
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 24 May 2016
Sec. Molecular Recognition
Volume 3 - 2016 | https://doi.org/10.3389/fmolb.2016.00019
This article is part of the Research Topic Modulating prokaryotic lifestyle by DNA-binding proteins View all 25 articles
CspA, the most characterized member of the csp gene family of Escherichia coli, is highly expressed not only in response to cold stress, but also during the early phase of growth at 37°C. Here, we investigate at molecular level the antagonistic role played by the nucleoid proteins FIS and H-NS in the regulation of cspA expression under non-stress conditions. By means of both probing experiments and immunological detection, we demonstrate in vitro the existence of binding sites for these proteins on the cspA regulatory region, in which FIS and H-NS bind simultaneously to form composite DNA-protein complexes. While the in vitro promoter activity of cspA is stimulated by FIS and repressed by H-NS, a compensatory effect is observed when both proteins are added in the transcription assay. Consistently with these findings, inactivation of fis and hns genes reversely affect the in vivo amount of cspA mRNA. In addition, by means of strains expressing a high level of the alarmone guanosine tetraphosphate ((p)ppGpp) and in vitro transcription assays, we show that the cspA promoter is sensitive to (p)ppGpp inhibition. The (p)ppGpp-mediated expression of fis and hns genes is also analyzed, thus clarifying some aspects of the regulatory loop governing cspA transcription.
The cspA gene of Escherichia coli encodes a nucleic acid-binding protein of 70 amino acid residues (CspA) that interacts preferentially with single stranded RNA and DNA (Jiang et al., 1997; Bae et al., 2000). CspA belongs to the csp gene family, a group which includes in E. coli a total of nine paralogs, called from cspA to cspI. CspA is known as the “major cold-shock protein” (Goldstein et al., 1990; Jones and Inouye, 1994) by virtue of the original observation that cold-shock induces its synthesis ex novo. However, CspA is also synthesized at 37°C, particularly during the early phase of growth (Brandi et al., 1999; Brandi and Pon, 2012). In addition to cspA, also cspB, cspE, cspG, cspI are cold shock inducible, unlike cspD, cspC, cspF and cspH which are expressed only at 37°C. In particular, cspD is expressed exclusively during stationary phase or nutritional stress, cspC is constitutively synthesized at 37°C, and cspF and cspH, the most distant related genes, remain to be characterized (Jones and Inouye, 1994; Yamanaka et al., 1998; Ermolenko and Makhatadze, 2002).
During cold-shock, CspA was found to affect both transcription (La Teana et al., 1991; Jones et al., 1992b) and translation (Brandi et al., 1996; Giuliodori et al., 2004) of other genes and it was also suggested to function as an RNA chaperone (Jiang et al., 1997; Bae et al., 2000). On the other hand, little is known about the role played by CspA at 37°C. The expression of cspA displays a multilevel regulation (transcription, mRNA stability and translation) modulated by multiple factors that may differently contribute to ensure a rapid and precise response to nutritional or environmental changes (Gualerzi et al., 2011). Interestingly, the regulation of cspA expression is different under stress and non-stress conditions. In fact, while the elevated production of CspA following a temperature down-shift is due mainly to post-transcriptional events (i.e., increased stability and preferential translation of cspA mRNA) rather than to the transcriptional stimulation of cspA promoter (Brandi et al., 1996; Goldenberg et al., 1996; Giuliodori et al., 2010), the synthesis of CspA at 37°C is prevalently regulated at transcriptional level (Brandi et al., 1999; Brandi and Pon, 2012).
Bacteria contain a heterogeneous group of polypeptides, collectively known as nucleoid-associated proteins (NAPs), that are cataloged as a family on the basis of functional similarities. These proteins bind to nucleic acids, are basic and have low molecular mass (Azam and Ishihama, 1999). In addition to their common architectural role in the organization of bacterial chromosome, they are able to modulate transcription initiation and to control DNA replication, segregation and repair (Browning et al., 2010; Dillon and Dorman, 2010; Rimsky and Travers, 2011; Wang et al., 2011). H-NS, one of the most abundant NAPs, preferentially binds to tracts of intrinsically curved DNA (A/T-rich sequences) and/or actively induces bending (Yamada et al., 1990; Spurio et al., 1997; Gordon et al., 2011). Thus, by interacting with curved DNA, often found in upstream promoter regions, this nucleoid protein, referred to as a “universal repressor,” silences transcription of its target genes (Atlung and Ingmer, 1997; Hommais et al., 2001; Dorman, 2004, 2007; Bouffartigues et al., 2007; Lang et al., 2007). FIS (Factor for Inversion Stimulation) is another NAP that binds to DNA and modulates the topology of DNA in a growth-phase dependent manner (Schneider et al., 1997; Muskhelishvili and Travers, 2003). Unlike H-NS, FIS is a positive regulator activating transcription of genes and operons associated with primary metabolism as stable RNAs (Ross et al., 1990; Gonzalez-Gil et al., 1996). Thus, H-NS and FIS, through direct and indirect effects, control the expression of a large number of genes and are viewed as global regulators of transcription in response to growth phase and environmental changes (reviewed in Kahramanoglou et al., 2011).
Since cspA belongs to the set of genes controlled by FIS and H-NS under non-stress conditions (Brandi et al., 1999), the prime aim of this study was to elucidate the molecular aspects of cspA regulation by these two NAPs expanding and deepening our previous knowledge. Our results demonstrate the existence of a functional interplay between FIS and H-NS, which are able to bind separately or simultaneously the cspA promoter region. The composition of the DNA-protein complexes thus formed has a different impact on the transcription of cspA, that is inhibited by H-NS, stimulated by FIS, and unaltered when both factors are present, in fully agreement with the in vivo data (Brandi et al., 1999). Finally, seeking further factors which could participate to this regulatory circuit, we found that cspA promoter is sensitive to (p)ppGpp, the mediator molecule of stringent response.
E. coli strains used in this study were: MRE600 (F-, rna) (Cammack and Wade, 1965); DH5α (Sambrook and Russell, 2001); WM2482 (correspond to MG1655 reference strain K-12, F− λ− ilvG− rfb-50 rph-1), WM2648 (MG1655 hns::hyg), WM2649 (MG1655 fis::kan) and WM 2650 (hns/fis double mutant of MG1655) a kind gift of Walter Messer's laboratory (Berlin) (Afflerbach et al., 1998); E. coli KT793 carrying IPTG-inducible relA in pKT31 (Tedin et al., 1995). Cells were grown at 37°C in Luria-Bertani broth or in M9 minimal medium supplemented with 0.4% glucose (Sambrook and Russell, 2001) or in “Phosphates-free” medium (100 mM Tris-HCl, pH 7.7, 0.5% glucose, 0.5% peptone, 10 mM NH4Cl, 0.7 mM NaNO3, 1mM Na2SO4, 0.5 mM MgSO4·7H2O, 0.05 mM MnCl2·4H2O, 0.02 mM FeSO4·7H2O) where indicated.
The plasmid pTZ310 was constructed by cloning the HpaII-HpaII fragment, containing the cspA promoter region (from pos. −145 to pos. +165), into the AccI site of the pTZ19R polylinker. H-NS was purified as described in Falconi et al. (1988); FIS was a kind gift from the laboratory of Regine Kahmann. DNA isolation, agarose gel electrophoresis, polymerase chain reaction and other DNA manipulations were performed according to standard procedures (Sambrook and Russell, 2001). Radioactivity associated to DNA or RNA was detected and quantified by Molecular Imager (Bio-Rad, model FX).
Total RNA was purified by hot phenol extraction from cells harvested at the indicated times and levels of individual mRNAs were detected by Northern blots probed with specific 5′-end-labeled oligonucleotides (Brandi et al., 1999). The hybridization was performed in the range of temperature 37–48°C, depending on the oligonucleotide used. The oligonucleotides used as specific probes are: 5′-CTTTCGATGGTGAAGGACACT-3′ for cspA; 5′-GCGCACGAAGAGTACGG-3′ for hns and 5′-CAGGGGTTTTTGGGTTACCT-3′ for fis.
