
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 29 September 2016
Sec. Molecular Recognition
Volume 3 - 2016 | https://doi.org/10.3389/fmolb.2016.00061
This article is part of the Research Topic Modulating prokaryotic lifestyle by DNA-binding proteins View all 25 articles
Shigella is a highly adapted human pathogen, mainly found in the developing world and causing a severe enteric syndrome. The highly sophisticated infectious strategy of Shigella banks on the capacity to invade the intestinal epithelial barrier and cause its inflammatory destruction. The cellular pathogenesis and clinical presentation of shigellosis are the sum of the complex action of a large number of bacterial virulence factors mainly located on a large virulence plasmid (pINV). The expression of pINV genes is controlled by multiple environmental stimuli through a regulatory cascade involving proteins and sRNAs encoded by both the pINV and the chromosome. The primary regulator of the virulence phenotype is VirF, a DNA-binding protein belonging to the AraC family of transcriptional regulators. The virF gene, located on the pINV, is expressed only within the host, mainly in response to the temperature transition occurring when the bacterium transits from the outer environment to the intestinal milieu. VirF then acts as anti-H-NS protein and directly activates the icsA and virB genes, triggering the full expression of the invasion program of Shigella. In this review we will focus on the structure of VirF, on its sophisticated regulation, and on its role as major player in the path leading from the non-invasive to the invasive phenotype of Shigella. We will address also the involvement of VirF in mechanisms aimed at withstanding adverse conditions inside the host, indicating that this protein is emerging as a global regulator whose action is not limited to virulence systems. Finally, we will discuss recent observations conferring VirF the potential of a novel antibacterial target for shigellosis.
Bacterial pathogens must often survive within fundamentally diverse habitats. Dynamic adaptation to the surroundings depends on the ability to sense environmental variations and to respond in an appropriate manner. This involves drastic changes in the transcriptional program of the cell. The capability to swiftly modulate gene expression requires investment by the bacterium in numerous gene functions that not only allow adaptation to different milieus but also enable the cell to co-ordinately rework its response to mutable conditions (Cases and de Lorenzo, 2005). The complexity of gene expression in pathogenic bacteria can be viewed as an evolutionary response to the challenge of surviving in changing environments (McAdams et al., 2004). Many pathogenic E. coli, including Shigella, are able to live in complex environments and have evolved intricate control systems. The expression of their pathogenic phenotype is the result of a multifactorial process which requires the synthesis of a large set of virulence determinants that may not be simultaneously needed during all stages of the infection process. These determinants are controlled by global regulatory networks, integrating specific regulators with conserved housekeeping processes.
Shigella is a facultative intracellular pathogen causing human bacillary dysentery, also known as shigellosis, a highly infectious disease widespread in developing countries. Although usually self-limiting, shigellosis can be fatal, especially in children (Anderson et al., 2016; Njamkepo et al., 2016; The et al., 2016). The pathogenicity of Shigella relies on its capacity to kill macrophages and invade colonic epithelial cells. Bacteria multiply intracellularly and spread to adjacent cells, with consequent cell death and inflammatory destruction of the mucosa (Schroeder and Hilbi, 2008; Ashida et al., 2015). The invasive process requires the coordinated expression of several genes located on the chromosome as well as on a virulence plasmid (pINV) (Sansonetti et al., 1982; Parsot, 2005). The acquisition of the pINV, a large F-type plasmid, by horizontal gene transfer (HTG) constitutes one of the most critical events in the evolution of the pathogenic lifestyle of Shigella since it encodes crucial elements of the molecular machinery required for invasion and intracellular survival (Pupo et al., 2000; The et al., 2016). pINV plasmids isolated from different Shigella spp. share significant homology and carry about 100 genes and an equivalent number of IS sequences (Buchrieser et al., 2000; Yang et al., 2005). Most genes required for host cell invasion and macrophage killing are contained in a conserved 31 kb region which is arranged as a PAI-like structure (Maurelli et al., 1985; Sasakawa et al., 1988). This so-called “entry region” consists of 34 genes organized into two large, divergently transcribed clusters. It contains the genes encoding the Ipa proteins, their chaperons and a specific T3SS system (Parsot, 2005). Besides these structural genes, the entry region hosts two regulatory genes coding for VirB and MxiE, the transcriptional activators required for the sequential expression of most pINV virulence genes (Beloin et al., 2002; Mavris et al., 2002). Scattered along the pINV, outside the entry region, are other genes encoding proteins crucial for the invasive process: the IcsA protein, responsible for the recruitment and polymerization of the host actin at one pole of the bacterial cell (Bernardini et al., 1989); the PhoN2 protein, required for proper IcsA localization (Scribano et al., 2014); the OspG protein, involved in the modulation of the host innate immune response (Kim et al., 2005); the IpaH proteins, which interfere with the host ubiquitination-dependent protein degradation (Ashida and Sasakawa, 2015); and the VirF protein, the primary virulence regulator (Sakai et al., 1988; Prosseda et al., 1998).
In this review we will focus on VirF summarizing its structure, its sophisticated regulation, its role as major player in the cascade of events leading to the activation of the virulence program of Shigella, and its involvement in mechanisms aimed at withstanding adverse conditions inside the host. We will also address how these features confer VirF the potential of a novel antibacterial target for shigellosis.
VirF is a member of the AraC family of transcriptional regulators. This large family contains more than 800 different proteins, found mainly among gram negative bacteria and involved in the regulation of carbon metabolism, stress response, and virulence (Gallegos et al., 1997; Egan, 2002). The AraC proteins are characterized by two structural domains: a conserved C-terminal DNA binding domain and a more variable N-terminal signaling domain. The two domains are connected by an unstructured linker. DNA binding involves two helix-turn-helix (HTH) motifs and has been studied by analyzing the structure of the MarA (Rhee et al., 1998) and Rob (Kwon et al., 2000) proteins in complex with their target DNA, leading to the proposal that two different modes of DNA binding might exist involving either one (Rob) or both (MarA) HTH motifs. The N-terminal region is responsible for multimerization and/or binding of cofactors (Egan, 2002). The AraC family has been traditionally divided into three classes (Gallegos et al., 1997). The first one consists of proteins which, like AraC, act as regulators in response to a chemical signal (Schleif, 2010). Proteins involved in stress response, such as MarA and Rob, constitute the second class. VirF belongs to the third class, whose members control transcription in response to a physical signal (like temperature) and mostly serve as virulence gene regulators. While the proteins of the first and third class act as homodimers, proteins of the second class operate mainly as monomers. Members of the AraC family are frequently insoluble proteins. Precise molecular characterization of VirF and detailed information on its mechanism of action are still scarce as purification of VirF has been obtained only in a few cases (Porter et al., 2004; Tran et al., 2011). Most groups (Tobe et al., 1993; Porter and Dorman, 2002; Koppolu et al., 2013; Emanuele et al., 2014; Emanuele and Garcia, 2015) have reported difficulties in isolating VirF in quantities suitable to in vitro analysis and have therefore used a MalE—VirF fusion protein which still retains VirF functionality despite the lack of the first 10 N-terminal aminoacids (Tobe et al., 1993; Koppolu et al., 2013; Emanuele and Garcia, 2015).
VirF carries the two canonical AraC DNA-binding HTH motifs within its C-terminus. To acquire structural and functional information about the VirF protein, the entire virF gene has been subjected to both, random and site-directed mutagenesis and mutated proteins have been assayed for their ability to activate the plasmid-encoded invasive genes by analyzing the expression of a mxiC-lacZ fusion (Porter and Dorman, 2002). Mutating the key residues of the first HTH motif, in particular in the positioning helix (I180) or in the recognition helix (K193), inactivates VirF. The second HTH motif is essential as well, since mutating the key residues (Y239 and I241), which according to the MarA-DNA co-crystal (Rhee et al., 1998) form specific contacts with DNA, leads again to VirF inactivation. The functionality of VirF is hampered also by modifications in the hydrophobic core of HTH2 or by deletion of the C-terminal HTH2 region. This strongly suggests that VirF interacts with its target sequences via both HTH motifs. Additional evidence on the relevance of the two motifs to DNA binding stems from studies on another AraC regulator, PerA, which shares significant homology with VirF. This protein is required for the activation of the bundle-forming pili in ETEC and is able to fully substitute VirF in the activation of the Shigella virB promoter (Porter et al., 2004). Mutations affecting the DNA-interacting nucleotides from either of the two C-terminal HTH motifs of PerA have been shown to inactivate the protein, indicating that also PerA requires both HTH motifs to interact with its DNA targets (Porter et al., 2004). However, while PerA and Rns, another AraC-like regulator from ETEC (Porter et al., 1998), are able to complement Shigella virF mutants, VirF is unable to restore the expression of the PerA or Rns regulated genes, suggesting that a particular DNA structure which forms only at Shigella VirF-regulated promoters is required as signal for the activation of the VirF protein (Porter and Dorman, 2002; Porter et al., 2004).
