
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 March 2025
Sec. Virology
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1563186
 Dan Li1,2
Dan Li1,2 Laixu Zhu1,2
Laixu Zhu1,2 Chenchen Cui1,2
Chenchen Cui1,2 Zhenchun Wu1,2
Zhenchun Wu1,2 Pengkai Qing1,2
Pengkai Qing1,2 Qiongqiong Zhou1,2*
Qiongqiong Zhou1,2* Peng Gao1,2
Peng Gao1,2 Yongning Zhang1,2
Yongning Zhang1,2 Lei Zhou1,2
Lei Zhou1,2 Xinna Ge1,2
Xinna Ge1,2 Xin Guo1,2
Xin Guo1,2 Jun Han1,2*
Jun Han1,2* Hanchun Yang1,2
Hanchun Yang1,2Introduction: Porcine reproductive and respiratory syndrome virus (PRRSV), an economically significant threat to the world pork production, is notoriously known for its heterogeneity, and therefore the current vaccines often fail to provide efficient cross-protection against diverse PRRSV strains.
Methods: By making chimeric viruses using HP-PRRSV-2 lineage 8 (JXwn06) and lineage 1 NADC30-like strains (CHsx1401) as model organisms, the recently results have shown that the viral structural protein-coding region is critical for induction of homologous immunity. In this study, the chimeric viruses were further constructed by exchanging the region coding for the minor (GP2/3/4) or major (GP5/M) structural proteins of JXwn06 on the backbone of CHsx1401 to generate two mutants CHsx1401-GP234JX and CHsx1401-GP5MJX.
Results: The subsequent animal experiment showed that all three chimeras could confer good protective immunity against the lethal challenge by HP-PRRSV strain JXwn06, and the survived pigs had much lower lung lesions, faster viremia clearance, and lower viral tissue load. However, the exchange of SP region as a whole performed better than either GP2/3/4 or GP5/M region alone, as the pigs in the latter groups showed transient fever following challenge and higher viral load in certain tissues, highlighting a synergistic role. Interestingly, as compared to the group CHsx1401-GP234JX, the group CHsx1401-GP5MJX showed excellent viremia clearance, comparable to the SP group.
Discussion: Our results in this report revealed the important role of ORFs2-4 and ORFs5-6 regions in induction of protective immunity and have important implications in understanding viral pathogenesis and further vaccine development.
Porcine reproductive and respiratory syndrome virus (PRRSV) has remained one of the greatest challenges in modern times to the global pig industry (Zhou et al., 2014; Zhang et al., 2022). This virus emerged almost simultaneously for the first time in both North America and Europe in late 1990s, and the associated diseases in the field are typically manifested as reproductive disorder in sows, respiratory distresses in young piglets, and frequent secondary bacterial infections (Dea et al., 2000; Hanada et al., 2005; Sánchez-Carvajal et al., 2022). As of today, many countries have maintained PRRSV epidemic status, leading to enormous economic losses (Kuhn et al., 2016; Han et al., 2017). As a single-stranded, positive-sense RNA virus, PRRSV is a member of the family Arteriviridae within the order Nidovirales and has a genomic size of about 15 kb that harbors at least 10 open reading frames (ORFs) (Lunney et al., 2010; Zhang et al., 2017; Zhou et al., 2021). Of them, ORF1 encodes viral replicase polyproteins, while ORFs2-7 code for structural proteins (SP) (Kappes and Faaberg, 2015; Zhang et al., 2021). In particular, ORFs2-4 specify the minor structural proteins GP2a, GP3, GP4, and the ion channel protein E (Wissink et al., 2005; Tian et al., 2012; Sun et al., 2013; Luo et al., 2023), whereas ORFs5-6 specify the major structural proteins GP5 and M and a newly discovered protein GP5a with unknown function (Fang and Snijder, 2010; Firth et al., 2011; Johnson et al., 2011). The last ORF7 expresses the viral nucleocapsid protein N (Li et al., 2015; Liu et al., 2018). Among these proteins, GP5 interacts with M to form heterodimers that are necessary for viral attachment, whereas GP2, GP3 and GP4 assemble into heterotrimers that enable virus penetration via engagement with the entry receptor CD163 (Zhou et al., 2009; Lei et al., 2024; Bai et al., 2025; Lim et al., 2025).
The PRRSV is notoriously known for its rapid genetic variation and evolution in the field (Yu et al., 2020). This property has led to the emergence of numerous variants and the quick expansion of viral genetic diversity. For example, in the past 35 years, the PRRSV-2 strains (previous North American type) have evolved into at least 9 lineages via phylogenetic analyses (Han et al., 2006; Brockmeier et al., 2012; Cui et al., 2022). In China, the years 2006–2015 have seen the havoc epidemic of lineage strains, represented by the Chinese highly pathogenic strains (HP-PRRSV), a group of viruses (e.g., JXwn06, HuN4, JXA1, etc.) that had ever devastated the Chinese swine industry (Han et al., 2017). While in recent years, they gradually give away the center stage to the lineage 1 NADC30/34-like strains (e.g., CHsx1401, SD17-38, Anheal-1, etc.), which show a clear genetic divergence greater than 20% at nucleotide level as compared to HP-PRRSV (Bao and Li, 2021; Yuan et al., 2022). Not surprisingly, the current modified live vaccines (MLV) fail to deliver efficient cross-protection against these rising strains (Li et al., 2014). The widespread application of MLV has also led to the selection of frequent recombinants between HP-PRRSV and NADC30/34 strains that potentially enable the virus to escape the preexisting immunity (Sun et al., 2016; Tian et al., 2017; Wang et al., 2018; Cao et al., 2022). To dissect the mechanisms behind the clinical observations, our recent studies have shown that the genetic variation of PRRSV nsp2 replicase protein-coding region is a key factor for modulating viral virulence and persistence, thus providing an explanation on the rise of NADC30 strains in the field, whereas the viral structure protein region (SP) is necessary for inducing efficient homologous immunity via the approach of constructing chimeric viruses (Kong et al., 2023a,b).
This report is a follow-up of our previous work and aims to assess the role of the specific SP regions in induction of homologous immunity. This study started with constructing chimeric mutants by exchanging the region coding for minor glycoproteins (GP2/3/4) or the major glycoproteins (GP5/M) from lineage 8 HP-PRRSV strain JXwn06 in the backbone of lineage 1 NADC30-like strain CHsx1401, followed by animal experiments. Our findings showed that the GP2/3/4 or GP5/M regions work synergistically, while CHsx1401 carrying the whole SP region of JXwn06, but not wild-type CHsx1401, provides the best protection efficacy against the challenge by JXwn06. These findings provide further insight into the structural proteins of PRRSV and their role in providing homologous protection for pigs against HP-PRRSV infection, which has great implications for development of novel PRRSV vaccines. The details are described below.
Primary porcine alveolar macrophages (PAMs) derived from 28-day-old specific pathogen free (SPF) piglets were cultured in RPMI 1640 (Gibco, 61870044) containing 10% fetal bovine serum (FBS; Gibco, 16140071) and 1% penicillin (50 U/mL) and streptomycin (50 mg/mL) at 37°C with 5% CO2. MARC-145 cells (African green monkey kidney epithelial cells, ATCC, CRL-12231) were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, 12491015) containing 10% fetal bovine serum (FBS; Gibco, 16140071) and 1% penicillin (50 U/mL) and streptomycin (50 mg/mL) at 37°C with 5% CO2. The Chinese highly pathogenic PRRSV strain JXwn06 (GenBank accession number: EF641008.1) and the NADC30-like strain CHsx1401 (GenBank accession number: KP861625.1) used in this study have been described previously (Zhou et al., 2009; Zhou et al., 2015).
The full-length infectious genomic clone plasmids pWSK-CHsx1401 and pWSK-JXwn06, including a cytomegalovirus (CMV) promoter, a hepatitis delta virus ribozyme sequence, and an SV40 polyadenylation signal, was successfully constructed, respectively. To facilitate the exchange of the GP2, GP3, and GP4 or GP5 and M regions, the corresponding genomic region sequences from pWSK-JXwn06, which served as the donor, along with the up-and downstream sequences of this region, were individually amplified using PCR with KOD DNA polymerase (TOYOBO, #KFX-101). The resulting products were purified and then cloned into the backbone plasmid pWSK-CHsx1401 that are double digested with the AscI and PacI restriction enzymes using a homologous recombination strategy with the ClonExpress MultiS one-step cloning kit (Vazyme, C113-02). This process ultimately led to the creation of the infectious clone plasmids pCHsx1401-GP234JX and pCHsx1401-GP5MJX. The primers used for the construction of the chimeric clones are listed in Supplementary Table S1. All constructs underwent verification through DNA sequencing and were prepared using the PureYield plasmid miniprep system (Promega, A2492).
The infectious cDNA clone plasmids were transfected into MARC-145 cells utilizing the LTX transfection reagent (Invitrogen, 15338100) following to the manufacturer’s instructions. Briefly, MARC-145 cells were cultured and transfected with plasmid at a density of approximately 80% in 6-well plates. The virus-induced cytopathic effect (CPE) was monitored daily, and the whole cell culture was harvested at 7 days post-transfection approximately. The rescued viruses were confirmed by indirect immunofluorescence assay (IFA) using the PRRSV N protein monoclonal antibody SDOW17 (Rural Technologies, United States). Finally, the RNA samples of chimeric viruses were subsequently extracted, reversed transcribed, and subjected to PCR for further verification through sequencing.
