
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 31 January 2020
Sec. RNA Networks and Biology
Volume 7 - 2020 | https://doi.org/10.3389/fmolb.2020.00008
This article is part of the Research Topic Role of Ribonucleoprotein Complexes in Neurodevelopment and in the Physiopathology of Neurological Diseases View all 9 articles
Activity-dependent protein synthesis plays an important role during neuronal development by fine-tuning the formation and function of neuronal circuits. Recent studies have shown that miRNAs are integral to this regulation because of their ability to control protein synthesis in a rapid, specific and potentially reversible manner. miRNA mediated regulation is a multistep process that involves inhibition of translation before degradation of targeted mRNA, which provides the possibility to store and reverse the inhibition at multiple stages. This flexibility is primarily thought to be derived from the composition of miRNA induced silencing complex (miRISC). AGO2 is likely the only obligatory component of miRISC, while multiple RBPs are shown to be associated with this core miRISC to form diverse miRISC complexes. The formation of these heterogeneous miRISC complexes is intricately regulated by various extracellular signals and cell-specific contexts. In this review, we discuss the composition of miRISC and its functions during neuronal development. Neurodevelopment is guided by both internal programs and external cues. Neuronal activity and external signals play an important role in the formation and refining of the neuronal network. miRISC composition and diversity have a critical role at distinct stages of neurodevelopment. Even though there is a good amount of literature available on the role of miRNAs mediated regulation of neuronal development, surprisingly the role of miRISC composition and its functional dynamics in neuronal development is not much discussed. In this article, we review the available literature on the heterogeneity of the neuronal miRISC composition and how this may influence translation regulation in the context of neuronal development.
MicroRNAs (miRNAs) are among the most studied and discussed regulators of gene expression in the last two decades. The reason they have attracted so much attention is because of their specificity in targeting and versatility in their function. These small RNAs (~22 nt long) recognize their targets through a “seed sequence” (forming 5–8 base pair match on target mRNA) and lead to a rapid decrease in the corresponding protein level by translation repression and/or degradation of mRNA (Baek et al., 2008; Guo et al., 2010; Bartel, 2018; Duchaine and Fabian, 2019). Due to their sequence specificity and requirement of short nucleotide stretch for base pairing, single miRNA can target several mRNAs (Hashimoto et al., 2013). miRNAs control gene expression at multiple levels and affect every aspect of cellular functions in multicellular organisms (Janga and Vallabhaneni, 2011). More recently, there has been an explosion of studies on miRNAs due to their potential implication in diagnostic and therapeutic approaches in multiple human diseases ranging from cancer to neurodegenerative disorders (Peng and Croce, 2016; Ramakrishna and Muddashetty, 2019). The effect of miRNAs on translation depends on the recruitment of a complex which includes several protein components. This protein complex together with miRNA forms the microRNA induced silencing complex (miRISC) which is essential to execute the miRNA function (Krol et al., 2010; Fabian and Sonenberg, 2012). While our appreciation of the role of miRNAs in biology is ever-increasing, there is a significant gap in our understanding of the function of individual components of the miRISC and its compositional heterogeneity.
Unlike small interfering RNA (siRNA), miRNA mediated translation repression is a multistep process. Several studies have indicated a clear correlation between decreased protein levels and the degradation of mRNAs targeted by miRNAs (Baek et al., 2008; Guo et al., 2010). However, a less appreciated fact is that the kinetics of this process is highly variable for different sets of mRNAs (Baek et al., 2008; Hendrickson et al., 2009; Guo et al., 2010). In all cases, translation repression precedes mRNA degradation and now it has been established that in many instances, translation repression can be reversed (Bhattacharyya et al., 2006; Muddashetty and Bassell, 2009; Djuranovic et al., 2011; Kute et al., 2019). The whole logic of a complex multistep process to induce translation repression could be to facilitate the reversibility of this process. This makes miRNA mediated regulation a powerful tool for the spatiotemporal regulation of gene expression. The kinetics and reversibility of miRISC depend to a large extent on its composition. Hence, it is very important to identify and classify these proteins into the core miRISC components and the additional/ancillary ones. The core miRISC facilitates the default miRISC pathway which includes, translation repression followed by deadenylation, decapping, and degradation of mRNA (Huntzinger and Izaurralde, 2011; Fabian and Sonenberg, 2012). Additional factors generally regulate the kinetics of this pathway which could also result in stalling of the process at any of the stages of the default pathway. More importantly, these factors could provide reversibility to the miRISC mediated translation repression and make it a more dynamic process to regulate gene expression.
Reversibility of miRNA mediated translation repression is likely to happen in all kinds of cells during the growth, division, differentiation or in response to stress and external cues (Bhattacharyya et al., 2006; Jafarifar et al., 2011; Bellon et al., 2017; Patranabis and Bhattacharyya, 2018). This mechanism is particularly apt for neurons where compartmentalized and cue dependent translation is evolved to a great extent. There are many reviews that discuss the role of miRNAs in neuronal development and plasticity (Siegel et al., 2011; Ye et al., 2016). But the dynamic property of miRISC greatly depends on its protein composition (Huntzinger and Izaurralde, 2011; Fabian and Sonenberg, 2012). In this review, we explore the protein composition of miRISC and how it determines the function of miRNAs in neurons. We particularly focus on different developmental stages of the nervous system where the alteration of this composition could fine-tune the gene expression to equip the response of neurons to external cues.
The miRISC function has two primary components, target recognition and silencing of the targeted transcript. The miRNA-element confers specificity to the target, while the gene expression repression functions are carried out by different protein components of miRISC (Fabian and Sonenberg, 2012; Duchaine and Fabian, 2019) (Figure 1). The primary protein partner of miRISC is the Argonaute protein. Argonaute proteins are an indispensable part of miRISC complex as they bring together miRNAs and mRNAs to form the core of miRISC (Höck and Meister, 2008; Bartel, 2018) (Figure 1A). The rest of the miRISC comprises of a diverse set of proteins, such as mRNA degradation machinery components, scaffolding proteins, and many different RNA binding proteins (RBPs), which assemble with the core miRISC to form compositionally diverse miRISCs. This composition, in turn, appears to be a crucial determinant of the kinetics and the outcome of the miRISC function (Dallaire et al., 2018).
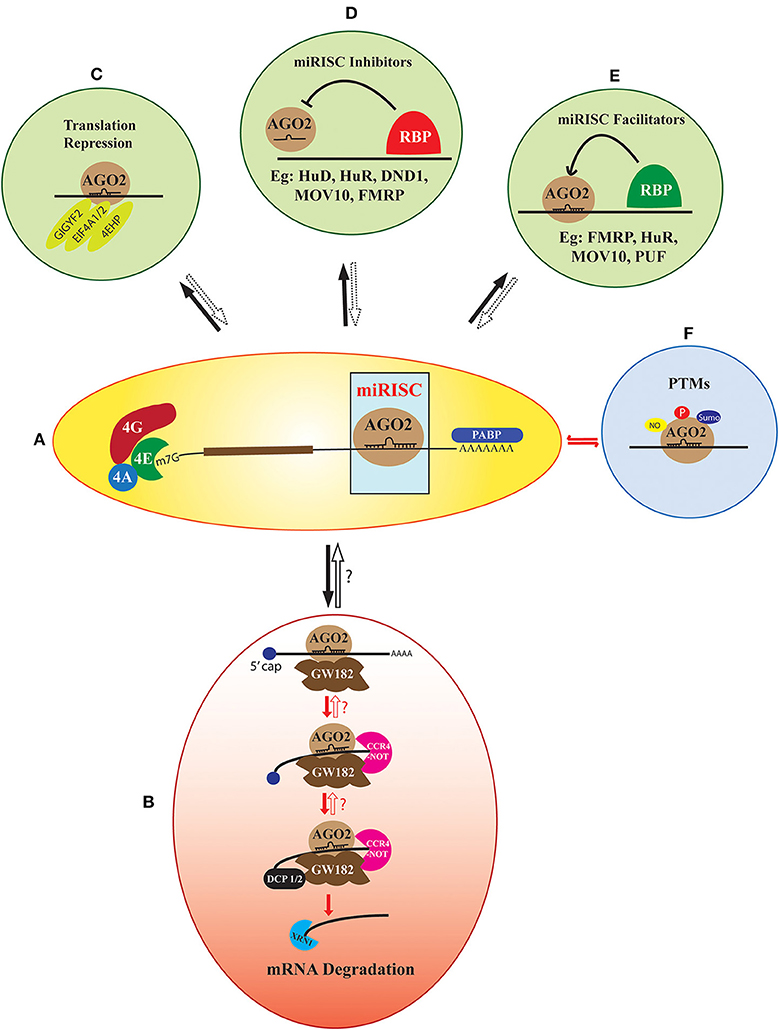
Figure 1. The heterogeneity in miRISC effector function. AGO2 recruited on a target mRNA along with its cognate miRNA form the core of miRISC (A). The default function of this complex is deadenylation and decapping-dependent degradation of the target mRNA. This pathway involves the sequential assembly of proteins like GW182, CCR4-NOT, DCP1/2, and XRN1 to the core miRISC (B). However, this multi-step assembly could be stalled and potentially reversed at several of these steps. Apart from the default pathway, the core miRISC can also associate with other translation regulators like GIGYF2, EIF4A1/2, and 4EHP, which lead to translation repression of the target mRNA without degradation (C). A large number of RNA Binding Proteins (RBPs) like FMRP, MOV10, Hu proteins, Pumilio, and DND1 either facilitate or inhibit miRISC assembly on an mRNA (D,E). Another added layer of heterogeneity is contributed by the post-translational modifications (PTMs) of the miRISC protein components. Modifications like phosphorylation, sumoylation, and nitrosylation are reported both on the core miRISC protein AGO2 or the accessory members (F). The fate of the target mRNA is determined by all the above-mentioned factors concerning miRISC along with the specific cellular context. However, the precise mechanisms of the reversible miRISC function, as well as its inhibitors and facilitators are not completely known.
As a default mechanism, the formation of miRISC on an mRNA leads to its degradation in a multistep pathway (Fabian et al., 2010; James et al., 2012; Jonas and Izaurralde, 2015) (Figure 1B). Integrated results from transcriptome-wide analysis coupled with ribosome profiling, and mass spectrometry techniques have revealed mRNA destabilization as the primary result of miRNA function in multiple cell lines (Baek et al., 2008; Hendrickson et al., 2009; Guo et al., 2010). This lead to the widely accepted (what we call “default”) model of miRISC function, which involves miRNA mediated inhibition of translation, followed by deadenylation, decapping, and degradation of target mRNA (Bazzini et al., 2012; Djuranovic et al., 2012). However, now there is sufficient evidence to suggest that this default pathway can be significantly altered by additional factors which can stall and reverse the miRNA mediated inhibition at multiple steps (Ashraf et al., 2006; Bhattacharyya et al., 2006; Kedde et al., 2007; Banerjee et al., 2009; Muddashetty et al., 2011; Kute et al., 2019). This reversible regulation provides flexibility to the system for rapid modulation of protein levels on different cues, bypassing the need for new mRNA transcription. In developmental contexts, such as oogenesis, early embryogenesis and in neurons, translational repression is preferred over irreversible mRNA decay. This helps to stably maintain a pool of specific mRNAs whose temporally controlled expression is critical in development or synaptic plasticity. Thus, we emphasize that the data from transcriptome-wide and ribosome-profiling studies should not be generalized, as this may ignore the importance of the structural and functional heterogeneity of miRISC in cell type and development specific contexts.
Argonaute (AGO) associates with miRNAs and load them on to their cognate target mRNAs to form the core of the miRISC (Figure 1A). Now there is good amount of data is available on the structure and function of distinct domains of Argonaute protein (Song et al., 2004; Hall, 2005; Peters and Meister, 2007; Höck and Meister, 2008; Hutvagner and Simard, 2008; Boland et al., 2011; Cenik and Zamore, 2011; Meister, 2013; Swarts et al., 2014; Dayeh et al., 2018). The Argonaute family of proteins are evolutionarily conserved with multiple paralogues present in diverse species (Swarts et al., 2014). The mammalian genome encodes four Argonaute proteins, of which only AGO2 possesses the endonucleolytic activity. AGO2 is the most abundant and is also best studied among its paralogues with respect to miRNA function (Carmell et al., 2004; Tuschl et al., 2004). However, all four Argonaute proteins can efficiently perform miRNA mediated translation repression and act in an overlapping and redundant manner (Su et al., 2009; Wang et al., 2012). Among the four AGO proteins, AGO3 and AGO4 are sparsely studied mostly due to the low abundance of these proteins in adult tissues (Su et al., 2009; Wang et al., 2012; Völler et al., 2016).
In addition to miRNA mediated post-transcriptional gene silencing, AGO proteins are reported to perform various moonlighting functions in other subcellular compartments. Nuclear localized AGO proteins regulate transcriptional silencing, epigenetic modulation, alternative splicing, and DNA repair (Huang and Li, 2014; Ross and Kassir, 2014). The function of nuclear-localized AGO complexes has been previously reviewed and hence will not be discussed further in this review (Huang and Li, 2014; Ross and Kassir, 2014; Kalantari et al., 2016; Sharma et al., 2016; Liu et al., 2018).
The imperfect complementarity of metazoan miRNA-mRNA pairs prevents the endonucleolytic activity of AGO2, leading to the requirement of additional factors for the effective degradation of target mRNA. In its default mode, the scaffolding protein GW182 is first recruited on to the core miRISC, which in turn facilitates the binding of deadenylation and decapping enzymes as well as exonucleases to form large miRISC complex (Fabian and Sonenberg, 2012). This complex primarily results in the degradation of targeted mRNA (Figure 1B). GW182, a critical component of this pathway, is a large Glycine Tryptophan (GW) repeats containing protein. The structure and function of different domains of GW182 have been critically reviewed (Ding and Han, 2007; Baillat and Shiekhattar, 2009; Chekulaeva et al., 2009; Eulalio et al., 2009; Bazzini et al., 2012; Zielezinski and Karlowski, 2015; Niaz and Hussain, 2018). GW182 binds to AGO2 through its N-terminal domain following which it interacts with Polyadenylate Binding Protein (PABP) on the 3′ end of mRNA. This interaction would interrupt the circularization of mRNA leading to translation repression (Zekri et al., 2009, 2013; Moretti et al., 2012). The C-terminal silencing domain of GW182 further recruits deadenylase complexes Poly(A)-Nuclease (PAN) deadenylation complex (PAN2-PAN3) and Carbon Catabolite Repressor 4 (CCR4)-Negative on TATA (NOT) to PABP free mRNA leading to deadenylation (Behm-Ansmant et al., 2006; Chekulaeva et al., 2011; Fabian et al., 2011). In most animal systems, the deadenylated mRNA is decapped by Decapping protein 1/2 (DCP1/2)-followed by mRNA degradation through 5′-3′ Exoribonuclease 1 (XRN1) (Rehwinkel et al., 2005; Behm-Ansmant et al., 2006).
