- 1Yunnan Provincial Engineering and Research Center for Sustainable Utilization of Honey Bee Resources, Eastern Bee Research Institute, College of Animal Science and Technology, Yunnan Agricultural University, Kunming, China
- 2Institute of Sericulture and Apiculture, Yunnan Academy of Agricultural Sciences, Mengzi, China
- 3College of Food Science and Technology, Yunnan Agricultural University, Kunming, China
Camellia reticulata, an ancient plant species endemic to Yunnan Province, China, remains underexplored in terms of its endophytic bacterial communities. The plant tissue pedicel serves as the connection between the flower and the stem, not only delivers nutrients but also transmits metabolic substances from endophytic bacteria to the nectar during long-term microbial colonization and probably improves the antagonistic activity of nectar against yeast. Hence, 138 isolates of endophytic bacteria have been isolated in this study from the pedicels of 12- and 60-year-old C. reticulata. Comparative analysis revealed significantly higher density of endophytic bacteria in older trees. Among these isolates, 29 exhibited inhibitory effects against nectar yeasts. Most of the isolates displayed positive results for Gram staining, catalase reaction, gelatin liquefaction, and motility. Additionally, the isolates demonstrated the ability to utilize diverse substrates, such as glucose, nitrate, and starch. Based on 16S rRNA molecular biology analysis, these isolates were identified to be 11 different species of 6 genera, with the majority belonging to Bacillus genus. Notably, C1 isolate, identified as Bacillus spizizenii, exhibited strongest antagonistic effect against three yeasts, i.e., Metschnikowia reukaufii, Cryptococcus laurentii, and Rhodotorula glutinis, with minimum inhibitory concentration values below 250 μg/mL. Major metabolites of B. spizizenii were aminoglycosides, beta-lactams, and quinolones, which possess antimicrobial activities. Furthermore, KEGG enrichment pathways primarily included the synthesis of plant secondary metabolites, phenylpropanoids, amino acids, alkaloids, flavonoids, neomycin, kanamycin, and gentamicin. Therefore, antagonistic activity of B. spizizenii against yeasts could be attributed to these antibiotics. The findings highlight the diverse endophytic bacteria associated with C. reticulata, indicating their potential as a valuable resource of bioactive metabolites. Additionally, this study provides new insights into the role of endophytic bacteria of pedicels in enhancing nectar resistance against yeasts.
1 Introduction
Nectar is a liquid secreted by the nectaries of flowering plants. It is rich in sugars, amino acids, organic acids, volatile substances, and various other compounds, attracting pollinators (bees, butterflies, birds, and other animals) and aiding in pollination (Canto and Herrera, 2012). However, the pollinators can pose a risk of nectar contamination by yeasts and bacteria (Russell et al., 2019; Zemenick et al., 2021). In particular, the nectar rich in various nutrients is highly conducive to microbial growth and proliferation. Yeasts belonging to phyla Ascomycota and Basidiomycota are found commonly in nectar, including genera Metschnikowia, Cryptococcus, Rhodotorula, and Saccharomyces (Canto and Herrera, 2012; Zhou et al., 2016). These yeasts have a unique ecological niche, as they live and proliferate in the nectar of flowers and can significantly alter the composition and concentration of sugars and amino acids, reducing the total sugar concentration and transforming the sucrose-rich nectar into fructose-rich nectar, thus affecting the interactions between plant and pollinators (Canto and Herrera, 2012; Zhou et al., 2016). It is obvious that the nectar does not decay due to microbial contamination and its quality remains superior, which indicates the presence of an effective antimicrobial mechanism (Herrera et al., 2008, 2009; Canto and Herrera, 2012). However, research on the antimicrobial substances in floral nectar is still in the exploratory stage. Furthermore, numerous factors may contribute to the efficacy of these substances, which need to be explored (Sasu et al., 2010).
Endophytic bacteria form a symbiotic relationship with the plant. They live inside plant tissues without causing any harm to host, providing resistance to plant against disease and environmental stress, promoting plant growth, and inhibiting the pathogens (Santoyo et al., 2016). In recent years, endophytic bacteria have been utilized as biocontrol agent to suppress plant pathogens. They can produce various antimicrobial metabolites, including alkaloids, peptides, steroids, and terpenoids, thereby providing resistance to host plants against microbial stress (Zhang et al., 2006; Pieterse et al., 2014). For instance, Bacillus velezensis KOF112 exhibited broad-spectrum antifungal activity and contributed to disease prevention and control in grape cultivation in a previous study (Hamaoka et al., 2021). Metabolites secreted by Sphingomonas melonis, isolated from rice seeds, exhibited significant antagonistic effects against the plant pathogen Burkholderia plantarii (Matsumoto et al., 2021). In addition, metabolites of Bacillus thuringiensis, isolated from Cordia dichotoma, also displayed diverse antimicrobial activities (Sharma and Mallubhotla, 2022). These reports highlight the significant role of endophytic bacteria in helping host plants respond to microbial infections. The colonization of endophytes can alter the composition of volatile metabolites and microbiota in nectar, which in turn attracts more pollinators and enhances the reproductive fitness of plants (González-Mas et al., 2023). These findings provide a new research avenue for endophytic bacteria to assist nectar resistance to yeasts.
Camellia reticulata (Theaceae family) is a rare and protected plant species in China. It is a unique woody oil tree species in Yunnan Province, which has a long history of cultivation by local people and holds great significance in economy (Feng, 1980). Observably, C. reticulata with the large quantity of nectar produced by its flowers over a long flowering period, experiences frequent visitation by birds, making the nectar highly susceptible to yeast contamination. However, its nectar is not fermented quickly in such a process. In previous studies, endophytic bacteria have been found in the roots, stems, and leaves of C. reticulata, exhibiting antagonistic effects against root rot, leaf blight, anthracnose, and soft rot (Li et al., 2009; Jiang et al., 2016). This shows that the plant tissue of C. reticulata is rich in endophytic bacteria, and improves the plant's fitness. Pedicels are crucial components of flowers, supporting not only their spatial distribution but also serving as the channels that connect the stems and flowers and enable the transport of nutrients from plant to flowers (Zhou, 2015). The sap, produced in the phloem sieve tubes of plants, is the primary component required for nectar formation, which is transported to the nectaries through the sieve tubes (Tejesvi et al., 2013). The endophytic bacteria are closely associated with this process, as they colonize the plant pedicels for extended periods and serve as valuable biological resources. However, there has been limited research on endophytic bacteria living in the pedicels of C. reticulata and their antagonistic activity against nectar yeasts. This research focuses on: (i) isolating endophytic bacteria from C. reticulata pedicels, (ii) screening and identifying antagonistic isolates against nectar yeasts, (iii) evaluating Bacillus spizizenii's anti-yeast potential, and analyzing its metabolite composition. The aim is to understand how heterospecific-pollinated plants maintain the stability and quality of nectar under yeast stress.
2 Materials and methods
2.1 Collection of plant samples
In February 2023, samples of C. reticulata were collected from Shaba Forest Farm in Tengchong, Yunnan Province (98°5′86″ E, 24°9′57″ N). This area has a tropical monsoon climate, with an average annual temperature of 14.6°C, average sunshine duration of ~2,167 h, average precipitation of 1,531 mm, and average altitude of 2,149 m. Based on the tree information management archival records of the forest farm, 12- and 60-year-old healthy C. reticulata plants were selected for sample collection. Specifically, fresh flower buds at the fully closed calyx stage were randomly collected from C. reticulata using sterilized scissors, placed in sterile sampling bags, and stored at 4°C (Figure 1).
2.2 Isolation of endophytic bacteria
2.2.1 Surface sterilization of samples
After washing the samples under flowing tap water, flower buds and stems were removed under sterile conditions, while preserving the pedicels. Following the method described by Cardoso et al. (2020), rigorous surface sterilization of pedicels was performed. Firstly, the pedicels were rinsed with sterile distilled water, and then immersed in 75% ethanol for 60 s. After four repeated rinsing steps, pedicels were dried using sterile filter paper, and then immersed in a 5% NaClO solution for 2 min. Subsequently, pedicels were rinsed again four times with sterile distilled water and dried with sterilized filter paper. To test the effectiveness of surface sterilization, 100 μL of the final rinse water was spread onto Luria Bertani (LB) and Tryptic Soy Agar (TSA) culture media. No microbial growth on media after 72 h incubation at 30°C indicated successful surface sterilization of the pedicels.
