- 1Dongzhimen Hospital Affiliated to Beijing University of Chinese Medicine, Beijing, China
- 2Department of TCM, Peking University People’s Hospital, Beijing, China
- 3Xiyuan Hospital Affiliated to China Academy of Chinese Medical Sciences, Beijing, China
- 4Department of TCM, Peking University International Hospital, Beijing, China
- 5Department of Integration of Chinese and Western Medicine, School of Basic Medical Sciences, Peking University, Beijing, China
Background: Asthma is a chronic inflammatory disease characterized by Th2-predominant inflammation and airway remodeling. Modified Guo Min decoction (MGMD) has been an extensive practical strategy for allergic disorders in China. Although its potential anti-asthmatic activity has been reported, the exact mechanism of action of MGMD in asthma remains unexplored.
Methods: Network pharmacology approach was employed to predict the active components, potential targets, and molecular mechanism of MGMD for asthma treatment, including drug-likeness evaluation, oral bioavailability prediction, protein–protein interaction (PPI) network construction and analysis, Gene Ontology (GO) terms, and Reactome pathway annotation. Molecular docking was carried out to investigate interactions between active compounds and potential targets.
Results: A total of 92 active compounds and 72 anti-asthma targets of MGMD were selected for analysis. The GO enrichment analysis results indicated that the anti-asthmatic targets of MGMD mainly participate in inflammatory and in airway remolding processes. The Reactome pathway analysis showed that MGMD prevents asthma mainly through regulation of the IL-4 and IL-13 signaling and the specialized pro-resolving mediators (SPMs) biosynthesis. Molecular docking results suggest that each bioactive compounds (quercetin, wogonin, luteolin, naringenin, and kaempferol) is capable to bind with STAT3, PTGS2, JUN, VEGFA, EGFR, and ALOX5.
Conclusion: This study revealed the active ingredients and potential molecular mechanism by which MGMD treatment is effective against airway inflammation and remodeling in asthma through regulating IL-4 and IL-13 signaling and SPMs biosynthesis.
Introduction
Asthma is a globally prevalent chronic inflammatory pulmonary disease, with an estimated 358 million affected individuals (Soriano et al., 2017). Persistent airway inflammation is the hallmark of asthma, which is infiltrated eosinophils, T lymphocytes, mast cells, and release of pro-inflammatory cytokines and lipid mediators. Lipid mediators, such as specialized pro-resolving mediators (SPMs), play a decisive role in the resolution of inflammation in asthma (Schett and Neurath, 2018; Kytikova et al., 2019). Actions of SPMs generated from polyunsaturated fatty acids (PUFAs) include inhibition of eosinophil and neutrophil migration, and decrease of T helper 2 (Th2) cytokine production (e.g., IL-4, IL-5, and IL-13) (Barnig et al., 2018). SPMs are originated via lipoxygenase (LOX)–catalyzed reaction. SPMs include omega-6 PUFA arachidonic acid derived lipoxins, as well as omega-3 PUFA eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) derived resolvins, protectins and maresins. Arachidonic acid also forms a range of pro-inflammatory mediators in asthma, such as prostaglandins via cyclooxygenase (COX)-2, and the leukotrienes via 5-LOX actions (Krishnamoorthy et al., 2018).
Airway remodeling is another key pathological features of asthma. It is characterized by thickening of basement membrane, goblet cell metaplasia, smooth muscle cell hyperplasia and hypertrophy, deposition of extracellular matrix proteins, vascular activation and angiogenesis (Jeffery, 2001; James and Wenzel, 2007). Synthesis of extracellular matrix components and collagen is promoted by epidermal growth factor (EGF) released by airway epithelial cells and TGF-β produced by myofibroblasts (Flood-Page et al., 2003; Le Cras et al., 2011). Under the influence of airway epithelial cells-derived endothelial growth factor (VEGF)-A, angiogenesis allow for extravasation of more inflammatory cells (Lambrecht and Hammad, 2012).
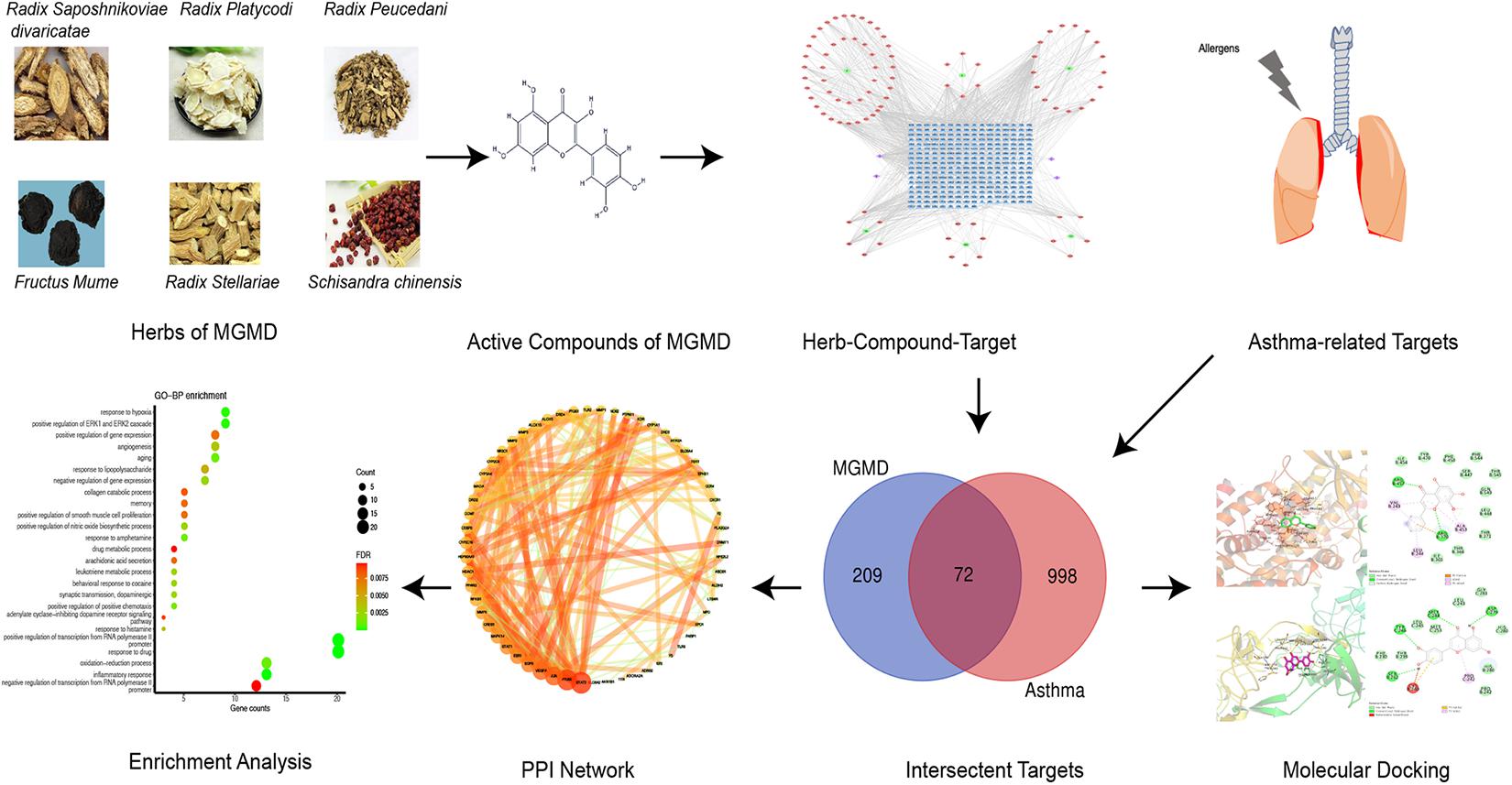
Traditional Chinese medicine (TCM) has been used to treat allergic diseases in Asia for hundreds of years (Liu L. et al., 2018). “Guo Min” means allergy in Chinese and Guo Min decoction (GMD) is a compound recipe composed by herbs, that raised by a well-known Chinese physician, Zhu Shenyu (Zhu and Liang, 1985). GMD has been widely used for allergic diseases treatment in China (Tao et al., 2018). The Modified Guo Min decoction (MGMD) comprises Fang Feng (Radix Saposhnikoviae divaricate, FF), Wu Mei (Fructus Mume, WM), Wu Wei Zi (Schisandra chinensis, WWZ), Yin Chai Hu (Radix Stellariae, YCH), Qian Hu (radix Peucedani, QH), and Jie Geng (Radix Platycodi, JG). Previous studies showed that prescriptions based on GMD increased the proportion of Th1 cells in the spleen tissue of asthmatic mice, reduced the Th2 subset of T cells, and balanced the number of Treg and Th17 cells (Qin et al., 2017). However, the therapeutic mechanisms of MGMD in asthma remain unclear. In this study, network pharmacology approach was used to explore the multi-components, multi-targets, and multi-pathways closely related to the anti-asthma effect of MGMD. The molecular docking method was also used to verify the binding of the active substance to the target of action to reveal and predict the efficacy of MGMD in asthma. The work scheme is shown in Figure 1.
Materials and Methods
Active Compounds Collection
The compounds of MGMD were extracted from the HERB database1 (Fang et al., 2020) and the encyclopedia of traditional Chinese medicine (ETCM)2 (Xu et al., 2019). HERB contains a comprehensive list of 7263 TCM herbs and 49258 ingredients by integrating multiple TCM databases included SymMap (Wu et al., 2019), TCMID 2.0 (Huang et al., 2018), TCMSP 2.3 (Ru et al., 2014), and TCM-ID (Chen et al., 2006). Oral bioavailability (OB) ≥ 30% and drug-likeness (DL) ≥ 0.18 were set as the thresholds to screen the active compounds from the TCMSP3. The properties of the other ingredients collected from the HERB were retrieved from the Swiss absorption, distribution, metabolism, and excretion (ADME) database4. The SMILES format files of the compounds were obtained from the PubChem5 database (Kim et al., 2019). The screening criterion for gastrointestinal GI absorption was set as high, and DL was satisfied with both “yes” at the same time (Daina et al., 2017). Active ingredients from ETCM were retrieved as DL Grading “Good” (Xu et al., 2019).
