- 1Department of Microbiology & Immunology, Medical University of South Carolina, Charleston, SC, United States
- 2Department of Regenerative Medicine & Cell Biology, Medical University of South Carolina, Charleston, SC, United States
- 3Hollings Cancer Center, Medical University of South Carolina, Charleston, SC, United States
- 4Department of Pathology and Laboratory Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States
- 5Department of Protective Immunity, Children’s Hospital of Philadelphia, Philadelphia, PA, United States
- 6Department Division of Gastroenterology, Cedars Sinai, Los Angeles, CA, United States
- 7Division of Gastroenterology and Hepatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States
- 8Dairy and Functional Foods Research Unit, United States Department of Agriculture, Washington, DC, United States
Clostridioides difficile is a bacterial pathogen that has been implicated in severe gastrointestinal infections. C. difficile has intrinsic green autofluorescence and the level of this autofluorescence is known to be increased by growth time and oxygen. Currently, it is unclear if dietary compounds or metabolites from the gut microbiota are able to enhance C. difficile autofluorescence. Here, we aimed to determine potential factors that affect C. difficile autofluorescence. After screening a large repertoire of compounds, we identified several substances, like L-lysine and pantothenate, that led to an increased C. difficile autofluorescence. We also found that several members of the gut microbiota, such as Enterococcus faecalis, Klebsiella aerogenes and K. pneumoniae, can increase C. difficile autofluorescence through their secreted compounds. We further focused on the effect of K. pneumoniae on C. difficile autofluorescence and found that multiple enteric strains of K. pneumoniae could enhance C. difficile’s autofluorescence. We used this enhanced autofluorescence to identify C. difficile in K. pneumoniae co-cultures by flow cytometry. Our findings shed light on the relationship between C. difficile and other members of the gut microbiota, as well as different factors that can affect C. difficile autofluorescence.
Introduction
Clostridioides difficile, formerly known as Clostridium difficile, is an anaerobic, spore-forming bacterium responsible for a spectrum of gastrointestinal illnesses ranging from mild diarrhea to severe pseudomembranous colitis and toxic megacolon (Guh and Kutty, 2018; Guh et al., 2020). C. difficile infection is a significant cause of antibiotic-associated diarrhea, and it has become a major public health concern globally due to its increasing incidence, severity, and recurrence rates (Guh et al., 2017; Singh et al., 2017; Dong et al., 2023). Risk factors for C. difficile infection include recent antibiotic exposure, advanced age, immunocompromised status, prolonged hospitalization, and residence in long-term care facilities (Eze et al., 2017; Davies et al., 2020). The intricate interplay between host factors, environmental factors, and the gut microbiota all contribute to the severity and recurrence of C. difficile infection. There is an urgent need to understand the pathogenic traits of this organism to safeguard public health.
Autofluorescence can be useful for detecting and studying bacteria like C. difficile in laboratory settings. Several researchers have noted that C. difficile generates a green autofluorescence when excited with ultraviolet light (Ransom Eric et al., 2015; Buckley et al., 2016; Donnelly et al., 2022; Oliveira Paiva et al., 2022). This has made fluorescently tagging C. difficile with GFP difficult, but it is also advantageous because the autofluorescent capacity of C. difficile allows researchers to visualize the presence and distribution of the bacterium without the need for staining or other labeling techniques. It has been documented that growth stage and the presence of oxygen increases C. difficile autofluorescence (Oliveira Paiva et al., 2022), but other compounds that influence the autofluorescence of C. difficile have not been identified. Additionally, it is not clear if other gut bacteria influence the autofluorescence capacity of C. difficile.
In this study we sought to identify compounds that regulate C. difficile autofluorescence. We screened >300 compounds and identified several novel candidates that increased autofluorescence in C. difficile. We also screened several commensal and pathobiont members of the gut microbiota and found Klebsiella species in particular enhanced C. difficile autofluorescence. When C. difficile was grown with clinical isolates of K. pneumoniae, they had enhanced autofluorescence and could be separated by flow cytometry. These data provide new insights into C. difficile autofluorescence.
Methods
Bacteria and culture conditions
The following bacteria were selected for the experiments: C. difficile R20291, Streptococcus mitis NCIMB 13770, Listeria monocytogenes BAA 751, Pseudomonas aeruginosa ATCC 27853, Enterococcus faecalis ATCC 35038, Morganella morganii ATCC25830, Klebsiella pneumoniae ATCC 35657, K. pneumoniae CB1, K. aerogenes ATCC 35029, Escherichia coli Nissle 1917, E. coli K12, Lactobacillus acidophilus ATCC 4356, Bifidobacterium breve ATCC 15698, and B. bifidum ATCC 29521. K. pneumoniae strains C015, A14 CFH, and A05E were isolated from the creeping mesenteric adipose of Crohn’s disease patients, identified with full-length 16S rRNA sequencing, and kindly provided by Suzanne Devkota. K. pneumoniae strains Dynamic 35, UNK34, and Dynamic 18 were isolated from the stool of patients with C. difficile infection, identified with MALDI, and kindly provided by Karley Mahalak.
C. difficile was streaked plated onto cycloserine-cefoxitin fructose agar (CCFA) and single colonies were inoculated into brain-heart infusion (BHI) broth supplemented with 2% yeast extract and 0.2% cysteine (BHIS; FisherScientific). Cultures were grown at 37°C anaerobically in an Anaerobe Systems AS-150 anaerobic chamber overnight before use. L. acidophilus was streak plated onto De Man, Rogosa, and Sharpe (MRS; FisherScientific) agar plates and single colonies were inoculated into MRS broth and grown at 37°C anaerobically overnight before use. B. breve and B. bifidum were streak plated onto Bifidobacterium selective agar (Anaerobe Systems, Cat # AS-6423) and single colonies were inoculated into MRS broth and grown at 37°C anaerobically overnight before use. S. mitis, L. monocytogenes, P. aeruginosa, E. faecalis, M. morganii, K. pneumoniae, K. aerogenes, and E. coli were streak plated onto BHI agar and single colonies were inoculated into BHI broth and grown aerobically, shaking at 37°C overnight before use.
Overnight cultures of bacteria were grown in their above indicated conditions before being subcultured at an OD600nm = 0.1 into 5 mL of a chemically defined media, ZMB1, supplemented with 100 mM glucose (Horvath et al., 2023). After 6 h of growth, the optical density of the bacterial cultures was examined at OD600nm on a ThermoFisher Nanodrop using the cuvette function as a proxy of growth (Table 1). Cultures were centrifuged at 4,450 x g for 10 min to pellet bacteria. The cell-free supernatant was filtered with a 0.22 μm syringe filter to collect sterile, log-phase conditioned media. This supernatant was used immediately for autofluorescence assays.
Autofluorescence measurements
To assess C. difficile autofluorescence over time, an overnight culture of C. difficile was subcultured into BHI at OD600mm = 0.1 and was grown at 37°C anaerobically. At 1, 3, 6, 9, 12, 18 and 24 h post-inoculation, 1 mL samples were removed and centrifuged at 5,000 x g for 5 min to pellet the bacteria. The bacterial pellet was washed twice with sterile phosphate-buffered saline (PBS) and 100 μL of the washed C. difficile in PBS was transferred to a 96-well plate and the fluorescence in the green channel (excitation 485 nm/emission 528 nm) was read on a Biotek Synergy H1 plate reader. Blank PBS without C. difficile was included as a negative control. Biolog© Phenotypic Microarray plates PM1, PM2a, PM9, and PM10 were used to measure C. difficile autofluorescence with various compounds. Overnight cultures of C. difficile were subcultured into BHI at OD600mm = 0.1 and grown at 37°C anaerobically for 6 h. C. difficile was then washed twice with PBS before being resuspended in PBS. To obtain baseline levels of autofluorescence in the Biolog microarray plates, we added 90 μL of PBS to the plate and read fluorescence in the green channel (485 nm/528 nm) on a Synergy H1 plate reader. We then added 10 μL C. difficile at OD600nm = 10 to the Biolog microarray plates containing 90 μL of PBS; resulting in a final concentration of OD600nm of 1.0 in PBS. The plates were incubated for five minutes before reading the green fluorescence (485 nm/528 nm) on a Synergy H1 plate reader. The reported fluorescence represents the values with the baseline fluorescence removed. To confirm the findings of the Biolog microarrays, we added concentrations of L-lysine (FisherSci) and pantothenate (FisherSci) ranging from 1–100 nM to 96-well plates and added C. difficile at an OD600nm of 1.0. Similar to the Biolog plate assay, autofluorescence was measured after 5 min of incubation at excitation 485 nm/528 nm on a Synergy H1 plate reader.
