- Guangdong Provincial Key Laboratory of Animal Molecular Design and Precise Breeding, School of Animal Science and Technology, Foshan University, Foshan, China
Introduction: Porcine reproductive and respiratory syndrome virus (PRRSV) is the causative agent 13 of porcine reproductive and respiratory syndrome (PRRS), which is one of the most economically 14 devastating viruses in the Vietnamese swine industry.
Methods: With a view toward determining the 15 genetic variation among PRRSV strains in Vietnam, we examined 271 PRRSV GP5 protein 16 sequences obtained from strains isolated in Vietnam from 2007 to 2023, for which we constructed 17 phylogenetic trees. Additionally, a collection of 52 PRRSV-1 strains and 80 PRRSV-2 strains 18 isolated in different years were specifically selected for nucleotide and amino acid homology analysis 19 and amino acid sequence alignment.
Results: The results revealed 76.1%–100.0% nucleotide and 20 75.2%–100.0% amino acid homologies for the PRRSV-1 GP5 gene, and 81.8%–100.0% nucleotide 21 and 81.1%–100.0% amino acid homologies for the PRRSV-2 GP5 gene. Amino acid mutation sites 22 in PRRSV-2 were found to be primarily distributed in the signal peptide region, antigenic sites, two 23 T-cell antigen regions, two highly variable regions (HVRs), and in the vicinity of the neutralizing 24 epitope, with a deletion mutation occurring in the neutralizing epitope, whereas amino acid mutations 25 in the PRRSV-1 sequences were found to occur predominantly in two T-cell epitopes. Genetic 26 analysis revealed that PRRSV-1 strains in Vietnam are of subtype 1 (Global), whereas PRRSV-2 27 strains are categorized into sublineages L1A, L5A, and L8E, with L8E being the predominantly 28 prevalent strain at present. Recombination analyses indicated that no significant recombination 29 events have occurred in any of the assessed 271 Vietnamese PRRSV strains.
Discussion: Our 30 analyses of 271 Vietnamese PRRSV strains have yielded valuable insights regarding the 31 epidemiological trends and genetic dynamics of PRRSV in Vietnam, and will provide a theoretical 32 basis for formulating prevention and control measures for PRRS and the development of PRRS 33 vaccines.
1 Introduction
The porcine reproductive and respiratory syndrome virus (PRRSV), the causal pathogen of porcine reproductive and respiratory syndrome (PRRS), primarily infects swine herds, causing reproductive disorders in pregnant sows, including miscarriage, the occurrence of stillbirths and mummified fetuses, as well as respiratory diseases in pigs of all ages (Sha et al., 2023). Since the initial isolation of PRRSV in Europe and the Americas in the early 1990s (Collins et al., 1992; Wensvoort et al., 1991), PRRS has become globally prevalent in many countries. In 2006, an outbreak of highly pathogenic porcine reproductive and respiratory syndrome (HP-PRRS), characterized by high fever and mortality in pigs of all ages, occurred in China and spread rapidly to many Asian countries (Guo et al., 2013). The incidence of HP-PRRS ranged from 50 to 100%, with mortality rates varying between 20 and 100%, resulting in the culling of approximately 40 million pigs in China in 2006–2007 (An et al., 2011), causing huge economic losses to the swine industry. However, given a range of complicating factors, including genetic diversity, immunosuppression, and secondary infections, the complete eradication of PRRS in the global swine industry presents significant challenge (Cai et al., 2023). Consequently, PRRS continues to pose a serious economic threat to the global swine industry.
PRRSV, a member of the viral genus Arterivirus, within the family Arteriviridae and order Nidovirales, is an enveloped, single-stranded positive-sense RNA virus (Adams et al., 2017). The PRRSV genome is approximately 15 kb in length and encodes 11 open reading frames (ORFs) (Zhang et al., 2023). ORF1a and ORF1b encode two polyproteins, namely, pp1a and pp1ab, respectively, which are subsequently cleaved by protein hydrolases to generate 16 non-structural proteins, whereas ORF2a, ORF2b, ORFs3–7, and ORF5a encode eight corresponding structural proteins (Chen et al., 2023). At present, PRRSV is divided into two distinct species, Betaarterivirus suid 1 (PRRSV-1) and Betaarterivirus suid 2 (PRRSV-2), which have approximately 60% nucleotide homology (Brinton et al., 2021; Lu et al., 2014). Among the viral proteins, the GP5 protein encoded by ORF5 is characterized by the highest structural diversity between PRRSV-1 and PRRSV-2 strains. At the amino acid level, the homology of PRRSV-1 and PRRSV-2 GP5 is between 50 and 55%, and this protein is thus commonly used in genetic sequence analysis (Lunney et al., 2010; Zhao et al., 2022). On the basis of ORF5 sequences, PRRSV-1 can be classified into four subtypes, namely, subtype 1 (Global), subtype 1 (Russia), subtype 2, and subtype 3, whereas PRRSV-2 can be classified into nine lineages and 37 sublineages (Li et al., 2022; Shi et al., 2010). However, Yim-Im et al. (2023) refined a phylogenetic classification system based on the PRRSV-2 ORF5, reclassifying PRRSV-2 into 11 lineages (L1-L11), 21 sublineages (L1A-L1F, L1H-L1J, L5A-L5B, L8A-L8E, and L9A-L9E), as well as the 5 taxa within L1C (L1C.1-L1C.5). Although partial sequences of PRRSV-1 strains isolated in Vietnam have been uploaded to the GenBank database, to the best of our knowledge, there have been no systematic studies on PRRSV-1 conducted to date, unlike PRRSV-2, for which there have been a number of studies. In 2007, an outbreak of highly pathogenic PRRS (HP-PRRS) was detected in Vietnam, which spread rapidly nationwide (Lee et al., 2019a). In the same year, Feng et al. (2008) isolated suspected PRRSV strains for the first time, and on the basis of bioinformatics analysis, revealed that these were strains of the PRRSV-2 species, with a 99% nucleotide homology to PRRSV-2 strains that were prevalent in China in 2006. However, as early as 2002, Kamakawa et al. (2006) had already detected PRRSV on free-range pig farms in Vietnam, thereby indicating that PRRS was prevalent in Vietnam prior to 2007. Do et al. (2016) found that among PRRSV field strains isolated from 2007 to 2015, all were of PRRSV-2 sub-lineages 8.7 and 5.1. Whereas for strains isolated in 2021, Nguyen et al. (2022) detected those of lineage 8, sublineage 8.7, lineage 1, and sublineage 1.4.
Since the 2007 PRRS outbreak, although numerous PRRS vaccines had been introduced in Vietnam (Lee et al., 2019b), until 2021, many swine farms in Vietnam on which pigs had for long been vaccinated with PRRS attenuated vaccines continued to experience PRRS outbreaks, and it was accordingly speculated that mutations in the GP5 were reducing the protective efficacy of these vaccines (Nguyen et al., 2022). To confirm this supposition and gain further insights into the genetic variations of the PRRSV GP5 gene in Vietnam from 2007 to 2023, we obtained 344 PRRSV GP5 sequences from the GenBank database and used these to construct phylogenetic trees. Among these sequences, we selected 52 PRRSV-1 and 80 PRRSV-2 GP5 sequences for amino acid and nucleotide homology analyses and amino acid sequence alignment, along with analyses of potential recombination events. Our findings in this study will provide a theoretical foundation for understanding future epidemiological trends in PRRS and for the development of vaccine-based prevention and control measures in Vietnam.
2 Materials and methods
2.1 Dataset used for sequence analysis
The GP5 sequences of 344 PRRSV isolates were obtained from the GenBank, NCBI database. Given the limited number of sequences of Vietnamese PRRSV-1 strains deposited in GenBank, we selected 52 PRRSV-1 strains (25 from Vietnam; 13 from China; 6 from the United States; 1 from Spain; 5 from Russia; 2 from Belarus) (Table 1) and 292 PRRSV-2 strains (246 from Vietnam; 21 from China; 25 from the United States) (Supplementary Table S1). Moreover, as the earliest database accession from Vietnam is a 2007 isolate, we selected PRRSV strains that were isolated from 2007 to 2023. To ensure the comprehensiveness of our genetic dynamics analysis of PRRSV in Vietnam, all 52 selected PRRSV-1 strains were analyzed for genetic variation (Table 1), whereas 80 strains of different lineages (including classical, vaccine and epidemic strains) isolated in different years were selected from the 292 PRRSV-2 GP5 sequences (Table 2).
