
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 11 March 2021
Sec. Molecular Diagnostics and Therapeutics
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.657161
This article is part of the Research Topic Tumor Microenvironment in Cancer Hallmarks and Therapeutics View all 57 articles
With high mortality and poor prognosis, hepatocellular carcinoma (LIHC) has become the fourth leading cause of cancer-related deaths worldwide. Most of the LIHC patients missed the best treatment period because of the untimely diagnosis. For others, even if they are temporarily cured, they have to face a very low prognostic survival rate and a very high risk of recurrence. Based on the characteristics of abnormal proliferation and uncontrolled growth of tumor cells. Cell Division Cycle Associated (CDCA) family genes, which are responsible for regulating the cell cycle and proliferation, were selected as our research object to explore the mechanism of hepatocarcinogenesis. To this end, we investigated the expression profiles of CDCA family genes in LIHC and corresponding normal tissues, and the effect of CDCAs expression on the survival of prognosis and immune cell infiltration through bioinformatics analysis methods and the publicly accessible online databases. In addition, we also analyzed the expression correlation of CDCAs and screened the neighboring genes related to functional CDCAs. The results revealed that the expression levels of CDCA1/3/5/8 were significantly increased in LIHC, regardless of stage, sex, race, drinking behavior, and other clinical factors. CDCAs expression was significantly correlated with poor prognosis and was positively correlated with the infiltration of dendritic cells, B cells, and macrophages. We also found that the most relevant neighboring genes to CDCAs in LIHC were SGO2, NDC80, BIRC5, INCENP, and PLOD1. In general, our work suggests that CDCA1/3/5/8 has the potential to be a diagnostic gene in hepatocarcinogenesis and prognostic biomarkers for LIHC patients.
Liver cancer is the fourth leading cause of cancer-related deaths worldwide, with an estimated incidence rate of over one million cases per year, which seriously endangers human health (Bray et al., 2018; Llovet et al., 2018). Hepatocellular carcinoma (LIHC), accounting for 75–85% of cases, is the main pathological type of primary liver cancer (Bray et al., 2018). The majority of LIHC cases arise from hepatitis B and hepatitis C in hepatocytes (Zhang et al., 2004; Makarova et al., 2016). Additionally, the rising incidence of LIHC is due to a high rate of alcohol consumption and the occurrence of non-alcoholic fatty liver disease (Schütte et al., 2009; Gish et al., 2016). Although the development of various treatment technologies, including surgical resection, liver transplantation surgery, interventional therapy, chemotherapy, and radiotherapy, may reduce mortality to some extent, LIHC patients still bear a poor five-year survival rate of only 20–30% (Pang et al., 2008; Yang et al., 2009). Thus, there is an urgent need to deepen the understanding of LIHC tumorigenesis and develop new treatment and monitoring methods for early detection and prolong the survival of LIHC patients.
Dysregulation in any process of cell division may lead to malignancy (Collins and Garrett, 2005). Cell Division Cycle Associated (CDCA) family proteins, including eight respective members of CDCA1-8, function to regulate the cell cycle and proliferation, which play an important role in biological process. Previous studies have shown that the expression levels of all or part of CDCA family genes are significantly upregulated in lung cancer, breast cancer, renal cell cancer, etc. This high expression is usually associated with a poor prognosis. For example, CDCA genes are highly expressed in ovarian cancer tissues and act on the PLK pathway to promote tumor invasion and metastasis (Chen et al., 2020a; Chen et al., 2020b). CDCA7 can accelerate the proliferation of lung adenocarcinoma and non-small cell lung cancer by regulating the cell cycle (Wang et al., 2019). High mRNA expression of CDCA3/5/7/8 in breast cancer significantly reduced the survival rate of patients (Phan et al., 2018). These findings strongly suggest the potential role of CDCAs in the tumorigenesis and prognosis of patients. It is necessary and meaningful to understand the mechanism of action of CDCAs in LIHC. To date, however, there is no systematic and comprehensive analysis of CDCAs in LIHC.
To reveal the mechanism of LIHC tumorigenesis and to identify diagnostic and prognostic markers or therapeutic targets for LIHC patients, in the current study, the transcriptional levels of CDCA genes were investigated by Oncomine and GEPIA, and we found that CDCA1/3/5/8 expression levels were significantly increased in LIHC. We also evaluated the impact of CDCA1/3/5/8 on the prognosis of LIHC patients. Specifically, we analyzed the effect of CDCAs expression on the prognostic survival rate of LIHC patients, the correlation between CDCAs transcription and various clinical factors by Kaplan-Meier plotter, and the effect of genetic alterations of CDCAs on prognosis by cBioPortal. The relationship between CDCA1/3/5/8 and immune cell infiltration was analyzed using TIMER 2.0. In addition, the interaction network between CDCAs and neighboring genes was mapped using GeneMANIA.
The Oncomine platform (https://www.oncomine.org/) is a publicly accessible online tumor-related gene microarray database that collects related gene expression profiles and relevant clinical information. The transcriptional levels of CDCA family genes (CDCA1–8) in different tumors and corresponding normal tissues were analyzed by Oncomine with approximately 200 samples. The expression levels were considered significantly different when fold change > 2.0 and p-value < 0.0001. We set the threshold value of gene rank to “top 10%” and the data type to “mRNA” (Rhodes et al., 2007).
GEPIA (http://gepia.cancer-pku.cn/index.html) was used to analyze the expression of CDCAs sequencing in liver cancer tissue based on the GTEx and TCGA databases (Tang et al., 2017). GEPIA was used to compare the CDCAs expression levels in HCC with the thresholds of |Log2 (Fold Change)| Cut-off: 1 and p-value Cut-off: 0.01. CDCAs expression in different HCC stages was analyzed using the default parameters.
