- 1Department of Chemistry, Biochemistry and Pharmaceutical Sciences, University of Bern, Bern, Switzerland
- 2Graduate School for Cellular and Biomedical Sciences, University of Bern, Bern, Switzerland
Ever since the “great oxidation event,” Earth’s cellular life forms had to cope with the danger of reactive oxygen species (ROS) affecting the integrity of biomolecules and hampering cellular metabolism circuits. Consequently, increasing ROS levels in the biosphere represented growing stress levels and thus shaped the evolution of species. Whether the ROS were produced endogenously or exogenously, different systems evolved to remove the ROS and repair the damage they inflicted. If ROS outweigh the cell’s capacity to remove the threat, we speak of oxidative stress. The injuries through oxidative stress in cells are diverse. This article reviews the damage oxidative stress imposes on the different steps of the central dogma of molecular biology in bacteria, focusing in particular on the RNA machines involved in transcription and translation.
Introduction
Sources of Oxidative Stress and the Bacterial Defense Mechanisms Against Them
The earliest single-celled life forms evolved on Earth in an anoxic environment around 4 billion years ago (Orgel, 1998). It was only after the “great oxidation event” approximately 2.4 billion years ago that, due to the emergence of photosynthesis, the oxygen level of Earth’s atmosphere rose significantly (Lyons et al., 2014). Consequently, cellular life forms had to adapt to the potential danger of harmful by-products of oxygen metabolism, better known as reactive oxygen species (ROS). ROS can damage all types of cellular components like nucleic acids, proteins and lipids (Imlay, 2013). Two of the most widely studied forms of ROS, superoxide (O2−) and hydrogen peroxide (H2O2), are constantly produced endogenously through the autoxidation of O2 on a range of both aerobic and non-aerobic respiratory flavoproteins (Imlay, 1995; Messner and Imlay, 1999; Kussmaul and Hirst, 2006; Korshunov and Imlay, 2010), as well as on non-respiratory flavoproteins (Massey et al., 1969; Geary and Meister, 1977; Grinblat et al., 1991). To protect the cells against these harmful compounds, bacteria evolved enzymes termed superoxide dismutases (SODs) to convert O2− into O2 and H2O2, as well as catalases and peroxidases to remove H2O2 to continuously neutralize the endogenously produced ROS (Imlay, 2008). In fully aerated Escherichia coli, the interplay between endogenous production and scavenging enzymes results in a steady-state intracellular concentration of ∼0.2 nM O2− and ∼50 nM H2O2 (Imlay, 2013). Internal concentrations can however be increased by exogenous sources. The membrane of bacteria is semipermeable for H2O2. Naturally produced H2O2 can therefore enter and potentially damage bacterial cells. Sources of H2O2 can be H2O2 generated through photochemistry in surface waters (Wilson et al., 2000b; Wilson et al., 2000a) (including in common lab media (Li and Imlay, 2018)), oxic-anoxic interfaces (e.g., near the intestinal epithelium) (Imlay, 2019), excretion from lactic acid bacteria (Tong et al., 2007; Liu et al., 2011; Bao et al., 2017), and potentially indirectly from phagocytes, which produce superoxide through an NADPH oxidase (Segal, 2008). O2− itself is not permeable through cytosolic membranes at neutral pH and can therefore not penetrate the bacterial cell enclosed in the phagosomes. One hypothesis is that the O2− dismutates spontaneously to H2O2 to exert the observed toxic effect on the captured bacteria in the phagocyte (Imlay, 2019). Due to the membranes’ impermeability to superoxide, other methods than simple diffusion must be used both in the lab and in nature to increase intracellular O2− concentration in bacteria. Redox-cycling antibiotics, such as the synthetic viologen paraquat or the naturally occurring phenazines and quinones, are able to penetrate bacterial cells. Inside the cell, they oxidize redox enzymes and produce O2− by transferring the electrons to oxygen (Turner and Messenger, 1986; Kuo et al., 1987; De Paiva et al., 2003; Inbaraj and Chignell, 2004). A unique source of ROS is found in phototrophic organisms. During photosynthesis, both O2− and H2O2 are produced at the photosystem I (Asada, 1999). Additionally, singlet oxygen (1O2) is generated at the photosystem II by a transfer of light energy to oxygen (Zolla and Rinalducci, 2002; Nishiyama et al., 2006; Glaeser et al., 2011). Singlet oxygen, another relevant ROS species, has a short half-life and diffuses freely through cells. Measurements of singlet oxygen travel distances in its lifetime determined that in microorganisms, 1O2 can theoretically be distributed throughout the whole bacterial cell once it is produced in the photosystems of the cytosolic membrane (Skovsen et al., 2005). In cyanobacteria, photosystems I and II are assisted by light-harvesting, antenna-like protein complexes called phycobilisomes (Singh et al., 2015). Close investigation of the ROS generated at phycobilisomes isolated from Synechocystis sp. PCC 6803 revealed that both a Type 1 and Type 2 photochemistry reaction (Foote, 1991) took place, resulting in O2− and 1O2 respectively (Rinalducci et al., 2008). While this list of ROS sources does not comprise the whole universe of potentially harmful ROS for bacteria (e.g., lipid peroxidation (Wang et al., 2004; Wang et al., 2006; Hong et al., 2019), ozone (Patil et al., 2011; Li et al., 2020)), this review focuses on the most widely studied forms of ROS to summarize our current understanding of oxidative stress damage and response in bacteria.
Oxidative stress can result in damage of both the backbone and bases of nucleic acids, both free and incorporated oxidized amino acids, as well as cofactors of proteins. To mitigate the damage of oxidative stress on cell biology, different stress response regulons are activated in bacteria, depending on the type of stressor (reviewed in (Imlay, 2015)). OxyR, a transcription factor widely distributed among gram-negative bacteria, is directly induced by H2O2. In most cases, the activated form of OxyR recruits RNA polymerase to transcribe about thirty different stress response genes (Seo et al., 2015), but there are also cases found in some bacteria where OxyR acts as a repressor to prevent the transcription of these genes under unstressed conditions (Loprasert et al., 2000; Luo et al., 2005; Chen et al., 2008; Ieva et al., 2008; Heo et al., 2010; Teramoto et al., 2013). PerR is an alternative transcription factor to OxyR, which is often found in gram-positive bacteria such as Bacillus subtilis (Jacquamet et al., 2009), but reports on PerR homologues in gram-negative bacteria, such as Campylobacter jejuni and Synechocystis sp. strain PCC 6803, have been published as well (Van Vliet et al., 1999; Li et al., 2004). The regulon of PerR contains most of the same stress response genes as the OxyR regulon (Helmann et al., 2003). Both induce enzymes to scavenge H2O2 and therefore mitigate further oxidative damage. However, only the OxyR regulon includes disulfide-reducing redoxins to repair already damaged proteins (Imlay, 2015; Seo et al., 2015). Two successive transcription factors are employed by E. coli to defend cells against the threat of increased O2− concentrations (Blanchard et al., 2007). SoxR is first activated by rising superoxide levels and induces the transcription of SoxS. SoxS in turn promotes the transcription of a defensive regulon encompassing 25 proteins to prevent the entry and accumulation of redox-active molecules in the cell (Blanchard et al., 2007; Seo et al., 2015). The SoxRS organization as found in E. coli is not a universal trait of all bacteria. In fact, performing a BLAST search for both SoxR and SoxS homologues in the bacterial domain revealed that SoxR is restricted to Proteobacteria and Actinobacteria, while SoxS was exclusively found in the family Enterobacteriaceae (Dietrich et al., 2008). The model bacterial system for the response to photooxidative stress through 1O2 is the anoxygenic anaerobic photosynthetic Rhodobacter sphaeroides. In R. sphaeroides, the 1O2 response regulon is activated through a cascade of transcription factors. First, the alternative sigma factor RpoE gets activated. While the RpoE regulon itself is rather small (Glaeser et al., 2007), it includes two additional sigma factors, RpoHI and RpoHII, which in turn activate the stress response regulon against singlet oxygen (Nuss et al., 2009; Nuss et al., 2010).
These defensive systems can be highly effective, allowing an exponentially growing E. coli culture to survive and overcome an extracellular H2O2 concentration that is 106 times the normal intracellular H2O2 concentration produced by endogenous activity (Zhu and Dai, 2019). Interestingly, cyanobacteria, which initially caused the great oxidation event (Schirrmeister et al., 2015), are much more susceptible to H2O2 as compared to eukaryotic aquatic microorganisms (Drábková et al., 2007; Mikula et al., 2012; Leunert et al., 2014; Liu et al., 2017). This higher susceptibility to H2O2 might potentially be exploited to combat increased cyanobacteria blooms caused by global warming (Chen et al., 2021). Notwithstanding the effectiveness of the response system, damage through oxidative stress can be acquired extremely quickly and acts non-discriminatory on all cellular compounds. In the next chapters, we will review how exactly oxidative damage influences different classes of biomolecules. In particular, we will focus on the damage that oxidative stress inflicts on the different steps of the extended central dogma of molecular biology in bacteria, which describes the flow of genetic information in a cell (Figure 1).
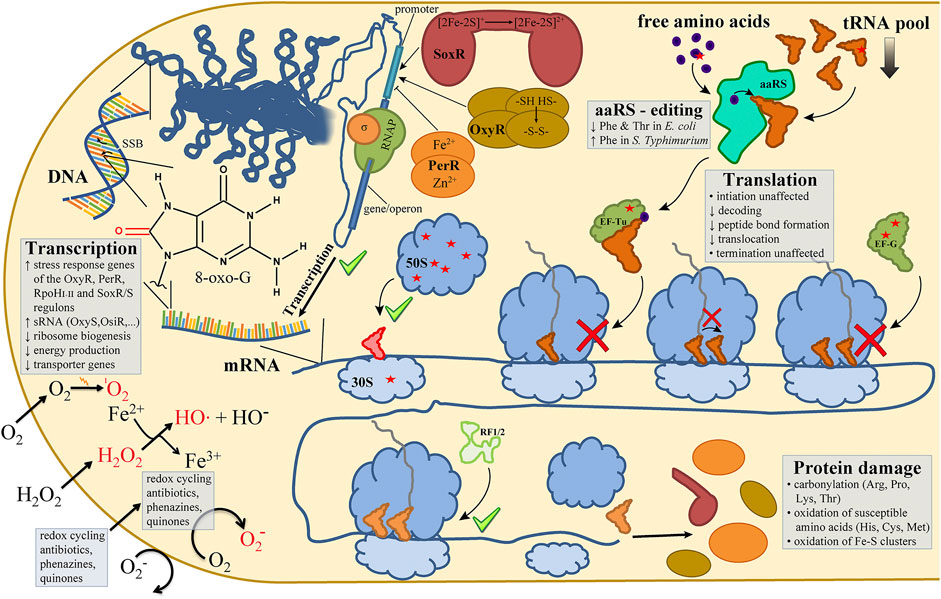
FIGURE 1. Oxidative stress in bacteria affects every sub step of the central dogma. In bacterial cells the most widely studied reactive oxygen species (ROS) are 1O2 (singlet oxygen), O2− (superoxide) and H2O2 (hydrogen peroxide), with the latter giving rise to HO• (hydroxyl radicals) via the Fenton reaction (lower left). ROS-dependent oxidations of DNA nucleobases (e.g., 8-oxo-G) or DNA backbone lesions (single-strand breaks; SSB) are recognized and trigger transcriptional responses mediated by oxidative stress-induced transcription factors (e.g., OxyR, PerR, SoxR) (upper left). Bacterial translation is affected in multiple ways by oxidative lesions (red asterisks) as observed in the pool of free amino acids, in elongation factors (EF-Tu, EF-G), ribosomal proteins, and in the main RNA species of the translation machinery (tRNA, rRNA) (upper right and center of the figure). Not necessarily expected, the sub steps of the ribosomal elongation cycle of protein biosynthesis are not affected by ROS to the same extent (center and lower right). While translation initiation and termination do not seem to be markedly inhibited (green check marks), A-site tRNA accommodation, peptide bond formation and EF-G-mediated tRNA translocation are inhibited by oxidative lesions (red cross signs). Please refer to the main text for more details on the ROS-mediated effects on the sub steps of the central dogma.
