
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 14 August 2020
Sec. Cellular Biochemistry
Volume 7 - 2020 | https://doi.org/10.3389/fmolb.2020.00200
Previous studies from this lab have determined that dedifferentiation of Müller glia occurs after eye drop application of an α7 nicotinic acetylcholine receptor (nAChR) agonist, PNU-282987, to the adult rodent eye. PNU-282987 acts on α7 nAChRs on retinal pigment epithelial cells to stimulate production of Müller-derived progenitor cells (MDPCs) and ultimately lead to neurogenesis. This current study was designed to test the hypothesis that the activation of genes involved in the HB-EGF/Ascl1/Lin28a signaling pathway in Müller glia leads to the genesis of MDPCs. RNA-seq was performed on a Müller glial cell line (rMC-1) following contact with supernatant collected from a retinal pigment epithelial (RPE) cell line treated with PNU-282987. Differentially regulated genes were compared with published literature of Müller glia dedifferentiation that occurs in lower vertebrate regeneration and early mammalian development. HB-EGF was significantly up-regulated by 8 h and expression increased through 12 h. By 48 h, up-regulation of Ascl1 and Lin28a was observed, two genes known to be rapidly induced in dedifferentiating zebrafish Müller glia. Up-regulation of other genes known to be involved in mammalian development and zebrafish regeneration were also observed, as well as down-regulation of some factors necessary for Müller glia cell identity. RNA-seq results were verified using qRT-PCR. Using immunocytochemistry, the presence of markers associated with MDCP identity, Otx2, Nestin, and Vsx2, were found to be expressed in the 48 h treatment group cultures. This study is novel in its demonstration that Müller glia in adult rodents can be induced into regenerative activity by stimulating genes involved in the HB-EGF/Ascl1/Lin28a pathway that leads to MDPCs after introducing conditioned media from PNU-282987 treated RPE. This study furthers our understanding of the mechanism by which Müller glia dedifferentiate in response to PNU-282987 in the adult mammalian retina.
Neurodegenerative diseases of the retina such as glaucoma, macular degeneration, retinal ischemia, and diabetic retinopathy lead to irreversible loss of retinal cells in adult mammals (Velez-Montoya et al., 2013; Weinreb et al., 2014; Duh et al., 2017; Dattilo et al., 2018). However, within the mammalian neuronal retina, neurogenesis does not typically occur after early development (Dhomen et al., 2006; Lamba et al., 2006; Sehgal et al., 2008). Decades of research has been performed in an effort to understand the inability of mammals to regenerate the central nervous system in adults, given the fact that many other animals are able to do so throughout adulthood (Dmitrieva et al., 2007; Gorsuch and Hyde, 2014). It is thought that the mechanism by which central nervous system regeneration occurs exists within mammals but has been inhibited through evolutionary time (Chen and Tonegawa, 1998; Nicholls et al., 1999; Tanaka and Ferretti, 2009; Echeverri, 2020).
Previous work from this lab has shown that administration of PNU-282987, an α7 nAChR-specific agonist (Bodnar et al., 2005; Iwamoto et al., 2013, 2014; Mata et al., 2015) via eye drops onto adult mice and rat eyes induced cell cycle reentry of Müller glial cells, which led to genesis of new mature retinal neurons. New cells first appeared in the inner nuclear layer and then migrated to the other layers (Webster et al., 2017, 2019). This observed inner nuclear migration is consistent with regenerative capacities in developing mice (Tackenberg et al., 2009; Webster et al., 2017). Cell cycle reentry was indicated by the presence of 5-bromo-2′-deoxyuridine (BrdU) positive cells observed after eye drop application of PNU-282987. The source of new BrdU positive cells was found to be Müller glia through lineage tracing with a transgenic TdTomato-Müller glia reporter line (Webster et al., 2017, 2019).
However, Müller glia do not express α7 nAChRs (Dmitrieva et al., 2007; Iwamoto et al., 2013, 2014; Webster et al., 2019). Of the cell types within the neural retina which contain α7 nAChRs, namely bipolar, some amacrine cells and retinal ganglion cells (Webster et al., 2017; Hall et al., 2019), acetylcholine (ACh) released from the starburst amacrine cells does not induce proliferation or interkinetic nuclear migration of Müller glia under physiological conditions. Instead, evidence was provided that PNU-282987 acts on the retinal pigment epithelium (RPE) when applied as eye drops (Webster et al., 2019). RPE cells contain α7 nACh receptors, are known to secret at least 20 different compounds under normal physiological conditions including growth factors and hormones, and are separated from ACh stimulation under normal physiological conditions by the outer limiting membrane (Grierson et al., 1994; Yoshii et al., 2007; Maneu et al., 2010). When PNU-282987 is administered as eyedrops into the eyes of adult mice and rats, it is hypothesized that PNU-282987 binds to alpha7 nAChRs on RPE directly outside the neural retina (Webster et al., 2019) to release signaling molecules onto the end feet of Müller glia, that then asymmetrically divide to produce Müller-derived progenitor cells (MDPCs). Evidence to support this hypothesis was provided by experiments in which RPE-J cell lines in culture were treated with 100 nM PNU-282987 and the resulting intraocular injection of supernatant was sufficient to produce cell cycle reentry of Müller glia in adult rodents in vivo (Webster et al., 2019).
This mechanism of generating new neurons from Müller glia is consistent with regeneration of the retina in adult zebrafish following injury. In Zebrafish, the HB-EGF/Ascl1/Lin28a pathway is found to be crucial in retinal regeneration following injury (Wan et al., 2012; Wan and Goldman, 2016). HB-EGF is one of many ligands which activate EGF receptors, others of which include EGF and TGFα (Wan et al., 2012). HB-EGF is localized to the point of damage in an injured zebrafish retina. It has been demonstrated that HB-EGF activates regeneration-associated gene Ascl1, which has been shown to modulate histone binding involved in dedifferentiation of neuronal cells (Vierbuchen et al., 2010). Ascl1 activates RNA binding protein Lin28a, which is known to inhibit microRNA Let-7 (Yao et al., 2016). Let-7 is involved in inhibition of proliferative abilities in adult Müller glia in zebrafish (Yao et al., 2016). When Let-7 is inhibited, the Wnt pathway is active, leading to dedifferentiation of Müller glia to produce MDPCs (Yao et al., 2016). In this current study, evidence will be presented that these pathways are also involved in the PNU-282987 response in mammalian Müller glia.
This study was designed to explore the gene expression profiles involved in dedifferentiation of adult Müller glia cells to MDPCs in mammals. These studies demonstrate that exposure of cultured Müller glia cells to transwells of RPE cells treated with PNU-282987, will induce genesis of MDPCs. In addition, the results from this study support the hypothesis that the HB-EGF/Ascl1/Lin28a pathway is activated in Müller glia after stimulation of supernatant from PNU-282987 treated RPE adult rat cell lines. The results from transcriptomic analysis were compared to published pathways involved in retina development and/or retinal regeneration. Furthermore, the transcriptomics were used to describe potential interactions involved in the dedifferentiation of mammalian Müller glia. Additionally, it was demonstrated that communication between PNU-282987-activated RPE and Müller glia can induce progenitor-like fate in Müller glia culture, a process that does not typically occurs in adult mammals. Understanding these mechanisms is an important step in learning to control neurogenesis and regeneration in the adult mammalian retina.
RPE cells derived from rats (RPE-J; 4-6 passages) were grown in Dulbecco’s modified Eagle’s medium containing 8% fetal bovine serum, 1% L-Glutamine, and 1% penicillin-streptomycin, using standard cell culture techniques and incubated in 33°C with 5% CO2. Propagation of RPE-J cell lines requires incubation lower than the standard 37°C. Cell culture reagents were obtained from Thermo Fisher Scientific (Waltham, MA, United States). RPE-J cell cultures were treated with dimethylsulfoxide (DMSO) (vehicle control) or PNU-282987 (100 nM; Sigma-Aldrich, St. Louis, MO, United States) for 24 h. Cells were plated in a T75 flask, 35 mm dish for immunocytochemistry or briefly in a 100 mm transwell dish (Corning, Inc., Corning, NY, United States) for co-culture with a cell line of Müller glia (rMC-1) and grown to 80% confluency. PNU-282987 or DMSO was applied to transwell or 35 mm dishes for 1 h. After thorough washing cells to remove any residual PNU-282987, cells were either placed in the co-culture system, or left for 72 h and “activated RPE supernatant” was collected. 100 nM PNU-282987 was used in this study, as previous dose-response studies from this lab have found 100 nM PNU-282987 to elicit the most robust proliferation response (Webster et al., 2017).
Müller glia cells derived from rats (rMC-1; 4-6 passages) were grown in Dulbecco’s modified Eagle’s medium containing 8% fetal bovine serum, 1% L-Glutamine, and 1% penicillin-streptomycin, using standard cell culture techniques and incubated at 37°C with 5% CO2. After 3 days, cells were switched to 33°C with 5% CO2 so they could be co-cultured with RPE cells. The change in temperature had no effect on Müller glia growth and morphology if they were first cultured in 37°C with 5% CO2. Müller glia were co-cultured with RPE-J in transwells at 33°C with 5% CO2. Cell culture reagents were obtained from Thermo Fisher Scientific (Waltham, MA, United States). rMC-1 cell cultures remained untreated and were co-cultured with RPE-J cells that had previously been treated with DMSO (vehicle control) or PNU-282987 (100 nM; Sigma-Aldrich, St. Louis, MO, United States). rMC-1 cells were plated in a T75 flask, 35 mm dish for immunocytochemistry, or briefly in a 100 mm transwell dish (Corning, Inc., Corning, NY, United States) and grown to 80% confluency for coculture with RPE-J. For RNA extraction, Müller glia were exposed to treated RPE via transwells for set times (8, 12, 24, or 48 h). When these co-culture exposure times were completed, cells were collected using 1:250 trypsin-EDTA and gently pelleted using centrifugation. For immunocytochemistry, Müller glia were grown to 30% confluency and treated with activated RPE-J supernatant after treatment with DMSO or PNU-282987 for 0, 8, or 48 h or were incubated in transwells containing treated RPE-J cells for the same amount of time.
Direct-zol RNA Miniprep kit (R2051; Zymo Research) was used for RNA extraction following the protocol provided. RNA from Müller glia cells in culture was collected in RNase-free water and stored at −80°C or on dry ice for overnight shipment to GeneWiz. GeneWiz performed RNA-seq next generation sequencing (NGS) using the Illumina NextSeq 550 high-output platform (Illumina, San Diego, CA, United States). Sequence reads were trimmed using Trimmomatic v.0.36. Trimmed sequence reads were mapped to the Rattus norvegicus Rnor6.0 reference genome available on ENSEMBL. Unique gene hit counts were calculated using featureCounts (subread package v.1.5.2). GeneWiz provided Deseq2 bioinformatics analysis. Pathway analysis was additionally performed in this lab using Reactome (Fabregat et al., 2014).
qRT-PCR was preformed using the SuperScript III Platinum One-Step kit with ROX (11745500, Thermo Fisher) with RNA collected from each of the five timepoints, as described previously. 50 ng of RNA was used for each reaction with a RIN ≥ 7 and set up in 10 μL reaction volumes, as per the manufacturers protocol. IDT and Thermo pre-designed primer/probe assays were used (Table 1) and efficiencies were validated for each primer/probe set to be between 90 and 110% (Supplementary Table S1). Each sample was run in triplicate and each assay was repeated in triplicate. For relative comparison of gene expression, the real-time results were analyzed using the comparative Ct method (2–ΔΔCt) normalized to a Gapdh housekeeping control. Negative controls were performed for each RNA sample and each primer/probe set in which no Reverse Transcriptase and no RNA template were added, respectively.
