
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci., 27 May 2021
Sec. Molecular Diagnostics and Therapeutics
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.646542
This article is part of the Research TopicTargetable RNA and Tumor HeterogeneityView all 4 articles
 Yiqiang Liu1,2†
Yiqiang Liu1,2† Hong Wu2†
Hong Wu2† Tao Luo1
Tao Luo1 Qiyu Luo2
Qiyu Luo2 Ziyu Meng2
Ziyu Meng2 Ying Shi2
Ying Shi2 Feifei Li1,2
Feifei Li1,2 Mingxin Liu2
Mingxin Liu2 Xinhao Peng2
Xinhao Peng2 Junjie Liu1
Junjie Liu1 Chuan Xu2*
Chuan Xu2* Weizhong Tang1*
Weizhong Tang1*Background: Colorectal cancer (CRC) is estimated to be one of the most common cancers and the leading cause of cancer-related death worldwide. SOX9 is commonly overexpressed in CRC and participates in drug resistance. In addition, DNA damage repair confers resistance to anticancer drugs. However, the correlation between DNA damage repair and high SOX9 expression is still unclear. In this study, we aimed to investigate the function and the specific underlying mechanism of the SOX9-dependent DNA damage repair pathway in CRC.
Methods: The expression levels of SOX9 and MMS22L in CRC were examined by immunohistochemistry (IHC) and TCGA analysis. RNA sequencing was conducted in RKO SOX9-deficient cells and RKO shControl cells. Mechanistic studies were performed in CRC cells by modulating SOX9 and MMS22L expression, and we evaluated drug sensitivity and DNA damage repair signaling events. In addition, we investigated the effect of oxaliplatin in tumors with SOX9 overexpression and low expression of MMS22L in vivo.
Results: Our study showed that SOX9 has a higher expression level in CRC tissues than in normal tissues and predicts poor prognosis in CRC patients. Overexpression and knockdown of SOX9 were associated with the efficacy of oxaliplatin. In addition, SOX9 activity was enriched in the DNA damage repair pathway via regulation of MMS22L expression and participation in DNA double-strand break repair. SOX9 was upregulated and formed a complex with MMS22L, which promoted the nuclear translocation of MMS22L upon oxaliplatin treatment. Moreover, the xenograft assay results showed that oxaliplatin abrogated tumor growth from cells with MMS22L downregulation in mice.
Conclusions: In CRC, activation of the SOX9-MMS22L-dependent DNA damage pathway is a core pathway regulating oxaliplatin sensitivity. Targeting this pathway in oxaliplatin-resistant CRC cells is a promising therapeutic option.
Colorectal cancer (CRC) is one of the most common cancers and the leading cause of cancer-related death worldwide (Siegel et al., 2020). CRC patients are generally diagnosed at an advanced stage, and death is usually caused by recurrence and metastasis to other organs (D. Chen et al., 2020). Drug resistance is not only the main cause of treatment failure but also the main factor that limits the survival of CRC patients (Carrasco-Garcia et al., 2016).
SOX9 is a transcription factor that has been reported to be linked with stem cell maintenance and is commonly overexpressed in solid tumors (Carrasco-Garcia et al., 2016; Christin et al., 2020; Kaushik et al., 2021). High expression of SOX9 in CRC correlates with advanced tumor stage and worse overall survival in patients (Lü et al., 2008). The oncogenic role of SOX9 is related to several cell signaling pathways, such as the TGFβ/Smad, Notch and Wnt/β-catenin pathways (Li and Neaves, 2006; Abdul Khalek et al., 2010). Studies have confirmed that SOX9-overexpressing cancer cells are resistant to oxaliplatin (Anuja et al., 2019; Qian et al., 2019; Abad et al., 2020; Das et al., 2020). However, the underlying mechanism of SOX9 in resistance to oxaliplatin is still unclear.
Oxaliplatin is a commonly used drug in the treatment of CRC and can inhibit DNA replication and transcription by promoting the formation of intra- and interstrand DNA breaks (Hochster and Sargent, 2016). Studies have shown that oxaliplatin adducts are less sensitive to the activity of DNA excision repair enzymes than native DNA and are equally damaging in DNA mismatch repair-proficient and DNA mismatch repair-deficient cells in vitro (Rothenberg, 2000). Understanding the molecular events controlling drug resistance is of paramount importance for cancer therapy. MMS22L is required for the repair of stalled/collapsed replication forks (O’Donnell et al., 2010). Studies have shown that human cells expressing low levels of MMS22L are sensitive to drugs that can induce replication fork reversal or Double-strand breaks (DSBs) at replication forks, such as camptothecin (CPT) (Duro et al., 2010; O’Connell et al., 2010). However, the regulatory role of MMS22L in SOX9-mediated chemotherapeutic resistance in CRC remains unclear. Therefore, identifying the role of the SOX9-DNA damage repair protein axis in anticancer drug resistance and aiming to elucidate its relevance to signaling pathway regulation may lead to the identification of new therapeutic targets.
The human colon cancer cell lines HCT116, HT-29, DLD1, RKO, and SW480 and the human normal colorectal mucosal cell line FHC were purchased from the American Type Culture Collection (ATCC) and cultured in Dulbecco’s modified Eagle’s medium (DMEM, CA, United States) supplemented with 10% fetal bovine serum (FBS, Jiangsu, China) and 100 U/ml penicillin-streptomycin. The culture conditions were a cell incubator with a temperature of 37°C and 5% carbon dioxide. Oxaliplatin was purchased from Selleck Chemical (Selleck, S1224, TX, United States).
CRC tumor and adjacent normal tissue slices were obtained from the Department of Pathology, Affiliated Tumor Hospital of Guangxi Medical University, Nanning, China, with approval from the Institutional Ethics Committee. All patients signed the informed consent form, and clinical data are provided in Supplementary Table 1.
The CMV-MCS-3FLAG-SV40-Neomycin lentiviral Vector expressing full-length human SOX9 (NM_000346.4) and the U6-MCS-Ubiquitin-cherry-IRES-puromycin shSOX9 lentiviral Vector were purchased from GeneChem (Shanghai, China). The lentiviral packaging procedure was performed as described previously (Wu et al., 2020). RKO and SW480 cells (1 × 106 cells/well) were infected with lentivirus. Stably transduced cells were selected and enriched with 4 μg/ml puromycin (Thermo Fisher Scientific, MA, United States). The sequences of the shRNA and the primers used to amplify the full-length sequence of human SOX9 are listed below:
SOX9-RNAi: forward: 5′-CCGGCTCCACCTTCACCTACAT GAACTCGAGTTCATGTAGGTGAAGGTGGAGTTTTTG-3′, reverse: 5′-GATCCAAAAACTCCACCTTCACCTACATGA ACTCGAGTTCATGTAGGTGAAGGTGGAG-3′.
