
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 11 April 2025
Sec. Microbiotechnology
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1559242
This article is part of the Research TopicRecent Insights on Thermophilic Anaerobic BacteriaView all 3 articles
 Yu-Ying Huang1,2,3,4†
Yu-Ying Huang1,2,3,4† Dan Zhu1,5,6†
Dan Zhu1,5,6† Li-Quan Yang1,3,5
Li-Quan Yang1,3,5 Maite Ortúzar3
Maite Ortúzar3 Zheng-Feng Yang1,6
Zheng-Feng Yang1,6 Zhi-Hua Lv1,5,6
Zhi-Hua Lv1,5,6 Kai-Qing Xie1
Kai-Qing Xie1 Hong-Chen Jiang2,7
Hong-Chen Jiang2,7 Wen-Jun Li1,3,4*
Wen-Jun Li1,3,4* Yi-Rui Yin1,3,4,5*
Yi-Rui Yin1,3,4,5*Introduction: As a renewable alternative to fossil fuels, second-generation bioethanol production relies heavily on efficient lignocellulose conversion, with β-glucosidase playing a critical role.
Methods: This study focused on the β-glucosidase gene y50bg4 discovered in the Tengchong Rehai metagenome. The recombinant enzyme Y50Bg4 was obtained through PCR amplification, cloning, and expression. It was subsequently separated and purified using a Ni-NTA affinity chromatography column, and its enzymatic properties were analyzed.
Results: Enzymatic characterization revealed that Y50Bg4 efficiently hydrolyzes substrates like cellobiose, pNPGlc, and lactose. Y50Bg4 achieved optimal activity at 60°C and pH 6.0, maintaining 100% stability after 2 h of incubation at 60°C. The residual activity remained above 60% after 24 h of incubation across a pH range of 4.0 to 10.0. Kinetic constants analysis showed Km values of 4.69 mg/mL for cellobiose and 0.53 mM for pNPGlc, with Vmax values of 39.71 μmol/min/mg and 20.39 μmol/min/mg, respectively. Furthermore, the enzyme exhibits exceptional glucose tolerance, with Y50Bg4 retaining over 80% of its activity even at a glucose concentration of 3,000 mM. In practical applications, Y50Bg4 can work synergistically to degrade corn straw when combined with commercial cellulase. When Y50Bg4 (0.05 mg/mL) was added to the commercial cellulase reaction system, the glucose yield from corn straw increased by 11.6% after a reaction period of 24 h at 50°C. The results indicate that Y50Bg4 exhibits the activities of both β-glucosidase and β-galactosidase. Molecular docking and kinetic simulations revealed that Y50Bg4 has a higher affinity for cellobiose than for lactose and identified structural regions (residues 325–350 and 390–410) that contribute to its thermal stability.
Discussion: These findings highlight the potential of Y50Bg4 for industrial applications in bioethanol production and cellulose hydrolysis. In summary, Y50Bg4, with its exceptional enzymatic properties, presents significant application value and market potential in industrial sectors such as bioethanol production and cellulose hydrolysis.
Lignocellulose is the most abundant and cost-effective renewable resource on Earth. Its main component is cellulose, which is a sustainable and environmentally friendly material for producing high-value industrial products, such as biofuels and chemicals. It plays a crucial role in addressing energy and environmental crises (Chimtong et al., 2011; Bilal et al., 2020) The complete hydrolysis of cellulose is driven by a combination of three main cellulases: endoglucanases (EC 3.2.1.4), exoglucanases (EC 3.2.1.91), and β-glucosidases (EC 3.2.1.21). Among these, β-glucosidase is the most important component of the cellulase system and acts as the key enzyme in cellulose hydrolysis (Bhatia et al., 2002). It degrades cellobiose and fibrooligosaccharide into glucose during cellulose saccharification (Bhatia et al., 2002; Lynd et al., 2002). However, insufficient catalytic activity of β-glucosidase can hinder glucose production, leading to the accumulation of cellobiose, which in turn impairs glucose formation. As a depolymerase inhibitor, cellobiose also suppresses the activity of endoglucanase and exoglucanase (Dekker, 1986). Therefore, β-glucosidase has become a key rate-limiting enzyme in the overall process and a significant bottleneck for the biodegradation of cellulose to glucose. Many β-glucosidases reported to date are very sensitive to glucose, with glucose inhibition constants (Ki) typically below 100 mM. In recent years, glucose-tolerant β-glucosidases have become a major research focus due to their potential to enhance the saccharification process of lignocellulosic materials (Fang et al., 2010). In addition, most natural enzymes can only react under mild conditions, which limits their industrial applicability. In contrast, thermostable β-glucosidases, known for their resistance to high temperatures, are gaining significant attention in industrial production. These enzymes are considered an ideal catalyst for natural glycoside conversion due to their robust properties, including heat stability (Yenenler and Sezerman, 2016; Zhang et al., 2017). For example, cellulose degradation typically occurs at temperatures above 50°C and can require extended reaction times (sometimes over 100 h). In these processes, the inclusion of thermostable β-glucosidases improves catalytic efficiency, reduces substrate degradation, and lowers both energy consumption and environmental impacts (Haki and Rakshit, 2003; Liu et al., 2012). Therefore, finding efficient, thermostable, and glucose-tolerant β-glucosidases is essential to advance both research and industrial applications of cellulase technology.
Recent studies have identified several β-glucosidases from hot spring environments that exhibit both thermostability and glucose tolerance (Kaushal et al., 2021; Sinha et al., 2021; Zhu et al., 2024). However, most microorganisms from natural environments cannot be easily cultured in pure culture in the laboratory. To overcome this limitation, metagenomic techniques, which enable the exploration of environmental microbial genetic resources, have been successfully used to clone enzymes of significant industrial value (Richardson et al., 2002; Green and Keller, 2006). With the increasing importance of β-glucosidases and their various biotechnological applications, many efforts have been made to isolate and characterize novel enzymes from different sources to improve their performance.
In this study, we screened a β-glucosidase gene (named y50bg4) from the metagenome of the Tengchong Rehai hot spring, based on its functional potential. The gene was induced and expressed in Escherichia coli DH5α, and the recombinant enzyme β-glucosidase Y50Bg4 was purified for further analysis. Y50Bg4, which belongs to the GH1 family, was characterized for its enzymatic properties, and its thermostable mechanism was investigated through molecular dynamics simulation and molecular docking studies. The results showed that Y50Bg4 exhibits appreciable thermal stability, a broad pH range, and glucose tolerance, positioning it as an ideal enzyme candidate for applications such as hemicellulose degradation and other industrial processes.
Sediment and spring water samples were collected from hot springs in the Rehai area, Tengchong County, Baoshan City, Yunnan Province, Southwest China (24.947063°N, 98.440565°E), and stored at room temperature in the laboratory for enrichment culture. A 5 g sediment sample and 100 mL of spring water were supplemented with 1 g of cellobiose to the MSM (Minimal Salt Medium) as a carbon source and incubated at 50°C for enrichment culture. Samples were collected every 24 h for determination of total sugar content until depletion, with enriched samples obtained by centrifugation at 4,000 × g. Metagenomic DNA was extracted using the MOBIO DNeasy PowerSoil Kit (USA) according to the manufacturer’s instructions, and metagenomic sequencing was performed on a HiSeq2500 platform at GENWIZ (Suzhou). The IMG server1 was used to analyze the sequences, and gene functions were predicted using KEGG (Nakaya et al., 2013), COG (Tatusov et al., 2001), and Pfam (Finn et al., 2014) databases.
Based on functional predictions, a gene encoding β-glucosidase (named y50bg4) was identified from metagenomic data. The nucleotide sequence was submitted to GenBank under the accession number PQ827568. The nucleotide and amino acid sequences of Y50Bg4 were aligned using the BLASTx and BLASTp programs2, respectively. Signal peptide prediction was done using SignalP3, and primary structure analysis and molecular weight prediction were determined with the ExPASy tool4.
Phylogenetic analysis was conducted using MEGA 7 software (Kumar et al., 2016). To determine the optimal substitution model for sequence alignment, a model selection test was performed, which identified the WANG model as the most appropriate based on its lowest Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC) scores. Maximum likelihood (ML) phylogenetic reconstruction was then carried out using this selected model, with nodal support assessed through 1,000 bootstrap replicates. To enhance the phylogenetic resolution of Y50Bg4 within the β-glucosidase family, we incorporated representative sequences from three glycoside hydrolase families (GH1, GH3, and related glycoside hydrolase families) in the phylogenetic tree construction. All reference sequences were obtained from the NCBI protein database5.
The 3D structure of Y50Bg4 was predicted using the AlphaFold2 server6 (Jumper et al., 2021; Varadi et al., 2022) and visualized in PyMOL (Delano, 2002). Model quality was assessed using the pLDDT score, which ranges from 0 to 100. The Y50Bg4 structure was compared with Thermomicrobium roseum β-glucosidase B9L147, and multiple sequence alignment was conducted using MEGA 7 and visualized and analyzed with ESPript7.
Specific primers (y50bg4)-F: (CATCATCATCATCATCATGAAAATAGATGGAGGGCTAAAATGT) and (y50bg4)-R: (GTGCTCGAGTGCGGCCGCAAGTTCTTTATACTTTTTAATTATTTC) containing restriction sites were designed abased on the y50bg4 DNA sequencing using Primer Premier 5.0 to amplify the full-length y50bg4 fragment. The underlined sequence corresponds to the homologous recombination fragment for insertion into the pSHY211 vector (Yin et al., 2023), which had been previously digested with EcoR I and Hind III. The complete open reading frame (ORF) of y50bg4 was amplified with the high-fidelity enzyme Gold Mix (Beijing Tsingke Biotech Co., Ltd., China) using the following PCR protocol: pre-denaturation at 95°C for 3 min, followed by 30 cycles of denaturation at 98°C for 20 s, annealing at 65°C for 30 s, extension at 72°C for 45 s, and a final extension at 72°C for 5 min. The resulting PCR product was inserted into the pSHY211 vector using the pEASY-Uni Seamless Cloning and Assembly Kit (TransGen Biotech, China), generating the expression plasmid pSHY211-y50bg4. E. coli DH5α was used for y50bg4 gene cloning and expression, with the bacteria cultured in LB medium containing 50 μg/mL kanamycin. DNA isolation and purification were performed using kits from Sangon (China).