Müller glia cells were grown to 30% confluency on 35 mm dishes (Thermo Fisher Scientific, Waltham, MA, United States) and treated for 0, 8, and 48 h with activated RPE supernatant or in transwells containing PNU-282987 treated RPE cells. After treatment, Müller glia cells were fixed using 4% paraformaldehyde (PFA) for 10 min at room temperature. PFA was then removed, and cells were washed 3x with 1X Phosphate Buffer Saline (PBS). To increase permeabilization, 0.5% Triton X-100 in 1X Tris Buffer Saline (TBSTr) was added and cells were incubated at room temperature for 15 min. TBSTr was removed and replaced with TBSTr with 10% Normal Goat Serum (NGS) for 4 h to minimize non-specific binding. Primary antibodies were diluted at a ratio of 1:300 in 1X TBS + 0.05% Triton X-100 with 1% NGS and incubated overnight at 4°C. Primary antibodies (Table 2) were then removed and cells were washed 3x with 1X TBS + 0.05% Triton X-100 for 10 min. Fluorescent dye labeled secondary antibodies (Table 2) were added for 2 h at room temperature in the dark, followed by Hoescht counterstain (1:10,000) for 10 min at room temperature. Cells were again washed 3x with 1X TBS. Fluorescently labeled cells were viewed using Nikon C2 + scanning confocal microscopy. Antibodies against OTX2, VSX2, and Nestin were used to identify the presence of retinal progenitor cells, and Vimentin was used to label Müller glial cytoskeleton as a positive control. All reagents were obtained from Thermo Fisher Scientific (Waltham, MA, United States) and antibodies were obtained from Abcam®, Inc. (Cambridge, MA, United States). In control studies, experiments were conducted to determine the specificity of all antibodies used. Antibodies used in this study originated from rabbit, mouse, goat, donkey, or sheep. Antibody processing was performed on untreated Müller glia rMC-1 cells as well as in experimentally treated cells of the same type to determine if positive labeling occurred in untreated controls. The number of immunostained cells in individual culture wells were normalized to the total number of cultured cells in each culture well that were counterstained with Hoescht. N’s of 5-10 were collected for each antibody used in this study and counts were averaged. In control studies, cells were processed with the primary antibody omitted. No significant epifluorescence was observed if the primary antibody was omitted.
Single comparisons involved a Student’s paired t-test for all culture experiments. For experiments requiring multiple comparisons, one-way ANOVA with post hoc analysis using the Holm-Bonferroni correction was performed. A Wald test was performed to generated P-values and log2 fold changes of RNA-seq results. Results were considered significant if P ≤ 0.05 and log2 fold changes were > 1 or < −1. Statistical analysis for RNA-seq was provided by GeneWiz. Heat map samples of significant genes were determined by systematic sampling using the equiprobability method to in order to accurately represent total sDEGs at each timepoint.
Bioinformatics performed by GeneWiz provided significant differentially expressed genes (sDEGs) and Volcano plots using the Wald test to determine DEGs with a P-value ≤ 0.05 and log2 fold changes > 1 or < −1. Heat maps were generated using Morpheus and Violin Plots using Plotly. Reactome was used to determine biological processes associated with pathways of identified genetic markers. Binary Alignment Sequences (BAMs) were generated by GeneWiz using STAR Aligner v.2.5.2b. All other tables and graphs were generated using Excel.
Previous studies from this lab found that supernatant collected from RPE cells treated with PNU-282987 (activated RPE) was sufficient to produce cell cycle reentry of the adult mammalian Müller glia in the retina when the supernatant was injected into the vitreal chamber of adult rodents in vivo. These same studies demonstrated that BrdU positive Müller glia dedifferentiate into MDPCs (Webster et al., 2017, 2019). To identify changes in Müller glia gene expression after exposure to activated RPE supernatant, RNA-seq was performed (GeneWiz Inc., NJ, United States) from total RNA extracted from Müller glia. rMC-1 Müller glial cell lines (Kerafast ENW001) were exposed to activated RPE via 75 mm Transwell (Corning 3419) dishes (Figure 1). RPE cells were cultured for 8, 12, 24, and 48 h in normal media (8% FBS DMEM with L-glutamine and Penicillin/Streptomyocin, Gibco 11971025) after exposure to 100 nM PNU-282987 for 24 h, followed by thorough rinsing with PBS. Previously, RPE culture was treated with PNU for 24 h, washed, and subsequently left in normal media for 72 h before injection in the mouse eye. This timeline induced robust BrdU incorporation (Webster et al., 2019). Subsequent experiments analyzed 12, 24, and 48 h cultures, which all showed some level of BrdU incorporation in the mouse retina (unpublished). This information was used to select the appropriate time points in order to see early changes in Müller glia transcript as well as changes leading up to the maximum response attainable while maintaining healthy cell cultures. In some 75 mm transwell dishes, RPE-J cells were treated with 1% DMSO for 24 h instead of 100 nM PNU-282987 to act as a vehicle control. Figure 1 summarizes the procedures used to perform these experiments.
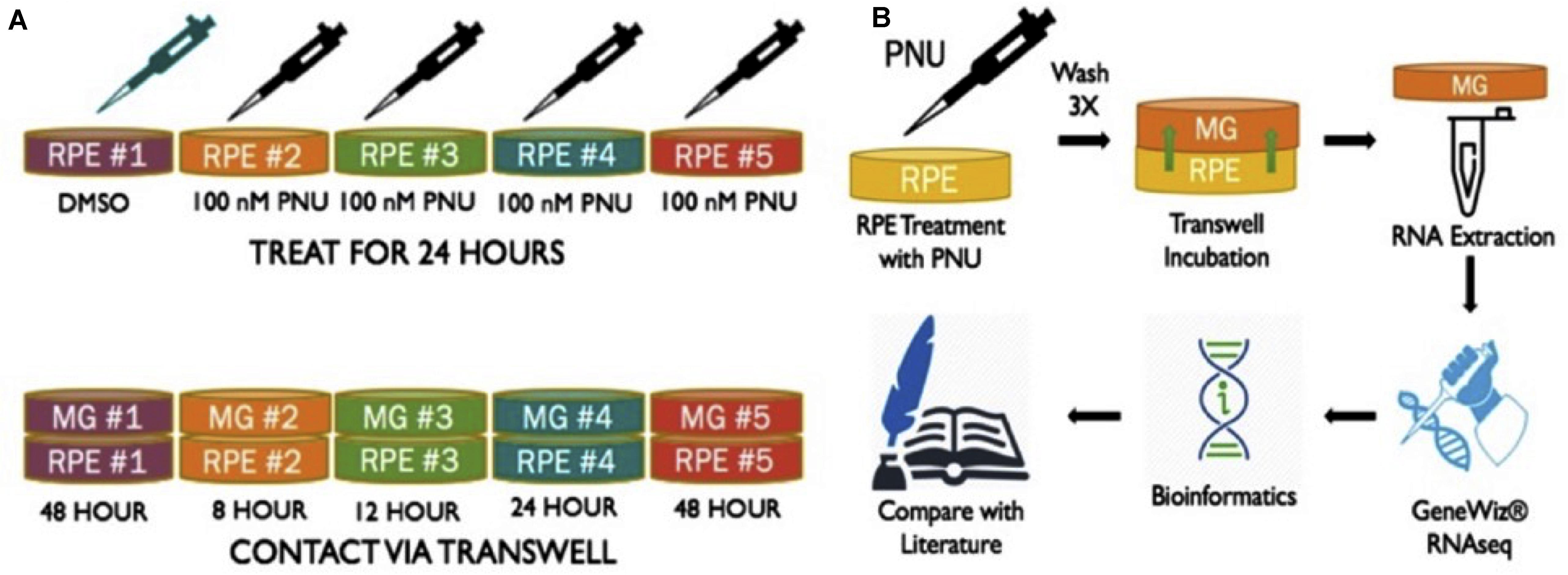
Figure 1. Schematic of methods performed to obtain RNA from Müller glia exposed to PNU-282987 treated RPE cells. (A) (1) RPE-J treated for 24 h with DMSO as a vehicle control. (2-5) RPE-J cells treated for 24 h with PNU-282987. PNU-282987 was removed and thoroughly washed away with PBS before being replace with normal media. (B) After PNU-282987 treatment and thorough rinsing, RPE were exposed to rMC-1 cells via transwells and allowed to incubate for 8, 12, 24, and 48 h. RNA was extracted from Müller glia and sent to GeneWiz for RNA sequencing. Some bioinformatics was performed to determine log2 fold changes of identified RNA sequences compared to DMSO controls.
In order to investigate transcriptional changes in Müller glia following the treatment with PNU-282987 described in Figure 1, RNA-seq analysis of a Müller glial cell line was performed. Each sample was sequenced by GeneWiz using the Illumina NextSeq 550 high-output platform (Illumina, San Diego, CA, United States). Log2 fold expression changes were obtained in approximately 15,000 identified genes total for the four timepoints. 1050 significantly differentially expressed genes (sDEGs) were identified, and were compared to literature identifying genes of interest known to be involved in dedifferentiation of Müller glia in teleost fish upon retinal injury (Hitchcock et al., 1996; Fischer and Reh, 2001; Ramachandran and Zhao, 2011; Meyers et al., 2012; Wan et al., 2012; Centanin and Wittbrodt, 2014; Goldman, 2014; Gorsuch and Hyde, 2014; Yao et al., 2016), as well as those involved in early development of the retina in vertebrate mammals such as mouse and rat (Bhattacharjee and Sanyal, 1975; Wang et al., 2001; Dhomen et al., 2006; Locker et al., 2006; Nelson et al., 2009; Phillips et al., 2011; Wright et al., 2015; Xia and Ahmad, 2016). Teleost fish, specifically zebrafish, were chosen as a comparative focal point in this study based on the zebrafish’s ability to regenerate the retina in adulthood following injury, the extensive availability of research published on this topic, and the finding that PNU-282987 induced neurogenesis in rats and mice are generated from Muller glia (Webster et al., 2017, 2019). For comparison, 100 molecules of interest per timepoint were chosen as a focus for this study, illustrated in the heat map in Figure 2A. The 100 genes per timepoint chosen to represent the sDEG data were identified by systematic sampling using the equiprobability method to accurately represent the distribution of log2 fold changes identified as significant at the timepoint.
