- 1School of Agricultural Engineering and Food Science, Shandong University of Technology, Zibo, China
- 2School of Resources and Environmental Engineering, Shandong University of Technology, Zibo, China
- 3School of Life Sciences and Medicine, Shandong University of Technology, Zibo, China
- 4Ministry of Education Key Laboratory of Pollution Processes and Environmental Criteria, College of Environmental Science and Engineering, Nankai University, Tianjin, China
Introduction: Soil acidification imperils soil health and hinders the agricultural sustainability. As being more tolerant than bacteria to soil acidification, so it would be more meaningful for agricultural management and crop yield to characterize fungal community in acidic soils and manifest its key drivers.
Method: This study investigated the composition and diversity of fungal communities and its key driving factors by collecting 90 soil samples from the acidic region of Jiaodong Peninsula China, spanning 3 × 104 km2.
Results: The results indicated that most soil pH values ranged from 5.01 to 6.42, and the exchangeable acidity (EA) content raised significantly (p < 0.01) along with soil acidic degree increasing. However, no significant differences were found in fungal community diversity and composition among various soil samples, which were all predominantly habited with the phyla of Ascomycota and Basidiomycota. Results of the linear discriminant analysis effect size (LEfSe) analysis revealed that saprophytic fungi were biomarkers of the slightly acidic soil (pH 6.0–6.5), including Nectriaceae, Thielavia, Nectria, Haematonectria, and unclassified Microascaceae, while plant pathogenic fungi, such as Didymellaceae, were biomarkers of the soils pH < 5.5. Similarly, the FUNGuild results also indicated that saprophytic fungi and pathogenic fungi were the dominant functional guilds in the investigated acidic soils, accounting for 66% of the total fungal communities. Redundancy analysis (RDA) revealed that soil pH as well as nitrate nitrogen (-N) and total nitrogen (TN) significantly associated with fungal community at the phylum level, whilst soil pH was the only factor significantly linked to individual fungal classes (p < 0.01 or 0.05). The further Mantel test analysis and structural equation modeling (SEM) suggested that, in contrast to the negative and directive driving of soil pH on fungal communities' variation, the specific plant pathogenic fungi, Gibberella and Didymellaceae, were significantly and positively associated with soil acidic characteristics (p < 0.05).
Discussion: These findings highlighted that, in addition to modulating the variation of soil fungal community, soil acidification might prime some plant pathogens development. So that, more attentions should be focused on impact of soil acidification on fungal ecology, as well as plant pathogenic fungi.
1 Introduction
Acidic soils widely distribute and occupy more than 50% of the world's potentially arable land (Kochian et al., 2005; Ma and Ryan, 2010). In China, the pH of farmland surface soil declined by 0.5 units on average due to excessive application of nitrogen (N) fertilizer during past three decades (Guo et al., 2010). Undoubtedly, soil acidification had become an serious issue in agricultural production in China (Li et al., 2017), as it could result in the significant losses of base cations and nutrients and decrease of cation exchange capacity (CEC), and inhibiting soil microbial activity as well as releasing of aluminum (Al) and manganese (Mn) (Bolan et al., 2003; Kunhikrishnan et al., 2016).
Soil fungi are massively abundantly distributed in soils, and play instrumental roles in the ecosystem, including pedogenesis, nutrient cycling, and disease suppression, etc. (Bardgett and van der Putten, 2014). Moreover, in contrast to soil bacteria, soil fungi exhibit a wider range in C:N and soil pH-growth optimum (Strickland and Rousk, 2010) and are more tolerant to acidic soil environments (Peñalva et al., 2008; Mosier et al., 2016). Recently, several studies investigated the composition and driving factors of soil fungal communities in acidic soils (Pan et al., 2015; Shi et al., 2018; Ye et al., 2020), and widely suggested that soil pH is the key factor modulating the fungal community structures in acidic soils (Muneer et al., 2021; Hu et al., 2024). However, given their prominent roles in key soil processes, more information about the fungal community composition and function in various acidic soils is of primary importance to prioritize ecosystem-level conservation and management efforts.
Previous studies showed that soil fungal communities not only facilitated the decomposition of soil organic matter (SOM) by secreting various degrading enzymes (Chen et al., 2014), but also could establish symbiotic or pathogenic relationships with plants (Pascale et al., 2020; Cao et al., 2024). So that, they were proved to be essential to maintain a variety of functions in above- and belowground ecosystems (Egidi et al., 2019; Yang et al., 2019; Pérez-Izquierdo et al., 2021). Within three trophic modes, FUNGuild classifies fungal community data into more manageable ecological guilds [e.g., arbuscular mycorrhizal (AM) fungi, ectomycorrhizal (ECM) fungi, plant pathogens, wood saprotrophs, etc.], totaling 12 guilds (Nguyen et al., 2016). For example, saprophytic fungi can decompose litter and promote nutrient cycling using their extracellular enzymes for providing essential nutrients for plant growth (Cao et al., 2022). Symbiotic fungi, such as AM fungi and ECM fungi, form associations with plants and help plants absorb nutrients, enhancing their ability to resist abiotic stresses (Cairney, 2012; Wahab et al., 2023). In contrast, plant pathogenic fungi, including Fusarium, Verticillium, and Ilyonectri, can cause plant diseases by inhibiting root development and causing root rot (De Coninck et al., 2015). Therefore, it is important to understand the structure and function of fungal communities in acidic soils and the potential risk to crop health, beside of their communities' diversity.
Several studies investigated soil microbial communities, including the composition and driving factors of soil fungal communities in acidic soil (Pan et al., 2015; Shi et al., 2018; Ye et al., 2020). Nonetheless, few reported studies have focused on the ecological functions of fungi, especially for those with potential pathogenicity in acidic soils. In this study, the composition and predicting functions of soil fungal communities were investigated in acidic soils of the Jiaodong Peninsula, a typical acidic region of Northern China. We aimed to explore the effect of soil acidification on soil fungal communities' composition and their ecological functions, and decipher their key driving factors, which is significant to assess the ecological risk of soil acidification to agroecosystems. It was hypothesized that: (1) as the similar agricultural practices, variation in the diversity and composition of fungal communities were not significant in various acidic soils of Jiaodong Peninsula, even with distinct acidification degree; (2) soil pH be the key factors driving changes on fungal community composition and ecological functions, as well as the fungal plant pathogens.
2 Materials and methods
2.1 Site description and soil sampling
Soil samples were collected from10 districts in Qingdao (QD), Yantai(YT), and Weihai (WH) of Jiaodong Peninsula, Shandong Province, China (35°35′-38°23′N, 119°59′-122°71′E, Figure 1). This region features a warm, temperate monsoon continental climate, with mean annual temperatures averaging 12.0–12.6°C. The annual mean rainfall ranges from 650 to 850 mm. The soil type was mainly classified as Alfisols, comprising over 50% (Soil Survey Staff, 2014; Zhao et al., 2023). A sampling point was set around per 7,000 hm2 according the area of the selected districts, and a total of 30 sampling points were set up spanning 3 × 104 km2 (Figure 1). Three sampling sites were chosen in each district, and at each sampling site, three 200 × 200 m quadrats with relatively similar conditions were selected. The distance between each quadrat was >25 km, thus the quadrats were considered independent from each other. A total of 90 topsoil (0–20 cm) samples were taken from fields in March to May, 2021. All of the soil samples were sieved (2 mm), after removing weeds, fine straw and other debris. A portion of each sample was taken for characterization of physical and chemical properties, while the other was frozen at −80°C for DNA extraction and PCR amplification analysis.
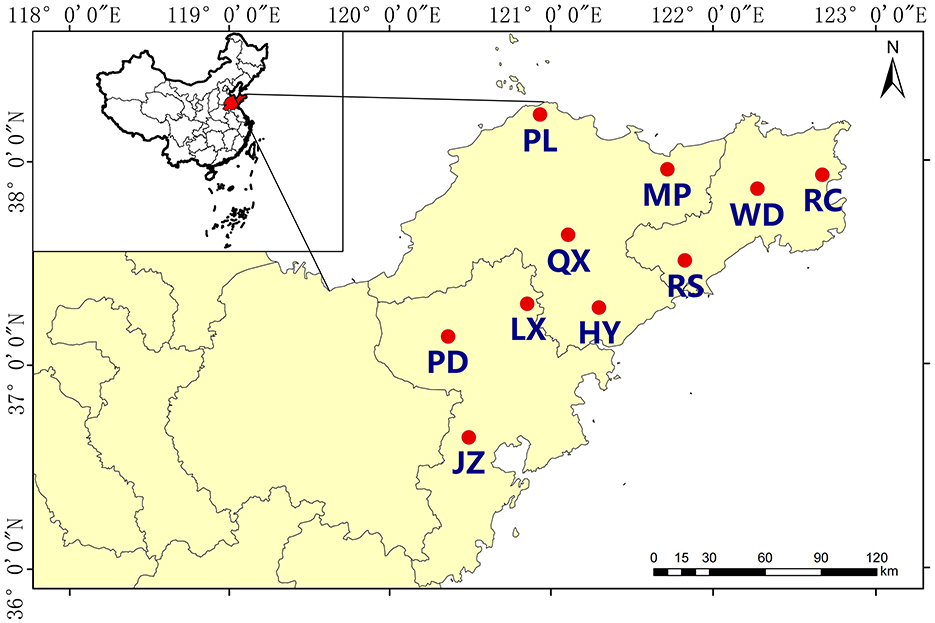
Figure 1. Sampling sites of acidic soils in Jiaodong Peninsula. The abbreviation in the figure refers to the site where samples were collected including Muping (MP), Haiyang (HY), Qixia (QX), Penglai (PL), Jiaozhou (JZ), Pingdu (PD), Laixi (LX), Rongcheng (RC), Rushan (RS), and Wendeng (WD).
