- 1Department of Biological and Ecological Sciences, University of Tuscia, Viterbo, Italy
- 2Laboratory of Ecology of Marine Fungi (CoNISMa), University of Tuscia, Viterbo, Italy
- 3Department of Biological, Chemical and Pharmaceutical Sciences and Technologies (STEBICEF), University of Palermo, Palermo, Italy
- 4Microbiology Section, Department of Health Promotion, Mother and Child Care, Internal Medicine and Medical Specialties “G. D’Alessandro”, University of Palermo, Palermo, Italy
- 5Department of Eco-Sustainable Marine Biotechnology, Stazione Zoologica Anton Dohrn, Naples, Italy
- 6Department of Pharmacy, University of Naples “Federico II”, Naples, Italy
- 7Laboratory of Applied Marine Microbiology (CoNISMa), University of Tuscia, Viterbo, Italy
Introduction: The marine environment is extremely complex and exerts strong evolutionary pressure often leading to the appearance of microbial strains with new metabolic competencies. Microorganisms in marine ecosystems are still largely unknown and should be explored and conserved for biodiversity preservation, possible ecosystem restoring, and other applications. Biodiversity conservation should become a basic ecological strategy of particular significance in relation to global change. In this context, the present research aimed at exploring the culturable mycobiota associated with the jellyfish Pelagia noctiluca, never studied before. In addition, the isolated strains were tested for potential application (antimicrobial activity and presence of genes related to the production of secondary metabolites).
Methods: Five jellyfishes were collected in the coastal area of Giglio Island and processed to isolate epizoic fungi. The strains were identified using a polyphasic approach (morphological, physiological, and molecular) and their salt preference was also investigated. The antifungal and antibacterial activity were tested for each strain with agar plug diffusion test. The presence of some key genes related to the main pathways for the production of secondary metabolites in fungi, polyketide synthases (PKSs), and non-ribosomal peptide synthase (NRPSs), was also assessed.
Results: A total of 164 isolates were obtained; after the dereplication, 40 morphotypes, and 23 species were identified. The phylogenetic analyses suggested the presence of new taxa belonging to Pleosporales: two new genera and species, and a new species of Tamaricicola. The detected mycobiota showed a relatively high diversity, if compared to other epizoic fungal communities. All isolated strains were marine fungi as confirmed by their salt preference and marked euryhalinism. The genes related to the two main pathways for the production of secondary metabolites in fungi, PKSs and NRPSs, were identified in four and nine strains, respectively. The antimicrobial activity was revealed in 70% of the strains, including the new taxa. The abundance of bioactive strains may be related to the potential involvement of epizoic fungi in host defense strategies. Moreover, these strains could show a high potential for further biotechnological applications particularly in the case of new taxa. All strains are maintained in culture collections.
1 Introduction
The marine environment is extremely complex showing several physio-chemical factors (salinity, low water activity, high concentrations of ions, etc.) exerting strong evolutionary pressure, often leading to the appearance of strains with new metabolic competencies (Raghukumar, 2008; Damare et al., 2011; Burgaud et al., 2022). Biodiversity in marine ecosystems is still largely unknown, in particular concerning the structure, composition, and functionality of the microbial communities. This is partly due to the technical hurdles of collecting accurate samples, primarily in open areas, across various marine ecosystems (Templado et al., 2010). In addition, there is still relatively little microbiological research effort, mainly considering the great heterogeneity of habitats, substrata, and ecological niches (Costello and Chaudhary, 2017). The effects of marine habitat constraints are particularly noticeable in organisms like fungi that, to survive, must adopt specific strategies to exert environmental “biochemical” control (bioactive compounds) to compete with other organisms (Jones et al., 2022; Pasqualetti et al., 2022). Over the past few decades, a number of novel and/or uncommon enzymes and metabolites from marine fungi have been discovered (Parte et al., 2017; Birolli et al., 2019; Carroll et al., 2019; Pasqualetti et al., 2019; Giovannini et al., 2019; Botta et al., 2020; Pasqualetti et al., 2022).
The search for bioactive molecules has been carried out by traditional culture-dependent approaches for many years. The advent of molecular methods allowed for improvement and sped out this process with a functional gene-based strategy of molecular screening overcoming some limitations of the traditional culture-dependent approach. In addition, the study of the targeted genes consents to find silent or cryptic gene clusters that potentially encode several bioactive metabolites (Bentley et al., 2002). The two main pathways for the production of secondary metabolites involve the non-ribosomal peptide synthase (NRPS) and polyketide synthase (PKS). It has been reported that fungi with PKS or NRPS genes showed valuable secondary bioactive metabolites (Cox and Simpson, 2009; Vassaux et al., 2019).
Furthermore, applied mycological investigations often do not provide taxonomic and ecological information regarding the studied organisms (Pang et al., 2018; Le et al., 2019; Shaker et al., 2021; Ren et al., 2022; Ye et al., 2022). In particular, the fungi are frequently described as “marine-derived” without providing any information to state the strain’s marine habitus (Pasqualetti et al., 2020). However, these inquiries are crucial for understanding the fungal biodiversity and the exact role of marine fungi in the sea ecosystems. Thus, despite the recent advancements in marine mycology, the scientific community is just beginning to understand the importance of fungi in the marine system and glimpse the complexity of their ecosystem services (Amend et al., 2019; Grossart et al., 2019). Despite the recent improvements in marine mycology, several taxonomical and ecological issues still need to be addressed. Among them, the main topics are: a deeper knowledge of fungal diversity and a formal description of new taxa, a broader characterization of the mycobiota associated with poorly studied or neglected substrata, and a wider comprehension of the ecological role of marine biotic fungi.
It is crucial to underline that the unknown biodiversity represents an important gap in understanding ecosystem functioning and a huge potential resource that could not be lost. One of the main instruments to address these topics is to promote the collection activities and best conservation practices, mainly regarding new taxa, and new marine strains of common terrestrial species. Biodiversity conservation, in situ and ex situ, is one of the main actions to contrast global change (Hagerman et al., 2010).
Recently, studies on the mycobiota associated with various substrata evidenced that marine fungi, like the terrestrial ones, exhibit some degree of specialization. This is more evident in epibiotic species since some of them are ubiquitous, others are specialized and strongly associated with a specific substratum, while others have intermediate patterns (Pasqualetti et al., 2014; Bovio et al., 2018; Nguyen and Thomas, 2018; Pasqualetti et al., 2020). Gao et al. (2008) showed great differences in the mycobiota composition associated with two coexisting Hawaiian marine sponges. Similar species-specificity has been observed in fungi associated with seagrass and seaweeds (Wainwright et al., 2017; Pasqualetti et al., 2020), in mycorrhizae-like associations in seagrass roots (Borovec and Vohnik, 2018), and in scleractinian corals (Williams et al., 2014). Although the nature of these interactions remains unclear, bioactive molecules may play significant roles in fungal interactions with marine hosts. Zhou et al. (2011) suggested that the presence of PKS or NRPS genes in epi-/endo-zoic fungi could have a potential role in the chemical host defense (Raghukumar and Ravindran, 2012).
Fungi associated with marine animals have received special attention since they are frequently found to be a valuable biotechnological resource, mainly due to the production of bioactive molecules (Marchese et al., 2020). Only a limited number of studies considered jellyfish mycobiota, and in general, the investigations did not focus on communities but only on the production of secondary metabolites by single strains (Yue et al., 2022; Li et al., 2023). For instance, some of the strains isolated from Nemopilema nomurai produced several new bioactive compounds and a new molecule of great interest (epicoccamide) was isolated from an epi-biont of Aurelia aurita (Wright et al., 2003; La Kim et al., 2012a,b). Although jellyfish-associated marine fungi have shown to be an unexploited source of novel molecules of biotechnological importance, the role of these metabolites in the interactions with their host remains almost unknown. Therefore, much more effort should be made to study the fungal community associated with different species of jellyfish and to understand the nature of these interactions.
Pelagia noctiluca (Cnidaria: Scyphozoa), known as the “mauve stinger,” is considered one of the most common jellyfish in the Mediterranean Sea (Mariottini et al., 2008; Canepa et al., 2014). Pelagia noctiluca is a pelagic organism generally pink-, mauve-, or light brown-colored, with a phosphorescent bell characterized by a thick jelly hemispherical umbrella, up to 12 cm in diameter with an exoumbrella with several nematocyst warts. It presents eight adradial marginal tentacles alternated with eight marginal rhopalia, four inter-radial gonads, and oral arms (Tibballs, 2006; Durgham et al., 2016). Pelagia noctiluca is a predator that feeds on several zooplankters, including eggs and larvae of nektonic and benthic organisms. This jellyfish performs daily vertical migrations, staying at the surface at night and sinking in deeper water during the day. This vertical distribution pattern coincides with the migration of zooplankton, which represents its main prey (Tilves et al., 2016).
To the best of our knowledge, there are no studies regarding the P. noctiluca mycobiota.
This work was aimed at studying the culturable assemblages of fungi related to P. noctiluca collected in a Posidonia meadow located at the Giglio Island (Tyrrhenian Sea, Italy). Marine fungi were isolated, identified, and taxonomically characterized with a polyphasic approach including morphological, physiological, molecular, and phylogenetic analysis. In addition, for a better comprehension of the P. noctiluca mycobiota ecological role and its possible biotechnological applications, presence of NRPS and PKS genes and preliminary antimicrobial activity was studied.
2 Materials and methods
2.1 Strain isolation
Samples of the jellyfish Pelagia noctiluca were collected from the “Cala Cupa” cove (4222008.0300 N, 1055004.0900E), Giglio Island (Tuscan Archipelago, North Tyrrhenian Sea) at 10 m depth near a previously studied meadow in May 2019. After capture by scuba divers, five entire living jellyfishes were placed separately in sterile containers and then stored in a refrigerator (icebox), and brought to the laboratory. The five animals were asymptomatic and well-developed, with equivalent size (umbrella range 9–11 cm in diameter). The samples were prepared for the isolation of fungi as follows. The samples were washed (five time) with sterile seawater to remove sediments, debris, and transient microorganisms (i.e., propagules not strictly associated with jellyfish) and gently blotted with sterile filter paper to remove water (Yue et al., 2015). To evaluate the effectiveness of this procedure in removing all the transient propagules, all washing solutions were plated on Potato Dextrose Agar Sea water (PDAs: PDA 39 g—Sigma-Aldrich, St. Louis, MO, United States—dissolved in 1 L of filtered seawater) and incubated at 20°C; to assess possible propagule growth (no growth was observed plating the last rinsing solution). Samples of the umbrella (U) and oral arms (OA) were excised manually from each animal and separately placed into sterile glass Petri dishes (80 mm × 15 mm). The inner tissues (IT) of the animals, including gonads, were also removed and placed in sterile Falcon tubes (50 mL).
