- 1Chiro National Sorghum Research and Training Center, Ethiopian Institute of Agricultural Research, Addis Ababa, Ethiopia
- 2Institute of Biotechnology, Addis Ababa University, Addis Ababa, Ethiopia
- 3Bio and Emerging Technology Institute, Addis Ababa, Ethiopia
- 4Department of Botany and Plant Pathology, Purdue University, West Lafayette, IN, United States
- 5College of Agriculture, Hawassa University, Hawassa, Ethiopia
- 6Department of Plant Breeding, Swedish University of Agricultural Sciences (SLU), Alnarp, Sweden
Introduction: Sorghum anthracnose, caused by Colletotrichum sublineola, is the most destructive disease of sorghum, which causes up to 80% grain yield loss in susceptible varieties. The use of resistance varieties is an effective, durable, and eco-friendly strategy for anthracnose control. Knowledge of the phenotypic and genetic variation in C. sublineola is vital for designing appropriate anthracnose management strategies.
Methods: The present study examined the morphology and virulence of 25 C. sublineola isolates recovered from various sorghum-producing regions of Ethiopia against 18 known sorghum anthracnose differentials, 6 Ethiopian sorghum landraces, and a variety of Bonsa.
Results: Analysis of variance (ANOVA) revealed significant differences among sorghum genotypes, C. sublineola isolates, and their interactions. There was a significant difference between the isolates in virulence, with each isolate exhibiting virulence in 8–72% of the sorghum genotypes tested. Among the 25 tested isolates, the top four most virulent isolates were from Pawe, suggesting that this area is suitable for pathogen diversity studies and host plant resistance screening. The sorghum genotypes IS_18760, Brandes, and Bonsa showed resistance to all tested isolates. Consequently, they may provide potential sources of resistance genes for sorghum breeding programs to develop cultivars resistant to different C. sublineola pathotypes. However, the resistant check SC748-5 was susceptible to isolates NK73_F37, while another resistant check SC112-14 was susceptible to isolates PW123_F47 and PW122_F47. Cluster analysis grouped 22 isolates into seven clusters based on their morphological characters, whereas 24 pathotypes were identified among 25 isolates that were tested on 25 sorghum genotypes.
Discussion: Hence, this study revealed high variation in C. sublineola in Ethiopia suggesting the need for broad-spectrum resistance to control the disease. Sorghum genotypes resistant to various C. sublineola isolates were identified in this study, which can be used in sorghum breeding programs aiming to develop resistant cultivars to anthracnose. Highly virulent C. sublineola isolates were also identified which could be used in sorghum germplasm resistance screening. The report is the first to show the existence of C. sublineola pathotypes in Ethiopia.
Introduction
Sorghum (Sorghum bicolor (L.) Moench) is among the major cereal crops grown in several regions of the world due to its versatility and adaptability to diverse climatic conditions (Charles et al., 2009). The crop performs well at altitudes ranging from 500 to 1700 m above sea levels (masl) with seasonal rainfall of 300 mm and above. More than 500 million people in over 30 countries depend on sorghum, making it the world’s fifth most important crop for human consumption after rice, wheat, maize, and potatoes (Acquaah, 2012; Martin, 2016; Bunker et al., 2019; Ahn, 2020). It has diverse uses worldwide with all plant parts used for different purposes (Charles et al., 2009). The global production of sorghum increased from 55 million metric tons (MMT) in 2000 to 62.12 MMT in 2021 (FAOSTAT, 2023). Sorghum is one of Ethiopia’s major staple and strategic food security crops (Bezabeh, 2015) ranking third after teff and maize in area coverage and fourth after maize, teff, and wheat in total production (ESS, 2022). A total of 4.45 MMT of sorghum is estimated to be produced on 1.65 million hectares of land in the country (ESS, 2022). However, the productivity of sorghum in Ethiopia is at 2.6 tons per hectare (tha−1) (ESS, 2022), which is below the average grain yield in the United States, China, and the European Union, which is 4.4, 4.76, and 5.l tha−1, respectively (USDA, 2021).
Drought, diseases, low soil fertility (nutrient deficiencies), insects (stem borer and weevil), quelea birds, striga, and weeds have been recognized as major sorghum production constraints across the world (Charles et al., 2009; Girma et al., 2020). Of these, anthracnose caused by the fungus Colletotrichum sublineola is an important disease causing a substantial loss in grain, forage, and stover yields in the hot humid regions of the world (Thakur et al., 2007; Chala et al., 2010). Anthracnose severity of up to 87% was reported in Ethiopia (Tsedaley et al., 2016). Under maximum disease severity, grain loss could reach 86% in Brazil (Cota et al., 2017) and 80% in the USA (Prom et al., 2023).