2.2 Measurement of soil properties
Soil pH was measured in a soil:deionized water mixture (1:5) by a pH meter. Soil CEC was determined based on the hexaamminecobalt trichloride method (Ciesielski and Sterckeman, 1997). SOM was analyzed by the potassium dichromate colorimetric method (Yang F. et al., 2017). Exchangeable acidity (EA), exchangeable hydrogen (EH+) and exchangeable aluminum (EAl) of the soil samples were performed using a titrator to adjust pH to the objective value in 1.0 M KCl extractions (Chen et al., 2011). Soil leachable ammonium nitrogen (-N) and nitrate nitrogen (-N) were measured by a UV-visible spectrophotometer in the same extracts (Li C. Y. et al., 2020). Total nitrogen (TN) was digested with the potassium persulfate catalyst and determined in the same analyzer (Cheng et al., 2021). Total phosphorus (TP) was measured by molybdenum-blue colorimetry (Wang et al., 2009). Available phosphorus (AP) content was determined based on the NaHCO3 extraction-molybdenum antimony anti-colorimetric method (Shen et al., 2018).
2.3 DNA extraction
Soil DNA from different samples were extracted with a FastDNA Spin Kit (MP Biomedicals, Santa Ana, CA) according to the manufacturer's instructions. Extractions were performed in triplicates for each of the selected 26 samples, and the extracts were then pooled for further analysis. The extracted DNA was eluted in 50 μL of Elution buffer and stored at −80°C until measurement in the PCR.
2.4 Sequencing and data processing
The fungal ITS2 region was amplified with the primes ITS1FI2 (5′-GTGARTCATCGAATCTTTG-3′) and ITS2 (5′-TCCTCCGCTTATTGATATGC-3′) (Karlsson et al., 2014). PCR amplification was performed in a total volume of 25 μL reaction mixture containing 25 ng of template DNA, 12.5 μL PCR Premix, 2.5 μL of each primer, and PCR-grade water to adjust the volume. The conditions for amplification were 98°C for 30 s; 32 cycles of denaturation at 98°C for 10 s, annealing at 54°C for 30 s, and extension at 72°C for 45 s; and then final extension at 72°C for 10 min. The PCR products were confirmed with 2% agarose gel electrophoresis. Then the PCR products were purified by AMPure XT beads (Beckman Coulter Genomics, Danvers, MA, USA) and quantified by Qubit (Invitrogen, USA). The amplicon pools were prepared for sequencing and the size and quantity of the amplicon library were assessed on Agilent 2100 Bioanalyzer (Agilent, USA) and with the Library Quantification Kit for Illumina (Kapa Biosciences, Woburn, MA, USA), respectively. The libraries were sequenced on NovaSeq PE250 platform at LC-Bio Technology (Hang Zhou, China). The sequencing data have been archived in the NCBI Sequence Read Archive under accession number PRJNA1045351.
Quality filtering on the raw reads were performed under specific filtering conditions to obtain the high-quality clean tags according to the fqtrim (v0.94). Chimeric sequences were filtered using Vsearch software (v2.3.4). After de-replication using DADA2, we obtained amplicon sequence variants (ASVs). The α-diversity and β-diversity were calculated by QIIME2, and the relative abundance (X fungi count/total count) is used in fungi taxonomy. The rarefaction curves of α-diversity indices, including Chao richness, Shannon index and Good's coverage, were saturated based on the ASV information (Supplementary Figure S1). According to the coverage values were 1.00, the majority of soil microbial associations in soil samples were covered. ASVs functional groups were predicted using FUNGuild (https://github.com/UMNFuN/FUNGuild) (Nguyen et al., 2016).
2.5 Statistical analysis
Mean and standard deviation of soil properties were calculated by Excel. Soil properties and the α-diversity indices were tested by one-way analysis of variance (ANOVA). Their differences among the soil samples were examined by the least significant difference (LSD) test (p < 0.05). These analyses were performed in SPSS 22.0. Liner regression model, columns, and box-plots were created by Origin 2022. Heatmap and correlation network, circos and sankey diagram were performed at the OmicStudio platform (https://www.omicstudio.cn/tool). The β-diversity was assessed through the non-metric multidimensional scaling (NMDS) analysis and the analysis of similarities (ANOSIM) based on Bray-Curtis distances, conducted with the R (version 4.3.2) package “vegan” and visualized using “ggplot2”. To conduct an analysis of differences in microbial communities, we used ALDEx2 analysis and LEfSe analysis for comparison. The ALDEx2 tool was used to determine significant differences at ASV level in terms of relative abundance between pairwise comparisons of fungal communities in soils with different soil pH gradients. Which tool performed the Wilcoxon's rank-sum tests, followed by the Benjamini-Hochberg FDR correction on p-value for each feature (Nearing et al., 2022). The linear discriminant analysis (LDA) together with effect size analysis (LEfSe) to identify fungal biomarkers characterizing the different pH range soil sample groups. The redundancy analysis (RDA) was performed using the same package in R, to identify the relationship between fungal community structure and physicochemical properties of the soils.
Based on maximum likelihood estimation, structural equation model (SEM) used IBM SPSS AMOS 24.0 to analyze the direct or indirect effects of soil properties on fungal communities. Several model fit summaries were used to evaluate our model: non-significant chi-square test (P > 0.05), low χ2/df (< 2), high goodness-of-fit index GFI > 0.9, and low root mean square errors of approximation RMSEA < 0.05.
3 Results
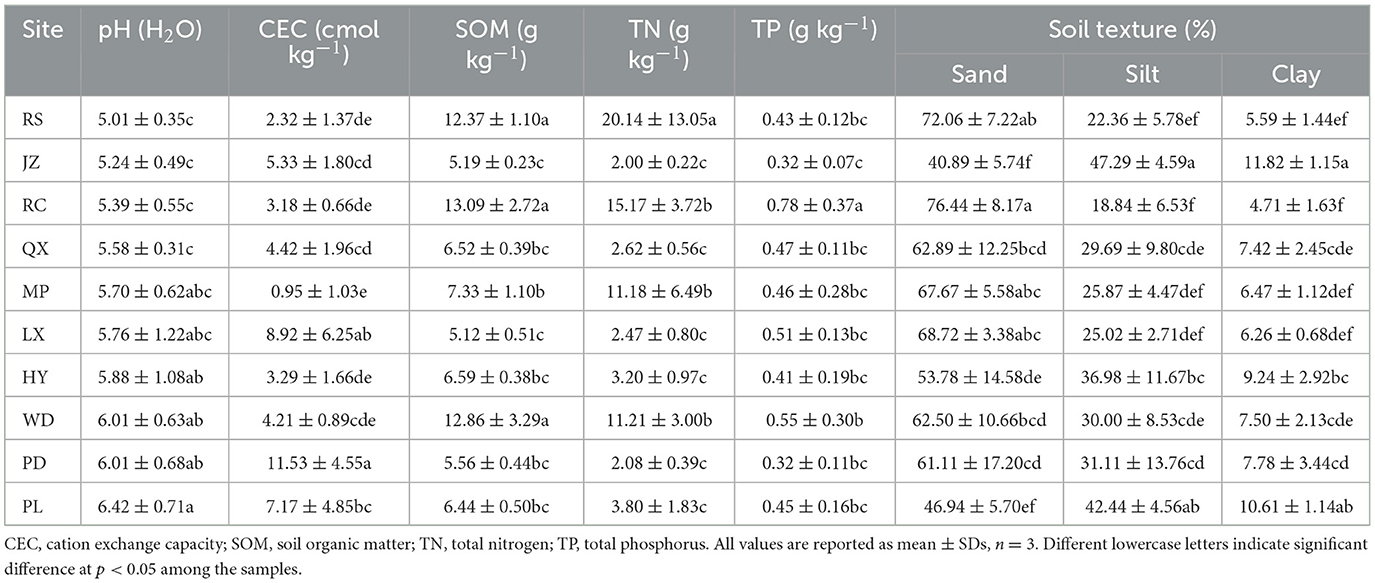
3.1 Soil physicochemical properties
The characteristics of soil acidification in Jiaodong Peninsula were clearly exhilarated in Table 1 and Figure 2A. RS soils had the lowest soil pH value of 5.01, while PL had the highest value (6.42), with the greatest difference of 1.41 units. Accordingly, the contents of EA and in soil were significantly higher at RS than the other sites (p < 0.05), whilst the EAl contents in JZ soils were the highest among all the soil samples (p < 0.05). Whatever, the significantly negative correlation (p < 0.01) between the content of EA and soil pH was also corroborated in the present study (Supplementary Figure S2).
