- 1Department of Epidemiology and Health Statistics, Xiangya School of Public Health, Central South University, Changsha, China
- 2School of Mathematics and Statistics, Hunan Normal University, Changsha, China
- 3Department of Pathology and Pathophysiology, Hunan Normal University School of Medicine, Changsha, China
- 4Department of Pathophysiology, Jishou University School of Medicine, Jishou, China
Due to the difficulty in predicting the prognosis of endometrial carcinoma (EC) patients by clinical variables alone, this study aims to build a new EC prognosis model integrating clinical and molecular information, so as to improve the accuracy of predicting the prognosis of EC. The clinical and gene expression data of 496 EC patients in the TCGA database were used to establish and validate this model. General Cox regression was applied to analyze clinical variables and RNAs. Elastic net-penalized Cox proportional hazard regression was employed to select the best EC prognosis-related RNAs, and ridge regression was used to construct the EC prognostic model. The predictive ability of the prognostic model was evaluated by the Kaplan–Meier curve and the area under the receiver operating characteristic curve (AUC-ROC). A clinical-RNA prognostic model integrating two clinical variables and 28 RNAs was established. The 5-year AUC of the clinical-RNA prognostic model was 0.932, which is higher than that of the clinical-alone (0.897) or RNA-alone prognostic model (0.836). This clinical-RNA prognostic model can better classify the prognosis risk of EC patients. In the training group (396 patients), the overall survival of EC patients was lower in the high-risk group than in the low-risk group [HR = 32.263, (95% CI, 7.707–135.058), P = 8e-14]. The same comparison result was also observed for the validation group. A novel EC prognosis model integrating clinical variables and RNAs was established, which can better predict the prognosis and help to improve the clinical management of EC patients.
Introduction
Endometrial carcinoma (EC) is a common malignant tumor of the female reproductive system, and its incidence is increasing (Chen W. et al., 2016; Siegel et al., 2020). Metastasis or recurrence often occurs in EC patients after surgery, and the median survival time of patients with recurrence or metastasis is generally <12 months (Obel et al., 2006). Chemotherapy and radiation therapy fail to kill tumor cells with high specificity. The 5-year overall survival rate of EC patients without metastasis is between 74 and 91% (Morice et al., 2016), while the rate is reduced to 68 or 17% for EC patients with local or distant metastases, respectively (Colombo et al., 2016). Therefore, it is urgent to study the factors and mechanisms that affect the prognosis of EC patients and improve the clinical management.
At present, the prognosis prediction of EC patients is mainly based on the age at diagnosis, FIGO stage, pathological classification, treatment method, and other clinical variables. Due to the strong individual differences in the stages of occurrence, development, and metastasis of EC, it is difficult to accurately predict the prognosis of EC patients through clinical variables only (Frederick and Straughn, 2009). Studies have shown that specific genes or molecular changes influence the prognosis of EC patients (Bell and Ellenson, 2019). Molecules such as ER, PR, p53, HER-2/neu, and Ki-67 have been used to predict EC recurrence or prognosis; nevertheless, the results are still controversial (Fanning et al., 2002; Jeon et al., 2006).
In recent years, a class of non-coding RNA (ncRNA), including microRNA (miRNA) and long non-coding RNA (lncRNA), which cannot encode proteins, has been found to play an important role in life regulation (Djebali et al., 2012). More and more studies show that abnormal expression of ncRNA is closely related to the prognosis of EC patients. For example, miRNA-200c, miR-944, HOTAIR, H19, and SRA are related to prognosis of EC patients or the malignant degree of EC tumors (Smolle et al., 2015; He et al., 2017; Wilczynski et al., 2018). The expression levels of miR-142-3p, miR-142-5p, and miR-15a-5p are higher in EC patients with progression-free survival (PFS)>21 months than in EC patients with PFS <21 months, suggesting that miR-142 and miR-15a may be useful for EC prognosis prediction (Jayaraman et al., 2017). Hsa-mir-15a.MIMAT0000068, hsa-mir-142.MIMAT0000433, hsa-mir-142.MIMAT0000434, hsa-mir-3170.MIMAT0015045, hsa-mir-1976.MIMAT0009451, and hsa-mir-146a.MIMAT0000449 are significantly related to EC overall survival (OS), and the six-microRNA signature is an independent prognostic factor of EC (Wang Y. et al., 2019). However, so far, there has been no report on EC prognostic model integrating mRNAs, miRNAs, lncRNAs, and clinical variables.
This study intends to analyze the clinical and genome-wide mRNA, miRNA, and lncRNA expression data of EC patients in The Cancer Genome Atlas (TCGA) database and screen RNAs and clinical variables that are related to EC prognosis, with the expectation of discovering new EC prognostic molecular markers and establishing the integrated clinical-mRNA–miRNA–lncRNA prognostic model, thus providing a theoretical basis for EC prognostic risk assessment and individualized treatment.
Materials and Methods
Data Acquisition and Selection
We searched the TCGA database and other open databases, including Gene Expression Omnibus (GEO), International Cancer Genome Consortium (ICGC), ArrayExpress, Oncomine, etc. Only the TCGA database has an EC-related dataset with both gene profiles and clinical survival information. Clinical data, RNAseq-HTSeq FPKM data (including mRNA and lncRNA profiles), and miRNAseq data of EC patients were downloaded from the TCGA database (https://gdc-portal.nci.nih.gov/) in June 2019, and the dataset obtained contains 548 EC patients. Then, 52 of 548 EC patients were excluded. The reasons for exclusion were as follows: (1) 14 EC patients had no clinical data or mRNA, miRNA, and lncRNA gene expression profiles, and (2) 38 EC patients survived <30 days after the first pathological diagnosis. Eventually, 496 EC patients were included in this study, and the data missing rates for each clinical variable and the expression of each gene were <10%. The missing data of the 496 EC patients included in this study were filled by predictive mean matching. Differential expression analysis was performed based on log2 transformation of RNA expression data. From among all the patients involved in this study, 100 EC patients were randomly selected as the validation group, while the remaining 396 EC patients were used as the training group for the construction of the EC prognostic model. Furthermore, 33 out of the 496 EC patients had mRNA, miRNA, and lncRNA gene expression profiles of paracancerous tissues available, which were used as the control group for differential expression analysis.
Construction of the Prognosis Model
The univariate Cox regression (the 2-sided log-rank test) was applied to analyze 12 clinical variables, including age at initial pathologic diagnosis, height, weight, histologic grade, clinical stage, histologic type, initial pathological diagnosis method, time since last menstruation, neoplasm status, race, surgical approach, and tissue indicator (prospective or retrospective). Clinical variables resulting in a univariate Cox regression P <0.05 were initially screened for inclusion multivariate Cox regression analysis (αin = 0.10, αout = 0.15). Then, the clinical variables selected by the multivariate Cox regression model were identified, and the prognosis clinical model (model 1) of EC patients based on the identified clinical variables was established. Clinical variables that had a hazard ratio (HR) for death>1 were considered to be risk-increasing clinical variables, and those with HR <1 were defined as protective clinical variables.
The RNA genes related to EC prognosis were screened by the following three steps: (1) A fold change (FC) and false discovery rate (FDR) were applied to identify RNAs with differential expression between the EC patient group (396 patients) and the control group (33 controls). mRNAs, miRNAs, and lncRNAs with a FC>2 or < -2 and FDR <0.05 were screened as differentially expressed RNAs. (2) Univariate Cox regression analysis was used to explore the relationship between the differentially expressed RNAs and the prognosis of EC patients, and the differentially expressed RNA with a univariate Cox regression P <0.05 is considered to be a prognosis-related RNA in EC patients. (3) The three types of RNAs (i.e., mRNA, miRNA, and lncRNA) with P <0.05 identified in the univariate Cox regression analysis were further subjected to elastic net-penalized Cox proportional hazards regression analysis with 10,000 iterations and 10 cross-validation (Zou and Hastie, 2005; Pak et al., 2020). Lastly, the mRNAs, miRNAs, and lncRNAs with a non-zero elastic net-penalized Cox proportional hazards regression coefficient were the final selected RNAs considered to be related to the OS of EC. Then, the ridge regression Cox model was used to fit the selected RNAs (mRNAs, miRNAs, and lncRNAs) to construct the prognosis models of mRNA (model 2), miRNA (model 3), and lncRNA (model 4), respectively. The integrated RNA molecular prognostic model (model 5) was then constructed by fitting the selected mRNAs, miRNAs, and lncRNAs with the ridge regression Cox model. Eventually, the integrated clinical-RNA prognostic model was established by fitting the screened prognosis-related clinical variables and RNAs with the ridge regression Cox model (model 6). RNAs that had a HR for death>1 were considered to be risk-increasing RNAs, and those with HR <1 were defined as protective RNAs.