PAMs or MARC-145 cells seeded in 12-well plates were infected with the indicated chimeric viruses at an MOI of 0.1. After incubation at 37°C for 1 h, the cells were washed three times with medium to remove unbound viruses and were then supplemented with either RPMI 1640 or DMEM containing 2% FBS, respectively. At the specified time points post-infection, the medium and cells were harvested for viral titration using an endpoint dilution assay, as described previously (Song et al., 2019). Virus titers were expressed as 50% tissue culture infective dose per mL (TCID50/mL).
A total of twenty-eight 28-day-old SPF piglets were procured from Shiji Pig Breeding & Management facility located in Shouyang County, China. All piglets were tested negative for PRRSV, pseudorabies virus, porcine circovirus type 2, classic swine fever virus, and African swine fever virus using reverse transcription polymerase chain reaction (RT-PCR) and commercial enzyme-linked immunosorbent assay (ELISA) kits for antibodies detection. Animals were housed in separate isolation rooms within the animal facility and were given a three-day acclimatization period prior to the commencement of the experiments. The piglets were randomly divided into 6 groups: CHsx1401 (n = 5), CHsx1401-SPJX (n = 5), CHsx1401-GP234JX (n = 5), CHsx1401-GP5MJX (n = 5), DMEM (n = 5) and a mock group (n = 3). Piglets in the experimental groups received intranasal administration of CHsx1401, CHsx1401-SPJX, CHsx1401-GP234JX, or CHsx1401-GP5MJX at a dosage of 2 × 105 TCID50, while those in the DMEM group were inoculated with DMEM. On the 42 DPI, except for mock group, all immunized piglets were challenged with wild-type JXwn06 strain at a dosage of 2 × 106 TCID50.
Animals were monitored for clinical signs and rectal temperatures daily. The detailed scoring system was implemented according to the method described previously (Li et al., 2014). Briefly, the clinical sign scoring system included gross clinical score (GCS), respiratory clinical score (RCS), and nervous signs score (NSS). The piglets’ weights were recorded weekly to calculate the average daily gain (ADG). Blood samples were collected at specified intervals to evaluate viremia and isolate peripheral blood mononuclear cells (PBMCs). On the 21 DPC (days post challenge), animals were euthanized, and the lungs, tonsils, inguinal lymph nodes (ILNs), and submandibular lymph nodes (SLNs) were collected and stained with hematoxylin and eosin (H&E) for histopathological examination. Gross lung lesions were blindly evaluated, with gross pathology scored on a scale of 100 based on the lesion area. The accessory lobes were assigned 5 points each and 27.5 points (15 for dorsal and 12.5 for ventral) for each of the right and left caudal lobes to reach a total of 100 points. According to the severity of the interstitial pneumonia, the lung microscopic lesions were blindly evaluated with a scale ranging from 0 to 4 (0, no microscopic lesions; 1, mild; 2, moderate multifocal interstitial pneumonia; 3, moderate diffusive interstitial pneumonia; 4, severe interstitial pneumonia).
Blood samples were collected using heparin anticoagulant tubes at specified time intervals post-infection, and PBMCs were isolated using Ficoll–Hypaque density gradient centrifugation according to the manufacturer’s instructions (TBD science, LTS1110). The cell deposit was resuspended in Cryostar freezing medium (CELLSAVINGTM, C40100) and stored in liquid nitrogen for future applications.
Heat inactivated serum samples were diluted at two-fold series with DMEM and subsequently incubated with an equal volume (50 μL) of the corresponding virus at a concentration of 100 TCID50 at 37°C for 1 h. Afterwards, the mixture was incubated with MARC-145 cells at 37°C for another 1 h. The cells were then washed three times with PBS, after which fresh DMEM media supplemented with 2% FBS was added. After a 24 h incubation period, PRRSV-positive cells were detected using IFA with PRRSV N protein antibody. The titer of neutralizing antibodies (NA) was calculated using the Reed-Muench method.
To quantify PRRSV chimeric viruses RNA loads in tissues, total RNAs were extracted with TRIzol (Thermo Fisher, 15596026) according to the manufacturer’s instructions. The cDNAs were synthesized by reverse transcription using the FastKing reverse transcription kit (Tiangen, KR116) with the indicated primers (Supplementary Table S2). To quantify the viral tissue load, a pair of specific primers and a corresponding probe were used to amplify JXwn06 nsp9 gene, and a standard curve was established for the nsp9 gene by plotting log10 copy number against the cycling threshold (C T ) value (γJXCT = −3.234x + 40.043).
GraphPad Prism version 7.0 (La Jolla, CA, United States) was used to perform the statistical analyses (two-tailed unpaired Student’s t-tests). Asterisks indicate statistical significance; ns, no significance; *p < 0.05, **0.001 < p < 0.01, ***p < 0.001, ****p < 0.0001. Error bars indicate ± standard deviation (SD).
Our previous study has demonstrated that the region coding for PRRSV structural proteins (SP) can provide effective homologous immune protection against the challenge by HP-PRRSV JXwn06 strain. To further delineate the specific region involved inducing protective immunity, the NADC30-like strain CHsx1401 was used as a receptor to construct chimeric viral mutants using reverse genetics. The region encoding GP2/3/4 or GP5/M was replaced by that from JXwn06 to make respective chimeric mutants CHsx1401-GP234JX and CHsx1401-GP5MJX (Figure 1A). Additionally, a derivative (CHsx1401-SPJX) carrying only the structural protein (SP) region of the strain JXwn06 was used as a control (Kong et al., 2023a,b). All mutants were successfully recovered upon transfection of the infectious cDNA clone plasmids into MARC-145 cells, an in vitro supporting cell line, and the virus-induced CPE was monitored on a daily basis and characterized by cell detachment and clustering. These viruses were further confirmed via whole genome sequencing and IFA using anti-PRRSV N protein antibody in MARC-145 cells (Figure 1B). Next, the virus growth curve analysis was performed at an MOI of 0.1 in both MARC-145 cells and in porcine primary alveolar macrophages (PAMs). The results showed that the parental viruses JXwn06 and CHsx1401 grew differently in both cell types by that JXwn06 grew better than CHsx1401 in MARC-145 cells while the situation was reversed in PAMs. Exchange of the whole SP region (CHsx1401-SPJX) slightly decreased the replication of CHsx1401 in MARC-145 cells but significantly in PAMs. It seems the region for GP2/3/4 is the determinant as its substitution (CHsx1401-GP234JX), but not GP5/M (CHsx1401-GP5MJX), reduced the virus growth (Figures 1C,D).
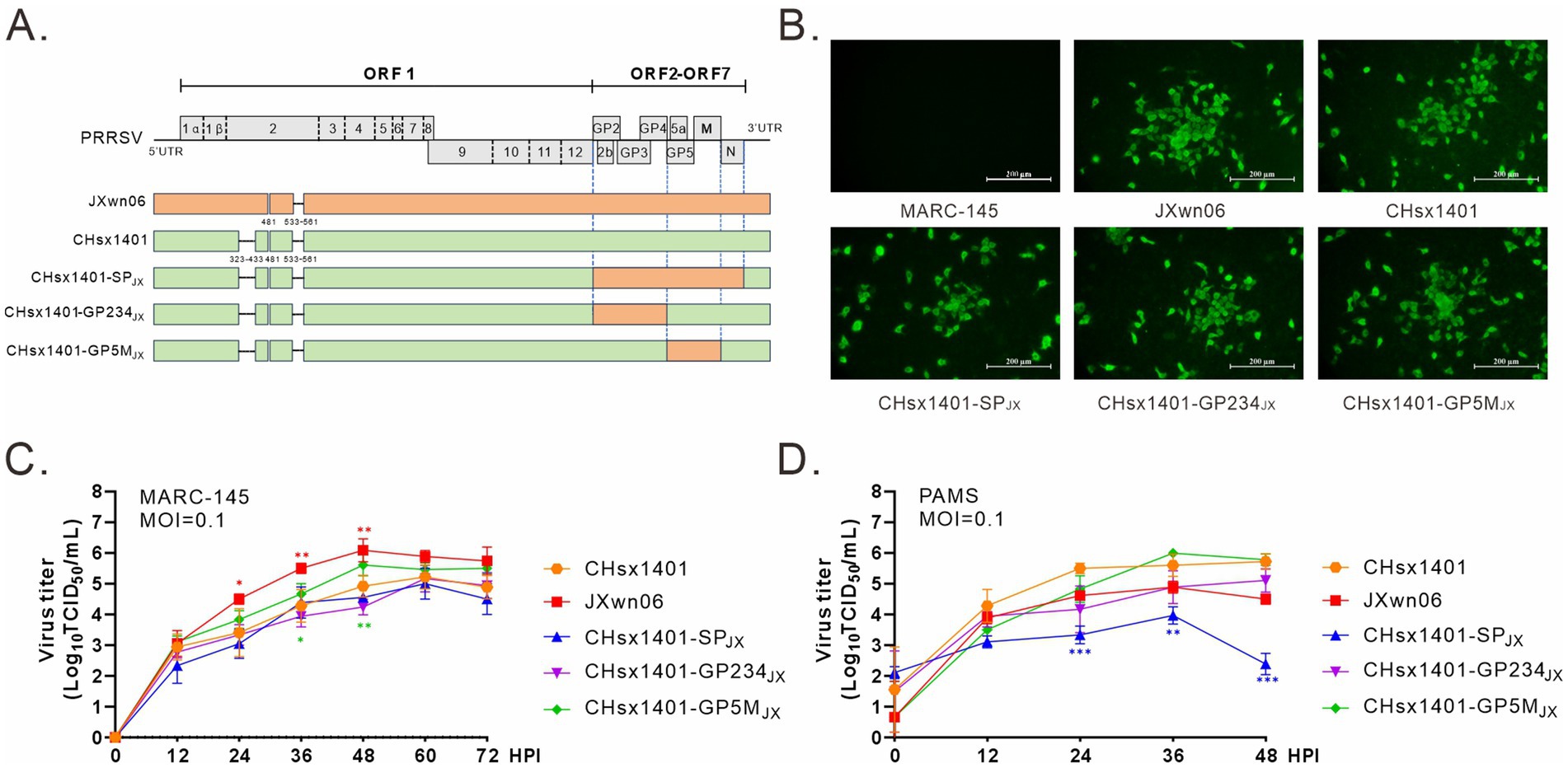
Figure 1. Construction and recovery of PRRSV chimeric mutants. (A) Genome organization and the constructs of PRRSV mutants; UTR, untranslated region. (B) Visualization of cytopathic effects (CPE) using IFA. MARC-145 cells in six-well plates were infected with the indicated viruses at an MOI of 0.1. At 24 h post infection (HPI), the cells were fixed and stained with PRRSV N protein antibody. CPE were visualized using a microscope. (C,D) Multiple-step growth analysis. MARC-145 cells (C) and PAMs (D) were infected with the indicated viruses at an MOI of 0.1. At the specified times intervals, the cells were harvested and subjected to titration by endpoint dilution assay in MARC-145 cells. Statistical analysis was performed using a two-tailed Student’s t-test, and error bars indicate means ± standard deviation (SD). Asterisks (*) indicate the statistical significance: *p < 0.05, **0.001 < p < 0.01, ***p < 0.001.