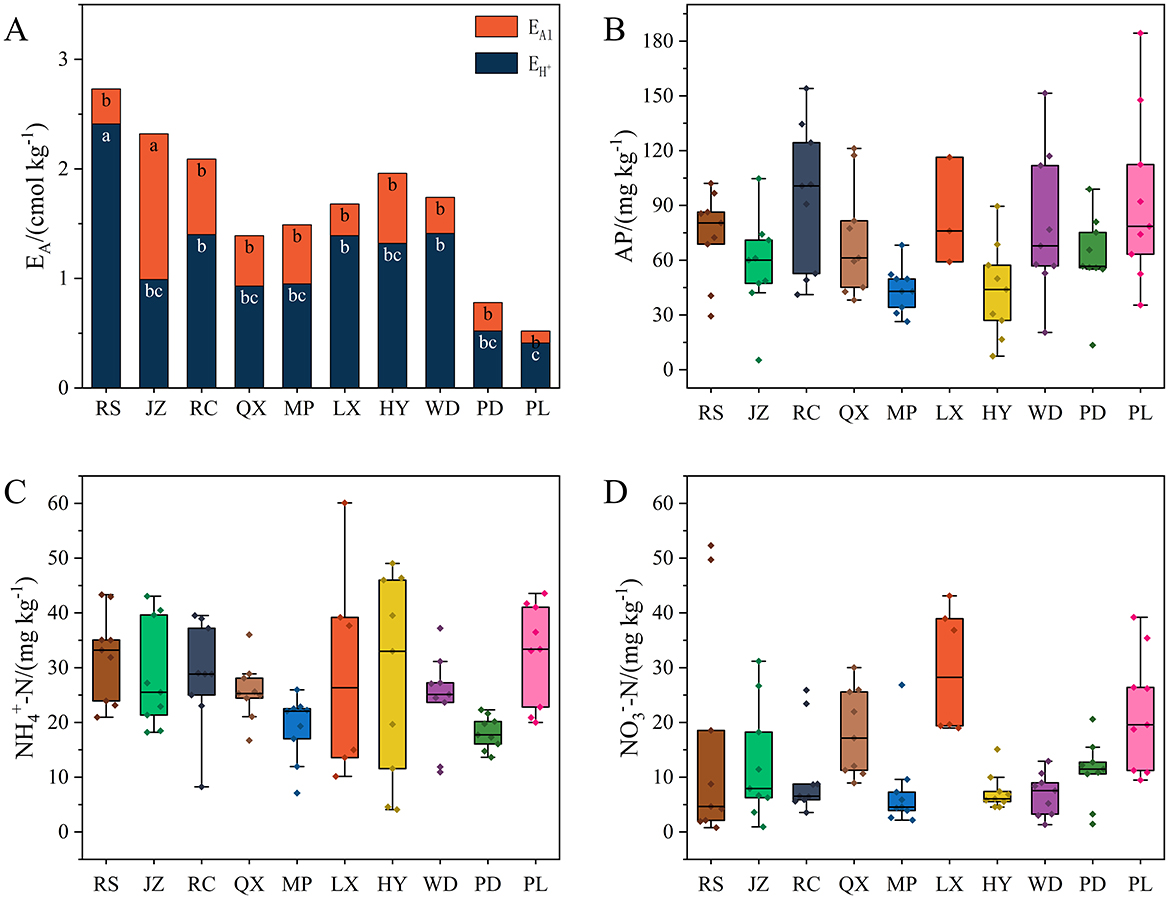
Figure 2. Contents of EA (A), AP (B), -N (C), and -N (D) in soil samples. Different lowercase letters denote significant difference among soil sampling sites (p < 0.05).
In most of soil samples, the nitrogen availability was usually lower with the increase of soil acidity (Figures 2C, D), while the contents of TN followed the opposite trend. The AP and TP contents were the highest in soils sampled from RC, which was consistent to the serious acidic conditions of this site. However, no significant associations were found between soil acidification gradient and the other soil properties, such as the SOM contents were significantly higher in soils from RC, RS and WD than the other sites (p < 0.05) (Figure 2B), whilst the contents of CEC in MP soils were the lowest among all the soil samples (p < 0.05), indicating the soils in MP had lower supply capacity of nutrients than the other sampling sites. There was no obvious fluctuation in the soil texture for most soil samples which predominant component was all of sand.
3.2 Characteristics of fungal community structure
Among all the investigated soils, no significant difference was found in fungal diversity, including Chao richness and Shannon index, even they were under various soil acidification degree (Figures 3A, B). Moreover, the NMDS analysis revealed that fungal communities did not exhibit a distinct separation along the soil pH gradient on the NMDS axis 1 (Figure 4A, Stress = 0.17). The NMDS results based on Bray-Curtis distance showed that the β-diversity of soil fungal communities was similar in Jiaodong Peninsula (Figure 4A), which were also substantiated by the results of ANOSIM (p > 0.05) (Figure 4B). The ALDEx2 analysis revealed that no significant difference was found in fungal community composition under various soil pH gradients (Supplementary Figure S6). This result was consistent with the fungal community diversity with increased soil acidity. All of these results suggested that fungal communities were every tolerant to acidic soils.
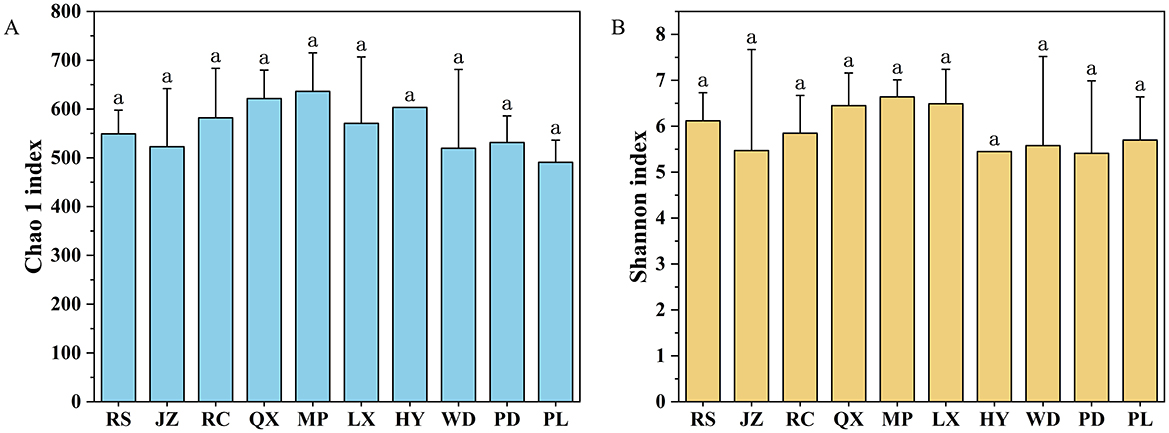
Figure 3. α-diversity indices of fungal communities in farmland: (A) Chao richness, and (B) Shannon index.
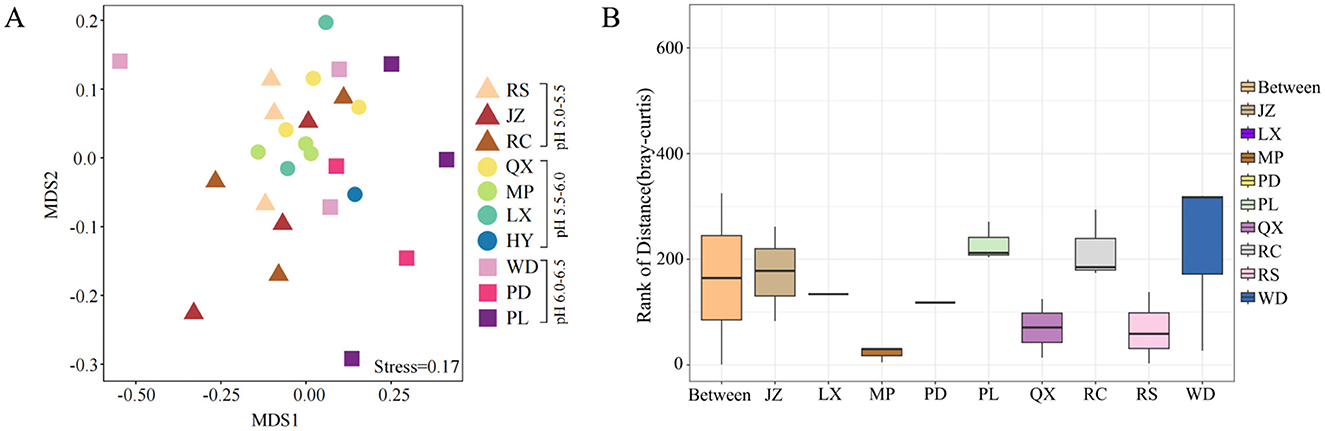
Figure 4. β-diversity of fungal communities in farmland: (A) non-metric multidimensional scaling (NMDS) plot, and (B) the analysis of similarities (ANOSIM) at the ASV level. The box corresponding to the “between” represents the distance value of the difference among groups, while the other boxes represent the distance value of the difference within each group.
