- 1Department of Plant Physiology, Institute of Biosciences, University of Rostock, Rostock, Germany
- 2Department of Plant Biochemistry, Faculty of Biology and Biotechnology, Ruhr-University Bochum, Bochum, Germany
- 3Science for Life Laboratory, School of Engineering Sciences in Chemistry, Biotechnology and Health, KTH—Royal Institute of Technology, Stockholm, Sweden
- 4Department of Biochemistry, Institute of Biosciences, University of Rostock, Rostock, Germany
- 5Department of Life, Light and Matter, Interdisciplinary Faculty, University of Rostock, Rostock, Germany
Fixation of CO2 into the organic compound formate by formate dehydrogenases (FDHs) is regarded as the oldest autotrophic process on Earth. It has been proposed that an FDH-dependent CO2 fixation module could support CO2 assimilation even in photoautotrophic organisms. In the present study, we characterized FDH from Clostridium carboxidivorans (ccFDH) due to its ability to reduce CO2 under aerobic conditions. During the production of recombinant ccFDH, in which the selenocysteine codon was replaced by Cys, we were able to replace the W with Mo as the transition metal in the ccFDH metal cofactor, resulting in a two-fold increase of 6 μmol formate min−1 in enzyme activity. Then, we generated ccFDH variants in which the strict NADH preference of the enzyme was changed to NADPH, as this reducing agent is produced in high amounts during the photosynthetic light process. Finally, we showed that the native ccFDH can also directly use ferredoxin as a reducing agent, which is produced by the photosynthetic light reactions at photosystem I. These data collectively suggest that ccFDH and, particularly, its optimized variants can be regarded as suitable enzymes to couple formate production to photosynthesis in photoautotroph organisms, which could potentially support CO2 assimilation via the Calvin–Benson–Bassham (CBB) cycle and minimize CO2 losses due to photorespiration.
Introduction
Formate is one of the simplest organic compounds and serves as a crucial source of carbon and reducing power for living cells. It can be produced through various routes, including the electrochemical reduction of CO2, photoreduction of CO2, hydrogenation of CO2, selective oxidation of biomass, partial oxidation of natural gas, and hydration of syngas (i.e., a mixture of hydrogen and carbon monoxide) (Bar-Even, 2016). Formate dehydrogenases (FDHs, EC1.17.1.9) are a heterogeneous group of proteins found in eukaryotes and prokaryotes (Jormakka et al., 2003; Yu et al., 2014), with some of them having the ability to catalyze the reversible reduction of CO2 to formate. In nature, many anaerobic bacteria and archaea can efficiently fix CO2 (or CO) into formate via the Wood–Ljungdahl pathway (Ragsdale and Pierce, 2008), which is the oldest autotrophic CO2 assimilation route on Earth (Moody et al., 2024). In recent years, the increased release of CO2 into the atmosphere due to human activities, mostly from burning fossil carbon sources for energy production, has been identified as the main driver of climate change (Lee et al., 2023). To mitigate this impact, carbon-neutral processes using FDHs to convert CO2 into organic compounds have become a focus of biotechnology and synthetic biology (Alissandratos et al., 2013a; Bar-Even, 2016).
FDHs can be divided into two major classes, based on their metal content and the subsequent chemical strategy employed by the active site to carry out formate oxidation (Maia et al., 2017). One class, metal-center-independent FDH proteins, belongs to the superfamily of D-specific dehydrogenases of 2-hydroxy acids (Tishkov and Popov, 2006; Maia et al., 2017). FDHs from this class usually catalyze formate oxidation coupled with the reduction of NAD+ to NADH. The other class, metal-center-dependent FDH proteins, contains a complex inventory of redox-active centers and is usually sensitive to oxygen (Maia et al., 2015). CO2-fixing FDHs in anaerobic bacteria and archaea performing the Wood–Ljungdahl pathway belong to the class of metal-center-dependent enzymes. These proteins contain molybdenum, tungsten, or other transition metals in their active site, coordinated by pyranopterin cofactors, to facilitate formate degradation or CO2 reduction. Additionally, to promote electron transport, they usually contain several iron–sulfur clusters. This group of FDHs is of special interest as they provide enzymes that could catalyze efficient CO2 fixation when coupled with naturally occurring formate assimilation. In the case of the Wood–Ljungdahl pathway, formate assimilation occurs via a formate tetrahydrofolate ligase that produces the C1 component formyl-tetrahydrofolate (formyl-THF). Formyl-THF is found ubiquitously in all living organisms within C1 metabolism, and thus, in terms of synthetic biology, it represents a potential entry point to various metabolic networks.
Recently, the establishment of FDH-mediated CO2 reduction into formate in photoautotrophic organisms was proposed to support CO2 fixation via the Calvin–Benson–Bassham (CBB) cycle (Bar-Even, 2018). In all organisms that perform oxygenic photosynthesis—cyanobacteria, algae, and plants—photosynthetic CO2 fixation is catalyzed by the key enzyme ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO). However, RubisCO has been identified as a limiting factor for efficient CO2 fixation under current atmospheric conditions because the enzyme is rather slow and has a low CO2 affinity. To offset these limitations, organisms rely on high amounts of RubisCO. The CO2 affinity of RubisCO enzymes varies between different organisms. For example, it is approximately 10 times lower in land plants than in most cyanobacteria and algae, which is compensated for by the evolution of a CO2-concentrating mechanism in the latter photoautotrophs (For a review, see Badger and Sharwood, 2023). Furthermore, RubisCO shows the oxygenase side reaction, where oxygen (O2) is fixed instead of CO2, leading to the formation of the toxic intermediate 2-phosphoglycolate, which needs to be salvaged into CBB intermediates at the expense of CO2 and energy losses via photorespiration (Tcherkez et al., 2006; Bar-Even, 2018; Kern et al., 2020). The salvaging of 2-phosphoglycolate in the photorespiratory cycle includes steps of the C1 metabolism. Hence, in photoautotrophs, the formyl-THF resulting from formate assimilation can be introduced into plant metabolism via photorespiration, after stepwise reduction to methylene-THF, which, together with glycine, serves for serine biosynthesis at serine hydroxymethyltransferase (SHMT). It has been discussed that an increased pool of methylene-THF due to efficient formate assimilation could turn photorespiration into less CO2-releasing or even CO2-fixing when the glycine-decarboxylase reaction is reversed. However, this has been only observed in synthetic biology studies involving Escherichia coli (e.g., Yishai et al., 2018) and not in photoautotrophs. Recently, the formate-assimilation pathway, including a reversed glycine decarboxylase flux, was successively established in E. coli, proving the previously designed CO2-fixing shunt as kinetically feasible and functional (Yishai et al., 2017; Bang and Lee, 2018; Döring et al., 2018; Kim et al., 2020). These synthetic biology approaches confirm that artificial formate assimilation pathways are compatible with cellular metabolism, but they have not yet seen widespread practical application.