2.2 Phylogenetic analysis
The 344 selected PRRSV GP5 sequences were used to perform phylogenetic analysis, with phylogenetic trees being constructed using MEGA software (Version 11.0.13) based on the maximum likelihood (ML) (substitution model: Nucleotide; Model/Method: Tamura-Nei model; Rates among Sites: Gamma Distributed With Invariant Sites (G + I); ML Heuristic Method: Nearest-Neighbor-Interchange (NNI); Initial Tree for ML: Make initial tree automatically (Default - NJ/BioNJ)) and neighbor-joining (NJ) (substitution model: Nucleotide; Model/Method: p-distance; Substitutions to Include: d: Transitions + Transversions; Rates among Sites: Uniform Rates) methods with 100 and 1,000 bootstrap replicates, respectively. Default parameter values were utilized for other parameters. The phylogenetic tree data thus obtained were imported into online software (The Interactive Tree Of Life1) for subsequent sorting and editing.
2.3 GP5 nucleotide and amino acid homology analysis and sequence alignment
For analysis of PRRSV GP5 nucleotide and amino acid homologies, we used the GraphPad Prism (Version 9.0.0) and the Clustal W method in the MegaAlign function of DNASTAR software (Version 7.0). DNASTAR software was also used for amino acid sequence comparisons.
2.4 Recombination analysis
To facilitate preliminary identification of potential recombination events in the 344 assessed PRRSV GP5 sequences, we used the RDP, GENECONV, BootScan, MaxChi, Chimera, SiScan, and 3Seq algorithms in RDP software (Version 4.0). Strains for which four or more positive (+) results and p < 0.05 were obtained were considered recombinant strains.
3 Results
3.1 Phylogenetic analysis
To determine the genetic variation of the PRRSV GP5 gene in Vietnam, we analyzed the evolution of this viral gene by constructing phylogenetic trees using gene sequences for 52 PRRSV-1 and 80 PRRSV-2 strains, which were selected based on the availability of GP5 sequence information in the global PRRSV classification system and GenBank database. The results revealed that all PRRSV-1 strains prevalent in Vietnam are subtype 1 (Global) (Figure 1). Contrastingly, the PRRSV-2 strains were grouped into three lineages (1, 5, and 8), among which, lineage 8 strains were identified as being predominant in Vietnam. The most recent genetic classification of PRRSV-2 (Yim-Im et al., 2023) indicates that the lineage 1 strains in Vietnam were part of sublineage L1A, while the lineage 5 strains were classified as sublineage L5A and the lineage 8 strains were identified as sublineage L8E. PRRSV-1 subtype 1 (Global) strains were genetically distant from the subtype 1 (Russia) strains and exhibited a closer genetic relationship with subtype 3. PRRSV-2 L1A and L5A are the most closely related, while they are more distantly related to L8E. In addition, PRRSV-2 strains recently detected in Vietnam in 2023 were established to be lineage 8 strains.
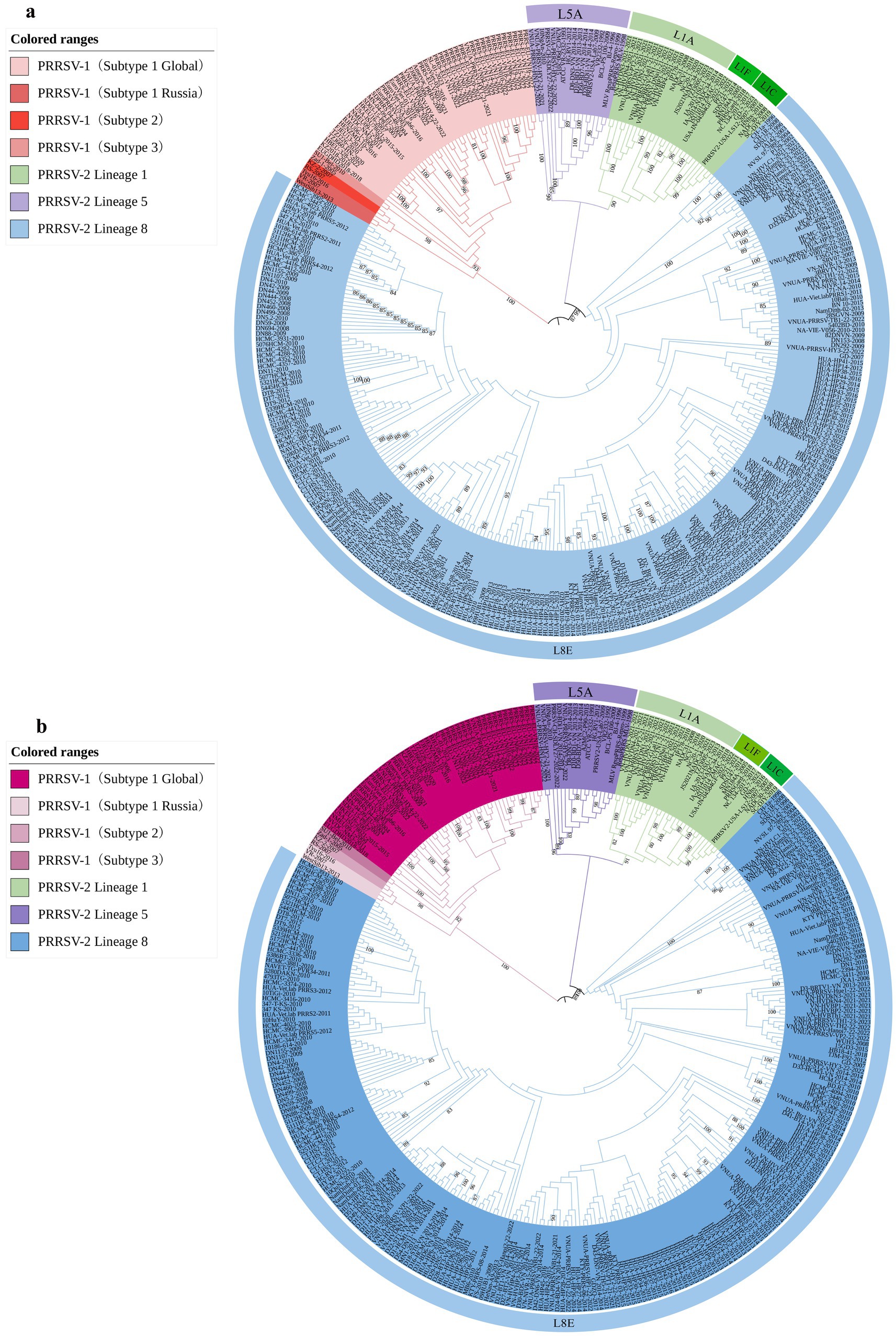
Figure 1. Phylogenetic analysis was conducted for 344 PRRSV GP5 sequences. (A) The construction of a phylogenetic tree was performed using the neighbor-joining method in MEGA software with 1,000 bootstrap replicates. (B) The construction of a phylogenetic tree was performed using the maximum likelihood method in MEGA software with 100 bootstrap replicates.
