
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 19 May 2017
Sec. Protein Folding, Misfolding and Degradation
Volume 4 - 2017 | https://doi.org/10.3389/fmolb.2017.00031
This article is part of the Research Topic The Role of AAA+ Proteins in Protein Repair and Degradation View all 22 articles
Gaseous carbon dioxide enters the biosphere almost exclusively via the active site of the enzyme ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco). This highly conserved catalyst has an almost universal propensity to non-productively interact with its substrate ribulose 1,5-bisphosphate, leading to the formation of dead-end inhibited complexes. In diverse autotrophic organisms this tendency has been counteracted by the recruitment of dedicated AAA+ (ATPases associated with various cellular activities) proteins that all use the energy of ATP hydrolysis to remodel inhibited Rubisco active sites leading to release of the inhibitor. Three evolutionarily distinct classes of these Rubisco activases (Rcas) have been discovered so far. Green and red-type Rca are mostly found in photosynthetic eukaryotes of the green and red plastid lineage respectively, whereas CbbQO is associated with chemoautotrophic bacteria. Ongoing mechanistic studies are elucidating how the various motors are utilizing both similar and contrasting strategies to ultimately perform their common function of cracking the inhibited Rubisco active site. The best studied mechanism utilized by red-type Rca appears to involve transient threading of the Rubisco large subunit C-terminal peptide, reminiscent of the action performed by Clp proteases. As well as providing a fascinating example of convergent molecular evolution, Rca proteins can be considered promising crop-improvement targets. Approaches aiming to replace Rubisco in plants with improved enzymes will need to ensure the presence of a compatible Rca protein. The thermolability of the Rca protein found in crop plants provides an opportunity to fortify photosynthesis against high temperature stress. Photosynthesis also appears to be limited by Rca when light conditions are fluctuating. Synthetic biology strategies aiming to enhance the autotrophic CO2 fixation machinery will need to take into consideration the requirement for Rubisco activases as well as their properties.
The vast majority of carbon dioxide entering the living world does so via the slow and non-specific enzyme ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) (Spreitzer and Salvucci, 2002). The realization that this enzyme often represents the rate-limiting step of photosynthesis has made it a long-standing target for crop improvement strategies (Parry et al., 2007; Whitney et al., 2011a; Ort et al., 2015; Sharwood et al., 2016b). The peculiar properties of Rubisco can be understood as an accident of natural history. A highly complex reaction mechanism for ribulose 1,5-bisphosphate (RuBP) carboxylation evolved once in a high CO2 atmosphere lacking O2 (Andrews and Lorimer, 1987, Figure 1A). The unprecedented increase in atmospheric oxygen following the evolution of oxygenic photosynthesis increased the propensity of RuBP oxygenation, making it physiologically relevant (Andrews et al., 1973; Tcherkez, 2016). This resulted in massive metabolite damage (Linster et al., 2013) in the form of a build-up of 2-phosphoglycolate, which in contemporary plants is repaired by photorespiration (Bauwe et al., 2010). In C3 plants exposed to the current atmospheric environment, photorespiration operates at ~20% of photosynthesis (Cegelski and Schaefer, 2006), making it the second highest flux pathway. Operation of the photorespiratory pathway is energetically wasteful, resulting in a high selection pressure to reduce its flux. However, Rubisco's extensive adaptive walks through sequence space were not rewarded by the discovery of catalytic solutions that eliminated oxygenation (Maynard Smith, 1970; Mueller-Cajar and Whitney, 2008a). Instead it appeared easier to evolve a myriad of diverse syndromes that concentrate CO2 at the active site of the carboxylase (Badger et al., 1998; Rae et al., 2013; Sage, 2013). However, all of these mechanisms involve active transport, and thus increase the metabolic cost per CO2 fixed. Therefore, there was a concomitant pressure to enhance the catalytic fidelity of the enzyme by increasing its CO2/O2 specificity, as manifested most strongly in C3 plants and red algae (Tcherkez et al., 2006).
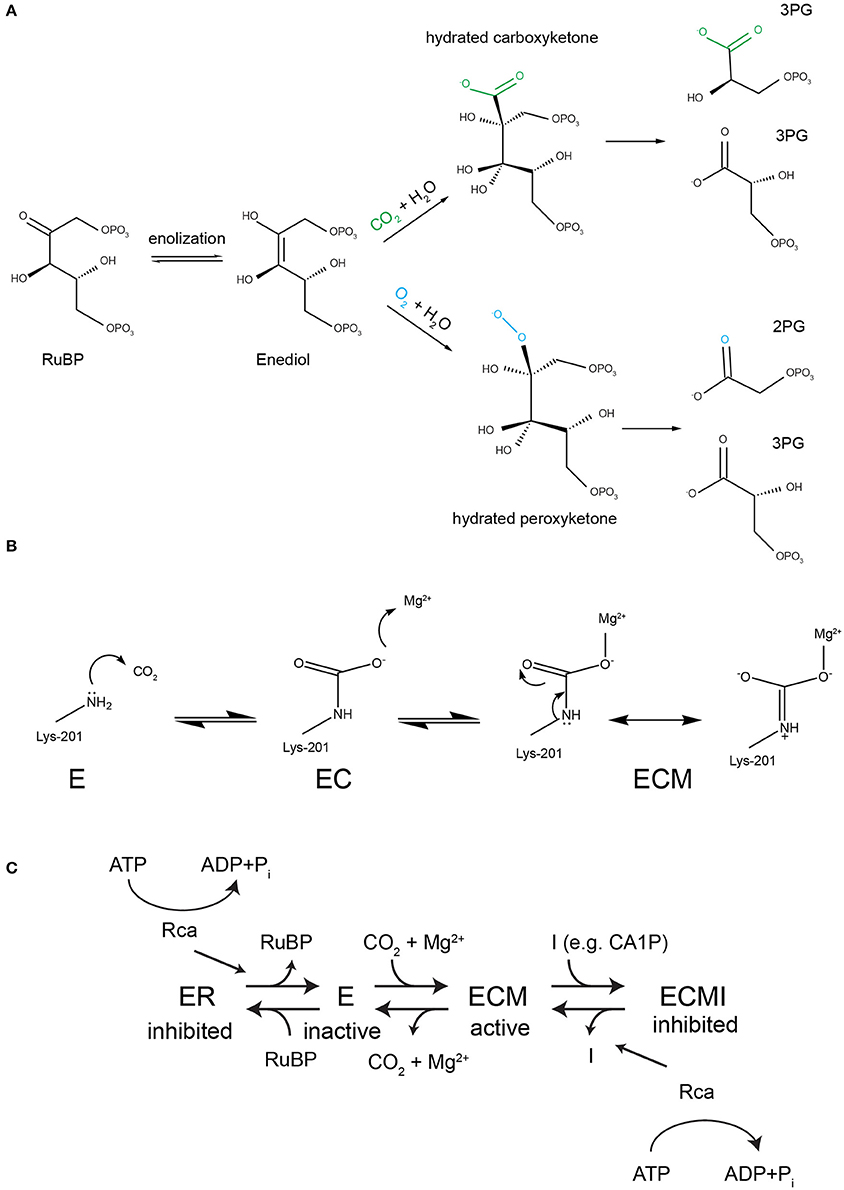
Figure 1. Rubisco's reaction mechanism and its inhibition properties. (A) A complex conserved reaction mechanism evolved to carboxylate ribulose 1,5-bisphosphate. The enediol intermediate can react with both oxygen and carbon dioxide. If oxygenation occurs the toxic metabolite 2-phosphoglycolate (2PG) is generated, which must be subjected to metabolite repair. (B) To perform the carboxylase reaction a conserved active site lysine (Lys-201 in spinach RbcL) must react with a non-substrate CO2 to form a carbamate (EC), followed by the binding of a Mg2+ ion to form the catalytically competent holoenzyme ECM. (C) Both the inactive apo (E) and the active holoenzyme (ECM) are prone to dead-end inhibition by sugar phosphates such as RuBP, which binds to E and CA1P (2-carboxy-D-arabinitol 1-phosphate), which binds to ECM. Rubisco activases (Rca) recognize inhibited active sites and use the energy of ATP hydrolysis to cause a conformational change that releases the inhibitor.
Catalysis by all Rubiscos requires two cofactors to bind at the active site permitting the functional holoenzyme to form (Figure 1B). A non-substrate CO2 reacts with the amine group of the conserved Lys-201 residue (spinach RbcL numbering) to form a carbamate. A Mg2+ ion is then bound to complete the activation process, forming the holoenzyme termed ECM (Lorimer et al., 1976; Cleland et al., 1998). The activated enzyme then binds the substrate RuBP, which is processed via a series of five partial reactions to eventually yield two molecules of 3-phosphoglycerate (3-PG) if carboxylated (Tcherkez, 2013, Figure 1A). The similarity in size and electrostatic potential of the gases CO2 and O2 (Kannappan and Gready, 2008) has culminated in a situation where the enzyme is unable to perfectly discriminate between the carboxylation substrate CO2 and the competing O2. The critical step at which the enzyme can influence the partitioning between carboxylation and oxygenation is during attack of the gaseous substrate by the enolized RuBP (Chen and Spreitzer, 1992). An analysis of decades of kinetic and isotope-fractionation data suggested that this task is achieved by a relative stabilization of the transition state for CO2, compared to O2 addition (Tcherkez et al., 2006). This stabilization manifests itself in both reduced flexibility of the active site and tighter binding of the carboxylated product (Pearce and Andrews, 2003). A well-documented outcome of this strategy is the trade-off where faster enzymes tend to exhibit higher Michaelis constants for CO2 and are less able to discriminate between CO2 and O2 (Bainbridge et al., 1995; Tcherkez et al., 2006; Savir et al., 2010). However, it is important to note that new Rubisco kinetic data is highlighting exceptions to these rules, at least regarding some algal enzymes exhibiting relatively low carboxylase efficiencies (Young et al., 2016).
A consequence of the described strategy, which tends to be less well popularized, relates to the tendency of the enzyme to become irreversibly inhibited by sugar phosphates. Since the unactivated apo-enzyme (E) already possesses all of the features required to bind the substrate RuBP, the active site will close when it encounters the substrate (Jordan et al., 1983; Duff et al., 2000). In the absence of the co-factors required to catalyze carboxylation or oxygenation, RuBP cannot be processed and is now bound unproductively, or “caught in the Rubisco mousetrap” (Andrews, 1996), to form Enzyme-RuBP (ER) (Figure 1C). At the same time, losing a valuable active site has reduced the capacity for carbon fixation of the host organism. RuBP is not the only inhibitory substrate, a palette of other sugar phosphates, including some generated by misfire-reactions of Rubisco itself, also tightly bind to the active site (Parry et al., 2008; Andralojc et al., 2012; Bracher et al., 2015). The affinity of the inhibitors is correlated with the enzyme's catalytic parameters, and based on the data available “superior” high specificity Rubiscos bind RuBP and other sugar phosphates more tightly than the low specificity enzymes with more flexible active sites (Pearce and Andrews, 2003; Pearce, 2006).
Over time, as Rubisco active sites became more and more adept at tightly binding the carboxylation-intermediate, the propensity for the apo-Enzyme to bind the substrate non-productively also increased (Pearce and Andrews, 2003). This led to a temporary removal of significant proportions of active sites from the pool of the enzyme. This problem could be alleviated by the action of molecular chaperones that would selectively engage inhibited Rubisco, and by performing a “chiropractic” maneuver (Carmo-Silva and Salvucci, 2011) conformationally reset the active site.
Earlier articles have comprehensively reviewed our knowledge on biochemical and physiological aspects of both the green-type (Portis, 1995, 2003; Portis et al., 2008; Carmo-Silva et al., 2015) and the red-type activase (Mueller-Cajar et al., 2014; Hauser et al., 2015b). Here we aim to direct attention toward the recent realization that in different autotrophic lineages multiple activase classes have converged on the same biochemical function. We attempt to integrate our understanding regarding mechanistic similarities and differences toward a framework regarding the chaperone-mediated rearrangement of the highly conserved inhibited Rubisco active site.
In spite of the single phylogenetic origin and highly conserved reaction chemistry of Rubisco, a number of highly distinct clades of Rubisco can be observed today (Tabita et al., 2008). All Rubiscos are comprised of ~55 kDa large subunits that assemble as anti-parallel dimers. Each dimer harbors two active sites formed by the β-barrel C-terminal domain of one subunit and the N-terminal domain (containing a 5-stranded mixed beta sheet) of the other (Knight et al., 1990). This basic functional unit is then often found to be assembled into higher oligomeric states.
