- 1University of Maryland Eastern Shore, Department of Agriculture, Food and Resource Science, Princess Anne, MD, United States
- 2United States Department of Agriculture, Agricultural Research Service, Dover, DE, United States
- 3National Oceanic and Atmospheric Administration, NOS, NCCOS, Cooperative Oxford Laboratory, Oxford, MD, United States
Shewanella is a genus of Gram-negative marine bacteria with some species associated with human and shellfish illnesses. This study evaluated the abundance of Shewanella species in oysters and seawater from the Chesapeake and Maryland Coastal Bays at four sites between 2019 and 2021. Physicochemical parameters such as temperature, salinity, dissolved oxygen, turbidity, pH, chlorophyll-a, rainfall within the last 48 h, total dissolved solids, and atmospheric pressure were also recorded to evaluate if there was a correlation between environmental parameters and the level of Shewanella. The highest total Shewanella counts were 1.8 × 107 CFU/g in oysters and 4.0 × 102 CFU/mL in seawater. 16S rRNA sequencing was performed on 1,344 representative isolates of which 890 (713 oyster, 177 seawater) were confirmed as Shewanella within 16 species. The top four species isolated from oysters and seawater were S. khirikhana a known shrimp pathogen (49%), S. marisflavi (19%), S. loihica (11%), and S. algae (8%). Testing for alpha and beta hemolysis were performed on all confirmed Shewanella isolates. Beta hemolysis was observed in 405 (46%) of the isolates of which 313 were in oysters and 92 in seawater. In oysters, beta-hemolysis was most prevalent in S. khirikhana (233 of 344 isolates, 68%), while in seawater 64 of 92 isolates (70%) were beta-hemolytic strains. Beta-hemolysis suggests that these could be potentially pathogenic strains. Correlations were performed between physicochemical attributes of the seawater and Shewanella counts. Only seawater temperature and dissolved oxygen correlated with Shewanella counts (r = 0.45 and − 0.41), respectively. No correlations were observed between the physicochemical parameters and Shewanella abundances in oysters. Results suggest that virulent strains of Shewanella may be present in oysters and seawater from the Chesapeake and Maryland Coastal Bays, perhaps as a consequence of rising seawater temperatures.
Introduction
Shewanella spp. are Gram-negative, facultatively anaerobic, singly flagellated, oxidase and catalase positive, rod-shaped bacteria that can produce H2S (Goyal et al., 2011). Members of the family Shewanellaceae are naturally occurring marine bacteria, distributed worldwide (Hau and Gralnick, 2007; Lemaire et al., 2020). Over 70 species of Shewanellaceae have been identified (Lemaire et al., 2020); however, there is still uncertainty of exactly how many species are truly human pathogens. Currently, five species have been associated with human infection: S. algae, S. putrefaciens (Hau and Gralnick, 2007), S. xiamensis (Zong, 2011), S. haliotis (Poovorawan et al., 2013; Zhang et al., 2018; Liu et al., 2019) and S. upenei (Yousfi et al., 2017). One recently identified species, S. khirikhana, has been associated with disease in cultured shrimp (Prachumwat et al., 2020). Shewanella is a genus that contains species that are human and shellfish pathogens as well as species that are widely recognized as seafood spoilage organisms.
Shewanella isolates can often exhibit hemolytic activity on blood agar plates, which is indicative of their ability to lyse red blood cells. Beta-hemolytic activity is a common feature associated with pathogenicity in various bacterial species. For instance, in Staphylococcus aureus, beta hemolysins are key virulence factors that contribute to tissue invasion and immune evasion, and their activity can be significantly reduced by inhibiting the ClpP protease, central regulator of virulence factors (Böttcher and Sieber, 2008). Similarly, Shewanella species such as S. algae and S. baltica have been shown to possess beta-hemolytic activity, and this trait may be predictive of their potential virulence, especially given their ability to grow at human body temperature (Richards et al., 2008). Moreover, the presence of genes encoding hemolysins, like those found in the S. algae STY3 genome, further supports the potential for these species to be pathogenic under the right conditions. These genes include those similar to hlyA, which encodes a toxin capable of causing cell lysis, and other genes involved in a hemolysin operon (Wu et al., 2018; Tamez et al., 2020). While S. baltica has not been commonly reported as a cause of human infections (Myung et al., 2009), the increasing number of illnesses linked to Shewanella infections in the last decade underscores the importance of considering these bacteria as emerging pathogens (Ng et al., 2022). These findings highlight the need for further research to elucidate the specific virulence mechanisms in Shewanella and their implications for human health.
Alpha hemolysins function by partially breaking down hemoglobin into biliverdin, resulting in a characteristic green discoloration around colonies on blood agar (Tasnim, 2017). This activity can damage host cells through mechanisms such as pore formation in cell membranes, as seen with pneumolysin in Streptococcus pneumoniae (Tasnim, 2017), which disrupts cellular integrity and can lead to cell death. Alpha hemolysins can interfere with host cell signaling pathways, suppressing immune responses and promoting bacterial survival. For example, pneumolysin produced by S. pneumoniae has been shown to impair host immune cells, allowing the bacteria to colonize and spread more effectively (Tasnim, 2017). In Shewanella, genomic analyses suggest the presence of genes encoding toxins similar to alpha hemolysins found in other bacteria, such as RTX-like toxins (Alav et al., 2021). These RTX toxins, like pneumolysin in S. pneumoniae, play roles in immune evasion and persistence within the host by targeting cell membranes and disrupting immune signaling (Tasnim, 2017). In Shewanella, RTX-like toxins may play a similar role in damaging host tissues, contributing to bacterial survival and colonization (Tasnim, 2017; Alav et al., 2021).
Pathogenic species reported most frequently in the literature are S. algae and S. putrefaciens, however, in the past, there have been problems identifying the microbes at the species level using conventional microbiological culture and biochemical analyses (Gram et al., 1999). More recently, this problem has been resolved by incorporating 16S rRNA gene sequencing, as the DNA sequences provide an opportunity for more definitive identification (Hau and Gralnick, 2007; Zong, 2011; Liu et al., 2013). Humans can become infected from certain Shewanella species by exposing cuts and abrasions in the skin to contaminated seawater, and from various water activities such as swimming, fishing, and boating. The consumption of raw or undercooked seafood has also been associated with illness (Myung et al., 2009; Liu et al., 2019). Some species are pathogenic to shellfish, causing production losses in the seafood industry (Huang et al., 2018). In humans, S. algae can cause soft tissue infections; bone, joint, and ear infections; lower respiratory tract infection; and occasional gastroenteritis (Holt et al., 2005; Dey et al., 2015; Takata et al., 2017; Kitaoka et al., 2019). It can also cause necrotizing fasciitis and typically attacks individuals who are immunocompromised, or have underlying conditions such as renal failure, diabetes mellitus, liver disease, and malignancies (Holt et al., 2005; Dey et al., 2015; Takata et al., 2017; Kitaoka et al., 2019).
Shewanella spp. can be readily isolated from seafood and environmental samples (Richards et al., 2008; Kang et al., 2013; Calvert County Health Department, 2017). Potentially pathogenic species were identified in the Delaware Bay with high numbers present in oysters and seawater during the summer months (Richards et al., 2008). Kang et al. (2013) isolated antibiotic resistant S. putrefaciens from shellfish in the West Sea in Korea. In 2017, cases of Shewanella infections have also been reported around the Chesapeake Bay area (Calvert County Health Department, 2017). In addition, one case has been reported during 2018 in New York involved an 87-year-old patient diagnosed with an infection caused by Shewanella haliotis (Liu et al., 2019). These cases underscore the potential pathogenicity of Shewanella species, particularly those isolated from marine environments. Furthermore, information is limited concerning the prevalence of this bacterium in oysters and seawater from the Chesapeake Bay and the Maryland Coastal Bays.
The objectives of this study were to: (i) isolate, identify, and quantify species of Shewanella in the Chesapeake and Maryland Coastal Bays; (ii) determine the levels of hemolytic Shewanella spp. in seawater and oysters as a potential indicator of their pathogenic potential, (iii) evaluate the seasonal distribution of Shewanella spp. in oysters and seawater at various sampling sites, and (iv) correlate Shewanella levels in oyster and seawater samples with physicochemical parameters of the seawater.
Materials and methods
Sampling sites
Study sites were selected to represent the range of salinities where Shewanella spp. may be present as well as represent the optimum conditions for the survival of oysters. Three sites in the Chesapeake Bay (Horn Point, Honga River, Tangier Sound) and one site in the Maryland Coastal Bays (Sinepuxent Bay on the Atlantic Ocean) were selected based on salinities, water quality, accessibility, and availability of oysters (Figure 1). The sites in the Maryland Coastal Bays represent the higher salinity range (generally 27–35 ppt). Salinities at selected sites were representative of oyster-growing areas nationally. The Chesapeake Bay is located on the East coast of the US within the states of Maryland and Virginia. The Chesapeake Bay is the largest estuary in North America. It is 314 km (195 miles) long and between 6 and 48 km (4 and 30 miles) wide, covering an area of 8,384 km2 (3,237 miles2), with 4,470 km2 (1,726 miles2) in Maryland. Salinities increase from north to south as one moves downstream from Maryland’s portion of the Bay into Virginia where the Chesapeake Bay meets the Atlantic Ocean. The Maryland Coastal Bays encompass the Assawoman Bay, Chincoteague Bay, Isle of Wright Bay, Newport Bay, Sinepuxent Bay, and St. Martin River (Rodgers et al., 2014). For this current study, because of the scarcity of wild oysters in some locations, we utilized oysters cultured at a site in the Sinepuxent Bay.
