- 1Division of Infectious Diseases, Department of Medicine, Medical College of Wisconsin, Milwaukee, WI, United States
- 2Department of Microbiology and Immunology, Medical College of Wisconsin, Milwaukee, WI, United States
Leptospirosis is a significant zoonosis worldwide, with disease severity ranging from a mild non-specific illness to multi-organ dysfunction and hemorrhage. The disease is caused by pathogenic bacteria of the genus Leptospira, which are classified into pathogenic and saprophytic clades. Bacterial binding to host molecules and cells, coordinated by adhesin proteins, is an important step in pathogenesis. While many leptospiral adhesins have been identified, the vast majority have not been characterized in vivo. Herein, we present an overview of the current methodologies and successes in identifying adhesins in Leptospira, including known biological roles in vivo. We will also identify and discuss potential areas for future research.
1 Introduction
Leptospirosis, the most widespread zoonosis globally, is caused by gram-negative spirochetes of the genus Leptospira (Costa et al., 2015; European Centre for Disease Prevention and Control, 2017). Disease ranges from a non-specific febrile illness to multiorgan dysfunction and hemorrhage, causing significant morbidity and mortality worldwide with the greatest impact on resource-poor populations (Weil, 1886; Alston and Brown, 1937; Gouveia et al., 2008; Costa et al., 2015). Leptospirosis is estimated to cause 60,000 deaths and a loss of productivity of $50 billion annually (Costa et al., 2015; Torgerson et al., 2015; Agampodi et al., 2023). Leptospirosis also affects animals, both domestic and wild, leading to substantial costs to the agricultural industry (Matthews et al., 1987; Barwick et al., 1998; Bennett et al., 1999; Ramos et al., 2006; Lilenbaum et al., 2008; Miotto et al., 2018; Canton et al., 2022).
Leptospira spp. are categorized into more than 300 serovars (serological variants) based upon sera reactivity to cell surface antigens, and into two clades (pathogenic, P, and saprophytic, S) and four subclades (P1, P2, S1, and S2) based upon genomic sequencing (Thibeaux et al., 2018; Vincent et al., 2019). Leptospira species range from highly pathogenic organisms in the P1 subclade, to free-living saprophytes unable to infect hosts in the S clade (Vincent et al., 2019). The enzootic cycle of Leptospira involves colonization of the renal proximal tubules in reservoir hosts, followed by shedding in the urine and infection of new hosts; humans are incidental hosts (reviewed in Adler and de la Pena, 2010; Adler et al., 2014; Haake and Levett, 2015).
Leptospira spp. remain significantly understudied. To combat this disease, researchers must determine how the pathogen is able to adapt and survive in various hosts and environments, which likely requires an interplay of bacterial, host, and environmental factors. One step in the infection process is binding to host tissue components. Bacteria produce a wide range of proteins known as adhesins that are responsible for binding host cells and molecules (reviewed in Marra and Isberg, 1996; Niemann et al., 2004; Pizarro-Cerda and Cossart, 2006; Patel et al., 2017; Paulsson and Riesbeck, 2018; Daroz et al., 2021). The specific adhesins produced by an organism contribute to its tropism and allow for successful infection. Numerous leptospiral outer membrane proteins (OMPs) have been shown to bind host cells and molecules, but the role the candidate adhesins play in natural infection remains unclear. A major question in the field remains: what is the role of these adhesins during initial contact with the host, and how do they contribute to colonization and adhesion to the proximal tubules of the kidneys? A detailed understanding of the arsenal of adhesins used by Leptospira is needed. Understanding this complex process will set the foundation for development of novel vaccines and therapeutic strategies to combat leptospirosis.
The goal of this review is to summarize methods of adhesin identification, highlight the gaps in knowledge regarding adhesins, and discuss what is needed in future adhesin research in Leptospira (Figure 1).
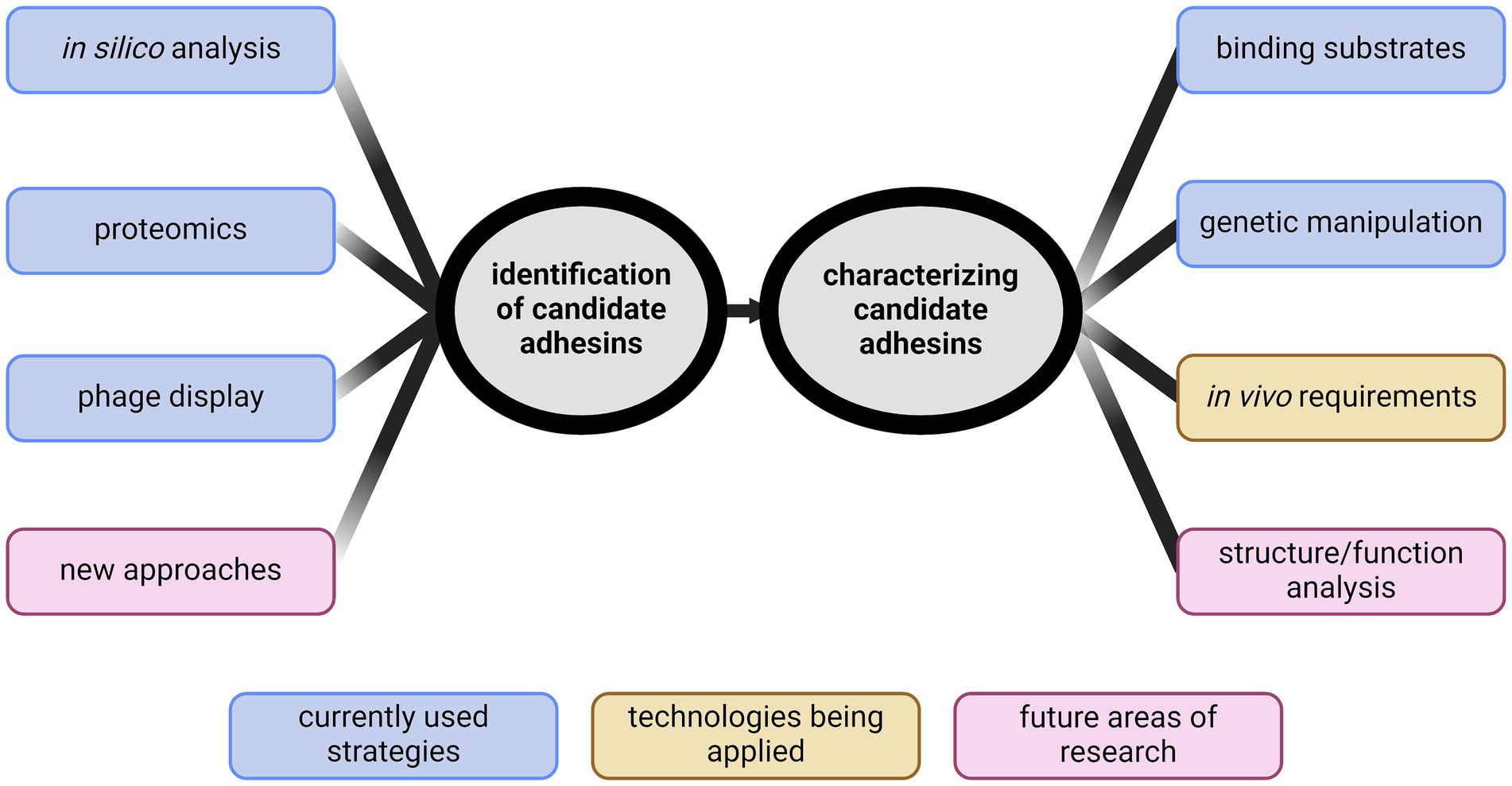
Figure 1. Identification of new directions for studying leptospiral adhesins. Identifying adhesins has primarily been performed through in silico analyses, phage display, or proteomics. New ideas for identification are needed. Characterization of candidate adhesins has typically been through demonstrating binding. Recent advancements in genetic manipulation of Leptospira spp. have provided new abilities to characterize candidates in vivo. Future research will require developing novel methods to identify biologically relevant adhesins and begin structure/function analyses to further define adhesin function. Created with BioRender.com.
2 Identification of Leptospira adhesins
2.1 The era of genomics
Whole genome sequencing revolutionized bacterial research. The first two bacterial genomes sequenced were those of Haemophilus influenzae and Mycoplasma genitalium in 1995 (Fleischmann et al., 1995; Fraser et al., 1995). By 2000, approximately 23 bacterial genomes had been described, with many more soon to follow (Schwartz, 2000). The first full Leptospira genome, L. interrogans serovar Lai, was reported by Ren et al. (2003). The genome consisted of two chromosomes of 4.33 megabases and 359 kilobases, encoding a total of 4,768 predicted genes (Ren et al., 2003). In 2004, the genome of L. interrogans sv. Copenhageni was described (Nascimento et al., 2004a,b). The non-pathogenic isolate L. biflexa sv. Patoc genome sequence was published by Picardeau et al. (2008). Pathogenic strains of Leptospira bind to human cells and ECM components more efficiently than do avirulent or saprophytic (S1) leptospires (Tsuchimoto et al., 1984; Ito and Yanagawa, 1987; Merien et al., 1997; Evangelista K. et al., 2014; Evangelista K. V. et al., 2014). This set the stage for identifying potential adhesins through comparative in silico analysis.