Figure 2 shows a phylogenetic tree of selected RbcL sequences relevant to the present discussion about Rca. The last common ancestor of all extant Rubiscos was probably the aforementioned dimer of large subunits, and this arrangement is still found in a subset of the so-called Form II enzymes, such as the well-studied enzyme from Rhodospirillum rubrum (Anderson and Fuller, 1969). Contemporary Form II enzymes are often found to occupy higher order oligomeric states with a hexameric arrangement recently found to be common (Satagopan et al., 2014; Tsai et al., 2015; Varaljay et al., 2016). A key early innovation in Rubisco evolution concerned the recruitment of the small subunit, a ~15 kDa scaffolding protein that stabilized tetramers of dimers resulting in a L8S8 stoichiometry. These enzymes constitute the Form I clade of Rubiscos (Spreitzer, 2003). This clade branched early into a red (Form IC and D) and green-type branch (Form IA and B), the large subunits of which today maintain about 50% sequence identity to each other. Form IA Rubiscos can be subdivided into Form IAQ and Form IAC sequences, the latter always being associated with carboxysomal gene clusters (Badger and Bek, 2008). It is interesting to note that the photosynthesizers dominating our planet's landmass, the higher plants, possess only a small slice of Rubisco's molecular diversity, all encoding a highly conserved Form IB enzyme derived from the ancestral cyanobacterial endosymbiont.
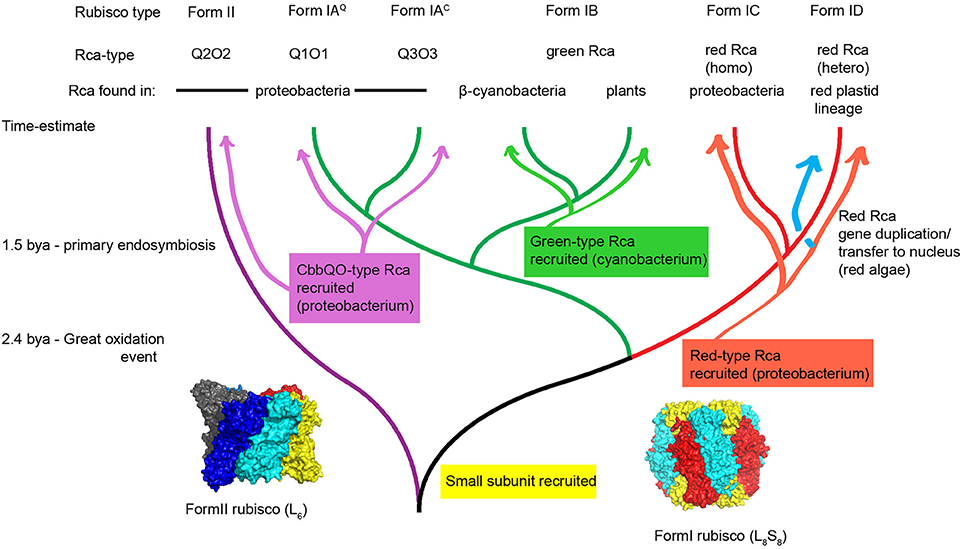
Figure 2. Hypothetical scheme for the evolution of Rubisco and its activases. Following the great oxidation event at least three different classes of Rubisco activase were recruited from the general molecular chaperone machinery toward a specialized Rubisco activase function in diverse autotrophic organisms. Green type and red-type Rca was maintained in eukaryotic phototrophs of the green and the red plastid lineage respectively. A phylogenetic tree was drawn using Rubisco large subunit sequences that are associated with activases. It is important to note that regarding non-red prokaryotic Rubisco sequences, many instances exist that do not have identifiable activase genes encoded in the same genome. Surface representations of a hexameric Form II Rubisco (pdb:4lf1) and spinach Form I Rubisco (pdb:8ruc) are shown. Structures shown in this paper were drawn using pymol.
Three distinct classes of Rubisco activase (green-, red-, and CbbQO-type) have now been identified (Salvucci et al., 1985; Mueller-Cajar et al., 2011; Tsai et al., 2015), permitting us to start dissecting the molecular underpinnings of how different organisms dealt with the outlined problem of blocked Rubisco active sites. The activases were recruited from highly distinct volumes of sequence space in the AAA+ protein universe (Ammelburg et al., 2006), and their AAA modules display less than 25% sequence identity between the groups. This vast and diverse group of molecular motors was clearly well suited for the task of active site rearrangement, as their unifying functional characteristic relates to conformationally remodeling macromolecular substrates using the energy of ATP hydrolysis (Hanson and Whiteheart, 2005; Sysoeva, 2016). The identified activases are not closely related to other well characterized extant molecular chaperones, which currently precludes the formulation of detailed hypotheses regarding their historical evolutionary trajectory.
Green-type Rcas represent the first discovered (Salvucci et al., 1985) and due to their presence in all higher plants, most extensively studied activase system (Portis, 2003; Carmo-Silva et al., 2015). They are evolutionarily derived from cyanobacteria, where homologs are found associated with carboxysomal green-type Form IB Rubisco (Li et al., 1993). Importantly, an experimental verification of the cyanobacterial activase's biochemical function is still elusive (Bracher et al., 2017). The distribution is not universal, but is associated with strains belonging to clade A and B1 according to the classification by Kerfeld and colleagues (Shih et al., 2013; Zarzycki et al., 2013). These are thought to form the sister group to the primary endosymbiont (Ochoa de Alda et al., 2014), which would indicate that Rca was transferred together with Form IB Rubisco during the primary endosymbiotic event about 1.5 billion years ago (Yoon et al., 2004).
On a structural level green-type Rcas show similarity to p97/CDC48 (Hasse et al., 2015) and classification of the C-terminal subdomain revealed a relationship to the D2 AAA+ module of N-ethylmaleimide-sensitive factor (NSF) (Ammelburg et al., 2006). Both of these belong to the classical clade of AAA proteins (Iyer et al., 2004). It is thus reasonable to conclude that specialization toward activase activity occurred using a general molecular chaperone in this clade in an ancient cyanobacterium as a starting point.
The gene encoding red-type Rca (also known as CbbX), is always found in an operon with the red-type (Form IC) Rubisco encoding genes in mixotrophic proteobacteria (Gibson and Tabita, 1997; Badger and Bek, 2008). It is also encountered in the chloroplast genomes of the red lineage (Oudot-Le Secq et al., 2007). A proposed explanation for this distribution involved horizontal gene transfer of the rbcL-rbcS-cbbX gene cluster from a proteobacterium to an ancestor of the primary endosymbiont (Delwiche and Palmer, 1996; Nisbet et al., 2004). Alternatively horizontal gene transfer occurred subsequent to the endosymbiotic event in the ancestor of the red algae, which subsequently lost the green Form IB Rubisco genes (Maier et al., 2000; Rice and Palmer, 2006). Where sequence data exists, it appears eukaryotes possessing red-type Rubisco always encode an additional CbbX isoform in the nuclear or nucleomorph genome (Hovde et al., 2015), and this is thought to be a consequence of gene duplication and migration of one copy to the nuclear genome in an early rhodophyte (Fujita et al., 2008). In the red algae Cyanidioschyzon merolae, the functional red-type Rca has been shown to be a 1:1 hetero-oligomer of the plastid and the nuclear encoded isoform (Loganathan et al., 2016), and we expect this scenario to hold true for red lineage phytoplankton in general.
The closest structural neighbors of red-type Rca, as determined by a DALI search are the helicase RuvB and protease-associated motors such as HslU and ClpX (Hasse et al., 2015). HslU and ClpX are powerful unfoldases that generally thread substrate proteins marked for degradation through their axial pore of the hexamer into a proteolytic chamber (Sauer and Baker, 2011). However, recently more gentle conformational rearrangements have been documented for the mitochondrial ClpX. In this case ClpX acts on an enzyme involved in heme biosynthesis and catalyzes the insertion of a cofactor (Kardon et al., 2015). Hence it is conceivable that subtle “pulling” on enzymes to bring about conformational transitions that favor inhibitor release or co-factor insertion is not an unusual scenario (Olivares et al., 2016). It is therefore a reasonable hypothesis that red-type Rca evolved in proteobacteria from a general molecular chaperone using the axial pore threading mechanism that was either involved in correcting protein conformations or protein complex maturation (including co-factor insertion).
The genes encoding the CbbQO-type activase system (Hayashi et al., 1997, 1999; Sutter et al., 2015; Tsai et al., 2015) are broadly distributed among proteobacteria, but associate strongly with chemolithoautotrophs that use sulfur oxidation as energy source (Badger and Bek, 2008). CbbQ belongs to the large, but relatively poorly characterized MoxR group of AAA+ proteins, which is often found encoded in operons together with a second protein containing a von Willebrand Factor A (VWA) domain (Snider and Houry, 2006; Wong and Houry, 2012).
Different isoforms of the AAA+ protein CbbQ and the VWA-domain containing CbbO assemble as hetero-oligomeric complexes in a Q6O1 stoichiometry (Sutter et al., 2015; Tsai et al., 2015). Two complexes encoded by Acidithiobacillus ferrooxidans activate phylogenetically remote Rubiscos (Q1O1 activates Form IAQ and Q2O2 activates Form II) that are encoded by the same genome (Tsai et al., 2015). In addition there is a third cbbQ-cbbO gene pair (termed Q3O3 in Figure 2) associated with a carboxysomal gene cluster, which contains genes encoding a Form IAC Rubisco (Heinhorst et al., 2002). The activase function of Q3O3, which is homologous to a complex recently purified and characterized for ATPase activity, has not yet been confirmed (Sutter et al., 2015). This work also pointed out that the presence of multiple Rubisco operons encoding different CbbQ and CbbO isoforms in the same organism is common. It is thus possible that the ancestor of the CbbQO complex became specialized for one Rubisco form, and then switched substrate following a gene duplication. Alternatively the ancestral CbbQO was a generalist Rca and already functional at remodeling both types of Rubisco. The feasibility to reconstruct ancestral proteins offers a tantalizing opportunity to illuminate these details experimentally (Shih et al., 2016).
Gene pairs highly homologous to CbbQ and CbbO that are not associated with Rubisco genes also exist in proteobacteria (Snider and Houry, 2006; Sutter et al., 2015). The genes encoding the AAA+ protein NirQ and VWA domain protein NorD, are associated with denitrification gene clusters. In the absence of either NirQ or NorD, nitric oxide reductase is produced in non-functional form, implicating NirQ-NorD in enzyme maturation or assembly (Jungst and Zumft, 1992; de Boer et al., 1996). The best biochemically characterized MoxR AAA+ ATPase chaperone system is RavA-ViaA, where RavA is the AAA+ motor, and ViaA is an interacting VWA-domain containing protein (Snider et al., 2006; Wong et al., 2017). Intriguingly one of a number of described function of RavA involves a reduction of the affinity of the allosteric inhibitor ppGpp to the enzyme lysine decarboxylase (albeit in a ViaA independent manner) (El Bakkouri et al., 2010; Kanjee et al., 2011). Therefore, it is likely that in this family many chaperones with functions related to the modulation of enzyme activity remain to be discovered. The CbbQO Rubisco activation system was likely derived from such an origin.
It is established that contemporary Rubisco enzymes all share a common ancestor (Tabita et al., 2007), and although there is significant diversity in quaternary structure, tertiary structure is essentially conserved (Andersson, 2008; Andersson and Backlund, 2008). The implication is thus that the different Rca motors will encounter a highly similar substrate, irrespective of its origin. It is therefore reasonable to expect that Rca mechanisms will display similarities. Consequently motor-substrate specificity should be exchangeable by targeted mutagenesis once the mechanisms are understood in sufficient detail.
Representative examples of Form I and Form II inhibited Rubisco complexes that function as Rca substrates are shown in Figure 3. The active site is located at the C-terminal face of the beta strands forming the αβ barrel. Residues contributing to the active site are mostly found in the loops connecting the beta strands of the barrel to the downstream helices, but a few are donated by the N-terminal domain of the opposing subunit. Once the substrate RuBP has bound, loop 6 of the beta barrel folds over the active site to form the closed state (Karkehabadi et al., 2007). Loop 6 contributes a critical lysine residue (Form I- K334, Form II- K330), which is thought to position the CO2 molecule for carboxylation. In Form I enzymes, closure of the active site is accompanied by the C-terminal strand of the large subunit folding over loop 6, with Asp-473 believed to act as a latch residue (Duff et al., 2000; Satagopan and Spreitzer, 2004). The thus secured C-terminus is envisaged to be under tension to push down on Loop 6 via Lys-128 (Bainbridge et al., 1998), providing rigidity to the carboxylation ready active site (Duff et al., 2000). In stark contrast to the C-terminal locking mechanism in Form I Rubisco, inspection of the closed form of the carboxy-arabinitol 1,5 bisphosphate (CABP) bound Form II hexamer from Rhodopseudomonas palustris reveals that the C-terminus does not fold over and lock down Loop 6, but is instead positioned at the apex of the complex (Satagopan et al., 2014) (Figure 3B). As a consequence Loop 6 is surface exposed in these structures (Satagopan et al., 2014; Varaljay et al., 2016). Instead of being held in place by the C-terminus, the structure reveals a salt-bridge between Glu-332 (R. palustris RbcL labeling) and Lys-33 on the opposite subunit. These residues are conserved in many Form II enzymes, and the interaction may thus be part of an alternative Loop 6 locking mechanism. Another important feature of active site closure concerns a 2° rotation of the N-terminal domain, resulting in a reduced distance between the phosphate binding sites of the active site (Taylor and Andersson, 1996; Duff et al., 2000).
