
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 11 February 2025
Sec. Microbiological Chemistry and Geomicrobiology
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1522265
This article is part of the Research TopicExploring Processes and Applications of Metal-Microbe InteractionsView all 11 articles
Cutting fluid is a type of fluid used in the metal-cutting process. It is prone to microbial growth during use, which can lead to the deterioration of its various useful properties; however, the mechanism underlying this deterioration remains unclear. This study analyzed the microbial diversity of field-sampled cutting fluids, and those with higher levels of diversity were used to inoculate other fluid samples in order to further study the effects of microbial growth on the properties of cutting fluids. The results show that the surface of cutting fluid sampled from the tank of a machining tool tank contained predominantly aerobic bacteria, while the bottom mainly harbored anaerobic and facultative microorganisms, with Yarrowia lipolytica representing the dominant fungus. Some obligate anaerobic bacteria were also present in the cutting fluid. Organic acids secreted by anaerobic microbial activity reduced the pH of the cutting fluid, as well as its resistance to corrosion. The metabolic activity of the aerobic microorganisms also consumed certain key components of the cutting fluid, which ultimately further lowered its pH and resistance to corrosion. Moreover, the number of fungi increased significantly during the later stages of the experiment. The rolling and bridging action of the resulting fungal mycelium caused flocculation of the effective components in the cutting fluid, resulting in reduced lubricity and poor stability. This study provides a theoretical basis for developing more effective measures to inhibit microbial growth and delay the deterioration of cutting fluid, thereby helping to improve the technical quality of the metal-cutting industry.
Cutting fluid (i.e., coolant) is a type of industrial liquid used for cooling, lubricating, cleaning, and preventing corrosion during the metal-cutting process. It comprises and is synthesized using a variety of functional chemical additives (Osama et al., 2017; Choudhury and Muaz, 2020; Chen et al., 2024). In recent years, as cutting speeds have increased in this industry, greater requirements are being placed concerning the safety and cooling properties of cutting fluid, leading to the gradual replacement of traditional oil-based cutting fluids with water-based ones. These water-based cutting fluids mainly contain water and mineral oil, along with anti-corrosion agents, defoamers, lubricants, and other additives with different chemical structures that ensure their performance and stability (Simpson et al., 2000; Lillienberg et al., 2008; Lillienberg et al., 2010). These additives are organic, and thus can represent nutritional sources for certain microorganisms (Trafny, 2013).
Microorganisms can enter cutting fluids in various ways. For example, tap water—which is often used to dilute concentrated cutting fluid—can represent a significant source of microbial contamination. Residues from used cutting fluids often remain even after pipelines or the dead spaces of machines have been cleaned, thus representing another source of microbial contamination when new fluids are added to such cutting systems. Microorganisms also enter cutting fluids through the equipment, or be inoculated by the equipment operators. Fresh cutting fluids are also susceptible to contamination by microorganisms from dust particles or aerosols. Such microorganisms not only threaten the health of workers in the industry, but also lead to spoiling of the cutting fluid, which can produce foul odors and affect its physicochemical properties (Liu et al., 2010; Schwarz et al., 2015). This, in turn, can lead to deterioration of the quality of the workpiece and reduce the lifespan of the tool (Rakić and Rakić, 2002).
Studies have shown that microorganisms are highly capable of growth in water-based cutting fluids from machining plants, leading to environmental malodor, fluid discoloration, and increased acidity (Gast et al., 2001; Grijalbo et al., 2015). After isolating fungi and bacteria from cutting fluids, Elansky et al. investigated their growth in cutting fluid, and hypothesized that microorganisms could be used to treat metal-working fluid waste through biodegradation (Elansky et al., 2022). Hill found that microbial growth consumes certain components of cutting fluid and increases the diameter of the droplets formed by its dispersion, which in turn leads to decreased performance that affects the machining accuracy of workpiece surfaces (Hill, 1992). Rabenstein et al. studied the microbial community in water-based metal-working fluids and the microbial degradation of their active ingredients, ultimately demonstrating that the active ingredients were not consumed simultaneously, but rather in a sequential order (Rabenstein et al., 2009). Zhang et al. investigated the effect of corrosion caused by microorganisms on cemented carbide (WC-30Co) cutting tools in an oil–water emulsion (Zhang et al., 2015). They found that preferential cobalt loss determined the corrosion failure in biodegraded emulsions, and that the sulfate-reducing bacterium Citrobacter sp. made the emulsion even more corrosive to the hard metal. To date, the safety hazards and economic losses caused by microorganisms have attracted significant attention in the fields of marine engineering, pipeline transportation, construction engineering, and nuclear power (Yazdi et al., 2022; Lu et al., 2023; Sun et al., 2023; Li et al., 2024). Conversely, microbial growth in cutting fluid, and the related deterioration of its performance, as well as the environmental pollution caused by the resultant waste (Sankaranarayanan et al., 2021; Korkmaz et al., 2023), has not yet attracted sufficient attention. This has resulted in a scarcity of research concerning the underlying mechanisms of its degradation.
The main objectives of this study were to: (1) examine the diversity and characteristics of the microbial communities in industrial cutting fluid; (2) study the effects of microbial growth and metabolism on the performance of industrial cutting fluid, based on parameters such as pH, anticorrosion, lubrication, and stability, under laboratory conditions; and (3) analyze the mechanism underlying the microbially-induced deterioration of industrial cutting fluid. The theoretical data provided herein will hopefully be of great significance to preventing microbial corrosion in industrial cutting fluid, as well as for maintaining and extending the life of cutting fluids used in metal-processing operations.
Cutting fluid samples (labeled “CF”), were collected from the surface and bottom of four equally-sized machine tool tanks (labeled 1, 2, 3, and 4), that had been filled with the same cutting fluid, used to cut the same metal, and received equal use at a research institute for aerospace materials in Shanghai, China. All of the cutting fluids had been in use for 4 months prior to the study, during which they had blackened and begun emitting a foul odor. Samples (50 mL) of each fluid were centrifuged in a high-speed refrigerated centrifuge (Multifuge X1R, Thermo Fisher Scientific, Waltham, MA, United States) at 44,720 × g and 4°C for 10 min. The supernatants were discarded, and the pellets were resuspended and diluted in physiological buffered saline (PBS), after which the resultant solutions were centrifuged again under the same conditions. The final pellets were then analyzed for biodiversity via high-throughput sequencing.
Microbial DNA from the cutting fluid samples was extracted using a FastDNA Spin Kit for Soil (MP Biomedicals LLC, Irvine, SC, United States), after which the concentration and purity of the extracted DNA were measured using a NanoDrop 2000 fluorometer (Thermo Fisher Scientific, Waltham, MA, United States).
The V3–V4 hypervariable region of the bacterial 16S rRNA gene was amplified using the universal primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′). The fungal ITS2 region was amplified using the universal primer sets ITS1 (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 (5′-TGCGTTCTTCATCGATGC-3′) (Abarenkov et al., 2010). Both the 16S rRNA and ITS rRNA amplicons were generated using the same protocol. The PCR mixtures comprised 5× buffer (10 μL), forward primer (1 μL), reverse primer (1 μL), deoxy-ribonucleoside triphosphates (dNTPs) (10 μL), double-distilled water (ddH2O), Phusion high-fidelity DNA polymerase (0.5 μL, New England Biolabs, Inc., Ipswich, MA, United States), and template DNA (30 ng), with total reaction volumes of 50 μL. The cycling parameters were: 94°C for 2 min; followed by 24 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s; before a final extension at 72°C for 5 min (Rognes et al., 2016). The generated PCR products were recovered via 2% agarose gel electrophoresis before being purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Silicon Valley, CA, United States). A Quantus™ fluorometer (Promega Corporation, Madison, WI, United States) was used for nucleic acid quantification of purified PCR products. Sequencing was then performed on an Illumina MiSeq PE3000 platform (Illumina, Inc., San Diego, CA, United States) at Shanghai Meiji Biomedical Technology Co., Ltd., China (Zhang et al., 2014). The data presented in the study are deposited in the National Center for Biotechnology Information (NCBI) repository, accession number SRP554559: PRJNA1204126.
