- 1The Robert H Smith Faculty of Agriculture, Food and Environment, The Institute of Biochemistry, Food and Nutrition, The Hebrew University, Rehovot, Israel
- 2The Fritz Haber Center for Molecular Dynamics, The Hebrew University, Jerusalem, Israel
- 3Department of Molecular Genetics, German Institute of Human Nutrition Potsdam-Rehbruecke, Nuthetal, Germany
- 4Center for Integrative Physiology and Molecular Medicine, Saarland University, Homburg, Germany
Chickens sense the bitter taste of structurally different molecules with merely three bitter taste receptors (Gallus gallus taste 2 receptors, ggTas2rs), representing a minimal case of bitter perception. Some bitter compounds like quinine, diphenidol and chlorpheniramine, activate all three ggTas2rs, while others selectively activate one or two of the receptors. We focus on bitter compounds with different selectivity profiles toward the three receptors, to shed light on the molecular recognition complexity in bitter taste. Using homology modeling and induced-fit docking simulations, we investigated the binding modes of ggTas2r agonists. Interestingly, promiscuous compounds are predicted to establish polar interactions with position 6.51 and hydrophobic interactions with positions 3.32 and 5.42 in all ggTas2rs; whereas certain residues are responsible for receptor selectivity. Lys3.29 and Asn3.36 are suggested as ggTas2r1-specificity-conferring residues; Gln6.55 as ggTas2r2-specificity-conferring residue; Ser5.38 and Gln7.42 as ggTas2r7-specificity conferring residues. The selectivity profile of quinine analogs, quinidine, epiquinidine and ethylhydrocupreine, was then characterized by combining calcium-imaging experiments and in silico approaches. ggTas2r models were used to virtually screen BitterDB compounds. ~50% of compounds known to be bitter to human are likely to be bitter to chicken, with 25, 20, 37% predicted to be ggTas2r1, ggTas2r2, ggTas2r7 agonists, respectively. Predicted ggTas2rs agonists can be tested with in vitro and in vivo experiments, contributing to our understanding of bitter taste in chicken and, consequently, to the improvement of chicken feed.
Introduction
Bitter taste is one of the basic taste modalities thought to protect organisms from consuming poisons that are often bitter. Bitter taste perception is mediated by bitter taste receptors (Tas2rs), a subfamily of Class A G-protein coupled receptors (GPCRs) (Di Pizio and Niv, 2014; Di Pizio et al., 2016). Recently, it has been shown that Tas2rs are expressed in tissues other than the mouth (Behrens and Meyerhof, 2010; Clark et al., 2012). These extra-oral Tas2rs have been implicated in diverse functions, including cellular responses to toxins (Lee and Cohen, 2014), bronchodilation (Ting-A-Kee et al., 2015), and regulation of thyroid hormones (Clark et al., 2015), suggesting that sensing bitter compounds is likely to have physiological roles beyond food evaluation.
The molecular recognition of bitter molecules by their receptors is rather complex. The structures of close to 700 bitter compounds have been gathered in the BitterDB database (http://bitterdb.agri.huji.ac.il; Wiener et al., 2012), which currently contains a special session dedicated to chicken (http://bitterdb.agri.huji.ac.il/dbbitter.php?mode_organism=Chicken). However, the number of bitter compounds is estimated to be thousands (Meyerhof et al., 2010) as recently confirmed by the BitterPredict protocol (Dagan-Wiener et al., 2017). In humans, these numerous compounds are perceived as bitter by 25 receptors (TAS2Rs); some of the receptors are still orphan, or have few known agonists, while others can be activated by numerous and structurally dissimilar compounds (Di Pizio and Niv, 2015). Similarly, the ligands vary in the repertoire of bitter receptors that they activate: some bitter compounds are selective toward a single TAS2R, while others activate multiple TAS2Rs (Di Pizio and Niv, 2015).
Interestingly, the number of Tas2rs varies by species. Chickens sense the bitter taste with merely three chicken (Gallus gallus) bitter taste receptors, ggTas2r1, ggTas2r2, ggTas2r7 (Go, 2006), which are expressed, together with their downstream signaling components, in both gustatory and extra-gustatory tissues (Cheled-Shoval et al., 2014, 2015). With their small Tas2r repertoire, chickens represent an optimal system to investigate the molecular mechanism of bitter taste perception. In 2014, a set of 46 bitter compounds, previously screened on human bitter taste receptors (Meyerhof et al., 2010), has been profiled against all chicken bitter taste receptors under the same assay conditions (Behrens et al., 2014), leading to the de-orphanization of ggTas2rs and furnishing a consistent dataset of ggTas2r agonists. The results of this screening indeed demonstrated that the smaller repertoire of ggTas2rs compared to human TAS2Rs is partially compensated by a broader repertoire of ligands. Even though ggTas2rs appear to be promiscuous and tend to accommodate many chemically diverse compounds, bitter compounds may exhibit different selectivity/promiscuity profiles toward chicken bitter taste receptors, and can be very selective for a specific subtype (Behrens et al., 2014). So far, a total of 25 bitter agonists for the three chicken Tas2rs and an antagonist for ggTas2r1 and ggTas2r7 have been unraveled (Behrens et al., 2014; Hirose et al., 2015; Dey et al., 2016; Cheled-Shoval et al., 2017). We took advantage of the simple ggTas2r system to compare in vitro vs. in vivo detection thresholds of selective and promiscuous ggTas2r agonists. In general, in vivo thresholds were similar or up to two orders of magnitude higher than the in vitro ones, but the in vivo:in vitro ratios were different for different ligands and ggTas2r-promiscuous ligands did not exhibit lower ratios than ggTas2r-selective ligands (Cheled-Shoval et al., 2017). Recently, integrating in silico and in vitro experiments on ggTas2r1 we investigated the binding modes of known agonists into the binding site and predicted additional ligands, providing a docking strategy for chemosensory receptors and other GPCRs, where the sequence identity between models and templates is very low (Di Pizio et al., 2017). Here, we use a similar approach to analyze the promiscuity/selectivity profile of bitter compounds in chicken, aiming to unravel what makes compounds active toward all ggTas2rs or selective for a particular subtype.