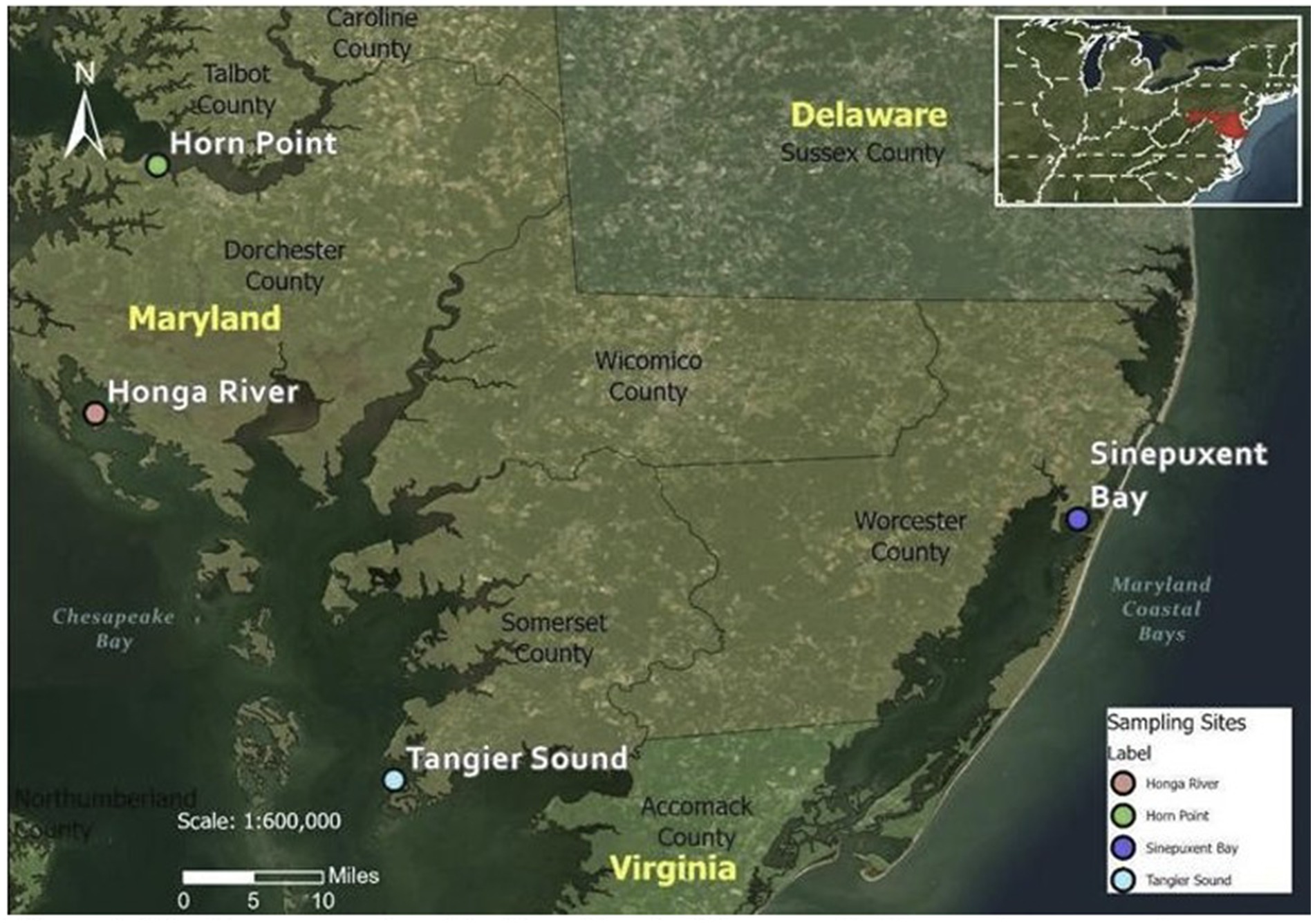
Figure 1. The three sample collection sites in Chesapeake Bay and one sample collection site in Maryland Coastal Bay. Coordinates for sampling sites: Horn Point (38°35′33”N 76°07′47”W); Honga River (38°20′03″N 76°11″W); Tangier Sound (37°57′09″N 75°53′01″W); Sinepuxent Bay (38°13′27″N 75°10′21″W).
Collection of samples
Eighty-one oyster samplings were performed, consisting of 12 oysters per sampling, and 82 seawater samplings (1-liter each) were collected from three study sites in the Chesapeake Bay and one site along the Maryland Coastal Bays monthly from June 2019 through October 2021 (Supplementary Table S1); except for 4 months (March–June 2020) as the university was closed due to the COVID-19 lockdown. At Horn Point, a total of 22 water and 22 oyster samplings were conducted. Similarly, 21 water and 21 oyster samplings were performed in the Honga River. In Tangier Sound, a total of 23 water and 23 oyster samplings were conducted. Lastly, at Maryland Coastal Bay, 16 water and 15 oyster samplings were performed. On a few occasions, we were not able to collect samples from the Maryland Coastal Bays site due to inclement weather and technical issues. Physicochemical parameters such as salinity, water and air temperature, turbidity, total dissolved solids, dissolved oxygen, pH, chlorophyll-a, precipitation, and atmospheric pressure were recorded using a YSI Pro Plus Multi-parameter Meter (Yellow Springs Instrument Co., Yellow Springs, OH). Upon harvesting, oysters were bagged and chilled in a cooler filled with ice and a sheet of bubble wrap was used to prevent direct contact of samples and ice. A Smart Button data logger (TEquipment, Long Branch, NJ) was used to monitor and verify that shipping temperatures of the oysters remained at or above 10°C.
Oysters—isolation and enumeration
Twelve oysters were divided into three sub-samples, using four oysters per subsample. The entire amount of oyster meat and liquor was homogenized in a small blender (Waring Commercial, 7010S, Wilmington, NC) for 1.5 min at high speed. Twenty-five grams of the oyster homogenate were added to a large blender (Waring Commercial, 7010S, Wilmington, NC) along with 225 mL of 0.1% peptone dilution buffer (weight/volume) and blended for 1 min at high speed. Serial dilutions of each homogenate (to a final dilution of 10−6) and water sample (to a final dilution of 10−4) were prepared in 0.1% peptone dilution buffer. Shewanella algae and E. coli were used as positive and negative controls for all experiments. One hundred microliters of each dilution were spread plated in duplicate onto Iron Agar (Condalab, Madrid, Spain) plates using an EddyJet 2 W- Spiral Plater (Neutec Group Inc., IUL S.A., Farmingdale, NY) followed by incubation of the plates for 48 h at 35°C. After incubation, the number of bacterial colonies on each plate were counted and recorded. Up to 10 (or 10% if CFU > 100) presumptive colonies were picked and streaked onto tryptic soy agar (TSA) (Difco™, Becton, Dickinson and Company, Sparks, MD) with 0.5% added NaCl (1% total NaCl) and incubated over night to ensure sufficient growth and isolation. The Bactidrop Oxidase Test Kit (Remel, Lenexa, KS) was used to determine if the isolates were oxidase positive since Shewanella are oxidase positive. Only oxidase positive colonies were inoculated into tryptic soy broth (TSB) (Difco™, Becton, Dickinson and Company, Sparks, MD) with 1% added NaCl (1.5% total NaCl) and incubated overnight at 35°C to create fresh culture. Glycerol stocks were prepared by combining equal volumes of the fresh culture and 50% sterile glycerol and storage at −80°C for subsequent hemolysis testing and 16S rRNA sequencing.
Seawater—isolation and enumeration
Seawater was divided into three sub-samples and was used to make 10-fold serial dilutions from 100 to 10−4 in 0.1% peptone buffer. One hundred microliters of the seawater dilutions as well as the undiluted seawater was spread plated onto iron agar using the EddyJet Spiral Plater. These plates were then incubated for 48 h at 35°C. After incubation, the number of bacterial colonies on each plate was counted and recorded. Up to 10 (or 10% if CFU > 100) presumptive colonies were picked and streaked for oxidase and hemolysis testing, as described above. Glycerol stocks were prepared from colonies on the TSA plates.