2.2.2 Isolation, purification, and preservation of endophytic bacteria
Firstly, 1 g surface-sterilized pedicels were transferred into a sterile mortar and grounded along with a small amount of quartz sand and 1 mL of PBS solution. After thorough grinding, 9 mL of PBS solution was added into the pedicel extracts and mixed well. Subsequently, 101-104-fold dilution gradients were prepared, and 100 μL of these diluted suspensions were plated onto both LB and TSA culture media. The culture plates were then incubated at 30°C for 24–72 h, with three replicates for each diluted suspension. LB agar medium, exhibiting superior results compared to TSA medium, was selected for further isolation and purification of endophytic bacteria. Based on colony morphology, isolates were purified through repeated streaking and sub-culturing. The purified isolates were cultured in LB broth media at 30°C and 130 rpm until they reached the exponential growth phase. Finally, pure isolates were preserved in 50% glycerol stocks and stored at −80°C for subsequent experiments.
2.3 Screening of endophytic bacteria with antagonistic activity against nectar yeasts
Three yeasts were used as indicators to screen the endophytic bacteria with antagonistic activity against nectar yeasts. These yeasts, Metschnikowia reukaufii (CGMCC2.2728), Cryptococcus laurentii (CGMCC2.2425), and Rhodotorula glutinis (CGMCC2.2945), were all procured from the China General Microbiological Culture Collection Center (CGMCC).
Firstly, the yeasts were recovered and cultured using Malt Extract Broth (MEB) medium. Their suspensions were prepared by diluting the cultures to a concentration of 0.5 McFarland standard (~108 CFU/mL) using PBS solution. A sterile cotton swab was used to evenly spread the prepared suspensions onto Potato Dextrose Agar (PDA) plates as the test media plates. Subsequently, single colonies of endophytic bacterial isolates isolated from the C. reticulata pedicels were picked with an inoculating needle and placed on the test media plates. After overnight incubation at 28°C, inhibition zones on the plates were observed. This step is a preliminary screening, which is simple to operate and can quickly find endophytic bacteria with antagonistic effect on nectar yeast. To evaluate their anti-yeast effects, a re-screening using equivalent quantities of fermentation broth is required. These bacteria were then cultured in LB broth media for 3 days at 34°C with shaking at 120 r/min. The supernatants were obtained by centrifugation of cultures at 12,000 rpm for 15 min, and used to determine the antagonistic activity against nectar yeasts. Hundred microliter of supernatant was added to the oxford cups, while LB medium without the supernatant of endophytic bacteria served as a negative control. LB medium with amphotericin B was used as a positive control. All media were sealed with parafilm and incubated at 28°C for 24 h. After incubation, diameter of inhibition zone produced by each endophytic bacterial isolate against yeasts were measured.
2.4 Identification of endophytic bacteria with antagonistic activity against nectar yeasts
The endophytic bacterial isolate showing antagonistic activity against yeasts were preliminarily identified by observing their morphological characteristics, Gram-reaction, motility, and activities of amylase, catalase, and gelatinase enzymes (Goodfellow et al., 2009). In addition, using the total DNA extraction kit (E.Z.N.A.® Soil DNA Kit), DNA of each endophytic bacterial isolate with anti-yeast activity was extracted. A NanoDrop spectrophotometer was used to determine the concentration of genomic DNA by measuring the absorbance at 260 nm. The primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) were used to amplify the 16S rRNA gene of isolated DNA samples through polymerase chain reaction (PCR). Due to their high conservativeness, broad applicability, moderate GC content and Tm values, as well as strong support in the literature, have become the preferred primer pair for the amplification and identification of bacterial 16S rRNA genes, and are widely used for bacterial identification. The reaction system (25 μL) consisted of: 1 μL of each primer (10 μmol/L), 2 μL of template (20 ng/μL), and 21 μL of PCR Mix. PCR reaction conditions were set as: 96°C for 5 min; followed by 35 cycles of 96°C for 20 s, 56°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 10 min. PCR products were stored at 4°C. The amplification results were verified by 1% agarose gel electrophoresis (150 V/100 mA for 20 min), and the target DNA fragments were recovered using the QIAquick Agarose Gel Recovery Kit (Item code: 28704). Purified DNA fragments were sent to Shanghai Majorbio Bio-Pharm Technology Co., Ltd. for sequencing. The obtained sequences were aligned using DNAMAN 6.0.3.99 (Lynnon Biosoft, San Ramon, CA, USA) and analyzed using BlastN program (http://www.ncbi.nlm.nih.gov/BLAST). The sequences were also cross-checked with the EzTaxon and RDP databases to select the highest scoring species for further analysis, based on the similarity to identified bacteria. All obtained bacterial 16S rRNA sequences have been deposited in GenBank (Supplementary Table 1).
2.5 Extraction of crude extract of B. spizizenii with ethyl acetate
The endophytic bacterium C1 B. spizizenii was activated and revitalized prior to inoculation into LB broth medium. It was then incubated at 34°C in a shaking incubator at 120 r/min for 12 h to obtain the seed bacterial culture. Subsequently, a 2% inoculum of the seed culture was added to fresh LB broth and cultured at 34°C with shaking at 120 r/min for 3 days to produce an abundant fermentation broth. The fermentation broth was then subjected to ultrasonic disruption for 1 h, followed by UV light irradiation for 45 min. Thereafter, it was centrifuged at 12,000 r/min for 15 min to collect the supernatant. Ethyl acetate was used to extract the supernatant in a 1:1 volume ratio. The resulting solution was rotary evaporated at 45°C to remove the ethyl acetate, yielding a crude extract that was stored at 4°C for future use.
2.6 Analysis of the anti-yeast properties of metabolites from B. spizizenii
The method described by Wang et al. (2023) was employed to analyze the antimicrobial properties of ethyl acetate extract obtained from the endophytic bacterium B. spizizenii. The minimum inhibitory concentration (MIC) values were determined using the broth dilution method in accordance with the guidelines of Clinical and Laboratory Standards Institute (CLSI), using amphotericin B as the positive control. Specifically, in each well of the first row of a 96-well plate, 180 μL of yeast suspension (1.25 × 106 CFU/mL) was added, while the remaining wells received only 100 μL of yeast suspension. A crude extract solution of B. spizizenii in DMSO (10 mg/mL) was prepared, and 20 μL of this solution was added into the wells of first row containing 180 μL of yeast suspension. Subsequently, a 2-fold serial dilution was performed within the 96-well plate, and the turbidity in each well was observed to determine the lowest concentration of B. spizizenii required to inhibit yeast growth. This lowest concentration was considered the MIC value.
2.7 Determination of the components of metabolites obtained from B. spizizenii
Non-targeted metabolomics liquid chromatography and mass spectroscopy (LC-MS/MS) was used to analyze the metabolites of B. spizizenii. A sample of crude extract weighing 20 mg (±1 mg) was added to 400 μL of a 70% methanol-water internal standard extraction solution. The mixture was vortexed for 3 min and subjected to ultrasonic extraction in an ice-water bath for 10 min. After removing the sample, it was vortexed for an additional 1 min and allowed to stand at −20°C for 30 min. Following this, the mixture was centrifuged at 12,000 r/min for 10 min at 4°C, and the resulting supernatant was filtered through a 0.22 μm membrane. The filtered solution was then transferred to a detection vial for LC-MS/MS analysis. This process was conducted at Shanghai Majorbio Bio-Pharm Technology Co., Ltd. The extracted metabolites were then subjected to analysis of pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The molecular weights of the metabolites were determined based on the m/z ratios of the parent ions in the MS1 spectra. Molecular formula predictions were made using mass deviation (ppm) and adduct ion information, followed by matching with the database for primary identification of the metabolites. Additionally, for quantitative analysis, the MS2 spectra of the detected metabolites were compared with fragment ion information from the database to achieve secondary identification.
2.8 Statistical analysis
SPSS 22 statistical software (SPSS Inc., Chicago, IL, USA) was used to calculate the mean and standard deviation of the bacterial densities and the size of inhibition zones produced due to anti-yeast activity of endophytic bacteria. P < 0.05 indicates a statistically significant difference.
3 Results
3.1 Isolation of endophytic bacteria from pedicels of C. reticulata
Disinfection is crucial to remove the unwanted microorganisms from the surface of plant samples. After 72 h tissue culturing, no microbial growth was observed on LB and TSA media. This confirmed the successful surface sterilization of collected pedicels of C. reticulata. Based on colony morphology, a total of 138 endophytic bacterial isolates were isolated and purified from the pedicels of 12- and 60-year-old C. reticulata, with bacterial densities of 3.9 × 103 ± 0.72 × 103 CFU/g and 16.5 × 103 ± 3.58 × 103 CFU/g, respectively. Among the 138 isolates, 52 were isolated from 12-year-old trees and 86 were isolated from 60-year-old trees.