The 310 bp DNA fragment, excised from pTZ310 by BamHI/HindIII digestion, was end-labeled with α-[32P]-dATP by fill-in reaction using the DNA polymerase Klenow fragment. About 5–10 ng of the radioactive DNA fragment were incubated with the indicated amounts of purified FIS and H-NS at 25°C in a reaction mixture (15 μl) containing 10 mM Tris HCl (pH 8), 10 mM MgCl2, 100 mM NaCl, 10 mM KCl, 1 mM spermidine, 0.5 mM dithiothreitol, 5% glycerol, 0.08 mg ml −1 BSA, and 50 ng Poly dI-dC as competitor DNA. After 15 min of incubation, samples were subjected to electrophoresis on 6% polyacrylamide gel in TBE buffer (Sambrook and Russell, 2001).
The combined EMSA-Western blot analysis was carried out essentially as described above except that each reaction mix contained 120 ng of a cold DNA fragment corresponding to cspA promoter. This fragment (340 bp) was obtained by PCR amplification using the primer pairs 5′-CAACCCGGCATTAAGTAAGC-3′ and 5′-CCATTTTACGATACCAGTCA-3′ on a 1200 bp DNA fragment cloned in pTZ19R (Brandi et al., 1996). Samples were loaded in duplicate on 6% polyacrylamide gel which was subsequently electro-transferred (35 min at 2.5 mA/cmq) into a nitrocellulose membrane. The filter was divided into the two duplicates: one half was incubated with polyclonal antibodies anti-FIS and the other half with antibodies anti-H-NS. Finally, proteins detection was carried out using Alkaline phosphatase Conjugated anti-rabbit IgG and NBT/BCIP as substrates.
The DNA fragment used in footprints was excised from pTZ310 with BamHI/PstI or HindIII/SmaI and end-labeled by fill-in reaction with α-[32P]-dATP using Klenow fragment of DNA polymerase. The radioactive DNA was pre-incubated with the indicated amounts of FIS or/and H-NS for 20 min at 25°C in 30 μl of in vitro transcription buffer (see below). After addition of 15 ng of DNase I, the reaction was prolonged for further 45 s and then stopped on ice with 1.5 μl of 0.5 M EDTA (pH 8) and 10 μl of 10 M NH4 acetate (pH 7.3). Partially digested DNA was ethanol precipitated in presence of 1 μg of tRNA as carrier and then loaded on a 7% polyacrylamide-urea gel (Sambrook and Russell, 2001).
In vitro transcription assays were programmed with pKKcspA310::cat, a pKK232-8 derivative, carrying a 310 bp DNA fragment of cspA promoter region (from pos. −145 to pos. +165; Goldenberg et al., 1997). The reactions, carried out at 37°C in 25 μl of transcription buffer (10 mM Tris HCl, pH 8, 10 mM MgCl2, 100 mM NaCl, 2 mM spermidine, 2 mM dithiothreitol, 0.1 mg ml−1 BSA), contain 0.15 U of E. coli RNA Polymerase (USB), 0.5 mM of each NTP, 5 U of human placental ribonuclease inhibitor and 100 ng of DNA template. At the indicated times, the reaction was stopped with 1.5 μl of 0.5 M EDTA (pH 8) and 10 μl of 10 M NH4 acetate (pH 7.5) and mRNA ethanol precipitated. The amount of the cat reporter gene transcribed in vitro was determined by Northern blotting, probed with a 32P-labeled cat fragment and quantified by Molecular Imager (Bio-Rad, model FX).
In previous studies, we have shown that cspA is highly expressed not only during cold-shock but also under non-stress conditions when cells grow at 37°C (Brandi et al., 1999; Brandi and Pon, 2012). Here, to understand how the physiological state of the cell could affect the expression of cspA, we monitored the cspA mRNA levels in cells escaping from stationary phase as a function of the availability of nutrients. To accomplish this goal, an overnight culture of E. coli grown at 37°C was diluted in rich (LB) or minimal (M9) fresh medium and bulk RNA, extracted from cells at increasing times after the nutritional up-shift, was used for Northern analysis (Figure 1). Transcription of cspA is promptly induced upon cell dilution, in both media, albeit to different extents (~40 fold in M9 and ~100 fold in LB). Furthermore, the level of transcript augments within the initial 80 min, a time preceding the first bacterial division as denoted by the constant number of cells (CFU), and drops off to the initial level after the cells start dividing. These observations are confirmed by detecting the cspA mRNA by RT-qPCR in cells growing in LB (Figure S6C). In light of this finding, we focused our study on those factors, likely affecting cspA regulation at 37°C, that are known to couple gene expression to growth conditions.
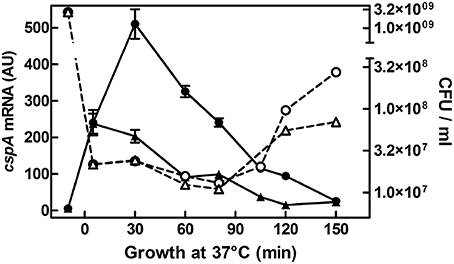
Figure 1. cspA mRNA steady state level following growth resumption at 37°C. An overnight culture of E. coli MRE600 grown at 37°C in M9 medium was diluted (1:100) in parallel with LB (●) or with M9 (▴) fresh medium and cell aliquots were harvested for RNA extraction at the indicated time after resumption from stationary phase. About 6 μg of total RNA were subjected to Northern analysis. The cellular level of cspA transcripts were evaluated by imager quantification of the radioactivity associated with the mRNA and expressed as Arbitrary Units (AU). Data represent the average of at least two independent experiments and standard deviation is indicated. The profile of Colony Forming Units (CFU) during growth in rich (LB,○) or minimal (M9, Δ) medium as a function of time after nutritional up-shift is also reported. Further details are provided in Materials and Methods.
Although, FIS and H-NS have been shown to influence cspA expression (Brandi et al., 1999), so far the evidence of a direct binding of these proteins to the promoter region of the target gene was lacking. Thus, we investigated the interaction of these two NAPs with the cspA DNA by electrophoretic mobility shift assay (EMSA). For this purpose, a fragment of 310 bp spanning from position −145 to position +165, was incubated with H-NS, FIS or with a mixture containing both proteins at different molar ratios. As seen in Figure 2A, when tested individually, H-NS produces an EMSA pattern typical of an all-or-none response, suggesting that this protein binds in a co-operative manner to the 310 bp DNA fragment containing the cspA promoter as described for other genetic systems (Falconi et al., 1993, 1998; Madrid et al., 2002; Giangrossi et al., 2005; Ulissi et al., 2014). In the absence of FIS, H-NS has little or no effect below the critical concentration of 360 nM, whereas it forms a stable nucleoprotein complex at 520 nM (Figure 2A). The addition of more H-NS (730 nM) causes the appearance of a new band with reduced mobility, suggesting that, at the maximum concentration tested, H-NS oligomerizes occupying all high and low affinity sites presents on this DNA fragment. Unlike H-NS, a discrete retardation band is detected even at very low FIS concentrations (7 nM) and additional bands with progressively lowered mobility appear as a function of increasing FIS concentrations (14 nM in Figure 2A, 28 and 56 nM in Figure S1). This pattern suggests that the 310 bp DNA fragment contains multiple sites for which FIS displays different affinities and that are saturated in a concentration-dependent manner by this protein. Furthermore, compared to H-NS, a relative little amount of FIS is sufficient to occupy, at least partially, all sites.
