
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 February 2025
Sec. Infectious Agents and Disease
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1540534
Upon recognizing bacterial lipopolysaccharide (LPS), human TLR4 initiates two distinct signaling pathways: the MyD88 pathway from the cell surface or the TRIF pathway following endocytosis. While the first is associated with strong pro-inflammatory responses, the latter is linked to dendritic cell maturation and T cell priming. Changes in LPS structure can influence the activation of either or both pathways. This study investigates the influence of specific structural features of Bordetella LPS on these pathways: the O antigen, the number of acyl chains in lipid A and the glucosamine modification of the phosphates of the lipid A diglucosamine backbone. Systematically engineered Bordetella LPS differing in one or more of these features were studied by quantifying NFκB and IRF3 activation—indicators of MyD88 and TRIF pathway activation, respectively. The findings reveal that the glucosamine modification of lipid A plays a dominant role in TLR4-mediated signaling, overriding the influence of the O antigen and lipid A acylation. The absence of glucosamine modification significantly reduced the activation of both MyD88 and TRIF pathways, underscoring its importance in promoting TLR4 dimerization. Furthermore, under-acylation of LPS (with 4 or 5 acyl chains) partially reduced NFκB activation, while completely abrogating TRIF pathway activation. In contrast, hexa-and hepta-acylated LPS equally and robustly activated both pathways. Lastly, the Bordetella O antigen selectively biased signaling towards the TRIF pathway without affecting the MyD88 pathway. This study provides valuable insights into how specific LPS structural modifications can be leveraged to tailor TLR4-mediated signaling.
Lipopolysaccharide (LPS) is the predominant component of the outer membrane of most Gram-negative bacteria. Its structure is typically composed of lipid A, core sugars and, in some bacteria, O antigen polysaccharide. The lipid A, in turn, consists of 4 to 7 acyl chains attached to a diglucosamine backbone. Together, this glycolipid entity is highly immunodominant and a key bacterial virulence factor.
The human innate immune system mainly detects LPS using the pattern recognition receptor complex, Toll-like receptor-4/Myeloid differentiation factor-2 (TLR4/MD-2) (Lu et al., 2008; Ciesielska et al., 2021). Its cofactors, LPS binding protein and CD14, relay LPS to TLR4/MD-2 from the bacterial surface or from solution. When lipid A of LPS binds to the pocket of MD-2, it triggers the dimerization of two TLR4/MD-2—LPS complexes and initiates one of two signaling cascades. The dimer can signal from the cell surface using the MyD88-dependant pathway to activate the transcription factors, NFκB and/or AP-1, and thereby induce a strong pro-inflammatory response. Additionally, the dimer can also be endocytosed where it interacts with adaptors, TRAM and TRIF, to induce IRF3-mediated type-I interferons, which thereby promote dendritic cell (DC) maturation and differentiation and consequently impact T cell priming. TRIF-mediated signaling also leads to the late-phase activation of NFκB and AP-1. Alternative to LPS recognition by TLR4/MD-2, endocytosed or cytosolic LPS can also be detected by caspases-4/11 to activate inflammasomes (Zamyatina and Heine, 2020).
Consequently, bacteria have evolved to evade or temper the immune response generated by altering their LPS structure. Modification to the LPS biosynthesis pathway itself can alter the lipid A backbone or acyl chain characteristics as well as change the properties of or even completely replace the O antigen. Bacteria also have mechanisms in place to alter their LPS post-synthesis largely under the regulation of two-component systems. Examples include the addition, removal or hydroxylation of acyl chains, altering the charge of lipid A or addition of groups to the core oligosaccharide (Bertani and Ruiz, 2018; Simpson and Trent, 2019). All these factors, put together, alter its interaction with TLR4/MD-2 and subsequently, the degrees to which the downstream pathways are activated.
The traditional hexa-acylated E. coli LPS is an agonist of TLR4, strongly activating both MyD88 and TRIF pathways. In contrast, under-acylated LPS usually act as antagonists as reducing the number of acyl chains alters their fit in the MD-2 pocket (Park et al., 2009). For example, penta-acylated R. sphaeroides LPS does not trigger TLR4-mediated signaling and, in fact, competitively inhibits the binding of other agonistic LPS (Qureshi et al., 1991; Kirikae et al., 1994; Stevens et al., 2013; Anwar et al., 2015). In the middle of these extremes lies monophosphoryl lipid A (MPLA), an FDA-approved adjuvant that is a chemically detoxified LPS isolated from Salmonella minnesota Re595. MPLA has been shown to greatly reduce the MyD88-dependent pro-inflammatory response while preserving signaling via the TRIF pathway (Mata-Haro et al., 2007).
LPS also forms a part of the outer membrane of Bordetella spp., a group of small, Gram-negative, coccobacilli that cause a highly contagious respiratory disease in a wide range of hosts, from humans to various mammals including sheep, pigs and mice, as a well as poultry and wild birds (Rivera et al., 2020). The Bordetella spp. have evolved, potentially due to the different selection pressures faced in the different hosts, to express a wide variety in LPS structures. B. pertussis is a penta-acylated member devoid of an O antigen in the otherwise predominantly hexa-acylated, O antigen-expressing Bordetella family. Some species (B. pertussis, B. bronchiseptica, B. parapertussis, and B. avium) modify their backbone phosphates with charged glucosamine (GlcN) moieties (Marr et al., 2008; Novikov et al., 2014; Novikov et al., 2019), while some (B. pertussis, B. bronchiseptica, B. avium, and B. hinzii) decorate their core with a distal trisaccharide (Preston et al., 2006; El Hamidi et al., 2009; Novikov et al., 2019). Additional diversity in the acyl chain length, distal trisaccharide and the O antigen composition is brought about by genetic diversity in the genes and loci involved in their biosynthesis and ligation (Novikov et al., 2019). Much remains unknown as to how these structural differences in Bordetella LPS influence their recognition by the host TLR4/MD-2 and the subsequent downstream signaling.
Of all Bordetella species, human respiratory illness (whooping cough) is mainly caused by B. pertussis and B. parapertussis, with B. pertussis causing the more severe disease. The number of cases of B. pertussis infection is resurging in many parts of the world, including USA, Canada, and Europe in 2024 with case numbers surpassing those in 2019 after a brief respite due to COVID-19 pandemic-related restrictions.1,2,3 In parallel, B. parapertussis is also reemerging, with a significant increase in its detection rates in PCR-tested samples in 2023 (Noble et al., 2024). Hence, these strains warrant further investigation, and this study focused on the recognition of B. pertussis and B. parapertussis LPS variants by human TLR4.
Despite infecting the same host, B. pertussis and B. parapertussis have evolved to exhibit significant differences in their LPS structure. B. parapertussis encodes a homopolymeric O antigen while B. pertussis does not (Di Fabio et al., 1992; Preston and Maskell, 2001). Studies using E. coli LPS implicate the O antigen in biasing TLR4-mediated signaling towards the TRIF pathway by interacting with the cofactor, CD14 (Gangloff et al., 2005; Jiang et al., 2005; Zanoni et al., 2012). Fedele et al., 2008 showed that purified B. parapertussis LPS (with an O antigen) induced significantly lower monocyte-derived DC (MDDC) maturation in the absence of CD14, while MDDC maturation triggered by purified B. pertussis LPS (without an O antigen) was not affected by the absence of CD14. Thus, the O antigen was implicated in playing a role in CD14-mediated LPS signaling (Fedele et al., 2008). Our study utilized systematically engineered Bordetella LPS with and without the O antigen to delineate its influence on the activation of TLR4-mediated signaling pathways: the MyD88 pathway and, particularly, the CD14-dependent TRIF pathway.
Another feature differentiating the strains is the number of acyl chains. B. pertussis is penta-acylated, while B. parapertussis is hexa-acylated. Under-acylation in Gram-negative bacteria has been associated with reduced TLR4 signaling (Meng et al., 2010; Herath et al., 2013; Anwar et al., 2015). Similarly, in Bordetella spp., hexa-acylated species (either B. parapertussis or B. pertussis modified to encode a hexa-acylated structure) were shown to activate NFκB mediated responses to a greater degree than penta-acylated B. pertussis (Geurtsen et al., 2009; Fathy Mohamed and Fernandez, 2024). This study expanded on these findings, investigating the activation of both TLR4-mediated signaling pathways by Bordetella LPS that expressed a wider variation in acyl chain numbers (from 4 to 7 acyl chains).
Lastly, both B. pertussis and B. parapertussis decorate the phosphates of the diglucosamine backbone with GlcN moieties (Marr et al., 2008). This modification has been shown to promote both the MyD88 (Geurtsen et al., 2009; Marr et al., 2010a; Marr et al., 2010b) and the TRIF pathway (Marr et al., 2010a) by influencing TLR4 dimerization (Maeshima et al., 2015). This study examined the influence of the GlcN moiety on TLR4-mediated signaling in combination with other structural modifications of Bordetella LPS, i.e., the presence or absence of the O antigen and the alteration in the number of acyl chains.
