
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 18 March 2021
Sec. Molecular Recognition
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.648468
This article is part of the Research Topic Prokaryotic Communications: From Macromolecular Interdomain to Intercellular Talks (Recognition) and Beyond View all 21 articles
 Wilfried J. J. Meijer1*
Wilfried J. J. Meijer1* D. Roeland Boer2
D. Roeland Boer2 Saúl Ares3
Saúl Ares3 Carlos Alfonso4
Carlos Alfonso4 Fernando Rojo5
Fernando Rojo5 Juan R. Luque-Ortega6
Juan R. Luque-Ortega6 Ling Juan Wu7*
Ling Juan Wu7*Bacterial conjugation is the main horizontal gene transfer route responsible for the spread of antibiotic resistance, virulence and toxin genes. During conjugation, DNA is transferred from a donor to a recipient cell via a sophisticated channel connecting the two cells. Conjugation not only affects many different aspects of the plasmid and the host, ranging from the properties of the membrane and the cell surface of the donor, to other developmental processes such as competence, it probably also poses a burden on the donor cell due to the expression of the large number of genes involved in the conjugation process. Therefore, expression of the conjugation genes must be strictly controlled. Over the past decade, the regulation of the conjugation genes present on the conjugative Bacillus subtilis plasmid pLS20 has been studied using a variety of methods including genetic, biochemical, biophysical and structural approaches. This review focuses on the interplay between RcopLS20, RappLS20 and Phr*pLS20, the proteins that control the activity of the main conjugation promoter Pc located upstream of the conjugation operon. Proper expression of the conjugation genes requires the following two fundamental elements. First, conjugation is repressed by default and an intercellular quorum-signaling system is used to sense conditions favorable for conjugation. Second, different layers of regulation act together to repress the Pc promoter in a strict manner but allowing rapid activation. During conjugation, ssDNA is exported from the cell by a membrane-embedded DNA translocation machine. Another membrane-embedded DNA translocation machine imports ssDNA in competent cells. Evidences are reviewed indicating that conjugation and competence are probably mutually exclusive processes. Some of the questions that remain unanswered are discussed.
Horizontal Gene Transfer (HGT) refers to the process by which a DNA element/region is transferred from one cell to another. HGT occurs at large scale in bacteria, and a single HGT event can result in the acquisition of several to many genes by a cell and therefore plays a major role in bacterial evolution (Soucy et al., 2015). Unfortunately, HGT also contributes importantly to the dissemination of antibiotic resistance, virulence and toxin genes. The spread of these genes has now become a major global problem, with serious consequences on both human lives and economy (Partridge et al., 2018; CDC, 2019; Lerminiaux and Cameron, 2019). A better understanding of the HGT processes is needed to design drugs or strategies to impede the spread of antibiotic resistance and other pernicious genes. There are three main mechanisms responsible for HGT: transformation, transduction and conjugation (Ochman et al., 2000; Frost et al., 2005; Thomas and Nielsen, 2005; Novick et al., 2010). Of these, conjugation is the mechanism that is majorly responsible for the spread of antibiotic resistance, toxin and virulence genes (Mazel and Davies, 1999; Davies and Davies, 2010).
During conjugation, a DNA element is transferred from a donor cell to a recipient cell; hence, it requires direct contact between the two cells. Conjugative elements are often present on plasmids, but they can also be integrated in a bacterial chromosome. These latter elements are named integrative and conjugative elements (ICE). Conjugation occurs in both Gram-positive (G+) and Gram-negative (G−) bacteria, and although there are important differences (see below), the basics of the conjugation process are conserved in G+ and G− bacteria. Conjugation can be divided into four discernable stages, which have been described in detail previously (Chen et al., 2005; Arutyunov and Frost, 2013; Cabezon et al., 2014; Goessweiner-Mohr et al., 2014; Koraimann, 2018; Li et al., 2019; Waksman, 2019). Below we briefly summarize each stage, to illustrate the large number of proteins involved in conjugation, and the complexity of the process in which the many proteins act together in a temporally orchestrated manner. During stage 1) transfer competent donor cells select and attach to a suitable recipient cell (also known as mating pair formation). This process involves surface-located proteins, some of which are adhesion proteins that establish contact with the recipient cell. The adhesion proteins encoded by conjugative elements present in G− bacteria are generally located at the tip of a pilus structure. Conjugative elements of G+ bacteria do not form so-called sex-pili but probably encode adhesions to fulfill this function. While some proteins are involved in establishing contact, other surface-located proteins serve to inhibit conjugation between two donor cells, in a process named exclusion. In stage 2) the conjugation element is processed to generate the DNA that is transferred to the recipient cell, which in most cases is single-stranded DNA (ssDNA). The DNA processing reaction requires a relaxase, which forms a nucleoprotein complex called the relaxosome that introduces a site- and strand-specific nick within the origin of transfer (oriT). Host and/or plasmid-encoded proteins, named auxiliary proteins, often assist in the DNA processing reaction. During stage 3) a sophisticated membrane-embedded translocation machinery, also called transferosome, is synthesized through which the DNA is transferred. The intercellular transferosome, which is a form of type IV secretion system, is composed of at least eight but often more different proteins (Christie et al., 2014; Christie, 2016; Li et al., 2019; Waksman, 2019). Moreover, most of these proteins are present in multiple copies in the transferosome. The plasmid is recruited to the transferosome through interactions of an ATPase, named coupling protein (CP) and which is located at the cytoplasmic entry side of the transferosome, and the relaxosome proteins. Next, the relaxase pilots transfer of the ssDNA into the recipient cell in an energy consuming process that involves, besides the CP, one or two additional ATPases. Stage 4) is the establishment of the DNA once it has entered the recipient cell. Once transferred, the ssDNA has to be converted into double-stranded DNA (dsDNA). Many conjugative elements encode a primase that is involved in the conversion of ss to dsDNA. Finally, as most bacteria have evolved defense systems that inactivate incoming foreign DNA, including conjugative elements, conjugative elements encode proteins that transiently inactivate these defense mechanisms. So far, little is known about this step and the proteins involved.
Expression of the conjugation genes of most systems are strictly controlled because of at least two reasons. First, expression of the many proteins involved in the conjugation process imposes an energetic burden on the cell. Second, activation of the conjugation process has major consequences for both the plasmid and the host. Conjugative plasmids replicate by the theta type of replication mechanism during vegetative growth of the host. However, replication switches to the rolling-circle mode during conjugation in order to generate the ssDNA molecule that is transferred to the recipient cell (Smillie et al., 2010; Cabezon et al., 2014). Moreover, probably all conjugative plasmids possess a partitioning system to ensure that each daughter cell receives a copy of the plasmid upon cell division (Baxter and Funnell, 2014; Brooks and Hwang, 2017; Bouet and Funnell, 2019). During conjugation, however, the plasmid is recruited to the entry site of the transferosome. The conjugation process also has consequences for the host cell, as it alters characteristics of the membrane and the cell surface.
Studies on conjugative plasmids replicating in G- bacteria have shown that the conjugation genes are strictly controlled by a combination of plasmid and host-encoded factors. In particular, the mechanism controlling conjugation genes of the IncF incompatibility group has been studied in depth (Frost and Koraimann, 2010; Koraimann, 2018). Compared to G− bacteria, conjugation in G+ bacteria is less understood, and little is known about the regulation of conjugation genes (Singh and Meijer, 2014; Kohler et al., 2019). Exceptions are the two related enterococcal plasmids pCF10 and pAD1. Conjugation of these plasmids is induced by pheromones that are produced by plasmid-free recipient cells (for review see, Clewell, 2011; Dunny and Johnson, 2011; Chatterjee et al., 2013; Dunny and Berntsson, 2016).
The 65 kb plasmid pLS20 was originally identified in the Bacillus subtilis natto strain IFO3335 (Tanaka and Koshikawa, 1977) and is known to be conjugative in liquid as well as on solid media (Koehler and Thorne, 1987; Meijer et al., 1998; Itaya et al., 2006). Although very little was known about conjugation systems present on B. subtilis plasmids, as a host B. subtilis is one of the best studied G+ bacteria (Sonenshein et al., 1993; sonenshein et al., 2001). There are also other reasons why B. subtilis is an interesting host to study conjugation. B. subtilis is closely related to fastidious and pathogenic bacilli, and more distantly related to the Gram+ pathogen Listeria. B. subtilis is a soil-dwelling bacterium that is found all over the world; therefore pLS20 may be a representative of a group of globally distributed conjugative plasmids. Finally, B. subtilis strains are also gut commensals in animals and humans (Cutting, 2011), so it is possible that B. subtilis plasmids play a role in HGT in the gut.
Earlier studies on pLS20 have determined the replication region of the plasmid (Meijer et al., 1995), and shown that faithful segregation of the plasmid during cell division is ensured by the actin-like Alp7A protein (Derman et al., 2009). However, little was known about the conjugation process and how it was regulated. In this review we describe the advances that have been made in recent years in understanding how expression of the pLS20 conjugation genes is regulated, and identify the similarities and differences with two other conjugative systems of Gram+ origin. We also discuss future directions to unanswered questions. The most important gaps of knowledge concern structural insights in 1) how repressor molecules generate DNA looping, 2) how the antirepressor interacts with the repressor, and 3) if and how genes involved in different steps of the conjugation process–all located in a single large operon-are differentially expressed. More general reviews of regulation of conjugation in G+ and G− can be found elsewhere (Grohmann et al., 2003; Frost and Koraimann, 2010; Koraimann and Wagner, 2014; Singh and Meijer, 2014; Kohler et al., 2019).