3.2 Screening of endophytic bacteria with antagonistic activity against nectar yeasts
These isolates were subjected to preliminarily screening for antagonistic activity against nectar yeasts. Around 21.01% isolates (29 isolates) showed inhibitory effects on at least one yeast, with 20 inhibiting M. reukaufii, 24 inhibiting C. laurentii, and 27 inhibiting R. glutinis. Meanwhile, 11.59% isolates (16 isolates) exhibited antagonistic activity against all three yeasts, including C1, C37, C38, C46, F1, F2, F6, F9, F15, F22, F37, F44, F50, F51, F53, and F56 (Table 1).
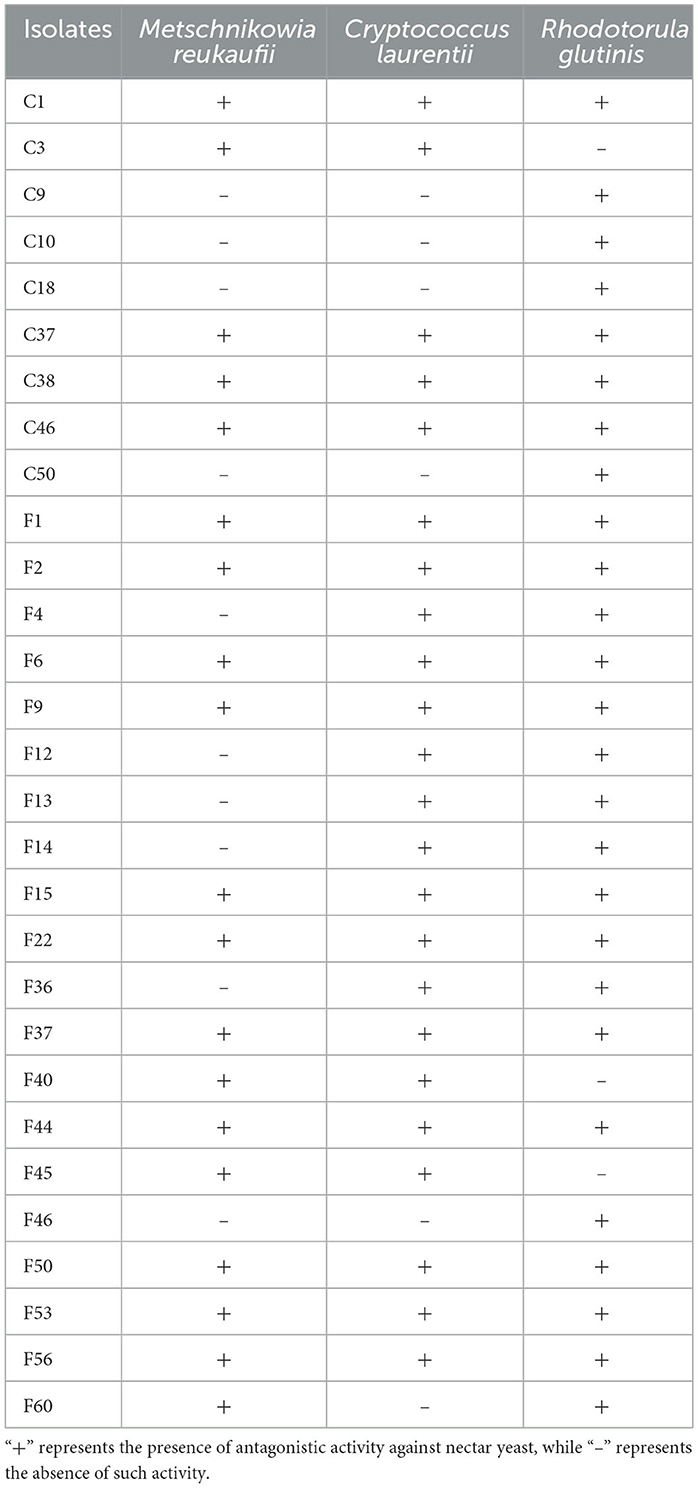
Table 1. Screening of endophytic bacteria isolated from the pedicels of C. reticulata with antagonistic activity against nectar yeasts.
Furthermore, the antagonistic activity of isolates against yeasts was further assessed by the diameter of their inhibition zones produced by their fermentation substances (Table 2). The screening results showed the antagonistic activity of 29 isolates against at least one yeast. Except for F45, all isolates demonstrated antagonistic effects against R. glutinis. Among these, C1 showed the best inhibitory effect against all three yeasts, similar to amphotericin B, highlighting its significant anti-yeast potential. The fermentation substances of C1 produced inhibition zones with diameters of 19.23 ± 0.95 mm, 19.93 ± 0.68 mm, and 21.53 ± 1.13 mm against M. reukaufii, C. laurentii, and R. glutinis, respectively.
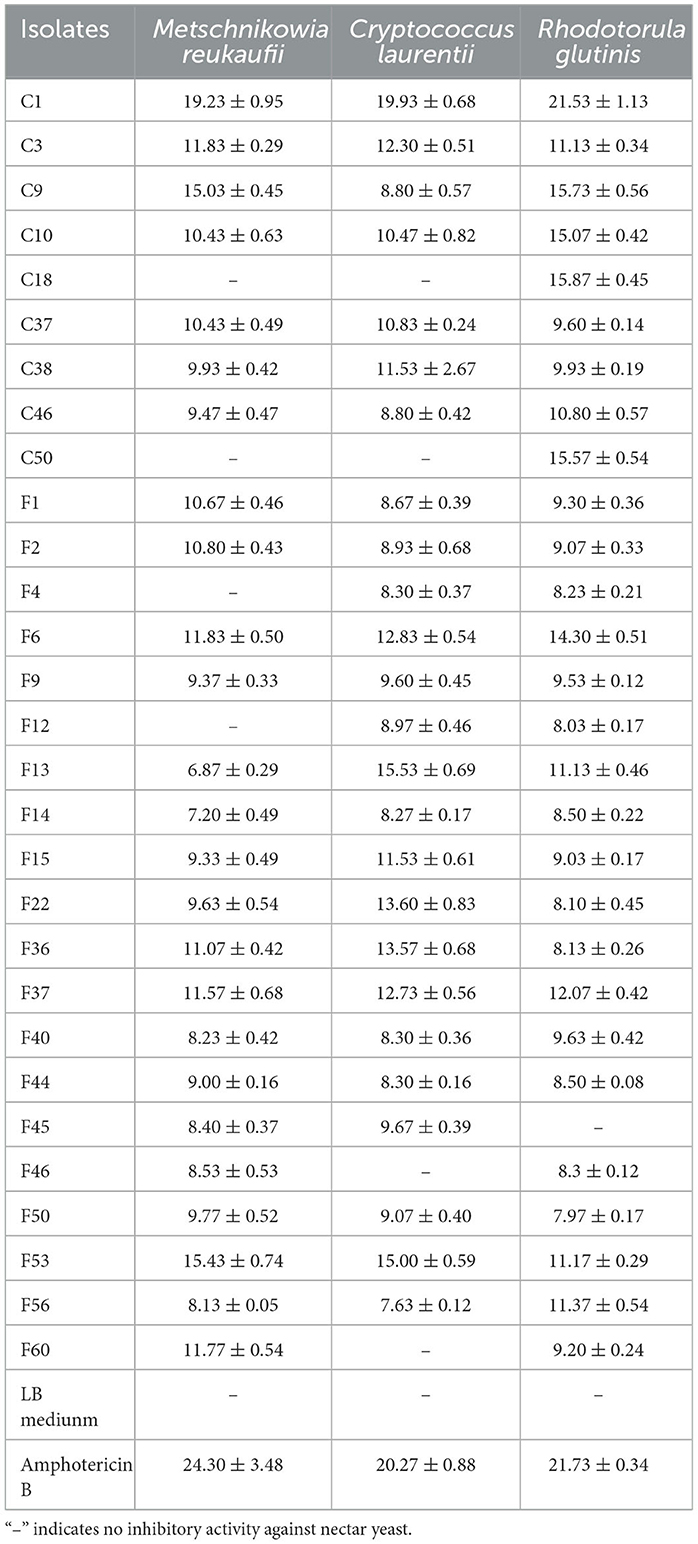
Table 2. Diameter of inhibition zones of endophytic bacteria with antagonistic activity against nectar yeasts (mean ± S.E., unit: mm).