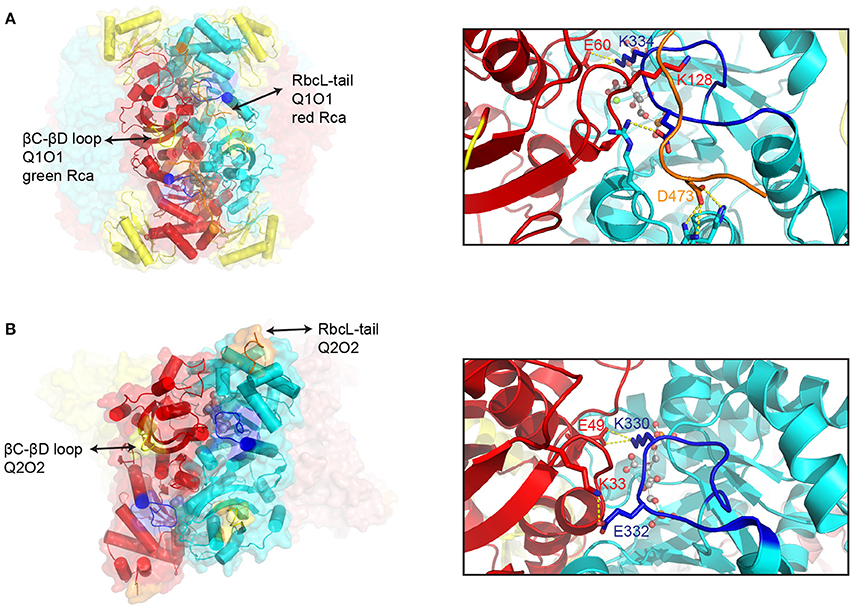
Figure 3. Structural features of inhibited Rubisco complexes. A comparison of structural elements involved in the Rca-mediated activation of Form I (A) and Form II (B) Rubisco. Left panels: Surface representation of CABP-bound spinach (pdb:8ruc) and R. palustris Rubisco (pdb:4lf1). One large subunit dimer pair (in red and cyan) is shown with helices represented by cylinders. Key segments are colored as follows: βC-βD loop, yellow; Loop 6, blue; C-terminal strand, orange. Right panels: Close-up of the active site highlighting differences in Loop 6 (in blue) closure between Form I and Form II Rubisco. Key residues and interactions are highlighted. Bound CABP is shown in ball and stick representation. The following indicated residues are conserved and functionally equivalent (Form I/Form II): E60/E49; K334/K330).
Based on these observations, the conformational changes to bring about an opening of the active site catalyzed by the Rca motors could either involve manipulation of the C-terminal domain, for instance by disruption of the latched C-terminus in Form I enzymes, or Rca-induced movement of the N-terminal domain. In fact both strategies appear to be utilized.
The three classes of Rca identified so far all belong to distantly related branches of the AAA+ protein superfamily and possess a single AAA+ domain. Experimentally determined atomic models of the AAA+ module of all activase classes are now available, and all exhibit the expected architecture of this protein family (Henderson et al., 2011; Mueller-Cajar et al., 2011; Stotz et al., 2011; Hasse et al., 2015; Sutter et al., 2015). A Rossmann fold forms the nucleotide binding domain, which is followed by a small α-helical subdomain (Erzberger and Berger, 2006, Figure 4A). AAA+ proteins commonly form hexameric rings, and this is certainly the functional form of both the red-type (Mueller-Cajar et al., 2011; Loganathan et al., 2016) and the CbbQO-type Rcas (Sutter et al., 2015; Tsai et al., 2015) as verified by negative-stain electron microscopy.
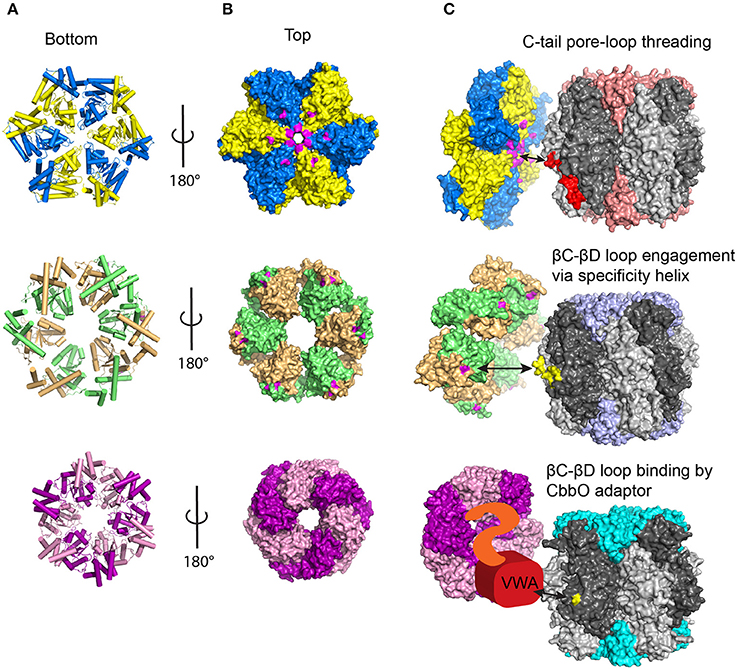
Figure 4. Current models of Rubisco activase function. (A) Bottom view of the different Rca hexameric models showing helices in cylinder view. Adjacent subunits are colored differently (B) Top view of the Rca models in surface representation. Residues known to be involved in protein-protein interactions with Rubisco are colored in magenta for red and green-type Rca. (C) Current mechanistic models for the different Rca systems. See text for details. Known Rca interacting segments on Rubisco are shown in red (RbcL C-tail) and yellow (interacting βC-βD loop residues). Red-type Rca /Rubisco, PDB:3ZUH/1BXN; Green-type Rca/Rubisco PDB:3ZW6/8RUC; CbbQ/Form IA Rubisco, PDB:5C3C/1SVD.
It is interesting to note that the proteobacterial red-type Rca forms an ATPase inactive fibril in the presence of Mg-ATP. Binding of Rubisco's substrate RuBP to a pocket located in the α-helical subdomain triggers an oligomeric transition to the ATPase and activase functional hexamer (Mueller-Cajar et al., 2011). In contrast the enzyme from the red algae Cyanidioschyzon merolae presents as a constitutive hexamer composed of alternately arranged nuclear and plastid-encoded isoforms (Loganathan et al., 2016). However, the RuBP-binding pocket is conserved in both isoforms and ATPase activity is stimulated by the addition of RuBP. Thus, in both prokaryotes and eukaryotes enzymatic activity of red-type Rca is allosterically regulated by the substrate of the remodeller's target. Nevertheless, mutational studies indicated that the two red-type Rca isoforms in red algae are functionally non-equivalent. For instance eliminating ATPase function of the plastid-encoded isoform by mutating the conserved Walker B glutamate to glutamine counterintuitively enhanced ATP hydrolysis of the hetero-oligomeric complex and resulted only in slight impairment of activase function. In contrast the equivalent substitution in the nuclear encoded isoform eliminated both Rca and ATPase function (Table 1). It remains to be seen whether these specializations have resulted in genuine enhancements in activase function or whether they are manifestations of molecular ratchet- type evolutionary trajectories (Gray et al., 2010; Finnigan et al., 2012).
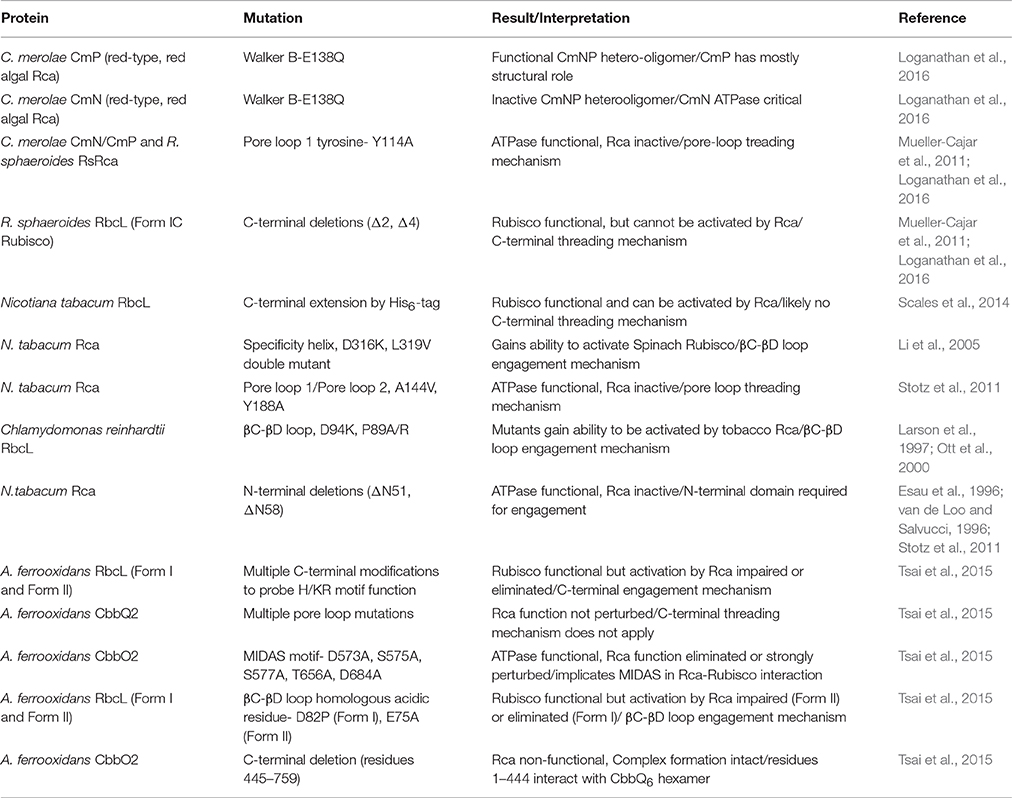
Table 1. Overview of selected key Rca and Rubisco mutants providing insights into the activation mechanism and listed in the order referred to in the text.
The in vitro oligomeric state of the green-type Rcas is highly polydisperse, possibly ranging from monomeric (Keown et al., 2013) to very large assemblies (Barta et al., 2010; Chakraborty et al., 2012; Kuriata et al., 2014). However, the existence of functional, stable hexamers (Blayney et al., 2011; Stotz et al., 2011; Keown and Pearce, 2014) suggest that this is also the functional species. It is possible that the oligomeric forms may be transitional to permit efficient movement of the activases through the extremely crowded chloroplast stroma (Harris and Koniger, 1997), permitting this less abundant helper protein to shuttle between inactive Rubisco active sites as required. Hexameric assemblies would then occur transiently to form the functional assembly at the inhibited substrate Rubisco. Consistent with this notion, green-type activases rapidly exchange subunits in vitro (Salvucci and Klein, 1994; van de Loo and Salvucci, 1998; Stotz et al., 2011). Regulation of the green-type Rca in higher plants is complex (Carmo-Silva and Salvucci, 2013; Hazra et al., 2015), with a number of mostly energy-related signals integrating. These include redox modulation by thioredoxin and inhibition by ADP (reviewed by Carmo-Silva et al., 2015 and Portis, 2003) and most recently reversible phosphorylation (Boex-Fontvieille et al., 2014; Kim et al., 2016).
CbbQO is unique among activases, in that the AAA+ hexamer CbbQ associates with a single adaptor protein CbbO, which is essential for activase function. The CbbQ6O1 complexes are monodisperse and do not disassemble as assessed by gel filtration chromatography (Tsai et al., 2015). Finally, both CbbQO and red-type Rubisco activases exhibit a strong stimulation of their ATPase activity when assayed in the presence of inhibited Rubisco complexes (Mueller-Cajar et al., 2011; Tsai et al., 2015; Loganathan et al., 2016). This type of regulation is not observed in the green-type Rcas (Robinson and Portis, 1989; Hazra et al., 2015).
