
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 January 2025
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1490293
Antifungal peptides are promising drug candidates to fight fungal infections in the clinics and agriculture. However, recent data suggest that antifungal peptides might also play a role within their own producing organism to survive nutrient limiting conditions. We have therefore studied the function of the antifungal AnAFP in Aspergillus niger in more detail. To achieve this, we established a Tet-on controlled anafp expression system, which allowed us to study a null and an overexpression phenotype in the same isolate. We observed that increased intracellular AnAFP expression reduces growth of A. niger and prematurely activates autophagy. Comparative transcriptome analyses of glucose-starving mycelium demonstrated that increased anafp expression strongly impacts expression of genes important for cell wall integrity and remodeling, as well as genes with a predicted function in metabolism and transport of carbohydrates, proteins, and lipids. Notably, genes encoding regulators of conidiophore development such as flbC and flbD became induced upon anafp overexpression. Fluorescent analyses of a Tet-on driven AnAFP::eGFP fusion protein congruently unraveled that AnAFP localizes to cell walls and septa of A. niger. Moreover, AnAFP::eGFP expression is spatially restricted to selected compartments only and affected cells displayed a sudden reduction in hyphal diameter. From these data we conclude that AnAFP is important to drive vegetative growth and sporulation in A. niger during nutrient limitation through autophagic recycling. We predict that AnAFP drives nutrient mobilization through selective cell lysis to ensure the survival of the whole colony during phases of starvation.
The estimated worldwide annual agricultural loss due to pathogenic fungi range from 14 to 18 percent (Savary et al., 2019). Even more threatening is the number of about 1.5 million humans who die each year because of fungal infections (Fisher et al., 2020). To fight the increasing numbers of fungal infections, antimicrobial peptides are discussed as an alternative to traditional drugs and antibiotics, as they have lower risks for resistance development (Cesare et al., 2020). Of specific interests are antifungal peptides from fungal origin which are currently understudied. About 1,400 antifungal peptides are listed in the Antimicrobial Peptide Database (APD3), but only a few dozen are from fungi (Wang et al., 2016; Feurstein et al., 2022; Paege et al., 2016). AFP, the first antifungal peptide from a fungus was isolated in 1965 from the supernatant of the filamentous fungus Aspergillus giganteus and served as eponym for other members of the family (Olson and Goerner, 1965; Meyer and Jung, 2018). All peptides of the AFP family are structurally highly conserved and are secreted by filamentous fungi, likely to fight competing fungi in their vicinity. Many of these peptides undergo processing after a signal peptide is cleaved, before maturing to their active form. They are small (about 50 amino acids), net positively charged, and enriched in the amino acids glycine, cysteine and lysine. They contain a specific cysteine spacing pattern that form 3–4 disulfide bridges thought to contribute to their resistance against denaturation or proteolytic degradation. Remarkably, all AFPs are amphipathic which enable them to interact with fungal plasma membranes, although it is thought that the integrity of the plasma membranes of the respective producing fungus remains unaffected (Paege et al., 2019; Hagen et al., 2007; Struyfs et al., 2021). Ultimately, this interaction can either lead to the loss of membrane integrity or the peptides are taken up by the cell to induce regulated cell death. AFP targets can be glucosylceramides, while binding sites for cell wall polymers, i.e., chitin were also reported (Paege et al., 2019; Hagen et al., 2007; Struyfs et al., 2021). However, the specific target(s) for many AFP members remain(s) unknown.
Despite the strong structural similarities among the AFP family members, attacked fungi activate different defense mechanisms to fight their growth inhibitory effect (Meyer and Jung, 2018). For example, the cell wall integrity and the calcium/calcineurin pathway that both lead to the synthesis of cell wall polymers such as glucans and chitin are important to survive attacks by AFP and AnAFP from A. giganteus and A. niger, respectively, whereas the G protein and cAMP/PKA pathway that leads to membrane polarization, rapid calcium influx, and apoptosis-like cell death is fundamental to survive the attack by PAF from Penicillium chrysogenum or NFAP from Neosartorya fischeri, respectively.
Notably, AFPs are under tight temporal and spatial expression control in their own producing strains, with a gene expression profile which is similar to that of autophagic genes. Furthermore, expression of AFPs becomes strongly upregulated during adverse conditions including nutrient limitation, osmotic, oxidative, or pH stress but not necessarily in the presence of a competing fungal organism (Meyer and Jung, 2018). The genes are expressed in axenic cultures and conceptually mirror the very tight expression pattern and growth inhibitory activity of cannibal-toxins in endospore forming bacteria (Meyer and Jung, 2018). We thus recently proposed that AFPs could play an endogenous role for their producing organisms comparable to bacterial cannibal toxins, i.e., they sacrifice a subpopulation of its producing mycelium to recycle nutrients under nutrient-limiting conditions (Meyer and Jung, 2018). In agreement, studies on PAF from P. chrysogenum and AFPB from P. digitatum demonstrated that these AFPs induce cell death of their host cells (Kovács et al., 2014; Bugeda et al., 2020). Also, two meta-transcriptomic analyses in A. niger uncovered that the anafp co-expression network, which consists of about 600 positively and 400 negatively co-expressed genes, is enriched in gene functions driving processes such as asexual development, polysaccharide catabolism, carbon starvation, nutrient recycling, autophagy, autolysis, cell death, and asexual development (Paege et al., 2016; Schäpe et al., 2023). Proven regulators of anafp expression in A. niger are so far CreA (carbon catabolite repressor), VelC (activator of secondary metabolism) and StuA (repressor of asexual development) (Paege et al., 2016; Schäpe et al., 2019). Interestingly, regulation of anafp expression was predicted to be downstream of flbA-E genes (the signaling cascade that activates the asexual developmental transcription factor BrlA) but upstream of the brlA gene (Meyer and Jung, 2018).
To better understand the endogenous function of AnAFP in A. niger, we harnessed in this study the doxycycline-responsive conditional Tet-on expression system, which is well established for A. niger (Meyer et al., 2011; Wanka et al., 2016a; Cairns et al., 2023). We generated a strain in which anafp expression is off (no doxycycline addition to the growth medium) or switched on (addition of doxycycline). The impact of AnAFP on the growth of A. niger was determined and autophagy quantified in the presence or absence of anafp expression. The effect of anafp expression was further investigated using RNA sequencing, differential gene expression and gene ontology analyses. Finally, we tracked the intracellular localization of natively expressed AnAFP fused to the green fluorescent protein eGFP using confocal microscopy and live cell imaging.
All strains used in this study are listed in Table 1. E. coli TOP 10 (Invitrogen) was used for cloning. A. niger was cultivated in minimal medium (MM) or in complete medium (CM) as described previously (Paege et al., 2016). All A. niger strains were stored at −70°C in 0.9% NaCl and 16.7% glycerol. For the generation of a spore solution, CM agar plates were inoculated from the cryo stock and spores were harvested after a 3-day cultivation at 30°C. Spores were suspended in 8 mL 0.9% sodium chloride solution which was filtered through a Miracloth filter. Spore solutions were always made fresh and were stored a maximum of 3 days at 4°C in darkness, prior to inoculation.
The A. niger strains STS9.2, STS19.3, LV1.1 and LV2.2 were generated via Cas9-mediated DNA integration; 5 μg of purified Cas9 was preassembled with 65 pmol of in vitro transcribed sgRNA (Kwon et al., 2019). Cas9 (fused N-terminally to a 6xHis tag and C-terminally to the nuclear localization sequence KKRKV) was expressed in E. coli CZ4.1, using the plasmid pET28a/Cas9-Cys (Addgene #53261). The donor DNAs for BBA22.6, STS4.10, STS9.2 and JW3.3 were assembled via Gibson cloning and the DNAs for STS19.3, LV1.1 and LV2.3 were generated via the modular cloning kit from Addgene (Weber et al., 2011). DNA integration in BBA22.6 was performed via single recombination into the pyrG locus of strain MA170.27, using hygromycin as a selection marker. For the generation of STS4.10, the Tet-on promotor was fused to the predicted 5’ untranslated region for anafp in A. niger CBS513.88 (46 bp) (Delmas et al., 2012; Hagiwara et al., 2016). Additionally, the trpC terminator used for conventional expression under the Tet-on promotor was replaced by the native anafp terminator (1 kb length) as it includes the 3´-UTR of anafp. A split marker approach was used for integration and selection was performed using a recyclable pyrG marker gene from A. oryzae (Arentshorst et al., 2015b). The anafp promotor region (1 kb length) was used as 5’ homology arm and the anafp terminator was used as the 3’ homolgy arm. In strain STS9.2, the promotor and the open reading frame (ORF) were deleted via replacement with a hygromycin resistance cassette, with 1 kb homology arms binding 1 kb upstream of the anafp promotor and to the anafp terminator, respectively. For the generation of strain STS19.3, the donor DNA was introduced in STS9.2 via double crossing over, using the same homology arms as used for STS9.2. The AnAFP::eGFP fusion protein is expressed under the Tet-on promotor fused to the 5´ UTR of anafp. For the generation of strain JW1.1, STS4.10 was cultivated on 5-FOA while strain JW3.3 was created through a single recombination event into the pyrG locus of strain JW1.1 as described earlier (Arentshorst et al., 2015a). For the development of knock-out cassettes used in strains LV1.1 and LV2.1, a terbinafine resistance cassette of PhttA::ergA::Thxk was amplified with 60 bp overhang primers homologous to the regions directly upstream of the start codon or downstream of the stop codon of atg1 or atg8. Standard techniques of molecular biology used in this work were described previously (Meyer et al., 2014; Meurant, 2012). All strains were confirmed by Southern blotting as described previously (Sambrook and Russell, 2001). Probes used for detection are listed in Additional File 1.
For growth quantification of A. niger strains on CM agar plates (90 mm), 105 spores in 5 μL physiological salt (0.9% NaCl) solution were point-inoculated in the center of mixed cellulose ester (MCE) membranes [Millipore S-Pak Filter 0.22 μM, 47 mm] which were placed on 20 mL of solidified CM agar. After 16 h of incubation at either 30°C or 37°C, the membranes were photographed and weighed and transferred to a fresh CM agar plate supplemented with 0, 5, or 20 μg/mL doxycycline. After further incubation for 48 h, membranes were weighed again. Biomass and the colony diameter difference was calculated by subtraction of corresponding values between 16 and 64 h of cultivation. Spores were harvested by mixing MCE membranes with 15 mL of physiological salt (0.9% NaCl) solution and a cotton stick was used to bring the spores into solution. The spore concentrations were counted using a Thoma cell counting chamber and the total amount of spores was divided by the colony diameter of the colony.
To measure the optical density of A. niger cultures, the Aquila Biolabs system was used, which records backscattered light in shake flask cultures (Bruder et al., 2016). Cell suspension measurements were taken at 60-s intervals using the CGQuant software (version 3.5) allowing for real-time, automated and non-invasive optical density measurements from the flasks in motion. Control flasks with uninoculated cultivation medium served to monitor background scatter. Given the variability of initial optical density values, including those in control flasks, the first 10 readings (10 min) were averaged and set to zero.
Aspergillus niger strain STS4.10 was cultivated at 30°C in 250 mL shake flasks containing 50 mL of CM and 20 μg/mL doxycycline inoculated with 5×106 spores/mL. After 24, 48, 72, and 96 h of cultivation, 30 mL of culture broth were centrifuged (4,000 g, 15 min) and filtered (0.2 μm). The supernatants were applied to an FPLC gradient cation-exchange chromatography with a SikiIIPak TOYOPEARL Sulfate – 650F 5 mL, 8×100 mm column from Tosoh Bioscience. The method did run for 78 min at 1 mL/min with buffer A (50 mM NaOAc, pH4) and buffer B (50 mM NaOAc +1.5 M NaCl, pH4). 0–8 min 100% buffer A, 8–70 min 0 to 100% B as a gradient, 72–78 min 100% B.
Elution fractions containing a single Peak at around 65% buffer B were applied to a gradient reversed phase HPLC using a MultoHigh®Bio-200-C18 5 μ column (125×4 mm), (A) milli-Q water and (B) acetonitrile (both solvents acidified with 0.1% TFA). The method run at a flow rate of 1 mL/min, starting with 2% solvent B for 2 min, increasing 2–49% B between 4–16 min and 49–95% B between 16–18 min, hold 95% B till 20 min and decrease 95 to 2% B till 34 min. Concentrations were calculated from the area under the curve using a previously purified and MS verified AnAFP standard (1 mg/mL) as reference. Mass spectrometry analyses was performed at the Institute of Chemistry, Technische Universität Berlin, with an Orbitrap XL from Thermo Fisher Scientific and electron spray ionization. Before ionization, samples were separated via reversed phase liquid chromatography (Grom-Sil 120 ODS-4- HE, 3 µm). Here, eluent 1 (H2O + 0.1% HCOOH) and eluent 2 (acetonitrile+0.1% HCOOH) were used at a flow rate of 0.3 mL/min.
For localization of eGFP to vacuoles in A. niger strains JW3.3, LV1.1, and LV2.3, respectively, microscopy coverslips were placed in MM supplemented with 0.003% yeast extract (Ohly) inoculated with 5 mL of 103 spores/mL inside a 6 well plate. Biological duplicates were generated for each strain. After 16 h of incubation at 30°C, starvation and anafp expression was induced through media exchange (MM lacking glucose and yeast extract but containing 0 or 20 μg/mL doxycycline). The coverslips were further incubated at 30°C and imaged 4 h after media exchange through differential interference contrast and fluorescence microscopy (green channel) on a Leica DM5000 CS system equipped with a DFC365 FX CCD camera. Six images from each biological sample were captured using a HC PL APO CS2 40x objective with a 1.30 aperture. A minimum of 100 vacuoles across all images were counted.