Kaplan-Meier plotter (http://kmplot.com/analysis/) was used to evaluate the influence of different expression levels of CDCA family genes on prognostic value, including overall survival (OS), disease-specific survival (DSS), relapse-free survival (RFS), progression-free survival (PFS), and OS of liver cancer patients with different clinical factors. We analyzed all samples in the database with the parameters of Group Cut-off: Median; Hazards Ratio: Yes; 95% Confidence Interval: Yes; Follow-up threshold: All.
cBioPortal (http://www.cbioportal.org/) was used to perform the interactive analysis of biomolecules in tumor tissues in the TCGA database (Gao et al., 2016). Here, we used it to analyze the alterations in the frequency of CDCAs genes change. Putative copy-number calls on 370 cases determined using GISTIC 2.0. In module Comparison/Survival, we analyzed the influence of alterations on prognostic survival in HCC patients by default parameters.
GeneMANIA (http://genemania.org/), based on many large, publicly available biological datasets, is used to identify intra-genomic associations and find co-expressing biomolecules. Here, GeneMANIA was used to identify the physical interactions and co-expression of CDCAs with 20 related genes in Homo sapiens datasets by default parameters.
Using TIMER 2.0 (http://timer.comp-genomics.org/), we analyzed the relationship between the expression of CDCAs and infiltration levels of immune cells in liver cancer tissue. The TIMER database was used to determine the abundance of tumor infiltrates based on biomarker gene expression analysis (Li et al., 2017). Here, we chose CDCA1/3/5/8 as input and, in turn, detected cancer cells under the Immune Association module. B cells, CD8+ T cells, CD4+ T cells, neutrophils, macrophages, and dendritic cells were selected as the test types according to Li et al. (Li et al., 2016; Danaher et al., 2017). Gene expression values were transformed to Log2 RSEM values.
Dysregulation in any process of cell division may lead to malignancy. It has been reported that there are eight respective members in the CDCA family genes, among which members may play an independent role or function cooperatively. Oncomine analysis revealed that most CDCA gene members were significantly upregulated in 15 cancer types (Figure 1). For LIHC, in particular, we found consistent results in both the Oncomine and GEPIA databases, i.e., the expression of CDCA1, CDCA3, CDCA5, and CDCA8 in LIHC was significantly higher than those in normal tissues (Figures 1, 2A). Therefore, these genes were selected for follow-up analysis and research objects.
Tumor development is usually subdivided into four stages to assess the extent of cancer cell proliferation and to determine the prognostic potential of patients. In this study, significant differences were observed in the above four members of CDCA genes in different stages of liver cancer, suggesting that these four genes may function in the whole course of LIHC tumorigenesis (Figure 2B).
Using the Kaplan-Meier plotter, we found that the transcription level of the four test CDCA genes was significantly correlated with the prognostic survival rate of liver cancer patients (Figure 3). Specifically, high expression of CDCAs in OS, DSS, RFS, and PFS represented poor prognosis, indicating that active transcription of CDCA1/3/5/8 might cause health risks, and these genes could be potential prognostic biomarkers for LIHC patients (Figure 4).
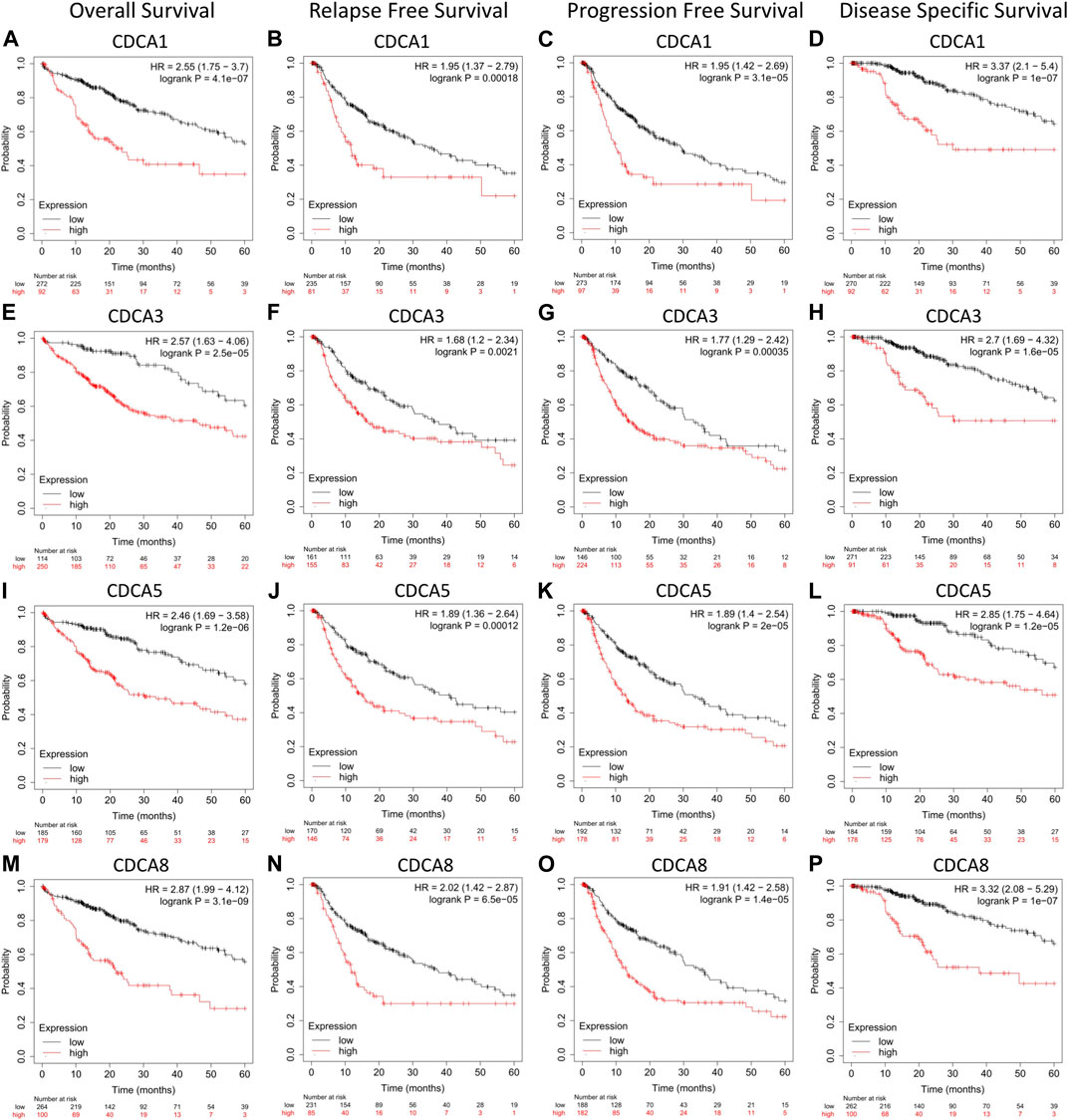
FIGURE 4. Correlation analysis between CDCAs expression and prognostic survival in LIHC patients via Kaplan-Meier plotter analysis. (A‐D) Correlation between expression of CDCA1 and survival curves. (E‐H) Correlation between expression of CDCA3 and survival curves. (I‐L) Correlation between expression of CDCA5 and survival curves. (M‐P) Correlation between expression of CDCA8 and survival curves.