To evaluate the pathogenic properties of these chimeric viruses, 28-day-old specific pathogen-free (SPF) piglets were randomly divided into 6 groups and intranasally immunized with indicated viruses at a dose of 2 × 105 TCID50 or with DMEM medium as a negative control, and no treatment for mock group. The rectal temperature and clinical symptoms of piglets were monitored daily (Figure 2A). Consistent with previous report, piglets infected with the parental virus CHsx1401 exhibited a transient fever exceeding 40°C on 3 DPI (days post infection) and maintained a good appetite, whereas the group CHsx1401-SPJX displayed elevated temperatures beginning from 1 DPI, peaking at an average of 40.6°C on 2 DPI, accompanied by depression and reduced appetite; however, their body temperature returned back to normal on day 4 DPI. Interestingly, the CHsx1401-GP234JX group showed increased temperature (40.4°C) from 5 DPI and last for about 1 week, while the CHsx1401-GP5MJX group developed slight fever on 6 DPI and, with a maximum average body temperature of approximately 40°C. Throughout the experimental process, individual piglets exhibited a loss of appetite during episodes of fever, which subsequently returned to normal levels (Figure 2B). Additionally, after chimeric viruses infection, all piglets exhibited a moderate coughing, sneezing, lethargic, and a decreased appetite. Among them, the CHsx1401-SPJX group showed a strongest clinical symptom than other groups; and subsequently, appetite returned to normal and respiratory symptoms gradually abated after the temperature dropped at 28 DPI (Figure 2C). In comparison, CHsx1401-GP234JX and CHsx1401-GP5MJX groups displayed a slightly lower average daily gain (ADG) than CHsx1401 and CHsx1401-SPJX groups during the 28 DPI period, whereas a higher ADG in CHsx1401-GP234JX and CHsx1401-GP5MJX groups than CHsx1401 and CHsx1401-SPJX groups was assay after the 28 DPI period (Figure 2D).
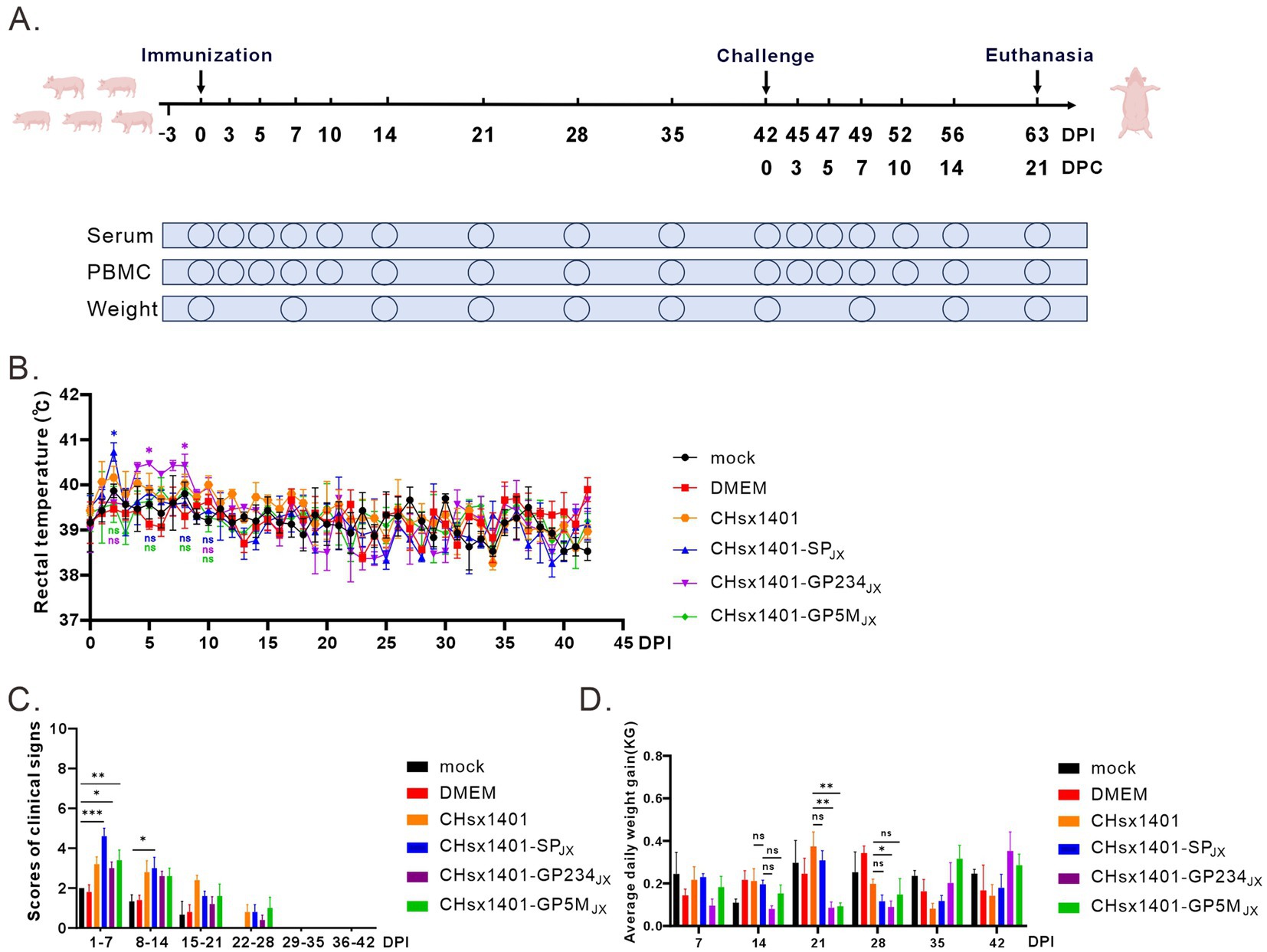
Figure 2. Pathogenicity analysis of PRRSV chimeras in piglets. (A) Scheme of the animal experiment protocol. (B) The rectal temperature curves of piglets after immunization with PRRSV chimeric mutants. (C) The clinical mental state scores of piglets. (D) Average daily weight gain of piglets. Total scores for each piglet represent the sum of the GCS, RCS, and NSS (C,D). Statistical analysis was performed using a two-tailed Student’s t test, and error bars indicate means ± standard deviation (SD). Asterisks (*) indicate the statistical significance: *p < 0.05, **0.001 < p < 0.01, ***p < 0.001, ns, no significance.
The turnover of inoculated chimeric viruses in the blood was investigated. Except of the CHsx1401-SPJX group, viremia was progressively detected in the other piglets at 3 DPI and became detectable for all piglets at 7 DPI. This trend persisted until the 14 DPI. Among these groups, the viral load in the blood of the CHsx1401 group displayed fastest rising, but was slowly and completely cleared until 42 DPI. The mutant CHsx1401-SPJX exhibited the fastest declining kinetics as compared to other groups (Figure 3). The groups of CHsx1401-GP234JX and CHsx1401-GP5MJX showed intermediate phenotype; they reached peaks around 5 DPI-7 DPI, with viral clearance occurring first at 14 DPI for these groups, except for one piglet that cleared the virus at 28 DPI. Nevertheless, the overall clearing trend of CHsx1401-GP234JX group was faster than that of the CHsx1401-GP5MJX group. Both CHsx1401-GP234JX and CHsx1401-GP5MJX were completely cleared at 35 DPI.