Confocal laser scanning microscopy (CLSM) was performed on A. niger strain STS19.3. 103 spores/mL were cultivated within an 8-well μ-slide (ibiTreat) with each well containing 300 μL of MM. After 16 h of incubation at 30°C, starvation and anafp expression was induced through media exchange (MM lacking glucose and yeast extract but containing 0 or 20 μg/mL doxycycline). Immediately after inducing carbon starvation by medium change, automated CLSM was performed hourly for a duration of up to 72 h. Imaging was conducted on a LEICA TCS SP8 microscope, utilizing a 20x objective (HC PL APO CS2 IMM, NA: 0.75) paired with a pump that supplied water as immersion fluid for imaging. Microscopy was performed within a temperature-controlled box at 30°C. The excitation laser was set at 488 nm and the detector was set to wavelengths between 495 nm and 545 nm. Z-stacks up to 250 nm were used to create focus stacks of the fluorescence channel using ImageJ (version 1.53 s), while Helicon Focus (version 8.1.0) was used for stacking bright field images.
Investigation of apoptosis-like cell death phenotype was performed as described recently (Semighini and Harris, 2010). In brief, glass coverslips were coated with 0.01% poly-L-lysine solution (Sigma) according to the instruction provided by the manufacturer. Coated coverslips were UV-sterilized for 30 min, sealed and stored at room temperature until use. They were transferred to 6-well plates and 5 mL of MM, inoculated with spores of STS4.10 and FW35.1 to a final concentration of 105 spores/mL. Samples were incubated at 30°C for 15 h. After incubation, the medium was replaced by fresh and preheated MM containing 0, 5 or 20 μg/mL doxycycline. For positive controls, 0, 5, 10, 100, 500, or 850 mM H2O2 were used instead of doxycycline. The samples were further incubated for 4 h at 30°C. Afterwards, medium was removed, and 5 mL of fixation solution [5% DMSO, 3.7% formaldehyde, 85% PEM buffer (25 mM EGTA, 5 mM MgSO4, 50 mM PIPES)] was added to each well, followed by incubation for 15 min at room temperature. After removing the fixation solution, the coverslips were transferred to new wells containing 5 mL PEM buffer and incubated for 5 min. Buffer was discarded and incubation was repeated twice with fresh 5 mL PEM buffer for each time. Next, coverslips were transferred to new 6-well plates with 5 mL of nuclear staining solution (1,000 ng/mL Hoechst 33258 in ultrapure water) and incubated for 5 min. Nuclear staining solution was protected from light throughout the whole experiment. Afterwards, the solution was removed, and 5 mL of PEM buffer was added, followed by an incubation for 3 min. The buffer was discarded, and PEM buffer incubation was repeated three times. Coverslips were carefully dried and transferred, with the hyphae facing down, onto a drop of 10 μL mounting solution (1× PBS buffer, 50% glycerol, 0.1% n-propyl-gallate) on a glass slide. After removing excess fluid, the slides were sealed with transparent nail polish and stored overnight at 4°C. For each sample, 10 images of different regions of a coverslip were taken. Brightfield and fluorescence images [using the CFP channel (460–500 nm) were recorded as z-stacks (10–30 images per stack)] on a Leica DM5000 CS with the CCD microscope camera Leica DFC365 FX, which was used with the 100x, 1.40 aperture HCX PL APO CS objective. For brightfield images, the exposure time was set to 34 ms, for fluorescence images the exposure time was set to 1 s, each setting with a gain of 1.0. Z stacks were processed in image J. All necessary image analysis steps can be found in the ImageJ macro code available in Additional File 6.
For anafp mRNA quantification, A. niger strains BBA22.6 and STS4.10 were cultivated in 250 mL shake flask cultures filled with 50 mL CM, containing 5×106 spores/mL, in the presence or absence of 20 μg/mL doxycycline, at 30°C and 250 rpm in a shaking incubator (Infors HT, Multitron II). Strains were grown until the mid-exponential growth phase (for 16 h), the biomass was harvested through filtration (Sartorius cotton filter 3 hw, 45 mm, 8–12 μm), shock frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Aspergillus niger strains STS4.10 and VG8.27 were cultivated in a 5 L bioreactor (BioStat A Plus Sartorius) at 30° C as we have described earlier (Nitsche et al., 2012). The strains were cultivated in four liters CM inoculated with 106 spores/mL, in the absence of doxycycline. The initial pH was set to 5.8 and not regulated throughout the cultivation. After 14 h of cultivation, when fungal growth reached the mid-exponential phase, 7 mL of culture broth were harvested in duplicates and filtered through nitrocellulose membranes (Sartorius 0.45 μM, 50 mm). The membranes were placed on glucose free MM agar plates with 20 mL medium containing 0 or 5 μg/mL doxycycline. An additional nitrocellulose membrane was placed on top of the biomass and pressed down tightly with a sterile pestle to avoid an air interface and thus repress sporulation. After 35 h of starvation at 30°C, biomass was scraped from the membranes and shock frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Biomass samples stored at −80°C were thawed on ice and approximately 200–300 mg of wet biomass was mixed with 1 mL of TRIzol solution (Thermo Scientific) and glass beads in a 2 mL screw cap tube. Cell disruption was performed using a VWR 4-Place Mini Bead Mill Homogeniser, applying three 30-s intervals at maximum speed (5 m/s). In-between, cells were put on ice for at least 1 min. Cell debris were centrifuged at 4 °C for 5 min at maximum speed. Total RNA was purified using the Zymo Research Direct-zol RNA Miniprep Kit (R2050) following the manufacturer’s instructions. To eliminate possible genomic DNA, the DNA-free™ DNA Removal Kit from Thermo Fischer (AM1906) was used. cDNA synthesis was carried out with the RevertAid cDNA Synthesis Kit from Thermo Fischer (K1632). Non reverse transcribed RNA was used to control for gDNA contamination in qPCR experiments. For qPCR, 100 ng of cDNA were used as a template with the Blue S’green qPCR master mix (Biozym). RNA quality for qPCR was verified by agarose gel electrophoresis. Actin was amplified using the forward primer: TGTACCCCGGTATCTCCGAC and the reverse primer: CTCGTCGTACTCCTGCTTGG, anafp was amplified using ATGCAGCTCACCAGCATTGCCATC (forward) and TCTGGCAATCGACCGTCTTA (reverse).
RNA samples were quantified using Qubit 4.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and RNA integrity was checked with RNA Kit on Agilent 5300 Fragment Analyzer (Agilent Technologies, Palo Alto, CA, USA). RNA sequencing libraries were prepared using the NEBNext Ultra RNA Library Prep Kit for Illumina following the manufacturer’s instructions (NEB, Ipswich, MA, USA). The libraries were sequenced using the NovaSeq instrument with a minimum of 10 million reads per sample. Sequencing quality was verified using MultiQC (Ewels et al., 2016) and untrimmed reads were mapped against the published genome of A. niger ATCC64974 (N402) using STAR (Dobin and Gingeras, 2015). For GO term analysis, differentially expressed genes with a log2 fold change >1 and a p-value <0.05 (1349 genes, FDR-corrected, Benjamini-Hochberg method (Benjamini and Hochberg, 1995)) were translated into syntenic orthologs of A. niger CBS513.88 (1229 genes) using FungiDB (Basenko et al., 2018). For non-syntenic orthologs, BLASTp was used against the genome of A. niger CBS513.88 and only the best hit with a score > 60 was considered as an ortholog (101 genes). To classify genes according to their function, existing gene lists were used to find overlaps among the differentially expressed, syntenic, and non-syntenic genes. Raw RNA Seq data has been deposited under: https://www.ncbi.nlm.nih.gov/geo/, accession code: GSE241846. GO term enrichment analysis for differentially expressed genes responding to afpB overexpression in P. digitatum was done using FungiFun2 in P. digitatum (strain PHI26 / CECT 20796) (Priebe et al., 2015).
Given that the primary function of AnAFP is to protect the extracellular space against nutrient competitors, one would expect highest anafp promoter activity at hyphal tips of foraging hyphae at the colony’s edge. To visualize the dynamics of the anafp promoter activity, we thus used our previously described A. niger strain PK2.9 in which the anafp gene has been deleted and the anafp promoter drives expression of the reporter gene luciferase anafp instead (Paege et al., 2016). This strain was cultivated on complete medium (CM) agar in the presence of luciferin to resolve the anafp promoter activity spatially and temporally through long-exposure photography. Supplementary Video S1 and Figure 1 illustrate that the luminescent signal forms a concentric ring along the outer rim of the colony that continuously moves with the same speed as the growth speed of the colony. Most notably, the concentric anafp expression ring has a thickness of only 1–2 mm and is continuously kept a few mm behind the growth front, i.e., exactly when the vegetative mycelium acquires the competence to form conidiophores (Schäpe et al., 2023). This demonstrates that transcriptional activation of the anafp promotor does not occur at the apical growth front but becomes induced when asexual development becomes initiated and shortly thereafter repressed.
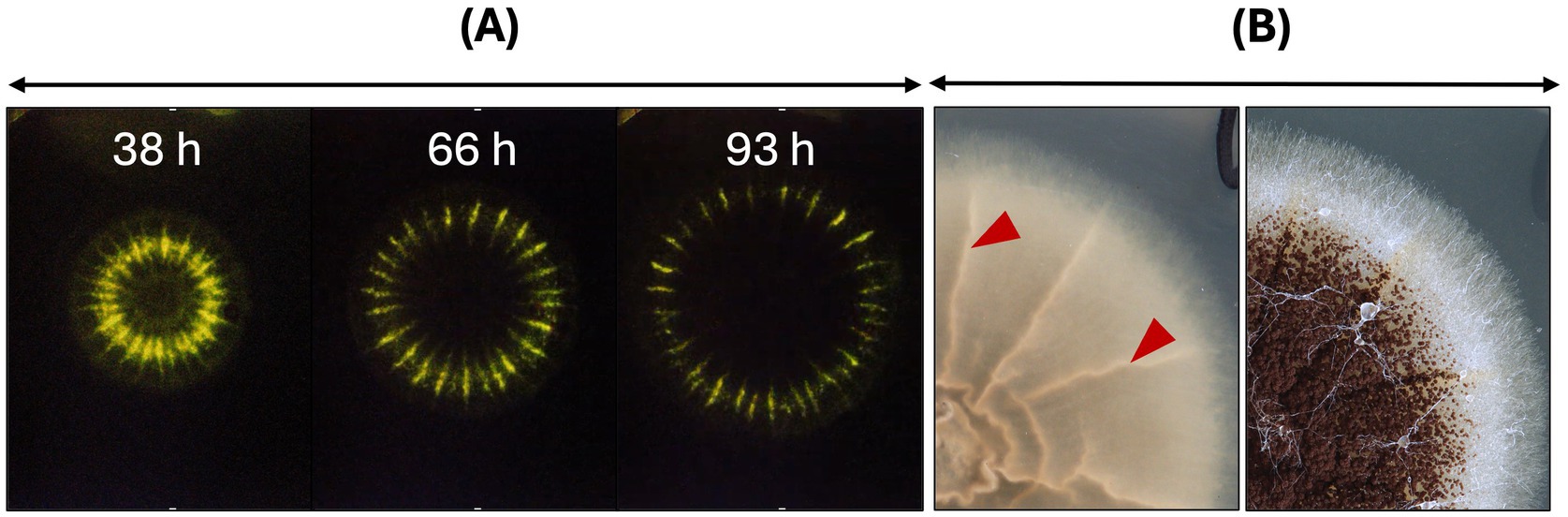
Figure 1. The anafp promotor activity within a growing colony of A. niger on solid CM agar. (A) The promotor activity of A. niger PK2.9 (Panafp-luciferase; Δanafp) is visualized through the luciferase reporter gene, which uses medium-supplemented luciferin as substrate [for cultivation and detection details see Schäpe et al. (2023)]. Three timepoints, derived from a time lapse video of long exposure photographs in darkness, photographed from the bottom (see Supplementary Video S1). The luminescence within the colony of A. niger appears as a greenish-yellow concentric ring and is enhanced in wrinkled areas of the colony likely due to more condensed biomass expressing the reporter gene. Note that the luminesce signal fades during prolonged incubation time, supposedly due to limited luciferin stability. (B) Colony of PK2.9 photographed from the bottom (left) and top (right). Red arrowheads mark wrinkled colony structures, where luminescence is detected as much brighter signal. An overlay of short exposed colonies under light conditions and luminescent long-exposed images can be seen in Schäpe et al. (2023).
A conditional Tet-on::anafp strain was harnessed to study the effect of anafp deletion and overexpression on the growth of A. niger. Therefore, the coding sequence of anafp was cloned into the Tet-on system (Meyer et al., 2011) and integrated in the previously described strain MA170.27 lacking the anafp gene (Wanka et al., 2016a). Integration via homologous recombination at the pyrG locus in the resulting strain BBA22.6 was verified by Southern blotting and its capability to transcribe anafp mRNA in response to doxycycline (Dox) via qPCR. For cloning details and Southern data see Materials and methods and Supplementary Figure S1. Figure 2A demonstrates that anafp mRNA is detectable in large amounts in the presence of 20 μg/mL Dox, while nearly absent when Dox was omitted. However, the AnAFP protein was unexpectedly not detected in the supernatant of shake flask cultures between 17 h and 96 h (Figure 2B). We thus suspected that the presence of the anafp 5’ and 3′ untranslated region in the Tet-on system might be of importance for post-transcriptional regulation of the anafp gene (Note that usually only the ORF of a gene of interest is cloned in the Tet-on system), where it becomes integrated directly after the minimal promoter of gpdA (Pmin) that follows by operator binding site tetO7 (Meyer et al., 2011). Untranslated regions (UTRs) are positioned in the promotor and terminator regions of eukaryotic genes and often play central roles in the translational regulation of eukaryotic genes (Leppek et al., 2018). We thus determined the UTR sequences of the anafp gene from previously published RNA sequencing data (Delmas et al., 2012; Hagiwara et al., 2016; Schäpe et al., 2023) and decided to integrate the Tet-on promoter at the native anafp locus, i.e., after the anafp promotor region and right before the predicted 5’ UTR sequence (−46 bp upstream of the start codon ATG). The terminator sequence (1 kb, including the 3’ UTR) was left unchanged. For cloning strategy and Southern blot verification, see Supplementary Figure S2. The resulting strain was named STS4.10. Through the integration of the Tet-on::anafp expression cassette at the native anafp genomic locus, we could detect both Dox-dependent transcription of anafp (Figure 2A) and secretion of AnAFP into the culture supernatant of STS4.10, as early as 17 h of cultivation (Figure 2B). The identity and full-length of AnAFP was verified via mass spectrometry (Supplementary Figure S3).
