- 1Department of Cardiology, Anesthesiology and Emergency Medicine, Henan Province People's Hospital and People's Hospital of Zhengzhou University, Zhengzhou, China
- 2Department of Cardiology, People's Hospital of Zhengzhou University, Zhengzhou, China
- 3Department of Cardiology, Henan Province People's Hospital and People's Hospital of Zhengzhou University, Zhengzhou, China
Cardiovascular disease is the main disease that affects human life span. In recent years, the disease has been increasingly addressed at the molecular levels, for example, pseudogenes are now known to be involved in the pathogenesis and development of cardiovascular diseases. Pseudogenes are non-coding homologs of protein-coding genes and were once called “junk gene.” Since they are highly homologous to their functional parental genes, it is somewhat difficult to distinguish them. With the development of sequencing technology and bioinformatics, pseudogenes have become readily identifiable. Recent studies indicate that pseudogenes are closely related to cardiovascular diseases. This review provides an overview of pseudogenes and their roles in the pathogenesis of cardiovascular diseases. This new knowledge adds to our understanding of cardiovascular disease at the molecular level and will help develop new biomarkers and therapeutic approaches designed to prevent and treat the disease.
Introduction
Cardiovascular diseases (CVDs) are the leading cause of death, taking an about 18 million lives globally every year (Goradel et al., 2018). CVDs are composed of coronary heart disease, cerebrovascular disease, rheumatic heart disease as well as other related conditions. A better understanding of CVDs at the molecular level is particularly important to develop preventive and therapeutic strategies. In this regard, tremendous effects have been made toward the elucidation of molecular mechanisms underlying various CVDs (Uchida and Dimmeler, 2015; Li S. et al., 2018). For example, expression of long non-coding RNAs (lncRNAs) has been detected under normal physiological conditions and in disease states, some lncRNAs are found to regulate acute myocardial infarction (such as Novlnc6) (Uchida and Dimmeler, 2015) and acute heart failure [such as Mhrt (Zhang et al., 2019), H19 (Greco et al., 2016), and growth arrest specific 5 (Wang G. et al., 2019)], and gene transcripts such as MALAT1 and Tie-1-AS are found to control the growth and functions of blood vessels (Uchida and Dimmeler, 2015). Recently, Spinraza (a novel antisense oligonucleotide therapy) is approved by FDA to treat spinal muscular atrophy resulted from pseudogene SMN2, in which the antisense oligonucleotide is used to increase SMN2 exon 7 inclusion, thus increasing levels of survival motor neuron (SMN) protein (Chiriboga, 2017; Paton, 2017; Berciano et al., 2020). This is an excellent example of how our understanding of molecular mechanism of pseudogene has been translated to important therapeutic solution.
Pseudogene was first identified in 1977 (Jacq et al., 1977), and generally refers to non-functional DNA sequences derived from non-sense or frameshift mutations on protein coding regions of
ancient functional genes. Therefore, it has high sequence homolog with the parental gene but does not encode any specific protein or peptide. Most pseudogenes in the human genome have not been characterized for biological functions. For a long time, they have been considered as “junk gene” as a result of ongoing evolution. However, data obtained during the past decade have indicated that this interpretation of the usefulness of pseudogenes is not entirely correct, and many pseudogenes have important biological and genetic functions (Jacq et al., 1977; Jingsi et al., 2015; Xie et al., 2019; Cheetham et al., 2020). With the advances in sequencing technology, more and more pseudogenes were identified (Dong et al., 2016). In the review, we address the classification, identification and role of pseudogenes in the pathogenesis of CVDs with emphasis on the mechanisms underlying gene expression regulation by pseudogenes.
Classification of Pseudogenes
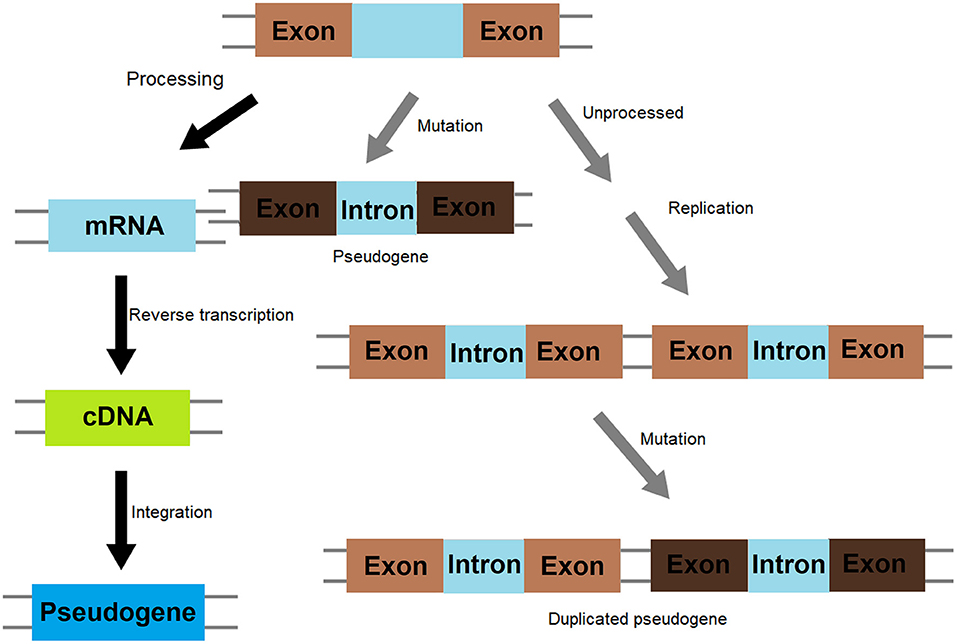
Pseudogenes were originally defined as non-functional genomic DNA sequences that are initially derived from genes. Therefore, it is assumed that various kinds of gene mutations occurred to generate pseudogenes are selectively neutral and the mutated genes are able to transmit to the next generations (Balakirev and Ayala, 2003). Generally speaking, pseudogenes are highly homologous with DNA sequences of ancestral functional genes but cannot be translated into protein due to the lack of critical regulatory elements such as promoters or the presence of premature stop codons resulted from frameshift mutations. They can be divided into non-processed or processed pseudogenes (Figure 1) (Milligan et al., 2016; Maranda et al., 2019). Non-processed pseudogenes are mainly derived from functional genes by processes such as unequal crossing over events during cell division. In these events, the homologous sequences are not paired precisely, a sequence is deleted in one strand and replaced with a duplication from its sister chromatid in mitosis or from its homologous chromosome during meiosis. They generally contain a promoter and other regulatory sequence elements such as enhancers and an intact or partial exon-intron structure. Unitary pseudogenes are a class of unprocessed pseudogenes that do not have functional counterparts in the genome (Zhang et al., 2010) and can be identified by analyzing the global inventory of orthologs between the human genome and its mammalian relatives (Zhang et al., 2010). On the other hand, processed pseudogenes result from the insertion of reversely-transcribed and mature RNA molecule (intron-less cDNA) into the genome, and the abundance of processed pseudogenes are therefore positively related to the expression level of genes from which the RNA molecules are derived (McDonell and Drouin, 2012). They usually do not have introns but a poly-A tail as cDNA sequences (Weiner et al., 1986) and are preferentially located in regions of low recombination rates in the human genome (Liu et al., 2010). They may be formed as a result of somatic reactivation of retrotransposons occurring during cancer development (Cooke et al., 2014). Different from processed pseudogenes, the presence of genetic structures in non-processed pseudogenes, such as exon-intron structures, may provide clues to trace the origin of these genes through bioinformatics analysis. Evolutionally, processed pseudogenes are derived from functional homologs and have distinct genetic features stemming from point mutations, deletions, and insertions. Once formed, pseudogenes can be inherited and transmitted to offspring and generate secondary pseudogenes through evolution.
