Corrigendum: Systematic Analysis of Expression Profiles and Prognostic Significance for FAM83 Family in Non-Small-Cell Lung Cancer
- 1Department of Medical Oncology, Harbin Medical University Cancer Hospital, Harbin, China
- 2Department of Gastrointestinal Oncology, Harbin Medical University Cancer Hospital, Harbin, China
Background: Lung cancer remains a common malignancy and the leading cause of cancer-related deaths in the world. Although dramatic progress made in multimodal therapies, it still has a poor prognosis. The Family with sequence similarity 83 (FAM83) of poorly characterized proteins are defined by the presence of the conserved DUF1669 domain of unknown function at their N-termini, most of which significantly elevated levels of expression in multiple cancers. However, the expression and prognostic values of different FAM83 family in lung cancer, especially in non-small-cell lung cancer (NSCLC), have not been clarified.
Methods: ONCOMINE, UALCAN, GEPIA, Kaplan–Meier Plotter, cBioPortal, and STRING databases were utilized in this study.
Results: The transcriptional levels of FAM83A/B/C/D/F/G/H were up-regulated in patients with NSCLC. A noticeable correlation was found between the over-expressions of FAM83A/B/D/F/H and clinical cancer stages in NSCLC patients. Besides, higher mRNA expressions of FAM83A/B/C/D/F/H were discovered to be expressively associated with overall survival (OS) in lung cancer patients, furthermore, FAM83A, FAM83C, and FAM83H in OS group achieved 0.9475/1, 0.971897/1, and 0.9454545/0.8974359 specificity/sensitivity in distinguishing short survivors from long survivors, respectively. Moreover, a high mutation rate of FAM83 family (51%) was also observed in lung adenocarcinoma (LUAD) patients, and genetic alteration in the FAM83 family was associated with shorter OS and disease-free survival (DFS) in LUAD patients.
Conclusion: Our results indicated that FAM83A/H might play important roles in NSCLC tumorigenesis and might be risk factor for the survival of NSCLC patients.
Introduction
In both sexes combined, lung cancer is the most commonly diagnosed cancer and the leading cause of cancer death (Bray et al., 2018). There are two main types of lung cancer: small cell lung cancer and non-small-cell lung cancer (NSCLC) (Alberg et al., 2005; Braicu et al., 2019), among which, NSCLC is mainly composed of lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), accounting for approximately 85% of new lung cancer cases (Wang Y. et al., 2019; Zheng et al., 2020). Efforts have been made in studying the mechanisms of the development, progression, and metastasis of NSCLC (Wang and Adjei, 2015; Popper, 2016; Inamura, 2018). However, the molecular characteristics of NSCLC, to date, remain unknown. Probing new highly specific and sensitive biomarkers and new molecular targets can not only help to elucidate the molecular mechanism of NSCLC patients, but also improve the prognosis of NSCLC patients.
The Family with sequence similarity 83 (FAM83) of proteins comprises of eight members of A–H, which have been categorized based on a highly conservative domain of the unknown function (DUF1669) at the N-terminus, while the C-terminal regions varied greatly among the different family members (Bartel et al., 2016; Snijders et al., 2017; Bozatzi and Sapkota, 2018). The first detailed analysis of the FAM83 proteins in transformation resulted from the separate identification of FAM83A and FAM83B in distinct genetic screens performed by the Bissell (Lee et al., 2012) and Jackson Laboratories (Cipriano et al., 2012), respectively. Subsequently, diverse studies have confirmed that members of FAM83 family with overexpression or dysregulation play important role in cell growth, proliferation, metastasis, and resistance to precision therapies (Bartel et al., 2016). Zhou et al. (2019) demonstrated that FAM83A accelerated NSCLC cell migration and invasion through the activation of epithelial-mesenchymal transition (EMT) via PI3K/ATK/Snail signaling. Moreover, a study verified that miR-143 restrained the proliferation, migration, and invasion of esophageal squamous cell carcinoma (ESCC) cells and also lead to G1/G0 phase arrest via down-regulation of FAM83F expression (Mao et al., 2016). A comprehensive study of FAM83 family members in NSCLC will help to uncover the molecular mechanisms involved in the development of NSCLC and could unveil novel prognostic and therapeutic targets for the intractable disease.
In the past few years, studies have demonstrated abnormal expression in some members of the FAM83 family and their prognostic value. For example, Fuziwara et al. (2019) reported that FAM83F, a novel oncogenic protein, was overexpressed in thyroid cancer and cross-regulated MAPK and TGF signaling pathways to promote the biological behavior and differentiation of thyroid follicular cells. FAM83B was recently identified as a novel oncogene involved in activating CRAF/MAPK signaling and driving epithelial cell transformation (Cipriano et al., 2012). Furthermore, evidence suggested that the upregulation of FAM83 members was remarkably correlated with an elevated breast tumor grade and decreased overall survival (OS) (Cipriano et al., 2014). Nevertheless, the role of distinct FAM83 family members remained unknown in the development and progression of NSCLC. In this present study, bioinformatics was performed initially to address this problem by analyzing the expression, prognosis, and mutations of different FAM83 family members and their relations with individual cancer stages in NSCLC patients. Furthermore, we also analyzed the predicted functions and pathways of the mutations in the FAM83 family as well as their 80 frequently altered genes.
Materials and Methods
Ethics Statement
The study has been admitted by the Institutional Review Board of The Harbin Medical University. Since all the data were retrieved from the online databases, it could be affirmed that all written informed consent had already been obtained.
ONCOMINE Database
ONCOMINE database1 is a publicly accessible online cancer microarray database, which provides a genome-wide expression analysis (Rhodes et al., 2004). It was utilized to analyze the transcription levels of FAM83 family between disparate cancer tissues and their corresponding adjacent normal control samples. In this study, the cell color is determined by the best gene rank percentile for the analysis within the cell, and Q–Q graph and histogram were used to detect whether the sample data obeyed normal distribution, then the Student’s t-test was applied to generate a p-value. A p-value of 0.05, a fold change of 2, and a gene rank in the top 10% were set as the significance thresholds.
UALCAN
UALCAN2 is an omnibus, user-friendly, and interactive web resource based on The Cancer Genome Atlas (TCGA) level 3 RNA-seq and clinical data from 31 cancer types (Chandrashekar et al., 2017). In our study, UALCAN was used to illustrate the distinct expression levels of tumor and normal tissues. Student’s t-test was used to generate a p-value and the p-value cutoff was 0.05.
GEPIA
GEPIA3 is a newly developed interactive web server for elaborating the RNA sequencing expression data of 9736 tumors and 8587 normal samples from the TCGA and Genotype-tissue Expression dataset, utilizing a standard processing pipeline. GEPIA offers customizable functions such as tumor/normal differential expression analysis, profiling according to cancer types or pathological stages, patient survival analysis, similar gene detection, correlation analysis, and dimensionality reduction analysis (Tang et al., 2017) For analysis, “Multiple Gene Comparison” was used to evaluate the multiple gene comparison analysis of FAM83 family. “Expression DIY” was utilized to compare eight FAM83 family members’ association with clinicopathologic parameters. The Student’s t-test was used to generate a p-value and the p-value cutoff was 0.05.
Kaplan–Meier Plotter
The Kaplan Meier plotter4 is able to assess the effect of 54 k genes on survival in 21 cancer types. The largest datasets include breast, ovarian, lung, and gastric cancer (Nagy et al., 2018). In this study, it was used to evaluate the prognostic value of FAM83 family mRNA expression in which cancer patients were split into high and low expression group based on median values of mRNA expression and validated by K-M survival curves, with the hazard ratio (HR) with 95% confidence intervals (CI) and log-rank p-value. The statically significant difference was considered when a p-value is <0.05.
TCGA Data and cBioPortal
The Cancer Genome Atlas had both sequencing and pathological data on 30 different cancers (2012). The LUAD (TCGA, Firehose Legacy) dataset including data from 515 cases with pathology reports was selected for further analyses of FAM83 family using the cBioPortal5), which is a comprehensive web resource for exploring, visualizing, and analyzing multidimensional cancer genomics data. In this study, we analyzed the genomic profiles of eight FAM83 family members, which contained mutations, putative copy-number alterations from GISTIC, and mRNA Expression z-Scores (RNASeq V2 RSEM) with a z-score threshold of ±1.8 (Cerami et al., 2012; Gao et al., 2013). Genetic mutations in FAM83 family and their association with OS and disease free survival (DFS) of lung cancer patients were displayed as Kaplan–Meier plots, and the log-rank test was performed to identify the significance of the diversity between the survival curves, and when a p-value is <0.05, the difference was considered statically significant. Co-expressed genes of FAM83 family was performed with the “Co-expression” module of cBioportal. Pearson’s correlation coefficient was used to investigate the correlation between FAM83 family and co-expressed genes and top ten co-expressed genes of each FAM83 family with the largest Pearson’s correlation coefficient were listed.
STRING
STRING6 is a database of known and predicted protein–protein interactions (PPI) (Szklarczyk et al., 2019). Herein, to detect the role of FAM83 family co-expressed genes, the online database of STRING was applied to analyze associations among the PPI network of FAM83 family co-expressed genes, and the species were set to Homo sapiens and a combined score of >0.7 was considered statistically significant. The nodes meant proteins; the edges meant the interaction of proteins and we hide disconnected nodes in the network.
