
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 14 March 2019
Sec. Structural Biology
Volume 6 - 2019 | https://doi.org/10.3389/fmolb.2019.00013
This article is part of the Research Topic Structural Biology of Membrane Transport and Signalling: New Insights and Developments View all 6 articles
 Thibault Viennet1,2†
Thibault Viennet1,2† Stefanie Bungert-Plümke3†
Stefanie Bungert-Plümke3† Shantha Elter1
Shantha Elter1 Aldino Viegas1
Aldino Viegas1 Christoph Fahlke3*
Christoph Fahlke3* Manuel Etzkorn1,2*
Manuel Etzkorn1,2*Barttin is an accessory subunit of ClC-K chloride channels expressed in the kidney and the inner ear. Main functions of ClC-K/barttin channels are the generation of the cortico-medullary osmotic gradients in the kidney and the endocochlear potential in the inner ear. Mutations in the gene encoding barttin, BSND, result in impaired urinary concentration and sensory deafness. Barttin is predicted to be a two helical integral membrane protein that directly interacts with its ion channel in the membrane bilayer where it stabilizes the channel complex, promotes its incorporation into the surface membrane and leads to channel activation. It therefore is an attractive target to address fundamental questions of intermolecular communication within the membrane. However, so far inherent challenges in protein expression and stabilization prevented comprehensive in vitro studies and structural characterization. Here we demonstrate that cell-free expression enables production of sufficient quantities of an isotope-labeled barttin variant (I72X Barttin, capable to promote surface membrane insertion and channel activation) for NMR-based structural studies. Additionally, we established purification protocols as well as reconstitution strategies in detergent micelles and phospholipid bilayer nanodiscs. Stability, folding, and NMR data quality are reported as well as a suitable assignment strategy, paving the way to its structural characterization.
ClC-K channels form a subgroup of chloride channels within the ClC family of chloride channels and chloride/proton antiporters. They are expressed in the kidney and in the inner ear, and are essential for NaCl re-absorption in the loop of Henle and for potassium secretion by the stria vascularis (Fahlke and Fischer, 2010; Stolting et al., 2014). Barttin is the accessory β-subunit of ClC-K channels. It is the gene product of BSND, the disease gene of a rare human disease, called Bartter syndrome IV. Patients suffer from a severe impairment of urinary concentration ability as well as sensorineural deafness (Landau et al., 1995; Rickheit et al., 2008; Riazuddin et al., 2009; Tan et al., 2017). Barttin has been shown to be necessary for the function of both chloride channels ClC-Ka and ClC-Kb (Estevez et al., 2001) present, respectively, in inner ears and in kidneys. Barttin modifies protein stability, promotes its insertion into the plasma membrane, and turns the channel into a conductive state (Hayama et al., 2003; Scholl et al., 2006).
The predicted transmembrane topology of barttin consists of a short cytoplasmic N-terminus of eight amino acids, two transmembrane helices encompassing the amino acids between 9 and 54 and a large cytoplasmic C-terminus. Whereas, the transmembrane core of barttin including the short cytoplasmic N-terminus is sufficient for fulfilling the effects on channel stability and intracellular trafficking, amino acids in the C-terminus are required for normal function of ClC-K/barttin channels (Scholl et al., 2006; Fischer et al., 2010; Steinke et al., 2015; Wojciechowski et al., 2018). Co-immunoprecipitation studies and confocal microscopy indicate binding of barttin to the B- and/or J-helix at the outer surface of the pore-forming subunit of ClC-K channel (Tajima et al., 2007). Despite this model and features, the structural characteristics of barttin remain elusive, pointing to the need for proper investigation of the protein, aiming to better understand the pathophysiology, the effect of BSND mutations (Steinke et al., 2015; Wojciechowski et al., 2018), and hoping to discover targets for drug development (Tajima et al., 2007).
While detergent micelles are most commonly used for structural studies of membrane proteins, they exhibit severe destabilization (Seddon et al., 2004) and denaturation effects (Zhou and Cross, 2013). Phospholipid bilayer nanodiscs (NDs) have proven to be a useful alternative membrane mimetic (Bayburt and Sligar, 2010; Nasr et al., 2017; Viennet et al., 2018). They have the advantage of allowing the use of numerous types of lipids, and have the power to mimic key properties of membranes thus providing potentially more native environments to membrane proteins. The nanodisc system has been successfully applied in numerous NMR-based studies (Viegas et al., 2016a), including dynamic and structural investigations of β-barrel proteins (Hagn et al., 2013; Fox et al., 2014) as well as the tumor necrosis factor receptor p75NTR (Mineev et al., 2015) which contains a single transmembrane domain or to the seven-transmembrane helices protein bacteriorhodopsin (Etzkorn et al., 2013). Although NMR-optimized constructs have been developed (Hagn et al., 2013), NDs are still a challenging environment for high resolution NMR studies due to their overall large size. As such, pursuing a combined approach using both detergent micelles and nanodiscs in order to exploit the good NMR properties of micelles and the presumably more native environment of the nanodiscs can be a valid approach. This was done in the case of Opa60, where restraints derived from NMR in both dodecylphosphocholine (DPC) and 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) bilayer NDs were used (Fox et al., 2014).
Here we report on expression, purification and reconstitution properties of barttin, which so far eluded a comprehensive in vitro characterization. The usage of an E. coli based cell-free protein expression system enabled production of larger quantities of barttin that we used for reconstitution procedures both in detergent micelles and in phospholipid bilayer nanodiscs. For the latter the influence of lipid and detergent types as well as detergent removal procedures on the quality of resulting nanodiscs was systematically evaluated. Using the optimized protocols, barttin could be successfully solubilized in detergent micelles as well as refolded into nanodiscs in sufficient quantities for subsequent structural and biophysical characterization. As expected NMR spectral quality is largely improved in detergent micelles, however our data show that barttin is considerably less stable in the used micelle system as compared to nanodiscs. While barttin adopts a secondary structure in detergent micelles consistent with its predicted behavior, the NMR spectra in micelles and nanodiscs differ more than expected for the changing environments, indicating that the two environments do not necessarily stabilize the same barttin structure.
A barttin construct containing the N-terminal 72 residues of barttin preceded by a His-tag (His-I72X) was cloned into a pET21a vector and plasmid DNA was produced. Barttin was expressed in an E. coli-based cell-free expression system following established protocols (Schwarz et al., 2007; Klammt et al., 2012). For NMR samples expression was carried out using deuterated buffers (>90% D2O) and either triple (2H, 13C, 15N) labeled Algal extract or custom-made mix with selected amino acids with different isotope labeling suitable for combinatorial assignments was used. Since the Algal isotope mixture lacks the four amino acids: Cys, Trp, Gln, and Asn, the missing amino acids were added in natural abundance at final concentrations of 1 mM. The combinatorial test sample contained the following labeling pattern: 15N-Val, 15N-Cys, 15N-Leu, 15N-Ser, 13C'-Met, u-(13C;15N)-Val and all other amino-acid types at natural abundance. Dialysis mode reactions were carried out at 28°C with or without the presence of nanodiscs. After 12–16 h, the reaction mix was centrifuged for 10 min at 12,000·g. The pellet was washed once with 5 to 10 volumes of buffer containing 50 mM NaH2PO4 pH 8.0, 300 mM NaCl (buffer A, all reagents from Sigma Aldrich if not stated otherwise) and Complete protease inhibitors (Roche). The resulting pellet was stored at −20°C until further use.