Genetic Information is Stored in DNA
DNA cannot be damaged directly neither by H2O2 nor by O2− alone (Winterbourn, 2013). Nevertheless, mutants lacking SODs (Farr et al., 1986) or catalases and peroxidases (Park et al., 2005) show increased mutagenesis rates. The mutagenic effect of H2O2 is easily explained by the Fenton reaction (Fenton, 1894) mediated by Fe2+-ions associated with the DNA:
The resulting hydroxyl radical is short-lived due to its high reactivity. It can react with most biomolecules at almost diffusion-limited rates. When produced in close proximity to DNA, hydroxyl radicals can damage both the nucleobase or the deoxyribose moieties, leading to mutations and strand breaks (Hutchinson, 1985; Dizdaroglu et al., 1991; Evans et al., 2004). Fittingly, Rai et al. found Fe2+ preferably associated to specific short DNA sequences (Rai et al., 2001) that had previously been found to be preferentially cleaved under oxidative stress conditions (Henle et al., 1999). A commonly used marker for oxidative stress damage is 8-oxo-7,8-dihydroguanosine (8-oxo-G), the most commonly observed oxidation product of a guanine base (Figure 1). Guanine has a lower reduction potential than the other DNA bases. Nearby oxidized bases can therefore readily be repaired by an electron transfer from the guanine base to the oxidized base (Candeias and Steenken, 1993), effectively transferring the oxidation site onto the guanine. 8-oxo-G can base-pair with both a cytosine or adenine base, thus affecting the coding potential of DNA (Cheng et al., 1992). Crystal structures of a Bacillus stearothermophilus DNA polymerase with 8-oxo-G pairing to both the cognate and mismatched base showed that under these conditions the proofreading mechanisms of the DNA polymerase are no longer effective (Hsu et al., 2004). In fact, Hsu et al. observed an inversion of the ordinary mismatch recognition. The in theory canonical 8-oxo-G:cytosine base-pair behaved as a mismatch, whereas the 8-oxo-G:adenine base-pair was recognized as cognate, which ultimately leads to a G to T transversion in the replicated DNA strand.
The impact of O2− on DNA is less obvious. In the end, the observed DNA damage is again due to the Fenton reaction described above. The involvement of superoxide is however only indirect. Keyer et al. showed that SOD deficient bacterial strains had increased free intracellular iron concentrations (Keyer and Imlay, 1996). The authors were also able to show that O2− excised the iron from a family of dehydratases that contain [4Fe-4S] clusters. Additionally, O2− was shown to reduce Fe3+ to Fe2+, regenerating ferrous iron for a continuous Fenton reaction (Fong et al., 1976). Finally, O2− is converted to H2O2 through the actions of SODs. The increase in both Fenton reaction educts results in the observed higher mutagenesis rate. Taken together, these findings highlight the need of bacterial cells to properly regulate their intracellular pool of free iron as well as iron associated and bound to macromolecular complexes. Unsurprisingly, both the transcription factors OxyR and PerR induce the transcription of an iron import repressor (fur) and a DNA-binding mini-ferritin (dps, mrgA in B. subtilis) to minimize free iron under conditions of oxidative stress (Imlay, 2015). The SoxRS system also activates Fur but has no effect on Dps. Instead it stimulates transcription of the endonuclease IV (nfo) gene for DNA repair (Seo et al., 2015). Similarly, iron-binding proteins are upregulated through the RpoE/RpoHI/II sigma factors cascade in R. sphaeroides, while uptake mechanisms for Fe2+ are minimized (Glaeser et al., 2007; Nuss et al., 2009).
In contrast to O2−, singlet oxygen can directly react with DNA. Unlike HO∙, which targets all nucleobases, 1O2 exclusively interacts with guanine, which contains conjugated, electron-rich double bonds (Ravanat et al., 2001). The resulting oxidation product of the base is however again 8-oxo-G (Cadet et al., 2006; Cadet et al., 2010).
DNA is Transcribed to RNA
In bacteria, the genetic information stored in DNA is transcribed to RNA by a single RNA polymerase (RNAP) structurally similar to eukaryotic RNA polymerase II (Ebright, 2000). Specificity of gene expression is modulated by the use of different sigma (σ) factors and an array of transcription factors such as the above mentioned OxyR, PerR, and SoxR/S (Figure 1). The fact that the stress response genes in their regulons are actively transcribed and translated as a response to oxidative stress is evidence that the RNAP is not incapacitated under these conditions. The aforementioned transcription factors themselves are activated by oxidation. For example, H2O2 oxidation of OxyR induces the formation of an intramolecular disulfide bond, which results in a significant conformational change of OxyR into the DNA binding form (Choi et al., 2001). PerR, which is often found in gram-positive bacteria, generally works as a repressor. Upon oxidation, it loses the ability to bind DNA and thus enables the stress regulons to be transcribed. PerR subunits have two metal ion binding sites; the structural site which irreversibly binds Zn2+ and the regulatory site which reversibly binds Fe2+ (Herbig and Helmann, 2001). PerR is deactivated by H2O2 through metal-catalyzed oxidation of histidine residues that coordinate the Fe2+ ion in the regulatory site, resulting in the dissociation of PerR from DNA (Lee and Helmann, 2006; Traoré et al., 2009). The homodimer SoxR, which is first triggered in the SoxRS cascade, is activated through the oxidation of [2Fe-2S]+ clusters when cells are exposed to redox-cycling compounds (Gaudu et al., 1997). Whether it is O2− that directly induces SoxR oxidation is however disputed (Imlay, 2015). Activation of the alternative transcription factor RpoE in R. sphaeroides by singlet oxygen is sensed by ChR, an anti-transcription factor in complex with RpoE (Campbell et al., 2007). ChR is targeted by 1O2, which results in the disassembly of the complex, releasing active RpoE (Campbell et al., 2007).
Additional evidence for the functionality of RNAP under oxidative stress is the transcription of small regulatory noncoding RNAs (sRNAs). One example found in E. coli is OxyS, which is induced by OxyR. This 109 nucleotides (nt) long RNA has been implicated in the regulation of as many as 40 genes and deletion of the oxyS locus results in higher mutagenesis rates compared to wildtype cells when treated with H2O2 (Altuvia et al., 1997). Barshishat et al. showed that OxyS induces a temporary cell cycle arrest, therefore allowing for proper DNA damage repair (Barshishat et al., 2018). The cell cycle arrest is prompted through the repression of the nusG gene upon binding of up to two OxyS molecules around the ribosome binding site on the nusG mRNA. NusG is a cofactor of Rho-dependent transcription and has been implicated in the suppression of prophages and other horizontally acquired portions of the genome (Li et al., 1993; Cardinale et al., 2008). The downregulation of NusG results in elevated expression of the rac prophage which encodes for the kilR gene. KilR prevents proper assembly of FtsZ, a ring-like structured protein essential for cell division (Bi and Lutkenhaus, 1991), contributing to the observed cell cycle arrest upon oxidative stress (Barshishat et al., 2018). Several additional sRNAs have been found to be induced by both 1O2 and O2− in R. sphaeroides (Berghoff et al., 2009). A subset of these sRNAs were shown to be directly regulated by the RpoE/RpoHI/II sigma factors cascade as a response to photooxidative stress (Berghoff et al., 2009; Nuss et al., 2010). Later reports confirmed the roles of these sRNAs in oxidative stress resistance (Adnan et al., 2015; Billenkamp et al., 2015; Peng et al., 2016).
An inverted role was described for the sRNA IsrR in the cyanobacterium Synechocystis sp. PCC 6803. IsrR was found to regulate the expression of the iron deficiency-stress induced protein A (IsiA), which was postulated to shade the photosystem II for oxidative stress prevention (Yeremenko et al., 2004). However, in this case, IsrR is not induced by oxidative stress but is constitutively transcribed, resulting in diminished isiA mRNA levels. Upon oxidative stress, IsrR levels are reduced, allowing for isiA mRNA accumulation and translation (Dühring et al., 2006).
The Transcribed mRNA is Translated to Proteins
Upon a short H2O2 burst, Liu et al. found that in E. coli 8-oxo-G levels were higher in RNA than in DNA, meaning that oxidative stress has a stronger effect on RNA (Liu et al., 2012). Additionally, they showed that highly structured RNA like tRNAs (transfer RNA) and rRNAs (ribosomal RNA) are not protected from oxidative damage. This is in line with findings of other groups that oxidative stress generally inhibits the translation process (Zhong et al., 2015; Zhu and Dai, 2019). This chapter will now discuss in more detail how the different cogwheels of the translation machinery are affected by oxidative stress (Figure 1).
Messenger RNA (mRNA)
mRNAs carry the information for the synthesis of a protein. Equal to its mutagenic effect in DNA replication, 8-oxo-G can affect the decoding process during translation. Using an E. coli in vitro translation system, Simms et al. showed that an oxidized base in the A-site codon can lead to the incorporation of a non-cognate amino acid (Simms et al., 2014). 8-oxo-G preferably adopts the syn conformation of the glycosidic bond in the ribosomal A-site, resulting in an 8-oxo-G:A base pair during the decoding step (Thomas et al., 2019). However, regardless of whether the incoming tRNA had a C or an A base pairing with the 8-oxo-G, translation rates at these codons in the in vitro system were decreased to an extent that suggests strong translational stalling in vivo instead (Simms et al., 2014). Incorporation of a non-cognate amino acid could lead to misfolding and protein aggregation (Drummond and Wilke, 2008). In agreement with the conclusion of Simms et al., a later study found no increase in protein aggregation under oxidative stress conditions (Zhong et al., 2015). Simms et al. additionally found that oxidation at the wobble position, where usually non-cognate base pairing is tolerated more freely, equally affected peptide bond formation rates as oxidation at the other two positions of the codon did. Summarizing their findings, Simms et al. concluded that oxidations of mRNA bases inhibit the decoding step by preventing the small subunit from adopting the active conformation required for proper tRNA selection regardless of the position of the oxidative lesion in the codon. This observation, however, seems to be exclusive to mRNA-tRNA interactions in the decoding center, since peptide release mediated by bacterial release factors (RF1 and RF2) was not affected by oxidized stop codons (Simms et al., 2014). The hypothesis that mostly the elongation step of translation seems to be affected by oxidative stress is supported by findings of other groups. Zhong et al. found an increase of polysomes under oxidative stress conditions (Zhong et al., 2015), an observation that can be explained by uninhibited translation initiation and stalling ribosomes on the mRNA. Similar results were found in a cyanobacterium (Nishiyama et al., 2001; Nishiyama et al., 2004). In a more recent publication, Zhu et al. quantified the elongation rate in E. coli upon addition of H2O2 (Zhu and Dai, 2019). They found an immediate and dose-dependent decline of the elongation rate upon stress induction (down to one amino acid per second at 5 mM H2O2), followed by a gradual recovery to a normal elongation rate of approximately 16 amino acids per second. Furthermore, the immediate drop in elongation rate and the H2O2 dose-dependent gradual incline of the rate to normal levels correlated well with an observed dose-dependent growth arrest phase.
Generally, stress response mechanisms and changes in growth rate can have diverse effects on mRNA stability in bacteria in both global and gene-specific ways (Vargas-Blanco and Shell, 2020). To our knowledge, there is no numerical data available on mRNA degradation rates in bacteria under oxidative stress conditions. Using a DNA microarray analysis of H2O2 stressed E. coli cells, Zheng et al. reported an upregulation of the OxyR regulon and other stress response genes, as well as a downregulation of many ribosomal protein genes, cold shock genes, ATP synthase genes, and transporter genes (Zheng et al., 2001). There are however some reports on sRNAs regulating mRNA stability upon oxidative stress conditions. As discussed above, sRNAs can positively affect oxidative stress resistance (Dühring et al., 2006; Adnan et al., 2015; Billenkamp et al., 2015; Peng et al., 2016; Barshishat et al., 2018). While in the case of IsrR the degradation of the sRNA was shown to stabilize isiA mRNA (Dühring et al., 2006), a different mechanism was described for the sRNA OsiR found in Deinococcus radiodurans. OsiR also positively regulates oxidative stress tolerance, but its transcription is increased upon oxidative stress (Gao et al., 2020). OsiR targets the mRNA of KatE2, one of two KatE-type catalases found in D. radiodurans (Jeong et al., 2016). Under oxidative stress conditions, OsiR base-pairs with the katE mRNA, which results in increased mRNA transcript levels and increased translation of KatE (Gao et al., 2020).