In order to optimize the isolation procedures for the umbrellas and the oral arms, some preliminary tests were carried out to compare the homogenization and the direct plating methods. The results demonstrated that the isolation yield using the homogenization method was definitely lower than that achieved by the direct plating. Moreover, all the fungal species isolated by the homogenization method were also found by direct plating. Accordingly, the isolation was carried out using two different techniques for the different districts of the animals: “direct plating” for OA and U, and “homogenization and plating” for IT, within 24 h from sampling.
Oral arms and U samples were cut into small pieces (10 pieces of ca. 5 mm2 for each substratum and sample) and directly placed on Petri dishes (five pieces for each plate 9 cm Ø) containing PDAs. Twenty Petri dishes were set up for OA and U.
For IT, 5 g (1 g from each animal) of tissues was added to 10 mL of sterile seawater and homogenized (ULTRA-TURRAX®-IKA, Staufen, Germany). The homogenate and 1:10 and 1:100 dilutions, in sterile sea water, were used to inoculate the plates: 0.5 mL of each dilution was spread in five Petri plates (9 cm Ø) containing PDAs. Five plates for each substratum and dilution were performed.
To prevent bacterial growth all media were supplemented with an antibiotic mix of 0.2 g/L streptomycin sulfate and 0.007 g/L penicillin G (Sigma-Aldrich, St. Louis, MO, United States). All procedures were carried out under sterile conditions.
A total of 50 plates were incubated at 20°C in the dark for up to 6 weeks and regularly monitored. All developed colonies were collected, transferred on appropriate media, and isolated in pure cultures. Rejection of duplicates (dereplication) was carried out by preliminary morphological, taxonomical and physiological analyses on different cultural media: PDAs, Corn Meal Agar Sea water (CMAs: 17 g CMA—Sigma-Aldrich dissolved in 1 L of filtered sea water), Malt Extract Agar Sea water (MEAs: 50 g MEA—Sigma-Aldrich dissolved in 1 L of filtered seawater), and Czapeck Sea water (CZs: 49 g CZ—Sigma-Aldrich, dissolved in 1 L of filtered seawater). However, every strain showing any morphological differences at macroscopic (colony) or microscopic (reproductive structures: asexual conidia, conidiphores, conidioma, or sexual structures) level were maintained as different morphotypes. The same dereplication approach was used considering physiological features revealed by the growth on different substrata (rate of growth, texture of the colonies, exudate, and pigment production).
All different morphotypes were cryo-conserved (Cryoinstant Mixed, VWR, Leuven, Belgium) and maintained in the culture collection of the “Laboratory of Ecology of Marine Fungi,” (CoNISMa) Department of Ecological and Biological Sciences (DEB), University of Tuscia, Viterbo, Italy. The strains of new taxa were also deposited at the international public institution, Mycotheca Universitatis Taurinensis (MUT).
2.2 Strain identification
A polyphasic approach that included morpho-physiological, molecular, and phylogenetic analyses was used to identify fungal strains.
2.2.1 Morpho-physiological identification
The strains were first identified at the genus level, and then at the species level when possible, using morpho-physiological identification based on macroscopic, microscopic, and physiological features (Ellis, 1971, 1976; Kohlmeyer and Kohlmeyer, 1979; Pitt, 1979; Sutton, 1980; Hyde et al., 2000; Klich, 2002; Domsch et al., 2007; Jones et al., 2015).
2.2.2 Molecular identification
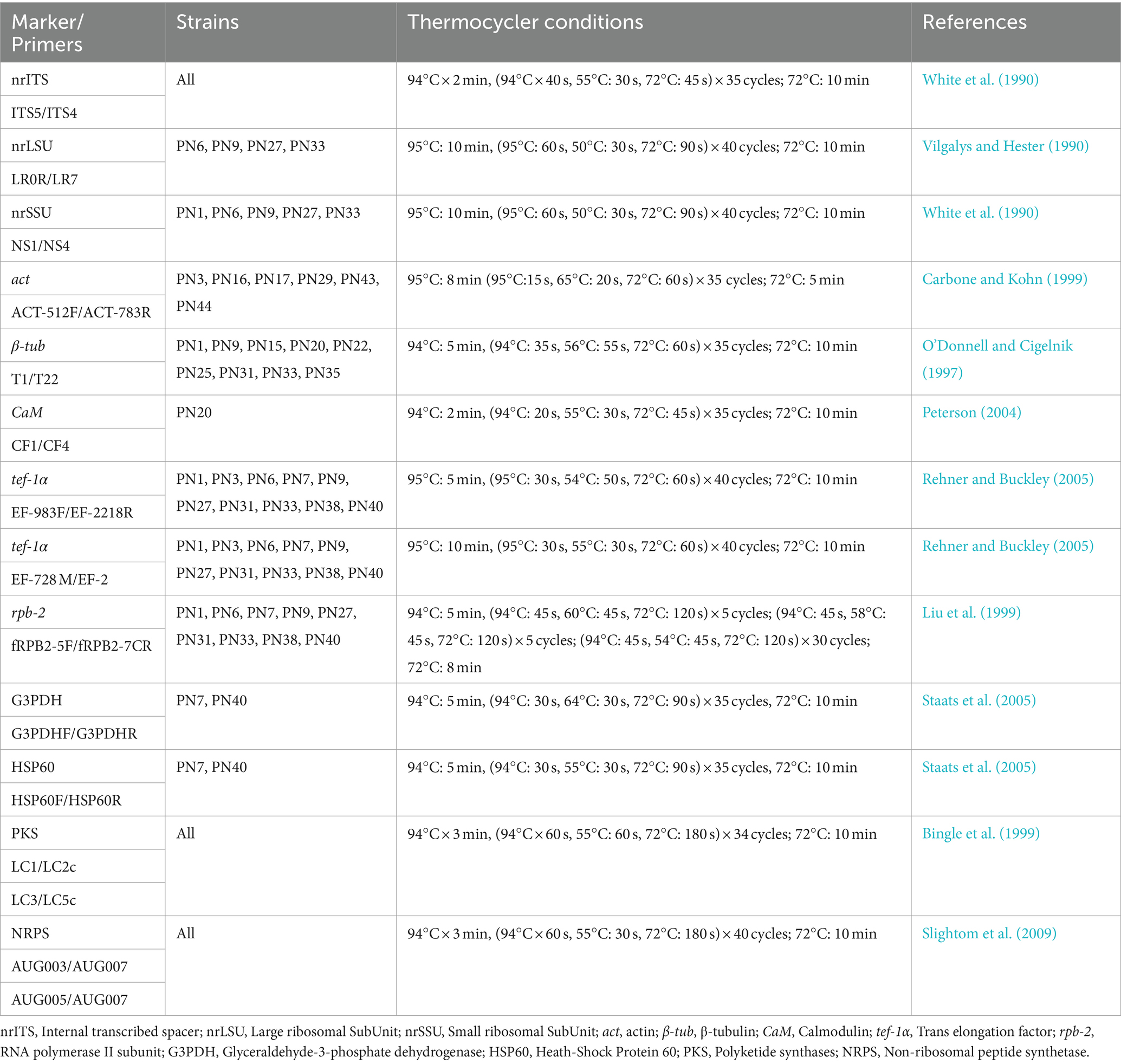
Genomic DNA was extracted using the ZR Fungal/Bacterial DNA MiniPrep Kit (Zymo Research, Irvine, CA, United States) according to the manufacturer’s instructions. The quantity of DNA was spectrophotometrically quantified (Qubit, Thermo Fisher Scientific, Waltham, MA, United States), and DNA samples were stored at −20°C. For each fungal strain, the ITS rDNA (ITS1-5.8S-ITS2) was amplified using the universal primers ITS5 and ITS4 (White et al., 1990). Based on the preliminary taxonomic assignment, other specific primers were selected as reported in Table 1. Amplifications were run in a 2720 Thermal Cycler (Applied Biosystem, Waltham, MA, United States) (Table 1).
PCRs were performed in a volume of 25 μL mixture containing: 0.5 μL of each primer (0.2 μM), 2.5 μL of MgCl2 (25 mM), 1.5 μL of 5 x buffer, 0.5 μL of dNTPs (0.2 mM), 0.2 μL of Go-Taq Polymerase (Promega, Madison, WI, United States), and 2 μL of genomic DNA; the final volume (25 μL) was reached adding ultrapure water. The PCR products were purified (E.Z.N.A. Cycle Pure kit Omega Bio-tek, Norcross, GA, United States) and sent to Eurofins Genomics (Ebersberg, Germany) for sequencing. Sequences obtained were checked and trimmed with Chromas Lite 2.1 program and then compared with those deposited in GenBank NCBI (National Center for Biotechnology Information, Bethesda, MD, United States). Newly generated sequences were deposited in GenBank, the accession numbers are reported in Table 2.