Even minor differences in LPS structure alter the LPS’ properties and its interaction with the host TLR4/MD-2 (Miller et al., 2005; Maeshima and Fernandez, 2013). Thus, the extent to which the MyD88 and the TRIF pathways are activated is affected, which consequently influences downstream adaptive immune responses (Fitzgerald and Kagan, 2020; Duan et al., 2022). In this study, we found that the GlcN modification of Bordetella LPS increases the activation of both MyD88 and TRIF pathways, irrespective of alterations to the other structural features studied. Additionally, while an extra acyl chain in Bordetella LPS did not alter TLR4 signaling, under-acylation partially reduced NFkB responses and failed to activate the TRIF pathway. Finally, the Bordetella O antigen biased signaling towards the TRIF pathway without affecting the MyD88 pathway activation. Altogether, this study highlights the nuanced interplay of these structural features in activating TLR4-mediated signaling pathways.
All strains and plasmids used in this study are listed in Table 1. Bacteria were grown as described before (Ifill et al., 2021). As needed, media were supplemented with nalidixic acid (Nal; 30 μg/mL), gentamicin (Gm; 15 μg/mL), kanamycin (Kan; 75 μg/mL), diaminopimelic acid (DAP; 250 μg/mL) and/or anhydrous tetracycline (aTC; 12.5 ng/mL).
A markerless clean deletion protocol was used to delete the genes of interest: lgmA-D, waaL, wbmA-E, pagP and pagL in B. parapertussis as described previously (Marr et al., 2010a; Shah et al., 2013; Ifill et al., 2021). Due to the sequence similarity in the lgm locus between B. pertussis and B. parapertussis, the same plasmid (pSS4245 ΔlgmA-D) used to delete the locus in B. pertussis (Shah et al., 2013), was used in B. parapertussis as well. pIG02 was used for the clean deletion of other genes by cloning ~600-bp of the upstream and downstream regions of the genes of interest [separated by an SpeI restriction enzyme (RE) site] between the KpnI and BamHI RE sites in its multiple cloning site (MCS). This construct was then transformed into E. coli RHO3. These allelic exchange plasmids were then conjugated into B. parapertussis using the di-parental mating protocol as previously published (Ifill et al., 2021) with the noted absence of nalidixic acid (B. parapertussis strain is not Nalr). In the case of double mutants, the allelic exchange protocol was repeated with the generated single gene deletion mutant and the RHO3 strain containing the pIG02 construct for the clean deletion of the second gene of interest.
The B. parapertussis waaL, wbmA-E, pagP and pagL deletion mutants were complemented with their respective genes using pIG10, an anhydrous tetracycline (aTC) inducible expression plasmid created for Bordetella species by Gyles Ifill (Ifill and Fernandez, manuscript in preparation). The gene of interest was amplified and cloned into the MCS of pIG10 between the SpeI and BamHI RE sites. In the case of B. parapertussis ΔpagP ΔpagL, both genes were cloned into pIG10 separated by a HindIII RE site and a ribosome-binding sequence, 5′-GGCAAGTCTAAAGCCATAGAAGGATAC-3′ to ensure the expression of both genes. The construct was introduced into the respective mutants using di-parental conjugation as described above.
The lgm locus, along with ~1,000-bp of the upstream region (to include its native promoter), was introduced into the chromosome using the mini-Tn7 transposon delivery plasmid, pUC18T-mini-Tn7T-Km-FRT, at the attTn7 site located downstream of the highly conserved and essential glmS genes (Choi et al., 2005). The construct, and its transposase vector, pTNS2, were introduced into the ΔlgmA-D mutants using tri-parental conjugation. This method, similar to di-parental mating described above, involved mixing E. coli RHO3 containing the pUC18-miniTn7T-lgmA-D construct, E. coli RHO3 carrying pTNS2 and the B. parapertussis mutant strain in the ratio of 1:1:2. Kanamycin was used as the selection antibiotic. Successful integration of the transposon was confirmed by PCR.
B. pertussis and B. parapertussis strains were grown in liquid culture inoculated at an initial OD600 of 0.001. They were grown to an OD600 of 0.6–0.8 (up to 72 h under agitation). 1.5 mL of bacterial suspension (concentrated to an OD600 of 2) was digested with DNase I, RNase and proteinase K, and the resulting lysate was separated using tricine-SDS-PAGE and visualized using silver staining (Marolda et al., 2006).
To prepare cells for MALDI-TOF analysis, 100 mL of B. pertussis or B. parapertussis liquid culture was grown as stated above. The bacteria were harvested, and lipid A was extracted using the ammonium-isobutyrate method (El Hamidi et al., 2005) and analyzed in the Applied Biosystems MALDI-TOF spectrometer as described previously (Fathy Mohamed and Fernandez, 2024). Data was acquired and analyzed using the Data Explorer software and graphed using GraphPad Prism 10 (RRID:SCR_002798).
HEK-Blue™ hTLR4 cells (InvivoGen Cat# hkb-htlr4) and HEK-Blue™ Null2 cells (InvivoGen Cat# hkb-null2) were cultured as described previously (Shah et al., 2013). HEK-Blue™ hTLR4 cells are engineered from HEK293 cell line to stably express human TLR4, MD2, CD14 and an inducible secreted embryonic alkaline phosphatase (SEAP) reporter gene to measure NFκB activation. HEK-Blue™ Null2 is the parental cell line of HEK-Blue™ hTLR4 expressing the SEAP reporter alone to exclude NFκB responses induced by the activation of endogenously expressed pattern recognition receptors, including TLR3, TLR5 and RIG-1-like receptors.
THP1-Dual™ cells (InvivoGen Cat# thpd-nfis) were cultured in RPMI 1640 (GIBCO) containing 10% heat-inactivated (30 min at 56°C) fetal bovine serum (Sigma), 2 mM GlutaMAX (GIBCO), 25 mM HEPES (GIBCO), 100 μg/mL Normocin (InvivoGen), and Penicillin-Streptomycin (100 U/mL-100 μg/mL; GIBCO) in the presence of selection antibiotics: 100 μg/mL zeocin (InvivoGen) and 10 μg/mL blastocidin (InvivoGen). They were passaged after reaching densities of 1–2 × 106 cells/mL. THP1-Dual™ cells are engineered from human THP-1 monocyte cell line to express two inducible reporter genes, SEAP to measure NFκB activation and Lucia luciferase to measure IRF3 activation.
THP-1 cells (ATCC) were cultured as described previously (Fathy Mohamed and Fernandez, 2024) and passaged after reaching a density of 1 × 106 cells/mL.
All cell lines were incubated in a CO2 incubator at 37°C with 5% CO2.
All Bordetella strains, and the positive control, E. coli DH5α, were grown to an OD600 of 0.6–0.8. They were concentrated to an OD600 of 5 in phosphate-buffered saline (PBS) and heat-killed at 60°C for 1 h. The lack of viability was confirmed by spotting a small aliquot (2 μL) on agar plates and checking for the lack of bacterial growth after incubating the plates for up to 5 days. The heat-killed samples were stored at −20°C.
E. coli K12 LPS (InvivoGen) was resuspended as recommended and stored in aliquots at −20°C. When needed, an aliquot was thawed, placed in a sonicating water bath for 10 min and then used to prepare required dilutions for the respective assays.
HEK-Blue™ hTLR4 cells and HEK-Blue™ Null2 cells were grown to ~70–80% confluency. Then the reporter assay was carried out as described previously (Shah et al., 2013) using the indicated dilution of heat-killed bacterial suspension as stimulants or media for negative control. The alkaline phosphatase reporter activity was quantified by measuring the absorbance after the indicated incubation period with the QUANTI-Blue reagent (Invivogen) at 650 nm in the Molecular Devices SpectraMax 190 microplate reader or the Thermo Scientific VarioSkan Flash multimode plate reader. Readings were converted as a percentage of B. parapertussis WT. One-way ANOVA with Tukey’s multiple comparison test was performed using GraphPad Prism 10.
One hundred and eighty microliters of THP-1 Dual™ cells (~100,000 cells/well) were aliquoted per well of 96-well flat-bottomed, tissue culture-treated plates (Corning, Cat# 353072). They were differentiated into macrophages by treating them with 50 ng/mL phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich) for 48 h at 37°C in a CO2 incubator. The cells were then washed with fresh media twice to remove traces of PMA and rested for 72 h. On day 4, the cells were washed again, and 180 μL of fresh media was added per well. Subsequently, 20 μL of stimulant prepared in media was added per well to obtain the desired final concentration of stimulant (1:100 dilution of heat-killed bacterial suspension; endotoxin-free water for negative control; 1 μg/mL E. coli K12 LPS as positive control). After incubation at 37°C for 24 h, 10 μL of the supernatant was used to determine the luciferase activity using the QUANTI-Luc 4 Lucia/Gaussia reagent (InvivoGen) as per manufacturer’s flash detection protocol in the Perkin-Elmer Victor X5 Multilabel reader. Readings were converted as a percentage of B. parapertussis WT. Mixed-effects analysis with Tukey’s multiple comparison test was performed using GraphPad Prism 10.
