
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Mol. Biosci. , 31 March 2021
Sec. RNA Networks and Biology
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.642732
This article is part of the Research Topic The Role of RNA in Genome Stability: To Wreck or Repair? View all 7 articles
Chromosome instability is a hallmark of cancer and is caused by inaccurate segregation of chromosomes. One cellular structure used to avoid this fate is the kinetochore, which binds to the centromere on the chromosome. Human centromeres are poorly understood, since sequencing and analyzing repeated alpha-satellite DNA regions, which can span a few megabases at the centromere, are particularly difficult. However, recent analyses revealed that these regions are actively transcribed and that transcription levels are tightly regulated, unveiling a possible role of RNA at the centromere. In this short review, we focus on the recent discovery of the function of human centromeric RNA in the regulation and structure of the centromere, and discuss the consequences of dysregulation of centromeric RNA in cancer.
Accurate chromosome segregation is fundamental for cell division. Errors in this process can lead to chromosome instability, leading to aneuploidy, which is correlated with cancer (Zhu et al., 2011; Santaguida and Amon, 2015). The centromere is a component of each chromosome used for accurate chromosome segregation. The kinetochore, the structure responsible for binding the chromosome to spindle microtubules and for chromosome movement during cell division, is assembled on the centromere (Van Hooser et al., 2001). The identity and inheritance of the centromere are thought to be determined epigenetically by the deposition of the species-specific histone 3 variant CENH3 (CENP-A in mammals, CID in D. melanogaster, and Cse4 in S. cerevisiae) nucleosomes interspersed with classical Histone 3 nucleosomes (Blower et al., 2002; Fukagawa and Earnshaw, 2014; Niikura et al., 2016). The centromere of the budding yeast S. cerevisiae consists of a 100 nucleotides DNA sequence motif and is referred to as a point centromere (Pluta et al., 1995). In all other eukaryotes, centromeres are composed of repetitive DNA sequences on several hundred kilobases, referred to as regional centromeres (Pluta et al., 1995). Furthermore, the DNA composition of each centromere presents a high variation between each chromosome (Eichler, 1999; Melters et al., 2013). The “centromere paradox” refers to how highly diverse centromere sequences are, even in closely related eukaryotes (Eichler, 1999). Human centromeres are composed of α-satellite repeated tandemly to form a block of satellites, called higher-order repeat (HOR) that are composed of a set number of monomers that vary from 2 to 34 (Willard, 1985; Willard et al., 1986; Alexandrov et al., 1993; McNulty et al., 2017). Despite the repetitive sequences composing the centromere, this region is transcriptionally active, with the transcription of genes in rice (Nagaki et al., 2004; Mizuno et al., 2011). For other organisms, centromeric DNA encodes for siRNA (Grishok et al., 2000; Volpe et al., 2002; Zilberman et al., 2003; Fukagawa et al., 2004; Pal-Bhadra et al., 2004) and long-non-coding RNA called cenRNA (Wong et al., 2007; Carone et al., 2009).
A dynamic balance between euchromatin and heterochromatin at the centromeric region is required for active kinetochore (Nakano et al., 2008) and transcription of these regions (Saffery et al., 2003). In Drosophila and human cells, the centromeric region presents a distinct set of histone modification, one being the H3 Lys4-diMe, a modification associated with open but not active euchromatin, and lack clear marker for both heterochromatin (such as H3 Lys9 diMe) or euchromatin (such as H3 Lys9 Ac) (Sullivan and Karpen, 2004). Centromeric DNA encodes mainly non-coding RNA which are transcribed by the RNA polymerase II (Chan et al., 2012; Rošić et al., 2014), and may participate in the assembly and function of the centromeres.
In S. cerevisiae, transcription at the centromere is regulated in an RNA polymerase II-dependent manner, and the level of transcription is critical to maintaining the centromere function (Ohkuni and Kitagawa, 2011; Hildebrand and Biggins, 2016). Higher-level transcription at the centromere leads to its inactivation of chromosome missegregation (Hill and Bloom, 1987), while a base level of transcriptional activity is required for centromere function (Ohkuni and Kitagawa, 2011). This regulation is realized by a competition between the transcription factors Cbf1 and Ste12 and the silencing factors Sir1, Hst1, or Cdc14 (Ohkuni and Kitagawa, 2011). Point centromere presents in S. pombe display similar behavior, with too much or too little centromeric non-coding RNA leading to centromere malfunction (Ling and Yuen, 2019). Similarly, in mice, forced accumulation of the centromeric minor satellite transcripts leads to defects in chromosome segregation (Bouzinba-Segard et al., 2006), while in human cells, the inhibition of the RNA polymerase II during mitosis leads to a decrease in centromeric α satellite transcription, anaphase lagging cells and abnormal chromosome splitting (Chan et al., 2012; McNulty et al., 2017). This suggests that, in mammals, the transcription at the centromere is tightly regulated, maybe by its interaction with the nucleolus that may induce its repression (Bury et al., 2020). In human cells, one of the regulators of cenRNA transcription is ZFAT, which binds to a specific DNA sequence present on each centromere (Ishikura et al., 2020). Its presence at the centromere increases the centromeric level of the histone acetylase KAT2B and its product the histone 4 acetylated at the lysine 8 (Ishikura et al., 2020). This leads to the recruitment of the RNA pol II through BRD4 to the centromere, thus allowing ZFAT to control cenRNA transcription (Ishikura et al., 2020).