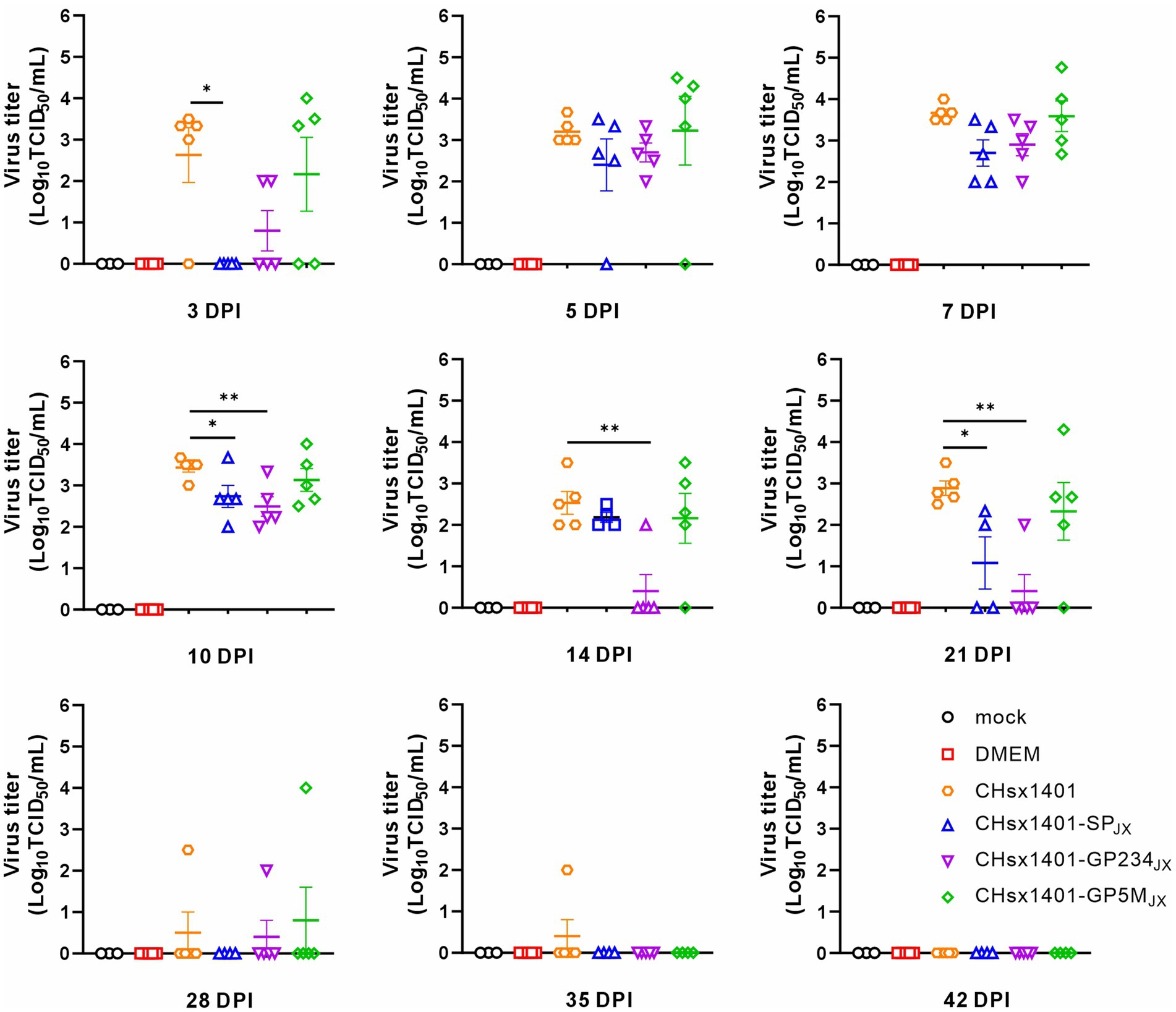
Figure 3. Viremia clearance analysis of chimeric mutants in piglets. The viral load in the blood harvested at various time intervals following infection was determined in MARC-145 cells using endpoint dilution assay. Statistical analysis was performed using a two-tailed Student’s t-test, and error bars indicate means ± standard deviation (SD). Asterisks (*) indicate the statistical significance: *p < 0.05, **0.001 < p < 0.01, ***p < 0.001.
To evaluate the immunoprotective effect of chimeric viruses, all piglets were challenged with the wild-type JXwn06 at 42 DPI, a time point when viremia of all infected piglets became undetectable. As expected, the groups immunized with CHsx1401 and DMEM exhibited the quickest and most notable increase in body temperature, reaching a peak of approximately 41.7°C. This significant rise was accompanied by severe respiratory symptoms, as evidenced by significant signs of coughing, nasal discharge, labored breathing, and a lack of appetite. The DMEM group experienced cyanotic ears, depression, and lost appetite at 7 DPC, and all piglets eventually died at 14 DPC after 10 days of elevated high temperatures. However, the body temperatures of 80% piglets in CHsx1401 immunization group returned to normal and 20% piglets also experienced death (Figures 4A,B). On the other hand, all the piglets in the groups CHsx1401-GP234JX and CHsx1401-GP5MJX survived the challenge, but some pigs experienced a transient increase of the body temperatures to 40.5°C for several days before returning back to baseline. Interestingly, the group CHsx1401-GP5MJX showed longer prolonger higher temperatures than the group CHsx1401-GP234JX (Figure 4A). In contrast, the group CHsx1401-SPJX showed normal temperature fluctuation during the course of challenge and all pigs survived. Overall, there was no significant difference in ADG among CHsx1401-GP234JX, and CHsx1401-GP5MJX immunization groups (Figure 4C), as evidenced by the piglets displaying good mental condition and normal appetites (Figure 4D).
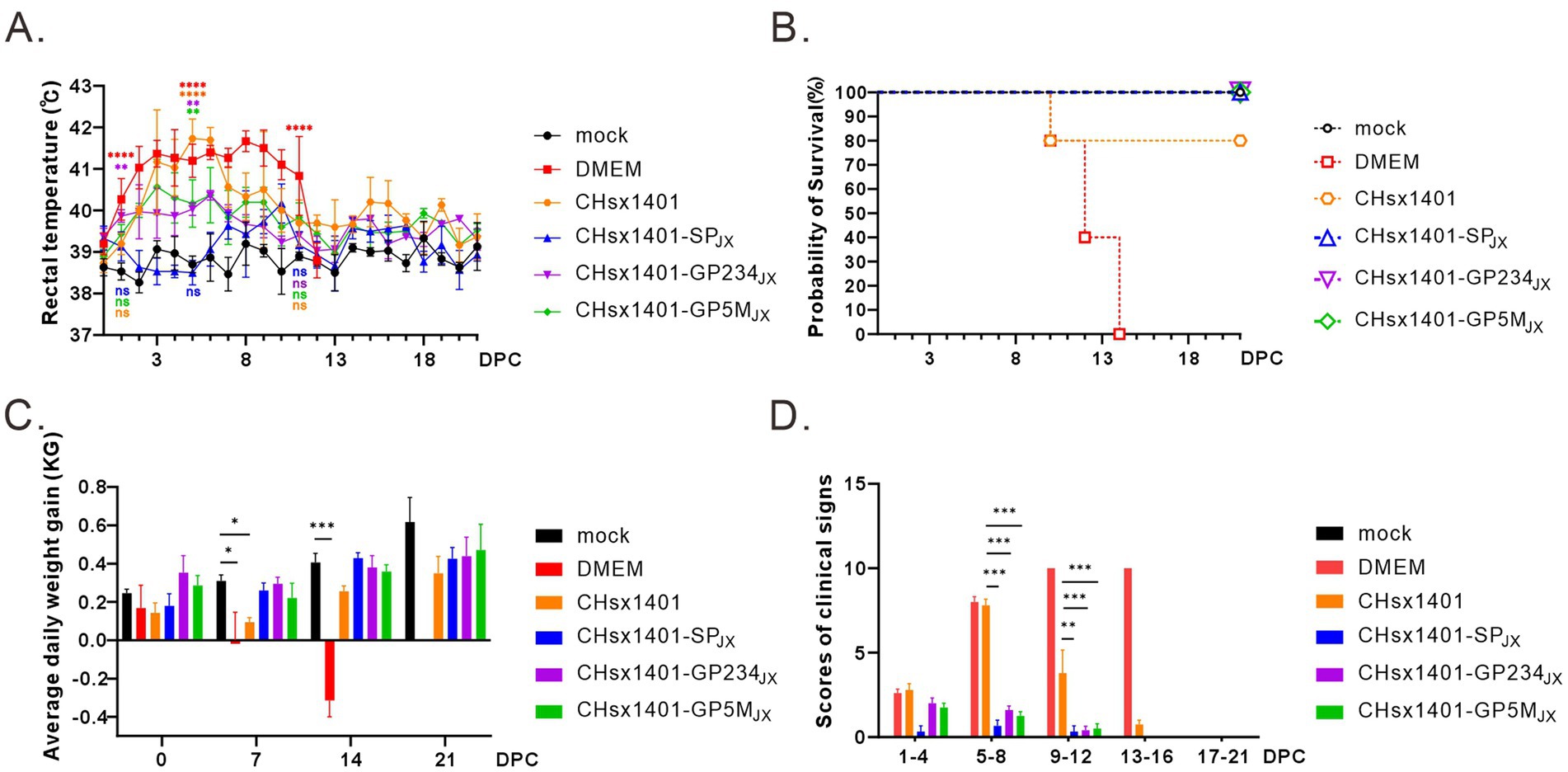
Figure 4. Structural proteins are crucial for the immune protection of HP-PRRSV JXwn06 challenge. (A) The rectal temperature fluctuates following JXwn06 challenge. (B) Survival curve. (C) The average daily weight gain of piglets after the JXwn06 challenge. (D) Clinical mental state scores of different groups. The clinical scoring included the gross clinical score (GCS), respiratory clinical score (RCS), and nervous signs score (NSS). Total scores for each piglet represent the sum of the GCS, RCS, and NSS. Statistical analysis was performed using a two-tailed Student’s t-test, and error bars indicate means ± standard deviation (SD). Asterisks (*) indicate the statistical significance: *p < 0.05, **0.001 < p < 0.01, ***p < 0.001, ****p < 0.0001; ns, no significance.