The THP-1 stimulation assay was performed as previously described (Marr et al., 2010a). THP-1 cells were differentiated using PMA and stimulated with the desired dilution of stimulant (1:10 dilution of heat-killed bacterial suspension in complete RPMI 1640; sterile media for negative “no stimulation” control; 10 μg/mL of E. coli K12 LPS) in duplicates. At 4 h post-stimulation, the supernatant was removed, and the THP-1 cells were washed with sterile PBS. The cells were then scraped in 100 μL PBS and stored in Eppendorf tubes at −20°C.
When performing western blot experiments, the cells were denatured and proteins were separated by 12% SDS-PAGE as described before (Fathy Mohamed and Fernandez, 2024). Three such gels were prepared for each stimulation assay. The first gel was stained with PageBlue™ protein staining solution (Thermo Scientific). Image Lab Software (RRID:SCR_014210) was used to detect total protein content of the sample lanes. Proteins from the second and third gels were transferred to Immobilon-P polyvinylidene difluoride (PVDF) membranes (Sigma) and immunoblotted for p-IRF3 and p-STAT1, respectively (Fathy Mohamed and Fernandez, 2024). Primary antibodies anti-IRF3 (phosphor S386) antibody EPR2346 (Abcam Cat# ab76493, RRID:AB_1523836) and phospho-Stat1 (Tyr701) (58D6) Rabbit mAb (Cell Signaling Technology Cat# 9167, RRID:AB_561284), as well as, secondary antibody Peroxidase AffiniPure™ Goat anti-Rabbit IgG (H + L) (Jackson ImmunoResearch Labs Cat# 111–035-144, RRID:AB_2307391) were used. The proteins were detected using chemiluminescence (ECL™ Prime Western Blotting Detection reagent; Cytivia) in the BioRad Imaging System. The membrane was exposed for 360 s, and the image obtained was analyzed using Image Lab Software to calculate band intensities. The band intensities, normalized to the total protein content, were converted as a percentage of B. parapertussis WT. One-way ANOVA with Tukey’s multiple comparison test was performed using GraphPad Prism 10.
To further understand how TLR4 differentiates structurally distinct Bordetella LPS, B. pertussis and B. parapertussis strains were genetically engineered to encode LPS with different structural features. First, to investigate if the O antigen biases signaling towards the TRIF pathway, LPS structures with and without the O antigen were engineered. Of the two parent strains, B. parapertussis expresses an O antigen due to the presence of an intact wbm biosynthesis locus, while, B. pertussis lacks an O antigen as the locus was replaced by an insertion sequence (Di Fabio et al., 1992; Preston and Maskell, 2001). The O antigen from B. parapertussis was removed by deleting either the ligase (WaaL) that attaches the pre-formed O antigen to the lipid A + core moiety (Mulford and Osborn, 1983; Kalynych et al., 2014) or by deleting the first five genes of the wbm locus (wbmA-E), which has been shown sufficient to prevent O antigen synthesis (Preston et al., 1999) as indicated in Figure 1A. Thus, hexa-acylated LPS with (B. parapertussis WT) and without (B. parapertussis ΔwaaL or ΔwbmA-E) an O antigen were generated. Additionally, by altering the number of acyl chains in B. parapertussis as outlined below, we generated a penta-acylated LPS expressing an O antigen (B. parapertussis ΔpagP ΔpagL) which could now be compared to the penta-acylated B. pertussis WT LPS that lacks the O antigen.
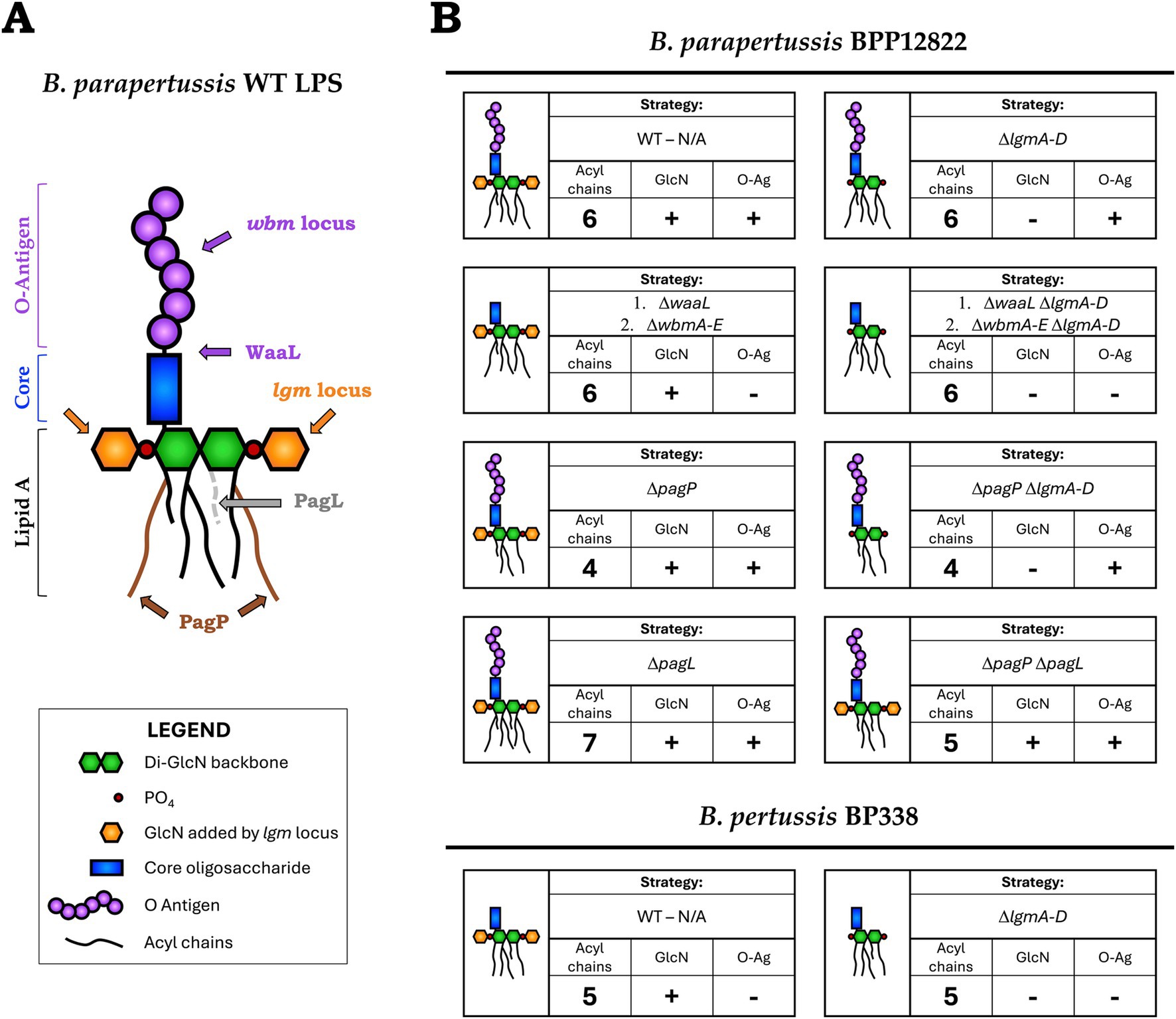
Figure 1. (A) Schematic of B. parapertussis WT LPS structure with the targets of deleted genes/loci indicated—lipid A of B. parapertussis consists of six acyl chains attached to a diglucosamine backbone (green) to which the core (blue) and the O antigen (purple) is attached. Deleting the ligase, WaaL, or the first five genes of the wbm locus creates an LPS species devoid of O antigen (purple). The number of acyl chains was modified by deleting PagP, which adds two palmitate groups (brown chains) to lipid A, or PagL, a deacylase that removes an acyl chain (grey dotted). The decoration of the phosphates (red) of the lipid A backbone with glucosamine moieties (orange) was prevented by deleting the lgm locus. (B) Bordetella WT and mutant strains used in the study—each block represents a Bordetella strain (WT or mutant), schematic of the encoded LPS structure and the strategy (genes deleted) to obtain said structure. It also indicates the number of acyl chains in the final structure and if it encodes the GlcN modifications and the O antigen.
To study the influence of the number of acyl chains on TLR4 signaling, strains encoding lipid A with 4 to 7 acyl chains were created by deleting pagP and/or pagL from hexa-acylated B. parapertussis (El Hamidi et al., 2009). PagP is a palmitoyl transferase that adds a secondary palmitate group to the acyl chains present at the C2 and C3′ positions (Hittle et al., 2015) (Figure 1A). On the other hand, PagL is a lipid A deacylase that removes the 3-hydroxydecanoic acid moiety from the C3 position (Geurtsen et al., 2006). Hence, deleting pagP, pagL or both together led to LPS species with 4, 7 or 5 acyl chains respectively, that could now be compared to B. parapertussis and B. pertussis LPS containing 6 and 5 acyl chains, respectively.
Both B. pertussis and B. parapertussis encode the lgm locus responsible for the GlcN modification of the phosphates of the lipid A backbone (Marr et al., 2008; Geurtsen et al., 2009). To study their role in TLR4 signaling in conjunction with other LPS structural features, the entire locus, lgmA-D, was deleted from both B. pertussis and B. parapertussis to create LPS with and without the GlcN modification.
