- 1Centre for Agricultural Resources Research, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Shijiazhuang, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Institute of Marine Sciences and Guangdong Provincial Key Laboratory of Marine Biotechnology, Shantou University, Shantou, China
- 4Agricultural Research Corporation (ARC), Ministry of Agriculture, Wad Madani, Sudan
- 5Xinjiang Key Laboratory of Desert Plant Roots Ecology and Vegetation Restoration, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences, Urumqi, China
Plants establish specific interactions with microorganisms, which are vital for promoting growth and resilience. Although advancements in microbiome modulation technologies show great potential for sustainable agriculture, several challenges have hindered the wider application of plant microbiomes in the field. These challenges may include inconsistent microbial colonization, competition with native microbiota, and environmental variability. Current strategies, while promising, often yield inconsistent results in real-world agricultural settings, highlighting the need for more refined approaches. Agricultural practices and plant genotypes significantly influence the composition and function of plant-associated microbiota. A data-driven strategy that incorporates genomic profiling, environmental assessments, and optimized delivery systems is essential for selecting effective microbial strains. Additionally, refining farming practices, such as crop rotation, intercropping, and reduced tillage, along with robust plant breeding programs, can greatly enhance crop health and productivity.
1 Introduction
Over the past few decades, research has unveiled the intricate and essential role of the plant microbiome in supporting plant growth, health, and resilience. The plant microbiome, encompassing diverse microbial communities associated with plant organs, includes bacteria, fungi, and archaea, with bacterial components being the most studied (Buée et al., 2009; Vorholt, 2012; Reinhold-Hurek et al., 2015). These microbial communities interact with the plant as a holobiont, which plays a critical role in maintaining plant productivity, particularly under changing environmental conditions (Berg et al., 2016; Brader et al., 2017; Lemanceau et al., 2017; Hassani et al., 2018).
As agriculture faces growing challenges such as climate change, resource depletion, and the increasing demand for sustainable practices, traditional reliance on chemical inputs like pesticides and fertilizers has raised concerns about their long-term environmental and economic impacts (Bhattacharyya and Jha, 2012). In response, the plant microbiome has emerged as a promising alternative to these inputs, offering a biologically driven approach to enhancing crop health and productivity. Microorganisms, particularly plant growth-promoting bacteria (PGPB), have demonstrated the capacity to improve nutrient uptake, stimulate plant growth, and enhance resistance to pathogens, positioning them as valuable tools for sustainable agriculture (Mitter et al., 2016). This review provides an up-to-date discussion on the potential of plant microbiome technology in addressing current challenges facing modern agriculture, with a focus on bacteria. To this end, we first summarize the knowledge on the diversity and functionality of the plant microbiome, and finally propose approaches to harness the microbiome for sustainable agriculture.
2 Plant microbiome: functionality and diversity
2.1 Belowground plant microbiome
Plants recruit their microbiome from various environmental reservoirs, including the rhizosphere, phyllosphere, spermosphere, anthosphere, and carposphere (Hardoim et al., 2015). The root microbiome is mainly horizontally transferred; that is, it comes from the soil environment, containing different microorganisms (Shade et al., 2017). Nevertheless, bacteria can also transmit vertically through seeds (Fierer, 2017). Microorganisms thrive in developing plant roots, and seeds are an important source of them (Truyens et al., 2015; Nelson, 2018). Plant root system presents a unique environmental niche for the soil microbial community, which settles in the root, rhizosphere and aerial part (Berendsen et al., 2012). One of the most significant mutualistic relationships in the root microbiome is the nodule formation between leguminous plants and nitrogen-fixing bacteria such as Rhizobium (Liu et al., 2021; Granada Agudelo et al., 2023). These nodules enhance nitrogen availability, which is fundamental to plant functionality and growth (Lindström and Mousavi, 2020), ultimately improving soil fertility and supporting sustainable agriculture (Ojija and Aloo, 2023). Nodule formation represents a classic example of a mutualistic interaction where both the plant and the microbes benefit (Haldar and Sengupta, 2015).
The rhizosphere, a thin layer of soil directly influenced by plant roots, is regarded as a hot zone of microbial activity and one of the most complicated ecosystems (Raaijmakers et al., 2009). Donn et al. (2015) demonstrated that bacterial composition of wheat rhizosphere changed across years and noticed the abundance of Pseudomonads and Actinobacteria was 10-fold greater in rhizosphere than that of bulk soil. Root exudates (e.g., amino acids, fatty acids, organic acids, phenolics, nucleotides, plant growth regulators, sugars, putrescine, vitamins, and sterols) are leading factors in terms of shifting microbial composition around roots, the so-called rhizosphere effect (Mendes et al., 2013; Rasmann and Turlings, 2016; Sasse et al., 2018; Zhalnina et al., 2018; Jiang et al., 2019). For example, benzoxazinoids (BXs), a class of defensive secondary metabolites attributed to maize roots, had a critical role in changing root-associated microbiota composition. Actinobacteria and Proteobacteria are shown to be affected by BXs compounds (Hu et al., 2018). Overall, phenotype and genotype variations in plant traits are the definitive explanation for this hypothesis, which can guide specific microbial communities.
A vast range of bacterial endophytes can successfully invade plant roots. Bacterial endophytes frequently enter root tissues via passive processes such as root fractures or lateral root emergence points, as well as active mechanisms (Kandel et al., 2017) (Figure 1). Endophytes’ colonization and transmission within plants are impacted by various factors, such as plant resource allocation and the endophytes’ colonization ability. Various bacterial taxa have been found to be able to colonize inside root tissues, such as Acidobacteria, Proteobacteria, Verrucomicrobia, Bacteroidetes, Chloroflexi, Planctomycetes, Gemmatimonadetes and Firmicutes which commonly occur in grapevine roots (Burns et al., 2015; Samad et al., 2017a; Correa-Galeote et al., 2018). Additionally, the study by Correa-Galeote et al. (2018) found that Proteobacteria, Firmicutes and Bacteroidetes are the predominant phyla within the maize roots and the history of soil cultivation substantially influenced the abundance of these phyla.
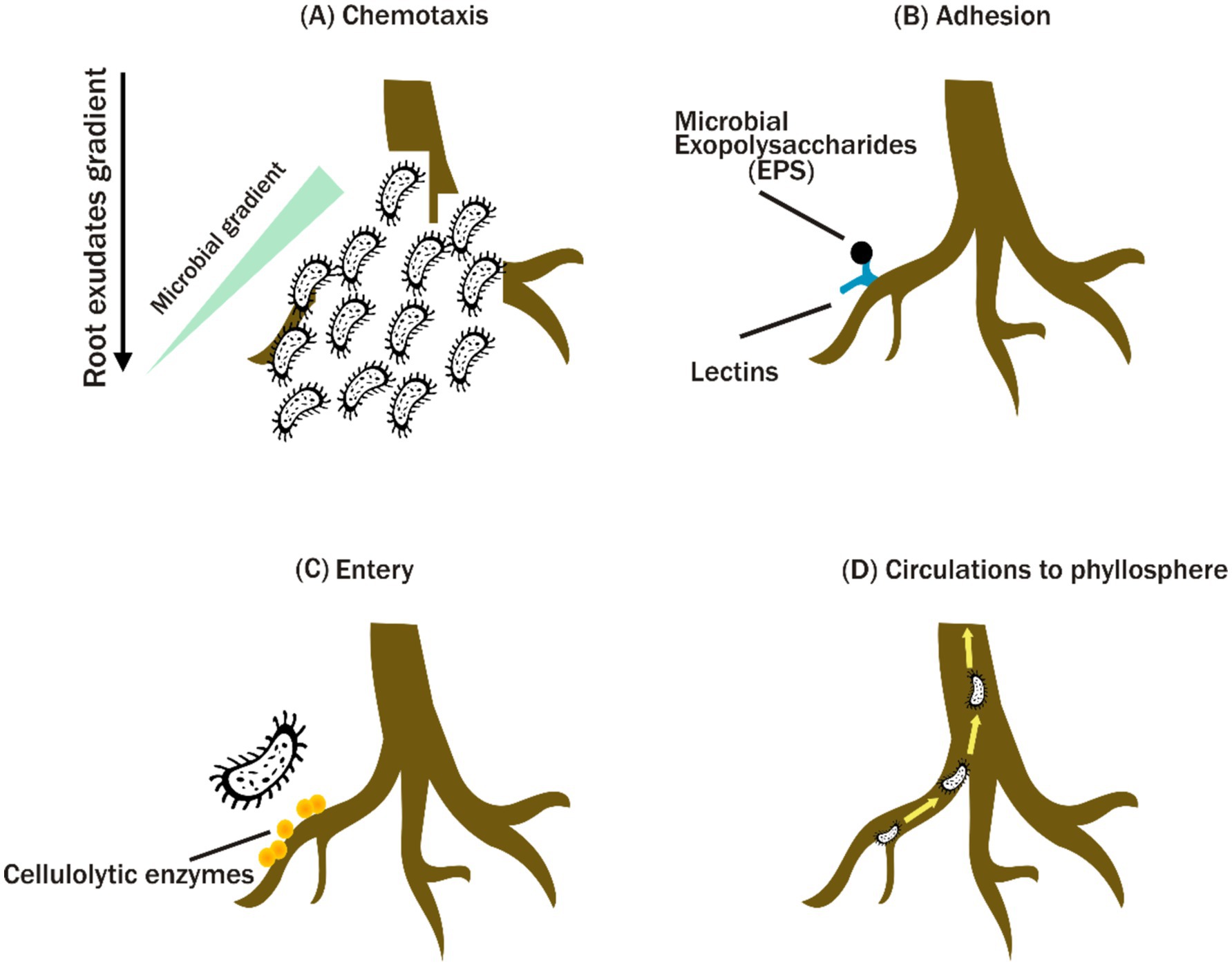
Figure 1. Mechanisms of plant microbiome colonization. Microbial recruitment in the rhizosphere is initiated by chemotaxis toward plant exudates (A). Microbes adhere to plant lectins via exopolysaccharides (EPS) (B), penetrate root tissues with the aid of cellulolytic enzymes (C), and disseminate throughout the plant system (D).
