
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Microbiol., 22 January 2025
Sec. Microorganisms in Vertebrate Digestive Systems
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1513253
Background: Intestinal dysbiosis was considered a pivotal pathological mechanism underlying sarcopenia. Despite the fervor surrounding research in this domain, substantial controversy persists regarding the obtained outcomes.
Objective: To systematically summarized the disparities in gut microbiota composition between the group afflicted by sarcopenia and non-sarcopenia controls.
Methods: PubMed, Medline, CINAHL, EMBASE, Scopus, Web of Science and Google Scholer, Cochrane Library and gray literature sources were systematically searched for in randomized controlled trials. Meta-analysis and random-effects meta-regression were conducted using Rev. Man 5.3. Overall effect was measured using Hedges’s g and determined using Z-statistics. Cochran’s Q test and I2 were used to investigate heterogeneity. The Newcastle-Ottawa Scale was used to assess overall quality of evidence.
Results: Ten studies, including 421 cases of sarcopenia and 1,642 cases of controls, were included in the meta-analysis. Patients with sarcopenia showed significantly reduced gut microbiota in α diversity, and β diversity was significantly different in 8/9 of included studies. We also found more abundance of phylum Proteobacteria and genus Escherichia-Shigella, and less abundance of phylum Firmicutes and genus Faecalibacterium, Prevotella 9, Blautia in the sarcopenia group.
Conclusion: The gut microbiota composition in patients with sarcopenia has undergone alterations, serving as a fundamental reference for further investigation into the potential pathogenic mechanisms and treatment strategies for sarcopenia.
Sarcopenia is a prevalent age-related skeletal muscle disorder characterized by the progressive loss of muscle mass and decline in muscular strength. Sarcopenia has an incidence of 36% in individuals <60 years, and 27% in individuals ≥60 years, the prevalence of severe sarcopenia was as high as 9%, and this percentage continues to increase with age (Petermann-Rocha et al., 2022). The number of people with sarcopenia is predicted to increase to 1.2 billion by 2025 and double by 2050 (Almohaisen et al., 2022). It is one of the leading health issues in the older adults, and it increases disability risk, falls as well as injuries related to falls, hospitalization, limitation of independence, and mortality, It has a certain burden on the social medical system (Cruz-Jentoft et al., 2019; Sayer and Cruz-Jentoft, 2022). Age-related mechanisms that contribute to the onset of sarcopenia encompass inflammation, immunosenescence, anabolic resistance, reduced levels of physical activity, and heightened oxidative stress (Chen et al., 2020; Cho et al., 2022).
The gut microbiota exerts a pivotal role in the aging process by regulating energy balance, metabolism, and inflammation to impact the progression of sarcopenia (Papadopoulou, 2020; Ling et al., 2022). The gut microbiota is recognized as an overlooked endocrine organ, exerting regulatory control over the host’s homeostasis through the fermentation of undigested food in the colon, thereby generating a wide array of bioactive molecules (Fan et al., 2023). For instance, short-chain fatty acids (SCFAs), such as acetate, propionate, and butyrate derived from dietary fiber, have demonstrated favorable effects on the host by enhancing skeletal muscle growth. The recently proposed concept of the gut-muscle axis suggests a potential correlation between gut microbiota and the quality and functionality of skeletal muscle (Liao et al., 2020). Yan et al. showed that the diversity and richness of gut microbiota were lower in sarcopenia patients than in controls. Among them, a decrease in the ratio of Prevotella to Bacteroidetes (P/B) and a decrease in the abundance of Coprococcus and Lachnospiraceae were significant indicators. Prevotella and Bacteroidetes are involved in dietary fiber fermentation and the production of short-chain fatty acids (SCFAs), which are essential for the maintenance of muscle mass and function. A lower P/B ratio indicates a reduced capacity for SCFA production, which may negatively affect muscle health. Furthermore, the reduction in the quantity of Faecalibacterium and Lachnospiraceae, which are the main producers of short-chain fatty acids, is closely related to the decrease in short-chain fatty acid levels, which may further lead to muscle atrophy and weakened muscle strength. In view of this, monitoring these specific bacterial markers can provide us with early warning signals of sarcopenia, enabling us to take timely intervention measures (Yan et al., 2023). Research into the association between gut microbiota and muscle frailty in elderly populations has demonstrated that the composition of intestinal flora undergoes significant alterations in sarcopenia patients. Specifically, the relative abundance of Lactobacillus, Bacteroides, and Prevotella decreases, whereas that of Leminoxella increases markedly (Grosicki et al., 2018). Through pyrosequencing analysis of 16S rRNA, it was found that the quantities of Ruminococcus and Brucella in the intestines of sarcopenia patients decreased significantly, while the abundance of Escherichia coli increased (Grosicki et al., 2018). It is worth noting that as people age, the composition of the human intestinal flora changes due to muscle weakness (Dao et al., 2020). Through the study of 35 community residents over 70 years old (including 18 sarcopenia patients and 17 healthy controls), it was shown that the quantities of Helicobacter and Ruminococcus in the intestines of sarcopenia patients increased, while the quantities of Pasteurellaceae and Christensenellaceae decreased (Wang X. et al., 2022; Wang Y. et al., 2022). In addition, the serum aspartate concentration was higher in sarcopenia patients, while the circulating levels of threonine and macrophage inflammatory protein 1α were lower.
In recent years, more research has focused on the relationship between sarcopenia and gut microbiota composition. At the genus level, Prevotella and Lacococcus faecalis have been identified as key markers and studies have shown their significantly reduced abundance in the sarcopenia population (Kang et al., 2021). At the family level, Trichospiraceae and Ruminoccaceae have received much attention for their important role in intestinal health and metabolism in patients with sarcopenia (Zhou et al., 2023). At the phylum level, the relative abundance of Firmicutes and Bacteroidetes was significantly decreased in patients with sarcopenia (Kang et al., 2021). The above taxa are believed to play an important role in the development of sarcopenia by affecting the inflammatory response and energy satisfaction.
