- 1Department of Zoonoses, Faculty of Veterinary Medicine, Sohag University, Sohag, Egypt
- 2Laboratory of Kafr El Sheikh Fever Hospital, Kafr El Sheikh Fever Hospital, Kafr El-Sheikh, Egypt
- 3Department of Pathology, Faculty of Veterinary Medicine, Kafrelsheikh University, Kafr El-Sheikh, Egypt
- 4Department of Hygiene and Zoonoses, Faculty of Veterinary Medicine, Mansoura University, Mansoura, Egypt
- 5Department of Nursing, College of Applied Medical Sciences, Jouf University, Sakaka, Saudi Arabia
- 6Department of Critical Care and Emergency Nursing, Faculty of Nursing, Mansoura University, Mansoura, Egypt
- 7Laboratory of Wildlife and Forensic Pathology/Biomedical Science Examination and Research Center, Department of Veterinary Medicine, Faculty of Veterinary Medicine, Okayama University of Science, Okayama, Japan
The recent emergence of COVID‐19 represents one of the biggest challenges facing the world today. Despite the recent attempts to understand the epidemiological pattern and pathogenesis of the disease, detailed data about the physiology and pathology of the disease is still out of reach. Moreover, the lack of a widespread vaccine prompts an urgent call for developing a proper intervention strategy against the virus. Importantly, identification of novel molecules that target replication of the virus represents one of the promising strategies for the control this pandemic crisis. Among others, honey bee products contain numerous bioactive compounds such as propolis and several phenolic compounds that possess a wide range of therapeutic properties for combating various pathological disorders and infectious agents. The intention of the present review is to highlight the stages of SARS-CoV-2 lifecycle, the molecular mechanisms explaining the health benefits of honey bee products on COVID‐19 physiology and pathology and the possible limitations. Further future research is suggested to explore more about bee natural bioactive compounds as potential candidates against SARS-CoV-2.
Introduction
Corona viruses (CoV) are a group of positive-sense single-stranded RNA viruses ranging between 26 and 32 kb in size (Graham et al., 2013; Song et al., 2019). This group of viruses belongs to genus β-Coronavirus, family Coronaviridae, and order Nidovirales (Paules et al., 2020). The epidemiological profile of Corona viruses’ infection involves a wide range of hosts that include humans, birds, and other mammals (Monchatre-Leroy et al., 2017; Cui et al., 2019). The clinical impact of these viruses ranges from asymptomatic cases to severe symptoms, affecting respiratory, digestive, and genital organs. In accordance with COVID‐19, the disease has been emerged and detected for the first time in patients with respiratory illness of unknown etiology in the urban center town, Hubei Province, Central China (Graham et al., 2013; Song et al., 2019; Paraskevis et al., 2020). The viral agent was defined as coronavirus illness 2019 (COVID-19) or SARS-CoV-2 (Li et al., 2020b; Sun et al., 2020; Xu et al., 2020). As of December 15, 2020, more than 73,212,302 confirmed cases of COVID-19 and 1,628,442 deaths have been reported worldwide (Worldometer, 2020). Interestingly, honeybee products have been used in treatment of many diseases including tumor and immune-related diseases (Yusuf et al., 2007; Wieckiewicz et al., 2013). In this regard, honey, propolis, Bee pollen and Bee venom created by bees possess many biological activities like antibiotic, antifungal, antioxidant, antiviral, inhibitor, anti-cancer, and immunomodulatory, hepatoprotective effects (Banskota et al., 2001; Tolba et al., 2013; Pasupuleti et al., 2017; Hashem, 2020; Shaldam et al., 2020). The following sections include an overview about structure, pathogenesis and mechanistic activities of SARS-CoV-2 and the potential application of Bee’s products in treatment of the disease.
Structure of SARS-CoV-2
As mentioned above, SARS-CoV-2 or CoV is an RNA virus belonging to the genus β-Coronavirus (Paules et al., 2020). This virus is a positive-sense RNA virus with a size of around 30 kb and about 74–99% identity with the coronavirus from the placental mammal (Manis javanica) and horseshoe bat (Rhinolophus sinicus) (Bat-CoVRaTG13), respectively (Zhu et al., 2020b). The typical CoV contains a minimum of six open reading frames (ORFs) (Consortium, 2004). The primary ORF (ORF1a/b) is a simple fraction that concerns the entire order length and encodes sixteen non-structural proteins (nsp1-16) (Consortium, 2004; Narayanan et al., 2015). Furthermore, the virus includes four main structural proteins; spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins which are shown in Figure 1 (Consortium, 2004; Peiris et al., 2004; Li et al., 2020a; Nadeem et al., 2020; Wrapp et al., 2020). It should be stressed that most of the non-structural proteins are known to play a significant role in virus replication while structural proteins are vital for particle assembly and for inflicting CoV infection (Consortium, 2004; McBride et al., 2014; Astuti and Ysrafil, 2020). Moreover, specific structural and accent proteins, like HE macromolecules are also encoded by CoV (Chen et al., 2020a; Naqvi et al., 2020).
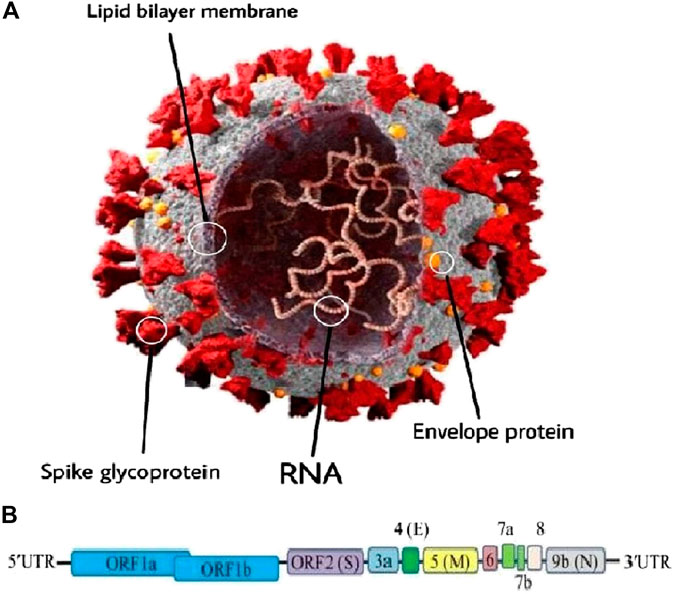
FIGURE 1. (A) Schematic representation of the genome organization and functional domains of S protein for COVID-19. The single-stranded RNA genomes of COVID-19 encode two large genes, the ORF1a and ORF1b genes, which encode 16 non-structural proteins (nsp1–nsp16). The structural genes encode the structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N), which are common features to all coronaviruses. (B) The SARS-CoV-2 genome is arranged in the order of 5′-replicase (ORF1a/b)–structural proteins [spike (S)–envelope (E)–membrane (M)–nucleocapsid (N)]−3′ (reproduced from Consortium, 2004; Peiris et al., 2004; Li et al., 2020a; Nadeem et al., 2020; Wrapp et al., 2020).