To establish CO2 reduction into formate in oxygenic phototrophs, a suitable FDH is needed, one that not only efficiently reduces CO2 but is also oxygen-tolerant and can use NADPH or ferredoxin as the electron source. A promising candidate for expression in phototrophs is the NAD-dependent FDH from acetogenic Clostridium carboxidivorans, ccFDH. It has been reported to preferentially reduce CO2 to formate even in the presence of O2 (Alissandratos et al., 2013b). In the present study, we aimed to biochemically characterize this enzyme and take the first steps in optimizing its acceptor capabilities for future application in oxygenic phototrophs.
Materials and methods
Gene synthesis and cloning
The nucleotide sequence of the ccFDH (Acc. No. UniProt E2IQB0) was synthesized by Baseclear (Leiden, Netherlands), in which codon 139 for selenocysteine (Sec) was changed to TGC encoding cysteine (139Cys; see Supplementary Figure S1). The gene was inserted via NheI/BglII into pET28a (Novagen) for subsequent expression with an N-terminal His6-tag. The resulting construct was confirmed by sequencing (Microsynth, Göttingen, Germany). Further amino acid substitutions were introduced by site-directed mutagenesis using primers listed in Supplementary Table S1.
Bioinformatic analysis
The homology model of the ccFDH was created using the SWISS-MODEL (Waterhouse et al., 2018; Studer et al., 2020) using FDH-H from E. coli as the template (64.01% sequence identity; QMEANDisCo Global: 0.83 ± 0.05). The conservation of amino acids was depicted using the ConSurf web server (Ashkenazy et al., 2016).
Protein expression and purification
Aerobic protein expression was performed in the strain E. coli BL21(DE3) using LB medium (Roth, Germany). Pre-cultures were diluted to an OD600 of 0.1 and incubated until they reached 0.6–0.8. The expression was induced by the addition of 1 mM IPTG. The cells were shaken at room temperature (RT) for 4 h. For co-factor determination, the E. coli cells were grown in M9 minimal medium either without any additions, with 1 mM Na2MoO4, or with 1 mM Na2WO4. For all other experiments, LB was supplemented with 1 mM Na2MoO4.
The cells were homogenized in a lysis buffer [50 mM HEPES pH 7.8, 100 mM NaCl] after incubation with lysozyme at RT, followed by sonication on ice. The lysate was cleared by centrifugation (30,000 × g, 30 min, 4°C), and the protein was immobilized via IMAC, washed two times with 10 batch volumes of a wash buffer (50 mM HEPES pH 7.8, 100 mM NaCl, 50 mM imidazole). Elution was performed with two batch volumes of an elution buffer (50 mM HEPES pH 7.8, 100 mM NaCl, 500 mM imidazole). Protein concentration was determined using amido black (Schulz et al., 1994).
Mass spectrometry
A total of 50 μg of the elution fraction of the ccFDH was prepared for mass spectrometry via filter-aided sample preparation. After tryptic digestion, the peptides were analyzed on a nanoACQUITY / SYNAPT GS-2 HDMS System, and the data were evaluated using Progenesis QI for Proteomics (Waters, Germany).
Enzyme activity
The NADH-dependent formate production was monitored by the decrease in NADH absorbance at 340 nm, as adopted from Alissandratos et al. (2013b). In brief, 5 μM of the ccFDH and 0.2 mM NADH in 50 mM Bis-Tris pH 6.8 were equilibrated at 30°C to a constant absorbance at 340 nm. The reaction was then started by the addition of 100 mM sodium bicarbonate. Initial velocity within the first 2 min after induction was used to calculate kinetic parameters. The formate oxidation was measured in the presence of 0.2 mM NAD+ and 100 mM sodium formate under the same buffer conditions.
Ferredoxin-dependent enzyme assay
A total of 10 μM of Fdx from Spinacia oleracea (soFdx) was pre-reduced by 1 mM sodium dithionite under anaerobic conditions with constant nitrogen bubbling in an Fdx buffer [50 mM Bis-Tris pH 6.8, 100 mM NaCl] for 5 min. The reaction started by the addition of 5 μM ccFDH and 100 mM sodium bicarbonate and stopped by heating the reaction mixture to 95°C. Protein aggregates were pelleted by centrifugation, and formate was detected in the clear supernatant.
Formate detection
Formate was colorimetrically detected using the formate detection kit provided by Merck (Darmstadt, Germany). The assay was performed according to the manufacturer’s protocol.