Identification of Pseudogene
With the development of computer technology, bioinformatics, and especially sequencing technology, the whole genome sequences of many species have been sequenced and published. Pseudogenes are being identified through sequence alignment and genome-wide transcriptomic and proteomic studies (Xu and Zhang, 2016). Using RNA sequencing (RNA-seq), new transcriptomes are becoming available quickly. The transcriptomes can be assessed to discover pseudogenes using bioinformatics tools to search, compare, and screen relevant genomic databases and proteomic databases, and estimate the types and abundance of pseudogenes. Several tools are currently available for the identification of pseudogenes, such as PseudoPipe (Zhang et al., 2006) (http://www.pseudogene.org/pseudopipe/), Pseudofinder (https://github.com/filip-husnik/pseudo-finder), Retrofinder, and REtrotransposed Gene EXPlorer (Regexp) (Molineris et al., 2010). PseudoPipe is a computational pipeline based on homology to screen and search a mammalian genome. It can identify both non-processed and processed pseudogene sequences. In this program, the sequences of candidate pseudogene sequences are compared with protein sequences to remove annotated gene sequences and repetitive redundant sequences, generating clusters of genes from a unique parent. Pseudogenes are also grouped based on their homology, intron-exon structures, and presence of stop codons and frameshifts in the sequences (Zhang et al., 2006). Different from PseudoPipe, Pseudofinder is designed to detect pseudogene candidates from bacterial and archaeal genomes.
Despite purposes and procedures, all these methods depend on genomic, transcriptomic and proteomic data, and are very efficient for species with expressed sequence tags (EST) or proteomic data. However, they are not effective for species whose information is not available or organisms lacking large EST collections. REGEXP was developed to systematically identify retrotransposition events. This method, unlike other existing approaches, does not depend on a prior knowledge of mRNA sequences. Using this pipeline, 2,288 processed pseudogenes have been identified in the human genome (Molineris et al., 2010). Recently, several new approaches have been developed to identify pseudogenes from various sources. For examples, competing endogenous RNAs (ceRNAs) networks are explored to link pseudogenes and circular RNAs (circRNAs) to identify pseudogenes, which also provide significant insights into gene regulatory network with implication of human diseases (Li et al., 2018a). In addition, a computational pipeline (CIRCpseudo) has been developed to identify potential pseudogenes derived from circRNAs based on the feature that a circRNA-derived pseudogene would have an exon-exon junction in a reversed orientation (Dong et al., 2016). After searching the mouse and human reference genomes, 42 pseudogenes originated from circRFWD2 in mouse and 24 circRNA-derived pseudogenes in human were identified (Dong et al., 2016). Several databases have been developed to provide information about the transcriptional regulation, expression, functions, and mechanisms of pseudogenes as well as their roles in biological processes and diseases (Khelifi et al., 2005; Zheng et al., 2018). In addition, circRNAs have also been implicated in many diseases, including tumor and heart disease (Kalyana-Sundaram et al., 2012; Wang et al., 2016; Li et al., 2018b).
Function of Pseudogene
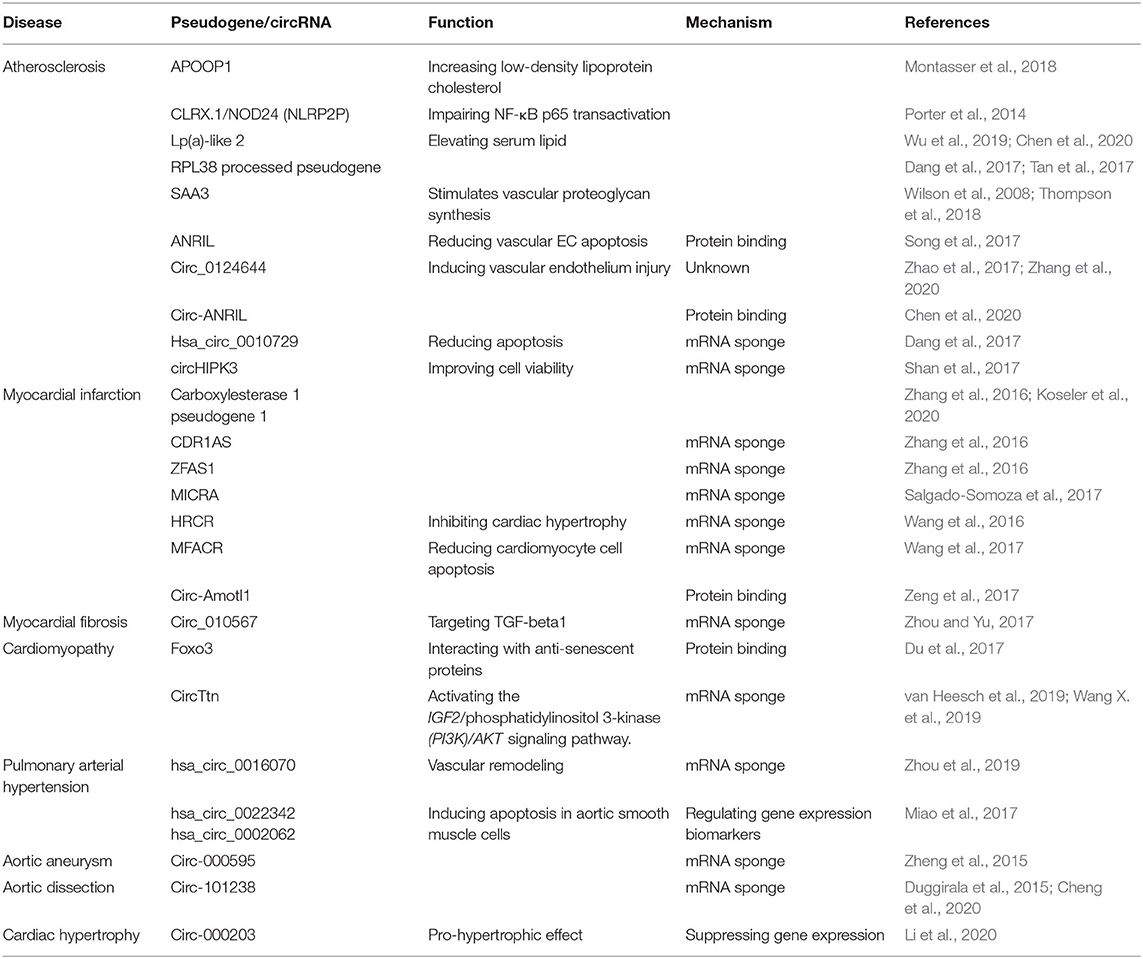
Pseudogenes cannot be translated into proteins, so they were once considered to be “dead genes” or “junk genes.” Although intensive efforts and studies have been made to elucidate and understand the function of pseudogenes, their biological and genetic roles still remain largely unknown. However, during the past decades, functional studies of pseudogenes have evolved from the initial discovery of single pseudogene to the modes and mechanisms of parental gene regulation by pseudogenes. It is found that pseudogenes regulate parental genes mainly via their transcripts. Expressed pseudogenes may be transcribed to form antisense RNAs, which may complement with the transcripts of parental genes to generate double strands or complemented stands, leading to change in expression of parent genes or resulting in destabilization of mRNA from the parental genes. For example, in an insertion-generated mouse mutant having polycystic kidneys and exhibiting bone deformity, the introduced transgene was found to have inserted in the vicinity of an expressed pseudogene Makorin1, termed Makorin1-p1. This transgenic insertion decreased the transcription level of Makorin1-p1, leading to destabilization of cellular Makorin1 mRNA (Hirotsune et al., 2003). Pseudogenes may form endogenous small interfering (SI) RNAs, when the double strand RNAs between pseudogene transcripts and their parent transcripts or repetitive sequences of pseudogene are sliced by endonuclease. These siRNAs may control and regulate the expression of their parental genes through mechanisms such as RNA interference (Abedini et al., 2018). For instance, it was found that many endogenous siRNAs (endo-siRNAs) are usually produced and formed from RNAs such as double-stranded RNAs generated by hybridization of transcripts spliced from genes that can code protein to antisense transcripts from homologous pseudogenes. Endo-siRNAs can interact with Piwi (P-element-induced wimpy testis)-interacting RNAs to repress the movement of transposable genetic elements (Tam et al., 2008). In addition, the competitive endogenous