- 1College of Public Health, Shanghai University of Medicine and Health Sciences, Shanghai, China
- 2Shanghai Center for Disease Control and Prevention, Shanghai, China
- 3Department of Food Science, University of Otago, Dunedin, New Zealand
- 4New Zealand Food Safety Science and Research Centre, Palmerston North, New Zealand
The emergence of multidrug-resistant (MDR) Salmonella Enteritidis has highlighted the importance of regularly monitoring for the occurrence of antibiotic-resistant strains. The current study combined phenotyping analysis and whole-genome-sequencing (WGS) to investigate the associations between the antibiotic-resistant phenotypes (ARPs) and genetic characteristics determinants in 95 Salmonella Enteritidis isolates from retail meat and environmental samples in China (2014–2019). Phenotypic analyses revealed that 70 isolates (73.68%) were MDR with 12 distinct resistance patterns. Most MDR strains (81.43%) had NAL-AMP-FIS-STR ± TET profiles, showing a fluctuating trend from 2015 to 2019, likely influenced by tetracycline withdrawal management. WGS identified four types of mutations in the gyrA gene were associated with nalidixic acid resistance. The co-carrying of blaTEM, sul2 and aph(6)-Id/aph(3″)-Ib was likely mediated by an X1-type plasmid, corresponding to resistance against ampicillin, sulfisoxazole, and streptomycin. Combining phenotypic analyses and WGS data, the 31 sequenced strains were primarily divided into two clusters, with most epidemic resistant strains in the largest cluster A. Identical ARP patterns observed across different sample types, regions, and isolation years but clustering together in cluster A suggested potential cross-contamination within the retail chain. Cluster B exhibited more diverse resistance patterns and genetic characteristics. Notably, three isolates in cluster B require special mention: a monophasic strain resistant to eight antibiotics, a strain exhibiting highly heteroresistance, and a strain with additional exotoxin genes. These results highlight the importance of ongoing surveillance and the utility of WGS to track and understand antibiotic resistance in Salmonella Enteritidis.
1 Introduction
Foodborne Salmonella Enteritidis is predominantly associated with eggs, chicken and related products (Hofer, 2021; Gu et al., 2020). However, this bacterium can occur throughout the food production chain, infecting other food animals and contaminating a wide range of foods (Guerrero et al., 2022; Bellil et al., 2023; Yang et al., 2020). Salmonella Enteritidis accounts for approximately 40–60% of global salmonellosis cases (Pearce et al., 2018) and is the most frequently detected serovar among patients with diarrhea in China (Wang et al., 2022). The main approach for treating salmonellosis relies on antibiotics, with third-generation cephalosporins and fluoroquinolones being the first-line clinical options (Konyali et al., 2023). However, due to the widespread use of antibiotics, the incidence of multi-drug resistant (MDR) Salmonella stains is increasing, with over 70% of isolates from patients with diarrhea, food animals, or retailed foods in China being reported as being MDR (Wang et al., 2020; Song et al., 2020). Previous investigations into Salmonella Enteritidis and other Salmonella serovars has demonstrated significant variations in the prevalence of antibiotic resistance among isolates from different sample types, geographic regions and years of isolation (Zakaria et al., 2022; Ksibi et al., 2022; Kang et al., 2022). Moreover, the antibiotic resistance patterns of this serovar often differ from those observed in other Salmonella serovars (Gu et al., 2020; Kang et al., 2022). Therefore, it is essential to continually update data on the resistance patterns of this prominent serovar across different time frames, regions and sample types to trace the origin of resistant strains and elucidate the development of their resistance profiles.
Antibiotic resistance in Salmonella can develop through point mutations in the bacterial genome or via the horizontal transfer of genetic elements carrying antibiotic resistant genes (ARGs) (Rakitin et al., 2021; Zhao et al., 2023). Whole genome sequencing (WGS) has become both a complementary and alternative approach to traditional methods for evaluating the genetic diversity of ARGs in Salmonella (Hu et al., 2020; Alzahrani et al., 2023). Additionally, WGS can provide valuable insights into other factors, such as virulence determinants, mobile genetic elements, and other genomic changes linked to pathogenicity and antibiotic resistance (Ksibi et al., 2022). Furthermore, WGS offers superior resolution and accuracy for correlation analyses based on the core genome compared to conventional molecular typing methods, such as pulsed-field gel electrophoresis (PFGE) (Edirmanasinghe et al., 2017; Keefer et al., 2019; Rounds et al., 2020). An enhanced ability to connect resistance and virulence phenotypes with genotypes, will facilitate epidemiological investigations, as detailed data on the genomic context of each isolate will help in the determination potential transmission routes (Edirmanasinghe et al., 2017; Sun et al., 2024; García-Soto et al., 2023).
The current study determined the occurrence of antibiotic resistance in 95 Salmonella Enteritidis isolates, primarily obtained from retail meat and environmental samples during 2014–2019 in China, and investigated the temporal and geographical distribution of antibiotic resistance patterns among these isolates. WGS analyses were conducted to determine the diversity of resistance and virulence genes and to analyze the genetic relatedness among these isolates. The findings provide a scientific basis for quantitatively assessing the public health risk posed by Salmonella Enteritidis.
2 Materials and methods
2.1 Salmonella Enteritidis isolates
