
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Microbiol., 03 March 2025
Sec. Virology
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1509890
Background and aims: Hepatitis viruses pose a significant global health challenge, necessitating accurate and efficient diagnostic methods. The CRISPR-Cas system, renowned for gene editing, shows potential tool in virus detection. This systematic review and meta-analysis aims to evaluate the diagnostic accuracy of CRISPR-Cas-based tests for hepatitis viruses, aiming to provide evidence for their effectiveness in clinical settings.
Methods: Studies from Web of Science, PubMed, and CNKI were analyzed. A bivariate random-effects model was employed to compute pooled estimates for sensitivity, specificity, and the area under the summary receiver operating characteristic (SROC) curve. Additionally, the methodological quality of the studies was evaluated using the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) tool.
Results: Following a rigorous screening process, 14 studies meeting our inclusion criteria were selected from an initial pool of 657 studies. The pooled sensitivity and specificity of the CRISPR-Cas system in hepatitis virus detection showed high sensitivity (0.99, 95% CI: 0.95–1.00) and specificity (0.99, 95% CI: 0.93–1.00) with SROC area 1.00 (95% CI: 0.99–1.00). However, considering the notable heterogeneity among the included studies, subgroup analyses and meta-regression were conducted. These analyses revealed that the type of hepatitis virus detected and the format of the final result presentation could be potential sources of this heterogeneity.
Conclusion: This systematic review and meta-analysis demonstrates the high diagnostic accuracy of CRISPR-Cas system in detecting hepatitis viruses. However, conclusions are limited by study number and quality. Therefore, more high-quality data are still needed to support this conclusion.
• The CRISPR-Cas system demonstrates very high sensitivity and specificity in detecting hepatitis viruses, suggesting it can accurately identify both infected and non-infected individuals.
• There is notable variability among the studies included in the analysis, which may be due to differences in the types of hepatitis viruses being detected and how the test results are presented.
• While the current evidence is promising, the number and quality of existing studies are limited, indicating a need for more high-quality research to confirm the effectiveness of CRISPR-Cas-based tests.
Hepatitis viruses mainly consist of five types: hepatitis A virus (HAV), hepatitis B virus (HBV), hepatitis C virus (HCV), hepatitis D virus (HDV) and hepatitis E virus (HEV) (Lanini et al., 2019). Globally, the number of HBV infection was estimated to be 296 million, with over 95% of cases in low-and middle-income countries. However, only 12–25% of patients have access to diagnosis and antiviral treatment (Yuen et al., 2018; Hsu et al., 2023). Approximately 58 million people worldwide suffer from chronic HCV infection, with 60–80% progressing to chronic hepatitis and potential risks of cirrhosis and liver cancer (Polaris Observatory HCV Collaborators, 2022). Nevertheless, due to the limited diagnostic capability, only 21% of HCV-infected individuals are diagnosed (Martinello et al., 2023; Pietschmann and Brown, 2019). Chronic HDV infection is considered the most severe form of viral hepatitis, and coinfection with HBV accelerates the progression of cirrhosis and hepatocellular carcinoma. Roughly 12 million people are infected with HDV globally, but this estimate remains controversial due to inadequate screening (Negro and Lok, 2023; Stockdale et al., 2020; European Association for the Study of the Liver, 2023). HEV is the leading cause of acute viral hepatitis worldwide, resulting in approximately 20 million infections and 70,000 deaths annually, posing a global public health concern (Aslan and Balaban, 2020; Kamar et al., 2017). In response, the World Health Organization has set a strategy to eliminate viral hepatitis as a public health threat by 2030, focusing on improving the efficiency and coverage of hepatitis virus testing (World Health Organization, 2017). This raises new and higher demands for hepatitis virus detection.
Currently, nucleic acid testing (NAT) is an important index to diagnose hepatitis virus, the most commonly used method of which is quantitative polymerase chain reaction (qPCR) (Pisano et al., 2021; Fu et al., 2023; Manso et al., 2024; Wedemeyer et al., 2023; Germer et al., 2017). Although significant advancements have been made in recent decades, this approach still faces several constraints: (a) it requires technically trained personnel with certified laboratory experience, (b) it often takes a long time to produce results, and (c) the high-sensitivity equipment required for RNA detection is expensive (Gupta et al., 2021). These limitations undermine the versatility of qPCR, especially in areas with limited medical resources. Therefore, cost-effective and highly sensitive NAT methods for hepatitis viruses are crucial for monitoring the occurrence and development of the disease and evaluating its treatment effect.
The CRISPR-Cas system, consisting of clustered regularly interspaced short palindromic repeats (CRISPR) and associated proteins (Cas), functions as a protective immune mechanism in prokaryotes, shielding them from external pathogens. This defense mechanism involves three key steps: first, inserting foreign nucleic acid fragments into the CRISPR sequence; second, initiating crRNA expression through transcription; third, guiding Cas nucleases to the target fragment through mature crRNA and then cleaving the homologous sequence of the invader’s genome (Barrangou and Marraffini, 2014; Hendriks et al., 2020). Due to its simplicity and programmability, the CRISPR-Cas system has rapidly become the preferred gene editing tool (Manghwar et al., 2019). Gootenberg et al. (2017) have established a molecular diagnostic technology platform using CRISPR-Cas system combined with recombinase polymerase amplification (RPA) for NAT, which has attracted widespread attention in the field of molecular diagnostics.
CRISPR-Cas12 and CRISPR-Cas13 exhibit targeted recognition and cleavage of double-stranded DNA (dsDNA) and RNA, respectively (Pickar-Oliver and Gersbach, 2019). When selectively targeting homologous sequences, Cas12 and Cas13 effectors undergo conformational changes, acting as signal amplifiers in NAT, resulting in significantly increased sensitivity. Additionally, this reaction process can be rapidly completed under mild conditions, which is highly beneficial for point-of-care testing (POCT) and large-scale screening of viral hepatitis (Kellner et al., 2019).