Results and Discussion
Tas2r-Ligand Relations
Figure 1 represents the ligand repertoire of ggTas2rs vs. that of TAS2Rs. Promiscuous compounds for chicken activate several human TAS2Rs, and the most selective compounds for chicken are selective for humans as well. Therefore, understanding how selectivity is achieved in chicken may provide insights about the selectivity of bitter compounds in humans.
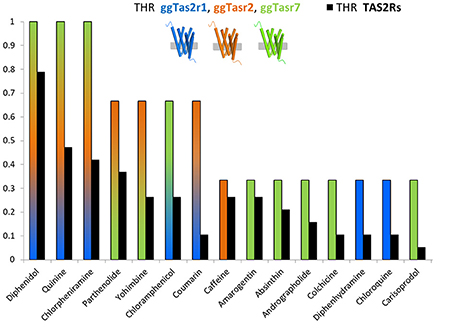
Figure 1. THR (target hit-rate) of bitter compounds toward human TAS2Rs (black bars) and chicken ggTas2r1, ggTas2r2, ggTas2r7 (blue, orange, and green bars, respectively). THR parameter is the number of targets hit at a specific concentration divided by the number of targets tested (Azzaoui et al., 2007; Di Pizio and Niv, 2015). Specifically, only compounds that elicited receptor activation of both human TAS2Rs and ggTas2rs at concentration of 300 μM or lower were used to generate this graph.
Among the compounds reported in Figure 1, we can observe ggTas2r1-selective molecules, i.e., diphenhydramine and chloroquine, ggTas2r2-selective molecules, i.e., caffeine, ggTas2r7-selective compounds, i.e., amarogentin, andrographolide, etc.; but also promiscuous compounds - diphenidol, quinine and chlorpheniramine activate all ggTas2rs; or ligands with an intermediate promiscuity toward the chicken receptors - parthenolide and yohimbine activate ggTas2r2 and ggTas2r7, while coumarin activate ggTas2r1 and ggTas2r2, and chloramphenicol responds to ggTas2r1 and ggTas2r7.
Binding Pocket of ggTas2rs
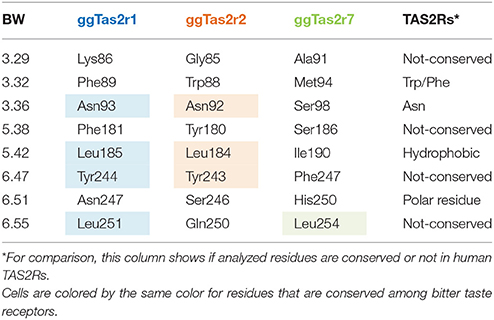
In order to identify the specific residues that may be responsible for the selectivity of each ggTas2r toward their agonists, we analyzed similarities and differences in the binding site. As previous works on human bitter taste receptors suggested (Brockhoff et al., 2010; Born et al., 2013; Karaman et al., 2016), the location of the binding site in ggTas2r1 coincides with the canonical binding site of Class A GPCRs (Di Pizio et al., 2017). Importantly, our recent investigation of the ggTas2r1 agonist-bound conformation led to identify Lys863.29, Phe893.32, Asn933.36, Phe1815.38, Leu1855.42, Tyr2446.47, Asn2476.51, and Leu2516.55 as the ggTas2r1 residues involved in agonist binding and recognition (Di Pizio et al., 2017). Transmembrane (TM) residues are identified throughout the text by a superscript numbering system according to the Ballesteros-Weinstein (BW) numbering method, where the residue corresponding to the Class A GPCRs most conserved residue in TM number X is assigned the index X.50, and the remaining residues are numbered relative to this position (Ballesteros and Weinstein, 1995).
Table 1 shows the different compositions of the agonist-interacting residues among the three chicken bitter taste receptors. Only three residues are conserved between the receptors ggTas2r1 and ggTas2r2 (Asn3.36, Leu5.42, Tyr6.47), one residue is conserved between ggTas2r1 and ggTas2r7 (Leu6.55), while no residues are conserved between ggTas2r2 and ggTas2r7. ggTas2r7 differs from the other two receptors: indeed, in the phylogenetic tree in the Figure 1 of the paper from Behrens et al. (2014), ggTas2r1 and ggTas2r2 are grouped together, while ggTas2r7 appears in a separate branch. Interestingly, Asn3.36 and Asn6.51, found to be relevant in agonist/ggTas2r1 binding (Di Pizio et al., 2017), are replaced by Asn3.36-Ser6.51, and Ser3.36-His6.51 combinations in ggTas2r2 and ggTas2r7, respectively. Phe3.32 and Phe5.38, involved in aromatic interactions in agonist-ggTas2r1 complexes (Di Pizio et al., 2017), are replaced by Trp3.32 and Tyr5.38, and Met3.32 and Ser5.38 in ggTas2r2 and ggTas2r7, respectively.
Binding Modes of Promiscuous vs. Selective Compounds
To investigate the influence of amino acid differences on the binding and the selectivity profile of bitter compounds, we analyzed the binding modes of both selective and promiscuous ligands in the cognate receptors. We built ggTas2r2 and ggTas2r7 models, analyzed the binding modes of their agonists with induced-fit docking simulations, and compared the previously predicted agonist/ggTas2r1 binding poses (Di Pizio et al., 2017) with those analyzed here. A matrix with all interactions found for each agonist/ggTas2r complex is reported in Supplementary Figure S1.