The temporal sequence and the relative contribution of translation repression and mRNA degradation in the canonical mode of miRISC function are not fully understood. However, majority of the studies suggest that translation repression is followed by mRNA degradation unless cellular degradation machinery is inhibited (Bazzini et al., 2012; Béthune et al., 2012; Djuranovic et al., 2012; Jonas and Izaurralde, 2015). The coupling between translation repression and mRNA degradation depends on relative kinetics of translation repression, deadenylation, and decapping process. Hence, we hypothesize that translation repression by miRISC can be effectively uncoupled from mRNA degradation thus providing an effective way for reversible translational regulation. Compared to the mRNA degradation function, the mechanisms contributing to translational repression by miRNAs are more heterogeneous and less understood. In the following sub-section, we describe several well-studied mechanisms of miRISC mediated translation repression.
Apart from the default mechanism, miRISC is shown to function via several alternate mechanisms (Figure 1C). These involve the interaction of core miRISC with components of the translation machinery and diverse RNA binding proteins. Translation machinery proteins shown to be central in such alternate pathways include Eukaryotic Initiation Factor 4F (eIF4F) complex subunit eIF4A (Meijer et al., 2013; Fukao et al., 2014; Fukaya et al., 2014), and the helicase DDX6 (Kamenska et al., 2016). The interaction of miRISC with these proteins either precludes the normal assembly of the translational machinery or recruits additional elements, such as eIF4E homolog 4EHP. miRISC is also shown to interact with the components of the ribosomal complex, such as Receptor for Activated C Kinase 1 (RACK1), Ribosomal Protein S14 (RPS14), Ribosomal Protein L5 (RPL5), and Ribosomal Protein L11 (RPL11) (Chan and Slack, 2009; Jannot et al., 2011; Liao et al., 2013).
The activity of miRISC is heavily modulated by its interactions with multiple RBPs (Fukao et al., 2015; Gardiner et al., 2015; Loffreda et al., 2015). These RBPs either associate directly with miRISC to influence its function, or they modulate miRISC activity by binding close to the miRNA binding site in the target mRNAs. Proteins like Fragile X Mental Retardation Protein (FMRP), MOV10 Moloney leukemia virus 10 (MOV10), Ataxin 2, and Fused in Sarcoma (FUS) directly associate with core miRISC to influence its downstream effector functions (Banerjee et al., 2009; McCann et al., 2011; Muddashetty et al., 2011; Sudhakaran et al., 2014; Zhang T. et al., 2018) (Figures 1D,E). The association of FMRP with miRISC was earlier contested as one of the report has shown that FMRP does not associate with miRISC and is an important component of stress granules (Didiot et al., 2008). However, further later reports showed the association of FMRP with core miRISC protein AGO2 (Jin et al., 2004; Muddashetty et al., 2011; Kute et al., 2019). RBP Pumilio positively modulates miRISC activity by unwinding mRNA 3′UTR to promote miRNA binding (Friend et al., 2012). Multiple members of TRIM-NHL family of proteins interact with AGO to positively modulate miRISC repressive activity (Hammell et al., 2009; Schwamborn et al., 2009) (Figure 1E). In contrast, proteins, such as Dead end protein homolog 1 (DND1), Heterogeneous Nuclear Ribonucleoprotein L (hnRNPL), APOBEC3G and Hu family proteins inhibit miRISC function either by competing with miRISC target site on mRNA or by preventing functional miRISC formation (Bhattacharyya et al., 2006; Kedde et al., 2007; Jafarifar et al., 2011; Liu et al., 2012) (Figure 1D). Notably, most of the miRISC repression modulated by different RBPs is shown to be reversible. Although many of these studies investigate the interaction of various RBPs with AGO2, the association of these RBPs with GW182 and how that may influence miRISC reversibility remains unclear.
The work described until now clearly establishes the wide compositional diversity of miRISC. The important question is how this diversity influences the effector function of miRISC? The broad mechanisms by which the composition and external cues modulate the miRISC function is shown in Figure 2. These factors include cellular proliferation status, specific spatiotemporal context, and different extracellular signals. External signals modify the miRISC function through different mechanisms. One such crucial mechanism is post-translational modifications (PTMs) (Figures 1F, 2C). The core miRISC component AGO2 is modified by phosphorylation, sumoylation, prolyl hydroxylation, and ubiquitination at multiple sites (Figure 1F). Such PTMs regulate divergent aspects of AGO2 function, such as stability, miRNA binding, mRNA association, and interaction with GW182 (Qi et al., 2008; Rüdel et al., 2011; Horman et al., 2013; Martinez and Gregory, 2013; Sahin et al., 2014; Bridge et al., 2017; Chinen and Lei, 2017; Golden et al., 2017). Another mechanism of regulating miRISC is via protein-protein interactions. AGO2-GW182 interaction is highly regulated by various extracellular cues (Olejniczak et al., 2013, 2016; Wu et al., 2013; La Rocca et al., 2015; Bridge et al., 2017; Rajgor et al., 2018). AGO2 interaction with RBPs, as well as the modulation of miRISC activity by RBPs are shown to be regulated by different signals like hypoxia, arsenite stress, mTOR signaling, and neuronal activity (Bhattacharyya et al., 2006; Banerjee et al., 2009; Jafarifar et al., 2011; Muddashetty et al., 2011; Sosanya et al., 2013). Cell-specific regulation of miRISC composition was recently demonstrated by work from Simard's lab (Dallaire et al., 2018). The structure of the miRNA binding site on mRNA also regulates the miRISC (Zhang K. et al., 2018). Additionally, specific subcellular localization of miRNA and miRISC machinery can also confer structural and functional dynamicity to miRISC (Trabucchi, 2019). These mechanisms generally have an overlapping regulatory function, as in, a post-translational modification can affect the level or localization of a particular miRISC protein.
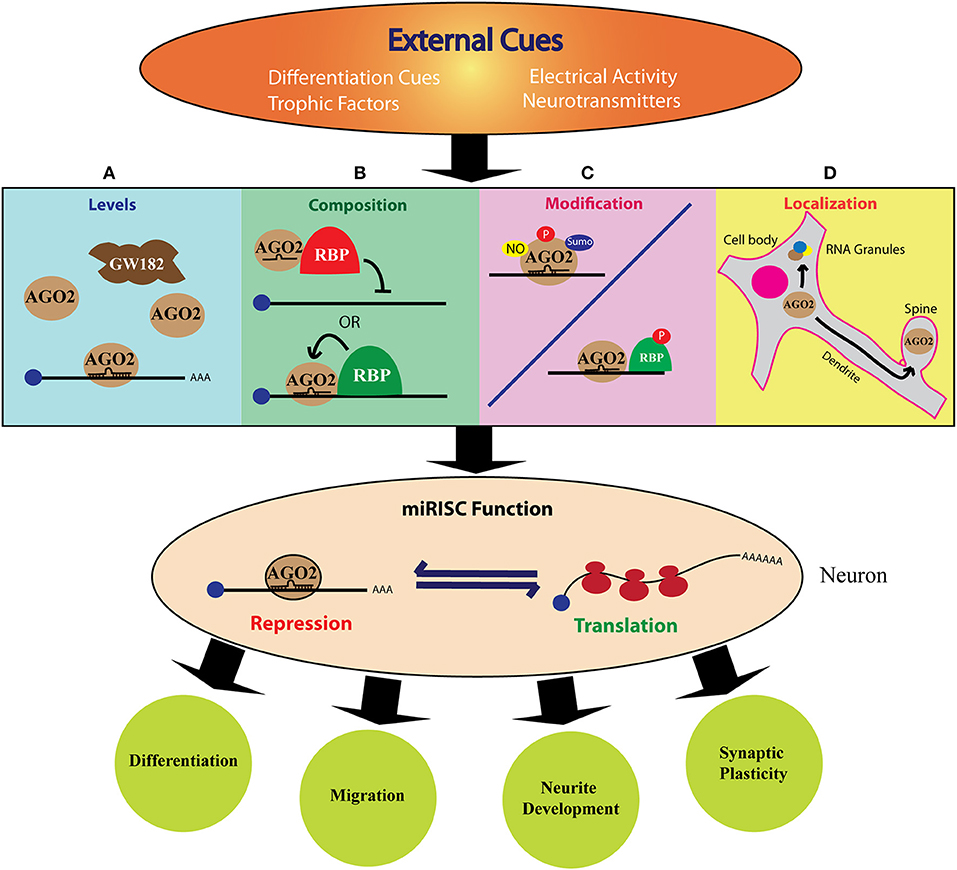
Figure 2. The role of miRISC in cue mediated protein synthesis during neurodevelopment. The above figure shows how different external cues can modulate miRISC function to fine-tune neurodevelopment. The specific, rapid, local, and reversible mode of miRNA action qualifies miRISC mediated mechanism as an excellent mode for extrinsic regulation of neuronal protein synthesis. External signals, such as trophic factors or neuronal activity can induce changes in the levels (A), composition (B), post-translational modification (C), or localization (D) of one or more of miRISC components. This can alter the fine balance between translation and repression/decay of the target mRNA. This, in turn, leads to specific changes in the cellular proteome either globally or locally and enable the cell to differentiate, migrate, grow neurites or form synapses.
Spatiotemporal regulation of the intracellular proteome is important for every cell in multicellular organisms. But, it is of critical significance to neurons due to their highly polarized organization, and the specificity of the connections. There is substantial de-centralization of protein synthesis in neurons which is well-established through the demonstration of the presence of the translational machinery, as well as, a wide repertoire of mRNAs, in various neuronal compartments (Zelená, 1970; Steward and Levy, 1982; Torre and Steward, 1992; Chicurel et al., 1993; Rao and Steward, 1993). Apart from that, neurons also have evolved mechanisms to tightly regulate protein synthesis in response to neuronal activity. There is now evidence to show the significant contribution of activity-mediated protein synthesis in neurodevelopment and synaptic plasticity (Kang and Schuman, 1996; Kauderer and Kandel, 2000; Campbell and Holt, 2001; Wu et al., 2005; Yao et al., 2006; Costa-Mattioli et al., 2009; Blair et al., 2017; Ravindran et al., 2019). Defects in this regulation are also thought to be the underlying pathophysiology of several neurodevelopmental disorders. There are several excellent recent reviews on these concepts (Steward and Schuman, 2003; Swanger and Bassell, 2013; Kim and Jung, 2015). Since this review is focused on the role of translational regulation during neurodevelopment, we have listed out different external cues affecting the stages of neuronal development as well as the different proteins reported to be translationally regulated during specific stages in Figure 3. However, it is to be noted that a signal to the target-protein link is not established for many of the cases.
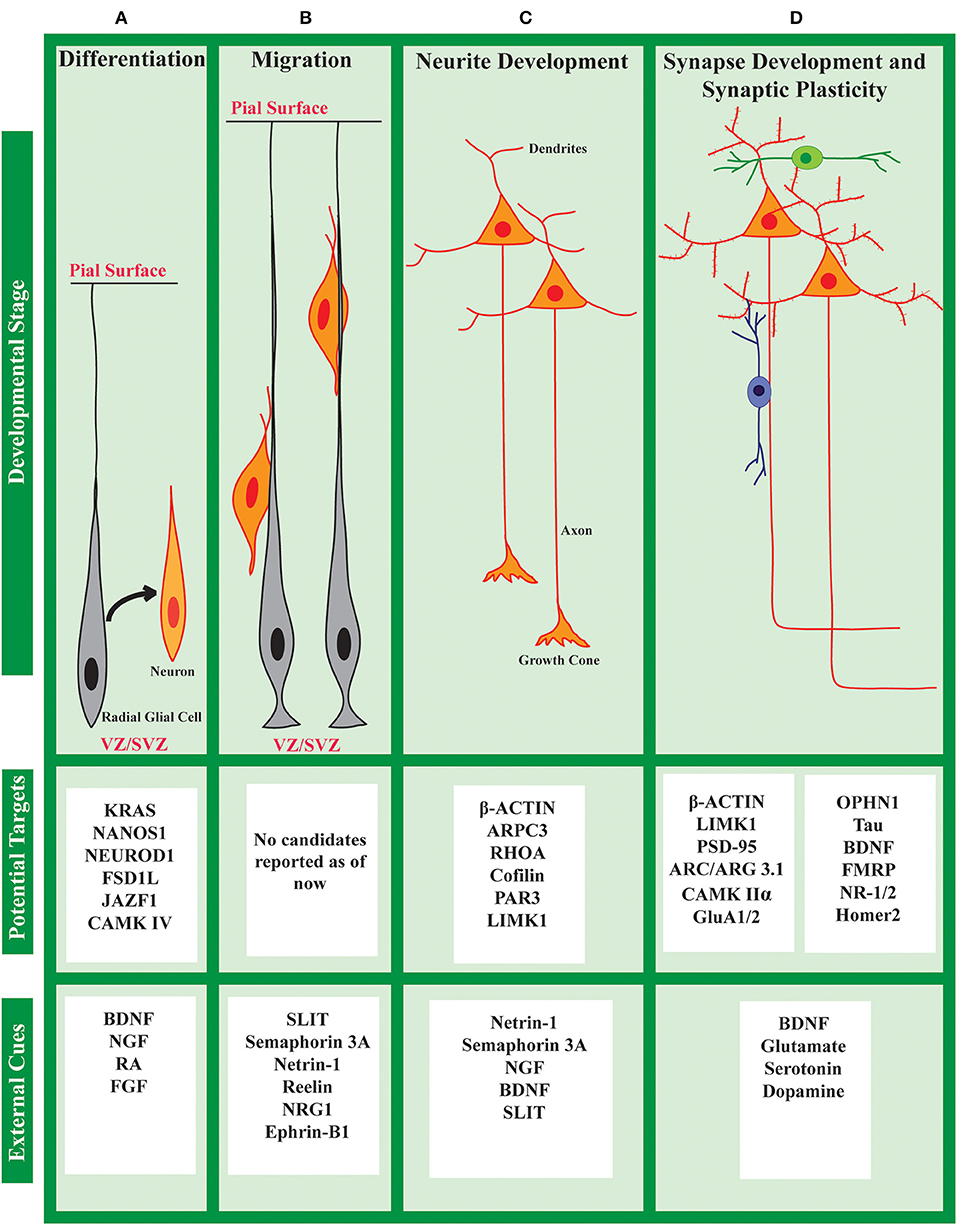
Figure 3. Cue induced protein synthesis in neuronal development: cues and targets. The development of a neuron can be divided into different stages (A–D) as shown above. At each stage, the neuron responds to a set of external cues that regulates its development. Along with activating transcription, these cues also affect the translational status of specific mRNAs to fine-tune the global/local proteome. Several such candidates are listed above, although specific links between cues and candidates are not known in many cases.