AraC and several members of the AraC family are known to form homodimers (Gallegos et al., 1997; Egan, 2002; Schleif, 2010) and the residues required for self-association are contained in the N-terminal domain. As yet the only evidence suggesting that VirF may act as dimer/oligomer stems from the observation that two mutants unable to bind DNA are dominant negative when co-expressed with the wild type VirF. However, the fact that other HTH1 or HTH2 mutations have no trans-dominant effect rises the possibility (Porter and Dorman, 2002) that VirF binds first as monomer and then, following DNA bending and DNA-DNA interactions, forms a multisubunit nucleoprotein complex, as previously observed in the case of the melibiose-dependent activator MelR (Bourgerie et al., 1997). This is consistent with the observation that a MalE—VirF fusion protein recognizes a single, large (about 100 bp) region on the virB promoter (Tobe et al., 1993). However, recent footprinting analyses using purified VirF and revealing the presence of four distinct VirF binding sites (each spanning 40–60 bp) within the icsA-RnaG regulatory region (Tran et al., 2011) indicate that, rather than interacting at a single large spot, VirF recognizes its target sites with different affinities and may give rise to a large nucleoprotein complex only at higher protein concentration.
The transcriptional activation of the invasive operons relies on the response to environmental stimuli like temperature, pH, and osmolarity, commonly encountered in the human intestine (Schroeder and Hilbi, 2008). This process requires the VirF protein (Sakai et al., 1988; Adler et al., 1989; Falconi et al., 1998; Durand et al., 2000) which, through a sophisticated regulatory cascade (Dorman and Porter, 1998; Prosseda et al., 2002; Parsot, 2005), leads to the full expression of the virulence phenotype (Figure 1). Temperature is a crucial factor since transcription of the pINV invasion genes is strongly repressed at 30°C by the chromosome-encoded protein H-NS (Maurelli and Sansonetti, 1988; Falconi et al., 1998). The primary event following the upshift of Shigella to the host temperature (37°C) is the synthesis of VirF which acts as an antisilencer, relieving H-NS mediated repression at the virB (Tobe et al., 1993) and icsA (Tran et al., 2011) loci. This is not uncommon: in many bacterial pathogens H-NS, one of the major components of the bacterial nucleoid, acts as transcriptional silencer of virulence genes located on mobile genetic elements (Dorman, 2004, 2007). As for the icsA gene, it encodes an outer membrane protein which promotes the intra- and intercellular spreading of Shigella among the epithelial cells of the colon (Bernardini et al., 1989; Lett et al., 1989). The other VirF-activated gene encodes a secondary transcriptional activator, VirB, which antagonizes the repressive activity of H-NS at the promoters of several pINV virulence genes by remodeling the DNA within H-NS-DNA nucleoprotein complexes (Beloin et al., 2002; McKenna et al., 2003). In particular, VirB activates, among others, the genes coding for the Shigella T3SS (Mxi and Spa proteins), for its early effectors and their chaperones (Ipa and Ipg proteins), and for MxiE, the last regulator in the cascade (Parsot, 2005; Schroeder and Hilbi, 2008). MxiE is another AraC-like protein (Kane et al., 2002; Mavris et al., 2002) which positively controls the expression of the late effectors but it becomes available as activator only when the IpgC chaperone facilitates its release from an anti-activator complex (OspD/Spa15).
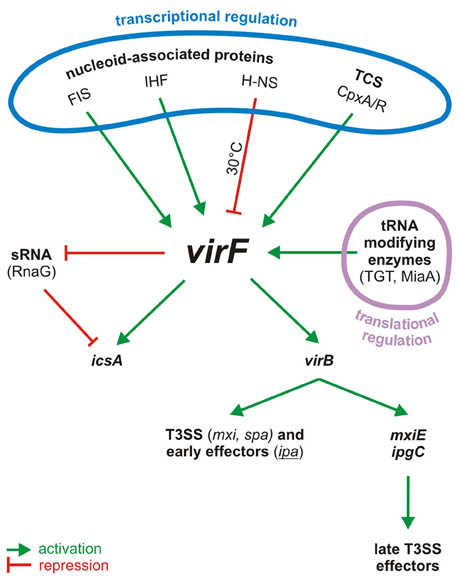
Figure 1. Centrality of VirF in the pINV regulatory cascade of Shigella. The control of VirF occurs at transcriptional and translation levels. The major proteins involved in the regulation of VirF in response to environmental stimuli and nutrient conditions are indicated. Once synthesized, VirF activates the icsA and virB genes and represses the synthesis of the sRNA RnaG. VirB, the second regulator, then activates several operons, including those for a T3SS system and for the early effectors. VirB activates also the last regulator, MxiE, which in association with IpgC activates the late effectors. TCS, two component system.
The role of VirF as positive activator was first evidenced by observing that in S. flexneri (Sakai et al., 1988) and in S. sonnei (Kato et al., 1989) the expression of the four Ipa antigen proteins and of IcsA (known also as VirG) is silenced in the absence of VirF and increased when VirF is over expressed. It was then demonstrated (Adler et al., 1989; Tobe et al., 1991) that the regulatory activity of VirF is exerted directly when it interacts with the icsA gene, while it occurs indirectly, i.e., through the mediation of VirB, on the ipa operons. This gave the first basis for the classic cascade model. Originally, the regulatory role of VirF remained unclear and the protein was simply regarded as the element responsible for the capacity of Shigella to bind Congo red, a phenotype (Pcr+) associated with the expression of virulence (Sakai et al., 1986a). Initial molecular analyses of the pINV had led to the identification of an about 1 kb region responsible for the Pcr+ phenotype in E. coli. This region turned out to be located within the F fragment of a SalI pINV restriction digests and contained a gene, hence named virF, which was shown to be essential but not sufficient for the full expression of virulence in S. flexneri (Sakai et al., 1986a,b). Additional support to the active role played by VirF as regulator banks on the observations that its intracellular concentration is strictly related to the expression of virulence genes, that a VirF threshold level is required for the activation of the second regulator, virB, and that overexpression of virF fully restores the invasive phenotype at non-permissive temperature or in pINV-integrated strains (Adler et al., 1989; Dagberg and Uhlin, 1992; Colonna et al., 1995; Prosseda et al., 1998; Durand et al., 2000).
Cloning and sequencing analyses of the pINV plasmid have revealed that, in contrast to virB which is localized within the 31 kb PAI, the virF gene constitutes a “desert island” located 60 kb outside the PAI and surrounded by a mosaic of IS sequences (Buchrieser et al., 2000; Venkatesan et al., 2001; Prosseda et al., 2006), suggesting that it might have been acquired by the plasmid genome independently from the “entry region” (Prosseda et al., 2002). Besides its position, one of the striking features of the virF sequence is its low GC content—about 30%; interestingly this is true also for other virulence-related genes in the 31 kb PAI—as compared to that of the whole pINV plasmid and of the Shigella chromosome, which are about 48 and 50%, respectively (Buchrieser et al., 2000; Venkatesan et al., 2001; Yang et al., 2005). The virF gene exhibits canonical −35 and −10 regulatory boxes, however no clear ribosome binding site is found. While attempting to purify the protein it has been observed that VirF exists in three different forms: 30, 27, and 21 kDa (Sakai et al., 1986b; Kato et al., 1989). The larger form has been considered the active one while as yet no functional activity has been attributed to the two minor forms.
