
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 12 February 2025
Sec. Food Microbiology
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1543686
This article is part of the Research TopicCritical- and High-Priority Pathogens in the Food ChainView all articles
Detection of Shiga toxin-producing Escherichia coli (STEC) presenting high risk of human infections is challenging. In France, the latest Anses opinion categorized STEC in four groups based on their association with severe forms of clinical infection. STEC strains carrying the eae gene, particularly those of serogroups O157, O26, O111, O103, O145, O121, O45 and more recently O80 (top 8 serogroups), are usually monitored worldwide, whereas eae-negative STEC strains that are less clinically significant are not surveyed. Screening food enrichment broths with classical genetic markers (stx, eae) can overestimate the presence of highly virulent STEC, causing needless disruption and costs for food producers. Recently the updated MLG5C reference method introduced additional genetic markers (espK, espV) in the detection scheme to improve specificity and effectiveness of priority STEC detection in foodstuffs. This study, conducted on beef samples with a new method supporting the regulatory USDA-FSIS MLG5C.04 method, showed that 92% of the stx-positive samples carry stx2 alone or in association with stx1. Among the stx2-positive samples, stx2a and/or stx2d subtypes dominate. Introduction of espK, espV markers on 868 stx+/eae+ beef enrichment broths reduced the number of presumptive positive results by 31%, compared to the ISO/TS 13136:2012 reference method. Subsequent analysis of the presumptive positives combining the O-group and the eae-subtype provided also a significant reduction of the number of the presumptive positive for the top 8 eae-positive STEC serogroups; and showed that O26, O103 and O157 were the most prevalent ones. Regarding the stx+/eae- samples, which are proportionally extremely predominant in beef as compared with the stx+/eae+ samples, 65% of them were positive for the serogroups monitored in this study (O91, O171, O174, O148, O146, O128 O113 and O104). The high occurrence of serogroup O113 observed in beef samples is not corroborated by the clinical data reported in France. Routine testing of beef samples should be revised to prioritize a hierarchical surveillance system based only on high risk STEC (STEC carrying the eae gene) and not on all STEC. This approach would provide Food Business Operators a significant improvement, saving time and costs while maintaining a high level of product safety.
Shiga toxin-producing Escherichia coli (STEC) are defined as strains that harbor the stx gene, which produces Shiga toxins, but may carry other virulence genes (FAO and WHO, 2018). The Shiga toxins belong to two major types stx1 further divided into three subtypes (a, c and d) and stx2 comprising 15 subtypes (a-o) (Lindsey et al., 2023). STEC strains do not carry the “Locus of Enterocyte Effacement” (LEE), a genomic island that includes the eae gene encoding the intimin, an adhesion factor to the intestinal mucosa first identified in enteropathogenic E. coli (EPEC), and which cause infant diarrhea in developing countries (Olayinka et al., 2024). STEC infections range from mild diarrhea to severe enterohemorrhagic diseases such as hemorrhagic colitis (HC), and hemolytic-uremic syndrome (HUS) (Boerlin et al., 1998; Kobayashi et al., 2013). Although more than 400 STEC serotypes have been isolated from human cases (sporadic and epidemic), only a minority are responsible for most infection cases (Beutin and Fach, 2014). A pathogenic subset called typical enterohemorrhagic E. coli (EHEC), defined by the presence of stx and eae genes, is responsible for enterohemorrhagic disease (Croxen and Finlay, 2010). The most frequent serotypes classified in this group are the top 7 serotypes O157:H7, O26:H11, O111:H8, O103:H2, O145:H28, O121:H19 and O45:H2 (Anses, 2023). Atypical EHEC strains encode stx genes but do not carry the LEE. The clinically significant ones belong mainly to O-groups O91, O171, O174, O148, O146, O128, O113, and O104 (Anses, 2023). However, as these are rarely associated with large foodborne outbreaks, they are not currently monitored in foods. While the top 7 serogroups of typical EHEC are closely monitored by U.S. food safety authorities, the EU prioritizes five EHEC serogroups (O157, O26, O111, O103, and O145). In recent years, O80 (eae-positive), has emerged as an important serogroup associated with STEC infections in France (Cointe et al., 2018; Cointe et al., 2020) and the French Agency for Food, Environmental and Occupational Health and Safety (Anses) recommends its routine monitoring in foods in the future.
The genetic markers aaiC and aggR are also used to target some cross-pathotype E. coli like the hybrid STEC/EAEC O104:H4 strain that caused a large epidemic in 2011 in both France and Germany, (Bielaszewska et al., 2011; Muniesa et al., 2012). Since then, it has been clearly evidenced that this epidemic strain was not zoonotic (Auvray et al., 2012). More recently, different clones of hybrid STEC of serotype O80:H2 caused HUS cases in Europe (Cointe et al., 2021). These hybrid strains were LEE- and stx2-positive (though a particular type of intimin (eae-xi) and different combinations of stx2a, stx2c and stx2d genes).
Based on French clinical epidemiological and microbiological data and international literature, a categorization of STEC strains has been recently proposed in the latest Anses opinion (Anses, 2023). Clinical isolates were classified in four groups. Group I [(stx2a and/or stx2d)+, (eae or aaiC and/or aggR)+] accounts for 84% of the notified HUS and 34% of notified bloody diarrhea. Group II [(stx2a and/or stx2d)+, (eae or aaiC/aggR)−] accounts for only 5% of the notified HUS and 6% of notified bloody diarrhea. Group III [(other stx-subtypes)+, (eae or aaiC/aggR)+] is associated with 7% of the notified HUS and 42% of notified bloody diarrhea. Group IV [(other stx-subtypes)+, (eae or aaiC/aggR)−] is reported in only 4% of the notified HUS and 18% of notified bloody diarrhea. French data obtained in Human showed clearly a clinical significance of STEC strains from groups I and III. According to the FAO, STEC strains stx2a and eae- or aggR-positive are at higher risk of causing HUS (FAO and WHO, 2018). In addition, the presence of stx2d –alone or in combination with eae or aggR– is also considered at risk for causing HUS. The National Advisory Committee on Microbiological Criteria for Foods (NACMCF) and the United States Department of Agriculture (USDA) classify STEC carrying stx2a with aggR or eae at high risk of causing severe forms (NACMCF, 2019). These different approaches for assessing STEC pathogenicity nowadays focus on virulence profiles rather than serogroups, moving away from the seropathotype classification (Karmali et al., 2003). Although the serogroup is epidemiologically important for tracking outbreaks, it is insufficient for predicting pathogenicity, especially for strains not fully serotyped.