A total of 95 Salmonella Enteritidis isolates were used in the study (Supplementary Table S1). Among the 95 isolates, 67 were isolated from chicken meat, 13 from duck meat, 5 from egg products, 4 from pork, 2 from river water environment, 2 from freshwater fish, and 1 from a food poisoning incident. Geographically, 48 of the isolates originated from Shanghai and Shandong in East China, 33 from Guangdong in South China, and 14 from other regions and cities across China. These isolates were collected over the period from 2014 to 2019. More detailed information of these isolates is listed in Supplementary Table S1. These isolates were stored in a − 80°C freezer in our laboratory and reactivated by overnight cultivation at 37°C prior to use. The serovar of every isolate was re-verified using a PCR method, as described in a previous study (Liu et al., 2012). In brief, the serotype-specific primer sen-1392 (Liu et al., 2012) for Salmonella Enteritidis was used for PCR, with slight adjustments to the PCR conditions. Specifically, the conditions included an initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 10 min, and a final hold at 4°C. In the subsequent agarose gel electrophoresis, a 656 bp band was detected in the PCR products of all isolates, indicating that they were all Salmonella Enteritidis.
2.2 Antimicrobial susceptibility testing
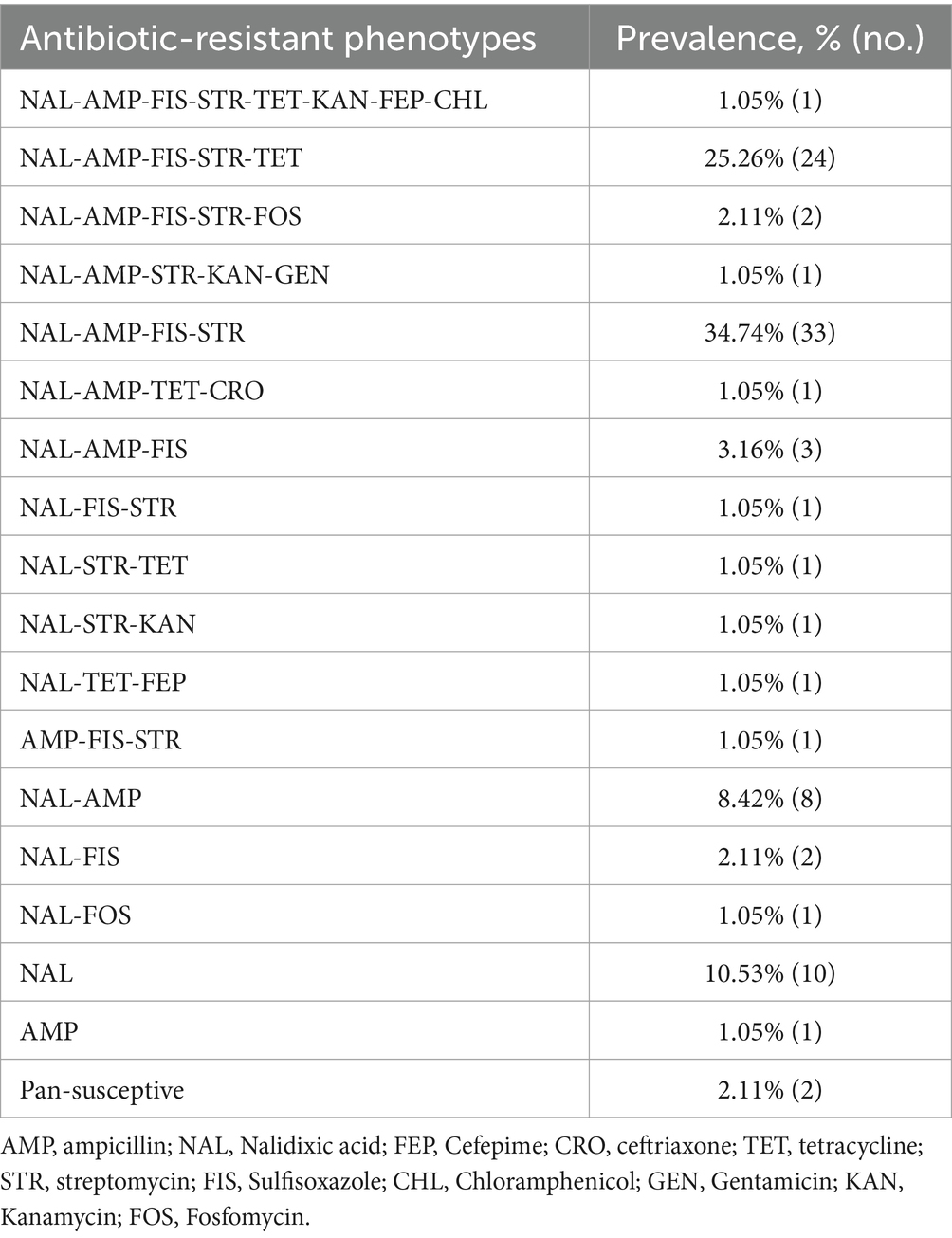
According to the guidelines recommended by the Clinical and Laboratory Standards Institute (CLSI) (CLSI, 2020), the minimum inhibitory concentration (MIC) for a range of antibiotics was determined using the agar dilution method. A total of 14 antibiotics were tested, including amikacin (AMK), gentamicin (GEN), kanamycin (KAN), streptomycin (STR), ampicillin (AMP), ceftriaxone (CRO), cefepime (FEP), sulfisoxazole (FIS), chloramphenicol (CHL), tetracycline (TET), ciprofloxacin (CIP), ofloxacin (OFX), nalidixic (NAL) and fosfomycin (FOS). Bacterial cultures were prepared by inoculating fresh colonies into Mueller Hinton broth (Beijing Landbridge Technology Co., Ltd., Beijing, China) and incubating at 37°C for 18–24 h. The inoculum density was adjusted to the McFarland 0.5 standard prior to testing. The agar dilution method was performed by preparing antibiotic-containing Mueller Hinton agar (Beijing Landbridge Technology Co., Ltd., Beijing, China) plates in accordance with CLSI guidelines (CLSI, 2020), and the inoculated plates were incubated at 37°C for 18–24 h. Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 25923 were used as quality control bacteria in the MIC determinations. Breakpoints for all the tested antibiotics were used according to the interpretive standards by CLSI (2020). Bar charts for analysis on the distribution of sample types, geographic regions and years of isolation across different antibiotic resistance phenotypes (ARPs) were created using ChiPlot.1
2.3 Whole genome sequencing and clustering tree construction
Of the 95 isolates, 31 were selected for WGS based on their resistance profiles, sample types, isolation dates, and geographic distribution to ensure representation of the broader dataset. WGS was carried out by Illumina platform Hiseq 2,500 in the Majorbio Corporation (Shanghai, China). Briefly, genomic DNA was extracted from each strain using the TIAN amp Bacterial DNA Kit (Tiangen, Beijing, China) following the manufacturer’s instructions. All genomes were constructed as fragments with an insert length of 500 bp to generate sequencing libraries using the NEB Next Ultra DNA library Prey Kit for Illumina (NEB, Beverly, MA, USA) according to the manufacturer’s recommendations. All genomes were assembled from scratch using SPAdes.2 The core genomes were aligned using MAFFT.3 To infer the clustering relationships among the bacterial isolates based on their core genomes, maximum-likelihood (ML) trees were constructed using IQ-TREE.4 Clustering trees were visualized using FigTree5 and edited for clarity in Adobe Illustrator. In Adobe Illustrator, the tree images were adjusted by repositioning branches, resizing labels, and adding color to improve readability and enhance the visual presentation of the clustering relationships.
2.4 In silico analysis of genetic information and plasmid typing
The detection of ARGs and point mutations was accomplished using ResFinder.6 The virulence factors (VFs) were investigated through the Virulence Factor Database (VFDB).7 A threshold of 90% was established to define a significant match between the identified gene sequences and those known to confer resistance and virulence, while a minimum length coverage criterion of 60% was applied to further validate the presence of these ARGs and VFs in the isolates examined. The PlasmidFinder database8 was employed to detect and characterize plasmid replicons in the bacterial samples. The 90% nucleotide identity cutoff was used to filter out the corresponding plasmid replication types in each isolate. All these bioinformatics analyses were based on the 31 sequenced representative isolates.
3 Results
3.1 Antibiotic resistance of 95 Salmonella Enteritidis isolates
Among the 95 Salmonella Enteritidis isolates analyzed, resistance was most frequently observed for NAL (91/95, 95.79%), followed by AMP (75/95, 78.95%), FIS (65/95, 68.42%), STR (57/95, 60.00%), and TET (28/95, 29.47%). All isolates were susceptible to AMK, CIP, and OFX, with low resistance rates (ranging from 1.05 to 3.16%) to other tested agents including GEN (1/95, 1.05%), CHL (1/95, 1.05%), CRO (1/95, 1.05%), EFP (2/95, 2.11%), KAN (3/95, 3.16%), and FOS (3/95, 3.16%). Notably, most isolates exhibited low MICs to third-generation cephalosporins (CRO and EFP) and fluoroquinolones (CIP and OFX), with MIC90 values below 0.125 μg/mL (Table 1).
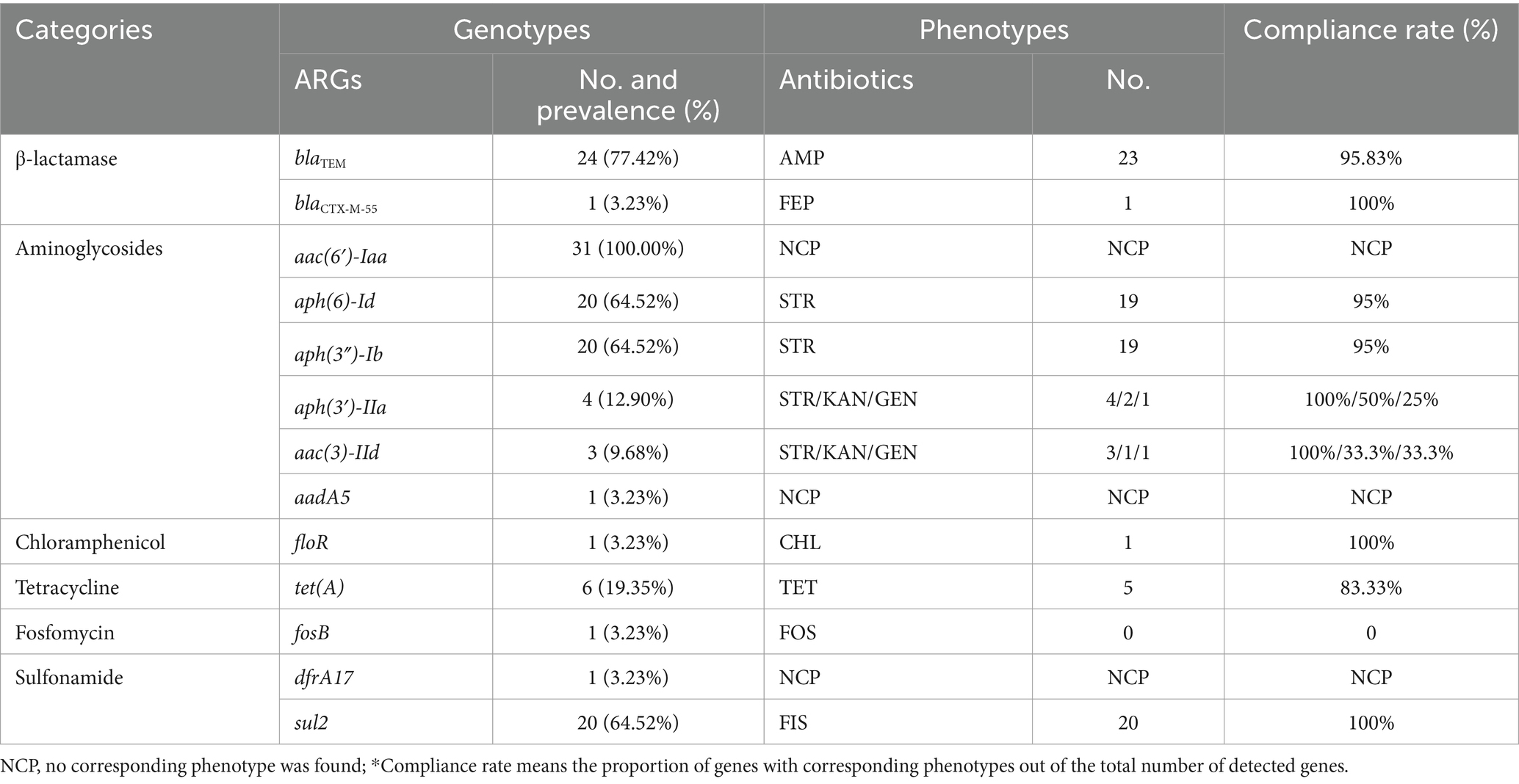
A total of 70 isolates (73.68%) were classified as MDR, defined as being resistant to three or more antibiotic agents. These isolates displayed 12 distinct MDR patterns (Table 2), including 1 isolate that showed resistance to 8 out of 14 antibiotics tested (NAL-AMP-FIS-STR-TET-KAN-FEP-CHL), with the underlined antibiotics forming the ACSSuT resistance pattern. The two most prevalent MDR profiles in this study were NAL-AMP-FIS-STR which was exhibited by 33 isolates and NAL-AMP-FIS-STR-TET which was exhibited by 24 isolates (Table 2). A third pattern (NAL-AMP-FIS) was exhibited by 3 isolates, and a fourth pattern (NAL-AMP-FIS-STR-FOS) by 2 isolates. Each of the other eight patterns was represented by a single isolate (Table 2). Along with 2 pan-sensitive strains, the remaining 23 strains which were resistant to one or two antibiotics were categorized into 5 different resistance profiles, with NAL resistance observed in 10 strains, and NAL-AMP resistance in 8 strains (Table 2).