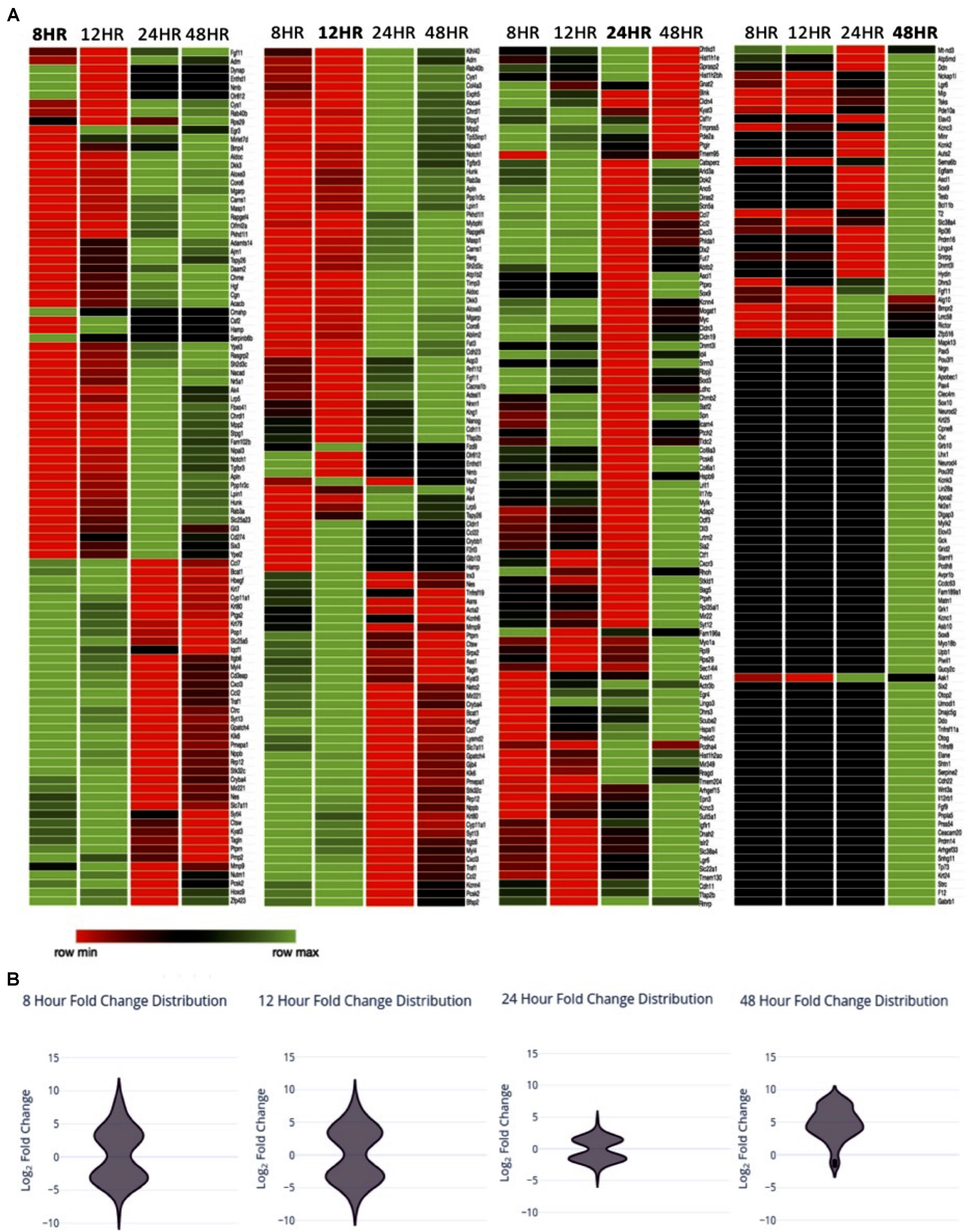
Figure 2. Expression changes over time (A) Heat map with of sDEGs from Müller glia RNA expression changes 8, 12, 24, and 48 h post-exposure to PNU-282987 treated RPE cells, compared to 48-h DMSO control conditions. sDEGs represented include a linear sample of total sDEGs per timepoint. Created using Morpheus. (B) Violin distribution plots of sDEGs per timepoint at the timepoint represented.
In Table 3, the number of sDEG at each collection time point is summarized as well as the direction of expression for the genes at each collection time point and any change in gene expression at later time points. For example, at the 8-h time point, 409 DEGs were found to be statistically significant (P-value < 0.05, log2 fold change > 1 or < −1) compared to DMSO control. At 8 h, 240 of the 409 DEGs were down-regulated compared to control (log2 fold change between −1.81 and −8.01), while 169 (41.3%) were up-regulated (log2 fold change between 1.78 and 8.56). The majority of sDEGs that were down-regulated at 8 h reversed and were up-regulated by 24 and 48 h post-treatment. Conversely, the majority of sDEGs that were up-regulated at 8 h were down-regulated by 24 and 48 h post-treatment. As seen in Table 3, the reversal of gene expression at later time points occurred for most genes, indicating these genes are expressed for a relatively narrow window of time and are then actively reversed. This “on/off” type of behavior for the genes is a typical pattern for genes involved in growth, development and dedifferentiation.
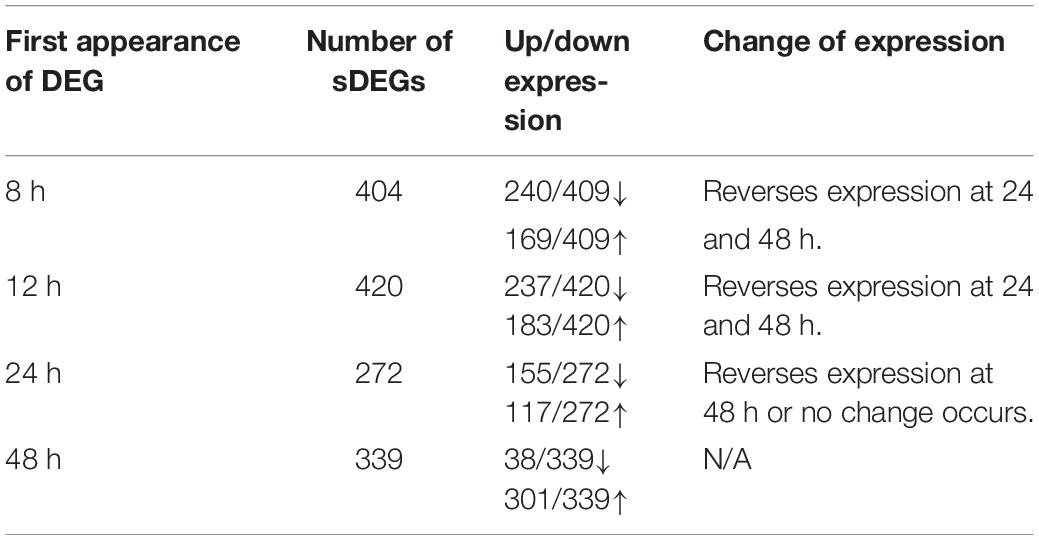
Table 3. Changes of significant differential gene expression (sDEG) that occurred when Muller glia cells were cultured in transwells with PNU-282987 treated RPE cells for various amount of time.
As seen in Table 3, most of the sDEGs first expressed 48 h post-treatment compared to the DMSO control were up-regulated and had much larger fold changes than genes expressed at other time points (log2 fold change between 1.0 and 8.71). These genes were specifically analyzed for their potential contribution toward dedifferentiation of Muller glia into MDPCs.
In total, there was a total of 1050 unique sDEGs when all timepoints were considered. This list was analyzed using Reactome pathway analysis in order to understand which biological processes in Rattus norvegicus these transcripts are known to be involved in Figure 3A. 120 genes involved in signal transduction were identified. Specifically, genes involved in signaling by TGF-β, Notch, Wnt, MAPK, Hippo, and Hedgehog pathways were identified in the sDEGs analyzed. These pathways are all known to be involved in retinal regeneration in vertebrates like zebrafish and chick (Hayes et al., 2007; Tackenberg et al., 2009; Meyers et al., 2012; Wan and Goldman, 2016; Yao et al., 2016). As well as signal transduction, genes involved in developmental pathways, metabolism and immunology were identified. The majority of genes involved in development pertain to pathways in nervous system development and nervous system function. Genes involved in early development were particularly relevant for this study as fully differentiated Müller glia do not typically express early neuronal development genes. 14 genes involved in mitosis were identified, as well as genes involved in G1/S DNA damage checkpoints. Though the analysis was performed on RNA extracted from mitotically active cell lines, normalization of values compared to the control condition accounted for the genes involved in immortalization.
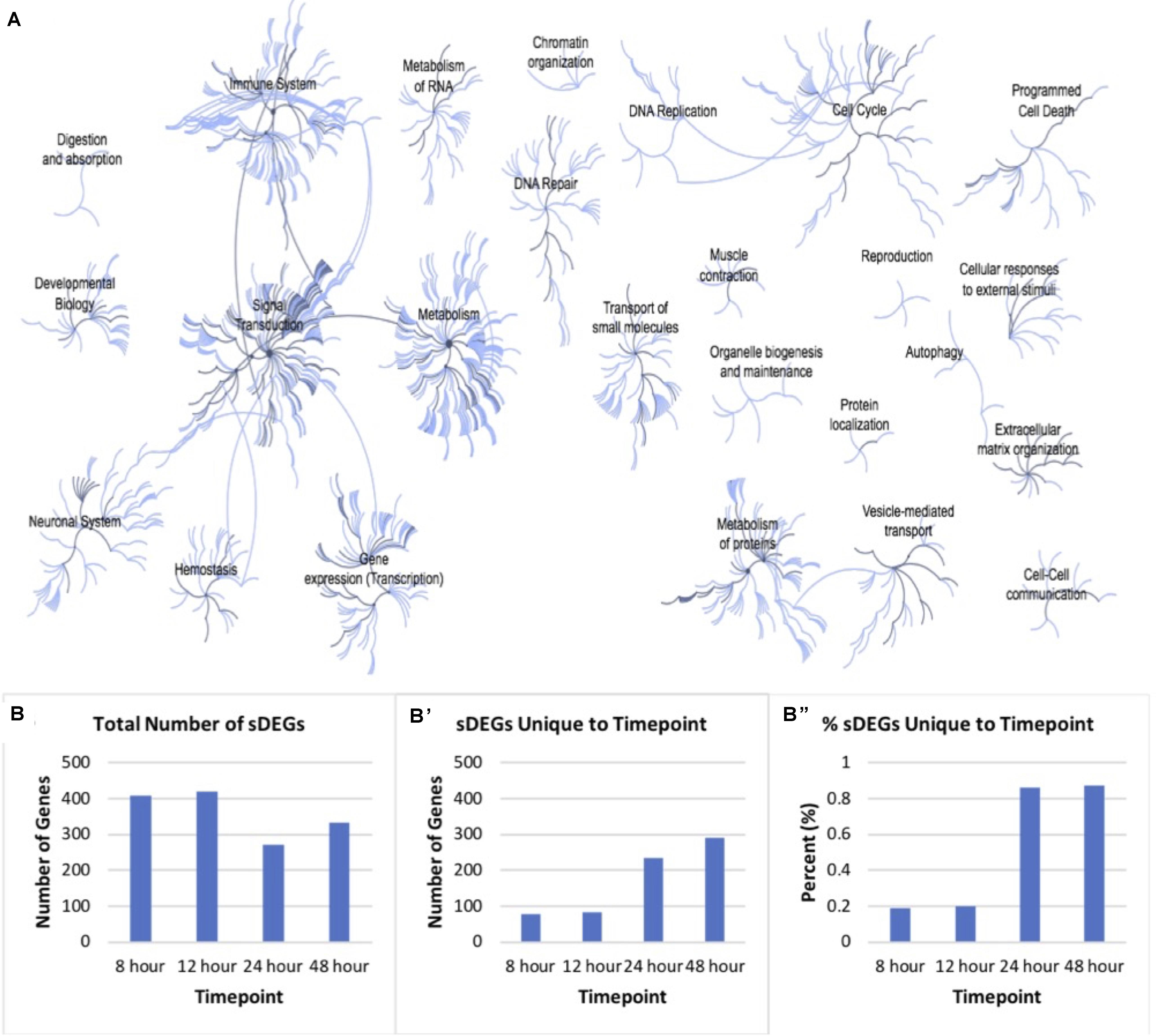
Figure 3. Pathway analysis of differentially expressed genes. (A) Reactome map display of the distribution of biological processes in which significant differentially expressed genes identified via mRNA sequencing of timepoints listed in Figure 2 are involved in rat. (B) Distribution of total significantly expressed genes 8, 12, 24, and 48-h post-exposure to PNU-282987 treated RPE supernatant compared to DMSO control. (B′) sDEGs which were only found to be significant at the corresponding timepoint. (B′′) Percent of sDEGs which were unique to a timepoint compared to the total number at that timepoint.
Although genes involved in development and differentiation was the focus of this study, genes involved in metabolism were also identified that are known to be involved in metabolic processes such as protein synthesis, regulation of insulin-like growth factor (IGF) transport, RNA processing, integration of energy metabolism, chromatin modification, gene silencing, and protein localization. In addition, a large immune response was also detected through Reactome pathway analysis, including significant expression changes of inflammatory genes like macrophage expressed 1 (mpeg1) and cytokine expressed Csf1R (Huynh et al., 2013; Mitchell et al., 2018). In zebrafish, acute inflammation occurs in the damaged tissue and is known to promote dedifferentiation and proliferation in response to injury in the retina (Silva et al., 2020). However, the change of gene expression pertaining to the immune response was not the focus of this study.