The ISO/TS 13136 (EU) and MLG5C.03 (US) reference methods (ISO, 2012; USDA-FSIS, 2021) bear the disadvantage to produce many false-positive as the virulence markers (stx and eae) are found on mobile genetic elements. Thus, other non-pathogenic bacteria and free bacteriophages can carry them as well. These false positives are a global challenge for the regulatory agencies and food producers, causing needless disruption and costs. The USDA Food Safety and Inspection Service (FSIS) has declared six non-O157 STEC serogroups (O26, O45, O103, O111, O121 and O145), known as the “big six,” as adulterants in raw beef products, in addition to E. coli O157:H7 (USDA-FSIS, 2012). In 2023, FSIS announced that it began testing all raw beef samples for non-O157 STEC, including raw ground beef, and other beef components (USDA-FSIS, 2023). The MLG5C reference method was updated in 2024 by incorporating recent advancements from the bioMérieux GENE-UP Pathogenic E. coli (PEC) kit (USDA-FSIS, 2024). This new approach of the MLG5C.04 includes additional virulence markers like the type III effectors espK and espV (Delannoy et al., 2013a) enhancing specificity for pathogenic EHEC strains.
This study aims to evaluate the proof of concept of a new method to predict, at the early screening step, beef enrichments broths that are presumptive positives for highly virulent STEC (i.e., strains that carry both the stx and eae genes, particularly those classified in groups I and III in the latest Anses opinion). This study is not an epidemiological survey and does not reflect the prevalence of STEC or EHEC in the French beef sectors.
Beef samples tested in this study were selected by Interbev (Association Nationale Inter-professionnelle du Bétail et des Viandes) from all production regions of France. They were composed of ground beef and carcasses that were processed by local laboratories with enrichment conditions supporting the growth of non-O157 and O157 STEC (Walker et al., 2024). Then, DNA was extracted from the beef enrichments and tested to determine the stx/eae status, as described by Delannoy et al. (2022). Extracted DNA were stored at −20°C until transportation to Anses for PCR analysis.
Sample selection was biased to get greater numbers of DNA samples positive for STEC, with a target of 868 samples positive for both stx and eae. Thus, this sample selection scheme does not reflect the real prevalence of STEC or EHEC in the French beef sectors. However, it is well designed to assess the contribution of the new genetic markers in the reduction in the number of presumptive positive samples after the first phase of screening with stx and eae. Samples positive for stx and eae were further PCR tested for the top 5 EHEC serogroups plus O121, O45 and O80, isolation of strains was attempted for confirmation of the top 5 EHEC serotypes regulated in France (O157, O145, O111, O103 and O26). Following the recommendation of the French ministry of agriculture, the appropriate sanitary measures were taken in positive cases of EHEC top 5. Moreover, regarding samples that were positive for stx only, they were further PCR tested for the following atypical EHEC O-groups: O91, O171, O174, O148, O146, O128, O113, and O104 (Figure 1).
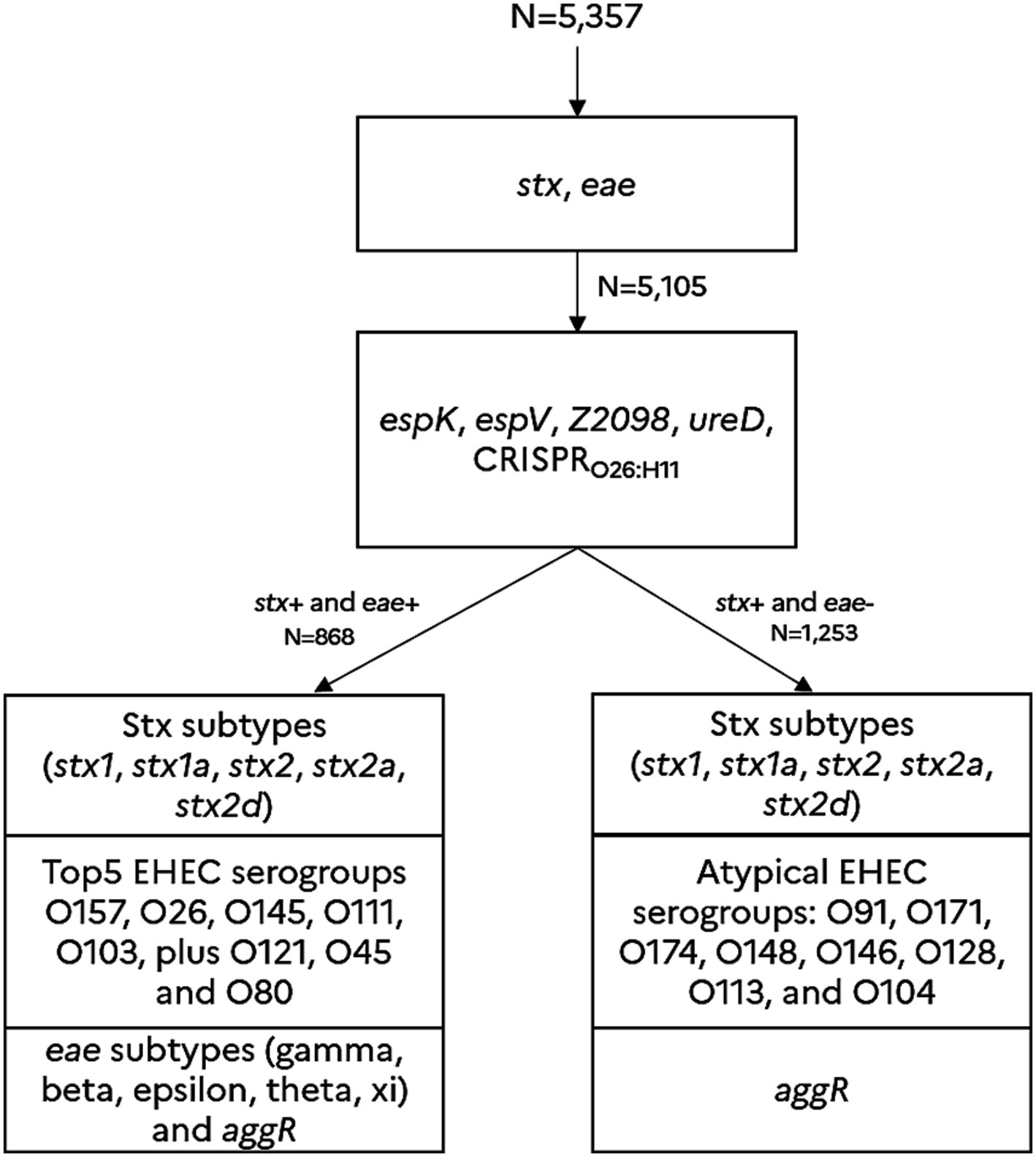
Figure 1. Screening approach for predicting the presence of typical EHEC and atypical EHEC in beef samples. The detection scheme (in line with the new MLG5C.04 reference method) is based on a sequential detection of various genetic markers such as the stx genes (including the significant stx2a and stx2d sub-types), the eae gene (including the eae sub-types gamma, beta, epsilon, theta and xi), and the molecular markers, espK, espV, ureD, Z2098, and CRISPRO26:H11. These genetic markers were previously described and validated as hallmarks of virulent STEC (Delannoy et al., 2013a, 2013b, 2015, 2016, 2022). The marker aggR is a hallmark of typical enteroaggregative E. coli. The genetic markers associated with the main common EHEC O-groups O157 (eae-gamma), O145 (eae-gamma), O26 (eae-beta) O111 (eae-thêta), O103 (eae-epsilon), O121 (eae-epsilon), O45 (eae-epsilon) and O80 (eae-xi), plus the uncommon EHEC O-groups (O91, O171, O174, O148, O146, O128, O113, and O104) were also tested.