To examine C. difficile autofluorescence in response to bacterial metabolites, we grew S. mitis NCIMB 13770, L. monocytogenes BAA 751, P. aeruginosa ATCC 27853, E. faecalis ATCC 35038, M. morganii ATCC25830, K. pneumoniae ATCC 35657, K. aerogenes ATCC 35029, E. coli Nissle 1917, E. coli K12, L. acidophilus ATCC 4356, B. breve ATCC 15698, and B. bifidum ATCC 29521 in 5 mL cultures of ZMB1 for 6 h and collected cell-free supernatant (see Bacteria and Culture Conditions). 100 μL of this cell-free supernatant was added to a 96-well DeepWell microplate (FisherSci #12-566-120) in quadruplicate and then 900 μL of C. difficile was added in ZMB1 at an OD600nm = 0.1. The plate was grown anaerobically for 9 h and then 100 μL of culture was transferred via a multi-channel pipette to a 96-well conical bottom microplate (FisherSci # 277143). The plate was centrifuged at 2,000 x g for 10 min to pellet the bacteria. The plate was washed twice with PBS and the C. difficile bacteria were resupended in PBS and transferred to a 96-well flat bottom microplate (FisherSci #12-565-501). This plate was then read on a Synergy H1 plate reader at 485 nm/528 nm. The pH of the C. difficile-supernatant cultures was examined using a liquid litmus assay, as previously described (Engevik et al., 2023). Briefly, 200 μL of the C. difficile culture was transferred from the 96-well DeepWell microplate to a 96-well conical bottom microplate and the plate was centrifuged at 2,000 x g for 10 min to pellet the bacteria. Then 150 μL of the cell-free supernatant was transferred to a 96-well filter plate containing a 0.2 μm PVDF filter plate (FisherSci # MSIPS4510) and the supernatant was sterile filtered. 90 μL of the sterile supernatant was transferred to a 96-well flat bottom microplate and 10 μL of litmus (10 mg/mL) was added to the solution. The plate was read at 520 nm and 590 nm and the ratio was used to calculate the pH via a standard curve.
As confirmation that bacterial products could influence C. difficile autofluorescence, E. faecalis ATCC 35038, K. pneumoniae ATCC 35657, and K. aerogenes ATCC 35029 were grown in ZMB1 for 6 h and 25 μL of the cell-free supernatant from these cultures were added to 96-well flat bottomed plate. As a negative control, uninoculated ZMB1 was added to the plate as well. We then added 65 μL of PBS and read the baseline levels of autofluorescence on a Synergy H1 plate reader. After getting baseline values, we added 10 μL of a 6 h culture of C. difficile in PBS to the plate, incubated for 5 min and then read fluorescence on a Synergy H1 plate reader. This assay was repeated just with K. pneumoniae ATCC 25657 supernatant at various concentrations (3.125–100% supernatant).
Clostridioides difficile and Klebsiella pneumoniae co-cultures
C. difficile and K. pneumoniae strains (ATCC 35657, CB1, C015, A14 CFH, A05E, Dynamic 35, UNK34, and Dynamic 18) were grown overnight as mono-cultures in BHI. The bacteria were then subcultured at an OD600nm = 0.1 in 5 mL of ZMB1 supplemented with 100 mM glucose for mono-cultures or an OD600nm = 0.05 in 5 mL of ZMB1 supplemented with glucose for co-cultures. Cultures were incubated anaerobically at 37°C for 6 h. After the incubation, cultures were centrifuged at 4,425 x g for 5 min to pellet the bacteria and the pellets were washed twice with PBS. The culture pellets were resuspended in PBS and read for green fluorescence (485 nm/528 nm) and OD600nm using a Synergy H1 plate reader.
Flow cytometry
C. difficile and K. pneumoniae were grown in mono- and co-cultures in ZMB1 anaerobically at 37°C for 20 h and then used for flow cytometry analysis. Bacteria were pelleted and resuspended in 4% paraformaldehyde (Alfa Aesar, Cat# J61899) and incubated at room temperature for one hour. Bacterial cultures were washed twice with PBS before being resuspended in 10 μg/mL of Hoechst. After 10 min of room-temperature incubation with Hoechst, the bacteria were washed with 0.2% BSA in PBS before being resuspended in 0.2% BSA in PBS. Bacteria were then run on a Beckman Coulter CytoFLEX LX flow cytometer. As bacteria are too small for the machine to detect unstained, the primary threshold was set to UV525 to detect the Hoechst stained bacteria. Autofluorescence was measured as B525-FITC. Analysis was performed using FlowJo version 10.
gDNA isolation and qPCR
To confirm the presence of both bacteria in co-culture conditions, C. difficile and K. pneumoniae strains were grown in mono- and co-cultures in ZMB1 anaerobically at 37°C for 20 h. After 20 h, 1 mL of the culture was transferred to a microfuge tube and centrifuged at 5,000 x g for 5 min to pellet the bacteria. We isolated gDNA from the bacterial pellet using the Zymo Quick-DNA Fecal/Soil Microbe Kits according to the manufacturer’s instructions. Quantitative real time PCR (qPCR) was performed using a Bio-Rad CFX96 Real Time qPCR machine (Bio-Rad). Forward and reverse primers for C. difficile and Klebsiella were added to SYBR Green mastermix (Genesee Scientific #17-501DP) and gDNA. Bacterial colony forming units (CFUs) were calculated from CT values based on standard curves of C. difficile and K. pneumoniae.
Fluorescent imaging
Images of C. difficile autofluorescence were obtained from cultures of C. dififcile in PBS incubated with 50% cell-free supernatant from K. pneumonia ATCC 35657 in ZMBI and co-cultures of C. difficile and K. pneumoniae ATCC 35657 grown together in ZMB1 for 20 h. After 5 min of incubation in a 1 mL microfuge tube, 100 μL of the solution was transferred to a glass slide and coverslipped. Images were obtained on a Zeiss Axio microscope and the images were analyzed with FIJI (Formerly Image J) software (NIH) with the relative fluorescent intensity obtained for each channel.
Results
Clostridioides difficile autofluorescence is influenced by various compounds
Since the autofluorescence in C. difficile 630 has been shown to be growth-dependent (Oliveira Paiva et al., 2022), we first sought to determine if the autofluorescence of C. difficile R20291 was also growth-phase dependent. C. difficile R20291 was grown in BHI at a starting OD600nm = 0.1 and samples were collected during lag phase (1 and 3 h), exponential phase (6 and 9 h), stationary phase (12 and 18 h), and death phase (24 h) (Figure 1). As expected, we found that C. difficile grew over time, with the highest growth observed after 18 h of incubation (Figure 1A). Mirroring the growth, we observed that C. difficile autofluorescence increased overtime, with the highest amount of autofluorescence occurring at 18 h (Figure 1B).