Despite the economic significance of the disease in Ethiopia, information on sorghum C. sublineola pathotypes and virulence patterns is highly limited. Knowledge of the pathogen virulence variability is essential for deploying durable and effective management strategies. The identification and deployment of resistance genes into improved varieties susceptible to C. sublineola is an effective approach to managing sorghum anthracnose (Prom et al., 2012). Anthracnose resistance genes are believed to be widespread due to the high diversity of both the crop and the pathogen in their primary centers of origin, Ethiopia and Sudan (Thakur et al., 2007; Chala et al., 2010) including some genes that provide broad-spectrum resistance (Cuevas et al., 2019). Furthermore, previous studies have hinted at the potential existence of resistant sorghum landraces in Ethiopia that could be utilized for the development of sorghum cultivars resistant to anthracnose (Cuevas et al., 2019; Prom et al., 2022). Hence, investigating the distribution of C. sublineola pathotypes and identifying resistance sources could greatly help to address the challenge of sorghum production due to anthracnose in Ethiopia and beyond. This study is, therefore, primarily aimed at determining the virulence variability of 25 C. sublineola isolates collected from major sorghum growing areas in Ethiopia. Additionally, the study aimed at identifying sorghum genotypes resistant to anthracnose for use in sorghum breeding programs.
Materials and methods
Sample collection and isolation
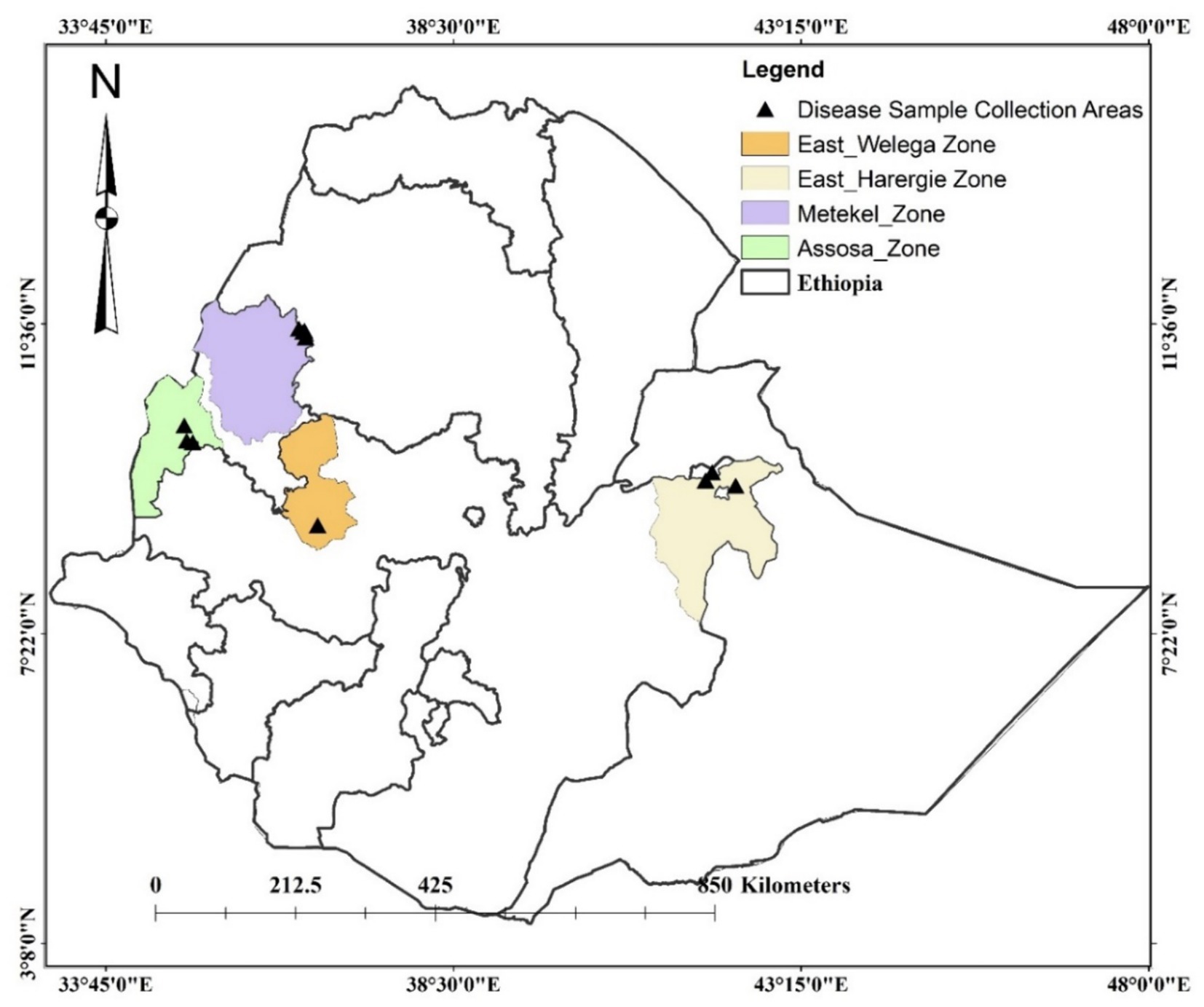
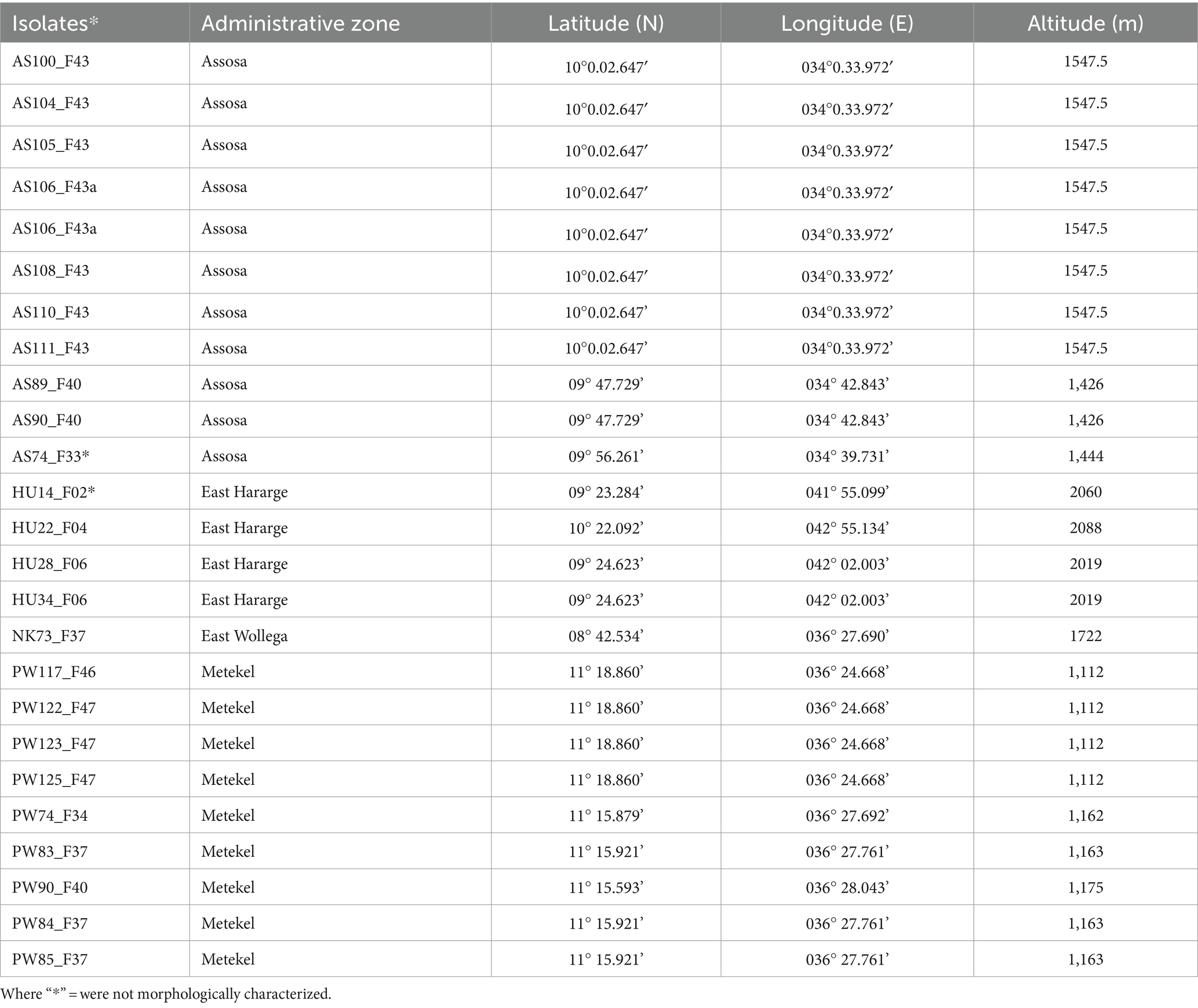
Sorghum leaf samples, with typical symptoms of anthracnose, were collected from Assosa, Haramaya, Nekamte, and Pawe districts in Ethiopia (Figure 1). These locations are categorized by hot and humid weather conditions, which are conducive to pathogen infection and proliferation. Assosa and Pawe districts are in the Assosa and Metekel zones, respectively, of the Benishangul-Gumuz Regional State. The sampling site in Assosa and Pawe had an altitude range of 1,426–1547.5 masl and 1,112–1,175 masl, respectively (Table 1). Assosa’s mean annual temperature and rainfall were 22.5°C and 1912.77 mm, respectively, whereas Pawe’s were 24.7°C and 1,254 mm. Haramaya and Nekamte districts are situated in East Hararge and East Wollega zones, respectively, of Oromia Regional State. The sampling sites in Haramaya had an altitude range of 2019–2088 masl, while the sampling site in Nekamte had an altitude of 1722 masl (Table 1). Harmaya’s mean annual temperature and rainfall were 20.8°C and 567.17 mm, respectively, while Nekamte’s were 21.44°C and 1306.16 mm.
A total of 372 diseased leaf samples of sorghum were collected during the 2020 and 2022 cropping seasons from 142 sorghum fields. The sampled sorghum fields in a district were at least 5 km apart. Sample collection was made at crop growth stages (GS) from complete anthesis to the milking stage. Sampling was done at one to three random points within a farmer’s field. In the Assosa, Haramaya, and Pawe research stations, 10–15 samples were collected because of the plants’ high phenotypic diversity and their diverse disease symptoms. A paper bag labeled with a sample code was used to collect anthracnose symptomatic leaves of each sorghum genotype and transport them to the plant pathology laboratory of Melkassa Agricultural Research Center (MARC) of the Ethiopian Institute of Agricultural Research (EIAR) for analysis. Table 1 presents sample passport data, such as administrative regions, geographical coordinates, and altitudes. For pathogen isolation, infected leaf samples were surface sterilized with 1% sodium hypochlorite for 30 s followed by three times rinsing with sterilized distilled water. Sterilized leaves were allowed to dry in the laminar flow cabinet and transferred to 9 cm Petri dishes with potato dextrose agar (PDA; potato 200 g/L, dextrose 20 g/L, agar 15 g/L) (Sigma-Aldrich-70139) amended with 100 mg of chloramphenicol (Sigma-Aldrich-C0378) and incubated at 26°C for 5–7 days under 12 h light period. Single spore isolates were obtained from a spread of conidial suspension on water agar. For this, a diluted spore suspension was transferred to water agar and incubated at 26°C for 16 h. Then a germinating and unbranched single spore was transferred to PDA for further analysis.
Morphological characterization
A total of 114 pathogen isolates were morphologically characterized using an OLYMPUS-SC50 digital compound microscope. The morphological characterization of the isolates was conducted on the 7th day of incubation except for colony size which was measured for four consecutive days at a 24 h interval starting at 48 h of incubation. Data on conidia, appressoria, acervuli, and setae were recorded as described by Crouch et al. (2009). For morphological characterization, a sample was taken 1 cm from the center of the colony and smeared in a drop of water. The conidia populations were determined by counting the number of conidia in suspension under a compound microscope with a 40x objective lens and were categorized into four (3 = high, when the number of conidia was more than 300; 2 = medium, when the number of conidia was 100–300; 1 = low, when the number of conidia was 1–100; and 0 = none, when there were no conidia) (Figure 2). The conidia length and width were recorded from the mean of three conidia, while the maximum number of setae was counted. Among the 114 isolates, 22 isolates representing the four administrative zones (9, 3, 1, and 9 from Assosa, East Hararge, East Wollega, and Metekel, respectively) were selected based on geographical origin and morphological characters for virulence variability analysis. In addition, three morphologically uncharacterized isolates (two from Assosa and one from East Hararge) were added for the virulence analysis.
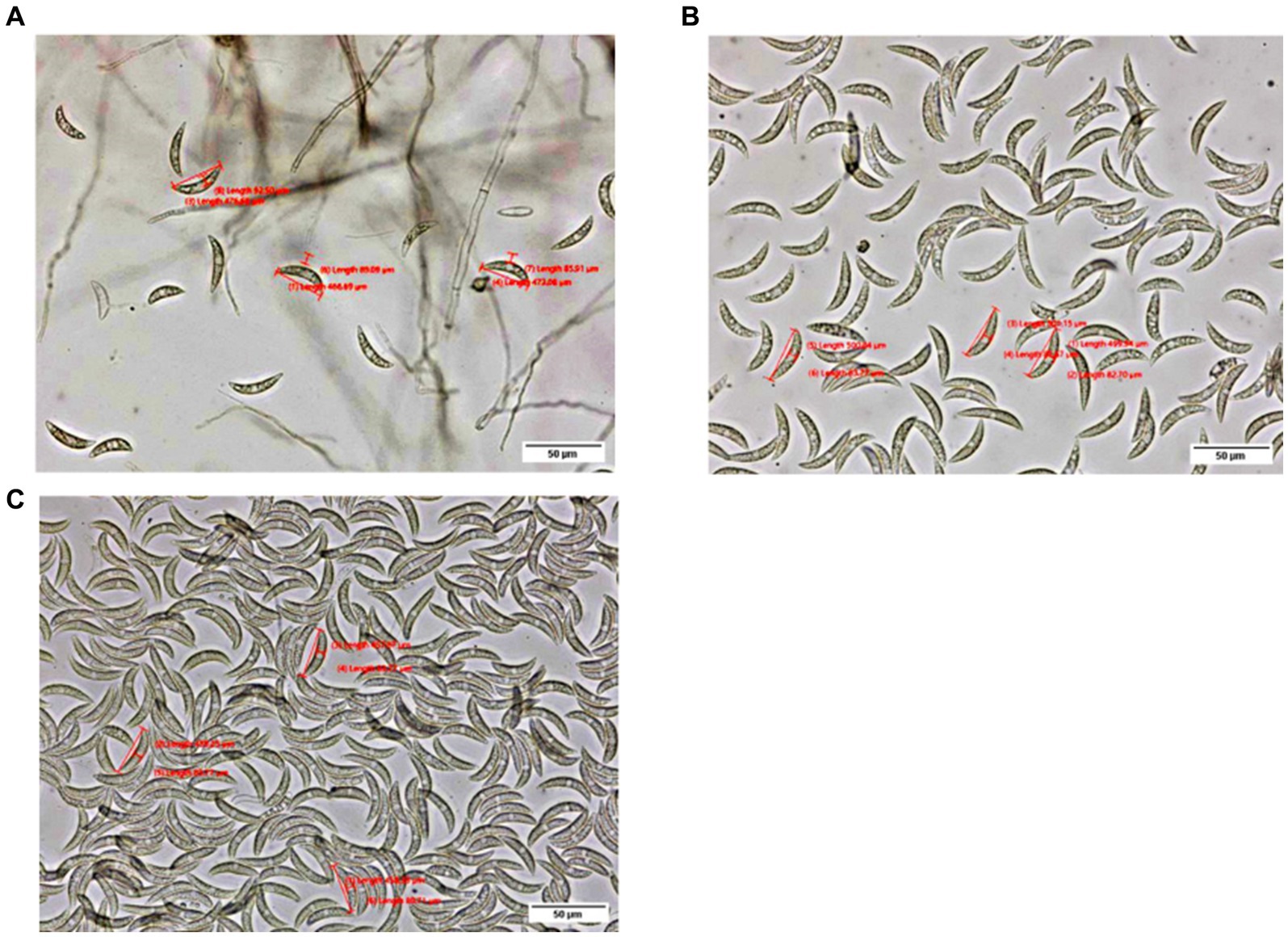
Figure 2. Pictures depict low (A), medium (B), and high (C) conidia populations, as observed under an OLYMPUS-SC50 digital compound microscope at a 40× objective lens.