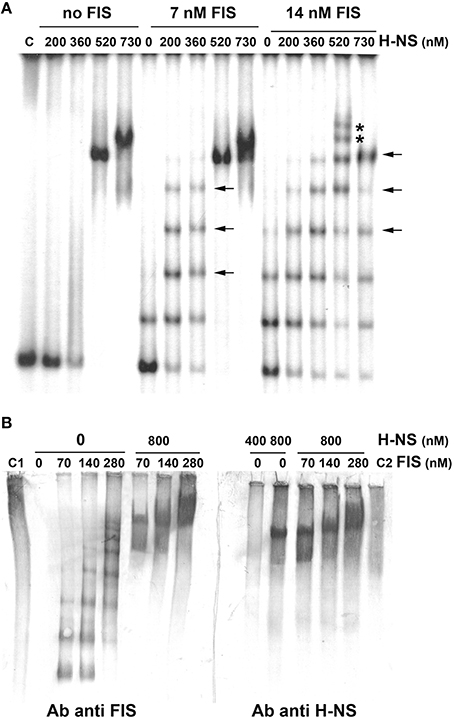
Figure 2. Band shift of a DNA fragment containing the cspA promoter by FIS and H-NS. (A) The 310 bp DNA fragment, containing the cspA promoter region including the entire 5′-UTR, labeled with 32P, was used in electrophoretic mobility shift assay (EMSA) with the indicated amounts of H-NS and/or FIS. DNA-protein complexes formed preferentially or exclusively in the presence of both FIS and H-NS are indicated by arrows or asterisks, respectively. (B) EMSA experiments, carried out in duplicate, were subjected to Western Blot. After electrophoresis and elettroblotting, membranes were alternatively developed with antibodies anti-FIS or anti-H-NS as indicated. Lanes C1 and C2 represent the free FIS and H-NS proteins without DNA, respectively. Proteins concentration is given assuming dimeric structure. For further details see Materials and Methods.
Interestingly, the addition of 200–300 nM of H-NS to low concentrations of FIS produces retarded bands (indicated by horizontal arrows in Figure 2A, Figure S1) with a mobility similar to those bands present when FIS alone is added at higher concentrations. Nevertheless, when both H-NS and FIS are added at a certain concentration ratio, additional bands (indicated by asterisks in Figure 2A, Figure S1), not found in the individual H-NS and FIS patterns, appear. On the other hand, when FIS and H-NS are added at low (≤14 nM) and high (>500 nM) concentrations, respectively, the most retarded complex seems to prevalently contain H-NS, since it displays a mobility similar to that obtained with H-NS alone at 520 nM (Figure 2A). All together, these observations indicate that FIS and H-NS might simultaneously bind to the same DNA molecule (the 310 bp DNA fragment) when are present in a given range of concentration ratios.
To verify this hypothesis, we carried out a band shift assay in which the DNA fragment was not radioactive and therefore nucleoprotein complexes were immunodetected using antibodies against FIS or H-NS (Figure 2B). Under the native conditions used for electrophoresis, unbound FIS and H-NS appear diffused throughout the lane (control samples C1 and C2) whereas discrete retarded bands are visible only in the presence of DNA. As expected, when FIS and H-NS are individually tested, DNA-protein complexes visualized by antibodies are superimposable to those obtained with labeled DNA (compare panels B and A of Figure 2). Nevertheless, when FIS (70–280 nM) and H-NS (800 nM) are combined, the same DNA-protein aggregates are revealed using either anti-FIS or anti-H-NS antibodies (Figure 2B). The control experiment shown in Figure S2 rules out the possibility that this result could be due to a cross-reaction between anti-FIS and H-NS or anti-H-NS and FIS. Therefore, all together, these data strongly suggest that both proteins can simultaneously interact with the 310 bp DNA fragment containing the cspA promoter to originate miscellaneous complexes.
EMSA experiments prompted us to localize FIS and H-NS binding sites on cspA promoter, in an attempt to characterize also the nature of the complexes containing both proteins. To this end, we carried out DNase I footprints and compared the digestion patterns obtained with single proteins to that observed with a mixture of FIS and H-NS (Figures 3A,B). When individually tested, FIS interacts with two sites of ~35 bp in length centered at positions −10 (site 2) and −60 (site 3) producing typical hypersensitive points, while H-NS covers a fairly wide region (~100 nucleotides) centered at position −40. When mixed together FIS and H-NS cover all the available sites, giving rise to an extended protection spanning from position −90 to position +20 on cspA promoter. Remarkably, the DNAse I digestion pattern observed at this cumulative site is essentially a merge of protected and hypersensitive positions characteristic of FIS and H-NS individual sites. Therefore, although FIS and H-NS protections are almost completely overlapping on both DNA strands (see scheme in Figure 3C), the two proteins apparently do not compete for binding to the same target DNA.
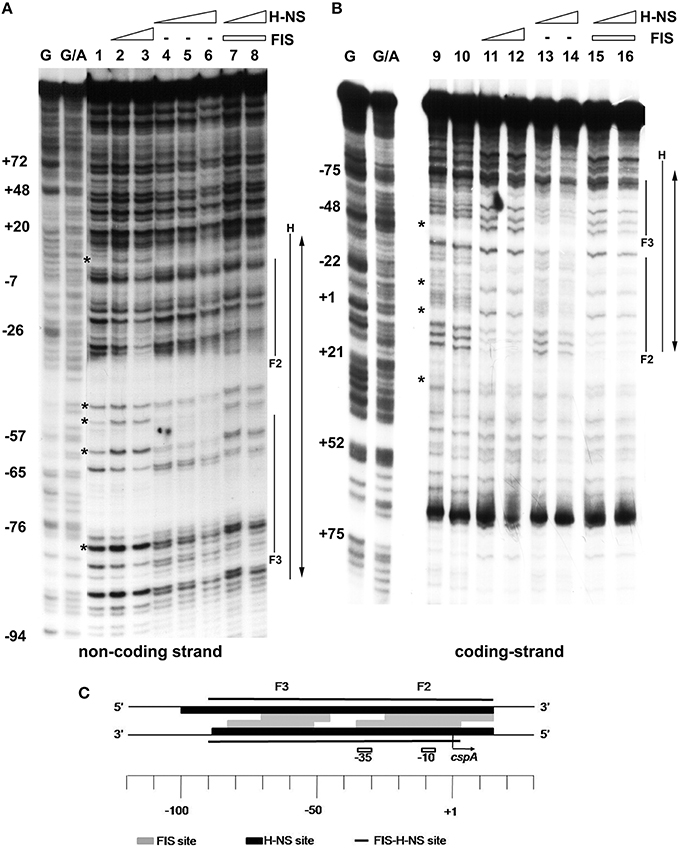
Figure 3. Mapping of FIS and H-NS binding sites on cspA promoter region by DNase I footprinting. Footprinting analysis was carried out on both non-coding strand (A) and coding strand (B) of a 310 bp DNA fragment corresponding to cspA promoter in the presence of the following FIS and HNS dimeric concentrations: no proteins, lanes 1, 9, and 10; 278 nM FIS, lanes 2 and 11; 555 nM FIS, lanes 3 and 12; 93 nM H-NS, lanes 4 and 13; 185 nM H-NS, lanes 5 and 14; 277 nM H-NS, lane 6; 278 nM FIS and 93 nM H-NS, lanes 7 and 15; 278 nM FIS and 185 nM H-NS, lanes 8 and 16. FIS and H-NS sites are indicated by solid lines while the double-headed arrows show protections resulting from the concomitant bond of FIS and H-NS. Sites hypersensitive to DNase I due to FIS-DNA interaction are indicated by asterisk. G and G+A represent the Maxam and Gilbert sequencing reactions. Localization of FIS (gray boxes), H-NS (black boxes), and FIS-H-NS (solid line) binding sites are schematically indicated on cspA promoter region (C).
An extensive scanning of the regions adjacent to the cspA minimum promoter (Figures S3, S4) reveals the existence of other positions recognized by these nucleoid proteins. FIS covers two other distinct sites numbered F1 and F4 and centered at positions +32 and −120, respectively, while H-NS extends its protection to basically the entire cspA promoter due to its extensive oligomerization (Spurio et al., 1997; Badaut et al., 2002; Stella et al., 2005; Giangrossi et al., 2014). According to this premise, we found several in silico predicted binding sites for H-NS (Figure 4A) that are all imperfect fits to its consensus sequences (Lang et al., 2007) and can be considered as nucleation sites where the protein initially binds before spreading to adjacent DNA tracts on cspA sequence. The same in silico approach allowed us to identify also five potential FIS binding sites matching the FIS Logo (Figure 4B) proposed by Shao et al. (2008), four of which fall in the regions shielded or exposed to DNase I cleavage by FIS. The overall H-NS and FIS protections on the sequence of cspA regulatory region along with their in silico predicted binding sites are summarized in Figure 4C.