In recent years, the emergence of high-throughput sequencing technologies such as 16S rRNA and metagenomics has sparked increasing interest in exploring the potential role of gut microbiota in the pathogenesis of muscular dystrophy (Wang X. et al., 2022; Wang Y. et al., 2022). However, the existing research results of 16S rRNA sequencing are inconsistent, and there is a lack of systematic summary, making it difficult to provide clear reference basis for the prevention and clinical treatment of sarcopenia. Therefore, this study aims to explore the differences in diversity and richness of the gut microbiota between sarcopenia and non-sarcopenia populations through systematic review and meta-analysis, in order to clarify the potential role of GM and its metabolites in the pathogenesis of sarcopenia, and provide new theoretical basis and practical methods for the prevention and treatment of sarcopenia.
The present systematic review and meta-analysis was conducted in accordance with the guidelines provided by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) (Page et al., 2021). Full-text original research articles on sarcopenia individual and non-sarcopenia individual were identified in a search of the PubMed, Medline, CINAHL, EMBASE, Scopus, Web of Science and Google Scholer, and Cochrane Library and gray literature sources, from the establishment of the database until February 2024. A combination of subject words and corresponding free words were used for the search, including “sarcopenia” OR “Gut-Muscle axis” OR “skeletal muscle,” “Muscle mass,” “microbiota,” “gut microbiota,” “microbiome,” “metabolomic,” “intestinal flora.” The study protocol was registered with PROSPERO (CRD42024523222).
The two authors (WGN and LYJ) independently conducted a thorough examination and selection of the complete texts that satisfy the specified criteria: (1) These articles are peer-reviewed and written in English; (2) They compare the diversity and abundance of gut microbiota between patients with sarcopenia and healthy or non-sarcopenia control groups; (3) The gut microbiota is derived from fecal samples; (4) Participants include adults aged 18 years and above, excluding studies focused on children as their microbial composition is less stable during development and cannot be compared to adults; (5) Sufficient statistical data should be provided, such as mean, standard deviation, interquartile range, p-value, maximum value, minimum value, etc., to estimate effect size; (6) Alpha diversity indices that can be correctly collected should be presented in the article or Supplementary material (e.g., tables, box plots, bar graphs). Case reports, systematic reviews, and animal studies are excluded.
The findings encompass the diversity of gut microbiota (including α-diversity and β-diversity) as well as variations in gut microbiota abundance between individuals with muscular dystrophy and healthy or non-muscular dystrophy control groups.
Two researchers (WGN and LYJ) independently screened eligible studies and excluded articles that did not meet the inclusion criteria. The following data were collected: article title and publication year, country of origin, age range of participants, gender distribution, sample size, method used for microbial assessment, region targeted for 16S rRNA sequencing analysis, as well as bacterial changes observed in patients with muscular dystrophy. To assess alterations in relative abundance at the phylum and genus levels of bacteria, trends indicating increased or decreased relative abundances were extracted for five and seven major bacterial taxa, respectively. For analyzing changes in relative abundance at the genus and species levels of bacteria, mean values along with their corresponding standard deviations (SD) were directly obtained from 10 included articles. In cases where direct mean values and SD were unavailable, quartile ranges along with maximum and minimum values were extracted from original figures to calculate mean values and SD.
The Newcastle-Ottawa Scale (Zeng et al., 2015) was employed to assess the quality of the literature included in the study. This scale has a maximum score of 7, and studies achieving a total score ≥ 5 are considered as high-quality. Two researchers independently conducted blinded assessments on the included studies, with any discrepancies that arose during this process being resolved through consultation with a third expert.
The effect size was calculated using a random-effects, inverse variance weighted model in RevMan 5.3 software. To estimate the mean and standard deviation of median, maximum, and minimum values, we used a previously reported transformation equation assuming mild departure from normal distribution (Hozo et al., 2005). Hedges’ g effect size was computed as the average difference between the sarcopenia group and non-sarcopenia group divided by their combined standard deviation. We assessed heterogeneity of each study using Q-statistics and I2. Funnel plot was employed for quantitative evaluation of potential publication bias. The significance level was set at p < 0.05.
Kindly request the primary author for access to the corresponding data.
After conducting a comprehensive literature search and eliminating duplicate articles, we obtained a total of 397 relevant papers. Following the review of titles and abstracts, 16 studies were identified as potentially meeting our inclusion criteria for meta-analysis (Figure 1). Upon meticulous examination of the full texts, 22 studies were subsequently excluded: two were review articles (Liu et al., 2021; Nikkhah et al., 2023), five were non-case–control studies (Jackson et al., 2016; Daily and Park, 2022; Lapauw et al., 2023; Yan et al., 2023; Zhao et al., 2023; Iwasaka et al., 2024), three involved animal experiments (Kim et al., 2022; Lee et al., 2023; Mo et al., 2023), and two were do not mention the diversity of OUT of the microbiota (Chen et al., 2023; Zhou et al., 2023). Consequently, the final meta-analysis comprised 10 remaining articles (Figure 1). All the 10 studies included 869 men and 1,148 women over 60 years old, and 46 people in one study (Yan et al., 2023) did not mention gender. Table 1 provides an overview of the clinical characteristics and demographic features observed in these 10 studies (Kang et al., 2021; Han et al., 2022; Lee et al., 2022; Wang X. et al., 2022; Wang Y. et al., 2022; He et al., 2023; Yan et al., 2023; Zhang et al., 2023; Zhou et al., 2023; Lou et al., 2024). These studies were conducted in Italy (Ticinesi et al., 2020) and China with a collective participation of 421 individuals diagnosed with sarcopenia and 1,642 non-sarcopenia controls (Table 1).
The alpha diversity index is a measure of the number of biotic species within a community as well as the relative abundance of biotic species among them. α diversity is mainly Chao1 richness estimator, Shannon diversity index, Simpson diversity index and Observed species were calculated. The results showed that sarcopenia group demonstrated significantly reduced α diversity as indexed by Chao 1 index (n = 7, SMD = −0.49, 95% CI: −0.81 to −0.17, I2 = 65%) (Figure 2A), Observed species index (n = 4, SMD = −0.64, 95% CI: −1.11 to −0.17, I2 = 65%) (Figure 2B), and Simpson index (n = 5, SMD = −1.62, 95% CI: −2.93 to −0.31, I2 = 95%) (Figure 2D). Shannon index showed a decreased trend (n = 7, SMD = −0.26, 95% CI: −0.61 to 0.09, I2 = 77%), although there was no statistical significance (Figure 2C).