During mitosis, most regions within the condensed chromosomes are transcriptionally inactive, while centromeric regions are not (Chan et al., 2012; Liu et al., 2015). During the cell cycle, there is a stable centromeric transcription to ensure a stable centromere and kinetochore cohesion (Liu et al., 2015), but there is still some peak of expression occurring at different phases of the cell cycle. For example, in budding yeast, the centromeric expression occurs mainly in the S phase (Ling and Yuen, 2019), while in mice, a peak in the G2/M phase is observed (Ferri et al., 2009). In human cells, the RNA pol II is transcriptionally active at the mitotic kinetochore (Chan et al., 2012). R-loops, the byproduct of DNA-RNA hybridization, have important physiological functions but are also contributing to defects in genome integrity and chromosome fragility (Aguilera and García-Muse, 2012). In budding yeast, the accumulation of R-loops at the centromere is associated with a defect in kinetochore integrity (Mishra et al., 2021). In human, DHX9 is one element generating centromeric R-loops (Chakraborty et al., 2018), and while R-loops causes genomic instability in S phase (Buisson et al., 2015), they are necessary for faithful mitosis through a mitotic specific ATR pathway, independent of DNA damage or replication, but dependent of Aurora B and R-loops, ensuring accurate chromosome segregation (Aze et al., 2016; Kabeche et al., 2018).
A unique set of non-coding RNA is produced from each human alpha satellite array, and kinetochore assembly requires this RNA at the active centromere site (McNulty et al., 2017). Also, active transcription at the centromere is required for de novo deposition and incorporation of CENP-A into chromatin, and as such, to keep the centromere position (Choi et al., 2011; Bobkov et al., 2018). The products of transcription are highly variable lengths of RNAs, ranging from 500 to 2000+ nucleotides (Bouzinba-Segard et al., 2006; McNulty et al., 2017); in mice, these undergo post-transcriptional processing to generate smaller RNAs from 120 to 150 nucleotides (Bouzinba-Segard et al., 2006). These long non-coding RNAs are referred to as cenRNA.
It is thought that the RNAi has an important role in chromosome function since the suppression of different RNAi proteins displays a chromosome lagging phenotype in many eukaryotes (Gutbrod and Martienssen, 2020). In Cryptococcus yeast, loss of the RNAi machinery triggers the attrition of the retrotransposon composing the centromere, resulting in the shortening of centromere length (Yadav et al., 2018). In S. Pombe, transcription of the centromeric sequence and their processing by the RNAi machinery through the ribonuclease Dicer is required for chromosome segregation and gene silencing (Provost et al., 2002; Volpe et al., 2002). Rpb7, a subunit of RNA Pol II, actively recognizes a centromeric promoter and transcribes a part of the centromere to a pre-siRNA required for RNAi-directed chromatin silencing (Djupedal et al., 2005).
In higher eukaryotes, the effect of the RNAi machinery is less clear. The depletion of Dicer in mouse embryonic stem cells leads to an accumulation of centromeric transcripts (Kanellopoulou et al., 2005; Murchison et al., 2005). However, these two studies obtain opposite results about the loss of heterochromatin modification around the centromere, possibly caused by a small amount of truncated Dicer produced by an allele that may retain or inhibit the cytosine methylation at the centromeres (Murchison et al., 2005). Dicer processes the double-strand RNA derived from the centromeric repeat into smaller RNAs ranging from 25 to 150 nucleotides, that may be incorporated into the mammalian complex RITS (Kanellopoulou et al., 2005). In chicken-human hybrid cells, the depletion of Dicer results in the accumulation of transcript from α-satellite sequences, without affecting the localization of CENP-A or CENP-C, but results in the diffusion of HP1 throughout the entire chromosome, opposed to focusing on the centromere (Fukagawa et al., 2004). In Drosophila cells, the RNAi machinery affects the localization of HP1 and the heterochromatic silencing (Pal-Bhadra et al., 2004). More recently, Huang et al. showed that in human cell lines, the level and distribution of chromosome-associated α-satellite RNA on the centromere is dependent on the presence of siRNA generated by Dicer, guided by AGO2, and direct the deposition of CENPC at the centromere (Huang et al., 2015).
Multiple proteins interact with these RNAs, and are involved in heterochromatin formation or are centromere-specific factors, such as the H3K9 methyltransferase SUV39H1 and 2 (Johnson et al., 2017; Velazquez Camacho et al., 2017), HP1 (Maison et al., 2011), CENP-A (Quénet and Dalal, 2014), CENP-A chaperone protein HJURP (Quénet and Dalal, 2014) CENP-C (Huang et al., 2015) and Aurora B (Blower, 2016).
The fact that centromeres contain RNA has been known for several decades (Heidemann et al., 1977; Rieder, 1979). In maize, products of transcription of centromeric retrotransposons and satellite repeat—small RNAs that range from 40 to 200 nucleotides—are tightly bound to the centromeric histone 3, the maize equivalent of CENP-A, and components of the kinetochore (Topp et al., 2004). This transcriptional activity may facilitate the replacement and maintenance of the centromere (Topp et al., 2004). Maize CENP-C protein presents a DNA and an RNA binding motif, both of them required for the recruitment of CENP-C at the centromere, and that binding to RNA highly increases CENP-C affinity to DNA (Du et al., 2010). In Drosophila, the cenRNA produced from the 359-bp repeat satellite III is required for the localization of both CENP-A and CENP-C at the centromere for all chromosomes (Rošić et al., 2014). In human cells, cenRNA transcription is required for the localization of CENP-A and HJURP at the centromere (Quénet and Dalal, 2014). Depletion of CENP-A does not change the transcription level of these RNAs, but the reduction of cenRNA reduces the loading of CENP-A at the centromere, suggesting that the cenRNA themselves recruit CENP-A (McNulty et al., 2017). Besides, treatment by an RNAse results in the de-colocalization of CENP-C from mature human kinetochore, suggesting that cenRNA may also have a major function in the recruitment of CENP-C and its localization in the centromere (Wong et al., 2007).
