- 1David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA, United States
- 2Cancer Center of Southern California, Santa Monica, CA, United States
- 3Aveni Foundation, Santa Monica, CA, United States
- 4Counterpoint Biomedica LLC, Santa Monica, CA, United States
- 5Delta Next-Gene, LLC, Santa Monica, CA, United States
Editorial on the Research Topic
Celebrating the 200th mendel’s anniversary: gene-targeted diagnostics and therapies for cancer
In a bicentennial celebration of the quantitative analytical investigations of Gregor Mendel (1822–1884)—which established the basic principles of heredity and the scientific foundations of classical genetics—we present a Special Research Topic entitled Gene-Targeted Diagnostics and Therapies for Cancer, which highlights the latest translations of molecular genetics, functional genomics, and applied proteomics into cancer therapy at the cutting-edge of precision medicine. Figure 1 shows an artist’s illustration of the chronology of events from Gregor Mendel’s experiments on pea plants to the discovery of the genetic code to genetic engineering of viral vectors for gene therapy applications (Jeffrey et al. 2023).
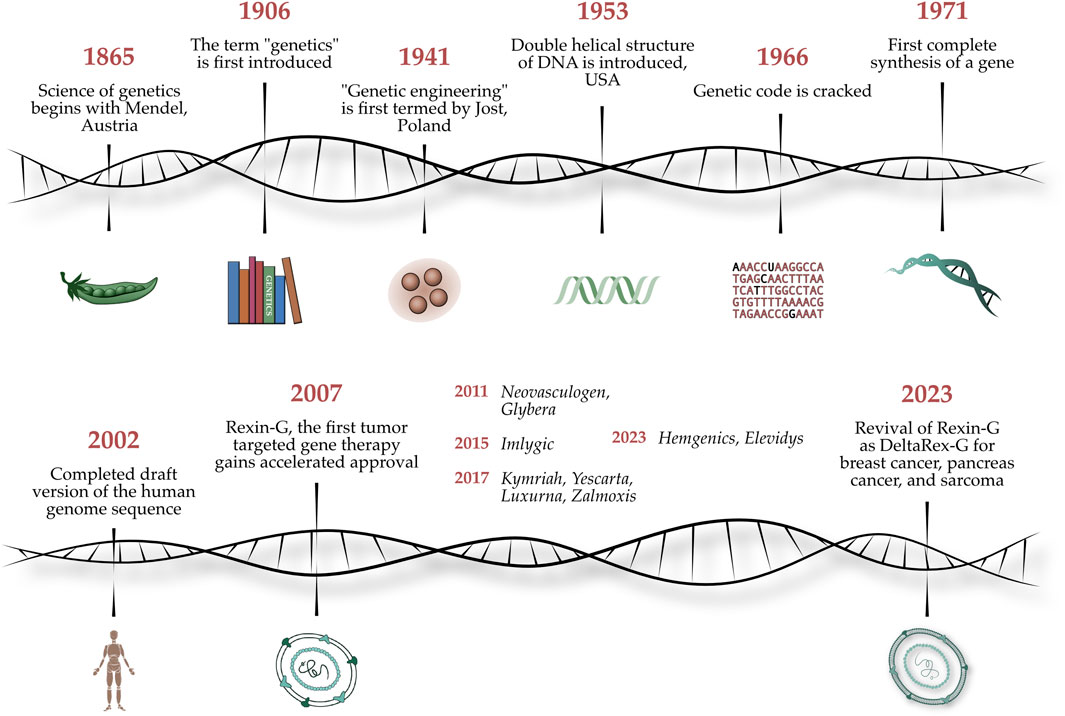
FIGURE 1. Legend: Celebrating Mendel’s 200th Anniversary. An artist’s illustration of the chronology of events from Gregor Mendel’s experiments on pea plants to the discovery of the genetic code to whole genome sequencing, to genetic engineering of viral vectors for gene therapy applications (Jeffrey et al. 2023).
Over the years, the concepts of classical genetics have necessarily been enlarged in scope to include the emergent concepts of chromosome pairs, the DNA double helix, the genetic code, epigenetics, the discovery of split-genes (introns and exons), the vast preponderance of non-coding/regulatory RNA/transcribed sequences within the human genome (Gayon, 2016), and the myriad of executive biochemical pathways (Yasutis and Kozminski, 2013) that regulate mammalian gene expression. Indeed, the availability of advanced DNA sequencing technologies has served to identify the functional gene regulatory elements (including promoters, enhancers, silencers, and insulators) which, along with the cognate transcription factors, regulate and coordinate gene transcription; and yet, when mutated or otherwise dysregulated, can become major drivers of human disease (Chatterjee and Ahituv, 2017).