Stages of SARS-CoV-2 Lifecycle and the Potential Inhibition Targets
It is noteworthy to state that SARS-CoV-2 targets cells through the infectious agent structural spike (S) supermolecule that binds to the angiotensin-converting enzyme two (ACE2) receptor (Wang et al., 2007; Magrone et al., 2020). Following its binding to the receptor, the virus uses host cell receptors and endosomes to enter cells while this action is facilitated by transmembrane protease/serine subfamily member 2 via the S supermolecule (Hoffmann et al., 2020b). Once within the cell, the infectious agent polyproteins area unit synthesized targets the assembly of replicase-transcriptase complex (Sawicki et al., 2005). The virus then synthesizes RNA via its RNA-dependent RNA enzyme and the structural proteins area unit synthesized, resulting in completion of assembly which is followed by the release of infectious agent particles (Fung and Liu, 2014; Fehr and Perlman, 2015; Chen et al., 2020a). These steps of the SARS-CoV-2 lifecycle represent potential drug targets (Figure 2) and there are other drug targets which trigger infectious agent entry and immune regulation pathways (Savarino et al., 2003; Al-Bari, 2017).
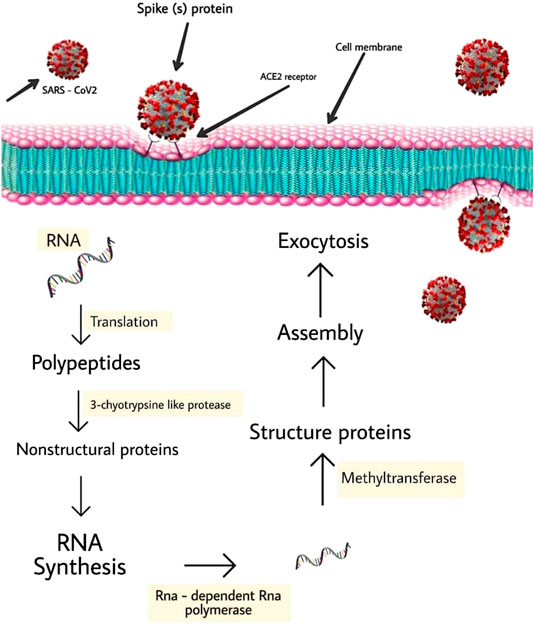
FIGURE 2. Schematic represents virus-induced host immune system response and viral processing within target cells.
The Role of Natural Therapy Strategy in SARS-CoV-2
Revising the available literature, several previous studies explored the promising role of some natural compounds and phytochemical extracts, e.g. Lycoris radiata (red spider lily), Lindera aggregata, Pyrrosia lingua (a fern), and Artemisia annua (sweet wormwood), in treatment of outbreaks of SARS (Wu et al., 2004; Hoever et al., 2005; Li et al., 2005; Kim et al., 2010). Taken into account, these previously mentioned extracts showed various degrees of activity against SARS-CoV ranged from moderate to potent and their antiviral actions were dose-dependent. Among others, Lycoris radiata (red spider lily) expressed the most potent antiviral activity (Li et al., 2005). Glycyrrhizin, which is an active compound contained in licorice roots, is another example for the herbal extracts that displayed potent antiviral activity against SARS-CoV by inhibiting the replication of the virus when tested on 10 different clinical strains of SARS-CoV (Cinatl et al., 2003; Hoever et al., 2005). Furthermore, Lycorine, a toxic crystalline alkaloid found in various Amaryllidaceae species and Baicalin (a constituent of the Baikal skullcap plant), has also shown potent antiviral effects against SARS-CoV (Fielding et al., 2020). Interestingly, myricetin, scutellarein, and phenolic compounds from dyer’s woad and Japanese nutmeg-yew have shown to be potent antagonists of SARS-CoV enzymes, including nsP13 helicase and 3CL protease (Lin et al., 2005; Ryu et al., 2010; Yu et al., 2012). In addition, some natural phytomedicines, e.g., the aqueous extract of fish mint, mediated several antiviral mechanisms in SARS-CoV (Lau et al., 2008). However, it should be borne in mind that some discrepancy in results has been reported as these in vitro data may not correlate with in vivo findings that renders the use of some of these natural compounds as an effective antiviral agent. The following subsections will highlight the potential inhibition targets in the different stages of SARS-CoV-2 lifecycle and the potential application of bee products in treatment of the disease.
Potential Repressive Properties Against ACE-2 Receptors
The intra- or inter-species transmission of β-coronaviruses (CoVs) requires an interaction between the infective agent and the host cell receptors that results in the invasion of the virus into host cells (Li, 2016). Some recent studies reveal that human, pig, and civet cell lines allowed SARS-CoV-2 infection and replication, indicating that the virus uses ACE2 receptor for infection (Hoffmann et al., 2020a; Zhu et al., 2020a; Zhu et al., 2020b; Letko et al., 2020; Zhou et al., 2020). ACE2 is extremely expressed within the respiratory organs that make the lung tissue highly vulnerable (Hamming et al., 2004). In addition, the ACE2 receptor is expressed within the epithelial tissue cells of gut, kidney, and heart cells (Zhang and Liu, 2020). It is therefore not surprising to state that ACE2 blockers are another choice to control the infection (Ton et al., 2020). Similarly, some molecules such as GSK1838705A (a small-molecule kinase inhibitor), KT203 (inhibitor of α/β-hydrolase domain), KT185 (brain-penetrant and selective ABHD6 inhibitor), and BMS195614 (selective RARα antagonist) showed strong binding affinities with receptor binding sites (RBD) of the infective agent S-protein. These molecules facilitate the management of fast infection by participating the virus at entry points (Choudhary et al., 2020). Therefore, ACE II enzyme inhibition seems an important target for treatment of these cases of infection caused by SARS-CoV-2.
Propolis, or bee glue, is defined as a natural resinous mixture produced by bees through its collection from nature (Sforcin, 2016; Drescher et al., 2019). The honey bee produces this mixture though mixing saliva and beeswax together with the collected exudate from several botanical sources such as tree and plants buds (Zabaiou et al., 2017). Interestingly, this mixture possesses a wide range of activity against various infections agents in addition to its role in wound healing (Pasupuleti et al., 2017; Oryan et al., 2018). In accordance with its texture, crude propolis could be extremely viscous and slightly soluble in water. Propolis has been an important element of apitherapy for hundreds of years. Recently, it has been used as an additive in the name of the ancient practice of medicine (Pobiega et al., 2018; Anjum et al., 2019). The bulk of the active ingredients of propolis comprise the family of polyphenols. In this concern, phenolic acids, flavonoids (flavanones, flavones, flavonols etc.), stilbenes, and tannins are considered the most active polyphenols of propolis (Graikou et al., 2015; Anjum et al., 2019). In addition, several previous in vitro and in vivo studies showed that flavonoids have high potential for inhibition of Angiotensin-Converting enzyme (ACE) (Hussain et al., 2018; Wang et al., 2018; Silveira et al., 2019). A recent study measured and checked the binding constants of 10 flavonoids, including caffeic acid, caffeic acid phenethyl ester, galangin, chrysin, rutin, hesperetin, myricetin, pinocembrin, quercetin, and luteolin, using the AutoDock 4.2 molecular arrival program and compared to a reference substance of MLN-4760 which is known as ACE2 inhibitor (Güler et al., 2020). The results showed that rutin has the simplest inhibition potential among the studied molecules. Clearly, the high potential of flavonoids extracts to bind to ACE II receptors indicates that this natural bee product might exhibit marked activity for Covid- 19 treatment (Güler et al., 2020; Shaldam et al., 2020). However, these findings must be supported by experimental studies.