SOX9-control: forward: 5′-CCGGTTCTCCGAACGTGTCA CGTCTCGAGTTCATGTAGGTGAAGGTGGAGTTTTTG-3′, reverse: 5′-GATCCAAAAATTCTCCGAACGTGTCACGTC TCGAGTTCATGTAGGTGAAGGTGGAG-3′.
MMS22L-RNAi: forward: 5′-CCGGCCAACTCTTCAAGGA AACTAACTCGAGTTAGTTTCCTTGAAGAGTTGGTTT TTG-3′, reverse: 5′-AATTCAAAAACCAACTCTTCAAGG AAACTAACTCGAGTTAGTTTCCTTGAAGAGTTGG-3′.
MMS22L-control: forward: 5′-CCGGTTCTCCGAACGTGT CACGTCTCGAGTTAGTTTCCTTGAAGAGTTGGTTTTTG-3′, reverse: 5′-AATTCAAAAATTCTCCGAACGTGTCACGTC TCGAGTTAGTTTCCTTGAAGAGTTGG-3′.
The primers used to amplify the full-length sequence of human SOX9 were as follows: forward: 5′-GAGGAAGTCG GTGAAGAACGG-3′, reverse: 5′-CTGAGCGGGGTTCATG TAGG-3′.
Total RNA was extracted from RKO shControl (shCtrl) and RKO shSOX9 cells. An RNA library was constructed using a TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, CA, United States) and subjected to 125 bp/150 bp paired-end sequencing on an Illumina platform (HiSeqTM 2500 or Illumina HiSeqTM X Ten) by Haplox (Jiangxi, China). The differentially expressed genes (DEGs) were identified using Cuffdiff with q value < 0.05 and fold change >2 as the criteria. Gene set enrichment analysis (GSEA) was performed on the software.broadinstitute.org/gsea website. Volcano plots, pathway enrichment analysis visualizations and heatmaps were generated with the R program. All details were described in our previous work (Wu et al., 2020).
Immunohistochemical staining was performed on tumor tissue slides as previously described (Wu et al., 2020). The slides were incubated with rabbit polyclonal anti-SOX9 (1:200, ABclonal, A19710, Hubei, China) and anti-MMS22L (1:200, Bioss, 17689R, Beijing, China) antibodies at 4°C overnight. SOX9 and MMS22L expression was evaluated by using a system considering the staining intensity (0 means negative; 1 means weak; 2 means moderate; and 3 means strong) and the percentage of positively stained cells (<5% = 0, 5% to <25% = 1, 25% to 50% = 2, > 50 to <75% = 3, ≥ 75% = 4). The final score was calculated by multiplying the extent score by the intensity score, and expression was accordingly classified as low or high with a score of four as the maximum value for low expression (Zou et al., 2019).
Total protein was obtained by the following steps. Cancer cells were lysed using radioimmunoprecipitation assay buffer (RIPA, Thermo Fisher Scientific, 89901, MA, United States):Phenylmethylsulfonyl Fluoride (PMSF, Beyotime, ST505, Shanghai, China) = 100:1 on ice for 30 min. Samples were centrifuged at 15,000 × g for 15 min; the supernatant was then collected, and 5 × loading buffer was added and heated at 95°C for 10 min. The nuclear fractions were separated using a Nuclear Extraction Kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions, and protein was obtained as described above. The samples were separated by SDS-PAGE, and proteins were transferred to Polyvinylidene Difluoride (PVDF) membranes (Millipore, IPVH00010, MA, United States). After blocking in 5% milk-PBST for 1 h at room temperature, the membranes were incubated with primary antibodies at 4°C overnight and with secondary antibodies at room temperature for 1 h. Positive bands were visualized by enhanced chemiluminescence (ECL) (Millipore, WBKLS0100, MA, United States). Information on the primary antibodies is provided in Supplementary Table 2.
Total RNA was extracted from cancer cells using TRIzol Reagent (Invitrogen, United States), and quantitative real-time polymerase chain reaction (qRT-PCR) was performed using a SYBR Prime Script RT-PCR kit (TaKaRa, Osaka, Japan) in a CFX Connect Real-Time System (Bio-Rad, CA, United States). GAPDH was used as the internal control. The primer sequences are listed as follows:
SOX9: forward: 5′-AGGAAGCTCGCGGACCAGTAC-3′, reverse: 5′-GGTGGTCCTTCTTGTGCTGCAC-3′; GAPDH: forward: 5′-GTTCGTCATGGGTGTGAAC-3′, reverse: 5′-ATGGCATGGACTGTGGTCAT-3′; MMS22L: forward: 5′- ATGTGAGCGGGAATCTCTT-3′, reverse: 5′-GAAACACT TGGGGTTCGTC-3′.
Cells were seeded on coverslips (5 × 104 cells per well) in a 24-well plate. 12 h after plating, cells were exposed to oxaliplatin at the indicated doses for 4 h. Then, cells were fixed with 4% paraformaldehyde and permeabilized with PBS containing 0.3% Triton X-100. After blocking with goat serum, cells were incubated with primary antibodies at 4°C overnight. Then, cells were incubated with fluorescent secondary antibodies (Molecular Probes, Eugene, OR, United States) (1:400) at room temperature for 30 min. Nuclei were stained with DAPI. A Nikon Eclipse Ti-S fluorescence microscope (Nikon, Tokyo, Japan) was used to acquire images. The paraffin sections required antigen repair prior to the above procedure.
CRC cells were seeded in 96-well plates at a density of 2 × 103 cells per well. Oxaliplatin was added to the culture medium at a concentration of 5 μM. Over the following 5 days, the CCK8 assay was performed daily. Cell Counting Kit-8 solution (Biosharp, BS350B, Anhui, China) was added to each well prior to incubation at 37°C for 2 h. Optical density was measured based on the absorbance at a wavelength of 450 nm.
CRC cells were treated with 5 μM oxaliplatin. According to the manufacturer’s instructions, the early apoptosis rate was analyzed by the Annexin-V staining method using a PE Annexin-V Apoptosis Detection Kit (Beyotime, C1065M, Shanghai, China) and a FITC Annexin-V Apoptosis Detection Kit (Beyotime, C1062M, Shanghai, China). We used FlowJo v.10 software for data analysis.