Determination of conjugation efficiencies as a function of growth revealed that maximum conjugation efficiencies, in the range of 1 × 10−3, could be obtained only during a short window of time near the end of the exponential growth phase. Conjugation efficiency decreases sharply when donor cells enter the stationary phase, and eventually declines below the detection level (<1 × 10−8) (Singh et al., 2013). These results indicate that the conjugation process is strictly regulated. Conjugation efficiencies, which are also low at early time points, increase during the exponential growth phase. However, this increase in efficiency was not due to recipient-produced pheromones though as occurs for the enterococcal pheromone-responsive conjugative plasmids (Dunny, 2013), because similar results were obtained when the growth medium was replaced by fresh medium before mating (Singh et al., 2013). This demonstrated that regulation of pLS20 conjugation is fundamentally different from the enterococcal pheromone-responsive plasmids. The narrow time window of efficient conjugation does not depend on the growth phase of the recipient cells. Thus, pLS20 conjugation is strictly repressed except for a small window of time near the end of the exponential growth phase.
Our annotation of the plasmid revealed a large putative operon spanning genes 28-74 in which some genes share similarity with conjugation genes (Singh et al., 2013; see Figure 1A for a schematic view of plasmid pLS20). This indicated that genes 28-74 constitute a large conjugation operon, which was later confirmed (see below). A single divergently oriented gene, 27c, is located immediately upstream of the conjugation operon. Its product, protein p27c, is a putative Xre-type transcriptional regulator predicted to contain a typical DNA binding Helix-Turn-Helix (HTH) domain near its N-terminus. Protein p27c has been shown to be the master regulator of pLS20 conjugation and was named RcopLS20 (regulator of conjugation of pLS20, see Figure 1B) (Singh et al., 2013). Ectopic overexpression of rcopLS20 (= gene 27c) from a chromosomal location blocked conjugation, and conjugation levels were constitutively high in a derivative of pLS20 lacking the gene (pLS20rco). Plasmid pLS20rco was very unstable in wild type B. subtilis cells and its integrity required the ectopic expression of gene rcopLS20 in the host, suggesting that constitutive expression of the conjugation operon is generating high levels of stress to the host. Evidence that RcopLS20 is a transcriptional regulator was obtained by RNAseq analyses (Singh et al., 2013). Thus, genes within the conjugation operon were expressed among the highest level of all plasmid-encoded genes when samples were taken from donor cell cultures at late exponential growth phase, but their levels were >16 fold lower for samples of cells in which rcopLS20 was ectopically overexpressed.
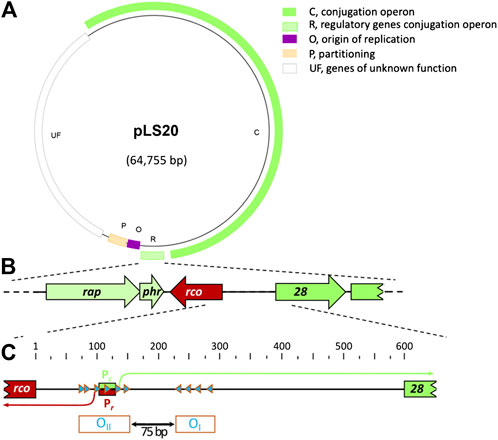
FIGURE 1. General characteristics of pLS20 and genetic organization of the pLS20 region involved in regulation of the conjugation genes. (A). Map of pLS20 (accession number AB615352.1). Different modules are indicated with colors. Light green box, region containing the three genes regulating the activity of the main conjugation promoter Pc (indicated with “R”); dark green box, conjugation operon (indicated with “C”). Purple box, origin of replication region (indicated with “O”); orange box, partitioning genes (indicated with “P”); and white box, module with unknown genes (indicated with “UF”). (B). Blow-up of the region encoding the genes rappLS20 (rap), phrpLS20 (phr) and rcopLS20 (rco) involved in regulating the expression of the conjugation genes, together with the 5’ region of the conjugation operon. (C). Blow-up of the gene 28-rcopLS20 intergenic region and its features involved in regulating expression of the conjugation genes. The position of the divergently oriented and overlapping promoters Pc and Pr are indicated with green and red boxes, respectively. Transcription initiation sites are indicated with bent arrows and the RcopLS20 binding sites with blue triangles. The numbers at the top correspond to bp of the almost 600 bp intergenic region. The operators OI and OII, which are separated by a 75 bp spacer, are indicated at the bottom.
Several analyses, including RNAseq, deletion analyses and primer extension experiments revealed that the promoters driving expression of the divergently oriented genes rcopLS20 and gene 28, the first gene in the conjugation operon, are located in the 595 bps intergenic region (Ramachandran et al., 2014; see also Figure 1). The promoter upstream of rcopLS20 is named Pr, and the one upstream of the conjugation operon Pc. The Pc promoter is located 462 bp upstream of gene 28 and, interestingly, overlaps with the divergently oriented Pr promoter. The strength of the two promoters has been determined using transcriptional lacZ fusions (Ramachandran et al., 2014). These analyses showed that the conjugation promoter Pc is a strong promoter, and promoter Pr, driving expression of rcopLS20, is a very weak promoter whose activity was barely detectable. Surprisingly, analyses of additional strains containing, besides a transcriptional lacZ fusion, either pLS20 or a copy of rcopLS20 under the control of an inducible promoter, revealed that RcopLS20 does not just repress the strong promoter Pc, but also regulates its own promoter, Pr. At low expression levels RcopLS20 stimulates the activity of Pr but at high expression level it represses its own promoter (Ramachandran et al., 2014). It is important to mention that the maximum strength of the Pr promoter is about 50–75-fold lower than that of the Pc promoter (Singh et al., 2020). In summary, RcopLS20 represses the strong promoter Pc and simultaneously regulates the activity of its own promoter Pr (Ramachandran et al., 2014).
The binding regions of RcopLS20 were initially determined by analyzing the response of Pr and Pc promoters to ectopically expressed rcopLS20, using strains containing lacZ fused to promoter-containing fragments of different lengths (Ramachandran et al., 2014). This approach revealed that proper regulation required the presence of two DNA regions. These two regions contain 10 octamer boxes whose sequences are (nearly) identical to CAGTGAAA (see Figure 1C for a schematic view). In the first region a cluster of six of these boxes, all in the same orientation, are located near, and overlapping with the Pr and Pc promoters. The second region contains four clustered boxes whose orientation is opposite to those present in the first region. The two clusters of boxes are separated by 75 bp. Binding of RcopLS20 to these boxes was demonstrated by DNase I footprinting assays, and also by gel retardation experiments using fragments with/out mutation in the octamers (Ramachandran et al., 2014). The regions containing the four and six octamers are named operator OI and OII, respectively (see Figure 1C).
The presence of two convergently oriented RcopLS20 operators raised the possibility that binding of RcopLS20 to these operators could result in DNA looping. However, the spacer region between the two operators is only 75 bp. Due to intrinsic stiffness of DNA spacer regions in DNA-loops are generally larger than 90 bp. DNA loops containing smaller spacer lengths are only possible when the DNA region is intrinsically bent. Circular permutation assays showed that the region containing the RcopLS20 operators is indeed intrinsically bent and that the center of the bent is located in approximately the middle of the 75 bp spacer region between operators OI and OII (Ramachandran et al., 2014). Several lines of evidence supported that binding of RcopLS20 to operators OI and OII results in DNA looping. First, ultracentrifugation analysis showed that RcopLS20 forms tetramers in solution (Ramachandran et al., 2014), which is in line with the observation that proteins generating DNA looping commonly form multimers in solution. Second, particularly for short spacer regions, DNA looping requires a certain phasing between the two operators so that operator-bound proteins are in the required configuration to generate the DNA loop. Enlarging the spacer region between the two operators with a half helical turn (i.e., 5 bp) disrupted RcopLS20 mediated regulation of the Pc promoter in vivo (Ramachandran et al., 2014). Finally, RcopLS20-mediated DNA looping was further supported by gel retardation and ultracentrifugation experiments, which also provided important insights into the mode of DNA binding (Singh et al., 2020). DNA looping may be generated as a consequence of one of the two fundamentally different modes by which tetrameric RcopLS20 units bind DNA, as schematically shown in Figure 2A. The difference between these two modes of DNA binding can be distinguished by gel retardation experiments using DNA fragments containing only one operator, as the second binding mode generates a maximum of two retarded species, while the first binding mode would allow the generation of more than two retarded species. Retardations using DNA fragments containing either operator OI or OII both generated a maximum of two retarded species (Ramachandran et al., 2014), indicating that DNA looping occurs through interaction between two RcopLS20 tetramers each bound to an operator. This conclusion was supported by additional gel retardation experiments using two DNA fragments, each containing one operator but having different lengths and distinct flanking sequences, allowing the fragments to be distinguished by Southern blotting. Gel retardation using a mixture of the small and large DNA fragment resulted in the appearance of an additional retarded species composed of a small and a large DNA fragment (Singh et al., 2020). Furthermore, SAXS analysis showed that RcopLS20 forms complexes of increased size at concentrations above 200 μM, indicating that RcopLS20 tetramers can interact with each other (Crespo et al., 2020), which would be expected if DNA binding occurs via the second mode. The combined evidences strongly indicate that efficient repression of the Pc promoter and simultaneous activation of the Pr promoter by RcopLS20 is achieved through DNA looping as a consequence of interaction between the RcopLS20 tetramers bound to operators OI and OII. We cannot fully excluded the possibility that, in addition to DNA looping, binding of RcopLS20 also introduces other effects on the DNA configuration or orientation.
