
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 January 2025
Sec. Extreme Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1505699
This article is part of the Research Topic Extremophiles: Environmental Adaptation Mechanisms, Modification to Synthetic Biology, and Industrial Application View all 5 articles
Nitrogen metabolism, related genes, and other stress-resistance genes are poorly understood in Bosea strain. To date, most of the research work in Bosea strains has been focused on thiosulfate oxidation and arsenic reduction. This work aimed to better understand and identify genomic features that enable thiosulfate-oxidizing lichen-associated Bosea sp. PAMC26642 from the Arctic region of Svalbard, Norway, to withstand harsh environments. Comparative genomic analysis was performed using various bioinformatics tools to compare Bosea sp. PAMC26642 with other strains of the same genus, emphasizing nitrogen metabolism and stress adaptability. During genomic analysis of Bosea sp. PAMC26642, assimilatory nitrogen metabolic pathway and its associated enzymes such as nitrate reductase, NAD(P)H-nitrite reductase, ferredoxin-nitrite reductase, glutamine synthetase, glutamine synthase, and glutamate dehydrogenase were identified. In addition, carbonic anhydrase, cyanate lyase, and nitronate monooxygenase were also identified. Furthermore, the strain demonstrated nitrate reduction at two different temperatures (15°C and 25°C). Enzymes associated with various stress adaptation pathways, including oxidative stress (superoxide dismutase, catalase, and thiol peroxidase), osmotic stress (OmpR), temperature stress (Csp and Hsp), and heavy metal resistance, were also identified. The average Nucleotide Identity (ANI) value is found to be below the threshold of 94-95%, indicating this bacterium might be a potential new species. This study is very helpful in determining the diversity of thiosulfate-oxidizing nitrate-reducing bacteria, as well as their ability to adapt to extreme environments. These bacteria can be used in the future for environmental, biotechnological, and agricultural purposes, particularly in processes involving sulfur and nitrogen transformation.
Nitrogen metabolism and its regulation have been studied across various ecosystems, including the polar regions (Arctic/Antarctic) by various microorganisms such as Cyanobacteria (Nitrosospira, Nitrosomonas, Nostoc, and Anabaena) (Hayashi et al., 2020; Magalhães et al., 2014; Makhalanyane et al., 2015; Fernández-Valiente et al., 2001), Actinobacteria (Streptomyces and Frankiniaceae) (De Scally et al., 2016; Papale et al., 2018), and Proteobacteria (Burkholderiales) (Garrido-Benavent et al., 2020). Nitrogen metabolism in prokaryotes involves a complex interplay between transporter proteins, signaling proteins, and transcriptional regulators. Furthermore, they also involve the coordinated expression of enzymes that utilize extracellular nitrogen sources and intracellular biosynthesis of nitrogen-containing compounds (Harper et al., 2008; Merrick and Edwards, 1995). Despite these insights, there remains a significant gap in understanding how bacteria meet their nitrogen requirements. This gap is particularly evident in extreme environments, such as the polar regions. One such bacterium is Bosea sp. (specifically, Bosea sp. PAMC26642) reported by Kim et al. (2013) whose ability to survive and thrive in cold and nutrient-poor conditions could provide valuable information on microbial adaptation.
The genus Bosea was first reported by Das et al. in 1996. It belongs to the order Hyphomicrobiales and the family Boseaceae (Das and Mishra, 1996; Hördt et al., 2020). In addition to that, Bosea species have been isolated from various environments including Arctic lichen (Bosea sp. PAMC26642) (Kim et al., 2013), agricultural soils (Das et al., 1996), hospital water systems (La Scola et al., 2003), lakes (Albert et al., 2019), anaerobic digester sludge (Ouattara et al., 2003), root nodules of legumes (De Meyer and Willems, 2012), and pyrite rock (Walczak et al., 2018). Bosea strains were reported to have arsenite-, sulfide-, and antimonite-oxidizing abilities (Walczak et al., 2018; Lu et al., 2018). Bosea strains are mostly studied for the remediation purpose of metalloids such as arsenic and antimony removal/transformation (Lu et al., 2018; Xiang et al., 2022). Although Bosea species have been studied across different environments and for various purposes, studies focusing on their nitrogen metabolism remain limited. In particular, the nitrogen metabolism of Bosea sp. PAMC26642 has not yet been explored.
The main aim of this study was to have a deeper understanding of the bacteria Bosea sp. PAMC26642; to conduct the genome analysis of Bosea sp. PAMC26642 strain; furthermore, to compare the strain with other Bosea species from the same genus with the use of various bioinformatics tools and software; to determine the unique genes/enzymes that allow them to adapt to the extreme environment; and to focus primarily on nitrogen metabolic enzymes and the associated nitrate assimilation pathway. The findings from the study will lay the foundation for leveraging cold-adapted microorganisms like Bosea sp. PAMC26642 to address environmental challenges and promote ecosystem resilience in a changing climate. Furthermore, the potential bacteria can be used in the future for environmental, agricultural, and biotechnological purposes.
Bosea sp. PAMC26642 was isolated from the Arctic Lichen Stereocaulon sp., collected in Svalbard, Norway (78°55′N, 11°56′E), by the Korea Polar Research Institute (KOPRI, Incheon, Republic of Korea). The detailed procedure of the isolation of Bosea sp. PAMC26642 was reported by Kim et al. (2013). After the collection of lichen, a segment of the lichen thallus was excised with sterile scissors or knife and subjected to vertexing for 10 min in sterilized 0.85% NaCl solution, which was subsequently discarded, and the procedure was repeated two times. Afterward, the tissue was further disrupted using a mortar in the same NaCl solution. After the washing steps, the disrupted tissue was spread onto Bennett’s vitamin agar, comprising 10.0 g D-glucose, 1.0 g yeast extract, 2.0 g peptone, 1.0 g beef extract, 1.0 mL vitamins, 1.0 L distilled water, and 16.0 g agar. The plates were incubated at 28°C for 15 to 21 days, allowing for the growth of bacterial colonies. Subsequently, the colonies were subcultured three times to isolate a pure culture. The final pure culture of Bosea sp. PAMC26642 was preserved at −80°C in 20% glycerol. After storage, the culture was utilized by selecting the appropriate medium and temperature according to the experimental design.
For complete genome sequencing, genomic DNA was extracted from Bosea sp. PAMC26642, using a QIAamp DNA Mini Kit (Qiagen Inc., Valencia, CA), determined the quantity and purity using an Agilent 2,100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Genome sequencing was performed using PacBio RS II single-molecule real-time (SMRT) sequencing technology (Pacific Biosciences, Menlo Park, CA). SMRTbell library inserts (10 kb) were sequenced using SMRT cells. Raw sequence data were generated from 98,259 reads and 1,308,437,307 bp that were assembled de novo by using the hierarchical genome-assembly process (HGAP) protocol (Chin et al., 2013) and RS HGAP Assembly 2 in SMRT analysis version 2.3 software (Pacific Biosciences; https://github.com/PacificBiosciences/SMRT-Analysis). The complete genome sequence has been deposited at GenBank/EMBL/DDBJ under the accession numbers CP014301 and CP014302, and the complete genome sequence of Bosea sp. PAMC26642 has already been reported by Kang et al. (2016).
A total of 11 strains of Bosea with a complete genome sequence, including strain of interest, Bosea sp. PAMC26642 were retrieved from the NCBI nucleotide database1 on 30 August 2023. Functional annotation of the genome of our strain and reference strains was carried out by Rapid Annotation using a Subsystem Technology (RAST) server. The RAST server enables the identification and annotation of genes along with their associated functions as well as coding DNA sequences (CDSs) (Aziz et al., 2008) available at https://rast.nmpdr.org on 14 February 2024. Furthermore, Bosea sp. PAMC26642 as well as other reference strains were mined for the presence of genes/proteins having roles in nitrogen metabolism, stress resistance, and cold adaptation.
A total of 11 strains of Bosea with a complete genome sequence, including our strain, were retrieved from the NCBI nucleotide database (see text footnote 1) on 30 August 2023. A phylogenetic tree was constructed using 16S rRNA sequences of the complete genomes of Bosea strains together with other Bosea strains obtained from the NCBI. These 16S rRNA sequences were aligned using MUSCLE, MEGA 11, and a neighbor-joining method (Edgar, 2004a; Edgar, 2004b; Tamura et al., 2021; Qi et al., 2004; Felsenstein, 1992; Tamura et al., 2004). A maximum composite likelihood model was used to construct the phylogenetic tree. Branch numbers represent percentages of bootstrap values in 1000 sampling replicates. Similarly, a phylogenetic tree based on housekeeping genes such as dnaK, recA, gyrB, and trpB was also prepared.
The genomic sequence of Bosea sp. PAMC26642 strain was uploaded to a free bioinformatics platform, TYGS2 for the genome-based taxonomic analysis, accessed on 4 January 2024 (Meier-Kolthoff and Göker, 2019). The phylogenetic tree was reconstructed using FastME 2.1.6.1, including SPR post-processing from the GBDP (Lefort et al., 2015). Branch support was inferred from 100 pseudo-bootstrap replicates each. ANI is used to compare the genomic similarity between two microbial genomes. It is very important for species identification as it provides a high-resolution metric for species delineation, especially in bacteria and archaea. Furthermore, ANI provides a more robust, objective measure and detects even small genomic differences. It allows for fine-scale distinctions between closely related species or strains. ANI values were derived from the ANI tool3 (Yoon et al., 2017). Moreover, the genome-to-genome distance calculation web server4 (Lee et al., 2016). OrthoANI values were calculated using the Orthologous Average Nucleotide Identity Tool (OAT) software (Lee et al., 2016).
From the assembled genome, sequences of Bosea sp. PAMC26642 were taken, and the nitrate assimilatory genes were predicted, and annotated by BLASTP. The protein sequences were searched against the Nr and Swiss-Prot databases to find the sequences with the most identities for multiple sequence alignments. Multiple sequence alignments of all the proteins were conducted using Clustal Omega (Sievers and Higgins, 2018) and were subjected to ESPript 35 (Robert and Gouet, 2014). To gain a better understanding of the gene’s function and evolution, the protein sequences were subjected to domain analysis using the InterPro database https://www.ebi.ac.uk/interpro/ (provides functional analysis of proteins by classifying them into families and predicting domains and important sites) (Blum et al., 2021).
The pathway for nitrogen metabolism in the strain was studied with the KEGG database. 3D structures of these analyzed proteins were predicted by the online program PHYRE 2.0 Server6 in the intensive mode (Kelley et al., 2015) accessed on 14 September 2023. Protein sequences were used to search against the ExPDB template library. Sequences with the most identities were used for model generation. All 3D images were generated and colored by rainbow from N to C terminals (Bienfait and Ertl, 2013).
For the wet-lab experiment standardized, high-quality culture media and reagents were used. All the instruments were calibrated regularly according to the manufacturer’s instructions and for precise measurement. The isolated strains were cultured in R2A broth (MB cell Ltd., Seoul, Republic of Korea) at three different temperatures at 15°C, 25°C, and 37°C to check their growth. In addition to that, the strain was also tested for their growth in nitrate broth containing peptone (5 gm), meat extract (3 gm), and KNO3 (1 gm) with pH 7.0. A nitrate reduction assay was performed based on the bacterial ability to reduce nitrate to nitrite by using a standard operating procedure. The presence of nitrite can be detected with specific reagents such as reagent A (sulfanilic acid 8 gm/l, glacial acetic acid 286 mL/L, and demineralized water 714 mL/L) and reagent B (glacial acetic acid 286 mL/L, N, N-dimethyl-1-napthaylamine 6 mL/L, and demineralized water 714 mL/L), which produce a color change. For confirmation of nitrate reduction, zinc dust was also added. For quality control measures, both positive and negative controls were used. The strain Bosea sp. PAMC26642 (test organism) was cultured in nitrate broth, including an abiotic control without any microorganisms and positive control with Escherichia coli strain at three different temperatures for 2 days at 15°C, 25°C, and 37°C; 1 mL of bacterial culture from different temperatures was measured by using a spectrophotometer (Biochrome, Libra S35PC, Cambridge, UK). A uniform OD600 having a consistent bacterial cell density was taken in test tubes. A few drops of reagents A and B were added. The change in color to red from a colorless solution was monitored. For confirmation, a pinch of zinc dust was added to the tube with reagents A and B. All the experiments were performed in triplicates and are reproducible.