The raw fastq files were demultiplexed based on the barcode. PE reads for all samples were run through Trimmomatic (version 0.35) to remove low quality base pairs using these parameters (SLIDINGWINDOW: 50:20; MINLEN: 50) (Pruesse et al., 2007). Trimmed reads were then cut primer and adaptors using cutadapt (version:1.16). And then further merged using FLASH program (version 1.2.11) with default parameters. The low quality contigs were removed based on screen.seqs command using the following filtering parameters, maxambig = 0, minlength = 200, maxlength =485, maxhomop = 8.
The 16S sequences were analyzed using a combination of software mothur (version 1.33.3),UPARSE (version v8.1.1756), and R (version 3.6.3). The demultiplexed reads were clustered at 97% sequence identity into operational taxonomic units (OTUs) and the chimera was removed and the singleton OTUs were deleted using the UPARSE pipeline (Edgar et al., 2011; Edgar, 2013). The OTU representative sequences were assignment for taxonomy against Silva 128 database with confidence score ≥ 0.6 by the classify.seqs command in mothur. As for the ITS taxonomic analysis, the UNITE database was used.
For the alpha-diversity analysis, Shannon, simpson, Chao1, ACE index and rarefaction curves were calculated and plotted by R. For the beta-diversity metrics, the weighted and unweighted UniFrac distance matrix were calculated and visualized with Principal Coordinate Analysis (PCoA) by ape package in R, NMDS by vegan package in R and tree by dendextend package in R (Harmon-Smith et al., 2010). The bray curits and jaccard metrics were calculated by vegan package in R and visualized also by R the same as Unifrac analysis.
In order to better analyze the effects of microorganisms on cutting fluid performance, the cutting fluid formulation was simplified and its fungicide was removed for this experiment. The fluid’s composition therefore comprised: 4.9% nonylphenol polyoxyethylene ether, 7.8% triethanolamine, 9.6% fatty alcohol polyoxyethylene ether, 9.6% dehydrated sorbitol fatty acid ester (i.e., Span), 9.6% polyethylene glycol dioleate, 50.3% mineral oil, 6% tall oil, 0.2% organic silicon, and 2% organic phosphate ester (all proportions in w/w). Before the formal experiments, the simplified cutting fluid was diluted to a volume fraction of 5% wt (the concentration recommended by the supplier) with deionized water, and dubbed “SCF.” This fluid contained sufficient organic components to supply ample nutrients for microbial growth, so it was not further supplemented with any additional nutrients. The total volume of this solution was 1 L.
To ensure high microbial diversity, CF samples with higher levels of biodiversity were selected as sources of the microbial strains. Appropriate amounts of spoiled CF were inoculated into 100 mL aliquots of sterilized SCF solution, to initial bacterial concentrations of ~106 CFU/mL. Since the cutting fluid was milky-white, it was difficult to accurately determine its microbial concentration using the absorbance method. Therefore, to plot the growth curve of the microorganisms in the fluid, microbial concentrations at different incubation times were quantified via the plate-count method. Three replicate solutions were prepared to increase the experiment’s accuracy. The main components of the solid medium used in the plates were: beef extract (5 g/L), peptone (5 g/L), triethanolamine borate and its derivatives (100 g/L), oleic acid (5 g/L), methanol (3 g/L), organophosphate ester and its derivatives (3 g/L), and agar (15 g/L) (Yin et al., 2019). The bacterial solution was appropriately diluted via step-wise dilution. Aliquots of the diluted microbial suspension (1 mL) were taken using sterile pipettes and inoculated onto the solid medium in Petri dishes, before being evenly spread using a sterilized glass scraper. The plates were then left face-up on the sterile experimental benchtop for 30 min to allow the solution to penetrate the medium, before being inverted for culturing in a controlled-temperature incubator at 37°C. The plates were removed at different time points, and Scan 300 colony-counters (Interscience, Puy Capel, Cantal Province, France) were used to count the resultant colonies.
The SCF solution was packed in two 1 L conical flasks that were plugged with cotton plugs, sterilized in an autoclave at 121°C for 20 min, then air-cooled in a biosafety cabinet. One of the conical flasks was inoculated with 200 μL of a CF sample with high microbial diversity, which was dubbed “Test-SCF.” The other conical flask was not inoculated, to serve as a control, and dubbed “Control-SCF.” These two conical bottles were incubated at 38°C for static culturing. After 1 month, samples from the middle regions of the solutions within the flasks were taken so that the compositions of their bacterial and fungal communities could be analyzed.
To study the effect of microbial growth on pH, he pH levels of the two SCF solutions were measured at 7, 14, 21, and 28 days using an FE20 pH meter (Mettler-Toledo International Inc., Zurich, Switzerland). The redox potential (ORP) of two solutions was measured at various stages throughout the experiment using an SX712 ORP meter (Shanghai San-Xin Instrumentation, Inc., Shanghai, China).
The corrosion inhibition ability of the cutting fluid was evaluated using the iron filings method. A section of qualitative filter paper was placed into a Petri dish, after which 2 g of iron filings were uniformly spread over its surface, with the weight confirmed using a precision balance. A 1 mL sample of SCF was then added to saturate the iron filings, and the dish was sealed using its lid. After incubating at room temperature for 2 h, the filter paper was removed and the iron filings were washed with running water. The number of rust spots was then counted, once the filter paper had dried completely.
Given the use of both onsite-sampled CF and the laboratory-prepared SCF, both needed to be analyzed regarding the impact of microbial contamination on their abilities to inhibit corrosion. Electrochemical analyses, including measurements of open circuit potential (OCP) and polarization curves, were conducted using samples of 7,050 aluminum alloy (Shanghai Hanglv Aviation Materials Co., Ltd., Shanghai, China) that were exposed to spoiled cutting fluids for varying periods. The aluminum alloy, sourced from an aerospace materials research institute in Shanghai, comprised 0.04% Cr, 0.12% Zr, 6.2% Zn, 0.11% Si, 0.08% Fe, 0.06% Mn, 2.3% Mg, 0.05% Ti, and 0.04% Cu, with the remaining composition being aluminum. Prior to testing, the alloy was sectioned into 10 × 10 × 2 mm samples that were sequentially polished under wet conditions with 800 grit silicon carbide paper, ultrasonically cleaned in anhydrous ethanol for 10 min, and air-dried. Lengths of copper wire were then soldered to the reverse sides of each sample. The soldered faces and remaining samples were encapsulated in epoxy resin, leaving only 10 × 10 mm working surfaces exposed on each. These working surfaces were further polished using 800 grit water-resistant sandpaper, sequentially cleaned with anhydrous ethanol and deionized water, and dried before being used for electrochemical evaluation.
Electrochemical evaluation was performed using a CHI 660 electrochemical workstation (Shanghai Chenhua Instrument Co., Ltd., Shanghai, China). The experimental configuration used a three-electrode system comprising a saturated calomel electrode (SCE) as the reference, a platinum plate (10 × 10 mm) as the counter electrode, and aluminum alloy samples immersed in experimental cutting fluid samples as the working electrodes. The sterilized SCF comprised the electrolyte used to assess the electrochemical behavior of the working electrode. After being immersed for varying lengths of time, samples were extracted from the conical flasks and tested in an electrochemical testing system. To ensure the system’s stability prior to the measurements, all of the samples were equilibrated at OCP for 30 min in a sterile environment at ambient temperature.
Polarization curve tests were carried out at a scan rate of 5 mV/s and range of −1 to +1 V relative to the stabilized OCP. The resultant polarization curve data were analyzed and fitted using the electrochemical workstation’s integrated analysis software.
Lubricity is commonly quantified by measuring tapping torque, with higher values signifying better lubricity. As the experiment progressed, the effects of microbial activity on the tapping torque values of the cutting fluid samples were quantified using a Labtap G8 threading torque meter (Microtap, Taufkirchen, BY, Germany). These measurements used aluminum alloy 7,075 for the standard extruded-hole test block, with a hole diameter and depth of 3.7 mm and 20 mm, respectively. A syringe was used to inject the solutions into the holes until they were fully filled. The tap used for extrusion molding had a diameter of 3.64 mm. The tests were conducted at a set rotational speed of 1,500 rpm, an applied torque of 400 N·cm, and a tapping depth of 12 cm.
