
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 28 May 2021
Sec. Molecular Diagnostics and Therapeutics
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.672040
This article is part of the Research Topic Engineering Signal Sensors Based on Reprogrammed CRISPR Technologies View all 23 articles
By targeting key genes, the CRISPR system can effectively exert its anti-cancer activity. The latest research suggests that the CRISPReader system that regulates gene transcription can effectively target and inhibit bladder cancer cells by sensing transcription factors such as c-Myc and Get-1 in the cell. An interesting question is whether the CRISPReader system can exert its anti-cancer ability against a variety of tumors by sensing the broad-spectrum transcription factor Ets-1. In this work, a CRISPReader system that senses the Ets-1 transcription factor has been constructed. It can effectively identify a variety of cancer cell lines, and specifically induce apoptosis in cancer cells. This study fully confirmed the effectiveness of Ets-1 as a broad-spectrum cancer related signal and provided a new anti-cancer tool based on the CRISPReader system.
CRISPR has become a popular gene editing technology in recent years, which can be used to modify species and investigate gene functions (Lino et al., 2018). The CRISPR nuclease, such as Cas9 protein, can induce DNA double-strand breaks and DNA recombination repair mediated by homologous templates (Doudna and Charpentier, 2014). The mutated dead Cas9, dCas9, can bind to the gene promoter region to regulate gene transcription levels (Gjaltema and Schulz, 2018). By modifying the dCas9 protein or sgRNA, an inducible dCas9 system can be constructed (Dai et al., 2018). The regulation of target genes by dCas protein is activated by sensing exogenous inducers or endogenous proteins within cells (Liu Y. et al., 2016; Liu et al., 2017). This is significant because it successfully realizes the systematic sensation of the key signals of tumor cells. The regulation of downstream gene expression through inducible dCas9 is not only conducive to the specific recognition of cancer cells but also can effectively inhibit the malignant growth of tumors. This provides new strategies for basic research and the clinical treatment of tumors.
A key aspect of the research progress made for the CRISPR transcriptional regulation system is to use the dCas9 transcription activator to bind to its transcription initiation element to replace the promoter to perform gene expression functions. This is a very important technological advancement. The related technology is called “CRISPReader” (Zhan H. et al., 2019), which was originally used to reduce the gene expression cassette of CRISPR so that it can be loaded into AAV vectors with limited capacity. The gene circuit based on CRISPReader can sense transcription factors such as c-Myc and Get-1 in cancer cells (Liu et al., 2020). It can not only effectively identify bladder cancer cells from non-bladder cancer cells, but also effectively inhibit the growth and metastasis of cancer cells. However, this gene circuit is only suitable for the gene therapy of bladder cancer because of the tissue expression specificity of the Get-1 transcription factor.
Ets-1 is a transcription factor that is widely expressed in a variety of tumors (Hahne et al., 2008). Previous studies have found that the TERT promoter of many tumors has high frequency mutations, and the mutated sequence increases the binding site of Ets-1 (Li et al., 2015; Liu L. et al., 2016). TERT is also a broad-spectrum tumor marker, which is conducive to the excessive division of tumor cells.
A more interesting idea is to use CRISPReader to sense Ets-1 and to develop a broad-spectrum anti-cancer tool. In this study, based on the CRISPReader technology, we successfully constructed a gene circuit that senses the endogenous Ets-1 transcription factor in cells. It can identify a variety of cancer cells including bladder cancer and gastric cancer, and can specifically and effectively induce apoptosis of cancer cells.
The cancer cell lines used in this study were purchased from the Institute of Cell Research, Chinese Academy of Sciences (Shanghai, China). Cells were grown in a high-sugar DMEM medium (Qingdao Jieshikang Biotechnology Co., Ltd.) supplemented with 10% fetal bovine serum (Invitrogen) at 37°C in a 5% carbon dioxide atmosphere.
The cDNA sequences for sgRNA binding regions were designed, synthesized, and inserted into the corresponding vector, which had been digested with restriction endonucleases. All vectors were transformed into One Shot TOP10 Chemically Competent E. coli cells, and the desired expression clones were identified using polymerase chain reaction (PCR) amplification and electrophoresis, and then confirmed with Sanger sequencing.