In time, the identification and characterization of cellular proto-oncogenes, the concept of oncogene addictions, and the loss of tumor suppressors observed in the processes of multistep carcinogenesis—that is, in the initiation, promotion, and progression of metastatic cancers—came to be viewed cytogenetically as heritable traits in the pathogenesis and evolution of solid tumors, seen as the evolving populations of metastatic cancer cells (Al-Shihabi et al., 2018; Grzes et al., 2020). The topical paper presented by Zou et al. demonstrates the power of Whole Genome Sequencing (WGS) and patient-matched samples in a rare, highly recurrent soft tissue sarcoma to examine genomic alterations, clonal evolution, and mutation rates in primary tumors, when compared with metastatic and recurrent tumor samples, to gain insights into the molecular pathogenesis, disease staging, and potential therapeutic strategies.
Among the Research Topic of topical papers presented herein is a timely retrospective review from Gordon and Hall who previously identified the executive components of a functional “proline-directed” protein kinase-mediated signal transduction cascade—from the human EGFR-associated/p38 Mapk14 kinase (Williams et al., 1993), to the human Cdk-activating kinase (Wu et al., 1994a), to the human Cdk2 (CDK2) kinase (Elledge et al., 1992), to the human Cyclin G1 (CYCG1) oncogene (Wu et al., 1994b), to the Cyclin G1-binding p18 (FX3) gene product, aka p18-Hamlet (Xu et al., 2000)—characterizing the Cyclin G1(CCNG1) oncogene as a pivotal and commanding locus: revealing the Cyclin G1 Axis of protein-protein interactions (PPIs) that mediate stem cell activation, genome stability, and sustained survival, or death, in somatic stem cells (Gordon et al., 2018). On the one hand, the mitogenic Cyclin G1/Cdk (Cdk2 or Cdk5)/Myc/Pin1 Axis of PPIs govern early stem cell activation and the biochemical Competence to proliferate, serving as a sustained and sustaining Survival Factor; while genomic stability and DNA-repair mechanisms are governed largely by the Cyclin G1/PP2A/MDM2/p53/p18-Hamlet Axis of interacting proteins, which ensure DNA fidelity at defined cell cycle checkpoints, in coordination with the p53 tumor suppressor (Vousden and Carol Prives, 2009) and with p18-Hamlet, a Cyclin G1-binding phosphoprotein which serves as a molecular sensor of genomic stress within the molecular checks and balances that determine cell fate. (Cuadrado et al., 2007; Lafarga et al., 2007).
The epic translation of gene-based research to applied clinical oncology was facilitated by the development of a broadly cytocidal dominant-negative construct of the Cyclin G1 oncogene (dnG1), along with the molecular engineering of synthetic lesion-targeted gene therapy expression vectors guided by pathotropic targeting (Hall et al., 2010); i.e., by affinity for the exposed anaplastic collagenous extracellular matrix proteins that are characteristic signatures of the tumor microenvironment (Su and Karin, 2023). A testament to perseverance and quantitative analytical investigations in clinical medicine, the first reports of long-term (>10 years) cancer-free survival in patients with advanced, previously intractable, chemo-resistant, metastatic cancers of the pancreas, bone, and breast were formally published in 2021 (Liu et al., 2021), which amounts to somewhat more than an academic milestone. Indeed, the topical papers presented herein by Chawla et al. and Bruckner et al., describe the recent clinical revival, restored cGMP production, the latest regulatory approvals, and current clinical trials of DeltaRex-G for the treatment of advanced pancreatic cancer and sarcoma, and carcinoma of breast, respectively. Finally, the topical article presented by Ticha et al., explores the limits and potential role of minimally invasive localized venous/tumor DNA sampling methodologies to improve the sensitivity and utility of liquid biopsies to monitor biological markers, disease progression, and treatment responses in complex clinical settings.
In summary, the field of genetics has experienced momentous growth from Mendel’s pea plant experiments to the advent of an increasing number of gene therapy products for cancer and genetic disorders.