In Xenopus eggs, the centromeric region is transcribed as cenRNA, and this cenRNA localizes to mitotic centromeres, chromatin, and spindle, through the chromosome passenger complex (CPC) allowing the activation of Aurora B (Blower, 2016). The transcription of these RNAs at the centromere region is required for attachment of the kinetochores to the mitotic spindle (Blower, 2016). Centromeric minor satellite RNAs from mice are also associated with proteins of the CPC, Aurora B, and Survivin (Ferri et al., 2009). Also, the kinase activity of Aurora B is dependent on RNA, and these satellite RNAs may be required (Ferri et al., 2009). In humans, α satellite RNA associates with Aurora B and INCENP, another component of the CPC, and their depletion from the cell result in the mislocalization of the CPC from the centromere region (Ideue et al., 2014).
There is some evidence of direct interaction between cenRNAs and centromeric protein, pointing out the critical roles of cenRNAs in multiple organisms. In Drosophila, the cenRNA produced from the 359-bp repeat satellite III binds to CENP-C (Rošić et al., 2014), while in human cells, cenRNAs are associated with CENP-A and CENP-C at the centromere (McNulty et al., 2017). Still, in human cells, CENP-C directly interacts in vitro with ∝ satellite RNAs through an RNA binding motif (Wong et al., 2007). In mammals, cenRNA interaction with Aurora B may play an important role in recruiting the CPC to the centromere (Ferri et al., 2009; Ideue et al., 2014).
Overall, cenRNA, as a component of the kinetochore, is involved in the recruitment of CENP-A, its chaperone HJURP, and CENP-C at the centromere, and is directly interacting with CENP-A and CENP-C. These components are involved in the recruitment of other parts of the centromere constitutive associated-network (CCAN) (Hori et al., 2008; Klare et al., 2015). Together with Aurora B, these components build the full kinetochore (Figure 1).
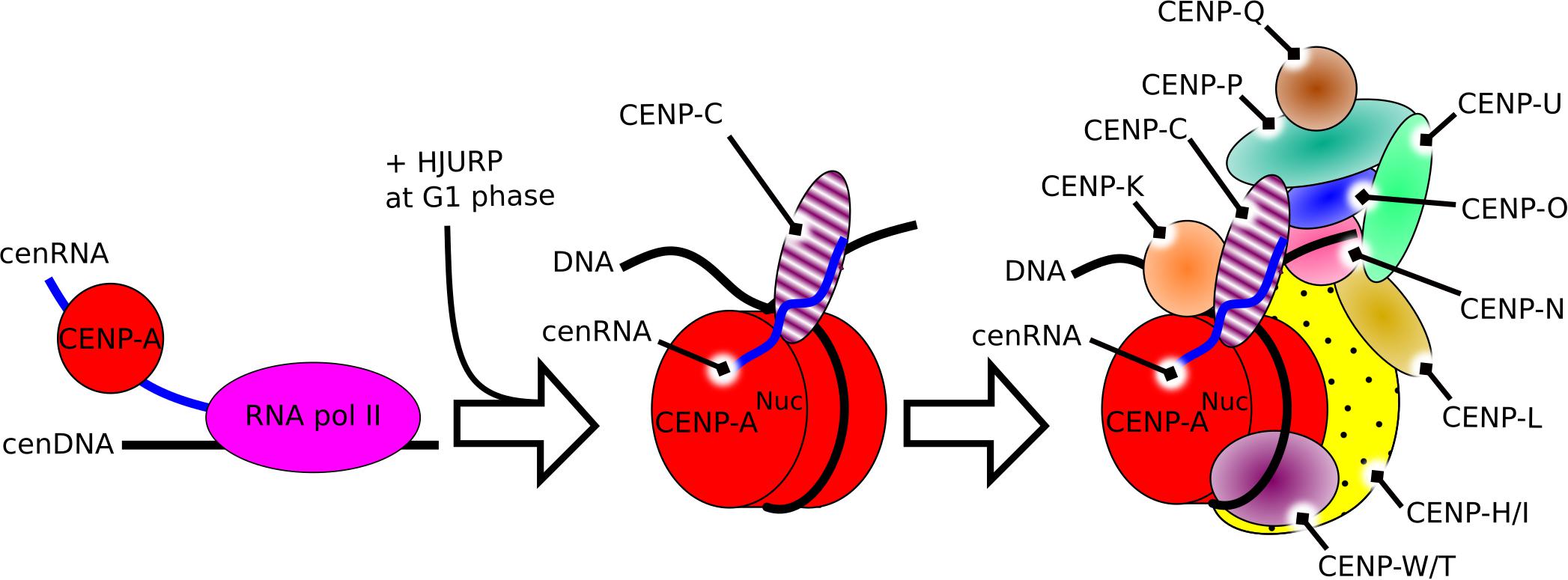
Figure 1. Schematic representation of the constitutive role of cenRNA in the structure of the centromere in humans. Left, the product of transcription of alpha-satellite DNA by the RNA polymerase II recruits CENP-A. Middle, after the constitution of the CENP-A nucleosome, cenRNA recruits CENP-C. Right, during mitosis, cenRNA may still be associated with the CCAN, and recruit the CPC via interaction with Aurora B to build the whole kinetochore.