The above plasmids were transfected into various cancer cells using Lipofectamine 2000 kit according to the operation instructions when the cell density reached 60 ∼70%.
After transfection for 48 h, cells were obtained. Next, the activities of firefly and ranilla luciferase were analyzed by double luciferase reporting assay (Promega) in line with the manufacturer’s specifications.
GFP reporter gene assay was also used for studying the regulation of gene expression. A specialized microscope was used to see GFP-expressing cells. The percentage of cells showing green fluorescence was quantified by the Image J software1.
Cancer cells were transiently transfected with plasmid vectors, and then a BD flow cytometer (Beijing Delica Biotechnology Co., Ltd.) was used to detect the cells that had been transfected for 48 h and stained by Annexin V and PI in a 6-hole plate, and the experiment was repeated three times.
Cancer cells were transiently transfected with plasmid vectors, and then cells were analyzed with CCK-8 assay after being cultured in DMEM medium for 24, 48, and 72 h. For the determination of cell proliferation, a Cell-Counting Kit 8 (Dojindo Laboratories) was used according to the instructions. Fluorescence (450 nm) was recorded using a microplate reader.
SPSS 21.0 (SPSS, Inc., Chicago, IL, United States) was used to conduct statistical analysis. T-test was applied to express the measurement data, which was expressed by the mean number ± standard deviation (x ± SD). P < 0.05 was considered statistically significant.
We inserted the binding sequence of transcription factor Ets-1 into the upstream of the transcription initiation element TATA box. Then, the sgRNA binding site was inserted between the two elements. The Cas9/VP64 and sgRNA expression framework were inserted downstream of the transcription start site. The sgRNA also targets the promoter region of luciferase or GFP. When the expression of the transcription factor Ets-1 in the cell is very low, neither Cas9/VP64 nor sgRNA is expressed. In cancer cells expressing Ets-1, Cas9/VP64 and sgRNA continue to drive their self-expression, and then transcriptionally activate luciferase (Figure 1A) or GFP (Figure 1B).
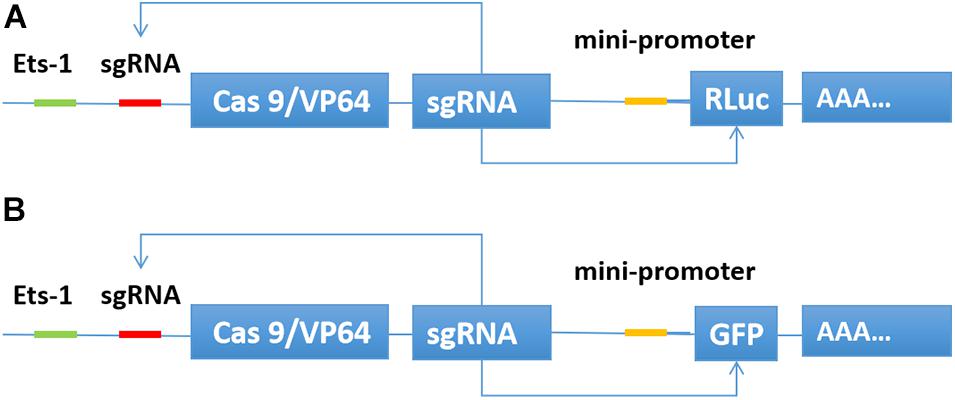
Figure 1. Construction of CRISPReader sensing the Ets-1 transcription factor. When Ets-1 binds to the promoter region, weak expression of Cas9/VP64 can be initiated. After Cas9/VP64 is combined with sgRNA, it can further bind its promoter and drive its own expression. In this way, the expression level of Cas9/VP64 is continuously increased through positive feedback, and then the transcription of luciferase (A) or GFP (B) is activated.
To verify whether the CRISPReader system can specifically recognize bladder cancer cells by sensing the expression of the transcription factor Ets-1, we transiently transferred the plasmid expressing the system into a series of cancer cells and corresponding normal cells. The luciferase test results suggest that the CRISPReader can significantly activate the expression of luciferase in T24, 5637, RT4 bladder cancer cell lines that highly express Ets-1, but cannot express luciferase in normal urothelial cells SV-HUC-1 (Figure 2A). The results were the same when the reporter gene was GFP (Figure 2B). The difference in the expression of reporter genes suggests that the CRISPReader system can effectively distinguish bladder tumor cells from normal bladder cells.
