
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 21 August 2020
Sec. Molecular Diagnostics and Therapeutics
Volume 7 - 2020 | https://doi.org/10.3389/fmolb.2020.00213
This article is part of the Research Topic Characterizing the Multi-faceted Dynamics of Tumor Cell Plasticity View all 21 articles
Among the transcription factors that are conserved across phylogeny, the grainyhead family holds vital roles in driving the epithelial cell fate. In Drosophila, the function of grainyhead (grh) gene is essential during developmental processes such as epithelial differentiation, tracheal tube formation, maintenance of wing and hair polarity, and epidermal barrier wound repair. Three main mammalian orthologs of grh: Grainyhead-like 1-3 (GRHL1, GRHL2, and GRHL3) are highly conserved in terms of their gene structures and functions. GRHL proteins are essentially associated with the development and maintenance of the epithelial phenotype across diverse physiological conditions such as epidermal differentiation and craniofacial development as well as pathological functions including hearing impairment and neural tube defects. More importantly, through direct chromatin binding and induction of epigenetic alterations, GRHL factors function as potent suppressors of oncogenic cellular dedifferentiation program – epithelial-mesenchymal transition and its associated tumor-promoting phenotypes such as tumor cell migration and invasion. On the contrary, GRHL factors also induce pro-tumorigenic effects such as increased migration and anchorage-independent growth in certain tumor types. Furthermore, investigations focusing on the epithelial-specific activation of grh and GRHL factors have revealed that these factors potentially act as a pioneer factor in establishing a cell-type/cell-state specific accessible chromatin landscape that is exclusive for epithelial gene transcription. In this review, we highlight the essential roles of grh and GRHL factors during embryogenesis and pathogenesis, with a special focus on its emerging pioneering function.
The grainyhead (grh) transcription factor is a member of the ancestral LSF/Grainyhead gene family. Originally identified in Drosophila (previously known as Elf-1 or NTF-1) as an embryonic lethal locus, grh mutant Drosophila embryos show immature cuticle development, patchy tracheal network and the most notable ‘granular’ head skeleton aberration (Nüsslein-Volhard et al., 1984; Bray and Kafatos, 1991). The LSF/Grainyhead family of proteins are functionally distinctive by their nature of oligomerization and mechanism of DNA-binding module on cognate regulatory sites (Traylor-Knowles et al., 2010). Therefore, the gene family is subdivided into two branches: the LSF/CP2 subfamily and the Grainyhead subfamily, resulting from a major gene duplication event dated more than 700 million years ago (Wilanowski et al., 2002; Venkatesan et al., 2003). These transcription factors bind to cis-regulatory elements and control the expression of crucial genes during early embryonic development and tissue homeostasis. The LSF/CP2 subfamily has three mammalian orthologs and evolved from the ancestral gene gemini (dCP2) in Drosophila. Recent critical reviews have discussed the role of LSF/CP2 subfamily members (TFCP2, TFCP2L1, and UBP1) in various aspects of development and human diseases including cancer (Kotarba et al., 2018; Taracha et al., 2018). In this review, we elaborate on the essential functions of grainyhead in Drosophila and the three mammalian orthologs: Grainyhead-like proteins (GRHL1, GRHL2, and GRHL3). Additionally, we focus on human GRHL2 as an important determinant of the epithelial phenotype during development and as a gatekeeper of epithelial differentiation in several human cancers. Finally, we also discuss in detail the novel pioneering role of grainyhead and Grainyhead-like proteins in contouring the chromatin landscape during embryonic development and cancer progression.
The single grh gene in Drosophila and Caenorhabditis elegans exists as multiple orthologs in the vertebrates, which are denoted as grainyhead-like (grhl) genes. These genes remain evolutionarily conserved from insects to humans. In addition, a varying number of splice variants generated from alternative splicing events and alternative transcriptional initiation sites further highlight the underlying complexity and their gene regulatory networks operating during development and disease progression (Uv et al., 1997; Miles et al., 2017). Of note, three orthologs of grainyhead exist in humans: Grainyhead-like 1 (GRHL1), Grainyhead-like 2 (GRHL2) and Grainyhead-like 3 (GRHL3). Each protein contains three annotated functional domains: a transcriptional activation domain (TAD) at the N-terminus; a central DNA-binding domain (DBD) structurally similar to the equivalent of p53, and a dimerization domain (DD) at the C-terminus with unique ubiquitin-like folds. The TAD is the least conserved domain between Drosophila and mammalian orthologs, which could be partially due to the presence of an isoleucine-rich segment that has poor conservation across phylogeny (Attardi et al., 1993; Werth et al., 2010). With respect to the human GRHL2 amino acid sequence homology, the DBD holds higher level of sequence identity in all GRHL proteins, when compared to all other domains across the model organisms (Table 1).
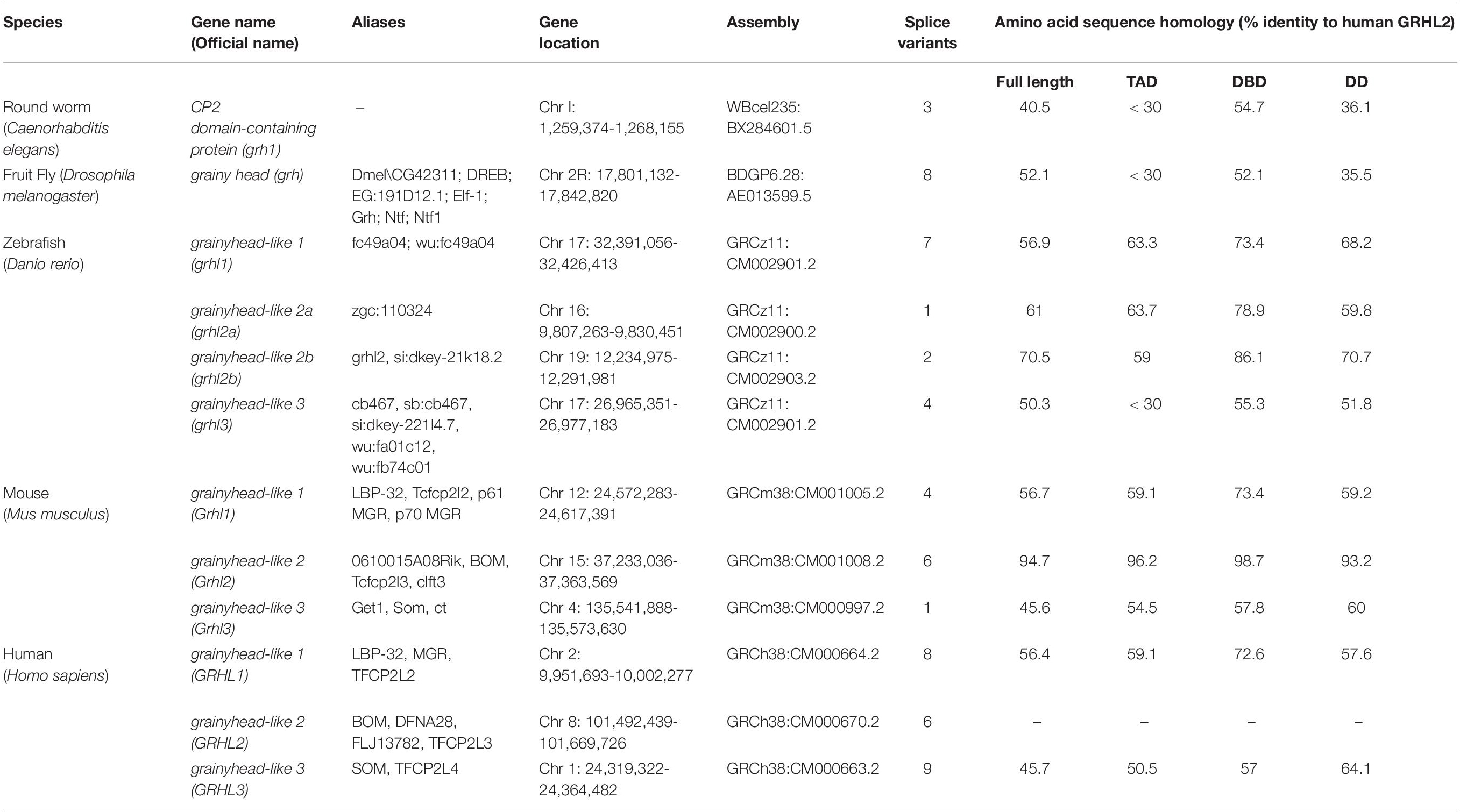
Table 1. Members of the grainyhead/Grainyhead-like transcription factor family and protein sequence homology in nematode, fruit fly, zebrafish, and mammals.
Members of the grh/GRHL family share a similar palindromic DNA-binding motif (AACCGGTT), with different levels of variability for some target genes (Table 2). In Drosophila, Grh recognizes DNA regulatory sequences upstream of genes Ddc (TGAACCGGTCCTGCGG) and en (GTGAGCCGGCGAAACCGGTT), whereas the binding motif on Ubx and ftz promoters is (T/C)NAAC(C/T)GGT(T/C) (Bray et al., 1988; Soeller et al., 1988; Dynlacht et al., 1989; Wilanowski et al., 2002; Venkatesan et al., 2003). In mammals, GRHL binding motifs display as two adjacent repeats of Grainyhead consensus sequences, with two tandem core CNNG motifs set apart by five bases. For example, the mouse Grhl2 binding site in intron 2 of Cdh1 (AAACCAGTCAAACCAGTT) and the promoter of Cldn4 (AATCCAGAGAAACTGGTC) are strikingly similar to the human GRHL2 binding motif on the intron 2 of the CDH1 (GCAAACCAGCCAAACCAGTTT) and the promoter of CLDN4 (GGAATCCAGAGAAACTGGTCAG) (Werth et al., 2010; Chung et al., 2016). The invariant CNNG tandem motifs share similarity with the binding motif of Tfcp2l1 from the CP2 family suggesting a close phylogenetic relationship with members of the p53 family (two CNNG set apart by six bases) based on protein folding and the binding of DNA (Kokoszynska et al., 2008). A recent study on the crystal structure of Grhl1/2 DBDs shows that these domains share a common fold with p53, substantiating earlier computational predictions (Ming et al., 2018).
In metazoans, two major cell types form the basis of organ development: epithelium and mesenchyme. Epithelial cells are generated first during the embryonic development, while mesenchymal cells are derived from the pre-existing epithelial cells through a process called epithelial-to-mesenchymal transition (EMT) (Hay, 1995; Thiery and Sleeman, 2006; Kalluri and Weinberg, 2009). Epithelial cells usually maintain a strict, aligned cellular polarity (apical and basal surfaces) and remain closely connected to adjacent cells through specialized transmembrane structures, such as tight junctions, adherens junctions, and desmosomes. In contrast, due to their lack of stable cell-cell attachment and apical-basal polarity, mesenchymal cells possess higher migratory abilities and interact extensively with the surrounding extracellular matrix. The earliest developmental EMT occurs during gastrulation, where mesenchymal cells are generated from epithelial epiblast cells. The mesenchyme further condenses to form mesoderm (middle layer of the embryo) and endoderm (inner layer of the embryo), which eventually form the vertebral column, bony appendages and connective tissues (Hay, 2005). However, the epithelia is the stable state of cellular organization that forms the epidermis, the primary layer covering the external surface of the body that provides protection against external physical and mechanical stress. The following sections describe the vital roles of Grainyhead and Grainyhead-like proteins in the epidermis and epithelia.
The recurrence of EMT is also observed during the early development of the nervous system to generate neural crest cells. During the embryonic process termed neurulation, epithelial neural plate (neural ectoderm) bends, invaginates and fuses along the dorsal midline to form a cylindrical structure called the neural tube. Subsequent closure of the neural tube in anterior and posterior directions guides formation of the future brain and spinal cord (Wilde et al., 2014). Meanwhile, the dorsal neuroepithelial cells located close to the neural tube lose intercellular connections, undergo EMT, and migrate away to become neural crest cells (Acloque et al., 2009). Further delamination and migration of neural crest cells navigate to populate multiple niches throughout the embryo, ultimately progress toward terminal differentiation derivatives such as ganglia of the peripheral and enteric nervous system, cardiac valves, bone and cartilage of the facial skeleton (Simões-Costa and Bronner, 2013; Muñoz and Trainor, 2015). Members of the Grainyhead-like family provide distinct regional signals during neurulation, neural crest migration, and neural tube closure that are discussed in detail in an upcoming section.
In Drosophila, the formation of protective exoskeleton called cuticle during the early stages of larval development serves many important functions during the adult life including the protection against water loss and the maintenance of structural framework for locomotion. grh remains to be essential for the development of the epidermal barrier and the repair of barriers after wounding. The embryonically lethal larval cuticles of grh mutants are multilayered and grossly inflated structures, generating the “blimp” phenotype that are functionally weaker when compared to wild-type cuticles (Uv et al., 1997; Ostrowski et al., 2002; Hemphälä et al., 2003). grh mutant embryos carrying induced aseptic epidermal wounds fail to restore the expression of Dopa decarboxylase (Ddc in the wound border), one of the two key enzymes contributing for the formation and hardening of larval and adult cuticles. This results in the defective phosphorylation of grh by ERK which is required for wound-dependent regeneration of the epidermal barrier (Mace et al., 2005; Geiger et al., 2011; Kim and McGinnis, 2011). Other studies focusing on Drosophila epidermal wound healing and amnioserosa (defects in dorsal closure) have identified the direct regulation of Grh on major targets including stit, msn, cora, sinu, and fas3 (Narasimha et al., 2008; Pearson et al., 2009; Wang and Samakovlis, 2012). In addition to controlling the epithelial and epidermal morphogenesis in Drosophila, Grh directly regulates the expression of key genes (Table 2) that are involved in the tracheal tube formation (Hemphälä et al., 2003), the maturation of central nervous system (Almeida and Bray, 2005; Khandelwal et al., 2017), and the maintenance of polarity in wing and hair (Lee and Adler, 2004).
The GRHL family members play crucial roles during the development of several epithelial tissues (Figure 1). During the development of mouse circumvallate papilla (specialized dome shaped region located at the back of a tongue), knockdown of Grhl3 significantly alters the epithelial structure and disrupts the epithelial integrity by having high proliferation, low apoptosis, and enhanced migration in epithelial tongues cells of embryonic mice (Adhikari et al., 2017). Several reports have claimed the prominent function of GRHL factors in epidermal integrity. Grhl1 directly controls the expression of the desmosomal cadherin, desmoglein 1 (Dsg1) and mice deficient of Grhl1 show an abnormal desmosome phenotype in the interfollicular epidermis, ultimately delaying the initial skin coat growth and poor hair anchoring to the follicle (Wilanowski et al., 2008).