For detergent solubilization tests, pellets were directly solubilized in 50 mM NaH2PO4 pH 8.0, 150 mM NaCl supplemented with either 20 mM sodium dodecylsulfate (SDS), 100 mM decylphosphocholine (FOS-10, Cube Biotech) 100 mM dodecylphosphocholine (FOS-12 or DPC, Cube Biotech), 100 mM N,N-dimethyldodecylamine N-oxide (LDAO, Cube Biotech), 42 mM lyso-myristoylphosphatidylglycerol (LMPG, Avanti polar lipids), 250 mM n-decyl-β-D-maltoside (DM, Anatrace) or 196 mM n-dodecyl-β-D-maltoside (DDM, Cube Biotech) in a thermomixer (Eppendorf) at 37°C, 800 rpm for 2 h, without further purification.
For small-scale NMR samples and combinatorial-labeled samples, pellets were directly solubilized with NMR buffer (20 mM NaPi pH 7.0, 100 mM NaCl, 2 mM TCEP, 0.2% (v/v) NaN3 and 10% (v/v) D2O) supplemented with either 100 mM DPC, 42 mM LMPG or 100 mM LDAO in a thermomixer (Eppendorf) at 37°C, 800 rpm for 2 h without further purification.
For large-scale triple-labeled (2H,13C,15N) samples, pellet was solubilized at room temperature for 30 min with 10 volumes of buffer A supplemented with 2 mM DTT, 50 mM LDAO, and Complete protease inhibitors, then centrifuged at room temperature at 16,000·g for 30 min. The supernatant was incubated with previously washed Ni-NTA agarose chemical beads (Macherey-Nagel) at room temperature for 1 h. The slurry was transferred to a column, washed with 10 column volumes of buffer A supplemented with 2 mM DTT and 10 mM LDAO. Barttin was eluted using buffer A supplemented with 2 mM DTT, 10 mM LDAO and 300 mM imidazole, fractions containing Barttin were pooled and applied to a desalting column equilibrated with NMR buffer supplemented with 10 mM LDAO. Finally, the eluate was concentrated in a 10 kDa cutoff Vivaspin concentrator.
E. coli BL21 (DE3) were transformed with the MSP1D1 or MSP1D1Δ5 plasmid DNA in a pET28a vector as reported in Ritchie et al. (2009); Hagn et al. (2013). In short, cells were grown in LB medium. Protein was resuspended with 6M Gdn-HCl and purified by IMAC (without denaturating agent). The elution fractions were pooled and dialyzed in order to remove imidazole. N-terminal His-tag was cleaved using TEV protease incubated overnight at 4°C. ΔHis-MSP1D1 or ΔHis-MSP1D1Δ5 was separated from MSP1D1 or MSP1D1Δ5 by IMAC.
Barttin was purified from washed CFE pellets to SDS following similar procedure as in LDAO. In short, pellets were solubilized in buffer containing 20 mM SDS, supernatant was diluted to 10 mM SDS previous to binding to Ni-NTA agarose beads. Either the slurry or the pooled elution fractions were used for nanodiscs assembly.
Barttin in SDS, ΔHis MSP1D1 or MSP1D1Δ5 (6-fold molar excess over barttin) and lipids (450-fold molar excess over barttin for MSP1D1 or 270-fold for MSP1D1Δ5) solubilized in 60 mM Na-cholate were mixed together in 20 mM Tris-HCl pH 7.5, 100 mM NaCl, 2 mM DTT, 10 mM SDS. Different lipids (all from Avanti polar lipids) were used including 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), a mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol (POPG) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) in the ratio 1:4 and a mixture of 1,2-distearoyl-sn-glycero-3-phospho-L-serine (DSPS), 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), and 1,2-diarachidoyl-sn-glycero-3-phosphocholine (DAPC) in ratio 1:3:3:3 (Mitchell et al., 2007; Kim et al., 2013). Lipids in chloroform were tried under nitrogen flow and stored under vacuum before usage.
Detergent removal procedure was empirically optimized, different approaches including usage of Biobeads SM-2 (Biorad) on-column or in the purified product, dialysis through a 10 kDa cutoff membrane (Thermo Scientific), on-column fast and stepwise wash of the detergent were tested. For more information see Results section.
For all off-column procedures an additional IMAC purification was done in order to separate nanodiscs containing Barttin from those which did not. Finally, Barttin-containing NDs were purified by SEC on a HiLoad 16/600 Superdex 200 pg (for large scale) or Superdex 10/300 gl (for small scale) column (GE Healthcare) equilibrated with 20 mM sodium phosphate pH 7.0, 100 mM NaCl, 2 mM DTT using a ÄKTA pure device running at 1 ml/min. The main peak corresponding to expected NDs was pooled and concentrated using a Vivaspin centrifugal device of 10 kDa MWCO. For NMR samples, final concentrations of 2 mM TCEP, 10% v/v D2O and 0.01% v/v NaN3 were added before measurements.
For co-translational incorporation of Barttin into NDs, empty nanodiscs assembled with DMPC according to established protocols (Denisov et al., 2004; Ritchie et al., 2009) were used in the cell-free system's reaction mix at a concentration of 58 μM.
Proteins were analyzed using denaturing, non-continuous tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis according to published methods (Schägger and von Jagow, 1987). The gel consists of three parts, a 17% acrylamide separation gel, a 10% intermediate gel and a 5% stacking gel. To separate the proteins in an electric field, the following anode buffer (200 mM Tris-HCl pH 8.9) and cathode buffer (200 mM Tris-HCl, 100 mM Tricine and 0.1% SDS) were used. The PageRuler Plus prestained protein ladder (ThermoFisher) was used as protein standard. Proteins were visualized after Coomassie blue staining.
A sample of 10 μM Barttin in LDAO micelles was investigated using circular dichroism (Jasco J-715) in 20 mM NaPi pH 6.8, 100 mM NaCl, 2 mM TCEP and at room temperature. Measurement from 260 to 200 nm was done at a constant bandwidth of 1 nm, with a data pitch of 0.2 nm and repeated 10 times for averaging. Data was converted to mean-residue molar ellipticity and convoluted using the K2d protocol.
NMR experiments were performed on Bruker Avance III HD+ spectrometers operating either at 600 or 700 MHz, both equipped with a triple resonance TCI (1H, 13C, 15N) cryoprobe. Data was collected at 35°C in 20 mM NaPi pH 6.8, 100 mM NaCl, 2 mM TCEP, 0.2% (v/v) NaN3, and 10% (v/v) D2O. For experiments using LDAO micelles, the detergent concentration after concentration was estimated to roughly 100 mM. All NMR experiments contained transverse relaxation optimized spectroscopy (TROSY) components (Pervushin et al., 1997; Salzmann et al., 1999). The 2D 13CO- and 13Cα-filtered TROSY-HSQC pulse sequences were designed from their respective 3D experiments. All NMR spectra were processed with TOPSPIN 3.2 (Bruker) and analyzed with CARA (Keller, 2004) and CCPN (Vranken et al., 2005).