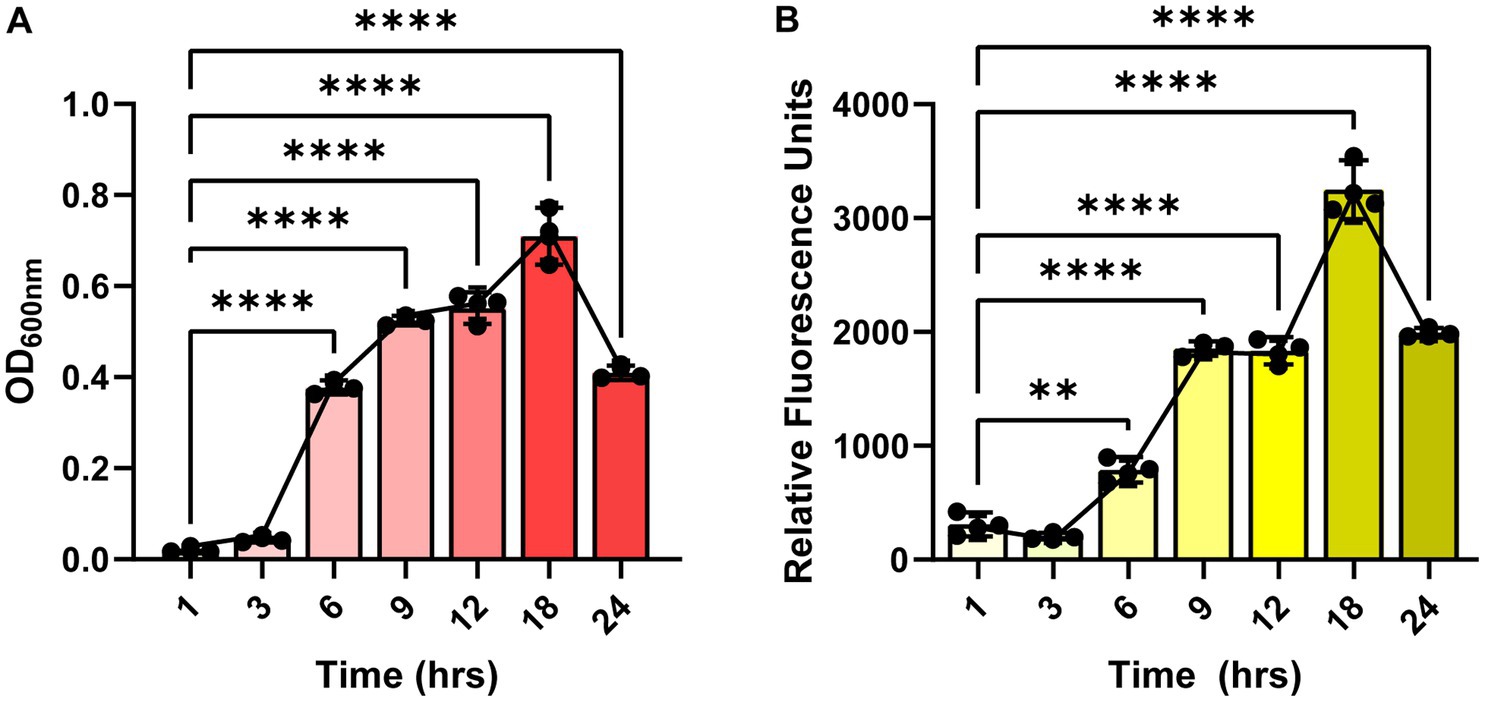
Figure 1. Clostridioides difficile autofluorescence is growth-phase dependent. C. difficile R20291 was grown in BHI at a starting OD600nm of 0.1 and growth and fluorescence was monitored after 1, 3, 6, 9, 12, 18 and 24 h post-inoculation. (A) Growth was quantified by measuring OD600 of the culture at various time points. (B) Autofluorescence was quantified by measuring green fluorescence (485 nm/528 nm) of washed C. difficile resuspended in PBS at various time points. Data are represented as mean ± stdev. Repeated Measures ANOVA, **p < 0.01, ***p < 0.001, ****p < 0.0001.
We next aimed to determine what compounds could potentially impact C. difficile autofluorescence. We utilized Biolog© Phenotypic Microarray plates to screen a large number of compounds efficiently (Figures 2A–G; Figures 3A–C). We collected baseline levels of autofluorescence of the compounds and then added a 6 h culture of C. difficile R29201 in PBS at a final OD600nm of 1 to the plates and monitored the fluorescence after a 5 min incubation. We observed that several compounds could increase C. difficile autofluorescence (Figures 2A,C–G; Figures 3A,C). The compounds that induced the highest autofluorescence in C. difficile were the monosaccharide 2-deoxy-ribose (2.4-fold increase) (Figure 2A), acid sorbic acid (2.5-fold increase) (Figure 3A), sodium lactate (3.1-fold increase) (Figure 3C), 2,3-butanedione (2.3-fold increase) (Figure 3C). We also found a ~ 1.5-fold increase in C. difficile autofluorescence in response to the disaccharide turnanose (Figure 2B), polysaccharide laminarin (Figure 2D), alcohol sugar xylitol (Figure 2E), amino sugar N-acetyl-D-Glucosaminitol (Figure 2F), amino acid L-Lysine (Figure 2G), acids α-keto-glutaric acid, D-amino valeric acid, and tartaric acid (Figure 3A). Interestingly, we also found some compounds that decreased C. difficile fluorescence. We found that C. difficile autofluorescence was lower with aspartic acid (1.8-fold decrease) (Figure 3A), dextrin (1.9-fold decrease) (Figure 3C), D-trehalose (2.2-fold decrease) (Figure 2B), D-alanine (2.4-fold decrease) (Figure 2G), N-Acetyl-D-glucosamine (2.6-fold decrease) (Figure 2F), acetoacetic acid (2.9-fold decrease) (Figure 3A), L-fucose (3.2-fold decrease) (Figure 2A), D-glucosamine (5.6-fold decrease) (Figure 2F) and pyruvic acid (6.2-fold decrease) (Figure 3A). We did not observe a pattern in the type of compounds which influenced C. difficile autofluorescence; we observed compounds that could increase or decrease autofluorescence in the same classes of compounds. This indicates that C. difficile autofluorescence is influenced by multiple types of compounds.
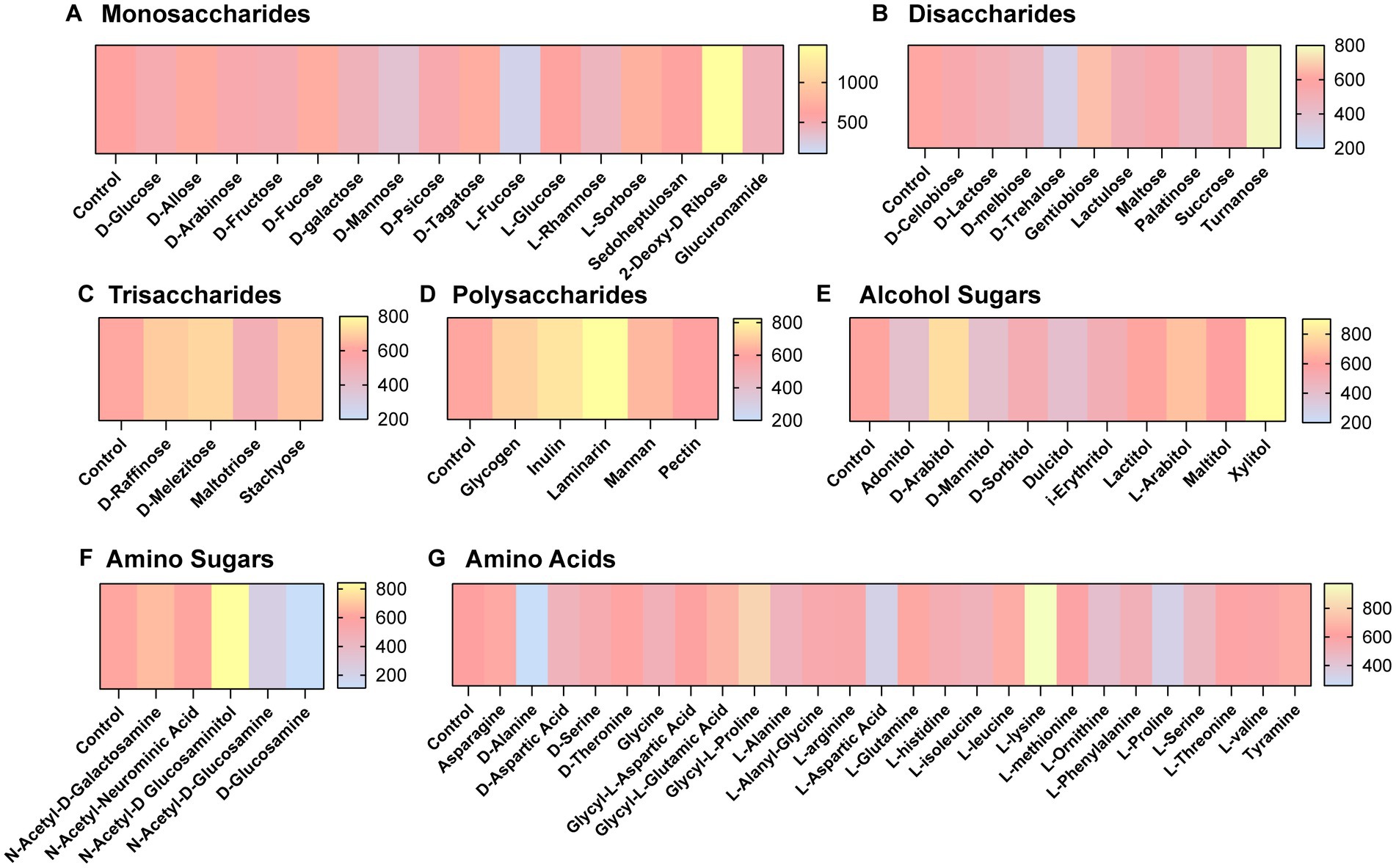
Figure 2. Clostridioides difficile autofluorescence is affected by certain sugars and amino acids. C. difficile was grown for 6 h in BHI anaerobically, washed with PBS and then added to Biolog© Phenotypic Microarray plates in PBS at an OD600nm = 1. Baseline line fluorescence and fluorescence with C. difficile was measured in the green channel (485 nm/528 nm). Compounds are grouped into (A) monosaccharides, (B) disaccharides, (C) trisaccharides, (D) polysaccharides, (E) alcohol sugars, (F) amino sugars, and (G) amino acids.
