
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Microbiol. , 20 March 2025
Sec. Microbiotechnology
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1551928
This article is a correction to:
Biostimulation of Indigenous Microbial Community for Bioremediation of Petroleum Refinery Sludge
 Jayeeta Sarkar1
Jayeeta Sarkar1 Sufia K. Kazy2*
Sufia K. Kazy2* Abhishek Gupta1
Abhishek Gupta1 Avishek Dutta3
Avishek Dutta3 Balaram Mohapatra1
Balaram Mohapatra1 Ajoy Roy2
Ajoy Roy2 Paramita Bera4
Paramita Bera4 Adinpunya Mitra4
Adinpunya Mitra4 Pinaki Sar1*
Pinaki Sar1*A corrigendum on
Biostimulation of indigenous microbial community for bioremediation of petroleum refinery sludge
by Sarkar, J., Kazy, S. K., Gupta, A., Dutta, A., Mohapatra, B., Roy, A., Bera, P., Mitra, A., and Sar, P. (2016). Front. Microbiol. 7:1407. doi: 10.3389/fmicb.2016.01407
In the published article, there was an error in the legend for Supplementary Figure S4 as published. There was a typographical error in the legend causing S4C to be referred to as S4G. Chromatograms shown earlier were rescaled, which are now replaced with original chromatograms. The legend has been updated accordingly.
In the published article, there was an error in Figure 2, with the legend “Gas chromatogram for GR3, U and N samples.” In the original figure, the chromatogram of N_30 was inadvertently duplicated in place of N_90. The correct chromatogram has been restored and updated in the revised Figure 2. The corrected Figure 2 and its caption are provided here.
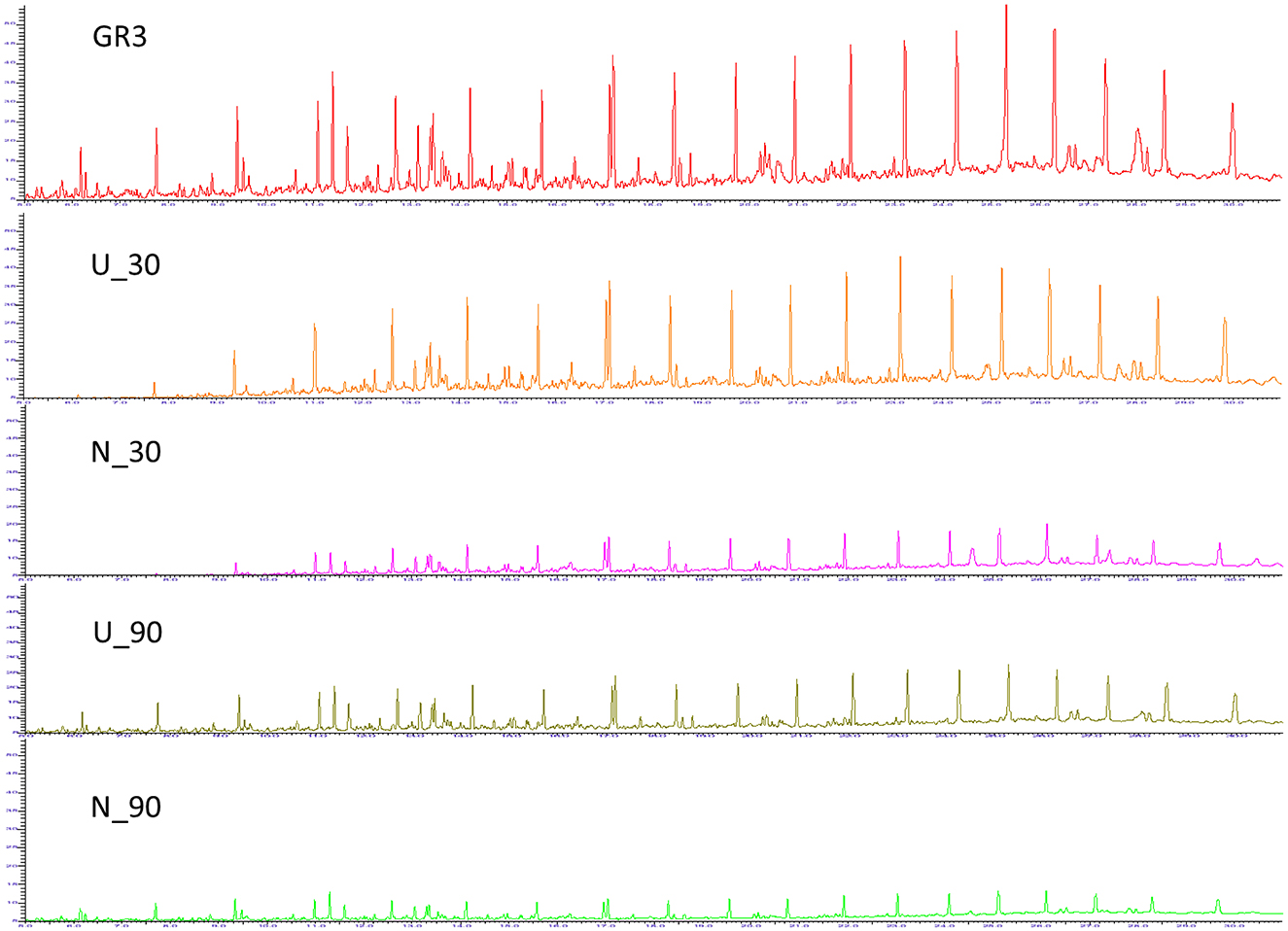
Figure 2. Gas chromatogram for GR3, U and N samples. GC-FID chromatograms showing constituent hydrocarbons after 30 and 90 days of microcosm incubation with or without nitrate amendment (U and N). Peaks have been assigned to carbon chain length corresponding to TPH standard (C10–28).”