To evaluate the role of the SOX9-MMS22L axis in vivo, 4-week-old BALB/c nude mice (DOSSY, Chengdu, China) were subcutaneously inoculated with RKO (5 × 106 cells/100 μl PBS) cells stably transfected with Vector+shCtrl, oeSOX9+shCtrl, Vector+shMMS22L, oeSOX9+shMMS22L plasmids. Mice were intraperitoneally injected with oxaliplatin (7.5 mg/kg) every 2 days for 10 days and sacrificed on Day 28 (Liu et al., 2020). The tumor volume was estimated every 4 days and calculated using the following formula: tumor volume = 0.5 × (length × width2). A portion of the xenograft tumors in the mice was taken for IHC staining. All animal experiments were performed in compliance with the guidelines of the Animal Ethics Committee of the Affiliated Tumor Hospital of Guangxi Medical University.
Statistical analysis was performed with GraphPad Prism 7.0 (CA, United States). One-way ANOVA or two-tailed unpaired t-tests were used to determine the level of significance. All data used in this study satisfied the assumptions of the statistical tests. All quantitative data are shown as the mean ± S.D. or mean ± S.E.M. values as indicated. P < 0.05 was considered statistically significant. All experiments were carried out at least three times.
To verify the expression status of SOX9 in CRC patients, we examined SOX9 expression in 80 pairs of CRC tissues by IHC staining. Our results confirmed the high expression of SOX9 in tumor tissues compared to the adjacent non-tumor tissues (Figures 1A,B). In addition, 65% of CRC patients had strong SOX9 expression (Figure 1C). Similar results were observed in CRC cells (HCT116, HT-29, DLD1, SW480, and RKO) compared with human normal colorectal mucosal cells (FHC) (Supplementary Figure 4). Furthermore, SOX9high CRC patients showed shorter overall survival, disease-specific survival and progression-free survival times than SOX9low patients in the TCGA database (Figures 1D–F). Conclusively, these data indicate that SOX9 is aberrantly highly expressed in tumor tissues and predicts poor prognosis in CRC patients.
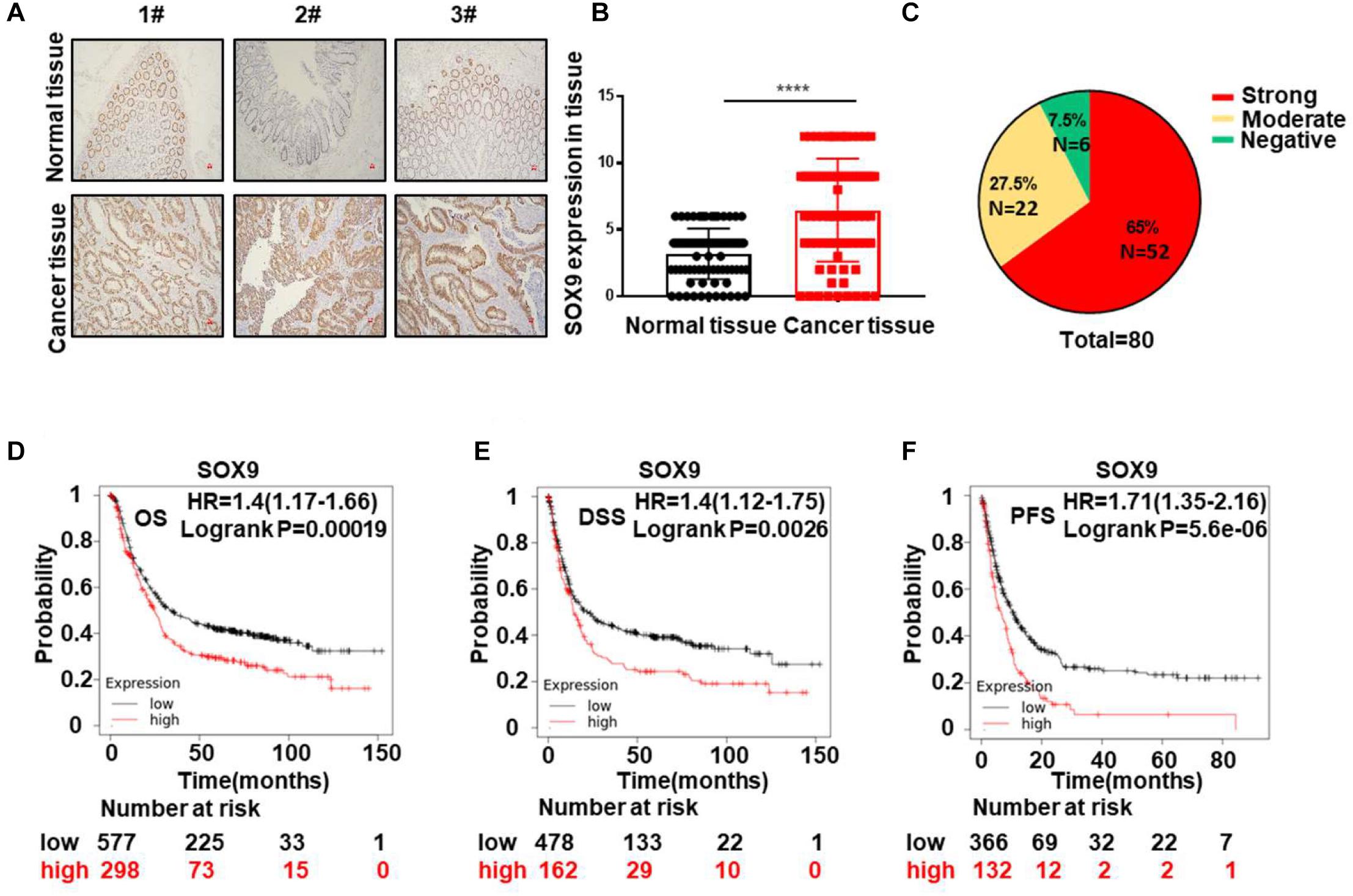
Figure 1. SOX9 is aberrantly highly expressed in tumor tissues and predicts poor prognosis in CRC. (A) Representative IHC staining image of the expression of SOX9 in three pairs of CRC tissues (T) and paired adjacent normal tissues (N). Scale bar = 50 μm. (B) Quantitative analysis of SOX9 expression in 80 CRC tumor tissues (T) and adjacent normal tissues (N). (C) Percentages of negative, moderate, and strong SOX9 expression in CRC tumor tissues (n = 80). (D–F) Kaplan–Meier analysis of the overall survival (OS) (D) and disease-specific survival (DSS) rates. (E) Progression-free survival (PFS) (F) in CRC patients with SOX9low and SOX9high tumors in the TCGA database. *P < 0.05, **P < 0.01, and ****P < 0.0001.