For expression of Y50Bg4, the recombinant plasmid was transformed into E. coli DH5α. The transformants were cultured in 200 mL of LB liquid medium with 50 μg/mL of kanamycin at 37°C with shaking at 200 rpm for 8 h. The culture was then incubated at 20°C with shaking at 200 rpm for 24 h. Cells were harvested by centrifugation at 10,000 × g for 15 min at 4°C, and then disrupted by sonication. The lysate was centrifuged at 12,000 × g for 30 min at 4°C, and the supernatant was purified using a Ni-chelating affinity column (Histrap, TransGen Biotech, China) as described by Yin et al. (2017). The purified recombinant β-glucosidase Y50Bg4 was analyzed by 12% SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis), with protein bands visualized by Coomassie Brilliant Blue dye R-250 staining. The protein concentration was determined using the Bradford Protein Assay Kit (order number C503031, Sangon Biotech, China), with bovine serum albumin as the standard (Bradford, 1976).
Enzyme activity was measured using the glucose oxidase-peroxidase assay kit with cellobiose as substrate (Yin et al., 2018). A 10 μL aliquot of 10-fold diluted enzyme solution after purification by Ni-NTA affinity chromatography was added to 90 μL of a buffer containing 1% (w/v) cellobiose. To minimize the occurrence of non-specific side reactions, such as substrate degradation or the acceleration of unintended chemical reactions, the enzymatic reaction was terminated immediately after 30 min under optimal conditions by transferring the samples to a pre-cooled −80°C environment and freezing them for 2–3 min. The transfer time was strictly limited to 30 s to reduce the impact of temperature fluctuations on enzyme activity. To quantify enzyme activity, 10 μL of the reaction mixture was added to a 96-well culture plate with 200 μL of glucose oxidase-peroxidase assay kit buffer. The mixture was incubated at 37°C for 10 min, and absorbance at OD490 nm was measured. One unit (U) of β-glucosidase activity was defined as the amount of enzyme required to release 2 μmol of glucose from cellobiose per minute. All experimental conditions were strictly controlled to ensure consistency across replicates.
The optimum temperature was determined by measuring enzyme activity across a temperature range of 30–85°C at the optimal pH. The optimal pH was assessed at the identified optimal temperature using sodium citrate buffer (pH 3.0–8.0) and glycine-NaOH buffer (pH 8.0–10.6). Results were expressed as the relative activity compared to the values obtained at the optimal temperature and pH. For thermal stability testing of Y50Bg4, the enzyme was pre-incubated at 50, 55, and 60°C for different time intervals (0-180 min). Residual enzyme activity was subsequently measured under optimal conditions. pH stability was assessed by incubating the enzyme in the specified buffers at 4°C for 12 and 24 h, and the residual activity was measured under standard conditions.
To assess the effect of glucose on the enzymatic activity of Y50Bg4, p-nitrophenyl-β-D-glucopyranoside (pNPGlc) was used as the substrate. A 10 μg protein was added to 200 μL of reaction mixture containing 2.5 mM pNPGlc (Sigma, St. Louis, MO, United States). The reaction was incubated for 5 min at the optimal temperature, then terminated by adding 450 μL of 1 M Na2CO3. The release of p-nitrophenol was measured by monitoring the absorbance at 405 nm, using p-nitrophenol (Sigma, St. Louis, Missouri, United States) as a standard. To evaluate the effect of β-glucose on enzyme activity, different concentrations of β-glucose (0–3 M) were added to the reaction mixture. Relative enzyme activity was determined by comparing it with a control reaction that did not include glucose.
The impact of metal ions and organic reagents on β-glucosidase activity was investigated by adding various metal ions (K+, Mg2+, Fe3+, Ca2+, Co2+, Ni2+, Mn2+, Pb2+, Cu2+, Zn2+, and Ag+) to the enzyme solution at final concentrations of 1 mM and 10 mM. Additionally, the effects of organic reagents, including Ethylenediaminetetraacetic acid (EDTA), Tween-80, Ethanol, Sodium Dodecyl Sulfate (SDS), and Phenyl Methyl Sulfonyl Fluoride (PMSF), were tested at final concentrations of 0.1 and 1%. The enzyme was incubated at 60°C, and the residual activity was measured after 30 min of treatment.
To determine the substrate specificity of Y50Bg4, a variety of substrates were tested, including cellobiose, p-nitrophenyl-β-D-glucopyranoside (pNPGlc), p-nitrophenyl-β-D-galactopyranoside (pNPGla), α-lactose, sucrose, maltose, gentiobiose, d-(+)-melibiose hydrate, d-(+)-trehalose dihydrate, raffinose, microcrystalline cellulose (Avicel), beechwood xylan, carboxymethylcellulose sodium (CMC-Na), corncob xylan, and bagasse xylan, all at a concentration of 1% (w/v). Cellulase and xylanase activities were quantified using the 3,5-dinitrosalicylic acid assay. One unit (U) of enzyme activity was defined as the amount of enzyme required to release 1 μmol of reducing sugar per minute.
The kinetic properties of recombinant Y50Bg4, including Km and Vmax, were evaluated in sodium citrate buffer (pH 6.0) at 60°C, using cellobiose and pNPGlc as substrates. Substrate concentrations ranged from 0 to 20 mg/mL for cellobiose and 0–2 mM for pNPGlc. The kinetic parameters were determined through non-linear regression analysis using the Michaelis–Menten equation in GraphPad Prism 5, yielding the Km and Vmax values. These results were further validated by Hanes-Woolf plots.
Ten gram of corn stover were ground and sieved through an 80-mesh sieve. The stover was then boiled in 100 mL of hot water for 30 min, filtered, and dried at 80°C. A 0.2 g aliquot of the pretreated corn was added to 1 mL of buffer (pH 5.6) along with 0.2 mg of cellulase (Sangon Biotechnology, China) and 0.03 mg of Y50Bg4. The mixture was incubated at 50°C, and samples were collected at 0, 1, 2, 3, 4, 5, 6, 7, 8, 18, and 24 h post-reaction. Glucose concentration in the reaction mixture was determined using a glucose oxidase assay kit. A control reaction without enzyme was also included.
A similarity search was performed using BLAST to identify an ambient β-glucosidase ThBGL1A with high homology to Y50Bg4, which was derived from Thermobifida halotolerans YIM 90462T. Molecular dynamics simulation was performed to analyze the structural stability and conformational flexibility of both Y50Bg4 and ThBGL1A. All simulations were conducted with identical steps and parameters. MD simulations were performed using GROMACS software, based on the protein structure and topology files obtained previously (Abraham et al., 2015). For the construction of the simulated environment, protein molecules were placed in a suitable solvent model to simulate their real state in the physiological environment. At the same time, an appropriate amount of counterions was added to make the simulation system electrically neutral. For the simulation parameter settings, we adopt a series of parameters that have been widely verified and optimized. The time step was set to 2 fs to ensure the simulation accuracy while effectively controlling the consumption of computational resources. The temperature is set at 300 K to simulate the environmental conditions close to the physiological temperature and to ensure that the structure and kinetic behavior of proteins are more realistic. The pressure was maintained at 1 bar to simulate the normal physiological pressure environment. The root means square deviation (RMSD) of the backbone and the root mean square fluctuation (RMSF) of Cα per residue during the simulations were calculated and compared using the GROMACS tools (gmx rms and gmx rmsf).
The 2D structures of the small molecule ligands, α-lactose and cellobiose, were downloaded from PubChem8, in SDF format and converted to 3D structures using ChemDraw 3D software, followed by energy minimization. The receptor Y50Bg4 and the ligands α-lactose and cellobiose were docked using AutoDock Vina software. The protein Y50Bg4 and the ligand molecules were pre-processed, and docking parameters were set with the following box dimensions: center_x = −0.133, center_y = −1.194, center_z = 0.019, size_x = 36.0, size_y = 28.5, size_z = 29.25. The docking simulations were executed, and optimized conformations for the different ligands were obtained.
All experiments were conducted in triplicate unless otherwise noted. Results were expressed as mean ± standard error of the mean (SEM). Statistical analyses were performed using SPSS version 20.0. One-way ANOVA followed by Tukey’s post hoc test was used for comparison among multiple experimental groups. A p-value of less than 0.05 was considered statistically significant, with a p-value less than 0.01 (**) indicating highly significant differences and p-values less than 0.05 (*) indicating significant differences.
Metagenomic sequencing of the hot spring enrichment from Tengchong Rehai yielded a comprehensive dataset comprising 6.72 Gbp and including 28,299 contigs longer than 1,000 bp. This dataset enabled the identification of 192,214 unique genes, among which 44 were predicted to be β-glucosidase genes. Based on gene integrity and sequence novelty, a novel gene y50bg4, encoding a GH1 family β-glucosidase, was identified from a Tengchong hot spring metagenomic data. Y50Bg4 is predicted to contain a glycoside hydrolase (GH1) structural domain (Pfam 00232; COG2723 β-glucosidase). The gene comprises a 1,338 bp open reading frame encoding a polypeptide of 446 amino acids. Based on EXPASY calculations, the predicted isoelectric point (pI) and molecular weight (Mw) are 6.4 and 52.7 kDa, respectively.
Sequence alignment analysis revealed that Y50Bg4 exhibits 80.65% sequence similarity with β-glucosidase from Caloramator quimbayensis (NCBI accession: WP278305990.1). Phylogenetic reconstruction demonstrated that these two enzymes form a distinct cluster, with Y50Bg4 showing close evolutionary relationships to other characterized GH1 family β-glucosidases (Figure 1). These findings provide strong evidence supporting the classification of Y50Bg4 within the glycoside hydrolase family 1 (GH1). Multiple sequence alignment confirmed conserved regions typical of GH1 family β-glucosidases (Supplementary Figure S1). NEP and TENG are typical conserved sites for GH1 family β-glucosidases, and these two motifs function in binding to glucosidase substrates. In addition, the glutamate residues E168 and E354 within these motifs are crucial for catalytic function, serving as a proton donor and a nucleophile, respectively (Supplementary Figure S1).
