
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 22 January 2025
Sec. Microbial Physiology and Metabolism
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1533918
 Li-Yan Zhang1
Li-Yan Zhang1 Tian-Tian Li1
Tian-Tian Li1 Hong-Xin Liao1
Hong-Xin Liao1 Jin-Rui Wen1
Jin-Rui Wen1 Hong-Yan Nie1
Hong-Yan Nie1 Fu-Rong Xu1
Fu-Rong Xu1 Xiao-Yun Liu2*
Xiao-Yun Liu2* Xian Dong1*
Xian Dong1*Background: The antifungal properties of essential oils (EOs) and their active constituents have been well documented. Histone acetylation is pivotal in modulating gene expression and influences biological processes in living organisms.
Results: This study demonstrated that menthone, the primary component of Mentha haplocalyx EO, exhibited notable antifungal activity against Fusarium proliferatum (EC50 = 6.099 mmol/L). The treatment significantly inhibited hyphal growth, reduced spore germination rates from 31.49 to 4.95%, decreased spore viability from 46.88 to 20.91%, and reduced spore production by a factor of 17.92 compared with the control group while simultaneously enhancing cell membrane permeability. However, the direct relationship between menthone and histone acetylation in inhibiting F. proliferatum remains nebulous. Our RNA sequencing (RNA-seq) analysis identified 7,332 differentially expressed genes (DEGs) between the control and menthone-treated groups, 3,442 upregulated and 3,880 downregulated, primarily enriched in pathways related to ribosome biogenesis and energy metabolism. Chromatin immunoprecipitation sequencing (ChIP-seq) analysis revealed that menthone inhibited the growth of F. proliferatum by decreasing H3K27ac levels and interfering with the transcription of energy metabolism-related genes. By integrating the RNA-seq data with the ChIP-seq results, we identified 110 DEGs associated with reduced H3K27ac modification primarily associated with ribosome biogenesis. Menthone affected the growth of F. proliferatum by reducing the expression of ribosome biogenesis-related genes (FPRO_06392, FPRO_01260, FPRO_10795, and FPRO_01372).
Conclusion: This study elucidated the mechanism by which menthone inhibits F. proliferatum growth from a histone acetylation modification perspective, providing insights into its application as an antifungal agent to prevent root rot in Panax notoginseng.
Panax notoginseng (burk) Chen is a perennial herbaceous plant belonging to the Araliaceae family and recognized for its therapeutic applications in cardiovascular and orthopedic diseases (Xu et al., 2019). This is attributable to their unique active compounds and high medicinal value. However, due to its specific habitat requirements and extended cultivation cycle, P. notoginseng is susceptible to various diseases, of which root rot is one of the most detrimental. This can lead to the cracking and decay of the primary medicinal parts and their roots, resulting in significant economic losses (Ning et al., 2021). Root rot disease in P. notoginseng is primarily caused by pathogenic fungi belonging to the genus Fusarium. These pathogens adhere to the surface of the root system via hyphae, establishing colonies on the roots, which subsequently invade the vascular system and cause root rot (Zheng et al., 2022). Our research group has identified that, alongside Fusarium oxysporum and Fusarium solani, another pathogen responsible for root rot in P. notoginseng is Fusarium proliferatum, which can infect its roots and facilitate disease progression (Ma et al., 2019).
F. proliferatum is a major pathogenic fungus with a broad host range capable of causing root rot in various crops, including peanuts, tomatoes, and rice (Sun et al., 2022; Ye et al., 2020; Wang et al., 2021). Additionally, F. proliferatum can secrete toxins that may pose health risks to humans and animals (Alshannaq and Yu, 2017). F. proliferatum is a necrotrophic fungus widely distributed in the soil, where it can inhabit plant tissues and persist for many years by producing large quantities of chlamydospores. Under favorable environmental conditions, the spores of F. proliferatum germinate and disperse through wind and rain, leading to disease outbreaks (Nguyen et al., 2016; Panth et al., 2020). Currently, the management of root rot caused by F. proliferatum relies heavily on chemical pesticides, which can mitigate fungal diseases to some extent; however, prolonged use raises concerns regarding pesticide residues and environmental pollution, thereby posing threats to human health (Panth et al., 2020; Zeng et al., 2021).
Essential oils (EOs) are complex mixtures of volatile plant-derived compounds. In addition to their notable antimicrobial properties, these oils offer several advantages, including easy biodegradability and the absence of residues, which complicate the development of resistance by pathogens. Consequently, EOs represent a significant source of novel plant-based antimicrobial agents (Alonso-Gato et al., 2021). Mentha haplocalyx is a notable medicinal plant that belongs to the Lamiaceae family and is widely distributed across China, Korea, and Japan (Tang et al., 2024). Its EOs exhibit potent antibacterial activity against various pathogenic microorganisms (Karpiński, 2020). Research has indicated that menthone, a primary component of M. haplocalyx EOs, can disrupt the cell membranes of pathogens, causing the leakage of cellular contents and inhibiting cellular respiration, ultimately disturbing the internal equilibrium of the cells and resulting in cell death (Kang et al., 2019). Furthermore, menthone is one of the main constituents responsible for biological activities, such as antibacterial effects against Helicobacter pylori, antifungal properties against Fusarium sambucinum, and antibiofilm activity against methicillin-resistant Staphylococcus aureus (Piasecki et al., 2023; Pérez-Vázquez et al., 2022; Zhao et al., 2023). However, data on the molecular mechanisms by which menthone inhibits Fusarium species remain relatively limited.
Epigenetic modifications, including histone acetylation and methylation, play pivotal roles in regulating chromatin activity and gene transcription (Berger, 2007). Specifically, H3 acetylation at lysine 27 is frequently associated with activation of gene transcription (Zhang et al., 2021). In this study, we investigated the inhibitory effects of menthone on F. proliferatum by measuring EC50 values, spore germination rates, spore viability, spore yield, and cell membrane permeability. Additionally, we employed RNA sequencing (RNA-seq) and chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) analyses to examine the overall changes in gene transcription and histone acetylation levels following menthone treatment in F. proliferatum, thereby exploring its antifungal mechanism from the perspective of histone acetylation.
Our results indicate that menthone can compromise the cell membrane integrity of F. proliferatum and modulate the expression of genes associated with metabolic pathways related to ribosome biogenesis by reducing histone acetylation levels within this fungal strain. This finding highlights the potential antifungal targets of menthone and provides theoretical support for the development of targeted biological pesticides.
The fungal strains used in this study were isolated from the roots of P. notoginseng exhibiting root rot. After isolating and purifying the strains, they were placed on potato dextrose agar (PDA) medium and incubated at a temperature of 28°C for a duration of 7 days. As previously described (Nie et al., 2024), DNA was extracted from the target strain and sequenced. Sequencing results were submitted to GenBank. A comparative analysis using NCBI BLAST revealed that the sequence exhibited 100% similarity to F. proliferatum (MH712158.1). The GenBank accession number is OP430570.1.
Menthone was procured from Shanghai Yuanye Biological Technology Co., Ltd. (CAS. 14073-97-3, purity ≥97%). The effect of menthone on inhibiting the mycelial growth of F. proliferatum was evaluated using the mycelial growth rate method (Zhou et al., 2017). In a sterile environment, menthone was dissolved in a suspension of 20/1,000 Dimethyl sulfoxide (DMSO) and 1/1,000 Tween-80 (2-DMSO-T) and filtered to obtain a sterile filtrate. The menthone solution was diluted to various concentrations using a two-fold dilution method. The prepared menthone was incorporated into the PDA culture medium to create menthone-containing plates. A PDA plate containing only the 2-DMSO-T suspension was used as the negative control. Fungal blocks with a diameter of 5 mm were inoculated onto the central area of PDA plates containing different concentrations of menthone (0 mM, 0.5 mM, 1 mM, 2 mM, 4 mM, 8 mM, and 16 mM). The plates were incubated at 28°C in an incubator at constant temperature for 4 days. Each treatment comprised five replicates. Colony diameters were measured, and the inhibition rate was calculated. A linear regression equation was established using the least squares method, resulting in a toxicity regression equation. The EC50 value was subsequently derived from this toxicity regression equation (Zou et al., 2021). The percentage of mycelial growth inhibition is determined using the following formula. Inhibition rate (%) = (C − B)/(C) × 100, where C is the diameter of the negative control group colony (mm) and B is the diameter of the treatment group colony (mm).
Spore viability was assessed using a fluorescent staining method involving acridine orange and propidium iodide (Manzo-Valencia et al., 2016). The experimental procedure for spore germination was modified from the method described by Tanapichatsakul et al. (2020). Menthone was prepared according to the procedure outlined in section 2.2. Subsequently, the solution was transferred to a sterile EP tube (5 mL) containing 2 mL 1/3 PDA liquid culture medium. The solutions were thoroughly mixed to achieve a menthone concentration of 1 × EC50. An appropriate amount of the spore suspension was then added, with 2-DMSO-T serving as a negative control. Each treatment was performed in triplicates. After incubating the mixture on a shaker at 28°C and 180 rpm in the dark for 24 h, The rate of spore germination is determined using the following formula:
Spore production was assessed as previously described with modification (Zheng et al., 2014). Menthone was added to Bilay’s culture medium at a concentration of 1 × EC50. Subsequently, 2 mL of the spore suspension, at a concentration of 1 × 106 spores/mL, was added to each treatment group. The control group was treated with an equivalent volume of 2-DMSO-T as a solvent control. All samples were incubated in a thermostatic shaker at 28°C and agitated at 180 rpm in darkness for 5 days to facilitate shaking culture. Following incubation, spores were filtered through four layers of sterile cellulose acetate membranes [Sigma-Aldrich (Shanghai) Trading, MO, United States]. The resulting filtrate was thoroughly mixed, and samples were collected for analysis. The number of spores was quantified under a microscope using a hemocytometer. Each treatment included three biological replicates.