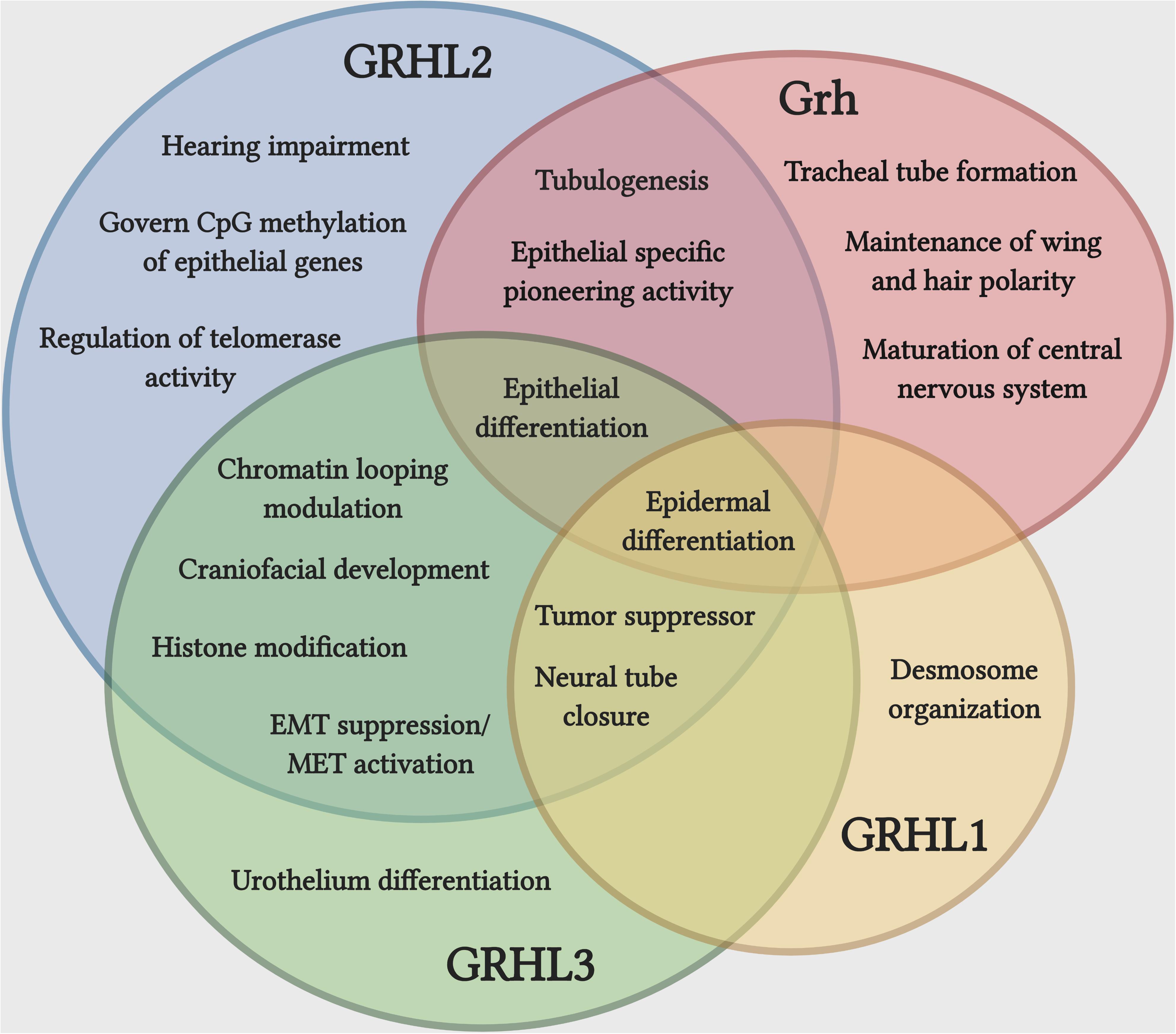
Figure 1. Unique and cooperative functions of Grh and GRHL family during development and disease. Epidermal differentiation is the most common function shared by Grh and GRHL family, while a set of functions associated with epithelial differentiation is carried out by more than one member of the family. The overlapping function does not indicate that they are functionally redundant. Illustration created with Biorender.com.
During early embryogenesis, Grhl2 expression is predominantly observed in several barrier-forming epithelial tissues, including the surface ectoderm, the otic ectoderm, and the gut tube (Auden et al., 2006; Werth et al., 2010). At the molecular level, Grhl2 binds to cis-regulatory elements and controls the timely expression of the apical junctional complex proteins such as the adherens junction component E-cadherin and the tight junction molecules claudin 3, claudin 4 (Cldn3/4), and an epithelial-specific member of small guanosine triphosphatase Rab25, which are crucial for epithelial differentiation (Werth et al., 2010; Senga et al., 2012; Tanimizu and Mitaka, 2013). GRHL2 is involved in the epithelial morphogenesis of the lung epithelium and essential for the establishment and maintenance of the epithelial barrier of mucociliary airways. In primary human bronchial epithelial cells, GRHL2 directly or indirectly regulates the expression of proteins that form apical junction assembly and cell polarity (CDH1, TJP1, RAB25) as well as essential proteins that are required to establish barrier function (PVRL4, VAV1, and ESRP1/2). Mutant cells carrying dominant–negative GRHL2 protein consequently fail to form polarized epithelium with barrier function (Gao et al., 2013). Conditional deletion of Grhl2 in mouse tracheal basal cells and in vitro CRISPR/Cas9 genome editing of GRHL2 in human basal cells have been shown to disrupt the differentiation of ciliated cells via targeting multiple genes in the Notch signaling pathway and ciliogenesis such as Mcidas, Rfx2, and Myb (Gao et al., 2015). In cooperation with Nkx2-1, a homeobox transcription factor, Grhl2 regulates the expression of cell-cell interaction genes such as semaphorins and their receptors, which are crucial for maintaining the lung epithelial identity (Varma et al., 2012). Using mouse lung epithelial cells, the same study shows that Grhl2 binds to the Nkx2-1 promoter regions and Nkx2-1 binds to the Grhl2 intronic region and generates a positive feedback loop to reinforce lung epithelial phenotypes. Furthermore, a recent study reports that the loss of Grhl2 in the developing mice lung epithelium reduces the expression of Elf5, an epithelial-specific transcription factor, and eventually leads to the impaired ciliated cell differentiation and the reduction of distal progenitor cells (Kersbergen et al., 2018).
Grhl3 controls epidermal differentiation and wound-repair by directly regulating the expression of two crucial genes: Transglutaminase-1, an enzyme that crosslinks structural components of the superficial epidermis (Ting et al., 2005), and RhoGEF19, a RhoA activator of the planar cell polarity (PCP) signaling pathway (Caddy et al., 2010). Grhl3 functionally interacts with the LIM-only protein LMO4 to regulate the differentiation of the epidermis, where mice lacking functional Grhl3 and LMO4 expression show severe defective skin barrier formation and failure of eyelid development affecting the expression of multiple genes linked to the epidermal terminal differentiation and F-actin cable formation (Yu et al., 2006, 2008; Hislop et al., 2008). Mice deleted for epidermal specific Grhl3 have digit fusion (syndactyly) due to abnormal adhesion of the periderm covering the developing digits (Kashgari et al., 2020). In autosomal-recessive ectodermal dysplasia syndrome, whole exome-sequencing from affected individuals revealed the presence of homozygous mutations in the GRHL2 locus. These mutant keratinocytes showed changes in the cellular phenotype and failure to form intact cell-cell junctions, partly due to the cytoplasmic translocation of GRHL2 (Petrof et al., 2014). Interestingly, cytoplasmic translocation of GRHL3 induces the activation of the non-canonical Wnt signaling pathway involving PCP genes, and eventually alters the mechanical properties essential for enduring tensile force during epithelial differentiation (Kimura-Yoshida et al., 2018). In additional to the pulmonary and epidermal epithelium, another major tissue that plays an important role in the barrier function is the urothelial membrane, which controls the selective movement of water and solutes between urine and tissues. Grhl3 is highly expressed in mature umbrella cells of the bladder epithelium. It directly targets the expression of uroplakin II, a major protein component of the asymmetric urothelial membrane plaques, thereby regulating the terminal differentiation and barrier function of the bladder epithelium (Yu et al., 2009).
During neurulation, the single-layered neurepithelium distinguishes into two different cell fates: neural ectoderm and surface (non-neural) ectoderm prior to neural tube closure. The fate choice between neural and surface ectoderm is highly regulated through signaling interplay of Wnt, FGF and BMP activity (Murry and Keller, 2008). At the molecular level, canonical Wnt signaling mediated Grhl3 expression is essential for the specification of surface ectoderm cell fate, whereas, repression of Grhl3 by Dickkopf1 (Dkk1), a canonical Wnt signaling antagonist, leads to the specification of neural ectoderm cell fate (Ting et al., 2003; Kimura-Yoshida et al., 2015). Thus, the balance between Wnt controlled Grhl3 activation or repression regulates the binary cell fate choice of neural and surface ectoderm identity, which is essential for subsequent neural tube closure. The remodeling of five or more polarized epithelial cells converging radially around a central point of fusion to form transient “rosette” like structures are identified during the formation of multiple organ systems, including surface ectodermal lineage specification (Afonso and Henrique, 2006; Nishimura and Takeichi, 2008; Harding et al., 2014). Genetic fate mapping using Grhl3Cre/+ mice have reported that the rosette forming cells of the surface ectoderm are Grhl3-expressing lineage cells, and Grhl3 mutants showed severe disruption of rosette formation, exhibiting fully penetrant spina bifida (Molè et al., 2020; Zhou et al., 2020). Single-cell RNA sequencing of neural rosettes in vitro, generated from human induced pluripotent stem cells, showed GRHL2 and GRHL3 were highly expressed in early rosettes highlighting the role for GRHL TFs in neurulation in humans (Shang et al., 2018). Therefore, these findings denote an earlier role for GRHL factors in neurulation, such as lineage specification, prior to its contribution in neural tube closure.
Delamination and migration of neural crest cell from the border of the surface and neural ectoderm involves activation of EMT. Although GRHL factors are potent suppressors of EMT and associated phenotypes, depletion of grhl3/Grhl3 in zebrafish and mouse embryos do not affect any stages of neural crest cell development and activity (Dworkin et al., 2014; Kimura-Yoshida et al., 2015). This might indicate that fate specification of surface ectoderm in the neural plate border is partly driven by GRHL factors. However, the involvement of other GRHL factors in neural crest cell migration remains to be explored.
Following the delineation of the neurepithelium, the developing neural tube converges and fuses along the midline to form complete the neural tube closure event. Among the GRHL family, disruption of GRHL2 and GRHL3 functions generated severe neural tube defects. Loss of Grhl2 expression in the surface ectoderm resulted in abnormal mesenchymal phenotypes, with increase in vimentin expression and downregulation of epithelial genes such as Fermt1, Esrp1, and Tmprss2, eventually resulting in neural tube closure defects (Pyrgaki et al., 2011; Ray and Niswander, 2016). In the Grhl2-null mutants, the expression levels of two Grhl2 direct targets, E-cadherin (Cdh1) and Claudin 4 (Cldn4), are significantly reduced in the surface ectoderm leading to neural tube defects (Werth et al., 2010). In contrast, overexpression of Grhl2 could also be the underlying cause of defective neural tube closure in Axial defects mutant mouse (Brouns et al., 2011). Similarly, Grhl3-null mutants exhibited fully penetrant spina bifida, and lack of Grhl3 expression in the hindgut caused curly tail phenotype, which occurs during the final stages of neural tube closure (Ting et al., 2003; Auden et al., 2006; Gustavsson et al., 2007). Moreover, Grhl2 and Grhl3 exhibit cooperative activity during neurulation closure at the forebrain/midbrain boundary and spinal closure from mid to lower thoracic region (Rifat et al., 2010). Taken together, members of the GRHL family play critical roles during several stages of neural development and dysregulation of these factors during neural development renders severe impact on neural tube closure.
An array of studies has reported the implication of GRHL members in multiple human diseases including cancers. Essential functioning of GRHL factors are implicated during carcinogenesis as well as during tumor suppression indicating that these factors play complex and controversial roles in regulating different cancer entities. In the following section we highlight several prominent findings that describe the essential roles of GRHL factors in pathophysiology.
Few recent studies have demonstrated the association of expression levels of GRHL members with patient outcomes during cancer progression. In colorectal cancer, higher expression levels of GRHL1 and GRHL3 are associated with worse disease-free survival, whereas low levels of all three members confer better overall survival of patients (Yuan et al., 2020). GRHL2 expression is enriched in human breast cancer stem cell-like subpopulation and is included in a 31-gene signature predictive of distant metastasis in estrogen receptor–negative breast cancer cohorts (Leth-Larsen et al., 2012). Gene correlation analysis of a breast cancer cohort showed that higher expression of GRHL2 is correlated with worse relapse-free survival in Luminal A, Luminal B, HER2+, and Basal-like subtypes (Mooney et al., 2017). Furthermore, overexpression of GRHL2 in breast cancer cell lines showed significant increase in migration, invasion potential and also correlated with unfavorable breast cancer patient characteristics – grade III tumors and large tumor size at the time of diagnosis (Yang et al., 2013). High expression of GRHL2 is also observed in pancreatic cancer patients with worsened overall survival (Wang et al., 2019).
At the molecular level, GRHL2 directly regulates the expression of the EGFR family member ERBB3 and the Wnt ligand Wnt7A, and overexpression of GRHL2 in metastatic breast cancer cells exhibit increased anchorage-independent growth, migratory and invasive potential (Xiang et al., 2012; Werner et al., 2013). Aberrant activation of ERBB3 potentially forms heterodimers with ERBB2 that directly contributes to decreased survival rate coupled with increased resistance to chemotherapy (Liu et al., 2007, 3; Sithanandam and Anderson, 2008; Garrett et al., 2011). Conditional deletion of Grhl2 prevents oral cancer development in a chronic, chemically induced carcinogen model, when compared to the aggressive tumor formation in Grhl2 wild-type mice (Chen et al., 2018b). In addition, the study also identifies that GRHL2 mediated activation of MAP kinase signaling and repression of TGF-β signaling in oral squamous cell carcinoma cell lines, dually render tumor promoting effects during the early stages of carcinogenesis. The role of GRHL2 in prostate cancer seems to be mainly oncogenic, as its expression is higher in prostate cancer tissue samples (Danila et al., 2014; Paltoglou et al., 2017). Paltoglou et al. showed that the loss of GRHL2 via silencing resulted in the loss of androgen receptor (AR) expression and demonstrated the presence of a positive feedback loop between GRHL2 and AR to promote prostate cancer growth (Paltoglou et al., 2017). In addition to driving AR expression, GRHL2 also acts as an AR transcriptional co-activator that enhances the oncogenic AR signaling pathway in prostate cancer progression. Furthermore, the oncogenic role of GRHL2 is observed in hepatocellular carcinoma (Tanaka et al., 2008), esophageal cancer (Shao et al., 2017), oral squamous cell carcinoma (Kang et al., 2009; Chen et al., 2016), and colorectal carcinoma (Quan et al., 2014, 2015).
In squamous cell carcinoma, GRHL3 functions as a strong tumor suppressor, where the deletion of Grhl3 in keratinocytes leads to hyper-proliferation epidermal keratinocytes that are more prone to chemical carcinogen induced spontaneous squamous cell carcinoma formation (Darido et al., 2011). Mechanistically, Grhl3 depletion in keratinocytes leads to the loss of tumor suppressor Pten expression, which induces the activation of PI3K/AKT/mTOR signaling and the oncogenic miR-21 expression, culminating in the formation of aggressive and poorly differentiated squamous cell carcinoma. Oncogenic Ras-mediated Grhl3–/– mouse epidermal keratinocytes were more prone to tumorigenesis by upregulating the miR-21 levels (Bhandari et al., 2013). Similarly, in a two-stage chemical skin carcinogenesis model, over 40% of benign papilloma developed into squamous cell carcinoma in Grhl1–/– mice, when compared to one-fourth of such tumor formation in Grhl1+/+ mice, due to the severe impairment of epidermal barrier and aberrant terminal differentiation of keratinocytes (Mlacki et al., 2014). In neuroblastoma, patients with high levels of GRHL1 expression show favorable prognosis, consistent with the suppressed tumor growth phenotype seen in the xenografts carrying forced GRHL1 expression in MYCN-amplified neuroblastoma cells (Fabian et al., 2014). At the molecular level, co-recruitment of MYCN and HDAC3 to the GRHL1 promoter represses its transcription. Esophageal squamous cell carcinoma with low expression of GRHL1 levels show poor differentiation and the patients are associated with reduced overall survival rate (Li M. et al., 2019). The tumor suppressive role of GRHL2 is largely mediated though the suppression of EMT which is summarized in the following section.