The complete genome sequence of all 11 strains, including our strain, after annotation from RAST analysis, was also compared in terms of genes that are involved in different stress adaptation mechanisms such as oxidative stress, heavy metal resistance, and salt stress.
A circular map of Bosea sp. PAMC26642 and a subsystem distribution based on the RAST SEED analysis of Bosea sp. PAMC26642 are shown in Figures 1, 2, respectively. Bosea sp. PAMC26642 and other strains of Bosea were compared. The general genome features and genomic information for all 11 strains are summarized in Tables 1, 2, respectively. All the compared strains have high GC content and are nearly similar.
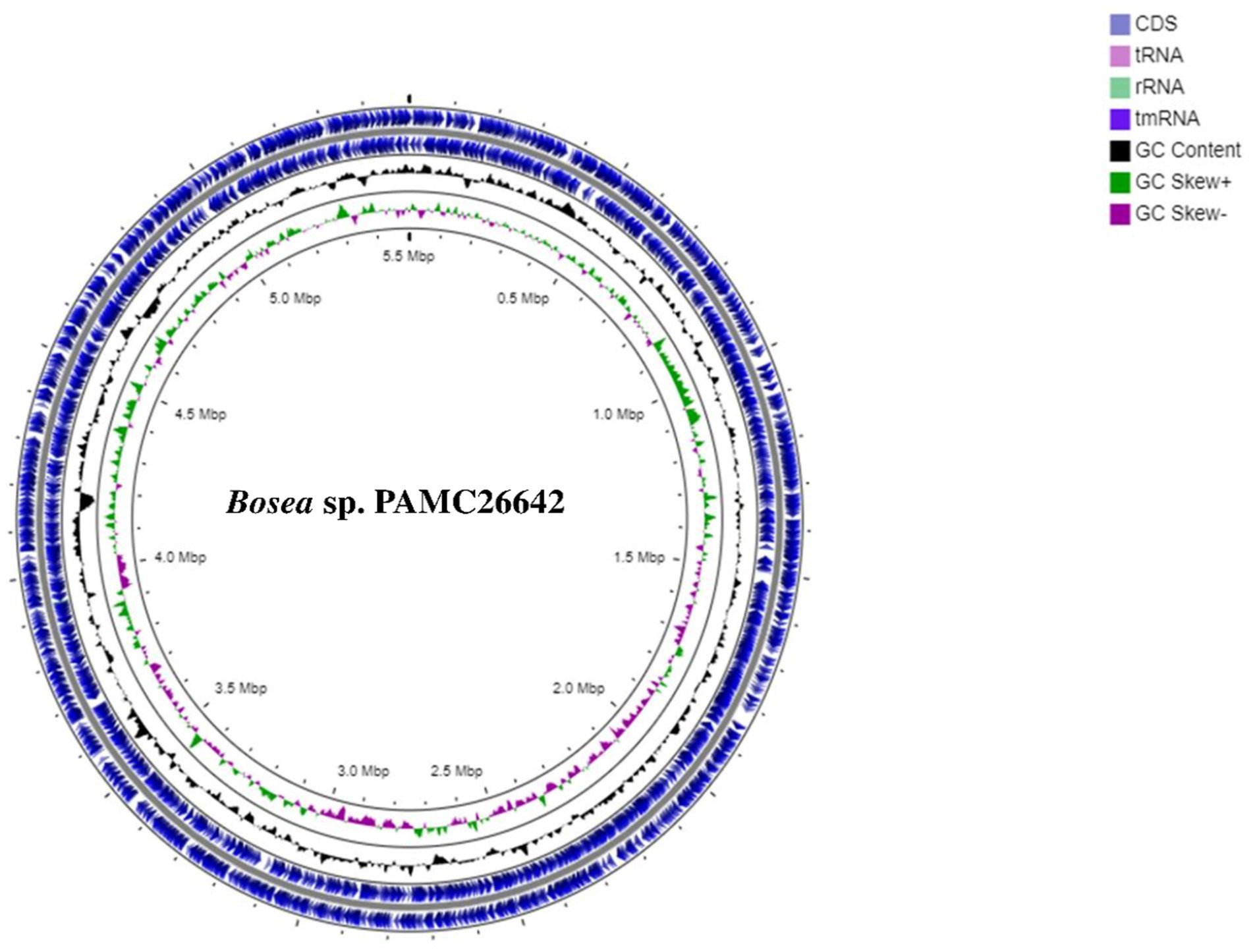
Figure 1. Circular map of Bosea sp. PAMC26642. Circular representation of genome with basic features including CDS and tRNA distributions, rRNA, tmRNA, GC content, GC Skew+, and GC Skew-from the outer circle to the inner circle.
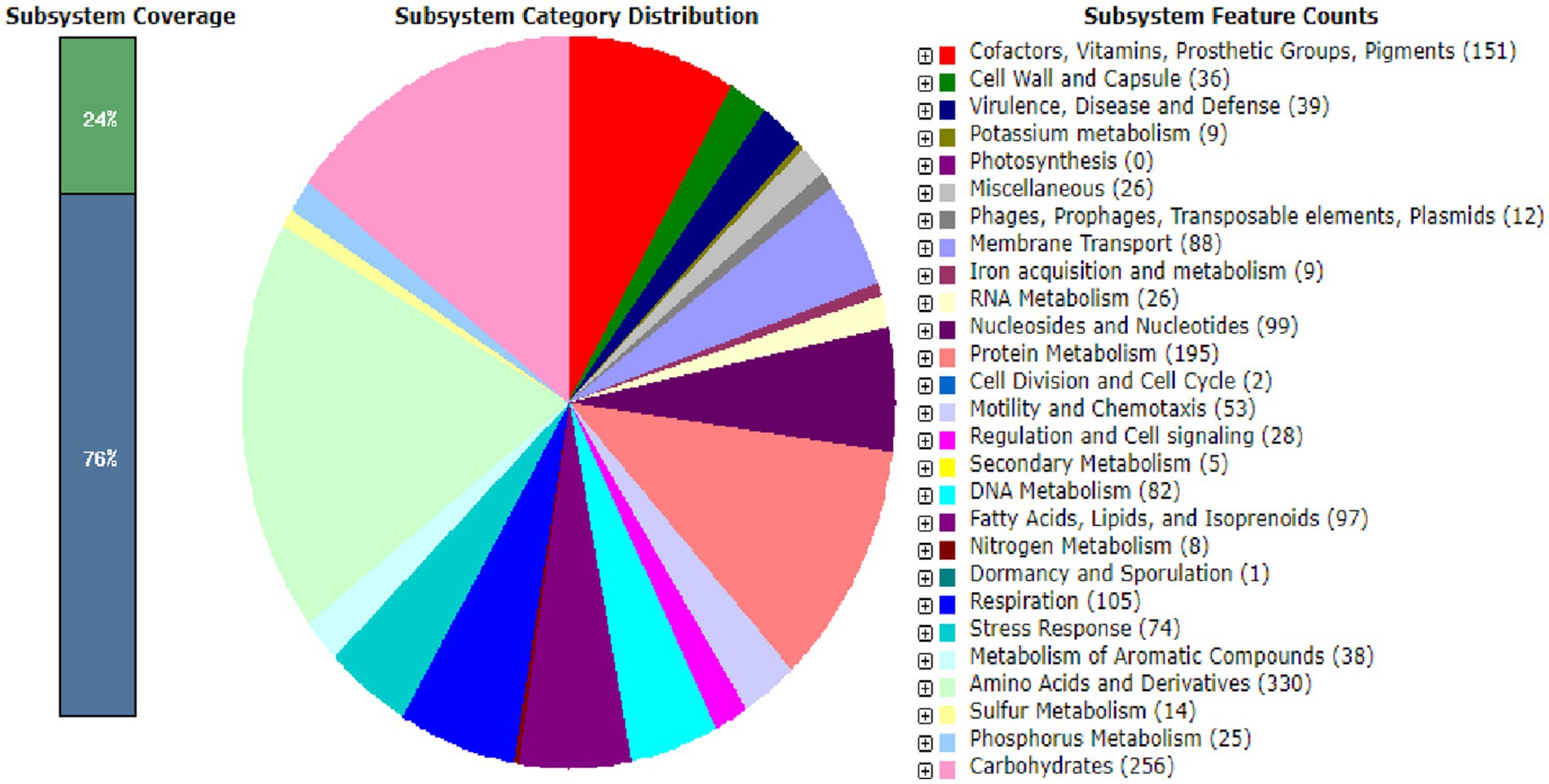
Figure 2. Subsystem distribution based on the RAST SEED analysis of Bosea sp. PAMC26642. The number of protein-coding genes (in parenthesis) are predicted to be involved in that cellular process.
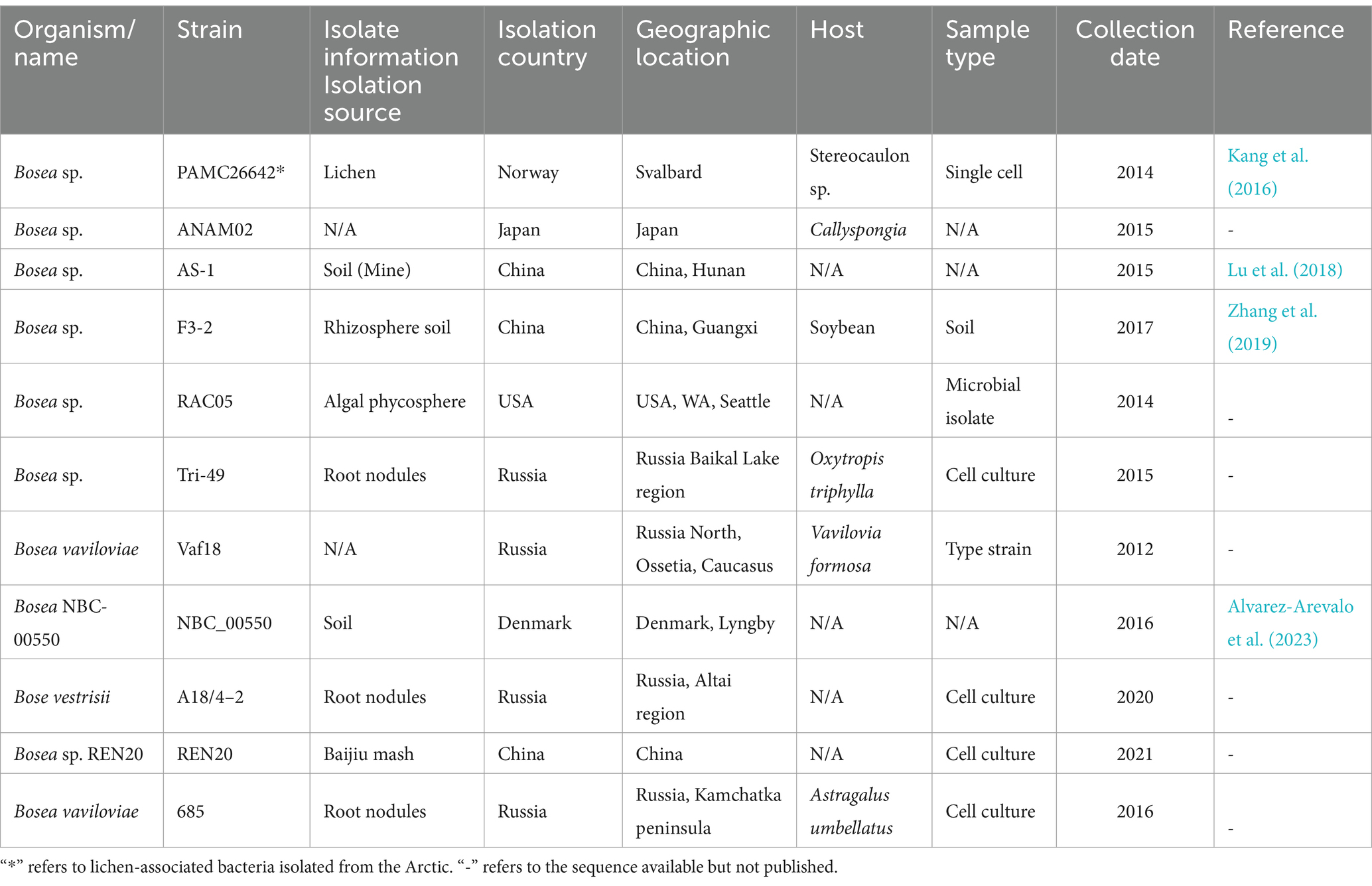
Table 1. General information and isolation sources of all the Bosea strains having a complete genome available in the NCBI database to date.