Binding modes of promiscuous compounds, e.g., diphenidol (Figure 2A and Supplementary Figure S2A): diphenidol in complex with ggTas2r1 forms H-bonds with Asn3.36 and Asn6.51, π-π stacking interaction with Phe5.38, and hydrophobic interactions with Leu5.42, Tyr6.47, and with Phe3.32 (Figure 2A1; Di Pizio et al., 2017). These interactions are well-conserved in diphenidol/ggTas2r2 and diphenidol/ggTas2r7 complexes (Figures 2A2,A3). In all cases we observe an H-bond interaction in position 6.51 (Ser in ggTas2r2 and His in ggTas2r7), and hydrophobic or aromatic interactions in positions 3.32, 5.42, 6.47. This allows diphenidol to assume a similar conformation in all three receptors with only slight differences: the π-π interaction with Phe5.38 in ggTas2r1 is replaced by a π-π stacking interaction with Tyr5.38 in ggTas2r2, but is lacking in ggTas2r7 where we have a polar residue, i.e., Ser, in this position; the H-bond between the ligand and ggTas2r1 Asn3.36 is not conserved in the other two receptors.
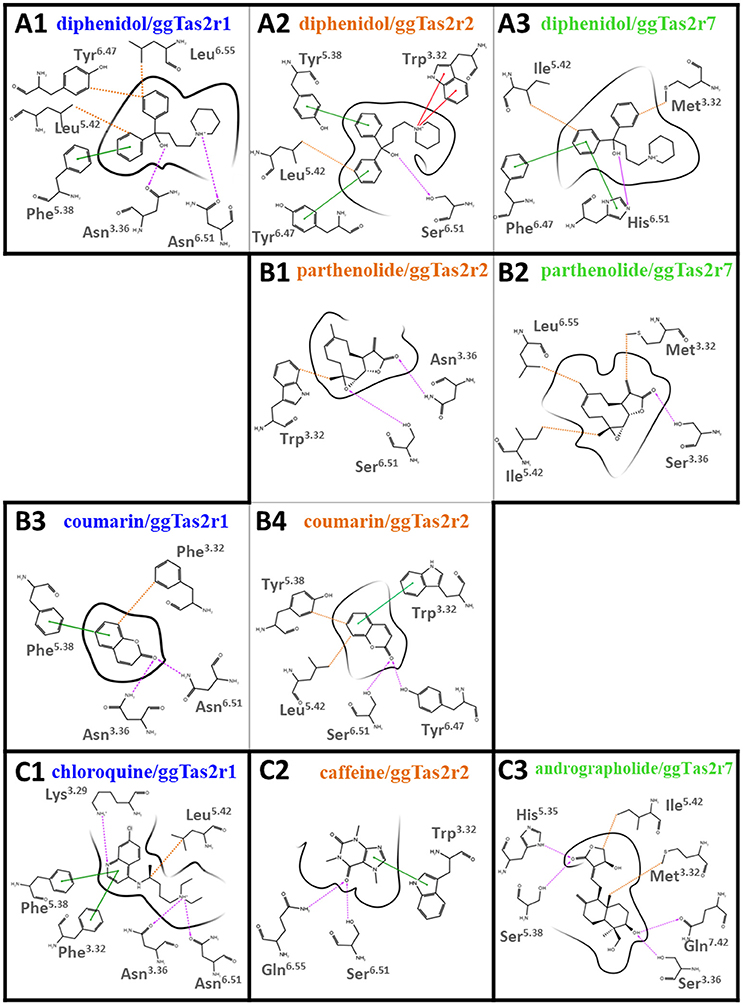
Figure 2. 2D representation of predicted binding modes of promiscuous (A), intermediate-promiscuous (B) and selective compounds (C) into the ggTas2r binding sites. H-bond interactions are shown as magenta dashed arrows, π-π interactions as green lines, cation-π interactions as red lines and hydrophobic interactions are represented as the distances between the closest heavy atoms of the ligand and the residue as orange dotted lines. Binding modes of diphenidol, coumarin, and chloroquine into ggTas2r1 have been described in our previous work (Di Pizio et al., 2017).
Binding modes of intermediate-promiscuous compounds, e.g., parthenolide and coumarin (Figure 2B and Supplementary Figure S2B). Parthenolide activates ggTas2r2 and ggTas2r7: it forms H-bonds with Asn3.36 and Ser6.51, and hydrophobic interactions with Trp3.32 when in complex with ggTas2r2 (Figure 2B1); similarly, it forms an H-bond with Ser3.36 and hydrophobic interactions with Met3.32, Ile5.42, and Leu6.55 in the ggTas2r7 binding site (Figure 2B2). Coumarin activates ggTas2r1 and ggTas2r2: in complex with ggTas2r1, it establishes H-bonds with Asn3.36 and Asn6.51, π-π stacking interaction with Phe5.38, and hydrophobic interactions with Phe3.32 (Figure 2B3; Di Pizio et al., 2017). The H-bond interaction between the ligand and the residue in position 6.51 is conserved in the ggTas2r2 complex as well, the H-bond in position 3.36 is lacking and replaced by an additional H-bond with Tyr6.47 (Figure 2B4). Moreover, in coumarin/ggTas2r2 binding pose, hydrophobic and π-π stacking interactions are observed with Leu5.42 and Tyr5.38, and Trp3.32, respectively (Figure 2B4).