For different types of clinical factors, we further analyzed the relationship between the expression levels of CDCA1/3/5/8 genes and prognostic OS (Table 1). Here, we concluded that the high expression levels of CDCAs led to poor prognosis regardless of sex, race, or alcohol consumption. Notably, when there was no hepatitis virus or vascular infection in LIHC patients, the expression of CDCA1/3/5/8 was significantly correlated with the survival rate. Interestingly, only CDCA8 was significantly associated with survival in patients with hepatitis virus or vascular infection (Table 1).
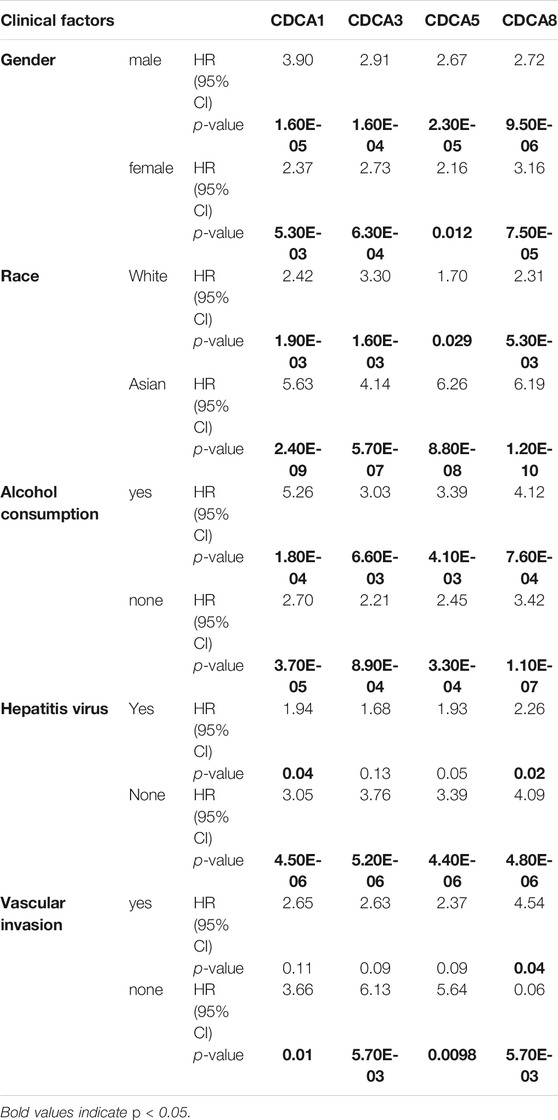
TABLE 1. Correlation analysis between CDCAs expression and prognostic overall survival in LIHC patients with different clinicopathological factors via Kaplan-Meier plotter analysis.
We extracted CDCA1/3/5/8 genes and investigated the percentages of genetic alterations in the TCGA dataset. In a total of 360 samples, the alteration frequency of CDCAs was as high as 29.44% in LIHC (106/360) (Figure 5A), with 27% of CDCA1, 0.8% of CDCA3, 7% of CDCA5, and 6% of CDCA8 (Figure 5B). Next, we performed a correlation analysis between cases with (or without) CDCAs genetic alterations and prognostic survival. There was a significant correlation between CDCA alterations and survival of both OS (4.348e-3) (Figure 5C) and DFS (7.932e-3) (Figure 5D), implying that these alterations will aggravate the hepatocarcinogenesis mediated by high expression of CDCAs, which is not conducive to the survival of patients.