RNA (ceRNA) may compete with miRNA generated by pseudogenes for target genes to regulate the expression of target genes (Thomson and Dinger, 2016). For example, pseudogene RNAs can function as “sponge” to competitively bind miRNAs, leading to release or attenuation of repression through separating targeted miRNAs away from parental mRNAs, since binding of miRNA to the target RNA is not 100% complementary, it is likely that one miRNA can bind to multiple target RNAs. Such interactions and binding generate a complex gene regulatory network that may affect the expression of functional genes. An increase in abundance of pseudogene RNA can strengthen the blocking of miRNA and eliminate suppression on the expression of parental genes, leading to an up-regulation of parental gene expression. Studies have shown that the regulation of gene expression by pseudogene in this manner needs the presence of RNAs that bear common and functional metal regulatory elements (MREs) (Poliseno et al., 2010). Furthermore, some pseudogenes can also encode short peptides and proteins, although this is contradictive to original definition of pseudogenes. Some pseudogenes may gain or retain function during evolution. For example, the processed pseudogene NLRP2P is related specifically to higher primate. It is found to be closely associated with the Pyrin-only protein 2 (POP2/PYDC2). The open-reading frame of the NLRP2P gene located on chromosome X has characteristics that are consistent with a processed pseudogene (retrotransposon). However, it encodes Pyrin-domain-related protein with 45-amino acids (Porter et al., 2014). Using a new bioinformatics method, it was found that 40% lncRNAs and pseudogene RNAs expressed in human cells are translated into peptides and 74% of pseudogene peptides have conserved ORFs in mouse transcripts, implying that these peptides are potentially functional (Ji et al., 2015). As a consequence of biomedicine research, large number of pseudogenes have been implicated in many human diseases (Pink et al., 2011; Grander and Johnsson, 2016), including cancer (Emadi-Baygi et al., 2017; Tutar et al., 2018) and CVDs (Table 1).
Pseudogenes and CVDs
Atherosclerosis
Statins are the most commonly prescribed CVD drugs. To identify biomarkers and determinants that are responsible for statin response, Kim et al. compared whole transcriptome sequence data of patients collected from simvastatin and control after simvastatin treatment. They found that one of the most differentially expressed genes is zinc finger protein 542 pseudogene (ZNF542P). It is considered as the signature gene because the changes of its expression are most correlated with statin-induced changes in the cellular level of cholesterol ester. They further showed that knock-down of the ZNF542P gene in a human hepatoma cell line increases the intracellular levels of cholesterol ester after the cells were exposed to simvastatin. These findings indicate that ZNF542P may have a role in low-density lipoprotein cholesterol (LDL-C) response to simvastatin (Kim et al., 2018). However, since these genes were identified from cell lines, not directly from tissues of patients treated with simvastatin, it is likely that they might not be the most responsive genes in vivo.
Elevated LDL-C level is a main risk factor for CVDs because it contributes to, and enhances the development and progression of atherosclerotic lesions. Currently, only 20% of the variation in LDL-C levels can be attributed to genetic variants. Through an array-based association analysis, it was found that a variant is strongly associated with LDL-C levels and the expression of a transcribed pseudogene, APOOP1 located on chromosome 5, increased LDL-C level and vascular plaque formation (Montasser et al., 2018), suggesting that this may be a novel mechanism of lipid homeostasis.
Serum amyloid-alpha (SAA) and high-sensitivity C-reactive protein (hs-CRP) are sensitive biomarkers of acute inflammation and are related to atherosclerosis. Recent studies show that there is a significant and independent relationship between SAA and development of potential cardiovascular events, suggesting that systemic inflammations, manifested as high SAA or hs-CRP levels, may accelerate the destabilization of existing atherosclerotic plaques (Johnson et al., 2004). SAA has three transcripts, SAA1/SAA2/SAA3. Among them, SAA3 is a human pseudogene. In the animal model, it is an expressed acute phase isoform in mice when acute inflammatory reaction occurs. Using ApoE(–/–) mouse model, it was shown that over-expression of SAA3 results in a 4-fold increase in atherosclerosis lesion size over control while knockdown of SAA3 decreases atherosclerosis (Johnson et al., 2004), implying that this gene may be a new therapeutic target for atherosclerosis.
Non-coding locus INK4 on chromosome 9p21.3 is closely related to atherosclerosis. The antisense non-coding RNA in INK4 locus ANRIL was confirmed to be involved in the pathological process of atherosclerosis, leading to carotid plaque, stroke, aneurysms, peripheral arterial disease, heart failure, and cardiovascular death events (Holdt and Teupser, 2012). However, ANRIL is not directly related to high risk factors such as hypertension and hyperlipidemia. Mechanistically, it forms cicrANRIL, which binds to a number of proteins such as the nucleolar protein pescadillo homolog 1 (PES1), leading to impairment of exonuclease-mediated pre-rRNA processing and ribosome biogenesis in vascular smooth muscle cells and macrophages, resulting in apoptosis and inhibition of proliferation (Holdt et al., 2016).