Testing the hemolysis
For hemolysis testing, the isolates were evaluated for alpha, or beta hemolytic activity on the TSA with 5% sheep’s blood agar plates. After isolates were incubated overnight on TSA with 1% NaCl, a 3-mm inoculum loop was used to pick a single colony from the TSA plate. This colony was mixed into TSB with 1% added NaCl and incubated overnight at 35°C to create a fresh culture. The fresh culture was then streaked onto TSA with 5% sheep’s blood agar plates (Thermo Fisher Scientific, Waltham, MA), and was incubated overnight at 35°C. Hemolytic activity was assessed the next day: alpha hemolysis was identified by dark green discoloration (indicative of partial hemoglobin breakdown), beta hemolysis by neon green zones of complete hemolysis, and gamma hemolysis by the absence of hemolytic activity.
16S rRNA sequencing
16S rRNA sequencing was employed for the identification of presumptive Shewanella isolates at Genewiz (2019–2020) and Azenta (2021) (South Plainfield, NJ). Genomic DNA was extracted from overnight cultures of the isolates grown in 1% tryptic soy agar using a DNeasy tissue kit (Qiagen, Valencia, CA). PCR amplification of a fragment of the 16S rRNA gene was performed on each genomic preparation using forward primer 63F (5-CAG GCC TAA CAC ATG CAA GTC-3) and reverse primer 1387R (5-GGG CGG WGT GTA CAA GGC-3), along with master mix and Taq DNA polymerase, following the protocol described by Marchesi et al. (1998). Sequencing was carried out in both forward and reverse directions using the 63F and 1387R primers, respectively, provided by SeqXcel Inc. (San Diego, CA) resulting in approximately 750 bp of sequence data in each direction. The sequences were viewed in Chromas (version 2.6.6, Technelysium Ply Ltd., South Brisbane, Australia) and trimmed. Identification was carried out by comparing the sequences to the GenBank database using the BLASTn program (Richards et al., 2008; Altschul et al., 1990). The sequences were submitted to NCBI/GenBank by EzBiome Inc. (Gaithersburg, MD). Finally, the presumptive Shewanella colonies were quantified by multiplying the percentage of isolates confirmed to be Shewanella via 16S rRNA sequencing by the total number of presumptive colonies, with the data converted to CFU/g or CFU/mL of sample.
Statistical analysis
Pearson’s correlations (Cor, RStudio 2022.07.1 + 554; R Studio Team, Vienna, Austria) were performed to assess similarities between CFU levels and physicochemical parameters. Correlations were considered weak when the r values were between 0.40 and 0.49, moderate when r values were between 0.50 and 0.69, and strong when r-values were ≥ 0.70. One-way ANOVA tests were used to assess differences between sampling sites, sampling months, and sampling years, with significance defined as p-value <0.05. Levene’s test was used to assess variances among oyster and water samples, with significance defined as p-value <0.05.
Results
Identification of Shewanella species
Out of 1,344 (1,099 oyster, and 245 seawater) presumptive Shewanella isolates, a total of 713 and 177 isolates were confirmed in oysters and seawater, respectively, using 16S rRNA sequencing. Sixteen different Shewanella spp. were identified, with three species that contain strains that are human pathogens (S. algae, S. xiamensis, and S. putrefaciens). The highest abundances of species detected in seawater and oysters were 436 S. khirikhana, 173 S. marisflavi, 102 S. loihica, and 73 S. algae (Table 1; Figure 2). These four species constitute 88% of the total Shewanella identified. Three species known to cause human illness were identified: S. algae (8%), S. xiamensis (2%), and S. putrefaciens (0.3%). Comparing the Chesapeake Bay and the Maryland Coastal Bays led to a higher frequency of isolating S. khirikhana in the Chesapeake Bay (54%), while over 60% of the Maryland Coastal Bays’ isolates were identified as S. marisflavi or S. loihica.
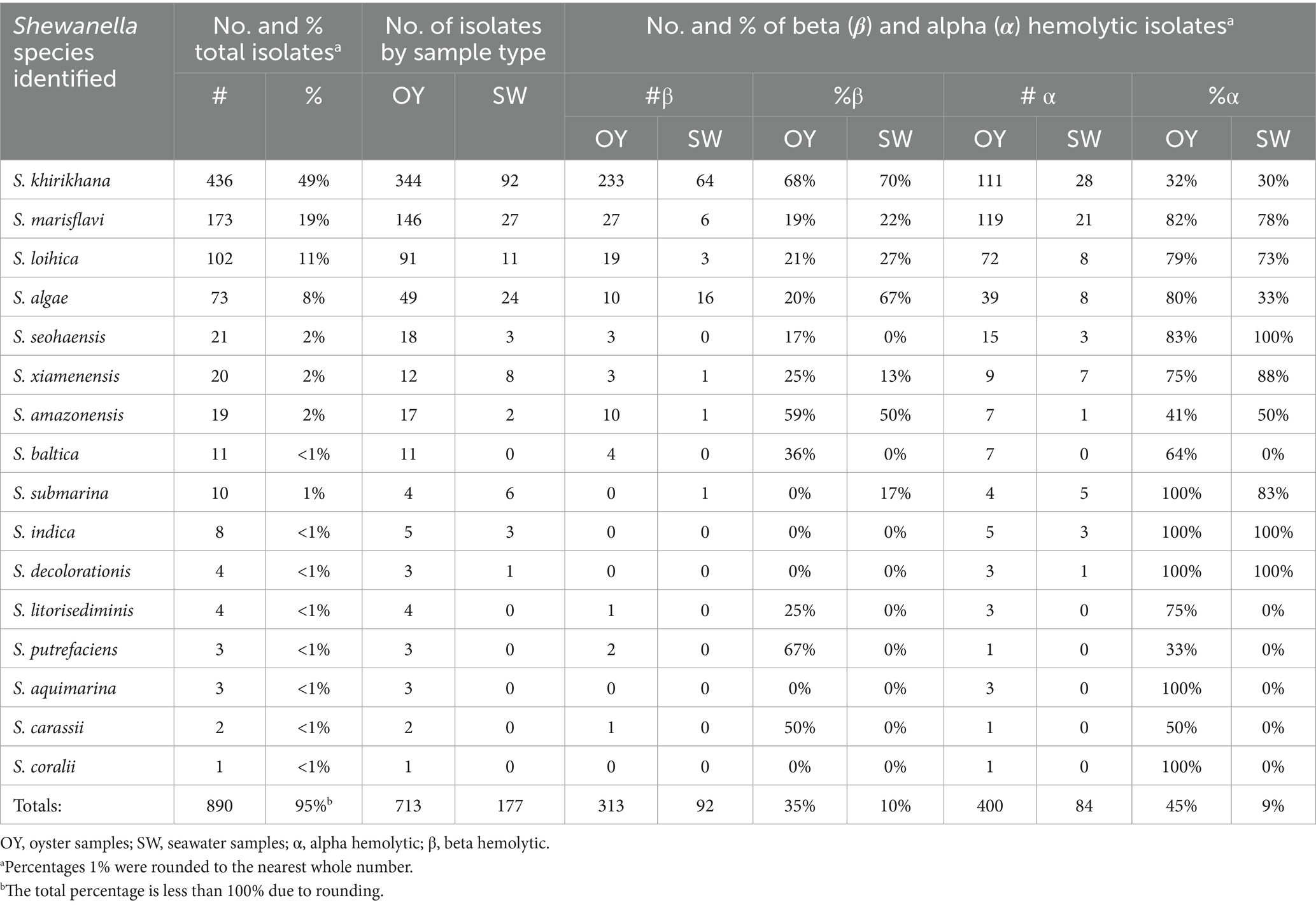
Table 1. Frequency and hemolytic activity of Shewanella species identified in oyster (OY) and seawater (SW) samples.
