
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 12 February 2025
Sec. Microorganisms in Vertebrate Digestive Systems
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1513541
Gut microbiota has been increasingly linked to metabolic health and diseases over the past few decades. Bile acids (BAs), the major components of bile, are bidirectionally linked to intestinal microbiota, also known as the gut microbiome-BA metabolic axis. Gut microbiota-derived bile salt hydrolase (BSH, EC 3.5.1.24), which catalyzes the “gateway” reaction in a wider pathway of bile acid modification, not only shapes the bile acid landscape, but also modulates the crosstalk between gut microbiota and host health. Therefore, microbial BSHs exhibit the potential to directly or indirectly influence microbial and host physiologies, and have been increasingly considered as promising targets for the modulation of gut microbiota to benefit animal and human health. However, their physiological functions in bacterial and host physiologies are still controversial and not clear. In this review, we mainly discuss the current evidence related to the physiological roles that BSHs played in gut microbiota and human health, and the possible underlying mechanisms. Meanwhile, we also present the potential applications of BSHs and BSH-producing probiotics in various fields. Finally, we describe several important questions that need to be addressed by further investigations. A detailed exploration of the physiological significance of BSHs will contribute to their future diagnostic and therapeutic applications in improving animal and human health.
Gut microbiota is a large population of commensal microorganisms inhabiting the gastrointestinal tract (GIT), including bacteria, viruses, archaea, protozoa and fungi (Parasar et al., 2019). Due to its ability to affect intestinal permeability, motility and sensitivity, mucosal immune function, and enteric nervous system, gut microbiome has been shown to play crucial roles in host health, while disruption of this population has been implicated in many diseases (Foley et al., 2021). For this reason, modulation of gut microbiota composition has become a potential alternative approach for preventing and treating diseases to maintain human health.
As the major components of bile, bile acids (BAs) represent an important class of metabolites that shape the gut microbiota, host physiology and metabolism. After de novo synthesis from cholesterol by host hepatocytes through cholesterol 7α-hydroxylase (CYP7A1), primary BAs, such as cholic acid (CA) and chenodeoxycholic acid (CDCA), are conjugated with either glycine or taurine to form conjugated bile acids, including glyco- and taurocholic acids (G/TCA) as well as glyco- and taurochenodeoxycholic acids (G/TCDCA, Figure 1). During the fasting state, these conjugated BAs are stored in the gallbladder. Upon ingestion of foods, they are excreted into the proximal small intestine via bile duct to promote the emulsification and absorption of dietary lipids and lipophilic vitamins (Yang et al., 2022). As they travel through the small intestine, the majority of conjugated BAs (>95%) are deconjugated by gut microbiota, and absorbed by active transport. The unabsorbed deconjugated BAs (CA or CDCA) are converted into secondary BAs [deoxycholic acid (DCA), lithocholic acid (LCA), etc.] through further microbial modifications, and absorbed into colon via passive diffusion. Both deconjugated and secondary BAs reabsorbed from the gut are taken up by hepatocytes through portal vein, reconjugated, and resecreted into bile for enterohepatic recirculation (Chiang and Ferrell, 2022), whereas remaining BAs are excreted into the feces. Recently, it has been found that BSHs from Lactiplantibacillus strains (Foley et al., 2023), B. longum NCTC 11818 (Rimal et al., 2024) and C. perfringens (Guzior et al., 2024) also exhibit amine N-acyl transferase activity that mediates the conjugation of amino acids to the acyl-site of deconjugated and secondary BAs, generating a diverse repertoire of microbial conjugated bile acids (MCBAs). This newly found function of BSHs expands BA diversity and complexity, and the compounds obtained also serve as substrates for BSHs and enter enterohepatic recirculation.
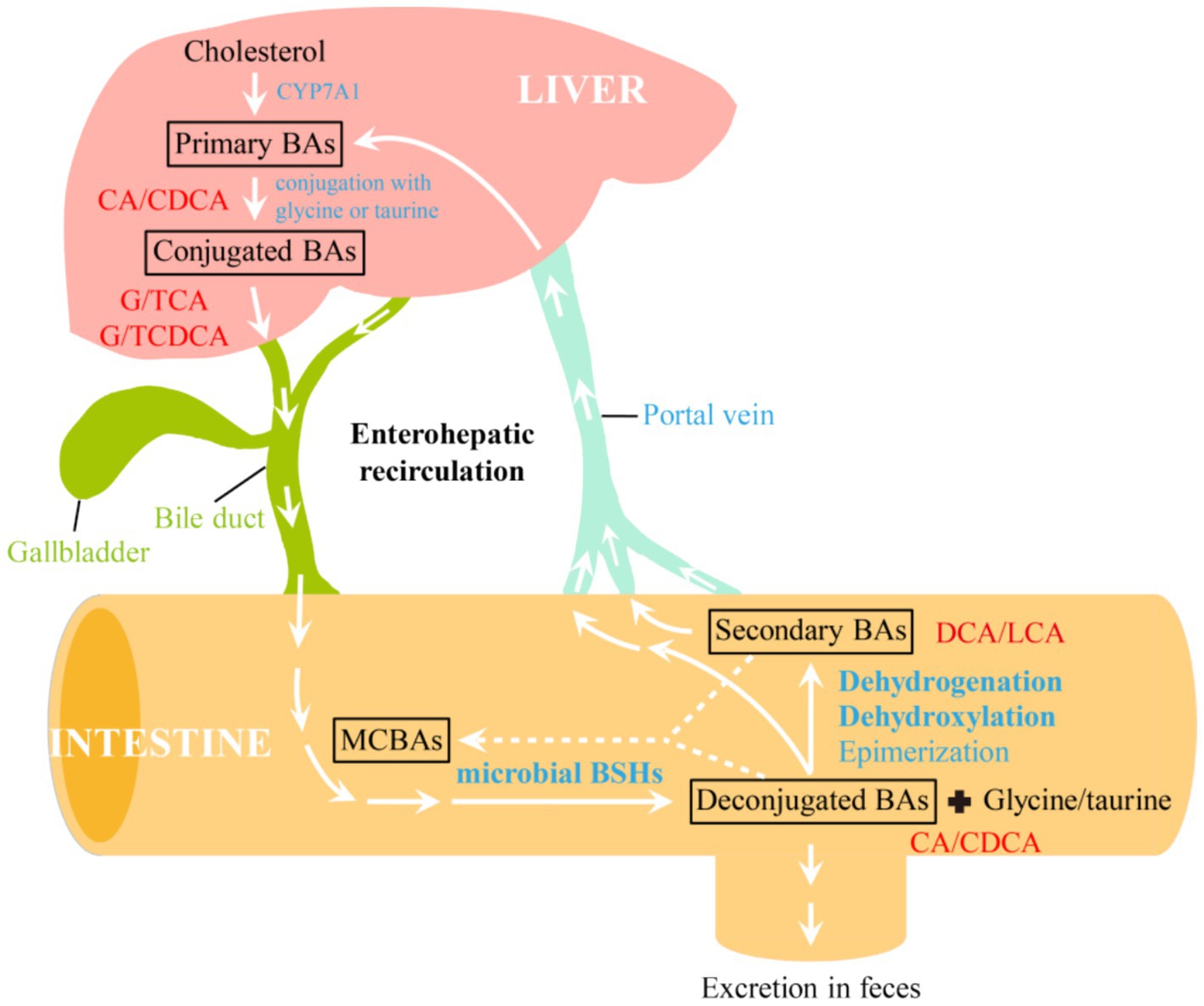
Figure 1. Enterohepatic circulation and modifications of bile acids by gut microbiome. CYP7A1, cholesterol 7α-hydroxylase; BSHs, bile salt hydrolases; BAs, bile acids; MCBAs, microbial conjugated bile acids; CA, cholic acid; CDCA, chenodeoxycholic acid; G/TCA, glyco- and taurocholic acids; G/TCDCA, glyco- and taurochenodeoxycholic acids; DCA, deoxycholic acid; LCA, lithocholic acid.
Intestinal microbiota and BAs are bidirectionally linked to form the well-known gut microbiome-BA metabolic axis (Figure 2), which has been increasingly recognized as a key regulator of host health (Wang et al., 2022). On the one hand, gut microbiota have evolved many strategies to modify BAs and change the BA pool, which in turn regulates BA metabolism by receptors and promotes bacterial survival (Guzior and Quinn, 2021). On the other hand, the amphipathic nature of BAs confer them detergent-like antimicrobial activities that inherently restrict the growth of some bacteria, thus reshaping the composition of gut microbiota (Peng et al., 2024). As metabolic integrators, BAs can also bind to a variety of nuclear and cell-surface receptors in enterocytes, liver and adipose tissues, including nuclear receptor farnesoid X receptor (FXR), liver X receptor (LXR), pregnane X receptor (PXR) or the G protein-coupled bile acid receptor 1 (GPBAR1, also known as TGR5) (Mohanty et al., 2024b; Zhang et al., 2024). By acting as agonists or antagonists for these receptors, BAs can regulate their own synthesis, immune homeostasis (Liu et al., 2022; Mohanty et al., 2024a), metabolic syndromes (Wang N. et al., 2021), lipid and energy metabolism (Govindarajan et al., 2016; Huang et al., 2019), cholesterol metabolism (Samson et al., 2024), neurological disorders (Sun et al., 2022; Wu et al., 2024), and drug absorption and bioavailability (Enright et al., 2018) (Figure 2). Therefore, bile acids act as key mediators to modulate the communications between gut microbiota and host health.
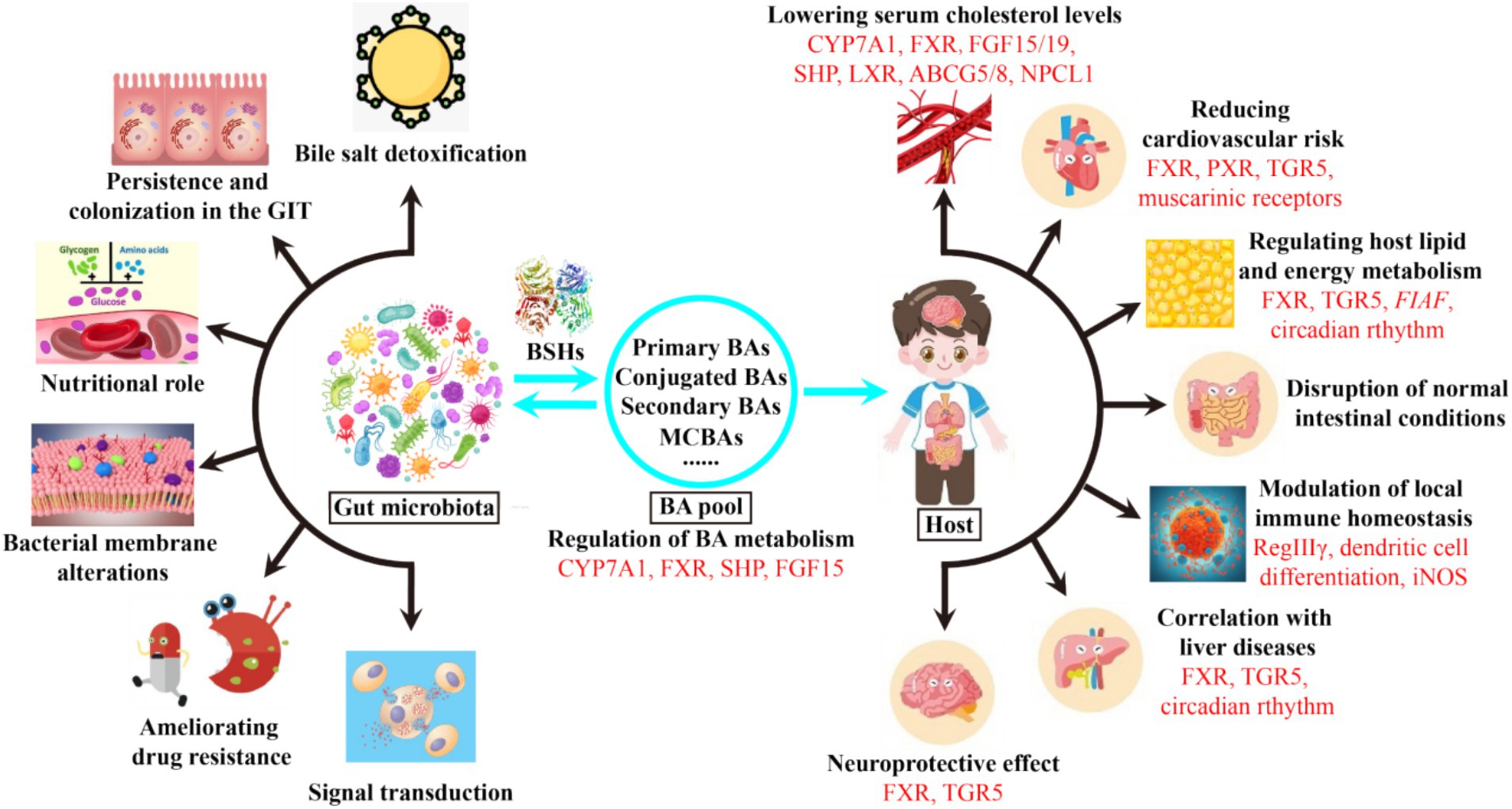
Figure 2. Effects of microbial BSHs on the bacterial and host physiologies. Bile acid receptors and related genes involved in each physiology are marked in red. GIT, gastrointestinal tract; MCBAs, microbial conjugated bile acids; FXR, farnesoid X receptor; FGF15/19, fibroblast growth factors 15 and 19; SHP, small heterodimer partner; LXR, liver X receptor; ABCG5/8, adenosine triphosphate-binding cassette transporters G5 and G8; NPCL1, Niemann-Pick C1-like 1; PXR, pregnane X receptor; TGR5, G protein-coupled bile acid receptor 1; FIAF, fasting-induced adipose factor; RegIIIγ, antimicrobial peptide RegIIIγ; iNOS, inducible nitric oxide synthase.
