
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci., 15 April 2021
Sec. Protein Folding, Misfolding and Degradation
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.664241
This article is part of the Research TopicFunctions and Mechanisms of Bacterial Protein Homeostasis and Stress ResponsesView all 24 articles
The spatial and temporal coordination of protein transport is an essential cornerstone of the bacterial adaptation to different environmental conditions. By adjusting the protein composition of extra-cytosolic compartments, like the inner and outer membranes or the periplasmic space, protein transport mechanisms help shaping protein homeostasis in response to various metabolic cues. The universally conserved SecYEG translocon acts at the center of bacterial protein transport and mediates the translocation of newly synthesized proteins into and across the cytoplasmic membrane. The ability of the SecYEG translocon to transport an enormous variety of different substrates is in part determined by its ability to interact with multiple targeting factors, chaperones and accessory proteins. These interactions are crucial for the assisted passage of newly synthesized proteins from the cytosol into the different bacterial compartments. In this review, we summarize the current knowledge about SecYEG-mediated protein transport, primarily in the model organism Escherichia coli, and describe the dynamic interaction of the SecYEG translocon with its multiple partner proteins. We furthermore highlight how protein transport is regulated and explore recent developments in using the SecYEG translocon as an antimicrobial target.
The dynamic control of protein synthesis, folding and degradation under different environmental conditions is essential for maintaining a functional proteome in eu- and prokaryotic cells (Mogk et al., 2011; Song et al., 2020). Protein trafficking pathways expand this proteostasis network and target proteins into subcellular compartments with specific folding conditions (Figure 1; Kudva et al., 2013; Tsirigotaki et al., 2017). Cell compartmentalization is a unifying principle in all cells and diversifies their metabolic activity by generating membrane-bordered reaction chambers. Prokaryotes lack the sophisticated intracellular organization that is usually observed in eukaryotes, but still maintain distinct compartments like the cytosol, the inner membrane, the periplasm and in Gram-negative bacteria also the outer membrane (Figure 1). Each extra-cytosolic compartment contains a dedicated protein composition which can only be maintained due to the presence of protein transport systems that export proteins out of the cytosol. The Gram-negative model organism Escherichia coli synthesizes approx. 4.400 different proteins1 and contains a predicted total number of 3–4 × 106 proteins per cell, calculated based on cell volume, average protein mass and average cellular protein concentration (Milo, 2013). Ribosome profiling studies suggest that roughly one third of these proteins, accounting to approx. 1.5 × 106 proteins per cell, execute their function outside of the cytosol (Li et al., 2014). The STEPdb databank of subcellular topologies of E. coli polypeptides2 lists approx. 1,000 different inner membrane proteins, approx. 400 periplasmic proteins and approx. 160 outer membrane proteins (OMPs) (Loos et al., 2019), all of which have in common the requirement for dedicated protein transport systems. N-terminal, cleavable signal sequences in secretory proteins and non-cleavable signal anchor sequences in inner membrane proteins provide the means to identify those proteins that have to be exported (Pugsley, 1990; von Heijne, 1994; Hegde and Bernstein, 2006; Steinberg et al., 2018).
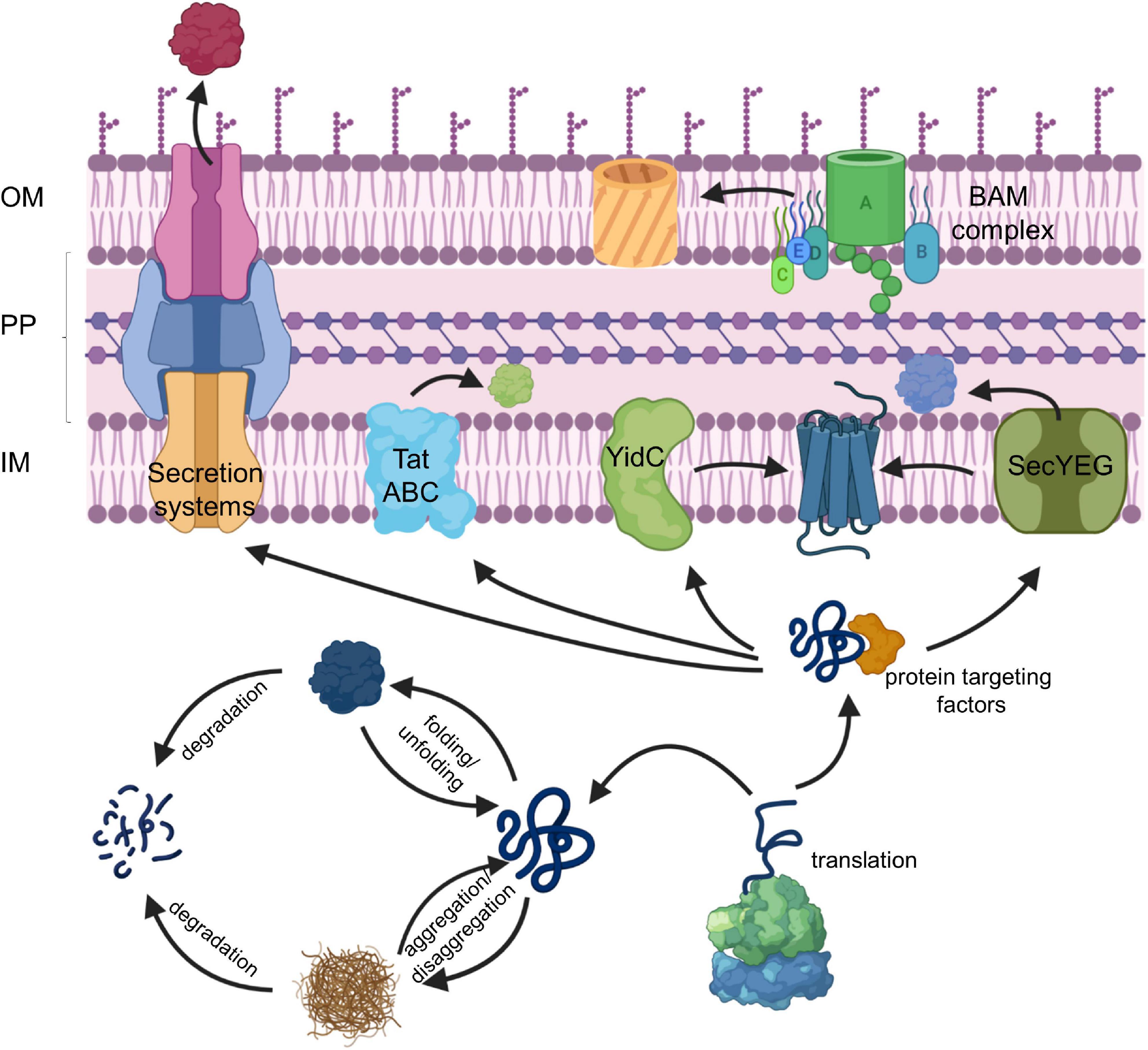
Figure 1. The proteostasis network in bacteria. For details see text. Secretion systems refer to the type I–IX protein secretion systems that have been identified in bacteria, although some of these secretion systems are only found in some species (Christie, 2019). IM, inner membrane; PP, periplasm; OM, outer membrane.
The majority of exported proteins engage the SecYEG translocon, a universally conserved protein transport channel that resides in the inner bacterial membrane and facilitates the insertion of membrane proteins into the inner membrane as well as the translocation of proteins across the inner membrane into the periplasm (Figure 1; Kudva et al., 2013; Denks et al., 2014). The heterotrimeric SecYEG translocon consists of SecY, SecE, and SecG as core proteins, but constitutes only a passive and sealed pore that connects the cytoplasm to the periplasm and the lipid phase of the membrane. For being active in protein transport, the SecYEG translocon depends on the coordinated interaction with multiple partner proteins that select potential SecYEG substrates (Lill et al., 1990; van der Does et al., 1996; Angelini et al., 2005), provide the driving force for protein transport (Tsukazaki et al., 2011; Knyazev et al., 2018), coordinate substrate release from the SecYEG channel (Beck et al., 2001; Houben et al., 2004; Sachelaru et al., 2017) and communicate with components of the proteostasis network (Kihara et al., 1996; Schäfer et al., 1999; Jauss et al., 2019). The SecYEG translocon also cooperates with additional protein transport systems (Figure 1), like the YidC insertase (Scotti et al., 2000; Sachelaru et al., 2015, 2017; Dalbey et al., 2017; Petriman et al., 2018), the Tat transport machinery (Keller et al., 2012; Kudva et al., 2013; Tooke et al., 2017) and the Bam complex (Wang et al., 2016; Alvira et al., 2020), which inserts β-barrel proteins into the outer membrane. Additional partner proteins of the SecYEG translocon have been recently identified by proteomic approaches (Chorev et al., 2018; Carlson et al., 2019; Jauss et al., 2019), further highlighting the dynamic nature of the SecYEG translocon, which is probably the basis for its ability to transport a large variety of highly different substrates.
The selective recognition of SecYEG substrates is achieved by two protein targeting systems that operate in parallel in bacterial cells (Koch et al., 2003; Rapoport, 2007; Driessen and Nouwen, 2008; Kudva et al., 2013; Smets et al., 2019). SecA-dependent protein targeting primarily acts on secretory proteins that contain a cleavable N-terminal signal sequence and this pathway is generally described as post-translational event (Figure 2). In contrast, inner membrane proteins with non-cleavable signal anchor sequences engage the signal recognition particle (SRP)-dependent targeting pathway, which operates primarily co-translationally and involves the ribosome-bound SRP (Pool et al., 2002; Gu et al., 2003; Halic et al., 2004; Schaffitzel et al., 2006) and the SecYEG-bound SRP receptor FtsY (Angelini et al., 2005, 2006; Kuhn et al., 2015; Draycheva et al., 2016; Steinberg et al., 2018; Figure 2). The SRP pathway can deliver membrane proteins also to the YidC insertase (Welte et al., 2012; Dalbey et al., 2017; McDowell et al., 2021), which can insert membrane proteins independently of SecYEG but also cooperates with the SecYEG translocon (Houben et al., 2000; Scotti et al., 2000; Serek et al., 2004; du Plessis et al., 2006; Yuan et al., 2007; Sachelaru et al., 2015, 2017; Dalbey et al., 2017). It is important to emphasize that the classification into post-translational targeting by SecA and co-translational targeting by SRP does not apply to all substrates. A co-translational targeting by SecA has been observed for the inner membrane protein RodZ, which contains a large cytosolic domain preceding its single transmembrane domain (Rawat et al., 2015; Wang et al., 2017), and for the periplasmic maltose binding protein MBP (Huber et al., 2017). This is in line with the ability of SecA to interact with translating and non-translating ribosomes (Eisner et al., 2003; Karamyshev and Johnson, 2005; Huber et al., 2011; Knupffer et al., 2019; Origi et al., 2019; Wang S. et al., 2019). On the other hand, a post-translational interaction of SRP has been shown for the small bacterial membrane proteins YohP and YkgR (Steinberg et al., 2020) and for the tail-anchored proteins DjlC, Flk, and SciP (Pross et al., 2016; Peschke et al., 2018).
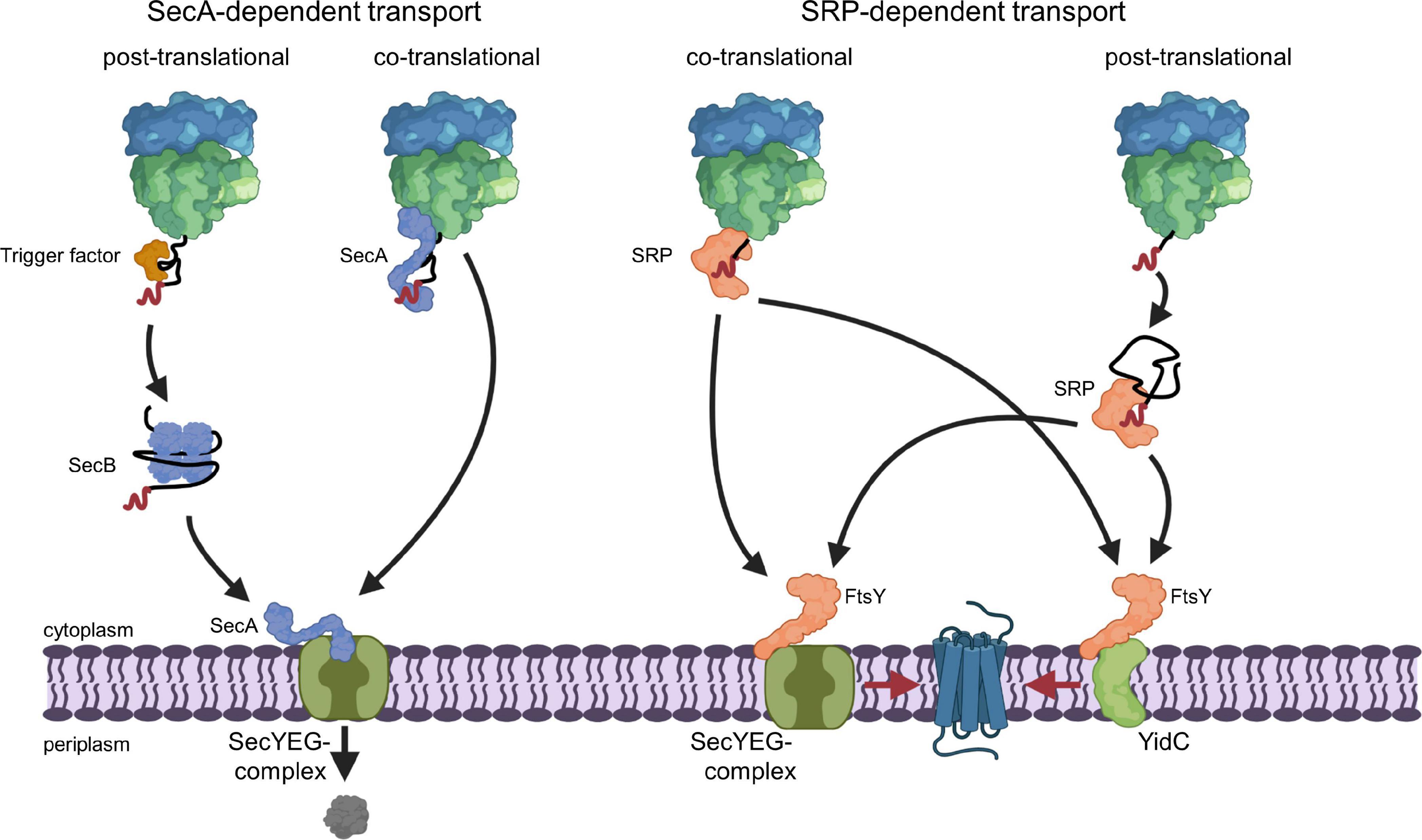
Figure 2. SecA- and SRP-dependent protein targeting in bacteria. The SecA- and SRP-dependent protein targeting pathways constitute the two main protein targeting pathways in bacteria and both can operate in a co- or post-translational mode. However, post-translational targeting of secretory proteins by SecA and co-translational targeting of membrane proteins by SRP are the preferred modes. Substrates of the post-translational SecA pathway are kept in a translocation competent state by chaperones, like the ribosome-bound TF or the cytosolic SecB. SecA serves as receptor for signal sequences (shown in red) of secretory proteins and is bound to the SecYEG translocon, which serves as main protein transport channel in bacteria. Repetitive ATP hydrolysis cycles by SecA allows for the translocation of the polypeptide across the SecY channel. SecA can also associate with the ribosome and target potential substrates co-translationally to the SecYEG translocon. The subsequent ATP-dependent translocation likely occurs then post-translationally, i.e., after the substrate is released from the ribosome. SRP binds with high affinity to translating ribosomes and traps the signal anchor sequence of a membrane protein when it emerges from the ribosomal peptide tunnel. SRP then delivers the translating ribosome (ribosome-associated nascent chain, RNC) to the SRP receptor FtsY. FtsY serves as SecYEG-bound receptor for nascent membrane proteins and engages similar binding sites as SecA on the SecYEG translocon. After SRP-FtsY contact, the translating ribosome docks onto the SecYEG translocon and ongoing translation inserts the protein into the lipid phase. FtsY can also associate with the YidC insertase and SRP can deliver less complex membrane proteins co-translationally to the YidC insertase for insertion. Small membrane proteins (<50 amino acids) and likely tail-anchored membrane proteins are post-translationally bound by SRP and targeted to SecYEG or YidC only after they have been released from the ribosome. This post-translational insertion by SRP is likely initiated by a so far largely uncharacterized mRNA-targeting step (Steinberg et al., 2020), which is not depicted in this cartoon.
The ATPase SecA is a multi-domain protein of 102 kDa that is found exclusively in bacteria and chloroplasts (Pohlschroder et al., 1997; Figure 3A). In E. coli it is present in about 2,000–5,000 copies per cell (Kudva et al., 2013; Smets et al., 2019) and therefore much more abundant than the SecYEG complex, which exists in about 500 copies (Kudva et al., 2013). SecA binds with high-affinity to the cytosolic loops of SecY (Douville et al., 1995; Mori and Ito, 2006; Kuhn et al., 2011) and to negatively charged phospholipids (Lill et al., 1990; Gold et al., 2010; Koch et al., 2016, 2019). In addition, a fraction of SecA is located in the cytosol (Chun and Randall, 1994; Hoffschulte et al., 1994), where it can exist as dimer (Woodbury et al., 2002; Banerjee et al., 2017a). The oligomeric state of membrane-bound SecA is controversially discussed. Liposome studies indicate that only the SecA monomer binds to phospholipids (Roussel and White, 2020), but a SecA dimer is functional in protein translocation (de Keyzer et al., 2005) and can function as receptor for preproteins (Gouridis et al., 2013). It has been suggested that one protomer is required for docking onto the SecYEG complex, while the second copy is involved in the downstream translocation upon ATP-dependent dissociation of the dimer (Or et al., 2002; Gouridis et al., 2013).
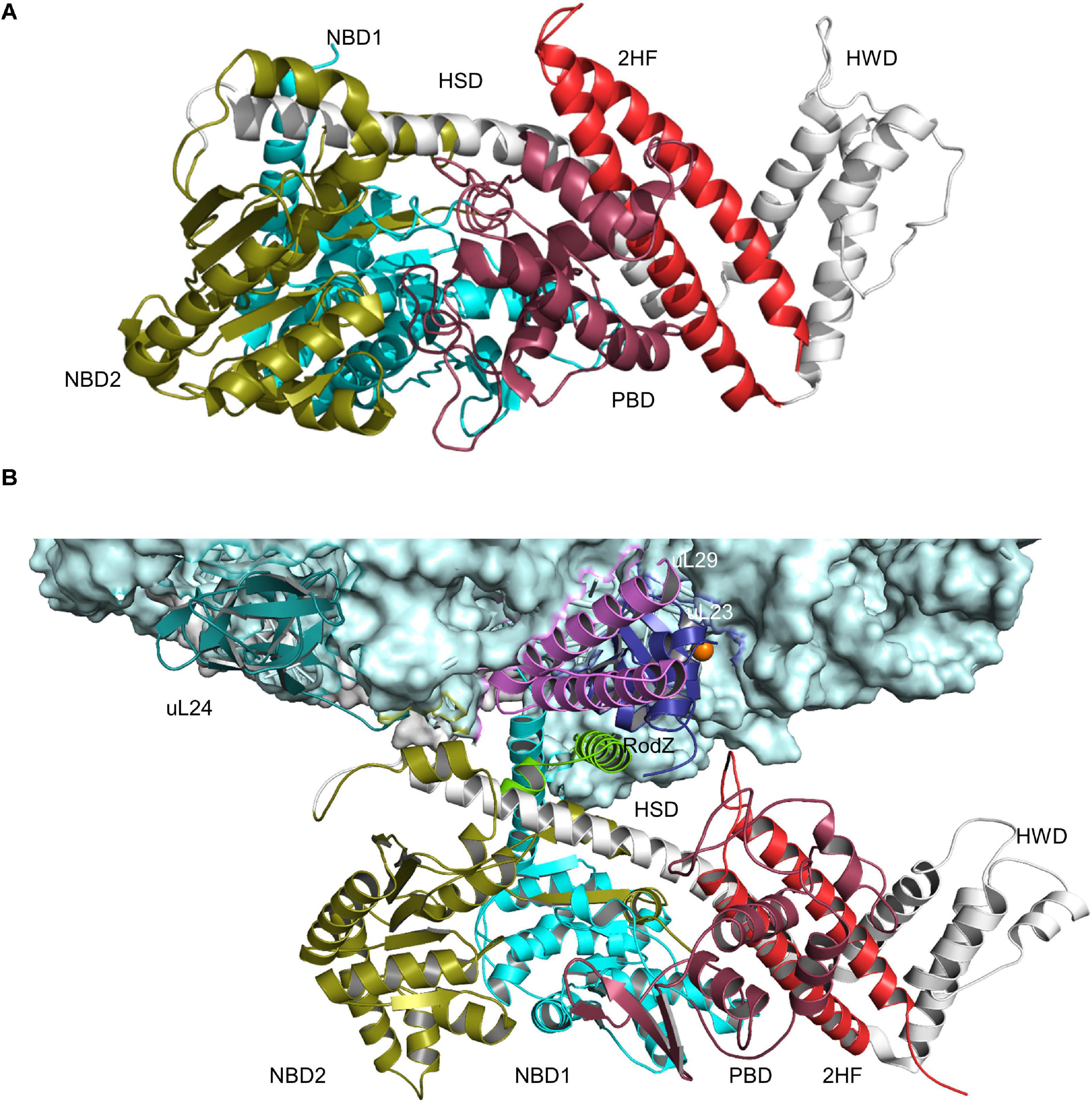
Figure 3. Structures of SecA and SecA bound to the ribosome. (A) Structure of B. subtilis SecA (PDB 5EUL), showing its multiple domains. The two nucleotide-binding domains NBD1 and NBD2 are shown in cyan and in olive, respectively. The peptide-binding domain (PBD) is shown in raspberry-red, the helical wing domain (HWD) and the helical scaffold domain (HSD) in gray and the two-helix finger (2HF) in red. (B) Structure of E. coli SecA bound to a translating ribosome (PDB 6S0K). The 50S ribosomal subunit is shown in light-blue and the nascent RodZ chain in green. Ribosomal proteins that are in contact with SecA [uL23 (blue), uL29 (pink), and uL24 (green)] and the different domains of SecA are labeled and shown in the same color-code as in (A).
SecYEG-bound SecA primarily recognizes its substrates after they have been released from the ribosome (Randall, 1983; Hartl et al., 1990; Swidersky et al., 1990; Chun and Randall, 1994; Fekkes et al., 1998). N-terminal signal sequences are bound via a shallow groove within the preprotein-binding domain (PBD) of SecA, also called preprotein cross-linking domain (PPXD) (Gelis et al., 2007; Grady et al., 2012). The PBD domain is located close to the two nucleotide binding domains (NBD1 and NBD2) and dynamic movements within the PBD link substrate recognition to ATP binding and hydrolysis (Karamanou et al., 2007; Gouridis et al., 2013; Figure 3A). Although signal sequences are probably the most important determinants for SecA-dependent targeting (Hegde and Bernstein, 2006), additional sequences within the mature domain of a secretory protein can also contribute to the specificity of the targeting reaction (Chatzi et al., 2017). Binding of SecA to sequences within the mature domain might be in particular important for keeping substrates in a translocation competent state, e.g., largely unfolded. Translocation competence is furthermore supported by chaperones like Trigger factor (TF) (Saio et al., 2014, 2018; Can et al., 2017; De Geyter et al., 2020) or SecB (Bechtluft et al., 2010; Huang et al., 2016; Figure 2). Due to its high affinity to ribosomes and its ability to bind to the ribosomal protein uL23 (Kramer et al., 2002), TF is one of the first contacts of the emerging nascent chain (Deuerling et al., 1999; Bornemann et al., 2014). Different to SecA, TF does not specifically bind to signal sequence-containing proteins but also binds to cytosolic proteins, although β-barrel OMPs appear to be the preferred target (Teter et al., 1999; Oh et al., 2011). It has been shown that protein translocation of some substrates is accelerated upon TF deletion and it was suggested that this reflects prolonged contact between TF and these outer membrane substrates (Lee and Bernstein, 2002). TF can also interact with SecB and the SecYEG-bound SecA, which probably helps to connect protein folding and protein transport (De Geyter et al., 2020). SecB is present in proteobacteria only and like TF not essential (Deuerling et al., 2003; Crane and Randall, 2017). It forms a tetramer with surface-exposed hydrophobic areas, which are involved in substrate binding (Knoblauch et al., 1999). SecB binds only to a small number of secretory proteins and releases its substrates upon binding to the C-terminus of SecA (Baars et al., 2006; Crane et al., 2006; Castanie-Cornet et al., 2014).
In addition to this post-translational substrate recognition, SecA can bind to its substrates also co-translationally (Eisner et al., 2003; Karamyshev and Johnson, 2005; Huber et al., 2011, 2017; Figure 2). This was observed for secretory proteins, like MBP (Chun and Randall, 1994; Huber et al., 2017), but also for the membrane protein RodZ (Rawat et al., 2015; Wang et al., 2017). SecA binds to the ribosome close to the ribosomal tunnel exit, which is formed by the ribosomal proteins uL23, uL24, and uL29 (Huber et al., 2011; Knupffer et al., 2019; Wang S. et al., 2019; Figure 3B). This is also the binding site for SRP and for many ribosome-associated chaperones and processing factors (Kramer et al., 2002, 2009; Denks et al., 2017; Knupffer et al., 2019). Importantly, it is the N-terminus of SecA that interacts with both the ribosome and with SecYEG or phospholipids (Knupffer et al., 2019; Origi et al., 2019) and thus SecA binding to ribosomes or to SecYEG appears to be mutual exclusive. This suggests that co-translational targeting by SecA is followed by a post-translational translocation across the SecYEG translocon. This assumption is also in line with the observation that SecA and ribosomes use almost identical binding sites on SecY (Prinz et al., 2000; Mori and Ito, 2006; Kuhn et al., 2011; Banerjee et al., 2017b) and that SecA and ribosomes compete for SecYEG binding (Wu et al., 2012).