Potential Repressing Properties Against Proteinase Enzyme
The inhibition of infectious agent proteinase is a crucial target in drug development. The 3C-like proteinase (3CLpro) might be a cysteine proteinase that hydrolyzes the polyproteins pp1a and pp1ab to supply purposeful proteins throughout the replication of the virus. As result of its extremely preserved sequence and essential properties, 3CLpro has been validated as a possible target for the treatment of respiratory illnesses such as MERS, and COVID-19 (Kumar et al., 2017; Dong et al., 2020; Jo et al., 2020). Recently, a wide range of natural and artificial inhibitors that focus on completely different sites and regions of 3CLpro have been developed (Muramatsu et al., 2016; He et al., 2020; Theerawatanasirikul et al., 2020; Ye et al., 2020). Because the extremely preserved process sites of 3CLpro area unit shared by CoVs (Kumar et al., 2017; Dong et al., 2020), tremendous efforts have been created to review this target in order to meet the urgent need for the development of anti-SARS-CoV-2 therapy (Ton et al., 2020). Their targets include antecedently approved medicine, run candidates, and bioactive agents that were known as potential treatments for respiratory illness and MERS (Ton et al., 2020). Most studies targeted the small-molecule compounds, through virtual screening, that supported the crystal structure of 3CLpro (Chen et al., 2020b). It should be stressed that the natural bee products such as flavonoids (herbacetin) and Chalcones are considered candidate compounds which are related to the protein activity or infectious agent load in vitro. In addition, Herbacetin (PubChem CID: 5280544) exerted outstanding repressing effects, with the IC50 values of 33.2 μM. An induced-fit docking tying up study with SARS-CoV 3CLpro (PDB ID: 4WY3) showed that herbacetin shaped four H-bonds at the S2 website besides the 8-hydroxyl cluster that was essential for the formation of H-bonds with Glu166 and Gln 189 (Jo et al., 2020).
Interestingly, several previous studies documented that galangin, kaempferol, chrysin, and pinocembrin were detected in Croatian Cystus incanus L. bee pollen (Saric et al., 2009). The presence of herbacetin, myricetin, tricetin, luteolin, and 3-O-methylquercetin was also documented (Campos et al., 2003). A series of alkylated chalcones isolated from Angelica keiskei, were evaluated for their repressing activities against SARS-CoV 3CLpro. Among these chalcones, compound, with a perhydroxyl cluster, showed the foremost potent repressing impact (IC50 = 11.4 ± 1.4 μM). Clearly, these previously mentioned results reveal that the perhydroxyl cluster can be crucial for the binding to SARS-CoV 3CLpro. Tying up studies of the compound with 3CLpro (PDB ID: 2ZU) showed that the carbonyl and hydroxyl group teams shaped H-bonds with His163 and Ser144, respectively. The perhydroxyl cluster shaped a powerful H-bond with the very important residue Cys145 (Park et al., 2016). It is noteworthy to state that honey contains various compounds besides water, sugars, free amino acids, proteins, enzymes, essential minerals, vitamins, and numerous phytochemicals (Escuredo et al., 2013). In addition, polyphenols are heterogeneous categories of chemical compounds which are divided into flavonoids (flavonols, flavones, flavanones, flavanols, chalcones, anthocyanidin, and isoflavones) and non-flavonoids (phenolic acids). The phenolic resin composition in honey mainly depends on its floral origin that can be used as a tool for classification and authentication, particularly within unifloral varieties (Kennedy and Wightman, 2011; Chan et al., 2013; Keckes et al., 2013; Campone et al., 2014). Importantly, two compounds of chalcones (2′, 4′- dihydroxychalcone and 2′,4′- dihydroxy 3′-methoxychalcone) were also found in propolis samples from the four-card monte phytogeographical region (Solórzano et al., 2012).
Potential Restrictive Properties Against Methyltransferase
It should be stressed that cap formation of an mRNAs infectious agent requires universally three sequent accelerator reactions. Firstly, RNA triphosphatase (TPase) removes the c-phosphate cluster from the 59-triphosphate finish (pppN) of the emergent RNA chain to come up with the diphosphate 5ʹ-ppN. Later on, RNA guanylyltransferase (GTase) transfers a Guanosine monophosphate (GMP) to the 59-diphosphate to yield the cap core structure (GpppN). Then, N7- MTase methylates the capping guanylate at the N7 position to supply a cap-0 structure (m7GpppN) (Furuichi and Shatkin, 2000). Taken into account, lower eukaryotes such as yeast use a cap-0 structure while higher eukaryotes and viruses sometimes markedly methylate the cap-0 structure at ribose 2ʹ-O position of the first and second nucleotide of the mRNA via a ribose 2ʹ-O MTase, which in turns create cap-1 and cap-2 structure, respectively (Furuichi and Shatkin, 2000). Taken into consideration, it was shown that ribose 2ʹ-O-methylation of viral RNA cap provided a mechanism for viruses to overcome the host immune recognition (Daffis et al., 2010; Zust et al., 2011). Furthermore, it was recently reported that SARS-CoV nsp16 acts as 2ʹ-O-MTase which together with nsp10 give the rise to cap-1 structure (Bouvet et al., 2010). Interestingly, some of the honey bee compounds showed high affinity on 2ʹ-O-methylates. Among the FDA-approved medications, paritaprevir and teniposide influence the conversion of spike macromolecule, 2′-o-ribose methyltransferase, dihydroergotamine and venetoclax to nucleocapsid macromolecule and 2′-o-ribose methyltransferase. Among the natural products, amyrin (triterpenes), procyanidin, and proanthocyanidin (category of flavonoids) influence the activity of 2′-o-ribose methyltransferase (Kadioglu et al., 2020). It is noteworthy to state that the major compounds of propolis are triterpenoids that have a relative concentration of 74%, steroids, and diterpenoids (Elnakady et al., 2017). Raw propolis contains over three hundred different compounds that largely consist of triterpenes (50% w/w), waxes (25–30%), and phenolics (5–10%), volatile mono- and sesquiterpenes (8–12%) and the last compound gives propolis its typical pitchy odor (Huang et al., 2014).
Potential Restrictive Properties Against Ribonucleic Acid-dependent RNA Enzyme
Ribonucleic acid-dependent RNA enzyme (RdRp) is considered a crucial enzyme for coronaviruses as it catalyzes the replication of ribonucleic acid from RNA templates. Importantly, a remarkable similarity in the sequences and cipher structures of RdRp were reported among the sequence of RdRp in severe acute respiratory syndrome coronavirus (SARS-CoV), SARS-CoV-2 and Middle East respiratory syndrome coronavirus (MERS-CoV) (Feng et al., 2020; Morse et al., 2020). Interestingly, Remdesivir (GS-5734) might represent a nucleoside ester analogue substance of RdRp since it showed broad-spectrum antiviral activity against many ribonucleic acid viruses, as well as filovirus, SARS-CoV and MERS-CoV(Tchesnokov et al., 2019; Gordon et al., 2020; Zhang and Liu, 2020). Furthermore, a recent report indicated that remdesivir improved the vital condition of a patient with COVID-19 that might represent a therapeutic target for SARS-CoV-2 (Holshue et al., 2020).
Interestingly, twelve completely different flavonoids were detected in propolis extracts namely, pinocembrin, acacetin, chrysin, rutin, luteolin, kaempferol, apigenin, myricetin, catechin, naringenin, galangin, and quercetin; 2 synthetic resin acids, caffeic acid and cinnamic acid (Volpi, 2004). Among others, myricetin has high binding affinity toward the RdRp of each SARS-CoV and SARS-CoV-2 with favorable materia medica properties. This compound has been consumed since a time long ago and does not possess any inherent toxicity besides exhibiting a broad range of therapeutic properties, suggesting the potential use of this natural compound as inhibitor for RdRp of SARS-CoV-2. However, additional in vitro and in vivo studies seem mandatory to validate its efficaciousness against SARS-CoV-2 (Singh et al., 2020).