As the high-throughput real-time PCR system relies on microfluidic PCR (PCR reaction taking on place on few nanoliters), a pre-amplification is required to guaranty the sensitivity of the method (Michelet et al., 2014). DNA extracts were processed by high-throughput real-time PCR using the Biomark™ real-time PCR system (Standard BioTools, USA) as described previously in an equivalent study performed on dairy products (Delannoy et al., 2022). Primers and probes used in high-throughput real-time PCR were those previously described by Delannoy et al. (2022) or were designed for this study. Table 1 reports all oligonucleotides used for this study.
Regarding the determination of presumptive positive samples for group I and III, three methods were explored and are defined as followed: method A (stx, eae, espK/Z2098/CRISPRO26:H11), method B (stx, eae, espK/espV/CRISPRO26:H11) and method C (stx, eae, espK/ureD/CRISPRO26:H11). A sample was considered presumptive positive if it tested positive for stx, eae, and at least one of the additional targets specified in each method. For example, a presumptive positive for method A was defined as a sample that tested stx+, eae+, and espK/Z2098/CRISPRO26:H11+ (meaning positive for at least one of the three targets espK, Z2098, CRISPRO26:H11). The same principle applies to methods B and C, where espV or ureD serve as the respective additional targets.
In a second step, all DNA samples stx+/eae+ (i.e., presumptive positives for group I and III) were further tested by real-time PCR for the main typical EHEC O-groups (O157, O26, O145, O111, O103, O145, O121, O45 and O80) and their corresponding eae-variants (eae-gamma, −beta, −epsilon, −theta, −xi subtypes). A presumptive positive recorded in this second screening was a sample stx+/eae+ that tested positive for the appropriate association O-group/eae-subtype.
The first screening with the new EHEC markers (methods A, B and C) reports the number of presumptive positives for all typical EHEC serotypes and not only for the top 7 EHEC serotypes. The second screening, as based on the combination of the O-group/eae type reports the number of presumptive positives for the top 7 EHEC serogroups plus O80, which are usually monitored worldwide. Data presented in this study showed the number of presumptive positives after the first screening (New EHEC markers) and the second screening step (O-groups/eae-types). Data were compared with the ISO/TS 13136 reference method (ISO, 2012).
Regarding the determination of presumptive positive samples for EHEC of groups II and IV, all DNA samples stx+/eae- were further PCR tested for the main atypical EHEC O-groups (O91, O171, O174, O148, O146, O128, O113, and O104).
Statistical analyses were performed on R Studio version 2021.09.0. Two-proportions z-tests (two-tailed or one tailed) were used to compare proportions of presumptive positives between the ISO reference method and the new approach (methods A, B, and C). The statistical tests were performed with α of 5%.
DNA extracts (n = 5,357) from beef were sent to Anses. The stx/eae status as determined by the local laboratories and Anses was identical for 5,105 DNA extracts. These samples having the same stx/eae status by conventional real-time PCR and by microfluidics real-time PCR were further used in this study. DNA extracts analysed in this study were either positive for stx alone (n = 1,253), positive for stx and eae (n = 868), positive for eae alone (n = 1,232), or negative for both stx and eae (n = 1,752). The study as a whole represents 225,504 real-time PCR determinations, which served to consolidate the results presented in this manuscript.
Among the DNA samples tested, a total of 2,121 DNA extracts were stx-positive (1,253 positive for stx only and 868 positive for both stx and eae). The data showed that the distribution of the stx1 and stx2 genes is quite different (Table 2). It is remarkable that only 8% of the stx-positive samples are positive for stx1 only. The stx2 gene was significantly more prevalent with 67% of the samples positive for stx2 only and 25% positive for stx1 and stx2 (p < 0.05). As a whole, 92% of the stx-positive samples are thus positive for stx2 alone or in association with stx1. Among the 1,955 stx2-positive samples, 93% are positive for at least stx2a and/or stx2d, showing the high prevalence of these two subtypes in beef samples. The other stx2 positive samples (approximately 6%) were positive for stx2-subtypes other than stx2a and stx2d.
All the 2,121 stx-positive samples were also categorized in groups I to IV based on the stx2a and stx2d real-time PCR results and the detection of eae and aggR. Overall, group I [(stx2a and/or stx2d)+, eae+] comprised 724 samples, group II [(stx2a and/or stx2d)+, eae-] comprised 1,102 samples, group III (other stx+, eae+) comprised 144 samples and group IV (other stx+, eae-) comprised 151 samples. It is remarkable to note that all stx+ samples tested negative for aggR (data not shown).
A total of 868 samples stx+/eae+ were investigated with methods A, B and C and the number of presumptive positives was reported for each method. In total, 597 presumptive positives were recorded according to method A, 596 with method B and 588 with method C. Alternate methods of pre-screening (methods A, B, and C) that include in addition to stx/eae, the detection of five novel markers (espK, espV, Z2098, ureD, and CRISPRO26:H11) allowed a significant reduction in the number of presumptive positives, with a reduction rate of approximately 31–32% with regard to the ISO/TS 13136:2012 reference method (Table 3). As shown in Figure 2, the reduction of the number of presumptive positives obtained with methods A, B and C is significantly higher for samples classified in group I (35–36% reduction) than in group III (12–15% reduction).
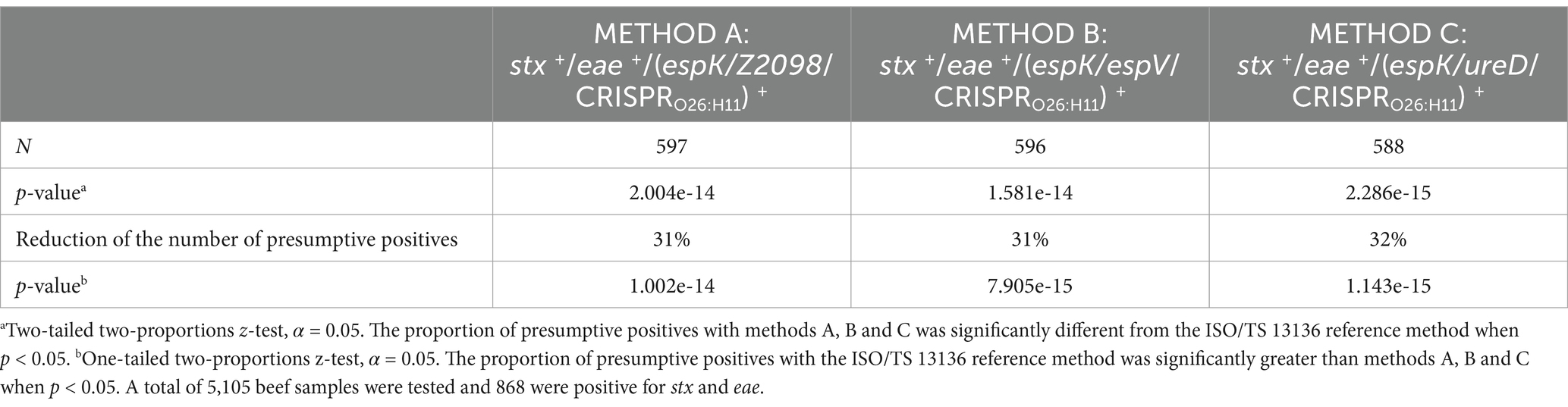
Table 3. Reduction of the number of presumptive positives by introducing new EHEC markers in the first screening phase of the ISO/TS 13136 reference method.
