- 1Laboratory of Anti-allergic Functional Molecules, College of Life Sciences and Medicine, Zhejiang Sci-Tech University, Hangzhou, China
- 2Hangzhou Zheda Dixun Biological Gene Engineering Co., Ltd., Hangzhou, China
- 3Beijing Key Laboratory of Precision Medicine for Diagnosis and Treatment of Allergic Diseases, Department of Allergy, National Clinical Research Center for Dermatologic and Immunologic Diseases, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing, China
Gut microbiota regulates the immune system, the development and progression of autoimmune diseases (AIDs) and overall health. Recent studies have played a crucial part in understanding the specific role of different gut bacterial strains and their metabolites in different AIDs. Microbial signatures in AIDs are revealed by advanced sequencing and metabolomics studies. Microbes such as Faecalibacterium prausnitzii, Akkermansia muciniphila, Anaerostipes caccae, Bacteroides sp., Roseburia sp., Blautia sp., Blautia faecis, Clostridium lavalense, Christensenellaceae sp., Coprococcus sp., Firmicutes sp., Ruminococcaceae sp., Lachnospiraceae sp., Megamonas sp., Monoglobus sp., Streptococcus pneumoniae and Bifidobacterium sp. help maintain immune homeostasis; whereas, Prevotella copri, Ruminococcus gnavus, Lactobacillus salivarius, Enterococcus gallinarum, Elizabeth menigoseptica, Collinsella sp., Escherichia sp., Fusobacterium sp., Enterobacter ludwigii, Enterobacteriaceae sp., Proteobacteria, Porphyromonas gingivalis, Porphyromonas nigrescens, Dorea sp., and Clostridium sp. cause immuno-pathogenesis. A complex web of interactions is revealed by understanding the influence of gut microbiota on immune cells and various T cell subsets such as CD4+ T cells, CD8+ T cells, natural killer T cells, γδ T cells, etc. Certain AIDs, including rheumatoid arthritis, diabetes mellitus, atopic asthma, inflammatory bowel disease and non-alcoholic fatty liver disease exhibit a state of dysbiosis, characterized by alterations in microbial diversity and relative abundance of specific taxa. This review summarizes recent developments in understanding the role of certain microbiota composition in specific AIDs, and the factors affecting specific regulatory T cells through certain microbial metabolites and also focuses the potential application and therapeutic significance of gut microbiota-based interventions as novel adjunctive therapies for AIDs. Further research to determine the precise association of each gut bacterial strain in specific diseases is required.
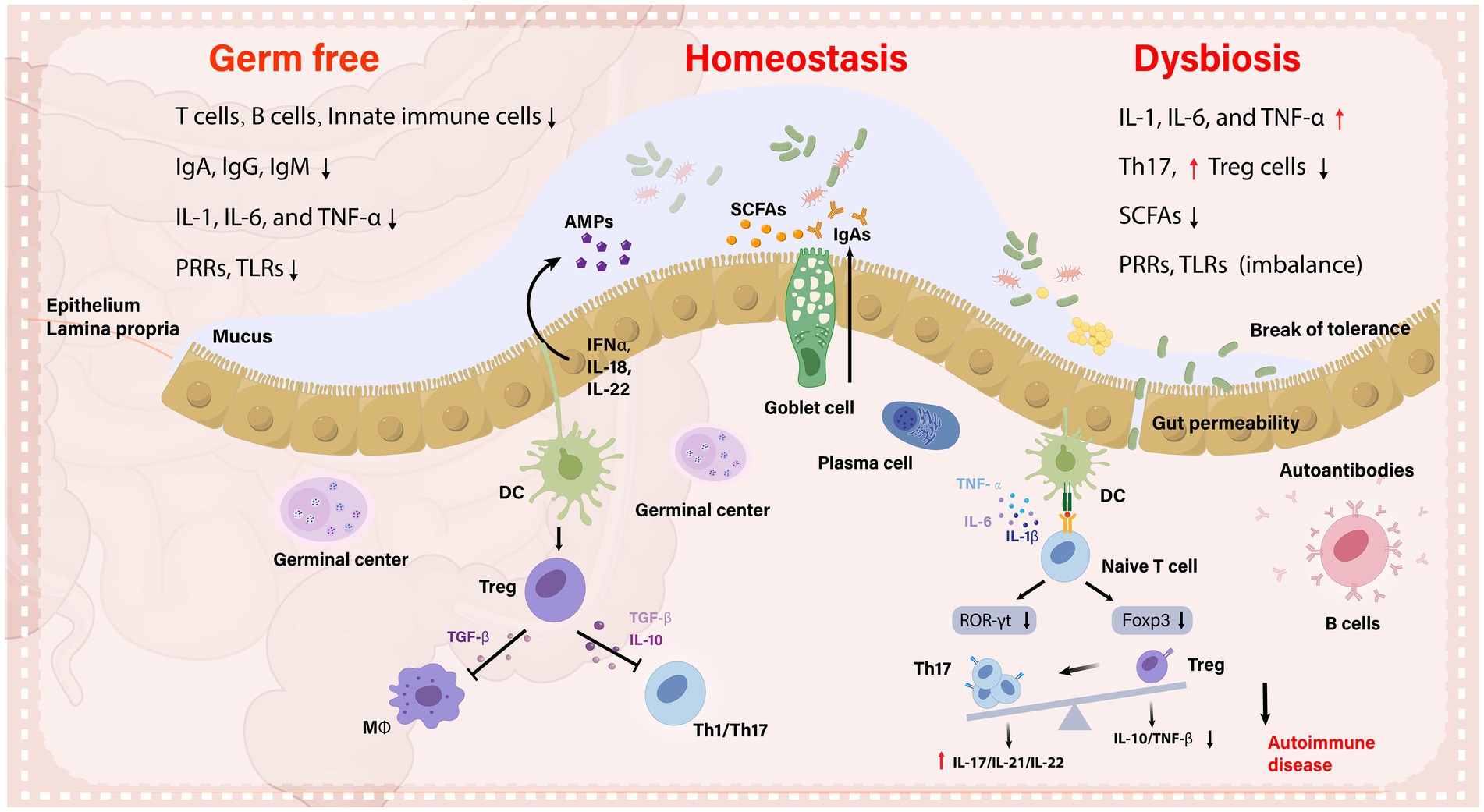
Graphical Abstract. The importance of the gut microbiota in immunological control and health is described in this graphical abstract, which contrasts its role in immune tolerance during homeostatic condition with that of autoimmune inflammation. It emphasizes the role of microbiota in T lymphocyte subpopulations, Gut-associated lymphoid tissue (GALT) development, and the synthesis of SCFAs and antimicrobial peptides. It also outlines how important dendritic cells (DCs) are in facilitating the synthesis of commensal-specific IgA via plasma cells, in order to maintain gut homeostasis. While, dysbiotic microbiota creates antigens in inflammatory conditions that lead to adverse immune responses, impairing mucosal integrity and exacerbating autoimmune diseases.
Introduction
It is well recognized that certain environmental factors cause autoimmune diseases (AIDs) in individuals having genetic predisposition, and there is evidence linking epigenetic dysregulation to AIDs pathogenesis (Araki and Mimura, 2018; Zhao et al., 2019). Numerous environmental factors, including pesticides, heavy metals and smoking have been linked to AIDs (Khan and Wang, 2018). In recent times, the gut microbiome is reported to be linked with AIDs due to its ability to cause immune dysregulatory and pro-inflammatory effects through microbiome dysbiosis (Dehner et al., 2019). The American gut project is recognized as largest crowdsourced citizen science project till now. The largest known human microbiome cohort was created from samples taken from thousands of participants via oral, skin, feces and other body sites. It is the PRJEB11419 project on NCBI (National Center for Biotechnology Information). In this project, a total of 1053 samples were related to the phenotype of AIDs. In the GMrepo database, 553 samples were selected from the 1053 samples for research, and 3095 species and 1016 genera were found (Wu et al., 2020). We sorted top 50 species/genera in descending order of median relative abundance and found that the species with higher median relative abundance include Elizabeth menigoseptica, Faecalibacterium sp. MC_41, etc., while the genera with higher median relative abundance include Bacteroides, Faecalibacterium, etc. (Figure 1). This large-scale, crowdsourced microbiome analysis offers insights about dysbiosis’ relation with the development of AIDs and may open avenues for personalized microbiome-based diagnostics and treatments.
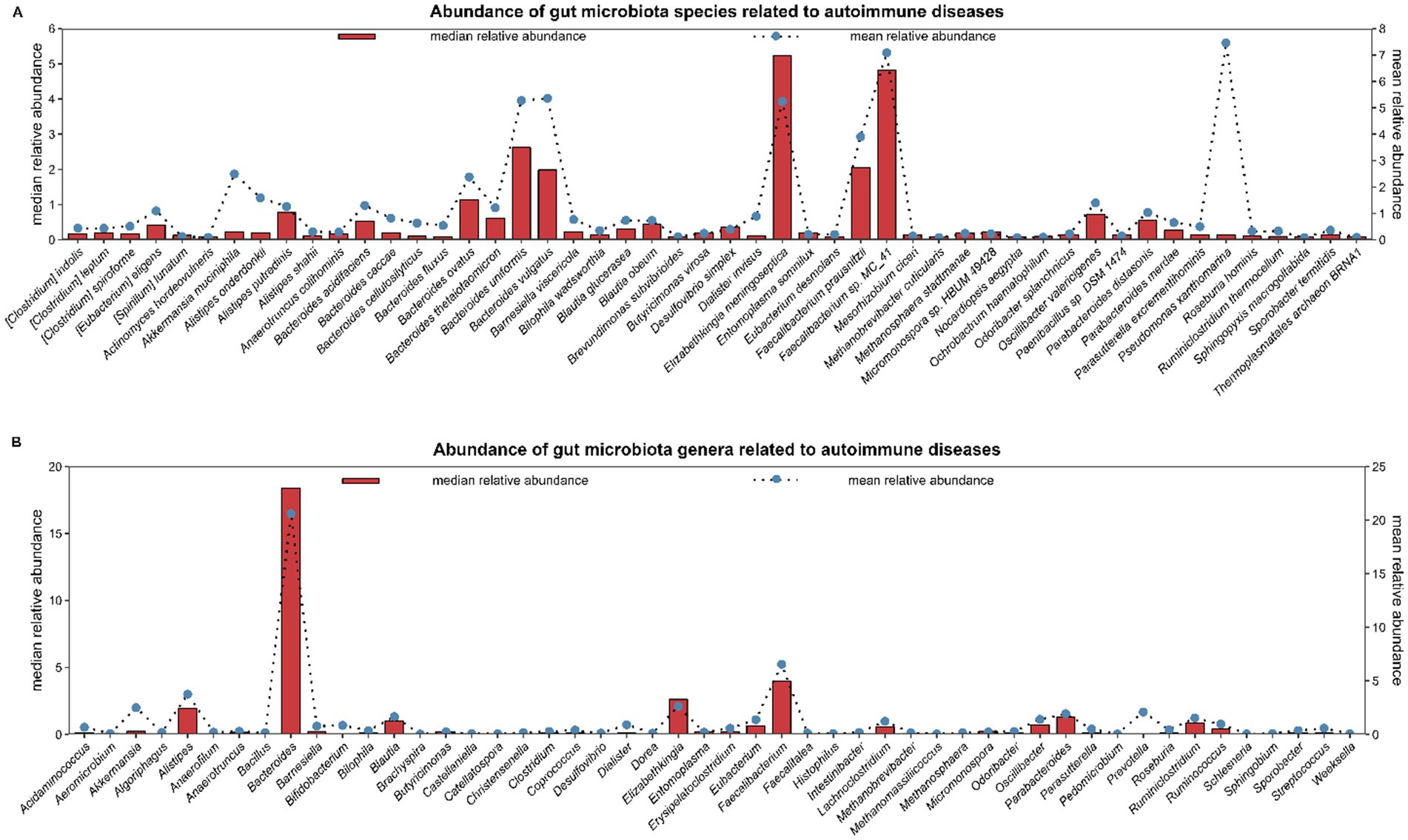
Figure 1. The American Gut project is the largest crowdsourced citizen science project to date. Fecal, oral, skin, and other body site samples collected from thousands of participants represent the largest human microbiome cohort in existence. It is the PRJEB11419 project on NCBI (National Center for Biotechnology Information). In the PRJEB11419 project, a total of 1,053 samples were related to the phenotype of autoimmune diseases. In the GMrepo database, 553 samples were selected from the 1,053 samples for research, and 3,095 species and 1,016 genera were found. In figure, the top 50 species/genera of gut microbiota are selected in descending order based on the median relative abundance. (A) shows 50 species; (B) shows 50 genera. Abundance: mean/median relative abundance of a species/genus in all samples of autoimmune diseases.
The immune homeostasis disruption leads to an increase in effector Th1, Th17, and plasma cell populations. Antigen-presenting cells (APCs), predominantly the DCs and macrophages, are significant because they sample antigens and initiate an inflammatory response. These APCs are able to transport toxins and antigens derived by the luminal microbiota to effector B and T cells, activating them in the process as part of the interaction of immune system with gut microbiota (Farache et al., 2013). Furthermore, antimicrobial peptides (AMPs), are essential for separating the gut bacteria from epithelium (Bevins and Salzman, 2011). AMPs are innate immuno-effectors that are released by epithelial cells in response to cytokines (IFNα, IL-18, and IL-22) produced by macrophages and DCs against microbial antigens (Mergaert, 2018). In a healthy state, when the immune system and gut microbiota work together, they encourage the immuno-modulatory T-reg cell differentiation and proliferation while suppressing the pro-inflammatory pathways (Mowat, 2018). However, abnormalities in the gut microbiota composition can cause harmful autoimmune reactions, especially in individuals having genetically predisposition. An illustration of this is the activation of Th17, which is linked to specific gut microbiota species such as Prevotella copri, Bifidobacterium adolescentis and Enterococcus gallinarum (Manfredo Vieira et al., 2018; Asquith et al., 2016; Tan et al., 2016).
Metabolites derived from the gut microbiota, like SCFAs, possess the ability to alter a cell's metabolic state, hence inducing regulatory B cells and preventing pentanoate-induced Th17 cell production (Luu et al., 2019). Furthermore, breakdown products from tryptophan can cause an increase in intraepithelial CD4+ CD8+ T cells, microbiota-generated ATP can stimulate Th17 cell proliferation, and polysaccharides derived from bacteria can stimulate regulatory T cells (Ahern and Maloy, 2020; Wiertsema et al., 2021). Moreover, the over-activation of plasma cells that produce antibodies is one way by which gut microbiota influence the immune system and contribute toward autoimmunity (Ma et al., 2019). Figure 2 shows that dysbiosis along with dysregulation of T cells leads to certain AIDs.
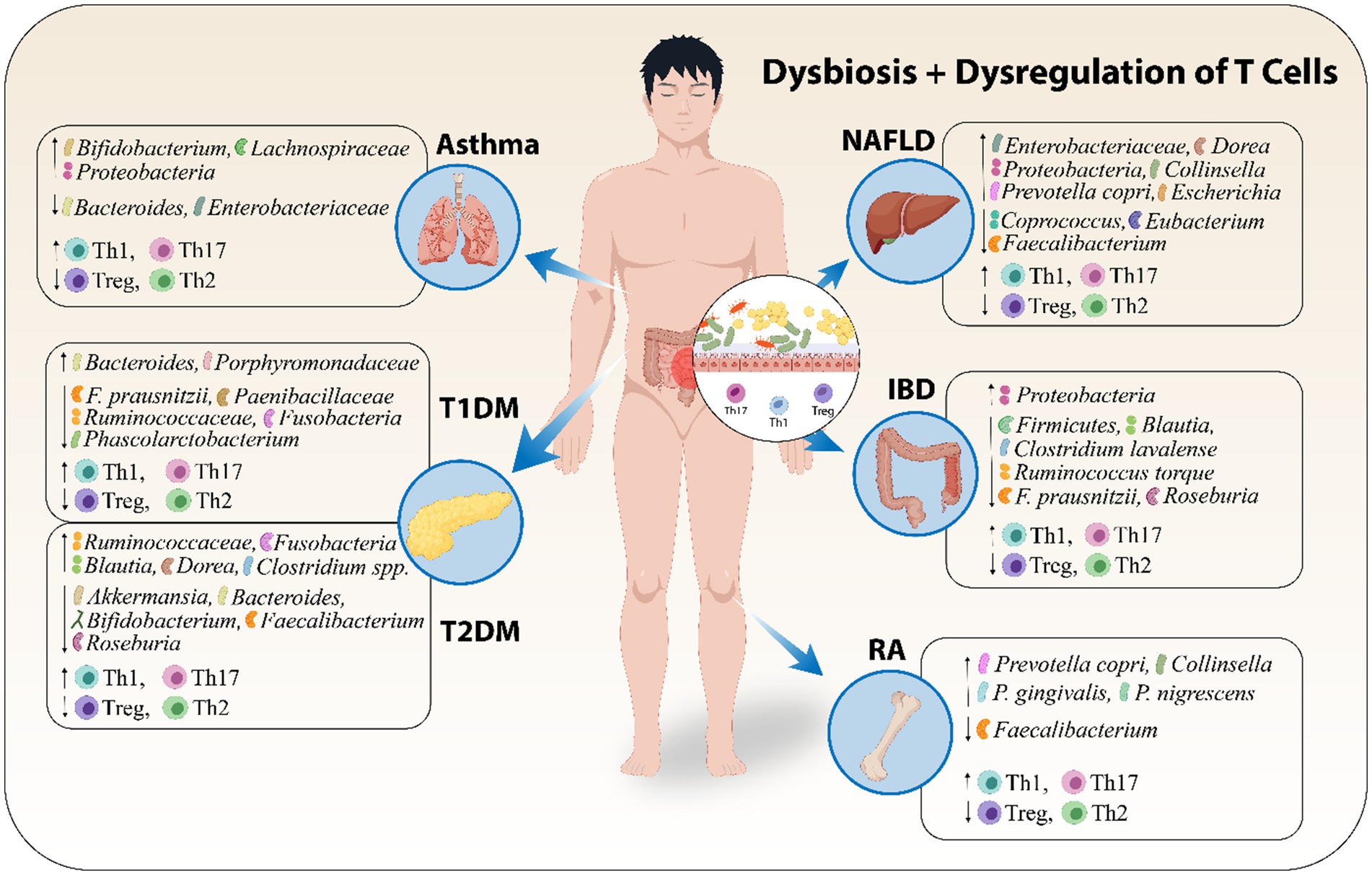
Figure 2. Dysbiosis along with dysregulation of T cells leads to certain AIDs. Abbreviations include Type 1 Diabetes Mellitus (T1DM), Type 2 Diabetes Mellitus (T2DM), and Non-Alcoholic Fatty Liver Disease (NAFLD), Inflammatory Bowel Disease (IBD), Rheumatoid Arthritis (RA).