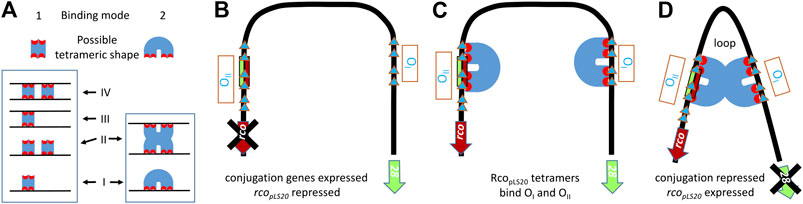
FIGURE 2. Probable mode of DNA binding by RcopLS20 to the intrinsically bent DNA region encompassing RcopLS20 operators OI and OII.(A). Schematic representation of two different binding modes by which tetrameric DNA binding proteins can bind a DNA fragment containing one operator. Binding modes 1 and 2 can generate up to four and two different protein-bound DNA configurations, respectively. (B–D). Schematic view of conjugation unrepressed state and repressed state involving DNA looping due to binding of RcopLS20 to operators OI and OII according to binding mode 2 explained in panel A. (B). Unrepressed state. The conjugation operon starting with gene 28 is derepressed when the operators OI and OII are not bound by RcopLS20 tetramers. (C). RcopLS20 tetramers can bind to operators OI and OII, resulting in a local high concentration and probably a specific configuration stimulating interaction between the two tetramers. (D). Repressed state. Interaction between the two RcopLS20 tetramers bound to operators OI and OII results in repression of the conjugation genes and simultaneous activation of rcopLS20. Blue triangles: RcopLS20 binding sites. Blue U shapes: RcopLS20 tetramers.
rcopLS20 is under the control of a very weak promoter (see above). Nonetheless, despite the low levels of RcopLS20 present in the cell, the conjugation process is strictly controlled as the number of transconjugants is below the detection level under conjugation-unfavorable conditions. Probably, this is achieved through DNA looping because it causes a high local concentration of the transcriptional regulator at the right location, which has been shown to be able to increase its specificity and affinity, and concomitantly controls the stochasticity of cellular processes (Vilar and Saiz, 2005; Oehler and Muller-Hill, 2010). Thus, DNA looping allows strict regulation at low protein concentration, while remaining sensitive for accurate activation of conjugation when appropriate conditions –as detailed in the following section-occur. Examples of DNA looping playing a crucial role in transcriptional regulation have been described before. One of the best-studied examples is DNA looping exerted by the CI protein and which is crucial for the genetic switch between the lysogenic and lytic state of the Escherichia coli phage λ (for review see, Dodd et al., 2005; Oppenheim et al., 2005). There are clear differences between the DNA looping mechanisms of λ and pLS20. Whereas RcopLS20 forms tetramers in solution, CI forms dimers. In addition, the region separating the operators is about 2.3 kb in phage λ, and 75 bp in pLS20.
Expression of the conjugation genes requires suspension of RcopLS20-mediated repression of the conjugation promoter Pc. In case of pLS20, the repressor RcopLS20 is inactivated by an antirepressor, RappLS20. Annotation of pLS20cat revealed that a two-gene rap-phr cassette is located downstream of rcopLS20, with direction of transcription convergent to that of rcopLS20 (see Figure 1B). Rap proteins belong to a large family of signal-peptide receptor proteins of the so-called RRNPP family, which is named after its prototypical members Rap, Rgg, NprR, PlcR and PrgX (for review see, Declerck et al., 2007; Rocha-Estrada et al., 2010; Dunny and Berntsson, 2016; Neiditch et al., 2017). RRNPP proteins, which are encoded by many bacteria belonging to the phylum of Firmicutes, are generally co-transcribed with a phr gene encoding the cognate signaling peptide. The Phr signaling peptides are synthesized as a pre-proprotein. After being secreted they are cleaved again to generate the mature peptide, which in most cases correspond to the C-terminal region of the pre-proprotein, and which can be imported into the cell by the oligopeptide permease system (Pottathil and Lazazzera, 2003; Monnet et al., 2016). The chromosome of B. subtilis contains eight rap-phr cassettes, and some Bacillus plasmids also contain such cassettes. Many of these rap-phr cassettes, including those on plasmids, function to interfere with developmental processes such as sporulation, competence and degradative enzyme production (Perego, 2001; Bongiorni et al., 2006; Smits et al., 2007; Parashar et al., 2013). Ectopically overexpressing the rap-phr cassette of pLS20 in B. subtilis did not seem to affect developmental processes of the host. However, it strongly affected the conjugation process, resulting in 1) increased maximum conjugation levels, 2) overexpression of genes located in the conjugation operon, and 3) allowing conjugation to occur during a much broader time window. Moreover, conjugation levels were severely affected for a pLS20 derivative lacking rap-phr (Singh et al., 2013). Addition of a synthetic peptide corresponding to the five C-terminal residues of the unprocessed Phr peptide also inhibited conjugation, showing that the activity of Rap is regulated by the mature signaling peptide (Singh et al., 2013). Based on these results the genes were named rappLS20 and phrpLS20 and the mature Phr peptide was referred to as Phr*pLS20.
The results outlined above suggest that RcopLS20, RappLS20 and Phr*pLS20 are the only pLS20-encoded proteins involved in regulation of the conjugation genes. This was confirmed using an in vivo system in which the regulatory components were uncoupled from their native setting in a B. subtilis strain where the rcopLS20 and rappLS20 genes are placed under different inducible promoters, and a lacZ reporter gene fused to the main conjugation promoter Pc. The main conjugation promoter Pc was active in the absence of both rcopLS20 and rappLS20 expression. Pc was repressed when rcopLS20 alone was induced, strongly supporting the view that RcopLS20 is directly responsible for repressing the Pc promoter, which is in line with in vitro results (see above). The Pc promoter became active when both rcopLS20 and rappLS20 were induced, showing that RappLS20 alone is sufficient to relieve RcopLS20-mediated repression of the Pc promoter. Finally, when both rcopLS20 and rappLS20 were induced in the presence of the mature peptide Phr*pLS20 (added to the growth medium), the Pc promoter remained inactive, demonstrating that Phr*pLS20 is required and sufficient to inactivate RappLS20 (Singh et al., 2020).
The interplay between RcopLS20, RappLS20 and Phr*pLS20 requires an understanding of the relative strengths of their promoters, as well as the activity of promoter Pc, which these proteins regulate. The strengths of Pc, Pr and the promoter driving expression of rappLS20 and phrpLS20, named Prap, were determined using strains containing gfp reporter cassettes in which either of these promoters was fused to the gfp gene (Singh et al., 2020). The results obtained with the gfp-fusion strains confirmed that Pc and Pr are a strong and a very weak promoter, respectively. Prap was found to be a weak promoter, whose strength was similar to the Pr promoter in cells containing pLS20, and about 50 to 75-fold lower than that of Pc (see Figure 3).
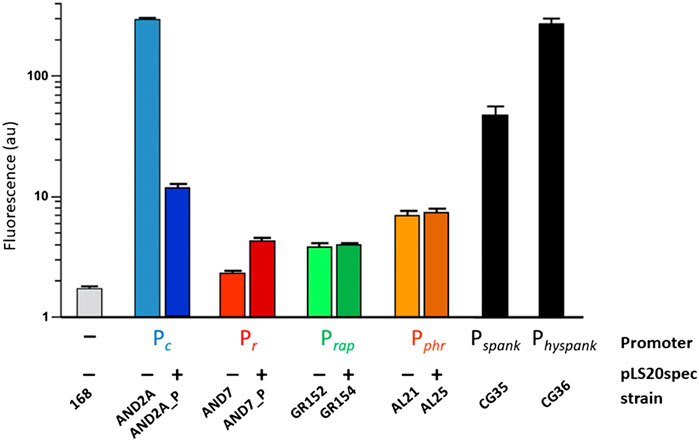
FIGURE 3. Relative strength of promoters Pc, Pr, Prap and Pphr determined by FACS analysis of strains containing transcriptional gfp fusions. Relative promoter strengths determined by cytometry using strains containing promoter Pc, Pr, Prap or Pphr transcriptionally fused to gfp. A negative control strain (-) and two positive control strains that contain gfp fused to the IPTG-inducible promoter Pspank or Physpank were included. Values correspond to late exponentially growing cultures (OD600 between 0.8-1). Light and dark colored bars reflect strains lacking or containing pLS20spec, respectively. The figure is adapted from (Singh et al., 2020).
Prap drives the expression of rappLS20 and phrpLS20. However, the concentration of PhrpLS20 will drop once it is exported from the cell and it is plausible that proper signaling of Phr*pLS20 requires higher expression levels than that of RappLS20. Analysis of an additional strain containing gfp fused to the upstream region of phrpLS20 revealed that the expression of phrpLS20 indeed is controlled by a second promoter, named Pphr, whose activity is about two-fold higher than that of Prap. Unlike the Pc and Pr promoters, the activities of Prap and Pphr were not affected by the presence of pLS20 in the cell, indicating that RcopLS20 does not regulate these promoters. The absence of RcopLS20 binding motifs in the vicinity of the Prap and Pphr promoters supports this conclusion (Singh et al., 2020). In summary, 1) the conjugation genes are under the control of the strong Pc promoter, 2) the rcopLS20 and rappLS20 genes, driving expression of the proteins regulating the activity of the Pc promoter, are preceded by very weak promoters, 3) expression of phr is controlled by Prap and additionally by the two-fold stronger promoter Pphr, presumably to accommodate for the dilution effect upon secretion, and 4) RcopLS20 represses Pc and regulates the Pr promoter, but not promoters Prap and Pphr.