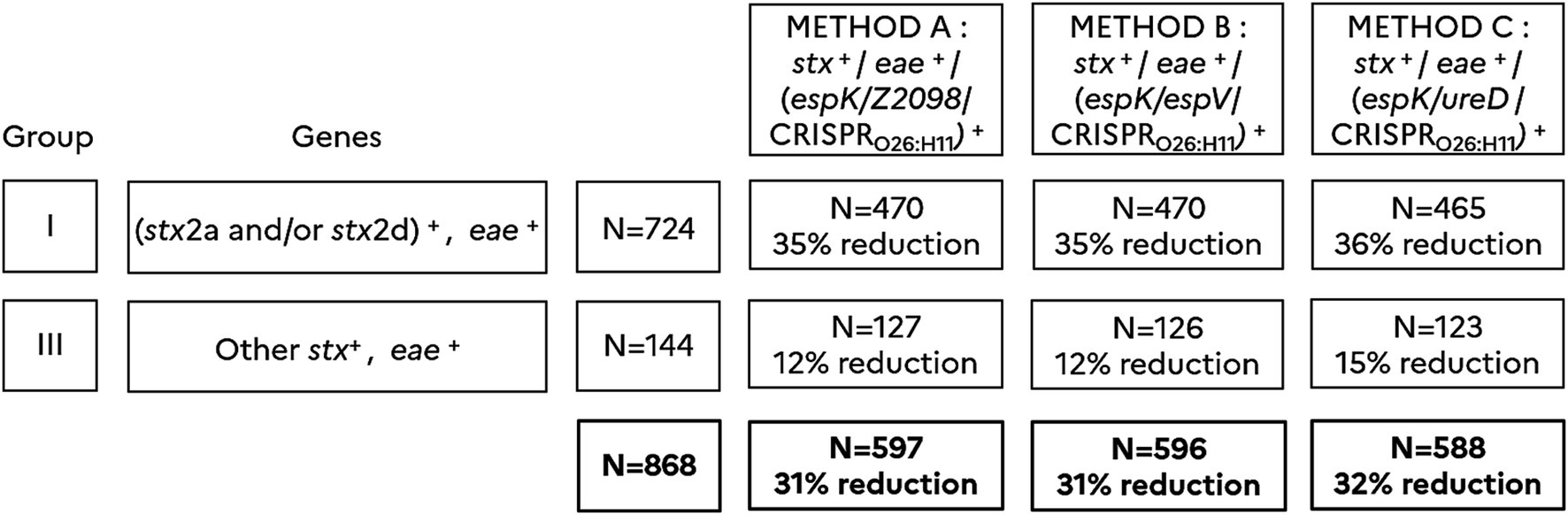
Figure 2. Classification of 2,121 stx-positive samples according to Anses opinion (2023) and comparison of methods A, B and C. (stx2a and/or stx2d)+ includes samples giving a positive result for stx2a and/or stx2d. Other stx+ includes samples giving a positive result for stx subtypes other than stx2a and stx2d. eae+ includes samples giving a positive result for eae. (espK/Z2098/CRISPRO26:H11)+ includes samples giving a positive result for at least one of the markers espK, Z2098, or CRISPRO26:H11. (espK/espV/CRISPRO26:H11)+ includes samples giving a positive result for at least one of the markers espK, espV, or CRISPRO26:H11. (espK/ureD/CRISPRO26:H11)+ includes samples giving a positive result for at least one of the markers espK, ureD, or CRISPRO26:H11. Samples are categorized in group I (N = 724) and III (N = 144) according to the Anses opinion. The percentage of reduction in the number of presumptive positives is calculated for each method A, B and C.
As methods A, B and C gave equivalent results, we chose method B, that is in line with the MLG5C.04, as an alternate method performing well to further analyze samples of groups I and III for the O-groups and the eae subtypes. Based on (1) the data obtained with the pre-screening method B, and (2) the data on the correlation between the presence of the O-antigen markers and the eae-subtypes, we could compare the results of this alternate method with the conventional ISO/TS 13136:2012 reference method which takes into account only the stx, eae and the O-antigen markers. Results were expressed for the top 7 EHEC serogroups plus serogroup O80 (top 8) that has recently emerged in Europe.
As shown in Figure 3 the number of presumptive positives for the top 8 typical EHEC serogroups identified with the ISO/TS 13136:2012 reference method is higher (628 samples) than those identified with the new approach including a pre-screening with method B and subsequent analysis with the combination of the O-group and the eae-subtype (397 samples). Thus, the new approach significantly reduced the number of the presumptive positive top 8 EHEC serogroups by 37% compared to the conventional ISO method (p < 0.05).