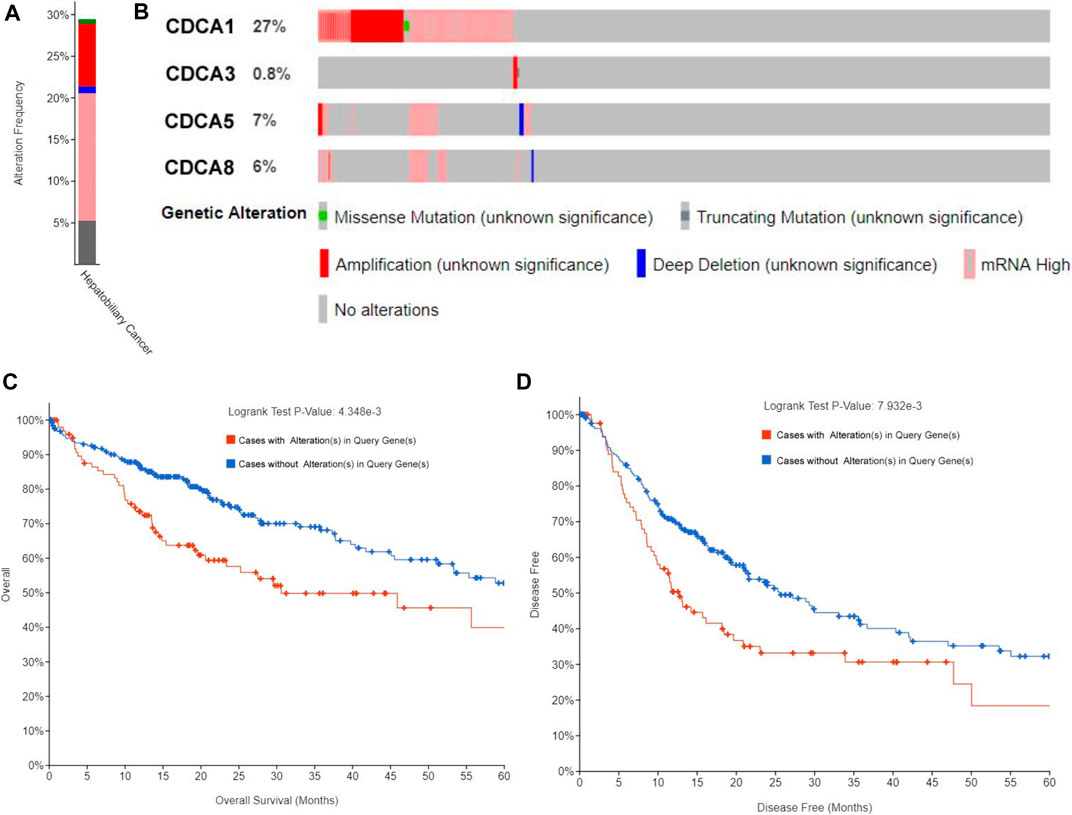
FIGURE 5. Alteration frequency analysis of CDCAs and its influence on prognosis in LIHC patients via cBioPortal analysis. (A,B) Alteration frequency of CDCAs based on the TCGA provisional dataset. (C,D) Kaplan-Meier plots comparing OS (C) and DFS (D) in cases with/without CDCAs genetic alterations.
TIMER 2.0 was used to investigate the correlation between the expression of CDCA1/3/5/8 and infiltration levels of immune cells (CD8+ T cells, CD4+ T cells, B cells, neutrophils, macrophages, and dendritic cells). Overall, there was a statistically significant positive correlation between CDCAs gene expression and most immune cell infiltration in LIHC. In detail, CDCAs showed a poor correlation with CD8+ T cells and neutrophils, but a stronger correlation with dendritic cells, B cells, and macrophages (Figure 6).
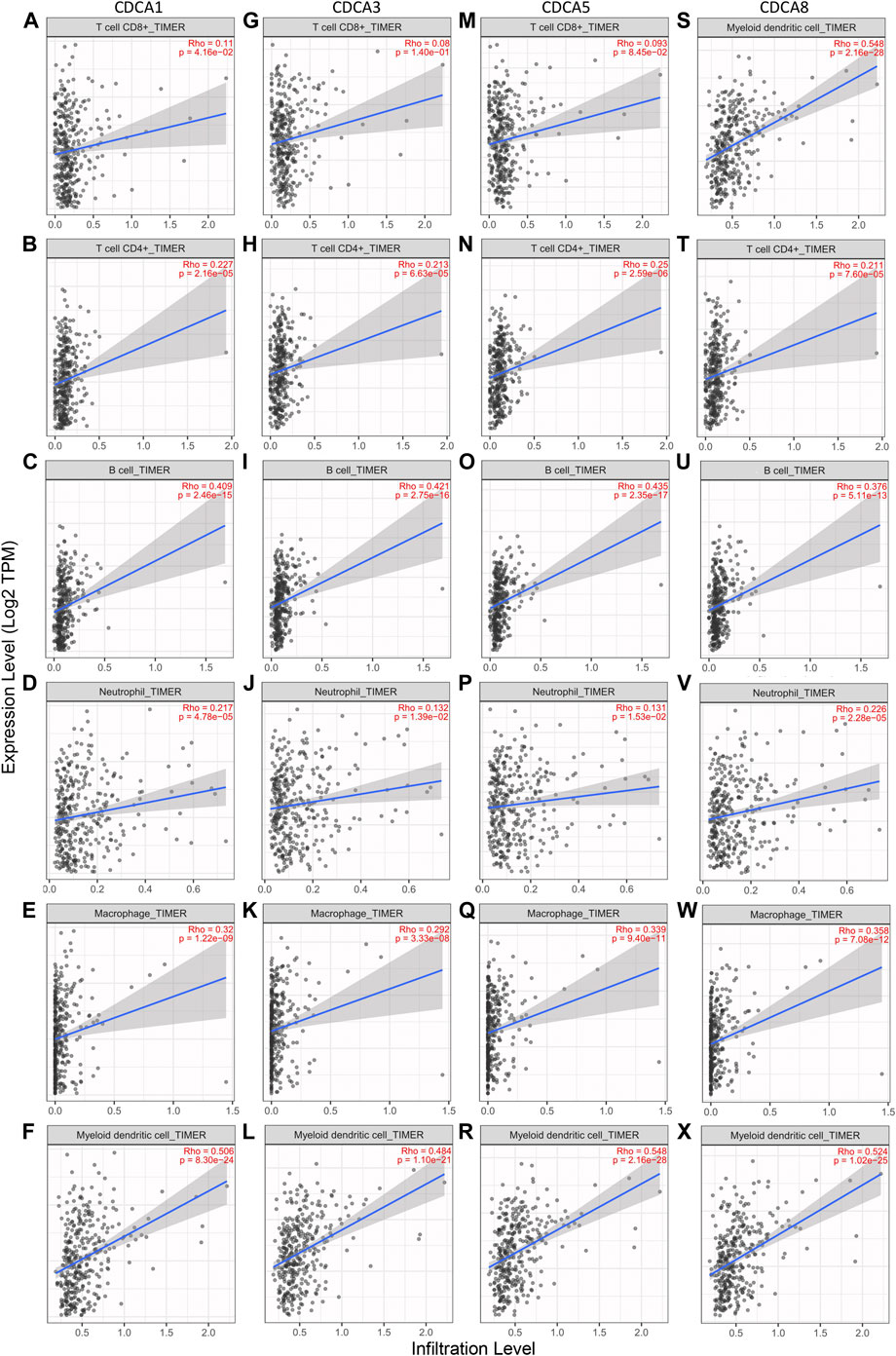
FIGURE 6. Correlation analysis between CDCAs expression and immune cell infiltration levels in LIHC patients via TIMER 2.0 analysis. (A‐F) Correlation analysis between CDCA1 expression and immune cell infiltration levels. (G‐L) Correlation analysis between CDCA3 expression and immune cell infiltration levels. (M‐R) Correlation analysis between CDCA5 expression and immune cell infiltration levels. (S‐X) Correlation analysis between CDCA8 expression and immune cell infiltration levels.