Additionally, spot inoculation with isolates C3, C9, C10, F13, F14, and F46 initially resulted in no antagonistic effects against some yeasts. However, the fermentation products of these isolates exhibited noticeable inhibitory effects on the corresponding yeasts. Therefore, it is necessary to use various methods to evaluate the antagonistic activity of endophytic bacteria against nectar yeasts.
3.3 Characterization of screened endophytic bacterial isolates
The 29 screened isolates of endophytic bacteria exhibited various morphological characteristics, such as distinct colors (including white, gray-white, and light yellow), margins (irregular, undulate, and entire), and shapes (rod and cocci) of colony (Figure 2). Gram staining results revealed that four isolates were Gram-negative, while the remaining isolates were all Gram-positive. All 29 isolates were positive for H2O2 and glucose metabolism. Nearly all isolates exhibited motility (except C18) and nitrate reductase activity (except for C10). Additionally, most isolates showed positive reactions for gelatin liquefaction (79.31%), methyl red (86.21%), and starch hydrolysis (86.21%). In contrast, only a few isolates showed positive responses for indole production (6.90%), V-P test (20.69%), and H2S production (20.69%) (Table 3).
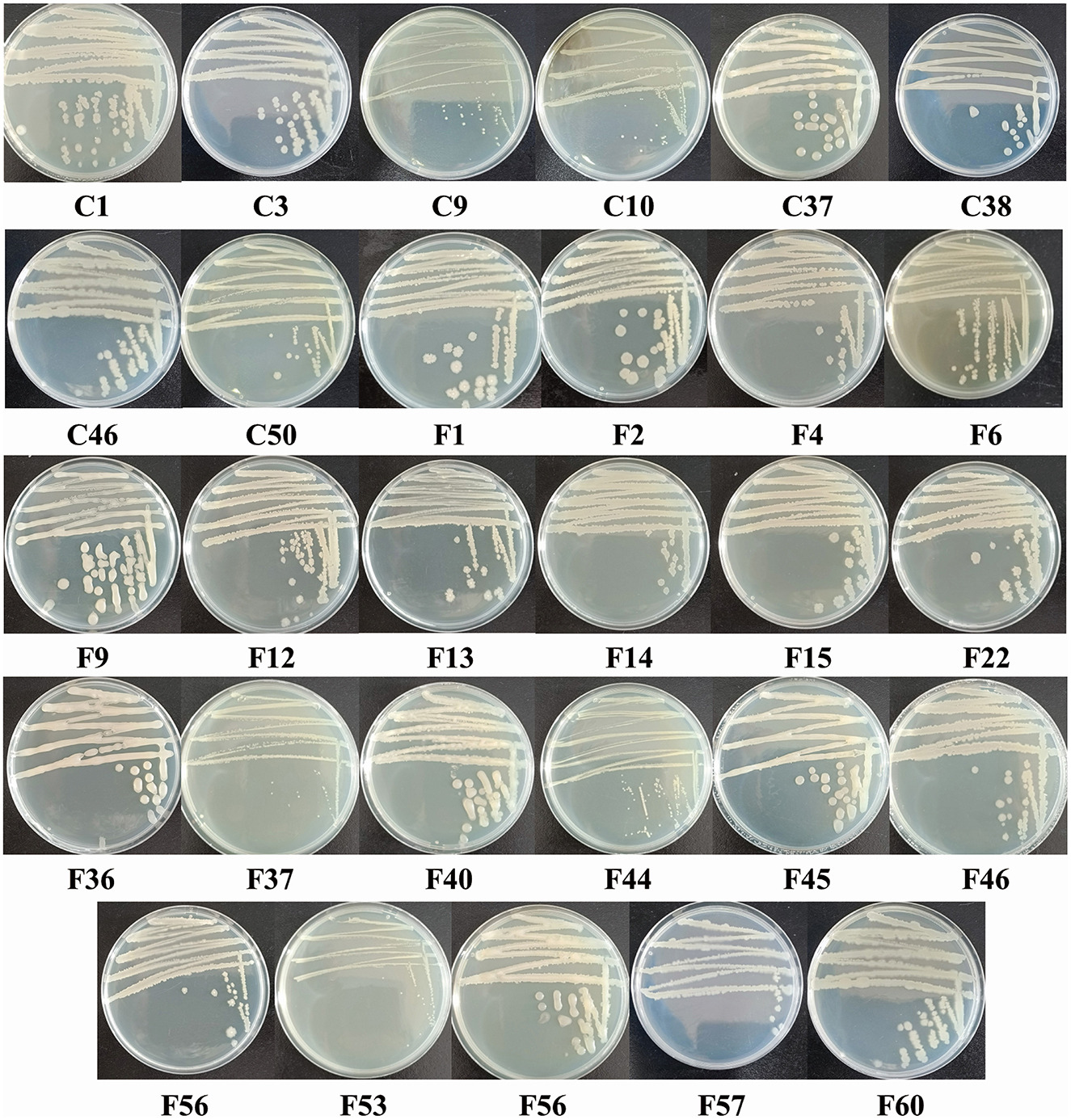
Figure 2. Colony morphology of endophytic bacterial isolates isolated from the pedicels of C. reticulata with antagonistic activity against nectar yeasts.
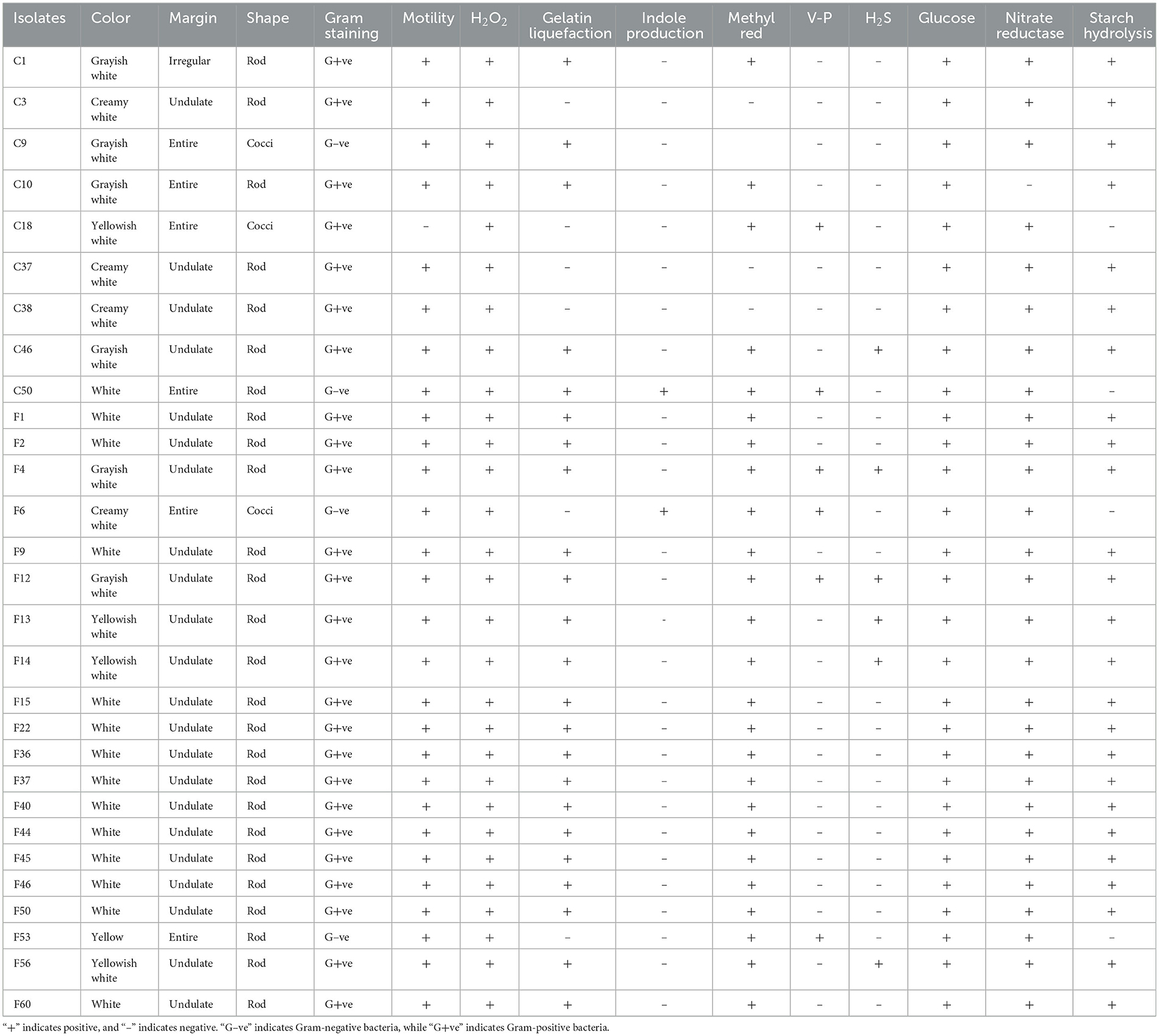
Table 3. Characterization of endophytic bacteria isolated from the pedicels of C. reticulata with antagonistic activity against nectar yeasts.