Figure 3B shows the number of sDEGs at each timepoint (B), as well as the number (B′) and percent (B′′) of those genes which were unique at each timepoint. For example, the 8 h timepoint shared 80.7% sDEGs of all with the 12 h timepoint, and only 2.2% with the 24 h timepoint and 3.9% with the 48 h timepoint. Overall, the 24 and 48 h timepoints had a more unique transcript than the other conditions. In vivo, these two timepoints produced the maximal number of BrdU + cells following PNU-282987 treatment and was the time when retinal progenitor markers began to be expressed (Webster et al., 2017, 2019). The patterns of gene expression changes in this data are consistent with what we would expect in cells that are proliferating (early) and then dedifferentiating (late).
To understand how the up- and down-regulation of these genes may contribute to dedifferentiation of the Müller glia, known pathways of retinal regeneration in zebrafish and chick, as well as those known to be involved in development of the mammalian retina were compared to the results obtained from RNA-seq results (Table 4). Table 4A shows sDEGs at the 8 h timepoint that are involved in genes associated with differentiation and proliferation. Up-regulation of β-catenin, a co-transcriptional regulator in the Wnt/β-Catenin pathway of proliferation (Gallina et al., 2016), HB-EGF, a ligand known to initiate retinal regeneration in zebrafish (Wan et al., 2012), and Mmp9, an initiator of Wnt signaling through the Hedgehog pathway of retinal regeneration in chick (Kaur et al., 2018) was observed. Down-regulation of Bmp4, a transcription factor associated with adult Müller cell identity as well as down-regulation of Wnt inhibitors such as transcription factor Dkk3, MicroRNA Let7d and hedgehog transcription factor Gli3 were observed at 8 h as well (Nakamura et al., 2007; Kaur et al., 2018).
Table 4B shows sDEGs that occurred at 12 h. The pattern of expression at 12 h was very similar to the 8 h timepoint, with up-regulation of β-catenin, HB-EGF, and Mmp9, as well as down-regulation of Dkk3. In addition, Fzd9, a frizzled receptor which Wnt ligands bind (Dijksterhuis et al., 2014), was up-regulated at 12 h, which could point to an increase in Wnt signaling at this time. In addition, early retinal progenitor markers Vsx2 and Nestin (Wright et al., 2010; Lee et al., 2012), were also upregulated at 12 h.
Table 4C shows sDEGs that occurred at 24 h and illustrates expression of genes for the first time. Specifically Ascl1 and the co-transcriptional regulator Lin28a, which are involved downstream of HB-EGF, activates Wnt signaling in zebrafish regeneration (Wan et al., 2012). In addition, the retinal progenitor marker Sox9 is expressed for the first time after 24 h of treatment (Poché et al., 2008).
Table 4D shows sDEGs that occurred at 48 h. There is up-regulation of Ascl1 again as well as Lin28a, Ascl1 recruiters Pou3f2 and Myt1 (Chanda et al., 2014), and the retinal progenitor marker Sox9. However, new up-regulation occurs as well, such as fibroblast growth factors Fgf9 and Fgf11. Fgf genes are involved in the regeneration response in zebrafish and chick retina (Fischer and Reh, 2001; Wan and Goldman, 2017) and have been shown to be regulators of Wnt signaling (Tang et al., 2019). These data highlight the conservation of gene expression profiles commonly observed in other models of retinal regeneration to the PNU-282987 activated mammalian system and demonstrate when expression occurs. Understanding the timing of these gene expression allowed us to hypothesis what signaling pathways are involved in Müller glia cell proliferation after exposure to PNU-282987 treated RPE cells.
Volcano plots (Figure 4) represent a distribution of the log2 fold changes seen at each timepoint. The black points represent differentially expressed genes, whereas blue points are those which were significantly up-regulated and red were significantly down-regulated. The volcano plots for 8 and 12 h show a consistent distribution of both significantly up- and down-regulated genes. The plot for 24 h shows fewer significant genes overall, with many genes showing a similarity in expression. The 48 h plot shows a striking disparity between up- and down-regulated differentially expressed genes, with the majority being up-regulated and to a higher degree than what is seen at other timepoints. This data overall points to up-regulation of genes early on which are related to proliferation and dedifferentiation of Müller glia and down-regulation of genes known to prevent proliferation. In later timepoints, up-regulation of genes associated with asymmetrical division of Müller glia that produce cells with a retinal progenitor-like fate are seen.
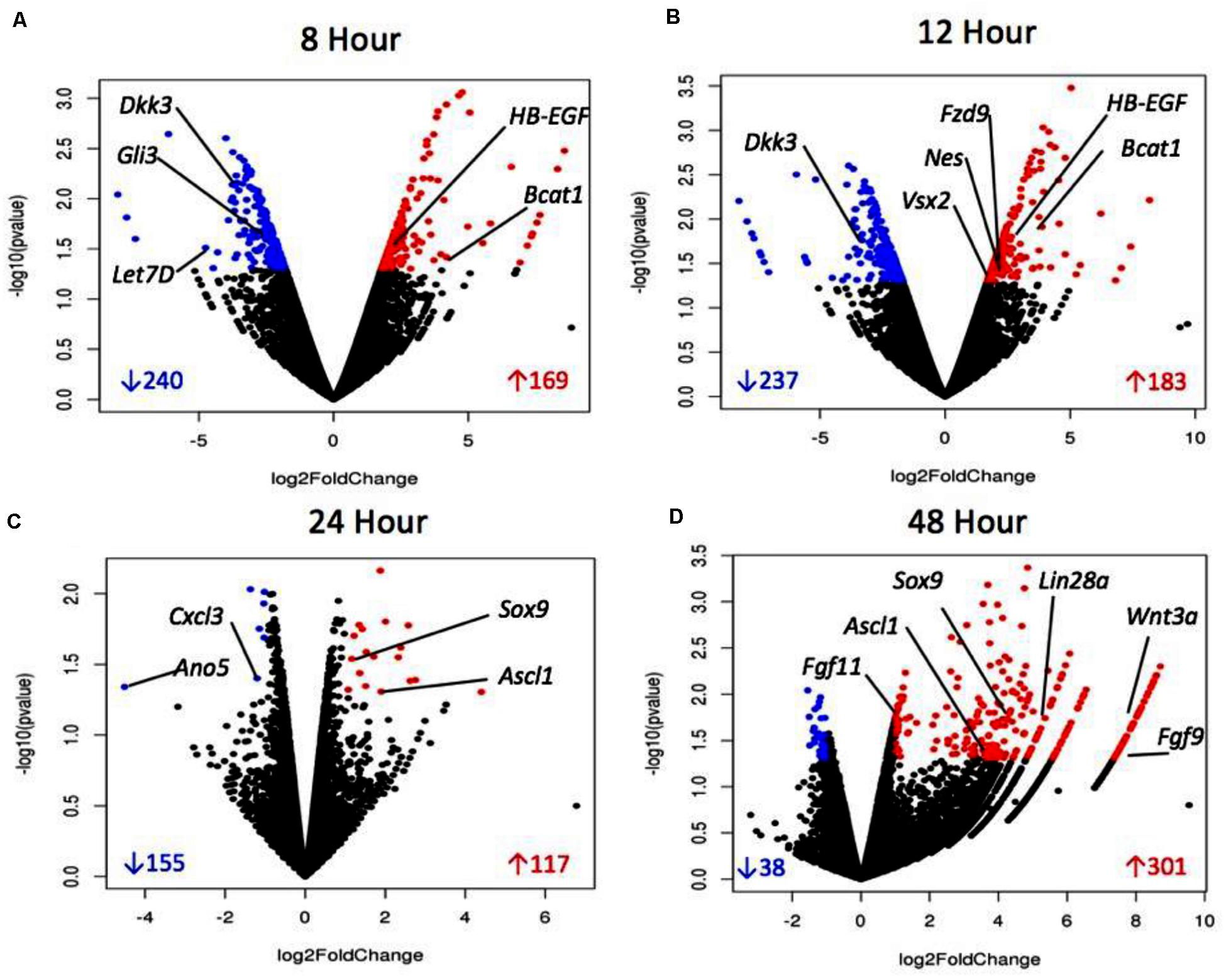
Figure 4. Volcano plots of sDEGs at 8 h vs. control (A), 12 h vs. control (B), 24 h vs. control, (C) and 48 h vs. control (D). Plots demonstrate all DEGs as dots based on their fold change, with down-regulated sDEGS in blue and up-regulated sDEGs in red.
qRT-PCR was performed to validate RNA sequencing results for several genes of interest thought to be involved in dedifferentiation pathways. RNA-seq data and qRT-PCR data were displayed for factors at differing treatment timepoints (Figure 5). BMP4, a protein coding gene in the TGFβ family, known to be down-regulated in chick MDPCs (Todd et al., 2017), is found to be down-regulated at 8 h in RNA seq results as well as all other timepoints in qRT-PCR. HB-EGF, found to be up-regulated at 8 and 12 h in RNA seq results, was confirmed to follow the same trend via qRT-PCR, as well as qRT-PCR confirmation of Ascl1 and Lin28a up-regulation at 48 and 24/48 h, respectively. These finding are consistent with the pathway of retinal regeneration in zebrafish in which HB-EGF activation at the site of injury leads to the activation of Ascl1 to then recruit Lin28a (Ramachandran and Zhao, 2011; Wan et al., 2012; Wan and Goldman, 2016). Bcat1 and Fzd9, up-regulated at 8 and 12 h, and Fgf9/Fgf11, up-regulated at 48 h in RNA seq, followed the same trend in qRT-PCR verification. Fzd9 is a receptor which binds Wnt ligands (Meyers et al., 2012; Wan et al., 2012; Tappeiner et al., 2016). FGF9 and FGF11 up-regulated in Müller-derived progenitor cells in other vertebrates (Fischer and Reh, 2001; Hochmann et al., 2012; Wan and Goldman, 2016). Mmp9 and Gli3, components of the sonic hedgehog (Shh) pathway that are up- and down-regulated respectively early in zebrafish regeneration (Wan and Goldman, 2016; Kaur et al., 2018) were up- and down-regulated at 8 and 12 h in RNA-seq, confirmed in qRT-PCR. Finally, retinal progenitor marker Sox9 (Young, 1985; Webster et al., 2017) was up-regulated at 24 and 24 h, which is confirmed in qRT-PCR results. This data provides evidence to support findings in the RNA-seq results.