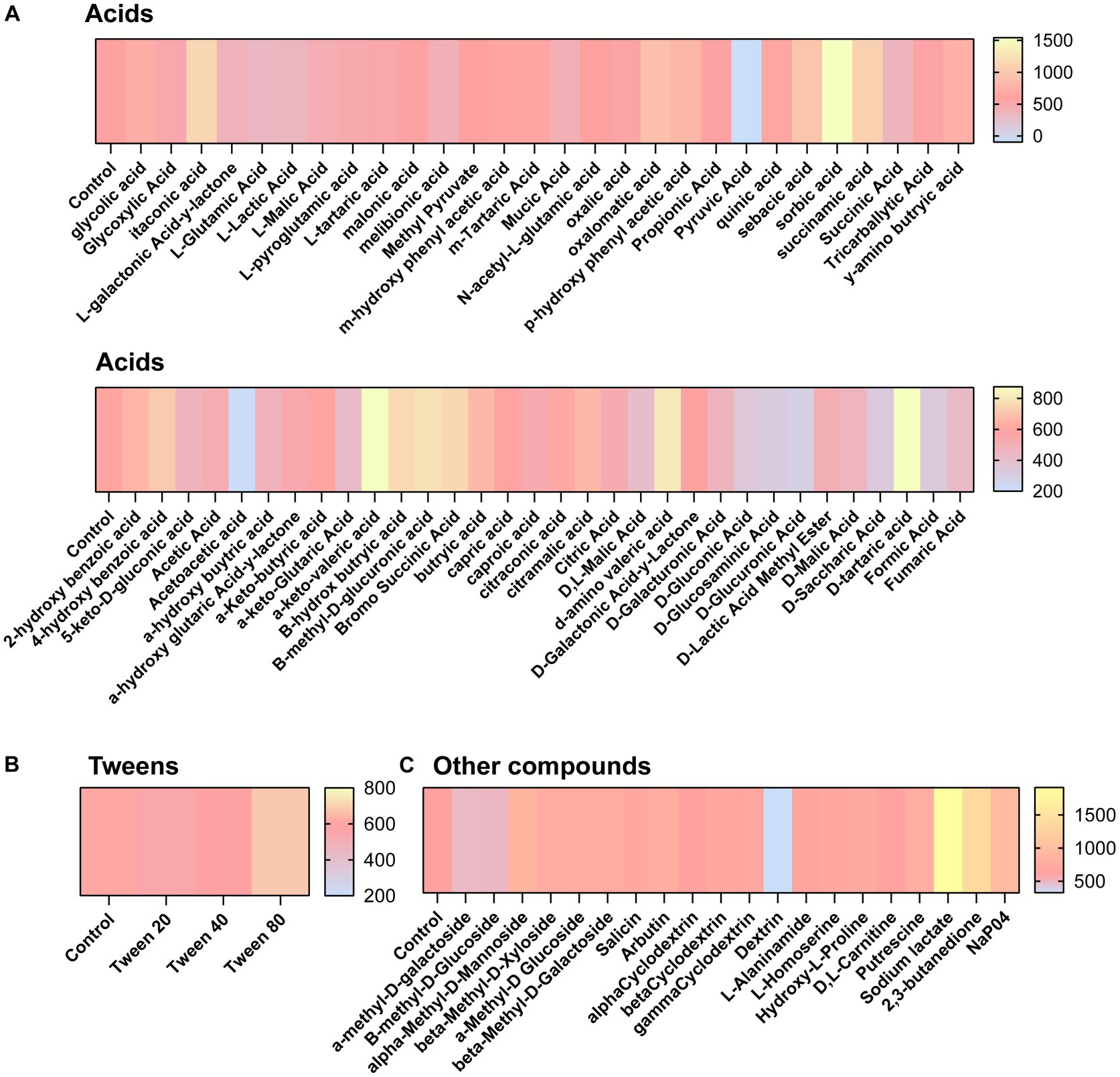
Figure 3. Clostridioides difficile autofluorescence is affected by various acids and other compounds. C. difficile was grown for 6 h in BHI anaerobically, washed with PBS and then added to Biolog© Phenotypic Microarray plates in PBS at an OD600nm = 1. Baseline line fluorescence and fluorescence with C. difficile was measured in the green channel (485 nm/528 nm). Compounds are grouped into (A) acids, (B) tweens, and (C) other compounds.
To confirm the findings from the Biolog© plates, we measured C. difficile autofluorescence with a range of concentrations from two of the compounds that showed an increase in C. difficile autofluorescence: L-lysine and pantothenate. Consistent with findings using the Biolog© microarrays, we saw a significant increase in C. difficile autofluorescence with L-lysine (Figure 4A) and pantothenate (Figure 4B) at concentrations ranging from 1 nM to 100 nM. We confirmed C. difficile’s autofluorescence by confocal microscopy (Supplementary Figure S2A). When we quantified the fluorescence of individual bacteria in confocal images, we observed cell to cell variation in the level of autofluorescence, with an overall increase in autofluorescence in the bacteria treated with 1 mM L-lysine and 1 mM pantothenate (Supplementary Figures S2B,C). These data support the finding that specific compounds can enhance C. difficile autofluorescence.