In pancreatic and other epithelial human cancers, overexpression of α-satellite transcripts (cenRNA) (Ting et al., 2011) is correlated with a poor clinical prognosis (Zhang et al., 2016). One function of BRCA1 is to facilitate the mono-ubiquitination of histones H2A associated with satellite DNA, which suppresses their transcription (Zhu et al., 2011). In some types of breast cancer, a lack of BRCA1 function leads to an overexpression of satellite RNA (Zhu et al., 2018). The over-expression of these RNAs destabilize the DNA replication forks, which results in the formation of R-loop and DNA damage at the centromere region (Reddy et al., 2011; Crossley et al., 2019), which is sufficient to induce aneuploidy (Zhu et al., 2018). Overexpression of alpha-satellite transcripts has been identified in both mouse and human pancreatic adenocarcinomas, within average a 40-fold increase compared to normal tissue (Ting et al., 2011). It is thought to be caused by the alteration of heterochromatin silencing, affecting both satellite DNA and LINE retrotransposons; the latter is linked with the expression of neuroendocrine genes proximal to LINE sequence (Ting et al., 2011). In general, high levels of cenRNA promote high CIN that correlates with tumor metastasis (Zhu et al., 2011; Turajlic and Swanton, 2016; Chan et al., 2017). CIN creates micronuclei caused by errors in chromosome segregation, resulting in the presence of cytosol DNA that activates the cGas-STING pathway (cyclic GMP-AMP synthase stimulator of interferon gene) (Bakhoum et al., 2018). Suppression of CIN reduces metastasis even in highly aneuploid cells while activating CIN promotes cellular invasion and metastasis (Bakhoum et al., 2018). A prolonged cell arrest before mitosis, caused by CIN, reduces the inflammatory signaling and the anti-tumor immunity in a cGas dependent manner (Bakhoum et al., 2018; Chen et al., 2020). Overexpression of Mad2 increases the karyotypes complexity through a CIN effect in Kras tumors, and if in the first step, the growth of the tumor is disadvantageous, in the second step a selective pressure on oncogene elimination leads to the development of persistent subclones that grow steadily (Rowald et al., 2016; Sheltzer et al., 2017). Similarly, in spontaneous lymphomas and lung tumors, an increased rate of CIN drives an elevated level of tumorigenesis, however, in the case of induced tumor formation, an increased rate of CIN is a more effective inhibitor than activator of tumorigenesis (Weaver et al., 2007).
Thus, detection of cenRNA might be a good predictor of cancer prognosis. In support of this concept, detection of pericentromeric SatII RNA, satellite RNAs present at specific heterochromatin located at the edges of the centromere, in biopsies is more tightly correlated with detection than the standard method (Ting et al., 2011; Bersani et al., 2015; Kishikawa et al., 2016). Sensing cytosol DNA to measure the amount of CIN also seems promising (Corrales et al., 2015; Jiang et al., 2020). Clinical trials are testing agonists of STING-nucleotide, with direct microinjection in the tumor of a synthetic cyclic dinucleotide (CDN) derivative that activates all human STING alleles (Corrales et al., 2015; Flood et al., 2019). Since CIN produces cytosol DNA that is detected by cGas-STING, this results in the activation of the STING pathway, involving the production of IFN-β and other cytokines (Ishikawa and Barber, 2008; Figure 2).
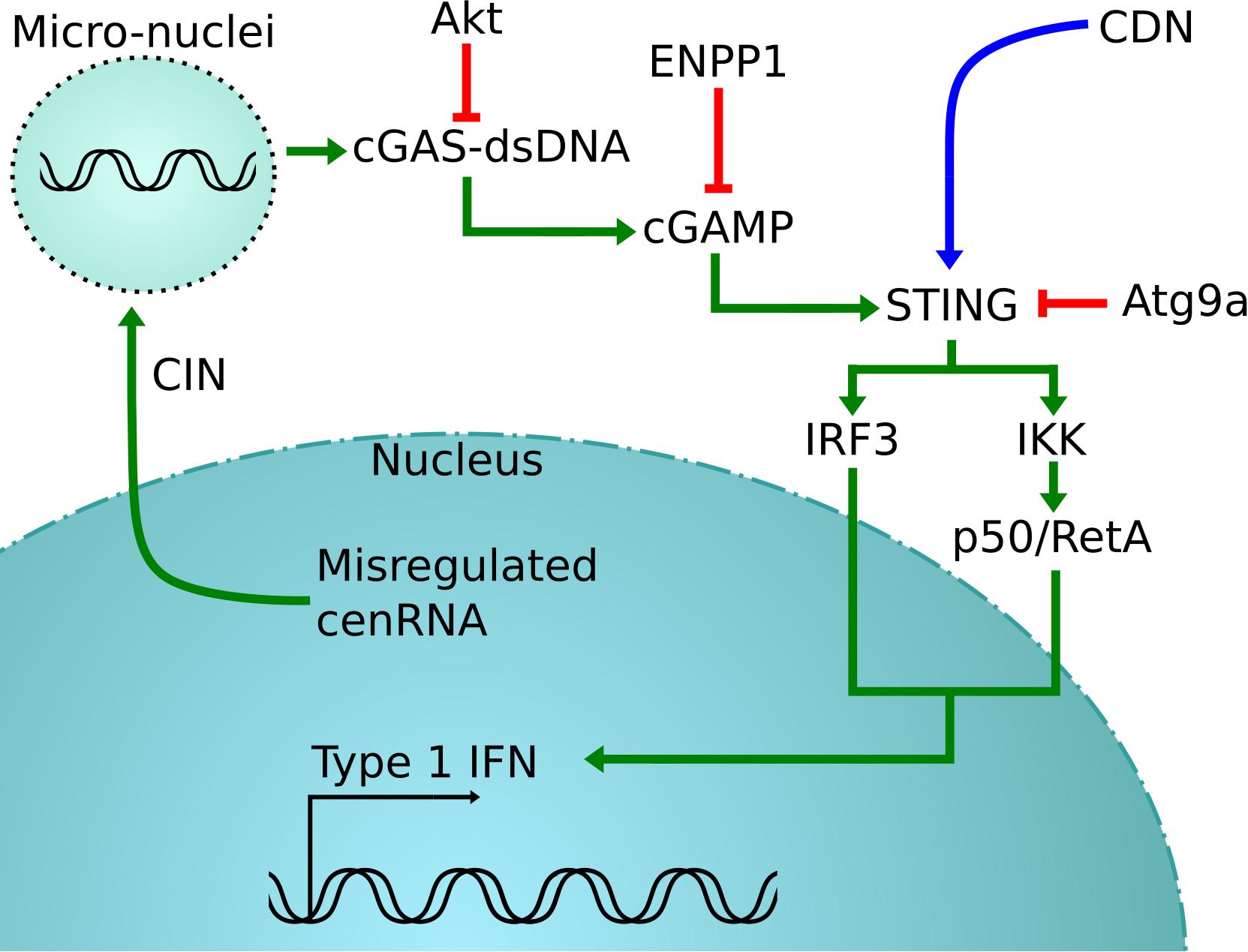
Figure 2. cGas-STING activation pathway. In green is the normal pathway, where cytosolic DNA created by the misregulation of cenRNA is detected by cGAS, which will cause the activation of STING (Corrales et al., 2015; Jiang et al., 2020). This is detrimental for cancer cells that will inactive STING using different strategies, in red (Saitoh et al., 2009; Xiong et al., 2018). One possible rescue is to inject synthetic CDN to force the activation of STING, shown in blue (Corrales et al., 2015).