In general, structural biology aspires to obtain data in a native environment of the target system in order to ensure relevant folding and necessary cofactors for function, without introducing bias due to in vitro handling procedures. While some techniques theoretically have the power to do so, such as in-cell NMR (Theillet et al., 2016) or dynamic nuclear polarization NMR (Viennet et al., 2016), they often face limitations in sensitivity and/or resolution, which is particularly true for membrane systems. For membrane proteins, the presence of a phospholipid bilayer with similar chemical composition and physical properties as the native membrane is evidently one of the most important environmental factors. When pursuing in vitro investigations, nanodiscs, as compared to other membrane mimetics such as detergent micelles, bicelles or, amphipols, exhibit several key advantages (Bayburt et al., 2002): (i) they provide homogenous lipid-bilayer particles, (ii) both sides of the bilayer are accessible in solution, (iii) they have high stability and low exchange rates of lipid and protein molecules between different NDs, (iv) the presence of the scaffold protein may mimic a crowded environment as often found in natural membranes, and (v) a large range of lipids (charge, length, unsaturations, non-phospholipids, etc.) can be incorporated allowing mimicking of properties of various types and states of membranes (Viegas et al., 2016a).
Structural studies of membrane proteins are often impeded by limitations in protein expression due to cytotoxic effects of the target protein in its heterologous overexpression systems. Unfortunately, barttin belongs to those membrane proteins that are difficult to produce in large quantities in conventional expression systems (unpublished observations). Therefore, so far, all published barttin data resulted from in cell experiments where protein amount is not critical (Scholl et al., 2006; Fahlke and Fischer, 2010; Fischer et al., 2010). Cell-free expression (CFE) offers a valuable alternative, which also has the advantage of supporting custom labeling strategies thanks to the direct use of amino acids and the very low level of metabolic scrambling (Katzen et al., 2009). We tested expression of barttin in an E. coli-based cell-free expression system. Following established protocols (Schwarz et al., 2007) we could obtain sufficient quantities of protein for subsequent characterization.
We initially carried out CFE in the absence of membrane mimetics. This strategy requires reconstitution of the resulting protein pellet into a suitable membrane mimetic. While canonical protocols for detergent reconstitution or incorporation into nanodiscs exist, they need to be empirically optimized for each target protein. For the latter factors such as the target protein to scaffold protein to lipid ratio, the detergent type and removal procedure may strongly differ for different target proteins (Viegas et al., 2016a).
To test barttin incorporation into nanodiscs we started with the following setup: SDS was used to solubilize barttin CFE pellets, MSP1D1 (D1) was used as scaffold protein, DMPC lipids were solubilized in sodium cholate, and adsorbent polystyrene beads were used to remove detergents. Although empty NDs assembly was detected, no detectable levels of barttin were incorporated with this setup (data not shown, see Table 1). Subsequently we changed the detergent in which lipids are solubilized to SDS, and the lipid mixture to DMPG:DMPC 1:4 (20% negatively charged head groups). While previously shown to largely improve quality of the data (Hagn et al., 2013), it did not lead to detectable improvements for barttin. The most probable explanation is that barttin, a rather hydrophobic and low molecular weight (10.1 kDa) protein, could adsorb to the polystyrene beads.
In the next step we therefore used dialysis to remove the detergent for ND assembly. The same conditions concerning lipids and detergents were tested and indeed successful incorporation of barttin in nanodiscs was detected (Figure 1A and Table 1). Unfortunately, the yield, defined as the recovery of barttin from the input to the purified NDs, remained very low (< 1%; see Table 1). Changing detergent for barttin solubilization to DPC micelles, which provided good results in other cases (Hagn et al., 2013; Fox et al., 2014), did not improve barttin incorporation. In addition (and possibly in line) with the low yield, the size exclusion profiles also reveal rather poor homogeneity (Figure 1A).
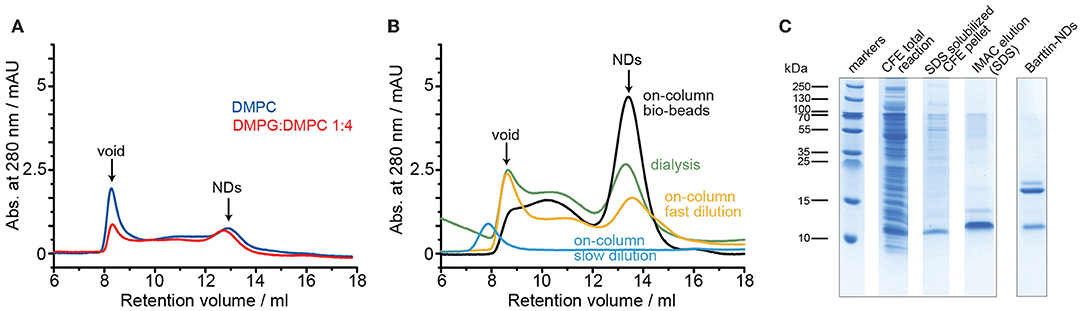
Figure 1. Optimization of barttin reconstitution into D1 NDs. (A) SEC profiles of barttin NDs (MSP1D1) assembled with DMPC (blue) and DMPG:DMPC 1:4 (red) upon sodium cholate dialysis. (B) SEC chromatograms of barttin NDs (MSP1D1) assembled with DMPC upon detergent dialysis (green), on-column biobeads adsorption (black), on-column fast dilution (yellow), and on-column step dilution (blue). (C) Coomassie blue-stained SDS-PAGE of CFE expressed barttin (total reaction), solubilized CFE pellet, SDS- purified barttin and purified (IMAC, before SEC) barttin NDs (MSP1D1) assembled with DMPC upon on-column biobeads adsorption.
Having found initial conditions allowing barttin incorporation into nanodiscs, additional parameters were screened in order to improve yields and homogeneity of the preparations. Since the detergent removal step seemed critical, three different approaches were tested using on-column procedures successfully reported before (Katayama et al., 2010). In short, Barttin in SDS was immobilized through its histidine tag to a Ni-NTA agarose column, incubated with MSP and lipids and the detergents were removed either by introduction of adsorbent beads, fast dilution of the slurry, or stepwise washing procedure. The latter did not allow any ND assembly, which we attribute to a too slow detergent removal rate, kinetically favoring lipid aggregation over nanodisc formation as seen from the SEC profile (Figure 1B). The other protocols were successful in producing substantial amounts of barttin-containing NDs, with an overall higher homogeneity and yield for the on-column polystyrene beads adsorption of detergents (Figure 1B and Table 1).
In addition to size homogeneity, another important factor in ND reconstitution is the resulting oligomeric state of the membrane protein, which is difficult to control as well as to measure accurately (Tsukamoto et al., 2010; Viegas et al., 2016a; Peetz et al., 2017). While, due to the lack of larger extra-membranous domains, incorporation of multiple copies of barttin into the NDs not necessarily changes the resulting SEC profile, the amount of barttin per ND can be estimated from the relative band intensities of MSP and barttin on a SDS-PAGE gel. The respective band intensities, after IMAC purification to remove empty NDs, are in line with a single copy of barttin in D1 NDs (Figure 1C).
Smaller nanodiscs, obtained by using shorter MSP variants (Hagn et al., 2013), are known to provide better NMR spectral quality and may provide better control of the oligomeric state for small to medium-sized membrane proteins. We therefore tested the membrane incorporation of barttin also using MSP1D1ΔH5 (Δ5)-based NDs. In this setup we additionally tested the possibility of reconstituting barttin in a more “native-like” phospholipid mixture based on a conserved aliphatic chain lengths distribution and negative charge content (Mitchell et al., 2007; Kim et al., 2013) using a lipid composition of DPPC:DSPC:DAPC:DSPS in a 3:3:3:1 molar ratio. For both standard DMPC and the “native-like lipid mix,” we tested the reconstitution using either dialysis or on-column adsorbent beads removal. In all conditions barttin incorporation was successful and provided sufficient homogeneity of the resulting NDs for NMR studies (Figure 2A). Usage of the “native-like lipid mix,” however, provided lower yields (Figure 2A and Table 1). The oligomeric state of barttin in Δ5 NDs was also estimated to one barttin per ND in all tested conditions (Figure 2B).