Potential Repressive Properties Against the Cytokine Storm
Revising the available literature, threatens of COVID-19 is partially related to cytokine storm which is defined as an exaggerated production of proinflammatory cytokines that results in multi organ system failure (Jose and Manuel, 2020; Tufan et al., 2020). It has been documented that COVID-19 infected patients with cytokine storm manifest high levels of cytokines, together with higher plasma levels of various interleukins (IL), including IL-2, IL-6, IL-7, IL-10, Granulocyte colony-stimulating factor (G-CSF), Interferon gamma (IFNγ), Microtubule-associated protein 1 alpha, and Tumor necrosis factor alpha (TNFα) (Costela-Ruiz et al., 2020; Mehta et al., 2020). Among others, IL6 is an inflammatory protein involved in cytokine storms that triggers the upregulation of T helper 1 (Th1) and T helper cell 2 (Th2) pathways (Kipar et al., 2006; Yan et al., 2018). In addition, whole bee venom down-regulates TNFα and IL-6 (Kim et al., 2011; Darwish et al., 2013; Shin et al., 2017). Moreover, bee venom is already utilized in some varieties of stylostixis for treatment of inflammatory arthritis (Lee et al., 2005). It was reported that cytokine IL10 down-regulates inflammatory cytokines like IL1 and TNF alpha in coronavirus infections (Cox, 1996). Bee venom (BV) was also reported to contain many enzymes, together with phospholipase A2 (PLA2), phospholipase B, spreading factor, acid enzyme and–glucosidase (Hossen et al., 2016). In a previous study, Park et al. (2015) showed that bee venom phospholipase A2 (BV PLA2) ameliorates allergic airway inflammation. This study found that BV PLA2 treatment causes diminished infiltration of neutrophils, eosinophils, lymphocytes, and macrophages in bronchoalveolar irrigation fluid (BLAF) (Park et al., 2015). Another study revealed that BV PLA2 includes a CD4+CD25 + Foxp3+ Treg cell-mediated protection against acute respiratory organ inflammation induced by actinotherapy (Shin et al., 2016). Interestingly, the exaggerated IL10 that was reported in beekeepers compared to the rest of population might be attributed to the chronic low level exposure to bee venom (Meiler et al., 2008). A recent study also proposed that beekeepers are often prevented from SARS-CoV-2 as results of this population developing a tolerance to bee stings (Yang et al., 2020). Clearly, this observation suggests that bee venom or product with active ingredients contained in bee venom can be used in people at high risk of serious COVID-19 to forestall or attenuate cytokine storm within the context of COVID-19.
Limitations of the Studies Carried out in Honey Bee Compounds
Given the above information, natural products are among the therapeutic options for SARS-CoV-2 infection (Serkedjieva et al., 1992; Calixto, 2005; Maruta and He, 2020). Propolis is an example for honey bee compounds that can reduce and alleviate the symptoms of inflammatory diseases by affecting various metabolic cycles (Machado et al., 2012; Hori et al., 2013; Piñeros et al., 2020). However, found many restrictions were found for the approval and acceptance of these substances as a health-promoting supplement in various countries since these compounds e.g., propolis products are not standardized and vary in their components and biological activity among countries and even at a regional level, and therefore, faced many relevant criticism (Bankova, 2005; Toreti et al., 2013; Miguel et al., 2014). However, it should be stressed that standardized propolis products e.g., standardized Brazilian propolis extract blend have recently become available to overcome this major drawback and showed higher safety profile and major effectiveness for treatment of many pathological conditions (Berretta, 2007; Berretta et al., 2012; Berretta et al., 2017; Silveira et al., 2019; Zaccaria et al., 2019). Therefore, standardized propolis is considered an example for natural products that can be used a nutraceutical or functional food resource that might provide a promising safe and easy to administer therapeutic for fighting COVID-19 pandemic (Fielding et al., 2020).
Conclusions, Limitations, and Future Perspectives
Given the above information, the current pandemic status of COVID-19 calls the urgent need to develop non-traditional novel drug targets and vaccines for combating the disease. Interestingly, several previous studies revealed the physiological and therapeutic actions of bee products (propolis, bee pollen, bee venom and honey) and their components, implicating their potential role in controlling various pathological conditions including COVID-19. However, it should be borne in mind that studies on bee bioactive compounds and their role in COVID-19 are limited and bee products may have different compositions. Taken together, the current review suggests further future studies on exploring the potential beneficial use of bee products besides investigation their detailed chemical analysis. This detailed information might provide clues for their use as potential drug targets for combating CoVID-19 either alone or in association with other drugs.
Author Contributions
EE and YM designed the idea of the review, drafted the manuscript and submitted the manuscript to the journal. WA, FE, SA, and TY involved in the conception of the idea of the manuscript and contributed his scientific advice and revision of the manuscript. The manuscript has been read and approved by all the authors.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Al-Bari, M. A. A. (2017). Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol Res Perspect 5 (1), e00293. doi:10.1002/prp2.293PRP2293
Anjum, S. I., Ullah, A., Khan, K. A., Attaullah, M., Khan, H., Ali, H., et al. (2019). Composition and functional properties of propolis (bee glue): a review. Saudi J. Biol. Sci. 26 (7), 1695–1703. doi:10.1016/j.sjbs.2018.08.013S1319-562X(18)30189-X
Astuti, I., and Ysrafil, (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab Syndr. 14 (4), 407–412. doi:10.1016/j.dsx.2020.04.020
Bankova, V. (2005). Recent trends and important developments in propolis research. Evid. base Compl. Alternative Med. 2, 29–32. doi:10.1093/ecam/neh059
Banskota, A. H., Tezuka, Y., and Kadota, S. (2001). Recent progress in pharmacological research of propolis. Phytother Res. 15 (7), 561–571. doi:10.1002/ptr.102910.1002/ptr.1029
Berretta, A. A., Nascimento, A. P., Bueno, P. C., Vaz, M. M., and Marchetti, J. M. (2012). Propolis standardized extract (EPP-AF®), an innovative chemically and biologically reproducible pharmaceutical compound for treating wounds. Int. J. Biol. Sci. 8 (4), 512. doi:10.7150/ijbs.3641
Berretta, A. A., Arruda, C., Miguel, F. G., Baptista, N., Nascimento, A. P., Marquele-Oliveira, F., et al. (2017). “Functional properties of Brazilian propolis: from chemical composition until the market,” in Superfood and functional food: an overview of their processing and utilization. (Norderstedt, Germany: BoD—Books on Demand), 55–98.
Berretta, A. A. (2007). Pesquisa pré-clínica e clínica de um gel termorreversível contendo extrato padronizado de própolis (EPP-AF) para a redução do tempo de cicatrização de lesões em pacientes queimados. São Paulo, Brazil: Universidade de São Paulo.