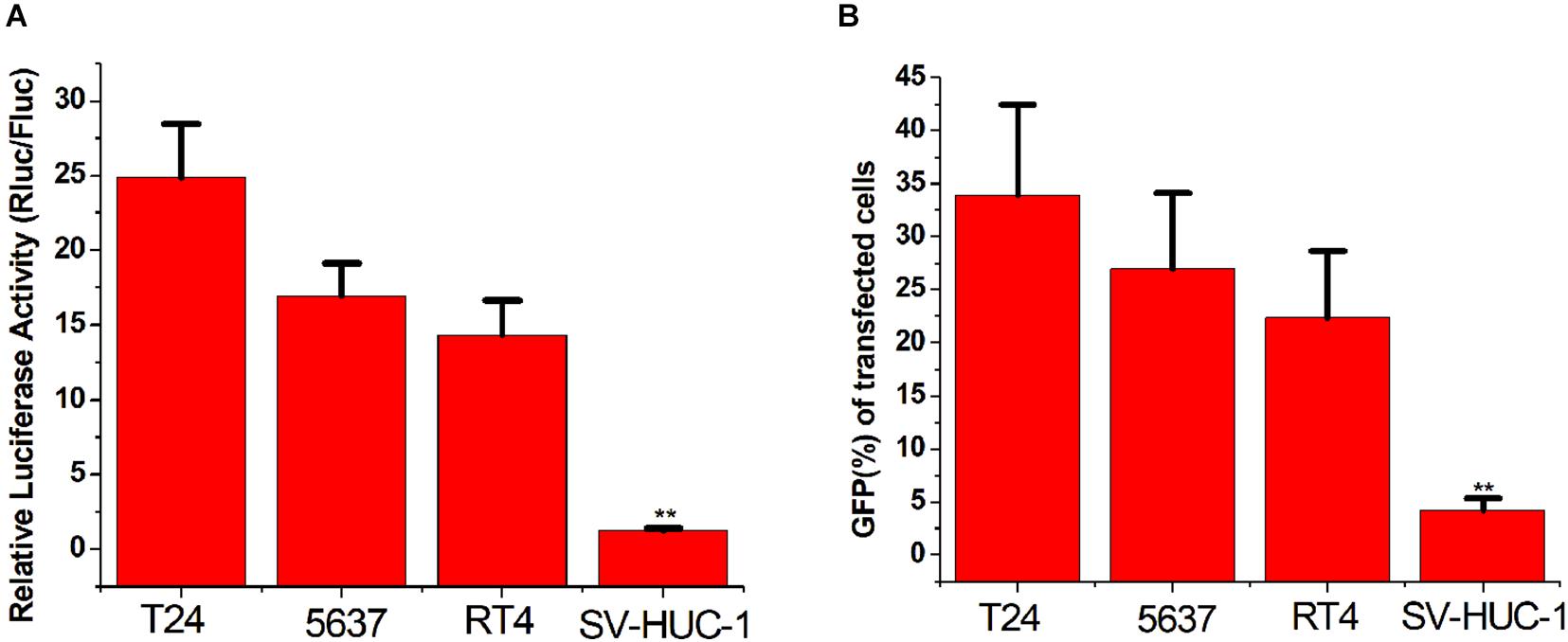
Figure 2. Distinguishing between bladder cancer cells and normal cells. (A) Cell luciferase activity was determined using the dual luciferase reporting assay (**p < 0.01). Data were expressed as mean ± SD. (B) Cell GFP expression was determined using the GFP reporter assay (**p < 0.01). Data were expressed as mean ± SD.
To verify the broad spectrum of the CRISPReader system in recognizing tumor cells, we transiently transferred the plasmid expressing the system into a series of gastric cancer cells and corresponding normal cells. The luciferase test results suggest that the CRISPReader can significantly activate the expression of luciferase in BGC-823, RPMI 1640, and HGC-27 gastric cancer cell lines that highly express Ets-1, while it cannot express luciferase in normal gastric epithelial cells GES-1 (Figure 3). The expression difference of the reporter gene suggests that the CRISPReader system can effectively distinguish gastric tumor cells from normal gastric cells, indicating that the Ets-1-sensing CRISPReader system has potential broad-spectrum anti-tumor capabilities.
