
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 15 February 2021
Sec. Molecular Diagnostics and Therapeutics
Volume 7 - 2020 | https://doi.org/10.3389/fmolb.2020.608447
During normal pregnancy, the placental trophoblast secretes a variety of steroid hormones and participates in the regulation of maternal physiological functions and fetal development. The CYP11A1 gene encodes the cholesterol side-chain cleavage enzyme P450scc, which catalyzes the production of pregnenolone from cholesterol, which is the first step in the synthesis of all steroid hormones. Under the influence of genetic susceptibility and certain environmental factors, such as drugs and toxins, the expression of CYP11A1 can be upregulated, thereby affecting steroid metabolism and physiological functions in trophoblast cells, as well as fetal development. Here, we demonstrate that upregulation of CYP11A1 in the BeWo cell line triggers excessive mitochondrial oxidative stress, leads to mitochondrial damage and interleukin-6 release, and contributes to the inhibition of proliferation and DNA damage in neuronal stem cells (NSCs). Furthermore, oxidative stress and inflammation can be ameliorated by vitamin D3 in a dose-dependent manner, thereby facilitating the rescue of NSC impairment. Our findings reveal the underlying mechanism in which upregulation of CYP11A1 is detrimental to the physiological function of trophoblasts and demonstrate the beneficial effects of vitamin D supplementation in preventing placental and neurodevelopmental damage associated with CYP11A1 upregulation during pregnancy.
In humans, the CYP11A1 gene is located on chromosome 15, and it encodes a cholesterol side-chain cleavage enzyme, P450scc, that is highly expressed in organs associated with the synthesis of steroid hormones, such as the adrenal gland, ovary, testis, and placenta (Hanukoglu, 1992; Strauss et al., 1996). P450scc catalyzes the production of pregnenolone from cholesterol, which is the rate-limiting step in steroidogenesis (Churchill and Kimura, 1979; Hanukoglu et al., 1990; Hanukoglu, 1992). Unlike the adrenal gland, ovary, and testis, the placenta does not express steroidogenic acute regulatory protein (StAR), which is responsible for transportation of cholesterol to the mitochondrial inner membrane; this is the real reason for the rate limitation in the afore mentioned organs (Stocco, 1999; Christenson and Strauss, 2000; Miller, 2007). Tuckey et al. pointed out that the supply of cholesterol to P450scc may approach saturation, and this rate limitation may be associated with the inhibition of P450scc activity by adrenodoxin reductase and its products (Tuckey, 2005). Thus, regulating CYP11A1 expression levels in the placental trophoblast can, theoretically, change steroidogenesis, thereby affecting the mitochondrial function and metabolic behavior of trophoblasts.
It has been demonstrated that a variety of factors in the intrauterine environment, such as drugs, toxins, hormones, and inflammation, can upregulate CYP11A1 expression in the trophoblast cells and impair pregnancy outcome (Beaudoin et al., 1997; Wang et al., 2014; Zhang et al., 2016b; Zhang et al., 2019). CYP11A1 has been identified as a candidate pathogenic gene for preeclampsia and shown to be hypomethylated and transcribed at higher levels in women with preeclampsia (Enquobahrie et al., 2008; Hogg et al., 2013). Our previous studies have demonstrated that upregulation of CYP11A1 induces trophoblast apoptosis and is positively correlated with liver damage during preeclampsia; however, the underlying mechanism remains elusive (He et al., 2013; Pan et al., 2017). In addition, due to the essential roles of steroid hormones in pregnancy, a valid question is whether or not the upregulation of this key gene has other negative effects.
Vitamin D is a vital nutrient for the maintenance of calcium homeostasis. In contrast to typical metabolism, in which vitamin D is first hydroxylated to calcidiol (25(OH)D) by CYP2R1 in the liver and then further hydroxylated to calcitriol (1,25(OH)D) by CYP27B1 in the kidney, Guryev (Guryev et al., 2003) and Zbytek et al (Zbytek et al., 2008). have uncovered a novel metabolic pathway, in which vitamin D is catalyzed to 20(OH)D by CYP11A1 (P450scc) in the placenta and showed anti-inflammatory activity and differential regulation (Guryev et al., 2003). In recent years, accumulating evidence from studies on pregnant women has demonstrated the preventive and therapeutic effects of vitamin D supplementation during pregnancy, especially in preventing preeclampsia. Fogacci et al. reviewed 27 clinical trials focusing on vitamin D supplementation and preeclampsia and concluded that although the underlying mechanism was unclear and the timing of dosage was controversial, the preventive effect of vitamin D supplementation on preeclampsia was firmly established (Fogacci et al., 2020). Based on these findings, we hypothesized that vitamin D could affect the impairment caused by CYP11A1 upregulation and that the underlying mechanisms may involve competitive binding to excess P450scc to decrease steroidogenesis and promote anti-inflammation via 20(OH)D production.
In this study, the BeWo cell line, a well-established in vitro trophoblast model for studying placental endocrine function, was used to investigate the signaling cascade triggered by CYP11A1 overexpression, including excessive oxidative stress, DNA damage, and apoptosis of mitochondria, leading to inflammatory cytokine release and contributing to inhibition of proliferation and DNA damage in neuronal stem cells (NSCs). In addition, we studied the putative role of vitamin D3 in preventing these negative effects. Overall, this study may provide a theoretical basis for the application of vitamin D supplementation during pregnancy.
After overexpression of CYP11A1 in BeWo cells and selection using puromycin, expression of the CYP11A1 gene was determined at the mRNA and protein level using qPCR and western blotting, respectively. Results showed significantly increased levels of CYP11A1 mRNA and protein compared to those in the vehicle or control cells (Figure 1). Interestingly, we also observed upregulation of CYP11A1 with low-dose bisphenol A (BPA) treatment (Supplemental Figure S1), indicating that both genetic variants and environmental factors, such as BPA, may lead to CYP11A1 upregulation in the placenta.
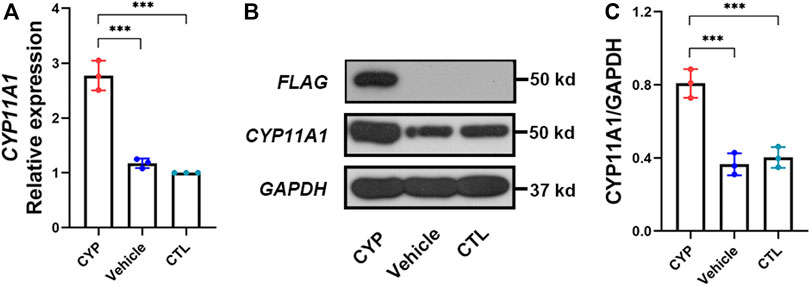
FIGURE 1. CYP11A1expression in BeWo cells was significantly increased after lentivirus transfection. (A) Relative mRNA expression levels and (B, C) protein expression levels of CYP11A1. CYP, BeWo cells overexpressing CYP11A1. Vehicle, BeWo cells transfected with vehicle. CTL, control. Statistical analysis was performed on the means from three independent experiments using one-way analysis of variance (ANOVA) with Tukey’s post hoc test. ***P < 0.001.
First, we analyzed the secretion of pregnenolone, a direct product of catalysis by P450scc. Pregnenolone production increased significantly with CYP11A1 overexpression and was reduced by the addition of 2 μM or 5 μM vitamin D3 (Figure 2A). Furthermore, the same trend was also observed for the secretion of progesterone, an indirect product of P450scc activity (Figure 2B). It should be noted that in trophoblast cells, progesterone is only catalyzed from pregnenolone by mitochondrial 3β-hydroxysteroid dehydrogenase type I (3βHSD-I), therefore, total levels of secreted pregnenolone should equal at least the sum of detected pregnenolone and progesterone (Figure 2C).