A change in our understanding of the etiology of different inflammatory disorders has resulted from the discovery of the gut microbiota involvement in the remodeling of immune cells' epigenome and it has opened new avenues for therapeutic mechanisms. The immune system's reaction to AIDs is complicated, and both B and T cells are essential to the pathophysiology of these conditions. While, gut microbiota has long been known to affect B cell functions and to regulate immune responses, there is mounting evidence that T cells, specifically T helper cells (Th1, Th2 and Th17) and regulatory T cells (Tregs) are key contributors to the immune dysregulation seen in AIDs. T cell subsets differentiation, including helper T cells (Th1, Th2 and Th17) and Treg cells, is associated with particular microbiota species (Mazmanian et al., 2005; Gaboriau-Routhiau et al., 2009; Atarashi et al., 2011). Furthermore, the gut microbiota's metabolic byproducts, e.g. short-chain fatty acids (SCFAs), have influence on the T cells activation and differentiation (Saade et al., 2022). Numerous factors, including metabolic activities, immune dysregulation and pro-inflammatory pathways can cause the gut microbiota dysbiosis. T cells-related disorders such as RA, type 1 and type 2 diabetes mellitus, atopic asthma, IBD and NAFLD are all influenced by dysbiosis (Valdes et al., 2018; Ananthakrishnan et al., 2018; Cenit et al., 2017). Hence, in this article, we have reviewed the gut microbiota relation with AIDs, how gut microbiota manipulates different T cells functions and contributes toward immuno-pathogenesis of different AIDs and also discussed the gut microbiota-based interventions as innovative therapeutic options for AIDs.
Material and methodology for literature survey
This review contains original research articles published in English language from 2004 to 2024, of which mostly were published after 2015. During the year of 2024, different database searches are performed on PubMed, Web of Science, Elsevier, MDPI and Springer Link etc., using keywords such as “gut microbiota” + “autoimmune diseases”, “inflammatory bowel disease”, “Treg cells function”, “CD4+ T cells”, “CD8+ T cells”, “γδ T cells”, “natural killer T cells”, “Type 1 diabetes mellitus”, “Type 2 diabetes mellitus”, “rheumatoid arthritis”, “atopic asthma”, and some other combined keyword searches. We only included studies that were published in well renowned journals of relevant topics and are completely consistent with the subject of this review. The purpose of this review article is to summarize the recent researches on autoimmune diseases and their relation with gut microbiota to provide enough information relevant to the topic of this review article. In order to facilitate readers, we have added mechanisms based figures for graphical presentation of concept and clear understanding.
Gut microbiota’s association with AIDs
The genetic susceptibilities of the individual cannot entirely describe the pathophysiology of human diseases, which emphasizes the need to investigate the roles played by environmental factors. Increasing research has demonstrated the link between aberrant changes in gut microbiota and a variety of disease types, including autoimmune disorders [such as spondyloarthritis (SpA), systemic lupus erythematosus (SLE), Sjögren’s syndrome (pSS), Multiple Sclerosis (MS), Inflammatory bowel disease (IBD), Rheumatoid arthritis (RA), etc] which is corroborated by our understanding of the pathogenic activities of gut microbiota (Yin et al., 2020; Thomas et al., 2019). The microbiota, which is found on the skin and mucosa of the host, coevolves with the host and is most prevalent in the gastrointestinal tract. These microbes contain roughly 150 times greater metagenome, or genomic contents in comparison to humans (Qin et al., 2010). The gut microbiota and immune system work together in a suitable manner to combine innate and adaptive immunity in such a way that selects, modifies, and terminates responses in the most appropriate manner. Immune system and gut bacterial communities correlate with each other in maintaining the healthy environment. Disturbance in either of these results in certain autoimmune disorders.
With time, the advancements in high-throughput DNA sequencing have made investigation of the complex gut microbes signature much easier, especially those microbes which are difficult to culture in vitro. Since metagenomic research was initially used to study the gut microbiota in gastrointestinal disorders such as colorectal cancer and IBD, now been extended to study the gut microbiota in AIDs such as SpA (Yin et al., 2020; Wen et al., 2017; Zhou et al., 2020), SLE (Li et al., 2019; Azzouz et al., 2019; Chen et al., 2019), MS (Jangi et al., 2016), RA (Chiang et al., 2019; Alpizar-Rodriguez et al., 2019; Zhang et al., 2015; Scher et al., 2013) and pSS (van der Meulen et al., 2019; Mandl et al., 2017). These techniques have uncovered dysbiosis patterns and linked it to immune regulation. However, limitations like insufficient strain-level resolution and the need for functional validation highlight the importance of integrating multi-omics approaches to better understand gut microbiota’s role in AIDs. The gut microbiota of the individuals having AIDs differs significantly in comparison to healthy people. For example, species such as Lactobacillus salivarius, Ruminococcus gnavus and Prevotella copri have been linked to the pathogenesis of AIDs. However, The complex relationships between the host and microbes make it difficult to translate data from animal models showing the immune-regulatory behavior of particular strains of bacteria and their metabolites into clinical implications (Hui et al., 2019; Evans-Marin et al., 2018; Zegarra-Ruiz et al., 2019; Manfredo Vieira et al., 2018). New biomarkers and treatment options can be obtained by examining temporal dynamics, transgenerational impacts, and microbial metabolite-induced epigenetic changes. Validating this hypothesis can provide information about tailored microbiota-based therapies for managing and preventing AIDs.
Gut microbiota disturbance and development of AIDs pathogenesis
One of the greatest interfaces (250–400 m2) in the human body exists between the host, antigens and environmental conditions in the gastrointestinal (GI) tract (Thursby and Juge, 2017). The human GI is home to greatest microbial community known as the "gut microbiota," that is made up of a wide variety of microorganisms, such as archaea, bacteria and eukarya, that have coevolved over millions of years with their host to form a complex and mutually beneficial symbiotic relationship (Neish, 2009). Various immune cells, including innate lymphocytes, macrophages, and DCs, make up the gut-associated lymphoid tissue (GALT), which is the first line of defense against pathogens (Bellocchi et al., 2019). The gut microbiota has been extensively studied in relation to human disease, and knowledge about its composition and functions has grown rapidly (Piccioni et al., 2022). The gut microbiota is mainly responsible for preserving the equilibrium between host defense and immune tolerance, and it is also a major factor in the development of the immune system (Zhang et al., 2023; Jiao et al., 2020; Piccioni et al., 2022). Interestingly, inadequate exposure to microbes during early life has been associated with an increased risk of AIDs. This highlights how important the early gut microbiota development is in influencing the immune system of their host (Stiemsma et al., 2015). The relationship between host immunity and gut microbiota suggests that AIDs are partly caused by dysbiosis of the gut microbiota.
The gut microbiota composition of individuals having AIDs differed significantly from that of healthy ones as demonstrated by advanced techniques like whole-genome sequencing and metatranscriptomic analysis, suggesting an imbalance in the gut ecosystem. The gut microbiota was found to be less diverse in cases of celiac disease, irritable bowel syndrome (Jackson et al., 2018) and several AIDs including RA, SpA, pSS, and SLE (Chiang et al., 2019; Azzouz et al., 2019; Wen et al., 2017; van der Meulen et al., 2019). It was indicated by a decrease in taxonomical richness and evenness in the microbiota. Therefore, there is need to explore whether specific microbial signatures, such as reduced levels of anti-inflammatory species or overabundance of pro-inflammatory taxa, correlate with disease activity and prognosis in different AIDs.
The presence of particular microbial taxa may serve as a biomarker for evaluating the severity of AIDs. For example, elevated R. gnavus was linked to lupus nephritis (Azzouz et al., 2019), while elevated L. salivarius was linked to higher disease activity in patients with SLE or RA (Zhang et al., 2015; Chen et al., 2019; Liu et al., 2013). Several studies highlight, R. gnavus, a bacteria known to degrade mucin, was in higher abundance in the feces of SLE, SpA, and RA patients. RA and ankylosing spondylitis (AS) patients had higher abundances of P. copri in their feces (Alpizar-Rodriguez et al., 2019; Scher et al., 2013; Wen et al., 2017; Zhou et al., 2020). On the other hand, a few bacteria, like Faecalibacterium prausnitzii, were found in lower amounts in AS and pSS patients (Yin et al., 2020; Stewart et al., 2018), indicating possible beneficial functions. In a recent study, Wang et al. reported, a significant decrease in the Firmicutes/Bacteroidetes (F/B) ratio in Lupus nephritis patients (Wang et al., 2023a). In a study reported, the Firmicutes in the microbial diversity was decreased in RA patients who had anti-citrullinated protein antibody (ACPA), while certain strains such as Blautia, Clostridiales and Akkermansia showed significant enrichment (Chiang et al., 2019).
Furthermore, Elevated number of pro-inflammatory bacteria like Akkermansia spp. and reduced numbers of Lachnospiraceae and Faecalibacterium (which produce anti-inflammatory chemicals) have been linked to the pathophysiology of MS (Jangi et al., 2016) A. muciniphila and Acinetobacter calcoaceticus, which were isolated from individuals suffering from MS and incubated in monocolonized mice, induced pro-inflammatory responses and Parabacteroides distasonis stimulated anti-inflammatory human IL-10+FoxP3+ cells and CD4+CD25+ T cells (Cekanaviciute et al., 2017; Sadeghpour Heravi, 2024). However, the predictive power of microbial species for disease severity is still unknown. Standardization of study methodologies and further research on the regulatory roles of different microbes across various AIDs is needed to address these inconsistencies.
Whereas the host immune system regulates microbial ecology, the microbiota also generates a wide range of biochemical active metabolites, including SCFAs and tryptophan metabolites, which impact the immune system's development and function. Studies on lupus-prone mice have supported the functional analysis found in patients with systemic lupus erythematosus (SLE), which indicated a rise in the biosynthesis of tryptophan, ornithine and arginine with a reduction in the biosynthesis of branched-chain amino acids (Chen et al., 2019). Abnormal pathways in patients with ankylosing spondylitis include degradation of glycosaminoglycan, lipopolysaccharide biosynthesis, and oxidative phosphorylation (Zhou et al., 2020).
Moreover, when several immune pathways were abnormally triggered due to dysbiosis, anti-inflammatory cytokines (like IL-4, IL-1ra, TGF-β, IL-10, IL-13, etc.) decreased and pro-inflammatory cytokines (like IL-6, IL-12, TNF-α, IFN-γ, IL-1β, IL-17, etc.) increased (Zhang et al., 2014). Therefore, it is crucial to conduct longitudinal research and functionally characterize gut microbial species. Translational research has to find a way to connect human and animal studies, find biomarkers based on microbiome, and develop focused treatments. Furthermore, investigating how lifestyle of an individual and environmental variables affect gut microbiota will help in developing preventative measures and improvement of the disease treatment. This will also clarify the complex interactions among host genetics, immunotypes and gut microbiota. Figure 3 describes the mechanism by which disturbed gut microbiota leads to AIDs and Table 1 highlights the increase or decrease in various bacterial species involved in different autoimmune diseases and their role toward diseases regulation.
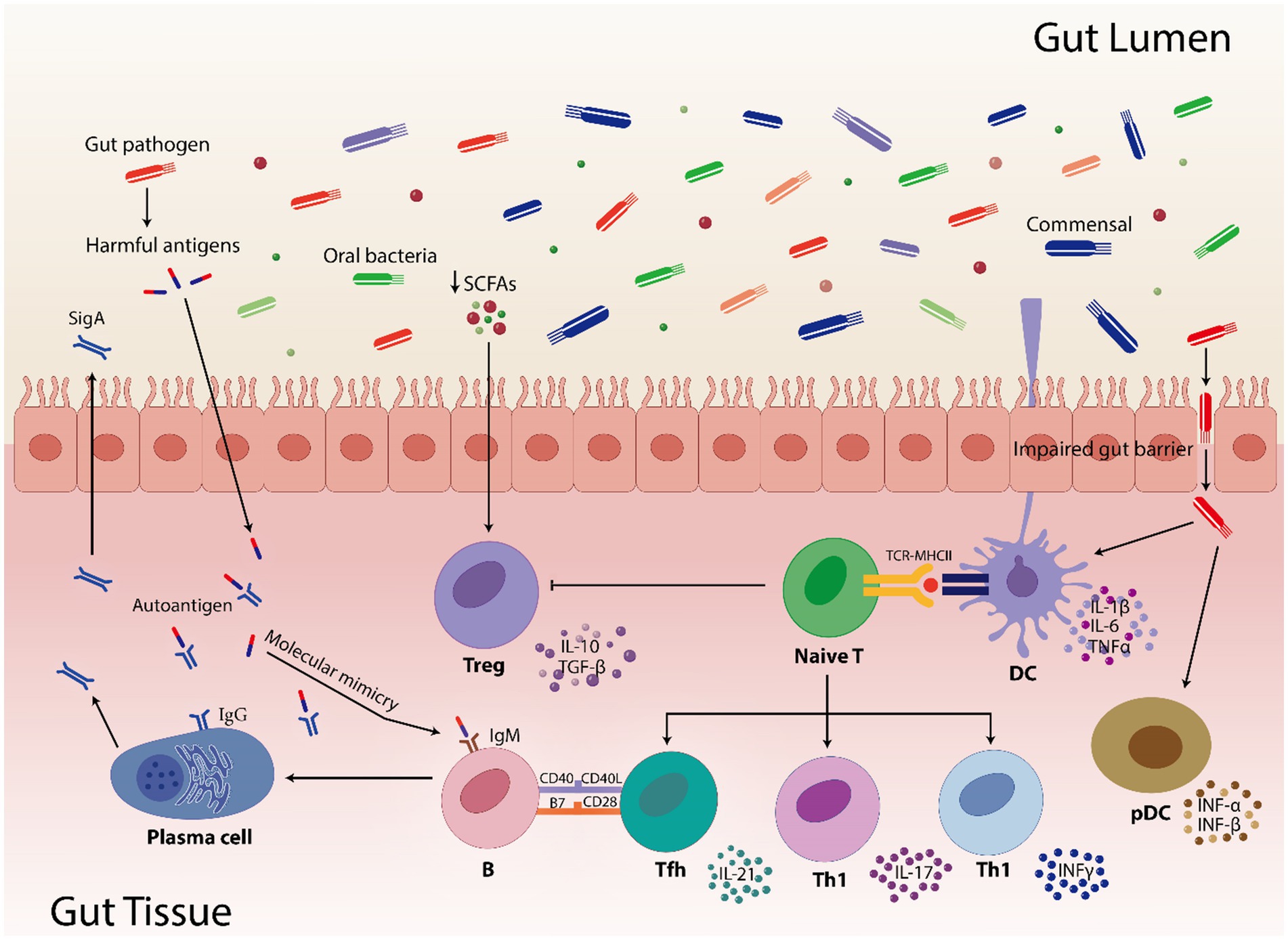
Figure 3. Highlights how gut pathogens and commensal bacteria can influence the gut immunity. Short-chain fatty acids (SCFAs) are involved in controlling immunological responses, including the activation of regulatory T cells (Treg). In pathogenic condition, SCFAs numbers are reduced which results in reduced activation of Treg cells. Furthermore, compromised gut barrier function facilitates the translocation of commensals and pathogens by inducing pro-inflammatory cytokines (IL-1β, IL-6, TNFα, INF-α, and INF-β) via dendritic cells (DC) and plasmacytoid dendritic cells (pDC). The TCR-MHCII contact on DCs activates naive T cells, which thereafter differentiate into diverse T helper cell subsets (Tfh, Th1, and Th17) and contribute to the gut’s immunological environment. Moreover, molecular mimicry is caused when microbial antigens structurally resemble the human autoantigens leading to B cell activation that use the assistance of Tfh cells to produces both protective sIgA and, in dysregulated states, pathogenic autoantibodies. In addition, impaired gut barrier facilitates the transfer of microbial antigens, which can result in immunological cross-reactions where T and B cells may mistakenly target host tissues by activating the T cell subsets and pro-inflammatory cytokines. These interactions highlight how the gut microbiome shapes autoimmune susceptibilities, especially in hosts who are genetically susceptible.
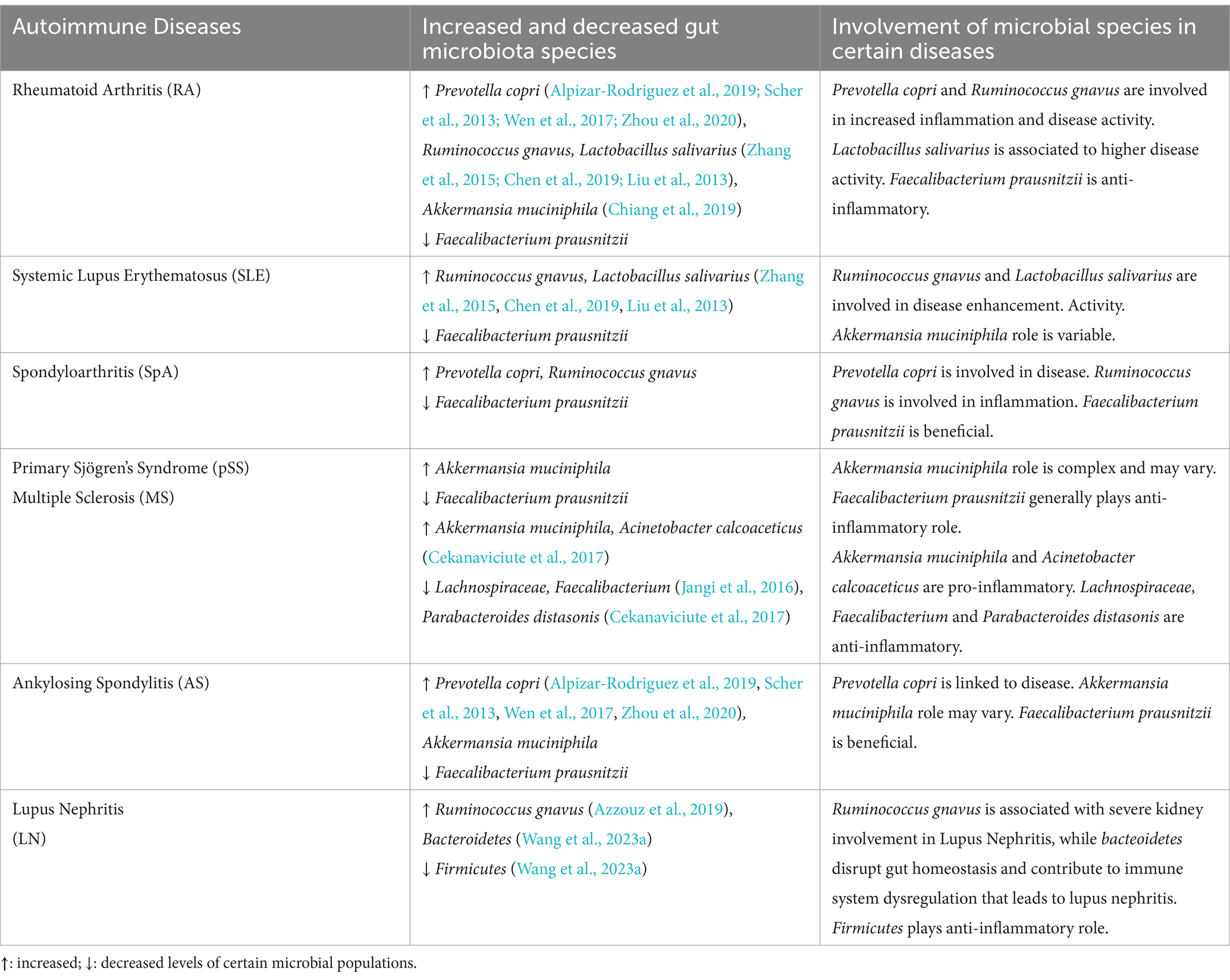
Table 1. Variations in population of gut microbes correlated with different autoimmune diseases and their involvement.
