- College of Plant Protection, Gansu Agricultural University, Lanzhou, China
Members of the genera Gymnopus and Mycena are vital for litter decomposition in tropical and humid temperate forests. In this study, the difference in morphological features among Gymnopus gansuensis, Gymnopus subsepiiconicus, and Mycena glabera was confirmed by DNA data. Gymnopus gansuensis and Gymnopus subsepiiconicus showed separate relationships with other species in the ITS and nLSU combined dataset utilized for the phylogeny of Gymnopus sect. Impudicae. In addition, Gymnopus gansuensis is characterized by pileus honey yellow at the center, margin pinkish buff to buff, stipe pinkish buff to fuscous, and basidiospores elliptic to briolette. Gymnopus subsepiiconicus is characterized by pileus clay buff to grayish brown at the center, margin pinkish buff to fawn, stipe dark brown to fuscous, and basidiospores elliptic. Based on the combined dataset of the ITS and TEF-1α, Mycena glabera has been detected as a separate lineage in the phylogenetic studies of Mycena sect. Calodontes. The ecological behaviors of the new species are described with illustrations.
1 Introduction
Gymnopus (Pers.) Roussel was classified as a member of the family Omphalotaceae by Antonín and Noordeloos (2010). There are 448 records of this genus in the Index Fungorum1 and 483 records in the MycoBank database.2 Approximately, 300 species of Gymnopus have been validly published (Coimbra et al., 2015; Kirk et al., 2008; Deng et al., 2016).
Antonín and Noordeloos (1997) proposed that Gymnopus should be classified based on white to cream-colored spore print, a non-insititious stipe, and different types of pileipellis. These authors divided the genus into three taxonomic groups (sect. Gymnopus, sect. Vestipedes, and sect. Levipedes) and four subgroups (subsect. Impudicae, subsect. Vestipedes, subsect. Levipedes, and subsect. Alkalivirentes). Further taxonomic revisions by Mata et al. (2006) proposed to rename Marasmius sect. Peronati to Gymnopus subsect., and Ovrebo’s (Mata and Ovrebo, 2009) establishment of Gymnopus sect. Androsacei.
Among the studies with molecular analysis, molecular research has been used to investigate the phylogenetic relationship within Gymnopus. Using nLSU sequences, Moncalvo et al. (2002) confirmed that Gymnopus, Lentinula, Rhodocollybia, and some other fungi belong to the Omphalotaceae clade. Mata et al. (2004) demonstrated a close relationship between Gymnopus and Marasmiellus Murrill, noting that Marasmiellus juniperinus Murrill, the type species of Marasmiellus, was included in Gymnopus sect. Levipedes. Wilson and Desjardin (2005) supported these findings, affirming that Gymnopus belongs to Omphalotaceae. Antonín and Noordeloos raised species of Gymnopus with a distinctly unpleasant smell to the sectional rank of Gymnopus sect. Impudicae. According to modern definition, species in Gymnopus sect. Impudicae are characterized by the pileipellis composed of cylindrical hyphae and a distinctive strong, unpleasant, onion, sewage, or rotten cabbage-like smell (Ryoo et al., 2016). Hu et al. (2024) conducted a comprehensive study about Gymnopus s. l. based on 527 gymnopoid collections from China. sect. Levipedes and sect. Impudicae were recognized as independent genera in Omphalotaceae, and six sections were accepted in Gymnopus depending on Melinda’s reagent. However, to be conservative, we still use the concept of sect. Impudicae due to this new classification framework, which needs further validation, and some species complexes still exist.
The genera Gymnopus and Mycena (Pers.) Roussel have also been studied in China. Teng (1963) reported the first (under Collybia) Gymnopus species in China as Collybia radicans P. Kumm. [= Hymenopellis radicata (Relhan) R.H. Petersen]. Overall, 19 taxa of Gymnopus were found in Southern China by Deng (2016). Ongoing research on Gymnopus has led to the identification of 57 species of Gymnopus s. l. based on morphological characteristics and molecular datasets and 32 species of Gymnopus s. str. described from China (Deng et al., 2016; Dai et al., 2010; Li et al., 2021a,b; Mešic et al., 2011; Wang et al., 2020; Sun et al., 2021; Hu et al., 2022; Hu et al., 2024). Mycena, one of the largest genera in the order Agaricales, comprises nearly 600 species distributed worldwide (He et al., 2020; Wei et al., 2024; Zhang et al., 2024). It has been separated into seven clades based on ITS+nLSU+SSU and 23 sections according to morphological feathers (Na et al., 2022; Cortés-Pérez et al., 2023). Mycena sect. Calodontes (Fr. ex Berk.) Quél. is characterized by its hygrophanous pileus exhibiting pinkish, reddish, purplish to brownish, intervenose lamellae, smooth cheilocystidia and pleurocystidia (if present), and frequently amyloid basidiospores (Cortés-Pérez et al., 2023). Pileipellis types and cheilocystidia characteristics may be vital for delimiting brownish Mycena species (Wei et al., 2024). As a significant group within the order Agaricales, Mycena encompasses at least 500 species globally, with most occurring in the northern temperate zone, primarily as saprophytes (Liu et al., 2022). These small mushrooms hold edible and medicinal value, or they may contain trace toxins and serve as germination fungi in the cultivation of Bl. With the application of multigene phylogeny in taxonomic studies, an increasing number of mushroom species are being reported from various regions around the world (Cortés-Pérez et al., 2023; Liu et al., 2022).
During our investigations of macrofungi in northwestern China, some specimens of Gymnopus and Mycena were collected. Gymnopus sect. Impudicae and Mycena sect. Calodontes phylogenetic analyses were performed in the present study using the combined datasets of ITS + nLSU and ITS + TEF-1α, respectively. Following morphological and molecular evidence, three previously unidentified species were discovered. Detailed descriptions and illustrations of these species are provided.
2 Materials and methods
2.1 Morphological examined
The specimens examined in this study were gathered in Northwest China, and their macroscopic features were recorded in fresh as the basis for the macromorphological descriptions, including host, ecological behaviors, geographical coordinates, location, altitude, collector, and date. Photographs of fresh basidiocarps and habitat information were taken in the field. All samples studied in this study were dehydrated and stored in the herbaria at Gansu Agricultural University (MHGAU, China). Micromorphological characters were observed from dried materials and examined with a light microscope (Nikon Eclipse E 80i microscope, Nikon, Tokyo, Japan) following the general techniques used in past research (Hu et al., 2022). In the descriptions, the abbreviations mean: IKI = Melzer’s reagent, IKI– = negative reaction in Melzer’s reagent, KOH = 5% potassium hydroxide, CB = Cotton Blue, CB+ = cyanophilous, CB− = acyanophilous, L = mean spore length, W = mean spore width, Q = variation in the L/W ratio between the specimens studied, and n = number of spores measured from the number of samples. The microscopic features were examined and described in 1% Congo red aqueous solution when necessary (Wei et al., 2024).
2.2 DNA extraction, PCR amplification, and sequencing
A CTAB plant genome rapid extraction kit-DN14 (Aidlab Biotechnologies Co., Ltd.) was utilized to extract DNA from dried specimens. With slight modifications, the manufacturer’s instructions for the PCR were carried out when employing the extracted DNA (Hu et al., 2022). The ITS gene region was amplified using the primer pairs ITS5/ITS4 and LR0R/LR7 for the nLSU gene region and 983F/1567R for the TEF-1α gene region (Gardes and Bruns, 1993; Cubeta et al., 1991). The PCR procedure for ITS and TEF-1α was as follows: initial denaturation at 95°C for 3 min, followed by 35 cycles at 94°C for 40 s, 56°C (ITS) or 55°C (TEF-1α) for 45 s and 72°C for 1 min, and a final extension of 72°C for 10 min (Li et al., 2022). The PCR procedure for nrLSU was as follows: initial denaturation at 94°C for 1 min, followed by 35 cycles at 94°C for 30 s, 50°C for 1 min, 72°C for 1.5 min, and a final extension of 72°C for 10 min (Li et al., 2022). The PCR products were purified and sequenced at the Xian Genomics Institute in China using the same pair of primers. Every newly produced sequence was uploaded to GenBank (Tables 1, 2).
