
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 08 January 2025
Sec. Microbial Symbioses
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1499315
This article is part of the Research Topic New Frontiers in Wolbachia Biology 2025 View all articles
Introduction: Phage WO represents the sole bacteriophage identified to infect Wolbachia, exerting a range of impacts on the ecological dynamics and evolutionary trajectories of its host. Given the extensive prevalence of Wolbachia across various species, phage WO is likely among the most prolific phage lineages within arthropod populations. To examine the diversity and evolutionary dynamics of phage WO, we conducted a screening for the presence of phage WO in Wolbachia-infected cricket species from China.
Methods: The presence of phage WO was detected using a PCR-based methodology. To elucidate the evolutionary forces driving phage WO diversity, analyses of intragenic recombination were conducted employing established recombination techniques, and horizontal transmission was investigated through comparative phylogenetic analysis of the phages and their hosts.
Results and discussion: Out of 19 cricket species infected with Wolbachia, 18 species were found to harbor phage WO. Notably, 13 of these 18 cricket species hosted multiple phage types, with the number of types ranging from two to 10, while the remaining five species harbored a single phage type. Twelve horizontal transmission events of phage WO were identified, wherein common phage WO types were shared among different Wolbachia strains. Notably, each phage WO horizontal transfer event was associated with distinct Wolbachia supergroups, specifically supergroups A, B, and F. Previous studies have found that four Wolbachia strains infect two to five species of crickets. However, among these cricket species, in addition to the shared phage WO types, all harbored species-specific phage WO types. This suggests that Wolbachia in crickets may acquire phage WO types through horizontal viral transfer between eukaryotes, independent of Wolbachia involvement. Furthermore, nine putative recombination events were identified across seven cricket species harboring multiple phage types. These findings suggest that horizontal transmission and intragenic recombination have played a significant role in the evolution of the phage WO genome, effectively enhancing the diversity of phage WO associated with crickets.
Wolbachia, members of the Anaplasmataceae family, are maternally inherited endosymbiotic bacteria that infect arthropods and filarial nematodes (Werren et al., 2008; Engelstädter and Hurst, 2009). These bacteria manipulate their host's reproduction by inducing various phenotypes, including cytoplasmic incompatibility (CI), parthenogenesis, feminization of genetic males, and male-killing (Werren et al., 2008).
Bacteriophages, the most abundant entities on Earth, play a pivotal role in the evolution of bacterial genomes (Hendrix et al., 1999; Bordenstein and Wernegreen, 2004). Wolbachia phage, a λ phage-like temperate double-stranded DNA (dsDNA) bacteriophage known as phage WO, was initially characterized from the Wolbachia strain wTai, which infects the cricket species Teleogryllus taiwanemma. Phage WO can exist in either a lysogenic state, integrated into the Wolbachia chromosome, or a lytic state, free in the cytoplasm (Masui et al., 2000, 2001). It is estimated that phage WO infects ~90% of Wolbachia supergroups A and B across various arthropod groups, but it is absent in the mutualistic C and D supergroups typically found in filarial nematodes (Bordenstein and Wernegreen, 2004; Gavotte et al., 2007; Gerth et al., 2014). Bordenstein and Bordenstein (2022) proposed that all phage WO should be classified within the genus Wovirus, belonging to the family Symbioviridae. Given the persistence of phage WO, it is hypothesized that phage WO confers certain advantages to its Wolbachia and arthropod hosts (Kent and Bordenstein, 2010). The transformation and integration of prophages are regarded as the primary mechanisms of lateral gene acquisition in Wolbachia, analogous to other prokaryotes (Bordenstein et al., 2006). Phage WO may facilitate lateral gene transfer in Wolbachia strains (Ishmael et al., 2009; Wang et al., 2016) and modulate Wolbachia density by inducing cell lysis or inhibiting replication (Bordenstein et al., 2006). Recent studies have identified the factors underlying CI as two genes, cifA and cifB, which are situated adjacent to one another within WO prophage regions of the Wolbachia genome. These genes have homologs in all known CI-inducing Wolbachia strains (Beckmann et al., 2017; LePage et al., 2017; Chen et al., 2019; Shropshire and Bordenstein, 2019).
The survey results of Gavotte et al. (2007) and Tanaka et al. (2009) indicated that the majority of phage-infected Wolbachia strains harbor a limited number of phage types, with ~85% containing only one or two distinct phage types. However, accumulating evidence suggests the occurrence of multiple phage infections in certain Wolbachia strains, with some strains exhibiting more than two phage types (Chauvatcharin et al., 2006; Gavotte et al., 2007; Zhu et al., 2021; Gao et al., 2022). For instance, 84% of the 19 butterflies collected from China, each infected with a single Wolbachia strain, were found to contain high levels of multiple phage types, ranging from 3 to 17 types (Gao et al., 2022). Additionally, Zhu et al. (2021) confirmed that the Wolbachia-infected gall wasp Andricus hakonensis harbored 27 distinct phage WO types.
Mutation, recombination, and genome segment reassortment are potential mediators of genetic alterations in viruses during replication (Domingo, 2010). The genome of a bacteriophage can be partitioned into functional units, or modules, each responsible for distinct processes such as head or tail formation, lysis, lysogeny, among others. Furthermore, within the shared gene pool, the primary evolutionary mechanism driving changes in double-stranded DNA bacteriophages is the rearrangement of genomic fragments through interactions with other bacteriophages (Hatfull, 2008). Phage WO is capable of evolving via mechanisms such as point mutation, intragenic recombination, deletion, and purifying selection (Kent et al., 2011). The nucleotide sequence of the minor capsid gene orf7 in the wKueA1 strain of Wolbachia exhibits a chimeric nature, with multiple intragenic recombination events identified through population genetic analysis (Bordenstein and Wernegreen, 2004). Previous research has offered molecular evidence of recombination within the orf7 gene, indicating that intragenic recombination serves as a significant evolutionary mechanism that substantially enhances the diversity of phage WO in gall wasps (Zhu et al., 2021) and butterflies (Gao et al., 2022). Additionally, base deletions occurring during replication have been shown to significantly drive the evolution of the phage genome, contributing to the diversity of phage WO associated with A. hakonensis (Su et al., 2021).
