
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 27 February 2025
Sec. Aquatic Microbiology
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1487584
This article is part of the Research Topic Progress in Microalgae Research, 2024: Freshwater Microalgae View all 6 articles
In eukaryotes, microRNAs (miRNAs) are generated by Dicer/Dicer-Like (DCL)-mediated cleavage. Previous studies identified three DCL genes (CrDCL1-3) in Chlamydomonas reinhardtii and indicated that CrDCL3 mediated the production of most miRNAs, while CrDCL1 protein was mainly involved in siRNA biogenesis. The role of CrDCL1 in miRNA biogenesis remains unclear. This study constructed a phylogenetic tree, performed structural analyses of Dicer/DCL proteins from multiple species and screened and verified dcl1 and dcl3 mutant strains. Using CC-5325 and dcl3 mutant as control groups, we performed sRNA-sequencing, RT-qPCR, and Northern blot verification experiments on dcl1 mutant to explore the involvement of CrDCL1 in miRNA biogenesis in C. reinhardtii. The results demonstrated that CrDCL1 was involved in the production of 22 miRNAs, including cre-miR910, novel-miR01, novel-miR03, novel-miR04, novel-miR05, and novel-miR06, whose production was depended not only on CrDCL1 but also on CrDCL3. The present findings highlight the production of some C. reinhardtii miRNAs that may be involved in multiple CrDCL proteins, which is different from animals and plants. The results of this study will enrich the knowledge of miRNA biogenesis in eukaryotes.
RNA-mediated silencing in eukaryotes involves highly conserved and specific gene expression regulatory mechanisms. Small RNAs (sRNAs) are incorporated into Piwi/Argonaute (AGO) proteins to form RNA-induced silencing complexes (RISC), which negatively regulate the expression of their target genes through complementary base pairing (Baulcombe, 2004; Meister, 2013). RNA-mediated silencing plays important roles in developmental regulation, genome stability maintenance in response to stress and viral infection (Baulcombe, 2004; Matranga and Zamore, 2007; Casas-Mollano et al., 2008). There are three major classes of sRNAs mediating RNA silencing in eukaryotes, including small interfering RNAs (siRNAs), microRNAs (miRNAs), and Piwi-interacting RNAs (piRNAs) (Matranga and Zamore, 2007; Ghildiyal and Zamore, 2009). Cleavage by the RNase III family enzyme Dicer or Dicer-like proteins produces siRNAs and miRNAs. The production of piRNAs is unclear, but it is independent from Dicer proteins (Iwasaki et al., 2015).
miRNAs are 18–24 nucleotide (nt) non-coding sRNAs that induce target mRNA cleavage or repress target mRNA translation at the post-transcriptional level. Following the initial discovery of miRNAs in Caenorhabditis elegans in 1993 (Lee et al., 1993), miRNAs were identified in other multicellular eukaryotes, such as Arabidopsis, Drosophila and humans. Subsequently, miRNAs were also identified in unicellular organisms later (Lou et al., 2018). In animals, miRNA biogenesis begins with the production of primary transcripts (pri-miRNAs) by RNA polymerase II. These pri-miRNAs are folded into imperfect stem-loop structures and processed to generate precursor-miRNAs (pre-miRNAs) in the nucleus by an RNase III enzyme called Drosha (Han et al., 2004). The pre-miRNAs are transported to the cytoplasm by Exportin 5, followed by cleavage of the loop by the RNase III enzyme Dicer to release the 18–24 nt miRNA/miRNA* duplex (Yi et al., 2003; Lund et al., 2004; Bartel, 2009). By contrast, the generation of plant miRNA/miRNA* duplexes occurs entirely within the nucleus, typically with two cleavage steps executed by an individual Dicer-like (DCL) RNase III enzyme. Plant miRNA/miRNA* duplexes are then transported to the cytoplasm and stabilized through HEN1 methylation (Kurihara and Watanabe, 2004; Kurihara et al., 2006; Eamens et al., 2009). In Arabidopsis thaliana, four Dicer homologs (AtDCL1-4) are discovered. AtDCL1 is mainly involved in miRNA biogenesis and AtDCL2-4 are involved in the biogenesis of various types of siRNAs. Notably, a few miRNAs such as miR822, miR839 and miR859 differ from other miRNAs in terms of their biogenesis: they are produced independently from AtDCL1 and rely on the cleavage by AtDCL4 (Rajagopalan et al., 2006; Ben et al., 2009; Tsuzuki et al., 2014; Yu et al., 2017).
miRNAs have been firstly reported in the single-cell alga Chlamydomonas reinhardtii in 2007 (Zhao et al., 2007; Molnár et al., 2007). Among the three CrDCLs (CrDCL1-3) in C. reinhardtii, CrDCL3 is involved in the production of most miRNAs with the assistance of CrDUS16 (Casas-Mollano et al., 2008; Merchant et al., 2007; Valli et al., 2016; Yamasaki et al., 2016). Casas-Mollano et al. demonstrated that CrDCL1 mediates siRNA accumulation and post-transcriptional silencing of the TOC1 retrotransposon (Casas-Mollano et al., 2008). The function of the CrDCL2 protein remains unknown. It is unclear whether the cleavage of pre-miRNAs occurs in the nucleus or cytoplasm, and whether the CrDCL1/CrDCL2 proteins play a role in the production of individual miRNAs similar to AtDCL4. This study aimed to verified whether CrDCL1 is also involved in miRNA processing, through sRNA high-throughput sequencing, using the dcl1 mutant. In comparison with the wild type CC-5325, differentially expressed miRNAs were identified and validated by real-time quantitative PCR (RT-qPCR) and Northern blot. By further evaluating the amount of differentially expressed miRNA in the dcl3 mutant, the results will figure out the role of CrDCL1 in miRNA biogenesis.
The wild type CC-5325, the dcl1 mutant strains (ID: LMJ.RY0402.124662; LMJ.RY0402.198146), and the dcl3 mutant strains (ID: LMJ.RY0402.253048; LMJ.RY0402.080558) were obtained from the Chlamydomonas Library Project (CLiP).1 These mutants were generated by random insertion of CIB1, a 2,223 bp double-stranded DNA transformation cassette, into the strain CC-5325 by electroporation (Zhang et al., 2014). The transformation cassette is composed of two random sequences, a PSAD promoter, a HSP70-RBCS2 promoter, a RBCS2 intron, the AphVIII gene (conferring paromomycin resistance), a PSAD terminator and the RPL12 terminator in the opposite direction to block transcription of the target gene.2 The dcl1 and dcl3 mutants exclusively harbored mutations in the CrDCL1 and CrDCL3 gene, respectively, without affecting other genes. Consequently, these mutants were chosen to investigate the functions of the CrDCL1 and CrDCL3 genes. C. reinhardtii cells were grown in Tris-acetate-phosphate (TAP) medium at 22°C under continuous illumination (100 μE m−2·s−1) and were aerated daily by shaking the bottles twice. For dcl1 and dcl3 mutants, the TAP medium was supplemented with 10 μg ml−1 paromomycin (Li et al., 2016; Zhang et al., 2014).
To confirm the mutants, colony PCR was performed. The genomic DNA was extracted from single colony obtained by streak cultivation, using Ultra DNA Isolation Kit (BEI-BEI BIOTECH). Primers used to verify the presence of 2,223 bp transformation cassette in mutants were listed in Supplementary Table S1.
The amino acid sequences of three C. reinhardtii DCL proteins (CrDCL1-3, gene IDs: Cre02.g141000, Cre16.g684715, Cre07.g345900), four Arabidopsis thaliana DCL proteins (AtDCL1-4, gene IDs: AT1G01040.2, AT3G03300.1, AT3G43920.2, AT5G20320.1), and two Oryza sativa putative DCL proteins (OsDCL2-3, gene IDs: LOC_Os09g14610.1, LOC_Os01g68120.1) were obtained from Phytozome (v12.1).3 The amino acid sequences of Dicer proteins from Mus musculus (Mm) (GenBank accession: NP_001398758.1), Homo sapiens (Hs) (GenBank accession: NP_001182502.1), Drosophila melanogaster (Dm) (GenBank accessions: AAF56056.1, AAF57830.2), Rhodotorula toruloides (Rt) (GenBank accession: XP_016275102.1), and Schizosaccharomyces pombe (Sp) (GenBank accession: NP_588215.2), as well as the amino acid sequences of Drosha protein from Cricetulus griseus (Cg) (GenBank accession: XP_035296509.1), Caenorhabditis elegans (Ce) (GenBank accession: O01326.2) and D. melanogaster (GenBank accession: AAF59169.1) were obtained from NCBI database.4 All sequences were aligned using ClustalW in MEGA X software and subsequently trimmed using trimAl v1.2rev57 with the “automated” parameter. The phylogenetic tree was constructed using the Maximum Likelihood method in the MEGA X software. The support for each node was tested with standard bootstrap analysis through 1,000 replications. The phylogenetic tree was visualized in iTOL v7.5
The feature of Dicer proteins was analyzed online at Pfam,6 SMART7 and NCBI (see text footnote 4). The secondary and tertiary structures of the CrDCL1-3 proteins were predicted using JPred: A Protein Secondary Structure Prediction Server8 and SWISS-MODEL,9 respectively.