EMT and MET (mesenchymal to epithelial transition, the reversal of EMT) functionally dictate the cellular dedifferentiation and differentiation status respectively and determine the nature of cellular behavior. Although EMT/MET processes occur spontaneously during fundamental events such as gastrulation, neural crest dissemination and organogenesis, execution of EMT/MET is also observed during wound healing, fibrosis and cancer (Thiery and Sleeman, 2006). Overcoming years of speculations about the reactivation of reversible EMT and MET program during cancer progression, compelling evidences from in vitro, in vivo and clinical findings support the crucial roles of EMT and MET during cancer progression (McInnes et al., 2015; Santamaria et al., 2017; Francart et al., 2018; Rios et al., 2019; Williams et al., 2019). In brief, acquisition of mesenchymal trait through EMT is regarded as an essential feature for epithelial-derived cancer cells to successfully metastasize the surrounding tissues and distant organs. Being a pivotal gatekeeper of epithelial integrity, GRHL2 suppresses EMT in a multipronged manner across cancer entities. Firstly, multiple independent investigations have shown that GRHL2 controls the ZEB1/miR-200 regulatory axis. The EMT inducer ZEB1 and epithelial-phenotype reinforcing microRNA-200 (miR-200) family members reciprocally control the expression of each other generating a double-negative feedback loop (Brabletz and Brabletz, 2010). The expression of ZEB1 drives the cancer cells to undergo EMT, whereas restoration of miR-200 expression is vital for cells to undergo epithelial differentiation or MET. On one hand, GRHL2 directly suppresses the expression of the EMT inducer ZEB1 in breast (Cieply et al., 2012; Werner et al., 2013), ovarian (Chung et al., 2016), bladder cancers (Shen et al., 2020), and sarcoma (Somarelli et al., 2016). On the other hand, GRHL2 activates the expression of miR-200 family members through direct promoter binding in oral (Chen et al., 2016), ovarian (Chung et al., 2016) cancers and sarcoma (Somarelli et al., 2016).
Secondly, GRHL2 suppresses TGF-β mediated migratory and invasive capabilities of gastric (Xiang et al., 2017), breast (Cieply et al., 2012; Werner et al., 2013), and oral (Chen et al., 2018b) cancer cells, where the activation of TGF-β signaling cascade is a significant inducer of EMT in tumor progression (Heldin et al., 2012). Thirdly, re-expression of GRHL2 in mesenchymal-like cells induces MET effects by restoring the expression of epithelial components such as E-cadherin, ZO-1 and downregulating mesenchymal markers including Vimentin, Snail, Slug and ZEB1 (Chung et al., 2016; Yang et al., 2019; Shen et al., 2020). Fourthly, GRHL2 expression suppresses stemness properties in CD44high/CD24low mesenchymal subpopulation cells of breast cancer cells and restores the anoikis sensitivity by altering intracellular H2O2 ROS levels (Cieply et al., 2012; Farris et al., 2016).
EMT-TFs Snail and ZEB1 are known to recruit epigenetic remodelers such as the DNA methyltransferases (DNMTs) and/or polycomb repressive complex 2 (PRC2) to generate a repressive chromatin around epithelial genes (Dong et al., 2012; Fukagawa et al., 2015). GRHL2 also interacts with multiple epigenetic regulators to dually suppress EMT and to induce MET phenotypes. GRHL2 significantly inhibits the histone acetyltransferase coactivator p300 and its activity on mesenchymal genes, which interfered with the branching morphogenesis and EMT of Madin–Darby canine kidney (MDCK) cells (Pifer et al., 2016). In addition, GRHL2 interaction with the histone methyltransferases KMT2C and KMT2D induces MET and ICAM-1 expression in cancer cells, which sensitizes these cells for optimal natural killer (NK) cells mediated activation and target cell killing, suggesting a potential link between the epithelial phenotype and cellular susceptibility to NK killing (MacFawn et al., 2019). Furthermore, using a set of ovarian cancer cell lines, we have shown that during the reactivation of epithelial genes, the presence of GRHL2 is essential for the modification of the epigenetic landscape into a permissive chromatin to allow the transcription of key epithelial genes such as E-cadherin, ESRP1 and OVOL2 (Chung et al., 2019).
Evidences have shown that GRHL members are the gatekeepers of early phenotype transition. Our assessment on a heterogeneous ovarian cancer cell line panel revealed the presence of intermediate cellular phenotypes that dually expressed epithelial and mesenchymal markers (Huang et al., 2013). Consequently, the concept of an ‘EMT spectrum’ has emerged, whereby EMT is regarded as a continuum consisting of multiple, transient intermediate phenotypes collectively referred as the EMT spectrum (Nieto et al., 2016; Yang et al., 2020). The prevalence of multiple intermediate EMT states is observed in breast (Grosse-Wilde et al., 2015; Yamashita et al., 2018), prostate (Ruscetti et al., 2015), and non-small lung cancer (Fustaino et al., 2017). GRHL2 knockdown in ovarian cancer cell lines harboring the epithelial phenotype results in a specific shift of subcellular E-cadherin localization with unaltered total E-cadherin protein abundance, generating a partial EMT phenotype (Chung et al., 2016). In lung cancer cells, GRHL2, OVOL2 and miR-145 play crucial roles in stabilizing the intermediate EMT phenotype (hybrid E/M), while transient knockdown of GRHL2 in cells with hybrid E/M phenotype switches to a complete EMT as evidenced by the disruption of partial EMT specific collective cell migration phenotype to a single cell migration phenotype (Jolly et al., 2016). Moreover, through computational modeling, the same study has predicted that GRHL2 promotes the association of hybrid E/M phenotype with high-tumor initiating stem-like traits, which might be helpful in stratifying patients with higher metastatic risk. Using a genetically engineered mouse model of squamous cell carcinoma, another recent study has identified spontaneous EMT and multiple intermediate EMT subpopulations with characteristic cell surface marker expressions (Pastushenko et al., 2018). Assay for transposase-accessible chromatin using sequencing (ATAC-seq) of these EMT subpopulations revealed the specific enrichment of a GRHL1 motif on the differentially expressed genes especially during the epithelial to early hybrid EMT state but not observed in the late hybrid EMT state or in mesenchymal subpopulations. Although tumor cells dynamically switch between epithelial, mesenchymal and intermediate E/M phenotypes through EMT and MET processes, mechanisms underlying the reversibility or irreversibility of such events are slowly emerging. In an inducible mammary EMT system (HMLE-Twist1-ER), epithelial clonal population (high GRHL2 expression) were susceptible to acquire a hybrid E/M phenotype and showed transient, reversible changes in chromatin accessibility when compared to the mesenchymal clonal population (low GRHL1/2 expression) (Eichelberger et al., 2020). In particular, ATAC-seq of the mesenchymal subpopulation revealed that the specific loss of chromatin accessibility along GRHL1/2 motifs governing loci of epithelial genes is crucial for these cells to enter an irreversible mesenchymal cell state or to resist trans-differentiation. Similarly, via mathematical modeling, another independent study has proposed two mechanisms that drive epithelial cells to resist undergoing EMT or enabling irreversible MET: (i) GRHL2 mediated epigenetic feedback on inhibition of ZEB1 and (ii) stochastic partitioning of biomolecules during cell division to generate different phenotypic subpopulations in regards to EMT (Jia et al., 2020). Altogether, these studies have unraveled that GRHL factors play crucial roles in establishing the EMT spectrum and moderating the EMT/MET dynamics during cancer progression. Importantly, comprehending the biology of such intermediate or hybrid trans-differentiation states is essential to combat clinically challenging issues such as metastatic aggressiveness and therapeutic resistance (Santamaria et al., 2017; Jolly et al., 2018, 2019; Williams et al., 2019).
Craniofacial development encompasses the patterning of bones, muscles and vasculatures of face, skull and jaws, governed by highly coordinated migration signals and spatiotemporal regulation of genetic and molecular factors. Owing to the gene nomenclature, Drosophila larvae carrying grh mutations have pronounced deformation in the chitinous head-skeleton morphology generating a granular head appearance (Bray and Kafatos, 1991). Similarly, deregulation of Grhl/GRHL factors are also heavily associated in the etiology of craniofacial malformations in mammals (Carpinelli et al., 2017). The failure of cranial neural tube closure in these mutants resulted in anterior spina bifida, prematurely apposed skull bones, split-face, defective neural fold elevation, cranioschisis and exencephaly, lumbosacral spina bifida (open neuropore) and a curled tail phenotype. Accordingly, multiple independent reports have observed that patients with craniofacial malformations are associated with microdeletions of a gene cluster at the chromosomal region 8q22.2-q22.3, comprising clinically relevant genes including GRHL2 (Kuechler et al., 2011; Kuroda et al., 2014; Sinajon et al., 2015; Chen et al., 2017; Hoebel et al., 2017; de Vries et al., 2020). Dominant-negative mutations in GRHL3 have been reported in the congenital disorder Van Der Woude syndrome, which is characterized by cleft lip and/or cleft palate (Peyrard-Janvid et al., 2014; Wang et al., 2016; Eshete et al., 2018).
Defects in neural tube closure generate severe congenital morbidity and mortality in human, which occurs at a high rate of 1 in every 1000 human pregnancies (Bhandari and Thada, 2020). Murine models carrying Grhl2 and Grhl3 conditional deletions are embryonically lethal with severe defects in organogenesis, dorso-lateral hinge point formation during neurulation and neural tube closure (Ting et al., 2003; Rifat et al., 2010; Werth et al., 2010; Pyrgaki et al., 2011; Menke et al., 2015; Goldie et al., 2016). Gene targeting in mice have demonstrated that the lack or surplus of Grhl2/Grhl3 expression could interfere with spinal neural tube closure (Gustavsson et al., 2007; Brouns et al., 2011; Nikolopoulou et al., 2017; De Castro et al., 2018). In particular, Grhl2 mediates the upregulation of cell-cell junction proteins via modulating the local actomyosin-dependent mechanical stress, which is essential for spinal neural tube closure (Nikolopoulou et al., 2019). The regulation of GRHL2 in the transactivation of OVOL1/2, ESRP1/2, miR-200 family and the suppression of ZEB1 expression during MET is also recapitulated during palate closure (Carpinelli et al., 2020). Furthermore, mouse models carrying Grhl3 dependent gene manipulation in the surface ectoderm showed severe defects in neural tube closure and open spina bifida (Camerer et al., 2010; Kimura-Yoshida et al., 2015; Molè et al., 2020).
Hearing impairment is another pathologic condition linked to GRHL2 mutation, where sequencing of the gene loci DFNA28 on chromosome 8q22 in a large American family is associated with progressive autosomal dominant hearing loss (Peters et al., 2002). The study initially identified a frameshift mutation 1609-1610insC generating the GRHL2 transcript with a premature stop codon in exon 14. A decade later, a second novel splice site mutation in GRHL2 – c.1258-1G > A resulting in p. Gly420Glufs0111 frameshift mutation in exon 10 was associated with age-related, post-lingual hearing loss (Vona et al., 2013). Subsequent studies have reported that mutations and/or gene polymorphisms in GRHL2 are implicated in hereditary and acquired hearing loss such as age-related hearing impairment, non-syndromic hearing loss, sudden sensorineural hearing loss and noise-induced hearing loss in Chinese, Korean, Roma and Hungarian populations (Kim et al., 2015; Zhang et al., 2015; Lin et al., 2016; Xu et al., 2016; Li X. et al., 2019; Matyas et al., 2019; Wu et al., 2020). Although mutations and gene polymorphisms of GRHL2 are associated with multiple hearing abnormalities, substantial association of GRHL2 in the development of the inner ear is not yet demonstrated. However, a recent examination of deafness genes in the non-human primate model marmoset (Callithrix jacchus) has revealed that GRHL2 expression is prevalent in cochlear duct lining cells, hair cells, and supporting cells of the inner ear (Hosoya et al., 2016), denoting that the definitive role of GRHL2 in the inner ear development needs further investigations.
On top of gene expression regulations through cis- and trans-regulatory elements, epigenetic modifications such as methylation of DNA cytosine and extensive post-translational modifications occur on the core octamer histone proteins collectively modulate chromatin landscape of underlying genes, and thereby regulate gene expression. Although methylation of DNA is a global phenomenon, concentrated methylation on short patches of CpG dinucleotide repeats (CpG islands) are often observed in genes that are suppressed at a particular cell state/type (Suzuki and Bird, 2008). A staggering number of histone modifications occur on the flexible N- and C-terminal ‘tail’ domains. These include prominent alterations such as lysine acetylation, lysine/arginine methylation, serine/threonine/tyrosine phosphorylation as well as many under-examined, minor changes such as lysine ubiquitination/sumoylation, citrullination, ADP-ribosylation, and proline isomerization (Bannister and Kouzarides, 2011). Alterations of CpG methylation patterns and histone modifications are observed in a variety of important cellular processes such as during cellular growth, differentiation, and cancer progression (Rothbart and Strahl, 2014; Audia and Campbell, 2016). GRHL factors have been shown to induce changes in CpG methylation levels and histone modifications during development.
In mouse kidney cells, depletion of active histone marks H3K4me3 and H3K9/14ac are observed at the E-cadherin promoter region exclusively in Grhl2-knockdown cells, denoting that Grhl2 expression is essential to sustain activating histone marks at the promoter (Werth et al., 2010). Selective expression of uroplakin II (UpkII) in mouse bladder epithelial cells is also regulated through Grhl3-mediated active H3K9ac mark enrichment on the UpkII promoter (Yu et al., 2009). During the development of kidney ureteric buds and collecting ductal epithelia, GRHL2 strongly associates with the active H3K4me3 mark of target genes (Cdh1, Rab25, Ovol2, and Cldn4) that are essential for lumen expansion and barrier formation (Aue et al., 2015). Besides direct transcription controls, GRHL2 imposes several epigenetic modifications on selected epidermal differentiation genes. GRHL2 overexpression in normal human epidermal keratinocytes leads to the inhibition of methylation at the CpG island of the hTERT promoter and restores hTERT expression, potentially by hindering the activity of DNA methyltransferase DNMT1 (Chen et al., 2010). In addition, GRHL2 overexpression in normal human epidermal keratinocytes also leads to the enrichment of H3K27me3 repressive mark, while simultaneously inhibiting the recruitment of histone demethylase Jmjd3 to the cognate promoters of epidermal differentiation genes such as involucrin (IVL), keratin 1 (KRT1), filaggrin (FLG) and cyclin dependent kinase inhibitor 2A (INK4A) (Chen et al., 2012). During epidermal differentiation, GRHL3 binds directly to the TGM1 promoter to control the expression of transglutaminase (TGM1) (Hopkin et al., 2012), a Ca2+-dependent enzyme is essential for the formation of cornified cell envelope (Eckert et al., 2005). GRHL3 further recruits the Trithorax complex components MLL2 and WDR5 to the target promoter and increases the active H3K4 methylation mark and drives TGM1 expression during epidermal differentiation (Hopkin et al., 2012). These studies substantiate the notion that GRHL factors have the potential to epigenetically modify chromatin states during cellular differentiation.
About two meters long, the double stranded DNA is condensed and packed into a typical eukaryotic interphase nucleus that only measures about six micrometers in diameter. This composite level of condensation starts primarily by wrapping the DNA around an octameric protein complex made of four core histones into a structure called nucleosome. Arrays of nucleosomes undergo further condensation into higher-order chromatin structures that functionally demarcate the chromatin boundaries into densely condensed heterochromatin and relatively less compressed euchromatin units. Although the high level of DNA packaging significantly deals with containing the genetic material in a miniscule space, DNA access to gene regulatory proteins during key cellular process such as transcription is greatly restricted. In this context, one crucial question is “How do TFs find their way to gene regulatory elements that are repressed or latent amidst these convoluted nucleosomal barriers in order to initiate transcription for diverse cellular processes?” A new set of regulatory proteins called ‘pioneer factors’ have been identified to accomplish this phenomenal task (Zaret and Carroll, 2011).
Pioneer factors belong to a unique class of TFs that can recognize and bind specific cis-regulatory units within permissive heterochromatin, and subsequently prime the chromatin for additional factors to bind, prior to transcription initiation (Zaret and Mango, 2016; Mayran and Drouin, 2018). The pioneer factors display few salient characteristics that are usually lacking in general TFs: (i) the ability to destabilize chromatin compaction (nucleosomes) and to bind to otherwise inaccessible heterochromatin regions, in a cell type/state-specific manner; (ii) the potential to alter pre-existing epigenetics modifications (such as DNA methylation and histone modification) to enhance DNA accessibility; and (iii) the remodeling of adjacent chromatin landscape to facilitate the binding of non-pioneer TFs prior to transcription initiation. In mammals, pioneer factors such as FOXA1, FOXD3, GATA-3, and PU.1 have crucial roles in development, cell fate conversions and deregulation of such factors in also implicated in cancer (Magnani et al., 2011; Iwafuchi-Doi and Zaret, 2016). Compelling evidences have accumulated to illustrate the novel role of Grainyhead and GRHL family proteins as pioneer factors in multiple cellular contexts.