The genes stated above were either deleted singly or in combination, deleting one gene at a time, using a markerless clean deletion protocol. In total, 10 Bordetella mutants were created that expressed eight different LPS structures that differed in the presence or absence of the O antigen, the number of acyl chains, and/or the presence or absence of the GlcN modification as indicated in Figure 1B. All Bordetella mutants were complemented with the respective deleted genes, either on an aTC inducible pIG10 plasmid (for waaL, wbmA-E, pagP and pagL) or the mini-Tn7 transposase system (for lgmA-D) (Choi et al., 2008). These strains, their respective WT strain as well as E. coli K12 LPS were used in subsequent experiments to study TLR4 recognition and signaling.
After confirming the deletion of the respective genes in the mutants using PCR, evidence for the presence or absence of the O antigen in the engineered LPS was obtained using tricine-SDS-PAGE and visualized using silver-staining. Bands corresponding to the lipid A + core moiety (bottom band) and the O antigen-containing LPS (top smear) were noted (Figure 2A). The homopolymeric O antigen-containing LPS of B. parapertussis WT is seen as a smear at the top of the gel (Lane 1). The mutants lacking the O antigen (ΔwaaL, ΔwbmA-E, ΔlgmA-D ΔwaaL and ΔlgmA-D ΔwbmA-E; Lanes 3 to 6) lack the smear at the top, confirming the loss of O antigen. All other mutants containing modifications to the lipid A alone show no change in the expression of the O antigen as expected (Lanes 2 and 7 to 10). B. pertussis WT and ΔlgmA-D mutants naturally do not express the O antigen as confirmed by the lack of the O antigen-containing LPS smear (Lanes 11 and 12). However, the lipid A + core band of B. pertussis migrates slower than that of B. parapertussis, presumably due to the addition of the distal trisaccharide unit to the core by the wlb locus, which is absent in B. parapertussis (Allen et al., 1998). The B. parapertussis O antigen mutants (ΔwaaL, ΔwbmA-E, ΔlgmA-D ΔwaaL and ΔlgmA-D ΔwbmA-E), when complemented with the respective deleted genes, demonstrate successful complementation of waaL or wbmA-E as indicated by the reappearance of the O antigen-containing LPS smear (Supplementary Figure S1).
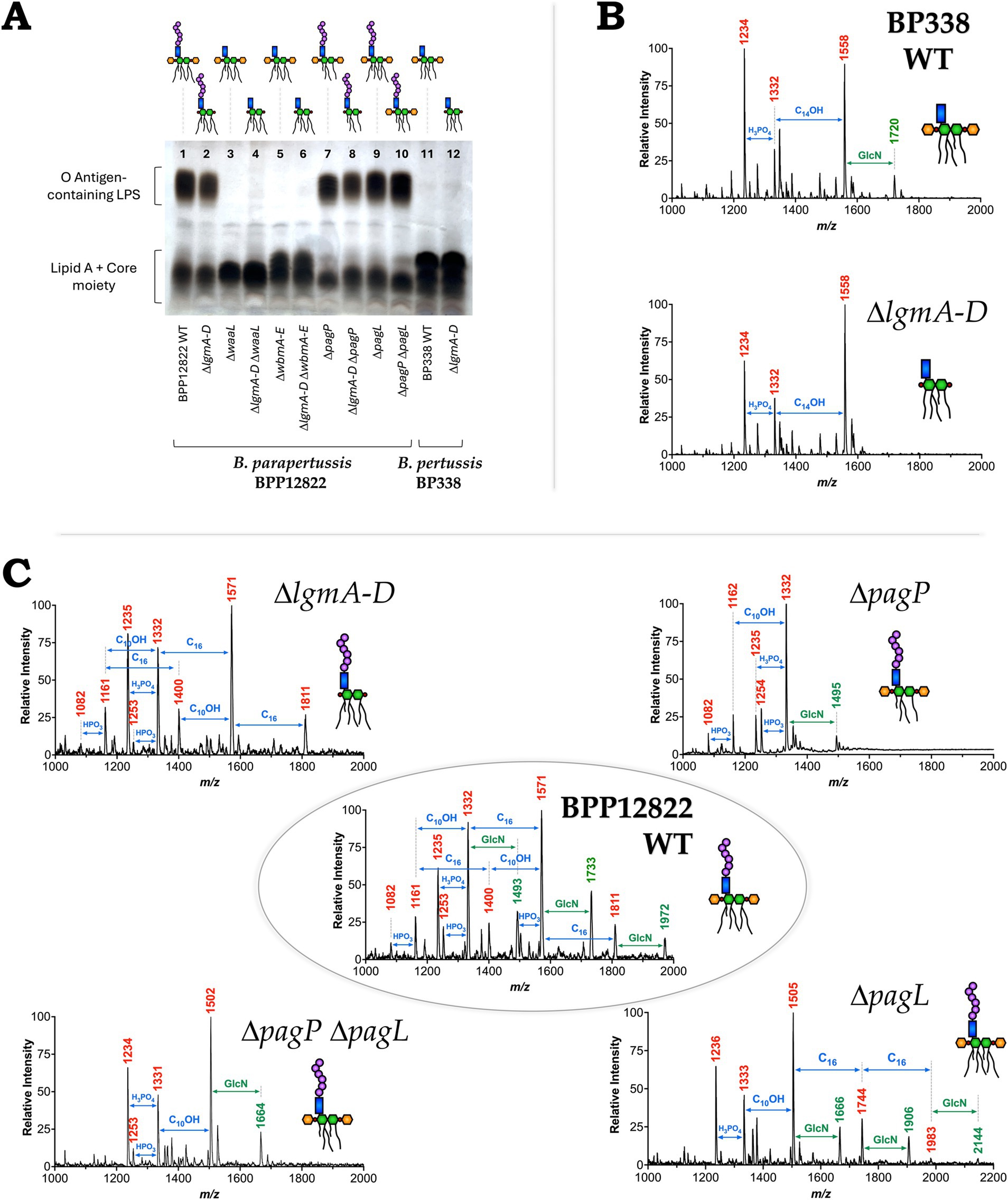
Figure 2. Validation of the structures of Bordetella LPS mutants. (A) Tricine-SDS-PAGE analysis of the variants—DNase I, RNase and proteinase K treated lysates were subjected to tricine-SDS-PAGE and stained using silver staining. (B) Negative-ion MALDI-TOF mass spectra of lipid A isolated from B. pertussis strains—WT (top) and ΔlgmA-D mutant (bottom). (C) Structural analysis of lipid A of select B. parapertussis variants—WT (center) and going clockwise: ΔlgmA-D mutant (top left), ΔpagP mutant (top right), ΔpagL mutant (bottom right) and ΔpagP ΔpagL mutant (bottom left). Predicted LPS structure is indicated beside each strain.
Evidence for the absence of the GlcN moiety (in the ΔlgmA-D mutants) and of the number of acyl chains present (in the ΔpagP and ΔpagL mutants) was acquired via MALDI-TOF. Lipid A was extracted using mild acid hydrolysis and analyzed using negative, linear ion-mode MALDI-TOF. While MALDI-TOF was performed for all mutants, Figures 2B,C illustrate the spectra of select mutants. Peaks of interest are labeled in red or green. Other m/z in the spectra correspond to micro-heterogeneity due to changes in hydroxylation and acyl chain length. The top panel of Figure 2B shows the spectra of B. pertussis WT LPS. The peak at m/z 1,558 represents the penta-acylated LPS species, followed by tetra-acylated species at m/z 1,332 and 1,234. The addition of a single GlcN moiety is observed as an addition of m/z 161 resulting in a peak at m/z 1,720 (green) (Marr et al., 2010a). The ΔlgmA-D mutant lacks this peak confirming the loss of this moiety (Figure 2B lower panel).
The central panel of Figure 2C shows the spectra of B. parapertussis WT LPS. Similar to what was previously reported for B. parapertussis (El Hamidi et al., 2009; Hittle et al., 2015), our data shows peaks at m/z 1,332, 1,571, and 1,811 that correspond to LPS species with 4, 5 and 6 acyl chains, respectively. The corresponding GlcN modified species, with m/z 161 higher, are highlighted in green (m/z 1,493, 1,733 and 1,972 respectively). The peaks corresponding to the GlcN modification are absent in the ΔlgmA-D mutant (top left). The ΔwaaL and ΔwbmA-E mutants, which did not have any modifications to their lipid A structure, had the same mass spectra as B. parapertussis WT (Supplementary Figure S2). Likewise, the ΔlgmA-D ΔwaaL and ΔlgmA-D ΔwbmA-E had mass spectra similar to the ΔlgmA-D mutant (Supplementary Figure S2). PagP adds two palmitate groups to B. parapertussis LPS, with m/z 238.4 each. Correspondingly, the ΔpagP mutant (top right) lacks the peaks at m/z 1,571 and 1,811 corresponding to the penta-and hexa-acylated species. Deletion of PagL prevents the deacylation of a C10-OH group with m/z 170.25. Hence, the ΔpagL mutant (bottom right) is seen to have an additional peak at m/z 1,983 indicative of a hepta-acylated species, followed by the addition of a single GlcN at m/z 2,144. Also, the ΔpagP ΔpagL double mutant (bottom left) corresponded to the loss of two palmitate groups at m/z 238.4 each and the addition of C10-OH group at m/z 170.25, resulting in a final loss of m/z 306.55. Thus, the double mutant has a major peak corresponding to a penta-acylated species with m/z 1,502, that is m/z 309 less than the hexa-acylated WT LPS at m/z 1,811.