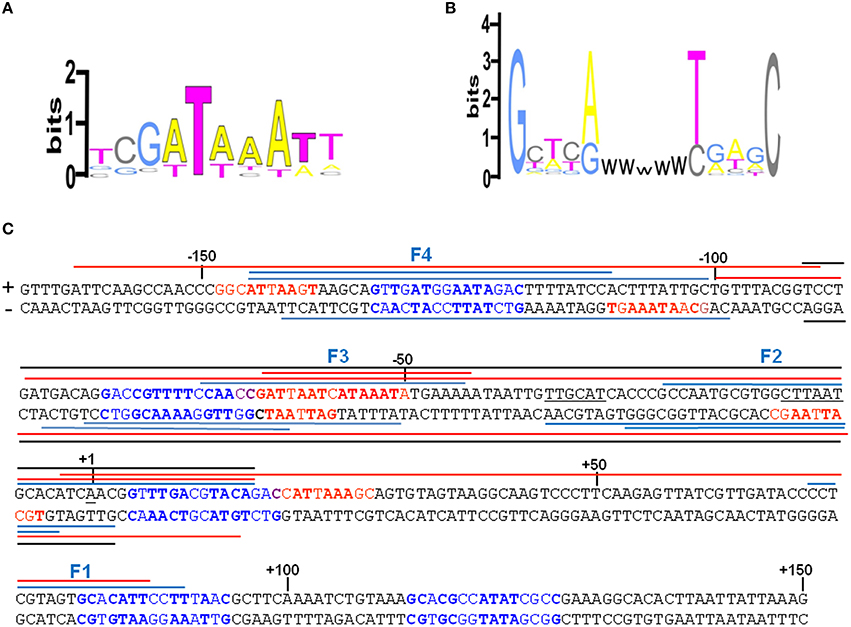
Figure 4. Localization of FIS and H-NS binding sites found within the cspA promoter region. (A) Logo representation of the H-NS binding motif (Lang et al., 2007). (B) Simplified version of the Logo for the FIS binding motif proposed by Shao et al. (2008). W = A or T. (C) The sequence of the promoter region of cspA (−167 to +150) is shown, with the transcriptional start point (+1), the −10 and −35 consensus promoter elements underlined in black. The bases in red represent the in silico predicted H-NS binding sites matching at least 4/6 bases of the core sequence of the H-NS binding motif and displaying less frequent bases in the other positions, while the bases in blue represent the in silico predicted FIS binding sites according to the FIS logo shown in (B). The bases in bold match those found more frequently in the H-NS and FIS binding sites, respectively. The colored lines placed above or below the + and − strand, respectively, indicate the protections from DNase I digestion found with H-NS alone (red), FIS alone (blue), or with a mixture of FIS and H-NS (black). Overlapping lines indicate DNase I footprintings found in independent experiments (Figure 3, Figures S3, S4 in Supplementary Material and other not shown). The protections were identified using either a 310 bp cspA fragment (for FIS, H-NS, and FIS+H-NS) or a 1200 bp cspA fragment (for FIS and H-NS alone).
In a previous paper, we provided evidence of a functional antagonism between FIS and H-NS on cspA expression (Brandi et al., 1999). This observation is consistent with the location, reported here, of H-NS and FIS binding sites, extending over the whole promoter region of cspA. Concerning the role of these two NAPs, while the inhibitory action of H-NS is commonly accepted, the function of FIS is still a point of debate since contradictory results have been reported about this protein that was also found to negatively regulate cspA (Yamanaka and Inouye, 2001). Thus, to address this issue, we assayed H-NS and FIS for their capability to affect cspA transcription in vitro. As seen in Figure 5A, the activity of cspA promoter is stimulated by FIS and repressed by H-NS, totally confirming our previous data. Under the experimental conditions used, the extents of FIS stimulation and H-NS inhibition are similar (~3-fold) as compared to transcription carried out in the absence of proteins. Consistently, when FIS and H-NS are added together, their opposed effects on transcription neutralize each other, thus restoring the basal activity of cspA promoter (compare C and FIS+H-NS curves in Figure 5A).
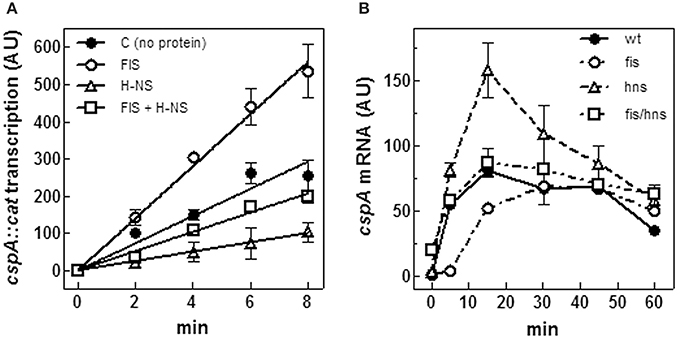
Figure 5. FIS and H-NS modulate cspA transcription. (A) In vitro transcription assay programmed with pKKcspA310::cat. DNA template was pre-incubated at 37°C in the absence of proteins (●), with 50 nM FIS (○), 375 nM H-NS (Δ) and with both FIS (50 nM) and H-NS (375 nM) (□) as dimers. The reaction was started by adding 0.15 units of RNA polymerase and the incubation was prolonged for the indicated times at 37°C, as described in Materials and Methods. (B) Steady-state levels of CspA mRNA were determined in wt E. coli cells (●), a fis null allele (○), an hns null allele (Δ), and a double mutant fis-hns (□). After a 10-fold dilution with fresh medium of saturated cultures grown in LB at 37°C, total RNA was extracted at the indicated times and subjected (10 μg) to Northern analysis. The radioactivity associated with cspA mRNA, normalized for the corresponding amounts of 16S rRNA, was quantified by Molecular Imager (Bio-Rad) and expressed as Arbitrary Units (AU). Data represent the average of at least two independent experiments and standard deviation is indicated.
The effect of H-NS and FIS on cspA expression was also studied in vivo. To this end, the steady state level of cspA mRNA was estimated at 37°C upon resumption from stationary phase of growth in wt, hns- and fis- strains and in hns/fis double deletion mutant. In agreement with Brandi et al. (1999) and Brandi and Pon (2012) in wt cells cspA expression is very high in early exponential growth and then progressively declines (Figure 5B). Interestingly, the lack of FIS causes a reduction of cspA transcript as compared to the wt within the initial 15 min, a time window that usually precedes the first cell division. However, at later stages, wt and fis- cells show comparable amounts of cspA mRNA (Figure 5B). On the contrary, inactivation of hns gene induces an increase of the cspA mRNA level that almost doubles in the first 20 min of growth with respect to the wt condition, and then declines. Finally, in agreement with the in vitro transcription assay (Figure 5A), the concomitant absence of FIS and H-NS results in a compensatory effect and ultimately has no significant consequences on the level of cspA mRNA (Figure 5B). Taken together, in vitro and in vivo data are fully consistent with each other and strongly indicate that H-NS and FIS, acting as negative and positive regulators, respectively, play an opposed role in modulating cspA transcription.
The alarmone guanosine tetraphosphate ((p)ppGpp) is a global regulator which is produced in most circumstances and modulates bacterial physiology (Hauryliuk et al., 2015). This small effector molecule is known to influence the expression of many genes thereby coupling the overall level of transcription to growth-rate (Potrykus and Cashel, 2008). Furthermore, overproduction of this unusual nucleotide prior to cold-shock was reported to lower the induction of most cold-shock genes, including cspA (Jones et al., 1992a).