A phylogenetic tree was constructed from the 16S rRNA sequences of the complete genomes of Bosea strains as shown in Figure 3A. Similarly, a phylogenetic tree based on different housekeeping genes such as rpoB, gyrB, atpD, dnaK, recA, and trpB is shown in Supplementary Figure S1. Phylogenetic studies using ANI values reflect more effectively the functional relationships involving strains as compared to 16S rRNA sequence studies (Chung et al., 2018).
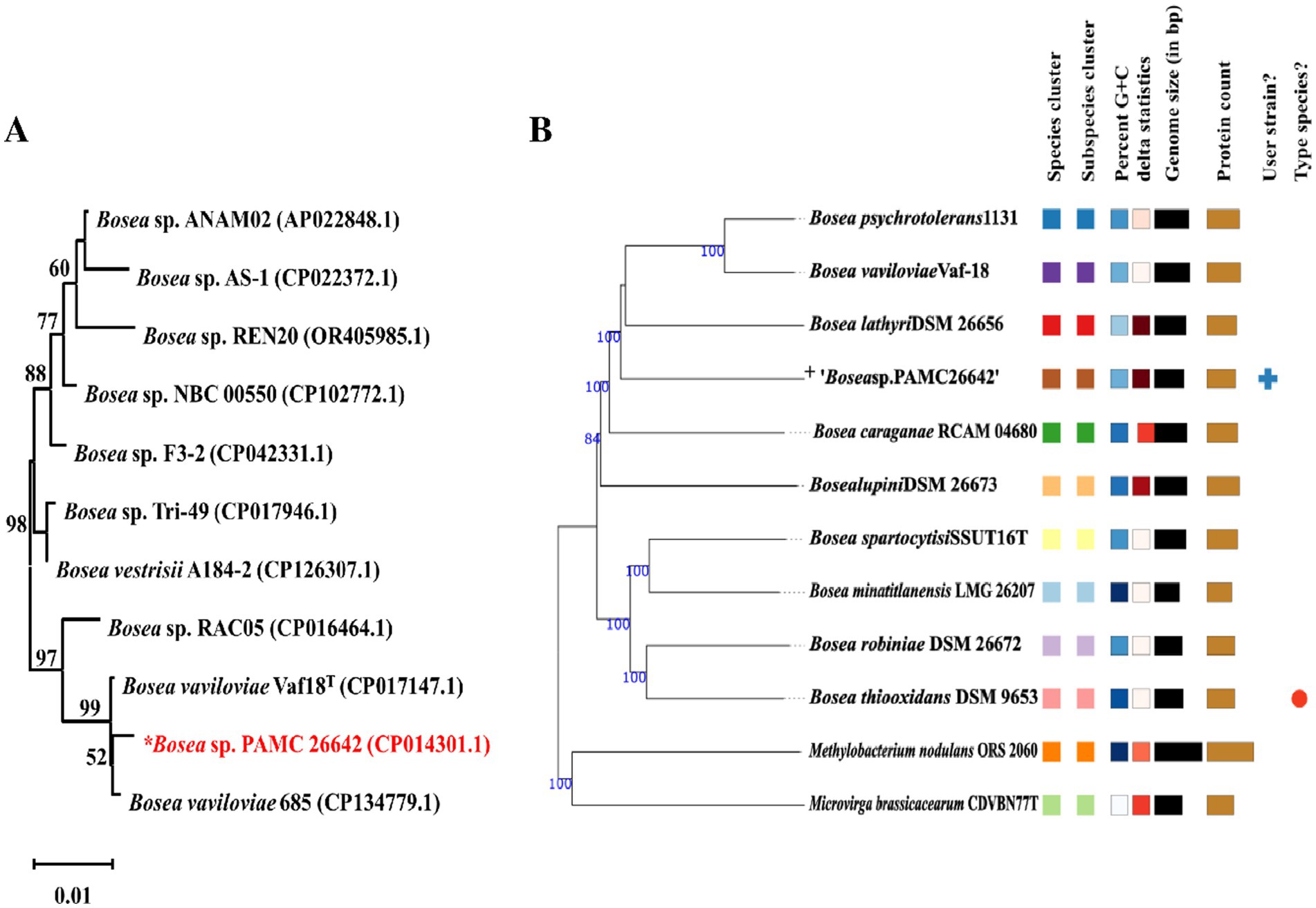
Figure 3. (A) Neighbor-joining phylogenetic trees of 16S rRNA gene of Bosea sp. without an outgroup. Percentages in the bootstrap test are from 1,000 replicates. (B) Phylogenomic tree based on genome sequences in the TYGS tree inferred with FastME 2.1.6.1 from GBDB. The branch lengths are scaled in terms of the GBDP distance formula d5. “+” denote Bosea sp. PAMC26642. Different colors are provided to indicate species and subspecies clusters. The same color denotes the same species cluster.
Genome Blast Distance Phylogeny (GBDP) analysis was calculated using the TYGS. Phylogenomic tree was shown in Figure 3B. TYGS showed that the ANI value obtained with the complete genome of Bosea sp. PAMC26642 is lower than the atypical 95–96% ANI value than the other closely related strain (Figure 4). This confirms that the Bosea sp. PAMC26642 might be a potential new species (Meier-Kolthoff and Göker, 2019; Goris et al., 2007; Richter and Rosselló-Móra, 2009; Meier-Kolthoff et al., 2013). In general, bacterial comparative genome analysis uses the ANI methods. As shown in Figure 4, each ANI value ranged from 79.26 to 100% between the bacteria genomes. Thus, we confirm that comparative genome results are lower than the common ANI values of 92–94. The ANI analysis shows the average nucleotide identity of all bacterial orthologous genes shared between any two genomes. It offers a robust resolution between bacterial strains of the same or closely related species (i.e., species showing 80–100% ANI) (Goris et al., 2007). However, ANI values do not represent genome evolution because orthologous genes can vary widely between the compared genomes. Nevertheless, ANI closely reflects the traditional microbiological concept of DNA–DNA hybridization relatedness for defining species, so many researchers use this method as it considers the fluid nature of the bacterial gene pool and implicitly considers shared functions (Jain et al., 2018). Thus, the ANI value is below the 90% threshold, indicating that the genome of Bosea sp. PAMC26642 has diverged significantly and provides insights into their evolutionary history and relationships.
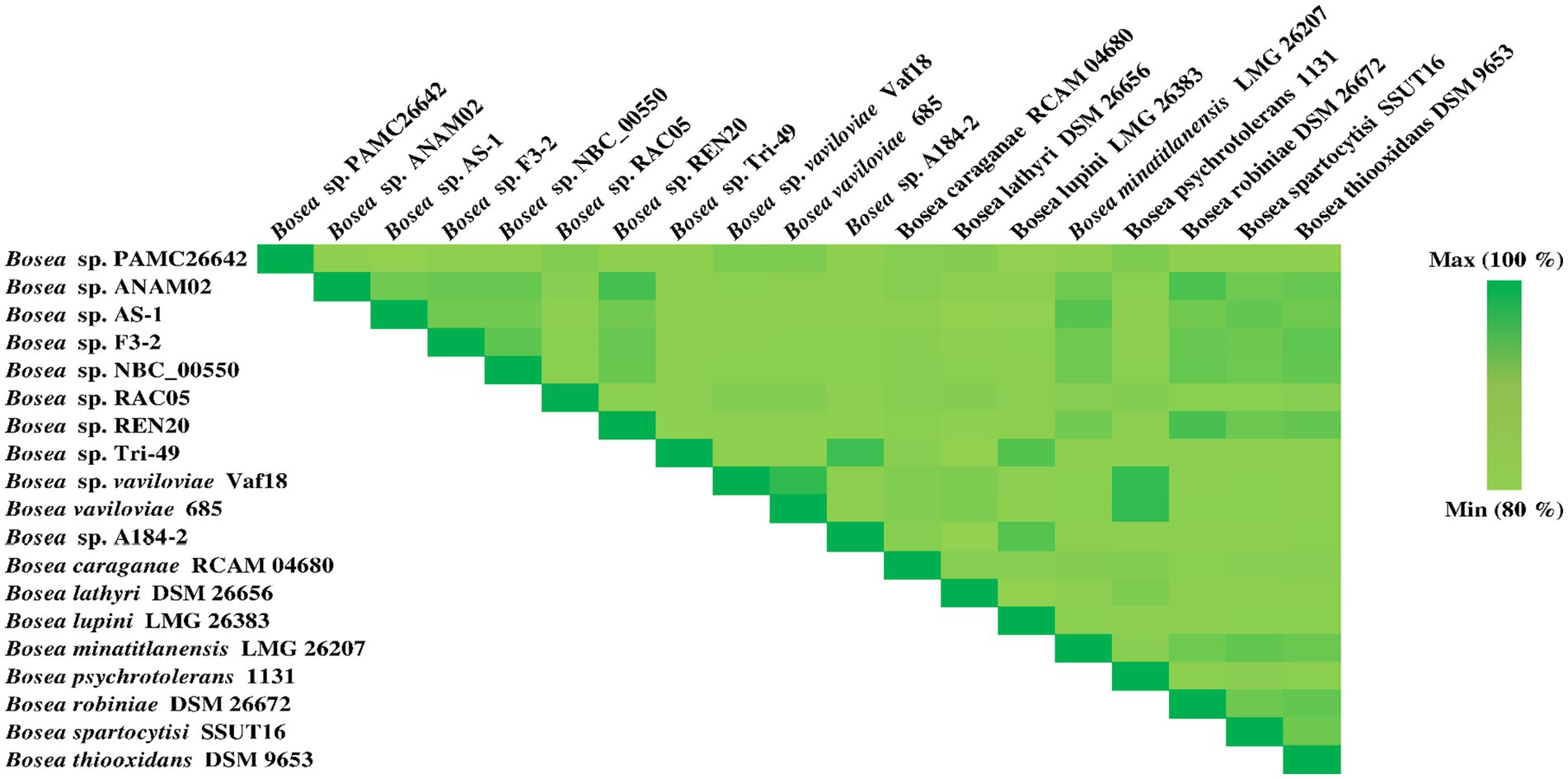
Figure 4. Heatmap generated using the OrthoANI values calculated from OAT software for Bosea sp. PAMC26642 with other closely related Bosea sp. including the type strain.