The program SpLip was designed to predict potential surface exposed lipoproteins that was based upon experimental data (Nascimento et al., 2004a; Setubal et al., 2006). Proteins predicted to be surface exposed were compared among the pathogenic isolates L. interrogans svv. Lai and Copenhageni (Ren et al., 2003; Nascimento et al., 2004a,b). Proteins of interest were screened for adhesive function, forming the basis for the identification and characterization of over 40 candidate adhesins by Nascimento et al. (2004a,b) (Gamberini et al., 2005; Barbosa et al., 2006; Vieira et al., 2007, 2010a,b; Atzingen et al., 2008, 2009; Gomez et al., 2008; Longhi et al., 2009; Oliveira et al., 2010, 2011, 2013; Mendes et al., 2011; Domingos et al., 2012, 2015; Fernandes et al., 2012, 2014; Souza et al., 2012; Siqueira et al., 2013, 2016, 2017; Silva et al., 2016; Pereira et al., 2017; Santos et al., 2018; Cavenague et al., 2019, 2023; Kochi et al., 2019, 2020; Rossini et al., 2020; Passalia et al., 2020a,b, 2021; Takahashi et al., 2021, 2022). Many of these proteins interact with multiple host substrates, often binding the same substrates as other candidates (Nascimento et al., 2004a,b; Gamberini et al., 2005; Barbosa et al., 2006; Vieira et al., 2007, 2010a,b; Atzingen et al., 2008, 2009; Gomez et al., 2008; Longhi et al., 2009; Oliveira et al., 2010, 2011, 2013; Mendes et al., 2011; Domingos et al., 2012, 2015; Fernandes et al., 2012, 2014; Souza et al., 2012; Siqueira et al., 2013, 2016, 2017; Silva et al., 2016; Pereira et al., 2017; Santos et al., 2018; Cavenague et al., 2019, 2023; Kochi et al., 2019, 2020; Rossini et al., 2020; Passalia et al., 2020a,b, 2021; Takahashi et al., 2021, 2022). It must be noted that non-specific binding in vitro cannot be ruled out. Taken together, this research suggests that Leptospira likely interact with numerous host proteins throughout infection and indicates that there is significant redundancy in the leptospiral adhesin proteome. The wide binding and redundancy likely aid Leptospira during infection, but also complicates research. Loss of a single adhesin is therefore less likely to cause a significant virulence defect, meaning therapeutic approaches targeting adhesion will need to include multiple proteins.
In silico analysis was also performed to identify genes in L. interrogans that had homology to known adhesin genes from other species. Proteins containing leucine-rich repeat (LRR) domains from other bacterial pathogens interact with host cells (Bierne et al., 2007; Ng and Xavier, 2011). Pathogenic Leptospira encode significantly more LRR proteins than do non-pathogenic Leptospira; LIC10831, an LRR protein from L. interrogans, was investigated and binds human cadherins (Eshghi et al., 2015; Fouts et al., 2016). In addition to comparisons between species, comparisons within a single genome have also identified adhesive proteins. LenA, also known as Lsa24 and LfhA, was known to have adhesive function (Barbosa et al., 2006). Stevenson et al. (2007) searched the L. interrogans genome for homology to lenA and identified five paralogs which they designated lenB-F. All Len proteins interact with laminin, but the newly identified paralogs also bind fibronectin, which can also be a component of the extracellular matrix (Barbosa et al., 2006; Stevenson et al., 2007).
Adhesins have also been identified in other Leptospira species. L. borgpetersenii is a species commonly carried in dairy cattle around the world (Hamond et al., 2022; Mazzanti et al., 2023), but can infect humans (Slack et al., 2006; Bourhy et al., 2012; Rajaonarivelo et al., 2023). Kamaruzaman et al. (2024) recently published an in silico analysis of L. borgpetersenii sv. Hardjo-Bovis looking for predicted surface exposed proteins present only in pathogens. They identified LBL0972 and LBL2618 that have homologs in the L. interrogans genome, and bind multiple substrates, including fibronectin, laminin, and fibrinogen (Kamaruzaman et al., 2024). A number of other candidate adhesins were identified from additional in silico approaches, including but not limited to LcpA (Barbosa et al., 2010; da Silva et al., 2015), LIC11966 (Ghosh et al., 2019), OmpL37 (Pinne and Haake, 2009; Pinne et al., 2010), MPL36 (Vieira et al., 2010a; Zhu et al., 2023), and LIC10271 (Sarma et al., 2023).
Taken together, in silico genomic analyses have been successful in identifying candidate adhesins. Today there are over 60 species and 300 serovars of Leptospira identified, with 72 reference genomes deposited in NCBI (Genome, 1988-2024; Thibeaux et al., 2018; Casanovas-Massana et al., 2019; Vincent et al., 2019; reviewed in Caimi and Ruybal, 2020; Dos Santos et al., 2023). This wealth of data will allow for extensive comparative analysis, helping to categorize strains by virulence and identify key differences. However, the in vivo role of the majority of the candidate adhesins is unknown. An additional complication is that many Leptospira adhesins bind extracellular matrix components, but specificity in biologically relevant conditions remains to be demonstrated. Future research must distinguish proteins that bind components in vitro from those with a biological and pathogenic role in vivo. Characterizing adhesins with in vivo roles will drive the development of future vaccine and therapeutic approaches.
2.2 Proteomics
As genomics advanced, so did proteomics. Pinne et al. (2012) combined genomics and proteomics to identify novel adhesins from L. interrogans sv. Copenhageni that bound fibronectin. In silico analysis was performed to determine a large set of genes that are predicted to encode surface proteins. A microarray containing these proteins was created and probed with host molecules to identify host-bacterial interactions. This method identified 15 fibronectin binding proteins, and Pinne et al. (2012) were able to confirm LIC10258, LIC10537, LIC10714, LIC11051, LIC11436, LIC11612, and LIC12631 bind fibronectin in vitro (Pinne et al., 2012).
Instead of predicting proteins on the outer surface, some groups have directly identified them on the basis of experimental data. One example is the leptospiral immunoglobulin-like proteins (Ligs), initially identified by searching for L. interrogans sv. Pomona proteins that reacted with sera from infected animals (Palaniappan et al., 2002). This study aimed to identify surface-exposed proteins to use as vaccine candidates, rather than identifying adhesins specifically. Lig proteins also contain domains similar to those of Y. pseudotuberculosis invasin and E. coli intimin, two known adhesins, thus suggesting the Lig proteins could play a role in Leptospira adhesion (Matsunaga et al., 2003). Later, other groups further characterized this three-protein family and confirmed the Lig proteins function as adhesins that bind laminin and fibrinogen (Choy et al., 2007; Castiblanco-Valencia et al., 2012; Hsieh et al., 2016). Lig proteins were of special interest because they were produced during infection, as evidenced by the host immune response, and had a known function. Despite efforts to develop a vaccine using recombinant Lig proteins, success has been limited. Multiple studies have shown that immunizing with one or a combination of Lig proteins, while protective against death, did not prevent infection and colonization (Palaniappan et al., 2006; Silva et al., 2007; Coutinho et al., 2011; Evangelista et al., 2017). Leptospires could still be shed and infect new hosts.
LipL32 was initially identified as an OMP produced during infection (Haake et al., 2000). Haake et al. (2000) identified the most prominent protein band in L. kirschneri by SDS-PAGE and, through further purification and sequencing, identified the lipL32 gene. Homologs of lipL32 were identified in pathogenic Leptospira but were not identified in non-pathogens (Haake et al., 2000). Later, LipL32 was confirmed by independent methods to be the most abundant protein on the outer membrane (Cullen et al., 2005; Malmstrom et al., 2009). It was subsequently characterized as an adhesin that binds fibronectin, collagens, laminin and human umbilical vein endothelial cells (HUVECs) (Hauk et al., 2008; Hoke et al., 2008; Sun et al., 2010). Thus, focusing on confirmed OMPs has also provided valuable information on candidate adhesins in Leptospira.
2.3 Phage display
Phage display, first reported by Smith (1985) as a method for purifying antibodies based upon affinity, has evolved significantly. A notable iteration is shotgun phage display, where random genome fragments are inserted into a phage coat gene, and therefore are present on the surface of the phage (Jacobsson et al., 2003). The resulting library is screened for fragments encoding peptides that bind to a chosen substrate. Enrichment of phage is performed multiple times in succession, and selected clones are isolated and sequenced to identify the encoded gene fragment. There are many advantages of using phage display: (1) adhesin identification relies on functional assays rather than predictions from in silico analysis, (2) as protein fragments are being analyzed, functional data is collected regarding specific domains and residues that are important in adhesion, and (3) identifying similar overlapping fragments increases the specificity and accuracy of data collected.
Lima et al. (2013) performed a shotgun phage display experiment in vitro and in vivo to identify proteins that bind kidney cells by creating a library from fragmented L. interrogans sv. Copenhageni DNA. Phage enrichment was performed based upon adherence to pig renal epithelial cells in vitro, or in vivo through injection into the heart of hamsters where phage was allowed to circulate for 5 min before harvesting the kidneys (Lima et al., 2013). LIC12976 was identified by both in vitro and in vivo strategies (Lima et al., 2013). To our knowledge, this is the only experiment identifying proteins based on function in a living host, and therefore provides clear biological relevance. An additional study by the same group enriched for phage that bound monkey renal epithelial cells (Lauretti-Ferreira et al., 2022). LIC10778 was identified to bind laminin, collagens I and IV, vitronectin and plasma, and cell fibronectins (Lauretti-Ferreira et al., 2022). The importance of LIC10778 in vivo remains unknown.
Another group identified proteins from L. interrogans sv. Copenhageni by phage display that bind Ea.hy926 endothelial cells in vitro (Evangelista et al., 2014). Evangelista K. V. et al. (2014) characterized LIC11574 and LIC13411 as candidate adhesins that bind VE-cadherin with nanomolar affinities and also bind other cadherins. Later work has followed up on this observation and demonstrated that heterologous production of LIC13411 increased binding of a non-pathogen to host tissues, which will be discussed in the next section of this review (Surdel et al., 2022b). Taken together, phage display is a valuable method for identifying candidate host-binding proteins in bacteria. The Leptospira field would greatly benefit from expanding this technique to identify additional biologically relevant adhesins in vivo. For example, in vivo phage display was used to find candidate Borrelia burgdorferi adhesins that bind the endothelium (Antonara et al., 2007). The known integrin-binding adhesin P66 was identified (Coburn et al., 1999; Coburn and Cugini, 2003; Kumar et al., 2015), as well as many novel adhesins that have all been validated by subsequent work (Antonara et al., 2007; Verma et al., 2009; Lin et al., 2015; Yang et al., 2016; Lin et al., 2020). Adapting this method to study Leptospira could provide novel insight into the arsenal of adhesins important during infection.