Currently known microbial modifications of BAs include deconjugation, dehydroxylation, epimerization, and dehydrogenation (Figure 1) (Guzior and Quinn, 2021; Mohanty et al., 2024b). Among them, deconjugation reaction which hydrolyzes conjugated BAs into less toxic unconjugated BAs and amino acids is considered as a “gatekeeper” for subsequent microbial modifications of BAs. This reaction is catalyzed by bile salt hydrolase (BSH, EC 3.5.1.24), an enzyme belonging to the N-terminal nucleophilic (Ntn) hydrolase superfamily. There are now more than 2,400 BSH enzymes widely distributed among gut-associated bacterial phyla inhabiting both the small intestine and large bowel of human, including Firmicutes, Bacteroidetes, and Actinobacteria (Jones et al., 2008; Mohanty et al., 2024a). It has been reported that Lactobacilli strains, which constitute an important part of the Firmicutes bacteria, are the main contributors to total BSH activity in the intestinal tracts of human (Song et al., 2023) and that they help to modulate the enterohepatic circulation. Gut microbial BSHs are therefore highly required for the formation and upkeep of the BA pool, and BSH-mediated BA metabolism modulates a microbe-host dialog that alters both local (gastrointestinal) and systematic (hepatic) host functions, including shaping gut microbiota (Govindarajan et al., 2016; Lynch et al., 2023) and regulation of host metabolism (Lin et al., 2014; Miremadi et al., 2014). Thus, gut microbiota-derived BSHs have become promising microbiome targets for the development of novel therapies to prevent and treat human diseases.
However, it is somewhat controversial whether the BSH activity of gut microbiome is beneficial or detrimental to the host. Although the potential positive aspects of BSH enzymes have been previously discussed (De Smet et al., 1998), other possible negative concerns about them have also been raised, such as disruption of lipid metabolism, colon carcinogenesis, gallstone formation, altered gut microbial populations and other gut associated disorders (Jones et al., 2008). We review here the available literature on the physiological roles of BSH activity for the enzyme-producing bacteria as well as for the hosts, including the contradictory findings obtained and the underlying mechanisms involved. We also present the potential applications of BSHs and BSH-harboring probiotics. Finally, we conclude with suggestions for future work. Knowledge gained here can provide an updated understanding of the physiological significance of microbial BSHs and support their wide applications in BSH-based biotherapeutics development.
Although the physiological roles of microbial BSHs are not yet fully understood, and may vary between bacterial species and genera, they have now been hypothesized to play multiple roles in the bacteria and hosts through several possible mechanisms, which are described in Figure 2 and briefly discussed below. Besides, they also have several potential applications in medical diagnosis, prevention and treatment of metabolic diseases, feed additives and other industries. Moreover, research is still ongoing to discern their exact functions.
Due to the lipophilic nature of the steroid ring, BAs are able to disorganize the cell membrane structure, acidify the cytoplasm and trigger DNA damage, displaying potent antimicrobial activities (Bustos et al., 2018). Therefore, bile represents a major challenge for bacteria to transit and survive in the GIT. Since BSH can convert more toxic conjugated BAs into less toxic free BAs, it has become a flexible response of gut microbes to BA stress and a desirable property when selecting a strain for use as a dietary adjunct (Urdaneta and Casadesús, 2017). In bacterial pathogens like Listeria innocua (Jones et al., 2008), L. monocytogenes (Begley et al., 2005) and Brucella abortus (Delpino et al., 2007), BSHs confer them with the ability to withstand the antimicrobial action of BAs in the gut, thus constituting a virulence factor for these bacteria, while bsh gene repression or disruption impairs the bile resistance of L. monocytogenes (Pombinho et al., 2020). Similarly, several studies have also established a connection between bile salt hydrolysis and bile tolerance of probiotic strains. For example, BSHs from Lactiplantibacillus plantarum WCFS1 (Lambert et al., 2008), L. gasseri JCM1131T (Kusada et al., 2021), Leptogranulimonas caecicola TOC12T and Granulimonas faecalis OPF53T (Morinaga et al., 2022) confer bile resistance on these bacteria, and the expression of bsh genes from L. plantarum Lp91 (Duary et al., 2012), B. longum NCC2705 (Yuan et al., 2008) and L. reuteri CRL1098 (Bustos et al., 2015) was also up-regulated in the presence of bile salts, whereas disruption of bsh genes in L. plantarum WCFS1 (Lambert et al., 2008), L. amylovorous (Grill et al., 2000), L. plantarum AR113 (Wang G. et al., 2021), and L. rhamnosus AMC010 (Arnold et al., 2018) renders these strains more susceptible to bile salts. Furthermore, several bsh genes have been heterologously expressed in BSH-negative strains L. innocua FH2333 (Jones et al., 2008), L. casei HX01 (Wang et al., 2011) and Leuconostoc citreum CB2567 (Seung Kee et al., 2015), which improves their survivability under bile stress or gut colonization in mice, contributing to their probiotic properties. Together, these findings lay the groundwork for engineering Lactobacilli with improved bile resistance to be used as antimicrobial agents or BA-altering therapeutics.
However, previous studies have demonstrated that bile tolerance of probiotics is not necessarily linearly dependent on their BSH activity, some of which have even failed to provide a direct link between them (Moser and Savage, 2001; Fang et al., 2009). BSHs from some Lactobacilli strains have been shown to confer no bile protection on them (Yang et al., 2019; Foley et al., 2021), and that L. monocytogenes relies upon other biomolecules rather than BSH activity for its growth in the murine gall bladder (Dowd et al., 2011). These conflicting findings can be partially attributed to the different concentrations, types and structures of BAs used in various studies, which determine their toxicity and differences in BSH substrate specificity (Bustos et al., 2018). It has been shown that the toxicity of glycoconjugated BAs is far greater than that of their tauroconjugated counterparts, and this may be the reason why most BSHs prefer to hydrolyze the former (Begley et al., 2005; Dong and Lee, 2018). Besides, intrinsic bile resistance is also a strain-specific trait and cannot be generalized within a species or a genus (Li and Chiang, 2014; Bustos et al., 2018). Given the complex and multifactorial effects of BA deconjugation, it is possible that such discrepancies are due to the use of different types of BAs, BSHs with various substrate specificities, and different strains and species among the studies.
Although the precise physiological function of BSHs in assisting probiotics to overcome the toxicity of conjugated BAs remains poorly understood, several possible mechanisms evolved by BSH-producing microorganisms have been proposed. Some studies have suggested that BSHs comprise part of a global response to acid and bile stresses (Dimarzio, 2016). Protonated forms of conjugated BAs are organic acids that exhibit toxicity toward the bacteria through intracellular acidification (Grill et al., 2000), while the unconjugated BAs released by BSHs are weaker acids and less soluble than their conjugated counterparts, thus recapturing the co-transported proton which may counteract the drop in internal pH under bile salts (Bustos et al., 2012). Unconjugated BAs produced by BSH hydrolysis can form biofilms on the cell surface under bile stress, which is another important bile resistance mechanism in several strains, including Lactiplantibacillus (Ambalam et al., 2012), Bifidobacteria (Jarocki et al., 2014), and Bacteroides fragilis (Pumbwe et al., 2007). Besides, BSH activity is also part of a broader strategy to alter cell membrane composition in response to environmental stress. In L. reuteri CRL 1098, BSHs change the fatty acid composition of the cell membrane, which can form complexes with precipitated BAs and cholesterol to maintain membrane fluidity, thus providing bile tolerance (Taranto et al., 2003). Recently, heterologous expression of the bsh gene from L. salivarius in Lactococcus lactis NZ9000 revealed that unconjugated BAs could form micelles around membrane surface of cells to combat the toxicity of conjugated BAs (Bi et al., 2016). Apart from the ability to deconjugated BAs, there are also other mechanisms involved in the bacterial bile tolerance, like PrfA-regulated bile exclusion system (Sleator et al., 2005) and Sigma B-regulated detoxification mechanism (Dowd et al., 2011) in L. monocytogenes, surface proteins, fatty acids and exopolysaccharides (Horáčková et al., 2018).
Adhesive property is an important factor that contributes to the colonization and survival of probiotics in the host gut, thus helping them exert health-promoting effects (Wu et al., 2022). For this reason, the ability to adhere to GIT has been proposed as one of the criteria for the selection of new probiotic strains. As BSHs can combat the deleterious effects of bile, BSH-mediated BA metabolism is an important function to assist microbial survival, gut persistence and colonization in the GIT (Pumbwe et al., 2007; Joyce et al., 2014b; Prete et al., 2020). In some pathogens, including Enterococcus faecalis (Shankar et al., 2002), L. monocytogenes (Begley et al., 2005) and B. abortus (Delpino et al., 2007), BSH activity has been identified as a virulence factor that influences bacterial persistence and dissemination in the host intestine. Accumulating evidence also suggests that BSHs improve mucosal adhesion and colonization abilities of probiotic strains in the GIT (Núñez-Sánchez et al., 2022), such as L. reuteri (Rodes et al., 2013), L. plantarum (Yang et al., 2019) and L. plantarum AR113 (Wu et al., 2022), further highlighting their significance as colonization factors (Jones et al., 2008; Grimm et al., 2014). Heterologous expression of bshA from L. acidophilus NCFM or bshB from L. johnsonii NCK88 in Escherichia coli C600 helped to significantly increase the colonization biomass of recombinant bacterium in the feces of germ-free mice compared with that of wide-type strain (Dimarzio, 2016). Conversely, BSHs from L. gasseri and Blautia obeum (Alavi et al., 2020) are even detrimental to the colonization of these strains and other pathogens in the GIT, contradicting their assumed roles (Moser and Savage, 2001). Therefore, the impact of BSH-catalyzed deconjugation on the adhesion and colonization of host strains is context-specific and depends on the BA core, the conjugated amino acid, and the BSHs (Foley et al., 2021). Further investigations are highly warranted to gain new insights into substrate-gene-performance, and to enable the engineering of probiotics to regulate their colonization abilities.
The mechanism underlying the contribution of BSHs to the fitness and colonization of probiotics is yet to be explored, which may include enhancing the adhesion abilities of host strains (Yang et al., 2019). As cell surface hydrophobicity is sometimes correlated with adhesion, BSH-positive L. plantarum PYPR1, which showed good surface hydrophobicity, could effectively adhere to epithelial cells and colonize (Yadav et al., 2016). The second mechanism could involve the incorporation of cholesterol moiety into bacterial membranes, which may change their fluidity, permeability, tensile strength and net charge, thus improving the colonization ability and survival of these microbes in the gut by enhancing bile tolerance and sensitivity to host defensins (Taranto et al., 2003). In vivo studies have demonstrated that the production of MCBAs positively correlated with the colonization of BSH-active bacteria (Rimal et al., 2024). It has also been shown that BSH activities of intestinal Bifidobacteria can increase the hydrogel-forming abilities of certain BAs, which may improve the colonization ability and survival of these bacteria in the GIT (Jarocki and Targoński, 2013; Jarocki et al., 2014). Recent evidence suggests that substrate specificities of BSHs from L. acidophilus and L. gasseri govern their stereospecific interaction mechanisms’ fitness and host colonization by limiting bacterial death when exposed to GCDCA (Foley et al., 2021), which is another strategy for Lactobacilli to adapt to their host niche (O'Flaherty et al., 2018). Besides BSHs, there are several other colonization-associated genes which can be divided into two categories: intestinal tissue-anchored elements and signaling molecules (Duary et al., 2012; Xiao et al., 2021). Thus, BSH-promoted strategy may work alongside with other mechanisms to help microbes to colonize the host GIT.
Bile acids can be utilized as nutrients, electron acceptors and environmental signals by the intestinal microbiota (Urdaneta and Casadesús, 2017). Amino acids released from BA deconjugation may be further converted into carbon dioxide, ammonium and sulfate, which serve as carbon and nitrogen sources for bacterial sustenance and survival (Tanaka et al., 2000; Ridlon et al., 2016). Huijghebaert and Hofmann (1986) and Van Eldere et al. (1988) found that BSH-positive strains of Clostridium utilized taurine as an electron acceptor and their growth rates were increased in the presence of tauroconjugated BAs and taurine. It has also been demonstrated that transcription of the B. longum bsh gene is coupled to a homolog of glnE that encodes a glutamine synthetase adenyltransferase that forms part of the nitrogen regulation cascade (Tanaka et al., 2000). Based on these findings, it was concluded that BA deconjugation may confer a nutritional advantage to hydrolytic strains. However, the sulfonic moiety of taurine can be dissimilated to hydrogen sulfide which is toxic to both bacteria and human host (Jarocki et al., 2014), which may refute this hypothesis. Consequently, the nutritional roles of BA deconjugation require further investigation.
The fluidity, composition, permeability, hydrophobicity, and net charge of bacterial membranes all determine the extent of damage by host defensins. Since BAs could disorganize the structure of cell membranes to display antimicrobial activity (Bustos et al., 2018), by changing the BA pool, BSH expression regulates gut bacterial membrane integrity and exposure to membrane-damaging BAs (Foley et al., 2021; McMillan et al., 2024), overall membrane composition and host cell internalization (Marchesini et al., 2011). Additionally, BSHs also facilitate the incorporation of BAs or cholesterol into bacterial membranes (Taranto et al., 2003; Bi et al., 2016), which may increase their tensile strength or change their fluidity and net charge, and ultimately improve the colonization and survival of these microbes in the gut (Taranto et al., 2003). Taken together, BSHs can directly and indirectly alter bacterial membranes, but the exact mechanism of membrane disruption likely varies in a substrate-dependent manner (Foley et al., 2021) and needs further extensive investigations.
Over the past few decades, the overuse and misuse of antibiotics have led to alterations in the intestinal microbiota balance and the rapid emergence of antibiotic-resistant pathogens (He et al., 2012). As a global public health concern, antibiotic resistance has become an increasingly serious obstacle in the treatment of infections (Korpela et al., 2016). It has been found that gut microbial BA metabolism influences the solubility and transport of poorly water-soluble drugs (Enright et al., 2018), and BA-activated receptors also play critical roles in the regulation of drug metabolism and detoxification (Li and Chiang, 2014). Under simulated gastric conditions, He et al. showed that deconjugated BAs produced by BSH hydrolysis could destruct cellular membrane permeability of selected foodborne pathogens, thus increasing their antibiotic susceptibility and eventually effectively inhibiting their growth (He et al., 2012). A more recent study demonstrated that use of macrolide in pre-school children could cause decrease in BSH activity and increase in macrolide resistance, also identifying a significant correlation between BSH enzyme and drug resistance (Korpela et al., 2016). Analogously, BSH was reported to be potentially involved in ameliorating multidrug resistance through the generation of unconjugated BAs with the capacity to access and inhibit P-gp ATPase (Enright et al., 2018). However, since BSHs are phylogenetically and structurally related to penicillin acylase, another member of Ntn hydrolase superfamily, some of them (like L. paragasseri JCM 5343T BSH) are bifunctional enzymes that degrade both BAs and β-lactam antibiotics (Kusada et al., 2022), thereby conferring the host gut bacteria antibiotic resistance and improving their survival in the mammalian intestine. More in vivo studies are therefore needed to decipher the exact roles that BSHs play in drug resistance. Anyway, these findings may provide guidance in the design of effective BSHs-based probiotic-antibiotic combination therapies for the improvement of intestinal microbiota balance.