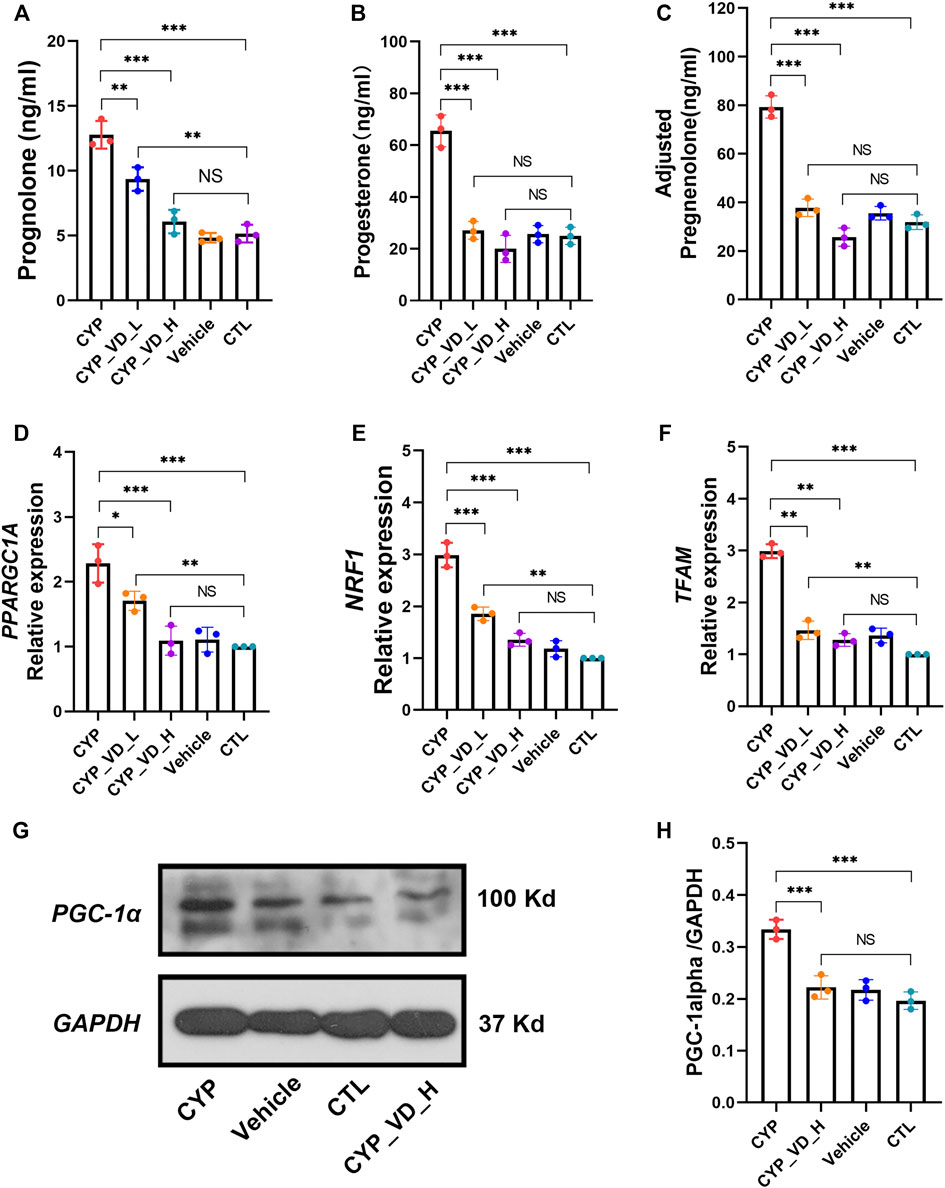
FIGURE 2. Excessive mitochondrial biosynthesis induced by upregulation of CYP11A1 in BeWo cells could be ameliorated by vitamin D3 addition. (A–C) Pregnenolone and progesterone secretion. (D–F) Relative expression of mitochondrial biosynthesis markers at the mRNA level. (G, H) Relative expression of PGC1-α. CYP, BeWo cells overexpressing CYP11A1. Vehicle, BeWo cells transfected with vehicle. CTL, control. CYP_VD_L, BeWo cells overexpressing CYP11A1 and treated with a low-dose of vitamin D3 (2 μM). CYP_VD_H, BeWo cells overexpressing CYP11A1 and treated with a high-dose of vitamin D3 (5 μM). Statistical analysis was performed on the means from three independent experiments using one-way analysis of variance (ANOVA) with Tukey’s post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001; NS, no significance.
Secretion of pregnenolone and progesterone requires the support of the mitochondrial biosynthesis system. Next, we observed a marked increase in the expression of the mitochondrial biosynthesis markers, namely PPARGC1A, NRF1, and TFAM at the mRNA level. PPARGC1A encodes PGC1-α, a member of the transcriptional coactivators and the master regulator of mitochondrial biogenesis. PGC1-α activates NRF1, which in turn co-activates TFAM, which is responsible for transcribing nuclear-encoded mitochondrial proteins. This alteration could be decreased and rescued by 2 μM and 5 μM of vitamin D3, respectively, (Figures 2D–F). Expression of these mitochondrial biosynthesis markers, represented by PPARGC1A, at the protein level was determined using western blotting, and displayed the same trend as described above (Figures 2G,H).
Together, these results indicated that the direct effect of CYP11A1 upregulation was excessive mitochondrial biosynthesis, which could be rescued by the addition of vitamin D3.
To evaluate whether excessive mitochondrial biosynthesis would lead to increased levels of reactive oxygen species, we analyzed levels of malonaldehyde (MDA), a product of lipid peroxidation generated by ROS. Results showed that overexpression of CYP11A1 induced a significant increase in MDA levels that could be decreased and rescued by 2 μM and 5 μM vitamin D3, respectively, (Figure 3A). Then, we searched for the origin of the excessive ROS. Although most intracellular ROS are produced in the mitochondria, they can also be detected in other cell organelles and in the cytoplasm. To determine the level of ROS specifically produced by mitochondria, we used a MitoSOX™ Red mitochondrial ROS probe, which has higher selectivity and specificity for ROS in mitochondria compared with other ROS probes such as DCFH-DA. As expected, higher ROS levels (higher red fluorescence signals) were observed in the CYP11A1 overexpression group compared to those in the vehicle or control groups (Figure 3B). In addition, treatment with 5 μM vitamin D3 significantly reduced ROS levels (Figure 3B).
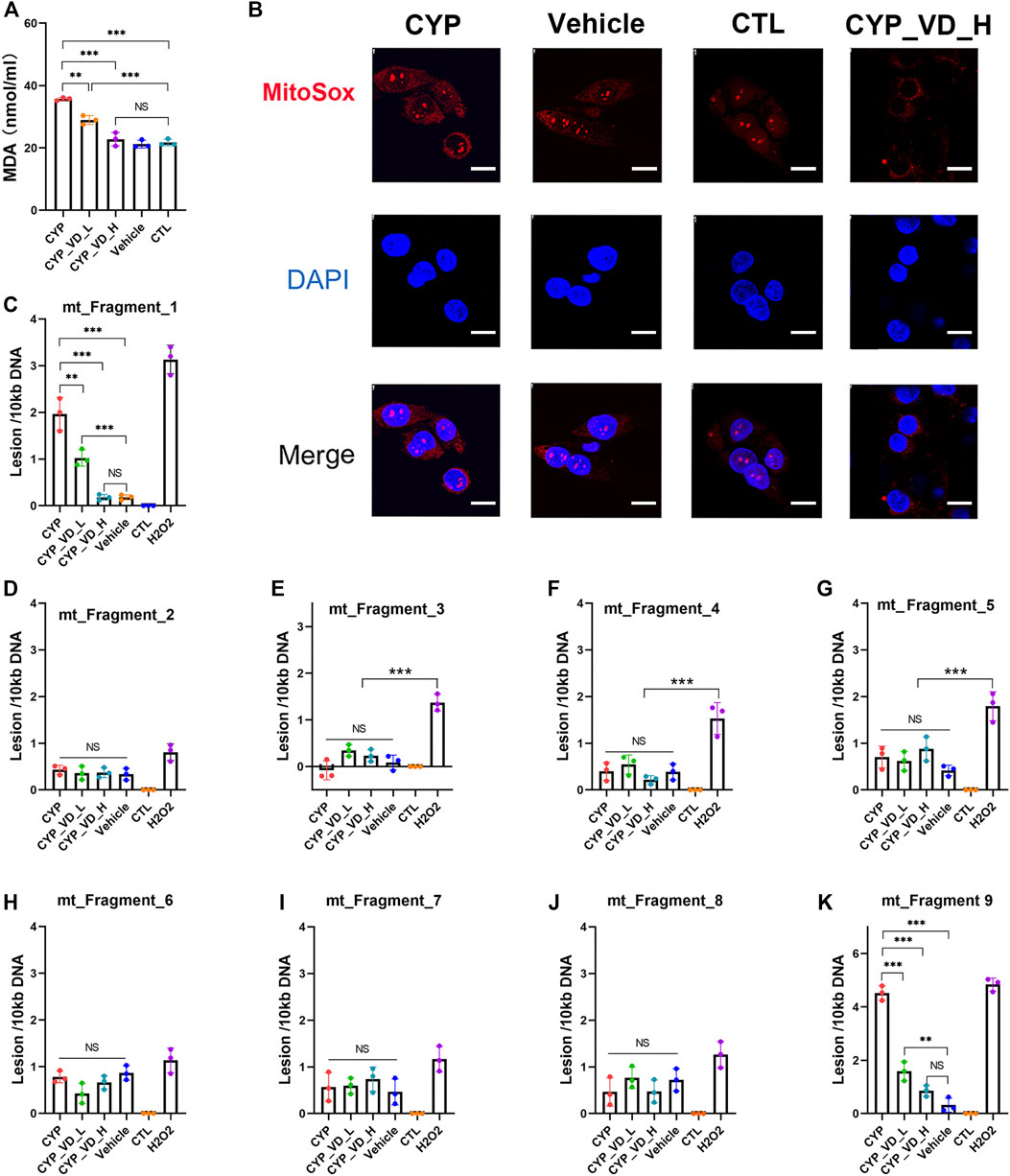
FIGURE 3. ROS accumulation and mtDNA damage induced by CYP11A1 overexpression could be rescued by vitamin D3in BeWo cells. (A) MDA production. (B) Mitochondrial ROS (red fluorescence). (C–K) mtDNA damage induced by ROS. CYP, BeWo cells overexpressing CYP11A1. Vehicle, BeWo cells transfected with vehicle. CTL, control. CYP_VD_L, BeWo cells overexpressing CYP11A1 and treated with a low-dose of vitamin D3 (2 μM). CYP_VD_H, BeWo cells overexpressing CYP11A1 and treated with a high-dose of vitamin D3 (5 μM). Statistical analysis was performed on the means from three independent experiments using one-way analysis of variance (ANOVA) with Tukey’s post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001; NS, no significance. Scale bars, 50 μm.