2.2 Microbiota in above-ground plant
Above-ground plant tissues (e.g., vegetative foliar portions, floral parts, and leaves) provide distinct habitats for epiphytes and endophytes; yet, the ecology of endosphere and episphere bacteria differs significantly. Most endophytes propagate systemically via the xylem to various plant compartments, but they can also reach plant tissues through aerial plant parts (Compant et al., 2010, 2011). Different above-ground plant compartments host distinct endophytic communities, influenced by the allocation of plant resources. According to observations, phyllobacteria also originated in the soil environment, affected by environmental and plant factors (Vorholt, 2012; Wallace et al., 2018).
Various microorganisms are found at the species and genus levels in the endosphere and phyllosphere. For example, the structural analysis of grapevine carposphere or phyllosphere microbiome revealed that the predominant genera were Sphingomonas, Frigoribacterium, Pseudomonas, Bacillus, Curtobacterium, Acinetobacter, Enterobacter, Citrobacter, Erwinia, Methylobacterium and Pantoea (Campisano et al., 2014; Kecskeméti et al., 2016; Wallace et al., 2018). Recently, Wallace et al. (2018) investigated the leaf microbiome of 300 various maize lines. They found that Methylobacteria and Sphingomonads were the predominant taxa. They also found that environmental variables mainly drove the microbial composition of the phyllosphere. The dominant taxa are found to be Pseudomonas and Enterobacteriaceae in apple flowers (Steven et al., 2018). Similarly, a number of studies have found Pseudomonas to be the most abundant genus in apples, almonds, grapefruit, tobacco and pumpkin flowers (Aleklett et al., 2014). In general, microbiota on and within the plant tissue originate mainly from seed, soil, and air, whose composition of communities was determined by several factors (e.g., environmental, farm management techniques, and soil). The unique assembly in host and compartments indicates that the plant has a strong functional interaction with its above-ground microbiota, but further research is needed to better understand the mechanisms governing this relationship. Above-ground microbiomes and endophytes are well known for their role in improving plant growth, stress mitigation, and disease resistance (Hassani et al., 2018; Stone et al., 2018).
2.3 Core and satellite microbiomes
Microorganisms that are consistently associated with specific plant species or genotypes, irrespective of environmental fluctuations, form the core plant microbiome (Toju et al., 2018). These core taxa are considered functionally critical due to their stable presence. Evolutionary selection has equipped them with essential genes that contribute to plant fitness. Notable examples include Bradyrhizobium, Sphingobium and Microvirga in potatoes (Pfeiffer et al., 2017), as well as Pseudomonadaceae, Micrococcaceae and Hyphomicrobiaceae in grapevines (Zarraonaindia et al., 2015). Keystone taxa within the core microbiome are pivotal in maintaining ecosystem stability and promoting plant health, primarily through nutrient cycling, pathogen suppression, and growth enhancement mechanisms (Lemanceau et al., 2017). Satellite taxa play critical roles in functions such as disease suppression and nutrient cycling, contributing disproportionately to the system’s overall stability (Hanski, 1982; Gera Hol et al., 2015). In contrast, satellite taxa are less abundant, displaying a transient presence across fewer environments. Although these taxa may not be as ubiquitous, their ecological contributions are significant, particularly in enhancing ecosystem resilience and stability (Jousset et al., 2017). Examples of satellite species include Lipomyces sp. (Kunito et al., 2012) and Cladorrhinum sp. (Tayyab et al., 2021), which contribute to aluminum toxicity reduction, plant growth promotion, and biological control.
Despite their transient and often rare nature, rare microorganisms are integral to the flexibility of the microbial community, enabling rapid responses to environmental perturbations. The concept of the “rare biosphere” is especially relevant here, as rare microbial taxa contribute to microbial diversity and the capacity of ecosystems to withstand biotic and abiotic stresses (Lynch and Neufeld, 2015).
The interplay between core and satellite microbiomes is essential in understanding the functional dynamics of plant health. Core taxa offer stability, providing critical support under standard environmental conditions. In contrast, satellite taxa introduce ecological flexibility, enabling the plant microbiome to adapt to environmental shifts. This synergistic interaction is fundamental to plant resilience, particularly in agriculture, where crops are exposed to diverse stresses. Case studies involving microbial inoculants have shown that targeting core taxa for long-term benefits, along with managing satellite taxa for short-term adaptations, can significantly enhance agricultural productivity and disease resistance (Compant et al., 2019). Understanding and leveraging the interactions between core and satellite microbiomes are vital for improving plant fitness and ecosystem health, especially in the face of environmental change and agricultural intensification (Figure 2).