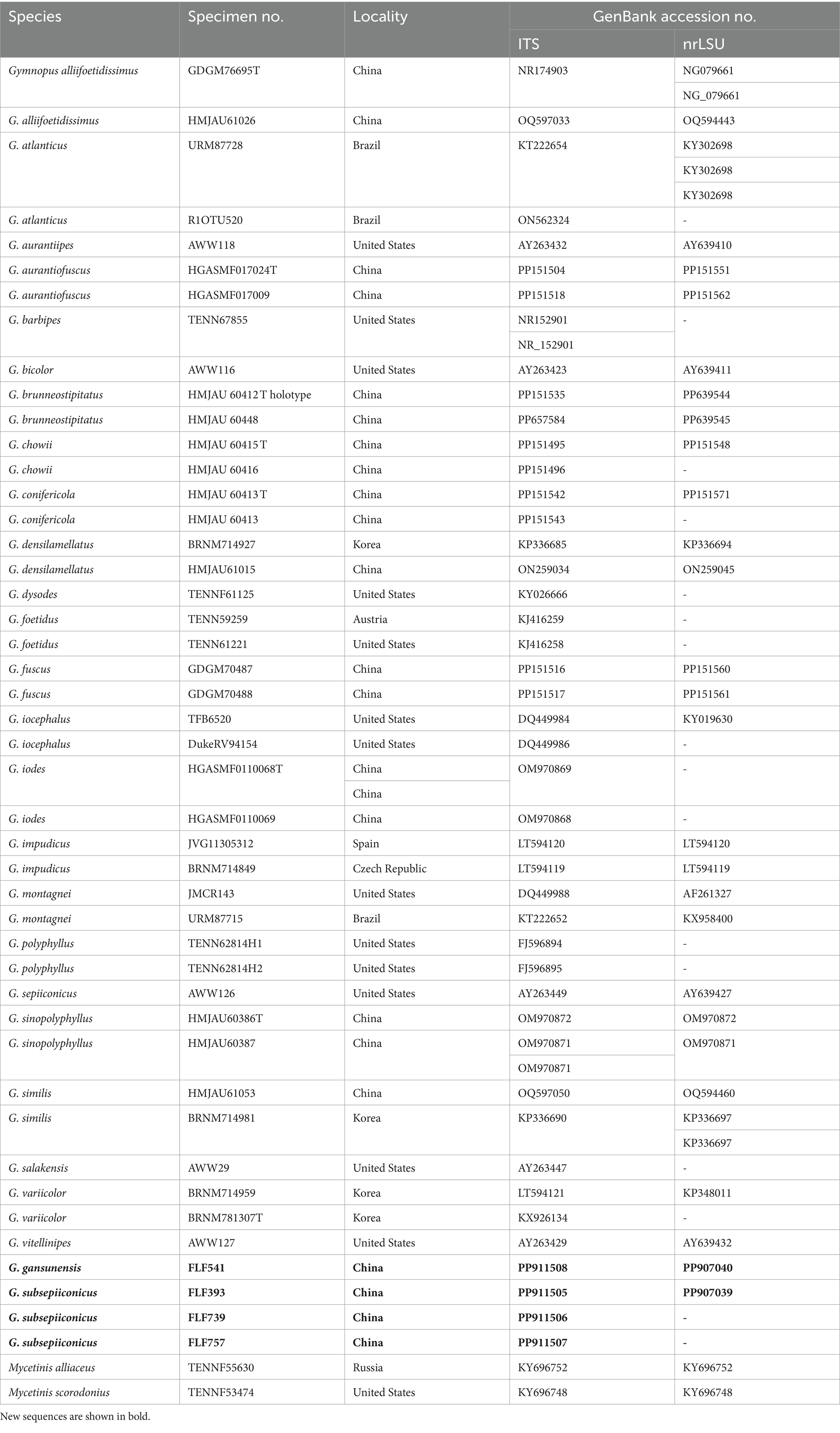
Table 1. A list of species, specimens, and GenBank accession numbers of sequences used in this study.
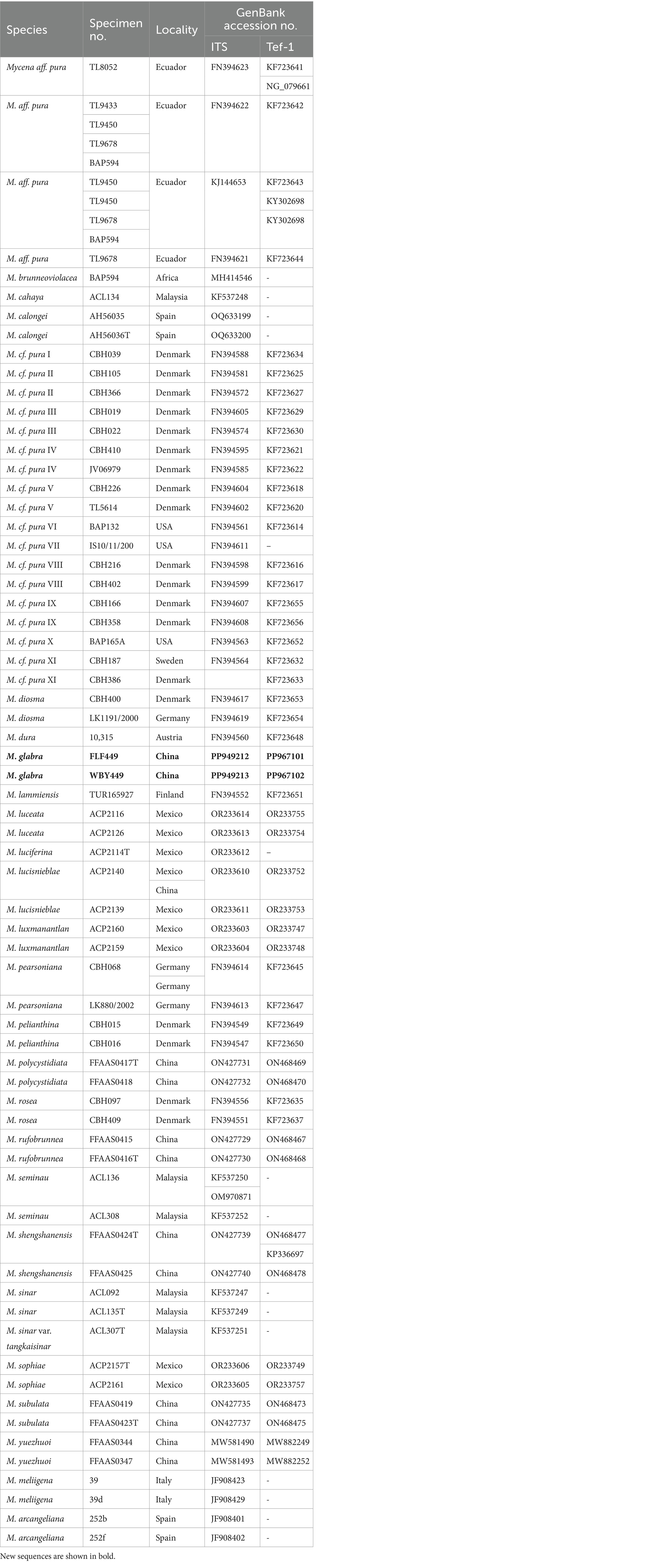
Table 2. A list of species, specimens, and GenBank accession numbers of sequences used in this study.
2.3 Phylogenetic analyses
A combined ITS + nLSU dataset was used to examine the phylogenetic relationship of Gymnopus, and for this, Mycetinis scorodonius (Fr.) A.W. Wilson & Desjardin and Mycetinis alliaceus (Jacq.) Earle ex A.W. Wilson & Desjardin were selected as the out-groups following the study by Li et al. (2022). The combined ITS + TEF-1α dataset was used to confirm the phylogenetic relationship of Mycena, and following the study by Na et al. (2022), M. meliigena (Berk. & Cooke) Sacc. and M. arcangeliana Bres were selected as the out-group. MAFFT 7 was used to align the datasets (Ryoo et al., 2016), and manual adjustments were made using BioEdit (Katoh and Standley, 2013). The partition homogeneity test (PHT) (Hall, 1999) of the two-gene dataset was tested by PAUP v. 4.0b10, respectively (Farris et al., 1994) under 1,000 homogeneity replicates. Alignments were spliced using Mesquite v.3.2. The best-fit model of nucleotide evolution for the datasets was selected, respectively, with Akaike’s information criterion (AIC) using MrModeltest 2.3 (Swofford, 2002; Guindon and Gascuel, 2003; Darriba et al., 2012). Phylogenetic analyses were conducted according to the previous study (Hu et al., 2022).