3.2 Nucleotide similarity
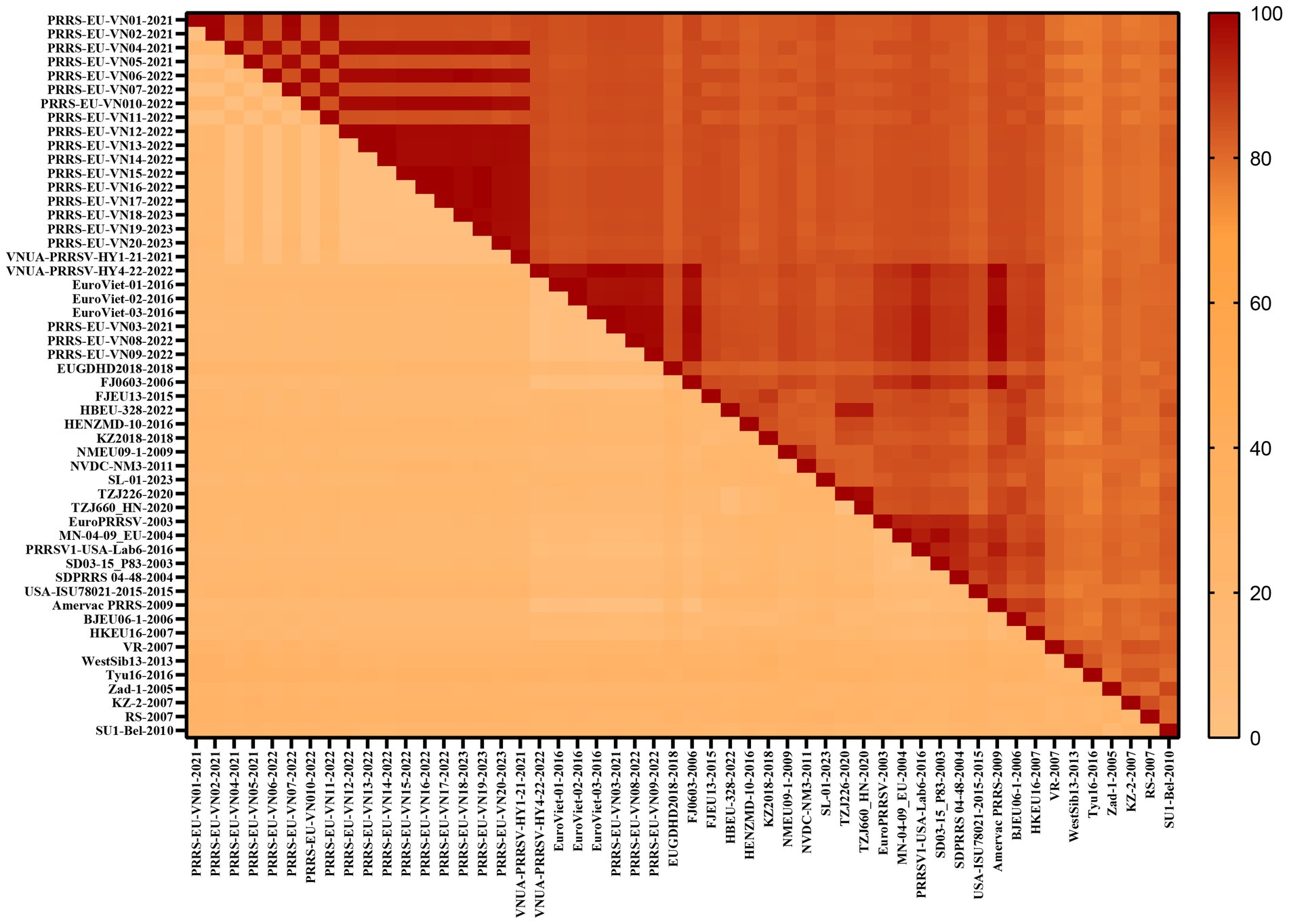
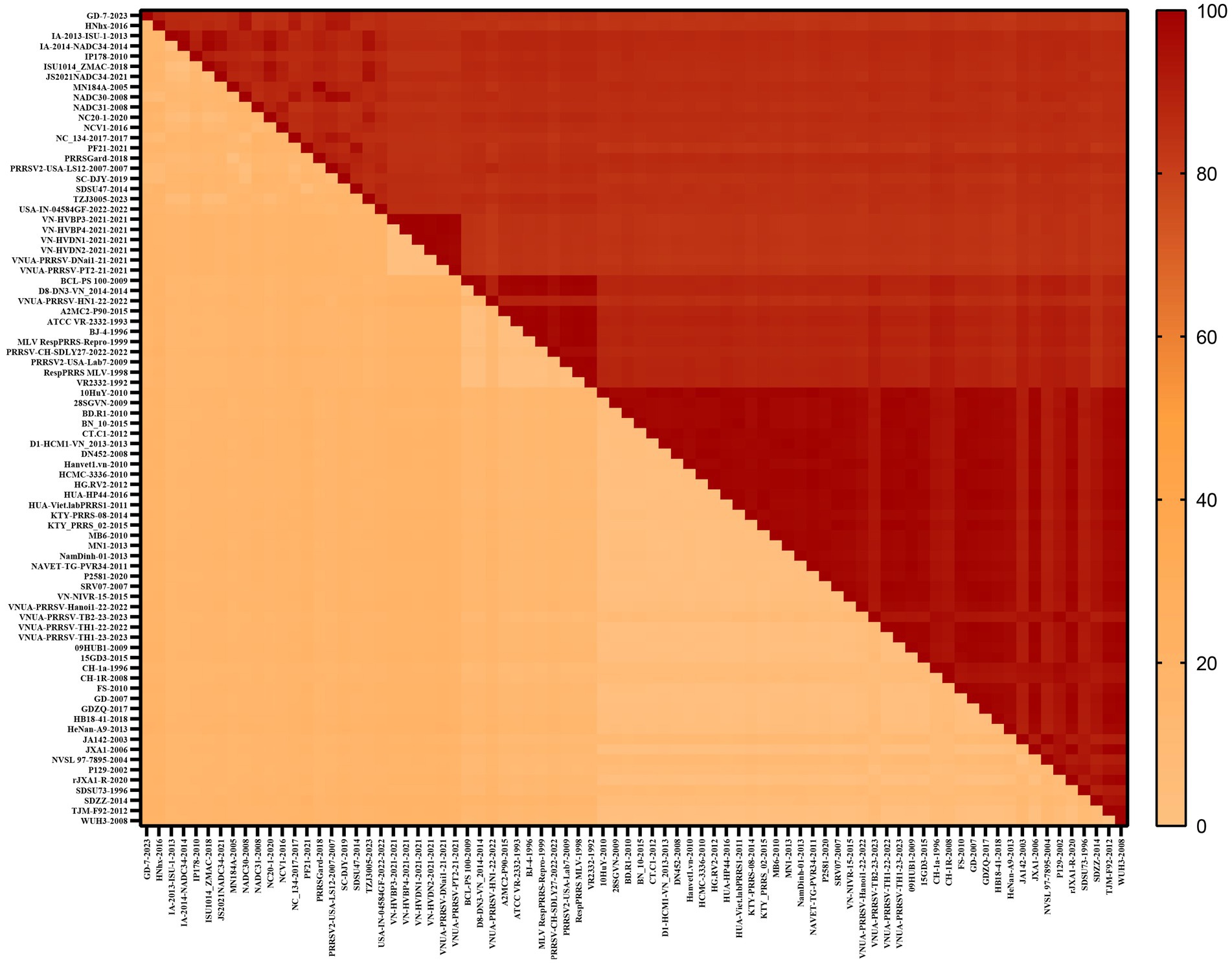
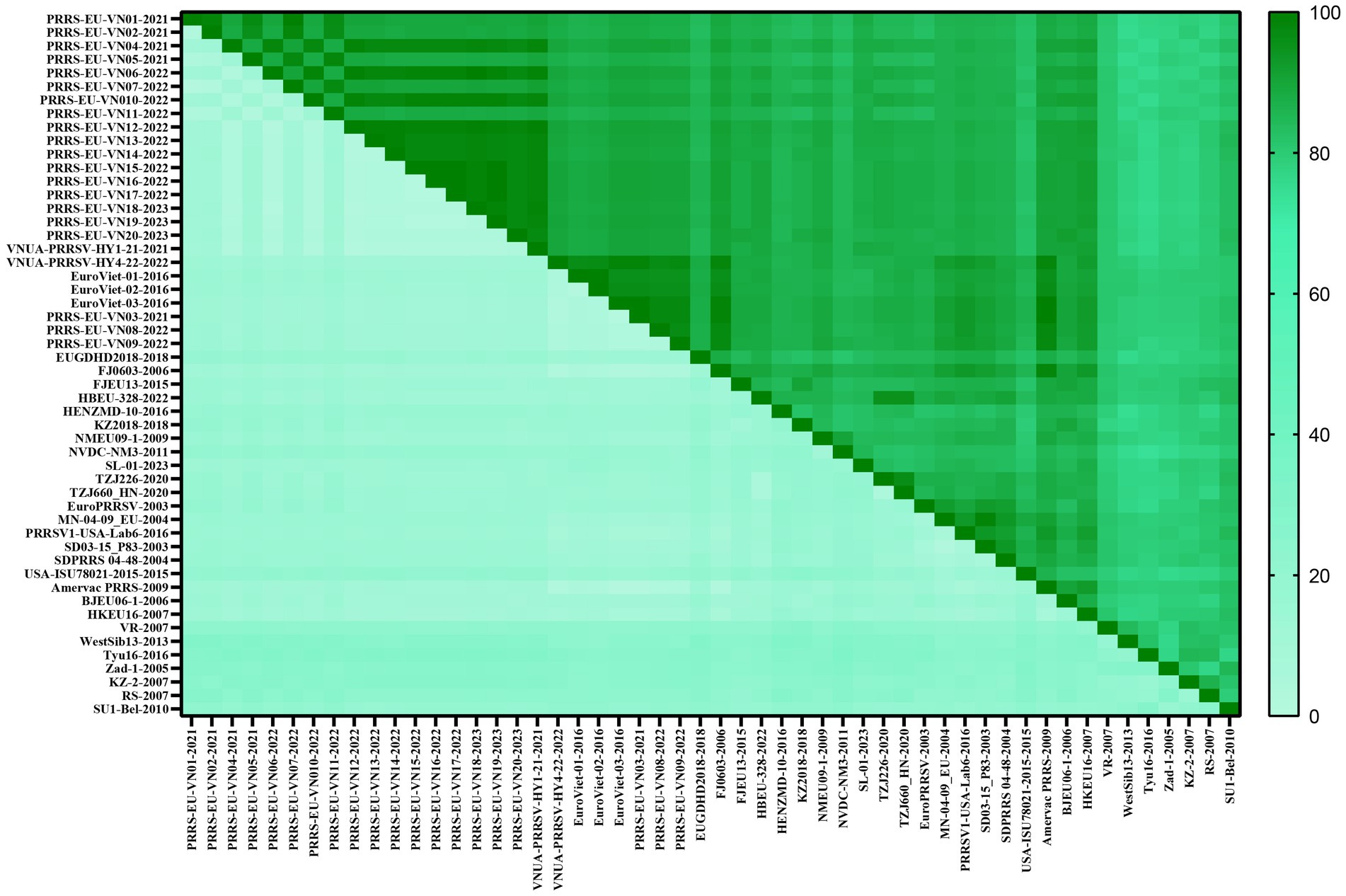
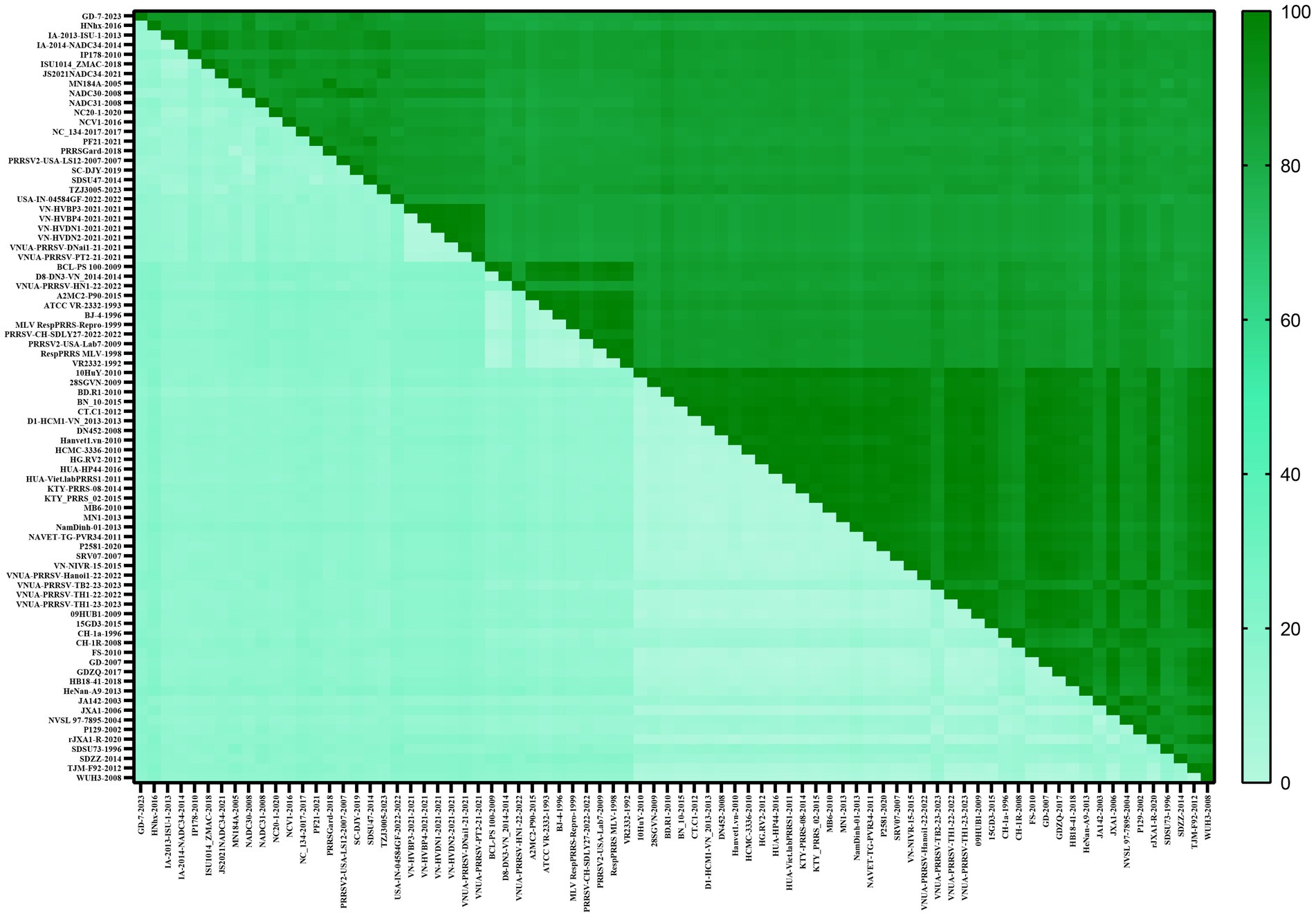
To gain a comprehensive understanding of the evolutionary genetic relationships among the different species and lineages of PRRSV, we performed nucleotide homology analyses for the PRRSV GP5 sequences. Analyses of the PRRSV-1 and PRRSV-2 GP5 sequences from Vietnam and other countries revealed nucleotide homologies 76.1–100.0% and 81.8–100.0%, respectively. Among the PRRSV-1 GP5 sequences (Figure 2; Supplementary Table S2), the lowest nucleotide homology (76.1%) was found between the strains of Vietnam and subtype 1 (Russia), whereas the PRRS-EU-VN06-2022 and PRRS-EU-VN18-2023 strains, and PRRS-EU-VN15-2022, PRRS-EU-VN16-2022, and PRRS-EU-VN17-2022 strains were characterized by the highest nucleotide homologies (100.0%). Among the PRRSV-2 GP5 sequences (Figure 3; Supplementary Table S3), the VNUA-PRRSV-DNai1-21–2021 and VNUA-PRRSV-HN1-22-2022 strains were found to have the lowest nucleotide homology (81.8%), whereas the VN-HVBP3-2021 and VN-HVBP4-2021 strains, the D1-HCM1-VN_2013 and MN1-2013 strains, and the HCMC-3336-2010 and NAVET-TG-PVR34-2011 strains had the highest nucleotide homologies (100.0%).