Due to the advantages of easy operation, cost-effectiveness, high sensitivity, and specificity in NAT, the use of the CRISPR-Cas system has been applied to the screening of various pathogens, including NAT of hepatitis viruses, but its diagnostic efficacy is still unclear. Therefore, this study aims to provide a brief meta-analysis of research on the diagnosis of hepatitis viruses based on the CRISPR-Cas system to evaluate its diagnostic efficacy (https://www.crd.york.ac.uk/PROSPERO/).
This systematic review and meta-analysis were conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines for DTA studies (McInnes et al., 2018). The study protocol was registered in the International Prospective Register of Systematic Reviews (PROSPERO) with registration ID CRD42024559017.
Database searches were conducted on Web of Science, PubMed, and CNKI without language restrictions to identify eligible studies on diagnostic test accuracy. Additionally, manual searches were performed on the reference lists of all studies identified by the search strategy. The search strategy included studies published in these databases up to August 1, 2024. Detailed search strategies are provided in the Supplementary File S1.
We defined the inclusion criteria as follows: (a) studies that used the CRISPR-Cas system for the detection of hepatitis virus nucleic acid; (b) studies that directly provided data on true positives (TP), false positives (FP), true negatives (TN), and false negatives (FN), or data that could be calculated to obtain these values; (c) studies that used quantitative PCR for nucleic acid on the included samples. However, studies were excluded if they met the following criteria: (a) the number of included samples was less than 15; (b) the study was a case report, review article, or meta-analysis article; (c) the study was a duplicate; (d) the gold standard was unclear or not used.
After removing duplicate studies, two authors (ZP and LX) independently screened all abstracts and full-texts. Using predetermined inclusion and exclusion criteria, the two authors extracted data from eligible studies. For studies that established multiple CRISPR-related NAT methods, data from different methods were extracted to maximize the amount of data. Disagreements were resolved by a third author (ZF).
Variables extracted from the selected studies included author names, study years, sample sizes, countries where the studies were conducted, types of Cas proteins used in the established methods, types of hepatitis viruses detected, and the presentation of results of the established methods. At the same time, data on TP (true positives), FP (false positives), FN (false negatives), and TN (true negatives) were extracted from the included studies.
We used the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) tool to evaluate the quality of the included studies (Whiting et al., 2011). This specifically includes the methodological quality of patient selection, index test, reference standard, as well as the flow and timing.
If any question is assessed as high risk, the bias risk for that item is considered high. If an item has two or more questions assessed as low risk, it is considered to have a low bias risk. If an item has two or more questions assessed as unclear, the bias risk for that item is considered unclear. Additionally, the applicability of patient selection, index tests, and reference tests was assessed. All assessments were independently conducted by two authors, and any disagreements were resolved by a third author.
By using the “Midas” package in Stata MP 16 software to perform a two-level random effects model analysis on the original research data, we obtained the pooled sensitivity, specificity, and area under the curve (AUC). The estimated values of sensitivity and specificity, along with their 95% confidence intervals (CIs), were presented in forest plot. Additionally, we calculated the summary receiver operating characteristic curve (SROC).
Heterogeneity was indicated by I2. Furthermore, we explored the potential sources of heterogeneity through subgroup analysis and meta-regression.
After searching three databases, a total of 657 relevant studies were retrieved, and after removing duplicates, there were 275 studies. We screened the 275 studies based on their titles and abstracts, excluding 218 studies. The remaining 57 studies were subject to full-text screening according to the inclusion criteria. Ultimately, this systematic review included 14 studies (Xu et al., 2024; Ho et al., 2023; Yue et al., 2021; He et al., 2024; Nguyen et al., 2023; Li et al., 2023; Kham-Kjing et al., 2022; Ding et al., 2021; Wang et al., 2021; Zhang et al., 2022; Chen et al., 2021; Tian et al., 2023a,b; Tian et al., 2022). A detailed flowchart of the study selection process and various reasons for exclusion is shown in Figure 1.