The stability of industrial cutting fluid is primarily assessed via two methods: oil droplet morphology analysis and zeta potential measurement. In general, smaller molecules or dispersed particles within the fluid correspond to greater absolute zeta potential values (positive or negative), signifying higher stability.
The methodology used to assess the oil droplet morphology in the cutting fluid samples involved staining with Oil Red O. A 1 g aliquot of Oil Red O was dissolved in 2 mL of anhydrous ethanol and mixed thoroughly to prepare the dye solution. A 100 μL aliquot of this dye was then added to 1 mL of the fluid being analyzed, followed by vigorous shaking to achieve uniform staining. The morphologies of the resultant oil droplets were then examined using a DM500 optical microscope (Leica, Wetzlar, HE, Germany).
Zeta potential was measured using a Litesizer 500 nanoscale particle analyzer (Anton Paar GmbH, Graz, ST, Austria) at various time points over the course of the experiment. Measurements were performed in an omega-shaped sample cell maintained at a target temperature of 25°C, with an equilibration time of 1 min. The Smoluchowski approximation was used, with Henry’s function set at 1.5. The adjustment mode was automatic, the running mode was manual (100 runs), and water was used as the solvent.
To examine the microbial degradation of the cutting fluid, 100 mg samples of three of its primary active ingredients—triethanolamine, tall oil, and organophosphate esters—were added separately to 150 mL aliquots of mineral medium without additional carbon sources. Nine such replicate samples were prepared for each ingredient. The composition of the mineral medium was 1.0 g/L K2HPO4, 1.0 g/L NaNO3, 0.5 g/L MgCl2 and 1.0 g/L MgSO4·7H2O in deionized water. The pH was adjusted to 8.5–9.0 using a 0.1 mol/L NaOH solution to mimic that of the cutting fluid used on-site. Samples of spoiled and sterile cutting fluid samples (2 mL each) were then inoculated into the test media and incubated at 25°C. The total organic carbon levels of each of these mixtures were tested after 0, 5, and 9 days of incubation, with three 10 mL samples tested from each mixture on a Multi N/C 3100 total organic carbon analyzer (Analytik Jena GmbH+Co., Jena, JN, Germany). The total volume was maintained at 150 mL with mineral medium. This approach facilitated precise quantification of which cutting fluid components were being actively metabolized by microorganisms.
Genus-level microbial community analysis revealed significant differences in the composition and relative abundances of the microorganisms retrieved from the surfaces and bottoms of the four machine tool tanks (Figure 1). The biodiversity was high in the B1 and B3 samples, and the relative abundances of the different microorganisms were relatively consistent. The microbial communities in the surface samples from the tanks of machines 1 and 4 were similar, being predominantly composed of Pseudomonas. Similarly, the microbial communities in the surface samples from the surface samples of machines 2 and 3 were also consistent, being predominantly composed of Pseudomonas, Acinetobacter, and Ochrobactrum. The bottom samples from all four machines mainly contained Brachymonas, Sebaldella, Corynebacterium, and Comamonas. Notably, the microbial communities in the surface samples had a high relative abundance of Pseudomonas, with percentages of 90.7, 57.5, 55.4, and 95.6% in samples 1–4, respectively. Pseudomonas is a facultative Gram-negative bacterium that thrives in aerobic conditions and can metabolize various carbon-, nitrogen-, and sulfur-containing compounds (Daniels et al., 2010; Babaei et al., 2014), thus facilitating the full degradation of various organic components (Caballero et al., 2005; Roca et al., 2009) and often leading to the deterioration of cutting fluid. The effective components in the cutting fluid were reduced by evaporation and splashing during the actual operational use of the cutting fluids. Microbial degradation represents a major factor that reduces the functional ingredients in cutting fluid (Rabenstein et al., 2009). Furthermore, the lower oxygen level at the bottom of the machine tool tank provides a favorable environment for the proliferation of anaerobic and facultative bacteria.
The Shannon index is used to measure the biodiversity of species. The higher the Shannon value is, the higher the microbial diversity will be (Lopes et al., 2024). The α-diversity of microbial communities in the four machine tool tanks were shown in Figure 2. The results showed that samples located bottom presented higher diversity and species richness in comparison with the samples located surface. The surface of the cutting fluid is usually in contact with air, and the oxygen content is relatively high. This oxygen-rich environment is conducive to the growth of aerobic microorganisms. For example, some bacteria of the genus Pseudomonas can utilize oxygen for respiration and decompose the organic components in the cutting fluid. However, at the bottom of the cutting fluid, oxygen diffuses relatively slowly, and it is easy to form an anoxic or even anaerobic environment. This provides living space for anaerobic microorganisms. These anaerobic microorganisms have unique metabolic pathways. As a result, aerobic, anaerobic and facultative anaerobic microorganisms coexist at the bottom, which increases the biodiversity. Based on the above results, it could be seen that sample B3 had a relatively high biodiversity and the relative abundances of each microorganism were relatively even. Therefore, sample B3 would be used as the source of strains and inoculated into the cutting fluid for experiments in the follow-up.
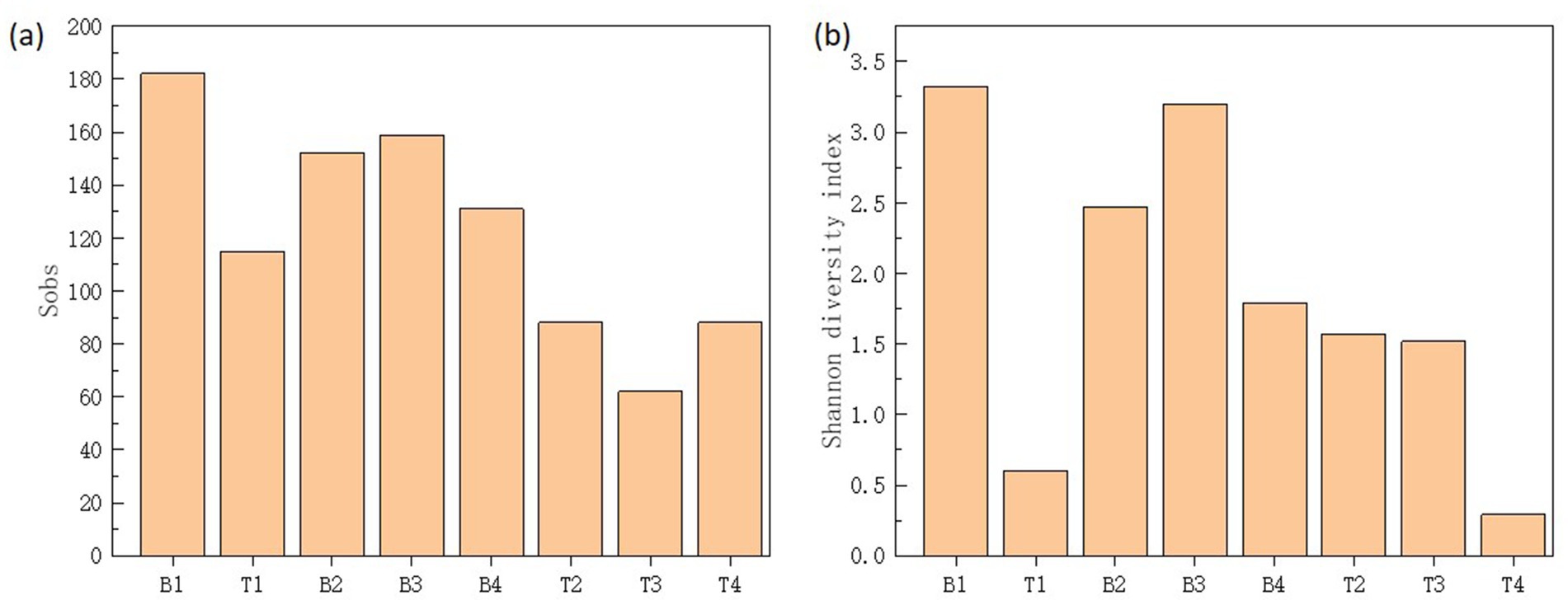
Figure 2. Sobs (a) and Shannon diversity index (b) of microbial communities in top (T1-4) and bottom (B1-4) cutting fluid samples taken from the four (1-4) machine tool tanks.