3.3 Amino acid sequence similarity
Analysis of the amino acid homologies of PRRSV GP5 sequences revealed homologies of 75.2–100.0% and 81.1–100.0% for the GP5 gene in PRRSV-1 and PRRSV-2 strains between Vietnam and other countries, respectively. Among the PRRSV-1 GP5 sequences (Figure 4; Supplementary Table S4), the lowest amino acid homology (75.2%) was detected between the strains of Vietnam and subtype 1 (Russia), whereas the highest homologies (100%) were between the PRRS-EU-VN03-2021 and EuroViet-03-2016 strains, PRRS-EU-VN06-2022 and PRRS-EU-VN18-2023 strains, and PRRS-EU-VN15-2022, PRRS-EU-VN16-2022, PRRS-EU-VN17-2022, and PRRS-EU-VN19-2023 strains. Among the 80 PRRSV-2 GP5 sequences (Figure 5; Supplementary Table S5), the HNhx-2016 and NamDinh-01–2013 strains, D8-DN3-VN_2014–2014 and NADC31-2008 strains had the lowest amino acid homology (81.1%), whereas the highest homologies (100%) were between the VN-HVBP3-2021 and VN-HVBP4-2021 strains, the D1-HCM1-VN_2013 and MN1-2013 strains, the CT.C1-2012, HG.RV2-2012 and 09HUB1-2009 strains, the HCMC-3336-2010 and NAVET-TG-PVR34-2011 strains, and the HUA-Viet.labPRRS1-2011 and VNUA-PRRSV-TH1-22-2022 strains.