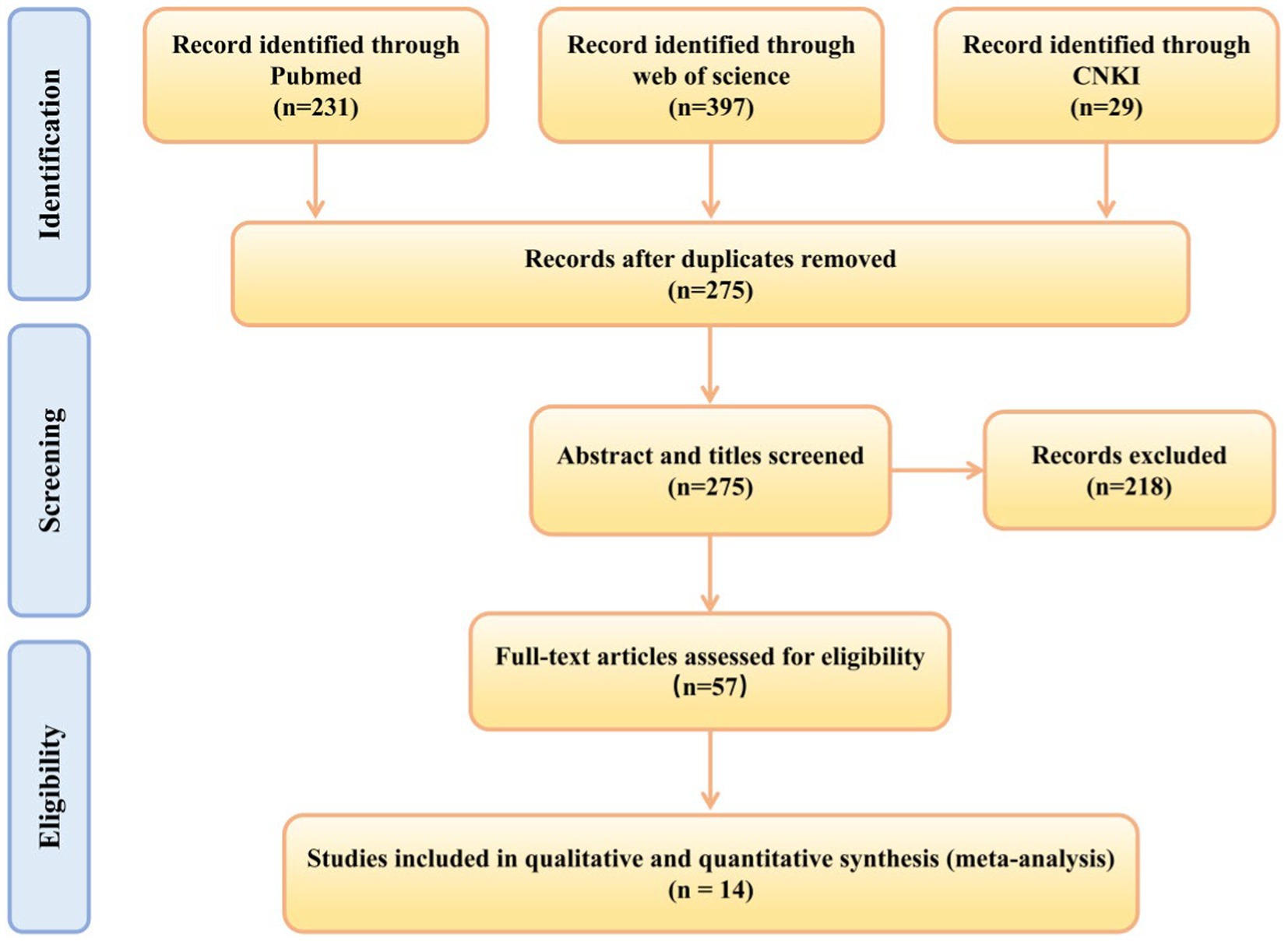
Figure 1. Preferred Reporting Items for Systematic Reviews and Meta-analysis flow diagram: search and study selection process for this review.
The 14 studies included were published from 2020 to 2024, with 12 studies from China and the remaining two from Thailand and the United States. The sample size of the included studies ranged from 17 to 312, and the types of Cas proteins involved Cas12 (8 studies) and Cas13 (6 studies). The hepatitis viruses detected included HBV, HCV, HDV, and HEV, and the results were presented in two forms: fluorescence and strips. Among them, four studies established both fluorescence and test strip methods. The main features of the included studies using CRISPR for hepatitis virus detection are shown in Table 1.
Figure 2A displays the individual QUADAS-2 assessment results for the 14 studies included, while Figure 2B presents the summary results. The risk of bias and applicability of all included studies were classified as “high,” “unknown”, and “low.” All 14 studies exhibited a low risk of bias in terms of flow and timing. In the patient selection domain, 42.9% (6/14) of studies had an unclear risk of bias due to the studies not specifying whether continuous or random samples were used, while the remaining studies had a low risk of bias. In the index test domain, 21.4% (3/14) of studies had an “unclear” risk of bias because they did not indicate whether samples were tested without knowledge of the gold standard, while the remaining studies had a low risk of bias. In the reference standard domain, 21.4% (3/14) of studies had a high risk of bias because it was uncertain in these studies whether the reference standard could accurately describe the sample situation. 14.3% (2/14) had an unclear risk of bias, and the remaining 64.3% (9/14) had a low risk of bias. In terms of applicability, 28.6% (4/14) of studies were classified as having an unclear risk in patient selection, while the remaining 71.4% (10/14) were classified as low risk. 7.1% (1/14) of studies were classified as having an unclear risk in the index test, while the remaining 92.9% (13/14) were classified as low risk. 28.6% (4/14) of studies were classified as having an unclear risk in the reference standard, while the remaining 71.4% (10/14) were classified as low risk.

Figure 2. Summary of the risk of bias of the included studies according to the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2). (A) Risk of bias and applicability concerns graph. (B) Risk of bias and applicability concerns summary.
Among the 14 studies included, the pooled sensitivity of the CRISPR-Cas system for detecting hepatitis viruses was 0.99 (95% CI 0.95–1.00), and the pooled specificity was 0.99 (95% CI 0.93–1.00). Figure 3 shows the forest plot of sensitivity and specificity for individual studies and the pooled results of the CRISPR-Cas system for detecting hepatitis viruses. The I2 test indicated significant heterogeneity among the studies. Figure 4 displays the SROC curve, and the area under curve (AUC) was 1.00 (95% CI 0.99–1.00). In addition, the funnel plot showed asymmetry, which was supported by the Egger test (p = 0.01) (Supplementary Figure S1).