Binding modes of selective compounds, e.g., chloroquine, caffeine, and andrographolide (Figure 2C and Supplementary Figure S2C). Chloroquine is selective for ggTas2r1 and binds Lys3.29, Asn3.36, and Asn6.51 through H-bonds, Phe3.32 and Phe5.38 through π-π stacking interactions, Leu5.42 with hydrophobic interactions. Lys3.29 is replaced by a glycine and an alanine in ggTas2r2 and ggTas2r7, respectively, and may be responsible for the selectivity of chloroquine for ggTas2r1. Indeed, it has been shown that mutating Lys3.29 to Ala causes severe impairments of the ggTas2r1 activation by chloroquine (Di Pizio et al., 2017). Caffeine is selective for ggTas2r2 and interacts with Trp3.32 through π-π stacking interaction, and with Ser6.51 and Gln6.55 through H-bonds. Gln6.55 is substituted with a leucine in both ggTas2r1 and ggTas2r7, and is not conserved in the TAS2R family. Andrographolide is selective for ggTas2r7 and forms several H-bonds (i.e., with His5.35, Ser5.38, Ser3.36, and Gln7.42) and hydrophobic interactions with Met3.32 and Ile5.42. Position 7.42, a non-conserved residue both in chicken and human, is not found to be involved in ligand-interaction in ggTas2r1 (Di Pizio et al., 2017) and ggTas2r2. Interestingly, the interaction pattern of selective compounds is different from what we observed for the promiscuous ligands.
The predicted binding poses of promiscuous compounds are compatible in all three ggTas2rs, with position 6.51 involved in polar interaction and positions 3.32 and 5.42 in hydrophobic interactions. Interestingly, Trp/Phe3.32 is conserved in TAS2Rs, as well as a polar residue is present in position 6.51 and a hydrophobic residue in 5.42 (Table 1). Comparing the binding modes of promiscuous ligands into the three receptors we could underline interesting differences due to: (1) differences in the amino acid sequence, such as the one of position 5.38 - the presence of an aromatic residue in this position in both ggTas2r1 and ggTas2r2 (Phe and Tyr, respectively) allows for aromatic interactions with ligands within those receptors, which are not possible with Ser5.58 of ggTas2r7; (2) different residue arrangements, such as the case of Asn3.36 - this position is conserved between ggTas2r1 and ggTas2r2, but because of the different environment around the residue, Asn3.36 is involved in direct H-bond interactions with the agonists in ggTas2r1, whereas it is involved in intramolecular interactions with Tyr6.47 and/or Trp3.32 in ggTas2r2, and is not always available for direct ligand-binding. On the contrary, selective compounds tend to interact with non-conserved residues, allowing us to identify ggTas2r1-, ggTas2r2-, and ggTas2r7-specific residues responsible for selectivity: Lys3.29 and Asn3.36 as ggTas2r1-specific residues; Gln6.55 as ggTas2r2-specific residue; Ser5.38 and Gln7.42 as ggTas2r7-specific residues.
ggTas2r Selectivity of Quinine Analogs
Quinine analogs, i.e., quinidine, epiquinidine and ethylhydrocupreine, are an example of structurally similar chemicals that elicit different receptor activations. They have been recently found to activate ggTas2r1 with different threshold concentrations (~0.3 μM quinidine, ~10 μM epiquinidine and ethylhydrocupreine) (Di Pizio et al., 2017). To characterize their selectivity profile toward the entire ggTas2r family, we carried out calcium-mobilization assays using these compounds toward ggTas2r2 and ggTas2r7 as well (Figure 3A). All tested compounds are more selective toward ggTas2r1 than toward ggTas2r2 and ggTas2r7, as shown by activation patterns. Epiquinidine and ethylhydrocupreine can still activate ggTas2r2 and ggTas2r7, whereas quinidine is a selective ggTas2r1 agonist. As described previously, quinidine binds ggTas2r1 through an H-bond with Asn3.36 and hydrophobic interactions with Phe3.32, Phe5.38, Leu5.42, Tyr6.47 (Di Pizio et al., 2017). This binding mode is not permitted in ggTas2r2 (where Asn3.36 assumes a different conformation) and in ggTas2r7 (where Asn3.36 is replaced with a shorter Ser). Instead, in complex with ggTas2r1, epiquinidine and ethylhydrocupreine form H-bonds with Asn6.51, π-π stacking interaction with Phe5.38, and hydrophobic interactions with Leu5.42. Indeed, the predicted binding modes of these compounds in ggTas2r2 show a π-π stacking interaction with Tyr5.38 that is not allowed in the ggTas2r7 binding site (Figure 3B).
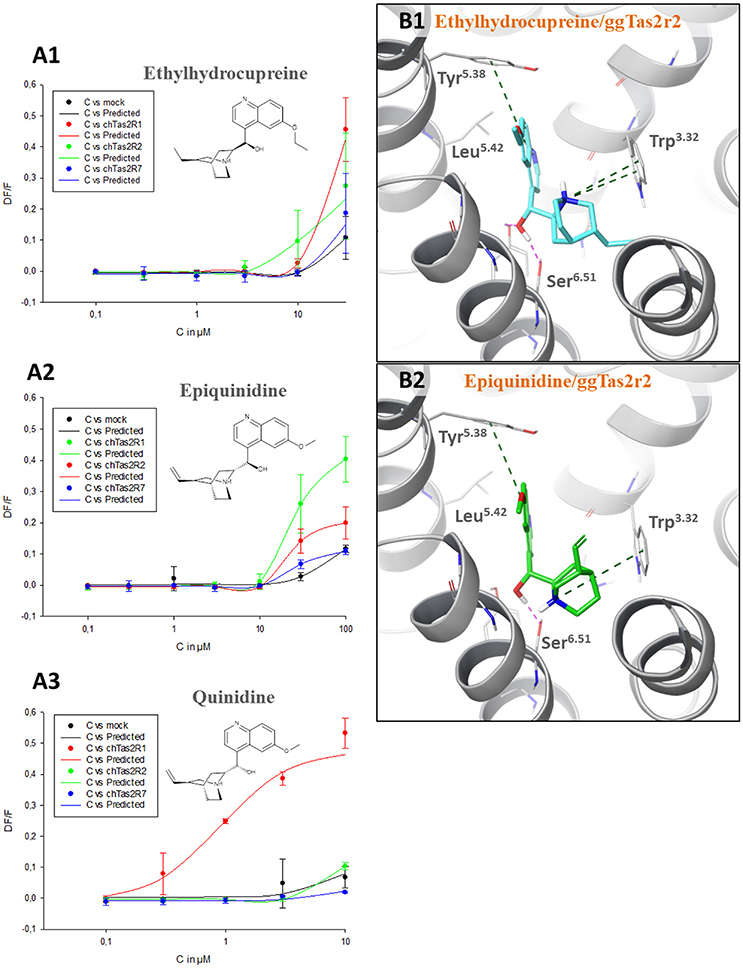
Figure 3. (A) Dose-response relations for ethylhydrocupreine (A1), epiquinidine (A2), and quinidine (A3) in cells expressing ggTas2r1, −2, and −7. The y-axis shows the relative fluorescence changes (ΔF/F), and the x axis the ligand concentration (C) in μM. Two experiments performed in duplicates were used for the calculations. The curves related to ggTas2r1 activation have been already published (Di Pizio et al., 2017). (B) Predicted binding modes of ethylhydrocupreine (in cyan, B1) and epiquinidine (in green, B2) into ggTas2r2 binding pocket. H-bond interactions are shown as magenta dotted lines, π-π interactions as green dashed lines.