The multi-layered regulation of the Pc promoter in combination with the Rap/Phr-based quorum sensing mechanism might be expected to cause heterogeneous activation of the Pc promoter in a population of donor cells. This seemed not to be the case though, because flow cytometry analyses of samples of B. subtilis cells having a chromosomal cassette containing the Pc-gfp fusion, or those harboring in addition pLS20, both showed homogeneous activity of the Pc promoter (Singh et al., 2020). In this set up, the Pc promoter was present on the bacterial chromosome whereas proteins regulating its activity were encoded by the resident plasmid containing its own Pc promoter. A homogeneous pattern of Pc promoter activity was also found for cells containing a derivative of pLS20cat in which a copy of the gfp gene was placed on pLS20 behind gene 28, the first gene of the conjugation operon. Together, these results strongly indicate that the multi-levels of regulation results in a sensitive genetic switch that activates the Pc promoter in a coordinated manner in most or all pLS20-containing cells (Singh et al., 2020). This does not automatically imply that activation of the Pc promoter guarantees successful plasmid transfer or even the initiation of the conjugation process in all cells. In fact, in most if not all conjugation systems studied, the full conjugation process appears to be activated only in a subpopulation of the donor cells (reviewed in, Koraimann and Wagner, 2014; Stingl and Koraimann, 2017). In the case of pLS20, checkpoints may be present downstream the Pc promoter. Moreover, successful transfer may not occur even when all conjugation genes are expressed due to, for example, unsuccessful mating pair formation or failure of the plasmid to establish itself in the new host. Moreover, it should be noted that the laboratory conditions under which conjugation is studied are very different from natural conditions in which environmental fluctuations at macro and microscale are likely to affect individual cells or subpopulations.
All members of the RRNPP family of proteins analyzed so far, are all-helical proteins composed of a large C-terminal domain (CTD) and a much smaller N-terminal domain (NTD). Whereas the large CTD interacts with its cognate signaling peptide resulting in conformational changes that affects the activity of the protein, the small NTD constitutes the effector domain that interacts with a target molecule ( D’Andrea and Regan, 2003; Rocha-Estrada et al., 2010; Neiditch et al., 2017). Although the effector domains of all RRNPP proteins are composed of three α-helices, they exert their regulatory functions in three fundamentally different ways. In the first subset of RRNPP proteins these three α-helices have evolved into Helix-Turn-Helix (HTH) DNA binding motifs, which allow them to negatively regulate protein expression by binding to DNA. Examples of such RRNPP proteins are PlcR from Bacillus thuringiensis (Declerck et al., 2007) and PrgX from Enterococcus faecalis (Shi et al., 2005; Dunny and Berntsson, 2016). Particularly the PrgX protein is interesting in the context of this review because it regulates the conjugation genes present on the plasmid pCF10 (see below). In the second subset of RRNPP proteins, the effector domain has phosphatase activity. In B. subtilis, the developmental process of sporulation is induced by the phosphorylated form of the master regulator Spo0A, Spo0A∼P. Spo0A phosphorylation is controlled by a phosphorelay consisting of several kinases and two phosphorelay proteins, Spo0F and Spo0B (Hoch, 1993; Grossman, 1995; Molle et al., 2003; Chastanet and Losick, 2011). Several Rap proteins, including RapA, RapB, RapE, RapH and RapJ, can interrupt the phosphorylation of Spo0A by dephosphorylating Spo0F∼P. RapA was the first Rap member discovered to have this activity and was named Rap, which stands for Response regulator Aspartate Phosphatase (Perego, 2001). Finally, in the third subset of RRNPP proteins, the effector domain binds to its cognate effector protein, which impedes the function of its cognate effector protein (in)directly by modulating the expression of the differentiation pathway involved. Examples of these are RapC, RapF, RapG, RapH and RapK (note that not all Rap proteins have phosphatase activity). It is also worth mentioning that the effector domain of some RRNPP proteins, for instance RapH and NprR, exhibit both activities.
Based on the features of the known RRNPP proteins, one possibility of how RappLS20 might activate the pLS20 conjugation genes is by competing with RcopLS20 for binding to DNA near the Pc promoter. However, both gel retardation and analytical ultracentrifugation (AUC) analyses showed that RappLS20 has no DNA binding activity, and the NTD of RappLS20 does not adopt a HTH configuration (Crespo et al., 2020; Singh et al., 2020). Instead, RappLS20 seems to exert its antirepressor activity by binding directly to RcopLS20, as bacterial two-hybrid analysis showed that RappLS20 and RcopLS20 interact in vivo (Singh et al., 2020). In vitro evidence that RappLS20 interacts directly with RcopLS20 has also been obtained by SAXS and analytical size exclusion chromatography (Crespo et al., 2020). Furthermore, AUC analyses provided conclusive evidence that RappLS20 forms predominantly dimers in solution. Sedimentation velocity experiments of mixtures of RappLS20 and RcopLS20 resulted in the appearance of a novel species whose deduced molecular weight was compatible with a complex of one RappLS20 dimer and one RcopLS20 tetramer (Singh et al., 2020). This conclusion was reinforced by multi-signal sedimentation velocity (MSSV) analysis from which it was determined that the heterocomplex was composed of a stoichiometry of 2.1 mol of RcopLS20 per mol of RappLS20. More importantly, besides demonstrating that RappLS20 interacts with free RcopLS20, AUC analyses also showed that RappLS20 is able to interact with DNA-bound RcopLS20, and that this results in the release of RcopLS20 from the DNA and the concomitant formation of RappLS20/RcopLS20 complexes. Complementary gel retardation experiments provided evidence that RappLS20 preferentially interacts with RcopLS20 tetramers involved in DNA looping (Singh et al., 2020). In summary, RappLS20 appears to activate the conjugation genes by preferentially interacting with RcopLS20 involved in DNA-looping, resulting in the detachment of RcopLS20 from its operators, and forming a heterocomplex in which one RappLS20 dimer binds one RcopLS20 tetramer. A schematic view of how RappLS20 activates the conjugation genes is shown in Figure 4.
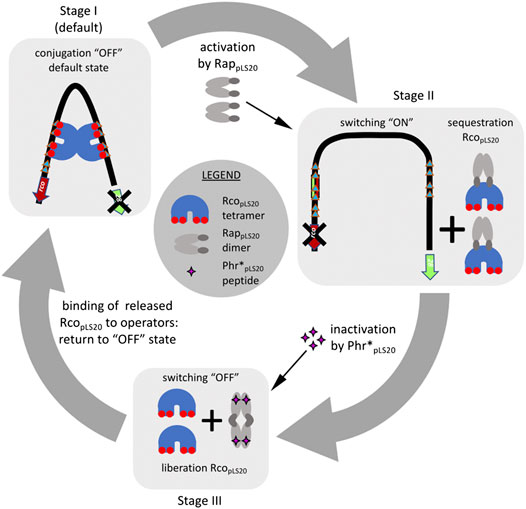
FIGURE 4. Overview of the current understanding of how the pLS20 conjugation genes are regulated by RcopLS20, RappLS20 and Phr*pLS20. Stage I, as default, the two RcopLS20 tetramers bound to operators OI and OII interact with each other resulting in DNA-looping. This configuration causes a steady state in which the strong conjugation promoter Pc is repressed and the weak Pr promoter driving expression of rcopLS20 is simultaneously stimulated. Stage II, the conjugation genes are activated when two apo-form of RappLS20 dimers detach two RcopLS20 tetramers from the DNA through direct interaction and generating RappLS20:RcopLS20 heterocomplexes in a stoichiometry of 1:2. Stage III, binding of the Phr*pLS20 signaling peptide provokes a conformational change of RappLS20 that stimulates tetramerization of RappLS20 via the so-called foot-2-foot configuration in which the N-terminal effector domains interact with each other and are, presumably, no longer available to interact with RcopLS20. The liberated RcopLS20 tetramers are then free to bind its operators thereby returning the system to the default conjugation repressed “OFF” state (stage I).
The presence of the mature signaling peptide Phr*pLS20 suppresses the RappLS20–mediated activation of the conjugation genes (see above). For other RRNPP members it has been shown that signaling peptides interact directly with the CTDs of their cognate RRNPP proteins, causing inactivation through peptide-induced conformational changes of the RRNPP protein. As outlined below, this is also the case for RappLS20 and Phr*pLS20. Phr*pLS20 binds directly to RappLS20 (see below) and the dissociation constant, Kd, for Phr*pLS20 was determined to be 2.1 and 7.4 μM by ultracentrifugation and fluorescence anisotropy approaches, respectively (Crespo et al., 2020; Singh et al., 2020). Although AUC analysis showed that RappLS20 forms predominantly dimers in solution (Crespo et al., 2020, see also above), it seems to have an intrinsic ability to form tetramers at higher concentrations, as 1.1% of RappLS20 was detected as tetramers in solution at 4.5 μM, and this percentage increased to 3.6% at 25 μM. Interestingly, AUC analysis showed that in the presence of Phr*pLS20, RappLS20 forms almost exclusively tetramers in solution (Crespo et al., 2020). The structures of the apo and peptide-bound forms of RappLS20 have been determined recently, which show that RappLS20 forms the typical structure of RRNPP family proteins (Crespo et al., 2020). RappLS20 is an all α-helical protein (17 antiparallel helices) that is composed of a large 14-helix C-terminal domain (CTD) that can bind the signal peptide, and a much smaller 3-helix N-terminal domain (NTD). The three helices of the NTD do not form a HTH topology, but have an approximate parallel configuration. The N- and CTDs are separated by a 13-residue long flexible loop that connects α-helices three and four. RappLS20 dimerizes through interactions between the CTDs, and these interactions involve helices H5-H7, H16-H17 and the C-terminal residues.