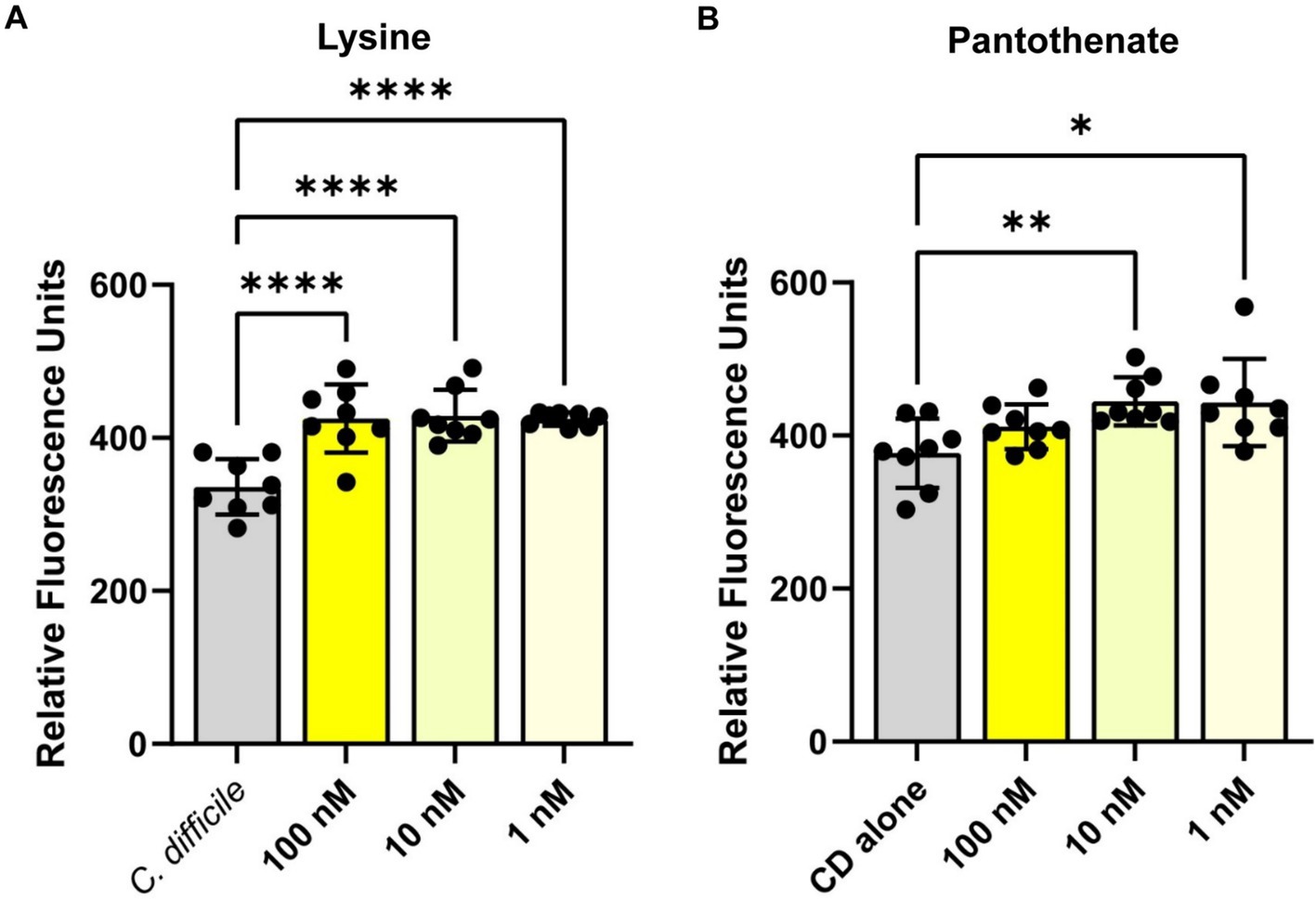
Figure 4. Clostridioides difficile autofluorescence increases in the presence of L-lysine and pantothenate. C. difficile was grown for 6 h in BHI anaerobically, washed with PBS and then added to various concentrations of (A) L-lysine or (B) pantothenate. Fluorescence was measured in the green channel (485 nm/528 nm). Data are represented as mean ± stdev. One Way ANOVA; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Gut bacteria enhance Clostridioides difficile autofluorescence
Several of the compounds we identified in the Biolog© microarrays that could induce C. difficile autofluorescence can be synthesized by bacteria. To investigate if bacterial metabolites could influence C. difficile autofluorescence, we grew C. difficile in cell-free bacterial conditioned media from various bacteria that can be found in the human gut (Figure 5). We found that that the metabolites from commensal E. coli, M. morganii, B. bifidum, B. breve and L. acidophilus did not affect C. difficile autofluorescence (Figure 5A). We also found that the classically pathogenic P. aeruginosa and L. monocytogenes metabolites did not influence autofluorescence in C. difficile. However, we did observe that metabolites from E. faecalis, S. mitis, K. aerogenes, and K. pneumoniae significantly increased overall C. difficile autofluorescence (Figure 5A). This effect was independent of C. difficile growth or culture pH, since C. difficile grew to a similar degree with all the bacterial supernatants and the final pH of C. difficile-supernatant culture was unchanged (Supplementary Figures S1A,B). To identify if C. difficile autofluorescence was increased rapidly by bacterial metabolites, we generated cell-free conditioned medium from E. faecalis, K. aerogenes and K. pneumoniae and examined C. difficile autofluorescence to these secreted metabolites. We found that all three of these cell-free supernatants could elevate C. difficile autofluorescence (Figure 5B). These results suggest that bacteria produce compounds that can increase C. difficile autofluorescence.
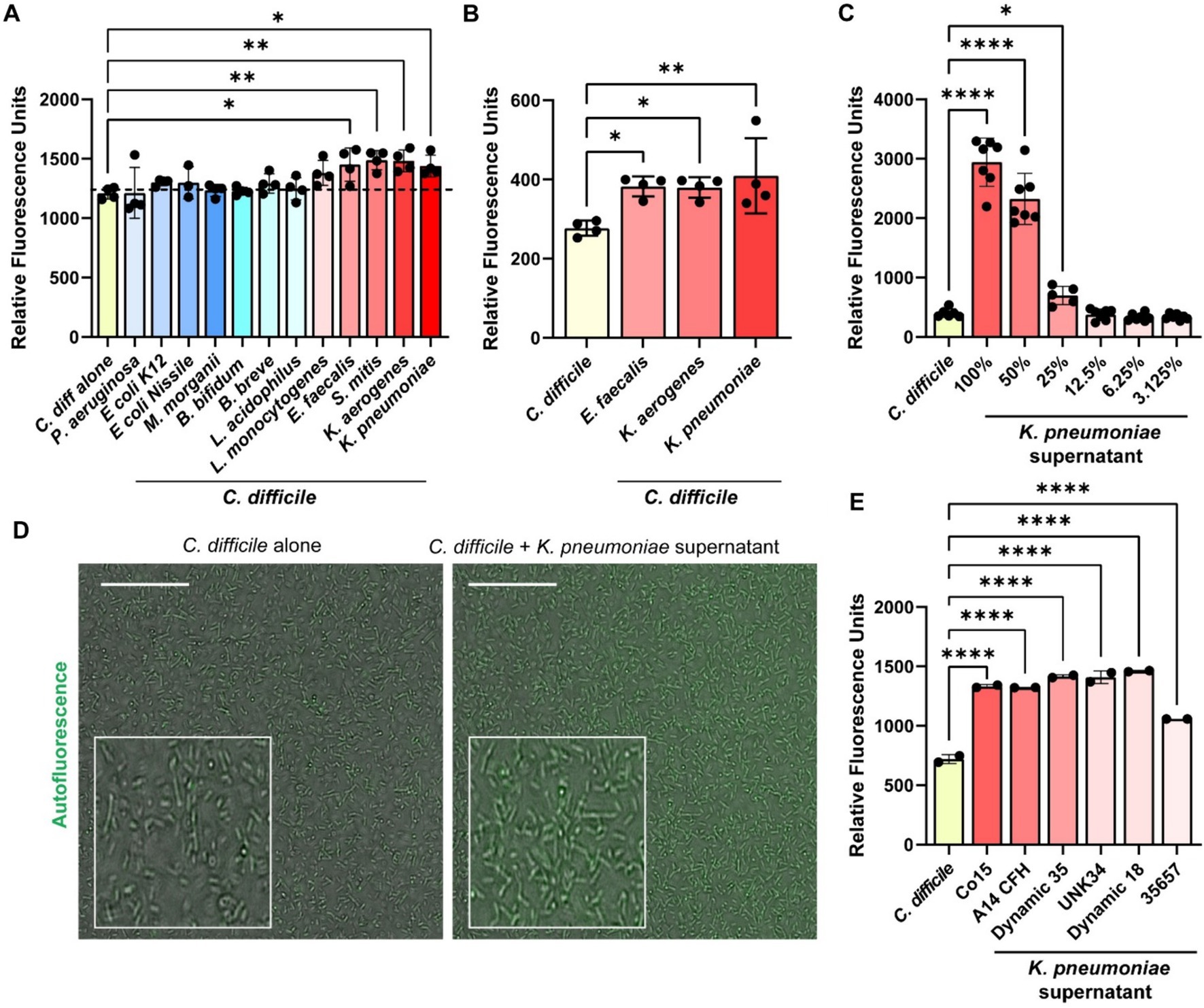
Figure 5. Bacterial metabolites increase C. difficile autofluorescence. (A) C. difficile was grown in a fully defined medium, ZMB1, supplemented with 10% of cell-free conditioned media from other bacteria. Green autofluorescence was measured in washed C. difficile cells after culturing for 20 h on a fluorescent plate reader (485 nm/528 nm). (B) C. difficile was grown for 6 h in BHI anaerobically, washed with PBS and then added to 25% of E. faecalis ATCC 35038, K. aerogenes ATCC 35029, and K. pneumoniae ATCC 35657 cell free conditioned media. Green autofluorescence was measured in washed C. difficile cells on a plate reader (485 nm/528 nm). (C) Green fluorescence was measured by a plate reader after C. difficile was incubated with various concentrations of K. pneumoniae ATCC 35657 conditioned media. (D) Representative fluorescent images of C. difficile alone or C. difficile after incubation with 50% K. pneumoniae ATCC 35657 cell free supernatant. Scale bar = 50 μm. (E) Green autofluorescence was measured after C. difficile by a plate reader was exposed to conditioned media from various K. pneumoniae strains after incubation. Data are represented as mean ± stdev. One Way ANOVA; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Klebsiella pneumoniae isolates enhance Clostridioides difficile autofluorescence
K. pneumoniae is an opportunistic bacterial species that is normally found in low abundance in the healthy human gut, but can increase substantially with antibiotics or in the setting of inflammation as one signature of a perturbed microbiota (Lepuschitz et al., 2020; Hudson Andrew et al., 2022). Studies have shown that K. pneumoniae relative abundance is increased in the feces of patients with C. difficile infection (Giuliano et al., 2014; Bruno et al., 2018; Hong et al., 2019; Golubovska et al., 2021). As a result, we decided to take a closer look at C. difficile autofluorescence with K. pneumoniae. We observed that C. difficile autofluorescence increased in a dose-dependent manner to the concentration of K. pneumoniae ATCC 35657 cell-free supernatant (Figure 5C). We also confirmed the increase in C. difficile’s autofluorescence in response to 50% of K. pneumoniae ATCC 35657 supernatant by microscopy (Figure 5D). When we quantified the fluorescence of individual C. difficile cells to K. pneumoniae metabolites by confocal microscopy, we found an overall increase in autofluorescence; although we observed a range of fluorescence in C. difficile bacteria (Supplementary Figures S3A,B). To determine if other K. pneumoniae strains could impact C. difficile autofluorescence, we obtained intestinal isolates of K. pneumoniae (C015, A14 CFH, A05E, Dynamic 35, UNK34, and Dynamic 18) and included another commercially available K. pneumoniae strain (CB1) in our assays. We generated cell-free conditioned media from these K. pneumoniae strains and found that C. difficile increased its overall autofluorescence when exposed to these K. pneumoniae supernatants for 5 min (Figure 5E). These results suggest that K. pneumoniae metabolites increase C. difficile autofluorescence, and that this effect is conserved across K. pneumoniae strains.