Next, the ability of the engineered Bordetella LPS to activate TLR4-mediated NFκB signaling was investigated. Heat-killed bacteria (at 1:100 dilution) from each strain were introduced to an NFκB reporter cell line, HEK-Blue™ hTLR4 cells, that expresses human TLR4/MD-2 and CD14. HEK-Blue™ Null2 was used as a control to rule out NFκB activation by endogenously expressed pattern recognition receptors. The degree of NFκB activation was measured 15 min post-mixing with the QUANTI-Blue reagent as an absorbance readout at 650 nm (Figure 3). Hexa-acylated E. coli DH5α was used as a control (Lane C1). B. parapertussis WT (Lane 1), with a hexa-acylated lipid A, did not induce as much NFκB as E. coli LPS (Lane C1), but it activated NFκB stronger than the penta-acylated B. pertussis LPS (Lane 11). The most prominent phenotype observed across the board was the significant reduction in NFκB activation upon the deletion of the lgm locus when compared to the WT or their respective single mutant parent in the case of a double mutant (Lanes 2, 4, 6, 8 and 12). Furthermore, the reduction in NFκB activation on deleting the lgm locus in B. parapertussis WT is much more striking than that observed in B. pertussis (Lanes 1 and 2 vs. Lanes 11 and 12).
Figure 3. Absence of GlcN modification and under-acylation of lipid A, but not the loss of the O antigen nor over-acylation of lipid A, significantly reduce NFκB activation via TLR4/MD-2—A 1:100 dilution of heat-killed bacteria was used to stimulate HEK-Blue™ hTLR4 reporter cells to measure NFκB activation 15 min post-mixing with the QUANTI-Blue reagent. The assay was repeated 5 times in total with 5 technical replicates each. The absolute absorbance readings were converted as a percentage of B. parapertussis WT and plotted. The histograms show the mean + standard deviation. One-way ANOVA with Tukey’s multiple comparison test was performed using GraphPad Prism 10. ns, not significant; ***p < 0.001 and ****p < 0.0001. Predicted LPS structure and lane number are indicated beside each strain. Dotted line represents average Null2 readings across samples.
Secondly, the O antigen had no impact on NFκB activation. B. parapertussis LPS lacking O antigen either by the deletion of the ligase (ΔwaaL; Lane 3) or the biosynthesis locus (ΔwbmA-E; Lane 5) showed equivalent NFκB activation as the WT strain (Lane 1). Also, penta-acylated Bordetella LPS that expresses the O antigen (B. parapertussis ΔpagP ΔpagL; Lane 10) and one that does not (B. pertussis WT; Lane 11) activated NFκB to equal degrees. Lastly, the number of acyl chains had a nuanced impact on NFκB signaling. Having an extra acyl chain (ΔpagL; Lane 9) compared to the B. parapertussis WT (Lane 1) was neither beneficial nor detrimental to NFκB activation. However, reducing the number of acyl chains (to 4 in ΔpagP mutant or 5 in ΔpagP ΔpagL double mutant; Lanes 7 and 10) reduced NFκB activation. Any reduction of NFκB activation was restored to WT levels upon complementation of the respective mutants (Supplementary Figure S3).
TLR4-mediated NFκB activation (as measured above) is triggered by both the MyD88 and the TRIF pathways. Of these, the MyD88 pathway is key for the immediate and strong activation of NFκB, while the TRIF pathway, though majorly involved in activating IRF3 and Type I interferons, triggers the late-phase activation of NFκB (Lu et al., 2008). Hence, we investigated if these structural changes in LPS biased the TLR4-mediated signaling toward the TRIF pathway.
To assess TRIF-mediated IRF3 activation, two techniques were employed. First, heat-killed bacteria were used to stimulate THP-1-Dual cells, which consist of a Lucia luciferase reporter under the control of the interferon-stimulated response element (IRSE) which is induced by phosphorylated IRF3 or Type 1 interferon-mediated STAT signaling. Luciferase activity was then measured at 24 h using the flash detection method and expressed as relative light units. E. coli K12 LPS was used as a positive control.
B. parapertussis WT showed high levels of IRF3 activation which was comparable to B. pertussis WT (Figure 4A; Lanes 1, 11). Both O antigen mutants (ΔwaaL and ΔwbmA-E; Lanes 3 and 5) showed a significant reduction in IRF3 activation. Altering the number of acyl chains down to 4 or 5 also abolished IRF3 activation (Lanes 7, 10), while increasing it to 7 had no impact (Lane 9). Intriguingly, the penta-acylated B. parapertussis ΔpagP ΔpagL mutant (Lane 10) activated IRF3 to a significantly lower extent than B. pertussis WT (Lane 11) despite it having an O antigen (whose presence seemed to increase IRF3 activation in B. parapertussis WT compared to its O antigen mutants). Upon the absence of the GlcN modification in respective single or double mutants, negligible IRF3 activation was observed (Lanes 2, 4, 6, 8, 12). Like that observed in NFκB activation, the drop in IRF3 activation in the B. parapertussis ΔlgmA-D mutant when compared to its WT is more dramatic compared to that observed in B. pertussis (Lanes 1 and 2 vs. Lanes 11 and 12).
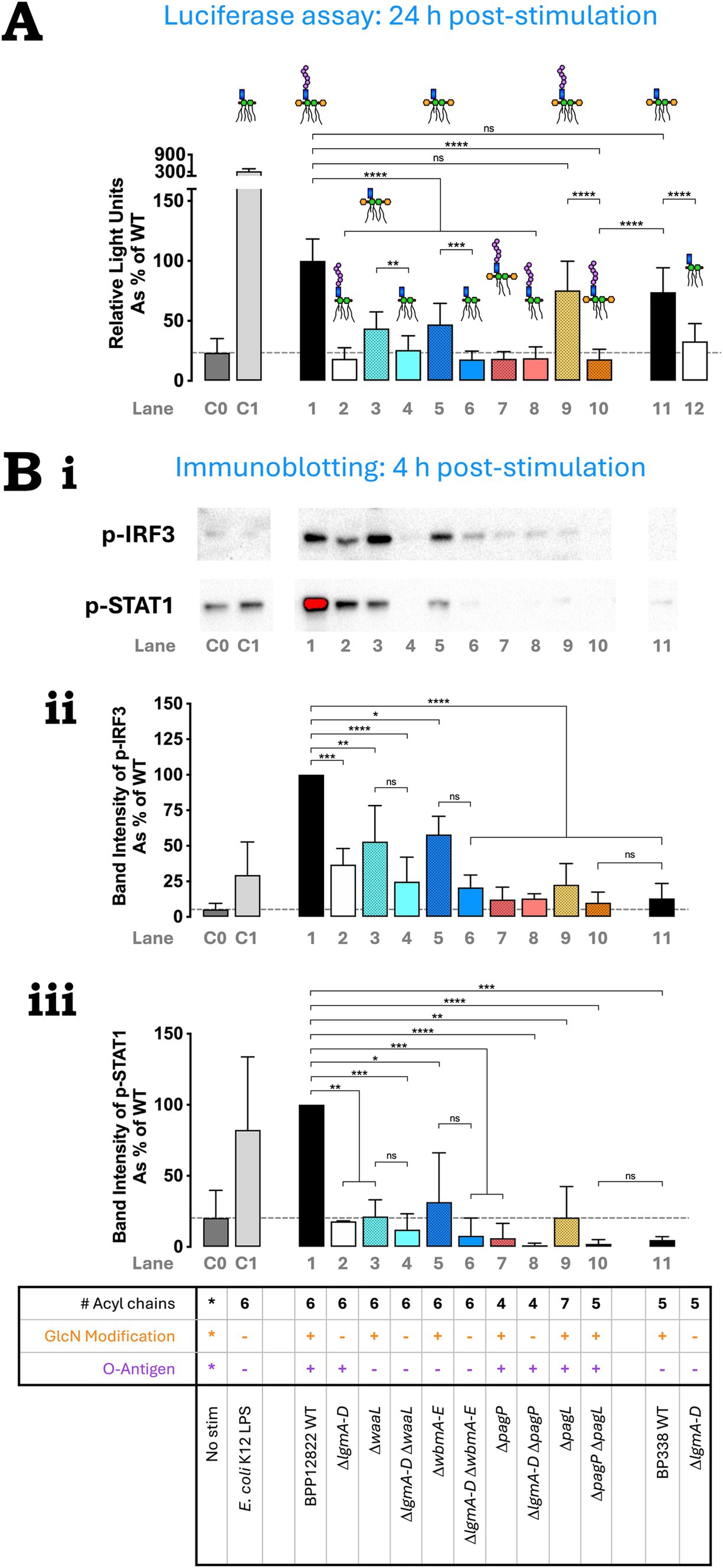
Figure 4. O antigen is important for TLR4-mediated TRIF pathway activation. (A) IRF3 activation by B. parapertussis strains using THP-1 Dual reporter assay—heat-killed bacteria were used to stimulate reporter THP-1 Dual™ cells for 24 h and the extent of IRF3 stimulation was measured as relative light units. The assay was repeated 4 times in total with 4 technical replicates each. The absolute luminescence readings were converted as a percentage of B. parapertussis WT readings and plotted. The histograms show the mean + standard deviation. Mixed-effects analysis with Tukey’s multiple comparison test was performed using GraphPad Prism 10. (B) p-IRF3 and p-STAT1 western blot indicate reduction in IRF3 and STAT1 phosphorylation upon the deletion of the lgm locus, O antigen and/or altering the number of acyl chains—heat-killed bacteria were used to stimulate PMA-differentiated THP-1 cells. Cells were collected at 4 h, lysed and immunoblotted for p-IRF3 and p-STAT1. (Bi) p-IRF3 and p-STAT1 western blot—a representative image of three repeats for the immunoblotting of p-IRF3 and p-STAT1 is shown. Quantification of p-IRF3 (Bii) and p-STAT-1 (Biii)—the band intensity for each repeat was calculated using Image Lab software, normalized to the total protein content and expressed as a percentage of B. parapertussis WT. Histograms show the mean + standard deviation. One-way ANOVA with Tukey’s multiple comparison test was performed using GraphPad Prism 10. ns, not significant; *p < 0.05, **, p < 0.01, ***p < 0.001, and ****p < 0.0001. Predicted LPS structure and lane numbers are indicated beside each Bordetella strain. Dotted line represents mean “no stimulation” reading.