Intracellular ROS is known to attack lipids and frequently damage mtDNA. The level of mtDNA damage was determined using qPCR, which demonstrated a significantly higher rate of DNA lesions within fragment 1 and fragment 9 (ChrM: 15361–2036) in the CYP11A1 overexpression group. The level of mtDNA damage was similar to that observed in cells treated with 500 μM H2O2. In contrast, no obvious damage was observed in fragments 3–8, indicating that the mtDNA damage was likely caused by ROS. Following treatment with 2 μM and 5 μM vitamin D3, the lesion rates in fragment 1 and fragment 9 gradually decreased (Figures 3C–K).
In brief, excessive mitochondrial biosynthesis in BeWo cells induced by CYP11A1 overexpression could lead to abnormally high mitochondrial ROS levels, manifested by increased MDA content and MitoSOX fluorescence signals, and hence damaged the mtDNA. Furthermore, addition of vitamin D3 decreased the ROS levels.
Mitochondrial apoptosis occurs frequently, along with ROS and mtDNA damage, and can be assessed by checking the membrane potential. Results of JC-1 staining revealed significantly decreased levels of JC-1 aggregates (red fluorescence) and increased levels of JC-1 monomers (green fluorescence) in the CYP11A1 overexpression group compared to those in vehicle and control groups, indicating a reversed mitochondrial membrane potential. This tendency to undergo apoptosis was rescued by treatment with 5 μM vitamin D3 (Figure 4A).
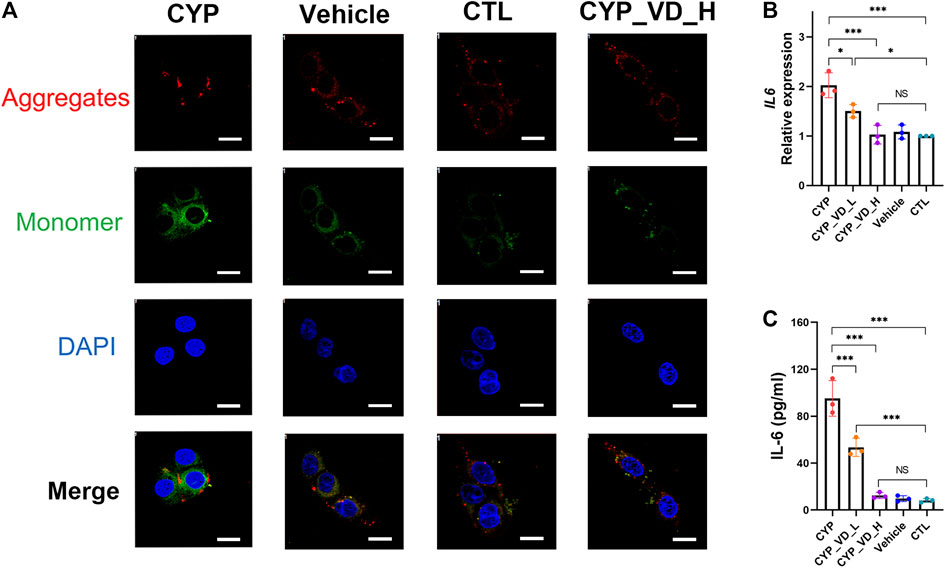
FIGURE 4. Trophoblast mitochondrial apoptosis and inflammation induced by upregulation of CYP11A1 could be ameliorated by vitamin D3addition. (A) JC-1 staining of BeWo mitochondria. Red fluorescence represents JC-1 aggregates, whereas green fluorescence represents JC-1 monomers. (B) Transcriptional levels of IL-6 in BeWo cells. (C) IL-6 released by BeWo cells. CYP, BeWo cells overexpressing CYP11A1. Vehicle, BeWo cells transfected with vehicle. CTL, control. CYP_VD_L, BeWo cells overexpressing CYP11A1 and treated with a low-dose of vitamin D3 (2 μM). CYP_VD_H, BeWo cells overexpressing CYP11A1 and treated with a high-dose of vitamin D3 (5 μM). Statistical analysis was performed on the means from three independent experiments using one-way analysis of variance (ANOVA) with Tukey’s post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001; NS, no significance. Scale bars, 50 μm.
To evaluate the effects of mitochondrial functional impairment on the trophoblast itself, we tested BeWo cells with an inflammatory cytokine panel that included interleukin (IL)-1β, IL-4, IL-6, IL-8, IL-10, IL-13, IL-17α, and IL-23, and tumor necrosis factor α (TNF-α) using a LuminexTM multiplex suspension array. Results showed that only IL-6 was present in the cells (data not shown). We further verified the production of IL-6 using qPCR and ELISA. Strikingly, the alteration in the inflammatory status of BeWo cells was verified. Furthermore, the expression and release of IL-6 was significantly increased in the CYP11A1 overexpression group compared to the vehicle or control groups. Moreover, after adding vitamin D3, the inflammation gradually decreased to regular levels (Figures 4B,C).
In this part, we examined adverse effects including mitochondrial apoptosis and cellular inflammation alterations, probably caused by increased mitochondrial ROS levels and subsequent mtDNA damage. The results showed that upregulation of CYP11A1 in BeWo cells may finally impair BeWo mitochondria and lead to inflammatory cytokines release. Addition of vitamin D3 rescued BeWo cells from these adverse effects.
To investigate the effects of the inhibition of proliferation and DNA damage on NSCs, NSCs were cultured for 12 h with neuronal maintenance medium (NMM) preconditioned by BeWo cells overexpressing CYP11A1 and with vitamin D3 treated. KI-67 immunostaining, which labels cells in the active phases of the cell cycle (G1, S, G2, and M phases), revealed a marked inhibition in proliferation of NSCs exposed to CYP11A1-upregulated BeWo cells; the inhibition of proliferation could be rescued by treatment with vitamin D3 (Figure 5A). This result was verified using the Cell Counting Kit-8 (CCK-8) assay (Figure 5B).