Strong evidence that VirF acts as a DNA binding protein capable to activate virB transcription by interacting with the upstream region of the virB promoter comes from the studies of Tobe et al. (1993). By deletion analysis of the virB promoter the authors have shown that the activation of virB requires a region of 110 bp upstream the transcription start site. The relevance of this region has been confirmed in the same study by footprinting with a MalE—VirF fusion protein. This allowed to identify a VirF binding site spanning from −117 to −17 which, by in vitro transcription, proved to be essential for the activation of virB. Considering the position of the VirF binding site and its role as activator, it has been proposed that VirF could act either by recruiting RNA polymerase at the virB promoter or by improving the ability of RNA polymerase to form an open complex (Tobe et al., 1993). The positive effect of VirF on virB transcription is counteracted by H-NS which has been shown to occupy a virB promoter region (spanning from −20 to +20) encompassing the RNA polymerase binding site (Tobe et al., 1993). Since the H-NS and VirF binding sites at the virB promoter are contiguous, it has been speculated that the binding of VirF may disrupt the H-NS-DNA repression complex, possibly being favored by thermally-induced changes in local supercoiling (Tobe et al., 1993). The relevance of superhelicity per se is supported by the finding that virB activation by VirF occurs much more efficiently on supercoiled templates than on relaxed ones (Tobe et al., 1993).
As mentioned before, also the icsA gene is controlled by VirF (Sakai et al., 1988). This gene encodes a protein responsible for the motility of Shigella and is located on the pINV plasmid outside the “entry region” (Bernardini et al., 1989; Lett et al., 1989). In contrast to all other structural genes involved in the invasive process, icsA does not require VirB (Figure 1). As is the case of virB, also icsA is repressed by the binding of H-NS to its regulatory region (Prosseda et al., 1998). This occurs at three sites (H-NS I, H-NS II, and H-NS III) and severely reduces icsA transcription at 30°C (Tran et al., 2011). Recent evidence shows that the regulation of icsA also depends on RnaG, a small antisense RNA transcribed on the complementary strand of icsA (Giangrossi et al., 2010). RnaG downregulates icsA transcription by means of two independent mechanisms, transcriptional interference, and transcriptional attenuation. In the first case the expression of RnaG decreases the activity of the icsA promoter. In the second case RnaG causes a premature termination of the icsA transcript (Giangrossi et al., 2010).
Experiments performed with purified VirF indicate that this protein is able to directly stimulate the expression of icsA and that it binds to four 40–60 bp sites, two of which overlap the icsA and RnaG promoters (Tran et al., 2011). This region hosts H-NS sites I and II. The relative position of binding sites for H-NS and VirF provides a physical basis for a possible functional competition between the two proteins. By monitoring the icsA promoter activity in the presence of both H-NS and VirF it has been shown that VirF is able to significantly counteract the H-NS-dependent inhibition at the icsA promoter, thus acting as an anti-H-NS protein. VirF expression increases rapidly when temperature is raised above 32°C (Falconi et al., 1998; Prosseda et al., 1998; Durand et al., 2000; Figure 2). It is possible to speculate that at the host temperature (37°C) the increased amount of VirF may facilitate the interaction with its sites and consequently disrupt the H-NS-DNA complexes by forming a putative H-NS-icsA-VirF intermediate able to promote the switch from a repressed to an active state (Tran et al., 2011). Besides inducing icsA activation, VirF is also able to repress the expression of RnaG (Figure 1) by binding to a specific site overlapping the RnaG promoter. Thus, VirF promotes icsA expression in two ways: by acting on icsA both directly and indirectly, i.e., via repression of RnaG transcription (Tran et al., 2011). Altogether the experimental evidence existing on the role of VirF as major regulator in the cascade of events regulating the virulence of Shigella highlights the antagonism between H-NS and AraC proteins, suggesting that this may be a common evolutionary strategy that pathogens adopt to control virulence genes (Egan, 2002; Dorman, 2004).
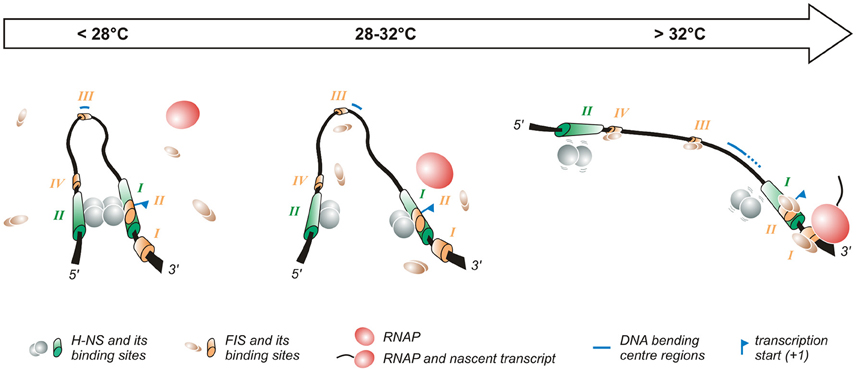
Figure 2. Temperature-dependent regulation of the promoter of the virF gene. The virF promoter contains two H-NS (indicated in green) and four FIS (indicated in orange) binding sites (Falconi et al., 1998, 2001). The arrow on the top of the figure indicates the transition from the outer environment (colder) to the intestinal host milieu (warmer). Left panel: At lower temperature a compact structure exists where H-NS represses virF by interacting with H-NS binding sites I and II which are separated by a DNA region whose sequence-mediated curvature is favored by the lower temperature. Middle panel: The intrinsic bend located between the two H-NS binding sites melts abruptly around 32°C and the bending center slides downstream unmasking a binding site for FIS. Right panel: By increasing the temperature the DNA bend relaxes further, H-NS interactions to binding sites I and II weaken and FIS gains easier access to its binding boxes, thus counteracting H-NS repression. Altogether these events lead to the formation of an active transcription complex.
The switch from the non-invasive to the invasive phenotype in Shigella is an elaborate process and the complexity of the regulatory mechanisms the virF gene undergoes is therefore not surprising. The expression of virF is affected by different environmental signals that act through highly diversified mechanisms. Nucleoid-associated proteins are known for their contribution to the transcriptional control of several genes including virulence genes (Rimsky and Travers, 2011). This is well exemplified in the case of virF (Prosseda et al., 2002) where H-NS (Falconi et al., 1998), FIS (Falconi et al., 2001), and IHF (Porter and Dorman, 1997b) are deeply involved as regulative elements.
The first studies aimed at understanding the temperature-regulated expression of the invasion phenotype of Shigella demonstrate that H-NS, originally designated virR (Maurelli and Sansonetti, 1988; Hromockyj et al., 1992), is responsible for silencing invasive genes at 30°C. Indeed, hns-defective mutants were shown to express the virulence determinants also at non-permissive temperature. Successive reports have revealed that the thermo-dependent expression of virF is lost in a Shigella hns-defective background, providing clear evidence that H-NS is able to affect the regulation of the virF gene (Dagberg and Uhlin, 1992; Colonna et al., 1995) and that this capacity is based on the ability of H-NS to bind the virF promoter with strong specificity (Prosseda et al., 1998). A detailed analysis (Falconi et al., 1998) of how H-NS interacts with the virF gene indicates that, both in vivo and in vitro, this protein is able to recognize and repress virF only below a critical temperature (32°C). This temperature-dependence relies on the interaction of H-NS with two binding sites within the virF promoter, spaced by a DNA linker region. The accessibility of the target DNA to H-NS varies significantly with temperature and H-NS is able to recognize its binding boxes only at lower temperature: with increasing temperature H-NS binding decreases, and at a temperature higher than 30°C the virF promoter becomes insensitive to H-NS repression. The position of the two H-NS binding sites on the virF promoter has been investigated in detail (Falconi et al., 1998). They have been mapped through in vivo and in vitro footprinting experiments and are localized around positions −250 (binding site II) and −1 (binding site I) with respect to the virF transcription start site. Site I overlaps the canonical −35 and −10 regulatory elements, suggesting that this region is involved in the transcriptional repression of the virF gene by H-NS: the interaction of H-NS with DNA may prevent the −35 and −10 boxes to be accessed by RNA polymerase (Figure 2). Binding site II maps far upstream the −35 and −10 sequences. In the absence of this site, H-NS forms a very unstable nucleoprotein complex, which cannot compete with RNA polymerase effectively. The DNA linker region separating the two H-NS binding sites is endowed with sequence-mediated curvature, a feature whose amplitude is inversely dependent on temperature (Prosseda et al., 2004, 2010). This DNA bend progressively relaxes with increasing temperature and is rapidly abolished above 32°C (Figure 2).