Numerous studies have identified Wolbachia infections in crickets. For instance, Giordano et al. (1997) documented the presence of Wolbachia in Gryllus assimilis, G. ovisopis, G. integer, G. rubens, and G. pennsylvanicus. In a significant contribution from China, Li et al. (2022) conducted the first comprehensive analyses of Wolbachia in crickets, revealing that 19 species across eight genera tested positive for the infection. Furthermore, Kamoda et al. (2000) confirmed the association between CI and Wolbachia infection in the cricket species Teleogryllus taiwanemma. Wolbachia may induce incomplete CI and enhance female fertility in Velarifictorus aspersus (Zhu and Liu, 2024). Additionally, Chafee et al. (2010) analyzed phage genes from natural sympatric field isolates in Gryllus pennsylvanicus. Kupritz et al. (2021) reported the isolation and characterization of a novel phage WO from Allonemobius socius. However, prior investigations into phage WO infections of Wolbachia in Chinese crickets have been lacking. Consequently, this study screened 19 species of Wolbachia-infected crickets for the presence of phage WO utilizing a PCR-based methodology. Additionally, to elucidate the evolutionary forces of phage WO diversity, we conducted analyses of intragenic recombination through established recombination techniques and examined horizontal transmission via comparative phylogenetic analysis of the phages and their hosts.
We used published DNA samples in this study. Insects collection, DNA extraction, and Wolbachia amplification using multi-locus sequence typing (MLST) gene primers have been described in Li et al. (2022).
Total DNA was extracted from each cricket individual. Phage WO infections were detected individually by using PCR to amplify a segment of the capsid protein gene orf7 with the primers WO-F (5′-CCCACATGAGCCAATGACGTCTG-3′) and WO-R (5′- CGTTCGCTCTGCAAGTAACTCCATTAAAAC-3′) (Masui et al., 2000). PCR amplification was carried out using a C1000 Touch thermal cycler (Bio-Rad, Hercules, CA, United States) in a reaction volume of 25 μL, consisting of 18.375 μL ddH2O, 2.5 μL PCR buffer, 2 μL dNTPs (10 mmol each), 1 μL of both forward and reverse primers (10 mM), 1 μL DNA extract, and 0.125 μL Taq polymerase (Takara, Dalian, China). The PCR cycling parameters included an initial denaturation step at 95°C for 3 min, followed by 35 cycles at 95°C for 30 s, 57°C for 40 s, and 72°C for 40 s, with a final extension step at 72°C for 5 min.
The orf7 fragment of each cricket species was sequenced from two to three individuals, with subsequent purification of the PCR products and direct sequencing of the orf7 gene fragments in both directions using PCR primers. The presence of multiple peaks during initial sequencing of each sample was interpreted as indicative of multiple infections. The PCR products underwent purification through the utilization of a DNA gene gel extraction kit and subsequent ligation into the pMD18-T cloning vector in adherence to the manufacturer's guidelines. Following this, 15 distinct positive colonies were isolated from each sample and cultivated in lysogeny broth medium containing ampicillin (100 mg/mL) and d-biotin (2 mM). Plasmids were then extracted and subjected to sequencing in both forward and reverse directions using M13F/R primers on an ABI 3730XL DNA sequencer (Applied Biosystems, Foster City, CA, United States).
Sequence analysis and homology comparisons were conducted utilizing the online BLAST program. Genetic distances between all sequence pairs were determined using the Kimura two-parameter distance model with the complete deletion option in MEGA software (version 4.0). Sequences exhibiting genetic distances below 1.5% nucleotide diversity in the orf7 gene were classified as identical haplotypes, as outlined by Chafee et al. (2010) and Zhu et al. (2021). The sequences have been deposited in GenBank under the following accession numbers: PQ674069–674193. The sequences of Wolbachia MLST genes (gatB, coxA, hcpA, fbpA, and ftsZ, were retrieved from Li et al., 2022, corresponding to GenBank MW680307–6803334) and the phage WO orf7 gene were aligned, respectively, using SEQMAN PRO v.11.2 (DNASTAR, Madison, WI, USA), followed by analysis using the neighbor joining method in PAUP v.4.0b (Swofford, 2003). The best evolutionary model was selected according to the corrected Akaike information criterion calculated using MEGA v.7.0. Bootstrap tests were conducted based on 1,000 replicates to assess branch support in the final maximum likelihood trees.
The individual segment alignments were examined for signs of intragenic recombination through various techniques within the Recombination Detection Program (RDP5) package (Heath et al., 2006). Six recombination detection methods available in the RDP5 program, including BootScan/rescan recombination test (Martin et al., 2005), 3Seq (Martin and Rybicki, 2000), Chimera (Posada and Crandall, 2001), GENECONV (Padidam et al., 1999), Siscan method (Gibbs et al., 2000), and MaxChi (Smith, 1992), were employed to identify recombinant sequences and breakpoints. Default settings were applied to all methods, with a maximum acceptable P-value cutoff of 0.05.
The reanalysis of Wolbachia MLST sequences from Li et al. (2022) allowed to identify 15 distinct sequence types (STs) belonging to supergroups A, B, and F in 19 out of the 22 cricket species under study. Phage WO was identified by PCR in 18 out of the 19 Wolbachia-infected cricket species (T. mitratus species did not show any positive orf7 PCR), rendering 127 phage WO orf7 sequences (see Table 1; Supplementary Table 1). Phage WO types with orf7 DNA sequences demonstrating a similarity > 98.5% were classified as identical types based on previous research (Chafee et al., 2010; Zhu et al., 2021). Each phage type was labeled as WO followed by the insect name and haplotype number. For instance, T. infernalis contained a singular phage type designated as WOTin, while Loxoblemmus taicoun from a specific geographical population in Changsha possessed two distinct phage types labeled as WOLta-1-CS and WOLta-2-CS. Thirteen Wolbachia-infected cricket species hosted multiple phage types and the other species harbored one type (Table 1; Supplementary Figures 1–3).