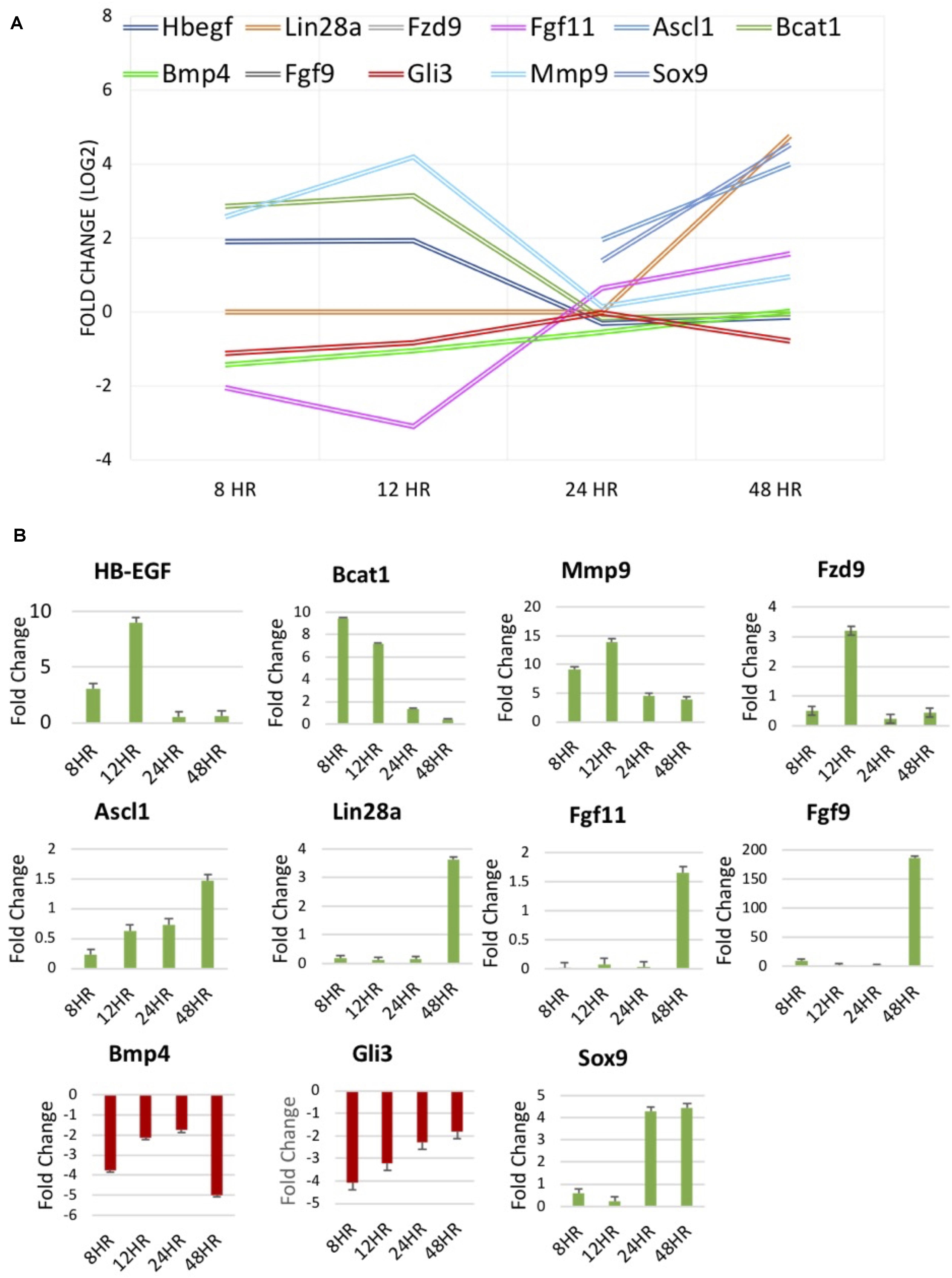
Figure 5. (A) RNA-seq results of molecules of interest identified in literature to be involved in regeneration of the zebrafish and chick retina and mammalian retinal development. (B) Fold change [2– –ΔΔCT] results of qPCR verification of molecules of interest in (A).
To provide evidence that MDPCs were produced as a result of signaling molecules released from the RPE, IHC was performed. To identify progenitor cell markers in Müller glia, rMC-1 cultures were exposed to activated RPE supernatant for 48 h in some studies, or allowed to incubate in transwells containing RPE cells that had previously been treated with PNU-282987. Both methods produced statistically similar changes in Müller glia. Following incubation with the supernatant or when with treated RPE cells in transwells, cells were labeled with antibodies against vimentin, VSX2, nestin and OTX2. Figure 6 illustrates confocal images obtained when the supernatant from PNU-282987 treated RPE cells were added to culture dishes of Müller glia, as the resolution and clarity of images obtained in culture dishes was higher than images of Müller glia attached to the mesh on transwells (Figure 6). Vimentin, a Müller glia marker found in the unaltered adult retina (Tackenberg et al., 2009), was used as a control and labeled all cells in both the treated or untreated conditions (Figures 6A,A′). Nestin, expressed in neural precursor cells in the developing retina (Božanić et al., 2006), labeled Müller glia when introduced to the supernatant from PNU282987 treated RPE cells to a significantly greater degree than that found in control cells (Figures 6B,B′). VSX2, found in prenatal eye cultures in neurospheres with neurogenic potential (Wright et al., 2010), immunostained significantly more Müller glia than in control cells (Figures 6C,C′). Finally, OTX2, another retinal progenitor marker widely accepted to represent stem-like qualities in the developing retina (Schmitt et al., 2009), immunostained significantly more treated Müller glia than in untreated cells (Figures 6D,D′). Müller cells with positive staining for retinal progenitor markers provide evidence that Müller glia dedifferentiate to a progenitor-like fate when exposed to signaling molecules released from PNU-282987 treated RPE cells. This result indicates that PNU-282987 induces a response in Müller glia that is not typically seen in adult mammalian neurons.
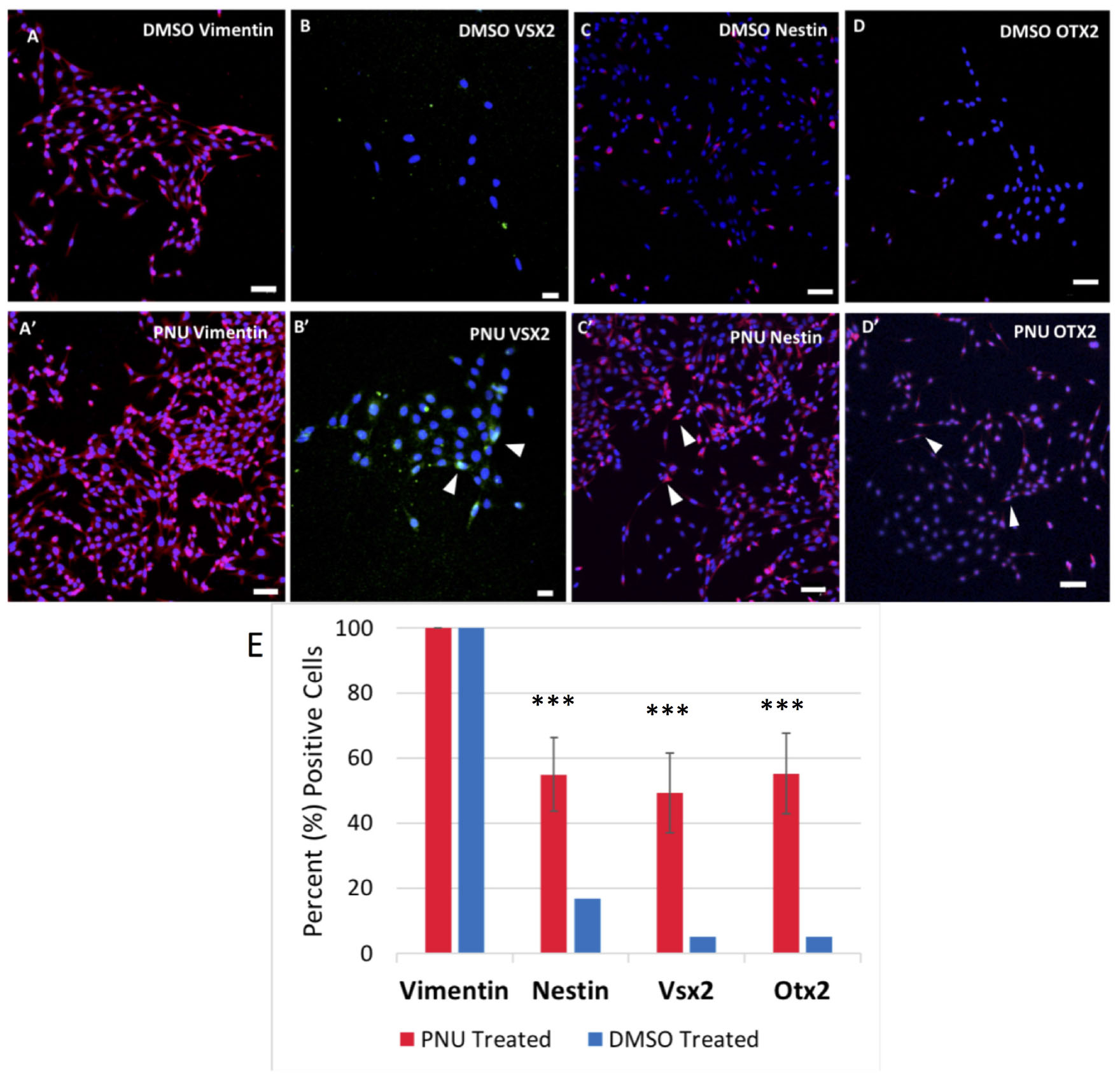
Figure 6. Immunocytochemistry of Müller glia after 48-h exposure to PNU-282987 treated RPE supernatant. Scale bars represent 100 microns. (A,A′) Müller glial cytoskeletal component, Vimentin, used as a control in DMSO and PNU-282987 treated rMC-1. (B,B′) Retinal progenitor marker VSX2 is identified in treated Müller glia. Arrows indicate VSX2 + single nuclei. (C,C′) Retinal progenitor marker Nestin labels cytoskeleton of treated Müller glia. Arrows indicate single Nestin + cells. (D,D′) Transcription factor OTX2 labels both nucleus and cytoskeleton of treated Müller glia. Arrows indicate OTX2 + cells. Scale bar represents 100 microns. (E) Quantification of cells positive for progenitor markers. A one-way ANOVA was performed to determine statistical significance. All three progenitor cell markers were found to be statistically significant when compared to DMSO controls (***P-value < 0.001) in PNU-282987 treated cells.
Based on the transcriptomics and IHC marker data, a summary of sDEGS involved in the dedifferentiation of Müller glia to retinal progenitor cells following activated RPE supernatant treatment is outlined in Figure 7. This figure was constructed using the expression patterns as well as the timing of expression. For example, HB-EGF, an EGFR ligand (8, 12 h up-regulation), is known to work upstream of Ascl1 (24, 48 h up-regulation), a basic helix-loop-helix transcription factor, which then activates RNA binding protein Lin28a (48 h up-regulation) to allow for dedifferentiation in the zebrafish retina (Wan and Goldman, 2016). MicroRNA Let-7 (Down-regulation at 8 and 12 h) is shown to inhibit dedifferentiation in the Müller glia in the adult zebrafish prior to injury (Wan et al., 2012; Kaur et al., 2018). Mmp9 (8, 12-h up-regulation) regulates Ascl1 and is found early in dedifferentiation (Locker et al., 2006). Also, Mmp9 along with Gli3 are Shh pathway regulators of Let-7 in Zebrafish regeneration (Wan and Goldman, 2016; Kaur et al., 2018) Fzd9 (12 h up-regulation) is a Wnt receptor known to be activated in HB-EGF/Ascl1 signaling (Ramachandran and Zhao, 2011). Finally, B cat1 signaling (Up-regulated at 8 and 12 h, later in qRT-PCR data) is both required in dedifferentiation to and maintenance of retinal progenitors produced in zebrafish (Meyers et al., 2012). β-Catenin also binds to Lin28a promotor to activate transcription (Yao et al., 2016). Wnt/β-catenin signaling is the pathway that drives the production of Müller-derived progenitor cells in chick (Gallina et al., 2016) as well as vertebrate eye development (Fujimura, 2016). The retinal progenitor marker, Sox9 (up-regulated at 24 and 48 h) as well as progenitor markers Nestin, OTX2 and VSX2, identified in IHC, provide evidence of a MDPC-like fate.