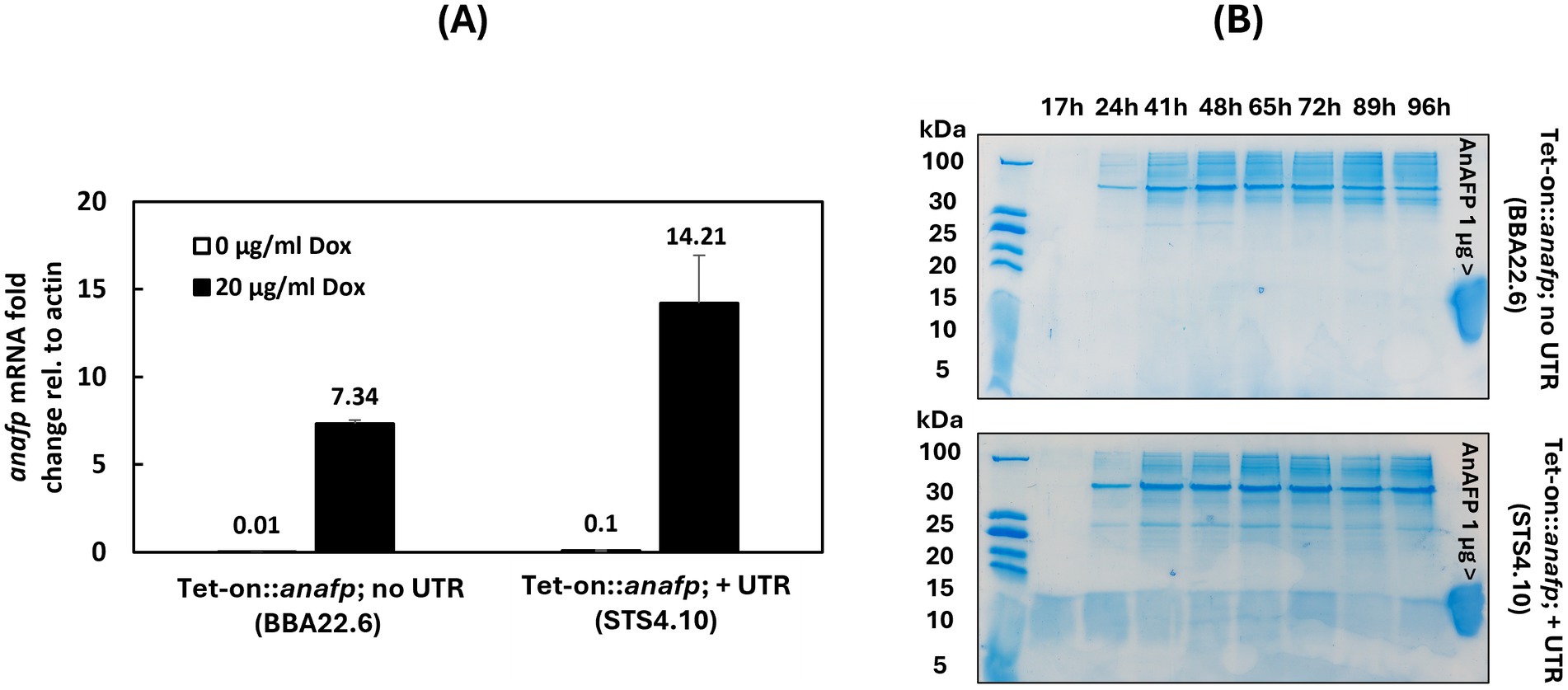
Figure 2. Tet-on driven anafp gene expression and the impact of 5′ and 3’ untranslated regions on its translation to AnAFP. (A) Quantification of anafp mRNA abundance in response to Dox for strain BBA 22.6 (Tet-on::anafp; no UTR) and strain STS4.10 (Tet-on::anafp, with UTR). (B) SDS-PAGE of a 50 mL culture supernatant of strains BBA22.6 (top) and STS4.10 (bottom). The strains were cultivated in CM + 20 μg/mL Dox at 30°C. Culture supernatants were sampled between 17 and 96 h, concentrated 4-times and 20 μL were used for protein gel electrophoresis. 1 μg purified AnAFP was used as positive control. Note that AnAFP protein bands often appear as a smear around 10 kDa.
To study the effects of AnAFP on the growth of A. niger, spores of strain STS4.10 (Tet-on::anafp) were point-inoculated on complete medium (CM) agar supplemented with 0, 5 or 20 μg/mL doxycycline (Dox) and biomass accumulation was monitored over time. As depicted in Figure 3, colony diameter reduced with increasing Dox concentrations, i.e., increased AnAFP production, an effect that was more pronounced when the cultivation temperature was set to 37°C compared to 30°C. Notably, there was a growth reduction measured in the overexpression control VG8.27 (Tet-on-luciferase). However, this reduction in growth was not as pronounced as it was observable in STS4.10 (Tet-on::anafp). Additionally, the number of spores produced remained mostly unaffected, except for a slight increase of spores per colony diameter at 20 μg/mL at 30°C. A similar reduction on growth through AnAFP was observed, when strain STS4.10 was cultivated in liquid shake flask cultures (Supplementary Figure S4). With this cultivation approach, we were able to determine the amount of secreted AnAFP by HPLC. A maximum of 1.4 μg/mL AnAFP was detectable after 24 h of cultivation, which declined to about 0.75 μg/mL AnAFP until the end of cultivation (96 h, Figure 4A).
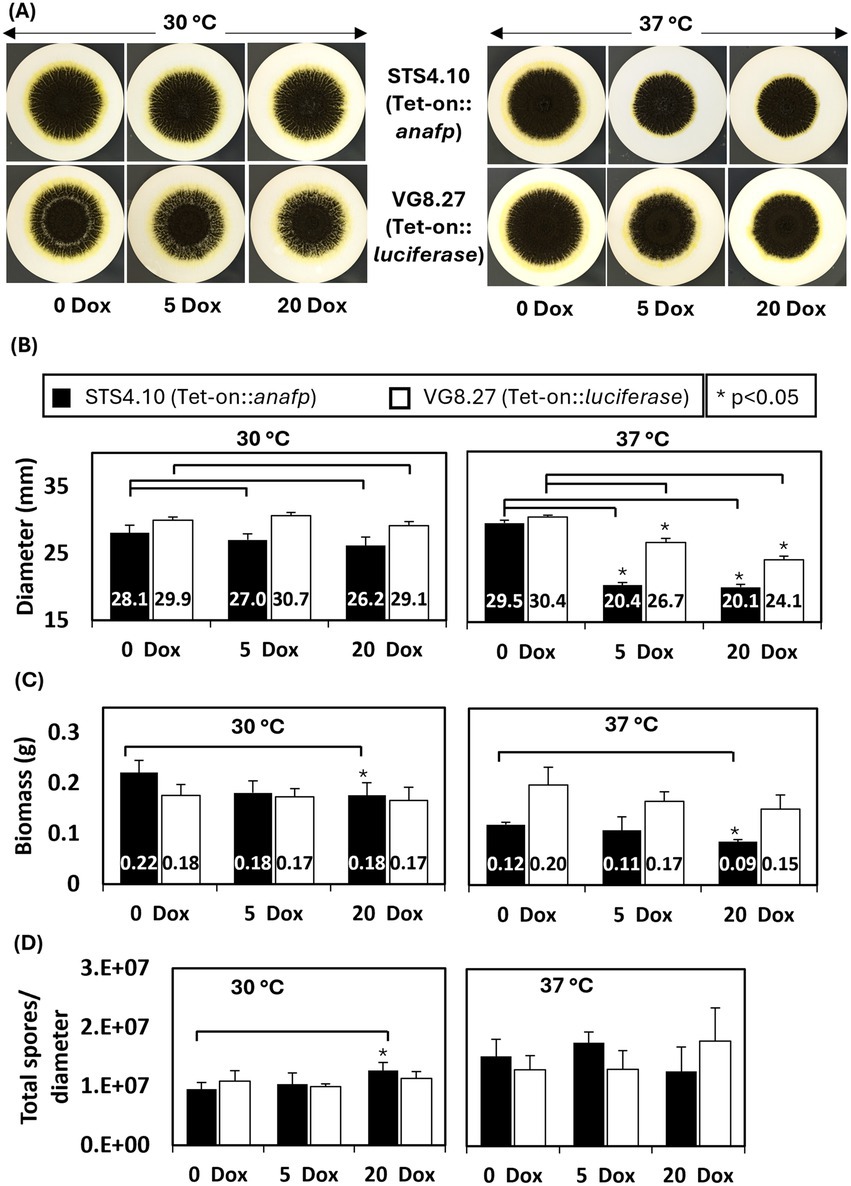
Figure 3. Overexpression of AnAFP reduces growth but not sporulation in A. niger. (A) Colony of strain STS4.10 (Tet-on::anafp) (top) and VG8.27 (Tet-on-luciferase) (bottom) on a mixed cellulose ester membrane (0.2 μm) on complete medium agar containing 0, 5 and 20 μg/mL Dox at 30°C or 37°C. After 64 h, colony diameter (B), biomass (C) and spore production normalized to the colony diameter (D) were determined as described in Materials and methods. Black bars refer to strain STS4.10 and white bars to a control strain VG8.27 which expresses the reporter gene luciferase instead of anafp under control of Tet-on (Meyer et al., 2011). Data derive from biological triplicate experiments (n = 3), p-value derived from paired, two tailed T-test.
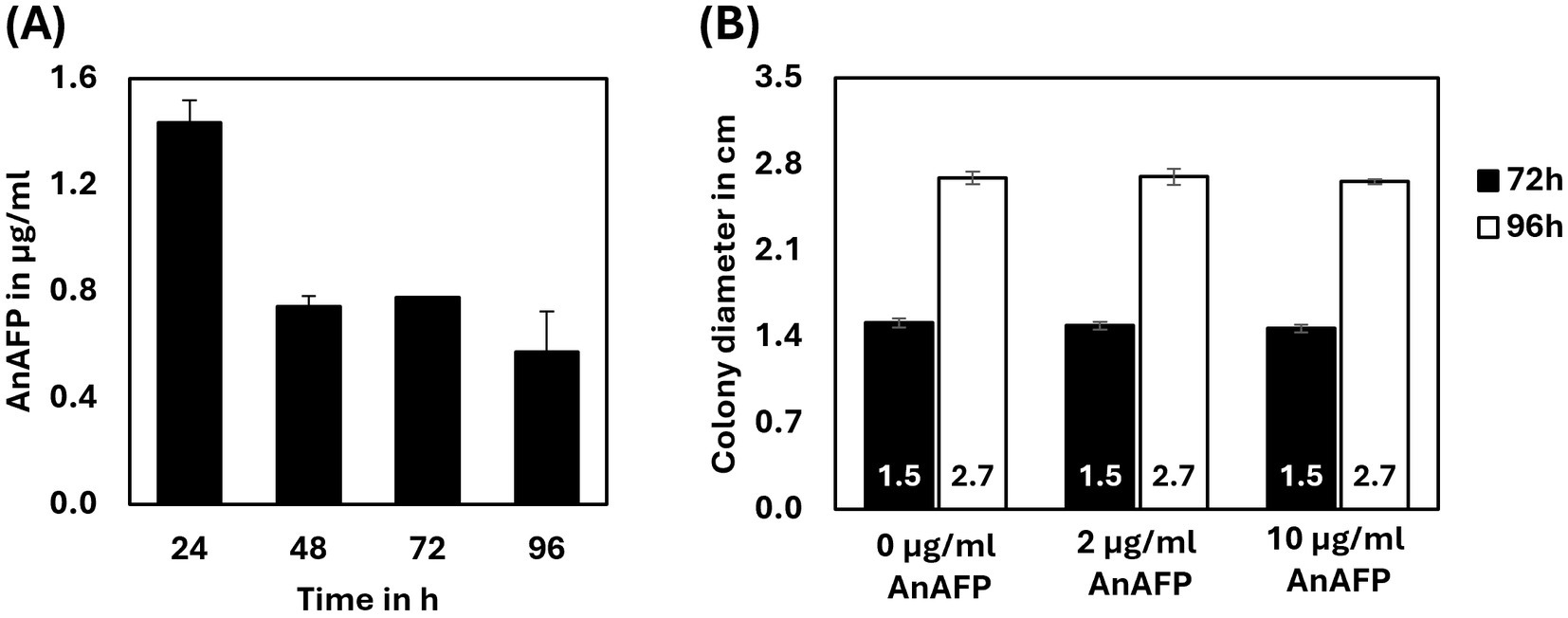
Figure 4. Externally applied AnAFP does not inhibit growth of A. niger. (A) The amount of AnAFP secreted by STS4.10 in shake flask cultures when cultivated up to 96 h at 30°C (n = 2). For cultivation and purification details see Materials and methods. (B) Colony diameter of strain STS4.10 after 72 h or 96 h of cultivation in the absence or presence of externally applied AnAFP (0, 2, and 10 μg/mL; n = 5).