Total RNA and Small RNA were extracted using RNAiso Plus (TaKaRa, Japan) and RNAiso for Small RNA (TaKaRa, Japan), respectively, following the manufacturer’s instruction. RNA quality was evaluated using a NanoDrop 2000 Ultra Microvolume Spectrophotometer (Thermo, MA, United States), with the 260/A280 ratios of total RNAs and sRNAs were approximately 2.0 and 1.75, respectively. For cDNA synthesis, genomic DNA was removed and reverse transcription was performed using the PrimeScript™ RT Reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Japan). Reverse transcription of sRNAs was performed with specific stem-loop primers (Supplementary Table S2). RT-qPCR was performed to detect RNA transcripts using PrimeScript™ RT-PCR (TaKaRa, Japan) on the ABI 7300 Real-Time PCR System (Framingham, MA, United States). The ACTIN gene and U4 snoRNA served as internal controls for normalizing mRNA and sRNA expression levels (Wang et al., 2017). All RT-qPCR primers are listed in Supplementary Table S3. The relative gene expression levels were calculated using the 2−△△CT method from three technical replicates.
The detected miRNA levels were further validated by Northern blot analysis. Total RNA was denatured at 70°C and separated by polyacrylamide gel electrophoresis using 0.5x TBE buffer. The RNA was then wet-transferred to a Hybond-NX nylon membrane (GE Healthcare) at a constant current of 0.3A for 50 min. The membrane was cross-linked with EDC cross linking solution at 65°C for 90 min. Following pre-hybridization with Hybridization buffer at 55°C for 40 min, 50 nmol/L miRNA probe and 25 nmol/L U4 probe were added for hybridization for 16 h. All reagents were prepared according to the methods described previously (Martinho et al., 2023). The membrane was washed with elution buffer (2x SSC and 0.1% SDS) and visualized using Procedure for Detection of Immobilized Nucleic Acids (Thermo, United States) and ChemiScope 3,300 Mini system (Clinx Science Instruments, China). The probes used for Northern blot were biotin-labeled at both the 5 “and 3” ends, and their sequences were listed in Supplementary Table S4. Integrated density of the band in images was measured three times using imageJ software.
All experiments were performed with more than three biological replicates to ensure repeatability.
After grown to the exponential phase (3.0–4.0 × 106 cells ml−1), the algal cells were collected by centrifugation, stored in liquid nitrogen and sent to Gene Denovo Biotechnology Company (Guangzhou, China) for sRNA-seq and analyses. In total, there were two groups of samples (CC-5325 and dcl1 mutant), each with three biological replicates.
Total RNAs were extracted by TRIzol, and 18–30 nt sRNAs were purified using polyacrylamide gel electrophoresis (PAGE), followed by 3′ and 5′ adapter ligation. The ligation products were reverse transcribed and amplified by PCR. The PCR products with the length of 140-160 bp were enriched to produce the cDNA libraries. Sequencing was performed using an Illumina HiSeq™ 2,500.
To obtain clean tags, the raw reads were filtered to remove the following: low quality reads (Q value ≤20 or containing unknown nucleotides N), reads without 3′ adapters or containing 5′ adapters, reads shorter than 18 nt (not including adapters) and reads containing polyA in the sRNA fragments. All clean tags were aligned with sRNAs in the GenBank (Release 209.0) and Pfam (11.0) databases to identify and eliminate rRNA, snoRNA, snRNA, and tRNA. They were also compared with the Chlamydomonas genome (v5.6) to remove repeat sequences and some tags that were potentially fragments from mRNA degradation. The remaining clean tags were queried against the miRBase database (Release 21) to identify the previously reported Chlamydomonas miRNAs (cre-miRNA), and the conservative miRNAs were obtained by alignment with miRNAs from other species. All unannotated tags were aligned with the Chlamydomonas genome, and novel miRNA candidates were predicted using Mireap_v0.2 software based on genome positions, hairpin structures, DCL cutting site, free energy and parameters between the miRNA and miRNA* strands (Ding et al., 2021; Kang et al., 2021).
miRNA expression levels in CC-5325 and dcl1 were normalized according to the following formula: Transcripts per million (TPM) = × 106. Furthermore, the fold change, the size of between-group differences and the associated p-values were calculated as following: Fold change (fc) = , and the size of between-group differences = log2(fc). Significant differences in miRNA expression level were identified based on a fold change ≥1.5 and p-value <0.05.
Target prediction for differentially expressed miRNAs and functional analysis.
Patmatch (v1.2) software was used to predict the targets of the differentially expressed miRNAs in the Chlamydomonas genome. To infer the potential functions of the differentially expressed miRNAs and their targets, we performed gene ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. For both analyses, significant enrichment was identified based on p < 0.05.
For RT-qPCR and Northern blot data, statistical significance was assessed using Dunnett’s multiple comparisons test and GraphPad Prism 9.1.2, at the levels of 0.01, 0.05, and 0.001. The data represents mean values from at least three biological replicates.
The Dicer proteins of C. reinhardtii, A. thaliana, O. sativa, M. musculus, H. sapiens, D. melanogaster, R. toruloides, and S. pombe were used to construct a phylogenetic tree in conjunction with the Drosha proteins of C. griseus, D. melanogaster, and C. elegans (Figure 1A). The analysis revealed that Drosha proteins of C. griseus, D. melanogaster, and C. elegans formed an outgroup, constituting a separate branch distinct from the Dicer proteins in each species. The Dicer/DCL proteins clustered according to their taxonomy categories: plants, animals, algae, and fungi. Within the plant branch, both A. thaliana and O. sativa exhibited multiple DCL proteins, which were grouped based on sequence similarity and function rather than species. For instance, AtDCL2 and OsDCL2 clustered together, as did AtDCL3 and OsDCL3. Three DCL proteins of C. reinhardtii formed an independent branch, evolutionarily distant from the Dicer/DCL proteins of other species.
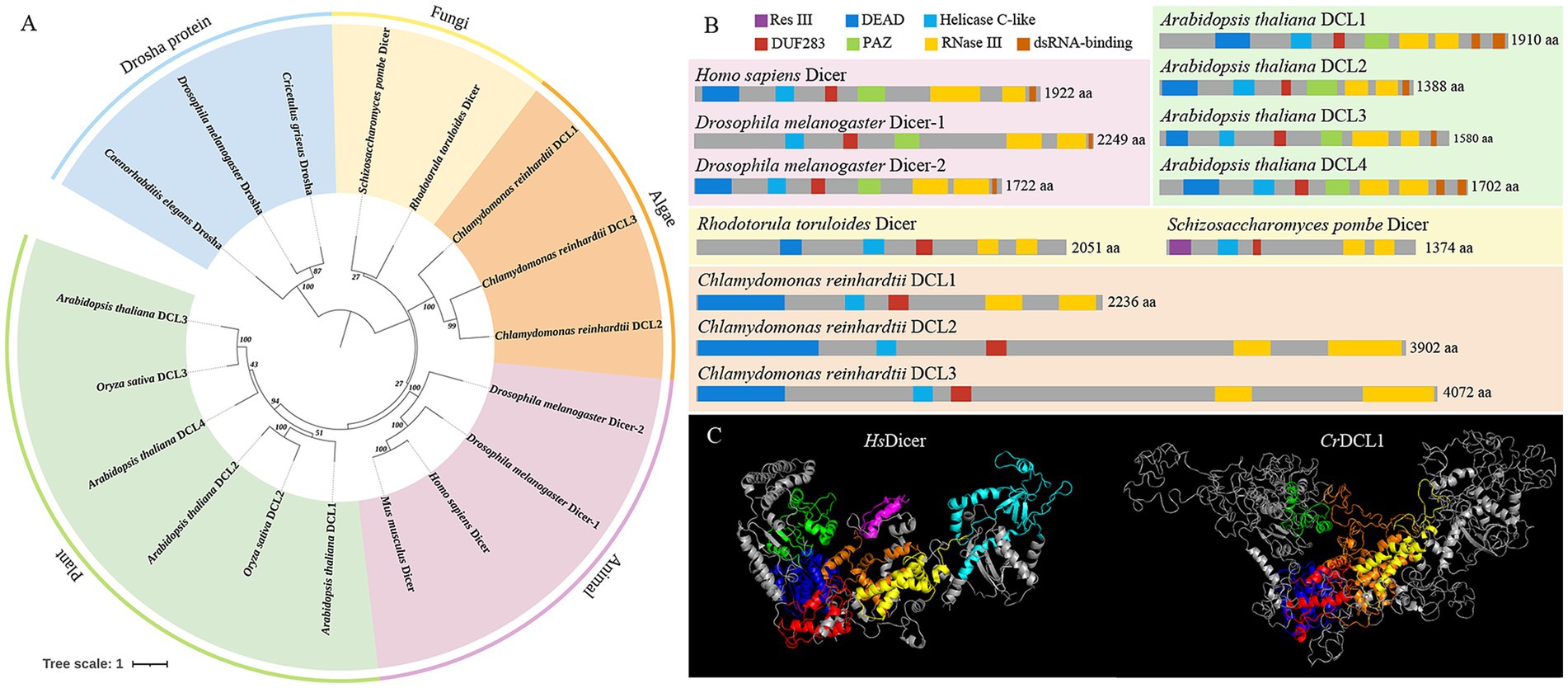
Figure 1. Feature of Dicer homologous proteins. (A) Phylogenetic tree analysis of Dicer and Drosha homologous proteins. (B) Structural domain analysis of Dicer homologous proteins. (C) Prediction of the three-dimensional structures of HsDicer and CrDCL1 proteins. In the three-dimensional structure diagram, the dark blue structures are the DEAD domains, the green structures are the Helicase C-like domains, the red structures are the DUF283 domains, the light blue structure is the PAZ domain, the yellow structures are the first RNase III domains, the orange structures are the second RNase III domains, and the purple structure is the dsRNA-binding domain.