Pioneer factors can remodel the chromatin landscape to expose functional cis-regulatory elements (such as enhancers) that recruit the binding of transcription factors, cofactors and collectively form a stable regulatory complex. Promoters are usually a minimal stretch of DNA sequences, located in proximity to the transcription start sites within a nucleosome-free chromatin landscape to enable easy access to the transcription machinery. In contrast, enhancers tend to be located far (either upstream or downstream) from the cognate promoters and enhance transcriptional outputs in a cell-type/state specific and spatiotemporal manner (Wittkopp and Kalay, 2012; Long et al., 2016). As a pioneer factor, Grainyhead and GRHL proteins bind to enhancers and regulate chromatin accessibility at the target genes during several developmental processes. During Drosophila eye development, the unbiased genome-wide characterization of direct TF interactions with enhancer regions of target genes have revealed a significant enrichment of Grainyhead in large fraction of active enhancer regions, which further elucidates the abundant Grh expression in the eye disc (Potier et al., 2014). The utilization of high-throughput genome-wide association methods such as the single-cell assay for transposable-accessible chromatin using sequencing (scATAC-seq) and quantitative trait loci for chromatin accessibility (caQTL) across a panel of Drosophila strains have revealed that the pioneer binding of Grh is essential for the opening and accessibility of epithelial cell enhancers (Jacobs et al., 2018). In these Grh binding sites, about 75% of Grh target sites are inaccessible due to the lack of Grh expression in the non-epithelial larval brain, whereas the ectopic overexpression of grh in the larval brain tissue profoundly increases the chromatin accessibility of these regions (Jacobs et al., 2018). These findings reiterate that, in Drosophila, Grh is the chief pioneer factor of the epithelial chromatin landscape. It potentially binds to the recognition sites and alters the closed chromatin landscape of non-epithelial tissues. In addition, the pioneering activity of Grh is also subjected to spatio-temporal and tissue specific regulation. Its pioneering activity has been reported to become essential during and after gastrulation but not during early embryogenesis (Nevil et al., 2020).
Investigations in mammalian GRHL factors also showed such pioneering roles in modulating the chromatin landscape of enhancers. During the early transition from the mouse naive embryonic stem cells (ESCs) to the primed pluripotent epiblast-like cells (EpiLCs), GRHL2 binds to latent enhancers (regions with low or no histone marks) and restores the epigenetic landscape toward an activation state (high levels of H3K4me1, H3K27ac histone marks and depletion of nucleosomes) of key genes that control the epithelial state and cell adhesion (Chen et al., 2018a; Figure 2). Importantly, in the ESC state, a different set of enhancers and TFs (such as KLF4, KLF5, and EKLF) controls the expression of the above mentioned genes, whereas in the EpiLCs, the control of gene expression switches toward the GRHL2-bound enhancers. Assaying across 47 human cell types, the positional distribution of TF binding motifs within the nucleosome-depleted enhancer sites have shown that GRHL1 is one of the six transcription factors that modulate DNA accessibility (Grossman et al., 2018). This study further elucidates that GRHL1 stably binds to the DNA with prolonged occupancy denoting that it may act in generating central anchor regions for potential transcription initiation. These results posit that in addition to pioneering the chromatin architecture, GRHL factors potentially mediate a major enhancer-switching phenomenon during cellular differentiation.
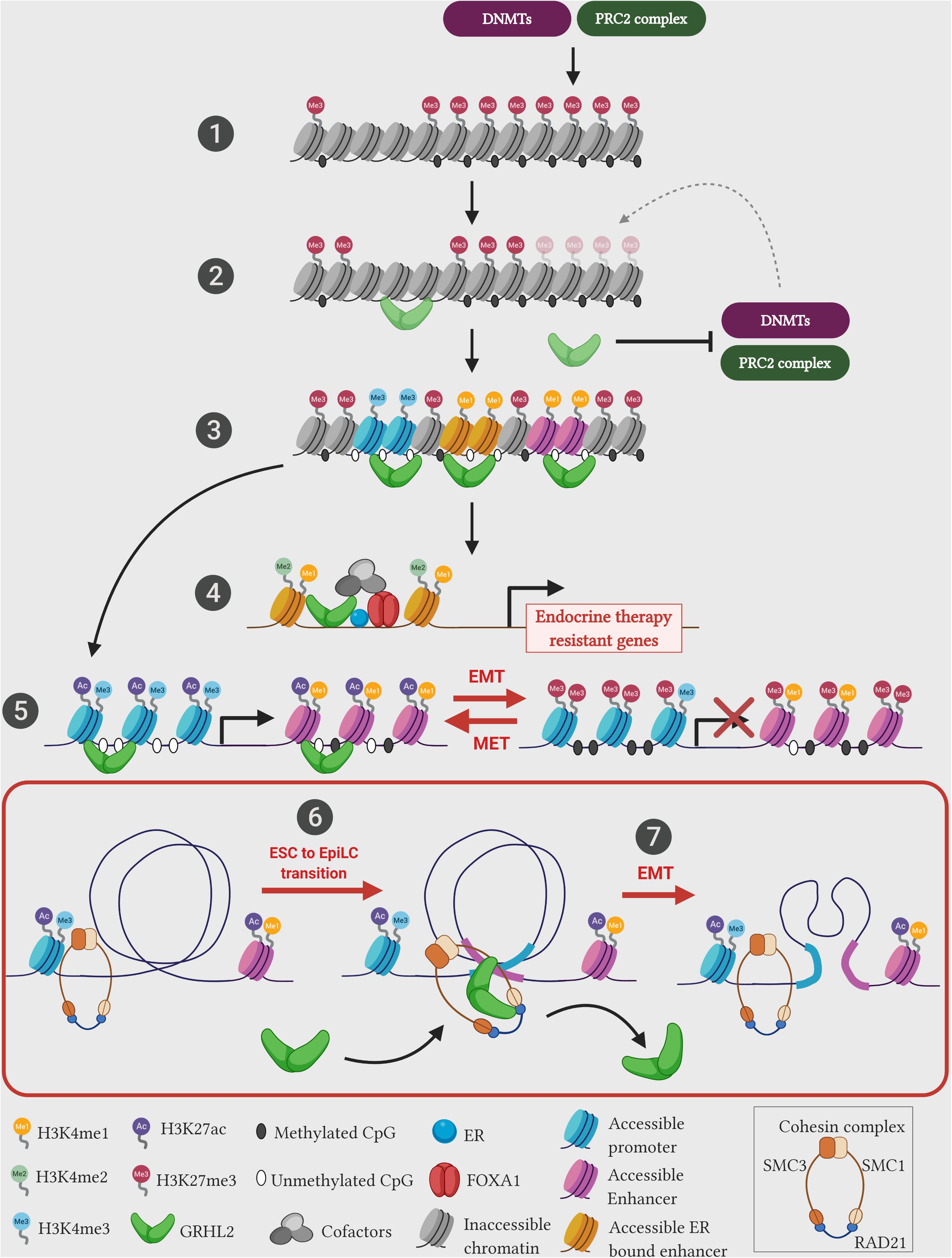
Figure 2. Emerging pioneering functions of GRHL2 during pathophysiology. Recent studies unearth the novel function of GRHL2 as a leading pioneer factor. (1) Typical chromatin landscape comprises cellular DNA wrapped around the core histone complex to form a nucleosome. Over half of the genome remains either in a latent/poised state with no histone marks/methylation status or in a repressed state studded with repressive chromatin marks (H3K27me3)/methylated CpG sites. These epigenetic marks are mediated through epigenetic repressors such as polycomb repressive complex 2 (PRC2) complex, and DNA methyltransferases (DNMTs). (2) Pioneer factors such as GRHL2 can potentially bind and activate latent chromatin in the context of ESC to EpiLC transition (6), or inhibit the activities of epigenetic repressors in the context of EMT (5). (3) GRHL2 primed regions generate accessible chromatin with unmethylated CpG islands and permissive histone marks (H3K4me3 – promoter; H3K4me1 – enhancer). (4) GRHL2 cooperates with pioneer factor FOXA1 at ER bound-active enhancer regions (studded with active histone H3K4me1/me2 marks) to drive transcription of endocrine therapy resistant genes. (5) In the presence of GRHL2, epigenetic landscape of epithelial genes are modified into a permissive chromatin (studded with H3K27ac and H3K4me3) at promoter and enhancer regions, whereas during cellular differentiation such as EMT, lack of GRHL2 expression results in change of chromatin setting of epithelial genes to a repressed state (hypermethylated promoters and enrichment of repressive H3K27me3 mark). (6) During ESC to EpiLC transition, GRHL2 associates with the cohesin component SMC1 on EpiLC-specific enhancer sites to facilitate transition. (7) In epithelial ovarian cancer cells, GRHL2 associates with the cohesin component RAD21 and brings distantly located gene regulatory elements in close proximity, to drive the expression of early epithelial genes such as ERBB3 and PERP. However, SNAI1-mediated EMT induction in these cells downregulates GRHL2 expression potentially disassembles cohesin structure, leading to reduced epithelial gene expression. Illustration created with Biorender.com.
GRHL proteins also exhibit the role of pioneering the enhancer landscape in human cancers. Chromatin states that denote the accessibility of the genomic region (such as active, repressed, heterochromatin, bivalent and poised) could be annotated using an automated ChromHMM algorithm (Ernst and Kellis, 2012). In ovarian cancer, using experimental ChIP-seq data derived from five major histone marks (H3K4me3, H3K4me1, H3K9me3, H3K27me3, and H3K27ac), our group has utilized this pipeline to show that upon the loss of GRHL2, a radical shift from an active chromatin state toward a latent, poised/bivalent, or repressed chromatin state occur across intronic and intergenic regions at the GRHL2 binding sites of epithelial genes such as MARVELD3, ESRP1, GRHL1, RAB25, OVOL2 and MUC20 (Chung et al., 2019). Upon re-expressing GRHL2 by using the inducible system, the chromatin changes of these GRHL2 binding sites located at the promoter and enhancer regions further shine the light on the pioneering function of GRHL2. GRHL2 is highly effective to induce MET in ovarian cancer cells with the intermediate phenotype. This is achieved via the suppression of PRC2 activity and to remove the repressive histone mark (H3K27me3) at the promoters with the corresponding suppression of HDAC activity at the enhancers to restore the H3K27ac mark (Chung et al., 2019). However, the pioneering capacity of GRHL2 might differ depending on cellular states along the EMT spectrum. The MET reversibility of GRHL2 in the highly mesenchymal cells has been quite limited suggesting that there would be state-specific pioneering reprogramming mechanisms. In human breast cancer cells, transient siRNA-mediated knockdown of all three GRHL orthologs show a reduced chromatin accessibility on GRHL-regulated enhancer elements that encode proteins required for cell-cell adhesion such as protocadherin-1 (PCDH1) and serine peptidase inhibitor 1 (SPINT1) (Jacobs et al., 2018). After priming the chromatin into an accessible regulatory landscape mediated by pioneer factors, tissue specific transcription factors and additional cofactors assemble to carryout gene expression. Such regulation is observed in the specific recruitment of GRHL factors to control steroid hormone-mediated gene expression in hormone-dependent cancers. Estrogen receptor α (ER) is a nuclear hormone receptor that drives over 70% of aggressive breast cancers. GRHL2 expression significantly correlates with ER-positive breast cancer tumors (Carroll et al., 2005; Xiang et al., 2012; Werner et al., 2013). ChIP-seq profiling of ER and phosphorylated ER at S118 (pS118-ER) occupancy sites shows a significant overlap with GRHL2 binding motifs (Helzer et al., 2018; Holding et al., 2019). This indicates that GRHL2 occupancy in the ER binding sites potentially drives ER transcription complex. The ER chromatin interaction and the subsequent gene expression changes are mediated through the pioneering activity of FOXA1 on cognate regulatory sites, independent of estrogen hormone signaling (Hurtado et al., 2011; Glont et al., 2019). Indeed, GRHL2 has been identified as the FOXA1 interaction partner at the ER bound-active enhancer regions demarcated with H3K4me1/me2 marks to promote tumor progression (Jozwik et al., 2016). This cooperation between FOXA1 and GRHL2 in ER driven breast cancer cells also contributes toward the resistance to endocrine therapy via the upregulation of LYPD/AGR2 (a receptor/ligand complex) making it a promising targetable frontier in endocrine therapy-resistant tumors (Cocce et al., 2019; Figure 2). These data clearly indicate the inevitable role of Grainyhead and GRHL proteins in the remodeling of gene regulatory units at the targeted sites.
As mentioned earlier, folding of the chromatin into three-dimensional structures are not only crucial for packaging DNA but also contribute toward fine-tuning of spatiotemporal gene regulation. For instance, in regulation of gene activity by cell-specific enhancers, distal enhancers are brought into close contact with its cognate promoters via DNA looping. Typically, DNA loops can occur between genomic loci which are tens to hundreds of kilobase pairs apart and are referred to as topological associated domains (TADs). The presence of TADs is evident across many species, ranging from Drosophila to mammals, and is a conserved feature of the three-dimensional chromatin architecture (Dixon et al., 2012; Hou et al., 2012; Nora et al., 2012; Sexton et al., 2012). These domains are demarcated by boundaries and often enriched in binding of architectural proteins: (i) sequence-specific CCTC-binding factor (CTCF) and (ii) cohesin protein complex, that serves to constrain the DNA loops within the TADs (Parelho et al., 2008; Stedman et al., 2008; Dixon et al., 2012; Rao et al., 2014). Therefore, TADs provide a structural and functional architecture across the genome, which permit short- and long-range chromatin interactions between regulatory elements within the same TADs (intradomain), while limiting interactions that span across the TAD boundaries (interdomain) to ensure proper gene regulation (Lupiáñez et al., 2015, 2016; Flavahan et al., 2016; Hnisz et al., 2016; Weischenfeldt et al., 2017; Beagan and Phillips-Cremins, 2020). The functional consequence of DNA loops in engaging promoters with its distal enhancers to drive transcriptional output of genes have been studied and reviewed (Sanyal et al., 2012; Long et al., 2016). In the context of the Grh family, a handful of studies indicate that GRHL factors have the potential to modulate chromatin looping structures and eventually affect gene expression. During mouse early embryonic development, intron 2 of the Cdh1 locus potentially functions as an enhancer element that control epithelium-specific E-cadherin expression (Stemmler et al., 2005). Using chromatin conformation capture-based techniques, it was revealed that the recruitment of Grhl2, Grhl3 and Hnf4α to multiple enhancers within intron 2 of Cdh1 resulted in functional DNA-loops to the Cdh1 promoter, thereby increasing the expression of the epithelial gene (Werth et al., 2010; Alotaibi et al., 2015). Formation of such DNA loops are essential for activating E-cadherin expression in mouse inner medullary collecting duct cells and thereby to induce epithelial differentiation in non-tumorigenic mouse mammary gland cells.
The formation of DNA loops is mainly mediated by the ring-shaped cohesin complex, which consists of four subunits – SMC1, SMC3, SCC1/RAD21, and SCC3/SA1/SA2 that topologically clasps chromatin into looping structures. As revealed by single-molecule imaging studies, the cohesin complex binds to DNA in a ring-shaped conformation and translocate along the chromatin in an ATPase-dependent fashion until it is impeded by CTCF (Davidson et al., 2016; Kanke et al., 2016; Stigler et al., 2016). It has been shown the transient degradation of cohesin resulted in the loss of DNA loops or loop domains, while CTCF degradation lead to the loss of DNA loops at the TAD boundaries, leading to subsequent loss of TAD insulation (Nora et al., 2017; Rao et al., 2017). Therefore, this highlights the role of cohesin complex lies heavily in the formation of loops linking two genomic loci and modulation of cohesin binding affects gene regulation during development and disease (Remeseiro et al., 2013). Recent studies have shown the direct interaction between GRHL factors and members of the cohesin complex. During the transition process of mouse ESCs to EpiLCs, Grhl2 predominantly associates with the cohesin subunit SMC1 to perform its pioneering function at enhancer regions of key epithelial genes to drive the transition process (Chen et al., 2018a). In the investigation on ovarian cancer cell lines, we also show that the co-occupancy of cohesin subunit RAD21 and GRHL2 on the promoter and enhancer elements of early epithelial genes (ERBB3 and PERP) is crucial for their expressions (Sundararajan et al., 2020; Figure 2). Moreover, we show that the recruitment of RAD21 on such enhancer regions are dependent on endogenous GRHL2 expression and the gradual loss of GRHL2 expression along the EMT spectrum of ovarian cancer cell lines might loosen up or alter the chromatin loop structures, which eventually lead to epithelial dedifferentiation. These observations highlight the identification of novel crosstalk between GRHL factors and the chromatin architectural complexes, where the pioneering activity of GRHL factors along the regulatory regions of epithelial genes potentially serve as loop anchors to mediate long-range functional chromatin interactions. Therefore, this mechanism appears to be an essential phenomenon in establishing the epithelial identity in development, while such interactions could be altered during cancer progression under conditions such as EMT. For example, in human sarcoma cells, ZEB1-associated chromatin remodeling factor BRG1 suppresses E-cadherin expression by blocking its promoter region (Somarelli et al., 2016). Depletion of BRG1 in the context of GRHL2 expression further upregulates E-cadherin expression, indicating that ZEB1-mediated chromatin remodeling interferes with GRHL factors associated pioneering function.