Photochronoamperometric measurements
Photochronoamperometric measurements were performed using a three-electrode system (Hartmann et al., 2018). A glassy carbon electrode, with a diameter of 4 mm, was used as the working electrode and was surrounded by a plastic tube containing the reaction medium. A Pt-wire, the counter electrode, and Ag/AgCl/3 M KCl, the reference electrode, were both placed in the reaction medium without touching the working electrode’s surface. The reaction medium included the following: 200 μL volume, 50 mM Bis-Tris pH 7.5, 30 mM NaCl, 0.03% n-Dodecyl-B-D-maltoside (β-DDM), 20 μM cytochrome C (Cyt c), 50 μg Chl photosystem I (PSI), 25, 50, or 100 μM soFdx, 25 μM ccFDH, and 200 mM sodium bicarbonate (HCO3−). After the deposition of the reaction medium on the electrode, the electrode was incubated in the dark for 3 min. Subsequently, a red light of 2,300 μE at 685 nm was switched on for 30 min, followed by dark incubation for 5 min. During the measurements, the electrode was applied with a potential of 0 V vs. Ag/AgCl/ 3 M KCL. Unless otherwise specified, all post-experimental procedures were conducted in the dark or under green light.
All photochronoamperometric measurements were performed using an AUTOLAB PGSTAT12 potentiostat/galvanostat. The light intensity was controlled by adapting the applied power-on LED with the computer-based control unit PXI-1033 with the DC precision power supply module PXI-4110 (National Instruments Germany GmbH, Munich, Germany).
The proteins used for the photochronoamperometric measurements, Cytc and PSI, were purified, as mentioned previously (El-Mohsnawy et al., 2010; Medipally et al., 2023). Both PSI and Cytc were obtained from Thermosynechococcus vestitus BP1.
Results and discussion
Protein purification and enzyme assay
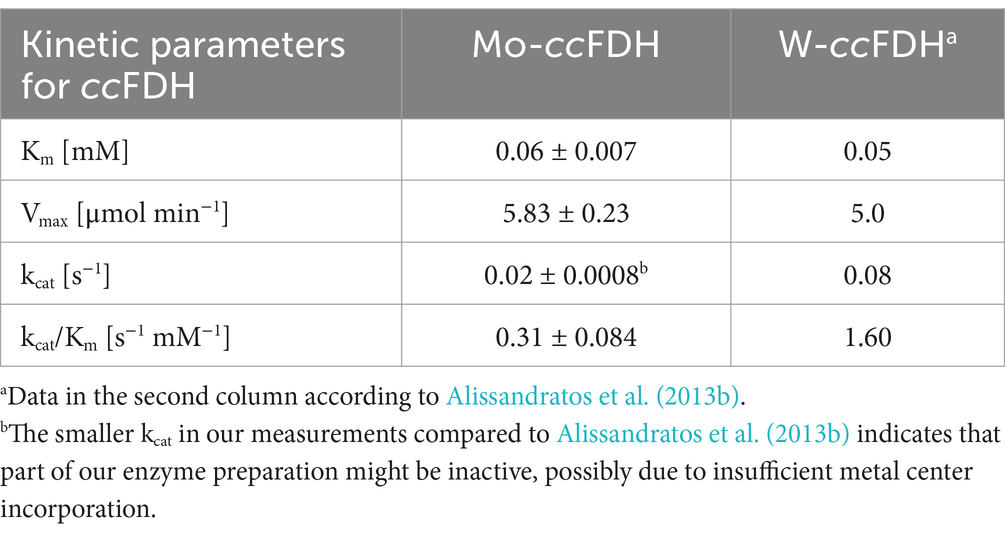
To assess whether the FDH from C. carboxidivorans (ccFDH) is a suitable target for expression in oxygenic phototrophs as an additional CO2-fixing enzyme, the biochemical features of the enzyme were investigated. For this purpose, a codon-optimized version of the ccFDH was cloned into the expression vector pET28a to obtain the recombinant protein with a N-terminal His6-tag. As reported before, the Sec codon 139Sec was exchanged for 139Cys (Alissandratos et al., 2013a) because the desired final hosts, such as cyanobacteria or plants, were not able to incorporate Sec into proteins (Zhang et al., 2006; Lopez Heras et al., 2011). The recombinant ccFDH was purified via IMAC as a soluble protein of approximately 80 kDa from the total protein extracts of E. coli. The elution fraction of the ccFDH revealed some proteinogenic contaminations, independent of the expression conditions (representative purification see Figure 1A). The mass spectrometry analysis revealed that these impurities did not interfere with our subsequent enzyme activity measurements because mostly chaperones and ribosomal proteins were detected (Supplementary Table S2).
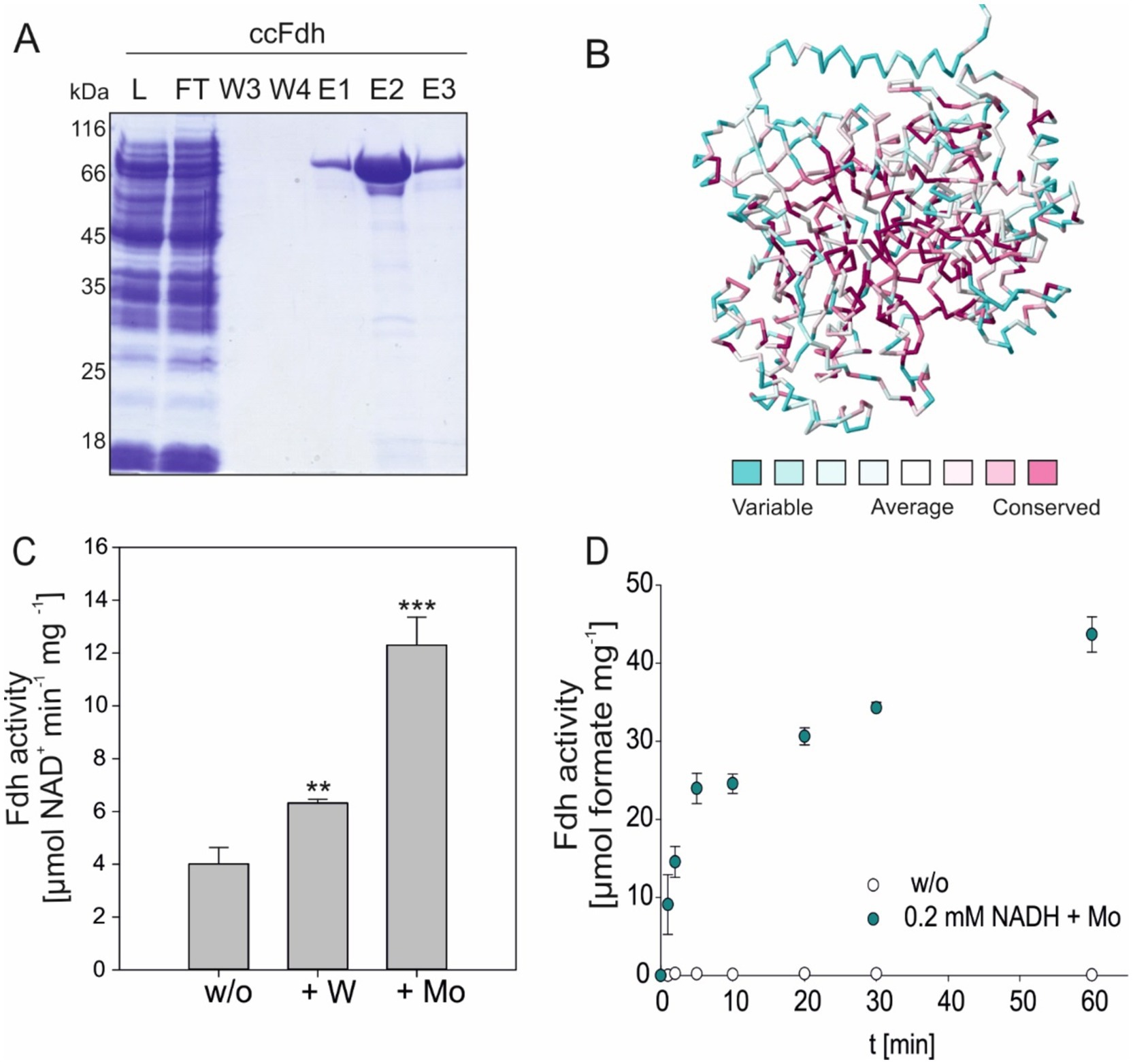
Figure 1. Molybdenum-dependent CO2 reduction by ccFDH. (A) Elution fraction after IMAC showing the degree of impurities. A representative purification is displayed. (B) Homology model showing the degree of conservation of the ccFDH based on the structure of ecFDH-H (PDB accession no. 1FDO). Modeling was carried out using SWISS-MODEL and the ConSurf web server. (C) NADH-depended CO2 reduction by the purified ccFDH obtained from the E. coli cultures either supplemented with 1 mM Na2MoO4, 1 mM Na2WO4, or without any addition. The FDH activity was monitored by the decrease in absorbance at 340 nm. The mean values of independent replicates (n = 6) are shown. ** > 0.05; *** > 0.01 (D) Time-dependent formate production by Mo-ccFDH measured at given time points using a colorimetric kit.