3.2 Distribution of sample sources across different ARPs
Bar charts were constructed to show the distribution of various sample types across different ARPs (Figure 1A). Retail meat samples exhibited almost all of the ARPs, with the phenotype AMP being the sole exception. The only strain from a food poisoning sample and two strains from freshwater fish all exhibited the phenotype NAL-AMP-FIS-STR, while the only strain from a ready-to-eat food sample exhibited the phenotype NAL-AMP-FIS-STR-TET. Both of these phenotypes were the top two most prevalent antibiotic resistance patterns in this study. Five strains from egg samples exhibited diverse ARPs. Among them, two strains were MDR: one exhibited the most prevalent resistance pattern (NAL-AMP-FIS-STR), while the other was also exhibited resistance to FOS. The remaining three strains were not extensively resistant, with each showing resistance to either AMP, NAL, or both of these antibiotics. Two strains from the river water environment showed significantly different ARPs to each other. One strain was fully sensitive to all the antibiotics tested, while the other stain exhibited the most prevalent resistance pattern (NAL-AMP-FIS-STR).
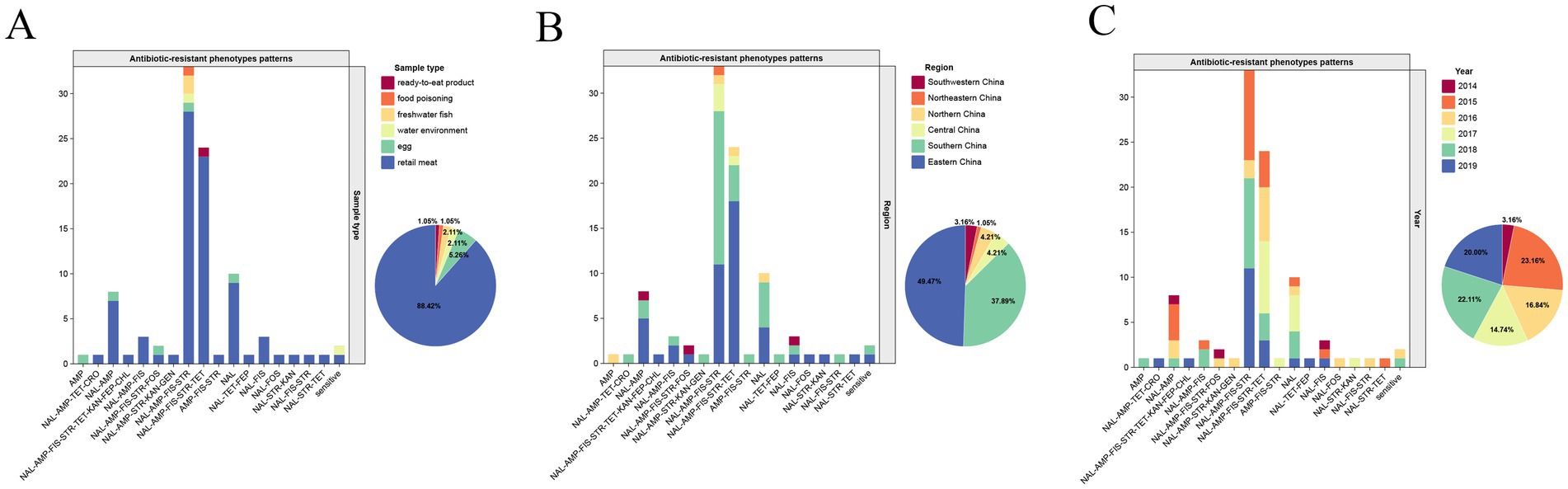
Figure 1. Distribution of sample types (A), geographic regions (B) and years of isolation (C) across different ARPs. Each bar represents a specific sample type, region and year, and its height corresponds to the number of samples that exhibit a certain ARP. The meanings of the abbreviations in the antibiotic-resistant phenotype patterns were shown in Table 1. The pie chart in each subgraph represents the proportion of strains in different categories.
From the perspective of geographic regions (Figure 1B), the three isolates from Southwestern China did not belong to either of the two most prevalent resistant patterns. The sole isolate from Northeastern China exhibited the most prevalent resistant pattern. The two most prevalent resistant patterns were also both detected in the other four regions of China. From the perspective of isolation years (Figure 1C), the three strains from 2014 did not belong to either of the two most prevalent resistant patterns, and interestingly, they were from Southwestern China. The most prevalent resistant pattern was not detected in isolates from 2017, while the second most prevalent resistant pattern was detected in isolates from 2015 to 2019. The annual distribution of the two most prevalent resistance patterns (NAL-AMP-FIS-STR ± TET) varied considerably from year to year (Figure 1C), meaning that in some years (2016 and 2017), strains resistant to TET were more common, while in others (2015, 2018 and 2019), strains lacking TET resistance were more prevalent.
3.3 Genetic analysis of 31 representative Salmonella Enteritidis isolates
A total of 31 Salmonella Enteritidis isolates were selected as representative strains for WGS. These representative strains encompassed all observed ARPs and two sensitive strains. Strains were selected based on a combination of ARPs, sample types, regions, and years. If a particular phenotype was only identified once in the 95 isolates studied, it was also included in the selection.
Using the ResFinder online prediction tool, a total of 16 antibiotic resistance genes were detected. These genes potentially corresponded to six different resistance categories (Table 3) which are described in detail below.
3.3.1 Aminoglycoside resistance genes
Six aminoglycoside resistance related genes were identified among the isolates (Table 3 and Figure 2). The three most prevalent genes were aac(6′)-Iaa, aph(6)-Id, and aph(3″)-Ib, which were detected in 100% (31/31), 64.52% (20/31), and 64.52% (20/31) of the isolates, respectively. Additionally, the aph(3′)-IIa gene was present in 12.90% (4/31) of the isolates, the aac(3)-IId gene was found in 9.68% (3/31), and the aadA5 gene was identified in 3.23% (1/31) of the isolates. The simultaneous or partial presence of the genes aph(6)-Id, aph(3″)-Ib, aph(3′)-IIa, and aac(3)-IId was linked to the STR phenotype, with compliance rates of 95 to 100% (Table 3). Additionally, the latter two genes (aph(3′)-IIa and aac(3)-IId) were also associated with the aminoglycosides GEN and KAN to some extent (Table 3).
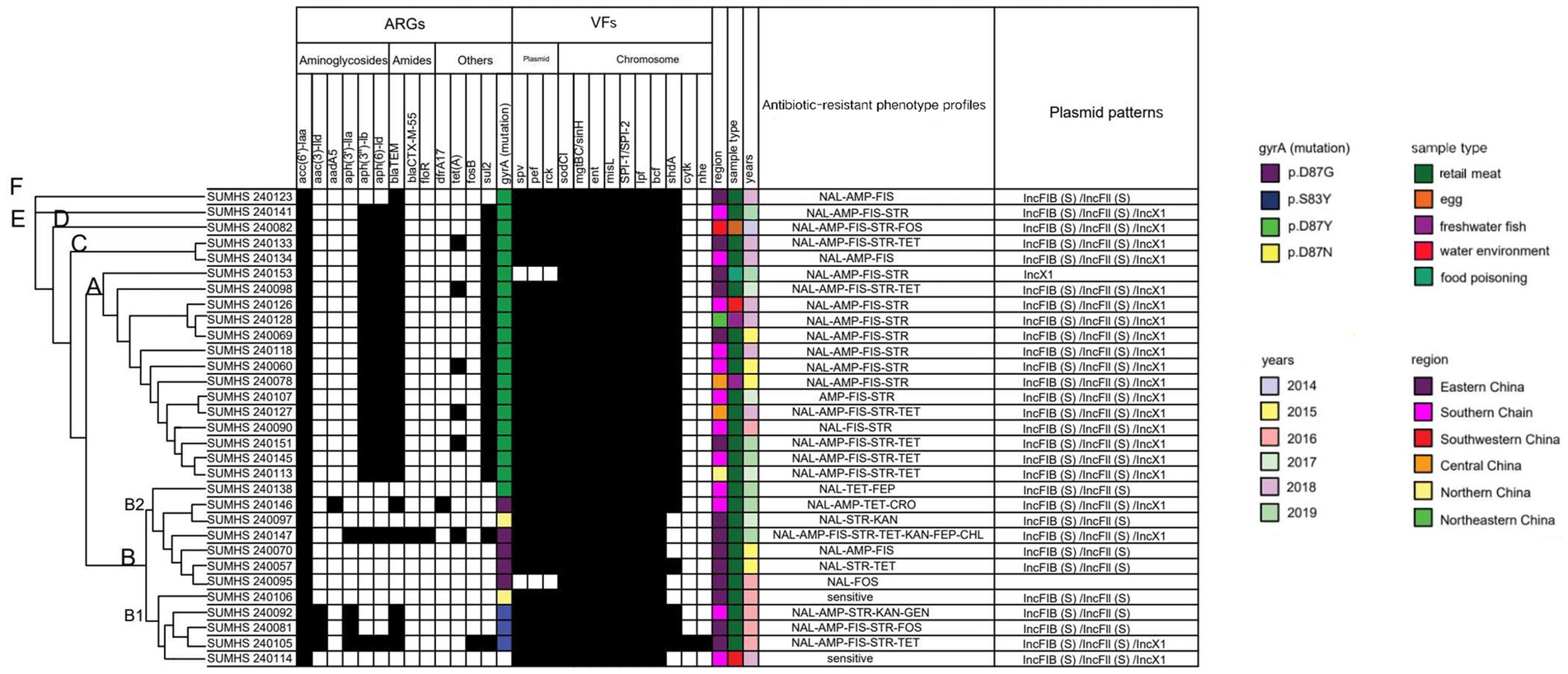
Figure 2. Clustering tree based on core genome and heatmap of 31 Salmonella Enteritidis strains: in silico analysis of virulence factors (VFs), antibiotic resistance genes (ARGs) and plasmid pattens.