The above assay was not sufficiently sensitive to detect immediate IRF3 activation at 4 h post-stimulation. Thus, western blot was used to detect the presence of phosphorylated IRF3 (p-IRF3) and phosphorylated STAT1 (p-STAT1) 4 h after the stimulation of PMA-differentiated THP-1 cells with heat-killed bacteria. A representative western blot image for p-IRF3 and p-STAT1 is shown in Figure 4B,i. The band intensity of p-IRF3 and p-STAT1 was quantified using Image Lab software and represented as a percentage of B. parapertussis WT in Figures 4B,ii,iii respectively. Except for B. parapertussis ΔpagL mutant (Lane 9) and B. pertussis WT (Lane 11), the western blot data at 4 h replicated the luciferase reporter assay results observed at 24 h. B. parapertussis had the highest intensity of p-IRF3 and p-STAT1 bands (Figures 4B,ii,iii; Lane 1). Deleting the O antigen caused a moderate reduction in p-IRF3 band intensity (Lanes 3, 5), while deleting the lgm locus (Lanes 2, 4, 6, 8, 12) or altering the number of acyl chains (Lanes 7, 9, 10) caused a significant reduction (Figure 4B,ii). Contrary to the luciferase assay, the hepta-acylated ΔpagL mutant (Lane 9) had significantly less p-IRF3 band intensity than the WT (Lane 1) at 4 h post-stimulation. Similarly, B. pertussis also had minimal IRF3 activation (Figure 4B,ii; Lane 11). Additionally, any structural changes to B. parapertussis WT LPS led to significantly lower STAT1 activation in all strains when compared to the WT (Figure 4B,iii; Lane 1). B. pertussis also had minimal STAT1 activation at 4 h (Figure 4B,iii; Lane 11).
Overall, all three LPS features studied: the GlcN modification of the backbone phosphates, the O antigen and the number of acyl chains, were observed to alter TLR4-mediated signaling, albeit in their own unique ways.
Previous studies on TLR4 signaling by B. pertussis LPS lacking GlcN modification showed a reduction in THP-1 macrophage-mediated cytokine production downstream of both the MyD88 pathway (e.g., IL-6, TNFα) and the TRIF pathway (e.g., IP-10, MCP-1, RANTES) (Marr et al., 2010a). Upon further investigation, a group of four negatively charged amino acid residues on human TLR4 were collectively shown to be important for its interaction with the positively charged GlcN moiety of B. pertussis LPS, thereby facilitating dimerization and subsequent NFκB activation (Maeshima et al., 2015). In this study, we were not only able to replicate the impact of GlcN modification on TLR4 signaling in B. pertussis but in hexa-acylated B. parapertussis as well. The lack of this modification was found to consistently and significantly reduce both NFκB and IRF3 activation in both strains. Additionally, deleting the lgm locus in waaL, wbmA-E or pagP mutants further reduced NFκB and IRF3 activity. Hence, this study provides compelling evidence that the GlcN modification had an overarching dominant influence on both TLR4-mediated signaling pathways, overriding the effects of the O antigen’s presence or absence and the number of acyl chains attached to lipid A. Furthermore, similar to trends reported by Geurtsen et al. (2009), whereby an insertional inactivation of arnT (i.e., lgmB) in B. parapertussis led to a greater drop in IL-6 production when compared to that in B. pertussis, we also observed a greater drop in both NFκB and IRF3 activation upon the deletion of the lgm locus in B. parapertussis compared to B. pertussis. These observations underscore the importance of the role of the GlcN moiety in the initial interaction and dimerization of TLR4/MD-2—LPS complexes which thereby dictates the overall activation of TLR4-mediated signaling, impacting the MyD88 and the TRIF pathway equally. While the presence of the GlcN moiety in the WT strains increases the visibility of the bacteria to the human immune system through TLR4, it has been proven beneficial to the bacteria by increasing resistance to cationic antimicrobial peptides and contributing to the integrity of the outer membrane (Shah et al., 2014). However, the benefits of the GlcN modification in B. parapertussis are unclear.
Secondary to the GlcN modification, the structural feature influencing TLR4 signaling the most was the number of acyl chains attached to lipid A. Hexa-acylated E. coli and B. parapertussis strongly activated NFκB and IRF3. In contrast, under-acylation to 4 or 5 acyl chains (in B. parapertussis ΔpagP and ΔpagP ΔpagL respectively) moderately reduced NFκB activation and completely abrogated signaling via the TRIF pathway. A study examining E. coli LPS and TLR4/MD-2 interaction showed that five of the acyl chains of LPS fit into the MD-2 pocket, while the sixth lay exposed and free to facilitate dimerization by interacting with hydrophobic residues on the second TLR4 (Park et al., 2009). Consequently, it stands to reason that under-acylation would change the fit of the LPS in the MD-2 pocket or prevent the exposure of an acyl chain and thereby weaken TLR4 dimerization and consequently, downstream signaling. This reasoning supports the results observed in our study as well as those seen with other under-acylated LPS like Lipid IVA, R. sphaeroides and LPS1435/1449 variant of P. gingivalis (Meng et al., 2010; Herath et al., 2013; Anwar et al., 2015). Thus, we deduce that under-acylation of B. parapertussis LPS could weaken TLR4 dimerization to an extent where it moderately signals via the cell surface-MyD88 pathway but hinders the endocytosis of the dimer and/or the activation of the TRIF pathway. On the other hand, increasing the number of acyl chains to 7 (B. parapertussis ΔpagL) did not impact both pathways at 24 h indicating that the extra acyl chain, presumably also exposed from the MD-2 pocket, does not hinder nor benefit TLR4-mediated signaling over time. However, it remains unclear why hepta-acylation affected early (4 h) TRIF pathway activation alone, unless explained by differences in experimental protocol.
Last of all, the presence or absence of the O antigen in either penta-or hexa-acylated Bordetella species did not alter NFκB activation. However, the loss of the O antigen in B. parapertussis led to a significant reduction in TRIF pathway activation at both 4 h and 24 h. This work corroborates studies by Fedele et al. (2008) and Zanoni et al. (2012) who showed that LPS with an O antigen induced superior DC maturation and IFN-β response respectively, when compared to its O antigen lacking LPS species, presumably due to its interaction with CD14. These studies, along with ours, support the theory that the O antigen interacts with the TLR4 cofactor, CD14, which is indispensable for TRIF pathway activation. This interaction thus promotes the endocytosis of the TLR4/MD-2—LPS dimer, biasing signaling towards the TRIF pathway without affecting the MyD88 pathway at the cell surface (Gangloff et al., 2005; Jiang et al., 2005; Zanoni et al., 2011; Zanoni et al., 2012). On the contrary, the penta-acylated species with and without the O antigen (B. parapertussis ΔpagP ΔpagL vs. B. pertussis WT) behaved differently. At 4 h post THP-1 stimulation, both species induced equally negligible levels of p-IRF3 and p-STAT1 despite one expressing the O antigen and the other not. Additionally, at 24 h post-stimulation, B. pertussis WT (without the O antigen) activated IRF3 significantly more than B. parapertussis ΔpagP ΔpagL expressing the O antigen, contrary to that observed in hexa-acylated LPS species. Inherent differences in LPS structure between B. pertussis and B. parapertussis such as acyl chain length and position, or the presence of the distal trisaccharide may contribute to the conflicting trends observed in IRF3 activation. Also, the influence of differences in experimental protocol, length of stimulation, or antigens encoded by B. pertussis and B. parapertussis on the observed results cannot be ruled out.