For FDH activity measurements, two principal assays were applied. In the majority of the cases, the oxidation of NAD(P)H during the conversion of CO2 into formate was followed spectrophotometrically, which permitted measurements in the presence of different substrate and cofactor concentrations. Alternatively, the formation of the reaction product, formate, was quantified using a colorimetric formate detection kit. The second assay confirmed that the NAD(P)H oxidation in our enzyme assays specifically produced the formate. The enzyme assays were conducted under ambient oxic conditions. Furthermore, the formate oxidation activity of the ccFDH in general was very low and could be considered undetectable under our experimental conditions (Supplementary Figure S2).
Molybdenum-dependent activity of ccFDH
Molybdenum (Mo) and tungsten (W) are the heaviest transition metals used by biological systems. They are present in the cofactors of oxidoreductases, including FDHs, due to their superior redox potential (Schwarz and Mendel, 2006). The comparison of the FDH sequences from different species revealed that the enzyme core, including the cofactor binding sites, is highly conserved (Figure 1B). According to the known crystal structures of FDH-H from E. coli with a verified Mo cofactor and FDH from Desulfovibrio vulgaris with a verified W cofactor, the Sec and neighboring His residues are thought to be involved in metal cation coordination and catalytic activity, respectively (Boyington et al., 1997; Raaijmakers et al., 2002). Alissandratos et al. (2013a) proved that the ccFDH harbors a W cofactor. However, the high degree of conservation in the metal cofactor-binding site made it difficult to obtain a clear structural or sequence-based prediction favoring one or other possible metals. Thus, we aimed to compare ccFDH-mediated CO2 reduction activity in the presence of either Mo or W. Furthermore, to apply the ccFDH to a broader host spectrum, the ability to accept Mo is of special interest.
For this purpose, the ccFDH was expressed in the E. coli cells grown in minimal M9 medium either supplemented with 1 mM Na2MoO4, 1 mM Na2WO4, or without any trace metal addition under aerobic conditions. The recombinant His-tagged protein was subsequently purified via IMAC. Using the spectrophotometric enzyme assay, the ccFDH expressed in the M9 medium without any supplementation revealed an activity of 4.0 ± 0.63 μmol NAD+ min−1 mg−1 (Figure 1C). This activity was equivalent to the initial production of approximately 4.4 μmol formate min−1 mg−1 during the first 5 min (Figure 1D). When expressed in the presence of 1 mM Na2WO4, the activity increased to 6.3 ± 0.14 μmol NAD+ min−1 mg−1, whereas the highest activity of 12.3 ± 1.05 μmol NAD+ min−1 mg−1 was measured with the 1 mM Na2MoO4 supplementation (Figure 1C). These results showed that CO2 reduction by the ccFDH did not solely rely on W, as reported previously (Alissandratos et al., 2013a), but was even enhanced in the presence of Mo. The increased catalytic rate with Mo was also observed in assays measuring the rise in the formate concentration over time. After a fast initial increase in the formate concentration, the velocity declined, but the formate was constantly produced for 1 h (Figure 1D). The detected formate concentration within the first 5 min was in good agreement with the NADH oxidation measured spectrophotometrically (Figure 1C). Hence, for all subsequent experiments, we used the ccFDH obtained from Mo-supplemented E. coli cultivations, referred to as Mo-ccFDH.