3 Expanding research beyond identification
Genomics, proteomics, and phage display have led to the identification of countless leptospiral candidate adhesins. As highlighted by this review, many lack functional data in a living host. To fully understand Leptospira pathogenesis, the field needs to move beyond identification and describe the function of these proteins in a living bacterium during infection. Additionally, the field needs higher-resolution analyses to identify specific residues and domains required for function to intelligently design future therapeutic and vaccine strategies.
3.1 The importance of adhesins in host-pathogen interactions
Historically, genetic manipulation of Leptospira has been difficult due to the limited genetic tools available and low rates of transformation. Recent advances are redefining the way we approach research on this genus (Bourhy et al., 2005; Picardeau, 2008; Murray et al., 2009; Alves et al., 2010; Aviat et al., 2010; Pappas et al., 2015; Pappas and Picardeau, 2015; Fernandes et al., 2021a,b,c, 2023a,b; Fernandes and Nascimento, 2022). Two approaches have been taken: heterologous expression of pathogen genes in a non-pathogen, or knockdown and knockouts in a pathogen.
Heterologous expression involves using a shuttle vector encoding a protein from a species different from its original source. In the case of Leptospira, genes from pathogenic species are cloned into a shuttle vector and expressed in a non-pathogen. Several proteins, including LIC11436, LIC11612, LIC11711, LIC12631, LIC13411, LigA, LigB, LMB216, and the mammalian cell entry protein (mce), have been shown to increase binding of the saprophyte to their respective ligands (Figueira et al., 2011; Pinne et al., 2012; Zhang et al., 2012; Toma et al., 2014; Kochi et al., 2020; Surdel et al., 2022a). This approach can be adapted for in vivo studies as well. A model of hematogenous dissemination, originally developed to measure bloodstream survival and tissue adhesion in Borrelia burgdorferi (Caine and Coburn, 2015), was adapted for use with Leptospira in 2022 (Surdel et al., 2022a). Surdel et al. (2022b) demonstrated that LIC13411, when produced in L. biflexa, was sufficient to enhance binding to host tissues, highlighting its potential role during infection. Pappas and Picardeau (2015) engineered a new shuttle vector that facilitates complementation and heterologous expression with increased success, making this strategy accessible for all laboratories (Picardeau, 2008). With this technology now widely successful, it is expected to become a standard baseline requirement for future publications identifying adhesins in Leptospira spp.
The classic method for demonstrating genetic requirements is through knockout experiments followed by complementation if there is a phenotype. In Leptospira, cloning difficulties have limited this approach. Early methods of creating knockouts relied on random transposon mutagenesis, a method typically used for screening (Bourhy et al., 2005; Murray et al., 2009). If a gene of interest were found to be interrupted by chance, this strain could then be used for additional studies such as validation of adhesins. One of the initial targeted methods used transcription activator-like effectors (TALEs) to knockdown genes of interest (Pappas and Picardeau, 2015). Pappas et al. (2015) employed TALEs to show that reducing both LigA and LigB production significantly decreased the virulence of L. interrogans, as measured by death and colonization (Pappas and Picardeau, 2015).
More recently, CRISPR/Cas technology has been adapted for use in Leptospira (Fernandes et al., 2021a,b,c, 2023a,b; Fernandes and Nascimento, 2022). Fernandes et al. (2021c, 2023b) used CRISPR/Cas to assess the in vivo requirements of several adhesins initially characterized in vitro, including LigAB, LipL32, LipL41, and LipL21. However, determining adhesin function with genetic mutants is challenging due to functional redundancy. For instance, only minimal changes were observed in the ability of these mutants to bind host molecules (Fernandes et al., 2021c, 2023b). Mutants of LigAB, LipL21, and LipL41 reduced death in hamsters, but leptospires still were able to colonize the kidneys (Fernandes et al., 2021c, 2023b). In contrast, a LipL32 mutant increased death in hamsters, potentially due to the altered proteome in this mutant (Fernandes et al., 2021c, 2023b). Another important consideration is the temporal requirements of each adhesin during infection. In B. burgdorferi, it is becoming increasingly clear that different adhesins are required for different steps of the endothelial transmigration process (Tan et al., 2023), and this could be true for various processes in Leptospira as well.
Advancements in genetic tools now enable testing of multiple mutations (Fernandes et al., 2023a), enhancing our ability to assign functions to proteins with redundant roles. Despite these strides, only a few adhesins have demonstrated importance in vivo, and none have been shown to be essential for virulence. With these genetic tools readily available, the adhesins previously identified in vitro can now be fully evaluated. This will be crucial in determining the role adhesins play in the pathogenesis of Leptospira.
3.2 Moving toward structure and function analysis
Understanding structure–function relationships of adhesins is essential for developing targeted vaccine and therapeutic strategies. By characterizing proteins at the sequence level, research can pinpoint regions that are critical for protein function. However, in Leptospira, there is currently very little information on candidate adhesin structure–function relationships. Eight complete structures of adhesins have been solved including Lp49 (Giuseppe et al., 2008), Lsa45 (Santos et al., 2023), LSS11580 (Hsu et al., 2020), LIC10831 (Miras et al., 2015), LIC11098 (Miras et al., 2015), LIC12759 (Miras et al., 2015), LIC12234 (Miras et al., 2015), and LipL32 (Hauk et al., 2009; Vivian et al., 2009; Tung et al., 2010). Two proteins, LigA (Mei et al., 2015a; Kumar et al., 2023) and LigB (Ptak et al., 2014; Mei et al., 2015b), only have specific domains solved.
Among these structures, only a couple have been investigated for structure–function relationships. Specific domains LigB, including LigB4 and LigB12, are important for binding human fibrinogen and tropoelastin (Hsieh et al., 2016, 2017). A few residues within LigB12 have been identified as important for binding, but a comprehensive structure–function analysis of LigB is still lacking (Hsieh et al., 2016, 2017). Mutation of residues within LipL32 that abolish calcium binding have shown that while calcium binding is not necessary for adhesion, it does influence the host immune response (Hauk et al., 2012; Lo et al., 2013).
Given this significant gap in knowledge, additional structure–function studies on leptospiral adhesins are crucial. This research is essential for designing vaccines and therapeutics targeting key epitopes recognized by the immune system. By separating distinct phenotypes in multi-functional adhesins, it may also be possible to engineer attenuated bacterial vaccines that are unable to bind and infect the host, while still displaying epitopes required to generate an adaptive immune response.
4 Conclusions
As new technologies continue to evolve in Leptospira research and other disciplines, methods for identifying and characterizing adhesins are also evolving. Techniques such as next-generation sequencing (NGS) offer more efficient characterization of genomic libraries than ever before. Experimental approaches that may have been under-used in the past, such as phage display or protein microarrays, may be combined with NGS to identify novel genes involved in host binding. These advancements are crucial for expanding our understanding of adhesin roles within a living host.
Using the new tools available in Leptospira, the field must now define which of the many candidates are in fact true adhesins. In the context of microbial pathogenesis, an adhesin must directly mediate the attachment of the bacteria to the host and be important during infection. Specifically, an adhesin must have a reasonable level of specificity and affinity (such as a KD < 1 μM). It must contain a definable binding domain, where the structure of the protein-ligand interaction can be delineated. The gene must be expressed during infection, and inactivation of adhesin genes should lead to a binding and ultimately virulence defect (either alone, or in combination with proteins of redundant function). This defect must be due to the adhesin-ligand interaction specifically (as opposed to unrelated functions the protein may have during pathogenesis). As these candidates are evaluated using state-of-the-art approaches, the full arsenal of Leptospira adhesins will be defined and can then be targeted by future vaccine and therapeutic strategies.
Anti-adhesin therapy is a promising antibacterial strategy that can be used in conjunction with, or in lieu of, classical antibiotics. Every step of the adhesion process has been targeted, including modulating host receptors (Svensson et al., 1994, 2006), synthesizing peptides to compete for binding (Okuda et al., 2010; Huebinger et al., 2016), and immunization against adhesins (Cook et al., 2007; McNeilly et al., 2010). Anti-adhesin therapy has many advantages to typical antibiotics, including long term protection imparted by immunization. Krachler and Orth (2013) hypothesize that since anti-adhesin therapy does not affect the fitness of the organism, it is less likely to select for resistance. Disrupting adhesion in sites with high natural clearance rates (such as the kidney and bladder), will have the greatest impact. By targeting leptospiral adherence to the kidney proximal tubules, the bacteria will be forced into their planktonic state, thus increasing the rate of clearance through the natural route (Ternent et al., 2015).
A detailed understanding of the arsenal of adhesins used by Leptospira is needed. Overall, it is evident that the tools available for studying adhesin function in Leptospira spp. are advancing rapidly. With these technological advancements, future research on functions of adhesins must incorporate these tools to fully understand their relevance and roles in pathogenesis.
Author contributions
MS: Conceptualization, Writing – original draft, Writing – review & editing. JC: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the NIAID P01 AI168148.
Acknowledgments
We thank Beth Hahn for her review of this manuscript prior to submission.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adler, B., Cameron, C., Haake, D., Hartskeerl, R., Ellis, W., Levitt, P., et al. (2014). Leptospira and leptospirosis. New York, NY: Springer.