The SRP pathway is a universally conserved targeting system that bacteria primarily use for inner membrane proteins (Figure 2) (Ulbrandt et al., 1997; de Gier et al., 1998; Valent et al., 1998; Cristobal et al., 1999; Koch et al., 1999, 2003; Koch and Muller, 2000). In E. coli, SRP consists of the protein Ffh and the 4.5S RNA (Figure 4A) and thus represents a basic version of the eukaryotic SRP, which consists of six protein subunits bound to the 7SL RNA (Koch et al., 2003). Still, the bacterial SRP and its receptor FtsY are sufficient to support protein targeting to mammalian endosomal membranes (Powers and Walter, 1997). The SRP pathway in bacteria not only targets the SecYEG translocon, but also the YidC insertase (Welte et al., 2012; Petriman et al., 2018), which inserts less-complex membrane proteins (Samuelson et al., 2000; Dalbey et al., 2017). Ffh and FtsY share a homologous NG domain with highly similar architecture and amino acid sequence (Freymann et al., 1997; Montoya et al., 1997). The respective N-domains form a four-helix bundle that is followed by the Ras-like GTPase domain (G-domain) (Figure 4A). The NG-domain of Ffh is C-terminally continued by the M-domain, which forms a flexible groove that is able to accommodate signal anchor sequences of different lengths and hydrophobicities. This flexibility explains why the bacterial SRP recognizes the hydrophobic signal anchor sequences of basically all inner membrane proteins and also the signal sequences of some secretory proteins and amphipathic helices of integral and membrane-associated proteins (Beha et al., 2003; Huber et al., 2005; Maier et al., 2008; Lim et al., 2013; Schibich et al., 2016). Substrate recognition by SRP is a multi-step process that is initiated by SRP binding to the ribosome, where it contacts primarily uL23, uL29, and the 23S rRNA close to the tunnel exit (Halic et al., 2006a, b; Schaffitzel et al., 2006; Figure 4B). SRP binds to vacant ribosomes with high affinity (Kd 50–60 nM) (Bornemann et al., 2008; Holtkamp et al., 2012) and the flexible C-terminus of Ffh protrudes into the ribosomal tunnel where it contacts the intra-tunnel loop of uL23 (Jomaa et al., 2016, 2017; Denks et al., 2017). This scanning mode allows SRP to screen ribosomes for potential substrates. When translation is initiated and the nascent chain reaches a length of approx. 25 amino acids, SRP is displaced from the intra-tunnel loop, which now contacts the nascent chain (Denks et al., 2017). However, SRP maintains contact to the surface-exposed domain of uL23 and this anticipatory or stand-by mode further increases the affinity (Kd 1 nM) and likely orients the M-domain for binding to the signal anchor sequence. When the nascent chain reaches a length of approx. 45–50 amino acids and the signal anchor sequence is exposed to the outside of the ribosome, SRP forms a stable complex with the ribosome-associated nascent chain (RNC) (Kd ≤ 1 nM) (Holtkamp et al., 2012; Schibich et al., 2016; Denks et al., 2017). The SRP-RNC complex is then targeted to the SRP receptor FtsY. Although some initial studies proposed that the SRP-RNC complex interacts with FtsY already in the cytosol (Shan et al., 2007; Saraogi et al., 2014), FtsY in Gram-positive and Gram-negative bacteria is almost exclusively membrane-bound (Mircheva et al., 2009).
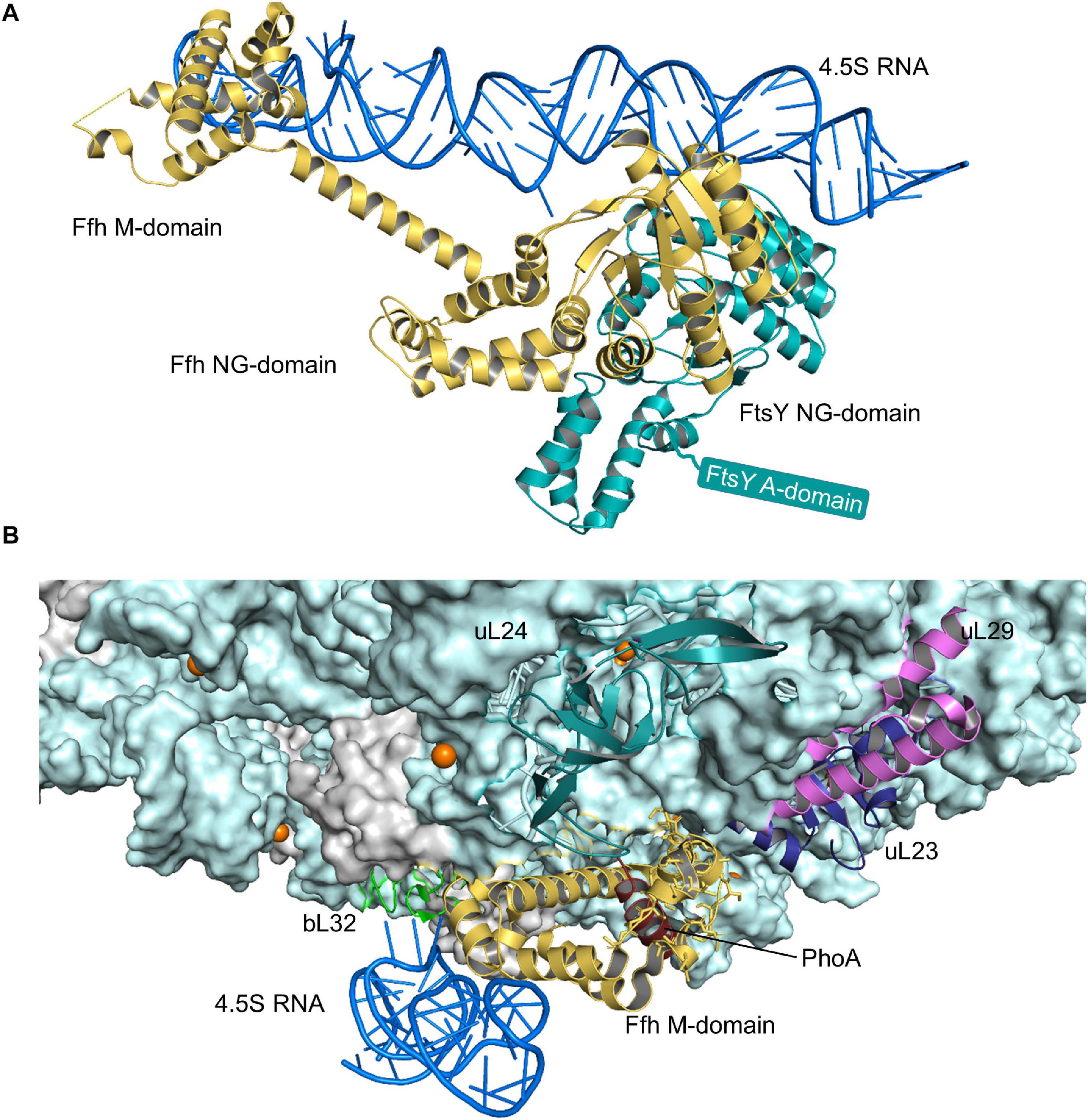
Figure 4. Structures of the SRP-FtsY-complex and the SRP-ribosome complex. (A) Structure of the E. coli SRP-FtsY complex (PDB 2XXA) (Ataide et al., 2011). Ffh, the protein component of the bacterial SRP is shown in yellow and the 4.5S RNA in dark-blue. The domains of Ffh are indicated. The NG-domain of FtsY is shown in green; the structure of the N-terminal A-domain of FtsY has not been solved yet and is shown as green box. (B) Structure of an SRP-RNC complex (PDB 5GAH). The 50S ribosomal subunit is shown in light-blue and the ribosomal proteins that provide the contact site for SRP are indicated, uL23 (blue), uL29 (pink), uL24 (green), and bL32 (light-green). Ffh is shown in yellow and the 4.5S RNA in dark-blue. The nascent PhoA chain is shown in dark red.
Membrane binding of FtsY is mediated by the A-domain, which precedes the NG-domain (Figure 4A), and by a membrane-targeting sequence at the interface of the A- and NG-domains (de Leeuw et al., 2000; Parlitz et al., 2007; Weiche et al., 2008; Braig et al., 2009; Erez et al., 2010; Kuhn et al., 2011). The A-domain is highly variable in length and sequence and so far no structural information is available, suggesting intrinsic flexibility (Montoya et al., 1997). The A-domain is not essential for protein targeting in E. coli (Eitan and Bibi, 2004), which is explained by the presence of additional binding sites for SecY and phospholipids in the N-domain of FtsY (Parlitz et al., 2007; Weiche et al., 2008; Braig et al., 2009; Erez et al., 2010; Kuhn et al., 2011). However, the A-domain is important for increasing the fidelity of the targeting reaction because it shields the SRP binding site when FtsY is not in contact with the SecYEG complex (Draycheva et al., 2016; Lakomek et al., 2016) and it thus prevents futile SRP-FtsY interactions. Binding of SRP-RNCs to the FtsY-SecYEG complex generates a transient quaternary complex (Kuhn et al., 2015; Jomaa et al., 2017; Draycheva et al., 2018; Figure 5). Subsequent movements of SRP expose the SecY binding site on the ribosome (Halic et al., 2006b) and simultaneous movements of FtsY expose the ribosome binding site on SecY (Halic et al., 2006b; Kuhn et al., 2015). This then allows for the docking of the RNC onto the SecYEG translocon and subsequent GTP hydrolysis by the FtsY-SRP complex (Egea et al., 2004; Focia et al., 2004; Saraogi et al., 2014). GTP-hydrolysis induces the dissociation of the FtsY-SRP complex and allows for the next round of targeting (Egea et al., 2004; Shan et al., 2004; Akopian et al., 2013a).
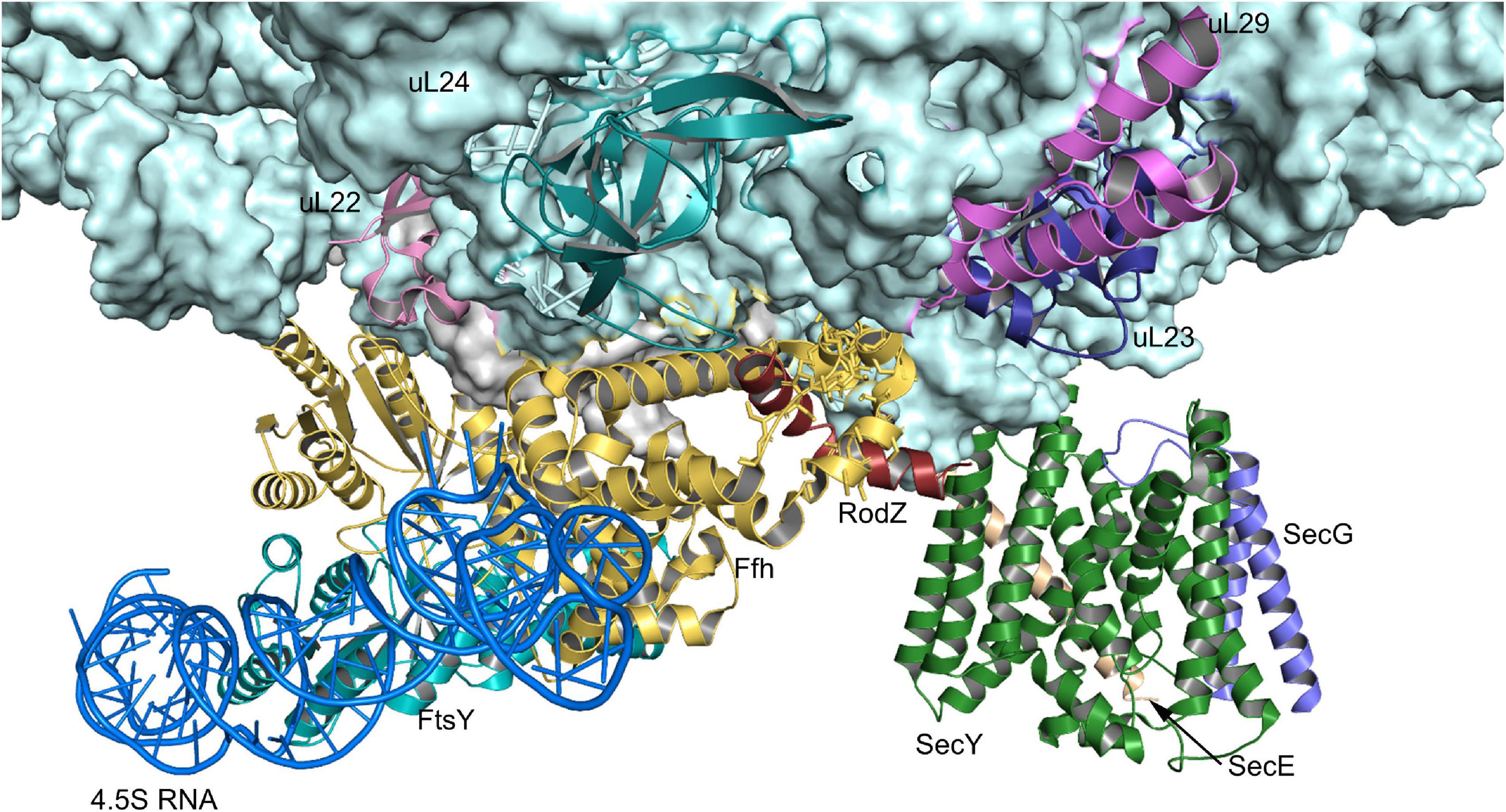
Figure 5. Structure of the quaternary RNC-SRP-FtsY-SecYEG complex. Structure of the quaternary complex (PDB 5NCO), depicting an early state of co-translational protein insertion. The subunits SecY, SecE and SecG of the SecYEG translocon are indicated by green, orange and blue color, respectively. The color code of the FtsY-SRP complex is as in Figure 4 and the nascent PhoA is shown in dark-red. Please note that in this structure, the SecYEG translocon is only tentatively fitted and would have to tilt by ∼20° to be accommodated within the membrane (Jomaa et al., 2017).
Importantly, the SecA and SRP pathways have several features in common: (1) SecA and SRP engage the same docking site on the ribosome and both protrude into the ribosomal tunnel (Denks et al., 2017; Knupffer et al., 2019; Wang S. et al., 2019). (2) FtsY and SecA are activated upon binding to anionic phospholipids and SecY (Mircheva et al., 2009; Kuhn et al., 2011; Stjepanovic et al., 2011; Draycheva et al., 2016; Koch et al., 2016). (3) FtsY, SecA and the ribosome use largely identical binding sites on SecY (Mori and Ito, 2006; Kuhn et al., 2011, 2015). A computational approach for investigating the early evolutionary history of protein transport systems indicates that the SRP/FtsY targeting pathway is the most ancient protein delivery system that probably even existed before the last universal common ancestor (LUCA) (Harris and Goldman, 2021). As protein transport is faster than translation (Pugsley, 1990; Rodnina and Wintermeyer, 2016), the evolution of a second targeting system in fast growing bacteria probably ensures that secretory proteins are kept in a translocation-competent state, when the limited number of SecYEG translocons are co-translationally engaged by SRP-substrates.
Finally, translation-independent membrane localization of some mRNAs encoding for membrane proteins has been observed in bacteria (Nevo-Dinur et al., 2011; Kannaiah and Amster-Choder, 2014; Kannaiah et al., 2019). One example is the small membrane protein YohP, which consists of just 27 amino acids and is predicted to be involved in the bacterial stress response (Hemm et al., 2010). The yohP mRNA was found to be almost exclusively membrane localized, but membrane insertion of the YohP protein by either the SecYEG complex or YidC still required SRP and FtsY (Steinberg et al., 2020). SRP contacts YohP post-translationally both in vivo and in vitro (Steinberg et al., 2020), questioning the paradigm that SRP has to be ribosome-bound for substrate recognition. For small membrane proteins, the post-translational recognition by SRP can be easily explained by the fact that they are already released from the peptidyl transferase domain of the ribosome before they are sufficiently exposed on the ribosomal surface for co-translational SRP recognition. Considering the rapidly increasing number of small membrane proteins discovered in bacteria (Storz et al., 2014; Weaver et al., 2019), the post-translational targeting by SRP could be as abundant as the co-translational targeting and might also be executed for C-tail anchored membrane proteins in bacteria (Abell et al., 2004; Pross et al., 2016; Peschke et al., 2018; Figure 2).
The first X-ray structure of the Sec translocon was obtained for the homologous SecYEβ complex from the archaeon Methanococcus janaschii and represented the resting state with a sealed pore (Van den Berg et al., 2004). In this resting conformation, which was later also obtained from other species (Li et al., 2007; Tsukazaki et al., 2008; Tanaka et al., 2015), SecY is organized in two halves formed by transmembrane helices (TMs) 1 to 5 and 6 to 10, respectively, which are connected by a loop between TM5 and 6, termed the hinge (Figure 6). In this clamshell-like structure, SecY forms two vestibules with a central constriction, called the pore ring, in the middle. The pore ring is formed by six bulky and hydrophobic isoleucine residues in E. coli and is sealed on the periplasmic side by a short helix (TM2a; the plug) (Figure 6B). The plug and the pore ring are important for maintaining the membrane barrier in the resting state and during translocation (Saparov et al., 2007; Park and Rapoport, 2011). This structural arrangement provided a first glimpse into how the SecY channel is able to translocate proteins across the membrane, but also to insert proteins into the membrane (Van den Berg et al., 2004). At the front of SecY, TMs 2/3, and 7/8 constitute a flexible crevice, called the lateral gate that allows access to the lipid phase (du Plessis et al., 2009; Hizlan et al., 2012; Bischoff et al., 2014; Gogala et al., 2014; Figure 6A). Cytosolically exposed loops of SecY provide the docking sites for SecA (Mori and Ito, 2006; Das and Oliver, 2011; Kuhn et al., 2011), FtsY (Angelini et al., 2005, 2006; Kuhn et al., 2011) and ribosomes (Prinz et al., 2000; Frauenfeld et al., 2011; Kuhn et al., 2011). Although these sites are not identical, they largely overlap (Kuhn et al., 2011), which indicates that SecA, FtsY and ribosomes compete for SecY binding (Wu et al., 2012; Kuhn et al., 2015). The tilted TM3 of SecE further stabilizes the hinge at the back of SecY and this appears to be crucial for its integrity because SecY is rapidly degraded by the membrane protease FtsH in the absence of SecE (Kihara et al., 1995; Lycklama a Nijeholt et al., 2013). SecG, the third subunit of the bacterial SecYEG complex, consists of two transmembrane domains, which are connected by a cytosolic loop (Figure 6). SecG is not essential for cell viability, but ΔsecG strains of E. coli exhibit protein transport defects in vivo (Nishiyama et al., 1994, 1996).
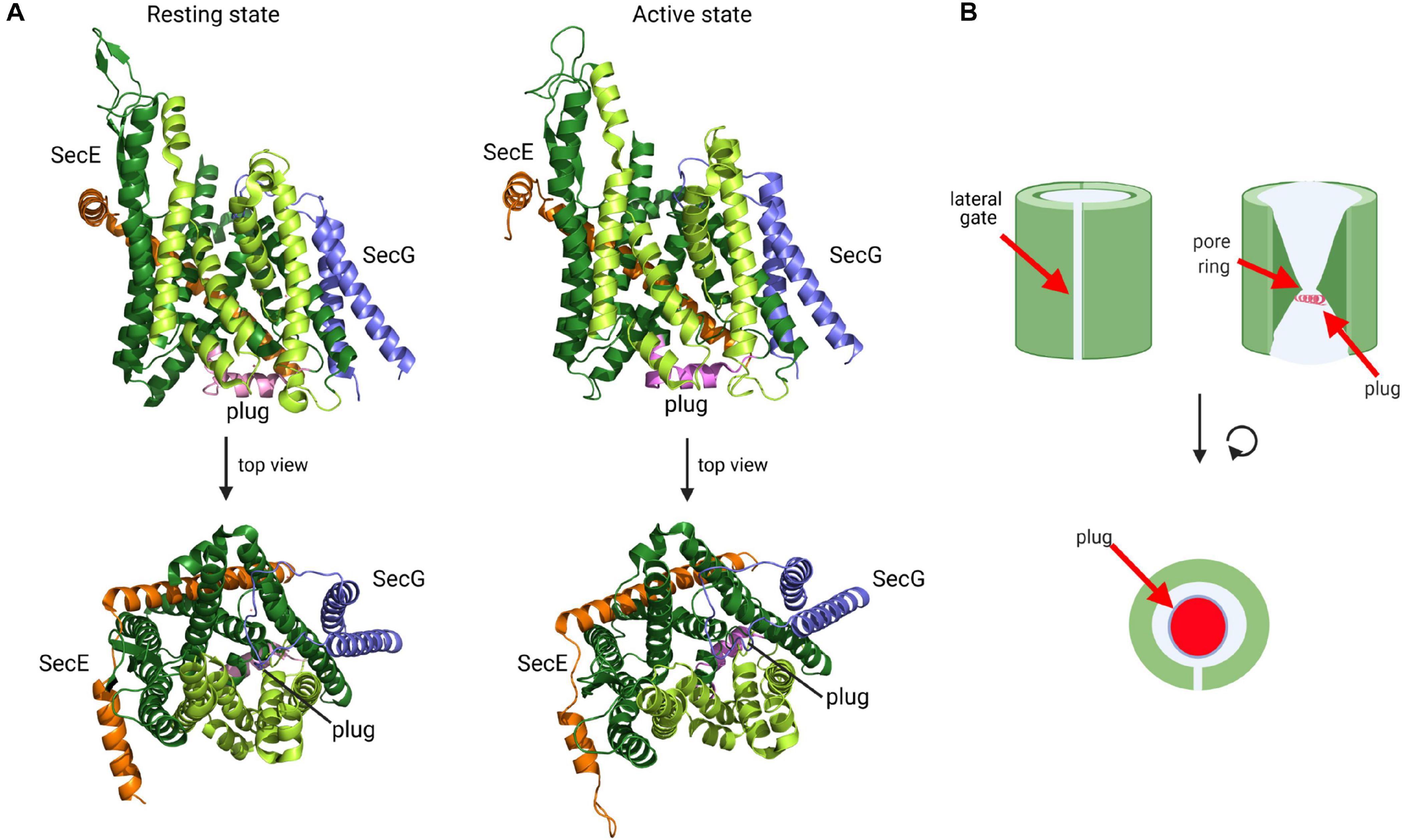
Figure 6. Structure of the SecYEG translocon in its resting state and active state. (A) Structure of T. thermophilus SecYEG in the resting state (PDB 5AWW) and the active state (PDB 5CH4). SecY is shown in green, SecE in orange and SecG in blue. The SecY transmembrane domains that constitute the lateral gate are shown in light green and the plug in magenta. The upper structures depict the front views of the SecYEG translocon and the lower structures the top view from the cytosol, respectively. (B) Schematic front view and view from the cytosol of the SecYEG translocon.
Activation of the SecYEG channel and subsequent protein transport requires opening of the lateral gate, expansion of the pore ring and movement of the plug (Collinson et al., 2015; Voorhees and Hegde, 2016b; Figure 6A). These movements have been documented by additional structures and a wealth of biochemical data. For the transport of secretory proteins, the SecYEG channel is activated by SecA, which serves a dual function: it acts as SecYEG bound receptor for proteins with cleavable signal sequences and provides the energy for translocation by multiple ATP-hydrolysis cycles (Douville et al., 1995; Manting et al., 1997; Tomkiewicz et al., 2006; Alami et al., 2007; Das and Oliver, 2011; Gold et al., 2013; Gouridis et al., 2013). A first structure of a SecYEG-SecA complex (Zimmer et al., 2008) revealed the insertion of the hairpin-like two-helix finger (2HF) of SecA into the cytoplasmic vestibule of SecY and a partial opening of the lateral gate. This opening is required for intercalation of the signal sequence within the lateral gate (du Plessis et al., 2009; Hizlan et al., 2012; Corey et al., 2016). This is depicted in the structure of the SecYEG-SecA complex with a covalently linked signal sequence (Li et al., 2016; Figure 7A). This structure shows that the hydrophobic segment of the signal sequence is located outside of the opened lateral gate. The segment following this hydrophobic part is trapped between TM3 and TM7 on the periplasmic part of the lateral gate and the signal sequence cleavage site is located within the periplasmic vestibule. Opening of the channel is further accompanied by movement of the plug to the back of the channel, where it resides close to SecE, validating previous cross-linking studies (Harris and Silhavy, 1999; Tam et al., 2005).
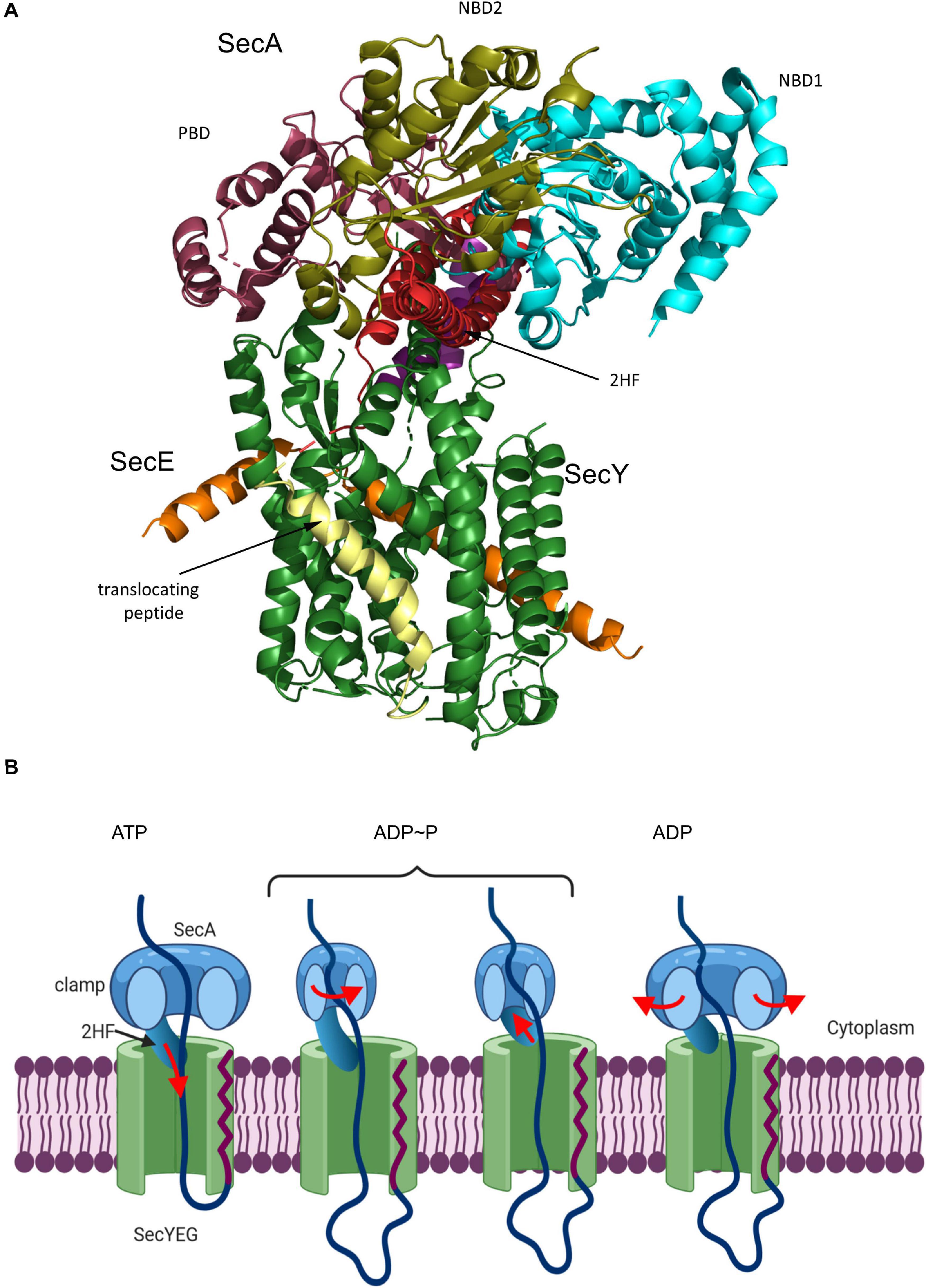
Figure 7. Structure of the substrate-engaged SecA-SecYEG complex and model for SecA-dependent translocation across the SecYEG-translocon. (A) Structure of the SecA-SecYEG complex from B. subtilis (PDB 5EUL). SecY and SecE are shown in green and orange, respectively, and the translocating peptide in yellow. The different domains of SecA are indicated. 2HF corresponds to the two-helix finger. (B) Upon ATP binding to SecA, the 2-helix-finger (2HF) inserts into the SecY channel and pushes the polypeptide into the channel. The signal sequence is depicted in red. For preventing back-sliding, the polypeptide binding domain (PBD) of SecA rotates toward the nucleotide-binding domain (NBD2) and forms a clamp that traps the polypeptide. This step likely occurs before or simultaneously with ATP-hydrolysis. Closing the clamp also leads to the retraction of the 2HF. After phosphate release, the clamp opens again and the polypeptide can slide deeper into the channel but in principle also backward. In vivo, backsliding at this stage could be prevented by contacts of the polypeptide to periplasmic chaperones, like Skp (Schäfer et al., 1999) or the PpiD/YfgM complex (Götzke et al., 2014; Jauss et al., 2019). In addition, the membrane potential is likely important for maintaining directionality of translocation (Driessen and Nouwen, 2008; Knyazev et al., 2018). Figure was modified after (Catipovic et al., 2019).