The microbial community structure in the cutting fluid from the Test-SCF sample is shown in Figure 3. The microbial communities were predominantly composed of Acinetobacter, Comamonas, and Pseudomonas at percentages of 60.0, 18.0, and 2.7%, respectively in Sample A; and 49.9, 6.9, and 9.0%, respectively, in Sample B.
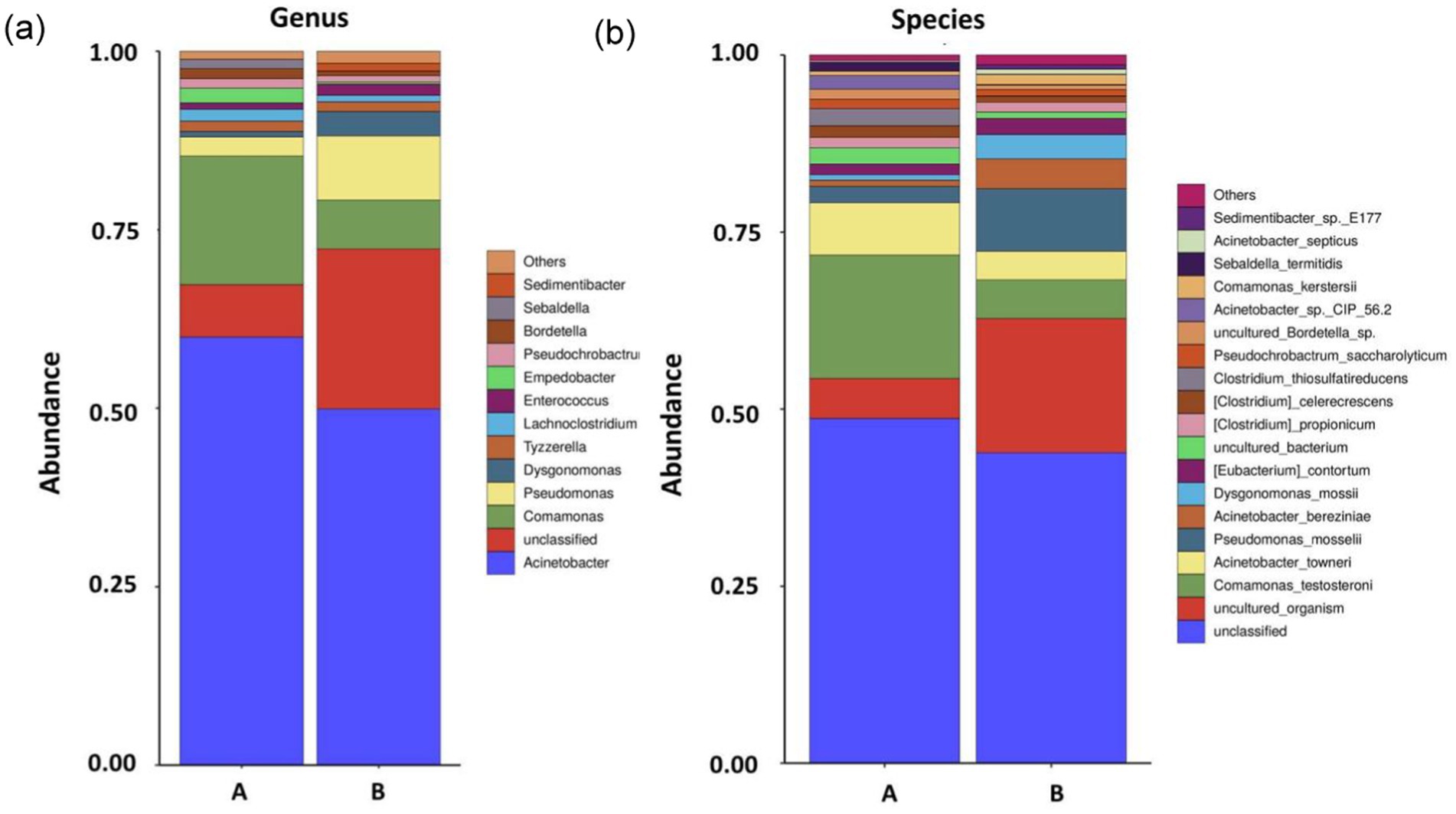
Figure 3. Microbial community structures and relative abundances at the conclusion of the enrichment cutting fluid culturing experiment at the genus (A) and species (B) levels (samples A and B are parallel samples).
Genus- and species-level analyses of the fungal communities showed that, although there were fewer types of fungi than bacteria in the Test-SCF sample, their relative abundances were higher (Figure 4).
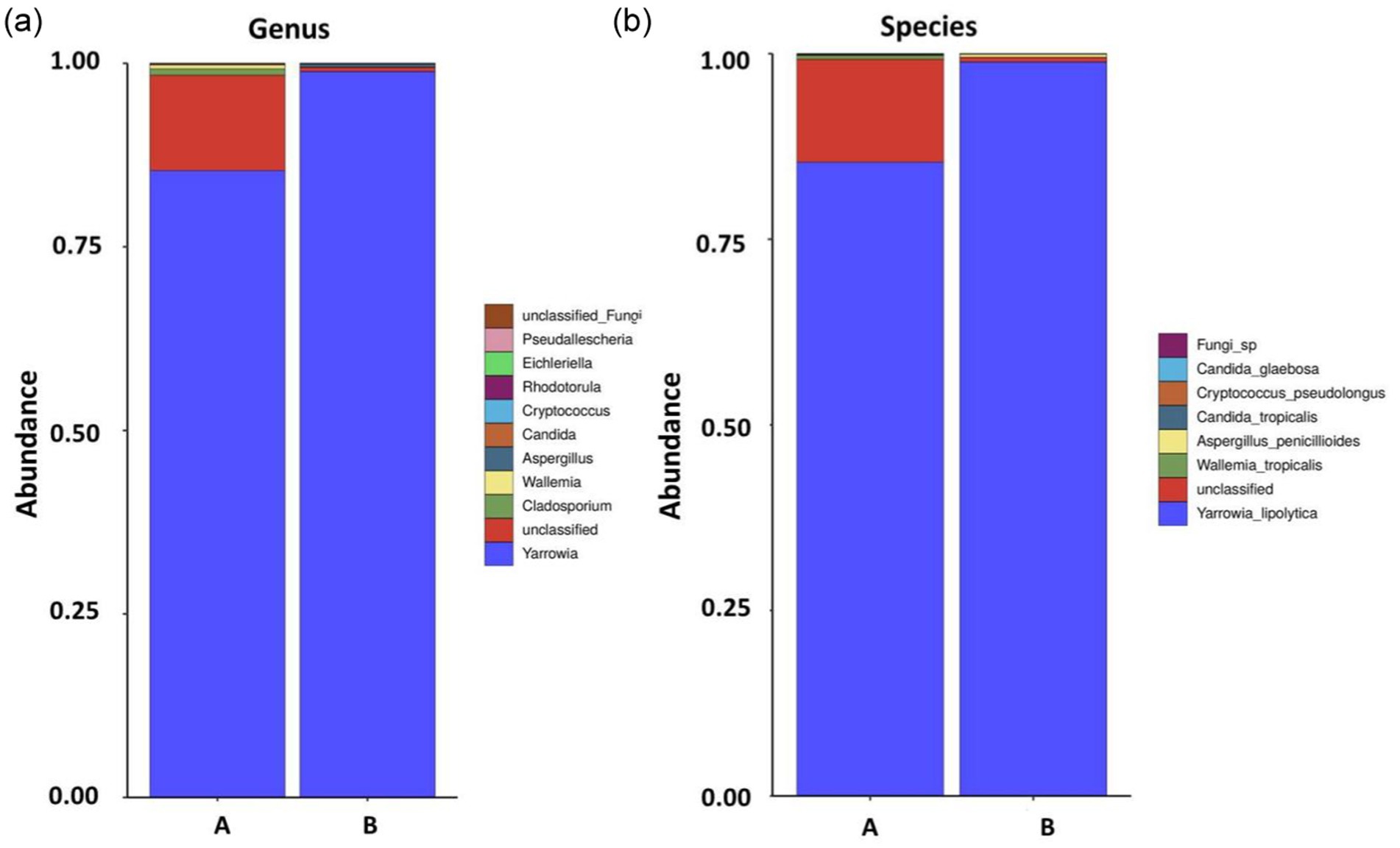
Figure 4. Fungal community structures and relative abundances in expanded experimental cutting fluids at the genus (A) and species (B) levels (samples A and B are parallel samples).