The fact that H-NS binding to the virF promoter, bending of the promoter region and repression of virF transcription are all sharply decreased by raising the temperature above a critical threshold suggests that the physical basis for the thermoregulated expression of virF resides in a temperature-dependent structural transition of the virF promoter, with the curved DNA tract within the promoter operating essentially as a thermosensor (Falconi et al., 1998). According to this model, with increasing temperature the transcriptionally inactive DNA architecture prevailing at low temperature would be replaced by a more relaxed geometry which no longer hinders the formation of a functional transcription complex (Falconi et al., 1998). This view has received further support by successive experiments (Prosseda et al., 2004) where the decreased DNA curvature of the virF promoter induced by raising the temperature is mimicked, at constant temperature, by templates (obtained by a targeted in vitro mutagenesis procedure) endowed with decreasing intrinsic DNA curvature. The results of footprinting and in vivo transcription assays using these templates highlight a strict correlation between intrinsic DNA curvature and the capability of H-NS to bind to its target sites and repress virF transcription. Moreover, the reciprocal rotational position of the two H-NS sites appears crucial for the thermoregulation of virF: moving the sites by about a half helix turn further apart produces templates which, despite a comparable overall bending level, are almost insensitive to H-NS repression, likely because the protein can no longer form a tight, transcription-blocking nucleoprotein complex. Temperature does not only affect the amplitude of the DNA bend within the virF promoter but also the position of the bending center, identified by means of circular permutation assays (Prosseda et al., 2004): at 4°C this spot maps at position −137 but with increasing temperature it slides downstream (up to position −55 at 60°C) within the region bounded by the two H-NS sites (Figure 2).
The complexity of the interaction between H-NS and bent DNA is increased further by the participation of FIS (Falconi et al., 2001). In agreement with the well-known growth phase dependent expression of FIS (Rimsky and Travers, 2011), the effect of this protein is higher in early exponential phase (Falconi et al., 2001), suggesting that it contributes to rapidly increase virF expression once bacteria have entered the host environment. FIS exerts, both in vivo and in vitro, a direct positive control on the transcription of virF and is able to bind to the virF promoter. Four FIS binding sites (I to IV), centered around positions +55 (site I), −1 (site II), −130 (site III, almost coinciding with the −137 position of the DNA bending center), and −200 (site IV), have been identified in the virF promoter region (Falconi et al., 2001). The interaction of FIS with site II likely hampers H-NS binding to its boxes. The downstream sliding of the DNA bending center and the concomitant curvature decrease occurring with increasing temperature favor the binding of FIS to the other sites inducing an adjustment to the geometry of the DNA that hinders long range H-NS/H-NS interaction, thus promoting the transcription of virF. Several reports have stressed the validity of this model—envisaging an environmentally induced structural collapse of a promoter's intrinsic bend as a regulative key—also among other pathogenic bacteria, like Y. enterocolitica virF (Rohde et al., 1999), and E. coli hly (Madrid et al., 2002).
Besides FIS also IHF is involved in the positive control of the pINV regulon by stimulating the transcription of the virF and virB genes (Porter and Dorman, 1997b). IHF is composed by two subunits encoded by the ihfA and ihfB genes (Rimsky and Travers, 2011). IHF binds to the virF, virB, and icsA promoters. In the case of virF it recognizes a 127 bp fragment spanning from the promoter-proximal sequence to the start of the ORF, with a putative binding site located between +45 and +57. In ihfA-defective strains entering stationary phase the expression of the Ipa proteins and of the T3SS components is decreased as a consequence of the reduced level of VirF and VirB (Porter and Dorman, 1997b). This growth-phase-dependent effect is consistent with the increased level of IHF during late growth phases (Rimsky and Travers, 2011). However, the observation that in a ihfA mutant virulence gene expression is reduced only two- to three-fold suggests that IHF, rather than constituting an absolute requirement, plays a modulatory role.
The expression of the Shigella invasive genes is repressed at low pH and low osmolarity (Porter and Dorman, 1997a). Also in these cases regulation occurs mainly at the level of virF expression. While it has been demonstrated that H-NS is involved, at least in part, in the repression of virF under low osmotic conditions (Mitobe et al., 2009), the pH-dependent regulation of virF requires the two component regulatory system CpxA/CpxR (Nakayama and Watanabe, 1995, 1998; Figure 1). The Cpx system responds to a broad range of stimuli including, besides pH, also salt, metals, lipids, and misfolded proteins that cause perturbations in the bacterial envelope (Raivio, 2014). In particular, it has been shown that the response regulator CpxR binds directly to a fragment containing the upstream promoter region of virF between positions −103 and −37. Phosphorylation of CpxR enhances its binding capacity and directly activates the transcription of virF (Nakayama and Watanabe, 1998). Due to the relevance of the CpxA/CpxR system in response to several stimuli connected to envelope stress, the integration of virF within the CpxR-controlled network represents a further regulatory layer acting on the fine control of VirF.
Besides being subject to a complex transcriptional control the virF gene is also regulated at the translational level. It has been shown that post-transcriptional modifications of tRNA affect the translation of VirF (Figure 1). The full expression of S. flexneri virulence genes depends on the presence of two modified tRNA nucleosides (queuosine, at position 34, and 2-methylthio-N6-isopentenyladenosine, adjacent to 3′-end of the anticodon). The synthesis of these nucleosides depends on the products of the tgt (encoding TGT, the tRNA-guanine transglycosylase) and miaA (coding for MiaA, a tRNA-N6-isopentyladenosine synthetase) genes, respectively. The intracellular concentration of VirF decreases in tgt mutants and in miaA mutants, inducing a low virulence phenotype and an avirulent phenotype, respectively (Durand et al., 1994, 1997). Overall, tRNA modifications are required for VirF to reach the threshold level necessary to activate the virulence cascade (Durand et al., 2000). Besides being involved in tRNA modification TGT also recognizes (Hurt et al., 2007) the virF mRNA in vitro and this recognition results in a site-specific modification at position 421 responsible for the change from guanine to adenine in the mRNA. As yet the physiological significance of this modification is unclear. A contribution of tRNA modification enzymes in virulence has been reported also in other systems: the GidA and MnmE enzymes, which act together as a complex, are both required for the expression of the full virulence phenotype in Salmonella enterica, Aeromonas hydrophila, and in the plant pathogen Pseudomonas syringae (Shippy and Fadle, 2014). Morevoer, the GidA/MnmE complex has been shown to be relevant for the stress response in Salmonella and MiaA is essential in E. coli for growth at higher temperature (Tsui et al., 1996).
Finally, a connection between bacterial metabolism and virulence gene expression has been found in S. flexneri, observing that the addition of ornithine to minimal medium reduces virF expression while the addition of putrescine, lysine and few other aminated metabolites counteract this inhibition (Durand and Björk, 2003, 2009). Moreover, it has been observed that proteins involved in the glycolytic pathway, most notably the carbon regulator CsrA, are required for the full expression of the Shigella virulence phenotype: indeed, the inactivation of csrA causes a significant reduction of virF and virB expression (Gore and Payne, 2010). As yet the molecular mechanism adopted by CsrA to control these genes is unknown.