Adler, B., and de la Pena, M. A. (2010). Leptospira and leptospirosis. Vet. Microbiol. 140, 287–296. doi: 10.1016/j.vetmic.2009.03.012
Agampodi, S., Gunarathna, S., Lee, J. S., and Excler, J. L. (2023). Global, regional, and country-level cost of leptospirosis due to loss of productivity in humans. PLoS Negl. Trop. Dis. 17:e0011291. doi: 10.1371/journal.pntd.0011291
Alston, J. M., and Brown, H. C. (1937). The epidemiology of Weil's disease (section of epidemiology and state medicine). Proc. R. Soc. Med. 30, 741–756. doi: 10.1177/003591573703000628
Alves, C. S., Melo, M. N., Franquelim, H. G., Ferre, R., Planas, M., Feliu, L., et al. (2010). Escherichia coli cell surface perturbation and disruption induced by antimicrobial peptides BP100 and pepR. J. Biol. Chem. 285, 27536–27544. doi: 10.1074/jbc.M110.130955
Antonara, S., Chafel, R. M., LaFrance, M., and Coburn, J. (2007). Borrelia burgdorferi adhesins identified using in vivo phage display. Mol. Microbiol. 66, 262–276. doi: 10.1111/j.1365-2958.2007.05924.x
Atzingen, M. V., Barbosa, A. S., De Brito, T., Vasconcellos, S. A., de Morais, Z. M., Lima, D. M., et al. (2008). Lsa21, a novel leptospiral protein binding adhesive matrix molecules and present during human infection. BMC Microbiol. 8:70. doi: 10.1186/1471-2180-8-70
Atzingen, M. V., Gomez, R. M., Schattner, M., Pretre, G., Goncales, A. P., de Morais, Z. M., et al. (2009). Lp95, a novel leptospiral protein that binds extracellular matrix components and activates e-selectin on endothelial cells. J. Infect. 59, 264–276. doi: 10.1016/j.jinf.2009.07.010
Aviat, F., Slamti, L., Cerqueira, G. M., Lourdault, K., and Picardeau, M. (2010). Expanding the genetic toolbox for Leptospira species by generation of fluorescent bacteria. Appl. Environ. Microbiol. 76, 8135–8142. doi: 10.1128/AEM.02199-10
Barbosa, A. S., Abreu, P. A., Neves, F. O., Atzingen, M. V., Watanabe, M. M., Vieira, M. L., et al. (2006). A newly identified leptospiral adhesin mediates attachment to laminin. Infect. Immun. 74, 6356–6364. doi: 10.1128/IAI.00460-06
Barbosa, A. S., Monaris, D., Silva, L. B., Morais, Z. M., Vasconcellos, S. A., Cianciarullo, A. M., et al. (2010). Functional characterization of LcpA, a surface-exposed protein of Leptospira spp. that binds the human complement regulator C4BP. Infect. Immun. 78, 3207–3216. doi: 10.1128/IAI.00279-10
Barwick, R. S., Mohammed, H. O., McDonough, P. L., and White, M. E. (1998). Epidemiologic features of equine Leptospira interrogans of human significance. Prev. Vet. Med. 36, 153–165. doi: 10.1016/S0167-5877(98)00069-5
Bennett, R. M., Christiansen, K., and Clifton-Hadley, R. S. (1999). Estimating the costs associated with endemic diseases of dairy cattle. J. Dairy Res. 66, 455–459. doi: 10.1017/S0022029999003684
Bierne, H., Sabet, C., Personnic, N., and Cossart, P. (2007). Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 9, 1156–1166. doi: 10.1016/j.micinf.2007.05.003
Bourhy, P., Collet, L., Lernout, T., Zinini, F., Hartskeerl, R. A., van der Linden, H., et al. (2012). Human Leptospira isolates circulating in Mayotte (Indian Ocean) have unique serological and molecular features. J. Clin. Microbiol. 50, 307–311. doi: 10.1128/JCM.05931-11
Bourhy, P., Louvel, H., Saint Girons, I., and Picardeau, M. (2005). Random insertional mutagenesis of Leptospira interrogans, the agent of leptospirosis, using a mariner transposon. J. Bacteriol. 187, 3255–3258. doi: 10.1128/JB.187.9.3255-3258.2005
Caimi, K., and Ruybal, P. (2020). Leptospira spp., a genus in the stage of diversity and genomic data expansion. Infect. Genet. Evol. 81:104241. doi: 10.1016/j.meegid.2020.104241
Caine, J. A., and Coburn, J. (2015). A short-term Borrelia burgdorferi infection model identifies tissue tropisms and bloodstream survival conferred by adhesion proteins. Infect. Immun. 83, 3184–3194. doi: 10.1128/IAI.00349-15
Canton, G. J., Moreno, F., Fiorentino, M. A., Hecker, Y. P., Spetter, M., Fiorani, F., et al. (2022). Spatial-temporal trends and economic losses associated with bovine abortifacients in Central Argentina. Trop. Anim. Health Prod. 54:242. doi: 10.1007/s11250-022-03237-0
Casanovas-Massana, A., Vincent, A. T., Bourhy, P., Neela, V. K., Veyrier, F. J., Picardeau, M., et al. (2019). Leptospira dzianensis and Leptospira putramalaysiae are later heterotypic synonyms of Leptospira yasudae and Leptospira stimsonii. Int. J. Syst. Evol. Microbiol. 71:004713. doi: 10.1099/ijsem.0.004713
Castiblanco-Valencia, M. M., Fraga, T. R., Silva, L. B., Monaris, D., Abreu, P. A., Strobel, S., et al. (2012). Leptospiral immunoglobulin-like proteins interact with human complement regulators factor H, FHL-1, FHR-1, and C4BP. J. Infect. Dis. 205, 995–1004. doi: 10.1093/infdis/jir875
Cavenague, M. F., Teixeira, A. F., Fernandes, L. G. V., and Nascimento, A. (2023). LIC12254 is a Leptospiral protein that interacts with Integrins via the RGD motif. Trop. Med. Infect. Dis. 8:249. doi: 10.3390/tropicalmed8050249
Cavenague, M. F., Teixeira, A. F., Filho, A. S., Souza, G. O., Vasconcellos, S. A., Heinemann, M. B., et al. (2019). Characterization of a novel protein of Leptospira interrogans exhibiting plasminogen, vitronectin and complement binding properties. Int. J. Med. Microbiol. 309, 116–129. doi: 10.1016/j.ijmm.2018.12.005
Choy, H. A., Kelley, M. M., Chen, T. L., Moller, A. K., Matsunaga, J., and Haake, D. A. (2007). Physiological osmotic induction of Leptospira interrogans adhesion: LigA and LigB bind extracellular matrix proteins and fibrinogen. Infect. Immun. 75, 2441–2450. doi: 10.1128/IAI.01635-06
Coburn, J., Chege, W., Magoun, L., Bodary, S. C., and Leong, J. M. (1999). Characterization of a candidate Borrelia burgdorferi beta3-chain integrin ligand identified using a phage display library. Mol. Microbiol. 34, 926–940. doi: 10.1046/j.1365-2958.1999.01654.x
Coburn, J., and Cugini, C. (2003). Targeted mutation of the outer membrane protein P66 disrupts attachment of the Lyme disease agent, Borrelia burgdorferi, to integrin alphavbeta3. Proc. Natl. Acad. Sci. U. S. A 100, 7301–7306. doi: 10.1073/pnas.1131117100
Cook, S. R., Maiti, P. K., DeVinney, R., Allen-Vercoe, E., Bach, S. J., and McAllister, T. A. (2007). Avian- and mammalian-derived antibodies against adherence-associated proteins inhibit host cell colonization by Escherichia coli O157:H7. J. Appl. Microbiol. 103, 1206–1219. doi: 10.1111/j.1365-2672.2007.03334.x
Costa, F., Hagan, J. E., Calcagno, J., Kane, M., Torgerson, P., Martinez-Silveira, M. S., et al. (2015). Global morbidity and mortality of leptospirosis: a systematic review. PLoS Negl. Trop. Dis. 9:e0003898. doi: 10.1371/journal.pntd.0003898
Coutinho, M. L., Choy, H. A., Kelley, M. M., Matsunaga, J., Babbitt, J. T., Lewis, M. S., et al. (2011). A LigA three-domain region protects hamsters from lethal infection by Leptospira interrogans. PLoS Negl. Trop. Dis. 5:e1422. doi: 10.1371/journal.pntd.0001422
Cullen, P. A., Xu, X., Matsunaga, J., Sanchez, Y., Ko, A. I., Haake, D. A., et al. (2005). Surfaceome of Leptospira spp. Infect. Immun. 73, 4853–4863. doi: 10.1128/IAI.73.8.4853-4863.2005
da Silva, L. B., Miragaia Ldos, S., Breda, L. C., Abe, C. M., Schmidt, M. C., Moro, A. M., et al. (2015). Pathogenic Leptospira species acquire factor H and vitronectin via the surface protein LcpA. Infect. Immun. 83, 888–897. doi: 10.1128/IAI.02844-14
Daroz, B. B., Fernandes, L. G. V., Cavenague, M. F., Kochi, L. T., Passalia, F. J., Takahashi, M. B., et al. (2021). A review on host-Leptospira interactions: what we know and future expectations. Front. Cell. Infect. Microbiol. 11:777709. doi: 10.3389/fcimb.2021.777709
Domingos, R. F., Fernandes, L. G., Romero, E. C., de Morais, Z. M., Vasconcellos, S. A., and Nascimento, A. L. (2015). Novel Leptospira interrogans protein Lsa32 is expressed during infection and binds laminin and plasminogen. Microbiology 161, 851–864. doi: 10.1099/mic.0.000041
Domingos, R. F., Vieira, M. L., Romero, E. C., Goncales, A. P., de Morais, Z. M., Vasconcellos, S. A., et al. (2012). Features of two proteins of Leptospira interrogans with potential role in host-pathogen interactions. BMC Microbiol. 12:50. doi: 10.1186/1471-2180-12-50
Dos Santos, R. P., Carvalho, N. B., Aburjaile, F., Sousa, T., Verissimo, G., Gomes, T., et al. (2023). Environmental biofilms from an Urban community in Salvador, Brazil, shelter previously uncharacterized saprophytic Leptospira. Microb. Ecol. 86, 2488–2501. doi: 10.1007/s00248-023-02253-3
Eshghi, A., Pappalardo, E., Hester, S., Thomas, B., Pretre, G., and Picardeau, M. (2015). Pathogenic Leptospira interrogans exoproteins are primarily involved in heterotrophic processes. Infect. Immun. 83, 3061–3073. doi: 10.1128/IAI.00427-15
European Centre for Disease Prevention and Control . (2017). Factsheet about leptospirosis. Available at: https://www.ecdc.europa.eu/en/leptospirosis/factsheet.