DAVID
Functions of FAM83 family mutations and 80 genes significantly related to FAM83 family mutations were analyzed by the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) in the Database for Annotation, Visualization, and Integrated Discovery (DAVID)7 (Huang da et al., 2009a,b). Gene ontology analyses focus on three domains: biological processes (BP), cellular components (CC), and molecular functions (MF), and such analyses are commonly used to predict the functional roles of FAM83 family mutations and 80 genes significantly associated with FAM83 family mutations, while KEGG analysis can define the pathways related to the FAM83 family mutations and 80 co-expressed genes associated with FAM83 family mutations. Only terms with p-value of <0.05 were considered as significant.
Results
Aberrant Expression of FAM83 Family in Patients With NSCLC
The FAM83 family genes located at definite genomic sites (Table 1; Bartel et al., 2016) and these proteins are described by the presence of the conserved DUF1669 domain of unknown function at their N-terminal, whereas the rest of the proteins vary in length and are not conserved between members (Figure 1A; Bozatzi and Sapkota, 2018). ONCOMINE database (see text footnote 1) and UALCAN (see text footnote 2) were utilized to explore expression levels of FAM83 family in cancers with those in normal samples. We used the Q–Q graph and histogram to detect whether the sample data obeyed normal distribution. As shown in Supplementary Figure 1, the sample data basically conforms to the normal distribution as we anticipated. Subsequently, we selected t-test as the main statistical method when analyzing the raw data in the ONCOMINE database. The results in Figure 1B showed that the mRNA expression levels of FAM83A/C/D/E/F/H were remarkably up-regulated in lung cancer tissues in multiple datasets. In the Okayama Lung dataset (Okayama et al., 2012), FAM83A overexpression was detected in LUAD (N = 226) compared with normal tissues (N = 20) with a fold change of 3.742 (p = 2.61E-22), while Hou found a 4.448-fold increase in FAM83A mRNA expression in 45 LUAD samples (p = 6.75E-13) (Hou et al., 2010), moreover, Selamat investigated a 3.928-fold increase in FAM83A mRNA expression in 58 LUAD tissues (p = 4.19E-17) (Selamat et al., 2012) and in the Garber dataset, the mRNA expression of FAM83A in LUAD (N = 40) was increased (by a fold change of 2.238, p = 1.00E-03) (Table 2; Garber et al., 2001). What is more, significant up-regulation of FAM83C was also found in LUAD tissues compared to normal tissues. The result from the Hou dataset unfolded that there was a 5.995-fold (p = 3.11E-09) increase in FAM83C mRNA expression in 27 LUAD tissues (Table 2; Hou et al., 2010). In the Garber dataset (Garber et al., 2001), FAM83D was overexpressed in all kinds of the lung cancer (N = 54) than in the normal tissues: by a fold change of 11.481, (p = 2.76-10) in LUSC, by a fold change of 6.635, (p = 4.07E-11) in LUAD, by a fold change of 7.297, (p = 2.00E-03) in large-cell lung carcinoma, by a fold change of 9.244 (p = 4.00E-03) in small cell lung carcinoma, respectively. Likewise, the transcription levels of FAM83D in 27 LUSC, 45 LUAD, and 19 large cell lung carcinoma (Hou et al., 2010) were higher than those in lung samples (N = 65), and their fold changes were 7.532 (p = 2.80E-20), 3.072 (p = 1.26E-11), 5.161 (p = 7.93E-06), respectively (Table 2). In the Selamat dataset (Selamat et al., 2012), the mRNA expression of FAM83E in LUAD (N = 58) increased with a fold change of 2.043 (p = 2.23E-14). A similar trend of FAM83F was shown in the Hou database (Hou et al., 2010). FAM83F was notably up-regulated in 27 LUSC and 19 Large Cell Lung Carcinoma, with a fold change of 3.110 (p = 3.25E-08) and 2.328 (p = 3.49E-05), respectively (Table 2). The transcriptional levels of FAM83H in 58 LUAD (fold change = 2.715, p = 1.07E-23) and in 27 LUSC (fold change = 2.493, p = 6.81E-14) considerably differed from those in the normal samples in the Selamat (Selamat et al., 2012) and Hou datasets (Hou et al., 2010), respectively (Table 2). Furthermore, using the UALCAN (see text footnote 2), we compared the mRNA expression of FAM83 family between 515 LUAD and 59 normal tissues, 503 LUSC and 52 normal tissues. The results in Figure 2 discovered that FAM83A/B/C/D/F/G/H were higher in LUAD and LUSC tissues than in lung tissues. Moreover, FAM83E was higher in LUAD than in lung tissues, nevertheless the expression level of FAM83E was lower in the LUSC tissues than in lung tissues. Besides, we also contrast the relative expression levels of FAM83 family in LUAD and LUSC tissues and determined that among all the factors we evaluated, FAM83A was the highest expression in LUAD and FAM83H was the highest in LUSC (Figure 3). Taken together, our results showed that transcriptional expressions of FAM83A/B/C/D/F/G/H were over-expressed in patients with NSCLC.

Figure 1. The expression of FAM83 family in distinct types of cancer diseases (ONCOMINE database). (A) The pattern diagram of the human FAM83 family of proteins and the conserved domain of unknown function, DUF1669 that characterizes them. (B) The figure shows the numbers of datasets with statistically significant mRNA up-regulation (red) or down-regulated expression (blue) of FAM83 family. Student’s t-test was used to compared the different transcriptional. Cutoff of p-value and fold change were as following: p-value: 0.01, fold change: 2, gene rank: 10%, data type: mRNA.
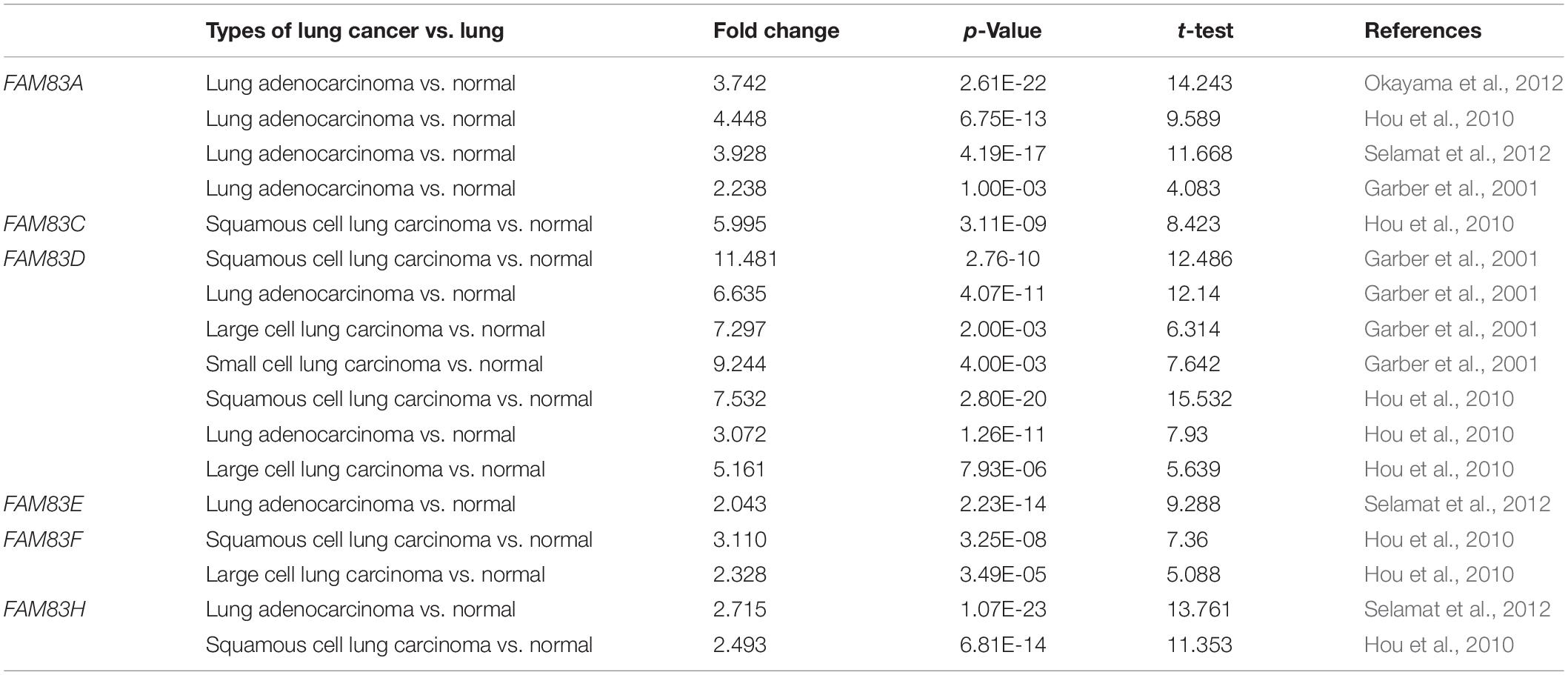
Table 2. Remarkable changes of FAM83 family expression in transcription level between lung cancer and normal lung tissues (ONCOMINE).