Next, enzyme assays with the Mo-ccFDH were performed in the presence of different NADH concentrations to estimate the biochemical properties of this enzyme. The obtained data were compared with the kinetic parameters for NADH oxidation in the W-cofactor-containing ccFDH (further referred to as W-ccFDH), as determined by Alissandratos et al. (2013b). The parameters for the Mo-ccFDH determined in our study are comparable to those determined by Alissandratos et al. (2013b) for the W-ccFDH (Table 1).
NADPH-dependent CO2 reduction by ccFDH
As mentioned above, the implementation of FDH-dependent CO2 fixation could support the CBB cycle and suppress photorespiration in photoautotrophs such as plants. To realize this strategy, an oxygen-tolerant FDH, such as ccFDH, which reduces CO2 to formate, needs to be expressed in photoautotrophs (Yishai et al., 2018; Song et al., 2020). However, in addition to oxygen tolerance, a well-suited FDH should be able to preferentially accept NADPH over NADH because photoautotrophs constantly produce NADPH in the photosynthetic light reactions. Hence, NADPH is the preferred reducing agent in most anabolic pathways, including CO2 assimilation (Park and Choi, 2017). Therefore, we aimed to change the cofactor specificity of ccFDH, which naturally did not show considerable activity in the presence of NADPH (Figure 2).
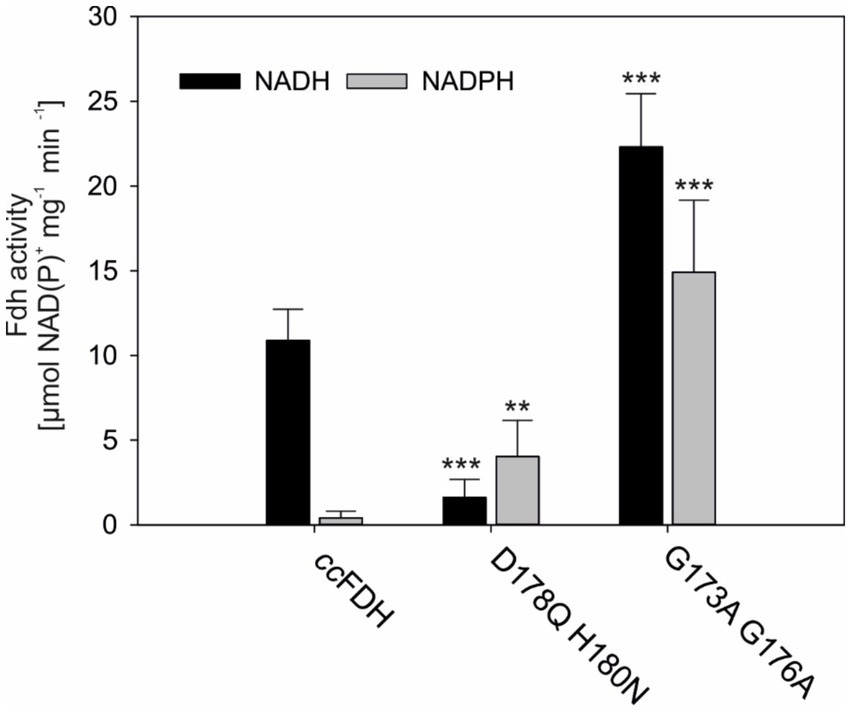
Figure 2. Cofactor specificity of Mo-ccFDH. The cofactor specificity was changed by introducing specific mutations in the nucleotide-binding site. All FDH variants were expressed in the presence of Mo. The mean values of three independent replicates and the standard deviation are shown. Significance was calculated according to the respective WT-FDH. ** > 0.05; *** > 0.01.
The nucleotide-binding site in the ccFDH was predicted using the Rossmann toolbox server (Kamiński et al., 2022). In ccFDH, it is located between amino acid residues 191 and 250 with a probability of 96.8%. The homology model revealed a typical βαβ fold within this region and a variation of the consensus nucleotide-binding sequence GxGxxxG located in a turn between β1 α1. In ccFDH, the consensus sequence lacks the first glycine and is composed of G173YNG178. To obtain weaker and more general cofactor binding, we introduced two glycine-to-alanine substitutions in this region, namely G173A and G178A. The variant G173A G176A showed a general increase in enzymatic activity compared to the native Mo-ccFDH and, as expected, was able to use NADPH with almost the same efficiency as NADH (Figure 2). In the presence of NADH, the activity of the variant G173A G176A increased two-fold compared to ccFDH, while its NADPH-dependent rate was 35-fold higher. Likely, the G173A G176A substitutions not only resulted in weaker and more general binding of the coenzymes but also caused faster cofactor release or exchange.
The preference of the FDH from Pseudomonas sp. 101 for NADH over NADPH was previously altered by exchanging D221 and H223, which are situated in close proximity to the GxGxxxG motif (Calzadiaz-Ramirez et al., 2020). Accordingly, we substituted D178 with glutamine (D178Q) and H180 with asparagine (H180N) in ccFDH. The amino acid substitutions in the D178Q H180N variant resulted in overall lower activity compared to the native Mo-ccFDH (Figure 2). However, the D178Q H180N variant showed the expected nearly two-fold higher specificity for NADPH over NADH.
Ferredoxin-dependent CO2 reduction by ccFDH
Electron transfer between iron–sulfur clusters has been shown to be primarily dependent on the intramolecular distance between the redox centers and should ideally be placed within the boundaries of 14 Å (Page et al., 1999). Assuming that the electron-accepting FeS cluster in the ccFDH is located at the N-terminal domain, in close proximity to the NAD(P)-binding domain, intramolecular electron transfer mediated by ferredoxin is likely and was tested in this study.