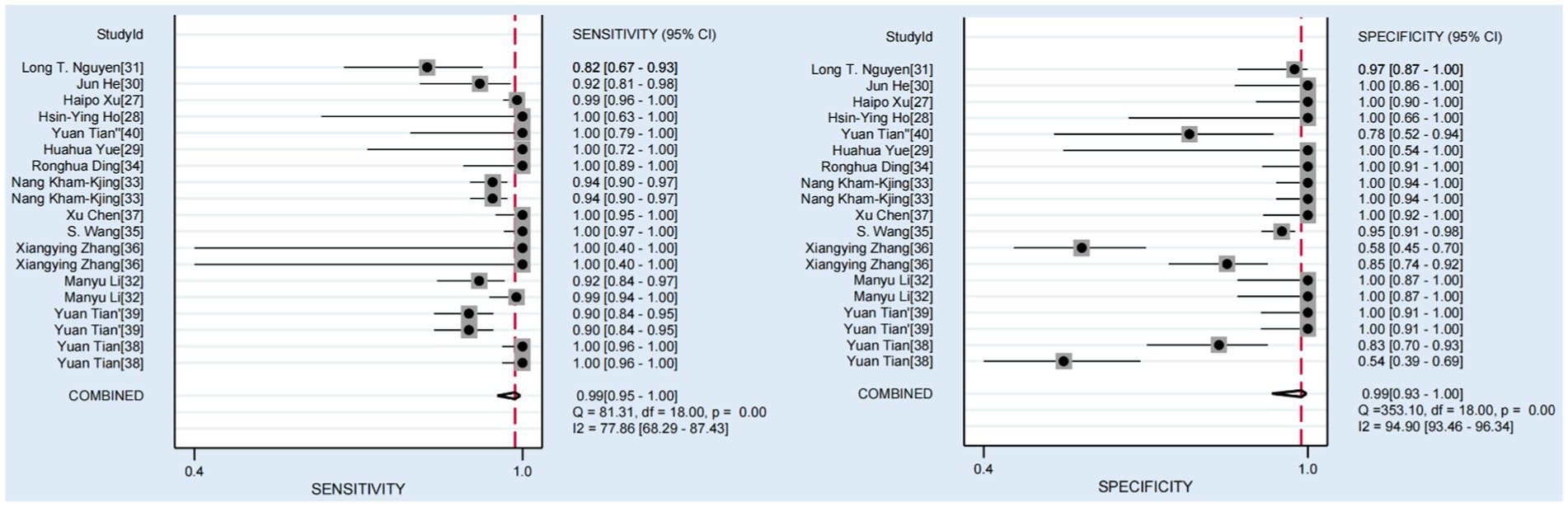
Figure 3. Forest plots of the pooled sensitivity and specificity for CRISPR-Cas system in diagnosis of hepatitis viruses.
To explore the sources of heterogeneity, we conducted subgroup analyses. First, we classified the studies based on the type of Cas protein used in the established NAT methods: 10 studies used Cas12 protein, and 10 studies used Cas13 protein. For studies using the Cas12 protein, the pooled sensitivity was 0.98 (95% CI 0.93–0.99), the specificity was 1.00 (95% CI 0.54–1.00), and the area under the SROC curve was 1.00 (95% CI 0.99–1.00). For studies using the Cas13 protein, the pooled sensitivity was 0.99 (95% CI 0.94–1.00), the specificity was 0.95 (95% CI 0.79–0.99), the area under the SROC curve was 1.00 (95% CI 0.99–1.00), and the results are shown in the Supplementary Figures S2, S3 (A: cas12/ B: cas13). And the forest plot is shown in the Supplementary Figures S4, S5 (A: cas12/ B: cas13). Detailed results of individual and pooled sensitivity and specificity are presented in Table 2. The pooled accuracy data of the CRISPR-Cas12/13 for detecting hepatitis viruses is presented in Table 3. Due to the limited number of studies included, other variables, including the type of hepatitis virus detected (DNA or RNA viruses), the result presentation format (fluorescence, strip) and sample nucleic acid amplification methods (PCR, recombinase polymerase amplification, RPA, recombinase-aid amplification, RAA/other amplification methods) were analyzed using meta-regression, and the results are shown in the Figure 5. In the sensitivity analysis, none of these four factors were statistically significant for sensitivity. In terms of specificity, both the format of result presentation and amplification methods exhibited statistical significance. This indicates that there were statistically significant differences in specificity based on whether the results were presented using fluorescence or test strips, as well as based on the sample nucleic acid amplification methods. These two factors may have contributed to the heterogeneity of specificity to some extent.