To provide a basis for exploring the immune mechanisms mediated by CDCAs and screening potential therapeutic targets, we further investigated the correlation of expression levels between CDCA1/3/5/8 and biomarker genes of immune cells and their subsets in LIHC. We found that the CDCA gene was positively correlated with all test biomarker genes of dendritic cells and B cells (Table 2). Although CDCAs showed a significant positive correlation with most of the biomarker genes of M1 macrophage and M2 macrophage, there was no correlation between CDCA1/3/5/8 and iNOS (macrophage biomarker). In addition, there was no association between CDCA3 and COX2 or CD163 (Table 2).
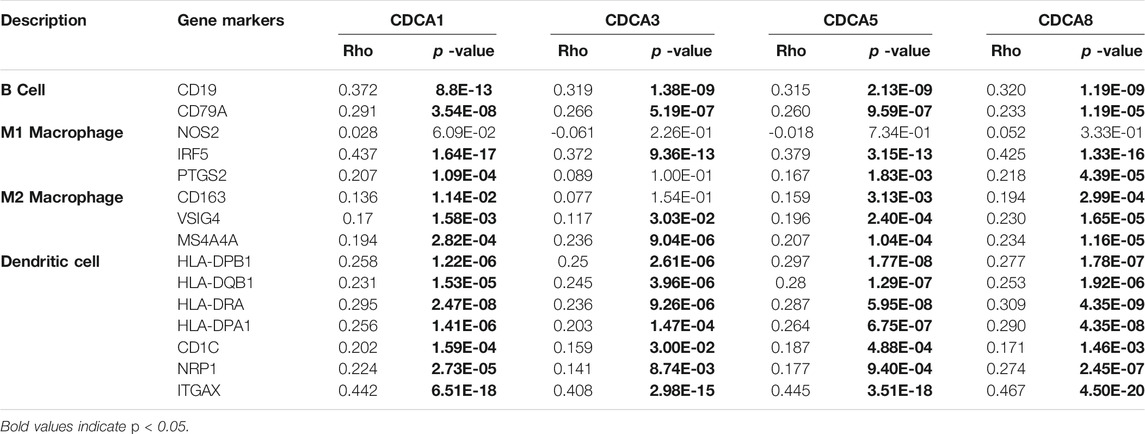
TABLE 2. Correlation analysis between the expression of CDCAs and immune cell biomarker genes in LIHC patients via TIMER 2.0 analysis.
At the genetic level, the expression correlation of the four member genes of CDCA1/3/5/8 in liver cancer patients was analyzed using cBioPortal. According to our results, the Pearson correlation coefficients ranged from 0.85–0.90, indicating a strong correlation between the expression patterns of these four CDCAs in LIHC (Figure 7A). The GeneMANIA dataset was used to explore the co-expression of CDCAs with other biomolecules. Here, we found that there are physical interactions between CDCA1, CDCA3, CDCA5, and CDCA8. The interaction network of CDCAs and the 20 most frequently neighboring genes are shown in Figure 7B. The top five CDCAs neighboring genes in LIHC were SGO2, NDC80, BIRC5, INCENP, and PLOD1.
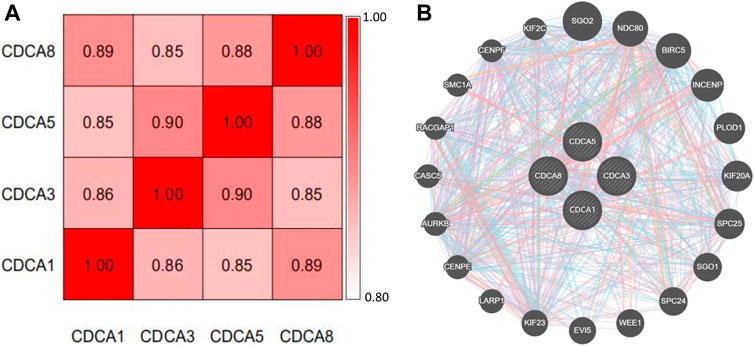
FIGURE 7. Co-expression and interaction of CDCAs at the gene and protein levels in LIHC patients. (A) Spearman’s correlation analysis of CDCAs. (B) Gene–gene interaction network among CDCAs in the GeneMANIA dataset.