RNA modifications have the potential to regulate mRNA stability. For example, N6-methyladenosine (m6A) can mark mRNAs for degradation in humans (Wang et al., 2014). Whereas m6A has been found to be an abundant mRNA modification in gram-negative bacteria, no change in the m6A/A ratio was observed under oxidative stress conditions in both E. coli and P. aeruginosa (Deng et al., 2015). Adenosine-to-inosine (A-to-I) RNA editing is another form of post-transcriptional RNA modification found in all domains of life (Torres et al., 2014). A recent report linked a specific A-to-I editing event on the mRNA of the flagellar filament protein fliC to an increased virulence in the bacterial plant pathogen Xanthomonas oryzae pv. oryzicola (Xoc) (Nie et al., 2020). Plants often generate ROS as a defensive reaction to either directly kill invading pathogens or, more importantly, work as signaling molecules for gene expression activation in the context of pathogen defense (Torres et al., 2006; Paiva and Bozza, 2014). The authors identified a serine to proline mutation (S128P) in FliC after exposing the bacteria to H2O2. The S128P editing event can potentially change filament structure and enhance biofilm formation, leading to an increased oxidative stress tolerance (Nie et al., 2020).
Transfer RNA (tRNA)
Aminoacylated tRNAs read the different codons on mRNAs with their anticodon to deliver the correct amino acid to the ribosome. Interestingly, Liu et al. found that tRNAs folded into their native structure displayed even slightly higher 8-oxo-G levels than denatured tRNAs when treated with H2O2in vitro (Liu et al., 2012). tRNAs are next to rRNA the second most abundant class of RNA and generally considered stable transcripts (Li et al., 2002). However, recent reports indicate that tRNA stability can be swiftly altered as a response to certain stress conditions and general demand (Svenningsen et al., 2017) In accordance, both Zhong et al. and Zhu et al. found a global reduction in full-length tRNA availability after the induction of oxidative stress by H2O2 in E. coli (Zhong et al., 2015; Zhu and Dai, 2019). The H2O2 dose-dependent reduction in full-length tRNAs could at least partially explain the strongly reduced elongation rate during oxidative stress conditions. Further evidence for this hypothesis is given by the observation that an overexpression of RNase D, resulting in an already initially reduced tRNA pool, lead to a hypersensitivity to H2O2. The opposite was true, when the tRNA pool was enriched by addition of a plasmid carrying extra copies of rare tRNA genes, cells became more resistant to oxidative stress (Zhu and Dai, 2019). Surprisingly, Leiva et al. published somewhat contradictory results to the two previous reports (Leiva et al., 2020). Instead of a global reduction of the tRNA pool, the authors found that from ten tested tRNA species only one specific tRNA (tRNAGly) was downregulated under both H2O2 and paraquat stress. One explanation for this discrepancy might be the fact that all three publications used different E. coli strains in their studies, hinting to the possibility that the response to oxidative stress could even be strain specific. Leiva et al. also investigated whether the tRNA modification pattern was altered in response to the oxidative stress. They found no change at all known modification sites for the tRNAGly isoacceptors before and after addition of the stress (Leiva et al., 2020). However, the authors also pointed out that abasic sites and unknown oxidation products of modified tRNAGly nucleotides might have escaped their analysis. Another study, using UV-induced oxidative stress, did describe degradation of many post-transcriptional modifications under oxidative stress conditions (Sun et al., 2018).
Aminoacyl-tRNA Synthetases (aaRS)
tRNAs are acylated with the correct amino acids by aaRSs. To ensure the cognate amino acid is loaded onto the 3′-CCA tail of the tRNA according to the genetic code, aaRS have evolved pretransfer and posttransfer proofreading functions (reviewed in (Ling et al., 2009)). While the aminoacylation active site of aaRSs exclude most non-cognate amino acids (pretransfer), especially structurally similar amino acids can escape this first proofreading sieve and be mistakenly added to a non-cognate tRNA. The posttransfer editing sites can correct these erroneous loadings. Oxidative stress has been shown to affect both the free amino acids (reviewed in (Stadtman and Levine, 2003)) and the editing sites of aaRSs. For example, the aromatic amino acid phenylalanine (Phe) is a prime target for oxidation through ROS. One of the potential oxidation products of Phe, meta-tyrosine (m-Tyr), has been shown to be incorporated into E. coli proteins at Phe codon positions if the editing site of PheRS was mutated. Additionally, the editing deficient PheRS E. coli strain was shown to be more sensitive to oxidative stress (Bullwinkle et al., 2014). Contradicting these findings, in Salmonella enterica serovar Typhimurium (S.Typhimurium) the PheRS displays increased proofreading activity of m-Tyr-tRNAPhe under oxidative stress conditions (Steiner et al., 2019). Salmonella have been shown to be able to survive inside of ROS producing macrophages (Steele-Mortimer, 2008). A restructuring of the S. Typhimurium PheRS under oxidative stress conditions to a hyperaccurate editing version can at least partially explain the observed resilience of Salmonella against human ROS-mediated immune defense mechanisms.
In the case of threonyl-tRNA synthetase (ThrRS), which is prone to incorporate the near-cognate serine (Ser), the editing site contains an essential cysteine residue (Dock-Bregeon et al., 2004). Surface cysteine can readily be oxidized by ROS. Consistently, oxidative stress has been found to deactivate the editing site of ThrRS and lead to the misincorporation of Ser at Thr codons (Ling and Söll, 2010; Wu et al., 2014). If protein mistranslation due to loading of non-cognate amino acids on tRNAs under oxidative stress happens in vivo, increased protein aggregates due to misfolded proteins might be expected. As mentioned above, no such aggregates were detected in a later study (Zhong et al., 2015). One explanation could be that the misfolded proteins are readily captured and degraded by proteases before they aggregate. Indeed, a protease-deficient E. coli strain was shown to be more susceptible to H2O2. An effect that was exaggerated when Ser was added in excess (Ling and Söll, 2010). These mistranslation events and the resulting need for chaperones show why genes encoding the HSP20 chaperones have been found to be induced by H2O2 in an OxyR independent manner (Zheng et al., 2001).
Ribosome
At the ribosome, proteins are formed by linking together the amino acids brought to the ribosome by tRNAs according to the codons of the mRNA present in the ribosomal decoding center. Bacterial ribosomes are enormous heterologous complexes consisting of a large (50S) and a small (30S) subunit. Both subunits contain RNA and protein components. Even though the ribosomal proteins outnumber the rRNA molecules (54 proteins to 3 rRNA molecules in E. coli), the rRNA content prevails and establishes the structural and functional core of the ribosome (Cech, 2000). As mentioned before, the high structural complexity of the ribosomes does not protect rRNA from oxidation (Liu et al., 2012). In a more recent study, we showed that the oxidation of the 50S subunit had more of a negative impact on in vitro translation than the oxidation of the 30S subunit (Willi et al., 2018; Figure 1). We observed the same trend with in vitro reconstituted 50S and 30S subunits using H2O2 treated 23S rRNA and 16S rRNA, respectively, in combination with ribosomal proteins isolated from unstressed Thermus aquaticus. In both cases, in vitro translation of a genuine mRNA was reduced. However, the reduction of translational activity was more severe using oxidized 23S rRNA as compared to 16S rRNA. The 23S rRNA of the 50S subunit harbors the peptidyl transferase center (PTC) of the ribosome. Interestingly, the investigation of the influence of oxidized PTC nucleotides using an atomic mutagenesis approach (Erlacher et al., 2011) showed varied results (Willi et al., 2018). Whereas the oxidation of 23S rRNA residues A2451 and U2585 resulted in a significant loss of translational activity in vitro, the oxidation of other active site nucleotides had no effect at all (U2506, A2602, G2447) or resulted in even slightly stimulated protein synthesis as in the case of 5-OH-C2063 (Willi et al., 2018). While these varied results highlight the complexity of oxidative stress damage, the question remains whether these particular nucleotides are actually prone to oxidation in vivo. Therefore, Willi et al. generated a crude oxidation map of the 23S, 16S, and 5S rRNAs using an 8-oxo-G antibody for immunoprecipitation of oxidized RNA followed by sequencing and alignment to rRNA genes. Fitting to the previous findings, we reported non-random occurrences of oxidation hotspots which were mostly found on the 23S rRNA, including in domain V which harbors the PTC nucleotides (Willi et al., 2018). Remarkably, 16S rRNA regions forming the decoding site and the anti-Shine-Dalgarno sequence of the small subunit appear to remain largely oxidation-damage free in vivo, which fits with previous reports that translation initiation was not affected by oxidative stress (Zhong et al., 2015). Efforts for a higher resolution oxidation map of the rRNAs comprising also other rRNA oxidative lesions are currently underway. As mentioned above, oxidation of rRNA inside the ribosome does not necessarily interfere with ribosome functions and oxidation of some residues even resulted in accelerated rates of protein synthesis in vitro (Willi et al., 2018). This illustrates that rRNA nucleobase oxidations can potentially be utilized by bacteria to fine-tune ribosome functions during different growth or stress conditions. Indeed, the natural occurrence of the bacterial rRNA modification 5-OH-C2501 gives further credence to this hypothesis. While in the polyextremophile Deinococcus radiodurans the modification is found ubiquitously on the bacterium’s ribosomes, it is completely absent in the rather closely related thermophile Thermus thermophilus (Havelund et al., 2011). In E. coli, the extent of the modification is growth phase dependent. While in early exponential phase only about 30% of ribosomes are modified, the frequency increase in stationary phase to about 70% (Havelund et al., 2011; Kimura et al., 2017). Recent research on the 5-OH-C2501 modification identified RlhA as the modifying enzyme, thereby completing the set of rRNA modifying enzymes in E. coli (Kimura et al., 2017). The exact biological role of this cytosine hydroxylation inside the ribosome is however so far unknown.
RNA molecules have a negatively charged phosphate backbone. Magnesium ions (Mg2+) are well known for their ability to counteract this negative charge and therefore stabilize the folding of large RNA molecules. Unsurprisingly, there are between 100–1,000 Mg2+ ions found on bacterial ribosomes (Zheng et al., 2015) that are essential for the structural integrity and functional activity of the ribosome (Klein et al., 2004). However, it has been shown that these Mg2+ ions can be replaced by Fe2+ and Mn2+ without significant loss of activity, mimicking the conditions of the early anoxic Earth (Athavale et al., 2012; Bray et al., 2018). While it has been shown before that exchange of Mg2+ ions in the ribosome can have a detrimental effect on rRNA stability (Winter et al., 1997; Polacek and Barta, 1998), research on the specific role of ribosome bound Fe2+ for oxidative lesions generated through localized Fenton reactions is scarce in bacteria. Most of the work has been focused on eukaryotes and human neurodegenerative diseases specifically (Ward et al., 2014; Dusek et al., 2015; Daglas and Adlard, 2018; Smethurst et al., 2020). However, since the core structures and functions of the ribosome are conserved (Melnikov et al., 2012), it is not far-fetched to assume that similar findings can be expected for bacteria. Indeed, our own unpublished data shows a significant increase of 8-oxo-G occurrences at nucleotides that are <2.4 Å from Mg2+ binding sites when ribosomes were treated with Fe2+ and H2O2.