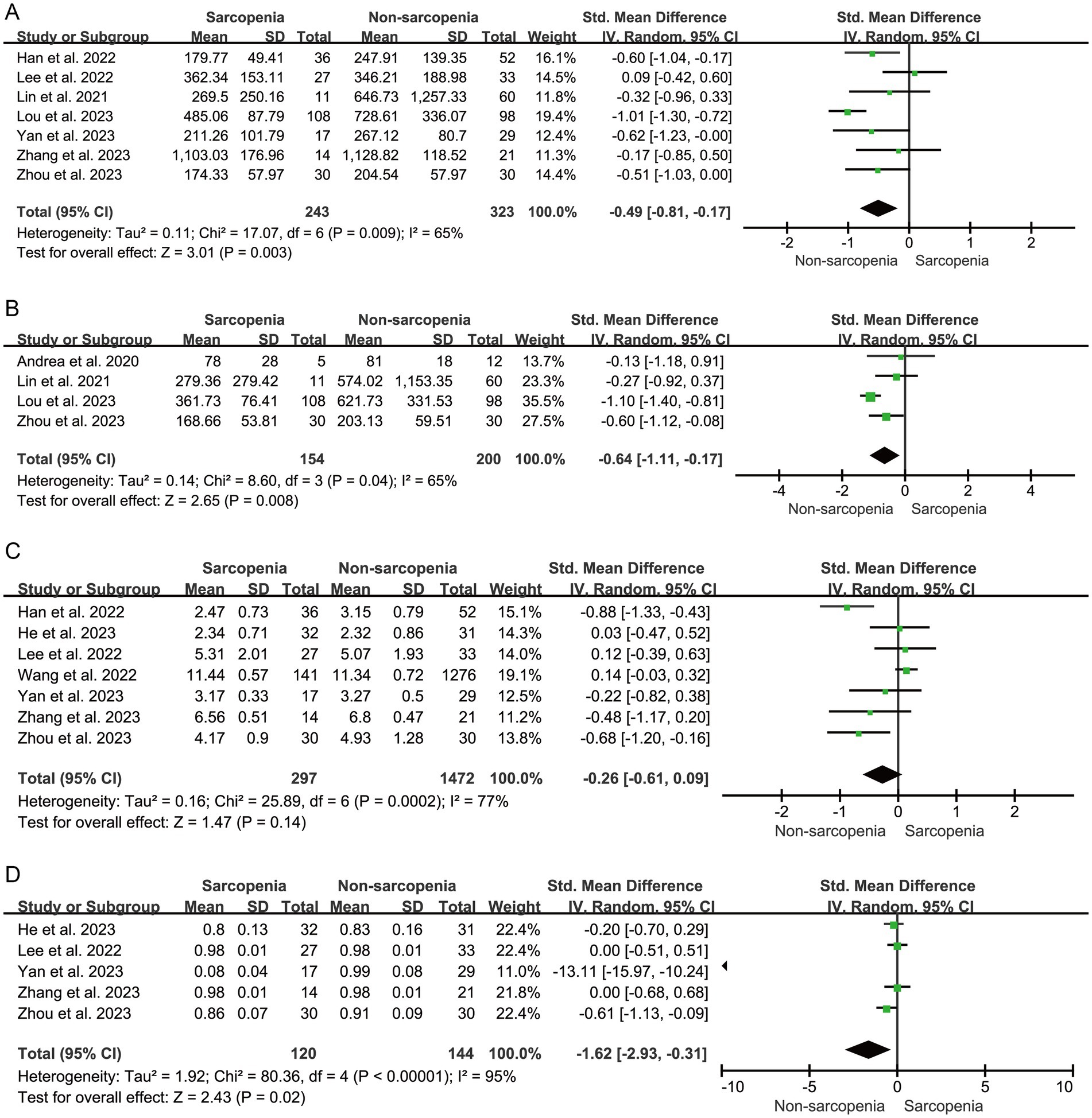
Figure 2. Forest plots of Chao 1 (A), Observed species (B), Shannon (C), and Simpson (D) between sarcopenia group and non-sarcopenia group.
The β diversity index is the species diversity between ecosystems, which contains a comparison of taxonomic units. That is to measure the differences between communities. The beta diversity index mainly includes PCoA analysis, and PLS-DA analysis. The evaluation of 13 β diversity indicators were assessed for nine studies, with the exception of one study (Lou et al., 2024) (Table 2). β diversity was significantly different in 8/9 of included studies. The principal coordinate analyses based on Bray–Curtis dissimilarity was most frequently measured, of which one study revealed no significant differences (Ticinesi et al., 2020), while seven studies revealed significant differences between sarcopenia and non-sarcopenia group (Han et al., 2022; Lee et al., 2022; Wang X. et al., 2022; Wang Y. et al., 2022; He et al., 2023; Lee et al., 2023; Yan et al., 2023; Zhang et al., 2023; Zhou et al., 2023). One study applied three methods to assess β diversity (Kang et al., 2021). The three methods are PLS-DA, Unweighted UniFrac distances matrix, and PCoA based on Unweighted UniFrac distances. The results of PLS-DA revealed significant differences, while the result of PCoA based on Unweighted UniFrac distances revealed no significances between sarcopenia and non-sarcopenia group.
Six studies were included to analyze the differences in the representative phyla of the microbiome in patients with sarcopenia and non-sarcopenia group. The trend of the relative abundance of phylum Proteobacteria (60%, 3/5) was increased, and the trends of Firmicutes (80%, 4/5) was decreased in the sarcopenia group (Figure 3A).