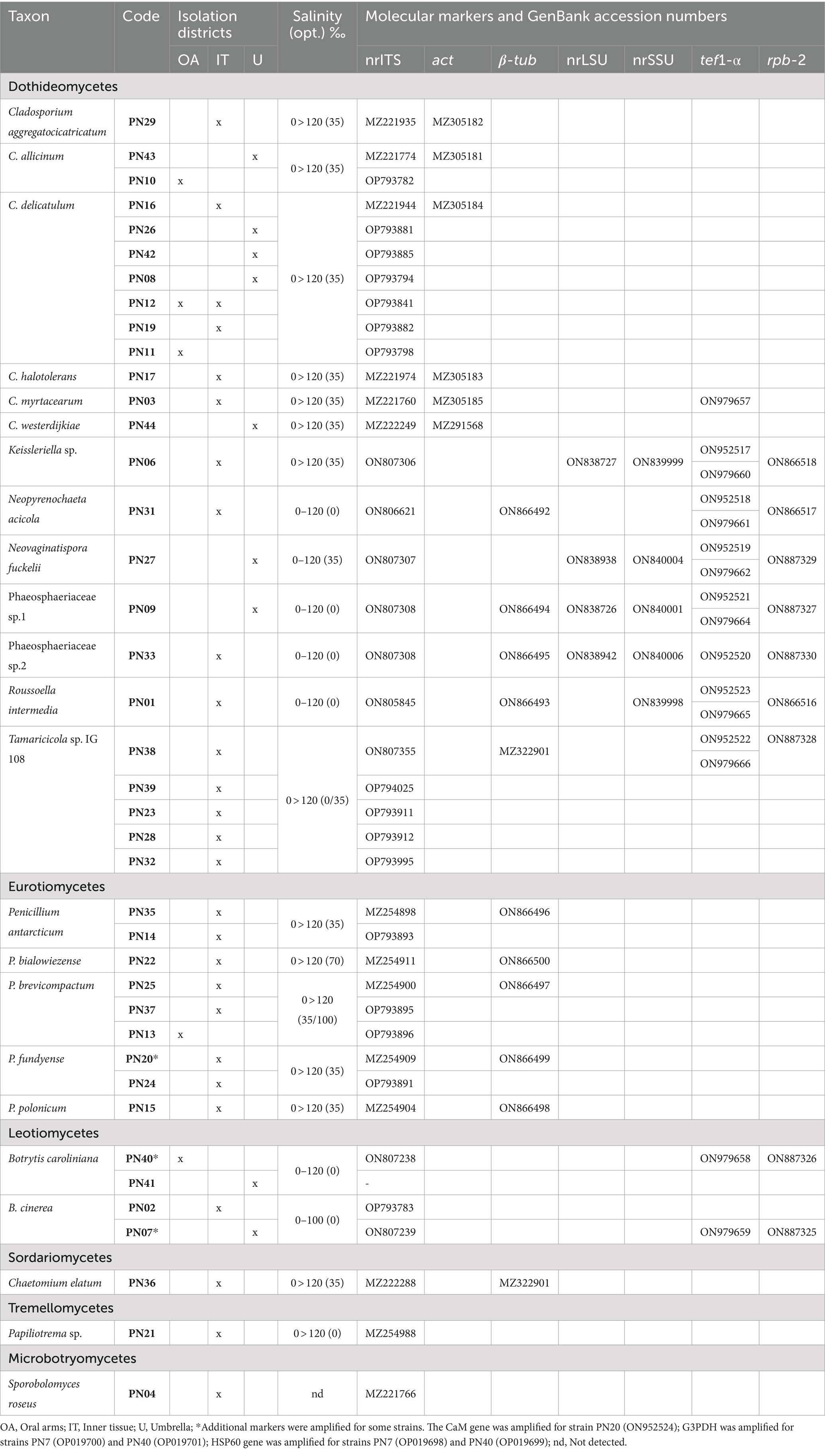
Table 2. Taxonomical attribution of the isolates from Pelagia noctiluca; strains and isolation districts, physiological characters (salinity tolerance, in brackets salinity optimum) and GenBank accession number of the obtained sequences.
Taxonomic assignment was based on similarity (NCBI-BLASTn algorithm) to reference sequences available at GenBank; similarity values higher than 98% (e-value > e_100) were considered reliable, attributions were confirmed by morphological and phylogenetic analyses.
Phylogenetic analysis was carried out on the nrITS region for all morphotypes; additional analyses, based on the actin (act) for Capnodiales, and β-tubulin (β-tub) for Eurotiales, were also performed.
Multi-locus phylogenetic analyses were performed, using concatenate datasets, based on nrITS region, nrLSU and nrSSU for Pleosporales and on rpb-2, G3PDH, and HSP60 for Leotiomycetes (Staats et al., 2005; Phookamsak et al., 2019). The references of all sequences included in the datasets are reported in Supplementary Tables S1–S5.
Sequences were aligned with the Clustal X 2.1 software (Thompson et al., 1997) using the default parameters. Alignments were checked and edited with BioEdit Alignment Editor 7.2.5 (Hall, 1999) and manually adjusted in MEGA X. For the multi-locus phylogenetic analyses, alignments of different markers were concatenated into a single data matrix with MEGA X. Phylogenetic inference was estimated using Maximum Likelihood (ML) and Bayesian inference (BI) as previously reported by Braconcini et al. (2024). ML analyses (10,000 bootstrap replicates) were run using the IQ-TREE web server, the best-fit evolution model was inferred using the ModelFinder in IQ-TREE; different models were used for each partition in the concatenated matrices (Trifinopoulos et al., 2016; Kalyaanamoorthy et al., 2017). The best-scoring trees were visualized using FigTree v.1.4 (FigTree (2024): Tree Figure Drawing Tool Version 1.4.4., 2024). The BI was performed with MrBayes 3.2.7 under different models for each partition as evaluated by jModelTest 2 (Posada, 2008) using Bayesian Information Criterion (Ronquist et al., 2012). The alignment was run for 1 million generations in two independent runs, each with four Markov Chains Monte Carlo (MCMC) and sampling every 100 iterations. As a “burn-in” measure, the first 25% of generated trees were discarded. MrBayes’ “sumt” function was used to generate a consensus tree, and Bayesian posterior probabilities (BYPP) were calculated.
Sequence alignment and phylogenetic tree were deposited in TreeBASE version 2 (2024) (submission ID: 31592). For each fungal species, the current name according to the “Index Fungorum” database (Index Fungorum database and web site, 2024) and recent literature was reported.
2.3 Determination of growth at different salinities
The effect of salinity on fungal growth was investigated on PDA modified with different NaCl concentrations: 0, 35, 70, 105, and 120‰. The plates (90 mm Ø) were inoculated with a single agar disk (diameter 3 mm) cut from the actively growing margin of 7-day-old colonies grown on PDAs. Plates were incubated at 25°C and colony diameter was measured daily for 7–30 days (in relation to the different growth rates) to estimate fungal growth. Experiments were carried out in triplicate.
2.4 Antimicrobial bioassay
2.4.1 Test microorganisms
All tests were performed on saline media. The halotolerant strains of Bacillus pumilus KB66, Pseudomonas fluorescens KB6 (Pesciaroli et al., 2012) and Penicillium griseofulvum TSF04 were used as test organisms to avoid interferences related to salt presence. The test strains were retrieved from the culture collection of microorganisms of the Laboratory of Ecology of Marine Fungi. Strains had been revitalized and sub-cultured on Luria Bertani Sea water (LBs) broth (25 g LB—Sigma-Aldrich dissolved in 1 L of filtered seawater) and MEAs for the bacterial and fungal strain, respectively.
To produce standardized inoculum, bacteria were grown in LBs at 30°C in an orbital shaker at 150 rpm. After overnight incubation, the bacteria were diluted by the addition of fresh media to an optical density (OD) of 0.8 at 680 nm. Penicillium griseofulvum was grown on MEAs for 7 days at 25°C. Conidia suspensions were prepared in sterile filtered seawater supplemented with 0.01% of Tween 80 and diluted to a final inoculum ranging from 0.5 × 105 to 1.0 × 105 cells/mL. Test plates were inoculated by spreading 100 μL of bacteria culture and 300 μL of spore suspension.
2.4.2 Agar plug diffusion method
The screening for antifungal and antibacterial activity was carried out with the agar plug diffusion method. Fungal strains were cultured on different cultural media (PDA, PDAs, MEA, MEAs, CYA, and CYAs; CYA medium is CZ supplemented with 5 g/L of Yeast Extract) for 90 days at 25°C. The fungal strains were grown for a relatively long period to enhance the production of secondary metabolites as evidenced by some preliminary tests. Agar plugs (3 cm2) were aseptically cut at the front of the colony and deposited on the agar surface of plates previously inoculated with the test organisms. Plugs of different cultural media were used as negative control. After incubation (24 h, 30°C for bacteria, and 48–72 h, 25°C for fungi) the antimicrobial activity was estimated by measuring the inhibitions halos.
2.5 PKS and NRPS genes screening
Seven PCR primers, designed for the highly conserved sequences of β-ketoacyl synthase (PKS) domains and the most conserved A domain of the NRPS gene, were used. PCR were performed as previously reported in section 2.2.2. PCR conditions and primers are reported in Table 1.
2.6 Statistical analysis
Principal Component Analysis (PCA) on salt growth preferences were made on the normalized colony diameters on the maximum growth (CANOCO Engine Version 5.2 2017; Šmilauer and Lepš, 2014).
3 Results and discussion
3.1 Strain isolation and identification
One hundred and sixty-four fungal isolates were obtained from samples of P. noctiluca. The fungal assemblages associated to umbrella and oral arms of each jellyfish were quite homogeneous, therefore they were merged to describe the P. noctiluca mycobiota. Considering the animals’ district, the inner tissues hosted the highest number of fungal colonizers (89 isolates), followed by umbrella (48 isolates), and oral arms (27 isolates). According to the morpho-physiological dereplication, 40 morphotypes were obtained. The morphotypes were characterized by molecular analysis using the nrITS region, the universal DNA barcode for fungi (Bergerow et al., 2010; Schoch et al., 2012). The analysis of the sequences (BLASTn) and subsequent phylogenetic analysis (Figure 1) allowed to group the 40 morphotypes in 23 distinct taxa. Nine morphotypes were identified at the species level, 26 at the genus level, and for the remaining five strains, all belonging to the Pleosporales order, no further attribution was possible with nrITS region. Considering these results, additional genes were analyzed to attain the attribution at the species level.
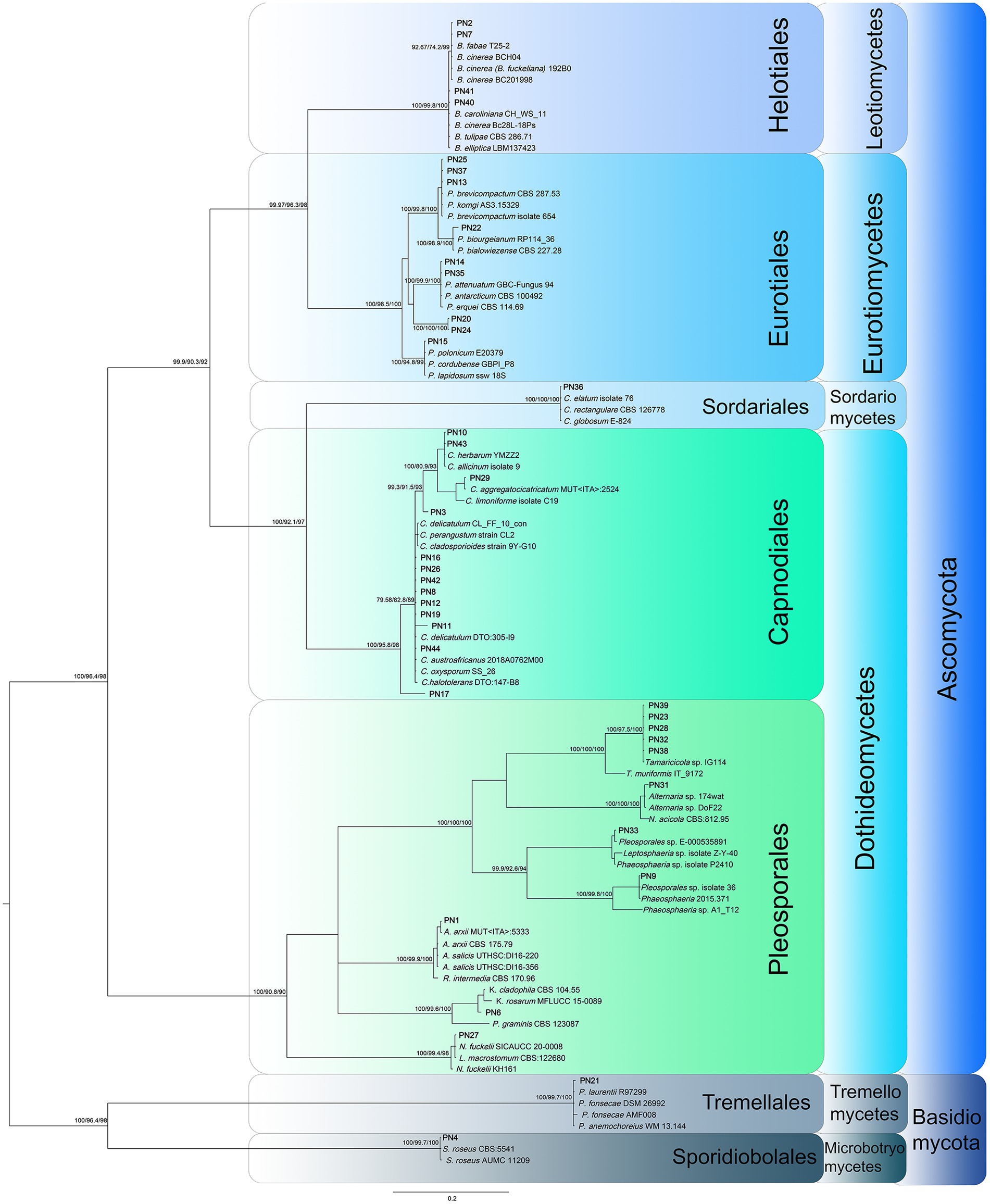
Figure 1. Phylogenetic inference based on ITS1, 5.8S rDNA, and ITS2 partitioned dataset was inferred using the Bayesian method. Branch numbers indicate BYPP values, and sH-aLRT and BP values from RA-ML analysis. The Bayesian Inference was performed under model as evaluated by jModelTest 2 using Bayesian Information Criterion (partition 1: TPM1uf + G, G = 4.445; partition 2: HKY + G, G = 0.140; partition 3: TrNef+G, G = 0.813).