The domain analyses of Dicer proteins revealed that AtDCL1-4, HsDicer and DmDicer1-2 all possess a DEAD/H-box helicase (hereinafter referred to as DEAD), Helicase C-like, DUF283, PAZ domain, two RNase III domains, and one or two dsRNA-binding domains. In contrast, RtDicer, SpDicer and CrDCL1-3 contain a DEAD, Helicase C-like, DUF283 and two RNase III domains but lack both PAZ and dsRNA-binding domains (Figure 1B). These findings suggested that DEAD, Helicase C-like, DUF283, and two RNase III domains are conserved across Dicer/DCL proteins, while PAZ and dsRNA-binding domains may have evolved later. Notably, SpDicer lacks the conserved DEAD domain but exhibits an additional unique Res III domain. There are not many reports on the Res III domain, and it remains unclear whether this domain compensates for the function of the DEAD domain or performs other unique functions.
Furthermore, CrDCL1 is comparable to other Dicer proteins in length, but CrDCL2 and CrDCL3 has twice number of amino acids as others. Notably, attempts to predict the secondary structure of CrDCL2 and CrDCL3 were unsuccessful. Similarly, predictions of their three-dimensional structures remained incomplete. Only the conserved domains (DEAD, Helicase C-like, DUF283 domains, and two RNase III domains) could be simulated. The three-dimensional structure of CrDCL1 and HsDicer was compared and results revealed an overall L-shaped structure for both proteins, with similar distribution patterns of their DEAD, Helicase C-like, DUF283 domains, and two RNase III domains. In this L-shaped structure, the Helicase C-like domains were located at the short arm, DEAD and DUF283 domains were located at the junction of the short and long arms, and two RNase III domains were located near DUF283 domain on the long arm. Additionally, the PAZ domain of HsDicer was located at the distal end of the long arm. It is important to note that CrDCL1 lacks the PAZ domain and dsRNA-binding domain but exhibits numerous irregular curls (Figure 1C).
The dcl mutants used in the present study were obtained from the Chlamydomonas Library Project (CLiP). Based on the recording, the dcl1 mutant strains LMJ.RY0402.124662 (dcl1-124662) and LMJ.RY0402.198146 (dcl1-198146) from CLiP harbored the cassette in a CDS region and intron region of the CrDCL1 gene, respectively. The dcl3 strains LMJ.RY0402.080558 (dcl3-080558) and LMJ.RY0402.253048 (dcl3-253048) harbored the cassette in an intron region and CDS region of the CrDCL3 gene, respectively.
We randomly selected four colonies of each mutant to confirm the DNA cassette insertion in the target gene, and then detected the mRNA transcript levels by RT-qPCR. The analysis showed that CrDCL1 transcript levels were significantly down-regulated in dcl1-124662 and dcl1-198146; CrDCL3 was significantly down-regulated in dcl3-253048 but increased in dcl3-080558 (Figure 2). Hence, for both genes, insertion of the cassette into the CDS region led to the significant inhibitory of CrDCL1 and CrDCL3 expression. CrDCL1 and CrDCL3 transcript levels were lowest in dcl1-124662 colony #3 and dcl3-253048 colony #3, respectively. Therefore, these two colonies were selected as the dcl1 and dcl3 mutants for the subsequent experiments.
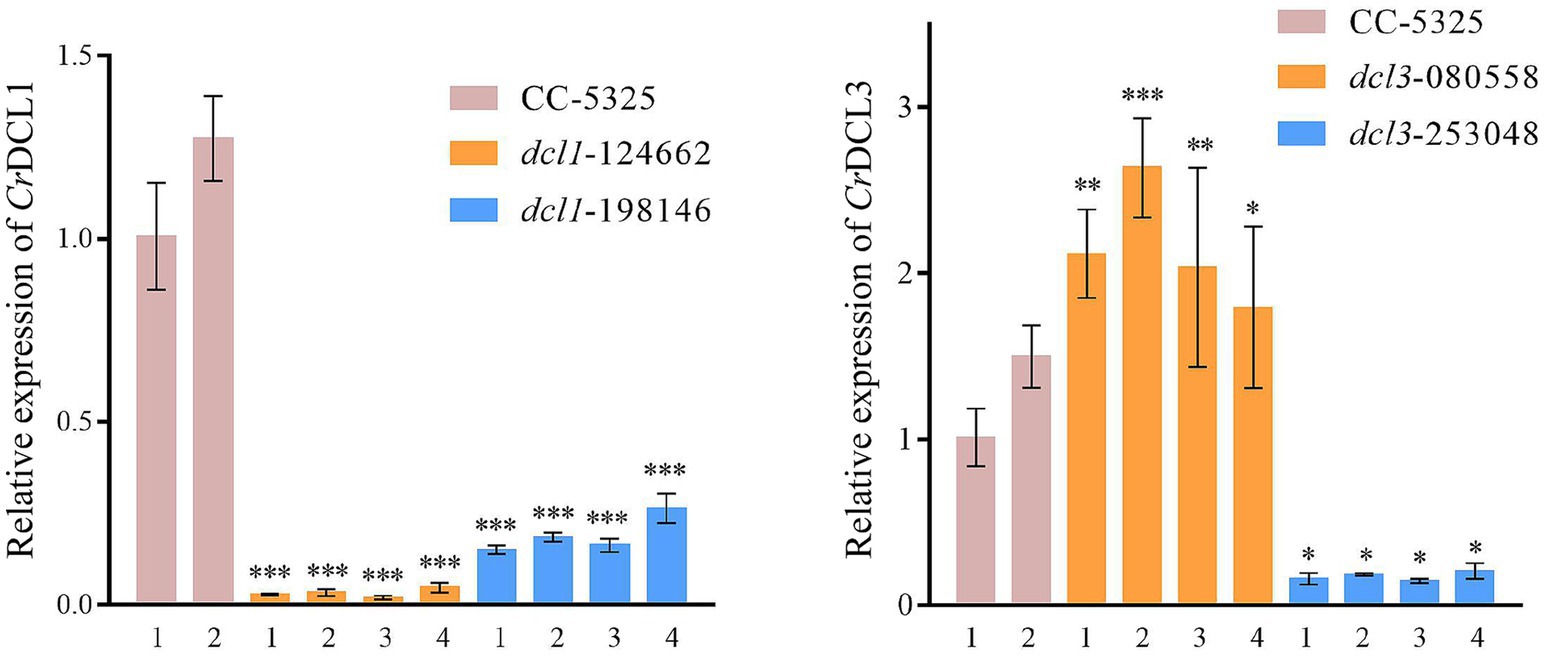
Figure 2. RT-qPCR analysis of CrDCL1 and CrDCL3 transcript levels in the mutants and control. ACTIN was used as an internal control for normalization. Bars indicate the standard error of the means (n = 3). *, **, and *** indicates the statistical significance between two means at the level of 0.05, 0.01, and 0.001, respectively.
Moreover, the expression of CrDCL1 gene in dcl3 mutant was no reduced, whereas CrDCL3 transcripts were significantly increased in dcl1 mutant (Supplementary Figure S1). This suggests that some functions of the CrDCL1 protein overlap with those of the CrDCL3 protein. In the dcl1 mutant, the loss of CrDCL1 protein function may lead to compensatory upregulation of CrDCL3 protein expression to perform related tasks.
The expression levels of previously reported miRNAs were analyzed by Northern blot in the dcl1 and dcl3 mutants. miR1162, miR1151b and miRB were undetectable in dcl3 (Supplementary Figure S2), indicating that CrDCL3 protein mediates the biogenesis of these three miRNAs. This conclusion aligns with previous report (Valli et al., 2016). Notably, density analysis of the Northern blot bands revealed a slight downregulation of miR1162 and miR1151b in dcl1 mutant relative to CC-5325, suggesting that CrDCL1 protein may also play a role in the production of some miRNAs.