The structure of the peptide-bound form of RappLS20 showed that Phr*pLS20 binds in a cavity of the CTD. Binding of the Phr*pLS20 peptide causes the N-terminal domains to swing outwards thereby favoring RappLS20 to form tetramers (Crespo et al., 2020). Interestingly, the dimer-dimer interface is principally formed between the NTDs of each dimer, resulting in a so-called foot-2-foot interaction (see stage III in Figure 4 for a schematic view). The small NTDs of RRNPP proteins form the effector domain and this is most likely also the case for RappLS20, implying that the NTD of RappLS20 interacts with RcopLS20. Due to the particular foot-2-foot configuration induced upon peptide binding, the N-terminal effector domain is no longer capable of interacting with RcopLS20 to relieve repression, providing the molecular mechanism by which Phr*pLS20 inactivates the antirepressor activity of RappLS20. Phr*pLS20 can also bind to RappLS20 complexed with RcopLS20. This conclusion is based on the results of size exclusion chromatography and SAXS experiments, which showed that the addition of Phr*pLS20 peptide to preformed RappLS20/RcopLS20 complexes resulted in disruption of these complexes and the appearance of elution and SAXS patterns that were similar to those of RcopLS20 alone and RappLS20-Phr*pLS20 complexes (i.e., RappLS20 tetramers) (Crespo et al., 2020).
Under certain growth conditions, B. subtilis can develop the state of natural competence in which cells can bind and adsorb extracellular DNA into the cytosol after which it can be integrated into the bacterial genome via homologous recombination (Dubnau, 1991; Hamoen et al., 2003). The processes of conjugation and competence have in common that they transport DNA via a sophisticated membrane-embedded DNA translocation machinery, but in opposite directions: DNA is exported in the case of conjugation, but imported during competence development. In both cases, only one of the two DNA strands (i.e., an ssDNA strand) is transported, and various other similarities exist between competence and conjugation related ssDNA transfer machines (Chen et al., 2005). However, simultaneous import and export of large ssDNA regions by the same cell may be incompatible for several reasons. For instance, simultaneous production and assembly of the ssDNA translocation machineries for competence and conjugation may interfere with each other and/or they may compete for the same cellular positions. In addition, the recombination enzymes synthesized during competence may act on ssDNA of the conjugative element. The following evidence indeed supports that conjugation and competence are incompatible processes and that at least two reciprocal mechanisms have evolved to avoid simultaneous activation of the competence and conjugation pathways in the same cell. Competence development is strongly suppressed in cells containing pLS20 by a plasmid-encoded protein called RokpLS20. RokpLS20 shares similarity with B. subtilis Rok, which is a repressor of comK that encodes the transcriptional activator of the recombination and the structural competence genes. As for Rok, RokpLS20 suppresses competence by repressing comK (Singh et al., 2012).
Like conjugation, competence is also induced by quorum sensing signals. The two-component ComP-ComA system plays a crucial role in the induction of competence: upon sensing the proper signals, the membrane-embedded kinase ComP activates ComA through phosphorylation which then activates competence genes, leading ultimately to the activation of the master regulator gene comK (Hamoen et al., 2003). RapF, encoded by the B. subtilis chromosome, inhibits stimulation of the competence pathway by interacting with ComA thereby preventing ComA from activating gene expression. RapF is cotranscribed with phrF that encodes the pre-proprotein which, after being processed, generates the signaling peptide PhrF*. High levels of PhrF* stimulate competence by inactivating RapF. Intriguingly, Phr*pLS20 and PhrF* are very similar: residues at positions 1, 3 and 4 are identical; position 2 concerns a conserved substitution of Lysine to Arginine, and position 5 a change from Tyrosine to Isoleucine. PhrF* has been shown to cross talk with RappLS20 (Singh et al., 2020). Thus, PhrF* can interact with RappLS20 with a Kd that is only 2.5-fold higher than that of Phr*pLS20 as determined by AUC (5.3 and 2.1 μM, respectively), and interaction with PhrF* results in tetramerization of RappLS20. More importantly, PhrF* is able to inactivate RappLS20in vivo when tested using the minimal regulatory system of pLS20 conjugation (see above).
All the evidences obtained so far show that the conjugation genes of pLS20 are regulated in a highly sophisticated manner ensuring that conjugation is only activated under conditions that favor a successful conjugation event, i.e. when potential recipient cells are present. A schematic view of the current knowledge of how the conjugation genes are regulated is presented in Figure 4. The main and strong conjugation promoter Pc located upstream of the conjugation operon is repressed by RcopLS20, which also controls the activity of its own weak promoter Pr to ensure that only low levels of RcopLS20 are produced. Despite these low levels, the tetrameric RcopLS20 efficiently represses the Pc promoter. This is most likely achieved through DNA looping that ensures high local concentration of RcopLS20 and simultaneous binding to multiple binding sites in the two operators. The low levels of RcopLS20 probably are also crucial to allow for a sensitive and rapid switch to activate the conjugation genes when favorable conditions occur. Activation of the conjugation genes occurs by the action of the dimeric form of RappLS20 that relieves RcopLS20-mediated repression of the Pc promoter. Binding of RappLS20 to RcopLS20 results in detachment of RcopLS20 from the DNA, and hence it is the apo form of RappLS20 that activates conjugation. Accumulating levels of Phr*pLS20 cause inactivation of RappLS20 by inducing a conformational change of RappLS20 that favors the formation of tetramers. Intriguingly, tetramerization occurs through interactions between the N-terminal effector domains with the consequence that these are presumably no longer available to interact with RcopLS20. Consequently, RappLS20 tetramerization results in the liberation of RcopLS20 from the RappLS20/RcopLS20 complexes, which are then available to bind its operators and thereby returning the system to the default “OFF” state. Therefore, as schematically shown in Figure 5, the activation state of the pLS20 conjugation genes is ultimately regulated by the concentration of the Phr*pLS20 signaling peptide. When all or a majority of the cells within a population contain the plasmid, Phr*pLS20 concentrations will be high and conjugation will be inhibited. The conjugation genes will become activated when Phr*pLS20 concentrations are low, which occurs when donor cells form a minority of the population and are surrounded by many potential recipient cells. In fact, the presence of recipient cells has a dual effect on lowering the Phr*pLS20 concentration: 1) the relative number of donor cells, which produce Phr*pLS20, will be lower in populations containing recipient cells, and 2) recipient cells can actively adsorb the signaling peptide. Finally, it is not known if the RappLS20-Phr*pLS20 complexes are destined for rapid degradation, or whether there exists a mechanism to recycle RappLS20 by removing Phr*pLS20 from the complex, thereby stimulating the activation of the conjugation pathway. It should be noted, however, that the promoter Prap driving expression of rappLS20 and phrpLS20 has been shown to be constitutive, thus resulting in de novo synthesis of cytosolic RappLS20, and PhrpLS20 that is destined for secretion (Singh et al., 2020).
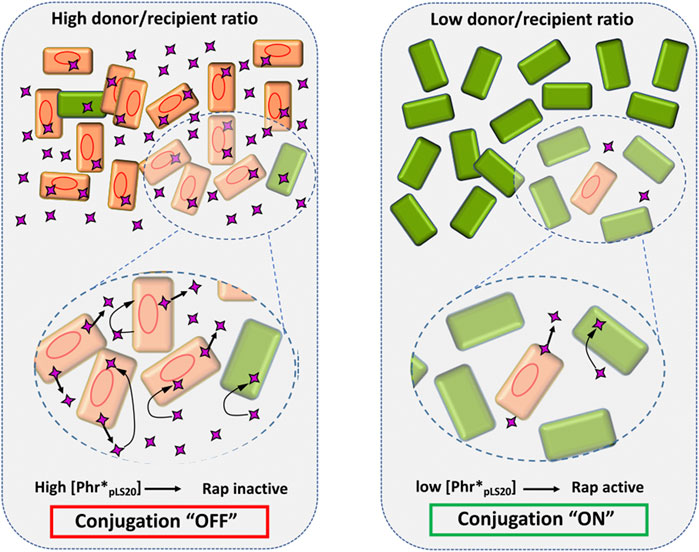
FIGURE 5. The concentration of the signaling peptide Phr*pLS20 determines the activation state of the conjugation genes. Schematic view of how a high and low donor/recipient ratio (left and right panel, respectively) affects the Phr*pLS20 concentration, and thereby the activation state of the conjugation process. Donor and recipient cells are indicated with orange and green rectangles, respectively. Red oval lines, pLS20; purple four-pointed star, signaling peptide. The amplified regions in the lower part illustrate secretion of the signaling peptide by donor cells, and its adsorption by donor and recipient cells as indicated by straight and bent arrows, respectively. Note that for simplicity only the mature form of the signaling peptide is shown. See text for details.