Evangelista, K., Franco, R., Schwab, A., and Coburn, J. (2014). Leptospira interrogans binds to cadherins. PLoS Negl. Trop. Dis. 8:e2672. doi: 10.1371/journal.pntd.0002672
Evangelista, K. V., Hahn, B., Wunder, E. A. Jr., Ko, A. I., Haake, D. A., and Coburn, J. (2014). Identification of cell-binding adhesins of Leptospira interrogans. PLoS Negl. Trop. Dis. 8:e3215. doi: 10.1371/journal.pntd.0003215
Evangelista, K. V., Lourdault, K., Matsunaga, J., and Haake, D. A. (2017). Immunoprotective properties of recombinant LigA and LigB in a hamster model of acute leptospirosis. PLoS One 12:e0180004. doi: 10.1371/journal.pone.0180004
Fernandes, L. G. V., Foltran, B. B., Teixeira, A. F., and Nascimento, A. (2023a). LipL41 and LigA/LigB gene silencing on a LipL32 knockout Leptospira interrogans reveals the impact of multiple mutations on virulence. Pathogens 12:1191. doi: 10.3390/pathogens12101191
Fernandes, L. G. V., Hornsby, R. L., Nascimento, A., and Nally, J. E. (2021a). Application of CRISPR interference (CRISPRi) for gene silencing in pathogenic species of Leptospira. J. Vis. Exp. 174:e62631. doi: 10.3791/62631-v
Fernandes, L. G. V., Hornsby, R. L., Nascimento, A., and Nally, J. E. (2021b). Genetic manipulation of pathogenic Leptospira: CRISPR interference (CRISPRi)-mediated gene silencing and rapid mutant recovery at 37 degrees C. Sci. Rep. 11:1768. doi: 10.1038/s41598-021-81400-7
Fernandes, L. G. V., and Nascimento, A. (2022). A novel breakthrough in Leptospira spp. mutagenesis: knockout by combination of CRISPR/Cas9 and non-homologous end-joining systems. Front. Microbiol. 13:915382. doi: 10.3389/fmicb.2022.915382
Fernandes, L. G. V., Putz, E. J., Stasko, J., Lippolis, J. D., Nascimento, A., and Nally, J. E. (2021c). Evaluation of LipL32 and LigA/LigB knockdown mutants in Leptospira interrogans serovar Copenhageni: impacts to proteome and virulence. Front. Microbiol. 12:799012. doi: 10.3389/fmicb.2021.799012
Fernandes, L. G. V., Teixeira, A. F., and Nascimento, A. (2023b). Evaluation of Leptospira interrogans knockdown mutants for LipL32, LipL41, LipL21, and OmpL1 proteins. Front. Microbiol. 14:1199660. doi: 10.3389/fmicb.2023.1199660
Fernandes, L. G. V., Vieira, M. L., Alves, I. J., de Morais, Z. M., Vasconcellos, S. A., Romero, E. C., et al. (2014). Functional and immunological evaluation of two novel proteins of Leptospira spp. Microbiology 160, 149–164. doi: 10.1099/mic.0.072074-0
Fernandes, L. G., Vieira, M. L., Kirchgatter, K., Alves, I. J., de Morais, Z. M., Vasconcellos, S. A., et al. (2012). OmpL1 is an extracellular matrix- and plasminogen-interacting protein of Leptospira spp. Infect. Immun. 80, 3679–3692. doi: 10.1128/IAI.00474-12
Figueira, C. P., Croda, J., Choy, H. A., Haake, D. A., Reis, M. G., Ko, A. I., et al. (2011). Heterologous expression of pathogen-specific genes ligA and ligB in the saprophyte Leptospira biflexa confers enhanced adhesion to cultured cells and fibronectin. BMC Microbiol. 11:129. doi: 10.1186/1471-2180-11-129
Fleischmann, R. D., Adams, M. D., White, O., Clayton, R. A., Kirkness, E. F., Kerlavage, A. R., et al. (1995). Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269, 496–512. doi: 10.1126/science.7542800
Fouts, D. E., Matthias, M. A., Adhikarla, H., Adler, B., Amorim-Santos, L., Berg, D. E., et al. (2016). What makes a bacterial species pathogenic?:comparative genomic analysis of the genus Leptospira. PLoS Negl. Trop. Dis. 10:e0004403. doi: 10.1371/journal.pntd.0004403
Fraser, C. M., Gocayne, J. D., White, O., Adams, M. D., Clayton, R. A., Fleischmann, R. D., et al. (1995). The minimal gene complement of Mycoplasma genitalium. Science 270, 397–404. doi: 10.1126/science.270.5235.397
Gamberini, M., Gomez, R. M., Atzingen, M. V., Martins, E. A., Vasconcellos, S. A., Romero, E. C., et al. (2005). Whole-genome analysis of Leptospira interrogans to identify potential vaccine candidates against leptospirosis. FEMS Microbiol. Lett. 244, 305–313. doi: 10.1016/j.femsle.2005.02.004
Genome . (1988-2024). Bethesda, MD: National Library of Medicine (US), National Center for Biotechnology Information. Available at: https://www.ncbi.nlm.nih.gov/datasets/genome/.
Ghosh, K. K., Prakash, A., Dhara, A., Hussain, M. S., Shrivastav, P., Kumar, P., et al. (2019). Role of Supramolecule ErpY-like lipoprotein of Leptospira in thrombin-catalyzed fibrin clot inhibition and binding to complement factors H and I, and its diagnostic potential. Infect. Immun. 87, 10–1128. doi: 10.1128/IAI.00536-19
Giuseppe, P. O., Neves, F. O., Nascimento, A. L., and Guimaraes, B. G. (2008). The leptospiral antigen Lp49 is a two-domain protein with putative protein binding function. J. Struct. Biol. 163, 53–60. doi: 10.1016/j.jsb.2008.04.003
Gomez, R. M., Vieira, M. L., Schattner, M., Malaver, E., Watanabe, M. M., Barbosa, A. S., et al. (2008). Putative outer membrane proteins of Leptospira interrogans stimulate human umbilical vein endothelial cells (HUVECS) and express during infection. Microb. Pathog. 45, 315–322. doi: 10.1016/j.micpath.2008.08.004
Gouveia, E. L., Metcalfe, J., de Carvalho, A. L., Aires, T. S., Villasboas-Bisneto, J. C., Queirroz, A., et al. (2008). Leptospirosis-associated severe pulmonary hemorrhagic syndrome, Salvador, Brazil. Emerg. Infect Dis. 14, 505–508. doi: 10.3201/eid1403.071064
Haake, D. A., Chao, G., Zuerner, R. L., Barnett, J. K., Barnett, D., Mazel, M., et al. (2000). The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect. Immun. 68, 2276–2285. doi: 10.1128/IAI.68.4.2276-2285.2000
Haake, D. A., and Levett, P. N. (2015). Leptospirosis in humans. Curr. Top. Microbiol. Immunol. 387, 65–97. doi: 10.1007/978-3-662-45059-8_5
Hamond, C., Dirsmith, K. L., LeCount, K., Soltero, F. V., Rivera-Garcia, S., Camp, P., et al. (2022). Leptospira borgpetersenii serovar Hardjo and Leptospira santarosai serogroup Pyrogenes isolated from bovine dairy herds in Puerto Rico. Front. Vet. Sci. 9:1025282. doi: 10.3389/fvets.2022.1025282
Hauk, P., Barbosa, A. S., Ho, P. L., and Farah, C. S. (2012). Calcium binding to Leptospira outer membrane antigen LipL32 is not necessary for its interaction with plasma fibronectin, collagen type IV, and plasminogen. J. Biol. Chem. 287, 4826–4834. doi: 10.1074/jbc.M111.277210
Hauk, P., Guzzo, C. R., Roman Ramos, H., Ho, P. L., and Farah, C. S. (2009). Structure and calcium-binding activity of LipL32, the major surface antigen of pathogenic Leptospira sp. J. Mol. Biol. 390, 722–736. doi: 10.1016/j.jmb.2009.05.034
Hauk, P., Macedo, F., Romero, E. C., Vasconcellos, S. A., de Morais, Z. M., Barbosa, A. S., et al. (2008). In LipL32, the major leptospiral lipoprotein, the C terminus is the primary immunogenic domain and mediates interaction with collagen IV and plasma fibronectin. Infect. Immun. 76, 2642–2650. doi: 10.1128/IAI.01639-07
Hoke, D. E., Egan, S., Cullen, P. A., and Adler, B. (2008). LipL32 is an extracellular matrix-interacting protein of Leptospira spp. and Pseudoalteromonas tunicata. Infect. Immun. 76, 2063–2069. doi: 10.1128/IAI.01643-07
Hsieh, C. L., Chang, E., Tseng, A., Ptak, C., Wu, L. C., Su, C. L., et al. (2016). Leptospira immunoglobulin-like protein B (LigB) binds to both the C-terminal 23 amino acids of fibrinogen alphaC domain and factor XIII: insight into the mechanism of LigB-mediated blockage of fibrinogen alpha chain cross-linking. PLoS Negl. Trop. Dis. 10:e0004974. doi: 10.1371/journal.pntd.0004974
Hsieh, C. L., Tseng, A., He, H., Kuo, C. J., Wang, X., and Chang, Y. F. (2017). Leptospira immunoglobulin-like protein B interacts with the 20th exon of human Tropoelastin contributing to Leptospiral adhesion to human lung cells. Front. Cell. Infect. Microbiol. 7:163. doi: 10.3389/fcimb.2017.00163
Hsu, S. H., Chu, C. H., Tian, Y. C., Chang, M. Y., Chou, L. F., Sun, Y. J., et al. (2020). Crystal structure of Leptospira leucine-rich repeat 20 reveals a novel E-cadherin binding protein to induce NGAL expression in HK2 cells. Biochem. J. 477, 4313–4326. doi: 10.1042/BCJ20200547