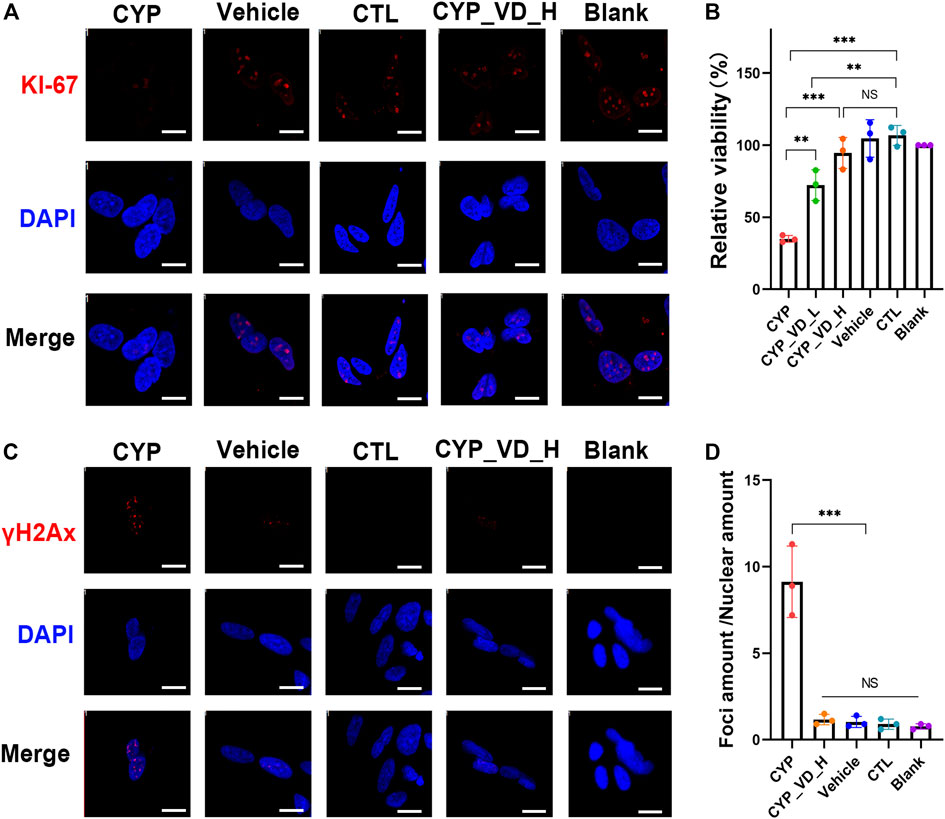
FIGURE 5. Inhibition of NSC proliferation and DNA damage induced by the conditioned medium of BeWo cells overexpressing CYP11A1 and intervention by vitamin D3. (A) KI-67 immunostaining in NSCs. (B) CCK-8 proliferation assay. (C, D) γH2AX immunostaining in NSCs. CYP, BeWo cells overexpressing CYP11A1. Vehicle, BeWo cells transfected with vehicle. CTL, control. CYP_VD_L, BeWo cells overexpressing CYP11A1 and treated with a low-dose of vitamin D3 (2 μM). CYP_VD_H, BeWo cells overexpressing CYP11A1 and treated with a high-dose of vitamin D3 (5 μM). Statistical analysis was performed on the means from three independent experiments using one-way analysis of variance (ANOVA) with Tukey’s post hoc test. ***P < 0.001. Scale bars, 20 μm.
To assess DNA damage in NSCs, we performed γH2AX immunofluorescence assays. When double-strand breaks (DSBs) occur in DNA, phosphorylation of H2AX at serine 139 is induced, resulting in the production of γH2AX. Immunofluorescence staining using anti-γH2AX antibodies reveals the formation of distinct foci around DSBs. Our results showed a highly significant increase in the quantity of γH2AX foci in NSCs exposed to BeWo cells expressing CYP11A1, indicating increased levels of DNA damage in these NSCs. Notably, vitamin D3 treatment effectively rescued this damage (Figures 5C,D).
In brief, here we try to extrapolate the adverse effects induced by overexpression of CYP11A1 in BeWo cells through culturing NSCs in BeWo conditioned medium. Results showed that the proliferation of NSCs was inhibited and DNA damage was observed. By adding vitamin D3 during medium conditioning, the damage to NSCs could be reduced.
In this study, we demonstrated that the overexpression of CYP11A1 in trophoblasts increased mitochondrial steroid synthesis resulting in a significant increase in the secretion of the direct product pregnenolone, as well as the indirect product progesterone. To support these metabolic changes, biosynthesis in mitochondria is passively increased. The observed increase in the expression levels of mitochondrial biosynthesis markers PPARGC1A, NRF1, and TFAM further supports this mechanism. Because of their vital role in transportation and hormone synthesis during pregnancy, the mitochondrial copy number and biosynthetic activities are originally higher in trophoblasts than in many other tissues (Miller, 2013; Martinez et al., 2015). Therefore, further increasing the mitochondrial activity inevitably leads to various impairments, such as ROS accumulation and consequent mtDNA damage. In this study, the damage to mtDNA caused by ROS was most distinct in ChrM: 15361–369, which harbors most of the mitochondrial D-loop (ChrM: 16024–576). This result is in agreement with those of previous studies (Rothfuss et al., 2010). The promoter of the heavy and light strands of the mitochondrial genome is located in this region (Sharma et al., 2005; Barshad et al., 2018), indicating that this damage can significantly affect mitochondrial function and cellular metabolic behavior, including mitochondrial genomic mutation, mitochondrial autophagy, and apoptosis. These findings further explain the underlying mechanism of trophoblast cell autophagy and apoptosis, triggered by the upregulation of CYP11A1 (Pan et al., 2017), and contribute to our understanding of the association between CYP11A1 and preeclampsia.
Although the pathogenesis of preeclampsia has not been fully elucidated, it is thought that inflammation plays a central role (Wolf et al., 2001; LaMarca et al., 2007; Ruma et al., 2008; Harmon et al., 2016). Previous studies have shown that autophagy, apoptosis, and necrosis occur in trophoblast cells and that secretion of inflammatory factors, especially IL-6, increases dramatically in preeclampsia (Mulla et al., 2009; Guinazu et al., 2012; Hawkins et al., 2018). In our study, although levels of other inflammatory cytokines were not altered, IL-6 increased over nine times in cells overexpressing CYP11A1 compared to the control cells, indicating that IL-6 expression is specifically induced by CYP11A1 overexpression in trophoblasts. In addition to preeclampsia, inflammation in trophoblasts has been associated with fetal developmental abnormalities like intrauterine growth restriction and neurodevelopmental disorders, including neurodevelopmental delay, cognitive impairment, and autism (Challis et al., 2009; Boutsikou et al., 2010; Goeden et al., 2016; Osokine and Erlebacher, 2017; Tang et al., 2017; Rudolph et al., 2018). A recent report indicated that inflammation was the mediator of the effect of any prenatal environmental adversity on fetal neurodevelopmental delay (Girchenko et al., 2020). In our study, the exposure of NSCs to medium conditioned with BeWo cells overexpressing CYP11A1 showed not only significant cell cycle arrest in the G0 phase but also remarkable amount of double-strand DNA damage. These results may reveal the indirect effects of CYP11A1 upregulation on fetal neurodevelopment (Figure 6).
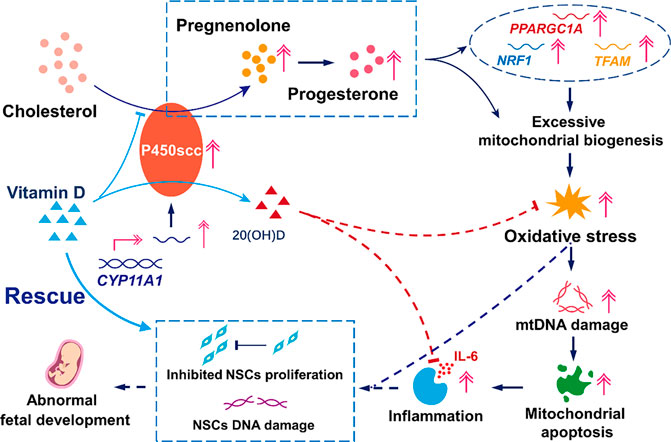
FIGURE 6. Model mechanism of the effects of upregulation of CYP11A1 in BeWo cells and amelioration of these effects by Vitamin D. The upregulation of CYP11A1 resulted in excessive mitochondrial biosynthesis, which triggered oxidative stress and led to trophoblast mitochondrial apoptosis and inflammation. This functional change in trophoblasts led to impaired NSCs characterized by inhibited proliferation and increased levels of DNA damage. Vitamin D supplementation proved to be an efficient intervention for the effects of CYP11A1 upregulation, mainly via its competitive binding to P450scc and the anti-inflammatory effect due to the subsequent production of 20(OH)D.