AAA+ proteins generally function by translating conformational changes brought about by ATP hydrolysis to a macromolecular substrate, and this principle applies to Rcas and Rubisco. The best described mechanisms so far involve the translocation of the substrate through the axial pore of the hexameric AAA+ ring. This involves a conserved pore loop 1 tyrosine in many well-studied systems, including ClpX (Siddiqui et al., 2004), ClpB/Hsp104 (Weibezahn et al., 2004) and the AAA+ unfoldase of the proteasome (Beckwith et al., 2013). In Table 1 I summarize biochemical evidence for the mechanistic models described in this section. The outlined threading mechanism appears to be utilized by red-type Rca in both photosynthetic bacteria and red algae. In this model, the activase transiently threads the C-terminus of the Rubisco large subunit into the pore (Figure 4C). Red-type Rubiscos all appear to possess a C-terminal extension of 11–12 residues following the critical latch residue Asp-473, which locks the C-terminus to its large subunit. Thus, by pulling on this peptide, the interaction of Asp-473 with its own subunit can be disrupted, releasing the lock and allowing loop-6 to retract, followed by release of the bound inhibitor. Substitutions with alanine of the conserved pore loop 1 tyrosine in both the bacterial and algal red-type Rca, as well as two and four amino-acid deletions of the RbcL C-terminus abolish activase function (Mueller-Cajar et al., 2011; Loganathan et al., 2016).
Interestingly this model, at least relating to transient threading of the Rubisco large subunit C-tail, is unlikely to apply to either of the other two Rca classes. In contrast to the red-type Rubiscos, the C-termini of green-type Rubiscos are of variable length, but often only have 2–4 residues following the latch residue (Satagopan and Spreitzer, 2004). Green-type Rca is thus unlikely to engage this short and variable motif. It was also found that an extension of the tobacco large subunit by six histidine residues did not affect Rca function (Scales et al., 2014). In addition the central pore of the green-type Rca hexamer has a larger diameter than that of red-type Rca, which lead to the hypothesis that a larger secondary structural element, such as a loop, could be threaded instead (Stotz et al., 2011). Consistent with the general theme of a poreloop threading mechanism mutational analysis of pore loop 1 and 2 resulted in the discovery of variants that maintained ATPase function but no longer activated Rubisco (Stotz et al., 2011). Notably, the AAA+ chaperone ClpB has been demonstrated to be capable of threading a looped segment (Haslberger et al., 2008), and the threading mechanism is therefore not limited to free N or C-termini.
The surface exposed βC-βD loop of the large subunit N-terminal domain has long been implicated in the interaction with green-type Rca (Figure 3A). Residues 89 and 94 (spinach numbering) in this loop are known to interact with residues 316 and 319 (tobacco Rca numbering) of the activase (Larson et al., 1997; Ott et al., 2000; Li et al., 2005), which are located on a helical insertion in the small subdomain of the AAA+ module (Stotz et al., 2011; Hasse et al., 2015). This interaction involves the same (top) face of the disc-shaped hexamer that is involved in red-type Rca function (Wachter et al., 2013, Figure 4B). In addition an N-terminal domain of ~70 amino acids is also involved in the Rubisco-Rca interaction (Esau et al., 1996; van de Loo and Salvucci, 1996; Stotz et al., 2011), however it is not resolved in current crystal structures. It is conceivable that following initial engagement by activase involving the mentioned structural elements (Figure 4C), a pulling force to the βC-βD loop could be brought about by Rca pore loop threading. Rigid body movement of the attached beta sheet would then result in the rotation of the N-terminal domain seen when comparing the closed and open form of the enzyme (Duff et al., 2000).
Mutational analysis of both CbbQO and the two different classes of substrate Rubisco revealed the basis of a common mechanism for CbbQO-type Rcas. More fascinatingly, the results revealed commonalities to both red- and green-type Rca function. It was noted that in spite of low (~30%) primary sequence identity of the Form I and Form II Rubisco large subunits, the C-termini of those enzymes encoded in cbbQ-cbbO containing gene clusters displayed a common C-terminal sequence motif (H/KR). Mutagenesis of this motif strongly impaired the ability of the target Rubiscos to be activated by their activases, drawing a strong mechanistic parallel to the pore-loop threading red-type Rcas (Tsai et al., 2015). However, experiments attempting to perturb the poorly conserved pore-loop region of CbbQ did not result in non-functional Rca, and I currently favor a model where the large subunit C-terminus is bound (and consequently immobilized) by the activase, rather than threaded. Here I am also considering the fact that in the Form II substrate the C-terminus does not occupy the same locked latch position as in the Form I complex (Figure 3, Satagopan et al., 2014), and thus exerting a pulling force on this motif would not have the same effect.
As is commonly observed for the MoxR class of AAA+ proteins, the CbbO adaptor encoded downstream of the cbbQ gene possesses a von Willebrand factor A (VWA) domain at its C-terminus (Whittaker and Hynes, 2002). This well-described protein-protein interaction module generally uses four residues that are part of a motif known as metal ion dependent adhesion site (MIDAS) to bind a divalent cation. Mutating conserved MIDAS residues mostly abolished CbbQO activase function (Tsai et al., 2015). A fifth ligand to the divalent cation is generally donated by an acidic residue of the interacting protein (Xiong et al., 2002; Santelli et al., 2004). It was discovered that mutating a conserved acidic residue in the previously mentioned surface exposed βC-βD loop of the Rubisco large subunit N-terminal domain to alanine abolished (Form I Rubisco) or greatly reduced (Form II) the ability of Rubisco to become activated by CbbQO (Tsai et al., 2015). Fascinatingly this residue is at the same position as the green-type Rca interacting residue 89 in higher plants Rubisco. We therefore predict that the ATP-hydrolysis powered conformational change brought about by CbbQO and green-type Rcas will emerge to be similar in nature (Figure 4C). The precise interaction between a CbbQ hexamer and the CbbO adaptor has not been resolved so far, but involves residues 1–444 of CbbO (Tsai et al., 2015). It is possible that the conformational changes of the hexamer generated by ATP hydrolysis are transmitted to the VWA domain via the CbbO N-terminal region (Figure 4C).
Disruption of the closed conformation of the Rubisco holoenzyme by Rca of all three classes will lead to release of the inhibitory sugar phosphate. The active site is thus reset either for cofactor binding, or acceptance of the substrate RuBP (if the inhibitor removed was already bound to ECM holoenzyme, Figure 1C).
A strong impetus regarding research into the detailed mechanisms underlying Rubisco repair in autotrophic organisms is provided by the realization that relatively poor Rubisco performance contributes to the low photosynthetic efficiency of plants, and enhancing its activity is predicted to significantly improve the yield of crops (Long et al., 2015). Given the tight coupling of carboxylase function to maintenance of its activation state by the described highly diverse Rca proteins, any modifications of Rubisco will need to keep in mind compatibilities and other properties of Rca.
A number of strategies regarding the enhancement of C3 photosynthesis rely on the concept of transplanting a Rubisco enzyme of choice into a target crop (Andrews and Whitney, 2003; Zhu et al., 2004). Such experiments need to ensure the presence of a suitable Rca, and technically this is not a difficult problem. Rca in higher plants is encoded by the nuclear genome, and thus Agrobacterium tumefaciens based transformation methods can successfully deliver a target Rca gene (Kurek et al., 2007; Kumar et al., 2009; Fukayama et al., 2012). Deletion or silencing of the endogenous Rca genes may be advantageous if hetero-oligomerization is likely to occur (for instance if a green-type Rca is to be transplanted). In particular the rapid development of CRISPR-Cas9 technology will facilitate this process further (Belhaj et al., 2015). However, the relative ease of Rca engineering does not extend to Rubisco. Since in higher plants the rubisco large subunit is encoded by the chloroplast (as opposed to the nuclear) genome, this achievement requires the replacement of the endogenous rbcL genes in multiple plastid genome copies. Following significant technical progress in the past decades it is now possible to routinely perform this experiment in tobacco plants using biolistic transformation. Here a particular boon has been the development of a marker-free tobacco-rubrum “master” line (Whitney and Sharwood, 2008), which has its endogenous hexadecameric Form IB Rubisco replaced by a bacterial dimeric Form II enzyme. Due to this Rubisco's low CO2/O2 specificity, it only permits plant growth at elevated levels of CO2 (Whitney and Andrews, 2001) and thus facilitates the isolation of transformants expressing more catalytically adept heterologous Form I enzymes. Key examples of successful rubisco transplantation experiments include various higher plant enzymes (Sharwood et al., 2008; Whitney et al., 2011b, 2015), a cyanobacterial Form I enzyme (Lin et al., 2014b) and an archaeal Form III enzyme from Methanococcus burtonii (Wilson et al., 2016). It is therefore technically feasible to produce functional heterologous Rubisco in tobacco plants, although expansion of the technology to other species has so far met with modest success and most crops cannot currently be modified in this manner (Bock, 2015). Current efforts in this area of research are aiming to identify better suited higher-plant Rubiscos (Orr et al., 2016; Sharwood et al., 2016a), or introducing single residue changes into the large subunit that result in desired catalytic switches (Whitney et al., 2011b). Here activase requirements should be easy to satisfy due to the wide level of compatibility between plant Rubiscos and green-type Rcas (Wang et al., 1992). Still a relative paucity of Rubisco-Rca compatibility data may require careful biochemical characterization on a case to case basis.
Although production of heterologous Rubisco in higher plants is currently feasible, a key limitation concerns our incomplete understanding of the enzyme's folding and assembly machinery, which results in either low Rubisco content, or a complete failure in functional Rubisco expression. Regarding the production of heterologous plant Rubisco, rapid progress is being made, for instance co-expression of the Rubisco assembly chaperone Raf1 (Feiz et al., 2012; Hauser et al., 2015a) permitted a doubling of correctly assembled Arabidopsis Rubisco large subunits in tobacco chloroplasts (Whitney et al., 2015).
Among the most tempting targets for transplantation are the red-type Form ID Rubiscos from red algae, some of which have evolved CO2/O2 specificity values that are twice as high than those found in the land plant Form 1B enzymes (Read and Tabita, 1994; Uemura et al., 1997). For instance functional production of the Rubisco from the red algae Griffithsia monilis (Whitney et al., 2001) in higher plant chloroplasts is predicted to result in a 27% increase in daily canopy carbon gain (Zhu et al., 2004). However, early experiments to produce these proteins in tobacco led to complete insolubility of the gene products (Whitney et al., 2001), consistent with an incompatibility of the folding and/or assembly chaperone machinery. Interestingly this apparent dependency on sophisticated chaperone machinery does not extend to the related bacterial Form IC red-type Rubiscos. The enzyme from Rhodobacter sphaeroides has no requirements for assembly chaperones, merely requiring the GroEL-ES chaperonin for productive folding of the large subunit in a reconstituted system (Joshi et al., 2015). Meeting the biogenesis requirements of Form ID Rubisco may thus be less complicated than that of the higher plant Form IB enzymes, which appear to require a plethora of assembly factors including Raf1, Raf2 and possibly RbcX (Liu et al., 2010; Feiz et al., 2014; Bracher et al., 2017). Once Form ID Rubisco transplantation has been achieved it will need to be supplemented with a red-type Rubisco activase. Based on the work with purified C. merolae proteins it is likely that the cognate algal Rca, a hetero-oligomer of nuclear and plastid encoded subunits, will be optimal for this purpose. However, the simpler homo-oligomeric bacterial red-type Rcas also presents with some activity toward the algal enzyme and thus may be sufficient (Loganathan et al., 2016).
A challenging goal that is currently being pursued by a number of groups involves the transplantation of the prokaryotic carboxysomal CO2-concentrating mechanism into the higher plant chloroplast (Price et al., 2008; Lin et al., 2014a,b). A combination of a high velocity Rubisco operating at very high CO2 concentrations achieved by carboxysomal Rubisco compartmentalization and active inorganic carbon transport should permit high carbon dioxide assimilation in the absence of photorespiration (Zarzycki et al., 2013). When considering this strategy it is important to realize that a subset of carboxysomal gene clusters include homologs of all three classes of Rca (Zarzycki et al., 2013; Sutter et al., 2015). Activase activity has not yet been demonstrated for any of the carboxysomally associated Rcas biochemically, and an inability to detect this function biochemically was reported in two cases (Li et al., 1999; Sutter et al., 2015). However, in my opinion the association of these Rca homologs with carboxysomal gene clusters is indicative that the associated Rubiscos have not escaped from the activase dependency. Progress here will likely require the use of Rubisco inhibitors other than RuBP, which binds only weakly to carboxysomal Rubiscos (Andrews and Abel, 1981; Pearce, 2006), as well as assay conditions that mimic the crowded carboxysomal interior. In order for Rca associated carboxysomes to function optimally, the relevant activase will likely also need to be supplied (Long et al., 2016).
It is intriguing that significant numbers of carboxysome-containing organisms do not appear to encode Rca proteins (Zarzycki et al., 2013), suggesting either a true activase independence or the existence of unidentified activase classes. Another enticing possibility would involve members of the general chaperone machinery functioning as activases, in a scenario resembling the situation prior to the evolutionary recruitment of specialized Rcas.