Bouvet, M., Debarnot, C., Imbert, I., Selisko, B., Snijder, E. J., Canard, B., et al. (2010). In vitro reconstitution of SARS-coronavirus mRNA cap methylation PLoS Pathog. 6 (4), e1000863. doi:10.1371/journal.ppat.1000863
Calixto, J. B. (2005). Twenty-five years of research on medicinal plants in Latin America: a personal view. J. Ethnopharmacol. 100 (1-2), 131–134. doi:10.1016/j.jep.2005.06.004
Campone, L., Piccinelli, A. L., Pagano, I., Carabetta, S., Di Sanzo, R., Russo, M., et al. (2014). Determination of phenolic compounds in honey using dispersive liquid-liquid microextraction. J. Chromatogr. A 1334, 9–15. doi:10.1016/j.chroma.2014.01.081
Campos, M. G., Webby, R. F., Markham, K. R., Mitchell, K. A., and Da Cunha, A. P. (2003). Age-induced diminution of free radical scavenging capacity in bee pollens and the contribution of constituent flavonoids. J. Agric. Food Chem. 51 (3), 742–745. doi:10.1021/jf0206466
Chan, C. W., Deadman, B. J., Manley-Harris, M., Wilkins, A. L., Alber, D. G., and Harry, E. (2013). Analysis of the flavonoid component of bioactive New Zealand mānuka (Leptospermum scoparium) honey and the isolation, characterisation and synthesis of an unusual pyrrole. Food Chem. 141 (3), 1772–1781. doi:10.1016/j.foodchem.2013.04.092
Chen, Y., Liu, Q., and Guo, D. (2020a). Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 92 (4), 418–423. doi:10.1002/jmv.25681
Chen, Y. W., Yiu, C. B., and Wong, K. Y. (2020b). Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL (pro)) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Res 9, 129. doi:10.12688/f1000research.22457.2
Choudhary, S., Malik, Y., and Tomar, S. (2020). Identification of SARS-CoV-2 cell entry inhibitors by drug repurposing using in silico structure-based virtual screening approach. Front. Immunol. 11, 1664. doi:10.3389/fimmu.2020.01664
Cinatl, J., Morgenstern, B., Bauer, G., Chandra, P., Rabenau, H., and Doerr, H. (2003). Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet 361 (9374), 2045–2046. doi:10.1016/s0140-6736(03)13615-x
Consortium, C. S. M. E. (2004). Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 303 (5664), 1666–1669. doi:10.1126/science.10920021092002
Costela-Ruiz, V. J., Illescas-Montes, R., Puerta-Puerta, J. M., Ruiz, C., and Melguizo-Rodríguez, L. (2020). SARS-CoV-2 infection: the role of cytokines in COVID-19 disease, Cytokine Growth Factor Rev., 54, 62–75. doi:10.1016/j.cytogfr.2020.06.001
Cox, G. (1996). IL-10 enhances resolution of pulmonary inflammation in vivo by promoting apoptosis of neutrophils. Am. J. Physiol. 271 (4 Pt 1), L566–L571. doi:10.1152/ajplung.1996.271.4.L566
Cui, J., Li, F., and Shi, Z. L. (2019). Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 17 (3), 181–192. doi:10.1038/s41579-018-0118-9
Daffis, S., Szretter, K. J., Schriewer, J., Li, J., Youn, S., Errett, J., et al. (2010). 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468 (7322), 452–456. doi:10.1038/nature09489nature09489
Darwish, S. F., El-Bakly, W. M., Arafa, H. M., and El-Demerdash, E. (2013). Targeting TNF-α and NF-κB activation by bee venom: role in suppressing adjuvant induced arthritis and methotrexate hepatotoxicity in rats. PloS One 8 (11), e79284. doi:10.1371/journal.pone.0079284PONE-D-13-26340
Dong, S., Sun, J., Mao, Z., Wang, L., Lu, Y. L., and Li, J. (2020). A guideline for homology modeling of the proteins from newly discovered betacoronavirus, 2019 novel coronavirus (2019‐nCoV). J. Med. Virol., 92, 1542. doi:10.1002/jmv.25768
Drescher, N., Klein, A. M., Schmitt, T., and Leonhardt, S. D. (2019). A clue on bee glue: new insight into the sources and factors driving resin intake in honeybees (Apis mellifera). PloS One 14 (2), e0210594. doi:10.1371/journal.pone.0210594
Elnakady, Y. A., Rushdi, A. I., Franke, R., Abutaha, N., Ebaid, H., Baabbad, M., et al. (2017). Characteristics, chemical compositions and biological activities of propolis from Al-Bahah, Saudi Arabia. Sci. Rep. 7, 41453. doi:10.1038/srep41453srep41453
Escuredo, O., Míguez, M., Fernández-González, M., and Carmen Seijo, M. (2013). Nutritional value and antioxidant activity of honeys produced in a European Atlantic area. Food Chem. 138 (2-3), 851–856. doi:10.1016/j.foodchem.2012.11.015S0308-8146(12)01742-6
Fehr, A. R., and Perlman, S. (2015). Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 1282, 1–23. doi:10.1007/978-1-4939-2438-7_1
Feng, W., Zong, W., Wang, F., and Ju, S. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a review. Mol. Canc. 19 (1), 1–14. doi:10.1186/s12943-020-01218-1
Fielding, B. C., da Silva Maia Bezerra Filho, C., Ismail, N. S. M., and Sousa, D. P. d. (2020). Alkaloids: therapeutic potential against human coronaviruses. Molecules 25 (23), 5496. doi:10.3390/molecules25235496
Fung, T. S., and Liu, D. X. (2014). Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 5, 296. doi:10.3389/fmicb.2014.00296
Furuichi, Y., and Shatkin, A. J. (2000). Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 55, 135–184. doi:10.1016/s0065-3527(00)55003-9
Gordon, C. J., Tchesnokov, E. P., Feng, J. Y., Porter, D. P., and Götte, M. (2020). The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 295, 013056. doi:10.1074/jbc.AC120.013056
Graham, R. L., Donaldson, E. F., and Baric, R. S. (2013). A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 11 (12), 836–848. doi:10.1038/nrmicro3143nrmicro3143
Graikou, K., Popova, M., Gortzi, O., Bankova, V., and Chinou, I. (2015). Characterization of biological evaluation of selected Mediterranean propolis samples. Is it a new type? LWT-Food Sci. Technol. 65, 261–267. doi:10.1016/j.lwt.2015.08.025
Güler, H. I., Tatar, G., Yildiz, O., Belduz, A. O., and Kolayli, S. (2020). An investigation of ethanolic propolis extracts: their potential inhibitor properties against ACE-II receptors for COVID-19 treatment by Molecular Docking Study. ScienceOpen Preprints. doi:10.35206/jan.762734
Hamming, I., Timens, W., Bulthuis, M. L., Lely, A. T., Navis, G., and van Goor, H. (2004). Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203 (2), 631–637. doi:10.1002/path.1570
Hashem, H. (2020). Silico approach of some selected honey constituents as SARS-CoV-2 main protease (COVID-19) inhibitors. EJMO 4 (3), 196–200. doi:10.14744/ejmo.2020.36102
He, J., Hu, L., Huang, X., Wang, C., Zhang, Z., Wang, Y., et al. (2020). Potential of coronavirus 3C-like protease inhibitors for the development of new anti-SARS-CoV-2 drugs: insights from structures of protease and inhibitors. Int. J. Antimicrob. Agents 56, 106055. doi:10.1016/j.ijantimicag.2020.106055