This study has given us a much deeper insight into how the GlcN modification, the number of acyl chains and the O antigen of Bordetella LPS influence the activation of TLR4-mediated MyD88 and TRIF pathways. In summary, the GlcN modification had an overarching effect over the O antigen and lipid A acylation, with its absence strongly reducing both MyD88 and TRIF pathway activation. Next, the under-acylation of LPS (to 4 or 5 acyl chains) partially reduced NFκB activation and abolished TRIF pathway activation while hexa-and hepta-acylated LPS equally and strongly activated NFκB and IRF3. Lastly, while not impacting the MyD88 pathway, the Bordetella O antigen biased signaling towards the TRIF pathway. This knowledge is not only helpful in understanding the interaction between LPS and TLR4 and the factors influencing downstream signaling, but also aids in creating engineered LPS species that can specifically modulate the immune response generated. Bacterial LPS structure can be tailored to delicately tune the MyD88 vs. TRIF response to enhance the generated immune response and memory, thereby informing vaccine design.
All data for this study are provided within the manuscript and supplementary information files, further inquiries can be directed to the corresponding author.
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
KM: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. YFM: Formal analysis, Investigation, Methodology, Writing – review & editing. RCF: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work is supported by the Canadian Institutes of Health Research (PG-53242) grant. KM is the recipient of the International Doctoral Fellowship from the University of British Columbia. YM is the recipient of a Michael Smith Foundation for Health Research Fellowship (RT-2021-1655).
We would like to thank Dr. Christopher Overall and his lab for providing us the opportunity to use the Bruker Autoflex MALDI-TOF spectrometer in his lab and Dr. Yoan Machado Hernandez for his technical assistance. We would like to express our deepest gratitude to Dr. Kenneth Harder for the use of the THP-1 Dual cells and his lab for their assistance in the THP-1 Dual luciferase reporter assay as well as the use of their Perkin-Elmer Victor X5 Multilabel reader for this assay. We would also like to thank Dr. Hugh Kim, Dr. Pauline Johnson, and Dr. Steven Hallam for the use of the BioRad Imaging System, the Molecular Devices SpectraMax 190 microplate reader and the Thermo Scientific VarioSkan Flash multimode plate reader, respectively. Finally, we would like to thank a member of our laboratory, Dr. Gyles Ifill for creating the pIG02 plasmid for the clean deletion protocol.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1540534/full#supplementary-material
1. ^Centers for Disease Control and Prevention (CDC).
2. ^European Centre for Disease Prevention and Control (ECDC).
3. ^Public Health Agency of Canada (PHAC).
Allen, A. G., Thomas, R. M., Cadisch, J. T., and Maskell, D. J. (1998). Molecular and functional analysis of the lipopolysaccharide biosynthesis locus wlb from Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Mol. Microbiol. 29, 27–38. doi: 10.1046/j.1365-2958.1998.00878.x
Anwar, M. A., Panneerselvam, S., Shah, M., and Choi, S. (2015). Insights into the species-specific TLR4 signaling mechanism in response to Rhodobacter sphaeroides lipid A detection. Sci. Rep. 5:7657. doi: 10.1038/srep07657
Bertani, B., and Ruiz, N. (2018). Function and biogenesis of lipopolysaccharides. EcoSal Plus 8. doi: 10.1128/ecosalplus.ESP-0001-2018
Chen, Q., Ng, V., Warfel, J. M., Merkel, T. J., and Stibitz, S. (2017). Activation of Bvg-repressed genes in Bordetella pertussis by RisA requires cross talk from noncooperonic histidine kinase RisK. J. Bacteriol. 199:e00475. doi: 10.1128/JB.00475-17
Choi, K. H., Gaynor, J. B., White, K. G., Lopez, C., Bosio, C. M., Karkhoff-Schweizer, R. R., et al. (2005). A Tn7-based broad-range bacterial cloning and expression system. Nat Methods. 2, 443–448. doi: 10.1038/nmeth765
Choi, K. H., Mima, T., Casart, Y., Rholl, D., Kumar, A., Beacham, I. R., et al. (2008). Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Appl. Environ. Microbiol. 74, 1064–1075. doi: 10.1128/AEM.02430-07
Ciesielska, A., Matyjek, M., and Kwiatkowska, K. (2021). TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell. Mol. Life Sci. 78, 1233–1261. doi: 10.1007/s00018-020-03656-y
de Jonge, E. F., Balhuizen, M. D., van Boxtel, R., Wu, J., Haagsman, H. P., and Tommassen, J. (2021). Heat shock enhances outer-membrane vesicle release in Bordetella spp. Curr. Res. Microb. Sci. 2:100009. doi: 10.1016/j.crmicr.2020.100009
Di Fabio, J. L., Caroff, M., Karibian, D., Richards, J. C., and Perry, M. B. (1992). Characterization of the common antigenic lipopolysaccharide O-chains produced by Bordetella bronchiseptica and Bordetella parapertussis. FEMS Microbiol. Lett. 97, 275–281. doi: 10.1016/0378-1097(92)90348-r
Duan, T., Du, Y., Xing, C., Wang, H. Y., and Wang, R. F. (2022). Toll-like receptor signaling and its role in cell-mediated immunity. Front. Immunol. 13:812774. doi: 10.3389/fimmu.2022.812774
El Hamidi, A., Novikov, A., Karibian, D., Perry, M. B., and Caroff, M. (2009). Structural characterization of Bordetella parapertussis lipid A. J. Lipid Res. 50, 854–859. doi: 10.1194/jlr.M800454-JLR200
El Hamidi, A., Tirsoaga, A., Novikov, A., Hussein, A., and Caroff, M. (2005). Microextraction of bacterial lipid A: easy and rapid method for mass spectrometric characterization. J. Lipid Res. 46, 1773–1778. doi: 10.1194/jlr.D500014-JLR200
Fathy Mohamed, Y., and Fernandez, R. C. (2024). Programming Bordetella pertussis lipid A to promote adjuvanticity. Microb. Cell Fact. 23:250. doi: 10.1186/s12934-024-02518-7
Fedele, G., Nasso, M., Spensieri, F., Palazzo, R., Frasca, L., Watanabe, M., et al. (2008). Lipopolysaccharides from Bordetella pertussis and Bordetella parapertussis differently modulate human dendritic cell functions resulting in divergent prevalence of Th17-polarized responses. J. Immunol. 181, 208–216. doi: 10.4049/jimmunol.181.1.208
Fitzgerald, K. A., and Kagan, J. C. (2020). Toll-like receptors and the control of immunity. Cell 180, 1044–1066. doi: 10.1016/j.cell.2020.02.041
Gangloff, S. C., Zahringer, U., Blondin, C., Guenounou, M., Silver, J., and Goyert, S. M. (2005). Influence of CD14 on ligand interactions between lipopolysaccharide and its receptor complex. J. Immunol. 175, 3940–3945. doi: 10.4049/jimmunol.175.6.3940
Geurtsen, J., Dzieciatkowska, M., Steeghs, L., Hamstra, H. J., Boleij, J., Broen, K., et al. (2009). Identification of a novel lipopolysaccharide core biosynthesis gene cluster in Bordetella pertussis, and influence of core structure and lipid A glucosamine substitution on endotoxic activity. Infect. Immun. 77, 2602–2611. doi: 10.1128/IAI.00033-09
Geurtsen, J., Steeghs, L., Hamstra, H. J., Ten Hove, J., de Haan, A., Kuipers, B., et al. (2006). Expression of the lipopolysaccharide-modifying enzymes PagP and PagL modulates the endotoxic activity of Bordetella pertussis. Infect. Immun. 74, 5574–5585. doi: 10.1128/IAI.00834-06
Herath, T. D., Darveau, R. P., Seneviratne, C. J., Wang, C. Y., Wang, Y., and Jin, L. (2013). Tetra-and penta-acylated lipid A structures of Porphyromonas gingivalis LPS differentially activate TLR4-mediated NF-kappaB signal transduction cascade and immuno-inflammatory response in human gingival fibroblasts. PLoS One 8:e58496. doi: 10.1371/journal.pone.0058496
Hittle, L. E., Jones, J. W., Hajjar, A. M., Ernst, R. K., and Preston, A. (2015). Bordetella parapertussis PagP mediates the addition of two palmitates to the lipopolysaccharide lipid A. J. Bacteriol. 197, 572–580. doi: 10.1128/JB.02236-14