Ribosomal proteins have no apparent shielding effect on proximate rRNA against oxidative stress lesions ((Willi et al., 2018), unpublished data). Nevertheless, they are also not spared from oxidative damage. In a global approach to detect redox sensitive cysteines before and after the addition of oxidative stress, Leichert et al. identified both small and large ribosomal subunit proteins that had at least one cysteine oxidized (Leichert et al., 2008). Another study found increased carbonylation of ribosomal proteins in viable but non-culturable E. coli cells found in a stationary phase culture (Desnues et al., 2003). A specific example of oxidative damage actually inhibiting the biological function of a ribosomal protein was described in two publications in 1978. In the stalk region of the ribosome, protein uL10 (nomenclature according to (Ban et al., 2014)) and multiple copies of the protein bL12 form one of the main interaction sites of the ribosome with translation factors, also known as the GTPase-associated center (Diaconu et al., 2005). The multiple copies of the bL12 protein have been shown to occur in two dimers on the uL10, whereas mutation of the uL10 to a single dimer-binding form showed strongly reduced translational initiation and elongation (Mandava et al., 2012). Oxidation of the three methionine residues found in bL12 to methionine sulfoxide resulted in a loss of dimerization. This leads to inhibited binding of bL12 to uL10 on the ribosome, which in turn leads to a decrease of EF-G dependent translation (see also below) (Caldwell et al., 1978; Koteliansky et al., 1978). Another publication shows an interesting connection between the ribosomal protein uS4, mistranslation, and the resistance to oxidative stress (Fan et al., 2015). The authors found that E. coli cells with error-prone ribosomes due to the I199N mutation of uS4 show a significantly lower H2O2 sensitivity than E. coli cells with wildtype ribosomes. Mistranslation of proteins leads to the induction of the general stress response transcription factor RpoS (Battesti et al., 2011). The regulon of RpoS includes the catalase KatE and the peroxidase OsmC, both of which were found to be upregulated in the error-prone E. coli strain, explaining the higher tolerance to H2O2 (Fan et al., 2015). The exact link between mistranslation and induction of RpoS is however not completely clear. The authors speculate that mistranslation of proteins buffers away the protease ClpP, which usually degrades RpoS. This would therefore provide a new pathway to activate the general stress response as a defense mechanism against H2O2 that is independent of the OxyR regulon, relying on protein mistranslation only. Additionally, the authors find that the deletion of the sRNA DsrA, which is known to positively regulate RpoS translation, results in the loss of the protective effect observed against H2O2. Mechanistic explanations of this observation remain to be elucidated.
Ribosome biogenesis is a particularly energy intensive process for the bacterial cell. Recycling the whole ribosome after oxidative damage would therefore appear very wasteful. To prevent the incorporation of oxidized free nucleotides in the first place, E. coli cells use the protein MutT to hydrolyze both 8-oxo-dGTP and 8-oxo-rGTP to the monophosphate forms, reducing mutagenic errors in DNA and RNA respectively (Taddei et al., 1997). While it is yet unclear whether the damaged rRNA can actually be repaired in cells, there is at least evidence that the polynucleotide phosphorylase (PNPase), which specifically binds oxidized RNA transcripts, also plays a role in proper ribosome assembly (Hayakawa et al., 2001; Cheng and Deutscher, 2003; Wu et al., 2009). In the case of damaged ribosomal proteins, there is evidence that a subset of ribosomal proteins can be exchanged in the fully assembled ribosome, including the above discussed uL10 and bL12 (Pulk et al., 2010).
Elongation Factors
Elongation factors (EF) are crucial for an efficient translation process. As was discussed above, mostly the elongation step of translation is inhibited by oxidative stress (Figure 1). It would therefore not be surprising, if also the essential elongation factors would be affected by ROS. Indeed, when Tamarit and co-workers analyzed the carbonylation pattern of proteins in E. coli after the treatment with H2O2, EF-G was found to be one of the primary targets (Tamarit et al., 1998). The same was true in Bacillus subtilis for both EF-Tu and EF-G (Mostertz and Hecker, 2003). EF-Tu assists the decoding step of elongation. It delivers aminoacylated tRNAs to the free A-site of the ribosome in a ternary complex with GTP. If correct base-pairing of the codon and anti-codon in the decoding center of the 16S rRNA is established, the GTP is hydrolyzed in the GTPase-associated center on the large subunit and EF-Tu is released (Rodnina, 2018). In the cyanobacterium Synechocystis sp. PCC 6803 (Synechocystis), H2O2 oxidizes a conserved cysteine of EF-Tu (Yutthanasirikul et al., 2016). The authors further showed that in an E. coli in vitro translation system the use of the oxidized EF-Tu resulted in strongly inhibited translation. Similar findings were described for EF-G in both Synechocystis and E. coli (Kojima et al., 2009; Nagano et al., 2015). EF-G catalyzes the tRNA/mRNA translocation step of elongation, where after a new peptide bond is formed between two amino acids, the ribosome moves along the mRNA to the next codon. Like EF-Tu, EF-G is a so-called G-protein. Its overall structure resembles the structure of the aa-tRNA/EF-Tu complex and it utilizes the hydrolysis of GTP triggered at the ribosomal GTPase-associated center as energy source (Koch et al., 2015; Rodnina, 2018). Upon oxidation with H2O2, two conserved cysteine residues formed a disulfide bridge which resulted in reduced GTPase activity and suppressed dissociation of EF-G from the ribosome (Kojima et al., 2009; Nagano et al., 2015). Interestingly, mutations of the two target cysteines to serines rendered the EF-G mutant resistant to oxidative stress and did not show significant loss of activity (Kojima et al., 2009). Therefore, the oxidation of these two conserved cysteines is likely a desired and expected mechanism for regulation of translation under adverse condition rather than random oxidative damage.
Translated Proteins Fulfill Diverse Functions in the Cell
Once translated and properly folded, proteins fulfill a wide array of cellular functions, ranging from structure to catalysis to metabolism and to reproduction. As discussed above in piecemeal, there are several ways how oxidative stress can damage proteins. Carbonylation represents an irreversible and unrepairable damage to proteins. It mostly affects the amino acids proline, arginine, lysine, and threonine and has been used in organisms of all domains of life as an indicator of oxidative damage (Tamarit et al., 1998; Nyström, 2005). Histidine can be oxidized to 2-oxo-histidine as has been shown in the deactivation of PerR through oxidative stress (Lee and Helmann, 2006; Traoré et al., 2009). The amino acids cysteine and methionine both contain an electron-rich sulfur atom in their side chains. Unsurprisingly, they are prone to oxidation through ROS as was seen in the case of OxyR activation (Choi et al., 2001), the loss of editing activity in the ThrRS (Dock-Bregeon et al., 2004), the loss of dimerization of bL12 (Caldwell et al., 1978; Koteliansky et al., 1978), and the loss of both EF-Tu and EF-G activity (Kojima et al., 2009; Nagano et al., 2015; Yutthanasirikul et al., 2016). Cysteine and methionine oxidation is further reviewed in (Ezraty et al., 2017). In the case of singlet oxygen, the impact on free amino acids has been studied extensively in vitro. 1O2 reacts selectively with only a subset of amino acids, namely cysteine, methionine, histidine, tyrosine, and tryptophan, and the interaction of 1O2 with these residues was found to be strongly solvent dependent (Di Mascio et al., 2019). Studies under more biological relevant conditions, however, confirmed these findings (Michaeli and Feitelson, 1994; Michaeli and Feitelson, 1995; Kim et al., 2008). Importantly, the accessibility of the five vulnerable amino acids in folded proteins (i.e., is the amino acid solvent exposed or buried inside of the protein) can alter the reactivity of a specific residue toward singlet oxygen (McDermott et al., 1991; Michaeli and Feitelson, 1994, Michaeli and Feitelson, 1995; Kim et al., 2008; Jensen et al., 2012; Sjöberg et al., 2016; Di Mascio et al., 2019). A final mechanism of ROS impact on proteins discussed above was the oxidation of iron-sulfur (Fe-S) clusters, as was seen in the case of SoxRS activation (Gaudu et al., 1997). Fe-S clusters are ancient prosthetic groups that could assemble spontaneously on polypeptides of the primordial, anoxic environment found during the first billion years of life on Earth (Imlay, 2006). Today, they are still one of the most ubiquitous cofactors found in nature. Their functions are extremely versatile, ranging from simple iron storage to the regulation of gene expression (Johnson et al., 2005). Both O2− and H2O2 can damage Fe-S clusters by either directly subtracting an electron or by generating reactive HO∙ through a Fenton reaction (Imlay, 2013). Additionally, most often cysteine is the coordinating amino acid of the polypeptide backbone forming the Fe-S cluster (Imlay, 2006). Oxidation of the coordinating cysteine by any generated ROS could lead to the inactivation of the Fe-S protein. Finally, H2O2 poisons the Isc system, which is responsible for Fe-S cluster synthesis. Therefore, in E. coli the OxyR regulon includes the activation of the Suf system, an alternative Fe-S cluster synthesis pathway (Seo et al., 2015).
Concluding Remarks
Once oxygen levels rose on Earth, bacterial cells had to develop systems to continuously remove endogenously generated ROS to keep oxidative damage to a minimum. Nonetheless, ROS levels inside cells can rise due to a variety of external reasons (Figure 1). Even though bacteria have efficient scavenging and repair systems, ever-emerging research in the field of oxidative stress has shown that all the steps of the central dogma of molecular biology are nonetheless susceptible to oxidative damage. In this review, we focused exclusively on bacterial systems. However, several parts of the central dogma of molecular biology are well conserved among all domains of life thus allowing extrapolation of insights gained in bacteria to other kingdoms. Especially the ribosome, the central hub of translation, has a highly conserved core found everywhere from bacteria to humans (Melnikov et al., 2012) and findings from bacteria can, to an extent and with reasonable caution, be extrapolated to more complex organisms. Nevertheless, there is a plethora of research on oxidative stress directly performed in mammalian systems to be found. Of the outmost interest is the apparent link between oxidative stress and neurodegenerative diseases such as Alzheimer’s and Parkinson’s disease (Barnham et al., 2004). Furthermore, there is an emerging view that these diseases and the associated oxidative stress are influenced by the interplay of the human host and the gut microbiome (Dumitrescu et al., 2018; Luca et al., 2019), highlighting once more the important role of bacteria in human health.
Author Contributions
MF wrote the entire first draft of the manuscript and designed the figure. NP supervised the work and wrote together with MF the final version of this article.
Funding
Research in our lab is funded by the Swiss National Science Foundation (grant 310030-188969 to NP).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Jessica Willi, Marina Cristodero, and Robert Rauscher for stimulating discussions.