In the initial approach, the formate production by the Mo-ccFDH was measured in the presence of 10 μM of ferredoxin from Spinacia oleracea (soFdx), reduced by sodium dithionite. Under our assay conditions, the enzyme was able to produce up to 42 μg of the formate within 1 h (Figure 3A). The initial reaction velocity, approximately 25.5 μmol min−1, exceeded that of the NADH-dependent formate production by 2.8-fold. However, the absolute formate concentration after 1 hour remained nearly the same. Most likely, the reduced soFdx was not stable for extended periods under our test conditions because oxygen could not be completely omitted from the assays, leading to soFdx oxidation.
Figure 3. Light-dependent ferredoxin-reduction can support formate production by Mo-FDH. (A) Time-resolved formate production by reduced spinach ferredoxin and Mo-ccFDH. The mean values of three independent replicates, along with the standard deviation, are shown. (B) Schematic representation of photosystem I (PSI)-based photocathodes for formate formation. CytC 6 : cytochrome c6, P700*/P700: excitation of the chlorophyll (Chl) molecule of PSI upon light excitation, FB−: reduced iron–sulfur cluster. Fd, ferredoxin; FDH, formate dehydrogenase. Midpoint potential values are according to literature references (Badura et al., 2011; Kothe et al., 2013; Sokol et al., 2018). The midpoint potential of all individual components of the cascade is defined relative to the standard hydrogen electrode. (C) Photochronoamperometry-based reduction of ferredoxin and subsequent formate production by FDH, shown by the development of photocurrent. Each sample contained CytC, PSI, and sodium bicarbonate. The additional components are provided in the figure legend. The mean value of three independent replicates from t 0 s (light on, yellow bulb) to 30 min (light off, black bulb) are shown.
Next, we aimed to investigate whether it is possible to couple ferredoxin-mediated electron transport from photosynthesis with Mo-ccFDH-mediated CO2 reduction. This was tested by the artificial reconstitution of the cathodic part of the photosynthetic electron transport chain, using a carbon electrode, CytC, photosystem I (PSI), soFdx, ccFDH, and CO2. The difference in the mid-point potentials between these components created a paradigm for electron transfer, as shown in Figure 3B. In this cascade, the initial electron donor was a carbon electrode with a mid-point potential of 0 V versus Ag/AgCl/ 3 M KCl and the final electron acceptor was CO2. The electron transfer within this cascade was monitored by changes in the photocurrent density. Upon light excitation, the photo-energized PSI initiated the rapid uptake of the electrons from the electrode via Cytc, which was visible by the formation of a negative current (Figure 3C). Then, the PSI-driven energized electrons were transferred via the soFdx to Mo-ccFDH, which further reduced CO2 to formate. When only the soFdx was added to this cascade (soFdx, green in Figure 3C), we observed a low photocurrent density of −29.15 nA cm−2 after 5 min of illumination, indicating a compromise in the electron consumption of any of the individual components in the cascade. When only the Mo-ccFDH was added, the current showed no significant decrease, with a value of −13.48 nA cm−2, after 5 min of illumination (ccFDH, blue in Figure 3C). However, when all components of the cascade were added, we observed the highest photocurrent density of −460.44 nA cm−2 after 5 min illumination, indicating electron consumption by the soFdx-dependent ccFDH. Here, the rate-limiting component was the soFdx (compared to Supplementary Figure S3), most likely due to the inefficient reduction of the plant Fdx by cyanobacterial PSI. These experiments clearly showed that the native ccFDH can convert CO2 into formate using a reduced Fdx, which is available in high amounts during the light period in photoautotrophs. However, the Fdx-dependent coupling of CO2 reduction via the ccFDH may have regulatory effects on the photosynthetic electron flow in vivo, which needs to be analyzed in future experiments once the enzyme is successfully expressed in a photoautotrophic cell.
Conclusion
The FDH from Clostridium carboxidivorans investigated in this study has the capability to reduce CO2 under aerobic conditions with high efficiency, while the reverse reaction is virtually absent. Moreover, in addition to W, it can also accept Mo as the transition metal in its pyranopterin cofactor, making it suitable for use in any organism that contains this cofactor family, such as most phototrophs, which use the Mo-cofactor in their nitrate reductase (Schwarz and Mendel, 2006). The cofactor preference of the ccFDH can be altered by a few amino acid substitutions in the Rossmann fold, enabling it to efficiently use NADPH in addition to NADH, which is produced in high amounts during photosynthetic light reactions. Finally, the native ccFDH can also directly use Fdx as a reducing agent, which is generated by the photosynthetic light reaction at PSI. These data collectively suggest that ccFDH and, particularly, slightly optimized variants can be regarded as suitable enzymes to couple formate production to photosynthesis in photoautotroph organisms, potentially supporting CO2 assimilation via the CBB cycle and minimizing CO2 losses due to photorespiration.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
E-MB: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft. HM: Data curation, Formal analysis, Investigation, Writing – review & editing. SaS: Data curation, Formal analysis, Investigation, Writing – review & editing. ShS: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. MN: Data curation, Formal analysis, Investigation, Resources, Supervision, Validation, Writing – review & editing. MH: Conceptualization, Funding acquisition, Supervision, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Shanshan Song was supported by the Chinese Scholarship Council (CSC). The project was supported by the German Federal Ministry for Education and Research (BMBF) through FormatPlant grant no. 031B0194B and the European Commission project Gain4Crops (proposal No. 862087) to MH. MMN and HM acknowledge the financial support received from the European Union Horizon 2020 research and innovation program (grant agreement no. 764920) as well as the Deutsche Forschungsgemeinschaft (DFG)-funded Research Training Group 2341 ‘Microbial Substrate Conversion (MiCon)’.