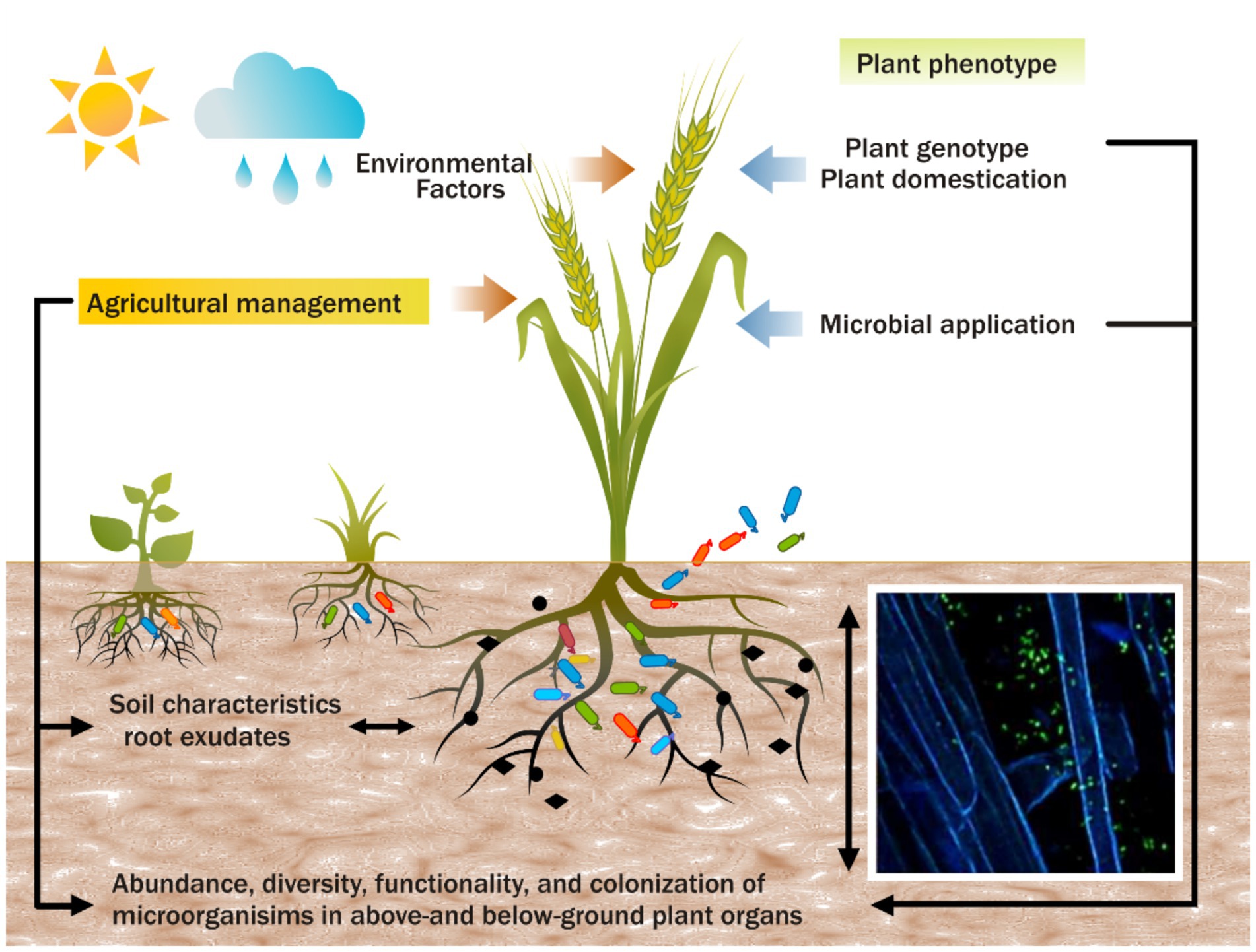
Figure 2. The complex interactions between plant genotypes, microbiota, and agricultural practices that influence plant phenotypes and traits.
3 Functions of plant microbiota
The plant microbiota encompasses a wide variety of microorganisms, including beneficial, neutral, and pathogenic species, each playing distinct roles in plant health and development. Plant growth-promoting bacteria (PGPB) form a crucial subset of the microbiota, promoting plant growth through various mechanisms.
3.1 Biological nitrogen fixation
Biological nitrogen fixation (BNF) is a key process wherein plant-associated bacteria convert atmospheric nitrogen into a usable form of ammonia for plants. This symbiotic relationship primarily occurs in leguminous plants through specialized root nodules that harbor nitrogen-fixing bacteria, such as Rhizobium and Bradyrhizobium (Irisarri et al., 2021; Prüß, 2022). Nitrogen fixation reduces the need for synthetic nitrogen fertilizers which are environmentally detrimental (Mahmud et al., 2020; Soumare et al., 2020). Furthermore, non-nodular bacteria like Gluconacetobacter diazotrophicus have expanded the scope of BNF to non-leguminous plants (Dent, 2018). Integrating nitrogen-fixing bacteria into sustainable agriculture reduces greenhouse gas emissions and enhances crop resilience, especially under drought conditions (Minamisawa, 2023).
3.2 Mineral solubilization
Microorganisms also facilitate the solubilization of key nutrients like phosphorus and potassium, which are often unavailable in their natural forms. Phosphate-solubilizing bacteria (PSB) convert insoluble phosphate into bioavailable forms through organic acid production (Tahir et al., 2018). Similarly, potassium-solubilizing bacteria (KSB) enhance the availability of potassium, critical for plant water regulation and enzyme activation (Balakrishnan et al., 2021; Mei et al., 2021). For example, Bacillus megaterium and Pseudomonas fluorescens have been shown to effectively solubilize phosphate, improving plant growth in phosphorus-deficient soils (Patel et al., 2016).
3.3 Phytohormone production
Plant-associated bacteria, particularly from the genera of Bacillus and Pseudomonas, are capable of producing phytohormones such as auxins, cytokinins, and gibberellins. These hormones regulate critical aspects of plant development, including cell division and elongation. Bacillus megaterium is a prolific producer of indole-3-acetic acid (IAA), a key auxin that enhances root growth and nutrient uptake (Ahmed and Hasnain, 2010; Tanveer and Ali, 2022). In addition, cytokinins produced by Pseudomonas species promote shoot development and delay leaf senescence, contributing to plant health under stress conditions (Shahraki et al., 2022).
3.4 Phytohormone regulation through ACC deaminase activity
Ethylene, a plant hormone produced in response to stress, can inhibit plant growth. Certain bacteria mitigate the negative effects of ethylene by producing the enzyme 1-aminocyclopropane-1-carboxylate (ACC) deaminase, which breaks down ACC, the ethylene precursor. For instance, Pseudomonas stutzeri A1501 facilitates rice growth under saline conditions by reducing ethylene levels (Han et al., 2015). These ACC deaminase-producing bacteria not only promote plant growth but also improve tolerance to environmental stresses like drought and salinity (Glick and Nascimento, 2021; Ali et al., 2023).
3.5 Siderophore activity
Siderophores are iron-chelating compounds produced by bacteria and fungi to solubilize and transport iron in iron-limited environments. P. fluorescens and B. subtilis produce siderophores such as pyoverdine and bacillibactin, which improve iron acquisition in plants (Nithyapriya et al., 2021; Zhang et al., 2022). This activity not only enhances plant growth but also provides biocontrol against pathogens by limiting their access to iron, an essential nutrient (Lyng et al., 2023; Celina et al., 2024).
3.6 Exopolysaccharide and biofilm formation
Exopolysaccharides are high-molecular-weight polysaccharides secreted by bacteria that form biofilms. EPS production facilitates bacterial adherence to plant roots, enhancing nutrient and water uptake. For example, Paenibacillus polymyxa produces EPS that aids in wheat root colonization and improves nutrient absorption (Yegorenkova et al., 2013). Biofilms protect the microbial community from environmental stresses, such as desiccation and salinity, and can also protect against pathogens by forming a physical barrier and competing for resources (Bokhari et al., 2021).
4 Factors governing microbial community composition
The composition of plant-associated microbial communities is shaped by a dynamic and multifaceted interplay of environmental factors, host-derived elements, and microbial interactions (Turner et al., 2013; Pang et al., 2019; Tayyab et al., 2022). Environmental conditions, including soil type, pH, temperature, moisture, and salinity, significantly influence microbial diversity (Hassani et al., 2018; Tayyab et al., 2021). The microbiome composition varies markedly between plant species, reflecting distinct microbial populations even in closely situated plants of phylogenetically distant species. For instance, grapevines and adjacent weeds exhibit species-specific microbial clades such as Pedobacter and Rhizobiaceae (Samad et al., 2017b). Experiments in controlled environments further demonstrate that the root microbiome composition in Ceanothus thyrsiflorus, Baccharis pilularis, and Pinus muricata reflects their respective native soils, underscoring the role of plant species identity in shaping microbial communities (Jones et al., 2019). The plant genotype plays a critical role in determining microbial community structure. Differential microbial abundances have been associated with specific plant genotypes, such as in wheat varieties where chromosomal substitutions affect rhizosphere microflora (Neal et al., 1970). Genetic modifications, such as the introduction of specific genes, can substantially alter microbial communities, as demonstrated in studies involving Sorghum bicolor and Arabidopsis thaliana (Bressan et al., 2009; Chaparro et al., 2014). Notably, certain sugarcane cultivars influence bacterial richness, with cultivars like “Haizhe 22” showing distinct beta diversity patterns and enriching bacterial genera (Tayyab et al., 2022). Plant exudates, such as amino acids, organic acids, sugars, and phenolic compounds, significantly influence microbial colonization and diversity (Sundaram et al., 2015; Olanrewaju et al., 2019; Arafat et al., 2020). Additionally, volatile compounds like methanol produced by plants contribute to the growth of phyllosphere microorganisms (Galbally and Kirstine, 2002; Uhlik et al., 2013). Plant health and immunity mechanisms, such as such as pattern recognition receptors (PRRs) responding to microbe-associated molecular patterns (MAMPs) and damage-associated molecular patterns (DAMPs) help regulate community composition through pathways like pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) (Boller and Felix, 2009; Dodds and Rathjen, 2010; Monaghan and Zipfel, 2012). Temporal shifts in microbial communities across plant developmental stages have been observed, with notable changes in the abundance of groups such as Actinobacteria, Acidobacteria, Bacteroidetes and Cyanobacteria (Chaparro et al., 2014; Chen et al., 2019). These shifts reflect the evolving requirements and interactions between plants and their microbial partners throughout the plant life cycle.