To further explore the role of SOX9 in CRC cells, we established CRC cells with SOX9 knockdown and overexpression, termed shSOX9 and oeSOX9 cells, respectively (Supplementary Figures 1A–D). Next, we analyzed shSOX9 and shCtrl RKO cells by transcriptome sequencing, and the results showed that drug sensitivity-related genes were significantly enriched in CRC cells with low SOX9 expression (Figures 2A–D). We detected the early apoptosis rate and cytotoxicity of colorectal cells treated with oxaliplatin. The results showed that loss of SOX9 significantly increased the sensitivity of cells to oxaliplatin, while gain of SOX9 decreased the drug sensitivity of CRC cells (Figures 2E–G and Supplementary Figure 2). To further explore the underlying mechanism of SOX9 in promoting oxaliplatin resistance in CRC cells, we analyzed the transcriptome sequencing results by GSEA and pathway enrichment analysis. The results showed that there were correlations between SOX9 and the DNA damage response (Figures 2H,I). Immunofluorescence and western blot analyses showed that the expression of γH2AX in shSOX9 RKO cells was higher than that in shCtrl cells. However, the expression of γH2AX in oeSOX9 RKO cells was lower than that in Vector cells (Figure 2J). Next, western blotting was conducted to detect the expression of the classical molecules in the DNA damage repair pathway. We found that loss or gain of SOX9 up- or downregulated MMS22L expression, respectively, but had no effect on the other molecules (Figure 2K). In summary, our data suggested that SOX9 affected the sensitivity of CRC cells to oxaliplatin by participating in DNA damage repair.

Figure 2. High expression of SOX9 promotes oxaliplatin resistance by inducing DNA repair in CRC cells. (A) Volcano plot showing the change in gene expression levels in shSOX9 RKO cells compared to control cells. (B) Heatmap showing the change in chemosensitivity and chemoresistance genes in shSOX9 RKO cells compared to control cells. (C,D) GSEA of drug sensitivity genes altered by SOX9 knockdown. (E,F) Representative images (E) and numbers (F) of early apoptotic shSOX9 and oeSOX9 CRC cells compared to control cells. (G) Cytotoxicity assay showing the proliferation of oeSOX9 and shSOX9 CRC cells treated with oxaliplatin compared to control cells. (H) GSEA of genes related to DNA damage altered by SOX9 knockdown in CRC cells. (I) Pathway enrichment analysis showing that the pathways related to DNA damage in shSOX9 CRC cells were extremely different from those in control cells. (J) Immunofluorescence showing representative images of γH2AX (green) in oeSOX9 and shSOX9 CRC cells treated with oxaliplatin compared to control cells. Nuclei were stained with DAPI (blue) (scale bar = 20 μm). (K) Western blotting was used to detect the expression of MMS22L, NBS1, and γH2AX in oeSOX9 and shSOX9 CRC cells treated with oxaliplatin compared to control cells. **p < 0.01; ***p < 0.001; and ****p < 0.0001.