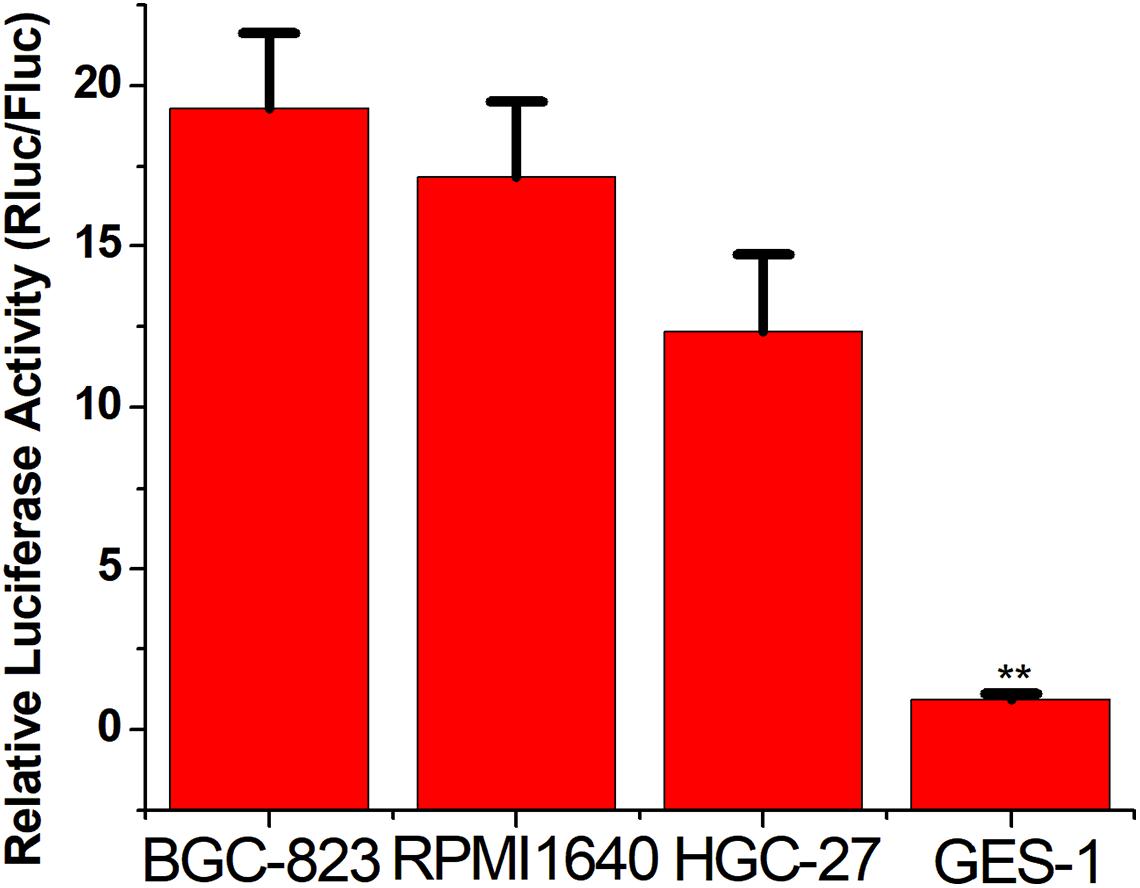
Figure 3. Distinguishing between gastric cancer cells and normal cells. Cell luciferase activity was determined using the dual luciferase reporting assay (**p < 0.01). Data were expressed as mean ± SD.
We then replaced the luciferase gene with the caspase-3 gene, which can induce apoptosis. The CRISPReader expression plasmid was transfected into bladder cancer and gastric cancer cell lines and the corresponding normal cell lines, respectively. The results of flow cytometry indicated that the apoptosis rate of tumor cell lines was significantly higher than that of normal cell lines (Figure 4), indicating that the Ets-1-sensitive CRISPReader system can effectively induce the apoptosis of different tumor cells and has broad-spectrum anti-tumor activity.