Previous research has identified that four strains of Wolbachia—ST-l (supergroup A), ST-543 (supergroup B), ST-j, and ST-k (supergroup F)—infect between two to five species of crickets (Li et al., 2022) (Table 1). To investigate the prevalence of phage WO infections within these Wolbachia strains, phylogenetic trees were constructed based on the orf7 sequences of phage WO types that infected Wolbachia strains across various insect species (Figure 1). Wolbachia strain ST-l was identified in five cricket species, including L. doenitzi, Loxoblemmus sp1., Loxoblemmus sp2., L. taicoun, and Mitius minor, while ST-k was present in four cricket species, namely Teleogryllus mitratus, L. jacobsoni, Velarifictorus micado, and Loxoblemmus sp4. Wolbachia strains ST-l and ST-k were found to be infected with 16 and 14 phage WO types, respectively, with no identical phage types observed among different cricket species (Figures 1A, D). Wolbachia strain ST-543 was identified in L. taicoun, L. montanus, and Loxoblemmus sp3, with these cricket species sharing two common phage WO types. Specifically, the sequences of WOLmo-6, WOLsp3-2, and WOLta-1-JS were identical, while those of WOLmo-5, WOLta-2-JS, and WOsp3-1 either matched or contained one base substitution. The remaining four phage WO types were exclusively present in L. montanus (Figure 1B). Wolbachia strain ST-j was found in V. micado and V. khasiensis. These two species of Velarifictorus shared a common phage WO type, as evidenced by the identical sequences of WOVmi-2-ZJJ and WOVkh-3 (Figure 1C).
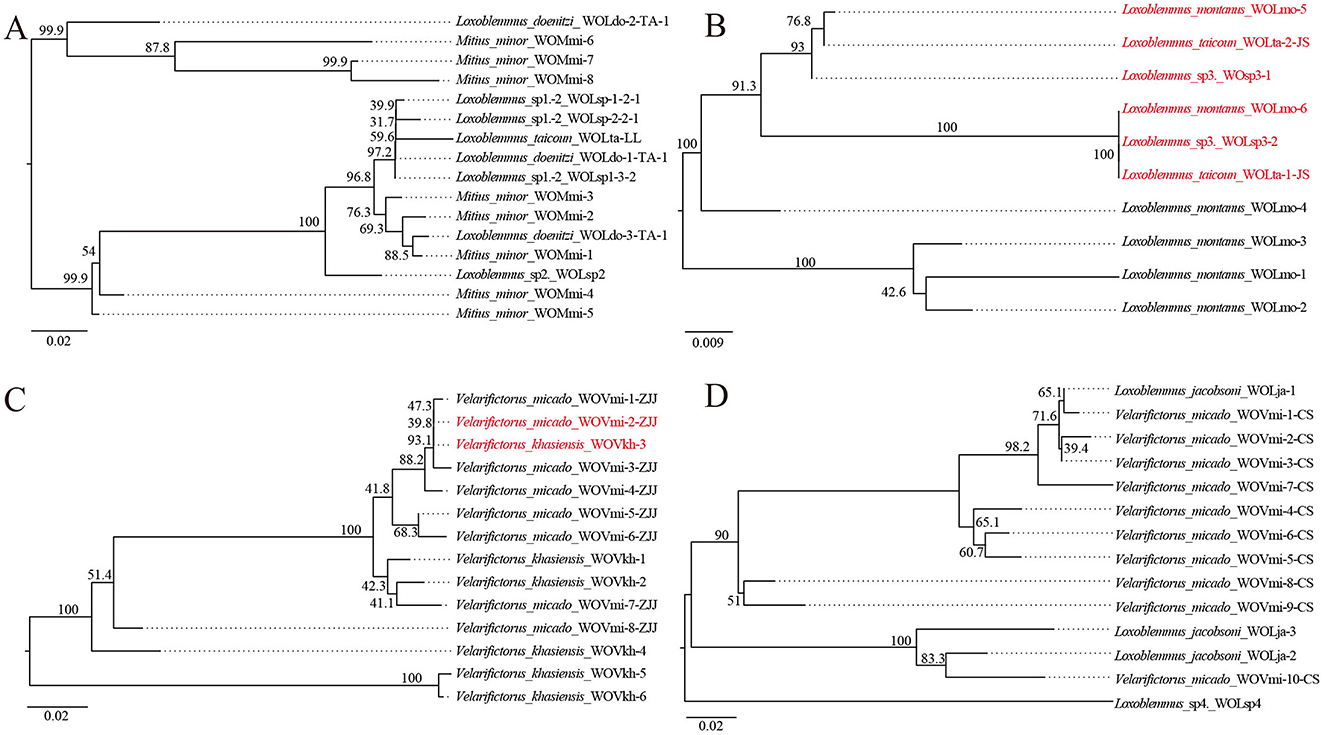
Figure 1. Neighbor joining phylogenetic tree obtained based on phage WO orf7 nucleotide sequences from Wolbachia strains ST-l (A), ST-543 (B), ST-j (C), and ST-k (D). Numbers above branches are bootstrap values based on 1,000 replicates. WO-1 refers to the serial number. Red font indicates identical orf7 sequences or those with similarity >98.5% to their respective sequences.
Phylogenetic analysis was performed on phage WO orf7 sequences and concatenated sequences of five Wolbachia MLST genes from crickets using neighbor joining methods (Figure 2). The results of the analysis revealed a lack of congruence between the phylogenies of phage WO and its host Wolbachia. Additionally, evidence was found for 12 instances of horizontal transmission of phage WO from crickets, where different Wolbachia strains shared common phage WO types; six involved four Wolbachia strains, four involved two Wolbachia strains, and one involved three Wolbachia strains (Figure 2). For instance, WOVmi-8-CS (derived from V. micado-CS), WOVkh-6 (derived from V. khasiensis), WOVmi-7-TA (derived from V. micado-TA), and WOVmi-3-LL (derived from V. micado-LL) exhibited identical orf7 sequences, indicating their classification within the same WO phage type. These phages were found to infect a minimum of four Wolbachia strains, including ST-a, ST-b, ST-j, and ST-k. The Wolbachia strains present in T. emma, L. doenitzi-TA-2, and Comidoblemmus nipponensis-CZ-2 could not be fully characterized due to difficulties in amplifying certain MLST loci (Li et al., 2022). All 12 instances of horizontal transmission involved distinct Wolbachia supergroups, with two instances occurring between supergroups A and B, one instance occurring between supergroups A and F, seven instances occurring between supergroups B and F, and two instances involving supergroups A, B, and F simultaneously.