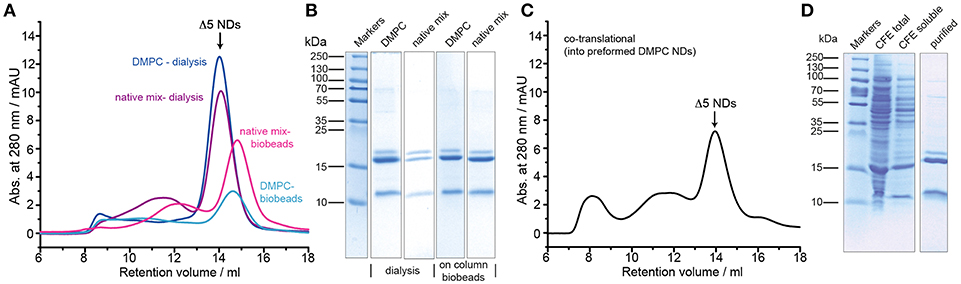
Figure 2. Optimization of barttin incorporation into Δ5 NDs. (A) SEC profiles of barttin NDs (MSP1Δ5) assembled with DMPC (blue) and “native-like lipid mix” (purple) upon detergent dialysis, and respective profiles for on-column biobeads adsorption for DMPC (cyan) and lipid mix (magenta). (B) Coomassie blue-stained SDS-PAGE gel of the corresponding barttin-NDs samples, purified via IMAC, before SEC. (C) SEC profile of barttin NDs formed via co-translational reconstitution of barttin into Δ5 NDs during CFE. (D) Coomassie blue stained gel of total CFE reaction in the presence of preformed NDs (MSP1Δ5, DMPC), soluble fraction and IMAC purified barttin NDs.
Noteworthy the usage of a cell-free expression system also allows to add preformed nanodiscs to the reaction mix to promote co-translational incorporation of the expressed protein directly into the NDs (Rues et al., 2016; Viegas et al., 2016a). We tested this approach using preformed DMPC Δ5 nanodiscs, which yielded fair quality and amount of barttin-containing NDs (Figures 2C,D). However, overall homogeneity and yield was lower as for other reconstitution protocols (see Table 1).
All previous experiments were carried out using a rather low amount of barttin (from two small scale 50 μl cell-free expressions). Since NMR studies require higher amounts of material (in the mg range), we tested up-scaling of the most promising protocols, i.e., “native-like lipid mix” together with on-column detergent adsorption and DMPC together with detergent dialysis. For both setups cell-free pellets from 3 ml CFE reactions were used. In line with general observations in upscaling, in both cases the yields dropped by 30 and 60%, respectively (Figures 3A,B and Table 1). Nevertheless, sufficient amounts of 15N-barttin in Δ5 NDs for NMR characterization could be obtained.
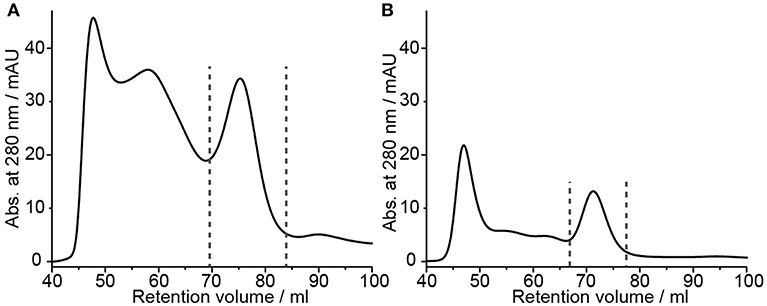
Figure 3. NMR sample preparation of barttin in Δ5 NDs. (A) SEC profile of up-scaled barttin-NDs (MSP1Δ5) assembled with “native-like lipid mix” upon on-column biobeads adsorption. (B) SEC profile of up-scaled barttin-NDs (MSP1Δ5) assembled with DMPC upon dialysis. Dotted line mark fractions collected in the corresponding NMR samples.
Summarizing the screening of the tested conditions, several key points in barttin expression and reconstitution can be concluded, including: (i) CFE offers a convenient approach to prepare high quantities of barttin (ii) cell-free expressed barttin can be easily purified (iii) barttin co-translationally inserts into preformed NDs during cell-free expression (iv) barttin is likely to adsorb into polystyrene beads (v) detergent removal setups and resulting removal rates during barttin-ND self-assembly are very critical parameters, and (vi) various types of phospholipid mixtures lead to successful incorporation of barttin into NDs.
Although providing clear advantages, nanodiscs can still be challenging for NMR-based studies due to their larger size as compared to detergent micelles (~30 kDa for barttin in LDAO micelles as compared to 110 kDa for Barttin in Δ5 NDs). Therefore, in cases where protein structure and function are not corrupted by the detergent, detergent micelles can facilitate NMR-based characterization.
To obtain barttin in detergent micelles, we initially performed detergent screening by directly solubilizing barttin from CFE protein pellets following previous approaches (Klammt et al., 2012). A range of detergents suitable for NMR studies were tested, including FOS-10, DPC, LDAO, LMPG, DM, and DDM (Figure 4A). Interestingly, all tested detergents with the exception of DM and DDM lead to similar solubilization levels as seen from the intensities of the corresponding bands in SDS-PAGE. Three of them were selected for initial NMR screening for their capacity to form small micelles (LDAO, LMPG) or their reported good NMR properties (LDAO, DPC) (Raschle et al., 2009; Hagn et al., 2013). 1D 1H NMR spectra of (not isotope labeled) barttin solubilized in the respective micelles without further purification were acquired and compared (Figure 4B). In general, the NMR spectra show an overall similar profile of the amide signals for barttin in the three different detergent micelles. However, the spectrum in LDAO shows several more intense and in particular also better resolved peaks on the flanking region of the amide signals. The latter is an important indicator for NMR-spectral quality (Viegas et al., 2016a). In addition, the appearance of tryptophan side-chain resonances was only observed in LDAO micelles (Figure 4B, Trp-label). We therefore decided to use LDAO micelles for the following steps.

Figure 4. Solubility and NMR properties of barttin in different detergent micelles. (A) Coomassie stained SDS-PAGE solubility screen using CFE pellets directly solubilized with indicated detergents (for different detergents only the soluble fraction after centrifugation at 20,000 g is loaded into the gel). (B) 1D 1H NMR screening of resulting most promising barttin containing micelles.
In order to further evaluate the characteristics of barttin in LDAO micelles we recorded circular dichroism (CD) data (Figure 5B). As expected for folded barttin the CD spectrum shows a double minimum around 225 and 215 nm, indicative for the presence of α-helical secondary structure. Deconvolution of the CD spectrum (K2d method) indicates a α-helical propensity of 37%. This value is in line with the proposed topology models of barttin (Fahlke and Fischer, 2010), which predict an α-helical content of 44% for the used construct (Figure 5A).