Therefore, we evaluated both in vivo and in vitro whether cspA promoter could respond to (p)ppGpp regulation also at 37°C (Figure 6). First of all, the intracellular (p)ppGpp level was artificially increased by IPTG induction of extra-chromosomal copies of relA gene placed under the control of the lacUV5 promoter in plasmid pTK31 (see Materials and Methods). The induction of (p)ppGpp synthesis from pTK31 was verified (not shown) by thin layer chromatography as previously described (Sarubbi et al., 1989). As expected, when cells in stationary phase were subjected to a nutritional up-shift, a sudden burst of cspA transcription was observed. By contrast, when the fresh medium was supplemented with IPTG, the high levels of the unusual nucleotide in induced cells significantly counteracted the characteristic promoter activation resulting in a substantial reduction of cspA messenger (Figure 6A). Thus, hypothesizing a direct action of (p)ppGpp, we investigated the effect of this molecule on cspA promoter activity in an in vitro purified system, following cspA transcription as a function of increasing reaction times. This experiment demonstrates that the level of cspA mRNA is decreased 2- and 4-fold in the presence of 200 and 400 μM of (p)ppGpp, respectively, as compared to the control curve obtained in the absence of the regulatory nucleotide (Figure 6B). According to the finding that (p)ppGpp-dependent inhibition of transcription of sensitive promoters results from the competition between the mediator molecule and NTPs substrates for access to the active center of RNA polymerase (Jöres and Wagner, 2003), the use of higher concentrations of NTPs (0.5 mM) alleviates the negative action of (p)ppGpp on cspA transcription (Figure S5). Altogether these results indicate that the stimulation of cspA expression, observed in early exponential growth at 37°C, is almost completely prevented by high levels of (p)ppGpp and that this effect reasonably resides on the ability of this molecule to directly repress the synthesis of mRNA from cspA promoter. Finally, to better understand the three components (FIS, H-NS, and (p)ppGpp) regulatory loop governing cspA expression, we analyzed, under our experimental conditions (cells carrying the pTK31 vector) fis and hns transcription as a function of increased intracellular concentrations of (p)ppGpp. As seen in Figure 7, activation of fis and hns promoters consequent to cell resumption from stationary phase is completely (panel A) and partially (panel B) abolished, respectively, by an elevated level of (p)ppGpp suggesting that transcription of both genes is negatively regulated by this alarmone.
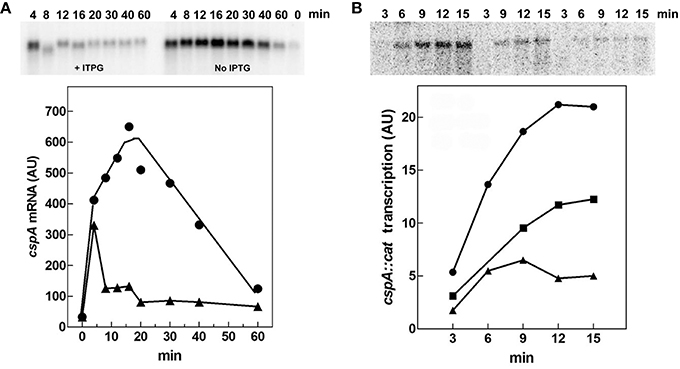
Figure 6. (p)ppGpp negatively affects cspA transcription. (A) The steady state levels of cspA transcript were determined during growth in Phosphates-free medium at 37°C in cells overproducing or not (p)ppGpp. Total RNA was extracted from cells in stationary phase (time zero) and at the indicated times after dilution with fresh medium alone (●) or supplemented with 400 μM IPTG (▴). The levels of cspA mRNA were estimated by Northern blotting analysis. (B) Effect of (p)ppGpp on the in vitro activity of cspA promoter. The transcriptional activity was estimated in the absence (●) or in presence of 200 (■) and 400 μM (▴) of (p)ppGpp. Reactions and processing of samples were performed as described in Materials and Methods. The supercoiled plasmid pKK310::cat used as DNA template was pre-incubated with (p)ppGpp and RNA polymerase for 5 min at 37°C. Then reactions were started by adding NTPs at a final concentration of 100 μM each. At the indicated incubation times, aliquots were withdrawn and transcription stopped with EDTA (f.c. 30 mM). Quantization of the Northern blots (upper panels), expressed as Arbitrary Units (AU), is plotted as a function of time (lower panels).
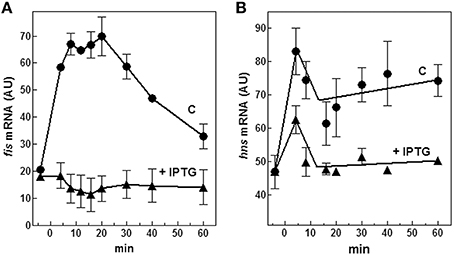
Figure 7. Effect of (p)ppGpp on fis and hns expression. Levels of fis (A) and hns (B) mRNAs were estimated by Northern blotting analysis of total RNA extracted from E. coli cells growing at 37°C and carrying the plasmid pTK31. Cells were collected in stationary phase (time zero) and at the indicated times after dilution with fresh medium. As described in Figure 6A, the control culture was grown in the absence of IPTG (▴) whereas the induced culture contained 400 μM IPTG (●) to activate the lacUV5::relA gene. Data represent the average of at least two independent experiments and standard deviation is indicated.
There is evidence that regulation of many bacterial genes is based on structural/functional interplays between two or more nucleoid-associated proteins which may play synergetic and/or antagonistic roles. In particular, FIS and H-NS cooperate to regulate several unrelated genetic systems in different bacteria. Some examples are rrnB (Afflerbach et al., 1999), oriC (Roth et al., 1994), nir (Browing et al., 2000), dps (Grainger et al., 2008), pel (Ouafa et al., 2012) as well as those systems that we have contributed to characterize, such as the FIS-H-NS-mediated regulation of the E. coli hns gene itself (Falconi et al., 1996), or the virulence gene virF of Shigella flexneri (Falconi et al., 2001).
In this study, in addition to confirming our earlier observations on the involvement of FIS and H-NS in controlling cspA (Brandi et al., 1999), we dissected their interplay at cspA promoter. Furthermore, we identified another factor, the (p)ppGpp, participating in this regulation, thus expanding our knowledge of the regulatory circuit governing the expression of cspA at 37°C.
Firstly, through both EMSA and footprinting assays, we have demonstrated the direct interaction of FIS and H-NS with the promoter region of cspA and identified their target sites as summarized in Figure 4. The appearance of discrete bands with shifted mobility indicates that FIS binds at least five sites on the cspA promoter region spanning from position −167 to position +150. This interaction is also confirmed by the FIS-dependent protections from DNase I cleavage of four discrete stretches of nucleotides (~30–40 bp). Notably, all these protected targets overlap a degenerated FIS consensus sequence predicted in silico using a software that allows identification of sequence motifs (Hu et al., 2003) based on the FIS binding site logo (Shao et al., 2008). With regard to H-NS, we found six sequences on cspA DNA partially matching the H-NS binding motif as identified by Lang et al. (2007). These sequences may represent high-affinity nucleation sites where H-NS initially binds before oligomerizating along DNA and interacting with adjacent lower affinity sites. Such binding property accounts for the extended regions protected by H-NS in footprinting experiments (Figure 3, Figure S4) and ultimately determines the complete coverage of cspA promoter region including the four mapped FIS sites (F1–F4). Both the standard EMSA and a modified EMSA, in which the DNA-protein complexes were localized by immunodetection (Figure 2), reveal that FIS and H-NS, instead of structurally competing for the binding to the DNA, at certain concentration ratios can contact simultaneously the region containing the cspA promoter each one recognizing their own targets. The formation of high-order aggregates containing both proteins is also evidenced by the occurrence of a single merged protection observed in footprints carried out with FIS and H-NS together. The coexistence of these two proteins is likely due to their different binding properties: while FIS interacts with the major groove (Osuna et al., 1991), H-NS contacts the minor groove of the DNA, as demonstrated by the finding that two DNA minor groove-binding molecules, distamycin and netropsin, effectively compete with H-NS for the binding to an AT-rich sequence (Yamada et al., 1990; Gordon et al., 2011). Furthermore, our EMSA experiments strongly suggest that the binding affinity of FIS and H-NS for cspA DNA increases when the proteins are combined (Figure 2, Figure S1). Two not mutually exclusive circumstances can explain this observation: FIS can bend DNA upon binding (Pan et al., 1996), thus facilitating the interaction of H-NS that is known to preferentially recognize curved DNA (Yamada et al., 1990) or H-NS can actively curve not curved DNA (Spurio et al., 1997), thus easing the positioning of FIS at its binding sites. Since the regions protected by FIS (site F2) and H-NS overlap the −35 and −10 elements of the promoter (Figure 4), their occupancy may allow the formation of contacts between the two nucleoid proteins and RNA polymerase thereby accounting for the stimulation/repression of cspA transcription by FIS and H-NS (Figure 5). This scenario shares many similarities with the models proposed for the regulation of hns and virF genes by H-NS and FIS in which the protein molar ratio reflects the nature of the hetero-complex formed and ultimately controls the switch between a transcriptionally active or repressed state (Falconi et al., 1996, 2001).
