- 1Department of Reproduction Management, Leibniz Institute for Zoo and Wildlife Research, Berlin, Germany
- 2Center for Precision Genome Editing and Genetic Technologies for Biomedicine, Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow, Russia
- 3Faculty of Biology, Lomonosov Moscow State University, Moscow, Russia
- 4Department of Biology, Washington University in St. Louis, St. Louis, MO, United States
- 5Biology Laboratory, Scuola Normale Superiore, Pisa, Italy
- 6Leibniz Institute on Aging – Fritz Lipmann Institute, Jena, Germany
- 7Department of General Zoology, Faculty of Biology, University of Duisburg-Essen, Essen, Germany
- 8Central Animal Laboratory, University Hospital Essen, Essen, Germany
- 9Faculty of Veterinary Medicine, Free University of Berlin, Berlin, Germany
- 10Division Signal Transduction, Institute for Genome Biology, Leibniz Institute for Farm Animal Biology, Dummerstorf, Germany
- 11Computational Biology Group, Leibniz Institute on Aging – Fritz Lipmann Institute, Jena, Germany
- 12Core Facility Life Science Computing, Leibniz Institute on Aging – Fritz Lipmann Institute, Jena, Germany
- 13Department of Biology and Evolution of Marine Organisms, Stazione Zoologica Anton Dohrn, Naples, Italy
- 14Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Moscow, Russia
Most research on mechanisms of aging is being conducted in a very limited number of classical model species, i.e., laboratory mouse (Mus musculus), rat (Rattus norvegicus domestica), the common fruit fly (Drosophila melanogaster) and roundworm (Caenorhabditis elegans). The obvious advantages of using these models are access to resources such as strains with known genetic properties, high-quality genomic and transcriptomic sequencing data, versatile experimental manipulation capabilities including well-established genome editing tools, as well as extensive experience in husbandry. However, this approach may introduce interpretation biases due to the specific characteristics of the investigated species, which may lead to inappropriate, or even false, generalization. For example, it is still unclear to what extent knowledge of aging mechanisms gained in short-lived model organisms is transferable to long-lived species such as humans. In addition, other specific adaptations favoring a long and healthy life from the immense evolutionary toolbox may be entirely missed. In this review, we summarize the specific characteristics of emerging animal models that have attracted the attention of gerontologists, we provide an overview of the available data and resources related to these models, and we summarize important insights gained from them in recent years. The models presented include short-lived ones such as killifish (Nothobranchius furzeri), long-lived ones such as primates (Callithrix jacchus, Cebus imitator, Macaca mulatta), bathyergid mole-rats (Heterocephalus glaber, Fukomys spp.), bats (Myotis spp.), birds, olms (Proteus anguinus), turtles, greenland sharks, bivalves (Arctica islandica), and potentially non-aging ones such as Hydra and Planaria.
Introduction
Most of our current knowledge on mechanisms of aging has been acquired using classical model systems such as mouse (Mus musculus) and rat (Rattus norvegicus domestica) as well as the common fruit fly (Drosophila melanogaster) and roundworm (Caenorhabditis elegans). The well-known advantages are easy handling, short generation times, availability of normed strains and standardized husbandry, a wealth of pre-existing information including high-quality and well-annotated genomic and transcriptomic sequencing data (for references see Table 1). The commercial availability of knock-in and knock-out models, cell lines comprising embryonic stem cells, and accessibility to genetic engineering tools such as CRISPR/Cas systems opens the possibility to investigate the underlying biological mechanisms of aging, e.g., by manipulation of related expression patterns (Conn, 2008; Roy et al., 2018).
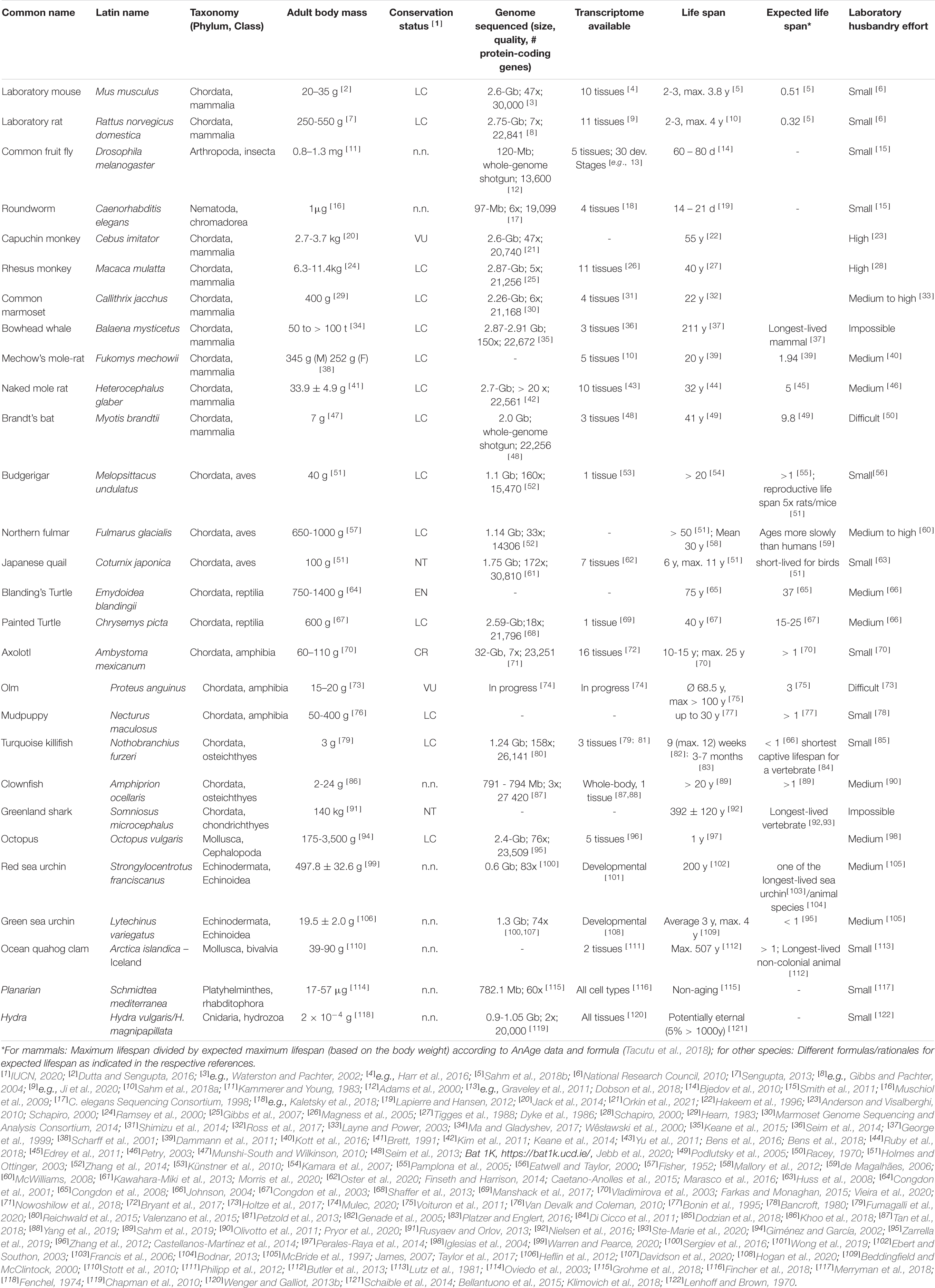
The drawback of narrowing the view to only a few model species is, however, that it impairs our efforts to gain a broad and general understanding of mechanisms underlying the fundamental biological processes that determine aging. Flies and nematodes with their maximum lifespans of up to a few months (Tacutu et al., 2018) are only to a limited extent representative of the entire animal kingdom in which aging rates vary 40,000-fold (Finch, 1994; Figure 1). Also, mice and rats, with a lifespan of up to 4 years and zebrafishes with up to 5.5 years (Tacutu et al., 2018), are considered short-lived. However, this does not only apply in absolute terms, but also with respect to the known positive correlation between lifespan and body size that explains 63% of the variation in maximum longevity across mammals, birds, amphibians and reptiles (de Magalhães et al., 2007; Fushan et al., 2015; Figure 1). With a life expectancy of only half that expected, mice and rats represent significant negative outliers (Prothero and Jürgens, 1987; de Magalhães et al., 2007). The correlation is highest in mammals and birds, slightly lower in reptiles, and even lower in amphibians, in which longevity further shows a relevant positive correlation with low ambient temperature, nocturnal lifestyle, poison defense, and captivity (Stark and Meiri, 2018). Data regarding fish and invertebrates are scarce, but suggest a similar positive allometric relationship of body weight and lifespan, e.g., among insects (Holm et al., 2016).
Figure 1. Overview of the body mass to life expectancy relation of canonical and alternative models of aging research. In mammals and many other species, lifespan generally correlates with body weight; therefore, larger species are expected to live longer (compare with Table 1). Note that the canonical models of aging research are all short-lived in relation to body mass. Selected remarkable traits of some of the mentioned species are highlighted.
It is still unclear to what extent knowledge about molecular aging mechanisms obtained in the canonical model species can be extrapolated to positive outliers such as humans who accordingly live more than 4.5 times as long as expected (Tacutu et al., 2018). For example, life-prolonging effects developed for short-lived species do not necessarily unfold the same beneficial effects in long-lived organisms (Miller and Nadon, 2000). Key components that have been successfully manipulated on a regular basis to extend lifespan in short-lived model organisms are, e.g., the growth hormone/insulin-like growth factor 1 (GH/IGF1) system or the mechanistic target of rapamycin (mTOR) pathway (Laplante and Sabatini, 2012; Junnila et al., 2013). Experimentally prolonged lifespans in short-lived species (Folch et al., 2018) are frequently associated with undesirable side-effects, e.g., decreased fertility or reduced pathogen resistance (Fontana et al., 2010). Finally, the classical model species themselves are equipped with possibly interfering, unique adaptations. For example, mice and rats display specific characteristics such as high fecundity which is associated with a vast range of physiological consequences (Hansen et al., 2014). In addition, research on traditional model organisms often focuses heavily on specific inbred lines, e.g., the mouse strain C57BL/6, which is used in about 90% of biomedical work on this species (Selman and Swindell, 2018). More natural populations show lifespans that in most cases far exceed those conferred by anti-aging interventions on conspecifics of such inbred strains. In addition, there is evidence that wild mouse populations, for instance, already have lower GH/IGF1 signaling - a condition that might thus merely be restored by many classical anti-aging interventions in inbred lines (Miller et al., 2002). The generalizability of findings from short-lived model organisms can and should be validated by studies in positive outliers of the lifespan to body mass correlation or by comparing these outliers with less long-lived species.
Based on those findings from short-lived, canonical model organisms three major categories for hallmarks of aging have been described (López-Otín et al., 2013). ‘Primary’ hallmarks are causally related to molecular damage during aging and comprise genomic instability, telomere attrition, epigenetic alteration, and loss of proteostasis. The ‘antagonistic’ hallmarks, deregulated nutrient sensing, mitochondrial dysfunction, and cellular senescence, have protective and beneficial hormetic effects at low, but detrimental effects at high levels. ‘Integrative hallmarks’ comprise stem cell exhaustion and altered intercellular communication, leading to loss of reserve capacity or resilience.
This review highlights some lesser known, non-canonical and emerging animal models of aging research by giving a short overview on their advantages and limitations, with the aim to facilitate the choice of model species, and to encourage further comparative approaches (Figures 1, 2). By exploring the greater picture, the variety of solutions for exceptional lifespans that originated over the course of evolution can help to identify relevant mechanisms, to gain a more holistic understanding of the processes involved, and eventually may contribute to efforts of extending human healthy aging. This besides underlines the importance of protecting these treasures of biodiversity.

Figure 2. Classical and alternative model species of aging research. In order from left to right, top to bottom: I Human (Homo sapiens); II Laboratory mouse (Mus musculus); III laboratory rat (Rattus norvegicus domestica); IV common fruit fly (Drosophila melanogaster); V roundworm (Caenorhabditis elegans); VI Rhesus monkey (Macaca mulatta); VII white-faced capuchin monkey (Cebus imitator); VIII bowhead whale (Balaena mysticetus); IX Mechow’s mole-rat (Fukomys mechowii); X naked mole-rat (Heterocephalus glaber); XI Brandt’s bat (Myotis brandtii); XII budgerigar (Melopsittacus undulatus); XIII northern fulmar (Fulmarus glacialis); XIV Japanese quail (Coturnix japonica); XV Blanding’s turtle (Emydoidea blandingii); XVI painted turtle (Chrysemys picta); XVII axolotl (Ambystoma mexicanum); XVIII olm (Proteus anguinus); XIX turquoise killifish (Nothobranchius furzeri); XX clownfish (Amphiprion ocellaris); XXI Greenland shark (Somniosus microcephalus); XXII octopus (Octopus vulgaris); XXIII lobster (Homarus americanus); XXIV red sea urchin (Strongylocentrotus franciscanus); XXV green sea urchin (Lytechinus variegatus); XXVI ocean quahog clam (Arctica islandica); XXVII Planarian (Schmidtea mediterranea); XVIII hydra (Hydra vulgaris). Attributions for used images: V Janine Kirstein; VIII A bowhead whale spyhops off the coast of western Sea of Okhotsk by Olga Shpak licensed under CC BY-SA 3.0; IX Philip Dammann; X, XIII, XVII, XVIII Susanne Holtze; XI Marcus Fritze; XIV; Male Japanese Quail by Ingrid Taylor licensed under CC BY 2.0; IXX Nadine Grimm on behalf of the FLI, Jena; XXI Close up image of a greenland shark taken at the floe edge of the Admiralty Inlet, Nunavut. by Hemming 1952 licensed under CC BY-SA 4.0; XXVI Arctica islandica (Ocean Quahog) by S. Rae licensed under CC BY 2.0; XXVII Anne Schroll on behalf of the FLI, Jena; XVIII Hydra oligactis by Marta Boroń licensed under CC BY 2.0; all other images were taken from Pixabay under the Simplified Pixabay License, links can be provided upon request. This figure was created by Susanne Holtze, IZW Berlin and is licensed under CC BY-SA 4.0.
Mammals
Primates
Due to their similar aging physiology, humans’ closest living relatives, chimpanzees (Pan troglodytes) and bonobos (P. paniscus) are of highest interest for research aiming at developing life-extending treatments for humans. However, this advantage is relativized by substantial financial, ethical (Gagneux et al., 2005), legal, and species conservation considerations, and by the impracticability imposed by lifespans of up to 60 years. New World marmosets and Old World macaques are also subject to strict ethical regulations (Mitchell et al., 2021), high cost of housing and long lives, but may embody a suitable compromise. They represent positive outliers from the positive allometric relationship between body weight and lifespan (Figure 1). Total metabolic energy per lifespan, per 1 kg body mass among terrestrial mammals in captivity was found to be highest in monkeys (Atanasov, 2006). The white-faced capuchin monkey (Cebus capucinus) has a maximal life expectancy of 55 years (Hakeem et al., 1996), the rhesus monkey (Macaca mulatta) of 40 years (Dyke et al., 1986; Tigges et al., 1988), and the common marmoset (Callithrix jacchus) of 22 years (Ross et al., 2017). Especially macaques and callitriche share many aging-related diseases with humans, affecting the skeletal, reproductive (Black and Lane, 2002; Colman et al., 2005) and vascular system (Clarkson and Mehaffey, 2009), sensory (Bito et al., 1982; Torre et al., 2004), and cognitive function (Voytko et al., 2009). They also develop diseases such as Alzheimer-like cerebral proteopathy (Heuer et al., 2012; Arnsten et al., 2019), cancer, amyloidosis, diabetes, and chronic renal disease (Tardif et al., 2011), as well as cellular aging (Shimizu et al., 2003). Aging interventions have shown increased longevity due to caloric restriction (Colman et al., 2014), and improved cognitive function due to estrogen therapy in postmenopausal monkeys (Voytko et al., 2009). The effect of rapamycin on non-human primates, a drug showing life-prolonging effects in many model organisms, show basic metabolic effects of long-term treatment on marmosets similar to that seen in short-lived species, with few adverse side effects (Ross et al., 2015; Colman, 2018).
State-of-the-art molecular techniques implemented for monkeys comprise embryonic stem cells of rhesus macaque and common marmoset (Thomson and Marshall, 1997), cloning (Chan et al., 2000) and genetic modification (Wolfgang et al., 2001; Sasaki et al., 2009). The genomes of both species were sequenced (Gibbs et al., 2007; Marmoset Genome Sequencing and Analysis Consortium, 2014) and induced pluripotent stem cells from somatic cells, capable of being differentiated into any cell type established (Liu et al., 2008; Wu et al., 2010). By genetic manipulation and incorporation into embryos, these may serve to generate genetically modified animals.
The disadvantages of using monkeys for aging research may be successfully addressed in the future. The risk associated with monkeys carrying zoonotic diseases, such as hepatitis A, herpes B virus, and tuberculosis (Rothschild, 2015) may be avoided by establishing specific pathogen free populations (Ross et al., 2017). Difficulties arising from the long developmental period and lifespan to perform studies of anti-aging interventions in these models may be solved by the application of modern precise methods of biological age measurements such as the Horvath epigenetic clock (Horvath et al., 2012) which allows for the detection of an intervention effect on a much shorter timescale. And finally, the ethical and conservation-related concerns of research on primates may be altogether circumvented by using the manifold and newly arising possibilities of in vitro and stem cell research.
Fukomys Mole-Rats
African mole-rats (family Bathyergidae) are subterranean rodents that have attracted the attention of gerontologists for more than 20 years due to their generally high longevity. This applies especially to the eusocial Fukomys and Heterocephalus genera. These genera typically live in extended family groups in which reproduction is monopolized by a few individuals (usually the founder pair). The other family members usually forego their own reproduction within the confines of their natal colonies (Jarvis and Bennett, 1993; Burda et al., 2000). Compared to their body size which lies between that of mice and rats, Fukomys species are very long-lived: maximum lifespans of > 20 years have been reported for the Ansell’s mole-rat (F. anselli; Dammann and Burda, 2006) and the giant mole-rat F. mechowii (Begall et al., 2021), respectively. Possible key factors are enhanced proteasome activity and that Fukomys mole-rats exhibit a comparatively high stability of gene expression during aging (Sahm et al., 2018a, 2021). Fukomys species are of particular interest to gerontologists because breeders of both sexes live on average approximately twice as long as their non-reproductive conspecifics (Dammann and Burda, 2006; Schmidt et al., 2013). Interestingly, it could be excluded that differences in food intake or behavior cause the different lifespans (Dammann et al., 2011). This feature, probably unique among mammals, offers researchers the opportunity to study highly divergent survival patterns within one genotype, without the inevitable shortcomings of inter-species comparisons. Furthermore, the higher longevity of the breeders is associated with several properties that are the opposite of what is often predicted based on findings from short-lived model species. For example, breeders, contrary to the predictions of the oxidative stress theory of aging, have more advanced glycation end products than age-matched non-breeders (Harman, 2001; Dammann et al., 2012). In addition, anabolic pathways such as insulin signaling and protein synthesis are up-regulated in breeders, which typically shortens the life of short-lived model organisms (Pan and Finkel, 2017; Sahm et al., 2021). Genome-wide comparisons of several long-lived, phylogenetically distinct subterranean rodents including Spalax galili, Heterocephalus glaber, and Fukomys damarensis have associated lifespan extensions with sequence changes in genes related to mitonuclear balance, protein synthesis, autophagy, inflammation as well as resistance to hypoxia, cytotoxins and cancer (Bennett and Faulkes, 2000; Begall et al., 2007; Davies et al., 2015; Sahm et al., 2018b).
Maintaining and breeding Fukomys species in captivity is relatively easy, yet time consuming due to generally long generation times and small litters. In fact, gestation and lactation times of ca. 3 month each, and mean litter sizes of 2-3 are typical, while full maturity is usually not reached before the second year of life. Maybe due to these difficulties, only a handful of academic laboratories currently breed Fukomys species. As ressources, for comparative studies on aging transcriptome assemblies are available for multiple species including F. anselli, F. mechowii and F. damarensis (Fang X. et al., 2014; Sahm et al., 2018b). A scaffold-level genome assembly is available for F. damarensis (Fang R. et al., 2014).
Naked Mole-Rats (Heterocephalus glaber)
Naked mole-rats, despite their small body mass (∼ 32 g; Brett, 1991) are the longest-lived rodents. Their maximal lifespan, exceeding 30 years (Buffenstein, 2005; Grimes et al., 2012; Ruby et al., 2018), is ten times longer than that of mice and five times longer than expected by body mass (Prothero and Jürgens, 1987). Only humans are similarly strong upward outliers among land-living mammals (Austad and Fischer, 1991). Phenotypical aging appears virtually absent for 80% of their lifespan (Edrey et al., 2011) including sarcopenia (Stoll et al., 2016), neurodegeneration (Edrey et al., 2013), disease, and cancer (Buffenstein, 2008; Delaney et al., 2013). Based on sustained fecundity and an apparently age-independent mortality rate, some authors even suggest that the naked mole-rat is the first known example of a non-aging mammal (Buffenstein, 2008; Ruby et al., 2018). Others consider this statement premature due to limited data of older cohorts (Dammann et al., 2019). At the molecular/physiological level, the naked mole-rat shows typical age-related decline, e.g., in activity levels, skin structure (Buffenstein and Jarvis, 2002), liver detoxification pathways (Heinze et al., 2018), lesion and lipofuscin accumulation in various organs (Edrey et al., 2011).
Various, partly overlapping mechanisms were suggested to contribute to longevity and cancer resistance (Shepard and Kissil, 2020), e.g., over-expression of alpha-2 macroglobulin (Thieme et al., 2015), efficient DNA damage repair (MacRae et al., 2015), apoptosis and autophagy of damaged cells (Zhao et al., 2014, 2016; Evdokimov et al., 2018), cytoprotective pathways (Lewis et al., 2015) and proteasome activity (Rodriguez et al., 2014). This list continues with translational fidelity (Azpurua et al., 2013), telomere and epigenome maintenance (Tan et al., 2017; Shekhidem et al., 2019), post-translational deimination (Pamenter et al., 2019), well-maintained splicing regulation (Lee et al., 2020), long non-coding RNAs (Jiang et al., 2016), and a strong innate immune response due to a highly developed myeloid compartment (Hilton et al., 2019; Shebzukhov et al., 2019). The surprisingly low incidence of cancer (Shepard and Kissil, 2020) was associated in particular with early contact inhibition (Seluanov et al., 2009). Accordingly, naked mole-rat cell division would arrest at a comparatively low cellular density mediated by ultra-high molecular mass hyaluronan (Tian et al., 2013, 2015). However, this explanation has been questioned due to non-replicability of central results (Braude et al., 2020; Hadi et al., 2020; del Marmol et al., 2021).