To investigate whether CrDCL1 is involved in the biogenesis of some miRNAs in C. reinhardtii, we performed sRNA-seq for CC-5325 and dcl1 mutant with three biological replicates. The sRNA characteristics were identical in the dcl1 mutant and CC-5325, as previously reported (Zhao et al., 2007; Valli et al., 2016). The sRNAs were 16–28 nt long with a normal distribution, and 30–40% of the sRNAs were 21 nt long (Figure 3A).
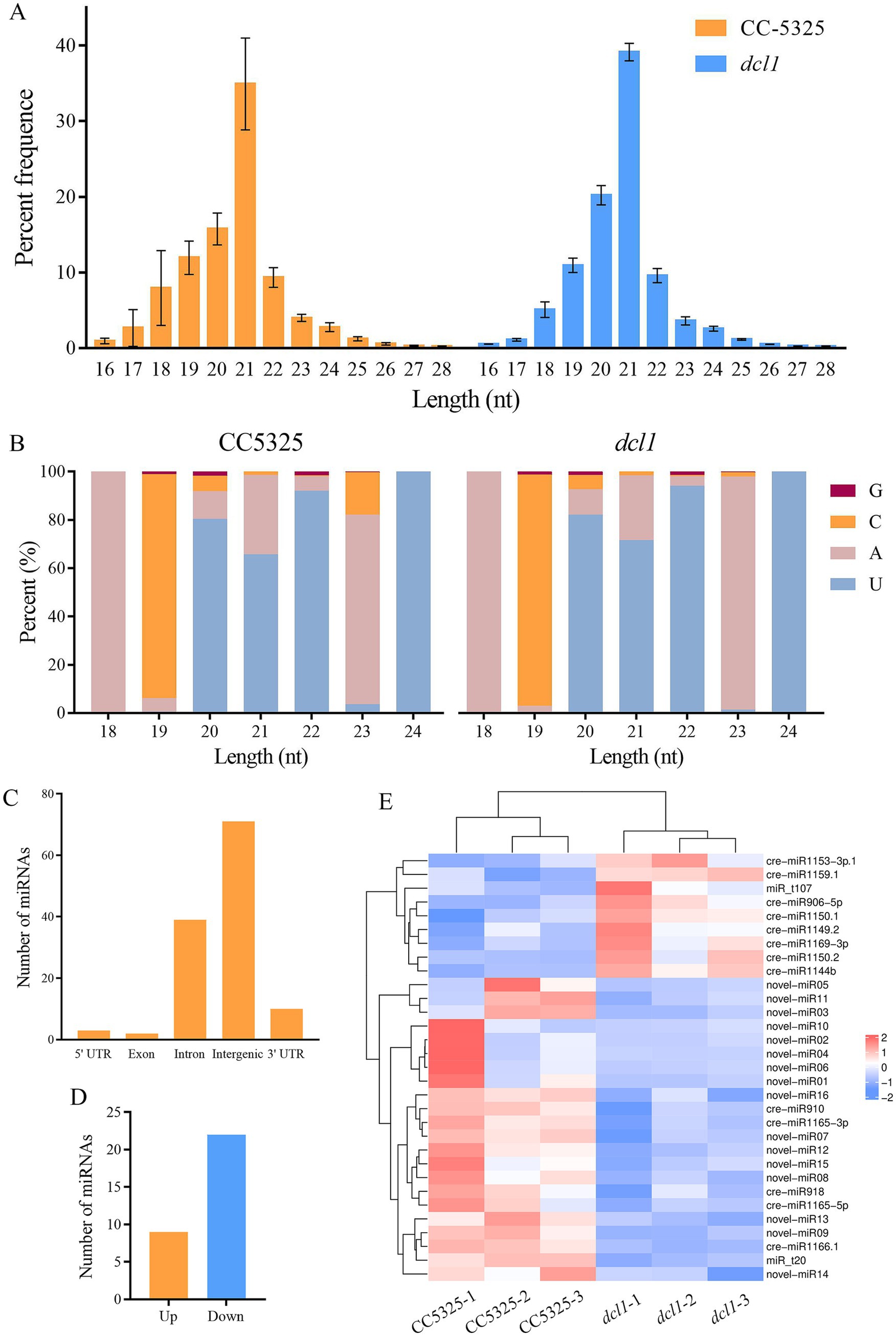
Figure 3. Characteristics of C. reinhardtii sRNAs from CC-5325 and the dcl1 mutant. (A) Length distribution of sRNAs. (B) Nucleotide frequency at the 5′ end of all identified miRNAs. (C) Genome locations of all identified miRNAs. (D) The number of miRNAs with significantly up-regulated or down-regulated expression levels in dcl1 compared to CC-5325. (E) Expression heat map of the differentially expressed miRNAs.
By comparing the sequences in the miRBase database and previous reports (Shu and Hu, 2012; Valli et al., 2016; Yamasaki and Cerutti, 2017; Voshall et al., 2017; Zhang et al., 2021), 117 previously reported cre-miRNAs were obtained. Among them, 19 cre-miRNAs only record mature sequences and precursor sequences in the miRBase database, lacking information on their chromosomal location. The precursors of these 19 cre-miRNAs were aligned with the updated Chlamydomonas genome (v5.6) to obtain their chromosomal location information, but the match failed. Considering them as novel miRNAs, we further predicted the precursors of these 19 unmatched miRNAs. Results were as follows: 9 miRNAs had no corresponding precursor sequences in C. reinhardtii, 7 miRNAs had a corresponding precursor, and the predicted precursor sequences of the remaining 3 miRNAs showed that their mature sequences completely complement their miRNA* (Table 1; Supplementary Figure S3). The miRNAs were mostly 20–22 nt in length with an obvious peak at 21 nt, and there was a bias toward U as the first nucleotide among the 20–22 nt miRNAs (Figure 3B). Analysis of the genomic location of these miRNAs showed that most of them derived from intergenic regions (Figure 3C).
To identify miRNAs associated with CrDCL1 from the sequencing data, we compared the miRNA expression levels in CC-5325 and dcl1 mutant. The analysis identified 9 miRNAs with significantly increased levels in dcl1 mutant [log2(fc) ≥ log21.5, p < 0.05] and 22 miRNAs with significantly decreased levels [log2(fc) ≤ −log21.5, p < 0.05], in comparison with CC-5325 (Figures 3D,E), suggesting that CrDCL1 may be involved in the biogenesis of these 22 down-regulated miRNAs. Based on their expression fold change, 14 of these 22 miRNAs were classified as “high-differential miRNAs” [log2(fc) ≤ −1], and the remaining 8 were referred to as “medium-differential miRNAs” [−1 ≤ log2(fc) ≤ −log21.5] (Table 2). These 14 high-differential miRNAs included one previously reported cre-miRNA (cre-miR1166.1) and 13 novel miRNAs. Among them, novel-miR02 had 3 precursors. The 8 medium-differential miRNAs included 4 previously reported cre-miRNAs and 4 novel miRNAs. Among them, the precursors of cre-miR1165-5p and cre-miR1165-3p were predicted again in this study (Table 1). To further investigate the potential involvement of CrDCL1 in the biogenesis of these miRNAs, we examined the expression levels of 10 high-differential miRNAs using RT-qPCR. The results showed that the expression levels of these 10 high-differential miRNAs were significantly down-regulated in dcl1 compared with CC-5325 (Figure 4). These results were consistent with the sRNA-seq data, which not only served as evidence for the accuracy of sRNA-seq data, but also proved that CrDCL1 protein affected the production of these 10 high-differential miRNAs. We also detected the expression levels of these miRNAs in the dcl3 mutant. Interestingly, the abundances of 5 novel miRNAs (novel-miR01, novel-miR03, novel-miR04, novel-miR05, and novel-miR06) in the dcl3 mutant were lower than that in CC-5325 but higher than that in dcl1, whereas three novel miRNAs (novel-miR07, novel-miR09, and novel-miR12) were up-regulated in dcl3 mutant relative to CC-5325 (Figure 4), indicating that the production of some miRNAs was mediated by CrDCL1 and CrDCL3 proteins.