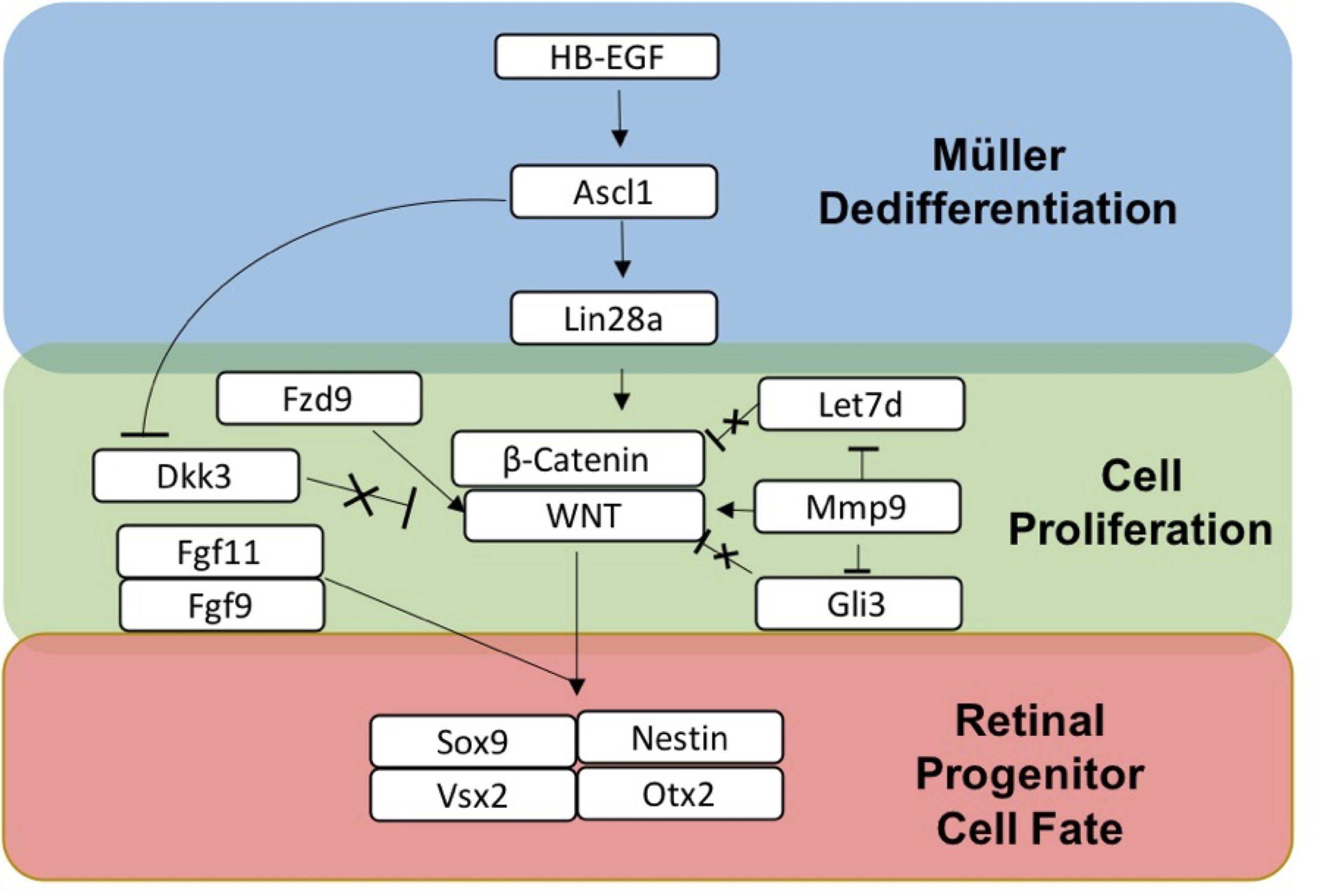
Figure 7. Summary of gene expression associated with dedifferentiation of Müller glia to Müller-derived progenitor cells in adult mammals. HB-EGF is up-regulated when Müller glia are exposed to RPE supernatant treated with PNU-282987.
In this study, we show that activated RPE supernatant initiates transcriptional changes in Müller glia that lead to gene expression profiles resembling that of dedifferentiated Müller glia and MDPCs including activation of genes involved the HB-EGF/Ascl1/Lin28a pathway. Activation of these genes were induced through signaling molecules released from PNU-282987 treated RPE cells, which has not been demonstrated before. Heat maps were generated comparing sDEG fold changes at experimental timepoints vs. control. Comparison of significant fold changes at 8/12 h reveals a distinct pattern of on/off behavior by sDEGs which were turned on at 8/12 h at off at 24/48, and vice versa. A vast change occurs at 48 h in which the majority of sDEGs are turned on with higher fold changes than seen at any other timepoint. Most sDEGs are unique to this timepoint. This pattern is similar to the change in gene expression pattern observed in the regenerating zebrafish and chick retina following injury. In zebrafish, proliferation inhibitors are turned off and signal transduction genes are turned on temporarily to allow for proliferation of Müller glia. Expression changes to upregulation of retinal progenitor markers later on in newly derived MDPCs (Fischer and Reh, 2001; Wan and Goldman, 2016; Todd et al., 2017).
Reactome analysis revealed significant changes in gene expression of sDEGs throughout the study in signal transduction pathways including Wnt/β-Catenin, TGF-β, Notch, MAPK, Hippo, and Hedgehog. Genes from these pathways were highlighted in Table 4 and in the volcano plots shown in Figure 4. Up-regulation of MAPK ligand HB-EGF was found at 8 and 12 h, followed by up-regulation of Ascl1 at 24 and 48 h and Lin28a at 48 h. HB-EGF is up-regulated at the injury site in Zebrafish, leading to activation of Ascl1 followed by Lin28a which stimulates the formation of multipotent Müller derived progenitor cells in the retina (Wan et al., 2012; Wan and Goldman, 2016). HB-EGF activation leads to Wnt/β-Catenin signaling in zebrafish, the pathway which allows for cell proliferation in the Zebrafish retina (Gallina et al., 2016; Wan and Goldman, 2016). HB-EGF is also known to act upstream of β-Catenin signaling (Wan et al., 2012). The β-catenin/Wnt pathway is essential in the production of Müller-derived progenitor cells in chick (Gallina et al., 2016) and in eye development (Fujimura, 2016). Bcat1 is up-regulated at 8 and 12 h according to sequencing data, as well as 24 and 48 h in qRT-PCR data. Dkk3, an inhibitor of the Wnt pathway found in adult mammalian Müller glia (Nakamura et al., 2007), is down-regulated at both 8 and 12 h in our data. Fzd9 is a Wnt receptor known to be activated in the HB-EGF/Ascl1 pathway by Wnt ligands (Ramachandran and Zhao, 2011; Gallina et al., 2016) and is up-regulated at 12 h with qRT-PCR confirmation. Wnt signaling occurs in the development of retinal progenitor cells (Meyers et al., 2012). We see down-regulation at 8 and 12 h of markers associated with fully differentiated Müller glia (Todd et al., 2017) (Bmp4), as well as inhibitors of cell cycle re-entry (Hes4, Hes6, Gli3, Let-7). Fibroblast growth factors are associated with proliferation following injury in the zebrafish retina (McCabe et al., 2006). FGF9 and FGF11 are released by retinal cells in Xenopus to initiate dedifferentiation and reprogramming (Fukui and Henry, 2012; Meyers et al., 2012). FGFR signaling also promotes retinal regeneration in chick and zebrafish (Fujimura, 2016; Gallina et al., 2016). Let-7 microRNA, a repressor of retinal regeneration-associated genes in zebrafish such as Ascl1, oct4 and myc, has been shown to be inhibited during retinal regeneration in zebrafish (Ramachandran et al., 2010). Let-7 is a regulator of sonic hedgehog (Shh) signaling (Locker et al., 2006) and is a marker for differentiation from retinal progenitor to adult cell in the developing retina (Xia and Ahmad, 2016). Shh component Mmp9 is found in injured retinal cells prior to progenitor cell markers and regulates Ascl1. Shh component Gli3 is a negative regulator of Ascl1 and is reduced in Müller glia progenitor cells (Kaur et al., 2018). Under normal physiological conditions, Gli3 physically interacts with the Ascl1 promotor to inhibit its expression (Kaur et al., 2018). Down-regulation of this gene allows the Ascl1 pathway to move forward and Müller glia to dedifferentiate into multipotent progenitor cells. These signal transduction genes are consistent with regeneration models in other vertebrates and suggest that the same genes and pathways found in animals that can regenerate can lead to dedifferentiation in adult mammals if they are activated.
qRT-PCR was able to verify expression and temporal trends of genes identified through RNA-seq. RNA-seq is highly accurate but represents n = 1. By validating with qRT-PCR, we were able to produce three biological replicates of 11 genes that verify the trend in expression seen with RNA-seq. This increased our sample size to n = 4 and provided evidence pertaining to the accuracy of results obtained in the RNA-seq data set.
The use of immortalized cell lines in this study could be viewed as a limitation for these studies. Immortalization is by definition an alteration of the genome of cells away from normal physiological conditions. However, results were normalized to the control condition. Therefore, all changes in gene expression were relative to the control in both cell lines. A standard quantity of RNA was used as well to normalize the comparative responses. Future studies will repeat these experiments in primary RPE and Müller glial cells to validate these findings.
Another limitation of this study is that only gene expression up to 48 h was analyzed as experiments were designed to look at early gene expression that led to MGPCs. In future experiments, it will be important to look at the RNA seq data for expression of the progenitor markers beyond 48 h. This data is absent or missing in the current study.
IHC experiments analyzing activated RPE treated Müller glia demonstrated that the conclusion of transcriptomics changes produce cells with a RPC-like fate. Previous studies in Zebrafish have demonstrated many pathways involved in Müller glial dedifferentiation and retinal regeneration that also appear to be involved in PNU-282987-induced dedifferentiation of Müller glia in adult mice. Given that Müller glia proliferate before production of MDPCs in other models, it is expected that progenitor cell markers would appear at later timepoints. At 48 h post-treatment in culture, we see Nestin, VSX2, and OTX2 expression. This provides evidence that these conditions can produce progenitor cell fate in culture. In mammals, damage typically induces proliferation of Müller glia in the form of reactive gliosis, which is not a beneficial outcome as no new neurons are generated. However, this study demonstrates the unique finding that PNU-282987 is able to generate MDPCs in adult mammals in the absence of injury, an outcome with more functional significance to the animals.
Given the data presented above, it was hypothesized that the sDEGs identified in this study are involved pathways of dedifferentiation within the adult mammalian retina following exposure to PNU-282987. The consistency of the data with injury-induced regeneration of the zebrafish retina points to the possibility that the mechanism may be partially conserved. Much work is left to be done to develop a defined pathway of proliferation to MDPCs in adult mammals in response to PNU-282987. However, this study provides a frame of reference to more fully understand the mechanism by which this phenomenon has been shown to occur.
The regenerative response to injury in the retina is common in lower vertebrates, such as teleost fish and chick (Stenkamp, 2015). Currently, the adult mammalian neuronal retina is not thought to, under normal physiological conditions, have the ability to restore loss of function caused by neuronal cell death past early development. However, this lab has found a robust neurogenic response in the adult mammalian retina after eye drop application of PNU-282987 (Webster et al., 2017, 2019) without inducing injury. Other research in this area has been met with success at a much more limited scale and requires external manipulations to generate few retinal precursors limited to a specific fate (Yao et al., 2018). The genetic profile and IHC of the MD generated as a result of PNU-282987 application indicate that multiple types of neurons can be generated. Also, these results demonstrate that the pathways involved in adult mice Müller glia dedifferentiation have many similarities with the regenerative capabilities of Müller glia in zebrafish. However, molecules released from PNU-282987 treated RPE cells can stimulate these pathways in adult mammals and bypass typical inhibition of mammalian Müller glia. The results from this study suggest that the inability of adult mammals to induce neurogenesis is not due to unique signaling cascades or genes in the mammalian system. Instead, it is possible that the mammalian Müller glia lack the signaling required to initiate cell division and should direct future research in this field. Ultimately, examining the mechanism by which this occurs will be useful in understanding which factors limit the mammalian system from regeneration in adulthood and how this can be manipulated to treat neurodegenerative diseases of various body systems, including the retina.