Naked mole-rats show remarkable hypoxia- and hypercapnia-resistance (Larson and Park, 2009) by switching their metabolism to fructose-driven glycolysis (Park et al., 2017) and induction of hypoxia inducible factor 1α and vascular endothelial growth factor A (Xiao et al., 2017). Hypoxia may reduce metabolic rate (Zhang and Lu, 2012) and cause hormetic effects (López-Otín et al., 2013).
Naked mole-rats challenge the free radical theory of aging, predicting reactive oxygen species (ROS) to result in cumulative, irreversible damage causing cellular senescence (Miwa et al., 2008). Despite low body temperature and resting metabolic rate, naked mole-rat mitochondrial oxygen consumption and ROS release are unexpectedly high (Munro et al., 2019). Accordingly, lipid (Andziak and Buffenstein, 2006; Edrey et al., 2014), protein, and DNA oxidation (Andziak et al., 2006) exceed levels of mice, but lack an age-related increase (Andziak and Buffenstein, 2006). Despite low glutathione levels, ROS damage-coping mechanisms such as superoxide dismutase, catalase (Andziak et al., 2005), α-tocopherol activity (Viltard et al., 2019), mitochondrial protein expression (Kim et al., 2011), function, and ROS production are maintained over their lifespan (Holtze et al., 2016; Skulachev et al., 2017; Vyssokikh et al., 2020). Isolated naked mole-rat mitochondria detoxify more ROS than mouse mitochondria via their antioxidative systems (Munro et al., 2019). Also, cell membranes (Hulbert et al., 2006) and arteries are ROS-insensitive (Labinskyy et al., 2006). Genes associated with oxidoreduction, detoxification and mitochondria are under positive selection (Sahm et al., 2018b), and differentially expressed (Yu et al., 2011; Stoll et al., 2016; Heinze et al., 2018).
Colonies of up to 300 individuals of the eusocial naked mole-rats usually contain one breeding pair (Brett, 1991) whose life expectancy exceeds that of non-breeders (Hochberg et al., 2016; Ruby et al., 2018), as in Fukomys spp., offering the unique opportunity to compare intraspecific, e.g., transcriptomic (Bens et al., 2018) differences in aging mechanisms.
The long lifespan, complex social system and lower reproductive rate render research on naked mole-rats more challenging compared to the similarly sized mice. The sacrifice or loss of a naked mole-rat queen may lead to long periods of social instability without reproduction or even the death of many individuals during fighting for successorship.
Bats (Chiroptera)
Bats can live at least 3 times longer than similarly sized non-flying eutherians (Brunet-Rossinni and Austad, 2004; Wilkinson and Adams, 2019), and Brandt’s bat (Myotis brandtii) can live > 41 years, which is up to 9.8 times longer (Podlutsky et al., 2005). Bats reconcile longevity with a high metabolic rate that results in a lifespan-energy expenditure that exceeds that of other mammals by as much as 2 times (Austad and Fischer, 1991). Vespertilionid bats show the greatest longevity-to-body-mass variation among mammals (Munshi-South and Wilkinson, 2010).
Bats’ efficient antiviral immune response (Huang et al., 2019) and tight inflammation control (Kacprzyk et al., 2017) may be primarily adaptations to flight-related rapid body temperature-surges and molecular damage as well as adaptations to crowding (O’Shea et al., 2014). Bats limit self-damaging inflammatory responses by altered interferon regulation (IRF3; Banerjee et al., 2020b), reduced TNF-α- expression (Banerjee et al., 2017), and dampened cellular damage, infection (Ahn et al., 2019) and DNA (Xie et al., 2018) sensing pathways, thereby preventing ‘cytokine storms’ and limiting sterile inflammation, linked to various age-related pathologies (‘inflammaging’; Franceschi et al., 2018). This enhances coexistence with viruses, rendering bats viral reservoir hosts (Banerjee et al., 2020a). Chiropteran cellular homeostatic adaptations may support antiviral mechanisms (O’Shea et al., 2014; Banerjee et al., 2020a). Elevated basal levels of autophagy in bats increase both with age (Huang et al., 2019) and in response to infection (Pteropus alecto; Laing et al., 2019). Notwithstanding high protein homeostasis, bat proteasome activity is surprisingly low (Salmon et al., 2009), probably compensated for by higher heat shock protein activity (Chionh et al., 2019) and macroautophagy (Pride et al., 2015). Calcium-dependent neutral proteases (“calpains”) implicated in degenerative processes show low activity in bats, consistent with decreased tissue degradation (Baudry et al., 1986).
Telomere length of long-lived Myotis species is telomerase-independent. Similar to the pattern found in fish (Debes et al., 2016), telomere length correlates with climatic conditions rather than with age or heritability in Myotis myotis (Foley et al., 2020; Seeker, 2020). Telomere-associated genes are under positive selection (Morgan et al., 2013) and differentially expressed (Foley et al., 2018). Bats maintain a stable microbiome (Hughes et al., 2018), and fruit eating species display low thyroxin levels (Ifuta et al., 1988). Growth hormone-related peculiarities (Seim et al., 2013) fail to explain longevity in bats (Davies et al., 2014).
In accordance with the oxidative stress theory of aging, bat mitochondrial ROS production is low given their high metabolic rates (M. lucifugus; Brunet-Rossinni, 2004). Bat mitochondrial maintenance is enhanced (Jebb et al., 2018; Vyssokikh et al., 2020) and mitochondrial DNA is under positive selection pressure (Nabholz et al., 2008). Bat cells are resistant to ROS-induced apoptosis (Ungvari et al., 2008), hydrogen peroxide (Harper et al., 2007) and to protein oxidation (Salmon et al., 2009). Antioxidant capacity is elevated and DNA-damage is reduced (Conde-Pérezprina et al., 2012) in long-lived (Desmodus rotundus) compared to short-lived species (Myotis velifer) and in torpid compared to active individuals (Sturnia lilium; Wilhelm Filho et al., 2007). Consistent with low cancer incidence (Kitsoulis et al., 2020), DNA repair and signaling pathways are maintained throughout the lifespan (Huang et al., 2019) and two long-lived species (M. myotis, M. lucifugus) possess additional copies of the tumor suppressor FBX031 (Seim et al., 2013). The genomes and transcriptomes published by Seim et al. together with further finished and ongoing genome sequencing projects of the approximately 1300 living bat species provide a solid basis for future comparative studies (Jebb et al., 2020).
Lack of established husbandry and captive breeding protocols for many species, as well as the long lifespans remain open challenges. The latter may be addressed e.g., by estimating chronological age of individual bats by using epigenetic signatures (Wilkinson et al., 2021).
Whales (Cetacea)
Whales are among the longest-lived mammals, with life expectancy comparable to, or exceeding, that of humans, as it is the case of the bowhead whale (Balaena mysticetus), which typically populates the cold seas of Greenland and represents the longest-lived extant mammal, being able to survive beyond 200 years of age (George et al., 1999; George and Bockstoce, 2008).
While presenting some age-associated physiological aspects common to other mammals in the senescent phase, such as the age-dependent reduction of fertility observed in humans (Tacutu et al., 2018), whales show an exceptionally low incidence of disease, even at old age. The natural resistance of bowhead whale cells to neoplastic degeneration in particular has been reported (Caulin and Maley, 2011; Magalhães, 2013) and is a classic example of solving Peto’s paradox, the observation that the incidence of cancer does not appear to correlate with the number of cells in an organism (Peto et al., 1975). However, the molecular mechanisms underlying these characteristics of longevity and resistance to age-related diseases (cancer, neurodegeneration, immunosenescence, metabolic dysfunctions) have not been clarified yet.
An interesting phenomenon observed in cetacean genomes is the duplication of genes related to cell cycle control and cancer protection, a phenomenon observed also in the elephant genome that contains multiple copies of the prototypical tumor suppressor TP53 (Sulak et al., 2016) and of further tumor suppressors (Vazquez and Lynch, 2021). Numerous duplicated genes have been described in the bowhead whale genome and some of these are of particular interest in the context of cell damage and survival: for example, the Proliferation Cell Nuclear Antigen plays a fundamental role in repairing DNA damage and is duplicated in the whale genome; both copies are expressed in different tissues and an amino acid substitution is present that alters its ability to interact with other effectors, and therefore its cellular function (Keane et al., 2015). Similarly, the coding sequence of some genes of the mTOR pathway such as LAMTOR1, directly involved in the regulation of various metabolic processes associated with cancer and aging, are modified (Keane et al., 2015). A subsequent study revealed duplication of cancer-related/DNA repair genes, a recurring theme in cetacean evolution. One notable example is UVRAG. This gene plays a dual role in autophagy and DNA repair and is present in multiple copies in the genomes of the sperm whale, North Atlantic right whale, and bowhead whale (Tollis et al., 2019). Consistently, quantitative transcriptome analysis revealed up-regulation of genes for DNA repair and autophagy in the gray whale (Toren et al., 2020). Finally, analysis of coding sequence variation independently identified positive selection on DNA repair/cancer suppression genes (Tollis et al., 2019).
In summary, the insights gained from genomic cetacean studies suggest that molecular adaptations in DNA repair genes played a key role in the evolution of cancer resistance and longevity in these species. Crucial resource requirements for these and presumably future findings were the initial sequencing of the B. mysticetus genome (Seim et al., 2014; Keane et al., 2015) which was followed by sequencing of the minke whale (Balaenoptera acutorostrata; Yim et al., 2014; Park et al., 2015; Malde et al., 2017), the gray whale (Eschrichtius robustus; Moskalev et al., 2017; Toren et al., 2020), the humpback whale (Megaptera novaeangliae; Tollis et al., 2019) and the largest mammal, the blue whale (Balaenoptera musculus; Árnason et al., 2018). As with other animals that cannot be kept for obvious reasons, a key challenge for the future is to identify experimental approaches that can be used to verify hypotheses derived from such comparative studies with regard to transferability to humans.
Birds
Birds (Aves)
Similar to bats, also birds have an increased life expectancy, e.g., more than 50 years in the northern fulmar (Fulmarus glacialis), despite higher body temperatures, glucose levels and metabolic rates – as compared to size-matched mammalian species (Holmes and Ottinger, 2003; Harrison and Lightfoot, 2006; Furness and Speakman, 2008; Munshi-South and Wilkinson, 2010). This may be partially explained by their lower reactive oxygen species generation per unit O2 consumption and lower oxidative damage levels (Lambert et al., 2007), which has been attributed to reduced activity of mitochondrial complex I (Barja et al., 1994) and their uricotelic metabolism, as elevated levels of blood urates serve as natural antioxidants (Cooper-Mullin and McWilliams, 2016). Among birds, parrots (order Psittaciformes) are especially longlived. A comparative genomic analysis of an endangered parrot species, the blue-fronted Amazon (Amazona aestiva) with 30 other bird species has identified longevity-associated genes under positive selection. These are involved in various cellular functions, including telomerase activity, DNA damage repair, cell proliferation control, cancer, immunity as well as anti-oxidative mechanisms (Wirthlin et al., 2018). Cells of another long-lived psittacine, the budgerigar (Melopsittacus undulatus) are more resistant to oxidative damage compared to short-lived Japanese quail (Coturnix japonica) (Ogburn et al., 2001) and compared to mice (Pamplona et al., 2005). Both budgerigar and Japanese quail are easy to keep and breed, and therefore are highly suitable laboratory species.
Although birds and mammals show similar age-related decline and pathology (Holmes and Ottinger, 2003), their reproductive senescence seems to differ. For example, in the short-lived whinchat, virtually no sign of reproductive senescence could be identified until the age of 4 years, although survival started to decline at an age of only 1 year (Fay et al., 2020). Furthermore, in contrast to mammals, female birds display similar or partially even shorter life expectancies than males (Orell and Belda, 2002; Fay et al., 2020). In a comprehensive study including 339 bird species, it was demonstrated that brain size, independently from body mass, correlates with longevity (Jiménez-Ortega et al., 2020). This supported the “cognitive buffer” hypothesis predicting reduced levels of extrinsic mortality due to improved behavioral flexibility by larger brains (Sol, 2009).
Birds as oviparous vertebrates are widely used for the study of hormone functions on growth and development of the embryo (Groothuis and von Engelhardt, 2005), because the egg represents a closed system almost independent from maternal control once laid. The effects of the hormonal axes on growth, development, and aging can be tested by direct injection, e.g., of thyroid hormones into eggs (Sarraude et al., 2020). The latter increased telomere lengths in collared flycatchers and it was hypothesized that prenatal hormone exposure by this mechanism might set the aging clock in birds (Stier et al., 2020). An age-related decline of telomerase activity was observed in short- but not in long-lived zebra-finches (Haussmann et al., 2004), and in different bird species a negative correlation between telomere shortening and lifespan was found (Sudyka et al., 2016). The annual rate of telomeric repeat shortening appears to be slower in long-lived birds compared to short-lived bird species (Tricola et al., 2018). The delayed senescence of many bird species represents a tremendous potential that can be exploited by comparative, descriptive or conclusive studies.
Despite the many similarities of birds with mammals in terms of cardiovascular anatomy, endothermy, high basal metabolism, and cognitive abilities, these traits have evolved convergently in both phylogenetically distant groups. Therefore, it is plausible to assume avian physiological mechanisms to substantially differ from those of mammals - which nevertheless does not necessarily render them less valuable.
Fish
Annual Killifishes (e.g., Nothobranchius)
Annual killifishes are adapted to the seasonal alternation of wet and rainy season and inhabit ephemeral ponds that last a few weeks or months (Myers, 1952; Vrtílek et al., 2018a). Duration of the ponds sets a pressure for rapid maturation and an upper limit to the post-hatch lifespan of these fishes. As a result, annual killifishes become sexually mature within a couple of weeks (Vrtílek et al., 2018b) and show rapid age-dependent physiological decline (Cellerino et al., 2016). The contribution of annual killifishes to aging research is two-fold: as a model taxon for comparative approaches based on high-throughput molecular analysis, and as an experimental model to investigate the effects of genetic and non-genetic interventions on lifespan and aging-associated phenotypes (Cellerino et al., 2016; Platzer and Englert, 2016; Poeschla and Valenzano, 2020).
African killifishes of the genus Nothobranchius represent a model for parallel evolution of lifespan. The distribution range of the genus overlaps with clines in aridity, and species originating from more humid habitats have a longer lifespan and slower accumulation of age-dependent cellular damage in captivity as compared to species originating from more arid habitats. This pattern is observed in multiple lineages (Tozzini et al., 2013) and was exploited to investigate the genetic architecture of natural lifespan variation (Sahm et al., 2017; Willemsen et al., 2020). Studies of molecular evolution clearly pointed to a prominent positive selection on genes coding for proteins involved in the transcription and translation of mitochondrially encoded members of the respiratory chain as well as proteins involved in the assembly of the respiratory complexes, including nuclearly encoded components of this complex (Sahm et al., 2017). The process that coordinates expression of nuclearly and mitochondrially encoded components of the respiratory chain is known as mitonuclear balance and it has been demonstrated that experimental interference with mitonuclear balance induces life-extension in C. elegans (Houtkooper et al., 2013). Notably, the treatment of annual killifish with low dosage of a complex I inhibitor also induces life-extension (Baumgart et al., 2016), demonstrating the complementarity of experimental and comparative approaches in killifishes.
Comparative approaches have also demonstrated that evolution of short lifespan is associated with a relaxation of selection on a large number of genes (Cui et al., 2020).
N. furzeri can be cultured in the laboratory with relative ease, in large numbers (Polačik et al., 2016) and with short generation times, as it is the vertebrate with the shortest documented captive lifespan (Di Cicco et al., 2011). Thus, it fills the gap between C. elegans and D. melanogaster, which are evolutionarily extremely distant from humans, on the one hand, and mice and rats, whose lifespan and housing requirements make life-long investigations unaffordable for many laboratories, on the other. The development of techniques for over-expression and gene knock-outs (Harel et al., 2016) enabled the identification and experimental validation of novel genetic mechanisms of aging. In particular, it was experimentally shown that activity of complex I of the respiratory chain can modulate lifespan (Baumgart et al., 2016), that the microRNA family miR-29 controls neuronal iron homeostasis (Ripa et al., 2017) and cardiac health (Heid et al., 2017) during aging and that decay of proteasome activity is an early event during aging causing loss of stoichiometry in protein complexes (Kelmer Sacramento et al., 2020). Finally, comparative studies of regeneration in killifish and zebrafish revealed evolutionarily conserved enhancers that represent an early response to amputation and are necessary for regeneration to occur (Wang et al., 2020).
In summary, annual killifishes and particularly N. furzeri, are the most diffused among the alternative models due to relative ease of housing and breeding. This has contributed to the availability of a number of key resources, such as high-quality genomes and transcriptomes (Reichwald et al., 2015; Valenzano et al., 2015), homo- and heterozygous laboratory strains (Valenzano et al., 2009), genetic manipulation tools (Harel et al., 2016) as well as an increasing knowledge about N. furzeri’s ecology and behavior (Thoré et al., 2020). An important challenge for the future is improved standardization, e.g., through the development of husbandry protocols, or time- and cost-efficient test frameworks to determine the effects of chemical compounds on lifespan (Thoré et al., 2020, 2021).
Long-Lived Fishes
Clownfishes (genus Amphiprion) are small (8-15 cm) marine fishes that have adapted to a symbiotic interaction with stinging sea anemones that protect them from predation (Pryor et al., 2020). Clownfishes form bonds with immobile anemones and it is possible to observe the same couple in consecutive seasons (Planes et al., 2009). It is therefore possible to estimate annual mortality which has been shown to be as low as 10% for males (Buston and García, 2007). Estimates of maximum natural lifespan for A. percula, based on recapture probability, indicated a lower bound of 22 years (Buston and García, 2007). This estimate is corroborated by observations of living aquarium specimens of A. ocellaris and A. melanopus, that are over 20 years old (Sahm et al., 2019).
Transcriptome sequencing of clownfishes revealed positive selection in genes related to redox, immunity and mitonuclear balance (Sahm et al., 2019). Further, a comparison with positive selection in naked mole-rats and killifish revealed convergent selection of the mitonuclear balance pathway indicating that selection on the same pathway can modulate lifespan in either direction (Sahm et al., 2019). The genomes of twelve different clownfish species are available and extensive transcriptome information is available for the species A. ocellaris (Pryor et al., 2020). Clownfishes are among the very few marine fishes that can be easily cultured in captivity and are currently used as a model to experimentally induce sex reversal (Casas et al., 2016) and pigmentation phenotypes (Salis et al., 2019). Clownfishes have the potential to become the first long-living experimental fish model.
Rockfishes are widely distributed in the Pacific Ocean (Love et al., 2002) and represent an example of adaptive radiation with over 100 species within the genus Sebastes (Hyde and Vetter, 2007). The biology and especially life-history of these fishes is known in detail because they are targeted by commercial fisheries and are therefore surveyed by the fishery authorities of United States and Canada. These fishes inhabit a variety of bathymetrics, from intertidal to around 1000 m (Love et al., 2002). A remarkable characteristic of this clade is the large variation in lifespan that ranges from around ten years to more than a century. Interestingly, there is a positive correlation between depth and longevity (Cailliet et al., 2001) and the longest-living species do not appear to undergo reproductive senescence, with fecundity increasing monotonically with age, that would qualify this clade as a model of negligible senescence. Due to the large number of species and large variation in life-history traits, rockfishes hold a great potential for comparative studies and extensive genomic investigations are ongoing (Xu et al., 2019).
The Greenland shark (Somniosus microcephalus) inhabits the cold environment of the North Atlantic and Arctic oceans. Paradoxically, this large (up to 5m) shark shows extremely slow annual growth. With a maximum reported lifespan of 392 ± 120 years, determined by radiocarbon measurements, it has been described as the longest-lived vertebrate species known (Nielsen et al., 2016). Remarkably, sexual maturity seems to be reached only after an age of over 100 years.
Although main aspects of Greenland shark biology and life-history are well described (MacNeil et al., 2012), only one molecular study has been conducted to date which tested a possible correlation between exceptional longevity of this species and its specific resistance to oxidative damage (Costantini et al., 2017). However, the assessment of glutathione peroxidase activity and protein carbonyls, as compared to other vertebrate species, did not lend any support for such a relationship.
To exploit the great potential of the Greenland shark for comparative studies on aging, the hopefully early publication of some key resources is required: these include not only the publication of the genome and transcriptome sequences, but also an expanded knowledge of its specific physiology.
Reptiles
Turtles (Cheloniidae)
Cheloniidae is a long-lived and successful reptilian family. Perhaps for the same reason, individual turtles protected by a hard carapace and plastron, tend to be long-lived as well. The giant tortoises often live past 100 years and many sea turtle species approach that mark. The genomes of giant Galapagos and Aldabra tortoises have unique genes for DNA repair, inflammatory mediators, telomerase, and nutrient sensing cell-cell inhibition (Quesada et al., 2019), and Leatherback turtles have highly active telomerase (Plot et al., 2012). However, none of these giants would be good candidates for a laboratory model organism because of the challenges of maintaining captive breeding colonies. Fortunately, Blanding’s turtle (Emydoidea blandingii) and Painted turtle (Chrysemys picta) are more modest sized, therefore more suitable for laboratory study and can live more than 60 years. Even more importantly, long-term field studies exist (since 1953) of Blanding’s Turtle and Painted Turtle (Congdon et al., 2001, 2003) which are especially useful in examining the demographic landscape under which their cellular and molecular adaptations have evolved. For example, long term monitoring of wild Painted Turtle populations has shown that earlier claims that they do not show any signs of senescence (via fertility decline or mortality increase with age) are untrue (Warner et al., 2016). Nonetheless, Cheloniidae do have a number of adaptations to extended hypoxia which may have led to their resistance to the oxidative damage associated with age. Many turtle species spend considerable time in hypoxia, either diving or hibernating in mud. During cycles of diving and resurfacing, mitochondria are forced to alternate between anaerobic and aerobic modes, resulting in production of more reactive oxygen species (Lutz and Prentice, 2002). Turtles deal with this challenge either by upregulating enzymes that clean up these reactive oxygen species, such as glutathione reductase, glutathione synthetase, and glutathione transferase (Willmore and Storey, 1997b), superoxide dismutase (Willmore and Storey, 1997a), necrosis factor kappa B (Krivoruchko and Storey, 2010) or by repairing the damage to lipids and nucleic acids (Willmore and Storey, 1997b). Turtles also have mechanisms for protecting ion channels from reactive oxygen species (Lutz et al., 2003) which may have particular application to age related neurodegenerative diseases in humans. While the genome of Blanding’s turtle has not yet been published, the painted turtle genome is available (Shaffer et al., 2013).