The specific, rapid, local, and reversible mode of miRNA action qualifies miRISC mediated mechanism as an excellent choice for regulating neuronal protein synthesis. Moreover, the reversibility of miRISC is a well-suited mechanism for neuronal functions because, in contrast to targeting the mRNA for degradation, neurons often require to store mRNA in a repressed state either for local transport or for cue mediated translation. As depicted in Figure 2, the dynamic nature of miRISC in neurons could be achieved in multiple ways. First, the expression of different miRISC components can be regulated during neuronal development and synaptic activity (Figure 2A). This can dictate the type of miRISC formed in a particular scenario. Additionally, the expression of the miRISC proteins can be regulated locally at specific neuronal compartments, such as dendrite, spine or axonal growth cone. Extracellular factors, such as axon guidance cues, neurotrophic factors, and neurotransmitter signals can also influence miRISC composition (Figure 2B) by altering the post-translational modification (Figure 2C) and sub-cellular localization (Figure 2D) of miRISC components. Not surprisingly, much of the literature investigating the role of RBPs in miRISC functions have utilized neuronal model systems.
Development of the nervous system in humans is a complex and prolonged phenomenon starting as early as 3 weeks of gestation and continuing until adolescence (Stiles and Jernigan, 2010; Elshazzly and Caban, 2019). It involves an orchestrated network of cell fate changes as well as cellular and tissue morphogenesis driven by cell-intrinsic and activity-dependent mechanisms (Katz et al., 1992). Development of the nervous system begins with neurulation—the process by which a segment of ectodermal cells specialize to form the neural plate which invaginates inside and fuses to form the neural tube. The neural tube is a hollow tube running in the anteroposterior axis of the embryo which gives rise to the central nervous system. The rostral end of the tube forms the brain and the caudal region forms the spinal cord. The peripheral nervous system is derived from the neural crest cells, which are formed near the dorsal line of fusion of the neural tube. Rodents also follow the same pattern of neurodevelopment. Although different kinds of cells are involved in the formation of the neuronal network, in this review, we have limited our area of interest to the development of the neurons. The development of a typical neuron in the central nervous system (CNS) can be broadly classified into the following overlapping phases: neurogenesis, subtype specification, neuronal migration, neurite growth, synaptogenesis, and synaptic pruning. In rodents, the first three phases are predominantly pre-natal and later phases continue into the post-natal period. In the next section, we will be discussing different miRISC complexes formed and their role in neuronal development.
In rodents, neurogenesis starts at around E10 and is complete by about E18 (Semple et al., 2013). A part of the neuronal population is derived directly from radial glial cells (RGC), which in turn is derived from neuroepithelial cells (NEC) (Figure 3A). Indirect neurogenesis occurs from RGCs through the production of intermediate progenitors (IP). The site of neurogenesis is close to the ventricular zone (VZ). The process of sub-type specification mostly coincides with neurogenesis. Early in the development, neural progenitor cells can give rise to a wide variety of neuronal populations. Later on, they become more sub-type specific (Taverna et al., 2014; Jiang and Nardelli, 2016).
The prevailing view in the field describes transcriptional regulation as the predominant gene regulatory program during neuronal differentiation. However, emerging studies have established the importance of post-transcriptional mechanisms in neuronal differentiation (Kraushar et al., 2014, 2015; Blair et al., 2017; Sugiyama et al., 2017; Hwang et al., 2018; Zahr et al., 2018; Mohammad et al., 2019; Tahmasebi et al., 2019). Multiple miRNAs have been shown to regulate distinct aspects of neuronal differentiation (Stappert et al., 2015). Studies from over a decade have established the pivotal role of miRISC proteins and their composition in regulating neuronal differentiation. To begin, the core miRISC protein AGO has been shown to undergo paralogue switching from AGO1 to AGO2 during nervous system development (Juvvuna et al., 2012). The study demonstrated a more potent regulation of miR-124 induced neuronal differentiation via AGO1-miRISC as compared to AGO2-miRISC. Additionally, the authors associated the increased fractional abundance of AGO2 with the progressive shortening of miRNA 3′ end during nervous system development. Thus, the authors suggested an interesting possibility where 3′ end trimming of miRNA regulates their target specificity across neuronal development. AGO1 is also reported to associate with TRIM-NHL protein Tripartite Motif-containing protein 32 (TRIM 32) to prevent neuronal differentiation in mouse cortical progenitor neurons. TRIM-32 associates with AGO1 to increase the activity of specific miRNAs, such as Let-7a. In C. elegans, Trim-NHL protein NHL-2 is shown to function as a cofactor for miRISC in regulating neuronal cell fate changes (Hammell et al., 2009). TRIM-32 was shown to associate with miRISC protein DDX6 for regulating neuronal differentiation (Nicklas et al., 2015). The mechanistic details of TRIM-NHL proteins interactions with AGO and its effect on the miRNA pathway are yet to be elucidated. However, findings across species suggest that they have a clear role in modulating the miRISC pathway during neuronal differentiation. Another recent study demonstrated that Nerve Growth Factor (NGF) induced differentiation of PC12 cells requires phosphorylation of AGO2 at Tyr-529 (Patranabis and Bhattacharyya, 2016). This results in the release of Let-7a miRNA from AGO2 and subsequent activation of Let-7a target KRAS leading to neuronal differentiation. Ectopic expression of P-body proteins, AGO2 or GW182 was sufficient to induce differentiation pointing to a direct link between miRISC activity and cell fate decisions (Patranabis and Bhattacharyya, 2018). An important role of miRISC was recently described in the regulation of the quiescent state in adult neuronal stem cells (Katz et al., 2016). This study demonstrated a non-canonical nuclear role of AGO and GW182 associated miR-9 complex in regulating neuronal differentiation. Alteration of the nuclear/cytoplasmic ratio of this miR-9-RISC resulted in deregulation of NSC quiescent state. An important future direction is to delineate whether such a mechanism also regulates neuronal differentiation during early development.
Multiple other miRISC interacting proteins, such as FMRP, Pumilio, and Hu family of proteins have been shown to play an indispensable role in regulation of neuronal differentiation (Kasashima et al., 1999; Anderson et al., 2000; Akamatsu et al., 2002; Castren et al., 2005; Tervonen et al., 2009). However, since all these proteins have been shown to regulate translation independent of miRISC, their relative contribution in the context of miRISC is yet to be elucidated. Thus, the current evidence indicates that even though transcriptional changes may be the predominant factor driving neurogenesis/differentiation, translational control through miRISC modulation also plays a critical role in driving these processes.
From the site of differentiation close to the VZ, neurons migrate to their final destination near the periphery of the neural tube by a process called radial migration. Initially in development, this occurs through somal translocation. Later, radial glial cells guide the neuronal migration (Figure 3B). As a general rule, neurons follow an “inside-out” migratory pattern, as the first formed neurons occupy the deeper layers and the later formed neurons migrate past them to form the superficial layers. In the developing neocortex, this migratory pattern leads to the formation of a six-layer architecture. Several external cues like Reelin, Ephrin, Semaphorin 3A, Netrin-1, etc. are known to regulate this process of migration (Jiang and Nardelli, 2016; Buchsbaum and Cappello, 2019). The importance of translation regulation in neuronal migration is sparsely studied. However, considering the directional migration of a polarized cell, it is very likely that local translation could be of importance. Indeed, some miRISC interactors are shown to play an important role in migration. For example, FMRP is shown to regulate neuronal migration and cortical positioning by affecting the multipolar to bipolar transition in migrating neurons (La Fata et al., 2014). Using mass spectrometry-based study, Weinmann et al. (2009) identified Importin-8 (IPO-8) as an AGO interacting protein. It interacts with AGO1-4 and is required for the recruitment of AGO complex onto a large set of target mRNAs. IPO-8 is also required for the radial migration of cortical projection neurons (Nganou et al., 2018). However, there is no direct evidence for the role of cue-induced miRISC modulation in neuronal migration. Technical limitations may be an important hurdle to study this. Developments in the field of organoid culture might bridge this gap in the future.
Neurons are heavily polarized cells with characteristic sub-type specific dendritic and axonal arbor. In most multipolar neurons in the cerebral cortex, several neurites outgrow from the soma and one of them is soon differentiated to be the axon. The rest of the neurites grow as dendrites. The axons generally grow faster and develop large growth cones at their tips which guide them to the putative targets (Figure 3C). Both axonal and dendritic growth is heavily dependent on various guidance cues and trophic factors (Dickson, 2002; Arikkath, 2012; Jiang and Nardelli, 2016).
Studies from the last three decades have established the essential role of cue mediated local translation in axonal growth and axonal regeneration. Subsequently, studies have also explored the role of miRISC composition in the regulation of the above-mentioned mechanism. Multiple components of miRISC machinery are shown to localize in developing axons and growth cones (Hengst et al., 2006; Murashov et al., 2007; Dajas-Bailador et al., 2012). A recent study has shown the co-localization of core miRISC (AGO2 and miRNAs) with mitochondria at axonal branch points and growth cone of peripheral nerve axons (Gershoni-Emek et al., 2018). miRISC protein FMRP has been shown to regulate Semaphorin 3A mediated growth cone collapse (Li, 2009). Another study has shown that FMRP associates with miR-181d and AGO2 to mediate axonal delivery of Map1b and Calm1 mRNAs (Wang et al., 2015). The authors further demonstrated that NGF mediated the local release of Map1b and Calm1 mRNA from FMRP containing repressive granules, resulting in axonal translation. This study provides an important proof of principle showing the importance of cue regulated miRISC composition in axonal development. It is important to determine whether miRISC also associates with other axonal transport proteins for regulating the transport of translationally repressed mRNA targets.
The importance of miRISC mediated translation regulation is not investigated extensively in mature axons. This is primarily due to the apparent absence of protein synthesis in mature axons (Kim and Jung, 2015; Biever et al., 2019). However, recent studies have demonstrated axonal translation and its importance in synaptic plasticity (Shigeoka et al., 2016; Younts et al., 2016; Scarnati et al., 2018), thereby providing a platform for future studies of miRISC involvement in translation regulation in mature axons.
miRISC is also shown to play an important role in regulating axonal regeneration upon injury. Expression of several miRISC components as AGO2, FMRP, GW182, and DCP1 is shown to be induced in axonal varicosities upon neuronal injury (Wu et al., 2011). This increased expression was accompanied by an increased co-localization of GW182 and DCP1 in axons. An interesting insight from these studies is that lesion-induced up-regulation of miRISC proteins was seen only in axons, leaving the possibility that the injury causes an axon-specific translation of miRISC proteins.
In contrast to its important role in axonal development, the importance of translation regulation in dendritic development is yet to be established. However, recent studies showing the essential requirement of ribosomes and different RBPs in dendrite morphogenesis, have hinted toward an important role of translation regulation in dendritic development (Vessey et al., 2010; Olesnicky et al., 2014; Antonacci et al., 2015; Slomnicki et al., 2016; Ravindran et al., 2019). Due to the apparent lack of studies on translation regulation in dendrite development, the function of miRISC and its compositional dynamics is very sparsely studied. Till date, only one study has shown the importance of cue dependent modulation of miRISC composition for the regulation of dendritic growth (Huang et al., 2012). The authors showed that BDNF increases the interaction of miRISC scaffolding protein GW182 with AGO2 and DDX6. This increased interaction regulates BDNF induced translation and dendritic growth (Huang et al., 2012). The authors also demonstrated that perturbation of GW182 function abolished BDNF induced dendritic growth. Interestingly, the authors did not see any effect of GW182 function on basal dendritic growth. However, this study was performed on DIV14 cultured neurons whereas most of the dendritic growth happens between DIV 3–12 in neuronal culture. Hence, it is important to determine whether a similar regulatory mechanism functions during the early stages of dendritic development.
Though the role of translation regulation in dendritic development is underexplored, several miRISC proteins are known to regulate dendrite development. In drosophila, dead box helicase Me31B (DDX6 homolog) interacts with FMRP and AGO to regulate dendritic elaboration in larval sensory neurons (Barbee et al., 2006). The study also showed that the effect of Me31B on dendritic growth is correlated with its ability to mediate translation repression via miRNA. Various miRISC components, such as DCP1A, AGO2, DDX6, and GW182 was shown to localize to dendrites to form dendritic P bodies like structures (dlP) (Barbee et al., 2006; Cougot et al., 2008). miRISC modulators, such as FMRP, MOV10, and Pumilio was also shown to regulate dendrite morphogenesis. However, this does not necessarily implicate their involvement with miRISC for the regulation of dendritic development, and this forms an exciting area for future studies.
Another correlative evidence suggesting miRISC as a regulator of dendrite morphogenesis is that various miRISC proteins show peak expression or activity during an intensive period of dendritogenesis. The decapping activator DCP1a shows development associated changes in its phosphorylation status (Blumenthal et al., 2009). The fractional abundance of the non-phosphorylated form of DCP1a increases in the adult brain. DCP1 phosphorylation is known to affect its activity as a decapping activator by regulating DCP1a-DCP2 interaction (Chiang et al., 2013). Thus, an interesting possibility is that the reduction in phosphorylated DCP1a could result in a reduced decapping and degradation efficiency of miRISC. Different subunits of the CCR4-NOT deadenylase complex also demonstrate a specific neurodevelopmental profile (Chen et al., 2011). The expression of subunits CNOT1, CNOT2, CNOT3, CNOT6, CNOT8, and CNOT9 drastically reduces during brain development. The functional implication of reduced expression of CCR4-NOT complex subunits at this developmental stage has not been determined. However, one can hypothesize that reduced CNOT complex expression along with reduced DCP1 phosphorylation might lead to a reduction in the proportion of degradatory miRISC in the adult brain. In summary, while the role of extrinsic modulation of miRISC is well-established during axonal development, its role in dendrite morphogenesis needs further explorations.