Relations and Overlaps between Predicted Bitter Compounds for Each ggTas2r
The ggTas2r models were next used to virtually screen molecules from BitterDB (Wiener et al., 2012) that are bitter for human, but have not yet been tested against ggTas2rs (642 compounds in total). To take the flexibility of the binding site residues into consideration, we used all agonist-bound models built for each ggTas2r subtype (the superimposition of all models is shown in Supplementary Figure S3). Pharmacophore modeling was employed as a filter prior to the docking screening. Pharmacophores are able to rationalize the receptor activity analyzing the ligand chemical structures (Martin et al., 2016), and pharmacophore-based virtual screening are generally very fast compared to high-throughput docking (Cheng et al., 2012). The pharmacophore filter allowed us to eliminate compounds that would have been obvious outliers and to submit to the docking analysis only compounds that have the molecular determinants responsible for receptor activity, speeding the screening process.
Pharmacophores built to predict ggTas2r1, ggTas2r2, and ggTas2r7 activity are represented in Figures 4A–C, respectively. ggTas2r1 pharmacophore, with chlorpheniramine as reference structure, is made of donor (D), hydrophobic (H) and aromatic (R) features, which correspond to the ligand groups that interact with Asn3.36, Phe3.32, and Phe5.38, respectively, in the chlorpheniramine/ggTas2r1 binding pose. Parthenolide is the reference compound of ggTas2r2 pharmacophore, where the acceptor (A) feature maps the carbonyl oxygen H-bonding with Asn3.36 and the hydrophobic (H) features match the aliphatic moieties of the ligands responsible for hydrophobic contacts with Trp883.32 and Tyr1805.38. Two acceptors and one hydrophobic feature were found as determinants for ggTas2r7 activation. These features map the carisoprodol groups interacting with Ser983.36, Gln2757.42, and Leu2546.55.
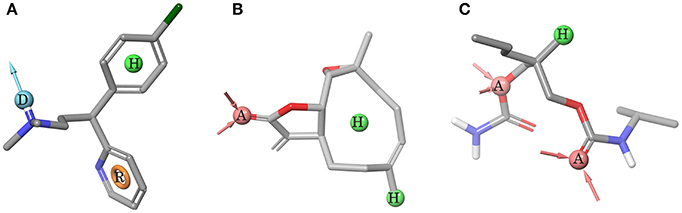
Figure 4. Pharmacophore models. (A) ggTas2r1 pharmacophore DHR (=H-Bond donor, hydrophobic feature, aromatic ring), reference structure chlorpheniramine, Sensitivity = 0.86, Specificity = 0.75. (B) ggTas2r2 pharmacophore AHH (=H-bond acceptor, hydrophobic feature, hydrophobic feature), reference structure quinine, Sensitivity = 0.67, Specificity = 0.62. (C) ggTas2r7 pharmacophore AAH (=H-bond acceptor, H-bond acceptor, hydrophobic feature), reference structure carisoprodol, Sensitivity = 0.78, Specificity = 0.68. Acceptor (red) and donor (cyan) features are shown as spheres and vectors, hydrophobic feature as green sphere, and aromatic feature as orange ring.
The screening of BitterDB against these pharmacophore models allowed for the selection of 165, 144, and 275 compounds as potential active compounds toward ggTas2r1,−2, and−7, respectively. Through docking, potential ggTas2r1 agonists were reduced to 162, equaling 25% of the database, the predicted ggTas2r2 agonists were reduced to 128, equaling 20% of screened library, and potential ggTas2r7 agonists were reduced to 236, equaling 37% of the screened compounds. In total, 332 of the 642 screened compounds are predicted to be bitter for chicken (52%). Interestingly, the percentage of the molecules selected with the virtual screening for each receptor is similar to that observed for the experimental screening, where ggTas2rs responded to 50% of tested molecules, ggTas2r1 and ggTas2r2 recognized ~20% of the compounds, and ggTas2r7 ~40% (Behrens et al., 2014). The relations between known and predicted compounds for each ggTas2r are represented in the Venn diagrams in Figure 5. Selected compounds, their pharmacophore and docking scores are reported in Supplementary Tables S1–S3.
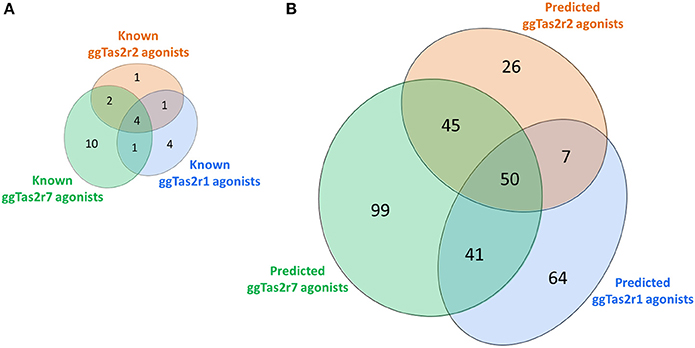
Figure 5. Venn diagrams of known (A) and predicted (B) ggTas2r agonists. The portions of ggTas2r1, −2, and −7 agonists are colored in blue, orange and green, respectively.