Hoever, G., Baltina, L., Michaelis, M., Kondratenko, R., Baltina, L., Tolstikov, G. A., et al. (2005). Antiviral activity of glycyrrhizic acid derivatives against SARS-coronavirus. J. Med. Chem. 48 (4), 1256–1259. doi:10.1021/jm0493008
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020b). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181 (2), 271–280.e8. doi:10.1016/j.cell.2020.02.052
Hoffmann, M., Kleine-Weber, H., Krüger, N., Müller, M., Drosten, C., and Pöhlmann, S. (2020a). The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv. doi:10.1101/2020.01.31.929042v1
Holshue, M. L., DeBolt, C., Lindquist, S., Lofy, K. H., Wiesman, J., Bruce, H., et al. (2020). First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 382 (10), 929–936. doi:10.1056/NEJMoa2001191
Hori, J. I., Zamboni, D. S., Carrão, D. B., Goldman, G. H., and Berretta, A. A. (2013). The inhibition of inflammasome by Brazilian propolis (EPP-AF). Evid. Based Complement Alternat. Med. 2013, 418508. doi:10.1155/2013/418508
Hossen, M. S., Shapla, U. M., Gan, S. H., and Khalil, M. I. (2016). Impact of bee venom enzymes on diseases and immune responses. Molecules 22 (1), E25. doi:10.3390/molecules22010025
Huang, S., Zhang, C. P., Wang, K., Li, G. Q., and Hu, F. L. (2014). Recent advances in the chemical composition of propolis. Molecules 19 (12), 19610–19632. doi:10.3390/molecules191219610molecules191219610
Hussain, F., Jahan, N., Rahman, K.-u., Sultana, B., and Jamil, S. (2018). Identification of hypotensive biofunctional compounds ofCoriandrum sativumand evaluation of their angiotensin-converting enzyme (ACE) inhibition potential. Oxid. Med. Cell. Longev., 2018, 1–11. doi:10.1155/2018/4643736
Jo, S., Kim, S., Shin, D. H., and Kim, M. S. (2020). Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzym. Inhib. Med. Chem. 35 (1), 145–151. doi:10.1080/14756366.2019.1690480
Jose, R. J., and Manuel, A. (2020). COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir Med 8 (6), e46–e47. doi:10.1016/S2213-2600(20)30216-2
Kadioglu, O., Saeed, M., Johannes Greten, H., and Efferth, T. (2020). Identification of novel compounds against three targets of SARS CoV-2 coronavirus by combined virtual screening and supervised machine learning. Bull. World Health. Organ. doi:10.2471/BLT.20.255943
Keckes, J., Trifkovic, J., Andric, F., Jovetic, M., Tesic, Z., and Milojkovic-Opsenica, D. (2013). Amino acids profile of Serbian unifloral honeys. J. Sci. Food Agric. 93 (13), 3368–3376. doi:10.1002/jsfa.6187
Kennedy, D. O., and Wightman, E. L. (2011). Herbal extracts and phytochemicals: plant secondary metabolites and the enhancement of human brain function. Adv. Nutr. 2 (1), 32–50. doi:10.3945/an.110.000117000117
Kim, H. Y., Eo, E. Y., Park, H., Kim, Y. C., Park, S., Shin, H. J., Kim, K., et al. (2010). Medicinal herbal extracts of Sophorae radix, Acanthopanacis cortex, Sanguisorbae radix and Torilis fructus inhibit coronavirus replication in vitro. Antivir. Ther. 15 (5), 697–709. doi:10.3851/IMP1615
Kim, S. K., Park, K. Y., Yoon, W. C., Park, S. H., Park, K. K., Yoo, D. H., et al. (2011). Melittin enhances apoptosis through suppression of IL-6/sIL-6R complex-induced NF-κB and STAT3 activation and Bcl-2 expression for human fibroblast-like synoviocytes in rheumatoid arthritis. Joint Bone Spine 78 (5), 471–477. doi:10.1016/j.jbspin.2011.01.004S1297-319X(11)00005-4
Kipar, A., Meli, M. L., Failing, K., Euler, T., Gomes-Keller, M. A., Schwartz, D., et al. (2006). Natural feline coronavirus infection: differences in cytokine patterns in association with the outcome of infection. Vet. Immunol. Immunopathol. 112 (3-4), 141–155. doi:10.1016/j.vetimm.2006.02.004
Kumar, V., Shin, J. S., Shie, J. J., Ku, K. B., Kim, C., Go, Y. Y., et al. (2017). Identification and evaluation of potent Middle East respiratory syndrome coronavirus (MERS-CoV) 3CL(Pro) inhibitors. Antivir. Res. 141, 101–106. doi:10.1016/j.antiviral.2017.02.007
Lau, K.-M., Lee, K.-M., Koon, C.-M., Cheung, C. S.-F., Lau, C.-P., Ho, H.-M., et al. (2008). Immunomodulatory and anti-SARS activities of Houttuynia cordata. J. Ethnopharmacol. 118 (1), 79–85. doi:10.1016/j.jep.2008.03.018
Lee, J. D., Park, H. J., Chae, Y., and Lim, S. (2005). An overview of bee venom acupuncture in the treatment of arthritis. Evid. Based Complement Alternat. Med. 2 (1), 79–84. doi:10.1093/ecam/neh070
Letko, M., Marzi, A., and Munster, V. (2020). Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5 (4), 562–569. doi:10.1038/s41564-020-0688-y
Li, F. (2016). Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 3 (1), 237–261. doi:10.1146/annurev-virology-110615-042301
Li, H., Liu, S. M., Yu, X. H., Tang, S. L., and Tang, C. K. (2020a). Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int. J. Antimicrob. Agents 55 (5), 105951. doi:10.1016/j.ijantimicag.2020.105951
Li, Q., Guan, X., Wu, P., Wang, X., Zhou, L., Tong, Y., et al. (2020b). Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 382 (13), 1199–1207. doi:10.1056/NEJMoa2001316
Li, S. Y., Chen, C., Zhang, H. Q., Guo, H. Y., Wang, H., Wang, L., et al. (2005). Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antivir. Res. 67 (1), 18–23. doi:10.1016/j.antiviral.2005.02.007
Lin, C. W., Tsai, F. J., Tsai, C. H., Lai, C. C., Wan, L., Ho, T. Y., et al. (2005). Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antivir. Res. 68 (1), 36–42. doi:10.1016/j.antiviral.2005.07.002
Machado, J. L., Assunção, A. K., da Silva, M. C., Dos Reis, A. S., Costa, G. C., Arruda, D. S., et al. (2012). Brazilian green propolis: anti-inflammatory property by an immunomodulatory activity. Evid. Based Complement Alternat. Med., 2012, 157652. doi:10.1155/2012/157652
Magrone, T., Magrone, M., and Jirillo, E. (2020). Focus on receptors for coronaviruses with special reference to angiotensin-converting enzyme 2 as a potential drug target - a perspective. Endocr. Metab Immune Disord. Drug Targets. 20 (6), 807–811. doi:10.2174/1871530320666200427112902
Maruta, H., and He, H. (2020). PAK1-blockers: potential therapeutics against COVID-19. Med. Drug Discov. 6, 100039. doi:10.1016/j.medidd.2020.100039
McBride, R., van Zyl, M., and Fielding, B. C. (2014). The coronavirus nucleocapsid is a multifunctional protein. Viruses 6 (8), 2991–3018. doi:10.3390/v6082991v6082991
Mehta, P., McAuley, D. F., Brown, M., Sanchez, E., Tattersall, R. S., and Manson, J. J. (2020). COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395 (10229), 1033–1034. doi:10.1016/S0140-6736(20)30628-0
Meiler, F., Zumkehr, J., Klunker, S., Rückert, B., Akdis, C. A., and Akdis, M. (2008). In vivo switch to IL-10-secreting T regulatory cells in high dose allergen exposure. J. Exp. Med. 205 (12), 2887–2898. doi:10.1084/jem.20080193jem.20080193