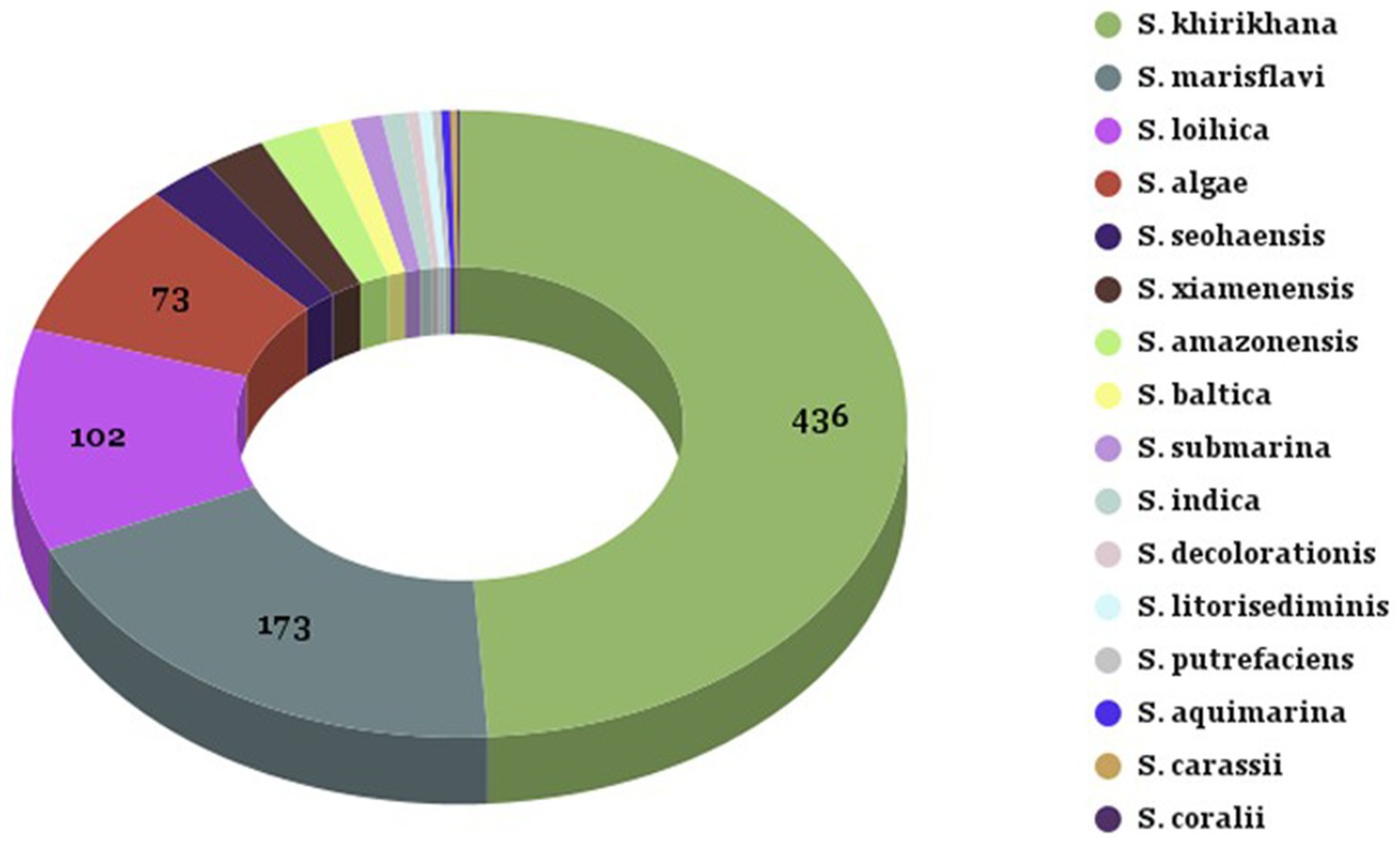
Figure 2. Summary of Shewanella species identified from oyster and seawater samples. Summary of Shewanella species identified from oyster and seawater samples collected from Chesapeake and Maryland Coastal Bays. The legend on the right corresponds to the color-coded sections of the pie chart, representing the different Shewanella species and their respective counts.
Identification of potentially pathogenic (hemolytic) Shewanella
Beta hemolysis was detected in 313 oyster isolates and 92 seawater isolates together representing 45% of the total confirmed Shewanella isolates from both sample types combined (Table 1). The majority were S. khirikhana with 233 out of a total of 436 beta hemolytic isolates (53%) from oysters and 64 out of 92 beta hemolytic isolates (70%) from seawater. Ten oyster isolates of S. algae (20%) were beta hemolytic while 16 beta hemolytic strains were observed in seawater isolates (67%) (Table 1). Alpha hemolysis was detected in 400 oyster isolates and 84 seawater isolates together representing 54% of the total confirmed Shewanella isolates from both sample types combined (Table 1). The majority were S. marisflavi with 119 out of a total of 400 alpha hemolytic isolates (30%) from oysters and 21 out of 84 alpha hemolytic isolates (25%) from seawater. Alpha hemolysis was also present in ≈ 30% of S. khirikhana isolates from oysters and seawater (Table 1). Thirty-nine oyster isolates of S. algae (80%) were alpha hemolytic while 8 alpha hemolytic strains were observed in seawater isolates (33%) (Table 1). It is unclear whether the pathogenicity of S. khirikhana toward shrimp is related to the presence of either alpha or beta hemolysis. Interestingly, S. marisflavi showed the highest number of alpha hemolytic strains in oyster samples, but a low number of beta hemolytic strains (Table 1).
Regional distribution of hemolytic strains
The regional distribution of hemolytic strains of Shewanella was analyzed over a three-year period (Table 2). All the isolates were either alpha or beta hemolytic or both. The number of beta hemolytic isolates in oysters from the four sites are as follows: Horn Point (160 isolates), Honga River (105 isolates), Tangier Sound (38 isolates), and the Maryland Coastal Bays (10 isolates) (Table 2). The beta hemolytic isolates obtained from seawater were Horn Point (53 isolates), followed by Tangier Sound (26 isolates), and Honga River (13 isolates). No isolates were reported from the Maryland Coastal Bay, perhaps due to the lower number of samples analyzed from that site. The number of alpha hemolytic isolates in oysters from the four sites were as follows: Horn Point (119 isolates), Honga River (96 isolates), Tangier Sound (134 isolates), and the Maryland Coastal Bays (61 isolates) (Table 2). The alpha hemolytic isolates obtained from seawater were Horn Point (39 isolates), Honga River (7 isolates), Tangier Sound (30 isolates), and the Maryland Coastal Bays (8 isolates) (Table 2). The percentage of total beta hemolytic isolates and alpha hemolytic isolates from oysters for each of the four sites are as follows: Horn Point (57% beta/43% alpha), Honga River (52% beta/48% alpha), Tangier Sound (23% beta/77% alpha), and Maryland Coastal Bays (14% beta/86% alpha). The Horn Point site contained the highest number and percentage of beta hemolytic isolates, and the Tangier Sound site contained the highest number of alpha hemolytic isolates; suggesting that these sites may have more strains potentially pathogenic to fish, shellfish, or humans. Similar comparisons performed for seawater revealed the following: Horn Point (58% beta/42% alpha), Honga River (65% beta/35% alpha), Tangier Sound (46% beta/54% alpha), and the Maryland Coastal Bays (0% beta/100% alpha).
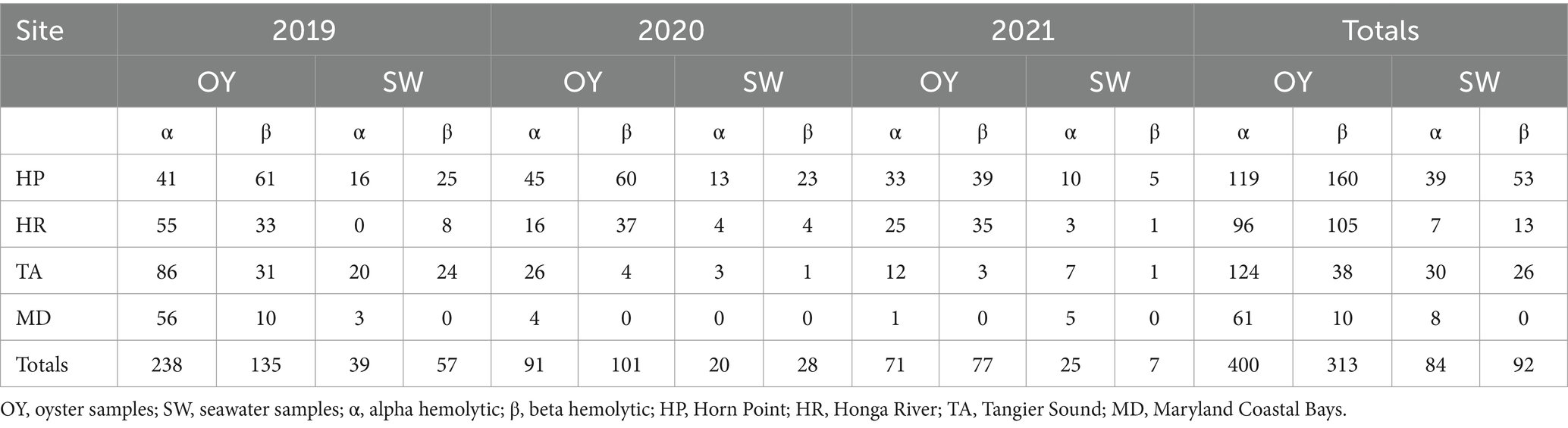
Table 2. Total number of alpha and beta hemolytic Shewanella strains isolated from oysters and seawater at four sites over 3 years.
Prevalence and seasonal distribution of total Shewanella
ANOVA tests were performed for Shewanella counts in seawater and oyster samples compared to sampling month, sampling site, and sampling year. For Shewanella counts in oyster samples, there were no significant differences between sampling site (p = 0.214), sampling month (p = 0.295), or sampling year (p = 0.280). For seawater, significant differences in Shewanella abundances were observed for sampling site (p < 0.01), and sampling month (p < 0.05), while sampling year showed no significant differences (p = 0.132) (Figure 3).