Plant materials, inoculation, and disease assessment
Plant materials
In total, 25 sorghum genotypes were used for virulence analysis of the pathogen isolates and resistance analysis of the genotypes against the isolates at the Swedish University of Agricultural Sciences (SLU), Department of Plant Breeding, Alnarp, Sweden. A total of 18 genotypes were sorghum differentials provided by Professor Tesfaye D. Mengiste (Purdue University, Department of Botany and Plant Pathology, United States), while 7 genotypes (six landraces and one variety, Bonsa) were obtained from the Ethiopian Institute of Agricultural Research, MARC, Ethiopia. The sorghum genotypes SC748-5 and BT × 623 were included as resistant and susceptible checks, respectively. In addition to the improved variety Bonsa, two landrace genotypes (TSL_100090 and ETSL_100267) were regarded as resistant, while two landrace genotypes (ETSL_100039 and ETSL_101249) and two landrace genotypes (ETSL_101388 and ETSL_100761) were regarded as moderately resistant and susceptible, respectively, based on data obtained from field trials conducted at four locations in Ethiopia during the 2022 cropping season (data not shown).
Experimental design and inoculation
In this study, detached leaf assay was used as it has been reported to give comparable results to direct inoculation of the whole plant under greenhouse conditions (Prom et al., 2016). For this, sorghum genotypes were planted in 3.5 L volume plastic pots filled with potting mix at SLU’s greenhouse in Alnarp. Forty days after planting, the sixth leaves of each seedling were cut into 5–6 cm long segments and transferred to a 9 cm Petri dish containing double filter papers for detached leaf assay. A split-plot design, with 25 sorghum genotypes as main plots and 25 fungal isolates as subplots, was used. In each Petri dish, three leaf segments from each plant were used. To fix the leaf segment to the filter papers and prevent drying, 5 mL sterilized distilled water was added to the filter papers, and leaf margins were covered with sterile tissue paper. A hemacytometer was used to adjust the spore concentration from 8 to 10 days-old culture to 1 × 106 spores/ml. Each leaf segment was inoculated at two locations with a drop of 30 μL conidia suspension. In total, 625 Petri dishes and 1875 leaf segments were used in this experiment.
Disease assessment and data collection
A 1 to 5 scoring scale modified from (Cuevas et al., 2021) was used to assess the infection level, where 1 = no disease symptom; 2 = red spots on the inoculated area but no acervuli formation hypersensitive reaction (HR) (1–10%); 3 = red lesions on the inoculated area and few spots with acervuli but not expanding (11–25%); 4 = red lesions and visible expanded acervuli spots on the inoculated area (26–50%), and 5 = abundant acervuli covered lesions beyond the inoculated area with dead tissues in the middle (more than 50%). The infection progress was photographed with a fixed Canon digital camera at 60, 84, 108, 132, 156, and 180 h post-inoculation (hpi) followed by image analysis. The genotype’s reaction scores (pathogen infection level) 1 and 2 are categorized as resistant, while scores 3–5 are categorized as susceptible. Pathogen re-isolation was conducted from 20 randomly selected infected leaf segments to confirm that the observed infection was due to the inoculated pathogen. For this purpose, infected leaf segments were assessed following the same method described earlier for pathogen isolation.
Determining hyphal growth of Colletotrichum sublineola isolates
Re-inoculation of the pathogen to new leaf segments was carried out to determine the structural growth of fungal hyphae and to evaluate the response of resistant and susceptible sorghum genotypes. For this, three pathogen isolates (AS106_F43a, NK73_F37, and PW123_F47) and three sorghum genotypes (BT × 623, SC748-5, and ETSL_1001249) were selected based on their reaction in the detached leaf assay. The analysis was conducted to determine the host–pathogen interaction and the structural growth of the fungal pathogen within the leaf. The assessment was conducted on the 7th day of inoculation. As a procedure, infected tissue was immersed in 100% ethanol for 2 days followed by 10% KOH for 2 h at 85°C to facilitate bleaching and complete chlorophyll removal. Subsequently, WGA-AF488 (green) was used to stain the fungal hyphae, while propidium iodide (red) was employed to stain dead host cells, providing a contrasting view as described in Redkar et al. (2018). The fungal hyphae and plant cell structure images were taken under a confocal microscope.
Data analysis
The morphological and disease severity data were subjected to ANOVA using PROC GLM (SAS version 9.4). Euclidean distance-based cluster analysis was used to determine the clustering pattern of the pathogen isolates based on the morphological data in the R package “hclust” (Savje, 2024). The Elbow method was used to determine the optimal number of clusters. The Corrplot package in R software was used to determine the correlation between the morphological traits of the pathogen isolates. ImageJ software was used to take images under a confocal microscope (Redkar et al., 2018).
Results
Morphological diversity of Colletotrichum sublineola isolates
Statistical analysis revealed significant differences (p < 0.05) among the C. Sublineola isolates in their conidia length, conidia width, colony size, and number of setae (Table 2). All the isolates had falcate conidia except PW84_F37, which had a mixture of falcate and flat conidia. The longest conidia (26.25 μm) was observed in the isolate PW90_F40, followed by PW74_F34 (26.13 μm), while the shortest conidia were found in isolates AS110_F43 (16.93 μm) and PW84_F37 (17.78 μm) (Table 2). The widest conidia (5.3 μm) was recorded in an isolate from Assosa (AS105_F43) followed by two isolates from Pawe (PW83_F37 = 5.28 μm and PW122_F47 = 5.1 μm). After 7 days of incubation, the largest colony (6.82 cm) was exhibited by isolates NK73_F37 and PW83_F37, whereas the smallest colony (2.2 cm) was recorded for isolate PW84_F37. Colony color was varied in all the isolates ranging from dirty white to gray and dark green. In general, isolates from Assosa and Nekamte showed a gray colony color at the center and dirty white at the margin, whereas isolates from Pawe showed dark green, light green, and salmon gray. However, all three isolates from Haramaya showed a dark green colony color. The maximum number of setae were recorded at Assosa from four isolates (AS100_F43 = 42.33, AS108_F43 = 39, AS90_F40 = 32, AS111_F43 = 21.67) (Table 2).
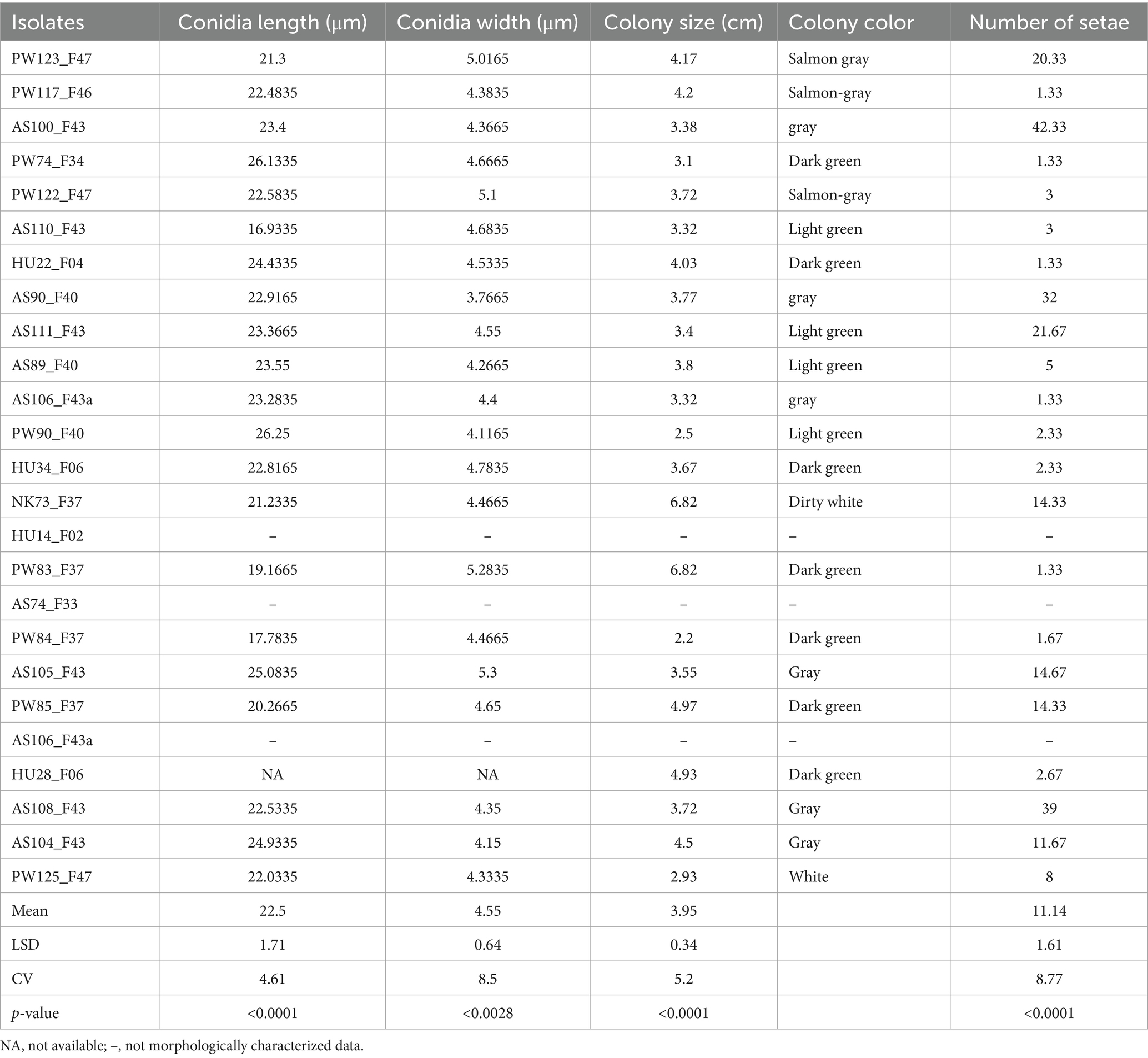
Table 2. Morphological description and variation of 22 C. sublineola isolates characterized in this study.
Clustering of Colletotrichum sublineola isolates
Euclidian distance-based cluster analysis was employed to visualize the relationships between the isolates (Figure 3). The analysis categorized the isolates into seven distinct morphological groups with low levels of admixture. Isolates from Assosa were predominantly grouped in clusters I and VI, while isolates from Pawe were distributed across clusters I, II, IV, V, and VI. Cluster III consisted of isolate HU28_F06 obtained from Haramaya while isolating HU34_F06 from the same area was assigned to cluster VII. Cluster IV comprised isolates from Nekamte (NK73_F37) and Pawe (PW83_F37). Isolates from Pawe were distributed across five clusters (I, II, IV, V, and VI), indicating their morphological diversity (Figure 3). The clustering analysis demonstrated that the isolates showed significant morphological variation with some degree of association with their geographical origin.