Idiopathic Pulmonary Arterial Hypertension
Idiopathic pulmonary arterial hypertension (IPAH) is a vasculopathy. It is characterized by increased pulmonary vascular resistance due to vasoconstriction and/or lung remodeling defects such as plexiform lesions, which are the hallmark of the PAH, as well as massive cell proliferation, remarkable, and irreversible vascular and angiogenic dysfunction. Octamer-binding transcription factor 4 (Oct4) is a transcription factor and is used as pluripotency markers in embryonic stem cell research. Six pseudogenes of the Oct4 family have been identified by using bioinformatics approach to analyze the genomic nucleotide sequences. Some of them are found involved in the excessive proliferation of pulmonary arterial smooth muscle cells (PASMC) in patients with IPAH. Therefore, they may have a role in the pathogenesis and development of IPAH. The expressions of Oct-4 isoforms are upregulated in IPAH-PASMC conditions. However, the mRNA levels of Oct-4 pseudogene Oct-4-psG1 and Oct-4-psG5 are significantly down-regulated in IPAH-PASMC (Figure 2) (Firth et al., 2010).
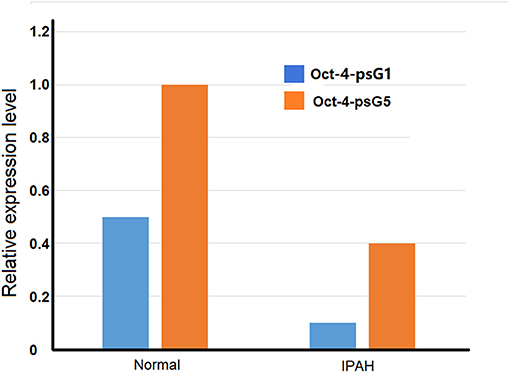
Figure 2. Comparison of expression of Oct-4 pseudogenes PSG1 and PSG5 in pulmonary arterial smooth muscle cells (PASMC) from idiopathic pulmonary arterial hypertension (IPAH) patients and normal individuals.
Heart Failure
Heart failure (HF) is a heterogeneous clinical syndrome stemming from cardiac overload and injury. The disease is expected to increase steadily in the upcoming years due to demographic changes such as aging. Therefore, a better understanding of molecular mechanisms underlying the disease is crucial for prevention and treatment of the disease. Using RNA-Seq, the expression of lncRNAs in 22 transplanted human models of HF hearts was studies. In HF heart models, a total of 84,793 mRNA coding and non-coding transcripts were identified using RNA-Seq, including 13,019 protein-coding genes, 2,085 lncRNA genes, and 1,064 pseudogenes. Based on the Ensembl release 73 (Genome Reference Consortium human genome build 37), 48 lncRNAs, 27 pseudogenes, and 30 antisense RNAs were identified for a total of 105 differentially expressed lncRNAs in the HF hearts. Compared with the donor hearts, HF hearts showed that 7.7% of protein-coding genes, 3.7% of lncRNAs, and 2.5% of pseudogenes are differentially expressed. In silico functional analyses suggested that many of these pseudogenes have possible regulatory roles and are of important for future mechanistic study (Di Salvo et al., 2015a). Based on RNA sequencing, 800–1,000 differentially expressed genes (DEGs) were identified between HF right ventricular (RV) and unused donor human heart RVs (DON RV), and many of them are likely pseudogenes. Since these DEGs had high sensitivities, specificities, predictive values and areas under the receiver operating characteristic curves, they might be novel HF myocardial biomarkers (Di Salvo et al., 2015b). In these studies, the sequenced transcripts were categorized based on the Ensembl release 73, which was released in 2013. More deferentially expressed transcripts could be identified with newer versions of the Ensembl release. Furthermore, annotation of these transcripts with other databases, such as GRCh38.p13 (Genome Reference Consortium Human Build 38 patch release 13) may offer more functional and classification information. When RNA-seq is used to profile the transcripts, the RNA sources have great impact on the transcriptome. In their work, end-stage human myocardium was used for RNA-seq, which may generate sequence bias due to medications and therapies (Di Salvo et al., 2015b), resulting in biomarkers less suitable for non-end stage patients. In addition, in vivo and in vitro experiments are needed to defined and validate the biological functions of the genes identified in silica. In general, parental and coding genes have high expression levels than corresponding pseudogenes. However, some pseudogenes have been found highly expressed. For example, the expression level of pseudogene FBXO43 is far more abundant than the corresponding parental gene (Shoji et al., 2014). Endogenous siRNA produced from pseudogene FBXO43 does not target the transcript of FBXO43, but the transcripts related to FBXO43 such as the transcripts of MAPK1, MDM2, OSMR, and IRAK3 and NMRA-like protein NMRAL1, which is a redox-sensitive transcriptional regulator. Reduced expression of NMRAL1 increases nitric oxide (NO) production and reduces cell viability. Compared with unused donor hearts, the expression level of NMRAL1 pseudogenes, but not NMRAL1 in right ventricle of HF was significantly reduced, indicating that NMRAL1 pseudogenes are involved in HF in right ventricle (Garciandia and Suarez, 2013). Identifications of genes including pseudogenes would provide targets for future molecular analysis on HF.
Other CVDs
The occurrence and prevalence of coronary artery disease are gender-depended. Men are more likely to be affected than women. Therefore, it is possible that the Y chromosome may contribute to this sexual difference. An analysis of 11 genetic markers positioned on the male-specific region of the Y chromosome in 3233 British men revealed nine haplogroups. The carriers of haplogroup I had a risk of coronary artery disease in about 50% higher likelihood than men with other Y chromosome lineages. The high risk possibly results from the interactions of immunity and inflammation in cardiac systems and associated with high blood pressure and myocardial infarction, but not with BMI, blood lipids, blood pressure, and C-reactive protein (Charchar et al., 2012). For men in the haplogroup I population, the elevated risk of coronary artery disease appears to be related to the down-regulations of several genes such as ubiquitously transcribed-tetratricopeptide repeat, the Y-linked gene (UTY) and protein kinases, and Y-linked, pseudogene (PRKY) in macrophages (Bloomer et al., 2013). PRKY is a transcribed pseudogene without exon 6 and a part of exon 5 (that encodes the functional domains of the kinase). However, the biological function of PRKY is largely unknown. Its functional homolog is located on the X chromosome and is involved in the maturation of macrophage and development of kidney (Li et al., 2002; Glesne and Huberman, 2006).