3.4 Identification of screened endophytic bacterial isolates
The 16S rRNA gene sequences were compared to known sequences in the GenBank database using BLAST. The results shown in Table 4 indicated 97-100% homology between the 16S rRNA sequences of 29 endophytic bacterial isolates and the known sequences. Among the 29 isolates, 23 belonged to the genus Bacillus, which accounted for the highest proportion. Specifically, the isolates were identified as B. subtilis (13 isolates), B. sp. (four isolates), B. velezensis (three isolates), B. licheniformis (two isolates), and B. spizizenii (one isolate). The remaining six isolates belonged to genus Erwinia (E. billingiae), Pseudomonas (P. fluorescens and P. tolaasii), Cytobacillus (C. firmus), Staphylococcus (S. haemolyticus), and Pantoea (P. agglomerans).
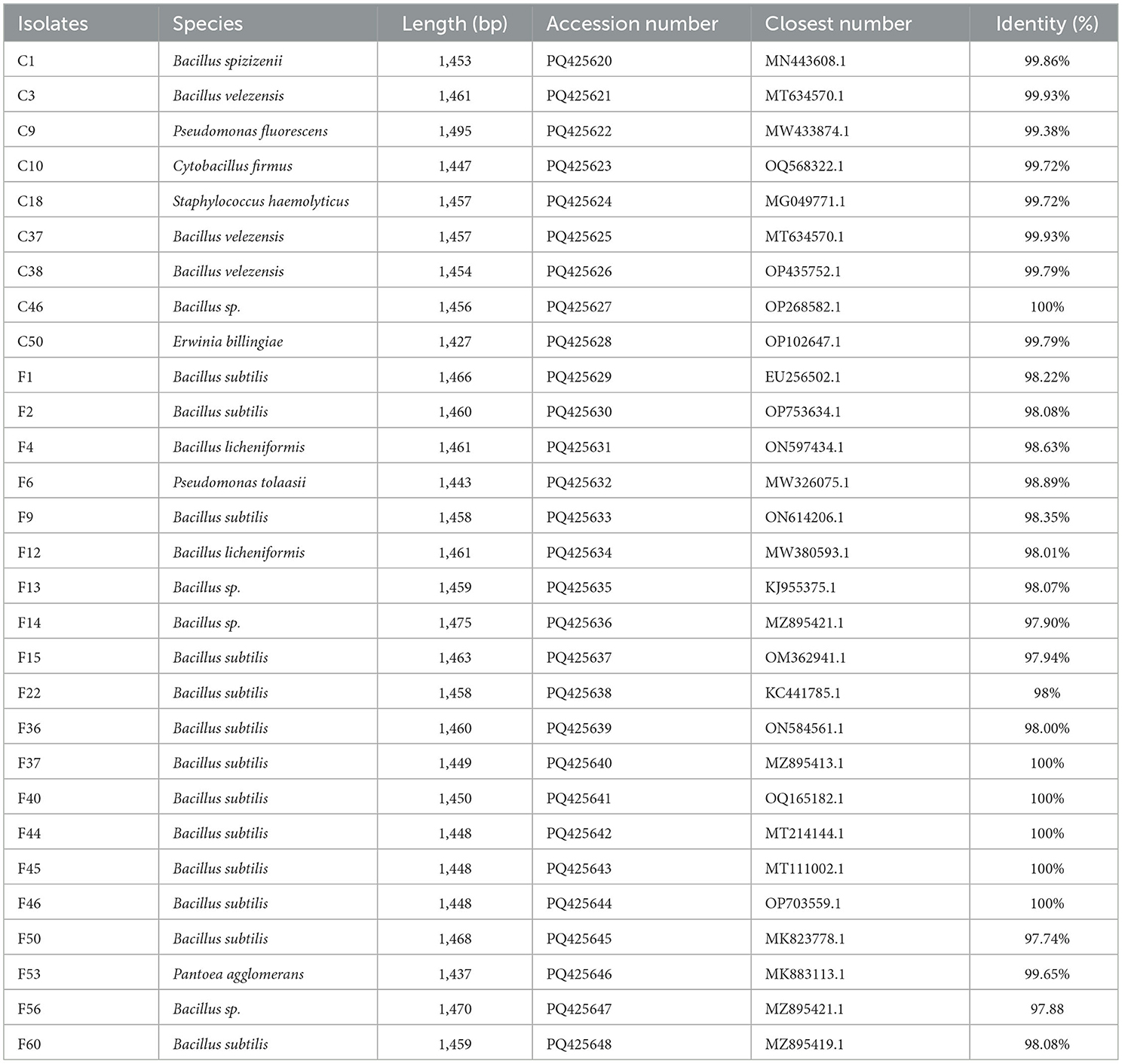
Table 4. Identification of endophytic bacterial isolates isolated from pedicels of C. reticulata with antagonistic activity against nectar yeasts.
3.5 Antagonistic effects of metabolites of B. spizizenii against nectar yeasts
The metabolites derived from B. spizizenii displayed potent antagonistic effects against M. reukaufii, C. laurentii, and R. glutinis, with MIC values of 62.5, 250, and 125 μg/mL, respectively. Notably, its antagonistic activity against these yeasts was comparable to that of the clinical drug amphotericin B (MIC values of 1.25, 0.63, and 1.25 μg/mL, respectively). These findings highlighted the significant potential of endophytic bacterium B. spizizenii to inhibit nectar yeasts.
3.6 Compositional analysis of the metabolites of B. spizizenii
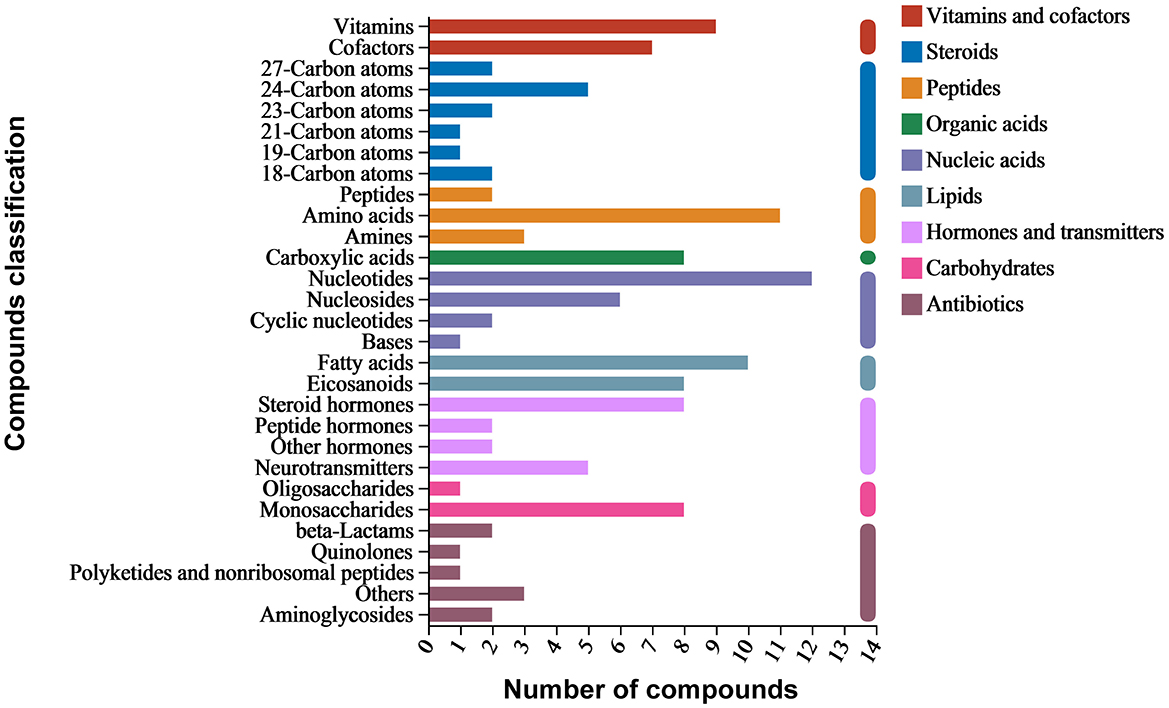
Around 3,563 substances were detected in the metabolites of B. spizizenii using LC-MS/MS. KEGG enrichment analysis revealed nine categories of enriched substances: vitamins and cofactors, steroids, peptides, organic acids, nucleic acids, lipids, hormones and transmitters, carbohydrates, and antibiotics. Specifically, nucleotides accounted for the highest proportion of total compounds, followed by amino acids, fatty acids, vitamins, and monosaccharides. Antibiotic families, such as aminoglycosides, beta-lactams, and quinolones were also detected (Figure 3).