We next sought to determine if K. pneumoniae could increase C. difficile autofluorescence when the bacteria are grown together. We grew C. difficile and K. pneumoniae in mono- and co-cultures in ZMB1 for 20 h. We found that C. difficile was present in all the co-cultures by qPCR (Supplementary Figure S4A). We also confirmed that C. difficile was autofluorescent in the co-cultures of C. difficile and K. pneumoniae ATCC 35657 by microscopy (Supplementary Figure S4B). We then performed flow cytometry on mono-cultures and co-cultures of C. difficile and K. pneumoniae. We found that all the K. pneumoniae strains had undetectable levels of autofluorescence (Supplementary Figure S5). When we grew C. difficile and K. pneumoniae together for 20 h, we could detect two populations of cells. Since K. pneumoniae does not exhibit green autofluorescence, any cells that were positive for green fluorescence were identified to be C. difficile. We observed that C. difficile had high green fluorescence when grown in co-culture with all strains of K. pneumoniae tested and this fluorescence could distinguish C. difficile from K. pneumoniae (Figure 6). Collectively these findings suggest that C. difficile’s autofluorescence is influenced by dietary compounds and bacterial metabolites and that the increase in C. difficile autofluorescence could potentially allow researchers to identify C. difficile in mixed bacterial populations.

Figure 6. Clostridioides difficile autofluorescence can distinguish C. difficile in co-culture with K. pneumoniae. C. difficile and K. pneumoniae strains were grown as a co-cultures in ZMB1 for 20 h, stained with Hoechst and examined by flow cytometry. The data depicts a representative gating strategy for identifying highly autofluorescent C. difficile in the green channel and histograms of the quantified fluorescence. (A) Flow analysis of C. difficile monocultures. (B) Flow cytometry analysis of C. difficile and K. pneumoniae co-cultures.
Discussion
C. difficile displays intrinsic green autofluorescence, but the factors that influence this autofluorescence have not been fully elucidated. In this study, we sought to identify compounds and conditions that could enhance C. difficile autofluorescence. We found that several compounds elevated C. difficile autofluorescence, including 2-deoxy-D-Ribose, turnanose, laminarin, L-lysine, N-acetyl-D-glucosaminitol, xylitol, sorbic acid, α-keto-valeric acid, D-amino valeric acid, D-tartaric acid, sodium lactate, 2,3-butanedione and pantothenate. We also found that bacteria can produce compounds which increase autofluorescence in C. difficile. Interestingly, we observed that K. pneumoniae strains were highly efficient at upregulating autofluorescence; so much so that we could grow C. difficile with K. pneumoniae and measure autofluorescence by flow cytometry. These findings expand our knowledge of C. difficile autofluorescence.
C. difficile autofluorescence has been reported by several groups. Ransom et al. first reported C. difficile’s green autofluorescence in 2015 while studying a method to fluorescently label C. difficile with minimal oxygen required to mature the chromophore (Ransom Eric et al., 2015). Since then, other groups have utilized this autofluorescence to visualize C. difficile in various capacities. Garcia-Garcia et al. paired C. difficile autofluorescence images with SNAP-PrkC staining (Garcia-Garcia et al., 2022). The widespread autofluorescence allowed for the localization of SNAP-PrkC to be visualized at both stationary growth phase, as well as sporulation phase of C. difficile. Kint et al. also used this autofluorescence as a tool to image C. difficile to then show localization of other targets (Kint et al., 2020). In this study, we expand the utility of using C. difficile autofluorescence to flow cytometry. We found that when C. difficile was grown with K. pneumoniae strains, it became highly autofluorescence and we could distinguish C. difficile and K. pneumoniae by flow cytometry. This method provides an alternative validation of C. difficile growth with other bacteria and could prove valuable for other studies involving C. difficile-microbe cross-talk. Previous studies have observed the beneficial relationship between C. difficile and other bacteria, such as Enterococci (Smith et al., 2022) and Fusobacteria (Engevik et al., 2021) so this technique offers another method to observe these relationships.