3.3.2 β-lactam resistance genes
Two distinct β-lactam resistance genes were identified (Table 3 and Figure 2). The blaTEM gene was present in 77.42% (24/31) of the isolates, most of which were associated with AMP, with the compliance rate at 95.83% (Table 3). The blaCTX-M-55 gene was detected in only 1 isolate (3.23%) (SUMHS 240147), which also exhibited resistance to FEP (Table 3 and Figure 2).
3.3.3 Quinolone resistance genes
Resistance to the quinolone antibiotic, NAL, was primarily linked to chromosomal mutations in the DNA gyrase subunit gene (gyrA). The mutation in gyrA gene was identified in 30 out of 31 isolates that classified into 4 types. The predominant mutation site was gyrA [87:D-Y] (66.67%, 20/30), followed by gyrA [87:D-G] (16.67%, 5/30), gyrA [83:S-Y] (10%, 3/30), and gyrA [87:D-N] (6.67%, 2/30) (Figure 2).
3.3.4 Other resistance genes
One or two resistance genes were identified in each of the following antibiotic resistance groups: TET (tetA gene), sulfonamides (sul2 and dfrA17 genes), CHL (floR gene) and FOS (fosB gene). The sul2 gene was found in 20/31 (64.52%) isolatess, while the tetA gene was detected in 6/31 (19.35%) isolate. The dfrA17 gene was identified in isolate SUMHS 240146, and the floR gene was found in isolate SUMHS 240147, in addition the fosB gene was identified in isolate SUMHS 240105 (Figure 2). The ARPs of most isolates generally aligned with their corresponding ARGs, such as floR to CHL, and sul2 to FIS, which had the 100% compliance rate (Table 3). However, some isolates exhibited resistance phenotypes for which a genetic basis could not be identified, while in other cases, ARGs were present in isolates that remained susceptible to the corresponding antibiotics. For example, although isolate SUMHS 240105 harbored the fosB gene, it did not show resistance to FOS; while three FOS resistance strains did not contain the fosB gene, which suggested that in these isolates resistance to FOS was due to yet to be identified gene(s). Similarly, six isolates were resistant to TET but lacked the tet(A) gene, which is typically responsible for TET resistance. Notably, among the 31 isolates, those (20/31) carrying the aph(6)-Id and aph(3″)-Ib genes were also found to possess the sul2 gene, and the blaTEM gene, associated with STR resistance, was frequently found in combination with both FIS and AMP (Figure 2).
Similarly, virulence genes and plasmid replicon types among the 31 isolates were detected using the VFDB online prediction tool and the PlasmidFinder database, respectively.
3.3.5 Virulence factors
Type III secretion systems (T3SS) encoded by Salmonella Pathogenicity Island (SPI-1 and SPI-2) were present in all 31 Salmonella Enteritidis isolates. The superoxide dismutase gene, sodC1, and fimbria related gene lpf, along with the metal ion-regulating genes mgtBC and misL, were also found in 100% of the isolates (Figure 2). Another fimbria related gene shdA was present in 80.65% (25/31) of the isolates. Additionally, plasmid mediated virulence genes were detected across most of strains (93.5%, 29/31), including fimbrial adhesins (pef), T3SS effectors (spv), and the complement resistance gene (rck). The remaining two virulence genes were less common, with nhe and cytK only being present in 3.23% (1/31) of the strains (Figure 2).
3.3.6 Plasmids assays
The plasmid profiles indicated that F-type (IncFIB/IncFII) and X1-type plasmids were common in Salmonella Enteritidis strains (Figure 2). F-type plasmids were present in 93.55% (29/31) of the strains, while X1-type plasmids were detected in 64.52% (20/31) of the strains. No plasmids were identified in SUMHS 240095, whereas only the X1-type plasmid was detected in SUMHS 240153. Notably, the presence of the X1-type plasmid corresponded with the co-occurrence of blaTEM, sul2 and aph(6)-Id/aph(3″)-Ib, as mentioned above, suggesting a high likelihood that these genes were located on the X1-type plasmid, conferring resistance to AMP-FIS-STR.
3.4 Genetic diversity and relatedness of 31 Salmonella Enteritidis isolates
For the purpose of illustrating the interrelationships among these Salmonella Enteritidis isolates, a clustering tree was constructed based on the core genome genes (Figure 2). This classified tree partitioned the 31 Salmonella Enteritidis isolates into 6 distinct clusters. Predominantly, two larger clusters were identified: cluster A, which encompassed 14 isolates, and cluster B, which was comprised of 12 isolates. Cluster B was distinctly subdivisible into B1, which encompasses 5 isolates, and B2, which contained the remaining 7 isolates. The remaining clusters, designated C, D, E and F, were notably smaller, each incorporating only 1 or 2 isolates.
The majority (7/8) of isolates carrying the most prevalent ARP (NAL-AMP-FIS-STR) (Figure 2) were classified into cluster A (Figure 2), which exhibited similar resistance and virulence genes, suggesting clonal transmission. However, the sample types, regions, and years of these 7 isolates differed, suggesting that strains with this ARP may be widespread in retail markets and the environment. This indicates that clonal transmission may be occurring due to cross-contamination during the sales process or food chain, likely involving the spread of bacteria between products, surfaces, or handlers, leading to the same bacterial clone being present in different locations or at different times. Notably, 6 closely clustered isolates were from 2015 to 2018, with the other 2 isolates, from 2019, having slightly longer clustering distances, which indicates that the genomes of the most recent isolates have slightly changed. Taking strain SUMHS 240153 as an example, the absence of three virulence genes, spv, pef and rcs, typically found on the virulence plasmid, was observed. This suggests that the strain might have diverged from the other 6 strains due to the potential loss of this specific virulence plasmid after infecting humans, as this was a food poisoning isolate. Similarly, the majority (5/7) of the isolates that possessed the second most prevalent ARP (NAL-AMP-FIS-STR-TET) (Table 2) were also classified into cluster A (Figure 2). These stains while all being isolated from retail meat, came from different regions and were isolated between 2017 and 2019, indicating that this prevalent resistance strain was most likely clonally transmitted through retail meat in various regions. Notably, one isolate (SUMHS 240105) exhibiting the second most prevalent ARP was classified into cluster B. This isolate differed significantly from the above 5 strains in terms of the resistance and virulence genes detected. For instance, it lacked the tet gene but exhibits a TET phenotype; 4 additional resistance genes [aac(3)-IId, aph(3″)-Ib, aph(3′)-IIa, fosB] were detected, yet they had no corresponding resistant phenotypes, referred to as heteroresistance; additionally, 2 virulence genes (ctyK and nhe) were uniquely found within this strain and the type of mutation in gyrA (S83Y) was different from others in cluster A. All of this data suggests that this strain may be a completely different clone.
Overall, both the ARGs and ARPs were more complicated in cluster B compared to for the other 5 clusters. Particularly, cluster B included four types of gyrA gene mutants (D87Y, D87N, D87G, and S83Y), whereas only the most typical mutation (gyrA: D87Y) existed in the other clusters (A, C, D, E, F). Similarly, the carriage of ARGs in the aminoglycoside class were uniform in the other 4 clusters (A, C, D, E, F), whereas this was not the case in cluster B. Additionally, the only two pan-susceptible strains among the tested isolates were also located within the B1 sub-branch of cluster B. With regard to geographic distribution and sample types; almost all the isolates in cluster B were from retail meat and all of them were isolated from Eastern or Southern China between 2015 and 2019. This suggested that these isolates in cluster B may have a wealth of transferable components which are carrying different ARGs or virulence genes.