Conclusions
In this study, we analyzed the current data on bitter compounds for chicken to unravel the molecular mechanism of ligand recognition and selectivity in chicken bitter taste receptors. Detailed structural models of the ligand-binding pockets were obtained with homology modeling and induced-fit docking techniques and allowed for the identification of residue positions that may confer subtype selectivity. Integrating in silico and in vitro approaches we characterized the selectivity profiles of quinidine, epiquinidine and ethylhydrocupreine toward ggTas2rs. We next used ggTas2r models to screen BitterDB compounds and we predicted that ~50% of compounds known to be bitter for human may activate chicken receptors. Predicted ggTas2rs agonists could be valuable for in vitro and in vivo experiments, contributing to our understanding of bitter perception in chickens and, consequently, leading to the improvement of chicken feed. Moreover, we provide a methodology that can be applied to identify residue positions that may confer bitter taste receptor selectivity in humans.
Methods
Structure-Based Analysis
ggTas2r1, ggTas2r2, and ggTas2r7 sequences were aligned using the webserver MAFFT (Katoh and Standley, 2013). ggTas2r1 has 58% sequence similarity (identity 35%) with ggTas2r2 and 46% sequence similarity (identity 25%) with ggTas2r7; ggTas2r2 has 41% sequence similarity (identity 23%) with ggTas2r7. Homology modeling was performed with Prime (version 4.2, Schrödinger, LLC, New York, NY, 2015) (Jacobson et al., 2002, 2004) using as template ggTas2r1 previously modeled and refined in complex with quinine (Di Pizio et al., 2017). Protein structures were then optimized with the Protein Preparation Wizard tool from Schrödinger at physiological pH. Induced-fit Docking simulations were run to investigate the binding modes of quinine, diphenidol, chlorpheniramine, parthenolide, yohimbine, coumarin, caffeine, epiquinidine and ethylhydrocupreine with ggTas2r2; and of quinine, diphenidol, chlorpheniramine, parthenolide, yohimbine, chloramphenicol, amarogentin, absinthin, andrographolide, colchicine and carisoprodol with ggTas2r7. Glide (version 6.9) and Prime (version 4.2) were used for the Induced-Fit Docking protocol available in the Schrödinger software suite 2015-4 (Friesner et al., 2004; Zhu et al., 2007). The grid box was built at 10 Å from quinine in the quinine/ggTas2r1 complex. The van der Waals scaling of receptors and ligands were 0.5. Side chains of residues within 4.0 Å of the ligand were refined. The Extended Sampling protocol and OPLS 2005 force field were used. The docking was performed with the Standard Precision (SP) mode of Glide. Glide score, combined with visual inspection, was used to select the docking poses. The Ligand Interaction Diagram available in Maestro (version 10.4, Schrödinger, LLC, New York, NY, 2015) was used to create the 2D representations of predicted binding modes shown in Figure 2.
Ligand-Based Analysis
Compounds and data activities reported in Table S2 of Behrens et al. (2014) were used to build the pharmacophore models using Phase (version 4.3, Schrödinger, LLC, New York, NY, 2015) (Dixon et al., 2006). Following features were modified: in donor (D) feature the setting [#1]([NH;X4]([#6;X4])([#6;X4])([#6;X4])) was set not to be excluded, the hydrophobic (H) feature was set to map both hydrophobic and aromatic moieties. The minimum intersite distance for features was set at 1 Å. 7 active (chloramphenicol, chloroquine, chlorpheniramine, diphenhydramin, diphenidol, nicotin, quinine) and 24 inactive molecules (absinthin, acetaminophen, alpha-thujone, amarogentin, andrographolide, brucine, caffeine, camphor, carisoprodol, colchicine, cycloheximide, denatonium, erythromycin, falcarindiol, ginkgolide A, limonin, noscapine, parthenolide, phenylthiocarbamide, quassin, saccharin, strychnine, thiamine, yohimbine) were used to build ggTas2r1 pharmacophore model DHR (one donor, one hydrophobic, and one aromatic feature). 6 actives (caffeine, chlorpheniramine, coumarin, diphenidol, parthenolide, quinine) and 26 inactives (absinthin, acetaminophen, alpha-thujone, amarogentin, andrographolide, azathioprine, brucine, camphor, carisoprodol, colchicine, cycloheximide, denatonium, diphenidramine, erythromycin, falcarindiol, ginkgolide A, limonin, nicotine, noscapine, phenylthiocarbamide, picrotoxinin, quassin, saccharin, strychnine, thiamine) were used to build the ggTas2r2 pharmacophore model AHH (one acceptor group and two hydrophobic features). 11 active (absinthin, amarogentin, andrographolide, carisoprodol, chloramphenicol, diphenidol, parthenolide, quinine, chlorpheniramine, colchicine, yohimbine) and 18 inactive molecules (acetaminophen, azathioprine, brucine, caffeine, camphor, chloroquine, coumarin, cycloheximide, denatonium, diphenidramine, falcarindiol, limonin, nicotine, noscapine, phenylthiocarbamide, saccharin, strychnine, thiamine) were used to build the ggTas2r7 pharmacophore model AAH (two acceptor and one hydrophobic features). Excluded volumes were added to each pharmacophore model around the reference structure (2 Å between the ligand and the shell). The sensitivity and specificity (SP) of the model were evaluated by the Equations (1) and (2):
where TP, TN, FP, and FN stand for the number of true positives, true negatives, false positives, and false negatives, respectively.