References
Adnan, F., Weber, L., and Klug, G. (2015). The sRNA SorY Confers Resistance during Photooxidative Stress by Affecting a Metabolite Transporter in Rhodobacter Sphaeroides. RNA Biol. 12, 569–577. doi:10.1080/15476286.2015.1031948
Altuvia, S., Weinstein-Fischer, D., Zhang, A., Postow, L., and Storz, G. (1997). A Small, Stable RNA Induced by Oxidative Stress: Role as a Pleiotropic Regulator and Antimutator. Cell 90, 43–53. doi:10.1016/S0092-8674(00)80312-8
Asada, K. (1999). The Water-Water Cycle in Chloroplasts: Scavenging of Active Oxygens and Dissipation of Excess Photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 601–639. doi:10.1146/annurev.arplant.50.1.601
Athavale, S. S., Petrov, A. S., Hsiao, C., Watkins, D., Prickett, C. D., Gossett, J. J., et al. (2012). RNA Folding and Catalysis Mediated by Iron (II). PLoS One 7, e38024. doi:10.1371/journal.pone.0038024
Ban, N., Beckmann, R., Cate, J. H. D., Dinman, J. D., Dragon, F., Ellis, S. R., et al. (2014). A New System for Naming Ribosomal Proteins. Curr. Opin. Struct. Biol. 24, 165–169. doi:10.1016/j.sbi.2014.01.002
Bao, X., Yang, J., De Soet, J. J., Liu, H., Gao, X., Van Loveren, C., et al. (2017). Factors Influencing the Competition between Streptococcus Oligofermentans and Streptococcus Mutans in Dual-Species Biofilms. Caries Res. 51, 507–514. doi:10.1159/000479044
Barnham, K. J., Masters, C. L., and Bush, A. I. (2004). Neurodegenerative Diseases and Oxidatives Stress. Nat. Rev. Drug Discov. 3, 205–214. doi:10.1038/nrd1330
Barshishat, S., Elgrably‐Weiss, M., Edelstein, J., Georg, J., Govindarajan, S., Haviv, M., et al. (2018). OxyS Small RNA Induces Cell Cycle Arrest to Allow DNA Damage Repair. EMBO J. 37, 413–426. doi:10.15252/embj.201797651
Battesti, A., Majdalani, N., and Gottesman, S. (2011). The RpoS-Mediated General Stress Response in Escherichia coli. Annu. Revie Microbiol. 65, 189–213. doi:10.1146/annurev-micro-090110-102946
Berghoff, B. A., Glaeser, J., Sharma, C. M., Vogel, J., and Klug, G. (2009). Photooxidative Stress-Induced and Abundant Small RNAs in Rhodobacter Sphaeroides. Mol. Microbiol. 74, 1497–1512. doi:10.1111/j.1365-2958.2009.06949.x
Bi, E., and Lutkenhaus, J. (1991). FtsZ Ring Structure Associated with Division in Escherichia coli. Nature 354, 161–164. doi:10.1038/354161a0
Billenkamp, F., Peng, T., Berghoff, B. A., and Klug, G. (2015). A Cluster of Four Homologous Small RNAs Modulates C1 Metabolism and the Pyruvate Dehydrogenase Complex in Rhodobacter Sphaeroides under Various Stress Conditions. J. Bacteriol. 197, 1839–1852. doi:10.1128/JB.02475-14
Blanchard, J. L., Wholey, W.-Y., Conlon, E. M., and Pomposiello, P. J. (2007). Rapid Changes in Gene Expression Dynamics in Response to Superoxide Reveal SoxRS-dependent and Independent Transcriptional Networks. PLoS One 2, e1186. doi:10.1371/journal.pone.0001186
Bray, M. S., Lenz, T. K., Haynes, J. W., Bowman, J. C., Petrov, A. S., Reddi, A. R., et al. (2018). Multiple Prebiotic Metals Mediate Translation. Proc. Natl. Acad. Sci. U. S. A. 115, 12164–12169. doi:10.1073/pnas.1803636115
Bullwinkle, T. J., Reynolds, N. M., Raina, M., Moghal, A., Matsa, E., Rajkovic, A., et al. (2014). Oxidation of Cellular Amino Acid Pools Leads to Cytotoxic Mistranslation of the Genetic Code. Elife 3, e02501. doi:10.7554/eLife.02501.001
Cadet, J., Douki, T., and Ravanat, J. L. (2010). Oxidatively Generated Base Damage to Cellular DNA. Free Radic. Biol. Med. 49, 9–21. doi:10.1016/j.freeradbiomed.2010.03.025
Cadet, J., Ravanat, J.-L., Martinez, G. R., Medeiros, M. H. G., and Di Mascio, P. (2006). Singlet Oxygen Oxidation of Isolated and Cellular DNA: Product Formation and Mechanistic Insights. Photochem. Photobiol. 82, 1219–1225. doi:10.1562/2006-06-09-IR-914
Caldwell, P., Luk, D. C., Weissbach, H., and Brot, N. (1978). Oxidation of the Methionine Residues of Escherichia coli Ribosomal Protein L12 Decreases the Protein’s Biological Activity. Proc. Natl. Acad. Sci. U. S. A. 75, 5349–5352. doi:10.1073/pnas.75.11.5349
Campbell, E. A., Greenwell, R., Anthony, J. R., Wang, S., Lim, L., Das, K., et al. (2007). A Conserved Structural Module Regulates Transcriptional Responses to Diverse Stress Signals in Bacteria. Mol. Cel 27, 793–805. doi:10.1016/j.molcel.2007.07.009
Candeias, L. P., and Steenken, S. (1993). Electron Transfer in Di(deoxy)nucleoside Phosphates in Aqueous Solution: Rapid Migration of Oxidative Damage (Via Adenine) to Guanine. J. Am. Chem. Soc. 115, 2437–2440. doi:10.1021/ja00059a044
Cardinale, C. J., Washburn, R. S., Tadigotla, V. R., Brown, L. M., Gottesman, M. E., and Nudler, E. (2008). Termination Factor Rho and its Cofactors NusA and NusG Silence Foreign DNA in E. coli. Science 320, 935–938. doi:10.1126/science.1152763
Cech, T. R. (2000). The Ribosome is a Ribozyme. Science 289, 878–879. doi:10.1126/science.289.5481.878
Chen, C., Wang, Y., Chen, K., Shi, X., and Yang, G. (2021). Using Hydrogen Peroxide to Control Cyanobacterial Blooms: A Mesocosm Study Focused on the Effects of Algal Density in Lake Chaohu, China. Environ. Pollut. 272, 115923. doi:10.1016/j.envpol.2020.115923
Chen, H., Xu, G., Zhao, Y., Tian, B., Lu, H., Yu, X., et al. (2008). A Novel OxyR Sensor and Regulator of Hydrogen Peroxide Stress with One Cysteine Residue in Deinococcus Radiodurans. PLoS One 3, e1602. doi:10.1371/journal.pone.0001602
Cheng, K. C., Cahill, D. S., Kasai, H., Nishimura, S., and Loeb, L. A. (1992). 8-Hydroxyguanine, an Abundant Form of Oxidative DNA Damage, Causes G → T and A → C Substitutions. J. Biol. Chem. 267, 166–172. doi:10.1016/s0021-9258(18)48474-8
Cheng, Z. F., and Deutscher, M. P. (2003). Quality Control of Ribosomal RNA Mediated by Polynucleotide Phosphorylase and RNase R. Proc. Natl. Acad. Sci. U. S. A. 100, 6388–6393. doi:10.1073/pnas.1231041100
Choi, H. J., Kim, S. J., Mukhopadhyay, P., Cho, S., Woo, J. R., Storz, G., et al. (2001). Structural Basis of the Redox Switch in the OxyR Transcription Factor. Cell 105, 103–113. doi:10.1016/S0092-8674(01)00300-2
Daglas, M., and Adlard, P. A. (2018). The Involvement of Iron in Traumatic Brain Injury and Neurodegenerative Disease. Front. Neurosci. 12, 981. doi:10.3389/fnins.2018.00981
De Paiva, S. R., Figueiredo, M. R., Aragão, T. V., and Coelho Kaplan, M. A. (2003). Antimicrobial Activity In Vitro of Plumbagin Isolated from Plumbago Species. Mem. Inst. Oswaldo Cruz 98, 959–961. doi:10.1590/s0074-02762003000700017
Deng, X., Chen, K., Luo, G.-Z., Weng, X., Ji, Q., Zhou, T., et al. (2015). Widespread Occurrence of N 6-methyladenosine in Bacterial mRNA. Nucleic Acids Res. 43, 6557–6567. doi:10.1093/nar/gkv596
Desnues, B., Cuny, C., Grégori, G., Dukan, S., Aguilaniu, H., and Nyström, T. (2003). Differential Oxidative Damage and Expression of Stress Defence Regulons in Culturable and Non‐culturable Escherichia Col Cells. EMBO Rep. 4, 400–404. doi:10.1038/sj.embor.embor799
Di Mascio, P., Martinez, G. R., Miyamoto, S., Ronsein, G. E., Medeiros, M. H. G., and Cadet, J. (2019). Singlet Molecular Oxygen Reactions with Nucleic Acids, Lipids, and Proteins. Chem. Rev. 119, 2043–2086. doi:10.1021/acs.chemrev.8b00554
Diaconu, M., Kothe, U., Schlünzen, F., Fischer, N., Harms, J. M., Tonevitsky, A. G., et al. (2005). Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTpase Activation. Cell 121, 991–1004. doi:10.1016/j.cell.2005.04.015
Dietrich, L. E. P., Teal, T. K., Price-Whelan, A., and Newman, D. K. (2008). Redox-active Antibiotics Control Gene Expression and Community Behavior in Divergent Bacteria. Science 321, 1203–1206. doi:10.1126/science.1160619
Dizdaroglu, M., Rao, G., Halliwell, B., and Gajewski, E. (1991). Damage to the DNA Bases in Mammalian Chromatin by Hydrogen Peroxide in the Presence of Ferric and Cupric Ions. Arch. Biochem. Biophys. 285, 317–324. doi:10.1016/0003-9861(91)90366-Q
Dock-Bregeon, A. C., Rees, B., Torres-Larios, A., Bey, G., Caillet, J., and Moras, D. (2004). Achieving Error-free Translation: The Mechanism of Proofreading of Threonyl-tRNA Synthetase at Atomic Resolution. Mol. Cel 16, 375–386. doi:10.1016/j.molcel.2004.10.002
Drábková, M., Matthijs, H. C. P., Admiraal, W., and Maršálek, B. (2007). Selective Effects of H2O2 on Cyanobacterial Photosynthesis. Photosynthetica 45, 363–369. doi:10.1007/s11099-007-0062-9
Drummond, D. A., and Wilke, C. O. (2008). Mistranslation-Induced Protein Misfolding as a Dominant Constraint on Coding-Sequence Evolution. Cell 134, 341–352. doi:10.1016/j.cell.2008.05.042
Dühring, U., Axmann, I. M., Hess, W. R., and Wilde, A. (2006). An Internal Antisense RNA Regulates Expression of the Photosynthesis Gene isiA. Proc. Natl. Acad. Sci. U. S. A. 103, 7054–7058. doi:10.1073/pnas.0600927103
Dumitrescu, L., Popescu-Olaru, I., Cozma, L., Tulbă, D., Hinescu, M. E., Ceafalan, L. C., et al. (2018). Oxidative Stress and the Microbiota-Gut-Brain Axis. Oxid. Med. Cel. Longev., 2406594. doi:10.1155/2018/2406594
Dusek, P., Roos, P. M., Litwin, T., Schneider, S. A., Flaten, T. P., and Aaseth, J. (2015). The Neurotoxicity of Iron, Copper and Manganese in Parkinson’s and Wilson’s Diseases. J. Trace Elem. Med. Biol. 31, 193–203. doi:10.1016/j.jtemb.2014.05.007
Ebright, R. H. (2000). RNA Polymerase: Structural Similarities between Bacterial RNA Polymerase and Eukaryotic RNA Polymerase II. J. Mol. Biol. 304, 687–698. doi:10.1006/jmbi.2000.4309
Erlacher, M. D., Chirkova, A., Voegele, P., and Polacek, N. (2011). Generation of Chemically Engineered Ribosomes for Atomic Mutagenesis Studies on Protein Biosynthesis. Nat. Protoc. 6, 580–592. doi:10.1038/nprot.2011.306
Evans, M. D., Dizdaroglu, M., and Cooke, M. S. (2004). Oxidative DNA Damage and Disease: Induction, Repair and Significance. Mutat. Res. - Rev. Mutat. Res. 567, 1–61. doi:10.1016/j.mrrev.2003.11.001
Ezraty, B., Gennaris, A., Barras, F., and Collet, J. F. (2017). Oxidative Stress, Protein Damage and Repair in Bacteria. Nat. Rev. Microbiol. 15, 385–396. doi:10.1038/nrmicro.2017.26
Fan, Y., Wu, J., Ung, M. H., De Lay, N., Cheng, C., and Ling, J. (2015). Protein Mistranslation Protects Bacteria against Oxidative Stress. Nucleic Acids Res. 43, 1740–1748. doi:10.1093/nar/gku1404
Farr, S. B., D’Ari, R., and Touati, D. (1986). Oxygen-dependent Mutagenesis in Escherichia coli Lacking Superoxide Dismutase. Proc. Natl. Acad. Sci. U. S. A. 83, 8268–8272. doi:10.1073/pnas.83.21.8268
Fenton, H. J. H. (1894). Oxidation of Tartaric Acid in Presence of Iron. J. Chem. Soc. Trans. 65, 899–910. doi:10.1039/CT8946500899
Fong, K. L., McCay, P. B., Poyer, J. L., Misra, H. P., and Keele, B. B. (1976). Evidence for Superoxide-dependent Reduction of Fe3+ and its Role in Enzyme-Generated Hydroxyl Radical Formation. Chem. Biol. Interact. 15, 77–89. doi:10.1016/0009-2797(76)90130-7
Foote, C. S. (1991). Definition of Type I and Type II Photosensitized Oxidation. Photochem. Photobiol. 54, 659. doi:10.1111/j.1751-1097.1991.tb02071.x
Gao, L., Chen, X., Tian, Y., Yan, Y., Zhan, Y., Zhou, Z., et al. (2020). The Novel ncRNA OsiR Positively Regulates Expression of katE2 and Is Required for Oxidative Stress Tolerance in Deinococcus Radiodurans. Int. J. Mol. Sci. 21, 3200. doi:10.3390/ijms21093200
Gaudu, P., Moon, N., and Weiss, B. (1997). Regulation of the soxRS Oxidative Stress Regulon: Reversible Oxidation of the Fe-S Centers of SoxR In Vivo. J. Biol. Chem. 272, 5082–5086. doi:10.1074/jbc.272.8.5082
Geary, L. E., and Meister, A. (1977). On the Mechanism of Glutamine Dependent Reductive Amination of α Ketoglutarate Catalyzed by Glutamate Synthase. J. Biol. Chem. 252, 3501–3508. doi:10.1016/s0021-9258(17)40419-4
Glaeser, J., Nuss, A. M., Berghoff, B. A., and Klug, G. (2011). Singlet Oxygen Stress in Microorganisms. Adv. Microb. Physiol. 58, 141–173. doi:10.1016/B978-0-12-381043-4.00004-0
Glaeser, J., Zobawa, M., Lottspeich, F., and Klug, G. (2007). Protein Synthesis Patterns Reveal a Complex Regulatory Response to Singlet Oxygen in Rhodobacter. J. Proteome Res. 6, 2460–2471. doi:10.1021/pr060624p
Grinblat, L., Sreider, C. M., and Stoppani, A. O. (1991). Superoxide Anion Production by Lipoamide Dehydrogenase Redox-Cycling: Effect of Enzyme Modifiers. Biochem. Int. 23, 83–92.