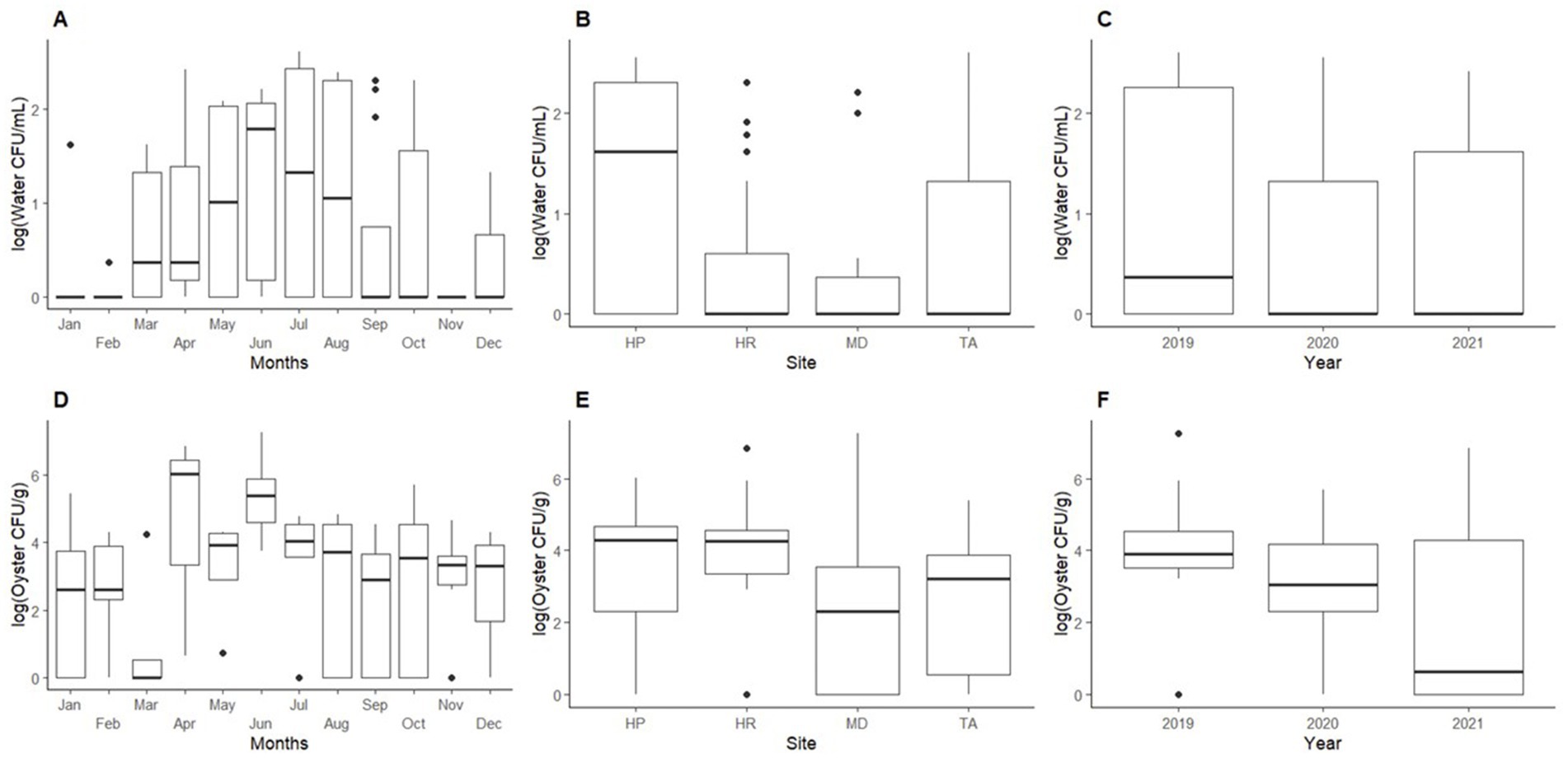
Figure 3. Mean abundance of Shewanella found in oyster and seawater. Top graphs show abundances of Shewanella in seawater by: (A) month, (B) sampling site, and (C) year. Bottom graphs show abundances of Shewanella in oysters by: (D) month, (E) sampling site, and (F) year. Abbreviations for graphs (B,E): HP, Horn Point; HR, Honga River; TA, Tangier Sound; and MD, Maryland Coastal Bay.
In water samples, 71% of the total isolates sequenced were identified as Shewanella. The highest counts of Shewanella were observed in May through August (Figure 3A). Horn Point had higher Shewanella occurrences from June through September with counts ranging from undetectable to 3.60 × 102 CFU/mL (July 2020). For the same period, Honga River, Tangier Sound, and Maryland Coastal Bay had counts from undetectable to 2.00 × 102 CFU/mL (August 2019), undetectable to 4.00 × 102 CFU/mL (July 2019); and undetectable to 2.60 × 101 CFU/mL (October 2019), respectively.
In oyster samples, 65% of the sequenced isolates were identified as Shewanella. The highest detection for oysters was observed from April through June (Figure 3D), oyster counts ranged from undetectable to 7.20 × 105 CFU/g at Horn Point (June 2021); undetectable to 8.40 × 105 CFU/g at Honga River (June 2019); undetectable to 2.40 × 105 CFU/g at Tangier Sound (June 2021); and undetectable to 1.80 × 107 CFU/g at Maryland Coastal Bay (June 2019) (Figure 3E).
Environmental influence on Shewanella
Seawater physiochemical parameters at the time of sample collections are given in Table 3. The seawater temperature ranged from 17.8–19.6°C with Horn Point averaging the lowest temperature and Maryland Coastal Bays averaging the highest water temperature. Salinity ranged from 12 to 31 ppt with Maryland Coastal Bays averaging the highest salinity levels and Horn Point averaging the lowest. Dissolved oxygen ranged from 7.5–8.6 mg/L with Tangier Sound averaging the highest levels and Maryland Coastal Bays averaging the lowest. Turbidity ranged from 5.8–10.6 Formazin Nephelometric Unit (FNU) with Horn Point and Honga River averaging the highest levels and Tangier Sound averaging the lowest. The pH was ≈ 8 for all sampling sites. Chlorophyll-a ranged from 6 to 8 μg/L with Horn Point averaging the highest measurements and Maryland Coastal Bay with the lowest. Precipitation ranged from about 3.0 to 7.4 mm with Horn Point receiving the least amount of average rainfall within 48 h prior to sampling and Tangier Sound averaging the most. Total dissolved solids ranged from 1,319 to 2,976 mg/L with Maryland Coastal Bay averaging the highest and Horn Point averaging the lowest. Atmospheric pressure consistently measured 765 mm Hg at all sampling sites (Table 3). Physicochemical water parameters such as water temperature, turbidity, and pH had lower averages in 2020 compared to 2019 and 2021.
Shewanella spp. were consistently isolated at higher concentrations in oysters compared to seawater (p < 0.05). This trend may be attributed to the filter-feeding behavior of Crassostrea virginica. Correlation coefficients (r-value) for various physicochemical parameters and total Shewanella abundances are shown in Table 4. For seawater, temperature had a low positive correlation (r = 0.45) with Shewanella abundance, while dissolved oxygen showed a low negative correlation (r = −0.41). For oysters, there were no notable correlations between the various parameters and abundances (r ≤ 0.35).
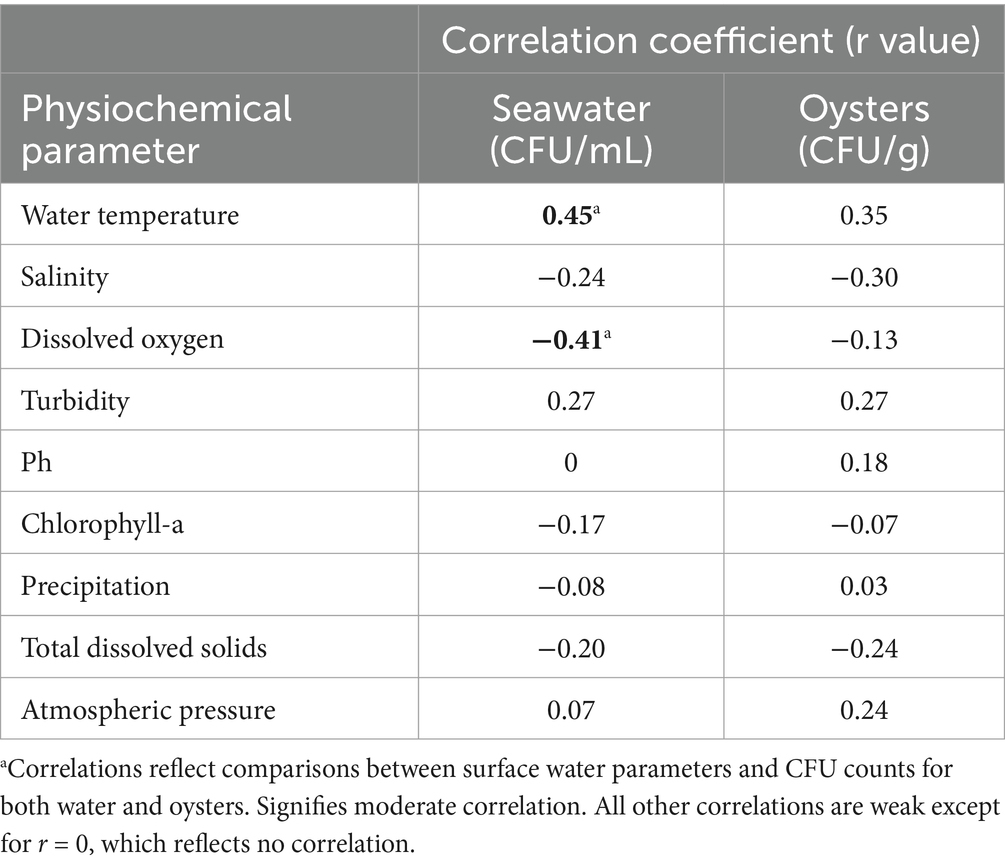
Table 4. Pearson correlation coefficients comparing the physiochemical parameters of the harvest water and oysters with the abundance of Shewanella isolation.