Evaluation of the Prognostic Model
The prognostic index (PI) is a weighted linear combination of various factors in the prognostic model. In the prognostic model, the PI value reflects the prognosis of the patient. PI is positively proportional to the risk function. A greater PI value indicates worse prognosis, and conversely, a smaller PI values means better prognosis. Standardization was carried out for PI to obtain a weighted prognostic index (WPI). The formulas used for calculating PI and WPI of each patient are as follows:
Where βi is the regression coefficient of the i-th factor in the model, Vi is the value of the i-th factor of EC patients, and mean (PI) and SD (PI) are the mean and standard deviation (SD) of the PI vector in EC patients, respectively. Applying WPI = 0 as the cutoff point, the patients were classified into two groups in terms of the predicted prognosis. Specifically, patients with WPI ≤ 0 were in the low-risk group, whereas those with WPI > 0 were in the high-risk group. The Kaplan–Meier curves of patients in the high-risk group and low-risk group were drawn and subjected to the log-rank test. P ≤ 0.05 indicates statistically significant difference in the OS between the two groups. The areas under the time-dependent ROC curves (AUC-ROC) of the six prognostic models were calculated. The model with the greatest AUC value was selected as the optimal prognostic model. An AUC value between 0.7 and 0.9 is generally believed to indicate medium predictive ability, while an AUC value greater than 0.9 indicates relatively ideal predictive ability. The larger is the AUC value, the stronger is the predictive ability of the model.
GO and KEGG Enrichment Analysis
The online tool DAVID (The Database for Annotation, Visualization and Integrated Discovery, version 6.8, http://david.abcc.ncifcrf.gov) was used to perform the GO and KEGG enrichment analysis for mRNAs, miRNA-targeted mRNAs (mRNAs with miRDB database-predicted scores higher than 90), and lncRNA-related mRNAs (Spearman's correlation coefficient rs>0.50 and P <0.05) in the EC prognostic molecular model. Fisher's exact test was employed to select terms with P <0.05 as significant GO and KEGG pathway terms. GO analysis annotates and classifies genes through biological process (BP), molecular function (MF), and cell composition (CC).
Statistical Analysis
Data analyses in this study were conducted by R, version 3.6.1. The missing data were filled by the “mice” R package (version 3.11.0), and differential expression analysis was performed with the “limma” R package (version 3.26.9). The “survival” R package (version 3.2-3) was applied for univariate Cox regression, multivariate Cox regression, and plotting Kaplan–Meier curves. The elastic net-penalized Cox proportional hazards regression model and the ridge regression Cox model were analyzed using the “glmnet” R package (version 3.0-2). The “timeROC” R package (version 0.4) was used to plot the time-dependent ROC curves and calculate the AUC values, and the “ggplot2” R package (version 3.3.1) was used to generate figures of GO and KEGG analysis.
Results
Workflow
Figure 1 shows the process of our Study. RNA expression data and corresponding clinical variable data from TCGA for EC were analyzed. Cox proportional hazards regression was used to analyze clinical variables related to EC prognosis. RNAs related to EC prognosis were screened by differential expression analysis, univariate Cox proportional hazards regression, and elastic net-penalized Cox proportional hazards regression. The EC prognostic model was constructed by using EC prognostic-related RNAs/clinical variables, and the performance of the prognostic model was evaluated.
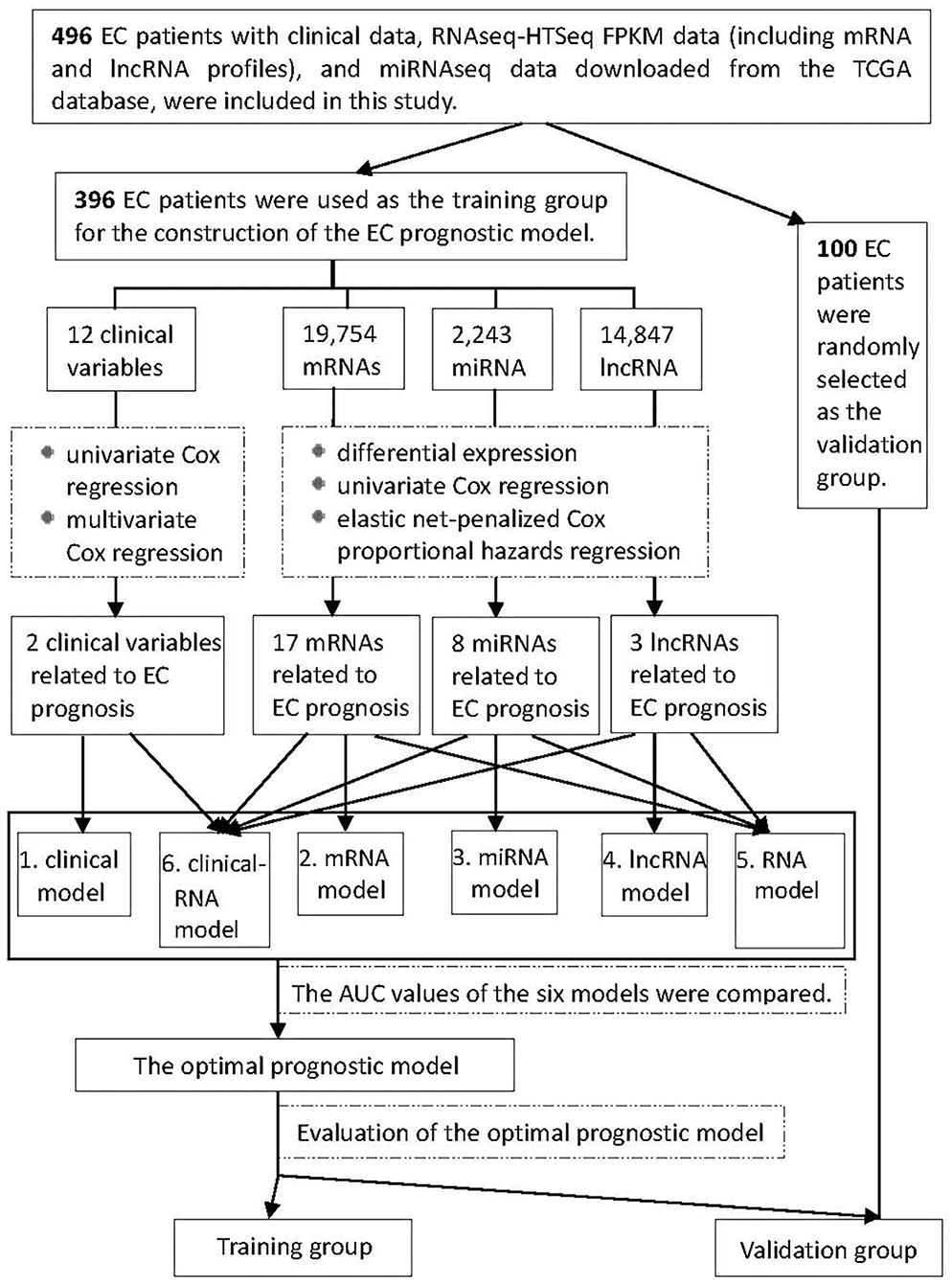
Figure 1. Flowchart of construction and evaluation of the EC prognostic model. AUC, the area under the receiver operating characteristic curve.
Clinical Characteristics and Prognosis Model of EC Patients
Among the 396 EC patients in the training group, 33 patients died by the follow-up deadline and 363 patients survived. The minimum and maximum ages of patients at initial pathological diagnosis were 33 years and 89 years, respectively, with the average age being 64.22 years (SD = 10.86 years). Univariate Cox regression analysis indicated that histological grade, clinical stage, and neoplasm status were statistically significant (P <0.05) among the 12 clinical variables. Further multivariate Cox regression analysis was performed for the three factors, and the results suggested that histological grade and neoplasm status are independent prognostic clinical variables of EC, and both of them are EC risk factors (HR > 1). Then, a clinical prognostic model of EC was established based on histological grade and neoplasm status (Table 1).