Ifill, G., Blimkie, T., Lee, A. H., Mackie, G. A., Chen, Q., Stibitz, S., et al. (2021). RNase III and RNase E influence posttranscriptional regulatory networks involved in virulence factor production, metabolism, and regulatory RNA processing in Bordetella pertussis. mSphere 6:e0065021. doi: 10.1128/mSphere.00650-21
Jiang, Z., Georgel, P., Du, X., Shamel, L., Sovath, S., Mudd, S., et al. (2005). CD14 is required for MyD88-independent LPS signaling. Nat. Immunol. 6, 565–570. doi: 10.1038/ni1207
Kalynych, S., Morona, R., and Cygler, M. (2014). Progress in understanding the assembly process of bacterial O-antigen. FEMS Microbiol. Rev. 38, 1048–1065. doi: 10.1111/1574-6976.12070
Kirikae, T., Schade, F. U., Kirikae, F., Qureshi, N., Takayama, K., and Rietschel, E. T. (1994). Diphosphoryl lipid A derived from the lipopolysaccharide (LPS) of Rhodobacter sphaeroides ATCC 17023 is a potent competitive LPS inhibitor in murine macrophage-like J774.1 cells. FEMS Immunol. Med. Microbiol. 9, 237–243. doi: 10.1111/j.1574-695X.1994.tb00499.x
Lopez, C. M., Rholl, D. A., Trunck, L. A., and Schweizer, H. P. (2009). Versatile dual-technology system for markerless allele replacement in Burkholderia pseudomallei. Appl. Environ. Microbiol. 75, 6496–6503. doi: 10.1128/AEM.01669-09
Lu, Y. C., Yeh, W. C., and Ohashi, P. S. (2008). LPS/TLR4 signal transduction pathway. Cytokine 42, 145–151. doi: 10.1016/j.cyto.2008.01.006
Maeshima, N., Evans-Atkinson, T., Hajjar, A. M., and Fernandez, R. C. (2015). Bordetella pertussis lipid A recognition by Toll-like receptor 4 and MD-2 is dependent on distinct charged and uncharged interfaces. J. Biol. Chem. 290, 13440–13453. doi: 10.1074/jbc.M115.653881
Maeshima, N., and Fernandez, R. C. (2013). Recognition of lipid A variants by the TLR4-MD-2 receptor complex. Front. Cell. Infect. Microbiol. 3:3. doi: 10.3389/fcimb.2013.00003
Marolda, C. L., Lahiry, P., Vines, E., Saldias, S., and Valvano, M. A. (2006). Micromethods for the characterization of lipid A-core and O-antigen lipopolysaccharide. Methods Mol. Biol. 347, 237–252. doi: 10.1385/1-59745-167-3:237
Marr, N., Hajjar, A. M., Shah, N. R., Novikov, A., Yam, C. S., Caroff, M., et al. (2010a). Substitution of the Bordetella pertussis lipid A phosphate groups with glucosamine is required for robust NF-kappaB activation and release of proinflammatory cytokines in cells expressing human but not murine Toll-like receptor 4-MD-2-CD14. Infect. Immun. 78, 2060–2069. doi: 10.1128/IAI.01346-09
Marr, N., Novikov, A., Hajjar, A. M., Caroff, M., and Fernandez, R. C. (2010b). Variability in the lipooligosaccharide structure and endotoxicity among Bordetella pertussis strains. J. Infect. Dis. 202, 1897–1906. doi: 10.1086/657409
Marr, N., Tirsoaga, A., Blanot, D., Fernandez, R., and Caroff, M. (2008). Glucosamine found as a substituent of both phosphate groups in Bordetella lipid A backbones: role of a BvgAS-activated ArnT ortholog. J. Bacteriol. 190, 4281–4290. doi: 10.1128/JB.01875-07
Mata-Haro, V., Cekic, C., Martin, M., Chilton, P. M., Casella, C. R., and Mitchell, T. C. (2007). The vaccine adjuvant monophosphoryl lipid A as a TRIF-biased agonist of TLR4. Science 316, 1628–1632. doi: 10.1126/science.1138963
Meng, J., Lien, E., and Golenbock, D. T. (2010). MD-2-mediated ionic interactions between lipid A and TLR4 are essential for receptor activation. J. Biol. Chem. 285, 8695–8702. doi: 10.1074/jbc.M109.075127
Miller, S. I., Ernst, R. K., and Bader, M. W. (2005). LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 3, 36–46. doi: 10.1038/nrmicro1068
Mulford, C. A., and Osborn, M. J. (1983). An intermediate step in translocation of lipopolysaccharide to the outer membrane of Salmonella typhimurium. Proc. Natl. Acad. Sci. U.S.A. 80, 1159–1163. doi: 10.1073/pnas.80.5.1159
Noble, B. A., Jiudice, S. S., Jones, J. D., and Timbrook, T. T. (2024). Reemergence of Bordetella parapertussis, United States, 2019–2023. Emerg. Infect. Dis. 30, 1058–1060. doi: 10.3201/eid3005.231278
Novikov, A., Marr, N., and Caroff, M. (2019). A comparative study of the complete lipopolysaccharide structures and biosynthesis loci of Bordetella avium, B. hinzii, and B. trematum. Biochimie 159, 81–92. doi: 10.1016/j.biochi.2018.12.011
Novikov, A., Shah, N. R., AlBitar-Nehme, S., Basheer, S. M., Trento, I., Tirsoaga, A., et al. (2014). Complete Bordetella avium, Bordetella hinzii and Bordetella trematum lipid A structures and genomic sequence analyses of the loci involved in their modifications. Innate Immun. 20, 659–672. doi: 10.1177/1753425913506950
Park, B. S., Song, D. H., Kim, H. M., Choi, B. S., Lee, H., and Lee, J. O. (2009). The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458, 1191–1195. doi: 10.1038/nature07830
Preston, A., Allen, A. G., Cadisch, J., Thomas, R., Stevens, K., Churcher, C. M., et al. (1999). Genetic basis for lipopolysaccharide O-antigen biosynthesis in Bordetellae. Infect. Immun. 67, 3763–3767. doi: 10.1128/IAI.67.8.3763-3767.1999
Preston, A., and Maskell, D. (2001). The molecular genetics and role in infection of lipopolysaccharide biosynthesis in the Bordetellae. J. Endotoxin Res. 7, 251–261. doi: 10.1179/096805101101532765
Preston, A., Petersen, B. O., Duus, J. O., Kubler-Kielb, J., Ben-Menachem, G., Li, J., et al. (2006). Complete structures of Bordetella bronchiseptica and Bordetella parapertussis lipopolysaccharides. J. Biol. Chem. 281, 18135–18144. doi: 10.1074/jbc.M513904200
Qureshi, N., Takayama, K., and Kurtz, R. (1991). Diphosphoryl lipid A obtained from the nontoxic lipopolysaccharide of Rhodopseudomonas sphaeroides is an endotoxin antagonist in mice. Infect. Immun. 59, 441–444. doi: 10.1128/iai.59.1.441-444.1991
Rivera, I., Linz, B., and Harvill, E. T. (2020). Evolution and conservation of Bordetella intracellular survival in eukaryotic host cells. Front. Microbiol. 11:557819. doi: 10.3389/fmicb.2020.557819
Shah, N. R., Albitar-Nehme, S., Kim, E., Marr, N., Novikov, A., Caroff, M., et al. (2013). Minor modifications to the phosphate groups and the C3′ acyl chain length of lipid A in two Bordetella pertussis strains, BP338 and 18-323, independently affect Toll-like receptor 4 protein activation. J. Biol. Chem. 288, 11751–11760. doi: 10.1074/jbc.M112.434365
Shah, N. R., Hancock, R. E., and Fernandez, R. C. (2014). Bordetella pertussis lipid A glucosamine modification confers resistance to cationic antimicrobial peptides and increases resistance to outer membrane perturbation. Antimicrob. Agents Chemother. 58, 4931–4934. doi: 10.1128/AAC.02590-14
Simpson, B. W., and Trent, M. S. (2019). Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol. 17, 403–416. doi: 10.1038/s41579-019-0201-x
Stevens, C. W., Aravind, S., Das, S., and Davis, R. L. (2013). Pharmacological characterization of LPS and opioid interactions at the Toll-like receptor 4. Br. J. Pharmacol. 168, 1421–1429. doi: 10.1111/bph.12028
Zamyatina, A., and Heine, H. (2020). Lipopolysaccharide recognition in the crossroads of TLR4 and caspase-4/11 mediated inflammatory pathways. Front. Immunol. 11:585146. doi: 10.3389/fimmu.2020.585146
Zanoni, I., Bodio, C., Broggi, A., Ostuni, R., Caccia, M., Collini, M., et al. (2012). Similarities and differences of innate immune responses elicited by smooth and rough LPS. Immunol. Lett. 142, 41–47. doi: 10.1016/j.imlet.2011.12.002
Keywords: Bordetella, LPS, TLR4, MyD88, NFκB, TRIF, IRF3
Citation: Manivannan K, Fathy Mohamed Y and Fernandez RC (2025) Determining the Bordetella LPS structural features that influence TLR4 downstream signaling. Front. Microbiol. 16:1540534. doi: 10.3389/fmicb.2025.1540534
Received: 06 December 2024; Accepted: 03 February 2025;
Published: 25 February 2025.
Edited by:
Axel Cloeckaert, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Yuhang Wang, The University of Iowa, United StatesCopyright © 2025 Manivannan, Fathy Mohamed and Fernandez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rachel C. Fernandez, cmFjaGVsZkBtYWlsLnViYy5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.