Target Fishing
Identification of Predicted Targets of MGMD
The active ingredients in drugs exert their biological effects via molecular targets. The targets related to the active compounds presented in MGMD were searched from ETCM (Xu et al., 2019) and Similarity ensemble approach (SEA)6 (Keiser et al., 2007). ETCM predicts targets by MedChem Studio and only outputs these targets with confidence score at least 0.8. The threshold of “human” and Tanimoto coefficients (Tc) > 0.57 were set to filter results predicted by SEA. The key targets corresponding to the compounds in such six herbs were obtained.
Building the Disease Target Database
Asthma-related targets were identified by integrating data from these databases: Comparative Toxicogenomics Database (CTD)7 (Grondin et al., 2018), Online Mendelian Inheritance in Man (OMIM)8 (Amberger and Hamosh, 2017), and the Human Gene Database (GeneCards)9 (Safran et al., 2010). For filtering the targets, “asthma” was defined as the key word and results were restricted to human genes/proteins. Targets with a relevance score above the median were selected in GeneCards database. The overlapping targets between MGMD and asthma were likely to be potential action targets of MGMD in the treatment of asthma.
PPI Network Construction and Analysis
To obtain the data of protein–protein interaction (PPI), the candidate targets were inputted to STRING10 with a minimum required interaction score > 0.7 and limited to “Homo sapiens” specie (von Mering et al., 2005). The visual network graphs were created by Cytoscape (version 3.7.1)11, an open-source software platform for visualizing complex networks (Shannon et al., 2003). The target interaction network parameters were calculated by NetworkAnalyzer.
Molecular Complex Detection (MCODE) of Cytoscape was used to searching the highly connected sub-networks in the PPI network. In the illustrations depicting MCODE results, a vertex-weighting-based scheme was used to identify local high-density areas in the graph. Density was generally defined based on the connection level of the graph (Bader and Hogue, 2003).
Protein Functional Enrichment Analysis
To reveal the potential biological functions of MGMD in the treatment of asthma, functional enrichment analyses were performed by the Database for Annotation, Visualization and Integrated Discovery (DAVID, v6.8)12 and Reactome13 (Fabregat et al., 2018). The results for Gene ontology (GO) biological processes (BPs) and Reactome pathway enrichment were saved and sorted by the adjusted P value corrected by the false discovery rate (FDR) algorithm for each term.
Network Construction
To demonstrate the multi-compound therapeutic features of MGMD, network constructions were performed as follows: (1) herb-compound-target Network (H-C-T network) was constructed to explore the active compounds and their potential targets. The core compounds were obtained through the H-C-T network. (2) PPI networks were built to analyze the target interactions. Hub targets involved in MGMD treatment of asthma were selected from the PPI network. (3) BP sub-networks were established for classification analysis of BPs in MGMD treatment for asthma. (4) Target pathway network (T-P network) was constructed to show the functional pathways of MGMD for the therapy of asthma.
Molecular Docking
Molecular docking was conducted to validate if MGMD’s compounds could bind to these targets. The 2D structures of the top five core compounds were downloaded from the TCMSP database (Ru et al., 2014). The structures were added charge and displayed rotatable keys by AutoDock Tools (version 1.5.6). The protein crystal structures corresponding to the core target genes were downloaded from the Protein Data Bank database (PDB)14 (Burley et al., 2017). Water and hetero molecules of the proteins were removed by Pymol. Hydrogen atoms and charge operations to the proteins was added by AutoDock Tools. The 3D Grid box for molecular docking simulation was also obtained by AutoDock tools was displayed by AutoDock Vina (version 1.1.2) (Trott and Olson, 2010). The results were analyzed and interpreted by PyMOL and Discovery Studio 2020.
Results
Construction of Herb-Compound-Target Network
In this study, 96 active compounds were screened from the six herbs in MGMD. Among them, 51, 19, 7, 6, 8, and 5 compounds were from FF, QH, JG, WM, WWZ, and YCH, respectively. MGMD contains a complex mixture of ingredients, some of them overlapped across 2 herbs, including decursinol, deoxygomisin A, nodakenetin, and naringenin. A total of 92 active compounds were identified after eliminating redundant entries.
Five hundred and twenty-three targets were associated with the 92 components identified in MGMD, of which 149 were associated with FF, 151 with QH, 83 with JG, 77 with WM, 23 with WWZ, and 40 with YCH. After eliminating overlapping targets, there were 281 targets remaining.
The H-C-T network of MGMD was visualized in Cytoscape (Figure 2). The network contained 379 nodes and 1021 edges. Quercetin showed the highest degree of connectivity in the network with 76 targets, followed by wogonin with 57, luteolin with 55, naringenin with 51, and kaempferol with 40. The properties of the H-C-T network were suitable for displaying complex ingredients, multiple targets, and close interactions between ingredients and targets. Detailed information about the active compounds and targets identified in MGMD is shown in Supplementary Table 1.
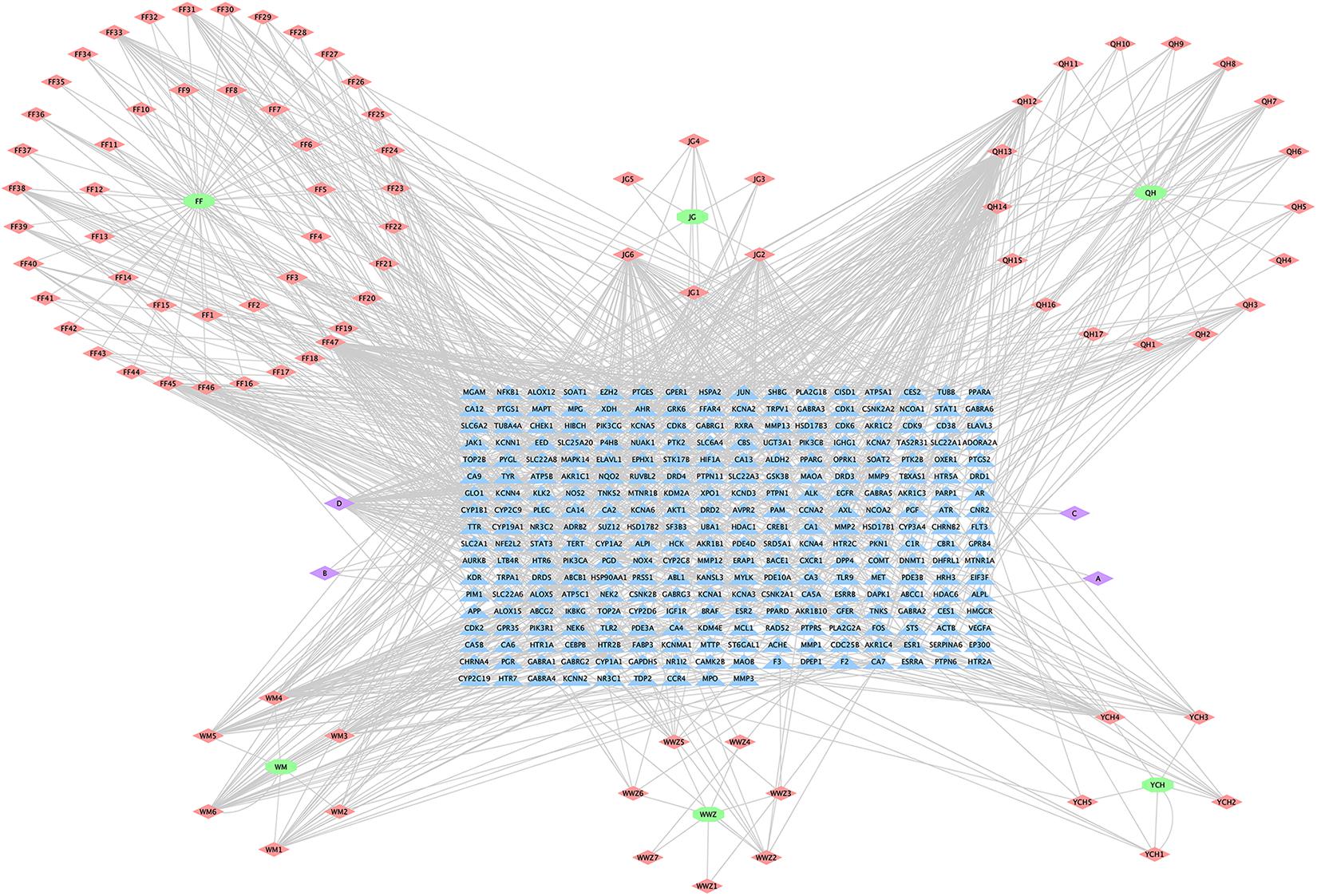
Figure 2. Herb-Compound-Target network (H-C-T network) of MGMD. Green ellipses represent the herbs present in MGMD; pink diamonds represent active compounds in each herb; purple diamonds represent active compounds shared by two herbs, and blue triangles correspond to related targets (The IDs of the components are described in Supplementary Table 1).
Potential Asthma Targets
The targets for asthma were integrated from multi-source databases and a final list of 1,070 disease-related targets obtained after eliminating duplicates (Supplementary Table 2). 72 overlapping targets were identified as the key targets for studying the anti-asthmatic activity of the MGMD compounds (Supplementary Table 3).