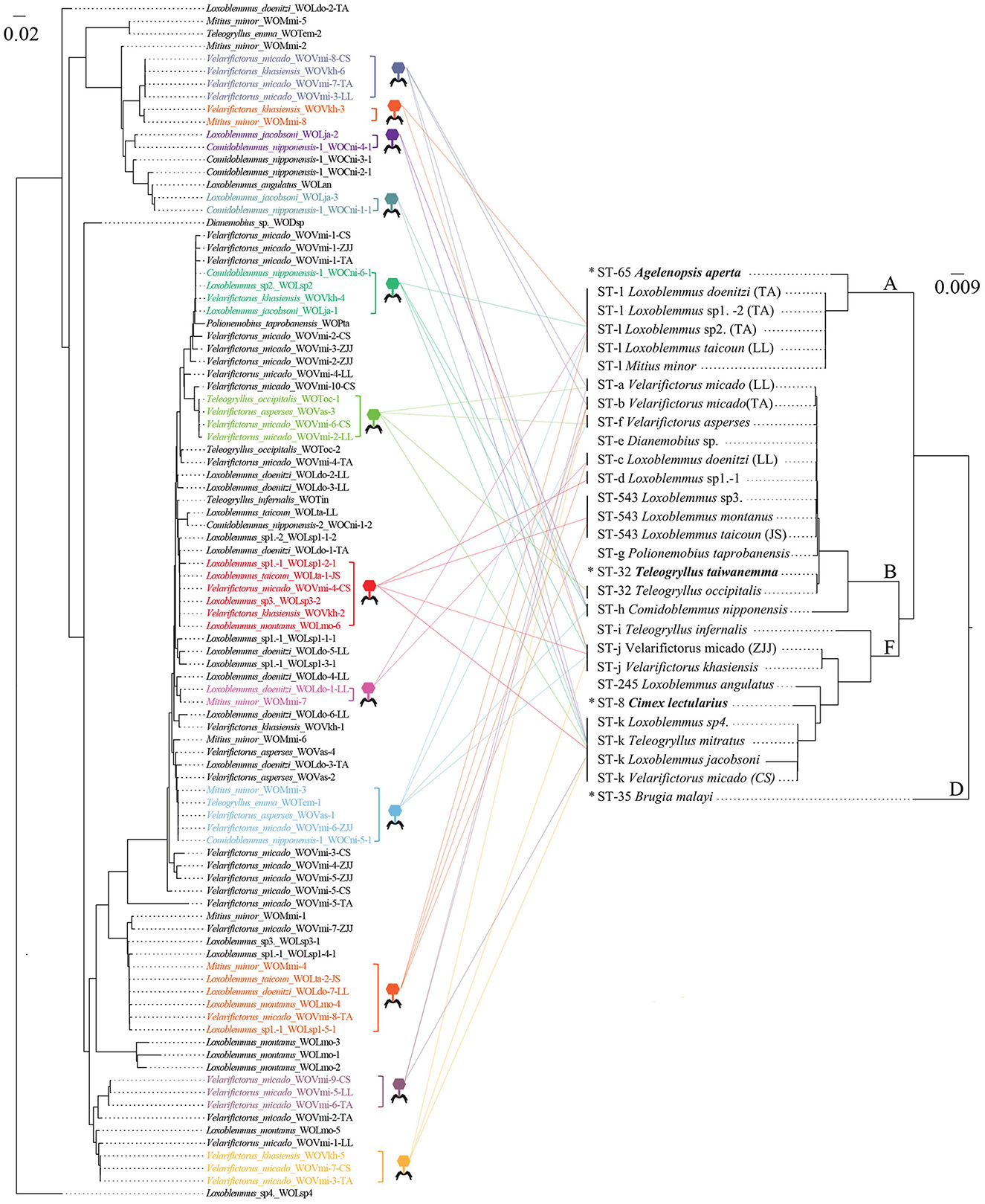
Figure 2. Comparisons of neighbor joining phylogenetic tree obtained for phage WO based on orf7 nucleotide sequences (Left) and Wolbachia based on concatenated sequences of multi-locus sequence type (MLST) genes (from Li et al., 2022) (Right). Colored fonts indicate identical orf7 sequences or those with similarity >98.5% to their respective sequences. The latter sequences are shown by icons with different colors and each icon represents a horizontal transfer event. The capital letters on the right indicate the Wolbachia supergroups. *Refers to known Wolbachia symbionts of various crickets retrieved from the MLST database.
To ascertain direct evidence of phage WO intragenic recombination, recombination analysis was conducted on aligned orf7 sequences sourced from the same cricket species utilizing the RDP5 software. Recombination events were exclusively included if identified by at least three detection methods within RDP5.
Nine putative recombination events were identified among seven cricket species that contained a variety of phage WO types (Table 2; Figure 3; Supplementary Figures 4, 5). These events involved a major and minor parent, both of which recombined at the same breakpoint to form a new phage WO lineage (Table 2). For instance, the phage WO type WOVmi-8-TA was found to be a recombinant using five of the six methods employed: 3Seq (P < 10−12), BootScan (P < 10−6), GENECONV (P < 10−5), Chimera (P < 10−7), and MaxChi (P < 10−9). The parental genotypes identified in this study were designated as WOVmi-7-TA and WOVmi-9-TA for the major and minor parents, respectively, with breakpoints located at positions 10 and 182 bp (Figure 3A). Furthermore, it was observed that a major parent in one recombinant could serve as a minor parent in another recombinant. For instance, WOVmi-7-TA acted as the major parent in the formation of recombinant WOVmi-8-TA, while simultaneously serving as the minor parent in the generation of WOVmi-5-TA (Figure 3B). It is important to acknowledge that major and minor parents have the ability to recombine through different breakpoints, resulting in the production of various recombinants. For instance, the recombinant WOLdo-6 and WOLdo-5 originated from the major parent WOLdo-3 and minor parent WOLdo-7, with breakpoints at 320 and 325 bp, respectively (Figures 3C, D). These findings indicate that intragenic recombination is prevalent and frequent in the phage WO types found in crickets.