The composition of the population of NAPs in the cell is not fixed and fluctuations of these proteins is thought to mediate global changes in nucleoid structure and transcriptional activity (Azam and Ishihama, 1999). In fact, growth phase and variations of other environmental parameters (i.e., temperature, pH, availability of nutrients and oxygen) produce a characteristic profile of nucleoid-associated proteins. While the intracellular level of H-NS is generally high and quite constant, the expression of FIS is strongly dependent on growth conditions, being elevated in early exponential phase and upon a nutrient up-shift (Ball et al., 1992; Dillon and Dorman, 2010).
Interestingly, the expression pattern of cspA at 37°C seems to mirror that of FIS, since cspA transcription is maximal before growth resumption from stationary phase (in the period of time immediately preceding the first cell division), and it is progressively reduced at later stages of growth. Since FIS is able to directly stimulate the transcription of cspA and to contrast the inhibitory effect of H-NS, as demonstrated by in vitro transcription assays (Figure 5A), it is likely that changes of FIS intracellular levels help to link the physiological state of cells and the environmental conditions to cspA expression. This model of regulation is confirmed also by in vivo experiments performed in different genetic backgrounds. In fact, with respect to wt strain, the raise in cspA mRNA level during the initial minutes after escape from stationary phase is reduced and delayed in fis- background and increased in hns- strain (Figure 5B). Furthermore, in line with our model, at increased culture age the cspA mRNA level in both wt and hns- strains becomes comparable to that of fis- strain. Therefore, FIS seems able to bind cspA promoter and sponsor the activation of cspA transcription in a dose-dependent manner so that when its level drops below a certain concentration, it can no longer contrast the inhibition by H-NS.
In addition to NAPs, cspA promoter is able to respond to changes of (p)ppGpp. Although, the intracellular concentration of this mediator molecule is quite stable under physiological growth conditions, its synthesis is affected by several types of nutritional limitations and other environmental stimuli, like an abrupt change of temperature (Magnusson et al., 2005). Here, we show that a high level of (p)ppGpp abolishes the in vivo induction of cspA after a nutritional up-shift and that this effect is due to a direct inhibition of cspA promoter activity (Figure 6). Numerous hypotheses have been made to explain the negative regulation of transcription by (p)ppGpp. These mechanisms, not mutually exclusive but possibly working in concert, rely on three main conditions: (i) the presence of particular features of promoters sensitive to (p)ppGpp; (ii) the direct interaction between (p)ppGpp and RNAP; (iii) the regulatory role of DksA protein (reviewed in Magnusson et al., 2005; Potrykus and Cashel, 2008; Hauryliuk et al., 2015). Interestingly, the cspA promoter contains, in the region between the TATA box (−10) and the transcriptional start site (+1), a GC-rich sequence termed discriminator (Figure 4), that is a key element of those promoters repressed by (p)ppGpp. Furthermore, in our genetic system, a direct interaction of (p)ppGpp with RNAP is supported by the fact that the in vitro inhibition of cspA transcription by this effector is dependent on the amount of NTPs added. In fact, an excess of NTPs (from 0.1 to 0.5 mM) causes a general decreased capability of the alarmone to negatively affect the promoter activity of cspA and makes the transcription repression independent from the concentration of (p)ppGpp (compare mRNA levels at 200 and 400 μM of (p)ppGpp in Figure 6B, Figure S5). Similarly, a sensitivity to the nature and concentration of initiating nucleotide was observed in the (p)ppGpp-mediated regulation of rRNA transcription (Jöres and Wagner, 2003; Kolmsee et al., 2011) where the major step affected by (p)ppGpp is the formation of the ternary transcription initiation complex. This finding strongly indicates that the unusual nucleotide and NTPs compete for access to the active center of RNAP and is consistent also with the (p)ppGpp-RNAP cocrystals that positioned (p)ppGpp in the secondary channel of the enzyme near the catalytic center (Artsimovitch et al., 2004). Notably, the amount of (p)ppGpp added in our in vitro transcription assays is very close to that used in similar studies (Heinemann and Wagner, 1997) and compatible with that estimated in vivo (~900 μM) in response to amino acid starvation (Traxler et al., 2008).
The cspA and fis genes are apparently regulated in parallel and this may be attributed to the role of the common regulatory molecule, (p)ppGpp. As cspA, also fis expression is controlled by this unusual nucleotide as demonstrated by the fact that the stringent response produced by the artificial induction of (p)ppGpp in early log cells causes a dramatic reduction of fis mRNA while transcription of hns, under the same experimental conditions, is less affected (Figure 7). Interestingly, also fis promoter contains a GC-rich discriminator sequence downstream the −10 position and its transcription peaks during the early logarithmic growth phase, a condition characterized by a low concentration of (p)ppGpp, to decrease soon thereafter, as the level of (p)ppGpp begins to rise in cells approaching the stationary phase (Mallik et al., 2004). Therefore, (p)ppGpp is a fundamental element of this regulatory circuit and is able to repress cspA transcription by a dual mechanism: one direct, exerting a negative action on the functionality of cspA promoter and one indirect, depriving the target gene of its natural transcriptional activator FIS. On the other hand, a low level of FIS favors the silencing of cspA by H-NS.
A recent expression analysis of csp genes based on quantitative RT-PCR (Czapski and Trun, 2014) has demonstrated that the levels of csp mRNAs change with growth phase and type of medium. In particular, in rich defined medium the transcripts of cspA, B, G, and I were found to accumulate preferentially in early log phase, those of cspC and cspD in mid-log phase and stationary phase, respectively, and cspE mRNA was found to be constitutively present. Our comparison of cspB, G, and I expression pattern using both Northern blotting and quantitative RT-qPCR (Figure S6) essentially confirms that these genes display a growth-dependent expression similar to that of cspA. Therefore, it is tempting to speculate that at 37°C also the fluctuations of other csp genes could be regulated by a network similar to that found for cspA and based on the antagonistic role of FIS/H-NS and the transcription inhibition by (p) ppGpp.
AB designed and performed most of the experiments giving an important contribution also to the analysis and interpretation of data. Additionally AB in collaboration with MG has been dealing with the preparation of figures, material and methods and references. MG and MF planned and carried out some experiments. MF and AG have mainly done the analysis and interpretation of results, drafting the work (including figures) and revising it critically.
Fondi di Ricerca di Ateneo (FAR), Università di Camerino, to MF.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmolb.2016.00019
Afflerbach, H., Schroder, O., and Wagner, R. (1999). Conformational changes of the upstream DNA mediated by H-NS and Fis regulate E.coli rrnB P1 promoter activity. J. Mol. Biol. 286, 339–353. doi: 10.1006/jmbi.1998.2494
Afflerbach, H., Schröder, O., and Wagner, R. (1998). Effects of the Escherichia coli DNA-binding protein H-NS on rRNA synthesis in vivo. Mol Microbiol. 28, 641–653. doi: 10.1046/j.1365-2958.1998.00829.x
Artsimovitch, I., Patlan, V., Sekine, S., Vassylyeva, M. N., Hosaka, T., Ochi, K., et al. (2004). Structural basis for transcriptional regulation by alarmone ppGpp. Cell 117, 299–310. doi: 10.1016/S0092-8674(04)00401-5
Atlung, T., and Ingmer, H. (1997). H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24, 7–17. doi: 10.1046/j.1365-2958.1997.3151679.x
Azam, T. A., and Ishihama, A. (1999). Twelve species of nucleoid-associate protein from Escherichia coli: sequence recognition specificity and binding affinity. J. Biol. Chem. 274, 33105–33113. doi: 10.1074/jbc.274.46.33105
Badaut, C., Williams, R., Arluison, V., Bouffartigues, E., Robert, B., Buc, H., et al. (2002). The degree of oligomerization of the H-NS nucleoid structuring protein is related to specific binding to DNA. J. Biol. Chem. 277, 41657–41666. doi: 10.1074/jbc.M206037200
Bae, W., Xia, B., Inouye, M., and Severinov, K. (2000). Escherichia coli CspA-family RNA chaperone are transcription antiterminators. Proc. Natl. Acad. Sci. U.S.A. 97, 7784–7789. doi: 10.1073/pnas.97.14.7784
Ball, C. A., Osuna, R., Ferguson, K. C., and Johnson, R. C. (1992). Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J. Bacteriol. 174, 8043–8056.