Gut microbiota-mediated manipulation of T cells function
Complex interactions between intestinal mucosal immunity and gut microbiota have a wide range of impacts on disease processes and homeostasis (Zhang and Pan, 2020). The human immune system and the microbiota mainly interact in GI tract. The innate immunity serves as first line of defense while the adaptive immunity precisely targets pathogens and creates immunological memory (Thakur et al., 2019). The gut microbiota has a major impact on disease states and immunological homeostasis by influencing the differentiation and function of different T cell subsets. Inflammation and immunological tolerance can be impacted by the promotion or inhibition of T helper (Th1, Th2, Th17), regulatory T (Treg), cytotoxic CD8+ T, natural killer T (NKT) and γδ T cells responses by particular bacterial species and their metabolites. These interactions are frequently upset by dysbiosis, which results in immunological abnormalities that fuel autoimmune disorders (AIDs).
CD4+ T cells
Adaptive immune responses mainly depend on CD4+ T helper cell differentiation into subsets (such as Tfhs, Th1s, Th2s, Th17s, Tregs, and other subtypes) with a variety of effector activities especially for the defense of host against pathogens (Saravia et al., 2019; Li et al., 2022b). Various studies highlight the influence of the gut microbiota on CD4+ T cells differentiation. Some species of Klebsiella genera, including K. pneumoniae and K. aeromobilis, cause Th1 cell responses in the gastrointestinal tract. Klebsiella colonization increases Th1 cell proliferation in the intestines of germ-free mice, thereby increasing the number of these cells (Atarashi et al., 2017) (Figure 4). Probiotic bacteria, especially Lactobacillus strains like L. salivarius (Ren et al., 2019) and L. plantarum (Matsusaki et al., 2016; Takeda et al., 2011), increase the Th1 cytokines [interferon-gamma (IFNγ) and tumor necrosis factor-alpha (TNFα)] production, while, inflammatory immune responses by Th1 cells were inhibited and antigen-specific oral tolerance was enhanced in arthritic mice treated with L. casei (So et al., 2008).
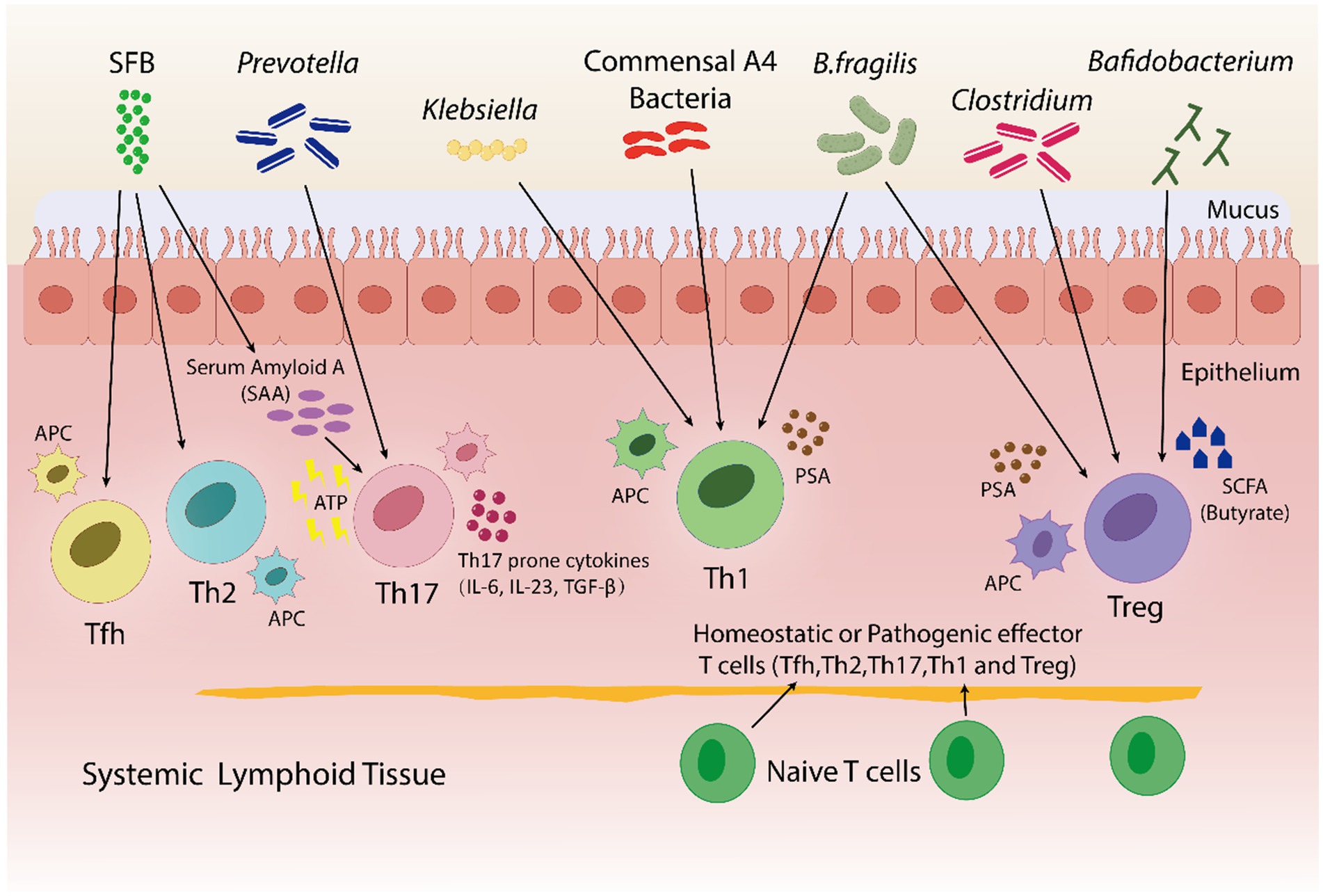
Figure 4. In both pathogenic and homeostatic settings, the microbiota is an essential factor in guiding T-cell differentiation. By using a bacterial product called PSA in a PSA-dependent pathway, B. fragilis promotes the growth of Th1-associated immune responses in germfree mice. Klebsiella and commensal A4 bacteria also induces Th1 cells. The proliferation of regulatory T (Treg) cells is encouraged by B. fragilis. By utilizing innate cells to produce either serum amyloid A (SAA) or adenosine 5′-triphosphate (ATP), SFB trigger a Th17 immune response. SFB also promotes the Tfh and Th2 cell production. Prevotella induces Th17 cells. Bafidobacterium produces SCFA, which stimulates the generation of Treg cells. Clostridium aids in the development of immunotolerance by promoting the production of Treg cells. Furthermore, within a particular tissue environment, systemic T cells can give rise to tissue-specific T cells through the activation of APCs by bacterial antigens.
A research indicated that germfree mice exhibits a preference for Th2 cell responses; however, the introduction of polysaccharide A (PSA) derived from Bacteroides fragilis corrects this imbalance by favoring Th1 cell responses (Mazmanian et al., 2005). PSA via the toll-like receptor (TLR2) interacts with DCs to stimulate the release of IL-12. IL-12 causes STAT 4 activation, which results in differentiation of CD4+ T cell into Th1 cells that produce IFN-γ (Wang et al., 2006). In healthy conditions, the differentiation of Th1 cells specific to Klebsiella can be controlled without causing severe inflammation in the gut. In contrast, during dysbiosis, Klebsiella dominance may induce severe gut inflammation through the induction of Th1 cell differentiation, as evidenced by more prevalence of Klebsiella species in fecal samples taken from IBD patients in comparison to healthy individuals (Lloyd-Price et al., 2019). Thus, the Th1/Th2 response can be balanced by the introduction of engineered B. fragilis strains that could overproduce polysaccharide A (PSA) and therefore, prevent Klebsiella-induced Th1 cell differentiation and severe gut inflammation in IBD patients.
Th2 cells generate antibodies to combat infections, proliferate and develop into plasma cells, and also stimulate B cells (Chen et al., 2021a). Certain types of bacteria, like A4 bacteria in the Lachnospiraceae family, prevent Th2 cells from differentiating and being active by making DCs based TGF-β (Wu et al., 2016). It has been determined that B. fragilis and lactobacillus strains inhibit Th2 activity, thereby enhancing Th1 activity (Mazmanian et al., 2005; Takeda et al., 2011; Won et al., 2011). Bamias et al. reported that the Th2 response was induced by commensal bacteria in the chronic phase of Crohn's disease (CD)-like ileitis in SAMP1/YitFc mouse models, and symptoms were also deteriorated (Bamias et al., 2007). A symbiotic combination of A4 bacteria, B. fragilis and Lactobacillus strains can be administered to Crohn’s disease patients, that may inhibit Th2 cell differentiation and activity, hence lowering chronic inflammation and enhancing symptom management.
Th17 cells stimulate neutrophils, produce and secrete a variety of inflammatory cytokines, including IL-17 and IL-22, and infiltrate lesions to increase the inflammation. Retinoic acid-related orphan receptor (RORγt), an important transcription factor, facilitates Th17 cell development and Th17 cytokine production (Nalbant and Eskier, 2016; Chen and Tang, 2021). Interestingly, Th17 cells are not present in germfree mice, rather, they are induced when bacteria colonize the mice (Lee and Kim, 2017). Certain bacteria, like gram-positive bacteria and SFB, have been found to be inducers of Th17 cell proliferation. Serum amyloid A (SAA), elevates when SFB typically penetrates the mucus layer and adheres to epithelial cells. SAA markedly enhances the Naive CD4+ T cell differentiation into Th17 cells (Atarashi et al., 2015; Li et al., 2022b).
Furthermore, the function of SFB such as Candidatus Arthromitus, and Prevotella has been reported in fostering strong Th17 cell proliferation along with the release of IL-17 and IL-22 (Atarashi et al., 2015; Farkas et al., 2015; Ivanov et al., 2009; Schnupf et al., 2015). In a study, P. gingivalis was introduced into a collagen-induced arthritis (CIA) mice model, found elevation in joint disease severity which was linked to systemic proinflammatory cytokine profiles that indicated Th17 pathway activation (Moen et al., 2006). A study reported a secondary bile acid produced by microbe to have a negative effect on Th17 cell differentiation. Th17 transcription factor RORγt (retinoic acid-related orphan receptor gamma t) was blocked by 3-oxolithocholic acid along with one of thirty diverse primary and secondary types of bile acids and the decrease in the differentiation of Th17 cells was observed in SPF mice (Hang et al., 2019). The exact mechanism by which certain bacteria induce intestinal Th17 differentiation is still unknown, thus, research must go into modifying gut microbiota to regulate the differentiation of Th17 cells and create innovative remedies for AIDs. Another promising therapeutic approach is to look into the interactions of bile acids with Th17 cells differentiation.
Treg cells are essential for the prevention of AIDs because they preserve immune homeostasis, control immune responses and promote tolerance to harmless antigens (Dominguez-Villar and Hafler, 2018). B. fragilis is able to trigger to generate large populations of Treg cells by producing PSA, which in turn causes Foxp3+ Treg cells to develop and produce IL-10 (Round and Mazmanian, 2010) as shown in Figure 4. Furthermore, adenosine and inosine, two bacterial metabolites, can bind with the adenosine A2A receptor (A2AR) on T cells, increasing the activity of Treg cell and suppressing the inflammatory responses of Th1 and Th17 (Liu et al., 2018; Wang et al., 2024). In a recent study Wang et al. found, Megamonas, Monoglobus, and Prevotella relative abundances were favorably connected with cytokine levels and CD4+ T cell counts; on the other hand, the T helper (Th17)/Treg ratio and the relative abundance of regulatory T cells (Tregs) were correlated negatively with RA disease activity (Wang et al., 2022b). Sun et al. reported the change in the composition of gut microbiota's by Bifidobacterium colonization. This improved the suppressive Treg cells function by encouraging their mitochondrial activity (Sun et al., 2020). Certain Lactobacillus strains, like Lacticaseibacillus casei, affect the differentiation and activity of T cells, resulting in the development of Treg cells and the release of IL-10 (Smits et al., 2005). L. acidophilus strain L-92 demonstrated related outcomes in BALB/c mice. L-92, when administered orally, led to a rise in the expression of Foxp3, TGF-β and IL-10 in mice under conditions of allergic contact dermatitis (ACD). This established the concept that L-92 mitigates ACD by increasing the proliferation of Treg cells, and Th1 and Th2 responses (Shah et al., 2012). A study conducted on in BALB/c mice, demonstrated that L. reuteri reduces allergic airway reactions (Forsythe et al., 2007) thereby, increases Foxp3 expression (Karimi et al., 2009), which positively affects the proliferation of Treg cell and reduces the intensity of AIDs. It has been shown that a different Lactobacillus strain, L. murinus regulates the small intestine's RORγt+ Treg cells, which reduces pulmonary inflammation caused by Mycobacterium tuberculosis infection (Bernard-Raichon et al., 2021).
Zhang et al. demonstrated that antibiotic-induced dysbiosis such as ampicillin, decreases Treg cell production and interferes with Th1 responses to infection caused by bacteria (Zhang and Bevan, 2011). Treg cell production and activities can also be affected by modifications in the microbiota, which can be caused by a variety of factors as Figure 4 illustrates, clostridium species are thought to have an effect on Treg cells, whose dysregulation can result in autoimmunity. These studies highlight, certain bacteria, such as B. fragilis and specific strains of Bifidobacterium and Lactobacillus, increase the production and activity of Treg cells, in result, reduce inflammatory responses and disease severity. Furthermore, antibiotics-induced dysbiosis can disrupt the production of Treg cells, that highlights the role healthy microbiota play in immune regulation.
CD8+ T cells
CD8+T cells also referred as cytotoxic T lymphocytes (CTLs), are essential components of the immune system because they specifically target and destroy malignant cells. Shimokawa et al. reported, Nematode-derived trehalose alters the gut microbiota and elevates the number of Ruminococcus species, which stimulates CD8+ T cells (Shimokawa et al., 2020). To enhance the effectiveness of immune-based therapies, there is need to understand how gut microbiota modulation, induced by dietary components like nematode-derived trehalose or probiotics, influences systemic immune responses like CD8+ T cell activation and infiltration of various tissues. A research discovered an association of commensal strains, predominantly Bacteroidetes, that might specifically produce CD8+ T cells unique to the microbiota having anticancer capabilities (Tanoue et al., 2019). Moreover, Bacteroidetes species produces integrase that contains a motif which raised the risk of systemic damage to the pancreas in Type 1 diabetes mellitus (T1DM) while simultaneously increasing CD8+ T cells in the gut (Nanjundappa et al., 2017). In a mice model with altered gut microbiota enriched in Fusobacteria expressed a magnesium transporter (Mgt), that accelerated the development of diabetes. An islet-specific glucose-6-phosphatase catalytic subunit–related protein (IGRP)-mimicking peptide carried by this transporter stimulated the CD8+ T cells specific to IGRP and caused diabetes in vivo (Tai et al., 2016).
Studies have suggested that the major SCFAs, like acetate, butyrate and propionate, which are metabolites of microbes, are involved in the mediation of CD8+ T cell function. A study demonstrated increased acetate levels in MS patients' blood, which were associated with CD8+ T cells that produce IL-17 (Pérez-Pérez et al., 2020). It is worth noting, the diet high in acetate and lesser butyrate level in non-obese diabetic (NOD) mice model of T1DM decreased the frequency of autoreactive CD8+ T cells and increased the number of CD4+Foxp3+ Treg cells in the spleen and colon, but there was no change in the peripheral lymph nodes (Mariño et al., 2017). Following this diet plan might decrease the frequency of pathogenic autoreactive T cells in MS and T1DM by shifting the balance toward a more regulatory T cell profile, thereby potentially mitigating disease progression. Through APCs, butyrate and propionate regulate the activation of CD8+ T cell and IL-12 production (Nastasi et al., 2017). According to another study, butyrate directly boosts the activity of CD8+ T cell by enhancing the expression of granzyme B and IFNγ (Luu et al., 2018). Thus, microbiome-immune axis could reveal new adjuvants for the treatment of certain AIDs, tailored to individual microbiota profiles.
IFNγ based CD8+ T cell induction might be linked with particular metabolites produced by bacterial strains, such as dimethylglycine, mevalonate and SCFAs (Tanoue et al., 2019; Bachem et al., 2019), which enter the bloodstream and cause CD8+ T cell systemic activation. As a result, the microbiota can influence the effectiveness of immunotherapy and CD8+ T cell function. Butyrate from Lachnospiraceae species was found to inhibit CD8+ T cells that secreted IFNγ. Butyrate inhibited DCs based stimulation of the IFN gene (STING), that is linked to responses of CD8+ T cell and reduces the effectiveness of radiation therapy (Yang et al., 2021). Pentanoate produced by Megasphaera malasiliensis stimulates the activity of effector CD8+ T cells. Higher levels of TNFα and IFNγ were found in the presence of M. massiliensis, and this had a favorable impact on adoptive T cell therapy's effectiveness (Luu et al., 2021). Future studies should focus on enhancing the efficacy of adoptive T cell therapies for controlling CD8+ T cell responses that may be done by engineering microbes to produce beneficial metabolites, such as acetate, butyrate and pentanoate.
Natural killer T cells (NKT)
NKT cells, resembles conventional T cells and can be influenced by the microbiota and have a role in autoimmune diseases (Simoni et al., 2013). NKT cells can sense the surrounding lipid environment by engaging their TCR with CD1-expressing cells, as opposed to CD4+ T cells' ability to sense protein antigens (Cheng et al., 2024). According to studies, gram-negative bacteria like Sphingomonas act as NKT cell stimulators, and commensal microbiota regulate the homeostasis of NKT cells. As microbial antigens, the glycosphingolipids and glycosylceramides derived from Sphingomonas induce NKT cell activation and secretion of IFNγ (Kinjo et al., 2005; Mattner et al., 2005). Moreover, 40–70% of the membrane phospholipids in the Bacteroides genus contain sphingolipids, which CD1d may present to NKT cells, the bacteria of this genus may be essential for controlling the population of NKT cells. B. fragilis synthesize an isoform of αGalCer. In a CD1d-dependent manner, this sphingolipid has been demonstrated to activate and stimulate the production of IFN-γ in both human and mouse NKT cells (Brown et al., 2021; Brailey et al., 2020). This approach may improve pathogen clearance and immune surveillance in chronic infections like cancer, with dosage and glycosphingolipid combinations personalized based on individual microbiome profiles.