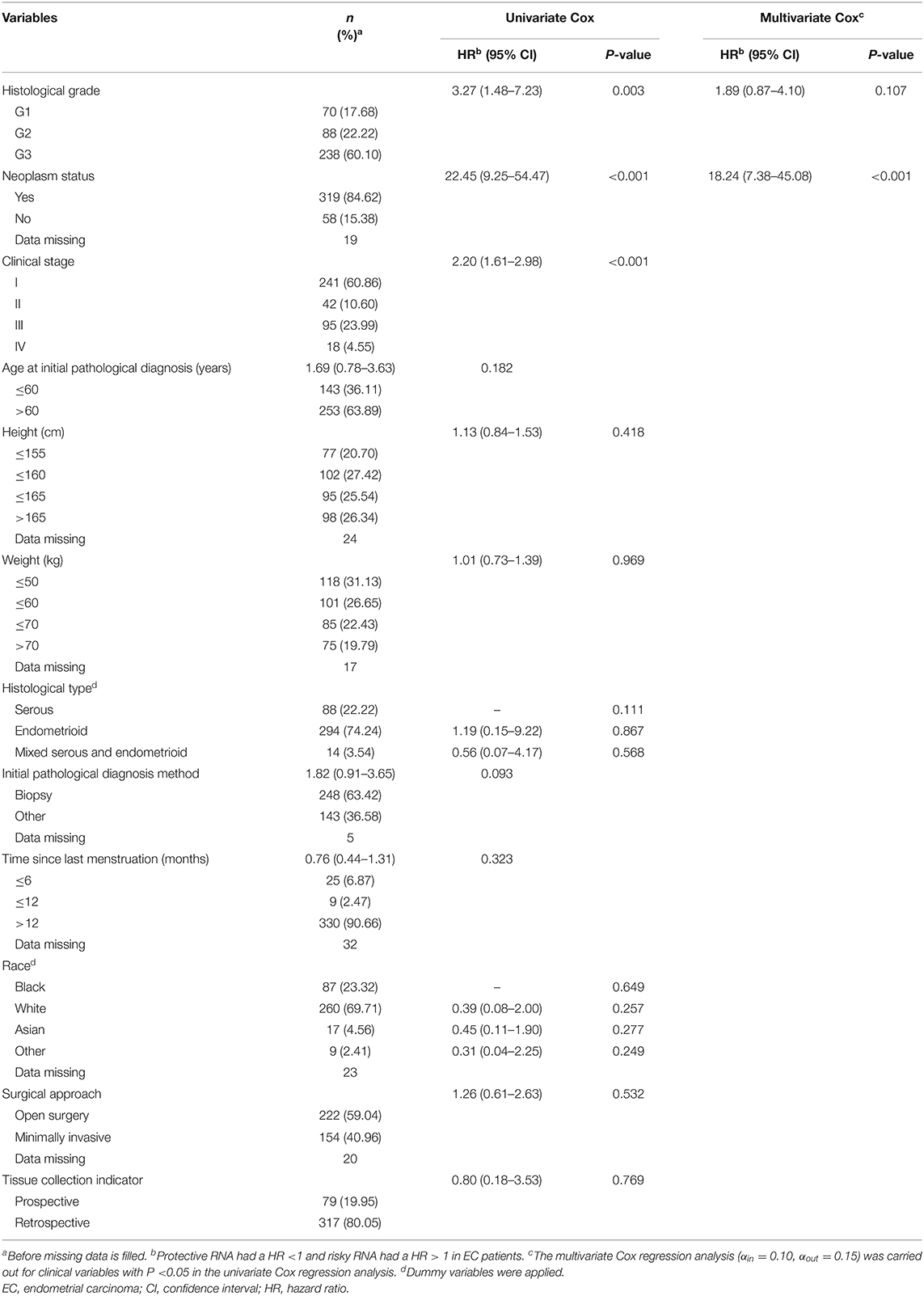
Table 1. Survival analysis results of demographic and clinical variables for EC patients in the prognostic model training group (396 patients).
Differentially Expressed and OS-Related RNAs of EC
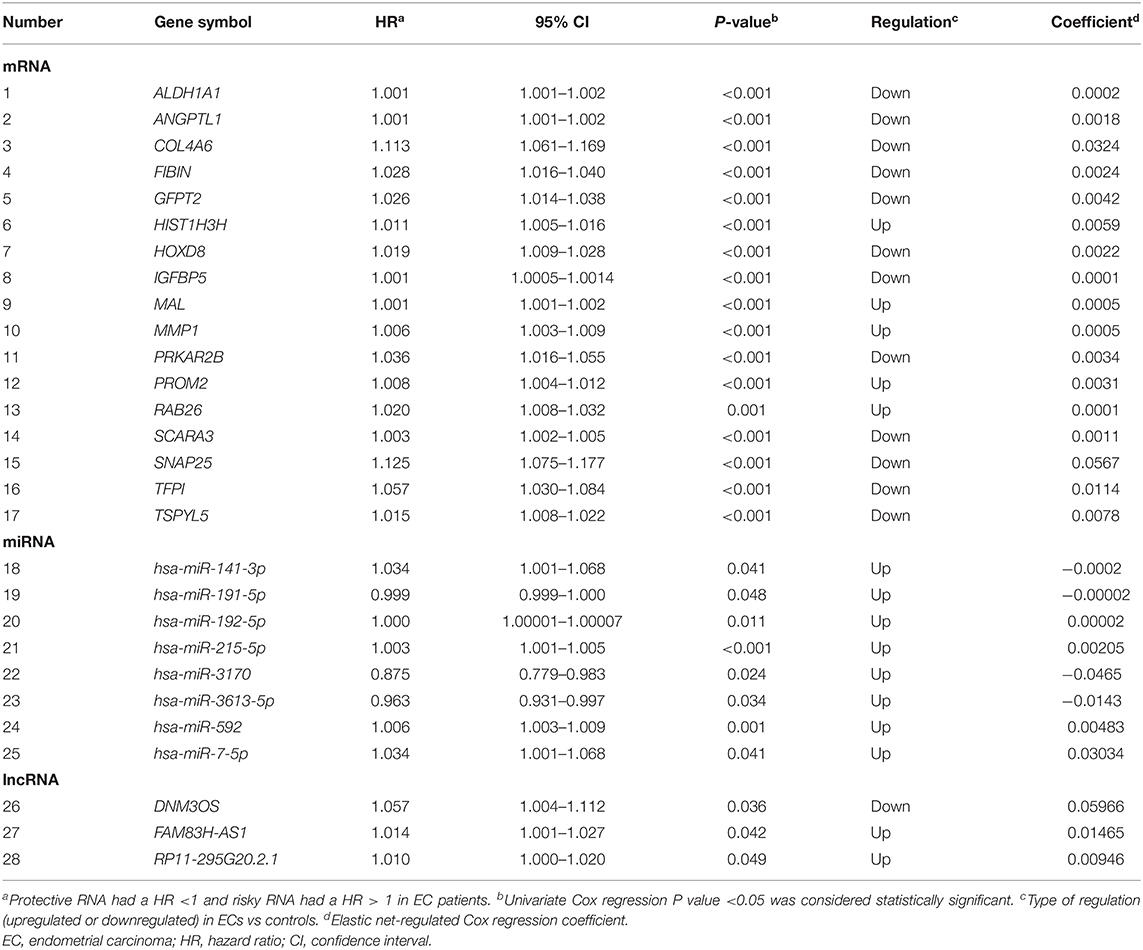
The EC RNA expression data acquired from TCGA database were preprocessed, and a total of 36,844 RNAs (19,754 mRNA, 2,243 miRNA, and 14,847 lncRNA) were included in this study. 1060 differential expression RNAs were screened by differential expression analysis, including 920 mRNAs (353 upregulation and 567 downregulation, Figure 2A), 100 miRNAs (83 upregulation and 17 downregulation, Figure 2B), and 40 lncRNAs (21 upregulation and 19 downregulation, Figure 2C). Univariate Cox regression was performed on these 1,060 differentially expressed RNAs individually, and there were 126 RNAs with P <0.05 (115 mRNAs, 8 miRNAs, and 3 lncRNAs). Subsequently, the whole 126 RNAs were subjected to elastic net-penalized Cox proportional hazard regression analysis, and 17 RNAs related to EC prognosis were selected, including 15 mRNAs (ANGPTL1, ALDH1A1, FIBIN, GFPT2, HIST1H3H, HOXD8, IGFBP5, MAL, MMP1, PRKAR2B, PROM2, SCARA3, SNAP25, TFPI, and TSPYL5), and 2 miRNAs (has-miR-215-5p and has-miR-592). There is no lncRNA in the 17 RNAs. We do not think it can reflect the whole picture of RNAs associated with EC prognosis. Recent studies have shown that although lncRNA does not encode proteins, lncRNA participates in gene expression regulation at various levels, such as transcriptional regulation and posttranscriptional regulation. The abnormal expression of lncRNA is usually associated with the occurrence, recurrence, and metastasis of tumors (Kopp and Mendell, 2018). Therefore, we used elastic net-penalized Cox proportional hazard regression to screen the three types of RNAs (mRNA, miRNA, and lncRNA), respectively. Eventually, 28 EC OS-related RNAs (17 mRNAs, 8 miRNAs, and 3 lncRNAs) were identified (Figure 3, Table 2). These 28 EC OS-related RNAs contain all 17 RNAs screened by elastic net-penalized Cox proportional hazard regression using whole genes.
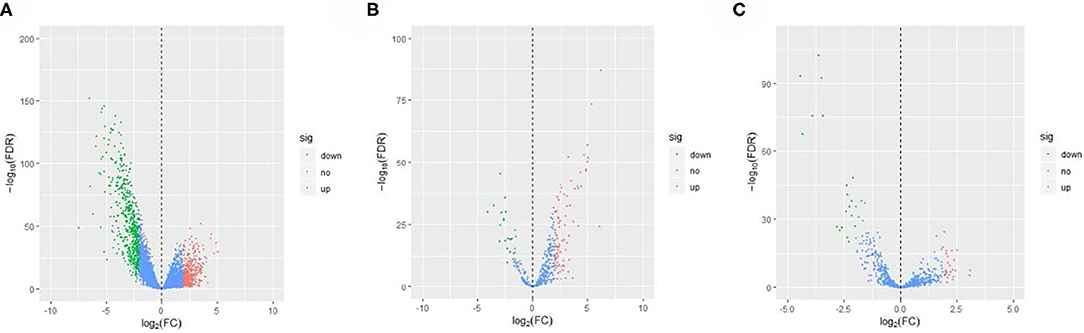
Figure 2. Volcano diagram of the differential expression RNAs in ECs and controls. (A) Volcano diagram of mRNA, (B) volcano diagram of miRNA, and (C) volcano diagram of lncRNA.