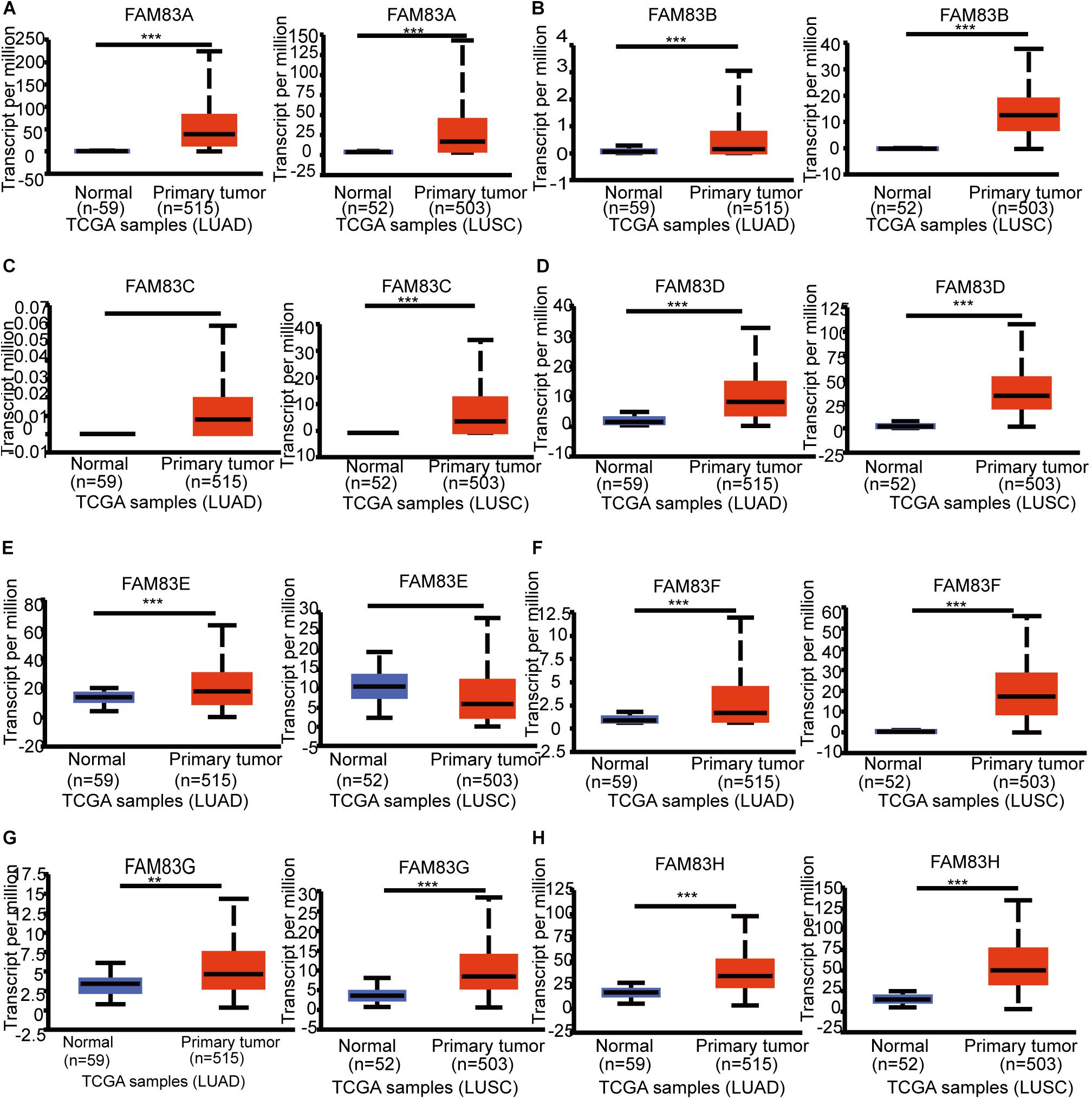
Figure 2. The mRNA expression of diverse FAM83 family members in NSCLC tissues and adjacent lung tissues (UALCAN). mRNA expressions of FAM83A/B/C/D/F/G/H were found to be over-expressed in LUAD and LUSC tissues compared to normal samples (A–D, F–H). FAM83E was higher in LUAD tissues than in lung tissues, whereas and the expression level of FAM83E was lower in the LUSC tissues than in lung tissues (E). ***p < 0.001, **p < 0.01.
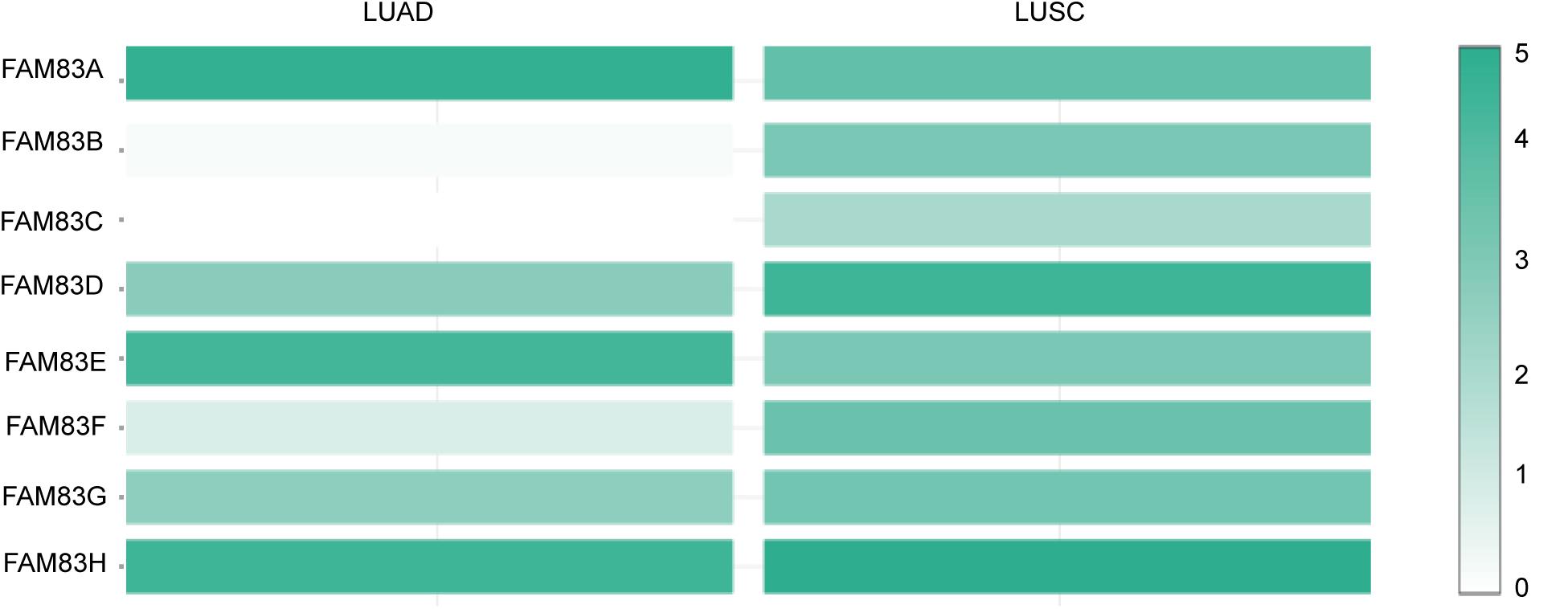
Figure 3. The relative level of FAM83 family in NSCLC. FAM83A was the highest expression in LUAD and FAM83H was the highest in LUSC.
Correlation Between mRNA Expression of Different FAM83 Family and Tumor Stages of NSCLC Patients
Next, we analyzed the relationship between the mRNA expression of different FAM83 family members with patients’ individual cancer stages of LUAD and LUSC patients by GEPIA (see text footnote 3). FAM83A, FAM83B, FAM83D, FAM83F, and FAM83H groups appreciably varied, whereas FAM83C, FAM83E, and FAM83G groups did not markedly differ (Figure 4). The reason why the mRNA expressions of FAM83C/E/G in NSCLC seemed to not significantly diverge may be as a result of small sample size. In short, the results above suggested that mRNA expressions of FAM83A/B/D/F/H were obviously related to patients’ individual cancer stages, and patients who were in more advanced cancer stages were inclined to express higher mRNA expression of FAM83A/B/D/F/H.