To determine whether the growth-inhibitory effect of AnAFP is due to an extracellular or intracellular mode of action, we added purified AnAFP to the agar medium of STS4.10. Remarkably, even 10 μg/mL externally supplied AnAFP did not reduce colony diameter of the strain (Figure 4B), suggesting that AnAFP produced by strain STS4.10 exerts its growth-inhibitory effect intracellularly.
As the data proposed an intracellular activity of AnAFP, we followed its localization by fusing it to the reporter gene egfp. Again, we decided to put anafp::egfp expression under the control of the Tet-on promoter, to be able to tune its expression from zero to overexpression. As described in Materials and methods and in Supplementary Figure S5, the Tet-on::anafp::egfp construct was integrated into the anafp locus and the resulting strain STS19.3 was verified through Southern blotting.
Strain STS19.3 was cultivated in 8-well ibiTreat microscopy slides for 16 h in minimal medium (MM) with glucose. After medium exchange to MM lacking glucose and in the absence or presence of 20 μg/mL Dox, growth and fluorescence were monitored hourly via confocal microscopy (see Materials and methods for details). Interestingly, despite being expressed by the Tet-on system in the presence of 20 μg/mL Dox, green fluorescence became only detectable after 30 h of carbon starvation (Figure 5). No fluorescence was measurable in the absence of Dox, in earlier hours after the medium shift or when medium change to MM with glucose was performed (Supplementary Figure S6). Thus AnAFP::eGFP fluorescence only occurred during carbon starvation. Notably, green fluorescence was mainly located at cell periphery and septa of vegetative hyphae, although cytosolic localization was measured as well (Figure 5). Green fluorescence was also detectable in conidiophores and hyphae in their vicinity, whereby neighboring cell compartments differed strongly in their fluorescent signal (Figure 5A). Furthermore, wave-like movement of the fluorescence signal from one cell compartment to the neighboring one was observed (Figure 5C; Supplementary Video S2). Between 33 h and 36 h post-starvation, we noted that cytosolic green fluorescence suddenly agglomerated in individual cell compartments, a phenomenon that coincided with a reduction of their hyphal diameter from about 4.5 μm to about 3.5–2.5 μm (Figures 5B,C). Thereafter, the AnAFP::eGFP signal slowly faded within 1 to 2 h of imaging.
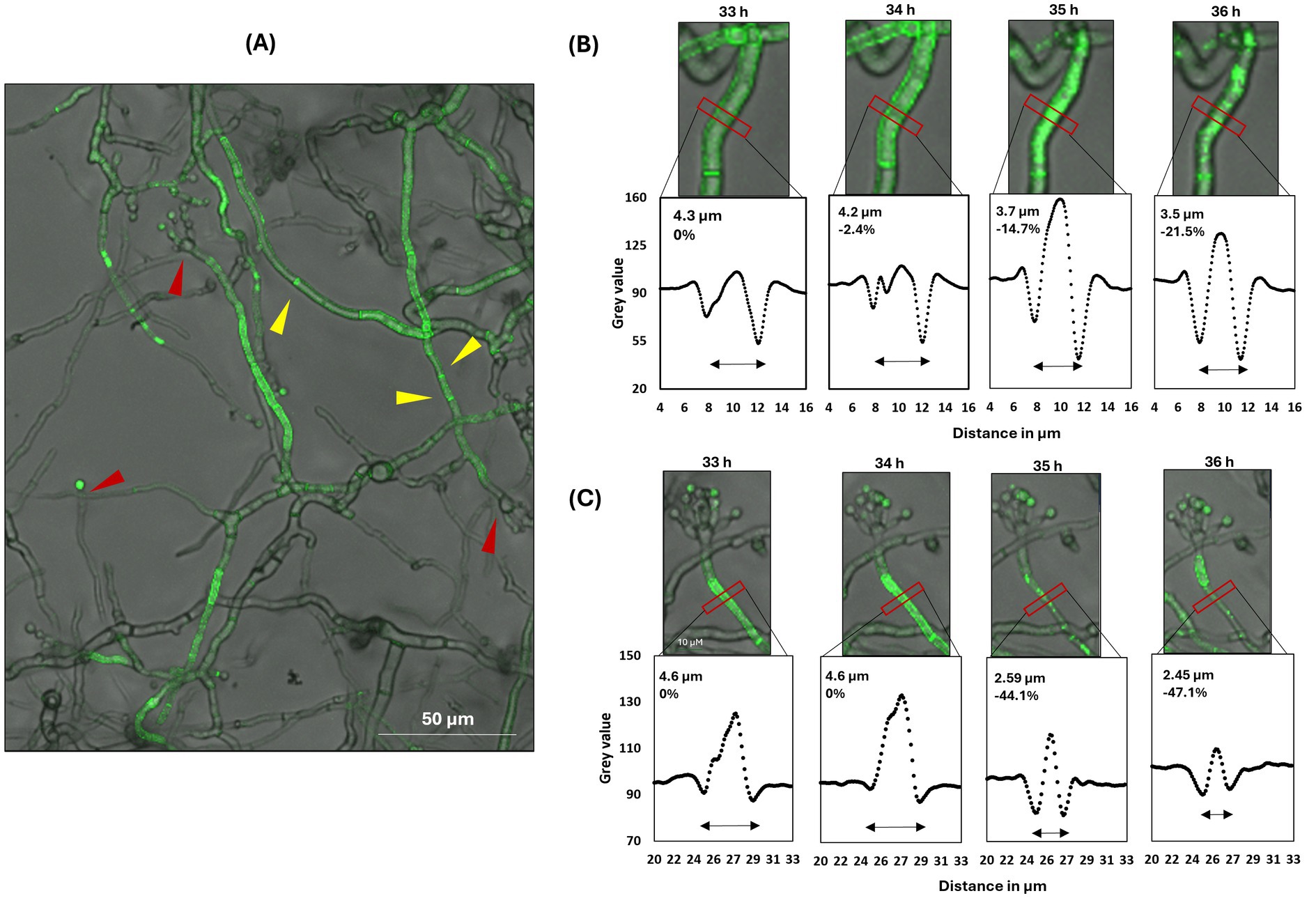
Figure 5. Spatial AnAFP::eGFP localization in strain STS19.3 expressing Tet-on::anafp::egfp. Hyphae were cultivated in minimal medium at the bottom of an 8-well microscopy slide. Green fluorescence became detectable only in the presence of 20 μg/mL Dox and after 30 h of carbon starvation. (A) The fluorescence signal of AnAFP::eGFP is primarily located in vegetative hyphae and in conidiophores and vegetative hyphae in their vicinity. Conidiophore heads are marked with a red arrowhead, septa with a yellow arrowhead. (B,C) show two representative close-ups visualizing the subcellular distribution of the AnAFP::eGFP signal between 33 and 36 h after carbon starvation. The grey values along the horizontal intersection with the hyphae, marked by the red box, is visualized below each micrograph. The hyphal diameter represents the distance between the 2 grey value minima, which estimates the center of the cell wall/ plasma membrane. Corresponding videos are presented in Supplementary Video S2.
The sudden reduction of hyphal diameter observed after expression of the anafp::egfp fusion protein indicated induced autolysis of selected cell compartments in response to AnAFP. The phenomenon of autolysis of selected cell compartments in response to carbon starvation and the emergence of conidiophores has been previously proposed to be linked to autophagy and asexual reproduction, based on transcriptional data from A. niger batch cultures (Nitsche et al., 2012). We thus speculated that induced expression of AnAFP provoked cell death of individual cell compartments under carbon starvation to fuel surviving ones.
In general, cell death in filamentous fungi can be classified into two main categories. The first category is a regulated form of cell death, which can be subdivided into (i) non-lytic (apoptosis, autophagy) (Gonçalves et al., 2017; Hamann et al., 2008; Pollack et al., 2009), (ii) lytic (necroptosis, pyroptosis, ferroptosis, autolysis) (Rico-Ramírez et al., 2022; White et al., 2002; Shen and Naqvi, 2021), and (iii) potentially lytic (HI/VI-mediated cell death, NLR-mediated cell death) (Rico-Ramírez et al., 2022; Uehling et al., 2017). The second category corresponds to a non-regulated form of cell death named necrosis (Gonçalves et al., 2017). Apoptosis and autophagy belonging to the first category are among the most studied cellular processes in fungi, for which a number of biomarkers are known (Sharon et al., 2009). We thus tested the possibility, that AnAFP could potentially provoke apoptosis in A. niger and performed a nuclear chromatin condensation assay known as gold standard for apoptosis. However, we could exclude apoptosis as mechanism provoked by AnAFP, as chromatin condensation was not observed upon Dox-induced anafp expression, whereas respective control samples did (Supplementary Figure S8).
We therefore tested whether Dox-induced anafp expression provoked an autophagic response. This was achieved via genetic modification of strain STS4.10 (Tet-on::anafp) that expressed eGFP constitutively in the cytoplasm. We generated this reporter strain as we could show earlier that cytoplasmatic eGFP fluorescence relocates to the vacuole during autophagy (Nitsche et al., 2013). Hence, we assumed that the proportion of fluorescently and non-fluorescently labeled vacuoles of A. niger could serve as quantitative proxy for autophagy activity. Cloning and integration of the respective PaopyrG::egfp::TtrpC construct into the STS4.10 background (Tet-on::anafp) resulted in strain JW3.3 (for details, see Supplementary Figure S9).
To determine the initiation and duration of autophagy, spores of strain JW3.3 were placed on microscopy coverslips and cultivated in MM including glucose at 30°C for 16 h, followed by a medium change to MM lacking glucose to induce carbon starvation. Initiation of autophagy in Dox-uninduced hyphae started after 6 h post starvation and already 100% of vacuoles became fluorescently labeled after 10 h post starvation (data not shown). This demonstrated that the autophagic response takes about 4 h in A. niger, under the conditions tested. Any impact of anafp gene expression on autophagy was thus evaluated after 4 h post starvation. As summarized in Figure 6, only 7.2% ± 4.1% of the vacuoles (n = 100) showed fluorescence in the absence of Dox, i.e., when the anafp gene was not expressed. In contrast, Dox-induced expression of anafp increased the amount of fluorescently labeled vacuoles to 80.6% ± 2.3%, suggesting that AnAFP can provoke autophagy. To verify this assumption, we generated two control strains deficient in: Atg1, a serine/threonine kinase and Atg8, an autophagosome-bound, ubiquitin-like protein. Both genes were shown to be essential for autophagy in A. niger (Nitsche et al., 2013). Control strain LV1.1 was a JW3.3 derivative, in which the gene atg1 was deleted, control strain LV2.3 a JW3.3 derivative, in which the gene atg8 was deleted. For cloning details and Southern proofs of successful deletion, see Materials and methods and Supplementary Figure S10. As in both strains LV1.1 and LV2.3 Dox-induced expression of the anafp gene did not provoke any translocation of the cytoplasmatic eGFP into the vacuoles in contrast to the autophagy-competent progenitor strain JW3.3 (Figure 6), we concluded that the intracellular mode of action of AnAFP is dependent on autophagy (at least of Atg1 and Atg8) and that its overexpression can even induce the onset of autophagy prematurely.
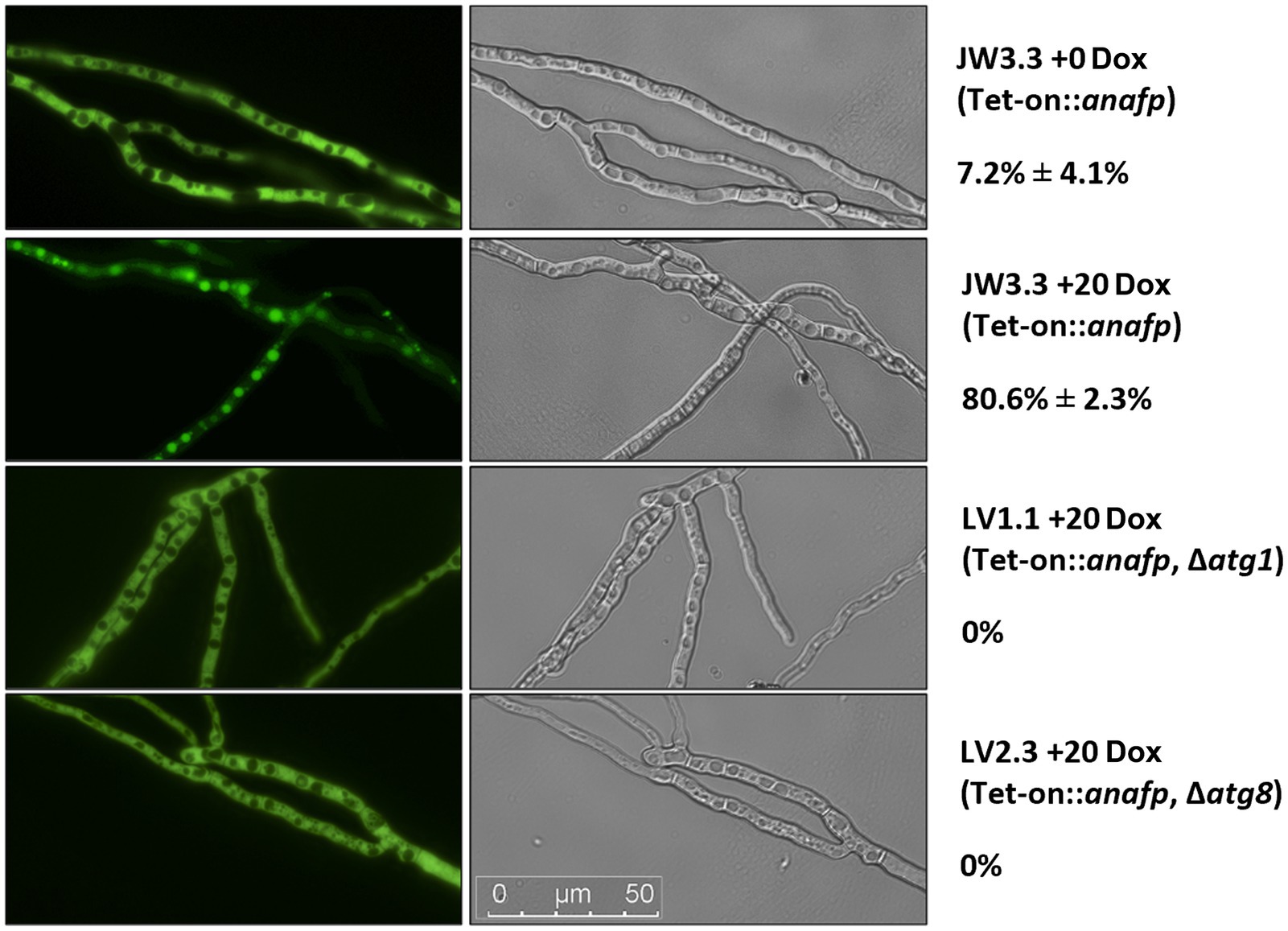
Figure 6. Quantification of autophagy through eGFP localization. Hyphae of strains JW3.3, LV1.1 and LV2.3 were grown on microscopy coverslips and subjected to carbon starvation as described in Materials and methods. Relevant genotype is given below the strain name. For at least 100 vacuoles, fluorescence was measured in the absence (Dox = 0) or presence (Dox = 20 μg/mL) of anafp expression. One representative picture is shown of the fluorescence channel (left) and the differential interference contrast channel (right). The percentage of filled vacuoles per total vacuoles counted is plotted next to the respective micrographs. Data refer to biological duplicate experiments.