All the piglets were euthanized and necropsied at 21 DPC. Postmortem examination revealed that, in comparison to the mock group, the DMEM and CHsx1401 group exhibited the most pronounced lung lesions were, displaying varying degrees of pulmonary damage; the CHsx1401-SPJX group displayed the normal or very minor lesion lesions, whereas the CHsx1401-GP5MJX and CHsx1401-GP234JX groups showed the intermediate phenotype (Figures 5A,C). A similar trend was also observed for the microscopic lesions by hematoxylin and eosin (H&E) staining of lungs (Figures 5B,D). The JXwn06 induced much more severe lung injury in DMEM and CHsx1401 immunization groups; with typical lesions that are often seen in HP-PRRSV-infected piglets, such as alveolar hemorrhage, inflammatory cell infiltration, and pulmonary congestion. However, these microscopic symptoms are rarely observed in CHsx1401-SPJX, CHsx1401-GP234JX, and CHsx1401-GP5MJX immunization groups (Figures 5B,D).
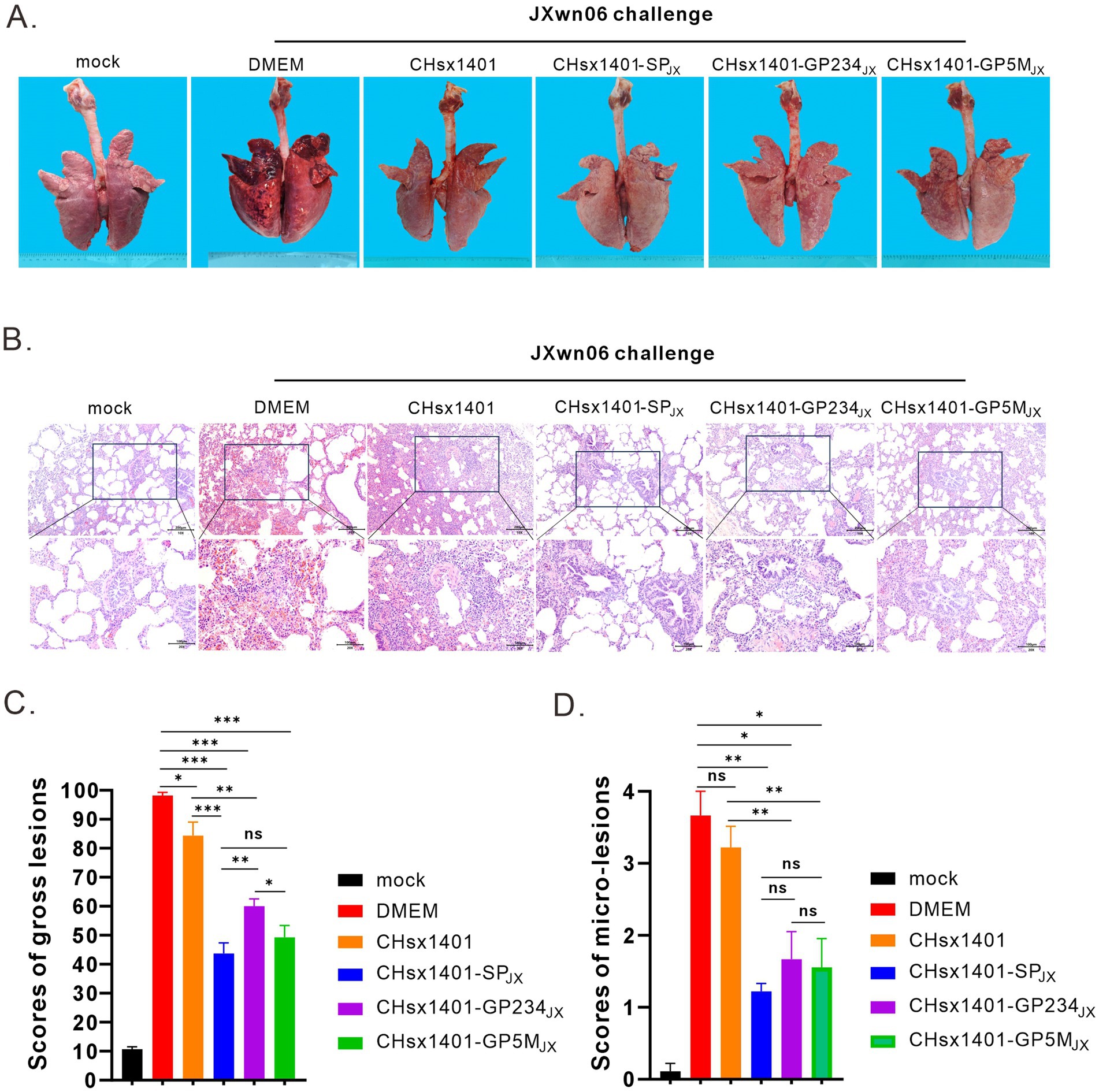
Figure 5. Evaluation of lung lesions and viral tissue load. (A) Representative gross lung lesions of each group at 21 DPC following JXwn06 challenge. (B) Representative images of microscopic lung lesions (H&E staining) at 21 DPC following JXwn06 challenge. (C,D) Mean scores of gross lesions (C) or microscopic lung lesions (D) of each group. Statistical analysis was performed using a two-tailed Student’s t-test, and error bars indicate means ± standard deviation (SD). Asterisks (*) indicate the statistical significance: *p < 0.05, **0.001 < p < 0.01, ***p < 0.001; ns, no significance.
Next, the effect on viral persistence in both blood and tissues was also assessed. The piglets in the groups DMEM and CHsx1401 mounted a significant viremia following challenge by JXwn06 that peaked at around 3 DPC-5 DPC, and the viremia for the group CHsx1401 were not cleared until 14 DPC. In contrast, all three chimera groups showed low level of viremia that were quickly cleared from the blood at 5 DPC-7 DPC. Specifically, the viremia was not detectable at 3 DPC for the groups CHsx1401-SPJX and CHsx1401-GP5MJX, suggesting that the region for GP5/M plays a significant role for viremia clearance (Figure 6A).
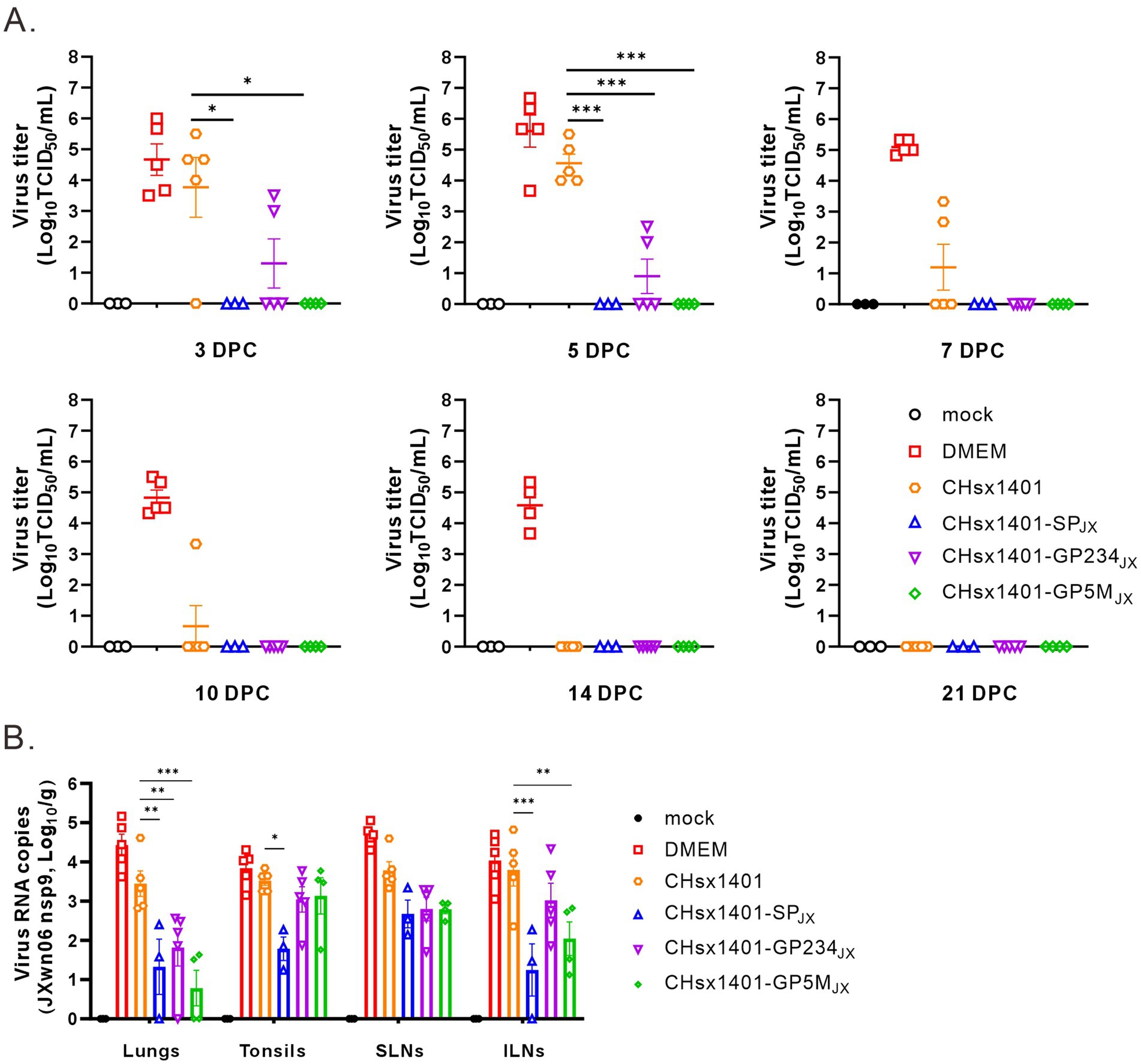
Figure 6. The structural proteins contribute to the clearance of the JXwn06 strain in the blood and the tissues of piglets. (A) Viremia status in the piglets following JXwn06 challenge. (B) Viral loads of JXwn06 strain in lungs and secondary lymphoid tissues were determined using RT-qPCR. Statistical analysis was performed using a two-tailed Student’s t-test, and error bars indicate means ± standard deviation (SD). Asterisks (*) indicate the statistical significance: *p < 0.05, **0.001 < p < 0.01, ***p < 0.001.