5 Harnessing the plant microbiome
5.1 Microbial inoculation
Microbial inoculation, central to agricultural microbiology, begins with the meticulous screening of strains for plant growth-promoting (PGP) traits under controlled conditions. Key traits include phosphate solubilization, nitrogen fixation, siderophore production, and plant hormone regulation. Fungal strains such as Purpureocillium sp. strain YZ1 have also been shown to modulate plant elemental uptake (Zheng et al., 2021). Strains that perform well in lab assays undergo greenhouse trials and subsequent field evaluations. Although microbial inoculants like Azospirillum brasilense Ab-V5 have demonstrated significant yield increases in maize and wheat under optimized conditions (Hungria et al., 2010), field efficacy remains variable, highlighting their potential when conditions are optimized (Fukami et al., 2016; Backer et al., 2018). Factors such as competition with native microflora, suboptimal strain selection, and environmental variables can undermine inoculant performance (Samad et al., 2017a). Formulation, including cell viability and niche compatibility, is crucial for sustained colonization and function (Chowdhury et al., 2015). Biocontrol agents like B. amyloliquefaciens FZB42 require early establishment and niche adaptation to trigger effective plant defenses (Chowdhury et al., 2015).
5.2 Leveraging microbial consortia
Emerging approaches advocate for using microbial consortia rather than single-strain inoculants to enhance reliability under diverse field conditions (Parnell et al., 2016; De Vrieze et al., 2018) Consortia offers complementary PGP effects and pathogen suppression, enhancing plant health and resilience (Berendsen et al., 2018). For example, P. consortia have demonstrated enhanced nutrient assimilation and stress resilience in plants (Guttieri et al., 2015; Hu et al., 2016). Tailoring consortia to specific environmental conditions and plant genotypes further enhances their efficacy, as shown by the host preference of Azospirillum (Chamam et al., 2013; Edwards et al., 2015; Santos-Medellín et al., 2017). Nonetheless, ongoing research is necessary to refine consortia selection and optimize their field performance (Herrera Paredes et al., 2018).
5.3 Innovations in formulation and delivery
Effective microbial formulations are essential for maintaining inoculant viability and efficacy in the field. Techniques such as encapsulation and surfactant addition improve adherence to plant surfaces, reducing environmental stresses like drift and UV exposure (Preininger et al., 2018; Timmusk et al., 2018). Innovations such as seed microbiome modulation through flower spray inoculation hold promise for improving microbial colonization and plant growth throughout the crop cycle (Pfeiffer et al., 2017). Immobilization technology can be promising to enhance field efficacy of microbial inoculants (Wu et al., 2021).
5.4 In situ microbiome engineering
The manipulation of existing natural microbial communities in situ is an important approach to improve plant health and productivity, which leverages the inherent diversity and functionality of microbial populations already present in the soil and plant environments. One effective strategy within this framework is the selective enhancement of beneficial microbial taxa through practices such as crop rotation, cover cropping, and organic amendments. These practices can promote the proliferation of beneficial microbes while suppressing pathogenic species, thereby enhancing plant resilience to stressors such as drought and disease (Carmen Orozco-Mosqueda et al., 2018; Wang and Haney, 2020).
In situ microbiome engineering also includes the application of microbial inoculants that are specifically tailored to enhance the native microbiome. For example, research has shown that inoculating crops with beneficial microbes can lead to significant increases in yield and stress tolerance (Lawson et al., 2019; Yang et al., 2023). Furthermore, the manipulation of root exudates through plant breeding or management practices can selectively recruit beneficial microbes, thereby optimizing the plant’s microbiome for enhanced performance (Clouse and Wagner, 2021; Nadarajah and Natasha Rahman, 2023).
5.5 Innovative systems
Design of novel microbial systems is an emerging approach in microbiome engineering. Two notable systems in this category are the Intraspecies Cross Environmental (ICE) system and the Combinatorial CRISPR Array-Guided Engineering (CRAGE) system.
The ICE system focuses on creating synthetic microbial consortia that can thrive across different environmental conditions. By selecting and combining microbial strains that exhibit complementary traits, researchers can develop robust microbial communities capable of enhancing plant growth under varying stress conditions. This approach allows for the fine-tuning of microbial interactions to optimize nutrient cycling and plant health (Uroz et al., 2019; Afridi et al., 2022). For instance, synthetic consortia can be engineered to improve nitrogen fixation or phosphorus solubilization, directly benefiting plant growth (Kaul et al., 2021).
The CRAGE system utilizes CRISPR technology to engineer microbial genomes for specific functions that benefit plant health. By targeting genes involved in beneficial traits such as stress tolerance, nutrient uptake, or pathogen resistance, researchers can create microbial strains with enhanced capabilities. This method not only allows for precise modifications but also facilitates the study of gene function in microbial communities, paving the way for more effective microbiome engineering strategies (Dastogeer et al., 2022; Malayejerdi and Pouresmaeil, 2020).
6 Harnessing plant microbiomes through agricultural approaches
6.1 Optimizing plant selection for enhanced microbial relationships
Incorporating plant microbiomes into crop breeding programs represents a crucial frontier in enhancing stress tolerance, nutrient uptake, and productivity (Haichar et al., 2008; Bakker et al., 2012; Ofek et al., 2014; Wei and Jousset, 2017) (Figure 2). Different genotypes foster distinct microbial associations, yet the genetic mechanisms driving these interactions remain poorly understood. Domestication has reduced plant genetic and microbial diversity, limiting the ability of modern crops to interact with beneficial microbes (Haudry et al., 2007; Bitocchi et al., 2013; Gopal and Gupta, 2016). However, recent studies reveal significant differences in microbial diversity and composition across sugarcane cultivars, highlighting the potential to select cultivars that promote beneficial microbial associations (Tayyab et al., 2022). Integrating microbiome considerations into breeding practices is pivotal for advancing sustainable agriculture and optimizing productivity (Li et al., 2024).
6.2 Effects of agricultural practices on plant-microbe dynamics
Agricultural practices, such as intercropping, organic farming, and reduced tillage, significantly influence plant-microbe interactions (Sánchez-Cañizares et al., 2017) (Figure 2). Practices like organic farming can enhance microbial diversity and abundance, improving ecosystem resilience and plant health (Hartmann et al., 2015; Hartman et al., 2018). For example, sugarcane–legume intercropping has been shown to enhance soil fertility and microbial diversity without compromising crop yields (Pang et al., 2022). The use of bacterial biological control agents also shows promise, with studies demonstrating improved soil chemistry, enriched beneficial microbial communities, and enhanced crop productivity in ratoon sugarcane (Fallah et al., 2023).
7 Conclusions and perspectives
Plant microbiomes play a crucial role in sustainable agriculture by fostering diverse and beneficial microbial communities that contribute to plant health and productivity. While the significance of plant-associated microbiomes has been recognized for centuries, it was not until the 1980s that substantial progress was made in understanding and harnessing their potential. Although microbial inoculants have been developed to enhance plant growth and resilience, their effectiveness in real-world agricultural settings has been inconsistent (Kimotho et al., 2024). This underscores the urgent need for targeted strategies to optimize microbial selection, formulation, and delivery methods to improve field performance.
Tailoring microbial consortia to specific crops and environmental conditions shows great promise in overcoming current challenges. Techniques like organic farming and intercropping can be employed to promote beneficial microbial interactions and enhance plant health (Wang and Li, 2019). Moreover, advancements in next-generation plant breeding that focus on facilitating favorable plant-microbe relationships offer exciting possibilities for boosting crop productivity and sustainability.