Signaling peptide receptor proteins belonging to the RRNPP family all have a similar structure that is composed of a small 3-helix N-terminal effector domain and a much larger CTD that, upon interaction with its cognate signal peptide, undergoes a conformational change affecting the functionality of the protein. Despite these conserved features, evolutionary adaptations have led to major effects on the mechanistic action of these proteins. This is exemplified here by comparing the three RRNPP members that regulate the conjugation genes of the conjugative plasmids pLS20 from B. subtilis, pCF10 from Enterococcus faecalis, and those of the ICEBs1 element that is present in many B. subtilis strains. The N-terminal effector domain of the pCF10-encoded RRNPP protein PrgX has evolved into a HTH DNA-binding domain. Remarkably, PrgX can interact with two signal peptides that cause opposing effects on its DNA binding. One of these signal peptides, iCF10, is encoded by the plasmid, and the other, cCF10 (the pheromone) by recipient cells. PrgX bound to the plasmid-encoded iCF10 adopts a conformation in which PrgX binds DNA resulting in repression of the conjugation genes. However, when PrgX binds to cCF10, DNA is released resulting in activation of the conjugation genes (for recent review see, Dunny and Berntsson, 2016). The regulatory system of pCF10 differs therefore in two important aspects from that of pLS20. First, in the case of pCF10 recipient cells are directly responsible for activation of the conjugation genes, and second, regulation of the conjugation genes is controlled by one instead of two regulatory proteins (PrgX vs. RappLS20 and RcopLS20).
The conjugation genes of ICEBs1 are repressed by a regulator named ImmR; an analogue of RcopLS20. Relieve of ImmR-mediated repression can occur in two ways. First, as a consequence of RecA-dependent SOS response to DNA damage, or second, by a mechanism that involves the ICE-encoded RRNPP protein RapI (Auchtung et al., 2007; Bose et al., 2008). However, activation of the conjugation genes by RapI occurs in a different way to RappLS20. RapI is required but not sufficient to activate the conjugation genes; a protease encoded by gene immA that degrades ImmR in a RapI-dependent manner is also needed. The exact mechanism of how RapI stimulates degradation of ImmR by ImmA is not yet known (Bose et al., 2008).
The presence and absence of a protease dedicated to degrade the repressor is likely to have important effects on reversibility of switching on and off of the conjugation pathway. Similar to RcopLS20, ImmR represses the conjugation promoter and simultaneously activates its own promoter (Auchtung et al., 2007). Degradation of ImmR will therefore result in activation of the conjugation genes and concomitantly inhibits de novo ImmR synthesis, causing conjugation of the ICEBs1 to be an irreversible process. In the case of pLS20, conjugation may be a reversible process, or at least it may be more flexible than the ICEBs1 system, because, rather than being degraded, RcopLS20 becomes temporally sequestered by RappLS20. Inactivation of RappLS20 by PhrpLS20 results in the release of RcopLS20, which can then bind to DNA and again repress the conjugation genes. Interaction of RappLS20 with RcopLS20 might inactivate RcopLS20 by altering its oligomerization state, in a similar way that RapF causes dissociation of its dimeric interacting partner ComA upon binding (Griffith and Grossman, 2008; Baker and Neiditch, 2011; Wolf et al., 2016). However, results of different analyses compellingly show that RcopLS20 is released as functional tetramers from the RappLS20-RcopLS20 complex upon addition of Phr*pLS20, lending further support for the view that activation of the pLS20 conjugation genes can be a reversible process.
Studies of the past years have resulted in major advances in understanding how the conjugation genes of pLS20 are regulated. Yet, multiple questions remain unanswered. Several of these concern multimerization of RcopLS20 and how RcopLS20 interacts with DNA and with RappLS20. In silico analysis indicates that RcopLS20 contains a HTH motif in its N-terminal region, which probably will be responsible for DNA binding. Therefore, the CTD of RcopLS20 is presumably involved in tetramerization, but how the RcopLS20 monomers interact to form a tetramer is unknown. Currently, it is also not exactly known how two tetramers interact when they are bound to DNA and induce DNA looping. Binding to DNA may generate a conformational change in the tetrameric structure of RcopLS20 that favors octamerization. Alternatively, octamerization may be favored by a local high concentration as the consequence of RcopLS20 binding to the intrinsically bent region containing operators OI and OII. The interacting surface between RcopLS20 and RappLS20 is also not known. RappLS20 may bind the DNA binding surface of RcopLS20 in an analogous way as RapF interacts with the DNA binding domain of ComA (Baker and Neiditch, 2011). However, the observation that RappLS20 appears to bind preferentially to RcopLS20 tetramers that are involved in DNA looping (see above) suggests that RappLS20 binds preferentially, but not exclusively, to the RcopLS20 surface generated upon dimerization of two RcopLS20 tetramers; this interface most likely does not involve the DNA binding domains. Various experimental approaches have shown that RcopLS20 does not bind DNA when it forms a complex with RappLS20 (Crespo et al., 2020; Singh et al., 2020). This may be because RappLS20 binds the DNA binding domain of RcopLS20. Alternatively, the formation of an RcopLS20/RappLS20 complex may induce a conformational change in RcopLS20 affecting its DNA binding ability. Answers to several or all of these questions may be obtained by unraveling the apo structure of RcopLS20 and the structures of RcopLS20 complexed with DNA and with RappLS20.
The activation of several developmental processes in B. subtilis such as sporulation, competence and motility depends on stochastic variability in expression of a master regulator that results in heterogeneic behavior of genetically identical cells within a population (Korobkova et al., 2004; Dubnau and Losick, 2006; Veening et al., 2008). A benefit of heterogeneity can be division of labor, where a subpopulation of cells produces products for the benefit of the entire community. Heterogeneity may also lead to the so-called bet-hedging strategies in which a subpopulation of differentiated cells is generated even in the absence of conditions favoring the differentiation process. Like division of labor, this strategy is beneficial for the community at the population level because the differentiated cells are adapted to adverse conditions that may arise suddenly. However, the process of conjugation is an energy consuming process and has major impacts on cell surface and membrane components, requiring tight repression at times when conditions for successful DNA transfer are not apt. Instead of a heterogeneic behavior resulting in a bet-hedging strategy, the conjugation process must be strictly controlled, allowing its activation to occur only under conditions that are favourable for successful transfer events, and this requires an efficient and sensitive genetic switch from an “OFF” to “ON” state only under conditions that favor successful plasmid transfer. Several features described above contribute to this efficient switch, such as overlapping divergent promoters, low levels of RcopLS20 and RappLS20, and DNA looping. We consider it possible that the multimerization state of RcopLS20, and particularly the apo and peptide bound forms of RappLS20 may also contribute to the sensitivity and efficiency of the switch.
Conjugation and competence appear to be incompatible processes. Although no evidence is currently available, one or two-way interference may also exist between conjugative plasmids and phage infection. Some Bacillus phages use a quorum sensing mechanism that plays a crucial role in entering the lytic or the lysogenic cycle (Erez et al., 2017). Perhaps, cross-talk exists between this quorum sensing system and the one regulating conjugation.
The work summarized in this review shows that conjugation is exquisitely controlled at the level of the Pc promoter. However, the conjugation operon is very large and contains many genes that are involved in different stages of the conjugation process, and it is very well possible that subsets of conjugation genes need to be expressed differentially or temporally. This may be achieved, at least in part, by different translation initiation signals, and protein/RNA stability, but we have recently obtained evidence that at least one other layer of expression is involved. pLS20cat gene 29 encodes a surface-located protein that exerts exclusion; i.e., it inhibits futile transfer of the plasmid between donor cells, thereby contributing to efficient transfer to plasmid-free recipient cells (Gago-Cordoba et al., 2019). Proper functioning of exclusion systems require that the gene(s) involved are expressed in all donor cells, but gene 29 is located inside the conjugation operon. This apparent paradox was solved by the identification of a weak promoter located upstream of gene 29, and a similar strategy is probably used in other conjugative systems as well (Gago-Cordoba et al., 2019). It is possible that the conjugation operon contains additional constitutive or regulated promoters that may modulate the expression of subsets of genes within the operon.
Contrary to most other conjugative systems, pLS20 conjugates efficiently in liquid medium and therefore it will have an important role in gene transfer under natural conditions. However, the predominant lifestyle of bacteria, including B. subtilis, is forming colonies or biofilms (Donlan, 2002). pLS20 also conjugates efficiently on solid medium (Koehler and Thorne, 1987; Meijer et al., 1998). As described above, the activation state of the conjugation process is ultimately determined by the concentration of the Phr*pLS20 signaling peptide. The results summarized in this review are based on conjugation experiments carried out in liquid medium, and particularly using shaking cultures. Under these conditions, the concentration of the exported signal peptide will be instantly homogenized, which is very different from the situation in which donor cells form part of a colony or biofilm where secretion of the signal peptide will lead to a concentration gradient. It will be interesting to see how the conjugation genes are regulated under such conditions.
WM and LW wrote the paper. DB, SA, FR, CA, and JRL-O edited the paper, WM and SA made Figures.
Ministry of Science and Innovation of the Spanish Government (PID2019 108778GB C21 (AEI/FEDER, EU)) to WM, (BIO2016-77883-C2-2-P and FIS2015-72574-EXP (AEI/FEDER, EU)), to DB, (PID2019-104544GBI00 (AEI/FEDER/EU)) to CA, (PID2019-109320GB-100/AEI/10.13039/501100011033) to SA, (PGC2018-094143-B-I00, (AEI/FEDER, EU)) to FR, and (FIS2016-78313-P) to SA. The Ministry of Science and Innovation of the Spanish Government also funded the Centro Nacional de Biotecnología (CNB) by means of the “Severo Ochoa” Center of Excellence grant SEV 2017-0712. A Wellcome Investigator grant (209500) to Jeff Errington supported LW. Institutional grants from the ‘Fundación Ramón Areces’ and ‘Banco de Santander’ supported the Centro de Biología Molecular ‘Severo Ochoa’.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank members of the different labs for useful discussions, and Daniel Zeigler of the Bacillus Genetic Stock Center for providing plasmids and strains essential for many experiments described in this review. We acknowledge the publication fee support of the CSIC Open Access Publication Support Initiative through its ‘Unit of Information Resources for Research’ (URICI).