Havelund, J. F., Michael, A., Giessing, B., Hansen, T., Rasmussen, A., Scott, L. G., et al. (2011). Identification of 5-Hydroxycytidine at Position 2501 Concludes Characterization of Modified Nucleotides in E. coli 23S rRNA. J. Mol. Biol. 411, 529–536. doi:10.1016/j.jmb.2011.06.036
Hayakawa, H., Kuwano, M., and Sekiguchi, M. (2001). Specific Binding of 8-Oxoguanine-Containing RNA to Polynucleotide Phosphorylase Protein. Biochemistry 40, 9977–9982. doi:10.1021/bi010595q
Helmann, J. D., Wu, M. F. W., Gaballa, A., Kobel, P. A., Morshedi, M. M., Fawcett, P., et al. (2003). The Global Transcriptional Response of Bacillus subtilis to Peroxide Stress is Coordinated by Three Transcription Factors. J. Bacteriol. 185, 243–253. doi:10.1128/JB.185.1.243-253.2003
Henle, E. S., Han, Z., Tang, N., Rai, P., Luo, Y., and Linn, S. (1999). Sequence-specific DNA Cleavage by Fe2+-Mediated Fenton Reactions Has Possible Biological Implications. J. Biol. Chem. 274, 962–971. doi:10.1074/jbc.274.2.962
Heo, Y. J., Chung, I. Y., Cho, W. J., Lee, B. Y., Kim, J. H., Choi, K. H., et al. (2010). The Major Catalase Gene (katA) of Pseudomonas aeruginosa PA14 is under Both Positive and Negative Control of the Global Transactivator OxyR in Response to Hydrogen Peroxide. J. Bacteriol. 192, 381–390. doi:10.1128/JB.00980-09
Herbig, A. F., and Helmann, J. D. (2001). Roles of Metal ions and Hydrogen Peroxide in Modulating the Interaction of the Bacillus subtilis PerR Peroxide Regulon Repressor with Operator DNA. Mol. Microbiol. 41, 849–859. doi:10.1046/j.1365-2958.2001.02543.x
Hong, Y., Zeng, J., Wang, X., Drlica, K., and Zhao, X. (2019). Post-Stress Bacterial Cell Death Mediated by Reactive Oxygen Species. Proc. Natl. Acad. Sci. U. S. A. 116, 10064–10071. doi:10.1073/pnas.1901730116
Hsu, G. W., Ober, M., Carell, T., and Beese, L. S. (2004). Error-Prone Replication of Oxidatively Damaged DNA by a High-Fidelity DNA Polymerase. Nature 431, 217–221. doi:10.1038/nature02908
Hutchinson, F. (1985). Chemical Changes Induced in DNA by Ionizing Radiation. Prog. Nucleic Acid Res. Mol. Biol. 32, 115–154. doi:10.1016/S0079-6603(08)60347-5
Ieva, R., Roncarati, D., Metruccio, M. M. E., Seib, K. L., Scarlato, V., and Delany, I. (2008). OxyR Tightly Regulates Catalase Expression in Neisseria Meningitidis through Both Repression and Activation Mechanisms. Mol. Microbiol. 70, 1152–1165. doi:10.1111/j.1365-2958.2008.06468.x
Imlay, J. A. (1995). A Metabolic Enzyme that Rapidly Produces Superoxide, Fumarate Reductase of Escherichia coli. J. Biol. Chem. 270, 19767–19777. doi:10.1016/s0021-9258(18)94548-5
Imlay, J. A. (2008). Cellular Defenses against Superoxide and Hydrogen Peroxide. Annu. Rev. Biochem. 77, 755–776. doi:10.1146/annurev.biochem.77.061606.161055
Imlay, J. A. (2006). Iron-sulphur Clusters and the Problem with Oxygen. Mol. Microbiol. 59, 1073–1082. doi:10.1111/j.1365-2958.2006.05028.x
Imlay, J. A. (2013). The Molecular Mechanisms and Physiological Consequences of Oxidative Stress: Lessons from a Model Bacterium. Nat. Publ. Gr. 11, 443–454. doi:10.1038/nrmicro3032
Imlay, J. A. (2015). Transcription Factors that Defend Bacteria against Reactive Oxygen Species. Annu. Rev. Microbiol. 69, 93–108. doi:10.1146/annurev-micro-091014-104322
Imlay, J. A. (2019). Where in the World Do Bacteria Experience Oxidative Stress? Environ. Microbiol. 21, 521–530. doi:10.1111/1462-2920.14445
Inbaraj, J. J., and Chignell, C. F. (2004). Cytotoxic Action of Juglone and Plumbagin: A Mechanistic Study Using HaCaT Keratinocytes. Chem. Res. Toxicol. 17, 55–62. doi:10.1021/tx034132s
Jacquamet, L., Traoré, D. A. K., Ferrer, J. L., Proux, O., Testemale, D., Hazemann, J. L., et al. (2009). Structural Characterization of the Active Form of PerR: Insights into the Metal-Induced Activation of PerR and Fur Proteins for DNA Binding. Mol. Microbiol. 73, 20–31. doi:10.1111/j.1365-2958.2009.06753.x
Jensen, R. L., Arnbjerg, J., and Ogilby, P. R. (2012). Reaction of Singlet Oxygen with Tryptophan in Proteins: A Pronounced Effect of the Local Environment on the Reaction Rate. J. Am. Chem. Soc. 134, 9820–9826. doi:10.1021/ja303710m
Jeong, S. W., Jung, J. H., Kim, M. K., Seo, H. S., Lim, H. M., and Lim, S. (2016). The Three Catalases in Deinococcus Radiodurans: Only Two Show Catalase Activity. Biochem. Biophys. Res. Commun. 469, 443–448. doi:10.1016/j.bbrc.2015.12.017
Johnson, D. C., Dean, D. R., Smith, A. D., and Johnson, M. K. (2005). Structure, Function, and Formation of Biological Iron-Sulfur Clusters. Annu. Rev. Biochem. 74, 247–281. doi:10.1146/annurev.biochem.74.082803.133518
Keyer, K., and Imlay, J. A. (1996). Superoxide Accelerates DNA Damage by Elevating Free-Iron Levels. Proc. Natl. Acad. Sci. U. S. A. 93, 13635–13640. doi:10.1073/pnas.93.24.13635
Kim, J., Rodriguez, M. E., Guo, M., Kenney, M. E., Oleinick, N. L., and Anderson, V. E. (2008). Oxidative Modification of Cytochrome c by Singlet Oxygen. Free Radic. Biol. Med. 44, 1700–1711. doi:10.1016/j.freeradbiomed.2007.12.031
Kimura, S., Sakai, Y., Ishiguro, K., and Suzuki, T. (2017). Biogenesis and iron-Dependency of Ribosomal RNA Hydroxylation. Nucleic Acids Res. 45, 12974–12986. doi:10.1093/nar/gkx969
Klein, D. J., Moore, P. B., and Steitz, T. A. (2004). The Contribution of Metal ions to the Structural Stability of the Large Ribosomal Subunit. RNA 10, 1366–1379. doi:10.1261/rna.7390804
Koch, M., Flür, S., Kreutz, C., Ennifar, E., Micura, R., and Polacek, N. (2015). Role of a Ribosomal RNA Phosphate Oxygen during the EF-G-Triggered GTP Hydrolysis. Proc. Natl. Acad. Sci. U. S. A. 112, E2561–E2568. doi:10.1073/pnas.1505231112
Kojima, K., Motohashi, K., Morota, T., Oshita, M., Hisabori, T., Hayashi, H., et al. (2009). Regulation of Translation by the Redox State of Elongation Factor G in the Cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 284, 18685–18691. doi:10.1074/jbc.M109.015131
Korshunov, S., and Imlay, J. A. (2010). Two Sources of Endogenous Hydrogen Peroxide in Escherichia coli. Mol. Microbiol. 75, 1389–1401. doi:10.1111/j.1365-2958.2010.07059.x
Koteliansky, V. E., Domogatsky, S. P., and Gudkov, A. T. (1978). Dimer State of Protein L7/L12 and EF-G-Dependent Reactions on Ribosomes. Eur. J. Biochem. 90, 319–323. doi:10.1111/j.1432-1033.1978.tb12607.x
Kuo, C. F., Mashino, T., and Fridovich, I. (1987). α,β-Dihydroxyisovalerate Dehydratase. A Superoxide-Sensitive Enzyme. J. Biol. Chem. 262, 4724–4727. doi:10.1016/S0021-9258(18)61255-4
Kussmaul, L., and Hirst, J. (2006). The Mechanism of Superoxide Production by NADH:ubiquinone Oxidoreductase (Complex I) from Bovine Heart Mitochondria. Proc. Natl. Acad. Sci. U. S. A. 103, 7607–7612. doi:10.1073/pnas.0510977103
Lee, J. W., and Helmann, J. D. (2006). The PerR Transcription Factor Senses H2O2 by Metal-Catalysed Histidine Oxidation. Nature 440, 363–367. doi:10.1038/nature04537
Leichert, L. I., Gehrke, F., Gudiseva, H. V., Blackwell, T., Ilbert, M., Walker, A. K., et al. (2008). Quantifying Changes in the Thiol Redox Proteome upon Oxidative Stress In Vivo. Proc. Natl. Acad. Sci. U. S. A. 105, 8197–8202. doi:10.1073/pnas.0707723105
Leiva, L. E., Pincheira, A., Elgamal, S., Kienast, S. D., Bravo, V., Leufken, J., et al. (2020). Modulation of Escherichia coli Translation by the Specific Inactivation of tRNAGly under Oxidative Stress. Front. Genet. 11, 856. doi:10.3389/fgene.2020.00856
Leunert, F., Eckert, W., Paul, A., Gerhardt, V., and Grossart, H.-P. (2014). Phytoplankton Response to UV-Generated Hydrogen Peroxide from Natural Organic Matter. J. Plankton Res. 36, 185–197. doi:10.1093/plankt/fbt096
Li, H., Singh, A. K., McIntyre, L. M., and Sherman, L. A. (2004). Differential Gene Expression in Response to Hydrogen Peroxide and the Putative PerR Regulon of Synechocystis Sp. Strain PCC 6803. J. Bacteriol. 186, 3331–3345. doi:10.1128/JB.186.11.3331-3345.2004
Li, J., Mason, S. W., and Greenblatt, J. (1993). Elongation Factor NusG Interacts with Termination Factor ρ to Regulate Termination and Antitermination of Transcription. Genes Dev. 7, 161–170. doi:10.1101/gad.7.1.161
Li, J., Rumancev, C., Lutze, H. V., Schmidt, T. C., Rosenhahn, A., and Schmitz, O. J. (2020). Effect of Ozone Stress on the Intracellular Metabolites from Cobetia Marina. Anal. Bioanal. Chem. 412, 5853–5861. doi:10.1007/s00216-020-02810-6
Li, X., and Imlay, J. A. (2018). Improved Measurements of Scant Hydrogen Peroxide Enable Experiments that Define its Threshold of Toxicity for Escherichia coli. Free Radic. Biol. Med. 120, 217–227. doi:10.1016/j.freeradbiomed.2018.03.025
Li, Z., Reimers, S., Pandit, S., and Deutscher, M. P. (2002). RNA Quality Control: Degradation of Defective Transfer RNA. EMBO J. 21, 1132–1138. doi:10.1093/emboj/21.5.1132
Ling, J., Reynolds, N., and Ibba, M. (2009). Aminoacyl-tRNA Synthesis and Translational Quality Control. Annu. Rev. Microbiol. 63, 61–78. doi:10.1146/annurev.micro.091208.073210
Ling, J., and Söll, D. (2010). Severe Oxidative Stress Induces Protein Mistranslation through Impairment of an Aminoacyl-tRNA Synthetase Editing Site. PNAS 107, 4028–4033. doi:10.1073/pnas.1000315107