For a long time it has been realized that plant photosynthesis is highly sensitive to temperature stress (Berry and Bjorkman, 1980), and that the reduction of this process was correlated with a loss in Rubisco activation state (Weis, 1981; Kobza and Edwards, 1987). The discovery that Rca is highly thermolabile, and undergoes heat denaturation at physiologically relevant temperatures provided a mechanistic basis to this observation (Feller et al., 1998; Crafts-Brandner and Salvucci, 2000; Salvucci and Crafts-Brandner, 2004a). This realization was followed by the critical demonstration that expression of more thermostable Rca proteins in Arabidopsis led to enhanced growth and biomass accumulation at moderately elevated growth temperatures (Kurek et al., 2007; Kumar et al., 2009). It is therefore imperative that these promising studies are followed by rigorous analyses of crop plants expressing more thermostable Rca proteins and such experiments have been reported to be taking place (Carmo-Silva et al., 2015). It will be most important to carefully analyse such plants for deleterious phenotypes at high temperatures, since Rca thermolability has been proposed to be regulatory (Sharkey, 2005). It may thus act as a thermal fuse to bring about Rubisco deactivation under stressful high temperature conditions.
In addressing these issues clearly opportunities exist in taking advantage of more thermostable Rca proteins that exist among natural variation (Salvucci and Crafts-Brandner, 2004b; Lawson et al., 2012; Scafaro et al., 2016). It is also worth pointing out that it may not be necessary to restrict oneself to green-type Rca. The characterized red-type Rca from the thermophilic rhodophyte C. merolae was a functional activase at 25°C, and able to hydrolyze ATP after incubation at 60°C (Loganathan et al., 2016). Protein engineering approaches that utilize both our mechanistic insights in combination with artificial evolution experiments that utilize an expanding suite of Rubisco dependent Escherichia coli (RDE) systems (Mueller-Cajar and Whitney, 2008b; Durao et al., 2015; Antonovsky et al., 2016; Wilson et al., 2016) will enable incompatibilities between specific Rubiscos and activases to be overcome.
An additional opportunity to enhance Rubisco function and photosynthesis by activase engineering relates to the naturally slow activation response of Rubisco under fluctuating light conditions (Mott and Woodrow, 2000; Lawson et al., 2012). Accordingly it was shown that Arabidopsis plants expressing less regulated Rubisco activase isoforms were able to activate Rubisco more rapidly than wild-type plants following a dark to light transition. This property translated to increased biomass accumulation when the plants were grown under a fluctuating light regimen (Carmo-Silva and Salvucci, 2013). Rice plants overexpressing an activase from maize also displayed faster induction of photosynthesis under fluctuating light conditions (Yamori et al., 2012). These results indicate that activases that are highly functional, and thus able to rapidly convert inhibited Rubisco complexes to the ECM holoenzyme, may be able to confer enhanced photosynthetic properties to plants exposed to fluctuating light conditions that may commonly be encountered in natural environments.
While considering the possibility of qualitatively superior activases it is also worth mentioning that the thus far described members of the red-type and CbbQO type Rca clades were all able to remove the extremely tight-binding inhibitor CABP from their cognate Rubiscos (Tsai et al., 2015; Loganathan et al., 2016), whereas the green-type Rca from higher plants is unable to do so (Robinson and Portis, 1988). Although more work is required regarding the relative affinity of CABP to various enzymes, these results indicate that different clades of Rca have evolved different levels of remodeling power that can potentially be utilized to advantage in heterologous contexts.
It appears likely that the crops of the future will possess a photosynthetic machinery consisting of carefully selected modules that will ensure maximum yield performance in their particular environment (Zhu et al., 2010; Kromdijk et al., 2016). The properties of Rubisco and its support cast will continue to play a critical role in this endeavor (Sharwood, 2017). In order to intelligently and effectively apply modifications to the photosynthesizers of our choice, a much denser network of Rubisco and activase related data is required (Hanson, 2016). This is critical because our dependence on Rubisco as key carbon fixation catalyst will be ongoing, at least until alternative and more efficient synthetic CO2 fixation pathways have been successfully and fully integrated into the metabolism of photoautotrophs (Bar-Even et al., 2010; Schwander et al., 2016).
The author confirms being the sole contributor of this work and approved it for publication.
My laboratory's research on rubisco activases was funded by Nanyang Technological University (startup grant) and the Ministry of Education of Singapore (MOE2013-T2-2-089).
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ammelburg, M., Frickey, T., and Lupas, A. N. (2006). Classification of AAA+ proteins. J. Struct. Biol. 156, 2–11. doi: 10.1016/j.jsb.2006.05.002
Anderson, L. E., and Fuller, R. C. (1969). Photosynthesis in Rhodospirillum rubrum. 4. Isolation and characterization of ribulose 1,5-Diphosphate Carboxylase. J. Biol. Chem. 244, 3105–3109.
Andersson, I. (2008). Catalysis and regulation in Rubisco. J. Exp. Bot. 59, 1555–1568. doi: 10.1093/jxb/ern091
Andersson, I., and Backlund, A. (2008). Structure and function of Rubisco. Plant Physiol. Biochem. 46, 275–291. doi: 10.1016/j.plaphy.2008.01.001
Andralojc, P. J., Madgwick, P. J., Tao, Y., Keys, A., Ward, J. L., Beale, M. H., et al. (2012). 2-Carboxy-D-arabinitol 1-phosphate (CA1P) phosphatase: evidence for a wider role in plant Rubisco regulation. Biochem. J. 442, 733–742. doi: 10.1042/BJ20111443
Andrews, T. J. (1996). The bait in the Rubisco mousetrap. Nat. Struct. Biol. 3, 3–7. doi: 10.1038/nsb0196-3
Andrews, T. J., and Abel, K. M. (1981). Kinetics and subunit interactions of ribulose bisphosphate carboxylase-oxygenase from the Cyanobacterium, Synechococcus Sp. J. Biol. Chem. 256, 8445–8451.
Andrews, T. J., and Lorimer, G. H. (1987). “Rubisco: Structure, mechanisms, and prospects for improvement,” in The Biochemistry of Plants: A Comprehensive Treatise, Vol. 10, Photosynthesis, eds M. D. Hatch and N. K. Boardman (New York, NY: Academic Press), 131–218.
Andrews, T. J., Lorimer, G. H., and Tolbert, N. E. (1973). Ribulose diphosphate oxygenase. I. Synthesis of phosphoglycolate by fraction-1 protein of leaves. Biochemistry 12, 11–18. doi: 10.1021/bi00725a003
Andrews, T. J., and Whitney, S. M. (2003). Manipulating ribulose bisphosphate carboxylase/oxygenase in the chloroplasts of higher plants. Arch. Biochem. Biophys. 414, 159–169. doi: 10.1016/S0003-9861(03)00100-0
Antonovsky, N., Gleizer, S., Noor, E., Zohar, Y., Herz, E., Barenholz, U., et al. (2016). Sugar Synthesis from CO2 in Escherichia coli. Cell 166, 115–125. doi: 10.1016/j.cell.2016.05.064
Badger, M. R., Andrews, T. J., Whitney, S. M., Ludwig, M., Yellowlees, D. C., Leggat, W., et al. (1998). The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast-based CO2-concentrating mechanisms in algae. Can. J. Bot Revue Can. Bot. 76, 1052–1071. doi: 10.1139/cjb-76-6-1052
Badger, M. R., and Bek, E. J. (2008). Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 59, 1525–1541. doi: 10.1093/jxb/erm297
Bainbridge, G., Anralojc, P. J., Madgwick, P. J., Pitts, J. E., and Parry, M. A. (1998). Effect of mutation of lysine-128 of the large subunit of ribulose bisphosphate carboxylase/oxygenase from Anacystis nidulans. Biochem. J. 336, 387–393.
Bainbridge, G., Madgwick, P., Parmar, S., Mitchell, R., Paul, M., Pitts, J., et al. (1995). Engineering Rubisco to change its catalytic properties. J. Exp. Bot. 46, 1269–1276. doi: 10.1093/jxb/46.special_issue.1269
Bar-Even, A., Noor, E., Lewis, N. E., and Milo, R. (2010). Design and analysis of synthetic carbon fixation pathways. Proc. Natl. Acad. Sci. U.S.A. 107, 8889–8894. doi: 10.1073/pnas.0907176107
Barta, C., Dunkle, A. M., Wachter, R. M., and Salvucci, M. E. (2010). Structural changes associated with the acute thermal instability of Rubisco activase. Arch. Biochem. Biophys. 499, 17–25. doi: 10.1016/j.abb.2010.04.022
Bauwe, H., Hagemann, M., and Fernie, A. R. (2010). Photorespiration: players, partners and origin. Trends Plant Sci. 15, 330–336. doi: 10.1016/j.tplants.2010.03.006
Beckwith, R., Estrin, E., Worden, E. J., and Martin, A. (2013). Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA plus unfoldase. Nat. Struct. Mol. Biol. 20, 1164. doi: 10.1038/nsmb.2659
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., Patron, N. J., and Nekrasov, V. (2015). Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 32, 76–84. doi: 10.1016/j.copbio.2014.11.007
Berry, J., and Bjorkman, O. (1980). Photosynthetic response and adaptation to temperature in higher-plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 31, 491–543. doi: 10.1146/annurev.pp.31.060180.002423
Blayney, M. J., Whitney, S. M., and Beck, J. L. (2011). NanoESI mass spectrometry of rubisco and rubisco activase structures and their interactions with nucleotides and sugar phosphates. J. Am. Soc. Mass Spectrom. 22, 1588–1601. doi: 10.1007/s13361-011-0187-8
Bock, R. (2015). Engineering plastid genomes: methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol. 66, 211–241. doi: 10.1146/annurev-arplant-050213-040212
Boex-Fontvieille, E., Daventure, M., Jossier, M., Hodges, M., Zivy, M., and Tcherkez, G. (2014). Phosphorylation pattern of Rubisco activase in Arabidopsis leaves. Plant Biol. 16, 550–557. doi: 10.1111/plb.12100
Bracher, A., Sharma, A., Starling-Windhof, A., Hartl, F. U., and Hayer-Hartl, M. (2015). Degradation of potent Rubisco inhibitor by selective sugar phosphatase. Nat. Plants 1:15006. doi: 10.1038/nplants.2015.6
Bracher, A., Whitney, S. M., Hartl, F. U., and Hayer-Hartl, M. (2017). Biogenesis and metabolic maintenance of Rubisco. Annu. Rev. Plant Biol. 68, 29–60. doi: 10.1146/annurev-arplant-043015-111633
Carmo-Silva, A. E., and Salvucci, M. E. (2011). The activity of Rubisco's molecular chaperone, Rubisco activase, in leaf extracts. Photosyn. Res. 108, 143–155. doi: 10.1007/s11120-011-9667-8
Carmo-Silva, A. E., and Salvucci, M. E. (2013). The regulatory properties of Rubisco activase differ among species and affect photosynthetic induction during light transitions. Plant Physiol. 161, 1645–1655. doi: 10.1104/pp.112.213348
Carmo-Silva, E., Scales, J. C., Madgwick, P. J., and Parry, M. A. (2015). Optimizing Rubisco and its regulation for greater resource use efficiency. Plant Cell Environ. 38, 1817–1832. doi: 10.1111/pce.12425
Cegelski, L., and Schaefer, J. (2006). NMR determination of photorespiration in intact leaves using in vivo 13CO2 labeling. J. Magn. Reson. 178, 1–10. doi: 10.1016/j.jmr.2005.10.010