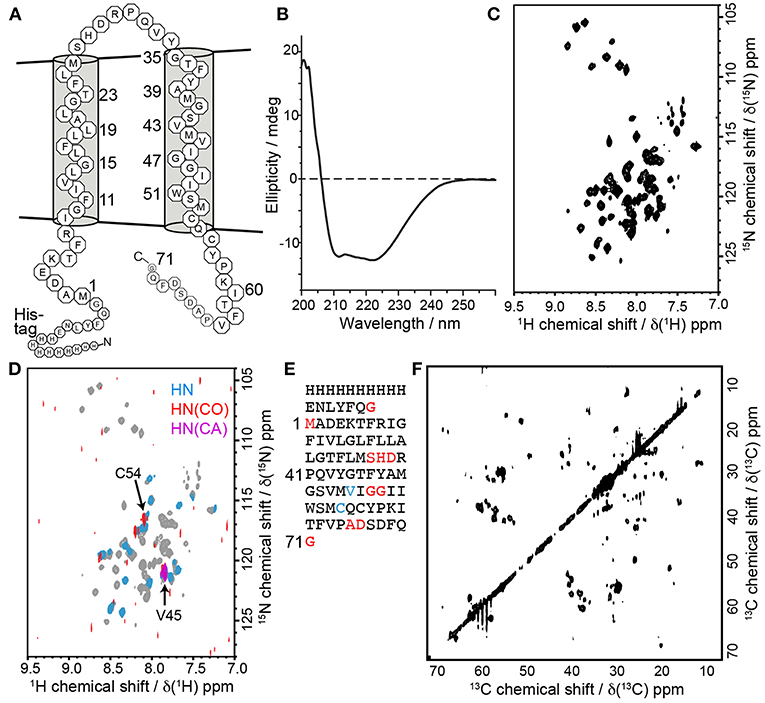
Figure 5. Barttin in LDAO micelles. (A) Expected topology model of barttin according to (Wojciechowski et al., 2015). (B) Circular dichroism spectrum of barttin in LDAO micelles. (C) [15N-1H]-HSQC spectrum of [U-2H-13C-15N]-labeled barttin in LDAO micelles (Cys, Asn and Trp are unlabeled). (D) Overlay of [15N-1H]-HSQC acquired on [2H-13C-15N]-labeled barttin in LDAO micelles (gray) and on a selectively labeled sample (see methods for labeling pattern, blue). In addition, the [15N-1H] dimension of a HNCO (red) and of a HNCA (purple) are shown. The labeling pattern directly allows assignments of indicated residues. (E) Primary sequence of the used barttin construct highlighting residues with assigned resonance frequencies from conventional 3D experiments (red) and using selective isotope labeling (blue). (F) 2D 13C-13C FLOPSY spectrum acquired using an UTOPIA NMR setup (Viegas et al., 2016b).
We additionally recorded 2D [15N-1H]-HSQC NMR data of isotopically labeled barttin in LDAO micelles. The spectrum shows dispersed and mostly resolved peaks (Figure 5C). From the 77 expected peaks about 65 are resolved. Note that cysteine, glutamine, asparagine and tryptophan are not 15N-labeled in the used sample. The overall peak positions and dispersion is also in line with the expected features of barttin. To obtain more comprehensive NMR insights into barttin in LDAO micelles we prepared a triple (2H, 13C, 15N) labeled sample of barttin at protein concentration of 250 μM. Note that amide proton back exchange was carried out during the reconstitution process to enable standard 1H-detected TROSY-based NMR experiments.
Since 1H and 15N chemical shift are less reliable secondary structure indicators as compared to 13C chemical shifts, we additionally recorded a 13C-13C correlation spectrum (Figure 5F). Note that the spectrum was obtained for free during the acquisition of a 3D NOESY spectrum using the UTOPIA setup (Viegas et al., 2016b). Evaluation of the amino acids specific 13Cα-13Cβ peak positions indicates that most residues predominantly occur in random coil and α-helical secondary structure, in line with the CD data and the expected topology model. Taken together these results suggest that barttin adopts a proper secondary structure in LDAO micelles.
Additionally, a set of 3D experiments for resonance assignment was carried out. Unfortunately and reproducibly, NMR spectra of barttin in LDAO micelles change their appearance rather fast (within a few days) indicative of protein degradation and/or aggregation (vide infra). The limited stability of barttin in micelles largely reduces the NMR spectral quality due to the alterations of the resonance frequencies of the protein over the time course of the 3D experiments. Consequently, the resulting 3D spectra are difficult to analyze and have lower signal-to-noise ratios than expected, especially when 13C-13C INEPT transfers are involved. This is often observed in large systems due the unfavorable relaxation properties and may indicate aggregation of barttin micelles. Albeit the spectral challenges, we could obtain resonance assignments for 10 residues (Figure 5E, red).
While the limited sample stability prevents usage of time-intensive 3D NMR experiments, in the case of barttin in LDAO it is still sufficient to obtain 2D spectra, which can be recorded sufficiently fast. Therefore, one possibility to obtain more information in this conditions is to use a combinatorial isotope labeling approach that only requires usage of simple 2D experiments (Parker et al., 2004). The principle is to produce several samples with different types of amino acids that are isotopically labeled with either only 15N, only 13CO or 13C and 15N. Comparison of 2D spectra from [15N-1H]-HSQC, 13CO-filtered HSQC, and 13Cα-filtered HSQC experiments lead to amino acid type assignment and ultimately to residue-specific assignments. This is made easy by the use of cell-free expression, which allows to introduce single amino acid types with the desired labeling (Lohr et al., 2012, 2015). While a full assignment is not attempted here, we investigated whether this strategy would in general be feasible for barttin in LDAO. We therefore prepared a selectively isotope labeled version of barttin in LDAO micelles and recorded in addition to the 2D 1H,15N-HSQC spectrum (Figure 5D, blue) also 2D versions of HNCO (Figure 5D, red) and HNCA (Figure 5D, purple) experiments. By using this single sample two additional unambiguous assignment can be made (Figure 5E, blue). We therefore anticipate that usage of a substantial number of differently isotope labeled samples should enable a near complete resonance assignment of the amide resonances. However, since it is not fully clear whether the LDAO embedded state resembles the functional relevant barttin structure (vide infra), it is at this point not clear whether this effort would be justified.
The lack of a reliable functional in vitro assay for barttin renders it difficult to judge, whether the protein adopts a relevant state after reconstitution in any membrane mimetic. In this respect it is only possible to compare structural integrity such as secondary structure with the expected behavior as well as to compare NMR-spectra obtained in different membrane mimetics. Under the (in general not necessarily valid) assumption that the NDs are more likely to induce a native-like protein fold, NMR offers the possibility to use spectra obtained in NDs as reference for the protein fold and compare it to the data obtained in detergent micelles (Shenkarev et al., 2009, 2010; Morgado et al., 2015).
One disadvantage of the nanodisc system is that the presence of the scaffold protein impedes light absorption-based experiments such as concentration determination at 280 nm or CD spectroscopy (Viegas et al., 2016a). Thus, the same characterization of barttin secondary structure in NDs is not feasible. However, for NMR measurements signal separation of target and scaffold protein is easily realizable via isotope enrichment of barttin in the CFE setup. We thus carried out an initial NMR characterization of isotope labeled barttin in nanodiscs. Two different ND preparations, i.e., using either 100 % DMPC lipids or using the “native-like lipid mixture” as described above, were used. 2D [15N-1H]-HSQC spectra of barttin in each of the two ND preparations show a rather small number of resolved peaks indicative of homogenous line broadening of bigger particles and/or heterogenous barttin conformations. Only about 15 out of the expected 77 peaks are visible. The most probable explanation for this is that only signals from residues situated in the protein termini or in the extra-membrane loop appear because of their NMR-favorable dynamic properties. However, without residue-specific assignments, only assumptions can be made.