The top five fungi with the largest relative abundance at the phylum level and class level were selected for the comparative analysis of fungal community composition (Figures 5A, B). Soil fungal communities in Jiaodong Peninsula primarily included members of phyla Ascomycota (65.24–91.60%), Basidiomycota (3.24–27.17%), Zygomycota (2.60–9.52%), unclassified Fungi (0.44–4.56%) and Chytridiomycota (0.16–1.55%), accounting for 99.88–100.00% of all the identified phyla. The first dominant phylum was Ascomycota, which presented a higher relative abundance in slightly acidic soils compared to strongly acidic soils, however Basidiomycota exhibited an inverse trend. The predominant classes in all soil samples were Dothideomycetes and Sordariomycetes, and in HY and PD soils, Sordariomycetes' relative abundance even exceeded 50% of all the designed classes. However, Dothideomycetes' relative abundance in WD and RS soils was 3.4–and 3.2 - fold higher than that in HY soil, respectively. As shown on Figures 5A, B, the relative abundance of the dominant phyla and classes in acidic soils exhibited spatial variations with increased soil acidity. Further analysis of the top 10 genera (relative abundance > 0.01%) revealed that members of Ascomycota, Basidiomycota, and Zygomycota were shared by all the soil samples (Supplementary Figure S3). The most abundant genera (relative abundance > 1%) were Pseudogymnoascus and Mortierella, followed by Gibberella, Humicola, and Podospora. The relative abundance of Humicola in HY soils had the highest value (43.94), significantly surpassing that of other soils, and this predominance may have been unrelated to soil acidity. In addition, the most representative pathotroph genera, including Gibberella, unclassified Didymellaceae, Oidiodendron and Pseudogymnoascus, were also determined among all soil samples. The RC soil exhibited the highest relative abundance of Pseudogymnoascus at 12.48%, which was classified as strongly acidic soil, whereas the PL soil, characterized as slightly acidic, showed the lowest relative abundance at 2.63%. Within the RS soil, the relative abundance of the unclassified Didymellaceae was the highest, reaching 15.72%, while the relative abundance of PL was the lowest, recorded at 0.21%. Therefore, the pathogenic fungi exhibited significant spatial distribution differences across soils with varying degrees of soil acidification, suggesting that soil pH likely played a pivotal role in shaping the composition and dominance of fungal communities within these ecosystems.
The LEfSe algorithm with an LDA score of 3.5 was used to characterize the differences in soil samples of different pH ranges. The results showed that 30 fungal clades exhibited significant differences in all soil samples (Supplementary Figure S4). Specifically, the less acidic soils (pH 6.0–6.5) were rich in Nectriaceae (family), Thielavia (genus), Nectria (genus), Haematonectria (genus) and unclassified Microascaceae (genus), which were belonged to the phylum Ascomycota. While in the higher acidic soil pH (pH 5.0–5.5), Didymellaceae with the highest abundance was only the biomarker taxa (family), yet belonging to Ascomycota. Besides, the enriched fungal taxa in pH 5.5–6.0 soils were all affiliated with the phyla Ascomycota (from class to genus) and Basidiomycota (from family to genus), but there were exhibited spatial variations in taxon abundance along soil pH gradients.
3.3 Relationship between environmental characteristics and fungal communities
Redundancy analysis (RDA) was applied to identify the effect of environmental characteristics on soil fungal structure in the acidic soils (Figures 6A, B). The eight environmental factors (i.e., pH, SOM, TN, TP, -N, -N, AP, and CEC) explained 31.25 and 31.54% of the variation degree of fungal communities at phylum and class level in the acidic soil samples. The results showed that soil pH, -N and TN were significantly related to the changes in fungal community at phylum level (p < 0.05) (Figure 6A). And at the class level, soil pH had a significant impact on the variation of fungal community structure across all soils (p < 0.01) (Figure 6B). Additionally, structural equation modeling (SEM) was used to fit the causal relationships between soil chemical properties and fungal communities, which explained 80% of the community composition (Figure 7). It was indicated that the soil acidification indicators, including soil pH and EA contents, had a negative direct influence on fungal community composition (p < 0.05). Overall, soil pH was the most important factor driving the soil fungal community composition.
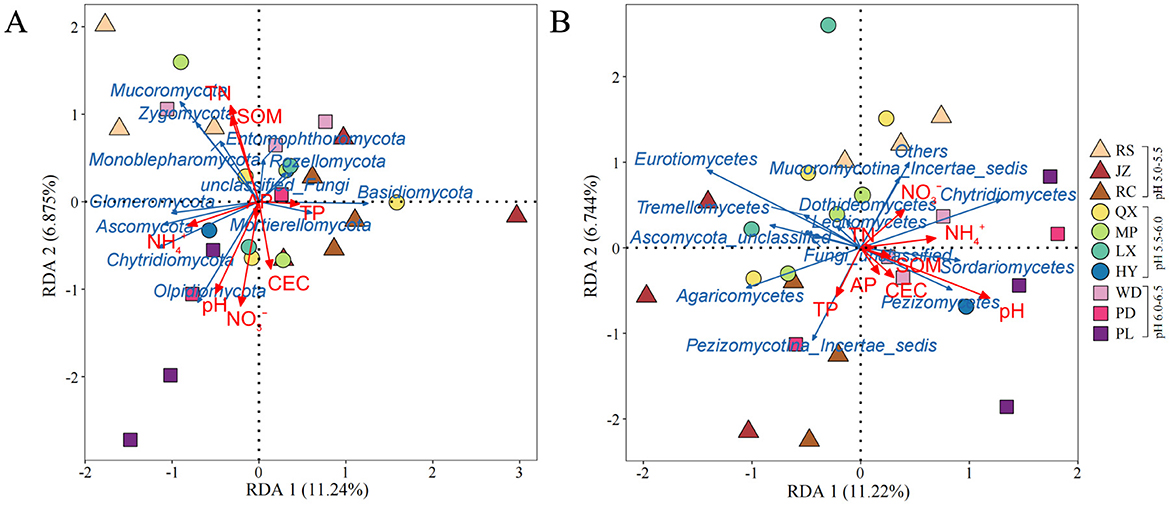
Figure 6. Redundancy analysis (RDA) of the relationships between fungal community structure at the (A) phylum level and (B) class level with soil environmental factors. The arrow length represents the strength of the correlation between the environmental factors and the fungi. And the acute angle (<90 degrees) represents a positive correlation, while the opposite indicates a negative correlation.
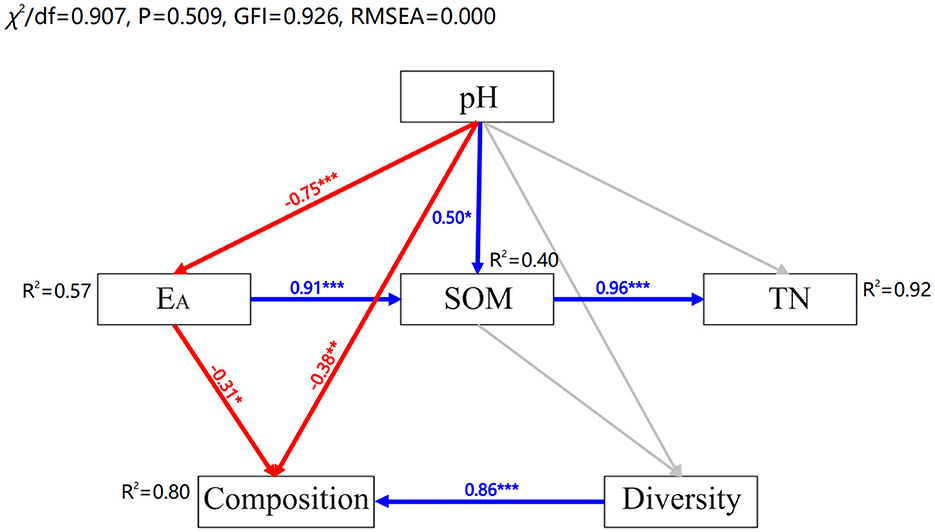
Figure 7. Structural equation modeling based on the effect of soil properties (pH, EA, SOM, and TN) on fungal diversity and composition. Blue and red arrows indicate significant positive and negative correlations, respectively. Gray arrows indicate non-significant relationships. Numbers at the arrows are standardized path coefficients. The width of the arrows indicates the strength of the relationship. R2 indicates the proportion of variance explained by the model. ***p < 0.001, **p < 0.01, *p < 0.05.
3.4 Fungal functional guilds in acidic soils
According to FUNGuild database, 69% of fungi could be categorized into three trophic modes, including saprophytic, pathogenic, symbiotic, or a combination of these. Saprophytic fungi and pathogenic fungi were the most diverse vegetative flora, accounting for 66% of the total fungal communities. There were no significant differences in the spatial distribution of ASVs belonging to saprotrophs and symbiotrophs with soil acidic degree increasing (Supplementary Figure S5). However, compared to the soils with 6 < pH < 6.5, more ASVs were attributed to pathotrophs in soils with a pH ranging from 5 to 6. This result was consistent with the composition of fungal communities at the genus level. Further analysis of the relationship between enriched ASVs and environmental factors at the genus level, we selected Gibberella and unclassified Didymellaceae for correlation analysis with environmental factors. The ASVs assigned to Gibberella were identified as pathotroph, and which allocated to unclassified Didymellaceae were classified as pathotroph-saprotroph. The abundance of Gibberella and unclassified Didymellaceae significantly correlated with the values of pH and contents of EA (p < 0.05) (Figure 8).