Discussion
This is the first comprehensive study that investigates the presence, diversity, influence of environmental parameters, and pathogenic potential of Shewanella species in oysters and seawater samples recovered from the Chesapeake Bay and the Maryland Coastal Bay in the Mid-Atlantic region of the USA. To maximize the range of collection conditions encountered during the study, low and high salinity oyster harvesting sites were selected. Compared to seawater, oyster samples contained more Shewanella at all sites. This was expected due to the ability of oysters to bioaccumulate large numbers of microorganisms as they filter the water (Center for Disease Control and Prevention, 2022). Our results are consistent with the finding of a previous study that reported higher levels of Photobacterium damselae and Shewanella in oysters in comparison to seawater (Richards et al., 2008). The diversity of Shewanella species identified in this study was unexpected and some of these species have not been previously reported in U.S. coastal waters. It is uncertain whether their presence was influenced by changes in climate, better identification methods, or the fact that Shewanella research has been very limited. Nearly half of the isolates identified in this study were S. khirikhana, which was only recently recommended as a new species (Prachumwat et al., 2020). It has a 16S rRNA sequence similar to that of S. amazonensis. S. khirikhana was associated with disease of cultivated shrimp in Thailand (Prachumwat et al., 2020). We were unable to identify previous reports of S. khirikhana or S. amazonensis species in the U.S. It is unclear whether S. khirikhana is responsible for disease in shrimp, or other shellfish, or fish species in U.S. aquaculture or in wild caught seafoods. Surveillance for this shellfish pathogen may be prudent because of its high prevalence in mid-Atlantic bays and the high numbers of alpha and beta hemolytic strains detected.
While the levels of total Shewanella varied among the bays, similar results were observed in the levels of total Shewanella between sample types (oyster and seawater) and the three sampling sites in the Chesapeake Bay. Although dissolved oxygen, chlorophyll-a levels, and seawater pH showed some correlation with Shewanella levels in seawater, no correlation was observed between the physicochemical parameters and Shewanella abundances in oysters. There can be many factors outside of environmental influences (Abirami et al., 2021; Marsay et al., 2023) that can lead to higher abundances of Shewanella and other bacteria. This can include host factors (Lemaire et al., 2020), nutrient availability, biofilm formation, genetic adaptations (Lemaire et al., 2020), anthropogenic influences, and enhanced metabolic activity of bacteria (Abirami et al., 2021).
The distribution of Shewanella species may be influenced by various factors, including environmental conditions and geographical location (Marsay et al., 2023). Different regions exhibit unique sets of physical and chemical parameters that shape the microbial communities thriving within them (Hanson et al., 2019). Specific environmental conditions, such as temperature, salinity, oxygen availability, and nutrient availability can shape the prevalence and adaptation of many microbial communities (Hanson et al., 2019; Marsay et al., 2023). The presence of geographical barriers, such as ocean currents or continental landmasses, can restrict the dispersal of certain species, resulting in regional variations in species composition (Hanson et al., 2019).
By comparing the Chesapeake Bay and the Maryland Coastal Bays, it was observed that isolation of S. khirikhana in the Chesapeake Bay was high, while over 90% of the Maryland Coastal Bays’ isolates were identified as S. marisflavi or S. loihica. Additionally, other studies have reported the isolation of several other Shewanella species, namely S. algae (Gram et al., 1999; Richards et al., 2008; Taherzadeh et al., 2014; Zago et al., 2020), S. putrefaciens (Richards et al., 2008; Kang and So, 2016), and S. baltica (Richards et al., 2008; Zago et al., 2020), which were also found in the Chesapeake Bay. These Shewanella species have been discussed and documented in studies conducted in the Delaware Bay (Richards et al., 2008), Danish Coastal waters (Gram et al., 1999), Persian Gulf (Taherzadeh et al., 2014), Adriatic Sea (Zago et al., 2020) and the West Sea (Kang and So, 2016).
Although all Shewanella isolates in this study were either alpha or beta hemolytic, the pathogenic potential of Shewanella species is not yet defined. If oysters become contaminated with hemolytic Shewanella, there could be potential safety concerns for shellfish consumers. The bacteria could cause health problems such as gastrointestinal illness or even more serious conditions like sepsis, particularly in individuals with weakened immune systems. Literature suggests that bacteria containing hemolysins may have higher antibiotic resistance and be more pathogenic toward humans (Jarraud et al., 2002; Nizet, 2002; Schroeder et al., 2017; Motamedi et al., 2018). We found that 45% of Shewanella isolates in this study had beta hemolytic activity. For example, S. algae is documented as pathogenic toward humans and exhibited a high percentage of beta-hemolytic activity in this study. With 45% of Shewanella isolates demonstrating beta hemolytic activity, including 20% of S. algae isolates from oysters and seawater, this finding raises concerns about potential pathogenicity.
S. algae has been increasingly recognized as an emerging human pathogen capable of causing serious infections, including those of the skin, soft tissue, bloodstream, and peritoneum (Kitaoka et al., 2019; Tamez et al., 2020). The presence of putative hemolysin genes in S. algae as discussed by Tamez et al. (2020), provides a molecular basis for its hemolytic activity and pathogenicity. Enzymes that are responsible for β-hemolysis have been established as a virulence factor for a variety of bacterial pathogens such as Enterococci (Nizet, 2002), Escherichia coli (Bhakdi et al., 1988), Listeria monocytogenes (Goebel et al., 1988; Raybourne, 2002), and Vibrio parahaemolyticus (Okuda and Nishibuchi, 1998). The presence of hemolytic activity in Shewanella has significant ramifications for the safety and quality of oysters and may well serve as an indicator of virulence in Shewanella spp. (Richards et al., 2008).
Shewanella algae demonstrates remarkable environmental adaptability and potential pathogenicity, as evidenced by its prevalence in marine ecosystems. Our findings on the prevalence and hemolytic activity of S. algae can be contextualized by comparing them with the study by Tseng et al. (2018). While Tseng et al. (2018) reported that S. algae was present in approximately 23% of shellfish samples and 28% of water samples, our study observed a higher prevalence with 67% in oysters and 33% in seawater. This difference may be attributed to geographical variations and/or sampling methods. However, the high prevalence suggests that oysters frequently harbor this bacterium, though its direct impact on oyster health has not been discussed. Notably, Tseng et al. (2018) examined hemolytic activity at both 25 and 37°C, finding that S. algae exhibited stronger hemolytic activity at human body temperature with 100% hemolysis after 72 h at 37°C compared to 50% at 25°C. In contrast, our study maintained a constant temperature of 35°C for all hemolytic testing and found beta hemolytic activity to be 20% in oysters and 67% in seawater, while alpha hemolytic activity was drastically higher in oysters at 80%, compared to 33% in seawater. Furthermore, Tseng et al. (2018) findings on S. algae’s adaptability to varying temperatures and salinities are comparable to our results; we noted a moderate correlation (r = 0.45) between water temperature and the abundance of Shewanella in our seawater samples (as noted in Table 4). This adaptability allows S. algae to survive in various marine environments, including oyster habitats, potentially affecting oyster microbiomes. These comparisons underscore the pathogenic potential of S. algae in both marine environments and human hosts, emphasizing the importance of continued monitoring for food safety implications.
A study by Richards et al. (2008) evaluated oysters and seawater from the Delaware Bay for the presence of potentially pathogenic Shewanella and Photobacterium species using the API 20E System (biMerieux Industries, Hazelwood, MO), which is a biochemical kit used to evaluate primarily human pathogens for clinical diagnoses. They found that out of 1,421 isolates tested, 12% (n = 170) of all picked colonies were identified as presumptive S. putrefaciens. The only Shewanella listed in the API database was S. putrefaciens of which some were subsequently identified as S. algae. 16S rRNA sequencing was performed and revealed a host of different Shewanella species, not present in the API database (Richards et al., 2008). Differences in assay methodologies between the Delaware study and the present study at the Maryland sites prevent direct comparison of results. Advances in scientific techniques and technologies have allowed researchers to uncover and distinguish between previously unknown or misidentified species. As a result, the number of recognized Shewanella species has expanded over time.