Centromeres are essential for chromosome segregation, and while this function is preserved across species, their structure and mechanisms underlying presents large differences. However, one common particularity is that the centromere DNA sequence is actively transcribed. The resulting RNA is then processed to result in RNAi in yeast and in mammals or cenRNA in mammals. These cenRNAs have been shown to be an indispensable part of the centromere by interacting with CENP-A and CENP-C, as well as actively recruiting other parts of the kinetochore. The dysregulation of the cenRNA expression leads to chromosome instability, which can result in some cases in metastasis of a cancer. One possible treatment to counter this effect is the injection of a synthetic CDN that activates the cGas-STING pathway, able to detect the presence of cytosolic DNA resulting from CIN and activating an immune-response. One point to further investigate is with which mechanism higher eukaryotes can finely tune the expression of cenRNAs, and if a drug can avoid the overexpression of cenRNA in cancer cells.
SL wrote the manuscript. KK directed how to write the manuscript and edited it. Both authors contributed to the article and approved the submitted version.
This work was supported by the National Science Foundation under Grant No. 1949653.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aguilera, A., and García-Muse, T. (2012). R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46, 115–124. doi: 10.1016/j.molcel.2012.04.009
Alexandrov, I. A., Medvedev, L. I., Mashkova, T. D., Kisselev, L. L., Romanova, L. Y., and Yurov, Y. B. (1993). Definition of a new alpha satellite suprachromosomal family characterized by monomeric organization. Nucleic Acids Res. 21, 2209–2215. doi: 10.1093/nar/21.9.2209
Aze, A., Sannino, V., Soffientini, P., Bachi, A., and Costanzo, V. (2016). Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression. Nat. Cell Biol. 18, 684–691. doi: 10.1038/ncb3344
Bakhoum, S. F., Ngo, B., Laughney, A. M., Cavallo, J.-A., Murphy, C. J., Ly, P., et al. (2018). Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472. doi: 10.1038/nature25432
Bersani, F., Lee, E., Kharchenko, P. V., Xu, A. W., Liu, M., Xega, K., et al. (2015). Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc. Natl. Acad. Sci. U.S.A. 112, 15148–15153. doi: 10.1073/pnas.1518008112
Blower, M. D. (2016). Centromeric transcription regulates Aurora-B localization and activation. Cell Rep. 15, 1624–1633. doi: 10.1016/j.celrep.2016.04.054
Blower, M. D., Sullivan, B. A., and Karpen, G. H. (2002). Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2, 319–330. doi: 10.1016/s1534-5807(02)00135-1
Bobkov, G. O. M., Gilbert, N., and Heun, P. (2018). Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation. J. Cell Biol. 217, 1957–1972. doi: 10.1083/jcb.201611087
Bouzinba-Segard, H., Guais, A., and Francastel, C. (2006). Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc. Natl. Acad. Sci. U.S.A. 103, 8709–8714. doi: 10.1073/pnas.0508006103
Buisson, R., Boisvert, J. L., Benes, C. H., and Zou, L. (2015). Distinct but concerted roles of ATR, DNA-PK, and Chk1 in countering replication stress during S phase. Mol. Cell 59, 1011–1024. doi: 10.1016/j.molcel.2015.07.029
Bury, L., Moodie, B., Ly, J., McKay, L. S., Miga, K. H., and Cheeseman, I. M. (2020). Alpha-satellite RNA transcripts are repressed by centromere-nucleolus associations. Elife 9:e59770. doi: 10.7554/eLife.59770
Carone, D. M., Longo, M. S., Ferreri, G. C., Hall, L., Harris, M., Shook, N., et al. (2009). A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma 118, 113–125. doi: 10.1007/s00412-008-0181-5
Chakraborty, P., Huang, J. T. J., and Hiom, K. (2018). DHX9 helicase promotes R-loop formation in cells with impaired RNA splicing. Nat. Commun. 9:4346.