The nitrate assimilatory proteins are summarized in Table 3. The protein sequences that were searched against the Nr database to find the top sequences with the most identities for the following multiple sequence alignments showed that nitrate reductase of Bosea sp. PAMC26642 showed high identities with Bosea vaviloviae (WP_069690485.1) of 86% (Safronova et al., 2015), Bosea sp. OK403 (WP_092176404.1) of 85% (Li et al., 2023), and Bosea lathyri (WP_244595725.1) of 85%. Similarly, for nitrite reductase, our strain showed the highest similarities with Bosea sp. Tri-44 (WP_129155718.1) of 91%, Bosea lathyri (WP_103874740.1) of 91%, and Bosea sp. Root483D1 (WP_057188657.1) of 88% (Karimi et al., 2020). For glutamine synthetase, our strain showed similarities of 98% with Bosea sp. 124 (WP_108049945.1), 98% with Bosea sp. lathyri (WP_103871513.1), and 97% with (WP_054207853.1), for glutamate synthase 93% similarities with Bosea sp. AAP35 (WP_197279720.1), 92% with Bosea sp. R86505 (WP_376984921), and 77% with Bradyrhizobium sp. LTSPM299 (WP_245322060.1), for glutamate dehydrogenase 88% similarities with Bosea vaviloviae (WP_069689058.1), 88% similarities with Bosea psychrotolerans (WP_103721262.1) (Albert et al., 2019), and 87% similarities with Bosea lathyri (WP_200828109.1) (De Meyer and Willems, 2012).
Furthermore, the protein sequences (nitrate reductase, nitrite reductase, glutamate synthase, glutamine synthetase, and glutamate dehydrogenase) were searched against the Swiss-Prot database to find the top sequences with the most identities are summarized in Supplementary Table S1. Multiple sequence alignment and InterProScan (software package that allows sequences to be scanned against InterPro’s member database signature) of glutamate dehydrogenase protein revealed the presence of ACT1 domain, ACT2 domain, ACT3, and catalytic domain found in all five bacteria (Bosea sp. PAMC26642, Mycolicibacterium smegmatis MC2 155, Halomonas elongata DSM 2581, Mycobacterium tuberculosis H37Rv, and Pseudomonas aeruginosa PAO1) compared, including Bosea sp. PAMC26642. The ACT domains have been reported to play a significant role in allosteric regulation and structural stability. The catalytic domain is involved in the activity characteristics of GDH, i.e., the reversible oxidative deamination of glutamate to α-ketoglutarate and ammonia, which is a central step in nitrogen metabolism (Lázaro et al., 2021; Sharkey et al., 2013; Kawakami et al., 2007; Plaitakis et al., 2017) (see Supplementary Tables S2.1, S5.1 for more detail). The presence of these domains in all bacteria analyzed implies these domains are essential for GDH functionality and likely share similar metabolic strategies. Similarly, a comparison of multiple sequence alignment of glutamate synthase protein in all five bacteria (Bosea sp. PAMC26642, Pyrococcus furiosus DSM 3638, Geobacter sulfurreducens KN400, Escherichia coli K-12, Halomonas elongata DSM 2581) revealed the presence 4Fe-4S ferredoxin-type iron–sulfur binding domain in Bosea sp. PAMC26642 and Escherichia coli K-12. This domain facilitates the transfer of electrons from reduced ferredoxin or NADPH to the enzyme active site, where the reductive conversion of glutamine-derived ammonia and 2-oxoglutarate into two molecules of glutamate occurs. The NADPH binding motif (conserved GXGXXG sequence) was identified in the glutamate synthase protein of all five compared bacteria including our strain. However, one amino acid was different in our strain. The details are summarized in Supplementary Table S2.2 and Supplementary Figure S5.2. The NADPH binding motif is important for protein function (Vanoni and Curti, 1999; Vanoni and Curti, 2008; Morandi et al., 2000).
Multiple sequence alignment and InterProScan of glutamine synthetase protein revealed the presence of glutamine synthetase (GS) beta-grasp domain and catalytic domain (see Supplementary Table S2.3; Supplementary Figure S5.1.3 for more detail), which are present in all compared bacteria (Bosea sp. PAMC26642, Bradyrhizobium diazoefficiens USDA 110, Azorhizobium caulinodans ORS 571, Sinorhizobium meliloti 1,021, Rhizobium leguminosarum bv. Viciae). The beta-grasp domain of glutamine synthetase plays an important role in protein stability, functionality, and protein–protein interactions (Burroughs et al., 2007). The preservation of the beta-grasp of glutamine synthetase domain among the bacteria including Bosea sp. PAMC26642 indicates they are evolutionarily conserved to maintain their functionality. The catalytic domain performs the enzyme core function. It catalyzes the ATP-dependent synthesis of glutamine from glutamate and ammonium ions.
Multiple sequence alignment and InterProScan of nitrate reductase protein of five bacteria compared (Bosea sp. PAMC26642, Klebsiella oxytoca, O33732.2, Shewanella frigidimarina NCIMB 400, Synechococcus elongatus PCC 7942 = FACHB-805, Synechocystis sp. PCC 6803 substr. Kazusa) reveals the presence of MopB-Nitrate-R-NapA-like domain in all compared bacteria. This domain is reported to be found typically in components of the bacterial nitrate reductase (Nap) complex, especially in the Nap A subunit, which is crucial for nitrate reduction. MopB-Nitrate-R-NapA-like domain facilitates interactions between MoCo and other cofactors, ensuring efficient electron flow to reduce nitrate. MopB-CT was identified in all except Klebsiella oxytoca, O33732.2. MopB-CT-Nitrate-R-NapA-like domain stabilizes the structure of the catalytic subunit. In addition, the Molybdop-Fe4S4–2 domain was also identified in all compared bacteria. The core function of this domain has been reported to transfer electrons and stabilize the incorporation of the 4Fe-4S cluster and molybdenum cofactor into the protein structure, ensuring proper enzyme functionality (Sparacino-Watkins et al., 2014; Coelho and Romao, 2015; Moreno-Vivián et al., 1999). The details of domain comparisons are summarized in Supplementary Table S2.4 and Supplementary Figure S5.4. Multiple sequence alignment and InterProScan of nitrite reductase protein reveal the presence of nitrite/sulfite reductase ferredoxin-like half domain present in all five compared bacteria (Bosea sp. PAMC26642, Synechococcus elongatus PCC 7942, Leptolyngbya laminosa, Mycobacterium tuberculosis CDC1551 and Mycobacterium avium subsp. paratuberculosis K-10). The details regarding the comparison of domains are summarized in Supplementary Table S2.5 and Supplementary Figure S5.5.
The genome analysis of Bosea sp. PAMC26642 showed the presence of nitrogen metabolic enzymes, transcription factors, and transporters. Among the three different nitrate-reducing systems (Nas, Nar, and Nap) reported in prokaryotes (Moreno-Vivián et al., 1999; González et al., 2006), the assimilatory pathway was identified in Bosea sp. PAMC26642. Nitrate reductase (EC 1.7.99.4), NAD(P)H-nitrite reductase (EC 1.7.1.4), ferrodoxin-nitrite reductase (EC 1.7.7.1), glutamine synthetase (EC 6.3.1.2), glutamate synthase (EC 1.4.1.13), and glutamate dehydrogenase (EC 1.4.1.2), which catalyzes the reversible conversion between 2-oxoglutarate/ammonium and glutamate using NAD(H) or NADP(H) as a coenzyme (Smith et al., 1975) was also identified. In addition to that, cyanate lyase (EC 4.2.1.104), an enzyme responsible for catalyzing the decomposition of cyanate in a bicarbonate-dependent reaction yielding carbamate, which spontaneously decarboxylates to ammonia and carbon dioxide (Johnson and Anderson, 1987), was also identified. Furthermore, carbonic anhydrase (EC 4.2.1.1) and nitronate monooxygenase (EC 1.13.12.16) were also identified. In addition, nitrate/nitrite transporter substrate-binding protein (Figure 5) was also identified. Nitrate is transported into a cell by an active transport system. Nitrate is converted to nitrite with the function of nitrate reductase, followed by the reduction in nitrite to ammonia and then the conversion of ammonia to glutamine through nitrite reductase and glutamine synthetase. Finally, glutamine is transformed into glutamate by glutamate synthase. Both glutamine and glutamate are essential substrates for protein synthesis and energy metabolism. Glutamate is metabolized into ammonia and α-ketoglutarate with glutamate dehydrogenase.
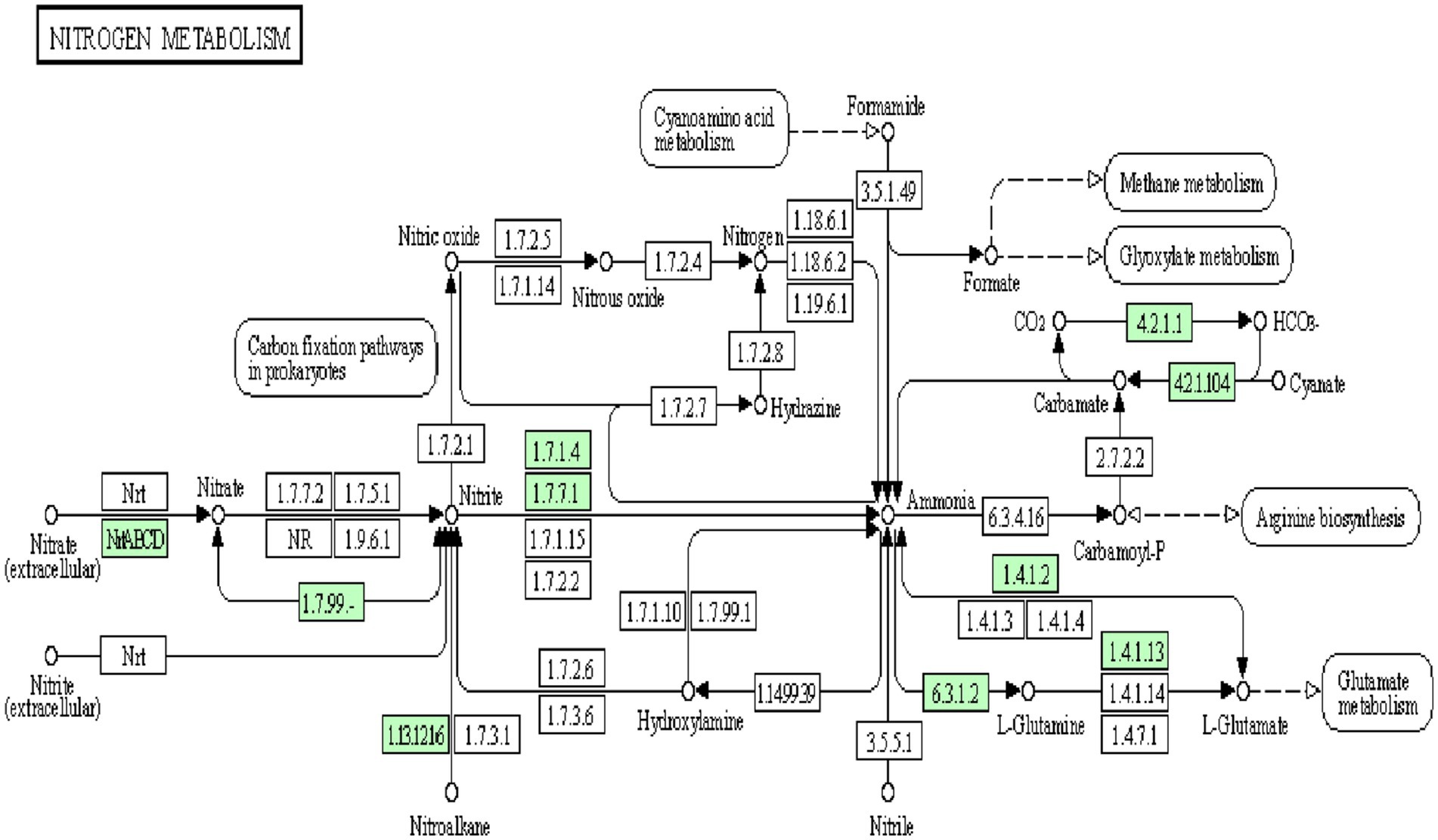
Figure 5. Snapshot of KEGG database showing nitrogen metabolism pathway in Bosea sp. PAMC26642. The enzymes available are highlighted in green.