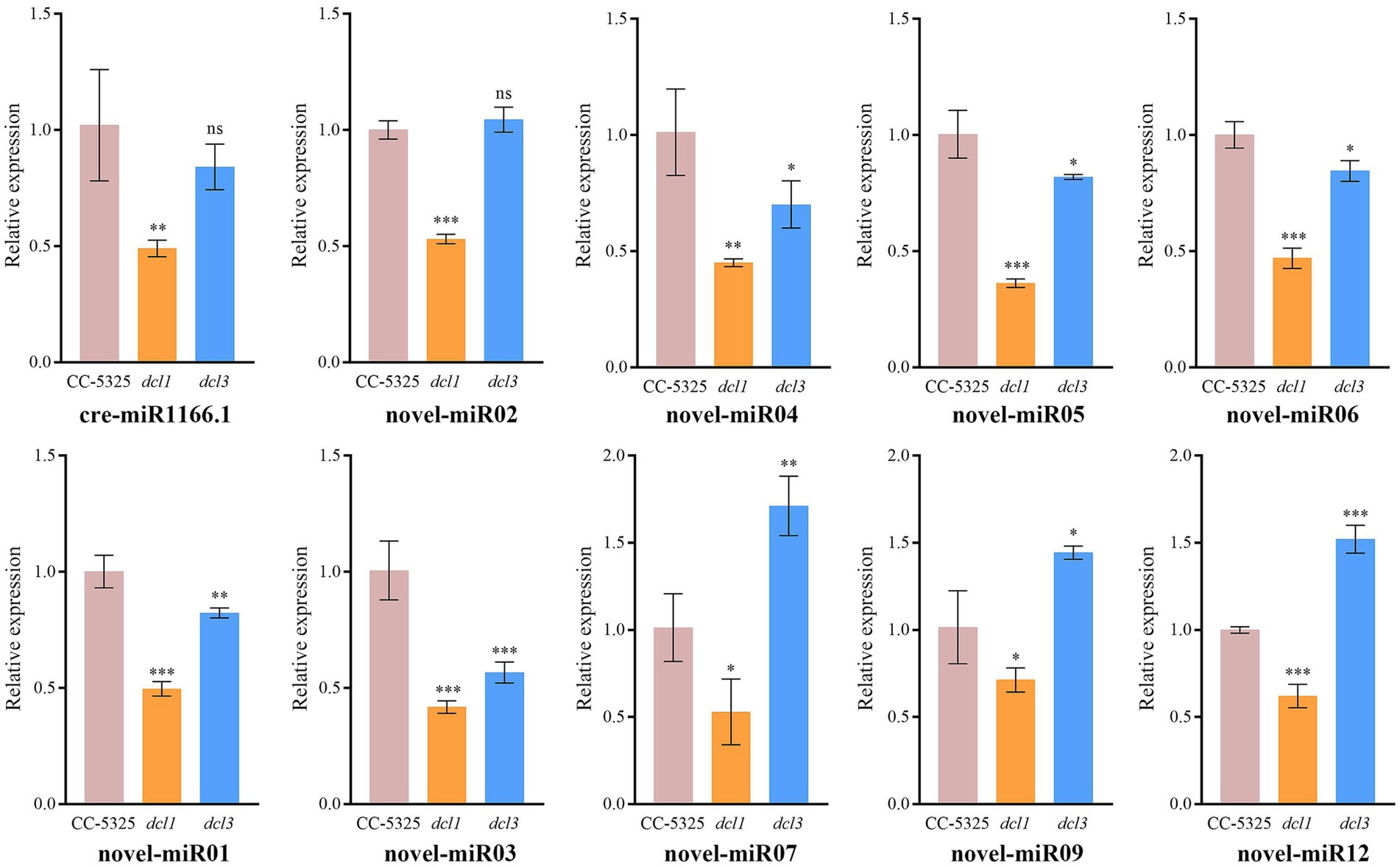
Figure 4. RT-qPCR analysis of 10 differentially expressed miRNAs in CC-5325, dcl1 and dcl3. The U4 gene was used for normalization. Error bars show the standard error from three technical replicates. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
We also performed Northern blot analysis to further validate the expression levels of the high-differential miRNAs in the mutants and control. However, due to their extremely low expression levels, only one of these high-differential miRNAs (novel-miR04) was detectable by Northern blot. Consisted with the RT-qPCR result, the abundance of novel-miR04 was slightly reduced in dcl3 and significantly reduced in dcl1 compared to the control (Supplementary Figure S4). Thereby, it strongly proposed that CrDCL1 is involved in the biogenesis of novel-miR04. We also analyzed the expression levels of cre-miR910, a confirmed cre-miRNA. Northern blot analysis indicated that cre-miR910 was significantly down-regulated in dcl1 and reduced to an undetectable level in dcl3 (Supplementary Figure S4). These results suggest that CrDCL1 and CrDCL3 are both involved in the biogenesis of some miRNAs (e.g., novel-miR04 and cre-miR910).
Finally, we performed target gene prediction and functional analysis for the 22 down-regulated miRNAs. A total of 384 target genes were obtained, 105 of which were target genes of previously reported cre-miRNAs. GO analysis was performed for all target genes, and the results indicated their involvement in various cellular components and molecular functions such as catalytic activity and binding. These target genes also have important roles in many biological processes including cellular processes, metabolic processes and single-organism processes (Figure 5A). The following cellular component terms were significantly enriched in the dataset, with 26 target genes associated with AP-type membrane coat adaptor complex, membrane coat, coated membrane, protein-DNA complex and cell projection. Forty-nine target genes were associated with lyase activity, transferase activity, oxidoreductase activity, catalytic activity, ion transmembrane transporter activity and monooxygenase activity (Figure 5B). KEGG pathway enrichment analysis was also performed. Among 40 target genes with functional descriptions associated with metabolic and signal transduction pathways, there was a significant enrichment of genes associated with fatty acid metabolism and the biosynthesis of unsaturated fatty acids (Figure 5C).
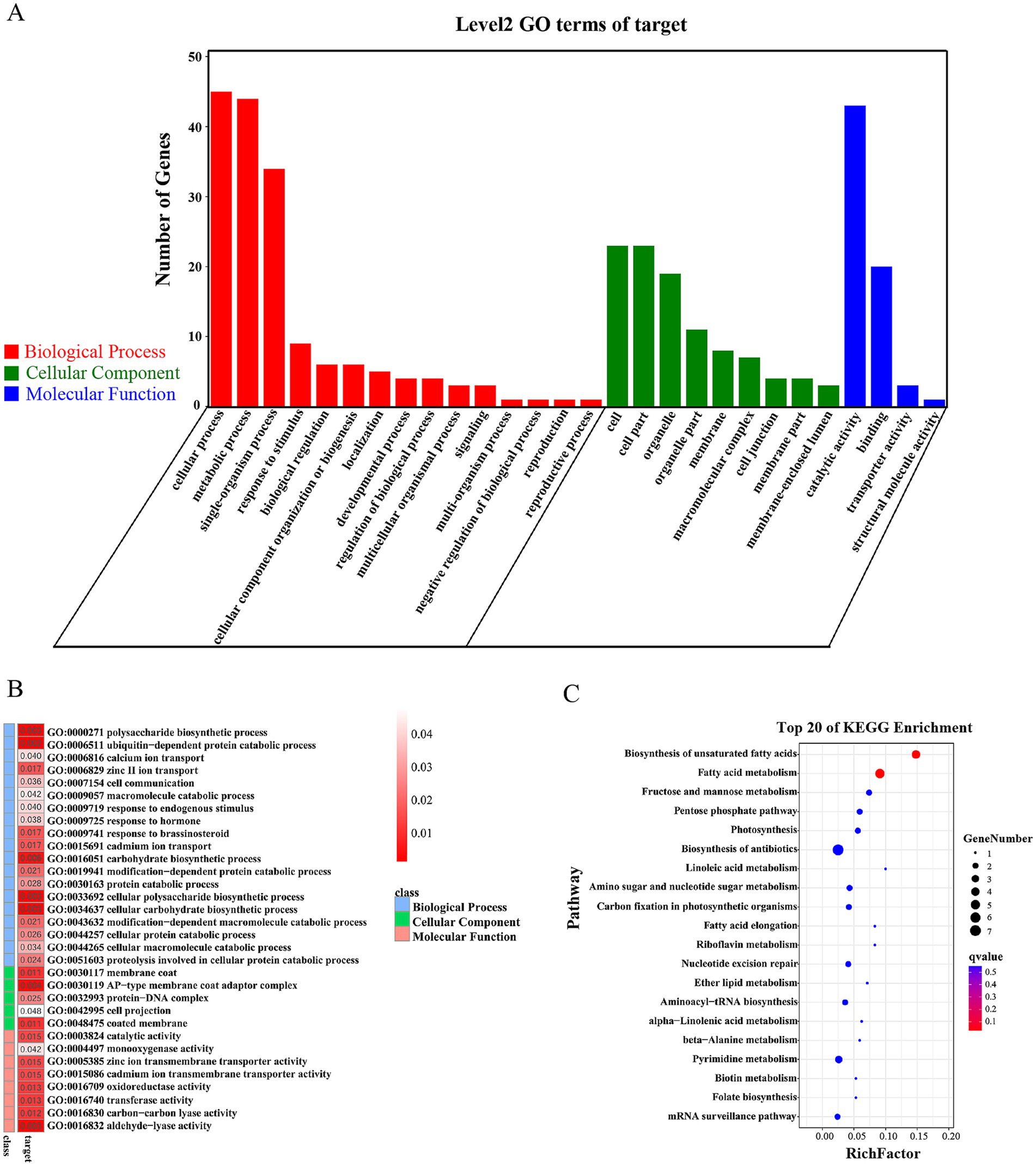
Figure 5. Functional analysis of the predicted targets of CrDCL1-related miRNAs. (A) Categorization of miRNA target genes by GO analysis. (B) Analysis of the miRNA target genes revealed a significant enrichment of various molecular function, cellular component and biological process terms. (C) KEGG pathway enrichment analysis of the predicted miRNA targets.