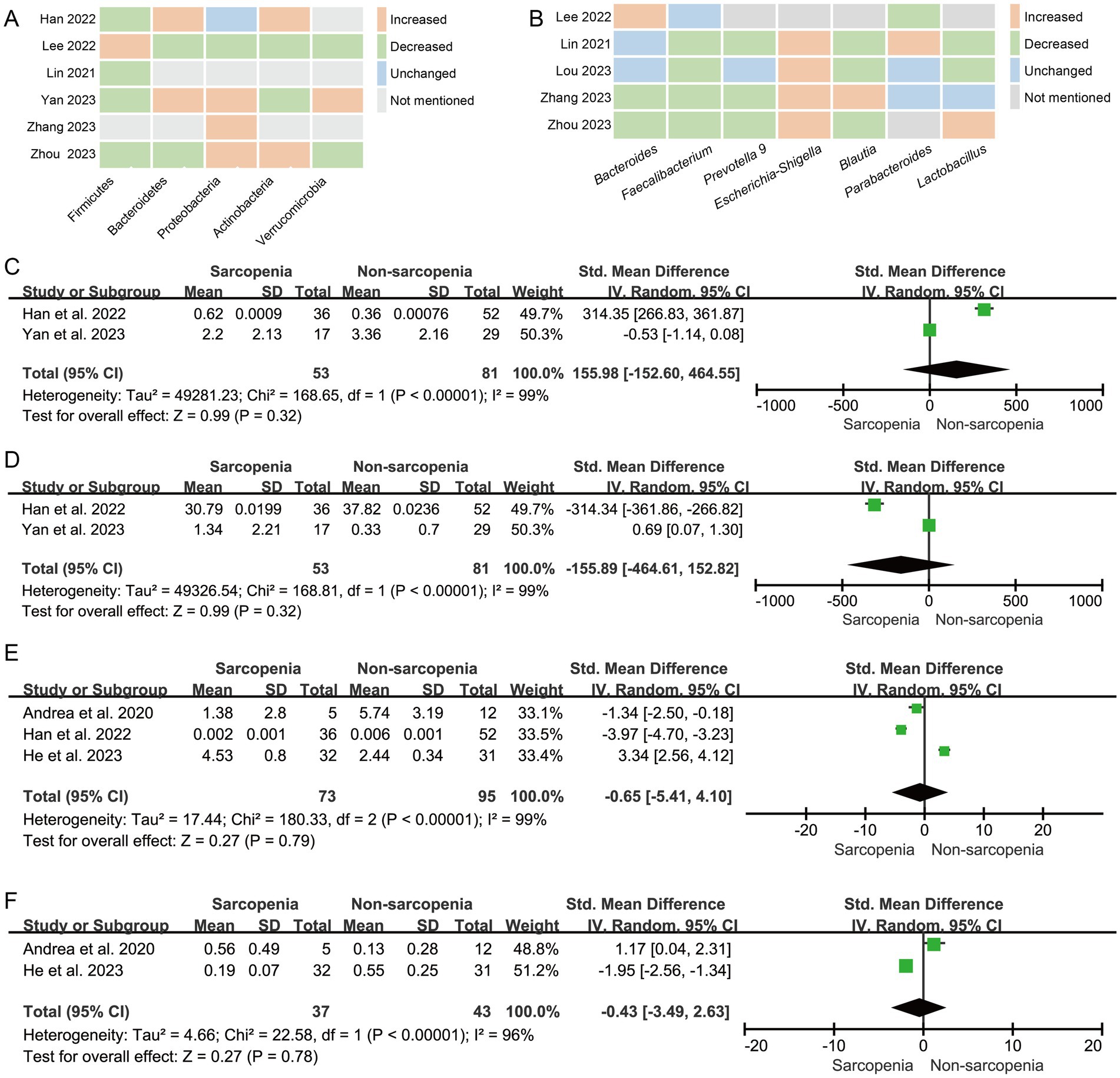
Figure 3. The changes in the relative abundanceof the microbiota in patients with sarcopenia compared with those of non-sarcopenia controls. (A) Heatmap analysis at the phylum level; (B) Heatmap analysis at the genus level; Forest plots of the relative abundance of (C) Dorea, (D) Bacteroides, (E) Faecalibacterium prausnitzii, and (F) Bifidobacterium longum between sarcopenia group and non-sarcopenia group.
Seven studies were included to analyze the differences in the representative genera of the microbiome in patients with sarcopenia and non-sarcopenia group. The trend of the relative abundance of genus Escherichia-Shigella (100%, 4/4) was increased, and the trends of Faecalibacterium (80%, 4/5), Prevotella 9 (75%, 3/4), and Blautia (75%, 3/4) were decreased in the sarcopenia group (Figure 3B).
In the meta-analysis of Dorea abundance, Two studies were included. The forest plot indicated that there was no statistically significant difference between sarcopenia group and non-sarcopenia group (n = 2, SMD = 155.98, 95% CI: −152.60 to 464.55, I2 = 99%) (Figure 3C). The results of Bacteroides (n = 2, SMD = −155.89, 95% CI: −464.61 to 152.82, I2 = 99%) (Figure 3D), Faecalibacterium prausnitzii (n = 3, SMD = −0.65, 95% CI: −5.41 to 4.10, I2 = 99%) (Figure 3E) and Bifidobacterium longum (n = 2, SMD = −0.43, 95% CI: −3.49 to 2.63, I2 = 96%) (Figure 3F) were consistent with Dorea.
The sensitivity analysis showed that excluding individual studies one by one had no significant impact on the standardized mean difference (SMD), which indicated that the SMD is not influenced by any individual study. Funnel plots (Supplementary Figure S1) indicated a lack of significant publication bias.
In recent years, the hypothesis of gut-muscle axis has been a popular topic of research at home and abroad (Liu et al., 2021). The current study found that the gut microbiota could affect host function in variety of ways and regulate the onset and progression of sarcopenia. For example, Studies have shown that intestinal microbiota could affect skeletal muscle by participating in the regulation of inflammation, immunity, endocrine system and protein synthesis (Li et al., 2022).