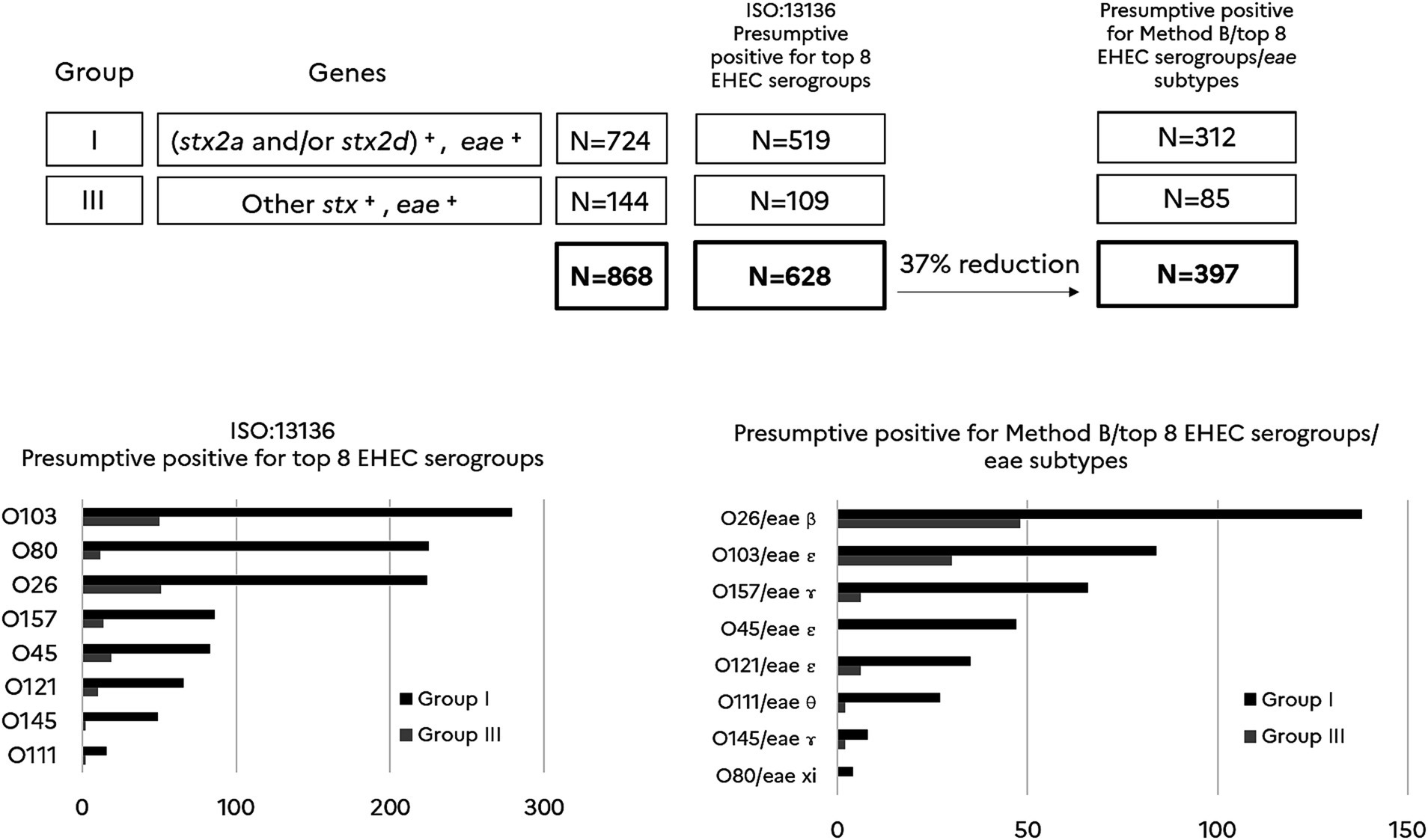
Figure 3. Comparison of presumptive positives for typical EHEC serogroups (top 8) using the ISO/TS13136:2012 reference method vs the new approach. (stx2a and/or stx2d)+ includes samples giving a positive result for stx2a and/or stx2d. Other stx+ includes samples giving a positive result for stx subtypes other than stx2a and stx2d. eae+ includes samples giving a positive result for eae. Samples are categorized in group I (N = 724) and III (N = 144) according to the Anses opinion. N = 628 is the number of presumptive positives for the top 8 EHEC serogroups according to the ISO:13136 reference method. N = 397 is the number of presumptive positives recorded by combining Method B, the top 8 EHEC serogroups and their corresponding eae subtype. The percentage of reduction in the number of presumptive positives is calculated for the new method when compared with the ISO reference method (p < 0.05). For each method, the histograms report the number of presumptive positives for each O-group and for samples categorized in groups I and III.
It is also noteworthy that the association of the O-groups with eae-subtypes affects the ranking of the top 8 typical EHEC serogroups with regard to the categorization obtained with the ISO reference method. The greatest selective effect was observed for O80. The association with eae-xi and O80 allowed discarding most of the presumptive samples for EHEC O80, showing that non-EHEC O80 are probably more prevalent in beef than EHEC O80.
With the objective to get an estimation of uncommon and atypical EHEC and their most prevalent serogroups, the stx+/eae- samples, which are categorized in groups II and IV, were tested for the French clinically significant top 6 atypical EHEC serogroups (O91, O171, O174, O148, O146, O128) plus serogroups O113 and O104. As shown in Figure 4, the data indicated that most of the atypical EHEC presumptive samples are categorized in group II (1,102 samples) and little in group IV (151 samples). Serogroup O113 highly dominates and is followed with a decreasing occurrence by serogroups O174, O91/O146, O148, O171, O104 and O128. One could notice in addition that 35% of the samples classified in groups II and IV tested negative for all the atypical EHEC serogroups monitored in this study (O91, O171, O174, O148, O146, O128, O113, and O104).
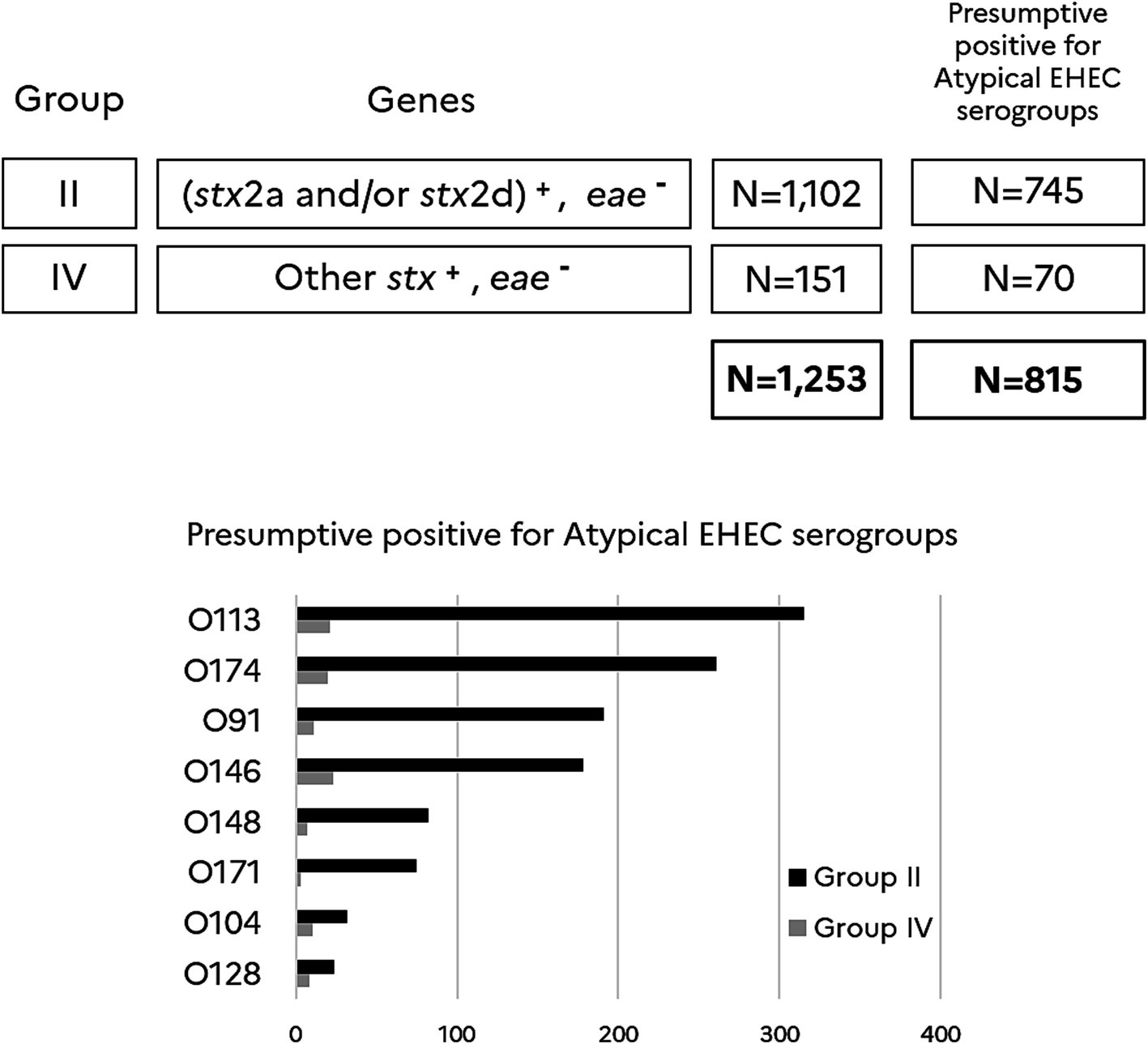
Figure 4. Atypical EHEC top 8 serogroups in stx+/eae- beef samples. (stx2a and/or stx2d)+ includes samples giving a positive result for stx2a and/or stx2d. Other stx+ includes samples giving a positive result for stx subtypes other than stx2a and stx2d. eae-includes samples giving a negative result for eae. Samples are categorized in group II (N = 1,102) and IV (N = 151) according to the Anses opinion. N = 815 is the number of presumptive positives for the top 8 atypical EHEC serogroups recorded in this study. The number of presumptive positives for each O-group and for samples categorized in groups II and IV is reported in the histogram.