To further verify the relationship between SOX9 and MMS22L, we performed IHC staining of sections from 80 CRC patients. The results showed that the expression of SOX9 and MMS22L had a positive correlation (Figures 3A,B). Moreover, immunofluorescence costaining of these sections showed that SOX9 and MMS22L were colocalized in CRC tissues and cells (Figures 3C,D). Co-IP analysis of SOX9 and MMS22L in CRC cells confirmed this finding (Figure 3E). Furthermore, the SOX9- MMS22L interaction increased with the exposure time to oxaliplatin treatment (Figure 3F). Gain of SOX9 expression promoted the nuclear translocation of MMS22L upon oxaliplatin treatment (Figure 3G). Furthermore, the absence of SOX9 in RKO cells led to the opposite effects (Figure 3G). In summary, our data showed that SOX9 participated in DNA damage repair by promoting the expression and nuclear entry of MMS22L upon oxaliplatin treatment, which induced oxaliplatin resistance in CRC cells.
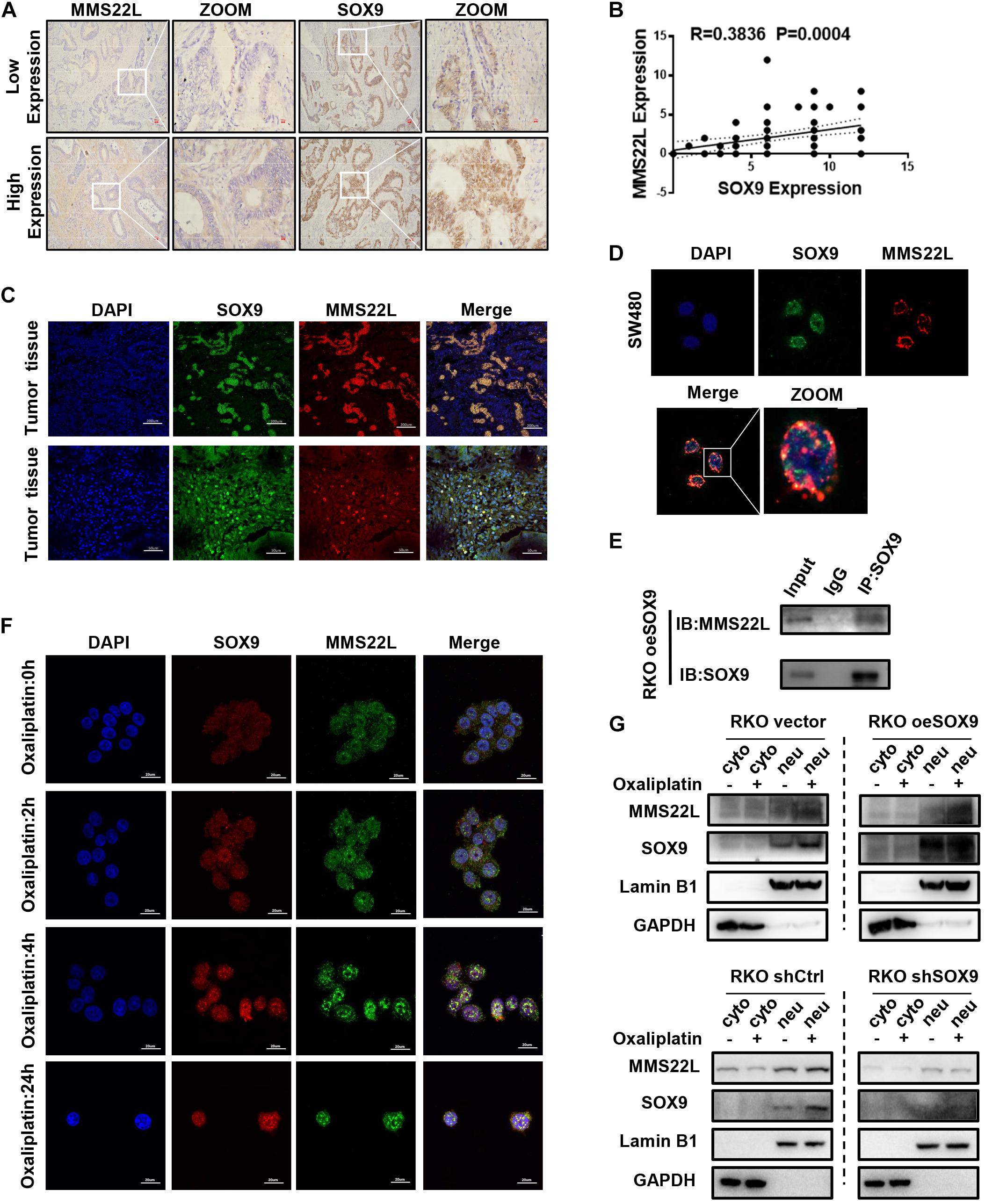
Figure 3. SOX9 enhances chemotherapeutic insensitivity in colorectal cancer by promoting MMS22L expression and nuclear translocation. (A) Representative images showing IHC staining of SOX9 and MMS22L in 2 CRC patients. Scale bar = 50 μm. (B) The correlation between SOX9 and MMS22L expression was analyzed in 80 CRC patients. (C,D) Representative immunofluorescence images showing SOX9 and MMS22L colocalization in CRC tissues and cell lines (scale bar = 20, 50, or 200 μm). (E) IP analysis of the interaction of MMS22L with SOX9 in SOX9-overexpressing RKO cells. (F) Immunofluorescence images showing the time-dependent variation in SOX9 and MMS22L colocalization in RKO cells treated with oxaliplatin. Scale bar = 20 μm. (G) Nuclear fractions were extracted from CRC cells, and the nuclear (neu) and cytoplasmic (without the nucleus; cyto) fractions were prepared. Immunoblot showing the protein levels of MMS22L and SOX9 expressed in Vector and oeSOX9 RKO cells and in shCtrl and shSOX9 RKO cells.
To explore the mechanism of oxaliplatin resistance in CRC cells with high SOX9 expression, we established shMMS22L and oeSOX9+shMMS22L RKO cells (Supplementary Figures 1E,F). Consistent with the previous results, immunofluorescence showed that overexpression of SOX9 augmented DNA repair under oxaliplatin treatment in CRC cells, but oeSOX9 RKO cells with loss of MMS22L showed less DNA repair activity than Vector RKO cells (Figures 4A,B). The results of early apoptosis detection by flow cytometry further demonstrated that MMS22L abolished the effects of SOX9 overexpression in cells treated with oxaliplatin (Figures 4C,D). In summary, our data indicated that SOX9 promoted oxaliplatin resistance in CRC cells through its interaction with MMS22L.
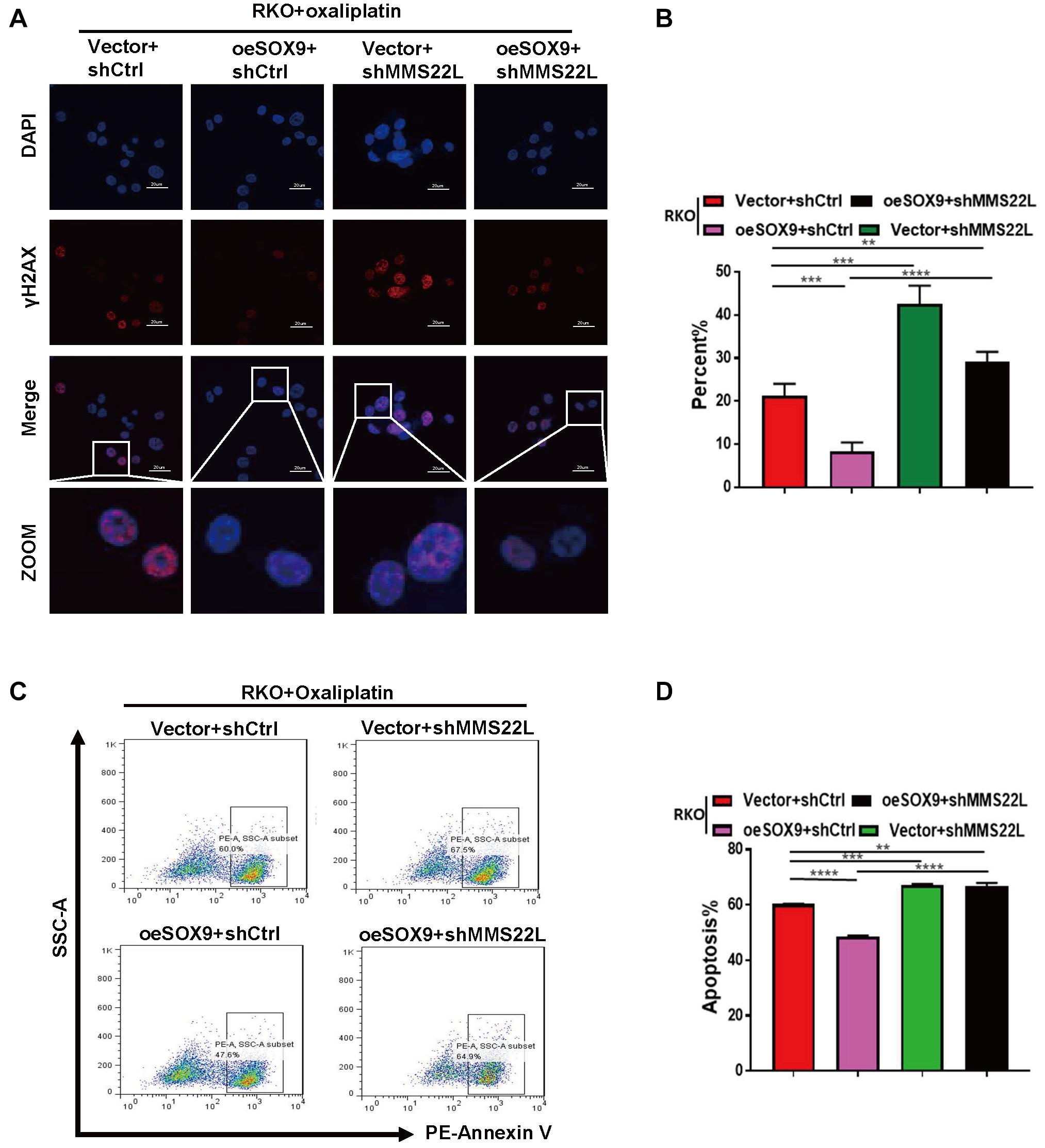
Figure 4. Loss of MMS22L enhances chemosensitivity in SOX9-overexpressing CRC cells. (A,B) Immunofluorescence images showing the typical appearance (A) and percentages (B) of γH2AX-positive cells in the indicated groups; nuclei were stained with DAPI (blue). Scale bar = 20 μm. (C,D) Flow cytometry results showing early apoptosis and the percentage of apoptotic cells among Vector+shCtrl, oeSOX9+shCtrl, Vector+shMMS22L, and oeSOX9+shMMS22L cells treated with oxaliplatin. **p < 0.01; ***p < 0.001; and ****p < 0.0001.