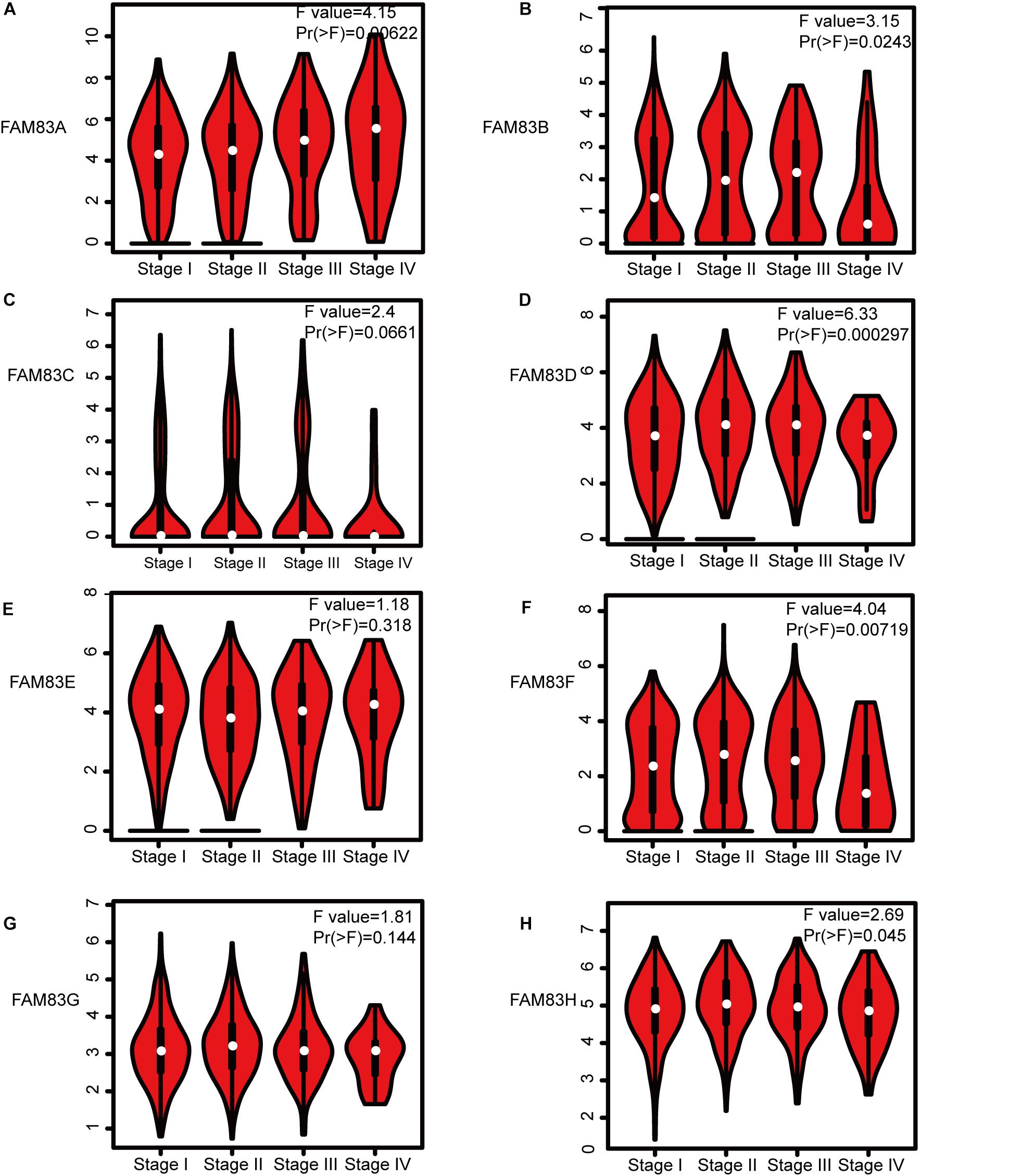
Figure 4. Correlation between FAM83 family expression and tumor stage in NSCLC patients (GEPIA). The mRNA expressions of FAM83A/B/D/F/H were significantly related to patients’ individual cancer stages (A,B,D,F,H), while mRNA expressions of FAM83C/E/G were not associated with patients’ individual cancer stages (C,E,G).
Prognostic Features of FAM83 Family in Patients With Lung Cancer
By means of data mining in the Kaplan–Meier plotter (2015 version; see text footnote 4), prognostic value of FAM83 family mRNA for lung cancer patients including OS, progression-free survival (FP), and post-progression survival (PPS) were investigated, respectively. It could be seen that, in each cohort, patients were divided into low and high risk group based on cutoff value (Figure 5 and Table 3). Reversed relationship was shown between OS and the mRNA levels of FAM83A/B/C/D/F/H; however, there was no significant association in lung cancer between OS and FAM83E. There was no significant correlation in lung cancer between FP and either FAM83D or FAM83H, whereas high FAM83A/B/C/E/F mRNA expression led to a reduced FP. In addition, increased FAM83A/D/F/H mRNA expression levels were associated with PPS. Unfortunately, by querying this public database, we did not find the relationship between the mRNA expression of FAM83G and prognosis. We also used scatter diagram to determine appropriate cutoff values of FAM83 family in OS group, FP group, and PPS group and to clarify their clinical significance (data from Supplementary Table 2). The scatter diagram in Supplementary Figure 2 revealed the statically significant difference (p-value < 0.05) when 362 and 1030 was adopted as the cutoff of FAM83A among all patients in OS group and FP group. And when p-value is <0.05, cutoff value of FAM83B (in FP group) is 13, FAM83C (in OS group and FP group) is 17 and 30, respectively, FAM83D (in PPS group) is 993, FAM83E (in FP group) is 16, FAM83F (in FP group) is 66, and FAM83H (in OS group) is 804. As shown in Supplementary Table 3, we described FAM83 family prognostic potential: FAM83A in OS group and PPS group achieved a 0.9475/1 and 0.952381/0.7586207 specificity/sensitivity in distinguishing short survivors from long survivors, accordingly. FAM83B in FP group achieved a 0.8235294/0.9910714 specificity/sensitivity in distinguishing short survivors from long survivors. FAM83C in OS group and FP group achieved a 0.971897/1 and 1/0.9810427 specificity/sensitivity in distinguishing short survivors from long survivors, respectively. FAM83D in PPS group achieved a 0.8333333/0.9661017 specificity/sensitivity. FAM83E in FP group achieved a 0.3424658/0.9504717 specificity/sensitivity in discriminating short survivors from long survivors. FAM83F in FP group achieved a 0.9148936/0.6 specificity/sensitivity in discriminating short survivors from long survivors. FAM83H in OS group achieved a 0.9454545/0.8974359 specificity/sensitivity in distinguishing poor survival.
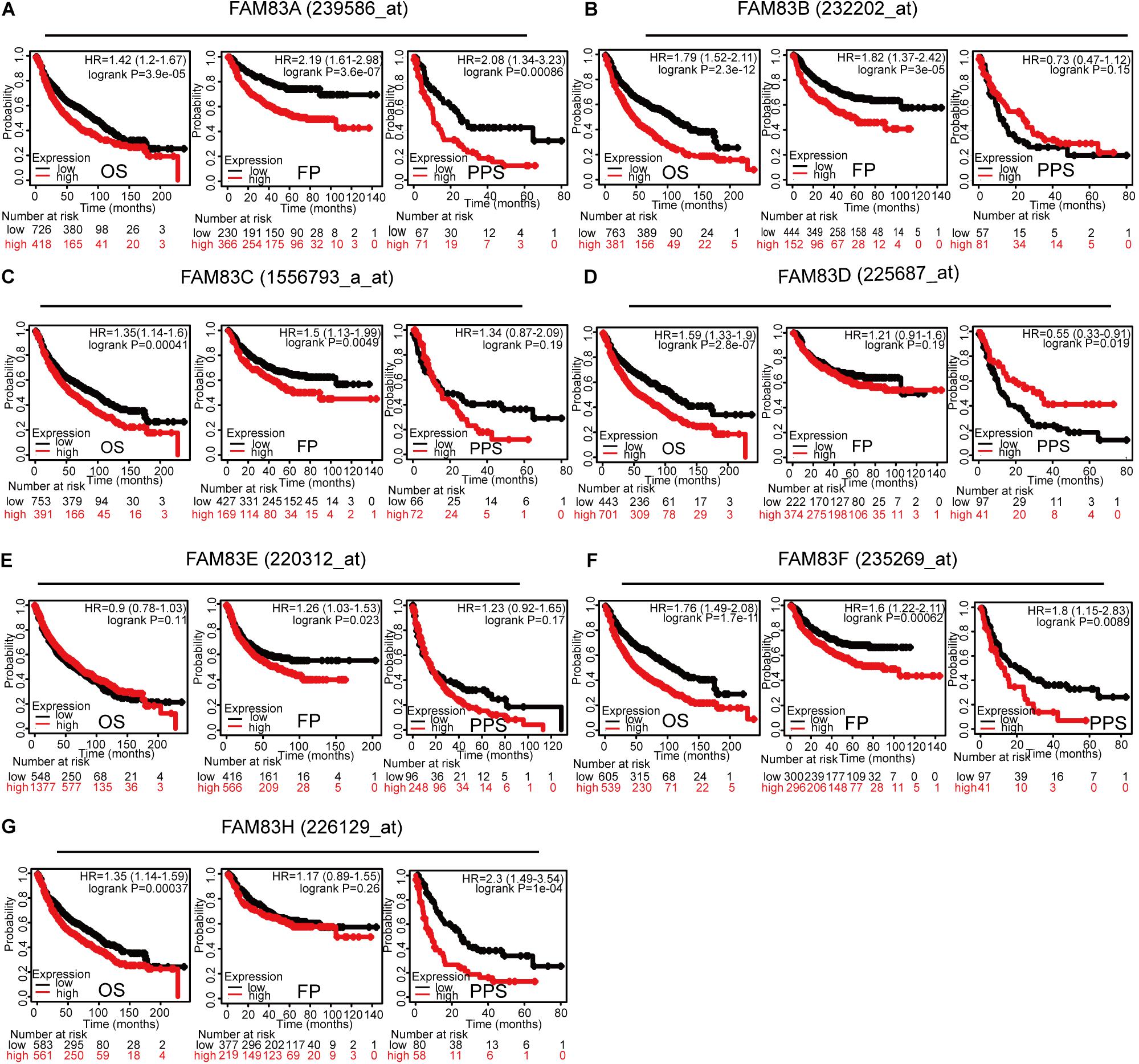
Figure 5. Prognostic feature of mRNA expression of distinct FAM83 family members in lung cancer patients (Kaplan–Meier plotter). The OS, FP, and PPS survival curves comparing patients with high (red) and low (black) FAM83 family member expression in lung cancer were plotted using the Kaplan–Meier plotter database at the threshold of p-value of <0.05. The association between Prognostic value and (A) FAM83A, (B) FAM83B, (C) FAM83C, (D) FAM83D, (E) FAM83E, (F) FAM83F, (G) FAM83H protein expression.