Since doxycycline induced expression of AnAFP lead to a reduction in growth and a premature autophagic response, the next step aimed to reveal the underlying transcriptomic network which is promoting these effects. Thus, we determined the genome-wide transcriptional response in strain STS4.10 (Tet-on::anafp) during carbon starvation and in the absence and presence of anafp. For this experiment, we cultivated this strain in submerged batch bioreactor cultures until the mid-exponential growth phase, harvested the biomass and transferred it to minimal medium agar plates lacking glucose but supplemented with either 0 or 5 μg/mL Dox. Following 35 h of carbon starvation, biomass was isolated from the agar plates, total RNA extracted and subjected to Illumina sequencing as described in Materials and methods. Strain VG8.27 expressing luciferase under control of the Tet-on promoter (Tet-on::luciferase) (Meyer et al., 2011) served as a control strain for this experiment and was thus cultivated with the same procedure. With this control we wished to ensure that any observed differential gene expression can be directly linked to anafp overexpression and not to an indirect cellular effect due to Dox addition and/or general cellular stress provoked by Tet-on driven protein expression. Differential gene expression analysis (see Materials and methods) of Tet-on driven luciferase expression in control strain VG8.27 (5 μg/mL Dox compared to 0 μg/mL Dox) indeed yielded only 7 genes, demonstrating that Dox addition and/or protein overexpression did not provoke any global transcriptional changes in A. niger. However, differential gene expression analysis of Tet-on driven anafp overexpression (5 μg/mL Dox compared to 0 μg/mL Dox) yielded a total of 1,349 genes (log 2-fold >1, false discovery rate (FDR) corrected p-value <0.05), whereby 599 were upregulated and 750 downregulated (Additional File 2). Notably, anafp is one of the most significantly, differentially expressed genes with a log 2-fold change of 8.1 and an adjusted p-value of 0.00011 (Additional File 2). As reads were mapped to the A. niger N402 genome which is insufficiently annotated, the differentially expressed genes were translated to syntenic orthologs of A. niger CBS513.88 which yielded 1,229 unique genes with a log2 fold change >1, which were thus used for GO term enrichment analysis. This analysis uncovered that anafp overexpression resulted in a severe up- and downregulation of genes, responsible for the metabolism, hydrolysis and transmembrane transport of carbohydrates, lipids, and proteins (GO:0005975; GO:0006520; GO:0006629; GO:0055085, Table 2; Additional File 3). Notably, 151 of 898 predicted A. niger transporters (16.9%) were differentially expressed. Similarly, between 14 and 19 percent of predicted hydrolases (carbohydrates, lipids, proteins) do respond to the presence of anafp, a selection of which has been highlighted in Table 3. The enrichment of the GO terms extracellular region (GO:0005576) and plasma membrane (GO:0005886) further underlined the involvement of proteins and enzymes for transport and metabolism upon anafp overexpression.
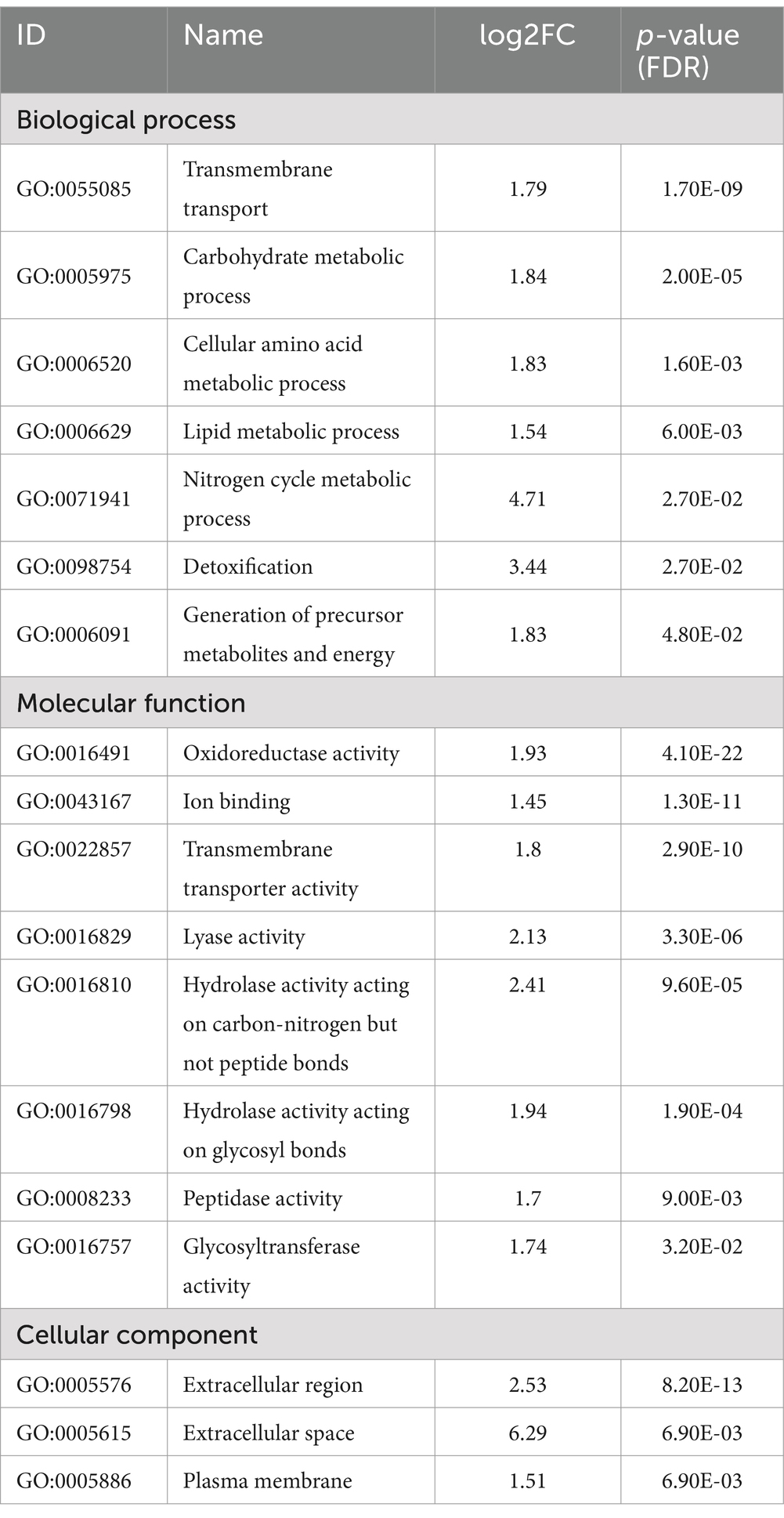
Table 2. Gene ontology enrichment of genes differentially expressed in response to anafp overexpression in A. niger log2 fold change >1, false discovery rate (FDR) corrected p-value <0.05; after Benjamini and Hochberg, 1995].
Notably, carbohydrate lyases and proteases were both up- or downregulated, while most lipases were upregulated only (Table 3). Interestingly, genes active in cell wall remodeling such as chitin and glucan synthases as well as chitinases were mostly downregulated, in agreement with downregulation of genes whose protein products are known as regulators of the cell wall integrity pathway (e.g., MkkA, BckA). The complete list of differentially expressed genes is summarized in Additional Files 2, 4, whereas Table 3 highlights some selected genes.
To understand, which regulators might drive these global changes, we searched among differentially expressed genes with an FDR adjusted p-value below 0.05 but with no log2 fold change restriction. This enlarged the differentially expressed gene set to 1,501 genes (Additional File 2). Previous studies reported that even minor changes in transcription factor gene expression (10–15%) can translate into large gene expression responses of individual target genes (Naqvi et al., 2023). The lowest fold change observed among transcription factors with an FDR adjusted p-value <0.05 was 0.63 which corresponds to a 55% change in expression. With this criterium, 91 out of 691 predicted transcription factor genes in A. niger (13%) were differentially expressed. Table 4 highlights selected transcription factors and other regulators for which a function has been verified in Aspergilli. Interestingly, transcription factor genes that responded to induced expression of anafp have a predicted function in asexual development (flbC, flbD, sfgA, ace2), DNA damage response (chkc, kusA, kueA), general stress response (msnA), fatty acid catabolism (farA), and calcium-, potassium- and iron ion homeostasis (aslA, sreA), respectively. One transcription factor which also responded to anafp expression is yap1, with mammalian orthologs important for apoptosis. Notably, also cell wall integrity regulators were downregulated (such as bckA, mkkA) including some of their effector genes (agsA/B, acuH; Table 2; Additional Files 2, 4).
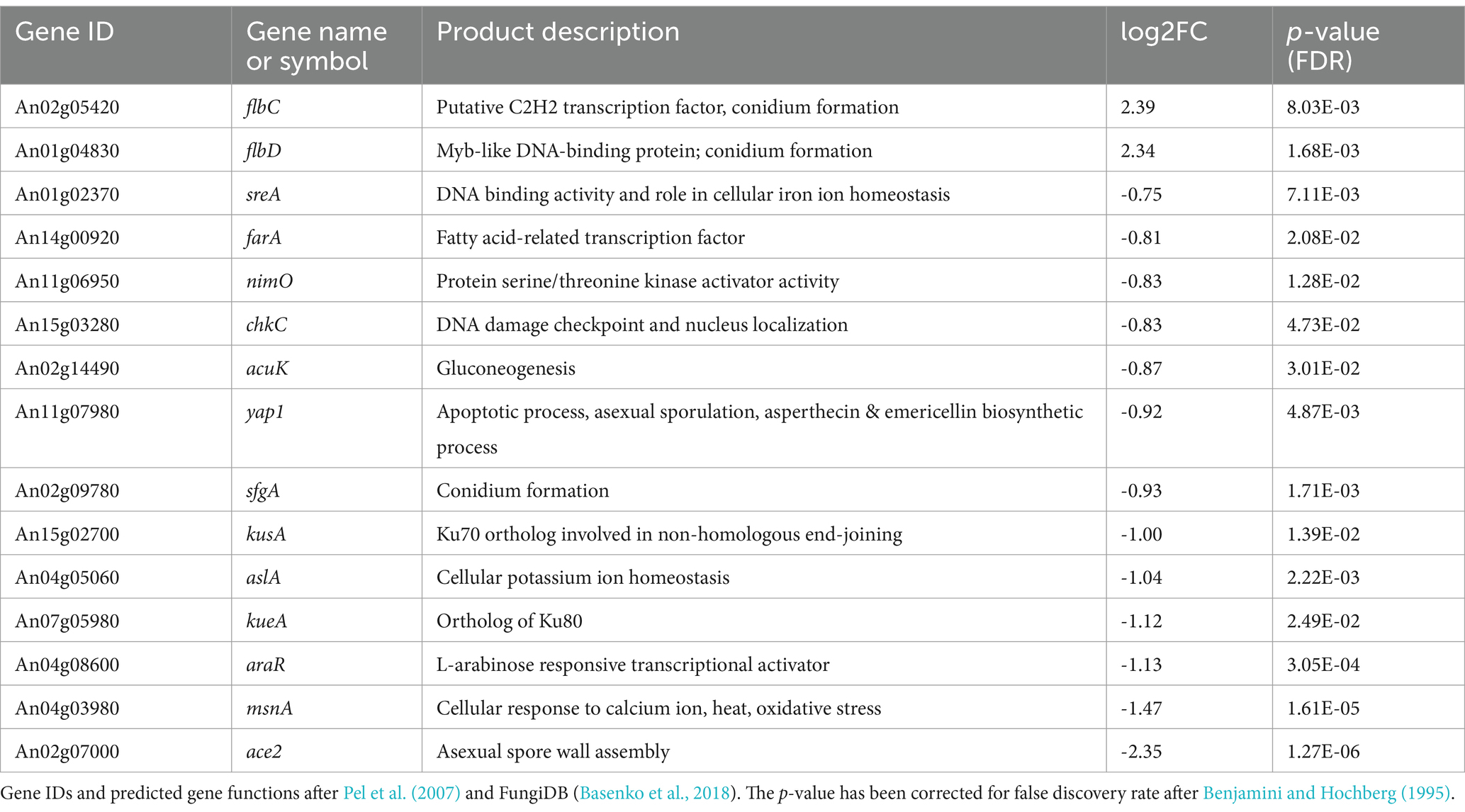
Table 4. Genes encoding regulators that were differentially expressed upon anafp overexpression in A. niger.