Amphibians
Long-Lived Amphibians
As with endothermic species, the lifespan of amphibians as ectothermic organisms correlates clearly with body mass and negatively correlates even more strongly than in endothermic species with body temperature (Keil et al., 2015; Stark et al., 2018). As an additional relevant factor, neoteny (juvenilization, decreased rate of development), which is known to be present in many long-lived species, is widespread among amphibians, especially in the order Caudata. The olm (Proteus anguinus) is a neotenic salamander that at a weight of 15-20 g has an average lifespan of 68 years and a predicted maximum lifespan of 102 years. It inhabits water cave habitats in Southern Europe with stable temperatures ranges of 9-11°C (Voituron et al., 2011). At this time, an ongoing olm genome project is not yet completed1 and little is known about the mechanisms that contribute to its longevity at the cellular level. It has been reported, however, that in line with its low metabolic rate the olm shows extremely low rates of cell proliferation, e.g., in blood cells (Gredar and Bizjak Mali, 2017). As resources, specific cell populations may be interesting in the context of long-term genome maintenance, for example, the so-called cell clusters of the intestine, which represent slowly proliferating stem cells of intestinal epithelium (Bizjak Mali and Bulog, 2004). Establishing captive Proteus anguinus populations is important not only for studies of the mechanisms of aging, but also for preservation of this endangered species (Holtze et al., 2017). Other neotenic amphibians that demonstrate a longer lifespan compared to endotherms of similar body mass include the common mudpuppy (Necturus maculosus), a species of the sister evolutionary branch (Zhang and Wake, 2009) that has a predicted lifespan of 34 years, and as a more distant relative, the axolotl (Ambystoma mexicanum) that lives for about 20 years (Tacutu et al., 2018). The axolotl itself can be also interesting for the studies on slow aging: researchers have been focused on their unique regeneration capacity for a long time, and now it is clear that the cellular mechanisms controlling epimorphic regeneration are also involved in the development of cancer and aging (McCusker and Gardiner, 2011; Vieira et al., 2020). For example, the expression of well-known oncogenes (Foxo1, Myc, Kazald1, etc.) is markedly increased in axolotl blastemas (tissue of regenerating limb) at the early stages of wound healing (Stewart et al., 2013; Bryant et al., 2017). At the same time, low incidence of cancer is characteristic for this species (Vieira et al., 2020). Recent work on single-cell sequencing of blastemas highlights the importance of different cell types involved in limb regeneration. For instance, highly represented regulatory myeloid and T cells possibly contribute pro-regenerative over oncogenic molecular context in growing blastemas (Leigh et al., 2018). Aside from abundant transcriptomic data, gene-editing methods are largely described in the axolotl model (Khattak and Tanaka, 2015; Kuo and Whited, 2015; Fei et al., 2017, 2018). Altogether it makes A. mexicanum a powerful system for searching for genetic or cellular factors which define differences between similarly built regeneration and tumorigenesis molecular programs (Boilly et al., 2017), possibly providing unexpected insights into aging research.
The assembly of salamander genomes is challenging because of its large size and excessive number of repetitive regions (axolotl genome; Nowoshilow et al., 2018). On the other hand, transcriptomic data is available for a number of salamanders (Joven et al., 2019), including axolotl (Stewart et al., 2013; Bryant et al., 2017) and the genome sequencing of further species such as the olm and Chinese giant salamander (Andrias davidianus) is currently on the way.
Invertebrates
Cephalopods
Coleoid cephalopods (octopuses, squids) have attracted the interest of gerontologists because of their highly variable lifespans (Schwarz et al., 2018). However, the significant variation in cephalopod lifespans remains little studied (Schwarz et al., 2018). For instance, littoral (O. minor, O. bimaculoides) and shallow water octopuses (O. vulgaris, O. maya) usually live for 1-2 years (Rodríguez-Domínguez et al., 2013; Perales-Raya et al., 2014; Kim et al., 2018), while the estimated lifespan of cold-water (Pareledone charcoti) or deep-sea octopuses (e.g., Graneledone boreopacifica) is 7-10 years (Schwarz et al., 2018). Both short-lived and long-lived octopods perform one cycle of reproduction, and die after hatching of the eggs, a phenomenon called phenoptosis – programmed death of the entire organism (Skulachev, 2012). Maximum lifespan in these species thus appears to be strictly tied to the duration of puberty and subsequent mating. Interestingly, parental behavior in octopods depends on a specific organ, the optic gland. During brooding the optic gland induces fasting by expression of steroids and insulin signaling regulators (Wang and Ragsdale, 2018). The removal of optic glands prolongs life and prevents parental care in octopuses (Wodinsky, 1977; Wang and Ragsdale, 2018). Furthermore, octopuses are the most highly organized invertebrates: their large neural and sensory systems as well as complex behavior keep gaining researchers’ attention (Huffard, 2013).
A large number of studies show a correlation between environmental temperature and growth rate in octopod embryos and larvae (Hoving and Robison, 2017; Schwarz et al., 2018; Braga et al., 2021). Thus, cold water species apparently have a prolonged maturation period. On the other hand, long-lived octopuses exhibit parental care and guard their egg-clutches throughout all embryonic stages; therefore, the period of brooding lasts longer (Robison et al., 2014). The molecular basis of these adaptations in long-lived octopuses remains largely unknown, but may be related to effective antioxidant systems. For instance, the short-lived O. vulgaris and O. tehuelchus demonstrate flexibility of antioxidant systems in different environments (Fassiano et al., 2017; Sillero-Ríos et al., 2018).
Laboratory housing conditions are well described for a variety of octopod species (Van Heukelem, 1977; Iglesias et al., 2004; Semmens et al., 2011), and keep improving. This opens the path to address novel biological questions, e.g., regarding evolutionary developmental biology (Maldonado et al., 2019). Genomic (Albertin et al., 2015; Kim et al., 2018; Zarrella et al., 2019) and transcriptomic (Zhang et al., 2012; Castellanos-Martínez et al., 2014) data so far are available only for short-lived octopuses. The combination of the rare semelparous reproductive strategy and their cognitive abilities render octopuses interesting alternative models of aging. To exploit this potential for comparative studies on aging, some key resources, such as genomes of longer-lived species and an expanded knowledge about their physiology need to be established.
Hydra
In demographic studies with species of the genus Hydra (freshwater polyps) it was shown that several of them have constant low mortality and high fertility rates over long periods of time (Martínez, 1998; Jones et al., 2014; Schaible et al., 2015) leading to the widespread view that these animals are non-senescent (Schaible et al., 2014; Bellantuono et al., 2015; Tomczyk et al., 2015; Klimovich et al., 2018). These studies predicted that at least 5% of the Hydra population would reach an age of more than 1000 years under laboratory conditions. It is assumed that this high potential for longevity has largely evolved as a by-product of Hydra’s regenerative capacity which allows these animals to fully restore any part of their body within days in the event of injury (Vogg et al., 2019b). When cut into pieces, a complete animal can regenerate from each piece, in extreme cases even from excisions corresponding to 1% of the original animal (Shimizu et al., 1993). The approximately 1 cm long body consists of essentially a digestive tube, with the mouth/anus on one side surrounded by the tentacles arranged in a circle, and a foot on the other. An adult polyp has at maximum about 100,000 cells of twelve cell types and three distinct stem cell lineages (Tomczyk et al., 2015). The regenerative ability is the result of a high proportion of stem cells, which ensures constant self-renewal with a cell cycle of 1-4 days depending on the stem cell lineage (Bosch and David, 1984; Bode, 1996). Driven by the continuous proliferation of stem cells, under optimal conditions Hydra usually reproduces asexually through budding. A reduction of the water temperature from 18° to 10°Celsius results in a time-dependent mortality increase among H. oligactis (Yoshida et al., 2006) suggesting that this measure is sufficient to induce aging. This offers the apparently unique opportunity to compare molecular signatures of non-aging and aging-like phenotypes within a single species (Bellantuono et al., 2015). Both annotated genomes (H. vulgaris, H. viridissima and H. oligactis) and a transcriptome (H. vulgaris) are available (Chapman et al., 2010; Wenger and Galliot, 2013a; Vogg et al., 2019a). Furthermore, high-resolution gene expression data sets are available, e.g., with respect to regeneration time series (Petersen et al., 2015) or on single cell level (Siebert et al., 2019). Despite being taxonomically more distant, Hydra shares as many genes with humans as D. melanogaster (both about 6000) and even more than C. elegans (about 4500) (Wenger and Galliot, 2013a). Particular attention has been paid, e.g., to the evolutionarily conserved transcription factor FoxO, which is believed to play a possible key role in maintaining the inexhaustible self-renewal capacity of Hydra stem cells and thus in apparent biological immortality (Boehm et al., 2012; Bellantuono et al., 2015; Martins et al., 2016). Generations of stable transgenic animals have been used several times both for overexpression and knockdown experiments (Wittlieb et al., 2006; Gee et al., 2010; Vogg et al., 2019a). Also, inducible RNA interference based gene silencing approaches can be applied (Watanabe et al., 2014; Vogg et al., 2019a). Hydra is easy to keep, both individually, e.g., in Petri dishes, and en masse, e.g., in glass bowls, at water temperatures of 18°Celsius and feeding them every one to three days. One of the key challenges for the future will be to transfer the knowledge gained from this evolutionarily distant species to humans, e.g., in the field of regenerative medicine.
Long-Lived Marine Non-colonial Invertebrates
Marine invertebrate species can be found in different invertebrate groups (Mollusca, Crustacea, Echinodermata), with different ecological niches, inhabiting a range of geographical zones at different temperatures and depths (Bodnar, 2009; Murthy and Ram, 2015; Ram and Costa, 2018). Among the features that unite these organisms are delayed growth and relatively low extrinsic mortality due to protective structures (shell, spines).
The most widely studied long-lived bivalve clam is the ocean quahog of the North Atlantic (Arctica islandica), the highest documented lifespan of which reaches at least 507 years. It should be noted that this abnormally high lifespan is characteristic for Icelandic population of A. islandica, while the Baltic Sea and White sea populations have a maximum lifespan of 30-50 years (Basova et al., 2012; Gruber et al., 2015). Ocean quahog is often mentioned as a candidate for negligible senescence: individuals develop rapidly until sexual maturation, then the growth ceases, and the organism exhibits no signs of senescence for decades. The same molecular dynamic was established for the antioxidant system: young ocean quahogs exhibit high antioxidant capacity compared to other bivalves, but then, at age of 30, the mitochondrial citrate synthase as well as the catalase and glutathione stabilize at lower post-maturation levels and show no tendency for further decline with age (Abele et al., 2008). A more recent study of mitochondrial membrane lipidome demonstrated lower peroxidation indexes in long-lived A. islandica than in short-lived bivalve species (Munro and Blier, 2012). However, an intraspecific study of six A. islandica populations suggests that lipid membrane composition does not correlate with longevity.
Highly variable mean telomere length and activity of telomerase, which is continuously expressed in different tissues, were investigated in the longest- versus shortest-lived populations of A. islandica. But the fact that equal telomere dynamics are shared between A. islandica populations with extremely different maximum lifespans indicates that constant telomerase activity apparently contributes to longevity but does not determine it (Gruber et al., 2014). Thus, unique factors defining the ocean quahog’s longevity remain to be discovered. A. islandica’s genome has not been published so far but a mitochondrial genome is available (Glöckner et al., 2013).
Genome sequencing of the red sea urchin (Strongylocentrotus franciscanus), which reaches a lifespan about 200 years (Tacutu et al., 2018), and of the green sea urchin (Lytechinus variegatus) allowed for a comparison between closely related long and short-living species. Mitochondrial proteins, lipid transport proteins (ApoB), proteins involved in amyloidogenesis (PSEN1) and the system of telomere maintenance (particularly, TERT) of S. franciscanus were enriched in amino acid substitutions which are specific for a long-lived species (Sergiev et al., 2016).
Homarus is a genus of lobsters whose members are estimated to reach lifespans of up to 50 years in the wild (Wolff, 1978) and up to 100 years in captivity (Bowden et al., 2020). They grow indefinitely, are able to regenerate limbs even at high ages and old animals may be more fertile than young ones (Klapper et al., 1998; Koopman et al., 2015; Tacutu et al., 2018). Unlike in most other animals, telomerase activity is high also in adults and in all tissues (Klapper et al., 1998).
Overall, marine invertebrates represent powerful models for aging research, combining ease of chronological age determination if captured in the wild and convenience of culturing in captivity. Further studies on longevity using these models require development of methods, such as cell culturing and gene manipulation, to be adapted for the marine invertebrate species (Ram and Costa, 2018).
Planaria (Tricladida)
Like Hydra, planarian flatworms are described as theoretically non-aging based on their regeneration ability (Sahu et al., 2017; Johnson et al., 2019). Although they are complex organisms with, for example, a central nervous system, a digestive tract and eyes, a complete individual can grow from small excised body fragments of almost any tissue. A high proportion of partly pluripotent adult stem cells (about 25-30% of all cells), called neoblasts, play a decisive role in this ability (Reddien and Sánchez Alvarado, 2004). The neoblasts incorporate positional signals into the processes of proliferation and differentiation and thus guarantee the almost perfect scaling of tissues and organs during regeneration (Reddien, 2018). Based on this, Planaria evolved asexual reproduction by fission as an alternative to sexual reproduction. The worm splits horizontally into a head and a tail part, each then restoring the missing part (Malinowski et al., 2017). Some species even reproduce exclusively this way. This means that in these species the neoblasts completely take over the function of the germline. Therefore, at least a part of the soma of these Planaria has to be non-aging. This “somatic immortality” can be illustrated by experiments in which the injection of a single neoblast was sufficient to save individuals in which all cycling cells were killed by irradiation and who then went on to establish new populations (Wagner et al., 2011). Stem cell aging is a major reason why organs and tissues in other species become less and less able to perform their tasks over time. It is therefore plausible that mechanisms associated with this process can be avoided in Planaria. In this context, it has been shown, e.g., that continuous telomere-shortening is avoided in the soma of asexual Planaria because telomerase is up-regulated in regeneration (Tan et al., 2012). On the other hand, the main biological function of cellular senescence is tumor suppression in tissues affected by somatic mutations (van Deursen, 2014). Therefore, it is often assumed that Planaria must have a high genomic stability and/or efficient DNA repair mechanisms (Sahu et al., 2017; Barghouth et al., 2019). In line with that, it was observed that the Piwi-piRNA pathway, which normally protects the germline from transposable element activity, is somatically expressed both in Planaria and Hydra (Sturm et al., 2017). More research is needed to find out whether activation of germline pathways can be a strategy to prevent stem cell aging. High-quality genome transcriptome assemblies are available for the frequently used species Schmidtea mediterranea (Grohme et al., 2018; Swapna et al., 2018) that contains both sexual and asexual strains. For the same species a single cell transcriptomics-based cell type atlas was published (Plass et al., 2018). Further genome and transcriptome assemblies are available for several additional species, including Dugesia japonica (Nishimura et al., 2012; An et al., 2018), Procotyla fluviatilis (Sikes and Newmark, 2013) and Dendrocoelum lacteum (Liu et al., 2013). Gene knock-down via RNA interference is well established and often used in Planaria (Rouhana et al., 2013). Further valuable tools are irradiation protocols for targeted or complete elimination of an individual’s neoblasts in combination with the possibility of transplantation of tissue or even single neoblasts (Wagner et al., 2011; Rojo-Laguna et al., 2019). As with Hydra, one of the great challenges for the future will be to explore applications for transferring the knowledge gained in these invertebrates to humans.
Discussion
Any selection of non-canonical model organisms is necessarily incomplete. Similar to African mole-rats, blind mole rats (Spalax), e.g., are strong positive outliers from the lifespan to body mass correlation (Tacutu et al., 2018) and also extremely cancer resistant – the latter possibly mediated by a concerted necrotic cell death mechanism (Gorbunova et al., 2012). Also, elephants exhibit strong cancer resistance, which was associated with a high number of copies of tumor suppressor TP53 (Sulak et al., 2016) and further tumor suppressors (Vazquez and Lynch, 2021). Ant queens reach extreme lifespans of up to 45 years unexpected for their size and, moreover, usually live many times longer than workers (Giraldo and Traniello, 2014). Deep-sea vestimentiferan tubeworms such as Lamellibrachia luymesi, Seepiophila jonesi and Escarpia laminata can reach lifespans of more than 250 years (Durkin et al., 2017). The “immortal jellyfish” Turritopsis dohrnii undergoes a reverse development from the medusa to the actually preceding, more juvenile polyp stage in case of injuries or age weakness due to transdifferentiation. It is assumed that this process can be repeated indefinitely and thus their lifespan, in contrast to most other species, is not intrinsically limited (Piraino et al., 1996). Chameleons of the genus Furcifer have highly variable life spans. For example, members of the southern population of the species F. labordi are considered the shortest-lived known tetrapods, with mean lifespans of 4-5 months, while parts of the northern population live twice as long and other species of the same genus even live more than 10 times as long (Karsten et al., 2008; Eckhardt et al., 2017; Tacutu et al., 2018). One criterion’ for selecting suitable models was the availability of scientific, age-relevant knowledge on a species or taxon. Nevertheless, we readily concede that the distinction is a matter of interpretation and that other species, as those briefly mentioned above, could possibly have been included.
Regarding potential molecular targets to slow down the aging process, unsurprisingly, the most promising targets from established (short-lived) model organisms also appear to some extent in the studies conducted in non-canonical species. As described above, this applies, e.g., to the GH/IGF1 axis, the mTOR pathway, and sirtuins (Fontana et al., 2010; Longo et al., 2015; Pan and Finkel, 2017; Duran-Ortiz et al., 2021). It is interesting to note, however, that especially in long-lived alternative model organisms other mechanisms seem to play an even more prominent role. This is particularly true for enhanced DNA repair, for which there is ample evidence from various extremely long-lived mammals, turtles, and Planaria that it may be a critical factor in longevity. The same applies to evolutionary adaptations that are associated with the coordination of protein synthesis of nuclear and mitochondrially coded components of the respiratory chain, called mitonuclear balance. Such adaptations have been seen so far in mole-rats, bats, killifishes and clownfishes. Moreover, findings in two long-lived social mole-rat genera (Heterocephalus and Fukomys) suggest that enhanced proteasome activity contributes to their long life and healthspan. There is also evidence that specific adaptations regarding the immune systems of mole-rats, bats and clownfish significantly contribute to their longevity (Figure 3A). Regarding the impact of oxidative stress and telomere maintenance on aging, however, findings from non-canonical model organisms are similarly ambiguous as those from canonical ones.
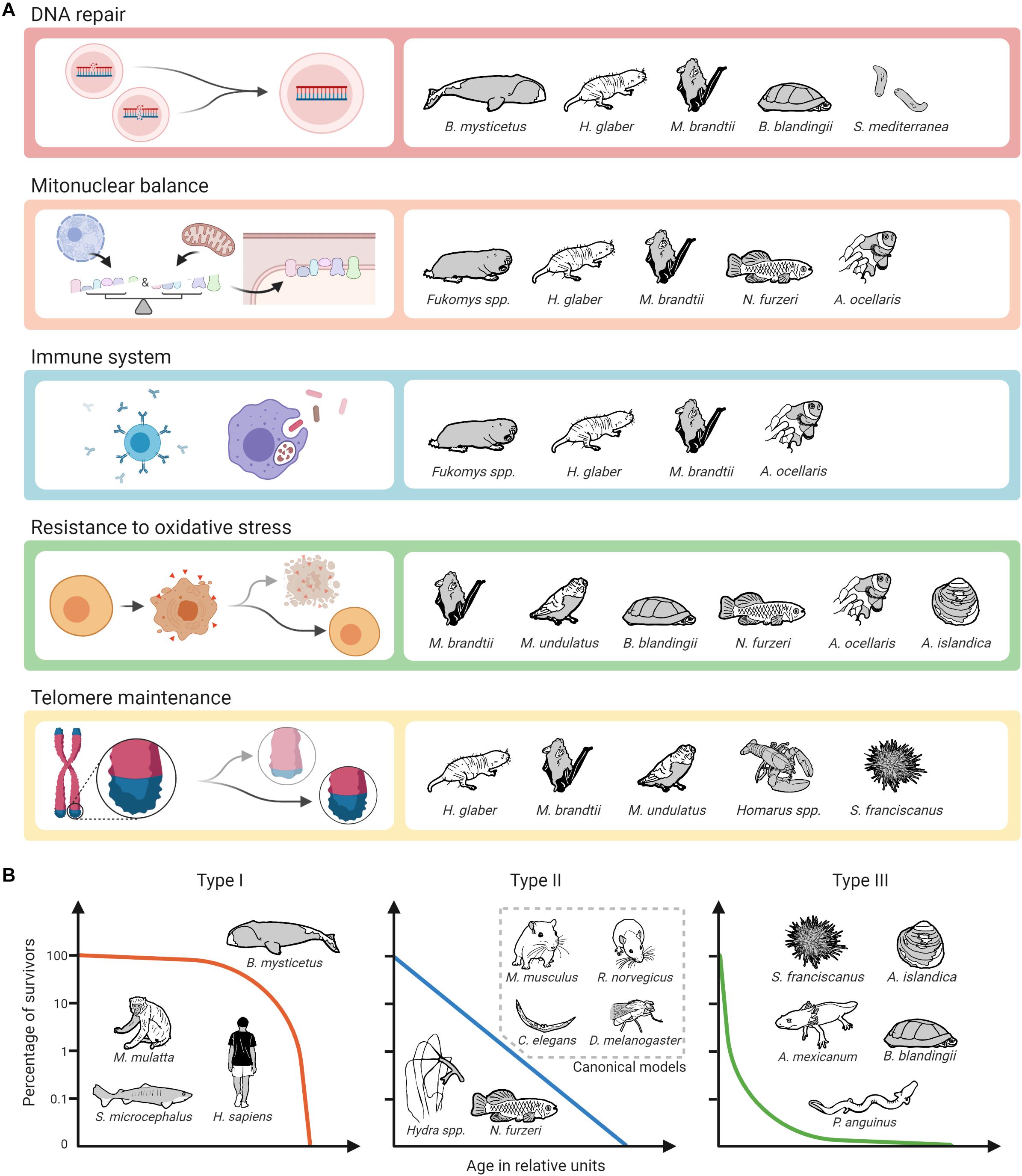
Figure 3. Biological pathways and mechanisms identified as potential contributors to longevity on alternative model organisms of aging and survival curve types. (A) The assignment of mechanisms and species does not claim to be complete, as alternative model organisms are not only less well studied than canonical ones, but also not all of the selected mechanisms have so far been addressed in all of the mentioned non-canonical species. Created with BioRender.com. (B) The assignment of survival curve types and species can only be done schematically.