The activation of SecYEG by SecA initiates the step-wise translocation of secretory proteins across the membrane. The reconstituted SecYEG-SecA complex was shown to generate a mechanical force of about 10pN (Robson et al., 2007; Gupta et al., 2020). Consequentially, several models were proposed on how the high conformational flexibility of SecA might be used for the ATP-dependent and stepwise translocation of a preprotein across the SecYEG channel (Erlandson et al., 2008a, b; Kusters et al., 2011; Gouridis et al., 2013; Ernst et al., 2018; Fessl et al., 2018; Corey et al., 2019; Komarudin and Driessen, 2019). Central to most models is the 2HF-domain of SecA (Erlandson et al., 2008a). The 2HF was shown to insert into the cytosolic vestibule of SecY, where it resides in close proximity to the preprotein (Zimmer et al., 2008). A highly conserved tyrosine residue at the tip of the loop is essential for SecA function, but immobilizing the 2HF on the SecYEG complex does not interfere with translocation (Whitehouse et al., 2012), suggesting that even restricted movements of the 2HF are sufficient to support translocation. Latest data support a push-and-slide mechanism of protein translocation that depends on a power stroke by SecA (Catipovic et al., 2019; Catipovic and Rapoport, 2020). In this model (Figure 7B), the 2HF moves toward the SecY channel upon ATP binding, thereby pushing the polypeptide into the channel. While the 2HF retracts during ATP hydrolysis from the channel, movement of the polypeptide-binding domain of SecA toward the nucleotide-binding domain generates a clamp that fixes the polypeptide in the channel. Phosphate release from SecA is suggested to open the clamp, which allows for some passive sliding of the polypeptide until the next ATP binds and the 2HF pushes the next segment of the polypeptide into the channel. The observation that cross-linking the 2HF to the cytosolic loop C4 of SecY does not impair protein translocation (Whitehouse et al., 2012) is possibly explained by the inherent flexibility of the large C4 loop which might still allow sufficient movements of the 2HF.
The 2HF is also central to an alternative model for SecA-dependent translocation, which suggests a Brownian ratchet mechanism (Collinson, 2019). In this model, SecA regulates channel opening via the 2HF, while substrate movement across the channel occurs via Brownian movement (Allen et al., 2016, 2020). ATP hydrolysis by SecA is suggested to prevent partial folding of substrates at the SecA-SecY interface, while the partial folding on the periplasmic side would prevent back-sliding and thus impose directionality to protein translocation (Fessl et al., 2018; Corey et al., 2019).
In both models, substrate translocation is further stimulated by the proton-motif-force (PMF), which probably adds to vectorial translocation (Brundage et al., 1990; Nouwen et al., 1996; Knyazev et al., 2018). Prior to completion of translocation, the signal sequence is cleaved off by signal peptidase and the mature domain is released into the periplasm (Josefsson and Randall, 1981a, b; Paetzel et al., 2002). This latter step is likely supported by periplasmic chaperones (Schäfer et al., 1999; Furst et al., 2018; Chum et al., 2019; Mas et al., 2019) (see below).
Inner membrane proteins are targeted to the SecYEG translocon co-translationally as RNCs by the SRP pathway (Figure 2; Koch et al., 1999; Beck et al., 2000; Neumann-Haefelin et al., 2000; Akopian et al., 2013b; Steinberg et al., 2018). The SRP receptor FtsY docks onto the SecYEG translocon and engages largely identical binding sites as SecA and the ribosome (Angelini et al., 2005, 2006; Kuhn et al., 2011, 2015). FtsY and SecA have comparable affinities for the SecYEG translocon and are present in comparable copy numbers in E. coli (Douville et al., 1995; Kudva et al., 2013; Kuhn et al., 2015) and it is currently unknown how access of either FtsY or SecA to the SecYEG translocon is regulated. Importantly, only SecY-bound FtsY exposes the SRP binding site and is thus able to direct the SRP-RNC complex to the SecYEG translocon (Mircheva et al., 2009; Draycheva et al., 2016). Structural information on the isolated FtsY-SecYEG complex is not available, but Cryo-EM structures of RNCs bound to the Sec translocon in the presence and absence of SRP and its receptor have been obtained from different species (Becker et al., 2009; Frauenfeld et al., 2011; Bischoff et al., 2014; Gogala et al., 2014; Voorhees et al., 2014; Jomaa et al., 2016, 2017; Voorhees and Hegde, 2016a; Kater et al., 2019). Binding of a non-translating ribosome to the Sec translocon, primarily via the cytosolic loop C5, results in small rearrangements which slightly open the cytosolic part of the lateral gate (Voorhees et al., 2014; Figure 8). The structure of a quaternary ribosome-SRP-FtsY-SecYEG complex revealed that FtsY aligns the ribosomal tunnel exit with the SecYEG channel (Jomaa et al., 2017; Figures 5, 8). The exposure of a short nascent membrane protein further opens the lateral gate on the cytosolic side (Kater et al., 2019) and full insertion of the signal anchor sequence leads to a rotation of helices 2–5 and 10 and allows trapping of the signal anchor sequence at the lateral gate (Voorhees and Hegde, 2016a; Figure 8). Simultaneously, the plug is displaced from its position at the pore ring and the channel is open to both the trans-side and the lipid side of the membrane. TMs downstream of the signal anchor sequence can exit the Sec translocon laterally one by one or in pairs (Heinrich and Rapoport, 2003; Houben et al., 2004; Sadlish et al., 2005). Lipid partitioning of TMs is largely determined by their hydrophobicity (Hessa et al., 2007; White and von Heijne, 2008) and moderately hydrophobic TMs possibly require the interaction with a more hydrophobic second TM to enter the lipid phase (Heinrich and Rapoport, 2003). These helix-helix interactions could occur within the Sec channel (Pitonzo et al., 2009), at the channel-lipid interface (Sadlish et al., 2005; Cross and High, 2009) or even before, at the end of the ribosomal tunnel (Tu et al., 2014; Holtkamp et al., 2015; Nilsson et al., 2015). Lateral release of transmembrane domains out of the SecY channel is further facilitated by YidC (Beck et al., 2001; Houben et al., 2002), which associates with the lateral gate of SecY to form a tetrameric protein channel (Sachelaru et al., 2015, 2017).
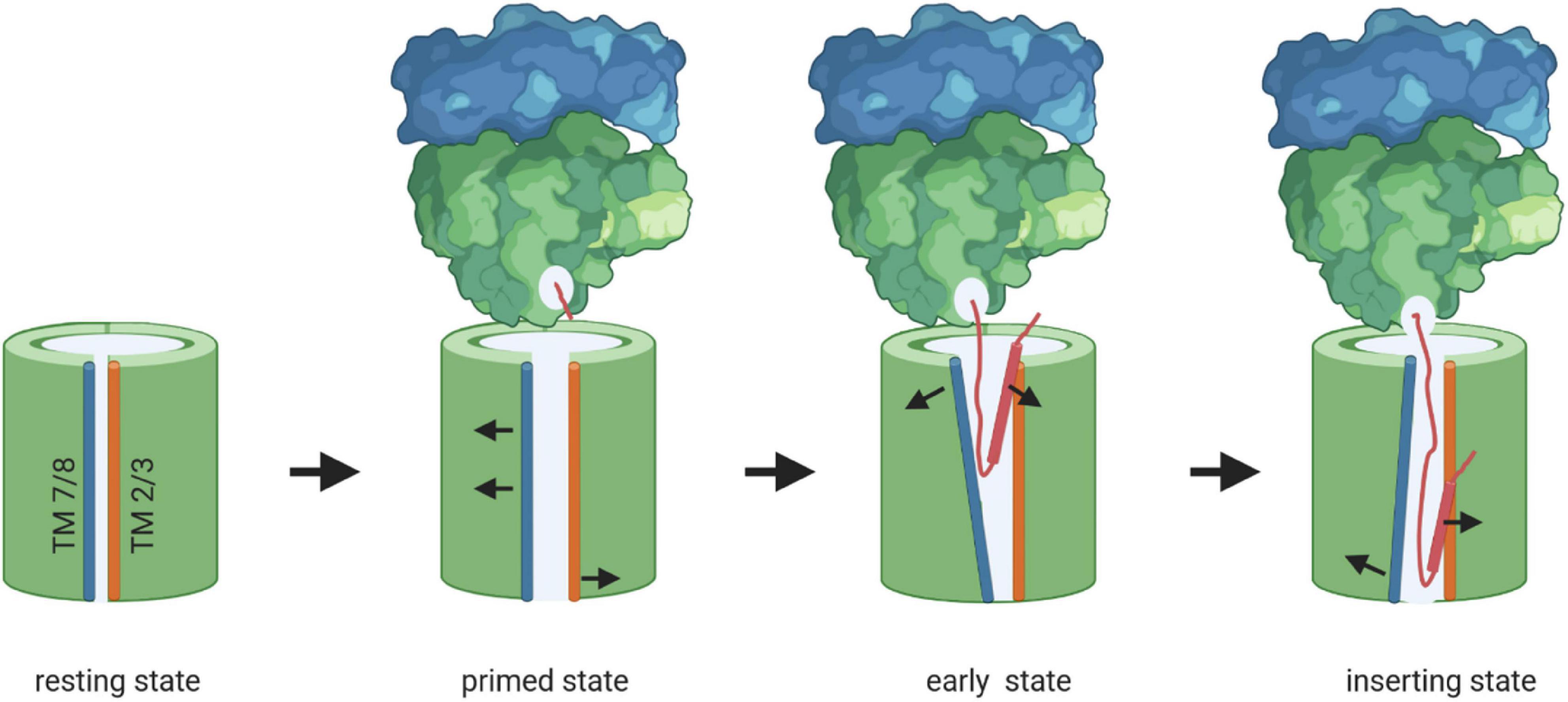
Figure 8. Model of membrane protein insertion via the SecYEG translocon. In the resting state of the SecYEG-translocon, the lateral gate, composed of transmembrane domains (TMs) 2/3 on one side (orange) and TMs 7/8 (blue) on the other side, is closed. Binding of the translating ribosome to the cytosolically exposed loop connecting TM 6 and 7 of SecY (C5-loop, not shown), causes the lateral gate to slightly open, which is then primed for the approaching nascent chain. The emerging nascent membrane protein (red) disrupts contacts between TM 2 and TM 7 on the cytosolic side of the membrane further, while TM 7 moves closer toward TM 3 on the periplasmic side. This creates a V-shaped crevice during the early state of insertion. This state is likely further stabilized by the two N-terminal TMs of SecE (not shown). Ongoing chain elongation positions the hydrophobic core of the signal peptide (red zylinder) at the lateral gate, where it occupies approx. the same position as TM 2 in the resting SecYEG channel, before it is released into the membrane.
Although there are some variations in the translocon structure when activated by SecA or the ribosome, the step-wise channel opening during post-translational translocation or co-translational insertion appears to be a conserved feature of the Sec translocon and is in line with multiple biochemical studies (du Plessis et al., 2009; Bonardi et al., 2011; Hizlan et al., 2012; Knyazev et al., 2013, 2014; Ge et al., 2014; Mercier et al., 2020). It is, however, currently unknown how channel opening and transport across the SecYEG translocon works for membrane proteins that are co-translationally targeted by SecA, like RodZ (Rawat et al., 2015; Wang et al., 2017; Figure 3). A simultaneous binding of SecA and the ribosome to SecY appears unlikely, considering that both engage overlapping binding sites on SecY (Kuhn et al., 2011). One possibility is that SecA starts inserting RodZ only after it is released from the ribosome. In this case, only targeting would occur co-translationally, while the actual insertion would be post-translationally. A similar situation is encountered during co-translational insertion of membrane proteins with large periplasmic loops, because their translocation requires SecA (Neumann-Haefelin et al., 2000; Deitermann et al., 2005). How the access of SecA to these loops during co-translational insertion is coordinated is currently unknown. Finally, how the SecYEG translocon handles small membrane proteins that are post-translationally targeted by SRP (Steinberg et al., 2020; Figure 2), i.e., when neither the ribosome nor SecA are involved, requires further analyses.
The Sec translocon in bacteria and eukaryotes is organized as a highly modular protein complex and multiple different entities have been structurally and biochemically characterized (Zimmer et al., 2008; Boy and Koch, 2009; Frauenfeld et al., 2011; Denks et al., 2014; Komar et al., 2016; Kater et al., 2019). The E. coli SecYEG translocon was found to exist as a functional monomer (Menetret et al., 2007; Kedrov et al., 2011; Park and Rapoport, 2012) and as a dimer stabilized by cardiolipin (Gold et al., 2010). SecYEG was furthermore found in heterotetrameric complexes with SecA (Zimmer et al., 2008) or YidC (Boy and Koch, 2009; Sachelaru et al., 2017), and as heterohexameric complexes with SecDFYajC (Duong and Wickner, 1997) or FtsY-SRP-RNCs (Jomaa et al., 2017). Finally, a heteroheptameric SecYEG-SecDFYajC-YidC complex was characterized and referred to as Holo-translocon (HTL) (Schulze et al., 2014; Komar et al., 2016). Several additional partner proteins have been identified, like the YfgM-PpiD chaperone complex (Antonoaea et al., 2008; Götzke et al., 2014; Sachelaru et al., 2014; Furst et al., 2018; Jauss et al., 2019), or the cytosolic protein Syd, which is suggested to serve together with the protease FtsH in quality control of the Sec translocon (Akiyama et al., 1996; Dalal et al., 2009; Table 1 and Figure 9). Non-proteinaceous partners are equally important for SecYEG function, like anionic phospholipids and cardiolipin (Prabudiansyah et al., 2015; Collinson, 2019; Bogdanov et al., 2020; Ryabichko et al., 2020) or the glycolipid MPiase, which was shown to support protein transport via the SecYEG translocon (Moser et al., 2013; Nishiyama and Shimamoto, 2014). The highly dynamic equilibrium between different SecYEG assemblies likely allows the SecYEG complex to adapt to a wide variety of different substrates and to different physiological conditions.
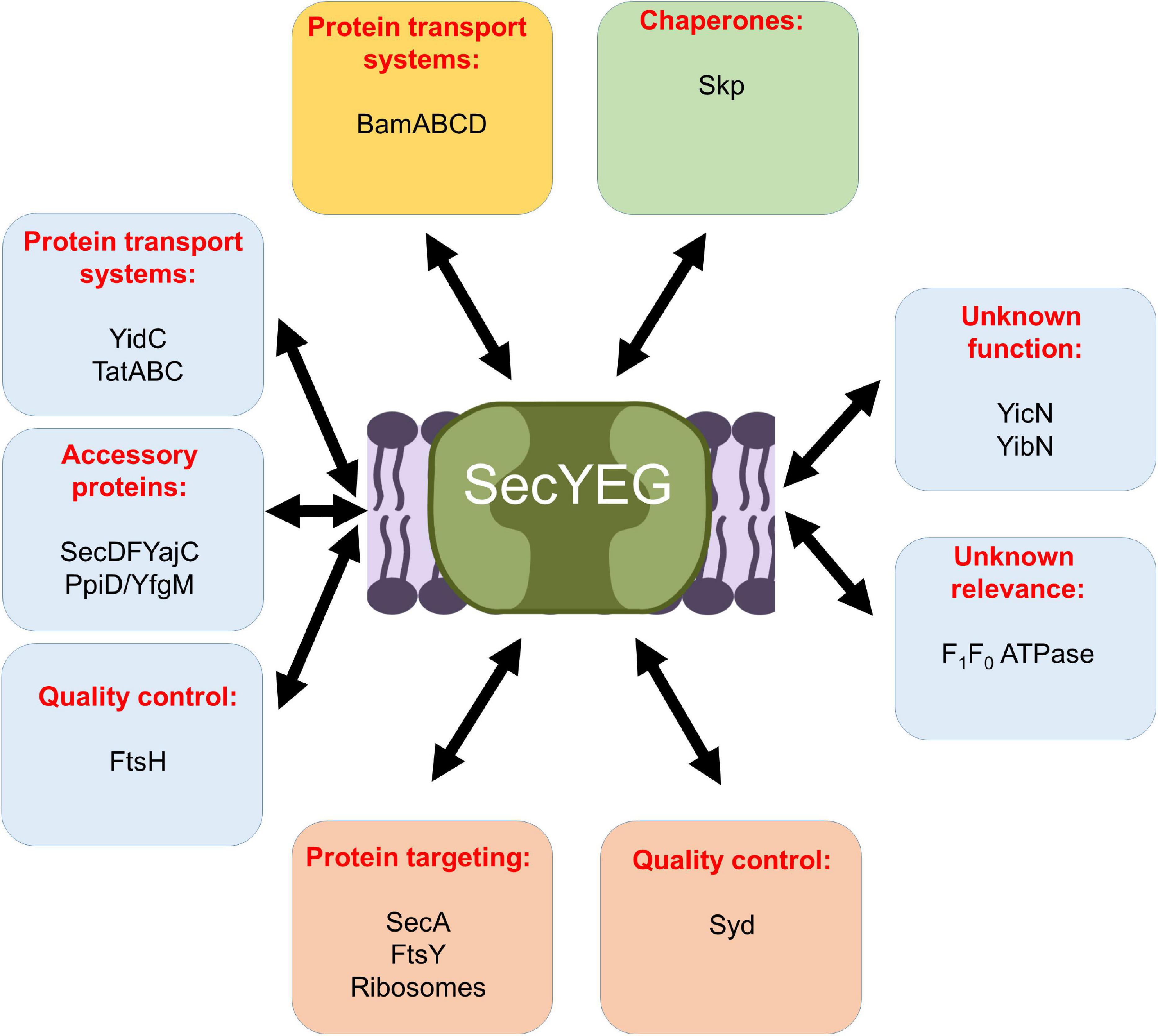
Figure 9. Schematic view on the protein interaction network of the E. coli SecYEG complex. Interactions within the inner membrane are shown in blue boxes, those that take place at the cytosolic phase of the inner membrane in orange boxes, those at the periplasmic side of the inner membrane in a green box, and those with the outer membrane are boxed in yellow. For details see text.
YidC is an inner membrane protein with six TMs in E. coli and a Nin-Cin-topology (Figure 10A). It belongs to a conserved group of proteins with homologues in mitochondria, chloroplasts, the endoplasmic reticulum and archaea (Borowska et al., 2015; Anghel et al., 2017; Kuhn and Kiefer, 2017; McDowell et al., 2021). Although YidC can act as SecYEG-independent insertase for some membrane proteins (Samuelson et al., 2000; Luirink et al., 2001; Serek et al., 2004; Welte et al., 2012), it also associates with the SecYEG complex (Scotti et al., 2000; Nouwen and Driessen, 2002; Li et al., 2013; Sachelaru et al., 2015, 2017).

Figure 10. Structures of YidC, SecDF and a model of the holo-translocon. (A) Structure of YidC from E. coli (PDB 6AL2). The conserved transmembrane domains (TMs) 2 to 6 of YidC are indicated (TM2, light blue; TM3, yellow; TM4, orange; TM5, light pink; TM6, red), while the structure of TM1 is still unknown. The short amphipathic helix EH1 is depicted in dark blue, the periplasmic loop P1 in dark green and the cytoplasmic loop C1 in light green. (B) Structure of the SecDF complex from Thermus thermophilus (PDB 5YHF). SecDF consists of 12 TMs, six each in SecD (TM1-6, pink) and SecF (TM7-12, green), and three periplasmic domains, termed P1-head (dark blue), P1-base (light blue) and P4 (yellow). (C) Modell of the holo-translocon based on the cryo-EM structure from E. coli (PDB 5MG3). SecY is shown in green, SecE in orange and SecG in blue, its ancillary subunits SecD in pink, SecF in green and YidC in yellow.
The conserved TMs 2 to 6 of YidC are organized as a globular helix bundle that forms a hydrophilic groove within the membrane, while the structure and position of TM1 is unknown (Kumazaki et al., 2014a, b; Figure 10A). The hydrophilic groove is blocked on the periplasmic side of the membrane by the short amphipathic EH1 helix, which is oriented in parallel to the membrane surface. The EH1 helix is part of the large P1-loop that connects TM1 and TM2 on the periplasmic side (Saaf et al., 1998; Oliver and Paetzel, 2008; Ravaud et al., 2008). On the cytosolic side of TM2, the C1-loop forms a helical coiled-coil domain that is essential for YidC function (Geng et al., 2015). The hydrophilic groove likely faces the TM domains of SecY and cross-link data demonstrate that TM1, TM3 and TM5 of YidC are in close contact to the lateral gate of SecY (Sachelaru et al., 2015; Petriman et al., 2018). YidC can even enter the SecY channel (Sachelaru et al., 2017) and this is achieved via the flexible TM1 and the P1-loop that reaches deep into the periplasmic cavity of SecY, where it makes contact to the plug domain of SecY (Jauss et al., 2019). TM1 was also found in contact with SecG, supporting its intrinsic flexibility (Petriman et al., 2018). Further contacts between YidC and SecY were observed for the C1-loop, while the P1-loop also contacts SecG, SecE and SecF. The C1-loop also provides the docking site for FtsY and is essential for the insertase function of YidC (Geng et al., 2015), but SecY-YidC contacts are maintained even in the absence of the C1-loop (Petriman et al., 2018). Crystallization and molecular dynamics simulations demonstrate that the C2 loop linking TM4 and TM5 is highly flexible (Tanaka et al., 2018). Together with the C-terminus of YidC, the C2-loop provides the ribosome binding site of YidC (Geng et al., 2015) and shields the hydrophilic groove on the cytosolic side (Tanaka et al., 2018). The intimate contact between the hydrophilic groove of YidC and the lateral gate of SecY provides further support for the concept that TMs leaving the SecY channel are first bound by YidC before they are released into the lipid phase (Beck et al., 2001; Houben et al., 2002). TMs exit the SecY channel in most cases sequentially (Serdiuk et al., 2019) and the hydrophilic groove of YidC probably reduces the hydrophobicity of the adjacent lateral gate of SecY and therefore further stimulates the release of the TMs into the inner membrane by a greasy slide. The amphipathic helix EH1 could act as a mechanical switch, tilting TM3 and supporting substrate release (Dalbey et al., 2017; He et al., 2020).
The inner membrane proteins SecD, SecF and YajC form a stable complex (Pogliano and Beckwith, 1994a, b) and were shown to interact with SecYEG and YidC (Duong and Wickner, 1997; Nouwen and Driessen, 2002). Depletion of SecDF causes cold sensitivity and the accumulation of precursor proteins in the cytosol, supporting their role in stimulating protein translocation across the membrane (Pogliano and Beckwith, 1994a). SecD mutants also lead to elevated levels of SecA (Rollo and Oliver, 1988), which is a typical sign of impaired protein translocation (Ito et al., 2018).
The crystal structure of the SecDF complex shows 12 TMs, six each in SecD and SecF, and three periplasmic domains, termed P1-head, P1-base and P4 (Tsukazaki et al., 2011; Figure 10B). The P1-head can undergo a large rotation, resulting in two distinct conformations, the F- and I-form. An amphiphilic cavity within the P1-head was proposed to bind precursor proteins (Furukawa et al., 2017, 2018). As protein translocation is strongly dependent on the PMF (Driessen and Wickner, 1991; Mori and Ito, 2003; Corey et al., 2018; Knyazev et al., 2018), PMF-driven conformational changes of the P1-head could help to pull substrates out of the SecYEG channel (Tsukazaki et al., 2011; Tsukazaki, 2018). This is in line with the assumption that the SecDF complex is necessary at a later stage of protein translocation (Pogliano and Beckwith, 1994a; Tsukazaki, 2018). The predicted low abundance of the SecDFYajC complex in E. coli (Pogliano and Beckwith, 1994a, b) suggests that such a pulling is only required for particular substrates or that other proteins execute a similar function, e.g., the YfgM-PpiD complex that also associates with the SecYEG translocon (Götzke et al., 2014; Sachelaru et al., 2014; Jauss et al., 2019).
SecF interacts with the P1-loop of YidC and the non-conserved residues 215–265 in the P1-loop are sufficient for SecF interaction (Xie et al., 2006), but these residues are not required for YidC function (Jiang et al., 2003). The phenotype of a secDF depletion strain can be rescued by YidC-overproduction, further supporting a cooperation between SecDF and YidC (Nouwen and Driessen, 2002; Li et al., 2013). The SecDF complex likely stabilizes the SecYEG-YidC interaction (Nouwen and Driessen, 2002; Tsukazaki, 2018), although the SecYEG-YidC interaction is also observed in the absence of the SecDFYajC complex (Boy and Koch, 2009; Sachelaru et al., 2015). Finally, SecDF might also play a role in efficient maturation and folding of OMPs (Alvira et al., 2020) and it was proposed that SecDF is part of an inter-membrane trafficking machinery that connects transport processes across the inner membrane with those at the outer membrane (Alvira et al., 2020) (see below).