As the axonal and dendritic processes meet their respective partners, further they mature to form pre- and post-synaptic compartments, respectively (Figure 3D). Neuronal activity and mutual chemical signaling between the two partners are critical for the establishment of a synapse. Typically, multiple weak connections are established initially between two partner neurons. Eventually, the network is refined by strengthening some connections and eliminating the rest (Yogev and Shen, 2014; Südhof, 2018). This process of synaptic pruning is a persistent feature of post-natal development until adulthood.
Studies have demonstrated the presence of entire protein synthesis machinery along with ribosomes at developing and mature synapses (Steward et al., 1988). Several studies have established the important function of protein synthesis in synapse formation, maintenance and pruning (Burry, 1985; Steward, 1987; Steward et al., 1988; Scheetz et al., 2000; Munno and Syed, 2003; McCann et al., 2007). Given the important role of cue regulated protein synthesis, it is very likely that miRISC mediated regulation has a critical role in synaptogenesis. The core miRISC component AGO2 has been shown to co-localize with miRNA processing enzyme dicer in the postsynaptic compartment (Lugli et al., 2005; Hanus and Schuman, 2013). FMRP-AGO miRISC was shown to play an important role in the regulation of synaptic growth at larval NMJ (Neuromuscular junction) in drosophila. Drosophila harboring mutations in dFMR1 or AGO1 (Drosophila homolog of mammalian AGO2) shows synaptic overgrowth phenotype along with an increased number of synaptic boutons (Jin et al., 2004). A recent study shows evidence for the important role of miRISC protein, decapping activator HPat1 in regulating synaptic growth at the glutamatergic NMJ in Drosophila larvae (Pradhan et al., 2012). The study demonstrated the presence of strong synaptic hyperplasia at the NMJ in HPat mutants. Further, the study showed that HPat genetically interacts with the catalytic subunit of the deadenylase complex (twin/CCR4) and the miRNA pathway (AGO1) to control bouton formation. These studies hint at the need to further explore the role of cue mediated miRISC modulation during synaptogenesis and pruning.
Synaptic plasticity is the mechanism by which specific patterns of synaptic activity results in changes in synaptic strength (Figure 3D). Different types of synaptic plasticity exist in the central nervous system which play an important role in the regulation of memory formation and consolidation. Multiple studies have established the unequivocal role of protein synthesis in different forms of synaptic plasticity (Kang and Schuman, 1996; Pfeiffer and Huber, 2006; Sutton and Schuman, 2006; McCann et al., 2007; Cajigas et al., 2010; Younts et al., 2016). Cue mediated modulation of miRISC composition and its importance in driving local protein synthesis is also widely studied in the context of synaptic plasticity (Ashraf et al., 2006; Banerjee et al., 2009; Muddashetty et al., 2011; Rajgor et al., 2018; Kute et al., 2019).
miRISC components have been shown to form heterogeneous and dynamic granules in neurons (Cougot et al., 2008; Zeitelhofer et al., 2008; Oh et al., 2013; Luchelli et al., 2015). These neuronal miRISC granules have been shown to re-localize to distal dendritic sites and post-synapse on synaptic activity (Cougot et al., 2008; Oh et al., 2013). Furthermore, synaptic stimulation has been shown to result in rapid loss of miRISC protein AGO2 from these neuronal miRISC granules (Cougot et al., 2008). A recent study demonstrated the important role of XRN1 containing synaptic granules in translation regulation downstream of different glutamate receptors (Luchelli et al., 2015). The authors characterized the presence of novel XRN1 granules at synapse named as synaptic XRN1 (SX) bodies, which did not contain any canonical P body components like DCP1a, EDC4, or RCK/P54. Further, it was shown that the synaptic concentration of SX bodies is affected by synaptic stimulation and is inversely correlated with the rate of local translation. Interestingly, disruption of SX bodies by XRN1 knockdown resulted in abrogation of NMDAR mediated translation repression response.
Compositionally distinct miRISC complexes have been shown to form in response to synaptic stimulation. These complexes play a crucial role in regulating the translation of specific mRNA downstream of synaptic activity. Drosophila miRISC protein AGO1, dFMR1, and Me31B were shown to interact with Ataxin-2 to regulate long term olfactory habituation and translation of CaMKII mRNA (McCann et al., 2011; Sudhakaran et al., 2014). Synaptic activation has been shown to modulate miRISC composition by affecting expression, post-translational modifications, subcellular localization and interactions of different miRISC constituents.
Work from Gary Bassell's lab established FMRP phosphorylation as a reversible switch in regulation miRISC mediated repression of specific target mRNAs (Muddashetty et al., 2011). This study demonstrated that phosphorylated FMRP associates with AGO2 and miR-125a to inhibit PSD-95 translation. mGluR activation causes FMRP phosphorylation, leading to dissociation of inhibitory miRISC complex from FMRP-bound PSD95 and its subsequent translation. Further studies suggested that mGluR induced dephosphorylation of FMRP facilitates its ubiquitination and proteasomal degradation (Nalavadi et al., 2012). Important questions like the interaction of FMRP with miRISC protein GW182 and how FMRP prevents miRISC mediated target degradation require further investigation. Surprisingly, the role of FMRP-miRISC mediated translation regulation downstream of NMDAR signaling remains relatively unexplored. Albeit, a recent study from our lab have suggested an important role of AGO2-MOV10-FMRP contain miRISC complexes in regulating NMDAR mediated translation response (Kute et al., 2019). This work shows that dissociation of MOV10 from AGO2 complexes is required for NMDAR mediated translation activation of a specific subset of mRNAs, such as PTEN and PSD95. NMDAR induced dissociation of MOV10 complexes from AGO2 requires phosphorylated FMRP. Consequentially, NMDAR induced translation of these specific target mRNAs is defective in FMR1 KO animals.
Modulating the phosphorylation of AGO2 is shown to be a mechanism employed for regulating NMDAR mediated translation response and spine dynamics (Antoniou et al., 2014; Rajgor et al., 2017, 2018). Hanley's lab has identified AGO2 as an interacting partner of PDZ and BAR domain-containing scaffolding protein: Protein Interacting with C Kinase 1 (PICK1) in neurons (Antoniou et al., 2014). PICK interaction with AGO2 has shown to localize AGO2 to recycling endosomes, and suppress AGO2 mediated miRNA repression (Antoniou et al., 2014). The authors showed that NMDAR induced long term depression (LTD) mediated increase in Ca2+ results in a reduction of PICK-AGO2 interaction, leading to AGO2 dissociation from endosomes. This led to a subsequent increase in AGO2 mediated repression by dendritically localized miRNAs like miR-134 and miR-138 (Rajgor et al., 2017). A later study from the same lab demonstrated that NMDAR stimulation results in increased interaction of AGO2 with GW182 and DDX6. The increased interaction was mediated by S387 phosphorylation of AGO2 via the AKT pathway and was essential for NMDAR induced spine shrinkage. On the contrary, another study has reported that NMDAR activation leads to the dephosphorylation of AGO2 at S387 (Paradis-Isler and Boehm, 2018). This dephosphorylation was shown to induce degradation of AGO2 resulting in de-repression of miRNA mediated silencing and subsequent spine growth and maturation. The explanation for such contradictory results could be the temporally distinct effect of NMDAR stimulation on synaptic translation and hence miRISC composition, an area needs to further explored. The above reports suggest that a difference in stimulation paradigm used and the time of experimentation after NMDAR stimulation (immediate vs. post-recovery experimentation), determines the phosphorylation response of AGO2 and resulting in distinct spine phenotypes.
Different miRISC components are degraded downstream of neuronal activity to mediate synaptic plasticity. For example, long term memory formation in drosophila NMJ is regulated by activity mediated rapid degradation of miRISC protein Armitage (Ashraf et al., 2006). Activity mediated Armitage degradation is required for the translation of synaptic mRNA CaMKII, which is further required for the induction of long-term potentiation (LTP). A similar mechanism has been described in hippocampal neurons where NMDAR mediated proteasomal degradation of miRISC protein MOV10 (Armitage homolog) leads to the translation of several synaptic mRNAs, such as LIMK1, CaMKIIα, and Lyp1a (Banerjee et al., 2009). These were among the first reports to show the importance of miRISC composition in reversible regulation of synaptic translation. The expression of deadenylase CCR4-NOT transcription complex subunit 7 (CNOT7) is regulated downstream of synaptic activity (Rajgor et al., 2017). The expression of CNOT7 drastically decreases upon glycine induced long-term potentiation of cultured hippocampal neurons (Rajgor et al., 2017). Furthermore, shRNA mediated knockdown of CNOT7 is shown to disrupt LTP, suggesting an important role of modulation of CNOT7 expression in LTP. Currently, it has not been established whether this reduction in CN0T7 also results in reduced activity of canonical miRISC. In short, miRISC reveals to be an important hub of regulation downstream of different synaptic signaling pathways and is crucial in mediating synaptic plasticity.
The work we discussed in this review highlight the importance of dynamic miRISC in normal neuronal development. Several miRISC proteins are shown to have a significant role during distinct stages of neuronal development as summarized in Table 1. But what is lacking is the understanding to link the role of these proteins in the context of miRISC function to their physiological impact. To get this insight, it is important to consider miRISC as a dynamic translation modulator rather than just as mRNA degradation machinery. In this review, we have analyzed how individual RBPs association with core miRISC may alter the kinetics and translation and degradation of a specific subset of mRNAs. Making a distinction between the “core” and “ancillary” components of miRISC helps us understand the role of many RBPs refereed to be part of miRISC as individual components or in combination. The key intention of this review is to reanalyze the role of the microRNA associated RNA binding proteins involved in neuronal development by their potential contribution to the dynamic nature of miRISC function.
Finally, it is important to highlight the role of the dynamic composition of miRISC in normal neuronal development as well as its potential implication in disorders of neuronal development. Indeed, defective translational regulation is a common feature of multiple neurodevelopmental and neuropsychiatric disorders (Kapur et al., 2017; Chen et al., 2019; Neelagandan et al., 2019; Zhou et al., 2019). The role of miRNA mediated regulation of gene expression has been widely studied in these disease conditions. Most of these studies have focused on the altered miRNA levels in neuronal disorders. While this is informative, we argue that it is also important to investigate the composition of miRISC under these disease conditions. Fragile X syndrome (FXS) could be an ideal case study, where the loss of FMRP (a well-studied miRISC associated RBP) leads to a significant defect in neuronal development (Jin et al., 2004; Bassell and Warren, 2008). Here, the role of FMRP regulating the miRISC function to the pathophysiology of FXS is tentatively established (Edbauer et al., 2010; Muddashetty et al., 2011; Kute et al., 2019; Ramakrishna and Muddashetty, 2019). FMRP and many other RBPs are likely to play a similar role in the pathophysiology of neurodevelopmental disorders due to their association with miRISC. Further studies are needed to elucidate this link which may help in the broader molecular understanding of neurodevelopmental disorders.
Recent SFARI study has identified missense mutations in miRISC proteins AGO1, TNRC6B, and CNOT3 among patients diagnosed with Autism Spectrum Disorders (Chen et al., 2019) Patients with microdeletions of chromosomal region 1p34.3 encompassing the AGO1 and AGO3 genes also show several neurodevelopmental defects (Tokita et al., 2015). Similarly, studies have also identified a crucial role of miRISC in neurodegenerative disorders. Reduced expression of AGO2 was observed in the brain of the rodent model for multiple sclerosis. This was further correlated with reduced AGO2-GW182 interaction and dysregulated miRISC assembly (Lewkowicz et al., 2015). In fact, miRISC protein GW182 was first identified from the serum auto-antibodies of patients with neuropathic symptoms (Eystathioy et al., 2002, 2003), and changes in its levels are implicated in Alzheimer's disease (Rouillard et al., 2016; Badhwar et al., 2017). Altered localization and trafficking of miRISC was recently reported in a cell culture model of amyotrophic lateral sclerosis (ALS) (Gershoni-Emek et al., 2018). Moreover, recent studies highlighting the interactions of miRISC components with proteins involved in neurodegenerative disorders, such as HTT, FUS, and Ataxin points toward an important role of miRISC composition in the pathology of these disease which should be investigated in detail (Savas et al., 2008; McCann et al., 2011; Zhang T. et al., 2018). Further, the above-mentioned studies showed that mutant versions of HTT and FUS impair miRNA mediated silencing, providing further evidence for the role of miRISC composition in driving neurodegenerative disorders (Savas et al., 2008; Zhang T. et al., 2018). All these studies indicate that it is not miRNA alone, but the composition of miRISC has a great significance in the pathophysiology of disorders of the nervous system. There is a great need and enormous potential for understanding the role of the dynamic composition and function of miRISC in health and disease-related to the development of the nervous system.
BN, SR, and RM reviewed the literature, conceptualized, and wrote the manuscript.