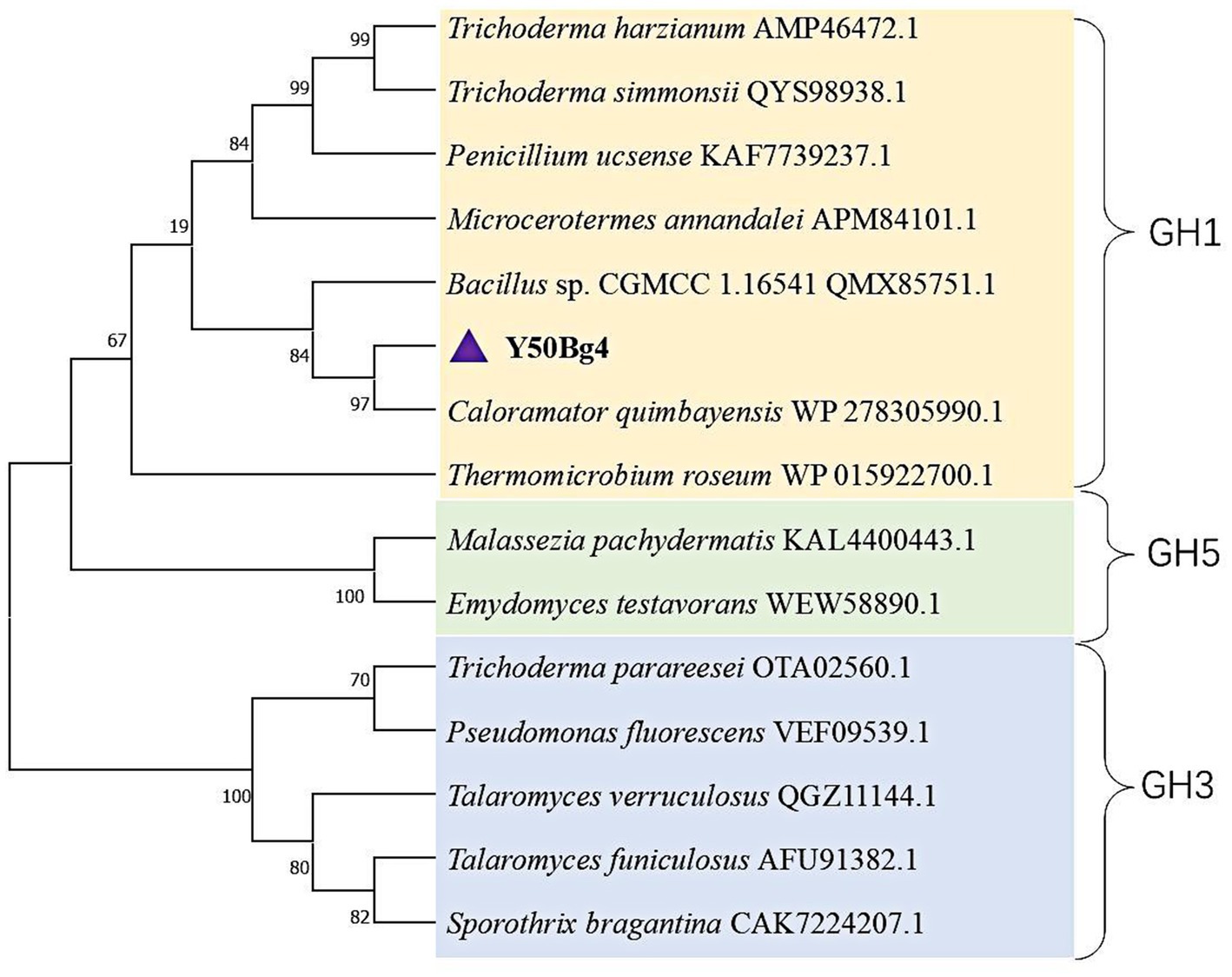
Figure 1. Phylogenetic analysis of Y50Bg4 and related β-glucosidases. The maximum likelihood phylogenetic tree was constructed using the WANG model with 1,000 bootstrap replicates. Branches are color-coded to represent different glycoside hydrolase families: yellow for GH1 family β-glucosidases, blue for GH3 family β-glucosidases, and green for GH5 family glycoside hydrolases. Y50Bg4 (highlighted in purple) clusters with β-glucosidase from Caloramator quimbayensis (80.65% sequence similarity) and is closely related to other GH1 family members. Bootstrap values (>50%) are shown at branch nodes.
The gene was successfully expressed in E. coli DH5α and the recombinant protein with an N-terminus His tag was purified using a Ni-NTA affinity chromatography column. SDS-PAGE analysis revealed a prominent 52 kDa band, consistent with the predicted molecular weight (Figure 2).
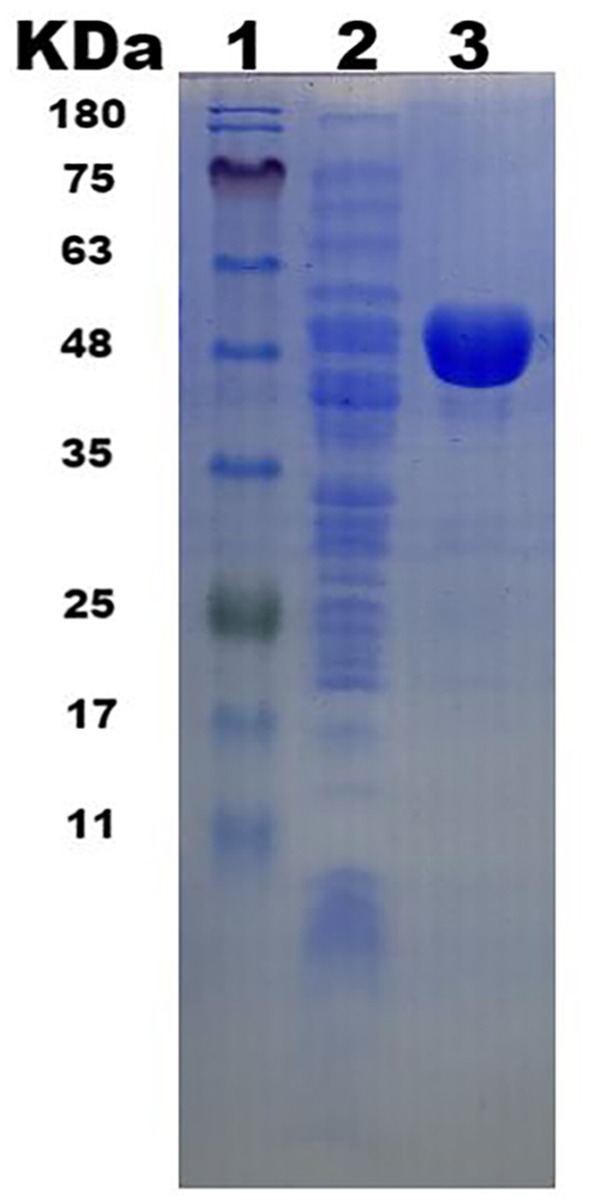
Figure 2. β-glucosidase Y50Bg4 SDS-PAGE electropherograms. (1. Protein Marker; 2. Total protein in E. coli DH5α/pSHY211-y50bg4 after expression; 3. Purified Y50Bg4 protein).
The 3D structural model of Y50Bg4 was predicted using AlphaFold, yielding a high-confidence model with a pLDDT score of 97.74 across 440 residues (Figure 3a). The structure displayed a classic 8 α/β TIM barrel, characteristic of GH1 enzymes. The predicted structural model of Y50Bg4 was superimposed onto B9L147 through comparative analysis with the three-dimensional structure of B9L147 (UniProt ID: B9L147), resulting in a root-mean-square deviation (RMSD) of 0.477 Å (Figure 3b). B9L147 is a GH1 β-glucosidase originating from Thermomicrobium roseum, a thermophilic bacterium known for its remarkable tolerance to glucose. Notably, the enzymatic activity of B9L147 was significantly enhanced at glucose concentrations of 1.25–1.5 M, showing an increase of more than 2.5-fold. Even at high glucose concentrations of up to 3 M, B9L147 retained over 100% of its specific activity. Both Y50Bg4 and B9L147 share the characteristic α/β TIM barrel domain, a hallmark of GH1 family enzymes.
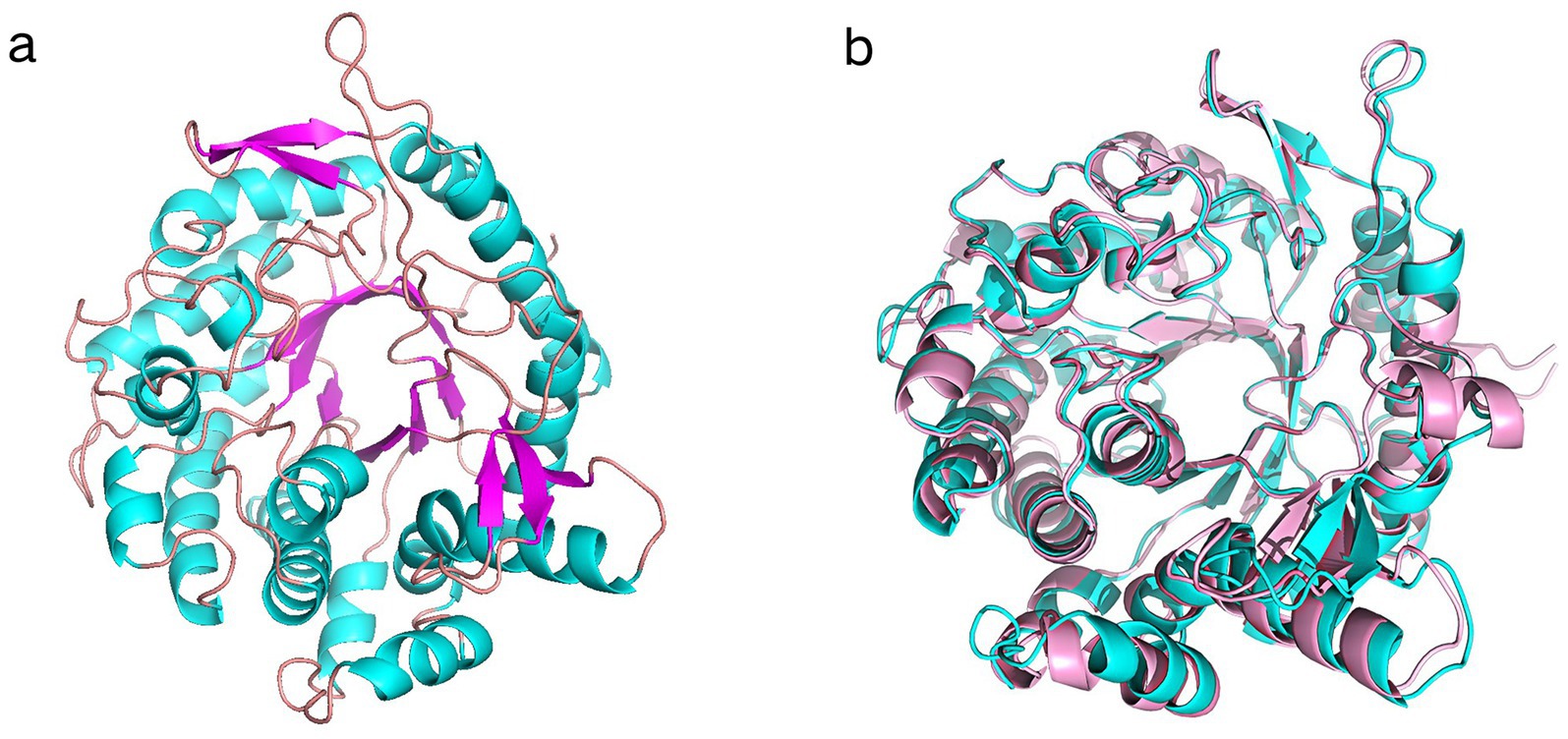
Figure 3. Superimposed results of Y50Bg4 3D structure prediction and B9L147 structure. (a) 3D structural model of Y50Bg4. (b) Superposition of the Y50Bg4 3D model generated by homology modeling (shown in cyan) onto the B9L147 structure (displayed in pink as a banded model). B9L147 (Uniprot ID) represents a GH1-family β-glucosidase originating from Thermomicrobium roseum.