Author contributions
NF: Writing–review and editing, Writing–original draft. EG: Writing–review and editing. SC: Writing–review and editing. FH: Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
SC, FH and EG are equity members of Counterpoint Biomedica LLC. FH and EG are equity members of Delta Next-Gene, LLC.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Al-Shihabi, A., Chawla, S. P., Hall, F. L., and Gordon, E. M. (2018). Exploiting oncogenic drivers along the CCNG1 pathway for cancer therapy and gene therapy. Mol. Ther. Oncolytics 11, 122–126. doi:10.1016/j.omto.2018.11.002
Chatterjee, S., and Ahituv, N. (2017). Gene regulatory elements, major drivers of human disease. Annu. Rev. Genom. Hum. Genet. 18, 45–63. doi:10.1146/annurev-genom-091416-035537
Cuadrado, A., Lafarga, V., Cheung, P. C., Dolado, I., Llanos, S., Cohen, P., et al. (2007). A new p38 MAP kinase-regulated transcriptional coactivator that stimulates p53-dependent apoptosis. EMBO J. 26, 2115–2126. doi:10.1038/sj.emboj.7601657
Elledge, S. J., Richman, R., Hall, F. L., Williams, R. T., Lodgson, N., and Harker, J. W. (1992). CDK2 encodes a 33-kDa cyclin A-associated protein kinase and is expressed before CDC2 in the cell cycle. Proc. Natl. Acad. Sci. USA. 89, 2907–2911. doi:10.1073/pnas.89.7.2907
Gayon, J. (2016). From Mendel to epigenetics: history of genetics. C.R Biol. 399, 225–230. doi:10.1016/j.crvi.2016.05.009
Gordon, E. M., Ravicz, J. R., Liu, S., Chawla, S. P., and Hall, F. L. (2018). Cell cycle checkpoint control: the cyclin G1/Mdm2/p53 axis emerges as a strategic target for broad-spectrum cancer gene therapy - a review of molecular mechanisms for oncologists. Mol. Clin. Oncol. 9, 115–134. doi:10.3892/mco.2018.1657
Grzes, M., Oron, M., Staszczak, Z., Jaiswar, A., Nowak-Niezgoda, M., and Walerych, D. (2020). A driver never works alone—interplay networks of mutant p53, MYC, RAS, and other universal oncogenic drivers in human cancer. Cancers 12, 1532. doi:10.3390/cancers12061532
Hall, F. L., Levy, J. P., Reed, R. A., Petchpud, W. N., Chua, V. S., Chawla, S. P., et al. (2010). Pathotropic targeting advances clinical oncology: tumor-targeted localization of therapeutic gene delivery. Oncol. Rep. 24, 829–833. doi:10.3892/or.2010.829
Jeffrey, S. L., Brigham, D. A., Chawla, S. P., Federman, N., Hall, F. L., and Gordon, E. M. (2023). From Mendel to gene therapy. Anticancer Res. 43, 4257–4261. doi:10.21873/anticanres.16620
Lafarga, V., Cuadrado, A., and Nebreda, A. R. (2007). p18 Hamlet mediates different p53-dependent responses to DNA damage-inducing agents. Cell Cycle 6, 2319–2322. doi:10.4161/cc.6.19.4741
Liu, S., Chawla, S. P., Bruckner, H., Morse, M. A., Federman, N., Srikureja, A., et al. (2021). Long term survival following DeltaRex-G/DeltaVax tumor-targeted gene therapy for advanced chemotherapy-resistant malignancies: an Academic Milestone. Clin. Oncol. 6, 1–7. Available at: https://clinicsinoncology.com/.
Su, H., and Karin, M. (2023). Multifaceted collagen-DDR1 signaling in cancer. Trends Cell Biol. doi:10.1016/j.tcb.2023.08.003
Vousden, K. H., and Carol Prives, C. (2009). Blinded by the light: the growing complexity of p53. Cell 137, 413–431. doi:10.1016/j.cell.2009.04.037
Williams, R., Sanghera, J., Wu, F., Carbonaro-Hall, D., Campbell, D. L., Warburton, D., et al. (1993). Identification of a human epidermal growth factor receptor-associated protein kinase as a new member of the Mitogen-activated Protein Kinase/Extracellular Signal-regulated Protein Kinase Family. J. Biol. Chem. 268, 18213–18217. doi:10.1016/s0021-9258(17)46832-3
Wu, L., Liu, L., Yee, A., Carbonaro-Hall, D., Tolo, V. T., and Hall, F. L. (1994b). Molecular-cloning of the human cycg1 gene encoding a g-type cyclin - overexpression in human osteosarcoma cells. Oncol. Rep. 1, 705–711. doi:10.3892/or.1.4.705
Wu, L., Yee, A., Liu, L., Carbonaro-Hall, D., Venkatesan, N., Tolo, V. T., et al. (1994a). Molecular cloning of the human CAK1 gene encoding a cyclin-dependent kinase-activating kinase. Oncogene 9, 2089–2096.
Xu, F., Wang, W., and Hall, F. L. (2000). Molecular cloning and characterization of FX3, a novel zinc-finger protein. Oncol. Rep. 7, 995–1001. doi:10.3892/or.7.5.995
Keywords: Cyclin G1, DeltaRex-G, sarcoma, pancreatic cancer, carcinoma of breast, whole genome sequencing, liquid biopsy, gene therapy
Citation: Federman N, Gordon EM, Chawla SP and Hall FL (2024) Editorial: Celebrating the 200th mendel’s anniversary: gene-targeted diagnostics and therapies for cancer. Front. Mol. Med. 4:1366963. doi: 10.3389/fmmed.2024.1366963
Received: 08 January 2024; Accepted: 15 January 2024;
Published: 27 February 2024.
Edited and reviewed by:
Masaru Katoh, National Cancer Center, JapanCopyright © 2024 Federman, Gordon, Chawla and Hall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erlinda M. Gordon, egordon@sarcomaoncology.com
 Noah Federman
Noah Federman