To the best of our knowledge, this current meta-analysis represents the first attempt to evaluate α-diversity and β-diversity in individuals diagnosed with sarcopenia. For the result of Shannon, excluding this study due to a significant difference in sample size (2658) compared to other studies (35–87), the findings become statistically significant (n = 6, SMD = −0.36, 95% CI: −0.70 to −0.013, I2 = 61%). Numerous studies have consistently reported a significant reduction in alpha diversity among patients with sarcopenia (Wang X. et al., 2022; Wang Y. et al., 2022; Lou et al., 2024). Additionally, Kang et al.’s investigation demonstrated that older adults exhibiting lower muscle mass exhibited significantly diminishedα diversity within their microbiota community (Kang et al., 2021). These findings align harmoniously with the outcomes of our meta-analysis. It is worth noting that muscle mass typically exhibits a positive correlation with muscle strength (function). Comparing the gut microbiota of women with and without sarcopenia, the women with sarcopenia showed low diversity, which predicted low health status (Sanz et al., 2018), and the reduced diversity of gut microbiota may impair the integrity of the intestinal barrier, allowing harmful substances including lipopolysaccharide to enter the bloodstream, which can not only trigger systemic inflammation, but also induce the up regulation of proinflammatory cytokines, ultimately stimulating protein catabolism and inhibiting muscle synthesis (Grosicki et al., 2018; Ashworth, 2021). Furthermore, this study suggests that there are notable disparities in gut microbiota composition between the sarcopenia group and non-sarcopenia group. Considering beta diversity, further investigation into disparities between healthy controls and individuals with sarcopenia appears necessary due to inconsistent findings.
In the colon of newborn healthy mammals, there is a slightly higher abundance of Proteobacteria, which primarily facilitate oxygen absorption and create a favorable environment for the colonization of obligate anaerobic bacteria. However, in adult healthy mammals, the abundance of Proteobacteria decreases to primarily support the host in maintaining an anaerobic intestinal environment (Bradley and Pollard, 2017). Common gastrointestinal tract bacteria belonging to the family Proteobacteria include Escherichia coli, Salmonella, Shigella, and Pseudomonas. The presence of reports suggests that an elevated proportion of Proteobacteria serves as a reliable indicator for dysbiosis in the gut microbiota (Fujisaka et al., 2023). The members of the phylum Proteobacteria are known for their complexity, however, the majority of Proteobacteria found in the gastrointestinal tract are facultative anaerobic and gram-negative bacteria. These bacteria have the ability to produce lipopolysaccharides (LPS) and stimulatory flagellar proteins, which can induce inflammatory reactions, thereby exhibiting a certain degree of pathogenicity. Research has demonstrated that LPS promotes inflammation primarily by entering the bloodstream via the intestinal tract and triggering an inflammatory response. Disruption of the intestinal microecological balance leads to a decrease in beneficial bacteria, increased intestinal permeability, and elevated expression of Gram-negative bacteria, which secrete endotoxins LPS. These endotoxins enter the bloodstream, bind to endotoxin-binding proteins, and are subsequently phagocytosed by macrophages expressing CD14. The activation of CD14 initiates a cascade of cellular metabolic reactions, leading to the secretion of various inflammatory factors from the cell nucleus, such as interleukin-6 (IL-6), tumor necrosis factor-alpha (TNF-α), and gamma interferon. This results in a persistent low-grade chronic inflammatory state within the body. Chronic inflammation further contributes to the development of chronic diseases and accelerates the aging process (Qin et al., 2024).
Escherichia-Shigella has been associated with a pro-in flammatory statusand (Zhao et al., 2022), persistent infection with adherent and invasive Escherichia led to chronic and persistent peripheral inflammation (Liang et al., 2022). Cattaneo found a positive correlation between changes in the abundance of Escherichia-Shigella and changes in the levels of the pro-inflammatory mole cules IL-6, CXCL2, and NLRP3, the genus Escherichia to induce the production of pro-inflammatory cytokines through NLRP3-dependent mechanism (Cattaneo et al., 2017). Moreover, changes in the abundance of inflammation-related bacteria, such as Shigella and Agathobacter, were observed in female sarcopenia patients (Norman et al., 2021). Inflammation is thought to underlie various physiological and pathological processes. Age-related chronic low-grade inflammation is one of the important factors of sarcopenia. Macrophages released pro-inflammatory factors, reduced proteosynthesis, and raised protein degradation of the skeletal muscle through a variety of ways (Nardone et al., 2021). Many bacterial pathogens, including Shigella, may increase skeletal muscle damage by provoking inflammasome activity and inducing inflammatory responses (Suzuki et al., 2014). The pathogenic mechanism of Escherichia-Shigella involves a series of critical steps: (1) traversing the gastrointestinal barrier to reach the colon; (2) being engulfed by resident colonic macrophages, which subsequently triggers inflammasome activation; (3) the activated inflammasome induces macrophage death, facilitating Shigella’s escape; and (4) invading colonic epithelial cells and spreading to neighboring cells, leading to epithelial cell death, ulceration, increased fluid accumulation in the colon, and the formation of pus, blood, and mucus. It is evident that Shigella’s capacity to evade immune surveillance and escape plays a crucial role in its pathogenicity. Previous research has demonstrated that Shigella employs the type III secretion system (T3SS) to inject effector proteins into host cells, thereby aiding the bacterium in evading host immune defenses. Specifically, natural killer (NK) cells secrete granzyme A, which targets gasdermin B (GSDMB) in epithelial cells, promoting its cleavage and activation. This process enables GSDMB to perforate bacterial cell membranes, resulting in bacterial death. However, wild-type Shigella can counteract this effect by using the effector protein IpaH7.8 to promote the proteasomal degradation of GSDMB, thereby avoiding the bactericidal action of NK cells (Hansen et al., 2021).