Almost three decades ago, foundation studies using Drosophila as a model organism have shown that the transcription factor Grainyhead is a crucial determinant of the epithelial phenotype and is involved in the development of vital fly organs such as epidermis, trachea, wings, and exoskeleton. Subsequent studies have identified that Grh potentially acts as a transcriptional activator and a repressor to regulate target gene expression, depending on the signaling events and its association with other transcription factors or co-factors. Interestingly, a recent study in the German cockroach (Blattella germanica) model has showed that Grh functions as a transcriptional repressor to regulate genes essential for the development of epithelium and molting of old integument (ecdysis) (Zhao et al., 2020). This indicates that the role of Grh in epithelial and epidermal differentiation is fundamental and evolutionarily conserved. Future Grh functional studies on other insect model systems might generate the possibility of Grh-mediated pest control management.
The three mammalian descendants of Grh – Grainyhead like 1-3 are also heavily implicated in the development of vital organs such as the neural tube, epidermis, and craniofacial skeleton. Although mutations and gene polymorphisms in Grhl genes were implicated in multiple human abnormalities such as hearing impairment, ectodermal dysplasia syndrome, and cleft palate formation, somatic mutations in GRHL genes in human cancer samples occur at a very low frequency (Kotarba et al., 2020). Moreover, the dual functioning of GRHL factors in carcinogenesis and tumor suppression indicate that GRHL factors impose a greater level of control over their target genes and miRNAs. In addition to directly controlling the target gene expression through promoter/enhancer binding, recent studies have shown that GRHL factors are potent modulators of the epigenetic landscape of target genes, which facilitate their spatiotemporal and cell type specific control. Such regulation is prevalent in the maintenance of epithelial barrier functions and the restoration of epithelial phenotypes during EMT/MET fluidity.
Recent studies underline how Grhl factors are essential during surface ectodermal neural lineage specification. Also, members of the GRHL family are potent repressors of the EMT program during development and cancer. These indicate that GRHL factors are at the crossroads of controlling epithelial, mesenchymal and neural-like phenotypes that determine cell lineage and transdifferentiation programs. It is therefore fair to hypothesize that tipping this balance during pathogenesis such as cancer might derail lineage specification, resulting in adverse phenotypes. Therefore, future studies on delineating GRHL factors-mediated of cellular and lineage plasticity through lineage tracing is worth exploring. Such investigations would shed light on the contribution of GRHL factors to the generation of neuroendocrine-like phenotypes, neuroendocrine differentiation observed in several cancers.
Genome-wide research progress in the last decade has brought a novel role of Grh/GRHL members as pioneer factors in limelight. Being pioneer factors, Grh/GRHL2 potentially gain access to the latent and repressed chromatin landscape of epithelial genes and prime such chromatin elements toward transcription initiation. Furthermore, GRHL2/3 associate with protein complexes that control chromatin 3D conformation structures (e.g., cohesins) and potentially regulate their access along the epithelial gene loci during cellular differentiation/dedifferentiation. Of note, chromatin conformational changes during EMT/MET programs during development and cancer progression are starting to emerge recently (Essafi et al., 2011; Yun et al., 2016; Sundararajan et al., 2020). Since GRHL2/3 are recently implicated in modulating the 3D chromatin architecture, future studies employing advanced sequencing techniques like ATAC-seq, FAIRE-seq, and Hi-C or Hi-ChIP would help us comprehend the interplay between GRHL factors and the chromatin accessibility during EMT/MET programs. Such investigations would also clarify our understanding on the dynamic changes of chromatin architecture along the EMT spectrum and eventually pave way toward improved cancer diagnostics and therapeutics.
VS and RH conceptualized and wrote the complete first version of the manuscript. All authors contributed to the manuscript revision, read, and approved the final version.
This work was supported by the National University Cancer Centre of Singapore (NCIS) Research Grant, Theme: EMT in Cancer and the Yushan Scholar Program by the Ministry of Education, Taiwan (NTU-109V0402) to RH.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acloque, H., Adams, M. S., Fishwick, K., Bronner-Fraser, M., and Nieto, M. A. (2009). Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J. Clin. Invest. 119, 1438–1449. doi: 10.1172/JCI38019
Adhikari, N., Neupane, S., Gwon, G.-J., Kim, J.-Y., An, C.-H., Lee, S., et al. (2017). Grhl3 modulates epithelial structure formation of the circumvallate papilla during mouse development. Histochem. Cell Biol. 147, 5–16. doi: 10.1007/s00418-016-1487-7
Afonso, C., and Henrique, D. (2006). PAR3 acts as a molecular organizer to define the apical domain of chick neuroepithelial cells. J. Cell Sci. 119, 4293–4304. doi: 10.1242/jcs.03170
Almeida, M. S., and Bray, S. J. (2005). Regulation of post-embryonic neuroblasts by Drosophila grainyhead. Mech. Dev. 122, 1282–1293. doi: 10.1016/j.mod.2005.08.004
Alotaibi, H., Basilicata, M. F., Shehwana, H., Kosowan, T., Schreck, I., Braeutigam, C., et al. (2015). Enhancer cooperativity as a novel mechanism underlying the transcriptional regulation of E-cadherin during mesenchymal to epithelial transition. Biochim. Biophys. Acta 1849, 731–742. doi: 10.1016/j.bbagrm.2015.01.005
Attardi, L. D., Seggern, D. V., and Tjian, R. (1993). Ectopic expression of wild-type or a dominant-negative mutant of transcription factor NTF-1 disrupts normal Drosophila development. PNAS 90, 10563–10567. doi: 10.1073/pnas.90.22.10563
Auden, A., Caddy, J., Wilanowski, T., Ting, S. B., Cunningham, J. M., and Jane, S. M. (2006). Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Express. Patterns 6, 964–970. doi: 10.1016/j.modgep.2006.03.011
Audia, J. E., and Campbell, R. M. (2016). Histone modifications and Cancer. Cold Spring Harb. Perspect. Biol. 8:a019521. doi: 10.1101/cshperspect.a019521
Aue, A., Hinze, C., Walentin, K., Ruffert, J., Yurtdas, Y., Werth, M., et al. (2015). A grainyhead-like 2/Ovo-Like 2 pathway regulates renal epithelial barrier function and lumen expansion. JASN 26, 2704–2715. doi: 10.1681/ASN.2014080759
Bannister, A. J., and Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21, 381–395. doi: 10.1038/cr.2011.22
Beagan, J. A., and Phillips-Cremins, J. E. (2020). On the existence and functionality of topologically associating domains. Nat. Genet. 52, 8–16. doi: 10.1038/s41588-019-0561-1
Bhandari, A., Gordon, W., Dizon, D., Hopkin, A. S., Gordon, E., Yu, Z., et al. (2013). The Grainyhead transcription factor Grhl3/Get1 suppresses miR-21 expression and tumorigenesis in skin: modulation of the miR-21 target MSH2 by RNA-binding protein DND1. Oncogene 32, 1497–1507. doi: 10.1038/onc.2012.168
Bhandari, J., and Thada, P. K. (2020). Neural Tube Disorders. Treasure Island, FL: StatPearls Publishing.
Boglev, Y., Wilanowski, T., Caddy, J., Parekh, V., Auden, A., Darido, C., et al. (2011). The unique and cooperative roles of the Grainy head-like transcription factors in epidermal development reflect unexpected target gene specificity. Dev. Biol. 349, 512–522. doi: 10.1016/j.ydbio.2010.11.011
Brabletz, S., and Brabletz, T. (2010). The ZEB/miR-200 feedback loop—a motor of cellular plasticity in development and cancer? EMBO Rep. 11, 670–677. doi: 10.1038/embor.2010.117
Bray, S. J., Johnson, W. A., Hirsh, J., Heberlein, U., and Tjian, R. (1988). A cis-acting element and associated binding factor required for CNS expression of the Drosophila melanogaster dopa decarboxylase gene. Embo J. 7, 177–188. doi: 10.1002/j.1460-2075.1988.tb02798.x
Bray, S. J., and Kafatos, F. C. (1991). Developmental function of Elf-1: an essential transcription factor during embryogenesis in Drosophila. Genes Dev. 5, 1672–1683. doi: 10.1101/gad.5.9.1672
Brouns, M. R., De Castro, S. C. P., Terwindt-Rouwenhorst, E. A., Massa, V., Hekking, J. W., Hirst, C. S., et al. (2011). Over-expression of Grhl2 causes spina bifida in the Axial defects mutant mouse. Hum. Mol. Genet. 20, 1536–1546. doi: 10.1093/hmg/ddr031
Caddy, J., Wilanowski, T., Darido, C., Dworkin, S., Ting, S. B., Zhao, Q., et al. (2010). Epidermal wound repair is regulated by the planar cell polarity signaling pathway. Dev. Cell 19, 138–147. doi: 10.1016/j.devcel.2010.06.008
Camerer, E., Barker, A., Duong, D. N., Ganesan, R., Kataoka, H., Cornelissen, I., et al. (2010). Local protease signaling contributes to neural tube closure in the mouse embryo. Dev. Cell 18, 25–38. doi: 10.1016/j.devcel.2009.11.014
Carpinelli, M. R., de Vries, M. E., Jane, S. M., and Dworkin, S. (2017). Grainyhead-like transcription factors in craniofacial development. J. Dent. Res. 96, 1200–1209. doi: 10.1177/0022034517719264
Carpinelli, M. R., Vries, M. E., de, Auden, A., Butt, T., Deng, Z., et al. (2020). Inactivation of Zeb1 in GRHL2-deficient mouse embryos rescues mid-gestation viability and secondary palate closure. Dis. Models Mech. 13:dmm042218. doi: 10.1242/dmm.042218
Carroll, J. S., Liu, X. S., Brodsky, A. S., Li, W., Meyer, C. A., Szary, A. J., et al. (2005). Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43. doi: 10.1016/j.cell.2005.05.008
Chen, A. F., Liu, A. J., Krishnakumar, R., Freimer, J. W., DeVeale, B., and Blelloch, R. (2018a). GRHL2-Dependent enhancer switching maintains a pluripotent stem cell transcriptional subnetwork after exit from naive pluripotency. Cell Stem Cell 23, 226.e4–238.e4. doi: 10.1016/j.stem.2018.06.005
Chen, W., Kang, K. L., Alshaikh, A., Varma, S., Lin, Y.-L., Shin, K.-H., et al. (2018b). Grainyhead-like 2 (GRHL2) knockout abolishes oral cancer development through reciprocal regulation of the MAP kinase and TGF-β signaling pathways. Oncogenesis 7:38. doi: 10.1038/s41389-018-0047-5
Chen, W., Shimane, T., Kawano, S., Alshaikh, A., Kim, S. Y., Chung, S. H., et al. (2018c). Human papillomavirus 16 E6 induces FoxM1B in oral keratinocytes through GRHL2. J. Dent. Res. 97, 795–802. doi: 10.1177/0022034518756071
Chen, C.-P., Chang, T.-Y., Hung, F.-Y., Chern, S.-R., Wu, P.-S., Chen, S.-W., et al. (2017). Prenatal diagnosis of an 8q22.2-q23.3 deletion associated with bilateral cleft lip and palate and intrauterine growth restriction on fetal ultrasound. Taiwanese J. Obstetr. Gynecol. 56, 843–846. doi: 10.1016/j.tjog.2017.10.026
Chen, W., Dong, Q., Shin, K.-H., Kim, R. H., Oh, J.-E., Park, N.-H., et al. (2010). Grainyhead-like 2 enhances the human telomerase reverse transcriptase gene expression by inhibiting DNA methylation at the 5’-CpG Island in normal human keratinocytes. J. Biol. Chem. 285, 40852–40863. doi: 10.1074/jbc.M110.103812
Chen, W., Xiao Liu, Z., Oh, J.-E., Shin, K.-H., Kim, R. H., Jiang, M., et al. (2012). Grainyhead-like 2 (GRHL2) inhibits keratinocyte differentiation through epigenetic mechanism. Cell Death Dis. 3:e450. doi: 10.1038/cddis.2012.190
Chen, W., Yi, J. K., Shimane, T., Mehrazarin, S., Lin, Y.-L., Shin, K.-H., et al. (2016). Grainyhead-like 2 regulates epithelial plasticity and stemness in oral cancer cells. Carcinogenesis 37, 500–510. doi: 10.1093/carcin/bgw027
Chung, V. Y., Tan, T. Z., Tan, M., Wong, M. K., Kuay, K. T., Yang, Z., et al. (2016). GRHL2-miR-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci. Rep. 6:19943. doi: 10.1038/srep19943
Chung, V. Y., Tan, T. Z., Ye, J., Huang, R.-L., Lai, H.-C., Kappei, D., et al. (2019). The role of GRHL2 and epigenetic remodeling in epithelial–mesenchymal plasticity in ovarian cancer cells. Commun. Biol. 2:272. doi: 10.1038/s42003-019-0506-3
Cieply, B., Riley, P., Pifer, P. M., Widmeyer, J., Addison, J. B., Ivanov, A. V., et al. (2012). Suppression of the epithelial–mesenchymal transition by grainyhead-like-2. Cancer Res. 72, 2440–2453. doi: 10.1158/0008-5472.CAN-11-4038
Cocce, K. J., Jasper, J. S., Desautels, T. K., Everett, L., Wardell, S., Westerling, T., et al. (2019). The lineage determining factor GRHL2 collaborates with FOXA1 to establish a targetable pathway in endocrine therapy-resistant breast cancer. Cell Rep. 29, 889.e10–903.e10. doi: 10.1016/j.celrep.2019.09.032
Danila, D. C., Anand, A., Schultz, N., Heller, G., Wan, M., Sung, C. C., et al. (2014). Analytic and clinical validation of a prostate cancer–enhanced messenger RNA detection assay in whole blood as a prognostic biomarker for survival. Eur. Urol. 65, 1191–1197. doi: 10.1016/j.eururo.2013.07.006
Darido, C., Georgy, S. R., Wilanowski, T., Dworkin, S., Auden, A., Zhao, Q., et al. (2011). Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell 20, 635–648. doi: 10.1016/j.ccr.2011.10.014
Davidson, I. F., Goetz, D., Zaczek, M. P., Molodtsov, M. I., Huis In ’t Veld, P. J., Weissmann, F., et al. (2016). Rapid movement and transcriptional re-localization of human cohesin on DNA. Embo J. 35, 2671–2685. doi: 10.15252/embj.201695402
De Castro, S. C. P., Hirst, C. S., Savery, D., Rolo, A., Lickert, H., Andersen, B., et al. (2018). Neural tube closure depends on expression of Grainyhead-like 3 in multiple tissues. Dev. Biol. 435, 130–137. doi: 10.1016/j.ydbio.2018.01.016