The evolution of bacterial pathogens from harmless ancestors mainly depends on the acquisition of virulence gene clusters on plasmids, phages, and pathogenicity islands by HGT. This process is complemented by the progressive adaptation to a specific niche determined by so-called pathoadaptive events such as mutations, rearrangements or deletions of genes unnecessary, or even deleterious, for optimal fitness to the new environment (Ochman and Moran, 2001). These events usually involve the concomitant acquisition or loss of regulatory factors which modify the transcriptional profile of the host to a significant extent. The evolution of Shigella from E. coli is a most studied exemplification of these events (Prosseda et al., 2012).
Global transcriptional analyses of E. coli cells expressing or lacking the virF gene have contributed to understand to what extent the arrival of VirF by the acquisition of the pINV plasmid in Shigella has altered the transcriptional program of the ancestor. These studies have revealed that the activity of VirF is not restricted to the regulation of plasmid-encoded virulence genes as icsA and virB but extends also to chromosomally-located genes (Barbagallo et al., 2011). Genes activated by VirF can be grouped into two categories: those which are functional both in Shigella and E. coli, and those that have been inactivated in Shigella. Among the more VirF-sensitive genes of the first group there are those encoding the heat shock proteins IbpA, GroESL, HtpG, DnaK, and Lon. Given the protective role exerted by these stress proteins, it can be hypothesized that the functional significance of VirF resides in activating genes which contribute to better withstand adverse conditions inside the host.
The existence of VirF-regulated genes silent in Shigella (i.e., the second group) suggests that some of them may encode factors perturbing the invasive process which likely have been silenced during evolution in order to optimize bacterial fitness within the host. Most of these genes encode yet unknown products. In this respect the speG gene, encoding the enzyme spermidine acetyl transferase (SAT), is an exception. In E. coli, SAT prevents the accumulation of spermidine (and the consequent toxic effects) by catalyzing the conversion of this polyamine into its physiologically inert form, acetylspermidine (Barbagallo et al., 2011). The speG gene belongs to the ynfB-speG operon (no function is as yet known for the ynfB product), whose induction by VirF has been observed only at 37°C (Barbagallo et al., 2011) in agreement with the thermodependency of virF expression (Falconi et al., 1998). The loss of speG constitutes a pathoadaptive mutation (Prosseda et al., 2012; Campilongo et al., 2014) since spermidine accumulation induced by the lack of SpeG activity increases Shigella survival inside macrophages and its resistance to oxidative stress conditions (Barbagallo et al., 2011). These data have led to hypothesize that, during the evolutionary transition from E. coli to Shigella, the acquisition of virF on the pINV plasmid might have caused an increased expression of speG, thus lowering the intracellular spermidine content (Di Martino et al., 2013). Evidence supporting this view stems from the observation that in Shigella speG expression is decreased in a VirF-defective background and that restoration of speG expression lowers the bacterial fitness within macrophages (Barbagallo et al., 2011).
Recently VirF has been shown to be involved in the regulation of mdtJI, another operon related to polyamine function in bacteria. In E. coli the mdtJI operon, which encodes an efflux pump belonging to the small multidrug resistance family of transporters, is almost silent because of a strong repression by H-NS (Leuzzi et al., 2015). Despite the high homology between Shigella and E. coli, in Shigella the mdtJI pump is expressed because the H-NS repressive effect is counteracted by VirF and favored by high levels of spermidine (Leuzzi et al., 2015). Intracellular spermidine does not affect the synthesis of VirF (Leuzzi et al., 2015) and the molecular mechanism by which this polyamine contributes towards the activation of mdtJI is still unclear. Genetic studies on the expression of mdtJI in Shigella indicate the presence of a VirF binding site around the promoter consensus boxes which partially overlaps an H-NS site, supporting the occurrence of a competition between the two proteins (Leuzzi et al., 2015), in close analogy with the observations on the virB and icsA promoters (Tobe et al., 1993; Tran et al., 2011). In Shigella MdtJI promotes the secretion of putrescine, the precursor of spermidine, and it has been proposed that VirF- and spermidine-mediated activation of the mdtJI operon represents a safety mechanism allowing spermidine to accumulate within the bacterial cell to a level optimally suited for survival within infected macrophages and, at the same time, avoid toxic side effects on bacterial viability due to spermidine excess (Leuzzi et al., 2015). Altogether, the observations on the ability of Shigella to activate chromosomal genes evidence the extent to which the acquisition of a new regulator by HGT represents a crucial event for reshaping the transcriptional profile of the core genome, facilitating bacterial adaptation to specific niches within infected hosts.
Each year Shigella is responsible for 125 million cases of illness, mainly in low income countries (The et al., 2016). Despite the enormous clinical relevance of these infections and the emergence of multiresistance strains, no vaccine has been as yet released for public use (Anderson et al., 2016). Several recent studies have focused on the development of novel treatment strategies targeting virulence instead of bacterial viability, since this is regarded as a highly effective approach to combat bacterial infections while minimizing the emergence of antibiotic resistances (Rasko and Sperandio, 2010). The expression of virulence factors is not required for cell viability and therefore bacterial pathogens should be subject to less selective pressure to develop resistance to inhibitors of virulence determinants. AraC proteins are considered very interesting candidate targets in anti-virulence strategies due to their critical role in controlling virulence in pathogenic bacteria (Rasko and Sperandio, 2010). Specific inhibitors can affect AraC-mediated processes at different stages, such as self-association, DNA binding, and recruitment of RNA polymerase.
As for shigellosis it is therefore not surprising that VirF has been considered a very good antivirulence target, since its silencing prevents host cell invasion and intercellular spreading without affecting bacterial viability (Sakai et al., 1988; Falconi et al., 1998). So far, two approaches have been adopted to identify potential VirF inhibitors: a high-throughput screening for small molecules and a targeted search among already characterized AraC inhibitors. In the first case (Hurt et al., 2010), the expression of virB in response to VirF has been analyzed using a pINV-cured S. flexneri strain harboring a recombinant plasmid containing the virF gene and a virB-lacZ fusion. The screening, performed initially on 42,000 compounds from several small-molecule libraries and then extended to an additional set of 100,000 compounds, has led to the identification of about 600 molecules meeting the selection criteria for VirF inhibition (Hurt et al., 2010; Emanuele et al., 2014). After selecting for candidates with favorable medicinal chemistry, low toxicity and a dose-dependent activity, five compounds able to inhibit VirF-driven transcriptional activation with very low IC50 values were considered for further studies. These compounds were characterized by the presence of aromatic or heterocyclic rings that could interact with DNA or with aromatic acids that typically associate with DNA (Hurt et al., 2010; Emanuele et al., 2014). In vivo assays on cell cultures have shown that only one polycyclic compound, named 19615 (methyl-[2-(2-phenyl-4a,9b-dihydro-benzo[4,5]furo[3,2-d]pyrimidin-4-γ1oxy9-ethyl]-amine), was able to severely affect intercellular spreading and induce a significant inhibition of cell invasion (Emanuele and Garcia, 2015). In particular this molecule has been demonstrated to be able to directly interact with a MalE—VirF fusion protein and inhibit its binding to the virB promoter. Further studies to structurally optimize the selected compound are required to fully clarify its effectiveness in an antivirulence therapy.
The second approach (Koppolu et al., 2013) adopted so far to identify possible VirF inhibitors has been centered on a small molecule, SE-1, a quinoline derivative [1-butyl-4-nitromethyl1-3-(quinolin-γl)-4Hquinoline] previously identified as an inhibitor of two AraC activators, RhaS, and RhaR (Skredenske et al., 2013). Since SE-1 interacts with the conserved DNA-binding domains of the AraC proteins (Skredenske et al., 2013), it has been considered able to potentially inhibit also other AraC activators. This turned out to be true: the data revealed that SE-1 induced a significant reduction in the expression of all VirF-controlled genes, consequently inhibiting the invasion of epithelial cells. SE-1 binds directly to a MalE—VirF fusion protein, a feature likely responsible for inhibiting VirF to interact with DNA and activate transcription. On account that SE-1 does not affect the growth of Shigella and does not have detectable toxicity in human cell cultures, it has been considered as another good candidate as novel antibacterial agent.