When comparing the NMR results obtained in the different NDs to the results in LDAO micelles, it is apparent that the positions of the visible peaks of barttin differ considerably between micelles and nanodiscs (Figure 6A). While the absence of peaks could be explained by the difference in particle sizes, the observed peak shifts indicate that the conformation of the protein is altered in the corresponding region of the protein. Under the assumption that only the extra-membrane residues are observed in the ND samples, this observation would be in line with the denaturation effect of detergents at the micelle-water interface as it was postulated in the case of OmpX (Hagn and Wagner, 2015). Since the conformation and dynamics of this region in general plays an important physiological role, it can at this point not be assumed that barttin in LDAO adopts the same conformation as barttin in nanodiscs.
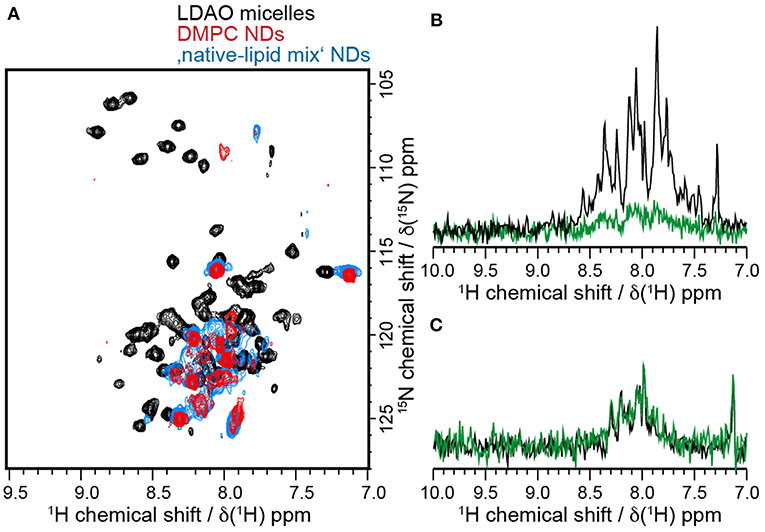
Figure 6. Barttin has different properties in LDAO and NDs. (A) [15N-1H]-HSQC spectra of 15N-labeled barttin in LDAO micelles (black), DMPC NDs (red) and “native-like lipid mix” NDs (blue). (B) 15N-filtered 1D 1H NMR spectra of 2H-13C-15N-labeled barttin in LDAO micelles at the beginning of the NMR measurements (black) and after 14 days (green). (C) 15N-filtered 1D 1H NMR spectra of 15N-labeled barttin in DMPC NDs at beginning of NMR measurements (black) and after 18 days (green).
Interestingly a large overlap between the NMR spectra obtained on barttin in pure DMPC and the “native-like lipid mix” NDs is visible, pointing to rather similar structures. Small changes do arise, which are in line with chemical shift perturbation induced by different membrane mimetic that do not alter protein function as e.g., observed for bacteriorhodopsin in detergent and nanodiscs (Etzkorn et al., 2013).
While NMR data acquired for barttin in LDAO micelles are clearly of much better quality, the stability of the sample is much lower than in NDs. Indeed, as discussed above we could observe gradual degradation of the HSQC signal over time when performing long-time measurements. Over the time course of 14 days a nearly complete disappearance of the signals was observed (Figure 6B). On the contrary barttin in nanodiscs showed no signal decay after 18 days (Figure 6C). This data demonstrates that barttin in the ND-system is considerably more stable as compared to LDAO micelles.
Our study could identify and overcome several key challenges in the in vitro characterization of barttin. We confirm the usefulness of cell-free expression systems for membrane proteins that are challenging to express in bacterial systems (Katzen et al., 2009). Solubilization of resulting CFE protein precipitates using detergent micelles was straightforward for nearly all tested detergents. Nevertheless, considering solubilization yield and NMR spectral quality, our detergent screening for barttin identified LDAO as best micelle environment for our study. CD and NMR data indicate that barttin in LDAO micelles adopts a similar secondary structure as predicted, it is however not possible to conclude that it adopts its native structure.
As an alternative to detergent micelles, we could identify conditions for which cell-free expressed barttin is successfully incorporated into nanodiscs. While the ND incorporation must be empirically optimized for each target, our screening of conditions reveals several features that may be of general relevance for other systems, in particular in respect to the used detergents, the detergent removal procedure and the used lipids. For barttin, usage of SDS for protein solubilization and Na-cholate for lipid solubilization provided best results. While we did not perform a thorough screening of other possible detergents for this step, in our experience and in line with several previous reports (Viegas et al., 2016a), this combination is rather robust and should also be applicable for many other systems. Detergent removal turned out to be the most important aspect of barttin reconstitution into nanodiscs. Our data suggest that barttin is entirely adsorbed to the most commonly used biobeads along with the detergents. This may be similar for other membrane proteins, in particular proteins with only one or two transmembrane helices and small hydrophilic domains. In the case of barttin, dialysis, or immobilization of the protein on a column (IMAC) while using biobeads, provided equally good strategies for detergent removal. Noteworthy, immobilization of the protein on a column during ND assembly can also reduce the number of steps and length of the reconstitution/purification procedure. In respect to the used lipids, we did not observe large effects in ND yield and homogeneity when using the most standard DMPC lipids or a more heterogenous native-like lipid mixture.
Comparison of NMR data of barttin in LDAO micelles or in lipid-bilayer nanodiscs reveals considerable differences reflecting most likely on variations in structure and dynamic of the extra-membrane residues. Interestingly, a comparison of NMR data obtained in nanodiscs formed by the model DMPC lipids are very similar to data obtained on the heterogeneous lipid mixture reflecting the most prominent lipids in barttin's cellular environment. This observation would be in line with the view that predominantly the extra-membranous regions are visible in the NMR spectra of barttin in the used NDs and that, unlike in the case of detergents, these regions are similar for barttin in the model lipid and in the native-like lipid bilayer.
Our data represent the first in vitro results of recombinantly expressed and purified barttin and reports on very initial structural results including insights into the effects of different membrane mimetics. While we have invested considerable efforts to overcome several of the major obstacles for the system, our results also suggest that structural characterization of barttin in nanodiscs is still substantially more challenging as expected from its size or as observed for other comparable systems. While barttin in detergent micelles is significantly better accessible via conventional solution NMR approaches, our data show that the protein is not stable in the used LDAO micelles. We demonstrate that combinatorial labeling could overcome this challenge, however our data also show that the barttin structure in micelles likely differs from the one in nanodiscs. Without a reliable in vitro functional assay, it can at this point not be decided, which membrane environment is suitable to support barttin's native structure. It is however clear that both tested systems, i.e., detergents and nanodisc, will present challenges in terms of stability or NMR spectral quality, respectively.
Overall several aspects of the presented approach should be transferable to other comparable systems where they may help to optimize sample preparation for structural studies. In respect to barttin we anticipate that our results enable future in vitro characterizations of the system, including interaction studies with the ClC-K channels.
All datasets generated for this study are included in the manuscript and/or the supplementary files.
TV, SB-P, SE, and AV conducted the experiments. TV and SB-P analyzed data. TV, SB-P, CF, and ME wrote the manuscript. All authors designed experiments and commented on the manuscript.