de Vries, M., Carpinelli, M., Rutland, E., Hatzipantelis, A., Partridge, D., Auden, A., et al. (2020). Interrogating the Grainyhead-like 2 (Grhl2) genomic locus identifies an enhancer element that regulates palatogenesis in mouse. Dev. Biol. 459, 194–203. doi: 10.1016/j.ydbio.2019.11.015
Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., et al. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. doi: 10.1038/nature11082
Dong, C., Wu, Y., Yao, J., Wang, Y., Yu, Y., Rychahou, P. G., et al. (2012). G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J. Clin. Invest. 122, 1469–1486. doi: 10.1172/JCI57349
Dworkin, S., Darido, C., Georgy, S. R., Wilanowski, T., Srivastava, S., Ellett, F., et al. (2012). Midbrain-hindbrain boundary patterning and morphogenesis are regulated by diverse grainy head-like 2-dependent pathways. Development 139, 525–536. doi: 10.1242/dev.066522
Dworkin, S., Simkin, J., Darido, C., Partridge, D. D., Georgy, S. R., Caddy, J., et al. (2014). Grainyhead-like 3 regulation of endothelin-1 in the pharyngeal endoderm is critical for growth and development of the craniofacial skeleton. Mech. Dev. 133, 77–90. doi: 10.1016/j.mod.2014.05.005
Dynlacht, B. D., Attardi, L. D., Admon, A., Freeman, M., and Tjian, R. (1989). Functional analysis of NTF-1, a developmentally regulated Drosophila transcription factor that binds neuronal cis elements. Genes Dev. 3, 1677–1688. doi: 10.1101/gad.3.11.1677
Eckert, R. L., Sturniolo, M. T., Broome, A.-M., Ruse, M., and Rorke, E. A. (2005). Transglutaminase function in epidermis. J. Invest. Dermatol. 124, 481–492. doi: 10.1111/j.0022-202X.2005.23627.x
Eichelberger, L., Saini, M., Moreno, H. D., Klein, C., Bartsch, J. M., Falcone, M., et al. (2020). Maintenance of epithelial traits and resistance to mesenchymal reprogramming promote proliferation in metastatic breast cancer. bioRxiv doi: 10.1101/2020.03.19.998823
Ernst, J., and Kellis, M. (2012). ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216. doi: 10.1038/nmeth.1906
Eshete, M. A., Liu, H., Li, M., Adeyemo, W. L., Gowans, L. J. J., Mossey, P. A., et al. (2018). Loss-of-function GRHL3 variants detected in african patients with isolated cleft palate. J. Dent. Res. 97, 41–48. doi: 10.1177/0022034517729819
Essafi, A., Webb, A., Berry, R. L., Slight, J., Burn, S. F., Spraggon, L., et al. (2011). A wt1-controlled chromatin switching mechanism underpins tissue-specific wnt4 activation and repression. Dev. Cell 21, 559–574. doi: 10.1016/j.devcel.2011.07.014
Fabian, J., Lodrini, M., Oehme, I., Schier, M. C., Thole, T. M., Hielscher, T., et al. (2014). GRHL1 acts as tumor suppressor in neuroblastoma and is negatively regulated by MYCN and HDAC3. Cancer Res. 74, 2604–2616. doi: 10.1158/0008-5472.CAN-13-1904
Farris, J. C., Pifer, P. M., Zheng, L., Gottlieb, E., Denvir, J., and Frisch, S. M. (2016). Grainyhead-like 2 reverses the metabolic changes induced by the oncogenic epithelial–mesenchymal transition: effects on anoikis. Mol. Cancer Res. 14, 528–538. doi: 10.1158/1541-7786.MCR-16-0050
Flavahan, W. A., Drier, Y., Liau, B. B., Gillespie, S. M., Venteicher, A. S., Stemmer-Rachamimov, A. O., et al. (2016). Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114. doi: 10.1038/nature16490
Francart, M.-E., Lambert, J., Vanwynsberghe, A. M., Thompson, E. W., Bourcy, M., Polette, M., et al. (2018). Epithelial-mesenchymal plasticity and circulating tumor cells: travel companions to metastases. Dev. Dyn. 247, 432–450. doi: 10.1002/dvdy.24506
Fukagawa, A., Ishii, H., Miyazawa, K., and Saitoh, M. (2015). δEF1 associates with DNMT1 and maintains DNA methylation of the E-cadherin promoter in breast cancer cells. Cancer Med. 4, 125–135. doi: 10.1002/cam4.347
Fustaino, V., Presutti, D., Colombo, T., Cardinali, B., Papoff, G., Brandi, R., et al. (2017). Characterization of epithelial-mesenchymal transition intermediate/hybrid phenotypes associated to resistance to EGFR inhibitors in non-small cell lung cancer cell lines. Oncotarget 8, 103340–103363. doi: 10.18632/oncotarget.21132
Gao, X., Bali, A. S., Randell, S. H., and Hogan, B. L. M. (2015). GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. J. Cell Biol. 211, 669–682. doi: 10.1083/jcb.201506014
Gao, X., Vockley, C. M., Pauli, F., Newberry, K. M., Xue, Y., Randell, S. H., et al. (2013). Evidence for multiple roles for grainyhead-like 2 in the establishment and maintenance of human mucociliary airway epithelium. PNAS 110, 9356–9361. doi: 10.1073/pnas.1307589110
Garrett, J. T., Olivares, M. G., Rinehart, C., Granja-Ingram, N. D., Sánchez, V., Chakrabarty, A., et al. (2011). Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. Proc. Natl. Acad. Sci. U.S.A. 108, 5021–5026. doi: 10.1073/pnas.1016140108
Geiger, J. A., Carvalho, L., Campos, I., Santos, A. C., and Jacinto, A. (2011). Hole-in-one mutant phenotypes link EGFR/ERK signaling to epithelial tissue repair in Drosophila. PLoS One 6:e28349. doi: 10.1371/journal.pone.0028349
Glont, S.-E., Chernukhin, I., and Carroll, J. S. (2019). Comprehensive genomic analysis reveals that the pioneering function of FOXA1 is independent of hormonal signaling. Cell Reports 26, 2558.e3–2565.e3. doi: 10.1016/j.celrep.2019.02.036
Goldie, S. J., Arhatari, B. D., Anderson, P., Auden, A., Partridge, D. D., Jane, S. M., et al. (2016). Mice lacking the conserved transcription factor Grainyhead-like 3 (Grhl3) display increased apposition of the frontal and parietal bones during embryonic development. BMC Dev. Biol. 16:37. doi: 10.1186/s12861-016-0136-7
Grosse-Wilde, A., Fouquier, d’Hérouël, A., McIntosh, E., Ertaylan, G., Skupin, A., et al. (2015). Stemness of the hybrid epithelial/mesenchymal state in breast cancer and its association with poor survival. PLoS One 10:e0126522. doi: 10.1371/journal.pone.0126522
Grossman, S. R., Engreitz, J., Ray, J. P., Nguyen, T. H., Hacohen, N., and Lander, E. S. (2018). Positional specificity of different transcription factor classes within enhancers. PNAS 115, E7222–E7230. doi: 10.1073/pnas.1804663115
Gustavsson, P., Greene, N. D. E., Lad, D., Pauws, E., de Castro, S. C. P., Stanier, P., et al. (2007). Increased expression of Grainyhead-like-3 rescues spina bifida in a folate-resistant mouse model. Hum. Mol. Genet. 16, 2640–2646. doi: 10.1093/hmg/ddm221
Han, Y., Mu, Y., Li, X., Xu, P., Tong, J., Liu, Z., et al. (2011). Grhl2 deficiency impairs otic development and hearing ability in a zebrafish model of the progressive dominant hearing loss DFNA28. Hum. Mol. Genet. 20, 3213–3226. doi: 10.1093/hmg/ddr234
Harding, M. J., McGraw, H. F., and Nechiporuk, A. (2014). The roles and regulation of multicellular rosette structures during morphogenesis. Development 141, 2549–2558. doi: 10.1242/dev.101444
Harrison, M. M., Botchan, M. R., and Cline, T. W. (2010). Grainyhead and Zelda compete for binding to the promoters of the earliest-expressed Drosophila genes. Dev. Biol. 345, 248–255. doi: 10.1016/j.ydbio.2010.06.026
Hay, E. D. (1995). An overview of epithelio-mesenchymal transformation. Acta Anat. 154, 8–20. doi: 10.1159/000147748
Hay, E. D. (2005). The mesenchymal cell, its role in the embryo, and the remarkable signaling mechanisms that create it. Dev. Dyn. 233, 706–720. doi: 10.1002/dvdy.20345
Heldin, C.-H., Vanlandewijck, M., and Moustakas, A. (2012). Regulation of EMT by TGFβ in cancer. FEBS Lett. 586, 1959–1970. doi: 10.1016/j.febslet.2012.02.037
Helzer, K. T., Szatkowski Ozers, M., Meyer, M. B., Benkusky, N. A., Solodin, N., Reese, R. M., et al. (2018). The phosphorylated estrogen receptor α (ER) cistrome identifies a subset of active enhancers enriched for direct ER-DNA binding and the transcription factor GRHL2. Mol. Cell Biol. 39, e417–e418. doi: 10.1128/MCB.00417-18
Hemphälä, J., Uv, A., Cantera, R., Bray, S., and Samakovlis, C. (2003). Grainy head controls apical membrane growth and tube elongation in response to Branchless/FGF signalling. Development 130, 249–258. doi: 10.1242/dev.00218
Hislop, N. R., Caddy, J., Ting, S. B., Auden, A., Vasudevan, S., King, S. L., et al. (2008). Grhl3 and Lmo4 play coordinate roles in epidermal migration. Dev. Biol. 321, 263–272. doi: 10.1016/j.ydbio.2008.06.026
Hnisz, D., Weintraub, A. S., Day, D. S., Valton, A.-L., Bak, R. O., Li, C. H., et al. (2016). Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458. doi: 10.1126/science.aad9024
Hoebel, A. K., Drichel, D., van de Vorst, M., Böhmer, A. C., Sivalingam, S., Ishorst, N., et al. (2017). Candidate genes for nonsyndromic cleft palate detected by exome sequencing. J. Dent. Res. 96, 1314–1321. doi: 10.1177/0022034517722761
Holding, A. N., Giorgi, F. M., Donnelly, A., Cullen, A. E., Nagarajan, S., Selth, L. A., et al. (2019). VULCAN integrates ChIP-seq with patient-derived co-expression networks to identify GRHL2 as a key co-regulator of ERa at enhancers in breast cancer. Genome Biol. 20:91. doi: 10.1186/s13059-019-1698-z
Hopkin, A. S., Gordon, W., Klein, R. H., Espitia, F., Daily, K., Zeller, M., et al. (2012). GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS Genet. 8:e1002829. doi: 10.1371/journal.pgen.1002829
Hosoya, M., Fujioka, M., Ogawa, K., and Okano, H. (2016). Distinct expression patterns of causative genes responsible for hereditary progressive hearing loss in non-human primate cochlea. Sci. Rep. 6:22250. doi: 10.1038/srep22250
Hou, C., Li, L., Qin, Z. S., and Corces, V. G. (2012). Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell 48, 471–484. doi: 10.1016/j.molcel.2012.08.031
Huang, R. Y.-J., Wong, M. K., Tan, T. Z., Kuay, K. T., Ng, A. H. C., Chung, V. Y., et al. (2013). An EMT spectrum defines an anoikis-resistant and spheroidogenic intermediate mesenchymal state that is sensitive to e-cadherin restoration by a src-kinase inhibitor, saracatinib (AZD0530). Cell Death Dis. 4:e915. doi: 10.1038/cddis.2013.442
Hurtado, A., Holmes, K. A., Ross-Innes, C. S., Schmidt, D., and Carroll, J. S. (2011). FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 43, 27–33. doi: 10.1038/ng.730
Iwafuchi-Doi, M., and Zaret, K. S. (2016). Cell fate control by pioneer transcription factors. Development 143, 1833–1837. doi: 10.1242/dev.133900
Jacobs, J., Atkins, M., Davie, K., Imrichova, H., Romanelli, L., Christiaens, V., et al. (2018). The transcription factor Grainy head primes epithelial enhancers for spatiotemporal activation by displacing nucleosomes. Nat. Genet. 50, 1011–1020. doi: 10.1038/s41588-018-0140-x
Jia, W., Tripathi, S., Chakraborty, P., Chedere, A., Rangarajan, A., Levine, H., et al. (2020). Epigenetic feedback and stochastic partitioning during cell division can drive resistance to EMT. Oncotarget 11, 2611–2624. doi: 10.18632/oncotarget.27651
Jolly, M. K., Mani, S. A., and Levine, H. (2018). Hybrid epithelial/mesenchymal phenotype(s): the ‘fittest’ for metastasis? Biochim. Biophys. Acta 1870, 151–157. doi: 10.1016/j.bbcan.2018.07.001
Jolly, M. K., Somarelli, J. A., Sheth, M., Biddle, A., Tripathi, S. C., Armstrong, A. J., et al. (2019). Hybrid epithelial/mesenchymal phenotypes promote metastasis and therapy resistance across carcinomas. Pharmacol. Therap. 194, 161–184. doi: 10.1016/j.pharmthera.2018.09.007
Jolly, M. K., Tripathi, S. C., Jia, D., Mooney, S. M., Celiktas, M., Hanash, S. M., et al. (2016). Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget 7, 27067–27084. doi: 10.18632/oncotarget.8166
Jozwik, K. M., Chernukhin, I., Serandour, A. A., Nagarajan, S., and Carroll, J. S. (2016). FOXA1 directs H3K4 monomethylation at enhancers via recruitment of the methyltransferase MLL3. Cell Rep. 17, 2715–2723. doi: 10.1016/j.celrep.2016.11.028
Kalluri, R., and Weinberg, R. A. (2009). The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428. doi: 10.1172/JCI39104
Kang, X., Chen, W., Kim, R. H., Kang, M. K., and Park, N.-H. (2009). Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. Oncogene 28, 565–574. doi: 10.1038/onc.2008.404
Kanke, M., Tahara, E., Huis In’t, Veld, P. J., and Nishiyama, T. (2016). Cohesin acetylation and Wapl-Pds5 oppositely regulate translocation of cohesin along DNA. Embo J. 35, 2686–2698. doi: 10.15252/embj.201695756
Kashgari, G., Meinecke, L., Gordon, W., Ruiz, B., Yang, J., Ma, A. L., et al. (2020). Epithelial migration and non-adhesive periderm are required for digit separation during mammalian development. Dev. Cell 52, 764.e–778.e. doi: 10.1016/j.devcel.2020.01.032
Kersbergen, A., Best, S. A., Dworkin, S., Ah-Cann, C., de Vries, M. E., Asselin-Labat, M.-L., et al. (2018). Lung morphogenesis is orchestrated through Grainyhead-like 2 (Grhl2) transcriptional programs. Dev. Biol. 443, 1–9. doi: 10.1016/j.ydbio.2018.09.002
Khandelwal, R., Sipani, R., Govinda Rajan, S., Kumar, R., and Joshi, R. (2017). Combinatorial action of grainyhead, extradenticle and notch in regulating Hox mediated apoptosis in Drosophila larval CNS. PLoS Genet. 13:e1007043. doi: 10.1371/journal.pgen.1007043
Kim, M., and McGinnis, W. (2011). Phosphorylation of grainy head by ERK is essential for wound-dependent regeneration but not for development of an epidermal barrier. Proc. Natl. Acad. Sci. U.S.A. 108, 650–655. doi: 10.1073/pnas.1016386108
Kim, Y.-R., Kim, M.-A., Sagong, B., Bae, S.-H., Lee, H.-J., Kim, H.-J., et al. (2015). Evaluation of the contribution of the EYA4 and GRHL2 genes in korean patients with autosomal dominant non-syndromic hearing loss. PLoS One 10:e0119443. doi: 10.1371/journal.pone.0119443
Kimura-Yoshida, C., Mochida, K., Ellwanger, K., Niehrs, C., and Matsuo, I. (2015). Fate specification of neural plate border by canonical Wnt signaling and Grhl3 is crucial for neural tube closure. Ebio Med. 2, 513–527. doi: 10.1016/j.ebiom.2015.04.012
Kimura-Yoshida, C., Mochida, K., Nakaya, M.-A., Mizutani, T., and Matsuo, I. (2018). Cytoplasmic localization of GRHL3 upon epidermal differentiation triggers cell shape change for epithelial morphogenesis. Nat. Commun. 9:4059. doi: 10.1038/s41467-018-06171-8