With high mortality and poor prognosis, LIHC has become the fourth leading cause of cancer-related deaths worldwide (Bray et al., 2018). Most of the LIHC patients missed the best treatment period because of the untimely diagnosis. For others, even if they are temporarily cured, they have to face a very low prognostic survival rate and a very high risk of recurrence. Based on the characteristics of abnormal proliferation and uncontrolled growth of tumor cells. CDCA genes, which are responsible for regulating the cell cycle and proliferation, were selected as our research object to explore the mechanism of hepatocarcinogenesis. Through a series of bioinformatics analysis methods and the use of publicly accessible online databases, including Oncomine, Kaplan-Meier plotter, GEPIA, GeneMANIA, cBioPortal, and TIMER 2.0, we investigated the expression profiles of CDCA family genes in LIHC and corresponding normal tissues, and the effect of CDCAs expression on the survival of prognosis and immune cell infiltration. In addition, we also analyzed the expression correlation of CDCAs and screened the neighboring genes related to functional CDCAs.
We found that the expression levels of CDCA1/3/5/8 were significantly increased in LIHC, regardless of stage, sex, race, drinking behavior, and other clinical factors. CDCAs expression was significantly correlated with poor prognosis of OS, DSS, RFS, and PFS and was positively correlated with the infiltration of dendritic cells, B cells, and macrophages. We also found that the most relevant neighboring genes to CDCAs in LIHC were SGO2, NDC80, BIRC5, INCENP, and PLOD1. In general, our work suggests that CDCA1/3/5/8 has the potential to be a diagnostic gene in hepatocarcinogenesis and prognostic biomarkers for LIHC patients.
The CDCA family consists of eight independent individuals named CDCA1–8. To clarify the role of CDCAs in LIHC, multiple database analysis was performed, and CDCA1/3/5/8 was found to be highly expressed in LIHC tissue. Moreover, this active expression of CDCAs was observed in all stages of LIHC, indicating that CDCA1/3/5/8 has potential as a diagnostic gene for the occurrence and development of LIHC. Similarly, CDCA has been shown to be highly expressed in many other cancer types, such as breast cancer and lung cancer, indicating its wide applicability and functional conservation. However, there was no significant upregulation of CDCAs in leukemia, suggesting that it is necessary to distinguish the types of cancer when they were used as diagnostic genes.
According to previous reports, CDCA1, also known as Nuf2, is mainly responsible for regulating cell mitosis (Zhang et al., 2015). Downregulation of CDCA1 expression can inhibit the proliferation of tumor cells, while overexpression of CDCA1 is associated with poor prognosis (Hu et al., 2015). CDCA3 has been shown to promote cell proliferation and invasion through activation of the Ras signaling pathway or hypomethylation in gastric cancer cells (Zhang et al., 2019). CDCA5 plays important roles in migration, proliferation, apoptosis, and invasion of tumor cells by regulating sister chromatid segregation and cohesion (Xu et al., 2019). CDCA8 is an integral part of the chromosomal passenger complex, which is involved in mitosis and contributes to distant metastasis of cancer cells (Jeon et al., 2017). It is worth noting that the high expression of these four genes has been proven to reduce the prognostic survival rate of many cancer patients. This is consistent with the results of our study. In particular, there is a significant negative correlation between CDCAs and four common prognostic indices of OS, DSS, RFS, and PFS, and this correlation is applicable to different clinical factors, suggesting that high expression of CDCAs may be one of the causes of poor prognosis. These biomolecules have the potential to be prognostic biomarkers for LIHC patients.
However, in the presence of pathological conditions, such as hepatitis virus or vascular invasion, there was no correlation between CDCAs and OS, except for CDCA8, suggesting that we should consider the patient’s clinical condition when selecting the diagnostic gene. In addition, we found that the alteration frequency of CDCAs is extremely high, and the genetic alteration will reduce the prognosis survival of LIHC patients. The factors causing gene alteration should be avoided in the process of treatment, and the degree of an alteration should be properly considered when detecting expression and evaluating prognosis.
Dendritic cells are special antigen-presenting cells that play a major role in activating T lymphocytes with anti-tumor effects (Wculek et al., 2020). B cells can produce a kind of IgG antibody to recognize a certain antigen in tumor tissue, which can inhibit tumor growth (Li et al., 2009). Macrophages were divided into M1 macrophages and M2 macrophages. M1 macrophages are mainly related to the recognition and attack of tumor cells, while M2 macrophages are related to tumor progression and immunosuppression (Yang and Zhang, 2017; Shapouri et al., 2018). CDCAs showed a significant positive correlation with dendritic cells, B cells, and macrophages, indicating that CDCA-mediated hepatocarcinogenesis might mobilize the activity of these immune cells and make them play an anti-tumor role. When we further analyzed the relationship between CDCA1/3/5/8 expression and the biomarker genes of immune cells, we found a significant positive correlation between CDCAs expression and all test biomarkers of B cells and dendritic cells. However, for M1 macrophages and M2 macrophages, CDCAs were only related to some of macrophages biomarker genes, indicating that there is a certain specificity and selectivity in this interaction, which also provides some basis for immunotherapy in the future.
Although these four genes have some differences in function and target genes (pathways), they jointly regulate the cell division cycle to promote the proliferation and invasion of tumor cells. The Pearson correlation coefficients of these four genes were all greater than 0.85. GeneMANIA analysis showed twenty neighboring genes to CDCAs, and the functions of these genes are also related to cell cycle regulation. For example, SGO1 is considered to play a major role in recruiting the chromosomal passenger complex to chromosomes (Bonner et al., 2020); SGO2 is a pericentromeric protein that associates with cohesin at centromeres and regulates chromosomal segregation during meiosis (El et al., 2017). SGO1/2 functions as an essential protector for centromeric cohesion and is required for accurate chromosome segregation during mitosis and meiosis (Dudas et al., 2011). PLOD1 has been reported to function in extracellular matrix formation and is involved in various diseases, including cancer (Wang et al., 2018). The NDC80 complex, including the main elements of NDC80, SPC24, and SPC25, is highly expressed in various tumors and cooperatively promotes the invasion and metastasis of tumor cells (Alushin et al., 2012; Umbreit et al., 2012). BIRC5 is a negative regulatory protein that inhibits tumor cell apoptosis and promotes cell proliferation (Cho et al., 2020; Shi et al., 2020).