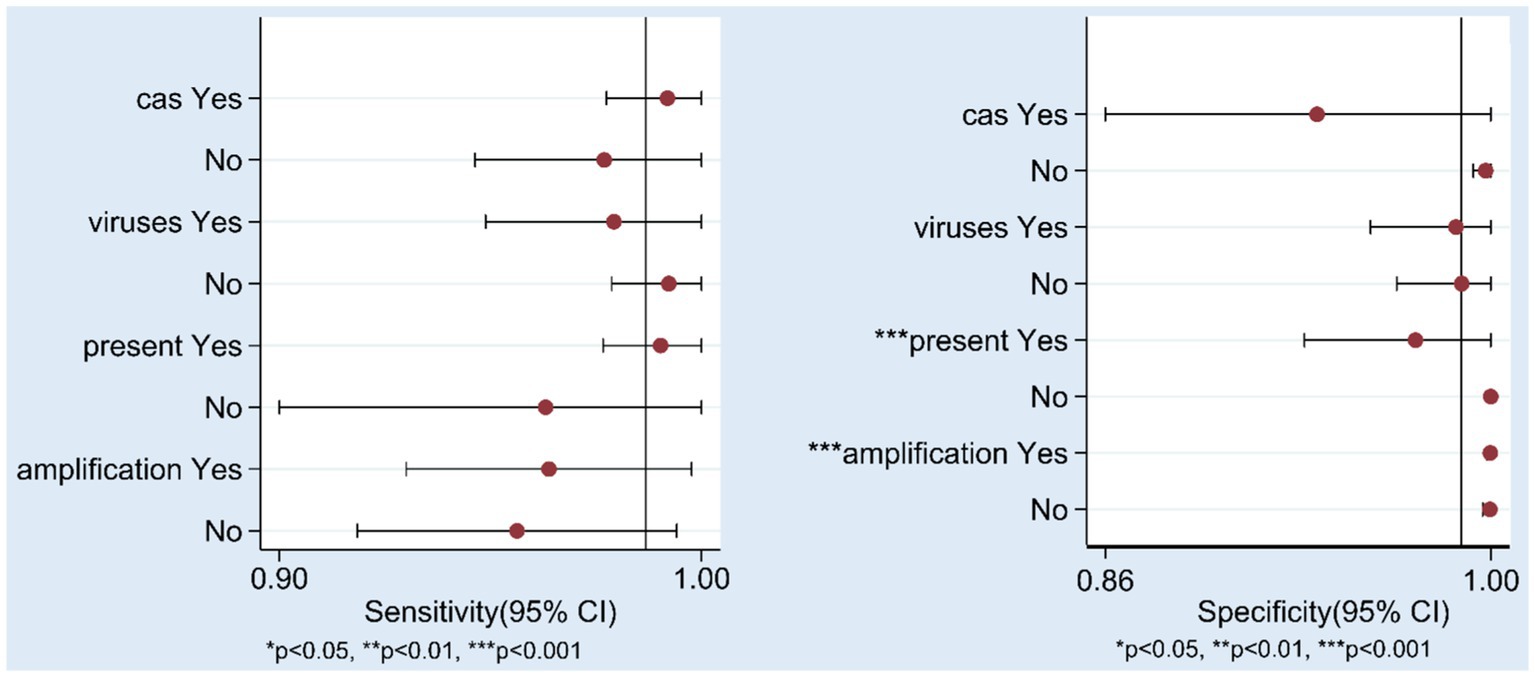
Figure 5. Results of Meta-regression. cas: CRISPR-Cas12/CRISPR-Cas13, virus: DNA/RNA viruses, present: fluorescence/test strip, amplification: PCR/RAA(RPA)/other amplification methods.
NAT results are widely accepted as a key indicator of hepatitis virus detection. However, the commonly employed qPCR for NAT faces several limitations that hinder large-scale screening, post-treatment monitoring, and utilization in regions with limited medical resources (Peeling et al., 2017). The CRISPR-Cas system, with its unique collateral cleavage activity of Cas proteins and efficient signal amplification mechanisms, offers a novel approach for ultra-sensitive and portable NAT.
In this systematic review and meta-analysis, we evaluated the accuracy of the CRISPR-Cas system in detecting hepatitis viruses. Our findings revealed a pooled sensitivity and specificity of 0.99 (95% CI: 0.95–1.00) and 0.99 (95% CI: 0.93–1.00), respectively, for hepatitis virus detection using the CRISPR-Cas system. The SROC curve demonstrated an AUC of 1.00 (95% CI: 0.99–1.00), indicating excellent diagnostic performance.
The I2 tests for sensitivity and specificity yielded values of 77.86 (95% CI: 68.29–87.43) and 94.90 (95% CI: 93.46–96.34), respectively, indicating substantial heterogeneity among the studies. To explore the sources of this heterogeneity, we stratified the included studies into Cas12 and Cas13 protein groups and constructed forest plots and SROC curves for each group. The results showed that for the Cas12 group, the pooled sensitivity was 0.98 (95% CI: 0.93–0.99) with an I2 test of 87.15 (95% CI: 80.45–93.85), while the pooled specificity was 1.00 (95% CI: 0.54–1.00) with an I2 test of 49.74 (95% CI: 13.33–86.15). For the Cas13 group, the pooled sensitivity was 0.99 (95% CI: 0.94–1.00) with an I2 test of 76.75 (95% CI: 62.53–90.97), and the pooled specificity was 0.95 (95% CI: 0.79–0.99) with an I2 test of 92.93 (95% CI: 89.83–96.03). Although a slight reduction in I2 values was observed after stratification, the differences were not statistically significant, suggesting that the use of different Cas proteins was not the primary source of heterogeneity. To further investigate the sources of heterogeneity, we performed meta-regression analysis, which revealed no statistically significant differences in sensitivity or specificity between the different Cas proteins, consistent with the subgroup analysis. It is noteworthy that there exist statistical differences in specificity between research on detecting DNA viruses and RNA viruses. This may be attributed to the fact that among the five common types of hepatitis viruses, only HBV is a DNA virus. Consequently, researchers have conducted more extensive studies on HBV, which has enhanced the accuracy of detection methods related to DNA viruses among hepatitis viruses. Furthermore, specific differences were also observed among studies based on the presentation of test results, which could be ascribed to the subjective nature of lateral flow analysis evaluation compared to fluorescence-based tests. Due to the existence of multiple genotypes within the same hepatitis virus, and a study has indicated the occurrence of recombination among different HBV genotypes (Liu et al., 2018). Consequently, the use of detection methods tailored to specific genotypes may be one of the reasons underlying the heterogeneity observed among various studies. Furthermore, the method of nucleic acid extraction from samples directly determines the test results, and differing nucleic acid extraction methods may also be one of the sources of heterogeneity among various studies. However, given the limited data included in this study, further research is still required to support these hypothesis.