A study reported that the commensal bacteria decreases the accumulation of mucosal NKT cells. The frequency and quantity of colon NKT cells were found to be higher in germfree mice compared to SPF mice, indicating a possible link between allergic asthma morbidity and IBD (Olszak et al., 2012). In neonatal mice, sphingolipid generated by B. fragilis suppresses NKT cell proliferation, regulating homeostasis (An et al., 2014). Pneumolysoid and type-3-polysaccharide (T3P) produced by Streptococcus pneumoniae stimulate Tregs, which reduce airway hyperreactivity dependent on NKT cell (Thorburn et al., 2012). Microbial bile acids regulate the accumulation of hepatic NKT cell through mediating CXCL16 (CXC-motif ligand 16) expression. Liver sinusoidal endothelial cells express more CXCL16 when exposed to bile acids modified by Clostridium species. Additionally, hepatic NKT cell recruitment has demonstrated remarkable anti-tumor responses against tumors of EL4 lymphoma (Ma et al., 2018). Therefore, it will be crucial to look into whether changes in microbiome of AIDs patients influence the pathophysiology of the disease by regulating NKT cells.
γδ T cells
γδ T cells are known to have semi-invariant TCRs consisting of γ and δ chains, and they are connected to many homeostatic functions such as wound healing and immunological monitoring (Ribot et al., 2021). In various disease models, γδ T cells play an important role as early responders, and frequently they produce the first cytokines like IFN-γ and IL-17 (Ribot et al., 2021; Chien et al., 2014). Although the amount of intestinal γδ intraepithelial lymphocytes (IELs) appears to be unaffected by the gut microbiota, some commensals can raise the frequency of γδ T cells that are positive for IL-17 and IL-1R1 by signaling via VAV1 (a guanine nucleotide exchange factor), which may provide protection against illness (Duan et al., 2010). Thus, there is need to investigate the type of commensals that increases IL-17 and IL-1R1-positive γδ T cells as it may result in the development of innovative probiotics for immune homeostasis. Moreover, the homeostasis of γδ T cells, which produce IL-17A, is also facilitated in the liver by the commensal microbiota. This disturbs the activation of these cells and their secretion of IL-17 cytokines, which in turn affects the advancement of NAFLD (Li et al., 2017a).
Li et al. (2017b) findings showed that via dose-dependent manner hepatic γδ T cell restoration was done by Escherichia coli. Additionally, immune surveillance was restored in mice treated with antibiotics when IL-17A and γδ T cells were added (Cheng et al., 2014). A study determined that microbial metabolite, propionate works as a major component in regulating γδ T cells based release of IL-17 (Dupraz et al., 2021). In the colon, Lactobacillus breves DM9218 expressed TLR2, which directly stimulated γδ T cells and had positive effects on colitis (Li et al., 2018). Certain beneficial bacteria, like Bacillus spp. and Bifidobacterium, increased TLR2 expression, which improved barrier functions via γδ T cells (Li et al., 2017b). Indeed, certain gut commensals can enhance IL-17 and IL-1R1-positive γδ T cells, that suggests using targeted probiotics might enhance immune defense. More specific insights related to these strains can lead to novel treatments for conditions like NAFLD and colitis by upregulating beneficial T cell activity. Table 2 presents the types of microbiota or metabolites involved in the manipulation of T cell subtypes functions and their effects.
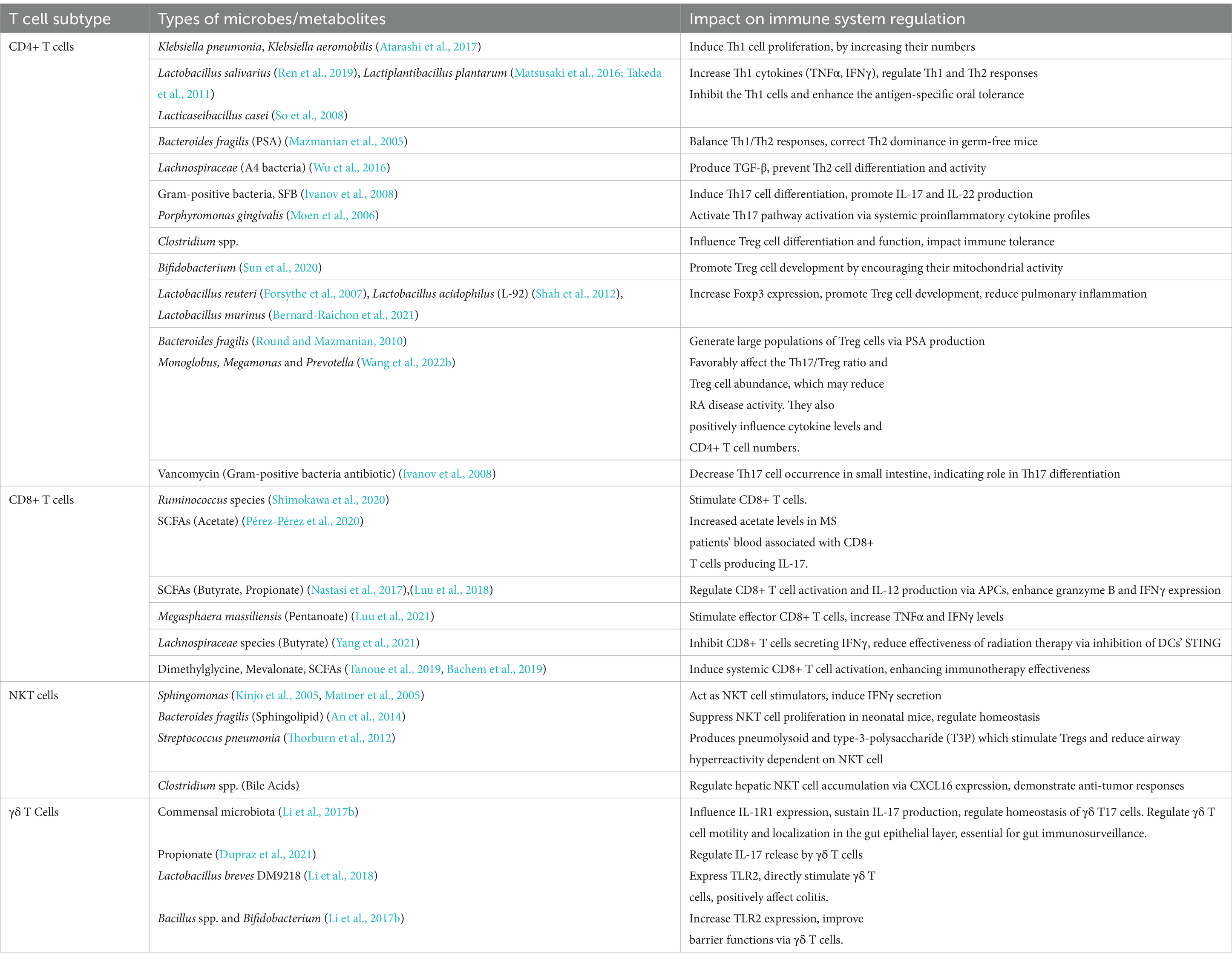
Table 2. Gut microbiota/metabolites mediated manipulation of T cell subtypes function and their outcomes.
Disease specific composition and functionality of different gut microbial species in major AIDs
Environmental factors and genetic predispositions are both known to enhance the development of AIDs, along with a disturbed gut microbiota emerging as a major area of interest. Research has indicated that several AIDs are influenced with deviations in the gut microbiota's composition and functionality. There is mounting evidence to suggest that the immunopathogenesis of AIDs is influenced by these disruptions in the gut microbiota. In the GMrepo database, we compared healthy phenotypes with six AIDs phenotypes (RA, T1DM, T2DM, Asthma, IBD, NAFLD) and identified gut microbiota significantly associated with the six AIDs, characterized by LDA scores. The higher the score, the more significant the relationship between gut microbiota and certain AIDs (Figure 5) (Wu et al., 2020).
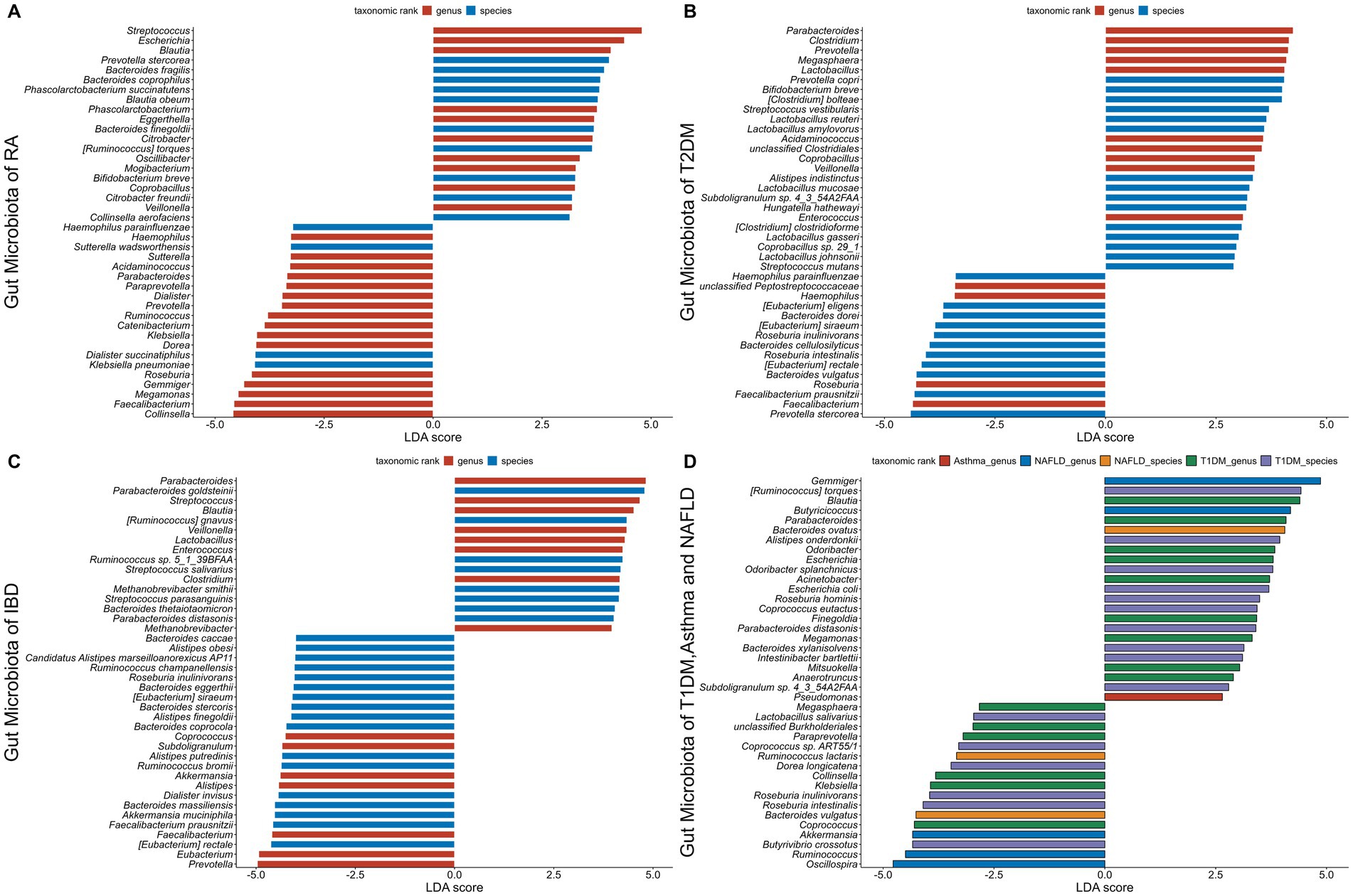
Figure 5. The gut microbiota associated with six autoimmune diseases (RA, T1DM, T2DM, Asthma, IBD, NAFLD). The LDA scores less than 0 indicate Health enriched taxa, while LDA scores larger than 0 indicate autoimmune diseases enriched taxa. The length of the bar chart represents the significance of differential phenotypes. (A–C) show the top 40 gut microbiota with the highest LDA scores for RA, T2DM, and IBD, respectively. (D) shows the gut microbiota with LDA scores greater than 0 for T1DM, asthma, and NAFLD. Nr. samples: In GMrepo, manually curation was performed for selected NCBI BioProjects in order to group samples according to their corresponding phenotypes to obtain Nr. samples, and identify marker taxa between a pair of phenotypes of interests, e.g., RA vs. Health. The total Nr. sample size involved in figure is Rheumatoid arthritis (RA,N = 233;HC,N = 174), Type 1 diabetes mellitus (T1DM,N = 113;HC,N = 153), Type 2 diabetes mellitus (T2DM,N = 240;HC,N = 264), Asthma (asthma,N = 145;HC,N = 1,451), Inflammatory bowel disease (IBD,N = 241;HC,N = 126), Non-alcoholic fatty liver disease (NAFLD,N = 81;HC,N = 62).
Rheumatoid arthritis (RA)
RA is a type of chronic AID that destroys joints and impairs its function. Recently, certain genetic factors along with gut dysbiosis has been proposed as the pathogenesis of RA (Maeda et al., 2016; Jubair et al., 2018). Microbiome research on germfree mice highlights the importance of instability in gut microbiota composition for the etiology of arthritis (Scher et al., 2013). Patients having RA exhibit markedly altered compositions of gut microbiota, with a decrease in Faecalibacterium, a beneficial microbe, and an increase in Prevotella species such as Prevotella copri (Wells et al., 2020). In a mouse model of arthritis, oral exposure to Prevotella nigrescens and Porphyromonas gingivalis exacerbated the inflammation by inducing IL-17 production by the immune system and promoting a Th17 cell response (de Aquino et al., 2014). In addition, Collinsella has been linked to cause severe arthritis in mouse models by promoting the expression of IL-17A and thereby increasing gut permeability. Collinsella has been found to be elevated in RA patients (Chen et al., 2016a). In early stages of RA, the patients have a dominant gut microbiota that includes Collinsella (Koh et al., 2023) and Prevotella copri (Zhang et al., 2015), indicating their potential roles in the etiology of disease. P. gingivalis has been found in the serum of RA patients as well as those who are at verge of developing RA (Mikuls et al., 2012).
The link between RA and SCFAs in the diet has been brought to light by many researches. Because of their function as histone deacetylase inhibitors, SCFAs especially butyrate, suppress the inflammation in RA (Chen et al., 2016b; Maslowski et al., 2009). Research has demonstrated that butyrate inhibits RA by suppressing the inflammatory cytokines secretion and by regulating Treg cells (Takahashi et al., 2020; Yang et al., 2024). By blocking histone deacetylase (HDAC)2 activity in osteoclasts and HDAC8 activity in T cells, butyrate decreased the inflammation in a CIA mice model, which in turn decreased the joint inflammation (Kim et al., 2018). In addition, in RA models, mice lacking SCFA receptors show increased inflammation (Maslowski et al., 2009). The influence of gut microbiota on RA is indicated by the presence of species such as Porphyromonas and the decrease in beneficial microbe like Faecalibacterium. Additionally, SCFAs such as butyrate have anti-inflammatory properties; this implies that dietary changes may help control RA by influencing immunological responses and gut health.
Type 1 diabetes mellitus (T1DM)
T1DM is a type of AIDs, caused when T lymphocytes attacks and destroys insulin-producing β-cells of pancreas. The link between T1DM and the gut microbiota is evident by the reduced numbers of intestinal Treg cells along with healthy microbiota in individuals suffering with T1DM (Anderson, 2023). Studies conducted on humans from a variety of ethnic backgrounds have consistently reported changes in gut microbiota in relation to T1DM (Kostic et al., 2015; Needell and Zipris, 2016; Mejía-León et al., 2014), marked by an increase in Bacteroides species and a decrease in the amount of bacteria that produce SCFA, namely F. prausnitzii (de Goffau et al., 2014; de Goffau et al., 2013; Chukhlovin et al., 2023). Children with T1DM, had lower levels of members of Clostridium clusters IV and XIVa (de Goffau et al., 2013). Moreover, the individuals with T1DM have been found to have decreased expression of intestinal FOXP3, an important transcription factor for the Treg cells activation (Badami et al., 2011). Additional data suggests that microbial diversity was lower (Kostic et al., 2015) and intestinal permeability was higher (Maffeis et al., 2016) prior to the T1DM diagnosis.
In a recent research, when LEfSe analysis was performed, 28 bacterial taxonomic clades showed statistically major alterations (13 elevated and 15 reduced) in T1DM patients in comparison to healthy controls. Porphyromonadaceae, a family of the Bacteroidetes phylum, was overrepresented in T1DM patients, but Paenibacillaceae, Veillonellaceae, Ruminococcaceae, and Phascolarctobacterium, all belonging to the Firmicutes, were most abundant in healthy individuals. Furthermore, the phylum Fusobacteria was differentially enriched in healthy participants (Abuqwider et al., 2023).
Studies on animals using NOD mouse models have clarified the SCFAs protective applications against T1DM. For instance, NOD mice of T1DM given specific diets that increased the amount of acetate and butyrate released by the bacteria which resulted in higher number of Treg cells and lower number of autoreactive T cells and protected them against the development of T1DM (Mariño et al., 2017). Furthermore, in a recent study conducted on T1DM model of NOD mice reported, decrease in the number of SCFAs (Wang et al., 2023b). Interestingly, gut permeability is shown to be a critical mediator between intestinal microbiota, inflammation, and the onset of T1DM, with implications for both human and animal models (Vaarala et al., 2008). Therefore, it is logical to hypothesize that SCFAs could preserve the integrity of the gut barrier by modifying the gut microbiota, encouraging tight junctions and thickening of the mucus layer, which would prevent the onset of T1DM.
Type 2 diabetes mellitus (T2DM)
The symptoms of T2DM include insufficient insulin secretion, increased insulin sensitivity and hepatic glucose production. According to recent studies, dysbiosis may have played a role in the development of T2DM (Arora et al., 2021). Numerous studies examining the gut microbiota of T2DM patients have discovered significant genus-level variations between the patients and healthy controls. These results suggest a connection between diabetes and modifications in the gut microbiota. Frequently reported results indicate that Ruminococcus, Fusobacterium, and Blautia have positive associations with T2DM, while the genera Akkermansia, Bacteroides, Bifidobacterium, Faecalibacterium and Roseburia have negative associations (Larsen et al., 2010; Gurung et al., 2020).
In comparison to healthy controls, the gut microbiota of individuals having T2DM from Northern China had lower levels of Akkermansia and Bifidobacteria species, but higher levels of Dorea (Li et al., 2020). In a metagenome-wide association study, adequate level of gut dysbiosis was found in patients having T2DM. Control samples had higher levels of Lactobacillus spp. and butyrate-producing bacteria, T2DM patients had higher levels of opportunistic pathogens like Clostridium spp. (Qin et al., 2012). Reduced levels of butyrate producing bacteria, which affect insulin sensitivity, are associated with T2DM (Vrieze et al., 2014). A diet that produces acetate and butyrate has been shown to improve gut integrity, stimulate Treg cells, secrete IL-21, and prevent T2DM (Puddu et al., 2014). Through particular G protein receptors (GPR41, GPR43), SCFAs cause intestinal L-cells to secrete GLP-1, which affects insulin release, pancreatic function, and central effects on appetite regulation (Fava, 2014).