The permeability of the mycelial membrane was evaluated in accordance with the previously described methodology (Liu et al., 2022). A total of 2 g of mycelium was added to a conical flask containing 40 mL of sterile water, followed by menthone to achieve a final concentration equivalent to 1 × EC50. The conductivity was measured at 0, 1, 2, 3, 4, 5, 6, 7, and 8 h and recorded as D1. Concurrently, the conductivity without mycelia for each treatment group was measured and denoted as D0. After boiling the mycelium for 10 min, their conductivity was measured again and recorded as D2. DMSO was used as a negative control, with three biological replicates per treatment group. The formula was employed to calculate the relative conductivity. Relative conductivity = (D1 − D0)/D2 × 100%.
The treatment method for the mycelia was consistent with that described in section 2.5. Samples were collected at time points of 0, 6, 12, 24, 48, and 72 h and subsequently centrifuged at 8,000 rpm for 5 min. The concentration of soluble proteins in the supernatant was determined using the Coomassie Brilliant Blue G-250 staining method (Hatada et al., 2006).
As previously described (Liu et al., 2022), the mycelia of the fungus were collected following treatment with menthone for 24 h, immediately frozen in liquid nitrogen, and stored at −80°C for subsequent experiments. Total RNA extraction was performed following the manufacturer’s guidelines using TRIzol reagent. The quality of the extracted total RNA was assessed with a NanoPhotometer spectrophotometer (IMPLEN, CA, United States), a Qubit 2.0 fluorometer (Life Technologies, CA, United States), and an Agilent 2100 bioanalyzer (Agilent Technologies, CA, United States). A cDNA library was constructed at Novogene Technology Co., Ltd. (Beijing, China) and RNA-seq sequencing was performed. After purifying the cDNA and constructing the library, it was sequenced on an Illumina Novaseq6000 platform. The sequencing data were analyzed using Illumina Casava software (version 1.8) to remove adapter sequences and low-quality reads. Subsequently, the filtered reads were aligned to the reference genome of F. proliferatum (NCBI Taxonomy ID. 1227346) using the Hisat2 (version 2.2.1) software with default parameters.
Gene expression levels were determined as the number of transcript fragments per kilobase of transcript per million fragments (FPKM). The DEseq2 (version 1.22.1) software was used to analyzed DEGs between the two groups, with a screening standard of |log2Fold Change| ≥1 and false discovery rate (FDR) <0.05 (Ling et al., 2024). Finally, the results of the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were visualized using the online platform available at https://cloud.metware.cn.
Histones were prepared according to the instructions provided in the Histone Extraction Kit (BBproExtra®). The proteins obtained were separated by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently transferred to a polyvinylidene fluoride (PVDF) membrane. The membrane was then incubated with specific antibodies for hybridization. After hybridization, the membrane was blocked with 4% BSA in PBST (PBS-0.1% Tween20) at room temperature for 2 h. The primary antibodies used were anti-H3K27ac (ABconal, A2771, dilution factor is 1:10,000), anti-H3K4me3 (ABconal, A22146, dilution factor is 1:10,000), anti-H3K9ac (ABconal, A21107, dilution factor is 1:1,000), and anti-H3 (ABconal, A2348, dilution factor is 1:10,000). The hybridized membrane was washed three times with PBST for 10 min each before incubation with the primary antibody in PBST containing 4% BSA for an additional 2 h. Subsequently, the membrane was washed six times with PBST for 10 min each. This was followed by incubation with the corresponding secondary antibody, goat anti-rabbit IgG (Abbkine, A23220, dilution factor is 1:1,000), for 2 h. Finally, excess secondary antibody was removed by washing the membrane six times with PBST prior to signal detection in a dark room. The intensity of the bands was quantitatively analyzed using ImageJ software (Perrella et al., 2024).
ChIP experiments were conducted as previously described (Zhou et al., 2023), with modifications. Initially, the hyphae of F. proliferatum were ground into powder using liquid nitrogen. The chromatin was extracted by cross-linking with 1% formaldehyde under vacuum for a duration of 10 min. Following the completion of the cross-linking process, 2.5 M glycine was added and subjected to vacuum for an additional 5 min to effectively terminate the cross-linking reaction. Genomic DNA was fragmented into approximately 500 bp fragments through physical sonication, and ChIP was performed using an anti-H3K27ac antibody (A2771). The sequencing library was constructed following the protocol provided by the Illumina TruSeq® ChIP Sample Prep Set A. Subsequently, sequencing was conducted on the Illumina HiSeq-PE150 platform, facilitated by Bioacme Biotechnology Co., Ltd.
Total RNA was extracted from the mycelia using TRIzol reagent. First-strand cDNA was synthesized from the total RNA using a reverse transcription kit (TaKaRa). Real-time fluorescence quantitative PCR (RT-qPCR) was conducted on the CFX Manager 3.1 system (Bio-Rad) employing the SYBR Premix ExTaq kit (TaKaRa). In the RT-qPCR and ChIP-qPCR experiments, QTUB or input (unimmunoprecipitated chromatin sample) served as normalization controls. The relative expression levels of genes were analyzed using the method. The primers used for RT-qPCR and ChIP-qPCR are listed in Supplementary Tables S1, S2, respectively.
All experiments conducted in this study were performed in triplicate for each sample, and the data were analyzed using one-way ANOVA with SPSS software version 19.0. Duncan’s multiple range test was used to assess the significance of differences (p < 0.05). All data are presented as mean ± standard deviation. The figures were generated using Adobe Illustrator 2022 and GraphPad Prism 9 software.
Menthone treatment significantly inhibited the hyphal growth of F. proliferatum. Under varying concentrations of menthone, a marked reduction in colony size was observed, with the inhibitory effect intensifying as the menthone concentration increased (Figure 1A). These findings suggest that menthone effectively suppresses hyphal growth. Additionally, the EC50 value of menthone against F. proliferatum was 6.099 mmol/L, indicating strong antifungal activity. Consequently, we selected 6.099 mmol/L (1 × EC50) as the menthone concentration for subsequent analyses.
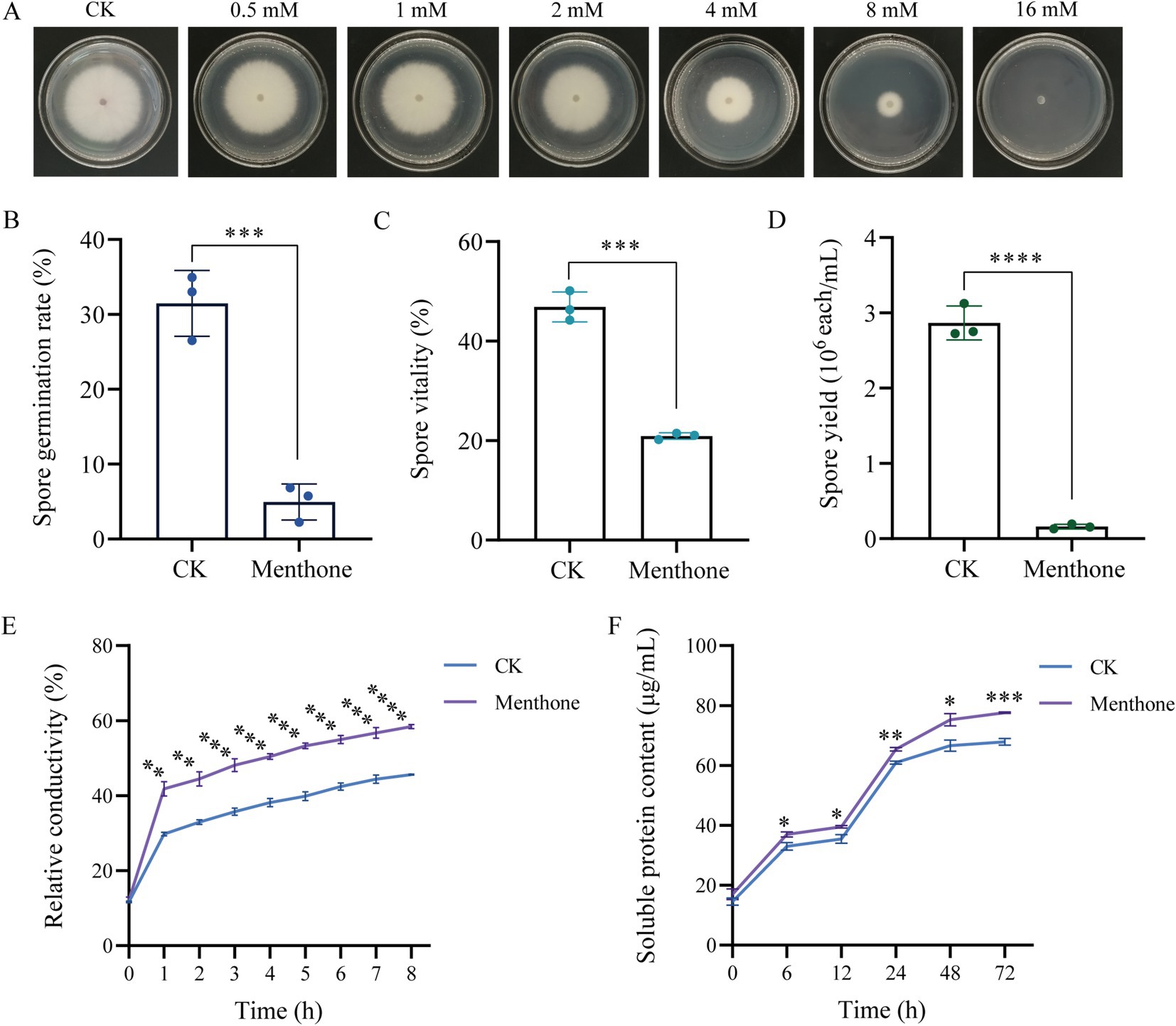
Figure 1. Antifungal effect of menthone against F. proliferatum. (A) Mycelial growth of F. proliferatum in PDA medium with various menthone concentrations. The effect of 1 × EC50 menthone on the spore germination rate (B), spore vitality (C), spore yield (D), relative electrical conductivity (E), and soluble protein content (F) of F. proliferatum. 2-DMSO-T (20/1,000 DMSO and 1/1,000 Tween-80 suspension) was used as a negative control (CK). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
The results of the spore germination assay indicated that menthone significantly inhibited the germination of F. proliferatum spores (Figure 1B). At a treatment concentration of 1 × EC50, the spore germination rate of F. proliferatum was only 4.95%, which contrasted the control group rate of 31.49%. This inhibition factor was approximately 6.36 times lower than that observed in the control group. Furthermore, under identical treatment conditions, the viability of F. proliferatum spores decreased from 46.88 to 20.91% (Figure 1C). These findings suggest that menthone exerts a pronounced inhibitory effect on spore germination and viability of F. proliferatum.