Apart from this, our review has not uncovered convergently evolved longevity patterns regarding cellular and molecular mechanisms yet. However, this is not very surprising given that high longevities evolved independently in very different phylogenetic lineages and equally different aquatic, fossorial, terrestrial, or airborne environments. Under so manyfold combinations of phylogenetic and ecological constraints that have shaped the gradual adaptation of the respective genomes, we should not a priori expect evolution to have always followed the same paths to reach converging solutions for this complex trait. Therefore, the recurrence of at least some patterns of genetic architecture, as described above, is far from trivial. Most surprising is that adaptations of processes such as mitonuclear balance or resistance to oxidative stress seem to play an important role both for longevity and extremely short lifespans. This suggests that evolutionary trade-offs exist along these processes that allow either adaptation toward selection advantages during the early stages of ontogenesis, e.g., rapid growth in N. furzeri, or modification toward longer lifespans. Furthermore, it is perhaps simply too early to expect more such patterns to emerge before these and other non-canonical species have been studied in more detail by a broader aging research community.
Canonical species did not become the workhorses of biological research by accident; their rapid maturity and reproduction make them economical to breed and rear in captivity. However, their rapid maturity and short lives are the result of millions of years of adaptation to a very different evolutionary landscape than that of the long-lived species we have reviewed above. So, although it made sense to first explore the cellular and molecular basis of aging in the already well studied, short-lived canonical species, it is time for us to move on if we are to progress.
Without doubt, non-canonical model organisms offer the possibility to study mechanisms relevant for an extended lifespan that do not occur in canonical ones. These include the species-specific causes of cancer resistance, e.g., in bathyergid mole-rats, blind mole-rats and elephants, the capacity of Hydra and Planaria stem cells to not exhaust over time, the physiological and molecular mechanisms linking longevity and neoteny in amphibians, and likely a wealth of further ones yet to be explored. Like humans, many of our non-canonical model species are adapted to relative safety from predators, either based on their subterranean environment, their special defenses, or their size. Unlike mice, rats, fruit flies and roundworms, with type II survivorship (sensu Pearl and Miner, 1935), many of our non-canonical species have evolved a type I or type III survivorship curve. Type I species typically include top predators or otherwise safe large individuals. We are long-lived because we are relatively safe from predation for our entire lives. Type III species on the other hand suffer much early mortality, but once they grow to a safe size, they can live very long lives. Type II species however are typically what we think of as “prey species” and suffer from predation at a relatively constant pace throughout their relatively short lives. Given that humans evolved under a type I survivorship curve, we might expect to find that we are better suited to exploit the longevity adaptations discovered in whales (such as DNA repair), but less with type III species such as giant clams (resistance to oxidative stress). On the other hand, perhaps we may be able to exploit a wide array of strategies to promote longevity as, e.g., bats apparently can (Figure 3B). One thing is certain, we will not discover the details of those strategies by restricting our work to the short-lived canonical species.
Conclusion
We summarized the specific characteristics of a broad taxonomic range of alternative animal models suitable for elucidating processes involved in delayed and accelerated senescence in terms of life expectancy, pre-existing discoveries, and available data and resources. The species presented have exceptional lifespans, an enormous regeneration potential or a remarkable resistance to aging-related diseases. Previous findings on the possible molecular causes confirm the mechanisms known from classical model organisms, but with a different weighting of the various known aging-related signaling pathways. In addition, there are some intriguing, life-prolonging mechanisms to which no equivalent in classical model organisms exists. Including this wealth of evolutionary adaptations into future research will most likely broaden our understanding of the aging process and may eventually contribute to the development of interventions for granting humans a longer and healthier lifespan.
Author Contributions
AS und SuH conceived and outlined the manuscript. SuH, AS, EG, SB, AC, PD, TH, AH, StH, PK, and ET wrote and edited the original draft. Graphs were prepared by SH and PK. SB and SuH did the final editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Leibniz Association (SAS-FLI-2016) and the Add-On Fellowship Program for Interdisciplinary Life Science of the Joachim Herz Foundation (to AS). EG was supported by grant # 075-15-2019-1660 from the Ministry of Science and Higher Education of the Russian Federation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Derek Ott for kind support with the figures.
Footnotes
References
Abele, D., Strahl, J., Brey, T., and Philipp, E. E. R. (2008). Imperceptible senescence: ageing in the ocean quahog Arctica islandica. Free Radical Res. 42, 474–480. doi: 10.1080/10715760802108849
Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. doi: 10.1126/science.287.5461.2185
Ahn, M., Anderson, D. E., Zhang, Q., Tan, C. W., Lim, B. L., Luko, K., et al. (2019). Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat. Microbiol. 4, 789–799. doi: 10.1038/s41564-019-0371-3
Albertin, C. B., Simakov, O., Mitros, T., Wang, Z. Y., Pungor, J. R., Edsinger-Gonzales, E., et al. (2015). The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 524, 220–224. doi: 10.1038/nature14668
An, Y., Kawaguchi, A., Zhao, C., Toyoda, A., Sharifi-Zarchi, A., Mousavi, S. A., et al. (2018). Draft genome of Dugesia japonica provides insights into conserved regulatory elements of the brain restriction gene nou-darake in planarians. Zool. Lett. 4:24. doi: 10.1186/s40851-018-0102-2
Anderson, J. R., and Visalberghi, E. (2010). “Capuchin monkeys,” in The UFAW Handbook on the Care and Management of Laboratory and Other Research Animals, Vol. 8, eds R. Hubrecht and J. Kirkwood (Hoboken, NJ: Wiley), 579–591.
Andziak, B., and Buffenstein, R. (2006). Disparate patterns of age-related changes in lipid peroxidation in long-lived naked mole-rats and shorter-lived mice. Aging Cell 5, 525–532. doi: 10.1111/j.1474-9726.2006.00246.x
Andziak, B., O’Connor, T. P., and Buffenstein, R. (2005). Antioxidants do not explain the disparate longevity between mice and the longest-living rodent, the naked mole-rat. Mech. Ageing Dev. 126, 1206–1212. doi: 10.1016/j.mad.2005.06.009
Andziak, B., O’Connor, T. P., Qi, W., DeWaal, E. M., Pierce, A., Chaudhuri, A. R., et al. (2006). High oxidative damage levels in the longest-living rodent, the naked mole-rat. Aging Cell 5, 463–471. doi: 10.1111/j.1474-9726.2006.00237.x
Árnason, Ú, Lammers, F., Kumar, V., Nilsson, M. A., and Janke, A. (2018). Whole-genome sequencing of the blue whale and other rorquals finds signatures for introgressive gene flow. Sci. Adv. 4:eaa9873. doi: 10.1126/sciadv.aap9873
Arnsten, A. F. T., Datta, D., Leslie, S., Yang, S.-T., Wang, M., and Nairn, A. C. (2019). Alzheimer’s-like pathology in aging rhesusmacaques: unique opportunity to study the etiology and treatment of Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 116, 26230–26238. doi: 10.1073/pnas.1903671116
Atanasov, A. T. (2006). Linear allometric relationship between total metabolic energy per life span and body mass of terrestrial mammals in captivity. Bulg. J. Vet. Med. 9, 159–174.
Austad, S. N., and Fischer, K. E. (1991). Mammalian aging, metabolism, and ecology: evidence from the bats and marsupials. J. Gerontol. 46, B47–B53. doi: 10.1093/geronj/46.2.B47
Azpurua, J., Ke, Z., Chen, I. X., Zhang, Q., Ermolenko, D. N., Zhang, Z. D., et al. (2013). Naked mole-rat has increased translational fidelity compared with the mouse, as well as a unique 28S ribosomal RNA cleavage. Proc. Natl. Acad. Sci. U.S.A. 110, 17350–17355. doi: 10.1073/pnas.1313473110
Bancroft, L. S. (1980). An introduction to the care and maintenance of Necturus spp. (mudpuppy), an aquatic salamander. J. Institute Anim. Tech. 31, 5–13.
Banerjee, A., Baker, M. L., Kulcsar, K., Misra, V., Plowright, R., and Mossman, K. (2020a). Novel insights into immune systems of bats. Front. Immunol. 11:26. doi: 10.3389/fimmu.2020.00026
Banerjee, A., Rapin, N., Bollinger, T., and Misra, V. (2017). Lack of inflammatory gene expression in bats: a unique role for a transcription repressor. Sci. Rep. 7:2232. doi: 10.1038/s41598-017-01513-w
Banerjee, A., Zhang, X., Yip, A., Schulz, K. S., Irving, A. T., Bowdish, D., et al. (2020b). Positive selection of a serine residue in Bat IRF3 confers enhanced antiviral protection. iScience 23:100958. doi: 10.1016/j.isci.2020.100958
Barghouth, P. G., Thiruvalluvan, M., LeGro, M., and Oviedo, N. J. (2019). DNA damage and tissue repair: What we can learn from planaria. Semin. Cell Dev. Biol. 87, 145–159. doi: 10.1016/j.semcdb.2018.04.013
Barja, G., Cadenas, S., Rojas, C., Pérez-Campo, R., and López-Torres, M. (1994). Low mitochondrial free radical production per unit O2 consumption can explain the simultaneous presence of high longevity and high aerobic metabolic rate in birds. Free Radical Res. 21, 317–327. doi: 10.3109/10715769409056584
Basova, L., Begum, S., Strahl, J., Sukhotin, A., Brey, T., Philipp, E., et al. (2012). Age-dependent patterns of antioxidants in Arctica islandica from six regionally separate populations with different lifespans. Aquat. Biol. 14, 141–152. doi: 10.3354/ab00387
Baudry, M., Dubrin, R., Beasley, L., Leon, M., and Lynch, G. (1986). Low levels of calpain activity in chiroptera brain: implications for mechanisms of aging. Neurobiol. Aging 7, 255–258. doi: 10.1016/0197-4580(86)90004-7
Baumgart, M., Priebe, S., Groth, M., Hartmann, N., Menzel, U., Pandolfini, L., et al. (2016). Longitudinal RNA-seq analysis of vertebrate aging identifies mitochondrial complex I as a small-molecule-sensitive modifier of lifespan. Cell Syst. 2, 122–132. doi: 10.1016/j.cels.2016.01.014
Beddingfield, S. D., and McClintock, J. B. (2000). Demographic characteristics of Lytechinus variegatus (Echinoidea: Echinodermata) from three habitats in a North Florida Bay, Gulf of Mexico. Mar. Ecol. 21, 17–40. doi: 10.1046/j.1439-0485.2000.00688.x
Begall, S., Burda, H., and Schleich, C. E. (2007). “Subterranean rodents: news from underground,” in Subterranean Rodents, eds S. Begall, H. Burda, and C. E. Schleich (New York, NY: Springer-Verlag Berlin Heidelberg), 3–9.
Begall, S., Nappe, R., Hohrenk, L., Schmidt, T. C., Burda, H., Sahm, A., et al. (2021). Life expectancy, family constellation and stress in giant mole-rats (Fukomys mechowii). Philos. Trans. R. Soc. B 376:20200207.
Bellantuono, A. J., Bridge, D., and Martínez, D. E. (2015). Hydra as a tractable, long-lived model system for senescence. Invert. Reprod. Dev. 59, 39–44. doi: 10.1080/07924259.2014.938196
Bennett, N. C., and Faulkes, C. G. (2000). African Mole-Rats: Ecology and Eusociality. Cambridge: Cambridge University Press.
Bens, M., Sahm, A., Groth, M., Jahn, N., Morhart, M., Holtze, S., et al. (2016). FRAMA: from RNA-seq data to annotated mRNA assemblies. BMC Genomics 17:54. doi: 10.1186/s12864-015-2349-8
Bens, M., Szafranski, K., Holtze, S., Sahm, A., Groth, M., Kestler, H. A., et al. (2018). Naked mole-rat transcriptome signatures of socially suppressed sexual maturation and links of reproduction to aging. BMC Biol 16:77. doi: 10.1186/s12915-018-0546-z
Bito, L. Z., DeRousseau, C. J., Kaufman, P. L., and Bito, J. W. (1982). Age-dependent loss of accommodative amplitude in rhesus monkeys: an animal model for presbyopia. Invest. Ophthalmol. Visual Sci. 23, 23–31.
Bizjak Mali, L., and Bulog, B. (2004). Histology and ultrastructure of the gut epithelium of the neotenic cave salamander, Proteus anguinus (Amphibia, Caudata). J. Morphol. 259, 82–89. doi: 10.1002/jmor.10171
Bjedov, I., Toivonen, J. M., Kerr, F., Slack, C., Jacobson, J., Foley, A., et al. (2010). Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 11, 35–46. doi: 10.1016/j.cmet.2009.11.010
Black, A., and Lane, M. A. (2002). Nonhuman primate models of skeletal and reproductive aging. Gerontology 48, 72–80. doi: 10.1159/000048930
Bode, H. R. (1996). The interstitial cell lineage of hydra: a stem cell system that arose early in evolution. J. Cell Sci. 109(Pt 6), 1155–1164.
Bodnar, A. (2013). Proteomic profiles reveal age-related changes in coelomic fluid of sea urchin species with different life spans. Exp. Gerontol. 48, 525–530. doi: 10.1016/j.exger.2013.01.014
Bodnar, A. G. (2009). Marine invertebrates as models for aging research. Exp. Gerontol. 44, 477–484. doi: 10.1016/j.exger.2009.05.001
Boehm, A.-M., Khalturin, K., Anton-Erxleben, F., Hemmrich, G., Klostermeier, U. C., Lopez-Quintero, J. A., et al. (2012). FoxO is a critical regulator of stem cell maintenance in immortal Hydra. Proc. Natl. Acad. Sci. U.S.A. 109, 19697–19702. doi: 10.1073/pnas.1209714109
Boilly, B., Faulkner, S., Jobling, P., and Hondermarck, H. (2017). Nerve dependence: from regeneration to cancer. Cancer Cell 31, 342–354. doi: 10.1016/j.ccell.2017.02.005
Bonin, J., DesGranges, J.-L., Bishop, C. A., Rodrigue, J., Gendron, A., and Elliott, J. E. (1995). Comparative study of contaminants in the mudpuppy (amphibia) and the common snapping turtle (reptilia), St. Lawrence River, Canada. Arch. Environ. Contamination Toxicol. 28, 184–194. doi: 10.1007/bf00217615
Bosch, T. C., and David, C. N. (1984). Growth regulation in hydra: relationship between epithelial cell cycle length and growth rate. Dev. Biol. 104, 161–171. doi: 10.1016/0012-1606(84)90045-9
Bowden, T. J., Kraev, I., and Lange, S. (2020). Extracellular vesicles and post-translational protein deimination signatures in mollusca—the blue mussel (Mytilus edulis), soft shell clam (Mya arenaria), Eastern Oyster (Crassostrea virginica) and Atlantic Jacknife Clam (Ensis leei). Biology 9:416.
Braga, R., Van der Molen, S., Pontones, J., and Ortiz, N. (2021). Embryonic development, hatching time and newborn juveniles of Octopus tehuelchus under two culture temperatures. Aquaculture 530:735778.
Braude, S., Holtze, S., Begall, S., Brenmoehl, J., Burda, H., Dammann, P., et al. (2020). Surprisingly long survival of premature conclusions about naked mole−rat biology. Biol. Rev. 96, 376–393. doi: 10.1111/brv.12660
Brett, R. A. (1991). The Ecology of Naked Mole-rat Colonies : Burrowing, Food and Limiting Factors. The Biology of the Naked Mole-Rat. Princeton, NJ: Princeton University Press, 137–184.
Brunet-Rossinni, A. K. (2004). Reduced free-radical production and extreme longevity in the little brown bat (Myotis lucifugus) versus two non-flying mammals. Mech. Ageing Dev. 125, 11–20. doi: 10.1016/j.mad.2003.09.003
Brunet-Rossinni, A. K., and Austad, S. N. (2004). Ageing studies on bats: a review. Biogerontology 5, 211–222. doi: 10.1023/B:BGEN.0000038022.65024.d8
Bryant, D. M., Johnson, K., DiTommaso, T., Tickle, T., Couger, M. B., Payzin-Dogru, D., et al. (2017). A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 18, 762–776. doi: 10.1016/j.celrep.2016.12.063
Buffenstein, R. (2005). The naked mole-rat: a new long-living model for human aging research. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 60, 1369–1377.
Buffenstein, R. (2008). Negligible senescence in the longest living rodent, the naked mole-rat: insights from a successfully aging species. J. Comp. Physiol. B 178, 439–445. doi: 10.1007/s00360-007-0237-5
Buffenstein, R., and Jarvis, J. U. M. (2002). The naked mole rat—a new record for the oldest living rodent. Sci. Aging Knowledge Environ. 2002:e7. doi: 10.1126/sageke.2002.21.pe7
Burda, H., Honeycutt, R. L., Begall, S., Locker-Grütjen, O., and Scharff, A. (2000). Are naked and common mole-rats eusocial and if so, why? Behav. Ecol. Sociobiol. 47, 293–303. doi: 10.1007/s002650050669
Buston, P. M., and García, M. B. (2007). An extraordinary life span estimate for the clown anemonefish Amphiprion percula. J. Fish Biol. 70, 1710–1719. doi: 10.1111/j.1095-8649.2007.01445.x
Butler, P. G., Wanamaker, A. D., Scourse, J. D., Richardson, C. A., and Reynolds, D. J. (2013). Variability of marine climate on the North Icelandic Shelf in a 1357-year proxy archive based on growth increments in the bivalve Arctica islandica. Palaeogeogr. Palaeoclimatol. Palaeoecol. 373, 141–151. doi: 10.1016/j.palaeo.2012.01.016
C. elegans Sequencing Consortium (1998). Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018. doi: 10.1126/science.282.5396.2012
Caetano-Anolles, K., Seo, M., Rodriguez-Zas, S., Oh, J.-D., Han, J. Y., Lee, K., et al. (2015). Comprehensive identification of sexual dimorphism-associated differentially expressed genes in two-way factorial designed RNA-seq data on Japanese Quail (Coturnix coturnix japonica). PLoS One 10:e0139324. doi: 10.1371/journal.pone.0139324
Cailliet, G. M., Andrews, A. H., Burton, E. J., Watters, D. L., Kline, D. E., and Ferry-Graham, L. A. (2001). Age determination and validation studies of marine fishes: do deep-dwellers live longer? Exp. Gerontol. 36, 739–764. doi: 10.1016/s0531-5565(00)00239-4
Casas, L., Saborido-Rey, F., Ryu, T., Michell, C., Ravasi, T., and Irigoien, X. (2016). Sex change in clownfish: molecular insights from transcriptome analysis. Sci. Rep. 6:35461. doi: 10.1038/srep35461
Castellanos-Martínez, S., Arteta, D., Catarino, S., and Gestal, C. (2014). De novo transcriptome sequencing of the Octopus vulgaris hemocytes using Illumina RNA-Seq technology: response to the infection by the gastrointestinal parasite Aggregata octopiana. PLoS One 9:e107873.
Caulin, A. F., and Maley, C. C. (2011). Peto’s Paradox: evolution’s prescription for cancer prevention. Trends Ecol. Evol. 26, 175–182. doi: 10.1016/j.tree.2011.01.002
Cellerino, A., Valenzano, D. R., and Reichard, M. (2016). From the bush to the bench: the annual Nothobranchius fishes as a new model system in biology. Biol. Rev. Camb. Philos. Soc. 91, 511–533. doi: 10.1111/brv.12183
Chan, A. W. S., Dominko, T., Luetjens, C. M., Neuber, E., Martinovich, C., Hewitson, L., et al. (2000). Clonal propagation of primate offspring by embryo splitting. Science 287, 317–319.
Chapman, J. A., Kirkness, E. F., Simakov, O., Hampson, S. E., Mitros, T., Weinmaier, T., et al. (2010). The dynamic genome of Hydra. Nature 464, 592–596. doi: 10.1038/nature08830
Chionh, Y. T., Cui, J., Koh, J., Mendenhall, I. H., Ng, J. H. J., Low, D., et al. (2019). High basal heat-shock protein expression in bats confers resistance to cellular heat/oxidative stress. Cell Stress Chaperones 24, 835–849. doi: 10.1007/s12192-019-01013-y
Clarkson, T. B., and Mehaffey, M. H. (2009). Coronary heart disease of females: lessons learned from nonhuman primates. Am. J. Primatol. 71, 785–793.
Colman, R. J. (2018). Non-human primates as a model for aging. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 2733–2741. doi: 10.1016/j.bbadis.2017.07.008
Colman, R. J., Beasley, T. M., Kemnitz, J. W., Johnson, S. C., Weindruch, R., and Anderson, R. M. (2014). Caloric restriction reduces age-related and all-cause mortality in rhesus monkeys. Nat. Commun. 5, 1–5.
Colman, R. J., McKiernan, S. H., Aiken, J. M., and Weindruch, R. (2005). Muscle mass loss in Rhesus monkeys: age of onset. Exp. Gerontol. 40, 573–581.
Conde-Pérezprina, J. C., Luna-López, A., González-Puertos, V. Y., Zenteno-Savín, T., and León-Galván, M. Á, and Königsberg, M. (2012). DNA MMR systems, microsatellite instability and antioxidant activity variations in two species of wild bats: Myotis velifer and Desmodus rotundus, as possible factors associated with longevity. AGE 34, 1473–1492. doi: 10.1007/s11357-012-9399-5
Congdon, J., Graham, T., Herman, T., and Lang, J. (2008). “Emydoidea blandingii (Holbrook 1838) – Blanding’s Turtle,” in Conservation Biology of Freshwater Turtles and Tortoises: A Compilation Project of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group. Chelonian Research Monographs No. 5, 015.1-015.12, eds A. G. J. Rhodin, P. C. H. Pritchard, P. P. van Dijk, R. A. Saumure, K. A. Buhlmann, and J. B. Iverson (Lunenburg, MA: Chelonian Research Foundation).