Liu, M., Gong, X., Alluri, R. K., Wu, J., Sablo, T., and Li, Z. (2012). Characterization of RNA Damage under Oxidative Stress in Escherichia coli. Biol. Chem. 393, 123–132. doi:10.1515/hsz-2011-0247
Liu, M., Shi, X., Chen, C., Yu, L., and Sun, C. (2017). Responses of Microcystis Colonies of Different Sizes to Hydrogen Peroxide Stress. Toxins (Basel). 9, 306. doi:10.3390/toxins9100306
Liu, X., Ramsey, M. M., Chen, X., Koley, D., Whiteley, M., and Bard, A. J. (2011). Real-time Mapping of a Hydrogen Peroxide Concentration Profile across a Polymicrobial Bacterial Biofilm Using Scanning Electrochemical Microscopy. Proc. Natl. Acad. Sci. U. S. A. 108, 2668–2673. doi:10.1073/pnas.1018391108
Loprasert, S., Fuangthong, M., Whangsuk, W., Atichartpongkul, S., and Mongkolsuk, S. (2000). Molecular and Physiological Analysis of an OxyR-Regulated ahpC Promoter in Xanthomonas Campestris Pv. Phaseoli. Mol. Microbiol. 37, 1504–1514. doi:10.1046/j.1365-2958.2000.02107.x
Luca, M., Di Mauro, M., Di Mauro, M., and Luca, A. (2019). Gut Microbiota in Alzheimer’s Disease, Depression, and Type 2 Diabetes Mellitus: The Role of Oxidative Stress. Oxid. Med. Cel. Longev. 2019, 4730539. doi:10.1155/2019/4730539
Luo, L., Qi, M. S., Yao, S. Y., Cheng, H. P., Zhu, J. B., and Yu, G. Q. (2005). Role of oxyR from Sinorhizobium Meliloti in Regulating the Expression of Catalases. Acta Biochim. Biophys. Sin. (Shanghai). 37, 421–428. doi:10.1111/j.1745-7270.2005.00055.x
Lyons, T. W., Reinhard, C. T., and Planavsky, N. J. (2014). The Rise of Oxygen in Earth’s Early Ocean and Atmosphere. Nature 506, 307–315. doi:10.1038/nature13068
Mandava, C. S., Peisker, K., Ederth, J., Kumar, R., Ge, X., Szaflarski, W., et al. (2012). Bacterial Ribosome Requires Multiple L12 Dimers for Efficient Initiation and Elongation of Protein Synthesis Involving IF2 and EF-G. Nucleic Acids Res. 40, 2054–2064. doi:10.1093/nar/gkr1031
Massey, V., Strickland, S., Mayhew, S. G., Howell, L. G., Engel, P. C., Matthews, R. G., et al. (1969). The Production of Superoxide Anion Radicals in the Reaction of Reduced Flavins and Flavoproteins with Molecular Oxygen. Biochem. Biophys. Res. Commun. 36, 891–897. doi:10.1016/0006-291X(69)90287-3
McDermott, M., Chiesa, R., Dillon, J., McDermott, M., and Roberts, J. E. (1991). Photooxidation of Specific Residues in α-Crystallin Polypeptides. Biochemistry 30, 8653–8660. doi:10.1021/bi00099a023
Melnikov, S., Ben-Shem, A., Garreau De Loubresse, N., Jenner, L., Yusupova, G., and Yusupov, M. (2012). One Core, Two Shells: Bacterial and Eukaryotic Ribosomes. Nat. Struct. Mol. Biol. 19, 560–567. doi:10.1038/nsmb.2313
Messner, K. R., and Imlay, J. A. (1999). The Identification of Primary Sites of Superoxide and Hydrogen Peroxide Formation in the Aerobic Respiratory Chain and Sulfite Reductase Complex of Escherichia coli. J. Biol. Chem. 274, 10119–10128. doi:10.1074/jbc.274.15.10119
Michaeli, A., and Feitelson, J. (1994). Reactivity of Singlet Oxygen toward Amino Acids and Peptides. Photochem. Photobiol. 59, 284–289. doi:10.1111/j.1751-1097.1994.tb05035.x
Michaeli, A., and Feitelson, J. (1995). Reactivity of Singlet Oxygen toward Large Peptides. Photochem. Photobiol. 61, 255–260. doi:10.1111/j.1751-1097.1995.tb03968.x
Mikula, P., Zezulka, S., Jancula, D., and Marsalek, B. (2012). Metabolic Activity and Membrane Integrity Changes in Microcystis Aeruginosa – New Findings on Hydrogen Peroxide Toxicity in Cyanobacteria. Eur. J. Phycol. 47, 195–206. doi:10.1080/09670262.2012.687144
Mostertz, J., and Hecker, M. (2003). Patterns of Protein Carbonylation Following Oxidative Stress in Wild-type and sigB Bacillus Subtilis Cells. Mol. Genet. Genomics 269, 640–648. doi:10.1007/s00438-003-0877-4
Nagano, T., Yutthanasirikul, R., Hihara, Y., Hisabori, T., Kanamori, T., Takeuchi, N., et al. (2015). Oxidation of Translation Factor EF-G Transiently Retards the Translational Elongation Cycle in Escherichia coli. J. Biochem. 158, 165–172. doi:10.1093/jb/mvv026
Nie, W., Wang, S., He, R., Xu, Q., Wang, P., Wu, Y., et al. (2020). A-to-I RNA Editing in Bacteria Increases Pathogenicity and Tolerance to Oxidative Stress. PLOS Pathog. 16, e1008740. doi:10.1371/journal.ppat.1008740
Nishiyama, Y., Allakhverdiev, S. I., and Murata, N. (2006). A New Paradigm for the Action of Reactive Oxygen Species in the Photoinhibition of Photosystem II. Biochim. Biophys. Acta - Bioenerg. 1757, 742–749. doi:10.1016/j.bbabio.2006.05.013
Nishiyama, Y., Allakhverdiev, S. I., Yamamoto, H., Hayashi, H., and Murata, N. (2004). Singlet Oxygen Inhibits the Repair of Photosystem II by Suppressing the Translation Elongation of the D1 Protein in Synechocystis sp. PCC 6803. Biochemistry 43, 11321–11330. doi:10.1021/bi036178q
Nishiyama, Y., Yamamoto, H., Allakhverdiev, S. I., Inaba, M., Yokota, A., and Murata, N. (2001). Oxidative Stress Inhibits the Repair of Photodamage to the Photosynthetic Machinery. EMBO J. 20, 5587–5594. doi:10.1093/emboj/20.20.5587
Nuss, A. M., Glaeser, J., Berghoff, B. A., and Klug, G. (2010). Overlapping Alternative Sigma Factor Regulons in the Response to Singlet Oxygen in Rhodobacter Sphaeroides. J. Bacteriol. 192, 2613–2623. doi:10.1128/JB.01605-09
Nuss, A. M., Glaeser, J., and Klug, G. (2009). RpoHII Activates Oxidative-Stress Defense Systems and is Controlled by RpoE in the Singlet Oxygen-dependent Response in Rhodobacter Sphaeroides. J. Bacteriol. 91, 220–230. doi:10.1128/JB.00925-08
Nyström, T. (2005). Role of Oxidative Carbonylation in Protein Quality Control and Senescence. EMBO J. 24, 1311–1317. doi:10.1038/sj.emboj.7600599
Orgel, L. E. (1998). The Origin of Life-a Review of Facts and Speculations. Trends Biochem. Sci. 23, 491–495. doi:10.1016/S0968-0004(98)01300-0
Paiva, C. N., and Bozza, M. T. (2014). Are Reactive Oxygen Species Always Detrimental to Pathogens? Antioxid. Redox Signal 20, 1000–1034. doi:10.1089/ars.2013.5447
Park, S., You, X., and Imlay, J. A. (2005). Substantial DNA Damage from Submicromolar Intracellular Hydrogen Peroxide Detected in Hpx- Mutants of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 102, 9317–9322. doi:10.1073/pnas.0502051102
Patil, S., Valdramidis, V. P., Karatzas, K. A. G., Cullen, P. J., and Bourke, P. (2011). Assessing the Microbial Oxidative Stress Mechanism of Ozone Treatment through the Responses of Escherichia coli Mutants. J. Appl. Microbiol. 111, 136–144. doi:10.1111/j.1365-2672.2011.05021.x
Peng, T., Berghoff, B. A., Oh, J. Il., Weber, L., Schirmer, J., Schwarz, J., et al. (2016). Regulation of a Polyamine Transporter by the Conserved 3′ UTR-Derived sRNA SorX Confers Resistance to Singlet Oxygen and Organic Hydroperoxides in Rhodobacter Sphaeroides. RNA Biol. 13, 988–999. doi:10.1080/15476286.2016.1212152
Polacek, N., and Barta, A. (1998). Metal ion Probing of rRNAs: Evidence for Evolutionarily Conserved Divalent Cation Binding Pockets. RNA 4, 1282–1294. doi:10.1017/S1355838298980347
Pulk, A., Liiv, A., Peil, L., Maiväli, Ü., Nierhaus, K., and Remme, J. (2010). Ribosome Reactivation by Replacement of Damaged Proteins. Mol. Microbiol. 75, 801–814. doi:10.1111/j.1365-2958.2009.07002.x
Rai, P., Cole, T. D., Wemmer, D. E., and Linn, S. (2001). Localization of Fe2+ at an RTGR Sequence within a DNA Duplex Explains Preferential Cleavage by Fe2+ and H2O2. J. Mol. Biol. 312, 1089–1101. doi:10.1006/jmbi.2001.5010
Ravanat, J.-L., Saint-Pierre, C., Di Mascio, P., Martinez, G. R., Medeiros, M. H. G., and Cadet, J. (2001). Damage to Isolated DNA Mediated by Singlet Oxygen. Helv. Chim. Acta 84, 3702–3709. doi:10.1002/1522-2675(20011219)84:12<3702::AID-HLCA3702>3.0
Rinalducci, S., Pedersen, J. Z., and Zolla, L. (2008). Generation of Reactive Oxygen Species upon Strong Visible Light Irradiation of Isolated Phycobilisomes from Synechocystis PCC 6803. Biochim. Biophys. Acta - Bioenerg. 1777, 417–424. doi:10.1016/j.bbabio.2008.02.005
Rodnina, M. V. (2018). Translation in Prokaryotes. Cold Spring Harb. Perspect. Biol. 10, 1–22. doi:10.1101/cshperspect.a032664
Schirrmeister, B. E., Gugger, M., and Donoghue, P. C. J. (2015). Cyanobacteria and the Great Oxidation Event: Evidence from Genes and Fossils. Palaeontol. 58, 769–785. doi:10.1111/pala.12178
Segal, A. W. (2008). The Function of the NADPH Oxidase of Phagocytes and its Relationship to Other NOXs in Plants, Invertebrates, and Mammals. Int. J. Biochem. Cel Biol. 40, 604–618. doi:10.1016/j.biocel.2007.10.003
Seo, S. W., Kim, D., Szubin, R., and Palsson, B. O. (2015). Genome-wide Reconstruction of OxyR and SoxRS Transcriptional Regulatory Networks under Oxidative Stress in Escherichia coli K-12 MG1655. Cell Rep. 12, 1289–1299. doi:10.1016/j.celrep.2015.07.043
Simms, C. L., Hudson, B. H., Mosior, J. W., Rangwala, A. S., and Zaher, H. S. (2014). An Active Role for the Ribosome in Determining the Fate of Oxidized mRNA. Cell Rep. 9, 1256–1264. doi:10.1016/j.celrep.2014.10.042