The existence of a HTL was first shown after co-expression and purification of its seven constituents (Schulze et al., 2014). The HTL comprises the core SecYEG translocon and its ancillary subunits SecDFYajC and YidC, forming a heteroheptameric complex (Schulze et al., 2014; Botte et al., 2016; Komar et al., 2016). The periplasmic domains of SecDF and YidC are localized on top of the SecY channel and are suggested to interact with emerging substrates, potentially preventing their backsliding (Botte et al., 2016; Figure 10C). The seven subunits of the HTL are arranged around a central lipid-filled chamber, which might provide a flexible and protected environment for TMs to fold, to acquire their final topology and to assemble (Goder et al., 1999; Dowhan et al., 2019; Martin et al., 2019). The presence of the lipid chamber could also promote the assembly of membrane protein complexes, a function that was assigned to YidC when in complex with SecYEG (Wagner et al., 2008). This concept would attribute the HTL a particular role during membrane protein insertion and indeed in vitro studies showed that the HTL was more efficient in protein insertion and less effective in SecA-dependent protein secretion than the SecYEG complex (Schulze et al., 2014). However, in these studies the HTL also increased the insertion of proteins that were classified as SecY-independent, like the phage protein Pf3 or subunit c of the F1F0 ATPase (Serek et al., 2004; van der Laan et al., 2004). The abundance of the HTL in the E. coli membrane is not entirely clear. Initial estimations suggested that the SecDF complex is present in only 30–100 copies per cell and thus about 10 times less abundant than SecYEG (Pogliano and Beckwith, 1994a, b). In contrast, ribosome profiling data indicated a 4:1 SecYEG:SecDF ratio (Li et al., 2014) and a recent proteomics study even proposed a 1:1 ratio (Schmidt et al., 2016). Considering that the HTL is only one of several SecYEG assemblies, it is important to emphasize that these absolute numbers would only predict the number of theoretically possible HTLs, but not the real number in the E. coli membrane.
The interaction of the SecYEG complex with periplasmic chaperones was first shown for Skp and it was suggested that Skp could facilitate substrate release from the SecY channel (Schäfer et al., 1999; Harms et al., 2001; Figure 9). A similar function was also proposed for the membrane-anchored periplasmic chaperone PpiD, which was found to contact a secretory protein exiting SecY (Antonoaea et al., 2008). PpiD forms a complex with YfgM, which contains like PpiD a single TM and a large periplasmic domain (Maddalo et al., 2011; Götzke et al., 2014). YfgM was also found as contact partner of SecYEG and the PpiD-YfgM complex was suggested to mediate substrate transfer from the SecYEG complex to other periplasmic chaperones, like SurA, Skp, or DegP (Götzke et al., 2014; Furst et al., 2018). PpiD contacts the lateral gate of SecY (Sachelaru et al., 2014) and its periplasmic domain deeply inserts into the periplasmic cavity of the SecY channel (Jauss et al., 2019). When the plug domain of SecY is deleted, the interaction between SecYEG and PpiD is enhanced both at the lateral gate as well as in the channel interior which suggests that channel opening controls the SecY-PpiD contact. These SecY-PpiD contacts as revealed by site-directed in vivo cross-linking are basically identical to the detected SecY-YidC contacts, which indicates that SecY can either interact with YidC or PpiD. However, PpiD and YidC show non-competitive binding to the SecYEG translocon in vivo (Jauss et al., 2019), pointing to the possible presence of two distinct SecYEG populations. This is also supported by Blue-Native PAGE analyses, which found SecYEG either in contact with YidC or PpiD/YfgM (Götzke et al., 2014) and by data showing that the SecY-PpiD contact is lost when SecY is engaged in inserting a membrane protein (Sachelaru et al., 2014). PpiD contains an inactive prolyl-isomerase domain in its periplasmic loop (Weininger et al., 2010) and does not seem to execute any pulling force on SecY substrates (Jauss et al., 2019). Still it improves translocation efficiency and the release of newly translocated substrates into the periplasm, possibly by preventing their backsliding into the periplasmic cavity of SecY (Furst et al., 2018). PpiD was also found to cross-link to the periplasmic chaperone SurA, providing further evidence for a role of PpiD in connecting the translocation machinery to the periplasmic folding machinery (Wang et al., 2016).
After their translocation across the inner membrane, β-barrel OMPs have to be inserted into the outer membrane (Bos et al., 2007; Konovalova et al., 2017). The β-barrel assembly machinery, the BAM complex, is localized in the outer membrane (OM) and facilitates the folding and insertion of OMPs into the OM (Ranava et al., 2018; Ricci and Silhavy, 2019). The complex has a molecular mass of around 203 kDa and comprises the core protein BamA and the four additional lipoprotein subunits BamBCDE (Noinaj et al., 2017; Figure 1). BamA contains a β-barrel domain and five polypeptide-transport-associated (POTRA) domains protruding into the periplasm. Even though only BamA and BamD are essential in vivo, all five subunits are necessary for unrestrained function of the complex (Iadanza et al., 2016).
The passage of OMPs from the SecYEG translocon to the BAM complex has been analyzed in multiple studies (reviewed in (Ricci and Silhavy, 2019). A direct interaction between the SecYEG translocon and the BAM complex was first suggested when a supercomplex consisting of BamA, BamB, SurA, PpiD, SecY, SecE, and SecA was found by native gel electrophoresis (Wang et al., 2016). Furthermore, cross-links between the periplasmic chaperone SurA and BamA consolidated the idea that translocation of OMPs across the inner membrane, passage through the periplasm and the insertion into the OM could be physically linked (Wang et al., 2016). BamA was furthermore found to co-purify with the Sec translocon (Jauss et al., 2019) and interactions between SecY and BamACD were identified in a peptidisc approach combined with affinity purification/mass-spectrometry (Carlson et al., 2019). The existence of connecting structures between the inner and outer membranes (so called Bayer’s patches) that could aid the biogenesis of OMPs were first postulated by Bayer (1968). However, they were controversially discussed since their discovery, although some biochemical evidence pointed to the existence of contact points between the outer and inner membrane (Ishidate et al., 1986; Kellenberger, 1990; Malinverni and Silhavy, 2011). This was recently verified by showing the interaction of the HTL with the BAM complex. This transient contact was shown to be conferred by the periplasmic loops of SecDF, YidC, and the BAM complex (Alvira et al., 2020). The periplasmic domain of SecD has multiple contact sites with BamBCD, while YidC interacts with BamABCD. Furthermore, there might be a potential interaction between YajC and BAM (Carlson et al., 2019). In contrast, the SecYEG complex alone is not able to directly interact with the BAM complex, probably due to the lack of large periplasmic domains. The HTL-BAM complex is further stabilized by cardiolipin (Alvira et al., 2020), which was already shown to be important for SecYEG complex stability (Gold et al., 2010; Ryabichko et al., 2020). A yet unsolved question is how OMPs are transported to and inserted into the OM without any apparent energy source due to the lack of ATP in the periplasm and the absence of an ion gradient across the outer membrane (Konovalova et al., 2017). The interaction between the HTL and BAM could facilitate the energetic coupling of inner membrane with outer membrane transport. Once OMP precursors are translocated across the SecYEG complex and the signal sequence is cleaved, the mature but yet unfolded protein is bound by periplasmic chaperons, such as PpiD (Antonoaea et al., 2008) and is then recognized by the BAM complex, forming a trans-periplasmic supercomplex with SecDF as potential energy supplier (Carlson et al., 2019; Alvira et al., 2020).
Functional and proteomic studies have identified several additional proteins as potential contact partners of the SecYEG complex (Kuhn et al., 2011; Carlson et al., 2019; Jauss et al., 2019), although the functional relevance of some of these interactions require further analyses (Table 1 and Figure 9).
The cytosolic protein Syd was shown to stabilize overexpressed SecY in E. coli (Shimoike et al., 1995) and to prevent access of SecA to an altered SecYEG translocon (Matsuo et al., 1998). Syd is suggested to bind to the C4 and C5 loops of SecY (Dalal et al., 2009), which are also part of the SecA binding site (Mori and Ito, 2006; Kuhn et al., 2011) and it appears that binding of SecA and Syd to SecY is mutually exclusive (Dalal et al., 2009). The SecY-Syd interaction could provide a quality control system for the correct assembly of the SecYEG complex, probably in conjunction with the essential zinc-metalloprotease FtsH (Kihara et al., 1995; Ito and Akiyama, 2005).
A cooperation between the SecYEG translocon and the Tat transport system for folded proteins (Kudva et al., 2013) was observed in Streptomyces coelicolor (Keller et al., 2012). Here, the first two TMs of the Rieske iron-sulfur protein are inserted via the SecYEG translocon, while TM3 is dependent on the Tat machinery. The dual requirement for the Sec- and Tat-machinery appears to be common for membrane proteins that contain globular, co-factor containing extracytoplasmic domains (Tooke et al., 2017), which are abundant in both Gram-positive and Gram-negative bacteria. TatA was also found co-purifying with the SecYEG translocon in E. coli, supporting the concept of a widespread cooperation between the Sec and Tat transport systems (Jauss et al., 2019).
The F1F0-ATPase was also shown to interact with the SecYEG complex (Chorev et al., 2018) and subunit b of F1F0-ATPase was enriched in a peptidisc approach (Young et al., 2020). The interaction of the protein translocation machinery with components of the respiratory chain has been extensively studied in the mitochondrial inner membrane (Pfanner et al., 2019), but the physiological importance of these interactions in the bacterial membrane requires further analyses.
YibN and YicN are two single-spanning membrane proteins of approx. 15 kDa that co-purify with SecYEG (Jauss et al., 2019; Young et al., 2020), but their functions have not been elucidated. A possible role of YibN in protein transport is supported by the observation that YibN is up-regulated when YidC is depleted (Wickstrom et al., 2011b) and in particular enriched when the SecYEG translocon is purified from secDF-depleted E. coli strains (Young et al., 2020). Nevertheless, the exact role of YibN/YicN in the translocation machinery and how they interact with the Sec translocon has still to be examined.
The SecYEG translocon, SecA, SRP, and FtsY are essential for cell viability, but conditional depletion strains have been generated for some of the respective genes and were analyzed for transcriptomic or proteomic responses. The depletion of SRP induces the σ32-response and leads to an up-regulation of several chaperones and proteases, like DnaK, GroEL, GroES, ClpB, IbpA, and FtsH (Bernstein and Hyndman, 2001; Wickstrom et al., 2011a; Figure 11). It furthermore induces the phage-shock protein A (PspA), which is generally associated with inner membrane damage (Manganelli and Gennaro, 2017). However, it does not lead to increased levels of stress-induced periplasmic proteins, like DegP or Skp (Wickstrom et al., 2011a), suggesting that the σE-dependent cell envelope stress response is not induced (Hews et al., 2019). This is rather surprising, because the insertion of SecY is dependent on the SRP/FtsY pathway (Koch and Muller, 2000) and SRP depletion should reduce the levels of SecY, which subsequently should impair the translocation of OMPs (Kudva et al., 2013). On the other hand, by promotor fusion experiments it was shown that impaired SecY activity is not strictly linked to the induction of the cell envelope stress response (Shimohata et al., 2007). It appears likely that the σE-dependent cell envelope stress response is only induced upon prolonged SRP-depletion or when SecYEG-dependent transport largely ceased. The up-regulation of chaperones and proteases is also observed in a conditional FtsY-depletion strain. However, FtsY-depletion additionally induced ribosome-inactivation via the ribosome-modulation factor (RMF) (Bürk et al., 2009). An up-regulation of chaperones/proteases and down-regulation of translation is also observed in eukaryotic cells upon SRP depletion (Mutka and Walter, 2001). Importantly, the depletion of the SRP pathway in either bacteria or eukaryotic cells does not cause a rapid decline in the membrane proteome (Ulbrandt et al., 1997; Wickstrom et al., 2011a; Costa et al., 2018). A possible explanation for this conundrum is the intrinsic affinity of ribosomes for the SecYEG complex (Prinz et al., 2000) and the presence of alternative targeting systems in eukaryotes (Ast et al., 2013).
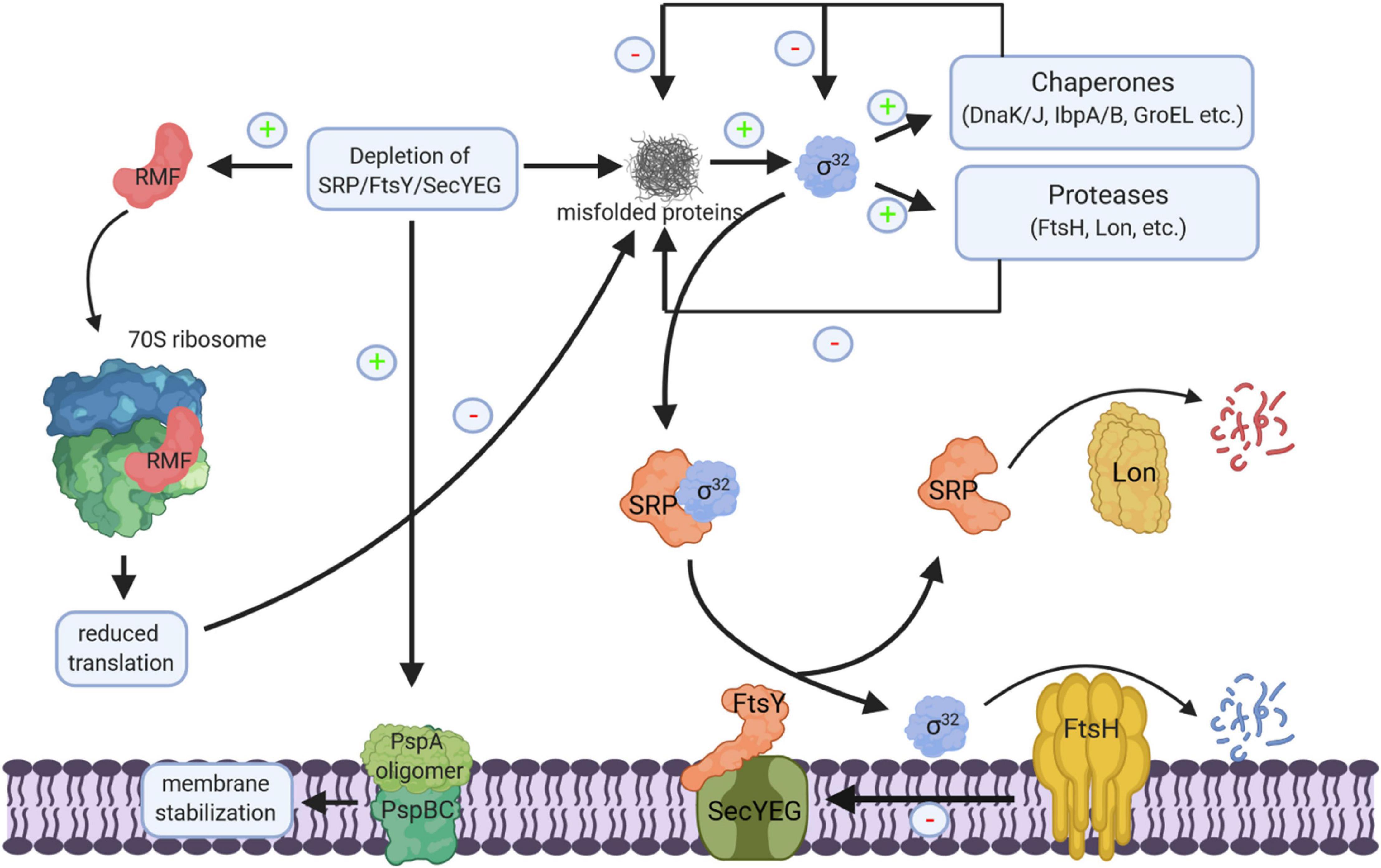
Figure 11. Cellular response to impaired protein transport. Depletion of SRP/FtsY or SecYEG induce a multifaceted response. This includes membrane stabilization via the induction of the phage-shock response (PspABC complex), the inhibition of translation via the induction of the ribosome-modulation factor (RMF) upon FtsY depletion and the induction of the σ32-response via the accumulation of misfolded proteins. Increased chaperone and protease production reduce the cellular concentration of misfolded proteins and provide a negative feedback loop for declining the σ32 response. Chaperones inhibit σ32 directly and the membrane bound protease FtsH degrades σ32. Membrane targeting of σ32 for degradation by FtsH is dependent on SRP/FtsY and SecYEG. Thus, upon SRP/FtsY or SecYEG depletion/saturation, elevated σ32 levels persist. FtsH also degrades misfolded/aggregated membrane proteins and SecY that is not in complex with its partner protein SecE. Ffh, the protein subunit of SRP is also a substrate of the Lon protease; in particular when Ffh is in excess over the 4.5S RNA, the RNA subunit of the bacterial SRP. “+” indicates increased production, “–” indicates reduced production, inhibition or degradation.
The cellular concentration of SRP is controlled by the Lon protease, which is induced upon stress conditions. However, Lon-dependent degradation of Ffh primarily occurs when the Ffh levels exceed the concentration of the 4.5S RNA (Sauerbrei et al., 2020) and it is unclear whether Lon also reduces the Ffh levels upon stress conditions. FtsY is encoded in the ftsYEX operon, upstream of the heat-shock sigma factor σ32 (Gill and Salmond, 1987, 1990; Weinreich et al., 1994), however, they seem not to be transcriptionally coupled (Gómez-Eichelmann and Helmstetter, 1999). FtsE and FtsX are involved in the control of peptidoglycan hydrolase activity and important for cell division (Pichoff et al., 2019), explaining the filamentous phenotype of ftsYEX mutations (Luirink et al., 1994). FtsY levels have been shown to increase at low temperature (Liu et al., 2016; Zhong and Zhao, 2019) and FtsY is subject to a proteolytic event, which degrades its N-terminal membrane targeting sequence (Weiche et al., 2008). However, the responsible protease and the physiological significance of this degradation are still unknown.
Mutants lacking SecB or depleted for SecA also show an up-regulation of the σ32-response due to the accumulation of secretory protein precursors in cytoplasm (Wild et al., 1992, 1993, 1996). SecB-deficient strains also show impaired growth on rich medium (Kumamoto and Beckwith, 1985; Wild et al., 1993), however, this is likely caused by a polar effect of the secB deletion on the downstream gpsA gene, which is involved in phospholipid biosynthesis (Shimizu et al., 1997).
The σ32-response and the formation of cytosolic aggregates containing many ribosomal proteins is also induced upon SecYE depletion (Wild et al., 1992, 1993, 1996; Baars et al., 2008). However, in comparison to SRP depletion, SecYE depletion has a more drastic effect on the steady-state levels of inner membrane proteins and secretory proteins (Baars et al., 2008). SecYE-depletion primarily reduces the levels of multi-spanning membrane proteins and the levels of membrane proteins with large periplasmic domains. Intriguingly, these membrane proteins cannot engage YidC as second integration site for membrane proteins (Samuelson et al., 2000; Serek et al., 2004) and are therefore strictly dependent on SecYEG. The levels of single spanning and short membrane proteins are less impaired by SecYE-depletion, because they can use YidC as alternative integration site when SecYEG is depleted. This is also in line with the observation that the SRP pathway can target both SecYEG and YidC (Welte et al., 2012; Petriman et al., 2018).
The σ32-response in E. coli is regulated by two feedback loops. Free chaperones, like DnaK or GroEL bind and inactivate σ32, while the inner membrane protease FtsH degrades σ32. It was recently shown that membrane targeting of σ32 is dependent on SRP, FtsY, and SecY (Lim et al., 2013; Miyazaki et al., 2016; Figure 11). Thus, depletion of SRP/FtsY increases the stability of σ32 by reducing its degradation via FtsH. This allows for increased chaperone and protease production when the SRP pathway or the SecYEG translocon are saturated and links protein transport directly to the proteostasis network.
The levels of SecY and SecE in E. coli are slightly higher on rich media compared to minimal media and are reduced in stationary phase (Yang et al., 2013; Crane and Randall, 2017). Thus, the expression of secY and secE seem to mimic the expression of house-keeping genes. A similar observation was made for secDF expression in S. coelicolor (Zhou et al., 2014). This is different for SecA; here an intriguing mechanism has been identified that allows E. coli to tailor SecA-levels to reduced translocation activity of the SecYEG translocon (Ito et al., 2010; Ito and Chiba, 2013). This was first recognized by studies showing that partial inactivation of SecYEG-dependent translocation by secY mutations or by adding the SecA-inhibitor sodium azide, led to an up-regulation of SecA (Oliver and Beckwith, 1982; Rollo and Oliver, 1988). This regulation is achieved by the product of the upstream secM gene, which is co-transcribed with secA. Both genes are separated on the mRNA by a stem-loop- like sequence that overlaps with the Shine-Dalgarno sequence of secA. SecM (secretion monitor) is a signal-sequence containing polypeptide that is translocated into the periplasm, where it is rapidly degraded. A particular feature of SecM is the presence of a stalling sequence at its C-terminus, which causes a transient translation arrest that is released during translocation. However, when translocation is compromised, translational arrest persists and the formation of the stem-loop is blocked, allowing the ribosome unhindered access to the Shine-Dalgarno sequence of the secA gene and increases the production of SecA (Ito et al., 2018). The use of monitoring substrates for adjusting the protein transport capacity has also been shown in Vibrio alginolyticus, where the substrate VemP controls the switch between a sodium-coupled SecDF2 complex and a proton-coupled SecDF1 complex in low Na+ environments (Ishii et al., 2015; Miyazaki et al., 2020). Similar systems are also active in Gram-positive bacteria, like B. subtilis. Here, the monitoring substrate MifM controls the expression of the alternative YidC2 when YidC1 is compromised (Chiba et al., 2011; Chiba and Ito, 2012, 2015).
Besides the minor growth-phase dependent regulation of SecY and SecE as described above, entries in the E. coli gene expression database do not reveal a strong transcriptional regulation of the respective genes in response to different growth or stress conditions (GenExpDB3). This is also validated by a proteomic approach, which demonstrated comparable levels of SecY, SecE and SecG over the entire growth phase of E. coli (Soufi et al., 2015). This is rather surprising, because secY is encoded in the spc operon together with genes for several ribosomal proteins (Lindahl et al., 1990; Ikegami et al., 2005). These genes are significantly down-regulated during stationary phase or when cells encounter stress conditions (Coenye and Vandamme, 2005; Ikegami et al., 2005; Starosta et al., 2014). The spc operon is under control of the rplN promotor and binding of RNA-polymerase is inhibited when cells enter stationary phase by the transcription factor DksA and the alarmone ppGpp, a hyper-phosphorylated guanosine derivative (Lemke et al., 2011; Haas et al., 2020). Thus, secY expression is obviously disconnected from the regulation of the other genes within the spc operon, probably by the presence of an internal promotor.
In E. coli, the levels of the two alarmones ppGpp and pppGpp are mainly controlled by the activity of two enzymes, RelA and SpoT (Atkinson et al., 2011; Potrykus and Cashel, 2018; Pausch et al., 2020). RelA primarily responds to stalled ribosomes upon amino acid starvation (Starosta et al., 2014; Steinchen et al., 2020), while SpoT activity increases upon fatty acid or carbon starvation (Battesti and Bouveret, 2009; Figure 12). High levels of (p)ppGpp induce a process called stringent response that is associated with a significant re-programming of cellular activities (Bennison et al., 2019; Irving et al., 2020). The (p)ppGpp levels raise from approx. 40 μM during exponential phase up to approx. 1 mM at the transition into stationary phase or upon amino acid starvation (Varik et al., 2017; Haas et al., 2020; Steinchen et al., 2020). Cellular re-programming is induced by two mechanisms: allosteric regulation of target proteins, like RNA polymerase, which leads to reduced expression of the spc-operon (Liang et al., 1999; Steinchen et al., 2020), and competitive inhibition of GTP-binding proteins, like the ribosome assembly factor ObgE (Sato et al., 2005; Persky et al., 2009; Feng et al., 2014), the initiation factor IF2 (Diez et al., 2020) or elongation factor EF-G (Mitkevich et al., 2010; Steinchen et al., 2020). As a result, ribosome biogenesis and translation are adjusted to substrate limitation.
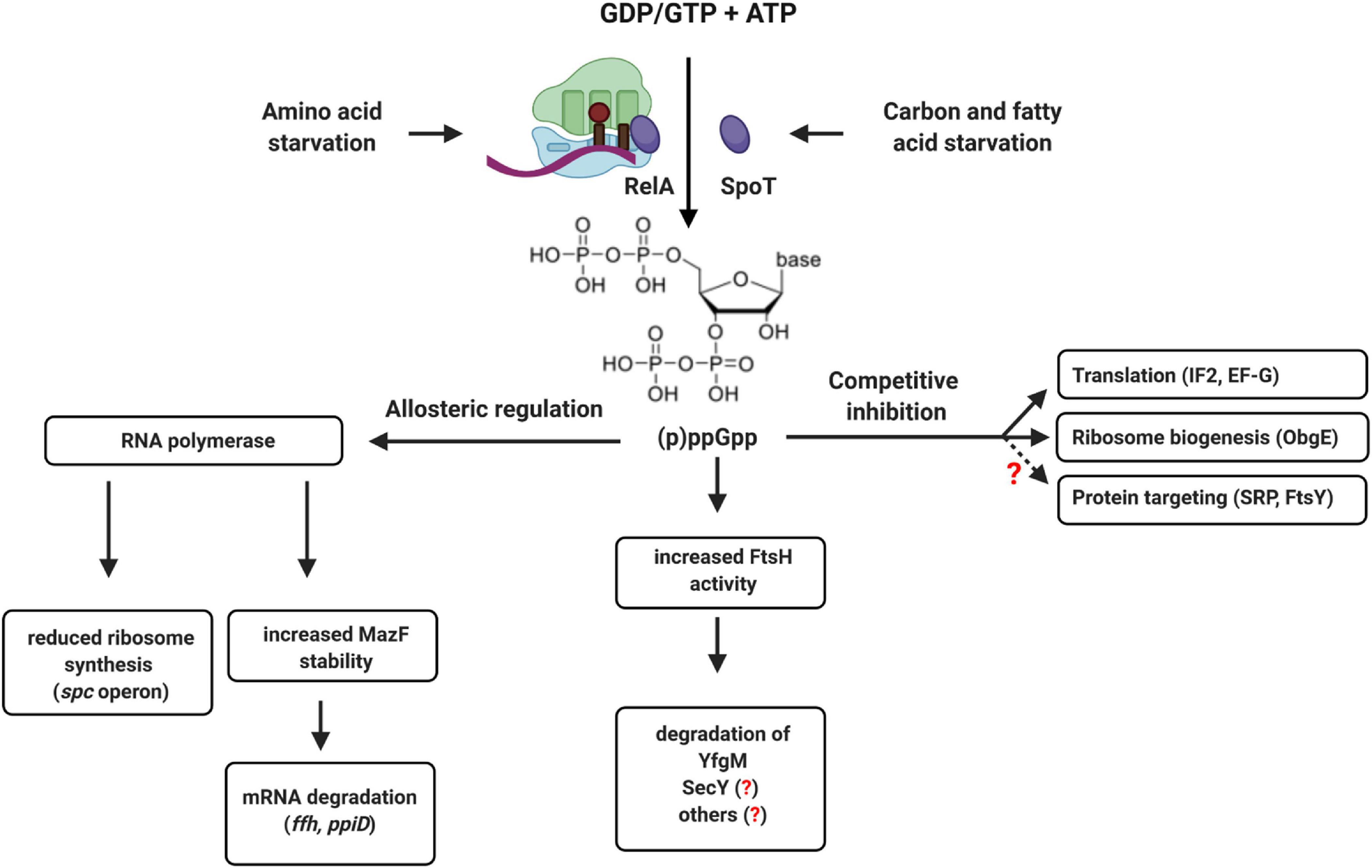
Figure 12. (p)ppGpp-dependent regulation of translation and protein transport in bacteria. The alarmones ppGpp and pppGpp are synthesized upon amino acid starvation by the ribosome-associated protein RelA or by the cytosolic protein SpoT upon carbon or fatty acid starvation. Allosteric regulation of RNA polymerase by (p)ppGpp reduces ribosome biogenesis and increases the stability of the ribonuclease MazF, which degrades multiple mRNAs. This includes the mRNA encoding for Ffh, the protein component of the bacterial SRP, or the ppiD mRNA, encoding for an accessory subunit of the SecYEG translocon. (p)ppGpp also increases the activity of FtsH, which can degrade SecY and YfgM. YfgM forms a complex with PpiD that associates with the SecYEG translocon. Whether SecY is specifically degraded by FtsH upon (p)ppGpp accumulation is not shown yet. (p)ppGpp also acts as competitive inhibitor of GTP-binding proteins like translation factors (IF2 and EF-G) or ribosome biogenesis proteins (ObgE). This leads to reduced ribosome biogenesis and reduced translation upon stress. Although not yet experimentally shown, it appears likely that increasing (p)ppGpp concentrations also inhibit the two GTPases SRP and FtsY, which would fine-tune the protein targeting machinery to the reduced translation rates.