The experiment identified 85.4% Yarrowia lipolytica in sample A, and 98.9% in sample B, with 13.9% unclassified fungi in sample A, and < 1% each of Wallemia_tropicalis, Candida_tropicalis, Cryptococcus, pseudolongus, and Candida_glaebosa. Yarrowia lipolytica exhibited the highest relative abundance, indicating its dominance in the cutting fluid. This yeast is characterized by rapid reproduction, a robust colonization capability, a high survival rate, and resilience in low pH environments.
The growth of microorganisms in the configured cutting fluid is presented in Figure 5. The graph indicates a slow microbial growth rate between 0 and 24 h, representing the lag phase. There was an exponential increase in biomass between 24 and 96 h, denoting rapid growth during the log phase. The biomass reached its peak at ~108 h, corresponding to the highest observed microbial concentration. After this point the biomass underwent a slight decline followed by a steady downward trend until 168 h, marking the onset of the death phase. Because of the high organic content in the cutting fluid that continuously supplied the necessary nutrients for microbial growth, the growth and death of the microorganisms eventually reached an equilibrium at a total count of ~1011 CFU/mL.
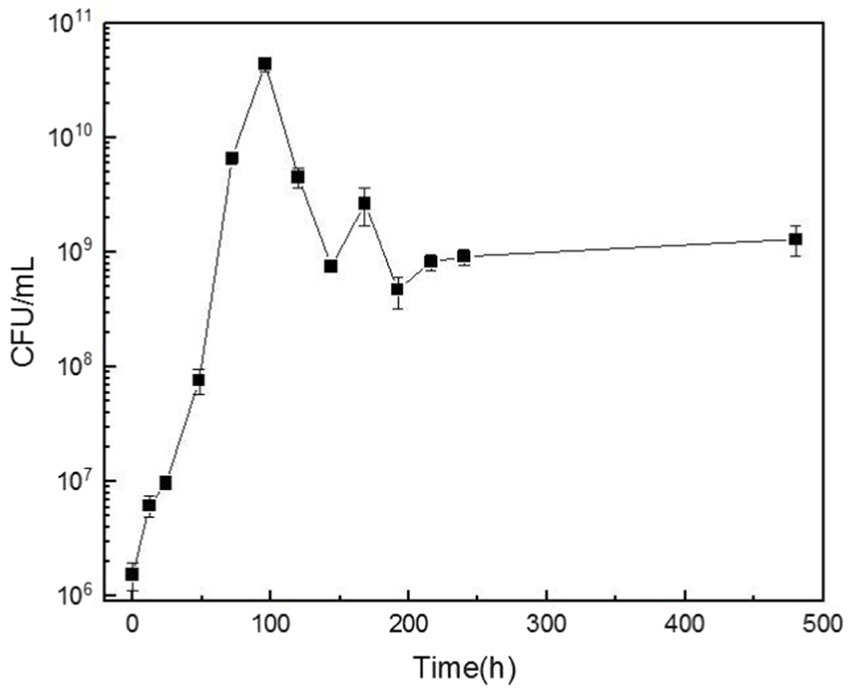
Figure 5. The variation curve of bacterial colony-forming units (CFU) per milliliter of cutting fluid over time.
The stability and corrosion-preventing physicochemical properties of cutting fluid are highly dependent on its pH, which is frequently used as a performance indicator in industrial testing. Cutting fluid pH should typically be maintained between 8.5–9.2 to ensure optimal performance.
Figure 6 displays the pH of cutting fluids over various time points, clearly showing that the pH of the cutting fluid sample that was inoculated with microorganisms decreased from an initial value of 8.81 to a final one of 7.15, whereas the pH of the uninoculated fluid did not change significantly. These results indicate that the presence of microorganisms significantly lowered the pH of the cutting fluid as the experiment progressed. The microbial growth curve indicates that microorganisms proliferate rapidly over time, which significantly impacts the pH of the cutting fluid. The microbial community analysis revealed the presence of various anaerobic bacteria, including the Sebaldella genus, at the bottom of the tool’s tank. These bacteria produce a range of organic acids such as acetic, lactic, and occasionally formic acid that reduce the pH of the cutting fluid (Li et al., 2023).
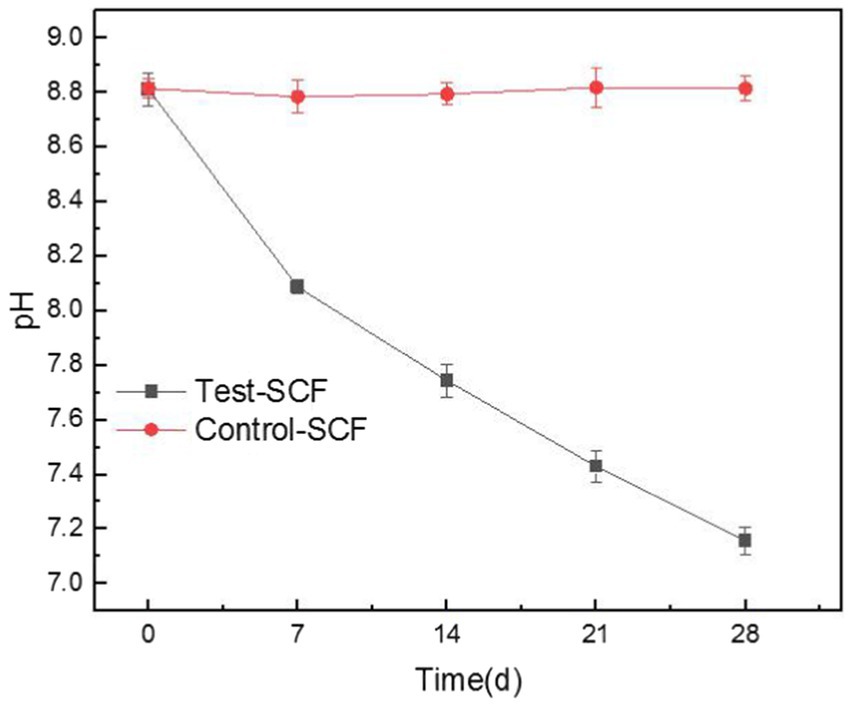
Figure 6. Trends in the pH values of the cutting fluids inoculated with bacteria (Test-SCF) and without bacteria (Control-SCF) over time.
After being used for extended periods, cutting fluid harbors aerobic, facultative, and anaerobic bacteria. Aerobic bacteria can degrade the active ingredients of the fluid, leading to decreased performance. The metabolism of facultative and anaerobic bacteria produces organic acids, resulting in a lower pH (Ma et al., 2018). Moreover, Y. lipolytica produces significant quantities of citric acid that accumulate over time (Ledesma-Amaro et al., 2016). This bacterium also produces other acidic substances such as alpha-ketoglutaric acid and unsaturated fatty acids (Guo et al., 2016).
The iron filings experimental results are shown in Figure 7. No rust spots were observed in the Control-SCF sample after 28 days, indicating no impact on the corrosion inhibition properties of the sterile SCF. Similarly, no rust spots were observed when iron filings were added to the fresh Test-SCF sample, indicating that the newly-prepared fluid effectively inhibited their corrosion. However, as the experiment progressed, more rust spots appeared on the filter paper, indicating a decrease in the corrosion-inhibiting properties of the fluid. This deterioration is likely attributable to a rapid increase in the microbial population and a corresponding intensification in their metabolic activity that significantly reduced the corrosion-inhibiting properties of the fluid.