3.4 Amino acid sequence alignment
On the basis of our multiple alignments of the GP5 sequences of 52 PRRSV-1 and 80 PRRSV-2 strains, we established that whereas there have been no insertion mutations in any of the assessed PRRSV GP5 sequences, and a deletion mutation has occurred in Vietnamese PRRSV-2 strains (L1A). In the GP5 sequence of PRRSV-1 subtype 1 (Global) (Figure 6), the sites of amino acid mutation were found to be concentrated primarily in two T-cell epitopes, with four mutations being detected in its N-glycosylation site. Of these mutations, an N37S alteration has occurred in EuroViet-03-2016 and PRRS-EU-VN03-2021, an N37D alteration has occurred in PRRS-EU-VN08-2022, an S38G alteration has occurred in PRRS-EU-VN09-2022, an N46D alteration was found in PRRS-EU-VN05-2021 and PRRS-EU-VN11-2022, whereas a N53K transition was observed in PRRS-EU-VN05-2022. In addition, some Vietnamese strains were found to be characterized by two specific mutations (H5C and A201V), one of which was located at the signal peptide. These analyses also revealed that the GP5 sequence of PRRSV-1 strain in Vietnam has fewer mutation sites and a lower degree of variation.
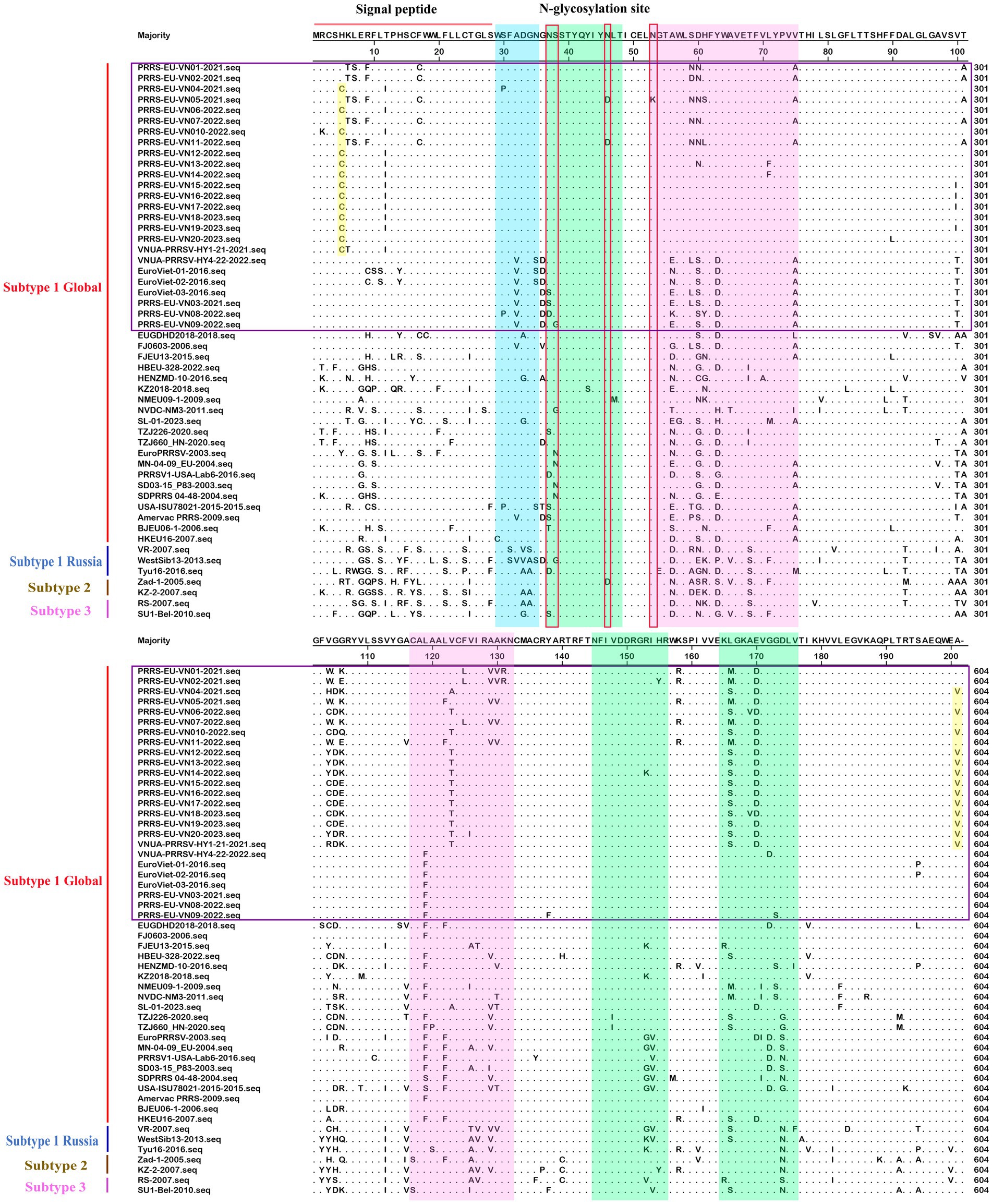
Figure 6. Alignment of 52 PRRSV-1 GP5 amino acid sequences. The Vietnamese strains are marked in purple. Potential neutralizing epitopes are marked. The Neutralizing epitope, B-cell epitope, T-cell epitope, and strain-specific mutations are represented by blue, green, pink, and yellow, respectively.
Compared with the PRRSV-1 strains, we found that amino acid sites in the PRRSV-2 GP5 gene appear to be more prone to mutation. These mutations we established to occur mainly in the signal peptide region, decoy epitope, two T-cell antigenic regions, two highly variable regions (HVRs), and in the vicinity of the neutralizing epitope (Figure 7). N-glycosylation site mutations were detected at amino acids 30, 32, 33, 34, 35, and 51, mainly in the HVR1 region and neutralizing epitope. Among these, a VNUA-PRRSV-HN1-22-2022 strain (L5A) has the most mutated N-glycosylation sites. We identified certain lineage-specific amino acid mutation sites, including a L47I mutation in the signal peptide region of GP5 in L1A; G9C, and C24Y mutations in L8E; and L93A, T66S, L127F, and S137A mutations in L5A. In addition, we detected some specific mutations in the L1A strains in Vietnam, including a deletion mutation at the 37th amino acid in the PNE region, and F26I, D61E, L145V, and E170I mutations (VNUA-PRRSV-DNAi1-21–2021, VN-HVDN2-2021, VN-HVBP3-2021, VN-HVBP4-2021, VN-HVDN1-2021, and VNUA-PRRSV-PT2-21–2021 strains). Among the three lineages, the strains of L1A and L5A were found to harbor the largest number of strain-specific mutations.
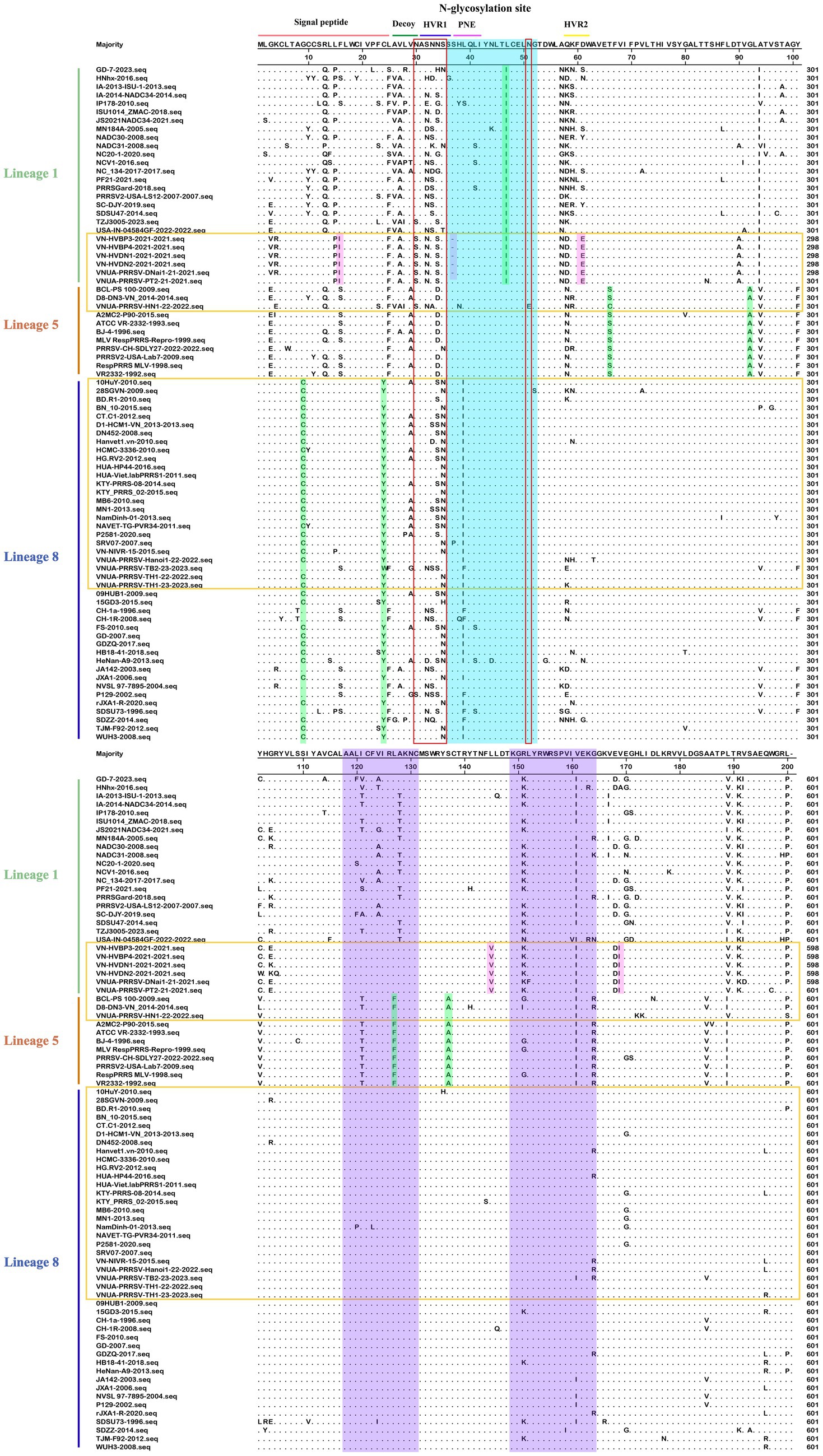
Figure 7. Alignment of 80 PRRSV-2 GP5 amino acid sequences. The Vietnamese strains are marked in yellow. The neutralizing epitope, two T-cell antigenic region, lineage-specific, and Vietnam strain specific mutations are represented by blue, purple, green, and pink, respectively.