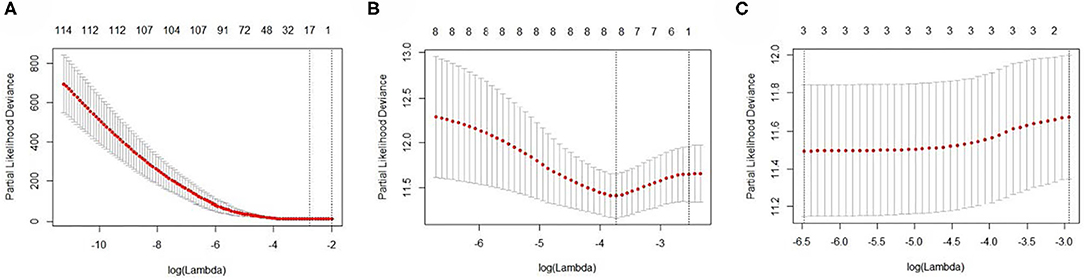
Figure 3. The cross-validation curves of the elastic net-penalized Cox proportional hazard regression. (A) Cross-validation curve for mRNA, (B) cross-validation curve for miRNA, and (C) cross-validation curve for lncRNA.
RNA Molecular Prognostic Models of EC
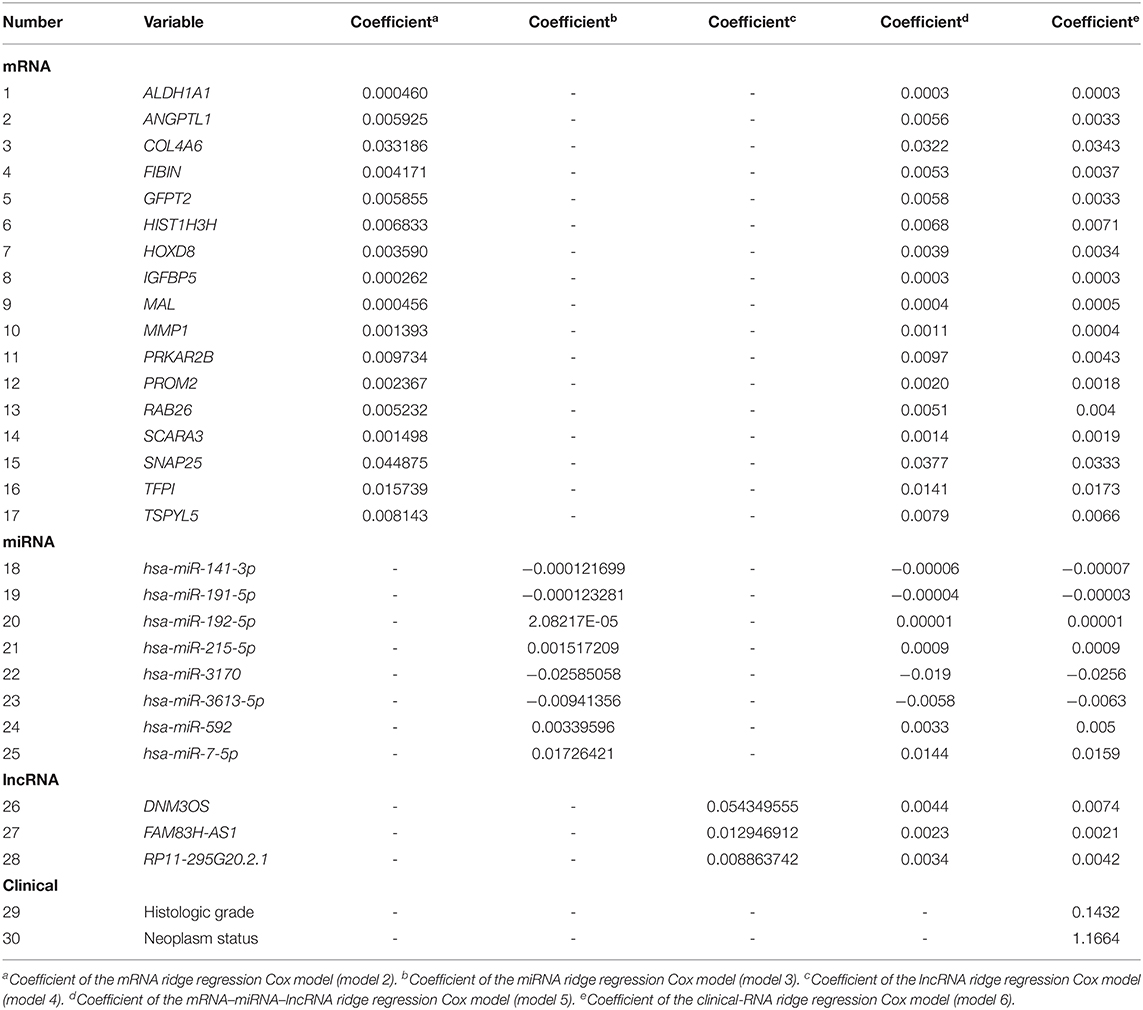
The EC OS-related 17 mRNAs, 8 miRNAs, and 3 lncRNAs were fitted with the ridge regression Cox model respectively to obtain the corresponding mRNA prognostic model, miRNA prognostic model, and lncRNA prognostic model. The 28 EC OS-related RNAs were fitted with the ridge regression Cox model, and an integrated mRNA–miRNA–lncRNA molecular prognostic model was established (Table 3).
Integrated Clinical-RNA Prognostic Model of EC
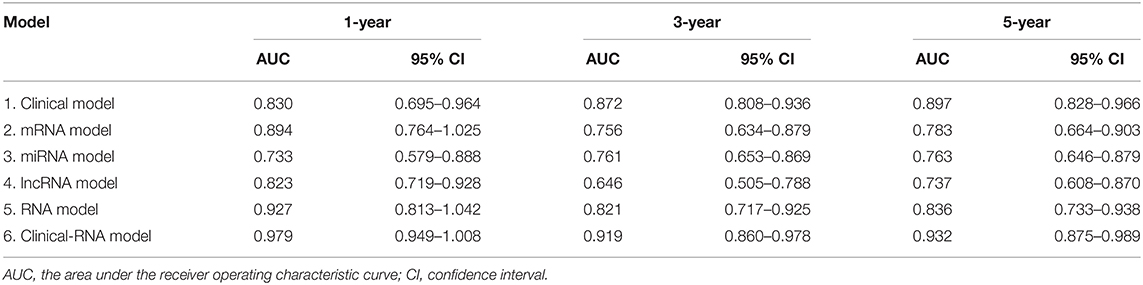
The EC prognosis-related 28 RNAs and 2 clinical variables were fitted with the ridge regression Cox model, and an integrated clinical-RNA prognostic model was established (Table 3). As listed in Table 4, the AUC (95% CI) values of the lncRNA model based on the selected three lncRNAs at 1, 3, and 5 years were 0.823 (0.719–0.928), 0.646 (0.505–0.788), and 0.737 (0.608–0.870), respectively. These results suggest that the three lncRNAs have a certain predictive effect on the prognosis of EC. The AUC-ROC showed that except for the lncRNA molecular model (model 4) with a 3-year prognosis AUC of 0.646 (<0.7), the minimum AUC value of other models was 0.733. The AUC of the integrated RNA molecular prognostic model (model 5) was ≥0.821, which is greater than the AUC of mRNA, miRNA, and lncRNA models, suggesting that the integrated RNA molecular prognostic model is superior to the mRNA or miRNA or lncRNA model in terms of predictive ability. As for the clinical prognostic model, the AUC value was ≥0.830, implying that the two clinical variables (i.e., histological grade and neoplasm status) screened in this study can predict the prognosis of EC. The 1-, 3-, and 5-year AUC values of the integrated clinical-RNA prognostic model (model 6) were ≥0.919, being greater than the AUC values of other models at the same time point. This indicates that the integrated clinical-RNA prognostic model has the best predictive ability among the six models (Figure 4).