Using the heuristic search, maximum parsimony (MP) analysis was carried out using PAUP*version 4.0b10. Gaps were considered as missing data, and all characters were given the same weight. This study chose the heuristic search option to determine the trees, which included TBR branch switching and 1,000 random sequence additions. All parsimonious trees were preserved, zero-length branches were collapsed, and the maximum number of trees was set to 5,000. A bootstrap analysis with 1,000 replicates was used to test the clade’s robustness (Posada and Crandall, 1998). For each maximum parsimonious tree (MPT), descriptive tree statistics including tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were computed. RAxmL v.7.2.8 was used to carry out maximum likelihood (ML) analysis with a GTR + G + I model (Felsenstein, 1985). The computer calculated all model parameters, but only the best maximum likelihood tree from each search was retained. The best-fit evolution model for Bayesian inference (BI) was used utilizing MrModeltest 2.3 (Stamatakis, 2006; Posada and Crandall, 1998). MrBayes 3.2.6 was used for BI, conducting two distinct runs, each starting from random trees with four independent chains running at the same time, doing 2 million repeats, and sampling a tree every 100 generations through the online platform CIPRES Science Gateway (www.phylo.org, accessed on 23 April 2024) (Nylander, 2004). The sequence alignment was deposited at TreeBase (Gymnopus submission ID: 31485; Mycena submission ID: 31513). A majority rule consensus tree of all the remaining trees was determined, and the first 25% of the sampled trees were eliminated as burn-in. Branches were considered significantly supported when the bootstrap supports for MP and ML were greater than or equal to 50%, and the Bayesian inference (BI) was greater than or equal to 0.95. FigTree v1.4.2 was used for the visualization of phylogenetic trees.
3 Results
3.1 Phylogenetic analyses
The sequences involved in the ITS + nLSU dataset of Gymnopus were obtained from 47 specimens representing 28 species. The dataset contains 47 ITS and 26 nLSU sequences; among them, four ITS and two nLSU sequences are newly generated. The dataset contained 1,217 characters, including gaps (418 characters for ITS, 799 characters for nLSU), with 930 characters constant, 55 variable and parsimonious uninformative, and 232 parsimonious informative. The maximum parsimony analysis of Gymnopus produced 4,953 equally parsimonious trees (TL = 578, CI = 0.633, RI = 0.822, RC = 0.521, HI = 0.367). The GTR + I + G models were selected as the most effective models for every region of the combined ITS + nLSU sequence dataset, and they were used in the Bayesian analysis. The topology obtained from MP and Bayesian analysis was similar to that of ML analysis. The Bayesian analysis yielded a topology that was concordant, with an average split frequency standard deviation of 0.005966. Only the ML tree is shown in Figure 2; bootstrap support values for MP and ML ≥50% and BI ≥0.95 are noted at the nodes, respectively.
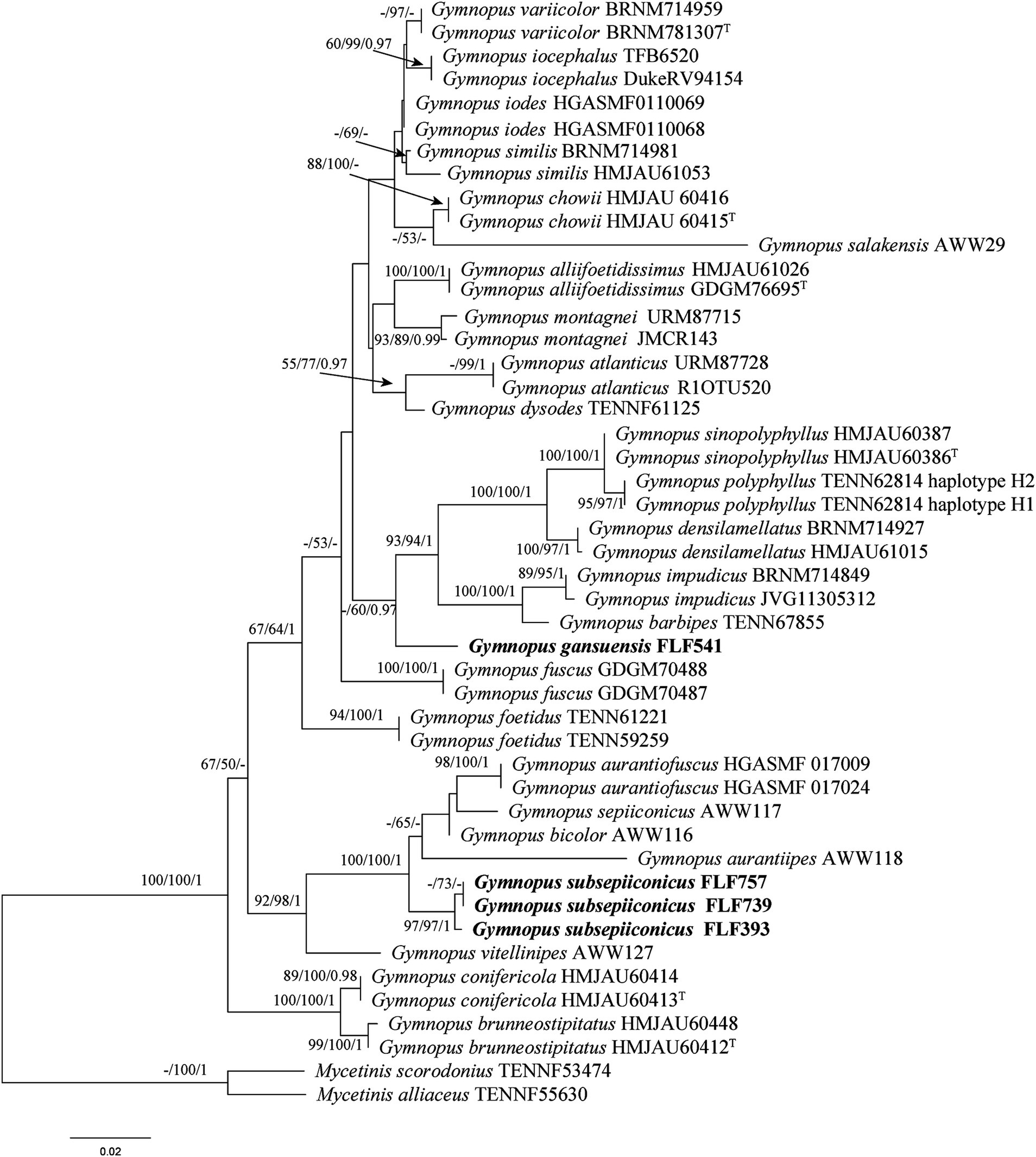
Figure 1. Maximum likelihood (ML) tree of species in Gymnopus based on the ITS + nLSU dataset. Bootstrap support values for MP and ML ≥50% and BI ≥0.95 are noted at each node, respectively. The new sequences are in bold.
The BLAST results show that the specimen FLF541 (Gymnopus gansuensis, holotype) is similar to Gymnopus sp. (taxon: 3341947), G. barbipes (taxon: 1460870), and G. impudicus (taxon: 206322), with a sequence similarity of 91.87%–95.73% from NCBI based on ITS with the top 10. Gymnopus subsepiiconicus (FLF393, Holotype) has a sequence similarity with G. erythropus (taxon: 230774), G. longisterigmaticus (taxon: 2935534), and G. longus (taxon: 2906493) ranging from 95.66% to 96.06% based on ITS with top 10.
The ITS + nLSU phylogenetic tree (Figure 2) confirmed the affinities of Gymnopus gansuensis and Gymnopus subsepiiconicus within Gymnopus, and both of them formed distinct, well-supported lineages (Figure 2).