The levels of total Shewanella in the Chesapeake and Maryland Coastal Bays can provide important information about the potential risks associated with consuming seafood from those areas. However, it is important to note that the presence of Shewanella alone does not necessarily indicate a safety concern, as not all species of Shewanella are pathogenic and for those that are, the levels of bacteria alone may not be sufficient to cause illness. Shewanella had the highest levels observed in 2019, while the least amount was observed in 2021. In 2019–2021, S. khirikhana was the most frequently isolated species from oyster and seawater samples. It is unclear if S. khirikhana outcompetes other species or if water conditions play some role in enhancing their growth or reproduction over other species. There was no significant variation between the years for the four most frequently isolated Shewanella species (S. khirikhana, S. marisflavi, S. loihica, and S. algae).
Knowledge is limited on the extent of disease caused by Shewanella species from the Chesapeake Bay and Maryland Coastal Bay or the origins of infections, such as from the consumption of contaminated seafood, by cuts or abrasions in the skin after exposure to a contaminated marine environment, or by recreational water activities. The detection of potentially pathogenic species in this study suggests the need for continued monitoring of Shewanella occurrences and further investigation of any association this bacterium may have with disease. Studies on factors that correlate with abundances of Shewanella to physicochemical parameters should be continued and expanded to other coastal regions. Monitoring the levels of pathogenic species should be continued, and investigations on antibiotic susceptibility should be performed. Enhanced reporting of Shewanella cases should be adopted to ascertain more accurate data on disease prevalence and its association with seafood consumption, particularly during the warmer months when Shewanella levels are the highest in both seawater and oysters.
However, the limitations of this study include limited sample size from the Maryland Coastal Bays due to logistical challenges, such as inclement weather and boating issues. In addition, the COVID-19 lockdown in 2020 caused the omission of critical sampling months (March–June). Furthermore, while hemolytic activity was assessed to infer pathogenic potential, direct evidence linking Shewanella species to seafood-related illnesses was not evaluated. Lastly, while correlations with physicochemical parameters were explored, the potential role of other environmental factors, such as pollution or nutrient inputs, remains to be evaluated, limited the number of samples collected from Maryland Coastal Bays, which may have impacted regional comparisons. Addressing these limitations in future studies will enhance our understanding of the ecological and clinical significance of Shewanella species in estuarine environments.
Conclusion
Shewanella species are of significant interest due to their pathogenic potential to humans and marine organisms, with multiple transmission routes including marine water, seafood, and wound infections. This study is the first comprehensive investigation into the diversity and pathogenic potential of Shewanella species recovered from oysters and seawater in the Chesapeake Bay and Maryland Coastal Bays. By filling critical data gaps on the abundance, diversity, and pathogenic potential of Shewanella, this research raises awareness of this emerging pathogen and lays the groundwork for developing intervention technologies to prevent or mitigate Shewanella-associated illnesses. Continued research in this area will be crucial to better understand the environmental factors driving the emergence of these pathogens and for informing public health strategies.
Author’s note
The use of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture (USDA). The USDA is an equal opportunity provider and employer.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, OR941802; https://www.ncbi.nlm.nih.gov/genbank/, OR943574.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
TJ: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. GR: Conceptualization, Data curation, Methodology, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing, Funding acquisition. JJ: Data curation, Formal analysis, Methodology, Software, Visualization, Writing – review & editing. HT: Data curation, Formal analysis, Software, Visualization, Writing – review & editing. EA: Investigation, Writing – review & editing, Data curation. DR: Data curation, Investigation, Writing – review & editing. PC: Conceptualization, Funding acquisition, Writing – review & editing. LD: Conceptualization, Funding acquisition, Validation, Writing – review & editing. SP: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. We thanked the United States Department of Agriculture CBG (award no. 2018–38821-27759) and Living Marine Resources Cooperative Science Center for funding. Tahirah Johnson was supported by the NOAA Office of Education Educational Partnership Program with Minority Serving Institutions (award nos. NA16SEC481007 and NA21SEC481005).
Acknowledgments
We are grateful to each of the oyster farmers who provided oyster samples and took us on their boats to gather physicochemical water parameters. Special thanks to Catherine Liu, Kelly Williams and Joan Meredith (deceased) for technical assistance, organizational aid and helpful suggestions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1502443/full#supplementary-material
References
Abirami, B., Radhakrishnan, M., Kumaran, S., and Wilson, A. (2021). Impacts of global warming on marine microbial communities. Sci. Total Environ. 791:147905. doi: 10.1016/j.scitotenv.2021.147905
Alav, I., Kobylka, J., Kuth, M. S., Pos, K. M., Picard, M., Blair, J. M. A., et al. (2021). Structure, assembly, and function of tripartite efflux and type 1 secretion systems in Gram-negative bacteria. Chem. Rev. 121, 5479–5596. doi: 10.1021/acs.chemrev.1c00055
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment 540 search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bhakdi, S., Mackman, N., Menestrina, G., Gray, L., Hugo, F., Seeger, W., et al. (1988). The hemolysin of Escherichia coli. Eur. J. Epidemiol. 4, 135–143. doi: 10.1007/BF00144740
Böttcher, T. S., and Sieber, S. A. (2008). β-Lactones as specific inhibitors of ClpP attenuate the production of extracellular virulence factors of Staphylococcus aureus. J. Am. Chem. Soc. 130, 14400–14401. doi: 10.1021/ja8051365
Calvert County Health Department (2017). Vibrio and Shewanella algae infections appear in waters around the Chesapeake. Available at: http://www.calverthealth.org/healththreats/diseases/vibrio.htm (Accessed April 22 2023).
Center for Disease Control and Prevention (2022). Oysters and vibriosis. Available at: https://www.cdc.gov/foodsafety/communication/oysters-and-vibriosis.html (Accessed April 22, 2023).
Dey, S., Bhattacharya, D., Roy, S., Nadgir, S. D., Patil, A., and Kholkute, S. D. (2015). Shewanella algae in acute gastroenteritis. Indian J. Med. Microbiol. 33, 172–175. doi: 10.4103/0255-0857.148442
Goebel, W., Chakraborty, T., and Kreft, J. (1988). Bacterial hemolysins as virulence factors. Antonie Van Leeuwenhoek 54, 453–463. doi: 10.1007/BF00461864
Goyal, R., Kaur, N., and Thakur, R. (2011). Human soft tissue infection by the emerging pathogen Shewanella algae. J. Infect. Dev. Ctries. 5, 310–312. doi: 10.3855/jidc.1436
Gram, L., Bundvad, A., Melchiorsen, J., Johansen, C., and Fonnesbech Vogel, B. (1999). Occurrence of Shewanella algae in Danish coastal water and effects of water temperature and culture conditions on its survival. Appl. Environ. Microbiol. 65, 3896–3900. doi: 10.1128/AEM.65.9.3896-3900.1999
Hanson, C. A., Muller, A. L., Loy, A., Dona, C., Appel, R., Jorgensen, B. B., et al. (2019). Historical factors associated with past environments influence the biogeography of thermophilic endospores in Arctic marine sediments. Front. Microbiol. 10:245. doi: 10.3389/fmicb.2019.00245
Hau, H. H., and Gralnick, J. A. (2007). Ecology and biotechnology of the genus Shewanella. Ann. Rev. Microbiol. 61, 237–258. doi: 10.1146/annurev.micro.61.080706.093257
Holt, H. M., Gahrn-Hansen, B., and Bruun, B. (2005). Shewanella algae and Shewanella putrefaciens: clinical and microbiological characteristics. Clin. Microbiol. Infect. 11, 347–352. doi: 10.1111/j.1469-0691.2005.01108.x