We next collected the tissues, including lungs, tonsils, inguinal lymph nodes (ILNs), and submandibular lymph nodes (SLNs) and processed individually for quantitative analyses of viral tissue load by absolute real-time reverse transcription PCR (RT-qPCR) by targeting the nsp9-coding region to differentiate CHsx1401 or its derivatives from JXwn06. Except for the mock control, viral RNAs could be detected in all tissues, but all four immunization groups had the ability to reduce to the viral tissue load of JXwn06 in tissues to varying extents (Figure 6B). Overall, the three chimeric groups showed a better reduction of viral tissue load than the CHsx1401 group, whereas the CHsx1401-SPJX and CHsx1401-GP5MJX groups exhibited the most pronounced reduction of viral load, achieving a decrease by approximate 100 ~ 1,000 fold in the lung and 100 fold in the ILNs compare to DMEM group. As analyzed for the three chimeric groups, CHsx1401-SPJX showed a slightly better or comparable effect for viral tissue load reduction than the groups CHsx1401-GP234JX, and CHsx1401-GP5MJX (Figure 6B). Together, the above findings suggest that the region for GP5 and M proteins has a greater efficiency in clearing the virus form the blood and tissues when compared to the region for GP2/3/4, but the best efficiency requires the whole region of SP.
To examine the humoral immune response induced by chimeric viruses, the serum samples from piglets were collected at indicated time points for analyzing the level of PRRSV antibodies by JNT Porcine reproductive and respiratory syndrome virus (PRRSV) AB ELISA kit. The PRRSV antibodies of all piglets became positive at 10 DPI. Although no significant differences were noted among the various groups, CHsx1401-GP5MJX induced a slightly lower humoral immune response than that of parental viruses CHsx1401. Notably, a discernible trend of antibody re-elevation was evident in all groups after wild-type JXwn06 challenge (Figure 7A).

Figure 7. The effect of exchange of SP on humoral immunity. (A) The levels of PRRSV N protein antibodies. (B,C) The neutralizing antibodies (NA) titer in the serum at 42 DPI (B) or 21 DPC (C) after immunization. Error bars represent standard deviation (SD). Asterisks (*) mean the statistical difference: ns, no significance; *p < 0.05, **0.001 < p < 0.01, ***p < 0.001, ns, no significance.
Subsequently, the neutralization abilities (NA) of antibodies against the CHsx1401 and JXwn06 viruses were evaluated at 42 DPI and 21 DPC in MARC-145 cells. At 42 DPI (before JXwn06 challenge), the neutralizing antibodies generated by CHsx1401 exhibit a pronounced neutralizing ability against the CHsx1401 strain, but almost no neutralizing ability against the JXwn06 strain; however, the neutralizing antibodies induced by CHsx1401-SPJX, CHsx1401-GP5MJX and CHsx1401-GP234JX showed substantial neutralizing efficacy against both CHsx1401 and JXwn06 strains. There was no statistically significant difference observed among CHsx1401-SPJX, CHsx1401-GP5MJX and CHsx1401-GP234JX in terms of their neutralizing antibody responses against either strain (p > 0.05) (Figure 7B). At the 21 DPC, the levels of neutralizing antibody against CHsx1401 elicited by the chimeric virus did not exhibit a significant increase compared to the levels observed at 42 DPI (before JXwn06 challenge); however, there was a slight increase in the neutralizing antibody titer against JXwn06 relative to the levels at 42 DPI. Among the three chimeric viruses, CHsx1401-SPJX elicited the highest level of neutralizing antibodies against JXwn06, followed by CHsx1401-GP5MJX, and CHsx1401-GP234JX was the weakest (p < 0.05) (Figure 7C).
Rapid evolution of PRRSV in the field represents a major challenge for PRRSV-associated disease control and prevention. The high genetic variability has rendered current MLV vaccines less efficient to provide cross-protection against the challenge of various field strains (Zhang et al., 2023). Hence, looking for the viral genetic determinants for inducing homologous immunity has been the focus for the PRRSV community. Our recent studies by making chimeric mutants have demonstrated that the viral structural protein-coding region (SP) is responsible for inducing homologous protective immunity. In this study, we went further to show that the regions for GP5/M and for GP2/3/4 play a synergistic role in inducing protective immunity and the best efficacy requires the whole SP region. The relevant significance and insights are discussed below.
The past 30 years have seen extensive efforts to understand the PRRSV-induced homologous and heterologous immunity or develop vaccines with broad cross-protection efficacy by making chimeric PRRSV mutants via exchange of certain specific SP regions or synthesized consensus sequences of different PRRSV strains (Tian et al., 2017; Park et al., 2021). So far, diverse viruses have been used as the acceptor (backbone), and they are from several PRRSV-2 lineages (L1, L5, and L8), and these include VR-2332 (Sun et al., 2016), FL12 (Vu et al., 2015; Shabir et al., 2016; Jeong et al., 2021; Choi et al., 2021), JSTZ1712-12 (HP-PRRSV, L8) (Chen et al., 2021; Li et al., 2022), GD (HP-PRRSV) (Li et al., 2024), Ingelvac PRRS MLV (Ellingson et al., 2010), VR2385 (Tian et al., 2017; Ni et al., 2013), and APRRS (very close to Primevac vaccine) (Li et al., 2008), and some from PRRSV-1 [SD01-08 (European PRRSV)] (Kimpston-Burkgren et al., 2017). Despite the result variations from different experiments from different laboratories, the results generate lots of useful information for understanding viral pathogenesis and cross-protection immunity. Overall, it is clear that the SP region plays an important role in inducing homologous immunity, although a definitive role of specific SP region is still under some debate, and the genetic variations often lead to reduce or poor cross protection efficiency or poor viral clearance, the consequence of which frequently results in generation of viral recombinants, leading to PRRS outbreaks (Li et al., 2017; van Geelen et al., 2018). However, there are two unfortunate caveats in most of these studies: either the viruses used for the challenge experiment are not virulent enough to cause the pig death, or the acceptor and challenge viruses are not heterologous enough. In this situation, it is necessary to use a highly virulent virus as a challenge model organism to further investigate the role of viral structural protein in induction of protective immunity. Hence, we used two genetically distant virus, NADC30-like strain CHsx1401 (lineage 1) and HP-PRRSV JXwn06 (lineage 8), as the respective acceptor and donor viruses. Genetically, their structural protein coding regions show a difference of about 28.25% at amino acid level. Moreover, we recently showed that immunization of piglets with CHsx1401 does not provide efficient protection against the challenge by HP-PRRSV strain JXwn06.
Our assessment started with a previous study by construction of chimeric viruses via substitution of SP of CHsx1401 with that of JXwn06 in the backbone of CHsx1401 to generate mutant CHsx1401-SPJX (Kong et al., 2023a). It has been showed that immunization of pigs with CHsx1401-SPJX can protect pigs from lethal challenge against JXwn06. The objective of this study was to further assess the individual role of the minor structural proteins GP2/3/4 and the major structural proteins GP5/M in induction of protective immunity. The challenge model was quite successful as the pigs in the group CHsx1401-SPJX all survived the lethal challenge by JXwn06 and exhibited normal temperate fluctuation whereas the death rate in the DMEM group was 100% and all the pigs died within 14 DPC. One significant finding from this experiment is that the exchange of either GP2/3/4 or GP5/M region alone is sufficient to provide clinical protection against the lethal challenge by JXwn06 in terms of the pig survival rate. In addition, immunization with the two chimeras significantly reduced clinical sign and tissue injury. It should be noted that some tissue damages in the lungs can still be seen in the chimera groups; however, these can be due to that the viruses used for immunization still retain virulence and could cause damage during the immunization stage. The second interesting finding is that GP5/M region appears to play a significant role in viremia clearance, and the pigs in this group did not mount an effective viremia. This result is also consistent with a previous observation (Sun et al., 2016). In contrast, two pigs in the GP2/3/4 group showed certain amount of viremia. Overall, the chimera groups exhibited a higher efficient and much faster viremia clearance than the CHsx1401 group. It should be noted that, despite great genetic difference, the immunization with CHsx1401 can provide partial protection as 80% of the pigs survived the lethal challenge and immunized pigs had a faster viremia clearance kinetics than the DMEM group, although much slower than the chimera groups.
In conclusion, the results in this report suggest that the SP region as a whole performs better than either GP2/3/4 or GP5/M region alone in induction of homologous immunity, whereas either of two regions can induce sufficient amount of protective immunity. These findings have important implications for understanding protective immunity and in further vaccine design and development.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
The animal study was approved by the Laboratory Animal Ethics Committee of China Agricultural University (license number: AW72903202-1-3). The study was conducted in accordance with the local legislation and institutional requirements.