In addition to SCFAs, gut microbiome alpha diversity has been linked to other serum metabolites. A microbiome-metabolite score that combined the levels of these metabolites in circulation showed stronger correlations with cardiometabolic traits. Significantly, this score was connected to the incidence and prevalence of T2DM, indicating that metabolites derived from the microbiome play a mechanistic role in the relationship between the composition of the microbiome and health (Menni et al., 2020). While considering the link between dysbiosis of the gut microbiota and T2DM, alteration of gut microbiota may prove to be a useful therapeutic approach. Insulin sensitivity and secretion may be improved by the development of personalized probiotic therapy to increase SCFA-producing bacteria. Moreover, a combined microbiome-metabolite score may also be utilized to mitigate the onset and progression of T2DM by enabling early diagnosis.
Atopic asthma
Reduced metabolic capacity and early-life dysbiosis of the gut microbiota can impair pulmonary and local immunity, making individuals more vulnerable to lung disorders like atopic asthma (Zhao et al., 2023). Microbes in the environment may have an impact on asthma risk even before birth. Numerous studies have demonstrated that exposure of mother to a farming environment protects her offspring from allergic reactions, such as asthma (Roduit et al., 2011; Loss et al., 2012; Ver Heul et al., 2019). Animal models used in experiments, especially newborn mice given antibiotics, showed reduced diversity of gut microbes, altered metabolite profiles, elevated response of immune cells and greater vulnerability to allergic inflammation of lungs (Russell et al., 2012; Cait et al., 2018). Moreover, the inflammation was reduced in these mice models when SCFAs were added in their diet. The mechanism for this improvement was linked to lower levels of circulating IgE and immune--modulating markers like T cells and IL-4-producing CD4+ T cells (Cait et al., 2018).
The relationship between the gut microbiota and asthma was investigated in a clinical research involving 58 patients having asthma and healthy controls. The findings showed a favorable relationship of the relative abundance of Bifidobacterium and Lachnospiraceae family with asthma. On the other hand, asthmatic participants had a decrease in the relative abundance of Bacteroides and Enterobacteriaceae family (Begley et al., 2018). Moreover, Proteobacteria was found to be the predominant phylum overrepresented in human observational studies (Wang et al., 2022a), which connected pro-inflammatory mechanisms to the pathogenesis and severity of asthma. Bifidobacterium administration has been demonstrated to increase IL-10–producing Treg cells, that aid in suppressing over reactive immune responses. Individuals with asthma who received a Bifidobacterium mixture in a randomized controlled experiment, reported better quality of life and clinical symptoms than those who received a placebo (Miraglia Del Giudice et al., 2017).
In human airway inflammation, SCFAs, known to be protective bacterial metabolites showed anti-inflammatory effects. The two most common bacterial phyla that produce SCFAs are Firmicutes and Bacteroidetes, and both are capable of producing acetate. In Firmicutes, (Coprococcus, Clostridium, and Ruminococcus) are the primary producers of butyrate. Bacteroidetes like Prevotella produce propionate (Zhao et al., 2023). Soluble fiber demonstrated anti-inflammatory properties through the binding of SCFAs to related G-protein-coupled receptors (GPCRs) (McLoughlin et al., 2019). Research also indicates the production of pro- and anti-inflammatory metabolites like histamine (Pugin et al., 2017) and oxylipins like 12,13-diHOME (Stewart, 2019) by gut bacteria, pointing a complex interaction between gut microbiota and asthma. Dietary fiber can be used by gut microbes to produce SCFAs, in order to control immunological response of the host (Silva and Bernardi, 2020). By modifying the development and activity of immune cells, SCFAs may be crucial in controlling asthma (Niu et al., 2023). The associated mechanisms that facilitate networking between the lungs and gut are still uncertain. More specifically, the pro-inflammatory responses in the lungs are inhibited by SCFAs derived from gut bacteria. Therefore, targeted focus on fostering SCFA-producing bacteria like Bifidobacterium and Prevotella might be a potential way to decrease asthma risk by promoting anti-inflammatory effects in the respiratory tract.
Inflammatory bowel disease (IBD)
IBD is a multifactorial disease, caused by environmental factors, genetic factors, and immune-mediated factors caused by microbiota and it consists of Crohn's disease (CD) and ulcerative colitis (UC) (Diez-Martin et al., 2024). Rather than a single causative organism, several microbes dysbiosis is linked to the onset of IBD (Lepage et al., 2011). Several studies show that gut microbes, which differ in composition and functionality from healthy controls, play a critical role in the manifestation of IBD (Thursby and Juge, 2017; Pandey et al., 2024). B. longum Bar 33 and L. acidophilus Bar 13 decrease the quantity of intraepithelial lymphocytes and increase the growth of Treg cells in mice models of intestinal inflammation caused by 2,4,6-trinitrobenzene sulphonic acid-induced colitis (Roselli et al., 2009). By stimulating and growing colonic CD4+ FoxP3+ Treg cells, L. casei DN-114 001 reduces the severity of the disease in a mouse colitis paradigm (2,4-dinitrobenzene sulphonic acid) (Hacini-Rachinel et al., 2009). Recently, a study on mice colonized with B. fragilis and a healthy human microbiota, highlighted that colitis induction resulted in a decrease in PSA in the "ON" orientation, which was reversed when inflammation decreased (Carasso et al., 2024). Similarly, a study employing colitis-prone mice shown that Enterobacter ludwigii was successful in reducing colitis symptoms. When compared to other antibiotic treatments, metronidazole had the greatest effect in reducing colitis in a colitis prone mouse model caused by dextran sulfate sodium (DSS). This effect was linked to an increase in the abundance of the gut microbiota species E. ludwigii. By using metabolites from E. ludwigii to stimulate Treg cells differentiation, the immunological tolerance response was boosted and mice's susceptibility to DSS-induced colitis was decreased (Li et al., 2022a).
Reduced microbial diversity, particularly a decline in Firmicutes and an up rise in Proteobacteria taxa, is indicative of IBD-associated dysbiosis. Reductions in the number of Firmicutes bacteria from the Lachnospiraceae and Ruminococcaceae families (Matsuoka and Kanai, 2015; Halfvarson et al., 2017), which are essential for the synthesis of butyrate, are frequently observed in active CD. Decreased butyrate production capacity is one of the functional disturbances associated with this depletion (Marchesi et al., 2007). Researches have reported reduced abundance of Blautia faecis, Clostridium lavalense, Roseburia inulinivorans and Ruminococcus torques in individuals having CD (Takahashi et al., 2016; Pandey et al., 2024). Findings in the ileal CD microbiota showed a reduced number of genes involved in the SCFAs synthesis, along with a decline in the butyrate-producing bacteria F. prausnitzii and Roseburia sp. (Liu et al., 2021).
Butyrate is known to treat IBD due to its ability to support colonocyte energy, inhibiting inflammation and improving the integrity of the epithelial barrier. A few studies applied a different strategy that uses probiotics to uplift in situ butyrate production by consuming butyrate-producing bacteria (Miquel et al., 2013; Tamanai-Shacoori et al., 2017). In a recent study, mice were protected against sorbitol-induced diarrhea by inoculation with butyrate-producing Anaerostipes caccae, even after the elimination of probiotic, which returned the abundance of Clostridia back to normal (Lee et al., 2024). This strategy suggests that focusing on microbial dysbiosis through supplementation of bacterial species which produce butyrate may be helpful in reestablishing the gut homeostasis and improving health of IBD patients.
Non-alcoholic fatty liver disease (NAFLD)
A rising global health concern, NAFLD is associated with an increased risk of cancer and liver disease. In this disease, there is excessive buildup of fat in the liver which is not linked with consumption of alcohol (Tang et al., 2024). Through a variety of immunological, epigenetic and metabolic processes, intestinal dysbiosis is a major contributor of NAFLD (Chen et al., 2020; Wolfe et al., 2023). The gut microbiota profile in individuals having NAFLD is strongly associated with systemic inflammation and can coexist with the development of hepatocellular carcinoma (Ponziani et al., 2019). Research indicates that in both NAFLD and non-alcoholic steatohepatitis (NASH), there is a reduction in alpha and beta diversities accompanied by modified microbial signatures (Caussy et al., 2019). These signatures include a rise in Enterobacteriaceae, Proteobacteria and genera like Collinsella, Dorea, Escherichia, and a decline in Coprococcus, Faecalibacterium, Eubacterium and Prevotella (Hoyles et al., 2018; Boursier et al., 2016). Despite the fact that these preliminary findings point to a detectable difference in microbial profiles between hepatic steatosis patients and controls, significant differences are recorded between studies with opposing findings in the literature. P. copri was found to be linked with a higher progression risk in both NAFLD and NASH, that may have connection with reduced capacity of butyrate production and elevated intestinal permeability (Moran-Ramos et al., 2023). Moreover, in the NASH, there is a reduction in the levels of the butyrate producer F. prausnitzii, a common microbial signature linked to other metabolic diseases (Caussy et al., 2019; Iebba et al., 2018).
Furthermore, it is possible that the increased Lactobacillus presence increases the amount of SCFAs in the gut, which have been demonstrated to reduce fat accumulation (Hou et al., 2022). According to another study, yinchen linggui zhugan decoction (YLZD) improved NAFLD treatment and raised Christensenellaceae abundance (Jiang et al., 2022). As for now, it is unknown how Christensenellaceae affects NAFLD. It is possible therefore, that this association is related to the improvement of fat metabolism (Tavella et al., 2021). There is a documented inverse relationship between insulin resistance and Christensenellaceae abundance. There may be a connection between Christensenellaceae and the onset of NAFLD because lower quantity of this bacterium is linked to more severe insulin resistance and consequent fat storage (Chen et al., 2021b). Through different mechanisms, the gut microbiota is linked to the onset and progression of NAFLD. When bacteria, like Collinsella sp. metabolize bile acids into oxo-bile acid intermediates, it can lead to increased intestinal permeability, which can exacerbate NAFLD (Doden et al., 2018; Purohit et al., 2024).
A large number of toxic metabolites that cause liver fibrosis and inflammation may be accessible to the liver due to dysbiosis and increased intestinal permeability. To shield liver cells from harmful or toxic metabolites the gut mucosal barriers must remain intact (Park et al., 2022). A. muciniphila not only improves the gut barrier integrity but also boosts Treg cell numbers and inhibits pro-inflammatory Th17 responses, both of which can help to prevent or lessen NAFLD condition (Nian et al., 2023). It is interesting to note that although butyrate and propionate were more common in mild to moderate NAFLD, the SCFAs especially acetate, were enriched in advanced NAFLD stages, suggesting different roles in disease severity (Aoki et al., 2021). Thus, the modulation of NAFLD progression is influenced by specific microbial metabolites that alter host metabolic and immune pathways. Moreover, more production of oxo-bile acids and SCFAs, by bacteria such as Collinsella sp. and Lactobacillus impact intestinal permeability and systemic inflammation. Targeting these microbial metabolic pathways could provide novel therapeutic strategies and personalized medicine methods for the control and therapy of NAFLD. Table 3 highlights the associations between the changes in gut microbiota, the role of SCFAs, and the mechanisms that leads to the development and progression of each autoimmune disease mentioned in this section.
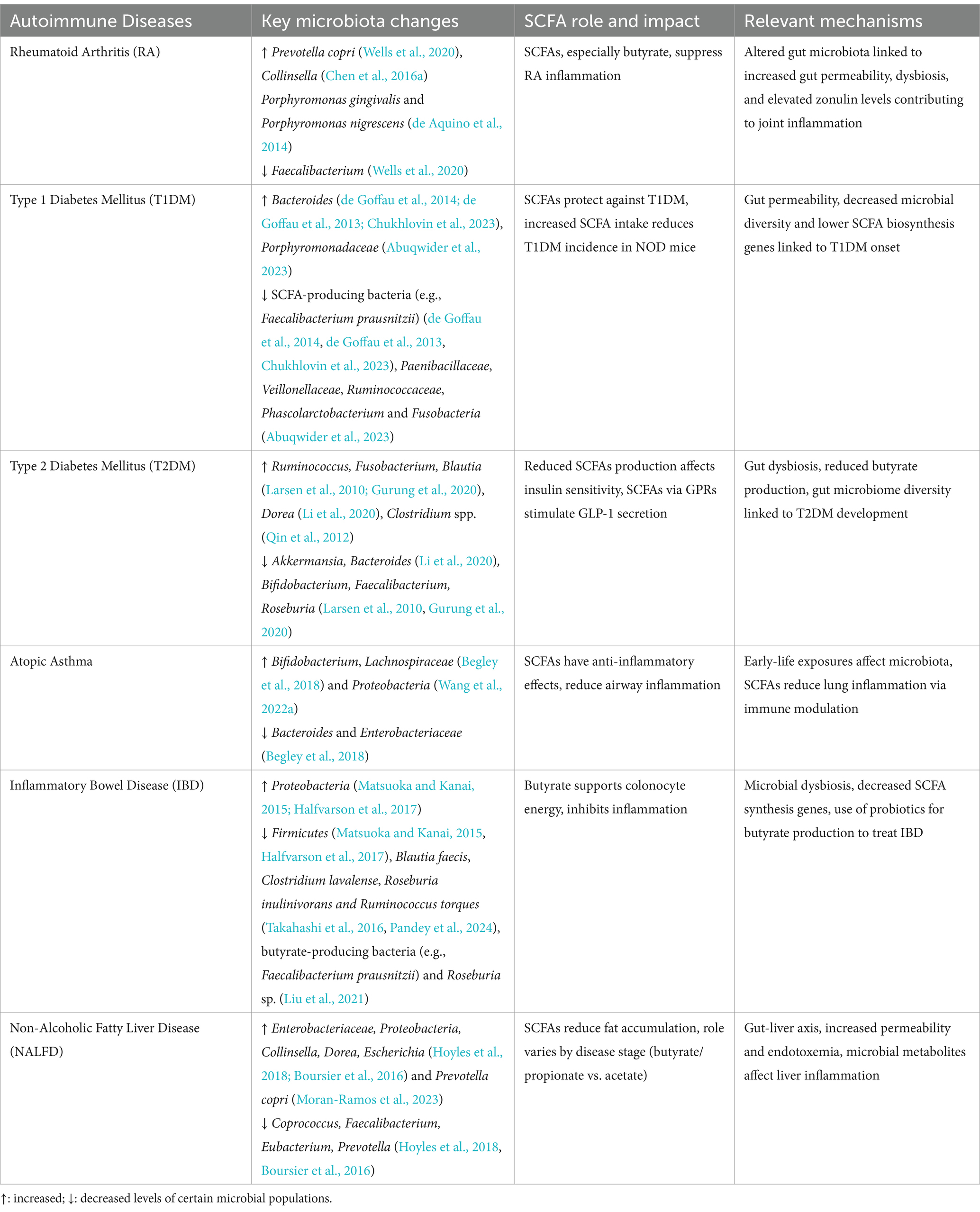
Table 3. The associations between gut microbiota changes, the role of SCFAs, and the mechanisms contributing to the development each autoimmune disease.
Conclusion and future perspectives
The etiology of autoimmune disorders is linked to gut dysbiosis. Therefore, in order to gain a deeper understanding of the effects of gut dysbiosis, we found the altered gut bacteria that were common in AIDs mentioned in this article and what effects they and their metabolites bring on immune system. It is interesting to note that the production of autoantibodies or the Th17 cells activation in reports of immune-related diseases are partly correlated with the common, changed gut bacteria that are abundant in AIDs. Notably, Beneficial gut microbiota strains like F. prausnitzii, A. muciniphila, and Roseburia sp. secrete immunomodulatory metabolites such as SCFAs, which support Treg cells production, while suppressing the inflammatory pathways in AIDs such as SLE, pSS, SpA, AS, Lupus nephritis, RA, IBD, T1DM, T2DM, Atopic asthma and NAFLD. Conversely, pathogenic strains, including P. copri and R. gnavus, disrupt gut barrier integrity by promoting pro-inflammatory Th17 polarization, and exacerbate immune dysregulation, contributing to disease progression.
The gut microbiota alterations have disease-specific implications. For example, in RA disease increased P. copri and decreased SCFA-producing bacteria correlate with heightened inflammation and disease severity. In SLE, elevated R. gnavus levels are associated with lupus nephritis, whereas beneficial species like Bacteroides sp. are depleted. IBD is marked by a loss of butyrate-producing bacteria like Roseburia and overrepresentation of pro-inflammatory Proteobacteria, contributing to chronic intestinal inflammation. It is acknowledged that different strains within the same species might differ significantly in terms of their pathogenicity and functionality (Conway and Cohen, 2015). For example, B. fragilis strains that are enterotoxigenic can stimulate Th17 responses through their enterotoxin (Wick et al., 2014), while non-toxigenic strains can trigger Treg cell responses through their capsular polysaccharide A (Wang et al., 2014). Therefore, it is worth understanding that the relationship between the gut microbiota and host immune system is so complicated and understanding the mechanism by which gut pathogens mediates immune-related disorders might be the solution to treat AIDs.
In this review, the relationship of the gut microbiota with immune system is covered along with the role of gut microbiota in certain AIDs. The pathogenic microbiota can induce Th1 and Th17 polarization, which can activate pro-inflammatory, self-reactive T cells that leads to onset of AIDs; Moreover, in AIDs, there is reduction in healthy microbiota that can provide regulatory, anti-inflammatory metabolites which promotes Treg induction. Restoring a healthy microbiota using targeted therapies holds promising avenue to manage AIDs. For example, probiotic treatments with Bifidobacterium and Lactobacillus strains can reduce the inflammation and increase Treg cells production. In addition, diet plan modification to produce SCFAs, like providing fiber-rich diet or supplementation with SCFA-producing bacteria has shown to decrease the disease severity. Fecal microbiota transplantation (FMT) has shown efficacy in mitigating the AIDs like IBD. Establishing causal relationships between specific microbial strains, their metabolites and AIDs through longitudinal studies like multi-omics approaches, including metagenomics, metabolomics, and transcriptomics should be the priority of future studies. Moreover, understanding every gene, metabolite, and protein in the microbiome that underlies intricate relationships with the immunological activities discussed here is still in its early stages. We have just begun to comprehend these relationships, and understanding every aspect of AIDs requires more comprehensive research. Above all, exploring this frontier in microbiota research not only promises to highlight the underpinnings of immune dysregulation but also to revolutionize the treatment paradigms for autoimmune diseases in future.
Author contributions
NB: Conceptualization, Writing – original draft, Writing – review & editing. XX: Formal analysis, Software, Writing – review & editing. CJ: Formal analysis, Visualization, Writing – review & editing. YW: Formal analysis, Software, Writing – review & editing. YL: Visualization, Writing – review & editing. JS: Writing – review & editing. BH: Conceptualization, Supervision, Writing – review & editing. SW: Resources, Writing – review & editing. AJ: Writing – review & editing, Visualization.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by Development and Application of Fully Automated Immunoassay System and Dust Mite Component Specific Antibody Detection Reagent (Protein Chip Method) (22040061-E); Postdoctoral Research Startup Funding from Zhejiang Sci-Tech University (22042342-Y); and The Cooperation Agreement between Zhejiang Sci-Tech-University and Hangzhou Zheda Dixun Biological Gene: Engineering Co, Ltd Anti-Allergy Functional Molecular Laboratory (agreement number 21040692-J).