Future research should focus on filling knowledge gaps related to how inoculants impact existing microbiomes, the functional dynamics within complex microbial communities, and the factors influencing microbial colonization in agricultural settings. A comprehensive understanding of these processes is essential for developing resilient microbial inoculants and consortia that can thrive in diverse farming environments. The development of tailored formulations and delivery systems suited to the specific requirements of crops and local conditions is vital for achieving success in the field. Innovative approaches such as encapsulation technologies and seed microbiome modulation show promise in enhancing microbial colonization and persistence, bridging the gap between laboratory research and practical application in agricultural settings.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
MH: Writing – original draft, Writing – review & editing. MT: Writing – review & editing. EB: Writing – review & editing. MI: Writing – review & editing. WI: Writing – review & editing. XL: Conceptualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (nos. 32250015 and U21A2024).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Afridi, M. S., Ali, S., Salam, A. K., Terra, W. C., Hafeez, A., Sumaira,, et al. (2022). Plant microbiome engineering: hopes or hypes. Biology (Basel) 11:1782. doi: 10.3390/biology11121782
Ahmed, A., and Hasnain, S. (2010). Auxin-producing Bacillus sp.: auxin quantification and effect on the growth of Solanum tuberosum. Pure Appl. Chem. 82, 313–319. doi: 10.1351/PAC-CON-09-02-06
Aleklett, K., Hart, M., and Shade, A. (2014). The microbial ecology of flowers: an emerging frontier in phyllosphere research1. Botany 92, 253–266. doi: 10.1139/cjb-2013-0166
Ali, S., Ahmad, M., Hamayun, M., and Alrefaei, A. F. (2023). Rhizobacteria containing ACC deaminase enhance plant growth and tolerance under abiotic stress conditions. J. Adv. Nutr. Sci. Technol. 3, 72–77. doi: 10.15228/anst.2023.v03.i03-4.p08
Arafat, Y., Ud Din, I., Tayyab, M., Jiang, Y., Chen, T., Cai, Z., et al. (2020). Soil sickness in aged tea plantation is associated with a shift in microbial communities as a result of plant polyphenol accumulation in the tea gardens. Front. Plant Sci. 11:601. doi: 10.3389/fpls.2020.00601
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9:402666. doi: 10.3389/fpls.2018.01473
Bakker, M. G., Manter, D. K., Sheflin, A. M., Weir, T. L., and Vivanco, J. M. (2012). Harnessing the rhizosphere microbiome through plant breeding and agricultural management. Plant Soil 360, 1–13. doi: 10.1007/s11104-012-1361-x
Balakrishnan, K., Thirumalairaj, J., Radhakrishnan, M., Gopikrishnan, V., and Balagurunathan, R. (2021). Phosphate Solubilization and plant growth promoting Actinobacteria from rhizosphere soil. Indian J. Agric. Res. 55, 87–92. doi: 10.18805/IJARe.A-5328
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berendsen, R. L., Vismans, G., Yu, K., Song, Y., De Jonge, R., Burgman, W. P., et al. (2018). Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 12, 1496–1507. doi: 10.1038/s41396-018-0093-1
Berg, G., Rybakova, D., Grube, M., and Köberl, M. (2016). The plant microbiome explored: implications for experimental botany. J. Exp. Bot. 67, 995–1002. doi: 10.1093/jxb/erv466
Bhattacharyya, P. N., and Jha, D. K. (2012). Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J. Microbiol. Biotechnol. 28, 1327–1350. doi: 10.1007/s11274-011-0979-9
Bitocchi, E., Bellucci, E., Giardini, A., Rau, D., Rodriguez, M., Biagetti, E., et al. (2013). Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol. 197, 300–313. doi: 10.1111/j.1469-8137.2012.04377.x
Bokhari, S. S., Qurashi, A. W., Yasmeen, R., Yasmeen, F., Nayab, N., and Raf, U. (2021). Analysis of cell surface properties of Exiguobacterium aurentiacum associated with Lumbricus terrestris and its potency as biological inoculants. Punjab Univ. J. Zool. 36, 197–204. doi: 10.17582/JOURNAL.PUJZ/2021.36.2.197.204
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Brader, G., Compant, S., Vescio, K., Mitter, B., Trognitz, F., Ma, L. J., et al. (2017). Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol. 55, 61–83. doi: 10.1146/annurev-phyto-080516-035641
Bressan, M., Roncato, M.-A., Bellvert, F., Comte, G., Haichar, F. E. Z., Achouak, W., et al. (2009). Exogenous glucosinolate produced by Arabidopsis thaliana has an impact on microbes in the rhizosphere and plant roots. ISME J. 3, 1243–1257. doi: 10.1038/ismej.2009.68
Buée, M., de Boer, W., Martin, F., van Overbeek, L., and Jurkevitch, E. (2009). The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321, 189–212. doi: 10.1007/s11104-009-9991-3
Burns, K. N., Kluepfel, D. A., Strauss, S. L., Bokulich, N. A., Cantu, D., and Steenwerth, K. L. (2015). Vineyard soil bacterial diversity and composition revealed by 16S rRNA genes: differentiation by geographic features. Soil Biol. Biochem. 91, 232–247. doi: 10.1016/j.soilbio.2015.09.002
Campisano, A., Antonielli, L., Pancher, M., Yousaf, S., Pindo, M., and Pertot, I. (2014). Bacterial endophytic communities in the grapevine depend on pest management. PLoS One 9:e112763. doi: 10.1371/journal.pone.0112763
Carmen Orozco-Mosqueda, M. D., Carmen Rocha-Granados, M. D., and Glick, B. R. (2018). Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol. Res. 208, 25–31. doi: 10.1016/j.micres.2018.01.005
Celina, R. A., Keshinee, K., and Elavarashi, E. (2024). Isolation and characterization of Bacillus species for the detection of siderophores. Res. J. Biotechnol. 19, 92–99. doi: 10.25303/1908rjbt092099
Chamam, A., Sanguin, H., Bellvert, F., Meiffren, G., Comte, G., Wisniewski-Dyé, F., et al. (2013). Plant secondary metabolite profiling evidences strain-dependent effect in the Azospirillum-Oryza sativa association. Phytochemistry 87, 65–77. doi: 10.1016/j.phytochem.2012.11.009
Chaparro, J. M., Badri, D. V., and Vivanco, J. M. (2014). Rhizosphere microbiome assemblage is affected by plant development. ISME J. 8, 790–803. doi: 10.1038/ismej.2013.196
Chen, S., Waghmode, T. R., Sun, R., Kuramae, E. E., Hu, C., and Liu, B. (2019). Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 7, 136–113. doi: 10.1186/s40168-019-0750-2
Chowdhury, S. P., Hartmann, A., Gao, X., and Borriss, R. (2015). Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42–a review. Front. Microbiol. 6:137829. doi: 10.3389/fmicb.2015.00780
Clouse, K. M., and Wagner, M. R. (2021). Plant genetics as a tool for manipulating crop microbiomes: opportunities and challenges. Front. Bioeng. Biotechnol. 9:567548. doi: 10.3389/fbioe.2021.567548
Compant, S., Clément, C., and Sessitsch, A. (2010). Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 42, 669–678. doi: 10.1016/j.soilbio.2009.11.024
Compant, S., Mitter, B., Colli-Mull, J. G., Gangl, H., and Sessitsch, A. (2011). Endophytes of grapevine flowers, berries, and seeds: identification of cultivable Bacteria, comparison with other plant parts, and visualization of niches of colonization. Microb. Ecol. 62, 188–197. doi: 10.1007/s00248-011-9883-y
Compant, S., Samad, A., Faist, H., and Sessitsch, A. (2019). A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19, 29–37. doi: 10.1016/j.jare.2019.03.004