Dilated cardiomyopathy (DCM) and hypertrophic cardiomyopathy (HCM) are two major cardiomyopathies. Khan et al. compared circRNA expression profiles in DCM, HCM, and normal cardiac tissues and found that RNA-binding motif protein 20 (RBM20), an essential protein for normal splicing of many cardiac genes, is also crucial to generate of a subset of circRNAs from the I-band of the titin gene. In RBM20-null mice, these titin circRNAs are down-regulated in DCM but not in HCM (Khan et al., 2016). These cardiac circRNAs are mostly (~90%) produced from constitutive exons and less (~10%) from alternatively spliced exons and are generated at the expense of their linear counterpart (Aufiero et al., 2018).
Arrhythmogenic right ventricular cardiomyopathy (ARVC) is a family-heritable disorder. It is characterized by progressive degeneration and destabilization of right ventricular myocardium, increased incidence of arrhythmias and an increased risk of sudden death at a young age. To identified gene associated with the disease, a number of genes located in the critical region of the microsatellite markers were screened, including laminin receptor 1 pseudogene 6 LAMR1P6, which is a processed transposable element within the ARVC6 critical interval (Asano et al., 2004) and a functional lamr1 retroposon gene is a cause of ARVC observed in the mouse via regulating mRNA stability of a homologous gene (Yonemura et al., 1998).
Marfan syndrome (MFS) is an autosomal dominant connective tissue disorder with an estimated incidence of 1/5,000 peoples. It affects the cardiovascular system as well as lung, skin, and dura. Since fibrillin-1 (FBN1) mRNA is indicator for MFS severity, identification of trans-acting regulators that controls FBN1 expression is important to elucidate the mechanism. Pseudogene, SNX7-ps1, is found to be associated with expression of a neighboring gene SLN (encoding sarcolipin) that plays role in skin fibroblasts. Since SNX7-ps1 expression is positively correlated with the SLN gene, it is likely that the two genes are controlled by regulatory elements for their expression (Benarroch et al., 2018).
CircRNAs have been shown to involve in a number of CVD conditions ranging from atherosclerosis to cardiac hypertrophy (Table 1) and they function as miRNA and protein sponges or transcriptional regulators to exert biological functions (Figure 3). For instance, cicrFoxO3 derived from the FoxO3 gene is highly expressed in the elderly and is positively corelated to senescence-associated β-galactosidase, SA-β-gal. It is proposed that cicrFoxO3 may be a protein sponge of inhibitor of differentiation-1, focal adhesion kinase, E2F transcription factor I, and hypoxia inducible factor 1, resulting in reduced availability of these proteins, thus reduced translocation of anti- senescence proteins into the nuclei and cellular senescence (Du et al., 2017). RBPs (RNA-binding proteins) have been found to be expressed and regulated in the heart. After treatment with doxorubicin, expression of circRNAs from Ttn (Titin), Fhod3 (Formin homology 2 domain containing 3), and Strn3 (Striatin, calmodulin-binding protein 3) and Qki5 was down-regulated, leading to reduced cell viability and cardiac senescence (Gupta et al., 2018).
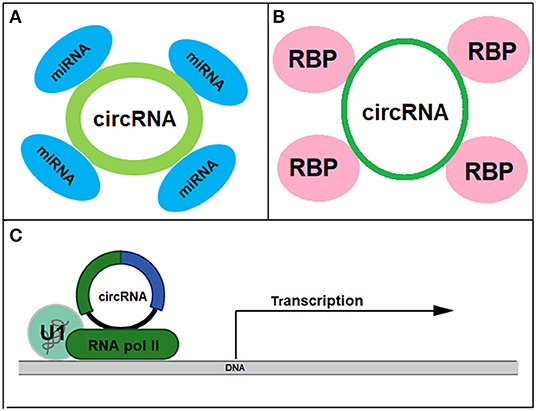
Figure 3. Biological functions of circRNAs as miRNA sponge (A), protein sponge (B), and transcriptional regulator (C).
Conclusion
With the continuous development of sequencing technology and bioinformatics, more and more pseudogenes are going to be discovered and studied. Current investigations have shown that pseudogenes can regulate gene expression and play important roles in the occurrence and development of CVDs. They may be used as new diagnostic markers or therapeutic targets to enhance our efforts to combat the diseases. However, compared to other molecular studies in human diseases, research on pseudogenes in CVDs is still in its infancy. More studies are needed to further our understanding of molecular mechanisms related to the role of pseudogenes in cardiovascular and other diseases.
Author Contributions
YQ, XW, and SA designed the study. YQ, XW, WL, DC, and HM collected the data and performed analysis. WL, DC, HM, and SA drafted the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by Nature Science Foundation of China (81970312) and Henan (182300410304).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abedini, F., Ebrahimi, M., and Hosseinkhani, H. (2018). Technology of RNA interference in advanced medicine. MicroRNA 7, 74–84. doi: 10.2174/2211536607666180129153307
Asano, Y., Takashima, S., Asakura, M., Shintani, Y., Liao, Y., Minamino, T., et al. (2004). Lamr1 functional retroposon causes right ventricular dysplasia in mice. Nat. Genet. 36, 123–130. doi: 10.1038/ng1294
Aufiero, S., van den Hoogenhof, M. M. G., Reckman, Y. J., Beqqali, A., van der Made, I., Kluin, J., et al. (2018). Cardiac circRNAs arise mainly from constitutive exons rather than alternatively spliced exons. RNA 24, 815–827. doi: 10.1261/rna.064394.117
Balakirev, E. S., and Ayala, F. J. (2003). Pseudogenes: are they “junk” or functional DNA? Annu. Rev. Genet. 37, 123–151. doi: 10.1146/annurev.genet.37.040103.103949
Benarroch, L., Aubart, M., Gross, M. S., Jacob, M. P., Arnaud, P., Hanna, N., et al. (2018). Marfan syndrome variability: investigation of the roles of sarcolipin and calcium as potential transregulator of FBN1 expression. Genes 9:421. doi: 10.3390/genes9090421
Berciano, M. T., Puente-Bedia, A., Medina-Samame, A., Rodriguez-Rey, J. C., Caldero, J., Lafarga, M., et al. (2020). Nusinersen ameliorates motor function and prevents motoneuron Cajal body disassembly and abnormal poly(A) RNA distribution in a SMA mouse model. Sci. Rep. 10:10738. doi: 10.1038/s41598-020-67569-3