To further demonstrate the effect of SOX9 on CRC cells in vivo, we established a subcutaneous xenograft model in nude mice. Mice were implanted with Vector +shCtrl, oeSOX9+shCtrl, Vector+shMMS22L or oeSOX9+shMMS22L RKO cells and treated with oxaliplatin (75 mg/kg) (Figure 5A). After 28 days, the xenografts were harvested. The results showed that oxaliplatin had better therapeutic efficacy against oeSOX9+shMMS22L RKO tumors than oeSOX9+shCtrl tumors (Figures 5A–D). IHC staining showed that the expression of γH2AX in the oeSOX9+shCtrl group was lower than that in the oeSOX9+shMMS22L group (Figure 5E). Therefore, these results indicate that SOX9 upregulation increases oxaliplatin resistance in CRC cells in vivo and that knocking down MMS22L reverses this effect.
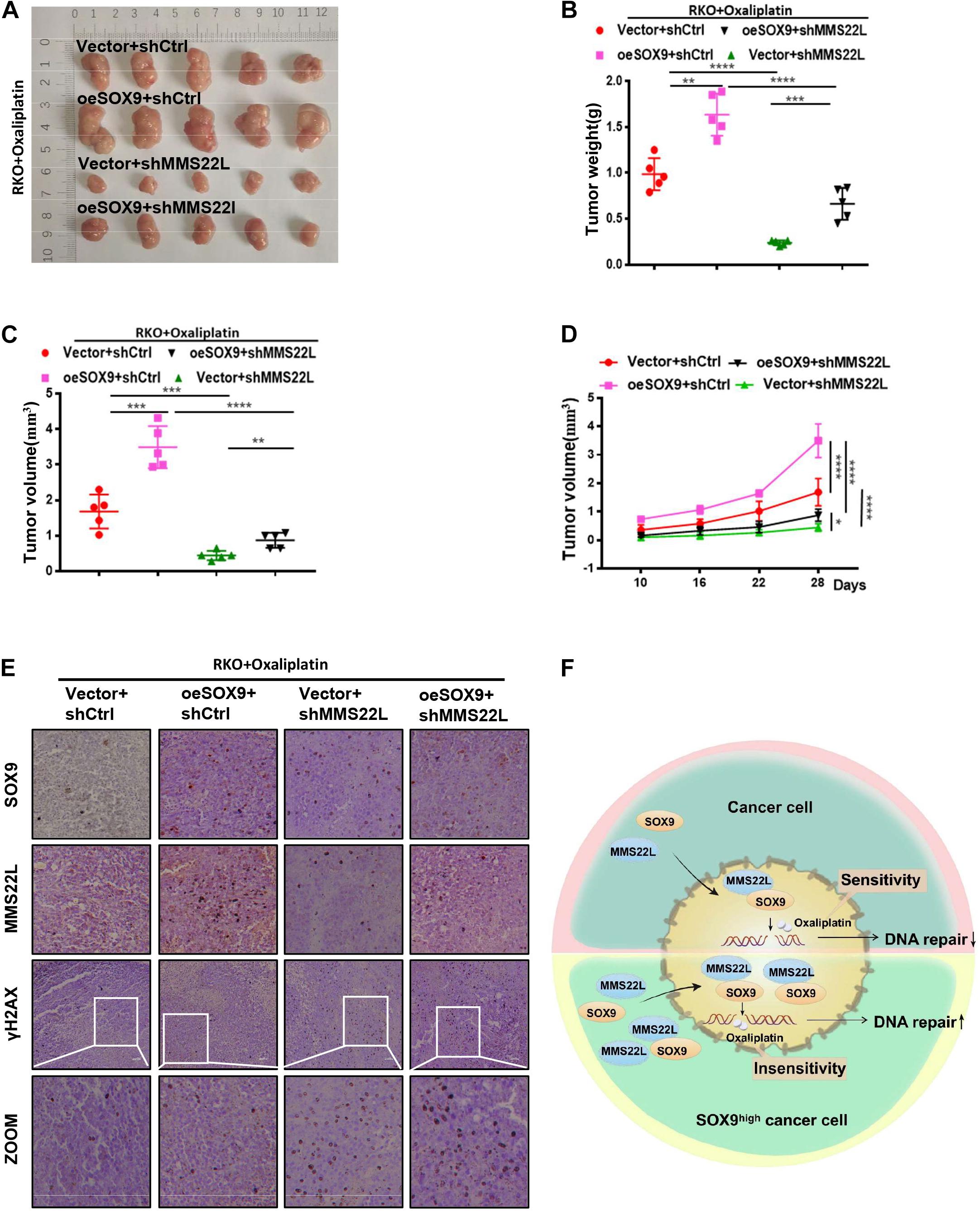
Figure 5. Loss of MMS22L enhances chemosensitivity in SOX9-overexpressing CRC cells in vivo. (A–D) Antitumor outcomes of RKO xenografts in male nude mice treated with oxaliplatin, including photographs of the tumors (A), average tumor weights (B), and tumor volumes (C), at the end of the experimental period, along with the time-dependent variation curves of tumor volume (D). (E) IHC staining images showing the expression of SOX9, MMS22L, and γH2AX in xenografts formed from Vector+shCtrl, oeSOX9+shCtrl, Vector+shMMS22L, and oeSOX9+shMMS22L cells. Scale bar = 50 μm. (F) Schematic diagram of the regulatory mechanism by which SOX9-mediated expression and nuclear translocation of MMS22L promotes DNA damage repair in CRC cells. **p < 0.01; ***p < 0.001; and ****p < 0.0001.