Arutyunov, D., and Frost, L. S. (2013). F conjugation: back to the beginning. Plasmid 70 (1), 18–32. doi:10.1016/j.plasmid.2013.03.010
Auchtung, J. M., Lee, C. A., Garrison, K. L., and Grossman, A. D. (2007). Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis. Mol. Microbiol. 64 (6), 1515–1528. doi:10.1111/j.1365-2958.2007.05748.x
Baker, M. D., and Neiditch, M. B. (2011). Structural basis of response regulator inhibition by a bacterial anti-activator protein. PLoS Biol. 9 (12), e1001226. doi:10.1371/journal.pbio.1001226
Baxter, J. C., and Funnell, B. E. (2014). Plasmid partition mechanisms. Microbiol. Spectr. 2 (6) 1–20. doi:10.1128/microbiolspec.PLAS-0023-2014
Bongiorni, C., Stoessel, R., Shoemaker, D., and Perego, M. (2006). Rap phosphatase of virulence plasmid pXO1 inhibits Bacillus anthracis sporulation. J. Bacteriol. 188 (2), 487–498. doi:10.1128/JB.188.2.487-498.2006
Bose, B., Auchtung, J. M., Lee, C. A., and Grossman, A. D. (2008). A conserved anti-repressor controls horizontal gene transfer by proteolysis. Mol. Microbiol. 70 (3), 570–582. doi:10.1111/j.1365-2958.2008.06414.x
Bouet, J. Y., and Funnell, B. E. (2019). Plasmid localization and partition in Enterobacteriaceae. EcoSal Plus 8 (2) 1–23. doi:10.1128/ecosalplus.ESP-0003-2019
Brooks, A. C., and Hwang, L. C. (2017). Reconstitutions of plasmid partition systems and their mechanisms. Plasmid 91, 37–41. doi:10.1016/j.plasmid.2017.03.004
Cabezón, E., Ripoll-Rozada, J., Peña, A., De la Cruz, F., and Arechaga, I. (2014). Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 39, 81–95. doi:10.1111/1574-6976.12085
CDC (2019). Antibiotic resistance threats in the United States. Atlanta, GA: U.S. Department of health and human services.
Chastanet, A., and Losick, R. (2011). Just-in-time control of Spo0A synthesis in Bacillus subtilis by multiple regulatory mechanisms. J. Bacteriol. 193 (22), 6366–6374. doi:10.1128/JB.06057-11
Chatterjee, A., Cook, L. C., Shu, C. C., Chen, Y., Manias, D. A., Ramkrishna, D., et al. (2013). Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer. Proc. Natl. Acad. Sci. U.S.A. 110 (17), 7086–7090. doi:10.1073/pnas.1212256110
Chen, I., Christie, P. J., and Dubnau, D. (2005). The ins and outs of DNA transfer in bacteria. Science 310 (5753), 1456–1460. doi:10.1126/science.114021
Christie, P. J. (2016). The mosaic type IV secretion systems. EcoSal Plus 7 (1), 10. doi:10.1128/ecosalplus.ESP-0020-2015
Christie, P. J., Whitaker, N., and González-Rivera, C. (2014). Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta 1843 (8), 1578–1591. doi:10.1016/j.bbamcr.2013.12.019
Clewell, D. B. (2011). Tales of conjugation and sex pheromones: a plasmid and enterococcal odyssey. Mob. Genet. Elem. 1 (1), 38–54. doi:10.4161/mge.1.1.15409
Crespo, I., Bernardo, N., Miguel-Arribas, A., Singh, P. K., Luque-Ortega, J. R., Alfonso, C., et al. (2020). Inactivation of the dimeric RappLS20 anti-repressor of the conjugation operon is mediated by peptide-induced tetramerization. Nucleic Acids Res. 48, 8113. doi:10.1093/nar/gkaa540
Cutting, S. M. (2011). Bacillus probiotics. Food Microbiol. 28 (2), 214–220. doi:10.1016/j.fm.2010.03.007
D’Andrea, L. D., and Regan, L. (2003). TPR proteins: the versatile helix. Trends Biochem. Sci. 28 (12), 655–662. doi:10.1016/j.tibs.2003.10.007
Davies, J., and Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74 (3), 417–433. doi:10.1128/MMBR.00016-10
Declerck, N., Bouillaut, L., Chaix, D., Rugani, N., Slamti, L., Hoh, F., et al. (2007). Structure of PlcR: insights into virulence regulation and evolution of quorum sensing in gram-positive bacteria. Proc. Natl. Acad. Sci. U.S.A. 104 (47), 18490–18495. doi:10.1073/pnas.0704501104
Derman, A. I., Becker, E. C., Truong, B. D., Fujioka, A., Tucey, T. M., Erb, M. L., et al. (2009). Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol. Microbiol. 73 (4), 534–552. doi:10.1111/j.1365-2958.2009.06771.x
Dodd, I. B., Shearwin, K. E., and Egan, J. B. (2005). Revisited gene regulation in bacteriophage lambda. Curr. Opin. Genet. Dev. 15 (2), 145–152. doi:10.1016/j.gde.2005.02.001
Donlan, R. M. (2002). Biofilms: microbial life on surfaces. Emer. Infect. Dis. 8 (9), 881–890. doi:10.3201/eid0809.020063
Dubnau, D. (1991). Genetic competence in Bacillus subtilis. Microbiol. Rev. 55, 395–424. doi:10.1128/mmbr.55.3.395-424.1991
Dubnau, D., and Losick, R. (2006). Bistability in bacteria. Mol. Microbiol. 61 (3), 564–572. doi:10.1111/j.1365-2958.2006.05249.x
Dunny, G. M. (2013). Enterococcal sex pheromones: signaling, social behavior, and evolution. Annu. Rev. Genet. 47, 457–482. doi:10.1146/annurev-genet-111212-133449
Dunny, G. M., and Berntsson, R. P. (2016). Enterococcal sex pheromones: evolutionary pathways to complex, two-signal systems. J. Bacteriol. 198 (11), 1556–1562. doi:10.1128/JB.00128-16
Dunny, G. M., and Johnson, C. M. (2011). Regulatory circuits controlling enterococcal conjugation: lessons for functional genomics. Curr. Opin. Microbiol. 14 (2), 174–180. doi:10.1016/j.mib.2011.01.008
Erez, Z., Steinberger-Levy, I., Shamir, M., Doron, S., Stokar-Avihail, A., Peleg, Y., et al. (2017). Communication between viruses guides lysis-lysogeny decisions. Nature 541 (7638), 488–493. doi:10.1038/nature21049
Frost, L. S., and Koraimann, G. (2010). Regulation of bacterial conjugation: balancing opportunity with adversity. Future Microbiol. 5 (7), 1057–1071. doi:10.2217/fmb.10.70
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi:10.1038/nrmicro1235
Gago-Córdoba, C., Val-Calvo, J., Miguel-Arribas, A., Serrano, E., Singh, P. K., Abia, D., et al. (2019). Surface exclusion revisited: function related to differential expression of the surface exclusion system of Bacillus subtilis plasmid pLS20. Front. Microbiol. 10, 1502. doi:10.3389/fmicb.2019.01502
Goessweiner-Mohr, N., Arends, K., Keller, W., and Grohmann, E. (2014). Conjugation in gram-positive bacteria. Microbiol. Spectr. 2 (4) 1–19. doi:10.1128/microbiolspec.PLAS-0004-2013
Griffith, K. L., and Grossman, A. D. (2008). A degenerate tripartite DNA-binding site required for activation of ComA-dependent quorum response gene expression in Bacillus subtilis. J. Mol. Biol. 381 (2), 261–275. doi:10.1016/j.jmb.2008.06.035
Grohmann, E., Muth, G., and Espinosa, M. (2003). Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67 (2), 277–contents. doi:10.1128/mmbr.67.2.277-301.2003
Grossman, A. D. (1995). Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29, 477–508. doi:10.1146/annurev.ge.29.120195.002401
Hamoen, L. W., Venema, G., and Kuipers, O. P. (2003). Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology (Reading, Engl) 149, 9–17. doi:10.1099/mic.0.26003-0
Hoch, J. A. (1993). “spo0 genes, the phosphorelay, and the initiation of sporulation,” in Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Editors A. L. Sonenshein, J. A. Hoch, and R. Losick (Washington, DC: American Society for Microbiology (ASM)), 747–755.