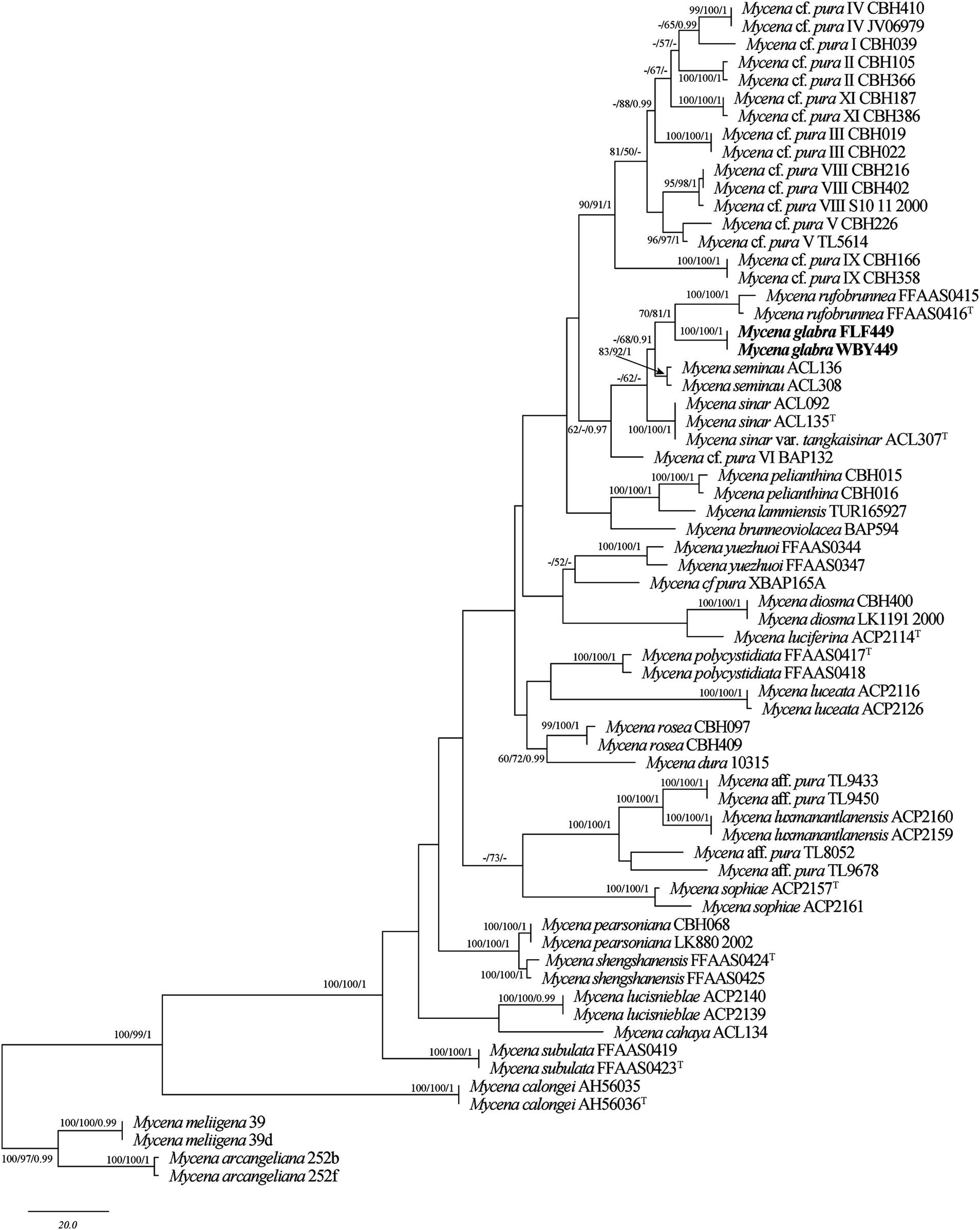
Figure 2. Maximum parsimony (MP) tree of species in Mycena based on the ITS + TEF-1α dataset. Bootstrap support values for MP and ML ≥50% and BI ≥0.95 are noted at each node, respectively. The new sequences are in bold.
The sequences involved in the ITS + TEF-1α dataset of Mycena were obtained from 66 specimens representing 37 taxa. The datasets contained 65 ITS and 51 TEF-1α sequences; among them, two ITS and two nLSU sequences are newly generated. The dataset contained 1,088 characters, including gaps (710 characters for ITS, 378 characters for TEF-1α), with 628 characters constant, 45 variable and parsimonious uninformative, and 415 parsimonious informative. The maximum parsimony analysis of Mycena produced 201 equally parsimonious trees (TL = 1,054, CI = 0.583, RI = 0.806, RC = 0.470, HI = 0.417). The GTR + I + G models were selected as the most effective models for every region of the combined ITS + TEF-1α sequence dataset, and it was used in the Bayesian analysis. The topology obtained from MP and Bayesian analysis was similar to that of ML analysis. The Bayesian analysis yielded a topology that was concordant, with an average split frequency standard deviation of 0.009123. Only the MP tree is shown in Figure 1; bootstrap support values for MP and ML ≥50% and BI ≥0.95 are noted at the nodes, respectively.
The BLAST results showed that the specimen FLF449 (Mycena glabera, Holotype) is similar to M. seminau (taxon: 1524328), M. rufobrunnea (taxon: 2942240), and M. sinar (taxon: 1524325), with a sequence similarity of 96.46%–97.83% from NCBI based on ITS with the top 10.
The ITS + TEF-1α phylogenetic tree (Figure 1) confirmed the affinities of Mycena glabera in Mycena sect. Calodontes, and two newly collected specimens formed distinct well-supported lineages (Figure 1).
3.2 Taxonomy
Gymnopus gansuensis, B.Y. Wang, T. F. Ma & L.F. Fan sp. nov. (Figures 3a,b, 4).

Figure 3. Basidiomata of Gymnopus species. Gymnopus gansuensis (a,b) and Gymnopus subsepiiconicus (c,d). Scale bars: 1 cm.
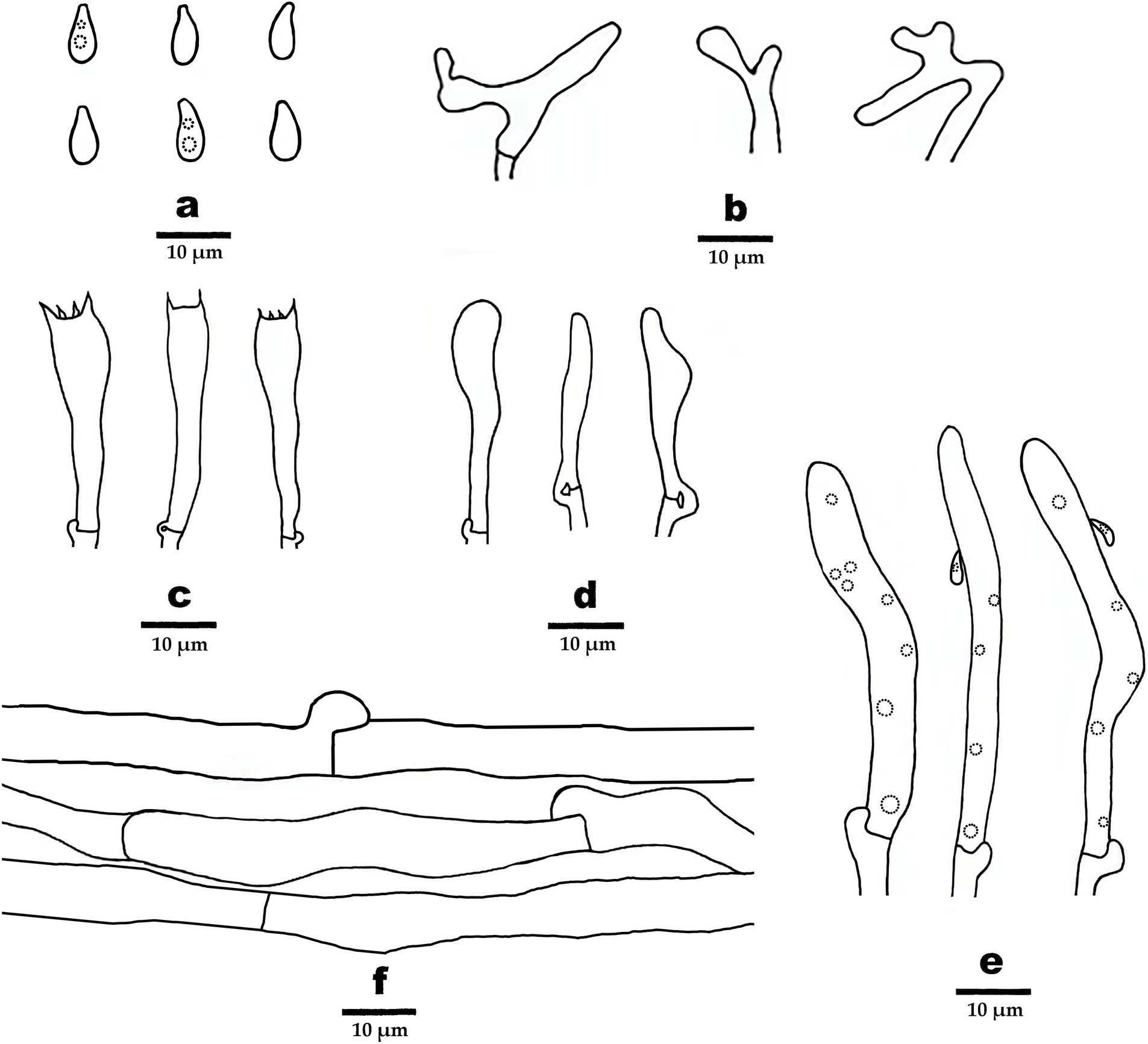
Figure 4. Microscopic structures of Gymnopus gansuensis (drawn from the holotype). (A) Basidiospores, (B) pileipellis, (C) basidia, (D) basidioles, (E) cheilocystidia, and (F) hyphae structure.