Huang, Y. T., Cheng, J. F., Chen, S. Y., Hong, Y. K., Wu, Z. Y., and Liu, P. Y. (2018). Draft genome sequence of carbapenem-resistant Shewanella algae strain AC isolated from small abalone (Haliotis diversicolor). J. Glob. Antimicrob. Resist. 14, 65–67. doi: 10.1016/j.jgar.2018.06.005
Jarraud, S., Mougel, C., Thioulouse, J., Lina, G., Meugnier, H., Forey, F., et al. (2002). Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70, 631–641. doi: 10.1128/IAI.70.2.631-641.2002
Kang, C. H., Shin, Y., Jeon, H., Choi, J. H., Jeong, S., and So, J. S. (2013). Antibiotic resistance of Shewanella putrefaciens isolated from shellfish collected from the West Sea in Korea. Mar. Pollut. Bull. 76, 85–88. doi: 10.1016/j.marpolbul.2013.09.025
Kang, C. H., and So, J. S. (2016). Antibiotic and heavy metal resistance in Shewanella putrefaciens strains isolated from shellfishes collected from West Sea, Korea. Mar. Pollut. Bull. 112, 111–116. doi: 10.1016/j.marpolbul.2016.08.033
Kitaoka, M., Ishida, M., and Sakaeda, H. (2019). Shewanella algae bacteremia in patients with biliary tract malignancy: report of three cases. Nihon Shokakibyo Gakkai Zasshi 116, 850–857. doi: 10.11405/nisshoshi.116.850
Lemaire, O. N., Mejean, V., and Iobbi-Nivol, C. (2020). The Shewanella genus: ubiquitous organisms sustaining and preserving aquatic ecosystems. FEMS Microbiol. Rev. 44, 155–170. doi: 10.1093/femsre/fuz031
Liu, D., Fiel, R. H., Cheng, L. S., Ogami, T., Wang, L., Singh, V., et al. (2019). Notes from the field: first reported case of Shewanella haliotis in the region of the Americas - New York, December 2018. MMWR 68, 1168–1169. doi: 10.15585/mmwr.mm6850a5
Liu, P. Y., Lin, C. F., Tung, K. C., Shyu, C. L., Wu, M. J., Liu, J. W., et al. (2013). Clinical and microbiological features of Shewanella bacteremia in patients with hepatobiliary disease. Intern. Med. 52, 431–438. doi: 10.2169/internalmedicine.52.8152
Marchesi, J. R., Sato, T., Weightman, A. J., Martin, T. A., Fry, J. C., Hiom, S. J., et al. (1998). Design and evaluation of useful bacterium-specific PCR primers that amplify 538 genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64, 795–799. doi: 10.1128/AEM.64.2.795-799.1998
Marsay, K. S., Ambrosino, A. C., Koucherov, Y., Davidov, K., Figueiredo, N., Yakovenko, I., et al. (2023). The geographical and seasonal effects on the composition of marine microplastic and its microbial communities: the case study of Israel and Portugal. Front. Microbiol. 14:1089926. doi: 10.3389/fmicb.2023.1089926
Motamedi, H., Asghari, B., Tahmasebi, H., and Arabestani, M. R. (2018). Identification of hemolysine genes and their association with antimicrobial resistance pattern among clinical isolates of Staphylococcus aureus in west of Iran. Adv. Biomed. Res. 7:153. doi: 10.4103/abr.abr_143_18
Myung, D. S., Jung, Y. S., Kang, S. J., Song, Y. A., Park, K. H., Jung, S. I., et al. (2009). Primary Shewanella algae bacteremia mimicking Vibrio septicemia. J. Korean Med. Sci. 24, 1192–1194. doi: 10.3346/jkms.2009.24.6.1192
Ng, W. W., Shum, H. P., and To, K.KSridhar, S. (2022). Emerging infections due to Shewanella spp.: a case series of 128 cases over 10 years. Front. Med. 9:850938. doi: 10.3389/fmed.2022.850938
Nizet, V. (2002). Streptococcal beta-hemolysins: genetics and role in disease pathogenesis. Trends Microbiol. 10, 575–580. doi: 10.1016/s0966-842x(02)02473-3
Okuda, J., and Nishibuchi, M. (1998). Manifestation of the Kanagawa phenomenon, the virulence-associated phenotype, of Vibrio parahaemolyticus depends on a particular single base change in the promoter of the thermostable direct haemolysin gene. Mol. Microbiol. 30, 499–511. doi: 10.1046/j.1365-2958.1998.01072.x
Poovorawan, K., Chatsuwan, T., Lakananurak, N., Chansaenroj, J., Komolmit, P., and Poovorawan, Y. (2013). Shewanella haliotis associated with severe soft tissue infection, Thailand, 2012. Emerg. Infect. Dis. 19, 1019–1021. doi: 10.3201/eid1906.121607
Prachumwat, A., Wechprasit, P., Srisala, J., Kriangsaksri, R., Flegel, T. W., Thitamadee, S., et al. (2020). Shewanella khirikhana sp. nov. - a shrimp pathogen isolated from a cultivation pond exhibiting early mortality syndrome. Microb. Biotechnol. 13, 781–795. doi: 10.1111/1751-7915.13538
Raybourne, R. B. (2002). Virulence testing of Listeria monocytogenes. J. AOAC Int. 85, 516–523. doi: 10.1093/jaoac/85.2.516
Richards, G. P., Watson, M. A., Crane, E. J. 3rd, Burt, I. G., and Bushek, D. (2008). Shewanella and Photobacterium spp. in oysters and seawater from the Delaware Bay. Appl. Environ. Microbiol. 74, 3323–3327. doi: 10.1128/AEM.00060-08
Rodgers, C., Parveen, S., Chigbu, P., Jacobs, J., Rhodes, M., and Harter-Dennis, J. (2014). Prevalence of Vibrio parahaemolyticus, and Vibrio vulnificus in blue crabs (Callinectes sapidus), seawater and sediments of the Maryland coastal bays. J. Appl. Microbiol. 117, 1198–1209. doi: 10.1111/jam.12608
Santos, A. C., Varughese, C. A., and Shankaran, S. (2021). Something odd and something new: a case of Shewanella putrefaciens peritonitis and bacteremia treated with intraperitoneal cefepime. Priv. Pract. Infect. Dis. 1:6. doi: 10.35995/ppid1020006
Schroeder, M., Brooks, B. D., and Brooks, A. E. (2017). The complex relationship between virulence and antibiotic resistance. Genes 8:39. doi: 10.3390/genes8010039
Taherzadeh, M., Katouli, M., Amirinejad, R., Farzaneh, M. R., and Gharibi, O. (2014). A case of wound infection caused by Shewanella algae in the south of Iran. New Microbes New Infect. 2, 29–30. doi: 10.1002/2052-2975.33
Takata, T., Chikumi, H., Morishita, S., Hamada, S., Hoi, S., Iyama, T., et al. (2017). Shewanella algae bacteremia in an end-stage renal disease patient: a case report and review of the literature. Intern. Med. 56, 729–732. doi: 10.2169/internalmedicine.56.7616
Tamez, A. M., McLaughlin, R. W., Li, J., Wan, X., and Zheng, J. (2020). Searching for putative virulence factors in the genomes of Shewanella indica and Shewanella algae. Arch. Microbiol. 203, 683–692. doi: 10.1007/s00203-020-02060-1
Tasnim, S. (2017). Rapid identification of Streptococcus pneumoniae strains using 16S rRNA gene based PCR method (Bachelor’s dissertation). Dhaka: BRAC University.
Tseng, S. Y., Liu, P. Y., Lee, Y. H., Wu, Z. Y., Huang, C. C., Cheng, C. C., et al. (2018). The pathogenicity of Shewanella algae and ability to tolerate a wide range of temperatures and salinities. Can. J. Infect. Dis. Med. Microbiol. 2018, 1–9. doi: 10.1155/2018/6976897
Wu, Z. Y., Ho, S. P., Cheng, J. F., Tung, K. C., Hong, T. K., Chen, S. Y., et al. (2018). Whole-genome characterization of Shewanella algae strain SYT3 isolated from seawater reveals insight into hemolysis. Future Microbiol. 13, 1709–1717. doi: 10.2217/fmb-2018-0267
Yousfi, K., Bekal, S., Usongo, V., and Touati, A. (2017). Current trends of human infections and antibiotic resistance of the genus Shewanella. Eur. J. Clin. Microbiol. Infect. Dis. 36, 1353–1362. doi: 10.1007/s10096-017-2962-3
Zago, V., Veschetti, L., Patuzzo, C., Malerba, G., and Lleo, M. M. (2020). Shewanella algae and Vibrio spp. strains isolated in Italian aquaculture farms are reservoirs of antibiotic resistant genes that might constitute a risk for human health. Mar. Pollut. Bull. 154:111057. doi: 10.1016/j.marpolbul.2020.111057
Zhang, F., Fang, Y., Pang, F., Liang, S., Lu, X., Kan, B., et al. (2018). Rare Shewanella spp. associated with pulmonary and bloodstream infections of cancer patients, China: a case report. BMC Infect. Dis. 18:454. doi: 10.1186/s12879-018-3354-8
Keywords: Shewanella species, oysters, seawater, pathogenicity, hemolytic, Chesapeake Bay, Maryland Coastal Bay, environmental influences
Citation: Johnson T, Richards GP, Jacobs J, Townsend H, Almuhaideb E, Rosales D, Chigbu P, Dasilva L and Parveen S (2025) Prevalence and pathogenic potential of Shewanella species in oysters and seawater collected from the Chesapeake Bay and Maryland Coastal Bays. Front. Microbiol. 16:1502443. doi: 10.3389/fmicb.2025.1502443
Edited by:
Nidia León-Sicairos, Autonomous University of Sinaloa, MexicoReviewed by:
Xiaolei Wang, Ocean University of China, ChinaShengqian Sun, Yantai Institute of Technology, China
Copyright © 2025 Johnson, Richards, Jacobs, Townsend, Almuhaideb, Rosales, Chigbu, Dasilva and Parveen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Salina Parveen, c3BhcnZlZW5AdW1lcy5lZHU=
 Tahirah Johnson1
Tahirah Johnson1 John Jacobs
John Jacobs Howard Townsend
Howard Townsend Paulinus Chigbu
Paulinus Chigbu Salina Parveen
Salina Parveen