Congdon, J. D., Nagle, R. D., Kinney, O. M., and van Loben Sels, R. C. (2001). Hypotheses of aging in a long-lived vertebrate, Blanding’s turtle (Emydoidea blandingii). Exp. Gerontol. 36, 813–827. doi: 10.1016/s0531-5565(00)00242-4
Congdon, J. D., Nagle, R. D., Kinney, O. M., van Loben Sels, R. C., Quinter, T., and Tinkle, D. W. (2003). Testing hypotheses of aging in long-lived painted turtles (Chrysemys picta). Exp. Gerontol. 38, 765–772. doi: 10.1016/s0531-5565(03)00106-2
Cooper-Mullin, C., and McWilliams, S. R. (2016). The role of the antioxidant system during intense endurance exercise: lessons from migrating birds. J. Exp. Biol. 219, 3684–3695. doi: 10.1242/jeb.123992
Costantini, D., Smith, S., Killen, S. S., Nielsen, J., and Steffensen, J. F. (2017). The Greenland shark: a new challenge for the oxidative stress theory of ageing? Comp. Biochem. Physiol. A Mol. Integr. Physiol. 203, 227–232. doi: 10.1016/j.cbpa.2016.09.026
Cui, R., Medeiros, T., Willemsen, D., Iasi, L. N. M., Collier, G. E., Graef, M., et al. (2020). Relaxed selection limits lifespan by increasing mutation load. Cell 180, 1272–1279. doi: 10.1016/j.cell.2020.02.038
Dammann, P., and Burda, H. (2006). Sexual activity and reproduction delay ageing in a mammal. Curr. Biol. 16, R117–R118. doi: 10.1016/j.cub.2006.02.012
Dammann, P., Scherag, A., Zak, N., Szafranski, K., Holtze, S., Begall, S., et al. (2019). Comment on ‘Naked mole-rat mortality rates defy Gompertzian laws by not increasing with age’. eLife 8:e45415. doi: 10.7554/eLife.45415
Dammann, P., Sell, D. R., Begall, S., Strauch, C., and Monnier, V. M. (2012). Advanced glycation end-products as markers of aging and longevity in the long-lived Ansell’s mole-rat (Fukomys anselli). J. Gerontol. Series A 67A, 573–583. doi: 10.1093/gerona/glr208
Dammann, P., Šumbera, R., Massmann, C., Scherag, A., and Burda, H. (2011). Extended longevity of reproductives appears to be common in Fukomys mole-rats (Rodentia. Bathyergidae). PLoS One 6, e18757. doi: 10.1371/journal.pone.0018757
Davidson, P. L., Guo, H., Wang, L., Berrio, A., Zhang, H., Chang, Y., et al. (2020). Chromosomal-level genome assembly of the sea urchin lytechinus variegatus substantially improves functional genomic analyses. Genome Biol. Evol. 12, 1080–1086. doi: 10.1093/gbe/evaa101
Davies, K. T. J., Bennett, N. C., Tsagkogeorga, G., Rossiter, S. J., and Faulkes, C. G. (2015). Family wide molecular adaptations to underground life in african mole-rats revealed by phylogenomic analysis. Mol. Biol. Evol. 32, 3089–3107. doi: 10.1093/molbev/msv175
Davies, K. T. J., Tsagkogeorga, G., Bennett, N. C., Dávalos, L. M., Faulkes, C. G., and Rossiter, S. J. (2014). Molecular evolution of growth hormone and insulin-like growth factor 1 receptors in long-lived, small-bodied mammals. Gene 549, 228–236. doi: 10.1016/j.gene.2014.07.061
de Magalhães, J. P. (2006). “Species selection in comparative studies of aging and antiaging research,” in Handbook of Models for Human Aging, ed. P. M. Conn (Valinhos: Elsevier), 9–20.
de Magalhães, J. P., Costa, J., and Church, G. M. (2007). An analysis of the relationship between metabolism, developmental schedules, and longevity using phylogenetic independent contrasts. J. Gerontol. A Biol. Sci. Med. Sci. 62, 149–160. doi: 10.1093/gerona/62.2.149
Debes, P. V., Visse, M., Panda, B., Ilmonen, P., and Vasemägi, A. (2016). Is telomere length a molecular marker of past thermal stress in wild fish? Mol. Ecol. 25, 5412–5424. doi: 10.1111/mec.13856
del Marmol, D., Holtze, S., Kichler, N., Sahm, A., Bihin, B., Bourguignon, V., et al. (2021). Abundance and size of hyaluronan in naked mole-rat tissues and plasma. Sci. Rep.
Delaney, M. A., Nagy, L., Kinsel, M. J., and Treuting, P. M. (2013). Spontaneous histologic lesions of the adult naked mole rat (Heterocephalus glaber): a retrospective survey of lesions in a zoo population. Vet. Pathol. 50, 607–621. doi: 10.1177/0300985812471543
Di Cicco, E., Tozzini, E. T., Rossi, G., and Cellerino, A. (2011). The short-lived annual fish Nothobranchius furzeri shows a typical teleost aging process reinforced by high incidence of age-dependent neoplasias. Exp. Gerontol. 46, 249–256. doi: 10.1016/j.exger.2010.10.011
Dobson, A. J., He, X., Blanc, E., Bolukbasi, E., Feseha, Y., Yang, M., et al. (2018). Tissue-specific transcriptome profiling of Drosophila reveals roles for GATA transcription factors in longevity by dietary restriction. NPJ Aging Mech. Dis. 4:5. doi: 10.1038/s41514-018-0024-4
Dodzian, J., Kean, S., Seidel, J., and Valenzano, D. R. (2018). A protocol for laboratory housing of Turquoise Killifish (Nothobranchius furzeri). J. Vis. Exp. 134:e57073. doi: 10.3791/57073
Duran-Ortiz, S., List, E. O., Basu, R., and Kopchick, J. J. (2021). Extending lifespan by modulating the growth hormone/insulin-like growth factor-1 axis: coming of age. Pituitary 1–19. doi: 10.1007/s11102-020-01117-0 [Epub ahead of print].
Durkin, A., Fisher, C. R., and Cordes, E. E. (2017). Extreme longevity in a deep-sea vestimentiferan tubeworm and its implications for the evolution of life history strategies. Sci. Nat. 104, 1–7.
Dutta, S., and Sengupta, P. (2016). Men and mice: relating their ages. Life Sci. 152, 244–248. doi: 10.1016/j.lfs.2015.10.025
Dyke, B., Gage, T. B., Mamelka, P. M., Goy, R. W., and Stone, W. H. (1986). A demographic analysis of the wisconsin regional primate center rhesus colony, 1962–1982. Am. J. Primatol. 10, 257–269.
Eatwell, K., and Taylor, N. (2000). Care and handling of the budgerigar. Veter. Nurs. J. 15, 227–230. doi: 10.1080/17415349.2000.11013048
Ebert, T. A., and Southon, J. R. (2003). Red sea urchins (Strongylocentrotus franciscanus) can live over 100 years: confirmation with A-bomb 14carbon. Fishery Bull. Natl. Oceanic Atmos. Admin. 101, 915–922.
Eckhardt, F., Kappeler, P. M., and Kraus, C. (2017). Highly variable lifespan in an annual reptile, Labord’s chameleon (Furcifer labordi). Sci. Rep. 7, 1–5.
Edrey, Y. H., Hanes, M., Pinto, M., Mele, J., and Buffenstein, R. (2011). Successful aging and sustained good health in the naked mole rat: a long-lived mammalian model for biogerontology and biomedical research. ILAR J. 52, 41–53. doi: 10.1093/ilar.52.1.41
Edrey, Y. H., Medina, D. X., Gaczynska, M., Osmulski, P. A., Oddo, S., Caccamo, A., et al. (2013). Amyloid beta and the longest-lived rodent: the naked mole-rat as a model for natural protection from Alzheimer’s disease. Neurobiol. Aging 34, 2352–2360. doi: 10.1016/j.neurobiolaging.2013.03.032
Edrey, Y. H., Oddo, S., Cornelius, C., Caccamo, A., Calabrese, V., and Buffenstein, R. (2014). Oxidative damage and amyloid-β metabolism in brain regions of the longest-lived rodents. J. Neurosci. Res. 92, 195–205. doi: 10.1002/jnr.23320
Evdokimov, A., Kutuzov, M., Petruseva, I., Lukjanchikova, N., Kashina, E., Kolova, E., et al. (2018). Naked mole rat cells display more efficient excision repair than mouse cells. Aging (Albany NY) 10, 1454–1473. doi: 10.18632/aging.101482
Fang, R., Liu, K., Zhao, Y., Li, H., Zhu, D., Du, Y., et al. (2014). Generation of naive induced pluripotent stem cells from rhesus monkey fibroblasts. Cell Stem Cell 15, 488–497.
Fang, X., Seim, I., Huang, Z., Gerashchenko, M. V., Xiong, Z., Turanov, A. A., et al. (2014). Adaptations to a subterranean environment and longevity revealed by the analysis of mole rat genomes. Cell Rep. 8, 1354–1364. doi: 10.1016/j.celrep.2014.07.030
Farkas, J. E., and Monaghan, J. R. (2015). Housing and maintenance of Ambystoma mexicanum, the Mexican axolotl. Methods Mol. Biol. 1290, 27–46. doi: 10.1007/978-1-4939-2495-0_3
Fassiano, A. V., Ortiz, N., and Ríos De Molina, M. C. (2017). Reproductive status, antioxidant defences and lipid peroxidation in Octopus tehuelchus (Cephalopoda: Octopodidae) females. J. Nat. History 51, 2645–2660. doi: 10.1080/00222933.2017.1329460
Fay, R., Schaub, M., Border, J. A., Henderson, I. G., Fahl, G., Feulner, J., et al. (2020). Evidence for senescence in survival but not in reproduction in a short-lived passerine. Ecol. Evol. 10, 5383–5390. doi: 10.1002/ece3.6281
Fei, J.-F., Lou, W. P.-K., Knapp, D., Murawala, P., Gerber, T., Taniguchi, Y., et al. (2018). Application and optimization of CRISPR-Cas9-mediated genome engineering in axolotl (Ambystoma mexicanum). Nat. Protoc. 13, 2908–2943. doi: 10.1038/s41596-018-0071-0
Fenchel, T. (1974). Intrinsic rate of natural increase: the relationship with body size. Oecologia 14, 317–326. doi: 10.1007/BF00384576
Finch, C. E. (1994). Longevity, Senescence, and the Genome. Chicago, IL: University of Chicago Press.
Fincher, C. T., Wurtzel, O., de Hoog, T., Kravarik, K. M., and Reddien, P. W. (2018). Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 360:eaaq1736. doi: 10.1126/science.aaq1736
Finseth, F. R., and Harrison, R. G. (2014). A comparison of next generation sequencing technologies for transcriptome assembly and utility for RNA-Seq in a non-model bird. PLoS One 9:e108550. doi: 10.1371/journal.pone.0108550
Folch, J., Busquets, O., Ettcheto, M., Sánchez-López, E., Pallàs, M., Beas-Zarate, C., et al. (2018). Experimental models for aging and their potential for novel drug discovery. Curr. Neuropharmacol. 16, 1466–1483.
Foley, N. M., Hughes, G. M., Huang, Z., Clarke, M., Jebb, D., Whelan, C. V., et al. (2018). Growing old, yet staying young: The role of telomeres in bats’ exceptional longevity. Sci. Adv. 4:eaao0926. doi: 10.1126/sciadv.aao0926
Foley, N. M., Petit, E. J., Brazier, T., Finarelli, J. A., Hughes, G. M., Touzalin, F., et al. (2020). Drivers of longitudinal telomere dynamics in a long-lived bat species, Myotis myotis. Mol. Ecol. 29, 2963–2977. doi: 10.1111/mec.15395
Fontana, L., Partridge, L., and Longo, V. D. (2010). Extending healthy life span—from yeast to humans. Science 328, 321–326. doi: 10.1126/science.1172539
Franceschi, C., Garagnani, P., Parini, P., Giuliani, C., and Santoro, A. (2018). Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 14, 576–590. doi: 10.1038/s41574-018-0059-4
Francis, N., Gregg, T., Owen, R., Ebert, T., and Bodnar, A. (2006). Lack of age-associated telomere shortening in long- and short-lived species of sea urchins. FEBS Lett. 580, 4713–4717. doi: 10.1016/j.febslet.2006.07.049
Fumagalli, M. R., Font-Clos, F., Milan, S., Zapperi, S., and La Porta, C. A. M. (2020). Comparative analysis of metabolic and transcriptomic features of Nothobranchius furzeri. J. R. Soc. Interface 17:20200217. doi: 10.1098/rsif.2020.0217
Furness, L. J., and Speakman, J. R. (2008). Energetics and longevity in birds. AGE 30, 75–87. doi: 10.1007/s11357-008-9054-3
Fushan, A. A., Turanov, A. A., Lee, S. G., Kim, E. B., Lobanov, A. V., Yim, S. H., et al. (2015). Gene expression defines natural changes in mammalian lifespan. Aging Cell 14, 352–365.
Gagneux, P., Moore, J. J., and Varki, A. (2005). The ethics of research on great apes. Nature 437, 27–29.
Gee, L., Hartig, J., Law, L., Wittlieb, J., Khalturin, K., Bosch, T. C. G., et al. (2010). beta-catenin plays a central role in setting up the head organizer in hydra. Dev. Biol. 340, 116–124. doi: 10.1016/j.ydbio.2009.12.036
Genade, T., Benedetti, M., Terzibasi, E., Roncaglia, P., Valenzano, D. R., Cattaneo, A., et al. (2005). Annual fishes of the genus Nothobranchius as a model system for aging research. Aging Cell 4, 223–233. doi: 10.1111/j.1474-9726.2005.00165.x
George, J. C., Bada, J., Zeh, J., Scott, L., Brown, S. E., O’Hara, T., et al. (1999). Age and growth estimates of bowhead whales (Balaena mysticetus) via aspartic acid racemization. Can. J. Zool. 77, 571–580. doi: 10.1139/z99-015
George, J. C., and Bockstoce, J. R. (2008). Two historical weapon fragments as an aid to estimating the longevity and movements of bowhead whales. Polar Biol 31, 751–754. doi: 10.1007/s00300-008-0407-2
Gibbs, R. A., and Pachter, L. (2004). Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428, 493–521.
Gibbs, R. A., Rogers, J., Katze, M. G., Bumgarner, R., Weinstock, G. M., Mardis, E. R., et al. (2007). Evolutionary and biomedical insights from the rhesus macaque genome. Science 316, 222–234.
Giménez, F. A., and García, B. G. (2002). Growth and food intake models in Octopus vulgaris Cuvier (1797): influence of body weight, temperature, sex and diet. Aqu. Int. 10, 361–377.
Giraldo, Y. M., and Traniello, J. F. A. (2014). Worker senescence and the sociobiology of aging in ants. Behav. Ecol. Sociobiol. 68, 1901–1919. doi: 10.1007/s00265-014-1826-4
Glöckner, G., Heinze, I., Platzer, M., Held, C., and Abele, D. (2013). The mitochondrial genome of Arctica islandica; Phylogeny and variation. PLoS One 8:e82857. doi: 10.1371/journal.pone.0082857
Gorbunova, V., Hine, C., Tian, X., Ablaeva, J., Gudkov, A. V., Nevo, E., et al. (2012). Cancer resistance in the blind mole rat is mediated by concerted necrotic cell death mechanism. Proc. Natl. Acad. Sci. U.S.A. 109, 19392–19396. doi: 10.1073/pnas.1217211109
Graveley, B. R., Brooks, A. N., Carlson, J. W., Duff, M. O., Landolin, J. M., Yang, L., et al. (2011). The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479. doi: 10.1038/nature09715
Gredar, T., and Bizjak Mali, L. (2017). Cultivation and morphology of blood cells of the olm Proteus anguinus. Cytogenet. Genome Res 148, 305–313.
Grimes, K. M., Lindsey, M. L., Gelfond, J. A. L., and Buffenstein, R. (2012). Getting to the heart of the matter: age-related changes in diastolic heart function in the longest-lived rodent, the naked mole rat. J. Gerontol. A Biol. Sci. Med. Sci. 67, 384–394. doi: 10.1093/gerona/glr222
Grohme, M. A., Schloissnig, S., Rozanski, A., Pippel, M., Young, G. R., Winkler, S., et al. (2018). The genome of Schmidtea mediterranea and the evolution of core cellular mechanisms. Nature 554, 56–61. doi: 10.1038/nature25473
Groothuis, T. G. G., and von Engelhardt, N. (2005). Investigating maternal hormones in avian eggs: measurement, manipulation, and interpretation. Ann. N. Y. Acad. Sci. 1046, 168–180. doi: 10.1196/annals.1343.014
Gruber, H., Schaible, R., Ridgway, I. D., Chow, T. T., Held, C., and Philipp, E. E. (2014). Telomere-independent ageing in the longest-lived non-colonial animal, Arctica islandica. Exp. Gerontol. 51, 38–45. doi: 10.1016/j.exger.2013.12.014
Gruber, H., Wessels, W., Boynton, P., Xu, J., Wohlgemuth, S., Leeuwenburgh, C., et al. (2015). Age-related cellular changes in the long-lived bivalve A. islandica. Age (Dordr) 37:90. doi: 10.1007/s11357-015-9831-8
Hadi, F., Kulaberoglu, Y., Lazarus, K. A., Bach, K., Ugur, R., Beattie, P., et al. (2020). Transformation of naked mole-rat cells. Nature 583, E1–E7. doi: 10.1038/s41586-020-2410-x
Hakeem, A., Sandoval, R. G., Jones, M., and Allman, J. (1996). “Brain and life span in primates,” in Handbook of the Psychology of Aging, 4th Edn, eds J. E. Birren and K. W. Schaie (San Diego, CA: Academic Press), 78–104.
Hansen, M., Flatt, T., and Aguilaniu, H. (2014). Reproduction, fat metabolism, and life span: what is the connection? Cell Metab. 19:1066. doi: 10.1016/j.cmet.2014.05.017
Harel, I., Valenzano, D. R., and Brunet, A. (2016). Efficient genome engineering approaches for the short-lived African turquoise killifish. Nat. Protoc. 11, 2010–2028. doi: 10.1038/nprot.2016.103
Harman, D. (2001). Aging: overview. Ann. N. Y. Acad. Sci. 928, 1–21. doi: 10.1111/j.1749-6632.2001.tb05631.x
Harper, J. M., Salmon, A. B., Leiser, S. F., Galecki, A. T., and Miller, R. A. (2007). Skin-derived fibroblasts from long-lived species are resistant to some, but not all, lethal stresses and to the mitochondrial inhibitor rotenone. Aging Cell 6, 1–13. doi: 10.1111/j.1474-9726.2006.00255.x
Harr, B., Karakoc, E., Neme, R., Teschke, M., Pfeifle, C., and Pezer, Ž, et al. (2016). Genomic resources for wild populations of the house mouse, Mus musculus and its close relative Mus spretus. Sci. Data 3:160075. doi: 10.1038/sdata.2016.75
Harrison, G. J., and Lightfoot, T. L. (2006). Clinical Avian Medicine (Vol. 1 and 2), ed. L. R. Harrison (Palm Beach, FL: Spix publishing).
Haussmann, M. F., Winkler, D. W., Huntington, C. E., Nisbet, I. C. T., and Vleck, C. M. (2004). Telomerase expression is differentially regulated in birds of differing life span. Ann. N. Y. Acad. Sci. 1019, 186–190. doi: 10.1196/annals.1297.029
Hearn, J. P. (1983). “The common marmoset (Callithrix jacchus),” in Reproduction in New World Primates, ed J. P. Hearn (Dordrecht: Springer), 181–215.
Heflin, L. E., Gibbs, V. K., Powell, M. L., Makowsky, R., Lawrence, J. M., Lawrence, A. L., et al. (2012). Effect of dietary protein and carbohydrate levels on weight gain and gonad production in the sea urchin Lytechinus variegatus. Aquaculture 35, 253–261. doi: 10.1016/j.aquaculture.2012.06.009
Heid, J., Cencioni, C., Ripa, R., Baumgart, M., Atlante, S., Milano, G., et al. (2017). Age-dependent increase of oxidative stress regulates microRNA-29 family preserving cardiac health. Sci. Rep. 7:16839. doi: 10.1038/s41598-017-16829-w
Heinze, I., Bens, M., Calzia, E., Holtze, S., Dakhovnik, O., Sahm, A., et al. (2018). Species comparison of liver proteomes reveals links to naked mole-rat longevity and human aging. BMC Biol. 16: 82. doi: 10.1186/s12915-018-0547-y
Heuer, E., Rosen, R. F., Cintron, A., and Walker, L. C. (2012). Nonhuman primate models of Alzheimer-like cerebral proteopathy. Curr. Pharma. Design 18, 1159–1169.
Hilton, H. G., Rubinstein, N. D., Janki, P., Ireland, A. T., Bernstein, N., Fong, N. L., et al. (2019). Single-cell transcriptomics of the naked mole-rat reveals unexpected features of mammalian immunity. PLoS Biol. 17:e3000528. doi: 10.1371/journal.pbio.3000528
Hochberg, M. E., Noble, R. J., and Braude, S. (2016). A hypothesis to explain cancers in confined colonies of naked mole rats. bioRxiv [Preprint]. doi: 10.1101/079012
Hogan, J. D., Keenan, J. L., Luo, L., Ibn-Salem, J., Lamba, A., Schatzberg, D., et al. (2020). The developmental transcriptome for Lytechinus variegatus exhibits temporally punctuated gene expression changes. Dev. Biol. 139–154. doi: 10.1016/j.ydbio.2019.12.002
Holm, S., Davis, R. B., Javoiš, J., Õunap, E., Kaasik, A., Molleman, F., et al. (2016). A comparative perspective on longevity: the effect of body size dominates over ecology in moths. J. Evol. Biol. 29, 2422–2435. doi: 10.1111/jeb.12966
Holmes, D. J., and Ottinger, M. A. (2003). Birds as long-lived animal models for the study of aging. Experimental Gerontology 38, 1365–1375. doi: 10.1016/j.exger.2003.10.018
Holtze, S., Eldarov, C. M., Vays, V. B., Vangeli, I. M., Vysokikh, M. Y., Bakeeva, L. E., et al. (2016). Study of age-dependent structural and functional changes of mitochondria in skeletal muscles and heart of naked mole rats (Heterocephalus glaber). Biochemistry (Mosc) 81, 1429–1437. doi: 10.1134/S000629791612004X
Holtze, S., Lukaè, M., Cizelj, I., Mutschmann, F., Szentiks, C. A., Jeliæ, D., et al. (2017). Monitoring health and reproductive status of olms (Proteus anguinus) by ultrasound. PLoS One 12:e0182209. doi: 10.1371/journal.pone.0182209
Horvath, S., Zhang, Y., Langfelder, P., Kahn, R. S., Boks, M. P., van Eijk, K., et al. (2012). Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 13, 1–18.
Houtkooper, R. H., Mouchiroud, L., Ryu, D., Moullan, N., Katsyuba, E., Knott, G., et al. (2013). Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 497, 451–457. doi: 10.1038/nature12188
Hoving, H. J. T., and Robison, B. H. (2017). The pace of life in deep-dwelling squids. Deep Sea Res. Part I Oceanogr. Res. Papers 126, 40–49.
Huang, Z., Whelan, C. V., Foley, N. M., Jebb, D., Touzalin, F., Petit, E. J., et al. (2019). Longitudinal comparative transcriptomics reveals unique mechanisms underlying extended healthspan in bats. Nat. Ecol. Evol. 3, 1110–1120. doi: 10.1038/s41559-019-0913-3
Huffard, C. L. (2013). Cephalopod neurobiology: an introduction for biologists working in other model systems. Invert. Neurosci. 13, 11–18.