Huebinger, R. M., Stones, D. H., de Souza, S. M., Carlson, D. L., Song, J., Vaz, D. P., et al. (2016). Targeting bacterial adherence inhibits multidrug-resistant Pseudomonas aeruginosa infection following burn injury. Sci. Rep. 6:39341. doi: 10.1038/srep39341
Ito, T., and Yanagawa, R. (1987). Leptospiral attachment to extracellular matrix of mouse fibroblast (L929) cells. Vet. Microbiol. 15, 89–96. doi: 10.1016/0378-1135(87)90133-7
Jacobsson, K., Rosander, A., Bjerketorp, J., and Frykberg, L. (2003). Shotgun phage display - selection for bacterial receptins or other exported proteins. Biol. Proced. Online 5, 123–135. doi: 10.1251/bpo54
Kamaruzaman, I. N. A., Staton, G. J., Ainsworth, S., Carter, S. D., and Evans, N. J. (2024). Characterisation of putative outer membrane proteins from Leptospira borgpetersenii serovar Hardjo-Bovis identifies novel adhesins and diversity in adhesion across genomospecies orthologs. Microorganisms 12:245. doi: 10.3390/microorganisms12020245
Kochi, L. T., Fernandes, L. G. V., and Nascimento, A. (2020). Heterologous expression of the pathogen-specific LIC11711 gene in the Saprophyte L. biflexa increases bacterial binding to laminin and plasminogen. Pathogens 9:599. doi: 10.3390/pathogens9080599
Kochi, L. T., Fernandes, L. G. V., Souza, G. O., Vasconcellos, S. A., Heinemann, M. B., Romero, E. C., et al. (2019). The interaction of two novel putative proteins of Leptospira interrogans with E-cadherin, plasminogen and complement components with potential role in bacterial infection. Virulence 10, 734–753. doi: 10.1080/21505594.2019.1650613
Krachler, A. M., and Orth, K. (2013). Targeting the bacteria-host interface: strategies in anti-adhesion therapy. Virulence 4, 284–294. doi: 10.4161/viru.24606
Kumar, D., Ristow, L. C., Shi, M., Mukherjee, P., Caine, J. A., Lee, W. Y., et al. (2015). Intravital imaging of vascular transmigration by the Lyme spirochete: requirement for the integrin binding residues of the B. burgdorferi P66 protein. PLoS Pathog 11:e1005333. doi: 10.1371/journal.ppat.1005333
Kumar, P., Vyas, P., Faisal, S. M., Chang, Y. F., and Akif, M. (2023). Crystal structure of a variable region segment of Leptospira host-interacting outer surface protein, LigA, reveals the orientation of Ig-like domains. Int. J. Biol. Macromol. 244:125445. doi: 10.1016/j.ijbiomac.2023.125445
Lauretti-Ferreira, F., Teixeira, A. A. R., Giordano, R. J., da Silva, J. B., Abreu, P. A. E., Barbosa, A. S., et al. (2022). Characterization of a virulence-modifying protein of Leptospira interrogans identified by shotgun phage display. Front. Microbiol. 13:1051698. doi: 10.3389/fmicb.2022.1051698
Lilenbaum, W., Varges, R., Medeiros, L., Cordeiro, A. G., Cavalcanti, A., Souza, G. N., et al. (2008). Risk factors associated with leptospirosis in dairy goats under tropical conditions in Brazil. Res. Vet. Sci. 84, 14–17. doi: 10.1016/j.rvsc.2007.03.011
Lima, S. S., Ching, A. T., Favaro, R. D., Da Silva, J. B., Oliveira, M. L., Carvalho, E., et al. (2013). Adhesin activity of Leptospira interrogans lipoprotein identified by in vivo and in vitro shotgun phage display. Biochem. Biophys. Res. Commun. 431, 342–347. doi: 10.1016/j.bbrc.2012.12.095
Lin, Y. P., Bhowmick, R., Coburn, J., and Leong, J. M. (2015). Host cell heparan sulfate glycosaminoglycans are ligands for OspF-related proteins of the Lyme disease spirochete. Cell. Microbiol. 17, 1464–1476. doi: 10.1111/cmi.12448
Lin, Y. P., Tan, X., Caine, J. A., Castellanos, M., Chaconas, G., Coburn, J., et al. (2020). Strain-specific joint invasion and colonization by Lyme disease spirochetes is promoted by outer surface protein C. PLoS Pathog. 16:e1008516. doi: 10.1371/journal.ppat.1008516
Lo, Y. Y., Hsu, S. H., Ko, Y. C., Hung, C. C., Chang, M. Y., Hsu, H. H., et al. (2013). Essential calcium-binding cluster of Leptospira LipL32 protein for inflammatory responses through the toll-like receptor 2 pathway. J. Biol. Chem. 288, 12335–12344. doi: 10.1074/jbc.M112.418699
Longhi, M. T., Oliveira, T. R., Romero, E. C., Goncales, A. P., de Morais, Z. M., Vasconcellos, S. A., et al. (2009). A newly identified protein of Leptospira interrogans mediates binding to laminin. J. Med. Microbiol. 58, 1275–1282. doi: 10.1099/jmm.0.011916-0
Malmstrom, J., Beck, M., Schmidt, A., Lange, V., Deutsch, E. W., and Aebersold, R. (2009). Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460, 762–765. doi: 10.1038/nature08184
Marra, A., and Isberg, R. R. (1996). Common entry mechanisms. Bacterial pathogenesis. Curr. Biol. 6, 1084–1086. doi: 10.1016/S0960-9822(02)70672-6
Matsunaga, J., Barocchi, M. A., Croda, J., Young, T. A., Sanchez, Y., Siqueira, I., et al. (2003). Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol. Microbiol. 49, 929–946. doi: 10.1046/j.1365-2958.2003.03619.x
Matthews, A. G., Waitkins, S. A., and Palmer, M. F. (1987). Serological study of leptospiral infections and endogenous uveitis among horses and ponies in the United Kingdom. Equine Vet. J. 19, 125–128. doi: 10.1111/j.2042-3306.1987.tb02605.x
Mazzanti, M., Scialfa, E., Rivero, M., and Passucci, J. (2023). Epidemiology of Leptospira spp. infection in a beef cattle area of Argentina. Front. Vet. Sci. 10:1083024. doi: 10.3389/fvets.2023.1083024
McNeilly, T. N., Mitchell, M. C., Rosser, T., McAteer, S., Low, J. C., Smith, D. G., et al. (2010). Immunization of cattle with a combination of purified intimin-531, EspA and Tir significantly reduces shedding of Escherichia coli O157:H7 following oral challenge. Vaccine 28, 1422–1428. doi: 10.1016/j.vaccine.2009.10.076
Mei, S., Zhang, J., Zhang, X., and Tu, X. (2015a). Solution structure of leptospiral LigA4 big domain. Biochem. Biophys. Res. Commun. 467, 288–292. doi: 10.1016/j.bbrc.2015.09.170
Mei, S., Zhang, J., Zhang, X., and Tu, X. (2015b). Solution structure of a bacterial immunoglobulin-like domain of the outer membrane protein (LigB) from Leptospira. Proteins 83, 195–200. doi: 10.1002/prot.24723
Mendes, R. S., Von Atzingen, M., de Morais, Z. M., Goncales, A. P., Serrano, S. M., Asega, A. F., et al. (2011). The novel leptospiral surface adhesin Lsa20 binds laminin and human plasminogen and is probably expressed during infection. Infect. Immun. 79, 4657–4667. doi: 10.1128/IAI.05583-11
Merien, F., Baranton, G., and Perolat, P. (1997). Invasion of Vero cells and induction of apoptosis in macrophages by pathogenic Leptospira interrogans are correlated with virulence. Infect. Immun. 65, 729–738. doi: 10.1128/iai.65.2.729-738.1997
Miotto, B. A., Guilloux, A. G. A., Tozzi, B. F., Moreno, L. Z., da Hora, A. S., Dias, R. A., et al. (2018). Prospective study of canine leptospirosis in shelter and stray dog populations: identification of chronic carriers and different Leptospira species infecting dogs. PLoS One 13:e0200384. doi: 10.1371/journal.pone.0200384
Miras, I., Saul, F., Nowakowski, M., Weber, P., Haouz, A., Shepard, W., et al. (2015). Structural characterization of a novel subfamily of leucine-rich repeat proteins from the human pathogen Leptospira interrogans. Acta Crystallogr. D Biol. Crystallogr. 71, 1351–1359. doi: 10.1107/S139900471500704X
Murray, G. L., Morel, V., Cerqueira, G. M., Croda, J., Srikram, A., Henry, R., et al. (2009). Genome-wide transposon mutagenesis in pathogenic Leptospira species. Infect. Immun. 77, 810–816. doi: 10.1128/IAI.01293-08
Nascimento, A. L., Ko, A. I., Martins, E. A., Monteiro-Vitorello, C. B., Ho, P. L., Haake, D. A., et al. (2004a). Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 186, 2164–2172. doi: 10.1128/JB.186.7.2164-2172.2004
Nascimento, A. L., Verjovski-Almeida, S., Van Sluys, M. A., Monteiro-Vitorello, C. B., Camargo, L. E., Digiampietri, L. A., et al. (2004b). Genome features of Leptospira interrogans serovar Copenhageni. Braz. J. Med. Biol. Res. 37, 459–477. doi: 10.1590/S0100-879X2004000400003
Ng, A., and Xavier, R. J. (2011). Leucine-rich repeat (LRR) proteins: integrators of pattern recognition and signaling in immunity. Autophagy 7, 1082–1084. doi: 10.4161/auto.7.9.16464