Y50Bg4 displayed the highest activity at 60°C, maintaining over 60% of its optimal activity between 30 and 60°C (Figure 4a). The optimal pH for activity was determined to be 6.0 and it was assessed at the optimal temperature (Figure 4b). After 2 h of incubation at 50°C, 55°C, and 60°C, the enzyme activity remained at 100%. However, after 3 h of incubation, the activity decreased to about 60% at 50°C and was completely lost at 55°C and 60°C (Figure 4c). Y50Bg4 retained over 60% of its activity across pH 4.0–10.0 after incubation at 4°C for 12 h and 24 h (Figure 4d).
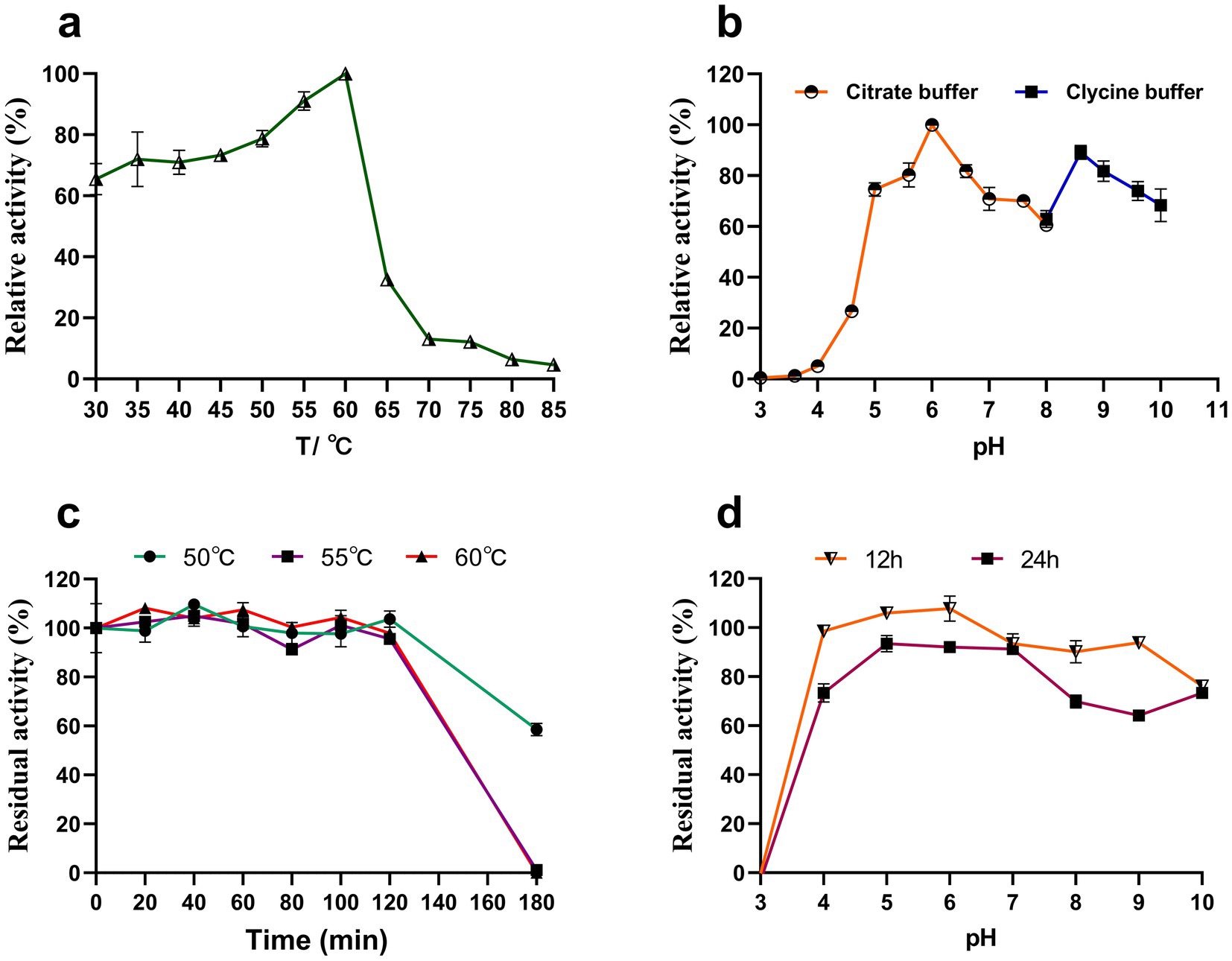
Figure 4. Effect of temperature and pH on the activity and stability of recombinant Y50Bg4. (a) Effect of temperature on Y50Bg4 activity. (b) Effect of pH on Y50Bg4 activity. (c) Effect of incubation at different temperatures (50, 55, and 60°C) for 0, 20, 40, 60, 80, 100, 120, 140, 160, and 180 min on the stability of Y50Bg4. (d) Effect of pH on the stability of Y50Bg4. The pH stability test was performed by pre-incubating the enzyme in different buffer systems at 4°C for 12 and 24 h. Values represent the mean of three biological replicates. Error bars represent the mean ± SEM of three biological replicates. The primary activity was taken as 100%.
Competitive inhibition by glucose is a common challenge for β-glucosidases and can limit their activity during enzyme-catalyzed hydrolysis. Therefore, high glucose tolerance provides a significant advantage for β-glucosidases in certain applications. In this study, the effect of glucose on Y50Bg4 activity was evaluated using 2.5 mM pNPGlc as the substrate. The results showed that Y50Bg4 retained 100% activity at glucose concentrations ranging from 0 to 500 mM and maintained over 80% activity even at 3,000 mM glucose (Figure 5). This remarkable resistance to high glucose concentrations highlights the potential of Y50Bg4 for applications in environments with elevated glucose levels.
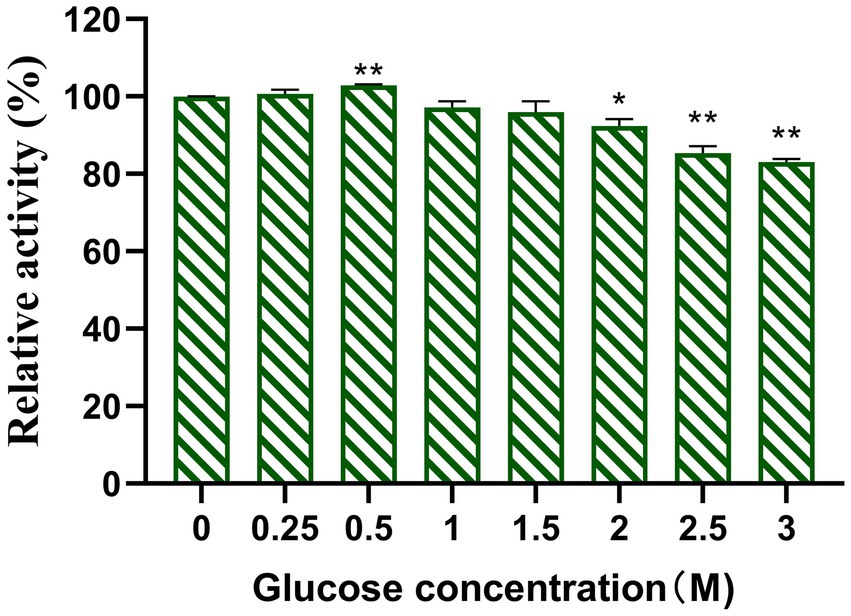
Figure 5. Effect of glucose on recombinant Y50Bg4. The final concentration was determined to be 1 mM using pNPGlc as the substrate. Values represent the mean of three biological replicates. Error bars represent the mean of three biological replicates ± SEM. Original viability was taken as 100%. p < 0.01 (**) means the difference is very significant, p < 0.05 (*) means the difference is significant.
The effects of metal ions and chemical reagents on the activity of Y50Bg4 were assessed at 1 and 10 mM (Table 1). At 1 mM, K+, Ca2+, Mg2+ and Fe3+ had minimal effects, while Cu2+ (11.79 ± 1.23), Mn2+ (7.28 ± 2.37), Pb2+ (26.15 ± 1.87) and Ag+ (1.94 ± 0.56) caused strong inhibition and Pb2+ (26.15 ± 0.56) and Ag+ (1.94 ± 0.56) had the highest inhibitory effect. At 10 mM, only K+, Ca2+, and Mg2+ remained non-inhibitory, while the remaining ions reduced activity to nearly 10%. Co2+ (0.07 ± 1.15) and Ag+ (0.21 ± 0.10) completely suppressed enzyme activity. Chemical reagents such as EDTA (11.86 ± 0.27), Tween-80 (48.83 ± 2.32), PMSF (12.96 ± 1.33), and SDS (9.75 ± 4.59) also significantly reduced activity at 1%.