Altogether the available data stress how Shigella, in its long route to becoming a successful pathogen, has evolved an elaborate regulatory system to ensure the coordinated activation of virulence determinants or prevent their wasteful expression, depending on environmental signals. The complexity of the circuitry regulating virulence in Shigella highlights the relevance of the firing of a major regulator, the virF gene, to the successful development of the invasive program. Through studies analyzing the interplay, either synergistic or antagonistic, among nucleoid-associated proteins, sRNA, global, and specific regulators, and intrinsic features of the DNA, the complex nature of the regulation of VirF and of the genes under its control is emerging with intriguing detail. However, while the master regulator has been identified almost three decades ago, several open questions still exist, such as the capacity of VirF to form dimers or more complex aggregates and the molecular mechanisms underlying its DNA binding specificity and its interactions with RNA polymerase. Understanding how the gene regulatory circuitry has evolved in bacterial pathogens represents a challenge. From the evolutionary standpoint this relates to the need to understand how genes acquired by HGT have integrated into existing regulatory networks and how newly acquired regulators have shaped the genome of the new bacterial host. In the long run a better understanding of the structure of the VirF protein can be expected to positively impact on the development of new therapeutic approaches based on the use of specific inhibitory compounds.
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication. In particular MLDM, BC and GP conceived the review and wrote the manuscript. MLDM and GM conceived the figures. MF and GM critically edited the final version of the review.
This research was supported by grants from Ministero della Ricerca e dell'Istruzione (PRIN 2012/WWJSX8K), Sapienza Università di Roma, Consiglio Nazionale delle Ricerche, and Institut Pasteur (PTR-24-16).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
FIS, factor for inversion stimulation; HGT, horizontal gene transfer; H-NS, heat stable nucleoid-structuring protein; HTH, helix-turn-helix; IC50, half maximal inhibitory concentration; IHF, integration host factor; Ipa, invasion plasmid antigen; IS, insertion sequence; MiaA, tRNA-N6-isopentyladenosine synthetase; PAI, pathogenicity island; PCr, Congo red phenotype; sRNA, small RNA; SAT, spermidine acetyl transferase; T3SS, type III secretion system; TCS, two component system; TGT, tRNA-guanine transglycosylase.
Adler, B., Sasakawa, C., Tobe, T., Makino, S., Komatsu, K., and Yoshikawa, M. (1989). A dual transcriptional activation system for the 230 kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol. Microbiol. 3, 627–635. doi: 10.1111/j.1365-2958.1989.tb00210.x
Anderson, M., Sansonetti, P. J., and Marteyn, B. S. (2016). Shigella diversity and changing landscape: insights for the twenty-first century. Front. Cell. Infect. Microbiol. 6:45. doi: 10.3389/fcimb.2016.00045
Ashida, H., Mimuro, H., and Sasakawa, C. (2015). Shigella manipulates host immune responses by delivering effector proteins with specific roles. Front. Immunol. 6:219. doi: 10.3389/fimmu.2015.00219
Ashida, H., and Sasakawa, C. (2015). Shigella IpaH family effectors as a versatile model for studying pathogenic bacteria. Front. Cell. Infect. Microbiol. 5:100. doi: 10.3389/fcimb.2015.00100
Barbagallo, M., Di Martino, M. L., Marcocci, L., Pietrangeli, P., De Carolis, E., Casalino, M., et al. (2011). A new piece of the Shigella Pathogenicity puzzle: spermidine accumulation by silencing of the speG gene. PLoS ONE 6:e27226. doi: 10.1371/journal.pone.0027226
Beloin, C., McKenna, S., and Dorman, C. J. (2002). Molecular dissection of VirB, a key regulator of the virulence cascade of Shigella flexneri. J. Biol. Chem. 277, 15333–15344. doi: 10.1074/jbc.M111429200
Bernardini, M. L., Mounier, J., D'Hauteville, H., Coquis-Rondon, M., and Sansonetti, P. J. (1989). Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl. Acad. Sci. U.S.A. 86, 3867–3871. doi: 10.1073/pnas.86.10.3867
Bourgerie, S. J., Michán, C. M., Thomas, M. S., Busby, S. J., and Hyde, E. I. (1997). DNA binding and DNA bending by the MelR transcription activator protein from Escherichia coli. Nucleic Acids Res. 25, 1685–1693. doi: 10.1093/nar/25.9.1685
Buchrieser, C., Glaser, P., Rusniok, C., Nedjari, H., D'Hauteville, H., Kunst, F., et al. (2000). The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38, 760–771. doi: 10.1046/j.1365-2958.2000.02179.x
Campilongo, R., Di Martino, M. L., Marcocci, L., Pietrangeli, P., Leuzzi, A., Grossi, M., et al. (2014). Molecular and functional profiling of the polyamine content in enteroinvasive E. coli: looking into the gap between commensal E. coli and harmful Shigella. PLoS ONE 9:e106589. doi: 10.1371/journal.pone.0106589
Cases, I., and de Lorenzo, V. (2005). Promoters in the environment: transcriptional regulation in its natural context. Nat. Rev. Microbiol. 3, 105–118. doi: 10.1038/nrmicro1084
Colonna, B., Casalino, M., Fradiani, P. A., Zagaglia, C., Naitza, S., Leoni, L., et al. (1995). H-NS regulation of virulence gene expression in enteroinvasive Escherichia coli harboring the virulence plasmid integrated into the host chromosome. J. Bacteriol. 177, 4703–4712.
Dagberg, B., and Uhlin, B. E. (1992). Regulation of virulence-associated plasmid genes in enteroinvasive Escherichia coli. J. Bacteriol. 174, 7606–7612.
Di Martino, M. L., Campilongo, R., Casalino, M., Micheli, G., Colonna, B., and Prosseda, G. (2013). Polyamines: emerging players in bacteria-host interactions. Int. J. Med. Microbiol. 303, 484–491. doi: 10.1016/j.ijmm.2013.06.008
Dorman, C. J. (2004). H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2, 391–400. doi: 10.1038/nrmicro883
Dorman, C. J. (2007). H-NS, the genome sentinel. Nat. Rev. Microbiol. 5, 157–161. doi: 10.1038/nrmicro1598
Dorman, C. J., and Porter, M. E. (1998). The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol. Microbiol. 29, 677–684.
Durand, J. M. B., and Björk, G. R. (2003). Putrescine or a combination of methionine and arginine restores virulence gene expression in a tRNA modification-deficient mutant of Shigella flexneri: a possible role in adaptation of virulence. Mol. Microbiol. 47, 519–527. doi: 10.1046/j.1365-2958.2003.03314.x
Durand, J. M. B., and Björk, G. R. (2009). Metabolic control through ornithine and uracil of epithelial cell invasion by Shigella flexneri. Microbiology 155(Pt 8), 2498–2508. doi: 10.1099/mic.0.028191-0
Durand, J. M., Björk, G. R., Kuwae, A., Yoshikawa, M., and Sasakawa, C. (1997). The modified nucleoside 2-methylthio-N6-isopentenyladenosine in tRNA of Shigella flexneri is required for expression of virulence genes. J. Bacteriol. 179, 5777–5782.
Durand, J. M., Dagberg, B., Uhlin, B. E., and Björk, G. R. (2000). Transfer RNA modification, temperature and DNA superhelicity have a common target in the regulatory network of the virulence of Shigella flexneri: the expression of the virF gene. Mol. Microbiol. 35, 924–935. doi: 10.1046/j.1365-2958.2000.01767.x
Durand, J. M., Okada, N., Tobe, T., Watarai, M., Fukuda, I., Suzuki, T., et al. (1994). vacC, a virulence-associated chromosomal locus of Shigella flexneri, is homologous to tgt, a gene encoding tRNA-guanine transglycosylase (Tgt) of Escherichia coli K-12. J. Bacteriol. 176, 4627–4634.