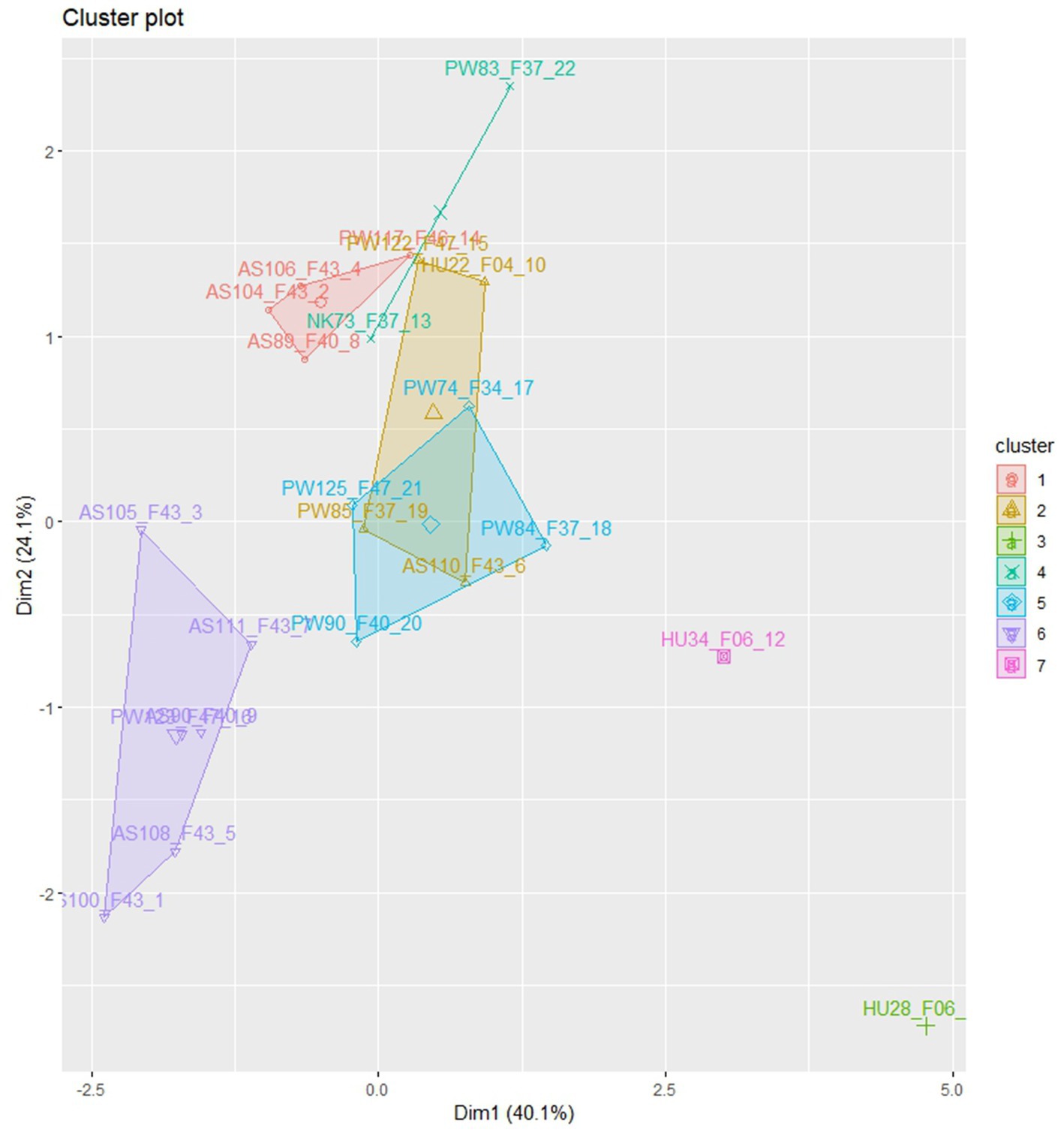
Figure 3. The clustering of 22 C. sublineola isolates in morphological variations, collected from four different administrative zones in Ethiopia.
Correlation analysis
Among the six morphological variables, the number of setae had a highly significant positive correlation with the acervuli population (p < 0.001) and a significant negative correlation with the appressoria population (p < 0.01) (Figure 4). Conidia width and length showed a highly significant positive correlation (r = 0.82, p < 0.001). However, they were not correlated with colony size, acervuli population, number of setae, and appressoria population. Acervuli population, number of setae, and conidia population showed significant positive correlations (r = 0.43–0.65) (Figure 4). In general, isolates with a high acervuli population had a high conidia population but were not correlated with the appressoria population.
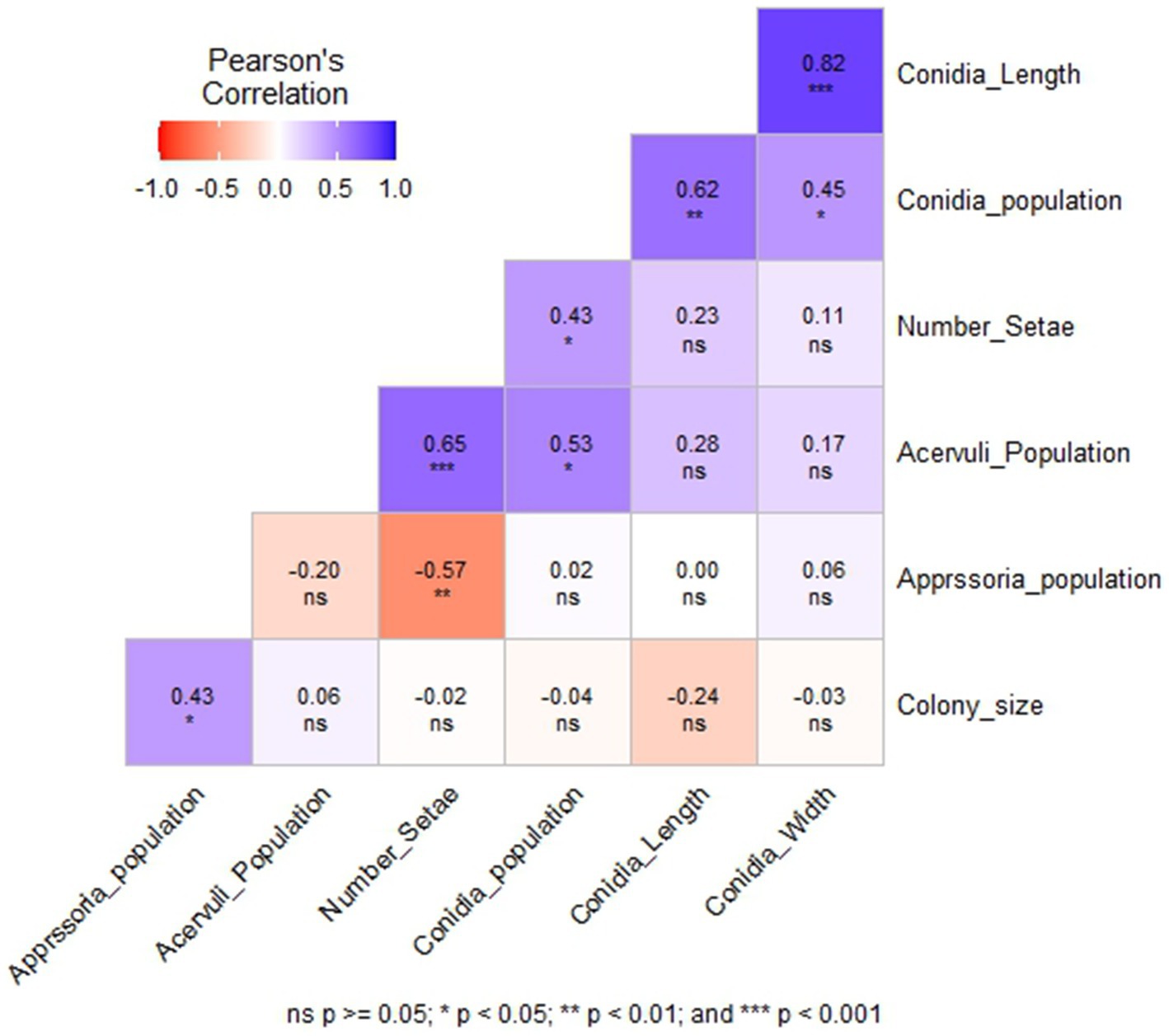
Figure 4. The correlation between six morphological variables of C. sublineola isolates collected from four administrative zones in Ethiopia.
Virulence level of Colletotrichum sublineola isolates and reaction of sorghum genotypes
The analysis of variance (ANOVA) revealed significant differences among sorghum genotypes, C. sublineola isolates, and their interactions (Table 3). Among the 25 tested isolates, the top 4 most virulent pathotypes were from Pawe (PW122_F47, PW117_F46, PW74_F34, and PW123_F47) followed by pathotypes from Assosa (Figure 5). Pathotype PW117_F46, which infected 18 (72%) of the sorghum genotypes, was the most virulent followed by pathotypes PW74_F34, PW122_F47, and PW123_F47, which infected 17 (68%) sorghum genotypes. Pathotype AS100_F43 infected 16 (64%) genotypes, including BT × 623 (Figure 5), whereas pathotypes PW125_F47, AS108_F43, and AS104_F43 were the least virulent, which infected only two, four, and five of the tested sorghum genotypes, respectively (Figure 5). Two pathotypes (HU28_F06 and HU34_F06) from Haramaya infected 28 and 32% of the sorghum genotypes tested, and hence, they are categorized as less virulent. Pathotypes HU14_F02 and HU22_F04 infected 52% of the sorghum genotypes tested; hence, their virulence is intermediate. Similarly, pathotype NK73_F37 collected from Nekamte showed an intermediate virulence level, even though it infected the sorghum genotype SC748-5, which was widely regarded as resistant to anthracnose. In total, 8 (AS100_F43, AS104_F43, AS105_F43, AS106_F43a, AS106_F43b, AS108_F43, AS110_F43, and AS111_F43) of the 11 isolates from Assosa were collected from the same field, and interestingly they exhibited different virulence levels. In addition, isolates isolated from the same leaf sample (AS106_F37a and AS106_F37b) showed different virulence pattern and colony color.
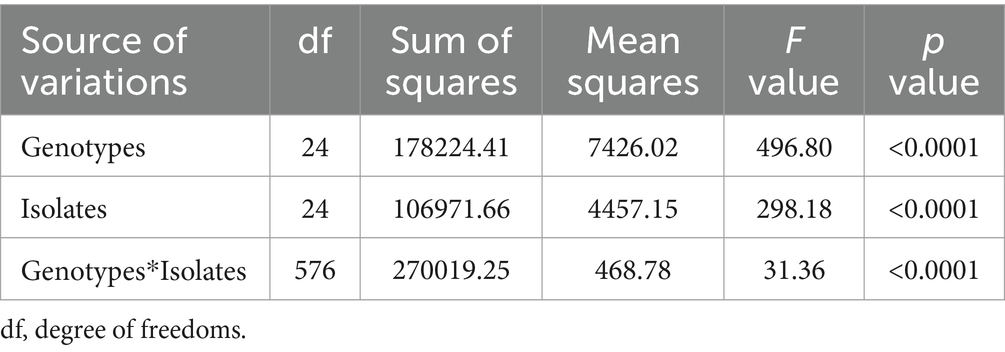
Table 3. ANOVA table with their F and p values for disease severity of 25 C. sublineola isolates on 25 sorghum genotypes.