Y50Bg4 hydrolyzed a variety of substrates, including cellobiose, lactose, pNPGlc, and gentiobiose, but was unable to hydrolyze maltose, melibiose, trehalose, raffinose, sucrose, beechwood xylan, corncob xylan, bagasse xylan, CMC-Na, and Avicel (Table 2). The highest activities were observed for cellobiose (31.06 ± 2.25 U/mg) and lactose (23.91 ± 1.74 U/mg), while lower activities were detected for pNPGlc (6.72 ± 0.76 U/mg), pNPGla (4.51 ± 0.02 U/mg), and gentiobiose (1.58 ± 0.16 U/mg).
The kinetic constants (Km and Vmax) were determined using Michaelis–Menten non-linear regression (Supplementary Figure S2), and the results have been validated by the Hanes-Woolf method (Supplementary Figure S3). With cellobiose as the substrate, Km was 4.69 mg/mL and Vmax was 39.71 μmol/min/mg. For pNPGlc as substrate Km was 0.53 mM and Vmax was 20.39 μmol/min/mg, yielding catalytic efficiencies (Kcat/Km) of 7.44 s−1/mg/ml and 33.80 s−1/mM, respectively (Table 3).
Enzymatic hydrolysis of corn stover was performed to evaluate the potential use of Y50Bg4 in lignocellulose degradation and revealed that Y50Bg4 alone showed no hydrolytic activity. However, the addition of Y50Bg4 to commercial cellulase increased the degradation rate by about 11.6% compared to the hydrolysis rate of the commercial cellulase-only group (Figure 6). This suggests that Y50Bg4 has no hydrolytic activity on corn stover alone but shows good synergistic hydrolysis in cooperation with other cellulases, suggesting its potential as an enzyme-promoted mixture component for converting lignocellulosic biomass into different chemicals.
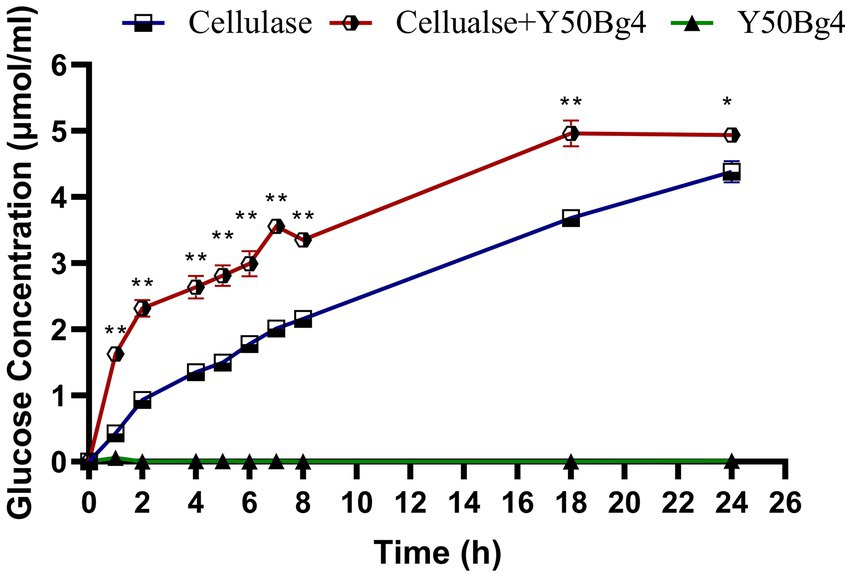
Figure 6. Synergistic degradation of corn stover by Y50Bg4 and commercial cellulases at 60°C. The amount of glucose released following the addition of Y50Bg4 was determined, with the group containing only commercial cellulase serving as the control. Values represent the mean of three biological replicates. Error bars represent the mean ± SEM of three biological replicates. Original viability was taken as 100%. p < 0.01 (**) means the difference is very significant, p < 0.05 (*) means the difference is significant.
The overall stability/equilibrium of Y50Bg4 and ThBGL1A during the molecular dynamics simulation process were evaluated by calculating the backbone RMSD as a function of time for both simulated systems. As shown in Figure 7a, the skeleton RMSD of the Y50Bg4 system remained relatively stable, fluctuating minimally throughout the simulation. In contrast, the ThBGL1A system exhibited greater fluctuations, requiring approximately 40 ns to stabilize before maintaining relatively steady values. Consequently, the MD trajectories between 10–50 ns for both systems were used as equilibrium trajectories for subsequent analysis. During the simulation, ThBGL1A consistently displayed higher RMSD values than Y50Bg4, indicating greater structural fluctuations and conformational changes compared to its initial structure. This increased flexibility of ThBGL1A might explain why Y50Bg4 demonstrates superior thermal stability. To investigate the conformational dynamics root-mean-square fluctuations (RMSF) of Cα atoms were calculated. As illustrated in Figure 7b, both proteins showed similar RMSF patterns in some regions, suggesting partial structural and interface similarity. However, Y50Bg4 exhibited lower RMSF values in most regions compared to ThBGL1A, aligning with the observed RMSD trends. Notably, two loop regions in ThBGL1A (residues 325–350 and 390–410) displayed significantly higher RMSF values than corresponding regions in Y50Bg4. These findings suggest that ThBGL1A possesses greater global conformational flexibility than Y50Bg4.

Figure 7. Root means square deviation (RMSD) and RMSF results for Y50Bg4, ThBGL1A. (a) RMSD value variation curves as a function of time for backbone atoms in MD simulations for Y50Bg4 (orange line) and ThBGL1A (blue line), respectively. (b) Root-mean-square fluctuation (RMSF) values of Cα atoms per residue for Y50Bg4 (orange line) and ThBGL1A (blue line).
Phylogenetic analysis revealed that the Y50Bg4 enzyme clusters closely with β-glucosidases from Caloramator quimbayensis. The study identified Caloramator quimbayensis as an anaerobic, moderately thermophilic bacterium isolated from terrestrial hot springs (Rubiano-Labrador et al., 2013). This bacterium grows within a temperature range of 37–55°C and pH 6.0–8.0, with optimal growth conditions at 50°C and pH 7.0, suggesting that Y50Bg4 may also exhibit thermophilic properties. It does not necessarily indicate that Caloramator quimbayensis is the direct origin of the Y50Bg4 enzyme. The actual source of Y50Bg4 remains unclear and may require further investigation, such as screening genomic or metagenomic data from environmental samples to identify its natural host. Y50Bg4 belongs to the GH1 family of β-glucosidases, which comprises most known β-glucosidases. GH1 enzymes are generally more glucose-tolerant than those from the GH3 family, playing a crucial role in cellulose saccharification (Shipkowski and Brenchley, 2005; Ketudat Cairns and Esen, 2010; de Almeida et al., 2020).
The conservation of the α/β TIM barrel fold within the GH1 family highlights its functional importance. This structural motif serves as a scaffold for the precise positioning of key catalytic residues, ensuring efficient substrate binding and hydrolysis (Zechel and Withers, 2000; Nagano et al., 2002). Specifically, the conserved glutamate residues E168 and E354, located within the active site, are known to function as proton donors and nucleophiles, respectively. The low RMSD value of 0.477 Å further emphasizes the close structural relationship between Y50Bg4 and B9L147. Such a small deviation suggests that the overall folding and architecture of the two enzymes are highly conserved, reflecting their shared functional mechanisms. The minor differences observed in the loop regions may contribute to variations in substrate specificity or thermostability, warranting further investigation.
Garbin et al. (2021) reported that β-glucosidase from the thermophilic fungus Thermoascus crustaceous remained stable at 30–55°C after 1 h of incubation, with about 60% residual activity at 60°C. In comparison, Toyama et al. (2018) found that the β-glucosidase AmBgl-LP from the Amazonian Lake Pollack metagenome maintained 60% residual activity after 30 min at 40°C but lost all activity after 10 min at 50°C. In contrast, as shown in Table 4, Y50Bg4 demonstrated better thermal stability, a broader pH tolerance, and higher efficiency in degrading naturally crystalline, structurally complex cellulose.
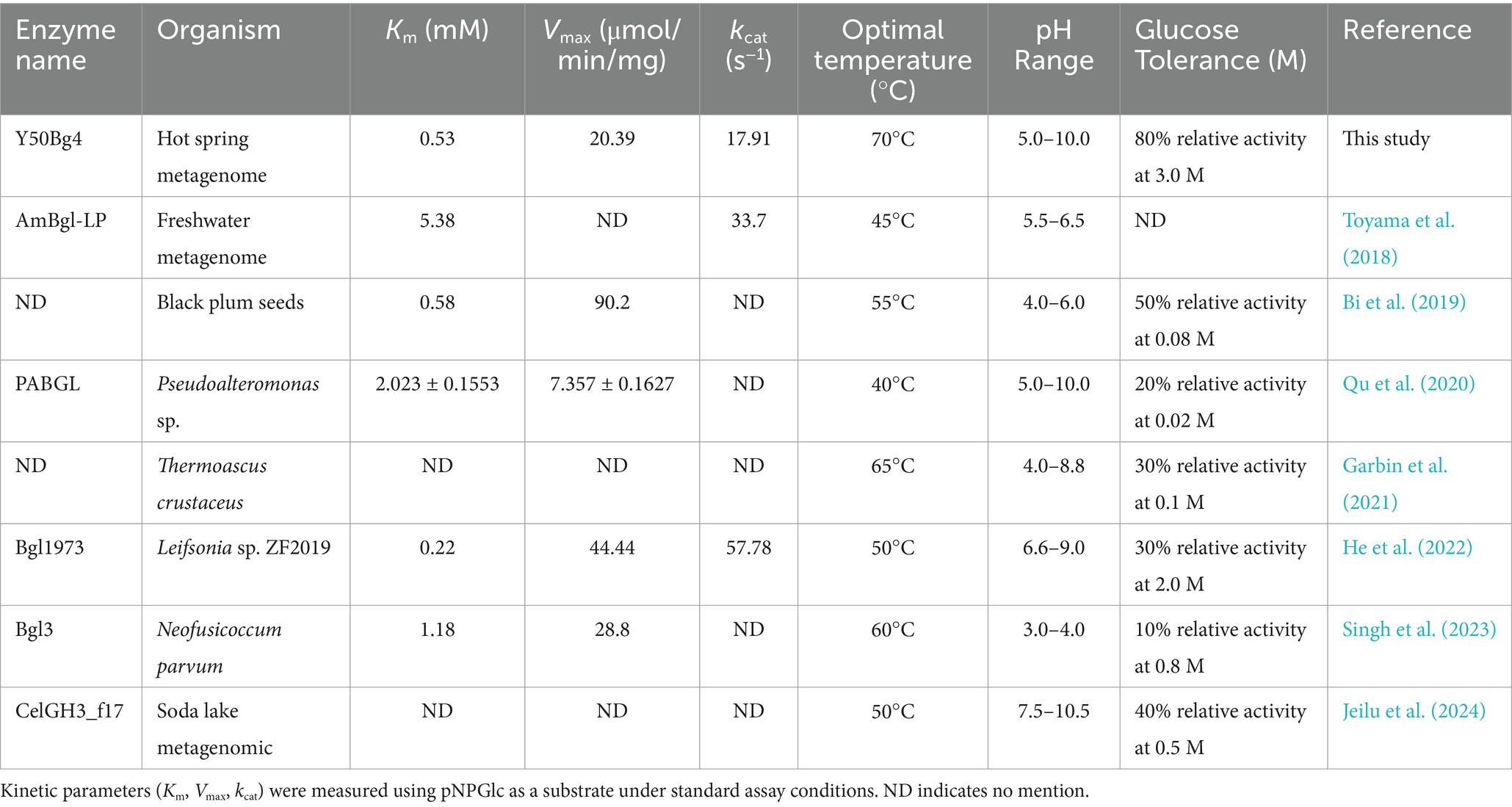
Table 4. Enzymatic properties as well as kinetic parameters of Y50Bg4 compared to other β-glucosidases.