All Cladosporium and Penicillium isolates were identified at the species level by the actin and β-tubulin genes, respectively (Supplementary Figures S1, S2; Bensch et al., 2012; Visagie et al., 2016), with the only exception of PN20. For this morphotype, the analysis of the CaM gene was also assessed for its species attribution to P. fundyense. The β-tubulin gene was also used to confirm the morphological identification of Chaetomium elatum PN36. The two Botrytis strains PN7 and PN40 were analyzed by a polyphasic approach including both molecular and morphological analyses. A multi-locus phylogenetic analysis was carried out using the rpb-2, HSP60, and G3PDH markers, as reported by Staats et al. (2005) to distinguish the most closely related neighbor species (Supplementary Figure S3). The strain PN7 is included in a well-supported group with three different species: B. cinerea, B. eucalypti, and B. pelargonii. Nevertheless, as already observed by Garfinkel (2021), these species must be considered conspecific, thus the strain PN7 was attributed to B. cinerea; this attribution was also confirmed by morphological analysis. Also, PN40 is grouped with three different species B. fabiopsis, B. galanthina, and B. caroliniana that have different morphological characters including presence/absence of sclerotia and conidial dimensions (Li et al., 2012). These features lead to the attribution of PN40 to B. caroliniana since it does not produce sclerotia, and conidia dimensions are 10–14 × 6–9 μm.
A multi-locus phylogenetic analysis based on the nrLSU, nrSSU, and nrITS was used to analyze the 11 morphotypes included in the Pleosporales in order to determine their taxonomic position within this large order (Figure 2). Subsequently, different molecular markers (β-tub, tef1-α, and/or rpb-2) were selected and analyzed for each family to reach specific attribution of the strains (Table 2). The pleosporelean morphotypes were included in six families. The strains included in Lentitheciaceae (PN6), Lophiostomataceae (PN27), Neopyrenochaetaceae (PN31), and Roussellaceae (PN1) were attributed to Keissleriella sp. (PN6), Neopyrenochaeta acicola (PN31), Neovaginatispora fuckelii (PN27), and Roussoella intermedia (PN1). For PN6, the analyses carried out on nrITS, nrLSU, and nrSSU did not allow to distinguish the morphotype from the related species K. cladophila, K. camporesi, K. sparticola, and K. rosarum. The tef1-α sequences, efficiently used to attribute some species in the genus Keissleriella, are not available for the species strictly related to PN6; in addition PN6 is a mycelia sterilia and morphological analysis is useless for species attribution. It is worth to note that some authors reported the inconsistency of some species included in the genus Keissleriella and suggested the necessity of a large revision of this genus (Hyde et al., 2020).

Figure 2. Pleosporales: Phylogenetic inference based on nrLSU, nrSSU, and nrITS (ITS1, 5.8S rDNA, and ITS2) combined dataset was inferred using Maximum Likelihood method (RA-ML), Hysterobrevium baoshanense and H. smilacis were used as outgroup. Branch numbers indicate BYPP values from Bayesian analysis, and sH-aLRT and BP values from RA-ML analysis. The Maximum Likelihood inference was performed under different models for each partition as evaluated by ModelFinder (IQ-TREE web server): K2P + I + G4 (partition 1), IM2 + F + I + G4 (partition 2), TIM2e + G4 (partition 3); K2P + I (partition 4); TIM2e + I + G4 (partition 5).
The remaining Pleosporales strains included in Pleosporaceae (PN38, PN39, PN23, PN28, and PN32) and Phaeosphaeriaceae (PN9 and PN33) families can be all ascribed to new taxa. The pleosporeacean strains were all included in a strong supported clade with a strain of Tamaricicola sp. IG108, previously collected in the same area (Pasqualetti et al., 2020). The clade is strictly related with the monospecific genus Tamaricicola and seem to represent a new lineage inside this genus (Figure 2).
The strains PN9 and PN33 represent two new lineages (genera) inside the family Phaeosphaeriaceae (Figure 2). This is also strongly supported by the analyses of genetic distances among PN9 and PN33 and the closest taxa of the family for all tested markers (Supplementary Tables S6, S7).
3.2 Growth at different salinities
According to Pasqualetti et al. (2020) analyses of salinity preferences were carried out to exclude, from the P. noctiluca fungal assemblage, the non-marine strains possibly present as propagules derived from terrestrial contamination and unable to growth at the sea salinity. All isolated strains are able to grow at sea salinity and included both halotolerant (36%) and facultative halophilic (64%) fungi (slight—and moderate–facultative halophiles, 56 and 8%, respectively; Gunde-Cimerman and Zalar, 2014; Raghukumar, 2017; Pasqualetti et al., 2020). However, the majority of strains showed their optimal growth at the sea salinity or higher (Table 2). Multivariate analysis carried out on growth data at different salinities, clearly identified four main groups (Figure 3). Strains characterized by optimal growth in the absence of salt or at salinities lower than that of seawater (halotolerant) were included in group I (PN7 and PN40) and group II (PN1, PN6, PN9, PN31, and PN33): Group I presented at sea salinity a growth reduction of 30% and Group II of 20% from the optimum. These strains showed a relatively low halotolerance with an 80–95% growth reduction (from the optimum) at salinities of 120‰. Isolates included in group III (PN16, PN36, PN29, PN43, PN44, PN3, PN35, PN15, PN20, and PN38) showed optimal growth at the sea salinity (35‰) and could be considered as facultative halophiles. Group IV included three strains, PN17, PN22, and PN25 presenting optimal growth at 70‰ (moderate-halophiles) and a remarkable euryhalinism, with strain PN25 showing an optimal growth in the range 35–105‰.
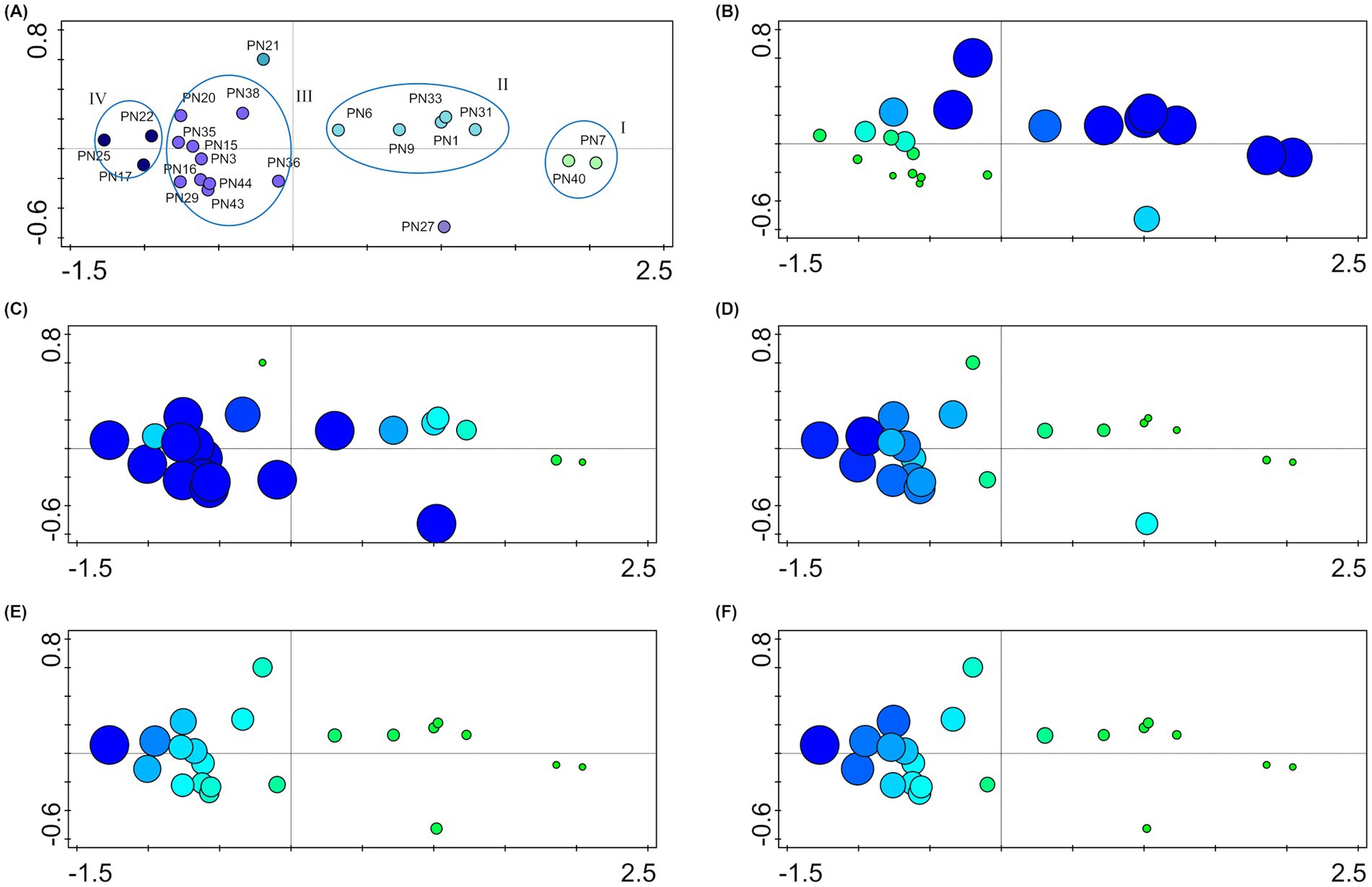
Figure 3. Salinity preferences of the studied strains. (A) Multivariate analysis (PCA) carried out on the growth data at different salinities. The plot shows strain distribution related to salinity preferences; four main groups (I, II, III, and IV) are identified from the ellipses. (B–F) Attribute plots in ordination space: dimensions and color gradient of the symbols indicate the percentage of growth of each strain at different salinities: 0‰ (B), 35‰ (C), 70‰ (D), 105‰ (E), and 120‰ (F).