Conflict of interest
YL and SW were employed by the Hangzhou Zheda Dixun Biological Gene Engineering Co., Ltd. The authors declare that this study received funding from Hangzhou Zheda Dixun Biological Gene Engineering Co, Ltd., Anti-Allergy Functional Molecular Laboratory. The funder had the following involvement in the study: reviewing and editing of the manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1477187/full#supplementary-material
Abbreviations
AIDs Autoimmune diseases RA Rheumatoid Arthritis IBD Inflammatory Bowel Disease NAFLD Non-alcoholic fatty liver disease T1DM Type 1 Diabetes Mellitus T2DM Type 2 Diabetes Mellitus GALT Gut-associated lymphoid tissue DCs Dendritic cells Th cells T helper cells T-reg cells T-regulatory cells APCs Antigen-presenting cells Tfh cells T follicular helper cells SPF Specific pathogen-free SCFAs Short-chain fatty acids SpA Spondyloarthritis SLE Systemic lupus erythematosus MS Multiple Sclerosis Pss Primary Sjögren’s syndrome LN Lupus nephritis BMI Body-mass-index AS Ankylosing spondylitis ACPA Anti-citrullinated protein antibody MHC Major histocompatibility complex IL Interleukin LPS Lipopolysaccharide TCR T cell receptor TGF Transforming growth factor TNF Tumor necrosis factor IFNs Type I interferons sIgA Secretory IgA SFB Segmented filamentous bacteria TNFα Tumor necrosis factor-alpha IFNγ Interferon-gamma PSA Polysaccharide A CD Crohn's disease RORγt Retinoid-related orphan receptor gamma t iNKT cells Invariant natural killer T cells CTLA-4 Cytotoxic T lymphocyte antigen-4 STING Stimulator of interferon genes CXCL16 CXC-motif ligand 16 IELs Intraepithelial lymphocytes SAA Serum amyloid A ATP Adenosine triphosphate CIA Collagen-induced arthritis ACD Allergic contact dermatitis NOD Non-obese diabetes GPR G protein receptors GPCR G-protein-coupled receptors UC Ulcerative colitis DSS Dextran sulfate sodium NASH Nonalcoholic steatohepatitis YLZD Yinchen Linggui Zhugan decoction TMAO Trimethylamine N-oxide
References
Abuqwider, J., Corrado, A., Scidà, G., Lupoli, R., Costabile, G., Mauriello, G., et al. (2023). Gut microbiome and blood glucose control in type 1 diabetes: a systematic review. Front. Endocrinol. 14. doi: 10.3389/fendo.2023.1265696
Ahern, P. P., and Maloy, K. J. (2020). Understanding immune–microbiota interactions in the intestine. Immunology 159, 4–14. doi: 10.1111/imm.13150
Alpizar-Rodriguez, D., Lesker, T. R., Gronow, A., Gilbert, B., Raemy, E., Lamacchia, C., et al. (2019). Prevotella copri in individuals at risk for rheumatoid arthritis. Ann. Rheum. Dis. 78, 590–593. doi: 10.1136/annrheumdis-2018-214514
An, D., Oh, S. F., Olszak, T., Neves, J. F., Avci, F. Y., Erturk-Hasdemir, D., et al. (2014). Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 156, 123–133. doi: 10.1016/j.cell.2013.11.042
Ananthakrishnan, A. N., Bernstein, C. N., Iliopoulos, D., Macpherson, A., Neurath, M. F., Ali, R. A. R., et al. (2018). Environmental triggers in IBD: a review of progress and evidence. Nat. Rev. Gastroenterol. Hepatol. 15, 39–49. doi: 10.1038/nrgastro.2017.136
Anderson, G. (2023). Type I diabetes pathoetiology and pathophysiology: roles of the gut microbiome, pancreatic cellular interactions, and the ‘bystander’ activation of memory CD8+ T cells. Int. J. Mol. Sci. 24:3300. doi: 10.3390/ijms24043300
Aoki, R., Onuki, M., Hattori, K., Ito, M., Yamada, T., Kamikado, K., et al. (2021). Commensal microbe-derived acetate suppresses NAFLD/NASH development via hepatic FFAR2 signalling in mice. Microbiome 9:188. doi: 10.1186/s40168-021-01125-7
Araki, Y., and Mimura, T. (2018). Epigenetic basis of autoimmune disorders in humans. Epigenetics Human Dis. 6, 353–385. doi: 10.1016/B978-0-12-812215-0.00012-1
Arora, A., Behl, T., Sehgal, A., Singh, S., Sharma, N., Bhatia, S., et al. (2021). Unravelling the involvement of gut microbiota in type 2 diabetes mellitus. Life Sci. 273:119311. doi: 10.1016/j.lfs.2021.119311
Asquith, M. J., Stauffer, P., Davin, S., Mitchell, C., Lin, P., and Rosenbaum, J. T. (2016). Perturbed mucosal immunity and Dysbiosis accompany clinical disease in a rat model of Spondyloarthritis. Arthritis Rheumatol. 68, 2151–2162. doi: 10.1002/art.39681
Atarashi, K., Suda, W., Luo, C., Kawaguchi, T., Motoo, I., Narushima, S., et al. (2017). Ectopic colonization of oral bacteria in the intestine drives T(H)1 cell induction and inflammation. Science 358, 359–365. doi: 10.1126/science.aan4526
Atarashi, K., Tanoue, T., Ando, M., Kamada, N., Nagano, Y., Narushima, S., et al. (2015). Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163, 367–380. doi: 10.1016/j.cell.2015.08.058
Atarashi, K., Tanoue, T., Shima, T., Imaoka, A., Kuwahara, T., Momose, Y., et al. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341. doi: 10.1126/science.1198469
Azzouz, D., Omarbekova, A., Heguy, A., Schwudke, D., Gisch, N., Rovin, B. H., et al. (2019). Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann. Rheum. Dis. 78, 947–956. doi: 10.1136/annrheumdis-2018-214856
Bachem, A., Makhlouf, C., Binger, K. J., De Souza, D. P., Tull, D., Hochheiser, K., et al. (2019). Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8(+) T cells. Immunity 51, 285–297.e5. doi: 10.1016/j.immuni.2019.06.002
Badami, E., Sorini, C., Coccia, M., Usuelli, V., Molteni, L., Bolla, A. M., et al. (2011). Defective differentiation of regulatory FoxP3+ T cells by small-intestinal dendritic cells in patients with type 1 diabetes. Diabetes 60, 2120–2124. doi: 10.2337/db10-1201
Bamias, G., Okazawa, A., Rivera-Nieves, J., Arseneau, K. O., De La Rue, S. A., Pizarro, T. T., et al. (2007). Commensal bacteria exacerbate intestinal inflammation but are not essential for the development of murine ileitis. J. Immunol. 178, 1809–1818. doi: 10.4049/jimmunol.178.3.1809
Begley, L., Madapoosi, S., Opron, K., Ndum, O., Baptist, A., Rysso, K., et al. (2018). Gut microbiota relationships to lung function and adult asthma phenotype: a pilot study. BMJ Open Respir. Res. 5:e000324. doi: 10.1136/bmjresp-2018-000324
Bellocchi, C., Fernández-Ochoa, Á., Montanelli, G., Vigone, B., Santaniello, A., Quirantes-Piné, R., et al. (2019). Identification of a shared microbiomic and metabolomic profile in systemic autoimmune diseases. J. Clin. Med. 8:1291. doi: 10.3390/jcm8091291
Bernard-Raichon, L., Colom, A., Monard, S. C., Namouchi, A., Cescato, M., Garnier, H., et al. (2021). A pulmonary Lactobacillus murinus strain induces Th17 and RORγt+ regulatory T cells and reduces lung inflammation. Front. Immunol. 12:727474. doi: 10.3389/fimmu.2021.727474
Bevins, C. L., and Salzman, N. H. (2011). Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9, 356–368. doi: 10.1038/nrmicro2546
Boursier, J., Mueller, O., Barret, M., Machado, M., Fizanne, L., Araujo-Perez, F., et al. (2016). The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 63, 764–775. doi: 10.1002/hep.28356
Brailey, P. M., Lebrusant-Fernandez, M., and Barral, P. (2020). NKT cells and the regulation of intestinal immunity: a two-way street. FEBS J. 287, 1686–1699. doi: 10.1111/febs.15238
Brown, L. W., Penaranda, C., and Kashyap, P. C. (2021). Production of α-galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol. 11:e1001610. doi: 10.1371/journal.pbio.1001610
Cait, A., Hughes, M. R., Antignano, F., Cait, J., Dimitriu, P. A., Maas, K. R., et al. (2018). Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 11, 785–795. doi: 10.1038/mi.2017.75
Carasso, S., Zaatry, R., Hajjo, H., Kadosh-Kariti, D., Ben-Assa, N., Naddaf, R., et al. (2024). Inflammation and bacteriophages affect DNA inversion states and functionality of the gut microbiota. Cell Host Microbe 32, 322–334.e9. doi: 10.1016/j.chom.2024.02.003
Caussy, C., Tripathi, A., Humphrey, G., Bassirian, S., Singh, S., Faulkner, C., et al. (2019). A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat. Commun. 10:1406. doi: 10.1038/s41467-019-09455-9
Cekanaviciute, E., Yoo, B. B., Runia, T. F., Debelius, J. W., Singh, S., Nelson, C. A., et al. (2017). Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. USA 114, 10713–10718. doi: 10.1073/pnas.1711235114
Cenit, M. C., Sanz, Y., and Codoñer-Franch, P. (2017). Influence of gut microbiota on neuropsychiatric disorders. World J. Gastroenterol. 23, 5486–5498. doi: 10.3748/wjg.v23.i30.5486
Chen, Z., Andreev, D., Oeser, K., Krljanac, B., Hueber, A., Kleyer, A., et al. (2016b). Th2 and eosinophil responses suppress inflammatory arthritis. Nat. Commun. 7:11596. doi: 10.1038/ncomms11596
Chen, Y., Cui, W., Li, X., and Yang, H. (2021a). Interaction between commensal bacteria, immune response and the intestinal barrier in inflammatory bowel disease. Front. Immunol. 12. doi: 10.3389/fimmu.2021.761981
Chen, H.-T., Huang, H.-L., Li, Y.-Q., Xu, H.-M., and Zhou, Y.-J. (2020). Therapeutic advances in non-alcoholic fatty liver disease: a microbiota-centered view. World J. Gastroenterol. 26, 1901–1911. doi: 10.3748/wjg.v26.i16.1901
Chen, B.-D., Jia, X.-M., Xu, J.-Y., Zhao, L.-D., Ji, J.-Y., Wu, B.-X., et al. (2019). Proinflammatory and autoimmunogenic gut microbiome in systemic lupus erythematosus. bioRxiv :621995. doi: 10.1101/621995
Chen, Z., Radjabzadeh, D., Chen, L., Kurilshikov, A., Kavousi, M., Ahmadizar, F., et al. (2021b). Association of Insulin Resistance and Type 2 diabetes with gut microbial diversity: a microbiome-wide analysis from population studies. JAMA Netw. Open 4:e2118811. doi: 10.1001/jamanetworkopen.2021.18811
Chen, P., and Tang, X. (2021). Gut microbiota as regulators of Th17/Treg balance in patients with myasthenia gravis. Front. Immunol. 12. doi: 10.3389/fimmu.2021.803101
Chen, J., Wright, K., Davis, J. M., Jeraldo, P., Marietta, E. V., and Taneja, V. (2016a). An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 8:43. doi: 10.1186/s13073-016-0299-7
Cheng, T. Y., Praveena, T., Govindarajan, S., Almeida, C. F., Pellicci, D. G., Arkins, W. C., et al. (2024). Lipidomic scanning of self-lipids identifies headless antigens for natural killer T cells. Proc. Natl. Acad. Sci. USA 121:e2321686121. doi: 10.1073/pnas.2321686121
Cheng, M., Qian, L., Shen, G., Bian, G., Xu, T., Xu, W., et al. (2014). Microbiota modulate tumoral immune surveillance in lung through a γδT17 immune cell-dependent mechanism. Cancer Res. 74, 4030–4041. doi: 10.1158/0008-5472.CAN-13-2462
Chiang, H. I., Li, J. R., Liu, C. C., Liu, P. Y., Chen, H. H., Chen, Y. M., et al. (2019). An Association of gut Microbiota with different phenotypes in Chinese patients with rheumatoid arthritis. J. Clin. Med. 8. doi: 10.3390/jcm8111770
Chien, Y.-H., Meyer, C., and Bonneville, M. (2014). γδ T cells: first line of defense and beyond. Annu. Rev. Immunol. 32, 121–155. doi: 10.1146/annurev-immunol-032713-120216
Chukhlovin, A. B., Dudurich, V. V., Kusakin, A. V., Polev, D. E., Ermachenko, E. D., Aseev, M. V., et al. (2023). Evaluation of gut microbiota in healthy persons and type 1 diabetes mellitus patients in North-Western Russia. Microorganisms 11:1813. doi: 10.3390/microorganisms11071813
Conway, T., and Cohen, P. S. (2015). Commensal and pathogenic Escherichia coli metabolism in the gut. Microbiol. Spectr. 3. doi: 10.1128/microbiolspec.MBP-0006-2014
de Aquino, S. G., Abdollahi-Roodsaz, S., Koenders, M. I., van de Loo, F. A., Pruijn, G. J., Marijnissen, R. J., et al. (2014). Periodontal pathogens directly promote autoimmune experimental arthritis by inducing a TLR2-and IL-1–driven Th17 response. J. Immunol. 192, 4103–4111. doi: 10.4049/jimmunol.1301970
de Goffau, M. C., Fuentes, S., Van Den Bogert, B., Honkanen, H., De Vos, W. M., Welling, G. W., et al. (2014). Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia 57, 1569–1577. doi: 10.1007/s00125-014-3274-0
de Goffau, M. C., Luopajärvi, K., Knip, M., Ilonen, J., Ruohtula, T., Härkönen, T., et al. (2013). Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes 62, 1238–1244. doi: 10.2337/db12-0526
Dehner, C., Fine, R., and Kriegel, M. A. (2019). The microbiome in systemic autoimmune disease: mechanistic insights from recent studies. Curr. Opin. Rheumatol. 31, 201–207. doi: 10.1097/BOR.0000000000000574
Diez-Martin, E., Hernandez-Suarez, L., Muñoz-Villafranca, C., Martin-Souto, L., Astigarraga, E., Ramirez-Garcia, A., et al. (2024). Inflammatory bowel disease: a comprehensive analysis of molecular bases, predictive biomarkers, diagnostic methods, and therapeutic options. Int. J. Mol. Sci. 25. doi: 10.3390/ijms25137062
Doden, H., Sallam, L. A., Devendran, S., Ly, L., Doden, G., Daniel, S. L., et al. (2018). Metabolism of Oxo-bile acids and characterization of recombinant 12α-Hydroxysteroid dehydrogenases from bile acid 7α-Dehydroxylating human gut Bacteria. Appl. Environ. Microbiol. 84. doi: 10.1128/aem.00235-18
Dominguez-Villar, M., and Hafler, D. A. (2018). Regulatory T cells in autoimmune disease. Nat. Immunol. 19, 665–673. doi: 10.1038/s41590-018-0120-4
Duan, J., Chung, H., Troy, E., and Kasper, D. L. (2010). Microbial colonization drives expansion of IL-1 receptor 1-expressing and IL-17-producing γ/δ T cells. Cell Host Microbe 7, 140–150. doi: 10.1016/j.chom.2010.01.005
Dupraz, L., Magniez, A., Rolhion, N., Richard, M. L., Da Costa, G., Touch, S., et al. (2021). Gut microbiota-derived short-chain fatty acids regulate IL-17 production by mouse and human intestinal γδ T cells. Cell Rep. 36:109332. doi: 10.1016/j.celrep.2021.109332
Evans-Marin, H., Rogier, R., Koralov, S. B., Manasson, J., Roeleveld, D., Van Der Kraan, P. M., et al. (2018). Microbiota-dependent involvement of Th17 cells in murine models of inflammatory arthritis. Arthritis Rheumatol. 70, 1971–1983. doi: 10.1002/art.40657
Farache, J., Koren, I., Milo, I., Gurevich, I., Kim, K. W., Zigmond, E., et al. (2013). Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity 38, 581–595. doi: 10.1016/j.immuni.2013.01.009
Farkas, A. M., Panea, C., Goto, Y., Nakato, G., Galan-Diez, M., Narushima, S., et al. (2015). Induction of Th17 cells by segmented filamentous bacteria in the murine intestine. J. Immunol. Methods 421, 104–111. doi: 10.1016/j.jim.2015.03.020
Fava, S. (2014). Glucagon-like peptide 1 and the cardiovascular system. Curr. Diabetes Rev. 10, 302–310. doi: 10.2174/1573399810666141030125830
Forsythe, P., Inman, M. D., and Bienenstock, J. (2007). Oral treatment with live Lactobacillus reuteri inhibits the allergic airway response in mice. Am. J. Respir. Crit. Care Med. 175, 561–569. doi: 10.1164/rccm.200606-821OC
Gaboriau-Routhiau, V., Rakotobe, S., Lécuyer, E., Mulder, I., Lan, A., Bridonneau, C., et al. (2009). The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689. doi: 10.1016/j.immuni.2009.08.020
Gurung, M., Li, Z., You, H., Rodrigues, R., Jump, D. B., Morgun, A., et al. (2020). Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 51:102590. doi: 10.1016/j.ebiom.2019.11.051
Hacini-Rachinel, F., Nancey, S., Boschetti, G., Sardi, F., Doucet-Ladeveze, R., Durand, P.-Y., et al. (2009). CD4+ T cells and Lactobacillus casei control relapsing colitis mediated by CD8+ T cells. J. Immunol. 183, 5477–5486. doi: 10.4049/jimmunol.0804267
Halfvarson, J., Brislawn, C. J., Lamendella, R., Vázquez-Baeza, Y., Walters, W. A., Bramer, L. M., et al. (2017). Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2:17004. doi: 10.1038/nmicrobiol.2017.4
Hang, S., Paik, D., Yao, L., Kim, E., Trinath, J., Lu, J., et al. (2019). Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature 576, 143–148. doi: 10.1038/s41586-019-1785-z
Hou, G., Yin, J., Wei, L., Li, R., Peng, W., Yuan, Y., et al. (2022). Lactobacillus delbrueckii might lower serum triglyceride levels via colonic microbiota modulation and SCFA-mediated fat metabolism in parenteral tissues of growing-finishing pigs. Front. Vet. Sci. 9:982349. doi: 10.3389/fvets.2022.982349
Hoyles, L., Fernández-Real, J. M., Federici, M., Serino, M., Abbott, J., Charpentier, J., et al. (2018). Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 24, 1070–1080. doi: 10.1038/s41591-018-0061-3