Considering the unique pathway of vitamin D metabolism by P450scc, we explored the effect of this nutrient factor in cells overexpressing CYP11A1. Interestingly, vitamin D intervened in the signaling cascade and rescued the function of NSCs. This ability may be attributed to two mechanisms: competitive binding to excess P450scc with cholesterol and the anti-inflammatory and antioxidant effects of its product, 20(OH)D (Janjetovic et al., 2009; Janjetovic et al., 2010; Slominski et al., 2014; Slominski et al., 2015), the detailed mechanisms of which warrant further investigation.
To date, several clinical trials have reported the preventive effects or correlation of vitamin D supplementation in gestational diabetes, premature delivery, neurodevelopmental disorders, and preeclampsia during pregnancy, although some results remain controversial (Mojibian et al., 2015; Perez-Lopez et al., 2015; Fort et al., 2016; Zhang et al., 2016a; Zhou et al., 2017; Curtis et al., 2018). Combined with our findings, we presume that although the pathogeneses of these diseases have not been fully elucidated, trophoblast cell damage and inflammation caused by upregulation of CYP11A1 expression might be involved and may be triggered by genetic factors or intrauterine exposure to adverse factors. It is assumed that vitamin D would be effective in treating these cases, but not in cases where CYP11A1 upregulation is not involved. Additionally, vitamin supplement dosage is an important factor, indicating that population background, environment, diet, and even vitamin D receptor genotypes should be considered for precise and individualized supplementation (Ferrari et al., 1998; Niimi et al., 2000; Touvier et al., 2015).
In summary, we elucidated the underlying mechanism of trophoblast damage caused by the upregulation of CYP11A1, which could be induced by toxins, drugs, hormones, or genetic variants. Furthermore, this upregulation of CYP11A1 induced trophoblast damage impaired NSCs in vitro. If extrapolated to a clinical situation, these effects may lead to neurodevelopmental disorders. Furthermore, we found that vitamin D supplementation was an efficient intervention method to rescue the impairment described above and provided additional evidence to support vitamin D supplementation during pregnancy for the prevention of diseases.
Our in vitro study has limitations for extrapolating cell line results to primary trophoblast cells or in vivo studies. In particular, the mechanism of NSC damage requires further investigation. Considering the major structural and functional differences in the placenta between human and rodent models, the “human placenta-on-a-chip” device may provide an important model for future studies (Cox et al., 2009; Robbins et al., 2010; Schmidt et al., 2015; Blundell et al., 2018).
BeWo cells (CCL-98, ATCC) were cultured in F12K medium (21127022, Gibco) containing 12% fetal bovine serum (FBS, 10270106, Gibco) and 1% penicillin/streptomycin (SV30010, Hyclone). Cells were divided into three groups: group 1 (CYP) was transfected with CYP11A1 (NM_000781) overexpression lentivirus (GENECHEM) with a multiplicity of infection (MOI) of 25, group 2 (vehicle) was transfected with the equivalent negative lentivirus as a vector, and group 3 was not transfected and was used as the control (CTL). After green fluorescent protein (GFP) was expressed, puromycin (2.5 μg/ml final concentration) was added to select the stable cell clones, and the expression of CYP11A1 at the mRNA and protein level was analyzed using qPCR and western blotting, respectively.
After overexpression of CYP11A1, BeWo cell syncytialization was induced by the addition of 25 μM forskolin (F127328, Aladdin) for 48 h. The medium was then replaced with neuronal maintenance medium (NMM), containing 50% DMEM/F12 GlutaMax (31331028, Gibco), 50% neurobasal medium (21103049, Gibco), 0.5% GlutaMAX supplement (35050038, Gibco), 1 × B27 supplement (17504044, Gibco), 1 × N2 supplement (17502048, Gibco), 1% penicillin/streptomycin (SV30010, Hyclone), 10 ng/ml b-FGF (PHG0263, Gibco), and 10 ng/ml EGF (PHG0311, Gibco). Vitamin D3 was added at concentrations of 0, 2, or 5 μM to the BeWo culture of group CYP, whereas NMM plus the equivalent volumes of DMSO were added to the vehicle and CTL groups for 24 h.
The cell culture supernatant, named conditioned NMM were harvested, carefully centrifuged at 600 x g for 5 min, filtered through a 0.22 μm syringe filter to remove cells and debris, and stored at −80°C until further use. Then, the levels of secreted pregnenolone, progesterone, and inflammatory cytokines were analyzed.
Meanwhile, the treated BeWo cells were harvested to extract DNA, RNA, and protein. The expression levels of PPARGC1A, NRF1, TFAM, and IL-6 were determined at the mRNA level using qPCR, and PGC-1α expression at the protein level was determined using western blotting. The level of mtDNA damage was assessed using qPCR, and mitochondrial ROS and apoptosis were assessed through MitoSox and JC-1 staining, respectively.
NSCs differentiated from human-induced pluripotent stem cells (hiPSCs) that were derived from human peripheral blood, were obtained from BioTalentum Ltd. The NSCs were seeded into laminin- (L2020, Sigma) and poly-L-ornithine- (P4957, Sigma) coated plates and cultured in NMM. After cell confluence reached 50%, the culture medium was replaced with conditioned NMM, as described above, or fresh NMM and cultured for 12 h. KI-67 immunofluorescence staining was performed to analyze NSC proliferation, and the results were confirmed using CCK-8 assays. γH2AX staining and immunofluorescence was performed to detect the amount of DSBs in the DNA.
The antibodies used in the western blotting experiments were anti-CYP11A1 (1:200, 13363-1-AP, Proteintech), anti-PGC1α (1:200, AF5395, Affinity), anti-DDDDK Tag (1:1000, 66008-3-Ig, Proteintech), anti-GAPDH (1:5000, EM1101, Huabio), HRP-conjugated goat anti-mouse IgG (1:5000, ZB-2305, Zsbio), and HRP-conjugated goat anti-rabbit IgG (1:5000, ZB-2301, Zsbio). The antibodies used for the immunofluorescence experiments were anti-KI-67 (1:50, 27309-1-AP, Proteintech), anti-γH2AX (1:400, ab26350, Abcam), goat anti-mouse antibody, Alexa Fluor 594 (1:1000, A11005, Invitrogen), and goat anti-rabbit antibody, Alexa Fluor 594 (1:1000, A11012, Invitrogen).
Cells were lysed using radioimmunoprecipitation assay (RIPA) buffer (P0013C, Beyotime) containing a protease and phosphatase inhibitor cocktail (B14011 and B15001, respectively, Bimake) to extract the total protein. Equal amounts of denatured protein samples were separated on 10% SDS-polyacrylamide gels and transferred to a polyvinylidene difluoride (PVDF) membrane (IPVH00010, Millipore) for immunoblot analysis. GAPDH was used as an internal control, and the intensity of bands was analyzed using Image J software (version 1.5).
For JC-1 (C2006, Beyotime) and MitoSox (M36008, Invitrogen) staining, BeWo cells were cultured on glass coverslips and washed with 1 × Hank's balanced salt solution (HBSS) (14065056, Gibco). JC-1 or MitoSox was loaded onto the cells and incubated for 15 min at 37°C, protected from light. The cells were then washed three times with warm 1 × HBSS and counterstained with 4,6-diamidino-2-phenylindole (DAPI, P0131, Beyotime) to label the nuclei. At least five quality images were acquired using a laser scanning confocal microscope (Olympus FV2000).
For immunofluorescence, NSCs were cultured on glass coverslips, washed with Dulbecco’s phosphate-buffered saline (DPBS) (14190144, Gibco), fixed with 4% paraformaldehyde, permeabilized with 0.3% Triton X-100 (T8200, Solarbio), blocked with 5% bovine serum albumin (BSA, 4240GR500, Biofroxx) in PBS, and incubated with primary antibodies overnight at 4°C. Then, the slides were washed in PBS, incubated with Alexa Fluor 594-labeled secondary antibodies for 1 h at room temperature, and counterstained with DAPI to label the nuclei. At least five quality images were acquired using a laser scanning confocal microscope (Olympus FV2000).