Correa-Galeote, D., Bedmar, E. J., and Arone, G. J. (2018). Maize endophytic bacterial diversity as affected by soil cultivation history. Front. Microbiol. 9:484. doi: 10.3389/fmicb.2018.00484
Dastogeer, G., Kao-Kniffin, J., and Okazaki, S. (2022). Editorial: plant microbiome: diversity, functions, and applications. Front. Microbiol. 13:1039212. doi: 10.3389/fmicb.2022.1039212
De Vrieze, M., Germanier, F., Vuille, N., and Weisskopf, L. (2018). Combining different potato-associated Pseudomonas strains for improved biocontrol of Phytophthora infestans. Front. Microbiol. 9:411905. doi: 10.3389/fmicb.2018.02573
Dent, D. (2018). Non-nodular endophytic bacterial Symbiosis and the nitrogen fixation of Gluconacetobacter diazotrophicus. Symbiosis 4, 53–81. doi: 10.5772/intechopen.75813
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Donn, S., Kirkegaard, J. A., Perera, G., Richardson, A. E., and Watt, M. (2015). Evolution of bacterial communities in the wheat crop rhizosphere. Environ. Microbiol. 17, 610–621. doi: 10.1111/1462-2920.12452
Edwards, J., Johnson, C., Santos-Medellín, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 112, E911–E920. doi: 10.1073/pnas.1414592112
Fallah, N., Pang, Z., Zhang, C., Tayyab, M., Yang, Z., Lin, Z., et al. (2023). Complementary effects of biochar, secondary metabolites, and bacteria biocontrol agents rejuvenate ratoon sugarcane traits and stimulate soil fertility. Ind. Crop. Prod. 202:117081. doi: 10.1016/j.indcrop.2023.117081
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Fukami, J., Nogueira, M. A., Araujo, R. S., and Hungria, M. (2016). Accessing inoculation methods of maize and wheat with Azospirillum brasilense. AMB Express 6, 3–13. doi: 10.1186/s13568-015-0171-y
Galbally, I. E., and Kirstine, W. (2002). The production of methanol by flowering plants and the global cycle of methanol. J. Atmos. Chem. 43, 195–229. doi: 10.1023/A:1020684815474
Gera Hol, W. H., de Boer, W., de Hollander, M., Kuramae, E. E., Meisner, A., and van der Putten, W. H. (2015). Context dependency and saturating effects of loss of rare soil microbes on plant productivity. Front. Plant Sci. 6:485. doi: 10.3389/fpls.2015.00485
Glick, B. R., and Nascimento, F. X. (2021). Pseudomonas 1-aminocyclopropane-1-carboxylate (Acc) deaminase and its role in beneficial plant-microbe interactions. Microorganisms 9:2467. doi: 10.3390/microorganisms9122467
Gopal, M., and Gupta, A. (2016). Microbiome selection could spur next-generation plant breeding strategies. Front. Microbiol. 7:1971. doi: 10.3389/fmicb.2016.01971
Granada Agudelo, M., Ruiz, B., Capela, D., and Remigi, P. (2023). The role of microbial interactions on rhizobial fitness. Front. Plant Sci. 14:1277262. doi: 10.3389/fpls.2023.1277262
Guttieri, M. J., Stephen Baenziger, P., Frels, K., Carver, B., Arnall, B., Wang, S., et al. (2015). Prospects for selecting wheat with increased zinc and decreased cadmium concentration in grain. Crop Sci. 55, 1712–1728. doi: 10.2135/cropsci2014.08.0559
Haichar, F. E. Z., Marol, C., Berge, O., Rangel-Castro, J. I., Prosser, J. I., Balesdent, J., et al. (2008). Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2, 1221–1230. doi: 10.1038/ismej.2008.80
Haldar, S., and Sengupta, S. (2015). Plant-microbe cross-talk in the rhizosphere: insight and biotechnological potential. Open Microbiol. J. 9, 1–7. doi: 10.2174/1874285801509010001
Han, Y., Wang, R., Yang, Z., Zhan, Y., Ma, Y., Ping, S., et al. (2015). 1-aminocyclopropane-1-carboxylate deaminase from Pseudomonas stutzeri A1501 facilitates the growth of rice in the presence of salt or heavy metals. J. Microbiol. Biotechnol. 25, 1119–1128. doi: 10.4014/jmb.1412.12053
Hanski, I. (1982). Dynamics of regional distribution: the Core and satellite species hypothesis. Oikos 38:210. doi: 10.2307/3544021
Hardoim, P. R., van Overbeek, L. S., Berg, G., Pirttilä, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/mmbr.00050-14
Hartman, K., van der Heijden, M. G. A., Wittwer, R. A., Banerjee, S., Walser, J. C., and Schlaeppi, K. (2018). Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 6:14. doi: 10.1186/s40168-017-0389-9
Hartmann, M., Frey, B., Mayer, J., Mäder, P., and Widmer, F. (2015). Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 9, 1177–1194. doi: 10.1038/ismej.2014.210
Hassani, M. A., Durán, P., and Hacquard, S. (2018). Microbial interactions within the plant holobiont. Microbiome 6:58. doi: 10.1186/s40168-018-0445-0
Haudry, A., Cenci, A., Ravel, C., Bataillon, T., Brunel, D., Poncet, C., et al. (2007). Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol. Biol. Evol. 24, 1506–1517. doi: 10.1093/molbev/msm077
Herrera Paredes, S., Gao, T., Law, T. F., Finkel, O. M., Mucyn, T., Teixeira, P. J. P. L., et al. (2018). Design of synthetic bacterial communities for predictable plant phenotypes. PLoS Biol. 16:e2003962. doi: 10.1371/journal.pbio.2003962
Hu, L., Robert, C. A. M., Cadot, S., Zhang, X., Ye, M., Li, B., et al. (2018). Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9:2738. doi: 10.1038/s41467-018-05122-7
Hu, J., Wei, Z., Friman, V.-P., Gu, S., Wang, X., Eisenhauer, N., et al. (2016). Probiotic diversity enhances rhizosphere microbiome function and plant disease suppression. MBio 7, 10–1128. doi: 10.1128/mBio.01790-16
Hungria, M., Campo, R. J., Souza, E. M., and Pedrosa, F. O. (2010). Inoculation with selected strains of Azospirillum brasilense and A. lipoferum improves yields of maize and wheat in Brazil. Plant Soil 331, 413–425. doi: 10.1007/s11104-009-0262-0
Irisarri, P., Imperial, J., Lattanzi, F. A., Monza, J., Palacios, J., Sanjuan, J., et al. (2021). Editorial: maximizing nitrogen fixation in legumes as a tool for sustainable agriculture intensification. Front. Agron. 3:796717. doi: 10.3389/fagro.2021.796717
Jiang, Y., Arafat, Y., Letuma, P., Ali, L., Tayyab, M., Waqas, M., et al. (2019). Restoration of long-term monoculture degraded tea orchard by green and goat manures applications system. Sustainability 11:1011. doi: 10.3390/su11041011
Jones, P., Garcia, B. J., Furches, A., Tuskan, G. A., and Jacobson, D. (2019). Plant host-associated mechanisms for microbial selection. Front. Plant Sci. 10:452782. doi: 10.3389/fpls.2019.00862
Jousset, A., Bienhold, C., Chatzinotas, A., Gallien, L., Gobet, A., Kurm, V., et al. (2017). Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J. 11, 853–862. doi: 10.1038/ismej.2016.174
Kandel, S. L., Joubert, P. M., and Doty, S. L. (2017). Bacterial endophyte colonization and distribution within plants. Microorganisms 5:77. doi: 10.3390/microorganisms5040077
Kaul, S., Choudhary, M., Gupta, S., and Dhar, M. K. (2021). Engineering host microbiome for crop improvement and sustainable agriculture. Front. Microbiol. 12:635917. doi: 10.3389/fmicb.2021.635917
Kecskeméti, E., Berkelmann-Löhnertz, B., and Reineke, A. (2016). Are epiphytic microbial communities in the carposphere of ripening grape clusters (Vitis vinifera L.) different between conventional, organic, and biodynamic grapes? PLoS One 11:e0160852. doi: 10.1371/journal.pone.0160852