Gut microbiome plays a prominent role in signal transduction via transforming primary BAs into secondary BAs by BSH deconjugation. In bacteria, many natural phenomena including bioluminescence, virulence, biofilm formation, and antibiotic production are now known to be regulated through quorum sensing. Acyl homoserine lactones (AHLs) serve as main signal molecules among many gram-negative bacteria, and quorum sensing mediated by these AHL molecules can be quenched by AHL acylases, which has been regarded as a new antimicrobial strategy (Mukherji and Prabhune, 2015). Because AHL acylases and BSHs both belong to the Ntn hydrolase superfamily, they show similar mode of action, and it has been recently demonstrated that BSH enzymes possibly participate in the disruption of quorum sensing and establishment of pathogenesis (Mukherji and Prabhune, 2015). Therefore, BSHs are involved in both pathogenic and probiotic roles, and have been recognized as “niche factors.”
There are now hundreds of known modifications to BAs and thousands of BA-associated genes, which greatly expand the chemical diversity of the BA pool (Mohanty et al., 2024a). Among them, increased BSH activity has been shown to influence BA composition through FXR, a ligand activated transcription factor highly expressed in the liver and intestine which controls hepatic BAs biosynthesis and enterohepatic circulation (Xie et al., 2020; Chiang and Ferrell, 2022). Reduced FXR activity increases the synthesis of BAs from cholesterol via the rate-limiting enzyme CYP7A1 and promotes the downregulation of the small heterodimer partner (SHP) which in turn activates the liver X receptor (LXR) and upregulates the adenosine triphosphate-binding cassette transporters G5 and G8 (ABCG5/8), thus promoting the conversion of cholesterol into BAs (Liu et al., 2021; Chiang and Ferrell, 2022). Most endogenous BAs bind and activate FXR, however, glycoursodeoxycholic acid in humans and tauro-β-muricholic acid in mice serve as FXR antagonists, which could counterbalance the homeostasis of BAs (Joyce et al., 2014a; Xie et al., 2020).
As predicted by WHO, cardiovascular diseases (CVDs) will remain the leading cause of death by 2030, affecting approximately 23.6 million people around the world (Ekong, 2021). Hypercholesterolemia is considered as one of the major risk factors for CVDs, and even a small reduction in serum cholesterol of 1% can reduce risk of coronary heart disease by 2–3%. Although pharmaceuticals are available to treat this disease, they are often expensive and suboptimal, and have unwanted side effects. In 1974, Mann and Spoerry (Mann, 1974) observed that the serum cholesterol levels of African Maasai men are lower than normal people after long-term consumption of fermented milk containing probiotics. Since then, oral administration of probiotics has increasingly been reported to have both preventive and therapeutical hypocholesterolemic effects (Jones et al., 2013), some of which can reduce serum cholesterol levels by as much as 22–33% (De Smet et al., 1998). Some in vivo clinical trials have also shown that oral consumption of probiotics or products containing them reduce both cholesterol and blood lipid concentrations in hypercholesterolemic subjects, while levels of triglycerides and high-density lipoprotein remain unchanged (Malpeli et al., 2015). Therefore, much effort has been devoted to developing BSH enzymes and BSH-producing probiotics as cholesterol-lowering agents.
With regard to the cholesterol removal effects of probiotics, several possible mechanisms have been proposed (reviewed in Jones et al., 2013; Horáčková et al., 2018): (1) co-precipitation of cholesterol with deconjugated BAs, on the one hand, deconjugated BAs produced by BSH hydrolysis are less-soluble under acidic conditions (pH < 0.6) and co-precipitate with cholesterol; on the other hand, deconjugated BAs cannot be reabsorbed by the intestines and are excreted in the feces, which will increase the de novo synthesis of BAs from cholesterol, thus leading to hypocholesterolemic levels (Horáčková et al., 2018); (2) cholesterol assimilation, cells directly bind cholesterol from the media to cellular surface under anaerobic conditions (Widodo et al., 2021), which makes less cholesterol available for absorption. Although several controversies exist, this mechanism has been shown to correlate with BSH activity and the parameters of bile resistance (Aswal et al., 2023); (3) synergistic effects of co-precipitation and bacterial assimilation (Kumar et al., 2012); (4) incorporation of cholesterol into cellular membrane (Miremadi et al., 2014), because co-precipitation of cholesterol with deconjugated BAs cannot occur in vivo since the intestinal pH is usually higher than 6.0 (Reis et al., 2017), probiotics may rely on this mechanism to remove cholesterol; (5) other mechanisms: conversion of cholesterol into coprostanol by cholesterol reductase of the strains (Horáčková et al., 2018), inhibition of Niemann-Pick C1-like 1 (NPCL1) protein (Jones et al., 2013), and production of short chain fatty acids, prebiotics that inhibit hepatic cholesterol synthesis (Ashaolu et al., 2021).
However, most of the hypotheses raised to date are based on in vitro experiments, and few attempts have been made to evaluate the possible hypocholesterolaemic mechanisms based on in vivo trials. Furthermore, the cholesterol-lowering efficacy of probiotics are influenced by many factors, such as the concentration of BAs, strains used and viable count, which complicates the elucidation of the exact mechanism underlying. But modification of BA metabolism through BSH activity is considered to be the core mechanism of the anti-cholesterolaemic effects (Agolino et al., 2024). To date, numerous studies have linked BSHs to hypercholesterolemia in mice and humans (reviewed in Jones et al., 2013; Reis et al., 2017). De Smet et al. were the first to explicitly demonstrate the efficacy of a BSH-active probiotic for lowering low-density lipoprotein (LDL) cholesterol levels in an animal model by increasing fecal BAs output (De Smet et al., 1998), which is the major pathway to remove cholesterol (Jones et al., 2012a). Subsequent trials in humans and mice have reported similar effects from oral administration of BSH-producing Lactobacilli strains (Jones et al., 2012a; Jones et al., 2012b; Wang et al., 2019) and immobilized BSH enzyme from L. buchneri ATCC 4005 (Sridevi et al., 2009). Importantly, the cholesterol-lowering effects of many BSH-positive Lactiplantibacillus strains were related to increased hepatic BA synthesis mediated by the FXR signaling pathway via upregulation of hepatic CYP7A1 in mice (Wang et al., 2019; Liu et al., 2021; Zhu et al., 2022; Zhao et al., 2024). More recently, follow-up trials reported that changes in BA composition and cholesterol synthesis were correlated with fibroblast growth factor 19 (FGF19, a key mediator of FXR signaling) in L. reuteri (Martoni et al., 2015), and gut-liver FXR-FGF19 axis in L. delbrueckii (Hou et al., 2020). BSH enzymes with different substrate specificities can modulate bile acid profiles differently and, consequently, have different mechanisms for cholesterol removal (Agolino et al., 2024). For instance, a BSH mutant F67A, which exclusively hydrolyzed TCA, reduced serum cholesterol levels in high-fat diet (HFD) mice through the modulation of intestinal FXR pathway, while another mutant YB81 preferred to hydrolyze GCA and reduced cholesterol levels by regulating changes in the intestinal flora and BSHs within the flora (Zhao et al., 2024). Therefore, mechanisms underlying the cholesterol-lowering effects of BSHs in vivo include elevated fecal excretion of deconjugated BAs, inhibition of hepatic cholesterol synthesis via FXR signaling, and induction of cellular LDL cholesterol uptake (Jones et al., 2012a). In addition to BSHs, gastrointestinal cholesterol efflux system encoded by ABCG5/8 (Jones et al., 2013) and S-layer protein (Guo et al., 2018) also contribute to the hypocholesterolemic activity of BSH-active probiotics.
CVDs are a group of disorders of the heart and blood vessels, and their evolution and progression depend on multiple factors, like hypercholesterolemia, dyslipidemia, diabetes mellitus, hypertension, smoking, etc. Among them, hypercholesterolemia caused by elevated levels of total and LDL cholesterol constitutes an important cardiovascular risk factor (Neverovskyi et al., 2020). Oral administration of L. plantarum 299v has already been shown to reduce CVD risk factors in smokers (Naruszewicz et al., 2002). However, there are limiting data regarding the clear-cut effects of BSHs on host cardiovascular risk. Recently, several in vivo studies with human beings have found that the total relative activity of gut bacterial BSH is negatively correlated with cardiovascular risk, total and LDL cholesterol levels, as well as the risk of dyslipidemia (Jia et al., 2020; Neverovskyi et al., 2020). The deconjugated BAs produced by BSH hydrolysis can regulate cardiovascular function through nuclear receptors (FXR, PXR and LXR) and G-protein-coupled receptors (TGR5 and muscarinic receptors) (Zhang et al., 2024).
Since deconjugated BAs are less efficient than their conjugated counterparts in the emulsification of dietary lipids, BSHs may cause disorders of lipid digestion in the hosts, thus leading to weight loss of the host (Huang et al., 2019). Numerous in vitro, in vivo and clinical trials have pointed to the fact that BSH-encoding strains and BSH enzymes directly reduced fat absorption, thus serving as functional foods for the treatment of obesity. An in vitro study found that BSH-producing L. plantarum AR113 and L. casei pWQH01 decreased hepatic lipid accumulation, while BSH-negative L. casei LC2W did not exert this beneficial effect (Huang et al., 2020). In mice, rats and hamsters, oral consumption of BSH enzymes, including recombinant BSH (Spratt et al., 2014), as well as BSH-active probiotics like L. plantarum H-87 (Liang et al., 2021) and L. acidophilus GOLDGUT-LA100 (Zheng et al., 2024) could inhibit their liver fat deposition, lipid digestion and body weight, eventually alleviating HFD-induced obesity. Using a controlled experimental system, Joyce et al. reported in vivo in animal models that increased BSH expression in the gut ecosystem led to lower body weight gain, lower adiposity and reduction of both circulating serum cholesterol and liver triglycerides (Joyce et al., 2014a). A systematic review and meta-analysis of randomized clinical trials showed that administration of probiotics counteracted some aspects of dislipidemia in hypercholesterolemic patients (Jiang et al., 2020). These findings ultimately suggest that BSH enzyme is a potential target for weight regulation and could be applied in the design of intervention strategies in humans and the agricultural sector. However, direct evidence supporting the significant role of intestinal BSHs in host lipid metabolism, energy harvest, as well as body weight change is still very limited. It has been shown that BSH from L. johnsonii LB1 (Dimarzio, 2016) does not affect the weight gain of mice, whereas BSH-deleted B. thetaiotaomicron causes significant weight loss in mice (Yao et al., 2018). Besides, the antioxidant tempol (Li et al., 2013), an Atlantic brown algae extract (Huebbe et al., 2017), theabrownin from Pu-erh tea (Kuang et al., 2020), and resistant starch (Li et al., 2024), which could reduce the BSH activities of gut microbiome, also remarkably ameliorate diet-induced obesity in mice or humans. These inconsistent findings thus call for more in-depth studies to decipher the BSH-mediated anti-obesity effect since it is more complex and influenced by many factors, such as diet, host, BSH activity, and bile acids.
The mechanism underlying weight regulation of microbial BSHs needs to be further elucidated. It has been reported that BSH activity in the gut regulates host lipid metabolism by impacting the expression of genes involved in cholesterol transport as well as lipid transport and synthesis in the duodenum, ileum and liver, such as Ppary and Angptl4 (Joyce et al., 2014a), and the fasting-induced adipose factor (FIAF) (Kersten, 2023). Some recent studies suggest that BSH activity alters gut microbiota and reduces obesity in mice through an FXR-mediated mechanism (Li et al., 2013; Liang et al., 2021), while unconjugated BAs affect TGR5-mediated adipose tissue development and weight loss (Svensson et al., 2013), and that there is a subtle interplay between FXR and TGR5 that warrants further investigations. Besides, BSH activity and unconjugated BAs also regulate the expression patterns of host circadian rhythm (e.g., Dbp) and other genes central to circadian clock (Joyce et al., 2014a; Joyce et al., 2014b; Govindarajan et al., 2016), which is clearly related to alterations in weight regulation and energy metabolism (Boege et al., 2021), and its disturbance causes the pathogenesis of obesity, inflammatory bowel diseases (IBD) and several liver diseases (Li and Chiang, 2014). Notably, obesity development is a complex physiological issue, and BSH-mediated bile salt metabolism is only one of several potential mechanisms by which microbiota affect host weight gain and energy harvest (Joyce et al., 2014a). Direct and controlled approaches are therefore required to obtain complete understanding of BSH-mediated regulation of host lipid metabolism and weight gain.
Due to the altered bile acid profile and abnormal lipid metabolism, deconjugation of bile salts by BSHs results in disruption of normal intestinal conditions and development of many intestinal diseases, such as short bowel syndrome. It has also been proposed that unconjugated BAs produced by BSHs are further modified into DCA, LCA and other secondary BAs by bacterial 7α-dehydroxylase (Figure 1), which may promote colorectal cancer, cause DNA damage and reactive oxygen species-associated oxidative stress in epithelial intestinal cells, or lead to impaired colonic mucosal function that would cause diarrhea, inflammation, and some other GIT diseases (Sun and Kato, 2016). Compared to its primary counterpart, DCA exhibits a 10-times higher antimicrobial activity, thus inhibiting the growth of many bacteria and altering gut microbial profiles (Liong et al., 2015). More hydrophobic secondary BAs also promote cholesterol crystallization, and the crystallized cholesterol together with bile pigments and calcium salts form gallstones (Thamer, 2022). Fortunately, the most commonly used probiotics (Lactobacilli and Bifidobacteria) cannot dehydroxylize deconjugated BAs, so they will mainly be precipitated and eliminated from body via feces, indicating no safety concern associated with these strains (Ahn et al., 2003). In contrast to these detrimental effects of BSHs on normal intestinal conditions, recent research indicated that B. fragilis BSH can alleviate necrotizing enterocolitis via restoring gut microbiota dysbiosis and BA metabolism balance (Chen et al., 2024), which necessitate an intense study into gut microbial BSHs to better understand their roles in the pathogenesis and treatment of intestinal disorders.