It is important to note that this systematic review and meta-analysis relied on qPCR as the gold standard. However, the gold standard may not always accurately reflect the true sample status. For example, in the study of Zhang et al. (2022), a CRISPR-Cas-based test for detecting HBV cccDNA was developed. Compared to qPCR, the new method has a higher false positive rate, but the results are closer to those of digital drop PCR (ddPCR), which may indicate greater sensitivity of the CRISPR-Cas system. Similarly, Wang et al. (2021) reported 10 “false positive” results inconsistent with qPCR, but confirmed as low viral load positive by ddPCR, further validating the potential of CRISPR-Cas to outperform qPCR in sensitivity, which may have led to an underestimation of the specificity of the CRISPR-Cas system for detecting hepatitis virus.
This study is subject to several limitations. Firstly, there exists significant heterogeneity among the included studies, which we addressed by adopting a random-effects model to mitigate its impact. Additionally, we attempted to conduct subgroup analyses to explore the sources of heterogeneity. However, due to an inadequate number of studies, we were unable to further analyze and draw definitive conclusions.
Secondly, a potential source of bias arises from the nature of the research paradigm employed. Since all the included studies adhered to a pattern of establishing a method followed by clinical sample validation, there is a tendency for authors to preferentially report positive findings. Conversely, studies yielding negative results may be less likely to be published, as researchers may lack the motivation to document them (Siddaway et al., 2019). This publication bias could inflate the apparent accuracy of the results beyond what is actually observed in practice. We also confirmed the existence of publication bias by drawing funnel plot (Supplementary Figure S2).
Thirdly, qPCR, which serves as the gold standard for comparison, is not without its own limitations. Firstly, qPCR may not always accurately reflect the true state of the samples due to various factors such as sample preparation, primer design, and amplification efficiency. Secondly, the lack of complete standardization of qPCR techniques across different laboratories hinders the direct comparability of results. This variability in techniques can introduce additional uncertainty into the interpretation of our study (Rizzetto and Hamid, 2021).
Finally, the novelty of CRISPR-Cas system for hepatitis virus detection is a factor that necessitates caution in interpreting our results. Given that this technology has only recently gained traction for this application, all the included studies in our analysis were published within the last 5 years. As such, our conclusions are based on a relatively limited body of evidence, and further research is warranted to refine this methodology and strengthen the support for our findings. More comprehensive and long-term studies are needed to fully evaluate the potential of CRISPR-Cas system in hepatitis virus detection.
The detection of hepatitis viruses remains a pivotal task within the realm of liver diseases and infectious diseases. This systematic review and meta-analysis, for the first time, presents diagnostic accuracy data for the utilization of the CRISPR-Cas system in detecting hepatitis viruses. However, due to the notable heterogeneity observed in the current studies, our findings necessitate further validation with more high-quality research data. Overall, the CRISPR-Cas system, with its excellent diagnostic accuracy, shows great potential in the identification of various hepatitis viruses and is expected to become a powerful alternative to traditional detection methods, leading a new chapter in future hepatitis virus diagnostic technology.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
ZP: Formal analysis, Investigation, Methodology, Writing – original draft. LX: Data curation, Project administration, Software, Writing – original draft. ZF: Resources, Software, Validation, Writing – review & editing. FR: Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (82002243 and 82100653), Key Projects of the Beijing Municipal Education Commission’s Science and Technology Plan (KZ202010025035), Chinese Institutes for Medical Research, Beijing (Grant No. CX24PY23), Beijing Hospitals Authority Youth Programme (QML20201702), Talent Cultivation Plan of Climbing the Peak of Beijing Municipal Hospital Administration (DFL20221503), Beijing Natural Science Foundation-Changping Innovation Joint Fund (L234046), Training Fund for Open Projects at Clinical Institutes and Departments of Capital Medical University (CCMU2023ZKYXZ003), High-Level Public Health Technical Talents Project of Beijing (Subject Leaders-02-13 and Xuekegugan-03-48).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1509890/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Funnel plot for included studies.
SUPPLEMENTARY FIGURE S2 | The SROC curves of the CRISPR-Cas12/13 system in diagnosis of hepatitis viruses. (A) CRISPR-Cas12. (B) CRISPR-Cas13.
SUPPLEMENTARY FIGURE S3 | Forest plots of the pooled sensitivity and specificity for CRISPR-Cas12/13 system in diagnosis of hepatitis viruses. (A) CRISPR-Cas12. (B) CRISPR-Cas13.