MycoBank: 855041.
Etymology: refers to the place of origin in Gansu Province, China.
Diagnosis: Differs from other Gymnopus species by pileus honey yellow at the center, margin pinkish buff to buff, stipe pinkish buff to fuscous, and basidiospores elliptic to briolette.
Holotype: CHINA. Gansu Province: Tibetan Autonomous Prefecture of Ganan, Taohe National Nature Reserve, N34°40′70″, E103°53′26″, in coniferous forest, 29 July 2023, MHGAU FLF541.
Basidiomata small-to-medium size, solitary. Pileus usually applanate or slightly convex, 1.0–2.5 cm in diameter, striated, hygrophanus, honey yellow at the center, margin pinkish buff to buff, entire, tomentose near margin. Context thin, fleshy, white to cinnamon buff, odorless. Lamellae subfree to adnate, pinkish buff to honey yellow, crowded. Stipe center, cylindrical, smooth in the upper part, covered with white hairs up to 2/3 (from the base upwards), fistulose, fibrous, 3.2–5.0 cm × 0.2–0.4 cm, pinkish buff near the pileus, fuscous at the bottom.
Hyphal system composed of generative hyphae with clamp connections, IKI–, CB–; tissues turn to black in KOH. Generative hyphae in context light brown to brown, thin-walled, occasionally branched, regularly arranged, 3–8 μm in diameter. Generative hyphae in lamella light brown, thin-walled, occasionally branched, regularly arranged, 5–11 μm in diameter.
Basidiospores elliptic to briolette, smooth, hyaline, IKI–, thin-walled, (6–) 6.5–8 (9) × 3–4 (−4.5) μm, L = 7.5 μm, W = 3.48 μm, Q = 2.16–2.25, (n = 60/2). Basidia clavate, smooth, hyaline, thin-walled, 27–35 μm × 6–7 μm, two- or four-spored, sterigmata 1.5–3.5 μm long. Cheilocystidia abundant, clavate, with obtuse at the center, smooth, hyaline, thin-walled, 51–63 μm × 5–6 μm. Pileipellis a cutis, made up of irregularly branched or weakly coralloid hyphae, inflated, smooth, hyaline to light brown, thin-walled, 4–8 μm wide.
Additional specimen (paratype) examined: CHINA. Gansu Province: Tibetan Autonomous Prefecture of Ganan, Taohe National Nature Reserve, N34°40′70″, E103°53′26″, in coniferous forest, 29 July 2023, MHGAU WBY541 (duplicate).
Gymnopus subsepiiconicus, B.Y. Wang, T. F. Ma & L.F. Fan sp. nov. (Figures 3c,d).
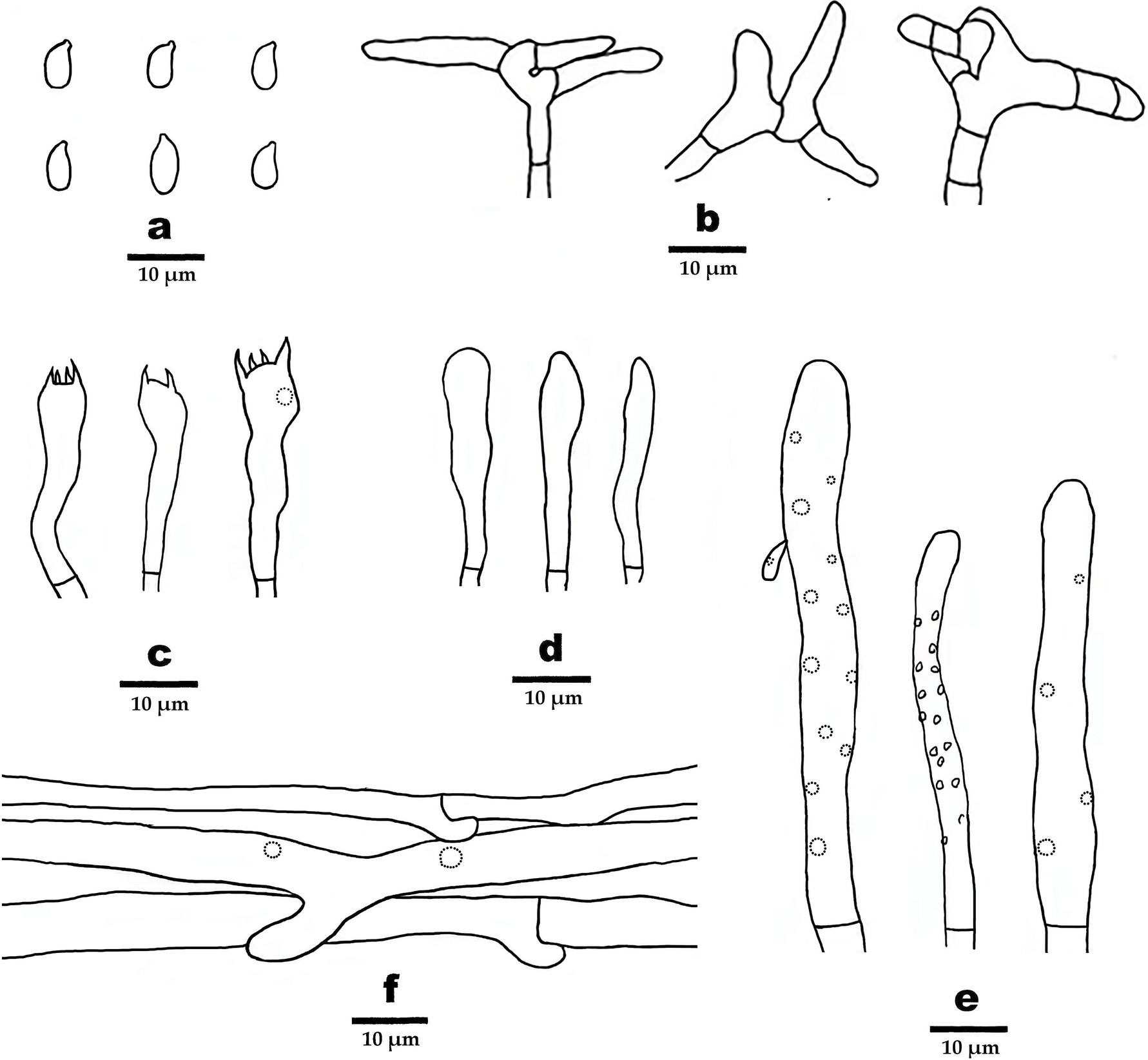
Figure 5. Microscopic structures of Gymnopus subsepiiconicus (drawn from the holotype). (A) Basidiospores, (B) pileipellis, (C) basidia, (D) basidioles, (E) cheilocystidia, and (F) hyphae structure.