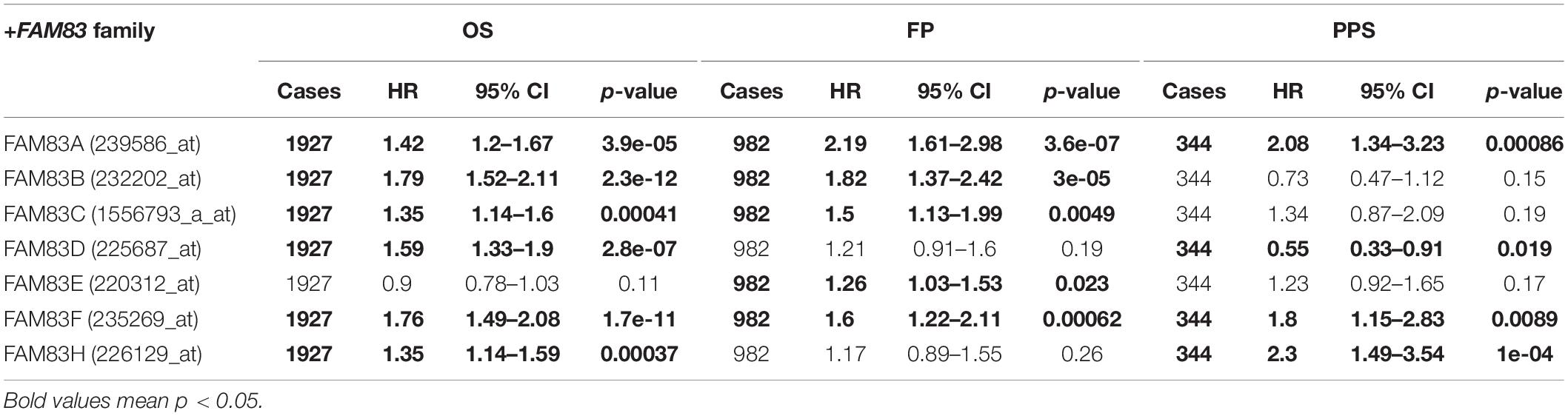
Table 3. The prognostic values of FAM83 family members in lung cancer patients (Kaplan–Meier plotter).
Genetic Mutations in FAM83 Family and Their Associations With OS and Disease-Free Survival of Lung Cancer Patients
Epigenetic alteration plays a vital role in early malignancies (Cancer Genome and Atlas Network, 2012). Then, we analyzed the FAM83 family alterations and correlations by using the cBioPortal online tool (see text footnote 5) for LUAD (TCGA, Firehose Legacy), in which FAM83 family was varied in 265 samples out of 515 patients with LUAD (51%) (Figure 6A). FAM83H, FAM83A, FAM83D, and FAM83B were the top four genes with genetic alterations, and their mutation rates were 23, 16, 14, and 10%, respectively. Besides, we also figured the correlations between FAM83 family by analyzing their mRNA expression (RNA Seq V2 RSEM) via the cBioPortal online tool for LUAD (TCGA, Firehose Legacy), which contained Pearson’s correction. The consequences exposed noteworthy and positive relationship in the following FAM83A with FAM83B and FAM83H (Figure 6B). Furthermore, we analyzed the relationship of genetic alteration in FAM83 family with OS and DFS of lung cancer patients. Results from the Kaplan–Meier plot and log-rank test uncovered that genetic alteration in FAM83 family was related to shorter OS (Figure 6C, p = 1.135E-3) and DFS (Figure 6D, p = 0.0381) of lung cancer patients. These discovery observed that the change of FAM83 family gene may also crucially affect the prognosis of lung cancer patients.
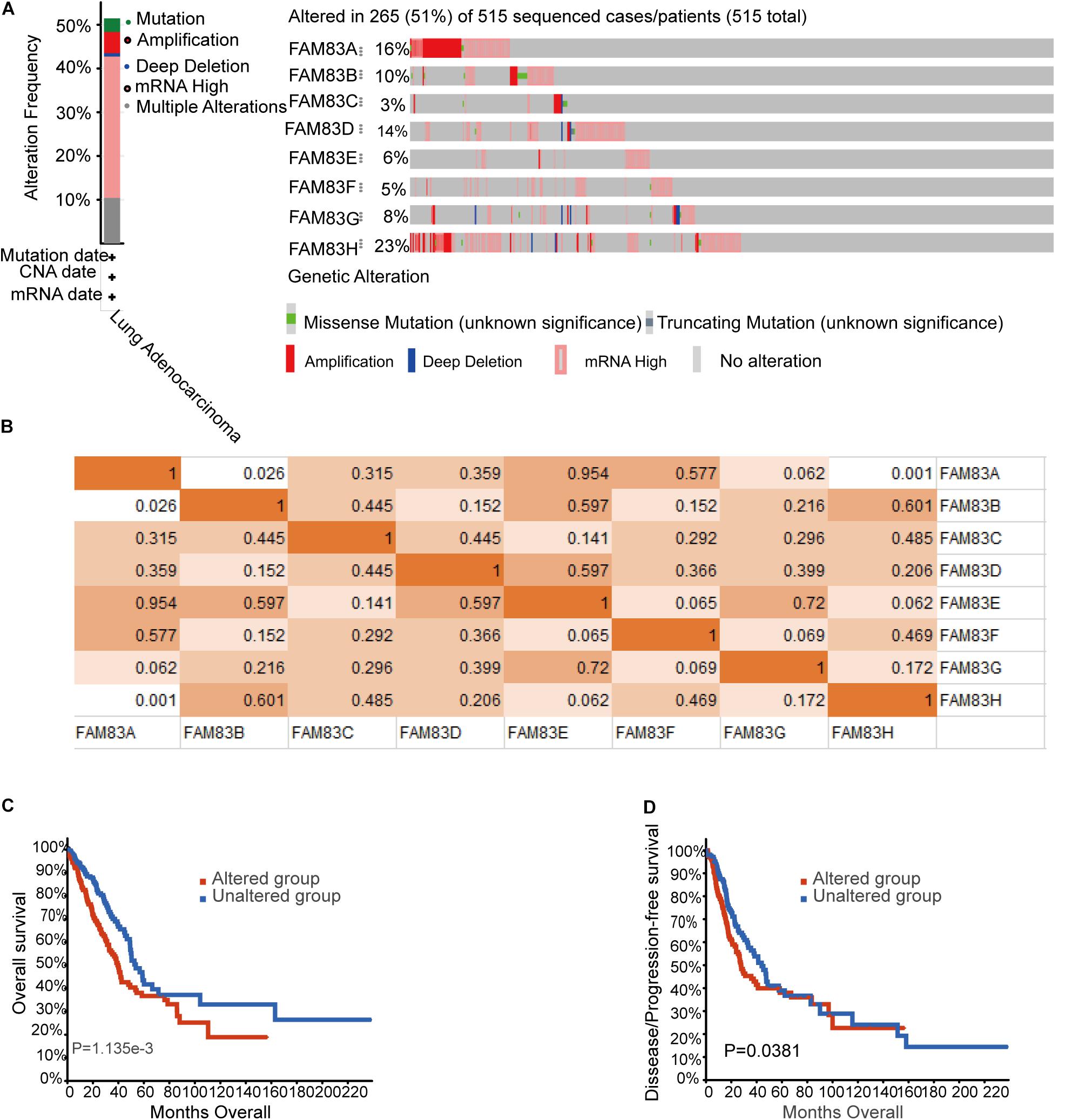
Figure 6. Genetic mutations in FAM83 family and their association with OS and DFS of NSCLC patients (cBioPortal). (A) Summary of alterations in different expressed FAM83 family in LUAD. (B) Correlations of different FAM83 family members with each other in LUAD. (C) Genetic alterations in FAM83 family were related to shorter OS and (D) DFS of LUAD patients.
GO and KEGG Enrichment Analysis of FAM83 Family and Their 80 Co-expression Genes in NSCLC Patients
After analyzing the genetic alterations in FAM83 family and their prognostic value in NSCLC patients, we then analyzed 80 co-expression genes that were significantly associated with FAM83 family mutations with the “Co-expression” module of cBiopotal and listed them in Supplementary Table 1.Subsequently, we constructed an integrated network by STRING (see text footnote 6). The results in Figure 7A exposed that the cell cycle-related genes, including CDCA5 and CDCA3, and chromosome segregation participant genes, such as KIF23 and KIF2C, were closely connected with FAM83 family alterations. In addition, GO and KEGG in DAVID (see text footnote 7) were employed to analyze the potential role of FAM83 family and their 80 co-expression genes. As shown in Figures 7B–E, we found that BP such as GO:0007062 (sister chromatid cohesion), GO:0030198 (extracellular matrix organization), GO:0007067 (mitotic nuclear division), GO:0051301 (cell division), and GO:0008283 (cell proliferation) were remarkably regulated by the FAM83 family mutations in NSCLC (Figure 7B). Moreover, CC, including GO:0005737 (cytoplasm), GO:0005829 (cytosol), GO:0005913 (cell–cell adherens junction), and GO:0005911 (cell–cell junction) were significantly associated with the FAM83 family alterations (Figure 7C). Furthermore, FAM83 family mutations also prominently affected the MF, such as GO:0005515 (protein binding), GO:0005524 (ATP binding), GO:0019901 (protein kinase binding), and GO:0098641 (cadherin binding involved in cell–cell adhesion). In the KEGG analysis, only hsa04530 (Tight junction) was greatly related to the functions of FAM83 family in NSCLC (Figure 7E).