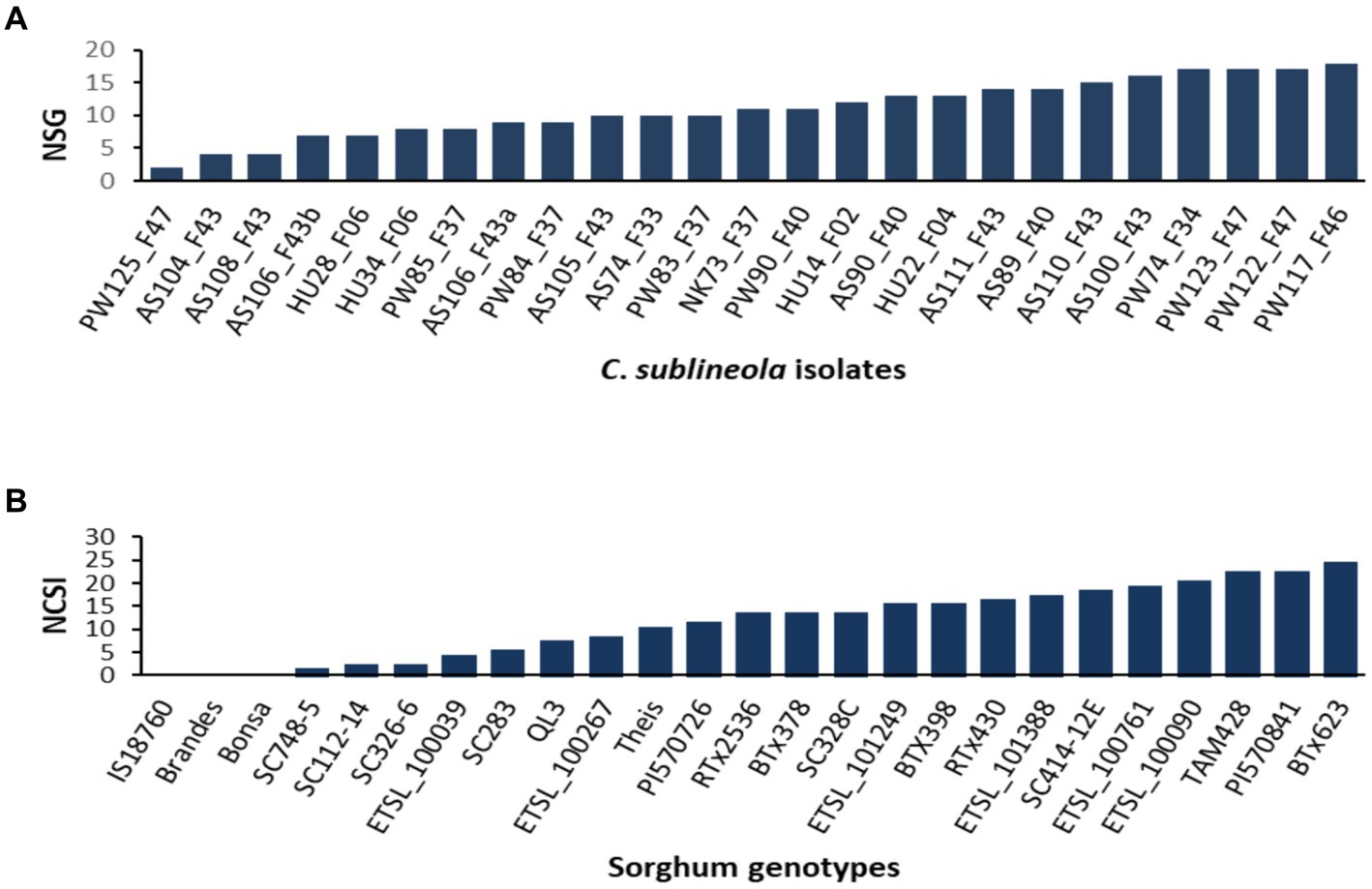
Figure 5. Bar graphs showing (A) the number of sorghum genotypes infected by each of the 25 C. sublineola isolates, and (B) the number of C. sublineola isolates that infected each of the 25 sorghum genotypes. NSG, number of sorghum genotypes; NCSI, number of C. sublineola isolates.
The ANOVA revealed significant differences (p < 0.05) between the C. sublineola isolates in anthracnose severity (Table 4). Among the tested isolates, PW123_F47 and PW117_F46 were the most aggressive, causing 31% anthracnose severity. AS100_F43 and PW74_F34 were the second-most aggressive pathotypes with 28% anthracnose severity (Table 4). C. sublineola Pathotypes vary considerably in their virulence and aggressiveness. For instance, AS106_F43a and AS106_F43b, which were isolated from the same leaf sample, showed differences in their virulence. Pathotype AS106_F43a was more aggressive than AS106_F43b, with severity scores of 17.87 and 8.87%, respectively. Moreover, AS106_F43a, which infected nine genotypes, was a bit more virulent than AS106_F43b, which infected seven genotypes. Pathotype PW125_F47 from Pawe was less virulent as it only infected two genotypes, BT × 623 and ETSL_100090. It was also the least aggressive pathotype with a 5.87% severity score.
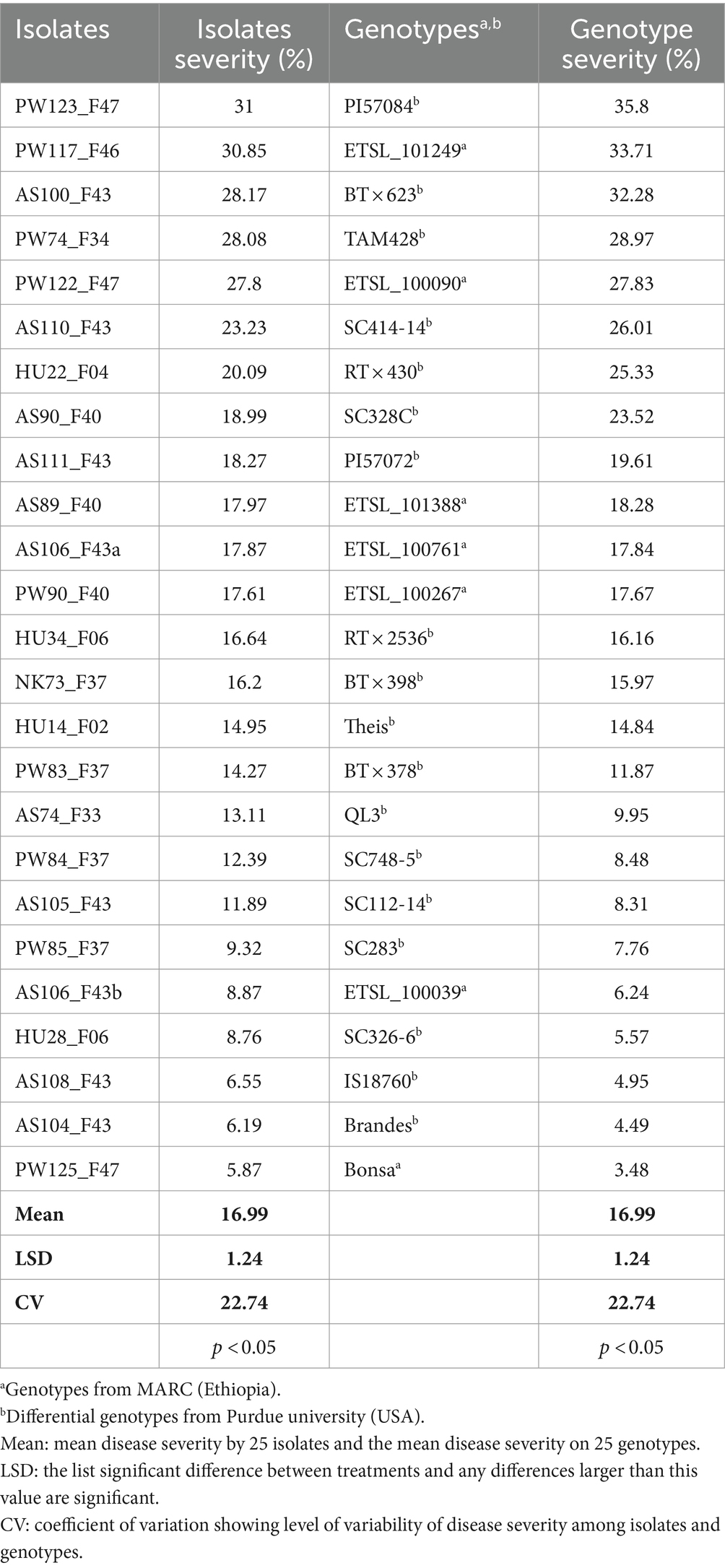
Table 4. Mean comparison of severity percentage (aggressiveness) of 25 C. sublineola isolates and severity level of 25 sorghum genotypes.
Generally, Ethiopian sorghum genotypes showed inconsistent reactions, except for the variety Bonsa, which is regarded as universally resistant, and genotype ETSL_100761, which was susceptible to 19 of the 25 isolates. Genotype PI57084 was found to be the most susceptible (disease severity = 35.8%) followed by the genotypes ETSL_101249 and BT × 623, which had severity scores of 33.71 and 32.28%, respectively (Table 4).
Virulence pattern and pathotype determination
The resistance screening revealed significantly different resistance levels among the tested sorghum genotypes. None of the C. sublineola isolates infected the sorghum genotypes IS_18760, Brandes, and Bonsa (Table 5). Conversely, genotype BT × 623 was susceptible to all isolates (Figure 6), except AS104_F43. Sorghum genotypes PI 570841 and TAM428 were susceptible to 22 (88%) of tested isolates. Genotype SC748-5 was susceptible to pathotype NK73_F37, while genotype SC112-14 was susceptible to pathotypes PW123_F47 and PW122_F47 (Table 5). Both SC748-5 and SC112-14 genotypes were previously reported as resistant genotypes (Prom et al., 2023). Among the Ethiopian genotypes, the variety Bonsa was the most resistant as it was not infected with any of the pathotypes. ETSL_100039 and ETSL_100267 were infected by four and eight pathotypes, respectively. ETSL_100090 was not infected and showed anthracnose symptoms under field conditions. However, it was susceptible to 80% of the pathotypes under greenhouse conditions and was the most susceptible among the Ethiopian sorghum genotypes. Genotype ETSL_100761 showed a susceptible reaction to most of the isolates in line with the field trial data. In general, the virulence pattern of 25 C. sublineola isolates tested against 25 sorghum genotypes revealed 24 pathotypes (Table 5).
Table 5. Pathotype determination and virulence pattern of 25 C. sublineola isolates on 25 sorghum genotypes.