Hui, W., Yu, D., Cao, Z., and Zhao, X. (2019). Butyrate inhibit collagen-induced arthritis via Treg/IL-10/Th17 axis. Int. Immunopharmacol. 68, 226–233. doi: 10.1016/j.intimp.2019.01.018
Iebba, V., Guerrieri, F., Di Gregorio, V., Levrero, M., Gagliardi, A., Santangelo, F., et al. (2018). Combining amplicon sequencing and metabolomics in cirrhotic patients highlights distinctive microbiota features involved in bacterial translocation, systemic inflammation and hepatic encephalopathy. Sci. Rep. 8:8210. doi: 10.1038/s41598-018-26509-y
Ivanov, I. I., Atarashi, K., Manel, N., Brodie, E. L., Shima, T., Karaoz, U., et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. doi: 10.1016/j.cell.2009.09.033
Ivanov, I. I., Frutos Rde, L., Manel, N., Yoshinaga, K., Rifkin, D. B., Sartor, R. B., et al. (2008). Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349. doi: 10.1016/j.chom.2008.09.009
Jackson, M. A., Verdi, S., Maxan, M. E., Shin, C. M., Zierer, J., Bowyer, R. C. E., et al. (2018). Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat. Commun. 9:2655. doi: 10.1038/s41467-018-05184-7
Jangi, S., Gandhi, R., Cox, L. M., Li, N., Von Glehn, F., Yan, R., et al. (2016). Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 7:12015. doi: 10.1038/ncomms12015
Jiang, H., Mao, T., Sun, Z., Shi, L., Han, X., Zhang, Y., et al. (2022). Yinchen Linggui Zhugan decoction ameliorates high fat diet-induced nonalcoholic fatty liver disease by modulation of SIRT1/Nrf2 signaling pathway and gut microbiota. Front. Microbiol. 13:1001778. doi: 10.3389/fmicb.2022.1001778
Jiao, Y., Wu, L., Huntington, N. D., and Zhang, X. (2020). Crosstalk between gut microbiota and innate immunity and its implication in autoimmune diseases. Front. Immunol. 11:282. doi: 10.3389/fimmu.2020.00282
Jubair, W. K., Hendrickson, J. D., Severs, E. L., Schulz, H. M., Adhikari, S., Ir, D., et al. (2018). Modulation of inflammatory arthritis in mice by gut microbiota through mucosal inflammation and autoantibody generation. Arthritis Rheumatol. 70, 1220–1233. doi: 10.1002/art.40490
Karimi, K., Inman, M. D., Bienenstock, J., and Forsythe, P. (2009). Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am. J. Respir. Crit. Care Med. 179, 186–193. doi: 10.1164/rccm.200806-951OC
Khan, M. F., and Wang, G. (2018). Environmental agents, oxidative stress and autoimmunity. Curr Opin Toxicol 7, 22–27. doi: 10.1016/j.cotox.2017.10.012
Kim, D. S., Kwon, J.-E., Lee, S. H., Kim, E. K., Ryu, J.-G., Jung, K.-A., et al. (2018). Attenuation of rheumatoid inflammation by sodium butyrate through reciprocal targeting of HDAC2 in osteoclasts and HDAC8 in T cells. Front. Immunol. 9:1525. doi: 10.3389/fimmu.2018.01525
Kinjo, Y., Wu, D., Kim, G., Xing, G. W., Poles, M. A., Ho, D. D., et al. (2005). Recognition of bacterial glycosphingolipids by natural killer T cells. Nature 434, 520–525. doi: 10.1038/nature03407
Koh, J. H., Lee, E. H., Cha, K. H., Pan, C.-H., Kim, D., and Kim, W.-U. (2023). Factors associated with the composition of the gut microbiome in patients with established rheumatoid arthritis and its value for predicting treatment responses. Arthritis Res. Ther. 25:32. doi: 10.1186/s13075-023-03013-x
Kostic, A. D., Gevers, D., Siljander, H., Vatanen, T., Hyötyläinen, T., Hämäläinen, A. M., et al. (2015). The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 17, 260–273. doi: 10.1016/j.chom.2015.01.001
Larsen, N., Vogensen, F. K., Van Den Berg, F. W., Nielsen, D. S., Andreasen, A. S., Pedersen, B. K., et al. (2010). Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 5:e9085. doi: 10.1371/journal.pone.0009085
Lee, N., and Kim, W.-U. (2017). Microbiota in T-cell homeostasis and inflammatory diseases. Exp. Mol. Med. 49:e340. doi: 10.1038/emm.2017.36
Lee, J. Y., Tiffany, C. R., Mahan, S. P., Kellom, M., Rogers, A. W. L., Nguyen, H., et al. (2024). High fat intake sustains sorbitol intolerance after antibiotic-mediated Clostridia depletion from the gut microbiota. Cell 187, 1191–1205.e15. doi: 10.1016/j.cell.2024.01.029
Lepage, P., Häsler, R., Spehlmann, M. E., Rehman, A., Zvirbliene, A., Begun, A., et al. (2011). Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 141, 227–236. doi: 10.1053/j.gastro.2011.04.011
Li, Q., Chang, Y., Zhang, K., Chen, H., Tao, S., and Zhang, Z. (2020). Implication of the gut microbiome composition of type 2 diabetic patients from northern China. Sci. Rep. 10:5450. doi: 10.1038/s41598-020-62224-3
Li, M., Gao, J., Tang, Y., Liu, M., Wu, S., Qu, K., et al. (2018). Traditional herbal medicine-derived sulforaphene LFS-01 reverses colitis in mice by selectively altering the gut microbiota and promoting intestinal gamma-delta T cells. Front. Pharmacol. 8:959. doi: 10.3389/fphar.2017.00959
Li, F., Hao, X., Chen, Y., Bai, L., Gao, X., Lian, Z., et al. (2017a). The microbiota maintain homeostasis of liver-resident γδT-17 cells in a lipid antigen/CD1d-dependent manner. Nat. Commun. 8:13839. doi: 10.1038/ncomms13839
Li, Q., Sun, X., Yu, K., Lv, J., Miao, C., Yang, J., et al. (2022a). Enterobacter ludwigii protects DSS-induced colitis through choline-mediated immune tolerance. Cell Rep. 40:111308. doi: 10.1016/j.celrep.2022.111308
Li, Y., Wang, H. F., Li, X., Li, H. X., Zhang, Q., Zhou, H. W., et al. (2019). Disordered intestinal microbes are associated with the activity of systemic lupus erythematosus. Clin. Sci. 133, 821–838. doi: 10.1042/CS20180841
Li, M., Wang, B., Sun, X., Tang, Y., Wei, X., Ge, B., et al. (2017b). Upregulation of intestinal barrier function in mice with DSS-induced colitis by a defined bacterial consortium is associated with expansion of IL-17A producing gamma delta T cells. Front. Immunol. 8:824. doi: 10.3389/fimmu.2017.00824
Li, Y., Ye, Z., Zhu, J., Fang, S., Meng, L., and Zhou, C. (2022b). Effects of gut microbiota on host adaptive immunity under immune homeostasis and tumor pathology state. Front. Immunol. 13. doi: 10.3389/fimmu.2022.844335
Liu, Y., Alookaran, J. J., and Rhoads, J. M. (2018). Probiotics in autoimmune and inflammatory disorders. Nutrients 10:1537. doi: 10.3390/nu10101537
Liu, S., Zhao, W., Lan, P., and Mou, X. (2021). The microbiome in inflammatory bowel diseases: from pathogenesis to therapy. Protein Cell 12, 331–345. doi: 10.1007/s13238-020-00745-3
Liu, X., Zou, Q., Zeng, B., Fang, Y., and Wei, H. (2013). Analysis of fecal Lactobacillus community structure in patients with early rheumatoid arthritis. Curr. Microbiol. 67, 170–176. doi: 10.1007/s00284-013-0338-1
Lloyd-Price, J., Arze, C., Ananthakrishnan, A. N., Schirmer, M., Avila-Pacheco, J., Poon, T. W., et al. (2019). Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662. doi: 10.1038/s41586-019-1237-9
Loss, G., Bitter, S., Wohlgensinger, J., Frei, R., Roduit, C., Genuneit, J., et al. (2012). Prenatal and early-life exposures alter expression of innate immunity genes: the PASTURE cohort study. J. Allergy Clin. Immunol. 130, 523–530.e9. doi: 10.1016/j.jaci.2012.05.049
Luu, M., Pautz, S., Kohl, V., Singh, R., Romero, R., Lucas, S., et al. (2019). The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat. Commun. 10:760. doi: 10.1038/s41467-019-08711-2
Luu, M., Riester, Z., Baldrich, A., Reichardt, N., Yuille, S., Busetti, A., et al. (2021). Microbial short-chain fatty acids modulate CD8(+) T cell responses and improve adoptive immunotherapy for cancer. Nat. Commun. 12:4077. doi: 10.1038/s41467-021-24331-1
Luu, M., Weigand, K., Wedi, F., Breidenbend, C., Leister, H., Pautz, S., et al. (2018). Regulation of the effector function of CD8+ T cells by gut microbiota-derived metabolite butyrate. Sci. Rep. 8:14430. doi: 10.1038/s41598-018-32860-x
Ma, C., Han, M., Heinrich, B., Fu, Q., Zhang, Q., Sandhu, M., et al. (2018). Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360. doi: 10.1126/science.aan5931
Ma, Y., Xu, X., Li, M., Cai, J., Wei, Q., and Niu, H. (2019). Gut microbiota promote the inflammatory response in the pathogenesis of systemic lupus erythematosus. Mol. Med. 25:35. doi: 10.1186/s10020-019-0102-5
Maeda, Y., Kurakawa, T., Umemoto, E., Motooka, D., Ito, Y., Gotoh, K., et al. (2016). Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 68, 2646–2661. doi: 10.1002/art.39783
Maffeis, C., Martina, A., Corradi, M., Quarella, S., Nori, N., Torriani, S., et al. (2016). Association between intestinal permeability and faecal microbiota composition in Italian children with beta cell autoimmunity at risk for type 1 diabetes. Diab. Metabol. Res Rev 32, 700–709. doi: 10.1002/dmrr.2790
Mandl, T., Marsal, J., Olsson, P., and Ohlsson, B. (2017). Severe intestinal dysbiosis is prevalent in primary Sjögren's syndrome and is associated with systemic disease activity. Arthritis Res. Ther. 19:237. doi: 10.1186/s13075-017-1446-2
Manfredo Vieira, S., Hiltensperger, M., Kumar, V., Zegarra-Ruiz, D., Dehner, C., Khan, N., et al. (2018). Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 359, 1156–1161. doi: 10.1126/science.aar7201
Marchesi, J. R., Holmes, E., Khan, F., Kochhar, S., Scanlan, P., Shanahan, F., et al. (2007). Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J. Proteome Res. 6, 546–551. doi: 10.1021/pr060470d
Mariño, E., Richards, J. L., McLeod, K. H., Stanley, D., Yap, Y. A., Knight, J., et al. (2017). Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat. Immunol. 18, 552–562. doi: 10.1038/ni.3713
Maslowski, K. M., Vieira, A. T., Ng, A., Kranich, J., Sierro, F., di Yu,, et al. (2009). Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286. doi: 10.1038/nature08530
Matsuoka, K., and Kanai, T. (2015). The gut microbiota and inflammatory bowel disease. Semin. Immunopathol. 37, 47–55. doi: 10.1007/s00281-014-0454-4
Matsusaki, T., Takeda, S., Takeshita, M., Arima, Y., Tsend-Ayush, C., Oyunsuren, T., et al. (2016). Augmentation of T helper type 1 immune response through intestinal immunity in murine cutaneous herpes simplex virus type 1 infection by probiotic Lactobacillus plantarum strain 06CC2. Int. Immunopharmacol. 39, 320–327. doi: 10.1016/j.intimp.2016.08.001
Mattner, J., Debord, K. L., Ismail, N., Goff, R. D., Cantu, C. III, Zhou, D., et al. (2005). Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature 434, 525–529. doi: 10.1038/nature03408
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O., and Kasper, D. L. (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118. doi: 10.1016/j.cell.2005.05.007
Mcloughlin, R., Berthon, B. S., Rogers, G. B., Baines, K. J., Leong, L. E. X., Gibson, P. G., et al. (2019). Soluble fibre supplementation with and without a probiotic in adults with asthma: a 7-day randomised, double blind, three way cross-over trial. EBioMedicine 46, 473–485. doi: 10.1016/j.ebiom.2019.07.048
Mejía-León, M. E., Petrosino, J. F., Ajami, N. J., Domínguez-Bello, M. G., and De La Barca, A. M. (2014). Fecal microbiota imbalance in Mexican children with type 1 diabetes. Sci. Rep. 4:3814. doi: 10.1038/srep03814
Menni, C., Zhu, J., Le Roy, C. I., Mompeo, O., Young, K., Rebholz, C. M., et al. (2020). Serum metabolites reflecting gut microbiome alpha diversity predict type 2 diabetes. Gut Microbes 11, 1632–1642. doi: 10.1080/19490976.2020.1778261
Mergaert, P. (2018). Role of antimicrobial peptides in controlling symbiotic bacterial populations. Nat. Prod. Rep. 35, 336–356. doi: 10.1039/c7np00056a
Mikuls, T. R., Thiele, G. M., Deane, K. D., Payne, J. B., O'Dell, J. R., Yu, F., et al. (2012). Porphyromonas gingivalis and disease-related autoantibodies in individuals at increased risk of rheumatoid arthritis. Arthritis Rheum. 64, 3522–3530. doi: 10.1002/art.34595
Miquel, S., Martín, R., Rossi, O., Bermúdez-Humarán, L. G., Chatel, J. M., Sokol, H., et al. (2013). Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 16, 255–261. doi: 10.1016/j.mib.2013.06.003
Miraglia Del Giudice, M., Indolfi, C., Capasso, M., Maiello, N., Decimo, F., and Ciprandi, G. (2017). Bifidobacterium mixture (B. longum BB536, B. infantis M-63, B. breve M-16V) treatment in children with seasonal allergic rhinitis and intermittent asthma. Ital. J. Pediatr. 43:25. doi: 10.1186/s13052-017-0340-5
Moen, K., Brun, J. G., Valen, M., Skartveit, L., Eribe, E. K., Olsen, I., et al. (2006). Synovial inflammation in active rheumatoid arthritis and psoriatic arthritis facilitates trapping of a variety of oral bacterial DNAs. Clin. Exp. Rheumatol. 24, 656–663
Moran-Ramos, S., Cerqueda-García, D., López-Contreras, B., Larrieta-Carrasco, E., Villamil-Ramírez, H., Molina-Cruz, S., et al. (2023). A metagenomic study identifies a Prevotella copri enriched microbial profile associated with non-alcoholic steatohepatitis in subjects with obesity. J. Gastroenterol. Hepatol. 38, 791–799. doi: 10.1111/jgh.16147
Mowat, A. M. (2018). To respond or not to respond - a personal perspective of intestinal tolerance. Nat. Rev. Immunol. 18, 405–415. doi: 10.1038/s41577-018-0002-x
Nalbant, A., and Eskier, D. (2016). Genes associated with T helper 17 cell differentiation and function. Frontiers in Bioscience 8, 427–435 doi: 10.2741/e777
Nanjundappa, R. H., Ronchi, F., Wang, J., Clemente-Casares, X., Yamanouchi, J., Umeshappa, C. S., et al. (2017). A gut microbial mimic that hijacks diabetogenic autoreactivity to suppress colitis. Cell 171, 655–667.e17. doi: 10.1016/j.cell.2017.09.022
Nastasi, C., Fredholm, S., Willerslev-Olsen, A., Hansen, M., Bonefeld, C. M., Geisler, C., et al. (2017). Butyrate and propionate inhibit antigen-specific CD8(+) T cell activation by suppressing IL-12 production by antigen-presenting cells. Sci. Rep. 7:14516. doi: 10.1038/s41598-017-15099-w
Needell, J. C., and Zipris, D. (2016). The role of the intestinal microbiome in type 1 diabetes pathogenesis. Curr. Diab. Rep. 16:89. doi: 10.1007/s11892-016-0781-z
Neish, A. S. (2009). Microbes in gastrointestinal health and disease. Gastroenterology. 136, 65–80. doi: 10.1053/j.gastro.2008.10.080
Nian, F., Wu, L., Xia, Q., Tian, P., Ding, C., and Lu, X. (2023). Akkermansia muciniphila and Bifidobacterium bifidum prevent NAFLD by regulating FXR expression and gut microbiota. J. Clin. Transl. Hepatol. 11, 763–776. doi: 10.14218/JCTH.2022.00415
Niu, J., Cui, M., Yang, X., Li, J., Yao, Y., Guo, Q., et al. (2023). Microbiota-derived acetate enhances host antiviral response via NLRP3. Nat. Commun. 14:642. doi: 10.1038/s41467-023-36323-4
Olszak, T., An, D., Zeissig, S., Vera, M. P., Richter, J., Franke, A., et al. (2012). Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493. doi: 10.1126/science.1219328
Pandey, H., Jain, D., Tang, D. W. T., Wong, S. H., and Lal, D. (2024). Gut microbiota in pathophysiology, diagnosis, and therapeutics of inflammatory bowel disease. Intest Res 22, 15–43. doi: 10.5217/ir.2023.00080
Park, J.-W., Kim, S.-E., Lee, N. Y., Kim, J.-H., Jung, J.-H., Jang, M.-K., et al. (2022). Role of microbiota-derived metabolites in alcoholic and non-alcoholic fatty liver diseases. Int. J. Mol. Sci. 23:426. doi: 10.3390/ijms23010426
Pérez-Pérez, S., Domínguez-Mozo, M. I., Alonso-Gómez, A., Medina, S., Villarrubia, N., Fernández-Velasco, J. I., et al. (2020). Acetate correlates with disability and immune response in multiple sclerosis. PeerJ 8:e10220. doi: 10.7717/peerj.10220
Piccioni, A., Cicchinelli, S., Valletta, F., De Luca, G., Longhitano, Y., Candelli, M., et al. (2022). Gut microbiota and autoimmune diseases: a charming real world together with probiotics. Curr. Med. Chem. 29, 3147–3159. doi: 10.2174/0929867328666210922161913
Ponziani, F. R., Bhoori, S., Castelli, C., Putignani, L., Rivoltini, L., Del Chierico, F., et al. (2019). Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease. Hepatology 69, 107–120. doi: 10.1002/hep.30036
Puddu, A., Sanguineti, R., Montecucco, F., and Viviani, G. L. (2014). Evidence for the gut microbiota short-chain fatty acids as key pathophysiological molecules improving diabetes. Mediat. Inflamm. 2014:162021. doi: 10.1155/2014/162021
Pugin, B., Barcik, W., Westermann, P., Heider, A., Wawrzyniak, M., Hellings, P., et al. (2017). A wide diversity of bacteria from the human gut produces and degrades biogenic amines. Microb. Ecol. Health Dis. 28:1353881. doi: 10.1080/16512235.2017.1353881