Chan, D. Y. L., Moralli, D., Khoja, S., and Monaco, Z. L. (2017). Noncoding centromeric RNA expression impairs chromosome stability in human and murine stem cells. Dis. Markers 2017:7506976. doi: 10.1155/2017/7506976
Chan, F. L., Marshall, O. J., Saffery, R., Kim, B. W., Earle, E., Choo, K. H. A., et al. (2012). Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc. Natl. Acad. Sci. U.S.A. 109, 1979–1984. doi: 10.1073/pnas.1108705109
Chen, J., Harding, S. M., Natesan, R., Tian, L., Benci, J. L., Li, W., et al. (2020). Cell cycle checkpoints cooperate to suppress DNA- and RNA-associated molecular pattern recognition and anti-tumor immune responses. Cell Rep. 32:108080. doi: 10.1016/j.celrep.2020.108080
Choi, E. S., Strålfors, A., Castillo, A. G., Durand-Dubief, M., Ekwall, K., and Allshire, R. C. (2011). Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J. Biol. Chem. 286, 23600–23607. doi: 10.1074/jbc.m111.228510
Corrales, L., Glickman, L. H., McWhirter, S. M., Kanne, D. B., Sivick, K. E., Katibah, G. E., et al. (2015). Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 11, 1018–1030. doi: 10.1016/j.celrep.2015.04.031
Crossley, M. P., Bocek, M., and Cimprich, K. A. (2019). R-loops as cellular regulators and genomic threats. Mol. Cell 73, 398–411. doi: 10.1016/j.molcel.2019.01.024
Djupedal, I., Portoso, M., Spåhr, H., Bonilla, C., Gustafsson, C. M., Allshire, R. C., et al. (2005). RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 19, 2301–2306. doi: 10.1101/gad.344205
Du, Y., Topp, C. N., and Dawe, R. K. (2010). DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 6:e1000835. doi: 10.1371/journal.pgen.1000835
Eichler, E. E. (1999). Repetitive conundrums of centromere structure and function. Hum. Mol. Genet. 8, 151–155. doi: 10.1093/hmg/8.2.151
Ferri, F., Bouzinba-Segard, H., Velasco, G., Hubé, F., and Francastel, C. (2009). Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 37, 5071–5080. doi: 10.1093/nar/gkp529
Flood, B. A., Higgs, E. F., Li, S., Luke, J. J., and Gajewski, T. F. (2019). STING pathway agonism as a cancer therapeutic. Immunol. Rev. 290, 24–38. doi: 10.1111/imr.12765
Fukagawa, T., and Earnshaw, W. C. (2014). The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell 30, 496–508. doi: 10.1016/j.devcel.2014.08.016
Fukagawa, T., Nogami, M., Yoshikawa, M., Ikeno, M., Okazaki, T., Takami, Y., et al. (2004). Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6, 784–791. doi: 10.1038/ncb1155
Grishok, A., Tabara, H., and Mello, C. C. (2000). Genetic requirements for inheritance of RNAi in C. elegans. Science 287, 2494–2497. doi: 10.1126/science.287.5462.2494
Gutbrod, M. J., and Martienssen, R. A. (2020). Conserved chromosomal functions of RNA interference. Nat. Rev. Genet. 21, 311–331. doi: 10.1038/s41576-019-0203-6
Heidemann, S. R., Sander, G., and Kirschner, M. W. (1977). Evidence for a functional role of RNA in centrioles. Cell 10, 337–350. doi: 10.1016/0092-8674(77)90021-6
Hildebrand, E. M., and Biggins, S. (2016). Regulation of budding yeast CENP-A levels prevents misincorporation at promoter nucleosomes and transcriptional defects. PLoS Genet. 12:e1005930. doi: 10.1371/journal.pgen.1005930
Hill, A., and Bloom, K. (1987). Genetic manipulation of centromere function. Mol. Cell. Biol. 7, 2397–2405. doi: 10.1128/mcb.7.7.2397
Hori, T., Amano, M., Suzuki, A., Backer, C. B., Welburn, J. P., Dong, Y., et al. (2008). CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135, 1039–1052. doi: 10.1016/j.cell.2008.10.019
Huang, C., Wang, X., Liu, X., Cao, S., and Shan, G. (2015). RNAi pathway participates in chromosome segregation in mammalian cells. Cell Discov. 1:15029.
Ideue, T., Cho, Y., Nishimura, K., and Tani, T. (2014). Involvement of satellite I noncoding RNA in regulation of chromosome segregation. Genes Cells 19, 528–538. doi: 10.1111/gtc.12149
Ishikawa, H., and Barber, G. N. (2008). STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678. doi: 10.1038/nature07317
Ishikura, S., Nakabayashi, K., Nagai, M., Tsunoda, T., and Shirasawa, S. (2020). ZFAT binds to centromeres to control noncoding RNA transcription through the KAT2B-H4K8ac-BRD4 axis. Nucleic Acids Res. 48, 10848–10866. doi: 10.1093/nar/gkaa815
Jiang, M., Chen, P., Wang, L., Li, W., Chen, B., Liu, Y., et al. (2020). cGAS-STING, an important pathway in cancer immunotherapy. J. Hematol. Oncol. 13:81.