Bloomer, L. D., Nelson, C. P., Eales, J., Denniff, M., Christofidou, P., Debiec, R., et al. (2013). Male-specific region of the Y chromosome and cardiovascular risk: phylogenetic analysis and gene expression studies. Arterioscler. Thromb. Vasc. Biol. 33, 1722–1727. doi: 10.1161/ATVBAHA.113.301608
Charchar, F. J., Bloomer, L. D., Barnes, T. A., Cowley, M. J., Nelson, C. P., Wang, Y., et al. (2012). Inheritance of coronary artery disease in men: an analysis of the role of the Y chromosome. Lancet 379, 915–922. doi: 10.1016/S0140-6736(11)61453-0
Cheetham, S. W., Faulkner, G. J., and Dinger, M. E. (2020). Overcoming challenges and dogmas to understand the functions of pseudogenes. Nat. Rev. Genet. 21, 191–201. doi: 10.1038/s41576-019-0196-1
Chen, L., Qu, H., Guo, M., Zhang, Y., Cui, Y., Yang, Q., et al. (2020). ANRIL and atherosclerosis. J. Clin. Pharm. Ther. 45, 240–248. doi: 10.1111/jcpt.13060
Cheng, M., Yang, Y., Xin, H., Li, M., Zong, T., He, X., et al. (2020). Non-coding RNAs in aortic dissection: from biomarkers to therapeutic targets. J. Cell. Mol. Med. 24, 11622–11637. doi: 10.1111/jcmm.15802
Chiriboga, C. A. (2017). Nusinersen for the treatment of spinal muscular atrophy. Expert Rev. Neurother. 17, 955–962. doi: 10.1080/14737175.2017.1364159
Cooke, S. L., Shlien, A., Marshall, J., Pipinikas, C. P., Martincorena, I., Tubio, J. M., et al. (2014). Processed pseudogenes acquired somatically during cancer development. Nat. Commun. 5:3644. doi: 10.1038/ncomms4644
Dang, R. Y., Liu, F. L., and Li, Y. (2017). Circular RNA hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186/HIF-1alpha axis. Biochem. Biophys. Res. Commun. 490, 104–110. doi: 10.1016/j.bbrc.2017.05.164
Di Salvo, T. G., Guo, Y., Su, Y. R., Clark, T., Brittain, E., Absi, T., et al. (2015a). Right ventricular long noncoding RNA expression in human heart failure. Pulm. Circ. 5, 135–161. doi: 10.1086/679721
Di Salvo, T. G., Yang, K. C., Brittain, E., Absi, T., Maltais, S., and Hemnes, A. (2015b). Right ventricular myocardial biomarkers in human heart failure. J. Card. Fail. 21, 398–411. doi: 10.1016/j.cardfail.2015.02.005
Dong, R., Zhang, X. O., Zhang, Y., Ma, X. K., Chen, L. L., and Yang, L. (2016). CircRNA-derived pseudogenes. Cell Res. 26, 747–750. doi: 10.1038/cr.2016.42
Du, W. W., Yang, W., Chen, Y., Wu, Z. K., Foster, F. S., Yang, Z., et al. (2017). Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 38, 1402–1412. doi: 10.1093/eurheartj/ehw001
Duggirala, A., Delogu, F., Angelini, T. G., Smith, T., Caputo, M., Rajakaruna, C., et al. (2015). Non coding RNAs in aortic aneurysmal disease. Front. Genet. 6:125. doi: 10.3389/fgene.2015.00125
Emadi-Baygi, M., Sedighi, R., Nourbakhsh, N., and Nikpour, P. (2017). Pseudogenes in gastric cancer pathogenesis: a review article. Brief. Funct. Genomics 16, 348–360. doi: 10.1093/bfgp/elx004
Firth, A. L., Yao, W., Remillard, C. V., Ogawa, A., and Yuan, J. X. (2010). Upregulation of Oct-4 isoforms in pulmonary artery smooth muscle cells from patients with pulmonary arterial hypertension. Am. J. Physiol. Lung Cell. Mol. Physiol. 298, L548–L557. doi: 10.1152/ajplung.00314.2009
Garciandia, A., and Suarez, T. (2013). The NMRA/NMRAL1 homologue PadA modulates the expression of extracellular cAMP relay genes during aggregation in Dictyostelium discoideum. Dev. Biol. 381, 411–422. doi: 10.1016/j.ydbio.2013.06.013
Glesne, D., and Huberman, E. (2006). Smad6 is a protein kinase X phosphorylation substrate and is required for HL-60 cell differentiation. Oncogene 25, 4086–4098. doi: 10.1038/sj.onc.1209436
Goradel, N. H., Hour, F. G., Negahdari, B., Malekshahi, Z. V., Hashemzehi, M., Masoudifar, A., et al. (2018). Stem cell therapy: a new therapeutic option for cardiovascular diseases. J. Cell. Biochem. 119, 95–104. doi: 10.1002/jcb.26169
Grander, D., and Johnsson, P. (2016). Pseudogene-expressed RNAs: emerging roles in gene regulation and disease. Curr. Top. Microbiol. Immunol. 394, 111–126. doi: 10.1007/82_2015_442
Greco, S., Zaccagnini, G., Perfetti, A., Fuschi, P., Valaperta, R., Voellenkle, C., et al. (2016). Long noncoding RNA dysregulation in ischemic heart failure. J. Transl. Med. 14:183. doi: 10.1186/s12967-016-0926-5
Gupta, S. K., Garg, A., Bar, C., Chatterjee, S., Foinquinos, A., Milting, H., et al. (2018). Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ. Res. 122, 246–254. doi: 10.1161/CIRCRESAHA.117.311335
Hirotsune, S., Yoshida, N., Chen, A., Garrett, L., Sugiyama, F., Takahashi, S., et al. (2003). An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature 423, 91–96. doi: 10.1038/nature01535
Holdt, L. M., Stahringer, A., Sass, K., Pichler, G., Kulak, N. A., Wilfert, W., et al. (2016). Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 7:12429. doi: 10.1038/ncomms12429
Holdt, L. M., and Teupser, D. (2012). Recent studies of the human chromosome 9p21 locus, which is associated with atherosclerosis in human populations. Arterioscler. Thromb. Vasc. Biol. 32, 196–206. doi: 10.1161/ATVBAHA.111.232678
Jacq, C., Miller, J. R., and Brownlee, G. G. (1977). A pseudogene structure in 5S DNA of Xenopus laevis. Cell 12, 109–120. doi: 10.1016/0092-8674(77)90189-1
Ji, Z., Song, R., Regev, A., and Struhl, K. (2015). Many lncRNAs, 5'UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife 4:e08890. doi: 10.7554/eLife.08890
Jingsi, T., Mingyao, Y., and Ying, L. (2015). Functional roles of pseudogenes in cancers. Yi Chuan 37, 8–16. doi: 10.16288/j.yczz.2015.01.002
Johnson, B. D., Kip, K. E., Marroquin, O. C., Ridker, P. M., Kelsey, S. F., Shaw, L. J., et al. (2004). Serum amyloid A as a predictor of coronary artery disease and cardiovascular outcome in women: the National Heart, Lung, and Blood Institute-Sponsored Women's Ischemia Syndrome Evaluation (WISE). Circulation 109, 726–732. doi: 10.1161/01.CIR.0000115516.54550.B1