This work was supported by grants from the German Research Foundation (DFG) (ET 103/2-1) to ME and from the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement N° 660258 to AV.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge access to the Jülich-Düsseldorf Biomolecular NMR Center. TV acknowledges support from the International North-Rhine-Westphalia Research School iGRASPseed.
Bayburt, T. H., Grinkova, Y. V., and Sligar, S. G. (2002). Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2, 853–856. doi: 10.1021/nl025623k
Bayburt, T. H., and Sligar, S. G. (2010). Membrane protein assembly into Nanodiscs. FEBS Lett. 584, 1721–1727. doi: 10.1016/j.febslet.2009.10.024
Denisov, I. G., Grinkova, Y. V., Lazarides, A. A., and Sligar, S. G. (2004). Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 126, 3477–3487. doi: 10.1021/ja0393574
Estevez, R., Boettger, T., Stein, V., Birkenhager, R., Otto, E., Hildebrandt, F., et al. (2001). Barttin is a Cl- channel beta-subunit crucial for renal Cl- reabsorption and inner ear K+ secretion. Nature 414, 558–561. doi: 10.1038/35107099
Etzkorn, M., Raschle, T., Hagn, F., Gelev, V., Rice, A. J., Walz, T., et al. (2013). Cell-free expressed bacteriorhodopsin in different soluble membrane mimetics: biophysical properties and NMR accessibility. Structure 21, 394–401. doi: 10.1016/j.str.2013.01.005
Fahlke, C., and Fischer, M. (2010). Physiology and pathophysiology of ClC-K/barttin channels. Front. Physiol. 1:155. doi: 10.3389/fphys.2010.00155
Fischer, M., Janssen, A. G., and Fahlke, C. (2010). Barttin activates ClC-K channel function by modulating gating. J. Am. Soc. Nephrol. 21, 1281–1289. doi: 10.1681/ASN.2009121274
Fox, D. A., Larsson, P., Lo, R. H., Kroncke, B. M., Kasson, P. M., and Columbus, L. (2014). Structure of the Neisserial outer membrane protein Opa(6)(0): loop flexibility essential to receptor recognition and bacterial engulfment. J. Am. Chem. Soc. 136, 9938–9946. doi: 10.1021/ja503093y
Hagn, F., Etzkorn, M., Raschle, T., and Wagner, G. (2013). Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J. Am. Chem. Soc. 135, 1919–1925. doi: 10.1021/ja310901f
Hagn, F., and Wagner, G. (2015). Structure refinement and membrane positioning of selectively labeled OmpX in phospholipid nanodiscs. J. Biomol. NMR 61, 249–260. doi: 10.1007/s10858-014-9883-6
Hayama, A., Rai, T., Sasaki, S., and Uchida, S. (2003). Molecular mechanisms of Bartter syndrome caused by mutations in the BSND gene. Histochem. Cell Biol. 119, 485–493. doi: 10.1007/s00418-003-0535-2
Katayama, H., Wang, J., Tama, F., Chollet, L., Gogol, E. P., Collier, R. J., et al. (2010). Three-dimensional structure of the anthrax toxin pore inserted into lipid nanodiscs and lipid vesicles. Proc. Natl. Acad. Sci. U.S.A. 107, 3453–3457. doi: 10.1073/pnas.1000100107
Katzen, F., Peterson, T. C., and Kudlicki, W. (2009). Membrane protein expression: no cells required. Trends Biotechnol. 27, 455–460. doi: 10.1016/j.tibtech.2009.05.005
Kim, O. Y., Jung, Y. S., Cho, Y., Chung, J. H., Hwang, G. S., and Shin, M. J. (2013). Altered heart and kidney phospholipid fatty acid composition are associated with cardiac hypertrophy in hypertensive rats. Clin. Biochem. 46, 1111–1117. doi: 10.1016/j.clinbiochem.2013.04.008
Klammt, C., Maslennikov, I., Bayrhuber, M., Eichmann, C., Vajpai, N., Chiu, E. J., et al. (2012). Facile backbone structure determination of human membrane proteins by NMR spectroscopy. Nat. Methods 9, 834–839. doi: 10.1038/nmeth.2033
Landau, D., Shalev, H., Ohaly, M., and Carmi, R. (1995). Infantile variant of Bartter syndrome and sensorineural deafness: a new autosomal recessive disorder. Am. J. Med. Genet. 59, 454–459. doi: 10.1002/ajmg.1320590411
Lohr, F., Reckel, S., Karbyshev, M., Connolly, P. J., Abdul-Manan, N., Bernhard, F., et al. (2012). Combinatorial triple-selective labeling as a tool to assist membrane protein backbone resonance assignment. J. Biomol. NMR 52, 197–210. doi: 10.1007/s10858-012-9601-1
Lohr, F., Tumulka, F., Bock, C., Abele, R., and Dotsch, V. (2015). An extended combinatorial 15N, 13Calpha, and 13C' labeling approach to protein backbone resonance assignment. J. Biomol. NMR 62, 263–279. doi: 10.1007/s10858-015-9941-8
Mineev, K. S., Goncharuk, S. A., Kuzmichev, P. K., Vilar, M., and Arseniev, A. S. (2015). NMR dynamics of transmembrane and intracellular domains of p75NTR in lipid-protein nanodiscs. Biophys. J. 109, 772–782. doi: 10.1016/j.bpj.2015.07.009
Mitchell, T. W., Ekroos, K., Blanksby, S. J., Hulbert, A. J., and Else, P. L. (2007). Differences in membrane acyl phospholipid composition between an endothermic mammal and an ectothermic reptile are not limited to any phospholipid class. J. Exp. Biol. 210, 3440–3450. doi: 10.1242/jeb.007286
Morgado, L., Zeth, K., Burmann, B. M., Maier, T., and Hiller, S. (2015). Characterization of the insertase BamA in three different membrane mimetics by solution NMR spectroscopy. J. Biomol. NMR 61, 333–345. doi: 10.1007/s10858-015-9906-y
Nasr, M. L., Baptista, D., Strauss, M., Sun, Z. J., Grigoriu, S., Huser, S., et al. (2017). Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat. Methods 14, 49–52. doi: 10.1038/nmeth.4079
Parker, M. J., Aulton-Jones, M., Hounslow, A. M., and Craven, C. J. (2004). A combinatorial selective labeling method for the assignment of backbone amide NMR resonances. J. Am. Chem. Soc. 126, 5020–5021. doi: 10.1021/ja039601r
Peetz, O., Henrich, E., Laguerre, A., Lohr, F., Hein, C., Dotsch, V., et al. (2017). Insights into cotranslational membrane protein insertion by combined LILBID-mass spectrometry and NMR spectroscopy. Anal. Chem. 89, 12314–12318. doi: 10.1021/acs.analchem.7b03309
Pervushin, K., Riek, R., Wider, G., and Wuthrich, K. (1997). Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U.S.A. 94, 12366–12371. doi: 10.1073/pnas.94.23.12366
Raschle, T., Hiller, S., Yu, T. Y., Rice, A. J., Walz, T., and Wagner, G. (2009). Structural and functional characterization of the integral membrane protein VDAC-1 in lipid bilayer nanodiscs. J. Am. Chem. Soc. 131, 17777–17779. doi: 10.1021/ja907918r
Riazuddin, S., Anwar, S., Fischer, M., Ahmed, Z. M., Khan, S. Y., Janssen, A. G., et al. (2009). Molecular basis of DFNB73: mutations of BSND can cause nonsyndromic deafness or Bartter syndrome. Am. J. Hum. Genet. 85, 273–280. doi: 10.1016/j.ajhg.2009.07.003
Rickheit, G., Maier, H., Strenzke, N., Andreescu, C. E., De Zeeuw, C. I., Muenscher, A., et al. (2008). Endocochlear potential depends on Cl- channels: mechanism underlying deafness in Bartter syndrome IV. EMBO J. 27, 2907–2917. doi: 10.1038/emboj.2008.203
Ritchie, T. K., Grinkova, Y. V., Bayburt, T. H., Denisov, I. G., Zolnerciks, J. K., Atkins, W. M., et al. (2009). “Chapter eleven - reconstitution of membrane proteins in phospholipid bilayer nanodiscs,” in Methods Enzymol, ed D. Nejat (New York, NY: Academic Press), 211–231.