Niemann, H. H., Schubert, W. D., and Heinz, D. W. (2004). Adhesins and invasins of pathogenic bacteria: a structural view. Microbes Infect. 6, 101–112. doi: 10.1016/j.micinf.2003.11.001
Okuda, K., Hanada, N., Usui, Y., Takeuchi, H., Koba, H., Nakao, R., et al. (2010). Inhibition of Streptococcus mutans adherence and biofilm formation using analogues of the SspB peptide. Arch. Oral Biol. 55, 754–762. doi: 10.1016/j.archoralbio.2010.06.014
Oliveira, R., de Morais, Z. M., Goncales, A. P., Romero, E. C., Vasconcellos, S. A., and Nascimento, A. L. (2011). Characterization of novel OmpA-like protein of Leptospira interrogans that binds extracellular matrix molecules and plasminogen. PLoS One 6:e21962. doi: 10.1371/journal.pone.0021962
Oliveira, R., Domingos, R. F., Siqueira, G. H., Fernandes, L. G., Souza, N. M., Vieira, M. L., et al. (2013). Adhesins of Leptospira interrogans mediate the interaction to fibrinogen and inhibit fibrin clot formation in vitro. PLoS Negl. Trop. Dis. 7:e2396. doi: 10.1371/journal.pntd.0002396
Oliveira, T. R., Longhi, M. T., Goncales, A. P., de Morais, Z. M., Vasconcellos, S. A., and Nascimento, A. L. (2010). LipL53, a temperature regulated protein from Leptospira interrogans that binds to extracellular matrix molecules. Microbes Infect. 12, 207–217. doi: 10.1016/j.micinf.2009.12.004
Palaniappan, R. U., Chang, Y. F., Jusuf, S. S., Artiushin, S., Timoney, J. F., McDonough, S. P., et al. (2002). Cloning and molecular characterization of an immunogenic LigA protein of Leptospira interrogans. Infect. Immun. 70, 5924–5930. doi: 10.1128/IAI.70.11.5924-5930.2002
Palaniappan, R. U., McDonough, S. P., Divers, T. J., Chen, C. S., Pan, M. J., Matsumoto, M., et al. (2006). Immunoprotection of recombinant leptospiral immunoglobulin-like protein a against Leptospira interrogans serovar Pomona infection. Infect. Immun. 74, 1745–1750. doi: 10.1128/IAI.74.3.1745-1750.2006
Pappas, C. J., Benaroudj, N., and Picardeau, M. (2015). A replicative plasmid vector allows efficient complementation of pathogenic Leptospira strains. Appl. Environ. Microbiol. 81, 3176–3181. doi: 10.1128/AEM.00173-15
Pappas, C. J., and Picardeau, M. (2015). Control of gene expression in Leptospira spp. by transcription activator-like effectors demonstrates a potential role for LigA and LigB in Leptospira interrogans virulence. Appl. Environ. Microbiol. 81, 7888–7892. doi: 10.1128/AEM.02202-15
Passalia, F. J., Carvalho, E., Heinemann, M. B., Vieira, M. L., and Nascimento, A. (2020a). The Leptospira interrogans LIC10774 is a multifunctional surface protein that binds calcium and interacts with host components. Microbiol. Res. 235:126470. doi: 10.1016/j.micres.2020.126470
Passalia, F. J., Heinemann, M. B., de Andrade, S. A., Nascimento, A., and Vieira, M. L. (2020b). Leptospira interrogans bat proteins impair host hemostasis by fibrinogen cleavage and platelet aggregation inhibition. Med. Microbiol. Immunol. 209, 201–213. doi: 10.1007/s00430-020-00664-4
Passalia, F. J., Heinemann, M. B., Vieira, M. L., and Nascimento, A. (2021). A novel Leptospira interrogans protein LIC13086 inhibits fibrin clot formation and interacts with host components. Front. Cell. Infect. Microbiol. 11:708739. doi: 10.3389/fcimb.2021.708739
Patel, S., Mathivanan, N., and Goyal, A. (2017). Bacterial adhesins, the pathogenic weapons to trick host defense arsenal. Biomed. Pharmacother. 93, 763–771. doi: 10.1016/j.biopha.2017.06.102
Paulsson, M., and Riesbeck, K. (2018). How bacteria hack the matrix and dodge the bullets of immunity. Eur. Respir. Rev. 27:180018. doi: 10.1183/16000617.0018-2018
Pereira, P. R. M., Fernandes, L. G. V., de Souza, G. O., Vasconcellos, S. A., Heinemann, M. B., Romero, E. C., et al. (2017). Multifunctional and redundant roles of Leptospira interrogans proteins in bacterial-adhesion and fibrin clotting inhibition. Int. J. Med. Microbiol. 307, 297–310. doi: 10.1016/j.ijmm.2017.05.006
Picardeau, M. (2008). Conjugative transfer between Escherichia coli and Leptospira spp. as a new genetic tool. Appl. Environ. Microbiol. 74, 319–322. doi: 10.1128/AEM.02172-07
Picardeau, M., Bulach, D. M., Bouchier, C., Zuerner, R. L., Zidane, N., Wilson, P. J., et al. (2008). Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One 3:e1607. doi: 10.1371/journal.pone.0001607
Pinne, M., Choy, H. A., and Haake, D. A. (2010). The OmpL37 surface-exposed protein is expressed by pathogenic Leptospira during infection and binds skin and vascular elastin. PLoS Negl. Trop. Dis. 4:e815. doi: 10.1371/journal.pntd.0000815
Pinne, M., and Haake, D. A. (2009). A comprehensive approach to identification of surface-exposed, outer membrane-spanning proteins of Leptospira interrogans. PLoS One 4:e6071. doi: 10.1371/journal.pone.0006071
Pinne, M., Matsunaga, J., and Haake, D. A. (2012). Leptospiral outer membrane protein microarray, a novel approach to identification of host ligand-binding proteins. J. Bacteriol. 194, 6074–6087. doi: 10.1128/JB.01119-12
Pizarro-Cerda, J., and Cossart, P. (2006). Bacterial adhesion and entry into host cells. Cell 124, 715–727. doi: 10.1016/j.cell.2006.02.012
Ptak, C. P., Hsieh, C. L., Lin, Y. P., Maltsev, A. S., Raman, R., Sharma, Y., et al. (2014). NMR solution structure of the terminal immunoglobulin-like domain from the Leptospira host-interacting outer membrane protein, LigB. Biochemistry 53, 5249–5260. doi: 10.1021/bi500669u
Rajaonarivelo, J. A., Desmoulin, A., Maillard, O., Collet, L., Baudino, F., Jaffar-Bandjee, M. C., et al. (2023). Clinical manifestations of human leptospirosis: bacteria matter. Front. Cell. Infect. Microbiol. 13:1259599. doi: 10.3389/fcimb.2023.1259599
Ramos, A. C., Souza, G. N., and Lilenbaum, W. (2006). Influence of leptospirosis on reproductive performance of sows in Brazil. Theriogenology 66, 1021–1025. doi: 10.1016/j.theriogenology.2005.08.028
Ren, S. X., Fu, G., Jiang, X. G., Zeng, R., Miao, Y. G., Xu, H., et al. (2003). Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422, 888–893. doi: 10.1038/nature01597
Rossini, A. D., Teixeira, A. F., Souza Filho, A., Souza, G. O., Vasconcellos, S. A., Heinemann, M. B., et al. (2020). Identification of a novel protein in the genome sequences of Leptospira interrogans with the ability to interact with host's components. J. Microbiol. Immunol. Infect. 53, 163–175. doi: 10.1016/j.jmii.2018.12.012
Santos, J. C., Handa, S., Fernandes, L. G. V., Bleicher, L., Gandin, C. A., de Oliveira-Neto, M., et al. (2023). Structural and biochemical characterization of Leptospira interrogans Lsa45 reveals a penicillin-binding protein with esterase activity. Process Biochem. 125, 141–153. doi: 10.1016/j.procbio.2022.12.010
Santos, J. V., Pereira, P. R. M., Fernandes, L. G. V., Siqueira, G. H., de Souza, G. O., Souza Filho, A., et al. (2018). Binding of human plasminogen by the lipoprotein LipL46 of Leptospira interrogans. Mol. Cell. Probes 37, 12–21. doi: 10.1016/j.mcp.2017.10.004
Sarma, A., Dhandapani, G., Phukan, H., Bhunia, P. K., De, A. K., Bhattacharya, D., et al. (2023). Leptospiral cell wall hydrolase (LIC_10271) binding peptidoglycan, lipopolysaccharide, and laminin and the protein show LysM and M23 domains are co-existing in pathogenic species. Res. Microbiol. 174:104107. doi: 10.1016/j.resmic.2023.104107
Schwartz, I. (2000). Microbial genomics: from sequence to function. Emerg. Infect. Dis. 6, 493–495. doi: 10.3201/eid0605.000508
Setubal, J. C., Reis, M., Matsunaga, J., and Haake, D. A. (2006). Lipoprotein computational prediction in spirochaetal genomes. Microbiology 152, 113–121. doi: 10.1099/mic.0.28317-0
Silva, L. P., Fernandes, L. G., Vieira, M. L., de Souza, G. O., Heinemann, M. B., Vasconcellos, S. A., et al. (2016). Evaluation of two novel leptospiral proteins for their interaction with human host components. Pathog. Dis. 74:ftw040. doi: 10.1093/femspd/ftw040
Silva, E. F., Medeiros, M. A., McBride, A. J., Matsunaga, J., Esteves, G. S., Ramos, J. G., et al. (2007). The terminal portion of leptospiral immunoglobulin-like protein LigA confers protective immunity against lethal infection in the hamster model of leptospirosis. Vaccine 25, 6277–6286. doi: 10.1016/j.vaccine.2007.05.053