4 Discussion
The current study analyzed the antibiotic resistance profiles of Salmonella Enteritidis isolates in China from 2014 to 2019 (Table 1 and Figure 1). Compared to data reported for Salmonella Enteritidis strains isolated from retail chicken samples from Shanghai between 2008 and 2012 (Zhou et al., 2018), resistance rates appear to have increased significantly: AMP from 50.70 to 78.95%, FIS from 49.32 to 68.42%, TET from 17.12 to 29.47%, and STR from 4.80% to a notable 60.00%. Further, compared to Salmonella Enteritidis strains isolated from clinical samples in Beijing between 2010 and 2014 (Qu et al., 2016), resistance rates also showed slight increases: AMP increased from 60.0 to 78.95%, FIS from 54.3 to 68.42%, and STR from 42.9 to 60.00%. These findings indicate a worrying increase in the resistance of Salmonella Enteritidis to commonly used antibiotics, which if not checked could limit treatment options and make infections harder to manage. Fortunately, current monitoring data from retail samples of Salmonella Enteritidis indicate that resistance to first-line clinical antibiotics, such as cephalosporins and fluoroquinolones, remains low (Yang et al., 2020; Zhou et al., 2018; Yin et al., 2022; Kanaan et al., 2023). The results of the current study support these observations, as all isolates were sensitive to fluoroquinolones and resistance to cephalosporins was limited, with the MIC90 values for both antibiotics being comparatively low (Table 1). In contrast, the situation is less optimistic in clinical settings for Salmonella Enteritidis (Li et al., 2021), as well as for other Salmonella serovars in retail samples, such as Salmonella Typhimurium (Yang et al., 2023) and Salmonella Indiana (Hu et al., 2022).
Although 12 different MDR profiles were identified in this study (Table 2), the majority of isolates (81.43%, 57/70) exhibited the NAL-AMP-FIS-STR ± TET profiles (Table 2 and Figure 1), which was consistent with the predominant resistance pattern reported for Salmonella Enteritidis in retail foods in Shanghai in 2020 (Yang et al., 2020) and in Shaanxi in 2021 (Dai et al., 2021). Interesting, the relative prevalence of the two most common resistance profiles differed from 2015 to 2019, owing to the presence or absence of TET (Figure 1C). TET was once widely used to promote growth and prevent animal diseases (Liang et al., 2023; Rincón-Gamboa et al., 2021). However, its non-therapeutic use, in China was strictly regulated during the years the isolates in the current study were obtained from, particularly in food products like meat, eggs, and milk, with strict adherence to withdrawal periods (Xu et al., 2020). This regulation may in part explain the alternating patterns of Salmonella Enteritidis resistance to TET and underscore the link between antibiotic resistance in retail meat strains and antibiotic use in farming. With new policies prohibiting TET as a growth-promoting feed additive since 2020, resistance to this antibiotic is expected to decline.
In the current study, a notable finding was the identification of strain SUMHS 240147, isolated from retail chicken in Shanghai in 2019, which exhibited resistance to eight antibiotics (Figure 2) and may be a derivative of the ACSSuT resistance pattern. The ACSSuT profile is typically associated with the main MDR type in Salmonella Typhimurium (Yang et al., 2020) but is rarely found in Salmonella Enteritidis. Recent studies have shown that the ACSSuT pattern is usually linked to high resistance against third-generation cephalosporins and quinolones in Salmonella, including Typhimurium (Yang et al., 2020), Newport and Dublin (Bhandari et al., 2023), and Enteritidis (Ma et al., 2018), which aligns with the findings of this study. These results highlight the need for regular monitoring to track changes in antibiotic resistance patterns, serving as a warning for other cities in China.
The egg-derived isolates in the current study exhibited five different ARP patterns, demonstrating a relatively rich resistance profile (Figure 1A). This is similar to a recent report on antibiotic resistance in Salmonella from poultry eggs (Bahramianfard et al., 2021), which showed that although the number of antibiotics Salmonella were resistant to was low, the resistance profiles were quite complex. A representative egg isolates (SUMHS 240082), which was sequenced was categorized into cluster D (Figure 2), exhibiting an additional FOS resistance compared to the second most prevalent resistant pattern (NAL-AMP-FIS-STR-TET). It shared the same resistance phenotype with a strain (SUMHS 240081) from the distantly related cluster B1, yet it had a significantly different genotype. These results suggested that the egg may not have been contaminated at the retail level and likely originated from chickens raised on farms that were infected with resistant Salmonella Enteritidis.
In silico analysis of antibiotic genes in the current study revealed complex relationships between the presence of specific genes and aminoglycoside phenotypes in Salmonella Enteritidis. While some genes, like aph(3″)-Ib and aph(6)-Id, showed a clear association with STR resistance, others, such as aac(6′)-Iaa and aac(3)-IId, did not demonstrate a clear correlation with resistance to certain aminoglycosides (Table 3). Previous studies have shown that the aph(3″)-Ib and aph(6)-Id genes are often detected together in STR-resistant strains (Srednik et al., 2022; Yue et al., 2022), which is consistent with the results obtained in the current study, where both genes were present in 20 out of 21 STR-resistant strains (concordance rate of 95%) (Table 3). Conversely, the gene aac(3)-IId which is sometimes associated with resistance to GEN (Davies et al., 2022; Cox et al., 2022) was detected in three strains of this study (Table 3 and Figure 2). However, among the three strains carrying the aac(3)-IId gene detected in this study, only one (SUMHS 240092) exhibited resistance to GEN (Figure 2). Furthermore, it was observed that for the aminoglycoside-resistant strains of the B cluster, the number of resistance genes did not correlate directly with the number of aminoglycoside resistance types (Figure 2). For instance, SUMHS 240105, which carried all four of the aforementioned genes, was only resistant to STR, while SUMHS 240092, which carried only aac(6′)-Iaa and aac(3)-IId, was resistant to STR, GEN, and KAN (Figure 2). This observation suggests that aminoglycoside resistance mechanisms in Salmonella Enteritidis are multifactorial and may involve additional factors beyond the presence and quantity of specific resistance genes. Further research is needed to fully elucidate these complexities.
Previous studies have indicated that blaTEM is the most common gene associated with AMP resistance (Zheng et al., 2021). Accordingly, the current study observed a 95.83% concordance rate (23/24) between the presence of this gene and AMP resistance (Table 3). Additionally, an association of the IncX1 plasmid with the blaTEM gene in Salmonella has previously been reported (Petrin et al., 2023). Consistent with this, 83.3% (20/24) of the AMP-resistant isolates in the current study carried the IncX1 plasmid (Figure 2), suggesting a high likelihood that the blaTEM gene in Salmonella Enteritidis is carried by the IncX1 plasmid. Moreover, the co-occurrence of blaTEM, sul2 and aph(6)-Id/aph(3″)-Ib, mediated by the X1 plasmid and resistance to AMP-FIS-STR, was also observed in a similar study by Li et al. (2022) for Salmonella isolated from dead poultry. This consistency suggests that the X1 plasmid plays a significant role in transmitting MDR among Salmonella strains from food animals to food products. Furthermore, the current study identified a strain (SUMHS 240147) carrying the blaCTX-M-55 gene that exhibited resistance to FEP (Figure 2). This finding aligns with previous reports that blaCTX-M encodes a plasmid-mediated enzyme that preferentially hydrolyzes CRO or FEP in Salmonella (Long et al., 2022). Interestingly, the current study detected one FEP-resistant strain (SUMHS 240138) and one CRO-resistant strain (SUMHS 240146) that lacked blaCTX-M, suggesting the presence of other genes encoding broad-spectrum cephalosporin resistance that we did not detect. Similarly, the tetA gene, which is associated with phenotypic resistance to TET (Petrin et al., 2023), was found in only 6 out of 11 TET-resistant strains. As these genes are plasmid-mediated, discrepancies may arise from variations in plasmid abundance or sequencing accuracy. The fosB gene, commonly found in Gram-positive bacteria such as Staphylococcus aureus (Hu et al., 2023), is associated with FOS resistance (Lamers et al., 2012). However, it is rarely detected in Gram-negative bacteria, such as Salmonella, and its association with resistance in this context is uncertain. This observation might explain why one strain in the current study which carried the fosB gene did not exhibit FOS resistance. In Salmonella, FOS resistance is typically linked to mutations in chromosomal genes glpT and murA (Couce et al., 2012; Takahata et al., 2010) or plasmid-mediated genes like fosA3 (Fang et al., 2020). However, the three FOS-resistant strains in the current study did not show the presence of these genes or mutations. This could be due to insufficient detection sensitivity or the involvement of other untested genes. Therefore, the aforementioned strains that exhibited resistance but lacked the corresponding genes, require more detailed sequencing and mechanistic studies.