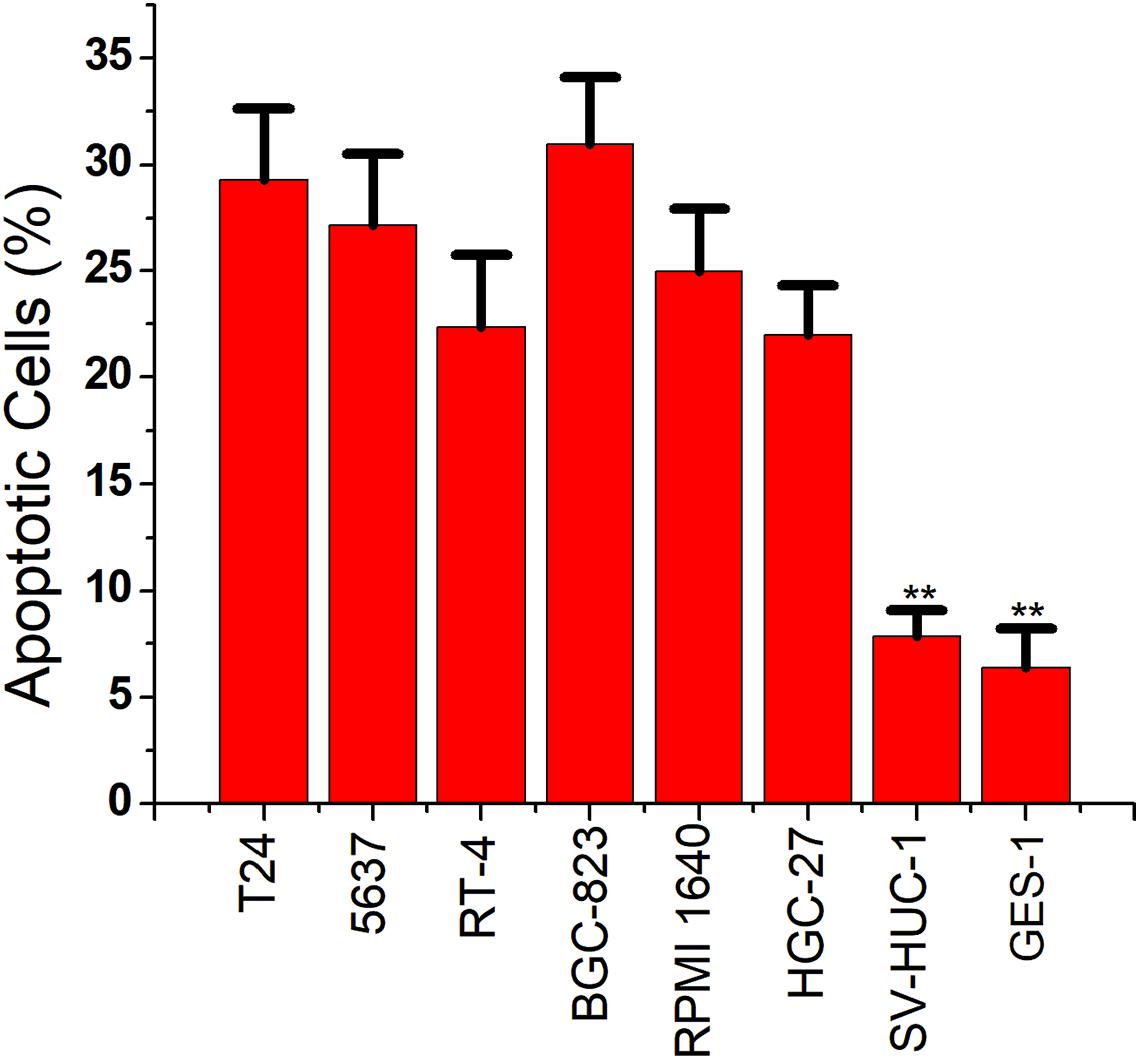
Figure 4. Specifically induced cancer cell apoptosis. Cell apoptosis activity was determined using the flow cytometry assay (**p < 0.01). Data were expressed as mean ± SD.