The sporulation of F. proliferatum was evaluated microscopically by counting the number of spores in a filtered mycelial suspension using a hemocytometer. The sporulation rate for the control group was 2.867 × 106 spores/mL. In contrast, the menthone-treated group demonstrated a significantly reduced sporulation rate of only 0.160 × 106 spores/mL, indicating an approximate reduction factor of 17.92 compared with the control group (Figure 1D). These results suggest that menthone exerts inhibitory effects on normal growth and spore development of F. proliferatum, leading to substantial impairment of its reproductive capacity.
The cell membrane plays a crucial role in preserving the structural integrity of the cell. When the cell membrane is compromised, normal physiological metabolism becomes impaired, and the leakage of cellular contents results in an increased conductance of the extracellular fluid. The effect of menthone on the membrane permeability of F. proliferatum was assessed by measuring its relative conductivity. Compared with the control group, a significant increase in the relative conductivity of F. proliferatum hyphae was observed with prolonged menthone treatment (Figure 1E). These findings indicate that menthone disrupts the membrane structure of F. proliferatum, resulting in electrolyte leakage and elevated extracellular conductivity.
The soluble protein content of F. proliferatum mycelia is shown in Figure 1F. Following menthone treatment, soluble protein content in the extracellular space of F. proliferatum mycelia gradually increased. Notably, protein levels exhibited a significant increase from 12 to 24 h, peaking at 72 h before stabilizing. Throughout the period from 6 to 72 h, the concentration of soluble protein in the treatment group was consistently and significantly higher than that observed in the control group. These findings suggest that menthone disrupts the cell membrane integrity of fungal cells, resulting in altered cell permeability and the subsequent leakage of cellular contents, inhibiting normal fungal growth.
To elucidate the antifungal mechanism of menthone against F. proliferatum, we performed transcriptome sequencing analysis of samples treated with menthone using the Illumina NovaSeq 6000 platform. After data cleaning and quality control, high-quality reads were generated for each independent sample, ranging from 21.25 to 31.77 million reads. These high-quality reads were successfully mapped to the reference genome of F. proliferatum, achieving mapping rates exceeding 90%, indicating the high level of reliability of our transcriptomic data (Supplementary Table S3). Principal component analysis (PCA) and correlation analyses demonstrated good biological repeatability within the sample groups, confirming the reliability of our results (Figures 2A,B).
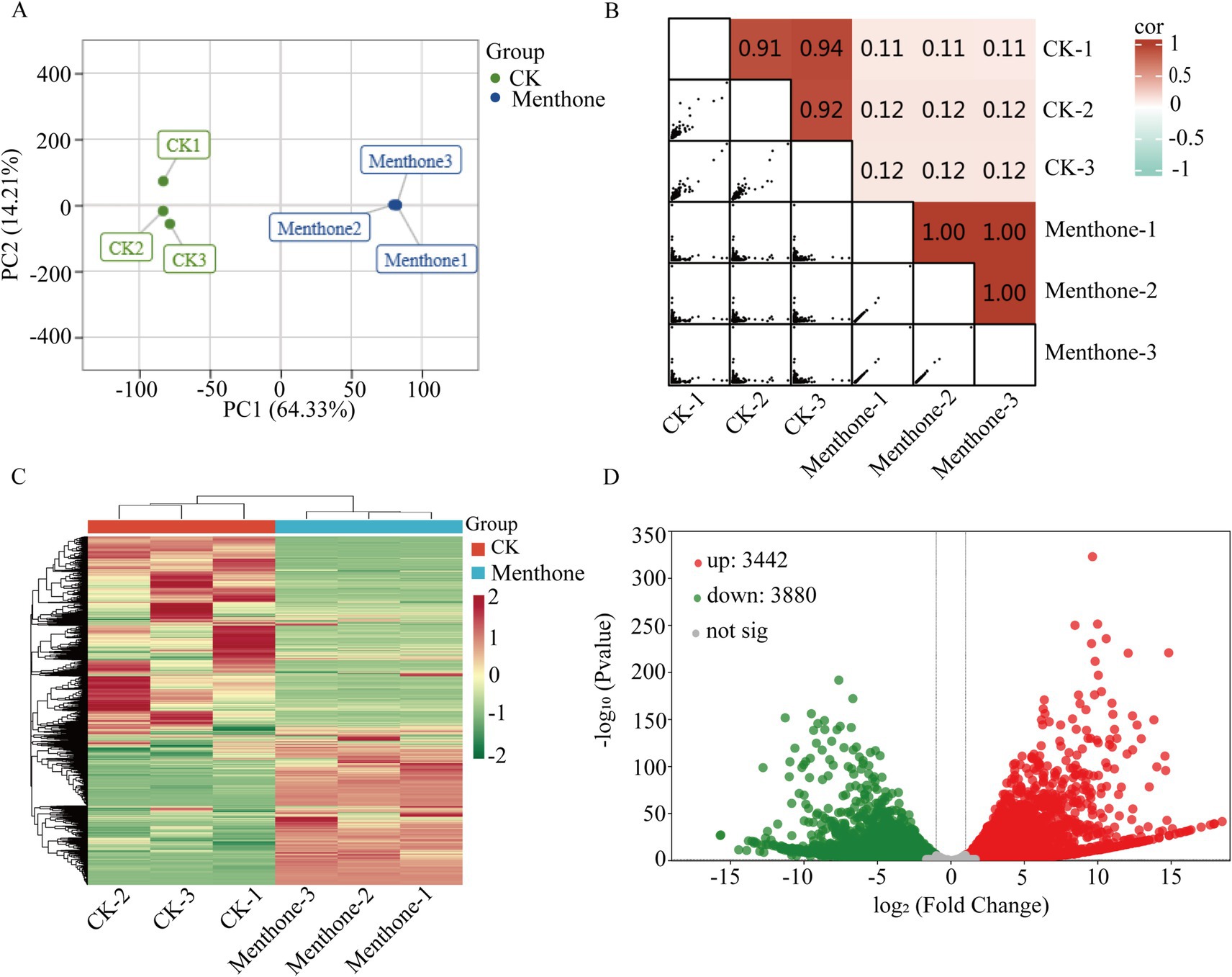
Figure 2. Preliminary analysis of transcriptomic data. (A) Principal component analysis score plot for all samples. The two treatments were control (CK) and menthol. (B) Heatmap of gene expression correlation. The color indicates the level of the correlation coefficient, and the number represents the correlation coefficient. (C) Clustering heat map of DEGs. (D) Volcano map of DEGs. The green and red colors represent downregulation and upregulation, respectively.
To further explore gene expression patterns, we identified DEGs using criteria of |log2Fold Change| ≥1 and false discovery rate (FDR) <0.05 for screening purposes. Our analysis revealed 3,442 upregulated genes and 3,880 downregulated genes in the menthone-treated group compared with the control group (Figures 2C,D). These findings suggest that menthone modulates the growth and development of F. proliferatum by inhibiting the expression of specific genes. Subsequently, the DEGs following menthone treatment were subjected to Gene Ontology (GO) functional enrichment analyses. The results indicated that upregulated DEGs were predominantly enriched in pathways related to the endoplasmic reticulum, cytoplasm, and lipid biosynthetic and metabolic processes. In contrast, downregulated DEGs were significantly enriched in pathways associated with ribonucleoprotein complex biosynthesis and nucleic acid-binding activities, including RNA- and snoRNA-binding pathways (Figures 3A,B).