Two specific strains classified in cluster B2 in the current study were also noteworthy. The strain SUMHS 240147 which exhibited an 8-fold resistant phenotype was isolated in 2019 (Figure 2), which was the most recent isolate among the batch of strains tested, making it worthy of attention at the current time. The ARPs were consistent with the ARGs, especially, with MDR-ACSSuT resistance commonly found on transferable components (e.g., plasmid) (Wottlin et al., 2022). However, MDR-ACSSuT resistance can also be located in chromosomes, as seen in Salmonella Typhimurium, where it is encoded within a pathogenic island (de Curraize et al., 2017). This suggests that the resistance might have originated from a common strain that underwent genetic mutation and acquired resistance genes. Interestingly, this strain was a monophasic variant (Supplementary Table S1). The emergence of such variants, which are frequently reported in Salmonella Typhimurium (Sun et al., 2020), could present certain challenges for the diagnosis and treatment of salmonellosis. Moreover, monophasic Salmonella Typhimurium often display increased resistance and pathogenicity (Sun et al., 2020; Qin et al., 2022). Therefore, this monophasic Salmonella Enteritidis strain, which exhibited high-quantity antibiotic resistance, warrants further toxicity testing. Another strain in cluster B2 that warrants attention was SUMHS 240146, which was extraordinarily unique in both phenotype and genotype. Specifically, it displayed resistance to CRO and TET, yet no corresponding ARGs were identified, indicating that it may have plasmid mediated unidentified genes. It was the only strain that carried the dfrA17 and aadA5 genes (Figure 2), which encode a dihydrofolate reductase enzyme for sulfamethoxazole/trimethoprim and an aminoglycoside-modifying enzyme (Ma et al., 2022), respectively. However, resistance to sulfonamides or aminoglycosides was not detected (Figure 2). Previous studies have shown that the dfrA17-aadA5 gene cassette is often found together with the aac(3)-Id gene in type I integrons (Meng et al., 2017). However, this strain did not carry the aac(3)-Id gene, suggesting it may possess a defective type I integron that contains genes related to sulfonamides or aminoglycosides, but these genes are not expressed. These discrepancies between the WGS-determined genotype and experimentally observed ARP, referred to as heteroresistance, may limit the ability of WGS to accurately predict true resistance rates (Zwe et al., 2020). Heteroresistance includes false negative errors (where phenotypic resistance is absent despite the presence of the genetic resistance determinant) and false positive errors (where resistance genes are present without corresponding phenotypic resistance). Unstable, temporary changes in genetic elements may be the main reasons that cause heteroresistance (Zwe et al., 2020). Unstable genetic features such as temporary loss of gene expression or lower copy number when resistance genes are located on mobile elements could not be reliably employed by WGS to predict phenotype, especially in next-generation sequencing (NGS) platforms utilized in this study due to its short length of the reads generated. Therefore, incorporating deep sequencing specifically targeting these mobile elements may offer a potential solution to alleviate the detection failures in silico.
From a virulence perspective, these strains also have a high pathogenic potential. All these resistant isolates possessed relatively abundant virulence genes as genes related to antioxidant (sodC1), metal ion transport (mgtBC and misL), bacteriocin production (ent), and major pathogenicity islands (SPI1 and SPI2), as well as genes related to adhesion that may be located within the pathogenicity islands (lpf and bcf), were present in all the strains (Figure 2). Strangely, half (6/12) of the isolates classified into cluster B lacked the shdA gene, which encodes a protein involved in bacterial flagella synthesis and assembly (Urrutia et al., 2014), and it is usually not located in pathogenicity islands. Since the loss of this flagellar gene was only detected in monophasic bacteria in this study (Supplementary Table S1), it is hypothesized that there may be a relationship between the two, which requires further confirmation. Genes related to cytotoxins (cytK) and enterotoxins (nhe) were only detected in the SUMHS 240105 strain, which is also the only strain classified into cluster B among those with the second epidemic resistance pattern (NAL-AMP-FIS-STR-TET) (Table 2). Therefore, this highly virulent and epidemic resistant bacteria (SUMHS 240105) is also very worthy of further study because these exotoxins, including cytotoxins and enterotoxins, are believed to be responsible for diarrheal food poisoning (Stenfors Arnesen et al., 2008). In the current study, strains that did not harbor the virulence genes pef, spv, and rck also did not carry F-type plasmids, and vice versa (Figure 2). This confirmed previous reports that these three virulence genes are mediated by F-type virulence plasmids specific in Salmonella Enteritidis (Ksibi et al., 2022; Silva et al., 2017; Villa et al., 2010).
A limitation of this study is that WGS was conducted on only 31 of the 95 isolates due to resource constraints. While these isolates were selected to represent diverse resistance profiles, future studies should aim to sequence all isolates or conduct in-depth sequencing (e.g., nanopore sequencing) on the three isolates that warrant special attention and compare them with similar strains in public databases to provide a more comprehensive understanding of the genomic determinants and resistance mechanisms.
5 Conclusion
This analysis of antibiotic susceptibility among 95 Salmonella Enteritidis isolates revealed a notable rise in resistance to AMP, FIS, STR, and TET. Fortunately, the resistance rate for first-line antibiotics remained comparatively low. The yearly fluctuations in TET resistance may have been influenced by the management of withdrawal periods. These findings highlight the importance of continuous monitoring. The prevalence of ARPs across various sample types, regions, and isolation years suggests the potential for cross-contamination through the retail chain. However, the complex resistance profiles of egg-derived isolates indicated that contamination might have occurred at the animal or farm level. Leveraging WGS data, we identified most ARGs that align with observed ARPs, while variations in VFs were also uncovered. Co-carrying of blaTEM, sul2 and aph(6)-Id/aph(3″)-Ib (64.52%) was likely mediated by an X1-type plasmid, while virulence genes including pef, spv, and rck were associated with an F-type plasmid. Clustering analysis, based on core genes from 31 representative strains, demonstrated the diversity in the development of resistance in Salmonella Enteritidis. Strains in cluster B exhibited distinct ARPs and ARGs. Notably, three strains in cluster B emerged with unique and potentially high-risk resistance phenotypes, as well as specific virulence and resistance genes, underscoring the need for vigilant monitoring of Salmonella Enteritidis. These findings provided new insights into the molecular epidemiology of antibiotic resistance and highlight critical intervention points for mitigating this public health threat.
Data availability statement
The complete sequences of 31 Salmonella Enteritidis have been deposited in the NCBI database 677 under PRJNA1161004.
Author contributions
LZ: Conceptualization, Formal analysis, Methodology, Writing – original draft. QD: Project administration, Software, Validation, Writing – original draft. XX: Investigation, Methodology, Writing – original draft. LL: Investigation, Writing – original draft. CQ: Validation, Writing – review & editing. PB: Supervision, Writing – review & editing. XZ: Conceptualization, Funding acquisition, Visualization, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (grant no. 32072320), Climbing Plan for Excellent Young Staffs at Shanghai University of Medicine & Health Sciences (grant no. A3-2601-24-311001), and Shanghai Municipal Health Commission Special Fund for Clinical Research in the Health Industry (grant no. 20234Y0058).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1502138/full#supplementary-material
Footnotes
1. ^https://www.chiplot.online/
2. ^http://cab.spbu.ru/software/spades/
3. ^https://mafft.cbrc.jp/alignment/software
5. ^http://tree.bio.ed.ac.uk/software/figtree
References
Alzahrani, K. O., Al-Reshoodi, F. M., Alshdokhi, E. A., Alhamed, A. S., Al Hadlaq, M. A., Mujallad, M. I., et al. (2023). Antimicrobial resistance and genomic characterization of Salmonella enterica isolates from chicken meat. Front. Microbiol. 14:1104164. doi: 10.3389/fmicb.2023.1104164
Bahramianfard, H., Derakhshandeh, A., Naziri, Z., and Khaltabadi Farahani, R. (2021). Prevalence, virulence factor and antimicrobial resistance analysis of Salmonella enteritidis from poultry and egg samples in Iran. BMC Vet. Res. 17:196. doi: 10.1186/s12917-021-02900-2
Bellil, Z., Mairi, A., Kendi, S., and Touati, A. (2023). Nontyphoid Salmonella in farm animals and food products in the Middle East and North Africa: a systematic review. Future Microbiol. 18, 521–534. doi: 10.2217/fmb-2022-0239
Bhandari, M., Poelstra, J. W., Kauffman, M., Varghese, B., Helmy, Y. A., Scaria, J., et al. (2023). Genomic diversity, antimicrobial resistance, plasmidome, and virulence profiles of Salmonella isolated from small specialty crop farms revealed by whole-genome sequencing. Antibiotics. 12:1637. doi: 10.3390/antibiotics12111637
CLSI (Ed.) (2020). Performance standards for antimicrobial susceptibility testing. 30th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute.