Analysis of the Network of Overlapping Targets
Protein–Protein Interaction (PPI) Network
The STRING database was used to acquire PPI relationships of 72 potential protein targets of MGMD as related to the treatment of asthma. The visualized PPI network was constructed by Cystoscape 3.7.1, constructing by 72 nodes represented proteins and 205 edges represented the interactions between the proteins (Figure 3A and Supplementary Table 4). The average node degree value of the PPI network was 6.508. 28 protein targets had significantly higher node degree than the average in the PPI network, including STAT3, PTGS2, JUN, vascular endothelial growth factor A (VEGFA), EGFR, ESR1, STAT1, CREB1, MAPK14, MMP9, NFKB1, PPARG, CYP2C19, HDAC1, HSP90AA1, CYP3A4, COMT, MAOA, CYP2C9, DRD2, CEBPB, ALOX15, PTGS1, arachidonate 5-lipoxygenase (ALOX5), DRD4, NR3C1, MMP3, and MMP2. These 28 targets showed the preliminary relationships between active compound and potential protein targets of MGMD treatment in asthma.
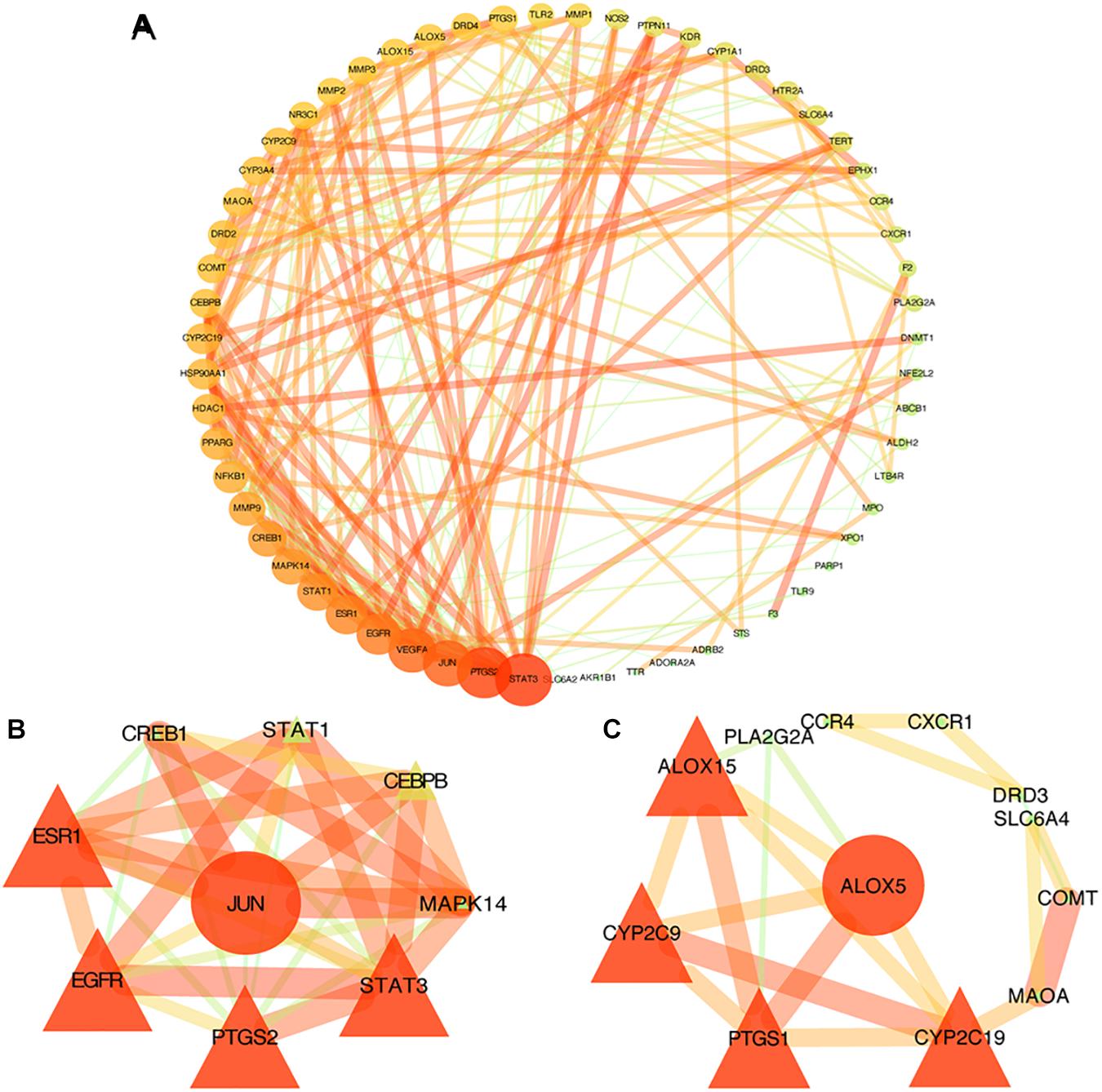
Figure 3. PPI networks of MGMD for asthma treatment. Each node represents a protein target and each line represents the interaction between two nodes. Nodes in red are important and nodes in green are less important in the network. Panel (A) is the PPI network diagram arranged according to the df. Two clusters detected in the MGMD-asthma PPI network. Panels (B,C) show clusters 1 and 2, respectively. The ellipses are seed nodes of each cluster.
Clustering Analysis of the PPI Network
Similar function clusters of the PPI network were selected by MCODE analysis based on topology to find densely interconnected regions using Cytoscape software. 2 clusters of functional modules were detected (K-core = 4) and the attribute values of the cluster were shown in Figures 3B,C.
Clusters in a PPI network are often protein complexes and parts of pathways, while clusters in a protein similarity network represent protein families. Cluster 1 comprised nine nodes and 33 edges with a score of 8.250 (Figure 3B). The seed node of this cluster was JUN (Jun proto-oncogene, also known as AP-1, cJUN, c-Jun), which involved in cell proliferation, differentiation, migration, transformation, and programmed cell death (Liu and Lin, 2007). Cluster 2 comprised 12 nodes and 21 sides with a score of 3.818 (Figure 3C). The seed node of this cluster was ALOX5 (arachidonate 5-lipoxygenase, also known as 5-LO, 5-LOX), an essential enzyme in the metabolism of arachidonic acid, which initiates the biosynthesis of leukotrienes (Bruno et al., 2018). Leukotrienes are powerful immune-regulating lipid mediators with established pathogenic roles in asthma. The connected proteins of ALOX5 in cluster 2 participating in the biosynthetic pathways of SPMs (Barnig et al., 2018; Kytikova et al., 2019), such as ALOX15 (15-LOX), cytochrome P450 proteins CYP2C9 and CYP2C19, are among the 28 protein targets which had higher node degree than the average in our PPI network. Thus, the constructed PPI network and the further functional clustering both indicate that MGMD intervene asthma by regulating SPMs biosynthesis.
GO Enrichment Analysis
To further explore the biological functions of the 72 potential targets of MGMD, GO enrichment analysis was constructed on DAVID website and obtained 76 terms (FDR < 0.05) of BPs. We found the top 25 most enriched BP terms represented in a bubble chart (Figure 4A and Supplementary Table 5). BP analysis revealed that the potential targets of MGMD were remarkably enriched in inflammation and airway remodeling.
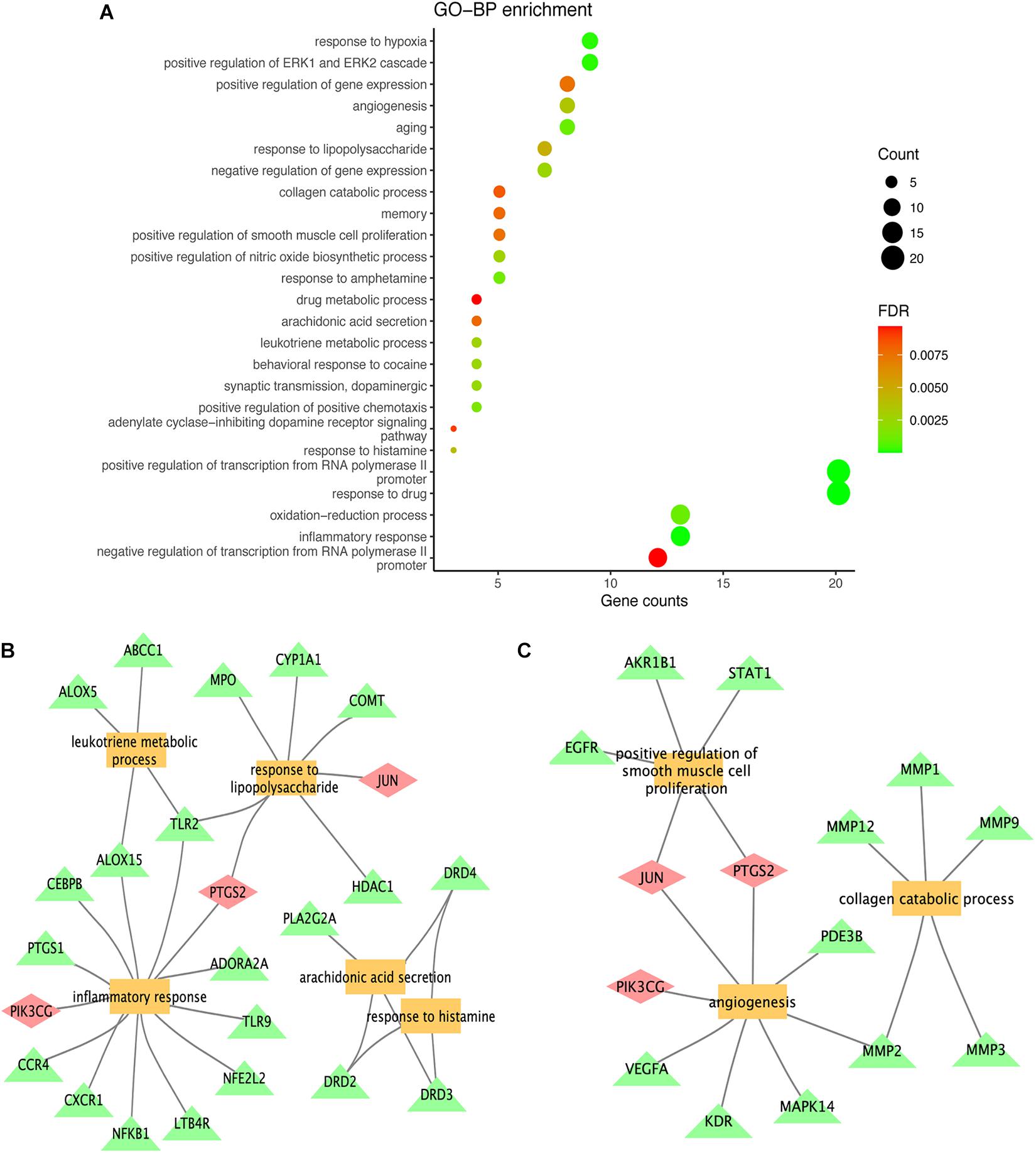
Figure 4. Results of GO term enrichment analysis of targets. The GO bar plot represents the top 25 biological (BP) process (A); inflammation and airway remodeling related to functional clusters represented by panels (B,C), respectively. The yellow squares represent biological processes, the green triangles represent genes, and the pink diamonds represent genes common to both categories.