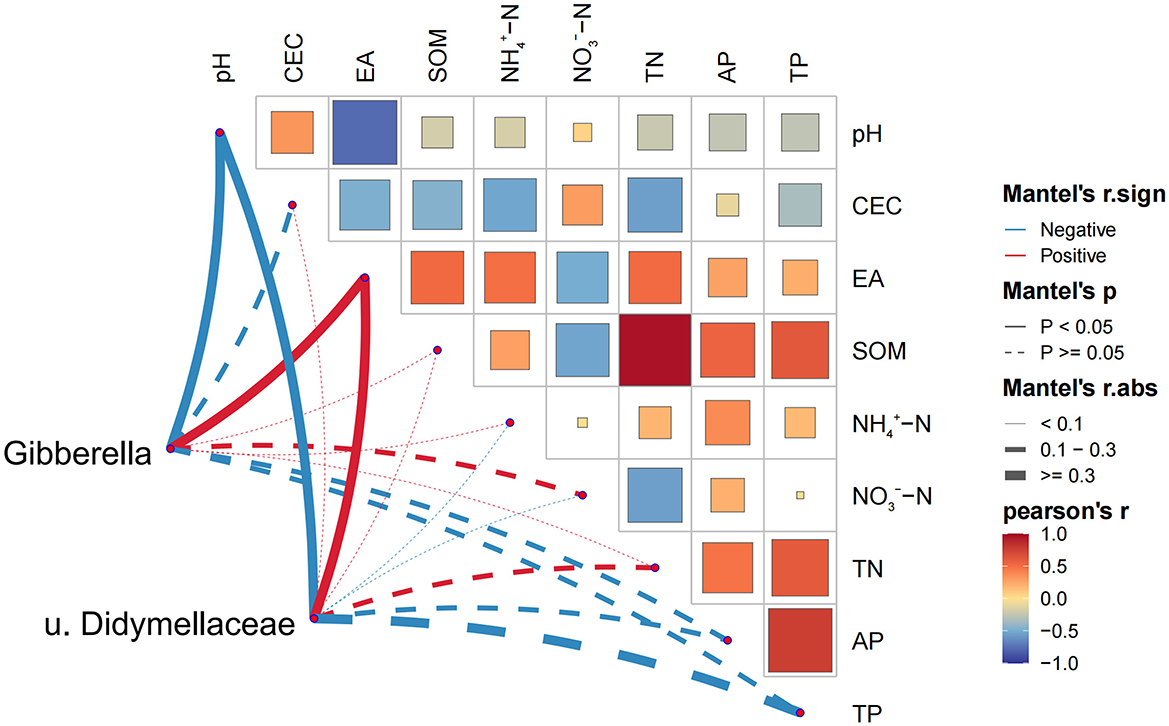
Figure 8. Correlation between Gibberella and unclassified Didymellaceae on soil environmental factors in Mantel test analysis.
4 Discussion
4.1 Distribution and influencing factors of fungi in acidic soils of Jiaodong Peninsula
Fungal species are better adapted and tolerant to acidic environments than bacteria, and can live normally in soils with a pH range from 5 to 9 pH (Nevarez et al., 2009; Liu et al., 2015; Ye et al., 2020). Whilst, it was also reported that soil fungal diversity significantly declined with the decrease of soil pH primarily caused by the long-term N and P fertilizer application (Zhou et al., 2016). However, in our study, little difference in fungal diversity was observed despite of significant variations across soil pH gradients (Figures 2A, 3; Table 1). NMDS and ANOSIM results indicated little difference in soil fungal communities' composition existed across all the soil samples (Figure 4). The opposite results to the prior researches should be related to the wide acclimation of fungi to acidic soils (Xiao et al., 2018; Turley et al., 2020). In addition, it may also be ascribed to their similar soil management practices and cropping systems (Schmidt et al., 2019). In further, the above results might indicate a relatively stable agroecosystem in the Jiaodong Peninsula. However, although the results did not show significant differences in fungal diversity and composition along soil acidic degree, there were still unidentified fungi that have not been specifically analyzed. Furthermore, fungi are only a part of the agroecosystem which cannot represent the whole microbial system. To extend the validity of the findings of this study, further investigation including soil bacteria is required in a larger scale of soil samples.
In contrast to the similar taxa composition, a significant variation of the relative abundance for each fungal taxon, whatever at phylum, class or genera level, were found across various soil samples (Figure 5; Supplementary Figure S3), indicating that the composition pattern of soil fungal communities varied spatially in Jiaodong Peninsula. Similar to previous studies (Ye et al., 2020; Wang et al., 2022), Ascomycota and Basidiomycota were also identified as the dominant fungal phyla in acidic soils of Jiaodong Peninsula (Figure 5A). Variations in the relative abundance of Basidiomycota across sampling sites were consistent with soils acidification degree, whereas Ascomycota being more prevalent in soils with slightly acid (6 < pH < 6.5). Ascomycota, which accounted over 65% of total fungal communities, had significant saprotrophic capacity (Xiong et al., 2014; Maharachchikumbura et al., 2015). An in-depth analysis showed that species of Pseudaleuria (Ascomycota, Pezizomycetes), which were identified as ectomycorrhizal-undefined saprotrophs, were abundant in slightly acidic soils (Supplementary Figure S3) and facilitated plant uptake of nutrients (Hansen et al., 2013; Ma et al., 2023). This indicates that slightly acidic degrees are beneficial for Pseudaleuria growth in Jiaodong Peninsula, which in turn enhances soil productivity (Xiang et al., 2020). Furthermore, the Podospora (Ascomycota, Sordariomycetes) genus are considered to be antifungal agents (Che et al., 2002; Ding et al., 2017), which can suppress the number of pathogenic fungi (e.g., Verticillium) on plant disease (Xu et al., 2012). The relative abundance of Podospora in slightly and moderately acidic soils sampled from LX, HY, PD and PL was clearly higher than that in strongly acidic soils (Supplementary Figure S3). The LEfSe analysis showed that the enrichment of Nectriaceae existed in the pH 6.0–6.5 soils, while the pH 5.0–5.5 soils were enriched with Didymellaceae, suggesting that these fungal taxa could be potential biomarkers for these two acidic soil ecosystems, respectively (Supplementary Figure S4). Most members of Didymellaceae were plant pathogens with a broad host range, primarily causing lesions of the leaves and stems (Chen et al., 2017). However, the typical genera biomarker of Nectriaceae were belonged to saprotroph, which commonly occurred on the soil-litter interface and promoted soil organic C and N turnover and accumulation (Crowther et al., 2012; Yang W. et al., 2017; Zeng and Zhuang, 2022). Therefore, with soil acidic stress increasing, the changes of soil fungal taxa would cause a potential risk on soil nutrient cycling and crop health.
More and more studies suggested that soil microbial communities were sensitive to changes in soil physicochemical properties, including soil pH, SOM and N and P nutrients (Liu et al., 2020; Li B. B. et al., 2021; Liu et al., 2021). In this study, we also found that the contents of -N and -N were quite low in the strongly and moderately acidic soils sampled from RS, JZ, RC, QX, and MP (Figures 2C, D). That is, limited available N in acidic soils, fungi might need to allocate more energy for N uptake and assimilation, resulting in reduced biomass and metabolic rates (Li J. et al., 2021). The following RDA results found that, soil TN, -N as well as pH were significantly associated with fungal communities' abundance (Figure 6), indicating these factors played an important role on fungal community shifting across the investigated soils. This finding is similar to the findings of Dong et al. (2022) who found that mineral-N (-N and -N) significantly affected the composition of the fungal communities. The further SEM analysis confirmed that soil pH played a crucial role in fungal community structure (Figure 7). This is consistent with the prior reports that soil pH was a master factor influencing the microbial community structure in acidic and neutral soils (Geisseler and Scow, 2014; Francioli et al., 2016; Ning et al., 2020). In general, the composition pattern and ecological function of soil fungal communities in agroecosystems of the Jiaodong Peninsula exhibited spatial variations, which mainly modulated by soil acidification.
4.2 Soil fungal functional groups in acidic soils and the potential ecological risk
The FUNGuild analysis revealed that the determined fungal communities in acidic soils of Jiaodong Peninsula mainly belonged to two functional groups of saprophytic and pathogenic fungi (Supplementary Figure S5). Among them, saprophytic fungi can convert complex SOM into available components (Francioli et al., 2021), while pathogenic fungi pose a serious threat to plant growth and health (Doehlemann et al., 2017), both of them are closely related to plant and soil productivity. Moreover, the distribution of saprophytic fungi showed no significant changes across three soil acidic degrees (strongly, moderately and slightly acid), indicating wide adaptations to the degraded environment (Boddy and Hiscox, 2016). Saprotrophic fungi preserved the capacity of soil nutrient supply under the stress of soil acidification (Pan et al., 2023). Whatever, the relative abundance of saprotrophs in slightly acidic soils were higher than that in strongly acidic soils, being consistent with the relative abundance of Ascomycota. It suggests that soil acidic stress might cause inhibition on the decomposition and transformation of organic material, yet affecting soil fertility in acidic soils (Ning et al., 2020).