3.3 Mycobiota of Pelagia noctiluca
Overall, the mycobiota of P. noctiluca was mainly composed of Ascomycota (21 taxa; 91.3%) with a small contribution of Basidiomycota (two taxa, 8.7%). Considering the phylum Ascomycota, Dothideomycetes and Eurotiomycetes were the most representative classes with 13 and 5 taxa, respectively. This agrees with previous reports describing these classes as the most representative in marine environments, considering both the number of taxa and abundance (Gonçalves et al., 2022; Wijayawardene et al., 2022).
As for the phylum Basidiomycota, only two taxa (one strain of Tremellomycetes and one of Microbotryomycetes) were identified. Although the result is in line with those previously reported in marine habitats/substrata, it must be also considered that the Basidiomycota could be underestimated due to the use of not selective techniques (Ding et al., 2011, Gonçalves et al., 2022). With respect to the isolation districts, the highest number of species (18) and colonies (89) were associated with the inner tissues, while seven and four species were recovered from the umbrella and oral arms, respectively. Oral arms showed a scarce colonization (27 colonies) and none of the four species found were exclusively associated with this district: B. caroliniana and C. allicinum were isolated from OA and U, P. brevicompactum from OA and IT and C. delicatulum was detected in all districts. On the contrary, the 43 and 83% of colonizers recovered in the umbrella and the inner tissues, respectively, were isolated only from one district (Table 2).
The most represented genera, in terms of species, were Cladosporium (six species) and Penicillium (five species). This result is not surprising since these genera are well adapted to marine environments and appear to be generally ubiquitous in the oceans (Raghukumar, 2017). Some of these species were frequently recorded in several marine habitats and substrata. For example, P. antarcticum and P. brevicompactum are well-known as saprotroph in seawater and as biont/saprotroph of algae, phanerogams, and animals (Paz et al., 2010; López-Legentil et al., 2015; Bovio et al., 2017, 2018, 2019; Marchese et al., 2020, 2021). Other species (i.e., C. aggregatocicatricatum, C. westerdijkiae, and P. bialowienzense) have been recorded on fewer marine substrata, even if they are widespread in terrestrial habitats (Bovio et al., 2017, 2018; Maamar et al., 2020; Ben-Dor Cohen et al., 2021).
Finally, the 22% of taxa (including the new genera and species) were detected for the first time in the marine environment. The high percentage of new strains detected emphasize the gap in our knowledge about real diversity of fungi in the sea. In the meantime, it underlines the great importance to focus on conservation strategies of biodiversity, considering also present and future prospectives linked to dramatic global changes. In this context, the maintaining of marine species in culture collections of microorganisms represents an important tool for their preservation, possible ecosystem restoring, and other applications. Under the ecological point of view, these species might be exclusively associated with P. noctiluca and considered as species-specific (“specialized”) organisms (Rambelli et al., 2004). However, the available information regarding the associations of these fungi with jellyfishes and other marine substrata is too scant to allow more than a hypothetical consideration. To the best of our knowledge, P. noctiluca was never studied before. Moreover, it is worth noting that most mycological studies on jellyfishes focused only on single strains and their biotechnological potential, without providing any ecological information on the single species or on the whole mycobiota (Wright et al., 2003; Liu et al., 2011, 2012; La Kim et al., 2012a,b; Liu et al., 2013; Yue et al., 2015; Tao et al., 2016; Liu et al., 2018; Regalado and Ramirez, 2019; Tan et al., 2019; Yue et al., 2022; Li et al., 2023). Globally only 14 species belonging to Cladosporium, Purpureocillium, Tilletiopsis, Aspergillus, Epicoccum, Paecilomyces, Penicillium, and Phoma were collected from the different jellyfish species studied (A. aurita, N. nomurai, and Catostylus sp.) (Supplementary Table S8). However, the only study focusing on a jellyfish mycobiota regarded N. nomurai, which showed a relatively low diversity in the fungal assemblage represented by just seven isolates and five species (Yue et al., 2015).
In the present study, 23 different fungal species were found on P. noctiluca and none of them was detected in the mycobiota of the other studied jellyfishes. On the contrary, some common genera such as Cladosporium and Penicillium were observed. Nevertheless, the significance of this genera in the mycobiota jellyfish characterization appears of low relevance considering, as already observed, that species of these genera are generally ubiquitous in the seas. Lastly, it is worth noting that the mycobiota of P. notiluca showed a relatively high diversity, if compared to other marine epizoic fungal communities (López-Legentil et al., 2015; Bovio et al., 2019).
3.4 Screening for potential bioactive molecules
The potential biotechnological interest of the isolated fungi, within a blue growth strategy perspective, has been tested by both plate screening and molecular survey of the target genes PKS and NRPS. These genes are commonly used to find species of interest for production of bioactive metabolites (Bentley et al., 2002; Molnár et al., 2010) since they are involved in the biosynthesis of a broad range of compounds such as antibiotics, antifungal, antiviral, anticancer, mycotoxins, antifouling, and pigments (Raimundo et al., 2018; Stroe et al., 2024). Moreover, the presence of these genes in fungi associated to marine macro-organisms (i.e., sponges, corals, algae, and mangrove plants) could suggest their potential roles in the host chemical defense process (Zhou et al., 2011; Hafez Ghoran et al., 2023).
A preliminary screening (agar plug diffusion method) was carried out to evaluate the potential production of antimicrobial molecules by the isolated species cultivated on different substrata (Table 3). Different media were utilized to promote different metabolic pathways and metabolites (Pinedo-Rivilla et al., 2022). Globally, the 70% of the strains exhibited inhibitory activity against one or two of the tested microorganisms (B. pumilus; P. fluorescens; P. griseofulvum). In general, no activity was observed against P. fluorescens, 11 strains showed antibacterial activity against B. pumilus and 11 strains antifungal activity. Five strains (PN16, PN31, PN33, PN38, and PN40), were active against both P. griseofulvum and B. pumilus gram-positive bacteria. Strains included in the order Pleosporales (PN9, PN33, PN27, and PN38) showed the strongest antibacterial activity, while the two Penicillium strains PN22 (P. bialowienzense) and PN25 (P. brevicompactum) showed the strongest activity against P. griseofulvum. The antifungal activity showed by various fungal species could be due to the production of cell-wall degrading enzymes (i.e., chitinases); however, PN22 and PN25 did not show any chitinolytic activity (data not shown). Considering the different substrata, in general, the most efficient to induce the production of bioactive molecules was PDAs (81% of active strain), only B. caroliniana showed activity when growing on all saline media, while only P. brevicompactum showed antifungal activity in a salt-free medium (CYA). Even if the antimicrobial activity tested by the plug diffusion method is not exhaustive, this preliminary result confirms that jellyfish-associated fungi can be a good source of active metabolites (Liu et al., 2013; Yue et al., 2015).
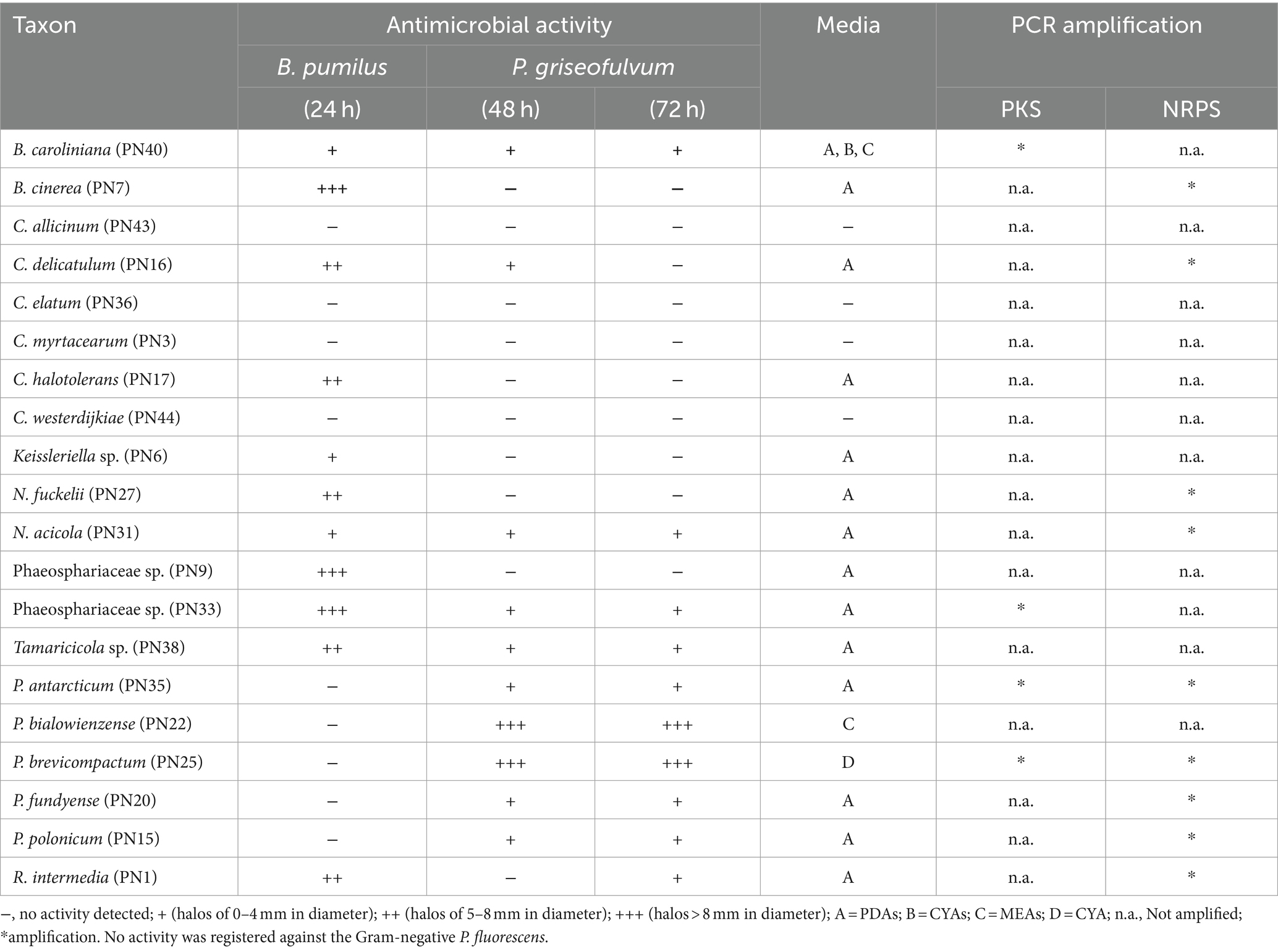
Table 3. Screening for antimicrobial activity: bioassay of antimicrobial activity of PN strains against Bacillus pumilus and Penicillium griseofulvum, Culture media, amplification of PKS, and NRPS genes.