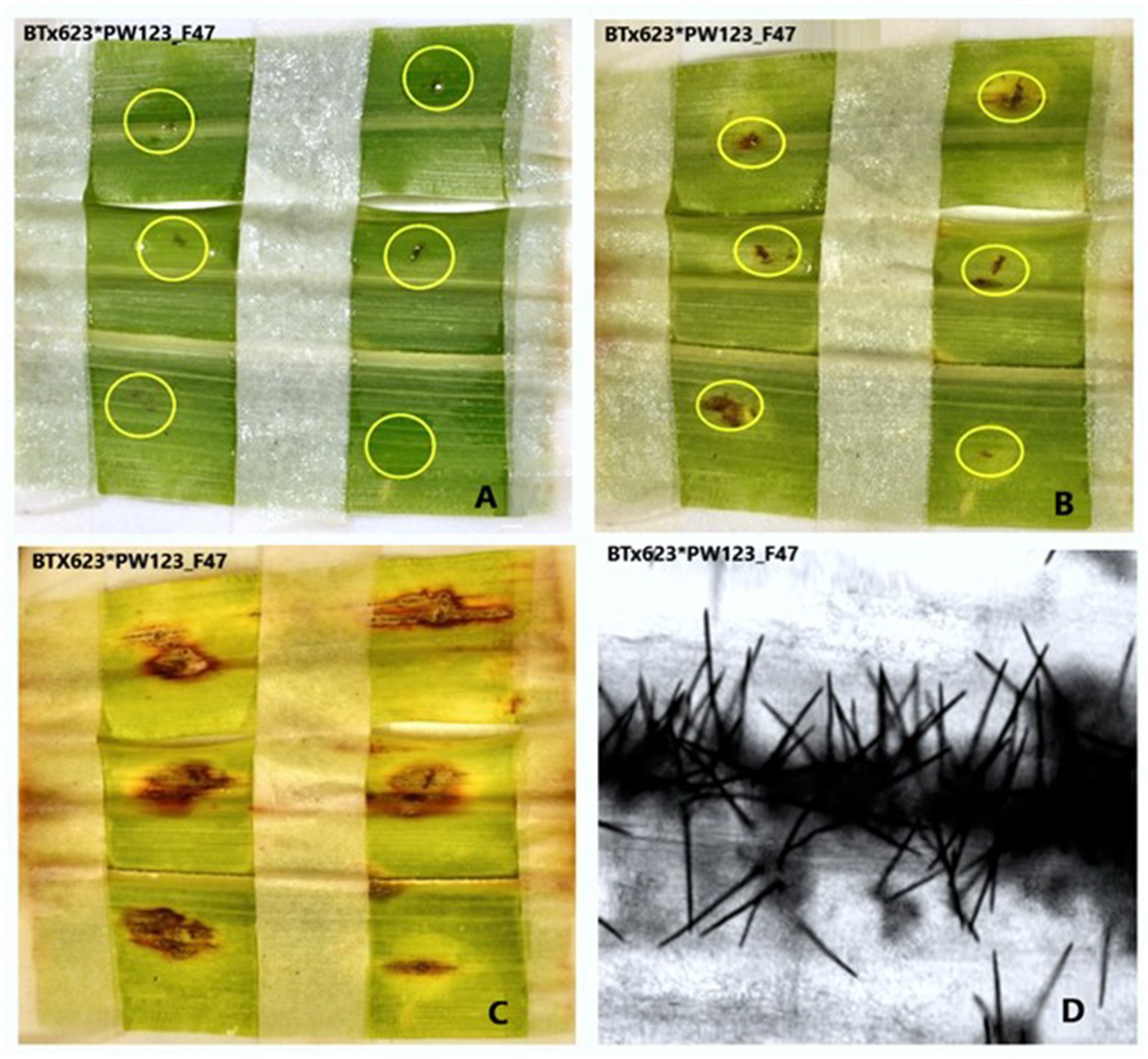
Figure 6. Pictures depicting the progress of anthracnose caused by C. sublineola isolate PW123_F47 to sorghum genotype BT × 623. (A) Second dpi, (B) fifth dpi, (C) eighth dpi, and (D) the development of Setae on the eighth day post inoculation (dpi) as observed under a confocal microscope.
Re-inoculation and evaluation of fungal hyphae growth
Figure 7 presents the reaction of SC748-5 sorghum genotype against NK73_F37 isolate. The symptom development was similar to the previous artificial inoculation for all three sorghum genotypes (BT × 623, SC748-5, and ETSL_1001249) to three C. sublineola isolates (AS106_F43a, NK73_F37, and PW123_F47) recorded on the seventh day of post inoculation (dpi). However, the hyphal growth was not clearly visible in genotype SC748-5 under a confocal microscope as in BT × 623 and ETSL_101249 (Figure 7C). Furthermore, the infected spots were not expanded to create coalescence in nearby spots. Such limited growth of symptoms could be an indication of a late response of SC748-5 to the inoculated isolate NK73_F37.
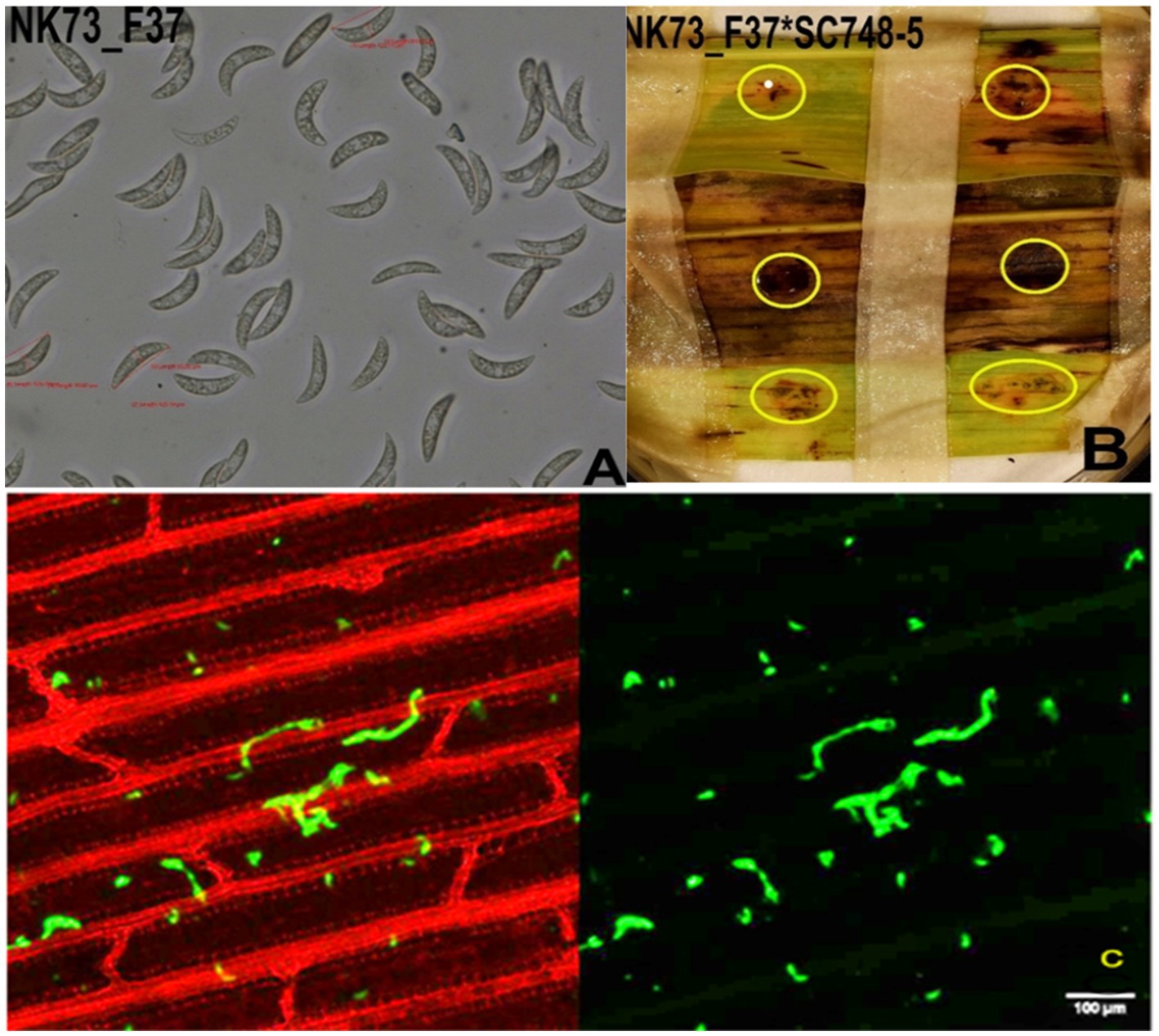
Figure 7. Infection of sorghum genotype SC748-5 by isolate NK73_F37. (A) Fungal conidia of NK73_F37; (B) Infection symptom development (acervuli circled with yellow rings), and (C) the growth of fungal hyphae of NK73_F37 in the leaf tissue of BT × 623 on the 7th dpi observed under a confocal microscope. Scale bars = 100 μm.
Discussion
Anthracnose of sorghum, caused by the fungus C. sublineola, poses a significant threat, leading to substantial losses in grain and biomass yields. The pathogen exhibits remarkable variation in symptoms, morphology, and pathogenicity, influenced by changes in environmental conditions and host factors (Valério et al., 2005; Chala, 2013). Understanding this variation within the pathogen is critical for developing durable and effective management strategies. This study employed morphological and virulence assessments to elucidate the interaction between C. sublineola isolates and sorghum genotypes. The methods employed in this study, constituting part of a polyphasic approach, facilitated the delineation of isolate variation. By utilizing a variety of sorghum anthracnose differentials, both qualitative (virulence) and quantitative (aggressiveness) distinctions among isolates were elucidated. The level of virulence provided insights into the presence of potential a virulence genes within the isolates and resistant genes within the host, contributing to a better understanding of the pathogen–host interaction.
The study revealed significant morphological and virulence variation among isolates, including between isolates recovered from the same leaf. Variation in virulence could be found within a C. sublineola population (Erpelding, 2011) in addition to the morphological variation, which could help to understand variation among isolates of the same species (Cannon et al., 2012). In Assosa, 8 of the 11 isolates were collected from the same research field and exhibited variation in virulence. In addition, pathotypes AS106_F37a and AS106_F37b were isolated from the same leaf sample and differed in virulence and colony morphology. Differentiation between isolates collected from a single research station elucidated the high variability of the pathogen, implying a lack of random mating in the pathogen population within a field (Prom et al., 2012). Similarly, high diversity among 22 isolates collected from a single field in Ethiopia has been reported (Chala, 2013), which is In line with the current study.