Miguel, M. G., Nunes, S., Dandlen, S. A., Cavaco, A. M., and Antunes, M. D. (2014). Phenols, flavonoids and antioxidant activity of aqueous and methanolic extracts of propolis (Apis mellifera L.) from Algarve, South Portugal. Food Sci. Technol (Campinas) 34 (1), 16–23. doi:10.1590/s0101-20612014000100002
Monchatre-Leroy, E., Boué, F., Boucher, J. M., Renault, C., Moutou, F., Ar Gouilh, M., et al. (2017). Identification of alpha and beta coronavirus in Wildlife species in France: bats, rodents, rabbits, and hedgehogs. Viruses 9 (12), E364. doi:10.3390/v9120364
Morse, J. S., Lalonde, T., Xu, S., and Liu, W. (2020). Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem. 21 (5), 730–738. doi:10.1002/cbic.202000047
Muramatsu, T., Takemoto, C., Kim, Y. T., Wang, H., Nishii, W., Terada, T., et al. (2016). SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proc. Natl. Acad. Sci. U.S.A. 113 (46), 12997–13002. doi:10.1073/pnas.1601327113
Nadeem, M. S., Zamzami, M. A., Choudhry, H., Murtaza, B. N., Kazmi, I., Ahmad, H., et al. (2020). Origin, potential therapeutic targets and treatment for coronavirus disease (COVID-19). Pathogens 9 (4), E307. doi:10.3390/pathogens9040307
Naqvi, A. A. T., Fatima, K., Mohammad, T., Fatima, U., Singh, I. K., Singh, A., et al. (2020). Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis., 1866, 165878. doi:10.1016/j.bbadis.2020.165878
Narayanan, K., Ramirez, S. I., Lokugamage, K. G., and Makino, S. (2015). Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res. 202, 89–100. doi:10.1016/j.virusres.2014.11.019S0168-1702(14)00494-8
Oryan, A., Alemzadeh, E., and Moshiri, A. (2018). Potential role of propolis in wound healing: biological properties and therapeutic activities. Biomed. Pharmacother. 98, 469–483. doi:10.1016/j.biopha.2017.12.069
Paraskevis, D., Kostaki, E. G., Magiorkinis, G., Panayiotakopoulos, G., Sourvinos, G., and Tsiodras, S. (2020). Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 79, 104212–111348. doi:10.1016/j.meegid.2020.104212
Park, J. Y., Ko, J. A., Kim, D. W., Kim, Y. M., Kwon, H. J., Jeong, H. J., et al. (2016). Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J. Enzym. Inhib. Med. Chem. 31 (1), 23–30. doi:10.3109/14756366.2014.1003215
Park, S., Baek, H., Jung, K. H., Lee, G., Lee, H., Kang, G. H., et al. (2015). Bee venom phospholipase A2 suppresses allergic airway inflammation in an ovalbumin-induced asthma model through the induction of regulatory T cells. Immun. Inflamm. Dis. 3 (4), 386–397. doi:10.1002/iid3.76IID376
Pasupuleti, V. R., Sammugam, L., Ramesh, N., and Gan, S. H. (2017). Honey, propolis, and royal jelly: a comprehensive review of their biological actions and health benefits. Oxid. Med. Cell. Longev. 2017, 1259510. doi:10.1155/2017/1259510
Paules, C. I., Marston, H. D., and Fauci, A. S. (2020). Coronavirus infections-more than just the common cold. Jama 323 (8), 707–708. doi:10.1001/jama.2020.0757
Peiris, J., Guan, Y., and Yuen, K. (2004). Severe acute respiratory syndrome. Nat. Med. 10 (12), S88–S97. doi:10.1038/nm1143
Piñeros, A. R., de Lima, M. H., Rodrigues, T., Gembre, A. F., Bertolini, T. B., Fonseca, M. D., et al. (2020). Green propolis increases myeloid suppressor cells and CD4+ Foxp3+ cells and reduces Th2 inflammation in the lungs after allergen exposure. J. Ethnopharmacol. 252, 112496. doi:10.1016/j.jep.2019.112496
Pobiega, K., Kraśniewska, K., and Gniewosz, M. (2018). Application of propolis in antimicrobial and antioxidative protection of food quality—a review. Trends Food Sci. Technol. 83, 53–62. doi:10.1016/j.tifs.2018.11.007
Ryu, Y. B., Jeong, H. J., Kim, J. H., Kim, Y. M., Park, J. Y., Kim, D., et al. (2010). Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL(pro) inhibition. Bioorg. Med. Chem. 18 (22), 7940–7947. doi:10.1016/j.bmc.2010.09.035
Saric, A., Balog, T., Sobocanec, S., Kusic, B., Sverko, V., Rusak, G., et al. (2009). Antioxidant effects of flavonoid from Croatian Cystus incanus L. rich bee pollen. Food Chem. Toxicol. 47 (3), 547–554. doi:10.1016/j.fct.2008.12.007S0278-6915(08)00705-9
Savarino, A., Boelaert, J. R., Cassone, A., Majori, G., and Cauda, R. (2003). Effects of chloroquine on viral infections: an old drug against today’s diseases? Lancet Infect. Dis. 3 (11), 722–727. doi:10.1016/s1473-3099(03)00806-5
Sawicki, S. G., Sawicki, D. L., Younker, D., Meyer, Y., Thiel, V., Stokes, H., et al. (2005). Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 1 (4), e39. doi:10.1371/journal.ppat.0010039
Serkedjieva, J., Manolova, N., and Bankova, V. (1992). Anti-influenza virus effect of some propolis constituents and their analogues (esters of substituted cinnamic acids). J. Nat. Prod. 55 (3), 294–302. doi:10.1021/np50081a003
Sforcin, J. (2016). Biological properties and therapeutic applications of propolis. Phytother Res. 30, 894. doi:10.1002/ptr.5605
Shaldam, M., Yahya, G., Mohamed, N., Abdel Daim, M., and Al Naggar, Y. (2020). Silico screening of potent bioactive compounds from honey bee products against COVID-19 target enzymes. chemRxiv. doi:10.26434/chemrxiv.12644102.v1
Shin, D., Lee, G., Sohn, S. H., Park, S., Jung, K. H., Lee, J. M., et al. (2016). Regulatory T cells contribute to the inhibition of radiation-induced acute lung inflammation via bee venom phospholipase A₂ in mice. Toxins 8 (5). doi:10.3390/toxins8050131E131
Shin, S. H., Ye, M. K., Choi, S. Y., and Park, K. K. (2017). The effects of melittin and apamin on airborne fungi-induced chemical mediator and extracellular matrix production from nasal polyp fibroblasts. Toxins 9 (11), E348. doi:10.3390/toxins9110348
Silveira, M. A. D., Teles, F., Berretta, A. A., Sanches, T. R., Rodrigues, C. E., Seguro, A. C., et al. (2019). Effects of Brazilian green propolis on proteinuria and renal function in patients with chronic kidney disease: a randomized, double-blind, placebo-controlled trial. BMC Nephrol. 20 (1), 140. doi:10.1186/s12882-019-1337-7
Singh, S., Sonawane, A., and Sadhukhan, S. (2020). Plant-derived natural polyphenols as potential antiviral drugs against SARS-CoV-2 via RNA‐dependent RNA polymerase (RdRp) inhibition: an in-silico analysis. J. Biomol. Struct. Dyn. 28, 1–16. doi:10.1080/07391102.2020.1796810
Solórzano, E., Vera, N., Cuello, S., Ordoñez, R., Zampini, C., Maldonado, L., et al. (2012). Chalcones in bioactive Argentine propolis collected in arid environments. Nat. Prod. Commun. 7, 879–882. doi:10.1177/1934578x1200700718
Song, Z., Xu, Y., Bao, L., Zhang, L., Yu, P., Qu, Y., et al. (2019). From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses 11 (1), E59. doi:10.3390/v11010059