BSH-mediated deconjugated BAs are recognized by host-encoded receptors as signaling molecules to regulate host immunity, since they can modulate a variety of intestinal effectors, including the dendritic cell differentiation and inducible nitric oxide synthase (Joyce et al., 2014b), and antimicrobial peptide RegIIIγ produced by intestinal paneth cells (Joyce et al., 2014a). An in vivo study showed that BSH-active strain L. mucosae DPC 6426 could modulate the immune system of mouse (Ryan et al., 2019). However, little information is available with regard to the effects of BSHs on regulating immune homeostasis, and further studies in this direction are necessary.
BA homesostasis modulates glucose and lipid metabolism via gut-liver axis and gut-adipose axis, respectively, and dysregulation of BA metabolism and circadian disturbance cause the pathogenesis of liver metabolic diseases including steatosis, type 2 diabetes, obesity, liver cirrhosis, hepatic cancer and non-alcoholic fatty liver disease (NAFLD) (Jia et al., 2020; Bourgin et al., 2021; Chiang and Ferrell, 2022). It has been shown that BSH enzymes participate in the pathogenesis of many liver diseases, including NAFLD, cholestasis, and colorectal cancer. In NAFLD patients, total fecal BA concentrations are generally elevated, whereas suppression of bacterial BSH activity by its inhibitor caffeic acid phenethyl ester can alleviate NAFLD through inhibiting intestinal FXR signaling (Zhong et al., 2023). Supplementation of BSH-producing L. plantarum Y15 could effectively ameliorate liver histopathological changes in mice, including relatively weak fat damage and unobserved small focal necrosis (Liu et al., 2021). Cholestasis refers to impaired bile flow from the liver to the intestine, and it significantly reduces BSH gene abundance and enzymatic activity in preterm neonates, indicating the requirement of this enzyme in early neonatal development (Lynch et al., 2023). Recently, it has been demonstrated that ursodeoxycholic acid, which is commonly used to treat cholestasis, enriches intestinal BSH-producing Bacteroidetes in intrahepatic cholestatic pregnancy (Ovadia et al., 2020). Therefore, BSH-active Bacteroidetes coupled with ursodeoxycholic acid can be used to cure this disease. As liver tumorigenesis is suppressed through an immune pathway which is stimulated by gut-derived primary BAs, but not secondary BAs (Ma et al., 2018), the overexpression of the bsh gene from B. fragilis NCTC 9343 in B. fragilis 638R, which led to the increase of unconjugated BAs in the colon, accelerated the progression of colorectal cancer under HFD treatment (Sun L. et al., 2023). Thus, modulating BSH activity to limit secondary BA metabolism would be a promising therapeutic for the prevention and treatment of colorectal and hepatic cancers.
As key modulators of the microbiota-gut-brain axis, BAs can enter the systematic circulation and across the blood–brain barriers, and subsequently display neuroprotective potential against several neurological diseases by targeting endogenous receptors, such as Alzheimer’s disease (Wu et al., 2024), major depressive disorder (Sun et al., 2022), etc. Besides, NAFLD also affects the development of neurological diseases via BA signaling, while inflammatory and systematic metabolic disorders in the brain are regulated by FXR and TGR5 (Ren et al., 2022). Recently, it has been found that the endogenous bile acid, tauroursodeoxycholic acid, may be an effective therapeutic to prevent and treat Alzheimer’s disease (Song et al., 2024). Besides, CDCA also exerts antidepressant effect through FXR signaling and it has become a biomarker and target potentially important for the diagnosis and treatment of major depressive disorder (Li et al., 2023). Therefore, BSHs may be linked to neurological diseases, and administration of BSH-active probiotics and BSH enzymes would become novel therapies to treat these neurological disorders by targeting gut microbiota.
Generally, BSH activity has been widely recognized as a functional probiotic biomarker due to its beneficial effects on GIT microbiota and host health, such as microbial bile tolerance, antimicrobial activity, colonization in the GIT, and cholesterol reduction (Miremadi et al., 2014). Bile salt hydrolase gene has been used as a potential food-grade selection marker for the construction of novel vectors for lactic acid bacteria (Yin et al., 2011). The TG motifs of BSH B and C from L. johnsonii PF01 can enhance the promoter strength, which makes them good candidate promoters for the construction of an E. coli-Lactobacilli shuttle vector (Chae et al., 2013). BSH-encoding genes also serve as valuable molecular markers for phylogenetic studies, and specific identification and selection of probiotics (Jarocki and Targoński, 2013), as well as detecting the contamination of E. faecalis in fermented foods (Yoon et al., 2020).
Gut microbiota-derived BSH activity act as a serum metabolomic biomarker, and its changes have been linked to many human diseases, such as hepatobiliary disease (Parasar et al., 2019), Crohn’s disease (Wang Y. et al., 2021), and non-obese liver fibrosis (Lee et al., 2020). Recently, chemoproteomic tools (Parasar et al., 2019), the non-invasive diagnostic tool (Khodakivskyi et al., 2021), and several BSH activity-based probes (Han et al., 2024) have been developed to quickly and cost-effectively assess and quantify BSH activity across a broad range of biological settings including pure enzymes and bacteria, intact fecal slurries, noninvasive imaging in live animals, and the entire gastrointestinal tract of mice and humans, which facilitate the prediction and diagnosis of the clinical status of these diseases. Taken together, BSHs can be used as biomarkers for the construction of food-grade vectors, selection of probiotics and medical diagnosis.
Partly due to their cholesterol-lowering effects through enterohepatic axis regulation, BSH-producing probiotics have now been largely used as hypocholesterolemic food supplements or “drugs” in this day and age (Palaniyandi et al., 2020). A growing number of people with hypercholesterolemia have used these products containing so-called “friendly” bacteria. In 2012, Cardioviva ™ (containing L. reuteri NCIMB 30242) was launched, the first and only probiotic on the market that has been clinically proven to naturally reduce total and LDL cholesterol levels by 9 and 11.6% in hypercholesterolaemic adults, respectively (Jones et al., 2012a; Jones et al., 2012b). More recently, administration of probiotic capsules containing the BSH-active L. plantarum ECGC 13110402 (LPLDL®), a commercialized probiotic bacterium, could significantly reduce total cholesterol levels in hypercholesterolemic subjects compared to the placebo group (Keleszade et al., 2022). In the future, more biotherapeutic agents, including fermented foods and additives of healthy diets such as yogurt harboring BSH-active Lactobacilli, will be developed to overcome hypercholesteremia.
Giardia duodenalis (also known as G. lamblia or G. intestinalis) is responsible for giardiasis, one of the most common and widely spread intestinal parasitic diseases worldwide, affecting both humans and animals. The spread of resistant parasite strains and the lack of appropriate medications urgently call for the development of novel therapeutic strategies. Recent findings suggested that BSH from the probiotic strain L. johnsonii La1 exhibited anti-giardial activities both in vitro and in vivo (Allain et al., 2017), and BSH activity has been included among the selection criteria for identifying anti-Giardia Lactobacilli strains. This effect is related to the generation of deconjugated BAs, which are toxic to the parasite in a dose-dependent manner, thus displaying a significant deleterious effect on the parasite. In another work, the anti-giardial effects of various Lactiplantibacillus strains were tested, and BSH-producing L. johnsonii La1 and L. gasseri CNCM I-4884 showed anti-parasitic abilities in vitro (Allain et al., 2018). However, in vivo studies demonstrated that L. gasseri CNCM I-4884 other than L. johnsonii La1 could significantly reduce G. duodenalis infection in a suckling mice model (Allain et al., 2018). Therefore, further experiments are highly warranted to investigate the potential of BSHs in treating giardiasis and the possible mechanisms underlying. Knowledge gained here represents a step toward the development of new prophylactic strategies to combat G. duodenalis in both humans and animals.
Clostridium difficile infection (CDI), which is caused by gut microbiota dysbiosis due to consumption of antibiotics, is one of the most common nosocomial gastrointestinal infections. CDI includes a broad range of disorders ranging from diarrhea to colitis and toxic megacolon, and their incidence, severity and costs are continuously increasing (Bourgin et al., 2021). The germination process of this bacterium is mediated by a host-derived molecule including BA sensing through the germinant receptor (Zhu et al., 2018). Matthew et al. found that BSHs with varying substrate preferences restricted C. difficile spore germination and growth in vitro, and colonization in pre-clinical in vivo models (Foley et al., 2023). Besides, B. ovatus SNUG 40239 also inhibit the growth of C. difficile by its BSH activity in vitro (Yoon et al., 2017). In vitro studies also showed that BSH-mediated hydrolysis of TCA effectively suppressed C. difficile germination, and administration of E. coli cells expressing highly active BSH contributed to the efficacy of fecal microbial transplantation in the treatment of CDI (Mullish et al., 2019). These results will facilitate the development of novel preventative or bacteriotherapy strategies targeting gut microbial BSHs to cure CDI. However, C. difficile also contains a BSH enzyme which favors taurine-conjugated BAs (Aguirre et al., 2022), highlighting the need for greater understanding the function of bacterial BSHs in the treatment of CDI.
IBD is a chronic inflammatory condition of the GIT characterized by a dysregulation of the gut mucosal immune functions and a dysbiotic gut microbiota occurring in genetically susceptible hosts. It encompasses two major phenotypes: ulcerative colitis and Crohn’s disease. Patients affected by IBD display low levels of gut microbiome-associated BSH activity (Jia et al., 2020), and many BSH-producing probiotics have been used as dietary supplements or drugs for the management of IBD (Gadaleta et al., 2022). In vivo studies showed that the probiotic candidate strain B. dorei BDX-01 (Sun X. et al., 2023), and BSH eznymes from L. plantarum AR113 (Feng et al., 2023) could ameliorate dextran sulfate sodium induced colitis in mice by regulating intestinal BSH activity and the FXR-NLRP3 signaling pathway, and antibiotic treatment could not abolish the protective effect of the former (Sun X. et al., 2023).
Obesity, one of the main causes of metabolic syndromes, is usually reflected as body fat accumulation, liver abnormalities, dyslipidemia and insulin resistance. It is also a group of inter-related metabolic conditions that greatly increase the risk of developing CVDs, diabetes and cancers (Gil-Rodríguez and Beresford, 2021). Nowadays, obesity has become a worldwide health problem, creating an urgent demand for its effective prevention and treatment. Although factors, such as diet and genetic background, contribute significantly to the prevalence of obesity, many studies have linked the gut microbiome to obesity and weight gain in humans and animals (Joyce et al., 2014b). Consequently, manipulation of gut microbiota might be a promising strategy to control obesity.
As unconjugated BAs are less efficient than conjugated molecules in the emulsification of dietary lipid and the formation of micelles (Huang et al., 2019), BSH enzymes and BSH-enriched microbiota are negatively related to the body weight gain and energy storage of the hosts (Korpela et al., 2016; Liang et al., 2021). Enhanced in situ BSH activity of gut microbiome not only causes weight loss in conventionally raised mice (Joyce et al., 2014a), but also lowers serum cholesterol levels in humans (Jones et al., 2008). Therefore, BSH enhancers have become one of the key mechanisms in the anti-hyperlipidemic and anti-hypercholesterolemic effects and have a potential application in human health. Thus, enhancing gut microbe-enriched BSH activity by dietary supplementation of highly BSH-active probiotics and BSH enhancers may offer potential as a biological alternative to pharmaceutical interventions to prevent and treat obesity and hypercholesterolemia.
Antibiotic growth promoters (AGPs) are a group of antibiotics used at subtherapeutic levels to improve the average daily weight gain and feed conversion efficiency in agricultural animals (Ayalew et al., 2022). Since the 1950s, AGPs have been successfully used by agricultural animal producers to improve growth performance. However, recent epidemiological studies strongly indicated that long-term and improper uses of AGPs resulted in the emergence of antimicrobial resistance and antibiotic residues in animal products, leading to food safety and public health threats (Ayalew et al., 2022). Consequently, there is a global trend to ban the use of AGPs in animal production, which necessitates the need to develop effective non-antibiotic alternatives to improve animal performance.
Numerous studies have shown that the growth-promoting effects of AGPs are inversely correlated with the decreased BSH activities of gut microbes (Smith et al., 2014; Rani et al., 2017) or the reduced abundance of BSH-producing bacteria in the small intestine (Robinson et al., 2019). As previously described, deconjugation of BAs by gut microbiota-derived BSHs results in lipid malabsorption and attenuates energy harvest, which may cause weight loss of the hosts (Joyce et al., 2014a; Korpela et al., 2016; Liang et al., 2021). Conversely, inhibiting the activities of intestinal BSHs can enhance lipid metabolism and energy harvest, thus improving the growth performance and feed efficiency of food animals (Bustos et al., 2018). Therefore, specific BSH inhibitors have been proposed as promising alternatives to AGPs for fattening food animals.
To date, various types of BSH inhibitors have been developed as novel alternatives to AGPs to enhance the productivity and sustainability of food animals. As conventional BSH inhibitors, high concentrations of cooper (CuCl2) and zinc (ZnSO4) have been reported to promote food digestion and body weight gain in different animal models (Liu et al., 2018). But long-term use of these metal ions will increase their accumulation in treated animals and the environment. Iodine reagents, such as sodium periodate, sodium iodate and potassium iodate, could also strongly inactivate BSHs (Smith, 2013), but their safety remains to be determined.
To discover more potent BSH inhibitors with satisfying safety profiles, many efforts have been devoted to screening from both natural and synthetic compounds. Several natural plant extracts, such as 2α-OH-protopanoxadiol (Xie et al., 2020), amentoflavone (Li et al., 2022a), licochalcone C and isobavachalcone (Li et al., 2022b), as well as green tea and B. vulgaris root extracts (Dibamehr et al., 2021) exhibit inhibitory effects against BSH enzymes or BSH-producing bacteria both in vitro and in vivo, thus serving as potential alternatives to AGPs. Using a high-throughput screening method, Smith et al. identified several potent inhibitors of L. salivarius NRRL B-30514 BSH from 2,240 various compounds, including riboflavin, caffeic acid phenethyl ester and carnosic acid (Smith et al., 2014). Thereafter, efficacies of these three inhibitors were evaluated using a chicken model (Geng et al., 2020). Among them, riboflavin and caffeic acid phenethyl ester could also inhibit the activities of BSHs from L. acidophilus PF01 (Lin et al., 2014) and L. gasseri FR4 (Rani et al., 2017) which have significantly different protein sequences and substrate spectrum. Besides, riboflavin has already been approved by the FDA of United States of America for the use as a feed additive to overcome vitamin B2 deficiencies in animals (Smith et al., 2014), and this compound alone or in conjunction with L. gasseri FR4 could also enhance the body weight of pigs and feed efficiency (Rani et al., 2017). Actually, there are many different subtypes of gut microbial BSHs with various substrate specificities (O'Flaherty et al., 2018), but the inhibitors obtained exhibit either moderate inhibitory or no inhibitory activities toward other BSHs, which greatly limits their practical usages.