Virtual Screening
BitterDB compounds were prepared with LigPrep (version 3.6, Schrödinger, LLC, New York, NY, 2015) through the generation of stereoisomers and protonation states at pH 7 ± 0.5 and then with Phase (version 4.3, Schrödinger, LLC, New York, NY, 2015) to generate the 3D Phase Database, where for each molecule, pharmacophore sites and 100 conformers were calculated. The database was screened against ggTas2r1, ggTas2r2, and ggTas2r7 pharmacophore models, setting at least two features to be mapped and applying a fitness threshold of 0.4 (Dixon et al., 2006). Molecules filtered with the pharmacophore screening were passed to the docking analysis with the Virtual Screening Workflow, which allowed us to use more receptors simultaneously. We used both the models of ggTas2r1 in complex with agonists built in our previous work and the models of ggTas2r2 and ggTas2r7 in complex with agonists built here, as detailed above. Glide score threshold of 4.0 kcal/mol was applied to select docking results.
Calcium-Mobilization Assays
ggTas2r1, ggTas2r2, and ggTas2r7 constructs, as well as empty expression vectors as controls were transiently transfected in HEK 293T cells stably expressing the chimeric Gα protein Gα16gust44, which were grown in 96-well plates exactly as described previously (Behrens et al., 2014). About 24 h after transfection, the cells were loaded with the calcium-sensitive dye Fluo4-AM in the presence of 2.5 mM probenecid, washed and placed in a fluorometric imaging plate reader (FLIPR-tetra, Molecular devices). Different concentrations of the bitter compounds (Sigma-Aldrich, Taufkirchen) were automatically applied and changes in fluorescence were monitored. A second application of 100 nM SST-14 stimulating endogenous somatostatin receptors was included as vitality control. Dose-response relations were calculated with SigmaPlot.
Author Contributions
ADP and MYN conceived the research. ADP and NS performed the ligand-based analysis; ADP conducted the structure-based experiments and wrote the manuscript. MB performed the in vitro experiments. MB and WM designed and analyzed the in vitro experiments. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Lady Davis Fellowship to ADP, Israel Science Foundation grants 432/12 and 494/16 and ISF-NSFC 2463/16 grants to MYN as well as support from the European Union's Seventh Framework Programme to WM (#613879; SynSignal) are gratefully acknowledged. MYN participates in the European COST Action CA15135 (MuTaLig). We thank Ulrike Redel for excellent technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2018.00006/full#supplementary-material
References
Azzaoui, K., Hamon, J., Faller, B., Whitebread, S., Jacoby, E., Bender, A., et al. (2007). Modeling promiscuity based on in vitro safety pharmacology profiling data. ChemMedChem 2, 874–880. doi: 10.1002/cmdc.200700036
Ballesteros, J. A., and Weinstein, H. (1995). [19] Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428. doi: 10.1016/S1043-9471(05)80049-7
Behrens, M., Korsching, S. I., and Meyerhof, W. (2014). Tuning properties of avian and frog bitter taste receptors dynamically fit gene repertoire sizes. Mol. Biol. Evol. 31, 3216–3227. doi: 10.1093/molbev/msu254
Behrens, M., and Meyerhof, W. (2010). Oral and extraoral bitter taste receptors. Results Probl. Cell Differ. 52, 87–99. doi: 10.1007/978-3-642-14426-4_8
Born, S., Levit, A., Niv, M. Y., Meyerhof, W., and Behrens, M. (2013). The human bitter taste receptor TAS2R10 is tailored to accommodate numerous diverse ligands. J. Neurosci. 33, 201–213. doi: 10.1523/JNEUROSCI.3248-12.2013
Brockhoff, A., Behrens, M., Niv, M. Y., and Meyerhof, W. (2010). Structural requirements of bitter taste receptor activation. Proc. Natl. Acad. Sci. U.S.A. 107, 24, 11110–11115. doi: 10.1073/pnas.0913862107
Cheled-Shoval, S. L., Behrens, M., Meyerhof, W., Niv, M. Y., and Uni, Z. (2014). Perinatal administration of a bitter tastant influences gene expression in chicken palate and duodenum. J. Agric. Food Chem. 62, 12512–12520. doi: 10.1021/jf502219a
Cheled-Shoval, S. L., Druyan, S., and Uni, Z. (2015). Bitter, sweet and umami taste receptors and downstream signaling effectors: expression in embryonic and growing chicken gastrointestinal tract. Poult. Sci. 94, 1928–1941. doi: 10.3382/ps/pev152
Cheled-Shoval, S., Behrens, M., Korb, A., Di Pizio, A., Meyerhof, W., Uni, Z., et al. (2017). From cell to beak: in vitro and in vivo characterization of chicken bitter taste thresholds. Molecules 22:E821. doi: 10.3390/molecules22050821
Cheng, T., Li, Q., Zhou, Z., Wang, Y., and Bryant, S. H. (2012). Structure-based virtual screening for drug discovery: a problem-centric review. AAPS J. 14, 133–141. doi: 10.1208/s12248-012-9322-0
Clark, A. A., Dotson, C. D., Elson, A. E., Voigt, A., Boehm, U., Munger, W., et al. (2015). TAS2R bitter taste receptors regulate thyroid function. FASEB J. 29, 164–172. doi: 10.1096/fj.14-262246