For the final step, we replaced the luciferase gene with the PTEN gene, which can inhibit cell proliferation. The CRISPReader expression plasmid was transfected into bladder cancer and gastric cancer cell lines and the corresponding normal cell lines, respectively. The results of the CCK-8 assay indicate that the proliferation rate of tumor cell lines was significantly lower than that of normal cell lines (Figure 5), indicating that the Ets-1-sensitive CRISPReader system can effectively inhibit the proliferation of different tumor cells and has broad-spectrum anti-tumor activity.
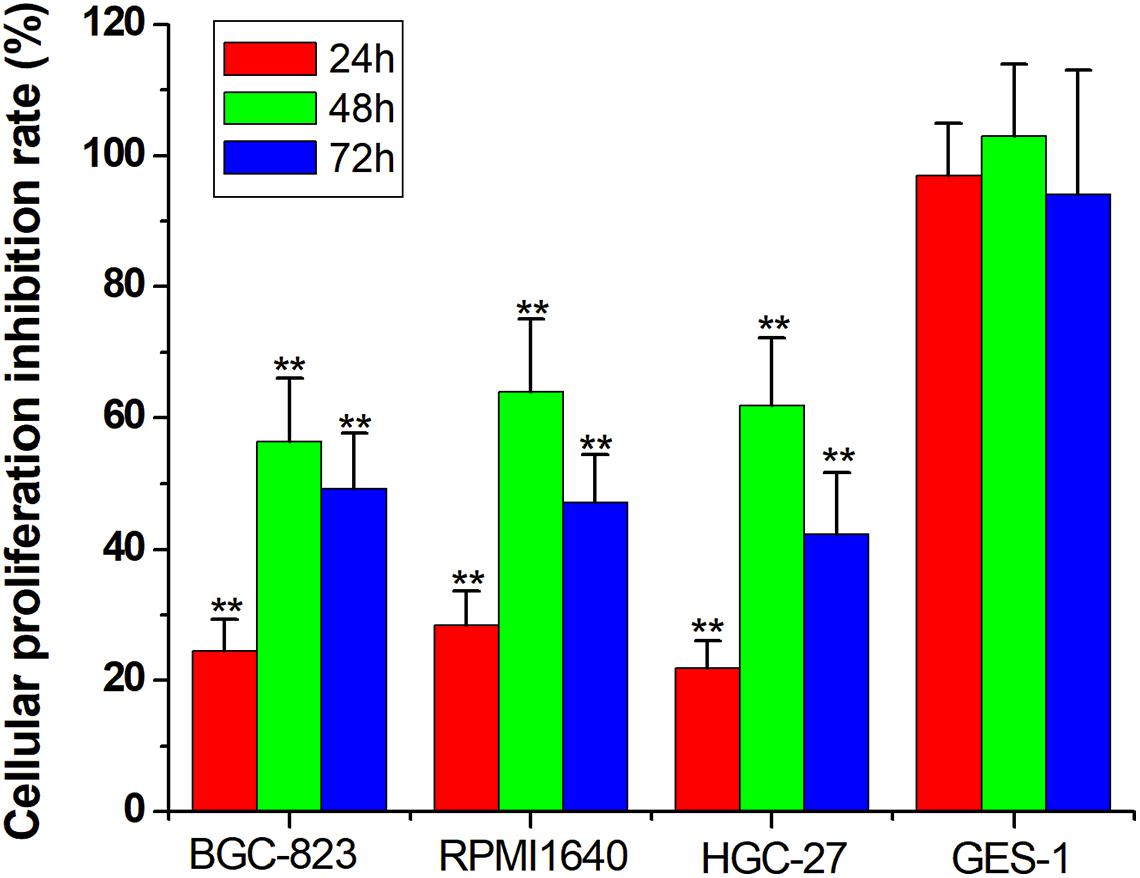
Figure 5. Specifically inhibited cancer cell proliferation. Cell proliferation rate was determined using the CCK-8 assay (**p < 0.01). Data were expressed as mean ± SD.
The application of the CRISPR system to the basic and therapeutic research of tumors is a major trend in current academic circles. An effective anti-cancer strategy is to use the CRISPR system to target fusion genes to exert anti-cancer effects (Zhan T. et al., 2019). However, the CRISPR system can not only regulate its downstream target genes but also sense a series of upstream signals, thus playing the role of the signal transmitter (Liu et al., 2018). It has obvious advantages when applied to tumor biotherapy, which can effectively identify and kill cancer cells. The biggest problem is how to choose the carcinogenic signal to be sensed. Ets-1 is a transcription factor that is generally highly expressed in tumors, so it is an ideal signal molecule for the CRISPR system.