Although BSH protein sequences vary largely among different gut strains, all BSHs possess six highly conserved active-site residues that include the catalytic cysteine (Cys2) (Dong and Lee, 2018). Based on this structural conservation, Adhikari et al. developed a covalent pan-inhibitor GR-7 which targeted both B. longum and B. thetaiotaomicron BSHs in a dose-dependent manner (Adhikari et al., 2020). This compound could effectively inhibit the deconjugation of BAs in vitro and in vivo, and did not significantly affect the viability of gut bacteria. To reduce off-target effects and enhance the potency and gut permeability, a LCA, the second-generation gut-restricted pan-BSH inhibitor, has been developed. As compared with the first generation compound, it displayed reduced toxicity toward mammalian cells and suppressed BSH activities in complex biological samples, including purified samples, bacterial cultures and conventional mouse fecal slurries (Adhikari et al., 2021). The usage of this inhibitor also prevented the development of hepatic inflammation and pathologic intestinal permeability in rats fed a choline-deficient, L-amino acid-defined HFD (Li et al., 2022c). Improved knowledge in the roles of BSHs and BSH-producing bacteria are highly warranted to design rational tailored pan-BSH inhibitors that would enhance animal health and performance. BSH pan-inhibitors could also be cooperated with certain BSH-positive probiotics to maximize the beneficial effects of these probiotics by alleviating their potential negative impacts on host fat digestion. Since noncovalent BSH inhibitors can avoid the development of resistance mutations (Adhikari et al., 2020), in the long term, they may prove to be the most effective tool for in vivo use. Therefore, further studies may be performed to confirm their growth-promoting effects.
Gut bacterial BSH has become a key target for the manipulation of gut microbiome and host health. Targeting this enzyme by dietary supplements, including crude extract, polysaccharides, phenols, saponins, alkaloids and dietary fiber, have been shown to effectively regulate gut microbiota and BAs-FXR signaling, thus modulating human health (Kastl et al., 2022). By modulating BSH activity, several natural products, like L-theanine, capsaicin and epigallocatechin-3-gallate, have been shown to reduce glucose levels (He et al., 2024), offering novel insights and strategies for type 2 diabetes prevention and treatment. Similar results have also been observed in medical constituents, such as caffeic acid phenethyl ester (Gonzalez et al., 2016) and gentamicin (Ma et al., 2023). In addition, recombinant BSH from B. longum could enhance the inhibition efficiency of TDCA toward C. perfringens virulence in chickens (Alenezi et al., 2024).
Many recent observations have indicated that gut microbial BSHs are involved in a multifaceted array of roles, directly or indirectly in the host and microbial physiologies, thus mediating a gut microbe-host dialog. It has been increasingly shown that BSH activity can reshape the landscape of BAs, which not only influences bacterial physiology, but also regulates a wide range of physiological processes in the hosts. As key gatekeepers and mediators of BA transformation, BSHs have been currently investigated as promising target enzymes in the manipulation of gut microbiota to benefit human and animal health. Although much effort has been undertaken to elucidate the physiological functions of gut microbiota-derived BSHs, detailed studies are still required to uncover their significance more clearly. Upcoming researches of BSHs should focus on their specific roles in the gut microbiome, and beneficial or detrimental effects on the host.
Microbial BSH activity has been considered as a beneficial property that may find importance for novel preventive and therapeutic strategies for conditions associated with BA dysbiosis. Despite the claimed benefits from human clinical studies carried out for the last few decades, controversies still exist in the roles of BSHs in bile detoxification, bacterial colonization, ameliorating drug resistance, lowering serum cholesterol levels, regulation of lipid metabolism and treatment of metabolic syndromes, owing to the fact that different clinical and methodological trials have complicated the use of probiotics in reaching a decisive outcome. Therefore, more properly-designed in vivo clinical trials are urgently warranted to investigate the exact function of BSHs to clarify conflicting findings in these fields and to explore the precise underlying mechanisms to have better formulations for animal and human consumption.
The substrate preferences of microbial BSHs have been extensively reviewed (Dong and Lee, 2018). Based on their kinetic parameters and specific activities toward various substrates, most BSH enzymes prefer to hydrolyze glycol-conjugated BAs, like GCA, GDCA and GCDCA, a few BSHs show high affinity for tauro-conjugates (TCA, TDCA and TCDCA), whereas others hydrolyze both glycol- and tauro-conjugated BAs. Although many structural analyses and amino acid substitution mutagenesis have been performed, the precise substrate recognition mechanism and the structural basis for the substrate preferences of BSHs remain to be elucidated. As summarized in this review, differences in the substrate specificity of BSHs not only influence the bile-detoxifying effects, colonization ability and growth of gut microbiota, but also affect host physiology, such as lowering serum cholesterol levels and modulating CDI. Overall, substrate preferences may underlie the protective vs. pathogenic effects of BSHs in the animal and human GIT. However, it is still unclear how differences in BSH substrate specificities affect BA pool, gut microbiota and host responses. In-depth analysis of the relationship between the functions and substrate specificities of BSHs would deepen our understanding of the BSH-mediated gut microbe-host dialog, and lay a solid foundation for the rational design of next generation BSH-active probiotics with improved colonization efficacy and persistence in the host as well as their usage as BA-altering biotherapeutics in the treatment of host specific metabolic diseases.
Extensive studies on gut microbiota have suggested that BSHs are key mechanistic microbiome targets for the development of BSH enhancers and inhibitors, which are promising measures for the control of diet-induced obesity in humans and novel non-antibiotic growth promoters in farm animals, respectively. To date, although a number of BSHs have been identified from different commensal bacteria, our understanding of the BSH structure–function relationship is still limited, since BSHs of various origins have different crystal structures and substrate specificities, and some strains even have more than one BSH homolog with different substrate preferences. With the development of artificial intelligence, structural biology, protein engineering, high throughput screening system and bioinformatics, structure-based computations would not only improve our understanding of the catalytic and substrate recognition mechanisms of BSHs, but also facilitate the rational design of BSH enhancers-based weight-reducing aids or BSH inhibitors-based alternatives to AGPs. Future research in this area would be really beneficial for human and animal health.
Although the therapeutic potential of gut microbiota-associated BSHs has been proposed, there are still knowledge gaps regarding the utility of BSHs as promoters of probiotic colonization and as treatments for diseases in human, which have hampered their development. Further studies are needed to better understand the function of BSHs in regulating host metabolism to develop BSH-based biotherapeutics. Extensive advances in the new “omics” technologies (e.g., functional metagenomics, the integrative microbiome-host transcriptome, the integrated transcriptomic and proteomic analyses) and systems biology approaches have allowed us to unravel the complex interactions between gut microbiome, BAs, BSH activity, the liver, GIT and host health. With the development of chemoproteomic tools and quantitative optical readout-based methods, we could also quickly and cost-effectively quantify BSH activity in complex biological samples, thus improving our understanding of altered BSH activities on important physiological processes and diseases. Thereafter, BSH-positive probiotics can be engineered by genome-editing tools such as CRISPR-Cas to be employed as biotherapeuticals for different needs.
ZD: Writing – original draft, Writing – review & editing. SY: Investigation, Writing – original draft. CT: Writing – review & editing. DL: Writing – review & editing. YK: Writing – review & editing. LY: Conceptualization, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the Specialized Research Fund for the Doctoral Program of Nanyang Normal University (grant No. 2020ZX004), Training Program for National Natural Science Foundation of China by Nanyang Normal University (grant No. 2023Y005), and the Henan Province Natural Science Foundation (grant No. 222300420251).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adhikari, A. A., Ramachandran, D., Chaudhari, S. N., Powell, C. E., Li, W., McCurry, M. D., et al. (2021). A gut-restricted lithocholic acid analog as an inhibitor of gut bacterial bile salt hydrolases. ACS Chem. Biol. 16, 1401–1412. doi: 10.1021/acschembio.1c00192
Adhikari, A. A., Seegar, T. C. M., Ficarro, S. B., McCurry, M. D., Ramachandran, D., Yao, L., et al. (2020). Development of a covalent inhibitor of gut bacterial bile salt hydrolases. Nat. Chem. Biol. 16, 318–326. doi: 10.1038/s41589-020-0467-3
Agolino, G., Pino, A., Vaccalluzzo, A., Cristofolini, M., Solieri, L., Caggia, C., et al. (2024). Bile salt hydrolase: the complexity behind its mechanism in relation to lowering-cholesterol lactobacilli probiotics. J. Funct. Foods 120:106357. doi: 10.1016/j.jff.2024.106357
Aguirre, A. M., Adegbite, A. O., and Sorg, J. A. (2022). Clostridioides difficile bile salt hydrolase activity has substrate specificity and affects biofilm formation. NPJ Biofilms Microbiomes 8:94. doi: 10.1038/s41522-022-00358-0
Ahn, Y. T., Kim, G. B., Lim, K. S., Baek, Y. J., and Kim, H. U. (2003). Deconjugation of bile salts by Lactobacillus acidophilus isolates. Int. Dairy J. 13, 303–311. doi: 10.1016/S0958-6946(02)00174-7
Alavi, S., Mitchell, J. D., Cho, J. Y., Liu, R., Macbeth, J. C., and Hsiao, A. (2020). Interpersonal gut microbiome variation drives susceptibility and resistance to cholera infection. Cell 181:e1513, 1533–1546.e13. doi: 10.1016/j.cell.2020.05.036
Alenezi, T., Alrubaye, B., Fu, Y., Shrestha, J., Algehani, S., Wang, H., et al. (2024). Recombinant bile salt hydrolase enhances the inhibition efficiency of taurodeoxycholic acid against Clostridium perfringens virulence. Pathogens 13:464. doi: 10.3390/pathogens13060464
Allain, T., Chaouch, S., Thomas, M., Travers, M. A., Valle, I., Langella, P., et al. (2018). Bile salt hydrolase activities: a novel target to screen anti-Giardia lactobacilli? Front. Microbiol. 9:89. doi: 10.3389/fmicb.2018.00089
Allain, T., Chaouch, S., Thomas, M., Vallee, I., Buret, A. G., Langella, P., et al. (2017). Bile-salt-hydrolases from the probiotic strain Lactobacillus johnsonii La1 mediate anti-giardial activity in vitro and in vivo. Front. Microbiol. 8:2707. doi: 10.3389/fmicb.2017.02707
Ambalam, P., Kondepudi, K. K., Nilsson, I., Wadström, T., and Ljungh, Å. (2012). Bile stimulates cell surface hydrophobicity, Congo red binding and biofilm formation of Lactobacillus strains. FEMS Microbiol. Lett. 333, 10–19. doi: 10.1111/j.1574-6968.2012.02590.x
Arnold, J. W., Simpson, J. B., Roach, J., Kwintkiewicz, J., and Azcarate-Peril, M. A. (2018). Intra-species genomic and physiological variability impact stress resistance in strains of probiotic potential. Front. Microbiol. 9:242. doi: 10.3389/fmicb.2018.00242
Ashaolu, T., Ashaolu, J., and Adeyeye, S. (2021). Fermentation of prebiotics by human colonic microbiota in vitro and short-chain fatty acids production: a critical review. J. Appl. Microbiol. 130, 677–687. doi: 10.1111/jam.14843
Aswal, M., Singhal, N., and Kumar, M. (2023). Comprehensive genomic analysis of hypocholesterolemic probiotic Enterococcus faecium LR13 reveals unique proteins involved in cholesterol-assimilation. Front. Nutr. 10:1082566. doi: 10.3389/fnut.2023.1082566
Ayalew, H., Zhang, H., Wang, J., Wu, S., Qiu, K., Qi, G., et al. (2022). Potential feed additives as antibiotic alternatives in broiler production. Front. Vet. Sci. 9:916473. doi: 10.3389/fvets.2022.916473
Begley, M., Sleator, R. D., Gahan, C. G. M., and Hill, C. (2005). Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. Immun. 73, 894–904. doi: 10.1128/IAI.73.2.894-904.2005
Bi, J., Liu, S., Du, G., and Chen, J. (2016). Bile salt tolerance of Lactococcus lactis is enhanced by expression of bile salt hydrolase thereby producing less bile acid in the cells. Biotechnol. Lett. 38, 659–665. doi: 10.1007/s10529-015-2018-7
Boege, H. L., Bhatti, M. Z., and St-Onge, M.-P. (2021). Circadian rhythms and meal timing: impact on energy balance and body weight. Curr. Opin. Biotech. 70, 1–6. doi: 10.1016/j.copbio.2020.08.009
Bourgin, M., Kriaa, A., Mkaouar, H., Mariaule, V., Jablaoui, A., Maguin, E., et al. (2021). Bile salt hydrolases: at the crossroads of microbiota and human health. Microorganisms 9:1122. doi: 10.3390/microorganisms9061122
Bustos, A. Y., de Valdez, G. F., Raya, R., de Almeida, A. M., Fadda, S., and Taranto, M. P. (2015). Proteomic analysis of the probiotic Lactobacillus reuteri CRL1098 reveals novel tolerance biomarkers to bile acid-induced stress. Food Res. Int. 77, 599–607. doi: 10.1016/j.foodres.2015.10.001
Bustos, A. Y., Font de Valdez, G., Fadda, S., and Taranto, M. P. (2018). New insights into bacterial bile resistance mechanisms: the role of bile salt hydrolase and its impact on human health. Food Res. Int. 112, 250–262. doi: 10.1016/j.foodres.2018.06.035
Bustos, A. Y., Saavedra, L., de Valdez, G. F., Raya, R. R., and Taranto, M. P. (2012). Relationship between bile salt hydrolase activity, changes in the internal pH and tolerance to bile acids in lactic acid bacteria. Biotechnol. Lett. 34, 1511–1518. doi: 10.1007/s10529-012-0932-5
Chae, J. P., Valeriano, V. D., Kim, G. B., and Kang, D. K. (2013). Molecular cloning, characterization and comparison of bile salt hydrolases from Lactobacillus johnsonii PF01. J. Appl. Microbiol. 114, 121–133. doi: 10.1111/jam.12027
Chen, Z., Chen, H., Huang, W., Guo, X., Yu, L., Shan, J., et al. (2024). Bacteroides fragilis alleviates necrotizing enterocolitis through restoring bile acid metabolism balance using bile salt hydrolase and inhibiting FXR-NLRP3 signaling pathway. Gut Microbes 16:2379566. doi: 10.1080/19490976.2024.2379566
Chiang, J. Y., and Ferrell, J. M. (2022). Discovery of farnesoid X receptor and its role in bile acid metabolism. Mol. Cell. Endocrinol. 548:111618. doi: 10.1016/j.mce.2022.111618
De Smet, I., De Boever, P., and Verstraete, W. (1998). Cholesterol lowering in pigs through enhanced bacterial bile salt hydrolase activity. Br. J. Nutr. 79, 185–194. doi: 10.1079/BJN19980030
Delpino, M. V., Marchesini, M. I., Estein, S. M., Comerci, D. J., Cassataro, J., Fossati, C. A., et al. (2007). A bile salt hydrolase of Brucella abortus contributes to the establishment of a successful infection through the oral route in mice. Infect. Immun. 75, 299–305. doi: 10.1128/IAI.00952-06
Dibamehr, A., Daneshyar, M., Tukmechi, A., and Froushani, S. (2021). The effects of different plant extracts on bile salt hydrolase activity of Lactobacillus strains isolated from the gastrointestinal tract of poultry. Vet. Arhiv. 91, 89–99. doi: 10.24099/vet.arhiv.0887
Dimarzio, M. J. (2016). Hijacking host metabolism with Lactobacillus—Understanding the implications of bile salt hydrolase diversity. Doctor of Philosophy, The Pennsylvania State University.