Analysis of genes and miRNAs related to the fatty acid metabolic pathway revealed that the Cre03.g213313, Cre04.g217945, and Cre01.g035400 genes had significant differences in the fatty acid metabolic pathway. Among them, Cre03.g213313 and Cre04.g217945 were involved in the unsaturated fatty acid biosynthesis pathway (Table 3). Target gene prediction revealed that Cre03.g213313, functioning as 3-oxoacyl-(fabG) is the target gene of novel-miR16; Cre04.g217945, functioning as a fatty acid desaturase (such as SCD, desC, stearoyl-CoA desaturase) is the target gene of novel-miR07; and Cre01.g035400, functioning as an E3 ubiquitin ligase (MECR, NRBF1; mitochondrial trans-2-enoyl-CoA reductase), is the target gene of novel-miR02 (Table 3).
In 2007, both Zhao et al. and Molnár et al. validated the miRNAs in C. reinhardtii identified in their studies, by alignment with the Chlamydomonas genome (v3.0) (Zhao et al., 2007; Molnár et al., 2007). Currently, 137 cre-miRNAs are listed in the miRBase database, most of which derived from their studies (Zhao et al., 2007; Molnár et al., 2007). According to several recent studies, an updated version of the Chlamydomonas genome was available (v5.6). Based on this updated v5.6 version, we found that 19 cre-miRNA precursors from miRBase were incorrect, and we re-predicted the precursors of these miRNAs. Our analysis indicates that 9 of these cre-miRNAs without precursors should be classified as non-miRNAs, 3 of these cre-miRNAs were completely complementary to their miRNA* and should be classified as siRNA, and the remaining 7 cre-miRNAs with a precursor are miRNA candidates.
Current studies have shown that fungi and most animals have only a single Dicer protein (Zhang et al., 2004; Drinnenberg et al., 2009; Makino et al., 2015), a few invertebrates have two Dicer proteins (Gao et al., 2014). Plants have multiple Dicer-like proteins (DCL) (Yu et al., 2017), and C. reinhardtii encodes three DCL proteins (CrDCL1-3) (Lou et al., 2018). We performed phylogenetic analysis using Dicer/DCL proteins from multiple species and found that the CrDCLs proteins formed an independently branch, and lacked PAZ and dsRNA-binding domains like RtDicer and SpDicer. In plants and animals, the PAZ and dsRNA-binding domains of Dicer/DCL proteins primarily influence their binding affinity for dsRNA. Moreover, the PAZ domain recognizes the 3′ end of pre-miRNA, serving as the starting point for Dicer/DCL cleavage, while the two RNase III domains function as a molecular ruler, enabling precise cutting of small RNAs into lengths of 21–25 nucleotides (Zhang et al., 2004; Macrae et al., 2006; Song and Rossi, 2017). Therefore, we speculate that Dicer/DCL proteins of animals and plants evolved from ancestral forms lacking PAZ and dsRNA-binding domains (such as CrDCL and RtDicer), leading to more accurate and specific RNA binding.
Previous studies have shown that the feature of the DUF283 domain of AtDCL4 and human Dicer protein are similar to that of the dsRNA-binding domain, and AtDCL4 DUF283 domain has a weak binding ability to dsRNA (Qin et al., 2010; Liu et al., 2018); human Dicer DUF283 domain can bind single-stranded nucleic acids in vitro (Kurzynska-Kokorniak et al., 2016). Our laboratory has also preliminarily proved that the DUF283 domain of the CrDCL protein can bind to dsRNA in vitro, through fluorescence anisotropy experiments (unpublished). On the other hand, DUF283 domain can recruit other dsRNA-binding domain proteins and participate in the binding of Dicer partner proteins (Dlakić, 2006). Therefore, we speculate that the DUF283 domain of the CrDCL protein may replace or recruit other proteins to replace partial functions of the PAZ and dsRNA-binding domains. Besides, previous studies have reconstructed the 3D structure of HsDicer protein, showing the overall L-shaped structure (Lau et al., 2009; Paturi and Deshmukh, 2021). The three-dimensional structure of the CrDCL1 protein predicted in this study is similar to that of the HsDicer protein. The distribution of various domains in CrDCL1 and HsDicer are similar. However, CrDCL1 has numerous irregular curls, whether they can replace the function of the PAZ and dsRNA-binding domain is still unclear and further verification needs to be conducted.
Since the coding sequences of CrDCL1 and CrDCL3 proteins are too long and have high GC content, it is a big challenge for full gene cloning. No signal peptide was predicted in CrDCL1 and CrDCL3, but the DEAD domains was presented and involved in various aspects of RNA metabolism, such as nuclear transcription, pre-mRNA splicing and nucleocytoplasmic transport (Schutz et al., 2010; Paysan-Lafosse et al., 2023). Therefore, we performed subcellular localization analysis using the 5′ partial (including the DEAD domain) of CrDCL1 and CrDCL3 proteins and found that they were both localized in the nucleus of onions (unpublished), which is consistent with the localization of CrDUS16 protein. It is preliminarily believed that, like plants, CrDCL proteins perform cleavage in the nucleus.
CrDCL3 is one of the three CrDCL proteins (CrDCL1-3) encoded in the C. reinhardtii genome, and it is mainly involved in miRNA biogenesis (Valli et al., 2016). CrDCL1 mediates siRNA biogenesis (Casas-Mollano et al., 2008), but it was not previously reported associating with miRNA biogenesis. CrDCL2 is not well characterized. In our analysis, most of the analyzed miRNAs were not detected in the dcl3 mutant by Northern blot. In addition, 9 miRNAs were detected to be significantly up-regulated in the dcl1 mutant by sRNA-seq. Combined with the result of up-regulated CrDCL3 protein expression in the dcl1 mutant, it can be speculated that the production of these 9 miRNAs may be affected by CrDCL3 protein rather than CrDCL1. This finding aligns with the established role of CrDCL3 in miRNA biogenesis. Interestingly, sRNA-seq identified 22 miRNAs significantly down-regulated in the dcl1 mutant compared to the control. Validation of the sRNA-seq data by RT-qPCR and Northern blot provided further evidence showing that CrDCL1 is involved in the biogenesis of these 22 miRNAs.
In animals, only one Dicer protein is expressed, and miRNAs are generated by two cleavage steps performed by Drosha and Dicer, respectively (Zhao et al., 2007). In plants, multiple DCL proteins are presented, but only one DCL protein involved in miRNA biogenesis. For instance, most miRNAs in Arabidopsis are generated only depend on AtDCL1 cleavage, while miR822, miR839 and miR859 only depend on AtDCL4 rather than AtDCL1 (Kurihara and Watanabe, 2004; Rajagopalan et al., 2006; Yu et al., 2017). Noticeably, 22 CrDCL1-related miRNAs (down-regulated miRNAs in dcl1 relative to the control) were still partially expressed in the dcl1 mutant. The RT-qPCR and Northern blot results of this study showed that cre-miR910, novel-miR01, novel-miR03, novel-miR04, novel-miR05 and novel-miR06 were down-regulated at different degrees in the dcl1 and dcl3 mutant, suggesting that along with CrDCL1, CrDCL3 plays a key role in the biogenesis of these miRNAs. It is concluded the biogenesis of miRNAs in C. reinhardtii may be performed by multiple CrDCL proteins, which is different from animals and plants. Nevertheless, the mechanisms of that remained to be explored.
Additionally, among the 10 high-differential miRNAs were detected by RT-qPCR, only 5 miRNAs were down-regulated in dcl3 mutant, and the down-regulation amplitude was smaller than that in the dcl1 mutant; compared with the control, the expression of CrDCL1 gene showed no difference in dcl3 mutants, whereas the CrDCL3 gene were significantly increased in dcl1 mutants. These results suggested that the production of these high-differential miRNAs mainly relied on the regulation of CrDCL0031 protein, but the function of CrDCL1 on the biogenesis of some miRNAs could be replaced by CrDCL3 in dcl1 mutants, thereby ensuring that algal cells can continue to produce these miRNAs. Whether this type of miRNA has important functions such as maintaining normal cell growth remains to be explored. Cre-miR910, as a medium-difference miRNA, was significantly down-regulated in dcl1 and undetectable in dcl3 compared to control. It is speculated that the medium-differential miRNAs were mainly relied on the cleavage of CrDCL3 protein, but this speculation still needs more verification.