Taken together, the differentially expressed gene set of this study confirms our previous comparative transcriptomic analyses and strongly points toward a connection between AnAFP, nutrient mobilization & transport, and asexual development in A. niger under carbon starvation conditions. It appears that AnAFP specifically stimulates upstream regulators of conidiophore development such as sfgA, flbC, and flbD, while expression of the regulatory genes executing sporulation such as brlA, wetA, and abaA remain unaffected. This is in excellent agreement with data from Figure 3 that demonstrated that sporulation is not affected by the presence of AnAFP in STS4.10.
AnAFP belongs to the antimicrobial peptide family with members across almost all kingdoms of live (Feurstein et al., 2022) and has thus withstood millions of years of evolutionary selection pressure. Some antimicrobial peptides have been reported to have acquired secondary functions next to their primary antimicrobial activity against nutrient competitors (Meyer and Jung, 2018; Yeaman and Yount, 2007; Yount and Yeaman, 2006). Examples include the ability of cathelicidins to contribute to wound healing in human cells (Lee et al., 2018; Arnett and Seveau, 2011), the function of hepcidin in human iron metabolism (Patel and Akhtar, 2017), the involvement of gloverins in insect development (Xu et al., 2012; Hwang and Kim, 2011), and the chitin binding and wound healing activity of AMPs in shrimps and horseshoe crabs (Otero-Gonzáiez et al., 2010).
We propose here a working model in which AnAFP also exerts a function for A. niger that goes far beyond its antifungal activity against nutrient competitors (Figure 7). Transcription of the anafp gene peaks in vegetative hyphae, conidiophores as well as hyphae in their vicinity but neither in the conidiophore vesicle nor in conidiospores (Figures 1, 5; Supplementary Video S1), demonstrating that transcription of the anafp gene is under tight spatial and temporal control. In general, conidiophore formation in Aspergilli follows a differentiation program which becomes initiated during carbon or nitrogen deprivation and is accompanied by a severe growth cessation of vegetative hyphae (Adams et al., 1998). Thus, anafp transcription is limited to vegetative hyphae competent for conidiophore formation, i.e., when their growth becomes repressed to allow differentiation into conidiophores. Notably, translated AnAFP provokes a sudden reduction in hyphal diameter of those hyphal compartments where it becomes expressed (Figure 5). From these data we propose that AnAFP is important to support the asexual differentiation program, i.e., asexual sporulation, in A. niger during nutrient, especially carbon limitation. We predict that AnAFP drives nutrient mobilization through autophagic recycling of vegetative hyphae to fuel growth of neighbouring compartments and conidiophores, and thus survival of the whole colony during phases of starvation (Figure 7).
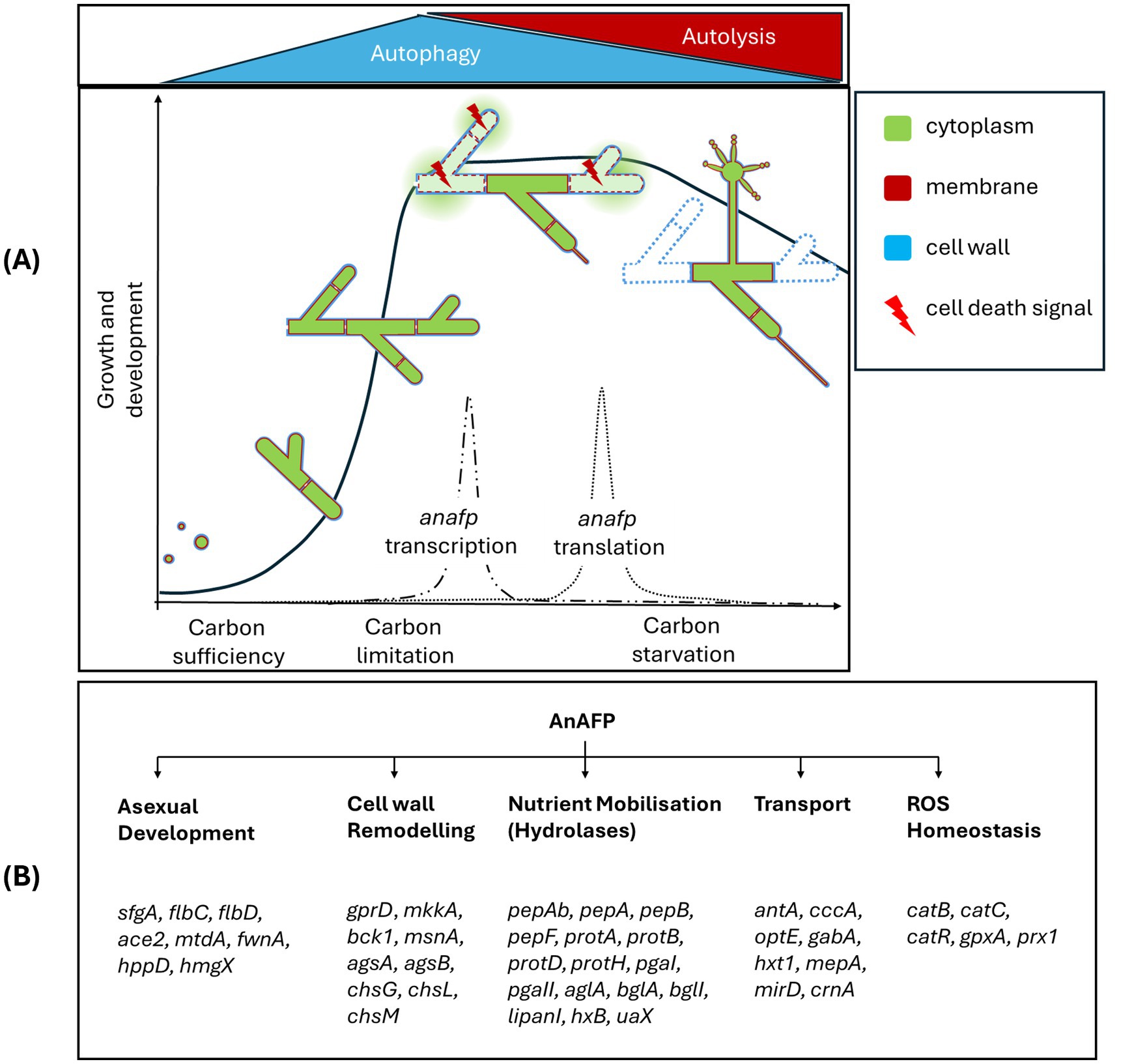
Figure 7. Working model for the intracellular function of AnAFP in A. niger based on data from this work and previous studies (Paege et al., 2016; Nitsche et al., 2012). (A) Depicted is the asexual life cycle of A. niger cells under solid-state and submerged growth conditions including spore germination, hyphal growth, vegetative mycelium formation and outgrowth of conidiophores. During exponential growth phase, hyphal compartments start to form visible vacuoles to recycle misfolded proteins. When nutrients in the environment become limiting, post-exponential growth starts and vacuolar size increases to provide nutrients and energy through autophagy. First conidiophores start to appear. During this narrow time window, the anafp gene becomes transcribed with a peak-like mRNA expression profile (Paege et al., 2016). Time-delayed (dependent on the cultivation condition), anafp mRNA becomes translated and intracellularly AnAFP is formed to support autophagic process. Throughout prolonged nutrient starvation, however, more nutrients need to become recycled to fuel conidiophore development. Hyphal compartments with higher AnAFP presence thus occur. This supports autolysis to become dominant over autophagy, i.e., some hyphal compartments with high amounts of AnAFP decrease their vacuole size, disintegrate their cell walls, so that eventually the cytoplasm disappears and such hyphal compartment significantly reduces its diameter. Finally, the vegetative mycelium population enters the death phase and autolysis of individual hyphal compartments provides further nutrients to support conidiophore formation and eventually sporulation. Altogether, this sequence ensures survival of the species. (B) AnAFP is embedded in many cellular regulatory processes that drive these different developmental stages. Selected genes are indicated that become differentially expressed as response to anafp overexpression. See Discussion for details.
We also report here that expression of the anafp gene is regulated at the post-transcriptional level. Although the anafp mRNA was produced in high amounts under the control of the Tet-on promotor, translation only occurred in the presence of its native anafp 5’ and 3’ UTRs (Figure 2). In agreement, the anafp::egfp fusion showed intracellular green fluorescence only after prolonged nutrient starvation despite being controlled under the Dox-induced Tet-on system and carrying its 5’ and 3’ UTRs (Figure 5). This data perfectly match our previous observations (Meyer and Jung, 2018) that expression of the anafp gene in wild type A. niger peaked 16 h after carbon starvation while extracellular AnAFP protein was detected only 160 h post starvation (van Munster et al., 2015). Note that such discrepancy between high level mRNA and lack of extracellular protein was also observed for two other members of the AFP family, namely the PAFB protein from P. chrysogenum (Huber et al., 2018) and the AfpB protein from P. digitatum (Garrigues et al., 2016). Here, the importance of native promotor and terminator sequence for AFP expression was also reported for the AfpA protein from P. expansum (Gandía et al., 2022; Sonderegger et al., 2016). Note that the choice of an optimal 5’ UTR has been demonstrated to increase the production of non-antifungal peptides, e.g., the reporter protein β-glucuronidase in A. oryzae (Koda et al., 2004).
Moreover, the regulation of AnAFP translation could be influenced by the cellular location of anafp mRNA. This impact on translation efficiency has been reported for cells displaying polarized growth such as filamentous fungi or neuronal cells (Das et al., 2021; Staples et al., 2023). It is thought that mRNAs travel within stress granules (a phase-separated accumulation of RNAs and ribosomes) within specific regions in the cell (i.e., tip, periphery, septa), while translation is repressed until the destination is reached or a certain environmental condition is encountered. Whether this is the case for the anafp mRNA in A. niger remains to be shown.
We therefore conclude that expression of the anafp gene in A. niger is under sophisticated transcriptional and post-transcriptional control and encodes a protein that exerts its function first intracellularly and then extracellularly. We furthermore propose that AnAFP exerts two different intracellular functions during growth of A. niger as summarized in Figure 7: (i) During early starvation (here 4 h after carbon depletion), it can induce the (premature) onset of autophagy to translocate cytoplasmatic proteins (here eGFP) into vacuoles in an Atg1 and Atg8 dependent manner for nutrient recycling (Figure 6). (ii) During prolonged starvation (here 30 h after carbon depletion), it assists in cellular lysis of cell compartments where it is expressed to fuel neighboring cell compartments and conidiophores with nutrients. This activity results in the sudden reduction of the hyphal diameter (Figure 5). Our transcriptomics analyses (Tables 3, 4) reinforce the autolytic role of AnAFP as hundreds of genes important for nutrient mobilization and metabolism become differentially expressed upon anafp overexpression. This also includes genes with a putative function in lytic cell death forms such as ferroptosis, heterokaryon incompatibility and NLR-associated cell death (Additional File 4).
We have previously analyzed the transcriptomic response of A. niger during carbon starvation in bioreactor cultures and could uncover that the mycelium responds with autophagy during early phases of nutrient deprivation but with autolysis during later stages of starvation (Nitsche et al., 2012). In parallel to the differential expression of autolytic genes, AnAFP also induced the expression of genes which respond to the presence of reactive oxygen species (ROS), as reflected by the enrichment of GO terms detoxification (GO:0098754) and hydrogen peroxide metabolic process (GO:0042743) (Additional File 3). Interestingly, an intertwined activity of antimicrobial peptides, reactive oxygen species (ROS) and compartmentalized cell death processes is a phenomenon described for the innate immune system of plants and animals and was most recently also proposed to exist in fungi (Gaspar and Pawlowska, 2022). ROS, a key characteristic of fungal apoptotic-like cell death is also often used as a way to mobilize nutrients (Gaspar and Pawlowska, 2022). Given the amphipathic nature of antimicrobial peptides and their involvement in immune responses across eukaryotic kingdoms, antifungal peptides could have evolved two functions: one to fight fungal nutrient competitors and one to act as cannibal-toxin to ensure species survival under nutrient scarcity. Regardless of the form of cell death which may be triggered by AnAFP, we argue that the liberated nutrients are used by hyphal compartments in the vicinity for survival and the establishment of conidiophores. The latter would explain why AnAFP::eGFP signatures can be found in conidiophores (Figure 5). Remarkably, a similar global transcriptional response was most recently published for AfpB in P. digitatum (Bugeda et al., 2020; Ropero-Pérez et al., 2023). We thus performed a comparative GO term analysis of differentially expressed genes in both strains A. niger and P. digitatum upon increased AnAFP/AfpB expression which uncovered an enrichment in hydrolase and transmembrane transporter expression (Additional File 5). Likewise, transcriptome data for PeAfpA from P. expansum also report significant changes in the expression of genes involved in cell wall remodeling, carbohydrate lysis, and nutrient transport (Ropero-Pérez et al., 2024), altogether suggesting that the observed role of AnAFP in nutrient mobilization in A. niger can also be extended to other AFP family members.