Hughes, G. M., Leech, J., Puechmaille, S. J., Lopez, J. V., and Teeling, E. C. (2018). Is there a link between aging and microbiome diversity in exceptional mammalian longevity? PeerJ 6:e4174. doi: 10.7717/peerj.4174
Hulbert, A. J., Faulks, S. C., and Buffenstein, R. (2006). Oxidation-resistant membrane phospholipids can explain longevity differences among the longest-living rodents and similarly-sized mice. J. Gerontol. A Biol. Sci. Med. Sci. 61, 1009–1018. doi: 10.1093/gerona/61.10.1009
Huss, D., Poynter, G., and Lansford, R. (2008). Japanese quail (Coturnix japonica) as a laboratory animal model. Lab. Anim. (NY) 37, 513–519. doi: 10.1038/laban1108-513
Hyde, J. R., and Vetter, R. D. (2007). The origin, evolution, and diversification of rockfishes of the genus Sebastes (Cuvier). Mol. Phylogenet. Evol. 790–811. doi: 10.1016/j.ympev.2006.12.026
Ifuta, N. B., Gevaerts, H., and Kühn, E. R. (1988). Thyroid hormones, testosterone, and estradiol-17β in plasma of Epomops franqueti (Tomes, 1860) (Chiroptera) in the rain forest of the equator. Gen. Comp. Endocrinol. 69, 378–380. doi: 10.1016/0016-6480(88)90028-7
Iglesias, J., Otero, J. J., Moxica, C., Fuentes, L., and Sánchez, F. J. (2004). The completed life cycle of the octopus (Octopus vulgaris, Cuvier) under culture conditions: paralarval rearing using Artemia and zoeae, and first data on juvenile growth up to 8 months of age. Aqua. Int. 12, 481–487.
IUCN (2020). The IUCN Red List of Threatened Species. Version 2020-2. Available online at: https://www.iucnredlist.org (accessed October 10, 2020).
Jack, K. M., Schoof, V. A., Sheller, C. R., Rich, C. I., Klingelhofer, P. P., Ziegler, T. E., et al. (2014). Hormonal correlates of male life history stages in wild white-faced capuchin monkeys (Cebus capucinus). Gen. Comp. Endocrinol. 195, 58–67.
James, P. (2007). The Effects of Environmental Factors and Husbandry Techniques on Roe Enhancement of the New Zealand Sea Urchin, Evechinus chloroticus. Wellington: Victoria University of Wellington.
Jarvis, J. U. M., and Bennett, N. C. (1993). Eusociality has evolved independently in 2 genera of bathyergid mole-rats - but occurs in no other subterranean mammal. Behav. Ecol. Sociobiol. 33, 253–260.
Jebb, D., Foley, N. M., Whelan, C. V., Touzalin, F., Puechmaille, S. J., and Teeling, E. C. (2018). Population level mitogenomics of long-lived bats reveals dynamic heteroplasmy and challenges the free radical theory of ageing. Sci. Rep. 8:13634. doi: 10.1038/s41598-018-31093-2
Jebb, D., Huang, Z., Pippel, M., Hughes, G. M., Lavrichenko, K., Devanna, P., et al. (2020). Six reference-quality genomes reveal evolution of bat adaptations. Nature 583, 578–584. doi: 10.1038/s41586-020-2486-3
Ji, X., Li, P., Fuscoe, J. C., Chen, G., Xiao, W., Shi, L., et al. (2020). A comprehensive rat transcriptome built from large scale RNA-seq-based annotation. Nucleic Acids Res. 48, 8320–8331. doi: 10.1093/nar/gkaa638
Jiang, Y.-Z., Liu, Y.-R., Xu, X.-E., Jin, X., Hu, X., Yu, K.-D., et al. (2016). Transcriptome analysis of triple-negative breast cancer reveals an integrated mRNA-lncRNA signature with predictive and prognostic value. Cancer Res. 76, 2105–2114. doi: 10.1158/0008-5472.CAN-15-3284
Jiménez-Ortega, D., Kolm, N., Immler, S., Maklakov, A. A., and Gonzalez-Voyer, A. (2020). Long life evolves in large-brained bird lineages. Evolution 74, 2617–2628. doi: 10.1111/evo.14087
Johnson, A. A., Shokhirev, M. N., and Shoshitaishvili, B. (2019). Revamping the evolutionary theories of aging. Ageing Res. Rev. 55:100947. doi: 10.1016/j.arr.2019.100947
Johnson, J. H. (2004). Husbandry and medicine of aquatic reptiles. Semin. Avian Exotic Pet. Med. 13, 223–228. doi: 10.1053/j.saep.2004.04.008
Jones, O. R., Scheuerlein, A., Salguero-Gómez, R., Camarda, C. G., Schaible, R., Casper, B. B., et al. (2014). Diversity of ageing across the tree of life. Nature 505, 169–173. doi: 10.1038/nature12789
Joven, A., Elewa, A., and Simon, A. (2019). Model systems for regeneration: salamanders. Development 146:dev167700. doi: 10.1242/dev.167700
Junnila, R. K., List, E. O., Berryman, D. E., Murrey, J. W., and Kopchick, J. J. (2013). The GH/IGF-1 axis in ageing and longevity. Nat. Rev. Endocrinol. 9, 366–376. doi: 10.1038/nrendo.2013.67
Kacprzyk, J., Hughes, G. M., Palsson-McDermott, E. M., Quinn, S. R., Puechmaille, S. J., O’Neill, L. A. J., et al. (2017). A potent anti-inflammatory response in bat macrophages may be linked to extended longevity and viral tolerance. Museum Institute Zool. Polish Acad. Sci. 19, 219–228.
Kaletsky, R., Yao, V., Williams, A., Runnels, A. M., Tadych, A., Zhou, S., et al. (2018). Transcriptome analysis of adult Caenorhabditis elegans cells reveals tissue-specific gene and isoform expression. PLoS Genet. 14:e1007559. doi: 10.1371/journal.pgen.1007559
Kamara, D., Geng, T., Xu, J., Guynn, S., Hopwood, K., and Smith, E. J. (2007). Primer note: isolation and characterization of microsatellite markers from the budgerigar, Melopsittacus undulatus. Mol. Ecol. Notes 7, 507–509. doi: 10.1111/j.1471-8286.2006.01636.x
Kammerer, C. M., and Young, S. S. Y. (1983). Directional selection for body weight in constant and variable environments in Drosophila melanogaster. Heredity 50, 237–251. doi: 10.1038/hdy.1983.27
Karsten, K. B., Andriamandimbiarisoa, L. N., Fox, S. F., and Raxworthy, C. J. (2008). A unique life history among tetrapods: an annual chameleon living mostly as an egg. Proc. Natl. Acad. Sci. U.S.A. 105, 8980–8984.
Kawahara-Miki, R., Sano, S., Nunome, M., Shimmura, T., Kuwayama, T., Takahashi, S., et al. (2013). Next-generation sequencing reveals genomic features in the Japanese quail. Genomics 345–353. doi: 10.1016/j.ygeno.2013.03.006
Keane, M., Craig, T., Alföldi, J., Berlin, A. M., Johnson, J., Seluanov, A., et al. (2014). The naked mole rat genome resource: facilitating analyses of cancer and longevity-related adaptations. Bioinformatics 30, 3558–3560. doi: 10.1093/bioinformatics/btu579
Keane, M., Semeiks, J., Webb, A. E., Li, Y. I., Quesada, V., Craig, T., et al. (2015). Insights into the evolution of longevity from the bowhead whale genome. Cell Rep. 10, 112–122. doi: 10.1016/j.celrep.2014.12.008
Keil, G., Cummings, E., and de Magalhães, J. P. (2015). Being cool: how body temperature influences ageing and longevity. Biogerontology 16, 383–397. doi: 10.1007/s10522-015-9571-2
Kelmer Sacramento, E., Kirkpatrick, J. M., Mazzetto, M., Baumgart, M., Bartolome, A., Di Sanzo, S., et al. (2020). Reduced proteasome activity in the aging brain results in ribosome stoichiometry loss and aggregation. Mol. Syst. Biol. 16:e9596. doi: 10.15252/msb.20209596
Khattak, S., Tanaka, E. M. (2015). Transgenesis in axolotl (Ambystoma mexicanum). Methods Mol. Biol. 1290, 269–277. doi: 10.1007/978-1-4939-2495-0_21
Khoo, M. L., Das, S. K., and Ghaffar, M. A. (2018). Growth pattern, diet and reproductive biology of the clownfish Amphiprion ocellaris in waters of Pulau Tioman, Malaysia. Egyp. J. Aquatic Res. 44, 233–239. doi: 10.1016/j.ejar.2018.07.003
Kim, B. M., Kang, S., Ahn, D. H., Jung, S. H., Rhee, H., Yoo, J. S., et al. (2018). The genome of common long-arm octopus Octopus minor. Gigascience 7:119. doi: 10.1093/gigascience/giy119
Kim, E. B., Fang, X., Fushan, A. A., Huang, Z., Lobanov, A. V., Han, L., et al. (2011). Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 479, 223–227. doi: 10.1038/nature10533
Kitsoulis, C. V., Baxevanis, A. D., and Abatzopoulos, T. J. (2020). The occurrence of cancer in vertebrates: a mini review. J. Biol. Res. (Thessalon) 27:9. doi: 10.1186/s40709-020-00119-0
Klapper, W., Kühne, K., Singh, K. K., Heidorn, K., Parwaresch, R., and Krupp, G. (1998). Longevity of lobsters is linked to ubiquitous telomerase expression. FEBS Lett. 439, 143–146.
Klimovich, A., Rehm, A., Wittlieb, J., Herbst, E.-M., Benavente, R., and Bosch, T. C. G. (2018). Non-senescent Hydra tolerates severe disturbances in the nuclear lamina. Aging (Albany NY) 10, 951–972. doi: 10.18632/aging.101440
Koopman, H. N., Westgate, A. J., and Siders, Z. A. (2015). Declining fecundity and factors affecting embryo quality in the American lobster (Homarus americanus) from the Bay of Fundy. Can. J. Fish. Aquatic Sci. 72, 352–363.
Kott, O., Nìmec, P., Fremlová, A., Mazoch, V., and Šumbera, R. (2016). Behavioural tests reveal severe visual deficits in the strictly Subterranean African mole-rats (Bathyergidae) but efficient vision in the fossorial rodent coruro (Spalacopus cyanus, Octodontidae). Ethology 122, 682–694. doi: 10.1111/eth.12515
Krivoruchko, A., and Storey, K. B. (2010). Molecular mechanisms of turtle anoxia tolerance: a role for NF-kappaB. Gene 450, 63–69. doi: 10.1016/j.gene.2009.10.005
Künstner, A., Wolf, J. B. W., Backström, N., Whitney, O., Balakrishnan, C. N., Day, L., et al. (2010). Comparative genomics based on massive parallel transcriptome sequencing reveals patterns of substitution and selection across 10 bird species. Mol. Ecol. 19(Suppl. 1), 266–276. doi: 10.1111/j.1365-294X.2009.04487.x
Kuo, T.-H., and Whited, J. L. (2015). Pseudotyped retroviruses for infecting axolotl. Methods Mol. Biol. 1290, 127–140. doi: 10.1007/978-1-4939-2495-0_10
Labinskyy, N., Csiszar, A., Orosz, Z., Smith, K., Rivera, A., Buffenstein, R., et al. (2006). Comparison of endothelial function, O2-∗ and H2O2 production, and vascular oxidative stress resistance between the longest-living rodent, the naked mole rat, and mice. Am. J. Physiol. Heart Circ. Physiol. 291, H2698–H2704. doi: 10.1152/ajpheart.00534.2006
Laing, E. D., Sterling, S. L., Weir, D. L., Beauregard, C. R., Smith, I. L., Larsen, S. E., et al. (2019). Enhanced autophagy contributes to reduced viral infection in black flying fox cells. Viruses 11:260. doi: 10.3390/v11030260
Lambert, A. J., Boysen, H. M., Buckingham, J. A., Yang, T., Podlutsky, A., Austad, S. N., et al. (2007). Low rates of hydrogen peroxide production by isolated heart mitochondria associate with long maximum lifespan in vertebrate homeotherms. Aging Cell 6, 607–618. doi: 10.1111/j.1474-9726.2007.00312.x
Lapierre, L. R., and Hansen, M. (2012). Lessons from C. elegans: signaling pathways for longevity. Trends Endocrinol. Metab. 23, 637–644. doi: 10.1016/j.tem.2012.07.007
Laplante, M., and Sabatini, D. M. (2012). mTOR signaling in growth control and disease. Cell 149, 274–293. doi: 10.1016/j.cell.2012.03.017
Larson, J., and Park, T. J. (2009). Extreme hypoxia tolerance of naked mole-rat brain. Neuroreport 20, 1634–1637. doi: 10.1097/WNR.0b013e32833370cf
Layne, D. G., and Power, R. A. (2003). Husbandry, handling, and nutrition for marmosets. Comp. Med. 53, 351–359.
Lee, B. P., Smith, M., Buffenstein, R., and Harries, L. W. (2020). Negligible senescence in naked mole rats may be a consequence of well-maintained splicing regulation. Geroscience 42, 633–651. doi: 10.1007/s11357-019-00150-7
Leigh, N. D., Dunlap, G. S., Johnson, K., Mariano, R., Oshiro, R., Wong, A. Y., et al. (2018). Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single-cell resolution. Nat. Commun. 9:5153. doi: 10.1038/s41467-018-07604-0
Lenhoff, H. M., and Brown, R. D. (1970). Mass culture of hydra: an improved method and its application to other aquatic invertebrates. Lab. Anim. 4, 139–154. doi: 10.1258/002367770781036463
Lewis, K. N., Wason, E., Edrey, Y. H., Kristan, D. M., Nevo, E., and Buffenstein, R. (2015). Regulation of Nrf2 signaling and longevity in naturally long-lived rodents. Proc. Natl. Acad. Sci. U.S.A. 112, 3722–3727. doi: 10.1073/pnas.1417566112
Liu, H., Zhu, F., Yong, J., Zhang, P., Hou, P., Li, H., et al. (2008). Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell 3, 587–590.
Liu, S.-Y., Selck, C., Friedrich, B., Lutz, R., Vila-Farré, M., Dahl, A., et al. (2013). Reactivating head regrowth in a regeneration-deficient planarian species. Nature 500, 81–84. doi: 10.1038/nature12414
Longo, V. D., Antebi, A., Bartke, A., Barzilai, N., Brown-Borg, H. M., Caruso, C., et al. (2015). Interventions to slow aging in humans: are we ready? Aging Cell 14, 497–510. doi: 10.1111/acel.12338
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M., and Kroemer, G. (2013). The hallmarks of aging. Cell 153, 1194–1217. doi: 10.1016/j.cell.2013.05.039
Love, M. S., Yoklavich, M., and Thorsteinson, L. K. (2002). The Rockfishes of the Northeast Pacific. Berkeley, CA: University of California Press.
Lutz, P. L., and Prentice, H. M. (2002). Sensing and responding to hypoxia, molecular and physiological mechanisms. Integr. Comp. Biol. 42, 463–468. doi: 10.1093/icb/42.3.463
Lutz, P. L., Prentice, H. M., and Milton, S. L. (2003). Is turtle longevity linked to enhanced mechanisms for surviving brain anoxia and reoxygenation? Exp. Gerontol. 38, 797–800. doi: 10.1016/s0531-5565(03)00111-6
Lutz, R. A., Goodsell, J. G., Mann, R., and Castagna, M. (1981). Experimental culture of the ocean quahog, Arctica islandica. J. World Maricult. Soc. 12, 196–205. doi: 10.1111/j.1749-7345.1981.tb00255.x
Ma, S., and Gladyshev, V. N. (2017). Molecular signatures of longevity: Insights from cross-species comparative studies. Semin. Cell Dev. Biol. 70, 190–203. doi: 10.1016/j.semcdb.2017.08.007
MacNeil, M. A., McMeans, B. C., Hussey, N. E., Vecsei, P., Svavarsson, J., Kovacs, K. M., et al. (2012). Biology of the Greenland shark Somniosus microcephalus. J. Fish. Biol. 80, 991–1018. doi: 10.1111/j.1095-8649.2012.03257.x
MacRae, S. L., Croken, M. M., Calder, R. B., Aliper, A., Milholland, B., White, R. R., et al. (2015). DNA repair in species with extreme lifespan differences. Aging (Albany NY) 7, 1171–1184. doi: 10.18632/aging.100866
Magness, C. L., Fellin, P. C., Thomas, M. J., Korth, M. J., Agy, M. B., Proll, S. C., et al. (2005). Analysis of the Macaca mulatta transcriptome and the sequence divergence between Macaca and human. Genome Biol. 6, 1–16.
Malde, K., Seliussen, B. B., Quintela, M., Dahle, G., Besnier, F., Skaug, H. J., et al. (2017). Whole genome resequencing reveals diagnostic markers for investigating global migration and hybridization between minke whale species. BMC Genomics 18:76. doi: 10.1186/s12864-016-3416-5
Maldonado, E., Rangel-Huerta, E., González-Gómez, R., Fajardo-Alvarado, G., and Morillo-Velarde, P. S. (2019). Octopus insularis as a new marine model for evolutionary developmental biology. Biol. Open 8:bio046086. doi: 10.1242/bio.046086
Malinowski, P. T., Cochet-Escartin, O., Kaj, K. J., Ronan, E., Groisman, A., Diamond, P. H., et al. (2017). Mechanics dictate where and how freshwater planarians fission. Proc. Natl. Acad. Sci. U.S.A. 114, 10888–10893. doi: 10.1073/pnas.1700762114
Mallory, M. L., Hatch, S. A., and Nettleship, D. N. (2012). Northern Fulmar (Fulmarus glacialis). Birds N. Am. doi: 10.2173/bna.361
Manshack, L. K., Conard, C. M., Bryan, S. J., Deem, S. L., Holliday, D. K., Bivens, N. J., et al. (2017). Transcriptomic alterations in the brain of painted turtles (Chrysemys picta) developmentally exposed to bisphenol A or ethinyl estradiol. Physiol. Genomics 49, 201–215. doi: 10.1152/physiolgenomics.00103.2016
Marasco, V., Herzyk, P., Robinson, J., and Spencer, K. A. (2016). Pre- and post-natal stress programming: developmental exposure to glucocorticoids causes long-term brain-region specific changes to transcriptome in the precocial Japanese quail. J. Neuroendocrinol. 28:10.1111/jne.12387. doi: 10.1111/jne.12387
Marmoset Genome Sequencing and Analysis Consortium (2014). The common marmoset genome provides insight into primate biology and evolution. Nat. Genet. 46:850.
Martínez, D. E. (1998). Mortality patterns suggest lack of senescence in hydra. Exp. Gerontol. 33, 217–225. doi: 10.1016/s0531-5565(97)00113-7
Martins, R., Lithgow, G. J., and Link, W. (2016). Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell 15, 196–207. doi: 10.1111/acel.12427
McBride, S. C., Pinnix, W. D., Lawrence, J. M., Lawrence, A. L., and Mulligan, T. M. (1997). The effect of temperature on production of gonads by the sea urchin strongylocentrotus franciscanus fed natural and prepared diets. J. World Aqua. Soc. 28, 357–365. doi: 10.1111/j.1749-7345.1997.tb00282.x
McCusker, C., and Gardiner, D. M. (2011). The axolotl model for regeneration and aging research: a mini-review. Gerontology 57, 565–571. doi: 10.1159/000323761
McWilliams, D. (2008). Nutritional considerations for captive Charadriiformes (shorebirds, gulls and alcids). CAB Rev. 3, 1–8. doi: 10.1079/pavsnnr20083028
Merryman, M. S., Alvarado, A. S., and Jenkin, J. C. (2018). Culturing Planarians in the Laboratory. Methods Mol. Biol. 1774, 241–258. doi: 10.1007/978-1-4939-7802-1_5
Miller, R. A., Harper, J. M., Dysko, R. C., Durkee, S. J., and Austad, S. N. (2002). Longer life spans and delayed maturation in wild-derived mice. Exp. Biol. Med. 227, 500–508.
Miller, R. A., and Nadon, N. L. (2000). Principles of animal use for gerontological research. J. Gerontol. A Biol. Sci. Med. Sci. 55, B117–B123. doi: 10.1093/gerona/55.3.b117
Mitchell, A. S., Hartig, R., Basso, M. A., Jarrett, W., Kastner, S., and Poirier, C. (2021). International primate neuroscience research regulation, public engagement and transparency opportunities. Neuroimage 117700.
Miwa, S., Beckman, K. B., and Muller, F. L. (2008). Oxidative Stress in Aging: From Model Systems to Human Diseases. New York, NY: Springer.
Morgan, C. C., Mc Cartney, A. M., Donoghue, M. T. A., Loughran, N. B., Spillane, C., Teeling, E. C., et al. (2013). Molecular adaptation of telomere associated genes in mammals. BMC Evol. Biol. 13:251. doi: 10.1186/1471-2148-13-251
Morris, K. M., Hindle, M. M., Boitard, S., Burt, D. W., Danner, A. F., Eory, L., et al. (2020). The quail genome: insights into social behaviour, seasonal biology and infectious disease response. BMC Biol. 18:14.
Moskalev, A. À, Kudryavtseva, A. V., Graphodatsky, A. S., Beklemisheva, V. R., Serdyukova, N. A., Krutovsky, K. V., et al. (2017). De novo assembling and primary analysis of genome and transcriptome of gray whale Eschrichtius robustus. BMC Evol. Biol. 17(Suppl. 2):258. doi: 10.1186/s12862-017-1103-z
Mulec, J. (2020). Welcome to the -omics era of the 21st century: will Proteus anguinus finally reveal all its mysteries? Acta Carsol. 1:49. doi: 10.3986/ac.v49i1.8009
Munro, D., Baldy, C., Pamenter, M. E., and Treberg, J. R. (2019). The exceptional longevity of the naked mole-rat may be explained by mitochondrial antioxidant defenses. Aging Cell 18:e12916. doi: 10.1111/acel.12916
Munro, D., and Blier, P. U. (2012). The extreme longevity of Arctica islandica is associated with increased peroxidation resistance in mitochondrial membranes. Aging Cell 11, 845–855. doi: 10.1111/j.1474-9726.2012.00847.x
Munshi-South, J., and Wilkinson, G. S. (2010). Bats and birds: exceptional longevity despite high metabolic rates. Ageing Res. Rev. 9, 12–19. doi: 10.1016/j.arr.2009.07.006
Murthy, M., and Ram, J. L. (2015). Invertebrates as model organisms for research on aging biology. Invert. Reprod. Dev. 59, 1–4. doi: 10.1080/07924259.2014.970002
Muschiol, D., Schroeder, F., and Traunspurger, W. (2009). Life cycle and population growth rate of Caenorhabditis elegans studied by a new method. BMC Ecol. 9:14. doi: 10.1186/1472-6785-9-14
Nabholz, B., Glémin, S., and Galtier, N. (2008). Strong variations of mitochondrial mutation rate across mammals—the longevity hypothesis. Mol. Biol. Evol. 25, 120–130. doi: 10.1093/molbev/msm248
National Research Council (2010). Guide for the Care and Use of Laboratory Animals. Washington, DC: National Academies Press.