MycoBank: 855412.
Etymology: refers to the species being similar to Gymnopus sepiiconicus.
Diagnosis: Differs from other Gymnopus species by pileus clay buff to grayish brown at the center, margin pinkish buff to fawn, stipe dark brown to fuscous, basidiospores elliptic.
Holotype: CHINA. Gansu Province: Lintan County Prefecture of Ganan, Yeliguan National Forest Park, N34°40′70″, E103°53′26″, in coniferous forest land, 27 July 2023, MHGAU FLF393.
Basidiomata in small-to-medium size, solitary to gregarious. Pileus usually applanate or slightly convex, 1.5 cm–2.2 cm in diameter, fluted, hygrophanus, clay buff to grayish brown at the center, margin pinkish buff to fawn, entire. Context thin, fleshy, light grayish brown, odorless. Lamellae subfree to adnate, honey yellow, crowded. Stipe central, cylindrical, 3.2–4.8 cm × 0.2–0.3 cm, grayish brown to dark brown near the pileus, fuscous at the bottom, smooth, fistulose, and fibrous.
Hyphal system composed of generative hyphae with clamp connections, IKI–, CB–; tissues turn to black in KOH. Generative hyphae in context light brown to brown, thin-walled, occasionally branched, regularly arranged, 3 μm –6 μm in diameter. Generative hyphae in lamella grayish brown to dark brown, thin-walled, occasionally branched, regularly arranged, 4 μm–8 μm in diameter.
Basidiospores elliptic, smooth, hyaline, inamyloid, thin-walled, 6–9 × 3–3.5 (−4) μm, L = 7.25 μm, W = 3.25 μm, Q = 2.05–2.4 (n = 60/2). Basidia clavate, smooth, hyaline, thin-walled, 19–40 μm × 5–7 μm, two- or four-spored sterigmata 2–5 μm long. Cheilocystidia abundant, clavate, smooth, hyaline, thin-walled, 52–111 μm× 5–6.5 μm. Pileipellis a cutis, made up of irregularly branched or weakly coralloid hyphae, inflated, smooth, hyaline to light brown, thin-walled, 5–15 μm wide.
Additional specimens (paratype) examined: CHINA. Gansu Province: Lintan County Prefecture of Ganan, Yeliguan National Forest Park, N34°56′58.924″, E103°35′47.026″, in coniferous forest, 12 September 2023, MHGAU FLF739, 13 September 2023, MHGAU FLF757.
Mycena glabera, B.Y. Wang, T. F. Ma & L.F. Fan sp. nov. (Figures 5, 6).
MycoBank: 855042.
Etymology: refers to the smooth and glabrous pileus.
Diagnosis: Mycena glabera is characterized by pileus convex or hemispherical, margin decurved, white to cream, slightly wavy, smooth, hygrophanus, surface glabrous, pale pink–yellow to fleshy pink at the center, basidiospores long ellipsoid to cylindrical, thin-walled, cheilocystidia fusiform, thin-walled.
Holotype: CHINA. Gansu Province: Tibetan Autonomous Prefecture of Ganan, Taohe National Nature Reserve, N34°40′70″, E103°53′26″, in coniferous forest, 28 July 2023, MHGAU FLF449.
Basidiomata thin and small, solitary, or scattered. Pileus convex or hemispherical to applanate, margin decurved, white to cream, slightly wavy, smooth, hygrophanus, with vertical stripes or grooves, surface glabrous, old lace to moccasin at the center, 10–20 mm in diameter. Context white, fleshy, very thin (< 1 mm). Lamellae subdecurrent, white to pale pink-yellow, distant. Stipe centrally attached, cylindrical, equal, smooth, fragile, hollow, 30–35 mm × 2–3 mm, white to cream near the pileus, pale wax yellow at the bottom.
Hyphal system composed of generative hyphae with clamp connections, IKI–, CB–; slightly inflated in KOH. Generative hyphae smooth, hyaline, thin-walled, regularly arranged, 3–9 μm in diameter.
Basidiospores long ellipsoid to cylindrical, smooth, hyaline, inamyloid, thin-walled, with obviously oil drop, (5.0–) 6.3–10.2 (−11.7) μm × 3.7–5.5 (−6.7) μm, L = 8.7 μm, W = 4.3 μm, Q = 2.0–2.15, (n = 60/2). Basidia clavate, smooth, hyaline, thin-walled, 29.0–45.0 μm × 6.3–12.0 μm, four-spored when mature, sterigmata up to 15.5 μm long. Cheilocystidia abundant, fusiform to subfusiform, with obtuse at the center, smooth, hyaline, thin-walled, 48.0–77.4 μm × 10.0–16.4 μm.
Additional specimen (paratype) examined: CHINA. Gansu Province: Tibetan Autonomous Prefecture of Ganan, Taohe National Nature Reserve, N34°40′70″, E103°53′26″, in coniferous forest, 28 July 2023, MHGAU WBY449 (duplicate) (Figure 7).
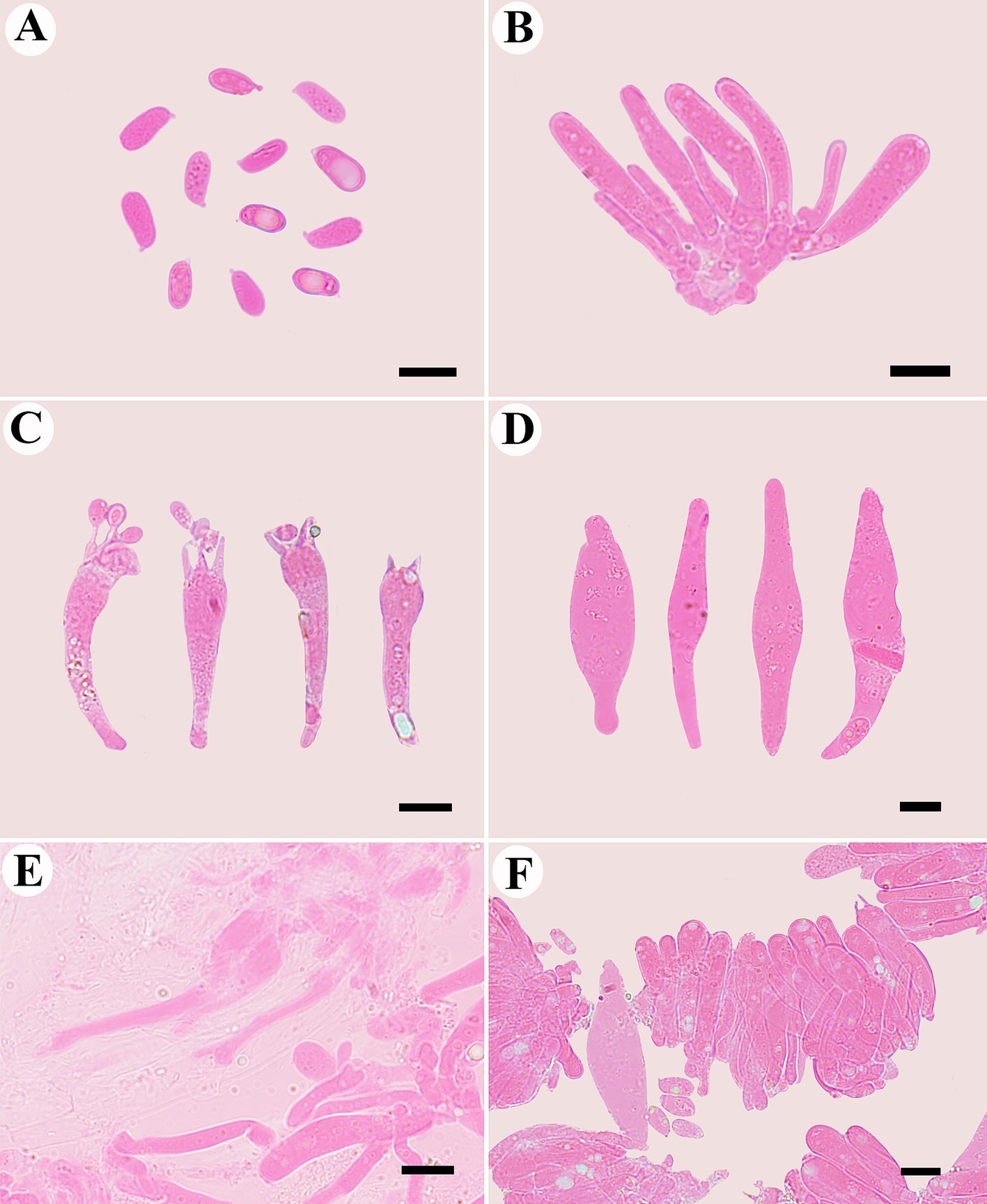
Figure 7. Microscopic structures of Mycena glabera (photographed from the holotype). (A) Basidiospores, (B) probasidia, (C) basidia, (D) cheilocystidia, (E) hyphae with clamp connections, and (F) hymenium.