As shown in Figure 4B, the potential targets of MGMD were involved in inflammatory response (GO ID: 0006954), response to lipopolysaccharide (GO ID: 0032496), leukotriene metabolic process (GO ID: 0006691), arachidonic acid secretion (GO ID: 0050482) and positive regulation of nitric oxide biosynthetic process (GO ID: 0045429). In addition, the BPs of airway remodeling were mainly enriched in positive regulation of smooth muscle cell proliferation (GO ID: 0048661), angiogenesis (GO ID: 0001525) and collagen catabolic process (GO ID: 0030574) (Figure 4C).
Pathway Enrichment Analysis
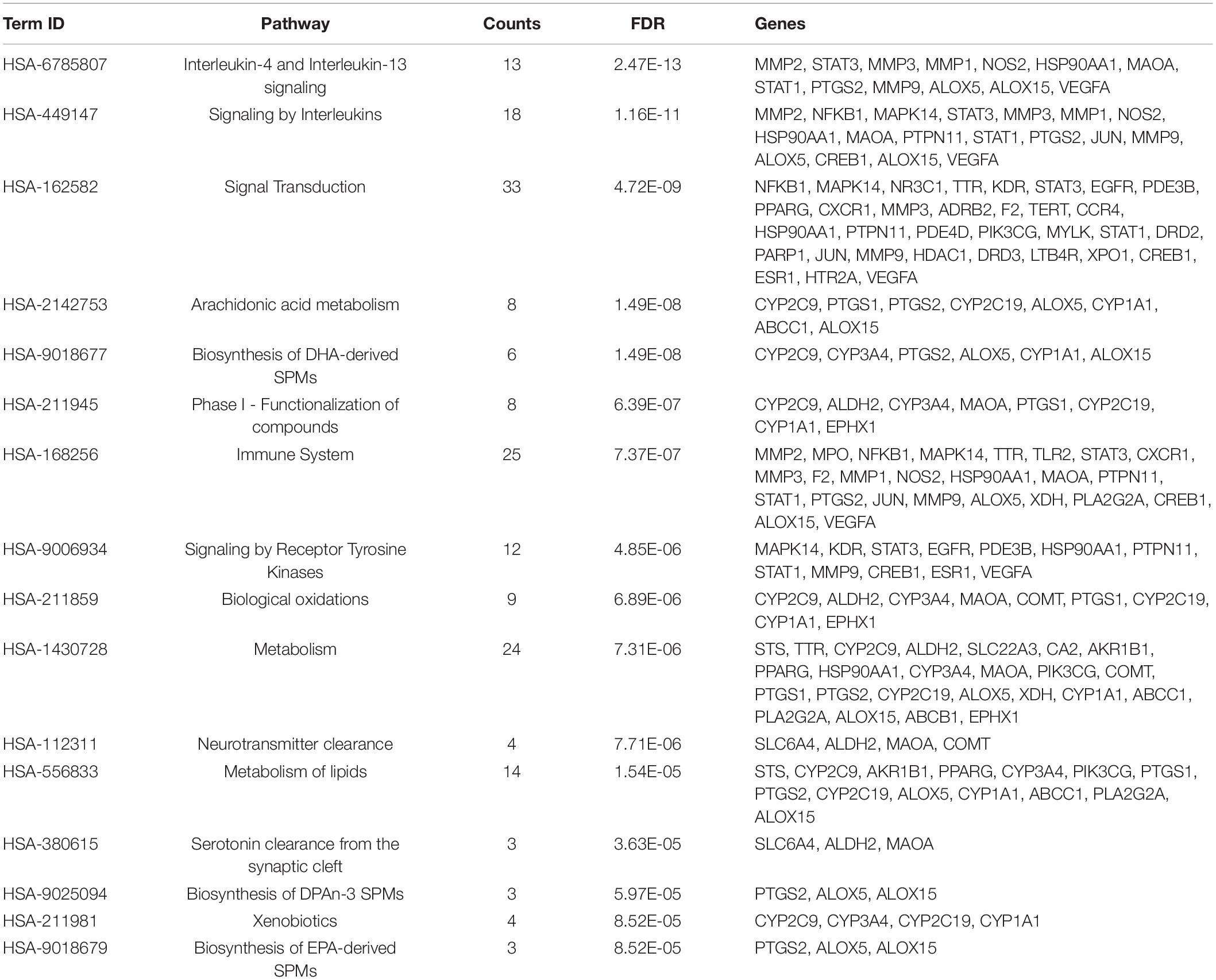
Reactome pathway enrichment analysis obtained 123 items (FDR < 0.05) and the top 16 pathways at a significance threshold of FDR < 10–5 were displayed (Figure 5A and Table 1). The pathways result was intensively enriched in SPMs biosynthesis and inflammatory and immune response, including arachidonic acid metabolism, metabolism of lipids, biosynthesis of EPA-derived SPMs, biosynthesis of DHA-derived SPMs, biosynthesis of DPAn-3 SPMs, interleukin-4 and interleukin-13 signaling, and signaling by interleukins and immune system.
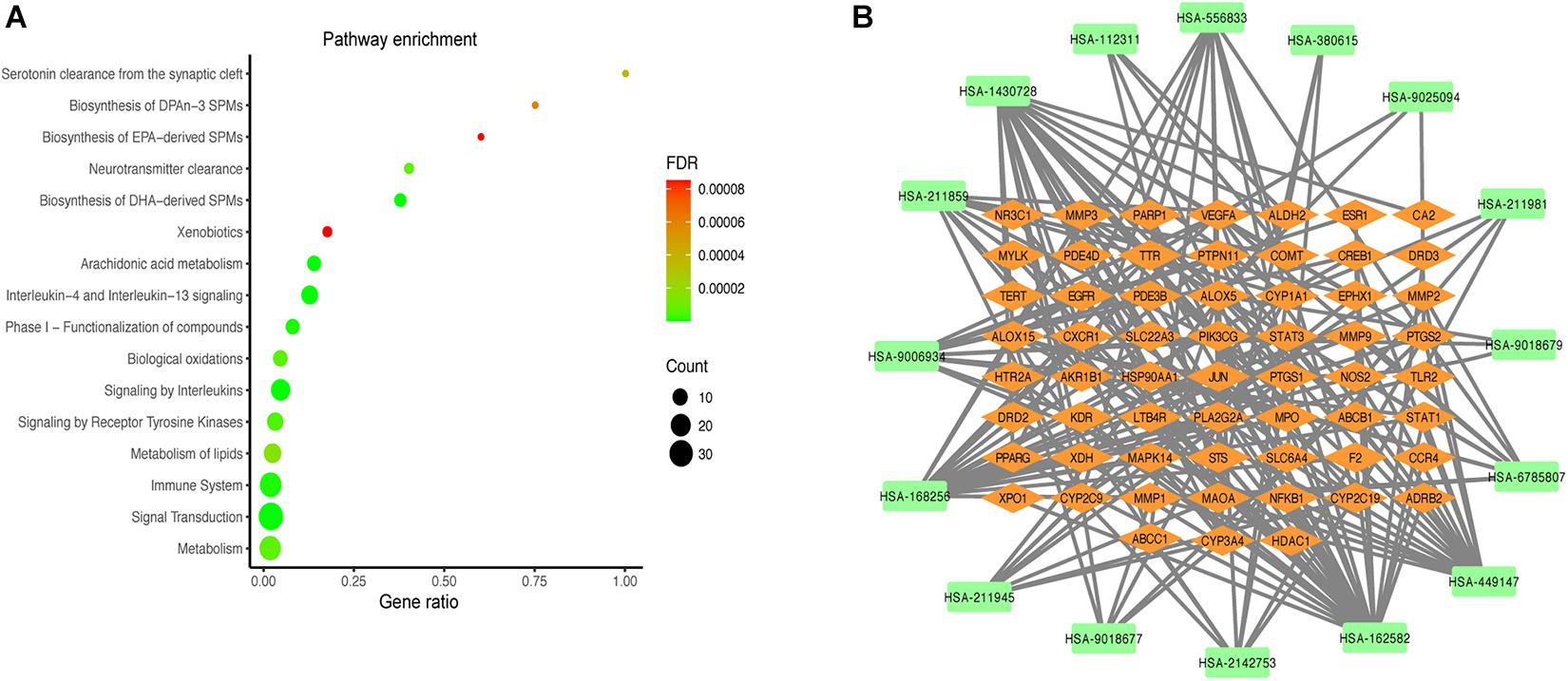
Figure 5. Results of the pathway analysis of the top 16 pathways: Bubble diagram of pathway (A) and T-P network diagram (B).