Sun, M. L., Yang, J. M., Sun, Y. P., and Su, G. H. (2020). Inhibitors of RAS might Be a good choice for the therapy of COVID-19 pneumonia. Zhonghua Jiehe He Huxi Zazhi 43, E014. doi:10.3760/cma.j.issn.1001-0939.2020.0014
Tchesnokov, E. P., Feng, J. Y., Porter, D. P., and Götte, M. (2019). Mechanism of inhibition of ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses 11 (4), E326. doi:10.3390/v11040326
Theerawatanasirikul, S., Kuo, C. J., Phetcharat, N., and Lekcharoensuk, P. (2020). In silico and in vitro analysis of small molecules and natural compounds targeting the 3CL protease of feline infectious peritonitis virus. Antivir. Res. 174, 104697. doi:10.1016/j.antiviral.2019.104697
Tolba, M. F., Azab, S. S., Khalifa, A. E., Abdel-Rahman, S. Z., and Abdel-Naim, A. B. (2013). Caffeic acid phenethyl ester, a promising component of propolis with a plethora of biological activities: a review on its anti-inflammatory, neuroprotective, hepatoprotective, and cardioprotective effects. IUBMB Life 65 (8), 699–709. doi:10.1002/iub.1189
Ton, A. T., Gentile, F., Hsing, M., Ban, F., and Cherkasov, A. (2020). Rapid identification of potential inhibitors of SARS‐CoV‐2 main protease by deep docking of 1.3 billion compounds. Mol. Inf., 39, 2000028. doi:10.1002/minf.202000028
Toreti, V. C., Sato, H. H., Pastore, G. M., and Park, Y. K. (2013). Recent progress of propolis for its biological and chemical compositions and its botanical origin. Evid. Based Complement Alternat. Med. 2013, 697390. doi:10.1155/2013/697390
Tufan, A., Avanoğlu Güler, A., and Matucci-Cerinic, M. (2020). COVID-19, immune system response, hyperinflammation and repurposing antirheumatic drugs. Turk. J. Med. Sci. 50 (SI-1), 620–632. doi:10.3906/sag-2004-168
Volpi, N. (2004). Separation of flavonoids and phenolic acids from propolis by capillary zone electrophoresis. Electrophoresis 25 (12), 1872–1878. doi:10.1002/elps.200405949
Wang, H., Rao, S., and Jiang, C. (2007). Molecular pathogenesis of severe acute respiratory syndrome. Microb. Infect. 9 (1), 119–126. doi:10.1016/j.micinf.2006.06.012
Wang, Q., Sui, X., Sui, D. J., and Yang, P. (2018). Flavonoid extract from propolis inhibits cardiac fibrosis triggered by myocardial infarction through upregulation of SIRT1. Evid. Based Complement Alternat. Med. 2018, 4957573. doi:10.1155/2018/4957573
Wieckiewicz, W., Miernik, M., Wieckiewicz, M., and Morawiec, T. (2013). Does propolis help to maintain oral health? Evid. Based Complement Alternat. Med. 2013, 351062. doi:10.1155/2013/351062
Worldometer (2020). Worldometer’s COVID-19 data. Available at: https://www.worldometers.info/coronavirus/ (Accessed December 15, 2020).
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367 (6483), 1260–1263. doi:10.1126/science.abb2507
Wu, C. Y., Jan, J. T., Ma, S. H., Kuo, C. J., Juan, H. F., Cheng, Y. S., et al. (2004). Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. U.S.A. 101 (27), 10012–10017. doi:10.1073/pnas.0403596101
Xu, X., Chen, P., Wang, J., Feng, J., Zhou, H., Li, X., et al. (2020). Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 63 (3), 457–460. doi:10.1007/s11427-020-1637-5
Yan, C., Deng, C., Liu, X., Chen, Y., Ye, J., Cai, R., et al. (2018). TNF-α induction of IL-6 in alveolar type II epithelial cells: contributions of JNK/c-Jun/AP-1 element, C/EBPδ/C/EBP binding site and IKK/NF-κB p65/κB site. Mol. Immunol. 101, 585–596. doi:10.1016/j.molimm.2018.05.004
Yang, W., Hu, F. L., and Xu, X. F. (2020). Bee venom and SARS-CoV-2. Toxicon 181, 69–70. doi:10.1016/j.toxicon.2020.04.105
Ye, G., Wang, X., Tong, X., Shi, Y., Fu, Z. F., and Peng, G. (2020). Structural basis for inhibiting porcine epidemic diarrhea virus replication with the 3C-like protease inhibitor GC376. Viruses 12 (2), E240. doi:10.3390/v12020240
Yu, M. S., Lee, J., Lee, J. M., Kim, Y., Chin, Y. W., Jee, J. G., et al. (2012). Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 22 (12), 4049–4054. doi:10.1016/j.bmcl.2012.04.081
Yusuf, N., Irby, C., Katiyar, S. K., and Elmets, C. A. (2007). Photoprotective effects of green tea polyphenols. Photodermatol. Photoimmunol. Photomed. 23 (1), 48–56. doi:10.1111/j.1600-0781.2007.00262.x
Zabaiou, N., Fouache, A., Trousson, A., Baron, S., Zellagui, A., Lahouel, M., et al. (2017). Biological properties of propolis extracts: something new from an ancient product. Chem. Phys. Lipids 207, 214. doi:10.1016/j.chemphyslip.2017.04.005
Zaccaria, V., Garzarella, E. U., Di Giovanni, C., Galeotti, F., Gisone, L., Campoccia, D., et al. (2019). Multi dynamic extraction: an innovative method to obtain a standardized chemically and biologically reproducible polyphenol extract from poplar-type propolis to Be used for its anti-infective properties. Materials 12 (22), 3746. doi:10.3390/ma12223746
Zhang, L., and Liu, Y. (2020). Potential interventions for novel coronavirus in China: a systematic review. J. Med. Virol. 92 (5), 479–490. doi:10.1002/jmv.25707
Zhou, P., Yang, X. L., Wang, X. G., Hu, B., Zhang, L., Zhang, W., et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579 (7798), 270–273. doi:10.1038/s41586-020-2012-7
Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., et al. (2020a). A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382 (8), 727–733. doi:10.1056/NEJMoa2001017
Zhu, Y., Yu, D., Yan, H., Chong, H., and He, Y. (2020b). Design of potent membrane fusion inhibitors against SARS-CoV-2, an emerging coronavirus with high fusogenic activity. J. Virol. 94 (14), e00635–20. doi:10.1128/JVI.00635-20
Keywords: COVID‐19, pathogenesis, honey bee, products, inhibitors
Citation: Elmahallawy EK, Mohamed Y, Abdo W, El-Gohary FA, Ahmed Awad Ali S and Yanai T (2021) New Insights Into Potential Benefits of Bioactive Compounds of Bee Products on COVID-19: A Review and Assessment of Recent Research. Front. Mol. Biosci. 7:618318. doi: 10.3389/fmolb.2020.618318
Received: 16 October 2020; Accepted: 29 December 2020;
Published: 08 February 2021.
Edited by:
Da’san Mahmoud Mousa Jaradat, Al-Balqa Applied University, JordanReviewed by:
Ana Cláudia Coelho, University of Trás-os-Montes and Alto Douro, PortugalMohammad Shahid, Children’s National Hospital, United States
Copyright © 2021 Elmahallawy, Mohamed, Abdo, El-Gohary, Ahmed Awad Ali and Yanai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ehab Kotb Elmahallawy, ZWVoYWFAdW5pbGVvbi5lcw==
†These authors have contributed equally to this work
 Ehab Kotb Elmahallawy
Ehab Kotb Elmahallawy Yasser Mohamed2†
Yasser Mohamed2† Fatma A. El-Gohary
Fatma A. El-Gohary