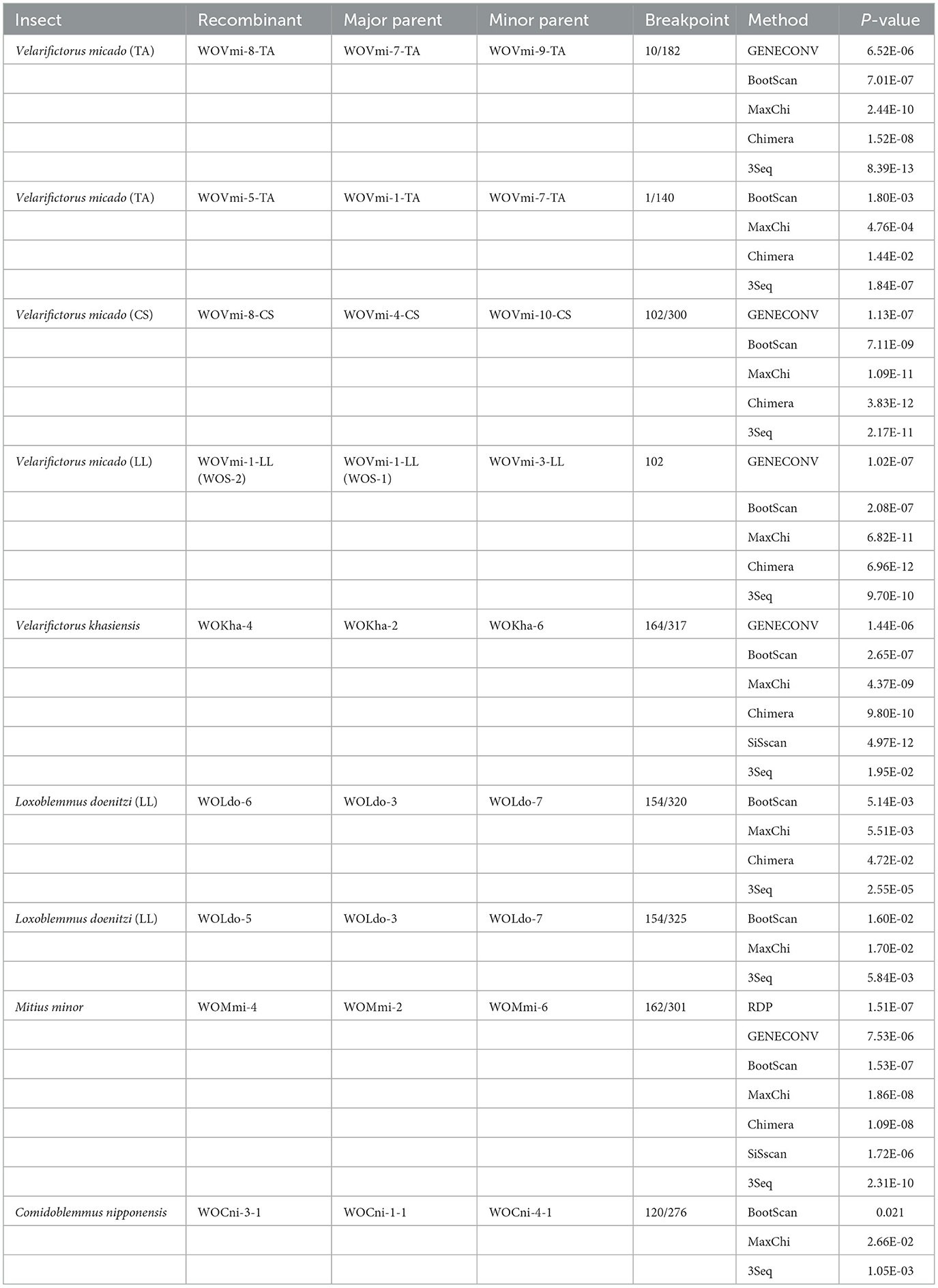
Table 2. Recombination analysis based on the phage WO orf7 gene in Gryllidae using six methods implemented in the RDP5 package.
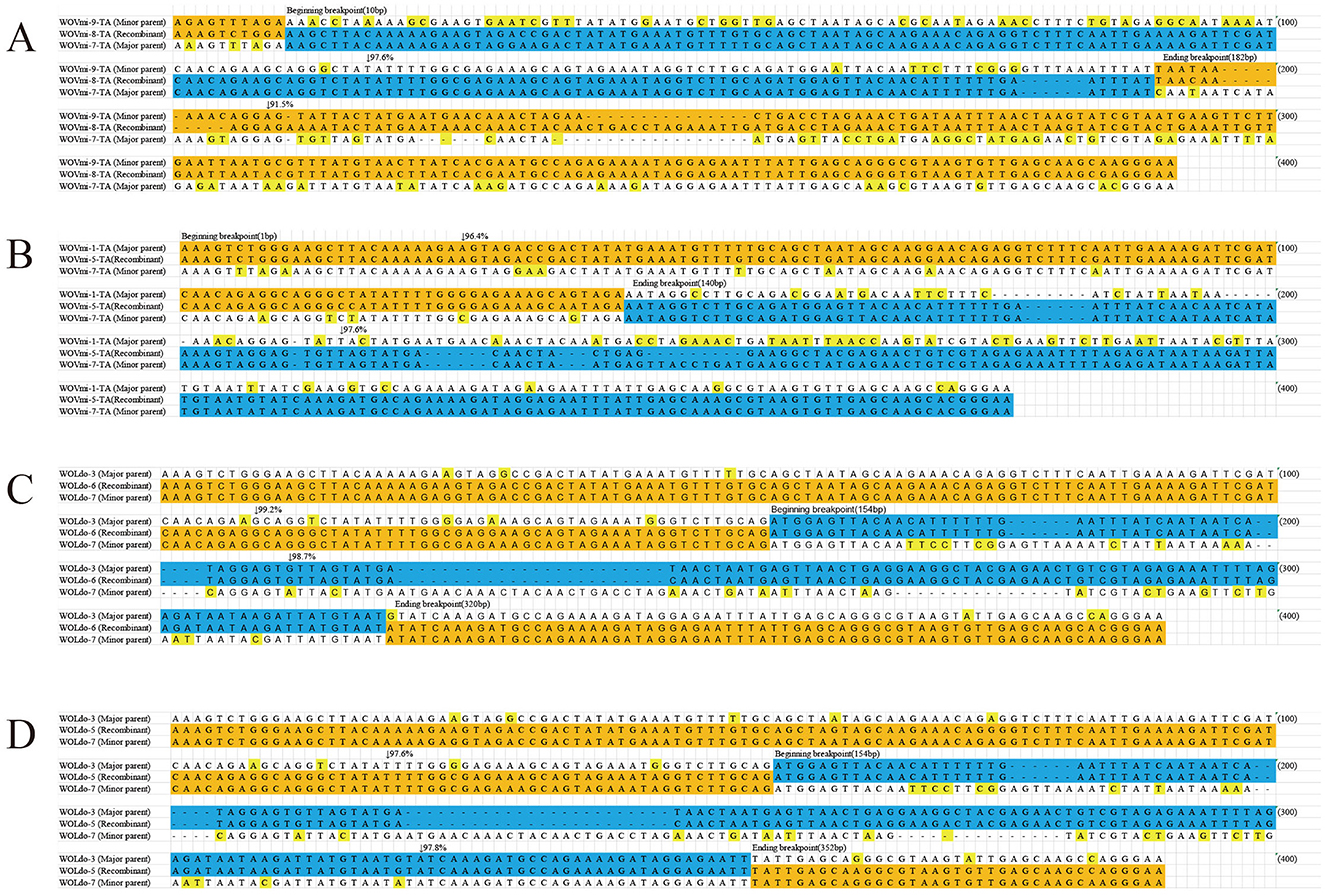
Figure 3. Recombination events identified for the orf7 gene between: WOVim-9-TA and WOVim-7-TA resulting in recombinant WOVim-8-TA (A); WOVim-1-TA and WOVim-7-TA resulting in recombinant WOVim-5-TA (B); WOLdo-3 and WOLdo-7 resulting in recombinant WOLdo-6 (C); and WOLdo-3 and WOLdo-7 resulting in recombinant WOLdo-5 (D).