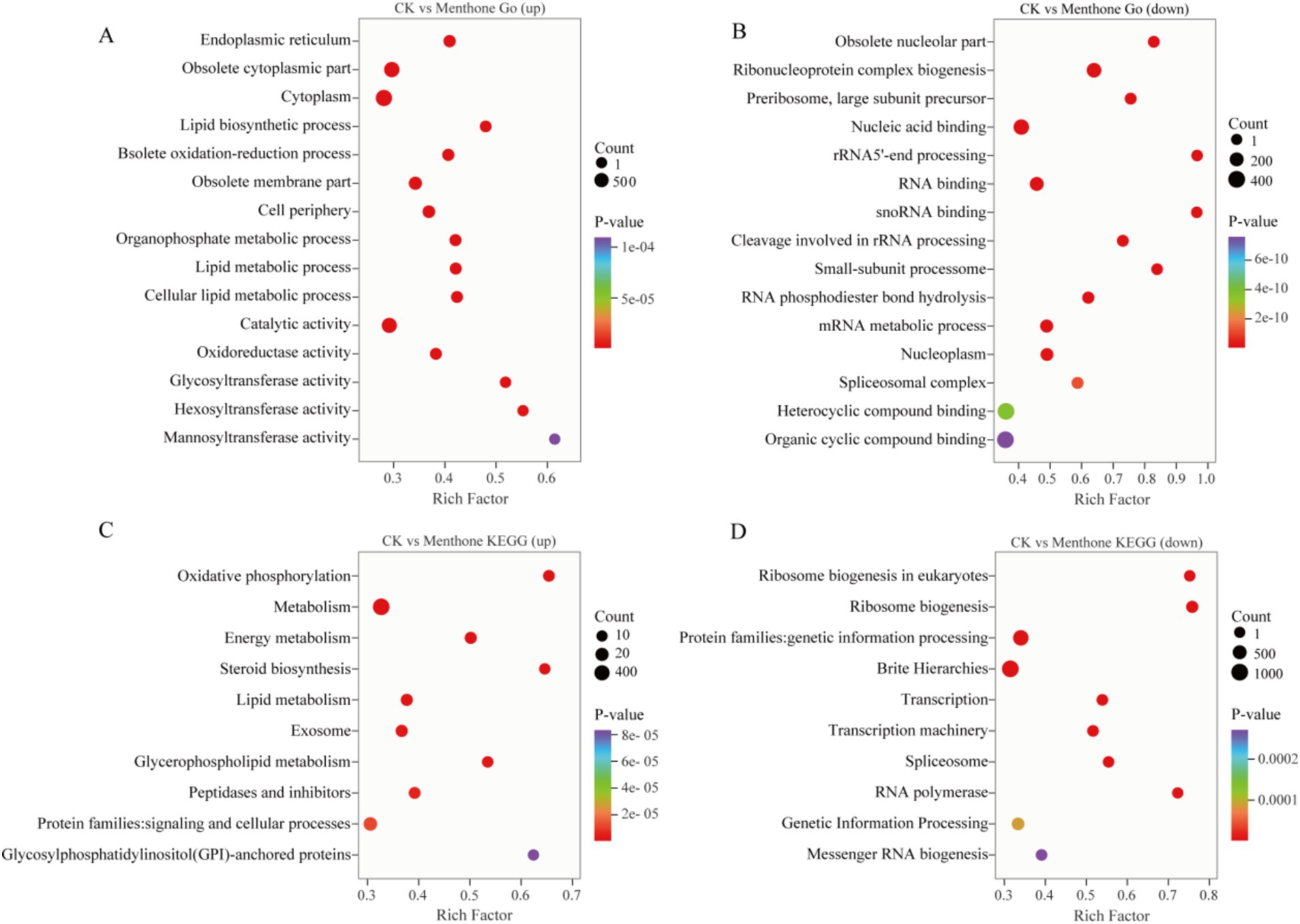
Figure 3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differentially expressed genes (DEGs). GO enrichment analysis of upregulated (A) and downregulated (B) DEGs between CK and menthone. KEGG enrichment analysis of upregulated (C) and downregulated (D) DEGs between CK and menthone. The x-axis represents the enrichment factor, while the y-axis indicates the pathway names. The color of the points reflects the p-value, with red indicating more significant enrichment and the size of the points corresponds to the number of DEGs.
To further investigate the biological functions of these DEGs in the growth and development of F. proliferatum, we conducted a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the DEGs, selecting the top 10 most significantly enriched pathways (Figures 3C,D).
DEGs upregulated by menthone treatment were predominantly enriched in oxidative phosphorylation, energy metabolism, steroid biosynthesis, and lipid metabolism. These findings suggest that menthone treatment enables F. proliferatum to mitigate membrane damage caused by menthone by enhancing steroid biosynthesis and lipid metabolism while simultaneously boosting oxidative phosphorylation and energy metabolism pathways to supply cellular energy. Conversely, the downregulated DEGs were primarily enriched in ribosome biogenesis, ribosome biogenesis in eukaryotes, transcription, and spliceosomes. The regulation of ribosomal biogenesis is a critical signal for cell growth and proliferation (Li et al., 2021). We hypothesized that menthone influences ribosome biogenesis in eukaryotes and other metabolic processes, affecting fungal protein synthesis and inhibiting the nutritional growth of F. proliferatum.
To elucidate the antifungal mechanism of menthone, we assessed the changes in histone modification levels using western blotting following menthone treatment. The results indicated that compared with the control group, the acetylation level of H3K27 was significantly reduced in F. proliferatum post-menthone treatment. No significant alterations were observed at other lysine sites of histone H3 (Figure 4; Supplementary Figure S1). This suggests that menthone primarily modulates the histone acetylation of H3K27 in F. proliferatum.

Figure 4. Menthone modulates the level of histone acetylation modification in F. proliferatum. (A) The levels of H3K9ac, H3K27ac, and H3K4me3 in F. proliferatum were detected using western blotting. Anti-H3 was used as the loading control. (B) Changes in relative gray values for the levels of histone acetylation and methylation modification in different treatment groups. *p < 0.05, ns, not significant.
Previous studies have established a close relationship between histone acetylation and gene activation. In this study, H3K27ac levels decreased following menthone treatment, which might have inhibited related gene expression. Therefore, we hypothesized that menthone inhibits the growth of F. proliferatum by suppressing histone acetylation.
To investigate the dynamic changes in H3K27 acetylation in F. proliferatum, we performed a ChIP-seq analysis. The results indicated that the number of genes enriched in high and low acetylation modifications affected by H3K27ac following menthone treatment was 488 and 362, respectively, when compared to the control (Figure 5B). The mapping rates for both ChIP-seq samples exceeded 80%, with approximately 40% of the modified binding sites located near the transcription start site (Figures 5A,D and Supplementary Table S4). Furthermore, H3K27 acetylation peaks were significantly enriched at transcription start sites across different treatment groups (Figure 5C). This finding aligns with the previously reported distribution patterns of histone markers (Papait et al., 2013), validating the authenticity of our sequencing data.
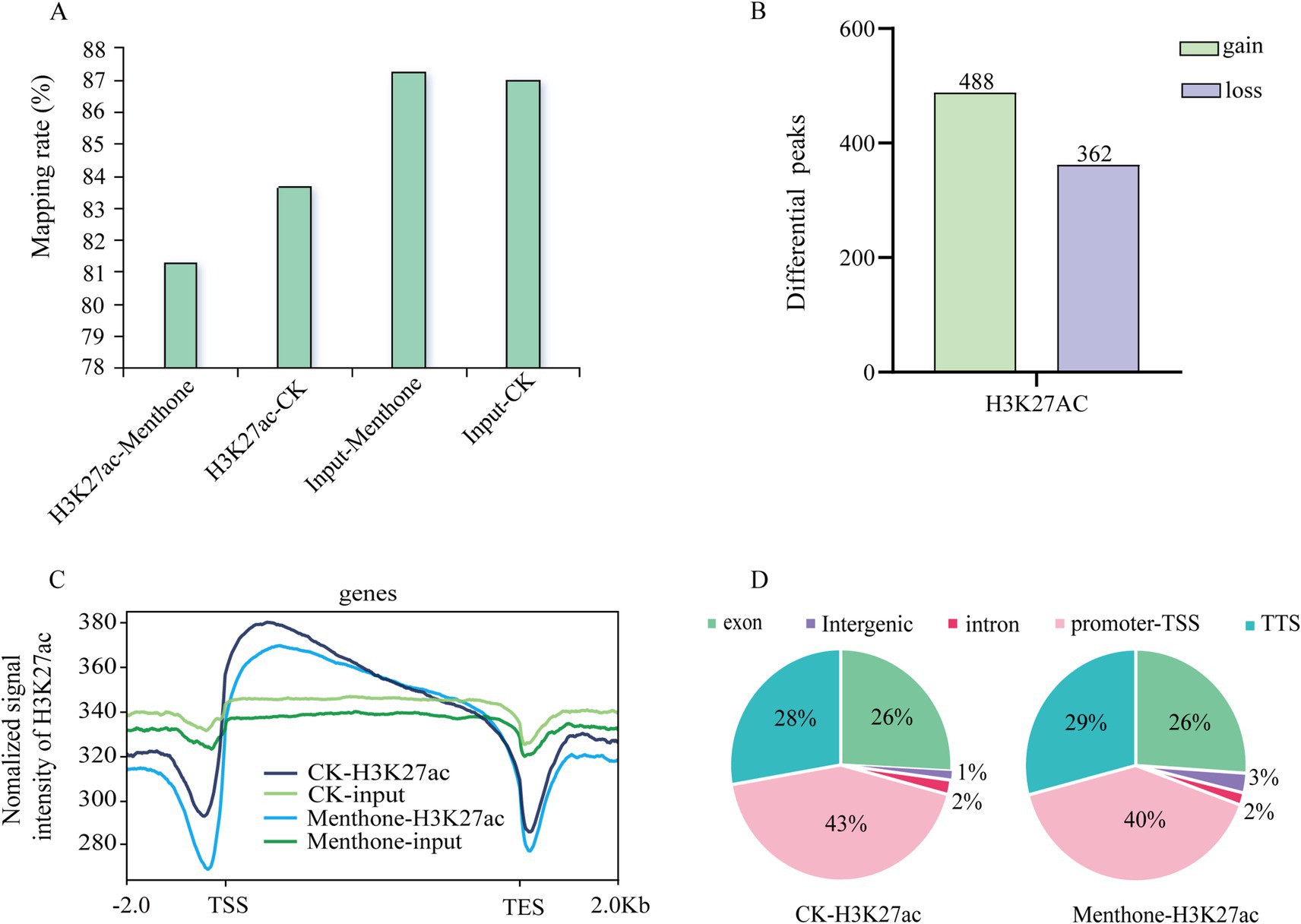
Figure 5. Preliminary analysis of ChIP-seq data. (A) Changes in H3K27ac levels in the menthone treatment group compared with the control group. (B) The number of genes associated with peaks that exhibit differences between the menthone treatment group and the control group. In comparison to the control group, genes within the menthone treatment group were classified as gain or loss genes based on the presence or absence of H3K27ac peaks. (C) Distribution of H3K27ac peaks at TSS-gene-TES across different treatment groups. TSS, transcriptional start site; TES, transcriptional end site. (D) The distribution of H3K27ac across various gene elements in different treatment groups.