Singh, N. K., Sonani, R. R., Prasad Rastogi, R., and Madamwar, D. (2015). The Phycobilisomes: An Early Requisite for Efficient Photosynthesis in Cyanobacteria. EXCLI J. 14, 268–289. doi:10.17179/excli2014-723
Sjöberg, B., Foley, S., Staicu, A., Pascu, A., Pascu, M., and Enescu, M. (2016). Protein Reactivity with Singlet Oxygen: Influence of the Solvent Exposure of the Reactive Amino Acid Residues. J. Photochem. Photobiol. B Biol. 159, 106–110. doi:10.1016/j.jphotobiol.2016.03.036
Skovsen, E., Snyder, J. W., Lambert, J. D. C., and Ogilby, P. R. (2005). Lifetime and Diffusion of Singlet Oxygen in a Cell. J. Phys. Chem. B 109, 8570–8573. doi:10.1021/jp051163i
Smethurst, D. G. J., Kovalev, N., McKenzie, E. R., Pestov, D. G., and Shcherbik, N. (2020). Iron-Mediated Degradation of Ribosomes under Oxidative Stress is Attenuated by Manganese. J. Biol. Chem. 295, 015025. doi:10.1074/jbc.ra120.015025
Stadtman, E. R., and Levine, R. L. (2003). Free Radical-Mediated Oxidation of Free Amino Acids and Amino Acid Residues in Proteins. Amino Acids 25, 207–218. doi:10.1007/s00726-003-0011-2
Steele-Mortimer, O. (2008). The Salmonella-Containing Vacuole—Moving with the Times. Curr. Opin. Microbiol. 11, 38–45. doi:10.1016/j.mib.2008.01.002
Steiner, R. E., Kyle, A. M., and Ibba, M. (2019). Oxidation of Phenylalanyl-tRNA Synthetase Positively Regulates Translational Quality Control. Proc. Natl. Acad. Sci. U. S. A. 116, 10058–10063. doi:10.1073/pnas.1901634116
Sun, C., Jora, M., Solivio, B., Limbach, P. A., and Addepalli, B. (2018). The Effects of Ultraviolet Radiation on Nucleoside Modifications in RNA. ACS Chem. Biol. 13, 567–572. doi:10.1021/acschembio.7b00898
Svenningsen, S. L., Kongstad, M., Stenum, T. S., Muñoz-Gómez, A. J., and Sørensen, M. A. (2017). Transfer RNA is Highly Unstable during Early Amino Acid Starvation in Escherichia coli. Nucleic Acids Res. 45, 793–804. doi:10.1093/nar/gkw1169
Taddei, F., Hayakawa, H., Bouton, M. F., Cirinesi, A. M., Matic, I., Sekiguchi, M., et al. (1997). Counteraction by MutT Protein of Transcriptional Errors Caused by Oxidative Damage. Science 278, 128–130. doi:10.1126/science.278.5335.128
Tamarit, J., Cabiscol, E., and Ros, J. (1998). Identification of the Major Oxidatively Damaged Proteins in Escherichia coli Cells Exposed to Oxidative Stress. J. Biol. Chem. 273, 3027–3032. doi:10.1074/jbc.273.5.3027
Teramoto, H., Inui, M., and Yukawa, H. (2013). OxyR Acts as a Transcriptional Repressor of Hydrogen Peroxide-Inducible Antioxidant Genes in Corynebacterium Glutamicum R. FEBS J. 280, 3298–3312. doi:10.1111/febs.12312
Thomas, E. N., Simms, C. L., Keedy, H. E., and Zaher, H. S. (2019). Insights into the Base-Pairing Preferences of 8-oxoguanosine on the Ribosome. Nucleic Acids Res. 47, 9857–9870. doi:10.1093/nar/gkz701
Tong, H., Chen, W., Merritt, J., Qi, F., Shi, W., and Dong, X. (2007). Streptococcus Oligofermentans Inhibits Streptococcus Mutans through Conversion of Lactic Acid into Inhibitory H2O2: A Possible Counteroffensive Strategy for Interspecies Competition. Mol. Microbiol. 63, 872–880. doi:10.1111/j.1365-2958.2006.05546.x
Torres, A. G., Piñeyro, D., Filonava, L., Stracker, T. H., Batlle, E., and Ribas De Pouplana, L. (2014). A-to-I Editing on tRNAs: Biochemical, Biological and Evolutionary Implications. FEBS Lett. 588, 4279–4286. doi:10.1016/j.febslet.2014.09.025
Torres, M. A., Jones, J. D. G., and Dangl, J. L. (2006). Reactive Oxygen Species Signaling in Response to Pathogens. Plant Physiol. 141, 373–378. doi:10.1104/pp.106.079467
Traoré, D. A. K., Ghazouani, A. El., Jacquamet, L., Borel, F., Ferrer, J. L., Lascoux, D., et al. (2009). Structural and Functional Characterization of 2-oxo-Histidine in Oxidized PerR Protein. Nat. Chem. Biol. 5, 53–59. doi:10.1038/nchembio.133
Turner, J. M., and Messenger, A. J. (1986). Occurrence, Biochemistry and Physiology of Phenazine Pigment Production. Adv. Microb. Physiol. 27, 211–275. doi:10.1016/s0065-2911(08)60306-9
Van Vliet, A. H. M., Baillon, M. L. A., Penn, C. W., and Ketley, J. M. (1999). Campylobacter Jejuni Contains Two Fur Homologs: Characterization of Iron-Responsive Regulation of Peroxide Stress Defense Genes by the PerR Repressor. J. Bacteriol. 181, 6371–6376. doi:10.1128/jb.181.20.6371-6376.1999
Vargas-Blanco, D. A., and Shell, S. S. (2020). Regulation of mRNA Stability during Bacterial Stress Responses. Front. Microbiol. 11, 2111. doi:10.3389/fmicb.2020.02111
Wang, G., Conover, R. C., Benoit, S., Olczak, A. A., Olson, J. W., Johnson, M. K., et al. (2004). Role of a Bacterial Organic Hydroperoxide Detoxification System in Preventing Catalase Inactivation. J. Biol. Chem. 279, 51908–51914. doi:10.1074/jbc.M408450200
Wang, G., Hong, Y., Johnson, M. K., and Maier, R. J. (2006). Lipid Peroxidation as a Source of Oxidative Damage in Helicobacter pylori: Protective Roles of Peroxiredoxins. Biochim. Biophys. Acta - Gen. Subj. 1760, 1596–1603. doi:10.1016/j.bbagen.2006.05.005
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N 6-methyladenosine-dependent Regulation of Messenger RNA Stability. Nature 505, 117–120. doi:10.1038/nature12730
Ward, R. J., Zucca, F. A., Duyn, J. H., Crichton, R. R., and Zecca, L. (2014). The Role of Iron in Brain Ageing and Neurodegenerative Disorders. Lancet Neurol. 13, 1045–1060. doi:10.1016/S1474-4422(14)70117-6
Willi, J., Küpfer, P., Evéquoz, D., Fernandez, G., Katz, A., Leumann, C., et al. (2018). Oxidative Stress Damages rRNA inside the Ribosome and Differentially Affects the Catalytic Center. Nucleic Acids Res. 46, 1945–1957. doi:10.1093/nar/gkx1308
Wilson, C. L., Hinman, N. W., Cooper, W. J., and Brown, C. F. (2000a). Hydrogen Peroxide Cycling in Surface Geothermal Waters of Yellowstone National Park. Environ. Sci. Technol. 34, 2655–2662. doi:10.1021/es9906397
Wilson, C. L., Hinman, N. W., and Sheridan, R. P. (2000b). Hydrogen Peroxide Formation and Decay in Iron-Rich Geothermal Waters: The Relative Roles of Abiotic and Biotic Mechanisms. Photochem. Photobiol. 71, 699. doi:10.1562/0031-8655(2000)071<0691:hpfadi>2.0.co.2
Winter, D., Polacek, N., Halama, I., Streicher, B., and Barta, A. (1997). Lead-Catalysed Specific Cleavage of Ribosomal RNAs. Nucleic Acids Res. 25, 1817–1824. doi:10.1093/nar/25.9.1817
Winterbourn, C. C. (2013). The biological chemistry of hydrogen peroxide. Meth. Enzymol. 528, 3–25. doi:10.1016/B978-0-12-405881-1.00001-X
Wu, J., Fan, Y., and Ling, J. (2014). Mechanism of Oxidant-Induced Mistranslation by Threonyl-tRNA Synthetase. Nucleic Acids Res. 42, 6523–6531. doi:10.1093/nar/gku271
Wu, J., Jiang, Z., Liu, M., Gong, X., Wu, S., Burns, C. M., et al. (2009). Polynucleotide Phosphorylase Protects Escherichia coli against Oxidative Stress. Biochemistry 48, 2012–2020. doi:10.1021/bi801752p
Yeremenko, N., Kouřil, R., Ihalainen, J. A., D’Haene, S., Van Oosterwijk, N., Andrizhiyevskaya, E. G., et al. (2004). Supramolecular Organization and Dual Function of the IsiA Chlorophyll-Binding Protein in Cyanobacteria. Biochemistry 43, 10308–10313. doi:10.1021/bi048772l
Yutthanasirikul, R., Nagano, T., Jimbo, H., Hihara, Y., Kanamori, T., Ueda, T., et al. (2016). Oxidation of a Cysteine Residue in Elongation Factor EF-Tu Reversibly Inhibits Translation in the Cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 291, 5860–5870. doi:10.1074/jbc.M115.706424
Zheng, H., Shabalin, I. G., Handing, K. B., Bujnicki, J. M., and Minor, W. (2015). Magnesium-Binding Architectures in RNA Crystal Structures: Validation, Binding Preferences, Classification and Motif Detection. Nucleic Acids Res. 43, 3789–3801. doi:10.1093/nar/gkv225
Zheng, M., Wang, X., Templeton, L. J., Smulski, D. R., LaRossa, R. A., and Storz, G. (2001). DNA Microarray-Mediated Transcriptional Profiling of the Escherichia coli Response to Hydrogen Peroxide. J. Bacteriol. 183, 4562–4570. doi:10.1128/JB.183.15.4562-4570.2001
Zhong, J., Xiao, C., Gu, W., Du, G., Sun, X., He, Q.-Y., et al. (2015). Transfer RNAs Mediate the Rapid Adaptation of Escherichia coli to Oxidative Stress. Plos Genet. 11, e1005302. doi:10.1371/journal.pgen.1005302
Zhu, M., and Dai, X. (2019). Maintenance of Translational Elongation Rate Underlies the Survival of Escherichia coli during Oxidative Stress. Nucleic Acids Res. 47, 7592–7604. doi:10.1093/nar/gkz467
Keywords: oxidative stress, ROS, oxidative damage, DNA damage, RNA damage, protein damage
Citation: Fasnacht M and Polacek N (2021) Oxidative Stress in Bacteria and the Central Dogma of Molecular Biology. Front. Mol. Biosci. 8:671037. doi: 10.3389/fmolb.2021.671037
Received: 22 February 2021; Accepted: 26 April 2021;
Published: 10 May 2021.
Edited by:
Claudio Scazzocchio, Imperial College London, United KingdomReviewed by:
Esa Tyystjärvi, University of Turku, FinlandSunny Sharma, Rutgers, The State University of New Jersey, United States
Copyright © 2021 Fasnacht and Polacek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Norbert Polacek, bm9yYmVydC5wb2xhY2VrQGRjYi51bmliZS5jaA==
 Michel Fasnacht
Michel Fasnacht Norbert Polacek
Norbert Polacek