Previous studies have demonstrated the occurrence of Wolbachia infections in various cricket genera, including Gryllus, Velarifictorus, Teleogryllus, Allonemobius, Loxoblemmus, Mitius, Dianemobius, Gryllodes, Polionemobius, and Comidoblemmus (Giordano et al., 1997; Kamoda et al., 2000; Mandel et al., 2001; Marshall, 2004; Jeong et al., 2012; Li et al., 2022). Wolbachia may induces CI in several cricket species, such as Teleogryllus taiwanemma (Kamoda et al., 2000) and Velarifictorus aspersus (Zhu and Liu, 2024). Nevertheless, the investigation of phage WO, which is associated with Wolbachia-infected crickets, has been limited to a relatively small number of studies (Masui et al., 2000; Chafee et al., 2010; Kupritz et al., 2021). In the present study, we report the first detection of phage WO infections associated with crickets in China. Our findings reveal that among the 19 Wolbachia-infected cricket species examined, phage WO was present in 18 species spanning the genera Velarifictorus, Teleogryllus, Loxoblemmus, Mitius, Dianemobius, Polionemobius, and Comidoblemmus, indicating a high prevalence of infection. Most Wolbachia strains infected by phage WO exhibited a limited diversity of phage types, with 85% containing only one or two distinct phage types (Gavotte et al., 2007; Tanaka et al., 2009). Nevertheless, instances of multiple phage WO infections have been documented in various Wolbachia strains (Gavotte et al., 2007; Zhu et al., 2021; Gao et al., 2022). Our findings revealed that 13 cricket species infected with Wolbachia harbored multiple phage types, with six of these species carrying more than five distinct phage types. For example, the Changsha population of V. micado infected with Wolbachia strain ST-k harbored 10 phage WO types, and V. khasiensis infected with Wolbachia strain ST-j harbored six phage WO types. These results indicate that Wolbachia-infected cricket species possess a high diversity of phage WO types, consistent with observations in gall wasps (Zhu et al., 2021) and butterflies (Gao et al., 2022).
Phage WO can disseminate among hosts through both vertical and horizontal transmissions. The absence of significant correlations between the evolutionary phylogenies of WO and Wolbachia (Bordenstein and Wernegreen, 2004; Gavotte et al., 2004; Zhu et al., 2021; Gao et al., 2022), along with the presence of divergent Wolbachia strains infecting either the same (Gavotte et al., 2004; Chauvatcharin et al., 2006) or different hosts (Wang et al., 2016; Zhu et al., 2021; Gao et al., 2022) that harbor identical phage WO types, indicates that numerous horizontal transfers of phage WO have likely occurred among various Wolbachia strains. In our study, we identified twelve instances of horizontal phage WO transmission, wherein distinct Wolbachia strains in crickets exhibited shared phage WO types. Remarkably, each of these horizontal transfer events was linked to different Wolbachia supergroups, specifically supergroups A, B, and F. During the lytic phase, prophages have the capacity to disrupt the cell membranes of both bacterial and eukaryotic cells. Subsequently, phage WO resides within the extracellular matrix of arthropods, enabling it to traverse the eukaryotic cell wall and initiate new infections (Masui et al., 2001; Bordenstein et al., 2006; Gavotte et al., 2007). Wolbachia strains ST-l, ST-543, ST-j, and ST-k were found to be shared among two to five cricket species (Li et al., 2022). Apart from the common phage WO types, nearly all of the cricket species harbored distinct, species-specific phage WO types. These findings strongly suggest that Wolbachia in crickets may acquire phage WO types through horizontal viral transfer between eukaryotes, independent of Wolbachia involvement, as has been previously reported in butterflies (Gao et al., 2022). Therefore, the horizontal transmission of phage WO—encompassing interactions across various supergroups of Wolbachia and among different insect hosts—may significantly contribute to the diversification of phage WO within Wolbachia-infected crickets.
Recombination and reassortment are recognized as mechanisms that enable RNA and DNA viruses to adapt to fluctuating environments. These processes can enhance viral genetic diversity and virulence, potentially leading to an expansion of host range (Domingo, 2010; Franzo et al., 2016; Chen et al., 2018). In the case of phage WO, the capsid protein gene orf7 undergoes frequent recombination, which substantially contributes to the genetic diversity of phage WO associated with Wolbachia-infected gall wasps (Zhu et al., 2021) and butterflies (Gao et al., 2022). Our research revealed that certain crickets were infected with a single Wolbachia strain harboring multiple phages. We identified nine recombination events within cricket species harboring diverse phage WO types. In certain phage WO lineages, a major parent in one recombinant could act as a mini parent in another recombinant; major and minor parental strains possess the capacity to recombine at different breakpoints and leading to the generation of various recombinant forms, thereby promoting active and frequent recombination. These findings indicate that intragenic recombination is a crucial evolutionary mechanism contributing to the high diversity of phage WO types associated with crickets.