The T-P network showed the associations of the top 16 pathways with 75 nodes and 187 edges (Figure 5B). The results revealed that the active compounds of the MGMD had effects on asthma through regulating various pathways, in particular through improving airway inflammatory infiltration resulted from Th2 cytokine production and lipid mediators.
Molecular Docking Verification of Core Compounds and Core Protein Targets
Furthermore, we verified the binding affinity between core compounds and protein targets of MGMD by molecular docking. As screened in H-C-T network, quercetin, wogonin, luteolin, naringenin, and kaempferol were the core compounds of MGMD. The hub protein targets were identified by top 5 node degree of PPI network and seed node of clusters, included STAT3, PTGS2, JUN, VEGFA, EGFR, and ALOX5.
The results obtained by the molecular docking software were shown in Table 2. The Grid box was centered to cover the active binding site and all essential residues. For the STAT3, grid box (126Å × 60Å × 126Å) centered at (0.233, 28.877, 33.555) Å, for the PTGS2, grid box (76Å × 94Å × 90Å) centered at (33.269, −4.946, 7.759) Å, for the JUN, grid box (90Å × 72Å × 70Å) centered at (−16.564, 23.05, 26.225) Å, for the VEGFA, grid box (58Å × 40Å × 40Å) centered at (2.965, −4.946, 7.759) Å, for the EGFR, grid box (126Å × 126Å × 126Å) centered at (80.339, 11.821, 66.594) Å, for the ALOX5, grid box (70Å × 90Å × 96Å) centered at (4.01, 45.829, 3.32) Å. As seen from Table 2, the binding affinity for the five core compounds and protein crystal structures corresponding to the core target genes were all greater than −5 kcal/mol, indicating that the compound had a certain affinity for the protein crystal structure. Wogonin showed the high binding affinity of −8.8 kcal/mol in ALOX5, −6.2 kcal/mol in JUN and −6.2 kcal/mol in VEGFA. Luteolin showed the high binding affinity of −8.3 kcal/mol in EGFR, −9.3 kcal/mol in PTGS2 and −7.9 kcal/mol in STAT3. The small-molecule compounds were tightly bound to the protein residues via various interactions (Figure 6 and Table 2).
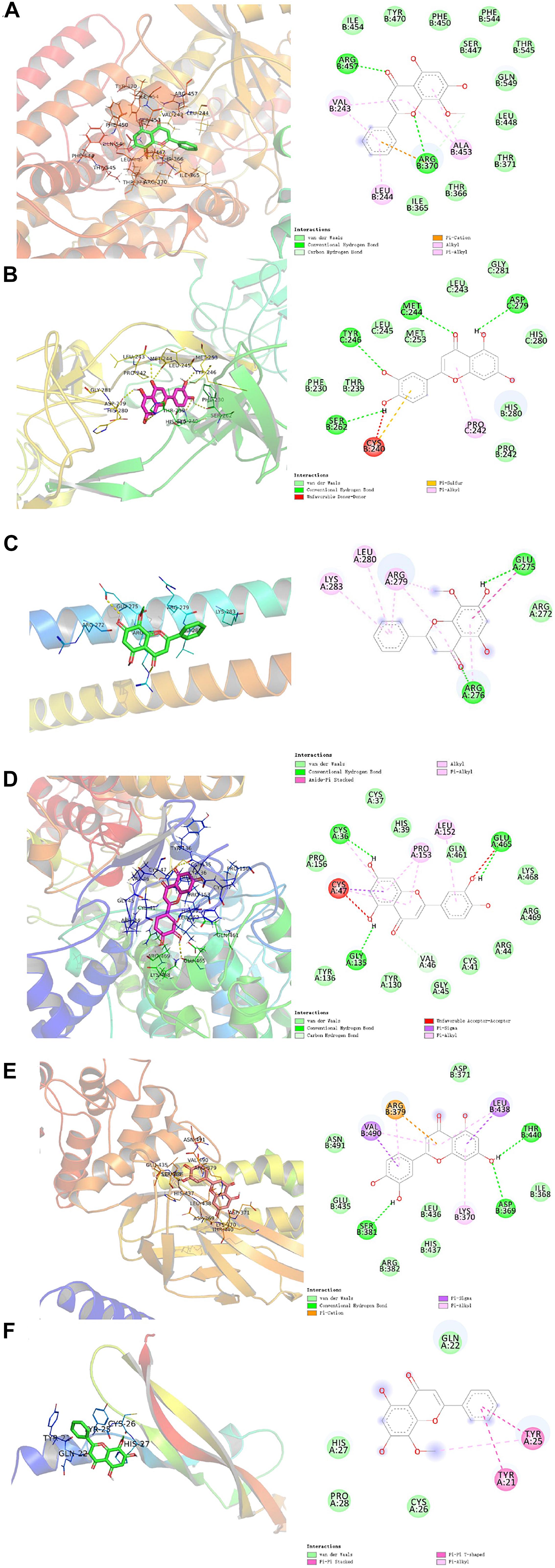
Figure 6. Molecular docking diagram of ALOX5 (A), JUN (B), VEGFA (C) complexed with wogonin; EGFR (D), PTGS2 (E), and STAT3 (F) complexed with luteolin.
Discussion
Asthma is a major public health challenge in China that affecting 45.7 million adults aged 20 years or older (Huang et al., 2019). TCM has a long history of clinical practice with available records, which provides a considerable number of classical herbal formulas widely used for disease treatment and represents a unique resource for drug development (Chao et al., 2017). Among the complex mixtures of compounds present in herbal medicines, unique combinations in the traditional complex formulas may exert major efficacy by targeting specific nodes of cellular signaling to intervene a specific chronic disease (Stermitz et al., 2000). Network pharmacology is used to study the intervention mechanism of drugs on diseases by constructing a biological interaction network (Yang et al., 2013). It systematically reveals the therapeutic effects of drugs on diseases from the interaction relationship between drugs, targets, and disease (Liu J. et al., 2018; Zhang et al., 2018). In a previous clinical study, we have found that MGMD had significant efficacy on respiratory symptoms (Zhang, 2016). To provide further insight of underlying mechanisms, we used the systematic pharmacology method and molecular docking to explore the potential molecular mechanism of the bioactive compounds.
We obtained 92 active compounds of MGMD from HERB, ECTM, SEA databases. A total of 72 putative anti-asthmatic targets of MGMD were selected by overlapping the targets between the MGMD-associated targets and the predicted asthma targets. According the H-C-T network topological analysis, we found the core compounds contained quercetin, wogonin, luteolin, naringenin, and kaempferol. Intriguingly, these compounds have all been reported to have therapeutic effects on inflammatory disease in previous studies. It had been demonstrated that all of the five core ingredients can reduce the allergic airway inflammation, inhibit levels of Th2 cytokine, including IL-4, IL-5, and IL-13, in OVA-sensitized and–challenged animals (Park et al., 2009; Shi et al., 2014; Bui et al., 2017; Jang et al., 2017; Molitorisova et al., 2021). Moreover, quercetin can inhibit human neutrophil elastase-induced MUC5AC expression in human airway epithelial cells through PKC/EGFR/ERK signal transduction pathway (Li et al., 2012). Both wogonin and luteolin had effect on reduction of airway mucus production (Lucas et al., 2015; Wang et al., 2021), meanwhile luteolin can also decrease collagen deposition in vovo (Wang et al., 2021). In addition, it had reported that kaempferol alleviated airway inflammation through modulating Tyk2-STAT1/3 signaling responsive (Gong et al., 2013), and suppressed COX2 expression level in lung tissues in asthmatic mice (Kang et al., 2018). These previous studies suggest that MGMD is likely to decrease Th2 cytokine production in asthma mainly through compound of quercetin, wogonin, luteolin, naringenin, and kaempferol.
The PPI network was constructed by 72 potential targets of MGMD which were involved in asthma, and the network was further analyzed by MCODE clustering to explore the asthma related pathways. Our results suggested that the core potential targets of MGMD were intensively enriched in the pathways related to SPMs biosynthesis, including ALOX5 (5-LOX), ALOX15 (15-LOX), PTGS1 (COX1), PTGS2 (COX2), cytochrome P450 proteins CYP2C9 and CYP2C19, which paly pivotal role in the resolution of inflammation in asthma (Schett and Neurath, 2018; Kytikova et al., 2019). 5-LOX is a crucial enzyme which helps in the conversion of arachidonic acid to leukotrienes. With the increasing number of indications for anti-leukotriene drugs, the development of 5-LOX inhibitor agents for asthma becomes increasingly important (Bruno et al., 2018; Sinha et al., 2019). Our results argue a potential that MGMD is a 5-LOX inhibitor for the treatment of asthma, in turn suggest a promising mechanism by which MGMD intervene asthma.