A putative 3D model for all the nitrate assimilatory pathway proteins (nitrate reductase, nitrite reductase, glutamine synthetase, glutamate synthase, and glutamate dehydrogenase) is shown in Figure 6, and its information is shown in Table 4. Putative 3D structure modeling of (1) nitrate reductase was generated based on the sequence residues from 13–716 with 77% coverage, 100% confidence, and 35% identity against C2v45A (2.80 Å). (2) nitrite reductase was generated based on the sequence residues from 51 to 588 with 89% coverage, 100% confidence, and 30% identity against C1Zj8B (2.80 Å). (3) glutamine synthetase was generated based on the sequence residues from 3 to 468 with 99% coverage, 100% confidence, and 62% identity against C1fpyE (2.89 Å). (4) glutamate synthase was generated based on the sequence residues from 15 to 534 with 96% coverage, 100% confidence, and 31% identity against C1gthD (2.25 Å). (5) glutamate dehydrogenase was generated based on the sequence residues from 39 to 1,616 with 97% coverage, 100% confidence, and 39% identity against C7jsrA (6.27 Å).
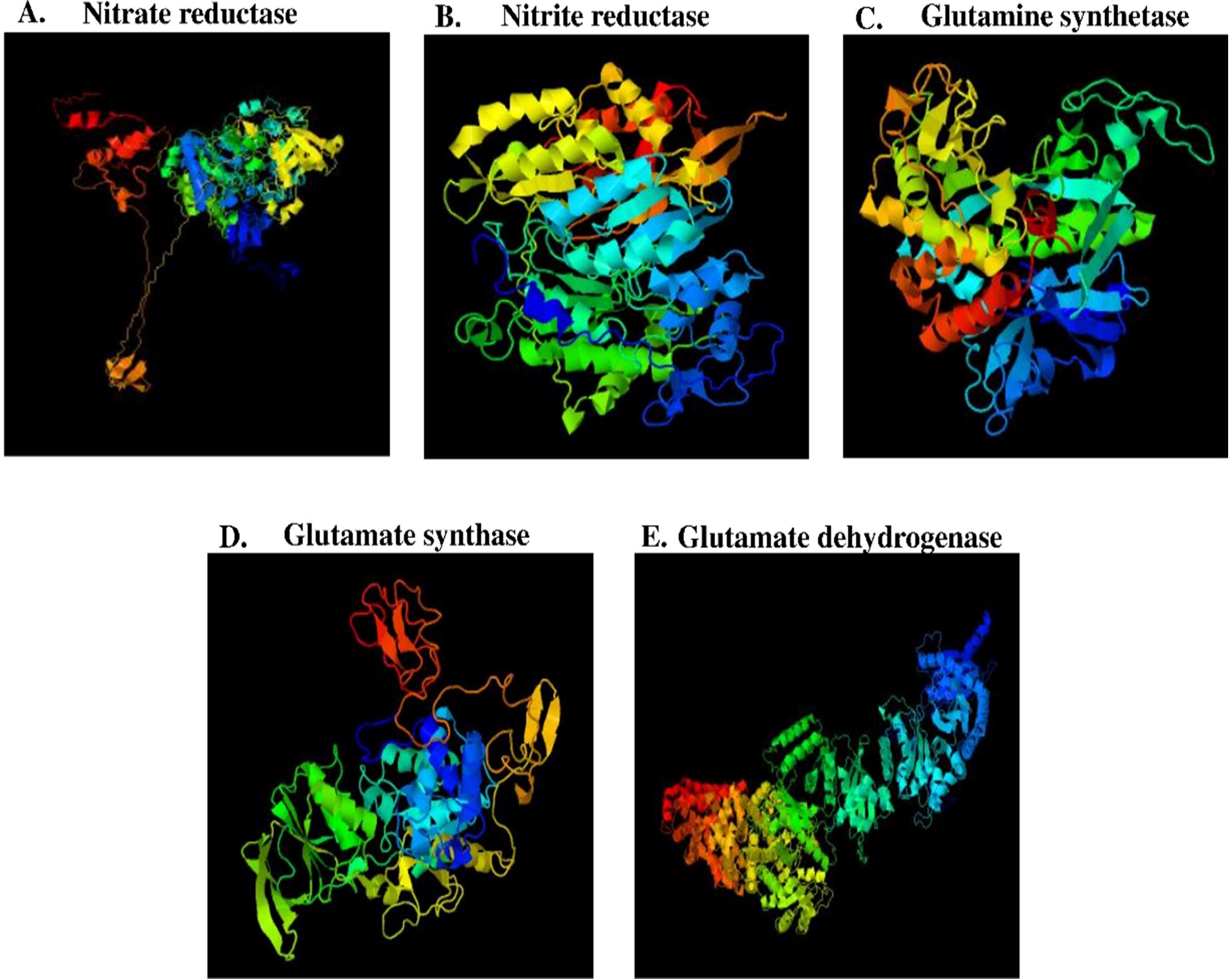
Figure 6. Putative 3D model of nitrate assimilatory proteins. (A) Nitrate reductase protein (B) Nitrite reductase protein (C) Glutamine synthetase protein, and (D) Glutamate synthase (E) Glutamate dehydrogenase protein.
The nitrate reduction assay was performed, which is based on the ability of bacteria to reduce nitrate to nitrite with the liberation of red color from a colorless solution. Bosea sp. PAMC26642 was tested for the nitrate reduction assay, including an abiotic control (without microorganisms) and E. coli as a positive control at temperatures of 15°C and 25°C. Both Bosea sp. PAMC26642 and E. coli showed reductions of nitrate at 15°C and 25°C. However, an abiotic control did not show any color change (Figure 7). The ability of bacteria to reduce nitrate at specific temperatures is a key adaptive trait, crucial for their survival across various environments. Furthermore, the enzymes required for nitrate reduction, primarily nitrate reductase, have temperature-dependent activity. These enzymes either work inefficiently or cease to function at temperatures outside their optimal range. The bacteria able to reduce nitrate at certain temperatures have implications such as ecological niche specialization, which minimizes the competition with other microorganisms. In addition to that, they also influence nutrient availability, overall ecosystem balance, survival, and adaptation to climate change. The result of nitrate reduction by Bosea sp. PAMC26642 at different temperatures (15°C and 25°C) revealed that the activity of nitrate reductase is functioning appropriately.
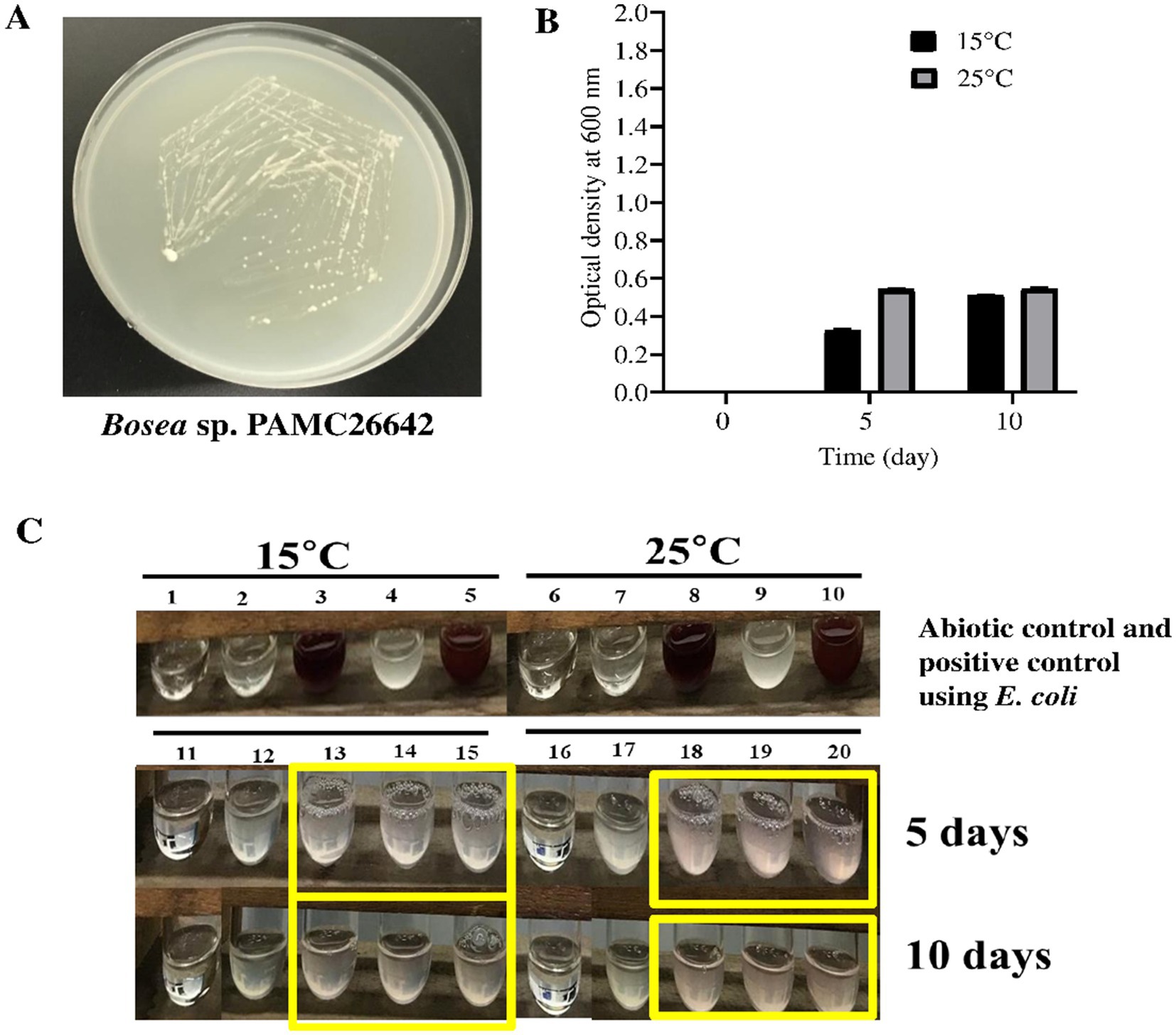
Figure 7. (A) Bosea sp. PAMC26642 in R2A media. (B) Growth of bacteria in nitrate media at different times and at different temperatures. The black box represents nitrate media containing cells at 15°C, and grey box represents nitrate media containing cells at 25°C. (C) Nitrate reduction assay at 15°C and 25°C. 1 and 6, abiotic control at 15°C and 25°C; 2 and 7, abiotic control with reagent A and reagent B at 15°C and 25°C; 3 and 8, abiotic control with reagent A, reagent B, and addition of zinc at 15°C and 25°C; 4 and 9, E. coli without reagent A and reagent B at 15°C and 25°C; 5 and 10, positive control using E. coli with reagent A and reagent B at 15°C and 25°C; 11 and 16, abiotic control at 15°C and 25°C; 12 and 17, Bosea culture without reagent A and reagent B; and 13, 14, 15, 18, 19, and 20, Bosea culture with reagent A and reagent B at 15°C and 25°C, respectively. Nitrate assays were performed at days 5 and 10. Experiments were performed in triplicates.