Bouffartigues, E., Buckle, M., Badaut, C., Travers, A., and Rimsky, S. (2007). H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 14, 441–448. doi: 10.1038/nsmb1233
Brandi, A., Pietroni, P., Gualerzi, C. O., and Pon, C. L. (1996). Post-transcriptional regulation of CspA expression in Escherichia coli. Mol. Microbiol. 19:231–240. doi: 10.1046/j.1365-2958.1996.362897.x
Brandi, A., and Pon, C. L. (2012). Expression of Escherichia coli cspA during early exponential growth at 37°C. Gene 492, 382–388. doi: 10.1016/j.gene.2011.10.047
Brandi, A., Spurio, R., Gualerzi, C. O., and Pon, C. L. (1999). Massive presence of the Escherichia coli “major cold-shock protein” CspA under non stress conditions. EMBO J. 18, 1653–1659. doi: 10.1093/emboj/18.6.1653
Browing, D. F., Cole, J. A., and Busby, S. (2000). Suppression of FNR-dependent transcription activation at the Escherichia coli nir promoter by FIS, IHF and H-NS: modulation of transcription initiation by a complex nucleo-protein assembly. Mol. Microbiol. 37, 1258–1269. doi: 10.1046/j.1365-2958.2000.02087.x
Browning, D. F., Grainger, D. C., and Busby, S. J. (2010). Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 13, 773–780. doi: 10.1016/j.mib.2010.09.013
Cammack, K. A., and Wade, H. E. (1965). The sedimentation behaviour of ribonuclease-active and -inactive ribosomes from bacteria. Biochem. J. 96, 671–680. doi: 10.1042/bj0960671
Czapski, T. R., and Trun, N. (2014). Expression of csp genes in E. coli K-12 in defined rich and defined minimal media during normal growth, and after cold-shock. Gene 547, 91–97. doi: 10.1016/j.gene.2014.06.033
Dillon, S. C., and Dorman, C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195. doi: 10.1038/nrmicro2261
Dorman, C. J. (2004). H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2, 391–400. doi: 10.1038/nrmicro883
Dorman, C. J. (2007). H-NS, the genome sentinel. Nat. Rev. Microbiol. 5, 157–161. doi: 10.1038/nrmicro1598
Ermolenko, D. N., and Makhatadze, G. I. (2002). Bacterial cold-shock proteins. Cell. Mol. Life Sci. 59, 1902–1913. doi: 10.1007/PL00012513
Falconi, M., Brandi, A., La Teana, A., Gualerzi, C. O., and Pon, C. L. (1996). Antagonistic involvement of FIS and H-NS proteins in the transcriptional control of hns expression. Mol. Microbiol. 19, 965–975. doi: 10.1046/j.1365-2958.1996.436961.x
Falconi, M., Colonna, B., Prosseda, G., Micheli, G., and Gualerzi, C. O. (1998). Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 17, 7033–7043. doi: 10.1093/emboj/17.23.7033
Falconi, M., Gualtieri, M. T., La Teana, A., Losso, M. A., and Pon, C. L. (1988). Proteins from prokaryotic nucleoid: primary and quaternary structure of the 15-KD Escherichia coli DNA binding protein H-NS. Mol. Microbiology 2, 323–329. doi: 10.1111/j.1365-2958.1988.tb00035.x
Falconi, M., Higgins, N. P., Spurio, R., Pon, C. L., and Gualerzi, C. O. (1993). Expression of the gene encoding the major bacterial nucleoid protein H-NS is subject to transcriptional auto-repression. Mol. Microbiol. 10, 273–282. doi: 10.1111/j.1365-2958.1993.tb01953.x
Falconi, M., Prosseda, G., Giangrossi, M., Beghetto, E., and Colonna, B. (2001). Involvement of FIS in the H-NS-mediated regulation of virF gene of Shigella and enteroinvasive Escherichia coli. Mol. Microbiol. 42, 439–452. doi: 10.1046/j.1365-2958.2001.02646.x
Giangrossi, M., Wintraecken, K., Spurio, R., and de Vries, R. (2014). Probing the relation between protein-protein interactions and DNA binding for a linker mutant of the bacterial nucleoid protein H-NS. Biochim. Biophys. Acta. 1844, 339–345. doi: 10.1016/j.bbapap.2013.11.010
Giangrossi, M., Zattoni, S., Tramonti, A., De Biase, D., and Falconi, M. (2005). Antagonistic role of H-NS and GadX in the regulation of the glutamate decarboxylase-dependent acid resistance system in Escherichia coli. J. Biol. Chem. 280, 21498–21505. doi: 10.1074/jbc.M413255200
Giuliodori, A. M., Brandi, A., Gualerzi, C. O., and Pon, C. L. (2004). Preferential translation of cold-shock mRNAs during cold adaptation. RNA 10, 265–276. doi: 10.1261/rna.5164904
Giuliodori, A. M., Di Pietro, F., Marzi, S., Masquida, B., Wagner, R., Romby, P., et al. (2010). The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol. Cell 37, 21–33. doi: 10.1016/j.molcel.2009.11.033
Goldenberg, D., Azar, I., and Oppenheim, A. B. (1996). Differential mRNA stability of the cspA gene in the cold-shock response of Escherichia coli. Mol. Microbiol. 19, 241–248. doi: 10.1046/j.1365-2958.1996.363898.x
Goldenberg, D., Azar, I., Oppenheim, A. B., Brandi, A., Pon, C. L., and Gualerzi, C. O. (1997). Role of Escherichia coli cspA promoter sequences and adaptation of translational apparatus in the cold shock response. Mol. Gen. Genet. 256, 282–290. doi: 10.1007/s004380050571
Goldstein, J., Pollitt, N. S., and Inouye, M. (1990). Major cold-shock protein of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 87, 283–287. doi: 10.1073/pnas.87.1.283
Gonzalez-Gil, G., Bringmann, P., and Kahmann, R. (1996). FIS a regulator of metabolism in Escherichia coli. Mol. Microbiol. 22, 21–29. doi: 10.1111/j.1365-2958.1996.tb02652.x
Gordon, B. R., Li, Y., Cote, A., Weirauch, M. T., Ding, P., Hughes, T. R., et al. (2011). Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. Proc. Natl. Acad. Sci. U.S.A. 108, 10690–10695. doi: 10.1073/pnas.1102544108
Grainger, D. C., Goldberg, M. D., Lee, D. J., and Busby, S. J. (2008). Selective repression by Fis and H-NS at the Escherichia coli dps promoter. Mol. Microbiol. 68, 1366–1377. doi: 10.1111/j.1365-2958.2008.06253.x
Gualerzi, C. O., Giuliodori, A. M., Brandi, A., Di Pietro, F., Piersimoni, L., Fabbretti, A., et al. (2011). “Translation Initiation at the root of the cold-shock translation bias,” in RIBOSOMES Structure, Function and Dynamics, eds M. V. Rodnina, W. Wintermeyer and R. Green (New York, NY: Springer Wien), 143–154.
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T., and Gerdes, K. (2015). Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309. doi: 10.1038/nrmicro3448
Heinemann, M., and Wagner, R. (1997). Guanosine 3′,5′-bis(diphosphate) (ppGpp)-dependent inhibition of transcription from stringently controlled Escherichia coli promoters can be explained by an altered initiation pathway that traps RNA polymerase. FEBS 247, 990–999. doi: 10.1111/j.1432-1033.1997.00990.x
Hommais, F., Krin, E., Laurent-Winter, C., Soutourina, O., Malpertuy, A., Le Caer, J. P., et al. (2001). Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40, 20–36. doi: 10.1046/j.1365-2958.2001.02358.x
Hu, Z., Frith, M., Niu, T., and Weng, Z. (2003). SeqVISTA: a graphical tool for sequence feature visualization and comparison. BMC Bioinformat. 4:1. doi: 10.1186/1471-2105-4-1
Jiang, W., Hou, Y., and Inouye, M. (1997). CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272, 196–202. doi: 10.1074/jbc.272.1.196
Jones, P. G., Cashel, M., Glaser, G., and Neidhardt, F. C. (1992a). Function of a relaxed-like state following temperature downshifts in Escherichia coli. J. Bacteriol. 174, 3903–3914.