This work was financially supported by the Science and Engineering Research Board (SERB), Department of Science & Technology (EMR/2016/006313) to RM. BN was funded by the CSIR-SRF fellowship [Award No: 09/860(0172)/2015-EMR-1]. SR was funded by the CSIR-SPM fellowship [Award No: SPM-07/860(0202)/2014].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Akamatsu, W., Fujihara, H., Mitsuhashi, T., Yano, M., Shibata, S., Hayakawa, Y., et al. (2005). The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc. Natl. Acad. Sci. U.S.A. 102, 4625–4630. doi: 10.1073/pnas.0407523102
Akamatsu, W., Okano, H. J., Osumi, N., Inoue, T., Nakamura, S., Sakakibara, S. I., et al. (2002). Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc. Natl. Acad. Sci. U.S.A. 96, 9885–9890. doi: 10.1073/pnas.96.17.9885
Anderson, K. D., Morin, M. A., Beckel-Mitchener, A., Mobarak, C. D., Neve, R. L., Furneaux, H. M., et al. (2000). Overexpression of HuD, but not of its truncated form HuD I+II, promotes GAP-43 gene expression and neurite outgrowth in PC12 cells in the absence of nerve growth factor. J. Neurochem. 75, 1103–1114. doi: 10.1046/j.1471-4159.2000.0751103.x
Antonacci, S., Forand, D., Wolf, M., Tyus, C., Barney, J., Kellogg, L., et al. (2015). Conserved RNA-binding proteins required for dendrite morphogenesis in Caenorhabditis elegans sensory neurons. G3 (Bethesda) 5, 639–653. doi: 10.1534/g3.115.017327
Antoniou, A., Baptista, M., Carney, N., and Hanley, J. G. (2014). PICK1 links Argonaute 2 to endosomes in neuronal dendrites and regulates miRNA activity. EMBO Rep. 15, 548–556. doi: 10.1002/embr.201337631
Arikkath, J. (2012). Molecular mechanisms of dendrite morphogenesis. Front. Cell. Neurosci. 6:61. doi: 10.3389/fncel.2012.00061
Ashraf, S. I., McLoon, A. L., Sclarsic, S. M., and Kunes, S. (2006). Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell 124, 191–205. doi: 10.1016/j.cell.2005.12.017
Badhwar, A., Brown, R., Stanimirovic, D. B., Haqqani, A. S., and Hamel, E. (2017). Proteomic differences in brain vessels of Alzheimer's disease mice: normalization by PPARI 3 agonist pioglitazone. J. Cereb. Blood Flow Metab. 37, 1120–1136. doi: 10.1177/0271678X16655172
Baek, D., Villén, J., Shin, C., Camargo, F. D., Gygi, S. P., and Bartel, D. P. (2008). The impact of microRNAs on protein output. Nature 455, 64–71. doi: 10.1038/nature07242
Baillat, D., and Shiekhattar, R. (2009). Functional dissection of the human TNRC6 (GW182-related) family of proteins. Mol. Cell. Biol. 29, 4144–4155. doi: 10.1128/MCB.00380-09
Banerjee, S., Neveu, P., and Kosik, K. S. (2009). A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron 64, 871–884. doi: 10.1016/j.neuron.2009.11.023
Barbee, S. A., Estes, P. S., Cziko, A. M., Hillebrand, J., Luedeman, R. A., Coller, J. M., et al. (2006). Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52, 997–1009. doi: 10.1016/j.neuron.2006.10.028
Bassell, G. J., and Warren, S. T. (2008). Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron 60, 201–214. doi: 10.1016/j.neuron.2008.10.004
Bazzini, A. A., Lee, M. T., and Giraldez, A. J. (2012). Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336, 233–237. doi: 10.1126/science.1215704
Behm-Ansmant, I., Rehwinkel, J., Doerks, T., Stark, A., Bork, P., and Izaurralde, E. (2006). mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 20, 1885–1898. doi: 10.1101/gad.1424106
Bello, B. (2006). The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development 133, 2639–2648. doi: 10.1242/dev.02429
Bellon, A., Iyer, A., Bridi, S., Lee, F. C. Y., Ovando-Vázquez, C., Corradi, E., et al. (2017). miR-182 regulates Slit2-mediated axon guidance by modulating the local translation of a specific mRNA. Cell Rep. 18, 1171–1186. doi: 10.1016/j.celrep.2016.12.093
Benitez, M. J., Sanchez-Ponce, D., Garrido, J. J., and Wandosell, F. (2014). Hsp90 activity is necessary to acquire a proper neuronal polarization. Biochim. Biophys. Acta Mol. Cell Res. 1843, 245–252. doi: 10.1016/j.bbamcr.2013.11.013
Béthune, J., Artus-Revel, C. G., and Filipowicz, W. (2012). Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 13, 716–723. doi: 10.1038/embor.2012.82
Betschinger, J., Mechtler, K., and Knoblich, J. A. (2006). Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124, 1241–1253. doi: 10.1016/j.cell.2006.01.038
Bhattacharyya, S. N., Habermacher, R., Martine, U., Closs, E. I., and Filipowicz, W. (2006). Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125, 1111–1124. doi: 10.1016/j.cell.2006.04.031
Biever, A., Donlin-Asp, P. G., and Schuman, E. M. (2019). Local translation in neuronal processes. Curr. Opin. Neurobiol. 57, 141–148. doi: 10.1016/j.conb.2019.02.008
Blair, J. D., Hockemeyer, D., Doudna, J. A., Bateup, H. S., and Floor, S. N. (2017). Widespread translational remodeling during human neuronal differentiation. Cell Rep. 21, 2005–2016. doi: 10.1016/j.celrep.2017.10.095
Blumenthal, J., Behar, L., Elliott, E., and Ginzburg, I. (2009). Dcp1a phosphorylation along neuronal development and stress. FEBS Lett. 583, 197–201. doi: 10.1016/j.febslet.2008.12.002
Boland, A., Huntzinger, E., Schmidt, S., Izaurralde, E., and Weichenrieder, O. (2011). Crystal structure of the MID-PIWI lobe of a eukaryotic argonaute protein. Proc. Natl. Acad. Sci. U.S.A. 108, 10466–10471. doi: 10.1073/pnas.1103946108
Bowman, S. K., Rolland, V., Betschinger, J., Kinsey, K. A., Emery, G., and Knoblich, J. A. (2008). The tumor suppressors brat and numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell 14, 535–546. doi: 10.1016/j.devcel.2008.03.004
Bridge, K. S., Shah, K. M., Li, Y., Foxler, D. E., Wong, S. C. K., Miller, D. C., et al. (2017). Argonaute utilization for miRNA silencing is determined by phosphorylation-dependent recruitment of LIM-domain-containing proteins. Cell Rep. 20, 173–187. doi: 10.1016/j.celrep.2017.06.027
Buchsbaum, I. Y., and Cappello, S. (2019). Neuronal migration in the CNS during development and disease: insights from in vivo and in vitro models. Development 146:dev163766. doi: 10.1242/dev.163766
Burow, D. A., Umeh-Garcia, M. C., True, M. B., Bakhaj, C. D., Ardell, D. H., and Cleary, M. D. (2015). Dynamic regulation of mRNA decay during neural development. Neural Dev. 10:11. doi: 10.1186/s13064-015-0038-6
Burry, R. W. (1985). Protein synthesis requirement for the formation of synaptic elements. Brain Res. 344, 109–119. doi: 10.1016/0006-8993(85)91194-1
Cajigas, I. J., Will, T., and Schuman, E. M. (2010). Protein homeostasis and synaptic plasticity. EMBO J. 29, 2746–2752. doi: 10.1038/emboj.2010.173
Callan, M. A., Cabernard, C., Heck, J., Luois, S., Doe, C. Q., and Zarnescu, D. C. (2010). Fragile X protein controls neural stem cell proliferation in the Drosophila brain. Hum. Mol. Genet. 19, 3068–3079. doi: 10.1093/hmg/ddq213
Campbell, D. S., and Holt, C. E. (2001). Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron 32, 1013–1026. doi: 10.1016/s0896-6273(01)00551-7
Carmell, M. A., Hannon, G. J., Marsden, C. G., Rivas, F. V., Thomson, J. M., Liu, J., et al. (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441. doi: 10.1126/science.1102513
Castren, M., Tervonen, T., Karkkainen, V., Heinonen, S., Castren, E., Larsson, K., et al. (2005). Altered differentiation of neural stem cells in fragile X syndrome. Proc. Natl. Acad. Sci. U.S.A. 102, 17834–17839. doi: 10.1073/pnas.0508995102
Cenik, E. S., and Zamore, P. D. (2011). Argonaute proteins. Curr. Biol. 21, R446–R449. doi: 10.1016/j.cub.2011.05.020
Chan, S. P., and Slack, F. J. (2009). Ribosomal protein RPS-14 modulates let-7 microRNA function in Caenorhabditis elegans. Dev. Biol. 334, 152–160. doi: 10.1016/j.ydbio.2009.07.011
Chekulaeva, M., Filipowicz, W., and Parker, R. (2009). Multiple independent domains of dGW182 function in miRNA- mediated repression in Drosophila. RNA 15, 794–803. doi: 10.1261/rna.1364909
Chekulaeva, M., Mathys, H., Zipprich, J. T., Attig, J., Colic, M., Parker, R., et al. (2011). MiRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 18, 1218–1226. doi: 10.1038/nsmb.2166
Chen, C., Ito, K., Takahashi, A., Wang, G., Suzuki, T., Nakazawa, T., et al. (2011). Distinct expression patterns of the subunits of the CCR4-NOT deadenylase complex during neural development. Biochem. Biophys. Res. Commun. 411, 360–364. doi: 10.1016/j.bbrc.2011.06.148
Chen, H. H., Yu, H. I., Chiang, W. C., Lin, Y. D., Shia, B. C., and Tarn, W. Y. (2012). hnRNP Q regulates Cdc42-mediated neuronal morphogenesis. Mol. Cell. Biol. 32, 2224–2238. doi: 10.1128/MCB.06550-11
Chen, Y. C., Chang, Y. W., and Huang, Y. S. (2019). Dysregulated translation in neurodevelopmental disorders: an overview of autism-risk genes involved in translation. Dev. Neurobiol. 79, 60–74. doi: 10.1002/dneu.22653
Chiang, P. Y., Shen, Y. F., Su, Y. L., Kao, C. H., Lin, N. Y., Hsu, P. H., et al. (2013). Phosphorylation of mRNA decapping protein Dcp1a by the ERK signaling pathway during early differentiation of 3T3-L1 preadipocytes. PLoS ONE 8:e61697. doi: 10.1371/journal.pone.0061697
Chicurel, M., Terrian, D., and Potter, H. (1993). mRNA at the synapse: analysis of a synaptosomal preparation enriched in hippocampal dendritic spines. J. Neurosci. 13, 4054–4063. doi: 10.1523/JNEUROSCI.13-09-04054.1993
Chinen, M., and Lei, E. P. (2017). Drosophila Argonaute2 turnover is regulated by the ubiquitin proteasome pathway. Biochem. Biophys. Res. Commun. 483, 951–957. doi: 10.1016/j.bbrc.2017.01.039
Costa-Mattioli, M., Sossin, W. S., Klann, E., and Sonenberg, N. (2009). Translational control of long-lasting synaptic plasticity and memory. Neuron 61, 10–26. doi: 10.1016/j.neuron.2008.10.055
Cougot, N., Bordonné, R., Bertrand, E., Rage, F., Bhattacharyya, S. N., Filipowicz, W., et al. (2008). Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J. Neurosci. 28, 13793–13804. doi: 10.1523/JNEUROSCI.4155-08.2008
Dajas-Bailador, F., Bonev, B., Garcez, P., Stanley, P., Guillemot, F., and Papalopulu, N. (2012). MicroRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat. Neurosci. 15, 697–699. doi: 10.1038/nn.3082
Dallaire, A., Frédérick, P. M., and Simard, M. J. (2018). Somatic and germline microRNAs form distinct silencing complexes to regulate their target mRNAs differently. Dev. Cell 47, 239–247.e4. doi: 10.1016/j.devcel.2018.08.022
Dayeh, D. M., Kruithoff, B. C., and Nakanishi, K. (2018). Structural and functional analyses reveal the contributions of the C- and N-lobes of Argonaute protein to selectivity of RNA target cleavage. J. Biol. Chem. 293, 6308–6325. doi: 10.1074/jbc.RA117.001051
Dear, M. L., Shilts, J., and Broadie, K. (2017). Neuronal activity drives FMRP- and HSPG-dependent matrix metalloproteinase function required for rapid synaptogenesis. Sci. Signal. 10, 36–39. doi: 10.1126/scisignal.aan3181
Dhananjaya D., Hung, K. Y., and Tarn, W. Y. (2018). RBM4 Modulates radial migration via alternative splicing of Dab1 during cortex development. Mol. Cell. Biol. 38, e00007–18. doi: 10.1128/MCB.00007-18
Dickson, B. J. (2002). Molecular mechanisms of axon guidance. Science 298, 1959–1964. doi: 10.1126/science.1072165
Didiot, M. C., Subramanian, M., Flatter, E., Mandel, J. L., and Moine, H. (2008). Cells lacking the fragile X mental retardation protein (FMRP) have normal RISC activity but exhibit altered stress granule assembly. Mol. Biol. Cell 20, 428–437. doi: 10.1091/mbc.e08-07-0737
Ding, L., and Han, M. (2007). GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 17, 411–416. doi: 10.1016/j.tcb.2007.06.003
Djuranovic, S., Nahvi, A., and Green, R. (2011). A parsimonious model for gene regulation by miRNAs. Science 331, 550–553. doi: 10.1126/science.1191138
Djuranovic, S., Nahvi, A., and Green, R. (2012). miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336, 237–241. doi: 10.1126/science.1215691
Dobashi, Y., Shoji, M., Wakata, Y., and Kameya, T. (1998). Expression of HuD protein is essential for initial phase of neuronal differentiation in rat pheachromocytoma PC12 cells. Biochem. Biophys. Res. Commun. 244, 226–229. doi: 10.1006/bbrc.1998.8247
Dockendorff, T. C., Su, H. S., McBride, S. M. J., Yang, Z., Choi, C. H., Siwicki, K. K., et al. (2002). Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron 34, 973–984. doi: 10.1016/s0896-6273(02)00724-9
Doll, C. A., and Broadie, K. (2015). Activity-dependent FMRP requirements in development of the neural circuitry of learning and memory. Development 142, 1346–1356. doi: 10.1242/dev.117127
Doll, C. A., Vita, D. J., and Broadie, K. (2017). Fragile X mental retardation protein requirements in activity-dependent critical period neural circuit refinement. Curr. Biol. 27, 2318–2330.e3. doi: 10.1016/j.cub.2017.06.046