Combining the results from the western blot analysis, we focused on the regulation of genes downregulated by histone modifications. Initially, we conducted GO enrichment analysis of the differentially acetylated peak genes to investigate the biological functions associated with these modified genes. Our findings revealed that these genes were predominantly enriched in tRNA modification, methylation, and processing (Figure 6A). Furthermore, KEGG enrichment analysis indicated that these genes were significantly enriched in cysteine and methionine metabolism and the spliceosome pathway (Figure 6B). These results imply that menthone treatment influences the functionality of the F. proliferatum spliceosome, resulting in disrupted gene expression, ultimately exerting antifungal effects and affecting the normal life processes of the fungus.
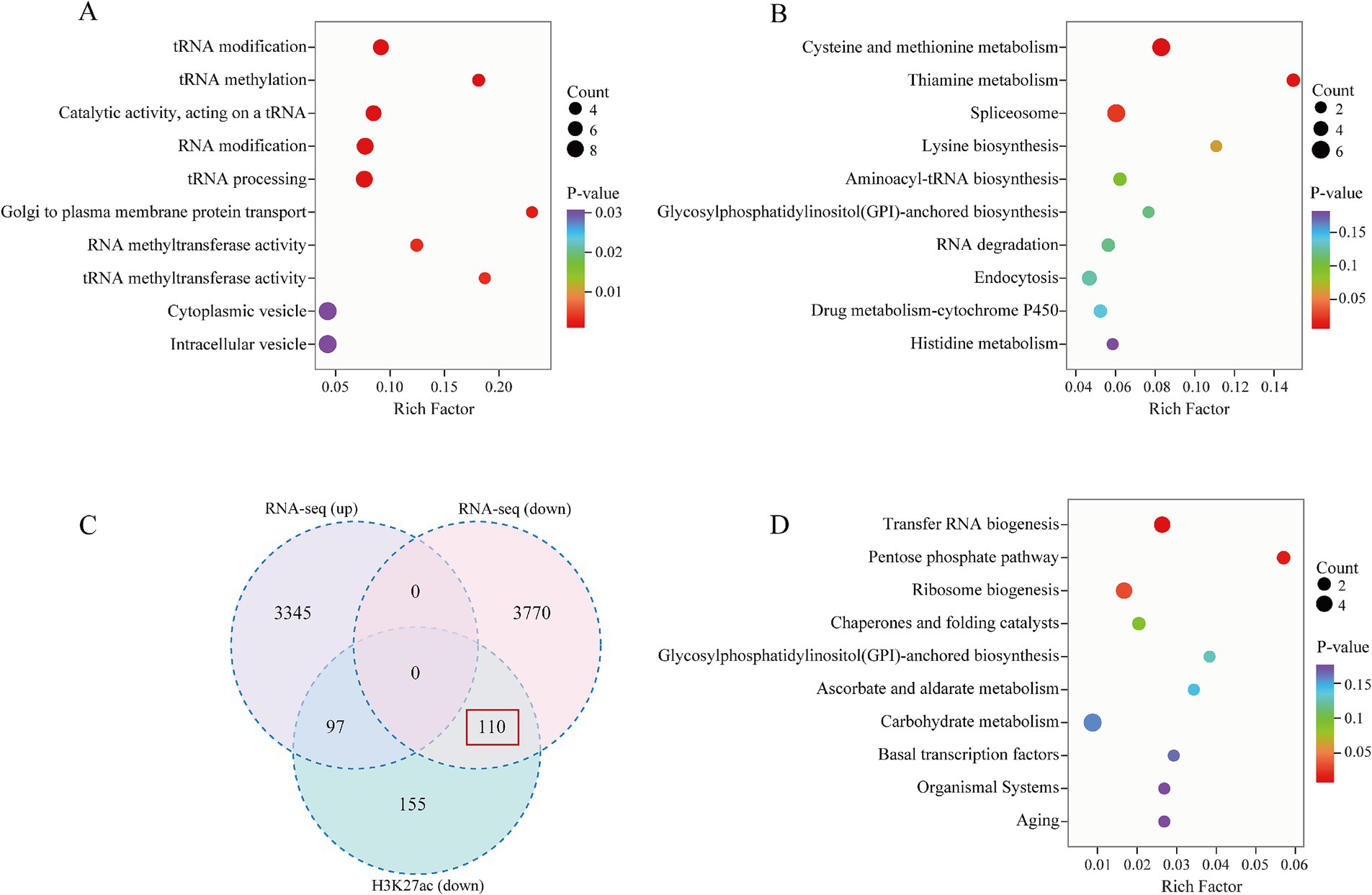
Figure 6. Analysis of genes associated with differential H3K27ac peaks following menthone treatment in F. proliferatum. (A) Gene Ontology (GO) enrichment analysis of low acetylation genes in the menthone treatment group compared with the control group. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of low acetylation genes in the menthone treatment group versus the control group. (C) Venn diagram illustrating low acetylation genes alongside transcriptionally upregulated and downregulated genes in the menthone treatment group relative to the control group. (D) KEGG enrichment analysis of low acetylation and transcriptionally downregulated genes (n = 110) within the menthone treatment cohort.
To investigate the regulatory mechanism of H3K27ac on gene transcription in F. proliferatum, we conducted a combined analysis of RNA-seq and ChIP-seq data to identify the transcripts associated with H3K27ac peaks following menthone treatment. Our comparative analysis revealed 362 low-acetylation peaks in the menthone-treated group, corresponding to 207 transcripts. Notably, the expression levels of 110 genes were significantly downregulated (Figure 6C). This finding suggests that after menthone treatment, H3K27ac primarily influences the downregulation of F. proliferatum genes, thereby inhibiting their growth and development. KEGG pathway enrichment analysis was performed to elucidate the pathways involved in the co-downregulated genes (n = 110) with low H3K27ac acetylation, indicating a significant enrichment within the ribosome biogenesis pathway (Figure 6D and Supplementary Tables S5, S6).
Several relevant genes were selected for RT-qPCR and ChIP-qPCR to validate the accuracy of the transcriptome and ChIP-seq data. The results indicated (Figure 7) that treatment with menthone significantly downregulated the expression levels of genes involved in ribosome biogenesis (FPRO_06392, FPRO_01260, and FPRO_01372), spliceosome-related genes (FPRO_01894 and FPRO_13101), and tRNA biosynthesis-related genes (FPRO_06284) in F. proliferatum compared to the control group. This finding is consistent with the transcriptome sequencing results, suggesting that the transcriptomic data are reliable.