NSC proliferation was measured using the CCK-8 (Abmole, M4839) assay. NSCs were seeded into laminin- and poly-L-ornithine-coated 96-well plates at a density of 5,000 cells per well and cultured with NMM for 24 h. The medium was then replaced with fresh or BeWo-conditioned NMM, as described above, and cultured further for 12 h. Then, 10 μl of CCK-8 solution was added to each well and incubated for another 4 h at 37°C. Absorbance was measured at 450 nm to determine cell viability, using a microplate reader (Cmax Plus, Molecular Devices).
BeWo-conditioned NMM, as described above, was collected, and stored at −80°C until further use. Pregnenolone levels were determined using an ELISA kit (FR E-2700, LDN) and readout wavelength of 450 nm on a microplate reader (Cmax Plus, Molecular Devices). Progesterone analysis was performed in-house, using the ADVIA Centaur XP Immunoassay System (Siemens, Germany). MDA levels were assessed using an assay kit (A003-1-2, Nanjing Jiancheng) based on thiobarbituric acid (TBA). IL-1β, IL-4, IL-6, IL-8, IL-10, IL-13, IL-17α, IL-23, and TNF-α levels were analyzed using a suspension array (MAGPX12157001, Luminex). IL-6 levels were further verified by using an ELISA kit (E-EL-H0102, Elabscience) and readout wavelength of 450 nm on a microplate reader (Cmax Plus, Molecular Devices). All analyses were performed according to the manufacturers’ instructions.
BeWo cells were cultured and treated as described earlier, and total RNA was extracted using TRIzol reagent (Invitrogen, 15596026). A NanoDrop 2000 (Thermo Scientific) was used to measure the amount of total RNA in each sample, and 500 ng of total RNA was subjected to reverse transcription (RT) using an RT reagent kit (Takara, RR037A) to form cDNA. Quantitative PCR for cDNA was performed in triplicate using SYBR Green qPCR Master Mix (Bimake, B21202) on an iCycler RT-PCR detection system (Bio-Rad Laboratories). GAPDH was used as an internal control. Delta-delta Ct value analysis was used to evaluate the relative expression of target genes. Primer pairs were designed using Primer3plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) and are listed in Table 1.
BeWo cells were cultured and treated as described earlier. Total genomic DNA was extracted using a specialized DNA extraction kit (DP304-03, TIANGEN) according to the manufacturer’s instructions. Treatment of normal BeWo cells with 500 μM H2O2 for 30 min was used as a positive control.
According to the method of Oliver (Rothfuss et al., 2010) and Artem et al. (Gureev et al., 2017), mtDNA damage was estimated based on semi-long run qPCR. Briefly, 9 primer pairs were used to amplify 9 long mtDNA fragments, ranging in size from 1,300 to 2,400 bp, whereas another pair of primers was used to amplify a short fragment ranging in size from 68 to 140 bp, located at the beginning of each long fragment (Figure 7). We used the primer pairs previously described by Artem et al. (Gureev et al., 2017) and listed in Table 2. For amplification, 5 ng of total DNA was utilized to perform qPCR using SYBR Green qPCR Master Mix (Bimake, B21202) on an iCycler RT-PCR Detection System (Bio-Rad Laboratories). The reaction conditions were: pre-incubation for 5 min at 95°C followed by 40 cycles of 95°C for 10 s, 61°C for 30 s, and 72°C for 10 s (small fragments) or 90–120 s (long fragments).
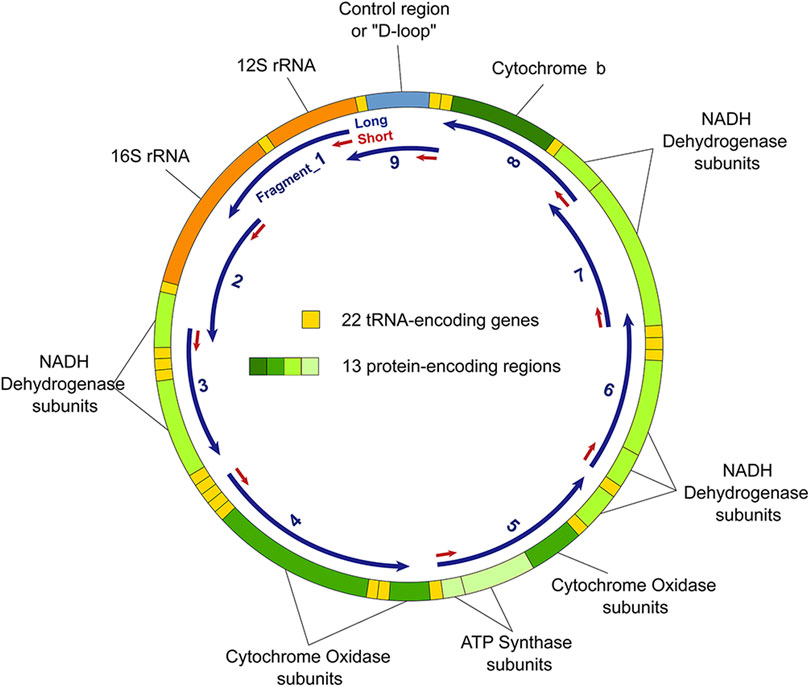
FIGURE 7. Mitochondrial DNA (mtDNA) and fragments used to estimate mtDNA damage. Fragment_1, ChrM: 298–2036; Fragment_2, ChrM: 2078–3403; Fragment_3, ChrM: 3318–4992; Fragment_4, ChrM: 5784–7517; Fragment_5, ChrM: 7858–9763; Fragment_6, ChrM: 9895–11856; Fragment_7, ChrM: 11775–13717; Fragment_8, ChrM: 13650–15381; and Fragment_9, ChrM: 15361–369.
To calculate the DNA lesions in each long fragment, delta Ct (every other group versus CTL) of the long fragment (Δlong) was compared to the delta Ct of its corresponding short fragment (Δshort):
Data were analyzed using GraphPad Prism software package (version 8.4; La Jolla, United States) and are presented as Mean ± SEM. One-way ANOVA test was used to assess differences among more than two groups and Tukey’s post hoc test was used to determine the difference between each two groups. A P-value <0.05 was considered statistically significant.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
XW designed the experiments, performed cell staining, and wrote the manuscript. ML cultured cells and collected data. XZ and YL performed the qPCR and western blot experiments. AD provided NSCs, guided cell culturing, and revised the manuscript. QS and GH revised the manuscript and supported the study. WX designed and supervised the experiments. All authors revised and approved the article.
This study was supported by National Key R&D Program of China (2018YFC1002804 and 2019YFE0103804), International Cooperation Project of Sichuan Science and Technology Department (2018HH0029), Sichuan Key Research and Development (2019YFS0411) and Chinese-Hungarian Bilateral Project (2018-2.1.14-TÉT-CN-2018-00011, Chinese No. 8-4).
Author AD was employed by the company BioTalentum Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2020.608447/full#supplementary-material.