Increasing (p)ppGpp concentrations likely also interfere with the activity of the GTPases FtsY and SRP and both proteins were identified as potential targets of (p)ppGpp (Wang B. et al., 2019). This would enable cells to adjust the protein targeting machinery to the reduced protein synthesis rate upon entry into stationary phase or during nutrient limitation. However, the consequences of (p)ppGpp on SRP-dependent protein targeting have not been studied so far. The accumulation of ppGpp also activates the MazEF toxin-antitoxin system (Moll and Engelberg-Kulka, 2012) and the mRNAs of both PpiD and Ffh were identified as potential targets of the riboendonuclease MazF (Sauert et al., 2016). This provides an additional link between stress conditions and the protein targeting and transport machinery that requires further analyses. Bacteria also produce hyper-phosphorylated adenosine derivatives, like (p)ppApp, although less is known about the conditions of synthesis and potential regulatory consequences (Travers, 1978; Bruhn-Olszewska et al., 2018; Ahmad et al., 2019). Still, it is tempting to speculate that by accumulating (p)ppGpp or (p)ppApp, bacteria can adjust protein transport by an allosteric or competitive mechanism, rather than by transcriptional or translational regulation. ppGpp also induces FtsH-dependent degradation of the SecYEG-interacting protein YfgM when cells enter stationary phase (Bittner et al., 2015). This is suggested to relieve the response regulator RcsB, thereby allowing cellular protection by the Rcs phosphorelay system (Lasserre et al., 2006; Wall et al., 2018). However, this would also reduce the levels of the PpiD-YfgM complex and thus impact on the SecYEG interactome under stress conditions. How stress conditions influence the steady-state levels of the protein transport machinery and the dynamic equilibrium between the different SecYEG assemblies is largely a terra incognita, but a promising area for future research.
The rapid rise of antibiotic resistance is a major problem for treating infections and novel antimicrobial strategies are of crucial importance (Rodríguez-Rojas et al., 2013; Sulaiman and Lam, 2021). Initial studies on exploring the protein transport machinery as potential target were focused on SecA inhibitors, because SecA homologues are absent in metazoans and SecA inhibition would affect most periplasmic and OMPs as well as some inner membrane proteins (Pohlschroder et al., 2005). Azide was the first described inhibitor of SecA (Oliver et al., 1990), but has no medical relevance due to its high toxicity (Chang and Lamm, 2003). Additional small molecule SecA inhibitors with broad-spectrum activity have been developed and include compounds like SEW-05929 and CD 09529, which inhibit the ATPase activity of SecA but are inactive on wild type E. coli strains (Li et al., 2008; Figure 13). Further studies identified 4-oxo-5-cyano thiouracils (Chaudhary et al., 2015), Fluorescein analogs (Huang et al., 2012) and triazole-pyrimidine analogs (Cui et al., 2016; Jin et al., 2016) as SecA inhibitors that are active against E. coli and S. aureus (Rao et al., 2014; De Waelheyns et al., 2015; Van Puyenbroeck and Vermeire, 2018).
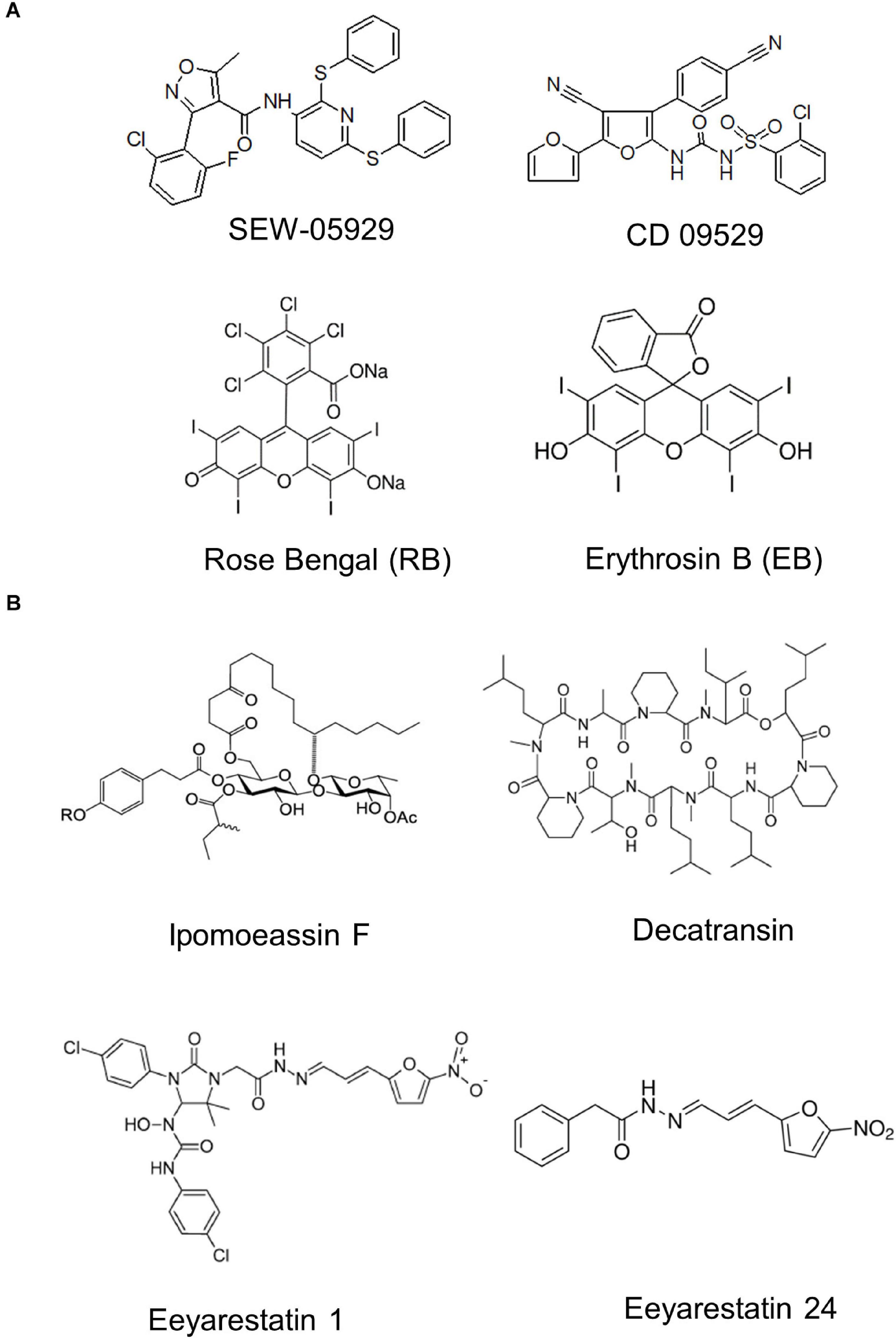
Figure 13. Inhibitors of bacterial protein translocation. (A) Inhibitors of the ATPase SecA. (B) Inhibitors of the SecYEG-translocon. Ipomeassin F, decatransin, eeyarastatin 1 and eeyarastatin 24 also act on the homologous Sec61 complex in eukaryotes. Chemical structures were retrieved from the Sigma Aldrich web resource (https://www.sigmaaldrich.com/) or adapted from (Li et al., 2008; Van Puyenbroeck and Vermeire, 2018; Zong et al., 2019).
The first characterized inhibitors of the Sec complex were synthetic signal peptides that have been shown to inhibit the eukaryotic Sec61 complex (Austen et al., 1984). The mammalian Sec61 complex is also inhibited by lanthanum ions, which stabilize the Sec61 channel in its open state (Erdmann et al., 2009). Components that inhibit both the eukaryotic Sec61 complex and the bacterial SecYEG complex are the glycoresin Ipomoeassin F (IpomF) (Zong et al., 2019; Steinberg et al., 2020), eeyarestatin (Cross et al., 2009; Steenhuis et al., 2021) and decatransin (Junne et al., 2015; Kalies and Römisch, 2015). IpomF was isolated from the morning glory Ipomea squamosa and shown to bind most likely near the lateral gate of Sec61α (Zong et al., 2019). IpomF also inhibits SecYEG-dependent transport in vitro, but this requires significantly higher concentrations than required for inhibition of Sec61-dependent transport (Zong et al., 2019; Steinberg et al., 2020). IpomF does not prevent the initial contact of substrate proteins with the SecYEG translocon, but rather blocks later stages of translocation (Steinberg et al., 2020).
Eeyarestatin I (ESI) was initially discovered as inhibitor of the retrograde protein transport into the endoplasmic reticulum and then shown to inhibit co-translational protein transport by the Sec61 complex (Cross et al., 2009). ESI does not inhibit growth of E. coli, but a smaller variant of ESI, ES24 (Gamayun et al., 2019), is active against E. coli and several clinically relevant pathogens (Steenhuis et al., 2021). ES24 likely binds to the cytosolic part of the lateral gate (Gamayun et al., 2019), but the antibacterial activity depends on the presence of the nitroreductases NfsA and NfsB, indicating that a specific reduction step is required to activate ES24 (Steenhuis et al., 2021). Decatransin is a naturally occurring fungal decadepsipeptide that was identified in a cancer drug screen and later shown to inhibit SecYEG/Sec61. Decatransin-resistant mutations mapped to the pore ring and to the plug of the Sec channel, suggesting that decatransin interferes with channel opening (Junne et al., 2015). However, whether these SecA- and SecY-inhibiting compounds also have clinical relevance requires further investigation.
The bacterial SecYEG translocon has been the focus of intense research for decades and served as a paradigm for genetic, biochemical and structural studies on protein transport mechanisms. The progress that has been made from the early genetic screens (Bassford et al., 1991; Beckwith, 2013) to the currently available structures is incredible (Smets et al., 2019; Tanaka and Tsukazaki, 2019). Snap-shots of the SecYEG translocon in contact with its most prominent partner proteins and of the SecYEG translocon in action during translocation or insertion of protein substrates have been attained and provide first insights into how these protein transport channels work. Still, structural information of substrate-engaged larger SecYEG assemblies, like the SecYEG-YidC complex, the SecYEG-PpiD/YfgM complex or the HTL, are needed for understanding how the SecYEG translocon handles the large variety of potential substrates. Equally needed are structures of the SecYEG translocon during the insertion of multi-spanning membrane proteins. It is also evident that the current picture of the SecYEG interactome is incomplete and includes only the most stable and abundant partner proteins. Many transient interactions only emerged upon improved mass spectrometry methods (Carlson et al., 2019; Jauss et al., 2019) and the functional characterization of these transient contacts will be a major challenge for the future. This will be particularly demanding if these contacts are only required for a specific subset of substrates, which are not in the tool box of frequently used model substrates. Analysing the transport of membrane proteins with large soluble domains at the N-terminus (Facey and Kuhn, 2003; Maier et al., 2008; Rawat et al., 2015; Wang S. et al., 2019) or very small membrane proteins, which basically consist of just a single transmembrane domain, has already revealed unexpected targeting and insertion requirements (Steinberg et al., 2018, 2020). Despite the increasing number of proteins interacting with the SecYEG translocon, the number of identified contact sites on SecY is rather low and mainly includes the cytosolic loop 5, the lateral gate and the periplasmic vestibule. This suggests that some proteins either compete for SecY binding, or interact with dedicated sub-populations of the SecYEG translocon and these subpopulations need to be further characterized. Our current view on bacterial protein transport pathways follows a rather strict dissection into multiple separate transport pathways, but recent data suggest that these pathways are intertwined. The best-studied example is of course the SecYEG-YidC interaction (Scotti et al., 2000), where YidC likely helps substrates to exit the SecY channel (Beck et al., 2001; Houben et al., 2002), although YidC can also act as SecYEG-independent insertase (Samuelson et al., 2000; Serek et al., 2004). But there are more examples, like the SecYEG-Tat interaction (Keller et al., 2012) or the SecYEG-Bam interaction (Alvira et al., 2020), and the collaboration between different transport systems needs to be further explored. Finally, it is largely unknown how the protein transport machinery responds to environmental changes or to stress conditions. Considering the multifaceted responses that down-regulate protein synthesis when cell encounter non-favorable conditions, it appears more than likely that similar, but so far unexplored mechanisms, also modulate the protein transport capacity of the cell. Thus, there is still a lot to learn about the SecYEG translocon or, to cite famous Isaac Newton: “What we know is a drop. What we don’t know is an ocean.”
JO, RN, AN, PS, and H-GK designed the review, prepared the figures, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) to HGK (DFG grants KO2184/8, KO2184/9 (SPP2002), SFB1381, Project-ID 403222702, and RTG 2202, Project-ID 278002225). Funding sources were not involved in the design of the article, writing the article or decision to submit the article for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures were created with BioRender.com and with PyMol.
Abell, B. M., Pool, M. R., Schlenker, O., Sinning, I., and High, S. (2004). Signal recognition particle mediates post-translational targeting in eukaryotes. EMBO J. 23, 2755–2764.
Ahmad, S., Wang, B., Walker, M. D., Tran, H. R., Stogios, P. J., Savchenko, A., et al. (2019). An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp. Nature 575, 674–678.
Akiyama, Y., Kihara, A., Tokuda, H., and Ito, K. (1996). FtsH (HflB) is an ATP-dependent protease selectively acting on SecY and some other membrane proteins. J. Biol. Chem. 271, 31196–31201.
Akopian, D., Dalal, K., Shen, K., Duong, F., and Shan, S. O. (2013a). SecYEG activates GTPases to drive the completion of cotranslational protein targeting. J. Cell Biol. 200, 397–405. doi: 10.1083/jcb.201208045
Akopian, D., Shen, K., Zhang, X., and Shan, S. O. (2013b). Signal recognition particle: an essential protein-targeting machine. Annu. Rev. Biochem. 82, 693–721.
Alami, M., Dalal, K., Lelj-Garolla, B., Sligar, S. G., and Duong, F. (2007). Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 26, 1995–2004. doi: 10.1038/sj.emboj.7601661
Allen, W. J., Corey, R. A., Oatley, P., Sessions, R. B., Baldwin, S. A., Radford, S. E., et al. (2016). Two-way communication between SecY and SecA suggests a Brownian ratchet mechanism for protein translocation. eLife 5:e15598. doi: 10.7554/eLife.15598
Allen, W. J., Watkins, D. W., Dillingham, M. S., and Collinson, I. (2020). Refined measurement of SecA-driven protein secretion reveals that translocation is indirectly coupled to ATP turnover. Proc. Natl. Acad. Sci. U.S.A. 117, 31808–31816. doi: 10.1073/pnas.2010906117
Alvira, S., Watkins, D. W., Troman, L., Allen, W. J., Lorriman, J. S., Degliesposti, G., et al. (2020). Inter-membrane association of the Sec and BAM translocons for bacterial outer-membrane biogenesis. eLife 9:e60669. doi: 10.7554/eLife.60669
Angelini, S., Boy, D., Schiltz, E., and Koch, H. G. (2006). Membrane binding of the bacterial signal recognition particle receptor involves two distinct binding sites. J. Cell Biol. 174, 715–724. doi: 10.1083/jcb.200606093
Angelini, S., Deitermann, S., and Koch, H. G. (2005). FtsY, the bacterial signal-recognition particle receptor, interacts functionally and physically with the SecYEG translocon. EMBO Rep. 6, 476–481. doi: 10.1038/sj.embor.7400385
Anghel, S. A., McGilvray, P. T., Hegde, R. S., and Keenan, R. J. (2017). Identification of Oxa1 homologs operating in the eukaryotic endoplasmic reticulum. Cell Rep. 21, 3708–3716. doi: 10.1016/j.celrep.2017.12.006
Antonoaea, R., Fürst, M., Nishiyama, K.-I., and Müller, M. (2008). The periplasmic chaperone PpiD interacts with secretory proteins exiting from the SecYEG translocon. Biochemistry 47, 5649–5656.
Ast, T., Cohen, G., and Schuldiner, M. (2013). A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell 152, 1134–1145. doi: 10.1016/j.cell.2013.02.003
Ataide, S. F., Schmitz, N., Shen, K., Ke, A., Shan, S. O., Doudna, J. A., et al. (2011). The crystal structure of the signal recognition particle in complex with its receptor. Science 331, 881–886. doi: 10.1126/science.1196473
Atkinson, G. C., Tenson, T., and Hauryliuk, V. (2011). The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6:e23479. doi: 10.1371/journal.pone.0023479
Austen, B. M., Hermon-Taylor, J., Kaderbhai, M. A., and Ridd, D. H. (1984). Design and synthesis of a consensus signal sequence that inhibits protein translocation into rough microsomal vesicles. Biochem. J. 224, 317–325. doi: 10.1042/bj2240317
Baars, L., Wagner, S., Wickstrom, D., Klepsch, M., Ytterberg, A. J., van Wijk, K. J., et al. (2008). Effects of SecE depletion on the inner and outer membrane proteomes of Escherichia coli. J. Bacteriol. 190, 3505–3525.
Baars, L., Ytterberg, A. J., Drew, D., Wagner, S., Thilo, C., van Wijk, K. J., et al. (2006). Defining the role of the Escherichia coli chaperone SecB using comparative proteomics. J. Biol. Chem. 281, 10024–10034. doi: 10.1074/jbc.M509929200
Banerjee, T., Lindenthal, C., and Oliver, D. (2017a). SecA functions in vivo as a discrete anti-parallel dimer to promote protein transport. Mol. Microbiol. 103, 439–451.
Banerjee, T., Zheng, Z., Abolafia, J., Harper, S., and Oliver, D. (2017b). The SecA protein deeply penetrates into the SecYEG channel during insertion, contacting most channel transmembrane helices and periplasmic regions. J. Biol. Chem. 292, 19693–19707.
Bassford, P., Beckwith, J., Ito, K., Kumamoto, C., Mizushima, S., Oliver, D., et al. (1991). The primary pathway of protein export in E. coli. Cell 65, 367–368.
Battesti, A., and Bouveret, E. (2009). Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein-SpoT interaction. J. Bacteriol. 191, 616–624.
Bayer, M. E. (1968). Areas of adhesion between wall and membrane of Escherichia coli. J. Gen. Microbiol. 53, 395–404.
Bechtluft, P., Kedrov, A., Slotboom, D. J., Nouwen, N., Tans, S. J., and Driessen, A. J. (2010). Tight hydrophobic contacts with the SecB chaperone prevent folding of substrate proteins. Biochemistry 49, 2380–2388. doi: 10.1021/bi902051e
Beck, K., Eisner, G., Trescher, D., Dalbey, R. E., Brunner, J., and Muller, M. (2001). YidC, an assembly site for polytopic Escherichia coli membrane proteins located in immediate proximity to the SecYE translocon and lipids. EMBO Rep. 2, 709–714. doi: 10.1093/embo-reports/kve154
Beck, K., Wu, L. F., Brunner, J., and Muller, M. (2000). Discrimination between SRP- and SecA/SecB-dependent substrates involves selective recognition of nascent chains by SRP and trigger factor. EMBO J. 19, 134–143.
Becker, T., Bhushan, S., Jarasch, A., Armache, J. P., Funes, S., Jossinet, F., et al. (2009). Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science 326, 1369–1373. doi: 10.1126/science.1178535
Beckwith, J. (2013). The Sec-dependent pathway. Res. Microbiol. 164, 497–504. doi: 10.1016/j.resmic.2013.03.007
Beha, D., Deitermann, S., Muller, M., and Koch, H. G. (2003). Export of beta-lactamase is independent of the signal recognition particle. J. Biol. Chem. 278, 22161–22167. doi: 10.1074/jbc.M300929200
Bennison, D. J., Irving, S. E., and Corrigan, R. M. (2019). The impact of the stringent response on TRAFAC GTPases and prokaryotic ribosome assembly. Cells 8:1313. doi: 10.3390/cells8111313
Bernstein, H. D., and Hyndman, J. B. (2001). Physiological basis for conservation of the signal recognition particle targeting pathway in Escherichia coli. J. Bacteriol. 183, 2187–2197. doi: 10.1128/jb.183.7.2187-2197.2001
Bischoff, L., Wickles, S., Berninghaus, O., van der Sluis, E. O., and Beckmann, R. (2014). Visualization of a polytopic membrane protein during SecY-mediated membrane insertion. Nat. Commun. 5:4103.
Bittner, L. M., Westphal, K., and Narberhaus, F. (2015). Conditional proteolysis of the membrane protein YfgM by the FtsH protease depends on a novel N-terminal degron. J. Biol. Chem. 290, 19367–19378. doi: 10.1074/jbc.M115.648550
Bogdanov, M., Pyrshev, K., Yesylevskyy, S., Ryabichko, S., Boiko, V., Ivanchenko, P., et al. (2020). Phospholipid distribution in the cytoplasmic membrane of Gram-negative bacteria is highly asymmetric, dynamic, and cell shape-dependent. Sci. Adv. 6:eaaz6333. doi: 10.1126/sciadv.aaz6333
Bonardi, F., Halza, E., Walko, M., Du Plessis, F., Nouwen, N., Feringa, B. L., et al. (2011). Probing the SecYEG translocation pore size with preproteins conjugated with sizable rigid spherical molecules. Proc. Natl. Acad. Sci. U.S.A. 108, 7775–7780. doi: 10.1073/pnas.1101705108
Bornemann, T., Holtkamp, W., and Wintermeyer, W. (2014). Interplay between trigger factor and other protein biogenesis factors on the ribosome. Nat. Commun. 5:4180.
Bornemann, T., Jockel, J., Rodnina, M. V., and Wintermeyer, W. (2008). Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat. Struct. Mol. Biol. 15, 494–499. doi: 10.1038/nsmb.1402
Borowska, M. T., Dominik, P. K., Anghel, S. A., Kossiakoff, A. A., and Keenan, R. J. (2015). A YidC-like protein in the archaeal plasma membrane. Structure 23, 1715–1724. doi: 10.1016/j.str.2015.06.025
Bos, M. P., Robert, V., and Tommassen, J. (2007). Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 61, 191–214. doi: 10.1146/annurev.micro.61.080706.093245
Botte, M., Zaccai, N. R., Nijeholt, J. L., Martin, R., Knoops, K., Papai, G., et al. (2016). A central cavity within the holo-translocon suggests a mechanism for membrane protein insertion. Sci. Rep. 6:38399.
Boy, D., and Koch, H. G. (2009). Visualization of distinct entities of the SecYEG translocon during translocation and integration of bacterial proteins. Mol. Biol. Cell 20, 1804–1815.
Braig, D., Bar, C., Thumfart, J. O., and Koch, H. G. (2009). Two cooperating helices constitute the lipid-binding domain of the bacterial SRP receptor. J. Mol. Biol. 390, 401–413. doi: 10.1016/j.jmb.2009.04.061
Bruhn-Olszewska, B., Molodtsov, V., Sobala, M., Dylewski, M., Murakami, K. S., Cashel, M., et al. (2018). Structure-function comparisons of (p)ppApp vs (p)ppGpp for Escherichia coli RNA polymerase binding sites and for rrnB P1 promoter regulatory responses in vitro. Biochim. Biophys. Acta Gene Regul. Mech. 1861, 731–742. doi: 10.1016/j.bbagrm.2018.07.005
Brundage, L., Hendrick, J. P., Schiebel, E., Driessen, A. J., and Wickner, W. (1990). The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell 62, 649–657. doi: 10.1016/0092-8674(90)90111-q
Bürk, J., Weiche, B., Wenk, M., Boy, D., Nestel, S., Heimrich, B., et al. (2009). Depletion of the signal recognition particle receptor inactivates ribosomes in Escherichia coli. J. Bacteriol. 191, 7017–7026.
Can, M. T., Kurkcuoglu, Z., Ezeroglu, G., Uyar, A., Kurkcuoglu, O., and Doruker, P. (2017). Conformational dynamics of bacterial trigger factor in apo and ribosome-bound states. PLoS One 12:e0176262. doi: 10.1371/journal.pone.0176262
Carlson, M. L., Stacey, R. G., Young, J. W., Wason, I. S., Zhao, Z., Rattray, D. G., et al. (2019). Profiling the E. coli membrane interactome captured in peptidisc libraries. eLife 8:e46615. doi: 10.7554/eLife.46615
Castanie-Cornet, M. P., Bruel, N., and Genevaux, P. (2014). Chaperone networking facilitates protein targeting to the bacterial cytoplasmic membrane. Biochim. Biophys. Acta. 1843, 1442–1456. doi: 10.1016/j.bbamcr.2013.11.007
Catipovic, M. A., and Rapoport, T. A. (2020). Protease protection assays show polypeptide movement into the SecY channel by power strokes of the SecA ATPase. EMBO Rep. 21:e50905. doi: 10.15252/embr.202050905
Catipovic, M. A., Bauer, B. W., Loparo, J. J., and Rapoport, T. A. (2019). Protein translocation by the SecA ATPase occurs by a power-stroke mechanism. EMBO J. 38:e101140. doi: 10.15252/embj.2018101140
Chang, S., and Lamm, S. H. (2003). Human health effects of sodium azide exposure: a literature review and analysis. Int. J. Toxicol. 22, 175–186. doi: 10.1080/10915810305109
Chatzi, K. E., Sardis, M. F., Tsirigotaki, A., Koukaki, M., Sostaric, N., Konijnenberg, A., et al. (2017). Preprotein mature domains contain translocase targeting signals that are essential for secretion. J. Cell Biol. 216, 1357–1369.
Chaudhary, A. S., Jin, J., Chen, W., Tai, P. C., and Wang, B. (2015). Design, syntheses and evaluation of 4-oxo-5-cyano thiouracils as SecA inhibitors. Bioorg. Med. Chem. 23, 105–117. doi: 10.1016/j.bmc.2014.11.017
Chiba, S., and Ito, K. (2012). Multisite ribosomal stalling: a unique mode of regulatory nascent chain action revealed for MifM. Mol. Cell 47, 863–872. doi: 10.1016/j.molcel.2012.06.034
Chiba, S., and Ito, K. (2015). MifM monitors total YidC activities of Bacillus subtilis, including that of YidC2, the target of regulation. J. Bacteriol. 197, 99–107.