Interestingly, the mobilization of genes important for hydrolases, transporters and programmed cell death is a phenomenon which has also been observed in more distantly related eukaryotic cells: namely in the process of germination in rice and barley. During germination thousands of genes are differentially expressed through the activity of specific transcription factors, one of which is a Myb-like DNA binding protein named MYBGA (Hong et al., 2012). The ortholog in Aspergilli is FlbD, a transcription factor with central role in asexual development in A. niger (Krijgsheld et al., 2013) and in (a) sexual differentiation and apoptosis in A. nidulans (Krijgsheld et al., 2013; Arratia-Quijada et al., 2012). Table 4 highlights that the flbD gene and its target gene, the flbC gene encoding the transcriptional activator FlbC [which itself activates expression of the brlA gene (Kwon et al., 2010)] are both upregulated upon anafp overexpression. In agreement, the sfgA gene encoding the repressor of both FlbD and FlbC is repressed. These genes play a crucial role in Aspergilli for the regulation of conidiophore and conidiospore formation, overall governed by the FluG-BrlA cascade (Park and Yu, 2012). Note that deletion of one of the key developmental regulators FlbA upstream of FlbD induces an autolysing phenotype in A. niger and increases anafp mRNA levels fivefold (Paege et al., 2016). On the other hand, anafp expression is independent of BrlA (Paege et al., 2016), which is the transcription factor that drives progression of the asexual developmental cascade toward sporulation, i.e., BrlA is key for the formation of phialides and conidia together with the transcription factors AbaA and WetA (Ojeda-López et al., 2018). In A. nidulans, it has been demonstrated that parts of this cascade not only regulate asexual reproduction but can also lead to the induction of autolysis through the production of proteinases and chitinases (Tamás et al., 2005). Similarly, the transcription factor FlbC was shown to play a role in the mobilization of hydrolase genes in A. oryzae (Tanaka and Gomi, 2021).
There is increasing evidence that the FluG-BrlA cascade has functions in Ascomycota (Pezizomycotina) that can go beyond asexual development and show their involvement in nutrient mobilisation through regulation of cell wall integrity and cell lysis (Ojeda-López et al., 2018; Guo et al., 2022). As discussed, our data show a strong differential regulation of genes involved in nutrient mobilization through autolysis and ROS.
Interestingly, genes active in autolysis have recently been reported to also contribute to conidiophore development and sporulation in A. fumigatus through the cell wall integrity pathway (CWI), whereby its central transcriptional factor RlmA directly regulates expression of the flbB, flbC, brlA, abaA genes (Rocha et al., 2020; Binder et al., 2011). Congruently, members of the CWI pathway in A. niger are consistently downregulated in response to anafp upregulation (Table 2). The capability of A. niger to detect and respond to cell wall stress is, however, not only dependent on the CWI pathway (transcription factor RlmA), but also on the general stress response pathway (transcription factor MsnA) and the calcium-calcineurin pathway (transcription factor CrzA) (Fiedler et al., 2014). All three transcription factors have also been shown to be important for asexual development in other Aspergilli (Rocha et al., 2020; Chang et al., 2011; Ren et al., 2021; Kovács et al., 2013). It is thus conceivable that anafp induced downregulation of the CWI pathway in A. niger reflects cell wall weakening, whereby depolymerized carbohydrates again serve nutrient mobilization for the generation of conidiophores.
This work uncovered that the antifungal peptide AnAFP, which was so far described to act exogenously as a secreted protein, exerts an important intracellular function for A. niger. We propose that the protein functions as a cannibal toxin that fine-tunes timing and efficiency of self-digestion during carbon starvation. We also conclude that AnAFP does not act as an ON/OFF switch for autophagy or autolysis as its deletion is neither lethal nor its down- or upregulation affects the general ability of A. niger to undergo asexual development and sporulation. We further show here that its expression is regulated in a very sophisticated manner in time and space at both the transcriptional and translational level. We propose that this ensures that only hyphal compartments competent for asexual development undergo autophagy and autolysis. Data from transcriptome analyses suggest that anafp expression is therefore embedded in many transcriptional regulatory programs which eventually ensure that the diameter of competent compartments become reduced to fuel neighbouring compartments and conidiophores. The intertwining of all subcellular and regulatory processes needs to be discovered in the future and – because of the entanglement of dying and outgrowing cells – will require new spatial and single cell omics approaches for fundamental understanding.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
SS: Writing – original draft, Writing – review & editing, Formal analysis, Investigation, Methodology, Visualization. LV: Writing – review & editing, Formal analysis, Investigation. BD: Writing – review & editing, Investigation. LS: Writing – review & editing, Investigation, Formal analysis, Methodology, Visualization. JW: Writing – review & editing, Investigation. SJ: Writing – review & editing, Conceptualization, Project administration, Supervision. VM: Writing – original draft, Writing – review & editing, Conceptualization, Project administration, Supervision, Funding acquisition, Resources.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The authors thank the Deutsche Forschungsgemeinschaft for funding this study (project grant 404869152 to VM). The authors also thank the Research Training Group SynPepBio funded by the Deutsche Forschungsgemeinschaft (DFG, GRK 2473 Bioactive Peptides, project number 392923329) for further support.
Special thanks go to Elena Babenko, Mathieu Gougeon, and Ramin Greckl for technical support. Open access funding was enabled and organized by Project DEAL.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1490293/full#supplementary-material
Adams, T. H., Wieser, J. K., and Yu, J.-H. (1998). Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62, 35–54. doi: 10.1128/MMBR.62.1.35-54.1998
Arentshorst, M., Lagendijk, E. L., and Ram, A. F. J. (2015a). A new vector for efficient gene targeting to the pyrG locus in Aspergillus niger. Fungal Biol. Biotechnol. 2, 2–5. doi: 10.1186/s40694-015-0012-4
Arentshorst, M., Niu, J., and Ram, A. F. J. (2015b). Efficient generation of Aspergillus niger knock out strains by combining NHEJ mutants and a split marker approach. Genet. Transforma. Syst. Fungi 1, 263–272. doi: 10.1007/978-3-319-10142-2_25
Arnett, E., and Seveau, S. (2011). The multifaceted activities of mammalian defensins. Curr. Pharm. Des. 17, 4254–4269. doi: 10.2174/138161211798999348
Arratia-Quijada, J., Sánchez, O., Scazzocchio, C., and Aguirre, J. (2012). FlbD, a Myb transcription factor of Aspergillus nidulans, is uniquely involved in both asexual and sexual differentiation. Eukaryot. Cell 11, 1132–1142. doi: 10.1128/EC.00101-12
Basenko, E. Y., Pulman, J. A., Shanmugasundram, A., Harb, O. S., Crouch, K., Starns, D., et al. (2018). FungiDB: an integrated bioinformatic resource for fungi and oomycetes. J. Fungi 4:39. doi: 10.3390/jof4010039
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Binder, U., Bencina, M., Eigentler, A., Meyer, V., and Marx, F. (2011). The Aspergillus gigenteus antifungal protein AFPNN5353 activates the cell wall integrity pathway and perturbs calcium homeostasis. BMC Microbiol. 11, 1–13. doi: 10.1186/1471-2180-11-209
Bos, C. J., Debets, A. J. M., Swart, K., Huybers, A., Kobus, G., and Slakhorst, S. M. (1988). Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr. Genet. 14, 437–443. doi: 10.1007/BF00521266
Bruder, S., Reifenrath, M., Thomik, T., Boles, E., and Herzog, K. (2016). Parallelised online biomass monitoring in shake flasks enables efficient strain and carbon source dependent growth characterisation of Saccharomyces cerevisiae. Microb. Cell Factories 15, 127–114. doi: 10.1186/s12934-016-0526-3
Bugeda, A., Garrigues, S., Gandía, M., Manzanares, P., Marcos, J. F., and Coca, M. (2020). The antifungal protein AfpB induces regulated cell death in its parental fungus Penicillium digitatum. Msphere 5, e00595–e00520. doi: 10.1128/msphere.00595-20
Cairns, T. C., de Kanter, T., Zheng, X. Z., Zheng, P., Sun, J., and Meyer, V. (2023). Regression modelling of conditional morphogene expression links and quantifies the impact of growth rate, fitness and macromorphology with protein secretion in Aspergillus niger. Biotechnol. Biofuels Bioprod. 16:95. doi: 10.1186/s13068-023-02345-9
Cesare, G. B. D., Cristy, S. A., Garsin, D. A., and Lorenz, M. C. (2020). Antimicrobial Peptides: a New Frontier in Antifungal Therapy. mBio 11:e02123-20. doi: 10.1128/mBio.02123-20
Chang, P.-K., Scharfenstein, L. L., Luo, M., Mahoney, N., Molyneux, R. J., Yu, J., et al. (2011). Loss of msnA, a putative stress regulatory gene, in Aspergillus parasiticus and Aspergillus flavus increased production of conidia, aflatoxins and kojic acid. Toxins 3, 82–104. doi: 10.3390/toxins3010082
Das, S., Vera, M., Gandin, V., Singer, R. H., and Tutucci, E. (2021). Intracellular mRNA transport and localized translation. Nat. Rev. Mol. Cell Biol. 22, 483–504. doi: 10.1038/s41580-021-00356-8
Delmas, S., Pullan, S. T., Gaddipati, S., Kokolski, M., Malla, S., Blythe, M. J., et al. (2012). Uncovering the genome-wide transcriptional responses of the filamentous fungus Aspergillus niger to lignocellulose using RNA sequencing. PLoS Genet. 8:e1002875. doi: 10.1371/journal.pgen.1002875
Dobin, A., and Gingeras, T. R. (2015). Mapping RNA-seq reads with STAR. Curr. Protoc. Bioinformatics 51, 11–14. doi: 10.1002/0471250953.bi1114s51
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Feurstein, C., Meyer, V., and Jung, S. (2022). Structure-activity predictions from computational mining of protein databases to assist modular design of antimicrobial peptides. Front. Microbiol. 13:812903. doi: 10.3389/fmicb.2022.812903
Fiedler, M. R., Lorenz, A., Nitsche, B. M., van den Hondel, C. A., Ram, A. F., Meyer, V., et al. (2014). The capacity of Aspergillus niger to sense and respond to cell wall stress requires at least three transcription factors: RlmA, MsnA and CrzA. Fungal Biol. Biotechnol. 1, 1–16. doi: 10.1186/s40694-014-0005-8
Fisher, M. C., Gurr, S. J., Cuomo, C. A., Blehert, D. S., Jin, H., Stukenbrock, E. H., et al. (2020). Threats Posed by the Fungal Kingdom to Humans, Wildlife, and Agriculture. mBio 11:e00449. doi: 10.1128/mBio.00449-20
Gandía, M., Moreno-Giménez, E., Giner-Llorca, M., Garrigues, S., Ropero-Pérez, C., Locascio, A., et al. (2022). Development of a FungalBraid Penicillium expansum-based expression system for the production of antifungal proteins in fungal biofactories. Microb. Biotechnol. 15, 630–647. doi: 10.1111/1751-7915.14006
Garrigues, S., Gandía, M., and Marcos, J. F. (2016). Occurrence and function of fungal antifungal proteins: a case study of the citrus postharvest pathogen Penicillium digitatum. Appl. Microbiol. Biotechnol. 100, 2243–2256. doi: 10.1007/s00253-015-7110-3
Gaspar, M. L., and Pawlowska, T. E. (2022). Innate immunity in fungi: is regulated cell death involved? PLoS Pathog. 18:e1010460. doi: 10.1371/journal.ppat.1010460
Gonçalves, A. P., Heller, J., Daskalov, A., Videira, A., and Glass, N. L. (2017). Regulated forms of cell death in fungi. Front. Microbiol. 8:1837. doi: 10.3389/fmicb.2017.01837
Guo, C.-T., Luo, X. C., Tong, S. M., Zhou, Y., Ying, S. H., and Feng, M. G. (2022). FluG and FluG-like FlrA coregulate manifold gene sets vital for fungal insect-pathogenic lifestyle but not involved in asexual development. Msystems 7, e00318–e00322. doi: 10.1128/msystems.00318-22
Hagen, S., Marx, F., Ram, A. F., and Meyer, V. (2007). The antifungal protein AFP from Aspergillus giganteus inhibits chitin synthesis in sensitive fungi. Appl. Environ. Microbiol. 73, 2128–2134. doi: 10.1128/AEM.02497-06
Hagiwara, D., Takahashi, H., Kusuya, Y., Kawamoto, S., Kamei, K., and Gonoi, T. (2016). Comparative transcriptome analysis revealing dormant conidia and germination associated genes in Aspergillus species: an essential role for AtfA in conidial dormancy. BMC Genomics 17, 1–18. doi: 10.1186/s12864-016-2689-z
Hamann, A., Brust, D., and Osiewacz, H. D. (2008). Apoptosis pathways in fungal growth, development and ageing. Trends Microbiol. 16, 276–283. doi: 10.1016/j.tim.2008.03.003
Hong, Y.-F., Ho, T. H. D., Wu, C. F., Ho, S. L., Yeh, R. H., Lu, C. A., et al. (2012). Convergent starvation signals and hormone crosstalk in regulating nutrient mobilization upon germination in cereals. Plant Cell 24, 2857–2873. doi: 10.1105/tpc.112.097741