Egan, S. M. (2002). Growing repertoire of AraC/XylS activators. J. Bacteriol. 184, 5529–5532. doi: 10.1128/JB.184.20.5529-5532.2002
Emanuele, A. A., Adams, N. E., Chen, Y.-C., Maurelli, A. T., and Garcia, G. A. (2014). Potential novel antibiotics from HTS targeting the virulence-regulating transcription factor, VirF, from Shigella flexneri. J. Antibiot. 67, 379–386. doi: 10.1038/ja.2014.10
Emanuele, A. A., and Garcia, G. A. (2015). Mechanism of action and initial, in vitro sar of an inhibitor of the Shigella flexneri virulence regulator VirF. PLoS ONE 10:e0137410. doi: 10.1371/journal.pone.0137410
Falconi, M., Colonna, B., Prosseda, G., Micheli, G., and Gualerzi, C. O. (1998). Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 17, 7033–7043. doi: 10.1093/emboj/17.23.7033
Falconi, M., Prosseda, G., Giangrossi, M., Beghetto, E., and Colonna, B. (2001). Involvement of FIS in the H-NS-mediated regulation of virF gene of Shigella and enteroinvasive Escherichia coli. Mol. Microbiol. 42, 439–452. doi: 10.1046/j.1365-2958.2001.02646.x
Gallegos, M. T., Schleif, R., Bairoch, A., Hofmann, K., and Ramos, J. L. (1997). AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61, 393–410.
Giangrossi, M., Prosseda, G., Tran, C. N., Brandi, A., Colonna, B., and Falconi, M. (2010). A novel antisense RNA regulates at transcriptional level the virulence gene icsA of Shigella flexneri. Nucleic Acids Res. 38, 3362–3375. doi: 10.1093/nar/gkq025
Gore, A. L., and Payne, S. M. (2010). CsrA and Cra influence Shigella flexneri pathogenesis. Infect. Immun. 78, 4674–4682. doi: 10.1128/IAI.00589-10
Hromockyj, A. E., Tucker, S. C., and Maurelli, A. T. (1992). Temperature regulation of Shigella virulence: identification of the repressor gene virR, an analogue of hns, and partial complementation by tyrosyl transfer RNA (tRNA1(Tyr)). Mol. Microbiol. 6, 2113–2124. doi: 10.1111/j.1365-2958.1992.tb01385.x
Hurt, J. K., McQuade, T. J., Emanuele, A., Larsen, M. J., and Garcia, G. A. (2010). High-throughput screening of the virulence regulator VirF: a novel antibacterial target for shigellosis. J. Biomol. Screen. 15, 379–387. doi: 10.1177/1087057110362101
Hurt, J. K., Olgen, S., and Garcia, G. A. (2007). Site-specific modification of Shigella flexneri virF mRNA by tRNA-guanine transglycosylase in vitro. Nucleic Acids Res. 35, 4905–4913. doi: 10.1093/nar/gkm473
Kane, C. D., Schuch, R., Day, W. A., and Maurelli, A. T. (2002). MxiE regulates intracellular expression of factors secreted by the Shigella flexneri 2a type III secretion system. J. Bacteriol. 184, 4409–4419. doi: 10.1128/JB.184.16.4409-4419.2002
Kato, J., Ito, K., Nakamura, A., and Watanabe, H. (1989). Cloning of regions required for contact hemolysis and entry into LLC-MK2 cells from Shigella sonnei form I plasmid: virF is a positive regulator gene for these phenotypes. Infect. Immun. 57, 1391–1398.
Kim, D. W., Lenzen, G., Page, A.-L., Legrain, P., Sansonetti, P. J., and Parsot, C. (2005). The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. U.S.A. 102, 14046–14051. doi: 10.1073/pnas.0504466102
Koppolu, V., Osaka, I., Skredenske, J. M., Kettle, B., Hefty, P. S., Li, J., et al. (2013). Small-molecule inhibitor of the Shigella flexneri master virulence regulator VirF. Infect. Immun. 81, 4220–4231. doi: 10.1128/IAI.00919-13
Kwon, H. J., Bennik, M. H., Demple, B., and Ellenberger, T. (2000). Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Biol. 7, 424–430. doi: 10.1038/75213
Lett, M. C., Sasakawa, C., Okada, N., Sakai, T., Makino, S., Yamada, M., et al. (1989). virG, a plasmid-coded virulence gene of Shigella flexneri: identification of the VirG protein and determination of the complete coding sequence. J. Bacteriol. 171, 353–359.
Leuzzi, A., Di Martino, M. L., Campilongo, R., Falconi, M., Barbagallo, M., Marcocci, L., et al. (2015). Multifactor regulation of the MdtJI polyamine transporter in Shigella. PLoS ONE 10:e0136744. doi: 10.1371/journal.pone.0136744
Madrid, C., Nieto, J. M., Paytubi, S., Falconi, M., Gualerzi, C. O., and Juárez, A. (2002). Temperature- and H-NS-dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J. Bacteriol. 184, 5058–5066. doi: 10.1128/JB.184.18.5058-5066.2002
Maurelli, A. T., Baudry, B., d'Hauteville, H., Hale, T. L., and Sansonetti, P. J. (1985). Cloning of plasmid DNA sequences involved in invasion of HeLa cells by Shigella flexneri. Infect. Immun. 49, 164–171.
Maurelli, A. T., and Sansonetti, P. J. (1988). Identification of a chromosomal gene controlling temperature-regulated expression of Shigella virulence. Proc. Natl. Acad. Sci. U.S.A. 85, 2820–2824. doi: 10.1073/pnas.85.8.2820
Mavris, M., Page, A.-L., Tournebize, R., Demers, B., Sansonetti, P., and Parsot, C. (2002). Regulation of transcription by the activity of the Shigella flexneri type III secretion apparatus. Mol. Microbiol. 43, 1543–1553. doi: 10.1046/j.1365-2958.2002.02836.x
McAdams, H. H., Srinivasan, B., and Arkin, A. P. (2004). The evolution of genetic regulatory systems in bacteria. Nat. Rev. Genet. 5, 169–178. doi: 10.1038/nrg1292
McKenna, S., Beloin, C., and Dorman, C. J. (2003). In vitro DNA-binding properties of VirB, the Shigella flexneri virulence regulatory protein. FEBS Lett. 545, 183–187. doi: 10.1016/S0014-5793(03)00524-6
Mitobe, J., Morita-Ishihara, T., Ishihama, A., and Watanabe, H. (2009). Involvement of RNA-binding protein Hfq in the osmotic-response regulation of invE gene expression in Shigella sonnei. BMC Microbiol. 9:110. doi: 10.1186/1471-2180-9-110
Nakayama, S., and Watanabe, H. (1995). Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J. Bacteriol. 177, 5062–5069.
Nakayama, S., and Watanabe, H. (1998). Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene. J. Bacteriol. 180, 3522–3528.
Njamkepo, E., Fawal, N., Tran-Dien, A., Hawkey, J., Strockbine, N., Jenkins, C., et al. (2016). Global phylogeography and evolutionary history of Shigella dysenteriae type 1. Nat. Microbiol. 1, 16027. doi: 10.1038/nmicrobiol.2016.27
Ochman, H., and Moran, N. A. (2001). Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292, 1096–1099. doi: 10.1126/science.1058543
Parsot, C. (2005). Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors. FEMS Microbiol. Lett. 252, 11–18. doi: 10.1016/j.femsle.2005.08.046
Porter, M. E., and Dorman, C. J. (1997a). Differential regulation of the plasmid-encoded genes in the Shigella flexneri virulence regulon. Mol. Gen. Genet. 256, 93–103.
Porter, M. E., and Dorman, C. J. (1997b). Positive regulation of Shigella flexneri virulence genes by integration host factor. J. Bacteriol. 179, 6537–6550.
Porter, M. E., and Dorman, C. J. (2002). In vivo DNA-binding and oligomerization properties of the Shigella flexneri AraC-like transcriptional regulator VirF as identified by random and site-specific mutagenesis. J. Bacteriol. 184, 531–539. doi: 10.1128/JB.184.2.531-539.2002
Porter, M. E., Mitchell, P., Roe, A. J., Free, A., Smith, D. G. E., and Gally, D. L. (2004). Direct and indirect transcriptional activation of virulence genes by an AraC-like protein, PerA from enteropathogenic Escherichia coli. Mol. Microbiol. 54, 1117–1133. doi: 10.1111/j.1365-2958.2004.04333.x
Porter, M. E., Smith, S. G., and Dorman, C. J. (1998). Two highly related regulatory proteins, Shigella flexneri VirF and enterotoxigenic Escherichia coli Rns, have common and distinct regulatory properties. FEMS Microbiol. Lett. 162, 303–309.