Chakraborty, M., Kuriata, A. M., Nathan Henderson, J., Salvucci, M. E., Wachter, R. M., and Levitus, M. (2012). Protein oligomerization monitored by fluorescence fluctuation spectroscopy: self-assembly of Rubisco activase. Biophys. J. 103, 949–958. doi: 10.1016/j.bpj.2012.07.034
Chen, Z., and Spreitzer, R. J. (1992). How various factors influence the CO2/O2 specificity of ribulose-1,5-bisphosphate carboxylase/oxygenase. Photosynth. Res. 31, 157–164. doi: 10.1007/BF00028792
Cleland, W. W., Andrews, T. J., Gutteridge, S., Hartman, F. C., and Lorimer, G. H. (1998). Mechanism of Rubisco - the carbamate as general base. Chem. Rev. 98, 549–561. doi: 10.1021/cr970010r
Crafts-Brandner, S. J., and Salvucci, M. E. (2000). Rubisco activase constrains the photosynthetic potential of leaves at high temperature and CO2. Proc. Natl. Acad. Sci. U.S.A. 97, 13430–13435. doi: 10.1073/pnas.230451497
de Boer, A. P., van der Oost, J., Reijnders, W. N., Westerhoff, H. V., Stouthamer, A. H., and van Spanning, R. J. (1996). Mutational analysis of the nor gene cluster which encodes nitric-oxide reductase from Paracoccus denitrificans. Eur. J. Biochem. 242, 592–600. doi: 10.1111/j.1432-1033.1996.0592r.x
Delwiche, C. F., and Palmer, J. D. (1996). Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol. Biol. Evol. 13, 873–882. doi: 10.1093/oxfordjournals.molbev.a025647
Duff, A. P., Andrews, T. J., and Curmi, P. M. G. (2000). The transition between the open and closed states of rubisco is triggered by the inter-phosphate distance of the bound bisphosphate. J. Mol. Biol. 298, 903–916. doi: 10.1006/jmbi.2000.3724
Durao, P., Aigner, H., Nagy, P., Mueller-Cajar, O., Hartl, F. U., and Hayer-Hartl, M. (2015). Opposing effects of folding and assembly chaperones on evolvability of Rubisco. Nat. Chem. Biol. 11, 148–155. doi: 10.1038/nchembio.1715
El Bakkouri, M., Gutsche, I., Kanjee, U., Zhao, B. Y., Yu, M. A., Goret, G., et al. (2010). Structure of RavA MoxR AAA plus protein reveals the design principles of a molecular cage modulating the inducible lysine decarboxylase activity. Proc. Natl. Acad. Sci. U.S.A. 107, 22499–22504. doi: 10.1073/pnas.1009092107
Erzberger, J. P., and Berger, J. M. (2006). Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35, 93–114. doi: 10.1146/annurev.biophys.35.040405.101933
Esau, B. D., Snyder, G. W., and Portis, A. R. (1996). Differential effects of N- and C-terminal deletions on the two activities of rubisco activase. Arch. Biochem. Biophys. 326, 100–105. doi: 10.1006/abbi.1996.0052
Feiz, L., Williams-Carrier, R., Belcher, S., Montano, M., Barkan, A., and Stern, D. B. (2014). A protein with an inactive pterin-4a-carbinolamine dehydratase domain is required for Rubisco biogenesis in plants. Plant J. 80, 862–869. doi: 10.1111/tpj.12686
Feiz, L., Williams-Carrier, R., Wostrikoff, K., Belcher, S., Barkan, A., and Stern, D. B. (2012). Ribulose-1,5-bis-phosphate carboxylase/oxygenase accumulation factor1 is required for holoenzyme assembly in maize. Plant Cell 24, 3435–3446. doi: 10.1105/tpc.112.102012
Feller, U., Crafts-Brandner, S. J., and Salvucci, M. E. (1998). Moderately high temperatures inhibit ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) activase-mediated activation of Rubisco. Plant Physiol. 116, 539–546. doi: 10.1104/pp.116.2.539
Finnigan, G. C., Hanson-Smith, V., Stevens, T. H., and Thornton, J. W. (2012). Evolution of increased complexity in a molecular machine. Nature 481, 360–364. doi: 10.1038/nature10724
Fujita, K., Ehira, S., Tanaka, K., Asai, K., and Ohta, N. (2008). Molecular phylogeny and evolution of the plastid and nuclear encoded cbbX genes in the unicellular red alga Cyanidioschyzon merolae. Genes Genet. Syst. 83, 127–133. doi: 10.1266/ggs.83.127
Fukayama, H., Ueguchi, C., Nishikawa, K., Katoh, N., Ishikawa, C., Masumoto, C., et al. (2012). Overexpression of rubisco activase decreases the photosynthetic CO2 assimilation rate by reducing rubisco content in rice leaves. Plant Cell Physiol. 53, 976–986. doi: 10.1093/pcp/pcs042
Gibson, J. L., and Tabita, F. R. (1997). Analysis of the cbbXYZ operon in Rhodobacter sphaeroides. J. Bact. 179, 663–669. doi: 10.1128/jb.179.3.663-669.1997
Gray, M. W., Lukes, J., Archibald, J. M., Keeling, P. J., and Doolittle, W. F. (2010). Cell biology. Irremediable complexity? Science 330, 920–921. doi: 10.1126/science.1198594
Hanson, D. T. (2016). Breaking the rules of Rubisco catalysis. J. Exp. Bot. 67, 3180–3182. doi: 10.1093/jxb/erw197
Hanson, P. I., and Whiteheart, S. W. (2005). AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6, 519–529. doi: 10.1038/nrm1684
Harris, G. C., and Koniger, M. (1997). The “high” concentrations of enzymes within the chloroplast. Photosyn. Res. 54, 5–23. doi: 10.1023/A:1005895213775
Haslberger, T., Zdanowicz, A., Brand, I., Kirstein, J., Turgay, K., Mogk, A., et al. (2008). Protein disaggregation by the AAA+ chaperone ClpB involves partial threading of looped polypeptide segments. Nat. Struct. Mol. Biol. 15, 641–650. doi: 10.1038/nsmb.1425
Hasse, D., Larsson, A. M., and Andersson, I. (2015). Structure of Arabidopsis thaliana Rubisco activase. Acta Crystallogr. D Biol. Crystallogr. 71(Pt 4), 800–808. doi: 10.1107/S1399004715001182
Hauser, T., Bhat, J. Y., Milicic, G., Wendler, P., Hartl, F. U., Bracher, A., et al. (2015a). Structure and mechanism of the Rubisco-assembly chaperone Raf1. Nat. Struct. Mol. Biol. 22, 720–728. doi: 10.1038/nsmb.3062
Hauser, T., Popilka, L., Hartl, F. U., and Hayer-Hartl, M. (2015b). Role of auxiliary proteins in Rubisco biogenesis and function. Nat. Plants 1, 15065. doi: 10.1038/nplants.2015.65
Hayashi, N. R., Arai, H., Kodama, T., and Igarashi, Y. (1997). The novel genes, cbbQ and cbbO, located downstream from the RubisCO genes of Pseudomonas hydrogenothermophila, affect the conformational states and activity of RubisCO. Biochem. Biophys. Res. Commun. 241, 565–569. doi: 10.1006/bbrc.1997.7853
Hayashi, N. R., Arai, H., Kodama, T., and Igarashi, Y. (1999). The cbbQ genes, located downstream of the form i and form II RubisCO genes, affect the activity of both RubisCOs. Biochem. Biophys. Res. Commun. 265, 177–183. doi: 10.1006/bbrc.1999.1103
Hazra, S., Henderson, J. N., Liles, K., Hilton, M. T., and Wachter, R. M. (2015). Regulation of ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) activase: product inhibition, cooperativity, and magnesium activation. J. Biol. Chem. 290, 24222–24236. doi: 10.1074/jbc.M115.651745
Heinhorst, S., Baker, S. H., Johnson, D. R., Davies, P. S., Cannon, G. C., and Shively, J. M. (2002). Two copies of form I RuBisCO genes in Acidithiobacillus ferrooxidans ATCC 23270. Curr. Microbiol. 45, 115–117. doi: 10.1007/s00284-001-0094-5
Henderson, J. N., Kuriata, A. M., Fromme, R., Salvucci, M. E., and Wachter, R. M. (2011). Atomic resolution x-ray structure of the substrate recognition domain of higher plant ribulose-bisphosphate carboxylase/oxygenase (Rubisco) activase. J. Biol. Chem. 286, 35683–35688. doi: 10.1074/jbc.C111.289595
Hovde, B. T., Deodato, C. R., Hunsperger, H. M., Ryken, S. A., Yost, W., Jha, R. K., et al. (2015). Genome sequence and transcriptome analyses of chrysochromulina tobin: metabolic tools for enhanced algal fitness in the prominent order prymnesiales (Haptophyceae). PLoS Genet. 11:e1005469. doi: 10.1371/journal.pgen.1005469
Iyer, L. M., Leipe, D. D., Koonin, E. V., and Aravind, L. (2004). Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146, 11–31. doi: 10.1016/j.jsb.2003.10.010
Jordan, D. B., Chollet, R., and Ogren, W. L. (1983). Binding of phosphorylated effectors by active and inactive forms of ribulose-1,5-bisphosphate carboxylase. Biochemistry 22, 3410–3418. doi: 10.1021/bi00283a017
Joshi, J., Mueller-Cajar, O., Tsai, Y. C., Hartl, F. U., and Hayer-Hartl, M. (2015). Role of small subunit in mediating assembly of red-type form I Rubisco. J. Biol. Chem. 290, 1066–1074. doi: 10.1074/jbc.M114.613091
Jungst, A., and Zumft, W. G. (1992). Interdependence of respiratory no reduction and nitrite reduction revealed by mutagenesis of Nirq, a novel gene in the denitrification gene-cluster of Pseudomonas-Stutzeri. FEBS Lett. 314, 308–314. doi: 10.1016/0014-5793(92)81495-8
Kanjee, U., Gutsche, I., Alexopoulos, E., Zhao, B., El Bakkouri, M., Thibault, G., et al. (2011). Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase. EMBO J. 30, 931–944. doi: 10.1038/emboj.2011.5
Kannappan, B., and Gready, J. E. (2008). Redefinition of Rubisco carboxylase reaction reveals origin of water for hydration and new roles for active-site residues. J. Am. Chem. Soc. 130, 15063–15080. doi: 10.1021/ja803464a
Kardon, J. R., Yien, Y. Y., Huston, N. C., Branco, D. S., Hildick-Smith, G. J., Rhee, K. Y., et al. (2015). Mitochondrial ClpX activates a key enzyme for heme biosynthesis and erythropoiesis. Cell 161, 858–867. doi: 10.1016/j.cell.2015.04.017
Karkehabadi, S., Satagopan, S., Taylor, T. C., Spreitzer, R. J., and Andersson, I. (2007). Structural analysis of altered large-subunit loop-6/carboxy-terminus interactions that influence catalytic efficiency and CO2/O2 specificity of ribulose-1,5-bisphosphate carboxylase/oxygenase. Biochemistry 46, 11080–11089. doi: 10.1021/bi701063f
Keown, J. R., Griffin, M. D., Mertens, H. D., and Pearce, F. G. (2013). Small oligomers of ribulose-bisphosphate carboxylase/oxygenase (Rubisco) activase are required for biological activity. J. Biol. Chem. 288, 20607–20615. doi: 10.1074/jbc.M113.466383
Keown, J. R., and Pearce, F. G. (2014). Characterization of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase activase isoforms reveals hexameric assemblies with increased thermal stability. Biochem. J. 464, 413–423. doi: 10.1042/BJ20140676
Kim, S. Y., Bender, K. W., Walker, B. J., Zielinski, R. E., Spalding, M. H., Ort, D. R., et al. (2016). The plastid casein kinase 2 phosphorylates Rubisco activase at the Thr-78 site but is not essential for regulation of Rubisco activation state. Front. Plant Sci. 7:404. doi: 10.3389/fpls.2016.00404
Knight, S., Andersson, I., and Brändén, C.-I. (1990). Crystallographic analysis of ribulose 1,5-bisphosphate carboxylase from spinach at 2.4 Å resolution. Subunit interactions and active site. J. Mol. Biol. 215, 113–160. doi: 10.1016/S0022-2836(05)80100-7
Kobza, J., and Edwards, G. E. (1987). Influences of leaf temperature on photosynthetic carbon metabolism in wheat. Plant Physiol. 83, 69–74. doi: 10.1104/pp.83.1.69
Kromdijk, J., Glowacka, K., Leonelli, L., Gabilly, S. T., Iwai, M., Niyogi, K. K., et al. (2016). Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 354, 857–861. doi: 10.1126/science.aai8878
Kumar, A., Li, C. S., and Portis, A. R. (2009). Arabidopsis thaliana expressing a thermostable chimeric Rubisco activase exhibits enhanced growth and higher rates of photosynthesis at moderately high temperatures. Photosyn. Res. 100, 143–153. doi: 10.1007/s11120-009-9438-y
Kurek, I., Chang, T. K., Bertain, S. M., Madrigal, A., Liu, L., Lassner, M. W., et al. (2007). Enhanced thermostability of arabidopsis Rubisco activase improves photosynthesis and growth rates under moderate heat stress. Plant Cell 19, 3230–3241. doi: 10.1105/tpc.107.054171
Kuriata, A. M., Chakraborty, M., Henderson, J. N., Hazra, S., Serban, A. J., Pham, T. V., et al. (2014). ATP and magnesium promote cotton short-form ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) activase hexamer formation at low micromolar concentrations. Biochemistry 53, 7232–7246. doi: 10.1021/bi500968h