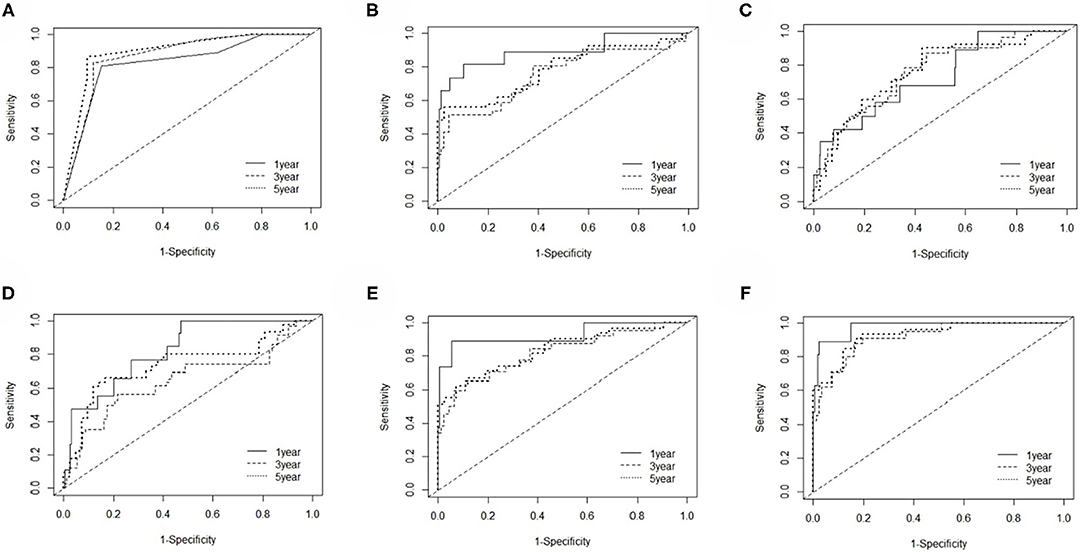
Figure 4. The time-dependent receiver operating characteristic curves. (A–F) The 1-, 3-, and 5-year ROC curves of the clinical model, mRNA model, miRNA model, lncRNA model, RNA model, and clinical-RNA model.
The integrated clinical-RNA prognostic model was used to calculate the WPI value of each EC patient in the training group (396 patients). The WPI value of EC patients ranged from −1.957 to 4.831. Taking WPI = 0 as the cutoff point, the EC patients were divided into the high-risk group (147 patients) and low-risk group (249 patients) (Figure 5A). The difference between the two groups' Kaplan–Meier curves was statistically significant (P = 8e-14). The prognosis of EC patients was worse in the high-risk group than in the low-risk group [HR = 32.263, (95% CI, 7.707–135.058)], suggesting that the integrated clinical-RNA prognostic model enables accurate prediction of the prognosis of EC patients (Figure 5C). Furthermore, this model was used to calculate the WPI value of each EC patient in the validation group (100 patients) to predict the prognosis of EC patients. The WPI value of EC patients in the validation group ranged from −1.455 to 3.822. Taking WPI = 0 as the cutoff point, the EC patients in the validation group were divided into the high-risk group (41 patients) and low-risk group (59 patients) (Figure 5B). The difference between the two groups' Kaplan–Meier curves was statistically significant (P = 0.0052). The prognosis of EC patients in the high-risk group was worse than that of patients in the low-risk group [HR = 6.674, (95% CI, 1.437–30.995)], showing that the integrated clinical-RNA prognostic model also has satisfactory accuracy in predicting the prognosis of EC patients in the validation group (Figure 5D).
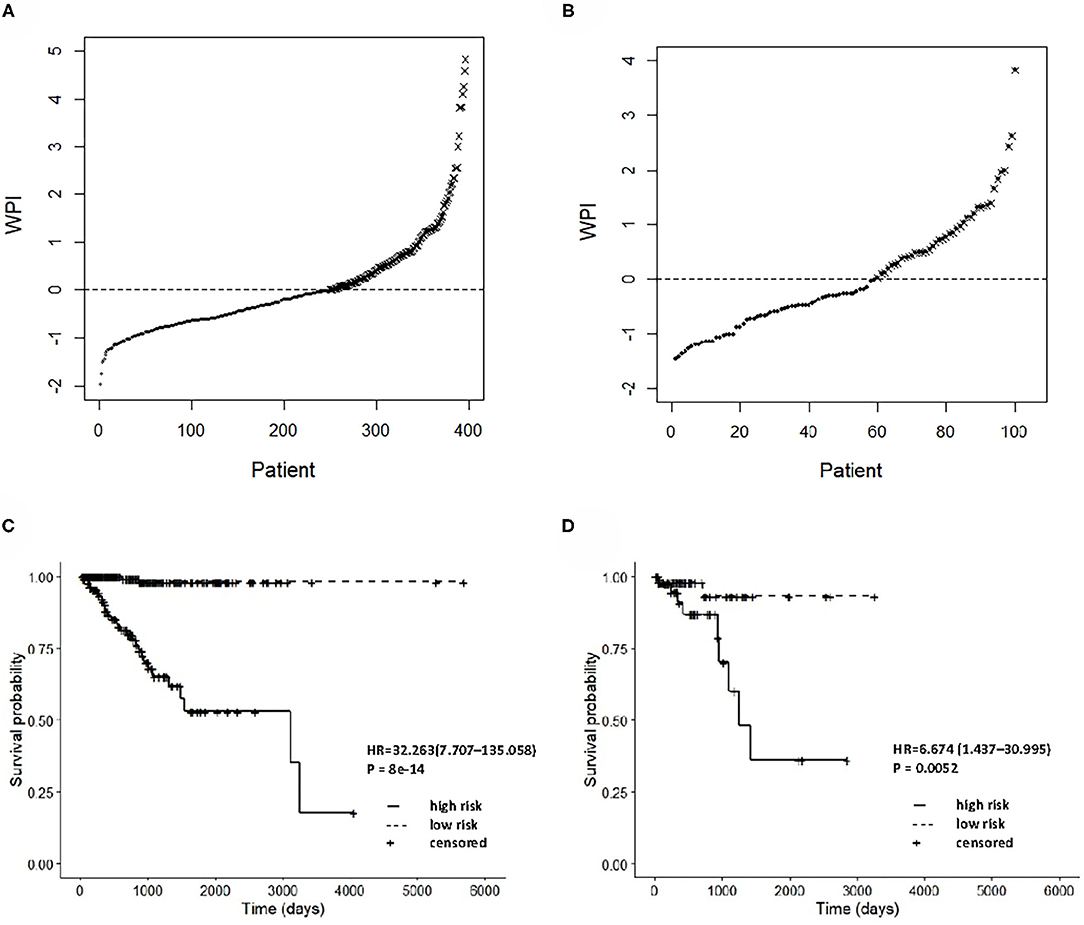
Figure 5. Prognostic performance assessment of integrated clinical-RNA prognostic model. WPI distributions and stratifications of EC patients in training set (A) and validating set (B), respectively. Kaplan–Meier curves for stratifications in training set (C) and validating set (D), respectively. WPI, weighed prognostic index; HR, hazard ratio.
Functional Analysis of EC Prognosis-Related Genes
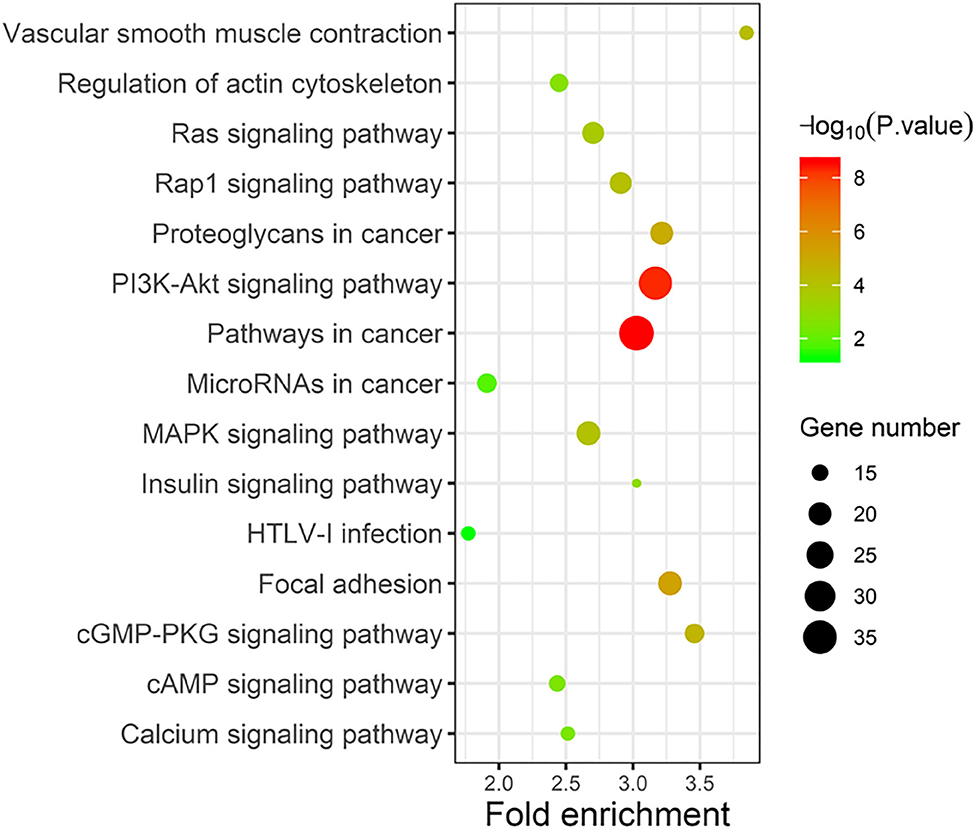
Taking the union of the 17 mRNAs, the 371 target mRNAs of the 8 miRNAs, and the 300 mRNAs related to the 3 lncRNAs in the RNA prognostic model, a gene set with 652 mRNAs was obtained and subjected to GO and KEGG analyses. The GO analysis results show that in biological processes, target genes are mainly enriched in “signaling,” “positive regulation of RNA polymerase II promoter transcription,” and “negative regulation of RNA polymerase II promoter transcription” (Figure 6A). In terms of cellular composition, the target genes are mainly located in the “cytoplasm” and “plasma membrane” (Figure 6B). As for the molecular function, “protein binding” is the most important mode (Figure 6C). According to the results of KEGG analysis, the top three pathways are pathways in cancer, the PI3K-Akt signaling pathway, and the focal adhesion pathway (Figure 7).