The results of the molecular survey are generally in accordance with the antimicrobial activity observed with cultural methods. The 70% of active strains presented PKS and/or NRPS genes: NRPS was present in nine strains, while PKS only in four strains and only P. brevicompactum and P. anctarticum presented both genes. Considering that Cladosporium and Penicillium were the most representative genera in the P. noctiluca fungal assemblage and that the primers used were designed on these taxa, it was quite surprising that the number of positive strains was relatively low. This is particularly true considering that most Cladosporium and Penicillium strains are known to produce various bioactive compounds related to these genes (El Hajj Assaf et al., 2020; Li et al., 2024). In particular, in this study for the Cladosporium genus only C. delicatulum was positive to NRPS and none of them to PKS. Finally, it is worth noting that P. bialowienzense, showing the strongest antifungal activity, was negative to both genes and, as already observed, to chitinolytic activity; indicating that its biological activity is probably related to others metabolic pathways.
Some new (Phaeosphaeriaceae sp. PN33) or neglected species (i.e., N. fuckelii and R. intermedia) that have never been studied for the presence of these genes, positive in this study, deserve further investigation.
In addition, the four strain (PN36, PN3, PN44, and PN43) that did not show activity were also negative to the gene amplification. It is possible that these results underestimated the real presence of these target genes, since the primers used for their amplification were designed for other specific genera, i.e., Penicillium, Aspergillus, and Cladosporium (Bingle et al., 1999). Hence, it is possible that they were unable to amplify homologous genes in other genera and species, in particular considering new taxa (Lee et al., 2001).
4 Conclusion
This work describes for the first time the culturable mycobiota associated with the jellyfish Pelagia noctiluca and highlights the relatively high biodiversity of marine fungi associated with this jellyfish. In addition, the study evidenced the presence of several marine strains, which were detected for the first time in the sea environment. Moreover, the identification of new taxa enphasize the gap in our knowledge about the real entity of the fungal impact in the sea. In the meantime, the work underlines the great importance to focus the research on conservation strategies of the fungal biodiversity, considering also the prospectives linked to rapidly increasing global changes. In this context, the conservation of marine strains in culture collections of microorganisms, according to the most recent good practices, represents an important strategy for their preservation, possible ecosystem restoring, and a number of potential applications also in the One Health framework. Here, the new marine strains appeared of particular interest for blue-biotechnological perspectives, since the screening results suggested that some of them could be used for the production of new drugs. However, further investigations need to be performed to evaluate the actual biotechnological value of the studied strains. This work evidenced the potential of the epizoic fungi related to P. noctiluca and lead the way to further investigations, not only to formally describe the new taxa, but also to get a deeper picture of the whole microbiota of this jellyfish, including molecular studies to depict the non-culturable moiety of the community. Studies in this sense are already in progress.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because the studied animals are not higher invertebrate and no ethical approval was required.
Author contributions
MP: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing, Data curation, Formal analysis, Investigation. MB: Data curation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing. PB: Data curation, Writing – review & editing, Methodology, Visualization. SG: Writing – review & editing, Formal analysis, Investigation, Writing – original draft. DS: Writing – review & editing, Data curation, Methodology, Visualization. DF: Data curation, Methodology, Visualization, Writing – review & editing. GD: Data curation, Methodology, Visualization, Writing – review & editing. SM: Data curation, Methodology, Visualization, Writing – review & editing. MF: Methodology, Writing – review & editing, Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The work was partially supported by the Programma Operativo Nazionale (PON-DOT1335703), Ricerca e Innovazione of the Italian Ministry of University and Research (MUR), and by the “Progetto di Ricerca di Interesse Nazionale” (PRIN)-Production and characterization of new bioactive molecules against emerging and/or multidrug-resistant pathogens by neglected polyextremophilic marine fungi (MYCOSEAS, no. 2022MPTT35).
Acknowledgments
The Research project implemented under the National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.4—Call for tender No. 3138 of 16 December 2021, rectified by Decree n.3175 of 18 December 2021 of Italian Ministry of University and Research funded by the European Union—Next Generation EU. Project code CN_00000033, Concession Decree No. 1034 of 17 June 2022 adopted by the Italian Ministry of University and Research, CUP J83C22000860007, Project title “National Biodiversity Future Center—NBFC.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1473269/full#supplementary-material
References
Amend, A., Burgaud, G., Cunliffe, M., Edgcomb, V. P., Ettinger, C. L., Gutiérrez, M. H., et al. (2019). Fungi in the marine environment: open questions and unsolved problems. MBio 10, 10–1128. doi: 10.1128/mBio.01189-18
Ben-Dor Cohen, E., Ilan, M., and Yarden, O. (2021). The culturable mycobiome of mesophotic Agelas oroides: constituents and changes following sponge transplantation to shallow water. J. Fungi 7:567. doi: 10.3390/jof7070567
Bensch, K., Braun, U., Groenewald, J. Z., and Crous, P. W. (2012). The genus Cladosporium. Stud. Mycol. 72, 1–401. doi: 10.3114/sim0003
Bentley, S. D., Chater, K. F., Cerdeño-Tárraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3 (2). Nature 417, 141–147. doi: 10.1038/417141a
Bergerow, D., Nilsson, H., Unterseher, M., and Maier, W. (2010). Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Appl. Microbiol. Biotechnol. 87, 99–108. doi: 10.1007/s00253-010-2585-4
Bingle, L. E., Simpson, T. J., and Lazarus, C. M. (1999). Ketosynthase domain probes identify two subclasses of fungal polyketide synthase genes. Fungal Genet. Biol. 26, 209–223. doi: 10.1006/fgbi.1999.1115
Birolli, W. G., Lima, R. N., and Porto, A. L. (2019). Applications of marine-derived microorganisms and their enzymes in biocatalysis and biotransformation, the underexplored potentials. Front. Microbiol. 10:1453. doi: 10.3389/fmicb.2019.01453
Borovec, O., and Vohnik, M. (2018). Ontogenetic transition from specialized root hairs to specific root-fungus symbiosis in the dominant Mediterranean seagrass Posidonia oceanica. Sci. Rep. 8:10773. doi: 10.1038/s41598-018-28989-4
Botta, L., Saladino, R., Barghini, P., Fenice, M., and Pasqualetti, M. (2020). Production and identification of two antifungal terpenoids from the Posidonia oceanica epiphytic Ascomycota Mariannaea humicola IG100. Microb. Cell Factories 19, 184–110. doi: 10.1186/s12934-020-01445-7
Bovio, E., Garzoli, L., Poli, A., Prigione, V., Firsova, D., McCormack, G. P., et al. (2018). The culturable mycobiota associated with three Atlantic sponges, including two new species: Thelebolus balaustiformis and T. spongiae. Fungal Syst. Evol. 1, 141–167. doi: 10.3114/fuse.2018.01.07
Bovio, E., Gnavi, G., Prigione, V., Spina, F., Denaro, R., Yakimov, M., et al. (2017). The culturable mycobiota of a Mediterranean marine site after an oil spill: isolation, identification and potential application in bioremediation. Sci. Total Environ. 576, 310–318. doi: 10.1016/j.scitotenv.2016.10.064
Bovio, E., Sfecci, E., Poli, A., Gnavi, G., Prigione, V., Lacour, T., et al. (2019). The culturable mycobiota associated with the Mediterranean sponges Aplysina cavernicola, Crambe crambe and Phorbas tenacior. FEMS Microbiol. Lett. 366:fnaa014. doi: 10.1093/femsle/fnaa014
Braconcini, M., Gorrasi, S., Fenice, M., Barghini, P., and Pasqualetti, M. (2024). Rambellisea gigliensis and Rambellisea halocynthiae, gen. Et spp. nov. (Lulworthiaceae) from the Marine Tunicate Halocynthia papillosa. J. Fungi 10:127. doi: 10.3390/jof10020127
Burgaud, G., Edgcomb, V., Hassett, B. T., Kumar, A., Li, W., Mara, P., et al. (2022). “Marine fungi” in The Marine Microbiome. eds. L. J. Stal and M. S. Cretoiu (Cham, Switzerland: Springer International Publishing), 243–295.
Canepa, A., Fuentes, V., Sabatés, A., Piraino, S., Boero, F., and Gili, J. M. (2014). “Pelagia noctiluca in the Mediterranean Sea” in Jellyfish Blooms. eds. K. A. Pitt and C. H. Lucas (Dordrecht: Netherland, Springer), 237–266.
Carbone, I., and Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91, 553–556. doi: 10.1080/00275514.1999.12061051
Carroll, A. R., Copp, B. R., Davis, R. A., Keyzers, R. A., and Prinsep, M. R. (2019). Marine natural products. Nat. Prod. Rep. 36, 122–173. doi: 10.1039/C8NP00092A
Costello, M. J., and Chaudhary, C. (2017). Marine biodiversity, biogeography, deep-sea gradients, and conservation. Curr. Biol. 27, R511–R527. doi: 10.1016/j.cub.2017.04.060
Cox, R. J., and Simpson, T. J. (2009). Fungal type I polyketide synthases. Methods Enzymol. 459, 49–78. doi: 10.1016/S0076-6879(09)04603-5
Damare, S., Singh, P., and Raghukumar, S. (2011). “Biotechnology of marine fungi” in Biology of Marine Fungi. ed. C. Raghukumar (Berlin, Germany: Springer), 277–297.
Ding, B., Yin, Y., Zhang, F., and Li, Z. (2011). Recovery and phylogenetic diversity of culturable fungi associated with marine sponges Clathrina luteoculcitella and Holoxea sp. in the South China Sea. Mar. Biotechnol. 13, 713–721. doi: 10.1007/s10126-010-9333-8
Domsch, K. H., Gams, W., and Anderson, T. (2007). Compendium of Soil Fungi. 2nd Edn. Eching: Academic Press.