As cutting fluid is primarily used to process aluminum alloys, this iron filings method does not comprehensively assess the impact of cutting fluid deterioration on its properties related to inhibiting the corrosion of aluminum alloy workpieces. Consequently, the effects of microbes on the ability of cutting fluid to inhibit corrosion were further investigated through electrochemical testing. Electrochemical tests were conducted using aluminum alloy samples that had been immersed in microorganism-inoculated cutting fluid for durations of 1, 7, 14, 21, and 28 days. Figure 8 presents the OCP of the aluminum alloy in the cutting fluid at various time points. Generally, a more negative OCP indicates that the alloy has a higher sensitivity to corrosion. The data revealed a noticeable downward trend in OCP, thereby suggesting that the sensitivity to corrosion of the aluminum alloy in the cutting fluid increased progressively. Cutting fluid includes corrosion inhibitors that are adsorbed onto metallic surfaces, exerting protective effects on the metal. Nevertheless, as the experiment progressed, the OCP of the aluminum alloy in the cutting fluid with microorganisms gradually declined. This was primarily attributed to the failure of the corrosion inhibitor to be adsorbed onto the surface of the aluminum alloy (Alobaid et al., 2024).
Figure 9 illustrates the tapping efficiency of the cutting fluid at different time points over the experiment. The results showed that the tapping torque value initially decreased gradually over time, before increasing again after 14 days. This suggests that in the early stage of the experiment, the presence of microorganisms slightly enhanced the lubricity of the cutting fluid (D’Addona et al., 2020). However, a biofilm was observed to have formed on the fluid’s surface. After stirring, clumpy substances appeared that resembled undissolved milk powder (Supplementary Figure S1). This observation was consistent with the characteristics of fungal growth, indicating that the number of fungi increased significantly. Fungal activity therefore likely became the determining factor, with Y. lipolytica representing the dominant fungus (Figure 4). In the cutting fluid environment with a high mineral oil content, Y. lipolytica follows a filamentous growth pattern (Ermakova and Morgunov, 1988). The growth and movement of fungal hyphae make the cutting fluid unable to form an effective lubricating film between the tool, chip, and machining surface (Elansky et al., 2022), resulting in reduced lubricity. Moreover, fungi produce various enzymes that can disrupt the active ingredients of cutting fluid, resulting in reduced lubricity (Buarque et al., 2023).
The impact of microorganisms on the stability of cutting fluid was assessed through droplet morphology and zeta potential. Figures 10, 11 show the morphology and diameter statistics of droplets in the cutting fluid at different time points over the experiment. The size of the oil droplets in the Test-SCF sample initially increased before increasing gradually after 14 days and stabilizing. Conversely, in the Control-SCF sample, the particle sizes of the oil droplets did not change appreciably, remaining stable at ~10 μm.
Zeta potential was used to show the stability of the cutting fluid system, with larger absolute values indicating a more stable system. Figure 12 shows the absolute value of the zeta potential of the freshly-prepared cutting fluid compared to the fluid that had progressed through 28 days of experimentation. The results indicated that the absolute zeta potential value decreased from 51.9 to 35.4 mV after 28 days. The significant reduction in the absolute value indicates a deterioration in the stability of the cutting fluid system.
This was primarily caused by the increased number of fungi after day 14 playing a dominant role. Fungal mycelia grew continuously in the cutting fluid, intertwining to form a network structure (Hotz et al., 2023). This led to the flocculation of oil droplets in the cutting fluid system, thereby reducing its stability. The effective components that provide lubrication also flocculated concurrently, resulting in the reduced lubricity observed in the tapping torque experiment results.
ORP has rarely been used to assess the performance of cutting fluid, but our study innovatively used this property to analyze the cutting fluid samples. In most biological systems, aerobic cells exhibit high cell potentials, whereas anaerobic ones have low potentials. Enzymatic activity, cellular assimilation capabilities, and microbial growth are also influenced by ORP. The ORP of the cutting fluid gradually decreased over the test month and eventually stabilized at a relatively low value (Figure 13). This indicated that the number of anaerobic bacteria in the cutting fluid increased as the experiment progressed. For example, there was a significant presence of bacteria such as S. termitidis and C. lubricantis, and even the obligate anaerobe C. celerecrescens was detected. This demonstrated a clear correlation between ORP and the dominant microorganisms in the cutting fluid.
In general, the growth and reproduction of microorganisms impacted the pH and corrosion inhibition capability of the cutting fluid in two ways. First, the microbial metabolic byproducts affected the pH and corrosion inhibition capability of the fluid. Second, the metabolic activity of the microorganisms themselves may have degraded the active ingredients of the cutting fluid.
Total organic carbon content was tested to investigate how microbial degradation affected the various components of industrial cutting fluid. Triethanolamine, which regulates pH, can cause pH imbalances in cutting fluid emulsions when degraded, thereby reducing performance (Pimenov et al., 2024). Tall oil, known for its excellent properties related to emulsification, lubrication, and corrosion inhibition, serves as a crucial emulsifying agent in cutting fluid (Kazeem et al., 2020). Organic phosphates, commonly added as corrosion inhibitors in cutting fluids for aluminum, help prevent corrosion during the machining process. These three substances were therefore selected to assess the effects of microbial degradation.
Figure 14 shows the effects of microbial degradation on these different components. Initially, the degradation rate of all three components was slow, owing to the lower microbial content and slower metabolism, thus resulting in a slower aerobic degradation rate. On day 5, during the logarithmic growth phase, the microbial population and metabolic activity increased substantially, accelerating the degradation of the organic components. By day 9, the degradation rates reached 26.82% for triethanolamine, 34.15% for tall oil, and 11.04% for organophosphate esters. The highest microbial degradation rates were observed for triethanolamine, followed by tall oil, with the organophosphate esters showing the lowest rates. Although their rate of microbial degradation was relatively slow, the organophosphate esters were nevertheless degraded. This indicates that, during metabolism, microorganisms can use corrosion-inhibiting organophosphate esters as a nutrient source, thus reducing the corrosion-inhibition properties of cutting fluid. Microorganisms also degraded triethanolamine, a pH regulator, ultimately leading to lowered pH in the cutting fluid.
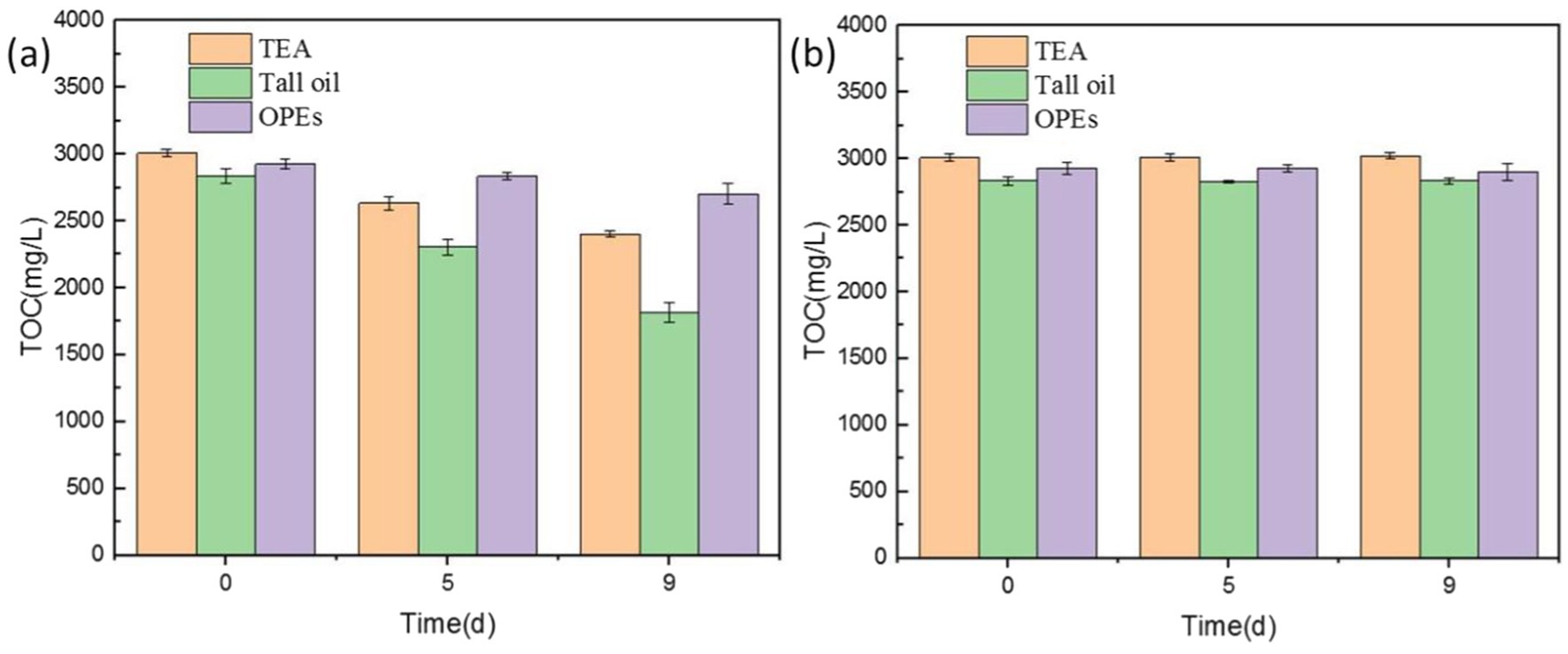
Figure 14. Total organic carbon (TOC) values of cutting fluid samples with (A) and without (B) microbial growth.