Alignment of the amino acid sequences of the PRRSV-1 and PRRSV-2 GP5 proteins revealed that PRRSV-2 L1A strains are characterized by the largest and most concentrated degree of variation, whereas PRRSV-1 subtype 1 (Global) strains have the lowest degree of amino acid variation. Moreover, compared with the PRRSV-1 strains, we detected a larger number of amino acid mutation sites in the neutralizing and T-cell epitopes of the GP5 sequence in PRRSV-2 strains, thereby indicating that in Vietnam, PRRSV-2 stains are more prone to mutation.
3.5 Recombination analysis
Based on multiple alignments of PRRSV GP5 sequences from the 344 PRRSV genomes, we established that there have been no significant recombination events with respect to GP5 protein.
4 Discussion
Since the first reported outbreak of HP-PRRS in China in 2006, this disease has spread rapidly throughout other Asian countries, including Vietnam, the Philippines, and Thailand (Do D. T. et al., 2015), and the continuous mutation and recombination of PRRSV present a considerable challenge to the global swine industry (Li et al., 2016). The GP5 protein of this virus, which is essential for virus assembly and infection, and also important for the induction of neutralizing antibodies, is characterized by a notable variability, thereby identifying it as a useful focus for studying the genetic diversity of PRRSV (Jantafong et al., 2015). On the basis of the GP5 gene of phylogenetic analyses revealed that in recent years, PRRSV-2 lineage 8 strains have been the prevalent strains in Vietnam. The strains of this lineage have been established to be more closely related to lineage 1 strains and most distantly related to those of lineage 5 strains, whereas PRRSV-1 subtype 1 (global) strains were closer to subtype 3 strains and distant from subtype 1 (Russia). A comparison of the evolutionary trees constructed using the ML and NJ methods revealed no differences in the genetic distances between PRRSV-1 and PRRSV-2, or among different PRRSV-2 lineages. This observation may be related to the selected PRRSV strain gene from Vietnam. The emergence of HP-PRRS was first reported in China in 2006, and in the following year, an outbreak of HP-PRRS was reported in northern Vietnam, which subsequently spread rapidly to central and southern regions (Do T. D. et al., 2015; Tian et al., 2007). Between 2008 and 2012, Thuy et al. (2013) isolated 32 PRRSV strains of the PRRSV-2 sublineage 8.7 in Vietnam, a majority of which were found to be closely related to the JXA1-2006 strain isolated from China. It has been proposed that PRRS outbreaks are also associated with climatic factors (Alkhamis et al., 2017). Given the close geographical proximity of northern Vietnam to China, it is speculated that these factors may have contributed to a close correlation in the PRRSV strains transmitted between Vietnam and China. In 2021, PRRSV-2 lineage 1 strains were reported in Vietnam for the first time, along with the isolation of a vaccine-like strain, VN-HVLC1-2021 (belonging to L8E) (Nguyen et al., 2022). Accordingly, it is speculated that there may have been variation or recombination in the PRRSV vaccine strains used on Vietnamese farms. In this regard, the phylogenetic trees constructed in this study reveal that the PRRSV-2 L8E strains are the most abundant among those isolated in Vietnam, whereas L1A strains are the least abundant. Although to date there have been no relevant systematic reports, we found that whereas PRRSV-1 strains were isolated in Vietnam in 2016, PRRSV-2 L1A strains appeared in 2021, the number of L5A strains exhibited a gradual increase in 2022, thereby highlighting the emerging complexity of the epidemiological and genetic evolution of PRRSV on Vietnamese swine farms. Although our analyses in this study were constrained to a certain extent by the limited number and variety of PRRSV sequences obtained from strains isolated in Vietnam, on the basis of the types of PRRSV strains isolated in recent years, it would be predicted that the prevalence of PRRSV-1 strains, along with PRRSV-2 lineage 1 and 5 strains may gradually increase in the future.
In this study, we accordingly assessed 52 PRRSV-1 and 80 PRRSV-2 strains to investigate the nucleotide and amino acid homologies and amino acid sequence alignments of the GP5 protein, which thereby enabled us to establish the genetic diversity of this protein in Vietnam and predict potential evolutionary trends. Our findings revealed nucleotide and amino acid homologies of 76.1–100.0% and 75.2–100.0%, respectively, for PRRSV-1 strain GP5 proteins and corresponding values of 81.8–100.0% and 81.1–100.0% for PRRSV-2 strain GP5 proteins. Among these, PRRSV-1 subtype 1 (Global) exhibited the lowest homology and the most distant phylogenetic relationship with subtype 1 (Russia). Furthermore, we established that the GP5 gene sequences of the Vietnamese PRRSV-2 strains CT.C1-2012 and HG.RV2-2012 are identical to that of the Chinese PRRSV-2 strain 09HUB1-2009 (100%), whereas we detected no close similarities regarding PRRSV-1 strains, thereby tending to indicate that the evolutionary trends of PRRSV strains in Vietnam and China are both similar and distinct.
N-glycosylation sites on the GP5 protein are essential for viral replication and play important roles in receptor binding, viral infectivity, and the induction of immune responses (Wei et al., 2012). It has been shown that different N-glycosylation site mutations have differing effects on PRRSV, including loss of polysaccharide residues in N34, N44, and N51 enhancing immunogenicity, whereas those at N33, N34, and N35 may enhance viral virulence; moreover, mutations in neutralizing epitopes can also lead to changes in PRRSV virulence and immune escape (Luo et al., 2023). Furthermore, specific mutations in N-glycosylation sites, such as D37N, D56N, S60N, and G63N, have been established to be closely associated with immune evasion, inhibition of host viral recognition, and loss of host neutralization capacity (Hsueh et al., 2023). In the present study, we identified ten N-glycosylation site mutations in the GP5 protein of Vietnamese PRRSV strains (4 in PRRSV-1, 6 in PRRSV-2), particularly at residues 33, 34, 35 and 51 of the PRRSV-2 GP5 protein, which may potentially play an important role to immune evasion or altered PRRSV virulence. All four glycosylation sites of PRRSV-1 were conserved, with mutations observed in only a few individual strains; whereas among the Vietnamese PRRSV-2 strains, the N51 site was relatively conserved, with mutations observed only in the strain VNUA-PRRSV-HN1-22-2022. Guo et al. (2019) have previously found that mutations to the 39th and 57th amino acids of GP5 contribute to virus evasion of host immune responses, and the 187–200 amino acid positions of the GP5 positions in both PRRSV-1 and PRRSV-2 were instrumental in the cross-neutralization reaction of antibody formation (de Lima et al., 2006). Compared with the findings reported by Nguyen et al. (2022), we identified certain identical amino acid mutation sites (aa 39, 52–61, 187–200) in the PRRSV-2 GP5 protein, which may confer the virus with the ability to evade host immunity; and certain different amino acid mutation sites were identified in sublineage L1A, including a novel deletion mutation (aa 37), a lineage-specific mutation (L47I) and four mutations specific (F26I, D61E, L145V, and E170I). In contrast, among the Vietnamese PRRSV-1 strains, with the exception of EuroViet-01-2016 and EuroViet-02-2016, which exhibited mutations at amino acid positions 187–200, no other strains demonstrated mutations at these sites. Furthermore, two specific mutations were identified in PRRSV-1 subtype 1 (Global): H5C and A201V. However, the association of these mutations with the epidemiological and genetic variation trends of PRRSV strain in Vietnam will necessitate further investigation.