This distinguishing feature of Bosea sp. PAMC26642 (in this study) is that they have a nitrogen assimilation pathway (nitrogen cycling) and its associated enzymes, which is important for adaptation in cold areas because nitrogen availability is a limiting factor in polar regions of the Arctic due to cold temperatures and nutrient-poor environment.
Aside from nitrogen metabolic enzymes, other enzymes involved in various stress adaptation mechanisms such as oxidative stress (superoxide dismutase, catalase, and thiol peroxidase), heavy metal resistance (arsenate reductase, ArsH), temperature stress (Csp and Hsp), and osmotic stress (OmpR) were identified in Bosea sp. PAMC26642. The comparison of this strain with other strains of the same genus in terms of genes predicted to be involved in different stress adaptation mechanisms after the annotation with the RAST server is shown in Table 5.
Among different types of stress temperature stress, either low or high temperature, bacteria have been reported to have evolved several mechanisms for coping with temperature stress and adapting to changing environmental conditions such as the production of cold shock protein (Csp) during low temperature. In addition, these Csps have been reported to contribute to osmotic, oxidative, starvation, pH, and ethanol stress tolerance (Keto-Timonen et al., 2016). In addition to Csps, there are other heat shock proteins (HSP) that are reported to be involved during bacterial high temperature-related environmental stress. HSP is found in various types of bacteria (Maleki et al., 2016), major Hsps are molecular chaperons GroEl-GroES, DNAJ, and GrpE, which were reported for regulation of folding as well as for heat shock response such as in E. coli (Nishihara et al., 1998; Arsène et al., 2000) and were identified in all the Bosea strains including HtpX (Kornitzer et al., 1991), whereas Hsp20 (Bakthisaran et al., 2015) was identified only in some Bosea strain. Furthermore, sigma factors play a crucial role in bacterial adaptability and survival, because of these factors the bacteria can swiftly change their gene expression profiles in response to environmental signals, optimizing their metabolic, growth, and stress responses. In addition, multiple sigma factors in bacteria have been reported to provide a mechanism for global coordinate regulation of classes of genes (Burgess, 2013). Different types of sigma factors such as RpoN, RpoE, RpoH, and RpoD have been reported in bacteria such as E. coli K-12, Pseudomonas aeruginosa, and Shewanella oneidensis MR-1 (Shimada et al., 2021; Potvin et al., 2008; Dai et al., 2015). Among these sigma factors, RpoD, RpoH, and RpoN were identified in all Bosea strains (Table 5). Osmotic stress-related protein, EnvZ/OmpR two-component system, which mediates osmotic stress response in several Gram-negative bacteria (Yuan et al., 2011), and EnvZ was identified in all Bosea strains. Superoxide dismutase (SOD), which converts superoxide radicals to the less toxic H2O2 and water. They were reported to be varied in microbes such as cytoplasmic Mn-SOD (encoded by sodA), Fe-SOD (encoded by sodB), and periplasmic Cu/Zn-SOD (encoded by sodC) (Seib et al., 2006; Sheehan et al., 2000; Najmuldeen et al., 2019). Among these three SODs, Mn-SOD was identified in all the Bosea strains and Cu/Zn-SOD was not identified in any of the Bosea strains including our strain. Catalases known for their protection against H2O2 (Katsuwon and Anderson, 1989) are classified into three groups, monofunctional heme-containing catalases (KatE), heme-containing catalase-peroxidase (KatG), and manganese-containing catalases. Among these catalases, KatE were identified in Bosea sp. PAMC26642, Bosea sp. NBC_00550, and Bosea sp. REN20. Manganese-catalase was identified in Bosea sp. NBC_00550 and Bosea vestrisii.
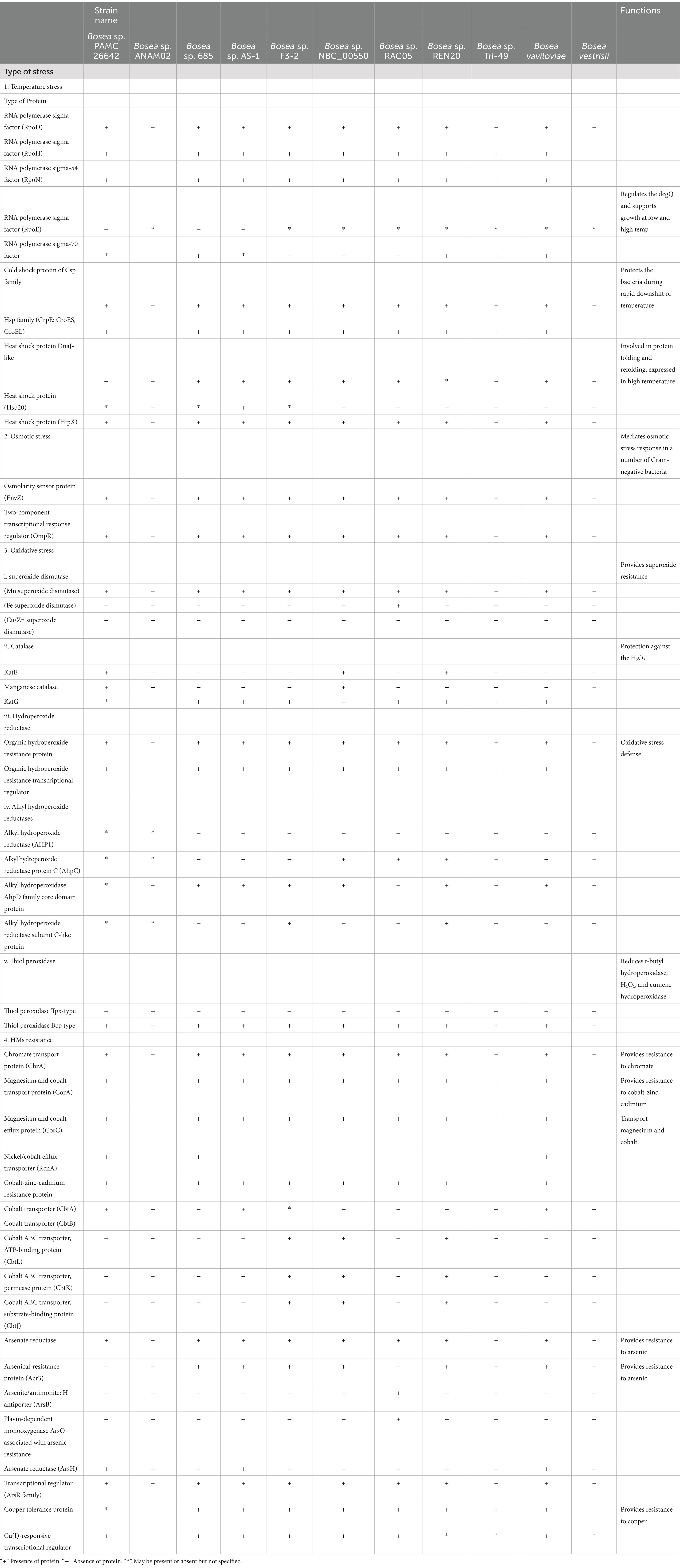
Table 5. Comparison of proteins that are involved in different stress adaptation mechanisms of all the Bosea strains having a complete genome.
Organic hydroperoxide resistance protein (Ohr) and organic hydroperoxide resistance regulator (OhrR), which are critical for organic peroxide resistance (Si et al., 2020) and could be involved in the detoxification of organic hydroperoxide (Shea and Mulks, 2002) were identified in all the Bosea strains. Peroxiredoxins are widespread in bacteria and are of two types of thiol peroxidase (Tpx), and the bacterioferritin co-migratory protein (Bcp) has been reported to play a role in the protection against oxidative stress, particularly that caused by excess oxygen and exogenous peroxidase as reported in Campylobacter jejuni (Atack et al., 2008). Furthermore, Tpx and Bcp appear to be able to use a wide variety of peroxide as substrates in vitro, such as hydrogen peroxide, organic peroxides, and lipid peroxides (Cha et al., 2004; Rho et al., 2006; Wang et al., 2005). Thiol peroxidase Bcp type was identified in all the Bosea strains. Heavy metal(loid)s toxicity has been considered as a global issue, and they have been considered as a serious environmental problem. Bacteria have been reported to have developed a resistance toward heavy metal(loid)s because of the presence of certain resistance genes. ArsH protein (organo arsenical oxidase) responsible for arsenic biotransformation (Chang et al., 2018) was identified in Bosea sp. AS-1, Bosea sp. Tri-49, and Bosea vaviloviae including our strain.
Similarly, the Acr3 protein belongs to the bile/arsenate/riboflavin transporter (BART) superfamily and is reported to be widespread in bacteria, archaea, fungi, and some plants (Mansour et al., 2007) was identified in all Bosea strains except Bosea sp. PAMC26642. Chromate transport protein (ChrA), membrane protein, and member of the chromate ion transporter protein (CHR) superfamily that confers resistance to the toxic ion chromate through the energy-dependent chromate efflux from the cytoplasm (Díaz-Pérez et al., 2007), was identified in all Bosea strains. RcnA, a nickel and cobalt-resistant protein reported in E. coli (Rodrigue et al., 2005), was identified in our strain, Bosea sp. 685, Bosea vaviloviae and Bosea vestrisii. HM-related protein (CorA) as reported by Afordoanyi et al. (2023) formerly known as magnesium and cobalt transport protein for mediating both influx and efflux of Mg2+ in Salmonella typhimurium and E. coli (Smith et al., 1998) were identified in all the Bosea strains. In addition, magnesium and cobalt transport protein and arsenate reductase were also identified in all the Bosea strains.
This study provides valuable insights into the nitrogen metabolic potential of lichen-associated Bosea sp. PAMC26642 from the polar region. It also sheds light on the strain’s stress adaptation mechanism. In this study, this strain has been compared to other species in the Bosea genus using comprehensive bioinformatics tools and wet-lab assays. The key enzymes of the assimilatory nitrogen metabolic pathway such as nitrate reductase, nitrite reductase, glutamine synthetase, glutamate synthase, and glutamate dehydrogenase were identified in Bosea sp. PAMC26642. In particular, the strain demonstrated nitrate reduction ability at 15°C and 25°C, highlighting its metabolic adaptability to cold environments. In addition, stress adaptation enzymes suggest resilience to oxidative stress, heavy metal resistance, temperature fluctuations, and osmotic stress. These findings not only expand our understanding of Bosea biodiversity and nitrogen metabolic capacity but also highlight its potential applications in ecosystem monitoring, nitrate bioremediation, and environment resilience strategies. This finding lays the foundation for leveraging cold-adapted microorganisms such as Bosea sp. PAMC26642 to address environmental challenges and promote ecosystem resilience in a changing climate. Overall, these findings will provide new knowledge gained in key areas like an enhanced understanding of nitrogen cycling in polar ecosystems, insight into cold-adapted metabolic adaptability, stress resilience mechanism in polar microorganisms, potential for bioremediation and environmental application and framework for ecosystem monitoring, and climate adaptation strategy.