The mechanism proposed as a result of this study could lead to the ability to control dedifferentiation in an effort to restore vision in retinal neurodegenerative disease models. At present, PNU-282987’s greatest effects would be in the retina, as it has been reported to inhibit a potassium channel in the heart if given systemically (Duris et al., 2011; Deng et al., 2019). However, understanding the mechanism of PNU-282987’s effect in the retina could allow for manipulation of these pathways to bring about a regenerative response. Ultimately, it is crucial to address the function of the retina after α7 nAChR agonist treatment. Even if new adult retinal neurons are generated after PNU-282987 treatment, one must ask if new cells make appropriate functional connections? Do they make synapses and function to affect visual processing? Can regeneration of new neurons reverse the functional effects of retinal disease, injury or age? One way to address these issues is to analyze electroretinogram (ERG) activity in the rodent eye before and after α7 nAChR agonist treatment with and without injury. These studies are currently underway.
The use of specific α7 nAChR agonists could potentially lead to important clinical treatments for other neurodegenerative diseases in the eye as well as in the brain where α7 nACh receptors have been shown to be involved; such as Alzheimer’s, Parkinson’s and Huntington’s diseases (Foucault-Fruchard and Antier, 2017). α7 nAChRs within the subventricular zone (SVZ), lining the lateral ventricle of the brain, have been found to be involved in neurogenesis in the SVZ when activated (Mudo et al., 2007; Narla et al., 2013; Shabani et al., 2020). As a result, understanding of the mechanism associated with cell cycle reentry of Müller glia in the retina after PNU-282987 treatment is crucial. Understanding the genes and signaling pathways that are involved in this dedifferentiation response may allow future treatments that directly stimulate proliferative pathways or genes without needing PNU-282987. For instance, the pathways identified in this study are known to act in similar ways in other areas of the mammalian central nervous system. HB-EGF is widely expressed in the brain, including the hippocampus and cerebral cortex, and is considered to multiple roles in the developing nervous system (Oyagi and Hara, 2012). Canonical Wnt/β-catenin signaling plays a big role in in tissue patterning in regulating rostral-caudal and medial-lateral patterning in the developing cortex (Noelanders and Vleminckx, 2017). Flushing out the mechanism by which neurogenesis is induced in the retina using PNU-282987 could allow for an understanding of inducible neurogenesis in adult mammals in the CNS systems.
The datasets generated for this study can be found in the NCBI GEO accessions GSE151477.
The animal study was reviewed and approved by IACUC Western Michigan University.
MS was responsible for generating all the data and figures for this study. SW was responsible for helping with analysis and intellectual qPCR and bioinformatic input as well as assisting with generating figures and editing writing. MW was responsible for helping to outline the project and for bioinformatic analysis. CL was the manager for this project and assisted with editing of the writing and analysis. All authors contributed to the article and approved the submitted version.
This study was funded by an NIH award to CL (#EY027970).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
A special thanks to Dr. Jeremy Duncan for his expert consultation and access to qRT-PCR equipment as well as to Dr. Pamela Hoppe for her editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2020.00200/full#supplementary-material
TABLE S1 | Summary of efficiency curves of all primer/probes used in qRT-PCR. All RNA Integrity Number (RIN) values for RNA used were greater than 6.0 and all efficiencies for primer/probes were between 90 and 110%.
Bhattacharjee, J., and Sanyal, S. (1975). Developmental origin and early differentiation of retinal Müller cells in mice. J. Anat. 120, 367–372.
Bodnar, A., Cortes-Burgos, L., Cook, K., Dinh, D., Groppi, V., Hajos, M., et al. (2005). Discovery and structure-activity relationship of quinuclidine benzamides as agonists of α7 nicotinic acetylcholine receptors. J. Med. Chem. 48, 905–908. doi: 10.1021/jm049363q
Božanić, D., Bocina, I., and Saraga-Babic, M. (2006). Involvement of cytoskelatal proteins and growth factor receptors during development of the human eye. Anat. Embry 211, 367–377. doi: 10.1007/s00429-006-0087-z
Centanin, L., and Wittbrodt, J. (2014). Retinal neurogenesis. Development 141, 241–244. doi: 10.1242/dev.083642
Chanda, S., Ang, C. E., Davila, J., Pak, C., Mall, M., Lee, Q. Y., et al. (2014). Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Rep. 3, 282–296. doi: 10.1016/j.stemcr.2014.05.020
Chen, D. F., and Tonegawa, S. (1998). Why do mature CNS neurons of mammals fail to re-establish connections following injury ± functions of Bcl-2. Cell Death Diff. 5, 816–822.
Dattilo, M., Newman, N., and Biousse, V. (2018). Acute retinal arterial ischemia. Ann. Eye Sci. 3:28. doi: 10.21037/aes.2018.05.04
Deng, Y., Guo, S.-L., Wei, B., Gao, X.-C., Zhou, Y.-C., and Li, J.-Q. (2019). Activation of nicotinic acetylcholine alpha7 receptor attenuates progression of monocrotaline-induced pulmonary hypertension in rats by downregulating the NLRP3 inflammasome. Front. Pharmacol. 10:128. doi: 10.3389/fphar.2019.00128
Dhomen, N., Balaggan, K., Pearson, R., Bainbridge, J., Levine, E., Ali, R., et al. (2006). Absence of chx10 causes neural progenitors to persist in the adult retina. IOVS 47, 386–396. doi: 10.1167/iovs.05-0428
Dijksterhuis, J. P., Peterson, J., and Schulte, G. (2014). WNT/Frizzled signaling; receptor -ligand selectivity with focus on FZD-G protein signaling and its physiological relevance: IUPHAR Review 3. Br. J. Pharmacol. 171, 1195–1209. doi: 10.1111/bph.12364
Dmitrieva, N., Strang, C., and Keyser, K. (2007). Expression of alpha 7 nicotinic acetylcholine receptors by bipolar, amacrine, and ganglion cells of the rabbit retina. J. Histochem. Cytochem. 55, 461–476. doi: 10.1369/jhc.6A7116.2006
Duh, E., Sun, J., and Stitt, A. (2017). Diabetic retinopathy: current understanding, mechanisms, and treatment strategies. JCI Insight 2:e93751. doi: 10.1172/jci.insight.93751
Duris, K., Manaenko, A., Suzuki, H., Rolland, W. B., Krafft, P. R., and Zhang, J. H. (2011). Alpha7 nicotinic acetylcholine receptor agonist, PNU-282987, attenuates early brain injury in a perforation model of subarachnoid hemorrhage in rats. Stroke 42, 3560–3566. doi: 10.1161/STROKEAHA.111.619965
Echeverri, K. (2020). The various routes to functional regeneration in the central nervous system. Comm. Biol. 3:47. doi: 10.1038/s42003-020-0773-z
Fabregat, A., Jupe, S., Matthews, L., Sidiropoulos, K., Gillespie, M., Garapati, P., et al. (2014). The reactome pathway knowledgebase. Nuc. Acids Res. 123, 141–150. doi: 10.1093/nar/gkt1102
Fischer, A., and Reh, T. (2001). Muller glia are a potential source of neural regeneration in the postnatal chicken retina. Nat. Neurosci. 4, 247–252. doi: 10.1038/85090
Foucault-Fruchard, L., and Antier, D. (2017). Therapeutic potential of α7 nicotinic receptor agonists to regulate neuroinflammation in neurodegenerative diseases. Neur. Regen. Res. 12, 1418–1421. doi: 10.4103/1673-5374.215244
Fujimura, N. (2016). Wnt/β-Catenin signaling in vertebrate eye development. Front. Cell and Dev. Biol. 4:138. doi: 10.3389/fcell.2016.00138
Fukui, L., and Henry, J. (2012). FGF signaling is required for lens regeneration in xenopus laevis. Biol. Bull. 221, 137–145. doi: 10.1086/BBLv221n1p137
Gallina, D., Palazzo, I., Steffenson, L., Todd, L., and Fischer, A. (2016). Wnt/β-catenin-signaling and the formation of Müller glia-derived progenitors in the chick retina. Dev. Neurobiol. 76, 983–1002. doi: 10.1002/dneu.22370
Goldman, D. (2014). Müller glia cell reprogramming and retina regeneration. Nat. Rev. Neurosci. 15, 431–442. doi: 10.1038/nrn3723
Gorsuch, R., and Hyde, D. (2014). Regulation of Müller glial dependent neuronal regeneration in the damaged adult zebrafish retina. Exper. Eye Res. 123, 131–140. doi: 10.1016/j.exer.2013.07.012
Grierson, I., Hiscott, P., Hogg, P., Robe, H., Mazure, A., and Larkin, G. (1994). Development, repair and regeneration of the retinal pigment epithelium. Eye 8, 255–262. doi: 10.1038/eye.1994.54
Hall, L. M., Hellmer, C. B., Koehler, C. C., and Ichinose, T. (2019). Bipolar cell type-specific expression and conductance of alpha-7 nicotinic acetylcholine receptors in the mouse retina. IOVS 60, 1353–1361. doi: 10.1167/iovs.18.2573
Hayes, S., Nelson, B., Buckingham, B., and Reh, T. (2007). Notch signaling regulates regeneration in the avian retina. Dev. Biol. 312, 300–311. doi: 10.1016/j.ydbio.2007.09.046
Hitchcock, P., Macdonald, R., VanDeRyt, J., and Wilson, S. (1996). Antibodies against Pax6 immunostain amacrine and ganglion cells and neuronal progenitors, but not rod precursors, in the normal and regenerating retina of the goldfish. J. Neurobio. 29, 399–413.