4 Discussion
Members of Gymnopus and Mycena are widely dispersed; however, their diversity is not well-recognized in China. In recent years, only three species of Gymnopus in China (G. ramulicola T.H. Li and S.F. Deng, G. alliifoetidissimus T.H. Li and J.P. Li, and G. pallipes J.P. Li and Chun Y. Deng) were originally described utilizing molecular evidence. Increased interest in the species variety of Mycena has resulted in the publication of numerous new species and significant advancements in scientific understanding. More Chinese scholars focus on the macrofungi in southwest and northeast China; however, the northwest region has received less attention. Phylogenetic analyses revealed that 23 species of Gymnopus sect. Impudicae (Figure 2) and 33 species of Mycena sect. Calodontes (Figure 1) were grouped together, respectively, including two new species of Gymnopus and one new species of Mycena from northwestern China. Our phylogenetic results are consistent with prior studies (Ryoo et al., 2016; Cubeta et al., 1991), and further information on the phylogeny and taxonomy of Gymnopus sect. Impudicae and Mycena sect. Calodontes is provided.
Based on our phylogenetic analysis, G. gansuensis formed a single branch that was separated from other Gymnopus species (Figure 2). Morphologically, G. similis is similar to G. gansuensis in having brownish orange pileal surface, brownish orange or light brown Lamellae, and similar size of basidiospores measuring 6.5–8.5 μm × 2.7–4 μm (Ryoo et al., 2016). In addition, G. gansuensis is characterized by pileus honey yellow at the center, margin pinkish buff to buff, stipe pinkish buff to fuscous, and basidiospores elliptic to briolette. However, G. similis differs from G. gansuensis by its light to reddish brown to darker (reddish) brown stipe and wider cheilocystidia (20–65 μm × 5–9 μm vs. 51–63 μm × 5–6 μm) (Ryoo et al., 2016).
Gymnopus subsepiiconicus is clustered together with G. aurantiipes (Corner) A.W. Wilson, Desjardin & E. Horak, Gymnopus bicolor A.W. Wilson, Desjardin & E. Horak, and G. sepiiconicus (Corner) A.W. Wilson, Desjardin, and E. Horak (Figure 2). Morphologically, G. aurantiipes is similar to G. subsepiiconicus in having solitary to gregarious basidiomata and convex to applanate pileus (Ronquist and Huelsenbeck, 2003). However, G. aurantiipes differs from G. subsepiiconicus by its orange pileal surface, yellow pileal margin, yellow to orange–brown to reddish brown stipe, and the smaller basidiospores (4.8–7.2 μm × 2.4–4 μm vs. 6–9 μm × 3–3.5 μm) (Ronquist and Huelsenbeck, 2003). Gymnopus bicolor is similar to G. subsepiiconicus in having fluted and hygrophanus pileus and central and cylindrical stipe. However, G. bicolor differs from G. subsepiiconicus by its disk brown pileus, pale orange brown to reddish brown stipe, smaller basidiospores (5.2–8 μm × 2.4–3.6 μm vs. 6–9 μm × 3–3.5 μm), shorter basidia (14.5–22.5 μm × 4–6 μm vs. 19–40 μm × 5–7 μm), and shorter cheilocystidia (17.5–25.5 μm × 6.5–9.5 μm vs. 52–111 μm × 5–6.5 μm) (Ronquist and Huelsenbeck, 2003). Gymnopus sepiiconicus is similar to G. subsepiiconicus in having convex to applanate pileus and cylindrical stipe. However, G. sepiiconicus differs from G. subsepiiconicus by its brown to dark brown pileal center, beige yellow to white pileal margin, and shorter basidiospores (4.8–6.4 μm × 2.4–4.4 μm vs. 6–9 μm × 3–3.5 μm) (Ronquist and Huelsenbeck, 2003; Wilson et al., 2004).
Mycena glabera, sistered with M. rufobrunnea Z.W. Liu, Y.P. Ge & Q., is grouped together with M. seminau A.L.C. Chew & Desjardin and M. sinar A.L.C. Chew & Desjardin (Figure 1) and indistinguishable in the shape or size of basidiospores. However, M. rufobrunnea can differ from M. glabera by having dark brown pileus at the center, grayish-magenta to dull violet stipe in the upper part, and utriform cheilocystidia (Cortés-Pérez et al., 2023; Liu et al., 2022). Mycena seminau is similar to M. glabera in having elongate to cylindrical, smooth, hyaline, and thin-walled basidiospores. However, M. seminau differs from M. glabera by its brown to dark brown pileus at the center, smaller basidia (20.8–25.6 μm × 4.8–8.0 μm vs. 29.0–45.0 μm × 6.3–12.0 μm) and basidiospores (7.6–8.8 μm × 3.6–4.4 μm vs. 6.3–10.2 μm × 3.7–5.5 μm) (Chew et al., 2014). Mycena glabera is similar to M. sinar in having elongate to cylindrical basidiospores and similar in size and subdecurrent lamellae. However, M. sinar can be differentiated from M. glabera by having brownish orange to yellowish brown pileus at the center, narrower basidia (4.8–8.8 vs. 6.3–12.0) and abundant, fusiform to subfusiform cheilocystidia (Chew et al., 2014).
The species diversity of Gymnopus and Mycena from China has been increased after the comprehensive research. This study provided a basis for further research on Gymnopus sect. Impudicae and Mycena sect. Calodontes. However, the systematic research of the genera was restricted because only a small number of Gymnopus sect. Impudicae and Mycena Sect. Calodontes species with multiple genes accessible could be used for the analysis. For the time being, the most effective DNA barcoding for the identification of Gymnopus and Mycena species is ITS, while more samples with multigene sequences, including mt-SSU, RPB1, and RPB2, are needed to further investigate the species diversity and phylogenetic relationships of mushroom-forming species.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
LF: Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. BW: Investigation, Methodology, Software, Visualization, Writing – review & editing. TM: Investigation, Writing – review & editing. BL: Investigation, Writing – review & editing. JM: Investigation, Writing – review & editing. XL: Investigation, Writing – review & editing. NB: Investigation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Research Foundation of Gansu Agricultural University for Advanced Talents (Nos. GAU-KYQD-2021-26), the National Natural Science Foundation of China (Nos. 32460005) and the Gansu Provincial University Foundation for Youth Doctors (Nos. 2022QB-080).
Acknowledgments
We express our gratitude to Bing Chen, Ying Yang, and Xiusheng Liu for their help during field collections. We would also like to thank the reviewers and the responsible editors whose corrections and suggestions have enabled our article to be published.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Antonín, V., and Noordeloos, M. E. (1997). A monograph of Marasmius, Collybia and related genera in Europe. Part 2: Collybia, Gymnopus, Rhodocollybia, Crinipellis, Chaetocalathus, and additions to Marasmiellus; Libri Botanici; IHW-Verlag: Eching, Germany, 17, pp. 1–256.
Antonín, V., and Noordeloos, M.E. (2010). A monograph of Marasmioid and Collybioid Fungi in Europe; IHW Verl: Eching, Germany.