Acknowledgments
We would like to thank Dr. Stefan Mikkat, Core Facility Proteome Analysis, Rostock University Medical Center, for the proteome analyses of our FDH elution fractions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1527626/full#supplementary-material
References
Alissandratos, A., Kim, H.-K., and Easton, C. J. (2013a). Formate production through biocatalysis. Bioengineered 4, 348–350. doi: 10.4161/bioe.25360
Alissandratos, A., Kim, H.-K., Matthews, H., Hennessy, J. E., Philbrook, A., and Easton, C. J. (2013b). Clostridium carboxidivorans strain P7T recombinant formate dehydrogenase catalyzes reduction of CO2 to formate. Appl. Environ. Microbiol. 79, 741–744. doi: 10.1128/AEM.02886-12
Ashkenazy, H., Abadi, S., Martz, E., Chay, O., Mayrose, I., Pupko, T., et al. (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350. doi: 10.1093/nar/gkw408
Badger, M. R., and Sharwood, R. E. (2023). Rubisco, the imperfect winner: it's all about the base. J. Exp. Bot. 74, 562–580. doi: 10.1093/jxb/erac458
Badura, A., Guschin, D., Kothe, T., Kopczak, M. J., Schuhmann, W., and Rögner, M. (2011). Photocurrent generation by photosystem 1 integrated in crosslinked redox hydrogels. Energy Environ. Sci. 4:2435. doi: 10.1039/c1ee01126j
Bang, J., and Lee, S. Y. (2018). Assimilation of formic acid and CO2 by engineered Escherichia coli equipped with reconstructed one-carbon assimilation pathways. Proc. Natl. Acad. Sci. 115, E9271–E9279. doi: 10.1073/pnas.1810386115
Bar-Even, A. (2016). Formate assimilation: the metabolic architecture of natural and synthetic pathways. Biochemistry 55, 3851–3863. doi: 10.1021/acs.biochem.6b00495
Bar-Even, A. (2018). Daring metabolic designs for enhanced plant carbon fixation. Plant Sci. 273, 71–83. doi: 10.1016/j.plantsci.2017.12.007
Boyington, J. C., Gladyshev, V. N., Khangulov, S. V., Stadtman, T. C., and Sun, P. D. (1997). Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science 275, 1305–1308. doi: 10.1126/science.275.5304.1305
Calzadiaz-Ramirez, L., Calvó-Tusell, C., Stoffel, G. M. M., Lindner, S. N., Osuna, S., Erb, T. J., et al. (2020). In vivo selection for Formate dehydrogenases with high efficiency and specificity toward NADP. ACS Catal. 10, 7512–7525. doi: 10.1021/acscatal.0c01487
Döring, V., Darii, E., Yishai, O., Bar-Even, A., and Bouzon, M. (2018). Implementation of a reductive route of one-carbon assimilation in Escherichia coli through directed evolution. ACS Synth. Biol. 7, 2029–2036. doi: 10.1021/acssynbio.8b00167
El-Mohsnawy, E., Kopczak, M. J., Schlodder, E., Nowaczyk, M., Meyer, H. E., Warscheid, B., et al. (2010). Structure and function of intact photosystem 1 monomers from the cyanobacterium Thermosynechococcus elongatus. Biochemistry 49, 4740–4751. doi: 10.1021/bi901807p
Hartmann, V., Ruff, A., Schuhmann, W., Rögner, M., and Nowaczyk, M. M. (2018). Analysis of photosystem II electron transfer with natural PsbA-variants by redox polymer/protein biophotoelectrochemistry. Photosynthetica 56, 229–235. doi: 10.1007/s11099-018-0775-y
Jormakka, M., Byrne, B., and Iwata, S. (2003). Formate dehydrogenase—a versatile enzyme in changing environments. Curr. Opin. Struct. Biol. 13, 418–423. doi: 10.1016/s0959-440x(03)00098-8
Kamiński, K., Ludwiczak, J., Jasiński, M., Bukala, A., Madaj, R., Szczepaniak, K., et al. (2022). Rossmann-toolbox: a deep learning-based protocol for the prediction and design of cofactor specificity in Rossmann fold proteins. Brief. Bioinform. 23:bbab371. doi: 10.1093/bib/bbab371
Kern, R., Facchinelli, F., Delwiche, C., Weber, A. P. M., Bauwe, H., and Hagemann, M. (2020). Evolution of Photorespiratory Glycolate oxidase among Archaeplastida. Plants (Basel) 9:106. doi: 10.3390/plants9010106
Kim, S., Lindner, S. N., Aslan, S., Yishai, O., Wenk, S., Schann, K., et al. (2020). Growth of E. coli on formate and methanol via the reductive glycine pathway. Nat. Chem. Biol. 16, 538–545. doi: 10.1038/s41589-020-0473-5
Kothe, T., Plumeré, N., Badura, A., Nowaczyk, M. M., Guschin, D. A., Rögner, M., et al. (2013). Combination of a photosystem 1-based photocathode and a photosystem 2-based photoanode to a Z-scheme mimic for biophotovoltaic applications. Angew. Chem. Int. Ed. Engl. 52, 14233–14236. doi: 10.1002/anie.201303671
Lee, H., Calvin, K., Dasgupta, D., Krinner, G., Mukherji, A., Thorne, P. W., et al. (2023). IPCC, 2023: Climate change 2023: Synthesis report. Contribution of working groups I, II and III to the sixth assessment report of the intergovernmental panel on climate change [Core writing team, H. Lee and J. Romero (eds.)]. Geneva, Switzerland: Intergovernmental Panel on Climate Change (IPCC).