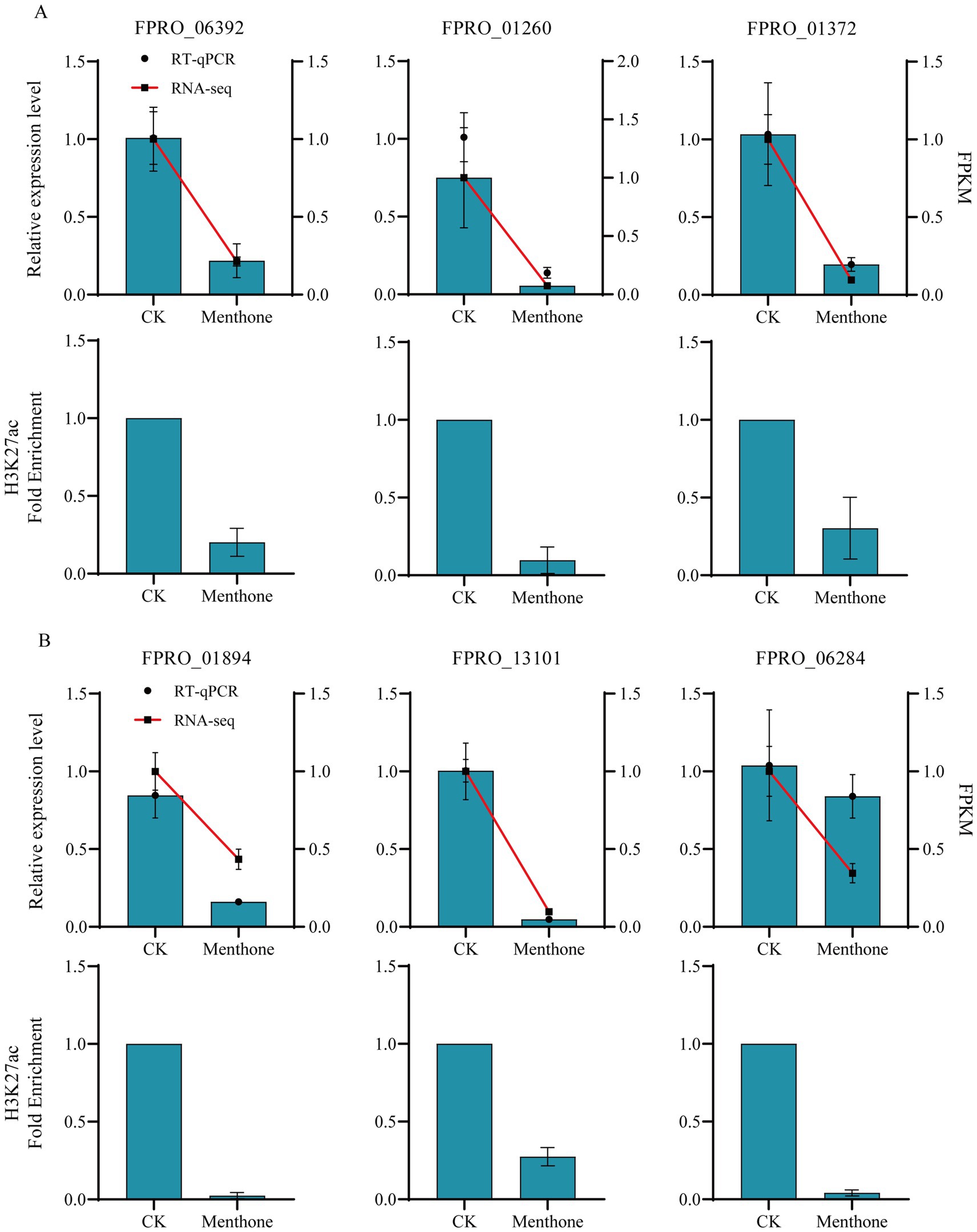
Figure 7. Validation of differential gene expression in F. proliferatum following menthol treatment assessed using RNA-seq and H3K27ac analysis. The RT-qPCR and ChIP-qPCR validation of ribosome biogenesis-related genes (A) and spliceosome and tRNA biosynthesis-related genes (B) in F. proliferatum following menthol treatment are presented.
Furthermore, after menthone treatment, there was a significant decrease in the H3K27ac modification levels of these genes relative to those in the control group. Consequently, reducing H3K27ac levels in the promoter regions of the relevant genes in F. proliferatum inhibited transcriptional activation. Our findings demonstrate that menthone treatment leads to decreased acetylation modifications at H3K27, suppressing gene transcription in F. proliferatum, subsequently affecting its growth and development.
Prolonged use of chemical pesticides poses a significant threat to human health and the environment, necessitating safe and effective new natural antimicrobial agents (Acheuk et al., 2022). Research has demonstrated that the essential oils (EOs) derived from M. haplocalyx exhibit a broad spectrum of antimicrobial properties against various pathogenic microorganisms, including F. oxysporum, F. proliferatum, and Rhizopus stolonifera (Soliman et al., 2022; Kumar et al., 2016; Yan et al., 2021). Menthone, which constitutes approximately 27.39% of the total content in M. haplocalyx EO, has strong inhibitory effects on pathogenic fungi such as Fusarium verticillioides, Aspergillus niger, and Botrytis cinerea (Felšöciová et al., 2020; Dambolena et al., 2008; Camele et al., 2021). In this study, we conducted in vitro antifungal experiments to demonstrate that menthone significantly inhibited hyphal growth, spore germination, spore viability, and yield of F. proliferatum (Figure 1). The cell membrane plays a crucial role in the maintenance of cellular integrity. When compromised, normal physiological activities are hindered, leading to the leakage of cellular contents (Andrade-Ochoa et al., 2021). In this study, menthone disrupted the integrity of the cell membrane in F. proliferatum while increasing its permeability (Figure 1), as evidenced by the elevated extracellular relative conductivity and soluble protein levels. These findings demonstrate the inhibitory function of menthone against the growth and development of F. proliferatum, which holds promise as a potential antifungal agent for controlling root rot associated with P. notoginseng.
Transcriptome sequencing has been used to elucidate the molecular mechanisms by which various organisms respond to stress at the molecular level. This approach provides insights into the DEGs involved in specific biological processes (Shi et al., 2023). In the present study, following menthone treatment, we identified 7,332 DEGs in F. proliferatum (Figures 2C,D). KEGG enrichment analysis revealed that these DEGs were significantly associated with oxidative phosphorylation, energy metabolism, lipid metabolism, ribosome biogenesis, and ribosome biogenesis in eukaryotic organisms, with a notable degree of enrichment (Figure 3). These findings indicate that menthone alters gene expression in F. graminearum, with most DEGs participating in fundamental physiological processes within the organism. In our previous study, we demonstrated that β-caryophyllene oxide, a constituent of M. haplocalyx EO, effectively reduced histone acetylation levels in F. proliferatum. This reduction subsequently inhibited ribosome biosynthesis and function, ultimately impairing the normal growth and pathogenicity of this pathogen (Liao et al., 2024).
Histone acetylation is crucial in epigenetic regulation that significantly influences various cellular processes, such as cell morphology, metabolic pathways, and protein synthesis (Yao et al., 2023; Wang et al., 2024). Dynamic regulation of histone acetylation primarily involves histone acetyltransferases (HATs) and histone deacetylases (HDACs), HATs facilitate the transfer of an acetyl group from acetyl-CoA to lysine residues on histones, whereas HDACs modulate gene expression by removing these acetyl groups from histones (Zhao et al., 2023). By modulating histone acetylation levels, HDACs can influence gene expression in critical biological processes. In the present study, menthone treatment reduced the level of H3K27 acetylation in F. oxysporum (Figure 4), inhibiting the expression of associated genes and adversely affecting normal growth. Typically, a decrease in histone acetylation correlates with increased HDAC activity. This shift results in a more compact chromatin structure, which inhibits gene transcription (Li et al., 2023). Previous studies have demonstrated that HDAC plays a vital role in the growth, development, and pathogenesis of plant pathogenic fungi (Brosch et al., 2008). In Magnaporthe oryzae, the histone deacetylase MoRpd3 plays a crucial regulatory role in fungal growth and pathogenicity. MoRpd3 overexpression in M. oryzae results in reduced hyphal production and loss of pathogenicity (Lin et al., 2021). This study showed that menthone promoted the expression of genes encoding the histone deacetylases FPRO_03415 and FPRO_13046 (Supplementary Table S7). Therefore, menthone can enhance the expression of these genes, thereby influencing histone acetylation and inhibiting the growth of F. proliferatum.
These findings suggest that menthone functions as an epigenetic modulator with the potential to inhibit fungal growth, potentially facilitating future research on HDAC activators. As documented in the existing literature, HDACs have been shown to modulate the acetylation of ribosomal proteins, thereby influencing the process of ribosome assembly (Xu et al., 2021). Furthermore, it has been demonstrated that the repression of genes involved in ribosome biogenesis contributes to the inhibition of mycelial growth and reduces the pathogenicity of the fungus Fusarium (Li et al., 2021). To elucidate the mechanism by which H3K27ac regulates gene transcription, we performed a combined analysis of RNA-seq and ChIP-seq data. This analysis revealed that 110 downregulated DEGs by low levels of H3K27ac modification (Figure 6C). Further examination indicated that most genes associated with ribosome biogenesis were downregulated, including FPRO_06392, FPRO_01260, FPRO_10795, and FPRO_01372, which encode heat shock proteins BUD20, NOC4, POP3, and HSP70, respectively. BUD20 plays a primary role in bud site selection. Research has demonstrated that this protein is involved in the epigenetic regulation of cell polarity during bipolar budding, and its reduced or absent expression leads to significant budding defects (Ni and Snyder, 2001). NOC4 belongs to the NOC family and collaborates with other NOC proteins in the processing and modification of preribosomal RNA, thereby playing a crucial role in ribosome biogenesis. The absence of NOC4 blocks ribosome synthesis, inhibiting cell growth (Milkereit et al., 2003). POP3 is a protein subunit of RNase MRP and RNase P primarily involved in pre-tRNA processing and is essential for cell viability. POP3 deletion results in cell death (Dichtl and Tollervey, 1997; Perederina et al., 2023). Ahamad et al. (2016) and Ahamad et al. (2021) demonstrated that in Schizosaccharomyces pombe, POP3 is critical for oxidative stress resistance; cells deficient in POP3 exhibit increased sensitivity to hydrogen peroxide and phenol red, adversely affecting spore formation and overall cell viability. HSP70 is a highly conserved molecular chaperone essential in fungal growth and development, secondary metabolite synthesis, and virulence. The deletion of HSP70 markedly affects hyphal growth and spore formation in Clonostachys rosea (Sun et al., 2019).
In summary, menthone regulates the expression of related genes by reducing the acetylation of H3K27ac in F. proliferatum, thereby influencing fungal growth. The decreased expression of BUD20, NOC4, POP3, and HSP70 may be crucial factors contributing to the inhibition of F. proliferatum spore germination, cell viability, hyphal growth, and spore production. Therefore, investigating histone acetylation modifications will provide novel insights into the antifungal mechanisms of menthone against the pathogen responsible for root rot in P. notoginseng. This study provides a theoretical foundation for developing targeted biological pesticides.