Furthermore, STAT3, JUN, EGFR, and VEGFA were also the core potential targets in our PPI network results. STAT3 and JUN are closely related to the pathological process of asthma (Simeone-Penney et al., 2007; García-Menaya et al., 2019). STAT3 can induce gene expression of cytokines, chemokines, and adhesion molecules with an important role in directing the inflammatory response and act as a novel epithelial regulator of the allergic response by altering Th2 cell recruitment and effector function (Schumann et al., 1996; Simeone-Penney et al., 2007). JUN activation appears to be implicated in airway inflammation by contributing to regulate the expression of Th2 cytokines, chemokines, growth factors, and adhesion molecules, which is associated with several inflammation related disorders such as asthma and allergy (Haeggström and Funk, 2011). There are extensive studies focusing on the relation among EGFR, VEGFA and asthma pathophysiology, which describe airway remodeling, airway hypermucus secretion, as well as immunological responses of airway inflammation (Hoshino et al., 2001; Lee et al., 2004; Burgel and Nadel, 2008; Inoue et al., 2020). MGMD compounds may exert anti-inflammatory and anti-remodeling activity through regulating STAT3, JUN, EGFR, and VEGFA.
The GO enrichment results strengthen the evidence that candidate protein targets of MGMD are involved in airway inflammation and remodeling. The enriched BPs of anti-inflammatory in our results included inflammatory response, leukotriene metabolic process and arachidonic acid secretion, suggesting that MGMD could influenced the lipid mediators, both pro-inflammatory mediators and SPMs, to ameliorate the airway inflammation in asthma patients. Moreover, the typical airway remodeling processes, including positive regulation of smooth muscle cell proliferation, angiogenesis and collagen catabolic process, were also enriched by MGMD related targets. Combined with the PPI network results, we propose that MGMD regulates SPMs biosynthesis by ALOX5 to reduce airway inflammation and relieve airway remodeling process by suppressing the EGFR and VEGFA signaling.
Our Reactome pathway enrichment results consistently argue that MGMD reduces allergic airway inflammation through the biosynthetic pathways of SPMs. Arachidonic acid metabolism, metabolism of lipids, biosynthesis of EPA-derived SPMs, biosynthesis of DHA-derived SPMs and biosynthesis of DPAn-3 SPMs were all enriched in Reactome pathway enrichment analysis. Additionally, interleukin-4 and interleukin-13 signaling, signaling by interleukins and immune system pathways were also enriched in our results. IL-4 and IL-13 are principal regulatory cytokines mainly secreted by activated Th2 cells, crucially important during the immune response in allergy and asthma (Nelms et al., 1999; Hershey, 2003). The receptors of allergic cytokines, including IL-4, IL-5, and IL-13, trigger the JAK/STAT pathway (Hsieh et al., 2011; Howell et al., 2018). Therefore, we propose that MGMD compounds targets the IL-4 and IL-13 pathway, which in turn activate transcription factors to modulate clinical symptoms of asthma (Figure 7).
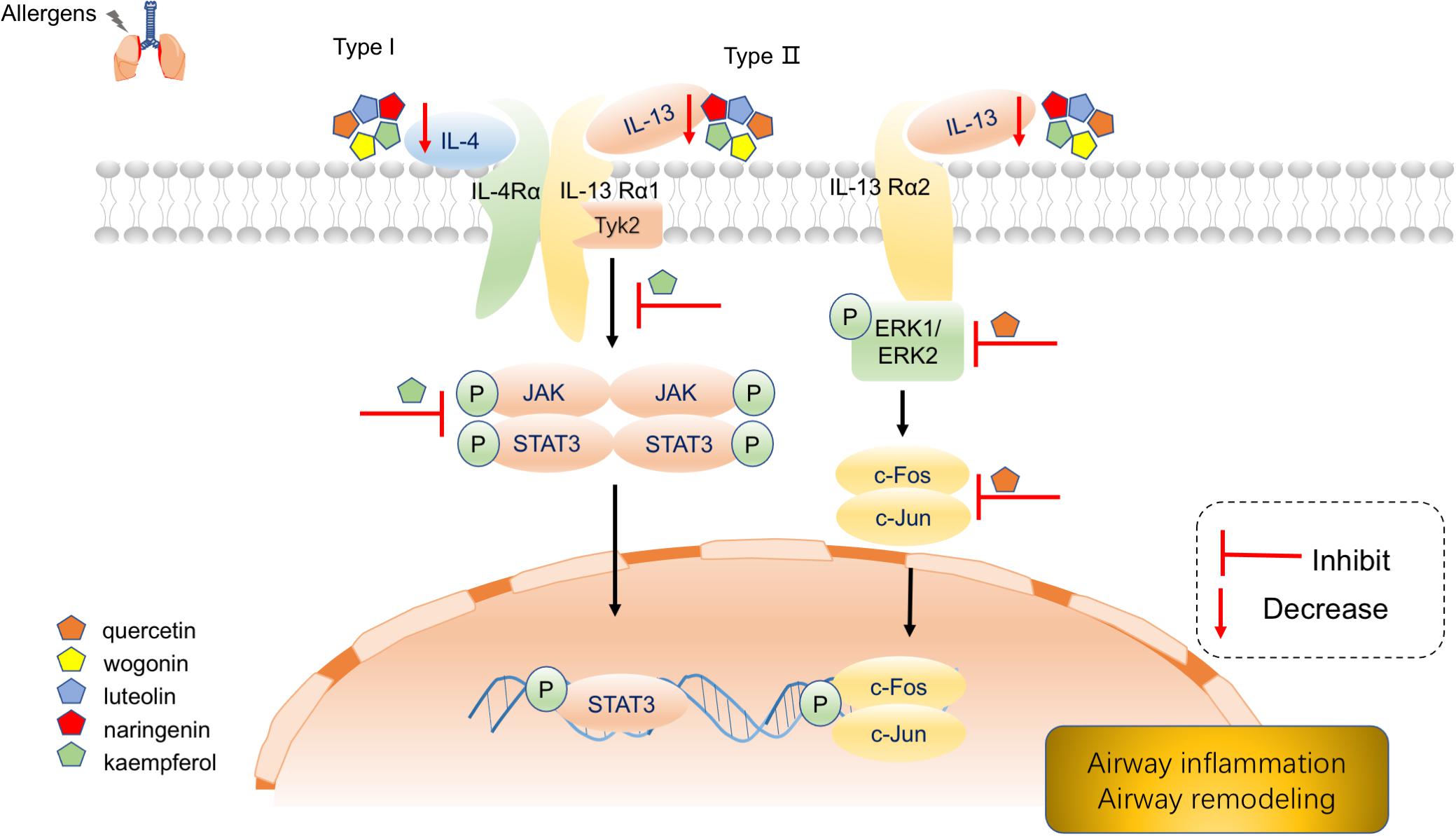
Figure 7. Schematic diagram depicting main components of MGMD inhibits IL-4 and IL-13 to mitigate airway allergic responses.
According to the compound-target interaction analysis, six targets with higher degrees on the PPI network (STAT3, PTGS2, JUN, VEGFA, EGFR, and ALOX5) were identified. In our molecular docking analysis, the binding mode with the best docking score was selected to analyze the interaction between the protein receptor and its predicted active compounds. The docking scores of all compound-target pairs are lower than −5 kcal/mol, indicating that each of the five core compounds (quercetin, wogonin, luteolin, naringenin, and kaempferol) has good binding affinity to all six targets. These results suggested that the core ingredients of MGMD could relieve airway inflammation and remodeling through binding STAT3, PTGS2, JUN, VEGFA, EGFR, and ALOX5.
Conclusion
On the ground of the above, network pharmacology strategy was presented to investigate the active compounds, potential anti-asthmatic targets and involved regulatory pathways in MGMD. Our results revealed the active ingredients and potential molecular mechanism by which MGMD treatment is effective against airway inflammation and remodeling in asthma through regulating IL-4 and IL-13 signaling and SPMs biosynthesis. These findings were partially validated previous studies. Our subsequent molecular docking showed that each bioactive compounds (quercetin, wogonin, luteolin, naringenin, and kaempferol) of MGMD has favorable binding abilities with STAT3, PTGS2, JUN, VEGFA, EGFR, and ALOX5, further arguing the potential molecular mechanism of action of MGMD in asthma. Our results will serve as a comprehensive reference for investigation of the mechanism by which MGMD intervene asthma.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
GW, BZ, and ZW: conceptualization. GW and BZ: methodology and visualization. BZ: software. GW and ZW: writing–original draft preparation. BZ, YM, YL, and XY: writing–review and editing. XY and CF: supervision and project administration. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (No. 81673921 to CF and No. 81804015 to XY).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.644561/full#supplementary-material
Abbreviations
AD, anti-allergic decoction; ADME, absorption distribution, metabolism and excretion; ALOX5, arachidonate 5-lipoxygenase; BP, biological process; H-C -T, herb-compound-target; DHA, docosahexaenoic acid; DL, drug-likeness; FF, Fangfeng (Saposhnikoviae Radix); EPA, Eicosapentaenoic acid; GO, Gene ontology; HL, Half-life; JG, Jiegeng (Platycodon grandiflorus); JUN, c-Jun NH2-terminal kinase; MGMD, Modified Guo Min decoction; MF, molecular function; OB, oral bioavailability; PPI, Protein–protein interaction; PUFA, polyunsaturated fatty acid; QH, Qianhu (Peucedanum praeruptorum); SPM, Specialized pro-resolving mediator; TCM, traditional Chinese medicine; TCMSP, traditional Chinese medicine system pharmacology database; VEGFA, vascular endothelial growth factor A; WM, Wumei (Prunus mume); WWZ, Wuweizi (Schisandra chinensis); YCH, Yinchaihu (Stellaria dichotoma L. var. lanceolata Bge).