It has been established that PRRSV spreads between farms via the transfer of infected animals and infected semen, aerosols, and items contaminated with the virus (Lee et al., 2019a), and Zimmerman et al. (1997) found that birds may be involved in the transmission of PRRSV. Accordingly, given that many small-scale farms in Vietnam operate mixed livestock farming systems (Lee et al., 2019b), the potential for virus transmission from avian species to pigs cannot be ruled out. Notably, such small-scale swine farms account for 70% of the total pig production in Vietnam, and farmers rearing pigs on these farms rarely use PRRSV vaccine (Lee et al., 2019a). Consequently, given the inadequate vaccine coverage and lack of effective biosecurity measures, these small-scale farms are at a heightened risk of PRRS outbreaks. Outbreaks are, however, by no means confined to these smallholdings, as PRRS outbreaks still occur on swine farms that have been conducting vaccination over prolonged periods, which would indicate that new mutations and antigen changes in PRRSV strains may have led to immune evasion. In addition, Vietnamese farmers continue to sell sick pigs in order to reduce the economic losses caused by PRRS, thereby making the prevention and control of PRRS even more challenging (Huong et al., 2016).
To summarize, in this study, we sought to examine nucleotide and amino acid homologies among different strains of PRRSV and characterized amino acid mutations in these strains based on analyses of the genetic dynamics of 271 PRRSV GP5 sequences obtained from Vietnamese strains isolated between 2007 to 2023. Although the GP5 protein of this virus has been established to show significant variability and is prone to recombination events, we detected no relevant recombination in this study, and we suspect that the recombination events may exist in other ORFs. During the course of PRRS epidemics, PRRSV strains undergo continuous mutation and evolution, thereby giving rise to numerous novel strains that can compromise the efficacy of vaccines. Moreover, as a consequence of economic imperatives and a lack of biosecurity awareness, small-scale farms in Vietnam tend to be characterized by low PRRSV vaccination rates and unrestricted pig trade, thereby presenting considerable challenges for ensuring effective PRRS prevention and control. On the basis of our analysis of PRRSV GP5 sequences obtained from strains isolated in different years, we provide a theoretical foundation for further research and the development of novel vaccines based on the GP5 gene.
5 Conclusions and outlook
In this study, we established that from 2007 to 2023, the PRRSV-1 strains prevalent in Vietnam were of subtype 1 (Global), along with sublineages L1A, L5A, and L8E PRRSV-2 strains, among which, L8E strains have tended to be the predominant type, followed by L5A, whereas L1A became prevalent only from 2021. Phylogenetic analysis based on sequences of the GP5 gene enabled us to assess the genetic diversity of PRRSV strains in Vietnam, and using such information, continuous monitoring of PRRSV GP5 genetic variation should be conducted to strengthen PRRSV prevention and control measures in this country.
On the basis of a survey of reports related to PRRSV in Vietnam, it has been established that the outbreaks of PRRS epidemics in the country can largely be traced to small-scale swine farms that account for the highest proportion of Vietnamese pig production. This can be ascribed to the fact that these farms tend to be based on mixed livestock and poultry breeding systems, combined with low vaccination rates against PRRSV and a lack of biosecurity awareness among pig farmers. Nevertheless, even on farms in which livestock have been routinely vaccinated against PRRS, herds still exhibit clinical symptoms related to PRRS, which provides evidence to indicate that PRRSV strains mutate and recombine to evade vaccine-promoted immunity, and thus highlights the inability of vaccines currently administered in Vietnam to effectively prevent and control PRRS. Given that the PRRSV GP5 protein shows high variability, and that mutations in the amino acid sequence can variously influence the virulence and immune evasion of PRRSV, as well as the neutralization capacity of the host, in-depth analyses of trends in the genetic variation of this protein could provide a theoretical basis for the development of novel vaccines and the establishment of effective prevention and control measures. Moreover, it is imperative that Vietnam further strengthens the cultivation of biosafety awareness among small-scale farmers and implement relevant biosafety measures, which, we anticipate, would contribute to the future control or even eradication of PRRS.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
GL: Writing – original draft, Writing – review & editing, Data curation, Formal analysis. YL: Formal analysis, Writing – original draft, Writing – review & editing. CH: Data curation, Writing – review & editing. XL: Methodology, Writing – review & editing. CL: Investigation, Writing – review & editing. KL: Investigation, Writing – review & editing. XY: Funding acquisition, Writing – review & editing. MZ: Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Guangdong Provincial Department of Agriculture and Rural Affairs-Guangdong Agricultural Technical Service “LightCavalry” Project2024 (NITG20240253), Guangdong Provincial Department of Education’s distinctive innovation initiative (2023KTSCX128), and the National Natural Science Foundation of China (31902279).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1475208/full#supplementary-material
Footnotes
References
Adams, M. J., Lefkowitz, E. J., King, A. M. Q., Harrach, B., Harrison, R. L., Knowles, N. J., et al. (2017). Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2017). Arch. Virol. 162, 2505–2538. doi: 10.1007/s00705-017-3358-5
Alkhamis, M. A., Arruda, A. G., Morrison, R. B., and Perez, A. M. (2017). Novel approaches for spatial and molecular surveillance of porcine reproductive and respiratory syndrome virus (PRRSv) in the United States. Sci. Rep. 7:4343. doi: 10.1038/s41598-017-04628-2
An, T. Q., Tian, Z. J., Leng, C. L., Peng, J. M., and Tong, G. Z. (2011). Highly pathogenic porcine reproductive and respiratory syndrome virus. Asia. Emerg. Infect. Dis. 17, 1782–1784. doi: 10.3201/eid1709.110411
Brinton, M. A., Gulyaeva, A. A., Balasuriya, U. B. R., Dunowska, M., Faaberg, K. S., Goldberg, T., et al. (2021). ICTV virus taxonomy profile: Arteriviridae 2021. J. Gen. Virol. 102:001632. doi: 10.1099/jgv.0.001632
Cai, H., Zhang, H., Cheng, H., Liu, M., Wen, S., and Ren, J. (2023). Progress in PRRSV infection and adaptive immune response mechanisms. Viruses 15:1442. doi: 10.3390/v15071442
Chen, X. X., Qiao, S., Li, R., Wang, J., Li, X., and Zhang, G. (2023). Evasion strategies of porcine reproductive and respiratory syndrome virus. Front. Microbiol. 14:1140449. doi: 10.3389/fmicb.2023.1140449
Collins, J. E., Benfield, D. A., Christianson, W. T., Harris, L., Hennings, J. C., Shaw, D. P., et al. (1992). Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J. Vet. Diagn. Invest. 4, 117–126. doi: 10.1177/104063879200400201
de Lima, M., Pattnaik, A. K., Flores, E. F., and Osorio, F. A. (2006). Serologic marker candidates identified among B-cell linear epitopes of Nsp 2 and structural proteins of a north American strain of porcine reproductive and respiratory syndrome virus. Virology 353, 410–421. doi: 10.1016/j.virol.2006.05.036