Clark, A. A., Liggett, S. B., and Munger, S. D. (2012). Extraoral bitter taste receptors as mediators of off-target drug effects. FASEB J. 26, 4827–4831. doi: 10.1096/fj.12-215087
Dagan-Wiener, A., Nissim, I., Ben Abu, N., Borgonovo, G., Bassoli, A., and Niv, M. Y. (2017). Bitter or not? BitterPredict, a tool for predicting taste from chemical structure. Sci. Rep. 7:12074. doi: 10.1038/s41598-017-12359-7
Dey, B., Kawabata, F., Kawabata, Y., Yoshida, Y., Nishimura, S., and Tabata, S. (2016). Identification of functional bitter taste receptors and their antagonist in chickens. Biochem. Biophys. Res. Commun. 482, 693–699. doi: 10.1016/j.bbrc.2016.11.096
Di Pizio, A., Levit, A., Slutzki, M., Behrens, M., Karaman, R., and Niv, M. Y. (2016). Comparing class a GPCRs to bitter taste receptors: structural motifs, ligand interactions and agonist-to-antagonist ratios. Methods Cell Biol. 132, 401–427. doi: 10.1016/bs.mcb.2015.10.005
Di Pizio, A., Kruetzfeldt, L. M., Cheled-Shoval, S., Meyerhof, W., Behrens, M., and Niv, M. Y. (2017). Ligand binding modes from low resolution GPCR models and mutagenesis: chicken bitter taste receptor as a test-case. Sci. Rep. 7:8223. doi: 10.1038/s41598-017-08344-9
Di Pizio, A., and Niv, M. Y. (2014). Computational studies of smell and taste receptors. Isr. J. Chem. 54, 1205–1218. doi: 10.1002/ijch.201400027
Di Pizio, A., and Niv, M. Y. (2015). Promiscuity and selectivity of bitter molecules and their receptors. Bioorg. Med. Chem. 23, 4082–4091. doi: 10.1016/j.bmc.2015.04.025
Dixon, S. L., Smondyrev, A. M., and Rao, S. N. (2006). PHASE: a novel approach to pharmacophore modeling and 3D database searching. Chem. Biol. Drug. Des. 67, 370–372. doi: 10.1111/j.1747-0285.2006.00384.x
Friesner, R. A., Banks, J. L., Murphy, R. B., Halgren, T. A., Klicic, J. J., Mainz, D. T., et al. (2004). Glide: a new approach for rapid, accurate docking and scoring. 1. method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749. doi: 10.1021/jm0306430
Go, Y. (2006). Lineage-specific expansions and contractions of the bitter taste receptor gene repertoire in vertebrates. Mol. Biol. Evol. 23, 964–972. doi: 10.1093/molbev/msj106
Hirose, N., Kawabata, Y., Kawabata, Y., Nishimura, S., and Tabata, S. (2015). Bitter taste receptor T2R1 activities were compatible with behavioral sensitivity to bitterness in chickens. Biochem. Biophys. Res. Commun. 460, 464–468. doi: 10.1016/j.bbrc.2015.03.056
Jacobson, M. P., Pincus, D. L., Rapp, C. S., Day, T. J., Honig, B., Shaw, D. E., et al. (2004). A hierarchical approach to all-atom protein loop prediction. Proteins 55, 351–367. doi: 10.1002/prot.10613
Jacobson, M. P., Friesner, R. A., Xiang, Z., and Honig, B. (2002). On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 320, 597–608. doi: 10.1016/S0022-2836(02)00470-9
Karaman, R., Nowak, S., Di Pizio, A., Kitaneh, H., Abu-Jaish, A., Behrens, W., et al. (2016). Probing the binding pocket of the broadly tuned human bitter taste receptor TAS2R14 by chemical modification of cognate agonists. Chem. Biol. Drug. Des. 88, 66–75. doi: 10.1111/cbdd.12734
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Lee, R., and Cohen, N. (2014). Bitter and sweet taste receptors in the respiratory epithelium in health and disease. J. Mol. Med. 92, 1235–1244. doi: 10.1007/s00109-014-1222-6
Martin, Y. C., Abagyan, R., Ferenczy, G. G., Gillet, V. J., Oprea, T. I., Zefirov, J., et al. (2016). Glossary of terms used in computational drug design, part II (IUPAC Recommendations 2015). Pure App. Chem. 88, 239–264. doi: 10.1515/pac-2012-1204
Meyerhof, W., Batram, C., Kuhn, C., Brockhoff, A., Chudoba, E., Bufe, B., et al. (2010). The molecular receptive ranges of human TAS2R bitter taste receptors. Chem. Senses 35, 157–170. doi: 10.1093/chemse/bjp092
Ting-A-Kee, R., Heinmiller, A., and van der Kooy, D. (2015). A proposed resolution to the paradox of drug reward: dopamine's evolution from an aversive signal to a facilitator of drug reward via negative reinforcement. Neurosci. Biobehav. Rev. 56, 50–61. doi: 10.1016/j.neubiorev.2015.06.016
Wiener, A., Shudler, M., Levit, A., and Niv, M. Y. (2012). BitterDB: a database of bitter compounds. Nucleic Acids Res. 40, D413–D419. doi: 10.1093/nar/gkr755
Keywords: bitter compounds, chicken Tas2rs, GPCRs, homology modeling, induced-fit docking, virtual screening, calcium-mobilization assays
Citation: Di Pizio A, Shy N, Behrens M, Meyerhof W and Niv MY (2018) Molecular Features Underlying Selectivity in Chicken Bitter Taste Receptors. Front. Mol. Biosci. 5:6. doi: 10.3389/fmolb.2018.00006
Received: 09 October 2017; Accepted: 12 January 2018;
Published: 31 January 2018.
Edited by:
Piero Andrea Temussi, University of Naples Federico II, ItalyReviewed by:
Anastassios C. Papageorgiou, University of Turku, FinlandPaolo Rovero, University of Florence, Italy
Copyright © 2018 Di Pizio, Shy, Behrens, Meyerhof and Niv. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonella Di Pizio, YWRpcGl6aW9AbWFpbC5odWppLmFjLmls
 Antonella Di Pizio
Antonella Di Pizio Nitzan Shy1,2
Nitzan Shy1,2 Maik Behrens
Maik Behrens Masha Y. Niv
Masha Y. Niv