Moreover, the supplementation of additional probiotics is recognized as a viable nutritional intervention that can enhance muscle mass and/or function while contributing to the prevention of muscle-degenerative diseases (Liu et al., 2021). Furthermore, Lee et al. observed that probiotics have the potential to enhance muscle mass, handgrip strength (HGS), gait speed, and balance while simultaneously mitigating sarcopenia, physical frailty, and fall incidence in elderly patients with primary osteoporosis (Karim et al., 2022). Another investigation further indicated that male athletes who consumed probiotics exhibited enhancements in muscle mass, strength, and exercise recovery (Przewlocka et al., 2020). The role of the gut microbiota in the development of muscle loss during aging is a crucial area that requires further studies for translation to patients.
Despite these interesting findings, our study is not without limitations. Firstly, the results of generalizability to other populations is questionable, because the vast majority of research into only from the two countries. Secondly, many studies are subject to significant bias. Statistically significant heterogeneity was observed among the included studies, which may be due to differences in dietary patterns, geographical background, and disease inclusion criteria (including different treatment regimens, medication doses, duration of disease, age, etc.). Nevertheless, we applied the random-model to estimate the effect sizes to reduce the influences of the heterogeneities on our results. Thirdly, it is important to consider that the utilization of diverse nucleic acid extraction methods and gene sequencing techniques (Table 1) may potentially introduce bias into the obtained results. For example, the differences ofαdiversity between sarcopenia and non-sarcopenia might be greater based on the V3-V4 region than those on the V4 region. However, the limited number of studies (one with V4 region and three with V3-V4 region) impeded us to perform additional analysis. Fourthly, in several studies, we manually extract the necessary data from the histogram, this may lead to another type of deviation. However, the procedure was carried out by two reviewers with full discussion and consensus. Therefore, we reasoned that the direction of statistical significance for between-group comparisons would not be materially affected because we performed this approach uniformly throughout the study. Fifthly, the effects in many occasions were assessed by very few studies and thus the current results should be interpreted with cautions. It merits future research to include more studies to provide stronger evidence on this issue.
In conclusion, we demonstrated that patients with sarcopenia showed significantly reduced α diversity in gut microbiota, and the composition of gut microbiota was significantly different with that of non-sarcopenia group. We also found more abundance of phylum Proteobacteria and genus Escherichia-Shigella, and less abundance of phylum Firmicutes and genus Faecalibacterium, Prevotella 9, Blautia in the sarcopenia group. In the future, a larger cohort study is needed to further examine the differences of gut microbiota in sarcopenia group.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
GW: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing, Software, Supervision, Validation, Visualization. YL: Data curation, Investigation, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing, Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources. HL: Writing – review & editing, Supervision. XY: Methodology, Supervision, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1513253/full#supplementary-material
Figure S1 | Funul plots of Chao 1 (A), Observed species (B), Shannon (C), and Simpson (D), Dorea (E), Bacteroides (F), Faecalibacterium prausnitzii (G), and Bifidobacterium longum (H).
Almohaisen, N., Gittins, M., Todd, C., Sremanakova, J., Sowerbutts, A. M., Aldossari, A., et al. (2022). Prevalence of undernutrition, frailty and sarcopenia in community-dwelling people aged 50 years and above: systematic review and meta-analysis. Nutrients 14:1537. doi: 10.3390/nu14081537
Ashworth, A. (2021). Sarcopenia and malnutrition: commonly occurring conditions in the older population. Br. J. Nurs. 30, S4–S10. doi: 10.12968/bjon.2021.30.21.S4
Bradley, P. H., and Pollard, K. S. (2017). Proteobacteria explain significant functional variability in the human gut microbiome. Microbiome 5:36. doi: 10.1186/s40168-017-0244-z
Cattaneo, A., Cattane, N., Galluzzi, S., Provasi, S., Lopizzo, N., Festari, C., et al. (2017). Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol. Aging 49, 60–68. doi: 10.1016/j.neurobiolaging.2016.08.019
Chen, S., Han, H., Sun, X., Zhou, G., Zhou, Q., and Li, Z. (2023). Causal effects of specific gut microbiota on musculoskeletal diseases: a bidirectional two-sample mendelian randomization study. Front. Microbiol. 14:1238800. doi: 10.3389/fmicb.2023.1238800
Chen, L. K., Woo, J., Assantachai, P., Auyeung, T. W., Chou, M. Y., Iijima, K., et al. (2020). Asian working group for sarcopenia: 2019 consensus update on sarcopenia diagnosis and treatment. J. Am. Med. Dir. Assoc. 21, 300–307.e2. doi: 10.1016/j.jamda.2019.12.012
Cho, M. R., Lee, S., and Song, S. K. (2022). A review of sarcopenia pathophysiology, diagnosis, treatment and future direction. J. Korean Med. Sci. 37:e146. doi: 10.3346/jkms.2022.37.e146