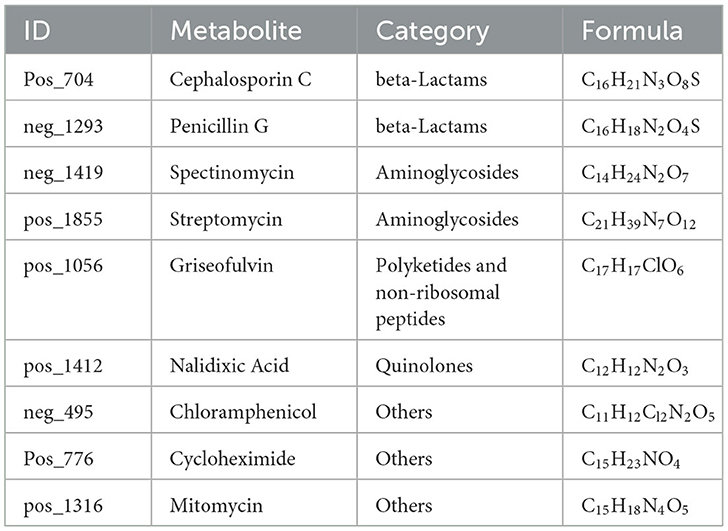
Active substances related to anti-yeast activity were detected in negative ion mode, including chloramphenicol, penicillin G, and spectinomycin. On the other hand, cephalosporin C, cycloheximide, griseofulvin, mitomycin, nalidixic acid, and streptomycin were detected in positive ion mode (Table 5). These antibiotics mainly belong to beta-lactams, polyketides and non-ribosomal peptides, quinolones, aminoglycosides, and other protein families. The results suggested that B. spizizenii inhibited the growth of nectar yeasts by producing multiple antibiotics, which indicates its great potential as a biocontrol bacterium.
KEGG database annotations revealed the major pathways involved in the production of metabolites in B. spizizenii. These pathways primarily included the biosynthesis of plant secondary metabolites, phenylpropanoid, and amino acid, as well as nucleotide metabolism, ABC transportation, and pyrimidine metabolism (Figure 4).
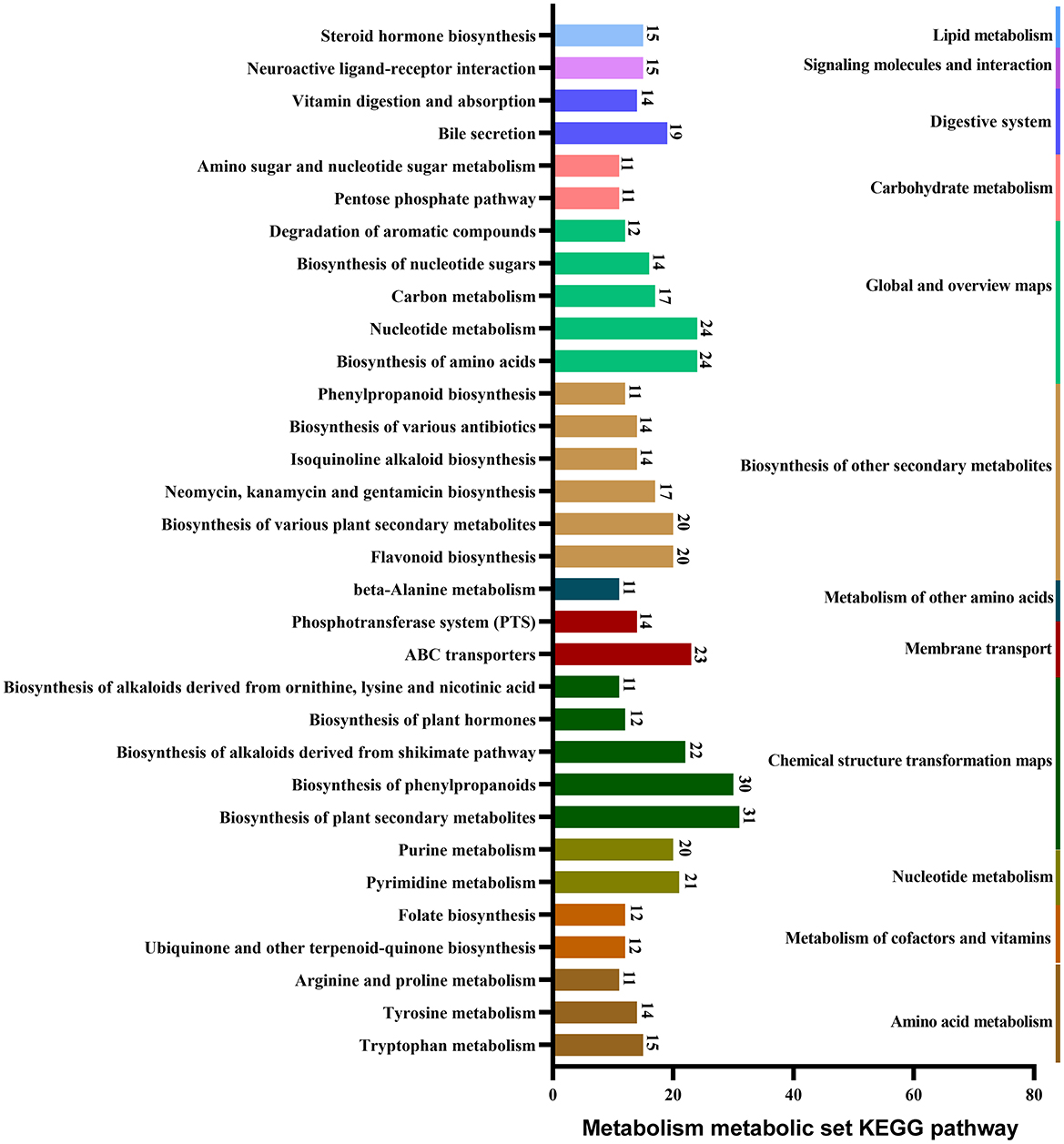
Figure 4. Major pathways involved in the metabolism of B. spizizenii, revealed by KEGG database annotations.
4 Discussion
Endophytic bacteria are commonly present in plants. As research continues to expand, endophytic bacteria have been increasingly isolated and identified from plant tissues (Hardoim et al., 2015). However, C. reticulata has been rarely explored, especially in its native Yunnan Province. Using surface sterilization, 138 endophytic bacterial isolates were isolated from the pedicels of 12- and 60-year-old C. reticulata trees. The density and diversity of endophytic bacterial isolates in the pedicels of 60-year-old C. reticulata were higher than those of the 12-year-old pedicel tissues. This finding was consistent with the results obtained through high-throughput sequencing (Huang et al., 2023). The invasion and colonization of host plant by endophytic bacteria represent dynamic processes in terms of time and space. Endophytic bacteria not only modify the colonization sites through transpiration, but also exhibit temporal changes in the extent of colonization and species diversity. As plants grow, density and diversity of endophytic bacteria increase (Wagner et al., 2016; Liu et al., 2017). Throughout the lifespan of plants, tissues with longer inoculation periods tend to exhibit higher richness of endophytic bacteria, which may be attributed to larger colonization amount and longer microbial growth period (Ercolani, 1991). In old trees, richness and diversity of endophytic bacteria are significantly higher due to long-term accumulation and co-evolutionary processes. These trees serve as natural microbial resource libraries, providing valuable resources for screening, investigating, and utilizing the endophytic bacteria living within plants.