Durgham, H., Ikhtiyar, S., and Ibraheem, R. (2016). First record of Pelagia noctiluca (Forsskål, 1775) on the coast of Syria. Mar. Biodivers. Rec. 9, 1–3. doi: 10.1186/s41200-016-0060-3
El Hajj Assaf, C., Zetina-Serrano, C., Tahtah, N., Khoury, A. E., Atoui, A., Oswald, I. P., et al. (2020). Regulation of secondary metabolism in the Penicillium genus. Int. J. Mol. Sci. 21:9462. doi: 10.3390/ijms21249462
Ellis, M.B. (1976). More dematiaceous Hyphomycete. Commonwealth Mycological Institute, Kew, Surrey, England.
FigTree (2024): Tree Figure Drawing Tool Version 1.4.4. (2024). Available online at: http://tree.bio.ed.ac.uk/software/Figtree/ (Accessed July 24, 2024).
Gao, Z., Li, B., Zheng, C., and Wang, G. (2008). Molecular detection of fungal communities in the Hawaiian marine sponges Suberites zeteki and Mycale armata. Appl. Environ. Microbiol. 74, 6091–6101. doi: 10.1128/AEM.01315-08
Garfinkel, A. R. (2021). The history of Botrytis taxonomy, the rise of phylogenetics, and implications for species recognition. Phytopathology 111, 437–454. doi: 10.1094/PHYTO-06-20-0211-IA
Giovannini, V., Barghini, P., Gorrasi, S., Massimiliano, F., and Pasqualetti, M. (2019). Marine fungi: a potential source of novel enzymes for environmental and biotechnological applications. J. Environ. Prot. Ecol. 20, 1214–1222.
Gonçalves, M. F., Esteves, A. C., and Alves, A. (2022). Marine fungi: opportunities and challenges. Encyclopedia 2, 559–577. doi: 10.3390/encyclopedia2010037
Grossart, H. P., Van den Wyngaert, S., Kagami, M., Wurzbacher, C., Cunliffe, M., and Rojas-Jimenez, K. (2019). Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 17, 339–354. doi: 10.1038/s41579-019-0175-8
Gunde-Cimerman, N., and Zalar, P. (2014). Extremely halotolerant and halophilic fungi inhabit brine in solar salterns around the globe. Food Technol. Biotechnol. 52, 170–179.
Hafez Ghoran, S., Taktaz, F., Sousa, E., Fernandes, C., and Kijjoa, A. (2023). Peptides from marine-derived Fungi: chemistry and biological activities. Mar. Drugs 21:510. doi: 10.3390/md21100510
Hagerman, S., Dowlatabadi, H., Satterfield, T., and McDaniels, T. (2010). Expert views on biodiversity conservation in an era of climate change. Glob. Environ. Chang. 20, 192–207. doi: 10.1016/j.gloenvcha.2009.10.005
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hyde, K. D., Dong, Y., Phookamsak, R., Jeewon, R., Bhat, D. J., Jones, E. G., et al. (2020). Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 100, 5–277. doi: 10.1007/s13225-020-00439-5
Hyde, K. D., Sarma, V. V., and Jones, E. B. G. (2000). “Morphology and taxonomy of higher marine fungi”, in Marine Mycology a practical approach. eds. K. D. Hyde and S. B. Pointing (Hong Kong: Fungal Diversity Press), 172–204.
Index Fungorum database and web site (2024). Available online at: https://www.indexfungorum.org (Accessed July 24, 2024).
Jones, E. G., Ramakrishna, S., Vikineswary, S., Das, D., Bahkali, A. H., Guo, S. Y., et al. (2022). How do fungi survive in the sea and respond to climate change? J. Fungi 8:291. doi: 10.3390/jof8030291
Jones, E. G., Suetrong, S., Sakayaroj, J., Bahkali, A. H., Abdel-Wahab, M. A., Boekhout, T., et al. (2015). Classification of marine Ascomycota, Basidiomycota, Blastocladiomycota and Chytridiomycota. Fungal Divers. 73, 1–72. doi: 10.1007/s13225-015-0339-4
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kohlmeyer, J., and Kohlmeyer, E. (1979). Marine Mycology—the Higher Fungi. New York: Academic Press.
La Kim, E., Li, J. L., Dang, H. T., Hong, J., Lee, C. O., Kim, D. K., et al. (2012a). Cytotoxic cytochalasins from the endozoic fungus Phoma sp. of the giant jellyfish Nemopilema nomurai. Bioorg. Med. Chem. Lett. 22, 3126–3129. doi: 10.1016/j.bmcl.2012.03.058
La Kim, E., Li, J. L., Xiao, B., Hong, J., Yoo, E. S., Yoon, W. D., et al. (2012b). A new cyclic tetrapeptide from the jellyfish-derived fungus Phoma sp. Chem. Pharm. Bull. 60, 1590–1593. doi: 10.1248/cpb.c12-00335
Le, H. M. T., Do, Q. T., Doan, M. H. T., Vu, Q. T., Nguyen, M. A., Vu, T. H. T., et al. (2019). Chemical composition and biological activities of metabolites from the marine fungi Penicillium sp. isolated from sediments of co to island, Vietnam. Molecules 24:3830. doi: 10.3390/molecules24213830
Lee, T., Yun, S. H., Hodge, K., Humber, R., Krasnoff, S., Turgeon, G., et al. (2001). Polyketide synthase genes in insect-and nematode-associated fungi. Appl. Microbiol. Biotechnol. 56, 181–187. doi: 10.1007/s002530100637
Li, X., Kerrigan, J., Chai, W., and Schnabel, G. (2012). Botrytis caroliniana, a new species isolated from blackberry on South Carolina. Mycologia 104, 650–658. doi: 10.3852/11-218
Li, D. D., Luo, X., Ying, W., La Kim, E., Hong, J., Lee, J. H., et al. (2023). Peroxisome proliferator activated receptor-γ agonistic compounds from the jellyfish-derived fungus Cladosporium oxysporum. Chem. Biodivers. 20:e202300851. doi: 10.1002/cbdv.202300851
Li, Y., Wang, Y., Wang, H., Shi, T., and Wang, B. (2024). The genus Cladosporium: a prospective producer of natural products. Int. J. Mol. Sci. 25:1652. doi: 10.3390/ijms25031652
Liu, J., Li, F., Kim, E. L., Hong, J., and Jung, J. H. (2013). Viriditoxin, from a jellyfish-derived fungus, is antibiotic to fish pathogens. Nat. Prod. Sci. 19, 61–65.
Liu, J., Li, F., Kim, E. L., Li, J. L., Hong, J., Bae, K. S., et al. (2011). Antibacterial polyketides from the jellyfish-derived fungus Paecilomyces variotii. J. Nat. Prod. 74, 1826–1829. doi: 10.1021/np200350b
Liu, J., Li, F., Lee, Y. M., Li, J. L., Hong, J. K., Yoon, W. D., et al. (2012). An anacardic acid analog from the jellyfish-derived fungus Paecilomyces variotii. Nat. Prod. Sci. 18, 8–12.
Liu, S., Su, M., Song, S. J., Hong, J., Chung, H. Y., and Jung, J. H. (2018). An anti-inflammatory PPAR-γ agonist from the jellyfish-derived fungus Penicillium chrysogenum J08NF-4. J. Nat. Prod. 81, 356–363. doi: 10.1021/acs.jnatprod.7b00846
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
López-Legentil, S., Erwin, P. M., Turon, M., and Yarden, O. (2015). Diversity of fungi isolated from three temperate ascidians. Symbiosis 66, 99–106. doi: 10.1007/s13199-015-0339-x
Maamar, A., Lucchesi, M. E., Debaets, S., Nguyen van Long, N., Quemener, M., Coton, E., et al. (2020). Highlighting the crude oil bioremediation potential of marine fungi isolated from the port of Oran (Algeria). Diversity 12:196. doi: 10.3390/d12050196
Marchese, P., Garzoli, L., Gnavi, G., O’Connell, E., Bouraoui, A., Mehiri, M., et al. (2020). Diversity and bioactivity of fungi associated with the marine sea cucumber Holothuria poli: disclosing the strains potential for biomedical applications. J. Appl. Microbiol. 129, 612–625. doi: 10.1111/jam.14659
Marchese, P., Garzoli, L., Young, R., Allcock, L., Barry, F., Tuohy, M., et al. (2021). Fungi populate deep-sea coral gardens as well as marine sediments in the Irish Atlantic Ocean. Environ. Microbiol. 23, 4168–4184. doi: 10.1111/1462-2920.15560
Mariottini, G. L., Giacco, E., and Pane, L. (2008). The mauve stinger Pelagia noctiluca (Forsskål, 1775). Distribution, ecology, toxicity and epidemiology of stings. Mar. Drugs 6, 496–513. doi: 10.3390/md6030496
Molnár, I., Gibson, D. M., and Krasnoff, S. B. (2010). Secondary metabolites from entomopathogenic Hypocrealean fungi. Nat. Prod. Rep. 27, 1241–1275. doi: 10.1039/c001459c
Nguyen, M. T., and Thomas, T. (2018). Diversity, host-specificity and stability of sponge-associated fungal communities of co-occurring sponges. PeerJ 6:e4965. doi: 10.7717/peerj.4965
O’Donnell, K., and Cigelnik, E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 7, 103–116. doi: 10.1006/mpev.1996.0376
Pang, X., Lin, X., Tian, Y., Liang, R., Wang, J., Yang, B., et al. (2018). Three new polyketides from the marine sponge-derived fungus Trichoderma sp. SCSIO41004. Nat. Prod. Res. 32, 105–111. doi: 10.1080/14786419.2017.1338286
Parte, S., Sirisha, V. L., and D’Souza, J. S. (2017). “Biotechnological applications of marine enzymes from algae, bacteria, fungi, and sponges” in Advances in Food and Nutrition Research. ed. F. Toldra (Cambridge: MA:Academic Press), 75–106.