DL: Data curation, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. LZhu: Writing – original draft, Methodology, Software. CC: Investigation, Methodology, Software, Writing – review & editing. ZW: Methodology, Resources, Software, Validation, Writing – original draft. PQ: Resources, Software, Writing – original draft. QZ: Data curation, Methodology, Supervision, Validation, Writing – original draft, Writing – review & editing. PG: Investigation, Methodology, Resources, Writing – review & editing. YZ: Formal analysis, Supervision, Writing – review & editing. LZho: Conceptualization, Data curation, Resources, Writing – review & editing. XGe: Data curation, Supervision, Validation, Writing – review & editing. XGu: Data curation, Resources, Supervision, Writing – review & editing. JH: Conceptualization, Data curation, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. HY: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Visualization, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32025035), the National Key Research and Development Program of China (2024YFD1800503), the Key Project of the National Natural Science Foundation of China (32330106), China Agriculture Research System of MOF and MARA (CARS-35), and 2115-Talent Development Program of China Agricultural University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1563186/full#supplementary-material
Bai, Y.-Z., Wang, S., Sun, Y., Liu, Y.-G., Zhang, H.-L., Wang, Q., et al. (2025). The full-length nsp2 replicase contributes to viral assembly in highly pathogenic PRRSV-2. J. Virol. 99:e0182124. doi: 10.1128/jvi.01821-24
Bao, H., and Li, X. (2021). Emergence and spread of NADC34-like PRRSV in China. Transbound. Emerg. Dis. 68, 3005–3008. doi: 10.1111/tbed.14316
Brockmeier, S. L., Loving, C. L., Vorwald, A. C., Kehrli, M. E., Baker, R. B., Nicholson, T. L., et al. (2012). Genomic sequence and virulence comparison of four type 2 porcine reproductive and respiratory syndrome virus strains. Virus Res. 169, 212–221. doi: 10.1016/j.virusres.2012.07.030
Cao, Z., Chen, J., Li, L., Liu, J., Tong, W., Zhou, Y., et al. (2022). A rescued NADC30-like virus by reverse genetic manipulation exhibits moderate virulence and a promising application perspective. Virus Res. 316:198801. doi: 10.1016/j.virusres.2022.198801
Chen, N., Li, S., Tian, Y., Li, X., Li, S., Li, J., et al. (2021). Chimeric HP-PRRSV2 containing an ORF2-6 consensus sequence induces antibodies with broadly neutralizing activity and confers cross protection against virulent NADC30-like isolate. Vet. Res. 52:74. doi: 10.1186/s13567-021-00944-8
Choi, H. Y., Lee, S. H., Ahn, S. H., Choi, J. C., Jeong, J. Y., Lee, B. J., et al. (2021). A chimeric porcine reproductive and respiratory syndrome virus (PRRSV)-2 vaccine is safe under international guidelines and effective both in experimental and field conditions. Res. Vet. Sci. 135, 143–152. doi: 10.1016/j.rvsc.2021.01.012
Cui, X., Xia, D., Huang, X., Sun, Y., Shi, M., Zhang, J., et al. (2022). Analysis of recombinant characteristics based on 949 PRRSV-2 genomic sequences obtained from 1991 to 2021 shows that viral multiplication ability contributes to dominant recombination. Microbiol. Spectr. 10:e0293422. doi: 10.1128/spectrum.02934-22
Dea, S., Gagnon, C. A., Mardassi, H., Pirzadeh, B., and Rogan, D. (2000). Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the north American and European isolates. Arch. Virol. 145, 659–688. doi: 10.1007/s007050050662
Ellingson, J. S., Wang, Y., Layton, S., Ciacci-Zanella, J., Roof, M. B., and Faaberg, K. S. (2010). Vaccine efficacy of porcine reproductive and respiratory syndrome virus chimeras. Vaccine 28, 2679–2686. doi: 10.1016/j.vaccine.2009.12.073
Fang, Y., and Snijder, E. J. (2010). The PRRSV replicase: exploring the multifunctionality of an intriguing set of non-structural proteins. Virus Res. 154, 61–76. doi: 10.1016/j.virusres.2010.07.030
Firth, A. E., Zevenhoven-Dobbe, J. C., Wills, N. M., Go, Y. Y., Balasuriya, U. B. R., Atkins, J. F., et al. (2011). Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. J. Gen. Virol. 92, 1097–1106. doi: 10.1099/vir.0.029264-0
Han, J., Wang, Y., and Faaberg, K. S. (2006). Complete genome analysis of RFLP 184 isolates of porcine reproductive and respiratory syndrome virus. Virus Res. 122, 175–182. doi: 10.1016/j.virusres.2006.06.003
Han, J., Zhou, L., Ge, X., Guo, X., and Yang, H. (2017). Pathogenesis and control of the Chinese highly pathogenic porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 209, 30–47. doi: 10.1016/j.vetmic.2017.02.020
Hanada, K., Suzuki, Y., Nakane, T., Hirose, O., and Gojobori, T. (2005). The origin and evolution of porcine reproductive and respiratory syndrome viruses. Mol. Biol. Evol. 22, 1024–1031. doi: 10.1093/molbev/msi089
Jeong, C. G., Khatun, A., Nazki, S., Kim, S. C., Noh, Y. H., Kang, S. C., et al. (2021). Evaluation of the cross-protective efficacy of a chimeric PRRSV vaccine against two genetically diverse PRRSV2 field strains in a reproductive model. Vaccine 9:1258. doi: 10.3390/vaccines9111258
Johnson, C. R., Griggs, T. F., Gnanandarajah, J., and Murtaugh, M. P. (2011). Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J. Gen. Virol. 92, 1107–1116. doi: 10.1099/vir.0.030213-0
Kappes, M. A., and Faaberg, K. S. (2015). PRRSV structure, replication and recombination: origin of phenotype and genotype diversity. Virology 479-480, 475–486. doi: 10.1016/j.virol.2015.02.012
Kimpston-Burkgren, K., Correas, I., Osorio, F. A., Steffen, D., Pattnaik, A. K., Fang, Y., et al. (2017). Relative contribution of porcine reproductive and respiratory syndrome virus open reading frames 2-4 to the induction of protective immunity. Vaccine 35, 4408–4413. doi: 10.1016/j.vaccine.2017.06.061
Kong, C., Li, D., Hu, Y., Gao, P., Zhang, Y., Zhou, L., et al. (2023a). The genetic variation of porcine reproductive and respiratory syndrome virus Replicase protein nsp2 modulates viral virulence and persistence. J. Virol. 97:e0168922. doi: 10.1128/jvi.01689-22
Kong, C., Li, D., Hu, Y., Gao, P., Zhang, Y., Zhou, L., et al. (2023b). The replicase protein nsp2 of Chinese highly pathogenic porcine reproductive and respiratory virus is involved in protective immunity by promoting viral clearance. One Health Adv. 1, 1–16. doi: 10.1186/s44280-023-00026-8
Kuhn, J. H., Lauck, M., Bailey, A. L., Shchetinin, A. M., Vishnevskaya, T. V., Bào, Y., et al. (2016). Reorganization and expansion of the nidoviral family Arteriviridae. Arch. Virol. 161, 755–768. doi: 10.1007/s00705-015-2672-z
Lei, X., Jiang, Y., Yu, W., Chen, X., Qin, Y., Wang, N., et al. (2024). Intermolecular disulfide bond of PRRSV GP5 and M facilitates VLPs secretion and cell binding. Vet. Microbiol. 298:110249. doi: 10.1016/j.vetmic.2024.110249
Li, X., Bao, H., Wang, Y., and Tian, K. (2017). Widespread of NADC30-like PRRSV in China: another Pandora's box for Chinese pig industry as the outbreak of highly pathogenic PRRSV in 2006? Infect. Genet. Evol 49, 12–13. doi: 10.1016/j.meegid.2016.12.021
Li, J., Li, S., Qiu, M., Li, X., Li, C., Feng, B., et al. (2022). Minor and major envelope proteins of PRRSV play synergistic roles in inducing heterologous neutralizing antibodies and conferring cross protection. Virus Res. 315:198789. doi: 10.1016/j.virusres.2022.198789
Li, Y., Tas, A., Sun, Z., Snijder, E. J., and Fang, Y. (2015). Proteolytic processing of the porcine reproductive and respiratory syndrome virus replicase. Virus Res. 202, 48–59. doi: 10.1016/j.virusres.2014.12.027
Li, Y., Wang, Y., Pei, X., Chen, S., Jing, Y., Wu, Y., et al. (2024). A chimeric strain of porcine reproductive and respiratory syndrome virus 2 derived from HP-PRRSV and NADC30-like PRRSV confers cross-protection against both strains. Vet. Res. 55:132. doi: 10.1186/s13567-024-01390-y
Li, X., Zhang, J., Lü, J., Yu, D., Yao, H., and Yuan, S. (2008). Construction and application of chimeric infectious clones of porcine reproductive and respiratory syndrome virus. Sheng Wu Gong Cheng Xue Bao 24, 1573–1581. (in Chinese).