Chiba, S., Kanamori, T., Ueda, T., Akiyama, Y., Pogliano, K., and Ito, K. (2011). Recruitment of a species-specific translational arrest module to monitor different cellular processes. Proc. Natl. Acad. Sci. U.S.A. 108, 6073–6078. doi: 10.1073/pnas.1018343108
Chorev, D. S., Baker, L. A., Wu, D., Beilsten-Edmands, V., Rouse, S. L., Zeev-Ben-Mordehai, T., et al. (2018). Protein assemblies ejected directly from native membranes yield complexes for mass spectrometry. Science 362, 829–834. doi: 10.1126/science.aau0976
Christie, P. J. (2019). The rich tapestry of bacterial protein translocation systems. Protein J. 38, 389–408.
Chum, A. P., Shoemaker, S. R., Fleming, P. J., and Fleming, K. G. (2019). Plasticity and transient binding are key ingredients of the periplasmic chaperone network. Protein Sci 28, 1340–1349. doi: 10.1002/pro.3641
Chun, S. Y., and Randall, L. L. (1994). In vivo studies of the role of SecA during protein export in Escherichia coli. J. Bacteriol. 176, 4197–4203.
Coenye, T., and Vandamme, P. (2005). Organisation of the S10, spc and alpha ribosomal protein gene clusters in prokaryotic genomes. FEMS Microbiol. Lett. 242, 117–126. doi: 10.1016/j.femsle.2004.10.050
Collinson, I. (2019). The dynamic ATP-driven mechanism of bacterial protein translocation and the critical role of phospholipids. Front. Microbiol. 10:1217. doi: 10.3389/fmicb.2019.01217
Collinson, I., Corey, R. A., and Allen, W. J. (2015). Channel crossing: How are proteins shipped across the bacterial plasma membrane? Philos. Trans. R. Soc. Lond. B Biol. Sci. 370:20150025. doi: 10.1098/rstb.2015.0025
Corey, R. A., Ahdash, Z., Shah, A., Pyle, E., Allen, W. J., Fessl, T., et al. (2019). ATP-induced asymmetric pre-protein folding as a driver of protein translocation through the Sec machinery. eLife 8:e41803. doi: 10.7554/eLife.41803
Corey, R. A., Allen, W. J., Komar, J., Masiulis, S., Menzies, S., Robson, A., et al. (2016). Unlocking the bacterial SecY Translocon. Structure 24, 518–527. doi: 10.1016/j.str.2016.02.001
Corey, R. A., Pyle, E., Allen, W. J., Watkins, D. W., Casiraghi, M., Miroux, B., et al. (2018). Specific cardiolipin-SecY interactions are required for proton-motive force stimulation of protein secretion. Proc. Natl. Acad. Sci. U.S.A. 115, 7967–7972. doi: 10.1073/pnas.1721536115
Costa, E. A., Subramanian, K., Nunnari, J., and Weissman, J. S. (2018). Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science 359, 689–692. doi: 10.1126/science.aar3607
Crane, J. M., and Randall, L. L. (2017). The Sec system: protein export in Escherichia coli. EcoSal Plus 7:10.1128/ecosalplus.ESP-0002-2017. doi: 10.1128/ecosalplus.ESP-0002-2017
Crane, J. M., Suo, Y., Lilly, A. A., Mao, C., Hubbell, W. L., and Randall, L. L. (2006). Sites of interaction of a precursor polypeptide on the export chaperone SecB mapped by site-directed spin labeling. J. Mol. Biol. 363, 63–74. doi: 10.1016/j.jmb.2006.07.021
Cristobal, S., Scotti, P., Luirink, J., von Heijne, G., and de Gier, J. W. (1999). The signal recognition particle-targeting pathway does not necessarily deliver proteins to the sec-translocase in Escherichia coli. J. Biol. Chem. 274, 20068–20070.
Cross, B. C., and High, S. (2009). Dissecting the physiological role of selective transmembrane-segment retention at the ER translocon. J. Cell Sci. 122(Pt 11), 1768–1777. doi: 10.1242/jcs.046094
Cross, B. C., McKibbin, C., Callan, A. C., Roboti, P., Piacenti, M., Rabu, C., et al. (2009). Eeyarestatin I inhibits Sec61-mediated protein translocation at the endoplasmic reticulum. J. Cell Sci. 122(Pt 23), 4393–4400.
Cui, J., Jin, J., Chaudhary, A. S., Hsieh, Y. H., Zhang, H., Dai, C., et al. (2016). Design, synthesis and evaluation of Triazole-Pyrimidine analogues as SecA inhibitors. ChemMedChem 11, 43–56. doi: 10.1016/j.bmc.2015.09.027
Dalal, K., Nguyen, N., Alami, M., Tan, J., Moraes, T. F., Lee, W. C., et al. (2009). Structure, binding, and activity of Syd, a SecY-interacting protein. J. Biol. Chem. 284, 7897–7902. doi: 10.1074/jbc.M808305200
Dalbey, R., Koch, H. G., and Kuhn, A. (2017). Targeting and insertion of membrane proteins. EcoSalPlus 7, 1–28.
Danese, P. N., Murphy, C. K., and Silhavy, T. J. (1995). Multicopy suppression of cold-sensitive sec mutations in Escherichia coli. J. Bacteriol. 177, 4969–4973. doi: 10.1128/jb.177.17.4969-4973.1995
Das, S., and Oliver, D. B. (2011). Mapping of the SecA. SecY and SecA. SecG interfaces by site-directed in vivo photocross-linking. J. Biol. Chem. 286, 12371–12380. doi: 10.1074/jbc.M110.182931
De Geyter, J., Portaliou, A. G., Srinivasu, B., Krishnamurthy, S., Economou, A., and Karamanou, S. (2020). Trigger factor is a bona fide secretory pathway chaperone that interacts with SecB and the translocase. EMBO Rep. 21:e49054. doi: 10.15252/embr.201949054
de Gier, J. W., Scotti, P. A., Saaf, A., Valent, Q. A., Kuhn, A., Luirink, J., et al. (1998). Differential use of the signal recognition particle translocase targeting pathway for inner membrane protein assembly in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 95, 14646–14651. doi: 10.1093/emboj/18.11.2982
de Keyzer, J., van der Sluis, E. O., Spelbrink, R. E., Nijstad, N., de Kruijff, B., Nouwen, N., et al. (2005). Covalently dimerized SecA is functional in protein translocation. J. Biol. Chem. 280, 35255–35260. doi: 10.1074/jbc.M506157200
de Leeuw, E., te Kaat, K., Moser, C., Menestrina, G., Demel, R., de Kruijff, B., et al. (2000). Anionic phospholipids are involved in membrane association of FtsY and stimulate its GTPase activity. EMBO J. 19, 531–541. doi: 10.1093/emboj/19.4.531
De Waelheyns, E., Segers, K., Sardis, M. F., Anne, J., Nicolaes, G. A., and Economou, A. (2015). Identification of small-molecule inhibitors against SecA by structure-based virtual ligand screening. J. Antibiot. 68, 666–673. doi: 10.1038/ja.2015.53
Deitermann, S., Sprie, G. S., and Koch, H. G. (2005). A dual function for SecA in the assembly of single spanning membrane proteins in Escherichia coli. J. Biol. Chem. 280, 39077–39085. doi: 10.1074/jbc.M509647200
Denks, K., Sliwinski, N., Erichsen, V., Borodkina, B., Origi, A., and Koch, H. G. (2017). The signal recognition particle contacts uL23 and scans substrate translation inside the ribosomal tunnel. Nat. Microbiol. 2:16265. doi: 10.1028/nmicrobiol.2016.265
Denks, K., Vogt, A., Sachelaru, I., Petriman, N. A., Kudva, R., and Koch, H. G. (2014). The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Mol. Membr. Biol. 31, 58–84.
Deuerling, E., Patzelt, H., Vorderwulbecke, S., Rauch, T., Kramer, G., Schaffitzel, E., et al. (2003). Trigger Factor and DnaK possess overlapping substrate pools and binding specificities. Mol. Microbiol. 47, 1317–1328.
Deuerling, E., Schulze-Specking, A., Tomoyasu, T., Mogk, A., and Bukau, B. (1999). Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400, 693–696. doi: 10.1038/23301
Diez, S., Ryu, J., Caban, K., Gonzalez, R. L. Jr., and Dworkin, J. (2020). The alarmones (p)ppGpp directly regulate translation initiation during entry into quiescence. Proc. Natl. Acad. Sci. U.S.A. 117, 15565–15572. doi: 10.1073/pnas.1920013117
Douville, K., Price, A., Eichler, J., Economou, A., and Wickner, W. (1995). SecYEG and SecA are the stoichiometric components of preprotein translocase. J. Biol. Chem. 270, 20106–20111.
Dowhan, W., Vitrac, H., and Bogdanov, M. (2019). Lipid-assisted membrane protein folding and topogenesis. Protein J. 38, 274–288.
Draycheva, A., Bornemann, T., Ryazanov, S., Lakomek, N. A., and Wintermeyer, W. (2016). The bacterial SRP receptor, FtsY, is activated on binding to the translocon. Mol. Microbiol. 102, 152–167. doi: 10.1111/mmi.13452
Draycheva, A., Lee, S., and Wintermeyer, W. (2018). Cotranslational protein targeting to the membrane: nascent-chain transfer in a quaternary complex formed at the translocon. Sci. Rep. 8:9922.
Driessen, A. J., and Nouwen, N. (2008). Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77, 643–667. doi: 10.1146/annurev.biochem.77.061606.160747
Driessen, A. J., and Wickner, W. (1991). Proton transfer is rate-limiting for translocation of precursor proteins by the Escherichia coli translocase. Proc. Natl. Acad. Sci. U.S.A. 88, 2471–2475.
du Plessis, D. J., Berrelkamp, G., Nouwen, N., and Driessen, A. J. (2009). The lateral gate of SecYEG opens during protein translocation. J. Biol. Chem. 284, 15805–15814. doi: 10.1074/jbc.M901855200
du Plessis, D. J., Nouwen, N., and Driessen, A. J. (2006). Subunit a of cytochrome o oxidase requires both YidC and SecYEG for membrane insertion. J. Biol. Chem. 281, 12248–12252. doi: 10.1074/jbc.M600048200
Duong, F., and Wickner, W. (1997). Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 16, 2756–2768.
Egea, P. F., Shan, S. O., Napetschnig, J., Savage, D. F., Walter, P., and Stroud, R. M. (2004). Substrate twinning activates the signal recognition particle and its receptor. Nature 427, 215–221. doi: 10.1038/nature02250
Eisner, G., Koch, H. G., Beck, K., Brunner, J., and Muller, M. (2003). Ligand crowding at a nascent signal sequence. J. Cell Biol. 163, 35–44. doi: 10.1083/jcb.200306069
Eitan, A., and Bibi, E. (2004). The core Escherichia coli signal recognition particle receptor contains only the N and G domains of FtsY. J. Bacteriol. 186, 2492–2494.
Erdmann, F., Jung, M., Eyrisch, S., Lang, S., Helms, V., Wagner, R., et al. (2009). Lanthanum ions inhibit the mammalian Sec61 complex in its channel dynamics and protein transport activity. FEBS Lett. 583, 2359–2364. doi: 10.1016/j.febslet.2009.06.032
Erez, E., Stjepanovic, G., Zelazny, A. M., Brugger, B., Sinning, I., and Bibi, E. (2010). Genetic evidence for functional interaction of the Escherichia coli signal recognition particle receptor with acidic lipids in vivo. J. Biol. Chem. 285, 40508–40514. doi: 10.1074/jbc.M110.140921
Erlandson, K. J., Miller, S. B., Nam, Y., Osborne, A. R., Zimmer, J., and Rapoport, T. A. (2008a). A role for the two-helix finger of the SecA ATPase in protein translocation. Nature 455, 984–987. doi: 10.1038/nature07439
Erlandson, K. J., Or, E., Osborne, A. R., and Rapoport, T. A. (2008b). Analysis of polypeptide movement in the SecY channel during SecA-mediated protein translocation. J. Biol. Chem. 283, 15709–15715. doi: 10.1074/jbc.M710356200
Ernst, I., Haase, M., Ernst, S., Yuan, S., Kuhn, A., and Leptihn, S. (2018). Large conformational changes of a highly dynamic pre-protein binding domain in SecA. Commun. Biol. 1:130.
Facey, S. J., and Kuhn, A. (2003). The sensor protein KdpD inserts into the Escherichia coli membrane independent of the Sec translocase and YidC. Eur. J. Biochem. 270, 1724–1734.
Fekkes, P., de Wit, J. G., van der Wolk, J. P., Kimsey, H. H., Kumamoto, C. A., and Driessen, A. J. (1998). Preprotein transfer to the Escherichia coli translocase requires the co-operative binding of SecB and the signal sequence to SecA. Mol. Microbiol. 29, 1179–1190. doi: 10.1046/j.1365-2958.1998.00997.x
Feng, B., Mandava, C. S., Guo, Q., Wang, J., Cao, W., Li, N., et al. (2014). Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol. 12:e1001866. doi: 10.1371/journal.pbio.1001866
Fessl, T., Watkins, D., Oatley, P., Allen, W. J., Corey, R. A., Horne, J., et al. (2018). Dynamic action of the Sec machinery during initiation, protein translocation and termination. eLife 7:e35112.
Focia, P. J., Shepotinovskaya, I. V., Seidler, J. A., and Freymann, D. M. (2004). Heterodimeric GTPase core of the SRP targeting complex. Science 303, 373–377. doi: 10.1126/science.1090827
Frauenfeld, J., Gumbart, J., Sluis, E. O., Funes, S., Gartmann, M., Beatrix, B., et al. (2011). Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 18, 614–621. doi: 10.1038/nsmb.2026
Freymann, D. M., Keenan, R. J., Stroud, R. M., and Walter, P. (1997). Structure of the conserved GTPase domain of the signal recognition particle. Nature 385, 361–364. doi: 10.1038/385361a0
Furst, M., Zhou, Y., Merfort, J., and Muller, M. (2018). Involvement of PpiD in Sec-dependent protein translocation. Biochim. Biophys. Acta Mol. Cell Res. 1865, 273–280. doi: 10.1016/j.bbamcr.2017.10.012
Furukawa, A., Nakayama, S., Yoshikaie, K., Tanaka, Y., and Tsukazaki, T. (2018). Remote coupled drastic β-Barrel to β-sheet transition of the protein translocation motor. Structure 26, 485–489.e2. doi: 10.1016/j.str.2018.01.002
Furukawa, A., Yoshikaie, K., Mori, T., Mori, H., Morimoto, Y. V., Sugano, Y., et al. (2017). Tunnel formation inferred from the I-form structures of the proton-driven protein secretion motor SecDF. Cell Rep. 19, 895–901. doi: 10.1016/j.celrep.2017.04.030
Gamayun, I., O’Keefe, S., Pick, T., Klein, M. C., Nguyen, D., McKibbin, C., et al. (2019). Eeyarestatin compounds selectively enhance Sec61-mediated Ca(2+) leakage from the endoplasmic reticulum. Cell Chem. Biol. 26, 571–583.e6. doi: 10.1016/j.chembiol.2019.01.010
Ge, Y., Draycheva, A., Bornemann, T., Rodnina, M. V., and Wintermeyer, W. (2014). Lateral Opening of the bacterial translocon on ribosome binding and signal peptide insertion. Nat. Commun. 5:5263.
Gelis, I., Bonvin, A. M., Keramisanou, D., Koukaki, M., Gouridis, G., Karamanou, S., et al. (2007). Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell 131, 756–769.
Geng, Y., Kedrov, A., Caumanns, J. J., Crevenna, A. H., Lamb, D. C., Beckmann, R., et al. (2015). Role of the cytosolic loop C2 and the C terminus of YidC in ribosome binding and insertion activity. J. Biol. Chem. 290, 17250–17261. doi: 10.1074/jbc.M115.650309
Gill, D. R., and Salmond, G. P. (1987). The Escherichia coli cell division proteins FtsY, FtsE and FtsX are inner membrane-associated. Mol. Gen. Genet. 210, 504–508. doi: 10.1007/bf00327204
Gill, D. R., and Salmond, G. P. (1990). The identification of the Escherichia coli ftsY gene product: an unusual protein. Mol. Microbiol. 4, 575–583. doi: 10.1111/j.1365-2958.1990.tb00626.x
Goder, V., Bieri, C., and Spiess, M. (1999). Glycosylation can influence topogenesis of membrane proteins and reveals dynamic reorientation of nascent polypeptides within the translocon. J. Cell Biol. 147, 257–266.
Gogala, M., Becker, T., Beatrix, B., Armache, J. P., Barrio-Garcia, C., Berninghausen, O., et al. (2014). Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature 506, 107–110.
Gold, V. A., Robson, A., Bao, H., Romantsov, T., Duong, F., and Collinson, I. (2010). The action of cardiolipin on the bacterial translocon. Proc. Natl. Acad. Sci. U.S.A. 107, 10044–10049. doi: 10.1073/pnas.0914680107
Gold, V. A., Whitehouse, S., Robson, A., and Collinson, I. (2013). The dynamic action of SecA during the initiation of protein translocation. Biochem. J. 449, 695–705. doi: 10.1042/bj20121314
Gómez-Eichelmann, M. C., and Helmstetter, C. E. (1999). Transcription level of operon ftsYEX and activity of promoter P1 of rpoH during the cell cycle in Escherichia coli. J. Basic Microbiol. 39, 237–242.
Götzke, H., Palombo, I., Muheim, C., Perrody, E., Genevaux, P., Kudva, R., et al. (2014). YfgM is an ancillary subunit of the SecYEG translocon in Escherichia coli. J. Biol. Chem. 289, 19089–19097.
Gouridis, G., Karamanou, S., Sardis, M. F., Scharer, M. A., Capitani, G., and Economou, A. (2013). Quaternary dynamics of the SecA motor drive translocase catalysis. Mol. Cell 52, 655–666. doi: 10.1016/j.molcel.2013.10.036
Grady, L. M., Michtavy, J., and Oliver, D. B. (2012). Characterization of the Escherichia coli SecA signal peptide-binding site. J. Bacteriol. 194, 307–316.
Gu, S. Q., Peske, F., Wieden, H. J., Rodnina, M. V., and Wintermeyer, W. (2003). The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA 9, 566–573.
Gupta, R., Toptygin, D., and Kaiser, C. M. (2020). The SecA motor generates mechanical force during protein translocation. Nat. Commun. 11:3802.
Haas, T. M., Qiu, D., Häner, M., Angebauer, L., Ripp, A., Singh, J., et al. (2020). Four phosphates at one blow: access to pentaphosphorylated magic spot nucleotides and their analysis by capillary electrophoresis. J. Org. Chem. 85, 14496–14506. doi: 10.1021/acs.joc.0c00841
Halic, M., Becker, T., Pool, M. R., Spahn, C. M., Grassucci, R. A., Frank, J., et al. (2004). Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427, 808–814. doi: 10.1038/nature02342
Halic, M., Blau, M., Becker, T., Mielke, T., Pool, M. R., Wild, K., et al. (2006a). Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature 444, 507–511. doi: 10.1038/nature05326
Halic, M., Gartmann, M., Schlenker, O., Mielke, T., Pool, M. R., Sinning, I., et al. (2006b). Signal recognition particle receptor exposes the ribosomal translocon binding site. Science 312, 745–747. doi: 10.1126/science.1124864
Harms, N., Konigstein, G., Dontje, W., Müller, M., Oudega, B., Luirink, J., et al. (2001). The early interaction of the outer membrane protein PhoE with the periplasmic chaperone Skp occurs at the cytoplasmic membrane. J. Biol. Chem. 276, 18804–18811.
Harris, A. J., and Goldman, A. D. (2021). The very early evolution of protein translocation across membranes. PLoS Comput. Biol. 17:e1008623. doi: 10.1371/journal.pcbi.1008623
Harris, C. R., and Silhavy, T. J. (1999). Mapping an interface of SecY (PrlA) and SecE (PrlG) by using synthetic phenotypes and in vivo cross-linking. J. Bacteriol. 181, 3438–3444. doi: 10.1128/jb.181.11.3438-3444.1999
Hartl, F. U., Lecker, S., Schiebel, E., Hendrick, J. P., and Wickner, W. (1990). The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell 63, 269–279. doi: 10.1016/0092-8674(90)90160-g
He, H., Kuhn, A., and Dalbey, R. E. (2020). Tracking the stepwise movement of a membrane-inserting protein in vivo. J. Mol. Biol. 432, 484–496. doi: 10.1016/j.jmb.2019.10.010
Hegde, R. S., and Bernstein, H. D. (2006). The surprising complexity of signal sequences. Trends Biochem. Sci. 31, 563–571. doi: 10.1016/j.tibs.2006.08.004
Heinrich, S. U., and Rapoport, T. A. (2003). Cooperation of transmembrane segments during the integration of a double-spanning protein into the ER membrane. EMBO J. 22, 3654–3663. doi: 10.1093/emboj/cdg346
Hemm, M. R., Paul, B. J., Miranda-Rios, J., Zhang, A., Soltanzad, N., and Storz, G. (2010). Small stress response proteins in Escherichia coli: proteins missed by classical proteomic studies. J. Bacteriol. 192, 46–58. doi: 10.1093/nar/gkq054
Hessa, T., Meindl-Beinker, N. M., Bernsel, A., Kim, H., Sato, Y., Lerch-Bader, M., et al. (2007). Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450, 1026–1030. doi: 10.1038/nature06387
Hews, C. L., Cho, T., Rowley, G., and Raivio, T. L. (2019). Maintaining integrity under stress: envelope stress response regulation of pathogenesis in gram-negative bacteria. Front. Cell. Infect. Microbiol. 9:313. doi: 10.3389/fcimb.2019.00313
Hizlan, D., Robson, A., Whitehouse, S., Gold, V. A., Vonck, J., Mills, D., et al. (2012). Structure of the SecY complex unlocked by a preprotein mimic. Cell Rep. 1, 21–28. doi: 10.1016/j.celrep.2011.11.003
Hoffschulte, H. K., Drees, B., and Muller, M. (1994). Identification of a soluble SecA/SecB complex by means of a subfractionated cell-free export system. J. Biol. Chem. 269, 12833–12839.
Holtkamp, W., Kokic, G., Jager, M., Mittelstaet, J., Komar, A. A., and Rodnina, M. V. (2015). Cotranslational protein folding on the ribosome monitored in real time. Science 350, 1104–1107. doi: 10.1126/science.aad0344
Holtkamp, W., Lee, S., Bornemann, T., Senyushkina, T., Rodnina, M. V., and Wintermeyer, W. (2012). Dynamic switch of the signal recognition particle from scanning to targeting. Nat. Struct. Mol. Biol. 19, 1332–1337. doi: 10.1038/nsmb.2421
Houben, E. N., Scotti, P. A., Valent, Q. A., Brunner, J., de Gier, J. L., Oudega, B., et al. (2000). Nascent Lep inserts into the Escherichia coli inner membrane in the vicinity of YidC, SecY and SecA. FEBS Lett. 476, 229–233.
Houben, E. N., ten Hagen-Jongman, C. M., Brunner, J., Oudega, B., and Luirink, J. (2004). The two membrane segments of leader peptidase partition one by one into the lipid bilayer via a Sec/YidC interface. EMBO Rep. 5, 970–975. doi: 10.1038/sj.embor.7400261
Houben, E. N., Urbanus, M. L., Van Der Laan, M., Ten Hagen-Jongman, C. M., Driessen, A. J., Brunner, J., et al. (2002). YidC and SecY mediate membrane insertion of a Type I transmembrane domain. J. Biol. Chem. 277, 35880–35886. doi: 10.1074/jbc.M205556200
Huang, C., Rossi, P., Saio, T., and Kalodimos, C. G. (2016). Structural basis for the antifolding activity of a molecular chaperone. Nature 537, 202–206. doi: 10.1038/nature18965
Huang, Y. J., Wang, H., Gao, F. B., Li, M., Yang, H., Wang, B., et al. (2012). Fluorescein analogues inhibit SecA ATPase: the first sub-micromolar inhibitor of bacterial protein translocation. ChemMedChem 7, 571–577. doi: 10.1002/cmdc.201100594
Huber, D., Boy, D., Xia, Y., Olma, M. H., Gerstein, M., and Beckwith, J. (2005). Use of thioredoxin as a reporter to identify a subset of Escherichia coli signal sequences that promote signal recognition particle-dependent translocation. J. Bacteriol. 187, 2983–2991.
Huber, D., Jamshad, M., Hanmer, R., Schibich, D., Doring, K., Marcomini, I., et al. (2017). SecA cotranslationally interacts with nascent substrate proteins in vivo. J. Bacteriol. 199:e00622-16.
Huber, D., Rajagopalan, N., Preissler, S., Rocco, M. A., Merz, F., Kramer, G., et al. (2011). SecA interacts with ribosomes in order to facilitate posttranslational translocation in bacteria. Mol. Cell 41, 343–353. doi: 10.1016/j.molcel.2010.12.028
Iadanza, M. G., Higgins, A. J., Schiffrin, B., Calabrese, A. N., Brockwell, D. J., Ashcroft, A. E., et al. (2016). Lateral opening in the intact β-barrel assembly machinery captured by cryo-EM. Nat. Commun. 7:12865. doi: 10.1038/ncomms12865
Ikegami, A., Nishiyama, K., Matsuyama, S., and Tokuda, H. (2005). Disruption of rpmJ encoding ribosomal protein L36 decreases the expression of secY upstream of the spc operon and inhibits protein translocation in Escherichia coli. Biosci. Biotechnol. Biochem. 69, 1595–1602. doi: 10.1271/bbb.69.1595
Irving, S. E., Choudhury, N. R., and Corrigan, R. M. (2020). The stringent response and physiological roles of (pp)pGpp in bacteria. Nat. Rev. Microbiol. 19, 256–271. doi: 10.1038/s41579-020-00470-y
Ishidate, K., Creeger, E. S., Zrike, J., Deb, S., Glauner, B., MacAlister, T. J., et al. (1986). Isolation of differentiated membrane domains from Escherichia coli and Salmonella typhimurium, including a fraction containing attachment sites between the inner and outer membranes and the murein skeleton of the cell envelope. J. Biol. Chem. 261, 428–443.
Ishii, E., Chiba, S., Hashimoto, N., Kojima, S., Homma, M., Ito, K., et al. (2015). Nascent chain-monitored remodeling of the Sec machinery for salinity adaptation of marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 112, E5513–E5522. doi: 10.1073/pnas.1513001112
Ito, K., and Akiyama, Y. (2005). Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59, 211–231. doi: 10.1146/annurev.micro.59.030804.121316
Ito, K., and Chiba, S. (2013). Arrest peptides: cis-acting modulators of translation. Annu. Rev. Biochem. 82, 171–202.