Dong, Z., and Lee, B. H. (2018). Bile salt hydrolases: structure and function, substrate preference and inhibitor development. Protein Sci. 27, 1742–1754. doi: 10.1002/pro.3484
Dowd, G. C., Joyce, S. A., Hill, C., and Gahan, C. G. (2011). Investigation of the mechanisms by which Listeria monocytogenes grows in porcine gallbladder bile. Infect. Immun. 79, 369–379. doi: 10.1128/iai.00330-10
Duary, R. K., Batish, V. K., and Grover, S. (2012). Relative gene expression of bile salt hydrolase and surface proteins in two putative indigenous Lactobacillus plantarum strains under in vitro gut conditions. Mol. Biol. Rep. 39, 2541–2552. doi: 10.1007/s11033-011-1006-9
Ekong, H. C. (2021). A critical assessment of global and regional trends in cardiovascular disease (CVD) burden and its factors. Intercont. J. End. Sci. Technol. 5, 9–15.
Enright, E. F., Govindarajan, K., Darrer, R., MacSharry, J., Joyce, S. A., and Gahan, C. G. M. (2018). Gut microbiota-mediated bile acid transformations alter the cellular response to multidrug resistant transporter substrates in vitro: focus on P-glycoprotein. Mol. Pharm. 15, 5711–5727. doi: 10.1021/acs.molpharmaceut.8b00875
Fang, F., Li, Y., Bumann, M., Raftis, E. J., Casey, P. G., Cooney, J. C., et al. (2009). Allelic variation of bile salt hydrolase genes in Lactobacillus salivarius does not determine bile resistance levels. J. Biotechnol. 191, 5743–5757. doi: 10.1128/JB.00506-09
Feng, X., Bu, Z., Tang, H., Xia, Y., Song, X., Ai, L., et al. (2023). Bile salt hydrolase of Lactiplantibacillus plantarum plays important roles in amelioration of DSS-induced colitis. iScience 26:106196. doi: 10.1016/j.isci.2023.106196
Foley, M. H., O'Flaherty, S., Allen, G., Rivera, A. J., Stewart, A. K., Barrangou, R., et al. (2021). Lactobacillus bile salt hydrolase substrate specificity governs bacterial fitness and host colonization. Proc. Natl. Acad. Sci. USA 118:e2017709118. doi: 10.1073/pnas.2017709118
Foley, M. H., Walker, M. E., Stewart, A. K., O’Flaherty, S., Gentry, E. C., Patel, S., et al. (2023). Bile salt hydrolases shape the bile acid landscape and restrict Clostridioides difficile growth in the murine gut. Nat. Microbiol. 8, 611–628. doi: 10.1038/s41564-023-01337-7
Gadaleta, R. M., Cariello, M., Crudele, L., and Moschetta, A. (2022). Bile salt hydrolase-competent probiotics in the management of IBD: unlocking the "bile acid code". Nutrients 14:3212. doi: 10.3390/nu14153212
Geng, W., Long, S. L., Chang, Y.-J., Saxton, A. M., Joyce, S. A., and Lin, J. (2020). Evaluation of bile salt hydrolase inhibitor efficacy for modulating host bile profile and physiology using a chicken model system. Sci. Rep. (UK) 10:4941. doi: 10.1038/s41598-020-61723-7
Gil-Rodríguez, A. M., and Beresford, T. (2021). Bile salt hydrolase and lipase inhibitory activity in reconstituted skim milk fermented with lactic acid bacteria. J. Funct. Foods 77:104342. doi: 10.1016/j.jff.2020.104342
Gonzalez, F. J., Jiang, C., and Patterson, A. D. (2016). An intestinal microbiota–farnesoid X receptor axis modulates metabolic disease. Gastroenterology 151, 845–859. doi: 10.1053/j.gastro.2016.08.057
Govindarajan, K., MacSharry, J., Casey, P. G., Shanahan, F., Joyce, S. A., and Gahan, C. G. (2016). Unconjugated bile acids influence expression of circadian genes: a potential mechanism for microbe-host crosstalk. PLoS One 11:e0167319. doi: 10.1371/journal.pone.0167319
Grill, J. P., Cayuela, C., Antoine, J. M., and Schneider, F. (2000). Isolation and characterization of a Lactobacillus amylovorus mutant depleted in conjugated bile salt hydrolase activity: relation between activity and bile salt resistance. J. Appl. Microbiol. 89, 553–563. doi: 10.1046/j.1365-2672.2000.01147.x
Grimm, V., Westermann, C., and Riedel, C. U. (2014). Bifidobacteria-host interactions—an update on colonisation factors. Biomed. Res. Int. 2014:960826. doi: 10.1155/2014/960826
Guo, C. F., Zhang, S., Yuan, Y. H., Li, J. Y., and Yue, T. L. (2018). Bile salt hydrolase and S-layer protein are the key factors affecting the hypocholesterolemic activity of Lactobacillus casei-fermented milk in hamsters. Mol. Nutr. Food Res. 62:1800728. doi: 10.1002/mnfr.201800728
Guzior, D. V., Okros, M., Shivel, M., Armwald, B., Bridges, C., Fu, Y., et al. (2024). Bile salt hydrolase acyltransferase activity expands bile acid diversity. Nature 626, 852–858. doi: 10.1038/s41586-024-07017-8
Guzior, D. V., and Quinn, R. A. (2021). Review: microbial transformations of human bile acids. Microbiome 9:140. doi: 10.1186/s40168-021-01101-1
Han, L., Xu, R., Conwell, A. N., Takahashi, S., Parasar, B., and Chang, P. V. (2024). Bile salt hydrolase activity-based probes for monitoring gut microbial bile acid metabolism. Chembiochem 25:e202300821. doi: 10.1002/cbic.202300821
He, J., Liu, X., Zhang, J., Wang, R., Cao, X., and Liu, G. (2024). Gut microbiome-derived hydrolases—an underrated target of natural product metabolism. Front. Cell. Infect. Microbiol. 14:1392249. doi: 10.3389/fcimb.2024.1392249
He, X., Zou, Y., Cho, Y., and Ahn, J. (2012). Effects of bile salt deconjugation by probiotic strains on the survival of antibiotic-resistant foodborne pathogens under simulated gastric conditions. J. Food Protect. 75, 1090–1098. doi: 10.4315/0362-028X.JFP-11-456
Horáčková, Š., Plocková, M., and Demnerová, K. (2018). Importance of microbial defence systems to bile salts and mechanisms of serum cholesterol reduction. Biotechnol. Adv. 36, 682–690. doi: 10.1016/j.biotechadv.2017.12.005
Hou, G., Peng, W., Wei, L., Li, R., Yuan, Y., Huang, X., et al. (2020). Lactobacillus delbrueckii interfere with bile acid enterohepatic circulation to regulate cholesterol metabolism of growing–finishing pigs via its bile salt hydrolase activity. Front. Nutr. 7:617676. doi: 10.3389/fnut.2020.617676
Huang, C. H., Ho, C. Y., Chen, C. T., Hsu, H. F., and Lin, Y. H. (2019). Probiotic BSH activity and anti-obesity potential of Lactobacillus plantarum strain TCI378 isolated from Korean kimchi. Prev. Nutr. Food Sci. 24, 434–441. doi: 10.3746/pnf.2019.24.4.434
Huang, W. L., Wang, G. Q., Xia, Y. J., Xiong, Z. Q., and Ai, L. Z. (2020). Bile salt hydrolase-overexpressing Lactobacillus strains can improve hepatic lipid accumulation in vitro in an NAFLD cell model. Food Nutr. Res. 64:3751. doi: 10.29219/fnr.v64.3751
Huebbe, P., Nikolai, S., Schloesser, A., Herebian, D., Campbell, G., Gluer, C. C., et al. (2017). An extract from the Atlantic brown algae Saccorhiza polyschides counteracts diet-induced obesity in mice via a gut related multi-factorial mechanisms. Oncotarget 8, 73501–73515. doi: 10.18632/oncotarget.18113
Huijghebaert, S. M., and Hofmann, A. F. (1986). Influence of the amino acid moiety on deconjugation of bile acid amidates by cholylglycine hydrolase or human fecal cultures. J. Lipid Res. 27, 742–752. doi: 10.1016/S0022-2275(20)38791-5
Jarocki, P., Podleśny, M., Glibowski, P., and Targoński, Z. (2014). A new insight into the physiological role of bile salt hydrolase among intestinal bacteria from the genus Bifidobacterium. PLoS One 9:e114379. doi: 10.1371/journal.pone.0114379
Jarocki, P., and Targoński, Z. (2013). Genetic diversity of bile salt hydrolases among human intestinal Bifidobacteria. Curr. Microbiol. 67, 286–292. doi: 10.1007/s00284-013-0362-1
Jia, B., Park, D., Hahn, Y., and Jeon, C. O. (2020). Metagenomic analysis of the human microbiome reveals the association between the abundance of gut bile salt hydrolases and host health. Gut Microbes 11, 1300–1313. doi: 10.1080/19490976.2020.1748261
Jiang, J., Wu, C., Zhang, C., Zhao, J., Yu, L., Zhang, H., et al. (2020). Effects of probiotic supplementation on cardiovascular risk factors in hypercholesterolemia: a systematic review and meta-analysis of randomized clinical trial. J. Funct. Foods 74:104177. doi: 10.1016/j.jff.2020.104177
Jones, B. V., Begley, M., Hill, C., Gahan, C. G., and Marchesi, J. R. (2008). Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA 105, 13580–13585. doi: 10.1073/pnas.0804437105
Jones, M. L., Martoni, C. J., Parent, M., and Prakash, S. (2012a). Cholesterol-lowering efficacy of a microencapsulated bile salt hydrolase-active Lactobacillus reuteri NCIMB 30242 yoghurt formulation in hypercholesterolaemic adults. Br. J. Nutr. 107, 1505–1513. doi: 10.1017/S0007114511004703
Jones, M. L., Martoni, C. J., and Prakash, S. (2012b). Cholesterol lowering and inhibition of sterol absorption by Lactobacillus reuteri NCIMB 30242: a randomized controlled trial. Eur. J. Clin. Nutr. 66, 1234–1241. doi: 10.1038/ejcn.2012.126
Jones, M. L., Tomaro-Duchesneau, C., Martoni, C. J., and Prakash, S. (2013). Cholesterol lowering with bile salt hydrolase-active probiotic bacteria, mechanism of action, clinical evidence, and future direction for heart health applications. Expert. Opin. Biol. Ther. 13, 631–642. doi: 10.1517/14712598.2013.758706
Joyce, S. A., MacSharry, J., Casey, P. G., Kinsella, M., Murphy, E. F., Shanahan, F., et al. (2014a). Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. USA 111, 7421–7426. doi: 10.1073/pnas.1323599111
Joyce, S. A., Shanahan, F., Hill, C., and Gahan, C. G. (2014b). Bacterial bile salt hydrolase in host metabolism: potential for influencing gastrointestinal microbe-host crosstalk. Gut Microbes 5, 669–674. doi: 10.4161/19490976.2014.969986
Kastl, A., Zong, W., Gershuni, V. M., Friedman, E. S., Tanes, C., Boateng, A., et al. (2022). Dietary fiber-based regulation of bile salt hydrolase activity in the gut microbiota and its relevance to human disease. Gut Microbes 14:2083417. doi: 10.1080/19490976.2022.2083417
Keleszade, E., Kolida, S., and Costabile, A. (2022). The cholesterol lowering efficacy of Lactobacillus plantarum ECGC 13110402 in hypercholesterolemic adults: a double-blind, randomized, placebo controlled, pilot human intervention study. J. Funct. Foods 89:104939. doi: 10.1016/j.jff.2022.104939
Kersten, S. (2023). The impact of fasting on adipose tissue metabolism. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 1868:159262. doi: 10.1016/j.bbalip.2022.159262
Khodakivskyi, P. V., Lauber, C. L., Yevtodiyenko, A., Bazhin, A. A., Bruce, S., Ringel-Kulka, T., et al. (2021). Noninvasive imaging and quantification of bile salt hydrolase activity: from bacteria to humans. Sci. Adv. 7:eaaz9857. doi: 10.1126/sciadv.aaz9857
Korpela, K., Salonen, A., Virta, L. J., Kekkonen, R. A., Forslund, K., Bork, P., et al. (2016). Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 7:10410. doi: 10.1038/ncomms10410
Kuang, J., Zheng, X., Huang, F., Wang, S., Li, M., Zhao, M., et al. (2020). Anti-adipogenic effect of theabrownin is mediated by bile acid alternative synthesis via gut microbiota remodeling. Meta 10:475. doi: 10.3390/metabo10110475
Kumar, R., Grover, S., and Batish, V. K. (2012). Bile salt hydrolase (Bsh) activity screening of lactobacilli: in vitro selection of indigenous Lactobacillus strains with potential bile salt hydrolysing and cholesterol-lowering ability. Probiotics Antimicrob. Proteins 4, 162–172. doi: 10.1007/s12602-012-9101-3
Kusada, H., Arita, M., Tohno, M., and Tamaki, H. (2022). Bile salt hydrolase degrades β-lactam antibiotics and confers antibiotic resistance on Lactobacillus paragasseri. Front. Microbiol. 13:858263. doi: 10.3389/fmicb.2022.858263
Kusada, H., Morinaga, K., and Tamaki, H. (2021). Identification of bile salt hydrolase and bile salt resistance in a probiotic bacterium Lactobacillus gasseri JCM1131T. Microorganisms 9:1011. doi: 10.3390/microorganisms9051011
Lambert, J. M., Bongers, R. S., de Vos, W. M., and Kleerebezem, M. (2008). Functional analysis of four bile salt hydrolase and penicillin acylase family members in Lactobacillus plantarum WCFS1. Appl. Environ. Microbiol. 74, 4719–4726. doi: 10.1128/AEM.00137-08
Lee, G., You, H. J., Bajaj, J. S., Joo, S. K., Yu, J., Park, S., et al. (2020). Distinct signatures of gut microbiome and metabolites associated with significant fibrosis in non-obese NAFLD. Nat. Commun. 11:4982. doi: 10.1038/s41467-020-18754-5
Li, D. K., Chaudhari, S. N., Lee, Y., Sojoodi, M., Adhikari, A. A., Zukerberg, L., et al. (2022c). Inhibition of microbial deconjugation of micellar bile acids protects against intestinal permeability and liver injury. Sci. Adv. 8:eabo2794. doi: 10.1126/sciadv.abo2794
Li, T., and Chiang, J. Y. (2014). Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 66, 948–983. doi: 10.1124/pr.113.008201
Li, F., Jiang, C., Krausz, K. W., Li, Y., Albert, I., Hao, H., et al. (2013). Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 4:2384. doi: 10.1038/ncomms3384
Li, C.-Y., Wang, H.-N., He, R.-J., Huang, J., Song, L.-L., Song, Y.-Q., et al. (2022a). Discovery and characterization of amentoflavone as a naturally occurring inhibitor against the bile salt hydrolase produced by Lactobacillus salivarius. Food Funct. 13, 3318–3328. doi: 10.1039/D1FO03277A
Li, C.-Y., Wang, H.-N., Zhu, G.-H., Song, L.-L., Hou, X.-D., Huo, P.-C., et al. (2022b). Discovery and characterization of naturally occurring chalcones as potent inhibitors of bile salt hydrolases. Acta Mat. Med. 1, 164–176. doi: 10.15212/AMM-2022-0003
Li, H., Zhang, L., Li, J., Wu, Q., Qian, L., He, J., et al. (2024). Resistant starch intake facilitates weight loss in humans by reshaping the gut microbiota. Nat. Metab. 6, 578–597. doi: 10.1038/s42255-024-00988-y
Li, H., Zhu, X., Xu, J., Li, L., Kan, W., Bao, H., et al. (2023). The FXR mediated anti-depression effect of CDCA underpinned its therapeutic potentiation for MDD. Int. Immunopharmacol. 115:109626. doi: 10.1016/j.intimp.2022.109626
Liang, C., Zhou, X. H., Gong, P. M., Niu, H. Y., Lyu, L. Z., Wu, Y. F., et al. (2021). Lactiplantibacillus plantarum H-87 prevents high-fat diet-induced obesity by regulating bile acid metabolism in C57BL/6J mice. Food Funct. 12, 4315–4324. doi: 10.1039/d1fo00260k
Lin, J., Negga, R., Zeng, X., and Smith, K. (2014). Effect of bile salt hydrolase inhibitors on a bile salt hydrolase from Lactobacillus acidophilus. Pathogens 3, 947–956. doi: 10.3390/pathogens3040947
Liong, M.-T., Lee, B., Choi, S. B., Lew, L. C., Lau, A., and Daliri, E. (2015). “Cholesterol-lowering effects of probiotics and prebiotics” in Probiotics and prebiotics: Current research and future trends, 429–446.