Itaya, M., Sakaya, N., Matsunaga, S., Fujita, K., and Kaneko, S. (2006). Conjugational transfer kinetics of pLS20 between Bacillus subtilis in liquid medium. Biosci. Biotechnol. Biochem. 70, 740–742. doi:10.1271/bbb.70.740
Koehler, T. M., and Thorne, C. B. (1987). Bacillus subtilis (natto) plasmid pLS20 mediates interspecies plasmid transfer. J. Bacteriol. 169 (11), 5271–5278. doi:10.1128/jb.169.11.5271-5278.1987
Kohler, V., Keller, W., and Grohmann, E. (2019). Regulation of gram-positive conjugation. Front. Microbiol. 10, 1134. doi:10.3389/fmicb.2019.01134
Koraimann, G. (2018). Spread and persistence of virulence and antibiotic resistance genes: a ride on the F plasmid conjugation module. EcoSal Plus 8 (1) 1–23. doi:10.1128/ecosalplus.ESP-0003-2018
Koraimann, G., and Wagner, M. A. (2014). Social behavior and decision making in bacterial conjugation. Front. Cell Infect. Microbiol. 4, 54. doi:10.3389/fcimb.2014.00054
Korobkova, E., Emonet, T., Vilar, J. M., Shimizu, T. S., and Cluzel, P. (2004). From molecular noise to behavioural variability in a single bacterium. Nature 428 (6982), 574–578. doi:10.1038/nature02404
Lerminiaux, N. A., and Cameron, A. D. S. (2019). Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 65 (1), 34–44. doi:10.1139/cjm-2018-0275
Li, Y. G., Hu, B., and Christie, P. J. (2019). Biological and structural diversity of type IV secretion systems. Microbiol. Spectr. 7 (2) 1–10. doi:10.1128/microbiolspec.PSIB-0012-2018
Mazel, D., and Davies, J. (1999). Antibiotic resistance in microbes. Cell Mol. Life Sci. 56 (9–10), 742–754. doi:10.1007/s000180050021
Meijer, W. J., de Boer, A., van Tongeren, S., Venema, G., and Bron, S. (1995). Characterization of the replication region of the Bacillus subtilis plasmid pLS20: a novel type of replicon. Nucleic Acids Res. 23, 3214–3223. doi:10.1093/nar/23.16.3214
Meijer, W. J., Wisman, G. B., Terpstra, P., Thorsted, P. B., Thomas, C. M., Holsappel, S., et al. (1998). Rolling-circle plasmids from Bacillus subtilis: complete nucleotide sequences and analyses of genes of pTA1015, pTA1040, pTA1050 and pTA1060, and comparisons with related plasmids from gram-positive bacteria. FEMS Microbiol. Rev. 21 (4), 337–368. doi:10.1111/j.1574-6976.1998.tb00357.x
Molle, V., Fujita, M., Jensen, S. T., Eichenberger, P., González-Pastor, J. E., Liu, J. S., et al. (2003). The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50 (5), 1683–1701. doi:10.1046/j.1365-2958.2003.03818.x
Monnet, V., Juillard, V., and Gardan, R. (2016). Peptide conversations in gram-positive bacteria. Crit. Rev. Microbiol. 42 (3), 339–351. doi:10.3109/1040841X.2014.948804
Neiditch, M. B., Capodagli, G. C., Prehna, G., and Federle, M. J. (2017). Genetic and structural analyses of RRNPP intercellular peptide signaling of gram-positive bacteria. Annu. Rev. Genet. 51, 311–333. doi:10.1146/annurev-genet-120116-023507
Novick, R. P., Christie, G. E., and Penadés, J. R. (2010). The phage-related chromosomal islands of gram-positive bacteria. Nat. Rev. Microbiol. 8 (8), 541–551. doi:10.1038/nrmicro2393
Ochman, H., Lawrence, J. G., and Groisman, E. A. (2000). Lateral gene transfer and the nature of bacterial innovation. Nature 405 (6784), 299–304. doi:10.1038/35012500
Oehler, S., and Müller-Hill, B. (2010). High local concentration: a fundamental strategy of life. J. Mol. Biol. 395 (2), 242–253. doi:10.1016/j.jmb.2009.10.056
Oppenheim, A. B., Kobiler, O., Stavans, J., Court, D. L., and Adhya, S. (2005). Switches in bacteriophage lambda development. Annu. Rev. Genet. 39, 409–429. doi:10.1146/annurev.genet.39.073003.113656
Parashar, V., Konkol, M. A., Kearns, D. B., and Neiditch, M. B. (2013). A plasmid-encoded phosphatase regulates Bacillus subtilis biofilm architecture, sporulation, and genetic competence. J. Bacteriol. 195 (10), 2437–2448. doi:10.1128/JB.02030-12
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31 (4), e00088-17. doi:10.1128/CMR.00088-17
Perego, M. (2001). A new family of aspartyl phosphate phosphatases targeting the sporulation transcription factor Spo0A of Bacillus subtilis. Mol. Microbiol. 42 (1), 133–143. doi:10.1046/j.1365-2958.2001.02611.x
Pottathil, M., and Lazazzera, B. A. (2003). The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis. Front. Biosci. 8, 32–45. doi:10.2741/913
Ramachandran, G., Singh, P. K., Luque-Ortega, J. R., Yuste, L., Alfonso, C., Rojo, F., et al. (2014). A complex genetic switch involving overlapping divergent promoters and DNA looping regulates expression of conjugation genes of a gram-positive plasmid. PLoS Genet. 10 (10), e1004733. doi:10.1371/journal.pgen.1004733
Rocha-Estrada, J., Aceves-Diez, A. E., Guarneros, G., and de la Torre, M. (2010). The RNPP family of quorum-sensing proteins in gram-positive bacteria. Appl. Microbiol. Biotechnol. 87 (3), 913–923. doi:10.1007/s00253-010-2651-y
Shi, K., Brown, C. K., Gu, Z. Y., Kozlowicz, B. K., Dunny, G. M., Ohlendorf, D. H., et al. (2005). Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc. Natl. Acad. Sci. U.S.A. 102 (51), 18596–18601. doi:10.1073/pnas10.1073/pnas.0506163102
Singh, P. K., and Meijer, W. J. (2014). Diverse regulatory circuits for transfer of conjugative elements. FEMS Microbiol. Lett. 358 (2), 119–128. doi:10.1111/1574-6968.12526
Singh, P. K., Ramachandran, G., Durán-Alcalde, L., Alonso, C., Wu, L. J., and Meijer, W. J. (2012). Inhibition of Bacillus subtilis natural competence by a native, conjugative plasmid-encoded comK repressor protein. Environ. Microbiol. 14 (10), 2812–2825. doi:10.1111/j.1462-2920.2012.02819.x
Singh, P. K., Ramachandran, G., Ramos-Ruiz, R., Peiró-Pastor, R., Abia, D., Wu, L. J., et al. (2013). Mobility of the native Bacillus subtilis conjugative plasmid pLS20 is regulated by intercellular signaling. PLoS Genet. 9 (10), e1003892. doi:10.1371/journal.pgen.1003892
Singh, P. K., Serrano, E., Ramachandran, G., Miguel-Arribas, A., Gago-Cordoba, C., Val-Calvo, J., et al. (2020). Reversible regulation of conjugation of Bacillus subtilis plasmid pLS20 by the quorum sensing peptide responsive anti-repressor RappLS20. Nucleic Acids Res. 48 (19), 10785–10801. doi:10.1093/nar/gkaa797
Smillie, C., Garcillán-Barcia, M. P., Francia, M. V., Rocha, E. P., and De la Cruz, F. (2010). Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74 (3), 434–452. doi:10.1128/MMBR.00020-10
Smits, W. K., Bongiorni, C., Veening, J. W., Hamoen, L. W., Kuipers, O. P., and Perego, M. (2007). Temporal separation of distinct differentiation pathways by a dual specificity Rap-Phr system in Bacillus subtilis. Mol. Microbiol. 65 (1), 103–120. doi:10.1111/j.1365-2958.2007.05776.x
Sonenshein, A. L., Hoch, J. A., and Losick, R. (1993). Bacillus subtilis and other Gram-positive bacteria; Biochemistry, physiology, and molecular genetics. Washington, DC: American Society for Microbiology (ASM).
Sonenshein, A. L., Hoch, J. A., and Losick, R. (2001). Bacillus subtilis and its closest relatives: from genes to cells. Washington, DC: ASM Press.
Soucy, S. M., Huang, J., and Gogarten, J. P. (2015). Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 16 (8), 472–482. doi:10.1038/nrg3962
Stingl, K., and Koraimann, G. (2017). Prokaryotic information games: how and when to take up and secrete DNA. Curr. Top. Microbiol. Immunol. 413, 61–92. doi:10.1007/978-3-319-75241-9_3
Tanaka, T., and Koshikawa, T. (1977). Isolation and characterization of four types of plasmids from Bacillus subtilis (natto). J. Bacteriol. 131, 699–701. doi:10.1128/JB.131.2.699-701.1977
Thomas, C. M., and Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3 (9), 711–721. doi:10.1038/nrmicro1234
Veening, J. W., Smits, W. K., and Kuipers, O. P. (2008). Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62, 193–210. doi:10.1146/annurev.micro.62.081307.163002
Vilar, J. M., and Saiz, L. (2005). DNA looping in gene regulation: from the assembly of macromolecular complexes to the control of transcriptional noise. Curr. Opin. Genet. Dev. 15 (2), 136–144. doi:10.1016/j.gde.2005.02.005
Waksman, G. (2019). From conjugation to T4S systems in gram-negative bacteria: a mechanistic biology perspective. EMBO Rep. 20 (2), e47012. doi:10.15252/embr.201847012
Keywords: quorum sensing, Bacillus subtilis, Gram-positive bacteria, antibiotic resistance, horizontal gene transfer, DNA looping, transcriptional regulation, conjugation
Citation: Meijer WJJ, Boer DR, Ares S, Alfonso C, Rojo F, Luque-Ortega JR and Wu LJ (2021) Multiple Layered Control of the Conjugation Process of the Bacillus subtilis Plasmid pLS20. Front. Mol. Biosci. 8:648468. doi: 10.3389/fmolb.2021.648468
Received: 31 December 2020; Accepted: 08 February 2021;
Published: 18 March 2021.
Edited by:
Chew Chieng Yeo, Sultan Zainal Abidin University, MalaysiaReviewed by:
Günther Koraimann, University of Graz, Graz, AustriaCopyright © 2021 Meijer, Boer, Ares, Alfonso, Luque-Ortega and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wilfried J. J. Meijer, d21laWplckBjYm0uY3NpYy5lcw==; Ling Juan Wu, bC5qLnd1QG5ld2Nhc3RsZS5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.