In the published article, there was an error in Figure 3, with the legend “Denaturing gradient gel electrophoresis of microcosm samples.” Figure 3A showed the DGGE banding patterns of the studied samples and is comprised of lanes of the same gel which were spliced from the original gel image and arranged to match the UPGMA clustering done subsequently. Unfortunately, during the compilation of this gel image, lane P was duplicated as NS, and there was some mistake in the lane N as well. We are sorry for the oversight. The image has now been recompiled and all lanes of Figure 3A have been corrected. Re-analysis of the revised Figure 3A produced a change in the UPGMA clustering pattern and Figure 3B has been replaced accordingly. Based on the outcome of UPGMA re-analysis, the concerned results have been updated. The corrected Figure 3 and its caption are provided here.
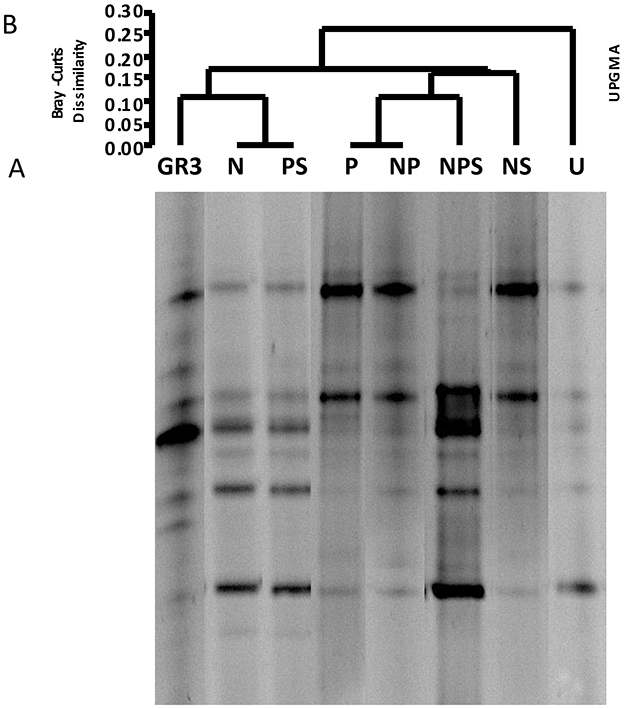
Figure 3. Denaturing gradient gel electrophoresis of microcosm samples. DGGE patterns of bacterial communities from microcosms with various amendments (A) and Bray–Curtis cluster analysis among the DGGE band patterns (B). No amendment control (U) was also included. For Bray–Curtis analysis presence or absence of bands at corresponding positions was considered”.
In the published article, there was an error in Figure 3, as explained previously. In light of the rectification of this figure, the results inferred from it have also been updated.
A correction has been made to Results, Shift in Microbial Community with Amendments paragraph number one.
This sentence previously stated:
“However, the UPGMA on presence and absence of bands showed a relationship between the treatments (Figure 3B). Presence of two distinct clades at a similarity index of 0.25 to 0.2 showed a clear clustering of band patterns among GR3, N, P, PS (clade A) and U, NP, NS, NPS (clade B) samples. The result also indicated a distinct change in community composition following incubation within the microcosm without any treatment (U) which was consistent even for NP, NS and NPS amendments. Interestingly, even after incubating with N or P or PS, the community compositions were more similar to the original one (Figure 3A).”
The corrected sentence appears below:
“However, the UPGMA on presence and absence of bands showed a relationship between the treatments (Figure 3B). Presence of two distinct clades at a dissimilarity index of 0.15 to 0.20 showed a clustering of band patterns among GR3, N, PS (clade A) and P, NP, NPS, NS (clade B) samples with U cladding separately. The result also indicated a distinct change in community composition following incubation within the microcosm without any treatment (U). Interestingly, even after incubating with N or PS, the community compositions were more similar to the original one, relative to the other treatments (Figure 3A)”
The authors apologize for these errors and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: petroleum refinery sludge, microbial community, bioremediation, biostimulation, next generation sequencing
Citation: Sarkar J, Kazy SK, Gupta A, Dutta A, Mohapatra B, Roy A, Bera P, Mitra A and Sar P (2025) Corrigendum: Biostimulation of indigenous microbial community for bioremediation of petroleum refinery sludge. Front. Microbiol. 16:1551928. doi: 10.3389/fmicb.2025.1551928
Received: 03 January 2025; Accepted: 29 January 2025;
Published: 20 March 2025.
Edited and reviewed by: Eric Altermann, Massey University, New Zealand
Copyright © 2025 Sarkar, Kazy, Gupta, Dutta, Mohapatra, Roy, Bera, Mitra and Sar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sufia K. Kazy, c3VmaWFfa2F6eUB5YWhvby5jb20=; Pinaki Sar, c2FycGluYWtpQHlhaG9vLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.