All supporting data and protocols have been provided within the article or through Supplementary files. The datasets analyzed during the current study are available in the NCBI repository, accession numbers: NZ_CP014301.1 for Bosea sp. PAMC26642, NZ_ AP022848.1 for Bosea sp. ANAM02, NZ_CP042331.1 for Bosea sp. F3-2, NZ_CP022372.1 for Bosea sp. AS-1, NZ_CP016464.1 for Bosea sp. RAC05, NZ_CP017946.1 for Bosea sp. Tri-49, NZ_CP017147.1 for Bosea vaviloviae Vaf18, NZ_CP126307.1 for Bosea vestrisii A18/4-2, NZ_CP102772.1 for Bosea sp. NBC_00550, NZ_ CP134779.1 for Bosea vaviloviae 685, and NZ_OR405985.1 for Bosea sp. REN20.
AK: Conceptualization, Formal analysis, Investigation, Methodology, Writing – original draft. S-RH: Investigation, Writing – original draft. JL: Investigation, Writing – original draft. T-JO: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the project titled “Development of potential antibiotic compounds using polar organism resources (20200610)”, funded by the Ministry of Oceans and Fisheries, Korea.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1505699/full#supplementary-material
PDB, protein data bank; NCBI, National Center for Biotechnology Information; SMRT, single-molecule real time; HGAP, hierarchical genome assembly process; RAST, rapid annotation using subsystem technology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
1. ^https://www.ncbi.nlm.nih.gov/
3. ^www.ezbiocloud.net/tools/ani
Afordoanyi, D. M., Akosah, Y. A., Shnakhova, L., Saparmyradov, K., Diabankana, R. G. C., and Validov, S. (2023). Biotechnological key genes of the Rhodococcus erythropolis MGMM8 genome: genes for bioremediation, antibiotics, plant protection, and growth stimulation. Microorganisms 12:88. doi: 10.3390/microorganisms12010088
Albert, R. A., McGuine, M., Pavlons, S. C., Roecker, J., Bruess, J., Mossman, S., et al. (2019). Bosea psychrotolerans sp. nov., a psychrotrophic alphaproteobacterium isolated from Lake Michigan water. Int. J. Syst. Evol. Microbiol. 69, 1376–1383. doi: 10.1099/ijsem.0.003319
Alvarez-Arevalo, M., Sterndorff, E. B., Faurdal, D., Mourched, A. S., Charusanti, P., Jørgensen, T. S., et al. (2023). Complete, circular genome sequence of a Bosea sp. isolate from soil. Microbiol. Resour. Announc. 12, e00360–e00323. doi: 10.1128/MRA.00360-23
Arsène, F., Tomoyasu, T., and Bukau, B. (2000). The heat shock response of Escherichia coli. Int. J. Food Microbiol. 55, 3–9. doi: 10.1016/S0168-1605(00)00206-3
Atack, J. M., Harvey, P., Jones, M. A., and Kelly, D. J. (2008). The Campylobacter jejuni thiol peroxidases Tpx and Bcp both contribute to aerotolerance and peroxide-mediated stress resistance but have distinct substrate specificities. J. Bacteriol. 190, 5279–5290. doi: 10.1128/jb.00100-08
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9, 1–15. doi: 10.1186/1471-2164-9-75
Bakthisaran, R., Tangirala, R., and Rao, C. M. (2015). Small heat shock proteins: role in cellular functions and pathology. Biochim. Biophys. Acta-Proteins Proteom. 1854, 291–319. doi: 10.1016/j.bbapap.2014.12.019
Bienfait, B., and Ertl, P. (2013). JSME: a free molecule editor in JavaScript. J. Cheminform. 5, 1–6. doi: 10.1186/1758-2946-5-24
Blum, M., Chang, H. Y., Chuguransky, S., Grego, T., Kandasaamy, S., Mitchell, A., et al. (2021). The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49, D344–D354. doi: 10.1093/nar/gkaa977
Burgess, R. R. (2013). “Sigma factors,” in Brenner’s encyclopedia of genetics, 2nd edn. eds. S. Maloy and K. Hughes (San Diego, CA: Academic Press).
Burroughs, A. M., Balaji, S., Iyer, L. M., and Aravind, L. (2007). Small but versatile: the extraordinary functional and structural diversity of the β-grasp fold. Biol. Direct 2, 1–28. doi: 10.1186/1745-6150-2-18
Cha, M. K., Kim, W. C., Lim, C. J., Kim, K., and Kim, I. H. (2004). Escherichia coli periplasmic thiol peroxidase acts as lipid hydroperoxide peroxidase and the principal antioxidative function during anaerobic growth. J. Biol. Chem. 279, 8769–8778. doi: 10.1074/jbc.M312388200
Chang, J. S., Yoon, I. H., and Kim, K. W. (2018). Arsenic biotransformation potential of microbial arsH responses in the biogeochemical cycling of arsenic-contaminated groundwater. Chemosphere 191, 729–737. doi: 10.1016/j.chemosphere.2017.10.044
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Chung, W. H., Kang, J., Lim, M. Y., Lim, T. J., Lim, S., Roh, S. W., et al. (2018). Complete genome sequence and genomic characterization of Lactobacillus acidophilus LA1 (11869BP). Front. Pharmacol. 9:83. doi: 10.3389/fphar.2018.00083
Coelho, C., and Romao, M. J. (2015). Structural and mechanistic insights on nitrate reductases. Protein Sci. 24, 1901–1911. doi: 10.1002/pro.2801
Dai, J., Wei, H., Tian, C., Damron, F. H., Zhou, J., and Qiu, D. (2015). An extracytoplasmic function sigma factor-dependent periplasmic glutathione peroxidase is involved in oxidative stress response of Shewanella oneidensis. BMC Microbiol. 15, 34–12. doi: 10.1186/s12866-015-0357-0
Das, S. K., and Mishra, A. K. (1996). Transposon mutagenesis affecting thiosulfate oxidation in Bosea thiooxidans, a new chemolithoheterotrophic bacterium. J. Bacteriol. 178, 3628–3633. doi: 10.1128/jb.178.12.3628-3633.1996
Das, S. K., Mishra, A. K., Tindall, B. J., Rainey, F. A., and Stackebrandt, E. (1996). Oxidation of thiosulfate by a new bacterium, Bosea thiooxidans. (strain BI-42) gen. Nov., sp. nov.: analysis of phylogeny based on chemotaxonomy and 16S ribosomal DNA sequencing. Int. J. Syst. Evol. Microbiol. 46, 981–987. doi: 10.1099/00207713-46-4-981
De Meyer, S. E., and Willems, A. (2012). Multilocus sequence analysis of Bosea species and description of Bosea lupini sp. nov., Bosea lathyri sp. nov. and Bosea robiniae sp. nov., isolated from legumes. Int. J. Syst. Evol. Microbiol. 62, 2505–2510. doi: 10.1099/ijs.0.035477-0
De Scally, S. Z., Makhalanyane, T. P., Frossard, A., Hogg, I. D., and Cowan, D. A. (2016). Antarctic microbial communities are functionally redundant, adapted and resistant to short term temperature perturbations. Soil Biol. Biochem. 103, 160–170. doi: 10.1016/j.soilbio.2016.08.013
Díaz-Pérez, C., Cervantes, C., Campos-García, J., Julián-Sánchez, A., and Riveros-Rosas, H. (2007). Phylogenetic analysis of the chromate ion transporter (CHR) superfamily. FEBS J. 274, 6215–6227. doi: 10.1111/j.1742-4658.2007.06141.x
Edgar, R. C. (2004a). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5, 1–19. doi: 10.1186/1471-2105-5-113
Edgar, R. C. (2004b). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Felsenstein, J. (1992). Phylogenies from restriction sites: a maximum-likelihood approach. Evolution 46, 159–173. doi: 10.1111/j.1558-5646.1992.tb01991.x
Fernández-Valiente, E., Quesada, A., Howard-Williams, C., and Hawes, I. (2001). N2-fixation in cyanobacterial mats from ponds on the McMurdo ice shelf. Antarctica. Microb. Ecol. 42, 338–349. doi: 10.1007/s00248-001-1010-z
Garrido-Benavent, I., Pérez-Ortega, S., Durán, J., Ascaso, C., Pointing, S. B., Rodríguez-Cielos, R., et al. (2020). Differential colonization and succession of microbial communities in rock and soil substrates on a maritime antarctic glacier forefield. Front. Microbiol. 11:126. doi: 10.3389/fmicb.2020.00126
González, P. J., Correia, C., Moura, I., Brondino, C. D., and Moura, J. J. G. (2006). Bacterial nitrate reductases: molecular and biological aspects of nitrate reduction. J. Inorg. Biochem. 100, 1015–1023. doi: 10.1016/j.jinorgbio.2005.11.024
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Harper, C., Hayward, D., Wiid, I., and van Helden, P. (2008). Regulation of nitrogen metabolism in Mycobacterium tuberculosis: a comparison with mechanisms in Corynebacterium glutamicum and Streptomyces coelicolor. IUBMB Life 60, 643–650. doi: 10.1002/iub.100
Hayashi, K., Tanabe, Y., Fujitake, N., Kida, M., Wang, Y., Hayatsu, M., et al. (2020). Ammonia oxidation potentials and ammonia oxidizers of lichen–moss vegetated soils at two ice-free areas in East Antarctica. Microbes Environ. 35:n/a. doi: 10.1264/jsme2.ME19126
Hördt, A., López, M. G., Meier-Kolthoff, J. P., Schleuning, M., Weinhold, L. M., Tindall, B. J., et al. (2020). Analysis of 1, 000+ type-strain genomes substantially improves taxonomic classification of Alphaproteobacteria. Front. Microbiol. 11:468. doi: 10.3389/fmicb.2020.00468
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Johnson, W. V., and Anderson, P. M. (1987). Bicarbonate is a recycling substrate for cyanase. J. Biol. Chem. 262, 9021–9025. doi: 10.1016/S0021-9258(18)48040-4
Kang, S., Han, S. R., Oh, T. J., and Park, H. (2016). Complete genome sequence of thiosulfate-oxidizing Bosea sp. strain PAMC26642 isolated from an Arctic lichen. J. Biotechnol. 223, 38–39. doi: 10.1016/j.jbiotec.2016.02.033
Karimi, E., Geslain, E., KleinJan, H., Tanguy, G., Legeay, E., Corre, E., et al. (2020). Genome sequences of 72 bacterial strains isolated from Ectocarpus subulatus: a resource for algal microbiology. Genome Biol. Evol. 12, 3647–3655. doi: 10.1093/gbe/evz278
Katsuwon, J., and Anderson, A. J. (1989). Response of plant-colonizing pseudomonads to hydrogen peroxide. Appl. Environ. Microbiol. 55, 2985–2989. doi: 10.1128/aem.55.11.2985-2989.1989
Kawakami, R., Sakuraba, H., and Ohshima, T. (2007). Gene cloning and characterization of the very large NAD-dependent l-glutamate dehydrogenase from the psychrophile Janthinobacterium lividum, isolated from cold soil. J. Bacteriol. 189, 5626–5633. doi: 10.1128/jb.00496-07
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Keto-Timonen, R., Hietala, N., Palonen, E., Hakakorpi, A., Lindström, M., and Korkeala, H. (2016). Cold shock proteins: a minireview with special emphasis on Csp-family of enteropathogenic Yersinia. Front. Microbiol. 7:1151. doi: 10.3389/fmicb.2016.01151
Kim, M. K., Oh, T. J., and Park, H. (2013). Antimicrobial properties of the bacterial associates of the Arctic lichen Stereocaulon sp. Afr. J. Microbiol. Res. 7, 3651–3657. doi: 10.5897/AJMR12.1771