In summary, 18 out of 19 cricket species infected with Wolbachia were observed to harbor phage WO, with 13 species hosting multiple phage types. This finding indicates a significant diversity of phage WO types associated with Wolbachia-infected crickets. Furthermore, twelve horizontal transmission events of phage WO were identified, and different Wolbachia strains within crickets shared common phage WO types. Additionally, nine recombination events were detected in cricket species harboring diverse phage WO types. These results suggest that intragenic recombination and horizontal transmission are pivotal evolutionary forces contributing to the observed diversity of phage WO in Wolbachia-infected crickets.
The data presented in this study are deposited in the GeneBank repository, accession number PQ674069-674193.
The manuscript presents research on animals that do not require ethical approval for their study.
Q-CL: Writing – original draft, Data curation, Investigation, Software. Y-YL: Conceptualization, Methodology, Writing – original draft, Data curation, Investigation, Software. Y-SR: Data curation, Methodology, Software, Writing – original draft, Formal analysis, Validation. X-HY: Formal analysis, Methodology, Software, Writing – original draft. D-HZ: Formal analysis, Methodology, Writing – original draft, Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research and Development Program of China (No. 2018YFE0127100).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1499315/full#supplementary-material
Beckmann, J. F., Ronau, J. A., and Hochstrasser, M. (2017). A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat. Microbiol. 2, 1–7. doi: 10.1038/nmicrobiol.2017.7
Bordenstein, S. R., and Bordenstein, S. R. (2022). Widespread phages of endosymbionts: phage WO genomics and the proposed taxonomic classification of Symbioviridae. PLoS Genet. 18:e1010227. doi: 10.1371/journal.pgen.1010227
Bordenstein, S. R., Marshall, M. L., Fry, A. J., Kim, U., and Wernegreen, J. J. (2006). The tripartite associations between bacteriophage, Wolbachia, and arthropods. PLoS Pathog. 2:e43. doi: 10.1371/journal.ppat.0020043
Bordenstein, S. R., and Wernegreen, J. J. (2004). Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol. Biol. Evol. 21, 1981–1991. doi: 10.1093/molbev/msh211
Chafee, M. E., Funk, D. J., Harrison, R. G., and Bordenstein, S. R. (2010). Lateral phage transfer in obligate intracellular bacteria (Wolbachia): verification from natural populations. Mol. Biol. Evol. 27, 501–505. doi: 10.1093/molbev/msp275
Chauvatcharin, N., Ahantarig, A., Baimai, V., and Kittayapong, P. (2006). Bacteriophage WO-B and Wolbachia in natural mosquito hosts: infection incidence, transmission mode and relative density. Mol. Ecol. 15, 2451–2461. doi: 10.1111/j.1365-294X.2006.02947.x
Chen, H., Ronau, J. A., Beckmann, J. F., and Hochstrasser, M. (2019). A Wolbachia nuclease and its binding partner provide a distinct mechanism for cytoplasmic incompatibility. Proc. Natl. Acad. Sci. U. S. A. 116, 22314–22321. doi: 10.1073/pnas.1914571116
Chen, Y., Trovao, N. S., Wang, G., Zhao, W., He, P., Zhou, H., et al. (2018). Emergence and evolution of novel Reassortant influenza a viruses in canines in southern China. MBio 9, e00909–e00918. doi: 10.1128/mBio.00909-18
Engelstädter, J., and Hurst, G. D. D. (2009). The ecology and evolution of microbes that manipulate host reproduction. Annu. Rev. Ecol. Evol. Syst. 40, 127–149. doi: 10.1146/annurev.ecolsys.110308.120206
Franzo, G., Cortey, M., Segalés, J., Hughes, J., and Drigo, M. (2016). Phylodynamic analysis of porcine circovirus type 2 reveals global waves of emerging genotypes and the circulation of recombinant forms. Mol. Phylogenet. Evol. 100, 269–280. doi: 10.1016/j.ympev.2016.04.028
Gao, S., Ren, Y. S., Su, C. Y., and Zhu, D. H. (2022). High levels of multiple phage WO infections and its evolutionary dynamics associated with Wolbachia-infected. butterflies. Front. Microbiol. 13:865227. doi: 10.3389/fmicb.2022.865227
Gavotte, L., Henri, H., Stouthamer, R., Charif, D., Charlat, S., Boulétreau, M., et al. (2007). A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol. Biol. Evol. 24, 427–435. doi: 10.1093/molbev/msl171
Gavotte, L., Vavre, F., Henri, H., Ravallec, M., Stouthamer, R., and Boulétreau, M. (2004). Diversity, distribution and specificity of WO phage infection in Wolbachia of four insect species. Insect Mol. Biol. 13, 147–153. doi: 10.1111/j.0962-1075.2004.00471.x
Gerth, M., Gansauge, M. T., Weigert, A., and Bleidorn, C. (2014). Phylogenomic analyses uncover origin and spread of the Wolbachia pandemic. Nat. Commun. 5:5117. doi: 10.1038/ncomms6117
Gibbs, M. J., Armstrong, J. S., and Gibbs, A. J. (2000). Sister-Scanning: a monte carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16, 573–582. doi: 10.1093/bioinformatics/16.7.573
Giordano, R., Jackson, J. J., and Robertson, H. M. (1997). The role of Wolbachia bacteria in reproductive incompatibilities and hybrid zones of Diabrotica beetles and Gryllus crickets. Proc. Natl. Acad. Sci. U. S. A. 94, 11439–11444. doi: 10.1073/pnas.94.21.11439
Hatfull, G. F. (2008). Bacteriophage genomics. Curr. Opin. Microbiol. 11, 447–453. doi: 10.1016/j.mib.2008.09.004
Heath, L., Van Walt, E., Varsani, A., and Martin, D. P. (2006). Recombination patterns in aphthoviruses mirror those found in other picornaviruses. J. Virol. 80, 11827–11832. doi: 10.1128/JVI.01100-06