Larson, E. M., O'Brien, C. M., Zhu, G., Spreitzer, R. J., and Portis, A. R. Jr. (1997). Specificity for activase is changed by a Pro-89 to Arg substitution in the large subunit of Ribulose-1,5-bisphosphate carboxylase/oxygenase. J. Biol. Chem. 272, 17033–17037. doi: 10.1074/jbc.272.27.17033
Lawson, T., Kramer, D. M., and Raines, C. A. (2012). Improving yield by exploiting mechanisms underlying natural variation of photosynthesis. Curr. Opin. Biotechnol. 23, 215–220. doi: 10.1016/j.copbio.2011.12.012
Li, C. H., Salvucci, M. E., and Portis, A. R. (2005). Two residues of rubisco activase involved in recognition of the rubisco substrate. J. Biol. Chem. 280, 24864–24869. doi: 10.1074/jbc.M503547200
Li, L. A., Gibson, J. L., and Tabita, F. R. (1993). The Rubisco activase (Rca) gene is located downstream from Rbcs in anabaena Sp strain Ca and is detected in other anabaena nostoc strains. Plant Mol. Biol. 21, 753–764. doi: 10.1007/BF00027109
Li, L.-A., Zianni, M., and Tabita, F. R. (1999). Inactivation of the monocistronic rca gene in Anabaena variabilis suggests a physiological ribulose bisphosphate carboxylase/oxygenase activase-like function in heterocystous cyanobacteria. Plant Mol. Biol. 40, 467–478. doi: 10.1023/A:1006251808625
Lin, M. T., Occhialini, A., Andralojc, P. J., Devonshire, J., Hines, K. M., Parry, M. A., et al. (2014a). β-carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J. 79, 1–12. doi: 10.1111/tpj.12536
Lin, M. T., Occhialini, A., Andralojc, P. J., Parry, M. A., and Hanson, M. R. (2014b). A faster Rubisco with potential to increase photosynthesis in crops. Nature 513, 547–550. doi: 10.1038/nature13776
Linster, C. L., Van Schaftingen, E., and Hanson, A. D. (2013). Metabolite damage and its repair or pre-emption. Nat. Chem. Biol. 9, 72–80. doi: 10.1038/nchembio.1141
Liu, C. M., Young, A. L., Starling-Windhof, A., Bracher, A., Saschenbrecker, S., Rao, B. V., et al. (2010). Coupled chaperone action in folding and assembly of hexadecameric Rubisco. Nature 463, 197–202. doi: 10.1038/nature08651
Loganathan, N., Tsai, Y.-C. C., and Mueller-Cajar, O. (2016). Characterization of the heterooligomeric red-type Rubisco activase from red algae. Proc. Natl. Acad. Sci. U.S.A. 113, 14019–14024. doi: 10.1073/pnas.1610758113
Long, B. M., Rae, B. D., Rolland, V., Forster, B., and Price, G. D. (2016). Cyanobacterial CO2-concentrating mechanism components: function and prospects for plant metabolic engineering. Curr. Opin. Plant Biol. 31, 1–8. doi: 10.1016/j.pbi.2016.03.002
Long, S. P., Marshall-Colon, A., and Zhu, X. G. (2015). Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161, 56–66. doi: 10.1016/j.cell.2015.03.019
Lorimer, G. H., Badger, M. R., and Andrews, T. J. (1976). The activation of ribulose-1,5-bisphosphate carboxylase by carbon dioxide and magnesium ions. Equilibria, kinetics, a suggested mechanism and physiological implications. Biochemistry 15, 529–536. doi: 10.1021/bi00648a012
Maier, U.-G., Fraunholz, M., Zauner, S., Penny, S., and Douglas, S. (2000). A nucleomorph-encoded CbbX and the phylogeny of RuBisCo regulators. Mol. Biol. Evol. 17, 576–583. doi: 10.1093/oxfordjournals.molbev.a026337
Maynard Smith, J. (1970). Natural selection and concept of a protein space. Nature 225, 563–564. doi: 10.1038/225563a0
Mott, K. A., and Woodrow, I. E. (2000). Modelling the role of Rubisco activase in limiting non-steady-state photosynthesis. J. Exp. Bot. 51, 399–406. doi: 10.1093/jexbot/51.suppl_1.399
Mueller-Cajar, O., Stotz, M., and Bracher, A. (2014). Maintaining photosynthetic CO2 fixation via protein remodelling: the Rubisco activases. Photosyn. Res. 119, 191–201. doi: 10.1007/s11120-013-9819-0
Mueller-Cajar, O., Stotz, M., Wendler, P., Hartl, F. U., Bracher, A., and Hayer-Hartl, M. (2011). Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase. Nature 479, 194–199. doi: 10.1038/nature10568
Mueller-Cajar, O., and Whitney, S. M. (2008a). Directing the evolution of Rubisco and Rubisco activase: first impressions of a new tool for photosynthesis research. Photosyn. Res. 98, 667–675. doi: 10.1007/s11120-008-9324-z
Mueller-Cajar, O., and Whitney, S. M. (2008b). Evolving improved Synechococcus Rubisco functional expression in Escherichia coli. Biochem. J. 414, 205–214. doi: 10.1042/BJ20080668
Nisbet, R. E. R., Kilian, O., and McFadden, G. I. (2004). Diatom genomics: genetic acquisitions and mergers. Curr. Biol. 14, R1048–R1050. doi: 10.1016/j.cub.2004.11.043
Ochoa de Alda, J. A., Esteban, R., Diago, M. L., and Houmard, J. (2014). The plastid ancestor originated among one of the major cyanobacterial lineages. Nat. Commun. 5, 4937. doi: 10.1038/ncomms5937
Olivares, A. O., Baker, T. A., and Sauer, R. T. (2016). Mechanistic insights into bacterial AAA plus proteases and protein-remodelling machines. Nat. Rev. Microbiol. 14, 33–44. doi: 10.1038/nrmicro.2015.4
Orr, D. J., Alcantara, A., Kapralov, M. V., Andralojc, P. J., Carmo-Silva, E., and Parry, M. A. (2016). Surveying Rubisco diversity and temperature response to improve crop photosynthetic efficiency. Plant Physiol. 172, 707–717. doi: 10.1104/pp.16.00750
Ort, D. R., Merchant, S. S., Alric, J., Barkan, A., Blankenship, R. E., Bock, R., et al. (2015). Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc. Natl. Acad. Sci. U.S.A. 112, 8529–8536. doi: 10.1073/pnas.1424031112
Ott, C. M., Smith, B. D., Portis, A. R. Jr., and Spreitzer, R. J. (2000). Activase region on chloroplast ribulose-1,5-bisphosphate carboxylase/oxygenase. Nonconservative substitution in the large subunit alters species specificity of protein interaction. J. Biol. Chem. 275, 26241–26244. doi: 10.1074/jbc.M004580200
Oudot-Le Secq, M. P., Grimwood, J., Shapiro, H., Armbrust, E. V., Bowler, C., and Green, B. R. (2007). Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: comparison with other plastid genomes of the red lineage. Mol. Genet. Genomics 277, 427–439. doi: 10.1007/s00438-006-0199-4
Parry, M. A. J., Keys, A. J., Madgwick, P. J., Carmo-Silva, A. E., and Andralojc, P. J. (2008). Rubisco regulation: a role for inhibitors. J. Exp. Bot. 59, 1569–1580. doi: 10.1093/jxb/ern084
Parry, M. A. J., Madgwick, P. J., Carvalho, J. F. C., and Andralojc, P. J. (2007). Prospects for increasing photosynthesis by overcoming the limitations of Rubisco. J. Agric. Sci. 145, 31–43. doi: 10.1017/S0021859606006666
Pearce, F. G. (2006). Catalytic by-product formation and ligand binding by ribulose bisphosphate carboxylases from different phylogenies. Biochem. J. 399, 525–534. doi: 10.1042/BJ20060430
Pearce, F. G., and Andrews, T. J. (2003). The relationship between side reactions and slow inhibition of ribulose-bisphosphate carboxylase revealed by a loop 6 mutant of the tobacco enzyme. J. Biol. Chem. 278, 32526–32536. doi: 10.1074/jbc.M305493200
Portis, A. R. Jr. (1995). The regulation of Rubisco by Rubisco activase. J. Exp. Bot. 46, 1285–1291. doi: 10.1093/jxb/46.special_issue.1285
Portis, A. R. (2003). Rubisco activase - Rubisco's catalytic chaperone. Photosyn. Res. 75, 11–27. doi: 10.1023/A:1022458108678
Portis, A. R., Li, C. S., Wang, D. F., and Salvucci, M. E. (2008). Regulation of Rubisco activase and its interaction with Rubisco. J. Exp. Bot. 59, 1597–1604. doi: 10.1093/jxb/erm240
Price, G. D., Badger, M. R., Woodger, F. J., and Long, B. M. (2008). Advances in understanding the cyanobacterial CO(2)-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J. Exp. Bot. 59, 1441–1461. doi: 10.1093/jxb/erm112
Rae, B. D., Long, B. M., Badger, M. R., and Price, G. D. (2013). Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol. Mol. Biol. Rev. 77, 357–379. doi: 10.1128/MMBR.00061-12
Read, B. A., and Tabita, F. R. (1994). High substrate specificity factor ribulose bisphosphate carboxylase/oxygenase from eukaryotic marine algae and properties of recombinant cyanobacterial rubisco containing “algal” residue modifications. Arch. Biochem. Biophys. 312, 210–218. doi: 10.1006/abbi.1994.1301
Rice, D. W., and Palmer, J. D. (2006). An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 4:31. doi: 10.1186/1741-7007-4-31
Robinson, S. P., and Portis, A. R. (1988). Release of the nocturnal inhibitor, carboxyarabinitol-1-phosphate, from ribulose bisphosphate carboxylase oxygenase by Rubisco activase. FEBS Lett. 233, 413–416. doi: 10.1016/0014-5793(88)80473-3
Robinson, S. P., and Portis, A. R. (1989). Adenosine-triphosphate hydrolysis by purified Rubisco activase. Arch. Biochem. Biophys. 268, 93–99. doi: 10.1016/0003-9861(89)90568-7
Sage, R. F. (2013). Photorespiratory compensation: a driver for biological diversity. Plant Biol. 15, 624–638. doi: 10.1111/plb.12024
Salvucci, M. E., and Crafts-Brandner, S. J. (2004a). Mechanism for deactivation of Rubisco under moderate heat stress. Physiol. Plant. 122, 513–519. doi: 10.1111/j.1399-3054.2004.00419.x
Salvucci, M. E., and Crafts-Brandner, S. J. (2004b). Relationship between the heat tolerance of photosynthesis and the thermal stability of rubisco activase in plants from contrasting thermal environments. Plant Physiol. 134, 1460–1470. doi: 10.1104/pp.103.038323
Salvucci, M. E., and Klein, R. R. (1994). Site-directed mutagenesis of a reactive lysyl residue (Lys- 247) of Rubisco activase. Arch. Biochem. Biophys. 314, 178–185. doi: 10.1006/abbi.1994.1427
Salvucci, M. E., Portis, A. R., and Ogren, W. L. (1985). A soluble chloroplast protein catalyzes ribulosebisphosphate carboxylase oxygenase activation in vivo. Photosyn. Res. 7, 193–201. doi: 10.1007/BF00037012
Santelli, E., Bankston, L. A., Leppla, S. H., and Liddington, R. C. (2004). Crystal structure of a complex between anthrax toxin and its host cell receptor. Nature 430, 905–908. doi: 10.1038/nature02763
Satagopan, S., Chan, S., Perry, L. J., and Tabita, F. R. (2014). Structure-function studies with the unique hexameric form II ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Rhodopseudomonas palustris. J. Biol. Chem. 289, 21433–21450. doi: 10.1074/jbc.M114.578625
Satagopan, S., and Spreitzer, R. J. (2004). Substitutions at the Asp-473 latch residue of chlamydomonas ribulosebisphosphate carboxylase/oxygenase cause decreases in carboxylation efficiency and CO2/O2 specificity. J. Biol. Chem. 279, 14240–14244. doi: 10.1074/jbc.M313215200
Sauer, R. T., and Baker, T. A. (2011). AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80, 587–612. doi: 10.1146/annurev-biochem-060408-172623
Savir, Y., Noor, E., Milo, R., and Tlusty, T. (2010). Cross-species analysis traces adaptation of Rubisco toward optimality in a low-dimensional landscape. Proc. Natl. Acad. Sci. U.S.A. 107, 3475–3480. doi: 10.1073/pnas.0911663107
Scafaro, A. P., Galle, A., Van Rie, J., Carmo-Silva, E., Salvucci, M. E., and Atwell, B. J. (2016). Heat tolerance in a wild Oryza species is attributed to maintenance of Rubisco activation by a thermally stable Rubisco activase ortholog. New Phytol. 211, 899–911. doi: 10.1111/nph.13963
Scales, J. C., Parry, M. A., and Salvucci, M. E. (2014). A non-radioactive method for measuring Rubisco activase activity in the presence of variable ATP: ADP ratios, including modifications for measuring the activity and activation state of Rubisco. Photosyn. Res. 119, 355–365. doi: 10.1007/s11120-013-9964-5