Hochmann, S., Kaslin, J., Hans, S., Weber, A., Machate, A., Geffarth, M., et al. (2012). Fgf signaling is required for photoreceptor maintenance in the adult zebrafish retina. PLoS One 7:e30365. doi: 10.1371/journal.pone.0030365
Huynh, D., Akcora, D., Malaterre, J., Chan, C. K., Dai, X. M., Bertoncello, I., et al. (2013). CSF-1 receptor-dependent colon development, homeostasis and inflammatory stress response. PLoS One 8:e56951. doi: 10.1371/journal.pone.0056951
Iwamoto, K., Birkholz, P., Schipper, A., Mata, D., Linn, D., and Linn, C. (2014). A nicotinic acetylcholine receptor agonist prevents loss of retinal ganglion cells in a glaucoma model. IOVS 55, 1078–1087. doi: 10.1167/iovs.13-12688
Iwamoto, K., Mata, D., Linn, D., and Linn, C. (2013). Neuroprotection of rat retinal ganglion cells mediated through alpha7 nicotinic acetylcholine receptors. Neuroscience 237, 184–198. doi: 10.1016/j.neuroscience.2013.02.003
Kaur, S., Gupta, S., Chaudhary, M., Khursheed, M. A., Mitra, S., Kurup, A. J., et al. (2018). Let-7 MicroRNA-mediated regulation of shh signaling and the gene regulatory network is essential for retina regeneration. Cell Rep. 23, 1409–1923. doi: 10.1016/j.celrep.2018.04.002
Lamba, D. A., Karl, M. O., Ware, C. B., and Reh, T. A. (2006). Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc. Nat. Acad. Sci. U.S.A. 103, 12769–12774. doi: 10.1073/pnas.0601990103
Lee, J. H., Park, H. S., Shin, J. M., Chun, M. H., and Oh, S. J. (2012). Nestin expressing progenitor cells during establishment of the neural retina and its vasculature. Anat. Cell Biol. 45, 38–46. doi: 10.5115/acb.2012.45.1.38
Locker, M. A., Agathocleous, M., Amato, M. A., Parain, K., Harris, W. A., and Perron, M. (2006). Hedgehog signaling and the retina: insights into the mechanisms controlling the proliferative properties of neural precursors. Genes Dev. 20, 3036–3048. doi: 10.1101/gad.391106
Maneu, V., Gerona, G., Fernandez, L., Cuenca, N., and Lax, P. (2010). Evidence of alpha7 nicotinic acetylcholine receptor expression in retinal pigment epithelial cells. Vis. Neurosci. 27, 139–147. doi: 10.1017/S0952523810000246
Mata, D., Linn, D., and Linn, C. (2015). Retinal ganglion cell neuroprotection induced by activation of alpha7 nicotinic acetylcholine receptors. Neuropharmacology 99, 337–346. doi: 10.1016/j.neuropharm.2015.07.036
McCabe, K., McGuire, C., and Reh, T. (2006). Pea3 expression is regulated by FGF signaling in developing retina. Dev. Dyn. 235, 327–335. doi: 10.1002/dvdy.20631
Meyers, J., Hu, L., Moses, A., Kaboli, K., Papandrea, A., and Raymond, P. A. (2012). β-catenin/Wnt signaling controls progenitor fate in the developing and regenerating zebrafish retina. Neur. Dev. 7:30. doi: 10.1186/1749-8104-7-30
Mitchell, D. M., Lovel, A. G., and Stenkamp, D. L. (2018). Dynamic changes in microglial and macrophage characteristics during degeneration and regeneration of the zebrafish retina. J. Neuroinflamm. 15:163. doi: 10.1186/s12974-018-1185-6
Mudo, G., Belluardo, N., and Fuxe, K. (2007). Nicotinic receptor agonists as neuroprotective/neurotrophic drugs. Neural Transmission. Prog. Mol. Mechan. 114, 135–147. doi: 10.1007/s00702-006-0561
Nakamura, R. E., Hunter, D. D., Yi, H., Brunken, W. J., and Hackam, A. S. (2007). Identification of two novel activities of the Wnt signaling regulator Dickkopf 3 and characterization of its expression in the mouse retina. BMC Cell Biol. 8:52. doi: 10.1186/1471-2121-8-52
Narla, S., Klejbor, I., Birkaya, B., Lee, Y. W., Morys, J., Stachowiak, E. K., et al. (2013). Alpha7 nicotinic receptor agonist reactivates neurogenesis in adult brain. Biochem. Pharmacol. 86, 1099–1104. doi: 10.1016/j.bcp.2013.07.028
Nelson, B., Hartman, B., Ray, C., Hayashi, T., Bermingham-McDonogh, O., and Reh, T. (2009). Acheate-scute like 1 (Ascl1) is required for normal delta-like (Dll) gene expression and notch signaling during retinal development. Dev. Dyn. 238, 2163–2178. doi: 10.1002/dvdy.21848
Nicholls, J., Adams, W., Eugenin, J., Geiser, R., Lepre, M., Luque, J., et al. (1999). Why does the central nervous system not regenerate after injury? Sur. Ophth 43, 136–141. doi: 10.1016/s0039-6257(99)00008-9
Noelanders, R., and Vleminckx, K. (2017). How Wnt signaling builds the brain: bridging development and disease. Neurosci. 23, 314–329. doi: 10.1177/1073858416667270
Oyagi, A., and Hara, H. (2012). Essential roles of heparin-binding epidermal growth factor-like growth factor in the brain. CNS Neurosci. Ther. 18, 803–810. doi: 10.1111/j.1755-5949.2012.00371.x
Phillips, J. B., Blanco-Sanchez, B., Lentz, J. J., Tallafuss, A., Khanobdee, K., Sampath, S., et al. (2011). Harmonin (Ush1c) is required in zebrafish Muller glial cells for photoreceptor synaptic development and function. Dis. Mod. Mech. 4, 786–800. doi: 10.1242/dmm.006429
Poché, R. A., Furuta, Y., Chaboissier, M.-C., Schedl, A., and Behringer, R. R. (2008). Sox9 is expressed in mouse multipotent retinal progenitor cells and functions in Muller glial cell development. J. Comp. Neurol. 510, 237–250. doi: 10.1002/cne.21746
Ramachandran, R., Fausett, B., and Goldman, D. (2010). Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat. Cell Biol. 12, 1101–1107. doi: 10.1038/ncb2115
Ramachandran, R., and Zhao, X. F. (2011). Ascl1a/Dkk/β-catenin signaling pathway is necessary and glycogen synthase kinase-3β inhibition is sufficient for zebrafish retina regeneration. PNAS 108, 15858–15863. doi: 10.1073/pnas.1107220108
Schmitt, S., Aftab, U., Jiang, C., Redenti, S., Klassen, H., Miljan, E., et al. (2009). Molecular characterization of human retinal progenitor cells. IOVS 50, 5901–5908. doi: 10.1167/iovs.08-3067
Sehgal, R., Karcavich, R., Carlson, S., and Belecky-Adams, T. L. (2008). Ectopic Pax2 expression in chick ventral optic cup phenocopies loss of Pax2 expression. Dev. Biol. 319, 23–33. doi: 10.1016/j.ydbio.2008.03.041
Shabani, Z., Mahmoudi, J., Farajdokht, F., and Sadish-Eteghad, S. (2020). An overview of nicotinic cholinergic system signaling in neurogenesis. Arch. Med. Res. 51, 287–296. doi: 10.1016/j.arcmed.2020.03.014
Silva, N. J., Nagashima, M., Li, J., Kakuk-Atkins, L., Ashrafzadeh, M., Hyde, D. R., et al. (2020). Inflammation and matrix metalloproteinase 9 (Mmp-9) regulate photoreceptor regeneration in adult zebrafish. Glia 68, 1445–1465. doi: 10.1002/glia.23792
Stenkamp, D. (2015). Development of the vertebrate eye and retina. Prog. Mol. Biol. Trans. Sci. 134, 397–414. doi: 10.1016/bs.pmbts.2015.06.006
Tackenberg, M., Tucker, B., Swift, J., Jiang, C., Redenti, S., Greenberg, K., et al. (2009). Müller cell activation, proliferation and migration following laser injury. Mol. Vis. 15, 1886–1896.
Tanaka, E., and Ferretti, P. (2009). Considering the evolution of regeneration in the central nervous system. Nat. Rev. Neurosci. 10, 713–723. doi: 10.1038/nrn2707
Tang, D., He, Y., Li, W., and Li, H. (2019). Wnt/beta-catenin interacts with the FGF pathway to promote proliferation and regenerative cell proliferation in the zebrafish lateral line neuromast. Exp. Mol. Med. 51, 1–16. doi: 10.1038/s12276-019-0247-x
Tappeiner, C., Maurer, E., Sallin, P., Bise, T., Enzmann, V., and Tschopp, M. (2016). Inhibition of the TGFβ Pathway enhances retinal regeneration in adult Zebrafish. PLoS One 11:e0167073. doi: 10.1371/journal.pone.0167073
Todd, L., Palazzo, I., Squires, N., Mendonca, N., and Fischer, A. J. (2017). BMP- and TGFβ-signaling regulate the formation of Müller glia-derived progenitor cells in the avian retina. Glia 65, 1640–1655. doi: 10.1002/glia.23185
Velez-Montoya, R., Oliver, S., Olson, J., Fine, S., Mandava, N., and Quiroz-Mercado, H. (2013). Current knowledge and trends in age-related macular degeneration: today’s and future treatments. Retina 33, 1487–1502.
Vierbuchen, T., Ostermeier, A., Pang, Z., Kokubu, Y., Südhof, T., and Wernig, M. (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041. doi: 10.1038/nature08797
Wan, J., and Goldman, D. (2016). Retina regeneration in zebrafish. Curr. Opin. Gen. Dev. 40, 41–47. doi: 10.1016/j.gde.2016.05.009
Wan, J., and Goldman, D. (2017). Opposing actions of Fgf8a on notch signaling distinguish two Muller glial cell populations that contribute to retina growth and regeneration. Cell Rep. 19, 849–862. doi: 10.1016/j.celrep.2017.04.009
Wan, J., Ramachandran, R., and Goldman, D. (2012). HB-EGF is necessary and sufficient for Müller glia dedifferentiation and retina regeneration. Dev. Cell 22, 334–347. doi: 10.1016/j.devcel.2011.11.020
Wang, S., Kim, B., Ding, K., Wang, H., Sun, D., Johnson, R., et al. (2001). Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 15, 24–29. doi: 10.1101/gad.855301
Webster, M., Barnett, B., Stanchfield, M., Paris, J., Webster, S., Cooley-Themm, C., et al. (2019). Stimulation of retinal pigment epithelium with an α7 nAChR agonist leads to Müller glia dependent neurogenesis in the adult mammalian retina. IOVS 60, 570–579. doi: 10.1167/iovs.18-25722
Webster, M., Cooley-Themm, C., Barnett, J., Bach, H., Vainner, J., Webster, S., et al. (2017). Evidence of BrdU-positive retinal neurons after application of an Alpha7 nicotinic acetylcholine receptor agonist. Neuroscience 346, 437–446. doi: 10.1016/j.neuroscience.2017.01.029
Weinreb, R., Aung, T., and Medeiros, F. (2014). The pathophysiology and treatment of glaucoma. J. Amer. Med. Assoc. 311, 1901–1911. doi: 10.1001/jama.2014.3192
Wright, E. M., Perveen, R., Bowers, N., Ramsden, S., McCann, E., O’Driscoll, M., et al. (2010). VSX2 in microphthalmia: a novel splice site mutation producing a severe microphthalmia phenotype. Br. J. Ophthalmol. 94, 386–388. doi: 10.1136/bjo.2009.159996
Wright, L. S., Pinilla, I., Saha, J., Clermont, J. M., Lien, J. S., Borys, K. D., et al. (2015). Vsx2 and ASCL1 are indicators of neurogenic compentence in human retinal progenitor cultures. PLoS One 10:e0135830. doi: 10.1371/journal.pone.0135830
Xia, X., and Ahmad, I. (2016). let-7 microRNA regulates neurogliogenesis in the mammalian retina through Hmga2. Dev. Biol. 410, 70–85. doi: 10.1016/j.ydbio.2015.12.010
Yao, K., Qiu, S., Tian, L., Snider, W., Flannery, J., Schaffer, D., et al. (2016). Wnt Regulates proliferation and neurogenic potential of Müller glial cells via a Lin28/let-7 miRNA-dependent pathway in adult mammalian retinas. Cell Rep. 17, 165–178.
Yao, K., Qui, S., Yanbin, V., Park, S. J. H., Mohns, E. J., Mehta, B., et al. (2018). Restoration of vision after de novo genesis of rod photoreceptors in mammalian retinas. Nature 560, 484–488. doi: 10.1038/s41586-018-0425-3
Yoshii, C., Ueda, Y., Okamoto, M., and Araki, M. (2007). Neural retinal regeneration in the anuran amphibian Xenopus laevis post-metamorphosis: transdifferentiation of retinal pigmented epithelium regenerates the neural retina. Dev. Biol. 303, 45–56. doi: 10.1016/j.ydbio.2006.11.024
Keywords: mammalian retinal regeneration, Müller glia, dedifferentiation, retinal pigment epithelium, α7 nicotinic acetylcholine receptor agonist
Citation: Stanchfield ML, Webster SE, Webster MK and Linn CL (2020) Involvement of HB-EGF/Ascl1/Lin28a Genes in Dedifferentiation of Adult Mammalian Müller Glia. Front. Mol. Biosci. 7:200. doi: 10.3389/fmolb.2020.00200
Received: 16 May 2020; Accepted: 24 July 2020;
Published: 14 August 2020.
Edited by:
Eleonora Napoli, University of California, Davis, United StatesReviewed by:
Monica Lamas, Center for Research and Advanced Studies, MexicoCopyright © 2020 Stanchfield, Webster, Webster and Linn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cindy L. Linn, Q2luZHkubGlubkB3bWljaC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.