Using beneficial microorganisms and their metabolites is an effective biocontrol method to prevent and control diseases in agricultural and forest crops (Abbasi et al., 2002). The biocontrol mechanisms of endophytic bacteria primarily involve: (1) competition for advantageous ecological niches and nutrients against pathogens, (2) the secretion of antimicrobial substances that provide antibacterial activity, (3) parasitism on pathogens, (4) the induction of systemic resistance in host plants to pathogens, and (5) the regulation of the microecological system of the host plant (Compant et al., 2005; Ansari and Mahmood, 2019; Kumar and Singh, 2020). Ultimately, these mechanisms promote host plant growth or enhance resistance, thereby enabling effective defense against plant pathogens. Thus, endophytic bacteria help host plants respond to microbial infestation, suggesting that these bacteria may also assist plant nectar in antagonizing nectar yeast. Due to the nectar is nutritious, it attracts colonization by yeasts. However, the active metabolism of these yeasts depletes the nutritional components of the nectar, significantly reducing the quality of the rewards for pollinators and ultimately weakening the reproductive fitness of cross-pollinated plants (Herrera et al., 2008; Canto and Herrera, 2012). Therefore, it was essential to isolate numerous yeast-inhibiting endophytic bacterial isolates from the pedicels of C. reticulata to study the role of endophytes in maintaining the stability and quality of nectar.
The endophytic bacteria isolated from the pedicel of C. reticulata were diverse. In the present study, 29 endophytic bacterial isolates exhibited inhibitory effects against at least one species of nectar yeast. 16S rRNA sequence analysis revealed that the isolates belonged to six different genera, including Bacillus, Erwinia, Pseudomonas, Cytobacillus, Staphylococcus, and Pantoea, with most isolates belonging to genus Bacillus. Endophytic bacteria belonging to the genera Bacillus, Pseudomonas, Enterobacter and Sporosarcina have been widely reported previously in many plants (Li et al., 2012; Cabanás et al., 2014). Bacillus are Gram-positive bacteria that have become a biocontrol research hotspot, as they can produce a variety of antimicrobial substances (Ongena and Jacques, 2008; Sumi et al., 2015). The metabolites of Bacillus include glucanase, protease, cellulase, chitinase, etc., which inhibit the growth of pathogens by degrading their cell walls. They can also produce lipopeptide substances that resist the invasion and harmful effects of plant pathogens. Furthermore, they promote plant growth by competing with pathogenic bacteria for nutrients, secreting antimicrobial peptides and antibiotics, and inducing the systemic disease resistance in plants (Pan et al., 2017). In this study, some endophytic bacteria of genus Bacillus showed at least one or more enzyme activities and could utilize various substrates, such as glucose, nitrate, and starch. This finding suggests that these bacteria can promote the utilization of energy in the host plant.
The fermentation substances of isolate C1, identified as B. spizizenii, exhibited the best antagonistic activity against yeasts. This antagonistic activity was comparable to the activity shown by positive control, i.e., amphotericin B. Furthermore, B. spizizenii could produce diverse antibiotics belonging to aminoglycosides, Macrolides, and quinolones classes, such as spectinomycin, bacillomycin D, and macrolactin (Chen et al., 2021; Han et al., 2021). Previous studies have also shown the ability of B. spizizenii to secrete the antibiotics of other classes, as well as several lipophilic membrane-active agents, such as subtilin, subtilosin, surfactin, and fengycin (Kim et al., 2017; Lim et al., 2017; Zhao et al., 2018). These findings suggest that the antagonistic activity of B. spizizenii against nectar yeasts can be attributed to its antibiotic-secreting properties. The main metabolic pathways of B. spizizenii included the biosynthesis of plant secondary metabolites, phenylpropanoid compounds, amino acids, nucleotide metabolism, and other compounds. Additionally, other metabolic pathways were also discovered, such as the biosynthesis of neomycin, kanamycin, kanosamine, etc. These metabolites and pathways provide new research directions to further explore the antimicrobial mechanism of B. spizizenii. Camellia reticulata secretes a large amount of nectar and relies on pollinators for reproduction. The antimicrobial substances in the nectar are generally believed to be derived from the active substances produced by the plant itself. In addition, the endophytic bacteria colonizing the pedicels also produce abundant secondary metabolites and antibiotics, which may be crucial for maintaining the nectar homeostasis. This finding may provide new ideas for the analysis of response mechanisms of heterologous pollinating plants to microbial infestation. However, the relevant conclusions need to be verified by further research. Exploring the potential of these metabolites and antibiotics in biological control and understanding the underlying mechanisms is also an important direction for future research.
5 Conclusion
The pedicels of C. reticulata are rich in endophytic bacteria. In this study, 29 yeast-resisting endophytic bacteria were isolated and screened using spot inoculation and oxford cup methods. Most of the isolates belonged to Bacillus genus, with a few isolates belonging to Erwinia, Pseudomonas, Cytobacillus, Staphylococcus, and Pantoea. Especially, the endophytic bacterium C1, identified as B. spizizenii, exhibited the strongest inhibitory effect on nectar yeasts. This isolate was found to be a source of structurally and functionally diverse natural compounds, exhibiting anti-yeast effects through the synthesis of antibiotics, such as chloramphenicol, penicillin G, spectinomycin, etc. This study highlighted the potential uses of endophytic bacteria in maintaining nectar quality and stability, as well as the reproductive fitness of host plant, providing new insights into the role of endophytic bacteria of pedicels in nectar resistance against yeasts.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
QM: Writing – original draft, Methodology. RH: Writing – original draft, Software, Visualization. LX: Methodology, Writing – original draft. XW: Investigation, Writing – original draft. SD: Software, Writing – original draft. DY: Investigation, Writing – original draft. WZ: Software, Writing – original draft. XD: Conceptualization, Writing – original draft. XG: Methodology, Writing – review & editing. KD: Data curation, Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by National Natural Science Foundation of China (grant numbers: 32060241 and 31572339), China Agriculture Research System of the Ministry of Finance and the Ministry of Agriculture and Rural Affairs (grant number: CARS-44-KXJ13), and Reserve Talents Training Program for Young and Middle-Aged Academic and Technical Leaders in Yunnan (grant number: 2018HB041).
Acknowledgments
We thank Zhongling Jiang and Junli Zhang for their help with preliminary treatments, as well as MJEditor (https://www.mjeditor.com) for providing English editing services during the preparation of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1459354/full#supplementary-material
References
Abbasi, P. A., Al-Dahmani, J., Sahin, F., Hoitink, H. A. J., and Miller, S. A. (2002). Effect of compost amendments on disease severity and yield of tomato in conventional and organic production systems. Plant Dis. 86, 156–161. doi: 10.1094/PDIS.2002.86.2.156
Ansari, R. A., and Mahmood, I. (2019). Plant Health Under Biotic Stress. Singapore: Springer Nature.
Cabanás, C. G.-L., Schilir,ò, E., Valverde-Corredor, A., and Mercado-Blanco, J. (2014). The biocontrol endophytic bacterium Pseudomonas fluorescens PICF7 induces systemic defense responses in aerial tissues upon colonization of olive roots. Front. Microbiol. 5:427. doi: 10.3389/fmicb.2014.00427
Canto, A., and Herrera, C. M. (2012). Micro-organisms behind the pollination scenes: microbial imprint on floral nectar sugar variation in a tropical plant community. Ann. Bot. 110, 1173–1183. doi: 10.1093/aob/mcs183
Cardoso, V. M., Campos, F. F., Santos, A. R. O., Ottoni, M. H. F., Rosa, C. A., Almeida, V. G., et al. (2020). Biotechnological applications of the medicinal plant Pseudobrickellia brasiliensis and its isolated endophytic bacteria. J. Appl. Microbiol. 129, 926–934. doi: 10.1111/jam.14666
Chen, L., Wang, X., and Liu, Y. (2021). Contribution of macrolactin in Bacillus velezensis CLA178 to the antagonistic activities against Agrobacterium tumefaciens C58. Arch. Microbiol. 203, 1743–1752. doi: 10.1007/s00203-020-02141-1
Compant, S., Duffy, B., Nowak, J., Clément, C., and Barka, E. A. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospect. Appl. Environ. Microbiol. 71, 4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005
Ercolani, G. L. (1991). Distribution of epiphytic bacteria on olive leaves and the influence of leaf age and sampling time. Microb. Ecol. 21, 35–48. doi: 10.1007/BF02539143
Feng, G. (1980). Species and Utilization of Camellia in Yunnan (in Chinese). Kunming: Yunnan people's Publishing House.
González-Mas, N., Cuenca-Medina, M., García-Mozo, H., Muñoz-Redondo, J. M., Moreno-Rojas, J. M., Padilla-Álvarez, F., et al. (2023). Endophytic Beauveria bassiana modifies flowering phenology, floral volatile profile and pollinator behaviour in melon. Entomol. Gener. 43, 961–969. doi: 10.1127/entomologia/2023/1991
Goodfellow, M., Kämpfer, P., Busse, H.-J., Trujillo, M. E., Suzuki, K., Ludwig, W., et al. (2009). Bergey's Manual of Systematic Bacteriology. New York, NY: Springer.