Footnotes
- ^ http://herb.ac.cn
- ^ http://www.tcmip.cn/ETCM/index.php/Home/Index/
- ^ http://tcmspw.com/tcmsp.php
- ^ http://www.swissadme.ch/
- ^ https://pubchem.ncbi.nlm.nih.gov/
- ^ http://sea.bkslab.org
- ^ http://ctdbase.org/
- ^ https://omim.org/
- ^ https://www.genecards.org/
- ^ http://string-db.org
- ^ http://www.cytoscape.org/
- ^ https://david.ncifcrf.gov/
- ^ https://reactome.org/
- ^ http://www1.rcsb.org/
References
Amberger, J. S., and Hamosh, A. (2017). Searching Online Mendelian Inheritance in Man (OMIM): a Knowledgebase of Human Genes and Genetic Phenotypes. Curr. Protoc. Bioinformatics 58, 1.2.1–1.2.12. doi: 10.1002/cpbi.27
Bader, G. D., and Hogue, C. W. V. (2003). An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4:2. doi: 10.1186/1471-2105-4-2
Barnig, C., Frossard, N., and Levy, B. D. (2018). Towards targeting resolution pathways of airway inflammation in asthma. Pharmacol. Ther. 186, 98–113. doi: 10.1016/j.pharmthera.2018.01.004
Bruno, F., Spaziano, G., Liparulo, A., Roviezzo, F., Nabavi, S. M., Sureda, A., et al. (2018). Recent advances in the search for novel 5-lipoxygenase inhibitors for the treatment of asthma. Eur. J. Med. Chem. 153, 65–72. doi: 10.1016/j.ejmech.2017.10.020
Bui, T. T., Piao, C. H., Song, C. H., Lee, C.-H., Shin, H. S., and Chai, O. H. (2017). Baicalein, wogonin, and Scutellaria baicalensis ethanol extract alleviate ovalbumin-induced allergic airway inflammation and mast cell-mediated anaphylactic shock by regulation of Th1/Th2 imbalance and histamine release. Anat. Cell Biol. 50, 124–134. doi: 10.5115/acb.2017.50.2.124
Burgel, P.-R., and Nadel, J. A. (2008). Epidermal growth factor receptor-mediated innate immune responses and their roles in airway diseases. Eur. Respir. J. 32, 1068–1081. doi: 10.1183/09031936.00172007
Burley, S. K., Berman, H. M., Kleywegt, G. J., Markley, J. L., Nakamura, H., and Velankar, S. (2017). Protein Data Bank (PDB): the Single Global Macromolecular Structure Archive. Methods Mol. Biol. 1607, 627–641. doi: 10.1007/978-1-4939-7000-1_26
Chao, J., Dai, Y., Verpoorte, R., Lam, W., Cheng, Y.-C., Pao, L.-H., et al. (2017). Major achievements of evidence-based traditional Chinese medicine in treating major diseases. Biochem. Pharmacol. 139, 94–104. doi: 10.1016/j.bcp.2017.06.123
Chen, X., Zhou, H., Liu, Y. B., Wang, J. F., Li, H., Ung, C. Y., et al. (2006). Database of traditional Chinese medicine and its application to studies of mechanism and to prescription validation. Br. J. Pharmacol. 149, 1092–1103. doi: 10.1038/sj.bjp.0706945
Daina, A., Michielin, O., and Zoete, V. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7:42717. doi: 10.1038/srep42717
Fabregat, A., Jupe, S., Matthews, L., Sidiropoulos, K., Gillespie, M., Garapati, P., et al. (2018). The Reactome Pathway Knowledgebase. Nucleic Acids Res. 46, D649–D655. doi: 10.1093/nar/gkx1132
Fang, S., Dong, L., Liu, L., Guo, J., Zhao, L., Zhang, J., et al. (2020). HERB: a high-throughput experiment- and reference-guided database of traditional Chinese medicine. Nucleic Acids Res. 49, D1197–D1206. doi: 10.1093/nar/gkaa1063
Flood-Page, P., Menzies-Gow, A., Phipps, S., Ying, S., Wangoo, A., Ludwig, M. S., et al. (2003). Anti-IL-5 treatment reduces deposition of ECM proteins in the bronchial subepithelial basement membrane of mild atopic asthmatics. J. Clin. Invest. 112, 1029–1036. doi: 10.1172/JCI17974
García-Menaya, J. M., Cordobés-Durán, C., García-Martín, E., and Agúndez, J. A. G. (2019). Pharmacogenetic Factors Affecting Asthma Treatment Response. Potential Implications for Drug Therapy. Front. Pharmacol. 10:520. doi: 10.3389/fphar.2019.00520
Gong, J.-H., Shin, D., Han, S.-Y., Park, S.-H., Kang, M.-K., Kim, J.-L., et al. (2013). Blockade of Airway Inflammation by Kaempferol via Disturbing Tyk-STAT Signaling in Airway Epithelial Cells and in Asthmatic Mice. Evid. Based Complement. Alternat. Med. 2013:250725. doi: 10.1155/2013/250725
Grondin, C. J., Davis, A. P., Wiegers, T. C., Wiegers, J. A., and Mattingly, C. J. (2018). Accessing an Expanded Exposure Science Module at the Comparative Toxicogenomics Database. Environ. Health Perspect. 126:014501. doi: 10.1289/EHP2873
Haeggström, J. Z., and Funk, C. D. (2011). Lipoxygenase and leukotriene pathways: biochemistry, biology, and roles in disease. Chem. Rev. 111, 5866–5898. doi: 10.1021/cr200246d
Hershey, G. K. K. (2003). IL-13 receptors and signaling pathways: an evolving web. J. Allergy Clin. Immunol. 111, 677–690. doi: 10.1067/mai.2003.1333
Hoshino, M., Takahashi, M., and Aoike, N. (2001). Expression of vascular endothelial growth factor, basic fibroblast growth factor, and angiogenin immunoreactivity in asthmatic airways and its relationship to angiogenesis. J. Allergy Clin. Immunol. 107, 295–301. doi: 10.1067/mai.2001.111928
Howell, M. D., Fitzsimons, C., and Smith, P. A. (2018). JAK/STAT inhibitors and other small molecule cytokine antagonists for the treatment of allergic disease. Ann. Allergy Asthma Immunol. 120, 367–375. doi: 10.1016/j.anai.2018.02.012
Hsieh, Y.-Y., Chang, C.-C., Hsu, C.-M., Wan, L., Chen, S.-Y., Lin, W.-H., et al. (2011). JAK-1 rs2780895 C-related genotype and allele but not JAK-1 rs10789166, rs4916008, rs2780885, rs17127114, and rs3806277 are associated with higher susceptibility to asthma. Genet. Test. Mol. Biomarkers 15, 841–847. doi: 10.1089/gtmb.2011.0002
Huang, K., Yang, T., Xu, J., Yang, L., Zhao, J., Zhang, X., et al. (2019). Prevalence, risk factors, and management of asthma in China: a national cross-sectional study. Lancet 394, 407–418. doi: 10.1016/S0140-6736(19)31147-X
Huang, L., Xie, D., Yu, Y., Liu, H., Shi, Y., Shi, T., et al. (2018). TCMID 2.0: a comprehensive resource for TCM. Nucleic Acids Res. 46, D1117–D1120. doi: 10.1093/nar/gkx1028
Inoue, H., Akimoto, K., Homma, T., Tanaka, A., and Sagara, H. (2020). Airway Epithelial Dysfunction in Asthma: relevant to Epidermal Growth Factor Receptors and Airway Epithelial Cells. J. Clin. Med. 9:3698. doi: 10.3390/jcm9113698
James, A. L., and Wenzel, S. (2007). Clinical relevance of airway remodelling in airway diseases. Eur. Respir. J. 30, 134–155. doi: 10.1183/09031936.00146905
Jang, T. Y., Jung, A.-Y., Kyung, T.-S., Kim, D.-Y., Hwang, J.-H., and Kim, Y. H. (2017). Anti-allergic effect of luteolin in mice with allergic asthma and rhinitis. Cent. Eur. J. Immunol. 42, 24–29. doi: 10.5114/ceji.2017.67315
Jeffery, P. K. (2001). Remodeling in Asthma and Chronic Obstructive Lung Disease. Am. J. Respir. Crit. Care Med. 164, S28–38. doi: 10.1164/ajrccm.164.supplement_2.2106061
Kang, D. R., Belal, S. A., Choe, H. S., Shin, D. K., and Shim, K. S. (2018). Effect of Kaempferol on Cyclooxygenase 2 (Cox2) and Cytosolic Phospholipase A2 (cPLA2) Protein Expression in BALB/c Mice. Iran. J. Allergy Asthma Immunol. 17, 428–435. doi: 10.18502/ijaai.v17i5.301
Keiser, M. J., Roth, B. L., Armbruster, B. N., Ernsberger, P., Irwin, J. J., and Shoichet, B. K. (2007). Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 25, 197–206. doi: 10.1038/nbt1284
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., et al. (2019). PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 47, D1102–D1109. doi: 10.1093/nar/gky1033
Krishnamoorthy, N., Abdulnour, R.-E. E., Walker, K. H., Engstrom, B. D., and Levy, B. D. (2018). Specialized Proresolving Mediators in Innate and Adaptive Immune Responses in Airway Diseases. Physiol. Rev. 98, 1335–1370. doi: 10.1152/physrev.00026.2017
Kytikova, O., Novgorodtseva, T., Denisenko, Y., Antonyuk, M., and Gvozdenko, T. (2019). Pro-Resolving Lipid Mediators in the Pathophysiology of Asthma. Medicina 55:284. doi: 10.3390/medicina55060284
Lambrecht, B. N., and Hammad, H. (2012). The airway epithelium in asthma. Nat. Med. 18, 684–692. doi: 10.1038/nm.2737