Schwander, T., Schada von Borzyskowski, L., Burgener, S., Cortina, N. S., and Erb, T. J. (2016). A synthetic pathway for the fixation of carbon dioxide in vitro. Science 354, 900–904. doi: 10.1126/science.aah5237
Sharkey, T. D. (2005). Effects of moderate heat stress on photosynthesis: importance of thylakoid reactions, rubisco deactivation, reactive oxygen species, and thermotolerance provided by isoprene. Plant Cell Environ. 28, 269–277. doi: 10.1111/j.1365-3040.2005.01324.x
Sharwood, R. E. (2017). Engineering chloroplasts to improve Rubisco catalysis: prospects for translating improvements into food and fiber crops. New Phytol. 213, 494–510. doi: 10.1111/nph.14351
Sharwood, R. E., Ghannoum, O., Kapralov, M. V., Gunn, L. H., and Whitney, S. M. (2016a). Temperature responses of Rubisco from Paniceae grasses provide opportunities for improving C3 photosynthesis. Nat. Plants 2:16186. doi: 10.1038/nplants.2016.186
Sharwood, R. E., Ghannoum, O., and Whitney, S. M. (2016b). Prospects for improving CO2 fixation in C3-crops through understanding C4-Rubisco biogenesis and catalytic diversity. Curr. Opin. Plant Biol. 31, 135–142. doi: 10.1016/j.pbi.2016.04.002
Sharwood, R. E., von Caemmerer, S., Maliga, P., and Whitney, S. M. (2008). The catalytic properties of hybrid Rubisco comprising tobacco small and sunflower large subunits mirror the kinetically equivalent source Rubiscos and can support tobacco growth. Plant Physiol. 146, 83–96. doi: 10.1104/pp.107.109058
Shih, P. M., Occhialini, A., Cameron, J. C., Andralojc, P. J., Parry, M. A., and Kerfeld, C. A. (2016). Biochemical characterization of predicted Precambrian RuBisCO. Nat. Commun. 7:10382. doi: 10.1038/ncomms10382
Shih, P. M., Wu, D., Latifi, A., Axen, S. D., Fewer, D. P., Talla, E., et al. (2013). Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 110, 1053–1058. doi: 10.1073/pnas.1217107110
Siddiqui, S. M., Sauer, R. T., and Baker, T. A. (2004). Role of the processing pore of the ClpX AAA+ ATPase in the recognition and engagement of specific protein substrates. Genes Dev. 18, 369–374. doi: 10.1101/gad.1170304
Snider, J., Gutsche, I., Lin, M., Baby, S., Cox, B., Butland, G., et al. (2006). Formation of a distinctive complex between the inducible bacterial lysine decarboxylase and a novel AAA+ ATPase. J. Biol. Chem. 281, 1532–1546. doi: 10.1074/jbc.M511172200
Snider, J., and Houry, W. A. (2006). MoxR AAA+ ATPases: a novel family of molecular chaperones? J. Struct. Biol. 156, 200–209. doi: 10.1016/j.jsb.2006.02.009
Spreitzer, R. J. (2003). Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch. Biochem. Biophys. 414, 141–149. doi: 10.1016/S0003-9861(03)00171-1
Spreitzer, R. J., and Salvucci, M. E. (2002). Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annu. Rev. Plant Biol. 53, 449–475. doi: 10.1146/annurev.arplant.53.100301.135233
Stotz, M., Mueller-Cajar, O., Ciniawsky, S., Wendler, P., Hartl, F. U., Bracher, A., et al. (2011). Structure of green-type Rubisco activase from tobacco. Nat. Struct. Mol. Biol. 18, 1366–1370. doi: 10.1038/nsmb.2171
Sutter, M., Roberts, E. W., Gonzalez, R. C., Bates, C., Dawoud, S., Landry, K., et al. (2015). Structural characterization of a newly identified component of α-carboxysomes: the AAA+ domain protein CsoCbbQ. Sci. Rep. 5:16243. doi: 10.1038/srep16243
Sysoeva, T. A. (2016). Assessing heterogeneity in oligomeric AAA+ machines. Cell. Mol. Life Sci. 74, 1001–1018. doi: 10.1007/s00018-016-2374-z
Tabita, F. R., Hanson, T. E., Li, H., Satagopan, S., Singh, J., and Chan, S. (2007). Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol. Mol. Biol. Rev. 71, 576–599. doi: 10.1128/MMBR.00015-07
Tabita, F. R., Satagopan, S., Hanson, T. E., Kreel, N. E., and Scott, S. S. (2008). Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J. Exp. Bot. 59, 1515–1524. doi: 10.1093/jxb/erm361
Taylor, T. C., and Andersson, I. (1996). Structural transitions during activation and ligand binding in hexadecameric Rubisco inferred from the crystal structure of the activated unliganded spinach enzyme. Nat. Struct. Biol. 3, 95–101. doi: 10.1038/nsb0196-95
Tcherkez, G. (2013). Modelling the reaction mechanism of ribulose-1,5-bisphosphate carboxylase/oxygenase and consequences for kinetic parameters. Plant Cell Environ. 36, 1586–1596. doi: 10.1111/pce.12066
Tcherkez, G. (2016). The mechanism of Rubisco-catalysed oxygenation. Plant Cell Environ. 39, 983–997. doi: 10.1111/pce.12629
Tcherkez, G. G. B., Farquhar, G. D., and Andrews, T. J. (2006). Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. PNAS 103, 7246–7251. doi: 10.1073/pnas.0600605103
Tsai, Y. C., Lapina, M. C., Bhushan, S., and Mueller-Cajar, O. (2015). Identification and characterization of multiple rubisco activases in chemoautotrophic bacteria. Nat. Commun. 6:8883. doi: 10.1038/ncomms9883
Uemura, K., Anwaruzzaman, M. S., and Yokota, A. (1997). Ribulose-1,5-bisphosphate carboxylase/oxygenase from thermophilic red algae with a strong specificity for CO2 fixation. Biochem. Biophys. Res. Commun. 233, 568–571. doi: 10.1006/bbrc.1997.6497
van de Loo, F. J., and Salvucci, M. E. (1996). Activation of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) involves Rubisco activase Trp16. Biochemistry 35, 8143–8148. doi: 10.1021/bi9604901
van de Loo, F. J., and Salvucci, M. E. (1998). Involvement of two aspartate residues of rubisco activase in coordination of the ATP γ-phosphate and subunit cooperativity. Biochemistry 37, 4621–4625.
Varaljay, V. A., Satagopan, S., North, J. A., Witte, B., Dourado, M. N., Anantharaman, K., et al. (2016). Functional metagenomic selection of ribulose 1, 5-bisphosphate carboxylase/oxygenase from uncultivated bacteria. Environ. Microbiol. 18, 1187–1199. doi: 10.1111/1462-2920.13138
Wachter, R. M., Salvucci, M. E., Carmo-Silva, A. E., Barta, C., Genkov, T., and Spreitzer, R. J. (2013). Activation of interspecies-hybrid Rubisco enzymes to assess different models for the Rubisco-Rubisco activase interaction. Photosyn. Res. 117, 557–566. doi: 10.1007/s11120-013-9827-0
Wang, Z.-Y., Snyder, G. W., Esau, B. D., Portis, A. R. Jr., and Ogren, W. L. (1992). Species-dependent variation in the interaction of substrate-bound ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) and Rubisco activase. Plant Physiol. 100, 1858–1862. doi: 10.1104/pp.100.4.1858
Weibezahn, J., Tessarz, P., Schlieker, C., Zahn, R., Maglica, Z., Lee, S., et al. (2004). Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119, 653–665. doi: 10.1016/j.cell.2004.11.027
Weis, E. (1981). Reversible heat-inactivation of the calvin cycle: a possible mechanism of the temperature regulation of photosynthesis. Planta 151, 33–39. doi: 10.1007/BF00384234
Whitney, S. M., and Andrews, T. J. (2001). Plastome-encoded bacterial ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) supports photosynthesis and growth in tobacco. Proc. Natl. Acad. Sci. U.S.A. 98, 14738–14743. doi: 10.1073/pnas.261417298
Whitney, S. M., Baldet, P., Hudson, G. S., and Andrews, T. J. (2001). Form I Rubiscos from non-green algae are expressed abundantly but not assembled in tobacco chloroplasts. Plant J. 26, 535–547. doi: 10.1046/j.1365-313x.2001.01056.x
Whitney, S. M., Birch, R., Kelso, C., Beck, J. L., and Kapralov, M. V. (2015). Improving recombinant Rubisco biogenesis, plant photosynthesis and growth by coexpressing its ancillary RAF1 chaperone. Proc. Natl. Acad. Sci. U.S.A. 112, 3564–3569. doi: 10.1073/pnas.1420536112
Whitney, S. M., Houtz, R. L., and Alonso, H. (2011a). Advancing our understanding and capacity to engineer nature's CO(2)-sequestering enzyme, Rubisco. Plant Physiol. 155, 27–35. doi: 10.1104/pp.110.164814
Whitney, S. M., and Sharwood, R. E. (2008). Construction of a tobacco master line to improve Rubisco engineering in chloroplasts. J. Exp. Bot. 59, 1909–1921. doi: 10.1093/jxb/erm311
Whitney, S. M., Sharwood, R. E., Orr, D., White, S. J., Alonso, H., and Galmes, J. (2011b). Isoleucine 309 acts as a C4 catalytic switch that increases ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) carboxylation rate in Flaveria. Proc. Natl. Acad. Sci. U.S.A. 108, 14688–14693. doi: 10.1073/pnas.1109503108
Whittaker, C. A., and Hynes, R. O. (2002). Distribution and evolution of von Willebrand/integrin a domains: widely dispersed adhesion and elsewhere. Mol. Biol. Cell 13, 3369–3387. doi: 10.1091/mbc.E02-05-0259
Wilson, R. H., Alonso, H., and Whitney, S. M. (2016). Evolving Methanococcoides burtonii archaeal Rubisco for improved photosynthesis and plant growth. Sci. Rep. 6:22284. doi: 10.1038/srep22284
Wong, K. S., Bhandari, V., Janga, S. C., and Houry, W. A. (2017). The RavA-ViaA chaperone-like system interacts with and modulates the activity of the fumarate reductase respiratory complex. J. Mol. Biol. 429, 324–344. doi: 10.1016/j.jmb.2016.12.008
Wong, K. S., and Houry, W. A. (2012). Novel structural and functional insights into the MoxR family of AAA+ ATPases. J. Struct. Biol. 179, 211–221. doi: 10.1016/j.jsb.2012.03.010
Xiong, J. P., Stehle, T., Zhang, R. G., Joachimiak, A., Frech, M., Goodman, S. L., et al. (2002). Crystal structure of the extracellular segment of integrin alpha V β 3 in complex with an Arg-Gly-Asp ligand. Science 296, 151–155. doi: 10.1126/science.1069040
Yamori, W., Masumoto, C., Fukayama, H., and Makino, A. (2012). Rubisco activase is a key regulator of non-steady-state photosynthesis at any leaf temperature and, to a lesser extent, of steady-state photosynthesis at high temperature. Plant J. Cell Mol. Biol. 71, 871–880. doi: 10.1111/j.1365-313X.2012.05041.x
Yoon, H. S., Hackett, J. D., Ciniglia, C., Pinto, G., and Bhattacharya, D. (2004). A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21, 809–818. doi: 10.1093/molbev/msh075
Young, J. N., Heureux, A. M., Sharwood, R. E., Rickaby, R. E., Morel, F. M., and Whitney, S. M. (2016). Large variation in the Rubisco kinetics of diatoms reveals diversity among their carbon-concentrating mechanisms. J. Exp. Bot. 67, 3445–3456. doi: 10.1093/jxb/erw163
Zarzycki, J., Axen, S. D., Kinney, J. N., and Kerfeld, C. A. (2013). Cyanobacterial-based approaches to improving photosynthesis in plants. J. Exp. Bot. 64, 787–798. doi: 10.1093/jxb/ers294
Zhu, X. G., Long, S. P., and Ort, D. R. (2010). “Improving photosynthetic efficiency for greater yield,” in Annual Review of Plant Biology, Vol. 61, eds S. Merchant, W. R. Briggs, and D. Ort (Palo Alto, CA: Annual Reviews), 235–261.
Keywords: Rubisco, activase, photosynthesis, AAA+ proteins, molecular chaperones, carbon fixation
Citation: Mueller-Cajar O (2017) The Diverse AAA+ Machines that Repair Inhibited Rubisco Active Sites. Front. Mol. Biosci. 4:31. doi: 10.3389/fmolb.2017.00031
Received: 17 March 2017; Accepted: 29 April 2017;
Published: 19 May 2017.
Edited by:
Walid A. Houry, University of Toronto, CanadaReviewed by:
Robert Edward Sharwood, Australian National University, AustraliaCopyright © 2017 Mueller-Cajar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oliver Mueller-Cajar, Y2FqYXJAbnR1LmVkdS5zZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.