It remains difficult to fully define human pathogenic STEC or identify virulence factors for STEC that absolutely predict the potential to cause human disease (EFSA and ECDC, 2021). However, data analysis of the French and European epidemiological studies underlines the relative importance of some virulence factors/markers. In the latest Anses opinion (Anses, 2023), the characteristics of strains isolated from cases of STEC infections are as follows: (i) the stx2 gene is found in 82% of cases of infections and 95% of cases of HUS; (ii) the eae gene is found in 80% of cases of infections and 90% of cases of HUS; (iii) the stx sub-types mostly associated with HUS are stx2a (58%) and stx2d (31%); (iv) three virulence profiles are responsible for more than 80% of HUS: [stx2a+/eae+], [stx2d+/eae+] and [stx1a+/stx2a+/eae+]; (v) the top 5 serogroups (O157, O26, O103, O145 and O111) and O80 represent 80% of cases of HUS in France. Regarding the clinical isolates that were eae-negative, the most frequently found serogroups are O146, O91 and O128.
Detection of EHEC in foods remains particularly challenging. In the present study, we designed and optimized a molecular risk assessment approach in beef samples that is in line with the MLG5C.04 reference method. This approach aims to provide a significant reduction in false positives while ensuring alignment with the latest Anses recommendations (Anses, 2023) that pointed out STEC of groups I and III as higher risk. For food business operators (FBO) it is important to improve the current detection methods with the aim of increasing the percentage of batches released at the end of the first screening step and of reducing the number of confirmations on isolated colonies while maintaining a high (or even higher) level of product safety.
In this study on beef samples collected in France, we showed that among the stx positive enriched beef samples the occurrence of stx2 is very high (p < 0.05). Indeed, it is remarkable that 92% of the stx positive enriched beef samples carry stx2 alone (67%) or in association with stx1 (25%). Moreover, the stx2 sub-typing scheme allowed to estimate that 93% of the stx2 positive samples are also positive for at least stx2a and/or stx2d. After testing the stx + samples for the eae gene, one could categorize them into groups I to IV according to the latest Anses opinion (Anses, 2023). Among the stx + beef samples, those categorized in group II dominate over the other groups. To refine the analysis of beef enrichments classified in groups I and III, additional biomarkers were tested in complement to the stx and eae genes. This new panel of genes included genetic markers identified previously (Delannoy et al., 2013a, 2013b) as highly associated with pathogenic STEC strains (stx+, eae+), detecting the top 7 EHEC together with many other serotypes than the top 7 of typical EHEC, offering then the possibility for identifying new emerging typical EHEC strains (Delannoy et al., 2013a, 2013b). Conversely, these genetic markers were very rarely (1.1–3.4%) associated with STEC and with non-pathogenic E. coli. Introduction of these markers in the detection scheme provided a significant reduction in the number of stx+/eae + beef samples that require a second screening step for serogroup determination. Moreover, as it relies on the detection of espK/espV it is also consistent with the updated MLG5C.04 reference method (USDA-FSIS, 2024).
Hence, a 31% reduction in the number of presumptive positive was achieved by using the alternate method A (stx/eae/espK/Z2098) or method B (stx/eae/espK/espV), and 32% with method C (stx/eae/espK/ureD) when they were combined with the CRISPRO26:H11 PCR assay. This CRISPR based PCR assay was introduced to detect a minor O26 lineage of EHEC that emerged 10 years ago in France but which declined and is no longer detected in humans (Delannoy et al., 2015). The three methods performed well and gave equivalent levels of selectivity to get a more precise risk assessment. As it is in line with MLG5C.04 reference method, we chose method B (stx/eae/espK/espV) in association with the CRISPRO26:H11 PCR assay, to narrow down the EHEC screening step in beef samples and to go further for EHEC serogroups determination. The new approach including a pre-screening with method B and subsequent analysis combining the O-group and the eae-subtype provided a 54% reduction in the number of the presumptive positive top 7 EHEC plus EHEC O80 (top 8 EHEC serogroups) as compared with a single screening for stx+/eae+. Presumptive positive samples for the top 8 EHEC serogroups accounted for 43% of samples of group I and 59% of group III. The first three serogroups of typical EHEC in presumptive positive samples are O26, O103 and O157.
Regarding beef samples (stx+, eae-) classified in groups II and IV, 88% were in group II and 12% in group IV. Group II, which appears extremely predominant in the beef industry, remains very rarely associated with STEC infections in France (Anses, 2023). Presumptive positive samples for the atypical top 8 EHEC serogroups accounted for 68% of samples of group II and 46% of group IV. The decreasing occurrence of atypical EHEC serogroups is the following: O113, O174, O91/O146, O148, O171, O104 and O128. Serogroup O113 is the most common, whereas serogroup O104 circulates poorly. The high occurrence of serogroup O113 in beef samples is not corroborated by the clinical data reported in the latest Anses opinion (Anses, 2023). As there is no correlation between the epidemiological data observed from the clinical side and the beef industry, we should rethink about the routine testing of raw meat to prioritize a hierarchical surveillance system, which would be based only on high risk STEC and not on all STEC. Given the data recorded in clinical samples in France and those obtained in this study, it makes sense for the routine surveillance of raw meat to focus only on highly virulent STEC of groups I and III (stx+, eae+). Here, we showed that the introduction of the new EHEC markers espK/espV in the detection scheme (at the first screening step on enriched samples) would certainly provide the beef industry a selective and reliable method for tracking the regulated top 7 EHEC serogroups together with other eae-positive STEC serotypes that may emerge in the future. The multiplication of genetic markers that must be included in the detection scheme may be one limitation of this approach as it requires optimization of multiplex PCRs for high throughput routine testing. Multiplication of genetic markers will also increase the cost of the routine analysis for screening highly virulent STEC in beef. Another limitation is that one could never exclude the emergence of new virulent STEC strains that would fail to be detected with these markers. The method developed as part of this project has nonetheless the potential to be used to prioritize STEC risk in raw meat, as although Shiga toxin is essential to STEC pathogenesis, not all STEC strains are systematically associated with disease in humans. The FBO need for their own controls an approach that focus on the higher risk STEC. The approach described here aligns with the updated USDA-FSIS MLG5C.04 method while adapting to the complexities of STEC virulence and epidemiology in the food industry.