Jones, P. G., Krah, R., Tafuri, S. R., and Wolffe, A. P. (1992b). DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J. Bacteriol. 174, 5798–5802.
Jones, P. J., and Inouye, M. (1994). The cold-shock response - a hot topic. Mol. Microbiol. 11, 811–818. doi: 10.1111/j.1365-2958.1994.tb00359.x
Jöres, L., and Wagner, R. (2003). Essential steps in the ppGpp-dependent regulation of bacterial ribosomal RNA promoters can be explained by substrate competition. J. Biol. Chem. 278, 16834–16843. doi: 10.1074/jbc.M300196200
Kahramanoglou, C., Seshasayee, A. S., Prieto, A. I., Ibberson, D., Schmidt, S., Zimmermann, J., et al. (2011). Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 39, 2073–2091. doi: 10.1093/nar/gkq934
Kolmsee, T., Delic, D., Agyenim, T., Calles, C., and Wagner, R. (2011). Differential stringent control of Escherichia coli rRNA promoters: effects of ppGpp, DksA and the initiating nucleotides. Microbiology 157, 2871–2879. doi: 10.1099/mic.0.052357-0
Lang, B., Blot, N., Bouffartigues, E., Buckle, M., Geertz, M., Gualerzi, C. O., et al. (2007). High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 35, 6330–6337. doi: 10.1093/nar/gkm712
La Teana, A., Brandi, A., Falconi, M., Spurio, R., Pon, C. L., and Gualerzi, C. O. (1991). Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H-NS. Proc. Natl. Acad. Sci. U.S.A. 88, 10907–10911. doi: 10.1073/pnas.88.23.10907
Madrid, C., Nieto, J. M., Paytubi, S., Falconi, M., Gualerzi, C. O., and Juárez, A. (2002). Temperature- and H-NS-dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J. Bacteriol. 184, 5058–5066. doi: 10.1128/JB.184.18.5058-5066.2002
Magnusson, L. U., Farewell, A., and Nyström, T. (2005). ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13, 236–242. doi: 10.1016/j.tim.2005.03.008
Mallik, P., Pratt, T. S., Beach, M. B., Bradley, M. D., Undamatla, J., and Osuna, R. (2004). Growth phase-dependent regulation and stringent control of fis are conserved processes in enteric bacteria and involve a single promoter (fisP) in Escherichia coli. J. Bacteriol. 186, 122–135. doi: 10.1128/JB.186.1.122-135.2004
Muskhelishvili, G., and Travers, A. (2003). Transcription factor as a topological homeostat. Front. Biosci. 8, d279–d285. doi: 10.2741/969
Osuna, R., Finkel, S. E., and Johnson, R. C. (1991). Identification of two functional regions in Fis: the N-terminus is required to promote Hin-mediated DNA inversion but not λ excision. EMBO J. 10, 1593–1603.
Ouafa, Z. A., Reverchon, S., Lautier, T., Muskhelishvili, G., and Nasser, W. (2012). The nucleoid-associated proteins H-NS and FIS modulate the DNA supercoiling response of the pel genes, the major virulence factors in the plant pathogen bacterium Dickeya dadantii. Nucleic Acids Res. 40, 4306–4319. doi: 10.1093/nar/gks014
Pan, C. Q., Finkel, S. E., Cramton, S. E., Feng, J. A., Sigman, D. S., and Johnson, R. C. (1996). Variable structures of Fis-DNA complexes determined by flanking DNA-protein contacts. J. Mol. Biol. 264, 675–695. doi: 10.1006/jmbi.1996.0669
Potrykus, K., and Cashel, M. (2008). (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51. doi: 10.1146/annurev.micro.62.081307.162903
Rimsky, S., and Travers, A. (2011). Pervasive regulation of nucleoid structure and function by nucleoid-associated proteins. Curr. Opin. Microbiol. 14, 136–141. doi: 10.1016/j.mib.2011.01.003
Ross, W., Thompson, J. F., Newlands, J. T., and Gourse, R. L. (1990). E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 9, 3733–3742.
Roth, A., Urmoneit, B., and Messer, W. (1994). Functions of histone-like proteins in the initiation of DNA replication at oriC of Escherichia coli. Biochimie 76, 917–923. doi: 10.1016/0300-9084(94)90016-7
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, NY: CSHL Press.
Sarubbi, E., Rudd, K. E., Xiao, H., Ikehara, K., Kalman, M., and Cashel, M. (1989). Characterization of the spoT gene of Escherichia coli. J. Biol. Chem. 264, 15074–15082.
Schneider, R., Travers, A., and Muskhelishvili, G. (1997). FIS modulates growth phase-dependent topological transitions of DNA in Escherichia coli. Mol. Microbiol. 26, 519–530. doi: 10.1046/j.1365-2958.1997.5951971.x
Shao, Y., Feldman-Cohen, L. S., and Osuna, R. (2008). Functional characterization of the Escherichia coli Fis-DNA binding sequence. J. Mol. Biol. 376, 771–785. doi: 10.1016/j.jmb.2007.11.101
Spurio, R., Falconi, M., Brandi, A., Pon, C. L., and Gualerzi, C. O. (1997). The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 16, 1795–1805. doi: 10.1093/emboj/16.7.1795
Stella, S., Spurio, R., Falconi, M., Pon, C. L., and Gualerzi, C. O. (2005). Nature and mechanism of the in vivo oligomerization of nucleoid protein H-NS. EMBO J. 24, 2896–2905. doi: 10.1038/sj.emboj.7600754
Tedin, K., Witte, A., Reisinger, G., Lubitz, W., and Bläsi, U. (1995). Evaluation of the E. coli rrnB P1 promoter and phage-derived lysis genes for the use in a biological containment system: a concept study. J. Biotechnol. 39, 137–148. doi: 10.1016/0168-1656(95)00003-9
Traxler, M. F., Summers, S. M., Nguyen, H. T., Zacharia, V. M., and Hightower, G. A. (2008). The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 68, 1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x
Ulissi, U., Fabbretti, A., Sette, M., Giuliodori, A. M., and Spurio, R. (2014). Time-resolved assembly of a nucleoprotein complex between Shigella flexneri virF promoter and its transcriptional repressor H-NS. Nucleic Acids Res. 42, 13039–13050. doi: 10.1093/nar/gku1052
Wang, W., Li, G. W., Chen, C., Xie, X. S., and Zhuang, X. (2011). Chromosome organization by a nucleoid-associated protein in live bacteria. Science 9, 1445–1449. doi: 10.1126/science.1204697
Yamada, H., Muramatsu, S., and Mizuno, T. (1990). An Escherichia coli protein that preferentially binds to sharply curved DNA. J. Biochem. 108, 420–425.
Yamanaka, K., Fang, L., and Inouye, M. (1998). The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol. Microbiol. 27, 247–255. doi: 10.1046/j.1365-2958.1998.00683.x
Keywords: cspA gene, FIS, H-NS, guanosine tetraphosphate, DNA-protein interaction, gene regulation in Bacteria, transcription
Citation: Brandi A, Giangrossi M, Giuliodori AM and Falconi M (2016) An Interplay among FIS, H-NS, and Guanosine Tetraphosphate Modulates Transcription of the Escherichia coli cspA Gene under Physiological Growth Conditions. Front. Mol. Biosci. 3:19. doi: 10.3389/fmolb.2016.00019
Received: 29 January 2016; Accepted: 01 May 2016;
Published: 24 May 2016.
Edited by:
Antonio Juárez, Universitat de Barcelona, SpainReviewed by:
Eduard Torrents, Institute for Bioengineering of Catalonia, SpainCopyright © 2016 Brandi, Giangrossi, Giuliodori and Falconi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maurizio Falconi, bWF1cml6aW8uZmFsY29uaUB1bmljYW0uaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.