Image 1: Supplementary Figure S1
Image 2: Supplementary Figure S2
Image 3: Supplementary Figure S3
Image 4: Supplementary Figure S4
Image 5: Supplementary Figure S5
Aslan, A. T., and Balaban, H. Y. (2020). Hepatitis E virus: epidemiology, diagnosis, clinical manifestations, and treatment. World J. Gastroenterol. 26, 5543–5560. doi: 10.3748/wjg.v26.i37.5543
Barrangou, R., and Marraffini, L. A. (2014). CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell 54, 234–244. doi: 10.1016/j.molcel.2014.03.011
Chen, X., Tan, Y., Wang, S., Wu, X., Liu, R., Yang, X., et al. (2021). A CRISPR-Cas12b-based platform for ultrasensitive, rapid, and highly specific detection of hepatitis B virus genotypes B and C in clinical application. Front. Bioeng. Biotechnol. 9:743322. doi: 10.3389/fbioe.2021.743322
Ding, R., Long, J., Yuan, M., Zheng, X., Shen, Y., Jin, Y., et al. (2021). CRISPR/Cas12-based ultra-sensitive and specific point-of-care detection of HBV. Int. J. Mol. Sci. 22:4842. doi: 10.3390/ijms22094842
European Association for the Study of the Liver (2023). EASL clinical practice guidelines on hepatitis delta virus. J. Hepatol. 79, 433–460. doi: 10.1016/j.jhep.2023.05.001
Fu, M. X., Simmonds, P., Andreani, J., Baklan, H., Webster, M., Asadi, R., et al. (2023). Ultrasensitive PCR system for HBV DNA detection: risk stratification for occult hepatitis B virus infection in English blood donors. J. Med. Virol. 95:e29144. doi: 10.1002/jmv.29144
Germer, J. J., Ankoudinova, I., Belousov, Y. S., Mahoney, W., Dong, C., Meng, J., et al. (2017). Hepatitis E virus (HEV) detection and quantification by a real-time reverse transcription-PCR assay calibrated to the World Health Organization standard for HEV RNA. J. Clin. Microbiol. 55, 1478–1487. doi: 10.1128/JCM.02334-16
Gootenberg, J. S., Abudayyeh, O. O., Lee, J. W., Essletzbichler, P., Joung, J., Verdine, V., et al. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442. doi: 10.1126/science.aam9321
Gupta, E., Khodare, A., Rani, N., Singh, G., Aggarwal, K., and Sharma, M. (2021). Performance evaluation of Xpert HBV viral load (VL) assay: point-of-care molecular test to strengthen and decentralize management of chronic hepatitis B (CHB) infection. J. Virol. Methods 290:114063. doi: 10.1016/j.jviromet.2021.114063
He, J., Hu, X., Weng, X., Wang, H., Yu, J., Jiang, T., et al. (2024). Efficient, specific and direct detection of double-stranded DNA targets using Cas12f1 nucleases and engineered guide RNAs. Biosens. Bioelectron. 260:116428. doi: 10.1016/j.bios.2024.116428
Hendriks, D., Clevers, H., and Artegiani, B. (2020). CRISPR-Cas tools and their application in genetic engineering of human stem cells and organoids. Cell Stem Cell 27, 705–731. doi: 10.1016/j.stem.2020.10.014
Ho, H. Y., Kao, W. S., Deval, P., Dai, C. Y., Chen, Y. H., Yu, M. L., et al. (2023). Rapid and sensitive LAMP/CRISPR-powered diagnostics to detect different hepatitis C virus genotypes using an ITO-based EG-FET biosensing platform. Sens. Actuators B 394:134278. doi: 10.1016/j.snb.2023.134278
Hsu, Y. C., Huang, D. Q., and Nguyen, M. H. (2023). Global burden of hepatitis B virus: current status, missed opportunities and a call for action. Nat. Rev. Gastroenterol. Hepatol. 20, 524–537. doi: 10.1038/s41575-023-00760-9
Kamar, N., Izopet, J., Pavio, N., Aggarwal, R., Labrique, A., Wedemeyer, H., et al. (2017). Hepatitis E virus infection. Nat. Rev. Dis. Primers 3:17086. doi: 10.1038/nrdp.2017.86
Kellner, M. J., Koob, J. G., Gootenberg, J. S., Abudayyeh, O. O., and Zhang, F. (2019). SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 14, 2986–3012. doi: 10.1038/s41596-019-0210-2
Kham-Kjing, N., Ngo-Giang-Huong, N., Tragoolpua, K., Khamduang, W., and Hongjaisee, S. (2022). Highly specific and rapid detection of hepatitis C virus using RT-LAMP-coupled CRISPR-Cas12 assay. Diagnostics 12:1524. doi: 10.3390/diagnostics12071524
Lanini, S., Ustianowski, A., Pisapia, R., Zumla, A., and Ippolito, G. (2019). Viral hepatitis: etiology, epidemiology, transmission, diagnostics, treatment, and prevention. Infect. Dis. Clin. N. Am. 33, 1045–1062. doi: 10.1016/j.idc.2019.08.004
Li, M., He, Q., Li, T., Wan, W., and Zhou, H. (2023). Development and evaluation of a CRISPR-Cas13a system-based diagnostic for hepatitis E virus. Clin. Chem. Lab. Med. 62, 1237–1247. doi: 10.1515/cclm-2023-1007
Liu, B., Yang, J. X., Yan, L., Zhuang, H., and Li, T. (2018). Novel HBV recombinants between genotypes B and C in 3′-terminal reverse transcriptase (RT) sequences are associated with enhanced viral DNA load, higher RT point mutation rates and place of birth among Chinese patients. Infect. Genet. Evol. 57, 26–35. doi: 10.1016/j.meegid.2017.10.023
Manghwar, H., Lindsey, K., Zhang, X., and Jin, S. (2019). CRISPR/Cas system: recent advances and future prospects for genome editing. Trends Plant Sci. 24, 1102–1125. doi: 10.1016/j.tplants.2019.09.006
Manso, T., de Salazar, A., Rodríguez-Velasco, M., García, F., and Aguilera, A. (2024). Detection and quantification of HBV and HCV in plasma samples by means of different molecular assays: comparative study. Enferm. Infecc. Microbiol. Clin. 42, 13–16. doi: 10.1016/j.eimc.2022.07.007