ANOVA revealed significant differences among the tested C. sublineola isolates collected from major sorghum growing areas in Ethiopia. Variation among isolates may arise from geographical isolation and specialized adaptation to hosts. In line with this, Erpelding and Prom (2009) stated that C. sublineola has a global distribution, with diverse pathotypes observed across various climatic regions. The present study also revealed a wide distribution of C. sublineola in Ethiopia as well as considerable variations in their virulence pattern and morphological characteristics. The differences in pathogenicity and virulence of C. sublineola in the country imply the need for continuous assessment of the pathogen genetic variation. As part of this approach, resistance genes could be identified and used to manage sorghum anthracnose effectively and sustainably.
The presence of 24 pathotypes in 25 isolates including those collected from the same field and leaf that showed different virulence levels in sorghum differentials indicates extreme pathogen variability. Cluster analysis revealed that isolates in five of the seven clusters were grouped according to their geographical origin, suggesting a significant contribution of collection sites to isolate differences. However, isolate grouping in terms of colony morphology did not correlate with virulence level. For instance, the most virulent pathotype PW74_F34 was grouped with the non-virulent pathotype PW125_F43 in cluster V, and the virulent pathotype PW117_F43 was grouped with the non-virulent pathotype AS104_F46 in cluster I. Pathotypes from Pawe, including the four top-ranked virulent pathotypes were distributed across five clusters, indicating high pathotype diversity in this part of the country. Cluster IV contained one pathotype from Pawe (PW83_F37) and one pathotype from Nekamte (NK73_F37), both showing similar pathogenicity and virulence against the genotypes and having similar morphological characters except the lower number of setae in pathotype PW83_F37. This may suggest a wide geographical distribution of some C. sublineola variants.
Although the infection of the sorghum genotype SC748-5 by pathotype NK73_F37 may suggest the emergence of a new virulent C. sublineola variant in Ethiopia, further study should be conducted for deeper insight including the gene-for-gene interaction. In the genus Colletotrichum, the emergence of genetic variants is common due to heterokaryosis, sexual cycle, transposable elements, and point mutation (Lopes et al., 2020). While the significance of the sexual cycle in generating genetic diversity is obvious and the role of heterokaryosis in the emergence of field variants remains uncertain, genetic differentiation of C. sublineola can result in resistance breakdown (Lopes et al., 2020). Isolation by distance and host diversification in the same field could contribute to the emergence of new pathogen variants. In sorghum anthracnose, many races have evolved to overcome LRR type R genes, and the host contains a large family comprising evolving LRR genes (Ahn et al., 2019). The present study revealed that the most virulent C. sublineola Pathotypes were collected from Pawe; this could be attributed to its favorable environment for the pathogen and a genetically diverse host cultivated in the area, which drives the pathogen toward increased pathogenicity and virulence compared to other areas. Similar observations were made regarding Puccinia species, where efforts to breed resistance in cereal crops introduced new selective pressures, resulting in the proliferation of virulent gene frequencies (Simon et al., 2020).
The tested sorghum genotypes showed different levels of resistance to the inoculated C. sublineola isolates ranging from no infection to being infected by multiple isolates. The sorghum genotypes IS_18760, Brandes, and Bonsa showed the highest resistance and were least affected by all 25 isolates, implying their broad-spectrum resistance to the C. sublineola variant. Hence, they could be a promising source of resistance genes for breeding sorghum cultivars resistant to anthracnose. Genotypes SC748-5 and SC112-14 were regarded as resistant to C. sublineola isolates, and genes on chromosome 5 contributed to their resistance (Valério et al., 2005; Prom et al., 2012; Prom et al., 2023). However, in the present study, genotype SC748-5 was susceptible to pathotype NK73_F37, and genotype SC112-14 was also susceptible to PW122_F47 and PW123_F47, indicating the presence of resistance-breaking C. sublineola pathotypes in Ethiopia. Conversely, sorghum genotype PI_57084 was the most susceptible followed by ETSL_101249, BT × 623, and TAM428. The present study also showed significant differences in aggressiveness among the tested C. sublineola isolates. Among the tested isolates, pathotypes PW123_F47 and PW117_F46 were the most aggressive causing anthracnose severity of 31%. This indicates that these isolates are useful for further sorghum germplasm screening for anthracnose resistance. Moreover, it indicates that Pawe is a hot spot for pathogen virulence and genetic variability analyses as well as for field evaluation of sorghum genetic resources for their response to C. sublineola infection.
The re-inoculation experiment involving known resistant and susceptible genotypes (SC748-5, BT × 623, and ETSL_1001249) with three isolates (NK73_F37, PW123_F47, and AS106_F43a) showed reproducibility of observed isolate–genotype interactions. However, proper structural growth of the fungi was observed only in genotypes BT × 623 and ETSL_1001249 (not SC748-5). Despite the fact that numerous anthracnose resistance loci have been reported in sorghum in recent years, resistance genes appeared to be not equally effective in different sorghum growing regions because of diverse C. sublineola pathotypes (Stutts and Vermerris, 2020). The limited fungal structural growth in SC748-5 could be because it possesses a dominant transcription factor gene conferring anthracnose resistance (Prom et al., 2012), and regulates gene expression patterns (Buiate et al., 2017). In general, a hypersensitivity reaction develops from plant-gene-producing pattern recognition receptors (PRRs) (Abreha et al., 2021), which recognize specific molecular patterns associated with pathogens (Zhang et al., 2014; Couto and Zipfel, 2016).
The infection symptom observed in the known resistant genotype (SC748-5) could be (1) late recognition of the pathogen by the host, which could lead to late response (Künstler et al., 2016), (2) the inoculated isolate NK73_F37 had evolved effectors that overcame the host resistance genes (Möller and Stukenbrock, 2017), which led to a change in the mechanism of infection, or (3) the pathogen might have initially colonized the host and remained in a latent or quiescent state for an extended period until it begins to macerate a plant tissue (Cannon et al., 2012) or combinations of these. In all cases, low or delayed host defense-related gene expression could occur. The dynamic nature of C. sublineola pathotypes, and resistant encoding genes associated with environmental factors could influence the host reaction time.
Cluster analysis conducted to visualize the relationships among the isolates revealed seven groups with some levels of admixture. None of the clusters exclusively contained isolates of a single geographical area indicating gene flow between isolates in different areas through various means like infected seed exchange and a step-by-step distribution of fungal spores over long distances. In line with this, Mathur et al. (2003) and Marley et al. (2005) stated that C. sublineola isolates obtained from different locations, the morphology of isolates extracted from a single-lesion culture, and even single-conidial derivatives of a single-lesion culture can vary considerably. Grouping the isolates based on their infection pattern and virulence level resulted in 24 pathotypes. Similar to the present finding, approximately 40 anthracnose pathotypes were identified among geographically diverse isolates tested under greenhouse and field conditions (Smith and Frederiksen, 2000; Mathur et al., 2003; Zanette et al., 2009). In Brazil, 22 pathotypes were identified among 37 isolates using 10 differentials (Valério et al., 2005). In the United States, Prom et al. (2012) reported 17 pathotypes among 18 isolates using 18 sorghum differentials, while Prom et al. (2023) reported 27 new pathotypes among 30 isolates using the same differential lines. This indicates that C. sublineola is a highly variable fungal pathogen.
Conclusion
This study revealed that C. sublineola isolates from major sorghum growing areas in Ethiopia were diverse in virulence patterns and aggressiveness. Variation in virulence was observed even between isolates extracted from plants in the same field in a single sample, indicating the pathogen’s extreme variation. C. sublineola isolates collected from Pawe were highly virulent and aggressive, indicating that the area is suitable for studies on pathogen diversity and host-resistance screening against anthracnose. For instance, four isolates (PW122_F47, PW117_F46, PW74_F34, and PW123_F47) collected from Pawe were the most virulent (on 68–72% of sorghum genotypes). Among the 25 tested sorghum genotypes, three genotypes (IS_18760, Brandes, and Bonsa) were resistant to all tested isolates, hence, they could serve as potential sources of resistance genes to be utilized in sorghum breeding programs aimed at developing sorghum cultivars resistant to a variety of C. sublineola pathotypes. On the other hand, some sorghum genotypes previously considered resistant to anthracnose, such as SC748-5 and SC112-14, were susceptible to some isolates. This suggests the need for continuous monitoring and further improvement of resistant germplasm through breeding. Overall, the present study confirmed the presence of a large number of C. sublineola pathotypes in Ethiopia which significantly differ in virulence. This study also documented the host resistance levels against the tested isolates. Sorghum genotypes resistant to various C. sublineola isolates were identified, so they can serve as sources of genes for developing cultivars to anthracnose through breeding. The current study only targeted areas highly prone to sorghum anthracnose. Thus, future studies should cover a broader geographic area where sorghum is grown in order to gain a more comprehensive understanding of the geographical distribution of C. sublineola pathotypes. Additionally, DNA marker-based genetic diversity analysis of C. sublineola populations in Ethiopia, as well as functional analysis of host-pathogen interactions, are highly recommended for effective control of sorghum anthracnose.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
MM: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. KT: Conceptualization, Investigation, Methodology, Resources, Supervision, Validation, Writing – review & editing. TeM: Methodology, Resources, Supervision, Validation, Writing – review & editing. AC: Methodology, Supervision, Validation, Visualization, Writing – review & editing. HN: Methodology, Supervision, Visualization, Writing – review & editing. TiM: Conceptualization, Methodology, Supervision, Validation, Visualization, Writing – review & editing. KA: Data curation, Funding acquisition, Supervision, Validation, Writing – review & editing. MG: Conceptualization, Data curation, Investigation, Methodology, Resources, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This experiment was funded by the Ethiopian Institute of Agricultural Research (EIAR) and the Swedish International Development Cooperation Agency (SIDA) project, AAU. This study was also supported by the Swedish Research Council grant (VR) grant #2019-05596 and Nilsson-Ehle Endowments grant #2020-41703. The contents are the sole responsibility of the authors and do not reflect the views of the funder.
Acknowledgments
The authors extend their heartfelt thanks to Mr. Abera Serbiessa for his invaluable support during laboratory and greenhouse research work and Dr. Salla Marttila for her assistance during imaging of the fungal hyphae under confocal microscope both from the Swedish University of Agricultural Sciences (SLU).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abreha, K. B., Ortiz, R., Carlsson, A. S., and Geleta, M. (2021). Understanding the Sorghum-Colletotrichum sublineola interactions for enhanced host resistance. Front. Plant Sci. 12:641969. doi: 10.3389/fpls.2021.641969
Acquaah, G. (2012). Principles of plant genetics and breeding. Second Edn. Maryland, USA: John Wiley & Sons, Ltd.
Ahn, E. J. S. (2020). Host defense responses of Sorghum and Johnsongrass against Colletotrichum Sublineola and identification of candidate defense related genes in Sorghum. USA: Texas A&M University.
Ahn, E., Hu, Z., Perumal, R., Prom, L. K., Odvody, G., Upadhyaya, D., et al. (2019). Genome wide association analysis of Sorghum Mini Core lines regarding anthracnose, downy mildew, and head smut. PLoS One 14:e0216671. doi: 10.1371/journal.pone.0216671
Bezabeh, E. (2015). Trends, growth and instability of Tefft production in Ethiopia. Res. J. Agric. Environ. Manag. 4, 78–081.