Li, Y., Zhou, L., Zhang, J., Ge, X., Zhou, R., Zheng, H., et al. (2014). Nsp9 and Nsp10 contribute to the fatal virulence of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China. PLoS Pathog. 10:e1004216. doi: 10.1371/journal.ppat.1004216
Lim, B., Kim, S. C., Kim, H. J., Kim, J. H., Seo, Y. J., Lim, C., et al. (2025). Single-cell transcriptomics of bronchoalveolar lavage during PRRSV infection with different virulence. Nat. Commun. 16:1112. doi: 10.1038/s41467-024-54676-2
Liu, Y., Hu, Y., Chai, Y., Liu, L., Song, J., Zhou, S., et al. (2018). Identification of non-structural protein 8 as the N-terminus of the RNA-dependent RNA polymerase of porcine reproductive and respiratory syndrome virus. Virol. Sin. 33, 429–439. doi: 10.1007/s12250-018-0054-x
Lunney, J. K., Benfield, D. A., and Rowland, R. R. (2010). Porcine reproductive and respiratory syndrome virus: an update on an emerging and re-emerging viral disease of swine. Virus Res. 154, 1–6. doi: 10.1016/j.virusres.2010.10.009
Luo, Q., Zheng, Y., He, Y., Li, G., Zhang, H., Sha, H., et al. (2023). Genetic variation and recombination analysis of the GP5 (GP5a) gene of PRRSV-2 strains in China from 1996 to 2022. Front. Microbiol. 14:1238766. doi: 10.3389/fmicb.2023.1238766
Ni, Y. Y., Opriessnig, T., Zhou, L., Cao, D., Huang, Y. W., Halbur, P. G., et al. (2013). Attenuation of porcine reproductive and respiratory syndrome virus by molecular breeding of virus envelope genes from genetically divergent strains. J. Virol. 87, 304–313. doi: 10.1128/JVI.01789-12
Park, C., Lee, M. S., Baek, J. H., Cho, S. H., Hyun, B. H., You, S. H., et al. (2021). Intradermal co-inoculation of codon pair deoptimization (CPD)-attenuated chimeric porcine reproductive and respiratory syndrome virus (PRRSV) with toll like receptor (TLR) agonists enhanced the protective effects in pigs against heterologous challenge. Vet. Microbiol. 256:109048. doi: 10.1016/j.vetmic.2021.109048
Sánchez-Carvajal, J. M., Rodríguez-Gómez, I. M., Ruedas-Torres, I., Zaldívar-López, S., Larenas-Muñoz, F., Bautista-Moreno, R., et al. (2022). Time series transcriptomic analysis of Bronchoalveolar lavage cells from piglets infected with virulent or low-virulent porcine reproductive and respiratory syndrome virus 1. J. Virol. 96:e0114021. doi: 10.1128/JVI.01140-21
Shabir, N., Khatun, A., Nazki, S., Kim, B., Choi, E. J., Sun, D., et al. (2016). Evaluation of the cross-protective efficacy of a chimeric porcine reproductive and respiratory syndrome virus constructed based on two field strains. Viruses 8:240. doi: 10.3390/v8080240
Song, J., Gao, P., Kong, C., Zhou, L., Ge, X., Guo, X., et al. (2019). The nsp2 hypervariable region of porcine reproductive and respiratory syndrome virus strain JXwn06 is associated with viral cellular tropism to primary porcine alveolar macrophages. J. Virol. 93, e01436–e01419. doi: 10.1128/JVI.01436-19
Sun, D., Khatun, A., Kim, W. I., Cooper, V., Cho, Y. I., Wang, C., et al. (2016). Attempts to enhance cross-protection against porcine reproductive and respiratory syndrome viruses using chimeric viruses containing structural genes from two antigenically distinct strains. Vaccine 34, 4335–4342. doi: 10.1016/j.vaccine.2016.06.069
Sun, L., Li, Y., Liu, R., Wang, X., Gao, F., Lin, T., et al. (2013). Porcine reproductive and respiratory syndrome virus ORF5a protein is essential for virus viability. Virus Res. 171, 178–185. doi: 10.1016/j.virusres.2012.11.005
Tian, D., Cao, D., Lynn Heffron, C., Yugo, D. M., Rogers, A. J., Overend, C., et al. (2017). Enhancing heterologous protection in pigs vaccinated with chimeric porcine reproductive and respiratory syndrome virus containing the full-length sequences of shuffled structural genes of multiple heterologous strains. Vaccine 35, 2427–2434. doi: 10.1016/j.vaccine.2017.03.046
Tian, D., Wei, Z., Zevenhoven-Dobbe, J. C., Liu, R., Tong, G., Snijder, E. J., et al. (2012). Arterivirus minor envelope proteins are a major determinant of viral tropism in cell culture. J. Virol. 86, 3701–3712. doi: 10.1128/JVI.06836-11
van Geelen, A. G. M., Anderson, T. K., Lager, K. M., Das, P. B., Otis, N. J., Montiel, N. A., et al. (2018). Porcine reproductive and respiratory disease virus: evolution and recombination yields distinct ORF5 RFLP 1-7-4 viruses with individual pathogenicity. Virology 513, 168–179. doi: 10.1016/j.virol.2017.10.002
Vu, H. L., Ma, F., Laegreid, W. W., Pattnaik, A. K., Steffen, D., Doster, A. R., et al. (2015). A synthetic porcine reproductive and respiratory syndrome virus strain confers unprecedented levels of heterologous protection. J. Virol. 89, 12070–12083. doi: 10.1128/JVI.01657-15
Wang, H. M., Liu, Y. G., Tang, Y. D., Liu, T. X., Zheng, L. L., Wang, T. Y., et al. (2018). A natural recombinant PRRSV between HP-PRRSV JXA1-like and NADC30-like strains. Transbound. Emerg. Dis. 65, 1078–1086. doi: 10.1111/tbed.12852
Wissink, E. H., Kroese, M. V., van Wijk, H. A., Rijsewijk, F. A., Meulenberg, J. J., and Rottier, P. J. (2005). Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. J. Virol. 79, 12495–12506. doi: 10.1128/JVI.79.19.12495-12506.2005
Yu, F., Yan, Y., Shi, M., Liu, H. Z., Zhang, H. L., Yang, Y. B., et al. (2020). Phylogenetics, genomic recombination, and NSP2 polymorphic patterns of porcine reproductive and respiratory syndrome virus in China and the United States in 2014-2018. J. Virol. 94, e01813–e01819. doi: 10.1128/JVI.01813-19
Yuan, L., Zhu, Z., Fan, J., Liu, P., Li, Y., Li, Q., et al. (2022). High pathogenicity of a Chinese NADC34-like PRRSV on pigs. Microbiol. Spectr. 10:e0154122. doi: 10.1128/spectrum.01541-22
Zhang, Q., Bai, J., Hou, H., Song, Z., Zhao, Y., and Jiang, P. (2017). A novel recombinant porcine reproductive and respiratory syndrome virus with significant variation in cell adaption and pathogenicity. Vet. Microbiol. 208, 150–158. doi: 10.1016/j.vetmic.2017.07.028
Zhang, J., Bai, J., Sun, Y., Liu, X., Gao, Y., Wang, X., et al. (2022). Comparison of pathogenicity of different subgenotype porcine reproductive and respiratory syndrome viruses isolated in China. Microb. Pathog. 168:105607. doi: 10.1016/j.micpath.2022.105607
Zhang, M., Han, X., Osterrieder, K., and Veit, M. (2021). Palmitoylation of the envelope membrane proteins GP5 and M of porcine reproductive and respiratory syndrome virus is essential for virus growth. PLoS Pathog. 17:e1009554. doi: 10.1371/journal.ppat.1009554
Zhang, H., Luo, Q., He, Y., Zheng, Y., Sha, H., Li, G., et al. (2023). Research Progress on the development of porcine reproductive and respiratory syndrome vaccines. Vet. Sci. 10:491. doi: 10.3390/vetsci10080491
Zhou, L., Chen, S., Zhang, J., Zeng, J., Guo, X., Ge, X., et al. (2009). Molecular variation analysis of porcine reproductive and respiratory syndrome virus in China. Virus Res. 145, 97–105. doi: 10.1016/j.virusres.2009.06.014
Zhou, L., Ge, X., and Yang, H. (2021). Porcine reproductive and respiratory syndrome modified live virus vaccine: a “leaky” vaccine with debatable efficacy and safety. Vaccine 9:362. doi: 10.3390/vaccines9040362
Zhou, L., Wang, Z., Ding, Y., Ge, X., Guo, X., and Yang, H. (2015). NADC30-like strain of porcine reproductive and respiratory syndrome virus, China. Emerg. Infect. Dis. 21, 2256–2257. doi: 10.3201/eid2112.150360
Zhou, L., Yang, X., Tian, Y., Yin, S., Geng, G., Ge, X., et al. (2014). Genetic diversity analysis of genotype 2 porcine reproductive and respiratory syndrome viruses emerging in recent years in China. Bio Med Res. Int. 2014:8068. doi: 10.1155/2014/748068
Keywords: porcine reproductive and respiratory syndrome virus, structural proteins, homologous protection, pathogenicity, vaccine development
Citation: Li D, Zhu L, Cui C, Wu Z, Qing P, Zhou Q, Gao P, Zhang Y, Zhou L, Ge X, Guo X, Han J and Yang H (2025) The role of major and minor structural proteins of porcine reproductive and respiratory syndrome virus in induction of protective immunity. Front. Microbiol. 16:1563186. doi: 10.3389/fmicb.2025.1563186
Received: 19 January 2025; Accepted: 05 March 2025;
Published: 19 March 2025.
Edited by:
Hongjun Chen, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Shao-Lun Zhai, Guangdong Academy of Agricultural Sciences, ChinaCopyright © 2025 Li, Zhu, Cui, Wu, Qing, Zhou, Gao, Zhang, Zhou, Ge, Guo, Han and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Han, aGFueDAxNThAY2F1LmVkdS5jbg==; Qiongqiong Zhou, WmhvdXFxQGNhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.