Purohit, A., Kandiyal, B., Kumar, S., Pragasam, A. K., Kamboj, P., Talukdar, D., et al. (2024). Collinsella aerofaciens linked with increased ethanol production and liver inflammation contribute to the pathophysiology of NAFLD. Science 27:108764. doi: 10.1016/j.isci.2023.108764
Qin, J., Li, Y., Cai, Z., Li, S., Zhu, J., Zhang, F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. doi: 10.1038/nature11450
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Ren, D., Wang, D., Liu, H., Shen, M., and Yu, H. (2019). Two strains of probiotic Lactobacillus enhance immune response and promote naive T cell polarization to Th1. Food Agric. Immunol. 30, 281–295. doi: 10.1080/09540105.2019.1579785
Ribot, J. C., Lopes, N., and Silva-Santos, B. (2021). γδ T cells in tissue physiology and surveillance. Nat. Rev. Immunol. 21, 221–232. doi: 10.1038/s41577-020-00452-4
Roduit, C., Wohlgensinger, J., Frei, R., Bitter, S., Bieli, C., Loeliger, S., et al. (2011). Prenatal animal contact and gene expression of innate immunity receptors at birth are associated with atopic dermatitis. J. Allergy Clin. Immunol. 127, 179–185.e1. doi: 10.1016/j.jaci.2010.10.010
Roselli, M., Finamore, A., Nuccitelli, S., Carnevali, P., Brigidi, P., Vitali, B., et al. (2009). Prevention of TNBS-induced colitis by different Lactobacillus and Bifidobacterium strains is associated with an expansion of γδT and regulatory T cells of intestinal intraepithelial lymphocytes. Inflamm. Bowel Dis. 15, 1526–1536. doi: 10.1002/ibd.20961
Round, J. L., and Mazmanian, S. K. (2010). Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA 107, 12204–12209. doi: 10.1073/pnas.0909122107
Russell, S. L., Gold, M. J., Hartmann, M., Willing, B. P., Thorson, L., Wlodarska, M., et al. (2012). Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 13, 440–447. doi: 10.1038/embor.2012.32
Saade, M., Andrés, W., Murtadha, A. M., Ghaidaa, J., Ali, M. S., Sajad, K., et al. (2022). The emerging role of microbiota-derived short-chain fatty acids in immunometabolism. Int. Immunopharmacol. 110:108983. doi: 10.1016/j.intimp.2022.108983
Sadeghpour Heravi, F. (2024). Gut microbiota and autoimmune diseases: mechanisms, treatment, challenges, and future recommendations. Curr. Clin. Microbiol. Rep. 11, 18–33. doi: 10.1007/s40588-023-00213-6
Saravia, J., Chapman, N. M., and Chi, H. (2019). Helper T cell differentiation. Cell. Mol. Immunol. 16, 634–643. doi: 10.1038/s41423-019-0220-6
Scher, J. U., Sczesnak, A., Longman, R. S., Segata, N., Ubeda, C., Bielski, C., et al. (2013). Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2:e01202. doi: 10.7554/eLife.01202
Schnupf, P., Gaboriau-Routhiau, V., Gros, M., Friedman, R., Moya-Nilges, M., Nigro, G., et al. (2015). Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature 520, 99–103. doi: 10.1038/nature14027
Shah, M. M., Saio, M., Yamashita, H., Tanaka, H., Takami, T., Ezaki, T., et al. (2012). Lactobacillus acidophilus strain L-92 induces CD4(+)CD25(+)Foxp3(+) regulatory T cells and suppresses allergic contact dermatitis. Biol. Pharm. Bull. 35, 612–616. doi: 10.1248/bpb.35.612
Shimokawa, C., Kato, T., Takeuchi, T., Ohshima, N., Furuki, T., Ohtsu, Y., et al. (2020). CD8+ regulatory T cells are critical in prevention of autoimmune-mediated diabetes. Nat. Commun. 11:1922. doi: 10.1038/s41467-020-15857-x
Silva, Y., and Bernardi, A. (2020). The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. 11:25. doi: 10.3389/fendo.2020.00025
Simoni, Y., Diana, J., Ghazarian, L., Beaudoin, L., and Lehuen, A. (2013). Therapeutic manipulation of natural killer (NK) T cells in autoimmunity: are we close to reality? Clin. Exper. Immunol. 171, 8–19. doi: 10.1111/j.1365-2249.2012.04625.x
Smits, H. H., Engering, A., Van Der Kleij, D., De Jong, E. C., Schipper, K., et al. (2005). Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin. J. Allergy Clin. Immunol. 115, 1260–1267. doi: 10.1016/j.jaci.2005.03.036
So, J. S., Kwon, H. K., Lee, C. G., Yi, H. J., Park, J. A., Lim, S. Y., et al. (2008). Lactobacillus casei suppresses experimental arthritis by down-regulating T helper 1 effector functions. Mol. Immunol. 45, 2690–2699. doi: 10.1016/j.molimm.2007.12.010
Stewart, C. J. (2019). Homing in on 12,13-diHOME in asthma. Nat. Microbiol. 4, 1774–1775. doi: 10.1038/s41564-019-0599-y
Stewart, C. J., Ajami, N. J., O’Brien, J. L., Hutchinson, D. S., Smith, D. P., Wong, M. C., et al. (2018). Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562, 583–588. doi: 10.1038/s41586-018-0617-x
Stiemsma, L. T., Reynolds, L. A., Turvey, S. E., and Finlay, B. B. (2015). The hygiene hypothesis: current perspectives and future therapies. Immunotargets Ther. 4, 143–157. doi: 10.2147/ITT.S61528
Sun, S., Luo, L., Liang, W., Yin, Q., Guo, J., Rush, A. M., et al. (2020). Bifidobacterium alters the gut microbiota and modulates the functional metabolism of T regulatory cells in the context of immune checkpoint blockade. Proc. Natl. Acad. Sci. 117, 27509–27515. doi: 10.1073/pnas.1921223117
Tai, N., Peng, J., Liu, F., Gulden, E., Hu, Y., Zhang, X., et al. (2016). Microbial antigen mimics activate diabetogenic CD8 T cells in NOD mice. J. Exp. Med. 213, 2129–2146. doi: 10.1084/jem.20160526
Takahashi, D., Hoshina, N., Kabumoto, Y., Maeda, Y., Suzuki, A., Tanabe, H., et al. (2020). Microbiota-derived butyrate limits the autoimmune response by promoting the differentiation of follicular regulatory T cells. EBioMedicine 58:102913. doi: 10.1016/j.ebiom.2020.102913
Takahashi, K., Nishida, A., Fujimoto, T., Fujii, M., Shioya, M., Imaeda, H., et al. (2016). Reduced abundance of butyrate-producing Bacteria species in the fecal microbial Community in Crohn's disease. Digestion 93, 59–65. doi: 10.1159/000441768
Takeda, S., Takeshita, M., Kikuchi, Y., Dashnyam, B., Kawahara, S., Yoshida, H., et al. (2011). Efficacy of oral administration of heat-killed probiotics from Mongolian dairy products against influenza infection in mice: alleviation of influenza infection by its immunomodulatory activity through intestinal immunity. Int. Immunopharmacol. 11, 1976–1983. doi: 10.1016/j.intimp.2011.08.007
Tamanai-Shacoori, Z., Smida, I., Bousarghin, L., Loreal, O., Meuric, V., Fong, S. B., et al. (2017). Roseburia spp.: a marker of health? Future Microbiol. 12, 157–170. doi: 10.2217/fmb-2016-0130
Tan, T. G., Sefik, E., Geva-Zatorsky, N., Kua, L., Naskar, D., Teng, F., et al. (2016). Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. USA 113, E8141–E8150. doi: 10.1073/pnas.1617460113
Tang, R., Liu, R., Zha, H., Cheng, Y., Ling, Z., and Li, L. (2024). Gut microbiota induced epigenetic modifications in the non-alcoholic fatty liver disease pathogenesis. Eng. Life Sci. 24:2300016. doi: 10.1002/elsc.202300016
Tanoue, T., Morita, S., Plichta, D. R., Skelly, A. N., Suda, W., Sugiura, Y., et al. (2019). A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565, 600–605. doi: 10.1038/s41586-019-0878-z
Tavella, T., Rampelli, S., Guidarelli, G., Bazzocchi, A., Gasperini, C., Pujos-Guillot, E., et al. (2021). Elevated gut microbiome abundance of Christensenellaceae, Porphyromonadaceae and Rikenellaceae is associated with reduced visceral adipose tissue and healthier metabolic profile in Italian elderly. Gut Microbes 13, 1–19. doi: 10.1080/19490976.2021.1880221
Thakur, A., Mikkelsen, H., and Jungersen, G. (2019). Intracellular pathogens: host immunity and microbial persistence strategies. J Immunol Res 2019, 1356540–1356524. doi: 10.1155/2019/1356540
Thomas, A. M., Manghi, P., Asnicar, F., Pasolli, E., Armanini, F., Zolfo, M., et al. (2019). Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 25, 667–678. doi: 10.1038/s41591-019-0405-7
Thorburn, A. N., Foster, P. S., Gibson, P. G., and Hansbro, P. M. (2012). Components of Streptococcus pneumoniae suppress allergic airways disease and NKT cells by inducing regulatory T cells. J. Immunol. 188, 4611–4620. doi: 10.4049/jimmunol.1101299
Thursby, E., and Juge, N. (2017). Introduction to the human gut microbiota. Biochem. J. 474, 1823–1836. doi: 10.1042/bcj20160510
Vaarala, O., Atkinson, M. A., and Neu, J. (2008). The "perfect storm" for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes 57, 2555–2562. doi: 10.2337/db08-0331
Valdes, A. M., Walter, J., Segal, E., and Spector, T. D. (2018). Role of the gut microbiota in nutrition and health. BMJ 361:k2179. doi: 10.1136/bmj.k2179
Van Der Meulen, T. A., Harmsen, H. J. M., Vila, A. V., Kurilshikov, A., Liefers, S. C., Zhernakova, A., et al. (2019). Shared gut, but distinct oral microbiota composition in primary Sjögren's syndrome and systemic lupus erythematosus. J. Autoimmun. 97, 77–87. doi: 10.1016/j.jaut.2018.10.009
Ver Heul, A., Planer, J., and Kau, A. L. (2019). The human microbiota and asthma. Clin. Rev. Allergy Immunol. 57, 350–363. doi: 10.1007/s12016-018-8719-7
Vrieze, A., Out, C., Fuentes, S., Jonker, L., Reuling, I., Kootte, R. S., et al. (2014). Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J. Hepatol. 60, 824–831. doi: 10.1016/j.jhep.2013.11.034
Wang, Y., Begum-Haque, S., Telesford, K. M., Ochoa-Repáraz, J., Christy, M., Kasper, E. J., et al. (2014). A commensal bacterial product elicits and modulates migratory capacity of CD39(+) CD4 T regulatory subsets in the suppression of neuroinflammation. Gut Microbes 5, 552–561. doi: 10.4161/gmic.29797
Wang, J., Chai, J., Zhang, L., Zhang, L., Yan, W., Sun, L., et al. (2022a). Microbiota associations with inflammatory pathways in asthma. Clin. Exp. Allergy 52, 697–705. doi: 10.1111/cea.14089
Wang, Q., McLoughlin, R. M., Cobb, B. A., Charrel-Dennis, M., Zaleski, K. J., Golenbock, D., et al. (2006). A bacterial carbohydrate links innate and adaptive responses through toll-like receptor 2. J. Exp. Med. 203, 2853–2863. doi: 10.1084/jem.20062008
Wang, Y., Shen, Y., Lu, S., and Wu, J. (2023b). EVOO supplement prevents type 1 diabetes by modulating gut microbiota and serum metabolites in NOD mice. Life Sci. 335:122274. doi: 10.1016/j.lfs.2023.122274
Wang, X., Yuan, W., Yang, C., Wang, Z., Zhang, J., Xu, D., et al. (2024). Emerging role of gut microbiota in autoimmune diseases. Front. Immunol. 15:1365554. doi: 10.3389/fimmu.2024.1365554
Wang, Q., Zhang, S.-X., Chang, M.-J., Qiao, J., Wang, C.-H., Li, X.-F., et al. (2022b). Characteristics of the gut microbiome and its relationship with peripheral CD4+ T cell subpopulations and cytokines in rheumatoid arthritis. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.799602
Wang, A., Zhao, J., Qin, Y., Zhang, Y., Xing, Y., Wang, Y., et al. (2023a). Alterations of the gut microbiota in the lupus nephritis: a systematic review. Ren. Fail. 45:2285877. doi: 10.1080/0886022X.2023.2285877
Wells, P. M., Adebayo, A. S., Bowyer, R. C. E., Freidin, M. B., Finckh, A., Strowig, T., et al. (2020). Associations between gut microbiota and genetic risk for rheumatoid arthritis in the absence of disease: a cross-sectional study. Lancet Rheumatol. 2, e418–e427. doi: 10.1016/S2665-9913(20)30064-3
Wen, C., Zheng, Z., Shao, T., Liu, L., Xie, Z., Le Chatelier, E., et al. (2017). Quantitative metagenomics reveals unique gut microbiome biomarkers in ankylosing spondylitis. Genome Biol. 18:142. doi: 10.1186/s13059-017-1271-6
Wick, E. C., Rabizadeh, S., Albesiano, E., Wu, X., Wu, S., Chan, J., et al. (2014). Stat3 activation in murine colitis induced by enterotoxigenic Bacteroides fragilis. Inflamm. Bowel Dis. 20, 821–834. doi: 10.1097/MIB.0000000000000019
Wiertsema, S. P., Van Bergenhenegouwen, J., Garssen, J., and Knippels, L. M. J. (2021). The interplay between the gut microbiome and the immune system in the context of infectious diseases throughout life and the role of nutrition in optimizing treatment strategies. Nutrients 13:886. doi: 10.3390/nu13030886
Wolfe, W., Xiang, Z., Yu, X., Li, P., Chen, H., Yao, M., et al. (2023). The challenge of applications of probiotics in gastrointestinal diseases. Adv. Gut Microbiome Res. 2023, 1–10. doi: 10.1155/2023/1984200
Won, T. J., Kim, B., Song, D. S., Lim, Y. T., Oh, E. S., Lee, D. I., et al. (2011). Modulation of Th1/Th2 balance by Lactobacillus strains isolated from kimchi via stimulation of macrophage cell line J774A.1 in vitro. J. Food Sci. 76, H55–H61. doi: 10.1111/j.1750-3841.2010.02031.x
Wu, W., Liu, H. P., Chen, F., Liu, H., Cao, A. T., Yao, S., et al. (2016). Commensal A4 bacteria inhibit intestinal Th2-cell responses through induction of dendritic cell TGF-β production. Eur. J. Immunol. 46, 1162–1167. doi: 10.1002/eji.201546160
Wu, S., Sun, C., Li, Y., Wang, T., Jia, L., Lai, S., et al. (2020). GMrepo: a database of curated and consistently annotated human gut metagenomes. Nucleic Acids Res. 48, D545–d553. doi: 10.1093/nar/gkz764
Yang, Y., Hong, Q., Zhang, X., and Liu, Z. (2024). Rheumatoid arthritis and the intestinal microbiome: probiotics as a potential therapy. Front. Immunol. 15. doi: 10.3389/fimmu.2024.1331486
Yang, C., Kwon, D. I., Kim, M., Im, S. H., and Lee, Y. J. (2021). Commensal microbiome expands Tγδ17 cells in the lung and promotes particulate matter-induced acute neutrophilia. Front. Immunol. 12:645741. doi: 10.3389/fimmu.2021.645741
Yin, J., Sternes, P. R., Wang, M., Song, J., Morrison, M., Li, T., et al. (2020). Shotgun metagenomics reveals an enrichment of potentially cross-reactive bacterial epitopes in ankylosing spondylitis patients, as well as the effects of TNFi therapy upon microbiome composition. Ann. Rheum. Dis. 79, 132–140. doi: 10.1136/annrheumdis-2019-215763
Zegarra-Ruiz, D. F., El Beidaq, A., Iñiguez, A. J., Lubrano Di Ricco, M., Manfredo Vieira, S., Kriegel, M. A., et al. (2019). A diet-sensitive commensal Lactobacillus strain mediates TLR7-dependent systemic autoimmunity. Cell Host Microbe 25, 113–127.e6. doi: 10.1016/j.chom.2018.11.009
Zhang, N., and Bevan, M. J. (2011). CD8(+) T cells: foot soldiers of the immune system. Immunity 35, 161–168. doi: 10.1016/j.immuni.2011.07.010
Zhang, H., Liao, X., Sparks, J. B., and Luo, X. M. (2014). Dynamics of gut microbiota in autoimmune lupus. Appl. Environ. Microbiol. 80, 7551–7560. doi: 10.1128/AEM.02676-14
Zhang, X., and Pan, Z. (2020). Influence of microbiota on immunity and immunotherapy for gastric and esophageal cancers. Gastroenterol. Rep. 8, 206–214. doi: 10.1093/gastro/goaa014
Zhang, Y., Peng, Y., and Xia, X. (2023). Autoimmune diseases and gut microbiota: a bibliometric and visual analysis from 2004 to 2022. Clin. Exp. Med. 23, 2813–2827. doi: 10.1007/s10238-023-01028-x
Zhang, X., Zhang, D., Jia, H., Feng, Q., Wang, D., Liang, D., et al. (2015). The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 21, 895–905. doi: 10.1038/nm.3914
Zhao, X., Hu, M., Zhou, H., Yang, Y., Shen, S., You, Y., et al. (2023). The role of gut microbiome in the complex relationship between respiratory tract infection and asthma. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1219942
Zhao, C. N., Xu, Z., Wu, G. C., Mao, Y. M., Liu, L. N., Qian, W., et al. (2019). Emerging role of air pollution in autoimmune diseases. Autoimmun. Rev. 18, 607–614. doi: 10.1016/j.autrev.2018.12.010
Keywords: muciniphila, Anaerostipes caccae, Bacteroides sp., Roseburia sp., Blautia sp., Blautia faecis, Clostridium lavalense, Christensenellaceae sp.
Citation: Bhutta NK, Xu X, Jian C, Wang Y, Liu Y, Sun J, Han B, Wu S and Javeed A (2024) Gut microbiota mediated T cells regulation and autoimmune diseases. Front. Microbiol. 15:1477187. doi: 10.3389/fmicb.2024.1477187
Edited by:
Zhangran Chen, Xiamen University, ChinaReviewed by:
Bin Wei, Zhejiang University of Technology, ChinaRongrong Cao, Soochow University, China
Copyright © 2024 Bhutta, Xu, Jian, Wang, Liu, Sun, Han, Wu and Javeed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingnan Han, aGFuYmluZ25hbkB6c3R1LmVkdS5jbg==; Shandong Wu, c2hhbmRvbmdyd3VAc2luYS5jb20=; Ansar Javeed, YWpjQHpzdHUuZWR1LmNu
 Nabeel Khalid Bhutta
Nabeel Khalid Bhutta Xiujin Xu1
Xiujin Xu1 Yi Liu
Yi Liu Bingnan Han
Bingnan Han Ansar Javeed
Ansar Javeed