Buiate, E. A. S., Xavier, K. V., Moore, N., Torres, M. F., Farman, M. L., Schardl, C. L., et al. (2017). A comparative genomic analysis of putative pathogenicity genes in the host-specific sibling species Colletotrichum Graminicola and Colletotrichum Sublineola. BMC Genomics 18, 1–24. doi: 10.1186/s12864-016-3457-9
Bunker, R. N., Tanwar, N. S., and Aggarwal, S. K. (2019). Management of sorghum anthracnose caused by colletotrichum graminicola (Ces.) Wilson. Int. J. Curr. Microbiol. App. Sci. 8:1371. doi: 10.20546/ijcmas.2019.810.159
Cannon, P. F., Damm, U., Johnston, P. R., and Weir, B. S. (2012). Colletotrichum-current status and future directions. Stud. Mycol. 73, 181–213. doi: 10.3114/sim0014
Chala, A. (2013). Genetic diversity of Colletotrichum Sublineolum isolates from a single field in southern Ethiopia and evidence for the existence of MAT2 genotypes in different parts of the country. SINET Ethiop. J. Sci. 36, 9–16.
Chala, A., Brurberg, M. B., and Tronsmo, A. M. (2010). Incidence and severity of Sorghum anthracnose in Ethiopia. Plant Pathol. J. 9, 23–30. doi: 10.3923/ppj.2010.23.30
Charles, W. S., Martha, M., Christopher, M., Elias, L., Girma, A., Kaizzi, K. C., et al. (2009). Atlas of Sorghum production in East and Southern Africa. University of Nebraska, Lincoln, 1–63.
Cota, L. V., Souza, A. G. C., Costa, R. V., Silva, D. D., Lanza, F. E., Aguiar, F. M., et al. (2017). Quantification of yield losses caused by leaf anthracnose on Sorghum in Brazil. J. Phytopathol. 165, 479–485. doi: 10.1111/jph.12582
Couto, D., and Zipfel, C. (2016). Regulation of pattern recognition receptor Signalling in plants. Nat. Rev. Immunol. 16, 537–552. doi: 10.1038/nri.2016.77
Crouch, J. A., Clarke, B. B., White, J. F., and Hillman, B. I. (2009). Systematic analysis of the falcate-Spored Graminicolous Colletotrichum and a description of six new species from warm-season grasses. Mycologia 101, 717–732. doi: 10.3852/08-230
Cuevas, H. E., Cruet Burgos, C. M., Prom, L. K., Knoll, J. E., Stutts, L. R., and Vermerris, W. (2021). The inheritance of anthracnose resistance in Sorghum differential lines QL3 and IS18760. Sci. Rep. 11:20525. doi: 10.1038/s41598-021-99994-3
Cuevas, E. H., Prom, L. K., and Cruet-Burgos, C. M. (2019). Genome-wide association mapping of anthracnose (Colletotrichum sublineolum) resistance in NPGS Ethiopian Sorghum germplasm. G3 (Bethesda) 9, 2879–2885. doi: 10.1534/g3.119.400350
Erpelding, J. E. (2011). Anthracnose field evaluation of Sorghum germplasm from Botswana. Plant Prot. Sci. 47, 149–156.
Erpelding, J. E., and Prom, L. K. (2009). Response to anthracnose infection for a subset of Ethiopian Sorghum germplasm. Plant Pathol. J. 6, 127–133. doi: 10.3923/ppj.2007.127.133
ESS (2022). “Area and production of major crops. Ethiopian statistics Servce, agricultural sample Surver” in Survey Report I, 132.
FAOSTAT. (2023). Agricultural production statistics 2000–2022. Rome: FAOSTAT Analytical Briefs, No. 79.
Girma, G., Nida, H., Tirfessa, A., Lule, D., Bejiga, T., Seyoum, A., et al. (2020). A comprehensive phenotypic and genomic characterization of Ethiopian Sorghum germplasm defines core collection and reveals rich genetic potential in adaptive traits. Plant Genome 13, 1–17. doi: 10.1002/tpg2.20055
Künstler, A., Bacsó, R., Gullner, G., Hafez, Y. M., and Király, L. (2016). Staying alive – is cell death dispensable for plant disease resistance during the hypersensitive response? Physiol. Mol. Plant Pathol. 93, 75–84. doi: 10.1016/j.pmpp.2016.01.003
Lopes, L., Lorena, H., Moreno, A., Lucas, H., and Correia, N. (2020). Colletotrichum: species complexes, lifestyle, and peculiarities of some sources of genetic variability. Appl. Microbiol. Biotechnol. 104, 1891–1904. doi: 10.1007/s00253-020-10363-y
Marley, P. S., Diourte, M., Neya, A., and Rattunde, F. W. (2005). Sorghum anthracnose and sustainable management strategies in west and Central Africa. J. Sustain. Agric. 25, 43–56. doi: 10.1300/J064v25n01_05
Martin, S. (2016). Breeding strategies for the adaptation of Sorghum (Sorghum Bicolor L. Moench) as a novel crop for temperate Europe. Germany: Justus-Liebig-University Giessen.
Mathur, K., Thakur, R. P., Neya, A., Marley, P. S., and Casela, C. R. (2003). “Sorghum anthracnose-problem and management Strategiesle” in Sorghum and millets diseases. ed. J. F. Leslie (Ames, Iowa, USA: Iowa State Press), 211–220.
Möller, M., and Stukenbrock, E. H. (2017). Evolution and genome architecture in fungal plant pathogens. Nat. Rev. Microbiol. 15, 756–771. doi: 10.1038/nrmicro.2017.76
Prom, L. K., Ahn, E. J. S., Perumal, R., Cuevas, H. E., Rooney, W. L., Isakeit, T. S., et al. (2023). Genetic diversity and classification of Colletotrichum Sublineola Pathotypes using a standard set of Sorghum differentials. J. Fungi 10:3. doi: 10.3390/jof10010003
Prom, L. K., Cuevas, H. E., Ahn, E., and Isakiet, T. (2022). Evaluation of a subset of Ethiopia Sorghum collection germplasm from the National Genetic Resources Program of the United States Department of Agriculture for anthracnose resistance. Am. J. Plant Sci. 13, 1403–1411. doi: 10.4236/ajps.2022.1312095
Prom, L. K., Cuevas, H., Isakeit, T., and Droleskey, R. (2016). Excised leaf method for high volume evaluation of Sorghum germplasm for resistance against Colletotrichum Sublineolum. Plant Pathol. J. 15, 11–16. doi: 10.3923/ppj.2016.11.16
Prom, L. K., Perumal, R., Erattaimuthu, S. R., Little, C. R., No, E. G., Erpelding, J. E., et al. (2012). Genetic diversity and Pathotype determination of Colletotrichum Sublineolum isolates causing anthracnose in Sorghum. Eur. J. Plant Pathol. 133, 671–685. doi: 10.1007/s10658-012-9946-z
Redkar, A., Jaeger, E., and Doehlemann, G. (2018). Visualization of growth and morphology of fungal hyphae in planta using WGA-AF488 and Propidium iodide co-staining. Bio Protoc. 8, 1–7. doi: 10.21769/bioprotoc.2942
Savje, F. (2024). Distances: tools for distance metrics (computer software manual). R Package Version 0.1.11. Available at: https://Github.Com/Fsavje/Distances (Accessed September 30, 2024).
Simon, S. J., Tschaplinski, T. J., LeBoldus, J. M., Keefover-Ring, K., Azeem, M., Chen, J. G., et al. (2020). Host plant genetic control of associated fungal and insect species in a Populus hybrid cross. Ecol. Evol. 10, 5119–5134. doi: 10.1002/ece3.6266
Smith, C. W., and Frederiksen, R. A. (2000). Sorghum origin, history, technology, and production. New York: John Wiley and Sons.
Stutts, L. R., and Vermerris, W. (2020). Elucidating anthracnose resistance mechanisms in Sorghum-a review. Phytopathology 110, 1863–1876. doi: 10.1094/PHYTO-04-20-0132-RVW
Thakur, R. P., Rao, V. P., Wu, B. M., Subbarao, K. V., Mathur, K., Tailor, H. C., et al. (2007). Genetic resistance to foliar anthracnose in Sorghum and pathogenic variability in Colletotrichum Graminicola. Indian Phytopathol. 60, 13–23.
Tsedaley, B., Adugna, G., and Fikre, L. (2016). Distribution and importance of Sorghum anthracnose (Colletotrichum Sublineolum) in southwestern and Western Ethiopia. Plant Pathol. J. 15, 75–85. doi: 10.3923/ppj.2016.75.85
USDA (2021). World agricultural production. United States Department of Agriculture, Foreign Agricultural Service. in Circular Series (WAP 7-21).
Valério, H. M., Resende, M. A., Weikert-Oliveira, R. C. B., and Casela, C. R. (2005). Virulence and molecular diversity in Colletotrichum Graminicola from Brazil virulence and molecular diversity in Colletotrichum Graminicola from Brazil. Mycopathologia 159, 449–459. doi: 10.1007/s11046-005-0373-y
Zanette, G. F., Nóbrega, G., and Luzia, M. (2009). Morphogenetic characterization of Colletotrichum Sublineolum strains, causal agent of anthracnose of Sorghum. Trop. Plant Pathol. 34, 146–151. doi: 10.1590/s1982-56762009000300002
Keywords: anthracnose, Colletotrichum sublineola , disease resistance, genotypes, landraces, virulence spectrum
Citation: Mekonen M, Tesfaye K, Mengiste T, Chala A, Nida H, Mekonnen T, Abreha KB and Geleta M (2024) Pathotype determination of sorghum anthracnose (Colletotrichum sublineola) isolates from Ethiopia using sorghum differentials. Front. Microbiol. 15:1458450. doi: 10.3389/fmicb.2024.1458450
Edited by:
Febri Doni, Padjadjaran University, IndonesiaReviewed by:
Angela T. Alleyne, The University of the West Indies, Cave Hill, BarbadosRasappa Viswanathan, Indian Council of Agricultural Research (ICAR), India
Copyright © 2024 Mekonen, Tesfaye, Mengiste, Chala, Nida, Mekonnen, Abreha and Geleta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mulatu Geleta, TXVsYXR1LkdlbGV0YS5EaWRhQHNsdS5zZQ==
†ORCID: Kassahun Tesfaye, orcid.org/0000-0002-4046-4657
Tesfaye Mengiste, orcid.org/0000-0001-8865-994X
Alemayehu Chala, orcid.org/0000-0001-5965-6336
 Moges Mekonen
Moges Mekonen Kassahun Tesfaye2,3†
Kassahun Tesfaye2,3† Habte Nida
Habte Nida Kibrom B. Abreha
Kibrom B. Abreha Mulatu Geleta
Mulatu Geleta