Kalyana-Sundaram, S., Kumar-Sinha, C., Shankar, S., Robinson, D. R., Wu, Y. M., Cao, X., et al. (2012). Expressed pseudogenes in the transcriptional landscape of human cancers. Cell 149, 1622–1634. doi: 10.1016/j.cell.2012.04.041
Khan, M. A., Reckman, Y. J., Aufiero, S., van den Hoogenhof, M. M., van der Made, I., Beqqali, A., et al. (2016). RBM20 regulates circular RNA production from the titin gene. Circ. Res. 119, 996–1003. doi: 10.1161/CIRCRESAHA.116.309568
Khelifi, A., Duret, L., and Mouchiroud, D. (2005). HOPPSIGEN: a database of human and mouse processed pseudogenes. Nucleic Acids Res. 33, D59–D66. doi: 10.1093/nar/gki084
Kim, K., Theusch, E., Kuang, Y. L., Dose, A., Mitchel, K., Cubitt, C., et al. (2018). ZNF542P is a pseudogene associated with LDL response to simvastatin treatment. Sci. Rep. 8:12443. doi: 10.1038/s41598-018-30859-y
Koseler, A., Ma, F., Kilic, I. D., Morselli, M., Kilic, O., and Pellegrini, M. (2020). Genome-wide DNA methylation profiling of blood from monozygotic twins discordant for myocardial infarction. In Vivo 34, 361–367. doi: 10.21873/invivo.11782
Li, H., Xu, J. D., Fang, X. H., Zhu, J. N., Yang, J., Pan, R., et al. (2020). Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc. Res. 116, 1323–1334. doi: 10.1093/cvr/cvz215
Li, S., Yang, B., Du, Y., Lin, Y., Liu, J., Huang, S., et al. (2018). Targeting PPARalpha for the treatment and understanding of cardiovascular diseases. Cell. Physiol. Biochem. 51, 2760–2775. doi: 10.1159/000495969
Li, X., Li, H. P., Amsler, K., Hyink, D., Wilson, P. D., and Burrow, C. R. (2002). PRKX, a phylogenetically and functionally distinct cAMP-dependent protein kinase, activates renal epithelial cell migration and morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 99, 9260–9265. doi: 10.1073/pnas.132051799
Li, Y., Huo, C., Lin, X., and Xu, J. (2018a). Computational identification of cross-talking ceRNAs. Adv. Exp. Med. Biol. 1094, 97–108. doi: 10.1007/978-981-13-0719-5_10
Li, Y., Liang, Y., Zhu, Y., Zhang, Y., and Bei, Y. (2018b). Noncoding RNAs in cardiac hypertrophy. J. Cardiovasc. Transl. Res. 11, 439–449. doi: 10.1007/s12265-018-9797-x
Liu, G., Li, H., and Cai, L. (2010). Processed pseudogenes are located preferentially in regions of low recombination rates in the human genome. J. Evol. Biol. 23, 1107–1115. doi: 10.1111/j.1420-9101.2010.01974.x
Maranda, V., Sunstrum, F. G., and Drouin, G. (2019). Both male and female gamete generating cells produce processed pseudogenes in the human genome. Gene 684, 70–75. doi: 10.1016/j.gene.2018.10.061
McDonell, L., and Drouin, G. (2012). The abundance of processed pseudogenes derived from glycolytic genes is correlated with their expression level. Genome 55, 147–151. doi: 10.1139/G2012-002
Miao, R., Wang, Y., Wan, J., Leng, D., Gong, J., Li, J., et al. (2017). Microarray expression profile of circular RNAs in chronic thromboembolic pulmonary hypertension. Medicine 96:e7354. doi: 10.1097/MD.0000000000007354
Milligan, M. J., Harvey, E., Yu, A., Morgan, A. L., Smith, D. L., Zhang, E., et al. (2016). Global intersection of long non-coding RNAs with processed and unprocessed pseudogenes in the human genome. Front. Genet. 7:26. doi: 10.3389/fgene.2016.00026
Molineris, I., Sales, G., Bianchi, F., Di Cunto, F., and Caselle, M. (2010). A new approach for the identification of processed pseudogenes. J. Comput. Biol. 17, 755–765. doi: 10.1089/cmb.2009.0027
Montasser, M. E., O'Hare, E. A., Wang, X., Howard, A. D., McFarland, R., Perry, J. A., et al. (2018). An APOO pseudogene on chromosome 5q is associated with low-density lipoprotein cholesterol levels. Circulation 138, 1343–1355. doi: 10.1161/CIRCULATIONAHA.118.034016
Paton, D. M. (2017). Nusinersen: antisense oligonucleotide to increase SMN protein production in spinal muscular atrophy. Drugs Today 53, 327–337. doi: 10.1358/dot.2017.53.6.2652413
Pink, R. C., Wicks, K., Caley, D. P., Punch, E. K., Jacobs, L., and Carter, D. R. (2011). Pseudogenes: pseudo-functional or key regulators in health and disease? RNA 17, 792–798. doi: 10.1261/rna.2658311
Poliseno, L., Salmena, L., Zhang, J., Carver, B., Haveman, W. J., and Pandolfi, P. P. (2010). A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038. doi: 10.1038/nature09144
Porter, K. A., Duffy, E. B., Nyland, P., Atianand, M. K., Sharifi, H., and Harton, J. A. (2014). The CLRX.1/NOD24 (NLRP2P) pseudogene codes a functional negative regulator of NF-kappaB, pyrin-only protein 4. Genes Immun. 15, 392–403. doi: 10.1038/gene.2014.30
Salgado-Somoza, A., Zhang, L., Vausort, M., and Devaux, Y. (2017). The circular RNA MICRA for risk stratification after myocardial infarction. Int. J. Cardiol. Heart Vasc. 17, 33–36. doi: 10.1016/j.ijcha.2017.11.001
Shan, K., Liu, C., Liu, B. H., Chen, X., Dong, R., Liu, X., et al. (2017). Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation 136, 1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004
Shoji, S., Muto, Y., Ikeda, M., He, F., Tsuda, K., Ohsawa, N., et al. (2014). The zinc-binding region (ZBR) fragment of Emi2 can inhibit APC/C by targeting its association with the coactivator Cdc20 and UBE2C-mediated ubiquitylation. FEBS Open Biol. 4, 689–703. doi: 10.1016/j.fob.2014.06.010
Song, C. L., Wang, J. P., Xue, X., Liu, N., Zhang, X. H., Zhao, Z., et al. (2017). Effect of circular ANRIL on the inflammatory response of vascular endothelial cells in a rat model of coronary atherosclerosis. Cell. Physiol. Biochem. 42, 1202–1212. doi: 10.1159/000478918
Tam, O. H., Aravin, A. A., Stein, P., Girard, A., Murchison, E. P., Cheloufi, S., et al. (2008). Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453, 534–538. doi: 10.1038/nature06904
Tan, X., Zhang, X., Pan, L., Tian, X., and Dong, P. (2017). Identification of key pathways and genes in advanced coronary atherosclerosis using bioinformatics analysis. Biomed Res. Int. 2017:4323496. doi: 10.1155/2017/4323496