Ito, K., Chiba, S., and Pogliano, K. (2010). Divergent stalling sequences sense and control cellular physiology. Biochem. Biophys. Res. Commun. 393, 1–5. doi: 10.1016/j.bbrc.2010.01.073
Ito, K., Mori, H., and Chiba, S. (2018). Monitoring substrate enables real-time regulation of a protein localization pathway. FEMS Microbiol. Lett. 365:fny109. doi: 10.1093/femsle/fny109
Jauss, B., Petriman, N. A., Drepper, F., Franz, L., Sachelaru, I., Welte, T., et al. (2019). Non-competitive binding of PpiD and YidC to the SecYEG translocon expands the global view on the SecYEG interactome in E. coli. J. Biol. Chem. 294, 19167–19183. doi: 10.1074/jbc.RA119.010686
Jiang, F., Chen, M., Yi, L., de Gier, J. W., Kuhn, A., and Dalbey, R. E. (2003). Defining the regions of Escherichia coli YidC that contribute to activity. J. Biol. Chem. 278, 48965–48972. doi: 10.1074/jbc.M307362200
Jin, F. (2020). Structural insights into the mechanism of a novel protein targeting pathway in Gram-negative bacteria. FEBS Open Bio 10, 561–579. doi: 10.1002/2211-5463.12813
Jin, J., Hsieh, Y. H., Cui, J., Damera, K., Dai, C., Chaudhary, A. S., et al. (2016). Using chemical probes to assess the feasibility of targeting SecA for developing antimicrobial agents against gram-negative bacteria. ChemMedChem 11, 2511–2521. doi: 10.1002/cmdc.201500447
Jomaa, A., Boehringer, D., Leibundgut, M., and Ban, N. (2016). Structure of the E. coli translating ribosome with SRP and its receptor and with the translocon. Nat. Comm. 7:10471.
Jomaa, A., Fu, Y. H., Boehringer, D., Leibundgut, M., Shan, S. O., and Ban, N. (2017). Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome. Nat. Commun. 8:15470.
Josefsson, L. G., and Randall, L. L. (1981a). Different exported proteins in E. coli show differences in the temporal mode of processing in vivo. Cell 25, 151–157.
Josefsson, L. G., and Randall, L. L. (1981b). Processing in vivo of precursor maltose-binding protein in Escherichia coli occurs post-translationally as well as co-translationally. J. Biol. Chem. 256, 2504–2507.
Junne, T., Wong, J., Studer, C., Aust, T., Bauer, B. W., Beibel, M., et al. (2015). Decatransin, a new natural product inhibiting protein translocation at the Sec61/SecYEG translocon. J. Cell Sci. 128, 1217–1229. doi: 10.1242/jcs.165746
Kalies, K. U., and Römisch, K. (2015). Inhibitors of protein translocation across the ER membrane. Traffic 16, 1027–1038. doi: 10.1111/tra.12308
Kannaiah, S., and Amster-Choder, O. (2014). Protein targeting via mRNA in bacteria. Biochim. Biophys. Acta 1843, 1457–1465. doi: 10.1016/j.bbamcr.2013.11.004
Kannaiah, S., Livny, J., and Amster-Choder, O. (2019). Spatiotemporal organization of the E. coli transcriptome: translation independence and engagement in regulation. Mol. Cell 76, 574–589.e7. doi: 10.1016/j.molcel.2019.08.013
Karamanou, S., Gouridis, G., Papanikou, E., Sianidis, G., Gelis, I., Keramisanou, D., et al. (2007). Preprotein-controlled catalysis in the helicase motor of SecA. EMBO J. 26, 2904–2914. doi: 10.1038/sj.emboj.7601721
Karamyshev, A. L., and Johnson, A. E. (2005). Selective SecA association with signal sequences in ribosome-bound nascent chains: a potential role for SecA in ribosome targeting to the bacterial membrane. J. Biol. Chem. 280, 37930–37940. doi: 10.1074/jbc.M509100200
Kater, L., Frieg, B., Berninghausen, O., Gohlke, H., Beckmann, R., and Kedrov, A. (2019). Partially inserted nascent chain unzips the lateral gate of the Sec translocon. EMBO Rep. 20:e48191.
Kedrov, A., Kusters, I., Krasnikov, V. V., and Driessen, A. J. (2011). A single copy of SecYEG is sufficient for preprotein translocation. EMBO J. 30, 4387–4397. doi: 10.1038/emboj.2011.314
Kellenberger, E. (1990). The ‘Bayer bridges’ confronted with results from improved electron microscopy methods. Mol. Microbiol. 4, 697–705. doi: 10.1111/j.1365-2958.1990.tb00640.x
Keller, R., de Keyzer, J., Driessen, A. J., and Palmer, T. (2012). Co-operation between different targeting pathways during integration of a membrane protein. J. Cell Biol. 199, 303–315. doi: 10.1083/jcb.201204149
Kihara, A., Akiyama, Y., and Ito, K. (1995). FtsH is required for proteolytic elimination of uncomplexed forms of SecY, an essential protein translocase subunit. Proc. Natl. Acad. Sci. U.S.A. 92, 4532–4536.
Kihara, A., Akiyama, Y., and Ito, K. (1996). A protease complex in the Escherichia coli plasma membrane: HflKC (HflA) forms a complex with FtsH (HflB), regulating its proteolytic activity against SecY. EMBO J. 15, 6122–6131.
Knoblauch, N. T., Rudiger, S., Schonfeld, H. J., Driessen, A. J., Schneider-Mergener, J., and Bukau, B. (1999). Substrate specificity of the SecB chaperone. J. Biol. Chem. 274, 34219–34225.
Knupffer, L., Fehrenbach, C., Denks, K., Erichsen, V., Petriman, N. A., and Koch, H. G. (2019). Molecular mimicry of SecA and signal recognition particle binding to the bacterial ribosome. mBio 10:e01317-19.
Knyazev, D. G., Kuttner, R., Zimmermann, M., Sobakinskaya, E., and Pohl, P. (2018). Driving forces of translocation through bacterial translocon SecYEG. J. Membr. Biol. 251, 329–343.
Knyazev, D. G., Lents, A., Krause, E., Ollinger, N., Siligan, C., Papinski, D., et al. (2013). The bacterial translocon SecYEG opens upon ribosome binding. J. Biol. Chem. 288, 17941–17946. doi: 10.1074/jbc.M113.477893
Knyazev, D. G., Winter, L., Bauer, B. W., Siligan, C., and Pohl, P. (2014). Ion conductivity of the bacterial translocation channel SecYEG engaged in translocation. J. Biol. Chem. 289, 24611–24616.
Koch, H. G., and Muller, M. (2000). Dissecting the translocase and integrase functions of the Escherichia coli SecYEG translocon. J. Cell Biol. 150, 689–694.
Koch, H. G., Hengelage, T., Neumann-Haefelin, C., MacFarlane, J., Hoffschulte, H. K., Schimz, K. L., et al. (1999). In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/SecB as constituents of two independent protein-targeting pathways of Escherichia coli. Mol. Biol. Cell 10, 2163–2173.
Koch, H. G., Moser, M., and Muller, M. (2003). Signal recognition particle-dependent protein targeting, universal to all kingdoms of life. Rev. Physiol. Biochem. Pharmacol. 146, 55–94.
Koch, S., de Wit, J. G., Vos, I., Birkner, J. P., Gordiichuk, P., Herrmann, A., et al. (2016). Lipids activate SecA for high affinity binding to the SecYEG complex. J. Biol. Chem. 291, 22534–22543.
Koch, S., Exterkate, M., López, C. A., Patro, M., Marrink, S. J., and Driessen, A. J. M. (2019). Two distinct anionic phospholipid-dependent events involved in SecA-mediated protein translocation. Biochim. Biophys. Acta Biomembr. 1861, 183035. doi: 10.1016/j.bbamem.2019.183035
Komar, J., Alvira, S., Schulze, R., Martin, R., Lycklama, A. N. J., Lee, S., et al. (2016). Membrane protein insertion and assembly by the bacterial holo-translocon SecYEG-SecDF-YajC-YidC. Biochem. J. 473, 3341–3354. doi: 10.1042/bcj20160545
Komarudin, A. G., and Driessen, A. J. M. (2019). SecA-mediated protein translocation through the SecYEG Channel. Microbiol. Spectr. 7:PSIB-0028-2019.
Konovalova, A., Kahne, D. E., and Silhavy, T. J. (2017). Outer membrane biogenesis. Annu. Rev. Microbiol. 71, 539–556.
Kramer, G., Boehringer, D., Ban, N., and Bukau, B. (2009). The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16, 589–597. doi: 10.1038/nsmb.1614
Kramer, G., Rauch, T., Rist, W., Vorderwulbecke, S., Patzelt, H., Schulze-Specking, A., et al. (2002). L23 protein functions as a chaperone docking site on the ribosome. Nature 419, 171–174. doi: 10.1038/nature01047
Kudva, R., Denks, K., Kuhn, P., Vogt, A., Muller, M., and Koch, H. G. (2013). Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res. Microbiol. 164, 505–534. doi: 10.1016/j.resmic.2013.03.016
Kuhn, A., and Kiefer, D. (2017). Membrane protein insertase YidC in bacteria and archaea. Mol. Microbiol. 103, 590–594. doi: 10.1111/mmi.13586
Kuhn, P., Draycheva, A., Vogt, A., Petriman, N. A., Sturm, L., Drepper, F., et al. (2015). Ribosome binding induces repositioning of the signal recognition particle receptor on the translocon. J. Cell Biol. 211, 91–104. doi: 10.1111/mmi.13321
Kuhn, P., Weiche, B., Sturm, L., Sommer, E., Drepper, F., Warscheid, B., et al. (2011). The bacterial SRP receptor, SecA and the ribosome use overlapping binding sites on the SecY translocon. Traffic 12, 563–578. doi: 10.1111/j.1600-0854.2011.01167.x
Kumamoto, C. A., and Beckwith, J. (1985). Evidence for specificity at an early step in protein export in Escherichia coli. J. Bacteriol. 163, 267–274. doi: 10.1128/jb.163.1.267-274.1985
Kumazaki, K., Chiba, S., Takemoto, M., Furukawa, A., Nishiyama, K., Sugano, Y., et al. (2014a). Structural basis of Sec-independent membrane protein insertion by YidC. Nature 509, 516–520. doi: 10.1038/nature13167
Kumazaki, K., Kishimoto, T., Furukawa, A., Mori, H., Tanaka, Y., Dohmae, N., et al. (2014b). Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase. Sci. Rep. 4:7299. doi: 10.1038/srep07299
Kusters, I., van den Bogaart, G., Kedrov, A., Krasnikov, V., Fulyani, F., Poolman, B., et al. (2011). Quaternary structure of SecA in solution and bound to SecYEG probed at the single molecule level. Structure 19, 430–439. doi: 10.1016/j.str.2010.12.016
Lakomek, N. A., Draycheva, A., Bornemann, T., and Wintermeyer, W. (2016). Electrostatics and intrinsic disorder drive translocon binding of the SRP Receptor FtsY. Angew. Chem. Int. Ed. Engl. 55, 9544–9547.
Lasserre, J. P., Beyne, E., Pyndiah, S., Lapaillerie, D., Claverol, S., and Bonneu, M. (2006). A complexomic study of Escherichia coli using two-dimensional blue native/SDS polyacrylamide gel electrophoresis. Electrophoresis 27, 3306–3321. doi: 10.1002/elps.200500912
Lee, H. C., and Bernstein, H. D. (2002). Trigger factor retards protein export in Escherichia coli. J. Biol. Chem. 277, 43527–43535. doi: 10.1074/jbc.M205950200
Lemke, J. J., Sanchez-Vazquez, P., Burgos, H. L., Hedberg, G., Ross, W., and Gourse, R. L. (2011). Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc. Natl. Acad. Sci. U.S.A. 108, 5712–5717.
Li, G. W., Burkhardt, D., Gross, C., and Weissman, J. S. (2014). Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635. doi: 10.1126/science.1251871
Li, L., Park, E., Ling, J., Ingram, J., Ploegh, H., and Rapoport, T. A. (2016). Crystal structure of a substrate-engaged SecY protein-translocation channel. Nature 531, 395–399.
Li, M., Huang, Y. J., Tai, P. C., and Wang, B. (2008). Discovery of the first SecA inhibitors using structure-based virtual screening. Biochem. Biophys. Res. Commun. 368, 839–845. doi: 10.1016/j.bbrc.2008.01.135
Li, W., Schulman, S., Boyd, D., Erlandson, K., Beckwith, J., and Rapoport, T. A. (2007). The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol. Cell 26, 511–521. doi: 10.1016/j.molcel.2007.05.002
Li, Z., Boyd, D., Reindl, M., and Goldberg, M. B. (2013). Identification of YidC residues that define interactions with the Sec apparatus. J. Bacteriol. 196, 367–377.
Liang, S., Bipatnath, M., Xu, Y., Chen, S., Dennis, P., Ehrenberg, M., et al. (1999). Activities of constitutive promoters in Escherichia coli. J. Mol. Biol. 292, 19–37. doi: 10.1006/jmbi.1999.3056
Lill, R., Dowhan, W., and Wickner, W. (1990). The ATPase activity of SecA is regulated by acidic phospholipids, SecY nd the leader and mature domain of precursor proteins. Cell 60, 271–280.
Lim, B., Miyazaki, R., Neher, S., Siegele, D. A., Ito, K., Walter, P., et al. (2013). Heat shock transcription factor sigma32 co-opts the signal recognition particle to regulate protein homeostasis in E. coli. PLoS Biol. 11:e1001735. doi: 10.1371/journal.pbio.1001735
Lindahl, L., Sor, F., Archer, R. H., Nomura, M., and Zengel, J. M. (1990). Transcriptional organization of the S10, spc and alpha operons of Escherichia coli. Biochim. Biophys. Acta 1050, 337–342.
Liu, R., Chen, H., Zhang, R., Zhou, Z., Hou, Z., Gao, D., et al. (2016). Comparative transcriptome analysis of Vibrio splendidus JZ6 reveals the mechanism of its pathogenicity at low temperatures. Appl. Environ. Microbiol. 82, 2050–2061.
Loos, M. S., Ramakrishnan, R., Vranken, W., Tsirigotaki, A., Tsare, E. P., Zorzini, V., et al. (2019). Structural basis of the subcellular topology landscape of Escherichia coli. Front. Microbiol. 10:1670. doi: 10.3389/fmicb.2019.01670
Luirink, J., Samuelsson, T., and de Gier, J. W. (2001). YidC/Oxa1p/Alb3: evolutionarily conserved mediators of membrane protein assembly. FEBS Lett. 501, 1–5.
Luirink, J., ten Hagen-Jongman, C. M., van der Weijden, C. C., Oudega, B., High, S., Dobberstein, B., et al. (1994). An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J. 13, 2289–2296.
Lycklama a Nijeholt, J. A., de Keyzer, J., Prabudiansyah, I., and Driessen, A. J. (2013). Characterization of the supporting role of SecE in protein translocation. FEBS Lett. 587, 3083–3088.
Maddalo, G., Stenberg-Bruzell, F., Götzke, H., Toddo, S., Björrkholm, P., Eriksson, H., et al. (2011). Systematic analysis of native membrane protein complexes in Escherichia coli. J. Proteome. Res. 10, 1848–1859. doi: 10.1021/pr101105c
Maier, K. S., Hubich, S., Liebhart, H., Krauss, S., Kuhn, A., and Facey, S. J. (2008). An amphiphilic region in the cytoplasmic domain of KdpD is recognized by the signal recognition particle and targeted to the Escherichia coli membrane. Mol. Microbiol. 68, 1471–1484.
Malinverni, J. C., and Silhavy, T. J. (2011). Assembly of outer membrane β-barrel proteins: the bam complex. EcoSal Plus 4:10.1128/ecosalplus.4.3.8. doi: 10.1128/ecosalplus.4.3.8
Manganelli, R., and Gennaro, M. L. (2017). Protecting from envelope stress: variations on the phage-shock-protein theme. Trends Microbiol. 25, 205–216. doi: 10.1016/j.tim.2016.10.001
Manting, E. H., van der Does, C., and Driessen, A. J. (1997). In vivo cross-linking of the SecA and SecY subunits of the Escherichia coli preprotein translocase. J. Bacteriol. 179, 5699–5704.
Martin, R., Larsen, A. H., Corey, R. A., Midtgaard, S. R., Frielinghaus, H., Schaffitzel, C., et al. (2019). Structure and dynamics of the central lipid pool and proteins of the bacterial Holo-Translocon. Biophys. J. 116, 1931–1940. doi: 10.1016/j.bpj.2019.04.002
Mas, G., Thoma, J., and Hiller, S. (2019). The periplasmic chaperones Skp and SurA. Subcell. Biochem. 92, 169–186. doi: 10.1007/978-3-030-18768-2_6
Matsuo, E., Mori, H., Shimoike, T., and Ito, K. (1998). Syd, a SecY-interacting protein, excludes SecA from the SecYE complex with an altered SecY24 subunit. J. Biol. Chem. 273, 18835–18840. doi: 10.1074/jbc.273.30.18835
McDowell, M. A., Heimes, M., and Sinning, I. (2021). Structural and molecular mechanisms for membrane protein biogenesis by the Oxa1 superfamily. Nat. Struct. Mol. Biol. 28, 234–239.
Menetret, J. F., Schaletzky, J., Clemons, W. M. Jr., Osborne, A. R., Skanland, S. S., Denison, C., et al. (2007). Ribosome binding of a single copy of the SecY complex: implications for protein translocation. Mol. Cell 28, 1083–1092. doi: 10.1016/j.molcel.2007.10.034
Mercier, E., Wintermeyer, W., and Rodnina, M. V. (2020). Co-translational insertion and topogenesis of bacterial membrane proteins monitored in real time. EMBO J. 39:e104054. doi: 10.15252/embj.2019104054
Milo, R. (2013). What is the total number of protein molecules per cell volume? A call to rethink some published values. Bioessays 35, 1050–1055. doi: 10.1002/bies.201300066
Mircheva, M., Boy, D., Weiche, B., Hucke, F., Graumann, P., and Koch, H. G. (2009). Predominant membrane localization is an essential feature of the bacterial signal recognition particle receptor. BMC Biol. 7:76. doi: 10.1186/1741-7007-7-76
Mitkevich, V. A., Ermakov, A., Kulikova, A. A., Tankov, S., Shyp, V., Soosaar, A., et al. (2010). Thermodynamic characterization of ppGpp binding to EF-G or IF2 and of initiator tRNA binding to free IF2 in the presence of GDP, GTP, or ppGpp. J. Mol. Biol. 402, 838–846. doi: 10.1016/j.jmb.2010.08.016
Miyazaki, R., Akiyama, Y., and Mori, H. (2020). Fine interaction profiling of VemP and mechanisms responsible for its translocation-coupled arrest-cancelation. eLife 9:e62623. doi: 10.7554/eLife.62623
Miyazaki, R., Yura, T., Suzuki, T., Dohmae, N., Mori, H., and Akiyama, Y. (2016). A novel SRP recognition sequence in the homeostatic control region of heat shock transcription Factor σ32. Sci. Rep. 6:24147. doi: 10.1038/srep24147
Mogk, A., Huber, D., and Bukau, B. (2011). Integrating protein homeostasis strategies in prokaryotes. Cold Spring Harb. Perspect. Biol. 3:a004366. doi: 10.1101/cshperspect.a004366
Moll, I., and Engelberg-Kulka, H. (2012). Selective translation during stress in Escherichia coli. Trends Biochem. Sci. 37, 493–498.
Montoya, G., Svensson, C., Luirink, J., and Sinning, I. (1997). Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature 385, 365–368. doi: 10.1038/385365a0
Mori, H., and Ito, K. (2003). Biochemical characterization of a mutationally altered protein translocase: proton motive force stimulation of the initiation phase of translocation. J. Bacteriol. 185, 405–412.
Mori, H., and Ito, K. (2006). Different modes of SecY-SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc. Natl. Acad. Sci. U.S.A. 103, 16159–16164. doi: 10.1073/pnas.0606390103
Moser, M., Nagamori, S., Huber, M., Tokuda, H., and Nishiyama, K. (2013). Glycolipozyme MPIase is essential for topology inversion of SecG during preprotein translocation. Proc. Natl. Acad. Sci. U.S.A. 110, 9734–9739. doi: 10.1073/pnas.1303160110
Mutka, S. C., and Walter, P. (2001). Multifaceted physiological response allows yeast to adapt to the loss of the signal recognition particle-dependent protein-targeting pathway. Mol. Biol. Cell 12, 577–588. doi: 10.1091/mbc.12.3.577
Neumann-Haefelin, C., Schafer, U., Muller, M., and Koch, H. G. (2000). SRP-dependent co-translational targeting and SecA-dependent translocation analyzed as individual steps in the export of a bacterial protein. EMBO J. 19, 6419–6426. doi: 10.1093/emboj/19.23.6419
Nevo-Dinur, K., Nussbaum-Shochat, A., Ben-Yehuda, S., and Amster-Choder, O. (2011). Translation-independent localization of mRNA in E. coli. Science 331, 1081–1084.
Nilsson, O. B., Hedman, R., Marino, J., Wickles, S., Bischoff, L., Johansson, M., et al. (2015). Cotranslational protein folding inside the ribosome exit tunnel. Cell Rep. 12, 1533–1540. doi: 10.1016/j.celrep.2015.07.065
Nishiyama, K., and Shimamoto, K. (2014). Glycolipozyme membrane protein integrase (MPIase): recent data. Biomol. Concepts 5, 429–438.
Nishiyama, K., Hanada, M., and Tokuda, H. (1994). Disruption of the gene encoding p12 (SecG) reveals the direct involvement and important function of SecG in the protein translocation of Escherichia coli at low temperature. EMBO J. 13, 3272–3277.
Nishiyama, K., Suzuki, T., and Tokuda, H. (1996). Inversion of the membrane topology of SecG coupled with SecA-dependent preprotein translocation. Cell 85, 71–81.
Noinaj, N., Gumbart, J. C., and Buchanan, S. K. (2017). The β-barrel assembly machinery in motion. Nat. Rev. Microbiol. 15, 197–204. doi: 10.1038/nrmicro.2016.191
Nouwen, N., and Driessen, A. J. (2002). SecDFyajC forms a heterotetrameric complex with YidC. Mol. Microbiol. 44, 1397–1405.
Nouwen, N., de Kruijff, B., and Tommassen, J. (1996). prlA suppressors in Escherichia coli relieve the proton electrochemical gradient dependency of translocation of wild-type precursors. Proc. Natl. Acad. Sci. U.S.A. 93, 5953–5957.
Oh, E., Becker, A. H., Sandikci, A., Huber, D., Chaba, R., Gloge, F., et al. (2011). Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147, 1295–1308. doi: 10.1038/nature10912
Oliver, D. B., and Beckwith, J. (1982). Identification of a new gene (secA) and gene product involved in the secretion of envelope proteins in Escherichia coli. J. Bacteriol. 150, 686–691. doi: 10.1128/jb.150.2.686-691.1982
Oliver, D. B., Cabelli, R. J., Dolan, K. M., and Jarosik, G. P. (1990). Azide-resistant mutants of Escherichia coli alter the SecA protein, an azide-sensitive component of the protein export machinery. Proc. Natl. Acad. Sci. U.S.A. 87, 8227–8231. doi: 10.1073/pnas.87.21.8227
Oliver, D. C., and Paetzel, M. (2008). Crystal structure of the major periplasmic domain of the bacterial membrane protein assembly facilitator YidC. J. Biol. Chem. 283, 5208–5216. doi: 10.1074/jbc.M708936200
Or, E., Navon, A., and Rapoport, T. (2002). Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J. 21, 4470–4479.
Origi, A., Natriashivili, A., Knupffer, L., Fehrenbach, C., Denks, K., Asti, R., et al. (2019). Yet another job for the bacterial ribosome. Microb. Cell 6, 524–526.
Paetzel, M., Karla, A., Strynadka, N. C., and Dalbey, R. E. (2002). Signal peptidases. Chem. Rev. 102, 4549–4580. doi: 10.1021/cr010166y
Park, E., and Rapoport, T. A. (2011). Preserving the membrane barrier for small molecules during bacterial protein translocation. Nature 473, 239–242.
Park, E., and Rapoport, T. A. (2012). Bacterial protein translocation requires only one copy of the SecY complex in vivo. J. Cell Biol. 198, 881–893. doi: 10.1083/jcb.201205140
Parlitz, R., Eitan, A., Stjepanovic, G., Bahari, L., Bange, G., Bibi, E., et al. (2007). Escherichia coli signal recognition particle receptor FtsY contains an essential and autonomous membrane-binding amphipathic helix. J. Biol. Chem. 282, 32176–32184. doi: 10.1074/jbc.M705430200
Pausch, P., Abdelshahid, M., Steinchen, W., Schäfer, H., Gratani, F. L., Freibert, S. A., et al. (2020). Structural basis for regulation of the opposing (p)ppGpp synthetase and hydrolase within the stringent response Orchestrator Rel. Cell Rep. 32:108157. doi: 10.1016/j.celrep.2020.108157
Persky, N. S., Ferullo, D. J., Cooper, D. L., Moore, H. R., and Lovett, S. T. (2009). The ObgE/CgtA GTPase influences the stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 73, 253–266. doi: 10.1111/j.1365-2958.2009.06767.x
Peschke, M., Le Goff, M., Koningstein, G. M., Karyolaimos, A., de Gier, J. W., van Ulsen, P., et al. (2018). SRP, FtsY, DnaK and YidC Are Required for the Biogenesis of the E. coli Tail-Anchored Membrane Proteins DjlC and Flk. J. Mol. Biol. 430, 389–403. doi: 10.1016/j.jmb.2017.12.004
Petriman, N. A., Jauss, B., Hufnagel, A., Franz, L., Sachelaru, I., Drepper, F., et al. (2018). The interaction network of the YidC insertase with the SecYEG translocon, SRP and the SRP receptor FtsY. Sci. Rep. 8:578. doi: 10.1038/s41598-017-19019-w
Pfanner, N., Warscheid, B., and Wiedemann, N. (2019). Mitochondrial proteins: from biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 20, 267–284.
Pichoff, S., Du, S., and Lutkenhaus, J. (2019). Roles of FtsEX in cell division. Res. Microbiol. 170, 374–380. doi: 10.1016/j.resmic.2019.07.003
Pitonzo, D., Yang, Z., Matsumura, Y., Johnson, A. E., and Skach, W. R. (2009). Sequence-specific retention and regulated integration of a nascent membrane protein by the endoplasmic reticulum Sec61 translocon. Mol. Biol. Cell 20, 685–698.
Pogliano, J. A., and Beckwith, J. (1994a). SecD and SecF facilitate protein export in Escherichia coli. EMBO J. 13, 554–561.
Pogliano, K. J., and Beckwith, J. (1994b). Genetic and molecular characterization of the Escherichia coli secD operon and its products. J. Bacteriol. 176, 804–814.
Pohlschroder, M., Hartmann, E., Hand, N. J., Dilks, K., and Haddad, A. (2005). Diversity and evolution of protein translocation. Annu. Rev. Microbiol. 59, 91–111. doi: 10.1146/annurev.micro.59.030804.121353
Pohlschroder, M., Prinz, W. A., Hartmann, E., and Beckwith, J. (1997). Protein translocation in the three domains of life: variations on a theme. Cell 91, 563–566.
Pool, M. R., Stumm, J., Fulga, T. A., Sinning, I., and Dobberstein, B. (2002). Distinct modes of signal recognition particle interaction with the ribosome. Science 297, 1345–1348.