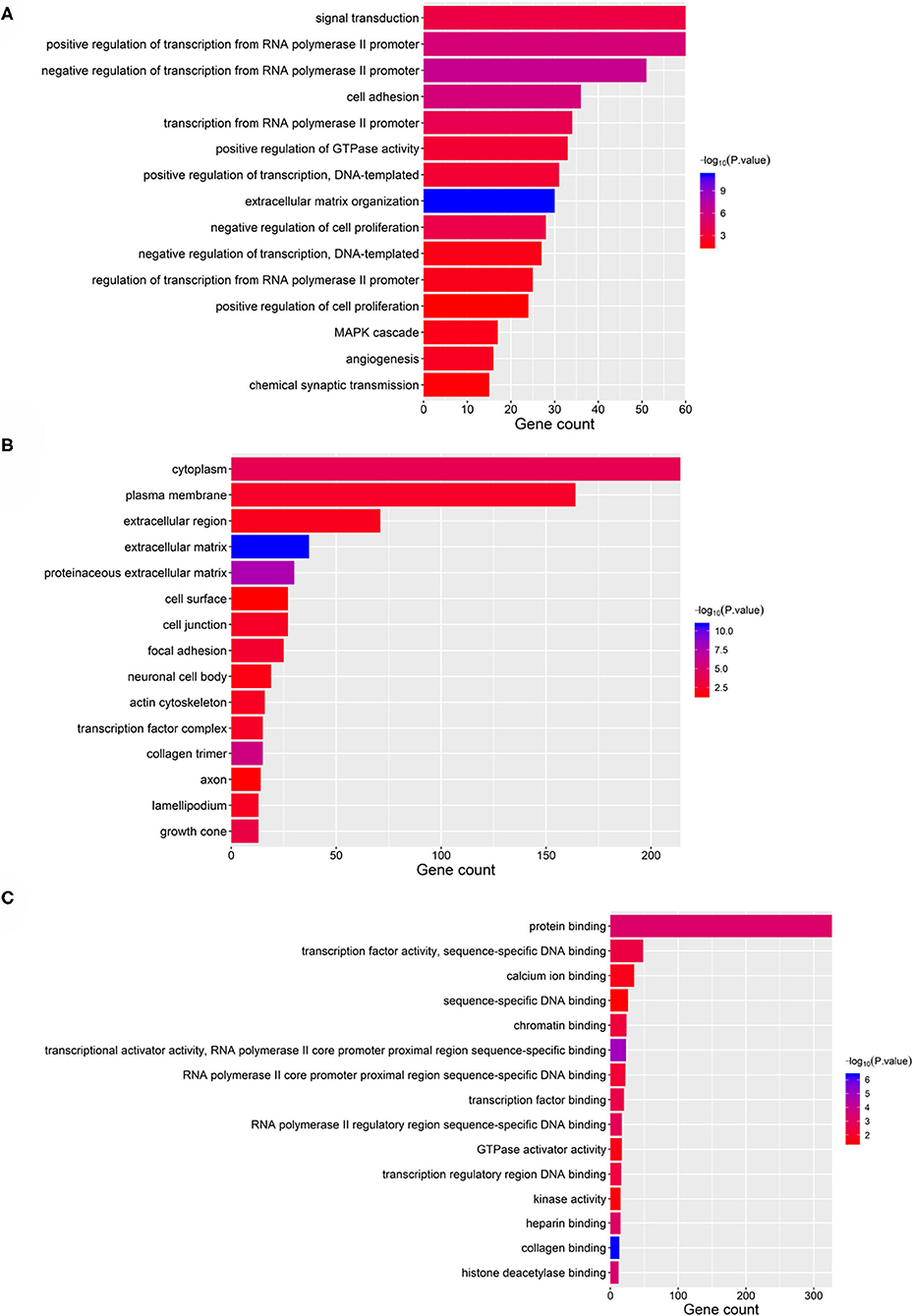
Figure 6. GO analysis of the 28 validated genes. (A) Biological process (BP), (B) cellular component (CC), and (C) molecular function (MF).
Discussion
At present, researchers have been searching for EC prognosis-related biomarkers and establishing EC prognostic prediction model with higher accuracy to provide better clues for formulating reasonable individualized treatment plans, thereby improving patients' prognostic quality of life. After acquiring the clinical data of EC patients from the TCGA database and the full mRNA, miRNA, and lncRNA genome expression profiles, 30 factors related to EC prognosis, including two clinical variables and 28 RNAs, were identified in this study. Based on the 30 EC prognosis-related factors, 6 EC prognosis models were established: clinical model, mRNA model, miRNA model, lncRNA model, integrated RNA model, and integrated clinical-RNA model. The clinical-RNA model displayed the highest AUC (≥ 0.919), indicating the strongest predictive ability among the six prognostic models.
Previous studies have shown that the clinical variables related to the prognosis of EC include pathological grade, pathological stage, FIGO stage, age at initial pathological diagnosis, degree of muscular invasion, vascular tumor thrombus, and lymph node metastasis (Braun et al., 2016; Morice et al., 2016). Our study suggests that histological grade and neoplasm status are independent prognostic factors for EC overall survival.
There were 17 prognosis-related mRNAs with HR>1, indicating that an increased expression level of these mRNAs will increase the risk of death in EC patients. Most of these genes are reportedly related to the occurrence, development, or prognosis of cancer. The expression of ALDH1A1 is upregulated in endometrial carcinoma cells (Shiba et al., 2019), and ALDH1A1 is a confirmed oncogene for lung cancer (Gao et al., 2015). As a member of angiopoietin-like protein genes, ANGPTL1 acts as a tumor-suppressor gene in various tumors (Chen H. A. et al., 2016). The absence of COL4A6 may cause familial hemorrhagic nephritis (Murata et al., 2016). GFPT2 is highly expressed in lung cancer (Zhang et al., 2018). HIST1H3H is a histone gene, and its high expression is related to the OS, relapse-free survival (RFS), and distant metastasis-free survival (DMFS) of breast cancer patients (Xie et al., 2019). HOXD8 belongs to a homeobox gene family and is closely related to cell proliferation, apoptosis, and cell cycle. Studies have found that HOXD8 is a downstream target gene of miR-5692a. MiR-5692a plays the role of an oncogene in the occurrence and development of liver cancer by regulating the expression of HOXD8 (Sun et al., 2019). IGFBP5 is a tumor-suppressor gene for leukemia, osteosarcoma, breast cancer, and pancreatic cancer and participates in cell biological functions, such as cell metastasis and apoptosis (Baxter, 2014). Hypermethylation of MAL in cervical intraepithelial neoplasia accelerates cervical lesions (Meršaková et al., 2018). High expression of MMP1 in cancer tissues leads to accelerated angiogenesis, thus promoting the proliferation and migration of cancer cells (Pahwa et al., 2014). It has been clarified that the PRKAR2B gene is overexpressed in castration-resistant prostate cancer (CRPC), which mainly promotes cell-cycle biological processes, accelerates CRPC cell proliferation and invasion, and inhibits CRPC cell apoptosis (Sha et al., 2017). The expression of PROM2 (prominin 2) is upregulated in kidney cancer and melanoma (Rohan et al., 2006; Winnepenninckx et al., 2006). The function of RAB26 is mainly related to membrane transport and cell autophagy. Some studies have found that RAB26 can affect the metastasis and invasion of breast cancer (Schwartz et al., 2007). SCARA3 inhibits the lethal effect of dexamethasone and bortezomib on myeloma cells (Brown et al., 2013). SNAP25 is mainly involved in the occurrence and development of mental diseases (González-Giraldo and Forero, 2020). TFPI reduces tumor cell-induced coagulation activation and lung metastasis, and it has shown inhibitory effect on primary and metastatic tumors in mice (Hembrough et al., 2003; Amirkhosravi et al., 2007). Some studies suggest that the methylation of the tumor-suppressor gene TSPYL5 will cause its expression silencing and, thereby, gastric cancer (Jung et al., 2008).