Consistent with other groups, we found that C. difficile autofluorescence increased with growth (Ransom Eric et al., 2015). Growth phase-dependent autofluorescence has also been observed in other bacteria, including Escherichia coli or Bacillus pumilus (Manzo et al., 2013; Mihalcescu et al., 2015). We opted to examine autofluorescence in C. difficile after 6 h of growth because this point had a lower baseline level of autofluorescence. This allowed us to monitor increases in autofluorescence in response to various factors. To the best of our knowledge, no group to date has shown that different bacteria and their metabolites can influence C. difficile’s autofluorescence. In this study we demonstrated that compounds potentially produced by bacteria and bacterial secreted metabolites can influence C. difficile autofluorescence. One of the interesting compounds we identified that elevated C. difficile autofluorescence of L-lysine. We found that several concentrations of L-lysine increased C. difficile’s autofluorescence. Certain bacteria are able to produce L-lysine, including Corynebacterium glutamicum (Nærdal et al., 2017), Bacillus methanolicus (Nærdal et al., 2017), various rumen bacteria (Styriak et al., 1992), and K. pneumoniae (Gilani et al., 2023). We speculate that L-lysine produced by K. pneumoniae could be responsible for the elevated levels of C. difficile autofluorescence that we observed with K. pneumoniae supernatant and co-cultures.
In addition to L-lysine, we also observed that pantothenate could increase C. difficile’s fluorescence. Pantothenate has been found in other bacteria to elevate glutathione and promote antibiotic resistance (Yan et al., 2023). It is not clear how pantothenate enhances autofluorescence, but it could be due to alterations in cell stress. A link between autofluorescence and oxidative stress has been proposed in Bacillus (Manzo et al., 2013) and E. coli (Mihalcescu et al., 2015). Others have found that autofluorescence can be enhanced in bacteria under antibiotic stress (Renggli et al., 2013) and has been correlated with an increase in expression of flavin biosynthesis pathways (Surre et al., 2018). In future, it would be interesting to examine C. difficile stress upon exposure to the autofluorescence inducing compounds identified in our study.
In this study we observed an overall increase in fluorescence intensity. This was observed by assessing the population on a fluorescent plate reader and by quantifying the fluorescence of individual bacteria by confocal microscopy. Interestingly we did identify variation among individual C. difficile cells in response to L-lysine, pantothenate and K. pneumoniae metabolites. Populations of genetically identical cells growing under uniform conditions can exhibit cell-to-cell heterogeneity in gene expression. Oliveira Paiva et al., 2022, used a similar single-cell autofluorescence analysis to examine C. difficile fluorescence and found that C. difficile had an increase in the average fluorescence intensity during the stationary growth phase, but there was significant heterogeneity between cells (Oliveira Paiva et al., 2022). Our data suggests an overall increase in autofluorescence intensity across all cells, although some cells did respond more than others. It is possible that a sub-population of C. difficile cells were better poised to respond to certain compounds and these cells exhibited a more rapid increase in autofluorescence. In the future it would be interesting to examine individual bacterial cells to further identify potential sub-populations of C. difficile.
A limitation of this study was that we were limited in the bacteria we could grow with C. difficile in co-culture to observe the effect on C. difficile autofluorescence. Bacteria such as P. aeruginosa (Zhang et al., 2024) and E. coli (Renggli et al., 2013) display their own autofluorescence. As such, we avoided using these bacteria to focus solely on C. difficile. We were then further limited by bacteria that would allow both itself and C. difficile to grow in co-culture. In a pilot experiment, we observed that several commensal bacteria out-competed C. difficile. As a result, we chose to focus on K. pneumoniae since the co-cultures supported the growth of both C. difficile and K. pneumonia. Future studies could potentially look at other bacterial co-culture pairs to see if other species enhance or diminish C. difficile autofluorescence.
In conclusion, our data sheds light on different factors that influence C. difficile autofluorescence, such as growth time in culture and compounds found in the human diet or produced by other gut bacteria. We specifically identified that L-lysine and K. pneumoniae could upregulate C. difficile autofluorescence. More in-depth studies will be needed to further understand the pathways being affected that lead to C. difficile autofluorescence, however these findings highlight a facet of the complexity of bacterial interactions.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
TT: Conceptualization, Data curation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing. JG: Data curation, Formal analysis, Investigation, Writing – review & editing. SaD: Data curation, Investigation, Writing – review & editing. JK: Data curation, Resources, Writing – review & editing. JZ: Investigation, Resources, Writing – review & editing. SuD: Investigation, Resources, Writing – review & editing. GW: Investigation, Resources, Writing – review & editing. KM: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing. AE: Formal analysis, Investigation, Resources, Writing – review & editing. ME: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Resources, Writing – review & editing. AT: Writing – review & editing, Data curation, Formal analysis.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Institute of Health T32DK124191-01A1 (TT), T32GM132055-01 (JSG), T32GM132055, Histochemical Society Cornerstone Grant (SD), K01DK123195 (MAE), K01DK121869 (ACE), P30 DK123704 (ACE), P20 GM120457 (MAE), U19AI174998 (JPZ), USDA In-House Project 8072-41000-108-00-D (KM), P30 DK050306 (GDW), and supported by MUSC startup funds to MAE and ACE. Klebsiella strain isolation was partially supported by the USDA In-House Project 8072-41000-108-00-D, “In Vitro human intestinal microbial ecosystems: effect of diet”.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1459795/full#supplementary-material
References
Bruno, G., Colangelo, L., Badiali, D., Minisola, S., Corazziari, E. S., and Gianni, W. (2018). Concomitant resolution through fecal microbiota transplantation of Clostridium difficile and OXA-48-producing Klebsiella pneumoniae. Infect. Dis. 50, 565–566. doi: 10.1080/23744235.2018.1442019
Buckley, A. M., Jukes, C., Candlish, D., Irvine, J. J., Spencer, J., Fagan, R. P., et al. (2016). Lighting up Clostridium Difficile: reporting gene expression using fluorescent Lov domains. Sci. Rep. 6:23463. doi: 10.1038/srep23463
Davies, K., Lawrence, J., Berry, C., Davis, G., Yu, H., Cai, B., et al. (2020). Risk factors for primary Clostridium difficile infection; results from the observational study of risk factors for Clostridium difficile infection in hospitalized patients with infective diarrhea (ORCHID). Front. Public Health 8:293. doi: 10.3389/fpubh.2020.00293
Dong, D., Su, T., Chen, W., Wang, D., Xue, Y., Lu, Q., et al. (2023). Clostridioides difficile aggravates dextran sulfate solution (DSS)-induced colitis by shaping the gut microbiota and promoting neutrophil recruitment. Gut Microbes 15:2192478. doi: 10.1080/19490976.2023.2192478