Finally, we performed preliminarily prediction and analysis of the biological metabolic pathways potentially mediated by CrDCL1 related miRNAs. This study aims to provide a reference for further research on the mechanisms and biotechnological applications of CrDCL1 protein and its related miRNA-mediated metabolic pathways. GO enrichment analysis indicated that the predicted target genes of these 22 miRNAs are associated with various molecular functions, cellular components and biological processes. Pathway enrichment analysis of their predicted target genes showed a significant enrichment of genes involved infatty acid metabolism. However, further analysis of significantly enriched target genes revealed only three miRNAs (novel-miR16, novel-miR07, novel-miR02) were associated with fatty acid metabolism. Notably, the predicted target gene of novel-miR02 is MECR. Previous study has demonstrated that the expression of the MECR gene in C. reinhardtii can enhance the production of total lipids and astaxanthin heterologous content (Sun et al., 2023). Our RT-qPCR experiment indicated that the expression of novel-miR02 primarily depends on CrDCL1 protein but not on CrDCL3 protein. We conducted fatty acid content detection in dcl1, dcl3 mutants as well as the control strain CC-5325 using the method described by Jia et al. (2019). By comparing with standards, 11 credible peaks were identified. The relative fatty acids contents were obtained using peak area normalization, and the average values from two biological replicates were calculated to obtain the data presented in Supplementary Table S5. Results showed that the contents of 9,12,15-octadecatrienoic acid, methyl ester, (Z,Z,Z-) and methyl 4,7,10,13-hexadecatetraenoate in dcl1 were slightly altered compared with CC-5325 (Supplementary Table S5). Therefore, whether novel-miR16, novel-miR07 and novel-miR02 are involved in the regulation of fatty acid metabolism, and whether the functional loss of CrDCL1 protein leading to the downregulation of novel-miR02 increases MECR gene expression, thereby affecting total lipids and astaxanthin heterologous content, requires further verified.
In summary, the evolutionary trends and structural predictions of Dicer/DCL proteins across various species indicated that DEAD, Helicase C-like, DUF283, and two RNase III domains are conserved domains in Dicer/DCL proteins. In contrast, the PAZ and dsRNA-binding domains, absent in CrDCL proteins, may have evolved later, contributing to the increased precision and complexity of small RNA generation in plants and animals. Moreover, based on the updated Chlamydomonas genome (v5.6), we excluded 12 miRNAs and re-predicted the precursors of 7 cre-miRNAs in miRBase. By analyzing miRNA expression levels, we demonstrated that CrDCL1 is involved in the biogenesis of 22 miRNAs. Notably, the production of cre-miR910, novel-miR01, novel-miR03, novel-miR04, novel-miR05 and novel-miR06 were regulated not only by CrDCL1 but also by CrDCL3. These findings highlight that the production of some C. reinhardtii miRNAs may involve multiple CrDCL proteins, differing from pathways observed in animals and plants. Furthermore, we performed target gene prediction analysis on 22 miRNAs and found that novel-miR02 may target the MECR gene, which is involved in the regulation of lipids and astaxanthin heterologous production. Despite these insights, there are still have many unresolved questions regarding the function of CrDCL proteins and the biogenesis of miRNAs in C. reinhardtii, necessitating further investigation.
Small RNA-seq datasets generated during this study have been submitted to the Genome Sequence Archive (GSA; https://ngdc.cncb.ac.cn/gsa/) under the accession number CRA023114.
TS: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. MT: Data curation, Formal analysis, Validation, Writing – review & editing. QD: Validation, Writing – original draft. ZH: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing. HL: Conceptualization, Methodology, Resources, Supervision, Writing – review & editing. SL: Methodology, Resources, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Natural Science Foundation of China (32273118, 32303034), China National Key Research and Development Project (2023YFF1001200, 2018YFA0902500), Guangxi Major Program for Science and Technology(GuikeAA24263042), Shenzhen Special Fund for Sustainable Development (KCXFZ20211020164013021), Guangdong Province Key Research and Development Project (2022B1111070005), The Project of DEGP (2023ZDZX4047), The Engineering Research Center Support Program from Development and Reform Commission of Shenzhen Municipality (XMHT20220104019) and Shenzhen University 2035 Program for Excellent Research (2022B010).
The authors gratefully acknowledge the supports from Instrumental Analysis Center of Shenzhen University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1487584/full#supplementary-material
Supplementary Figure S1 | RT-qPCR analysis of CrDCL1 and CrDCL3 genes in CC-5325, dcl1 and dcl3. ACTIN was used as an internal control for normalization. Bars indicate the standard error of the means (n = 3). ns, not significant; *p < 0.05; **p < 0.01.
Supplementary Figure S2 | Northern blot detection of miR1162, miR1151-b and miRB in the mutants and control. U4 was used as an internal control.
Supplementary Figure S3 | Predicted precursor structures of cre-miR1159.2, cre-miR1148.2, and cre-miR907.
Supplementary Figure S4 | Northern blot detection of novel-miR04 and cre-miR910 in the mutants and control. U4 gene was used as an internal control.
Supplementary Table S1 | PCR primers for the CrDCL genes.
Supplementary Table S2 | Specific stem-loop primers for reverse transcription.
Supplementary Table S3 | RT-qPCR primers for miRNAs, CrDCLs, ACTIN and U4.
Supplementary Table S4 | Northern blot probes for miRNAs and the U4 gene.
Supplementary Table S5 | Determination of the fatty acid content.
1. ^https://www.chlamylibrary.org/index
2. ^https://www.chlamylibrary.org/showCassette?cassette=CIB1
3. ^https://phytozome.jgi.doe.gov/pz/portal.html#
4. ^https://www.ncbi.nlm.nih.gov/
5. ^https://itol.embl.de/login.cgi?logout=1
7. ^http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Ben, A. B., Wirth, S., Merchan, F., Laporte, P., d’Aubenton-Carafa, Y., Hirsch, J., et al. (2009). Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 19, 57–69. doi: 10.1101/gr.080275.108
Casas-Mollano, J. A., Rohr, J., Kim, E. J., Balassa, E., Dijk, K., and Cerutti, H. (2008). Diversification of the core RNA interference machinery in Chlamydomonas reinhardtii and the role of DCL1 in transposon silencing. Genetics 179, 69–81. doi: 10.1534/genetics.107.086546
Ding, X., Guo, J., Zhang, Q., Yu, L., Zhao, T., and Yang, S. (2021). Heat-responsive miRNAs participate in the regulation of male fertility stability in soybean CMS-based F1 under high temperature stress. Int. J. Mol. Sci. 22:2446. doi: 10.3390/ijms22052446
Dlakić, M. (2006). DUF283 domain of dicer proteins has a double-stranded RNA-binding fold. Bioinformatics 22, 2711–2714. doi: 10.1093/bioinformatics/btl468
Drinnenberg, I. A., Weinberg, D. E., Xie, K. T., Mower, J. P., Wolfe, K. H., Fink, G. R., et al. (2009). RNAi in budding yeast. Science 326, 544–550. doi: 10.1126/science.1176945
Eamens, A. L., Smith, N. A., Curtin, S. J., Wang, M. B., and Waterhouse, P. M. (2009). The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15, 2219–2235. doi: 10.1261/rna.1646909
Gao, Z., Wang, M., Blair, D., Zheng, Y., and Dou, Y. (2014). Phylogenetic analysis of the endoribonuclease dicer family. PLoS One 9:e95350. doi: 10.1371/journal.pone.0095350
Ghildiyal, M., and Zamore, P. D. (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10, 94–108. doi: 10.1038/nrg2504
Han, J. J., Lee, Y., Yeom, K. H., Kim, Y. K., Jin, H., and Kim, V. N. (2004). The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 18, 3016–3027. doi: 10.1101/gad.1262504
Iwasaki, Y. W., Siomi, M. C., and Siomi, H. (2015). PIWI-interacting RNA: its biogenesis and functions. Annu. Rev. Biochem. 84, 405–433. doi: 10.1146/annurev-biochem-060614-034258
Jia, B., Xie, X., Wu, M., Lin, Z., Yin, J., lou, S., et al. (2019). Understanding the functions of endogenous DOF transcript factor in Chlamydomonas reinhardtii. Biotechnol. Biofuels 12:67. doi: 10.1186/s13068-019-1403-1
Kang, Y., Yang, X., Liu, Y., Shi, M., Zhang, W., Fan, Y., et al. (2021). Integration of mRNA and miRNA analysis reveals the molecular mechanism of potato (Solanum tuberosum L.) response to alkali stress. Int. J. Biol. Macromol. 182, 938–949. doi: 10.1016/j.ijbiomac.2021.04.094
Kurihara, Y., Takashi, Y., and Watanabe, Y. (2006). The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12, 206–212. doi: 10.1261/rna.2146906
Kurihara, Y., and Watanabe, Y. (2004). Arabidopsis micro-RNA biogenesis through dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 101, 12753–12758. doi: 10.1073/pnas.0403115101
Kurzynska-Kokorniak, A., Pokornowska, M., Koralewska, N., Hoffmann, W., Bienkowska-Szewczyk, K., and Figlerowicz, M. (2016). Revealing a new activity of the human dicer DUF283 domain in vitro. Sci. Rep. 6:23989. doi: 10.1038/srep23989
Lau, P., Potter, C. S., Carragher, B., and Macrae, I. J. (2009). Structure of the human dicer-TRBP complex by electron microscopy. Structure 17, 1326–1332. doi: 10.1016/j.str.2009.08.013
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. doi: 10.1016/0092-8674(93)90529-y
Li, X., Zhang, R., Patena, W., Gang, S. S., Blum, S. R., Ivanova, N., et al. (2016). An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell 28, 367–387. doi: 10.1105/tpc.15.00465
Liu, Z., Wang, J., Cheng, H., Ke, X., Sun, L., Zhang, Q. C., et al. (2018). Cryo-EM structure of human dicer and its complexes with a pre-miRNA substrate. Cell 173, 1549–1550. doi: 10.1016/j.cell.2018.05.031
Lou, S., Sun, T., Li, H., and Hu, Z. (2018). Mechanisms of microRNA-mediated gene regulation in unicellular model alga Chlamydomonas reinhardtii. Biotechnol. Biofuels 11:244. doi: 10.1186/s13068-018-1249-y
Lund, E., Guttinger, S., Calado, A., Dahlberg, J. E., and Kutay, U. (2004). Nuclear export of microRNA precursors. Science 303, 95–98. doi: 10.1126/science.1090599
Macrae, I. J., Zhou, K., Li, F., Repic, A., Brooks, A. N., Cande, W. Z., et al. (2006). Structural basis for double-stranded RNA processing by dicer. Science 311, 195–198. doi: 10.1126/science.1121638
Makino, S., Mishima, Y., Inoue, K., and Inada, T. (2015). Roles of mRNA fate modulators Dhh1 and Pat1 in TNRC6-dependent gene silencing recapitulated in yeast. J. Biol. Chem. 290, 8331–8347. doi: 10.1074/jbc.M114.615088
Martinho, C., and Lopez-Gomollon, S. (2023). “Detection of microRNAs by northern blot,” in MicroRNA detection and target identification: Methods and protocols. (New York, NY: Springer US), 47–66.