In this study, eight EC prognosis-related miRNAs were identified. The HR values of miR-141-3p, miR-192-5p, miR-215-5p, miR-592, and miR-7-5p were greater than 1, indicating that the five miRNAs are highly expressed in EC, acting as tumor genes and prognostic risk factors. MiR-141-3p acts as a tumor-suppressor gene in colorectal cancer and enhances the sensitivity of colorectal cancer cells to cetuximab by inhibiting EGFR (Xing et al., 2020). MiR-192-5p plays different roles in different cancers, e.g., it is highly expressed in gastric cancer and pancreatic ductal cancer, while the expression is low in lung cancer (Feng et al., 2011; Zhao et al., 2013; Chen et al., 2014). Overexpression of miR-215-5p in colorectal cancer leads to G2/M phase cell-cycle arrest and p53-dependent apoptosis induction, thus reducing the proliferation and migration of colorectal cancer cells (Vychytilova-Faltejskova et al., 2017). The biological function of miR-592 varies according to the cancer type. Its overexpression in liver cancer inhibits the proliferation and metastasis of cancer cells, while the opposite effect is observed in prostate cancer (Wang et al., 2012; Lv et al., 2015). Studies have shown that miR-7-5p can inhibit tumor development by regulating the PI3K/Akt pathway and the expression of the target gene KLF4 (Fang et al., 2012; Okuda et al., 2013). The other three miRNAs (i.e., miR-191-5p, miR-3170, and miR-3613-5p) have HR values lower than 1, indicating that these three genes are protective factors for EC prognosis, i.e., their high expression reduces the risk of death in EC patients. The overexpression of miR-191-5p in lung adenocarcinoma downregulates Wnt signaling via the target gene SATB1, thus blocking lung cancer cell migration and proliferation (Zhou et al., 2020). Studies have shown that the prognosis is better in EC patients with high expression of miR-3170 than in those with low expression of miR-3170 (Wang Y. et al., 2019). The expression level of miR-3613-5p in the serum of patients with endometriosis is significantly reduced (Cosar et al., 2019).
The HR values of the three identified EC prognosis-related lncRNAs (e.g., DNM3OS, FAM83H-AS1, and RP11-295G20.2.1) are all greater than 1, indicating that they are prognostic risk factors for EC. Studies have observed that the expression of DNM3OS is upregulated in gastric cancer tissues and cell lines. Knocking out DNM3OS hinders snail-mediated epithelial-to-mesenchymal transition, thereby inhibiting the proliferation, migration, and invasion of gastric cancer cells (Wang S. et al., 2019). FAM83H-AS1 promotes radiation resistance and metastasis of ovarian cancer via targeted HuR protein (Dou et al., 2019). At present, the specific biological function of RP11-295G20.2.1 is not clear, and its relationship with the occurrence, development, and prognosis of EC needs to be confirmed by further experimental research.
Conclusions
In summary, 28 RNAs that are related to the prognosis of EC patients were identified in this study, and a clinical-mRNA–miRNA–lncRNA prognostic model for EC patients was established. The predictive ability of this clinical-RNA model is significantly better than the clinical-alone model and RNA-alone model in terms of prognosis prediction for EC patients. This study provides a scientific basis for discovering new prognostic markers for EC patients, clarifying the molecular mechanism of EC prognosis, and improving prognosis and clinical management of EC patients.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
Conceptualization: XZ and XP. Data curation: FD. Formal analysis: FD and JM. Methodology: CQ. Software: FD and CQ. Writing—original draft: FD, JM, CQ, FY, XL, and XZ. Writing—review and editing: XP.
Funding
This research was funded by the National Natural Science Foundation of China (Grant Number 81472860).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank The Cancer Genome Atlas project for contributing the data for analyses of our study.
References
Amirkhosravi, A., Meyer, T., Amaya, M., Davila, M., Mousa, S. A., Robson, T., et al. (2007). The role of tissue factor pathway inhibitor in tumor growth and metastasis. Semin. Thromb Hemost. 33, 643–652. doi: 10.1055/s-2007-991531
Baxter, R. C. (2014). IGF binding proteins in cancer: mechanistic and clinical insights. Nat. Rev. Cancer 14, 329–341. doi: 10.1038/nrc3720
Bell, D. W., and Ellenson, L. H. (2019). Molecular genetics of endometrial carcinoma. Annu. Rev. Pathol. 14, 339–367. doi: 10.1146/annurev-pathol-020117-043609
Braun, M. M., Overbeek-Wager, E. A., and Grumbo, R. J. (2016). Diagnosis and management of endometrial cancer. Am. Fam. Phys. 93, 468–474.
Brown, C. O., Schibler, J., Fitzgerald, M. P., Singh, N., Salem, K., Zhan, F., et al. (2013). Scavenger receptor class A member 3 (SCARA3) in disease progression and therapy resistance in multiple myeloma. Leuk. Res. 37, 963–969. doi: 10.1016/j.leukres.2013.03.004
Chen, H. A., Kuo, T. C., Tseng, C. F., Ma, J. T., Yang, S. T., Yen, C. J., et al. (2016). Angiopoietin-like protein 1 antagonizes MET receptor activity to repress sorafenib resistance and cancer stemness in hepatocellular carcinoma. Hepatology 64, 1637–1651. doi: 10.1002/hep.28773
Chen, Q., Ge, X., Zhang, Y., Xia, H., Yuan, D., Tang, Q., et al. (2014). Plasma miR-122 and miR-192 as potential novel biomarkers for the early detection of distant metastasis of gastric cancer. Oncol. Rep. 31, 1863–1870. doi: 10.3892/or.2014.3004
Chen, W., Zheng, R., Baade, P. D., Zhang, S., Zeng, H., Bray, F., et al. (2016). Cancer statistics in China, 2015. CA. Cancer J. Clin. 66, 115–132. doi: 10.3322/caac.21338
Colombo, N., Creutzberg, C., Amant, F., Bosse, T., González-Martín, A., Ledermann, J., et al. (2016). ESMO-ESGO-ESTRO Consensus conference on endometrial cancer: diagnosis, treatment and follow-up. Ann. Oncol. 27, 16–41. doi: 10.1093/annonc/mdv484
Cosar, E., Mamillapalli, R., Moridi, I., Duleba, A., and Taylor, H. S. (2019). Serum microRNA biomarkers regulated by simvastatin in a primate model of endometriosis. Reprod. Sci. 26, 1343–1350. doi: 10.1177/1933719118765971
Djebali, S., Davis, C. A., Merkel, A., Dobin, A., Lassmann, T., Mortazavi, A., et al. (2012). Landscape of transcription in human cells. Nature 489, 101–108. doi: 10.1038/nature11233
Dou, Q., Xu, Y., Zhu, Y., Hu, Y., Yan, Y., and Yan, H. (2019). LncRNA FAM83H-AS1 contributes to the radioresistance, proliferation, and metastasis in ovarian cancer through stabilizing HuR protein. Eur. J. Pharmacol. 852, 134–141. doi: 10.1016/j.ejphar.2019.03.002
Fang, Y., Xue, J. L., Shen, Q., Chen, J., and Tian, L. (2012). MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology 55, 1852–1862. doi: 10.1002/hep.25576
Fanning, J., Brown, S., Phibbs, G., Kramer, T., and Zaher, A. (2002). Immunohistochemical evaluation is not prognostic for recurrence in fully staged high-risk endometrial cancer. Int. J. Gynecol. Cancer 12, 286–289. doi: 10.1136/ijgc-00009577-200205000-00008
Feng, S., Cong, S., Zhang, X., Bao, X., Wang, W., Li, H., et al. (2011). MicroRNA-192 targeting retinoblastoma 1 inhibits cell proliferation and induces cell apoptosis in lung cancer cells. Nucleic Acids Res. 39, 6669–6678. doi: 10.1093/nar/gkr232
Frederick, P. J., and Straughn, J. M. (2009). The role of comprehensive surgical staging in patients with endometrial cancer. Cancer Control 16, 23–29. doi: 10.1177/107327480901600104
Gao, F., Zhou, B., Xu, J. C., Gao, X., Li, S. X., Zhu, G. C., et al. (2015). The role of LGR5 and ALDH1 Al in non-small cell lung cancer: cancer progression and prognosis. Biochem. Biophys. Res. Commun. 462, 91–98. doi: 10.1016/j.bbrc.2015.04.029
González-Giraldo, Y., and Forero, D. A. (2020). A functional SNP in the synaptic SNAP25 gene is associated with impulsivity in a Colombian sample. 3 Biotech 10:134. doi: 10.1007/s13205-020-2110-0
He, Z., Xu, H., Meng, Y., and Kuang, Y. (2017). miR-944 acts as a prognostic marker and promotes the tumor progression in endometrial cancer. Biomed. Pharmacother. 88, 902–910. doi: 10.1016/j.biopha.2017.01.117
Hembrough, T. A., Swartz, G. M., Papathanassiu, A., Vlasuk, G. P., Rote, W. E., Green, S. J., et al. (2003). Tissue factor/factor VIIa inhibitors block angiogenesis and tumor growth through a non-hemostatic mechanism. Cancer Res. 63, 2997–3000.