Here we propose an optimized testing scheme that includes the detection of additional virulence markers characterizing virulent STEC. By providing a reduction of more than 30% in the number of presumptive positives, this method improves the specificity and effectiveness of STEC detection in foodstuffs, supporting regulatory protocols while reducing operational interruptions in food production.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
M-LT: Data curation, Formal analysis, Methodology, Validation, Writing – original draft. SD: Conceptualization, Formal analysis, Investigation, Methodology, Resources, Supervision, Visualization, Writing – original draft. PF: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Visualization, Writing – original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by Interbev (Association Nationale Inter-professionnelle du Bétail et des Viandes), Grant number SECU-21-02 and by the French Agency for Food, Environmental and Occupational Health and Safety (Anses) supported by the French Ministry of Agriculture and Food.
We are grateful to the “STEC working group” of Interbev for supervising the study and for organizing the shipment of the DNA extracts to Anses for the realization of this study. In particular, we thank CHARDON Hélène, VEAUCLIN Nathalie, ANTOINE-VAUDIN Garance, MELON-WERLY Flore, BILLET Isabelle, LE GOFF Olivier, and GOBERT Christel.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Anses. (2023). Avis relatif à la définition des souches pathogènes D’escherichia coli productrices de Shiga-toxines (saisine n°2020-SA-0095). Available at: https://www.anses.fr/fr/system/files/BIORISK2020SA0095.pdf (Accessed July 11, 2024).
Auvray, F., Dilasser, F., Bibbal, D., Kérourédan, M., Oswald, E., and Brugère, H. (2012). French cattle is not a reservoir of the highly virulent enteroaggregative Shiga toxin-producing Escherichia coli of serotype O104:H4. Vet. Microbiol. 158, 443–445. doi: 10.1016/j.vetmic.2012.02.029
Beutin, L., and Fach, P. (2014). Detection of Shiga toxin-producing Escherichia coli from nonhuman sources and strain typing. Microbiol. Spectr. 2:2013. doi: 10.1128/microbiolspec.EHEC-0001-2013
Bielaszewska, M., Mellmann, A., Zhang, W., Köck, R., Fruth, A., Bauwens, A., et al. (2011). Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 11, 671–676. doi: 10.1016/S1473-3099(11)70165-7
Boerlin, P., Chen, S., Colbourne, J. K., Johnson, R., De Grandis, S., and Gyles, C. (1998). Evolution of enterohemorrhagic Escherichia coli hemolysin plasmids and the locus for enterocyte effacement in Shiga toxin-producing E. coli. Infect. Immun. 66, 2553–2561. doi: 10.1128/IAI.66.6.2553-2561.1998
Bugarel, M., Beutin, L., Martin, A., Gill, A., and Fach, P. (2010). Micro-array for the identification of Shiga toxin-producing Escherichia coli (STEC) seropathotypes associated with hemorrhagic colitis and hemolytic uremic syndrome in humans. Int. J. Food Microbiol. 142, 318–329. doi: 10.1016/j.ijfoodmicro.2010.07.010
Cointe, A., Birgy, A., Bridier-Nahmias, A., Mariani-Kurkdjian, P., Walewski, V., Lévy, C., et al. (2020). Escherichia coli O80 hybrid pathotype strains producing Shiga toxin and ESBL: molecular characterization and potential therapeutic options. J. Antimicrob. Chemother. 75, 537–542. doi: 10.1093/jac/dkz484
Cointe, A., Birgy, A., Mariani-Kurkdjian, P., Liguori, S., Courroux, C., Blanco, J., et al. (2018). Emerging multidrug-resistant hybrid Pathotype Shiga toxin-producing Escherichia coli O80 and related strains of clonal complex 165, Europe. Emerg. Infect. Dis. 24, 2262–2269. doi: 10.3201/eid2412.180272
Cointe, A., Bizot, E., Delannoy, S., Fach, P., Bidet, P., Birgy, A., et al. (2021). Emergence of new ST301 Shiga toxin-producing Escherichia coli clones harboring extra-intestinal virulence traits in Europe. Toxins 13:686. doi: 10.3390/toxins13100686
Croxen, M. A., and Finlay, B. B. (2010). Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8, 26–38. doi: 10.1038/nrmicro2265
Delannoy, S., Beutin, L., Burgos, Y., and Fach, P. (2012b). Specific detection of enteroaggregative hemorrhagic Escherichia coli O104:H4 strains by use of the CRISPR locus as a target for a diagnostic real-time PCR. J. Clin. Microbiol. 50, 3485–3492. doi: 10.1128/JCM.01656-12
Delannoy, S., Beutin, L., and Fach, P. (2012a). Use of clustered regularly interspaced short palindromic repeat sequence polymorphisms for specific detection of enterohemorrhagic Escherichia coli strains of serotypes O26:H11, O45:H2, O103:H2, O111:H8, O121:H19, O145:H28, and O157:H7 by real-time PCR. J. Clin. Microbiol. 50, 4035–4040. doi: 10.1128/JCM.02097-12
Delannoy, S., Beutin, L., and Fach, P. (2013a). Discrimination of enterohemorrhagic Escherichia coli (EHEC) from non-EHEC strains based on detection of various combinations of type III effector genes. J. Clin. Microbiol. 51, 3257–3262. doi: 10.1128/JCM.01471-13
Delannoy, S., Beutin, L., and Fach, P. (2013b). Towards a molecular definition of enterohemorrhagic Escherichia coli (EHEC): detection of genes located on O island 57 as markers to distinguish EHEC from closely related enteropathogenic E. coli strains. J. Clin. Microbiol. 51, 1083–1088. doi: 10.1128/JCM.02864-12
Delannoy, S., Chaves, B. D., Ison, S. A., Webb, H. E., Beutin, L., Delaval, J., et al. (2016). Revisiting the STEC testing approach: using esp K and espV to make Enterohemorrhagic Escherichia coli (EHEC) detection more reliable in beef. Front. Microbiol. 7:1. doi: 10.3389/fmicb.2016.00001
Delannoy, S., Mariani-Kurkdjian, P., Bonacorsi, S., Liguori, S., and Fach, P. (2015). Characteristics of emerging human-pathogenic Escherichia coli O26:H11 strains isolated in France between 2010 and 2013 and carrying the stx 2d gene only. J. Clin. Microbiol. 53, 486–492. doi: 10.1128/JCM.02290-14
Delannoy, S., Tran, M. L., and Fach, P. (2022). Insights into the assessment of highly pathogenic Shiga toxin-producing Escherichia coli in raw milk and raw milk cheeses by high throughput real-time PCR. Int. J. Food Microbiol. 366:109564. doi: 10.1016/j.ijfoodmicro.2022.109564
EFSA and ECDC (2021). The European Union one health 2020 Zoonoses report. EFSA J. 19:e06971. doi: 10.2903/j.efsa.2021.6971
FAO and WHO. (2018). Shiga toxin-producing Escherichia coli (STEC) and food: Attribution, characterization, and monitoring. Available at: https://www.who.int/publications/i/item/9789241514279 (Accessed July 11, 2024).