Lopez Heras, I., Palomo, M., and Madrid, Y. (2011). Selenoproteins: the key factor in selenium essentiality. State of the art analytical techniques for selenoprotein studies. Anal. Bioanal. Chem. 400, 1717–1727. doi: 10.1007/s00216-011-4916-4
Maia, L. B., Moura, J. J. G., and Moura, I. (2015). Molybdenum and tungsten-dependent formate dehydrogenases. J. Biol. Inorg. Chem. 20, 287–309. doi: 10.1007/s00775-014-1218-2
Maia, L. B., Moura, I., and Moura, J. J. G. (2017). Molybdenum and tungsten-containing formate dehydrogenases: aiming to inspire a catalyst for carbon dioxide utilization. Inorg. Chim. Acta 455, 350–363. doi: 10.1016/j.ica.2016.07.010
Medipally, H., Mann, M., Kötting, C., van Berkel, W. J. H., and Nowaczyk, M. M. (2023). A clickable photosystem I, ferredoxin, and ferredoxin NADP+ reductase fusion system for light-driven NADPH regeneration. Chembiochem 24:e202300025. doi: 10.1002/cbic.202300025
Moody, E. R. R., Álvarez-Carretero, S., Mahendrarajah, T. A., Clark, J. W., Betts, H. C., Dombrowski, N., et al. (2024). The nature of the last universal common ancestor and its impact on the early earth system. Nat. Ecol. Evol. 8, 1654–1666. doi: 10.1038/s41559-024-02461-1
Page, C. C., Moser, C. C., Chen, X., and Dutton, P. L. (1999). Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature 402, 47–52. doi: 10.1038/46972
Park, J., and Choi, Y. (2017). Cofactor engineering in cyanobacteria to overcome imbalance between NADPH and NADH: a mini review. Front. Chem. Sci. Eng. 11, 66–71. doi: 10.1007/s11705-016-1591-1
Raaijmakers, H. C. A., Macieira, S., Dias, J., Teixeira, S., Bursakov, S., Huber, R., et al. (2002). Gene sequence and the 1.8 a crystal structure of the tungsten-containing Formate dehydrogenase from Desulfovibrio gigas. Structure 10, 1261–1272. doi: 10.1016/S0969-2126(02)00826-2
Ragsdale, S. W., and Pierce, E. (2008). Acetogenesis and the wood-Ljungdahl pathway of CO(2) fixation. Biochim. Biophys. Acta 1784, 1873–1898. doi: 10.1016/j.bbapap.2008.08.012
Schulz, J., Dettlaff, S., Fritzsche, U., Harms, U., Schiebel, H., Derer, W., et al. (1994). The amido black assay: a simple and quantitative multipurpose test of adhesion, proliferation, and cytotoxicity in microplate cultures of keratinocytes (HaCaT) and other cell types growing adherently or in suspension. J. Immunol. Methods 167, 1–13. doi: 10.1016/0022-1759(94)90069-8
Schwarz, G., and Mendel, R. R. (2006). Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu. Rev. Plant Biol. 57, 623–647. doi: 10.1146/annurev.arplant.57.032905.105437
Sokol, K. P., Robinson, W. E., Oliveira, A. R., Warnan, J., Nowaczyk, M. M., Ruff, A., et al. (2018). Photoreduction of CO2 with a Formate dehydrogenase driven by photosystem II using a semi-artificial Z-scheme architecture. J. Am. Chem. Soc. 140, 16418–16422. doi: 10.1021/jacs.8b10247
Song, S., Timm, S., Lindner, S. N., Reimann, V., Hess, W. R., Hagemann, M., et al. (2020). Expression of Formate-tetrahydrofolate ligase did not improve growth but interferes with nitrogen and carbon metabolism of Synechocystis sp. PCC 6803. Front. Microbiol. 11:1650. doi: 10.3389/fmicb.2020.01650
Studer, G., Rempfer, C., Waterhouse, A. M., Gumienny, R., Haas, J., and Schwede, T. (2020). QMEANDisCo-distance constraints applied on model quality estimation. Bioinformatics 36, 1765–1771. doi: 10.1093/bioinformatics/btz828
Tcherkez, G. G. B., Farquhar, G. D., and Andrews, T. J. (2006). Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl. Acad. Sci. 103, 7246–7251. doi: 10.1073/pnas.0600605103
Tishkov, V. I., and Popov, V. O. (2006). Protein engineering of formate dehydrogenase. Biomol. Eng. 23, 89–110. doi: 10.1016/j.bioeng.2006.02.003
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Yishai, O., Bouzon, M., Döring, V., and Bar-Even, A. (2018). In vivo assimilation of one-carbon via a synthetic reductive Glycine pathway in Escherichia coli. ACS Synth. Biol. 7, 2023–2028. doi: 10.1021/acssynbio.8b00131
Yishai, O., Goldbach, L., Tenenboim, H., Lindner, S. N., and Bar-Even, A. (2017). Engineered assimilation of exogenous and endogenous Formate in Escherichia coli. ACS Synth. Biol. 6, 1722–1731. doi: 10.1021/acssynbio.7b00086
Yu, S., Zhu, L., Zhou, C., An, T., Zhang, T., Jiang, B., et al. (2014). Promising properties of a formate dehydrogenase from a methanol-assimilating yeast Ogataea parapolymorpha DL-1 in his-tagged form. Appl. Microbiol. Biotechnol. 98, 1621–1630. doi: 10.1007/s00253-013-4996-5
Keywords: enzyme, photosynthesis, redox, site-specific mutants, carbon fixation
Citation: Brouwer E-M, Medipally HKR, Schwab S, Song S, Nowaczyk MM and Hagemann M (2025) Characterization of the oxygen-tolerant formate dehydrogenase from Clostridium carboxidivorans. Front. Microbiol. 15:1527626. doi: 10.3389/fmicb.2024.1527626
Edited by:
Ivan A. Berg, University of Münster, GermanyReviewed by:
Harvey J. M. Hou, Alabama State University, United StatesAmit Srivastava, The Czech Academy of Sciences, Czechia
Copyright © 2025 Brouwer, Medipally, Schwab, Song, Nowaczyk and Hagemann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eva-Maria Brouwer, ZXZhLW0uYnJvdXdlckBnbXguZGU=; Martin Hagemann, bWFydGluLmhhZ2VtYW5uQHVuaS1yb3N0b2NrLmRl
 Eva-Maria Brouwer
Eva-Maria Brouwer Hitesh K. R. Medipally2
,3
Hitesh K. R. Medipally2
,3
 Martin Hagemann
Martin Hagemann