The development and propagation of fungal plant pathogens in soil is a key factor stressing plant health and food security globally (Jones and Medina, 2020). The FUNGuild analysis indicated that plant pathogenic fungi constituted ~24% of the entire fungal community determined in soils of Jiaodong Peninsula, which might pose a potential threat to crop production as they include most of plant pathogens (Li Y. C. et al., 2020; Zhang et al., 2022). Moreover, the number of pathogenic ASVs in slightly acidic soils were lower than that in strongly acidic soils (Supplementary Figure S5). For example, Gibberella and unclassified Didymellaceae, the representative plant pathogenic fungi, were observed in soil samples of Jiaodong Peninsula, which had significantly higher abundance in strongly and moderately acidic soils (Supplementary Figure S3). Gibberella can easily lead to diseases such as root rot, crown rot, and wilting in crops (Gange et al., 1999; Kazan et al., 2012), indicating that the risk of root rot infection by Gibberella is higher in QX, MP, LX and HY than in other regions of the Jiaodong Peninsula (Tian et al., 2021). Interestingly, we found the highest relative abundance of unclassified Didymellaceae in strongly acidic soils sampled from RS, which could be affect photosynthesis and water transport in plants, thereby increasing the risk of disease in plants in RS. Moreover, the further analysis indicated that the abundance of Gibberella and unclassified Didymellaceae were positively correlated with soil acidification characteristics (Figure 8). Li et al. (2017) also showed that strongly acidic conditions favored the growth of the bacterial wilt in South China. Soil acidification greatly suppressed the capacity of soil microbiome to fight soil-borne pathogens (Li et al., 2023), seriously threating crop health. However, since some other factors (e.g., cropping systems, and MAT) were reported, which also affected the development of plant pathogens in agroecosystems (Thompson et al., 2014; Lu et al., 2022; Song et al., 2022), hence, the risk of infection by plant pathogens should be further evaluated in Jiaodong Peninsula. Taken together, functional guilds in the soils of Jiaodong Peninsula varied with soil acidic degree increasing, and these changes exerted adverse effects both on nutrient cycling and crop productivity.
5 Conclusion
In conclusion, our study demonstrated that in the Jiaodong Peninsula, soil fungal diversity exhibited no significant differences, but there is spatial variation in fungal community composition along the soil acidification gradient, yet there were all dominantly enriched with Ascomycota and Basidiomycota in acidic soils. Soil pH was identified to be the critical factor driving fungal community composition in acidic soils. LEfSe analysis in further revealed that the functional fungi contributing to soil organic matter degradation had a relative higher enrichment in the slightly acidic soils (6.0–6.5), while the fungi interacting with plant pathogens were well-enriched in the acidic soils (5.0–5.5). Therefore, soil acidification could cause more seriously potential risk on agroecosystem in addition to acidic soil environment. So that, in the future, more researches about acidic soils should be concentrated on the impact of microbes on the fertility and crop growth.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. All the DNA sequence data in this manuscript are deposited in the NCBI Sequence Read Archive, accession number PRJNA1045351.
Author contributions
JL: Data curation, Writing – original draft. ZW: Visualization, Writing – review & editing. GH: Data curation, Investigation, Writing – review & editing. GW: Formal analysis, Investigation, Writing – review & editing. FR: Formal analysis, Investigation, Writing – review & editing. YX: Visualization, Writing – review & editing. KL: Visualization, Writing – review & editing. ML: Visualization, Writing – review & editing. AL: Conceptualization, Methodology, Writing – review & editing. HL: Validation, Writing – review & editing. HS: Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The study was supported by the National Natural Science Foundation of China (NSFC) (Nos. 42077129 and 42177403), SDUT & Zibo City Integration Development Project (No. 2021SNPT0012), Shandong Province Natural Science Foundation under Grant No. ZR2020ZD19 and Shandong Provincial Natural Science Foundation for Outstanding Young Scholars (Grant No. ZR2022YQ34).
Acknowledgments
The authors would like to thank Lian-Chuan BioTechnology Co., Ltd for their helps in high throughput DNA sequencing data analysis and the Tutools platform (http://www.cloudtut.com) offering a free platform for data analysis and graphing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1496268/full#supplementary-material
References
Bardgett, R. D., and van der Putten, W. H. (2014). Belowground biodiversity and ecosystem functioning. Nature 515, 505–511. doi: 10.1038/nature13855
Boddy, L., and Hiscox, J. (2016). Fungal ecology: principles and mechanisms of colonization and competition by saprotrophic fungi. Microbiol. Spectr. 4, FUNK-0019-2016. doi: 10.1128/microbiolspec.FUNK-0019-2016
Bolan, N. S., Adriano, D. C., and Curtin, D. (2003). Soil acidification and liming interactions with nutrientand heavy metal transformation and bioavailability. Adv. Agron. 78, 215–272. doi: 10.1016/S0065-2113(02)78006-1
Cairney, J. W. G. (2012). Extramatrical mycelia of ectomycorrhizal fungi as moderators of carbon dynamics in forest soil. Soil Biol. Biochem. 47, 198–208. doi: 10.1016/j.soilbio.2011.12.029
Cao, T. T., Fang, Y., Chen, Y. R., Kong, X. S., Yang, J. B., Alharbi, H., et al. (2022). Synergy of saprotrophs with mycorrhiza for litter decomposition and hotspot formation depends on nutrient availability in the rhizosphere. Geoderma 410:115662. doi: 10.1016/j.geoderma.2021.115662
Cao, T. T., Luo, Y. C., Shi, M., Tian, X. J., and Kuzyakov, Y. (2024). Microbial interactions for nutrient acquisition in soil: miners, scavengers, and carriers. Soil Biol. Biochem. 188:109215. doi: 10.1016/j.soilbio.2023.109215
Che, Y. S., Gloer, J. B., Koster, B., and Malloch, D. (2002). Decipinin A and decipienolides A and B: new bioactive metabolites from the coprophilous fungus podospora decipiens. J. Nat. Prod. 65, 916–919. doi: 10.1021/np010575p
Chen, L., Zhang, J. B., Zhao, B. Z., Yan, P., Zhou, G. X., and Xin, X. L. (2014). Effects of straw amendment and moisture on microbial communities in Chinese fluvo-aquic soil. J. Soil Sedim. 14, 1829–1840. doi: 10.1007/s11368-014-0924-2
Chen, L. X., Duan, W. B., and Qiao, L. (2011). Study on nutrition characteristics and acidity in rhizosphere and non-rhizosphere soils in larch plantations. J. Soil Water Conserv. 25, 131–135. doi: 10.13870/j.cnki.stbcxb.2011.03.052
Chen, Q., Hou, L. W., Duan, W. J., Crous, P. W., and Cai, L. (2017). Didymellaceae revisited. Stud. Mycol. 87, 105–159. doi: 10.1016/j.simyco.2017.06.002
Cheng, Y. T., Li, P., Xu, G. C., Wang, X. K., Li, Z. B., Cheng, S. D., et al. (2021). Effects of dynamic factors of erosion on soil nitrogen and phosphorus loss under freeze-thaw conditions. Geoderma 390:114972. doi: 10.1016/j.geoderma.2021.114972
Ciesielski, H., and Sterckeman, T. (1997). Determination of cation exchange capacity and exchangeable cations in soils by means of cobalt hexamine trichloride. Effects of experimental conditions. Agronomie. 17, 1–7. doi: 10.1051/agro:19970101
Crowther, T. W., Boddy, L., and Hefin Jones, T. (2012). Functional and ecological consequences of saprotrophic fungus–grazer interactions. ISME J. 6, 1992–2001. doi: 10.1038/ismej.2012.53
De Coninck, B., Timmermans, P., Vos, C., Cammue, B. P. A., and Kazan, K. (2015). What lies beneath: belowground defense strategies in plants. Trends Plant Sci. 20, 91–101. doi: 10.1016/j.tplants.2014.09.007
Ding, J. L., Jiang, X., Guan, D. W., Zhao, B. S., Ma, M. C., Zhou, B. K., et al. (2017). Influence of inorganic fertilizer and organic manure application on fungal communities in a long-term field experiment of Chinese Mollisols. Appl. Soil Ecol. 111, 114–122. doi: 10.1016/j.apsoil.2016.12.003
Doehlemann, G., Ökmen, B., Zhu, W. J., and Sharon, A. (2017). Plant pathogenic fungi. Microbiol. Spectr. 5, FUNK-0023-2016. doi: 10.1128/microbiolspec.FUNK-0023-2016
Dong, H. Y., Shao, S. A., Liang, C. F., Xu, Q. F., Chen, J. H., and Qin, H. (2022). Impacts of litter composition on the structure and functional pathways of soil microbial community during phyllostachys edulis expansion. Agronomy 12:220. doi: 10.3390/agronomy12010220
Egidi, E., Delgado-Baquerizo, M., Plett, J. M., Wang, J., Eldridge, D. J., Bardgett, R. D., et al. (2019). A few Ascomycota taxa dominate soil fungal communities worldwide. Nat. Commun. 10:2369. doi: 10.1038/s41467-019-10373-z
Francioli, D., Schulz, E., Lentendu, G., Wubet, T., Buscot, F., and Reitz, T. (2016). Mineral vs. organic amendments: microbial community structure, activity and abundance of agriculturally relevant microbes are driven by long-term fertilization strategies. Front. Microbiol. 7:1446. doi: 10.3389/fmicb.2016.01446