Cruz-Jentoft, A. J., Bahat, G., Bauer, J., Boirie, Y., Bruyere, O., Cederholm, T., et al. (2019). Sarcopenia: revised european consensus on definition and diagnosis. Age Ageing 48, 16–31. doi: 10.1093/ageing/afy169
Daily, J. W., and Park, S. (2022). Sarcopenia is a cause and consequence of metabolic dysregulation in aging humans: effects of gut dysbiosis, glucose dysregulation, diet and lifestyle. Cells 11:338. doi: 10.3390/cells11030338
Dao, T., Green, A. E., Kim, Y. A., Bae, S. J., Ha, K. T., Gariani, K., et al. (2020). Sarcopenia and muscle aging: a brief overview. Endocrinol. Metab. 35, 716–732. doi: 10.3803/EnM.2020.405
Fan, L., Xia, Y., Wang, Y., Han, D., Liu, Y., Li, J., et al. (2023). Gut microbiota bridges dietary nutrients and host immunity. Sci. China Life Sci. 66, 2466–2514. doi: 10.1007/s11427-023-2346-1
Fujisaka, S., Watanabe, Y., and Tobe, K. (2023). The gut microbiome: a core regulator of metabolism. J. Endocrinol. 256:e220111. doi: 10.1530/JOE-22-0111
Grosicki, G. J., Fielding, R. A., and Lustgarten, M. S. (2018). Gut microbiota contribute to age-related changes in skeletal muscle size, composition, and function: biological basis for a gut-muscle axis. Calcif. Tissue Int. 102, 433–442. doi: 10.1007/s00223-017-0345-5
Han, D. S., Wu, W. K., Liu, P. Y., Yang, Y. T., Hsu, H. C., Kuo, C. H., et al. (2022). Differences in the gut microbiome and reduced fecal butyrate in elders with low skeletal muscle mass. Clin. Nutr. 41, 1491–1500. doi: 10.1016/j.clnu.2022.05.008
Hansen, J. M., de Jong, M. F., Wu, Q., Zhang, L. S., Heisler, D. B., Alto, L. T., et al. (2021). Pathogenic ubiquitination of gsdmb inhibits nk cell bactericidal functions. Cell 184, 3178–3191.e18. doi: 10.1016/j.cell.2021.04.036
He, Y., Cui, W., Fang, T., Zhang, Z., and Zeng, M. (2023). Metabolites of the gut microbiota may serve as precise diagnostic markers for sarcopenia in the elderly. Front. Microbiol. 14:1301805. doi: 10.3389/fmicb.2023.1301805
Hozo, S. P., Djulbegovic, B., and Hozo, I. (2005). Estimating the mean and variance from the median, range, and the size of a sample. BMC Med. Res. Methodol. 5:13. doi: 10.1186/1471-2288-5-13
Iwasaka, C., Nanri, H., Nakagata, T., Ohno, H., Tanisawa, K., Konishi, K., et al. (2024). Association of skeletal muscle function, quantity, and quality with gut microbiota in japanese adults: a cross-sectional study. Geriatr Gerontol Int 24, 53–60. doi: 10.1111/ggi.14751
Jackson, M. A., Jeffery, I. B., Beaumont, M., Bell, J. T., Clark, A. G., Ley, R. E., et al. (2016). Signatures of early frailty in the gut microbiota. Genome Med. 8:8. doi: 10.1186/s13073-016-0262-7
Kang, L., Li, P., Wang, D., Wang, T., Hao, D., and Qu, X. (2021). Alterations in intestinal microbiota diversity, composition, and function in patients with sarcopenia. Sci. Rep. 11:4628. doi: 10.1038/s41598-021-84031-0
Karim, A., Muhammad, T., Shahid, I. M., and Qaisar, R. (2022). A multistrain probiotic improves handgrip strength and functional capacity in patients with copd: a randomized controlled trial. Arch. Gerontol. Geriatr. 102:104721. doi: 10.1016/j.archger.2022.104721
Kim, K. H., Chung, Y., Huh, J. W., Park, D. J., Cho, Y., Oh, Y., et al. (2022). Gut microbiota of the young ameliorates physical fitness of the aged in mice. Microbiome 10:238. doi: 10.1186/s40168-022-01386-w
Lapauw, L., Dupont, J., Amini, N., Vercauteren, L., Verschueren, S., Tournoy, J., et al. (2023). Trial in elderly with musculoskeletal problems due to underlying sarcopenia-faeces to unravel the gut and inflammation translationally (tempus-fugit): protocol of a cross-sequential study to explore the gut-muscle axis in the development and treatment of sarcopenia in community-dwelling older adults. BMC Geriatr. 23:599. doi: 10.1186/s12877-023-04291-5
Lee, S. Y., Kim, J. H., Lee, D. Y., and Hur, S. J. (2023). Characterization of gut microbiota in mouse models of aging and sarcopenia. Microbiol. Res. 275:127462. doi: 10.1016/j.micres.2023.127462
Lee, Y., Song, S., Jung, S. Y., Bae, J., Hwang, N., and Kim, H. (2022). Sarcopenia in community-dwelling older adults is associated with the diversity and composition of the gut microbiota. Exp. Gerontol. 167:111927. doi: 10.1016/j.exger.2022.111927
Li, G., Jin, B., and Fan, Z. (2022). Mechanisms involved in gut microbiota regulation of skeletal muscle. Oxidative Med. Cell. Longev. 2022:2151191. doi: 10.1155/2022/2151191
Liang, H., Song, H., Zhang, X., Song, G., Wang, Y., Ding, X., et al. (2022). Metformin attenuated sepsis-related liver injury by modulating gut microbiota. Emerg. Microbes Infect. 11, 815–828. doi: 10.1080/22221751.2022.2045876
Liao, X., Wu, M., Hao, Y., and Deng, H. (2020). Exploring the preventive effect and mechanism of senile sarcopenia based on "gut-muscle axis". Front. Bioeng. Biotechnol. 8:590869. doi: 10.3389/fbioe.2020.590869
Ling, Z., Liu, X., Cheng, Y., Yan, X., and Wu, S. (2022). Gut microbiota and aging. Crit. Rev. Food Sci. Nutr. 62, 3509–3534. doi: 10.1080/10408398.2020.1867054
Liu, C., Cheung, W. H., Li, J., Chow, S. K., Yu, J., Wong, S. H., et al. (2021). Understanding the gut microbiota and sarcopenia: a systematic review. J. Cachexia Sarcopenia Muscle 12, 1393–1407. doi: 10.1002/jcsm.12784
Lou, J., Wang, Q., Wan, X., and Cheng, J. (2024). Changes and correlation analysis of intestinal microflora composition, inflammatory index, and skeletal muscle mass in elderly patients with sarcopenia. Geriatr Gerontol Int 24, 140–146. doi: 10.1111/ggi.14661