Donnelly, M. L., Shrestha, S., Ribis, J. W., Kuhn, P., Krasilnikov, M., Alves Feliciano, C., et al. (2022). Development of a dual-fluorescent-reporter system in Clostridioides difficile reveals a division of labor between virulence and transmission. Gene Expr. 7:e0013222. doi: 10.1128/msphere.00132-22
Engevik, M. A., Danhof, H. A., Auchtung, J., Endres, B. T., Ruan, W., Bassères, E., et al. (2021). Fusobacteriumnucleatum adheres to Clostridioides difficile via the RadD Adhesin to enhance biofilm formation in intestinal mucus. Gastroenterology 160, 1301–1314.e8. doi: 10.1053/j.gastro.2020.11.034
Engevik, K. A., Gonzalez, H., Daniels, C., Stavert, R. K., Oezguen, N., Engevik, M. A., et al. (2023). A high-throughput protocol for measuring solution pH of bacterial cultures using UV-vis absorption spectrophotometry. STAR Protoc 4:102540. doi: 10.1016/j.xpro.2023.102540
Eze, P., Balsells, E., Kyaw, M. H., and Nair, H. (2017). Risk factors for Clostridium difficile infectionsan overview of the evidence base and challenges in data synthesis. J. Glob. Health 7:010417. doi: 10.7189/jogh.07.010417
Garcia-Garcia, T., Douché, T., Giai Gianetto, Q., Poncet, S., El Omrani, N., Smits, W. K., et al. (2022). In-depth characterization of the Clostridioides difficile Phosphoproteome to identify Ser/Thr kinase substrates. Mol. Cell. Proteomics 21:100428. doi: 10.1016/j.mcpro.2022.100428
Gilani, N., Maqbool, T., Muhammad, M., Ume, H., Anwar, T., and Huma, Q. (2023). Screening of lysine production by Escherichia coli and Klebsiella isolates at different carbon sources. Biosci. J. 39:e39004. doi: 10.14393/BJ-v39n0a2023-60568
Giuliano, S., Guastalegname, M., Jenco, M., Morelli, A., Falcone, M., and Venditti, M. (2014). Severe community onset healthcare-associated Clostridium difficile infection complicated by carbapenemase producing Klebsiella pneumoniae bloodstream infection. BMC Infect. Dis. 14:475. doi: 10.1186/1471-2334-14-475
Golubovska, I., Vigante, D., Malzubris, M., Raga, L., Isajevs, S., and Miscuks, A. (2021). Severe Clostridium difficile infection with extremely high leucocytosis complicated by a concomitant bloodstream infection caused by Klebsiella pneumoniae after osteomyelitis surgery: a case report. Int. J. Surg. Case Rep. 78, 155–158. doi: 10.1016/j.ijscr.2020.12.018
Guh, A. Y., Adkins, S. H., Li, Q., Bulens, S. N., Farley, M. M., Smith, Z., et al. (2017). Risk factors for community-associated Clostridium difficile infection in adults: a case-control study. Open forum. Infect. Dis. 4:ofx171. doi: 10.1093/ofid/ofx171
Guh, A. Y., and Kutty, P. K. (2018). Clostridioides difficile Infection. Ann. Intern. Med. 169:Itc49-itc64. doi: 10.7326/AITC201810020
Guh, A. Y., Mu, Y., Winston, L. G., Johnston, H., Olson, D., Farley, M. M., et al. (2020). Trends in U.S. burden of Clostridioides difficile infection and outcomes. N. Engl. J. Med. 382, 1320–1330. doi: 10.1056/NEJMoa1910215
Hong, W., Yang, J., Cheng, Y., Huang, X., Rao, F., Zhang, T., et al. (2019). Bacteria co-colonizing with Clostridioides difficile in two asymptomatic patients. Open Life Sciences 14, 628–637. doi: 10.1515/biol-2019-0071
Horvath, T. D., Haidacher, S. J., Engevik, M. A., Luck, B., Ruan, W., Ihekweazu, F., et al. (2023). Interrogation of the mammalian gut-brain axis using LC-MS/MS-based targeted metabolomics with in vitro bacterial and organoid cultures and in vivo gnotobiotic mouse models. Nat. Protoc. 18, 490–529. doi: 10.1038/s41596-022-00767-7
Hudson Andrew, W., Barnes Andrew, J., Bray Andrew, S., Ornelles David, A., and Zafar, M. A. (2022). Klebsiella pneumoniae l-Fucose metabolism promotes gastrointestinal colonization and modulates its virulence determinants. Infect. Immun. 90, e00206–e00222. doi: 10.1128/iai.00206-22
Kint, N., Alves Feliciano, C., Martins, M. C., Morvan, C., Fernandes, S. F., Folgosa, F., et al. (2020). How the anaerobic Enteropathogen Clostridioides difficile tolerates low O(2) tensions. MBio 11, e01559–e01520. doi: 10.1128/mBio.01559-20
Lepuschitz, S., Hauser, K., Schriebl, A., Schlagenhaufen, C., Stöger, A., Chakeri, A., et al. (2020). Fecal Klebsiella pneumoniae carriage is intermittent and of high clonal diversity. Front. Microbiol. 11:581081. doi: 10.3389/fmicb.2020.581081
Manzo, N., Di Luccia, B., Isticato, R., D'apuzzo, E., De Felice, M., and Ricca, E. (2013). Pigmentation and sporulation are alternative cell fates in Bacillus pumilus SF214. PLoS One 8:e62093. doi: 10.1371/journal.pone.0062093
Mihalcescu, I., Van-Melle Gateau, M., Chelli, B., Pinel, C., and Ravanat, J. L. (2015). Green autofluorescence, a double edged monitoring tool for bacterial growth and activity in micro-plates. Phys. Biol. 12:066016. doi: 10.1088/1478-3975/12/6/066016
Nærdal, I., Netzer, R., Irla, M., Krog, A., Heggeset, T. M. B., Wendisch, V. F., et al. (2017). L-lysine production by Bacillus methanolicus: genome-based mutational analysis and l-lysine secretion engineering. J. Biotechnol. 244, 25–33. doi: 10.1016/j.jbiotec.2017.02.001
Oliveira Paiva, A. M., Friggen, A. H., Douwes, R., Wittekoek, B., and Smits, W. K. (2022). Practical observations on the use of fluorescent reporter systems in Clostridioides difficile. Antonie Van Leeuwenhoek 115, 297–323. doi: 10.1007/s10482-021-01691-8
Ransom Eric, M., Ellermeier Craig, D., and Weiss David, S. (2015). Use of mCherry red fluorescent protein for studies of protein localization and gene expression in Clostridium difficile. Appl. Environ. Microbiol. 81, 1652–1660. doi: 10.1128/AEM.03446-14
Renggli, S., Keck, W., Jenal, U., and Ritz, D. (2013). Role of autofluorescence in flow cytometric analysis of Escherichia coli treated with bactericidal antibiotics. J. Bacteriol. 195, 4067–4073. doi: 10.1128/JB.00393-13
Singh, H., Nugent, Z., Yu, B. N., Lix, L. M., Targownik, L. E., and Bernstein, C. N. (2017). Higher incidence of Clostridium difficile infection among individuals with inflammatory bowel disease. Gastroenterology 153:e432, 430–438.e2. doi: 10.1053/j.gastro.2017.04.044
Smith, A. B., Jenior, M. L., Keenan, O., Hart, J. L., Specker, J., Abbas, A., et al. (2022). Enterococci enhance Clostridioides difficile pathogenesis. Nature 611, 780–786. doi: 10.1038/s41586-022-05438-x
Styriak, I., Timashova-Kalcheva, E. O., Kmeí, V., and Maljuta, S. S. (1992). DAP-decarboxylase activity and lysine production by rumen bacteria. Arch. Tierernahr. 42, 71–77. doi: 10.1080/17450399209428531
Surre, J., Saint-Ruf, C., Collin, V., Orenga, S., Ramjeet, M., and Matic, I. (2018). Strong increase in the autofluorescence of cells signals struggle for survival. Sci. Rep. 8:12088. doi: 10.1038/s41598-018-30623-2
Yan, B. B., Dong, X. S., Wang, J. P., Li, X. Y., An, L., Wang, X. R., et al. (2023). Glutamate-pantothenate pathway promotes antibiotic resistance of Edwardsiella tarda. Front. Microbiol. 14:1264602. doi: 10.3389/fmicb.2023.1264602
Keywords: Clostridioides difficile, Klebsiella pneumoniae, autofluorescence, metabolites, intestine
Citation: Ticer TD, Tingler AM, Glover JS, Dooley SA, Kendrick J, Zackular JP, Devkota S, Wu GD, Mahalak K, Engevik A and Engevik MA (2024) Bacterial metabolites influence the autofluorescence of Clostridioides difficile. Front. Microbiol. 15:1459795. doi: 10.3389/fmicb.2024.1459795
Edited by:
Gregg Pettis, Louisiana State University, United StatesReviewed by:
Monica Serrano, Universidade Nova de Lisboa, PortugalFernando Gil, Andres Bello University, Chile
Copyright © 2024 Ticer, Tingler, Glover, Dooley, Kendrick, Zackular, Devkota, Wu, Mahalak, Engevik and Engevik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Melinda A. Engevik, ZW5nZXZpa0BtdXNjLmVkdQ==
 Taylor D. Ticer1
Taylor D. Ticer1 Janiece S. Glover
Janiece S. Glover Joseph P. Zackular
Joseph P. Zackular Suzanne Devkota
Suzanne Devkota Karley Mahalak
Karley Mahalak Melinda A. Engevik
Melinda A. Engevik