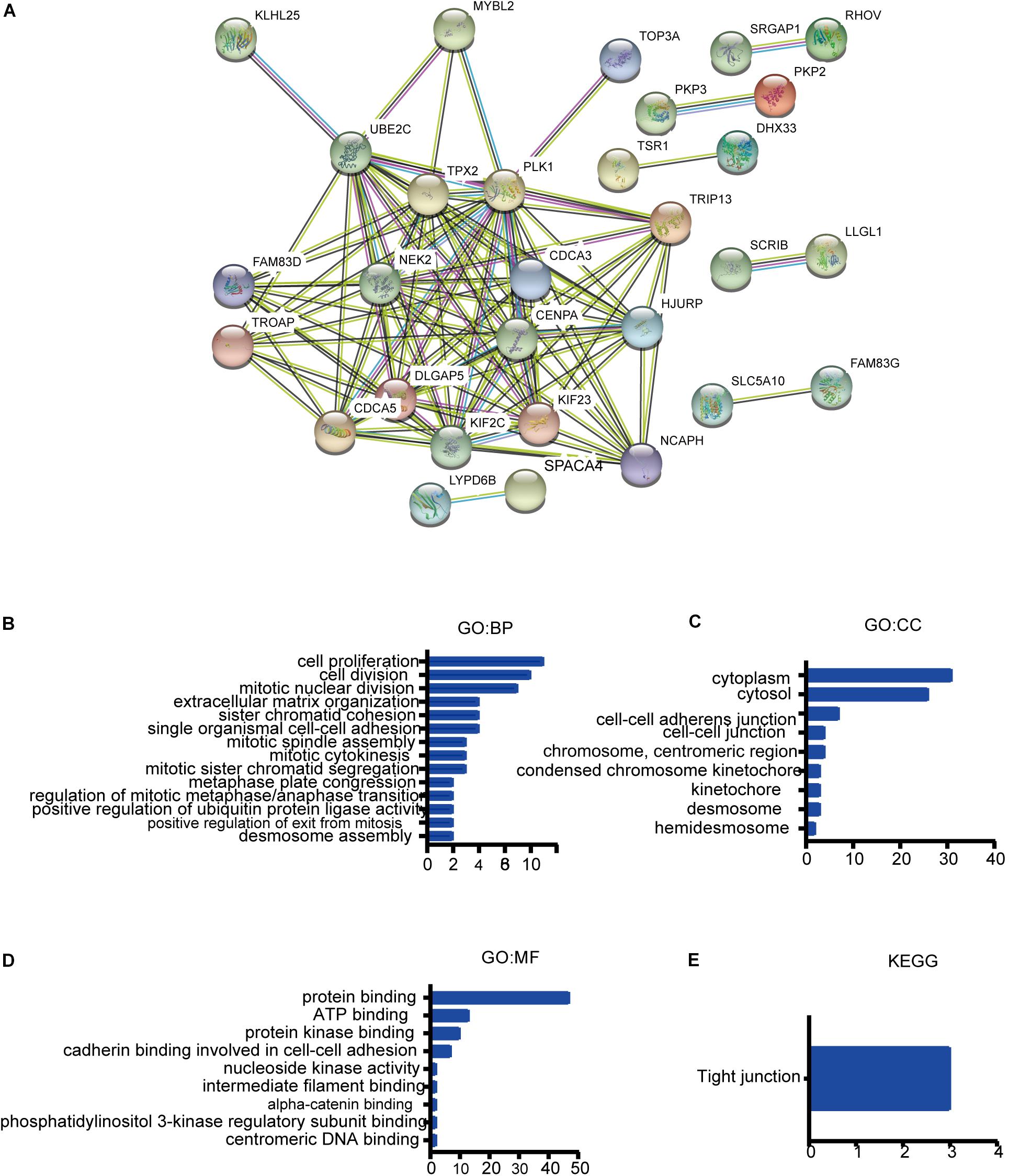
Figure 7. GO and KEGG enrichment analysis of FAM83 family and their 80 co-expression genes in NSCLC patients. (STRING and DAVID). (A) PPI network. The nodes meant proteins; the edges meant the interaction of proteins (B) BP. (C) CC. (D) MF. (E) KEGG.
Discussion
While all eight FAM83 members share the highly conserved DUF1669, they also possess unique C-terminus of variable length and otherwise lack any significant homology beyond the DUF1669 (Fulcher et al., 2018), explaining the reason of poor enrichment function. Thus, we consider that the DUF1669 is the carcinogenetic part of the FAM83 proteins (Cipriano et al., 2014), and therefore the most attractive therapeutic target. Based on the divergent sizes of the FAM83 proteins (ranging from 434–1179 amino acids), each protein likely has additional DUF1669-independent cellular functions (Bartel et al., 2016). Therefore, the present study is the first time to analyze the mRNA expression, mutation, and prognostic roles of different FAM83 factors in NSCLC.
FAM83A has been reported to play a role in promoting cancer and reducing drug sensitivity in a variety of cancer (Liu et al., 2020). Parameswaran et al. (2019) indicated that FAM83A was overexpressed in human and murine pancreatic cancers and protected from cell death in pancreatic cancer cells by activating essential MEK/ERK survival signaling. Similarly, a study on breast cancer found that overexpression of FAM83A also enhanced cell proliferation and invasiveness, while increasing resistance to tyrosine kinase inhibitors (TKIs). Conversely, down-regulation of FAM83A restrained the malignant phenotype, reduced tumor growth in immunocompromised mice, and increased TKI responsiveness (Grant, 2012). In the past few years, the expression of FAM83A was found to be noticeably over-expressed in lung cancer, moreover, FAM83A was related to an advanced TNM stage and poor prognosis, and regulated the Wnt and Hippo signaling pathways and EMT process to accelerate lung cancer cells’ proliferation and invasion (Zheng et al., 2020). In our present study, we found that the up-expression of FAM83A in NSCLC tissues and that the mRNA expression of FAM83A was dramatically related to patients’ individual cancer stages. What was more, higher mRNA expression of FAM83A led to a reduced OS, FP, and PPS of lung cancer patients, which were similar to the findings of Yu et al.’s (2020) studies. Further analysis determined that the patients were categorized by cutoffs of 362 for FAM83A low expression and high expression and achieved a 0.8333333/0.9661017 specificity/sensitivity in distinguishing poor survival from OS. These discoveries raise the possibility that this gene may play a remarkable role in the tumorigenesis of NSCLC and might become a new therapeutic target in the future.
Studies from Okabe et al. (2015) showed that FAM83B was a novel biomarker for the diagnosis and prognosis of LUSC. Furthermore, Shen et al. (2017) discovered that the expression of FAM83B was observably enhanced both in pancreatic ductal adenocarcinoma cell lines and tumor tissues. FAM83B expression was positively related with advanced clinical stage and poor vital status. Higher FAM83B expression led to a reduced OS in pancreatic ductal adenocarcinoma patients, regardless of the lymphatic metastasis status. What was more, down-regulation of FAM83B contributed to G0/G1 phase arrest and inhibition of cell proliferation (Shen et al., 2017). Similarly, Cipriano et al. (2012) found that FAM83B expression was dramatically elevated in cancer, increased tumor grade, and decreased OS. In our study, FAM83B expression levels in NSCLC tumor tissues were markedly higher than that in non-tumor tissues. We also certified that FAM83B expression was dramatically related to tumor stage in patients with NSCLC. Reduced OS and FP with high FAM83B expression, indicating that FAM83B may be a prognosis signature and potential oncogene of NSCLC.
In recent years, significant overexpression of FAM83D has been found in a variety of cancers, including lung cancer, hepatocellular carcinoma, gastric cancer, invasive ovarian cancer, and colorectal cancer (Liao et al., 2015; Mu et al., 2017; Wang F. et al., 2019; Zhang et al., 2019; Yin et al., 2020). Studies from Mu et al. (2017) showed that the FAM83D mRNA expression level was markedly up-regulated in colorectal cancerous tissues and cell lines. Mechanistically, FAM83D suppression promoted colorectal cancer cell apoptosis, inhibited cell proliferation, cell migration, and invasion through inhibiting the FBXW7/Notch1 signal pathway (Mu et al., 2017). Moreover, the up-regulation of FAM83D mRNA and protein levels were detected in human gastric cancer tumor tissues and cell lines, comparing with the adjacent normal tissues and non-malignant gastric epithelial cell lines, respectively, and that reversed relationship shown between OS, DFS, and FAM83D in gastric cancer patients (Wang F. et al., 2019). In our present research, FAM83D expression was higher in NSCLC tissues than in normal ones and was obviously and negatively related to tumor stage in NSCLC patients. Furthermore, an elevated level of FAM83D was markedly associated with worse OS and PPS in NSCLC patients. These discoveries raise the possibility that FAM83D may be a prognosis signature and potential oncogene of NSCLC.