Mo, X., Shen, L., Cheng, R., Wang, P., Wen, L., Sun, Y., et al. (2023). Faecal microbiota transplantation from young rats attenuates age-related sarcopenia revealed by multiomics analysis. J. Cachexia Sarcopenia Muscle 14, 2168–2183. doi: 10.1002/jcsm.13294
Nardone, O. M., de Sire, R., Petito, V., Testa, A., Villani, G., Scaldaferri, F., et al. (2021). Inflammatory bowel diseases and sarcopenia: the role of inflammation and gut microbiota in the development of muscle failure. Front. Immunol. 12:694217. doi: 10.3389/fimmu.2021.694217
Nikkhah, A., Ejtahed, H. S., Ettehad, M. F., Taghavi, M., Pakmehr, A., Hajipour, F., et al. (2023). The critical role of gut microbiota dysbiosis in skeletal muscle wasting: a systematic review. J. Appl. Microbiol. 134:lxac014. doi: 10.1093/jambio/lxac014
Norman, K., Hass, U., and Pirlich, M. (2021). Malnutrition in older adults-recent advances and remaining challenges. Nutrients 13:2764. doi: 10.3390/nu13082764
Page, M. J., Moher, D., Bossuyt, P. M., Boutron, I., Hoffmann, T. C., Mulrow, C. D., et al. (2021). Prisma 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews. BMJ 372:n160. doi: 10.1136/bmj.n160
Papadopoulou, S. K. (2020). Sarcopenia: a contemporary health problem among older adult populations. Nutrients 12:1293. doi: 10.3390/nu12051293
Petermann-Rocha, F., Balntzi, V., Gray, S. R., Lara, J., Ho, F. K., Pell, J. P., et al. (2022). Global prevalence of sarcopenia and severe sarcopenia: a systematic review and meta-analysis. J. Cachexia Sarcopenia Muscle 13, 86–99. doi: 10.1002/jcsm.12783
Przewlocka, K., Folwarski, M., Kazmierczak-Siedlecka, K., Skonieczna-Zydecka, K., and Kaczor, J. J. (2020). Gut-muscle axisexists and may affect skeletal muscle adaptation to training. Nutrients 12:1451. doi: 10.3390/nu12051451
Qin, C., Tian, G., Hu, J., Zou, X., and Yin, J. (2024). Recent chemical synthesis and immunological evaluation of glycans related to bacterial lipopolysaccharides. Curr. Opin. Chem. Biol. 78:102424. doi: 10.1016/j.cbpa.2023.102424
Sanz, Y., Romani-Perez, M., Benitez-Paez, A., Portune, K. J., Brigidi, P., Rampelli, S., et al. (2018). Towards microbiome-informed dietary recommendations for promoting metabolic and mental health: opinion papers of the mynewgut project. Clin. Nutr. 37, 2191–2197. doi: 10.1016/j.clnu.2018.07.007
Sayer, A. A., and Cruz-Jentoft, A. (2022). Sarcopenia definition, diagnosis and treatment: consensus is growing. Age Ageing 51:afac220. doi: 10.1093/ageing/afac220
Suzuki, S., Mimuro, H., Kim, M., Ogawa, M., Ashida, H., Toyotome, T., et al. (2014). Shigella ipah7.8 e3 ubiquitin ligase targets glomulin and activates inflammasomes to demolish macrophages. Proc. Natl. Acad. Sci. USA 111, E4254–E4263. doi: 10.1073/pnas.1324021111
Ticinesi, A., Mancabelli, L., Tagliaferri, S., Nouvenne, A., Milani, C., Del, R. D., et al. (2020). The gut-muscle axis in older subjects with low muscle mass and performance: a proof of concept study exploring fecal microbiota composition and function with shotgun metagenomics sequencing. Int. J. Mol. Sci. 21:8946. doi: 10.3390/ijms21238946
Wang, X., Chi, F., Wang, Y., and Zhang, W. (2022). Research progress on the relationship between gut microbiota and sarcopenia and its clinical significance. Chin. J. Microecol. 34, 1107–1111. doi: 10.13381/j.cnki.cjm.202209023
Wang, Y., Zhang, Y., Lane, N. E., Wu, J., Yang, T., Li, J., et al. (2022). Population-based metagenomics analysis reveals altered gut microbiome in sarcopenia: data from the xiangya sarcopenia study. J. Cachexia Sarcopenia Muscle 13, 2340–2351. doi: 10.1002/jcsm.13037
Yan, X., Li, H., Xie, R., Lin, L., Ding, L., Cheng, X., et al. (2023). Relationships between sarcopenia, nutrient intake, and gut microbiota in chinese community-dwelling older women. Arch. Gerontol. Geriatr. 113:105063. doi: 10.1016/j.archger.2023.105063
Zeng, X., Zhang, Y., Kwong, J. S., Zhang, C., Li, S., Sun, F., et al. (2015). The methodological quality assessment tools for preclinical and clinical studies, systematic review and meta-analysis, and clinical practice guideline: a systematic review. J. Evid. Based Med. 8, 2–10. doi: 10.1111/jebm.12141
Zhang, Y., Zhu, Y., Guo, Q., Wang, W., and Zhang, L. (2023). High-throughput sequencing analysis of the characteristics of the gut microbiota in aged patients with sarcopenia. Exp. Gerontol. 182:112287. doi: 10.1016/j.exger.2023.112287
Zhao, J., Bai, M., Ning, X., Qin, Y., Wang, Y., Yu, Z., et al. (2022). Expansion of Escherichia-Shigella in gut is associated with the onset and response to immunosuppressive therapy of Iga nephropathy. J. Am. Soc. Nephrol. 33, 2276–2292. doi: 10.1681/ASN.2022020189
Zhao, J., Liang, R., Song, Q., Song, S., Yue, J., and Wu, C. (2023). Investigating association between gut microbiota and sarcopenia-related traits: a mendelian randomization study. Precision. Clin. Med. 6:pbad10. doi: 10.1093/pcmedi/pbad010
Keywords: gut microbiota, sarcopenia, musculoskeletal diseases, effects, meta-analysis
Citation: Wang G, Li Y, Liu H and Yu X (2025) Gut microbiota in patients with sarcopenia: a systematic review and meta-analysis. Front. Microbiol. 16:1513253. doi: 10.3389/fmicb.2025.1513253
Received: 30 October 2024; Accepted: 10 January 2025;
Published: 22 January 2025.
Edited by:
Yongsheng Chen, Jinan University, ChinaReviewed by:
Yuchang Wang, Huazhong University of Science and Technology, ChinaCopyright © 2025 Wang, Li, Liu and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinjuan Yu, eXhqNDUwMUAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.