Kokoszynska, K., Ostrowski, J., Rychlewski, L., and Wyrwicz, L. S. (2008). The fold recognition of CP2 transcription factors gives new insights into the function and evolution of tumor suppressor protein p53. Cell Cycle 7, 2907–2915. doi: 10.4161/cc.7.18.6680
Kotarba, G., Krzywinska, E., Grabowska, A. I., Taracha, A., and Wilanowski, T. (2018). TFCP2/TFCP2L1/UBP1 transcription factors in cancer. Cancer Lett. 420, 72–79. doi: 10.1016/j.canlet.2018.01.078
Kotarba, G., Taracha-Wisniewska, A., and Wilanowski, T. (2020). Grainyhead-like transcription factors in cancer – Focus on recent developments. Exp. Biol. Med. 245, 402–410. doi: 10.1177/1535370220903009
Kuechler, A., Buysse, K., Clayton-Smith, J., Caignec, C. L., David, A., Engels, H., et al. (2011). Five patients with novel overlapping interstitial deletions in 8q22.2q22.3. Am. J. Med. Genet. Part A 155, 1857–1864. doi: 10.1002/ajmg.a.34072
Kuroda, Y., Ohashi, I., Saito, T., Nagai, J., Ida, K., Naruto, T., et al. (2014). Refinement of the deletion in 8q22.2–q22.3: the minimum deletion size at 8q22.3 related to intellectual disability and epilepsy. Am. J. Med. Genet. Part A 164, 2104–2108. doi: 10.1002/ajmg.a.36604
Lee, H., and Adler, P. N. (2004). The grainy head transcription factor is essential for the function of the frizzled pathway in the Drosophila wing. Mech. Dev. 121, 37–49. doi: 10.1016/j.mod.2003.11.002
Leth-Larsen, R., Terp, M. G., Christensen, A. G., Elias, D., Kühlwein, T., Jensen, O. N., et al. (2012). Functional heterogeneity within the CD44 high human breast cancer stem cell-like compartment reveals a gene signature predictive of distant metastasis. Mol. Med. 18, 1109–1121. doi: 10.2119/molmed.2012.00091
Li, M., Li, Z., Guan, X., and Qin, Y. (2019). Suppressor gene GRHL1 is associated with prognosis in patients with oesophageal squamous cell carcinoma. Oncol. Lett. 17, 4313–4320. doi: 10.3892/ol.2019.10072
Li, X., Zhu, Z., Li, W., Wei, L., Zhao, B., and Hao, Z. (2019). Polymorphism in GRHL2 gene may contribute to noise-induced hearing loss susceptibility: a meta-analysis. Braz. J. Otorhinolaryngol. 86, 370–375. doi: 10.1016/j.bjorl.2019.01.003
Lin, X., Teng, Y., Lan, J., He, B., Sun, H., and Xu, F. (2016). GRHL2 genetic polymorphisms may confer a protective effect against sudden sensorineural hearing loss. Mol. Med. Rep. 13, 2857–2863. doi: 10.3892/mmr.2016.4871
Liu, B., Ordonez-Ercan, D., Fan, Z., Edgerton, S. M., Yang, X., and Thor, A. D. (2007). Downregulation of erbB3 abrogates erbB2-mediated tamoxifen resistance in breast cancer cells. Int. J. Cancer 120, 1874–1882. doi: 10.1002/ijc.22423
Long, H. K., Prescott, S. L., and Wysocka, J. (2016). Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167, 1170–1187. doi: 10.1016/j.cell.2016.09.018
Lupiáñez, D. G., Kraft, K., Heinrich, V., Krawitz, P., Brancati, F., Klopocki, E., et al. (2015). Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025. doi: 10.1016/j.cell.2015.04.004
Lupiáñez, D. G., Spielmann, M., and Mundlos, S. (2016). Breaking TADs: how alterations of chromatin domains result in disease. Trends Genet. 32, 225–237. doi: 10.1016/j.tig.2016.01.003
Mace, K. A., Pearson, J. C., and McGinnis, W. (2005). An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science 308, 381–385. doi: 10.1126/science.1107573
MacFawn, I., Wilson, H., Selth, L. A., Leighton, I., Serebriiskii, I., Bleackley, R. C., et al. (2019). Grainyhead-like-2 confers NK-sensitivity through interactions with epigenetic modifiers. Mol. Immunol. 105, 137–149. doi: 10.1016/j.molimm.2018.11.006
Magnani, L., Eeckhoute, J., and Lupien, M. (2011). Pioneer factors: directing transcriptional regulators within the chromatin environment. Trends Genet. 27, 465–474. doi: 10.1016/j.tig.2011.07.002
Matsushita, T., Sakai, M., Yoshida, H., Morita, S., Hieda, Y., and Sakai, T. (2018). Grhl2 regulation of SPINT1 expression controls salivary gland development. Biochem. Biophys. Res. Commun. 504, 263–269. doi: 10.1016/j.bbrc.2018.08.166
Matyas, P., Postyeni, E., Komlosi, K., Szalai, R., Bene, J., Magyari, L., et al. (2019). Age-related hearing impairment associated NAT2, GRM7, GRHL2 susceptibility gene polymorphisms and haplotypes in roma and hungarian populations. Pathol. Oncol. Res. 25, 1349–1355. doi: 10.1007/s12253-018-0388-6
Mayran, A., and Drouin, J. (2018). Pioneer transcription factors shape the epigenetic landscape. J. Biol. Chem. 293, 13795–13804. doi: 10.1074/jbc.R117.001232
McInnes, L. M., Jacobson, N., Redfern, A., Dowling, A., Thompson, E. W., and Saunders, C. M. (2015). Clinical implications of circulating tumor cells of breast cancer patients: role of epithelial-mesenchymal plasticity. Front. Oncol. 5:42. doi: 10.3389/fonc.2015.00042
Mehrazarin, S., Chen, W., Oh, J.-E., Liu, Z. X., Kang, K. L., Yi, J. K., et al. (2015). The p63 gene is regulated by Grainyhead-like 2 (GRHL2) through reciprocal feedback and determines the epithelial phenotype in human keratinocytes. J. Biol. Chem. 290, 19999–20008. doi: 10.1074/jbc.M115.659144
Menke, C., Cionni, M., Siggers, T., Bulyk, M. L., Beier, D. R., and Stottmann, R. W. (2015). Grhl2 is required in nonneural tissues for neural progenitor survival and forebrain development. Genesis 53, 573–582. doi: 10.1002/dvg.22875
Miles, L. B., Dworkin, S., and Darido, C. (2017). Alternative splicing and start sites: lessons from the Grainyhead-like family. Dev. Biol. 429, 12–19. doi: 10.1016/j.ydbio.2017.06.018
Ming, Q., Roske, Y., Schuetz, A., Walentin, K., Ibraimi, I., Schmidt-Ott, K. M., et al. (2018). Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family. Nucl. Acids Res. 46, 2082–2095. doi: 10.1093/nar/gkx1299
Mlacki, M., Darido, C., Jane, S. M., and Wilanowski, T. (2014). Loss of Grainy head-like 1 is associated with disruption of the epidermal barrier and squamous cell carcinoma of the skin. PLoS One 9:e89247. doi: 10.1371/journal.pone.0089247
Molè, M. A., Galea, G. L., Rolo, A., Weberling, A., Nychyk, O., De Castro, S. C., et al. (2020). Integrin-mediated focal anchorage drives epithelial zippering during mouse neural tube closure. Dev. Cell 52, 321.e6–334.e6. doi: 10.1016/j.devcel.2020.01.012
Mooney, S. M., Talebian, V., Jolly, M. K., Jia, D., Gromala, M., Levine, H., et al. (2017). The GRHL2/zeb feedback loop—a key axis in the regulation of EMT in breast cancer. J. Cell. Biochem. 118, 2559–2570. doi: 10.1002/jcb.25974
Muñoz, W. A., and Trainor, P. A. (2015). “Chapter One - Neural crest cell evolution: how and when did a neural crest cell become a neural crest cell,” in Current Topics in Developmental Biology Neural Crest and Placodes, ed. P. A. Trainor (London: Academic Press), 3–26. doi: 10.1016/bs.ctdb.2014.11.001
Murry, C. E., and Keller, G. (2008). Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132, 661–680. doi: 10.1016/j.cell.2008.02.008
Narasimha, M., Uv, A., Krejci, A., Brown, N. H., and Bray, S. J. (2008). Grainy head promotes expression of septate junction proteins and influences epithelial morphogenesis. J. Cell. Sci. 121, 747–752. doi: 10.1242/jcs.019422
Nevil, M., Gibson, T. J., Bartolutti, C., Iyengar, A., and Harrison, M. M. (2020). Establishment of chromatin accessibility by the conserved transcription factor Grainy head is developmentally regulated. Development 147:dev185009. doi: 10.1242/dev.185009
Nieto, M. A., Huang, R. Y.-J., Jackson, R. A., and Thiery, J. P. (2016). EMT: 2016. Cell 166, 21–45. doi: 10.1016/j.cell.2016.06.028
Nikolopoulou, E., Galea, G. L., Rolo, A., Greene, N. D. E., and Copp, A. J. (2017). Neural tube closure: cellular, molecular and biomechanical mechanisms. Development 144, 552–566. doi: 10.1242/dev.145904
Nikolopoulou, E., Hirst, C. S., Galea, G., Venturini, C., Moulding, D., Marshall, A. R., et al. (2019). Spinal neural tube closure depends on regulation of surface ectoderm identity and biomechanics by Grhl2. Nat. Commun. 10, 1–17. doi: 10.1038/s41467-019-10164-6
Nishimura, T., and Takeichi, M. (2008). Shroom3-mediated recruitment of Rho kinases to the apical cell junctions regulates epithelial and neuroepithelial planar remodeling. Development 135, 1493–1502. doi: 10.1242/dev.019646
Nora, E. P., Goloborodko, A., Valton, A.-L., Gibcus, J. H., Uebersohn, A., Abdennur, N., et al. (2017). Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930.e22–944.e22. doi: 10.1016/j.cell.2017.05.004
Nora, E. P., Lajoie, B. R., Schulz, E. G., Giorgetti, L., Okamoto, I., Servant, N., et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385. doi: 10.1038/nature11049
Nüsslein-Volhard, C., Wieschaus, E., and Kluding, H. (1984). Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. Wilhelm. Roux’. Archiv. 193, 267–282. doi: 10.1007/BF00848156
Ostrowski, S., Dierick, H. A., and Bejsovec, A. (2002). Genetic control of cuticle formation during embryonic development of Drosophila melanogaster. Genetics 161, 171–182.
Paltoglou, S., Das, R., Townley, S. L., Hickey, T. E., Tarulli, G. A., Coutinho, I., et al. (2017). Novel androgen receptor coregulator GRHL2 exerts both oncogenic and antimetastatic functions in prostate cancer. Cancer Res. 77, 3417–3430. doi: 10.1158/0008-5472.CAN-16-1616
Parelho, V., Hadjur, S., Spivakov, M., Leleu, M., Sauer, S., Gregson, H. C., et al. (2008). Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132, 422–433. doi: 10.1016/j.cell.2008.01.011
Pastushenko, I., Brisebarre, A., Sifrim, A., Fioramonti, M., Revenco, T., Boumahdi, S., et al. (2018). Identification of the tumour transition states occurring during EMT. Nature 556, 463–468. doi: 10.1038/s41586-018-0040-3
Pearson, J. C., Juarez, M. T., Kim, M., Drivenes, Ø., and McGinnis, W. (2009). Multiple transcription factor codes activate epidermal wound-response genes in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 106, 2224–2229. doi: 10.1073/pnas.0810219106
Peters, L. M., Anderson, D. W., Griffith, A. J., Grundfast, K. M., San Agustin, T. B., Madeo, A. C., et al. (2002). Mutation of a transcription factor, TFCP2L3, causes progressive autosomal dominant hearing loss, DFNA28. Hum. Mol. Genet. 11, 2877–2885. doi: 10.1093/hmg/11.23.2877
Petrof, G., Nanda, A., Howden, J., Takeichi, T., McMillan, J. R., Aristodemou, S., et al. (2014). Mutations in GRHL2 result in an autosomal-recessive ectodermal dysplasia syndrome. Am. J. Hum. Genet. 95, 308–314. doi: 10.1016/j.ajhg.2014.08.001
Peyrard-Janvid, M., Leslie, E. J., Kousa, Y. A., Smith, T. L., Dunnwald, M., Magnusson, M., et al. (2014). Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. Am. J. Hum. Genet. 94, 23–32. doi: 10.1016/j.ajhg.2013.11.009
Pifer, P. M., Farris, J. C., Thomas, A. L., Stoilov, P., Denvir, J., Smith, D. M., et al. (2016). Grainyhead-like 2 inhibits the coactivator p300, suppressing tubulogenesis and the epithelial–mesenchymal transition. MBoC 27, 2479–2492. doi: 10.1091/mbc.E16-04-0249
Potier, D., Davie, K., Hulselmans, G., Naval Sanchez, M., Haagen, L., Huynh-Thu, V. A., et al. (2014). Mapping gene regulatory networks in Drosophila eye development by large-scale transcriptome perturbations and motif inference. Cell Rep. 9, 2290–2303. doi: 10.1016/j.celrep.2014.11.038
Pyrgaki, C., Liu, A., and Niswander, L. (2011). Grainyhead-like 2 regulates neural tube closure and adhesion molecule expression during neural fold fusion. Dev. Biol. 353, 38–49. doi: 10.1016/j.ydbio.2011.02.027
Quan, Y., Jin, R., Huang, A., Zhao, H., Feng, B., Zang, L., et al. (2014). Downregulation of GRHL2 inhibits the proliferation of colorectal cancer cells by targeting ZEB1. Cancer Biol. Ther. 15, 878–887. doi: 10.4161/cbt.28877
Quan, Y., Xu, M., Cui, P., Ye, M., Zhuang, B., and Min, Z. (2015). Grainyhead-like 2 promotes tumor growth and is associated with poor prognosis in colorectal cancer. J. Cancer 6, 342–350. doi: 10.7150/jca.10969
Rao, S. S. P., Huang, S.-C., Glenn, St Hilaire, B., Engreitz, J. M., Perez, E. M., et al. (2017). Cohesin loss eliminates all loop domains. Cell 171, 305.e24–320.e24. doi: 10.1016/j.cell.2017.09.026
Rao, S. S. P., Huntley, M. H., Durand, N. C., Stamenova, E. K., Bochkov, I. D., Robinson, J. T., et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. doi: 10.1016/j.cell.2014.11.021
Ray, H. J., and Niswander, L. A. (2016). Grainyhead-like 2 downstream targets act to suppress epithelial-to-mesenchymal transition during neural tube closure. Development 143, 1192–1204. doi: 10.1242/dev.129825
Remeseiro, S., Cuadrado, A., and Losada, A. (2013). Cohesin in development and disease. Development 140, 3715–3718. doi: 10.1242/dev.090605
Rifat, Y., Parekh, V., Wilanowski, T., Hislop, N. R., Auden, A., Ting, S. B., et al. (2010). Regional neural tube closure defined by the Grainy head-like transcription factors. Dev. Biol. 345, 237–245. doi: 10.1016/j.ydbio.2010.07.017
Rios, A. C., Capaldo, B. D., Vaillant, F., Pal, B., van Ineveld, R., Dawson, C. A., et al. (2019). Intraclonal plasticity in mammary tumors revealed through large-scale single-cell resolution 3D imaging. Cancer Cell 35:953. doi: 10.1016/j.ccell.2019.02.010
Rothbart, S. B., and Strahl, B. D. (2014). Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta 1839, 627–643. doi: 10.1016/j.bbagrm.2014.03.001
Ruscetti, M., Quach, B., Dadashian, E. L., Mulholland, D. J., and Wu, H. (2015). Tracking and functional characterization of epithelial-mesenchymal transition and mesenchymal tumor cells during prostate cancer metastasis. Cancer Res. 75, 2749–2759. doi: 10.1158/0008-5472.CAN-14-3476
Santamaria, P. G., Moreno-Bueno, G., Portillo, F., and Cano, A. (2017). EMT: present and future in clinical oncology. Mol. Oncol. 11, 718–738. doi: 10.1002/1878-0261.12091