Matranga, C., and Zamore, P. D. (2007). Small silencing RNAs. Curr. Biol. 17, R789–R793. doi: 10.1016/j.cub.2007.07.014
Meister, G. (2013). Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 14, 447–459. doi: 10.1038/nrg3462
Merchant, S. S., Prochnik, S. E., Vallon, O., Harris, E. H., Karpowicz, S. J., Witman, G. B., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250. doi: 10.1126/science.1143609
Molnár, A., Schwach, F., Studholme, D. J., Thuenemann, E. C., and Baulcombe, D. C. (2007). miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature 447, 1126–1129. doi: 10.1038/nature05903
Paturi, S., and Deshmukh, M. V. (2021). A glimpse of “dicer biology” through the structural and functional perspective. Front. Mol. Biosci. 8:643657. doi: 10.3389/fmolb.2021.643657
Paysan-Lafosse, T., Blum, M., Chuguransky, S., Grego, T., Pinto, B. L., Salazar, G. A., et al. (2023). InterPro in 2022. Nucleic Acids Res. 51, D418–D427. doi: 10.1093/nar/gkac993
Qin, H., Chen, F., Huan, X., Machida, S., Song, J., and Yuan, Y. A. (2010). Structure of the Arabidopsis thaliana DCL4 DUF283 domain reveals a noncanonical double-stranded RNA-binding fold for protein-protein interaction. RNA 16, 474–481. doi: 10.1261/rna.1965310
Rajagopalan, R., Vaucheret, H., Trejo, J., and Bartel, D. P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20, 3407–3425. doi: 10.1101/gad.1476406
Schutz, P., Karlberg, T., van den Berg, S., Collins, R., Lehtiö, L., Högbom, M., et al. (2010). Comparative structural analysis of human DEAD-box RNA helicases. PLoS One 5:e12791. doi: 10.1371/journal.pone.0012791
Shu, L., and Hu, Z. (2012). Characterization and differential expression of microRNAs elicited by sulfur deprivation in Chlamydomonas reinhardtii. BMC Genomics 13:108. doi: 10.1186/1471-2164-13-108
Song, M. S., and Rossi, J. J. (2017). Molecular mechanisms of dicer: endonuclease and enzymatic activity. Biochem. J. 474, 1603–1618. doi: 10.1042/BCJ20160759
Sun, J. P., Wei, X. H., Cong, X. M., Zhang, W. H., Qiu, L. X., and Zang, X. N. (2023). Expression of fatty acid related gene promotes astaxanthin heterologous production in Chlamydomonas reinhardtii. Front. Nutr. 10:1130065. doi: 10.3389/fnut.2023.1130065
Tsuzuki, M., Takeda, A., and Watanabe, Y. (2014). Recovery of dicer-like 1-late flowering phenotype by miR172 expressed by the noncanonical DCL4-dependent biogenesis pathway. RNA 20, 1320–1327. doi: 10.1261/rna.044966.114
Valli, A. A., Santos, B. A. C. M., Hnatova, S., Bassett, A. R., Molnar, A., Chung, B. Y., et al. (2016). Most microRNAs in the single-cell alga Chlamydomonas reinhardtii are produced by dicer-like 3-mediated cleavage of introns and untranslated regions of coding RNAs. Genome Res. 26, 519–529. doi: 10.1101/gr.199703.115
Voshall, A., Kim, E. J., Ma, X., Yamasaki, T., Moriyama, E. N., and Cerutti, H. (2017). miRNAs in the alga Chlamydomonas reinhardtii are not phylogenetically conserved and play a limited role in responses to nutrient deprivation. Sci. Rep. 7:5462. doi: 10.1038/s41598-017-05561-0
Wang, Y., Jiang, X., Hu, C., Sun, T., Zeng, Z., Cai, X., et al. (2017). Optogenetic regulation of artificial microRNA improves H2 production in green alga Chlamydomonas reinhardtii. Biotechnol. Biofuels 10:257. doi: 10.1186/s13068-017-0941-7
Yamasaki, T., and Cerutti, H. (2017). Cooperative processing of primary miRNAs by DUS16 and DCL3 in the unicellular green alga Chlamydomonas reinhardtii. Commun. Integr. Biol. 10:e1280208. doi: 10.1080/19420889.2017.1280208
Yamasaki, T., Onishi, M., Kim, E. J., Cerutti, H., and Ohama, T. (2016). RNA-binding protein DUS16 plays an essential role in primary miRNA processing in the unicellular alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 113, 10720–10725. doi: 10.1073/pnas.1523230113
Yi, R., Qin, Y., Macara, I. G., and Cullen, B. R. (2003). Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17, 3011–3016. doi: 10.1101/gad.1158803
Yu, Y., Jia, T., and Chen, X. (2017). The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 216, 1002–1017. doi: 10.1111/nph.14834
Zhang, H., Kolb, F. A., Jaskiewicz, L., Westhof, E., and Filipowicz, W. (2004). Single processing center models for human dicer and bacterial RNase III. Cell 118, 57–68. doi: 10.1016/j.cell.2004.06.017
Zhang, R., Patena, W., Armbruster, U., Gang, S. S., Blum, S. R., and Jonikas, M. C. (2014). High-throughput genotyping of green algal mutants reveals random distribution of mutagenic insertion sites and endonucleolytic cleavage of transforming DNA. Plant Cell 26, 1398–1409. doi: 10.1105/tpc.114.124099
Zhang, J., Shi, J., Yuan, C., Liu, X., Du, G., Fan, R., et al. (2021). MicroRNA expression profile analysis of Chlamydomonas reinhardtii during lipid accumulation process under nitrogen deprivation stresses. Bioengineering 9:6. doi: 10.3390/bioengineering9010006
Keywords: CrDCL1, microRNA, biogenesis, small RNA-sequencing, Chlamydomonas reinhardtii
Citation: Sun T, Tao M, Di Q, Hu Z, Li H and Lou S (2025) Identification of CrDCL1-mediated microRNA biogenesis in green alga Chlamydomonas reinhardtii. Front. Microbiol. 16:1487584. doi: 10.3389/fmicb.2025.1487584
Received: 28 August 2024; Accepted: 10 February 2025;
Published: 27 February 2025.
Edited by:
Yonghong Bi, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Adrian Alejandro Valli, Spanish National Research Council (CSIC), SpainCopyright © 2025 Sun, Tao, Di, Hu, Li and Lou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Li, bGlodWk4MEBzenUuZWR1LmNu; Sulin Lou, c2xsb3VAc3p1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.