Jayaraman, M., Radhakrishnan, R., Mathews, C. A., Yan, M., Husain, S., Moxley, K. M., et al. (2017). Identification of novel diagnostic and prognostic miRNA signatures in endometrial cancer. Genes Cancer 8, 566–576. doi: 10.18632/genesandcancer.144
Jeon, Y. T., Park, I. A., Kim, Y. B., Kim, J. W., Park, N. H., Kang, S. B., et al. (2006). Steroid receptor expressions in endometrial cancer: clinical significance and epidemiological implication. Cancer Lett. 239, 198–204. doi: 10.1016/j.canlet.2005.08.001
Jung, Y., Park, J., Bang, Y. J., and Kim, T. Y. (2008). Gene silencing of TSPYL5 mediated by aberrant promoter methylation in gastric cancers. Lab. Invest. 88, 153–160. doi: 10.1038/labinvest.3700706
Kopp, F., and Mendell, J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407. doi: 10.1016/j.cell.2018.01.011
Lv, Z. H., Rao, P., and Li, W. (2015). MiR-592 represses FOXO3 expression and promotes the proliferation of prostate cancer cells. Int. J. Clin. Exp. Med. 8, 15246–15253.
Meršaková, S., Holubeková, V., Grendár, M., Višnovský, J., Nachajová, M., Kalman, M., et al. (2018). Methylation of CADM1 and MAL together with HPV status in cytological cervical specimens serves an important role in the progression of cervical intraepithelial neoplasia. Oncol. Lett. 16, 7166–7174. doi: 10.3892/ol.2018.9505
Morice, P., Leary, A., Creutzberg, C., Abu-Rustum, N., and Darai, E. (2016). Endometrial cancer. Lancet 387, 1094–1108. doi: 10.1016/S0140-6736(15)00130-0
Murata, T., Katayama, K., Oohashi, T., Jahnukainen, T., Yonezawa, T., Sado, Y., et al. (2016). COL4A6 is dispensable for autosomal recessive Alport syndrome. Sci. Rep. 6:29450. doi: 10.1038/srep29450
Obel, J. C., Friberg, G., and Fleming, G. F. (2006). Chemotherapy in endometrial cancer. Clin. Adv. Hematol. Oncol. 4, 459–468.
Okuda, H., Xing, F., Pandey, P. R., Sharma, S., Watabe, M., Pai, S. K., et al. (2013). miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 73, 1434–1444. doi: 10.1158/0008-5472.CAN-12-2037
Pahwa, S., Stawikowski, M. J., and Fields, G. B. (2014). Monitoring and inhibiting MT1-MMP during cancer initiation and progression. Cancers 6, 416–435. doi: 10.3390/cancers6010416
Pak, K., Oh, S., Goh, T. S., Heo, H. J., Han, M., Jeong, D. C., et al. (2020). A user-friendly, web-based integrative tool (ESurv) for survival analysis: development and validation study. J. Med. Internet Res. 22:e16084. doi: 10.2196/16084
Rohan, S., Tu, J. J., Kao, J., Mukherjee, P., Campagne, F., Zhou, X. K., et al. (2006). Gene expression profiling separates chromophobe renal cell carcinoma from oncocytoma and identifies vesicular transport and cell junction proteins as differentially expressed genes. Clin. Cancer Res. 12, 6937–6945. doi: 10.1158/1078-0432.CCR-06-1268
Schwartz, S. L., Cao, C., Pylypenko, O., Rak, A., and Wandinger-Ness, A. (2007). RabGTPases at a glance. J. Cell Sci. 120, 3905–3910. doi: 10.1242/jcs.015909
Sha, J., Xue, W., Dong, B., Pan, J., Wu, X., Li, D., et al. (2017). PRKAR2B plays an oncogenic role in the castration-resistant prostate cancer. Oncotarget 8, 6114–6129. doi: 10.18632/oncotarget.14044
Shiba, S., Ikeda, K., Suzuki, T., Shintani, D., Okamoto, K., Horie-Inoue, K., et al. (2019). Hormonal regulation of patient-derived endometrial cancer stem-like cells generated by Three-dimensional culture. Endocrinology 160, 1895–1906. doi: 10.1210/en.2019-00362
Siegel, R. L., Miller, K. D., and Jemal, A. (2020). Cancer statistics, 2020. CA. Cancer J. Clin. 70, 7–30. doi: 10.3322/caac.21590
Smolle, M. A., Bullock, M. D., Ling, H., Pichler, M., and Haybaeck, J. (2015). Long non-coding RNAs in endometrial carcinoma. Int. J. Mol. Sci. 16, 26463–26472. doi: 10.3390/ijms161125962
Sun, S., Wang, N., Sun, Z., Wang, X., and Cui, H. (2019). MiR-5692a promotes proliferation and inhibits apoptosis by targeting HOXD8 in hepatocellular carcinoma. J. BUON. 24, 178–186.
Vychytilova-Faltejskova, P., Merhautova, J., Machackova, T., Gutierrez-Garcia, I., Garcia-Solano, J., Radova, L., et al. (2017). MiR-215-5p is a tumor suppressor in colorectal cancer targeting EGFR ligand epiregulin and its transcriptional inducer HOXB9. Oncogenesis 6, 399–406. doi: 10.1038/s41389-017-0006-6
Wang, S., Ni, B., Zhang, Z., Wang, C., Wo, L., Zhou, C., et al. (2019). Long non-coding RNA DNM3OS promotes tumor progression and EMT in gastric cancer by associating with Snail. Biochem. Biophys. Res. Commun. 511, 57–62. doi: 10.1016/j.bbrc.2019.02.030
Wang, W., Zhao, L. J., Tan, Y. X., Ren, H., and Qi, Z. T. (2012). Identification of deregulated miRNAs and their targets in hepatitis B virusassociatedhepato-cellular carcinoma. World J. Gastroenterol. 18, 5442–5453. doi: 10.3748/wjg.v18.i38.5442
Wang, Y., Xu, M., and Yang, Q. (2019). A six-microRNA signature predicts survival of patients with uterine corpusendometrial carcinoma. Curr. Probl. Cancer 43, 167–176. doi: 10.1016/j.currproblcancer.2018.02.002
Wilczynski, M., Danielska, J., Domanska-Senderowska, D., Dzieniecka, M., Szymanska, B., and Malinowski, A. (2018). Association of microRNA-200c expression levels with clinicopathological factors and prognosis in endometrioid endometrial cancer. Acta Obstet. Gynecol. Scand. 97, 560–569. doi: 10.1111/aogs.13306
Winnepenninckx, V., Lazar, V., Michiels, S., Dessen, P., Stas, M., Alonso, S. R., et al. (2006). Gene expression profiling of primary cutaneous melanoma and clinical outcome. J. Natl. Cancer. Inst. 98, 472–482. doi: 10.1093/jnci/djj103
Xie, W., Zhang, J., Zhong, P., Qin, S., Zhang, H., and Fan, X. (2019). Expression and potential prognostic value of histone family gene signature in breast cancer. Exp. Ther. Med. 18, 4893–4903. doi: 10.3892/etm.2019.8131
Xing, Y., Jing, H., Zhang, Y., Suo, J., and Qian, M. (2020). MicroRNA-141-3p affected proliferation, chemosensitivity, migration and invasion of colorectal cancer cells by targeting EGFR. Int. J. Biochem. Cell Biol. 118:105643. doi: 10.1016/j.biocel.2019.105643
Zhang, W., Bouchard, G., Yu, A., Shafiq, M., Jamali, M., Shrager, J. B., et al. (2018). GFPT2-expressing cancer-associated fibroblasts mediate metabolic reprogramming in human lung adenocarcinoma. Cancer Res. 78, 3445–3457. doi: 10.1158/0008-5472.CAN-17-2928
Zhao, C., Zhang, J., Zhang, S., Yu, D., Chen, Y., Liu, Q., et al. (2013). Diagnostic and biological significance of microRNA-192 in pancreatic ductal adenocarcinoma. Oncol. Rep. 30, 276–284. doi: 10.3892/or.2013.2420
Zhou, L. Y., Zhang, F. W., Tong, J., and Liu, F. (2020). MiR-191-5p inhibits lung adenocarcinoma by repressing SATB1 to inhibit Wnt pathway. Mol. Genet. Genomic Med. 8:e1043. doi: 10.1002/mgg3.1043
Keywords: endometrial carcinoma, cancer genomics, the Cancer Genome Atlas (TCGA), integrative model, prognosis
Citation: Deng F, Mu J, Qu C, Yang F, Liu X, Zeng X and Peng X (2021) A Novel Prognostic Model of Endometrial Carcinoma Based on Clinical Variables and Oncogenomic Gene Signature. Front. Mol. Biosci. 7:587822. doi: 10.3389/fmolb.2020.587822
Received: 27 July 2020; Accepted: 23 November 2020;
Published: 07 January 2021.
Edited by:
Peng Zhang, University of Maryland, United StatesReviewed by:
Siddharth Shukla, Howard Hughes Medical Institute (HHMI), United StatesJunjiang Fu, Southwest Medical University, China
Copyright © 2021 Deng, Mu, Qu, Yang, Liu, Zeng and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaomin Zeng, enhpYW9taW5AY3N1LmVkdS5jbg==; Xiaoning Peng, cHhpYW9uaW5nQGh1bm51LmVkdS5jbg==
 Fang Deng1
Fang Deng1 Xiaomin Zeng
Xiaomin Zeng