Martinello, M., Solomon, S. S., Terrault, N. A., and Dore, G. J. (2023). Hepatitis C. Lancet 402, 1085–1096. doi: 10.1016/S0140-6736(23)01320-X
McInnes, M. D. F., Moher, D., Thombs, B. D., TA, M. G., Clifford, T., Cohen, J. F., et al. (2018). Preferred reporting items for a systematic review and meta-analysis of diagnostic test accuracy studies: the PRISMA-DTA statement. JAMA 319, 388–396. doi: 10.1001/jama.2017.19163
Negro, F., and Lok, A. S. (2023). Hepatitis D: a review. JAMA. 330, 2376–2387. doi: 10.1001/jama.2023.23242
Nguyen, L. T., Rananaware, S. R., Yang, L. G., Macaluso, N. C., Ocana-Ortiz, J. E., Meister, K. S., et al. (2023). Engineering highly thermostable Cas12b via de novo structural analyses for one-pot detection of nucleic acids. Cell Rep Med. 4:101037. doi: 10.1016/j.xcrm.2023.101037
Peeling, R. W., Boeras, D. I., Marinucci, F., and Easterbrook, P. (2017). The future of viral hepatitis testing: innovations in testing technologies and approaches. BMC Infect. Dis. 17:699. doi: 10.1186/s12879-017-2775-0
Pickar-Oliver, A., and Gersbach, C. A. (2019). The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 20, 490–507. doi: 10.1038/s41580-019-0131-5
Pietschmann, T., and RJP, B. (2019). Hepatitis C virus. Trends Microbiol. 27, 379–380. doi: 10.1016/j.tim.2019.01.001
Pisano, M. B., Giadans, C. G., Flichman, D. M., Ré, V. E., Preciado, M. V., and Valva, P. (2021). Viral hepatitis update: progress and perspectives. World J. Gastroenterol. 27, 4018–4044. doi: 10.3748/wjg.v27.i26.4018
Polaris Observatory HCV Collaborators (2022). Global change in hepatitis C virus prevalence and cascade of care between 2015 and 2020: a modelling study. Lancet Gastroenterol. Hepatol. 7, 396–415. doi: 10.1016/S2468-1253(21)00472-6
Rizzetto, M., and Hamid, S. (2021). The medical impact of hepatitis D virus infection in Asia and Africa; time for a reappraisal. Liver Int. 41, 16–19. doi: 10.1111/liv.14729
Siddaway, A. P., Wood, A. M., and Hedges, L. V. (2019). How to do a systematic review: a best practice guide for conducting and reporting narrative reviews, meta-analyses, and meta-syntheses. Annu. Rev. Psychol. 70, 747–770. doi: 10.1146/annurev-psych-010418-102803
Stockdale, A. J., Kreuels, B., Henrion, M. Y. R., Giorgi, E., Kyomuhangi, I., de Martel, C., et al. (2020). The global prevalence of hepatitis D virus infection: systematic review and meta-analysis. J. Hepatol. 73, 523–532. doi: 10.1016/j.jhep.2020.04.008
Tian, Y., Fan, Z., Xu, L., Cao, Y., Chen, S., Pan, Z., et al. (2023a). CRISPR/Cas13a-assisted rapid and portable HBV DNA detection for low-level viremia patients. Emerg. Microbes Infect. 12:e2177088. doi: 10.1080/22221751.2023.2177088
Tian, Y., Fan, Z., Zhang, X., Xu, L., Cao, Y., Pan, Z., et al. (2023b). CRISPR/Cas13a-assisted accurate and portable hepatitis D virus RNA detection. Emerg. Microbes Infect. 12:2276337. doi: 10.1080/22221751.2023.2276337
Tian, Y., Wan, Y., Xu, L., Zhang, X. Y., Ren, F., and Wu, J. (2022). Clinical application evaluation of hepatitis B virus cccDNA detection method based on CRISPR-Cas13a technology. Hepatol Int. 16:306–315.
Wang, S., Li, H., Kou, Z., Ren, F., Jin, Y., Yang, L., et al. (2021). Highly sensitive and specific detection of hepatitis B virus DNA and drug resistance mutations utilizing the PCR-based CRISPR-Cas13a system. Clin. Microbiol. Infect. 27, 443–450. doi: 10.1016/j.cmi.2020.04.018
Wedemeyer, H., Leus, M., Glenn, J., Gordien, E., Kamili, S., Kapoor, H., et al. (2023). HDV RNA assays: performance characteristics, clinical utility, and challenges. Hepatology 81, 637–650. doi: 10.1097/HEP.0000000000000584
Whiting, P. F., AWS, R., Westwood, M. E., Mallett, S., Deeks, J. J., Reitsma, J. B., et al. (2011). QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 155, 529–536. doi: 10.7326/0003-4819-155-8-201110180-00009
Xu, H., Lin, G., Chen, R., Cai, Z., Sun, Y., Zhang, X., et al. (2024). CRISPR/Cas12b assisted loop-mediated isothermal amplification for easy, rapid and sensitive quantification of chronic HBV DNA in one-pot. Anal. Chim. Acta 1310:342702. doi: 10.1016/j.aca.2024.342702
Yue, H., Shu, B., Tian, T., Xiong, E., Huang, M., Zhu, D., et al. (2021). Droplet Cas12a assay enables DNA quantification from unamplified samples at the single-molecule level. Nano Lett. 21, 4643–4653. doi: 10.1021/acs.nanolett.1c00715
Yuen, M. F., Chen, D. S., Dusheiko, G. M., Janssen, H. L. A., Lau, D. T. Y., Locarnini, S. A., et al. (2018). Hepatitis B virus infection. Nat. Rev. Dis. Primers 4:18035. doi: 10.1038/nrdp.2018.35
Keywords: CRISPR-Cas, hepatitis virus, nucleic acid testing, diagnostic, meta-analysis
Citation: Pan Z, Xu L, Fan Z and Ren F (2025) CRISPR-Cas for hepatitis virus: a systematic review and meta-analysis of diagnostic test accuracy studies. Front. Microbiol. 16:1509890. doi: 10.3389/fmicb.2025.1509890
Received: 29 October 2024; Accepted: 05 February 2025;
Published: 03 March 2025.
Edited by:
Murugan Kasi, Manonmaniam Sundaranar University, IndiaReviewed by:
Puey Ounjai, Mahidol University, ThailandCopyright © 2025 Pan, Xu, Fan and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Ren, cmVuZmVuZzc1MTJAY2NtdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.