These proteins are more or less involved in the development of tumors, which explains the mechanism of CDCA-mediated hepatocarcinogenesis to a certain extent. This also screened out some targets for future research on new therapeutic methods. However, the specific roles of CDCAs in tumorigenesis and development, as well as their interaction with target proteins need to be further verified. The same shortcomings run through our research because the results were based on big data mining and analysis, which inevitably led to some false-positive results. An important purpose was to provide researchers with a more instructive research idea and tried to make our findings point out a direction for future research. Although we mined the data as comprehensively as possible and had obtained some meaningful conclusions, the relevant results still require further experimental and clinical verification.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. The patients/participants provided their written informed consent to participate in this study.
XL and X-WX contributed to the concept and wrote the manuscript. S-SJ, S-JK, Z-LK, and JL designed experiments, performed experiments, and analyzed data. S-SJ and S-JK contributed equally to this study.
This work was supported by 2019 Doctor Initiation Fund of Guizhou University of Chinese Medicine (3043-043190019) and Research Initiation Foundation for Doctor of Henan Agricultural University (30602107).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Alushin, G. M., Musinipally, V., Matson, D., Tooley, J., Stukenberg, P. T., and Nogales, E. (2012). Multimodal microtubule binding by the Ndc80 kinetochore complex. Nat. Struct. Mol. Biol. 19, 1161–1180. doi:10.1038/nsmb.2411
Bonner, M. K., Haase, J., Saunders, H., Gupta, H., Li, B., and Kelly, A. E. (2020). The borealin dimerization domain interacts with Sgo1 to drive aurora B-mediated spindle assembly. Mol. Biol. Cell. 31, 2207–2218. doi:10.1091/mbc.E20-05-0341
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi:10.3322/caac.21492
Chen, C., Chen, S., Luo, M., Yan, H., Pang, L., Zhu, C., et al. (2020a). The role of the CDCA gene family in ovarian cancer. Ann. Transl. Med. 8, 190. doi:10.21037/atm.2020.01.99
Chen, C., Chen, S., Pang, L., Yan, H., Luo, M., Zhao, Q., et al. (2020b). Analysis of the expression of cell division cycle-associated genes and its prognostic significance in human lung carcinoma: a review of the literature databases. Biomed. Res. Int. 2020, 1–14. doi:10.1155/2020/6412593
Cho, M., Lee, O.-H., Chang, E. M., Lee, S., Moon, S., Lee, J., et al. (2020). BIRC5 expression is regulated in uterine epithelium during the estrous cycle. Genes 11, 282–295. doi:10.3390/genes11030282
Collins, I., and Garrett, M. D. (2005). Targeting the cell division cycle in cancer: CDK and cell cycle checkpoint kinase inhibitors. Curr. Opin. Pharmacol. 5, 366–373. doi:10.1016/j.coph.2005.04.009
Danaher, P., Warren, S., Dennis, L., D’Amico, L., White, A., Disis, M. L., et al. (2017). Gene expression markers of tumor infiltrating leukocytes. J. Immunother. Cancer 5, 18–32. doi:10.1186/s40425-017-0215-8
Dudas, A., Ahmad, S., and Gregan, J. (2011). Sgo1 is required for co-segregation of sister chromatids during achiasmate meiosis I. Cell cycle 10, 951–955. doi:10.4161/cc.10.6.15032
El, Y. W., Buffin, E., Cladière, D., Gryaznova, Y., Berenguer, I., Touati, S. A., et al. (2017). Mps1 kinase-dependent Sgo2 centromere localisation mediates cohesin protection in mouse oocyte meiosis I. Nat. Commun. 8, 1–15. doi:10.1038/s41467-017-00774-3
Gao, J., Lindsay, J., Watt, S., Bahceci, I., Lukasse, P., Abeshouse, A., et al. (2016). Abstract 5277: The cBioPortal for cancer genomics and its application in precision oncology. Cancer Res. 76, 5277. doi:10.1158/1538-7445.AM2016-5277
Gish, R. G., Lencioni, R., Di Bisceglie, A. M., Raoul, J. L., and Mazzaferro, V. (2016). Role of the multidisciplinary team in the diagnosis and treatment of hepatocellular carcinoma. Expert Rev. Gastroenterol. Hepatol. 6, 173–185. doi:10.1586/egh.11.105
Hu, P., Shangguan, J., and Zhang, L. (2015). Downregulation of NUF2 inhibits tumor growth and induces apoptosis by regulating lncRNA AF339813. Int. J. Clin. Exp. Pathol. 8, 2638–2648. doi:10.1287/orsc.12.5.599.10094
Jeon, T.-W., Ko, M. J., Seo, Y.-R., Baik, I. H., Hwang, I., Ryoo, H.-M., et al. (2017). Abstract 3113: Knockdown of cell division cycle-associated 8 (CDCA8) suppresses hepatocellular carcinoma growth via the upregulation of tumor suppressor ATF3. Cancer Res. 77, 3113. doi:10.1158/1538-7445.AM2017-3113
Li, Q., Song, H., Teitz-Tennenbaum, S., Donald, E. J., Li, M., and Chang, A. E. (2009). In vivo sensitized and in vitro activated B cells mediate tumor regression in cancer adoptive immunotherapy. J. Immunol. 183, 3195–3203. doi:10.4049/jimmunol.0803773