Powers, T., and Walter, P. (1997). Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 16, 4880–4886. doi: 10.1093/emboj/16.16.4880
Prabudiansyah, I., Kusters, I., Caforio, A., and Driessen, A. J. (2015). Characterization of the annular lipid shell of the Sec translocon. Biochim. Biophys. Acta 1848(10 Pt A), 2050–2056. doi: 10.1016/j.bbamem.2015.06.024
Prinz, A., Behrens, C., Rapoport, T. A., Hartmann, E., and Kalies, K. U. (2000). Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal RNA. EMBO J. 19, 1900–1906. doi: 10.1093/emboj/19.8.1900
Pross, E., Soussoula, L., Seitl, I., Lupo, D., and Kuhn, A. (2016). Membrane targeting and insertion of the C-tail protein SciP. J. Mol. Biol. 428, 4218–4227. doi: 10.1016/j.jmb.2016.09.001
Pugsley, A. P. (1990). Translocation of proteins with signal sequences across membranes. Curr. Opin. Cell Biol. 2, 609–616.
Ranava, D., Caumont-Sarcos, A., Albenne, C., and Ieva, R. (2018). Bacterial machineries for the assembly of membrane-embedded β-barrel proteins. FEMS Microbiol. Lett. 365:fny087. doi: 10.1093/femsle/fny087
Randall, L. L. (1983). Translocation of domains of nascent periplasmic proteins across the cytoplasmic membrane is independent of elongation. Cell 33, 231–240.
Rao, C. V. S., De Waelheyns, E., Economou, A., and Anne, J. (2014). Antibiotic targeting of the bacterial secretory pathway. Biochim. Biophys. Acta 1843, 1762–1783. doi: 10.1016/j.bbamcr.2014.02.004
Rapoport, T. A. (2007). Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450, 663–669. doi: 10.1038/nature06384
Ravaud, S., Stjepanovic, G., Wild, K., and Sinning, I. (2008). The crystal structure of the periplasmic domain of the Escherichia coli membrane protein insertase YidC contains a substrate binding cleft. J. Biol. Chem. 283, 9350–9358. doi: 10.1074/jbc.M710493200
Rawat, S., Zhu, L., Lindner, E., Dalbey, R. E., and White, S. H. (2015). SecA drives transmembrane insertion of RodZ, an unusual single-span membrane protein. J. Mol. Biol. 427, 1023–1037.
Ricci, D. P., and Silhavy, T. J. (2019). Outer membrane protein insertion by the β-barrel assembly machine. EcoSal Plus 8:10.1128/ecosalplus.ESP-0035-2018. doi: 10.1128/ecosalplus.ESP-0035-2018
Robson, A., Booth, A. E., Gold, V. A., Clarke, A. R., and Collinson, I. (2007). A large conformational change couples the ATP binding site of SecA to the SecY protein channel. J. Mol. Biol. 374, 965–976. doi: 10.1016/j.jmb.2007.09.086
Rodnina, M. V., and Wintermeyer, W. (2016). Protein elongation, Co-translational folding and targeting. J. Mol. Biol. 428(10 Pt B), 2165–2185. doi: 10.1016/j.jmb.2016.03.022
Rodríguez-Rojas, A., Rodríguez-Beltrán, J., Couce, A., and Blázquez, J. (2013). Antibiotics and antibiotic resistance: a bitter fight against evolution. Int. J. Med. Microbiol. 303, 293–297. doi: 10.1016/j.ijmm.2013.02.004
Rollo, E. E., and Oliver, D. B. (1988). Regulation of the Escherichia coli secA gene by protein secretion defects: analysis of secA, secB, secD, and secY mutants. J. Bacteriol. 170, 3281–3282. doi: 10.1128/jb.170.7.3281-3282.1988
Roussel, G., and White, S. H. (2020). The SecA ATPase motor protein binds to Escherichia coli liposomes only as monomers. Biochim. Biophys. Acta Biomembr. 1862, 183358. doi: 10.1016/j.bbamem.2020.183358
Ryabichko, S., Ferreira, V. M., Vitrac, H., Kiyamova, R., Dowhan, W., and Bogdanov, M. (2020). Cardiolipin is required in vivo for the stability of bacterial translocon : 6296.
Saaf, A., Monne, M., de Gier, J. W., and von Heijne, G. (1998). Membrane topology of the 60-kDa Oxa1p homologue from Escherichia coli. J. Biol. Chem. 273, 30415–30418.
Sachelaru, I., Petriman, N. A., Kudva, R., and Koch, H. G. (2014). Dynamic interaction of the Sec Translocon with the chaperone PpiD. J. Biol. Chem. 289, 21706–21715.
Sachelaru, I., Petriman, N. A., Kudva, R., Kuhn, P., Welte, T., Knapp, B., et al. (2015). YidC occupies the lateral gate of the SecYEG translocon and is sequentially displaced by a nascent membrane protein. J. Biol. Chem. 290:14492. doi: 10.1089/ars.2015.6272
Sachelaru, I., Winter, L., Knyazev, D. G., Zimmermann, M., Vogt, A., Kuttner, R., et al. (2017). YidC and SecYEG form a heterotetrameric protein translocation channel. Sci. Rep. 7:101.
Sadlish, H., Pitonzo, D., Johnson, A., and Skach, W. (2005). Sequential triage of transmembrane segments by Sec61alpha during biogenesis of a native mutlispanning membrane protein. Nat. Struct. Mol. Biol. 12, 870–878.
Saio, T., Guan, X., Rossi, P., Economou, A., and Kalodimos, C. G. (2014). Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science 344:1250494. doi: 10.1126/science.1250494
Saio, T., Kawagoe, S., Ishimori, K., and Kalodimos, C. G. (2018). Oligomerization of a molecular chaperone modulates its activity. eLife 7:e35731. doi: 10.7554/eLife.35731
Samuelson, J. C., Chen, M., Jiang, F., Moller, I., Wiedmann, M., Kuhn, A., et al. (2000). YidC mediates membrane protein insertion in bacteria. Nature 406, 637–641. doi: 10.1038/35020586
Saparov, S. M., Erlandson, K., Cannon, K., Schaletzky, J., Schulman, S., Rapoport, T. A., et al. (2007). Determining the conductance of the SecY protein translocation channel for small molecules. Mol. Cell 26, 501–509.
Saraogi, I., Akopian, D., and Shan, S. O. (2014). Regulation of cargo recognition, commitment, and unloading drives cotranslational protein targeting. J. Cell Biol. 205, 693–706.
Sato, A., Kobayashi, G., Hayashi, H., Wada, A., Maeda, M., Hiragi, S., et al. (2005). The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells 10, 393–408.
Sauerbrei, B., Arends, J., Schünemann, D., and Narberhaus, F. (2020). Lon protease removes excess signal recognition particle protein in Escherichia coli. J. Bacteriol. 202:e00161-20.
Sauert, M., Wolfinger, M. T., Vesper, O., Müller, C., Byrgazov, K., and Moll, I. (2016). The MazF-regulon: a toolbox for the post-transcriptional stress response in Escherichia coli. Nucleic Acids Res. 44, 6660–6675. doi: 10.1093/nar/gkw115
Schäfer, U., Beck, K., and Müller, M. (1999). Skp, a molecular chaperone of Gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J. Biol. Chem. 274, 24567–24574.
Schaffitzel, C., Oswald, M., Berger, I., Ishikawa, T., Abrahams, J. P., Koerten, H. K., et al. (2006). Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature 444, 503–506. doi: 10.1038/nature05182
Schibich, D., Gloge, F., Pohner, I., Bjorkholm, P., Wade, R. C., von Heijne, G., et al. (2016). Global profiling of SRP interaction with nascent polypeptides. Nature 536, 219–223. doi: 10.1038/nature19070
Schmidt, A., Kochanowski, K., Vedelaar, S., Ahrné, E., Volkmer, B., Callipo, L., et al. (2016). The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 34, 104–110. doi: 10.1038/nbt.3418
Schulze, R. J., Komar, J., Botte, M., Allen, W. J., Whitehouse, S., Gold, V. A., et al. (2014). Membrane protein insertion and proton-motive-force-dependent secretion through the bacterial holo-translocon SecYEG-SecDF-YajC-YidC. Proc. Natl. Acad. Sci. U.S.A. 111, 4844–4849. doi: 10.1073/pnas.1315901111
Scotti, P. A., Urbanus, M. L., Brunner, J., de Gier, J. W., von Heijne, G., van der Does, C., et al. (2000). YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19, 542–549. doi: 10.1093/emboj/19.4.542
Serdiuk, T., Steudle, A., Mari, S. A., Manioglu, S., Kaback, H. R., Kuhn, A., et al. (2019). Insertion and folding pathways of single membrane proteins guided by translocases and insertases. Sci. Adv. 5:eaau6824. doi: 10.1126/sciadv.aau6824
Serek, J., Bauer-Manz, G., Struhalla, G., van den Berg, L., Kiefer, D., Dalbey, R., et al. (2004). Escherichia coli YidC is a membrane insertase for Sec-independent proteins. EMBO J. 23, 294–301. doi: 10.1038/sj.emboj.7600063
Shan, S. O., Chandrasekar, S., and Walter, P. (2007). Conformational changes in the GTPase modules of the signal reception particle and its receptor drive initiation of protein translocation. J. Cell Biol. 178, 611–620. doi: 10.1083/jcb.200702018
Shan, S. O., Stroud, R. M., and Walter, P. (2004). Mechanism of association and reciprocal activation of two GTPases. PLoS Biol. 2:e320. doi: 10.1371/journal.pbio.0020320
Shimizu, H., Nishiyama, K., and Tokuda, H. (1997). Expression of gpsA encoding biosynthetic sn-glycerol 3-phosphate dehydrogenase suppresses both the LB- phenotype of a secB null mutant and the cold-sensitive phenotype of a secG null mutant. Mol. Microbiol. 26, 1013–1021. doi: 10.1046/j.1365-2958.1997.6392003.x
Shimohata, N., Nagamori, S., Akiyama, Y., Kaback, H. R., and Ito, K. (2007). SecY alterations that impair membrane protein folding and generate a membrane stress. J. Cell Biol. 176, 307–317. doi: 10.1083/jcb.200611121
Shimoike, T., Taura, T., Kihara, A., Yoshihisa, T., Akiyama, Y., Cannon, K., et al. (1995). Product of a new gene, syd, functionally interacts with SecY when overproduced in Escherichia coli. J. Biol. Chem. 270, 5519–5526. doi: 10.1074/jbc.270.10.5519
Smets, D., Loos, M. S., Karamanou, S., and Economou, A. (2019). Protein transport across the bacterial plasma membrane by the sec pathway. Protein J. 38, 262–273.
Song, J., Herrmann, J. M., and Becker, T. (2020). Quality control of the mitochondrial proteome. Nat. Rev. Mol. Cell. Biol. 22, 54–70.
Soufi, B., Krug, K., Harst, A., and Macek, B. (2015). Characterization of the E. coli proteome and its modifications during growth and ethanol stress. Front. Microbiol. 6:103. doi: 10.3389/fmicb.2015.00103
Starosta, A. L., Lassak, J., Jung, K., and Wilson, D. N. (2014). The bacterial translation stress response. FEMS Microbiol. Rev. 38, 1172–1201.
Steenhuis, M., Koningstein, G. M., Oswald, J., Pick, T., O’Keefe, S., Koch, H. G., et al. (2021). Eeyarestatin 24 impairs SecYEG-dependent protein trafficking and inhibits growth of clinically relevant pathogens. Mol. Microbiol. 115, 28–40. doi: 10.1111/mmi.14589
Steinberg, R., Knupffer, L., Origi, A., Asti, R., and Koch, H. G. (2018). Co-translational protein targeting in bacteria. FEMS Microbiol. Lett. 365:fny095. doi: 10.1093/femsle/fny095
Steinberg, R., Origi, A., Natriashvili, A., Sarmah, P., Licheva, M., Walker, P. M., et al. (2020). Posttranslational insertion of small membrane proteins by the bacterial signal recognition particle. PLoS Biol. 18:e3000874. doi: 10.1371/journal.pbio.3000874
Steinchen, W., Zegarra, V., and Bange, G. (2020). (p)ppGpp: magic modulators of bacterial physiology and metabolism. Front. Microbiol. 11:2072. doi: 10.3389/fmicb.2020.02072
Stenberg, F., Chovanec, P., Maslen, S. L., Robinson, C. V., Ilag, L. L., von Heijne, G., et al. (2005). Protein complexes of the Escherichia coli cell envelope. J. Biol. Chem. 280, 34409–34419. doi: 10.1074/jbc.M506479200
Stjepanovic, G., Kapp, K., Bange, G., Graf, C., Parlitz, R., Wild, K., et al. (2011). Lipids trigger a conformational switch that regulates signal recognition particle (SRP)-mediated protein targeting. J. Biol. Chem. 286, 23489–23497. doi: 10.1074/jbc.M110.212340
Storz, G., Wolf, Y. I., and Ramamurthi, K. S. (2014). Small proteins can no longer be ignored. Annu. Rev. Biochem. 83, 753–777. doi: 10.1186/s12915-014-0096-y
Sulaiman, J. E., and Lam, H. (2021). Evolution of bacterial tolerance under antibiotic treatment and its implications on the development of resistance. Front. Microbiol. 12:617412. doi: 10.3389/fmicb.2021.617412
Swidersky, U. E., Hoffschulte, H. K., and Müller, M. (1990). Determinants of membrane-targeting and transmembrane translocation during bacterial protein export. EMBO J. 9, 1777–1785.
Tam, P. C., Maillard, A. P., Chan, K. K., and Duong, F. (2005). Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 24, 3380–3388. doi: 10.1038/sj.emboj.7600804
Tanaka, Y., and Tsukazaki, T. (2019). A snapshot of membrane protein insertion. EMBO Rep. 20:e49034. doi: 10.15252/embr.201949034
Tanaka, Y., Izumioka, A., Abdul Hamid, A., Fujii, A., Haruyama, T., Furukawa, A., et al. (2018). 2.8-Å crystal structure of Escherichia coli YidC revealing all core regions, including flexible C2 loop. Biochem. Biophys. Res. Commun. 505, 141–145. doi: 10.1016/j.bbrc.2018.09.043
Tanaka, Y., Sugano, Y., Takemoto, M., Mori, T., Furukawa, A., Kusakizako, T., et al. (2015). Crystal structures of SecYEG in lipidic cubic phase elucidate a precise resting and a peptide-bound state. Cell Rep. 13, 1561–1568. doi: 10.1016/j.celrep.2015.10.025
Teter, S. A., Houry, W. A., Ang, D., Tradler, T., Rockabrand, D., Fischer, G., et al. (1999). Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97, 755–765.
Tomkiewicz, D., Nouwen, N., van Leeuwen, R., Tans, S., and Driessen, A. J. (2006). SecA supports a constant rate of preprotein translocation. J. Biol. Chem. 281, 15709–15713. doi: 10.1074/jbc.M600205200
Tooke, F. J., Babot, M., Chandra, G., Buchanan, G., and Palmer, T. (2017). A unifying mechanism for the biogenesis of membrane proteins co-operatively integrated by the Sec and Tat pathways. eLife 6:e26577. doi: 10.7554/eLife.26577
Travers, A. A. (1978). ppApp alters transcriptional selectivity of Escherichia coli RNA polymerase. FEBS Lett. 94, 345–348.
Tsirigotaki, A., De Geyter, J., Sostaric, N., Economou, A., and Karamanou, S. (2017). Protein export through the bacterial Sec pathway. Nat. Rev. Microbiol. 15, 21–36. doi: 10.1038/nrmicro.2016.161
Tsukazaki, T. (2018). Structure-based working model of SecDF, a proton-driven bacterial protein translocation factor. FEMS Microbiol. Lett. 365:fny112.
Tsukazaki, T., Mori, H., Echizen, Y., Ishitani, R., Fukai, S., Tanaka, T., et al. (2011). Structure and function of a membrane component SecDF that enhances protein export. Nature 474, 235–238. doi: 10.1038/nature09980
Tsukazaki, T., Mori, H., Fukai, S., Ishitani, R., Mori, T., Dohmae, N., et al. (2008). Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 455, 988–991. doi: 10.1038/nature07421
Tu, L., Khanna, P., and Deutsch, C. (2014). Transmembrane segments form tertiary hairpins in the folding vestibule of the ribosome. J. Mol. Biol. 426, 185–198. doi: 10.1016/j.jmb.2013.09.013
Ulbrandt, N. D., Newitt, J. A., and Bernstein, H. D. (1997). The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88, 187–196.
Valent, Q. A., Scotti, P. A., High, S., de Gier, J. W., von Heijne, G., Lentzen, G., et al. (1998). The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 17, 2504–2512. doi: 10.1093/emboj/17.9.2504
Van den Berg, B., Clemons, W. M. Jr., Collinson, I., Modis, Y., Hartmann, E., et al. (2004). X-ray structure of a protein-conducting channel. Nature 427, 36–44. doi: 10.1038/nature02218
van der Does, C., den Blaauwen, T., de Wit, J. G., Manting, E. H., Groot, N. A., Fekkes, P., et al. (1996). SecA is an intrinsic subunit of the Escherichia coli preprotein translocase and exposes its carboxyl terminus to the periplasm. Mol. Microbiol. 22, 619–629.
van der Laan, M., Bechtluft, P., Kol, S., Nouwen, N., and Driessen, A. J. (2004). F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J. Cell Biol. 165, 213–222. doi: 10.1083/jcb.200402100
Van Puyenbroeck, V., and Vermeire, K. (2018). Inhibitors of protein translocation across membranes of the secretory pathway: novel antimicrobial and anticancer agents. Cell. Mol. Life Sci. 75, 1541–1558.
van Stelten, J., Silva, F., Belin, D., and Silhavy, T. J. (2009). Effects of antibiotics and a proto-oncogene homolog on destruction of protein translocator SecY. Science 325, 753–756. doi: 10.1126/science.1172221
Varik, V., Oliveira, S. R. A., Hauryliuk, V., and Tenson, T. (2017). HPLC-based quantification of bacterial housekeeping nucleotides and alarmone messengers ppGpp and pppGpp. Sci. Rep. 7:11022.
von Heijne, G. (1994). Signals for protein targeting into and across membranes. Subcell. Biochem. 22, 1–19.
Voorhees, R. M., and Hegde, R. S. (2016a). Structure of the Sec61 channel opened by a signal sequence. Science 351, 88–91. doi: 10.1126/science.aad4992
Voorhees, R. M., and Hegde, R. S. (2016b). Toward a structural understanding of co-translational protein translocation. Curr. Opin. Cell Biol. 41, 91–99. doi: 10.1016/j.ceb.2016.04.009
Voorhees, R. M., Fernández, I. S., Scheres, S. H., and Hegde, R. S. (2014). Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell 157, 1632–1643. doi: 10.1016/j.cell.2014.05.024
Wagner, S., Pop, O. I., Haan, G. J., Baars, L., Koningstein, G., Klepsch, M. M., et al. (2008). Biogenesis of MalF and the MalFGK(2) maltose transport complex in Escherichia coli requires YidC. J. Biol. Chem. 283, 17881–17890. doi: 10.1074/jbc.M801481200
Wall, E., Majdalani, N., and Gottesman, S. (2018). The complex rcs regulatory cascade. Annu. Rev. Microbiol. 72, 111–139.
Wang, B., Dai, P., Ding, D., Del Rosario, A., Grant, R. A., Pentelute, B. L., et al. (2019). Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 15, 141–150.
Wang, S., Jomaa, A., Jaskolowski, M., Yang, C. I., Ban, N., and Shan, S. O. (2019). The molecular mechanism of cotranslational membrane protein recognition and targeting by SecA. Nat. Struct. Mol. Biol. 26, 919–929.
Wang, S., Yang, C. I., and Shan, S. O. (2017). SecA mediates cotranslational targeting and translocation of an inner membrane protein. J. Cell Biol. 216, 3639–3653.
Wang, Y., Wang, R., Jin, F., Liu, Y., Yu, J., Fu, X., et al. (2016). A supercomplex spanning the inner and outer membranes mediates the biogenesis of β-barrel outer membrane proteins in bacteria. J. Biol. Chem. 291, 16720–16729. doi: 10.1074/jbc.M115.710715
Weaver, J., Mohammad, F., Buskirk, A. R., and Storz, G. (2019). Identifying small proteins by ribosome profiling with stalled initiation complexes. mBio 10:e02819-18. doi: 10.1128/mBio.02819-18
Weiche, B., Burk, J., Angelini, S., Schiltz, E., Thumfart, J. O., and Koch, H. G. (2008). A cleavable N-terminal membrane anchor is involved in membrane binding of the Escherichia coli SRP receptor. J. Mol. Biol. 377, 761–773. doi: 10.1016/j.jmb.2008.01.040
Weininger, U., Jakob, R. P., Kovermann, M., Balbach, J., and Schmid, F. X. (2010). The prolyl isomerase domain of PpiD from Escherichia coli shows a parvulin fold but is devoid of catalytic activity. Protein Sci. 19, 6–18. doi: 10.1002/pro.277
Weinreich, M. D., Yigit, H., and Reznikoff, W. S. (1994). Overexpression of the Tn5 transposase in Escherichia coli results in filamentation, aberrant nucleoid segregation, and cell death: analysis of E. coli and transposase suppressor mutations. J. Bacteriol. 176, 5494–5504. doi: 10.1128/jb.176.17.5494-5504.1994
Welte, T., Kudva, R., Kuhn, P., Sturm, L., Braig, D., Muller, M., et al. (2012). Promiscuous targeting of polytopic membrane proteins to SecYEG or YidC by the Escherichia coli signal recognition particle. Mol. Biol. Cell 23, 464–479.
White, S. H., and von Heijne, G. (2008). How translocons select transmembrane helices. Annu. Rev. Biophys. 37, 23–42. doi: 10.1146/annurev.biophys.37.032807.125904
Whitehouse, S., Gold, V. A., Robson, A., Allen, W. J., Sessions, R. B., and Collinson, I. (2012). Mobility of the SecA 2-helix-finger is not essential for polypeptide translocation via the SecYEG complex. J. Cell Biol. 199, 919–929. doi: 10.1083/jcb.201205191
Wickstrom, D., Wagner, S., Baars, L., Ytterberg, A. J., Klepsch, M., van Wijk, K. J., et al. (2011a). Consequences of depletion of the signal recognition particle in Escherichia coli. J. Biol. Chem. 286, 4598–4609. doi: 10.1111/j.1751-7915.2009.00148.x
Wickstrom, D., Wagner, S., Simonsson, P., Pop, O., Baars, L., Ytterberg, A. J., et al. (2011b). Characterization of the consequences of YidC depletion on the inner membrane proteome of E. coli using 2D blue native/SDS-PAGE. J. Mol. Biol. 409, 124–135. doi: 10.1016/j.jmb.2011.03.068
Wild, J., Altman, E., Yura, T., and Gross, C. A. (1992). DnaK and DnaJ heat shock proteins participate in protein export in Escherichia coli. Genes Dev. 6, 1165–1172. doi: 10.1101/gad.6.7.1165
Wild, J., Rossmeissl, P., Walter, W. A., and Gross, C. A. (1996). Involvement of the DnaK-DnaJ-GrpE chaperone team in protein secretion in Escherichia coli. J. Bacteriol. 178, 3608–3613. doi: 10.1128/jb.178.12.3608-3613.1996
Wild, J., Walter, W. A., Gross, C. A., and Altman, E. (1993). Accumulation of secretory protein precursors in Escherichia coli induces the heat shock response. J. Bacteriol. 175, 3992–3997. doi: 10.1128/jb.175.13.3992-3997.1993
Woodbury, R. L., Hardy, S. J., and Randall, L. L. (2002). Complex behavior in solution of homodimeric SecA. Protein Sci. 11, 875–882. doi: 10.1110/ps.4090102
Wu, Z. C., de Keyzer, J., Kedrov, A., and Driessen, A. J. (2012). Competitive binding of the SecA ATPase and ribosomes to the SecYEG translocon. J. Biol. Chem. 287, 7885–7895. doi: 10.1074/jbc.M111.297911
Xie, K., Kiefer, D., Nagler, G., Dalbey, R. E., and Kuhn, A. (2006). Different regions of the nonconserved large periplasmic domain of Escherichia coli YidC are involved in the SecF interaction and membrane insertase activity. Biochemistry 45, 13401–13408. doi: 10.1021/bi060826z
Yang, C. K., Lu, C. D., and Tai, P. C. (2013). Differential expression of secretion machinery during bacterial growth: SecY and SecF decrease while SecA increases during transition from exponential phase to stationary phase. Curr. Microbiol. 67, 682–687.
Young, J. W., Wason, I. S., Zhao, Z., Rattray, D. G., Foster, L. J., and Duong Van Hoa, F. (2020). His-tagged peptidiscs enable affinity purification of the membrane proteome for downstream mass spectrometry analysis. J. Proteome Res. 19, 2553–2562. doi: 10.1021/acs.jproteome.0c00022
Yu, Z., Laven, M., Klepsch, M., de Gier, J. W., Bitter, W., van Ulsen, P., et al. (2011). Role for Escherichia coli YidD in membrane protein insertion. J. Bacteriol. 193, 5242–5251.
Yuan, J., Phillips, G. J., and Dalbey, R. E. (2007). Isolation of cold-sensitive yidC mutants provides insights into the substrate profile of the YidC insertase and the importance of transmembrane 3 in YidC function. J. Bacteriol. 189, 8961–8972.
Zhang, X. X., Chan, C. S., Bao, H., Fang, Y., Foster, L. J., and Duong, F. (2012). Nanodiscs and SILAC-based mass spectrometry to identify a membrane protein interactome. J. Proteome Res. 11, 1454–1459. doi: 10.1021/pr200846y
Zhong, J., and Zhao, X. (2019). Transcriptomic analysis of viable but non-culturable Escherichia coli O157:H7 formation induced by low temperature. Microorganisms 7:634. doi: 10.3390/microorganisms7120634
Zhou, Z., Li, Y., Sun, N., Sun, Z., Lv, L., Wang, Y., et al. (2014). Function and evolution of two forms of SecDF homologs in Streptomyces coelicolor. PLoS One 9:e105237. doi: 10.1371/journal.pone.0105237
Zimmer, J., Nam, Y., and Rapoport, T. A. (2008). Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455, 936–943. doi: 10.1038/nature07335
Keywords: SecYEG translocon, protein transport, YidC, signal recognition particle, SecA, PpiD, FtsY, stress response
Citation: Oswald J, Njenga R, Natriashvili A, Sarmah P and Koch H-G (2021) The Dynamic SecYEG Translocon. Front. Mol. Biosci. 8:664241. doi: 10.3389/fmolb.2021.664241
Received: 04 February 2021; Accepted: 24 March 2021;
Published: 15 April 2021.
Edited by:
Kürşad Turgay, Max Planck Unit for the Science of Pathogens, Max-Planck-Gesellschaft (MPG), GermanyReviewed by:
Damon Huber, University of Birmingham, United KingdomCopyright © 2021 Oswald, Njenga, Natriashvili, Sarmah and Koch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hans-Georg Koch, SGFucy1HZW9yZy5Lb2NoQGJpb2NoZW1pZS51bmktZnJlaWJ1cmcuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.