Our study demonstrates that menthone significantly inhibits F. proliferatum by reducing mycelial growth, spore germination rates, viability, and yield while increasing cell membrane permeability. Transcriptome sequencing and ChIP-seq analyses revealed that menthone treatment reduced H3K27 acetylation levels in F. proliferatum and inhibited the expression of genes related to ribosome biogenesis pathways, ultimately suppressing growth. Our study highlights the potential antifungal targets of menthone through histone acetylation modification, provides a theoretical basis for its use as an antimicrobial agent against root rot disease caused by P. notoginseng, and offers insights into developing targeted biopesticides.
The RNA-seq data and ChIP-seq data presented in the study are deposited in the NCBI repository, accession number PRJNA1209135 and SAMN46191626.
L-YZ: Investigation, Methodology, Software, Writing – original draft. T-TL: Data curation, Formal analysis, Methodology, Writing – original draft. H-XL: Software, Validation, Visualization, Writing – original draft. J-RW: Data curation, Formal analysis, Writing – original draft. H-YN: Writing – original draft. F-RX: Resources, Writing – original draft. X-YL: Project administration, Supervision, Writing – review & editing. XD: Funding acquisition, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (82060683), Yunnan Provincial Science and Technology Plan-Basic Research Project (202301AW070008), Wang Yuan Chao Expert Workstation in Yunnan Province (202305AF150018), Natural Science Foundation of Hubei Province (2023AFB982), and Yunnan Provincial Department of Education Scientific Research Fund Project (2024Y365).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1533918/full#supplementary-material
EOs, Essential oils; DEGs, Differentially expressed genes; PDA, Potato dextrose agar; DMSO, Dimethyl sulfoxide; SDS-PAGE, Sodium dodecyl sulfate-polyacrylamide gel electrophoresis; ChIP, Chromatin immunoprecipitation; PCA, Principal component analysis; FDR, False discovery rate; HATs, Histone acetyltransferases; HDACs, Histone deacetylases.
Acheuk, F., Basiouni, S., Shehata, A. A., Dick, K., Hajri, H., Lasram, S., et al. (2022). Status and prospects of botanical biopesticides in Europe and Mediterranean countries. Biomol. Ther. 12:311. doi: 10.3390/biom12020311
Ahamad, N., Anjum, S., and Ahmed, S. (2021). Pyrogallol induces oxidative stress defects in the fission yeast S. pombe. MicroPubl. Biol. doi: 10.17912/micropub.biology.000348
Ahamad, N., Verma, S. K., and Ahmed, S. (2016). Activation of checkpoint kinase Chk1 by reactive oxygen species resulting from disruption of wat1/pop3 in Schizosaccharomyces pombe. Genetics 204, 1397–1406. doi: 10.1534/genetics.116.193896
Alonso-Gato, M., Astray, G., Mejuto, J. C., and Simal-Gandara, J. (2021). Essential oils as antimicrobials in crop protection. Antibiotics 10:34. doi: 10.3390/antibiotics10010034
Alshannaq, A., and Yu, J. H. (2017). Occurrence, toxicity, and analysis of major mycotoxins in food. Int. J. Environ. Res. Public Health 14:841. doi: 10.3390/ijerph14060632
Andrade-Ochoa, S., Chacón-Vargas, K. F., Sánchez-Torres, L. E., Rivera-Chavira, B. E., Nogueda-Torres, B., and Nevárez-Moorillón, G. V. (2021). Differential antimicrobial effect of essential oils and their main components: insights based on the cell membrane and external structure. Membranes 11:405. doi: 10.3390/membranes11060405
Berger, S. L. (2007). The complex language of chromatin regulation during transcription. Nature 447, 407–412. doi: 10.1038/nature05915
Brosch, G., Loidl, P., and Graessle, S. (2008). Histone modifications and chromatin dynamics: a focus on filamentous fungi. FEMS Microbiol. Rev. 32, 409–439. doi: 10.1111/j.1574-6976.2007.00100.x
Camele, I., Gruľová, D., and Elshafie, H. S. (2021). Chemical composition and antimicrobial properties of Mentha × piperita cv. ‘Kristinka’ essential oil. Plants 10:1567. doi: 10.3390/plants10081567
Dambolena, J. S., López, A. G., Cánepa, M. C., Theumer, M. G., Zygadlo, J. A., and Rubinstein, H. R. (2008). Inhibitory effect of cyclic terpenes (limonene, menthol, menthone and thymol) on Fusarium verticillioides MRC 826 growth and fumonisin B1 biosynthesis. Toxicon 51, 37–44. doi: 10.1016/j.toxicon.2007.07.005
Dichtl, B., and Tollervey, D. (1997). Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO J. 16, 417–429. doi: 10.1093/emboj/16.2.417
Felšöciová, S., Vukovic, N., Jeżowski, P., and Kačániová, M. (2020). Antifungal activity of selected volatile essential oils against Penicillium sp. Open life Sci. 15, 511–521. doi: 10.1515/biol-2020-0045
Hatada, Y., Ohta, Y., and Horikoshi, K. (2006). Hyperproduction and application of alpha-agarase to enzymatic enhancement of antioxidant activity of porphyran. J. Agric. Food Chem. 54, 9895–9900. doi: 10.1021/jf0613684
Kang, J. M., Jin, W. Y., Wang, J. F., Sun, Y. Y., Wu, X. X., and Liu, L. (2019). Antibacterial and anti-biofilm activities of peppermint essential oil against Staphylococcus aureus. LWT 101, 639–645. doi: 10.1016/j.lwt.2018.11.093
Karpiński, T. M. (2020). Essential oils of Lamiaceae family plants as antifungals. Biomol. Ther. 10:168. doi: 10.3390/biom10010103
Kumar, P., Mishra, S., Kumar, A., and Sharma, A. K. (2016). Antifungal efficacy of plant essential oils against stored grain fungi of Fusarium spp. J. Food Sci. Technol. 53, 3725–3734. doi: 10.1007/s13197-016-2347-0
Li, X., Mei, Q., Yu, Q., Wang, M., He, F., Xiao, D., et al. (2023). The TORC1 activates Rpd3L complex to deacetylate Ino80 and H2A.Z and repress autophagy. Sci. Adv. 9:eade8312. doi: 10.1126/sciadv.ade8312
Li, L., Zhu, T., Song, Y., Luo, X., Datla, R., and Ren, M. (2021). Target of rapamycin controls hyphal growth and pathogenicity through FoTIP4 in Fusarium oxysporum. Mol. Plant Pathol. 22, 1239–1255. doi: 10.1111/mpp.13108
Liao, H. X., Yang, J., Wen, J. R., Nie, H. Y., Zhao, J., Xu, F. R., et al. (2024). β-caryophyllene oxide inhibits lysine acetylation of histones in Fusarium proliferatum to block ribosomal biosynthesis and function. Pestic. Biochem. Physiol. 206:106213. doi: 10.1016/j.pestbp.2024.106213
Lin, C., Cao, X., Qu, Z., Zhang, S., Naqvi, N. I., and Deng, Y. Z. (2021). The histone deacetylases MoRpd3 and MoHst4 regulate growth, conidiation, and pathogenicity in the rice blast fungus Magnaporthe oryzae. mSphere 6:e0011821. doi: 10.1128/mSphere.00118-21
Ling, C. Q., Liao, H. X., Wen, J. R., Nie, H. Y., Zhang, L. Y., Xu, F. R., et al. (2024). Investigation of the inhibitory effects of Illicium verum essential oil nanoemulsion on Fusarium proliferatum via combined transcriptomics and metabolomics analysis. Curr. Microbiol. 81:182. doi: 10.1007/s00284-024-03724-7
Liu, X. Y., Huo, Y. Y., Yang, J., Li, T. T., Xu, F. R., Wan, H. P., et al. (2022). Integrated physiological, metabolomic, and proteome analysis of Alpinia officinarum Hance essential oil inhibits the growth of Fusarium oxysporum of Panax notoginseng. Front. Microbiol. 13:1031474. doi: 10.3389/fmicb.2022.1031474
Ma, Y. N., Chen, C. J., Li, Q. Q., Xu, F. R., Cheng, Y. X., and Dong, X. (2019). Monitoring antifungal agents of Artemisia annua against Fusarium oxysporum and Fusarium solani, associated with Panax notoginseng root-rot disease. Molecules 24:213. doi: 10.3390/molecules24010213
Manzo-Valencia, M. K., Valdés-Santiago, L., Sánchez-Segura, L., and Guzmán-de-Peña, D. L. (2016). Naphthalene acetic acid potassium salt (NAA-K+) affects conidial germination, sporulation, mycelial growth, cell surface morphology, and viability of Fusarium oxysporum f. sp. radici-lycopersici and F. oxysporum f. sp. cubense in vitro. J. Agric. Food Chem. 64, 8315–8323. doi: 10.1021/acs.jafc.6b03105
Milkereit, P., Strauss, D., Bassler, J., Gadal, O., Kühn, H., Schütz, S., et al. (2003). A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 278, 4072–4081. doi: 10.1074/jbc.M208898200
Nguyen, T. T., Dehne, H. W., and Steiner, U. (2016). Histopathological assessment of the infection of maize leaves by Fusarium graminearum, F. proliferatum, and F. verticillioides. Fungal Biol. 120, 1094–1104. doi: 10.1016/j.funbio.2016.05.013