Barshad, G., Marom, S., Cohen, T., and Mishmar, D. (2018). Mitochondrial DNA transcription and its regulation: an evolutionary perspective. Trends Genet. 34 (9), 682–692. doi:10.1016/j.tig.2018.05.009
Beaudoin, C., Blomquist, C. H., Bonenfant, M., and Tremblay, Y. (1997). Expression of the genes for 3 beta-hydroxysteroid dehydrogenase type 1 and cytochrome P450scc during syncytium formation by human placental cytotrophoblast cells in culture and the regulation by progesterone and estradiol. J. Endocrinol. 154 (3), 379–387. doi:10.1677/joe.0.1540379
Blundell, C., Yi, Y. S., Ma, L., Tess, E. R., Farrell, M. J., Georgescu, A., et al. (2018). Placental drug transport-on-a-chip: a microengineered in vitro model of transporter-mediated drug efflux in the human placental barrier. Adv. Healthc. Mater 7 (2), 1700786. doi:10.1002/adhm.201700786
Boutsikou, T., Mastorakos, G., Kyriakakou, M., Margeli, A., Hassiakos, D., Papassotiriou, I., et al. (2010). Circulating levels of inflammatory markers in intrauterine growth restriction. Mediat. Inflamm. 2010, 790605. doi:10.1155/2010/790605
Challis, J. R., Lockwood, C. J., Myatt, L., Norman, J. E., Strauss, J. F., and Petraglia, F. (2009). Inflammation and pregnancy. Reprod. Sci. 16 (2), 206–215. doi:10.1038/s41593-018-0128-y
Christenson, L. K., and Strauss, J. F. (2000). Steroidogenic acute regulatory protein (StAR) and the intramitochondrial translocation of cholesterol. Biochim. Biophys. Acta 1529 (1–3), 175–187. doi:10.1016/s1388-1981(00)00147-5
Churchill, P. F., and Kimura, T. (1979). Topological studies of cytochromes P-450scc and P-45011 beta in bovine adrenocortical inner mitochondrial membranes. Effects of controlled tryptic digestion. J. Biol. Chem. 254 (20), 10443–10448. doi:10.1016/S0021-9258(19)86728-5
Cox, B., Kotlyar, M., Evangelou, A. I., Ignatchenko, V., Ignatchenko, A., Whiteley, K., et al. (2009). Comparative systems biology of human and mouse as a tool to guide the modeling of human placental pathology. Mol. Syst. Biol. 5 (1), 279. doi:10.1038/msb.2009.37
Curtis, E. M., Moon, R. J., Harvey, N. C., and Cooper, C. (2018). Maternal vitamin D supplementation during pregnancy. Br. Med. Bull. 126 (1), 57–77. doi:10.1093/bmb/ldy010
Enquobahrie, D. A., Meller, M., Rice, K., Psaty, B.M., Siscovick, D. S., and Williams, M. A. (2008). Differential placental gene expression in preeclampsia. Am. J. Obstet. Gynecol. 199(5), 566–511. doi:10.1016/j.ajog.2008.04.020
Ferrari, S., Bonjour, J. P., and Rizzoli, R. (1998). The vitamin D receptor gene and calcium metabolism. Trends Endocrinol. Metab. 9 (7), 259–265. doi:10.1016/s1043-2760(98)00065-4
Fogacci, S., Fogacci, F., Banach, M., Michos, E. D., Hernandez, A. V., Lip, G. Y. H., et al. (2020). Vitamin D supplementation and incident preeclampsia: a systematic review and meta-analysis of randomized clinical trials. Clin. Nutr. 39 (6), 1742–1752. doi:10.1016/j.clnu.2019.08.015
Fort, P., Salas, A. A., Nicola, T., Craig, C. M., Carlo, W. A., and Ambalavanan, N. (2016). A Comparison of 3 vitamin D dosing regimens in extremely preterm infants: a randomized controlled trial. J. Pediatr. 174, 132–138. doi:10.1016/j.jpeds.2016.03.028
Girchenko, P., Lahti-Pulkkinen, M., Heinonen, K., Reynolds, R. M., Laivuori, H., Lipsanen, J., et al. (2020). Persistently high levels of maternal antenatal inflammation are associated with and mediate the effect of prenatal environmental adversities on neurodevelopmental delay in the offspring. Biol. Psychiat. 87 (10), 898–907. doi:10.1016/j.biopsych.2019.12.004
Goeden, N., Velasquez, J., Arnold, K. A., Chan, Y., Lund, B. T., Anderson, G. M., et al. (2016). Maternal inflammation disrupts fetal neurodevelopment via increased placental output of serotonin to the fetal brain. J. Neurosci. 36 (22), 6041–6049. doi:10.1523/JNEUROSCI.2534-15.2016
Guinazu, N., Rena, V., Genti-Raimondi, S., Rivero, V., and Magnarelli, G. (2012). Effects of the organophosphate insecticides phosmet and chlorpyrifos on trophoblast JEG-3 cell death, proliferation and inflammatory molecule production. Toxicol. In Vitro 26 (3), 406–413. doi:10.1016/j.tiv.2012.01.003
Gureev, A. P., Shaforostova, E. A., Starkov, A. A., and Popov, V. N. (2017). Simplified qPCR method for detecting excessive mtDNA damage induced by exogenous factors. Toxicology 382, 67–74. doi:10.1016/j.tox.2017.03.010
Guryev, O., Carvalho, R. A., Usanov, S., Gilep, A., and Estabrook, R. W. (2003). A pathway for the metabolism of vitamin D3: unique hydroxylated metabolites formed during catalysis with cytochrome P450scc (CYP11A1). Proc. Natl. Acad. Sci. U S A 100 (25), 14754–14759. doi:10.1073/pnas.2336107100
Hanukoglu, I. (1992). Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis. J. Steroid. Biochem. Mol. Biol. 43 (8), 779–804. doi:10.1016/0960-0760(92)90307-5
Hanukoglu, I., Suh, B. S., Himmelhoch, S., and Amsterdam, A. (1990). Induction and mitochondrial localization of cytochrome P450scc system enzymes in normal and transformed ovarian granulosa cells. J. Cell. Biol. 111 (4), 1373–1381. doi:10.1083/jcb.111.4.1373
Harmon, A. C., Cornelius, D. C., Amaral, L. M., Faulkner, J. L., Cunningham, M. W., Wallace, K., et al. (2016). The role of inflammation in the pathology of preeclampsia. Clin. Sci. 130 (6), 409–419. doi:10.1042/CS20150702
Hawkins, S. J., Crompton, L. A., Sood, A., Saunders, M., Boyle, N. T., Buckley, A., et al. (2018). Nanoparticle-induced neuronal toxicity across placental barriers is mediated by autophagy and dependent on astrocytes. Nat. Nanotechno.l 13 (5), 427–433. doi:10.1038/s41565-018-0085-3
He, G., Xu, W., Chen, Y., Liu, X., and Xi, M. (2013). Abnormal apoptosis of trophoblastic cells is related to the up-regulation of CYP11A gene in placenta of preeclampsia patients. PloS One 8 (3), e59609. doi:10.1371/journal.pone.0059609
Hogg, K., Blair, J. D., McFadden, D. E., von Dadelszen, P., and Robinson, W. P. (2013). Early onset pre-eclampsia is associated with altered DNA methylation of cortisol-signalling and steroidogenic genes in the placenta. PLoS One 8 (5), e62969. doi:10.1371/journal.pone.0062969
Janjetovic, Z., Tuckey, R. C., Nguyen, M. N., Thorpe, E. M., and Slominski, A. T. (2010). 20,23-dihydroxyvitamin D3, novel P450scc product, stimulates differentiation and inhibits proliferation and NF-kappaB activity in human keratinocytes. J. Cell Physio.l 223 (1), 36–48. doi:10.1002/jcp.21992
Janjetovic, Z., Zmijewski, M. A., Tuckey, R. C., DeLeon, D. A., Nguyen, M. N., Pfeffer, L. M., et al. (2009). 20-Hydroxycholecalciferol, product of vitamin D3 hydroxylation by P450scc, decreases NF-kappaB activity by increasing IkappaB alpha levels in human keratinocytes. PLoS One 4 (6), e5988. doi:10.1371/journal.pone.0005988
LaMarca, B. D., Ryan, M. J., Gilbert, J. S., Murphy, S. R., and Granger, J. P. (2007). Inflammatory cytokines in the pathophysiology of hypertension during preeclampsia. Curr. Hypertens Rep. 9 (6), 480–485. doi:10.1007/s11906-007-0088-1
Martinez, F., Olvera-Sanchez, S., Esparza-Perusquia, M., Gomez-Chang, E., and Flores-Herrera, O. (2015). Multiple functions of syncytiotrophoblast mitochondria. Steroids 103, 11–22. doi:10.1016/j.steroids.2015.09.006
Miller, W. L. (2007). Steroidogenic acute regulatory protein (StAR), a novel mitochondrial cholesterol transporter. Biochim. Biophys. Acta 1771 (6), 663–676. doi:10.1016/j.bbalip.2007.02.012
Miller, W. L. (2013). Steroid hormone synthesis in mitochondria. Mol. Cell Endocrinol. 379 (1–2), 62–73. doi:10.1016/j.mce.2013.04.014
Mojibian, M., Soheilykhah, S., Fallah Zadeh, M. A., and Jannati Moghadam, M. (2015). The effects of vitamin D supplementation on maternal and neonatal outcome: a randomized clinical trial. Iran. J. Reprod. Med. 13 (11), 687–696
Mulla, M. J., Brosens, J. J., Chamley, L. W., Giles, I., Pericleous, C., Rahman, A., et al. (2009). Antiphospholipid antibodies induce a pro-inflammatory response in first trimester trophoblast via the TLR4/MyD88 pathway. Am. J. Reprod. Immunol. 62 (2), 96–111. doi:10.1111/j.1600-0897.2009.00717.x
Niimi, T., Tomita, H., Sato, S., Akita, K., Maeda, H., Kawaguchi, H., et al. (2000). Vitamin D receptor gene polymorphism and calcium metabolism in sarcoidosis patients. Sarcoid. Vasc. Diffuse Lung Dis. 17 (3), 266–269.