Siqueira, G. H., Atzingen, M. V., Alves, I. J., de Morais, Z. M., Vasconcellos, S. A., and Nascimento, A. L. (2013). Characterization of three novel adhesins of Leptospira interrogans. Am. J. Trop. Med. Hyg. 89, 1103–1116. doi: 10.4269/ajtmh.13-0205
Siqueira, G. H., Atzingen, M. V., de Souza, G. O., Vasconcellos, S. A., and Nascimento, A. (2016). Leptospira interrogans Lsa23 protein recruits plasminogen, factor H and C4BP from normal human serum and mediates C3b and C4b degradation. Microbiology 162, 295–308. doi: 10.1099/mic.0.000217
Siqueira, G. H., de Souza, G. O., Heinemann, M. B., Vasconcellos, S. A., and Nascimento, A. (2017). The role of Lsa23 to mediate the interaction of Leptospira interrogans with the terminal complement components pathway. Microb. Pathog. 112, 182–189. doi: 10.1016/j.micpath.2017.09.058
Slack, A. T., Symonds, M. L., Dohnt, M. F., and Smythe, L. D. (2006). The epidemiology of leptospirosis and the emergence of Leptospira borgpetersenii serovar Arborea in Queensland, Australia, 1998-2004. Epidemiol. Infect. 134, 1217–1225. doi: 10.1017/S0950268806006352
Smith, G. P. (1985). Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317. doi: 10.1126/science.4001944
Souza, N. M., Vieira, M. L., Alves, I. J., de Morais, Z. M., Vasconcellos, S. A., and Nascimento, A. L. (2012). Lsa30, a novel adhesin of Leptospira interrogans binds human plasminogen and the complement regulator C4bp. Microb. Pathog. 53, 125–134. doi: 10.1016/j.micpath.2012.06.001
Stevenson, B., Choy, H. A., Pinne, M., Rotondi, M. L., Miller, M. C., Demoll, E., et al. (2007). Leptospira interrogans endostatin-like outer membrane proteins bind host fibronectin, laminin and regulators of complement. PLoS One 2:e1188. doi: 10.1371/journal.pone.0001188
Sun, Z., Bao, L., Li, D., Huang, B., and Wu, B. (2010). Effect of Leptospira interrogans outer membrane proteins LipL32 on HUVEC. Microb. Pathog. 49, 116–121. doi: 10.1016/j.micpath.2010.05.006
Surdel, M. C., Anderson, P. N., Hahn, B. L., and Coburn, J. (2022a). Hematogenous dissemination of pathogenic and non-pathogenic Leptospira in a short-term murine model of infection. Front. Cell. Infect. Microbiol. 12:917962. doi: 10.3389/fcimb.2022.917962
Surdel, M. C., Hahn, B. L., Anderson, P. N., and Coburn, J. (2022b). Heterologous production of the adhesin LIC13411 from pathogenic Leptospira facilitates binding of non-pathogenic Leptospira in vitro and in vivo. Front. Cell. Infect. Microbiol. 12:917963. doi: 10.3389/fcimb.2022.917963
Svensson, M., Lindstedt, R., Radin, N. S., and Svanborg, C. (1994). Epithelial glucosphingolipid expression as a determinant of bacterial adherence and cytokine production. Infect. Immun. 62, 4404–4410. doi: 10.1128/iai.62.10.4404-4410.1994
Svensson, M., Platt, F. M., and Svanborg, C. (2006). Glycolipid receptor depletion as an approach to specific antimicrobial therapy. FEMS Microbiol. Lett. 258, 1–8. doi: 10.1111/j.1574-6968.2006.00175.x
Takahashi, M. B., Teixeira, A. F., and Nascimento, A. (2021). The leptospiral LipL21 and LipL41 proteins exhibit a broad spectrum of interactions with host cell components. Virulence 12, 2798–2813. doi: 10.1080/21505594.2021.1993427
Takahashi, M. B., Teixeira, A. F., and Nascimento, A. (2022). Host cell binding mediated by Leptospira interrogans adhesins. Int. J. Mol. Sci. 23:15550. doi: 10.3390/ijms232415550
Tan, X., Castellanos, M., and Chaconas, G. (2023). Choreography of Lyme disease spirochete Adhesins to promote vascular escape. Microbiol. Spectr. 11:e0125423. doi: 10.1128/spectrum.01254-23
Ternent, L., Dyson, R. J., Krachler, A. M., and Jabbari, S. (2015). Bacterial fitness shapes the population dynamics of antibiotic-resistant and -susceptible bacteria in a model of combined antibiotic and anti-virulence treatment. J. Theor. Biol. 372, 1–11. doi: 10.1016/j.jtbi.2015.02.011
Thibeaux, R., Iraola, G., Ferrés, I., Bierque, E., Girault, D., Soupé-Gilbert, M. E., et al. (2018). Deciphering the unexplored Leptospira diversity from soils uncovers genomic evolution to virulence. Microb. Genom. 4:e000144. doi: 10.1099/mgen.0.000144
Toma, C., Murray, G. L., Nohara, T., Mizuyama, M., Koizumi, N., Adler, B., et al. (2014). Leptospiral outer membrane protein LMB216 is involved in enhancement of phagocytic uptake by macrophages. Cell. Microbiol. 16, 1366–1377. doi: 10.1111/cmi.12296
Torgerson, P. R., Hagan, J. E., Costa, F., Calcagno, J., Kane, M., Martinez-Silveira, M. S., et al. (2015). Global burden of leptospirosis: estimated in terms of disability adjusted life years. PLoS Negl. Trop. Dis. 9:e0004122. doi: 10.1371/journal.pntd.0004122
Tsuchimoto, M., Niikura, M., Ono, E., Kida, H., and Yanagawa, R. (1984). Leptospiral attachment to cultured cells. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 258, 268–274. doi: 10.1016/S0176-6724(84)80044-9
Tung, J. Y., Yang, C. W., Chou, S. W., Lin, C. C., and Sun, Y. J. (2010). Calcium binds to LipL32, a lipoprotein from pathogenic Leptospira, and modulates fibronectin binding. J. Biol. Chem. 285, 3245–3252. doi: 10.1074/jbc.M109.006320
Verma, A., Brissette, C. A., Bowman, A., and Stevenson, B. (2009). Borrelia burgdorferi BmpA is a laminin-binding protein. Infect. Immun. 77, 4940–4946. doi: 10.1128/IAI.01420-08
Vieira, M. L., Atzingen, M. V., Oliveira, T. R., Oliveira, R., Andrade, D. M., Vasconcellos, S. A., et al. (2010a). In vitro identification of novel plasminogen-binding receptors of the pathogen Leptospira interrogans. PLoS One 5:e11259. doi: 10.1371/journal.pone.0011259
Vieira, M. L., D'Atri, L. P., Schattner, M., Habarta, A. M., Barbosa, A. S., de Morais, Z. M., et al. (2007). A novel leptospiral protein increases ICAM-1 and E-selectin expression in human umbilical vein endothelial cells. FEMS Microbiol. Lett. 276, 172–180. doi: 10.1111/j.1574-6968.2007.00924.x
Vieira, M. L., de Morais, Z. M., Goncales, A. P., Romero, E. C., Vasconcellos, S. A., and Nascimento, A. L. (2010b). Lsa63, a newly identified surface protein of Leptospira interrogans binds laminin and collagen IV. J. Infect. 60, 52–64. doi: 10.1016/j.jinf.2009.10.047
Vincent, A. T., Schiettekatte, O., Goarant, C., Neela, V. K., Bernet, E., Thibeaux, R., et al. (2019). Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 13:e0007270. doi: 10.1371/journal.pntd.0007270
Vivian, J. P., Beddoe, T., McAlister, A. D., Wilce, M. C., Zaker-Tabrizi, L., Troy, S., et al. (2009). Crystal structure of LipL32, the most abundant surface protein of pathogenic Leptospira spp. J. Mol. Biol. 387, 1229–1238. doi: 10.1016/j.jmb.2009.02.038
Weil, A. (1886). Ueber eine eigenthumliche, mit Milztumor, Icterus und Nephritis einhergehende acute Infectionskrankheit. Dtsch. Arch. Klin. Med. 39, 209–232.
Yang, X., Lin, Y. P., Heselpoth, R. D., Buyuktanir, O., Qin, J., Kung, F., et al. (2016). Middle region of the Borrelia burgdorferi surface-located protein 1 (Lmp1) interacts with host chondroitin-6-sulfate and independently facilitates infection. Cell. Microbiol. 18, 97–110. doi: 10.1111/cmi.12487
Zhang, L., Zhang, C., Ojcius, D. M., Sun, D., Zhao, J., Lin, X., et al. (2012). The mammalian cell entry (Mce) protein of pathogenic Leptospira species is responsible for RGD motif-dependent infection of cells and animals. Mol. Microbiol. 83, 1006–1023. doi: 10.1111/j.1365-2958.2012.07985.x
Keywords: Leptospira, leptospirosis, adhesion, adhesins, binding, spirochete
Citation: Surdel MC and Coburn J (2024) Leptospiral adhesins: from identification to future perspectives. Front. Microbiol. 15:1458655. doi: 10.3389/fmicb.2024.1458655
Edited by:
Christopher J. Pappas, Manhattanville College, United StatesReviewed by:
Angela Silva Barbosa, Butantan Institute, BrazilCopyright © 2024 Surdel and Coburn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthew C. Surdel, bXN1cmRlbEBtY3cuZWR1
 Matthew C. Surdel
Matthew C. Surdel Jenifer Coburn
Jenifer Coburn