Kornitzer, D., Teff, D., Altuvia, S., and Oppenheim, A. B. (1991). Isolation, characterization, and sequence of an Escherichia coli heat shock gene, htpX. J. Bacteriol. 173, 2944–2953. doi: 10.1128/jb.173.9.2944-2953.1991
La Scola, B., Mallet, M. N., Grimont, P. A., and Raoult, D. (2003). Bosea eneae sp. nov., Bosea massiliensis sp. nov. and Bosea vestrisii sp. nov., isolated from hospital water supplies, and emendation of the genus Bosea (Das et al. 1996). Int. J. Syst. Evol. Microbiol. 53, 15–20. doi: 10.1099/ijs.0.02127-0
Lázaro, M., Melero, R., Huet, C., López-Alonso, J. P., Delgado, S., Dodu, A., et al. (2021). 3D architecture and structural flexibility revealed in the subfamily of large glutamate dehydrogenases by a mycobacterial enzyme. Commun Biol. 4:684. doi: 10.1038/s42003-021-02222-x
Lee, I., Ouk Kim, Y., Park, S. C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Lefort, V., Desper, R., and Gascuel, O. (2015). FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 32, 2798–2800. doi: 10.1093/molbev/msv150
Li, X., Wang, J., Su, W., Li, C., Qu, G., Yuan, B., et al. (2023). Characterization and engineering of cephalosporin C acylases to produce 7-Aminocephalosporanic acid. Mol. Catal. 550:113595. doi: 10.1016/j.mcat.2023.113595
Lu, X., Zhang, Y., Liu, C., Wu, M., and Wang, H. (2018). Characterization of the antimonite-and arsenite-oxidizing bacterium Bosea sp. AS-1 and its potential application in arsenic removal. J. Hazard. Mater. 359, 527–534. doi: 10.1016/j.jhazmat.2018.07.112
Magalhães, C. M., Machado, A., Frank-Fahle, B., Lee, C. K., and Cary, S. C. (2014). The ecological dichotomy of ammonia-oxidizing archaea and bacteria in the hyper-arid soils of the Antarctic dry valleys. Front. Microbiol. 5:515. doi: 10.3389/fmicb.2014.00515
Makhalanyane, T. P., Valverde, A., Velázquez, D., Gunnigle, E., Van Goethem, M. W., Quesada, A., et al. (2015). Ecology and biogeochemistry of cyanobacteria in soils, permafrost, aquatic and cryptic polar habitats. Biodivers. Conserv. 24, 819–840. doi: 10.1007/s10531-015-0902-z
Maleki, F., Khosravi, A., Nasser, A., Taghinejad, H., and Azizian, M. (2016). Bacterial heat shock protein activity. J. Clin. Diagn. Res. 10, BE01–BE03. doi: 10.7860/JCDR/2016/14568.7444
Mansour, N. M., Sawhney, M., Tamang, D. G., Vogl, C., and Saier, M. H. (2007). The bile/arsenite/riboflavin transporter (BART) superfamily. FEBS J. 274, 612–629. doi: 10.1111/j.1742-4658.2006.05627.x
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 14, 1–14. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182. doi: 10.1038/s41467-019-10210-3
Merrick, M. J., and Edwards, R. (1995). Nitrogen control in bacteria. Microbiol. Rev. 59, 604–622. doi: 10.1128/mr.59.4.604-622.1995
Morandi, P., Valzasina, B., Colombo, C., Curti, B., and Vanoni, M. A. (2000). Glutamate synthase: identification of the NADPH-binding site by site-directed mutagenesis. Biochemistry 39, 727–735. doi: 10.1021/bi9920329
Moreno-Vivián, C., Cabello, P., Martínez-Luque, M., Blasco, R., and Castillo, F. (1999). Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J. Bacteriol. 181, 6573–6584. doi: 10.1128/jb.181.21.6573-6584.1999
Najmuldeen, H., Alghamdi, R., Alghofaili, F., and Yesilkaya, H. (2019). Functional assessment of microbial superoxide dismutase isozymes suggests a differential role for each isozyme. Free Radic. Biol. Med. 134, 215–228. doi: 10.1016/j.freeradbiomed.2019.01.018
Nishihara, K., Kanemori, M., Kitagawa, M., Yanagi, H., and Yura, T. (1998). Chaperone coexpression plasmids: differential and synergistic roles of Dna K-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl. Environ. Microbiol. 64, 1694–1699. doi: 10.1128/AEM.64.5.1694-1699.1998
Ouattara, A. S., Assih, E. A., Thierry, S., Cayol, J. L., Labat, M., Monroy, O., et al. (2003). Bosea minatitlanensis sp. nov., a strictly aerobic bacterium isolated from an anaerobic digester. Int. J. Syst. Evol. Microbiol. 53, 1247–1251. doi: 10.1099/ijs.0.02540-0
Papale, M., Conte, A., Mikkonen, A., Michaud, L., La Ferla, R., Azzaro, M., et al. (2018). Prokaryotic assemblages within permafrost active layer at Edmonson point (northern Victoria land, Antarctica). Soil Biol. Biochem. 123, 165–179. doi: 10.1016/j.soilbio.2018.05.004
Plaitakis, A., Kalef-Ezra, E., Kotzamani, D., Zaganas, I., and Spanaki, C. (2017). The glutamate dehydrogenase pathway and its roles in cell and tissue biology in health and disease. Biology 6:11. doi: 10.3390/biology6010011
Potvin, E., Sanschagrin, F., and Levesque, R. C. (2008). Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 32, 38–55. doi: 10.1111/j.1574-6976.2007.00092.x
Qi, J., Luo, H., and Hao, B. (2004). CVTree: a phylogenetic tree reconstruction tool based on whole genomes. Nucleic Acids Res. 32, W45–W47. doi: 10.1093/nar/gkh362
Rho, B. S., Hung, L. W., Holton, J. M., Vigil, D., Kim, S. I., Park, M. S., et al. (2006). Functional and structural characterization of a thiol peroxidase from Mycobacterium tuberculosis. J. Mol. Biol. 361, 850–863. doi: 10.1016/j.jmb.2006.05.076
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 106, 19126–19131. doi: 10.1073/pnas.0906412106
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Rodrigue, A., Effantin, G., and Mandrand-Berthelot, M. A. (2005). Identification of rcnA (yohM), a nickel and cobalt resistance gene in Escherichia coli. J. Bacteriol. 187, 2912–2916. doi: 10.1128/jb.187.8.2912-2916.2005
Safronova, V. I., Kuznetsova, I. G., Sazanova, A. L., Kimeklis, A. K., Belimov, A. A., Andronov, E. E., et al. (2015). Bosea vaviloviae sp. nov., a new species of slow-growing rhizobia isolated from nodules of the relict species Vavilovia formosa (Stev.) fed. Antonie Van Leeuwenhoek 107, 911–920. doi: 10.1007/s10482-015-0383-9
Seib, K. L., Wu, H. J., Kidd, S. P., Apicella, M. A., Jennings, M. P., and McEwan, A. G. (2006). Defenses against oxidative stress in Neisseria gonorrhoeae: a system tailored for a challenging environment. Microbiol. Mol. Biol. Rev. 70, 344–361. doi: 10.1128/mmbr.00044-05
Sharkey, M. A., Oliveira, T. F., Engel, P. C., and Khan, A. R. (2013). Structure of NADP+-dependent glutamate dehydrogenase from Escherichia coli–reflections on the basis of coenzyme specificity in the family of glutamate dehydrogenases. FEBS J. 280, 4681–4692. doi: 10.1111/febs.12439
Shea, R. J., and Mulks, M. H. (2002). Ohr, encoding an organic hydroperoxide reductase, is an in vivo-induced gene in Actinobacillus pleuropneumoniae. Infect. Immun. 70, 794–802. doi: 10.1128/iai.70.2.794-802.2002
Sheehan, B. J., Langford, P. R., Rycroft, A. N., and Kroll, J. S. (2000). [cu, Zn]-superoxide dismutase mutants of the swine pathogen Actinobacillus pleuropneumoniae are unattenuated in infections of the natural host. Infect. Immun. 68, 4778–4781. doi: 10.1128/iai.68.8.4778-4781.2000
Shimada, T., Furuhata, S., and Ishihama, A. (2021). Whole set of constitutive promoters for RpoN sigma factor and the regulatory role of its enhancer protein NtrC in Escherichia coli K-12. Microb. Genom. 7:000653. doi: 10.1099/mgen.0.000653
Si, Y., Guo, D., Deng, S., Lu, X., Zhu, J., Rao, B., et al. (2020). Ohr and OhrR are critical for organic peroxide resistance and symbiosis in Azorhizobium caulinodans ORS571. Genes 11:335. doi: 10.3390/genes11030335
Sievers, F., and Higgins, D. G. (2018). Clustal omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145. doi: 10.1002/pro.3290
Smith, E. L., Austen, B. M., Blumenthal, K. M., and Nyc, J. F. (1975). “5 Glutamate dehydrogenases,” in The enzymes. Vol. 11. eds. P. D. Boyer (New York: Academic Press), 293–367.
Smith, R. L., Gottlieb, E., Kucharski, L. M., and Maguire, M. E. (1998). Functional similarity between archaeal and bacterial CorA magnesium transporters. J. Bacteriol. 180, 2788–2791. doi: 10.1128/jb.180.10.2788-2791.1998
Sparacino-Watkins, C., Stolz, J. F., and Basu, P. (2014). Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 43, 676–706. doi: 10.1039/c3cs60249d
Tamura, K., Nei, M., and Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. 101, 11030–11035. doi: 10.1073/pnas.0404206101
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Vanoni, M. A., and Curti, B. (1999). Glutamate synthase: a complex iron-sulfur flavoprotein. Cell. Mol. Life Sci. 55, 617–638. doi: 10.1007/s000180050319
Vanoni, M. A., and Curti, B. (2008). Structure–function studies of glutamate synthases: a class of self-regulated iron-sulfur flavoenzymes essential for nitrogen assimilation. IUBMB Life 60, 287–300. doi: 10.1002/iub.52
Walczak, A. B., Yee, N., and Young, L. Y. (2018). Draft genome sequence of Bosea sp. WAO an arsenite and sulfide oxidizer isolated from a pyrite rock outcrop in New Jersey. Stand. Genomic Sci. 13, 6–12. doi: 10.1186/s40793-018-0312-4
Wang, G., Olczak, A. A., Walton, J. P., and Maier, R. J. (2005). Contribution of the Helicobacter pylori thiol peroxidase bacterioferritin comigratory protein to oxidative stress resistance and host colonization. Infect. Immun. 73, 378–384. doi: 10.1128/iai.73.1.378-384.2005
Xiang, L., Liu, C., Liu, D., Ma, L., Qiu, X., Wang, H., et al. (2022). Antimony transformation and mobilization from stibnite by an antimonite oxidizing bacterium Bosea sp. AS-1. J. Environ. Sci. 111, 273–281. doi: 10.1016/j.jes.2021.03.042
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Yuan, J., Wei, B., Shi, M., and Gao, H. (2011). Functional assessment of EnvZ/OmpR two-component system in Shewanella oneidensis. PLoS One 6:e23701. doi: 10.1371/journal.pone.0023701
Keywords: Bosea sp., cold adaptation, comparative genomics, nitrogen metabolism, Gram-negative bacteria
Citation: Khanal A, Han S-R, Lee JH and Oh T-J (2025) Unraveling nitrogen metabolism, cold and stress adaptation in polar Bosea sp. PAMC26642 through comparative genome analysis. Front. Microbiol. 15:1505699. doi: 10.3389/fmicb.2024.1505699
Received: 03 October 2024; Accepted: 04 December 2024;
Published: 24 January 2025.
Edited by:
Xuejiao An, Jiangxi Agricultural University, ChinaReviewed by:
Zheng Lu, Hainan University, ChinaCopyright © 2025 Khanal, Han, Lee and Oh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tae-Jin Oh, dGpvaDM3ODJAc3VubW9vbi5hYy5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.