Most microbial β-glucosidases are competitively inhibited by glucose, with inhibition constant (Ki) values typically between 100 and 500 mM (Mamma et al., 2004; Karnchanatat et al., 2007). However, some β-glucosidases, such as Unbgl1A from soil metagenomic source, tolerate glucose concentrations up to 1,500 mM, with activation observed at 300 mM (Lu et al., 2013). Y50Bg4 exhibits higher glucose tolerance than most of the β-glucosidases, making it an ideal candidate for industrial processes requiring robust enzyme activity under high glucose concentrations. The glucose tolerance of Y50Bg4 may be linked to its active site architecture and substrate binding mode. Beta-glucosidases from the GH1 family have been reported to be generally glucose-tolerant, with a narrower and deeper active site at the α/β 8 barrel, whereas the majority of glucose-intolerant beta-glucosidases have been studied under the GH3 family, which has a shallower lumen (de Giuseppe et al., 2014). The binding of glucose to the shallow lumen easily leads to competitive inhibition with the substrate, while a wider or more flexible substrate-binding pocket can reduce the steric hindrance caused by glucose binding (Sengupta et al., 2023). Y50Bg4 showed sensitivity to metal ions and chemical reagents, of which Ag+, Zn2+, and SDS act as strong inhibitors, similar to other β-glucosidases (Zollner, 1989). However, Ca2+ and Mg2+ stabilized its activity, aligning with findings for other GH1β-glucosidases. The inhibitory effects of EDTA suggest that Y50Bg4 depends on metal ions for activity. These results are consistent with studies on β-glucosidases such as BGLA from Alteromonas sp. L82 and its F171W mutant (Sun et al., 2020), where SDS disrupted the enzyme three-dimensional structure and catalytic domain integrity (Geiger et al., 1999).
To further determine the hydrolytic activity of Y50Bg4 on lactose, the results of the optimal temperature (60°C) and optimal pH (6.6) of Y50Bg4 determined using lactose as the substrate were almost the same as those determined when cellobiose was used as the substrate (Supplementary Figure S4). Y50Bg4 efficiently hydrolyzed β-(1 → 4) glycosidic bonds but failed to cleave α-glycosidic linkages in sucrose or maltose and complex polysaccharides such as xylan, CMC-Na, and Avicel, whose long chains of polysaccharides hinder the activity of BGLs due to the complexity of the substrates. In summary, Y50Bg4 is a multifunctional enzyme with the activity of both β-glucosidase and β-galactosidase pairs. This result is consistent with Xie et al. (2020) who reported that the β-glucosidase Igag_0940 derived from hyperthermophile Ignisphaera aggregans also showed activity toward cellobiose, lactose, and is also a multifunctional glycoside hydrolase of GH1. Furthermore, Deng et al. (2020) reported that from the deep-sea bacterium Bacillus sp. D1 also screened and cloned a novel β-glucosidase, BglD1, with high β-galactosidase and trans glycosidic activities. This is also a feature of the GH1 family of glucosidases, which mostly have both β-galactosidase activity and β-glucosidase activity (Cantarel et al., 2009). Additionally, Y50Bg4 exhibited high catalytic efficiency for the synthesis of the substrate pNPGlc, which is a key factor in the development of all β-glucosidases (Rani et al., 2015; Akram et al., 2016). However, the catalytic efficiency of cellobiose is crucial in determining the efficient degradation of biomass.
The molecular docking of Y50Bg4 with cellobiose and α-lactose revealed successful binding of both substrates. Ten docked conformations with negative binding free energies were obtained for each ligand, indicating favorable interactions. The lowest binding free energy conformation for cellobiose was −7.5 kcal/mol (Figure 8a), while for α-lactose, it was −7.4 kcal/mol (Figure 8b), suggesting strong binding affinity for both substrates. Intermolecular interactions, especially hydrogen bonding, played an important role in the formation of enzyme-substrate complexes. In the Y50Bg4-cellobiose complex, six hydrogen bonds were observed, involving residues Asn-294, Tyr-293, Glu-348, Trp-402, and Glu-162. In contrast, the Y50Bg4-lactose complex formed only four hydrogen bonds, involving residues Trp-402, Tyr-293, and His-117. The higher number of hydrogen bonds with cellobiose suggests that Y50Bg4 interacts more efficiently with this substrate, indicating a substrate preference likely due to better structural compatibility. Notably, residues Trp-402 and Tyr-293 were involved in binding both substrates, consistent with findings by Khairudin and Mazlan (2013), who reported Trp-402 as a conserved residue involved in the interaction of proteins with fibrous disaccharides and polysaccharides. This supports the notion that these residues are critical for substrate recognition and catalysis, highlighting Y50Bg4 potential in biodegradation and glycolysis. Y50Bg4 may hydrolyze both α-lactose and cellobiose, which is consistent with our experimental results, suggesting Y50Bg4 is more efficient at hydrolyzing cellobiose. This makes it a promising candidate for enzymatic hydrolysis of lignocellulose into fermentable sugars, a key step in biofuel production. Multifunctional glycoside hydrolases like Y50Bg4 are valuable for the saccharification of cellulose and hemicellulose (Gruninger et al., 2014).
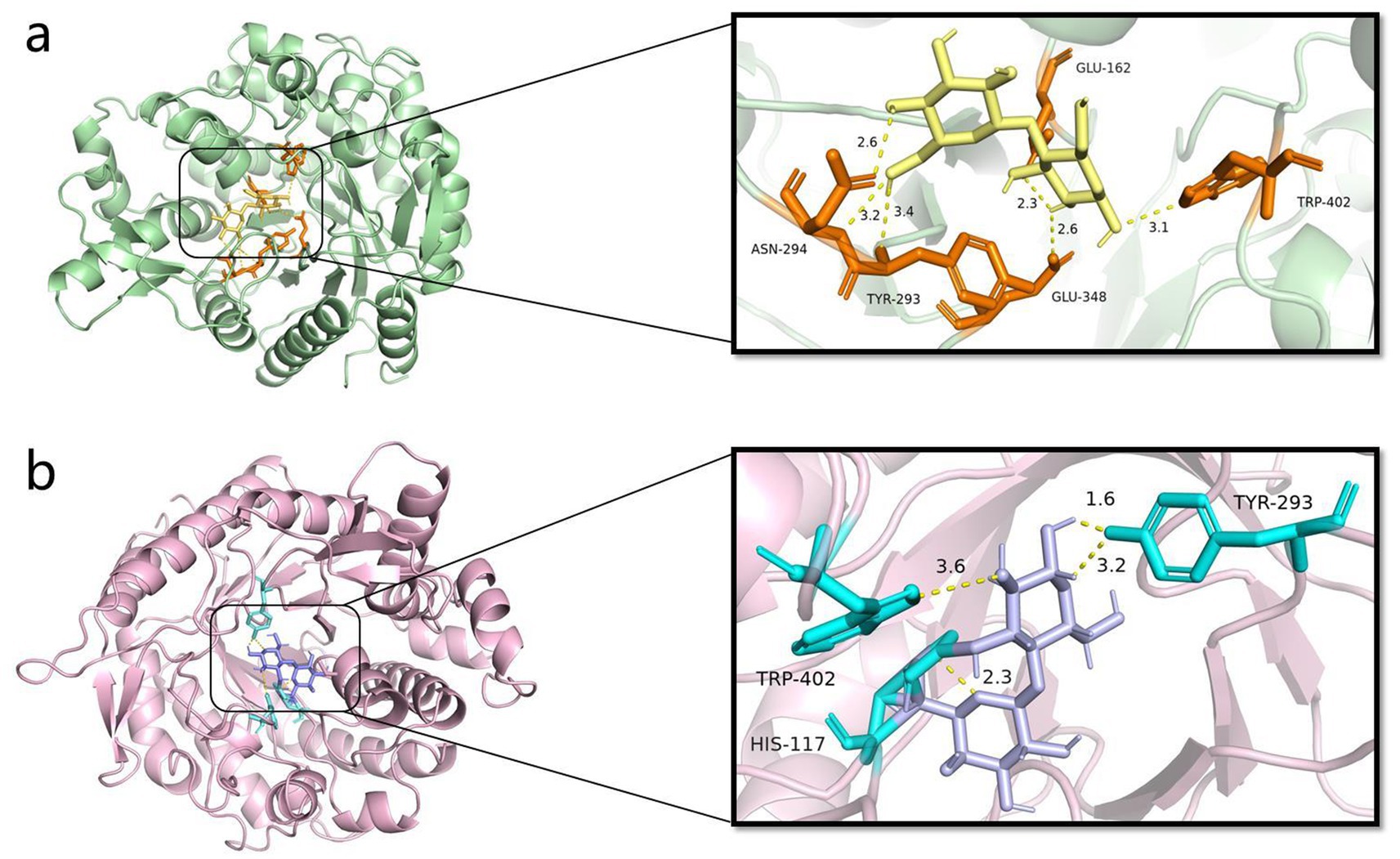
Figure 8. Y50Bg4 docking results with ligand. (a) Y50Bg4 docking results with cellobiose, yellow represents cellobiose, orange represents residues interacting with the ligand, and yellow dashed line represents hydrogen bonding. (b) Y50Bg4 docking results with lactose, purple represents lactose, cyan represents residues interacting with the ligand, and yellow dashed lines represent hydrogen bonds.