Cancer cells aquire drug resistance through different mechanisms (Chen and Shen, 2020; da Costa and Baiocchi, 2020; Qin et al., 2020; Zhu et al., 2020; Pastushenko et al., 2021), including enhancement of oxygen free radical scavenging, promotion of DNA damage repair, or inhibition of apoptotic signaling pathway activation (Lü et al., 2008; Bakkenist et al., 2018; Phi et al., 2018). In vitro studies suggest a role for SOX9 in regulating cell differentiation in the intestinal epithelium (Mori-Akiyama et al., 2007; Aguilar-Medina et al., 2019). A transcription factor related to stem cell maintenance, SOX9 is highly expressed in CRC and indicates poor prognosis (Aguilar-Medina et al., 2019). In human specimens, we confirmed that the expression of SOX9 in CRC was significantly upregulated compared to that in adjacent tissues and verified that SOX9 upregulation can promote the formation of CRC stem cells (Supplementary Figures 3A–C), similar to previous results. Recent studies on CRC have found that the overexpression of SOX9 in CRC is related to anticancer drug resistance, but the specific mechanism of drug resistance is not well studied. Considering the characteristics of cancer stem cells, we explored whether SOX9 participates in anticancer drug resistance by affecting DNA damage repair in cancer cells. Our results showed that SOX9 can affect the sensitivity of cancer cells to oxaliplatin by affecting the DNA damage repair. Overexpression of SOX9 significantly enhanced DNA damage repair, thus increasing the resistance of cancer cells to oxaliplatin. After knockdown of SOX9, this effect was significantly reversed. Moreover, we observed the expression of classical DNA damage repair-related molecules after knocking down or overexpressing SOX9 and found that SOX9 can affect MMS22L expression.
MMS22L is mainly localized in the nucleus and has been reported to be critical for protecting against DSBs formation in S-phase cells by dealing with replication fork stalling/collapse events (Piwko et al., 2011). Further research has shown that MMS22L participates in DNA repair by promoting the displacement of the single-stranded DNA binding protein RPA and assembly of RAD51 filaments at DNA damage sites in response to CPT (Piwko et al., 2016; Huang et al., 2018). It has been reported that SOX9 can usually influence related pathways by regulating the transcription of certain molecules. However, knockdown of SOX9 did not alter the mRNA level of MMS22L. In this study, we further confirmed an interaction between SOX9 and MMS22L by immunofluorescence and immunoprecipitation experiments. Furthermore, nucleocytoplasmic separation further confirmed that SOX9 increased the enrichment of MMS22L in the nucleus under treatment with oxaliplatin. Therefore, our results elucidated that SOX9 can participate in DNA damage repair by affecting the expression and nuclear translocation of MMS22L (Figure 5F).
In this manuscript, through overexpression or knockdown of SOX9 in CRC cells and a series of subsequent experiments, we elucidated that SOX9 can affect the sensitivity of CRC cells to oxaliplatin and preliminarily explored the mechanism of this phenomenon. However, there was a relative absence of SOX9 expression in drug-resistant CRC cells compared with wild-type cells. Therefore, we will screen oxaliplatin-resistant CRC cells in our next study and continue to perform more in-depth research. In conclusion, our study confirmed that SOX9, a transcription factor, is involved in the maintenance of stemness and the induction of drug resistance in CRC cells. Furthermore, we revealed a mechanism by which SOX9 regulates the expression and nuclear translocation of MMS22L and oxaliplatin resistance in CRC cells and provided a new focus for solving the problem of oxaliplatin resistance in CRC patients.
The data presented in the study are deposited in the SRA repository, accession number PRJNA707264.
The animal study was reviewed and approved by the Ethics Committee of Affiliated Tumor Hospital of Guangxi Medical University. All authors have consented to publication of the results presented in this manuscript.
YL, HW, and TL performed the experiments. YL, HW, TL, QL, ZM, YS, FL, ML, XP, and JL provided technical support. CX, WT, XP, and JL provided critical comments. YL, HW, TL, CX, WT, XP, and JL analyzed the data, designed the experiments, and wrote the manuscript. All authors read and approved the final manuscript.
The work was supported by research grants from the National Natural Science Foundation of China (no. 82004006 and 81873048) and Sichuan Provincial Science Fund for Distinguished Young Scholars of China (2020JDJQ0065).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.646542/full#supplementary-material
Abad, E., Graifer, D., and Lyakhovich, A. (2020). DNA damage response and resistance of cancer stem cells. Cancer Lett. 474, 106–117. doi: 10.1016/j.canlet.2020.01.008
Abdul Khalek, F. J., Gallicano, G. I., and Mishra, L. (2010). Colon cancer stem cells. Gastrointest Cancer Res. (Suppl 1), S16–S23.
Aguilar-Medina, M., Avendaño-Félix, M., Lizárraga-Verdugo, E., Bermúdez, M., Romero-Quintana, J. G., Ramos-Payan, R., et al. (2019). SOX9 stem-cell factor: clinical and functional relevance in cancer. J. Oncol. 2019:6754040. doi: 10.1155/2019/6754040
Anuja, K., Chowdhury, A. R., Saha, A., Roy, S., Rath, A. K., Kar, M., et al. (2019). Radiation-induced DNA damage response and resistance in colorectal cancer stem-like cells. Int. J. Radiat. Biol. 95, 667–679. doi: 10.1080/09553002.2019.1580401
Bakkenist, C. J., Lee, J. J., and Schmitz, J. C. (2018). ATM is required for the repair of oxaliplatin-induced DNA damage in colorectal cancer. Clin. Colorectal Cancer 17, 255–257. doi: 10.1016/j.clcc.2018.09.001
Carrasco-Garcia, E., Lopez, L., Aldaz, P., Arevalo, S., Aldaregia, J., Egaña, L., et al. (2016). SOX9-regulated cell plasticity in colorectal metastasis is attenuated by rapamycin. Sci. Rep. 6:32350. doi: 10.1038/srep32350