Hamaoka, K., Aoki, Y., and Suzuki, S. (2021). Isolation and characterization of endophyte Bacillus velezensis KOF112 from grapevine shoot xylem as biological control agent for fungal diseases. Plants 10:1815. doi: 10.3390/plants10091815
Han, X., Shen, D., Xiong, Q., Bao, B., Zhang, W., Dai, T., et al. (2021). The plant-beneficial rhizobacterium Bacillus velezensis FZB42 controls the soybean pathogen Phytophthora sojae due to bacilysin production. Appl. Environ. Microbiol. 87:e0160121. doi: 10.1128/AEM.01601-21
Hardoim, P. R., van Overbeek, L. S., Berg, G., Pirttilä, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/MMBR.00050-14
Herrera, C. M., de Vega, C., Canto, A., and Pozo, M. I. (2009). Yeasts in floral nectar: a quantitative survey. Ann. Bot. 103, 1415–1423. doi: 10.1093/aob/mcp026
Herrera, C. M., García, I. M., and Pérez, R. (2008). Invisible floral larcenies: microbial communities degrade floral nectar of bumble bee-pollinated plants. Ecology 89, 2369–2376. doi: 10.1890/08-0241.1
Huang, R., Wu, X., Xun, L., Meng, Q., Tian, Y., Dong, K., et al. (2023). Diversity of endophytic bacteria in pedicels of Camellia reticulate Lindl (in Chinese). J. South. Agric. 54, 1997–2004. doi: 10.3969/j.issn.2095-1191.2023.07.011
Jiang, W., Liu, X., Qiu, R., Wu, X., Wen, J., Luo, N., et al. (2016). Screening and identification of oil-tea endophytic antagonistic bacteria (in Chinese). J. Anhui Agric. Sci. 8, 125–127. doi: 10.13989/j.cnki.0517-6611.2016.08.045
Kim, S. Y., Lee, S. Y., Weon, H.-Y., Sang, M. K., and Song, J. (2017). Complete genome sequence of Bacillus velezensis M75, a biocontrol agent against fungal plant pathogens, isolated from cotton waste. J. Biotechnol. 241, 112–115. doi: 10.1016/j.jbiotec.2016.11.023
Li, H., Zhou, G., Lu, L., and Liu, J. (2009). Isolation and identification of endophytic bacteria antagonistic to Camellia oleifera anthracnose. Afr. J. Microbiol. Res. 3, 315–318. doi: 10.1109/BMEI.2009.5304770
Li, J., Zhao, G., Huang, H., Qin, S., Zhu, W., Zhao, L., et al. (2012). Isolation and characterization of culturable endophytic actinobacteria associated with Artemisia annua L. Antonie Van Leeuwenhoek 101, 515–527. doi: 10.1007/s10482-011-9661-3
Lim, S. M., Yoon, M.-Y., Choi, G. J., Choi, Y. H., Jang, K. S., Shin, T. S., et al. (2017). Diffusible and volatile antifungal compounds produced by an antagonistic Bacillus velezensis G341 against various phytopathogenic fungi. Plant Pathol. J. 33, 488–498. doi: 10.5423/PPJ.OA.04.2017.0073
Liu, H., Carvalhais, L. C., Crawford, M., Singh, E., Dennis, P. G., Pieterse, C. M. J., et al. (2017). Inner plant values: diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 8:2552. doi: 10.3389/fmicb.2017.02552
Matsumoto, H., Fan, X., Wang, Y., Kusstatscher, P., Duan, J., Wu, S., et al. (2021). Bacterial seed endophyte shapes disease resistance in rice. Nat. Plants 7, 60–72. doi: 10.1038/s41477-020-00826-5
Ongena, M., and Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Pan, H., Li, Q., and Hu, J. (2017). The complete genome sequence of Bacillus velezensis 9912D reveals its biocontrol mechanism as a novel commercial biological fungicide agent. J. Biotechnol. 247, 25–28. doi: 10.1016/j.jbiotec.2017.02.022
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., Wees, S. C. M. V., and Bakker, P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Russell, A. L., Rebolleda-Gómez, M., Shaible, T. M., and Ashman, T.-L. (2019). Movers and shakers: bumble bee foraging behavior shapes the dispersal of microbes among and within flowers. Ecosphere 10:e02714. doi: 10.1002/ecs2.2714
Santoyo, G., Moreno-Hagelsieb, G., del Carmen Orozco-Mosqueda, M., and Glick, B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99. doi: 10.1016/j.micres.2015.11.008
Sasu, M. A., Seidl-Adams, I., Wall, K., Winsor, J. A., and Stephenson, A. G. (2010). Floral transmission of erwinia tracheiphila by cucumber beetles in a wild Cucurbita pepo. Environ. Entomol. 39, 140–148. doi: 10.1603/EN09190
Sharma, M., and Mallubhotla, S. (2022). Diversity, antimicrobial activity, and antibiotic susceptibility pattern of endophytic bacteria sourced from Cordia dichotoma L. Front. Microbiol. 13:879386. doi: 10.3389/fmicb.2022.879386
Sumi, C. D., Yang, B. W., Yeo, I.-C., and Hahm, Y. T. (2015). Antimicrobial peptides of the genus Bacillus: a new era for antibiotics. Can. J. Microbiol. 61, 93–103. doi: 10.1139/cjm-2014-0613
Tejesvi, M. V., Segura, D. R., Schnorr, K. M., Sandvang, D., Mattila, S., Olsen, P. B., et al. (2013). An antimicrobial peptide from endophytic Fusarium tricinctum of Rhododendron tomentosum Harmaja. Fungal Divers. 60, 153–159. doi: 10.1007/s13225-013-0227-8
Wagner, M. R., Lundberg, D. S., del Rio, T. G., Tringe, S. G., Dangl, J. L., and Mitchell-Olds, T. (2016). Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 7:12151. doi: 10.1038/ncomms12151
Wang, H., Liu, Z., Duan, F., Chen, Y., Qiu, K., Xiong, Q., et al. (2023). Isolation, identification, and antibacterial evaluation of endophytic fungi from Gannan navel orange. Front. Microbiol. 14:1172629. doi: 10.3389/fmicb.2023.1172629
Zemenick, A. T., Vannette, R. L., and Rosenheim, J. A. (2021). Linked networks reveal dual roles of insect dispersal and species sorting for bacterial communities in flowers. Oikos 130, 697–707. doi: 10.1111/oik.06818
Zhang, H., Song, Y., and Tan, R. (2006). Biology and chemistry of endophytes. Nat. Prod. Rep. 23, 753–771. doi: 10.1039/b609472b
Zhao, P., Xue, Y., Gao, W., Li, J., Zu, X., Fu, D., et al. (2018). Bacillaceae-derived peptide antibiotics since 2000. Peptides 101, 10–16. doi: 10.1016/j.peptides.2017.12.018
Zhou, H. (2015). Concise Tutorial in Botany (in Chinese). Wuhan: Central China Normal University Press.
Keywords: tree age, pedicel endophytic bacteria, anti-yeast potential, bioactive metabolites, antibiotics, nectar homeostasis
Citation: Meng Q, Huang R, Xun L, Wu X, Deng S, Yue D, Zhao W, Dong X, Gong X and Dong K (2024) Endophytic bacteria in Camellia reticulata pedicels: isolation, screening and analysis of antagonistic activity against nectar yeasts. Front. Microbiol. 15:1459354. doi: 10.3389/fmicb.2024.1459354
Received: 04 July 2024; Accepted: 07 October 2024;
Published: 21 October 2024.
Edited by:
Iftikhar Ahmed, National Agricultural Research Center, PakistanReviewed by:
Orlando Borras-Hidalgo, Qilu University of Technology, ChinaLaith Khalil Tawfeeq Al-Ani, Universiti Sains Malaysia, Malaysia
Copyright © 2024 Meng, Huang, Xun, Wu, Deng, Yue, Zhao, Dong, Gong and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueyang Gong, eHVleWFuZ2cxMUAxMjYuY29t; Kun Dong, ZG9uZ2t1bjE5NzIyMDA0QGFsaXl1bi5jb20=
†These authors have contributed equally to this work and share first authorship
 Qingxin Meng
Qingxin Meng Rong Huang
Rong Huang Lijie Xun1,2
Lijie Xun1,2 Kun Dong
Kun Dong