The overexpression of FAM83F protein and mRNA had been found in ESCC tissues in recent years. Moreover, miR-143 restrained the expression of FAM83F and thereby refrained the proliferation, migration, and invasion and induced G1/G0 phase arrest of ESCC cells (Mao et al., 2016). Besides, FAM83F was also found high expression in papillary thyroid cancer (Fuziwara et al., 2019). Furthermore, the functional study had shown that MiR-650 expression in glioma tissues was greatly decreased, while the expression of FAM83F was remarkably up-regulated and MiR-650 could promote cell proliferation through facilitating the expression of FAM83F (Xu et al., 2018). In our present study, we verified that the expression of FAM83F in NSCLC tissues was higher than that in normal tissues. We also confirmed that FAM83F expression was distinctly correlated with tumor stage in patients with NSCLC patients. A high FAM83F expression was prominent correlated with the poor OS, FP, and PPS in all of the patients with lung cancer. These discoveries raise the possibility that FAM83F may be a prognosis signature and potential oncogene of NSCLC.
A study has discovered that the expression of FAM83H was increased in osteosarcomas tissues and tended to reduce the survival of osteosarcoma patients by univariate analysis. This study also discovered that FAM83H regulated the progression of osteosarcomas via a mechanism involving the stabilization of β-catenin and the promotion of proliferation and invasiveness of osteosarcomas (Kim et al., 2019). Functional study had shown that FAM83H was up-regulated in liver cancer cells, and nuclear expression of FAM83H signed shorter survival of HCC patients. They also considered FAM83H as an oncogene in liver cancer, which is closely associated with the MYC (Kim et al., 2017). Likewise, in our investigation, markedly higher mRNA expression of FAM83H was also found in NSCLC tissues, mRNA expression of FAM83H was remarkably correlated with patients’ individual cancer stages. Accordingly, higher mRNA expression of FAM83H was also considerably related with shorter OS and PPS of lung cancer patients. Consistent with our results, Chen et al. (2019) demonstrated that FAM83H is increased in cervical cancer tissues and that high FAM83H expression tends to worsen the OS. In addition, suppression of FAM83H refrained proliferation, colony formation, migration, and invasion of CC cells through inactivating PI3K/AKT pathway (Chen et al., 2019). Further analysis demonstrated that the patients were categorized by cutoffs of 804 for FAM83H low expression and high expression and achieved a 0.9454545/0.8974359 specificity/sensitivity in distinguishing poor survival from OS. Together with the other findings discussed above, our results suggested that FAM83H might play an oncogenic role in NSCLC and those findings might provide theoretical basis and wide possibilities to develop the combination of FAM83H and NSCLC.
There are a few studies on the role of FAM83 family members C, E, and G. The up-regulation of FAM83C and FAM83E was reported in bladder cancer specimens and ovarian cancer (Cipriano et al., 2014; Snijders et al., 2017). Our study disclosed that FAM83C was overexpressed in human NSCLC than normal tissues. What was more, higher mRNA expression of FAM83C was also significantly related with a shorter OS and FP of lung cancer patients, further analysis showed that the patients were categorized by cutoffs of 17 for FAM83C low expression and high expression and achieved a 0.971897/1 specificity/sensitivity in distinguishing poor survival from OS. Furthermore, the expression of FAM83E was increased in human LUAD than in normal tissues, while the expression of FAM83E was lower in LUSC tissues than in normal tissues. FAM83C/E expression were not related to tumor stage in patients with NSCLC and there was no obvious correlation in lung cancer between OS, PPS, and FAM83E expression. All in all, this finding was in contradiction with the role of FAM83C and FAM83E as an oncogene. Our study detected that the expression of FAM83G was up-regulated in human NSCLC, which was not related to the clinical characteristics of NSCLC patients and we could not find any literature as far as the role of FAM83G in cancer progression and data on prognostic factors in the Kaplan-Meier plotter.
There were some limitations in our study. On the one hand, all the data analyzed in our study were obtained from different online databases, which might cause background heterogeneity, thus further studies of larger sample sizes are required to confirm our findings. On the other hand, the study did not conduct experiments to verify the results obtained from bioinformatics analysis. Subsequently, further in vitro and in vivo studies should be performed to affirm our results, and might provide some desirable results to us.
Conclusion
In this study, we formulated the expression and prognostic value of FAM83 family in NSCLC. Besides, we also provided a thorough understanding of the heterogeneity and complexity of the molecular biology of NSCLC, which put forward a new direction for the diagnosis and treatment of NSCLC. Additionally, our results also revealed that overexpression of FAM83A/B/D/F/H was memorably related to clinical cancer stages in NSCLC patients. Besides, higher mRNA expressions of FAM83A/B/C/D/F/H were found to be notably associated with OS in lung cancer patients, furthermore, FAM83A, FAM83C, and FAM83H in OS group achieved a 0.9475/1, 0.971897/1, and 0.9454545/0.8974359 specificity/sensitivity in distinguishing short survivors from long survivors, respectively. Moreover, a high mutation rate of FAM83 family (51%) was also discovered in LUAD patients, and genetic alteration in FAM83 family was associated with a shorter OS and DFS in LUAD patients. Our results indicated that FAM83A/H might play an important role in NSCLC oncogenesis and might be a risk factor for survivals of NSCLC patients. To sum up, it is a charming hypothesis that targeting the FAM83A/H might be a correlatedly unexplored field in the future repertoire of tumor oncogenesis and provide a promising potential in tumor.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JG and QM developed the idea, designed the research, and performed the data analysis work. JG drafted the manuscript. YL and QM reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81672931).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2020.572406/full#supplementary-material
Supplementary Figure 1 | Q–Q graph and histogram were used to detect whether the sample data obeyed normal distribution. In Q–Q graph, X-axis is the normal distribution quantile and the Y-axis is the sample quantile (A–K left). In the histogram, the X-axis is the equidistance segmentation range of gene expression value, the Y-axis is the sample proportion located in this region, and the red curve is the density fitting curve, and the purple curve is the normal distribution fitting curve (A–K right).
Supplementary Figure 2 | The scatter diagram is used to describe the relationship between p-value and cutoff. The X-axis represents the cutoff, the Y-axis represents the p-value, and the position shown in the arrow is the cutoff value corresponding to the p < 0.05.
Abbreviations
NSCLC, non-small-cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; TCGA, The Cancer Genome Atlas; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological processes; CC, cellular components; MF, molecular functions; OS, over survival; DFS, disease free survival.
Footnotes
- ^ http://www.oncomine.org
- ^ http://ualcan.path.uab.edu/
- ^ http://gepia.cancer-pku.cn/
- ^ http://kmplot.com/analysis/
- ^ http://www.cbioportal.org
- ^ http://www.string-db.org
- ^ https://david.ncifcrf.gov/summary.jsp
References
Alberg, A. J., Brock, M. V., and Samet, J. M. (2005). Epidemiology of lung cancer: looking to the future. J. Clin. Oncol. 23, 3175–3185. doi: 10.1200/jco.2005.10.462
Bartel, C. A., Parameswaran, N., Cipriano, R., and Jackson, M. W. (2016). FAM83 proteins: fostering new interactions to drive oncogenic signaling and therapeutic resistance. Oncotarget 7, 52597–52612. doi: 10.18632/oncotarget.9544
Bozatzi, P., and Sapkota, G. P. (2018). The FAM83 family of proteins: from pseudo-PLDs to anchors for CK1 isoforms. Biochem. Soc. Trans. 46, 761–771. doi: 10.1042/bst20160277
Braicu, C., Zimta, A. A., Harangus, A., Iurca, I., Irimie, A., Coza, O., et al. (2019). The function of non-coding RNAs in lung cancer tumorigenesis. Cancers 11:605. doi: 10.3390/cancers11050605
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Cancer Genome and Atlas Network (2012). Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70. doi: 10.1038/nature11412
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. doi: 10.1158/2159-8290.cd-12-0095
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B., et al. (2017). UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19, 649–658. doi: 10.1016/j.neo.2017.05.002
Chen, C., Li, H. F., Hu, Y. J., Jiang, M. J., Liu, Q. S., and Zhou, J. (2019). Family with sequence similarity 83 Member H promotes the viability and metastasis of cervical cancer cells and indicates a poor prognosis. Yonsei Med. J. 60, 611–618. doi: 10.3349/ymj.2019.60.7.611
Cipriano, R., Graham, J., Miskimen, K. L., Bryson, B. L., Bruntz, R. C., Scott, S. A., et al. (2012). FAM83B mediates EGFR- and RAS-driven oncogenic transformation. J. Clin. Invest. 122, 3197–3210. doi: 10.1172/jci60517
Cipriano, R., Miskimen, K. L., Bryson, B. L., Foy, C. R., Bartel, C. A., and Jackson, M. W. (2014). Conserved oncogenic behavior of the FAM83 family regulates MAPK signaling in human cancer. Mol. Cancer Res. 12, 1156–1165. doi: 10.1158/1541-7786.mcr-13-0289
Fulcher, L. J., Bozatzi, P., Tachie-Menson, T., Wu, K. Z. L., Cummins, T. D., Bufton, J. C., et al. (2018). The DUF1669 domain of FAM83 family proteins anchor casein kinase 1 isoforms. Sci. Signal. 11:eaao2341. doi: 10.1126/scisignal.aao2341
Fuziwara, C. S., Saito, K. C., Leoni, S. G., Waitzberg, ÂF. L., and Kimura, E. T. (2019). The highly expressed FAM83F protein in papillary thyroid cancer exerts a pro-oncogenic role in thyroid follicular cells. Front. Endocrinol. 10:134. doi: 10.3389/fonc.2020.00134
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:Pl1. doi: 10.1126/scisignal.2004088
Garber, M. E., Troyanskaya, O. G., Schluens, K., Petersen, S., Thaesler, Z., Pacyna-Gengelbach, M., et al. (2001). Diversity of gene expression in adenocarcinoma of the lung. Proc. Natl. Acad. Sci. U.S.A. 98, 13784–13789.