Duchaine, T. F., and Fabian, M. R. (2019). Mechanistic insights into microrna-mediated gene silencing. Cold Spring Harb. Perspect. Biol. 11, 1–22. doi: 10.1101/cshperspect.a032771
Edbauer, D., Neilson, J. R., Foster, K. A., Wang, C. F., Seeburg, D. P., Batterton, M. N., et al. (2010). Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65, 373–384. doi: 10.1016/j.neuron.2010.01.005
Eulalio, A., Tritschler, F., and Izaurralde, E. (2009). The GW182 protein family in animal cells: new insights into domains required for miRNA-mediated gene silencing. RNA 15, 1433–1442. doi: 10.1261/rna.1703809
Eystathioy, T., Chan, E. K. L., Tenenbaum, S. A., Keene, J. D., Griffith, K., and Fritzler, M. J. (2002). Associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell 13, 1338–1351. doi: 10.1091/mbc.01-11-0544
Eystathioy, T., Fritzler, M. J., Jakymiw, A., Chan, E. K. L., Séraphin, B., and Cougot, N. (2003). The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9, 1171–1173. doi: 10.1261/rna.5810203
Fabian, M. R., Cieplak, M. K., Frank, F., Morita, M., Green, J., Srikumar, T., et al. (2011). MiRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat. Struct. Mol. Biol. 18, 1211–1217. doi: 10.1038/nsmb.2149
Fabian, M. R., and Sonenberg, N. (2012). The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 19, 586–593. doi: 10.1038/nsmb.2296
Fabian, M. R., Sonenberg, N., and Filipowicz, W. (2010). Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379. doi: 10.1146/annurev-biochem-060308-103103
Friend, K., Campbell, Z. T., Cooke, A., Kroll-Conner, P., Wickens, M. P., and Kimble, J. (2012). A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat. Struct. Mol. Biol. 19, 176–184. doi: 10.1038/nsmb.2214
Fukao, A., Aoyama, T., and Fujiwara, T. (2015). The molecular mechanism of translational control via the communication between the microRNA pathway and RNA-binding proteins. RNA Biol. 12, 922–926. doi: 10.1080/15476286.2015.1073436
Fukao, A., Mishima, Y., Takizawa, N., Oka, S., Imataka, H., Pelletier, J., et al. (2014). MicroRNAs trigger dissociation of eIF4AI and eIF4AII from target mRNAs in humans. Mol. Cell 56, 79–89. doi: 10.1016/j.molcel.2014.09.005
Fukaya, T., Iwakawa, H. oki, and Tomari, Y. (2014). MicroRNAs block assembly of eIF4F translation initiation complex in Drosophila. Mol. Cell 56, 67–78. doi: 10.1016/j.molcel.2014.09.004
Gardiner, A. S., Twiss, J. L., and Perrone-Bizzozero, N. I. (2015). Competing interactions of RNA-binding proteins, MicroRNAs, and their targets control neuronal development and function. Biomolecules 5, 2903–2918. doi: 10.3390/biom5042903
Gershoni-Emek, N., Altman, T., Ionescu, A., Costa, C. J., Gradus-Pery, T., Willis, D. E., et al. (2018). Localization of RNAi machinery to axonal branch points and growth cones is facilitated by mitochondria and is disrupted in ALS. Front. Mol. Neurosci. 11, 1–17. doi: 10.3389/fnmol.2018.00311
Golden, R. J., Chen, B., Li, T., Braun, J., Manjunath, H., Chen, X., et al. (2017). An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature 542, 197–202. doi: 10.1038/nature21025
Guo, H., Ingolia, N. T., Weissman, J. S., and Bartel, D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840. doi: 10.1038/nature09267
Hall, T. M. T. (2005). Structure and function of argonaute proteins. Structure 13, 1403–1408. doi: 10.1016/j.str.2005.08.005
Hammell, C. M., Lubin, I., Boag, P. R., Blackwell, T. K., and Ambros, V. (2009). nhl-2 Modulates MicroRNA Activity in Caenorhabditis elegans. Cell 136, 926–938. doi: 10.1016/j.cell.2009.01.053
Hanus, C., and Schuman, E. M. (2013). Proteostasis in complex dendrites. Nat. Rev. Neurosci. 14, 638–648. doi: 10.1038/nrn3546
Hashimoto, Y., Akiyama, Y., and Yuasa, Y. (2013). Multiple-to-multiple relationships between microRNAs and target genes in gastric cancer. PLoS ONE 8:e62589. doi: 10.1371/journal.pone.0062589
Hendrickson, D. G., Hogan, D. J., McCullough, H. L., Myers, J. W., Herschlag, D., Ferrell, J. E., et al. (2009). Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 7, 25–29. doi: 10.1371/journal.pbio.1000238
Hengst, U., Cox, L. J., Macosko, E. Z., and Jaffrey, S. R. (2006). Functional and selective RNA interference in developing axons and growth cones. J. Neurosci. 26, 5727–5732. doi: 10.1523/JNEUROSCI.5229-05.2006
Höck, J., and Meister, G. (2008). The Argonaute protein family. Genome Biol. 9:210. doi: 10.1186/gb-2008-9-2-210
Horman, S. R., Janas, M. M., Litterst, C., Wang, B., MacRae, I. J., Sever, M. J., et al. (2013). Akt-mediated phosphorylation of argonaute 2 downregulates cleavage and upregulates translational repression of microRNA targets. Mol. Cell 50, 356–367. doi: 10.1016/j.molcel.2013.03.015
Huang, V., and Li, L. C. (2014). Demystifying the nuclear function of Argonaute proteins. RNA Biol. 11, 18–24. doi: 10.4161/rna.27604
Huang, Y. W. A., Ruiz, C. R., Eyler, E. C. H., Lin, K., and Meffert, M. K. (2012). Dual regulation of miRNA biogenesis generates target specificity in neurotrophin-induced protein synthesis. Cell 148, 933–946. doi: 10.1016/j.cell.2012.01.036
Huntzinger, E., and Izaurralde, E. (2011). Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12, 99–110. doi: 10.1038/nrg2936
Hutvagner, G., and Simard, M. J. (2008). Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9, 22–32. doi: 10.1038/nrm2321
Hwang, D. W., Jaganathan, A., Shrestha, P., Jin, Y., El-Amine, N., Wang, S. H., et al. (2018). Chromatin-mediated translational control is essential for neural cell fate specification. Life Sci. Alliance 1, 1–13. doi: 10.26508/lsa.201700016
Ivanco, T. L., and Greenough, W. T. (2002). Altered mossy fiber distributions in adult Fmr1 (FVB) knockout mice. Hippocampus 12, 47–54. doi: 10.1002/hipo.10004
Jafarifar, F., Yao, P., Eswarappa, S. M., and Fox, P. L. (2011). Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J. 30, 1324–1334. doi: 10.1038/emboj.2011.38
James, V., Wong, S. C. K., and Sharp, T. V. (2012). MicroRNA-mediated gene silencing: are we close to a unifying model? Biomol. Concepts 3, 29–40. doi: 10.1515/bmc.2011.047
Janga, S. C., and Vallabhaneni, S. (2011). “MicroRNAs as post-transcriptional machines and their interplay with cellular networks,” in RNA Infrastructure and Networks, ed L. J. Collins (New York, NY: Springer New York), 59–74. doi: 10.1007/978-1-4614-0332-6_4
Jannot, G., Bajan, S., Giguère, N. J., Bouasker, S., Banville, I. H., Piquet, S., et al. (2011). The ribosomal protein RACK1 is required for microRNA function in both C. elegans and humans. EMBO Rep. 12, 581–586. doi: 10.1038/embor.2011.66
Jeon, S. J., Kim, J. W., Kim, K. C., Han, S. M., Go, H. S., Seo, J. E., et al. (2014). Translational regulation of NeuroD1 expression by FMRP: Involvement in glutamatergic neuronal differentiation of cultured rat primary neural progenitor cells. Cell. Mol. Neurobiol. 34, 297–305. doi: 10.1007/s10571-013-0014-9
Jiang, X., and Nardelli, J. (2016). Cellular and molecular introduction to brain development. Neurobiol. Dis. 92, 3–17. doi: 10.1016/j.nbd.2015.07.007
Jin, P., Zarnescu, D. C., Ceman, S., Nakamoto, M., Mowrey, J., Jongens, T. A., et al. (2004). Biochemical and genetic interaction between the fragile X mental retardation protein 3nd the microRNA pathway. Nat. Neurosci. 7, 113–117. doi: 10.1038/nn1174
Jonas, S., and Izaurralde, E. (2015). Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 16, 421–433. doi: 10.1038/nrg3965
Juvvuna, P. K., Khandelia, P., Lee, L. M., and Makeyev, E. V. (2012). Argonaute identity defines the length of mature mammalian microRNAs. Nucleic Acids Res. 40, 6808–6820. doi: 10.1093/nar/gks293
Kalantari, R., Chiang, C. M., and Corey, D. R. (2016). Regulation of mammalian transcription and splicing by nuclear RNAi. Nucleic Acids Res. 44, 524–537. doi: 10.1093/nar/gkv1305
Kamenska, A., Simpson, C., Vindry, C., Broomhead, H., Bénard, M., Ernoult-Lange, M., et al. (2016). The DDX6-4E-T interaction mediates translational repression and P-body assembly. Nucleic Acids Res. 44, 6318–6334. doi: 10.1093/nar/gkw565
Kang, H., and Schuman, E. M. (1996). A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science 273, 1402–1406. doi: 10.1126/science.273.5280.1402
Kapur, M., Monaghan, C. E., and Ackerman, S. L. (2017). Regulation of mRNA translation in neurons—a matter of life and death. Neuron 96, 616–637. doi: 10.1016/j.neuron.2017.09.057
Kasashima, K., Terashima, K., Yamamoto, K., Sakashita, E., and Sakamoto, H. (1999). Cytoplasmic localization is required for the mammalian ELAV-like protein HuD to induce neuronal differentiation. Genes Cells 4, 667–683. doi: 10.1046/j.1365-2443.1999.00292.x
Katz, L. C., Driscoll, M., Zimmermann, K., and Wiesel, T. N. (1992). Neural development. Brain Res. Rev. 17, 171–181. doi: 10.1016/0165-0173(92)90013-C
Katz, S., Cussigh, D., Urbán, N., Blomfield, I., Guillemot, F., Bally-Cuif, L., et al. (2016). A nuclear role for miR-9 and Argonaute proteins in balancing quiescent and activated neural stem cell states. Cell Rep. 17, 1383–1398. doi: 10.1016/j.celrep.2016.09.088
Kauderer, B. S., and Kandel, E. R. (2000). Capture of a protein synthesis-dependent component of long-term depression. Proc. Natl. Acad. Sci. U.S.A. 97, 13342–13347. doi: 10.1073/pnas.97.24.13342
Kedde, M., Strasser, M. J., Boldajipour, B., Vrielink, J. A. F. O., Slanchev, K., le Sage, C., et al. (2007). RNA-binding protein dnd1 inhibits microRNA access to target mRNA. Cell 131, 1273–1286. doi: 10.1016/j.cell.2007.11.034
Kim, E., and Jung, H. (2015). Local protein synthesis in neuronal axons: why and how we study. BMB Rep. 48, 139–146. doi: 10.5483/bmbrep.2015.48.3.010
Koch, P., Löhr, H. B., and Driever, W. (2014). A mutation in cnot8, component of the Ccr4-not complex regulating transcript stability, affects expression levels of developmental regulators and reveals a role of Fgf3 in development of caudal hypothalamic dopaminergic neurons. PLoS ONE 9:e113829. doi: 10.1371/journal.pone.0113829
Konopacki, F. A., Wong, H. H. W., Dwivedy, A., Bellon, A., Blower, M. D., and Holt, C. E. (2016). ESCRT-II controls retinal axon growth by regulating DCC receptor levels and local protein synthesis. Open Biol. 6:150218. doi: 10.1098/rsob.150218
Kraushar, M. L., Thompson, K., Wijeratne, H. R. S., Viljetic, B., Sakers, K., Marson, J. W., et al. (2014). Temporally defined neocortical translation and polysome assembly are determined by the RNA-binding protein Hu antigen R. Proc. Natl. Acad. Sci. U.S.A. 111, E3815–E3824. doi: 10.1073/pnas.1408305111
Kraushar, M. L., Viljetic, B., Wijeratne, H. R. S., Thompson, K., Jiao, X., Pike, J. W., et al. (2015). Thalamic WNT3 secretion spatiotemporally regulates the neocortical ribosome signature and mRNA translation to specify neocortical cell subtypes. J. Neurosci. 35, 10911–10926. doi: 10.1523/JNEUROSCI.0601-15.2015
Krol, J., Loedige, I., and Filipowicz, W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610. doi: 10.1038/nrg2843
Kute, P. M., Ramakrishna, S., Neelagandan, N., Chattarji, S., and Muddashetty, R. S. (2019). NMDAR mediated translation at the synapse is regulated by MOV10 and FMRP. Mol. Brain 12:65. doi: 10.1186/s13041-019-0473-0
La Fata, G., Gärtner, A., Domínguez-Iturza, N., Dresselaers, T., Dawitz, J., Poorthuis, R. B., et al. (2014). FMRP regulates multipolar to bipolar transition affecting neuronal migration and cortical circuitry. Nat. Neurosci. 17, 1693–1700. doi: 10.1038/nn.3870
La Rocca, G., Olejniczak, S. H., González, A. J., Briskin, D., Vidigal, J. A., Spraggon, L., et al. (2015). In vivo, Argonaute-bound microRNAs exist predominantly in a reservoir of low molecular weight complexes not associated with mRNA. Proc. Natl. Acad. Sci. U.S.A. 112, 767–772. doi: 10.1073/pnas.1424217112
Lee, C. Y., Wilkinson, B. D., Siegrist, S. E., Wharton, R. P., and Doe, C. Q. (2006). Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell 10, 441–449. doi: 10.1016/j.devcel.2006.01.017
Lewkowicz, P., Cwiklinska, H., Mycko, M. P., Cichalewska, M., Domowicz, M., Lewkowicz, N., et al. (2015). Dysregulated RNA-induced silencing complex (RISC) assembly within CNS corresponds with abnormal miRNA expression during autoimmune demyelination. J. Neurosci. 35, 7521–7537. doi: 10.1523/JNEUROSCI.4794-14.2015
Li, C. (2009). Fragile X mental retardation protein is involved in protein synthesis-dependent collapse of growth cones induced by Semaphorin-3A. Front. Neural Circuits 3, 1–10. doi: 10.3389/neuro.04.011.2009
Liao, J. M., Zhou, X., Gatignol, A., and Lu, H. (2013). Ribosomal proteins L5 and L11 co-operatively inactivate c-Myc via RNA-induced silencing complex. Oncogene 33, 4916–4923. doi: 10.1038/onc.2013.430