Ni, L., and Snyder, M. (2001). A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 12, 2147–2170. doi: 10.1091/mbc.12.7.2147
Nie, H., Liao, H., Wen, J., Ling, C., Zhang, L., Xu, F., et al. (2024). Foeniculum vulgare essential oil nanoemulsion inhibits Fusarium oxysporum causing Panax notoginseng root-rot disease. J. Ginseng Res. 48, 236–244. doi: 10.1016/j.jgr.2023.12.002
Ning, K., Li, M., Wei, G., Zhou, Y., Zhang, G., Huai, H., et al. (2021). Genomic and transcriptomic analysis provide insights into root rot resistance in Panax notoginseng. Front. Plant Sci. 12:775019. doi: 10.3389/fpls.2021.775019
Panth, M., Hassler, S. C., and Baysal-Gurel, F. (2020). Methods for management of soilborne diseases in crop production. Agriculture 10:16. doi: 10.3390/agriculture10010016
Papait, R., Cattaneo, P., Kunderfranco, P., Greco, C., Carullo, P., Guffanti, A., et al. (2013). Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc. Natl. Acad. Sci. U.S.A. 110, 20164–20169. doi: 10.1073/pnas.1315155110
Perederina, A., Berezin, I., and Krasilnikov, A. S. (2023). Proteins Rpr2 and Pop3 increase the activity and thermal stability of yeast RNase P. RNA Biol. 20, 149–153. doi: 10.1080/15476286.2023.2201110
Pérez-Vázquez, M. A. K., Pacheco-Hernández, Y., Lozoya-Gloria, E., Mosso-González, C., Ramírez-García, S. A., Romero-Arenas, O., et al. (2022). Peppermint essential oil and its major volatiles as protective agents against soft rot caused by Fusarium sambucinum in cera pepper (Capsicum pubescens). Chem. Biodivers. 19:e202100835. doi: 10.1002/cbdv.202100835
Perrella, G., Fasano, C., Donald, N. A., Daddiego, L., Fang, W., Martignago, D., et al. (2024). Histone deacetylase complex 1 and histone 1 epigenetically moderate stress responsiveness of Arabidopsis thaliana seedlings. New Phytol. 241, 166–179. doi: 10.1111/nph.19165
Piasecki, B., Korona-Głowniak, I., Kiełtyka-Dadasiewicz, A., and Ludwiczuk, A. (2023). Composition and anti-Helicobacter pylori properties of essential oils obtained from selected Mentha cultivars. Molecules 28:5690. doi: 10.3390/molecules28155690
Shi, J., Cui, Y., Zhang, J., Sun, L., and Tang, X. (2023). Transcriptomics reveals the molecular basis for methyl jasmonate to promote the synthesis of monoterpenoids in Schizonepeta tenuifolia Briq. Curr. Issues Mol. Biol. 45, 2738–2756. doi: 10.3390/cimb45040179
Soliman, S. A., Hafez, E. E., Al-Kolaibe, A. M. G., Abdel Razik, E. S., Abd-Ellatif, S., Ibrahim, A. A., et al. (2022). Biochemical characterization, antifungal activity, and relative gene expression of two Mentha essential oils controlling Fusarium oxysporum, the causal agent of Lycopersicon esculentum root rot. Plants 11:189. doi: 10.3390/plants11020189
Sun, W., Lei, T., Yuan, H., and Chen, S. (2022). Occurrence of root rot caused by Fusarium fujikuroi and Fusarium proliferatum on peanut in China. Plant Dis. 107:940. doi: 10.1094/PDIS-02-22-0438-PDN
Sun, Z. B., Wang, Q., Sun, M. H., and Li, S. D. (2019). The heat shock protein 70 gene is involved for colony morphology, sporulation and mycoparasitism of Clonostachys rosea. FEMS Microbiol. Lett. 366:FNZ188. doi: 10.1093/femsle/fnz188
Tanapichatsakul, C., Khruengsai, S., and Pripdeevech, P. (2020). In vitro and in vivo antifungal activity of Cuminum cyminum essential oil against aspergillus aculeatus causing bunch rot of postharvest grapes. PLoS One 15:e0242862. doi: 10.1371/journal.pone.0242862
Tang, H. P., Zhu, E. L., Bai, Q. X., Wang, S., Wang, Z. B., Wang, M., et al. (2024). Mentha haplocalyx Briq. (Mint): a comprehensive review on the botany, traditional uses, nutritional value, phytochemistry, health benefits, and applications. Chin. Med. 19:168. doi: 10.1186/s13020-024-01037-2
Wang, B., Duan, G., Liu, L., Long, Z., Bai, X., Ou, M., et al. (2024). UvHOS3-mediated histone deacetylation is essential for virulence and negatively regulates ustilaginoidin biosynthesis in Ustilaginoidea virens. Mol. Plant Pathol. 25:e13429. doi: 10.1111/mpp.13429
Wang, L., Ge, S. L., Zhao, K., and Shiwen, H. (2021). First report of Fusarium incarnatum causing spikelet rot on rice in China. Plant Dis. 105:3306. doi: 10.1094/PDIS-12-20-2660-PDN
Xu, Q., Liu, Q., Chen, Z., Yue, Y., Liu, Y., Zhao, Y., et al. (2021). Histone deacetylases control lysine acetylation of ribosomal proteins in rice. Nucleic Acids Res. 49, 4613–4628. doi: 10.1093/nar/gkab244
Xu, C., Wang, W., Wang, B., Zhang, T., Cui, X., Pu, Y., et al. (2019). Analytical methods and biological activities of Panax notoginseng saponins: recent trends. J. Ethnopharmacol. 236, 443–465. doi: 10.1016/j.jep.2019.02.035
Yan, J., Wu, H., Shi, F., Wang, H., Chen, K., Feng, J., et al. (2021). Antifungal activity screening for mint and thyme essential oils against Rhizopus stolonifer and their application in postharvest preservation of strawberry and peach fruits. J. Appl. Microbiol. 130, 1993–2007. doi: 10.1111/jam.14932
Yao, G., Han, N., Zheng, H., and Wang, L. (2023). The histone deacetylase HstD regulates fungal growth, development and secondary metabolite biosynthesis in Aspergillus terreus. Int. J. Mol. Sci. 24:12569. doi: 10.3390/ijms241612569
Ye, Q., Wang, R., Ruan, M., Yao, Z., Cheng, Y., Wan, H., et al. (2020). Genetic diversity and identification of wilt and root rot pathogens of tomato in China. Plant Dis. 104, 1715–1724. doi: 10.1094/PDIS-09-19-1873-RE
Zeng, Z. Y., Li, Q. Q., Huo, Y. Y., Chen, C. J., Duan, S. S., Xu, F. R., et al. (2021). Inhibitory effects of essential oils from Asteraceae plant against pathogenic fungi of Panax notoginseng. J. Appl. Microbiol. 130, 592–603. doi: 10.1111/jam.14606
Zhang, W., Huang, J., and Cook, D. E. (2021). Histone modification dynamics at H3K27 are associated with altered transcription of in planta induced genes in Magnaporthe oryzae. PLoS Genet. 17:e1009376. doi: 10.1371/journal.pgen.1009376
Zhao, H., Ge, Z., Zhou, M., Zeng, H., Wei, Y., Liu, G., et al. (2023). Histone deacetylase 9 regulates disease resistance through fine-tuning histone deacetylation of melatonin biosynthetic genes and melatonin accumulation in cassava. J. Pineal Res. 74:e12861. doi: 10.1111/jpi.12861
Zhao, W., Yang, C., Zhang, N., Peng, Y., Ma, Y., Gu, K., et al. (2023). Menthone exerts its antimicrobial activity against methicillin resistant Staphylococcus aureus by affecting cell membrane properties and lipid profile. Drug Des. Devel. Ther. 17, 219–236. doi: 10.2147/DDDT.S384716
Zheng, Z., Gao, T., Zhang, Y., Hou, Y., Wang, J., and Zhou, M. (2014). FgFim, a key protein regulating resistance to the fungicide JS399-19, asexual and sexual development, stress responses and virulence in Fusarium graminearum. Mol. Plant Pathol. 15, 488–499. doi: 10.1111/mpp.12108
Zheng, L., Qiu, B., Su, L., Wang, H., Cui, X., Ge, F., et al. (2022). Panax notoginseng WRKY transcription factor 9 is a positive regulator in responding to root rot pathogen Fusarium solani. Front. Plant Sci. 13:930644. doi: 10.3389/fpls.2022.930644
Zhou, K., Chen, D., Li, B., Zhang, B., Miao, F., and Zhou, L. (2017). Bioactivity and structure-activity relationship of cinnamic acid esters and their derivatives as potential antifungal agents for plant protection. PLoS One 12:e0176189. doi: 10.1371/journal.pone.0176189
Zhou, H., Yuan, Z., Han, S., He, H., Rong, J., Guo, D., et al. (2023). Global decrease in H3K9 acetylation in sorghum seed postgermination stages. J. Agric. Food Chem. 71, 5836–5850. doi: 10.1021/acs.jafc.2c08863
Keywords: antifungal activity, ribosomes, histone deacetylases, root rot disease, Panax notoginseng
Citation: Zhang L-Y, Li T-T, Liao H-X, Wen J-R, Nie H-Y, Xu F-R, Liu X-Y and Dong X (2025) Menthone lowers H3K27ac levels to inhibit Fusarium proliferatum growth. Front. Microbiol. 16:1533918. doi: 10.3389/fmicb.2025.1533918
Received: 25 November 2024; Accepted: 03 January 2025;
Published: 22 January 2025.
Edited by:
Minu Chaudhuri, Meharry Medical College, United StatesReviewed by:
Wenxing Liang, Qingdao Agricultural University, ChinaCopyright © 2025 Zhang, Li, Liao, Wen, Nie, Xu, Liu and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Yun Liu, bGl1eGlhb3l1bkBqaHVuLmVkdS5jbg==; Xian Dong, ZG9uZ3hpYW5fMTY1NTEyOUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.