Couce, A., Briales, A., Rodríguez-Rojas, A., Costas, C., Pascual, A., and Blázquez, J. (2012). Genome wide overexpression screen for fosfomycin resistance in Escherichia coli: MurA confers clinical resistance at low fitness cost. Antimicrob. Agents Chemother. 56, 2767–2769. doi: 10.1128/AAC.06122-11
Cox, G. W., Avery, B. P., Parmley, E. J., Irwin, R. J., Reid-Smith, R. J., Deckert, A. E., et al. (2022). A one health genomic investigation of gentamicin resistance in Escherichia coli from human and chicken sources in Canada, 2014 to 2017. Antimicrob. Agents Chemother. 66:e0067722. doi: 10.1128/aac.00677-22
Dai, W., Zhang, Y., Zhang, J., Xue, C., Yan, J., Li, X., et al. (2021). Analysis of antibiotic-induced drug resistance of Salmonella enteritidis and its biofilm formation mechanism. Bioengineered. 12, 10254–10263. doi: 10.1080/21655979.2021.1988251
Davies, N., Jørgensen, F., Willis, C., McLauchlin, J., and Chattaway, M. A. (2022). Whole genome sequencing reveals antimicrobial resistance determinants (AMR genes) of Salmonella enterica recovered from raw chicken and ready-to-eat leaves imported into England between 2014 and 2019. J. Appl. Microbiol. 133, 2569–2582. doi: 10.1111/jam.15728
de Curraize, C., Amoureux, L., Bador, J., Chapuis, A., Siebor, E., Clément, C., et al. (2017). Does the Salmonella Genomic Island 1 (SGI1) confer invasiveness properties to human isolates? BMC Infect. Dis. 17:741. doi: 10.1186/s12879-017-2847-1
Edirmanasinghe, R., Finley, R., Parmley, E. J., Avery, B. P., Carson, C., Bekal, S., et al. (2017). A whole-genome sequencing approach to study cefoxitin-resistant Salmonella enterica serovar Heidelberg isolates from various sources. Antimicrob. Agents Chemother. 61, e01919–e01916. doi: 10.1128/AAC.01919-16
Fang, L. X., Jiang, Q., Deng, G. H., He, B., Sun, R. Y., Zhang, J. F., et al. (2020). Diverse and flexible transmission of fosA3 associated with heterogeneous multidrug resistance regions in Salmonella enterica serovar typhimurium and Indiana isolates. Antimicrob. Agents Chemother. 64, e02001–e02019. doi: 10.1128/AAC.02001-19
García-Soto, S., Linde, J., and Methner, U. (2023). Epidemiological analysis on the occurrence of Salmonella enterica subspecies enterica serovar Dublin in the German federal state Schleswig-Holstein using whole-genome sequencing. Microorganisms 11:122. doi: 10.3390/microorganisms11010122
Gu, D., Wang, Z., Tian, Y., Kang, X., Meng, C., Chen, X., et al. (2020). Prevalence of Salmonella isolates and their distribution based on whole-genome sequence in a chicken slaughterhouse in Jiangsu, China. Front. Vet. Sci. 7:29. doi: 10.3389/fvets.2020.00029
Guerrero, T., Bayas-Rea, R., Erazo, E., and Zapata Mena, S. (2022). Nontyphoidal Salmonella in food from Latin America: a systematic review. Foodborne Pathog. Dis. 19, 85–103. doi: 10.1089/fpd.2020.2925
Hofer, U. (2021). Salmonella enteritidis: chicken or egg? Nat. Rev. Microbiol. 19:682. doi: 10.1038/s41579-021-00632-6
Hu, J., Chen, L., Li, G., Pan, Y., Lu, Y., Chen, J., et al. (2023). Prevalence and genetic characteristics of fosB-positive Staphylococcus aureus in duck farms in Guangdong, China in 2020. J. Antimicrob. Chemother. 78, 802–809. doi: 10.1093/jac/dkad014
Hu, L., Cao, G., Brown, E. W., Allard, M. W., Ma, L. M., Khan, A. A., et al. (2020). Antimicrobial resistance and related gene analysis of Salmonella from egg and chicken sources by whole-genome sequencing. Poult. Sci. 99, 7076–7083. doi: 10.1016/j.psj.2020.10.011
Hu, Y., He, Y., Nguyen, S. V., Liu, C., Liu, C., Gan, X., et al. (2022). Antimicrobial resistance of Salmonella Indiana from retail chickens in China and emergence of an mcr-1-harboring isolate with concurrent resistance to ciprofloxacin, cefotaxime, and colistin. Front. Microbiol. 13:955827. doi: 10.3389/fmicb.2022.955827
Kanaan, M. (2023). Prevalence and antimicrobial resistance of Salmonella enterica serovars Enteritidis and typhimurium isolated from retail chicken meat in Wasit markets, Iraq. Vet. World. 16, 455–463. doi: 10.14202/vetworld.2023.455-463
Kang, X., Wang, M., Meng, C., Li, A., Jiao, X., and Pan, Z. (2022). Prevalence and whole-genome sequencing analysis of Salmonella reveal its spread along the duck production chain. Poult. Sci. 101:101993. doi: 10.1016/j.psj.2022.101993
Keefer, A. B., Xiaoli, L., M'ikanatha, N. M., Yao, K., Hoffmann, M., and Dudley, E. G. (2019). Retrospective whole-genome sequencing analysis distinguished PFGE and drug-resistance-matched retail meat and clinical Salmonella isolates. Microbiology 165, 270–286. doi: 10.1099/mic.0.000768
Konyali, D., Guzel, M., and Soyer, Y. (2023). Genomic characterization of Salmonella enterica resistant to cephalosporin, quinolones, and macrolides. Curr. Microbiol. 80:344. doi: 10.1007/s00284-023-03458-y
Ksibi, B., Ktari, S., Ghedira, K., Othman, H., Maalej, S., Mnif, B., et al. (2022). Antimicrobial resistance genes, virulence markers and prophage sequences in Salmonella enterica serovar Enteritidis isolated in Tunisia using whole genome sequencing. Curr Res Microb Sci. 3:100151. doi: 10.1016/j.crmicr.2022.100151
Lamers, A. P., Keithly, M. E., Kim, K., Cook, P. D., Stec, D. F., Hines, K. M., et al. (2012). Synthesis of bacillithiol and the catalytic selectivity of FosB-type fosfomycin resistance proteins. Org. Lett. 14, 5207–5209. doi: 10.1021/ol302327t
Li, C., Zhang, Z., Xu, X., He, S., Zhao, X., Cui, Y., et al. (2021). Molecular characterization of cephalosporin-resistant Salmonella enteritidis ST11 isolates carrying Bla(CTX-M) from children with diarrhea. Foodborne Pathog. Dis. 18, 702–711. doi: 10.1089/fpd.2020.2878
Li, Y., Kang, X., Ed-Dra, A., Zhou, X., Jia, C., Müller, A., et al. (2022). Genome-based assessment of antimicrobial resistance and virulence potential of isolates of non-Pullorum/Gallinarum Salmonella serovars recovered from dead poultry in China. Microbiol. Spectr. 10:e0096522. doi: 10.1128/spectrum.00965-22
Liang, C., Wei, Y., Wang, X., Gao, J., Cui, H., Zhang, C., et al. (2023). Analysis of resistance gene diversity in the intestinal microbiome of broilers from two types of broiler garms in Hebei province, China. Antibiotics 12:1664. doi: 10.3390/antibiotics12121664
Liu, B., Zhou, X., Zhang, L., Liu, W., Dan, X., Shi, C., et al. (2012). Development of a novel multiplex PCR assay for the identification of Salmonella enterica typhimurium and Enteritidis. Food Control 27, 87–93. doi: 10.1016/j.foodcont.2012.01.062
Long, L., You, L., Wang, D., Wang, M., Wang, J., Bai, G., et al. (2022). Highly prevalent MDR, frequently carrying virulence genes and antimicrobial resistance genes in Salmonella enterica serovar 4,[5],12: i: - isolates from Guizhou province, China. PLoS One. 17:e0266443. doi: 10.1371/journal.pone.0266443
Ma, J., An, N., Li, W., Liu, M., and Li, S. (2022). Antimicrobial resistance and molecular characterization of gene cassettes from class 1 integrons in Salmonella strains. J. Med. Microbiol. 71:001574. doi: 10.1099/jmm.0.001574
Ma, Y., Li, M., Xu, X., Fu, Y., Xiong, Z., Zhang, L., et al. (2018). High-levels of resistance to quinolone and cephalosporin antibiotics in MDR-ACSSuT Salmonella enterica serovar Enteritidis mainly isolated from patients and foods in Shanghai, China. Int. J. Food Microbiol. 286, 190–196. doi: 10.1016/j.ijfoodmicro.2018.09.022
Meng, X., Zhang, Z., Li, K., Wang, Y., Xia, X., Wang, X., et al. (2017). Antibiotic susceptibility and molecular screening of class I integron in Salmonella isolates recovered from retail raw chicken carcasses in China. Microb. Drug Resist. 23, 230–235. doi: 10.1089/mdr.2015.0359
Pearce, M. E., Alikhan, N. F., Dallman, T. J., Zhou, Z., Grant, K., and Maiden, M. C. J. (2018). Comparative analysis of core genome MLST and SNP typing within a European Salmonella serovar Enteritidis outbreak. Int. J. Food Microbiol. 274, 1–11. doi: 10.1016/j.ijfoodmicro.2018.02.023