Liu, Y., Azad, M. A. K., Kong, X., Zhu, Q., and Yu, Z. (2022). Dietary bile acids supplementation modulates immune response, antioxidant capacity, glucose, and lipid metabolism in normal and intrauterine growth retardation piglets. Front. Nutr. 9:991812. doi: 10.3389/fnut.2022.991812
Liu, Y., Espinosa, C. D., Abelilla, J. J., Casas, G. A., Lagos, L. V., Lee, S. A., et al. (2018). Non-antibiotic feed additives in diets for pigs: a review. Anim. Nutr. 4, 113–125. doi: 10.1016/j.aninu.2018.01.007
Liu, Y., Zheng, S., Cui, J., Guo, T., and Zhang, J. (2021). Effect of bile salt hydrolase-active Lactobacillus plantarum Y15 on high cholesterol diet induced hypercholesterolemic mice. CyTA J. Food 19, 408–417. doi: 10.1080/19476337.2021.1914176
Lynch, L. E., Hair, A. B., Soni, K. G., Yang, H., Gollins, L. A., Narvaez-Rivas, M., et al. (2023). Cholestasis impairs gut microbiota development and bile salt hydrolase activity in preterm neonates. Gut Microbes 15:2183690. doi: 10.1080/19490976.2023.2183690
Ma, C., Han, M., Heinrich, B., Fu, Q., Zhang, Q., Sandhu, M., et al. (2018). Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360:eaan5931. doi: 10.1126/science.aan5931
Ma, Y., Wang, H., Yang, J., Xin, M., and Wu, X. (2023). Gentamicin alleviates cholestatic liver injury by decreasing gut microbiota-associated bile salt hydrolase activity in rats. Eur. J. Pharmacol. 951:175790. doi: 10.1016/j.ejphar.2023.175790
Malpeli, A., Taranto, M. P., Cravero, R. C., Tavella, M., Fasano, M. V., Vicentin, D., et al. (2015). Effect of daily consumption of Lactobacillus reuteri CRL 1098 on cholesterol reduction in hypercholesterolemic subjects. Food Nutr. Sci. 6, 1583–1590. doi: 10.4236/fns.2015.617163
Mann, G. V. (1974). Studies of a surfactant and cholesteremia in the Maasai. Am. J. Clin. Nutr. 27, 464–469. doi: 10.1093/ajcn/27.5.464
Marchesini, M. I., Connolly, J., Delpino, M. V., Baldi, P. C., Mujer, C. V., DelVecchio, V. G., et al. (2011). Brucella abortus choloylglycine hydrolase affects cell envelope composition and host cell internalization. PLoS One 6:e28480. doi: 10.1371/journal.pone.0028480
Martoni, C. J., Labbe, A., Ganopolsky, J. G., Prakash, S., and Jones, M. L. (2015). Changes in bile acids, FGF-19 and sterol absorption in response to bile salt hydrolase active L. reuteri NCIMB 30242. Gut Microbes 6, 57–65. doi: 10.1080/19490976.2015.1005474
McMillan, A. S., Foley, M. H., Perkins, C. E., and Theriot, C. M. (2024). Loss of Bacteroides thetaiotaomicron bile acid-altering enzymes impacts bacterial fitness and the global metabolic transcriptome. Microbiol. Spectr. 12:e0357623. doi: 10.1128/spectrum.03576-23
Miremadi, F., Ayyash, M., Sherkat, F., and Stojanovska, L. (2014). Cholesterol reduction mechanisms and fatty acid composition of cellular membranes of probiotic lactobacilli and Bifidobacteria. J. Funct. Foods 9, 295–305. doi: 10.1016/j.jff.2014.05.002
Mohanty, I., Allaband, C., Mannochio-Russo, H., El Abiead, Y., Hagey, L. R., Knight, R., et al. (2024a). The changing metabolic landscape of bile acids–keys to metabolism and immune regulation. Nat. Rev. Gastroenterol. Hepatol. 21, 493–516. doi: 10.1038/s41575-024-00914-3
Mohanty, I., Mannochio-Russo, H., Schweer, J. V., El Abiead, Y., Bittremieux, W., Xing, S., et al. (2024b). The underappreciated diversity of bile acid modifications. Cell 187:e1820, 1801–1818.e20. doi: 10.1016/j.cell.2024.02.019
Morinaga, K., Kusada, H., and Tamaki, H. (2022). Bile salt hydrolases with extended substrate specificity confer a high level of resistance to bile toxicity on Atopobiaceae bacteria. Int. J. Mol. Sci. 23:10980. doi: 10.3390/ijms231810980
Moser, S. A., and Savage, D. C. (2001). Bile salt hydrolase activity and resistance to toxicity of conjugated bile salts are unrelated properties in lactobacilli. Appl. Environ. Microbiol. 67, 3476–3480. doi: 10.1128/AEM.67.8.3476-3480.2001
Mukherji, R., and Prabhune, A. (2015). Possible correlation between bile salt hydrolysis and AHL deamidation: Staphylococcus epidermidis RM1, a potent quorum quencher and bile salt hydrolase producer. Appl. Biochem. Biotechnol. 176, 140–150. doi: 10.1007/s12010-015-1563-9
Mullish, B. H., McDonald, J. A. K., Pechlivanis, A., Allegretti, J. R., Kao, D., Barker, G. F., et al. (2019). Microbial bile salt hydrolases mediate the efficacy of faecal microbiota transplant in the treatment of recurrent Clostridioides difficile infection. Gut 68, 1791–1800. doi: 10.1136/gutjnl-2018-317842
Naruszewicz, M., Johansson, M.-L., Zapolska-Downar, D., and Bukowska, H. (2002). Effect of Lactobacillus plantarum 299v on cardiovascular disease risk factors in smokers. Am. J. Clin. Nutr. 76, 1249–1255. doi: 10.1093/ajcn/76.6.1249
Neverovskyi, A., Chernyavskyi, V., Shypulin, V., Gvozdecka, L., and Mikhnova, N. (2020). Gut bacterial bile salt hydrolase activity correlates with cardiovascular risk: a case-control study. Gastroenterologia :228:54. doi: 10.22141/2308-2097.54.4.2020.216711
Núñez-Sánchez, M. A., Herisson, F. M., Keane, J. M., García-González, N., Rossini, V., Pinhiero, J., et al. (2022). Microbial bile salt hydrolase activity influences gene expression profiles and gastrointestinal maturation in infant mice. Gut Microbes 14:2149023. doi: 10.1080/19490976.2022.2149023
O'Flaherty, S., Briner Crawley, A., Theriot, C. M., and Barrangou, R. (2018). The Lactobacillus bile salt hydrolase repertoire reveals niche-specific adaptation. mSphere 3, e00140–e00118. doi: 10.1128/mSphere.00140-18
Ovadia, C., Perdones-Montero, A., Fan, H. M., Mullish, B. H., McDonald, J. A., Papacleovoulou, G., et al. (2020). Ursodeoxycholic acid enriches intestinal bile salt hydrolase-expressing Bacteroidetes in cholestatic pregnancy. Sci. Rep. (UK) 10:3895. doi: 10.1038/s41598-020-60821-w
Palaniyandi, S. A., Damodharan, K., Suh, J.-W., and Yang, S. H. (2020). Probiotic characterization of cholesterol-lowering Lactobacillus fermentum MJM60397. Probiotics Antimicrob. Proteins 12, 1161–1172. doi: 10.1007/s12602-019-09585-y
Parasar, B., Zhou, H., Xiao, X., Shi, Q., Brito, I. L., and Chang, P. V. (2019). Chemoproteomic profiling of gut microbiota-associated bile salt hydrolase activity. ACS Cent. Sci. 5, 867–873. doi: 10.1021/acscentsci.9b00147
Peng, Y.-L., Wang, S.-H., Zhang, Y.-L., Chen, M.-Y., He, K., Li, Q., et al. (2024). Effects of bile acids on the growth, composition and metabolism of gut bacteria. NPJ Biofilms Microbiol. 10:112. doi: 10.1038/s41522-024-00566-w
Pombinho, R., Vieira, A., Camejo, A., Archambaud, C., Cossart, P., Sousa, S., et al. (2020). Virulence gene repression promotes Listeria monocytogenes systemic infection. Gut Microbes 11, 868–881. doi: 10.1080/19490976.2020.1712983
Prete, R., Long, S. L., Gallardo, A. L., Gahan, C. G., Corsetti, A., and Joyce, S. A. (2020). Beneficial bile acid metabolism from Lactobacillus plantarum of food origin. Sci. Rep. 10:1165. doi: 10.1038/s41598-020-58069-5
Pumbwe, L., Skilbeck, C. A., Nakano, V., Avila-Campos, M. J., Piazza, R. M. F., and Wexler, H. M. (2007). Bile salts enhance bacterial co-aggregation, bacterial-intestinal epithelial cell adhesion, biofilm formation and antimicrobial resistance of Bacteroides fragilis. Microb. Pathog. 43, 78–87. doi: 10.1016/j.micpath.2007.04.002
Rani, R. P., Anandharaj, M., and Ravindran, A. D. (2017). Characterization of bile salt hydrolase from Lactobacillus gasseri FR4 and demonstration of its substrate specificity and inhibitory mechanism using molecular docking analysis. Front. Microbiol. 8:1004. doi: 10.3389/fmicb.2017.01004
Reis, S. A., Conceição, L. L., Rosa, D. D., Siqueira, N. P., and Peluzio, M. C. G. (2017). Mechanisms responsible for the hypocholesterolaemic effect of regular consumption of probiotics. Nutr. Res. Rev. 30, 36–49. doi: 10.1017/s0954422416000226
Ren, Z.-L., Li, C.-X., Ma, C.-Y., Chen, D., Chen, J.-H., Xu, W.-X., et al. (2022). Linking nonalcoholic fatty liver disease and brain disease: focusing on bile acid signaling. Int. J. Mol. Sci. 23:13045. doi: 10.3390/ijms232113045
Ridlon, J. M., Harris, S. C., Bhowmik, S., Kang, D.-J., and Hylemon, P. B. (2016). Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7, 22–39. doi: 10.1080/19490976.2015.1127483
Rimal, B., Collins, S. L., Tanes, C. E., Rocha, E. R., Granda, M. A., Solanki, S., et al. (2024). Bile salt hydrolase catalyses formation of amine-conjugated bile acids. Nature 626, 859–863. doi: 10.1038/s41586-023-06990-w
Robinson, K., Becker, S., Xiao, Y., Lyu, W., Yang, Q., Zhu, H., et al. (2019). Differential impact of subtherapeutic antibiotics and ionophores on intestinal microbiota of broilers. Microorganisms 7:282. doi: 10.3390/microorganisms7090282
Rodes, L., Coussa-Charley, M., Marinescu, D., Paul, A., Fakhoury, M., Abbasi, S., et al. (2013). Design of a novel gut bacterial adhesion model for probiotic applications. Artif. Cells Nanomed. Biotechnol. 41, 116–124. doi: 10.3109/10731199.2012.712047
Ryan, P. M., Stolte, E. H., London, L. E., Wells, J. M., Long, S. L., Joyce, S. A., et al. (2019). Lactobacillus mucosae DPC 6426 as a bile-modifying and immunomodulatory microbe. BMC Microbiol. 19, 33–13. doi: 10.1186/s12866-019-1403-0
Samson, N., Bosoi, C. R., Roy, C., Turcotte, L., Tribouillard, L., Mouchiroud, M., et al. (2024). HSDL2 links nutritional cues to bile acid and cholesterol homeostasis. Sci. Adv. 10:eadk9681. doi: 10.1126/sciadv.adk9681
Seung Kee, C., Soo Jin, L., So-Yeon, S., Jin Seok, M., Ling, L., Wooha, J., et al. (2015). Development of bile salt-resistant Leuconostoc citreum by expression of bile salt hydrolase gene. J. Microbiol. Biotechnol. 25, 2100–2105. doi: 10.4014/jmb.1505.05072
Shankar, N., Baghdayan, A. S., and Gilmore, M. S. (2002). Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417, 746–750. doi: 10.1038/nature00802
Sleator, R. D., Wemekamp-Kamphuis, H. H., Gahan, C. G. M., Abee, T., and Hill, C. (2005). A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol. Microbiol. 55, 1183–1195. doi: 10.1111/j.1365-2958.2004.04454.x
Smith, K. R. (2013). Identification of bile salt hydrolase inhibitors, the promising alternative to antibiotic growth promoters to enhance animal production. Masters Theses, University of Tennessee.