Francioli, D., van Rijssel, S. Q., van Ruijven, J., Termorshuizen, A. J., Cotton, T. E. A., Dumbrell, A. J., et al. (2021). Plant functional group drives the community structure of saprophytic fungi in a grassland biodiversity experiment. Plant Soil 461, 91–105. doi: 10.1007/s11104-020-04454-y
Gange, A. C., Lindsay, D. E., and Ellis, L. S. (1999). Can arbuscular mycorrhizal fungi be used to control the undesirable grass Poa annua on golf courses? J. Appl. Ecol. 36, 909–919. doi: 10.1046/j.1365-2664.1999.00456.x
Geisseler, D., and Scow, K. M. (2014). Long-term effects of mineral fertilizers on soil microorganisms – A review. Soil Biol. Biochem. 75, 54–63. doi: 10.1016/j.soilbio.2014.03.023
Guo, J. H., Liu, X. J., Zhang, Y., Shen, J. L., Han, W. X., Zhang, W. F., et al. (2010). Significant acidification in major Chinese croplands. Science 327, 1008–1010. doi: 10.1126/science.1182570
Hansen, K., Perry, B. A., Dranginis, A. W., and Pfister, D. H. (2013). A phylogeny of the highly diverse cup-fungus family Pyronemataceae (Pezizomycetes, Ascomycota) clarifies relationships and evolution of selected life history traits. Mol. Phylogenet. Evol. 67, 311–335. doi: 10.1016/j.ympev.2013.01.014
Hu, Z. K., Delgado-Baquerizo, M., Fanin, N., Chen, X. Y., Zhou, Y., Du, G. Z., et al. (2024). Nutrient-induced acidification modulates soil biodiversity-function relationships. Nat. Commun. 15:2858. doi: 10.1038/s41467-024-47323-3
Jones, T.-K. L., and Medina, R. F. (2020). Corn stunt disease: an ideal insect–microbial–plant pathosystem for comprehensive studies of vector-borne plant diseases of corn. Plants 9:747. doi: 10.3390/plants9060747
Karlsson, I., Friberg, H., Steinberg, C., and Persson, P. (2014). Fungicide effects on fungal community composition in the wheat phyllosphere. PLoS ONE 9:e111786. doi: 10.1371/journal.pone.0111786
Kazan, K., Gardiner, D. M., and Manners, J. M. (2012). On the trail of a cereal killer: recent advances in Fusarium graminearum pathogenomics and host resistance. Mol. Plant Pathol. 13, 399–413. doi: 10.1111/j.1364-3703.2011.00762.x
Kochian, L. V., Piñeros, M. A., and Hoekenga, O. A. (2005). The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil 274, 175–195. doi: 10.1007/s11104-004-1158-7
Kunhikrishnan, A., Thangarajan, R., Bolan, N. S., Xu, Y., and Naidu, R. (2016). Functional relationships of soil acidification, liming, and greenhouse gas flux. Adv. Agron. 139, 1–71. doi: 10.1016/bs.agron.2016.05.001
Li, B. B., Roley, S. S., Duncan, D. S., Guo, J., and Tiedje, J. M. (2021). Long-term excess nitrogen fertilizer increases sensitivity of soil microbial community to seasonal change revealed by ecological network and metagenome analyses. Soil Biol. Biochem. 160:108349. doi: 10.1016/j.soilbio.2021.108349
Li, C. Y., Kong, X. Q., and Dong, H. Z. (2020). Nitrate uptake, transport and signaling regulation pathways. J. Nucl. Agric. Sci. 34, 982–993. doi: 10.11869/j.issn.100-8551.2020.05.0982
Li, J., Sang, C. P., Yang, J. Y., Qu, L. R., Xia, Z. W., Sun, H., et al. (2021). Stoichiometric imbalance and microbial community regulate microbial elements use efficiencies under nitrogen addition. Soil Biol. Biochem. 156:108207. doi: 10.1016/j.soilbio.2021.108207
Li, S. L., Liu, Y. Q., Wang, J., Yang, L., Zhang, S. T., Xu, C., et al. (2017). Soil acidification aggravates the occurrence of bacterial wilt in South China. Front. Microbiol. 8:703. doi: 10.3389/fmicb.2017.00703
Li, X., Chen, D., Carrión, V. J., Revillini, D., Yin, S., Dong, Y., et al. (2023). Acidification suppresses the natural capacity of soil microbiome to fight pathogenic Fusarium infections. Nat. Commun. 14:5090. doi: 10.1038/s41467-023-40810-z
Li, Y. C., Li, Z. W., Arafat, Y., and Lin, W. X. (2020). Studies on fungal communities and functional guilds shift in tea continuous cropping soils by high-throughput sequencing. Ann. Microbiol. 70:7. doi: 10.1186/s13213-020-01555-y
Liu, J., Sui, Y., Yu, Z., Shi, Y., Chu, H., Jin, J., et al. (2015). Soil carbon content drives the biogeographical distribution of fungal communities in the black soil zone of northeast China. Soil Biol. Biochem. 83, 29–39. doi: 10.1016/j.soilbio.2015.01.009
Liu, J. L., Li, S. Q., Yue, S. C., Tian, J. Q., Chen, H., Jiang, H. B., et al. (2021). Soil microbial community and network changes after long-term use of plastic mulch and nitrogen fertilization on semiarid farmland. Geoderma 396:115086. doi: 10.1016/j.geoderma.2021.115086
Liu, J. S., Ma, Q., Hui, X. L., Ran, J. Y., Ma, Q. X., Wang, X. S., et al. (2020). Long-term high-P fertilizer input decreased the total bacterial diversity but not phoD-harboring bacteria in wheat rhizosphere soil with available-P deficiency. Soil Biol. Biochem. 149:107918. doi: 10.1016/j.soilbio.2020.107918
Lu, J., Li, W. X., Yang, Y. H., Ye, F., Lu, H. Y., Chen, X. Y., et al. (2022). The impact of different rotation regime on the soil bacterial and fungal communities in an intensively managed agricultural region. Arch. Microbiol. 204:142. doi: 10.1007/s00203-021-02615-w
Ma, J. F., and Ryan, P. R. (2010). Foreword: understanding how plants cope with acid soils. Funct. Plant Biol. 37, iii–vi. doi: 10.1071/FPv37n4_FO
Ma, Y. J., Shen, S. Z., Wan, C., Wang, S. Q., Yang, F. X., Zhang, K. Q., et al. (2023). Organic fertilizer substitution over six years improves the productivity of garlic, bacterial diversity, and microbial communities network complexity. Appl. Soil Ecol. 182:104718. doi: 10.1016/j.apsoil.2022.104718
Maharachchikumbura, S. S. N., Hyde, K. D., Jones, E. B. G., McKenzie, E. H. C., Huang, S.-K., Abdel-Wahab, M. A., et al. (2015). Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers. 72, 199–301. doi: 10.1007/s13225-015-0331-z
Mosier, A. C., Miller, C. S., Frischkorn, K. R., Ohm, R. A., Li, Z., LaButti, K., et al. (2016). Fungi contribute critical but spatially varying roles in nitrogen and carbon cycling in acid mine drainage. Front. Microbiol. 7:238. doi: 10.3389/fmicb.2016.00238
Muneer, M. A., Huang, X. M., Hou, W., Zhang, Y. D., Cai, Y. Y., Munir, M. Z., et al. (2021). Response of fungal diversity, community composition, and functions to nutrients management in red soil. J. Fungi 7:554. doi: 10.3390/jof7070554
Nearing, J. T., Douglas, G. M., Hayes, M. G., MacDonald, J., Desai, D. K., Allward, N., et al. (2022). Microbiome differential abundance methods produce different results across 38 datasets. Nat. Commun. 13:342. doi: 10.1038/s41467-022-28034-z
Nevarez, L., Vasseur, V., Madec, A. L., Bras, M. A. L., Coroller, L., Leguérinel, I., et al. (2009). Physiological traits of Penicillium glabrum strain LCP 08.5568, a filamentous fungus isolated from bottled aromatised mineral water. Int. J. Food Microbiol. 130, 166–171. doi: 10.1016/j.ijfoodmicro.2009.01.013
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Ning, Q., Chen, L., Jia, Z. J., Zhang, C. Z., Ma, D. H., Li, F., et al. (2020). Multiple long-term observations reveal a strategy for soil pH-dependent fertilization and fungal communities in support of agricultural production - ScienceDirect. Agric. Ecosyst. Environ. 293:106837. doi: 10.1016/j.agee.2020.106837
Pan, C., Yu, W., Sun, C. C., Guo, J. H., Yu, Y. C., and Li, X. G. (2023). Saprotrophic fungi buffer the adverse effects of soil acidification on the soil nutrient supply ability of Chinese fir (Cunninghamia lanceolata) plantations. Eur. J. Soil Biol. 114:103462. doi: 10.1016/j.ejsobi.2022.103462
Pan, F. X., Li, Y. Y., Chapman, S. J., and Yao, H. Y. (2015). Effect of rice straw application on microbial community and activity in paddy soil under different water status. Environ. Sci. Pollut. Res. 23, 5941–5948. doi: 10.1007/s11356-015-5832-5
Pascale, A., Proietti, S., Pantelides, I. S., and Stringlis, I. A. (2020). Modulation of the root microbiome by plant molecules: the basis for targeted disease suppression and plant growth promotion. Front. Plant Sci. 10:1741. doi: 10.3389/fpls.2019.01741
Peñalva, M. A., Tilburn, J., Bignell, E., and Arst, H. N. (2008). Ambient pH gene regulation in fungi: making connections. Trends Microbiol. 16, 291–300. doi: 10.1016/j.tim.2008.03.006
Pérez-Izquierdo, L., Rincón, A., Lindahl, B. D., and Buée, M. (2021). Fungal community of forest soil: diversity, functions, and services. Forest Microbiol. 1, 231–255. doi: 10.1016/B978-0-12-822542-4.00022-X
Schmidt, R., Mitchell, J., and Scow, K. (2019). Cover cropping and no-till increase diversity and symbiotroph:saprotroph ratios of soil fungal communities. Soil Biol. Biochem. 129, 99–109. doi: 10.1016/j.soilbio.2018.11.010
Shen, F. F., Wu, J. P., Fan, H. B., Guo, X. M., and Wo, Q. D. (2018). Litterfall ecological stoichiometry and soil available nutrients under long-term nitrogen deposition in a Chinese fir plantation. Acta Ecol. Sinica 38, 7477–7487. doi: 10.5846/stxb201711242095
Shi, Y., Li, Y. T., Xiang, X. J., Sun, R. B., Yang, T., He, D., et al. (2018). Spatial scale affects the relative role of stochasticity versus determinism in soil bacterial communities in wheat fields across the North China Plain. Microbiome 6:27. doi: 10.1186/s40168-018-0409-4
Soil Survey Staff (2014). Keyto Soil Taxonomy. Washington, DC: United States Department of Agriculture (USDA).