Thompson, J. C., Wilson, P. G., Shridas, P., Ji, A., de Beer, M., de Beer, F. C., et al. (2018). Serum amyloid A3 is pro-atherogenic. Atherosclerosis 268, 32–35. doi: 10.1016/j.atherosclerosis.2017.11.011
Thomson, D. W., and Dinger, M. E. (2016). Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 17, 272–283. doi: 10.1038/nrg.2016.20
Tutar, L., Ozgur, A., and Tutar, Y. (2018). Involvement of miRNAs and pseudogenes in cancer. Methods Mol. Biol. 1699, 45–66. doi: 10.1007/978-1-4939-7435-1_3
Uchida, S., and Dimmeler, S. (2015). Long noncoding RNAs in cardiovascular diseases. Circ. Res. 116, 737–750. doi: 10.1161/CIRCRESAHA.116.302521
van Heesch, S., Witte, F., Schneider-Lunitz, V., Schulz, J. F., Adami, E., Faber, A. B., et al. (2019). The translational landscape of the human heart. Cell 178, 242–260 e229. doi: 10.1016/j.cell.2019.05.010
Wang, G., Zheng, X., Zheng, Y., Cao, R., Zhang, M., Sun, Y., et al. (2019). Construction and analysis of the lncRNAmiRNAmRNA network based on competitive endogenous RNA reveals functional genes in heart failure. Mol. Med. Rep. 19, 994–1003. doi: 10.3892/mmr.2018.9734
Wang, K., Gan, T. Y., Li, N., Liu, C. Y., Zhou, L. Y., Gao, J. N., et al. (2017). Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 24, 1111–1120. doi: 10.1038/cdd.2017.61
Wang, K., Long, B., Liu, F., Wang, J. X., Liu, C. Y., Zhao, B., et al. (2016). A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 37, 2602–2611. doi: 10.1093/eurheartj/ehv713
Wang, X., Cao, X., Dong, D., Shen, X., Cheng, J., Jiang, R., et al. (2019). Circular RNA TTN acts as a miR-432 sponge to facilitate proliferation and differentiation of myoblasts via the IGF2/PI3K/AKT signaling pathway. Mol. Ther. Nucleic Acids 18, 966–980. doi: 10.1016/j.omtn.2019.10.019
Weiner, A. M., Deininger, P. L., and Efstratiadis, A. (1986). Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu. Rev. Biochem. 55, 631–661. doi: 10.1146/annurev.bi.55.070186.003215
Wilson, P. G., Thompson, J. C., Webb, N. R., de Beer, F. C., King, V. L., and Tannock, L. R. (2008). Serum amyloid A, but not C-reactive protein, stimulates vascular proteoglycan synthesis in a pro-atherogenic manner. Am. J. Pathol. 173, 1902–1910. doi: 10.2353/ajpath.2008.080201
Wu, M. F., Xu, K. Z., Guo, Y. G., Yu, J., Wu, Y., and Lin, L. M. (2019). Lipoprotein(a) and atherosclerotic cardiovascular disease: current understanding and future perspectives. Cardiovasc. Drugs Ther. 33, 739–748. doi: 10.1007/s10557-019-06906-9
Xie, J., Chen, S., Xu, W., Zhao, Y., and Zhang, D. (2019). Origination and function of plant pseudogenes. Plant Signal. Behav. 14:1625698. doi: 10.1080/15592324.2019.1625698
Xu, J., and Zhang, J. (2016). Are human translated pseudogenes functional? Mol. Biol. Evol. 33, 755–760. doi: 10.1093/molbev/msv268
Yonemura, S., Hirao, M., Doi, Y., Takahashi, N., Kondo, T., Tsukita, S., et al. (1998). Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J. Cell Biol. 140, 885–895. doi: 10.1083/jcb.140.4.885
Zeng, Y., Du, W. W., Wu, Y., Yang, Z., Awan, F. M., Li, X., et al. (2017). A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics 7, 3842–3855. doi: 10.7150/thno.19764
Zhang, L., Wu, Y. J., and Zhang, S. L. (2019). Circulating lncRNA MHRT predicts survival of patients with chronic heart failure. J. Geriatr. Cardiol. 16, 818–821. doi: 10.11909/j.issn.1671-5411.2019.11.006
Zhang, S., Wang, W., Wu, X., and Zhou, X. (2020). Regulatory roles of circular RNAs in coronary artery disease. Mol. Ther. Nucleic Acids 21, 172–179. doi: 10.1016/j.omtn.2020.05.024
Zhang, Y., Sun, L., Xuan, L., Pan, Z., Li, K., Liu, S., et al. (2016). Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci. Rep. 6:22384. doi: 10.1038/srep22384
Zhang, Z., Carriero, N., Zheng, D., Karro, J., Harrison, P. M., and Gerstein, M. (2006). PseudoPipe: an automated pseudogene identification pipeline. Bioinformatics 22, 1437–1439. doi: 10.1093/bioinformatics/btl116
Zhang, Z. D., Frankish, A., Hunt, T., Harrow, J., and Gerstein, M. (2010). Identification and analysis of unitary pseudogenes: historic and contemporary gene losses in humans and other primates. Genome Biol. 11:R26. doi: 10.1186/gb-2010-11-3-r26
Zhao, Z., Li, X., Gao, C., Jian, D., Hao, P., Rao, L., et al. (2017). Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci. Rep. 7:39918. doi: 10.1038/srep39918
Zheng, C., Niu, H., Li, M., Zhang, H., Yang, Z., Tian, L., et al. (2015). Cyclic RNA hsacirc000595 regulates apoptosis of aortic smooth muscle cells. Mol. Med. Rep. 12, 6656–6662. doi: 10.3892/mmr.2015.4264
Zheng, L. L., Zhou, K. R., Liu, S., Zhang, D. Y., Wang, Z. L., Chen, Z. R., et al. (2018). dreamBase: DNA modification, RNA regulation and protein binding of expressed pseudogenes in human health and disease. Nucleic Acids Res. 46, D85–D91. doi: 10.1093/nar/gkx972
Zhou, B., and Yu, J. W. (2017). A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem. Biophys. Res. Commun 487, 769–775. doi: 10.1016/j.bbrc.2017.04.044
Keywords: cardiovascular disease, pseudogenes, non-coding gene, mutation, protein-coding, gene regulation
Citation: Qi Y, Wang X, Li W, Chen D, Meng H and An S (2021) Pseudogenes in Cardiovascular Disease. Front. Mol. Biosci. 7:622540. doi: 10.3389/fmolb.2020.622540
Received: 28 October 2020; Accepted: 07 December 2020;
Published: 10 February 2021.
Edited by:
Matteo Becatti, University of Florence, ItalyReviewed by:
Nicholas Delihas, Stony Brook Medicine, United StatesSteve Wilton, Murdoch University, Australia
Francesco Esposito, Consiglio Nazionale Delle Ricerche (CNR), Italy
Copyright © 2021 Qi, Wang, Li, Chen, Meng and An. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Songtao An, YW5zb25ndGFvMjAxMEAxNjMuY29t
 Yanyan Qi1
Yanyan Qi1 Songtao An
Songtao An