The microbial compositions of the spoiled cutting fluid samples varied significantly between those taken from the surface and the bottom of the onsite instrument’s liquid tank. The surface layer predominantly contained aerobic bacteria such as Pseudomonas, Acinetobacter, and Ochrobactrum. Conversely, the bottom layer was primarily composed of anaerobic and facultative bacteria such as Brachymonas, Sebaldella, Corynebacterium, and Comamonas. The results of laboratory microbial culturing experiments showed that the fungal communities in these samples mainly consisted of the yeast Yarrowia lipolytica. Microbial metabolic activity inhibited the formation of passivation films on the aluminum alloys, through the secretion of various organic acids. These activities also consumed the primary active ingredients of the cutting fluid, leading to a significant decrease in its pH and ability to inhibit corrosion. The presence of microorganisms slightly enhanced the lubricity of the cutting fluid; however, in the later stages of the experiment, an increase in fungi, as well as the corresponding sweeping and bridging actions of the fungal hyphae, caused the active ingredients in the cutting fluid system to flocculate, reducing both its lubricity and stability. The results of this study showed that microorganisms impact all properties of industrial cutting fluid, with the most pronounced effects being related to corrosion inhibition, pH, and stability. The experimental conditions were restricted to a laboratory environment, so there may be many other potential microbial species and influencing factors related to this phenomenon that have remained unexplored. A deeper understanding of the impact of microorganisms on the deterioration of cutting fluid performance plays a key role in improving the quality, efficiency, and sustainability of the cutting process. It will also hopefully lay a theoretical foundation for the future development and application of new cutting fluids.
The data presented in the study are deposited in the National Center for Biotechnology Information (NCBI) repository, accession numbers SRP554559 and PRJNA1204126.
YS: Formal analysis, Project administration, Writing – original draft. WZ: Data curation, Formal analysis, Investigation, Writing – original draft. LW: Writing – review & editing, Supervision, Visualization. YD: Conceptualization, Data curation, Methodology, Writing – review & editing. GG: Writing – review & editing, Supervision, Methodology. LD: Conceptualization, Funding acquisition, Methodology, Writing – review & editing. ZG: Conceptualization, Formal analysis, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The authors acknowledge the financial support of the Shanghai Engineering Technology Research Center of Deep Offshore Material (no. 19DZ2253100).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1522265/full#supplementary-material
Abarenkov, K., Nilsson, R. H., Larsson, K. H., Alexander, I. J., Eberhardt, U., Erland, S., et al. (2010). The UNITE database for molecular identification of fungi-recent updates and future perspectives. New Phytol. 186, 281–285. doi: 10.1111/j.1469-8137.2009.03160.x
Alobaid, A. A., Allah, A., El, M. A., Dahmani, K., Aribou, Z., Kharbouch, O., et al. (2024). Comprehensive investigation of novel synthesized Thiophenytoin derivative (antiepileptic drug) as an environmentally friendly corrosion inhibitor for mild steel in 1 M HCl: theoretical, electrochemical, and surface analysis perspectives. Mater. Today Commun. 41:110698. doi: 10.1016/j.mtcomm.2024.110698
Babaei, P., Ghasemi-Kahrizsangi, T., and Marashi, S. A. (2014). Modeling the differences in biochemical capabilities of pseudomonas species by flux balance analysis: how good are genome-scale metabolic networks at predicting the differences? The Scientific World J. 2014:289. doi: 10.1155/2014/416289
Buarque, F. S., Carniel, A., Ribeiro, B. D., and Coelho, M. A. Z. (2023). Selective enzymes separation from the fermentation broth of Yarrowia lipolytica using aqueous two-phase system based on quaternary ammonium compounds. Sep. Purif. Technol. 324:124539. doi: 10.1016/j.seppur.2023.124539
Caballero, A., Esteve-Núñez, A., Zylstra, G. J., and Ramos, J. L. (2005). Assimilation of nitrogen from nitrite and trinitrotoluene in Pseudomonas putida JLR11. J. Bacteriol. 187, 396–399. doi: 10.1128/JB.187.1.396-399.2005
Chen, H. M., Liu, N. C., Lu, H., Li, X. F., and Zheng, Y. Y. (2024). Evaluation of new nano-cutting fluids for the processing of carbon fiber-reinforced composite materials. J. Clean. Prod. 437:140771. doi: 10.1016/j.jclepro.2024.140771
Choudhury, S. K., and Muaz, M. (2020). Renewable metal working fluids for aluminum and heavy duty machining. Encycl. Renew. Sustain. Mater. 5, 242–248. doi: 10.1016/B978-0-12-803581-8.11546-2
D’Addona, D. M., Conte, S., Teti, R., Marzocchella, A., and Raganati, F. (2020). Feasibility study of using microorganisms as lubricant component in cutting fluids. Proc. CIRP 88, 606–611. doi: 10.1016/j.procir.2020.05.106
Daniels, C., Godoy, P., Duque, E., Molina-Henares, M. A., de la Torre, J., del Arco, J. M., et al. (2010). Global regulation of food supply by Pseudomonas putida DOT-T1E. J. Bacteriol. 192, 2169–2181. doi: 10.1128/JB.01129-09
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Elansky, S. N., Chudinova, E. M., Elansky, A. S., Kah, M. O., Sandzhieva, D. A., Mukabenova, B. A., et al. (2022). Microorganisms in spent water-miscible metalworking fluids as a resource of strains for their disposal. J. Clean. Prod. 350:131438. doi: 10.1016/j.jclepro.2022.131438
Ermakova, I. T., and Morgunov, I. G. (1988). Pathways of metabolism in Yarrowia (candida) Lipolytica yeasts. Microbiology 57, 429–432.