Song, X. L., Huang, L., Li, Y. Q., Zhao, C. Z., Tao, B., and Zhang, W. (2022). Characteristics of soil fungal communities in soybean rotations. Front. Plant Sci. 13:926731. doi: 10.3389/fpls.2022.926731
Strickland, M. S., and Rousk, J. (2010). Considering fungal:bacterial dominance in soils – methods, controls, and ecosystem implications. Soil Biol. Biochem. 42, 1385–1395. doi: 10.1016/j.soilbio.2010.05.007
Thompson, S. E., Levin, S., and Rodriguez-Iturbe, I. (2014). Rainfall and temperatures changes have confounding impacts on Phytophthora cinnamomi occurrence risk in the southwestern USA under climate change scenarios. Glob. Chang. Biol. 20, 1299–1312. doi: 10.1111/gcb.12463
Tian, L. X., Ou, J. R., Sun, X., Miao, Y. J., Pei, J., Zhao, L., et al. (2021). The discovery of pivotal fungus and major determinant factor shaping soil microbial community composition associated with rot root of American ginseng. Plant Signal. Behav. 16:1952372. doi: 10.1080/15592324.2021.1952372
Turley, N. E., Bell-Dereske, L., Evans, S. E., Brudvig, L. A., and Yang, G. (2020). Agricultural land-use history and restoration impact soil microbial biodiversity. J. Appl. Ecol. 57, 852–863. doi: 10.1111/1365-2664.13591
Wahab, A., Muhammad, M., Munir, A., Abdi, G., Zaman, W., Ayaz, A., et al. (2023). Role of arbuscular mycorrhizal fungi in regulating growth, enhancing productivity, and potentially influencing ecosystems under abiotic and biotic stresses. Plants 12:3102. doi: 10.3390/plants12173102
Wang, H. F., Li, C. Y., and Liu, X. X. (2009). Improvement on digestion methods in determination of total phosphorus in water by ammonium molybdate spectrophotometric method. China Water Wastewater 25, 81–83. doi: 10.1016/S1874-8651(10)60073-7
Wang, T. T., Cao, X. X., Chen, M. M., Lou, Y. H., Wang, H., Yang, Q. G., et al. (2022). Effects of soil acidification on bacterial and fungal communities in the Jiaodong Peninsula, Northern China. Agronomy 12:927. doi: 10.3390/agronomy12040927
Xiang, X. J., Liu, J., Zhang, J., Li, D. M., Xu, C. X., and Kuzyakov, Y. (2020). Divergence in fungal abundance and community structure between soils under long-term mineral and organic fertilization. Soil Till. Res. 196:104491. doi: 10.1016/j.still.2019.104491
Xiao, D., Huang, Y., Feng, S. Z., Ge, Y. H., Zhang, W., He, X. Y., et al. (2018). Soil organic carbon mineralization with fresh organic substrate and inorganic carbon additions in a red soil is controlled by fungal diversity along a pH gradient. Geoderma 321, 79–89. doi: 10.1016/j.geoderma.2018.02.003
Xiong, J. B., Peng, F., Sun, H. B., Xue, X., and Chu, H. Y. (2014). Divergent responses of soil fungi functional groups to short-term warming. Microb. Ecol. 68, 708–715. doi: 10.1007/s00248-014-0385-6
Xu, L. H., Ravnskov, S., Larsen, J., Nilsson, R. H., and Nicolaisen, M. (2012). Soil fungal community structure along a soil health gradient in pea fields examined using deep amplicon sequencing. Soil Biol. Biochem. 46, 26–32. doi: 10.1016/j.soilbio.2011.11.010
Yang, C., Li, J. J., Liu, N., and Zhang, Y. J. (2019). Effects of fairy ring fungi on plants and soil in the alpine and temperate grasslands of China. Plant Soil 441, 499–510. doi: 10.1007/s11104-019-04141-7
Yang, F., Xu, Y., Cui, Y., Meng, Y. D., Dong, Y., Li, R., et al. (2017). Variation of soil organic matter content in croplands of china over the last three decades. Acta Pedol. Sin. 54, 1047–1056.
Yang, W., Zhao, H., Leng, X., Cheng, X. L., and An, S. Q. (2017). Soil organic carbon and nitrogen dynamics following Spartina alterniflora invasion in a coastal wetland of eastern China. Catena 156, 281–289. doi: 10.1016/j.catena.2017.03.021
Ye, G. P., Lin, Y. X., Luo, J. F., Di, H. J., Lindsey, S., Liu, D. Y., et al. (2020). Responses of soil fungal diversity and community composition to long-term fertilization: field experiment in an acidic Ultisol and literature synthesis. Appl. Soil Ecol. 145:103305. doi: 10.1016/j.apsoil.2019.06.008
Zeng, Z. Q., and Zhuang, W. Y. (2022). New species of nectriaceae (hypocreales) from China. J. Fungi 8:1075. doi: 10.3390/jof8101075
Zhang, X., Zhao, W., Liu, Y., He, H., Kou, Y., and Liu, Q. (2022). Dominant plant species and soil properties drive differential responses of fungal communities and functions in the soils and roots during secondary forest succession in the subalpine region. Rhizosphere 21:100483. doi: 10.1016/j.rhisph.2022.100483
Zhao, X., Zhang, Z., Zhao, M., Song, X., Liu, X., and Zhang, X. (2023). The quantified and major influencing factors on spatial distribution of soil organic matter in provincial-scale farmland—a case study of Shandong Province in Eastern China. Appl. Sci. 13:3738. doi: 10.3390/app13063738
Keywords: acidic soils, fungal community composition, fungal functional guilds, soil pH, plant pathogens
Citation: Liu J, Wazir ZG, Hou G, Wang G, Rong F, Xu Y, Liu K, Li M, Liu A, Liu H and Sun H (2025) Composition and the predicted functions of fungal communities and the key drivers in acidic soils of Jiaodong Peninsula, China. Front. Microbiol. 15:1496268. doi: 10.3389/fmicb.2024.1496268
Received: 14 September 2024; Accepted: 09 December 2024;
Published: 06 January 2025.
Edited by:
Marika Pellegrini, University of L'Aquila, ItalyReviewed by:
Setu Bazie Tagele, University of California, Riverside, United StatesShengtao Xu, Yunnan Academy of Agricultural Sciences, China
Copyright © 2025 Liu, Wazir, Hou, Wang, Rong, Xu, Liu, Li, Liu, Liu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aiju Liu, YWlqdmxpdUBzZHV0LmVkdS5jbg==; Hongliang Liu, ZW1pY3JvYmVAc2R1dC5lZHUuY24=
 Jing Liu1
Jing Liu1 Guoqin Hou
Guoqin Hou Fangxu Rong
Fangxu Rong Kai Liu
Kai Liu Aiju Liu
Aiju Liu Hongliang Liu
Hongliang Liu Hongwen Sun
Hongwen Sun