Grant, S. (2012). FAM83A and FAM83B: candidate oncogenes and TKI resistance mediators. J. Clin. Invest. 122, 3048–3051. doi: 10.1172/jci64412
Hou, J., Aerts, J., den Hamer, B., van Ijcken, W., den Bakker, M., Riegman, P., et al. (2010). Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One 5:e10312. doi: 10.1371/journal.pone.0010312
Huang da, W., Sherman, B. T., and Lempicki, R. A. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. doi: 10.1093/nar/gkn923
Huang da, W., Sherman, B. T., and Lempicki, R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Inamura, K. (2018). Clinicopathological characteristics and mutations driving development of early lung adenocarcinoma: tumor initiation and progression. Int. J. Mol. Sci. 19:1259. doi: 10.3390/ijms19041259
Kim, K. M., Hussein, U. K., Park, S. H., Kang, M. A., Moon, Y. J., Zhang, Z., et al. (2019). FAM83H is involved in stabilization of β-catenin and progression of osteosarcomas. J. Exp. Clin. Cancer Res. 38:267.
Kim, K. M., Park, S. H., Bae, J. S., Noh, S. J., Tao, G. Z., Kim, J. R., et al. (2017). FAM83H is involved in the progression of hepatocellular carcinoma and is regulated by MYC. Sci. Rep. 7:3274.
Lee, S. Y., Meier, R., Furuta, S., Lenburg, M. E., Kenny, P. A., Xu, R., et al. (2012). FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice. J. Clin. Invest. 122, 3211–3220. doi: 10.1172/jci60498
Liao, W., Liu, W., Liu, X., Yuan, Q., Ou, Y., Qi, Y., et al. (2015). Upregulation of FAM83D affects the proliferation and invasion of hepatocellular carcinoma. Oncotarget 6, 24132–24147. doi: 10.18632/oncotarget.4432
Liu, C., Peng, X., Li, Y., Liu, S., Hou, R., Zhang, Y., et al. (2020). Positive feedback loop of FAM83A/PI3K/AKT/c-Jun induces migration, invasion and metastasis in hepatocellular carcinoma. Biomed. Pharmacother. 123:109780. doi: 10.1016/j.biopha.2019.109780
Mao, Y., Liu, J., Zhang, D., and Li, B. (2016). miR-143 inhibits tumor progression by targeting FAM83F in esophageal squamous cell carcinoma. Tumour Biol. 37, 9009–9022. doi: 10.1007/s13277-015-4760-9
Mu, Y., Zou, H., Chen, B., Fan, Y., and Luo, S. (2017). FAM83D knockdown regulates proliferation, migration and invasion of colorectal cancer through inhibiting FBXW7/Notch-1 signalling pathway. Biomed. Pharmacother. 90, 548–554. doi: 10.1016/j.biopha.2017.03.073
Nagy, Á, Lánczky, A., Menyhárt, O., and Gyõrffy, B. (2018). Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 8:9227.
Okabe, N., Ezaki, J., Yamaura, T., Muto, S., Osugi, J., Tamura, H., et al. (2015). FAM83B is a novel biomarker for diagnosis and prognosis of lung squamous cell carcinoma. Int. J. Oncol. 46, 999–1006. doi: 10.3892/ijo.2015.2817
Okayama, H., Kohno, T., Ishii, Y., Shimada, Y., Shiraishi, K., Iwakawa, R., et al. (2012). Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 72, 100–111. doi: 10.1158/0008-5472.can-11-1403
Parameswaran, N., Bartel, C. A., Hernandez-Sanchez, W., Miskimen, K. L., Smigiel, J. M., Khalil, A. M., et al. (2019). A FAM83A positive feed-back loop drives survival and tumorigenicity of pancreatic ductal adenocarcinomas. Sci. Rep. 9:13396.
Popper, H. H. (2016). Progression and metastasis of lung cancer. Cancer Metast. Rev. 35, 75–91. doi: 10.1007/s10555-016-9618-0
Rhodes, D. R., Yu, J., Shanker, K., Deshpande, N., Varambally, R., Ghosh, D., et al. (2004). ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6. doi: 10.1016/s1476-5586(04)80047-2
Selamat, S. A., Chung, B. S., Girard, L., Zhang, W., Zhang, Y., Campan, M., et al. (2012). Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 22, 1197–1211. doi: 10.1101/gr.132662.111
Shen, C. Q., Yan, T. T., Liu, W., Zhu, X. Q., Tian, X. L., Fu, X. L., et al. (2017). High Expression of FAM83B predicts poor prognosis in patients with pancreatic ductal adenocarcinoma and correlates with cell cycle and cell proliferation. J. Cancer 8, 3154–3165. doi: 10.7150/jca.20086
Snijders, A. M., Lee, S. Y., Hang, B., Hao, W., Bissell, M. J., and Mao, J. H. (2017). FAM83 family oncogenes are broadly involved in human cancers: an integrative multi-omics approach. Mol. Oncol. 11, 167–179. doi: 10.1002/1878-0261.12016
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613.
Tang, Z., Li, C., Kang, B., Gao, G., Li, C., and Zhang, Z. (2017). GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102.
Wang, F., Zhang, S., Wei, Y., Chen, H., Jiao, Z., and Li, Y. (2019). Upregulation of family with sequence similarity 83 member D expression enhances cell proliferation and motility via activation of Wnt/β-catenin signaling and predicts poor prognosis in gastric cancer. Cancer Manag. Res. 11, 6775–6791. doi: 10.2147/cmar.s203082
Wang, X., and Adjei, A. A. (2015). Lung cancer and metastasis: new opportunities and challenges. Cancer Metast. Rev. 34, 169–171. doi: 10.1007/s10555-015-9562-4
Wang, Y., Xu, R., Zhang, D., Lu, T., Yu, W., Wo, Y., et al. (2019). Circ-ZKSCAN1 regulates FAM83A expression and inactivates MAPK signaling by targeting miR-330-5p to promote non-small cell lung cancer progression. Transl. Lung Cancer Res. 8, 862–875. doi: 10.21037/tlcr.2019.11.04
Xu, L., Yu, Q. W., Fang, S. Q., Zheng, Y. K., and Qi, J. C. (2018). MiR-650 inhibits the progression of glioma by targeting FAM83F. Eur. Rev. Med. Pharmacol. Sci. 22, 8391–8398.
Yin, C., Lin, X., Wang, Y., Liu, X., Xiao, Y., Liu, J., et al. (2020). FAM83D promotes epithelial-mesenchymal transition, invasion and cisplatin resistance through regulating the AKT/mTOR pathway in non-small-cell lung cancer. Cell Oncol. 43, 395–407. doi: 10.1007/s13402-020-00494-9
Yu, J., Hou, M., and Pei, T. (2020). FAM83A is a prognosis signature and potential oncogene of lung adenocarcinoma. DNA Cell Biol. 39, 890–899. doi: 10.1089/dna.2019.4970
Zhang, Q., Yu, S., Lok, S. I. S., Wong, A. S. T., Jiao, Y., and Lee, L. T. O. (2019). FAM83D promotes ovarian cancer progression and its potential application in diagnosis of invasive ovarian cancer. J. Cell Mol. Med. 23, 4569–4581. doi: 10.1111/jcmm.14360
Zheng, Y. W., Li, Z. H., Lei, L., Liu, C. C., Wang, Z., Fei, L. R., et al. (2020). FAM83A promotes lung cancer progression by regulating the Wnt and hippo signaling pathways and indicates poor prognosis. Front. Oncol. 10:180. doi: 10.3389/fonc.2020.00180
Keywords: non-small-cell lung cancer, FAM83 family, bioinformatics analysis, biomarker, prognostic value
Citation: Gan J, Meng Q and Li Y (2020) Systematic Analysis of Expression Profiles and Prognostic Significance for FAM83 Family in Non-small-Cell Lung Cancer. Front. Mol. Biosci. 7:572406. doi: 10.3389/fmolb.2020.572406
Received: 16 July 2020; Accepted: 29 October 2020;
Published: 10 December 2020.
Edited by:
Matteo Becatti, University of Florence, ItalyReviewed by:
Natascia Marino, Indiana University, United StatesCornelia Braicu, Iuliu Hat̨ieganu University of Medicine and Pharmacy, Romania
Lasse Dahl Ejby Jensen, Linköping University, Sweden
Copyright © 2020 Gan, Meng and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingwei Meng, bXF3ZWlAMTI2LmNvbQ==; Yanjing Li, bGl5YW5qaW5nX2htdUAxMjYuY29t
 Junqing Gan1
Junqing Gan1 Qingwei Meng
Qingwei Meng