Petrin, S., Orsini, M., Massaro, A., Olsen, J., Barco, L., and Losasso, C. (2023). Phenotypic and genotypic antimicrobial resistance correlation and plasmid characterization in Salmonella spp. isolates from Italy reveal high heterogeneity among serovars. Front. Public Health 11:1221351. doi: 10.3389/fpubh.2023.1221351
Qin, X., Yang, M., Cai, H., Liu, Y., Gorris, L., Aslam, M. Z., et al. (2022). Antibiotic resistance of Salmonella Typhimurium monophasic variant 1,4,[5],12: i: - in China: a systematic review and meta-analysis. Antibiotics. 11:532. doi: 10.3390/antibiotics11040532
Qu, M., Lv, B., Zhang, X., Yan, H., Huang, Y., Qian, H., et al. (2016). Prevalence and antibiotic resistance of bacterial pathogens isolated from childhood diarrhea in Beijing, China (2010-2014). Gut Pathog. 8:31. doi: 10.1186/s13099-016-0116-2
Rakitin, A. L., Yushina, Y. K., Zaiko, E. V., Bataeva, D. S., Kuznetsova, O. A., Semenova, A. A., et al. (2021). Evaluation of antibiotic resistance of Salmonella serotypes and whole-genome sequencing of multi-resistant strains isolated from food products in Russia. Antibiotics 11:1. doi: 10.3390/antibiotics11010001
Rincón-Gamboa, S. M., Poutou-Piñales, R. A., and Carrascal-Camacho, A. K. (2021). Antimicrobial resistance of non-typhoid Salmonella in meat and meat products. Food Secur. 10:1731. doi: 10.3390/foods10081731
Rounds, J. M., Taylor, A. J., Eikmeier, D., Nichols, M. M., Lappi, V., Wirth, S. E., et al. (2020). Prospective Salmonella enteritidis surveillance and outbreak detection using whole genome sequencing, Minnesota 2015-2017. Epidemiol. Infect. 148:e254. doi: 10.1017/S0950268820001272
Silva, C., Puente, J. L., and Calva, E. (2017). Salmonella virulence plasmid: pathogenesis and ecology. Pathog. Dis. 75:ftx070. doi: 10.1093/femspd/ftx070
Song, Y., Wang, F., Liu, Y., Song, Y., Zhang, L., Zhang, F., et al. (2020). Occurrence and characterization of Salmonella isolated from chicken breeder flocks in nine Chinese provinces. Front. Vet. Sci. 7:479. doi: 10.3389/fvets.2020.00479
Srednik, M. E., Morningstar-Shaw, B. R., Hicks, J. A., Mackie, T. A., and Schlater, L. K. (2022). Antimicrobial resistance and genomic characterization of Salmonella enterica serovar Senftenberg isolates in production animals from the United States. Front. Microbiol. 13:979790. doi: 10.3389/fmicb.2022.979790
Stenfors Arnesen, L. P., Fagerlund, A., and Granum, P. E. (2008). From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32, 579–606. doi: 10.1111/j.1574-6976.2008.00112.x
Sun, H., Wan, Y., Du, P., and Bai, L. (2020). The epidemiology of monophasic Salmonella Typhimurium. Foodborne Pathog. Dis. 17, 87–97. doi: 10.1089/fpd.2019.2676
Sun, R. Y., Fang, L. X., Dai, J. J., Chen, K. C., Ke, B. X., Sun, J., et al. (2024). Antimicrobial resistance and population genomics of emerging multidrug-resistant Salmonella 4,[5], 12: i: - in Guangdong, China. mSystems 9:e0116423. doi: 10.1128/msystems.01164-23
Takahata, S., Ida, T., Hiraishi, T., Sakakibara, S., Maebashi, K., Terada, S., et al. (2010). Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int. J. Antimicrob. Agents 35, 333–337. doi: 10.1016/j.ijantimicag.2009.11.011
Urrutia, M. I, Fuentes, J. A., Valenzuela, L. M., Ortega, A. P., Hidalgo, A. A., and Mora, G. C. (2014). Salmonella Typhi shdA: pseudogene or allelic variant? Infect. Genet. Evol. 26, 146–152. doi: 10.1016/j.meegid.2014.05.013
Villa, L., García-Fernández, A., Fortini, D., and Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Wang, W., Zhao, L., Hu, Y., Dottorini, T., Fanning, S., Xu, J., et al. (2020). Epidemiological study on prevalence, serovar diversity, multidrug resistance, and CTX-M-type extended-spectrum β-lactamases of Salmonella spp. from patients with diarrhea, food of animal origin, and pets in several provinces of China. Antimicrob. Agents Chemother. 64, e00092–e00020. doi: 10.1128/AAC.00092-20
Wang, Y., Liu, Y., Lyu, N., Li, Z., Ma, S., Cao, D., et al. (2022). The temporal dynamics of antimicrobial-resistant Salmonella enterica and predominant serovars in China. Natl. Sci. Rev. 10:nwac269. doi: 10.1093/nsr/nwac269
Wottlin, L. R., Harvey, R. B., Norman, K. N., Burciaga, S., Loneragan, G. H., Droleskey, R. E., et al. (2022). Prevalence and antimicrobial resistance of nontyphoidal Salmonella enterica from head meat and trim for ground product at pork processing facilities. J. Food Prot. 85, 1008–1016. doi: 10.4315/JFP-22-049
Xu, J., Sangthong, R., McNeil, E., Tang, R., and Chongsuvivatwong, V. (2020). Antibiotic use in chicken farms in northwestern China. Antimicrob. Resist. Infect. Control 9:10. doi: 10.1186/s13756-019-0672-6
Yang, C., Chen, K., Ye, L., Heng, H., Yang, X., Wai-Chi Chan, E., et al. (2023). Prevalence and molecular characterization of cefotaxime-resistant Salmonella strains recovered from retail meat samples in Shenzhen, China, during 2014-2017. Microbiol Spectr. 11:e0488622. doi: 10.1128/spectrum.04886-22
Yang, J., Zhang, Z., Zhou, X., Cui, Y., Shi, C., and Shi, X. (2020). Prevalence and characterization of antimicrobial resistance in Salmonella enterica isolates from retail foods in Shanghai, China. Foodborne Pathog. Dis. 17, 35–43. doi: 10.1089/fpd.2019.2671
Yin, X., Dudley, E. G., Pinto, C. N., and M'ikanatha, N. M. (2022). Fluoroquinolone sales in food animals and quinolone resistance in non-typhoidal Salmonella from retail meats: United States, 2009-2018. J. Glob. Antimicrob. Resist. 29, 163–167. doi: 10.1016/j.jgar.2022.03.005
Yue, Y., Shen, M., Liu, X., Hao, Q., Kang, Y., Che, Y., et al. (2022). Whole-genome sequencing-based prediction and analysis of antimicrobial resistance in Yersinia enterocolitica from Ningxia, China. Front. Microbiol. 13:936425. doi: 10.3389/fmicb.2022.936425
Zakaria, Z., Hassan, L., Sharif, Z., Ahmad, N., Mohd, A. R., Amir, H. S., et al. (2022). Virulence gene profile, antimicrobial resistance and multilocus sequence typing of Salmonella enterica subsp. enterica serovar Enteritidis from chickens and chicken products. Animals 12:97. doi: 10.3390/ani12010097
Zhao, L., Liu, G., Tang, W., Song, X., Zhao, X., Wang, C., et al. (2023). Antimicrobial resistance and genomic characteristics of Salmonella from broilers in Shandong Province. Front. Vet. Sci. 10:1292401. doi: 10.3389/fvets.2023.1292401
Zheng, D., Ma, K., Du, J., Zhou, Y., Wu, G., Qiao, X., et al. (2021). Characterization of human origin Salmonella serovar 1,4,[5],12: i: - in eastern China, 2014 to 2018. Foodborne Pathog. Dis. 18, 790–797. doi: 10.1089/fpd.2021.0008
Zhou, X., Xu, L., Xu, X., Zhu, Y., Suo, Y., Shi, C., et al. (2018). Antimicrobial resistance and molecular characterization of Salmonella enterica serovar Enteritidis from retail chicken products in Shanghai, China. Foodborne Pathog. Dis. 15, 346–352. doi: 10.1089/fpd.2017.2387
Keywords: Salmonella Enteritidis, whole-genome-sequencing, multidrug-resistant, clustering analysis, genetic determinants
Citation: Zheng L, Di Q, Xu X, Liu L, Qu C, Bremer P and Zhou X (2025) Phenotypic and WGS-derived antibiotic resistance patterns of Salmonella Enteritidis isolates from retail meat and environment during 2014 to 2019 in China. Front. Microbiol. 16:1502138. doi: 10.3389/fmicb.2025.1502138
Edited by:
Fang He, Zhejiang Provincial People’s Hospital, ChinaReviewed by:
Guodong Zhang, United States Food and Drug Administration, United StatesPoonam Sharma, Oklahoma State University, United States
Copyright © 2025 Zheng, Di, Xu, Liu, Qu, Bremer and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiujuan Zhou, emhvdXhqNTU0QDE2My5jb20=; Phil Bremer, cGhpbC5icmVtZXJAb3RhZ28uYWMubno=
†These authors have contributed equally to this work and share first authorship
 Liya Zheng1†
Liya Zheng1† Xuebin Xu
Xuebin Xu Phil Bremer
Phil Bremer Xiujuan Zhou
Xiujuan Zhou