In this study, we successfully designed a CRISPReader system that senses the Ets-1 transcription factor. It can effectively identify a variety of tumor cell lines including bladder cancer and gastric cancer, and specifically induce apoptosis and inhibit the proliferation of cancer cells. This study fully confirmed the effectiveness of Ets-1 as a broad-spectrum tumor recognition signal and provided a new anti-cancer tool based on the CRISPR system.
The limitation of this study is that only in vitro cell experiments were performed, and animal experiments will be needed in the future to further prove the anti-cancer effect of this tool.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
KY provided the idea and designed the study details. All authors finished the experiments and wrote the manuscript, contributed to the article, and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Dai, X., Chen, X., Fang, Q., Li, J., and Bai, Z. (2018). Inducible CRISPR genome-editing tool: classifications and future trends. Crit. Rev. Biotechnol. 38, 573–586. doi: 10.1080/07388551.2017.1378999
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096.
Gjaltema, R. A. F., and Schulz, E. G. (2018). CRISPR/dCas9 switch systems for temporal transcriptional control. Methods Mol. Biol. 1767, 167–185. doi: 10.1007/978-1-4939-7774-1_8
Hahne, J. C., Okuducu, A. F., Sahin, A., Fafeur, V., Kiriakidis, S., and Wernert, N. (2008). The transcription factor ETS-1: its role in tumour development and strategies for its inhibition. Mini Rev. Med. Chem. 8, 1095–1105. doi: 10.2174/138955708785909934
Li, C., Wu, S., Wang, H., Bi, X., Yang, Z., Du, Y., et al. (2015). The C228T mutation of TERT promoter frequently occurs in bladder cancer stem cells and contributes to tumorigenesis of bladder cancer. Oncotarget 6, 19542–19551. doi: 10.18632/oncotarget.4295
Lino, C. A., Harper, J. C., Carney, J. P., and Timlin, J. A. (2018). Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 25, 1234–1257. doi: 10.1080/10717544.2018.1474964
Liu, L., Liu, Y., Zhang, T., Wu, H., Lin, M., Wang, C., et al. (2016). Synthetic Bax-Anti Bcl2 combination module actuated by super artificial hTERT promoter selectively inhibits malignant phenotypes of bladder cancer. J. Exp. Clin. Cancer Res. 8:3.
Liu, Y., Han, J., Chen, Z., Wu, H., Dong, H., and Nie, G. (2017). Engineering cell signaling using tunable CRISPR-Cpf1-based transcription factors. Nat. Commun. 8:2095.
Liu, Y., Huang, W., and Cai, Z. (2020). Synthesizing AND gate minigene circuits based on CRISPReader for identification of bladder cancer cells. Nat. Commun. 11:5486.
Liu, Y., Li, J., Chen, Z., Huang, W., and Cai, Z. (2018). Synthesizing artificial devices that redirect cellular information at will. Elife 10:e31936.
Liu, Y., Zhan, Y., Chen, Z., He, A., Li, J., and Wu, H. (2016). Directing cellular information flow via CRISPR signal conductors. Nat. Methods 13, 938–944. doi: 10.1038/nmeth.3994
Zhan, H., Zhou, Q., Gao, Q., Li, J., Huang, W., and Liu, Y. (2019). Multiplexed promoterless gene expression with CRISPReader. Genome Biol. 20, 113.
Keywords: Ets-1, CRISPReader, cancer, cancer related signal, gene therapy
Citation: Yang K, Zhou Y and Zhong H (2021) CRISPReader System Sensing the Ets-1 Transcription Factor Can Effectively Identify Cancer Cells. Front. Mol. Biosci. 8:672040. doi: 10.3389/fmolb.2021.672040
Received: 25 February 2021; Accepted: 16 April 2021;
Published: 28 May 2021.
Edited by:
Yonghao Zhan, Zhengzhou University, ChinaReviewed by:
Xiujuan Wu, Shanghai Xuhui Central Hospital, ChinaCopyright © 2021 Yang, Zhou and Zhong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kang Yang, NDkyNTUyOTMyQHFxLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.