Kimotho, R. N., Zheng, X., Li, F., Chen, Y., and Li, X. (2024). A potent endophytic fungus Purpureocillium lilacinum YZ1 protects against fusarium infection in field-grown wheat. New Phytol. 243, 1899–1916. doi: 10.1111/nph.19935
Kunito, T., Owaki, M., Ihyo, Y., Sumi, H., Toda, H., Fukuda, D., et al. (2012). Genera Burkholderia and Lipomyces are predominant aluminum-resistant microorganisms isolated from acidic forest soils using cycloheximide-amended growth media. Ann. Microbiol. 62, 1339–1344. doi: 10.1007/s13213-011-0393-4
Lawson, C. E., Harcombe, W. R., Hatzenpichler, R., Lindemann, S. R., Löffler, F. E., O’Malley, M., et al. (2019). Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol. 17, 725–741. doi: 10.1038/s41579-019-0255-9
Lemanceau, P., Blouin, M., Muller, D., and Moënne-Loccoz, Y. (2017). Let the Core microbiota be functional. Trends Plant Sci. 22, 583–595. doi: 10.1016/j.tplants.2017.04.008
Li, X., Zheng, X., Yadav, N., Saha, S., Salama, E. S., Li, X., et al. (2024). Rational management of the plant microbiome for the second green revolution. Plant Commun. 5:100812. doi: 10.1016/j.xplc.2024.100812
Lindström, K., and Mousavi, S. A. (2020). Effectiveness of nitrogen fixation in rhizobia. Microb. Biotechnol. 13, 1314–1335. doi: 10.1111/1751-7915.13517
Liu, Y., Ma, B., Chen, W., Schlaeppi, K., Erb, M., Stirling, E., et al. (2021). Rhizobium symbiotic capacity shapes root-associated microbiomes in soybean. Front. Microbiol. 12:709012. doi: 10.3389/fmicb.2021.709012
Lynch, M. D. J., and Neufeld, J. D. (2015). Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 13, 217–229. doi: 10.1038/nrmicro3400
Lyng, M., Jørgensen, J. P. B., Schostag, M. D., Jarmusch, S. A., Aguilar, D. K. C., Lozano-Andrade, C.-L. N., et al. (2023). Competition for iron shapes metabolic antagonism between Bacillus subtilis and Pseudomonas. BioRxiv 18, 1–13. doi: 10.1101/2023.06.12.544649
Mahmud, K., Makaju, S., Ibrahim, R., and Missaoui, A. (2020). Current progress in nitrogen fixing plants and microbiome research. Plan. Theory 9:97. doi: 10.3390/plants9010097
Malayejerdi, Z., and Pouresmaeil, O. (2020). Current applications of the microbiome engineering and its future: a brief review. J. Curr. Biomed. Rep. 1, 48–51. doi: 10.52547/jcbior.1.2.48
Mei, C., Chretien, R. L., Amaradasa, B. S., He, Y., Turner, A., and Lowman, S. (2021). Characterization of phosphate solubilizing bacterial endophytes and plant growth promotion in vitro and in greenhouse. Microorganisms 9:1935. doi: 10.3390/microorganisms9091935
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Minamisawa, K. (2023). Mitigation of greenhouse gas emission by nitrogen-fixing bacteria. Biosci. Biotechnol. Biochem. 87, 7–12. doi: 10.1093/bbb/zbac177
Mitter, B., Pfaffenbichler, N., and Sessitsch, A. (2016). Plant–microbe partnerships in 2020. Microb. Biotechnol. 9, 635–640. doi: 10.1111/1751-7915.12382
Monaghan, J., and Zipfel, C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349–357. doi: 10.1016/j.pbi.2012.05.006
Nadarajah, K. K., and Natasha Rahman, N. S. (2023). The microbial connection to sustainable agriculture. Plan. Theory 12:2307. doi: 10.3390/plants12122307
Neal, J. L., Atkinson, T. G., and Larson, R. I. (1970). Changes in the rhizosphere microflora of spring wheat induced by disomic substitution of a chromosome. Can. J. Microbiol. 16, 153–158. doi: 10.1139/m70-027
Nelson, E. B. (2018). The seed microbiome: origins, interactions, and impacts. Plant Soil 422, 7–34. doi: 10.1007/s11104-017-3289-7
Nithyapriya, S., Lalitha, S., Sayyed, R. Z., Reddy, M. S., Dailin, D. J., El Enshasy, H. A., et al. (2021). Production, purification, and characterization of bacillibactin siderophore of bacillus subtilis and its application for improvement in plant growth and oil content in sesame. Sustainability 13:5394. doi: 10.3390/su13105394
Ofek, M., Voronov-Goldman, M., Hadar, Y., and Minz, D. (2014). Host signature effect on plant root-associated microbiomes revealed through analyses of resident vs. active communities. Environ. Microbiol. 16, 2157–2167. doi: 10.1111/1462-2920.12228
Ojija, F., and Aloo, B. N. (2023). The role of rhizobia toward food production, food and soil security through microbial agro-input utilization in developing countries. Case Stud. Chem. Environ. Eng. 8:100404. doi: 10.1016/j.cscee.2023.100404
Olanrewaju, O. S., Ayangbenro, A. S., Glick, B. R., and Babalola, O. O. (2019). Plant health: feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol. 103, 1155–1166. doi: 10.1007/s00253-018-9556-6
Pang, Z., Fallah, N., Weng, P., Zhou, Y., Tang, X., Tayyab, M., et al. (2022). Sugarcane–peanut intercropping system enhances bacteria abundance, diversity, and sugarcane parameters in rhizospheric and bulk soils. Front. Microbiol. 12:815129. doi: 10.3389/fmicb.2021.815129
Pang, Z., Tayyab, M., Kong, C., Hu, C., Zhu, Z., Wei, X., et al. (2019). Liming positively modulates microbial community composition and function of sugarcane fields. Agronomy 9:808. doi: 10.3390/agronomy9120808
Parnell, J. J., Berka, R., Young, H. A., Sturino, J. M., Kang, Y., Barnhart, D. M., et al. (2016). From the lab to the farm: an industrial perspective of plant beneficial microorganisms. Front. Plant Sci. 7:209596. doi: 10.3389/fpls.2016.01110
Patel, G., Singh, S., Saxena, D. S. K., and Kaur, D. K. J. (2016). Isolation, biochemical characterization and production of biofertilizer from Bacillus megaterium. Int. J. Life Sci. Scienti. Res. 2, 749–752. doi: 10.21276/ijlssr.2016.2.6.16
Pfeiffer, S., Mitter, B., Oswald, A., Schloter-Hai, B., Schloter, M., Declerck, S., et al. (2017). Rhizosphere microbiomes of potato cultivated in the high Andes show stable and dynamic core microbiomes with different responses to plant development. FEMS Microbiol. Ecol. 93:fiw242. doi: 10.1093/femsec/fiw242
Preininger, C., Sauer, U., Bejarano, A., and Berninger, T. (2018). Concepts and applications of foliar spray for microbial inoculants. Appl. Microbiol. Biotechnol. 102, 7265–7282. doi: 10.1007/s00253-018-9173-4
Prüß, B. M. (2022). The three sisters of agriculture: an active learning activity on symbiotic nitrogen fixation. CourseSource 9, 1–10. doi: 10.24918/cs.2022.40
Raaijmakers, J. M., Paulitz, T. C., Steinberg, C., Alabouvette, C., and Moënne-Loccoz, Y. (2009). The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321, 341–361. doi: 10.1007/s11104-008-9568-6
Rasmann, S., and Turlings, T. C. J. (2016). Root signals that mediate mutualistic interactions in the rhizosphere. Curr. Opin. Plant Biol. 32, 62–68. doi: 10.1016/j.pbi.2016.06.017
Reinhold-Hurek, B., Bünger, W., Burbano, C. S., Sabale, M., and Hurek, T. (2015). Roots shaping their microbiome: global hotspots for microbial activity. Annu. Rev. Phytopathol. 53, 403–424. doi: 10.1146/annurev-phyto-082712-102342
Samad, A., Antonielli, L., Sessitsch, A., Compant, S., and Trognitz, F. (2017a). Comparative genome analysis of the vineyard weed endophyte Pseudomonas viridiflava CDRTc14 showing selective herbicidal activity. Sci. Rep. 7:17336. doi: 10.1038/s41598-017-16495-y
Samad, A., Trognitz, F., Compant, S., Antonielli, L., and Sessitsch, A. (2017b). Shared and host-specific microbiome diversity and functioning of grapevine and accompanying weed plants. Environ. Microbiol 19, 1407–1424. doi: 10.1111/1462-2920.13618