Hendrix, R. W., Smith, M. C., Burns, R. N., Ford, M. E., and Hatfull, G. F. (1999). Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. U. S. A. 96, 2192–2197. doi: 10.1073/pnas.96.5.2192
Ishmael, N., Hotopp, J. C. D., Ioannidis, P., Biber, S., Sakomoto, J., Siozios, S., et al. (2009). Extensive genomic diversity of closely related Wolbachia strains. Microbiology 155, 2211–2222. doi: 10.1099/mic.0.027581-0
Jeong, G., Ahn, J., Jang, Y., Choe, J. C., and Choi, H. (2012). Wolbachia infection in the Loxoblemmus complex (Orthoptera: Gryllidae) in Korea. J. Asia Pac. Entomol. 15, 563–566. doi: 10.1016/j.aspen.2012.07.002
Kamoda, S., Masui, S., Ishikawa, H., and Sasaki, T. (2000). Wolbachia infection and cytoplasmic incompatibility in the cricket Teleogryllus taiwanemma. J. Exp. Biol. 203, 2503–2509. doi: 10.1242/jeb.203.16.2503
Kent, B. N., and Bordenstein, S. R. (2010). Phage WO of Wolbachia: lambda of the endosymbiont world. Trends Microbiol. 18, 173–181. doi: 10.1016/j.tim.2009.12.011
Kent, B. N., Funkhouser, L. J., Setia, S., and Bordenstein, S. R. (2011). Evolutionary genomics of a temperate bacteriophage in an obligate intracellular bacteria (Wolbachia). PLoS ONE 6:e24984. doi: 10.1371/journal.pone.0024984
Kupritz, J., Martin, J., Fischer, K., Curtis, K. C., Fauver, J. R., Huang, Y., et al. (2021). Isolation and characterization of a novel bacteriophage WO from Allonemobius socius crickets in Missouri. PLoS ONE 16:e0250051. doi: 10.1371/journal.pone.0250051
LePage, D. P., Metcalf, J. A., Bordenstein, S. R., On, J., Perlmutter, J. I., Shropshire, J. D., et al. (2017). Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543, 243–247. doi: 10.1038/nature21391
Li, Y. Y., Zeng, Y., and Zhu, D. H. (2022). Determination of Wolbachia diversity in 23 cricket species (Gryllidae) from China. Ann. Entomol. Soc. Am. 115, 148–155 doi: 10.1093/aesa/saab032
Mandel, M. J., Ross, C. L., and Harrison, R. G. (2001). Do Wolbachia infections play a role in unidirectional incompatibilities in a field cricket hybrid zone? Mol. Ecol. 10, 703–709. doi: 10.1046/j.1365-294x.2001.01213.x
Marshall, J. L. (2004). The Allonemobius-Wolbachia host-endosymbiont system: evidence for rapid speciation and against reproductive isolation driven by cytoplasmic incompatibility. Evolution 58, 2409–2425. doi: 10.1111/j.0014-3820.2004.tb00871.x
Martin, D., and Rybicki, E. (2000). RDP: detection of recombination amongst aligned sequences. Bioinformatics 16, 562–563. doi: 10.1093/bioinformatics/16.6.562
Martin, D. P., Posada, D., Crandall, K. A., and Williamson, C. (2005). A modified BOOTSCAN algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retrov. 21, 98–102. doi: 10.1089/aid.2005.21.98
Masui, S., Kamoda, S., Sasaki, T., and Ishikawa, H. (2000). Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J. Mol. Evol. 51, 491–497. doi: 10.1007/s002390010112
Masui, S., Kuroiwa, H., Sasaki, T., Inui, M., Kuroiwa, T., and Ishikawa, H. (2001). Bacteriophage WO and virus-like particles in Wolbachia, an endosymbiont of arthropods. Biochem. Biophys. Res. Commun. 283, 1099–1104. doi: 10.1006/bbrc.2001.4906
Padidam, M., Sawye, S., and Fauquet, C. M. (1999). Possible emergence of new geminiviruses by frequent recombination. Virology 265, 218–225. doi: 10.1006/viro.1999.0056
Posada, D., and Crandall, K. A. (2001). Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. U. S. A. 98, 13757–13762. doi: 10.1073/pnas.241370698
Shropshire, J. D., and Bordenstein, S. R. (2019). Two-By-One model of cytoplasmic incompatibility: synthetic recapitulation by transgenic expression of cifA and cifB in Drosophila. PLoS Genet. 15:e1008221. doi: 10.1371/journal.pgen.1008221
Smith, J. M. (1992). Analyzing the mosaic structure of genes. J. Mol. Evol. 34, 126–129. doi: 10.1007/BF00182389
Su, C. Y., Zhu, D. H., and Yang, X. H. (2021). Design and testing of effective primers for amplification of the orf7 gene of phage WO associated with Andricus hakonensis. Insects 12:713. doi: 10.3390/insects12080713
Swofford, D. L. (2003). PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods), Version 4. Sunderland, MA: Sinauer Associates.
Tanaka, K., Furukawa, S., Nikoh, N., Sasaki, T., and Fukatsu, T. (2009). Complete WO phage sequences reveal their dynamic evolutionary trajectories and putative functional elements required for integrationin to the Wolbachia genome. Appl. Environ. Microbiol. 75, 5676–5686. doi: 10.1128/AEM.01172-09
Wang, G. H., Sun, B. F., Xiong, T. L., Wang, Y. K., Murfin, K. E., Xiao, J. H., et al. (2016). Bacteriophage WO can mediate horizontal gene transfer in endosymbiotic Wolbachia genomes. Front. Microbiol. 7:1867. doi: 10.3389/fmicb.2016.01867
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Zhu, D. H., and Liu, L. T. (2024). Cytoplasmic incompatibility and female fecundity associated with Wolbachia infection in a cricket species. Ecol. Entomol. 49, 67–76. doi: 10.1111/een.13282
Keywords: phage WO, multiple infections, recombination, horizontal transfer, Wolbachia, cricket
Citation: Luo Q-C, Li Y-Y, Ren Y-S, Yang X-H and Zhu D-H (2025) Phage WO diversity and evolutionary forces associated with Wolbachia-infected crickets. Front. Microbiol. 15:1499315. doi: 10.3389/fmicb.2024.1499315
Received: 20 September 2024; Accepted: 20 December 2024;
Published: 08 January 2025.
Edited by:
Rosario Gil, University of Valencia, SpainReviewed by:
Beatriz Sabater-Munoz, University of Valencia (UV), SpainCopyright © 2025 Luo, Li, Ren, Yang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dao-Hong Zhu, ZGFvaG9uZ3podUB5ZWFoLm5ldA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.