Nielsen, J., Hedeholm, R. B., Heinemeier, J., Bushnell, P. G., Christiansen, J. S., Olsen, J., et al. (2016). Eye lens radiocarbon reveals centuries of longevity in the Greenland shark (Somniosus microcephalus). Science 702–704. doi: 10.1126/science.aaf1703
Nishimura, O., Hirao, Y., Tarui, H., and Agata, K. (2012). Comparative transcriptome analysis between planarian Dugesia japonica and other platyhelminth species. BMC Genomics 13:289. doi: 10.1186/1471-2164-13-289
Nowoshilow, S., Schloissnig, S., Fei, J.-F., Dahl, A., Pang, A. W. C., Pippel, M., et al. (2018). The axolotl genome and the evolution of key tissue formation regulators. Nature 554, 50–55. doi: 10.1038/nature25458
Ogburn, C. E., Carlberg, K., Ottinger, M. A., Holmes, D. J., Martin, G. M., and Austad, S. N. (2001). Exceptional cellular resistance to oxidative damage in long-lived birds requires active gene expression. J. Gerontol. Ser. A 56, B468–B474. doi: 10.1093/gerona/56.11.b468
Olivotto, I., Planas, M., Simões, N., Holt, G. J., Avella, M. A., and Calado, R. (2011). Advances in breeding and rearing marine ornamentals. J. World Aqua. Soc. 42, 135–166. doi: 10.1111/j.1749-7345.2011.00453.x
Orell, M., and Belda, E. J. (2002). Delayed cost of reproduction and senescence in the willow tit Parus montanus. J. Anim. Ecol. 71, 55–64. doi: 10.1046/j.0021-8790.2001.00575.x
Orkin, J. D., Montague, M. J., Tejada-Martinez, D., de Manuel, M., Del Campo, J., Hernandez, S. C., et al. (2021). The genomics of ecological flexibility, large brains, and long lives in capuchin monkeys revealed with fecalFACS. Proc. Natl. Acad. Sci. U.S.A. 118::e2010632118. doi: 10.1073/pnas.2010632118
O’Shea, T. J., Cryan, P. M., Cunningham, A. A., Fooks, A. R., Hayman, D. T. S., Luis, A. D., et al. (2014). Bat flight and zoonotic viruses. Emerg. Infect. Dis. 20, 741–745. doi: 10.3201/eid2005.130539
Oster, M., Reyer, H., Trakooljul, N., Weber, F. M., Xi, L., Muráni, E., et al. (2020). Ileal transcriptome profiles of Japanese quail divergent in phosphorus utilization. Int. J. Mol. Sci. 21:2762. doi: 10.3390/ijms21082762
Oviedo, N. J., Newmark, P. A., and Sánchez Alvarado, A. (2003). Allometric scaling and proportion regulation in the freshwater planarian Schmidtea mediterranea. Dev. Dyn. 226, 326–333. doi: 10.1002/dvdy.10228
Pamenter, M. E., Uysal-Onganer, P., Huynh, K. W., Kraev, I., and Lange, S. (2019). Post-translational deimination of immunological and metabolic protein markers in plasma and extracellular vesicles of naked mole-RAT (Heterocephalus glaber). Int. J. Mol. Sci. 20:5378. doi: 10.3390/ijms20215378
Pamplona, R., Portero-Otín, M., Sanz, A., Ayala, V., Vasileva, E., and Barja, G. (2005). Protein and lipid oxidative damage and complex I content are lower in the brain of budgerigar and canaries than in mice, relation to aging rate. Age (Dordr) 267–280. doi: 10.1007/s11357-005-4562-x
Pan, H., and Finkel, T. (2017). Key proteins and pathways that regulate lifespan. J. Biol. Chem. 292, 6452–6460. doi: 10.1074/jbc.R116.771915
Park, J. Y., An, Y.-R., Kanda, N., An, C.-M., An, H. S., Kang, J.-H., et al. (2015). Cetaceans evolution: insights from the genome sequences of common minke whales. BMC Genomics 16:13. doi: 10.1186/s12864-015-1213-1
Park, T. J., Reznick, J., Peterson, B. L., Blass, G., Omerbašiæ, D., Bennett, N. C., et al. (2017). Fructose-driven glycolysis supports anoxia resistance in the naked mole-rat. Science 356, 307–311. doi: 10.1126/science.aab3896
Pearl, R., and Miner, J. R. (1935). Experimental studies on the duration of life. XIV. The comparative mortality of certain lower organisms. Q. Rev. Biol. 10, 60–79.
Perales-Raya, C., Jurado-Ruzafa, A., Bartolomé, A., Duque, V., Carrasco, M. N., and Fraile-Nuez, E. (2014). Age of spent Octopus vulgaris and stress mark analysis using beaks of wild individuals. Hydrobiologia 725, 105–114.
Petersen, H. O., Höger, S. K., Looso, M., Lengfeld, T., Kuhn, A., Warnken, U., et al. (2015). A comprehensive transcriptomic and proteomic analysis of hydra head regeneration. Mol. Biol. Evol. 32, 1928–1947. doi: 10.1093/molbev/msv079
Peto, R., Roe, F. J., Lee, P. N., Levy, L., and Clack, J. (1975). Cancer and ageing in mice and men. Br. J. Cancer 32, 411–426.
Petry, H. (2003). Zur artgerechten Haltung von afrikanischen Nacktmullen (Heterocephalus glaber). J. Anim. Physiol. Anim. Nutr. 87, 421–432. doi: 10.1046/j.0931-2439.2003.00453.x
Petzold, A., Reichwald, K., Groth, M., Taudien, S., Hartmann, N., Priebe, S., et al. (2013). The transcript catalogue of the short-lived fish Nothobranchius furzeri provides insights into age-dependent changes of mRNA levels. BMC Genomics 14:185. doi: 10.1186/1471-2164-14-185
Philipp, E. E. R., Wessels, W., Gruber, H., Strahl, J., Wagner, A. E., Ernst, I. M. A., et al. (2012). Gene expression and physiological changes of different populations of the long-lived bivalve Arctica islandica under low oxygen conditions. PLoS One 7:e44621. doi: 10.1371/journal.pone.0044621
Piraino, S., Boero, F., Aeschbach, B., and Schmid, V. (1996). Reversing the life cycle: medusae transforming into polyps and cell transdifferentiation in Turritopsis nutricula (Cnidaria, Hydrozoa). Biol. Bull. 190, 302–312. doi: 10.2307/1543022
Planes, S., Jones, G. P., and Thorrold, S. R. (2009). Larval dispersal connects fish populations in a network of marine protected areas. Proc. Natl. Acad. Sci. U.S.A. 106, 5693–5697. doi: 10.1073/pnas.0808007106
Plass, M., Solana, J., Wolf, F. A., Ayoub, S., Misios, A., Glažar, P., et al. (2018). Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 360:eaaq1723. doi: 10.1126/science.aaq1723
Platzer, M., and Englert, C. (2016). Nothobranchius furzeri: a model for aging research and more. Trends Genet. 32, 543–552. doi: 10.1016/j.tig.2016.06.006
Plot, V., Criscuolo, F., Zahn, S., and Georges, J.-Y. (2012). Telomeres, age and reproduction in a long-lived reptile. PLoS One 7:e40855. doi: 10.1371/journal.pone.0040855
Podlutsky, A. J., Khritankov, A. M., Ovodov, N. D., and Austad, S. N. (2005). A new field record for bat longevity. J. Gerontol. A Biol. Sci. Med. Sci. 60, 1366–1368. doi: 10.1093/gerona/60.11.1366
Poeschla, M., and Valenzano, D. R. (2020). The turquoise killifish: a genetically tractable model for the study of aging. J. Exp. Biol. 223(Suppl 1):jeb209296. doi: 10.1242/jeb.209296
Polačik, M., Blažek, R., and Reichard, M. (2016). Laboratory breeding of the short-lived annual killifish Nothobranchius furzeri. Nat. Protoc. 11, 1396–1413. doi: 10.1038/nprot.2016.080
Pride, H., Yu, Z., Sunchu, B., Mochnick, J., Coles, A., Zhang, Y., et al. (2015). Long-lived species have improved proteostasis compared to phylogenetically-related shorter-lived species. Biochem. Biophys. Res. Commun. 457, 669–675. doi: 10.1016/j.bbrc.2015.01.046
Prothero, J., and Jürgens, K. D. (1987). Scaling of maximal lifespan in mammals: a review. Evolution of Longevity in Animals 42, 49–74. doi: 10.1007/978-1-4613-1939-9_4
Pryor, S. H., Hill, R., Dixson, D. L., Fraser, N. J., Kelaher, B. P., and Scott, A. (2020). Anemonefish facilitate bleaching recovery in a host sea anemone. Sci. Rep. 10:18586. doi: 10.1038/s41598-020-75585-6
Quesada, V., Freitas-Rodríguez, S., Miller, J., Pérez-Silva, J. G., Jiang, Z.-F., Tapia, W., et al. (2019). Giant tortoise genomes provide insights into longevity and age-related disease. Nat. Ecol. Evol. 3, 87–95. doi: 10.1038/s41559-018-0733-x
Racey, P. A. (1970). The breeding, care and management of vespertilionid bats in the laboratory. Lab. Anim. 4, 171–183. doi: 10.1258/002367770781071635
Ram, J. L., and Costa, A. J. (eds). (2018). “Invertebrates as model organisms for research on aging biology,” in Conn’s Handbook of Models for Human Aging ([Place of Publication Not Identified], (Cambridge, MA: Elsevier Academic Press), 445–452.
Ramsey, J. J., Laatsch, J. L., and Kemnitz, J. W. (2000). Age and gender differences in body composition, energy expenditure, and glucoregulation of adult rhesus monkeys. J. Med. Primatol. 29, 11–19.
Reddien, P. W. (2018). The cellular and molecular basis for planarian regeneration. Cell 175, 327–345. doi: 10.1016/j.cell.2018.09.021
Reddien, P. W., and Sánchez Alvarado, A. (2004). Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 20, 725–757. doi: 10.1146/annurev.cellbio.20.010403.095114
Reichwald, K., Petzold, A., Koch, P., Downie, B. R., Hartmann, N., Pietsch, S., et al. (2015). Insights into sex chromosome evolution and aging from the genome of a short-lived fish. Cell 163, 1527–1538. doi: 10.1016/j.cell.2015.10.071
Ripa, R., Dolfi, L., Terrigno, M., Pandolfini, L., Savino, A., Arcucci, V., et al. (2017). MicroRNA miR-29 controls a compensatory response to limit neuronal iron accumulation during adult life and aging. BMC Biol. 15:9. doi: 10.1186/s12915-017-0354-x
Robison, B., Seibel, B., and Drazen, J. (2014). Deep-sea octopus (Graneledone boreopacifica) conducts the longest-known egg-brooding period of any animal. PLoS One 9:e103437.
Rodriguez, K. A., Osmulski, P. A., Pierce, A., Weintraub, S. T., Gaczynska, M., and Buffenstein, R. (2014). A cytosolic protein factor from the naked mole-rat activates proteasomes of other species and protects these from inhibition. Biochim. Biophys. Acta 1842, 2060–2072. doi: 10.1016/j.bbadis.2014.07.005
Rodríguez-Domínguez, A., Rosas, C., Méndez-Loeza, I., and Markaida, U. (2013). Validation of growth increments in stylets, beaks and lenses as ageing tools in Octopus maya. J. Exp. Mar. Biol. Ecol. 449, 194–199.
Rojo-Laguna, J. I., Garcia-Cabot, S., and Saló, E. (2019). Tissue transplantation in planarians: A useful tool for molecular analysis of pattern formation. Semin. Cell Dev. Biol. 87, 116–124. doi: 10.1016/j.semcdb.2018.05.022
Ross, C. N., Austad, S., Brasky, K., Brown, C. J., Forney, L. J., Gelfond, J. A., et al. (2017). The development of a specific pathogen free (SPF) barrier colony of marmosets (Callithrix jacchus) for aging research. Aging (Albany NY) 9:2544.
Ross, C., Salmon, A., Strong, R., Fernandez, E., Javors, M., Richardson, A., et al. (2015). Metabolic consequences of long-term rapamycin exposure on common marmoset monkeys (Callithrix jacchus). Aging (Albany NY) 7, 964–973. doi: 10.18632/aging.100843
Rouhana, L., Weiss, J. A., Forsthoefel, D. J., Lee, H., King, R. S., Inoue, T., et al. (2013). RNA interference by feeding in vitro-synthesized double-stranded RNA to planarians: methodology and dynamics. Dev. Dyn. 242, 718–730. doi: 10.1002/dvdy.23950
Roy, B., Zhao, J., Yang, C., Luo, W., Xiong, T., Li, Y., et al. (2018). CRISPR/Cascade 9-mediated genome editing-challenges and opportunities. Front. Genet. 9:240. doi: 10.3389/fgene.2018.00240
Ruby, J. G., Smith, M., and Buffenstein, R. (2018). Naked mole-rat mortality rates defy gompertzian laws by not increasing with age. elife 7:e31157. doi: 10.7554/eLife.31157
Rusyaev, S. M., and Orlov, A. M. (2013). Bycatches of the greenland shark Somniosus microcephalus (Squaliformes, Chondrichthyes) in the barents sea and the adjacent waters under bottom trawling data. J. Ichthyol. 53, 111–115. doi: 10.1134/S0032945213010128
Sahm, A., Almaida-Pagán, P., Bens, M., Mutalipassi, M., Lucas-Sánchez, A., de Costa Ruiz, J., et al. (2019). Analysis of the coding sequences of clownfish reveals molecular convergence in the evolution of lifespan. BMC Evol. Biol. 19:89. doi: 10.1186/s12862-019-1409-0
Sahm, A., Bens, M., Henning, Y., Vole, C., Groth, M., Schwab, M., et al. (2018a). Higher gene expression stability during aging in long-lived giant mole-rats than in short-lived rats. Aging (Albany NY) 10, 3938–3956. doi: 10.18632/aging.101683
Sahm, A., Bens, M., Platzer, M., and Cellerino, A. (2017). Parallel evolution of genes controlling mitonuclear balance in short-lived annual fishes. Aging Cell 16, 488–496. doi: 10.1111/acel.12577
Sahm, A., Bens, M., Szafranski, K., Holtze, S., Groth, M., Görlach, M., et al. (2018b). Long-lived rodents reveal signatures of positive selection in genes associated with lifespan. PLoS Genet. 14:e1007272. doi: 10.1371/journal.pgen.1007272
Sahm, A., Platzer, M., Koch, P., Henning, Y., Bens, M., Groth, M., et al. (2021). Increased longevity due to sexual activity in mole-rats is associated with transcriptional changes in HPA stress axis. eLife 10:e57843.
Sahu, S., Dattani, A., and Aboobaker, A. A. (2017). Secrets from immortal worms: what can we learn about biological ageing from the planarian model system? Semin. Cell Dev. Biol. 70, 108–121. doi: 10.1016/j.semcdb.2017.08.028
Salis, P., Lorin, T., Lewis, V., Rey, C., Marcionetti, A., Escande, M.-L., et al. (2019). Developmental and comparative transcriptomic identification of iridophore contribution to white barring in clownfish. Pigment Cell Melanoma Res. 23, 391–402. doi: 10.1111/pcmr.12766
Salmon, A. B., Leonard, S., Masamsetti, V., Pierce, A., Podlutsky, A. J., Podlutskaya, N., et al. (2009). The long lifespan of two bat species is correlated with resistance to protein oxidation and enhanced protein homeostasis. FASEB J. 23, 2317–2326. doi: 10.1096/fj.08-122523
Sarraude, T., Hsu, B.-Y., Groothuis, T., and Ruuskanen, S. (2020). Testing the short-and long-term effects of elevated prenatal exposure to different forms of thyroid hormones. PeerJ 8:e10175. doi: 10.7717/peerj.10175
Sasaki, E., Suemizu, H., Shimada, A., Hanazawa, K., Oiwa, R., Kamioka, M., et al. (2009). Generation of transgenic non-human primates with germline transmission. Nature 459, 523–527.
Schaible, R., Scheuerlein, A., Dańko, M. J., Gampe, J., Martínez, D. E., and Vaupel, J. W. (2015). Constant mortality and fertility over age in Hydra. Proc. Natl. Acad. Sci. U.S.A. 112, 15701–15706. doi: 10.1073/pnas.1521002112
Schaible, R., Sussman, M., and Kramer, B. H. (2014). Aging and potential for self-renewal: hydra living in the age of aging - a mini-review. Gerontology 60, 548–556. doi: 10.1159/000360397
Schapiro, S. J. (2000). A few new developments in primate housing and husbandry. Scand. J. Lab. Anim. Sci. 27, 103–110.
Scharff, A., Locker-Grütjen, O., Kawalika, M., and Burda, H. (2001). NATURAL HISTORY OF THE GIANT MOLE-RAT, CRYPTOMYS MECHOWI (RODENTIA: BATHYERGIDAE), FROM ZAMBIA. J. Mammal. 82, 1003–1015. doi: 10.1644/1545-15422001082<1003:NHOTGM<2.0.CO;2
Schmidt, C. M., Jarvis, J. U. M., and Bennett, N. C. (2013). The long-lived queen: reproduction and longevity in female eusocial Damaraland mole-rats (Fukomys damarensis). African Zool. 48, 193–196. doi: 10.1080/15627020.2013.11407583
Schwarz, R., Piatkowski, U., and Hoving, H. J. T. (2018). Impact of environmental temperature on the lifespan of octopods. Mar. Ecol. Prog. Ser. 605, 151–164.
Seeker, L. A. (2020). Telomere shortening correlates with harsh weather conditions in the bat species Myotis myotis. Mol. Ecol. 29, 2951–2953. doi: 10.1111/mec.15580
Seim, I., Fang, X., Xiong, Z., Lobanov, A. V., Huang, Z., Ma, S., et al. (2013). Genome analysis reveals insights into physiology and longevity of the Brandt’s bat Myotis brandtii. Nat. Commun. 4:2212. doi: 10.1038/ncomms3212
Seim, I., Ma, S., Zhou, X., Gerashchenko, M. V., Lee, S.-G., Suydam, R., et al. (2014). The transcriptome of the bowhead whale Balaena mysticetus reveals adaptations of the longest-lived mammal. Aging (Albany NY) 6, 879–899. doi: 10.18632/aging.100699
Seluanov, A., Hine, C., Azpurua, J., Feigenson, M., Bozzella, M., Mao, Z., et al. (2009). Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc. Natl. Acad. Sci. U.S.A. 106, 19352–19357.
Semmens, J., Doubleday, Z., Hoyle, K., and Pecl, G. (2011). A multilevel approach to examining cephalopod growth using Octopus pallidus as a model. J. Exp. Biol. 214, 2799–2807.
Sengupta, P. (2013). The laboratory rat: relating its age with human’s. Int. J. Prev. Med. 4, 624–630.
Sergiev, P. V., Artemov, A. A., Prokhortchouk, E. B., Dontsova, O. A., and Berezkin, G. V. (2016). Genomes of Strongylocentrotus franciscanus and Lytechinus variegatus: are there any genomic explanations for the two order of magnitude difference in the lifespan of sea urchins? Aging (Albany NY) 8, 260–271. doi: 10.18632/aging.100889
Shaffer, H. B., Minx, P., Warren, D. E., Shedlock, A. M., Thomson, R. C., Valenzuela, N., et al. (2013). The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol. 14:R28. doi: 10.1186/gb-2013-14-3-r28
Shebzukhov, Y., Holtze, S., Hirseland, H., Schäfer, H., Radbruch, A., Hildebrandt, T., et al. (2019). Identification of cross-reactive antibodies for the detection of lymphocytes, myeloid cells and haematopoietic precursors in the naked mole rat. Eur. J. Immunol. 49, 2103–2110. doi: 10.1002/eji.201948124
Shekhidem, A. H., Sharvit, L., Leman, E., Manov, I., Roichman, A., Holtze, S., et al. (2019). Telomeres and longevity: a cause or an effect? Int. J. Mol. Sci. 20:3233. doi: 10.3390/ijms20133233
Shepard, A., and Kissil, J. L. (2020). The use of non-traditional models in the study of cancer resistance-the case of the naked mole rat. Oncogene 39, 5083–5097. doi: 10.1038/s41388-020-1355-8
Shimizu, H., Sawada, Y., and Sugiyama, T. (1993). Minimum tissue size required for hydra regeneration. Dev. Biol. 155, 287–296. doi: 10.1006/dbio.1993.1028
Shimizu, M., Iwano, S., Uno, Y., Uehara, S., Inoue, T., Murayama, N., et al. (2014). Qualitative de novo analysis of full length cDNA and quantitative analysis of gene expression for common marmoset (Callithrix jacchus) transcriptomes using parallel long-read technology and short-read sequencing. PLoS One 9:e100936.
Shimizu, Y., Suzuki, J., Terao, K., and Ishida, T. (2003). In vitro aging of macaque adherent cells: similar pattern of cellular aging between human and macaque. Mech. Ageing Dev. 124, 237–244.