Sánchez-Cañizares, C., Jorrín, B., Poole, P. S., and Tkacz, A. (2017). Understanding the holobiont: the interdependence of plants and their microbiome. Curr. Opin. Microbiol. 38, 188–196. doi: 10.1016/j.mib.2017.07.001
Santos-Medellín, C., Edwards, J., Liechty, Z., Nguyen, B., and Sundaresan, V. (2017). Drought stress results in a compartment-specific restructuring of the rice root-associated microbiomes. MBio 8, 10–1128. doi: 10.1128/mBio.00764-17
Sasse, J., Martinoia, E., and Northen, T. (2018). Feed your friends: do Plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41. doi: 10.1016/j.tplants.2017.09.003
Shade, A., Jacques, M. A., and Barret, M. (2017). Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr. Opin. Microbiol. 37, 15–22. doi: 10.1016/j.mib.2017.03.010
Shahraki, A., Mohammadi-Sichani, M., and Ranjbar, M. (2022). The effect of rhizospheric bacteria on the physiological and biochemical characteristics of safflower (Carthamus tinctorius). Nova Biologica Reperta 9, 213–221. doi: 10.52547/nbr.9.3.213
Soumare, A., Diedhiou, A. G., Thuita, M., Hafidi, M., Ouhdouch, Y., Gopalakrishnan, S., et al. (2020). Exploiting biological nitrogen fixation: a route towards a sustainable agriculture. Plan. Theory 9, 1–22. doi: 10.3390/plants9081011
Steven, B., Huntley, R. B., and Zeng, Q. (2018). The influence of flower anatomy and apple cultivar on the apple flower phytobiome. Phytobiomes J. 2, 171–179. doi: 10.1094/PBIOMES-03-18-0015-R
Stone, B. W. G., Weingarten, E. A., and Jackson, C. R. (2018). The role of the phyllosphere microbiome in plant health and function. Annu. Plant Rev. Online 1, 533–556. doi: 10.1002/9781119312994.apr0614
Sundaram, B., Hiremath, A. J., and Krishnaswamy, J. (2015). Factors influencing the local scale colonisation and change in density of a widespread invasive plant species, Lantana camara, in South India. NeoBiota 25, 27–46. doi: 10.3897/neobiota.25.8354
Tahir, M., Khalid, U., Ijaz, M., Shah, G. M., Naeem, M. A., Shahid, M., et al. (2018). Combined application of bio-organic phosphate and phosphorus solubilizing bacteria (Bacillus strain MWT 14) improve the performance of bread wheat with low fertilizer input under an arid climate. Brazilian J. Microbiol. 49, 15–24. doi: 10.1016/j.bjm.2017.11.005
Tanveer, S., and Ali, B. (2022). Evaluation of Bacillus and Rhizobium strains to enhance the growth of Vigna radiata (L.) under drought stress. Pak-Euro J. Med. Life Sci. 5, 101–112. doi: 10.31580/pjmls.v5i1.2433
Tayyab, M., Islam, W., Noman, A., Pang, Z., Li, S., Lin, S., et al. (2022). Sugarcane cultivars manipulate rhizosphere bacterial communities' structure and composition of agriculturally important keystone taxa. 3 Biotech 12:32. doi: 10.1007/s13205-021-03091-1
Tayyab, M., Yang, Z., Zhang, C., Islam, W., Lin, W., and Zhang, H. (2021). Sugarcane monoculture drives microbial community composition, activity and abundance of agricultural-related microorganisms. Environ. Sci. Pollut. Res. 28, 48080–48096. doi: 10.1007/s11356-021-14033-y
Timmusk, S., Seisenbaeva, G., and Behers, L. (2018). Titania (TiO2) nanoparticles enhance the performance of growth-promoting rhizobacteria. Sci. Rep. 8:617. doi: 10.1038/s41598-017-18939-x
Toju, H., Peay, K. G., Yamamichi, M., Narisawa, K., Hiruma, K., Naito, K., et al. (2018). Core microbiomes for sustainable agroecosystems. Nat. Plants 4, 247–257. doi: 10.1038/s41477-018-0139-4
Truyens, S., Weyens, N., Cuypers, A., and Vangronsveld, J. (2015). Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ. Microbiol. Rep. 7, 40–50. doi: 10.1111/1758-2229.12181
Turner, T. R., Ramakrishnan, K., Walshaw, J., Heavens, D., Alston, M., Swarbreck, D., et al. (2013). Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 7, 2248–2258. doi: 10.1038/ismej.2013.119
Uhlik, O., Musilova, L., Ridl, J., Hroudova, M., Vlcek, C., Koubek, J., et al. (2013). Plant secondary metabolite-induced shifts in bacterial community structure and degradative ability in contaminated soil. Appl. Microbiol. Biotechnol. 97, 9245–9256. doi: 10.1007/s00253-012-4627-6
Uroz, S., Courty, P., and Oger, P. (2019). Plant symbionts are engineers of the plant-associated microbiome. Trends Plant Sci. 24, 905–916. doi: 10.1016/j.tplants.2019.06.008
Vorholt, J. A. (2012). Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840. doi: 10.1038/nrmicro2910
Wallace, J. G., Kremling, K. A., Kovar, L. L., and Buckler, E. S. (2018). Quantitative genetics of the maize leaf microbiome. Phytobiomes J. 2, 208–224. doi: 10.1094/PBIOMES-02-18-0008-R
Wang, N. R., and Haney, C. H. (2020). Harnessing the genetic potential of the plant microbiome. Biochem. (Lond) 42, 20–25. doi: 10.1042/bio20200042
Wang, L., and Li, X. (2019). Steering soil microbiome to enhance soil system resilience. Crit. Rev. Microbiol. 45, 743–753. doi: 10.1080/1040841X.2019.1700906
Wei, Z., and Jousset, A. (2017). Plant breeding Goes microbial. Trends Plant Sci. 22, 555–558. doi: 10.1016/j.tplants.2017.05.009
Wu, P., Wang, Z., Bhatnagar, A., Jeyakumar, P., Wang, H., Wang, Y., et al. (2021). Microorganisms-carbonaceous materials immobilized complexes: synthesis, adaptability and environmental applications. J. Hazard. Mater. 416:125915. doi: 10.1016/j.jhazmat.2021.125915
Yang, S., Liu, H., Xie, P., Wen, T., Shen, Q., and Yuan, J. (2023). Emerging pathways for engineering the rhizosphere microbiome for optimal plant health. J. Agric. Food Chem. 71, 4441–4449. doi: 10.1021/acs.jafc.2c08758
Yegorenkova, I. V., Tregubova, K. V., and Ignatov, V. V. (2013). Paenibacillus polymyxa rhizobacteria and their synthesized exoglycans in interaction with wheat roots: colonization and root hair deformation. Curr. Microbiol. 66, 481–486. doi: 10.1007/s00284-012-0297-y
Zarraonaindia, I., Owens, S. M., Weisenhorn, P., West, K., Hampton-Marcell, J., Lax, S., et al. (2015). The soil microbiome influences grapevine-associated microbiota. MBio 6, 10–1128. doi: 10.1128/mBio.02527-14
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., Da Rocha, U. N., Shi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Zhang, Q., Lin, Y., Shen, G., Zhang, H., and Lyu, S. (2022). Siderophores of Bacillus pumilus promote 2-keto-L-gulonic acid production in a vitamin C microbial fermentation system. J. Basic Microbiol. 62, 833–842. doi: 10.1002/jobm.202200237
Keywords: plant microbiome, microbial diversity, growth promotion, microbiome-based farming approaches, sustainable agriculture
Citation: Hanif MS, Tayyab M, Baillo EH, Islam MM, Islam W and Li X (2024) Plant microbiome technology for sustainable agriculture. Front. Microbiol. 15:1500260. doi: 10.3389/fmicb.2024.1500260
Edited by:
Bin Zhou, Chinese Academy of Tropical Agricultural Sciences, ChinaReviewed by:
Saira Ali, McMaster University, CanadaSudipta Sankar Bora, Assam Agricultural University, India
Copyright © 2024 Hanif, Tayyab, Baillo, Islam, Islam and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofang Li, eGZsaUBzanppYW0uYWMuY24=
 Muhammad Sajid Hanif
Muhammad Sajid Hanif Muhammad Tayyab
Muhammad Tayyab Elamin Hafiz Baillo
Elamin Hafiz Baillo M. Mominul Islam
M. Mominul Islam Waqar Islam
Waqar Islam Xiaofang Li
Xiaofang Li