Johnson, W. L., Yewdell, W. T., Bell, J. C., McNulty, S. M., Duda, Z., O’Neill, R. J., et al. (2017). RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. Elife 6:e25299. doi: 10.7554/eLife.25299
Kabeche, L., Nguyen, H. D., Buisson, R., and Zou, L. (2018). A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science 359, 108–114. doi: 10.1126/science.aan6490
Kanellopoulou, C., Muljo, S. A., Kung, A. L., Ganesan, S., Drapkin, R., Jenuwein, T., et al. (2005). Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19, 489–501. doi: 10.1101/gad.1248505
Kishikawa, T., Otsuka, M., Yoshikawa, T., Ohno, M., Yamamoto, K., Yamamoto, N., et al. (2016). Quantitation of circulating satellite RNAs in pancreatic cancer patients. JCI Insight 1:e86646.
Klare, K., Weir, J. R., Basilico, F., Zimniak, T., Massimiliano, L., Ludwigs, N., et al. (2015). CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J. Cell Biol. 210, 11–22. doi: 10.1083/jcb.201412028
Ling, Y. H., and Yuen, K. W. Y. (2019). Point centromere activity requires an optimal level of centromeric noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 116, 6270–6279. doi: 10.1073/pnas.1821384116
Liu, H., Qu, Q., Warrington, R., Rice, A., Cheng, N., and Yu, H. (2015). Mitotic transcription installs Sgo1 at centromeres to coordinate chromosome segregation. Mol. Cell 59, 426–436. doi: 10.1016/j.molcel.2015.06.018
Maison, C., Bailly, D., Roche, D., Montes de Oca, R., Probst, A. V., Vassias, I., et al. (2011). SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat. Genet. 43, 220–227. doi: 10.1038/ng.765
McNulty, S. M., Sullivan, L. L., and Sullivan, B. A. (2017). Human centromeres produce chromosome-specific and array-specific alpha satellite transcripts that are complexed with CENP-A and CENP-C. Dev. Cell 42, 226–240.e6.
Melters, D. P., Bradnam, K. R., Young, H. A., Telis, N., May, M. R., Ruby, J. G., et al. (2013). Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 14:R10.
Mishra, P. K., Chakraborty, A., Yeh, E., Feng, W., Bloom, K. S., and Basrai, M. A. (2021). R-loops at centromeric chromatin contribute to defects in kinetochore integrity and chromosomal instability in budding yeast. Mol. Biol. Cell 32, 74–89. doi: 10.1091/mbc.e20-06-0379
Mizuno, H., Kawahara, Y., Wu, J., Katayose, Y., Kanamori, H., Ikawa, H., et al. (2011). Asymmetric distribution of gene expression in the centromeric region of rice chromosome 5. Front. Plant Sci. 2:16. doi: 10.3389/fpls.2011.00016
Murchison, E. P., Partridge, J. F., Tam, O. H., Cheloufi, S., and Hannon, G. J. (2005). Characterization of dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 102, 12135–12140. doi: 10.1073/pnas.0505479102
Nagaki, K., Cheng, Z., Ouyang, S., Talbert, P. B., Kim, M., Jones, K. M., et al. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36, 138–145. doi: 10.1038/ng1289
Nakano, M., Cardinale, S., Noskov, V. N., Gassmann, R., Vagnarelli, P., Kandels-Lewis, S., et al. (2008). Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev. Cell 14, 507–522. doi: 10.1016/j.devcel.2008.02.001
Niikura, Y., Kitagawa, R., and Kitagawa, K. (2016). CENP-A ubiquitylation is inherited through dimerization between cell divisions. Cell Rep. 15, 61–76. doi: 10.1016/j.celrep.2016.03.010
Ohkuni, K., and Kitagawa, K. (2011). Endogenous transcription at the centromere facilitates centromere activity in budding yeast. Curr. Biol. 21, 1695–1703. doi: 10.1016/j.cub.2011.08.056
Pal-Bhadra, M., Leibovitch, B. A., Gandhi, S. G., Chikka, M. R., Bhadra, U., Birchler, J. A., et al. (2004). Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303, 669–672. doi: 10.1126/science.1092653
Pluta, A. F., Mackay, A. M., Ainsztein, A. M., Goldberg, I. G., and Earnshaw, W. C. (1995). The centromere: hub of chromosomal activities. Science 270, 1591–1594. doi: 10.1126/science.270.5242.1591
Provost, P., Silverstein, R. A., Dishart, D., Walfridsson, J., Djupedal, I., Kniola, B., et al. (2002). Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc. Natl. Acad. Sci. U.S.A. 99, 16648–16653. doi: 10.1073/pnas.212633199
Quénet, D., and Dalal, Y. (2014). A long non-coding RNA is required for targeting centromeric protein A to the human centromere. Elife 3:e03254.