Sanyal, A., Lajoie, B. R., Jain, G., and Dekker, J. (2012). The long-range interaction landscape of gene promoters. Nature 489, 109–113. doi: 10.1038/nature11279
Satishchandran, A., Ambade, A., Rao, S., Hsueh, Y.-C., Iracheta-Vellve, A., Tornai, D., et al. (2018). MicroRNA 122, regulated by GRLH2, protects livers of mice and patients from ethanol-induced liver disease. Gastroenterology 154, 238.e–252.e. doi: 10.1053/j.gastro.2017.09.022
Senga, K., Mostov, K. E., Mitaka, T., Miyajima, A., and Tanimizu, N. (2012). Grainyhead-like 2 regulates epithelial morphogenesis by establishing functional tight junctions through the organization of a molecular network among claudin3, claudin4, and Rab25. Mol. Biol. Cell 23, 2845–2855. doi: 10.1091/mbc.E12-02-0097
Sexton, T., Yaffe, E., Kenigsberg, E., Bantignies, F., Leblanc, B., Hoichman, M., et al. (2012). Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472. doi: 10.1016/j.cell.2012.01.010
Shang, Z., Chen, D., Wang, Q., Wang, S., Deng, Q., Wu, L., et al. (2018). Single-cell RNA-seq reveals dynamic transcriptome profiling in human early neural differentiation. Gigascience 7:giy117. doi: 10.1093/gigascience/giy117
Shao, C., Ji, C., Wang, X., and Hu, B. (2017). Expression and significance of GRHL2 in esophageal cancer. Onco Targets Ther. 10, 2025–2031. doi: 10.2147/OTT.S128294
Shen, J., Lv, X., and Zhang, L. (2020). GRHL2 acts as an anti-oncogene in bladder cancer by regulating ZEB1 in Epithelial-Mesenchymal Transition (EMT) process. Onco Targets Ther. 13, 2511–2522. doi: 10.2147/OTT.S239120
Simões-Costa, M., and Bronner, M. E. (2013). Insights into neural crest development and evolution from genomic analysis. Genome Res. 23, 1069–1080. doi: 10.1101/gr.157586.113
Sinajon, P., Gofine, T., Ingram, J., and So, J. (2015). Microdeletion 8q22.2-q22.3 in a 40-year-old male. Eur. J. Med. Genet. 58, 569–572. doi: 10.1016/j.ejmg.2015.10.004
Sithanandam, G., and Anderson, L. M. (2008). The ERBB3 receptor in cancer and cancer gene therapy. Cancer Gene Ther. 15, 413–448. doi: 10.1038/cgt.2008.15
Soeller, W. C., Poole, S. J., and Kornberg, T. (1988). In vitro transcription of the Drosophila engrailed gene. Genes Dev. 2, 68–81. doi: 10.1101/gad.2.1.68
Somarelli, J. A., Shetler, S., Jolly, M. K., Wang, X., Dewitt, S. B., Hish, A. J., et al. (2016). Mesenchymal-epithelial transition in sarcomas is controlled by the combinatorial expression of MicroRNA 200s and GRHL2. Mol. Cell. Biol. 36, 2503–2513. doi: 10.1128/MCB.00373-16
Stedman, W., Kang, H., Lin, S., Kissil, J. L., Bartolomei, M. S., and Lieberman, P. M. (2008). Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. Embo J. 27, 654–666. doi: 10.1038/emboj.2008.1
Stemmler, M. P., Hecht, A., and Kemler, R. (2005). E-cadherin intron 2 contains cis-regulatory elements essential for gene expression. Development 132, 965–976. doi: 10.1242/dev.01662
Stigler, J., Çamdere, G. Ö, Koshland, D. E., and Greene, E. C. (2016). Single-molecule imaging reveals a collapsed conformational state for DNA-bound cohesin. Cell Rep. 15, 988–998. doi: 10.1016/j.celrep.2016.04.003
Sundararajan, V., Tan, M., Zea Tan, T., Pang, Q. Y., Ye, J., Chung, V. Y., et al. (2020). SNAI1-driven sequential EMT changes attributed by selective chromatin enrichment of RAD21 and GRHL2. Cancers 12:1140. doi: 10.3390/cancers12051140
Suzuki, M. M., and Bird, A. (2008). DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 9, 465–476. doi: 10.1038/nrg2341
Tanaka, Y., Kanai, F., Tada, M., Tateishi, R., Sanada, M., Nannya, Y., et al. (2008). Gain of GRHL2 is associated with early recurrence of hepatocellular carcinoma. J. Hepatol. 49, 746–757. doi: 10.1016/j.jhep.2008.06.019
Tanimizu, N., and Mitaka, T. (2013). Role of grainyhead-like 2 in the formation of functional tight junctions. Tissue Barr. 1:e23495. doi: 10.4161/tisb.23495
Tanimizu, N., Nakamura, Y., Ichinohe, N., Mizuguchi, T., Hirata, K., and Mitaka, T. (2013). Hepatic biliary epithelial cells acquire epithelial integrity but lose plasticity to differentiate into hepatocytes in vitro during development. J. Cell Sci. 126, 5239–5246. doi: 10.1242/jcs.133082
Taracha, A., Kotarba, G., and Wilanowski, T. (2018). Neglected functions of TFCP2/TFCP2L1/UBP1 transcription factors may offer valuable insights into their mechanisms of action. Int. J. Mol. Sci. 19:2852. doi: 10.3390/ijms19102852
Thiery, J. P., and Sleeman, J. P. (2006). Complex networks orchestrate epithelial–mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 7, 131–142. doi: 10.1038/nrm1835
Ting, S. B., Caddy, J., Hislop, N., Wilanowski, T., Auden, A., Zhao, L., et al. (2005). A homolog of drosophila grainy head is essential for epidermal integrity in mice. Science 308, 411–413. doi: 10.1126/science.1107511
Ting, S. B., Wilanowski, T., Auden, A., Hall, M., Voss, A. K., Thomas, T., et al. (2003). Inositol- and folate-resistant neural tube defects in mice lacking the epithelial-specific factor Grhl-3. Nat. Med. 9, 1513–1519. doi: 10.1038/nm961
Traylor-Knowles, N., Hansen, U., Dubuc, T. Q., Martindale, M. Q., Kaufman, L., and Finnerty, J. R. (2010). The evolutionary diversification of LSF and Grainyhead transcription factors preceded the radiation of basal animal lineages. BMC Evol. Biol. 10:101. doi: 10.1186/1471-2148-10-101
Uv, A. E., Harrison, E. J., and Bray, S. J. (1997). Tissue-specific splicing and functions of the Drosophila transcription factor Grainyhead. Mol. Cell. Biol. 17, 6727–6735. doi: 10.1128/MCB.17.11.6727
Varma, S., Cao, Y., Tagne, J.-B., Lakshminarayanan, M., Li, J., Friedman, T. B., et al. (2012). The transcription factors Grainyhead-like 2 and NK2-Homeobox 1 form a regulatory loop that coordinates lung epithelial cell morphogenesis and differentiation. J. Biol. Chem. 287, 37282–37295. doi: 10.1074/jbc.M112.408401
Venkatesan, K., McManus, H., and Mello, C. (2003). Functional conservation between members of an ancient duplicated transcription factor family, LSF/Grainyhead. Nucl. Acids Res. 31, 4304–4316. doi: 10.1093/nar/gkg644
Vona, B., Nanda, I., Neuner, C., Müller, T., and Haaf, T. (2013). Confirmation of GRHL2 as the gene for the DFNA28 locus. Am. J. Med. Genet. A 161A, 2060–2065. doi: 10.1002/ajmg.a.36017
Walentin, K., Hinze, C., Werth, M., Haase, N., Varma, S., Morell, R., et al. (2015). A Grhl2-dependent gene network controls trophoblast branching morphogenesis. Development 142, 1125–1136. doi: 10.1242/dev.113829
Wang, G., Pan, J., Zhang, L., and Wang, C. (2019). Overexpression of grainyhead-like transcription factor 2 is associated with poor prognosis in human pancreatic carcinoma. Oncol. Lett. 17, 1491–1496. doi: 10.3892/ol.2018.9741
Wang, S., and Samakovlis, C. (2012). Grainy head and its target genes in epithelial morphogenesis and wound healing. Curr. Top. Dev. Biol. 98, 35–63. doi: 10.1016/B978-0-12-386499-4.00002-1
Wang, S., Tsarouhas, V., Xylourgidis, N., Sabri, N., Tiklová, K., Nautiyal, N., et al. (2009). The tyrosine kinase stitcher activates grainy head and epidermal wound healing in Drosophila. Nat. Cell Biol. 11, 890–895. doi: 10.1038/ncb1898
Wang, Y., Sun, Y., Huang, Y., Pan, Y., Jia, Z., Ma, L., et al. (2016). Association study between Van der Woude Syndrome causative gene GRHL3 and nonsyndromic cleft lip with or without cleft palate in a Chinese cohort. Gene 588, 69–73. doi: 10.1016/j.gene.2016.04.045
Watanabe, K., Villarreal-Ponce, A., Sun, P., Salmans, M. L., Fallahi, M., Andersen, B., et al. (2014). Mammary morphogenesis and regeneration require the inhibition of EMT at terminal end buds by Ovol2 transcriptional repressor. Dev. Cell 29, 59–74. doi: 10.1016/j.devcel.2014.03.006
Weischenfeldt, J., Dubash, T., Drainas, A. P., Mardin, B. R., Chen, Y., Stütz, A. M., et al. (2017). Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat. Genet. 49, 65–74. doi: 10.1038/ng.3722
Werner, S., Frey, S., Riethdorf, S., Schulze, C., Alawi, M., Kling, L., et al. (2013). Dual roles of the transcription factor Grainyhead-like 2 (GRHL2) in breast cancer. J. Biol. Chem. 288, 22993–23008. doi: 10.1074/jbc.M113.456293
Werth, M., Walentin, K., Aue, A., Schönheit, J., Wuebken, A., Pode-Shakked, N., et al. (2010). The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development 137, 3835–3845. doi: 10.1242/dev.055483
Wilanowski, T., Caddy, J., Ting, S. B., Hislop, N. R., Cerruti, L., Auden, A., et al. (2008). Perturbed desmosomal cadherin expression in grainy head-like 1-null mice. Embo J. 27, 886–897. doi: 10.1038/emboj.2008.24
Wilanowski, T., Tuckfield, A., Cerruti, L., O’Connell, S., Saint, R., Parekh, V., et al. (2002). A highly conserved novel family of mammalian developmental transcription factors related to Drosophila grainyhead. Mech. Dev. 114, 37–50. doi: 10.1016/S0925-4773(02)00046-1
Wilde, J. J., Petersen, J. R., and Niswander, L. (2014). Genetic, epigenetic, and environmental contributions to neural tube closure. Annu. Rev. Genet. 48, 583–611. doi: 10.1146/annurev-genet-120213-092208
Williams, E. D., Gao, D., Redfern, A., and Thompson, E. W. (2019). Controversies around epithelial–mesenchymal plasticity in cancer metastasis. Nat. Rev. Cancer 19, 716–732. doi: 10.1038/s41568-019-0213-x
Wittkopp, P. J., and Kalay, G. (2012). Cis -regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69. doi: 10.1038/nrg3095
Wu, D., Huang, W., Xu, Z., Li, S., Zhang, J., Chen, X., et al. (2020). Clinical and genetic study of 12 Chinese Han families with nonsyndromic deafness. Mol. Genet. Genomic Med. 8:e1177. doi: 10.1002/mgg3.1177
Xiang, J., Fu, X., Ran, W., and Wang, Z. (2017). Grhl2 reduces invasion and migration through inhibition of TGFβ-induced EMT in gastric cancer. Oncogenesis 6:e284. doi: 10.1038/oncsis.2016.83
Xiang, X., Deng, Z., Zhuang, X., Ju, S., Mu, J., Jiang, H., et al. (2012). Grhl2 determines the epithelial phenotype of breast cancers and promotes tumor progression. PLoS One 7:e50781. doi: 10.1371/journal.pone.0050781
Xu, X., Yang, Q., Jiao, J., He, L., Yu, S., Wang, J., et al. (2016). Genetic variation in POU4F3 and GRHL2 associated with noise-induced hearing loss in Chinese population: a case-control study. Int. J. Environ. Res. Public Health 13:561. doi: 10.3390/ijerph13060561
Yamashita, N., Tokunaga, E., Iimori, M., Inoue, Y., Tanaka, K., Kitao, H., et al. (2018). Epithelial paradox: clinical significance of coexpression of E-cadherin and vimentin with regard to invasion and metastasis of breast cancer. Clin. Breast Cancer 18, e1003–e1009. doi: 10.1016/j.clbc.2018.02.002
Yang, J., Antin, P., Berx, G., Blanpain, C., Brabletz, T., Bronner, M., et al. (2020). Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352. doi: 10.1038/s41580-020-0237-9
Yang, X., Vasudevan, P., Parekh, V., Penev, A., and Cunningham, J. M. (2013). Bridging cancer biology with the clinic: relative expression of a GRHL2-mediated gene-set pair predicts breast cancer metastasis. PLoS One 8:e56195. doi: 10.1371/journal.pone.0056195
Yang, Z., Wu, D., Chen, Y., Min, Z., and Quan, Y. (2019). GRHL2 inhibits colorectal cancer progression and metastasis via oppressing epithelial-mesenchymal transition. Cancer Biol. Ther. 20, 1195–1205. doi: 10.1080/15384047.2019.1599664
Yu, Z., Bhandari, A., Mannik, J., Pham, T., Xu, X., and Andersen, B. (2008). Grainyhead-like factor Get1/Grhl3 regulates formation of the epidermal leading edge during eyelid closure. Dev. Biol. 319, 56–67. doi: 10.1016/j.ydbio.2008.04.001
Yu, Z., Lin, K. K., Bhandari, A., Spencer, J. A., Xu, X., Wang, N., et al. (2006). The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev. Biol. 299, 122–136. doi: 10.1016/j.ydbio.2006.07.015
Yu, Z., Mannik, J., Soto, A., Lin, K. K., and Andersen, B. (2009). The epidermal differentiation-associated Grainyhead gene Get1/Grhl3 also regulates urothelial differentiation. Embo J. 28, 1890–1903. doi: 10.1038/emboj.2009.142
Yuan, M., Wang, J., and Fang, F. (2020). Grainyhead-like genes family may act as novel biomarkers in colon cancer. Onco Targets Ther. 13, 3237–3245. doi: 10.2147/OTT.S242763
Yun, J., Song, S.-H., Kim, H.-P., Han, S.-W., Yi, E. C., and Kim, T.-Y. (2016). Dynamic cohesin-mediated chromatin architecture controls epithelial–mesenchymal plasticity in cancer. Embo Rep. 17, 1343–1359. doi: 10.15252/embr.201541852
Zaret, K. S., and Carroll, J. S. (2011). Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241. doi: 10.1101/gad.176826.111
Zaret, K. S., and Mango, S. (2016). Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr. Opin. Genet. Dev. 37, 76–81. doi: 10.1016/j.gde.2015.12.003
Zhang, X., Liu, Y., Zhang, L., Yang, Z., Yang, L., Wang, X., et al. (2015). Associations of genetic variations in EYA4, GRHL2 and DFNA5 with noise-induced hearing loss in Chinese population: a case- control study. Environ. Health 14:77. doi: 10.1186/s12940-015-0063-2
Zhao, Z., Li, L., Cheng, M., Jing, A.-D., Liu, S.-N., Zhu, S.-M., et al. (2020). Grh signaling regulates epithelium development and ecdysis in Blattella germanica. Insect Sci. doi: 10.1111/1744-7917.12780
Keywords: Grainyhead, Grh, Grainyhead-like 2, GRHL2, epithelial-to-mesenchymal transition, pioneer factor, epithelial differentiation, epigenetics
Citation: Sundararajan V, Pang QY, Choolani M and Huang RY-J (2020) Spotlight on the Granules (Grainyhead-Like Proteins) – From an Evolutionary Conserved Controller of Epithelial Trait to Pioneering the Chromatin Landscape. Front. Mol. Biosci. 7:213. doi: 10.3389/fmolb.2020.00213
Received: 07 June 2020; Accepted: 03 August 2020;
Published: 21 August 2020.
Edited by:
Herbert Levine, Rice University, United StatesReviewed by:
Jason Somarelli, Duke University, United StatesCopyright © 2020 Sundararajan, Pang, Choolani and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruby Yun-Ju Huang, cnVieWh1YW5nQG50dS5lZHUudHc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.