Prosseda, G., Di Martino, M. L., Campilongo, R., Fioravanti, R., Micheli, G., Casalino, M., et al. (2012). Shedding of genes that interfere with the pathogenic lifestyle: the Shigella model. Res. Microbiol. 163, 399–406. doi: 10.1016/j.resmic.2012.07.004
Prosseda, G., Falconi, M., Giangrossi, M., Gualerzi, C. O., Micheli, G., and Colonna, B. (2004). The virF promoter in Shigella: more than just a curved DNA stretch. Mol. Microbiol. 51, 523–537. doi: 10.1046/j.1365-2958.2003.03848.x
Prosseda, G., Falconi, M., Nicoletti, M., Casalino, M., Micheli, G., and Colonna, B. (2002). Histone-like proteins and the Shigella invasivity regulon. Res. Microbiol. 153, 461–468. doi: 10.1016/S0923-2508(02)01346-3
Prosseda, G., Fradiani, P. A., Di Lorenzo, M., Falconi, M., Micheli, G., Casalino, M., et al. (1998). A role for H-NS in the regulation of the virF gene of Shigella and enteroinvasive Escherichia coli. Res. Microbiol. 149, 15–25. doi: 10.1016/S0923-2508(97)83619-4
Prosseda, G., Latella, M. C., Casalino, M., Nicoletti, M., Michienzi, S., and Colonna, B. (2006). Plasticity of the P junc promoter of ISEc11, a new insertion sequence of the IS1111 family. J. Bacteriol. 188, 4681–4689. doi: 10.1128/JB.00332-06
Prosseda, G., Mazzola, A., Di Martino, M. L., Tielker, D., Micheli, G., and Colonna, B. (2010). A temperature-induced narrow DNA curvature range sustains the maximum activity of a bacterial promoter in vitro. Biochemistry 49, 2778–2785. doi: 10.1021/bi902003g
Pupo, G. M., Lan, R., and Reeves, P. R. (2000). Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. U.S.A. 97, 10567–10572. doi: 10.1073/pnas.180094797
Raivio, T. L. (2014). Everything old is new again: an update on current research on the Cpx envelope stress response. Biochim. Biophys. Acta 1843, 1529–1541. doi: 10.1016/j.bbamcr.2013.10.018
Rasko, D. A., and Sperandio, V. (2010). Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 9, 117–128. doi: 10.1038/nrd3013
Rhee, S., Martin, R. G., Rosner, J. L., and Davies, D. R. (1998). A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. U.S.A. 95, 10413–10418.
Rimsky, S., and Travers, A. (2011). Pervasive regulation of nucleoid structure and function by nucleoid-associated proteins. Curr. Opin. Microbiol. 14, 136–141. doi: 10.1016/j.mib.2011.01.003
Rohde, J. R., Luan, X. S., Rohde, H., Fox, J. M., and Minnich, S. A. (1999). The Yersinia enterocolitica pYV virulence plasmid contains multiple intrinsic DNA bends which melt at 37 degrees C. J. Bacteriol. 181, 4198–4204.
Sakai, T., Sasakawa, C., Makino, S., Kamata, K., and Yoshikawa, M. (1986a). Molecular cloning of a genetic determinant for Congo red binding ability which is essential for the virulence of Shigella flexneri. Infect. Immun. 51, 476–482.
Sakai, T., Sasakawa, C., Makino, S., and Yoshikawa, M. (1986b). DNA sequence and product analysis of the virF locus responsible for congo red binding and cell invasion in Shigella flexneri 2a. Infect. Immun. 54, 395–402.
Sakai, T., Sasakawa, C., and Yoshikawa, M. (1988). Expression of four virulence antigens of Shigella flexneri is positively regulated at the transcriptional level by the 30 kiloDalton virF protein. Mol. Microbiol. 2, 589–597.
Sansonetti, P. J., Kopecko, D. J., and Formal, S. B. (1982). Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect. Immun. 35, 852–860.
Sasakawa, C., Kamata, K., Sakai, T., Makino, S., Yamada, M., Okada, N., et al. (1988). Virulence-associated genetic regions comprising 31 kilobases of the 230-kilobase plasmid in Shigella flexneri 2a. J. Bacteriol. 170, 2480–2484.
Schleif, R. (2010). AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol. Rev. 34, 779–796. doi: 10.1111/j.1574-6976.2010.00226.x
Schroeder, G. N., and Hilbi, H. (2008). Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin. Microbiol. Rev. 21, 134–156. doi: 10.1128/CMR.00032-07
Scribano, D., Petrucca, A., Pompili, M., Ambrosi, C., Bruni, E., Zagaglia, C., et al. (2014). Polar localization of PhoN2, a periplasmic virulence-associated factor of Shigella flexneri, is required for proper IcsA exposition at the old bacterial pole. PLoS ONE 9:e90230. doi: 10.1371/journal.pone.0090230
Shippy, D. C., and Fadle, A. A. (2014). tRNA modification enzymes GidA and MnmE: potential role in virulence of bacterial pathogens. Int. J. Mol. Sci. 15, 18267–18280. doi: 10.3390/ijms151018267
Skredenske, J. M., Koppolu, V., Kolin, A., Deng, J., Kettle, B., Taylor, B., et al. (2013). Identification of a small-molecule inhibitor of bacterial AraC family activators. J. Biomol. Screen. 18, 588–598. doi: 10.1177/1087057112474690
The, H. C., Thanh, D. P., Holt, K. E., Thomson, N. R., and Baker, S. (2016). The genomic signatures of Shigella evolution, adaptation and geographical spread. Nat. Rev. Microbiol. 14, 235-250. doi: 10.1038/nrmicro.2016.10
Tobe, T., Nagai, S., Okada, N., Adler, B., Yoshikawa, M., and Sasakawa, C. (1991). Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol. Microbiol. 5, 887–893.
Tobe, T., Yoshikawa, M., Mizuno, T., and Sasakawa, C. (1993). Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by virF and repression by H-NS. J. Bacteriol. 175, 6142–6149.
Tran, C. N., Giangrossi, M., Prosseda, G., Brandi, A., Di Martino, M. L., Colonna, B., et al. (2011). A multifactor regulatory circuit involving H-NS, VirF and an antisense RNA modulates transcription of the virulence gene icsA of Shigella flexneri. Nucleic Acids Res. 39, 8122–8134. doi: 10.1093/nar/gkr521
Tsui, H. C., Feng, G., and Winkler, M. E. (1996). Transcription of the mutL repair, miaA tRNA modification, hfq pleiotropic regulator, and hflA region protease genes of Escherichia coli K-12 from clustered Esigma32-specific promoters during heat shock. J. Bacteriol. 178, 5719–5731.
Venkatesan, M. M., Goldberg, M. B., Rose, D. J., Grotbeck, E. J., Burland, V., and Blattner, F. R. (2001). Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect. Immun. 69, 3271–3285. doi: 10.1128/IAI.69.5.3271-3285.2001
Keywords: VirF, Shigella, pathogenic E. coli, AraC proteins, transcriptional regulators, DNA binding proteins, bacterial virulence, antivirulence therapy
Citation: Di Martino ML, Falconi M, Micheli G, Colonna B and Prosseda G (2016) The Multifaceted Activity of the VirF Regulatory Protein in the Shigella Lifestyle. Front. Mol. Biosci. 3:61. doi: 10.3389/fmolb.2016.00061
Received: 28 July 2016; Accepted: 15 September 2016;
Published: 29 September 2016.
Edited by:
Tatiana Venkova, University of Texas Medical Branch-Galveston, USAReviewed by:
Alessandra Polissi, University of Milan, ItalyCopyright © 2016 Di Martino, Falconi, Micheli, Colonna and Prosseda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bianca Colonna, YmlhbmNhLmNvbG9ubmFAdW5pcm9tYTEuaXQ=
Gianni Prosseda, Z2lhbm5pLnByb3NzZWRhQHVuaXJvbWExLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.