Li, B., Severson, E., Pignon, J. C., Zhao, H., Li, T., Novak, J., et al. (2016). Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 17, 174–189. doi:10.1186/s13059-016-1028-7
Li, T., Fan, J., Wang, B., Traugh, N., Chen, Q., Liu, J. S., et al. (2017). TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77, 108–110. doi:10.1158/0008-5472.CAN-17-0307
Llovet, J. M., Montal, R., Sia, D., and Finn, R. S. (2018). Molecular therapies and precision medicine for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 15, 599–616. doi:10.1038/s41571-018-0073-4
Makarova, R. O. V., Altekruse, S. F., McNeel, T. S., Ulahannan, S., Duffy, A. G., Graubard, B. I., et al. (2016). Population attributable fractions of risk factors for hepatocellular carcinoma in the United States. Cancer 122, 1757–1765. doi:10.1002/cncr.29971
Pang, R. W. C., Joh, J. W., Johnson, P. J., Monden, M., Pawlik, T. M., Poon, R. T. P., et al. (2008). Biology of hepatocellular carcinoma. Ann. Surg. Oncol. 15, 962–971. doi:10.1245/s10434-007-9730-z
Phan, N. N., Wang, C. Y., Li, K. L., Chen, C. F., Chiao, C. C., Yu, H. G., et al. (2018). Distinct expression of CDCA3, CDCA5, and CDCA8 leads to shorter relapse free survival in breast cancer patient. Oncotarget 9, 6977–6992. doi:10.18632/oncotarget.24059
Rhodes, D. R., Kalyana-Sundaram, S., Mahavisno, V., Varambally, R., Yu, J., Briggs, B. B., et al. (2007). Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9, 166–180. doi:10.1593/neo.07112
Schütte, K., Bornschein, J., and Malfertheiner, P. (2009). Hepatocellular carcinoma-epidemiological trends and risk factors. Dig. Dis. 27, 80–92. doi:10.1159/000218339
Shapouri, M. A., Mohammadian, S., Vazini, H., Taghadosi, M., Esmaeili, S. A., Mardani, F., et al. (2018). Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 233, 6425–6440. doi:10.1002/jcp.26429
Shi, J., Tan, S. Y., Lee, A., Zhang, S., Sasidharan, S. L., Wong, B., et al. (2020). Restoring apoptosis dysregulation using survivin inhibitor in nasopharyngeal cancer. Head Neck 42, 913–923. doi:10.1002/hed.26068
Tang, Z., Li, C., Kang, B., Gao, G., Li, C., and Zhang, Z. (2017). GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, 98–102. doi:10.1093/nar/gkx247
Umbreit, N. T., Gestaut, D. R., Tien, J. F., Vollmar, B. S., Gonen, T., Asbury, C. L., et al. (2012). From the Cover: The Ndc80 kinetochore complex directly modulates microtubule dynamics. Proc. Natl. Acad. Sci. U.S.A. 109, 16113–16118. doi:10.1073/pnas.1209615109
Wang, D., Zhang, S., and Chen, F. (2018). High expression of PLOD1 drives tumorigenesis and affects clinical outcome in gastrointestinal carcinoma. Genet. Test. Mol. Biomarkers 22, 366–373. doi:10.1089/gtmb.2018.0009
Wang, H. Y., Ye, L., Xing, Z., Li, H. Q., Lv, T. F., Liu, H. B., et al. (2019). CDCA7 promotes lung adenocarcinoma proliferation via regulating the cell cycle. Pathol. Res. Pract. 215, 152559. doi:10.1016/j.prp.2019.152559
Wculek, S. K., Cueto, F. J., Mujal, A. M., Melero, I., Krummel, M. F., and Sancho, D. (2020). Dendritic cells in cancer immunology and immunotherapy. Nat. Rev. Immunol. 20, 7–24. doi:10.1038/s41577-019-0210-z
Xu, J., Zhu, C., Yu, Y., Wu, W., Cao, J., Li, Z., et al. (2019). Systematic cancer-testis gene expression analysis identified CDCA5 as a potential therapeutic target in esophageal squamous cell carcinoma. EBioMedicine 46, 54–65. doi:10.1016/j.ebiom.07.03010.1016/j.ebiom.2019.07.030
Yang, L. Y., Fang, F., Ou, D. P., Wu, W., Zeng, Z. J., and Wu, F. (2009). Solitary large hepatocellular carcinoma: a specific subtype of hepatocellular carcinoma with good outcome after hepatic resection. Ann. Surg. 249, 118–123. doi:10.1097/SLA.0b013e3181904988
Yang, L., and Zhang, Y. (2017). Tumor-associated macrophages: from basic research to clinical application. J. Hematol. Oncol. 10, 1–12. doi:10.1186/s13045-017-0430-2
Zhang, B. H., Yang, B. H., and Tang, Z. Y. (2004). Randomized controlled trial of screening for hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 130, 417–422. doi:10.1007/s00432-004-0552-0
Zhang, T., Zhou, Y., Qi, S. T., Wang, Z. B., Qian, W. P., Ouyang, Y. C., et al. (2015). Nuf2 is required for chromosome segregation during mouse oocyte meiotic maturation. Cell cycle 14, 2701–2710. doi:10.1080/15384101.2015.1058677
Keywords: CDCA, hepatocellular carcinoma, biomarkers, prognosis, tumor immunity
Citation: Jiang S-S, Ke S-J, Ke Z-L, Li J, Li X and Xie X-W (2021) Cell Division Cycle Associated Genes as Diagnostic and Prognostic Biomarkers in Hepatocellular Carcinoma. Front. Mol. Biosci. 8:657161. doi: 10.3389/fmolb.2021.657161
Received: 22 January 2021; Accepted: 08 February 2021;
Published: 11 March 2021.
Edited by:
Hongming Miao, Army Medical University, ChinaReviewed by:
Huai-Qiang Ju, Sun Yat-Sen University, ChinaCopyright © 2021 Jiang, Ke, Ke, Li, Li and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang Li, bGl4aWFuZzAyMTdAMTI2LmNvbQ==; Xing-Wei Xie, eGlleGluZ3dlaTEyM0AxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.