Chew, A. L. C., Tan, Y. S., Desjardin, D. E., Musa, M. Y., and Sabaratnam, S. (2014). Four new bioluminescent taxa of Mycena sect. Calodontes from peninsular Malaysia. Mycologia 106, 976–988. doi: 10.3852/13-274
Coimbra, V. R. M., Pinheiro, F. G. B., Wartchow, F., and Gibertoni, T. B. (2015). Studies on Gymnopus sect. Impudicae (Omphalotaceae, Agaricales) from northern Brazil: two new species and notes on G. montagnei. Mycol. Prog. 14:110. doi: 10.1007/s11557-015-1131-2
Cortés-Pérez, A., Guzmán-Dávalos, L., Ramírez-Cruz, V., Villalobos-Arámbula, A. R., Ruiz-Sanchez, E., and Ramírez-Guillén, F. (2023). New species of bioluminescent Mycena sect. Calodontes (Agaricales, Mycenaceae) from Mexico. J. Fungi 9:902. doi: 10.3390/jof9090902
Cubeta, M. A., Echandi, E., Abernethy, T., and Vilgalys, R. (1991). Characterization of anastomosis groups of Binucleate Rhizoctonia species using restriction analysis of an amplified ribosomal RNA gene. Phytopathology 81, 1395–1400. doi: 10.1094/Phyto-81-1395
Dai, Y. C., Zhou, L. W., Yang, Z. L., Wen, H. A., BAU, T., and Li, T. H. (2010). A revised checklist of edible of China. Mycosystema 29, 1–21.
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Deng, S.F. (2016). Taxonomy of Gymnopus and preliminary study of Marasmiaceae resource in South China. College of Agriculture, South China Agricultural University, China.
Deng, S. F., Li, T. H., Jiang, Z. D., and Song, B. (2016). Gymnopus Ramulicola sp. nov., a pinkish species from southern China. Mycotaxon 131, 663–670. doi: 10.5248/131.663
Farris, J. S., Källersjö, M., Kluge, A. G., and Bult, C. (1994). Testing significance of incongruence. Cladistics 10, 315–319. doi: 10.1111/j.1096-0031.1994.tb00181.x
Felsenstein, J. (1985). Confidence intervals on Phylogenetics: an approach using bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Gardes, M., and Bruns, T. D. (1993). Its primers with enhanced specificity for Basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Hall, T. A. (1999). Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
He, M. Q., Zhao, R. L., Mwanga, Z., and Dutta, A. K. (2020). 2019-notes, outline and divergence times of Basidiomycota. Fungal Divers. 99, 105–367. doi: 10.1007/s13225-019-00435-4
Hu, J. J., Tuo, Y. L., Yue, L., Zhao, G. P., He, X. L., Song, L. R., et al. (2024). New insights into Gymnopus s.l.: its systematic rearrangements and novel taxa. Mycosphere 15, 365–472. doi: 10.5943/mycosphere/15/1/3
Hu, J., Zhao, G., Tuo, Y., Rao, G., Zhang, Z., Qi, Z., et al. (2022). Morphological and molecular evidence reveal eight new species of Gymnopus from Northeast China. J. Fungi 8:349. doi: 10.3390/jof8040349
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kirk, P., Cannon, P., Minter, D., and Stalpers, J. (2008). Dictionary of the Fungi. 10th Edn. Wallingford, UK: CABI International.
Li, J. P., Li, Y., Li, T. H., Antonín, V., Hosen, I., Song, B., et al. (2021a). A preliminary report of Gymnopus sect. Impudicae (Omphalotaceae) from China. Phytotaxa 497, 263–276. doi: 10.11646/phytotaxa.497.3.5
Li, J. P., Pan, M. C., Li, Y., Deng, C. Y., Wang, X. M., Zhang, B. X., et al. (2022). Morpho-molecular evidence reveals four novel species of Gymnopus (Agaricales, Omphalotaceae) from China. J. Fungi 8:398. doi: 10.3390/jof8040398
Li, J. P., Song, B., Feng, Z., Wang, J., Deng, C. Y., and Yang, Y. H. (2021b). A new species of Gymnopus sect. Androsacei (Omphalotaceae, Agaricales) from China. Phytotaxa 521, 1–14. doi: 10.11646/phytotaxa.521.1.1
Liu, Z., Ge, Y., Zeng, H., Cheng, X., and Na, Q. (2022). Four new species of Mycena sect. Calodontes (Agaricales, Mycenaceae) from Northeast China. MycoKeys 93, 23–56. doi: 10.3897/mycokeys.93.86580
Mata, J. L., Hughes, K. W., and Petersen, R. H. (2004). Phylogenetic placement of Marasmiellus juniperinus. Mycoscience 45, 214–221. doi: 10.1007/S10267-004-0170-3
Mata, J. L., Hughes, K. W., and Petersen, R. H. (2006). An investigation of/Omphalotaceae (Fungi: Euagarics) with emphasis on the genus Gymnopus. Sydowia 58, 191–289.
Mata, J. L., and Ovrebo, C. L. (2009). New reports and illustrations of Gymnopus for Costa Rica and Panama. Fungal Divers. 38, 125–131. doi: 10.1002/yea.1704
Mešic, A., Tkalcec, Z., Deng, C. Y., Li, T. H., Pleše, B., and Cetkovic, H. (2011). Gymnopus Fuscotramus (Agaricales), a new species from southern China. Mycotaxon 117, 321–330. doi: 10.5248/117.321
Moncalvo, J. M., Vilgalys, R., Redhead, S. A., Johnson, J. E., James, T. Y., Catherine Aime, M., et al. (2002). One hundred and seventeen clades of Euagarics. Mol. Phylogenet. Evol. 23, 357–400. doi: 10.1016/S1055-7903(02)00027-1
Na, Q., Liu, Z., and Zeng, X. G. Y. (2022). Taxonomic studies of bluish Mycena (Mycenaceae, Agaricales) with two new species from northern China. Mycokeys 90, 119–145. doi: 10.3897/mycokeys.90.78880
Nylander, J.A.A. (2004). MrModeltest v2. Program. Distributed by the author; Evolutionary Biology Center, Uppsala University: Uppsala, Sweden.
Posada, D., and Crandall, K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Ryoo, R., Antonín, V., Ka, K.-H., and Tomšovský, M. (2016). Marasmioid and Gymnopoid Fungi of the Republic of Korea. 8. Gymnopus section Impudicae. Phytotaxa 286, 75–88. doi: 10.11646/phytotaxa.286.2.2
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Sun, Y. L., Luo, Y., and Bau, T. (2021). Notes on Basidiomycetes of Jilin Province (X). J. Fungal Res. 19, 139–147.
Swofford, D.L. (2002). PAUP*: phylogenetic analysis using parsimony (*and other methods), Version 4.0b10; Sinauer associates: Sunderland, MA, USA.
Wang, X. S., Bao, J. S., Bao, H., and Feng, J. (2020). Macrofungal diversity in Hanwula National Nature Reserve, Inner Mongolia. Mycosystema 39, 695–706. doi: 10.13346/j.mycosystema.190409
Wei, R., Ge, Y., Qi, L., Han, M., Zeng, H., Hu, Y., et al. (2024). Revealing brownish Mycena diversity in China: new discoveries and taxonomic insights. J. Fungi 10:439. doi: 10.3390/jof10060439
Wilson, A. W., and Desjardin, D. E. (2005). Phylogenetic relationships in the Gymnopoid and Marasmioid Fungi (Basidiomycetes, Euagarics clade). Mycologia 97, 667–679. doi: 10.1080/15572536.2006.11832797
Wilson, A. W., Desjardin, D. E., and Horak, E. (2004). Agaricales of Indonesia. 5. The genus Gymnopus from Java and Bali. Sydowia 56, 137–210. doi: 10.1023/B:MYCO.0000041833.41085.6f
Keywords: Gymnopus sect. Impudicae, Mycena, macrofungi, morphology, phylogenetic analyses
Citation: Fan L, Wang B, Ma T, Li B, Ma J, Lei X and Bao N (2024) Three new species of Gymnopus and Mycena (Agaricales, Basidiomycota) from Northwestern China. Front. Microbiol. 15:1487598. doi: 10.3389/fmicb.2024.1487598
Edited by:
Zhou Shi, Gladstone Institutes, United StatesReviewed by:
Victor Manuel Bandala, Instituto de Ecología (INECOL), MexicoChang-lin Zhao, Southwest Forestry University, China
Copyright © 2024 Fan, Wang, Ma, Li, Ma, Lei and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: LongFei Fan, ZmFubGZAZ3NhdS5lZHUuY24=
 LongFei Fan
LongFei Fan BiYue Wang
BiYue Wang Bin Li
Bin Li XuTao Lei
XuTao Lei