Do, D. T., Park, C., Choi, K., Jeong, J., Nguyen, T. T., Nguyen, K. D., et al. (2015). Comparison of two genetically distant type 2 porcine reproductive and respiratory syndrome virus (PRRSV) modified live vaccines against Vietnamese highly pathogenic PRRSV. Vet. Microbiol. 179, 233–241. doi: 10.1016/j.vetmic.2015.06.013
Do, T. D., Park, C., Choi, K., Jeong, J., Nguyen, T. T., Nguyen, D. Q., et al. (2015). Comparison of experimental infection with northern and southern Vietnamese strains of highly pathogenic porcine reproductive and respiratory syndrome virus. J. Comp. Pathol. 152, 227–237. doi: 10.1016/j.jcpa.2014.12.002
Do, H. Q., Trinh, D. T., Nguyen, T. L., Vu, T. T., Than, D. D., Van Lo, T., et al. (2016). Molecular evolution of type 2 porcine reproductive and respiratory syndrome viruses circulating in Vietnam from 2007 to 2015. BMC Vet. Res. 12:256. doi: 10.1186/s12917-016-0885-3
Feng, Y., Zhao, T., Nguyen, T., Inui, K., Ma, Y., Nguyen, T. H., et al. (2008). Porcine respiratory and reproductive syndrome virus variants, Vietnam and China, 2007. Emerg. Infect. Dis. 14, 1774–1776. doi: 10.3201/eid1411.071676
Guo, Z., Chen, X. X., Li, X., Qiao, S., Deng, R., and Zhang, G. (2019). Prevalence and genetic characteristics of porcine reproductive and respiratory syndrome virus in Central China during 2016-2017: NADC30-like PRRSVs are predominant. Microb. Pathog. 135:103657. doi: 10.1016/j.micpath.2019.103657
Guo, B., Lager, K. M., Schlink, S. N., Kehrli, M. E. Jr., Brockmeier, S. L., Miller, L. C., et al. (2013). Chinese and Vietnamese strains of HP-PRRSV cause different pathogenic outcomes in United States high health swine. Virology 446, 238–250. doi: 10.1016/j.virol.2013.08.008
Hsueh, F. C., Kuo, K. L., Hsu, F. Y., Wang, S. Y., Chiu, H. J., Wu, M. T., et al. (2023). Molecular characteristics and pathogenicity of porcine reproductive and respiratory syndrome virus (PRRSV) 1 in Taiwan during 2019-2020. Life 13:843. doi: 10.3390/life13030843
Huong, V. T., Thanh, L. V., Phu, V. D., Trinh, D. T., Inui, K., Tung, N., et al. (2016). Temporal and spatial association of Streptococcus suis infection in humans and porcine reproductive and respiratory syndrome outbreaks in pigs in northern Vietnam. Epidemiol. Infect. 144, 35–44. doi: 10.1017/S0950268815000990
Jantafong, T., Sangtong, P., Saenglub, W., Mungkundar, C., Romlamduan, N., Lekchareonsuk, C., et al. (2015). Genetic diversity of porcine reproductive and respiratory syndrome virus in Thailand and Southeast Asia from 2008 to 2013. Vet. Microbiol. 176, 229–238. doi: 10.1016/j.vetmic.2015.01.017
Kamakawa, A., Ho, T. V., and Yamada, S. (2006). Epidemiological survey of viral diseases of pigs in the Mekong delta of Vietnam between 1999 and 2003. Vet. Microbiol. 118, 47–56. doi: 10.1016/j.vetmic.2006.07.003
Lee, H. S., Pham, T. L., Nguyen, T. N., Lee, M., and Wieland, B. (2019a). Seasonal patterns and space-time clustering of porcine reproductive and respiratory syndrome (PRRS) cases from 2008 to 2016 in Vietnam. Transbound. Emerg. Dis. 66, 986–994. doi: 10.1111/tbed.13122
Lee, H. S., Thakur, K. K., Bui, V. N., Bui, A. N., Dang, M. V., and Wieland, B. (2019b). Simulation of control scenarios of porcine reproductive and respiratory syndrome in Nghe An Province in Vietnam. Transbound. Emerg. Dis. 66, 2279–2287. doi: 10.1111/tbed.13278
Li, X., Wu, J., Tan, F., Li, Y., Ji, G., Zhuang, J., et al. (2016). Genome characterization of two NADC30-like porcine reproductive and respiratory syndrome viruses in China. Springerplus 5:1677. doi: 10.1186/s40064-016-3336-5
Li, C., Xu, H., Zhao, J., Gong, B., Sun, Q., Xiang, L., et al. (2022). Epidemiological investigation and genetic evolutionary analysis of PRRSV-1 on a pig farm in China. Front. Microbiol. 13:1067173. doi: 10.3389/fmicb.2022.1067173
Lu, Z. H., Archibald, A. L., and Ait-Ali, T. (2014). Beyond the whole genome consensus: unravelling of PRRSV phylogenomics using next generation sequencing technologies. Virus Res. 194, 167–174. doi: 10.1016/j.virusres.2014.10.004
Lunney, J. K., Benfield, D. A., and Rowland, R. R. (2010). Porcine reproductive and respiratory syndrome virus: an update on an emerging and re-emerging viral disease of swine. Virus Res. 154, 1–6. doi: 10.1016/j.virusres.2010.10.009
Luo, Q., Zheng, Y., Zhang, H., Yang, Z., Sha, H., Kong, W., et al. (2023). Research Progress on glycoprotein 5 of porcine reproductive and respiratory syndrome virus. Animals 13:813. doi: 10.3390/ani13050813
Nguyen, N. H., Tran, H. A. T., Nguyen, T. Q., Nguyen, P. B. T., Le, T. H. T., Lai, D. C., et al. (2022). Phylogenetic analysis of porcine reproductive and respiratory syndrome virus in Vietnam, 2021. Virus Genes 58, 361–366. doi: 10.1007/s11262-022-01912-w
Sha, H., Zhang, H., Luo, Q., Zheng, Y., Zhu, Q., Wang, N., et al. (2023). Variations in the NSP4 gene of the type 2 porcine reproductive and respiratory syndrome virus isolated in China from 1996 to 2021. Virus Genes 59, 109–120. doi: 10.1007/s11262-022-01957-x
Shi, M., Lam, T. T.-Y., Hon, C.-C., Murtaugh, M. P., Davies, P. R., Hui, R. K.-H., et al. (2010). Phylogeny-based evolutionary, demographical, and geographical dissection of north American type 2 porcine reproductive and respiratory syndrome viruses. J. Virol. 84, 8700–8711. doi: 10.1128/jvi.02551-09
Thuy, N. T., Thu, N. T., Son, N. G., Ha le, T., Hung, V. K., Nguyen, N. T., et al. (2013). Genetic analysis of ORF5 porcine reproductive and respiratory syndrome virus isolated in Vietnam. Microbiol. Immunol. 57, 518–526. doi: 10.1111/1348-0421.12067
Tian, K., Yu, X., Zhao, T., Feng, Y., Cao, Z., Wang, C., et al. (2007). Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526. doi: 10.1371/journal.pone.0000526
Wei, Z., Lin, T., Sun, L., Li, Y., Wang, X., Gao, F., et al. (2012). N-linked glycosylation of GP5 of porcine reproductive and respiratory syndrome virus is critically important for virus replication in vivo. J. Virol. 86, 9941–9951. doi: 10.1128/jvi.07067-11
Wensvoort, G., Terpstra, C., Pol, J. M. A., Terlaak, E. A., and Braamskamp, J. (1991). Mystery swine disease in the Netherlands: the isolation of Lelystad virus. Vet. Q. 13, 121–130. doi: 10.1080/01652176.1991.9694296
Yim-Im, W., Anderson, T. K., Paploski, I. A. D., Vander Waal, K., Gauger, P., Krueger, K., et al. (2023). Refining PRRSV-2 genetic classification based on global ORF5 sequences and investigation of their geographic distributions and temporal changes. Microbiol. Spectr. 11:e0291623. doi: 10.1128/spectrum.02916-23
Zhang, H., Luo, Q., He, Y., Zheng, Y., Sha, H., Li, G., et al. (2023). Research Progress on the development of porcine reproductive and respiratory syndrome vaccines. Vet. Sci. 10:491. doi: 10.3390/vetsci10080491
Zhao, J., Xu, Z., Xu, T., Zhou, Y., Li, J., Deng, H., et al. (2022). Molecular characterization of the Nsp 2 and ORF5s of PRRSV strains in Sichuan China during 2012-2020. Animals 12:3309. doi: 10.3390/ani12233309
Keywords: PRRSV, GP5 gene, homology, phylogeny, genetic variation
Citation: Li G, Li Y, He C, Liu X, Lv C, Liu K, Yu X and Zhao M (2024) Sequence analysis of the GP5 protein of porcine reproductive and respiratory syndrome virus in Vietnam from 2007 to 2023. Front. Microbiol. 15:1475208. doi: 10.3389/fmicb.2024.1475208
Edited by:
Xin Yin, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Chaoliang Leng, Nanyang Normal University, ChinaMotohiro Akashi, Seikei University, Japan
Copyright © 2024 Li, Li, He, Liu, Lv, Liu, Yu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingang Yu, eXV4aW5nYW5nNDUyNUAxNjMuY29t; Mengmeng Zhao, bWVuZ21lbmd6aGFvMjAyMUBmb3N1LmVkdS5jbg==
†These authors have contributed equally to this work
 Gan Li
Gan Li Yilong Li†
Yilong Li† Chen Lv
Chen Lv Xingang Yu
Xingang Yu Mengmeng Zhao
Mengmeng Zhao