Fratamico, P. M., DebRoy, C., Miyamoto, T., and Liu, Y. (2009). PCR detection of enterohemorrhagic Escherichia coli O145 in food by targeting genes in the E. coli O145 O-antigen gene cluster and the Shiga toxin 1 and Shiga toxin 2 genes. Foodborne Pathog. Dis. 6, 605–611. doi: 10.1089/fpd.2008.0254
ISO (2012). ISO/TS 13136:2012, microbiology of food and animal feed—Real-time polymerase chain reaction (PCR) –based method for the detection of food-borne pathogens—Horizontal method for the detection of Shiga toxin-producing Escherichia coli (STEC) and the determination of O157, O111, O26, O103, and O145 Serogroups. Geneva: ISO.
Karmali, M. A., Mascarenhas, M., Shen, S., Ziebell, K., Johnson, S., Reid-Smith, R., et al. (2003). Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41, 4930–4940. doi: 10.1128/JCM.41.11.4930-4940.2003
Kobayashi, N., Lee, K., Yamazaki, A., Saito, S., Furukawa, I., Kono, T., et al. (2013). Virulence gene profiles and population genetic analysis for exploration of pathogenic serogroups of Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 51, 4022–4028. doi: 10.1128/JCM.01598-13
Lindsey, R. L., Prasad, A., Feldgarden, M., Gonzalez-Escalona, N., Kapsak, C., Klimke, W., et al. (2023). Identification and characterization of ten Escherichia coli strains encoding novel Shiga toxin 2 subtypes, Stx2n as well as Stx 2j, Stx2m, and Stx2o, in the United States. Microorganisms 11:2561. doi: 10.3390/microorganisms11102561
Michelet, L., Delannoy, S., Devillers, E., Umhang, G., Aspan, A., Juremalm, M., et al. (2014). High-throughput screening of tick-borne pathogens in Europe. Front. Cell. Infect. Microbiol. 4:103. doi: 10.3389/fcimb.2014.00103
Muniesa, M., Hammerl, J. A., Hertwig, S., Appel, B., and Brüssow, H. (2012). Shiga toxin-producing Escherichia coli O104:H4: a new challenge for microbiology. Appl. Environ. Microbiol. 78, 4065–4073. doi: 10.1128/AEM.00217-12
NACMCF (2019). Response to questions posed by the Food and Drug Administration regarding virulence factors and attributes that define foodborne Shiga toxin-producing Escherichia coli (STEC) as severe human pathogens (dagger). J. Food Prot. 82, 724–767. doi: 10.4315/0362-028X.JFP-18-479
Nielsen, E. M., and Andersen, M. T. (2003). Detection and characterization of verocytotoxinproducing Escherichia coli by automated 5′ nuclease PCR assay. J. Clin. Microbiol. 41, 2884–2893. doi: 10.1128/JCM.41.7.2884-2893.2003
Olayinka, A. A., Oginni-Falajiki, I. O., Okeke, I. N., and Aboderin, A. O. (2024). Diarrhoeagenic Escherichia coli associated with childhood diarrhoea in Osun state, Nigeria. BMC Inf. Dis. 24:928. doi: 10.1186/s12879-024-09793-0
Perelle, S., Dilasser, F., Grout, J., and Fach, P. (2004). Detection by 5′-nuclease PCR of Shiga-toxin producing Escherichia coli O26, O55, O91, O103, O111, O113, O145 and O157:H7, associated with the world’s most frequent clinical cases. Mol. Cell. Probes 18, 185–192. doi: 10.1016/j.mcp.2003.12.004
Perelle, S., Dilasser, F., Grout, J., and Fach, P. (2005). Detection of Escherichia coli serogroup O103 by real-time polymerase chain reaction. J. Appl. Microbiol. 98, 1162–1168. doi: 10.1111/j.1365-2672.2005.02545.x
USDA-FSIS. (2012). Risk Profile for Pathogenic Non-O157 Shiga Toxin-Producing Escherichia coli (non-O157 STEC). Available at: https://www.fsis.usda.gov/sites/default/files/media_file/2020-07/Non_O157_STEC_Risk_Profile_May2012.pdf (Accessed January 10, 2025).
USDA-FSIS. (2021). MLG5C.02 detection, isolation and identification of top seven Shiga toxin-producing Escherichia coli (STECs) from meat products and carcass and environmental sponges. Available at: https://www.fsis.usda.gov/sites/default/files/media_file/2021-08/MLG-5C.02.pdf (Accessed July 11, 2024).
USDA-FSIS. (2023). Expansion of FSIS Non-O157 Shiga Toxin-Producing Escherichia coli (STEC) Testing to Additional Raw Beef Products. Available at: https://www.fsis.usda.gov/sites/default/files/media_file/documents/06-23.pdf (Accessed January 10, 2025).
USDA-FSIS. (2024). MLG 5C.04 detection, isolation, and identification of top seven Shiga toxin-producing Escherichia coli (STEC) from meat products, carcass, and environmental sponges. Available at: https://www.fsis.usda.gov/sites/default/files/media_file/documents/MLG-5C.04.pdf (Accessed July 11, 2024).
Keywords: Shiga toxin (Stx) producing Escherichia coli (STEC), EHEC (Enterohaemorrhagic E. coli), STX, EAE, espK, espV, non-O157, MLG5C.04
Citation: Tran M-L, Delannoy S and Fach P (2025) Enhancing detection of STEC in the meat industry: insights into virulence of priority STEC. Front. Microbiol. 16:1543686. doi: 10.3389/fmicb.2025.1543686
Received: 11 December 2024; Accepted: 28 January 2025;
Published: 12 February 2025.
Edited by:
Daniel F. M. Monte, North Carolina State University, United StatesReviewed by:
Mabel Kamweli Aworh, North Carolina State University, United StatesCopyright © 2025 Tran, Delannoy and Fach. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mai-Lan Tran, bWFpLWxhbi50cmFuQGFuc2VzLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.