Liu, C., Zhang, X., Huang, F., Yang, B., Li, J., Liu, B., et al. (2012). APOBEC3G inhibits microRNA-mediated repression of translation by interfering with the interaction between Argonaute-2 and MOV10. J. Biol. Chem. 287, 29373–29383. doi: 10.1074/jbc.M112.354001
Liu, H., Lei, C., He, Q., Pan, Z., Xiao, D., and Tao, Y. (2018). Nuclear functions of mammalian microRNAs in gene regulation, immunity and cancer. Mol. Cancer 17, 1–14. doi: 10.1186/s12943-018-0765-5
Loewen, C., Boekhoff-Falk, G., Ganetzky, B., and Chtarbanova, S. (2018). A novel mutation in brain tumor causes both neural over-proliferation and neurodegeneration in adult Drosophila. G3 (Bethesda) 8, 3331–3346. doi: 10.1534/g3.118.200627
Loffreda, A., Rigamonti, A., Barabino, S. M. L., and Lenzken, S. C. (2015). RNA-binding proteins in the regulation of miRNA activity: a focus on neuronal functions. Biomolecules 5, 2363–2387. doi: 10.3390/biom5042363
Luchelli, L., Thomas, M. G., and Boccaccio, G. L. (2015). Synaptic control of mRNA translation by reversible assembly of XRN1 bodies. J. Cell Sci. 128, 1542–1554. doi: 10.1242/jcs.163295
Lugli, G., Larson, J., Martone, M. E., Jones, Y., and Smalheiser, N. R. (2005). Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 94, 896–905. doi: 10.1111/j.1471-4159.2005.03224.x
Martinez, N. J., and Gregory, R. I. (2013). Argonaute2 expression is post-transcriptionally coupled to microRNA abundance. RNA 19, 605–612. doi: 10.1261/rna.036434.112
Marusich, M. F., Furneaux, H. M., Henion, P. D., and Weston, J. A. (1994). Hu neuronal proteins are expressed in proliferating neurogenic cells. J. Neurobiol. 25, 143–155. doi: 10.1002/neu.480250206
McCann, C., Holohan, E. E., Das, S., Dervan, A., Larkin, A., Lee, J. A., et al. (2011). The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc. Natl. Acad. Sci. U.S.A. 108, E655–E662. doi: 10.1073/pnas.1107198108
McCann, C. M., Nguyen, Q. T., Neto, H. S., and Lichtman, J. W. (2007). Rapid synapse elimination after postsynaptic protein synthesis inhibition in vivo. J. Neurosci. 27, 6064–6067. doi: 10.1523/JNEUROSCI.0627-07.2007
McFleder, R. L., Mansur, F., and Richter, J. D. (2017). Dynamic control of dendritic mRNA expression by CNOT7 regulates synaptic efficacy and higher cognitive function. Cell Rep. 20, 683–696. doi: 10.1016/j.celrep.2017.06.078
Meijer, H. A., Kong, Y. W., Lu, W. T., Wilczynska, A., Spriggs, R. V., Robinson, S. W., et al. (2013). Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science 340, 82–85. doi: 10.1126/science.1231197
Meister, G. (2013). Argonaute proteins: Functional insights and emerging roles. Nat. Rev. Genet. 14, 447–459. doi: 10.1038/nrg3462
Menon, K. P., Andrews, S., Murthy, M., Gavis, E. R., and Zinn, K. (2009). The translational repressors nanos and pumilio have divergent effects on presynaptic terminal growth and postsynaptic glutamate receptor subunit composition. J. Neurosci. 29, 5558–5572. doi: 10.1523/JNEUROSCI.0520-09.2009
Menon, K. P., Sanyal, S., Habara, Y., Sanchez, R., Wharton, R. P., Ramaswami, M., et al. (2004). The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron 44, 663–676. doi: 10.1016/j.neuron.2004.10.028
Michel, C. I. (2004). Defective neuronal development in the mushroom bodies of Drosophila fragile X mental retardation 1 mutants. J. Neurosci. 24, 5798–5809. doi: 10.1523/JNEUROSCI.1102-04.2004
Mochida, G. H., Ganesh, V. S., de Michelena, M. I., Dias, H., Atabay, K. D., Kathrein, K. L., et al. (2012). CHMP1A encodes an essential regulator of BMI1-INK4A in cerebellar development. Nat. Genet. 44, 1260–1264. doi: 10.1038/ng.2425
Mohammad, L., Wiseman, J., Erickson, S., Yang, G., Mohammad, L., Wiseman, J., et al. (2019). Protein synthesis and translational control in neural stem cell development and neurogenesis. Brain 140, 582–598. doi: 10.1093/oxfordhb/9780190686307.013.21
Morales, J., Hiesinger, P. R., Schroeder, A. J., Kume, K., Verstreken, P., Jackson, F. R., et al. (2002). Drosophila fragile X protein DFXR regulates neuronal morphology and function in the brain. Neuron 34, 961–972. doi: 10.1016/s0896-6273(02)00731-6
Moretti, F., Kaiser, C., Zdanowicz-Specht, A., and Hentze, M. W. (2012). PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat. Struct. Mol. Biol. 19, 603–608. doi: 10.1038/nsmb.2309
Muddashetty, R., and Bassell, G. J. (2009). A boost in microRNAs shapes up the neuron. EMBO J. 28, 617–618. doi: 10.1038/emboj.2009.51
Muddashetty, R. S., Nalavadi, V. C., Gross, C., Yao, X., Xing, L., Laur, O., et al. (2011). Reversible Inhibition of PSD-95 mRNA Translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol. Cell 42, 673–688. doi: 10.1016/j.molcel.2011.05.006
Munno, D. W., and Syed, N. I. (2003). Synaptogenesis in the CNS: an odyssey from wiring together to firing together. J. Physiol. 552, 1–11. doi: 10.1113/jphysiol.2003.045062
Murashov, A. K., Chintalgattu, V., Islamov, R. R., Lever, T. E., Pak, E. S., Sierpinski, P. L., et al. (2007). RNAi pathway is functional in peripheral nerve axons. FASEB J. 21, 656–670. doi: 10.1096/fj.06-6155com
Nalavadi, V. C., Muddashetty, R. S., Gross, C., and Bassell, G. J. (2012). Dephosphorylation-induced ubiquitination and degradation of FMRP in dendrites: a role in immediate early mGluR-stimulated translation. J. Neurosci. 32, 2582–2587. doi: 10.1523/JNEUROSCI.5057-11.2012
Neelagandan, N., Gonnella, G., Dang, S., Janiesch, P. C., Miller, K. K., Küchler, K., et al. (2019). TDP-43 enhances translation of specific mRNAs linked to neurodegenerative disease. Nucleic Acids Res. 47, 341–361. doi: 10.1093/nar/gky972
Neumüller, R. A., Betschinger, J., Fischer, A., Bushati, N., Poernbacher, I., Mechtler, K., et al. (2008). Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454, 241–245. doi: 10.1038/nature07014
Nganou, G., Silva, C. G., Gladwyn-Ng, I., Engel, D., Coumans, B., Delgado-Escueta, A. V., et al. (2018). Importin-8 modulates division of apical progenitors, dendritogenesis and tangential migration during development of mouse cortex. Front. Mol. Neurosci. 11, 1–14. doi: 10.3389/fnmol.2018.00234
Niaz, S., and Hussain, M. U. (2018). Role of GW182 protein in the cell. Int. J. Biochem. Cell Biol. 101, 29–38. doi: 10.1016/j.biocel.2018.05.009
Nicklas, S., Okawa, S., Hillje, A. L., González-Cano, L., Del Sol, A., and Schwamborn, J. C. (2015). The RNA helicase DDX6 regulates cell-fate specification in neural stem cells via miRNAs. Nucleic Acids Res. 43, 2638–2654. doi: 10.1093/nar/gkv138
Oh, J. Y., Kwon, A., Jo, A., Kim, H., Goo, Y. S., Lee, J. A., et al. (2013). Activity-dependent synaptic localization of processing bodies and their role in dendritic structural plasticity. J. Cell Sci. 126, 2114–2123. doi: 10.1242/jcs.125690
Olejniczak, S. H., La Rocca, G., Gruber, J. J., and Thompson, C. B. (2013). Long-lived microRNA-Argonaute complexes in quiescent cells can be activated to regulate mitogenic responses. Proc. Natl. Acad. Sci. U.S.A. 110, 157–162. doi: 10.1073/pnas.1219958110
Olejniczak, S. H., La Rocca, G., Radler, M. R., Egan, S. M., Xiang, Q., Garippa, R., et al. (2016). Coordinated regulation of cap-dependent translation and microRNA function by convergent signaling pathways. Mol. Cell. Biol. 36, 2360–2373. doi: 10.1128/MCB.01011-15
Olesnicky, E. C., Bhogal, B., and Gavis, E. R. (2012). Combinatorial use of translational co-factors for cell type-specific regulation during neuronal morphogenesis in Drosophila. Dev. Biol. 365, 208–218. doi: 10.1016/j.ydbio.2012.02.028
Olesnicky, E. C., Killian, D. J., Garcia, E., Morton, M. C., Rathjen, A. R., Sola, I. E., et al. (2014). Extensive use of RNA-binding proteins in Drosophila sensory neuron dendrite morphogenesis. G3 (Bethesda) 4, 297–306. doi: 10.1534/g3.113.009795
Pan, L., Zhang, Y. Q., Woodruff, E., and Broadie, K. (2004). The Drosophila fragile X gene negatively regulates neuronal elaboration and synaptic differentiation. Curr. Biol. 14, 1863–1870. doi: 10.1016/j.cub.2004.09.085
Paradis-Isler, N., and Boehm, J. (2018). NMDA receptor-dependent dephosphorylation of serine 387 in Argonaute 2 increases its degradation and affects dendritic spine density and maturation. J. Biol. Chem. 293, 9311–9325. doi: 10.1074/jbc.RA117.001007
Patranabis, S., and Bhattacharyya, S. N. (2016). Phosphorylation of Ago2 and Subsequent Inactivation of let-7a RNP- Specific MicroRNAs Control Differentiation of Mammalian Sympathetic Neurons. Mol Cell Biol. 36, 1260–1271. doi: 10.1128/MCB.00054-16
Patranabis, S., and Bhattacharyya, S. N. (2018). P-body – induced inactivation of let-7a miRNP prevents the death of growth factor – deprived neuronal cells. FASEB J. 32, 1493–1509. doi: 10.1096/fj.201700633R
Peng, Y., and Croce, C. M. (2016). The role of microRNAs in human cancer. Signal Transduct. Target. Ther. 1:15004. doi: 10.1038/sigtrans.2015.4
Perrone-Bizzozero, N., and Bolognani, F. (2002). Role of HuD and other RNA-Binding proteins in neural development and plasticity. J. Neurosci. Res. 68, 121–126. doi: 10.1002/jnr.10175
Peters, L., and Meister, G. (2007). Argonaute proteins: mediators of RNA silencing. Mol. Cell 26, 611–623. doi: 10.1016/j.molcel.2007.05.001
Pfeiffer, B. E., and Huber, K. M. (2006). Current advances in local protein synthesis and synaptic plasticity. J. Neurosci. 26, 7147–7150. doi: 10.1523/JNEUROSCI.1797-06.2006
Pradhan, S. J., Nesler, K. R., Rosen, S. F., Kato, Y., Nakamura, A., Ramaswami, M., et al. (2012). The conserved P body component HPat/Pat1 negatively regulates synaptic terminal growth at the larval Drosophila neuromuscular junction. J. Cell Sci. 125, 6105–6116. doi: 10.1242/jcs.113043
Qi, H. H., Ongusaha, P. P., Myllyharju, J., Cheng, D., Pakkanen, O., Shi, Y., et al. (2008). Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature 455, 421–424. doi: 10.1038/nature07186
Quintá, H. R., Maschi, D., Gomez-Sanchez, C., Piwien-Pilipuk, G., and Galigniana, M. D. (2010). Subcellular rearrangement of hsp90-binding immunophilins accompanies neuronal differentiation and neurite outgrowth. J. Neurochem. 115, 716–734. doi: 10.1111/j.1471-4159.2010.06970.x
Rajgor, D., Fiuza, M., Parkinson, G. T., and Hanley, J. G. (2017). The PICK1 Ca2+ sensor modulates N-methyl-D-aspartate (NMDA) receptor-dependent microRNA-mediated translational repression in neurons. J. Biol. Chem. 292, 9774–9786. doi: 10.1074/jbc.M117.776302
Rajgor, D., Sanderson, T. M., Amici, M., Collingridge, G. L., and Hanley, J. G. (2018). NMDAR-dependent Argonaute 2 phosphorylation regulates miRNA activity and dendritic spine plasticity. EMBO J. 37:e97943. doi: 10.15252/embj.201797943
Ramakrishna, S., and Muddashetty, R. S. (2019). Emerging role of microRNAs in dementia. J. Mol. Biol. 431, 1743–1762. doi: 10.1016/j.jmb.2019.01.046
Rao, A., and Steward, O. (1993). Evaluation of RNAs present in synaptodendrosomes: dendritic, glial, and neuronal cell body contribution. J. Neurochem. 61, 835–844. doi: 10.1111/j.1471-4159.1993.tb03594.x
Ravindran, S., Nalavadi, V. C., and Muddashetty, R. S. (2019). BDNF induced translation of limk1 in developing neurons regulates dendrite growth by fine-tuning cofilin1 activity. Front. Mol. Neurosci. 12, 1–14. doi: 10.3389/fnmol.2019.00064
Rehwinkel, J., Behm-Ansmant, I., Gatfield, D., and Izaurralde, E. (2005). A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11, 1640–1647. doi: 10.1261/rna.2191905
Ross, J. P., and Kassir, Z. (2014). The varied roles of nuclear Argonaute-small RNA complexes and avenues for therapy. Mol. Ther. Nucleic Acids 3:e203. doi: 10.1038/mtna.2014.54
Rouillard, A. D., Gundersen, G. W., Fernandez, N. F., Wang, Z., Monteiro, C. D., McDermott, M. G., et al. (2016). The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, 1–16. doi: 10.1093/database/baw100
Rüdel, S., Wang, Y., Lenobel, R., Körner, R., Hsiao, H. H., Urlaub, H., et al. (2011). Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res. 39, 2330–2343. doi: 10.1093/nar/gkq1032
Saffary, R., and Xie, Z. (2011). FMRP Regulates the transition from radial glial cells to intermediate progenitor cells during neocortical development. J. Neurosci. 31, 1427–1439. doi: 10.1523/JNEUROSCI.4854-10.2011
Sahin, U., Lapaquette, P., Andrieux, A., Faure, G., and Dejean, A. (2014). Sumoylation of human Argonaute 2 at lysine-402 regulates its stability. PLoS ONE 9:e102957. doi: 10.1371/journal.pone.0102957
Savas, J. N., Makusky, A., Ottosen, S., Baillat, D., Then, F., Krainc, D., et al. (2008). Huntington's disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proc. Natl. Acad. Sci. U.S.A. 105, 10820–10825. doi: 10.1073/pnas.0800658105
Scarnati, M. S., Kataria, R., Biswas, M., and Paradiso, K. G. (2018). Active presynaptic ribosomes in the mammalian brain, and altered transmitter release after protein synthesis inhibition. Elife 7, 1–28. doi: 10.7554/eLife.36697