Rues, R. B., Dotsch, V., and Bernhard, F. (2016). Co-translational formation and pharmacological characterization of beta1-adrenergic receptor/nanodisc complexes with different lipid environments. Biochim. Biophys. Acta 1858, 1306–1316. doi: 10.1016/j.bbamem.2016.02.031
Salzmann, M., Wider, G., Pervushin, K., Senn, H., and Wüthrich, K. (1999). TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. J. Am. Chem. Soc. 121, 844–848. doi: 10.1021/ja9834226
Schägger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379. doi: 10.1016/0003-2697(87)90587-2
Scholl, U., Hebeisen, S., Janssen, A. G., Muller-Newen, G., Alekov, A., and Fahlke, C. (2006). Barttin modulates trafficking and function of ClC-K channels. Proc. Natl. Acad. Sci. U.S.A 103, 11411–11416. doi: 10.1073/pnas.0601631103
Schwarz, D., Junge, F., Durst, F., Frolich, N., Schneider, B., Reckel, S., et al. (2007). Preparative scale expression of membrane proteins in Escherichia coli-based continuous exchange cell-free systems. Nat. Protoc. 2, 2945–2957. doi: 10.1038/nprot.2007.426
Seddon, A. M., Curnow, P., and Booth, P. J. (2004). Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta 1666, 105–117. doi: 10.1016/j.bbamem.2004.04.011
Shenkarev, Z. O., Lyukmanova, E. N., Paramonov, A. S., Shingarova, L. N., Chupin, V. V., Kirpichnikov, M. P., et al. (2010). Lipid-protein nanodiscs as reference medium in detergent screening for high-resolution NMR studies of integral membrane proteins. J. Am. Chem. Soc. 132, 5628–5629. doi: 10.1021/ja9097498
Shenkarev, Z. O., Lyukmanova, E. N., Solozhenkin, O. I., Gagnidze, I. E., Nekrasova, O. V., Chupin, V. V., et al. (2009). Lipid-protein nanodiscs: possible application in high-resolution NMR investigations of membrane proteins and membrane-active peptides. Biochemistry Mosc. 74, 756–765. doi: 10.1134/S0006297909070086
Steinke, K. V., Gorinski, N., Wojciechowski, D., Todorov, V., Guseva, D., Ponimaskin, E., et al. (2015). Human CLC-K channels require palmitoylation of their accessory subunit barttin to be functional. J. Biol. Chem. 290, 17390–17400. doi: 10.1074/jbc.M114.631705
Stolting, G., Fischer, M., and Fahlke, C. (2014). CLC channel function and dysfunction in health and disease. Front. Physiol. 5:378. doi: 10.3389/fphys.2014.00378
Tajima, M., Hayama, A., Rai, T., Sasaki, S., and Uchida, S. (2007). Barttin binds to the outer lateral surface of the ClC-K2 chloride channel. Biochem. Biophys. Res. Commun. 362, 858–864. doi: 10.1016/j.bbrc.2007.08.097
Tan, H., Bungert-Plumke, S., Fahlke, C., and Stolting, G. (2017). Reduced membrane insertion of CLC-K by V33L barttin results in loss of hearing, but leaves kidney function intact. Front. Physiol. 8:269. doi: 10.3389/fphys.2017.00269
Theillet, F. X., Binolfi, A., Bekei, B., Martorana, A., Rose, H. M., Stuiver, M., et al. (2016). Structural disorder of monomeric alpha-synuclein persists in mammalian cells. Nature 530, 45–50. doi: 10.1038/nature16531
Tsukamoto, H., Sinha, A., DeWitt, M., and Farrens, D. L. (2010). Monomeric rhodopsin is the minimal functional unit required for arrestin binding. J. Mol. Biol. 399, 501–511. doi: 10.1016/j.jmb.2010.04.029
Viegas, A., Viennet, T., and Etzkorn, M. (2016a). The power, pitfalls and potential of the nanodisc system for NMR-based studies. Biol. Chem. 397, 1335–1354.
Viegas, A., Viennet, T., Yu, T. Y., Schumann, F., Bermel, W., Wagner, G., et al. (2016b). UTOPIA NMR: activating unexploited magnetization using interleaved low-gamma detection. J. Biomol. NMR 64, 9–15. doi: 10.1007/s10858-015-0008-7
Viennet, T., Viegas, A., Kuepper, A., Arens, S., Gelev, V., Petrov, O., et al. (2016). Selective protein hyperpolarization in cell lysates using targeted dynamic nuclear polarization. Angew. Chem. Int. Ed. Engl. 55, 10746–10750. doi: 10.1002/anie.201603205
Viennet, T., Wordehoff, M. M., Uluca, B., Poojari, C., Shaykhalishahi, H., Willbold, D., et al. (2018). Structural insights from lipid-bilayer nanodiscs link alpha-Synuclein membrane-binding modes to amyloid fibril formation. Commun Biol 1:44.
Vranken, W. F., Boucher, W., Stevens, T. J., Fogh, R. H., Pajon, A., Llinas, M., et al. (2005). The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696. doi: 10.1002/prot.20449
Wojciechowski, D., Fischer, M., and Fahlke, C. (2015). Tryptophan scanning mutagenesis identifies the molecular determinants of distinct barttin functions. J. Biol. Chem. 290, 18732–18743. doi: 10.1074/jbc.M114.625376
Wojciechowski, D., Thiemann, S., Schaal, C., Rahtz, A., de la Roche, J., Begemann, B., et al. (2018). Activation of renal ClC-K chloride channels depends on an intact N terminus of their accessory subunit barttin. J. Biol. Chem. 293, 8626–8637. doi: 10.1074/jbc.RA117.000860
Keywords: barttin, ion channel, lipid bilayer nanodisc, detergent micelle, nuclear magnetic resonance
Citation: Viennet T, Bungert-Plümke S, Elter S, Viegas A, Fahlke C and Etzkorn M (2019) Reconstitution and NMR Characterization of the Ion-Channel Accessory Subunit Barttin in Detergents and Lipid-Bilayer Nanodiscs. Front. Mol. Biosci. 6:13. doi: 10.3389/fmolb.2019.00013
Received: 07 January 2019; Accepted: 19 February 2019;
Published: 14 March 2019.
Edited by:
Anja Böckmann, UMS3760 Institut de Biologie et Chimie des Protéines (IBCP), FranceReviewed by:
Daniel Huster, Leipzig University, GermanyCopyright © 2019 Viennet, Bungert-Plümke, Elter, Viegas, Fahlke and Etzkorn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christoph Fahlke, Yy5mYWhsa2VAZnotanVlbGljaC5kZQ==
Manuel Etzkorn, bWFudWVsLmV0emtvcm5AaGh1LmRl
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.