Kinetic simulations revealed that Y50Bg4 exhibited lower structural fluctuations and greater conformational stability compared to the mesophilic enzyme ThBGL1A. It is well established that protein thermal stability is often associated with reduced conformational flexibility. The thermal stability of proteins is usually associated with their reduced flexibility (or increased rigidity), and rigidity protects proteins from unfolding, degradation by proteolytic hydrolysis, and chemical denaturation at high temperatures, implying that inhibition of fluctuations within the protein and the mobility may be responsible for its ability to resist thermal denaturation (Kamerzell and Middaugh, 2008; Karshikoff et al., 2015; Quezada et al., 2017). In the present study, we observed that the overall fluctuation of Y50Bg4 was lower than that of its homologous counterpart ThBGL1A. Supporting the theory that reduced flexibility enhances thermostability. Therefore, the greater thermal stability of Y50Bg4 can be attributed to its higher rigidity and less conformational flexibility than mesothermal enzyme. In addition to the above, the thermostability of Y50Bg4 is also likely attributed to several structural features commonly observed in thermostable enzymes. First, sequence analysis reveals a high proportion of hydrophobic residues, which may contribute to the formation of a stable hydrophobic core. This core helps maintain the enzyme’s tertiary structure under high temperatures. Second, the presence of salt bridges and hydrogen bonds in key regions of the protein could enhance its resistance to thermal denaturation. These interactions are known to stabilize proteins by reducing conformational flexibility at elevated temperatures. In summary, with its unique stability, high substrate specificity, and thermal adaptability, Y50Bg4 holds significant promise for industrial biocatalysis applications. Its efficient catalytic mechanism and resilience under harsh conditions make it an ideal candidate for processes such as biofuel production and biomass conversion.
The β-glucosidase gene y50bg4 from the Tengchong Thermal Sea metagenomic source was successfully cloned and heterologously expressed in E. coli. Detailed characterization of the recombinant β-glucosidase Y50Bg4 revealed notable enzymatic properties and provided insights into its thermostable mechanism. The results showed that Y50Bg4 has excellent thermal stability, making it a promising candidate for applications in high-temperature industrial processes. Structural analysis showed that Y50Bg4 has greater rigidity and more compact conformation compared to ambient enzymes4. Notably, Y50Bg4 is a bifunctional enzyme with both β-glucosidase and β-galactosidase activities, confirmed through molecular docking studies. In addition, its exceptional glucose tolerance surpasses that of most GH1-family β-glucosidases, making it an ideal candidate enzyme for use in cellulose degradation saccharification processes as well as biofuel production.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found: https://www.ncbi.nlm.nih.gov/genbank/, PQ827568.
Y-YH: Writing – original draft. DZ: Writing – review & editing. L-QY: Writing – review & editing. MO: Writing – review & editing. Z-FY: Writing – review & editing. Z-HL: Writing – review & editing. K-QX: Writing – review & editing. HJ: Writing – review & editing. W-JL: Writing – review & editing. Y-RY: Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the Yunnan Applied Basic Research Project (Nos. 202101AU070138 and 202501AT070411), the Science and Technology Major Project of Xizang (Program No. XZ2025), and Xingdian Talent Support Program of Yunnan Province (No. 230212528080).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1559242/full#supplementary-material
1. ^https://img.jgi.doe.gov/cgi-bin/mer/main.cgi
2. ^http://blast.ncbi.nlm.nih.gov/Blast.cgi
3. ^https://services.healthtech.dtu.dk/services/SignalP-5.0/
4. ^http://web.expasy.org/protparam/
5. ^https://www.ncbi.nlm.nih.gov/protein
Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., et al. (2015). GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25. doi: 10.1016/j.softx.2015.06.001
Akram, F., Haq, I. U., Khan, M. A., Hussain, Z., Mukhtar, H., and Iqbal, K. (2016). Cloning with kinetic and thermodynamic insight of a novel hyperthermostable β-glucosidase from Thermotoga naphthophila RKU-10T with excellent glucose tolerance. J. Mol. Catal. B Enzym. 124, 92–104. doi: 10.1016/j.molcatb.2015.12.005
Bhatia, Y., Mishra, S., and Bisaria, V. S. (2002). Microbial beta-glucosidases: cloning, properties, and applications. Crit. Rev. Biotechnol. 22, 375–407. doi: 10.1080/07388550290789568
Bi, Y., Zhu, C., Wang, Z., Luo, H., Fu, R., Zhao, X., et al. (2019). Purification and characterization of a glucose-tolerant β-glucosidase from black plum seed and its structural changes in ionic liquids. Food Chem. 274, 422–428. doi: 10.1016/j.foodchem.2018.09.007
Bilal, M., Wang, Z., Cui, J., Ferreira, L. F. R., Bharagava, R. N., and Iqbal, H. M. N. (2020). Environmental impact of lignocellulosic wastes and their effective exploitation as smart carriers – a drive towards greener and eco-friendlier biocatalytic systems. Sci. Total Environ. 722:137903. doi: 10.1016/j.scitotenv.2020.137903
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The carbohydrate-active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–D238. doi: 10.1093/nar/gkn663
Chimtong, S., Tachaapaikoon, C., Pason, P., Kyu, K. L., Kosugi, A., Mori, Y., et al. (2011). Isolation and characterization of endocellulase-free multienzyme complex from newly isolated Thermoanaerobacterium thermosaccharolyticum strain NOI-1. J. Microbiol. Biotechnol. 21, 284–292. doi: 10.4014/jmb.1009.09032
de Almeida, P. Z., de Oliveira, T. B., de Lucas, R. C., Salgado, J. C. S., Pérez, M. M., Gálan, B., et al. (2020). Heterologous production and biochemical characterization of a new highly glucose tolerant GH1 β-glucosidase from Anoxybacillus thermarum. Process Biochem. 99, 1–8. doi: 10.1016/j.procbio.2020.08.013
de Giuseppe, P. O., Souza, T. A. C. B., Souza, F. H. M., Zanphorlin, L. M., Machado, C. B., Ward, R. J., et al. (2014). Structural basis for glucose tolerance in GH1 β-glucosidases. Acta Cryst D 70, 1631–1639. doi: 10.1107/S1399004714006920
Dekker, R. F. (1986). Kinetic, inhibition, and stability properties of a commercial beta-D-glucosidase (cellobiase) preparation from aspergillus Niger and its suitability in the hydrolysis of lignocellulose. Biotechnol. Bioeng. 28, 1438–1442. doi: 10.1002/bit.260280918
Delano, W. L. (2002). The PyMol molecular graphics system. Prot. Struct. Funct. Bioinf. 30, 442–454.