Smith, K., Zeng, X. M., and Lin, J. (2014). Discovery of bile salt hydrolase inhibitors using an efficient high-throughput screening system. PLoS One 9:e85344. doi: 10.1371/journal.pone.0085344
Song, Z., Feng, S., Zhou, X., Song, Z., Li, J., and Li, P. (2023). Taxonomic identification of bile salt hydrolase-encoding lactobacilli: modulation of the enterohepatic bile acid profile. iMeta 2:e128. doi: 10.1002/imt2.128
Song, H., Liu, J., Wang, L., Hu, X., Li, J., Zhu, L., et al. (2024). Tauroursodeoxycholic acid: a bile acid that may be used for the prevention and treatment of Alzheimer’s disease. Front. Neurosci. 18:1348844. doi: 10.3389/fnins.2024.1348844
Spratt, E., Mo, Y., Li, Y., Lin, J., and Chen, G. (2014). The effects of oral administration of bile salt hydrolase on the partition of fatty acid and glucose oxidation in Zucker fatty rats (585.8). FASEB J. 28. doi: 10.1096/fasebj.28.1_supplement.585.8
Sridevi, N., Vishwe, P., and Prabhune, A. (2009). Hypocholesteremic effect of bile salt hydrolase from Lactobacillus buchneri ATCC 4005. Food Res. Int. 42, 516–520. doi: 10.1016/j.foodres.2009.02.016
Sun, X., Chen, Z., Yu, L., Zeng, W., Sun, B., Fan, H., et al. (2023). Bacteroides dorei BDX-01 alleviates DSS-induced experimental colitis in mice by regulating intestinal bile salt hydrolase activity and the FXR-NLRP3 signaling pathway. Front. Pharmacol. 14:1205323. doi: 10.3389/fphar.2023.1205323
Sun, J., and Kato, I. (2016). Gut microbiota, inflammation and colorectal cancer. Genes Dis. 3, 130–143. doi: 10.1016/j.gendis.2016.03.004
Sun, L., Zhang, Y., Cai, J., Rimal, B., Rocha, E. R., Coleman, J. P., et al. (2023). Bile salt hydrolase in non-enterotoxigenic Bacteroides potentiates colorectal cancer. Nat. Commun. 14:755. doi: 10.1038/s41467-023-36089-9
Sun, N., Zhang, J., Wang, J., Liu, Z., Wang, X., Kang, P., et al. (2022). Abnormal gut microbiota and bile acids in patients with first-episode major depressive disorder and correlation analysis. Psychiat. Clin. Neurosci. 76, 321–328. doi: 10.1111/pcn.13368
Svensson, P.-A., Olsson, M., Andersson-Assarsson, J. C., Taube, M., Pereira, M. J., Froguel, P., et al. (2013). The TGR5 gene is expressed in human subcutaneous adipose tissue and is associated with obesity, weight loss and resting metabolic rate. Biochem. Biophys. Res. Commun. 433, 563–566. doi: 10.1016/j.bbrc.2013.03.031
Tanaka, H., Hashiba, H., Kok, J., and Mierau, I. (2000). Bile salt hydrolase of Bifidobacterium longum-biochemical and genetic characterization. Appl. Environ. Microbiol. 66, 2502–2512. doi: 10.1128/AEM.66.6.2502-2512.2000
Taranto, M., Fernandez Murga, M., Lorca, G., and Valdez, G. D. (2003). Bile salts and cholesterol induce changes in the lipid cell membrane of Lactobacillus reuteri. J. Appl. Microbiol. 95, 86–91. doi: 10.1046/j.1365-2672.2003.01962.x
Thamer, S. J. (2022). Pathogenesis, diagnosis and treatment of gallstone disease: a brief review. Biomed. Chem. Sci. 1, 70–77. doi: 10.48112/bcs.v1i2.99
Urdaneta, V., and Casadesús, J. (2017). Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front. Med. 4:163. doi: 10.3389/fmed.2017.00163
Van Eldere, J., Robben, J., De Pauw, G., Merckx, R., and Eyssen, H. (1988). Isolation and identification of intestinal steroid-desulfating bacteria from rats and humans. Appl. Environ. Microbiol. 54, 2112–2117. doi: 10.1128/aem.54.8.2112-2117.1988
Wang, Y., Gao, X., Zhang, X., Xiao, F., Hu, H., Li, X., et al. (2021). Microbial and metabolic features associated with outcome of infliximab therapy in pediatric Crohn’s disease. Gut Microbes 13, 1–18. doi: 10.1080/19490976.2020.1865708
Wang, G., Huang, W., Xia, Y., Xiong, Z., and Ai, L. (2019). Cholesterol-lowering potentials of Lactobacillus strain overexpression of bile salt hydrolase on high cholesterol diet-induced hypercholesterolemic mice. Food Funct. 10, 1684–1695. doi: 10.1039/c8fo02181c
Wang, N., Wang, J., Zhang, T., Huang, L., Yan, W., Lu, L., et al. (2021). Alterations of gut microbiota and serum bile acids are associated with parenteral nutrition-associated liver disease. J. Pediatr. Surg. 56, 738–744. doi: 10.1016/j.jpedsurg.2020.06.035
Wang, G., Yin, S., An, H., Chen, S., and Hao, Y. (2011). Coexpression of bile salt hydrolase gene and catalase gene remarkably improves oxidative stress and bile salt resistance in Lactobacillus casei. J. Ind. Microbiol. Biotechnol. 38, 985–990. doi: 10.1007/s10295-010-0871-x
Wang, G., Yu, H., Feng, X., Tang, H., Xiong, Z., Xia, Y., et al. (2021). Specific bile salt hydrolase genes in Lactobacillus plantarum AR113 and relationship with bile salt resistance. LWT 145:111208. doi: 10.1016/j.lwt.2021.111208
Wang, Y., Zheng, L., Zhou, Z., Yao, D., Huang, Y., Liu, B., et al. (2022). Review article: insights into the bile acid-gut microbiota axis in intestinal failure-associated liver disease-redefining the treatment approach. Aliment. Pharmacol. Ther. 55, 49–63. doi: 10.1111/apt.16676
Widodo, W., Fanani, T. H., Fahreza, M. I., and Sukarno, A. S. (2021). Cholesterol assimilation of two probiotic strains of Lactobacillus casei used as dairy starter cultures. Appl. Food Biotechnol. 8, 103–112. doi: 10.22037/afb.v8i2.30661
Wu, M., Cheng, Y., Zhang, R., Han, W., Jiang, H., Bi, C., et al. (2024). Molecular mechanism and therapeutic strategy of bile acids in Alzheimer’s disease from the emerging perspective of the microbiota–gut–brain axis. Biomed. Pharmacother. 178:117228. doi: 10.1016/j.biopha.2024.117228
Wu, T., Wang, G., Tang, H., Xiong, Z., Song, X., Xia, Y., et al. (2022). Genes encoding bile salt hydrolase differentially affect adhesion of Lactiplantibacillus plantarum AR113. J. Sci. Food Agr. 102, 1522–1530. doi: 10.1002/jsfa.11487
Xiao, Y., Zhao, J., Zhang, H., Zhai, Q., and Chen, W. (2021). Mining genome traits that determine the different gut colonization potential of Lactobacillus and Bifidobacterium species. Microb. Genom. 7:000581. doi: 10.1099/mgen.0.000581
Xie, Z., Jiang, H., Liu, W., Zhang, X., Chen, D., Sun, S., et al. (2020). The triterpenoid sapogenin (2α-OH-protopanoxadiol) ameliorates metabolic syndrome via the intestinal FXR/GLP-1 axis through gut microbiota remodelling. Cell Death Dis. 11:770. doi: 10.1038/s41419-020-02974-0
Yadav, R., Puniya, A. K., and Shukla, P. (2016). Probiotic properties of Lactobacillus plantarum RYPR1 from an indigenous fermented beverage raabadi. Front. Microbiol. 7:1683. doi: 10.3389/fmicb.2016.01683
Yang, B., Huang, S., Yang, N., Cao, A., Zhao, L., Zhang, J., et al. (2022). Porcine bile acids promote the utilization of fat and vitamin a under low-fat diets. Front. Nutr. 9:1005195. doi: 10.3389/fnut.2022.1005195
Yang, Y., Liu, Y., Zhou, S., Huang, L., Chen, Y., and Huan, H. (2019). Bile salt hydrolase can improve Lactobacillus plantarum survival in gastrointestinal tract by enhancing their adhesion ability. FEMS Microbiol. Lett. 366:fnz100. doi: 10.1093/femsle/fnz100
Yao, L., Seaton, S. C., Ndousse-Fetter, S., Adhikari, A. A., DiBenedetto, N., Mina, A. I., et al. (2018). A selective gut bacterial bile salt hydrolase alters host metabolism. eLife 7:e37182. doi: 10.7554/eLife.37182
Yin, S., Zhai, Z., Wang, G., An, H., Luo, Y., and Hao, Y. (2011). A novel vector for lactic acid bacteria that uses a bile salt hydrolase gene as a potential food-grade selection marker. J. Biotechnol. 152, 49–53. doi: 10.1016/j.jbiotec.2011.01.018
Yoon, H. M., Song, S. J., Ko, J. S., Yoon, H. K., Kim, S. J., Kim, S. J., et al. (2020). PCR screening of Enterococcus faecalis from mixture of lactic acid bacteria using bile salt hydrolase gene as a selective marker. Indian J. Microbiol. 60, 251–253. doi: 10.1007/s12088-019-00851-9
Yoon, S., Yu, J., McDowell, A., Kim, S. H., You, H. J., and Ko, G. (2017). Bile salt hydrolase-mediated inhibitory effect of Bacteroides ovatus on growth of Clostridium difficile. J. Microbiol. 55, 892–899. doi: 10.1007/s12275-017-7340-4
Yuan, J., Wang, B., Sun, Z., Bo, X., Yuan, X., He, X., et al. (2008). Analysis of host-inducing proteome changes in Bifidobacterium longum NCC2705 grown in vivo. J. Proteome Res. 7, 375–385. doi: 10.1021/pr0704940
Zhang, Z., Lv, T., Wang, X., Wu, M., Zhang, R., Yang, X., et al. (2024). Role of the microbiota–gut–heart axis between bile acids and cardiovascular disease. Biomed. Pharmacother. 174:116567. doi: 10.1016/j.biopha.2024.116567
Zhao, M., Kuang, W., Yang, J., Liu, Y., Yang, M., Chen, Y., et al. (2024). Cholesterol lowering in diet-induced hypercholesterolemic mice using Lactobacillus bile salt hydrolases with different substrate specificities. Food Funct. 15, 1340–1354. doi: 10.1039/d3fo04871c
Zheng, Y., Zhang, S., Zhang, Z., Zhang, T., Teng, X., Xiao, G., et al. (2024). Isolation of Lactobacillus acidophilus strain and its anti-obesity effect in a diet induced obese murine model. Lett. Appl. Microbiol. 77:ovae021. doi: 10.1093/lambio/ovae021
Zhong, X.-C., Liu, Y.-M., Gao, X.-X., Krausz, K. W., Niu, B., Gonzalez, F. J., et al. (2023). Caffeic acid phenethyl ester suppresses intestinal FXR signaling and ameliorates nonalcoholic fatty liver disease by inhibiting bacterial bile salt hydrolase activity. Acta Pharmacol. Sin. 44, 145–156. doi: 10.1038/s41401-022-00921-7
Zhu, D., Sorg, J. A., and Sun, X. (2018). Clostridioides difficile biology: sporulation, germination, and corresponding therapies for C. difficile infection. Front. Cell. Infect. Microbiol. 8:29. doi: 10.3389/fcimb.2018.00029
Keywords: gut microbiota, host health, bile acids, bile salt hydrolase, physiological roles, potential applications
Citation: Dong Z, Yang S, Tang C, Li D, Kan Y and Yao L (2025) New insights into microbial bile salt hydrolases: from physiological roles to potential applications. Front. Microbiol. 16:1513541. doi: 10.3389/fmicb.2025.1513541
Received: 18 October 2024; Accepted: 03 February 2025;
Published: 12 February 2025.
Edited by:
Laurent Dufossé, Université de la Réunion, FranceReviewed by:
Jai K. Kaushik, National Dairy Research Institute (ICAR), IndiaCopyright © 2025 Dong, Yang, Tang, Li, Kan and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zixing Dong, c3RhcjE5ODcuY29tQDE2My5jb20=; Lunguang Yao, bHVuZ3Vhbmd5YW9AMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.