Gast, C. J. V. D., Knowles, C. J., Wright, M. A., and Thompson, I. P. (2001). Identification and characterisation of bacterial populations of an in-use metal-working fluid by phenotypic and genotypic methodology. Int. Biodeterior. Biodegradation 47, 113–123. doi: 10.1016/S0964-8305(01)00036-1
Grijalbo, L., Garbisu, C., Martin, I., Etxebarria, J., Gutierrez-Mañero, F. J., and Garcia, J. A. L. (2015). Functional diversity and dynamics of bacterial communities in a membrane bioreactor for the treatment of metal-working fluid wastewater. J. Water Health 13, 1006–1019. doi: 10.2166/wh.2015.079
Guo, H. W., Su, S. J., Madzak, C., Zhou, J. W., Chen, H. W., and Chen, G. (2016). Applying pathway engineering to enhance production of alpha-ketoglutarate in Yarrowia lipolytica. Appl. Microbiol. Biot. 100, 9875–9884. doi: 10.1007/s00253-016-7913-x
Harmon-Smith, M., Celia, L., Chertkov, O., Lapidus, A., Copeland, A., Del Rio, T. G., et al. (2010). Complete genome sequence of Sebaldella termitidis type strain (NCTC 11300T). Stand. Genomic Sci. 2, 220–227. doi: 10.4056/sigs.811799
Hill, E. C. (1992). COSHH regulations and microbial hazards associated with metal-working fluids. Tribol. Int. 25, 141–143. doi: 10.1016/0301-679X(92)90091-Z
Hotz, E. C., Bradshaw, A. J., Elliott, C., Carlson, K., Dentinger, B. T. M., and Naleway, S. E. (2023). Effect of agar concentration on structure and physiology of fungal hyphal systems. J. Mater. Res. Technol. 24, 7614–7623. doi: 10.1016/j.jmrt.2023.05.013
Kazeem, R. A., Fadare, D. A., Abutu, J., Lawal, S. A., and Adesina, O. S. (2020). Performance evaluation of jatropha oil-based cutting fluid in turning AISI 1525 steel alloy. CIRP J. Manuf. Sci. Technol. 31, 418–430. doi: 10.1016/j.cirpj.2020.07.004
Korkmaz, M. E., Gupta, M. K., Ross, N. S., and Sivalingam, V. (2023). Implementation of green cooling/lubrication strategies in metal cutting industries: a state of the art towards sustainable future and challenges. Sustain. Mater. Technol. 36:e00641. doi: 10.1016/j.susmat.2023.e00641
Ledesma-Amaro, R., Lazar, Z., Rakicka, M., Guo, Z. P., Fouchard, F., Crutz-Le, C. A. M., et al. (2016). Metabolic engineering of Yarrowia lipolytica to produce chemicals and fuels from xylose. Metab. Eng. 38, 115–124. doi: 10.1016/j.ymben.2016.07.001
Li, J. L., Chen, L. J., Wei, B., Xu, J., Wei, B. X., and Sun, C. (2024). Microbiologically influenced corrosion of circulating cooling systems in power plants – a review. Arab. J. Chem. 17:105529. doi: 10.1016/j.arabjc.2023.105529
Li, B. H., Huang, J. H., Li, X. L., Ma, Q., Zhao, P. F., and Huang, W. (2023). The microbial degradation of AAOO based cutting fluid wastewater. J. Water Process Eng. 55:104167. doi: 10.1016/j.jwpe.2023.104167
Lillienberg, L., Andersson, E. M., Jarvholm, B., and Torén, K. (2010). Respiratory symptoms and exposure-response relations in workers exposed to metalworking fluid aerosols. Ann. Occup. Hyg. 54, 403–411. doi: 10.1093/annhyg/meq009
Lillienberg, L., Burdorf, A., Mathiasson, L., and Thorneby, L. (2008). Exposure to metalworking fluid aerosols and determinants of exposure. Ann. Occup. Hyg. 52, 597–605. doi: 10.1093/annhyg/men043
Liu, H. M., Lin, Y. H., Tsai, M. Y., and Lin, W. H. (2010). Occurrence and characterization of culturable bacteria and fungi in metalworking environments. Aerobiologia 26, 339–350. doi: 10.1007/s10453-010-9169-8
Lopes, E. L., Semedo, M., Tomasino, M. P., Mendes, R., Sousa, J. B., and Magalhães, C. (2024). Horizontal distribution of marine microbial communities in the North Pacific subtropical front. Front. Microbiol. 15:1455196. doi: 10.3389/fmicb.2024.1455196
Lu, S. H., He, Y., Xu, R. C., Wang, N. X., Chen, S. Q., Dou, W. W., et al. (2023). Inhibition of microbial extracellular electron transfer corrosion of marine structural steel with multiple alloy elements. Bioelectrochemistry 151:108377. doi: 10.1016/j.bioelechem.2023.108377
Ma, S., Kim, K., Huh, J., Kim, D. E., Lee, S., and Hong, Y. (2018). Regeneration and purification of water-soluble cutting fluid through ozone treatment using an air dielectric barrier discharge. Sep. Purif. Technol. 199, 289–297. doi: 10.1016/j.seppur.2018.02.005
Osama, M., Singh, A., Walvekar, R., Khalid, M., Gupta, T. C., and Yin, W. W. (2017). Recent developments and performance review of metal working fluids. Tribol. Int. 114, 389–401. doi: 10.1016/j.triboint.2017.04.050
Pimenov, D. Y., Silva, L. R. R., Machado, A. R., França, P. H. P., Pintaude, G., Unune, D. R., et al. (2024). A comprehensive review of machinability of difficult-to-machine alloys with advanced lubricating and cooling techniques. Tribol. Int. 196:109677. doi: 10.1016/j.triboint.2024.109677
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., and Peplies, J. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Rabenstein, A., Koch, T., Remesch, M., Brinksmeier, E., and Kuever, J. (2009). Microbial degradation of water miscible metal working fluids. Int. Biodeterior. Biodegradation 63, 1023–1029. doi: 10.1016/j.ibiod.2009.07.005
Rakić, R., and Rakić, Z. (2002). The influence of the metal working fluids on machine tool failures. Wear 252, 438–444. doi: 10.1016/S0043-1648(01)00890-0
Roca, A., Rodriguez-Herva, J. J., and Ramos, J. L. (2009). Redundancy of enzymes for formaldehyde detoxification in Pseudomonas putida. J. Bacteriol. 191, 3367–3374. doi: 10.1128/JB.00076-09
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahe, F. (2016). VSEARCH: a versatile open source tool for metagenomics. Peer J 4:e2584. doi: 10.7717/peerj.2584
Sankaranarayanan, R., Hynes, N. R. J., Kumar, J. S., and Krolczyk, G. M. (2021). A comprehensive review on research developments of vegetable-oil based cutting fluids for sustainable machining challenges. J. Manuf. Process. 67, 286–313. doi: 10.1016/j.jmapro.2021.05.002
Schwarz, M., Dado, M., Hnilica, R., and Veverková, D. (2015). Environmental and health aspects of metalworking fluid use. Pol. J. Environ. Stud. 24, 37–45.
Simpson, A. T., Groves, J. A., Unwin, J., and Piney, M. (2000). Mineral oil metal working fluids (MWFs)-development of practical criteria for mist sampling. Ann. Occup. Hyg. 44, 165–172. doi: 10.1016/S0003-4878(99)00085-X
Sun, M., Xu, W. W., Rong, H., Chen, J. T., and Yu, C. L. (2023). Effects of dissolved oxygen (DO) in seawater on microbial corrosion of concrete: morphology, composition, compression analysis and transportation evaluation. Constr. Build. Mater. 367:130290. doi: 10.1016/j.conbuildmat.2023.130290
Trafny, E. A. (2013). Microorganisms in metalworking fluids: current issues in research and management. Int. J. Occup. Med. Environ. Health 26, 4–15. doi: 10.2478/S13382-013-0075-5
Yazdi, M., Khan, F., Abbassi, R., Quddus, N., and Castaneda-Lopez, H. (2022). A review of risk-based decision-making models for microbiologically influenced corrosion (MIC) in offshore pipelines. Reliab. Eng. Syst. Safe. 223:108474. doi: 10.1016/j.ress.2022.108474
Yin, Y. S., Guo, Z. W., Liu, T., Guo, N., and Shen, Y. Y. (2019). A culture medium used for culturing and isolating microorganisms in cutting fluid, as well as its preparation method and application. CN 110846247 a [patent]. China: State Intellectual Property Office of the People’s Republic of China. Available at: https://www.iprdb.com.
Zhang, Q. K., He, Y. H., Wang, W., Lin, N., Wu, C. H., and Li, N. F. (2015). Corrosion behavior of WC–co hardmetals in the oil-in-water emulsions containing sulfate reducing Citrobacter sp. Corros. Sci. 94, 48–60. doi: 10.1016/j.corsci.2015.01.036
Keywords: microbial reproduction, deterioration of cutting fluid performance, corrosion inhibition capability, lubricity, stability, microbial metabolism
Citation: Shen Y, Zhang W, Wu L, Dong Y, Guo G, Dong L and Guo Z (2025) Microbial proliferation deteriorates the corrosion inhibition capability, lubricity, and stability of cutting fluid. Front. Microbiol. 16:1522265. doi: 10.3389/fmicb.2025.1522265
Received: 04 November 2024; Accepted: 20 January 2025;
Published: 11 February 2025.
Edited by:
Eva Pakostova, Laurentian University, CanadaReviewed by:
Ruiyong Zhang, Chinese Academy of Sciences, ChinaCopyright © 2025 Shen, Zhang, Wu, Dong, Guo, Dong and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanyuan Shen, eXlzaGVuQHNobXR1LmVkdS5jbg==; Yaohua Dong, eWhkb25nQHNobXR1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.