Scheetz, A. J., Nairn, A. C., and Constantine-Paton, M. (2000). NMDA receptor-mediated control of protein synthesis at developing synapses. Nat. Neurosci. 3, 211–216. doi: 10.1038/72915
Schwamborn, J. C., Berezikov, E., and Knoblich, J. A. (2009). The TRIM-NHL protein TRIM32 activates micrornas and prevents self-renewal in mouse neural progenitors. Cell 136, 913–925. doi: 10.1016/j.cell.2008.12.024
Semple, B. D., Blomgren, K., Gimlin, K., Ferriero, D. M., and Noble-Haeusslein, L. J. (2013). Brain development in rodents and humans: identifying benchmarks of maturation and vulnerability to injury across species. Prog. Neurobiol. 106–107, 1–16. doi: 10.1016/j.pneurobio.2013.04.001
Sharma, N. R., Wang, X., Majerciak, V., Ajiro, M., Kruhlak, M., Meyers, C., et al. (2016). Cell type- and tissue contextdependent nuclear distribution of human Ago2. J. Biol. Chem. 291, 2302–2309. doi: 10.1074/jbc.C115.695049
Shen, M., Wang, F., Li, M., Sah, N., Stockton, M. E., Tidei, J. J., et al. (2019). Reduced mitochondrial fusion and Huntingtin levels contribute to impaired dendritic maturation and behavioral deficits in Fmr1-mutant mice. Nat. Neurosci. 22, 386–400. doi: 10.1038/s41593-019-0338-y
Shigeoka, T., Jung, H., Jung, J., Turner-Bridger, B., Ohk, J., Lin, J. Q., et al. (2016). Dynamic axonal translation in developing and mature visual circuits. Cell 166, 181–192. doi: 10.1016/j.cell.2016.05.029
Siegel, G., Saba, R., and Schratt, G. (2011). MicroRNAs in neurons: manifold regulatory roles at the synapse. Curr. Opin. Genet. Dev. 21, 491–497. doi: 10.1016/j.gde.2011.04.008
Skariah, G., Seimetz, J., Norsworthy, M., Lannom, M. C., Kenny, P. J., Elrakhawy, M., et al. (2017). Mov10 suppresses retroelements and regulates neuronal development and function in the developing brain. BMC Biol. 15, 1–19. doi: 10.1186/s12915-017-0387-1
Slomnicki, L. P., Pietrzak, M., Vashishta, A., Jones, J., Lynch, N., Elliot, S., et al. (2016). Requirement of neuronal ribosome synthesis for growth and maintenance of the dendritic tree. J. Biol. Chem. 291, 5721–5739. doi: 10.1074/jbc.M115.682161
Song, J. J., Smith, S. K., Hannon, G. J., and Joshua-Tor, L. (2004). Crystal structure of argonaute and its implications for RISC slicer activity. Science 305, 1434–1437. doi: 10.1126/science.1102514
Sosanya, N. M., Huang, P. P. C., Cacheaux, L. P., Chen, C. J., Nguyen, K., Perrone-Bizzozero, N. I., et al. (2013). Degradation of high affinity HuD targets releases Kv1.1 mRNA from miR-129 repression by mTORC1. J. Cell Biol. 202, 53–69. doi: 10.1083/jcb.201212089
Stappert, L., Roese-Koerner, B., and Brüstle, O. (2015). The role of microRNAs in human neural stem cells, neuronal differentiation and subtype specification. Cell Tissue Res. 359, 47–64. doi: 10.1007/s00441-014-1981-y
Steward, O. (1987). Regulation of synaptogenesis through the local synthesis of protein at the postsynaptic site. Prog. Brain Res. 71, 267–279. doi: 10.1016/s0079-6123(08)61830-0
Steward, O., Davis, L., Dotti, C., Phillips, L. L., Rao, A., and Banker, G. (1988). Protein synthesis and processing in cytoplasmic microdomains beneath postsynaptic sites on CNS neurons. Mol. Neurobiol. 2, 227–261. doi: 10.1007/BF02935634
Steward, O., and Levy, W. B. (1982). Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J. Neurosci. 2, 284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982
Steward, O., and Schuman, E. M. (2003). Compartmentalized synthesis and degradation of proteins in neurons. Neuron 40, 347–359. doi: 10.1016/s0896-6273(03)00635-4
Stiles, J., and Jernigan, T. L. (2010). The basics of brain development. Neuropsychol. Rev. 20, 327–348. doi: 10.1007/s11065-010-9148-4
Su, C. H., Hung, K. Y., Hung, S. C., and Tarn, W. Y. (2017). RBM4 Regulates neuronal differentiation of mesenchymal stem cells by modulating alternative splicing of Pyruvate kinase M. Mol. Cell. Biol. 37, e00466–16. doi: 10.1128/MCB.00466-16
Su, H., Trombly, M. I., Chen, J., and Wang, X. (2009). Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 23, 304–317. doi: 10.1101/gad.1749809
Sudhakaran, I. P., Hillebrand, J., Dervan, A., Das, S., Holohan, E. E., Hulsmeier, J., et al. (2014). FMRP and Ataxin-2 function together in long-term olfactory habituation and neuronal translational control. Proc. Natl. Acad. Sci. U.S.A. 111, E99–E108. doi: 10.1073/pnas.1309543111
Südhof, T. C. (2018). Towards an understanding of synapse formation. Neuron 100, 276–293. doi: 10.1016/j.neuron.2018.09.040
Sugiyama, H., Takahashi, K., Yamamoto, T., Iwasaki, M., Narita, M., Nakamura, M., et al. (2017). Nat1 promotes translation of specific proteins that induce differentiation of mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 114, 340–345. doi: 10.1073/pnas.1617234114
Sutton, M. A., and Schuman, E. M. (2006). Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127, 49–58. doi: 10.1016/j.cell.2006.09.014
Swanger, S. A., and Bassell, G. J. (2013). Dendritic protein synthesis in the normal and diseased brain. Neuroscience 232, 106–127. doi: 10.1016/j.neuroscience.2012.12.003
Swarts, D. C., Makarova, K., Wang, Y., Nakanishi, K., Ketting, R. F., Koonin, E. V., et al. (2014). The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 21, 743–753. doi: 10.1038/nsmb.2879
Sweeney, N. T., Brenman, J. E., Jan, Y. N., and Gao, F. B. (2006). The Coiled-coil protein shrub controls neuronal morphogenesis in Drosophila. Curr. Biol. 16, 1006–1011. doi: 10.1016/j.cub.2006.03.067
Tahmasebi, S., Amiri, M., and Sonenberg, N. (2019). Translational control in stem cells. Front. Genet. 9, 709. doi: 10.3389/fgene.2018.00709
Tarn, W. Y., Kuo, H. C., Yu, H. I., Liu, S. W., Tseng, C. T., Dhananjaya, D., et al. (2016). RBM4 promotes neuronal differentiation and neurite outgrowth by modulating Numb isoform expression. Mol. Biol. Cell 27, 1676–1683. doi: 10.1091/mbc.E15-11-0798
Taverna, E., Götz, M., and Huttner, W. B. (2014). The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 30, 465–502. doi: 10.1146/annurev-cellbio-101011-155801
Tervonen, T. A., Louhivuori, V., Sun, X., Hokkanen, M. E., Kratochwil, C. F., Zebryk, P., et al. (2009). Aberrant differentiation of glutamatergic cells in neocortex of mouse model for fragile X syndrome. Neurobiol. Dis. 33, 250–259. doi: 10.1016/j.nbd.2008.10.010
Tokita, M. J., Chow, P. M., Mirzaa, G., Dikow, N., Maas, B., Isidor, B., et al. (2015). Five children with deletions of 1p34.3 encompassing AGO1 and AGO3. Eur. J. Hum. Genet. 23, 761–765. doi: 10.1038/ejhg.2014.202
Torre, E. R., and Steward, O. (1992). Demonstration of local protein synthesis within dendrites using a new cell culture system that permits the isolation of living axons and dendrites from their cell bodies. J. Neurosci. 12, 762–772. doi: 10.1523/JNEUROSCI.12-03-00762.1992
Trabucchi, M. (2019). Subcellular Heterogeneity of the microRNA Machinery. Trends Genet. 35, 15–28. doi: 10.1016/j.tig.2018.10.006
Tuschl, T., Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., and Teng, G. (2004). Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15, 185–197. doi: 10.1016/j.molcel.2004.07.007
Vessey, J. P., Schoderboeck, L., Gingl, E., Luzi, E., Riefler, J., Di Leva, F., et al. (2010). Mammalian Pumilio 2 regulates dendrite morphogenesis and synaptic function. Proc. Natl. Acad. Sci. U.S.A. 107, 3222–3227. doi: 10.1073/pnas.0907128107
Völler, D., Linck, L., Bruckmann, A., Hauptmann, J., Deutzmann, R., Meister, G., et al. (2016). Argonaute family protein expression in normal tissue and cancer entities. PLoS ONE 11:e0161165. doi: 10.1371/journal.pone.0161165
Wang, B., Pan, L., Wei, M., Wang, Q., Liu, W. W., Wang, N., et al. (2015). FMRP-mediated axonal delivery of miR-181d regulates axon elongation by locally targeting Map1b and Calm1. Cell Rep. 13, 2794–2807. doi: 10.1016/j.celrep.2015.11.057
Wang, D., Zhang, Z., O'Loughlin, E., Lee, T., Houel, S., O'Carroll, D., et al. (2012). Quantitative functions of Argonaute proteins in mammalian development. Genes Dev. 26, 693–704. doi: 10.1101/gad.182758.111
Wang, X., Zorio, D. A. R., Schecterson, L., Lu, Y., and Wang, Y. (2018). Postsynaptic FMRP Regulates synaptogenesis in vivo in the developing cochlear nucleus. J. Neurosci. 38, 6445–6460. doi: 10.1523/JNEUROSCI.0665-18.2018
Weinmann, L., Höck, J., Ivacevic, T., Ohrt, T., Mütze, J., Schwille, P., et al. (2009). Importin 8 Is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 136, 496–507. doi: 10.1016/j.cell.2008.12.023
Wu, D., Raafat, M., Pak, E., Hammond, S., and Murashov, A. K. (2011). MicroRNA machinery responds to peripheral nerve lesion in an injury-regulated pattern. Neuroscience 190, 386–397. doi: 10.1016/j.neuroscience.2011.06.017
Wu, K. Y., Hengst, U., Cox, L. J., Macosko, E. Z., Jeromin, A., Urquhart, E. R., et al. (2005). Local translation of RhoA regulates growth cone collapse. Nature 436, 1020–1024. doi: 10.1038/nature03885
Wu, P. H., Isaji, M., and Carthew, R. W. (2013). Functionally diverse microRNA effector complexes are regulated by extracellular signaling. Mol. Cell 52, 113–123. doi: 10.1016/j.molcel.2013.08.023
Yao, J., Sasaki, Y., Wen, Z., Bassell, G. J., and Zheng, J. Q. (2006). An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat. Neurosci. 9, 1265–1273. doi: 10.1038/nn1773
Ye, B., Petritsch, C., Clark, I. E., Gavis, E. R., Jan, L. Y., and Jan, Y. N. (2004). nanos and pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr. Biol. 14, 314–321. doi: 10.1016/j.cub.2004.01.052
Ye, Y., Xu, H., Su, X., and He, X. (2016). Role of microRNA in governing synaptic plasticity. Neural Plast. 2016:4959523. doi: 10.1155/2016/4959523
Yogev, S., and Shen, K. (2014). Cellular and molecular mechanisms of synaptic specificity. Annu. Rev. Cell Dev. Biol. 30, 417–437. doi: 10.1146/annurev-cellbio-100913-012953
Younts, T. J., Monday, H. R., Dudok, B., Klein, M. E., Jordan, B. A., Katona, I., et al. (2016). Presynaptic protein synthesis is required for long-term plasticity of GABA release. Neuron 92, 479–492. doi: 10.1016/j.neuron.2016.09.040
Zahr, S. K., Yang, G., Kazan, H., Borrett, M. J., Yuzwa, S. A., Voronova, A., et al. (2018). A translational repression complex in developing mammalian neural stem cells that regulates neuronal specification. Neuron 97, 520–537.e6. doi: 10.1016/j.neuron.2017.12.045
Zeitelhofer, M., Karra, D., Macchi, P., Tolino, M., Thomas, S., Schwarz, M., et al. (2008). Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J. Neurosci. 28, 7555–7562. doi: 10.1523/JNEUROSCI.0104-08.2008
Zekri, L., Huntzinger, E., Heimstadt, S., and Izaurralde, E. (2009). The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol. Cell. Biol. 29, 6220–6231. doi: 10.1128/MCB.01081-09
Zekri, L., Kuzuoglu-Öztürk, D., and Izaurralde, E. (2013). GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. EMBO J. 32, 1052–1065. doi: 10.1038/emboj.2013.44
Zelená, J. (1970). Ribosome-like particles in myelinated axons of the rat. Brain Res. 24, 359–363. doi: 10.1016/0006-8993(70)90120-4
Zhang, K., Zhang, X., Cai, Z., Zhou, J., Cao, R., Zhao, Y., et al. (2018). A novel class of microRNA-recognition elements that function only within open reading frames. Nat. Struct. Mol. Biol. 25, 1019–1027. doi: 10.1038/s41594-018-0136-3
Zhang, T., Wu, Y. C., Mullane, P., Ji, Y. J., Liu, H., He, L., et al. (2018). FUS regulates activity of microRNA-mediated gene silencing. Mol. Cell 69, 787–801.e8. doi: 10.1016/j.molcel.2018.02.001
Zhou, Z. D., Selvaratnam, T., Lee, J. C. T., Chao, Y. X., and Tan, E. K. (2019). Molecular targets for modulating the protein translation vital to proteostasis and neuron degeneration in Parkinson's disease. Transl. Neurodegener. 8, 1–14. doi: 10.1186/s40035-019-0145-0
Zielezinski, A., and Karlowski, W. M. (2015). Early origin and adaptive evolution of the GW182 protein family, the key component of RNA silencing in animals. RNA Biol. 12, 761–770. doi: 10.1080/15476286.2015.1051302
Keywords: miRNA, miRISC, neuronal development, translation regulation, neuronal activity
Citation: Nawalpuri B, Ravindran S and Muddashetty RS (2020) The Role of Dynamic miRISC During Neuronal Development. Front. Mol. Biosci. 7:8. doi: 10.3389/fmolb.2020.00008
Received: 27 September 2019; Accepted: 10 January 2020;
Published: 31 January 2020.
Edited by:
Barbara Bardoni, UMR7275 Institut de Pharmacologie Moléculaire et Cellulaire (IPMC), FranceReviewed by:
Enzo Lalli, UMR7275 Institut de Pharmacologie Moléculaire et Cellulaire (IPMC), FranceCopyright © 2020 Nawalpuri, Ravindran and Muddashetty. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ravi S. Muddashetty, cmF2aXNtQGluc3RlbS5yZXMuaW4=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.