Chen, D., Zhang, M., Ruan, J., Li, X., Wang, S., Cheng, X., et al. (2020). The long non-coding RNA HOXA11-AS promotes epithelial mesenchymal transition by sponging miR-149-3p in colorectal cancer. J. Cancer 11, 6050–6058. doi: 10.7150/jca.49809
Chen, S., and Shen, X. (2020). Long noncoding RNAs: functions and mechanisms in colon cancer. Mol. Cancer 19:167. doi: 10.1186/s12943-020-01287-2
Christin, J. R., Wang, C., Chung, C. Y., Liu, Y., Dravis, C., Tang, W., et al. (2020). Stem cell determinant SOX9 promotes lineage plasticity and progression in basal-like breast cancer. Cell Rep. 31:107742. doi: 10.1016/j.celrep.2020.107742
da Costa, A., and Baiocchi, G. (2020). Genomic profiling of platinum-resistant ovarian cancer: the road into druggable targets. Semin. Cancer Biol. 20:30221-2: doi: 10.1016/j.semcancer.2020.10.016
Das, P. K., Islam, F., and Lam, A. K. (2020). The roles of cancer stem cells and therapy resistance in colorectal carcinoma. Cells 9:1392. doi: 10.3390/cells9061392
Duro, E., Lundin, C., Ask, K., Sanchez-Pulido, L., MacArtney, T. J., Toth, R., et al. (2010). Identification of the MMS22L-TONSL complex that promotes homologous recombination. Mol. Cell 40, 632–644. doi: 10.1016/j.molcel.2010.10.023
Hochster, H. S., and Sargent, D. J. (2016). One good DNA-damage deserves another: oxaliplatin in MSI-high colon cancer. J. Natl. Cancer Inst. 108:djw011. doi: 10.1093/jnci/djw011
Huang, T. H., Fowler, F., Chen, C. C., Shen, Z. J., Sleckman, B., and Tyler, J. K. (2018). The histone chaperones ASF1 and CAF-1 promote MMS22L-TONSL-mediated Rad51 loading onto ssDNA during homologous recombination in human cells. Mol. Cell 69, 879.e–892.e. doi: 10.1016/j.molcel.2018.01.031
Kaushik, G., Seshacharyulu, P., Rauth, S., Nallasamy, P., Rachagani, S., Nimmakayala, R. K., et al. (2021). Selective inhibition of stemness through EGFR/FOXA2/SOX9 axis reduces pancreatic cancer metastasis. Oncogene 40, 848–862. doi: 10.1038/s41388-020-01564-w
Li, L., and Neaves, W. B. (2006). Normal stem cells and cancer stem cells: the niche matters. Cancer Res. 66, 4553–4557. doi: 10.1158/0008-5472.Can-05-3986
Liu, Z., Xie, Y., Xiong, Y., Liu, S., Qiu, C., Zhu, Z., et al. (2020). TLR 7/8 agonist reverses oxaliplatin resistance in colorectal cancer via directing the myeloid-derived suppressor cells to tumoricidal M1-macrophages. Cancer Lett. 469, 173–185. doi: 10.1016/j.canlet.2019.10.020
Lü, B., Fang, Y., Xu, J., Wang, L., Xu, F., Xu, E., et al. (2008). Analysis of SOX9 expression in colorectal cancer. Am. J. Clin. Pathol. 130, 897–904. doi: 10.1309/ajcpw1w8gjbqgcni
Mori-Akiyama, Y., van den Born, M., van Es, J. H., Hamilton, S. R., Adams, H. P., Zhang, J., et al. (2007). SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology 133, 539–546. doi: 10.1053/j.gastro.2007.05.020
O’Connell, B. C., Adamson, B., Lydeard, J. R., Sowa, M. E., Ciccia, A., Bredemeyer, A. L., et al. (2010). A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L-NFKBIL2 complex required for genomic stability. Mol. Cell 40, 645–657. doi: 10.1016/j.molcel.2010.10.022
O’Donnell, L., Panier, S., Wildenhain, J., Tkach, J. M., Al-Hakim, A., Landry, M. C., et al. (2010). The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination. Mol. Cell 40, 619–631. doi: 10.1016/j.molcel.2010.10.024
Pastushenko, I., Mauri, F., Song, Y., de Cock, F., Meeusen, B., Swedlund, B., et al. (2021). Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis. Nature 589, 448–455. doi: 10.1038/s41586-020-03046-1
Phi, L. T. H. I, Sari, N., Yang, Y. G., Lee, S. H., Jun, N., Kim, K. S., et al. (2018). Cancer Stem Cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int. 2018:5416923. doi: 10.1155/2018/5416923
Piwko, W., Buser, R., and Peter, M. (2011). Rescuing stalled replication forks: MMS22L-TONSL, a novel complex for DNA replication fork repair in human cells. Cell Cycle 10, 1703–1705. doi: 10.4161/cc.10.11.15557
Piwko, W., Mlejnkova, L. J., Mutreja, K., Ranjha, L., Stafa, D., Smirnov, A., et al. (2016). The MMS22L-TONSL heterodimer directly promotes RAD51-dependent recombination upon replication stress. Embo J. 35, 2584–2601. doi: 10.15252/embj.201593132
Qian, Y., Wu, X., Yokoyama, Y., Okuzaki, D., Taguchi, M., Hirose, H., et al. (2019). E-cadherin-Fc chimera protein matrix enhances cancer stem-like properties and induces mesenchymal features in colon cancer cells. Cancer Sci. 110, 3520–3532. doi: 10.1111/cas.14193
Qin, S., Jiang, J., Lu, Y., Nice, E. C., Huang, C., Zhang, J., et al. (2020). Emerging role of tumor cell plasticity in modifying therapeutic response. Signal Transduct. Target. Ther. 5:228. doi: 10.1038/s41392-020-00313-5
Rothenberg, M. L. (2000). Efficacy of oxaliplatin in the treatment of colorectal cancer. Oncology (Williston Park) 14(12 Suppl 11), 9–14.
Siegel, R. L., Miller, K. D., Goding Sauer, A., Fedewa, S. A., Butterly, L. F., Anderson, J. C., et al. (2020). Colorectal cancer statistics, 2020. CA Cancer J. Clin. 70, 145–164. doi: 10.3322/caac.21601
Wu, H., Wang, T., Liu, Y., Li, X., Xu, S., Wu, C., et al. (2020). Mitophagy promotes sorafenib resistance through hypoxia-inducible ATAD3A dependent Axis. J. Exp. Clin. Cancer Res. 39:274. doi: 10.1186/s13046-020-01768-8
Zhu, L., Sun, H. T., Wang, S., Huang, S. L., Zheng, Y., Wang, C. Q., et al. (2020). Isolation and characterization of exosomes for cancer research. J. Hematol. Oncol. 13:152. doi: 10.1186/s13045-020-00987-y
Keywords: colorectal cancer, SOX9, MMS22L, DNA damage repair, oxaliplatin resistance
Citation: Liu Y, Wu H, Luo T, Luo Q, Meng Z, Shi Y, Li F, Liu M, Peng X, Liu J, Xu C and Tang W (2021) The SOX9-MMS22L Axis Promotes Oxaliplatin Resistance in Colorectal Cancer. Front. Mol. Biosci. 8:646542. doi: 10.3389/fmolb.2021.646542
Received: 27 December 2020; Accepted: 20 April 2021;
Published: 27 May 2021.
Edited by:
Xin Zhang, Jiangmen Central Hospital, ChinaCopyright © 2021 Liu, Wu, Luo, Luo, Meng, Shi, Li, Liu, Peng, Liu, Xu and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuan Xu, eHVjaHVhbjEwMEAxNjMuY29t; Weizhong Tang, dGFuZ3dlaXpob25nQGd4bXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.