Reddy, K., Tam, M., Bowater, R. P., Barber, M., Tomlinson, M., Nichol Edamura, K., et al. (2011). Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 39, 1749–1762. doi: 10.1093/nar/gkq935
Rieder, C. L. (1979). Ribonucleoprotein staining of centrioles and kinetochores in newt lung cell spindles. J. Cell Biol. 80, 1–9. doi: 10.1083/jcb.80.1.1
Rošić, S., Köhler, F., and Erhardt, S. (2014). Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell Biol. 207, 335–349. doi: 10.1083/jcb.201404097
Rowald, K., Mantovan, M., Passos, J., Buccitelli, C., Mardin, B. R., Korbel, J. O., et al. (2016). Negative selection and chromosome instability induced by Mad2 overexpression delay breast cancer but facilitate oncogene-independent outgrowth. Cell Rep. 15, 2679–2691. doi: 10.1016/j.celrep.2016.05.048
Saffery, R., Sumer, H., Hassan, S., Wong, L. H., Craig, J. M., Todokoro, K., et al. (2003). Transcription within a functional human centromere. Mol. Cell 12, 509–516. doi: 10.1016/s1097-2765(03)00279-x
Saitoh, T., Fujita, N., Hayashi, T., Takahara, K., Satoh, T., Lee, H., et al. (2009). Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. U.S.A. 106, 20842–20846. doi: 10.1073/pnas.0911267106
Santaguida, S., and Amon, A. (2015). Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 16, 473–485. doi: 10.1038/nrm4025
Sheltzer, J. M., Ko, J. H., Replogle, J. M., Habibe Burgos, N. C., Chung, E. S., Meehl, C. M., et al. (2017). Single-chromosome gains commonly function as tumor suppressors. Cancer Cell 31, 240–255. doi: 10.1016/j.ccell.2016.12.004
Sullivan, B. A., and Karpen, G. H. (2004). Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11, 1076–1083. doi: 10.1038/nsmb845
Ting, D. T., Lipson, D., Paul, S., Brannigan, B. W., Akhavanfard, S., Coffman, E. J., et al. (2011). Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331, 593–596. doi: 10.1126/science.1200801
Topp, C. N., Zhong, C. X., and Dawe, R. K. (2004). Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl. Acad. Sci. U.S.A. 101, 15986–15991. doi: 10.1073/pnas.0407154101
Turajlic, S., and Swanton, C. (2016). Metastasis as an evolutionary process. Science 352, 169–175. doi: 10.1126/science.aaf2784
Van Hooser, A. A., Ouspenski, I. I., Gregson, H. C., Starr, D. A., Yen, T. J., Goldberg, M. L., et al. (2001). Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114(Pt 19), 3529–3542.
Velazquez Camacho, O., Galan, C., Swist-Rosowska, K., Ching, R., Gamalinda, M., Karabiber, F., et al. (2017). Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation. Elife 6:e25293. doi: 10.7554/eLife.25293
Volpe, T. A., Kidner, C., Hall, I. M., Teng, G., Grewal, S. I. S., and Martienssen, R. A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837. doi: 10.1126/science.1074973
Weaver, B. A. A., Silk, A. D., Montagna, C., Verdier-Pinard, P., and Cleveland, D. W. (2007). Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11, 25–36. doi: 10.1016/j.ccr.2006.12.003
Willard, H. F. (1985). Chromosome-specific organization of human alpha satellite DNA. Am. J. Hum. Genet. 37, 524–532.
Willard, H. F., Waye, J. S., Skolnick, M. H., Schwartz, C. E., Powers, V. E., and England, S. B. (1986). Detection of restriction fragment length polymorphisms at the centromeres of human chromosomes by using chromosome-specific alpha satellite DNA probes: implications for development of centromere-based genetic linkage maps. Proc. Natl. Acad. Sci. U.S.A. 83, 5611–5615. doi: 10.1073/pnas.83.15.5611
Wong, L. H., Brettingham-Moore, K. H., Chan, L., Quach, J. M., Anderson, M. A., Northrop, E. L., et al. (2007). Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 17, 1146–1160. doi: 10.1101/gr.6022807
Xiong, M., Wang, S., Wang, Y.-Y., and Ran, Y. (2018). The regulation of cGAS. Virol. Sin. 33, 117–124. doi: 10.1007/s12250-018-0005-6
Yadav, V., Sun, S., Billmyre, R. B., Thimmappa, B. C., Shea, T., Lintner, R., et al. (2018). RNAi is a critical determinant of centromere evolution in closely related fungi. Proc. Natl. Acad. Sci. U.S.A. 115, 3108–3113. doi: 10.1073/pnas.1713725115
Zhang, W., Mao, J.-H., Zhu, W., Jain, A. K., Liu, K., Brown, J. B., et al. (2016). Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat. Commun. 7:12619.
Zhu, Q., Hoong, N., Aslanian, A., Hara, T., Benner, C., Heinz, S., et al. (2018). Heterochromatin-encoded satellite RNAs induce breast cancer. Mol. Cell 70, 842–853.e7.
Zhu, Q., Pao, G. M., Huynh, A. M., Suh, H., Tonnu, N., Nederlof, P. M., et al. (2011). BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477, 179–184. doi: 10.1038/nature10371
Keywords: cancer, cenRNA, lncRNA, kinetochore, centromere, chromosome instability
Citation: Leclerc S and Kitagawa K (2021) The Role of Human Centromeric RNA in Chromosome Stability. Front. Mol. Biosci. 8:642732. doi: 10.3389/fmolb.2021.642732
Received: 16 December 2020; Accepted: 26 February 2021;
Published: 31 March 2021.
Edited by:
Belén Gomez-Gonzalez, Andalusian Center of Molecular Biology and Regenerative Medicine (CABIMER), SpainReviewed by:
Lee Zou, Montreal General Hospital, CanadaCopyright © 2021 Leclerc and Kitagawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katsumi Kitagawa, a2l0YWdhd2FrQHV0aHNjc2EuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.