Deng, P., Meng, C., Wu, Y., Xu, J., Tang, X., Zhang, X., et al. (2020). An unusual GH1 β-glucosidase from marine sediment with β-galactosidase and transglycosidation activities for superior galacto-oligosaccharide synthesis. Appl. Microbiol. Biotechnol. 104, 4927–4943. doi: 10.1007/s00253-020-10578-z
Fang, Z., Fang, W., Liu, J., Hong, Y., Peng, H., Zhang, X., et al. (2010). Cloning and characterization of a beta-glucosidase from marine microbial metagenome with excellent glucose tolerance. J. Microbiol. Biotechnol. 20, 1351–1358. doi: 10.4014/jmb.1003.03011
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Garbin, A. P., Garcia, N. F. L., Cavalheiro, G. F., Silvestre, M. A., Rodrigues, A., Paz, M. F. D., et al. (2021). β-Glucosidase from thermophilic fungus Thermoascus crustaceus: production and industrial potential. An. Acad. Bras. Cienc. 93:e20191349. doi: 10.1590/0001-3765202120191349
Geiger, G., Furrer, G., Funk, F., Brandl, H., and Schulin, R. (1999). Heavy metal effects on beta-glucosidase activity influenced by pH and buffer systems. J. Enzym. Inhib. 14, 365–379. doi: 10.3109/14756369909030329
Green, B. D., and Keller, M. (2006). Capturing the uncultivated majority. Curr. Opin. Biotechnol. 17, 236–240. doi: 10.1016/j.copbio.2006.05.004
Gruninger, R. J., Gong, X., Forster, R. J., and McAllister, T. A. (2014). Biochemical and kinetic characterization of the multifunctional β-glucosidase/β-xylosidase/α-arabinosidase, Bgxa1. Appl. Microbiol. Biotechnol. 98, 3003–3012. doi: 10.1007/s00253-013-5191-4
Haki, G. D., and Rakshit, S. K. (2003). Developments in industrially important thermostable enzymes: a review. Bioresour. Technol. 89, 17–34. doi: 10.1016/S0960-8524(03)00033-6
He, Y., Wang, C., Jiao, R., Ni, Q., Wang, Y., Gao, Q., et al. (2022). Biochemical characterization of a novel glucose-tolerant GH3 β-glucosidase (Bgl1973) from Leifsonia sp. ZF2019. Appl. Microbiol. Biotechnol. 106, 5063–5079. doi: 10.1007/s00253-022-12064-0
Jeilu, O., Alexandersson, E., Johansson, E., Simachew, A., and Gessesse, A. (2024). A novel GH3-β-glucosidase from soda lake metagenomic libraries with desirable properties for biomass degradation. Sci. Rep. 14:10012. doi: 10.1038/s41598-024-60645-y
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kamerzell, T. J., and Middaugh, C. R. (2008). The complex inter-relationships between protein flexibility and stability. J. Pharm. Sci. 97, 3494–3517. doi: 10.1002/jps.21269
Karnchanatat, A., Petsom, A., Sangvanich, P., Piaphukiew, J., Whalley, A. J. S., Reynolds, C. D., et al. (2007). Purification and biochemical characterization of an extracellular beta-glucosidase from the wood-decaying fungus Daldinia eschscholzii (Ehrenb.:Fr.) Rehm. FEMS Microbiol. Lett. 270, 162–170. doi: 10.1111/j.1574-6968.2007.00662.x
Karshikoff, A., Nilsson, L., and Ladenstein, R. (2015). Rigidity versus flexibility: the dilemma of understanding protein thermal stability. FEBS J. 282, 3899–3917. doi: 10.1111/febs.13343
Kaushal, G., Rai, A. K., and Singh, S. P. (2021). A novel β-glucosidase from a hot-spring metagenome shows elevated thermal stability and tolerance to glucose and ethanol. Enzym. Microb. Technol. 145:109764. doi: 10.1016/j.enzmictec.2021.109764
Ketudat Cairns, J. R., and Esen, A. (2010). β-Glucosidases. Cell. Mol. Life Sci. 67, 3389–3405. doi: 10.1007/s00018-010-0399-2
Khairudin, N. B. A., and Mazlan, N. S. F. (2013). Molecular docking study of Beta-glucosidase with cellobiose, cellotetraose and cellotetriose. Bioinformation 9, 813–817. doi: 10.6026/97320630009813
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Liu, D., Zhang, R., Yang, X., Zhang, Z., Song, S., Miao, Y., et al. (2012). Characterization of a thermostable β-glucosidase from Aspergillus fumigatus Z5, and its functional expression in Pichia pastoris X33. Microb. Cell Factories 11:25. doi: 10.1186/1475-2859-11-25
Lu, J., Du, L., Wei, Y., Hu, Y., and Huang, R. (2013). Expression and characterization of a novel highly glucose-tolerant β-glucosidase from a soil metagenome. Acta Biochim. Biophys. Sin. 45, 664–673. doi: 10.1093/abbs/gmt061
Lynd, L. R., Weimer, P. J., van Zyl, W. H., and Pretorius, I. S. (2002). Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66, 506–577. doi: 10.1128/MMBR.66.3.506-577.2002
Mamma, D., Hatzinikolaou, D. G., and Christakopoulos, P. (2004). Biochemical and catalytic properties of two intracellular β-glucosidases from the fungus Penicillium decumbens active on flavonoid glucosides. J. Mol. Catal. B Enzym. 27, 183–190. doi: 10.1016/j.molcatb.2003.11.011
Nagano, N., Orengo, C. A., and Thornton, J. M. (2002). One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 321, 741–765. doi: 10.1016/S0022-2836(02)00649-6
Nakaya, A., Katayama, T., Itoh, M., Hiranuka, K., Kawashima, S., Moriya, Y., et al. (2013). KEGG OC: a large-scale automatic construction of taxonomy-based ortholog clusters. Nucleic Acids Res. 41, D353–D357. doi: 10.1093/nar/gks1239
Qu, X., Ding, B., Li, J., Liang, M., Du, L., Wei, Y., et al. (2020). Characterization of a GH3 halophilic β-glucosidase from Pseudoalteromonas and its NaCl-induced activity toward isoflavones. Int. J. Biol. Macromol. 164, 1392–1398. doi: 10.1016/j.ijbiomac.2020.07.300
Quezada, A. G., Díaz-Salazar, A. J., Cabrera, N., Pérez-Montfort, R., Piñeiro, Á., and Costas, M. (2017). Interplay between protein thermal flexibility and kinetic stability. Structure 25, 167–179. doi: 10.1016/j.str.2016.11.018
Rani, V., Mohanram, S., Tiwari, R., Nain, L., and Arora, A. (2015). Beta-glucosidase: key enzyme in determining efficiency of Cellulase and biomass hydrolysis. Journal of bioprocessing & biotechniques. Available online at: https://www.semanticscholar.org/paper/Beta-Glucosidase%3A-Key-Enzyme-in-Determining-of-and-Rani-Mohanram/c4711ace1f6a2e38ee9b020c7bc420572ce0aa3a (accessed January 3, 2025).
Richardson, T. H., Tan, X., Frey, G., Callen, W., Cabell, M., Lam, D., et al. (2002). A novel, high performance enzyme for starch liquefaction: Discovery and optimization of a low pH, thermostable α-amylase*. J. Biol. Chem. 277, 26501–26507. doi: 10.1074/jbc.M203183200
Rubiano-Labrador, C., Baena, S., Díaz-Cárdenas, C., and Patel, B. K. C. (2013). Caloramator quimbayensis sp. nov., an anaerobic, moderately thermophilic bacterium isolated from a terrestrial hot spring. Int. J. Syst. Evol. Microbiol. 63, 1396–1402. doi: 10.1099/ijs.0.037937-0
Sengupta, S., Datta, M., and Datta, S. (2023). “Chapter 5 – β-glucosidase: structure, function and industrial applications” in Glycoside Hydrolases. eds. A. Goyal and K. Sharma (Cambridge, MA, USA: Academic Press), 97–120.
Shipkowski, S., and Brenchley, J. E. (2005). Characterization of an unusual cold-active beta-glucosidase belonging to family 3 of the glycoside hydrolases from the psychrophilic isolate Paenibacillus sp. strain C7. Appl. Environ. Microbiol. 71, 4225–4232. doi: 10.1128/AEM.71.8.4225-4232.2005
Singh, N., Sithole, B., Kumar, A., and Govinden, R. (2023). A glucose tolerant β-glucosidase from a newly isolated Neofusicoccum parvum strain F7: production, purification, and characterization. Sci. Rep. 13:5134. doi: 10.1038/s41598-023-32353-6
Sinha, S. K., Datta, M., and Datta, S. (2021). A glucose tolerant β-glucosidase from Thermomicrobium roseum that can hydrolyze biomass in seawater. Green Chem. 23, 7299–7311. doi: 10.1039/D1GC01357B
Sun, J., Wang, W., Ying, Y., and Hao, J. (2020). A novel glucose-tolerant GH1 β-glucosidase and improvement of its glucose tolerance using site-directed mutation. Appl. Biochem. Biotechnol. 192, 999–1015. doi: 10.1007/s12010-020-03373-z
Tatusov, R. L., Natale, D. A., Garkavtsev, I. V., Tatusova, T. A., Shankavaram, U. T., Rao, B. S., et al. (2001). The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29, 22–28. doi: 10.1093/nar/29.1.22
Toyama, D., de Morais, M. A. B., Ramos, F. C., Zanphorlin, L. M., Tonoli, C. C. C., Balula, A. F., et al. (2018). A novel β-glucosidase isolated from the microbial metagenome of Lake Poraquê (Amazon, Brazil). Biochim. Biophys. Acta Proteins Proteom. 1866, 569–579. doi: 10.1016/j.bbapap.2018.02.001
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., et al. (2022). AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444. doi: 10.1093/nar/gkab1061
Xie, J., Xu, H., Jiang, J., Zhang, N., Yang, J., Zhao, J., et al. (2020). Characterization of a novel thermostable glucose-tolerant GH1 β-glucosidase from the hyperthermophile Ignisphaera aggregans and its application in the efficient production of baohuoside I from icariin and total epimedium flavonoids. Bioorg. Chem. 104:104296. doi: 10.1016/j.bioorg.2020.104296
Yenenler, A., and Sezerman, O. U. (2016). Design and characterizations of two novel cellulases through single-gene shuffling of Cel12A (EG3) gene from Trichoderma reseei. Protein Eng. Des. Sel. 29, 219–229. doi: 10.1093/protein/gzw011
Yin, Y. R., Li, X. W., Long, C. H., Li, L., Hang, Y. Y., Rao, M. D., et al. (2023). Characterization of a GH10 extremely thermophilic xylanase from the metagenome of hot spring for prebiotic production. Sci. Rep. 13:16053. doi: 10.1038/s41598-023-42920-6
Yin, Y. R., Meng, Z. H., Hu, Q. W., Jiang, Z., Xian, W. D., Li, L. H., et al. (2017). The hybrid strategy of Thermoactinospora rubra YIM 77501T for utilizing cellulose as a carbon source at different temperatures. Front. Microbiol. 8:942. doi: 10.3389/fmicb.2017.00942
Yin, Y. R., Sang, P., Xian, W. D., Li, X., Jiao, J. Y., Liu, L., et al. (2018). Expression and characteristics of two glucose-tolerant GH1 β-glucosidases from Actinomadura amylolytica YIM 77502T for promoting cellulose degradation. Front. Microbiol. 9:3149. doi: 10.3389/fmicb.2018.03149
Zechel, D. L., and Withers, S. G. (2000). Glycosidase mechanisms: anatomy of a finely tuned catalyst. Acc. Chem. Res. 33, 11–18. doi: 10.1021/ar970172+
Zhang, Z., Wang, M., Gao, R., Yu, X., and Chen, G. (2017). Synergistic effect of thermostable β-glucosidase TN0602 and cellulase on cellulose hydrolysis. 3 Biotech. 7, 54. doi: 10.1007/s13205-017-0672-2
Zhu, Q., Huang, Y., Yang, Z., Wu, X., Zhu, Q., Zheng, H., et al. (2024). A recombinant thermophilic and glucose-tolerant GH1 β-glucosidase derived from Hehua hot spring. Molecules 29:1017. doi: 10.3390/molecules29051017
Keywords: Tengchong Rehai, thermostable, glucotolerant, β-glucosidase, β-galactosidase
Citation: Huang Y-Y, Zhu D, Yang L-Q, Ortúzar M, Yang Z-F, Lv Z-H, Xie K-Q, Jiang H-C, Li W-J and Yin Y-R (2025) Characterization, thermostable mechanism, and molecular docking of a novel glucose-tolerant β-glucosidase/β-galactosidase from the GH1 family isolated from Rehai hot spring. Front. Microbiol. 16:1559242. doi: 10.3389/fmicb.2025.1559242
Received: 12 January 2025; Accepted: 20 March 2025;
Published: 11 April 2025.
Edited by:
Johann Orlygsson, University of Akureyri, IcelandReviewed by:
Cesar Diaz Perez, University of Guanajuato, MexicoCopyright © 2025 Huang, Zhu, Yang, Ortúzar, Yang, Lv, Xie, Jiang, Li and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi-Rui Yin, eWlueWlydWlAZGFsaS5lZHUuY24=; Wen-Jun Li, bGl3ZW5qdW4zQG1haWwuc3lzdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.