Pasqualetti, M., Barghini, P., Giovannini, V., and Fenice, M. (2019). High production of chitinolytic activity in halophilic conditions by a new marine strain of Clonostachys rosea. Molecules 24:1880. doi: 10.3390/molecules24101880
Pasqualetti, M., Giovannini, V., Barghini, P., Gorrasi, S., and Fenice, M. (2020). Diversity and ecology of culturable marine fungi associated with Posidonia oceanica leaves and their epiphytic algae Dictyota dichotoma and Sphaerococcus coronopifolius. Fungal Ecol. 44:100906. doi: 10.1016/j.funeco.2019.100906
Pasqualetti, M., Gorrasi, S., Giovannini, V., Braconcini, M., and Fenice, M. (2022). Polyextremophilic chitinolytic activity by a marine strain (IG119) of Clonostachys rosea. Molecules 27:688. doi: 10.3390/molecules27030688
Pasqualetti, M., Mulas, B., Rambelli, A., and Tempesta, S. (2014). Saprotrophic litter fungi in a Mediterranean ecosystem: behaviour on different substrata. Plant Biosyst. 148, 342–356. doi: 10.1080/11263504.2013.877534
Paz, Z., Komon-Zelazowska, M., Druzhinina, I. S., Aveskamp, M. M., Shnaiderman, A., Aluma, Y., et al. (2010). Diversity and potential antifungal properties of fungi associated with a Mediterranean sponge. Fungal Divers. 42, 17–26. doi: 10.1007/s13225-010-0020-x
Pesciaroli, C., Cupini, F., Selbmann, L., Barghini, P., and Fenice, M. (2012). Temperature preferences of bacteria isolated from seawater collected in Kandalaksha Bay, White Sea, Russia. Polar Biol. 35, 435–445. doi: 10.1007/s00300-011-1091-1
Peterson, S. W. (2004). Multilocus DNA sequence analysis shows that Penicillium biourgeianum is a distinct species closely related to P. brevicompactum and P. olsonii. Mycol. Res. 108, 434–440. doi: 10.1017/S0953756204009761
Phookamsak, R., Hyde, K. D., Jeewon, R., Bhat, D. J., Jones, E. G., Maharachchikumbura, S. S., et al. (2019). Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 95, 1–273. doi: 10.1007/s13225-019-00421-w
Pinedo-Rivilla, C., Aleu, J., and Durán-Patrón, R. (2022). Cryptic metabolites from marine-derived microorganisms using OSMAC and epigenetic approaches. Mar. Drugs 20:84. doi: 10.3390/md20020084
Pitt, J. I. (1979). The Genus Penicillium and its Teleomorphic States Eupenicillium and Talaromyces. London: Academic Press Inc.
Posada, D. (2008). jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Raghukumar, C. (2008). Marine fungal biotechnology: an ecological perspective. Fungal Divers. 31, 19–36.
Raghukumar, C., and Ravindran, J. (2012). “Fungi and their role in corals and coral reef ecosystems” in Biology of Marine Fungi, ed. C. Raghukumar (Berlin, Germany: Springer), 89–113.
Raimundo, I., Silva, S. G., Costa, R., and Keller-Costa, T. (2018). Bioactive secondary metabolites from octocoral-associated microbes—new chances for blue growth. Mar. Drugs 16:485. doi: 10.3390/md16120485
Rambelli, A., Mulas, B., and Pasqualetti, M. (2004). Comparative studies on microfungi in tropical ecosystems in Ivory Coast forest litter: behaviour on different substrata. Mycol. Res. 108, 325–336. doi: 10.1017/S0953756204009396
Regalado, R. R. H., and Ramirez, V. (2019). Molecular identification and hemolytic activity of four jellyfish-associated marine fungi from Cagbatano Bay, Pio Duran, Philippines. Int. J. Biosci. 15, 531–538. doi: 10.12692/ijb/15.4.531-538
Rehner, S. A., and Buckley, E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97, 84–98. doi: 10.3852/mycologia.97.1.84
Ren, Z., Yang, L., Ma, Q., Xie, Q., Dai, H., Sun, K., et al. (2022). Meroterpenoids and steroids from the marine-derived fungus Trametes sp. ZYX-Z-16. Molecules 27:8782. doi: 10.3390/molecules27248782
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schoch, C. L., Seifert, K. A., Huhndorf, S., Robert, V., Spouge, J. L., Levesque, C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. PNAS 109, 6241–6246. doi: 10.1073/pnas.1117018109
Shaker, S., Fan, R. Z., Li, H. J., and Lan, W. J. (2021). A pair of novel bisindole alkaloid enantiomers from marine fungus Fusarium sp. XBB-9. Nat. Prod. Res. 35, 1497–1503. doi: 10.1080/14786419.2019.1655416
Slightom, J. L., Metzger, B. P., Luu, H. T., and Elhammer, A. P. (2009). Cloning and molecular characterization of the gene encoding the Aureobasidin a biosynthesis complex in Aureobasidium pullulans BP-1938. Gene 431, 67–79. doi: 10.1016/j.gene.2008.11.011
Šmilauer, P., and Lepš, J. (2014). Multivariate Analysis of Ecological Data Using CANOCO 5. Cambridge, UK: Cambridge University Press.
Staats, M., van Baarlen, P., and van Kan, J. A. (2005). Molecular phylogeny of the plant pathogenic genus Botrytis and the evolution of host specificity. MBE 22, 333–346. doi: 10.1093/molbev/msi020
Stroe, M. C., Gao, J., Pitz, M., and Fischer, R. (2024). Complexity of fungal polyketide biosynthesis and function. Mol. Microbiol. 121, 18–25. doi: 10.1111/mmi.15192
Sutton, B. C. (1980). The Coelomycetes Fungi Imperfecti With Pycnidia, Acervuli and Stromata. London: CABI Publisher.
Tan, S., Yang, B., Liu, J., Xun, T., Liu, Y., and Zhou, X. (2019). Penicillixanthone a, a marine-derived dual-coreceptor antagonist as anti-HIV-1 agent. Nat. Prod. Res. 33, 1467–1471. doi: 10.1080/14786419.2017.1416376
Tao, G. Y., Liu, J., Jung, J. H., Guo, W., Wen, X. Q., and Liu, Y. (2016). Compounds from a jellyfish-derived fungus Aspergillus fumigates. Nat. Prod. Res. 22, 82–86. doi: 10.20307/nps.2016.22.2.82
Templado, J., Paulay, G., Gittenberger, A., and Meyer, C. (2010). Sampling the marine realm. ABC Taxa 8, 273–307.
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tibballs, J. (2006). Australian venomous jellyfish, envenomation syndromes, toxins and therapy. Toxicon 48, 830–859. doi: 10.1016/j.toxicon.2006.07.020
Tilves, U., Purcell, J. E., Fuentes, V. L., Torrents, A., Pascual, M., Raya, V., et al. (2016). Natural diet and predation impacts of Pelagia noctiluca on fish eggs and larvae in the NW Mediterranean. JPR 38, 1243–1254. doi: 10.1093/plankt/fbw059
TreeBASE version 2 (2024). Available online at: https://www.treebase.org/treebase-web/home.html (Accessed July 27, 2024).
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Vassaux, A., Meunier, L., Vandenbol, M., Baurain, D., Fickers, P., Jacques, P., et al. (2019). Nonribosomal peptides in fungal cell factories: from genome mining to optimized heterologous production. Biotechnol. Adv. 37:107449. doi: 10.1016/j.biotechadv.2019.107449
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Visagie, C. M., Seifert, K. A., Houbraken, J., Samson, R. A., and Jacobs, K. (2016). A phylogenetic revision of Penicillium sect. Exilicaulis, including nine new species from fynbos in South Africa. IMA Fungus 7, 75–117. doi: 10.5598/imafungus.2016.07.01.06
Wainwright, B. J., Zahn, G. L., Spalding, H. L., Sherwood, A. R., Smith, C. M., and Amend, A. S. (2017). Fungi associated with mesophotic macroalgae from the 'Au'au channel, west Maui are differentiated by host and overlap terrestrial communities. PeerJ 5:e3532. doi: 10.7717/peerj.3532
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR Protocols a Guide to Methods and Applications. eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York, USA: Academic Press), 315–322.
Wijayawardene, N. N., Dai, D. Q., Jayasinghe, P. K., Gunasekara, S. S., Nagano, Y., Tibpromma, S., et al. (2022). Ecological and oceanographic perspectives in future marine fungal taxonomy. J. Fungi 8:1141. doi: 10.3390/jof8111141
Williams, G. J., Price, N. N., Ushijima, B., Aeby, G. S., Callahan, S., Davy, S. K., et al. (2014). Ocean warming and acidification have complex interactive effects on the dynamics of a marine fungal disease. Proc. Biol. Sci. 281:20133069. doi: 10.1098/rspb.2013.3069
Wright, A. D., Osterhage, C., and König, G. M. (2003). Epicoccamide, a novel secondary metabolite from a jellyfish-derived culture of Epicoccum purpurascens. OBC 1, 507–510.
Ye, Y., Liang, J., She, J., Lin, X., Wang, J., Liu, Y., et al. (2022). Two new alkaloids and a new butenolide derivative from the beibu gulf sponge-derived fungus Penicillium sp. SCSIO 41413. Mar. Drugs 21:27. doi: 10.3390/md21010027
Yue, Y., Yu, H., Li, R., Xing, R., Liu, S., and Li, P. (2015). Exploring the antibacterial and antifungal potential of jellyfish-associated marine fungi by cultivation-dependent approaches. PLoS One 10:e0144394. doi: 10.1371/journal.pone.0144394
Yue, Y., Yu, H., Suo, Q., Li, R., Liu, S., Xing, R., et al. (2022). Discovery of a novel jellyfish venom metalloproteinase inhibitor from secondary metabolites isolated from jellyfish-derived fungus aspergillus versicolor SmT07. Chem. Biol. Interact. 365:110113. doi: 10.1016/j.cbi.2022.110113
Keywords: marine fungi, epizoic mycobiota, Pelagia noctiluca , antimicrobial activity, Mediterranean Sea, NRPS and PKS genes
Citation: Pasqualetti M, Braconcini M, Barghini P, Gorrasi S, Schillaci D, Ferraro D, Della Sala G, De Marino S and Fenice M (2024) From marine neglected substrata new fungal taxa of potential biotechnological interest: the case of Pelagia noctiluca. Front. Microbiol. 15:1473269. doi: 10.3389/fmicb.2024.1473269
Edited by:
Anna Maria Puglia, University of Palermo, ItalyReviewed by:
Giovanna Cristina Varese, University of Turin, ItalyElisabet Aranda, University of Granada, Spain
Copyright © 2024 Pasqualetti, Braconcini, Barghini, Gorrasi, Schillaci, Ferraro, Della Sala, De Marino and Fenice. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marcella Pasqualetti, bXBhc3F1YWxAdW5pdHVzLml0; Massimiliano Fenice, ZmVuaWNlQHVuaXR1cy5pdA==
†These authors share first authorship
 Marcella Pasqualetti
Marcella Pasqualetti Martina Braconcini
Martina Braconcini Paolo Barghini
Paolo Barghini Susanna Gorrasi
Susanna Gorrasi Domenico Schillaci
Domenico Schillaci Donatella Ferraro
Donatella Ferraro Gerardo Della Sala
Gerardo Della Sala Simona De Marino
Simona De Marino Massimiliano Fenice
Massimiliano Fenice