Siebert, S., Farrell, J. A., Cazet, J. F., Abeykoon, Y., Primack, A. S., Schnitzler, C. E., et al. (2019). Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science 365: eaav9314. doi: 10.1126/science.aav9314
Sikes, J. M., and Newmark, P. A. (2013). Restoration of anterior regeneration in a planarian with limited regenerative ability. Nature 500, 77–80. doi: 10.1038/nature12403
Sillero-Ríos, J., Sureda, A., Capó, X., Oliver-Codorniú, M., and Arechavala-Lopez, P. (2018). Biomarkers of physiological responses of Octopus vulgaris to different coastal environments in the western Mediterranean Sea. Mar. Pollut. Bull. 128, 240–247. doi: 10.1016/j.marpolbul.2018.01.032
Skulachev, V. P. (2012). What is “phenoptosis” and how to fight it? Biochemistry (Mosc) 77, 689–706. doi: 10.1134/S0006297912070012
Skulachev, V. P., Holtze, S., Vyssokikh, M. Y., Bakeeva, L. E., Skulachev, M. V., Markov, A. V., et al. (2017). Neoteny, prolongation of youth: from naked mole rats to “naked apes” (humans). Physiol. Rev. 97, 699–720. doi: 10.1152/physrev.00040.2015
Smith, S. A., Scimeca, J. M., and Mainous, M. E. (2011). Culture and maintenance of selected invertebrates in the laboratory and classroom. ILAR J. 52, 153–164. doi: 10.1093/ilar.52.2.153
Sol, D. (2009). Revisiting the cognitive buffer hypothesis for the evolution of large brains. Biol. Lett. 5, 130–133. doi: 10.1098/rsbl.2008.0621
Stark, G., and Meiri, S. (2018). Cold and dark captivity: drivers of amphibian longevity. Glob. Ecol. Biogeogr. 27, 1384–1397. doi: 10.1111/geb.12804
Stark, G., Tamar, K., Itescu, Y., Feldman, A., and Meiri, S. (2018). Cold and isolated ectotherms: drivers of reptilian longevity. Biol. J. Linn. Soc. 125, 730–740. doi: 10.1093/biolinnean/bly153
Ste-Marie, E., Watanabe, Y. Y., Semmens, J. M., Marcoux, M., and Hussey, N. E. (2020). A first look at the metabolic rate of Greenland sharks (Somniosus microcephalus) in the Canadian Arctic. Sci. Rep. 10:19297. doi: 10.1038/s41598-020-76371-0
Stewart, R., Rascón, C. A., Tian, S., Nie, J., Barry, C., Chu, L.-F., et al. (2013). Comparative RNA-seq analysis in the unsequenced axolotl: the oncogene burst highlights early gene expression in the blastema. PLoS Comput. Biol. 9:e1002936. doi: 10.1371/journal.pcbi.1002936
Stier, A., Hsu, B.-Y., Marciau, C., Doligez, B., Gustafsson, L., Bize, P., et al. (2020). Born to be young? Prenatal thyroid hormones increase early-life telomere length in wild collared flycatchers. Biol. Lett. 16:20200364. doi: 10.1098/rsbl.2020.0364
Stoll, E. A., Karapavlovic, N., Rosa, H., Woodmass, M., Rygiel, K., White, K., et al. (2016). Naked mole-rats maintain healthy skeletal muscle and Complex IV mitochondrial enzyme function into old age. Aging (Albany NY) 8, 3468–3485. doi: 10.18632/aging.101140
Stott, K. J., Austin, W., Sayer, M., Weidman, C. R., Cage, A. G., and Wilson, R. (2010). The potential of Arctica islandica growth records to reconstruct coastal climate in north west Scotland, UK. Quaternary Sci. Rev. 29, 1602–1613. doi: 10.1016/j.quascirev.2009.06.016
Sturm, Á, Perczel, A., Ivics, Z., and Vellai, T. (2017). The Piwi-piRNA pathway: road to immortality. Aging Cell 16, 906–911. doi: 10.1111/acel.12630
Sudyka, J., Arct, A., Drobniak, S., Gustafsson, L., and Cichoń, M. (2016). Longitudinal studies confirm faster telomere erosion in short-lived bird species. J. Ornithol. 157, 373–375. doi: 10.1007/s10336-015-1304-4
Sulak, M., Fong, L., Mika, K., Chigurupati, S., Yon, L., Mongan, N. P., et al. (2016). TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. eLife 5:e11994. doi: 10.7554/eLife.11994
Swapna, L. S., Molinaro, A. M., Lindsay-Mosher, N., Pearson, B. J., and Parkinson, J. (2018). Comparative transcriptomic analyses and single-cell RNA sequencing of the freshwater planarian Schmidtea mediterranea identify major cell types and pathway conservation. Genome Biol. 19:124. doi: 10.1186/s13059-018-1498-x
Tacutu, R., Thornton, D., Johnson, E., Budovsky, A., Barardo, D., Craig, T., et al. (2018). Human ageing genomic resources: new and updated databases. Nucleic Acids Res. 46, D1083–D1090. doi: 10.1093/nar/gkx1042
Tan, L., Ke, Z., Tombline, G., Macoretta, N., Hayes, K., Tian, X., et al. (2017). Naked mole rat cells have a stable epigenome that resists iPSC reprogramming. Stem Cell Rep. 9, 1721–1734. doi: 10.1016/j.stemcr.2017.10.001
Tan, M. H., Austin, C. M., Hammer, M. P., Lee, Y. P., Croft, L. J., and Gan, H. M. (2018). Finding nemo: hybrid assembly with oxford nanopore and illumina reads greatly improves the clownfish (Amphiprion ocellaris) genome assembly. Gigascience 7, 1–6. doi: 10.1093/gigascience/gix137
Tan, T. C. J., Rahman, R., Jaber-Hijazi, F., Felix, D. A., Chen, C., Louis, E. J., et al. (2012). Telomere maintenance and telomerase activity are differentially regulated in asexual and sexual worms. Proc. Natl. Acad. Sci. U.S.A. 109, 4209–4214. doi: 10.1073/pnas.1118885109
Tardif, S. D., Mansfield, K. G., Ratnam, R., Ross, C. N., and Ziegler, T. E. (2011). The marmoset as a model of aging and age-related diseases. ILAR J. 52, 54–65.
Taylor, A. M., Heflin, L. E., Powell, M. L., Lawrence, A. L., and Watts, S. A. (2017). Effects of dietary carbohydrate on weight gain and gonad production in small sea urchins, Lytechinus variegatus. Aquac. Nutr. 23, 375–386. doi: 10.1111/anu.12403
Thieme, R., Kurz, S., Kolb, M., Debebe, T., Holtze, S., Morhart, M., et al. (2015). Analysis of Alpha-2 macroglobulin from the long-lived and cancer-resistant naked mole-rat and human plasma. PLoS One 10:e0130470. doi: 10.1371/journal.pone.0130470
Thomson, J. A., and Marshall, V. S. (1997). 4 primate embryonic stem cells. Curr. Top. Dev. Biol. 38, 133–165.
Thoré, E. S., Brendonck, L., and Pinceel, T. (2020). Conspecific density and environmental complexity impact behaviour of turquoise killifish (Nothobranchius furzeri). J. Fish Biol. 97, 1448–1461.
Thoré, E. S., Philippe, C., Brendonck, L., and Pinceel, T. (2021). Towards improved fish tests in ecotoxicology-efficient chronic and multi-generational testing with the killifish Nothobranchius furzeri. Chemosphere 273:129697.
Tian, X., Azpurua, J., Hine, C., Vaidya, A., Myakishev-Rempel, M., Ablaeva, J., et al. (2013). High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 499, 346–349. doi: 10.1038/nature12234
Tian, X., Azpurua, J., Ke, Z., Augereau, A., Zhang, Z. D., Vijg, J., et al. (2015). INK4 locus of the tumor-resistant rodent, the naked mole rat, expresses a functional p15/p16 hybrid isoform. Proc. Natl. Acad. Sci. U.S.A. 112, 1053–1058. doi: 10.1073/pnas.1418203112
Tigges, J., Gordon, T. P., McClure, H. M., Hall, E. C., and Peters, A. (1988). Survival rate and life span of rhesus monkeys at the yerkes regional primate research center. Am. J. Primatol. 15, 263–273.
Tollis, M., Robbins, J., Webb, A. E., Kuderna, L. F. K., Caulin, A. F., Garcia, J. D., et al. (2019). Return to the sea, get huge, beat cancer: an analysis of cetacean genomes including an assembly for the humpback whale (Megaptera novaeangliae). Mol. Biol. Evol. 36, 1746–1763. doi: 10.1093/molbev/msz099
Tomczyk, S., Fischer, K., Austad, S., and Galliot, B. (2015). Hydra, a powerful model for aging studies. Invert. Reprod. Dev. 59, 11–16. doi: 10.1080/07924259.2014.927805
Toren, D., Kulaga, A., Jethva, M., Rubin, E., Snezhkina, A. V., Kudryavtseva, A. V., et al. (2020). Gray whale transcriptome reveals longevity adaptations associated with DNA repair and ubiquitination. Aging Cell 19:e13158. doi: 10.1111/acel.13158
Torre, I. I. I., Mattison, J. A., Fowler, C. G., Lane, M. A., Roth, G. S., and Ingram, D. K. (2004). Assessment of auditory function in rhesus monkeys (Macaca mulatta): effects of age and calorie restriction. Neurobiol. Aging 25, 945–954.
Tozzini, E. T., Dorn, A., Ng’oma, E., Polaèik, M., Blažek, R., Reichwald, K., et al. (2013). Parallel evolution of senescence in annual fishes in response to extrinsic mortality. BMC Evol. Biol. 13:77. doi: 10.1186/1471-2148-13-77
Tricola, G. M., Simons, M. J. P., Atema, E., Boughton, R. K., Brown, J. L., Dearborn, D. C., et al. (2018). The rate of telomere loss is related to maximum lifespan in birds. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 373:20160445. doi: 10.1098/rstb.2016.0445
Ungvari, Z., Buffenstein, R., Austad, S. N., Podlutsky, A., Kaley, G., and Csiszar, A. (2008). Oxidative stress in vascular senescence: lessons from successfully aging species. Front. Biosci. 13, 5056–5070. doi: 10.2741/3064
Valenzano, D. R., Benayoun, B. A., Singh, P. P., Zhang, E., Etter, P. D., Hu, C. K., et al. (2015). The African turquoise killifish genome provides insights into evolution and genetic architecture of lifespan. Cell 163, 1539–1554.
Valenzano, D. R., Kirschner, J., Kamber, R. A., Zhang, E., Weber, D., Cellerino, A., et al. (2009). Mapping loci associated with tail color and sex determination in the short-lived fish Nothobranchius furzeri. Genetics 183, 1385–1395.
van Deursen, J. M. (2014). The role of senescent cells in ageing. Nature 509, 439–446. doi: 10.1038/nature13193
Van Devalk, A. J., and Coleman, J. T. H. (2010). A proposed weight-length relationship for the Common Mudpuppy. Herpetol. Rev. 41, 29–31.
Van Heukelem, W. F. (1977). Laboratory maintenance, breeding, rearing, and biomedical research potential of the Yucatan octopus (Octopus maya). Lab. Anim. Sci. 27, 852–859.
Vazquez, J. M., and Lynch, V. J. (2021). Pervasive duplication of tumor suppressors in Afrotherians during the evolution of large bodies and reduced cancer risk. eLife 10:e65041.
Vieira, W. A., Wells, K. M., and McCusker, C. D. (2020). Advancements to the axolotl model for regeneration and aging. Gerontology 212–222. doi: 10.1159/000504294
Viltard, M., Durand, S., Pérez-Lanzón, M., Aprahamian, F., Lefevre, D., Leroy, C., et al. (2019). The metabolomic signature of extreme longevity: naked mole rats versus mice. Aging (Albany NY) 11, 4783–4800. doi: 10.18632/aging.102116
Vladimirova, I. G., Kleimenov, S. Y., Alekseeva, T. A., and Radzinskaya, L. I. (2003). Specific growth rate and the level of energy metabolism in the ontogeny of axolotl, Ambystoma mexicanum (Amphibia: Ambystomatidae). Biol. Bull. 30, 591–596. doi: 10.1023/B:BIBU.0000007716.17865.c8
Vogg, M. C., Beccari, L., Iglesias Ollé, L., Rampon, C., Vriz, S., Perruchoud, C., et al. (2019a). An evolutionarily-conserved Wnt3/β-catenin/Sp5 feedback loop restricts head organizer activity in Hydra. Nat. Commun. 10:312. doi: 10.1038/s41467-018-08242-2
Vogg, M. C., Galliot, B., and Tsiairis, C. D. (2019b). Model systems for regeneration: Hydra. Development 146:dev177212. doi: 10.1242/dev.177212
Voituron, Y., de Fraipont, M., Issartel, J., Guillaume, O., and Clobert, J. (2011). Extreme lifespan of the human fish (Proteus anguinus): a challenge for ageing mechanisms. Biol. Lett. 7, 105–107. doi: 10.1098/rsbl.2010.0539
Voytko, M. L., Tinkler, G. P., Browne, C., and Tobin, J. R. (2009). Neuroprotective effects of estrogen therapy for cognitive and neurobiological profiles of monkey models of menopause. Am. J. Primatol. 71, 794–801.
Vrtílek, M., Žák, J., Polaèik, M., Blažek, R., and Reichard, M. (2018a). Longitudinal demographic study of wild populations of African annual killifish. Sci. Rep. 8:4774. doi: 10.1038/s41598-018-22878-6
Vrtílek, M., Žák, J., Pšenièka, M., and Reichard, M. (2018b). Extremely rapid maturation of a wild African annual fish. Curr. Biol. 28, R822–R824. doi: 10.1016/j.cub.2018.06.031
Vyssokikh, M. Y., Holtze, S., Averina, O. A., Lyamzaev, K. G., Panteleeva, A. A., Marey, M. V., et al. (2020). Mild depolarization of the inner mitochondrial membrane is a crucial component of an anti-aging program. Proc. Natl. Acad. Sci. U.S.A. 117, 6491–6501. doi: 10.1073/pnas.1916414117
Wagner, D. E., Wang, I. E., and Reddien, P. W. (2011). Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811–816. doi: 10.1126/science.1203983
Wang, W., Hu, C.-K., Zeng, A., Alegre, D., Hu, D., Gotting, K., et al. (2020). Changes in regeneration-responsive enhancers shape regenerative capacities in vertebrates. Science 369:eaaz3090. doi: 10.1126/science.aaz3090
Wang, Z. Y., and Ragsdale, C. W. (2018). Multiple optic gland signaling pathways implicated in octopus maternal behaviors and death. J. Exp. Biol. 221:185751. doi: 10.1242/jeb.185751
Warner, D. A., Miller, D. A. W., Bronikowski, A. M., and Janzen, F. J. (2016). Decades of field data reveal that turtles senesce in the wild. Proc. Natl. Acad. Sci. U.S.A. 113, 6502–6507. doi: 10.1073/pnas.1600035113
Warren, E. M., and Pearce, C. M. (2020). Effect of transport method on subsequent survivorship and gonad yield/quality in the red sea urchin Mesocentrotus franciscanus. North Am. J. Aquaculture 82, 371–376. doi: 10.1002/naaq.10153
Watanabe, H., Schmidt, H. A., Kuhn, A., Höger, S. K., Kocagöz, Y., Laumann-Lipp, N., et al. (2014). Nodal signalling determines biradial asymmetry in Hydra. Nature 515, 112–115. doi: 10.1038/nature13666
Waterston, R. H., and Pachter, L. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562.
Wenger, Y., and Galliot, B. (2013a). Punctuated emergences of genetic and phenotypic innovations in eumetazoan, bilaterian, euteleostome, and hominidae ancestors. Genome Biol. Evol. 5, 1949–1968. doi: 10.1093/gbe/evt142
Wenger, Y., and Galliot, B. (2013b). RNAseq versus genome-predicted transcriptomes: a large population of novel transcripts identified in an Illumina-454 Hydra transcriptome. BMC Genomics 14:204. doi: 10.1186/1471-2164-14-204
Wêsławski, J. M., Hacquebord, L., Stempniewicz, L., and Malinga, M. (2000). Greenland whales and walruses in the Svalbard food web before and after exploitation. Oceanologia 42, 37–56.
Wilhelm Filho, D., Althoff, S. L., Dafré, A. L., and Boveris, A. (2007). Antioxidant defenses, longevity and ecophysiology of South American bats. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 146, 214–220. doi: 10.1016/j.cbpc.2006.11.015
Wilkinson, G. S., and Adams, D. M. (2019). Recurrent evolution of extreme longevity in bats. Biol. Lett. 15:20180860. doi: 10.1098/rsbl.2018.0860
Wilkinson, G. S., Adams, D. M., Haghani, A., Lu, A. T., Zoller, J., Breeze, C. E., et al. (2021). DNA methylation predicts age and provides insight into exceptional longevity of bats. Nat. Commun. 12, 1–13.
Willemsen, D., Cui, R., Reichard, M., and Valenzano, D. R. (2020). Intra-species differences in population size shape life history and genome evolution. eLife 9:e55794. doi: 10.7554/eLife.55794
Willmore, W. G., and Storey, K. B. (1997b). Glutathione systems and anoxia tolerance in turtles. Am. J. Physiol. 273, R219–R225. doi: 10.1152/ajpregu.1997.273.1.R219
Willmore, W. G., and Storey, K. B. (1997a). Antioxidant systems and anoxia tolerance in a freshwater turtle Trachemys scripta elegans. Mol. Cell Biochem. 170, 177–185. doi: 10.1023/a:1006817806010
Wirthlin, M., Lima, N. C., Guedes, R. L. M., Soares, A. E., Almeida, L. G. P., Cavaleiro, N. P., et al. (2018). Parrot genomes and the evolution of heightened longevity and cognition. Curr. Biol. 28, 4001–4008.
Wittlieb, J., Khalturin, K., Lohmann, J. U., Anton-Erxleben, F., and Bosch, T. C. G. (2006). Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 6208–6211. doi: 10.1073/pnas.0510163103
Wodinsky, J. (1977). Hormonal inhibition of feeding and death in octopus: control by optic gland secretion. Science 198, 948–951.
Wolfgang, M. J., Eisele, S. G., Browne, M. A., Schotzko, M. L., Garthwaite, M. A., Durning, M., et al. (2001). Rhesus monkey placental transgene expression after lentiviral gene transfer into preimplantation embryos. Proc. Natl. Acad. Sci. U.S.A. 98, 10728–10732.
Wong, J. M., Gaitán-Espitia, J. D., and Hofmann, G. E. (2019). Transcriptional profiles of early stage red sea urchins (Mesocentrotus franciscanus) reveal differential regulation of gene expression across development. Mar. Genomics 48:100692. doi: 10.1016/j.margen.2019.05.007
Wu, Y., Zhang, Y., Mishra, A., Tardif, S. D., and Hornsby, P. J. (2010). Generation of induced pluripotent stem cells from newborn marmoset skin fibroblasts. Stem Cell Res. 4, 180–188.
Xiao, B., Wang, S., Yang, G., Sun, X., Zhao, S., Lin, L., et al. (2017). HIF-1α contributes to hypoxia adaptation of the naked mole rat. Oncotarget 8, 109941–109951. doi: 10.18632/oncotarget.22767
Xie, J., Li, Y., Shen, X., Goh, G., Zhu, Y., Cui, J., et al. (2018). Dampened STING-dependent interferon activation in bats. Cell Host Microbe 23, 297–301.e4. doi: 10.1016/j.chom.2018.01.006
Xu, S., Zhao, L., Xiao, S., and Gao, T. (2019). Whole genome resequencing data for three rockfish species of Sebastes. Sci. Data 6:97. doi: 10.1038/s41597-019-0100-z
Yang, W., Lin, B., Li, G., Chen, H., and Liu, M. (2019). Sequencing and transcriptome analysis for reproduction-related genes identification and SSRs discovery in sequential hermaphrodite Amphiprion ocellaris. TrJFAS 19, 1049–1059.
Yim, H.-S., Cho, Y. S., Guang, X., Kang, S. G., Jeong, J.-Y., Cha, S.-S., et al. (2014). Minke whale genome and aquatic adaptation in cetaceans. Nat. Genet. 46, 88–92. doi: 10.1038/ng.2835
Yoshida, K., Fujisawa, T., Hwang, J. S., Ikeo, K., and Gojobori, T. (2006). Degeneration after sexual differentiation in hydra and its relevance to the evolution of aging. Gene 385, 64–70. doi: 10.1016/j.gene.2006.06.031
Yu, C., Li, Y., Holmes, A., Szafranski, K., Faulkes, C. G., Coen, C. W., et al. (2011). RNA sequencing reveals differential expression of mitochondrial and oxidation reduction genes in the long-lived naked mole-rat when compared to mice. PLoS One 6:e26729. doi: 10.1371/journal.pone.0026729
Zarrella, I., Herten, K., Maes, G. E., Tai, S., Yang, M., Seuntjens, E., et al. (2019). The survey and reference assisted assembly of the Octopus vulgaris genome. Sci. Data 6:13. doi: 10.1038/s41597-019-0017-6
Zhang, G., Li, B., Li, C., Gilbert, M. T. P., Jarvis, E. D., and Wang, J. (2014). Comparative genomic data of the avian phylogenomics project. Gigascience 3:26. doi: 10.1186/2047-217X-3-26
Zhang, L., and Lu, X. I. (2012). Amphibians live longer at higher altitudes but not at higher latitudes. Biol. J. Linn. Soc. 106, 623–632. doi: 10.1111/j.1095-8312.2012.01876.x
Zhang, P., and Wake, D. B. (2009). Higher-level salamander relationships and divergence dates inferred from complete mitochondrial genomes. Mol. Phylogenet. Evol. 53, 492–508. doi: 10.1016/j.ympev.2009.07.010
Zhang, X., Mao, Y., Huang, Z., Qu, M., Chen, J., Ding, S., et al. (2012). Transcriptome analysis of the Octopus vulgaris central nervous system. PLoS One 7:e40320.
Zhao, S., Li, L., Wang, S., Yu, C., Xiao, B., Lin, L., et al. (2016). H2O2 treatment or serum deprivation induces autophagy and apoptosis in naked mole-rat skin fibroblasts by inhibiting the PI3K/Akt signaling pathway. Oncotarget 7, 84839–84850. doi: 10.18632/oncotarget.13321
Keywords: Senescence, Heterocephalus glaber, Myotis, Nothobranchius furzeri, Proteus anguinus, Hydra oligactis, Greenland shark, resistance to cancer
Citation: Holtze S, Gorshkova E, Braude S, Cellerino A, Dammann P, Hildebrandt TB, Hoeflich A, Hoffmann S, Koch P, Terzibasi Tozzini E, Skulachev M, Skulachev VP and Sahm A (2021) Alternative Animal Models of Aging Research. Front. Mol. Biosci. 8:660959. doi: 10.3389/fmolb.2021.660959
Received: 30 January 2021; Accepted: 08 April 2021;
Published: 17 May 2021.
Edited by:
Venkaiah Betapudi, Chemical and Biological Defense Division, United States Department of Homeland Security, United StatesReviewed by:
Alexey Moskalev, Institute of Biology, Komi Scientific Center (RAS), RussiaEli Thoré, KU Leuven Research & Development, Belgium
Parvin Shahrestani, California State University, Fullerton, United States
Enoch Ng’Oma, University of Missouri, United States
Copyright © 2021 Holtze, Gorshkova, Braude, Cellerino, Dammann, Hildebrandt, Hoeflich, Hoffmann, Koch, Terzibasi Tozzini, Skulachev, Skulachev and Sahm. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Susanne Holtze, aG9sdHplQGl6dy1iZXJsaW4uZGU=; Arne Sahm, YXJuZS5zYWhtQGxlaWJuaXotZmxpLmRl
 Susanne Holtze
Susanne Holtze Ekaterina Gorshkova
Ekaterina Gorshkova Stan Braude
Stan Braude Alessandro Cellerino5,6
Alessandro Cellerino5,6 Philip Dammann
Philip Dammann Andreas Hoeflich
Andreas Hoeflich Philipp Koch
Philipp Koch Eva Terzibasi Tozzini
Eva Terzibasi Tozzini Arne Sahm
Arne Sahm