Le Cras, T. D., Acciani, T. H., Mushaben, E. M., Kramer, E. L., Pastura, P. A., Hardie, W. D., et al. (2011). Epithelial EGF receptor signaling mediates airway hyperreactivity and remodeling in a mouse model of chronic asthma. Am. J. Physiol. Lung Cell. Mol. Physiol. 300, L414–21. doi: 10.1152/ajplung.00346.2010
Lee, C. G., Link, H., Baluk, P., Homer, R. J., Chapoval, S., Bhandari, V., et al. (2004). Vascular endothelial growth factor (VEGF) induces remodeling and enhances TH2-mediated sensitization and inflammation in the lung. Nat. Med. 10, 1095–1103. doi: 10.1038/nm1105
Li, N., Li, Q., Zhou, X. D., Kolosov, V. P., and Perelman, J. M. (2012). The effect of quercetin on human neutrophil elastase-induced mucin5AC expression in human airway epithelial cells. Int. Immunopharmacol. 14, 195–201. doi: 10.1016/j.intimp.2012.07.008
Liu, J., and Lin, A. (2007). Wiring the cell signaling circuitry by the NF-κB and JNK1 crosstalk and its applications in human diseases. Oncogene 26, 3267–3278. doi: 10.1038/sj.onc.1210417
Liu, J., Liu, J., Shen, F., Qin, Z., Jiang, M., Zhu, J., et al. (2018). Systems pharmacology analysis of synergy of TCM: an example using saffron formula. Sci. Rep. 8:380. doi: 10.1038/s41598-017-18764-2
Liu, L., Wang, L., He, S., and Ma, Y. (2018). Immune Homeostasis: effects of Chinese Herbal Formulae and Herb-Derived Compounds on Allergic Asthma in Different Experimental Models. Chin. J. Integr. Med. 24, 390–398. doi: 10.1007/s11655-018-2836-2
Lucas, C. D., Dorward, D. A., Sharma, S., Rennie, J., Felton, J. M., Alessandri, A. L., et al. (2015). Wogonin Induces Eosinophil Apoptosis and Attenuates Allergic Airway Inflammation. Am. J. Respir. Crit. Care Med. 191, 626–636. doi: 10.1164/rccm.201408-1565OC
Molitorisova, M., Sutovska, M., Kazimierova, I., Barborikova, J., Joskova, M., Novakova, E., et al. (2021). The anti-asthmatic potential of flavonol kaempferol in an experimental model of allergic airway inflammation. Eur. J. Pharmacol. 891:173698. doi: 10.1016/j.ejphar.2020.173698
Nelms, K., Keegan, A. D., Zamorano, J., Ryan, J. J., and Paul, W. E. (1999). The IL-4 receptor: signaling mechanisms and biologic functions. Annu. Rev. Immunol. 17, 701–738. doi: 10.1146/annurev.immunol.17.1.701
Park, H., Lee, C.-M., Jung, I. D., Lee, J. S., Jeong, Y., Chang, J. H., et al. (2009). Quercetin regulates Th1/Th2 balance in a murine model of asthma. Int. Immunopharmacol. 9, 261–267. doi: 10.1016/j.intimp.2008.10.021
Qin, L., Lan, Y., and Ma, S. (2017). Effects of Modified Guomin Decoction on Treg Cell Specific Transcription Factor in Mice with Allergic Asthma. Tradit. Chin. Drug Res. Clin. Pharmacol. 28, 332–335.
Ru, J., Li, P., Wang, J., Zhou, W., Li, B., Huang, C., et al. (2014). TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 6:13. doi: 10.1186/1758-2946-6-13
Safran, M., Dalah, I., Alexander, J., Rosen, N., Iny Stein, T., Shmoish, M., et al. (2010). GeneCards Version 3: the human gene integrator. Database 2010:baq020. doi: 10.1093/database/baq020
Schett, G., and Neurath, M. F. (2018). Resolution of chronic inflammatory disease: universal and tissue-specific concepts. Nat. Commun. 9:3261. doi: 10.1038/s41467-018-05800-6
Schumann, R. R., Kirschning, C. J., Unbehaun, A., Aberle, H. P., Knope, H. P., Lamping, N., et al. (1996). The lipopolysaccharide-binding protein is a secretory class 1 acute-phase protein whose gene is transcriptionally activated by APRF/STAT/3 and other cytokine-inducible nuclear proteins. Mol. Cell. Biol. 16, 3490–3503. doi: 10.1128/mcb.16.7.3490
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shi, Y., Tan, Y., Mao, S., and Gu, W. (2014). Naringenin inhibits allergen-induced airway remodeling in a murine model of asthma. Mol. Med. Rep. 9, 1204–1208. doi: 10.3892/mmr.2014.1940
Simeone-Penney, M. C., Severgnini, M., Tu, P., Homer, R. J., Mariani, T. J., Cohn, L., et al. (2007). Airway epithelial STAT3 is required for allergic inflammation in a murine model of asthma. J. Immunol. 1950, 6191–6199. doi: 10.4049/jimmunol.178.10.6191
Sinha, S., Doble, M., and Manju, S. L. (2019). 5-Lipoxygenase as a drug target: a review on trends in inhibitors structural design, SAR and mechanism based approach. Bioorg. Med. Chem. 27, 3745–3759. doi: 10.1016/j.bmc.2019.06.040
Soriano, J. B., Abajobir, A. A., Abate, K. H., Abera, S. F., Agrawal, A., Ahmed, M. B., et al. (2017). Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Respir. Med. 5, 691–706. doi: 10.1016/S2213-2600(17)30293-X
Stermitz, F. R., Lorenz, P., Tawara, J. N., Zenewicz, L. A., and Lewis, K. (2000). Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc. Natl. Acad. Sci. U. S. A. 97, 1433–1437. doi: 10.1073/pnas.030540597
Tao, T., Zhang, Y., Li, J., and Mei, C. (2018). Review of experiments and clinical application research on Guomin Decoction. China J. Tradit. Chin. Med. Pharm. 33, 242–244.
Trott, O., and Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. doi: 10.1002/jcc.21334
von Mering, C., Jensen, L. J., Snel, B., Hooper, S. D., Krupp, M., Foglierini, M., et al. (2005). STRING: known and predicted protein–protein associations, integrated and transferred across organisms. Nucleic Acids Res. 33, D433–7. doi: 10.1093/nar/gki005
Wang, S., Wuniqiemu, T., Tang, W., Teng, F., Bian, Q., Yi, L., et al. (2021). Luteolin inhibits autophagy in allergic asthma by activating PI3K/Akt/mTOR signaling and inhibiting Beclin-1-PI3KC3 complex. Int. Immunopharmacol. 94:107460. doi: 10.1016/j.intimp.2021.107460
Wu, Y., Zhang, F., Yang, K., Fang, S., Bu, D., Li, H., et al. (2019). SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Res. 47, D1110–D1117. doi: 10.1093/nar/gky1021
Xu, H.-Y., Zhang, Y.-Q., Liu, Z.-M., Chen, T., Lv, C.-Y., Tang, S.-H., et al. (2019). ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 47, D976–D982. doi: 10.1093/nar/gky987
Yang, M., Chen, J.-L., Xu, L.-W., and Ji, G. (2013). Navigating traditional chinese medicine network pharmacology and computational tools. Evid. Based Complement. Alternat. Med. 2013:731969. doi: 10.1155/2013/731969
Zhang, G.-X., Zhang, Y.-Y., Zhang, X.-X., Wang, P.-Q., Liu, J., Liu, Q., et al. (2018). Different network pharmacology mechanisms of Danshen-based Fangjis in the treatment of stable angina. Acta Pharmacol. Sin. 39, 952–960. doi: 10.1038/aps.2017.191
Zhang, X. (2016). Clinical And Animal Experimental Study Of Allergy Decoction Combined With Cough Suppressant To Interfere With Pm2.5. Available online at: https://kns.cnki.net/kcms/detail/detail.aspx?dbcode=CMFD&dbname=CMFD201602&filename=1016071173.nh&v=THPQPTs5AmVKWk3lTRAFLU2OZN63qbJsXUW1RC1V3QQsD67MWJgqLm9VNc7mE1mF (accessed March 21, 2021).
Zhu, S., and Liang, X. (1985). Discussion on disease differentiation and medication based on syndrome differentiation from the application of Guomin Decoction. Chin. J. Med. 59:1985,Google Scholar
Keywords: asthma, airway inflammation, airway remodeling, Chinese medicine, network pharmacology
Citation: Wang G, Zhou B, Wang Z, Meng Y, Liu Y, Yao X and Feng C (2021) Pharmacological Mechanisms Underlying the Anti-asthmatic Effects of Modified Guomin Decoction Determined by Network Pharmacology and Molecular Docking. Front. Mol. Biosci. 8:644561. doi: 10.3389/fmolb.2021.644561
Received: 21 December 2020; Accepted: 29 March 2021;
Published: 22 April 2021.
Edited by:
Huahao Shen, Zhejiang University, ChinaReviewed by:
Zhihua Chen, Zhejiang University, ChinaKai Yang, State Key Laboratory of Respiratory Diseases, First Affiliated Hospital of Guangzhou Medical University, China
Copyright © 2021 Wang, Zhou, Wang, Meng, Liu, Yao and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cuiling Feng, ZmVuZ2N1aWxpbmdAc2luYS5jb20=; Xiaoqin Yao, eWFveGlhb3FpbkBwa3VpaC5lZHUuY24=
†These authors have contributed equally to this work
 Guishu Wang1,2†
Guishu Wang1,2† Zheyi Wang
Zheyi Wang Yufeng Meng
Yufeng Meng Yaqian Liu
Yaqian Liu Xiaoqin Yao
Xiaoqin Yao Cuiling Feng
Cuiling Feng