Osokine, I., and Erlebacher, A. (2017). Inflammation and autism: from maternal gut to fetal brain. Trends Mol. Med. 23 (12), 1070–1071. doi:10.1016/j.molmed.2017.10.008
Pan, T., He, G., Chen, M., Bao, C., Chen, Y., Liu, G., et al. (2017). Abnormal CYP11A1 gene expression induces excessive autophagy, contributing to the pathogenesis of preeclampsia. Oncotarget 8 (52), 89824–89836. doi:10.18632/oncotarget.21158
Perez-Lopez, F. R., Pasupuleti, V., Mezones-Holguin, E., Benites-Zapata, V. A., Thota, P., Deshpande, A., et al. (2015). Effect of vitamin D supplementation during pregnancy on maternal and neonatal outcomes: a systematic review and meta-analysis of randomized controlled trials. Fertil. Steril. 103 (5), 1278–1288. doi:10.1016/j.fertnstert.2015.02.019
Robbins, J. R., Skrzypczynska, K. M., Zeldovich, V. B., Kapidzic, M., and Bakardjiev, A. I. (2010). Placental syncytiotrophoblast constitutes a major barrier to vertical transmission of Listeria monocytogenes. PLoS Pathog. 6 (1), e1000732. doi:10.1371/journal.ppat.1000732
Rothfuss, O., Gasser, T., and Patenge, N. (2010). Analysis of differential DNA damage in the mitochondrial genome employing a semi-long run real-time PCR approach. Nucleic. Acids Res. 38 (4), e24. doi:10.1093/nar/gkp1082
Rudolph, M. D., Graham, A. M., Feczko, E., Miranda-Dominguez, O., Rasmussen, J. M., Nardos, R., et al. (2018). Maternal IL-6 during pregnancy can be estimated from newborn brain connectivity and predicts future working memory in offspring. Nat. Neurosci. 21 (5), 765–772. doi:10.1038/s41593-018-0128-y
Ruma, M., Boggess, K., Moss, K., Jared, H., Murtha, A., Beck, J., et al. (2008). Maternal periodontal disease, systemic inflammation, and risk for preeclampsia. Am. J. Obstet. Gynecol. 198 (4), 389–385. doi:10.1016/j.ajog.2007.12.002
Schmidt, A., Morales-Prieto, D. M., Pastuschek, J., Fröhlich, K., and Markert, U. R. (2015). Only humans have human placentas: molecular differences between mice and humans. J. Reprod. Immunol. 108, 65–71. doi:10.1016/j.jri.2015.03.001
Sharma, H., Singh, A., Sharma, C., Jain, S. K., and Singh, N. (2005). Mutations in the mitochondrial DNA D-loop region are frequent in cervical cancer. Canc. Cell Int. 5, 34. doi:10.1186/1475-2867-5-34
Slominski, A. T., Janjetovic, Z., Kim, T.-K., Wasilewski, P., Rosas, S., Hanna, S., et al. (2015). Novel non-calcemic secosteroids that are produced by human epidermal keratinocytes protect against solar radiation. J. Steroid Biochem. Mol. Biol. 148, 52–63. doi:10.1016/j.jsbmb.2015.01.014
Slominski, A. T., Kim, T. K., Li, W., Yi, A.K., Postlethwaite, A., and Tuckey, R. C. (2014). The role of CYP11A1 in the production of vitamin D metabolites and their role in the regulation of epidermal functions. J. Steroid Biochem. Mol. Biol. 144 (Pt A), 28–39. doi:10.1016/j.jsbmb.2013.10.012
Stocco, D. M. (1999). Steroidogenic acute regulatory (StAR) protein: what's new? Bioessays, 21, 768–775. doi:10.1002/(SICI)1521-1878(9199909)21:9<768::AID-BIES8>3.0.CO;2-2
Strauss, J. F., Martinez, F., and Kiriakidou, M. (1996). Placental steroid hormone synthesis: unique features and unanswered questions. Biol. Reprod. 54 (2), 303–311. doi:10.1095/biolreprod54.2.303
Tang, L., He, G., Liu, X., and Xu, W. (2017). Progress in the understanding of the etiology and predictability of fetal growth restriction. Reproduction 153 (6), R227–R240. doi:10.1530/REP-16-0287
Touvier, M., Deschasaux, M., Montourcy, M., Sutton, A., Charnaux, N., Kesse-Guyot, E., et al. (2015). Determinants of vitamin D status in Caucasian adults: influence of sun exposure, dietary intake, sociodemographic, lifestyle, anthropometric, and genetic factors. J. Invest. Dermatol. 135 (2), 378–388. doi:10.1038/jid.2014.400
Tuckey, R. C. (2005). Progesterone synthesis by the human placenta. Placenta 26 (4), 273–281. doi:10.1016/j.placenta.2004.06.012
Wang, F., Zhang, Q., Zhang, X., Luo, S., Ye, D., Guo, Y., et al. (2014). Preeclampsia induced by cadmium in rats is related to abnormal local glucocorticoid synthesis in placenta. Reprod. Biol. Endocrinol. 12, 77. doi:10.1186/1477-7827-12-77
Wolf, M., Kettyle, E., Sandler, L., Ecker, J. L., Roberts, J., and Thadhani, R. (2001). Obesity and preeclampsia: the potential role of inflammation. Obstet. Gynecol. 98 (5 Pt 1), 757–762. doi:10.1016/s0029-7844(01)01551-4
Zbytek, B., Janjetovic, Z., Tuckey, R. C., Zmijewski, M. A., Sweatman, T. W., Jones, E., et al. (2008). 20-Hydroxyvitamin D3, a product of vitamin D3 hydroxylation by cytochrome P450scc, stimulates keratinocyte differentiation. J. Invest. Dermatol. 128 (9), 2271–2280. doi:10.1038/jid.2008.62
Zhang, K., Hu, M., Zhang, L., Zhang, Q., and Huang, Y. (2019). The effect of BML-111 in preeclampsia rat model induced by the low dose of cadmium chloride. AJP Rep. 9 (3), e201–e208. doi:10.1055/s-0039-1693016
Zhang, Q., Cheng, Y., He, M., Li, T., Ma, Z., and Cheng, H. (2016a). Effect of various doses of vitamin D supplementation on pregnant women with gestational diabetes mellitus: a randomized controlled trial. Exp. Therap. Med. 12 (3), 1889–1895. doi:10.3892/etm.2016.3515
Zhang, Q., Huang, Y., Zhang, K., Huang, Y., Yan, Y., Wang, F., et al. (2016b). Cadmium-induced immune abnormality is a key pathogenic event in human and rat models of preeclampsia. Environ. Pollut. 218, 770–782. doi:10.1016/j.envpol.2016.07.073
Keywords: CYP11A1, P450scc, neuronal stem cells, trophoblast, vitamin D
Citation: Wang X, Li M, Zhang X, Li Y, He G, Dinnyés A, Sun Q and Xu W (2021) CYP11A1 Upregulation Leads to Trophoblast Oxidative Stress and Fetal Neurodevelopmental Toxicity That can be Rescued by Vitamin D. Front. Mol. Biosci. 7:608447. doi: 10.3389/fmolb.2020.608447
Received: 06 October 2020; Accepted: 30 December 2020;
Published: 15 February 2021.
Edited by:
Michal Mielcarek, Imperial College London, United KingdomReviewed by:
Cleo Smeets, Other, Paris, FranceCopyright © 2021 Wang, Li, Zhang, Li, He, Dinnyés, Sun and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenming Xu, eHV3ZW5taW5nQHNjdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.