Huber, A., Hajdu, D., Bratschun-Khan, D., Gáspári, Z., Varbanov, M., Philippot, S., et al. (2018). New antimicrobial potential and structural properties of PAFB: a cationic, cysteine-rich protein from Penicillium chrysogenum Q176. Sci. Rep. 8, 1–16. doi: 10.1038/s41598-018-20002-2
Hwang, J., and Kim, Y. (2011). RNA interference of an antimicrobial peptide, gloverin, of the beet armyworm, Spodoptera exigua, enhances susceptibility to Bacillus thuringiensis. J. Invertebr. Pathol. 108, 194–200. doi: 10.1016/j.jip.2011.09.003
Koda, A., Minetoki, T., Ozeki, K., and Hirotsune, M. (2004). Translation efficiency mediated by the 5′ untranslated region greatly affects protein production in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 66, 291–296. doi: 10.1007/s00253-004-1681-8
Kovács, B., Hegedűs, N., Bálint, M., Szabó, Z., Emri, T., Kiss, G., et al. (2014). Penicillium antifungal protein (PAF) is involved in the apoptotic and autophagic processes of the producer Penicillium chrysogenum. Acta Microbiol. Immunol. Hung. 61, 379–388. doi: 10.1556/amicr.61.2014.3.10
Kovács, Z., Szarka, M., Kovács, S., Boczonádi, I., Emri, T., Abe, K., et al. (2013). Effect of cell wall integrity stress and RlmA transcription factor on asexual development and autolysis in Aspergillus nidulans. Fungal Genet. Biol. 54, 1–14. doi: 10.1016/j.fgb.2013.02.004
Krijgsheld, P., Bleichrodt, R., van Veluw, G. J., Wang, F., Müller, W. H., Dijksterhuis, J., et al. (2013). Development in Aspergillus. Stud. Mycol. 74, 1–29. doi: 10.3114/sim0006
Kwon, N.-J., Garzia, A., Espeso, E. A., Ugalde, U., and Yu, J.-H. (2010). FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol. Microbiol. 77, 1203–1219. doi: 10.1111/j.1365-2958.2010.07282.x
Kwon, M. J., Schütze, T., Spohner, S., Haefner, S., and Meyer, V. (2019). Practical guidance for the implementation of the CRISPR genome editing tool in filamentous fungi. Fungal Biol. Biotechnol. 6, 1–11. doi: 10.1186/s40694-019-0079-4
Lee, M. W., Lee, E. Y., and Wong, G. C. L. (2018). What can pleiotropic proteins in innate immunity teach us about bioconjugation and molecular design? Bioconjug. Chem. 29, 2127–2139. doi: 10.1021/acs.bioconjchem.8b00176
Leppek, K., Das, R., and Barna, M. (2018). Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 19, 158–174. doi: 10.1038/nrm.2017.103
Meyer, V., and Jung, S. (2018). Antifungal peptides of the AFP family revisited: are these cannibal toxins? Microorganisms 6:50. doi: 10.3390/microorganisms6020050
Meyer, V., Ram, A. F. J., and Punt, P. J. (2014). Genetics, genetic manipulation, and approaches to strain improvement of filamentous fungi. Biol. Environ. Sci. Eng., 318–329. doi: 10.1128/9781555816827.ch22
Meyer, V., Wanka, F., van Gent, J., Arentshorst, M., van den Hondel, C. A. M. J. J., and Ram, A. F. J. (2011). Fungal gene expression on demand: an inducible, tunable, and metabolism-independent expression system for Aspergillus niger. Appl. Environ. Microbiol. 77, 2975–2983. doi: 10.1128/AEM.02740-10
Naqvi, S., Kim, S., Hoskens, H., Matthews, H. S., Spritz, R. A., Klein, O. D., et al. (2023). Precise modulation of transcription factor levels identifies features underlying dosage sensitivity. Nat. Genet. 55, 841–851. doi: 10.1038/s41588-023-01366-2
Nitsche, B. M., Burggraaf-van Welzen, A. M., Lamers, G., Meyer, V., and Ram, A. F. J. (2013). Autophagy promotes survival in aging submerged cultures of the filamentous fungus Aspergillus niger. Appl. Microbiol. Biotechnol. 97, 8205–8218. doi: 10.1007/s00253-013-4971-1
Nitsche, B. M., Jørgensen, T. R., Akeroyd, M., Meyer, V., and Ram, A. F. J. (2012). The carbon starvation response of Aspergillus niger during submerged cultivation: insights from the transcriptome and secretome. BMC Genomics 13, 380–322. doi: 10.1186/1471-2164-13-380
Ojeda-López, M., Chen, W., Eagle, C. E., Gutiérrez, G., Jia, W. L., Swilaiman, S. S., et al. (2018). Evolution of asexual and sexual reproduction in the aspergilli. Stud. Mycol. 91, 37–59. doi: 10.1016/j.simyco.2018.10.002
Olson, B. H., and Goerner, G. L. (1965). Alpha Sarcin, a new antitumor agent. Appl. Microbiol. 13, 314–321. doi: 10.1128/am.13.3.314-321.1965
Otero-Gonzáiez, A. J., Magalhães, B. S., Garcia-Villarino, M., López-Abarrategui, C., Sousa, D. A., Dias, S. C., et al. (2010). Antimicrobial peptides from marine invertebrates as a new frontier for microbial infection control. FASEB J. 24, 1320–1334. doi: 10.1096/fj.09-143388
Paege, N., Jung, S., Schäpe, P., Müller-Hagen, D., Ouedraogo, J. P., Heiderich, C., et al. (2016). A transcriptome meta-analysis proposes novel biological roles for the antifungal protein AnAFP in Aspergillus niger. PLoS One 11:e0165755. doi: 10.1371/journal.pone.0165755
Paege, N., Warnecke, D., Zäuner, S., Hagen, S., Rodrigues, A., Baumann, B., et al. (2019). Species-specific differences in the susceptibility of fungi to the antifungal protein AFP depend on C-3 saturation of glycosylceramides. MSphere 4, e00741–e00719. doi: 10.1128/mSphere.00741-19
Park, H.-S., and Yu, J.-H. (2012). Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 15, 669–677. doi: 10.1016/j.mib.2012.09.006
Patel, S., and Akhtar, N. (2017). Antimicrobial peptides (AMPs): the quintessential ‘offense and defense’ molecules are more than antimicrobials. Biomed. Pharmacother. 95, 1276–1283. doi: 10.1016/j.biopha.2017.09.042
Pel, H. J., de Winde, J. H., Archer, D. B., Dyer, P. S., Hofmann, G., Schaap, P. J., et al. (2007). Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25, 221–231. doi: 10.1038/nbt1282
Pollack, J., Harris, S., and Marten, M. (2009). Autophagy in filamentous fungi. Fungal Genet. Biol. 46, 1–8. doi: 10.1016/j.fgb.2008.10.010
Priebe, S., Kreisel, C., Horn, F., Guthke, R., and Linde, J. (2015). FungiFun2: a comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics 31, 445–446. doi: 10.1093/bioinformatics/btu627
Ren, Y., Zhang, C., Chen, Z., and Lu, L. (2021). The heterotrimeric transcription factor CCAAT-binding complex and Ca2+-CrzA signaling reversely regulate the transition between fungal hyphal growth and asexual reproduction. MBio 12, e03007–e03021. doi: 10.1128/mBio.03007-21
Rico-Ramírez, A. M., Gonçalves, A. P., and Louise Glass, N. (2022). Fungal cell death: the beginning of the end. Fungal Genet. Biol. 159:103671. doi: 10.1016/j.fgb.2022.103671
Rocha, M. C., Fabri, J. H. T. M., Simões, I. T., Silva-Rocha, R., Hagiwara, D., da Cunha, A. F., et al. (2020). The cell wall integrity pathway contributes to the early stages of Aspergillus fumigatus asexual development. Appl. Environ. Microbiol. 86, e02347–e02319. doi: 10.1128/AEM.02347-19
Ropero-Pérez, C., Bolós, B., Giner-Llorca, M., Locascio, A., Garrigues, S., Gandía, M., et al. (2023). Transcriptomic profile of Penicillium digitatum reveals novel aspects of the mode of action of the antifungal protein AfpB. Microbiol. Spectr. 11:e0484622. doi: 10.1128/spectrum.04846-22
Ropero-Pérez, C., Moreno-Giménez, E., Marcos, J. F., Manzanares, P., and Gandía, M. (2024). Studies on the biological role of the antifungal protein PeAfpA from Penicillium expansum by functional gene characterization and transcriptomic profiling. Int. J. Biol. Macromol. 266:131236. doi: 10.1016/j.ijbiomac.2024.131236
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., and Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Schäpe, P., Kwon, M. J., Baumann, B., Gutschmann, B., Jung, S., Lenz, S., et al. (2019). Updating genome annotation for the microbial cell factory Aspergillus niger using gene co-expression networks. Nucleic Acids Res. 47, 559–569. doi: 10.1093/nar/gky1183
Schäpe, P., Starke, S., Schuetze, T., Basenko, P. E., Jung, S., Cairns, T., et al. (2023). High-quality co-expression networks for accurate gene function predictions in the fungal cell factory Aspergillus niger and beyond. bioRxiv. doi: 10.1101/2023.07.28.550800
Semighini, C. P., and Harris, S. D. (2010). Methods to detect apoptotic-like cell death in filamentous fungi. Mol. Cell Biol. Methods Fungi 638:269. doi: 10.1007/978-1-60761-611-5_20
Sharon, A., Finkelstein, A., Shlezinger, N., and Hatam, I. (2009). Fungal apoptosis: function, genes and gene function. FEMS Microbiol. Rev. 33, 833–854. doi: 10.1111/j.1574-6976.2009.00180.x
Shen, Q., and Naqvi, N. I. (2021). Ferroptosis and microbial pathogenesis. PLoS Pathog. 17:e1009298. doi: 10.1371/journal.ppat.1009298
Sonderegger, C., Galgóczy, L., Garrigues, S., Fizil, Á., Borics, A., Manzanares, P., et al. (2016). A Penicillium chrysogenum-based expression system for the production of small, cysteine-rich antifungal proteins for structural and functional analyses. Microb. Cell Factories 15, 1–14. doi: 10.1186/s12934-016-0586-4
Staples, M. I., Frazer, C., Fawzi, N. L., and Bennett, R. J. (2023). Phase separation in fungi. Nat. Microbiol. 8, 375–386. doi: 10.1038/s41564-022-01314-6
Struyfs, C., Cammue, B. P. A., and Thevissen, K. (2021). Membrane-interacting antifungal peptides. Front. Cell Dev. Biol. 9:649875. doi: 10.3389/fcell.2021.649875
Tamás, E., Molnár, Z., Pusztahelyi, T., Varecza, Z., and Pócsi, I. (2005). The FluG-BrlA pathway contributes to the initialisation of autolysis in submerged Aspergillus nidulans cultures. Mycol. Res. 109, 757–763. doi: 10.1017/S0953756205003023
Tanaka, M., and Gomi, K. (2021). Induction and repression of hydrolase genes in Aspergillus oryzae. Front. Microbiol. 12:677603. doi: 10.3389/fmicb.2021.677603
Uehling, J., Deveau, A., and Paoletti, M. (2017). Do fungi have an innate immune response? An NLR-based comparison to plant and animal immune systems. PLoS Pathog. 13:e1006578. doi: 10.1371/journal.ppat.1006578
van Hartingsveldt, W., Mattern, I. E., van Zeijl, C. M. J., Pouwels, P. H., van den Hondel, C. A. M. J. J., and den, v. (1987). Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol. Gen. Genet. MGG 206, 71–75. doi: 10.1007/BF00326538
van Munster, J. M., Nitsche, B. M., Akeroyd, M., Dijkhuizen, L., van der Maarel, M. J. E. C., and Ram, A. F. J. (2015). Systems approaches to predict the functions of glycoside hydrolases during the life cycle of Aspergillus niger using developmental mutants ΔbrlA and ΔflbA. PLoS One 10:e0116269. doi: 10.1371/journal.pone.0116269
Wang, G., Li, X., and Wang, Z. (2016). APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44, D1087–D1093. doi: 10.1093/nar/gkv1278
Wanka, F., Cairns, T., Boecker, S., Berens, C., Happel, A., Zheng, X., et al. (2016b). Tet-on, or Tet-off, that is the question: advanced conditional gene expression in Aspergillus. Fungal Genet. Biol. 89, 72–83. doi: 10.1016/j.fgb.2015.11.003
Wanka, F., Arentshorst, M., Cairns, T. C., Jørgensen, T., Ram, A. F. J., Meyer, V., et al. (2016a). Highly active promoters and native secretion signals for protein production during extremely low growth rates in Aspergillus niger. Microb. Cell Factories 15, 1–12.
Weber, E., Engler, C., Gruetzner, R., Werner, S., and Marillonnet, S. (2011). A modular cloning system for standardized assembly of multigene constructs. PLoS One 6:e16765. doi: 10.1371/journal.pone.0016765
White, S., McIntyre, M., Berry, D. R., and McNeil, B. (2002). The autolysis of industrial filamentous fungi. Crit. Rev. Biotechnol. 22, 1–14. doi: 10.1080/07388550290789432
Xu, X.-X., Zhong, X., Yi, H.-Y., and Yu, X.-Q. (2012). Manduca sexta gloverin binds microbial components and is active against bacteria and fungi. Dev. Comp. Immunol. 38, 275–284. doi: 10.1016/j.dci.2012.06.012
Yeaman, M. R., and Yount, N. Y. (2007). Unifying themes in host defence effector polypeptides. Nat. Rev. Microbiol. 5, 727–740. doi: 10.1038/nrmicro1744
Keywords: Aspergillus niger , antifungal protein, nutrient mobilization, asexual development, survival, apoptosis, autophagy, AnAFP
Citation: Starke S, Velleman L, Dobbert B, Seibert L, Witte J, Jung S and Meyer V (2025) The antifungal peptide AnAFP from Aspergillus niger promotes nutrient mobilization through autophagic recycling during asexual development. Front. Microbiol. 15:1490293. doi: 10.3389/fmicb.2024.1490293
Received: 02 September 2024; Accepted: 05 November 2024;
Published: 24 January 2025.
Edited by:
Martin Gruhlke, Gesellschaft für Natur- und Wirkstoffforschung e.V., GermanyReviewed by:
Shikha Joon, National Cancer Institute at Frederick (NIH), United StatesCopyright © 2025 Starke, Velleman, Dobbert, Seibert, Witte, Jung and Meyer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sascha Jung, cy5qdW5nQHR1LWJlcmxpbi5kZQ==; Vera Meyer, dmVyYS5tZXllckB0dS1iZXJsaW4uZGU=
†ORCID: Stephan Starke, https://orcid.org/0009-0008-0128-0223
Laura Velleman, https://orcid.org/0009-0002-7347-8067
Luis Seibert, https://orcid.org/0009-0002-0464-8720
Jordi Witte, https://orcid.org/0009-0004-7239-9193
Sascha Jung, https://orcid.org/0000-0003-4970-6904
Vera Meyer, https://orcid.org/0000-0002-2298-2258
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.